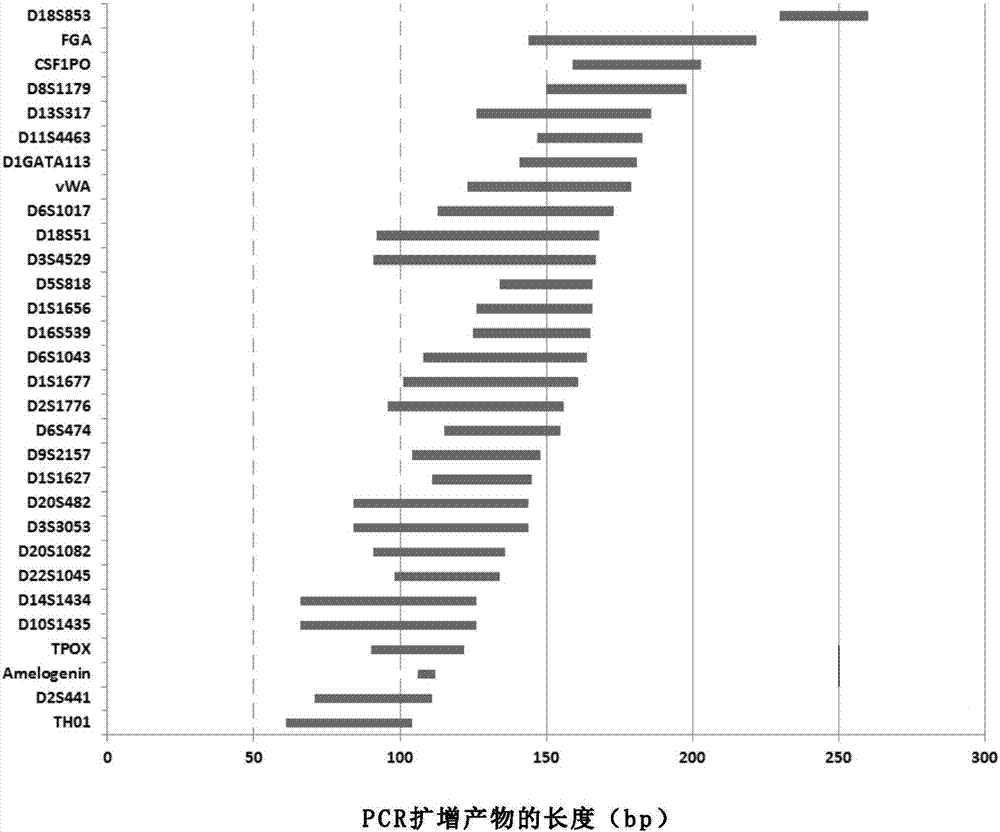
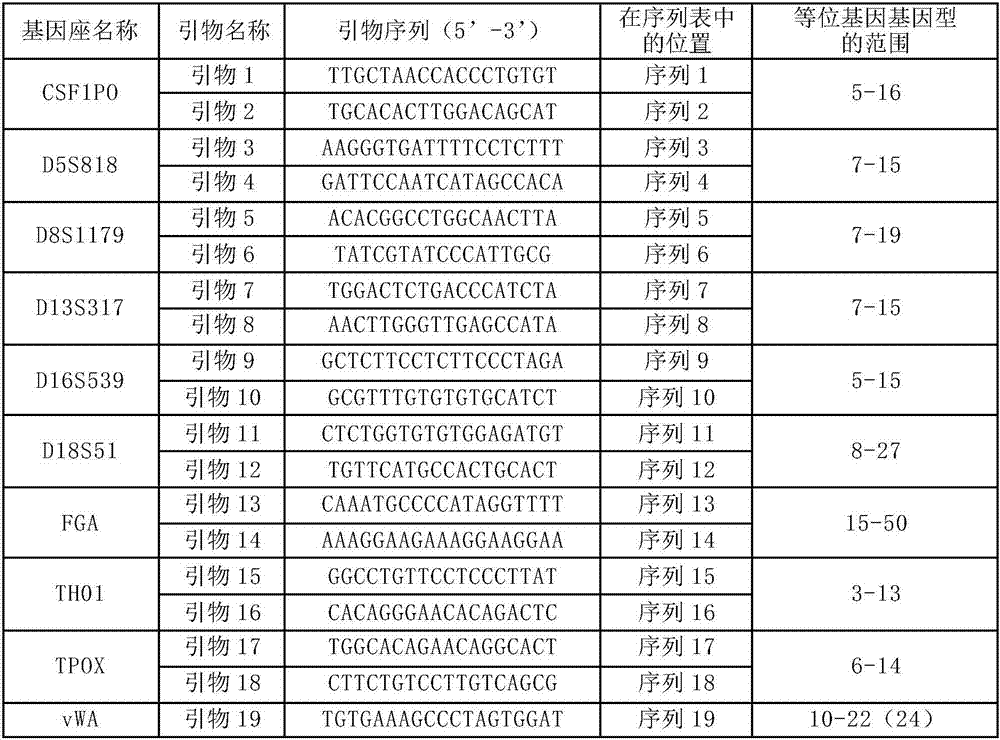
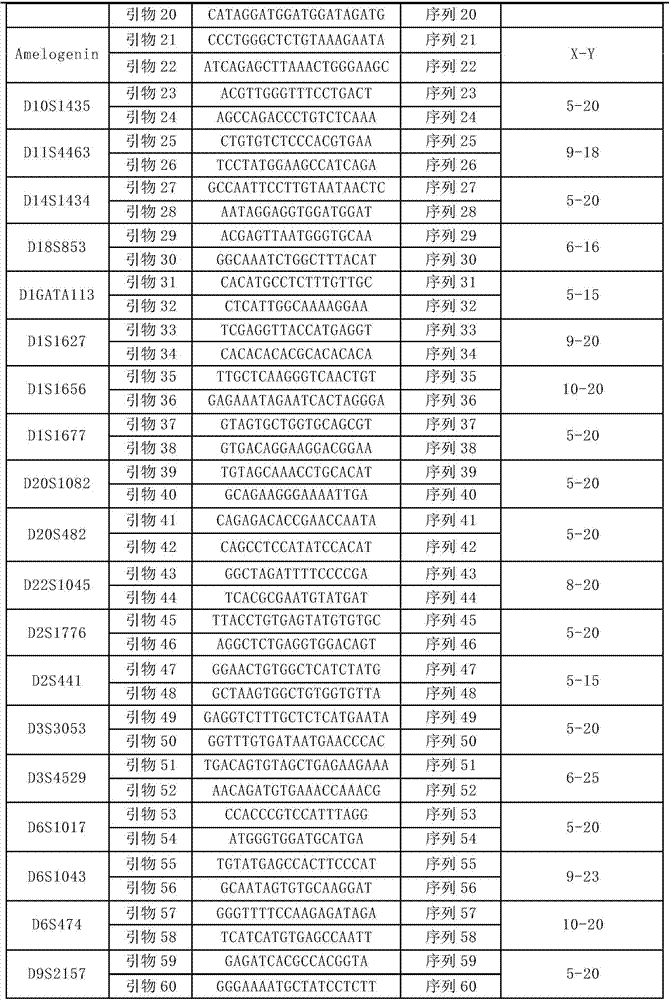
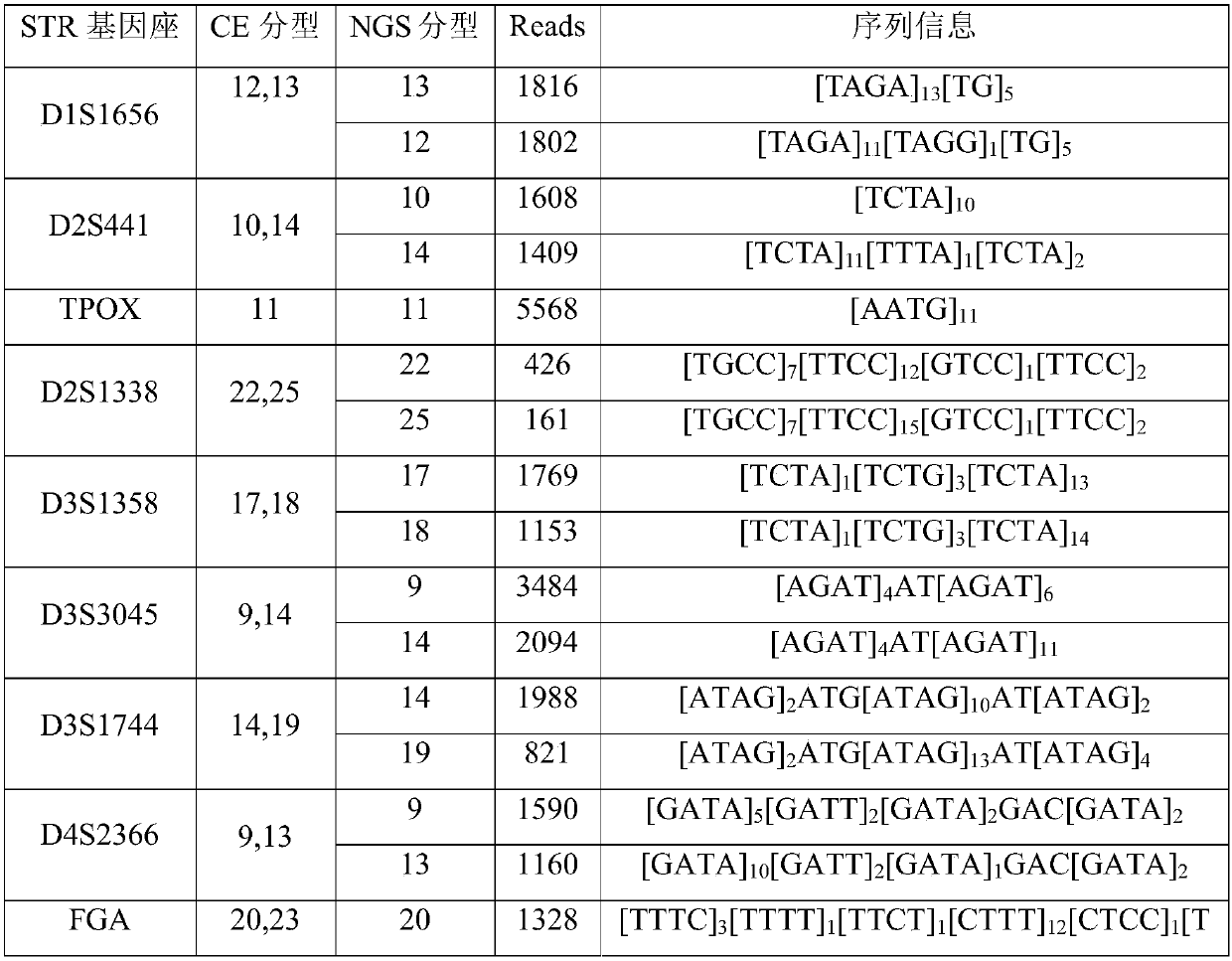
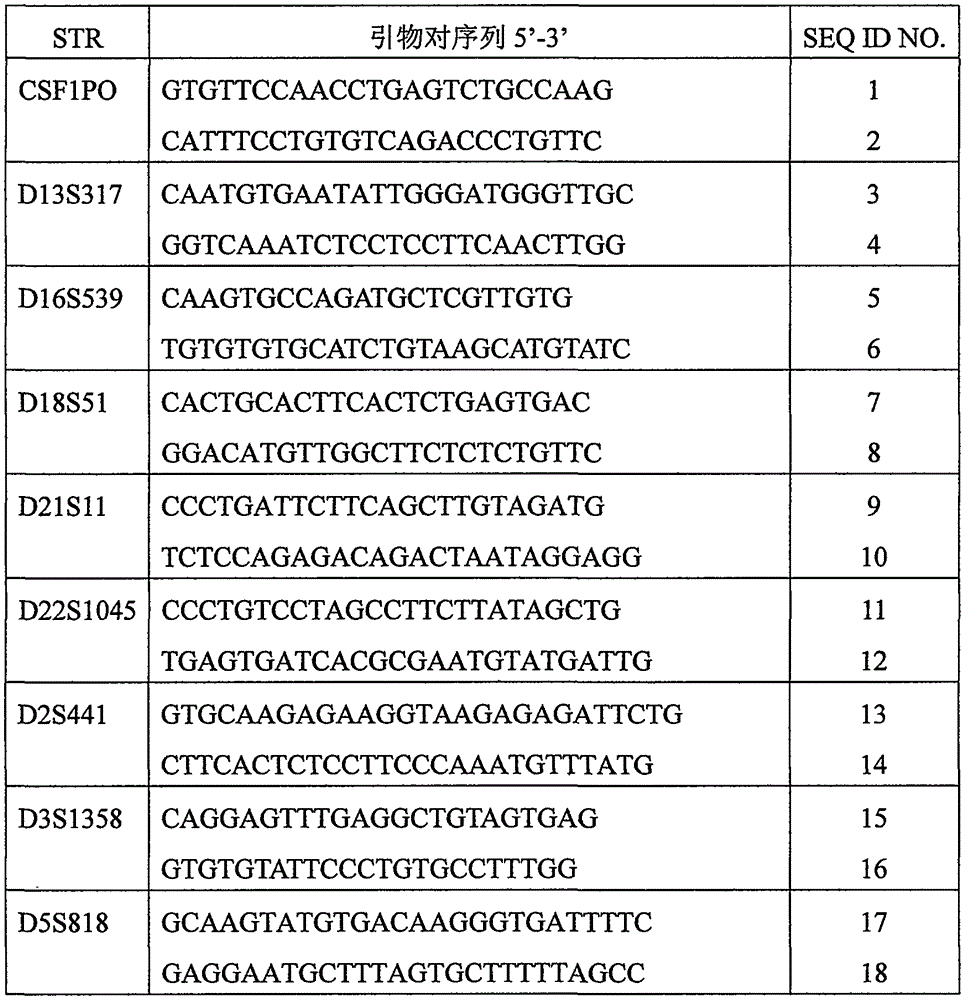
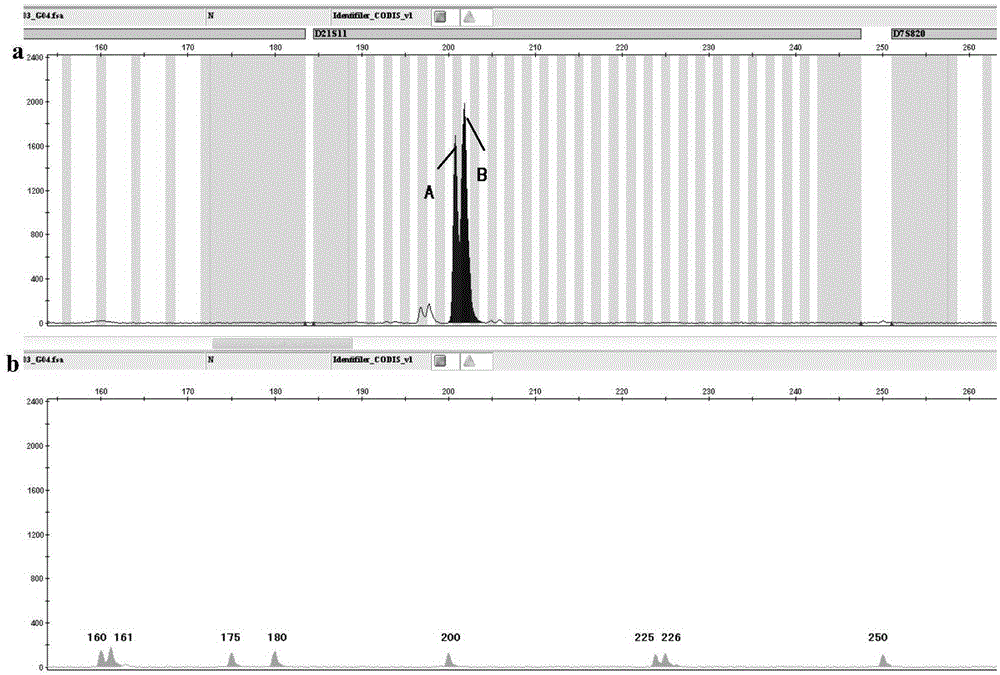
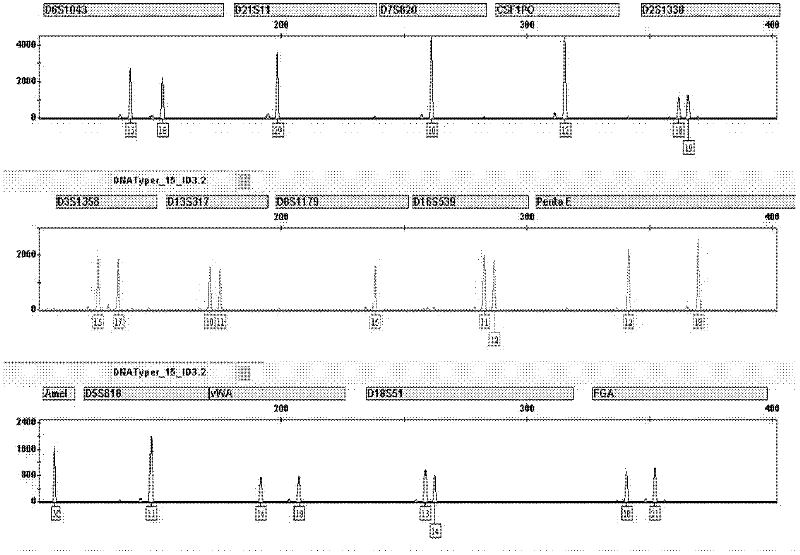
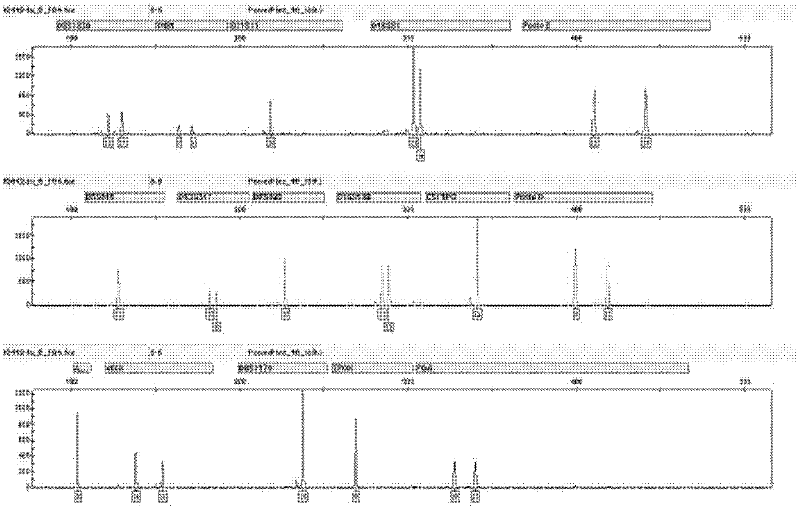
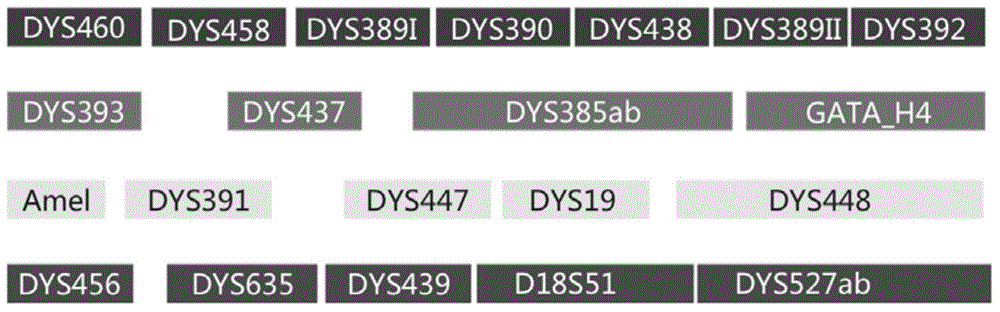
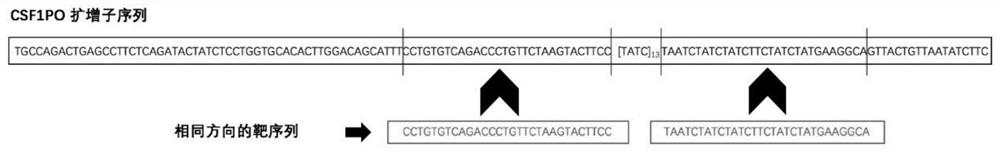
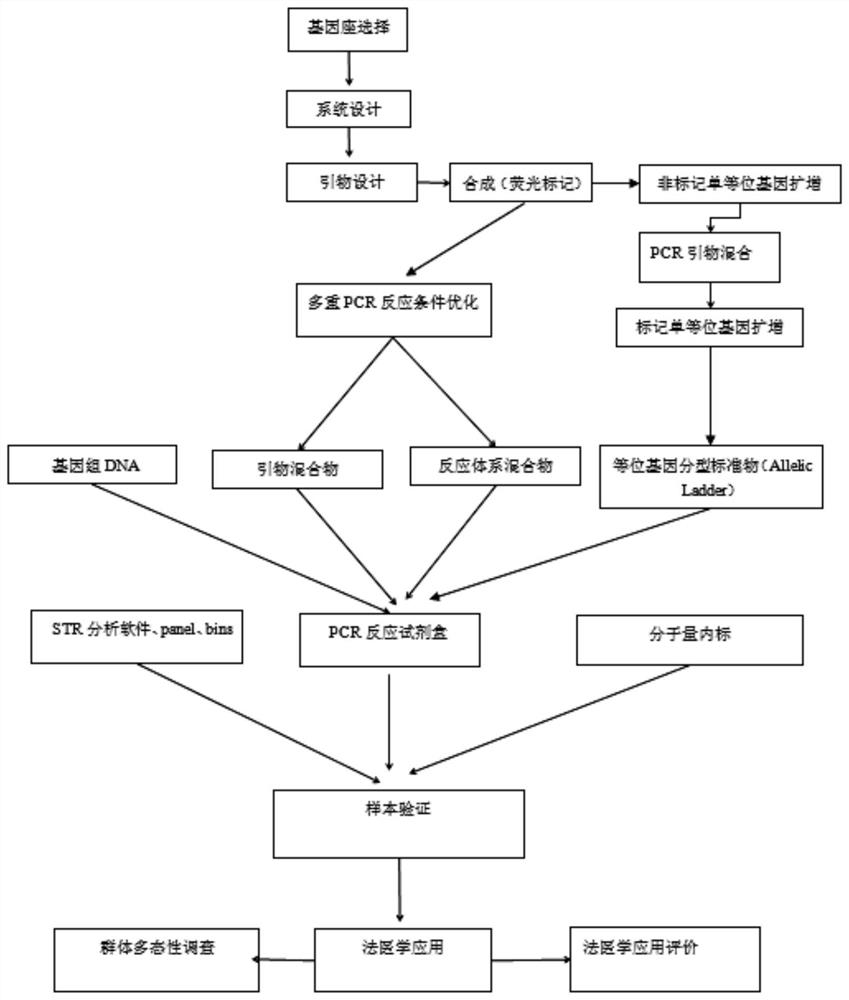
Patents
Literature
63 results about "Str typing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for Typing an Individual Using Short Tandem Repeat (Str) Loci of the Genomic Dna
The present invention relates to a novel STR typing strategy, which allows the simultaneous amplification and subsequent analysis of several (e.g. eleven) polymorphic systems with amplicon sizes of less than 270 bp. Thereby, after a PCR amplification the multiplex reaction is divided into two sets of STR multiplexes and analyzed separately. This multiplex system was particularly developed and tested for use in forensic investigations, where only limited amounts of DNA or only highly degraded DNA is available, for example, when the DNA is isolated from the roots of telogen hair.
Owner:JOHANNES GUTENBERG UNIV
Method utilizing magnetic particles to purify deoxyribonucleic acid (DNA) from samples containing trace nucleic acid
The invention provides a method utilizing magnetic nanocomposite to conveniently, quickly and efficiently purify deoxyribonucleic acid (DNA) from medical samples and other samples containing trace nucleic acid. The method comprises the following steps of: (1) taking a sample to be purified, adding the guanidinium lysis solution with the final concentration of 3-6M and the dithiothreitol with the final concentration of 0.01-0.05M, uniformly mixing, and then carrying out full pyrolysis in a water bath for 5-30min; (2) forming a mixture solution containing magnetic particle-DNA