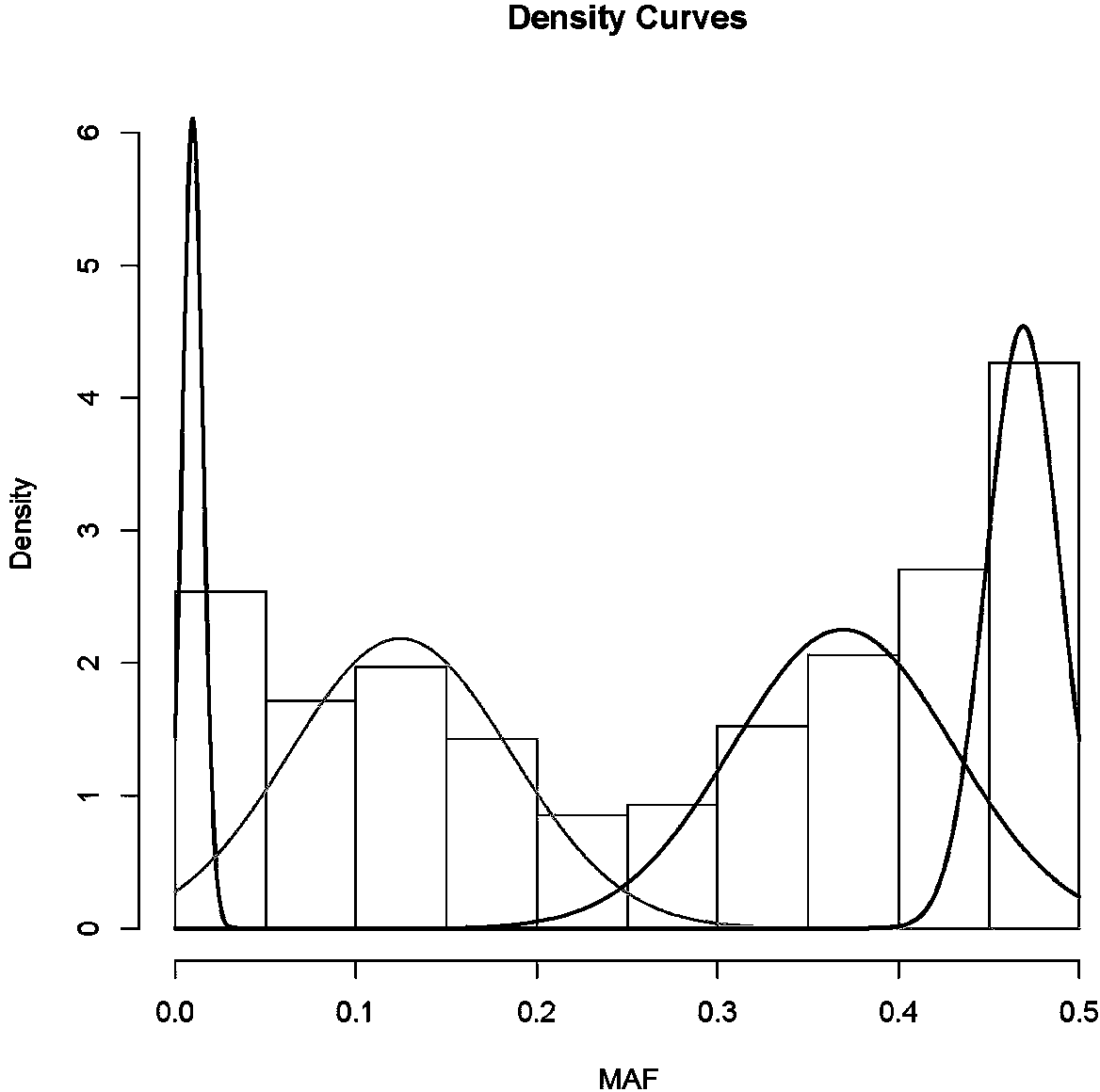
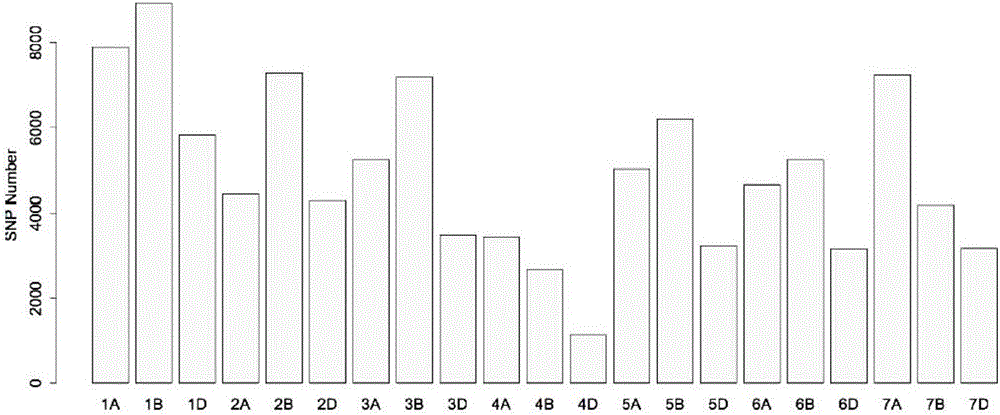
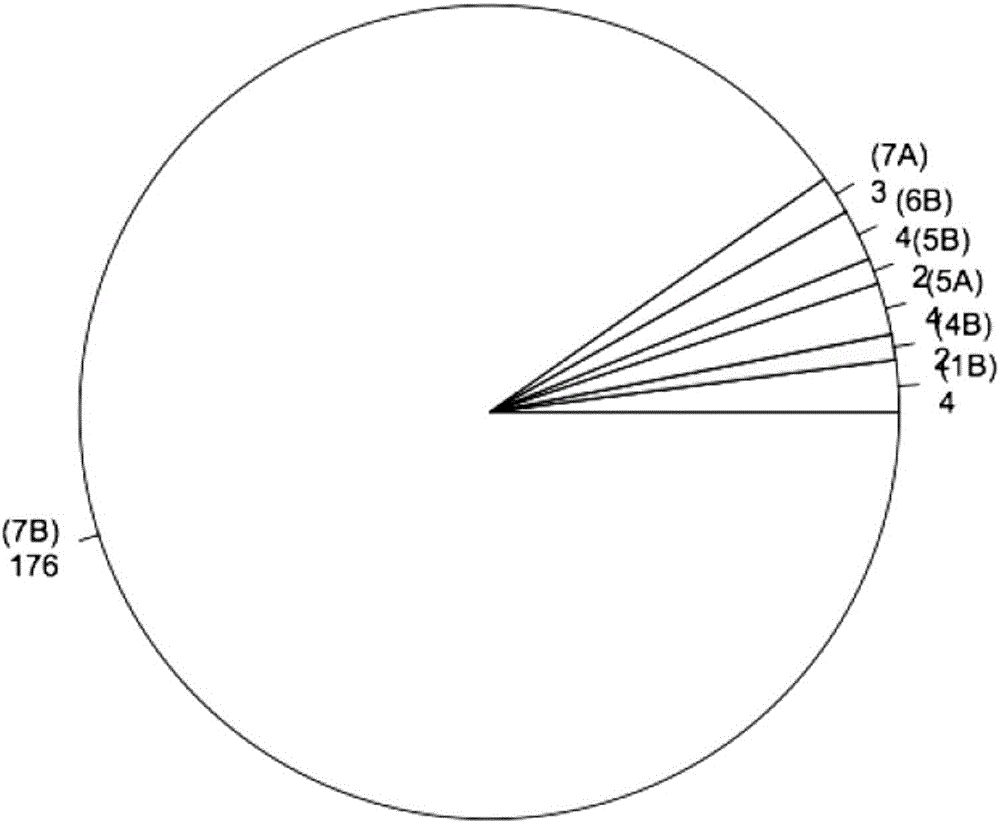
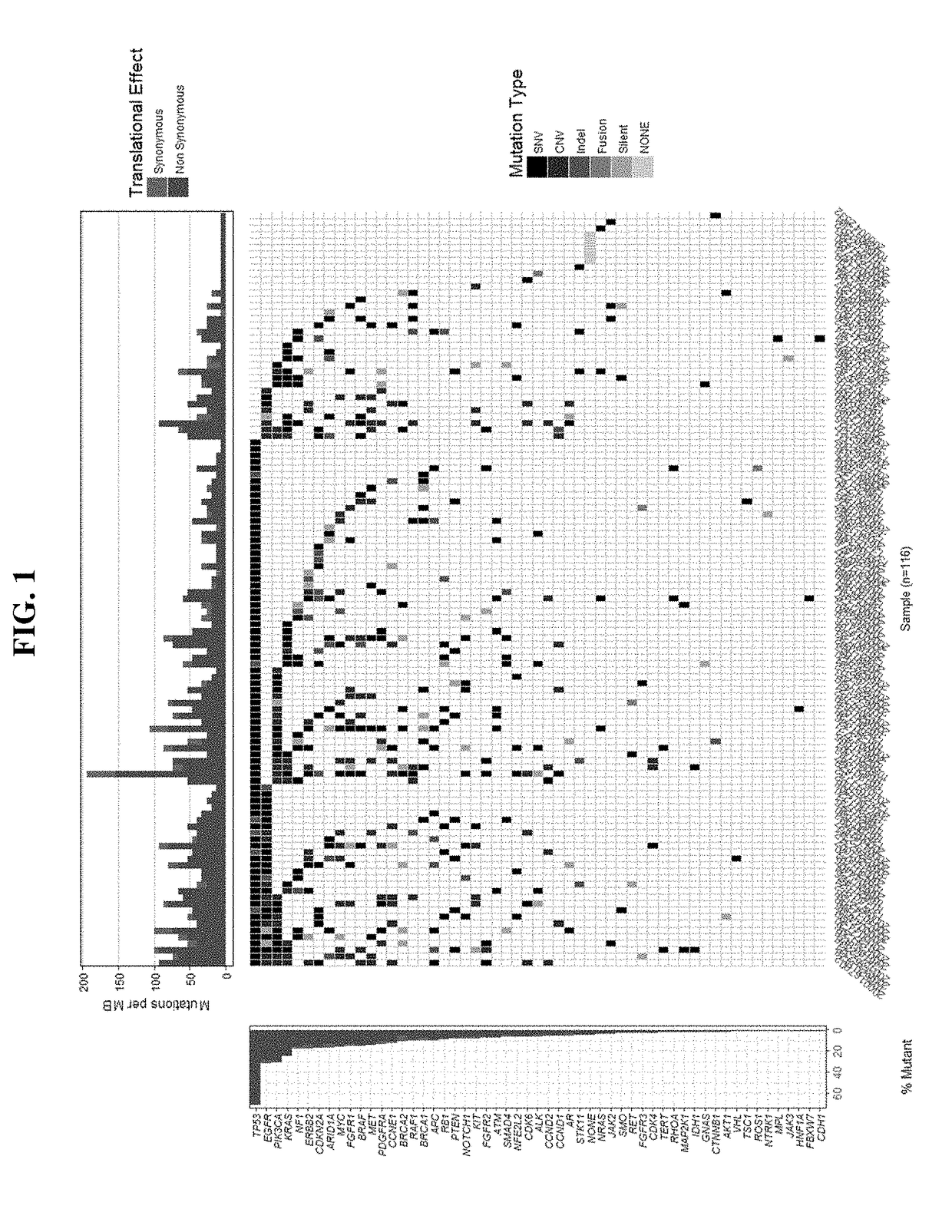
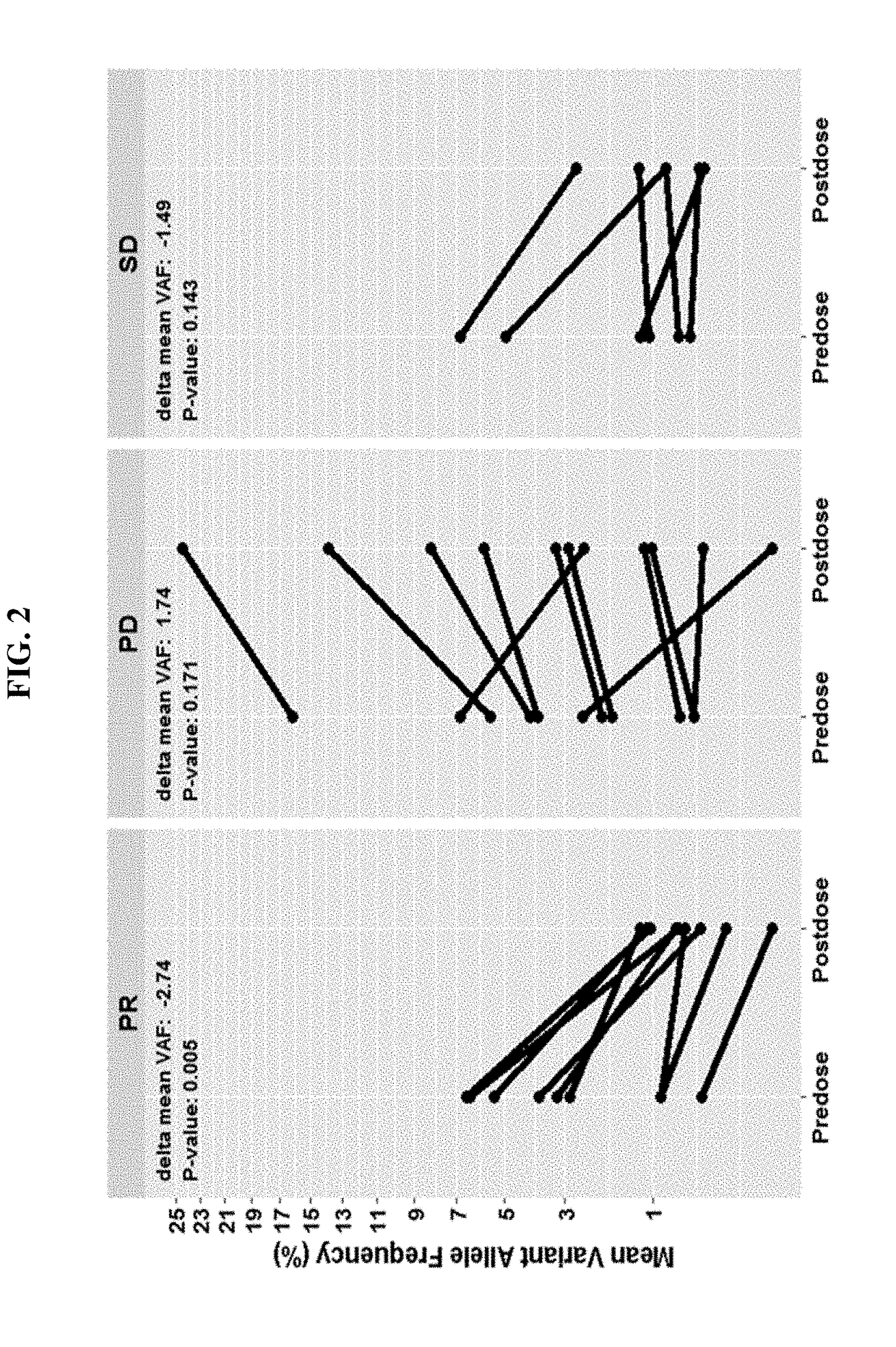
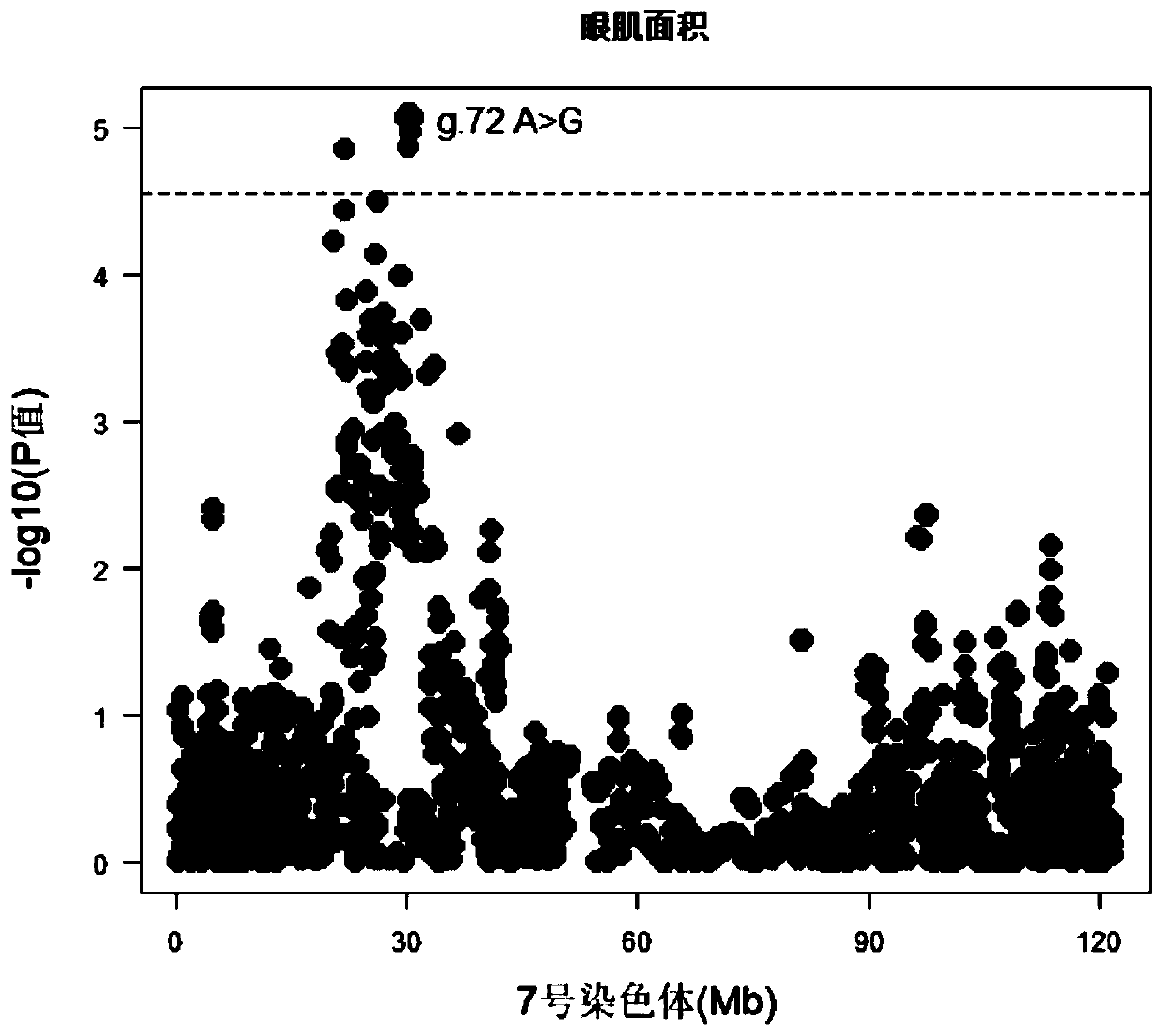
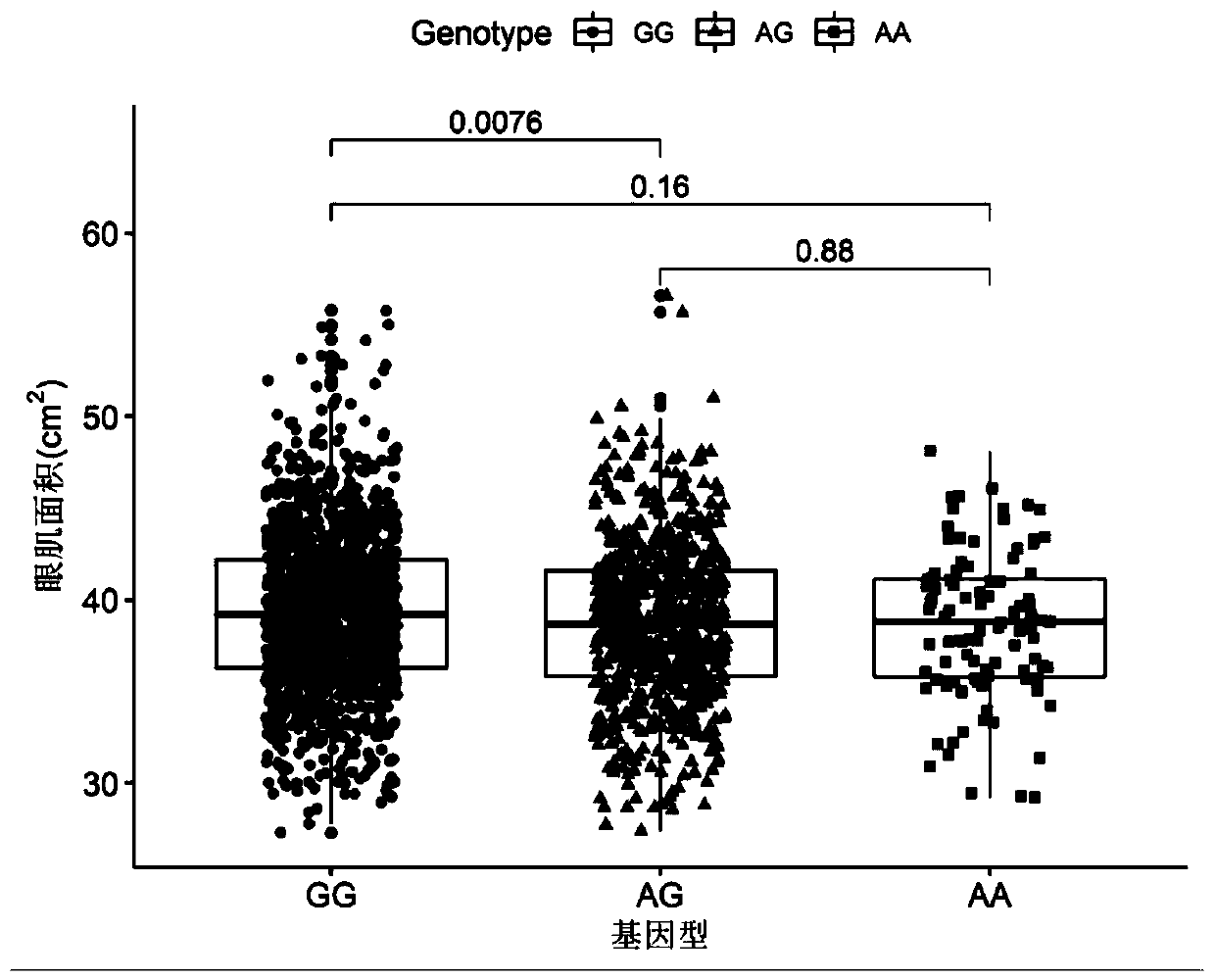
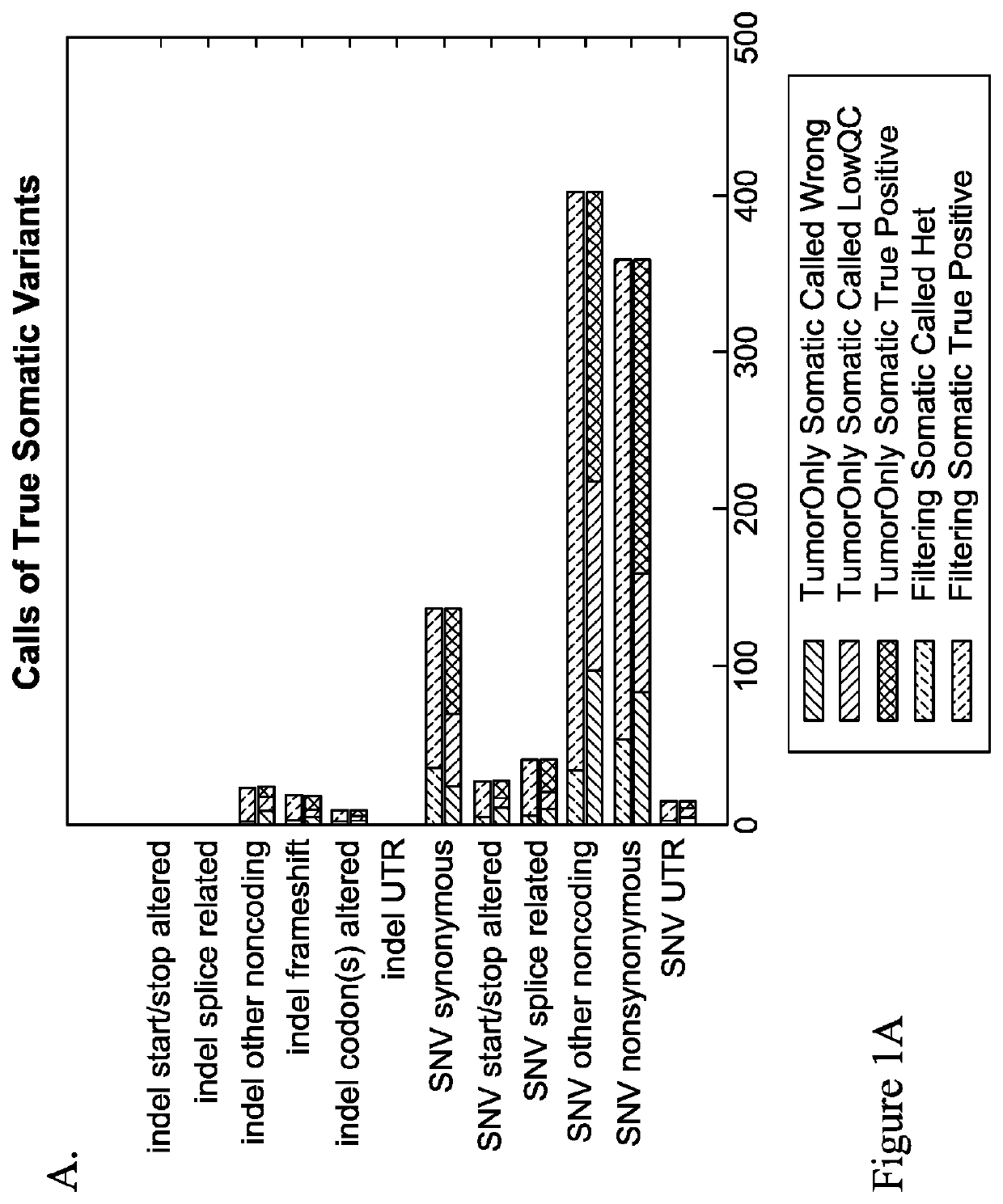
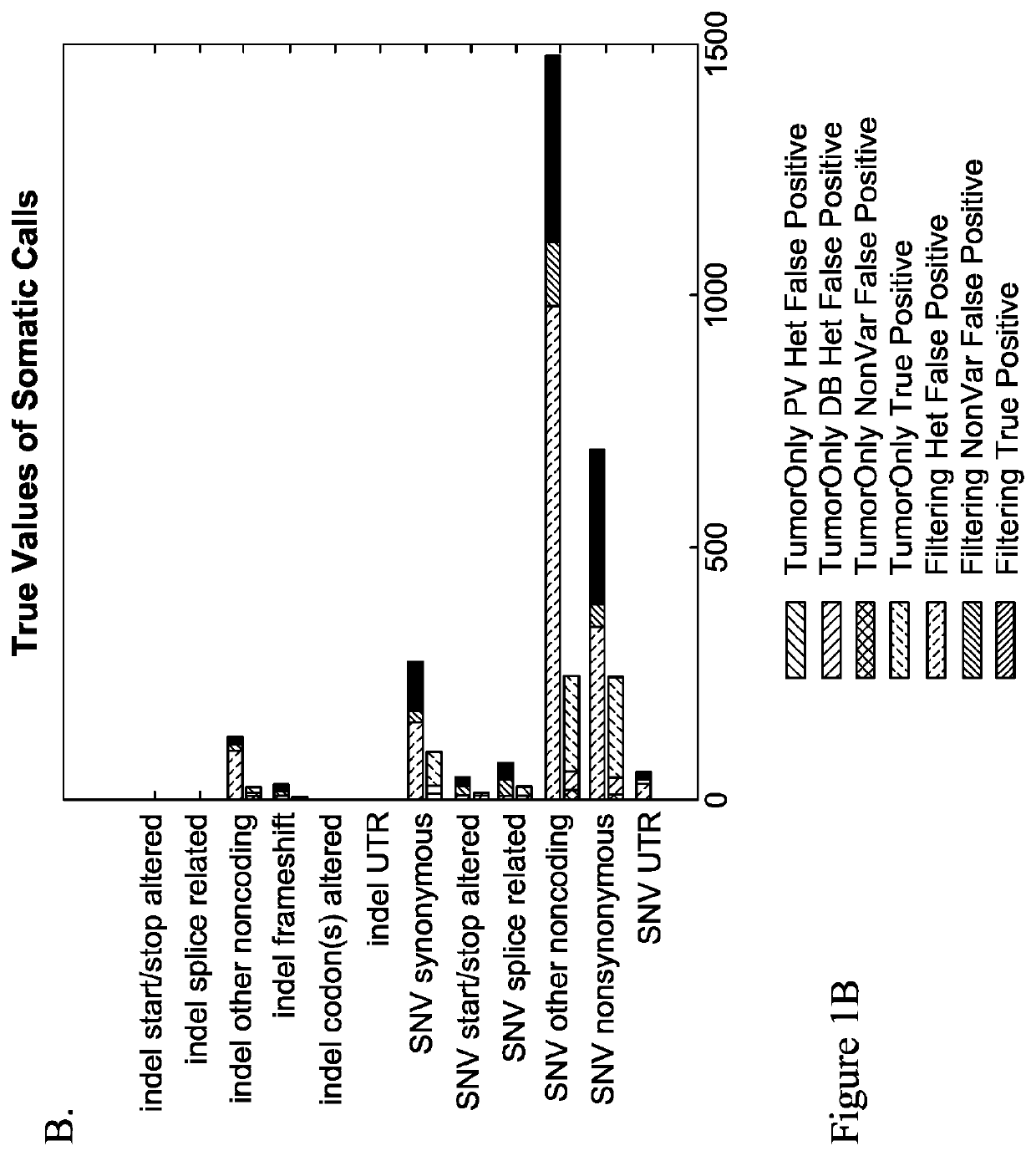
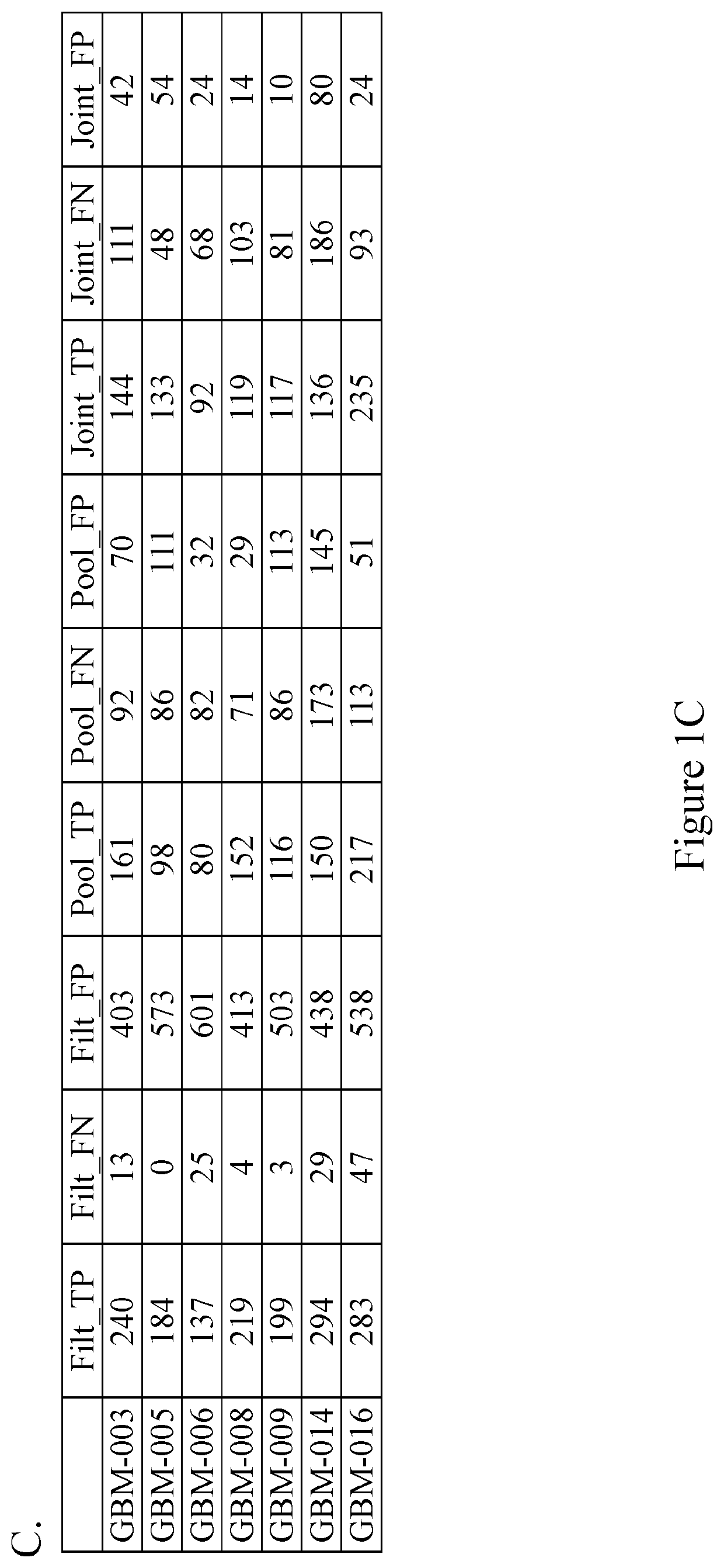
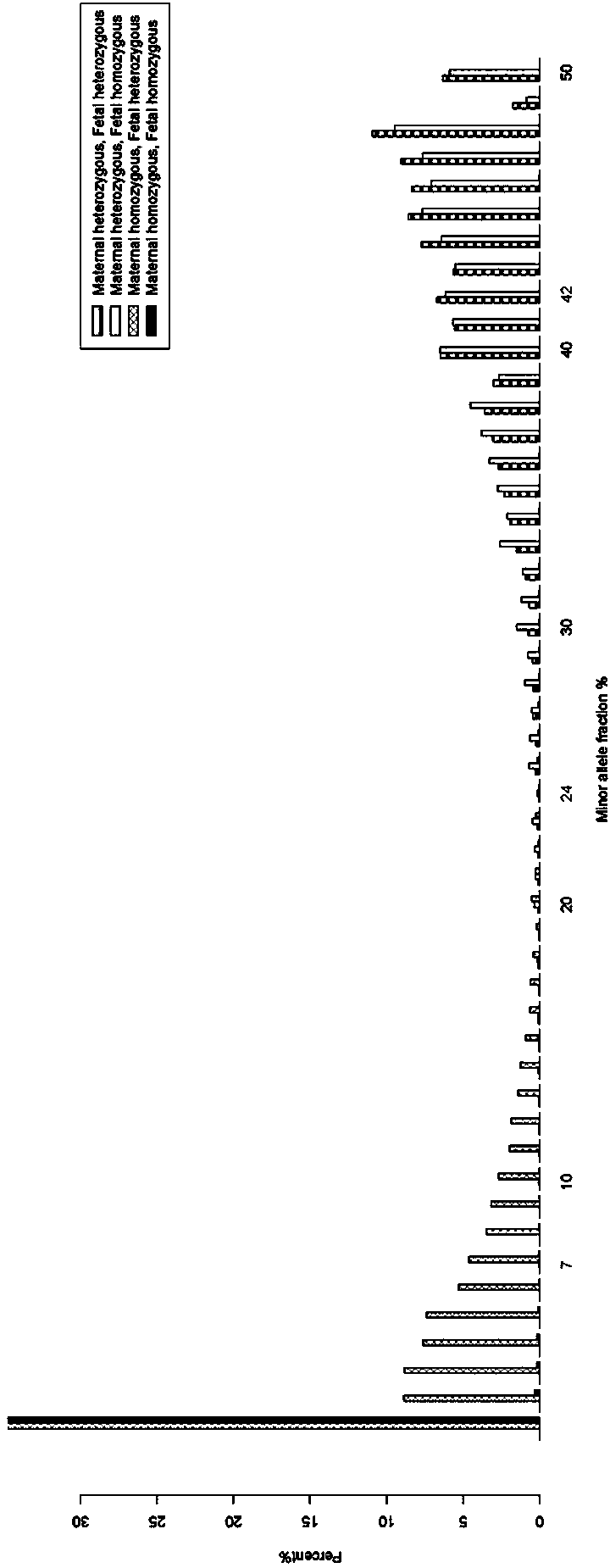Patents
Literature
140 results about "Allele frequency" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
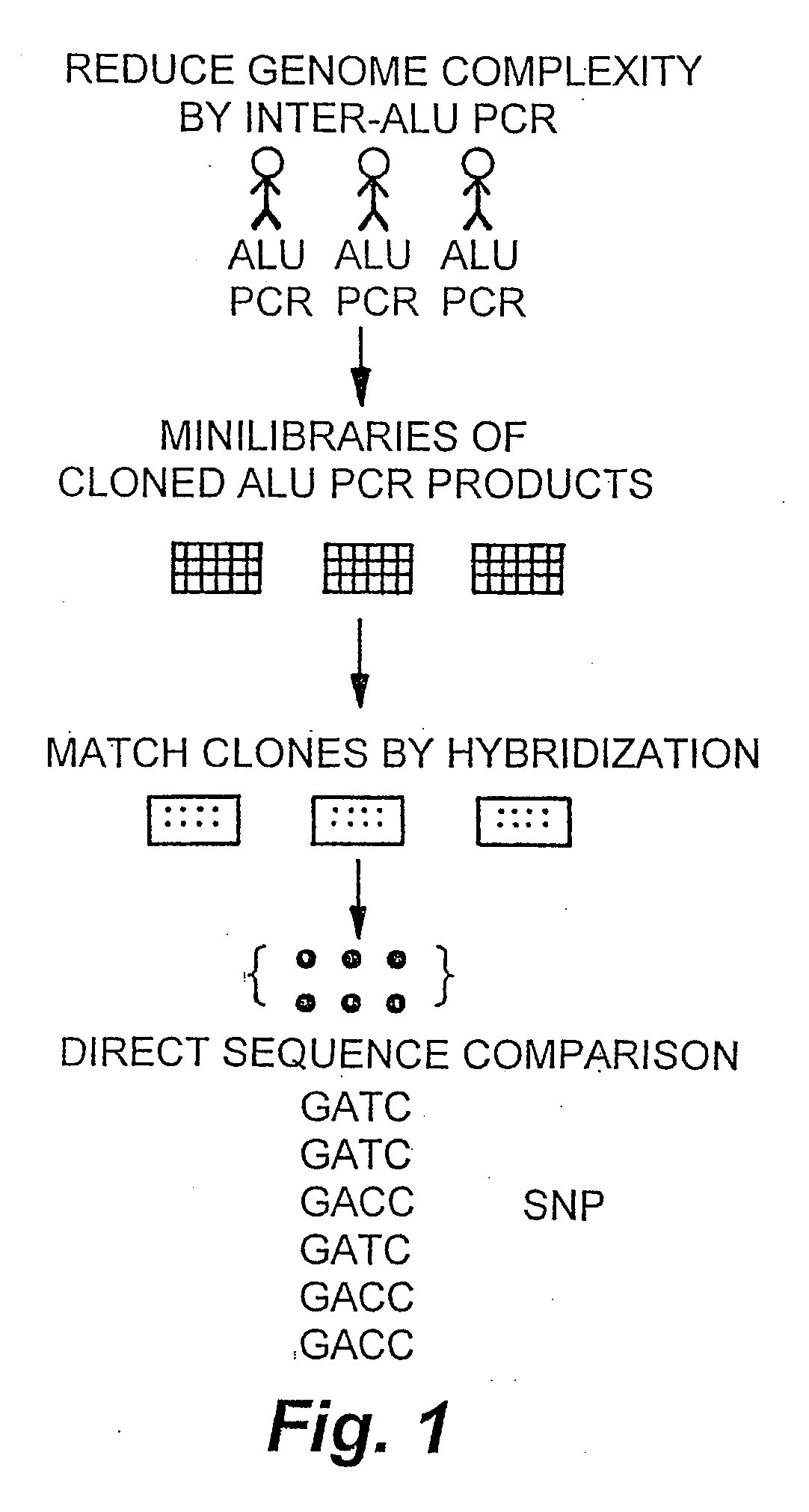
Allele frequency, or gene frequency, is the relative frequency of an allele (variant of a gene) at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele. Microevolution is the change in allele frequencies that occurs over time within a population.
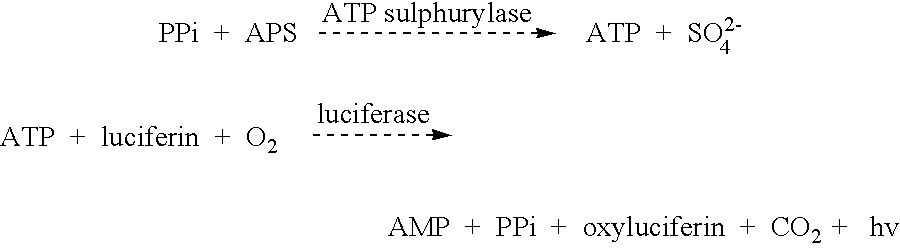
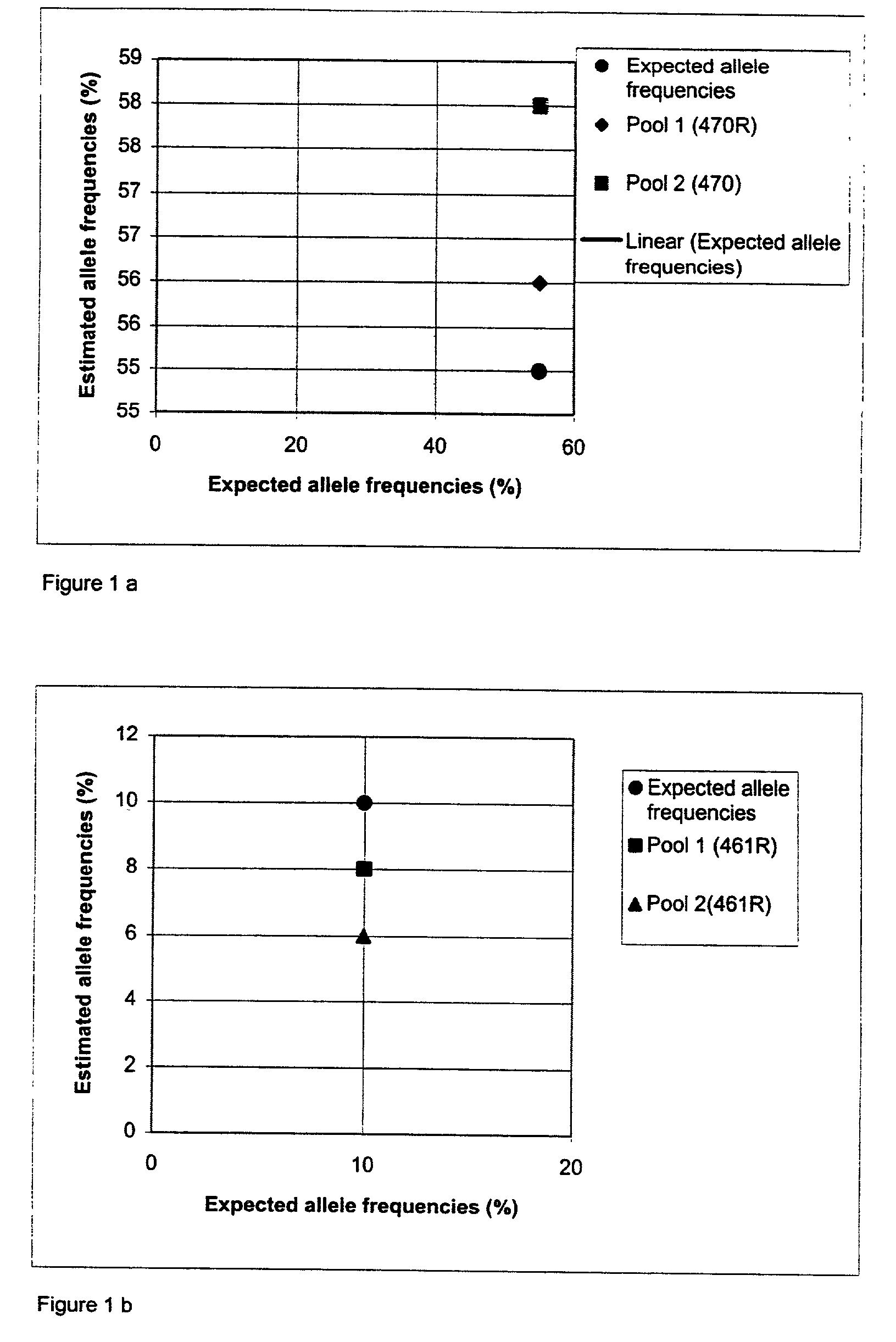
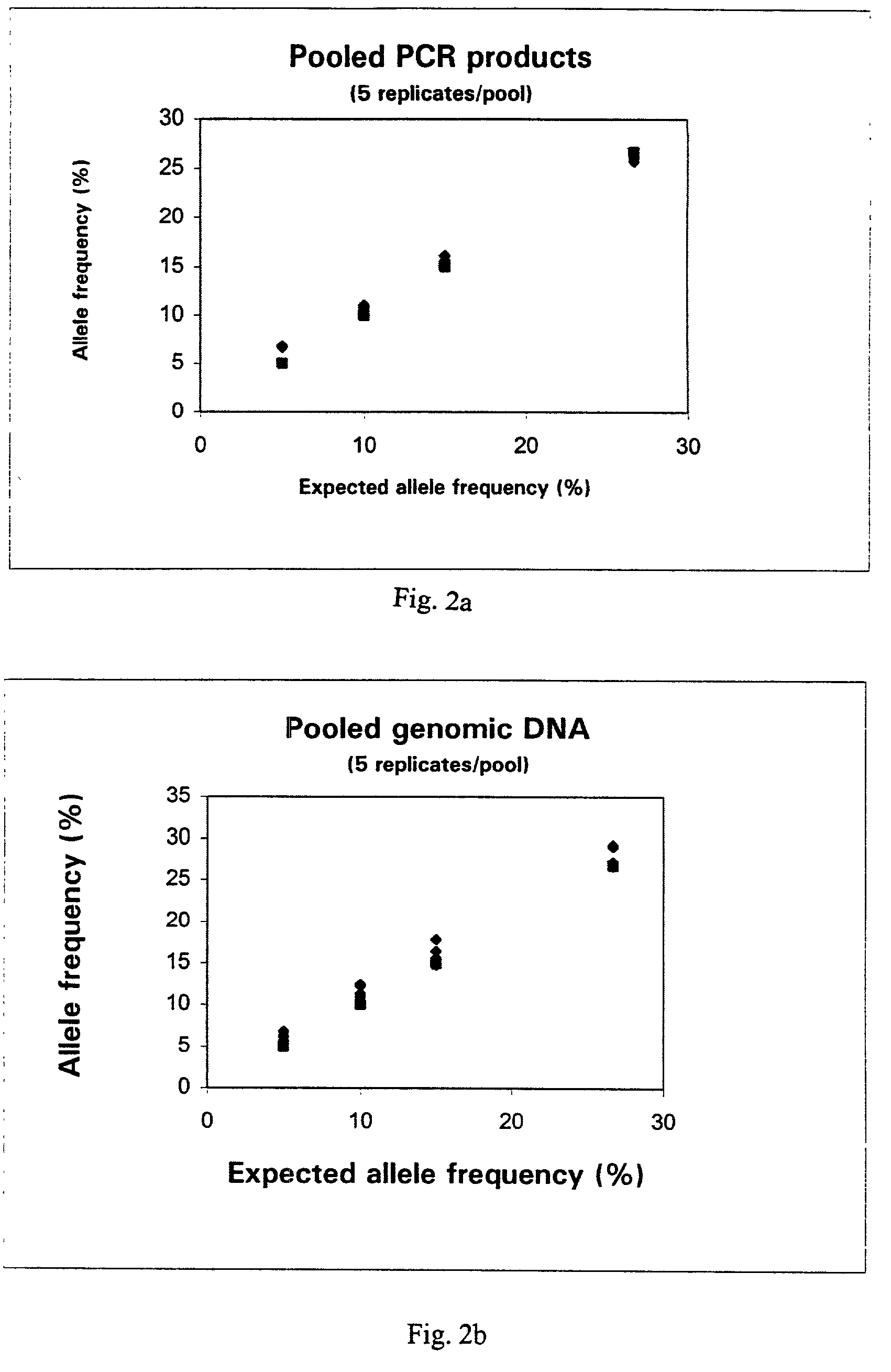
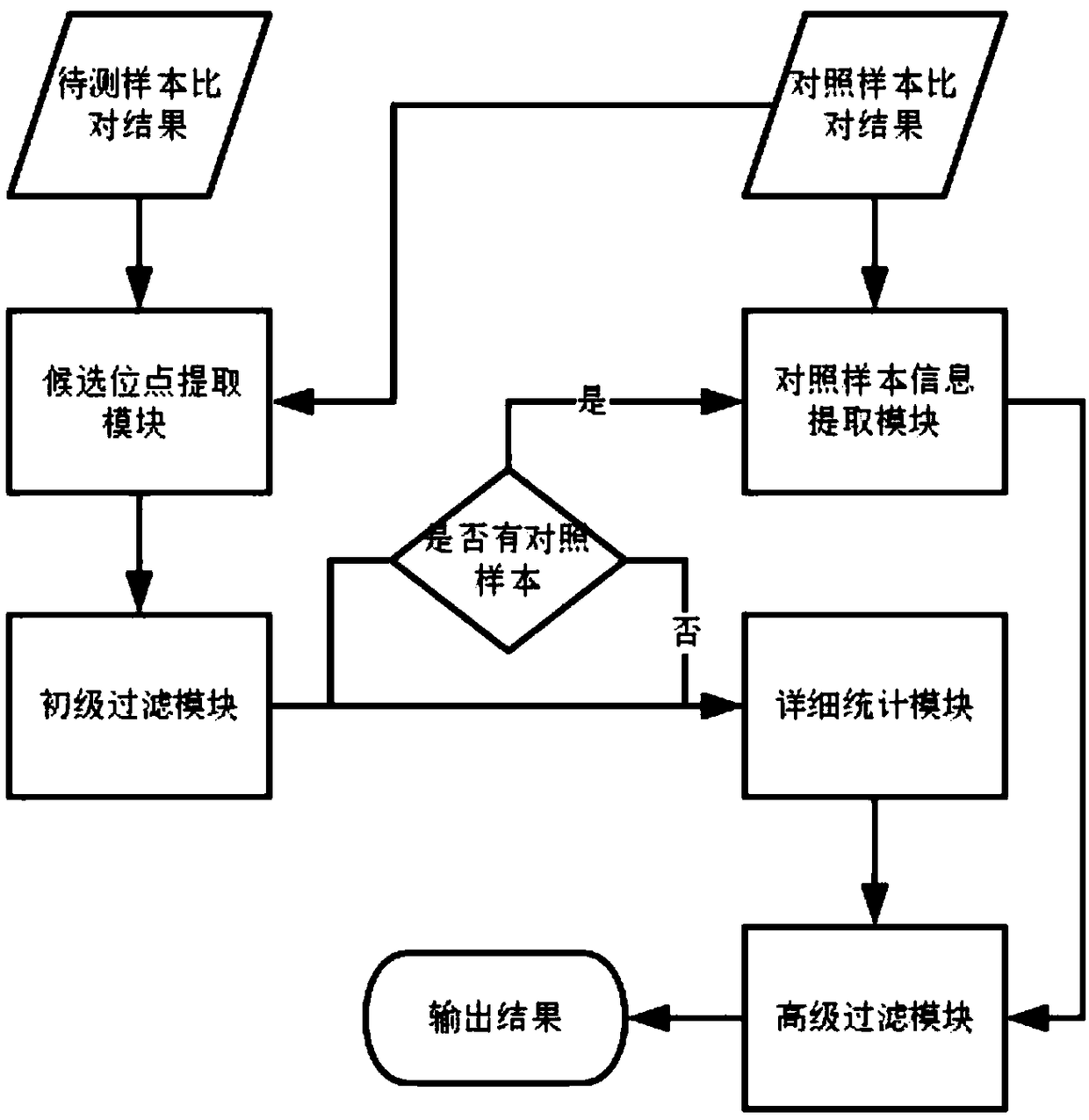
Method for determining allele frequencies
InactiveUS7078168B2Simple and reliable processSugar derivativesMicrobiological testing/measurementNucleotideAllele frequency
The present invention related to a method of determining the frequency of an allele in a population of nucleic acid molecules. The method comprises pooling the nucleic acid molecules of a population of nucleic acids, performing primer extension reactions using a primer which binds at a predetermined site located in nucleic acid molecules, and obtaining a pattern of nucleotide incorporation.