composite; (3) separating the magnetic particle-DNA composite from the mixture solution; (4) cleaning; and (5) eluting to obtain the purified DNA. The method can satisfy the requirement of extracting high-quality DNA from the medical samples, and is applied in downstream polymerase chain reaction (PCR) amplification, submarine thermal reactor (STR) somatotyping and other applications; and the method does not need centrifugation, and has quick and simple operation steps. According to the method, a kit which not only is applicable to manual operation but also can be mated with an automated instrument can be produced, so the medical sample DNA can be obtained through purification.
Owner:XIAN GOLDMAG NANOBIOTECH
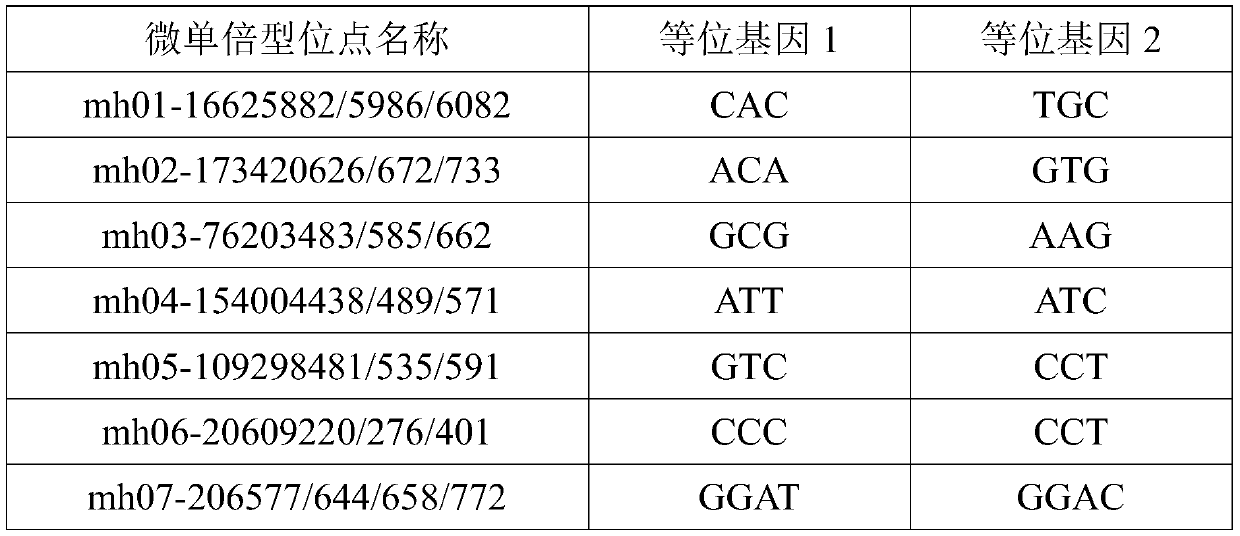
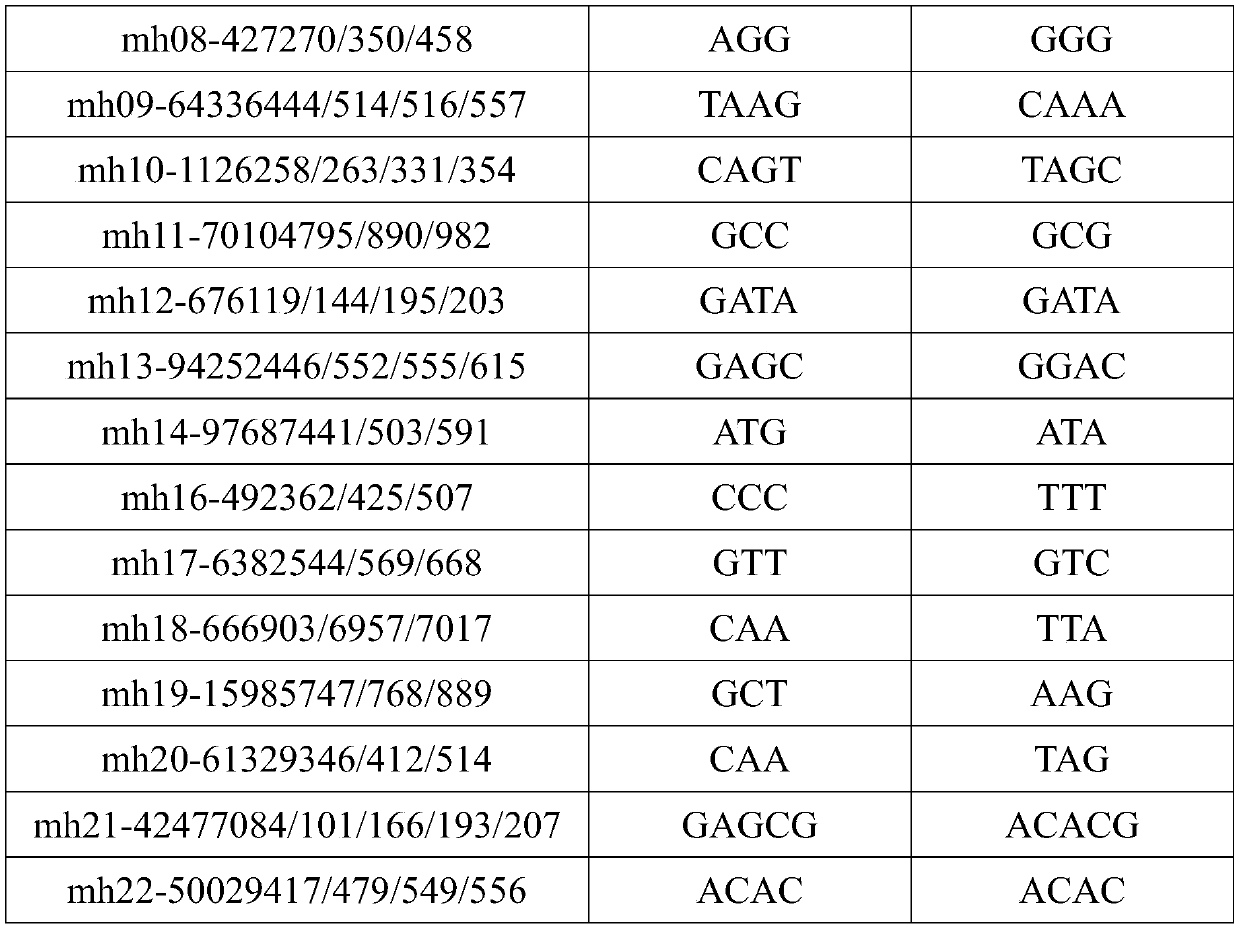
Multiplex amplification system, and next generation sequencing typing kit and typing method for 21 micro-haplotype sites
ActiveCN110218781AShorten the lengthLow mutation rateMicrobiological testing/measurementSequence analysisStr typingAllele frequency
The invention relates to the technical field of molecular biology, and especially relates to a multiplex amplification system, and a next generation sequencing typing kit and typing method for 21 micro-haplotype sites. The 21 micro-haplotype sites in the multiplex amplification system are derived from 21 autosomes, are balanced in allele frequency, are mutually independent between the sites, and have the advantages of polymorphism and low mutation rate. The kit contains 53 PCR-amplified single-end specific primers shown in SEQ ID NO. 1-SEQ ID NO.53, and can detect the 21 microhaplotype sites in a same reaction system. The multiplex amplification system, the kit and the typing method provided by the invention can solve the technical problem that a current STR typing technology cannot obtainan ideal typing result, and allele loss may occur due to excessive number of SNP sites.
Owner:HEBEI MEDICAL UNIVERSITY
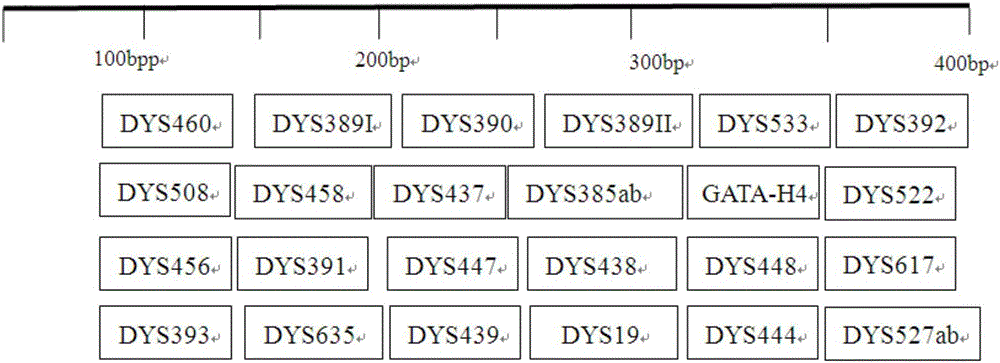
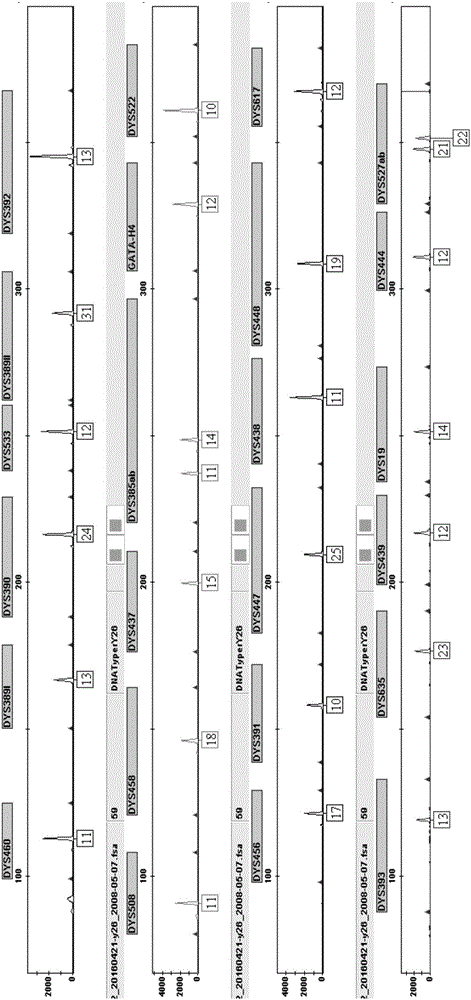
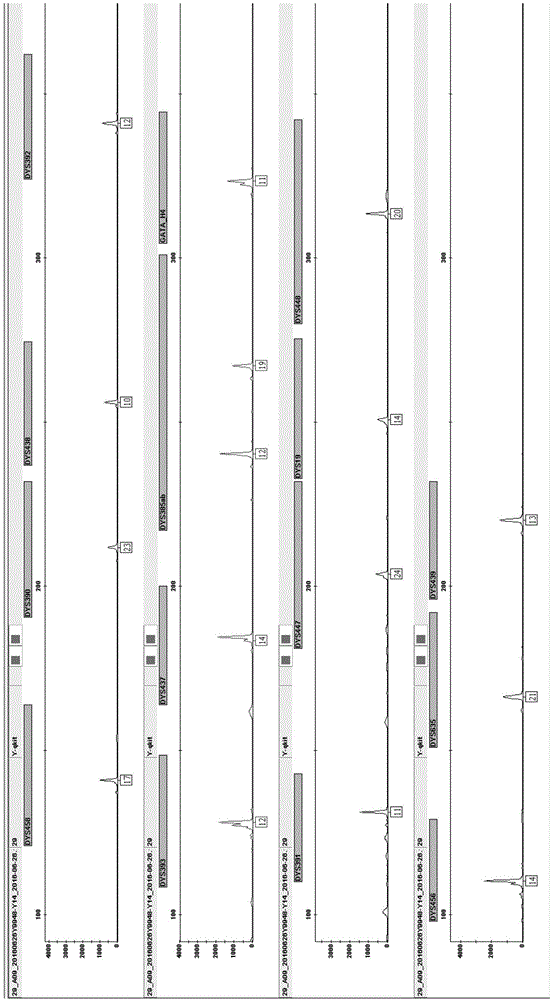
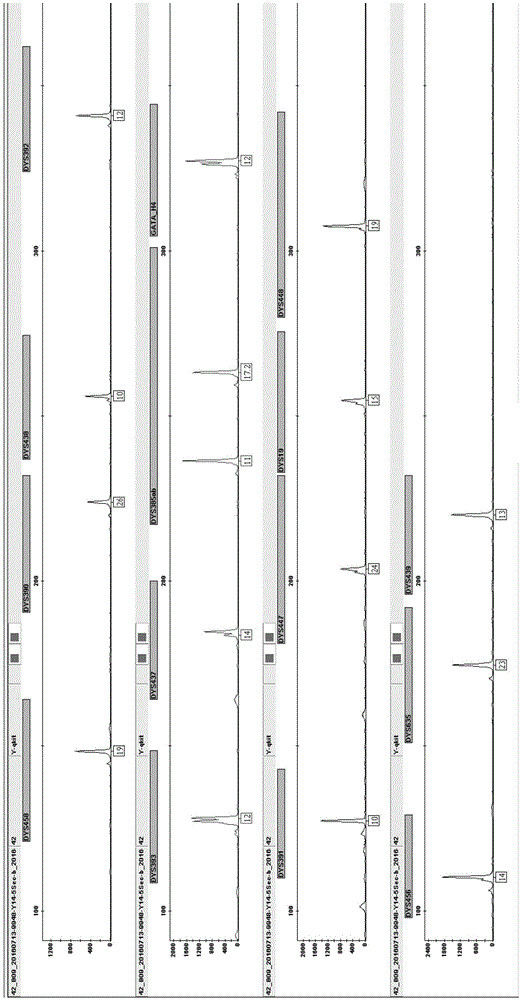
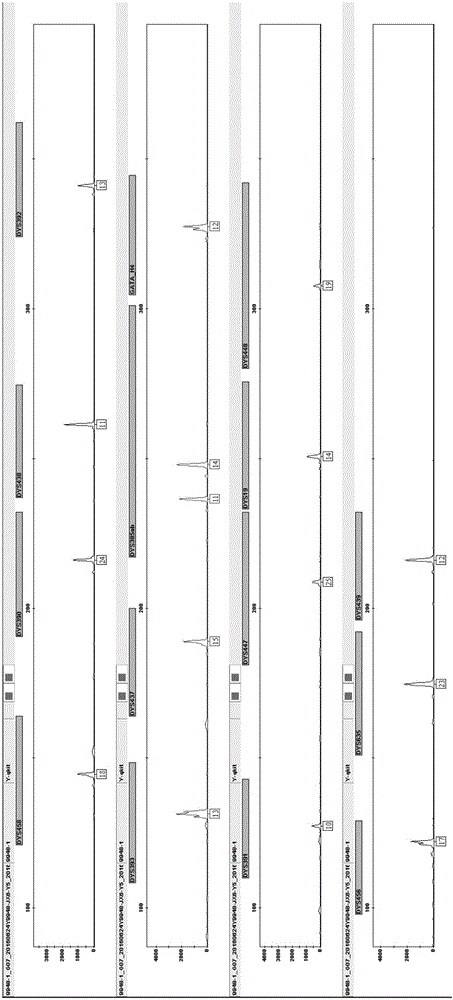
Method and system for realizing Y-STR typing on male individual by adopting 26 Y-STR genetic loci
ActiveCN106755340AEvenly distributedMeet the inspection requirementsMicrobiological testing/measurementStr typingTyping
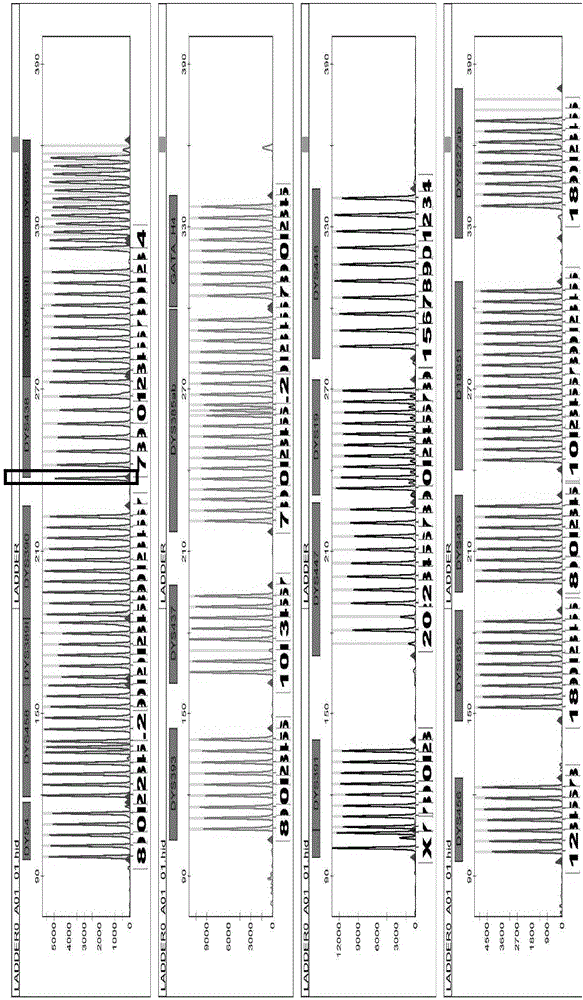
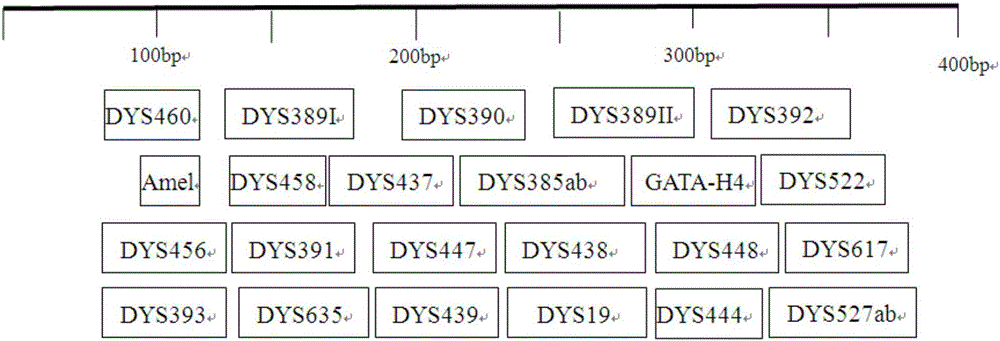
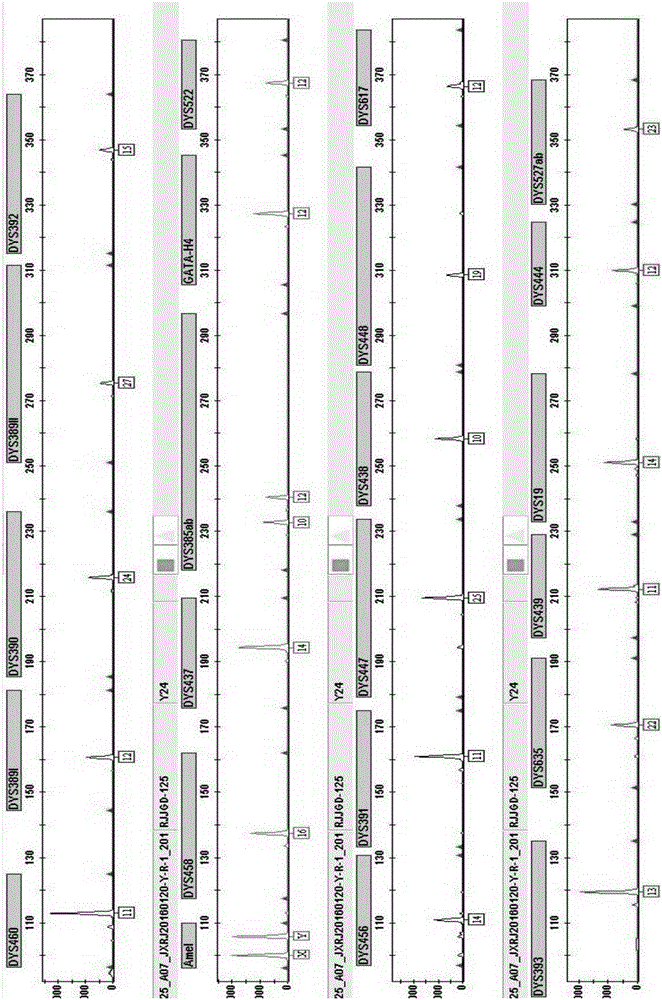
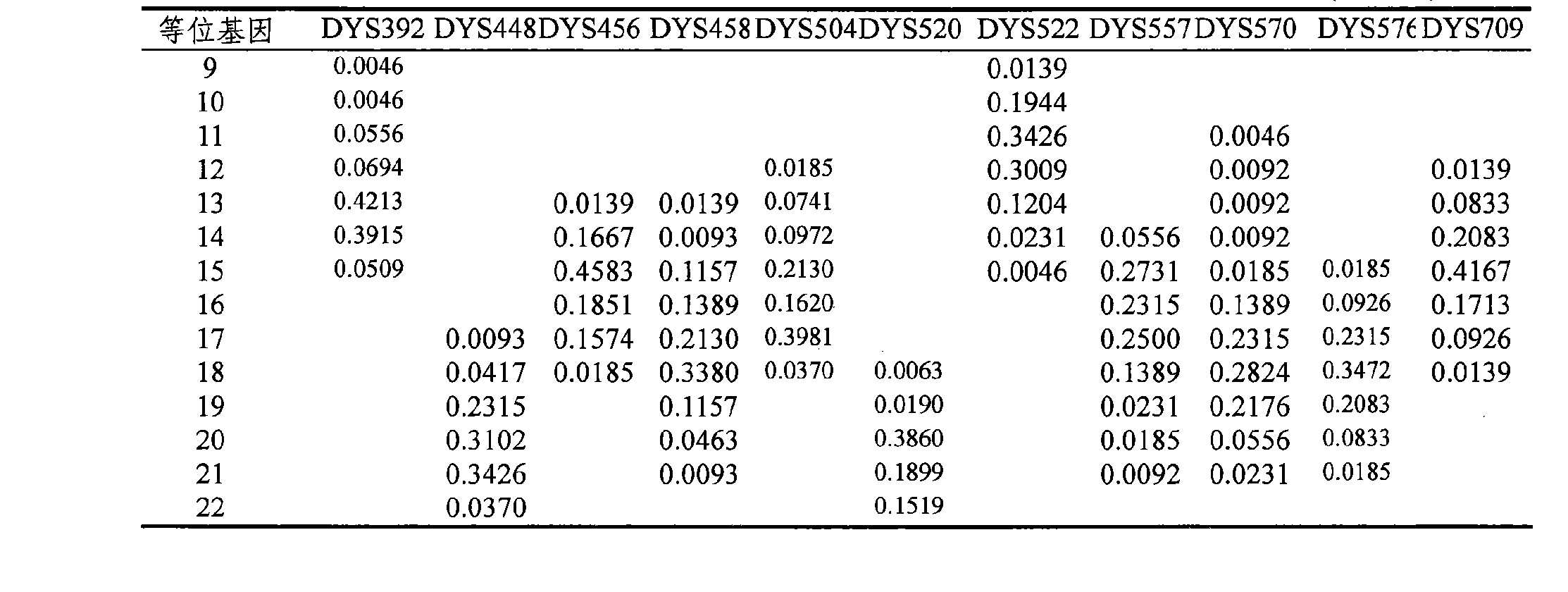
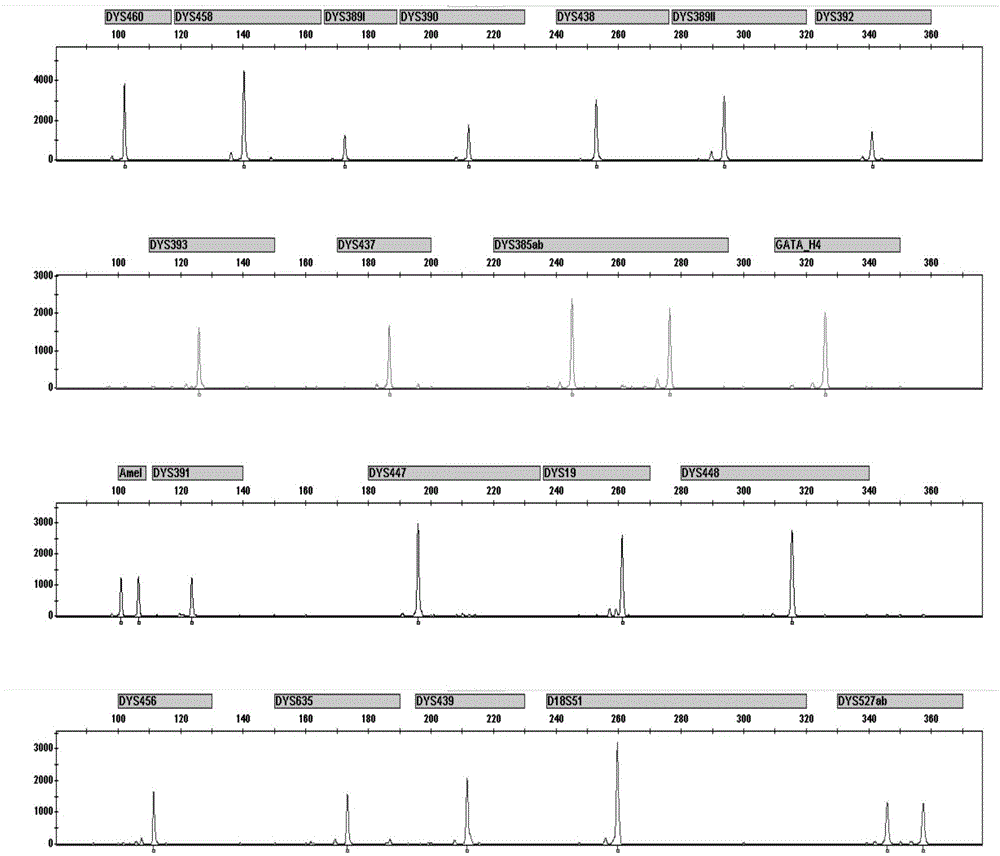
The invention provides a method and a system for realizing Y-STR typing on the male individual by adopting 26 Y-STR genetic loci. The method comprises the following steps: acquiring DNA of the male individual, and acquiring the genotypes of the 26 Y-STR genetic loci of the DNA, wherein the 26 Y-STR genetic loci are as follows: DYS19, DYS385a / b, DYS389I / II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS448, DYS456, DYS458, DYS635, GATA H4, DYS438, DYS439, DYS460, DYS447, DYS527a / b, DYS617, DYS522, DYS444, DYS508 and DYS533; and acquiring the Y-STR typing result of the male individual according to the genotypes of the 26 genetic loci of the male individual. With the typing result, the operations including family checking, paternity identification, detection for the male element in male and female mixed samples, and the detection for the male element in a plurality of male mixed samples can be realized on the male individual.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
STR kit for micro-amount degradation sample and application of STR kit
ActiveCN106521013AIncrease the number of detectionsHigh sensitivityMicrobiological testing/measurementStr typingAmelogenin
The invention discloses an STR kit for a micro-amount degradation sample and application of the STR kit. The kit comprises 17 pairs of specific amplification primers of STR gene loci, the 17 pairs of specific amplification primers are divided into 5 groups, and fluorescent labels with 6 colors are related. The STR kit has the advantages of rapidness, high sensitiveness and high inhibition resistance on detection of micro-amount degradation samples. A mini type amplification system of the 17 gene loci established by the invention is short in amplified fragment, and is suitable for micro-amount, highly degraded or high-inhibitor-content difficult samples; and during actual application, STR types of special biological samples such as skeletons, contact cast-off cells and blood samples in soil can be obtained successively. DYS391 gene loci are introduced in a system, and the auxiliary judgment effect on sex genes Amelogenin can be achieved; and the kit can rapidly detect the samples, and the whole PCR process does not exceed 50 minutes.