Owner:QIAGEN GMBH
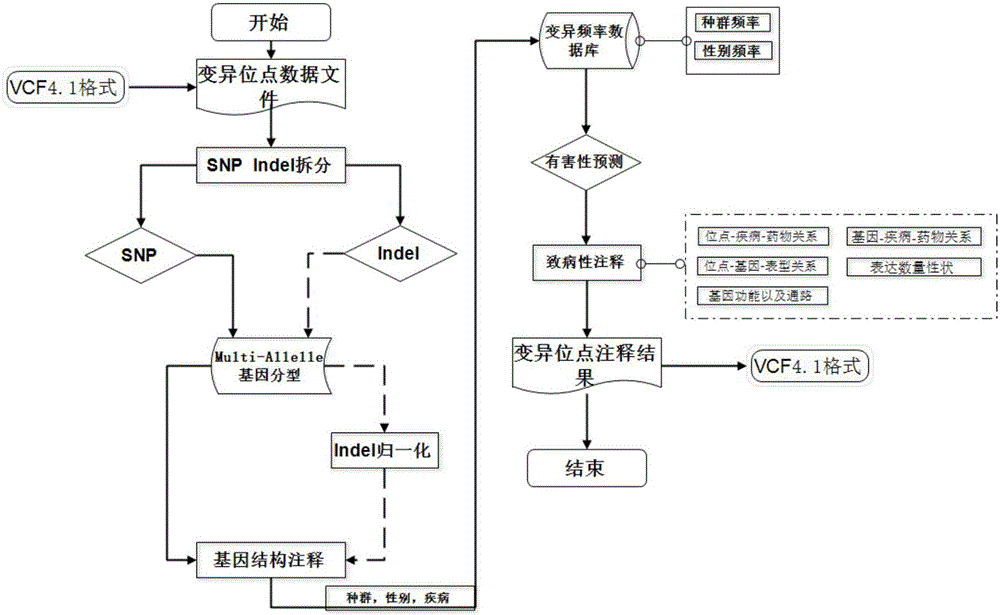
Annotation method and annotation system of whole-genome variant data
InactiveCN106156538AAccurate NotesSolve the screening puzzleProteomicsGenomicsReference genesAllele frequency
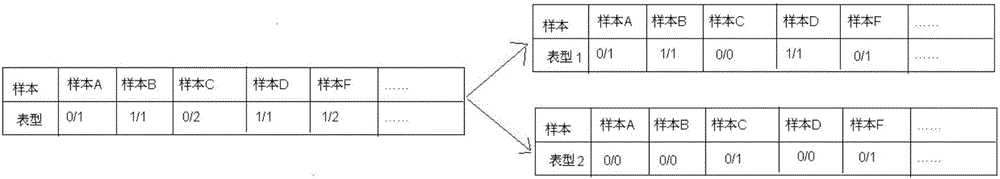
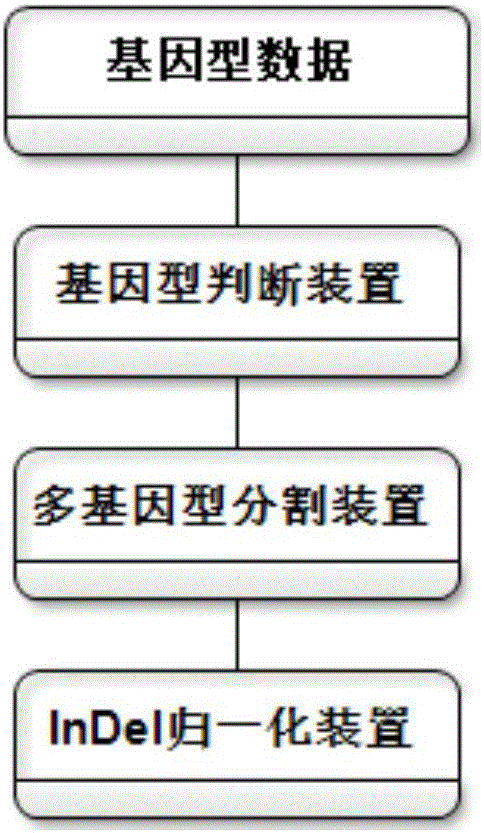
The invention discloses an annotation method and an annotation system of whole-genome variant data. The method comprises the following steps of S1, creating a variant data file, wherein the variant data are stored according to a national standard VCF format as an input file; S2, performing multi-allele genotyping, firstly performing genotype judgment, representing a basic group which is consistent with a reference genome by zero, and representing the basic groups which are inconsistent with the reference gene group by 1, 2, 3,..., then performing SNP and InDel multi-allele type resolution so that the allele type is represented by zero and one; S3, causing InDel generation position normalization, namely performing InDel generation position normalization according to a left justification and simplification normalization method; and S4, performing annotation, namely performing gene structure annotation, allele frequency annotation, variable site harm prediction and pathogenicity annotation. The annotation method and the annotation system improve integrity and accuracy of annotation information.
Owner:天津诺禾医学检验所有限公司
Methods and products related to genotyping and DNA analysis
InactiveUS20090098551A1Improve throughputReduce complexityMicrobiological testing/measurementProteomicsGenome ScanAllele-specific oligonucleotide
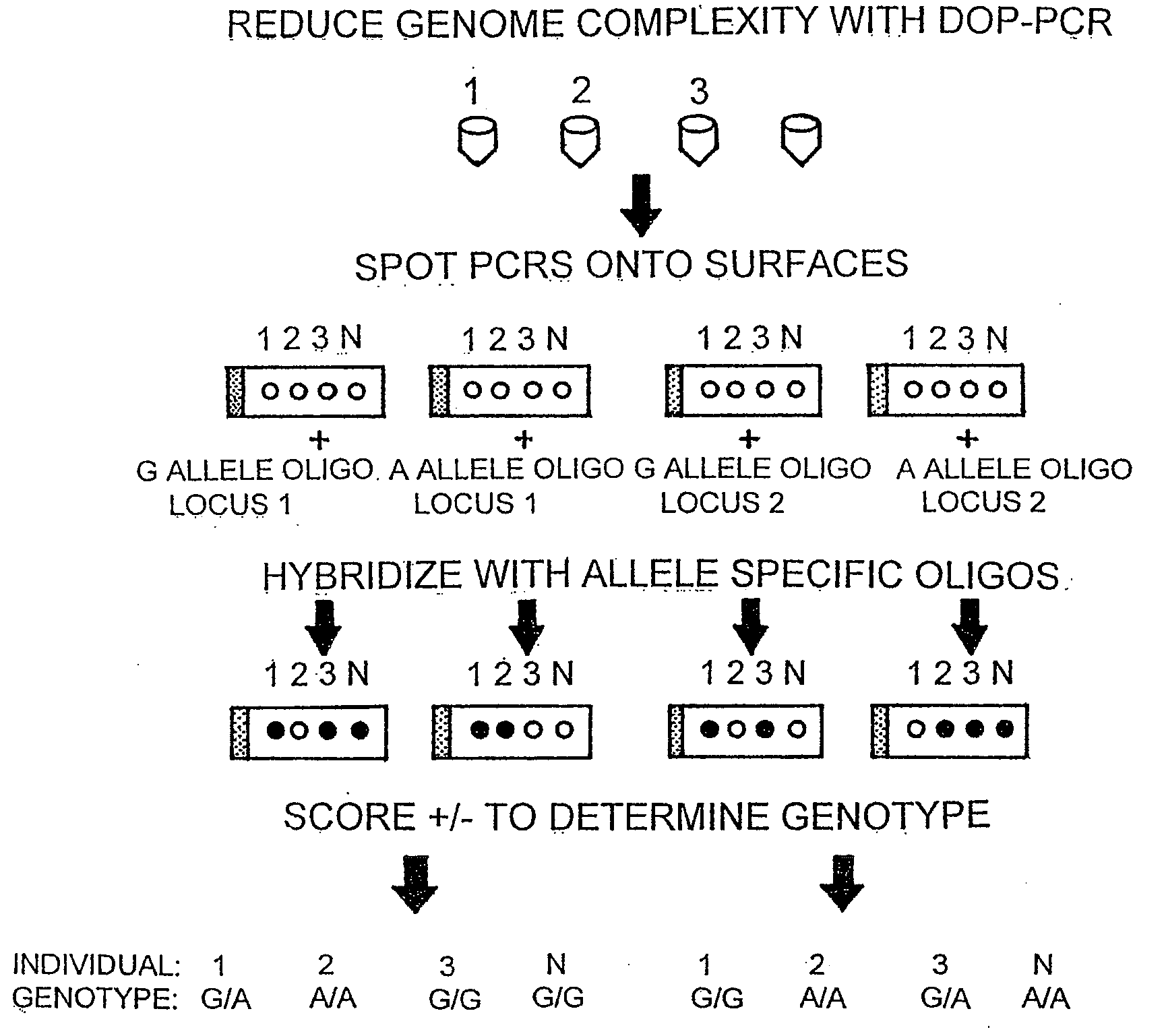
The invention encompasses methods and products related to genotyping. The method of genotyping of the invention is based on the use of single nucleotide polymorphisms (SNPs) to perform high throughput genome scans. The high throughput method can be performed by hybridizing SNP allele-specific oligonucleotides and a reduced complexity genome (RCG). The invention also relates to methods of preparing the SNP specific oligonucleotides and RCGs, methods of fingerprinting, determining allele frequency for a SNP, characterizing tumors, generating a genomic classification code for a genome, identifying previously unknown SNPs, and related compositions and kits.
Owner:MASSACHUSETTS INST OF TECH
Detection method and system for copy number variation in genome
The invention provides a method for analyzing copy number variation in a genome of a testee. According to the method, mathematical manipulation is creatively performed on variables extracted from sequencing data, new two-dimensional variables, namely a sequencing depth ratio (SDR) and alternate allele frequency (AAF) or alternate allelic haploid frequency (AHF) are derived, the signal / noise ratio is further increased, detection sensitivity and precision are improved, and information contained in the high-throughput DNA sequencing data is utilized more fully. Besides, the method is used for processing free DNA in a body fluid sample of the testee, wherein when a CNV feature signal of a certain locus of the genome is extracted from the sequencing data, the information close to the locus is introduced, so that signal strength is greatly improved, and the copy number variation in the free DNA in urine or serum can be effectively detected. The invention furthermore provides a system for implementing the method.
Owner:郝柯 +1
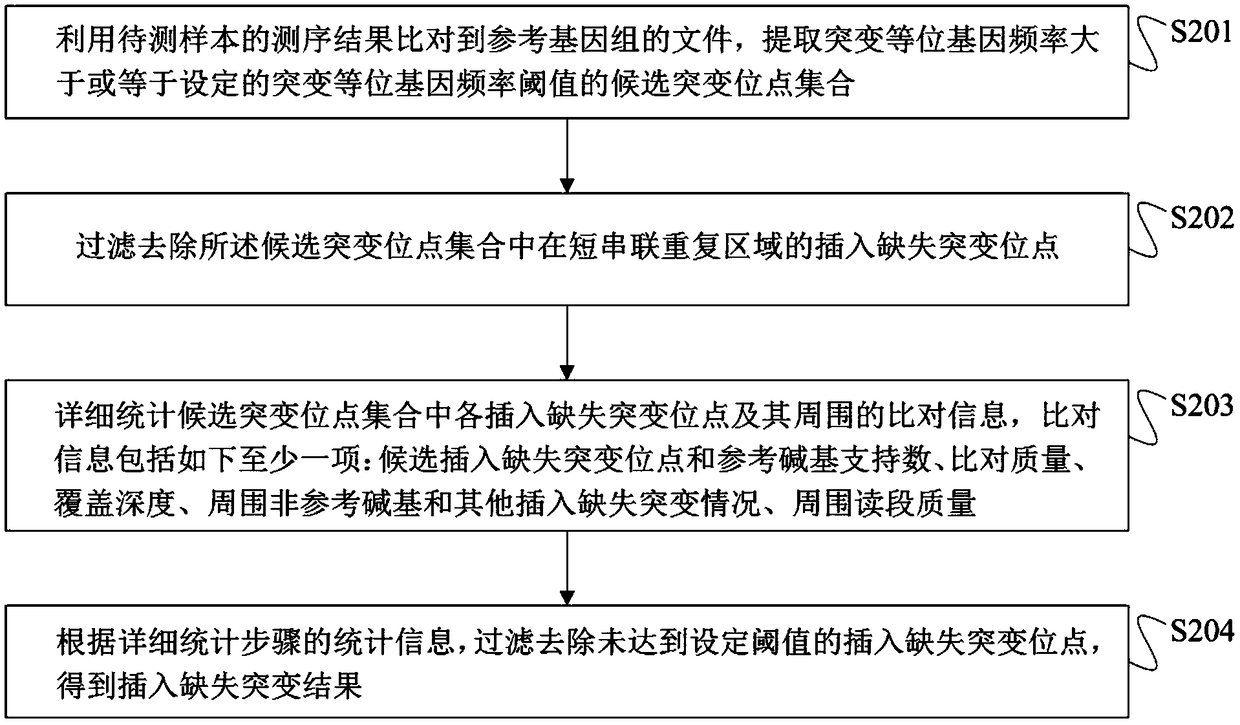
Method for detection of insertion deletion mutation based on second generation sequencing, device and storage medium
ActiveCN108690871AIncreased sensitivityStrong specificityMicrobiological testing/measurementProteomicsInsertion deletionAllele frequency
The present application discloses a method for detection of insertion deletion mutation based on second generation sequencing, a device and a storage medium. The method comprises the following steps:comparing a sample to be tested with a file of a reference genome to extract a set of candidate mutation sites with mutation allele frequency being greater than or equal to a threshold; filtering to remove sites in a short tandem repeat region; making detail statistics of comparison information of the mutation sites and comparison information surrounding the mutation sites, wherein the comparisoninformation includes InDel site and reference base support number, comparison quality, coverage depth, surrounding non-reference base and other insertion deletion mutations, surrounding read quality;and filtering to remove sites that do not reach the set threshold according to statistical information to obtain mutation results. The method does not require partial assembly, and filters second-generation sequencing data in advance to quickly eliminate most of false positive results caused by the comparison, reduces detection running time and computing resources, improves detection efficiency, has strong sensitivity and specificity, and can quickly and accurately detect InDel mutations.
Owner:深圳裕策生物科技有限公司
Method of reducing insect resistant pests in transgenic crops
The present invention discloses Resistance Management (RM) practices that are critical to safeguard Bacillus thuringiensis as a natural resource and sustain genetically modified corn expressing Bt toxins as a suitable method for ECB and WCRW management. A useful tool in developing RM strategies is to develop laboratory selected colonies that exhibit high levels of resistance to a particular toxin. The availability of selected strains allows determination of the genetic expression of resistance (i.e., dominant vs. recessive, autosomal vs. sex-linked) and whether or not the resistance mechanism is specific for a given toxin. In addition, the availability of resistant strains will allow estimation of the particular resistance allele frequency in the field, and provides a tool to identify the biochemical and physiological basis of resistance and a means to develop molecular probes to monitor the evolution of resistance in the field.
Owner:PIONEER HI BRED INT INC
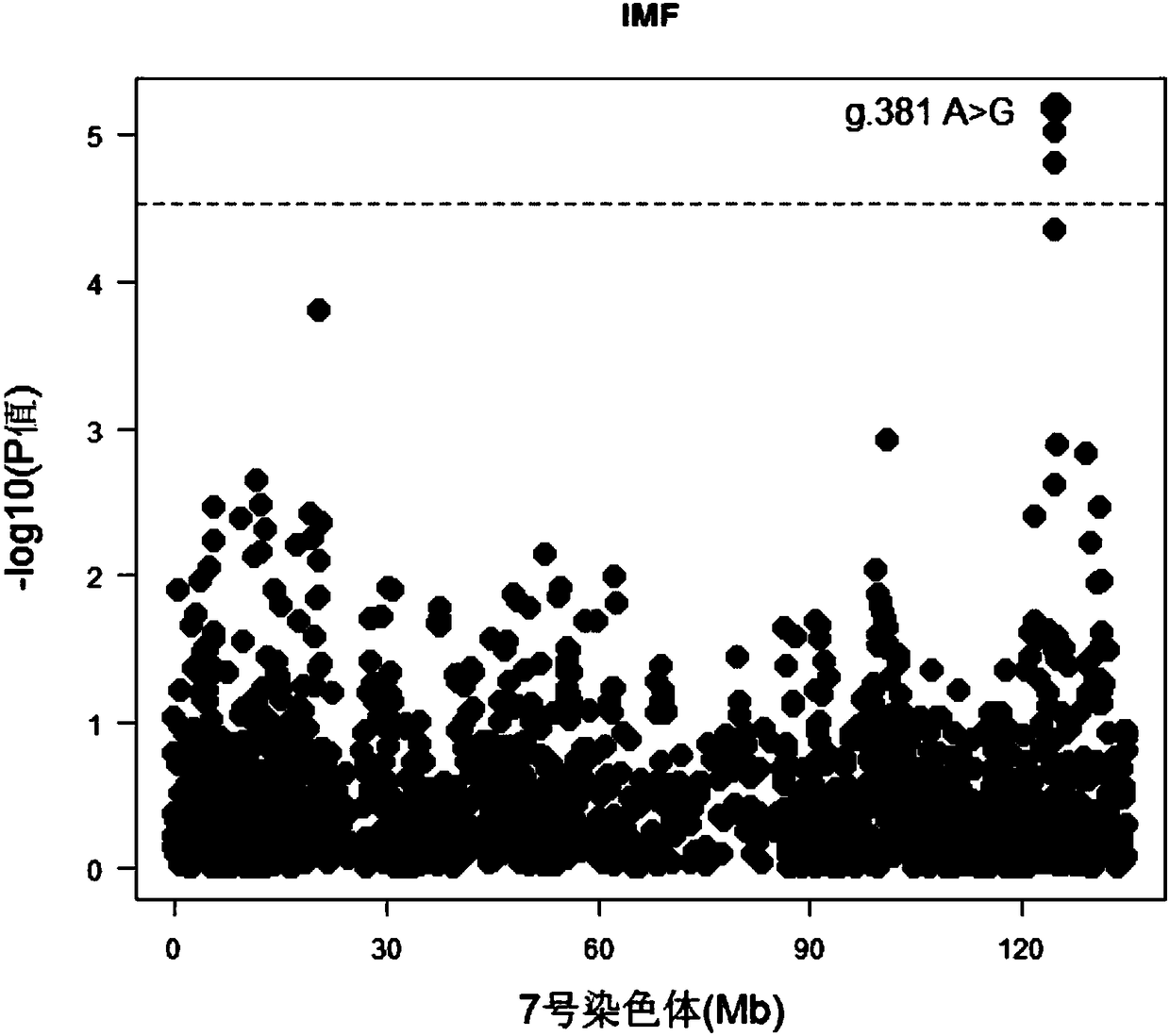
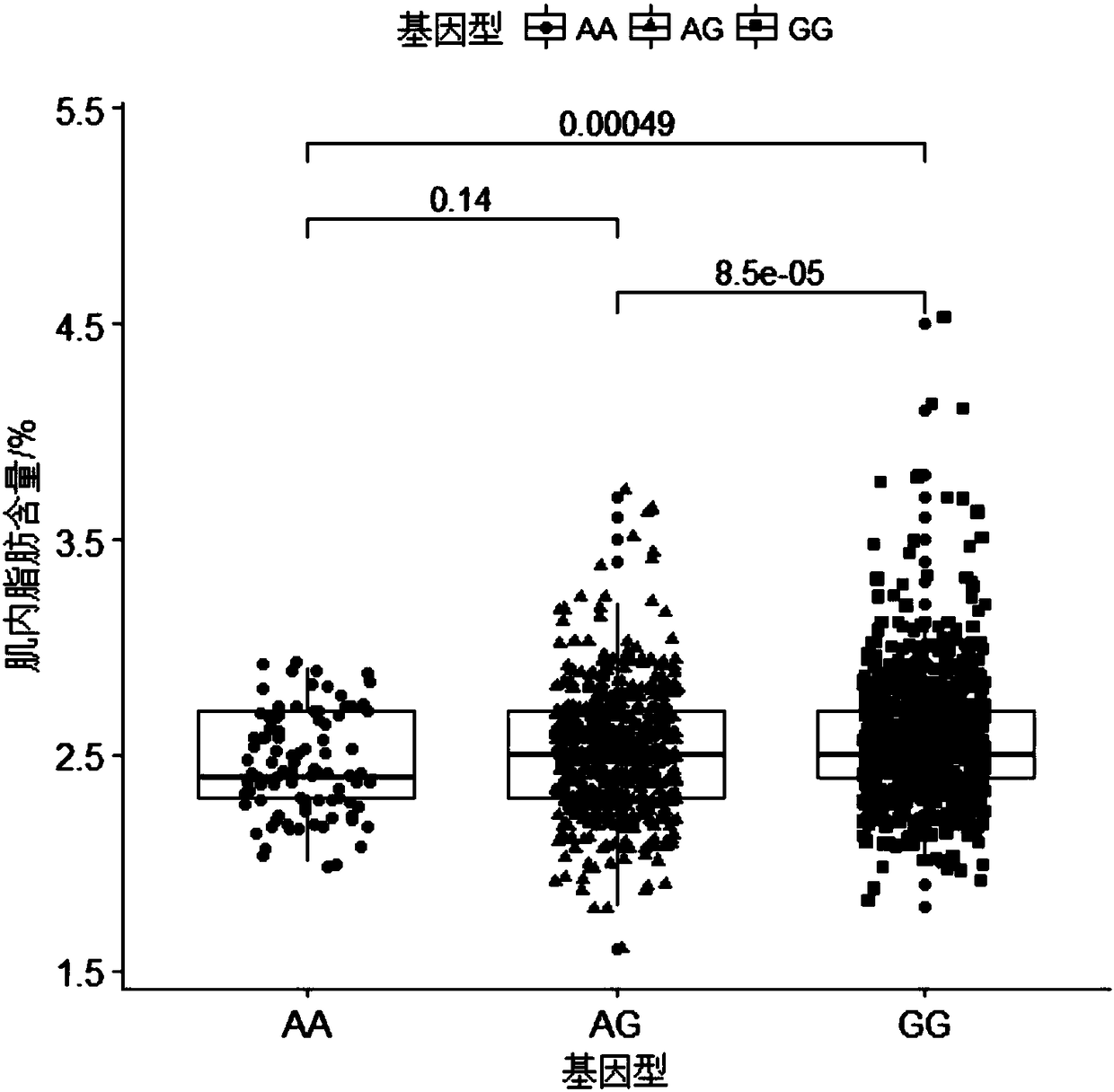
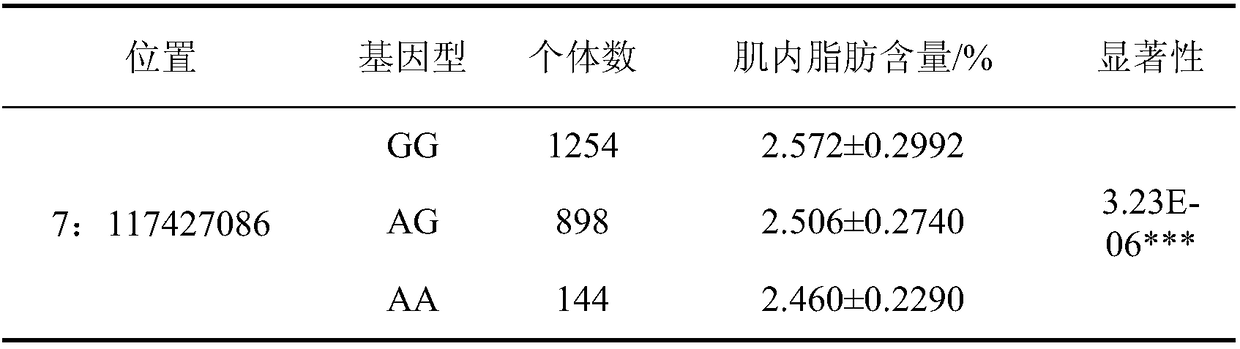
Molecular marker capable of influencing intramuscular fat content of Duroc boar and application
ActiveCN108315433AImprove meat qualityIncrease profitMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyIntramuscular fat
The invention belongs to the fields of molecular biological technology and molecular marker technology and relates to an SNP molecular marker related to intramuscular fat content of a Duroc boar. Thesite of the SNP marker is the 117427086th nucleotide site on the chromosome 7 of the International Pig Reference Genome 11.1 version, and the basic group of the site is G or A. By preferably selectingthe dominant allele of the SNP, the frequency of the dominant allele can be increased generation by generation, the intramuscular fat content of the boar can be increased, and the genetic improvementprogress of the boar can be accelerated, thereby effectively improving the economic benefit of boar breeding.