Owner:AGCU SCIENTECH +1
Reagent kit for detecting gene loci on basis of next generation sequencing technologies and special primer combination of reagent kit
ActiveCN107099529AThe typing result is accurateMicrobiological testing/measurementDNA/RNA fragmentationStr typingTyping
The invention discloses a reagent kit for detecting gene loci on the basis of next generation sequencing technologies and a special primer combination of the reagent kit. The primer combination comprises 60 types of DNA (deoxyribonucleic acid) molecules shown from sequences 1 to sequences 60 in sequence tables. The reagent kit for detecting the gene loci on the basis of the next generation sequencing technologies and the primer combination have the advantages that as proved by experiments, accurate typing results can be obtained when the reagent kit is used for detecting STR (short tandem repeat) typing of the 30 gene loci in standard substances 2800M; the reagent kit has important application value.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Next generation sequencing typing reagent kit of 43 STR sites and method
The invention relates to a next generation sequencing typing reagent kit of 43 STR sites and a method. The reagent kit comprises a PCR amplification reagent kit, wherein the PCR amplification reagentkit comprises 79 PCR amplification single-end specific primers of the 43 STR sites, and the nucleotide sequence of the PCR amplification single-end specific primers is as shown in SEQ ID NO.1-SEQ ID NO.79. The primers in the reagent kit can enable the polymorphism of each gene locus to be increased, so that probability of exclusion and system effectiveness can be improved, and the testing result is accurate and reliable; and testing results can provide new data for development of a NGS-STR typing technique, and a new detection scheme is provided for cases of target missing person survey, complex genetic relationship identification and the like through united application of a plurality of STR sites of the reagent kits in the prior art.
Owner:HEBEI MEDICAL UNIVERSITY
Processing method for high-throughput sequencing information of human short tandem repeat
The invention discloses a processing method for high-throughput sequencing information of human short tandem repeat and belongs to the field of biological detection. The method comprises the steps that sequences with a preset sequencing length in STR high-throughput sequencing information of a single chip are reserved, and first sequences are formed; the first sequences are classified into different sample folders according to sample label information, the first sequences are reclassified into different STR locus folders according to STR target fragment specific primer information, and second sequences are formed; stepped reference sequences for different STR loci are established, the second sequences are compared with sequences of corresponding STR loci in the second sequences, and third sequences with the sequence similarity greater than or equal to 90% are reserved; and the threshold value of the number of sample sequencing entries is set to be 1,000, the threshold value of the number of locus sequencing entries is set to be 50, the threshold value of the number of typing sequencing entries in the loci is set to be 5, the threshold value of the number of the typing sequencing entries in the loci and the number of the locus sequencing entries is set to be 40%, and sequences with threshold values greater than or equal to the threshold values in the third sequences are screened to obtain an STR typing result.
Owner:IPE BIOTECHNOLOGY CO LTD
High-throughput STR typing system capable of identifying complex consanguinity and kit thereof
ActiveCN110499373AImprove balanceAchieve single-tube amplificationMicrobiological testing/measurementDNA/RNA fragmentationStr typingNucleic acid detection
The invention discloses a high-throughput STR typing system capable of identifying complex consanguinity and a kit thereof, and relates to the technical field of nucleic acid detection in vitro. The high-throughput STR typing system capable of identifying complex consanguinity comprises PCR primers used for amplifying 60, 118 or 179 linked autosomal STR loci; and the kit comprises a PCR primer composition, an Index connector sequence, an IGT-EM707 polymerase mixture, an amplification buffer enhancer NB, a YF buffer solution B, a database setting-up reagent and the like. The high-throughput STRtyping system capable of identifying complex consanguinity and the kit thereof provided by the invention are capable of realizing single-tube amplification of 60, 118 or 179 linked autosomal STR loci; and moreover, the STR typing system and the kit thereof are good in balance, high in sensitivity, good in specificity, and accurate in typing result. Thus, the high-throughput STR typing system capable of identifying complex consanguinity and the kit thereof can be used for identifying complex relationships of different consanguinity at the same level.
Owner:SHANXI MEDICAL UNIV
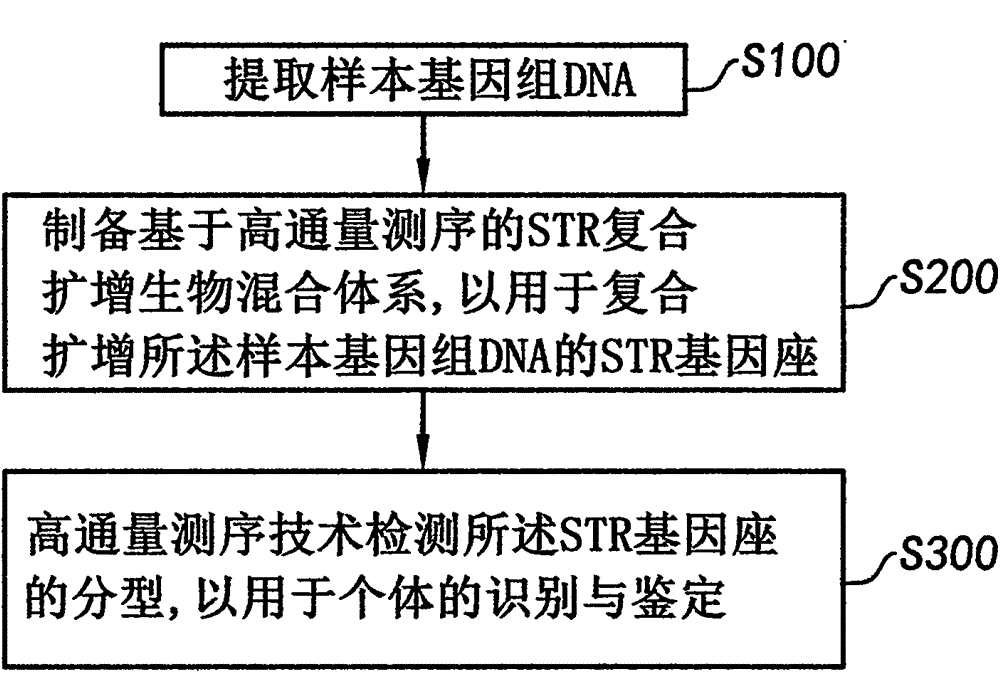
System and method for detecting STR subtype at high throughput
ActiveCN104673907AImprove detection accuracyImprove visibilityMicrobiological testing/measurementStr typingGenotype
The invention discloses a system and a method for detecting an STR subtype at high throughput. The method comprises the steps of (S100) extracting a sample genome DNA; (S200) preparing an STR multiplex amplification bio-mixed system based on high throughput sequencing to carry out multiplex amplification on an STR gene locus of the sample genome DNA; and (S300) detecting the genotype of the STR gene locus by using a high throughput sequencing technology to recognize and authenticate individuals. The method is suitable for detecting the STR subtype in the field of forensic science.
Owner:SHANGHAI CRIMINAL SCI TECH RES INST
STR typing method for loca D8S1179 based on next generation sequencing
The invention relates to the field of molecular biology and particularly discloses a specific primer pair for amplifying a gene D8S1179. The specific primer pair comprises D8S1179-FP:5'-TTTGTATTTCATGTGTACATTCGTAATTC-3' and D8S1179-RP:5'-ACCTATCCTGTAGATTATTTTCACTGATTG-3'. The invention further provides an STR typing method for a loca D8S1179 based on next generation sequencing and particularly provides a primer combination for being connected with a proper adaptor. The STR typing method is applied to the STR typing of the loca D8S1179. According to the primer pair capable of amplifying the gene D8S1179, after concentrations of different sample products subjected to PCR amplification, the equivalent mixing is carried out, and a genome library is constructed according to a BIOO RAPID DNA kit; the library is input into a computer by virtue of a next generation sequencer, data is subjected to bioinformatics analysis, and the typing results such as times of repetition of NDA molecules with same length, SNP of the loca and the difference among side wings of the loca of D8S1179 can be found.
Owner:BEIJING CENT FOR PHYSICAL & CHEM ANALYSIS
Method and system for Y-STR typing of individual man
ActiveCN106520980AEvenly distributedMeet the inspection requirementsMicrobiological testing/measurementStr typingTyping
The invention provides method and system for Y-STR typing of an individual man. The method comprises the following steps: acquiring the DNAs of the individual man, wherein acquired DNAs comprise genotypes of 25 gene loci including 24 Y-STR gene loci and a gender determination locus Amelogenin, and the 24 Y-STR gene loci are DYS460, DYS389I / II, DYS390, DYS392, DYS458, DYS437, DYS385a / b, GATA_H4, DYS522, DYS456, DYS391, DYS447, DYS438, DYS448, DYS393, DYS635, DYS439, DYS19, DYS444, DYS527a / b, DYS617; and acquiring the Y-STR typing results of the individual man according to the 25 gene loci of the individual man. The Y-STR typing results can be used for family checking, male paternity testing, detection of male components in a hybrid male and female sample, detection of male components in a mixed sample of a plurality of males and the like for the individual man.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Y chromosome MiniSTR typing reagent kit, preparation and use thereof
InactiveCN101475994AHigh multiplexing efficiencyGood for typingMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceX chromosome
The invention discloses a Y chromosome parting kit and its preparation method and application. The present invention designs three pairs of public primer pairs, optimizedly select 12 X chromosome STR loci suitable for the genetic distribution of Chinese population and designs MiniSTR primers fro part of loci, and respectively adds the three pairs of public primers to the 5' end of the MiniSTR primers to obtain 24 additional primers; the use of the inventive kit can simultaneously amplify 12 X chromosome STR loci at a single-tube, avoids the competition between these primers and the formation of heterozygosity dimers, increases the amplification efficiency of each locus, wherein, the length of amplification products can be less than the 280bp, and has a high sensitivity (as low as 30pg), a higher STR parting success rate can be obtained and the amplification efficiency of medical examiner DNA degradation samples can be improved. The inventive kit has advantages of high sensitivity, high specificity, high detection efficiency, low preparation cost and the like.