Owner:SOUTH CHINA AGRI UNIV
SNP mark combination for traceability and identification of beef cattle individuals and meat products and application of SNP mark combination
ActiveCN107201409ARich in polymorphic sitesHigh polymorphismMicrobiological testing/measurementAgricultural scienceFood safety
The invention discloses an SNP mark combination for traceability and identification of beef cattle individuals and meat products and application of the SNP mark combination. The SNP mark combination disclosed by the invention comprises 32 highly polymorphic loci distributed on 29 autosomes. The minimum allele frequencies of the combination loci are all greater than 0.35. The experiments show that the combination can be successfully applied to the identification of the beef cattle individuals except for yak breeds in the market and the traceability of the meat products and is conducive to the realization of the whole chain traceability of breeding-slaughtering-selling to really ensure food safety.
Owner:INST OF QUALITY STANDARD & TESTING TECH FOR AGRO PROD OF CAAS
29 micro-haplotype sites, screening method, multiplex amplification system and application
ActiveCN108504749AReduce dosageReduce difficultyMicrobiological testing/measurementDNA/RNA fragmentationTypingAllele frequency
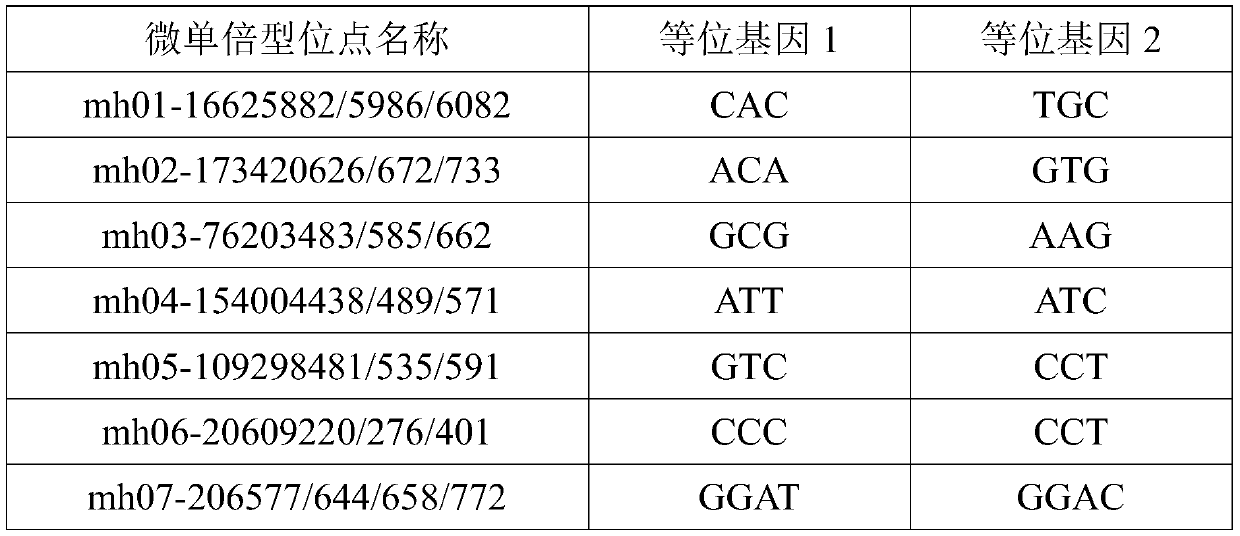
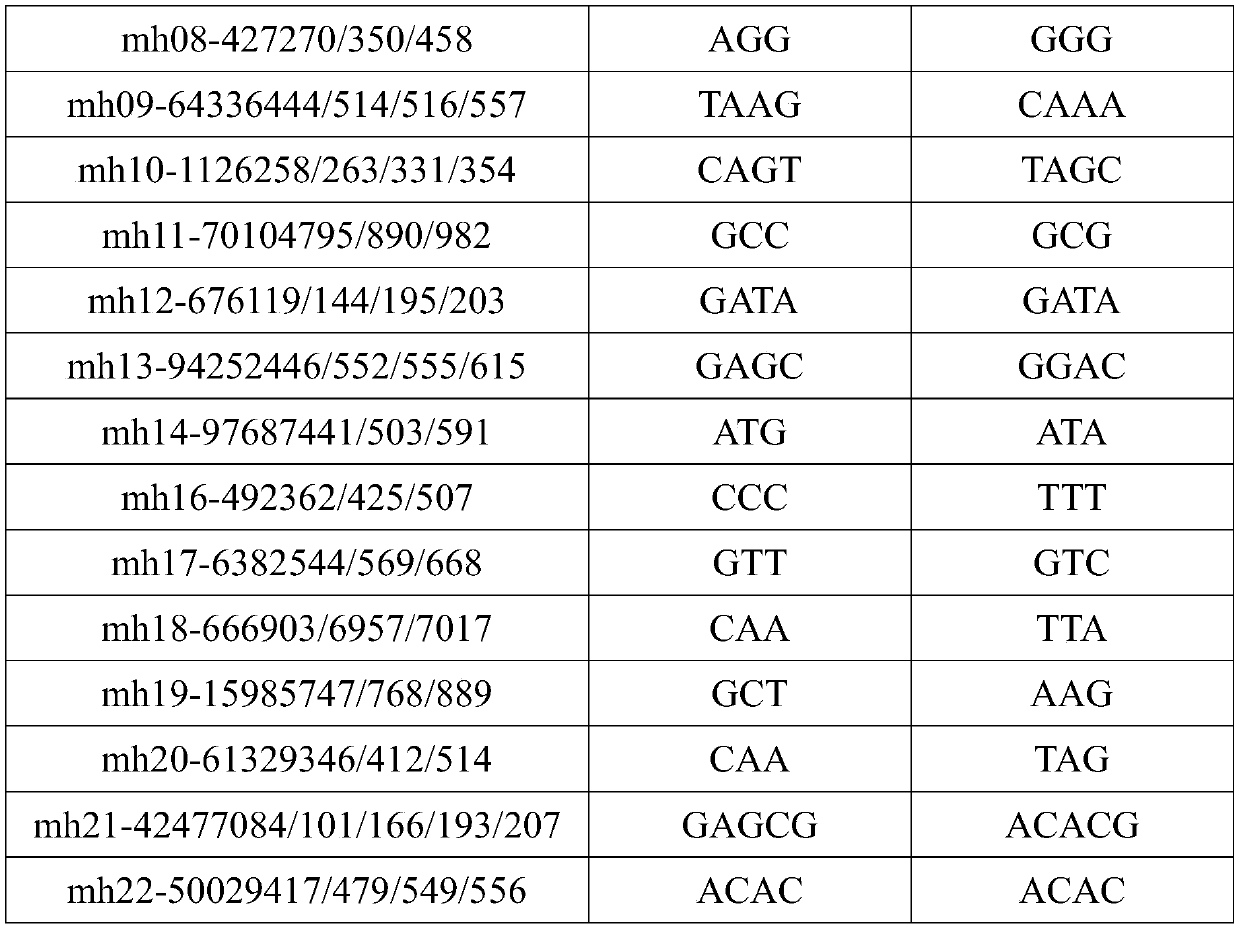
The invention discloses 29 micro-haplotype sites, a screening method, a multiplex amplification system and an application. The screening method comprises the following steps: acquiring population data, acquiring micro-haplotype genotypes and allele frequencies from the population data, calculating forensic related parameters of candidate sites in different populations and excluding sites affectingsubsequent sequencing typing to obtain the 29 micro-haplotype sites; designing primers and amplification conditions according to the 29 micro-haplotype sites, and establishing the multiplex amplification system based on three-round PCR amplification systems. According to the multiplex amplification system, DNA templates extracted from mixed biological samples are amplified, amplification resultsare subjected to large-scale parallel sequencing, value of the sequencing depth of each micro-haplotype site in the DNA templates is obtained, and finally, the purpose of typing the mixed biological samples is achieved. The multiplex amplification system is suitable for large-scale parallel sequencing technology and has the advantages of high detection flux, high detection speed, large quantity ofobtained data, more accurate detection results and the like.
Owner:NANJING MEDICAL UNIV
Method for judging gene type of fetus
ActiveCN104182655ALow costDeep sequencingSpecial data processing applicationsRelationship - FatherOriginal data
The invention relates to a method for judging the gene type of a fetus. The method comprises the following specific steps: A, extracting a DNA (deoxyribonucleic acid) sample of peripheral blood of a pregnant woman, and carrying out exon sequencing to obtain original data; B, carrying out quality control on the original data, comparing with a reference sequence, detecting SNP (Single Nucleotide Polymorphism), annotating and counting; C, based on four combination modes of the gene types of the pregnant woman and the fetus, calculating to obtain the Gaussian mixture model of the minimum gene frequency of SNP sites with the four combination modes by using a maximum expected value algorithm; D, calculating to obtain three critical values of the Gaussian mixture model; E, judging the gene type of the fetus by comparing the minimum gene frequency of each SNP site with the three critical values. According to the method for judging the gene type of the fetus disclosed by the invention, the mutation site of fetal gene can be calculated by means of the peripheral blood of the pregnant woman, the method is accurate and safe, DNAs from the father do not need, the sequencing depth is deeper, the cost is reduced, and the method is economic and practical.
Owner:SHANGHAI MAJORBIO BIO PHARM TECH
Wheat BSR-Seq gene locating method
ActiveCN106202995ALow costHigh precisionSequence analysisSpecial data processing applicationsReference genome sequenceAllele frequency
The invention discloses a wheat BSR-Seq gene locating method. The wheat BSR-Seq gene locating method comprises the following steps of: constructing and sequencing mixing pools, performing quality variation mining, screening a transcript tightly linked to a target gene, developing and locating a molecular marker, etc. The next-generation transcriptome sequencing technology (transcriptome sequencing, RNA-Seq) and a mixing pool technology (Bulked Segregant Analysis BSA) are combined; a wheat sequencing draft sequence is used as a reference sequence at first; then, a lot of high-quality SNP heritable variations on the transcript is mined at high throughput by adopting the next-generation sequencing technology; the allele frequency is precisely calculated in combination with the mixing pool technology, so that the transcript tightly linked to the target gene possibly can be rapidly screened out; false positive is precisely checked and controlled through Fish; the method is independent of a reference genomic sequence, low in cost, rapid and high in precision; the wheat gene locating efficiency and precision are increased; the wheat polymorphic molecular marker developing cost is reduced; therefore, the wheat gene fine locating working time is reduced to several months from several years; the locating precision is reduced to a fraction or 0 cM from several cM; and the fine locating cost is reduced to several thousands from several ten thousands.
Owner:北京麦美瑞生物科技有限公司
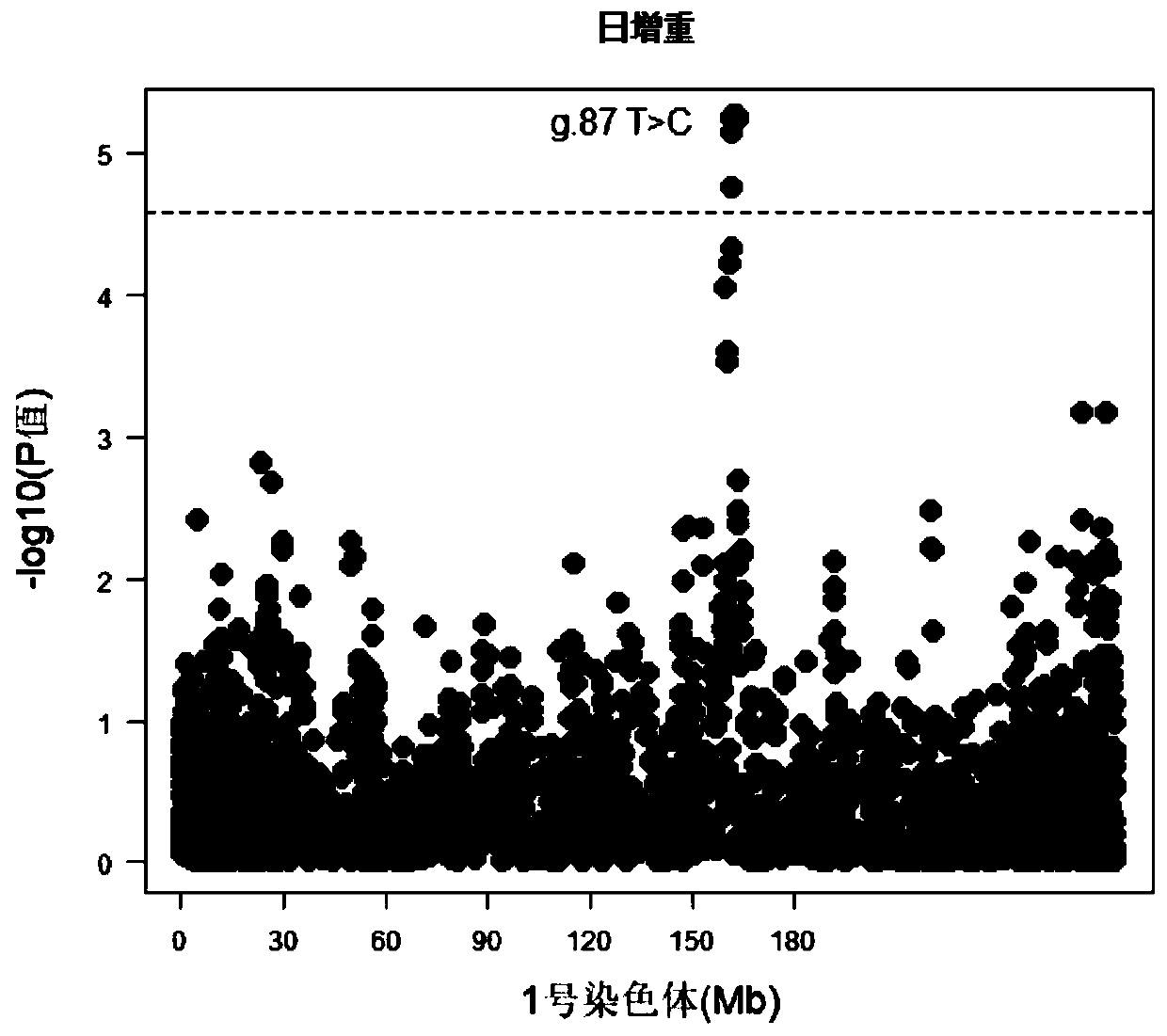
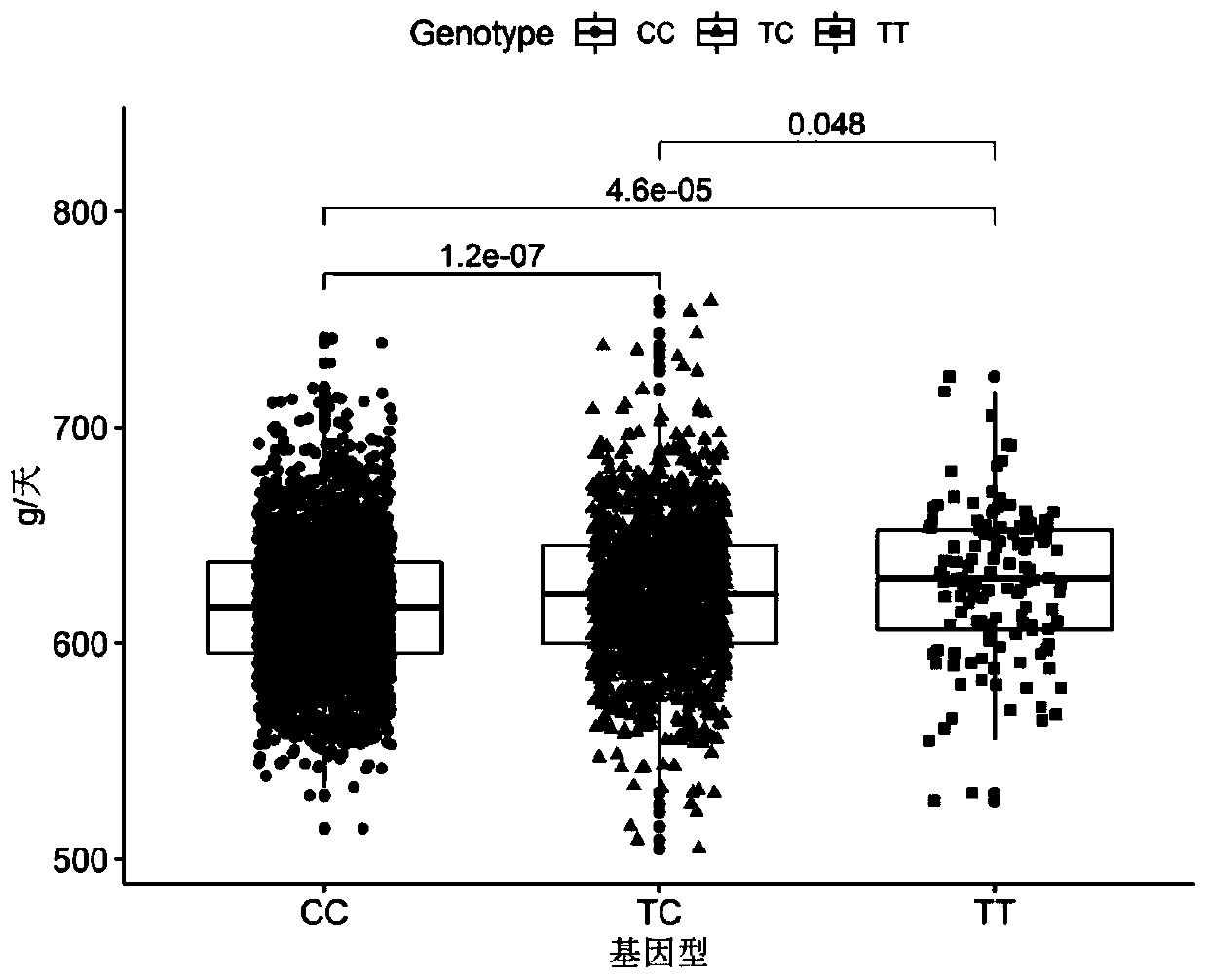
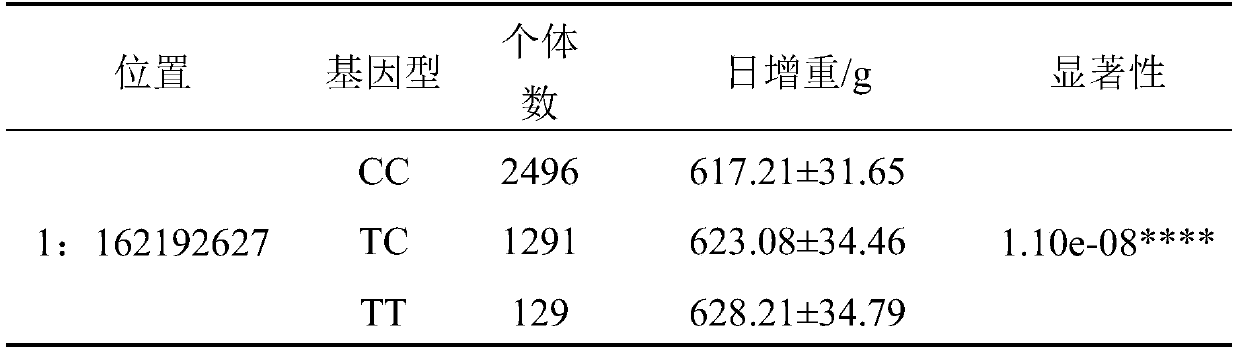
Single nucleotide polymorphisms (SNP) molecular marker located on porcine chromosome 1 and related to daily gain of pig and application of SNP molecular marker
InactiveCN110541038AIncrease daily weight gainEfficient and accurate breedingMicrobiological testing/measurementDNA/RNA fragmentationNucleotideAllele frequency
The invention relates to the fields of molecular biotechnologies and molecular marker technologies, in particular to a single nucleotide polymorphisms (SNP) molecular marker located on a porcine chromosome 1 and related to daily gain of a pig and application of the SNP molecular marker. The SNP site marked by the SNP molecular marker located on the porcine chromosome 1 and related to the daily gain of the pig corresponds to 162192627 T>C mutation on the chromosome 1 of an international porcine genome 11.1 version reference sequence. By optimizing a dominant allele of the SNP, the dominantallele frequency can be increased by generations, the daily gain of the pig is increased, pig genetic improvement development is accelerated, and thus economic benefits of pig breeding are effectivelyincreased.
Owner:SOUTH CHINA AGRI UNIV
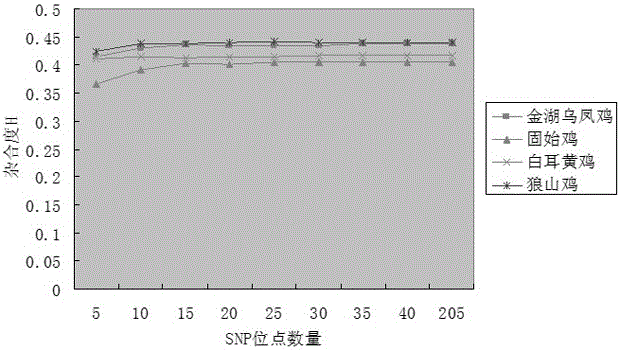
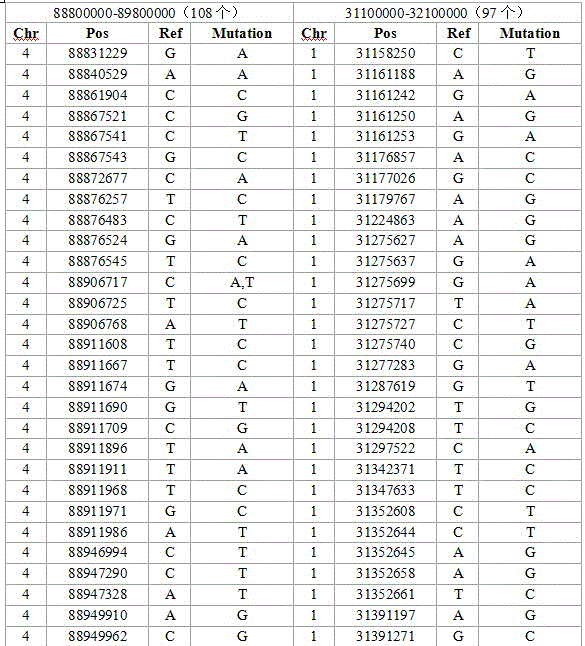
SNP mark screening method for monitoring breed conservation effect of poultry, application thereof to chicken breed conservation and authentication method of SNP mark
PendingCN106434867AImprove scienceImprove accuracyMicrobiological testing/measurementHigh densityAllele frequency
The invention relates to a high-density SNP molecular mark screening method for monitoring the breed conservation effect of poultry, application there to chicken breed conservation and an authentication method of an SNP molecular mark. Multiple breeds of poultry of the same kind are selected, multiple individuals are selected for each breed of poultry, and SNP screening includes the following steps that 1, all the breeds of poultry are shared, and each SNP site at least has two alleles in any breed; 2, distribution is relatively concentrated on a gene group, and the number of SNP sites within the interval of the gene group 1M is larger than 90; 3, genetic diversity is abundant, and the minimum allele frequency of each SNP site in any breed is larger than 0.1, and the SNP sites meeting the requirements of the step1, step2 and step3 are obtained through screening. A simplified gene group RAD-seq sequencing technology is utilized for sequencing chicken breeds at different places, and the uniform SNP sites which are distributed on different chicken gene groups in a concentrated mode and have abundant genetic diversity are authenticated through sequence comparison.