Owner:CHINA UNIVERSITY OF POLITICAL SCIENCE AND LAW
Multiplex system, method for detecting unbalanced mixed samples through compound system and application of method
ActiveCN109576378AAchieving Simultaneous AmplificationImprove analytical performanceMicrobiological testing/measurementDNA/RNA fragmentationForward primerStr typing
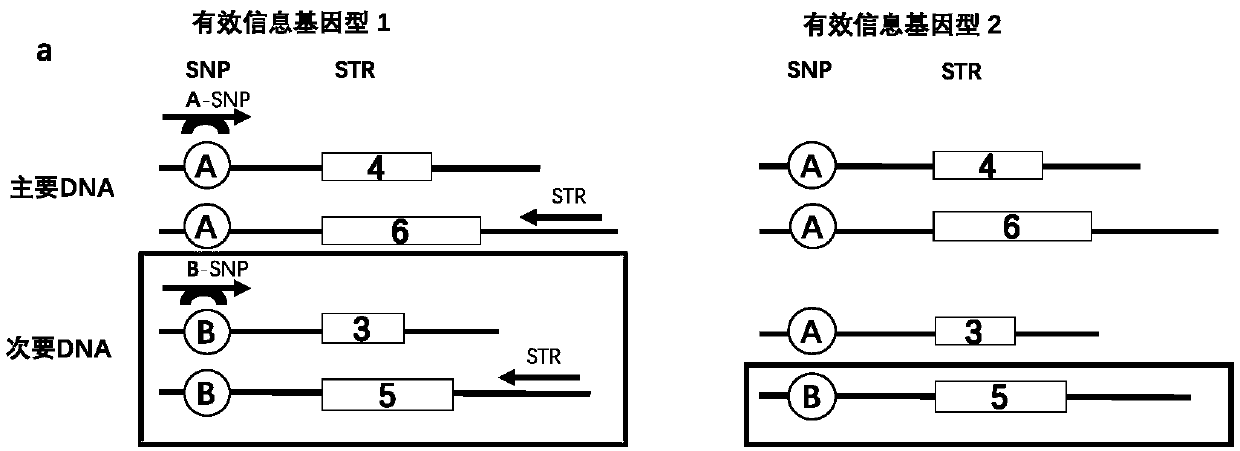
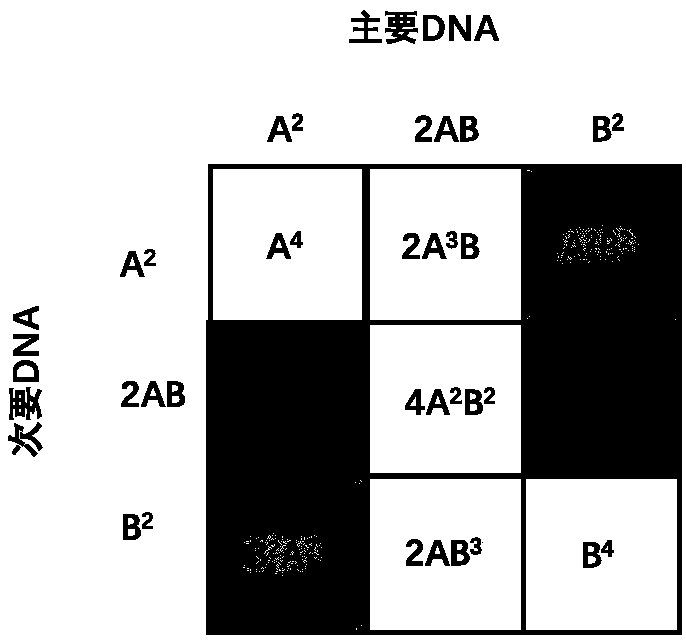
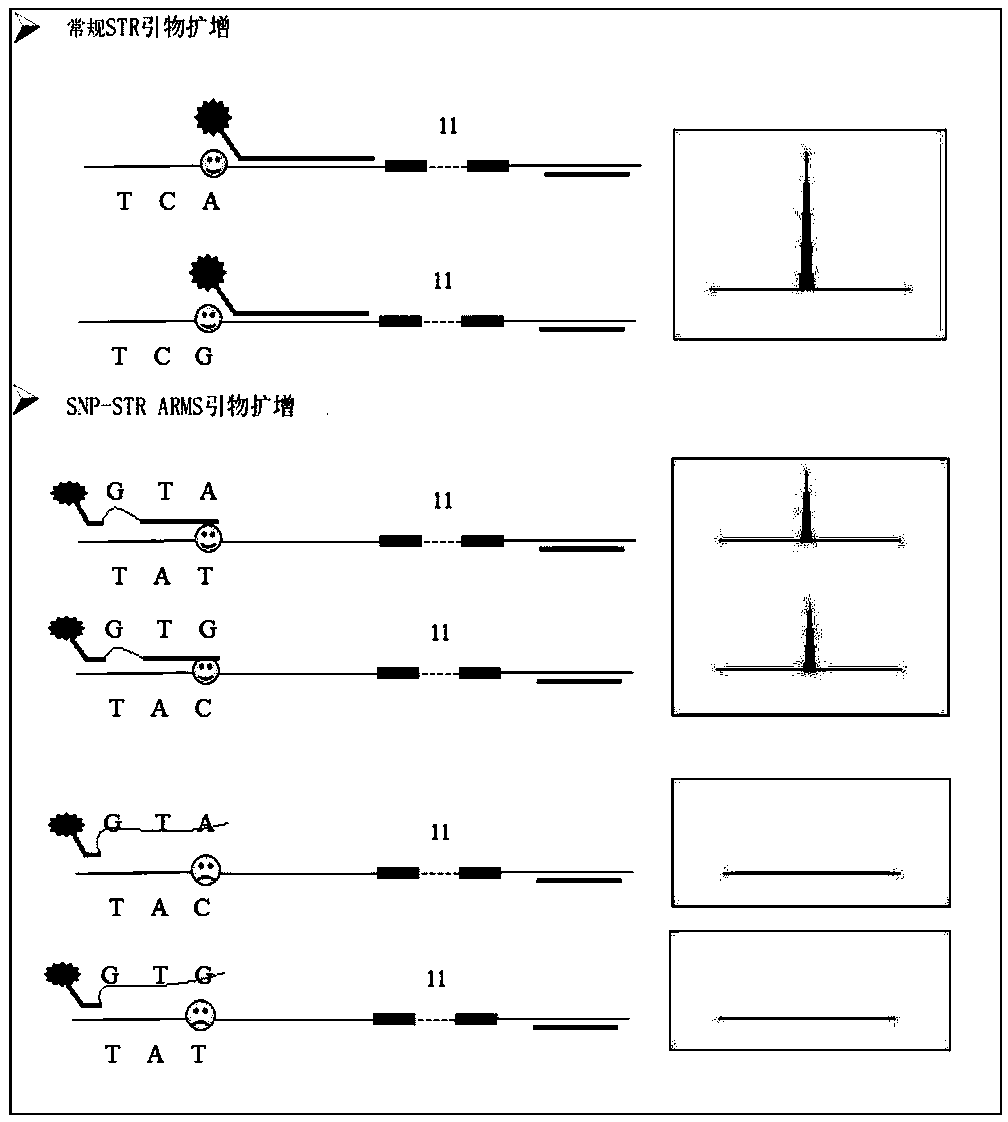
The invention discloses a multiplex system, a method for detecting unbalanced mixed samples through the multiplex system and application of the method. The multiplex system comprises primers for amplifying 18 SNP-STR (single nucleotide polymorphism-short tandem repeat) sites, each site comprises a common reverse primer and two specific forward primers for increasing mutation, and the specific sequences of the primers are as shown in the SEQ ID NO.1-54 in the specification. The SNP-STR (single nucleotide polymorphism-short tandem repeat) multiplex amplification system has the advantages that the operation process is simplified, the system can be widely used on the basis of existing forensic evidence conventional inspection equipment, mixed samples of two persons at a forensic scene are detected, the same STR (short tandem repeat) typing samples are distinguished, non-invasive prenatal diagnosis and non-invasive prenatal paternity test are realized, a new technical means is provided, andclues and evidence are provided for solving a case.
Owner:SICHUAN UNIV
Five-color fluorescence STR parting method for synchronously detecting 23 gene loci and special kit of method
PendingCN110066792AImprove individual recognitionImprove stabilityMicrobiological testing/measurementDNA/RNA fragmentationCapillary electrophoresisStr typing
The invention discloses a five-color fluorescence STR parting method for synchronously detecting 23 gene loci and a special kit of the method. The method comprises the following steps that a genome DNA of a to-be-detected individual is used as a template, the kit is adopted for PCR amplification, and a PCR amplification product is obtained; the PCR amplification product is taken, subjected to degeneration and then subjected to capillary electrophoresis detection, and genotypes of the 23 gene loci of the genome DNA of the to-be-detected individual are obtained; then an STR parting result of theto-be-detected individual is obtained. The kit comprises a primer pair combination, nucleotide sequences of 46 primers forming the primer pair combination are shown in the sequences 1-46 in a sequence table in sequence, and one primer in each primer pair is marked with fluorescence. The kit has high individual identification capacity and stability, is compatible with basic standard allele loci ofcaucasian, xanthoderm, melanoderm and brown race and is suitable for individual identification and affinity authentication of the Chinese nation population. The kit has high application value.
Owner:BGI FORENSIC TECH (SHENZHEN) CO LTD
Fluorescence labelled multiplex amplification primer set containing human genome DNA 22 loci, kit and application thereof
InactiveCN106591463AHigh sensitivityImprove scalabilityMicrobiological testing/measurementDNA/RNA fragmentationHuman DNA sequencingStr typing
The invention provides a fluorescence labelled multiplex amplification primer set containing human genome DNA 22 loci, a kit and application thereof. The kit cooperates with a new hot start Taq enzyme and a corresponding premixed system of a PCR buffer solution to enhance the resistant ability of a PCR system to an inhibitor. At the same time, the PCR sensitivity is enhanced, all the 22 genome loci can be detected under a DNA template amount of 50pg, and the kit can be stored at 4DEG C for one year. The kit is suitable for forensic STR typing detection, parentage identification, individual recognition and other multiplex PCR application fields.
Owner:AGCU SCIENTECH
Method and system for acquiring autosomal STR gene locus typing result of single individual from mixed seminal stains
ActiveCN104232751AGood repeatabilityIntuitive test resultsMicrobiological testing/measurementStr typingTyping
The invention provides a method and a system for acquiring an autosomal STR gene locus typing result of a single individual from mixed seminal stains. The method comprises the following steps of: acquiring different Y-STR gene loci; acquiring autosomal STR gene locus haplotype typing results of a sufficient amount of Y type spermoblast; merging the autosomal STR gene locus haplotype typing results of the Y type spermoblast, which have same typing results of the different Y-STR gene loci to obtain the autosomal STR gene locus typing result of the single individual from the mixed seminal stains. The invention also provides the system for acquiring the autosomal STR gene locus typing result of the single individual from the mixed seminal stains. The method and system which are provided by the invention can acquire the typing result of the single individual from the mixed seminal stains by comprehensively analyzing autosomal STR and Y-STR typing results, thereby providing a powerful technical support for a public security organ to fast determine a criminal suspect.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Multiple PCR targeted capture typing system and kit based on high-throughput sequencing technology
ActiveCN110499372AImprove balanceAchieve single-tube amplificationMicrobiological testing/measurementPolymerase LConsanguinity
The present invention discloses a multiple PCR targeted capture typing system and kit based on a high-throughput sequencing technology, and relates to the technical field of nucleic acid in-vitro detection. The STR typing system comprises PCR primers for amplification of 58, 119, or 179 linked autosomal STR gene loci. The kit comprises PCR primer combination, Index linker sequences, an IGT-EM707 polymerase mixture, an amplification buffer enhancer NB, a YF buffer B, library building reagents, etc. The provided STR typing system and kit can achieve single-tube amplification of the 58, 119, or 179 linked autosomal STR gene loci, are good in balance, high in sensitivity, good in specificity, and accurate in typing results, and can be used to identify complex consanguinity relationships of different consanguinity relationships at a same level.
Owner:SHANXI MEDICAL UNIV
Composite amplification system based on Y-STR locus and specific primer combination thereof
ActiveCN108866201AMeet the inspection requirementsEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationStr typingAgricultural science
The invention discloses a composite amplification system based on a Y-STR locus and a specific primer combination thereof. The primer combination provided by the invention is formed by 30 kinds of DNAmolecules shown as sequence 1 to sequence 30 in a sequence table. The primer combination is used for building the composite amplification system based on the Y-STR locus; then, male individual STR typing is performed; the detected polymorphism is high; the population screening and individual recognition capability of a Y chromosome genetic marker can be enhanced; particularly important identification values are realized on the distant relatives of the same family. Therefore when the primer combination provided by the invention is used for sample screening, the cost is low; the efficiency is high; important application values are realized.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Llinked autosomal (STR typing system based on high-throughput sequencing technology and kit
ActiveCN110734982AImprove balanceAchieve single-tube amplificationMicrobiological testing/measurementDNA/RNA fragmentationStr typingTyping
The invention claims a linked autosomal short tandem repeat (STR) typing system based on a high-throughput sequencing technology and a kit, and relates to the technical field of nucleic acid in-vitrodetection. The STR typing system includes PCR primers for amplification of 61, 121 or 179 linked autosomal STR loci, and the kit includes a PCR primer combination, an Index linker sequence, an IGT-EM707 polymerase mixture, an amplification buffer enhancing agent NB, a YF buffer liquid B and a library building reagent. The STR typing system and kit provided by the invention can realize single-tubeamplification of the 61, 121 or 179 linked autosomal STR loci, have good balance, high sensitivity, good specificity, and accurate typing results, and can be used to identify complex genetic relationships of different genetic relationships at a same level.
Owner:SHANXI MEDICAL UNIV
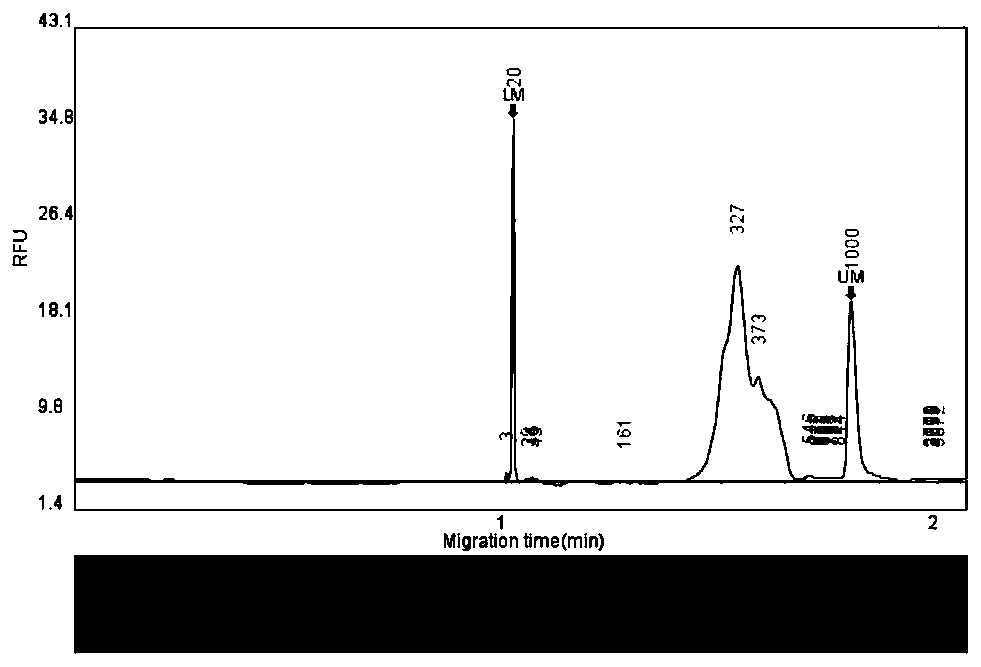
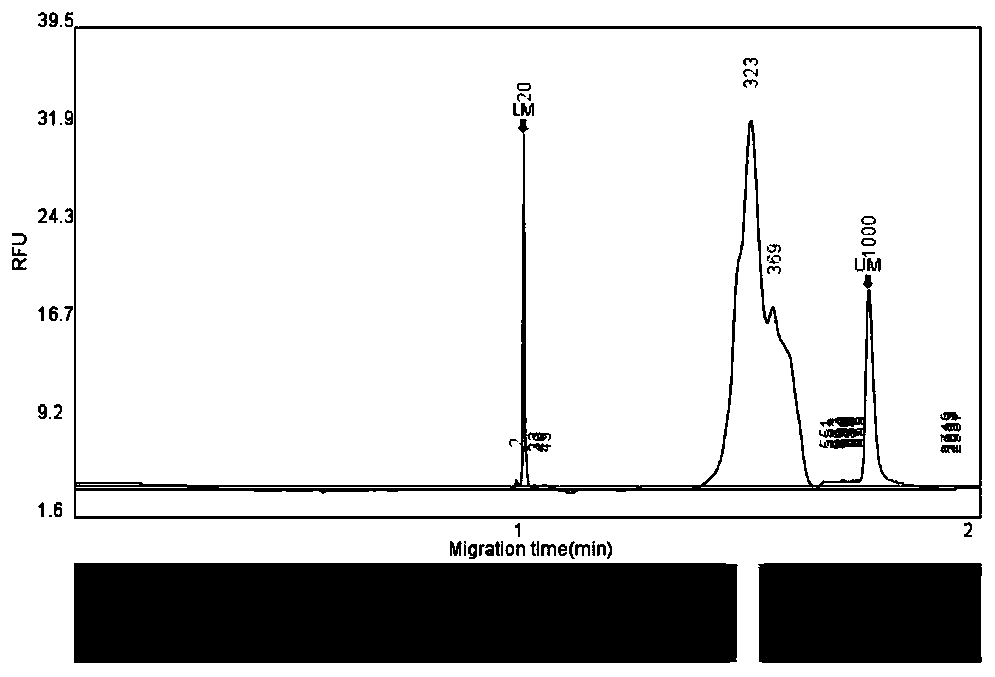
Molecular weight internal lane standard and application thereof
InactiveCN105296473AEasy to distinguishEasy to readMicrobiological testing/measurementDNA/RNA fragmentationStr typingInternal standard
The invention belongs to the technical field of biology and discloses a molecular weight internal lane standard. The molecular weight internal lane standard comprises n nucleotide fragments with recognizable markers and different molecular weights, wherein the n nucleotide fragments include at least p nucleotide fragment pairs differing by one basic group respectively, in other words, the basic group number of one fragment of any nucleotide fragment pair differing by one basic group is m, and the basic group number of the other fragment is m+1 or m-1. Compared with existing commercial molecular weight internal lane standards, the molecular weight internal lane standard has the advantages that the molecular weight internal lane standard is capable of distinguishing single basic group, effectively differentiating PCR (polymerase chain reaction) product fragments differing by one basic group and guaranteeing correct typing to facilitate interpretation of detection personnel, thereby being widely applicable to STR typing, sequencing, large-fragment analysis and genome analysis.
Owner:SUZHOU NUHIGH BIOTECH
str type standard substance
ActiveCN102286476AMicrobiological testing/measurementVector-based foreign material introductionNucleotideGenetic engineering
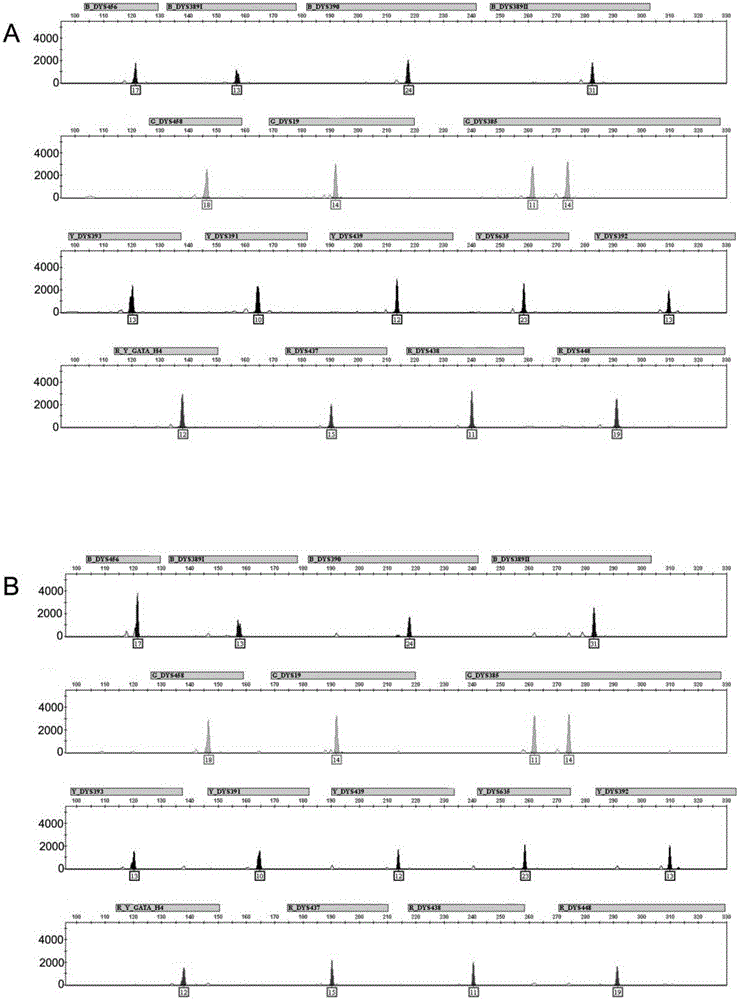
The invention discloses a short tandem repeat (STR) parting standard substance, which is a deoxyribonucleic acid (DNA) mixture and comprises 31 DNA segments with the nucleotide sequences as 1) to 31). In the invention, all allelic genes in the bits of Chinese people groups are researched and studied through focusing on the discovered STR gene bits in the existing forensic molecular genetics, and in addition, the biostatistics technology is used for studying the gene frequency distribution of each allelic gene. On the basis, the experiments techniques such as molecular genetics, molecule biology, cell biology, genetic engineering and the like are integrally utilized, the allelic gene DNA information base is developed and built for the first time, all of the allelic gene DNA segments such as STR bits are included, further, the forensic DNA standard reference system independently developed by China is further built, and the special artificially synthesized STR standard substance is formed.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
DNA examination reagent for sample examination, and special primer thereof
ActiveCN104911247AComplete typingQuick checkMicrobiological testing/measurementDNA/RNA fragmentationStr typingForensic dna
The invention discloses a DNA examination reagent for sample examination, and a special primer thereof. A DNA examination primer group provided by the invention is composed of a primer pair group used for amplifying 21 Y-STR sites, a primer pair for amplifying a sex site Amelogenin, and a primer pair for amplifying an autosome SRT site. Experiments prove that the DNA examination reagent can simultaneously examine the 21 Y-STR sites and the autosome SRT site, and the primer specificity is high, so complete SRT typing can be obtained, peak patterns are sharp and intelligible, the balance is good, no Pull-up peaks or stutter bands appear, no non-specific artificial products appear, and legal medical experts' DNA large scale sample examination requirements are completely met.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
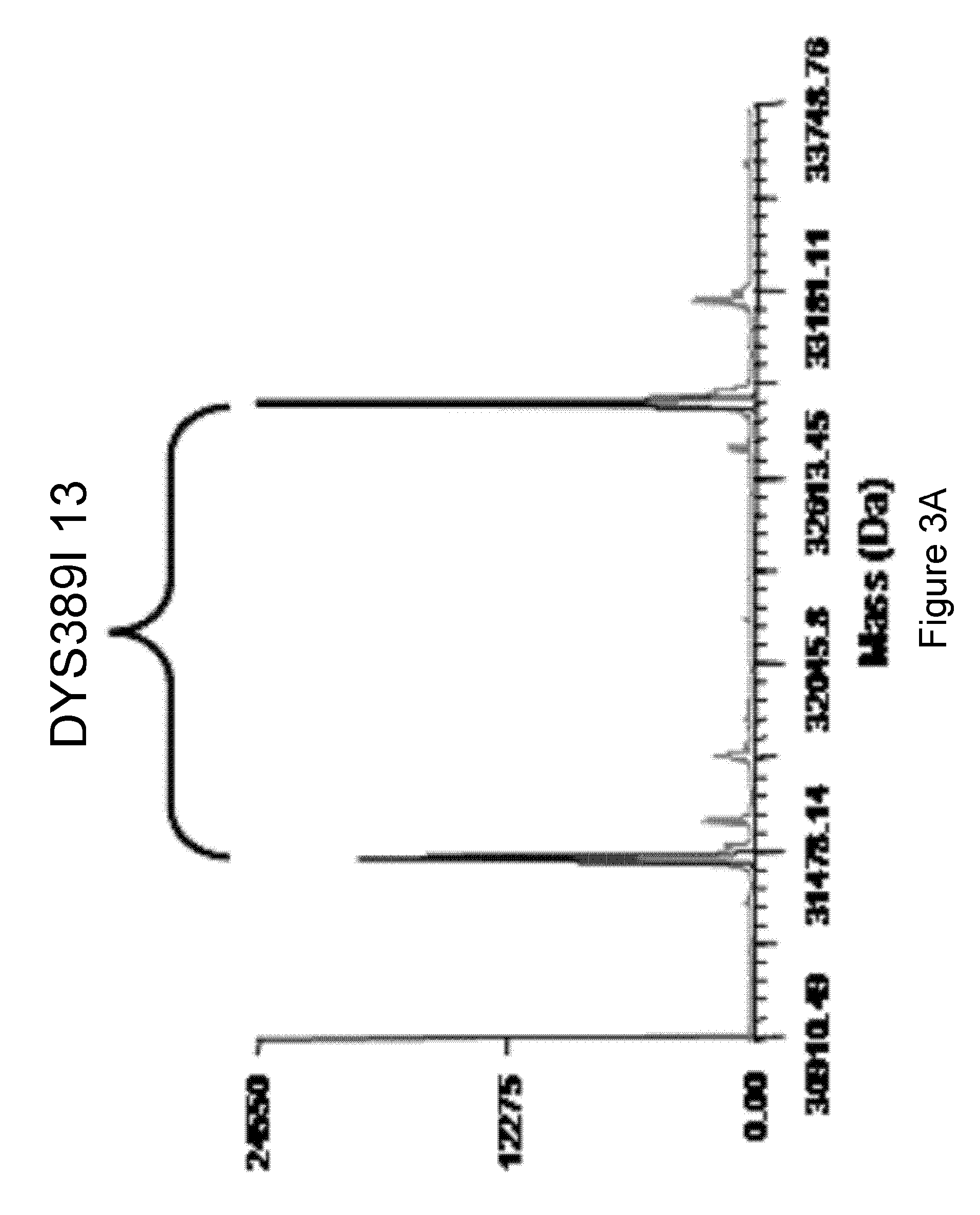
Methods For Rapid Forensic DNA Analysis
InactiveUS20120021427A1Minimizing non-templated adenylationSugar derivativesMicrobiological testing/measurementStr typingForensic dna
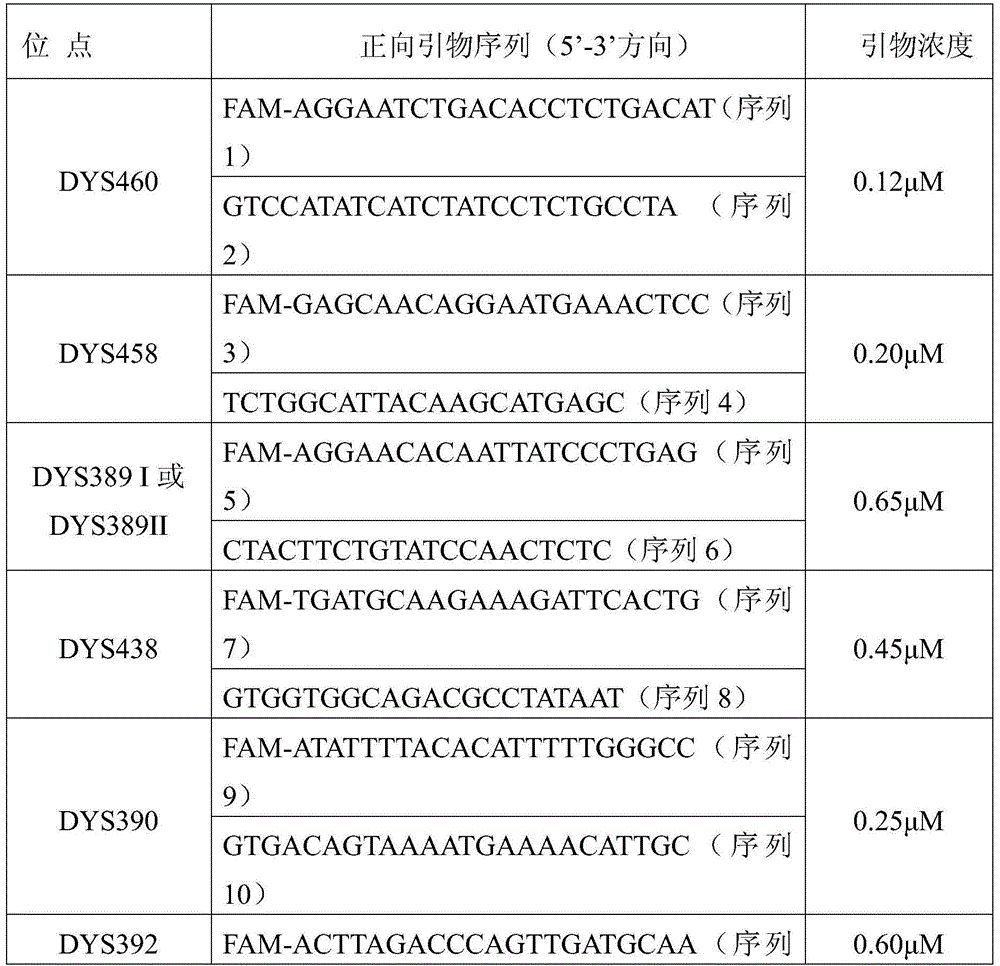
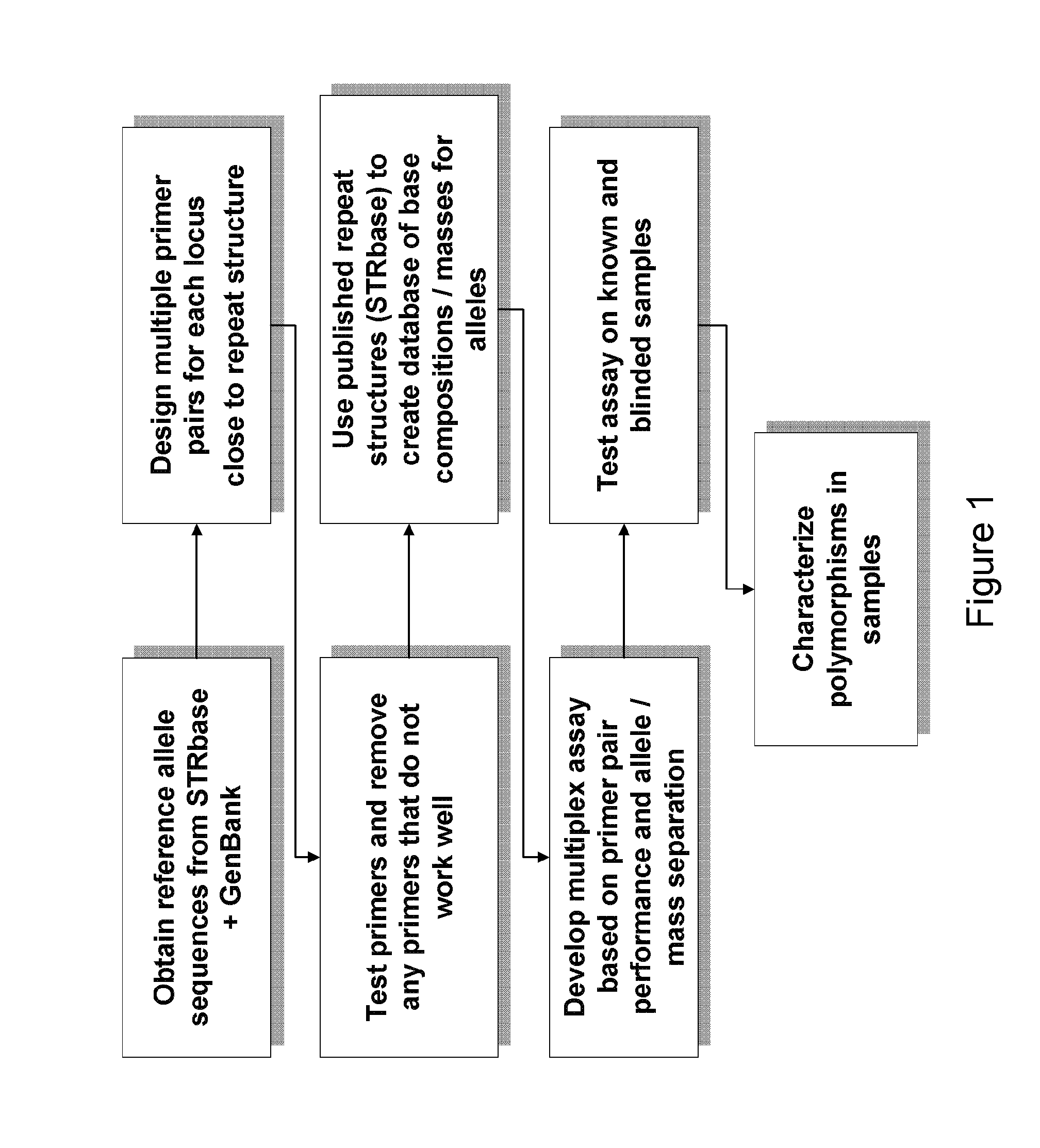
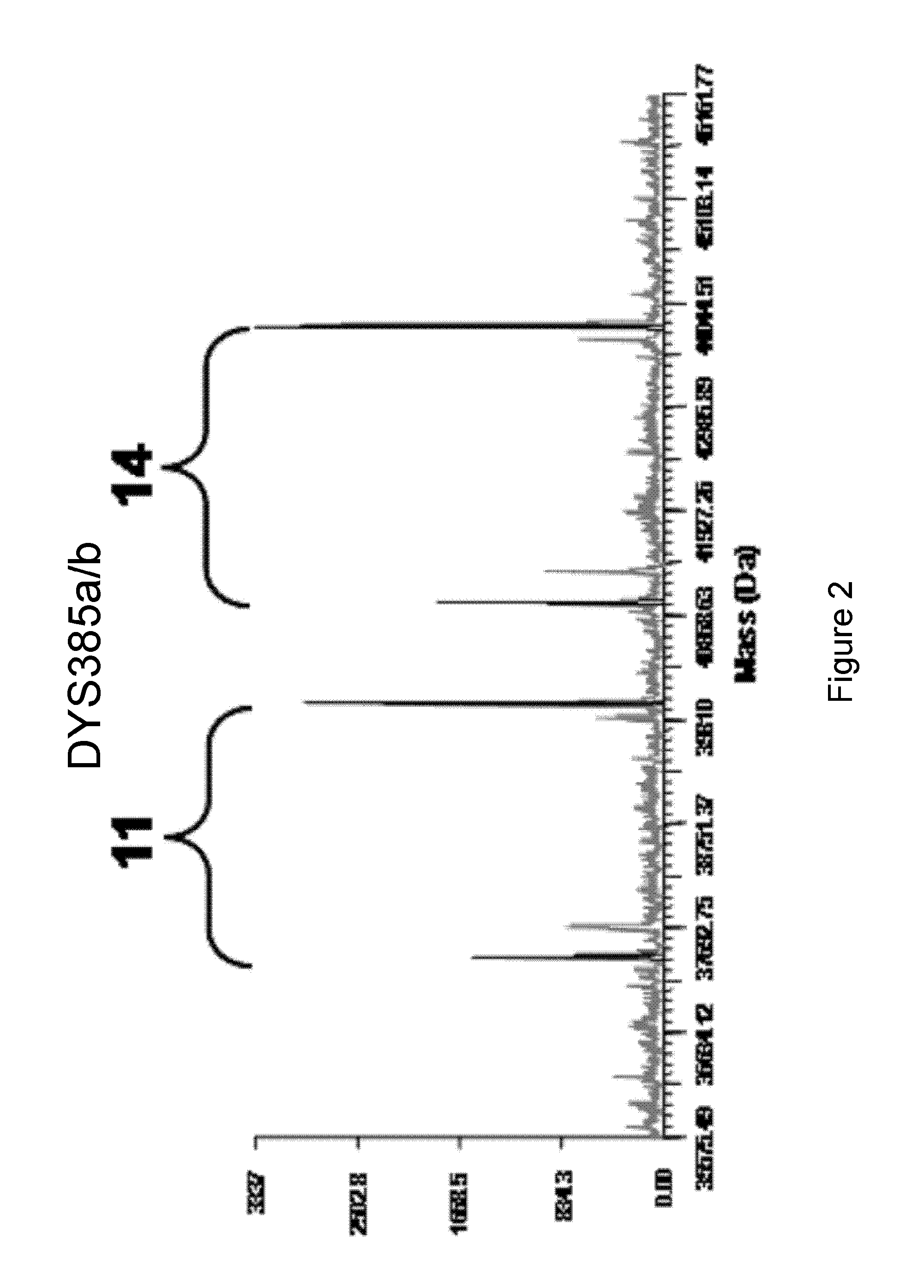
The present invention provides methods and primer pairs for rapid, high-resolution forensic analysis of DNA and STR-typing by using amplification and mass spectrometry, determining the molecular masses and calculating base compositions of amplification products and comparing the molecular masses with the molecular masses of theoretical amplicons indexed in a database.