Owner:JIANGSU INST OF POULTRY SCI
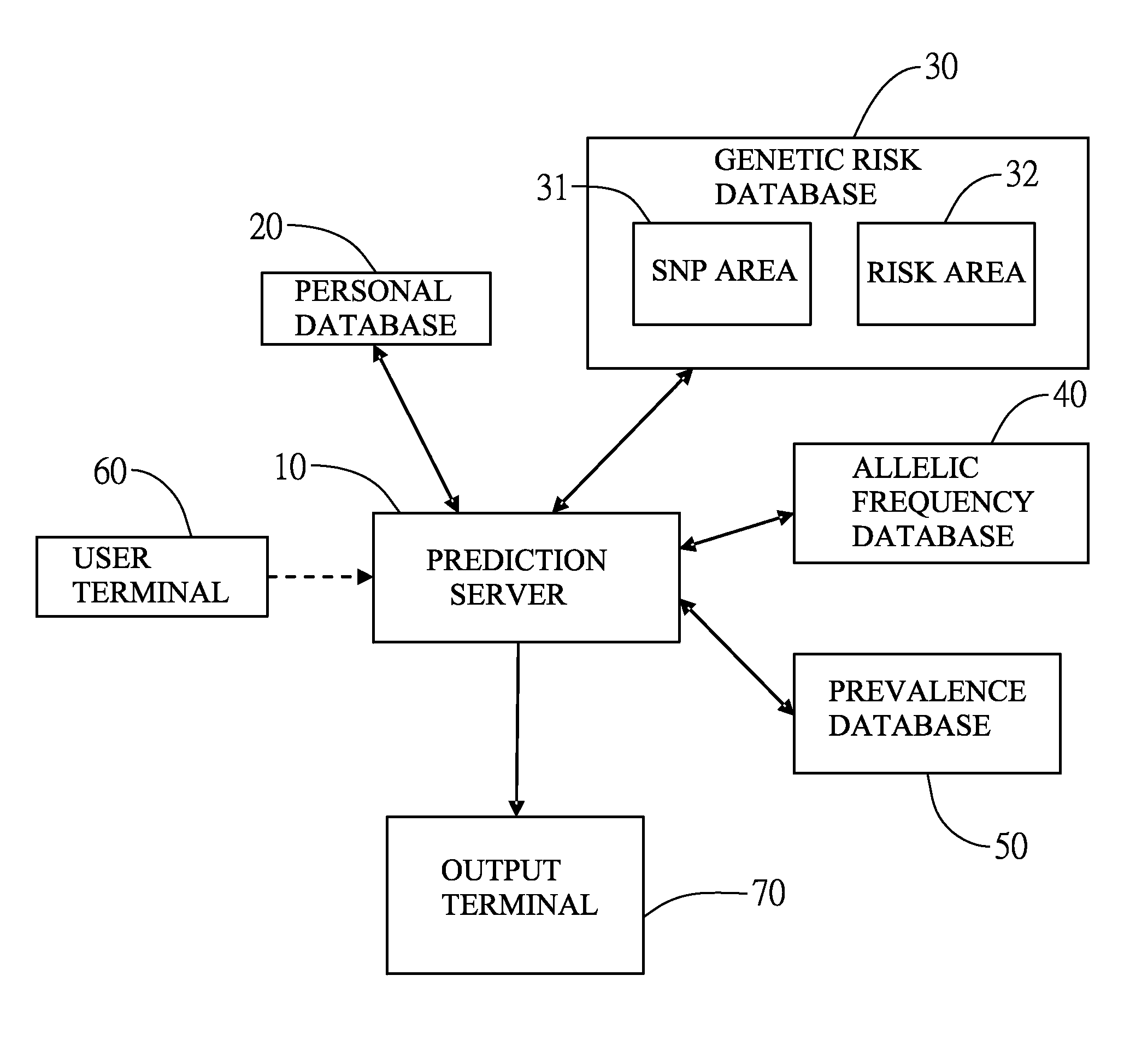
Method for forming personal nutrition complex according to incidence of disease and genetic polymorphism by a prediction system
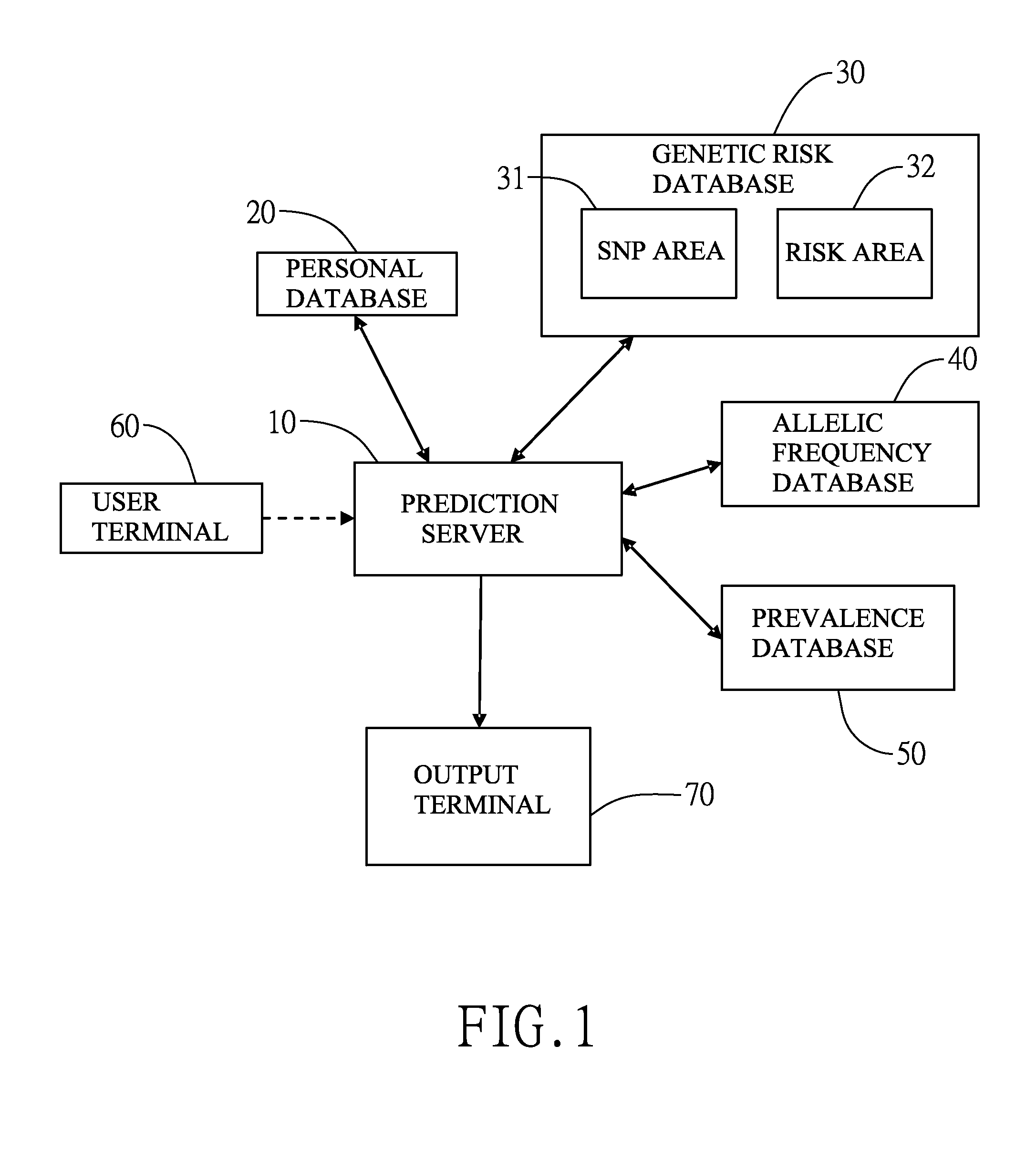
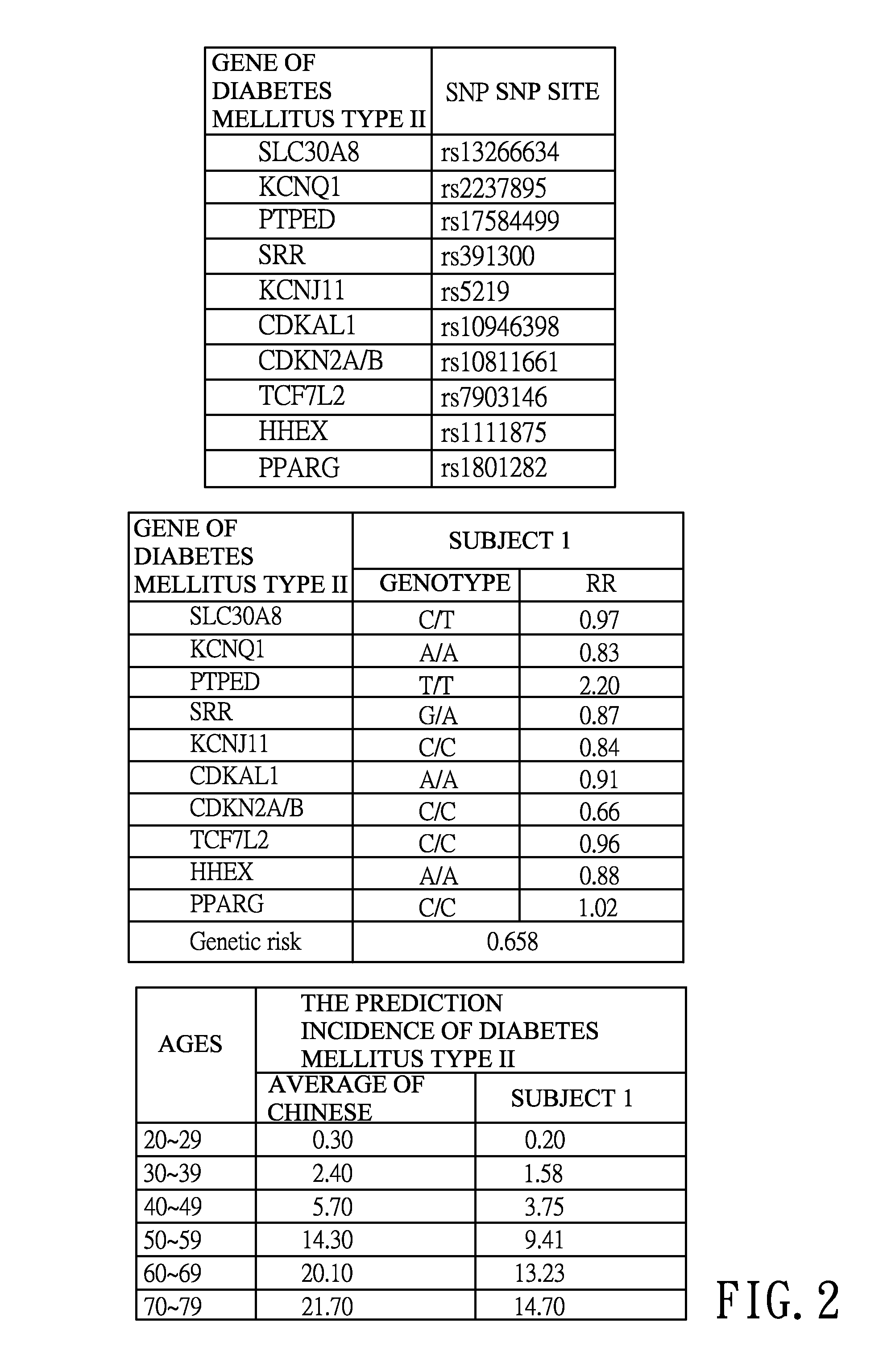
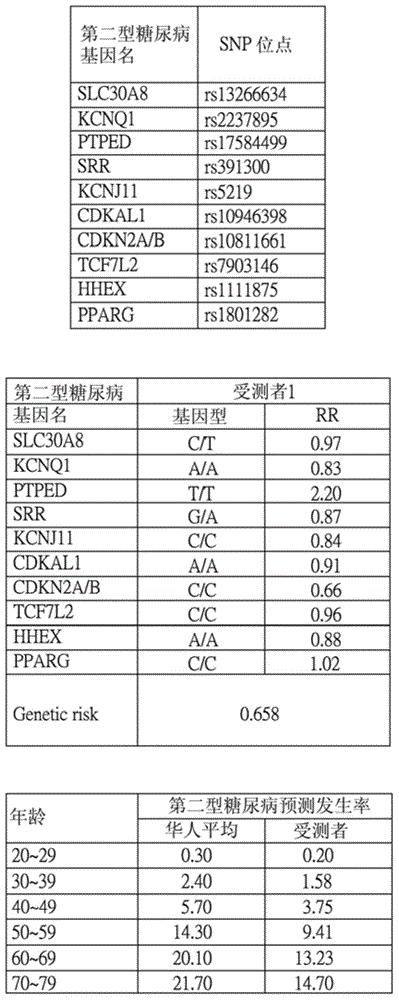
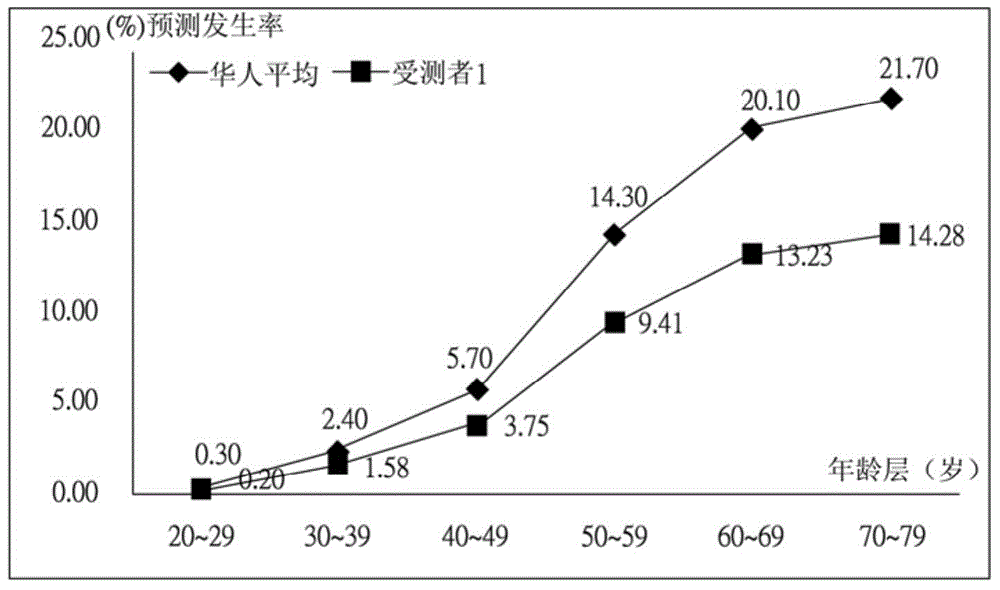
InactiveUS20160140288A1Convenient and efficientHealth-index calculationProteomicsAllele frequencyAdditive ingredient
The present invention relates to a system for predicting an incidence of disease from genetic polymorphism and uses the prediction result to form a personal nutrition complex. The system collects at least one personal information and single nucleotide polymorphism (SNP) information then exchanges the above information with databases including a personal database, a genetic risk database, an allelic frequency database, and a prevalence database. Finally, the system will output a prediction report and indicates a risk of specific disease and a plurality of abnormal genes. According to the prediction results, the system also can provide a plurality of nutritional supplement ingredients to form a personal nutrition complex. Users can receive a comprehensive and an effective nutritional supplement countermeasure about abnormal genes for prevention of the specific disease.
Owner:TCI GENE
Molecular marker identification method related to pig birth litter weight and fetal weight and application of molecular marker identification method
ActiveCN106011245AMicrobiological testing/measurementDNA/RNA fragmentationAllele frequencyFetal weight
The invention discloses a molecular marker identification method related to pig birth litter weight and fetal weight and an application of the molecular marker identification method. The identification method comprises the following steps: collecting a sample; extracting and detecting pig genome DNA; then, conducting SNP (single nucleotide polymorphism) chip genotype judgment and genotype data quality control: adding a PCR amplification primer pair and conducting a PCR amplification reaction by taking DNA of the sample as a template, purifying a reaction product and co-crystallizing the purified reaction product with a mass spectrum chip, conducting detection and typing by virtue of a flight mass spectrum method, detecting an SNP site of a target gene, computing an SNP genotype and an allele frequency by virtue of PopGene3.2, and conducting association analysis on the SNP and characters by virtue of SAS9.2 software; and finally, conducting data collating and analysis. The identification method disclosed by the invention, by determining the genotype of a pig individual, can assist identification of multiparous litter weight and fetal weight.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI +1
Preparation method of tumor sample sequencing reference
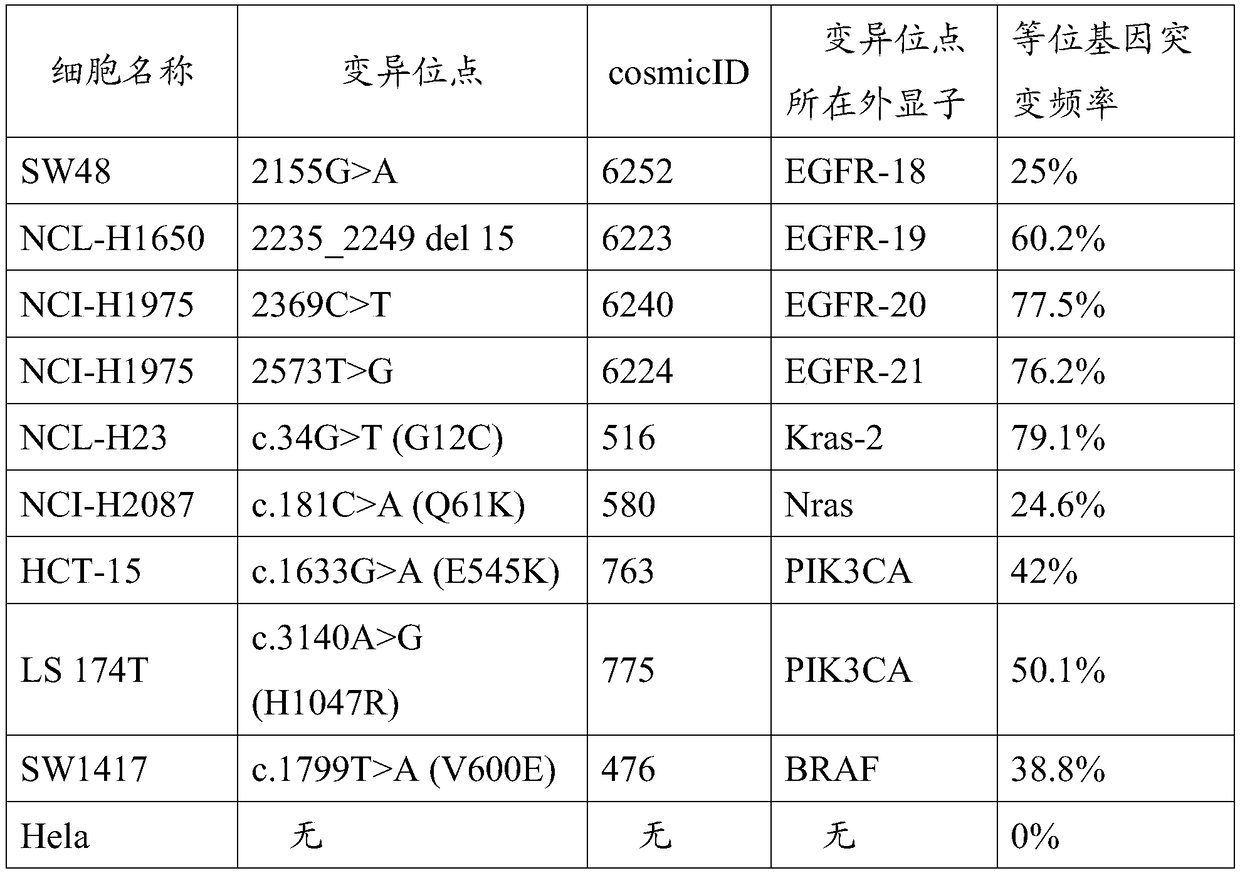
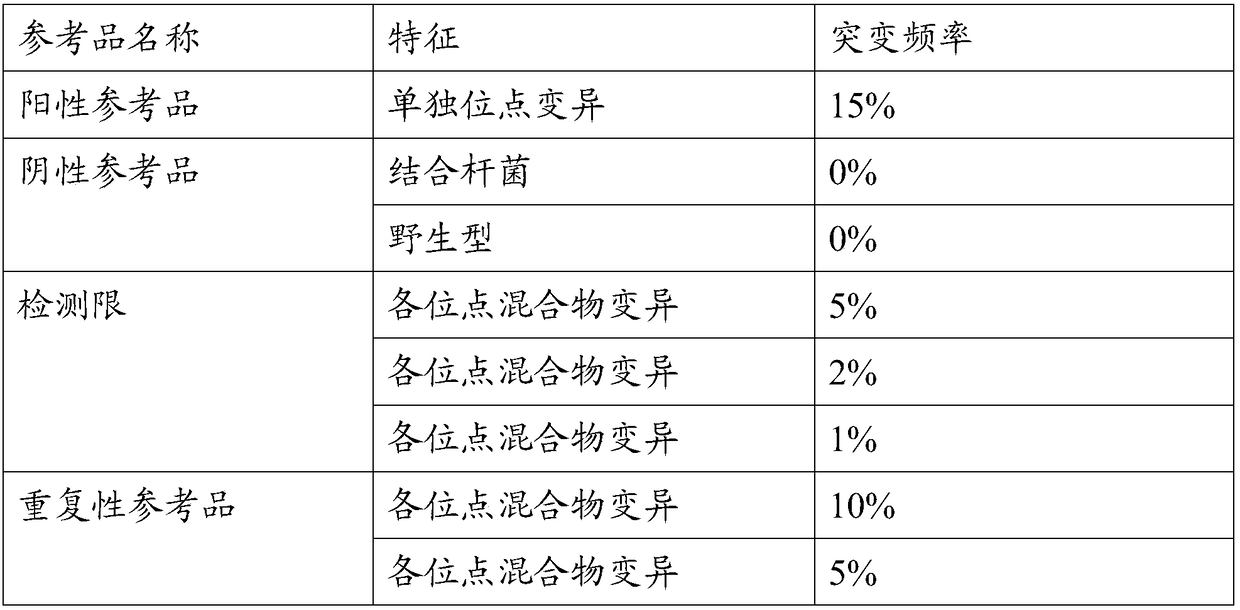
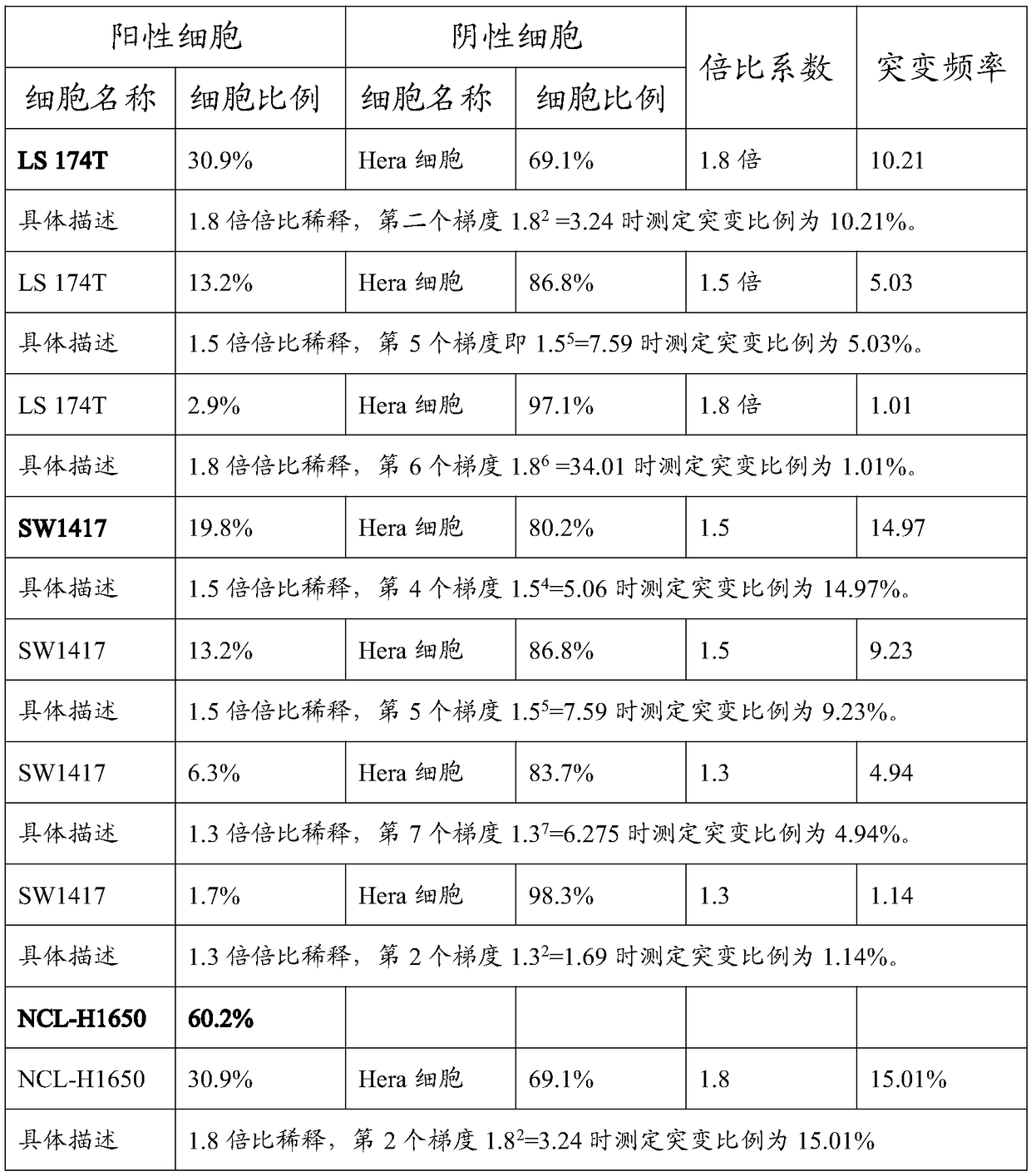
ActiveCN108728516AClear genetic backgroundAccurate countMicrobiological testing/measurementAllele frequencyMutation frequency
The invention provides a preparation method of a tumor sample sequencing reference, and belongs to the technical field of high-throughput sequencing. The method comprises the following steps: performing monoclonal culture on a tumor cell line to obtain a monoclonal cell line and extracting a whole genome; determining an allele frequency; determining a dilution multiple according to an allele mutation frequency and a target allele mutation frequency; diluting the monoclonal cell line with negative cells in multiple proportion to obtain mixed cells; extracting a whole genome of the mixed cells,and determining an allele mutation frequency; and according to the comparison between the allele mutation frequency and the target allele mutation frequency of the mixed cells, refining the multiple of multiple-proportion dilution, and diluting the monoclonal cell line with negative cells to obtain the reference. According to the method disclosed by the invention, the gene background of a cell line is determined by combination of cell strain monoclone and high-throughput sequencing; and a multiple-proportion dilution method with multi-step gradual refinement is combined with a flow cytometry to accurately count cells so as to ensure high repeatability of reference preparation.