Owner:IBIS BIOSCI
Method and system for Y-STR typing of individual man
ActiveCN106520973AShort detection timeAchieve paternal kinship identificationMicrobiological testing/measurementStr typingTyping
The invention provides method and system for Y-STR typing of an individual man. The method comprises the following steps: extracting the DNAs of the individual man to acquire the genotypes of 16 Y-chromosome STR gene loci of the DNAs, wherein the STR gene loci are DYS458, DYS390, DYS438, DYS392, DYS393, DYS437, DYS385a / b, GATA_H4, DYS391, DYS447, DYS19, DYS448, DYS456, DYS393, DYS635 and DYS439; and acquiring the Y-STR typing results of the individual man according to the 16 STR gene loci of the individual man. The invention also provides a system for rapid Y-STR typing of the individual man. The method and system provided by the invention can realize rapid, stable and high-quality Y-chromosome STR typing and apply the Y-STR typing results to rapid identification of patriarchal relatives, family checking and the like.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
A method for data transmission of allele of a locus by two-dimensional code video
InactiveCN109146036AOvercoming Human Transcription ProblemsSolve data transmissionCo-operative working arrangementsStr typingIsolated system
The invention discloses a method for gene locus allele data transmission by using two-dimensional code video. The method for data transmission of allele of a gene locus by using a two-dimensional codevideo comprises that follow steps of: by coding the locus allele data of each sample generated by STR typing software identified by different DNA kits, the two-dimensional code video frame is successively obtained from the two-dimensional code video frame; after the two-dimensional code frame image of one frame data in the two-dimensional code video frame is successfully read, a response identification two-dimensional code image is formed, and the data two-dimensional code frame image of the next frame is played. The method for visually exchanging data between two physically isolated system terminals by using two-dimensional code as a medium overcomes the problem that the data in the prior art is artificially copied, and overcomes the problem that a single bar code and two-dimensional code cannot transmit multiple identification samples of a kit at one time.
Owner:ANHUI UNIVERSITY OF TECHNOLOGY AND SCIENCE
Analysis method for multiplex amplification STR data
PendingCN112786107AExact typing methodImprove accuracySequence analysisInstrumentsMultiplexWhole genome sequence analysis
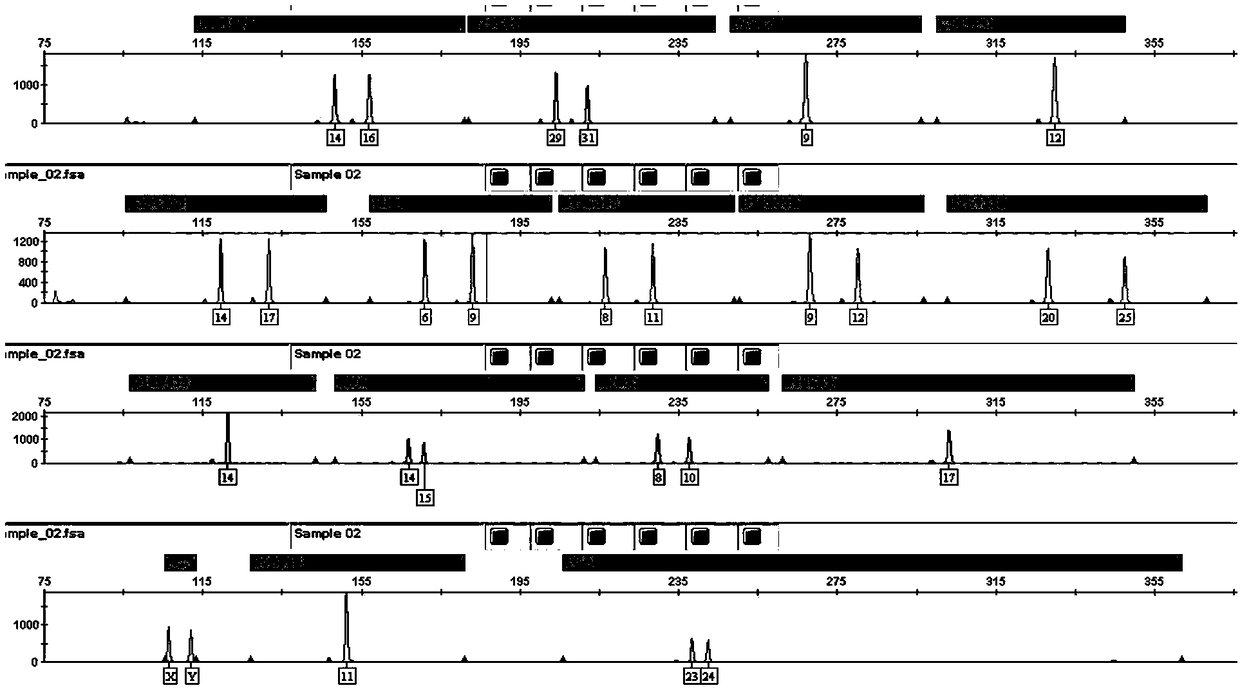
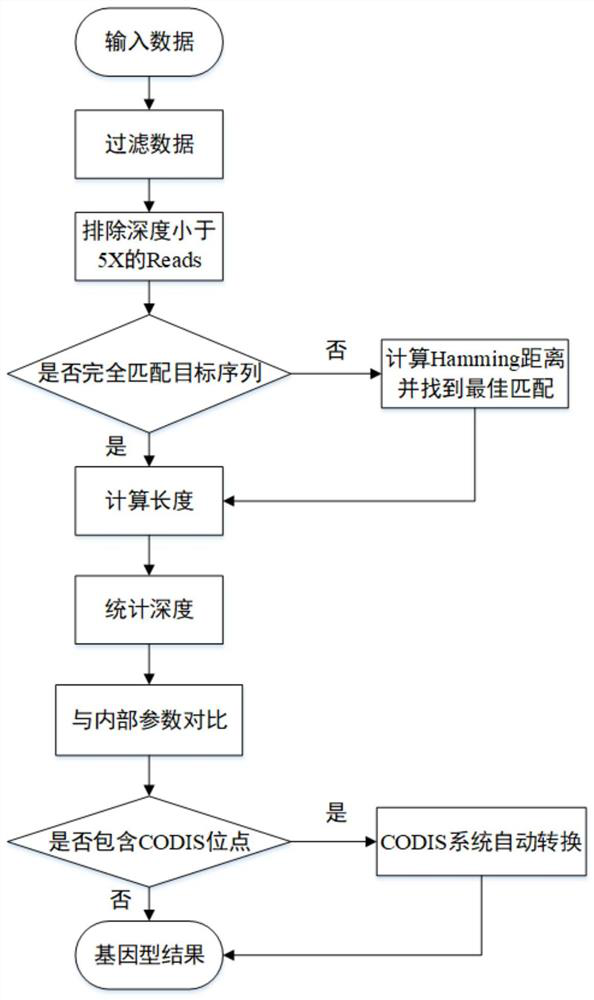
The invention discloses an analysis method for multiplex amplification STR data. The analysis method comprises the following steps: 1, site reference information: inputting data; 2, quality control (preprocessing): filtering data; 3, an STR typing method: a) searching for optimal matching, and b) referencing internal reference data. According to the method, high typing accuracy and timeliness can be guaranteed, CODIS system automatic conversion of STR loci can be achieved, the consistency of the STR loci and capillary electrophoresis data reaches up to 100% in the aspect of accuracy, and the analysis time is obviously shorter than that of mainstream analysis software in the market in the aspect of timeliness. Therefore, as an important supplementary method for whole genome sequence analysis, the process has great practical significance to forensic genetics.
Owner:深圳百人科技有限公司
29-plex Y-STR typing system for family search and paternal biogeographic ancestor inference
PendingCN112143816AEasy to operateType easy to readMicrobiological testing/measurementStr typingMedicine
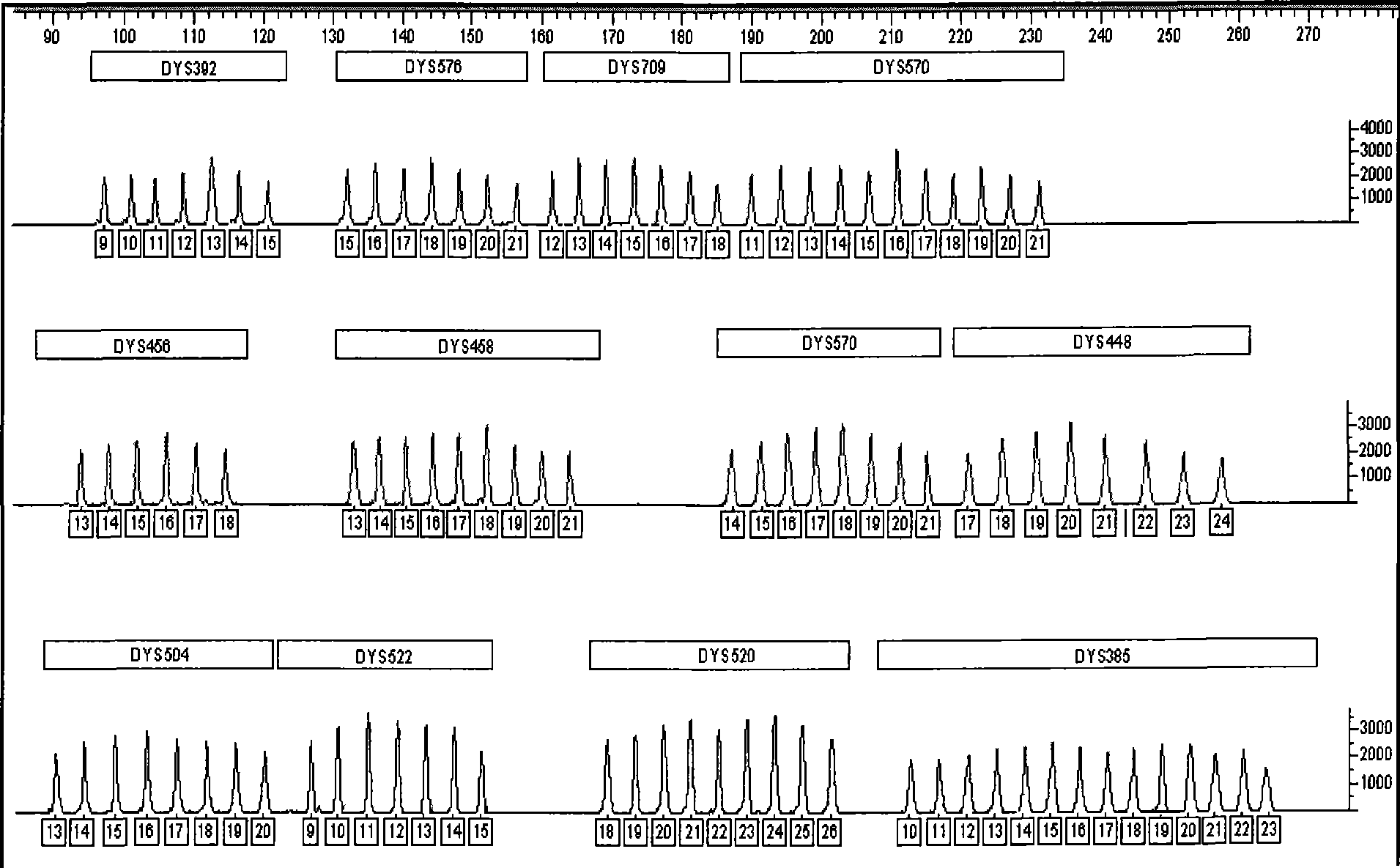
The invention provides a 29-plex Y-STR typing system for family search and paternal biogeographic ancestor inference. Specifically, a Y chromosome short tandem repeat typing kit provided by the invention contains detection reagents for specifically detecting Y chromosome STR loci in the following groups (a), (b) and (c) that: the group (a) includes DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392,DYS393, DYS385, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and Y-GATA-H4; the group (b) includes DYS526a, DYS526b, DYS570, DYS576 and DYS626; and the group (c) includes DYS388, DYS460, DYS481, DYS593, DYS596, DYS643 and DYS645. The typing system disclosed by the invention has the advantages of simplicity in operation, easiness in typing reading, high amplification specificity, balanced typing result peak value, reliable and stable performance and the like.