Owner:安徽鼎晶生物科技有限公司 +1
Tumor burden as measured by cell free DNA
InactiveUS20180282417A1Good chanceLong survivalMicrobiological testing/measurementImmunoglobulins against cell receptors/antigens/surface-determinantsCell freeAllele frequency
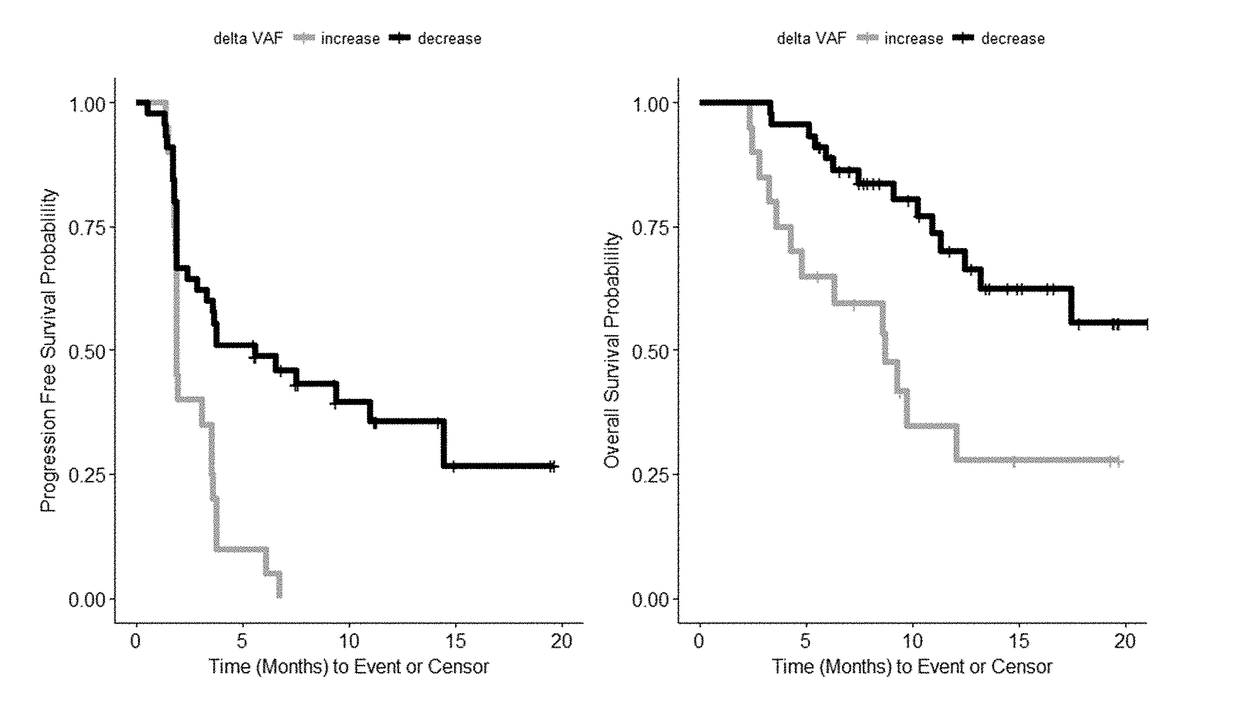
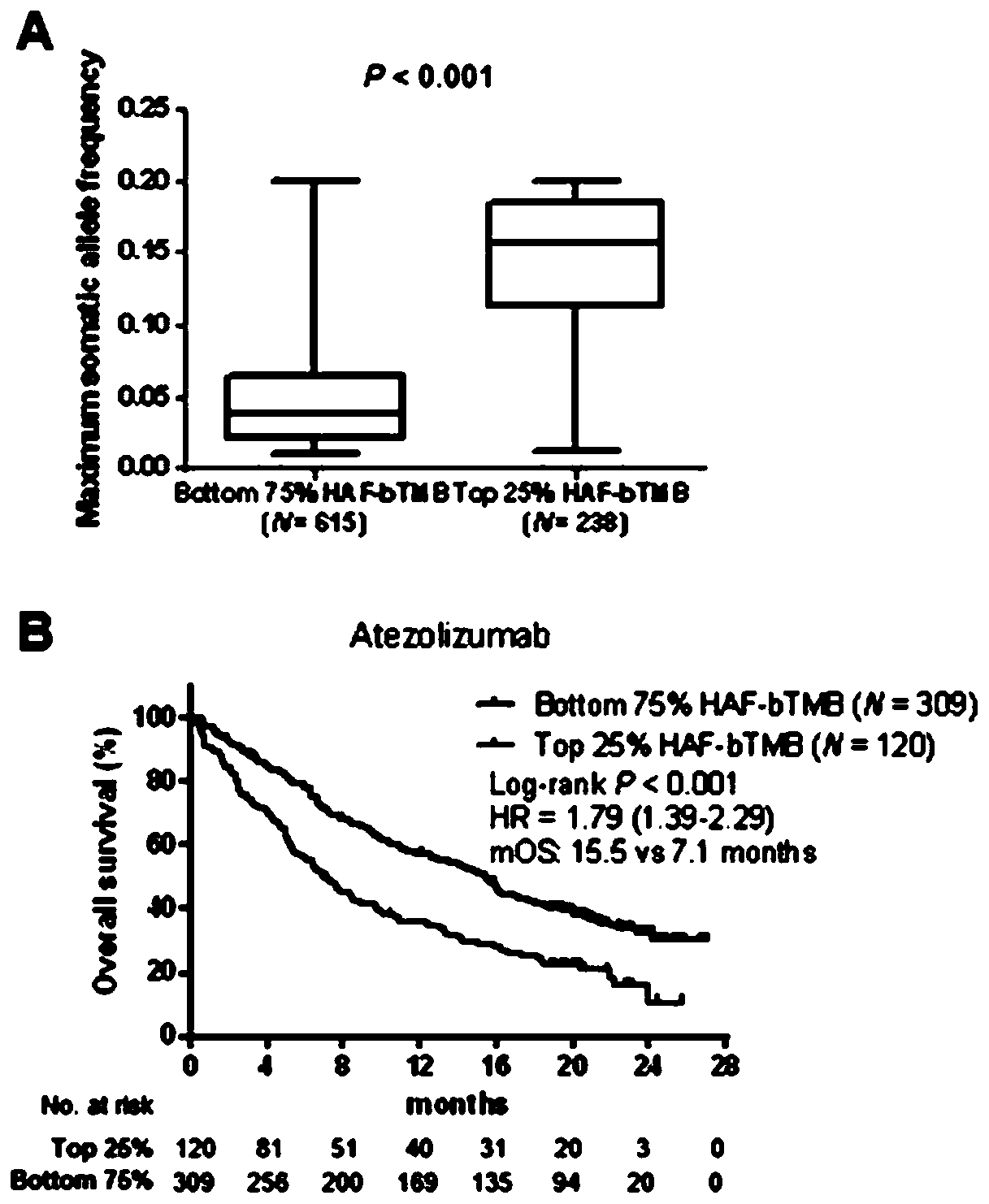
Disclosed are methods for treating cancer (e.g., solid tumor cancers, lung cancer, bladder head and neck cancer) with an anti-PD-L1 antibody in a patient identified as being responsive to anti-PD-L1 antibody therapy by detecting a mutation in one or more disclosed circulating tumor DNA (ctDNA) markers. Also disclosed are methods for determining the efficacy of anti-PD-L1 therapeutic antibody treatment in a patient having lung cancer or bladder cancer comprising detecting variant allele frequency in ctDNA in plasma samples and determining the difference of the variant allele frequency in ctDNA between the first and at least second plasma samples, wherein a decrease in the variant allele frequency in the at least second plasma sample relative to the first plasma sample identifies the anti-PD-L1 antibody treatment as effective. The disclosure also provides methods of identifying a subject having a cancer responsive to a therapy comprising an anti-PD-L1 antibody by detecting the expression of a mutation in one or more circulating tumor DNA (ctDNA) markers.
Owner:MEDIMMUNE LLC
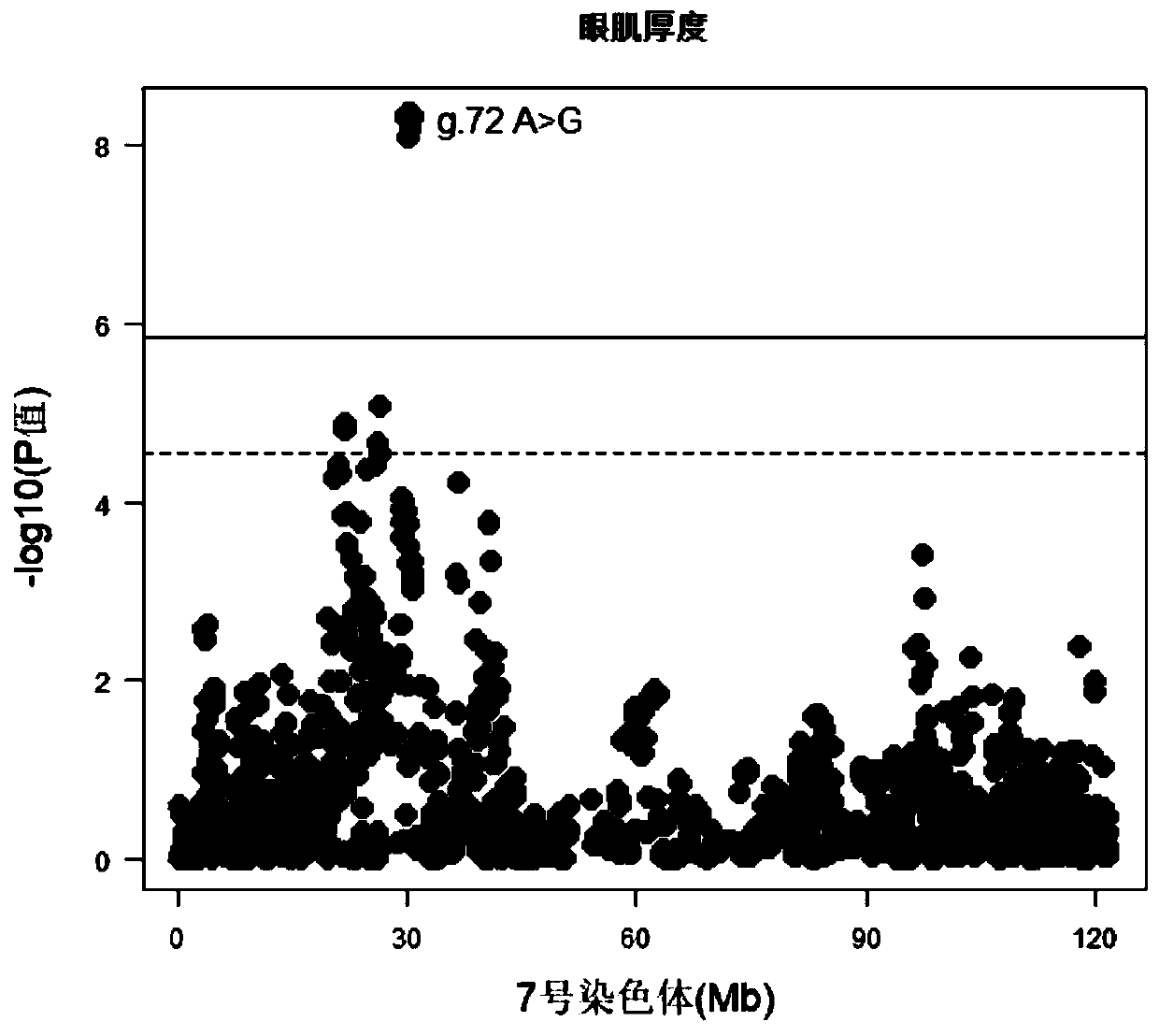
SNP molecular marker which is positioned on No.7 porcine chromosome and related to area and thickness of eye muscles and application
ActiveCN110218798AIncrease the areaIncreasing the thicknessMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAllele frequency
The invention belongs to the technical field of molecular biotechnology and molecular markers and particularly relates to an SNP molecular marker which is positioned on a No.7 porcine chromosome and related to the area and thickness of the eye muscles and application. An SNP locus of the SNP molecular marker positioned on the No.7 porcine chromosome and related to the area and thickness of the eyemuscles corresponds to a mutation (A>G) of the 30342161st site of the No.7 chromosome of an international porcine genome 11.1 version reference sequence. The invention also provides a primer pair foridentifying the molecular marker. Through the molecular marker and the primer pair, dominant alleles of SNP can be screened generation by generation, the frequency of the dominant alleles is increased, the area and thickness of the eye muscles of a pig are increased, the economic benefit of pork is finally increased, and the molecular marker and the application have very important significance inthe progress of genetic improvement of pigs.
Owner:SOUTH CHINA AGRI UNIV
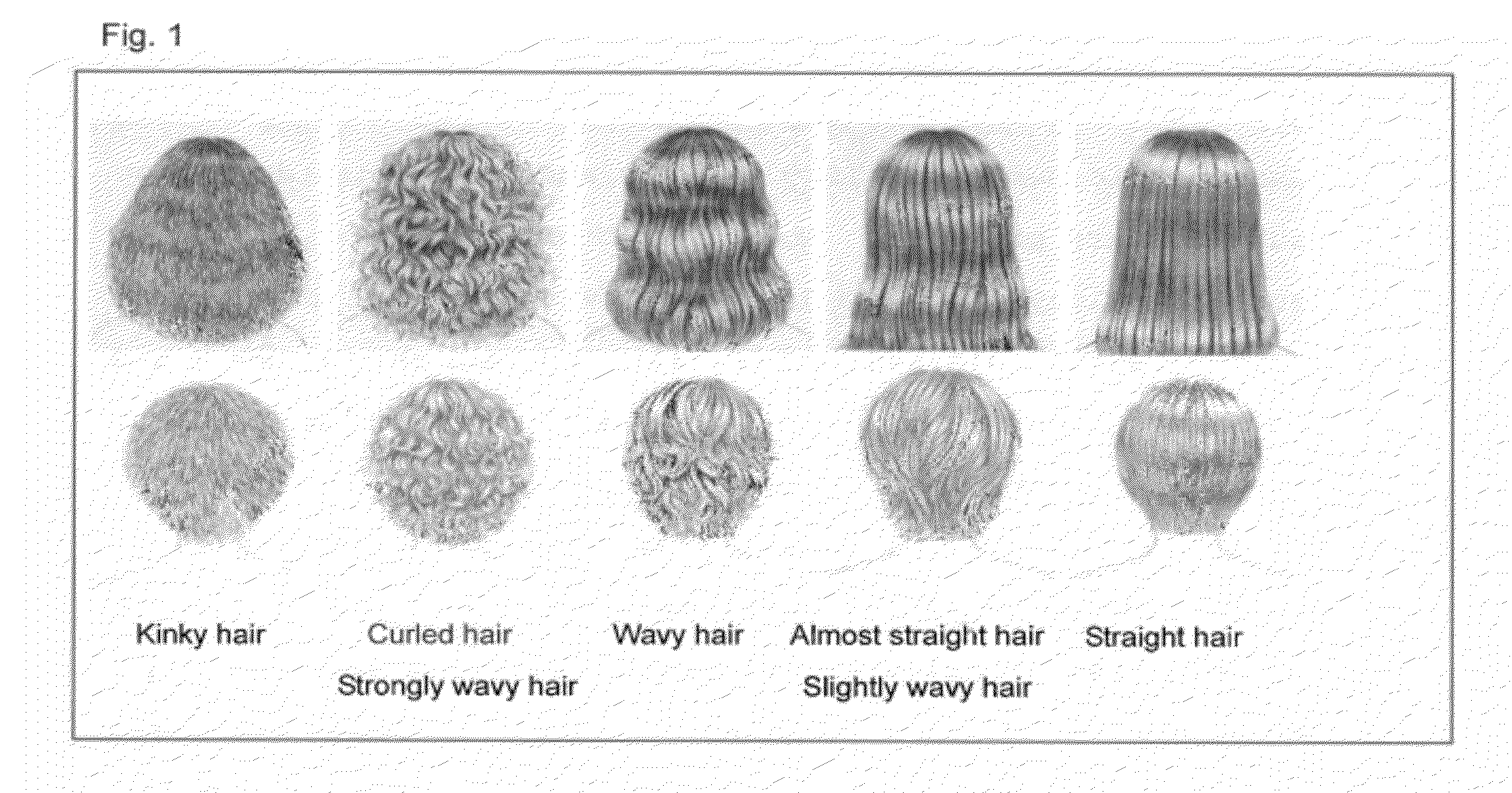
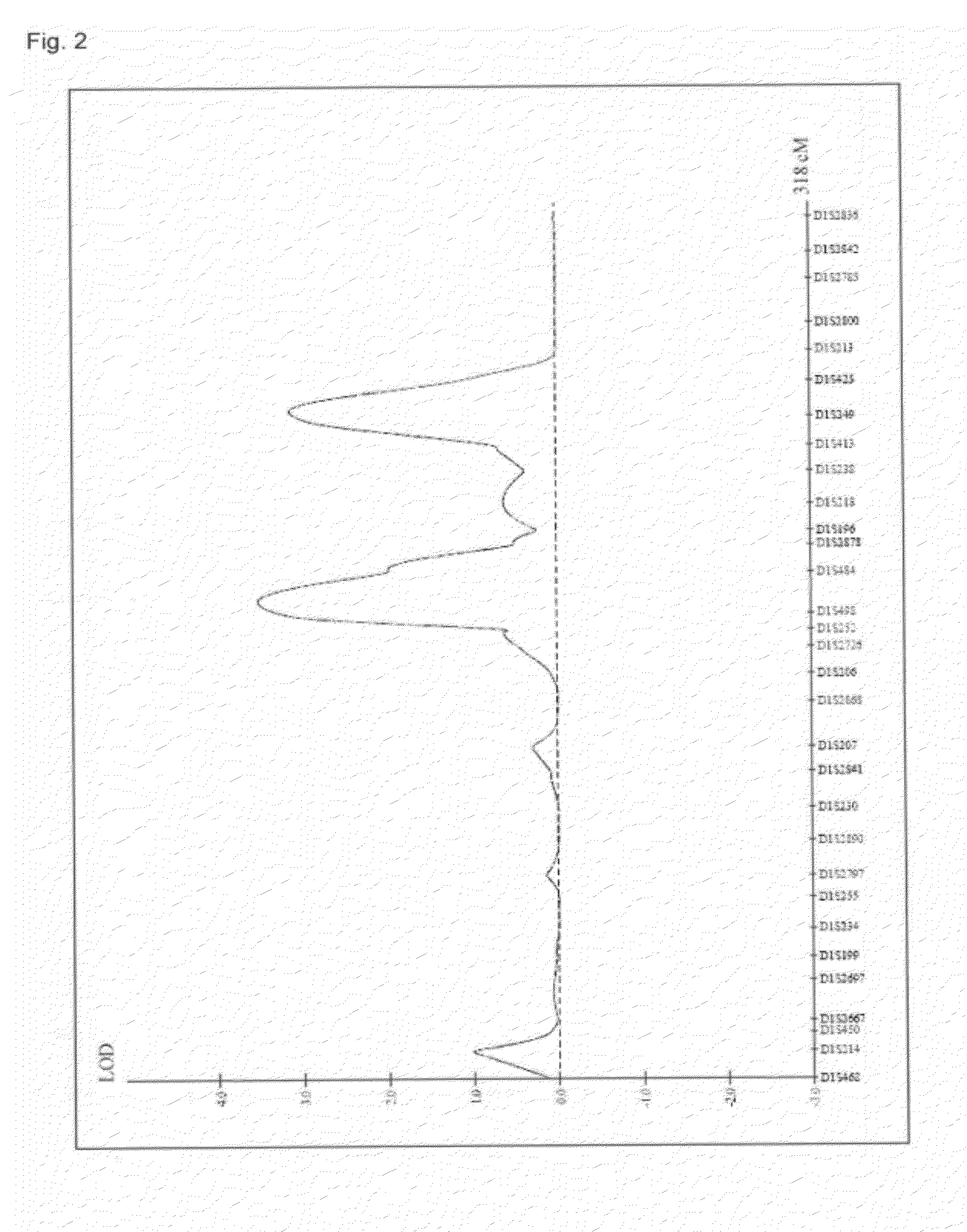
Hair Shape Susceptibility Gene
InactiveUS20120329726A1Cosmetic preparationsPeptide/protein ingredientsAllele frequencyGenetic linkage disequilibrium
A genetic polymorphism and a hair shape susceptibility gene that are related to hair shape, and a method for determining the genetic susceptibility to hair shape in individual test subjects are provided. Disclosed is a hair shape susceptibility gene, which overlaps with a haplotype block in 1q21.3 region (D1S2696 to D1S2346) of human chromosome 1 and comprises a portion or the entirety of the base sequence of the haplotype block, wherein the haplotype block is determined by a linkage disequilibrium analysis conducted on a single nucleotide polymorphism (SNP) marker whose allele frequency differs statistically significantly between a group having a curly hair trait and a group having a non-curly hair trait, and consists of a base sequence set forth in any one of SEQ ID NO: 1 to NO: 5.
Owner:KAO CORP
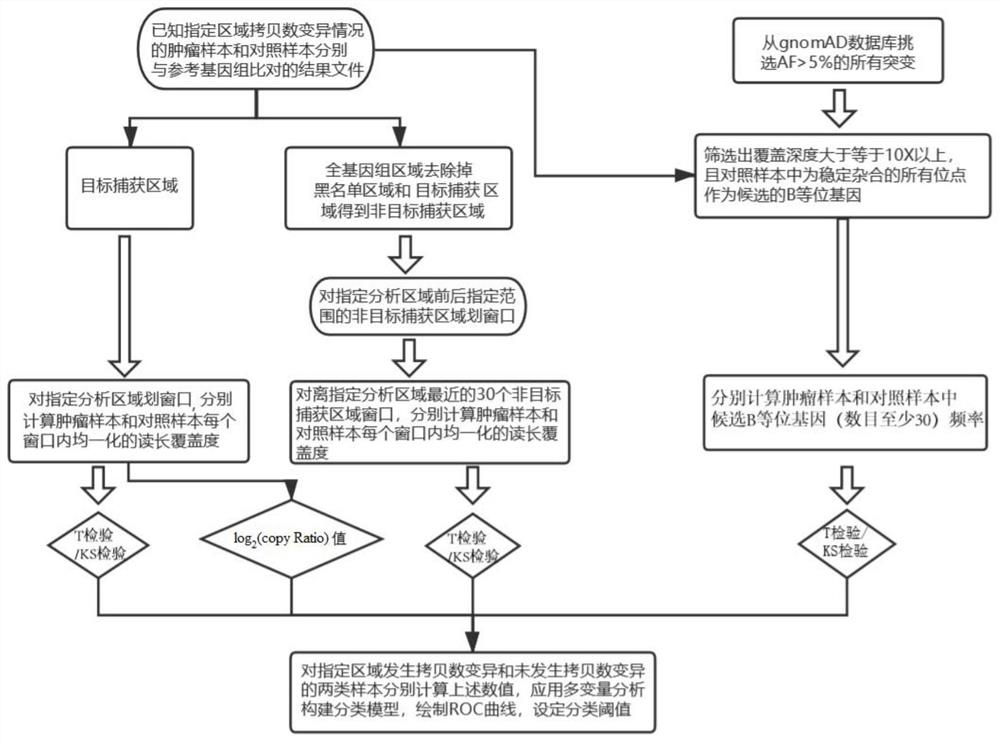
Method and device for detecting somatic cell copy number variation in specified genome region
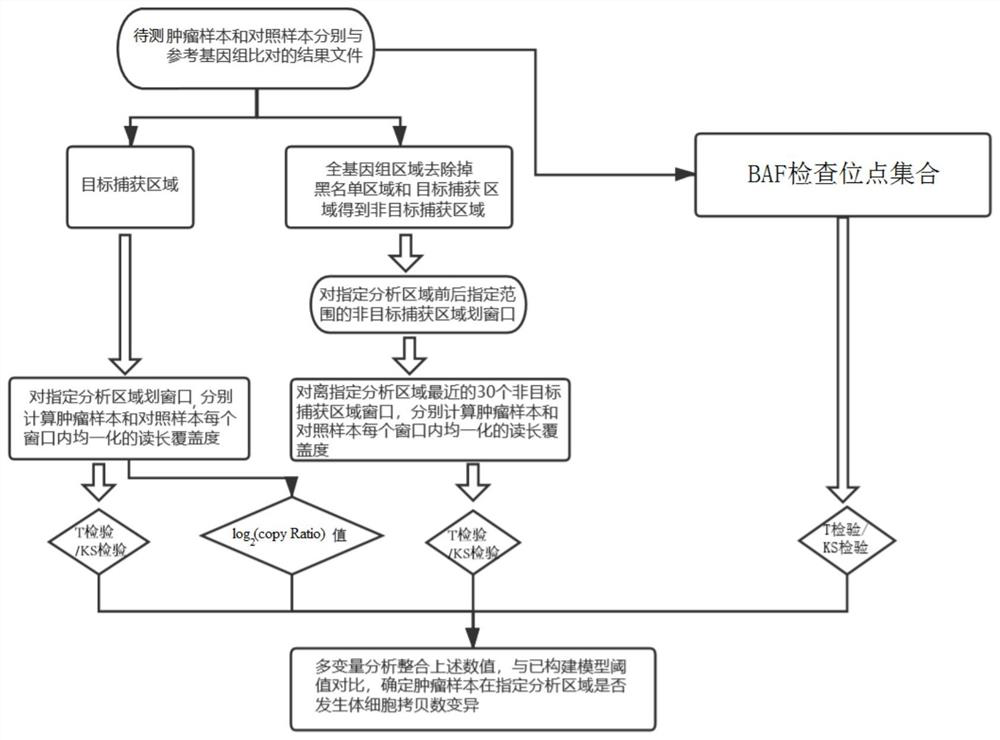
ActiveCN111968701AThe test result is accurateEfficient detectionProteomicsGenomicsTarget captureAllele frequency
The invention relates to the field of genes, in particular to a method and a device for detecting somatic cell copy number variation in a specified genome region. According to the method, a pluralityof indexes including a log2 (copy Ratio) value, B allele frequency (BAF) difference significance, difference significance of homogenized read length coverage in a target capture area and difference significance of homogenized read length coverage in a non-target capture area are integrated, and multivariable analysis is applied to synthesize a plurality of index analysis results, so efficient andaccurate copy number variation detection is carried out on a specified gene or interval.
Owner:BEIJING GENEPLUS TECH +1
Multiplex amplification system, and next generation sequencing typing kit and typing method for 21 micro-haplotype sites
ActiveCN110218781AShorten the lengthLow mutation rateMicrobiological testing/measurementSequence analysisStr typingAllele frequency
The invention relates to the technical field of molecular biology, and especially relates to a multiplex amplification system, and a next generation sequencing typing kit and typing method for 21 micro-haplotype sites. The 21 micro-haplotype sites in the multiplex amplification system are derived from 21 autosomes, are balanced in allele frequency, are mutually independent between the sites, and have the advantages of polymorphism and low mutation rate. The kit contains 53 PCR-amplified single-end specific primers shown in SEQ ID NO. 1-SEQ ID NO.53, and can detect the 21 microhaplotype sites in a same reaction system. The multiplex amplification system, the kit and the typing method provided by the invention can solve the technical problem that a current STR typing technology cannot obtainan ideal typing result, and allele loss may occur due to excessive number of SNP sites.
Owner:HEBEI MEDICAL UNIVERSITY
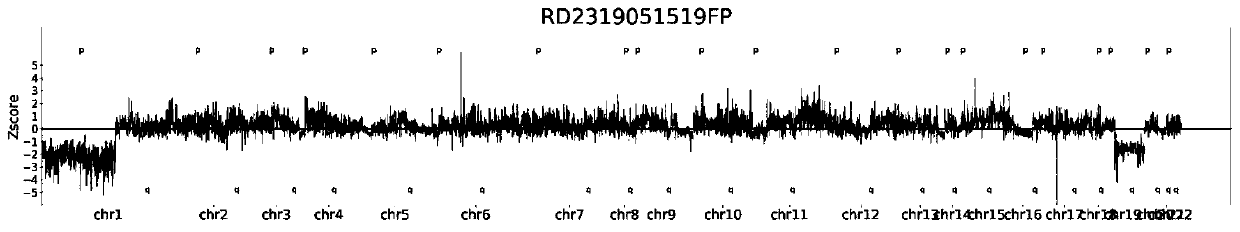
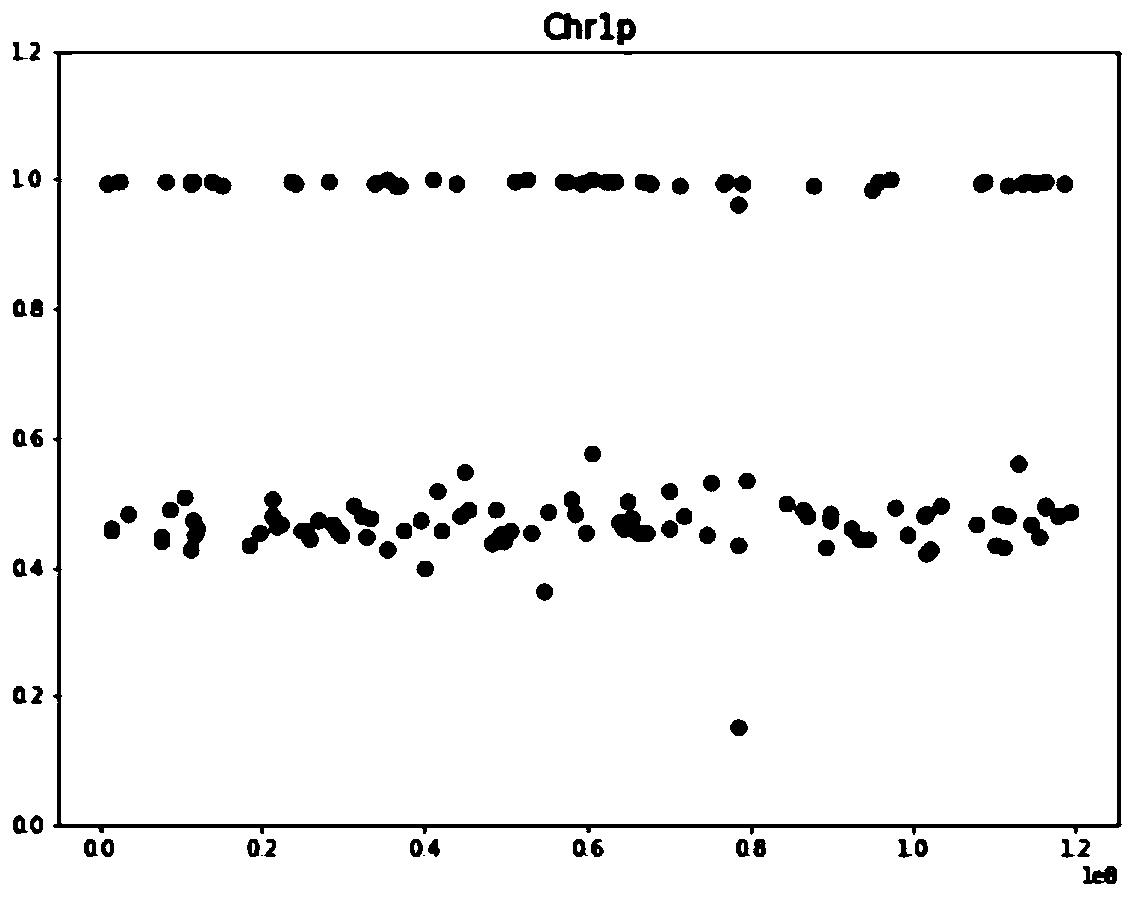
Chromosome aneuploidy analysis method and chromosome aneuploidy analysis system
ActiveCN111091868AImprove detection accuracyReduce the impact of the result judgmentBiostatisticsProteomicsStatistical analysisAllele frequency
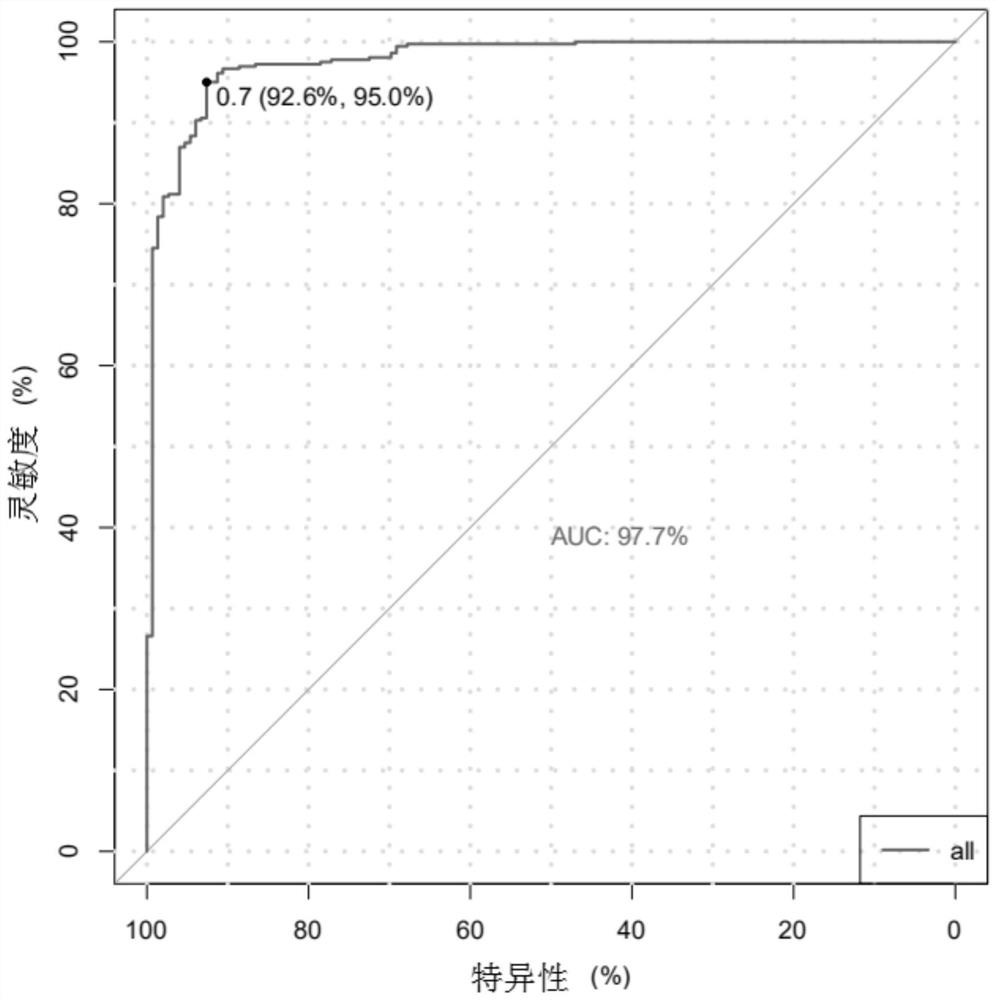
The invention provides a chromosome aneuploidy analysis method and a chromosome aneuploidy analysis system. According to the method, sample sequencing depth is subjected to statistical analysis basedon a Zscore algorithm, and the condition of chromosome aneuploidy is comprehensively judged in combination with statistical analysis of SNP allele frequency of samples. The method provided by the invention is high in accuracy, is not influenced by the SNP locus number of a sample to be detected, has high specificity and sensitivity (up to 100%), and can also be used for judging whether abnormal chromosomes are deleted or amplified.
Owner:SIMCERE DIAGNOSTICS CO LTD +2
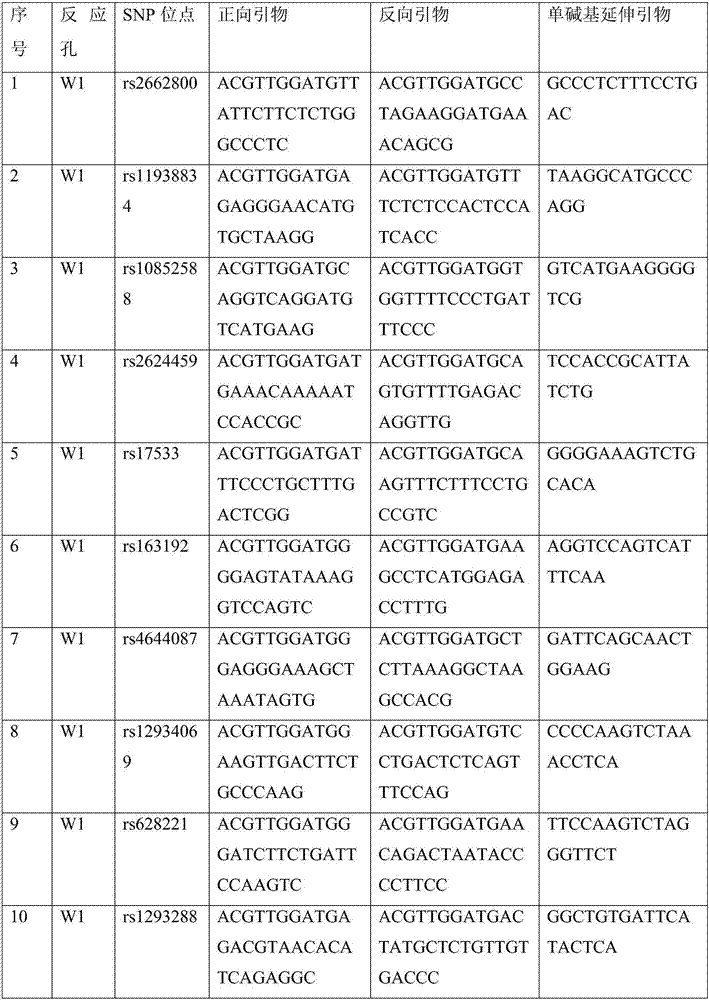
Genetic marker for human individual recognition and/or paternity identification and detection method thereof and kit
ActiveCN107541554ABroad multi-racial adaptabilityThe test result is accurateMicrobiological testing/measurementDNA/RNA fragmentationAllele frequencyCommercial kit
The invention discloses a genetic marker combination for human individual recognition and / or paternity identification and a detection method thereof. By conducting whole-genome SNPs unbiased scanningon multiple races, a SNPs site combination widely applicable to the multiple races is screened, and 116 autosome SNPs sites which come from 37 groups in different regions in the whole world, have thehypermorph allel frequency and low difference and achieve independent inheritance and 12 X chromosome SNPs sites which come from 37 groups in different regions in the whole world, have the hypermorphallel frequency and low difference and achieve independent inheritance are screened from 25,580,678 SNPs sites in a genome-wide scale. Compared with an existing commercial kit, the SNPs site combination has higher and wider multiracial adaptation, the cumulative individual recognition probability and the cumulative probability of exclusion are both significantly superior to those of the commercialkit, the detection result is more accurate, and a great application prospect is achieved.
Owner:SUN YAT SEN UNIV
Method for generating SSR molecular identification numbers of fruit trees based on SSR genotypes
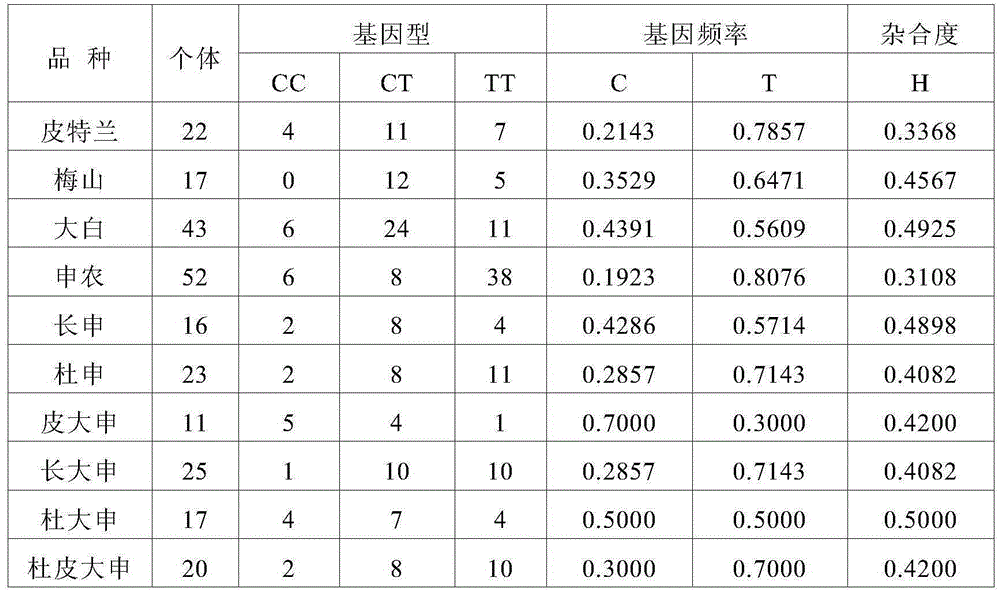
The invention provides a method for generating SSR molecular identification numbers of fruit trees based on SSR genotypes. The method comprises the following steps: screening SSR primers for fruit tree variety amplification and performing PCR amplification on the fruit tree varieties; after performing electrophoretic detection on the PCR amplification product, counting the genotypes of the fruit tree varieties at various SSR sites, arranging all the genotypes at various SSR sites in an ascending order before assignment, marking the non-deficient genotype as 00, and numbering the genotypes in order from 01 to 99, and then sequentially connecting the genotype numbers of each variety at various SSR sites in a certain SSR site order based on the standard, thereby forming the SSR molecular identification number of the fruit tree variety. The method takes the difference of the allele frequencies of various SSR sites due to such factors as selection, mutation and drift into full account.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
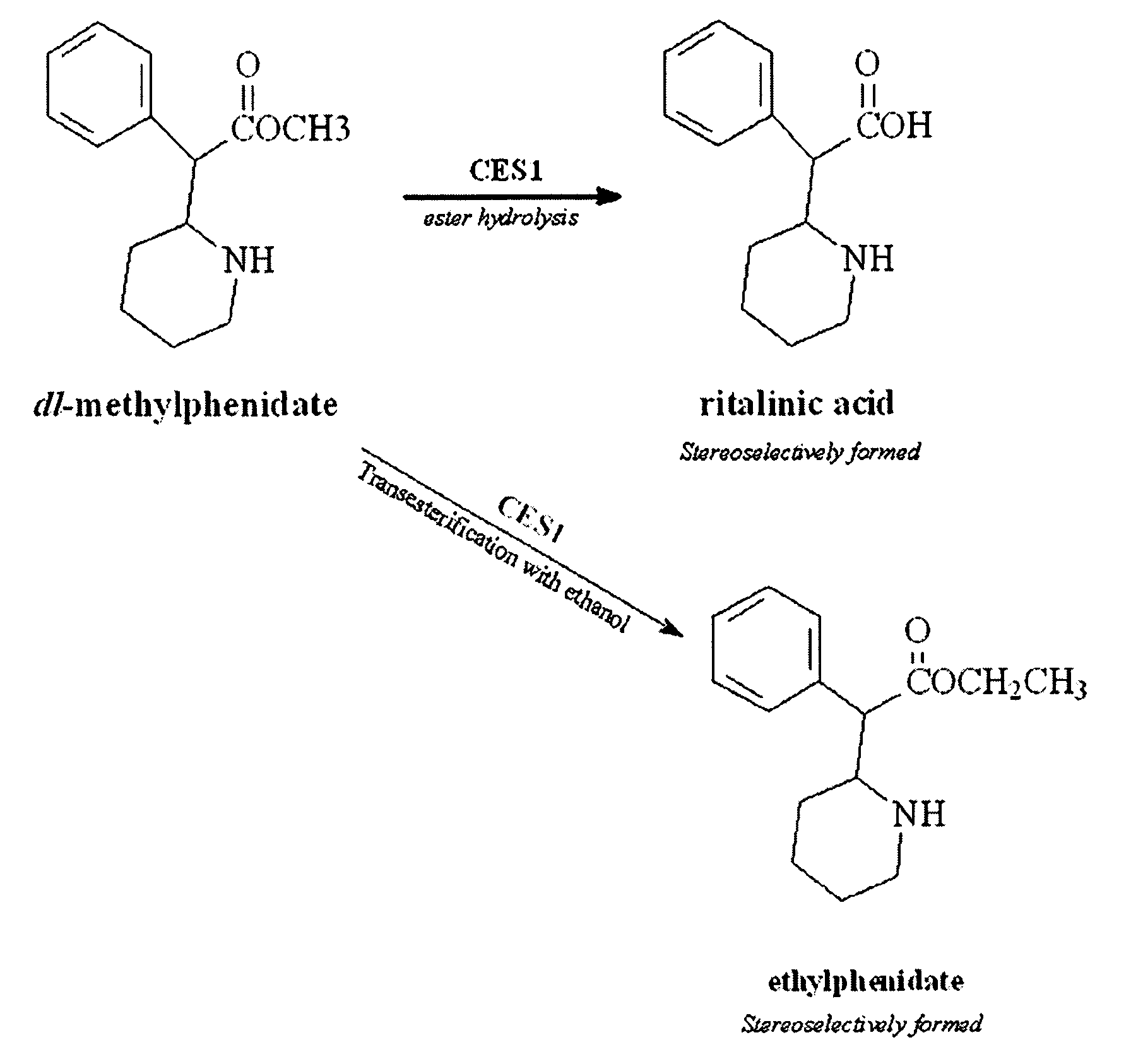
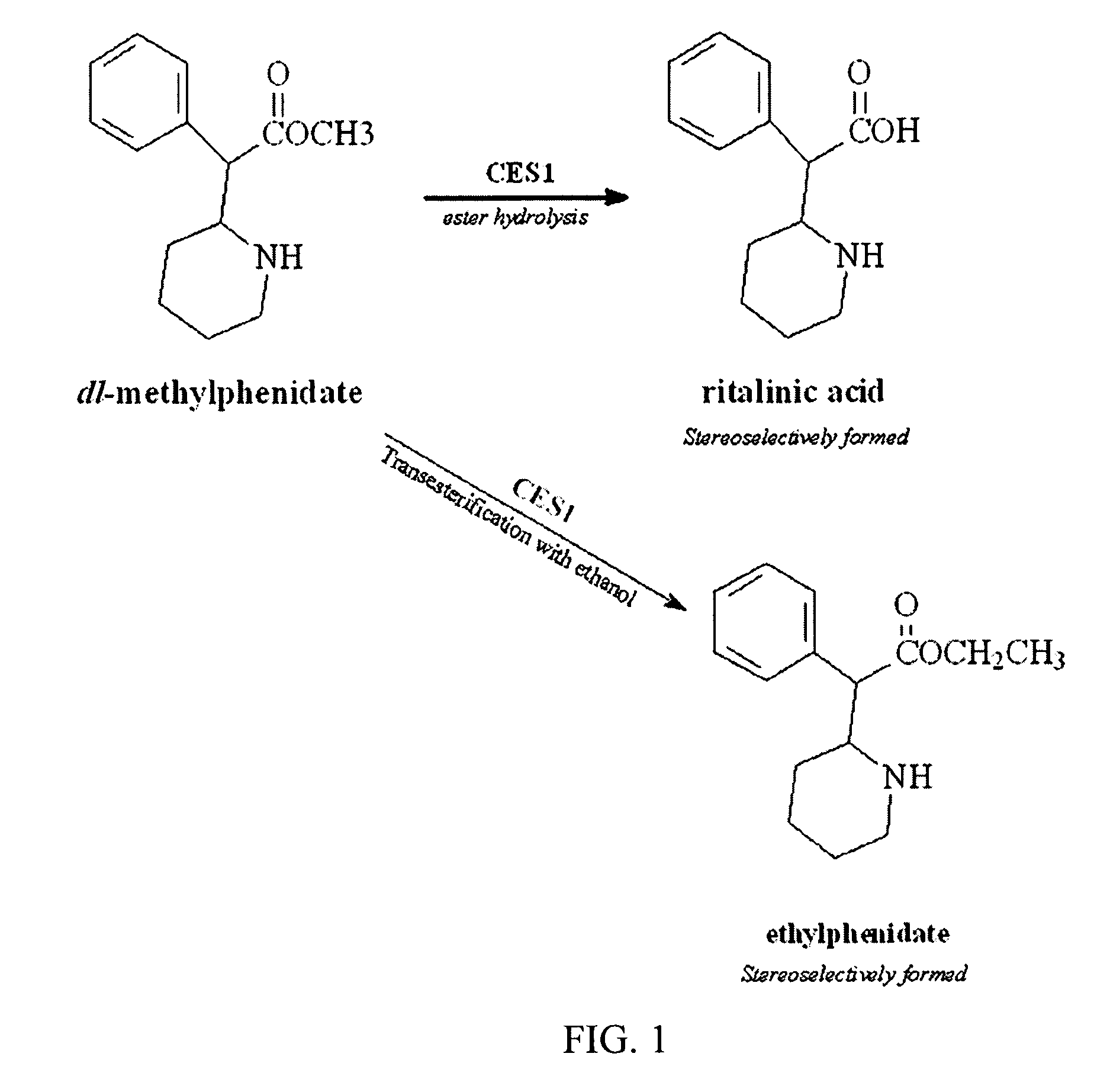
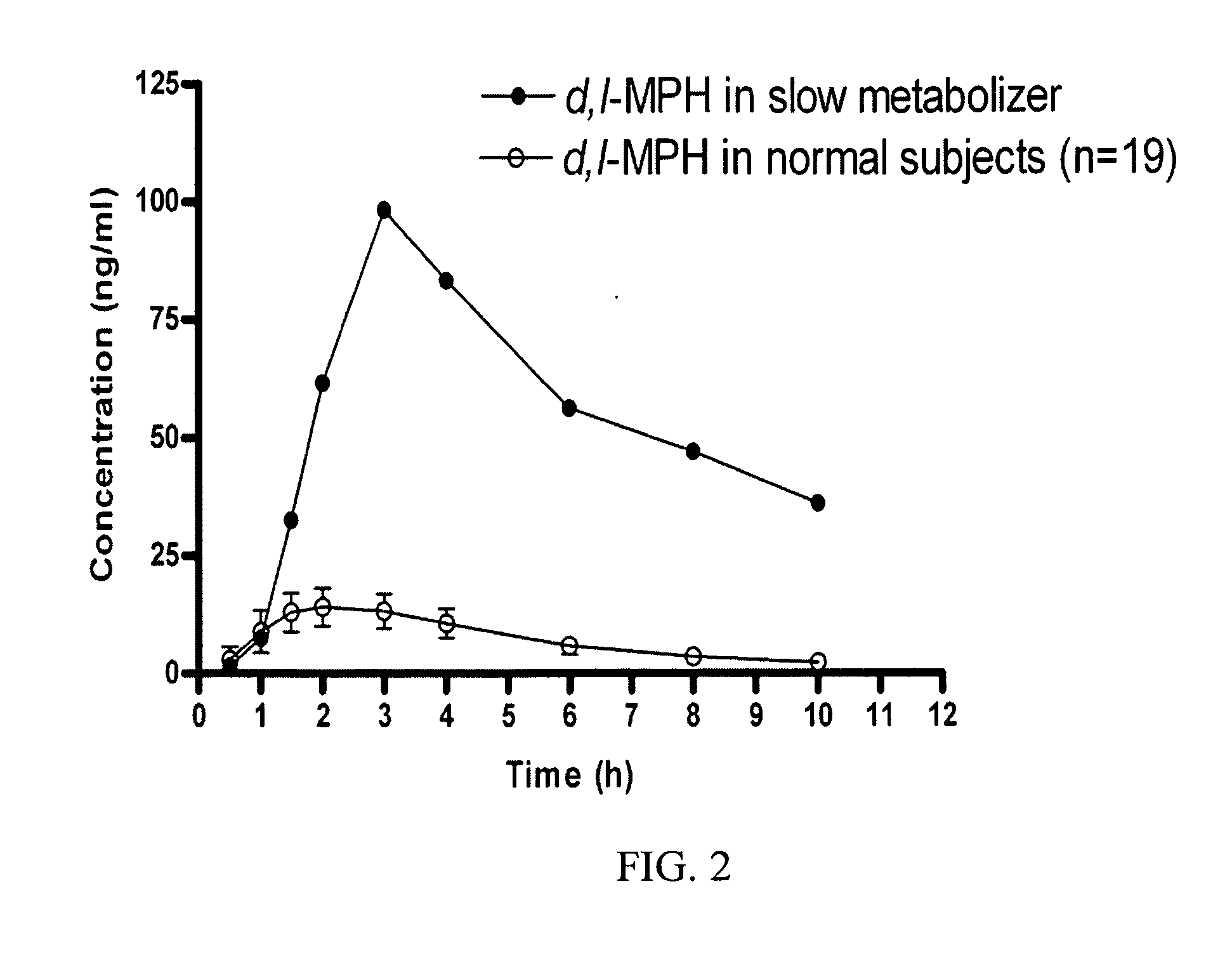
Carboxylesterase-1 Polymorphisms and Methods of Use Therefor
InactiveUS20110020801A1Improve throughputEfficient CatalysisMicrobiological testing/measurementBiological material analysisCaucasian populationNucleotide
Methods and kits are provided for detecting polymorphisms in carboxylesterase-1 (CES1). Several single nucleotide polymorphisms (SNPs) in CES1 in humans, and methods for detecting the same, are provided (e.g., Gly143Glu, 12754T>del). Results indicate that the Gly143Glu (9486G>A) polymorphism has an allelic frequency of 1.5% in the Caucasian population. Polymorphisms of the present invention may alter the function of the carboxylesterase-1 enzyme (hCES1). Thus, the methods and kits of the present invention may be used to personalize a therapy and / or avoid adverse consequences of altered metabolism of a therapeutic or compound (e.g., enalapril, methylphenidate, etc.) which may result due to a CES1 polymorphism. In addition, recombinant cells lines overexpressing wild-type CES1 or expressing CES1 mutants are provided. Such cell lines may be used to assess the effects of candidate compounds on CES1, and the action of CES1 on these candidate compounds.
Owner:MARKOWITZ JOHN S +1
Blood-based tumor mutation burden (bTMB) biomarker, and measuring method and uses thereof
ActiveCN110643703AEffectively predict the efficacy of immunotherapyMicrobiological testing/measurementBase JHematological Tumor
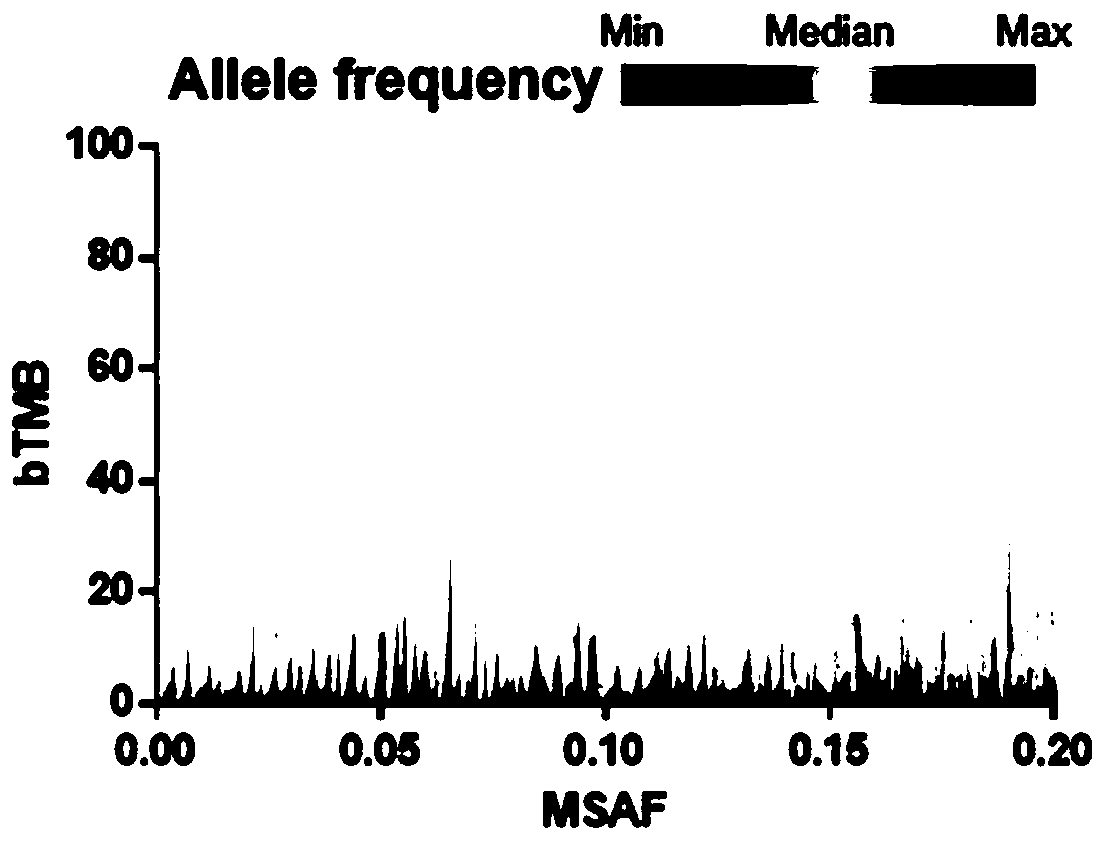
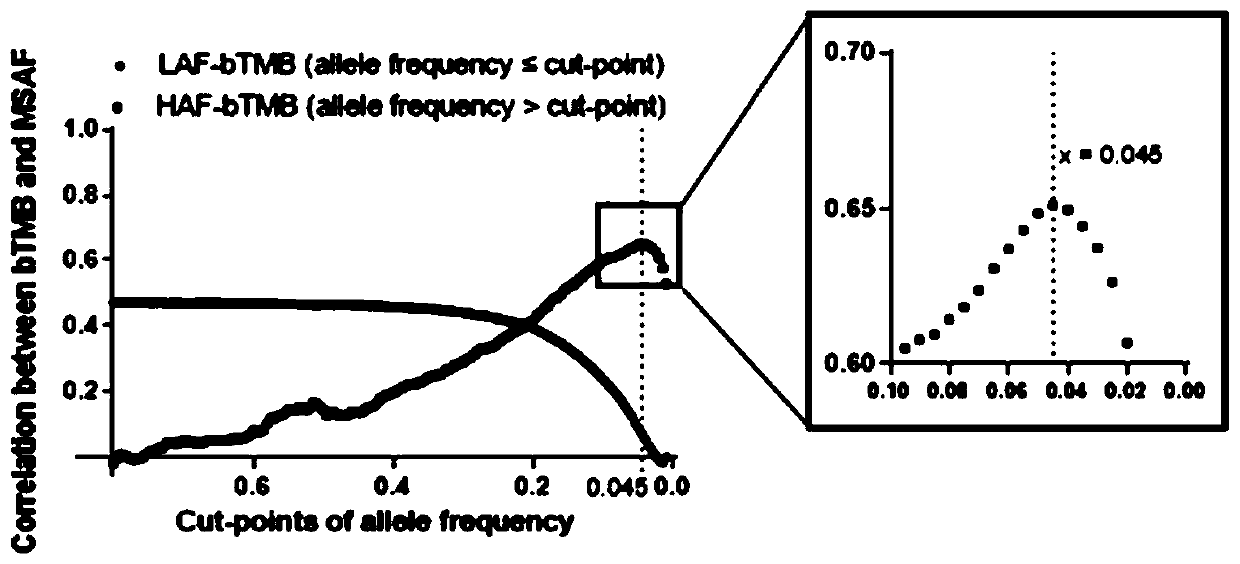
The invention relates to a blood-based tumor mutation burden (bTMB) biomarker, and an measuring method and uses thereof. The bTMB biomarker is obtained from the following steps that cell-free DNA (cfDNA) is obtained from a blood sample of a subject, the number of somatic mutations on sequencing bases is measured, the number of somatic mutations of low allele abundance is taken as the bTMB biomarker and expressed as LAF-bTMB, wherein the low allele abundance means that the allele frequency is less than, for example, 25%, 24%, 23%, 22%, 21%, 20%, 19%, 18%, 17%, 16%, 15%, 14%, 13%, 12%, 11%, 10%,9%, 8%, 7%, 6%, 5%, 4 %, 3%, 2%, 1%, for example, between 0.3% to 25%, preferably less than, for example, 13%, 12%, 11%, 10%, 9%, 8%, 7%, 6%, 5%, 4%, 3%, 2%, 1%, for example, between 0.5% to 13%, andparticularly preferably less than 6.5%, 6%, 5.5%, 5%, 4.5%, 4 %, 3.5%, 3%, 2.5%, and the LAF-bTMB is calculated according to the total number of mutations in a sample measuring area.
Owner:CANCER INST & HOSPITAL CHINESE ACADEMY OF MEDICAL SCI +1
Single nucleotide polymorphism disease incidence prediction system
ActiveCN104573408AEasy accessAchieve efficiencySpecial data processing applicationsAllele frequencyDisease cause
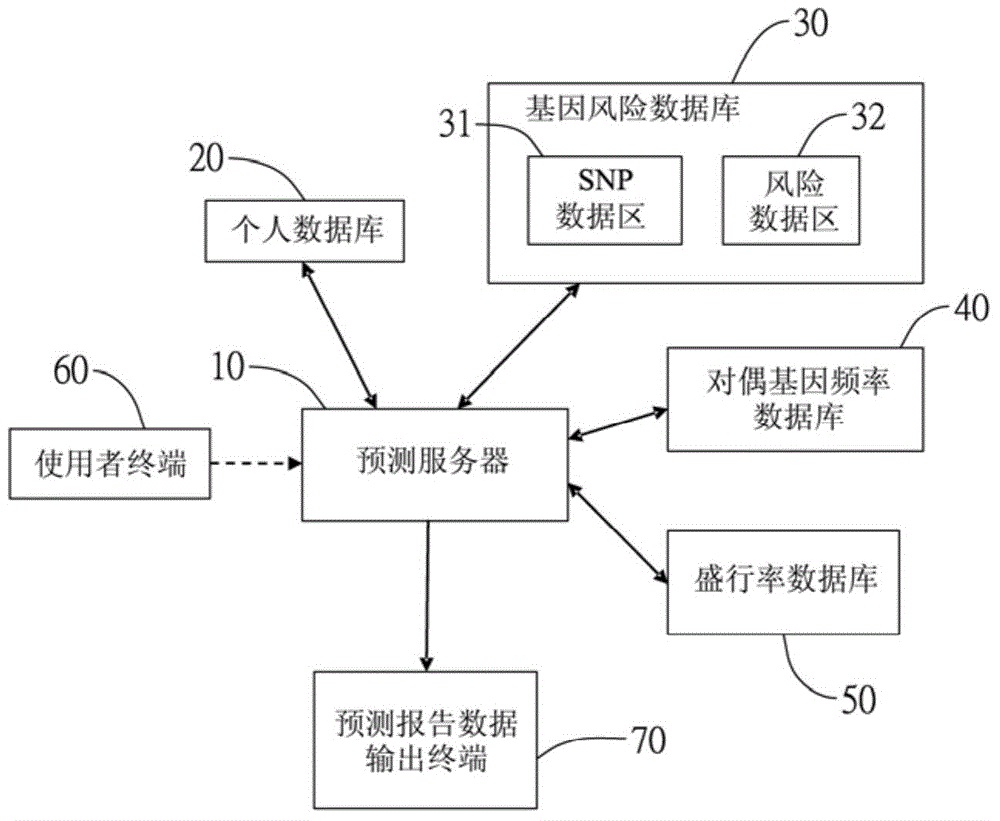
The invention provides a single nucleotide polymorphism disease incidence prediction system. According to the system, a prediction server collects more than one group of personal data and gene data and respectively transmits the data to a personal database for storage and a gene risk database for data exchange, the prediction server obtains a single nucleotide polymorphism data and a risk data via the gene risk database and respectively exchanges a frequency data with an allele frequency database, and the prediction server generates a genetic risk data according to the operation of the above single nucleotide polymorphism data, the risk data, and the frequency data, obtains a plurality of prevalence data via a prevalence database, and rapidly generates a prediction report according to the prevalence data and the genetic risk data. Therefore, via the above effective method, users can conveniently obtain reference data of disease incidence related to self genes.
Owner:TCI GENE
SNP molecular markers used in chromosome 6 of pig for traceability and application thereof
ActiveCN105255877AImprove testing coverageImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceAllele frequency
The invention discloses SNP molecular markers used in chromosome 6 of a pig for traceability and application thereof. The SNP molecular markers are obtained by means of NCBI database sequence alignment, and peculiarly recognized by Eco72I restrictive endonuclease, the total number is 670 bp, a base substitution exists in a base group of the 284th position and is 284 C or 284 T, and the sequence of the molecular markers is shown as SEQ ID NO.1; distribution conditions alleles of the SNP molecular markers in test groups are analyzed in the test groups including ten pig varieties or strains, it is found that allele frequencies of the SNP molecular markers in the different varieties or strains are approximate, polymorphism is rich, the allele frequency distribution difference among the varieties or the strains is small, heterozygosity is larger than 0.3, and the SNP molecular markers can be used for DNA traceability of pork products.
Owner:SHANGHAI ACAD OF AGRI SCI
Methods of detecting somatic and germline variants in impure tumors
PendingUS20190362808A1Maximizes sumMicrobiological testing/measurementBiostatisticsAllele frequencySomatic cell
A system is provided that considers allele fraction shifts as a function of copy number and clonal heterogeneity. The system leverages differences between allele frequencies to differentiate between somatic and normal variants in impure tumor samples. In solid tumors, stromal cells and infiltrating lymphocytes are typically interspersed among the tumor cells. The normal cell contamination in tumors can be leveraged to differentiate somatic from germline variants. We explicitly model allelic copy number and clonal sample fractions so that we can examine how these factors impact the power to detect somatic variants. The system models the copy number alterations, which can also affect the allele frequencies of both somatic and germline variants. The expected allele frequencies can be calculated. The expected allele frequencies for somatic and germline differ with tumor content for different copy number alterations.
Owner:TRANSLATIONAL GENOMICS RESEARCH INSTITUTE
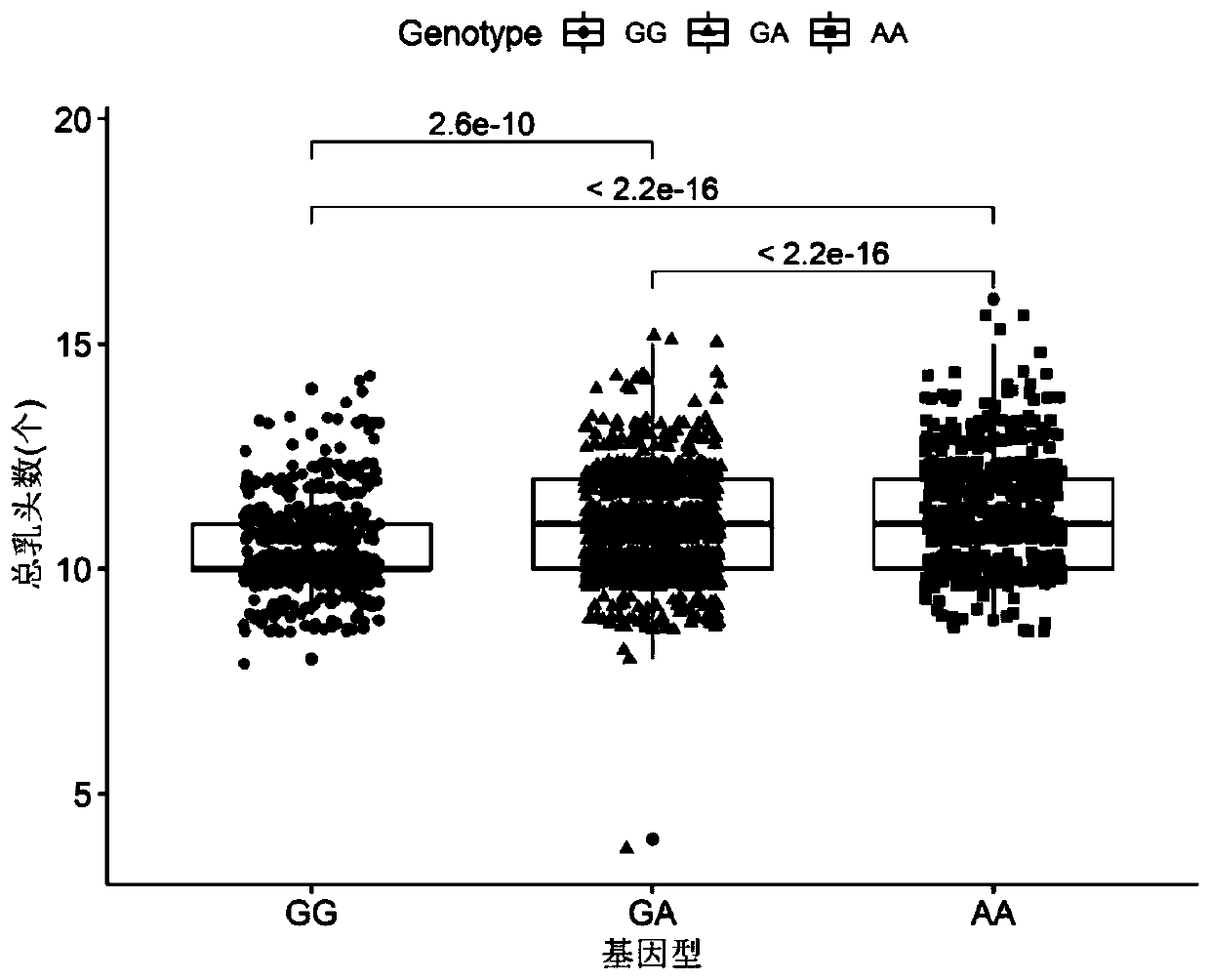
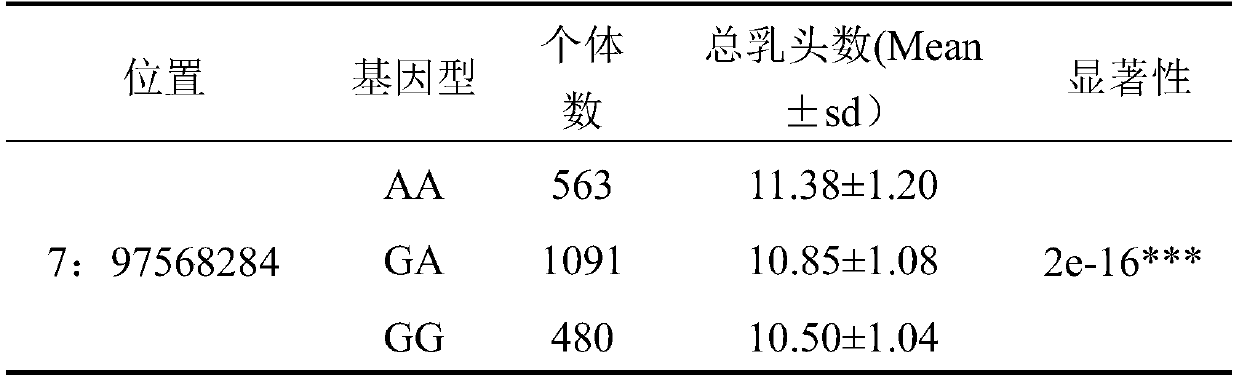
SNP molecular marker located on porcine No. 7 chromosome and related to total teat number and application of SNP molecular marker
ActiveCN110144408AFast and Accurate BreedingImprove reproductive performanceMicrobiological testing/measurementDNA/RNA fragmentationMedicineAllele frequency
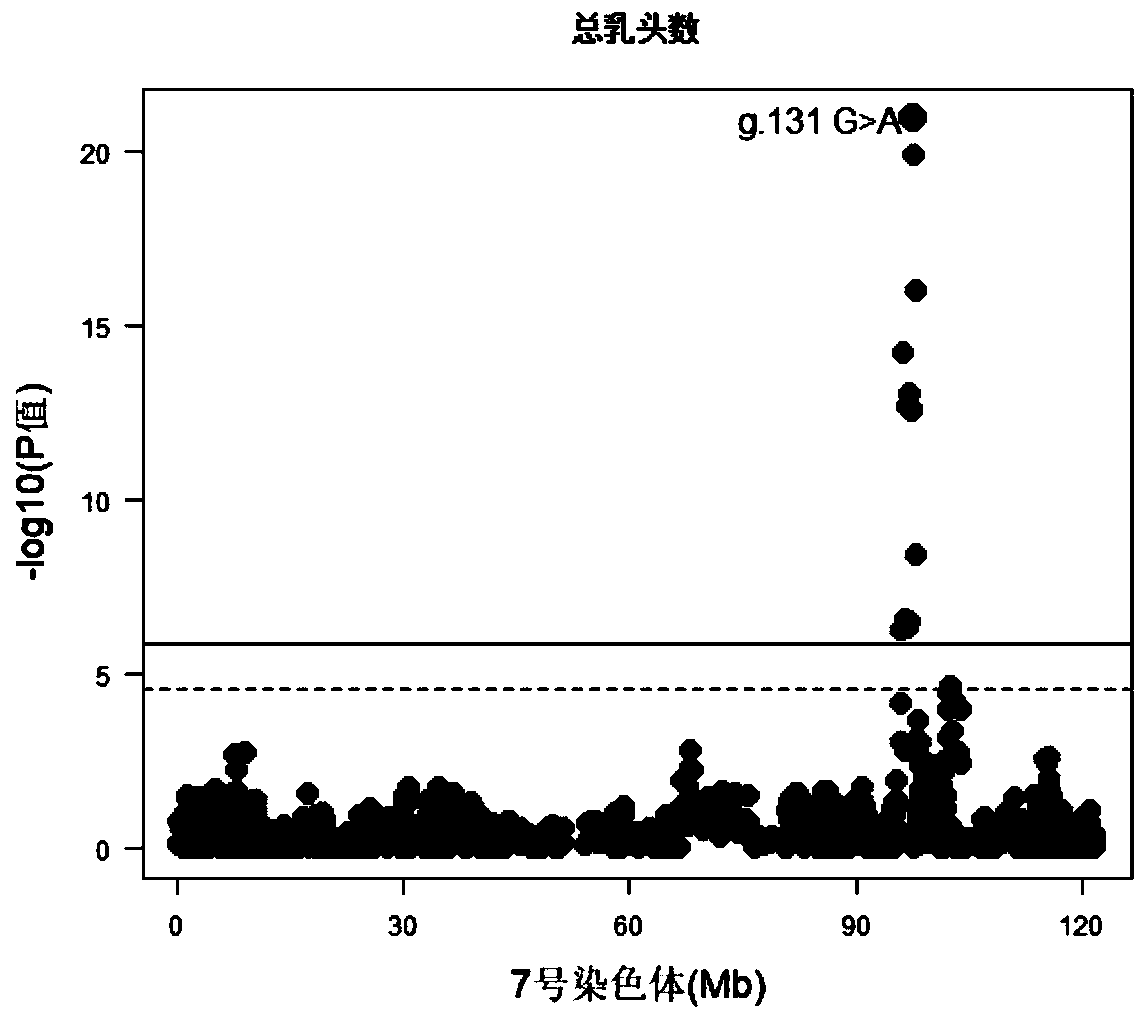
The invention belongs to the technical field of a molecule biology technique and molecular markers, and particularly relates to an SNP molecular marker located on a porcine No. 7 chromosome and related to total teat number and an application of the SNP molecular marker. The SNP site of the SNP molecular marker located on a porcine No. 7 chromosome and related to total teat number corresponds to the 97568284th G>A mutation on the No. 7 chromosome of a reference sequence of an international porcine genome 11.1 edition. The invention also provides a primer pair of the SNP molecular marker locatedon a porcine No. 7 chromosome and related to total teat number. According to the SNP molecular marker disclosed by the invention, the dominant allele of the SNP is preferably selected, so that the dominant allele frequency can be increased by generation, the reproductive performance of pigs can be increased, the heredity improvement progress of the pigs can be accelerated, and economic benefits can be effectively increased.
Owner:SOUTH CHINA AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com