Owner:ACADEMY OF FORENSIC SCIENCE
A kind of str kit and its application for trace and degraded samples
ActiveCN106521013BIncrease the number of detectionsHigh sensitivityMicrobiological testing/measurementStr typingGenetics
Owner:AGCU SCIENTECH +1
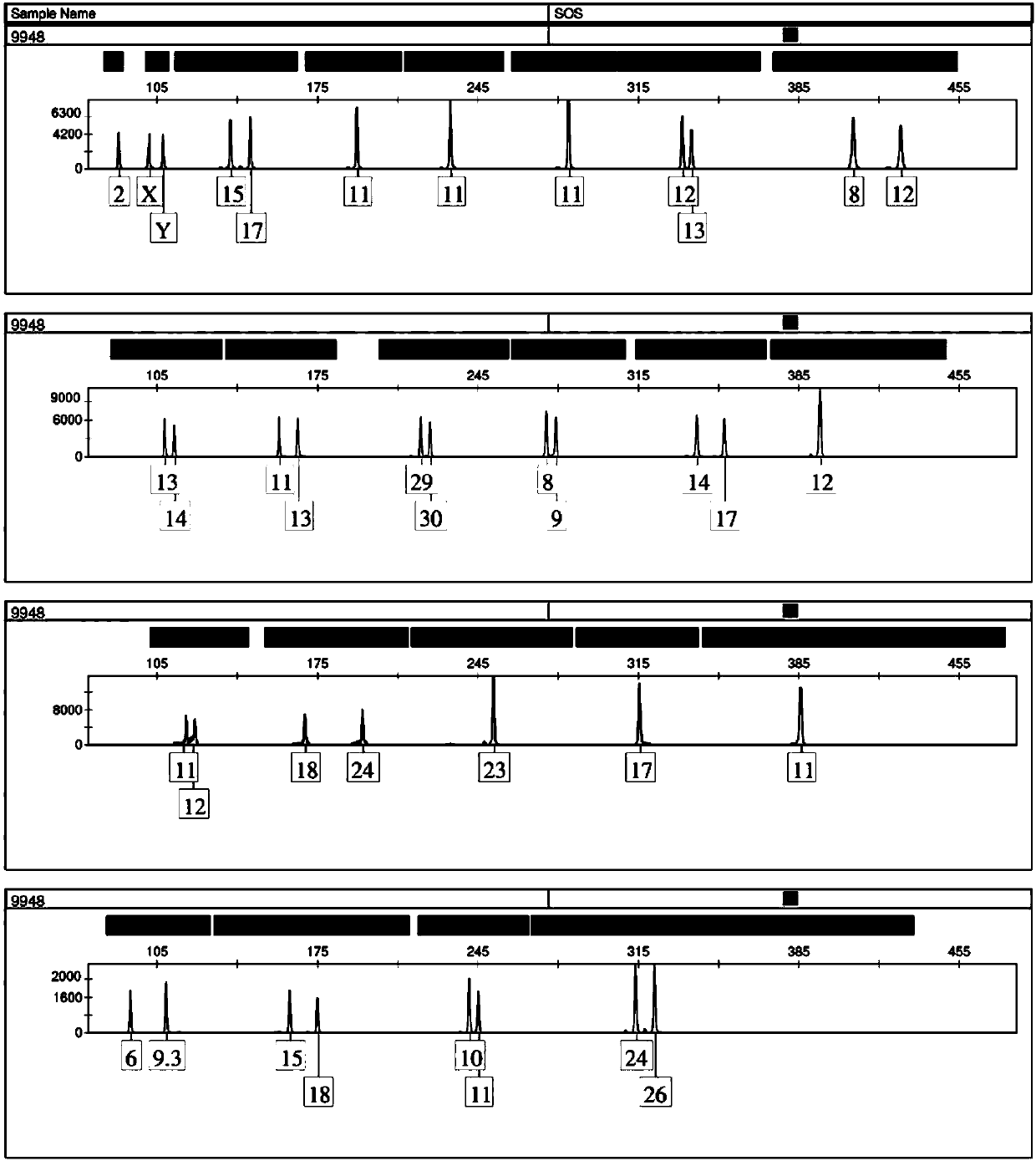
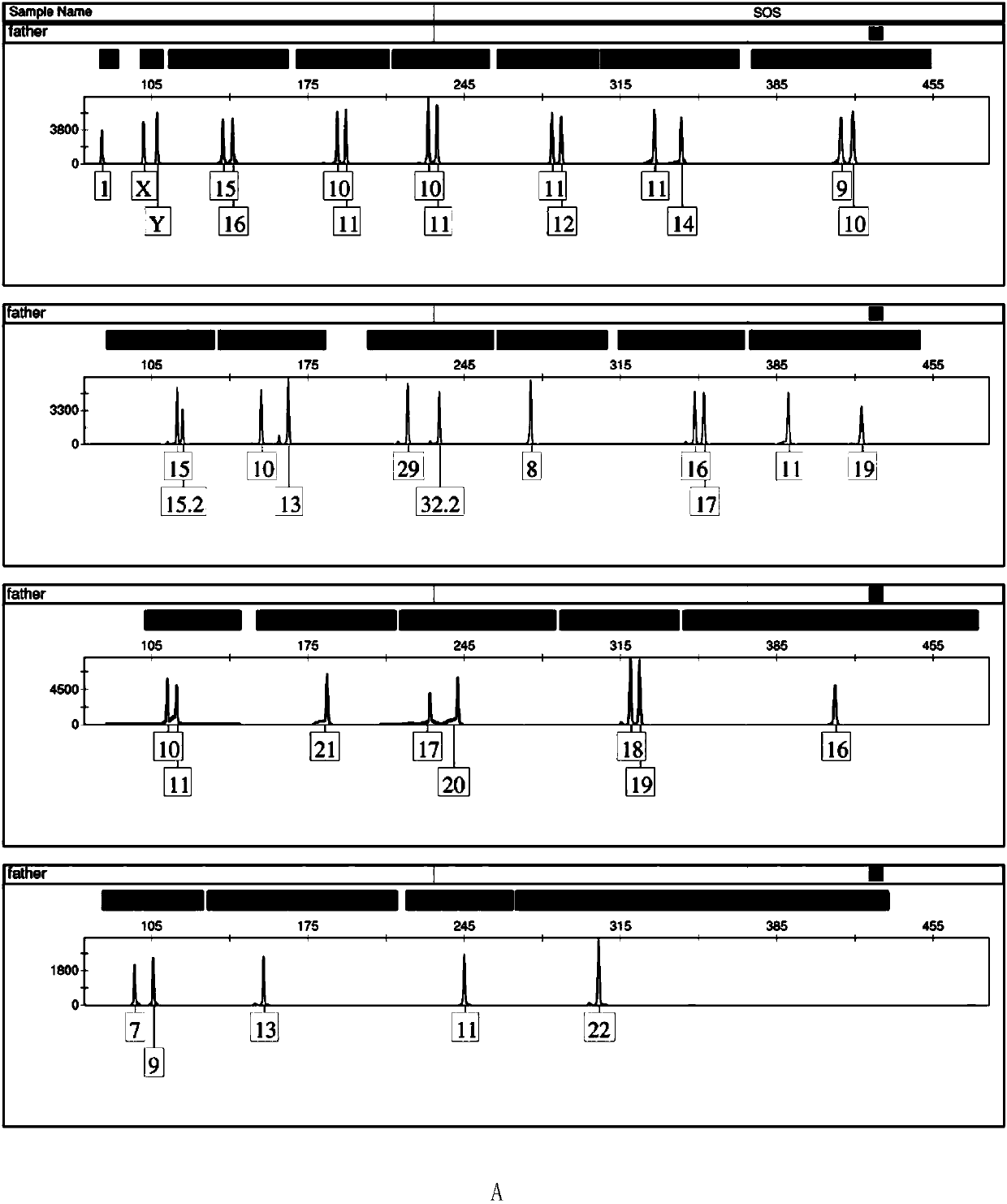
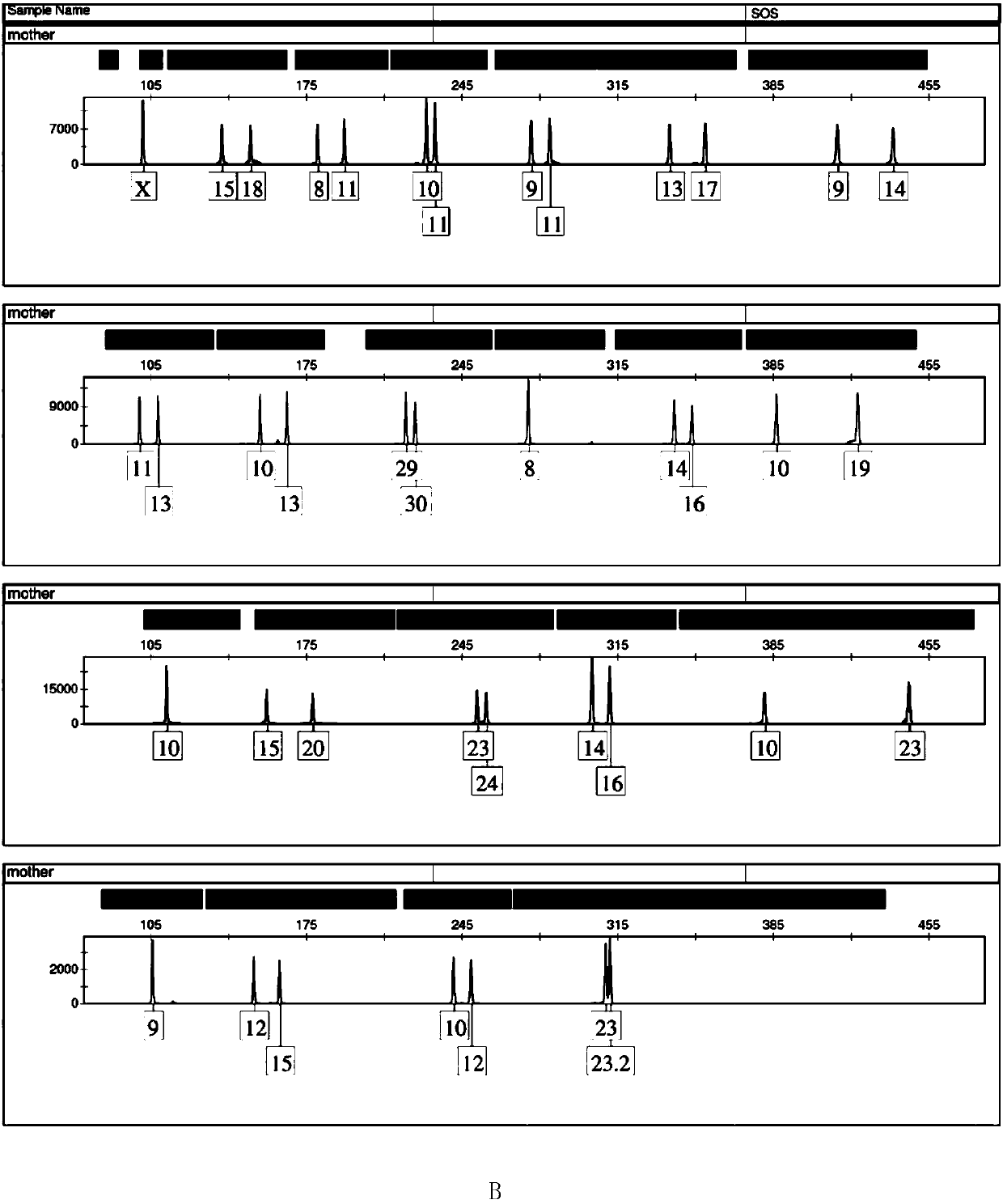
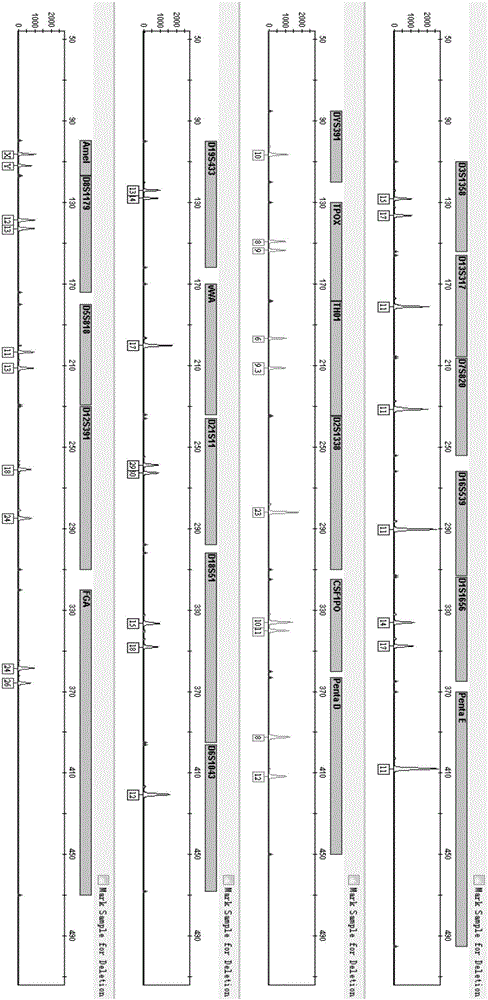
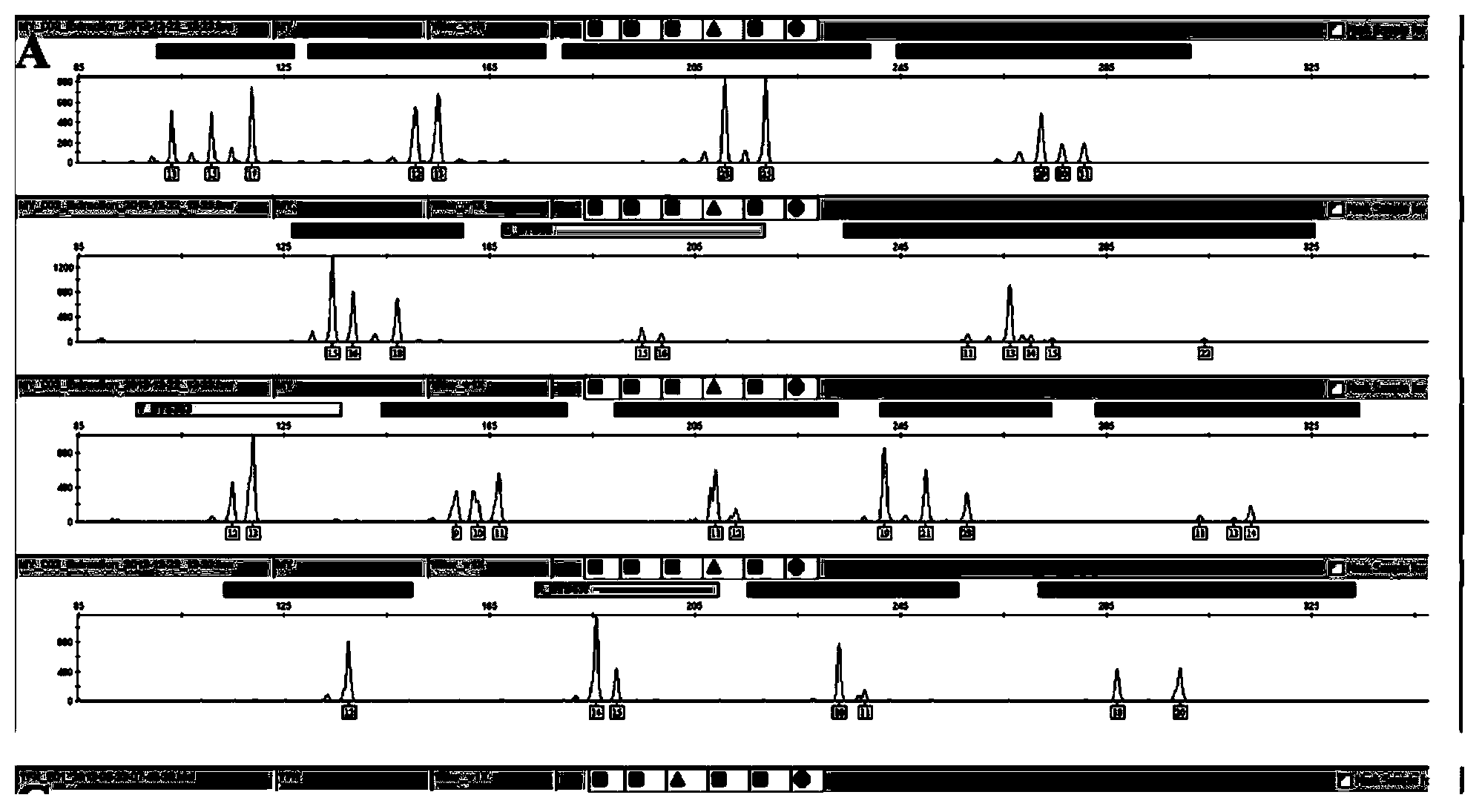
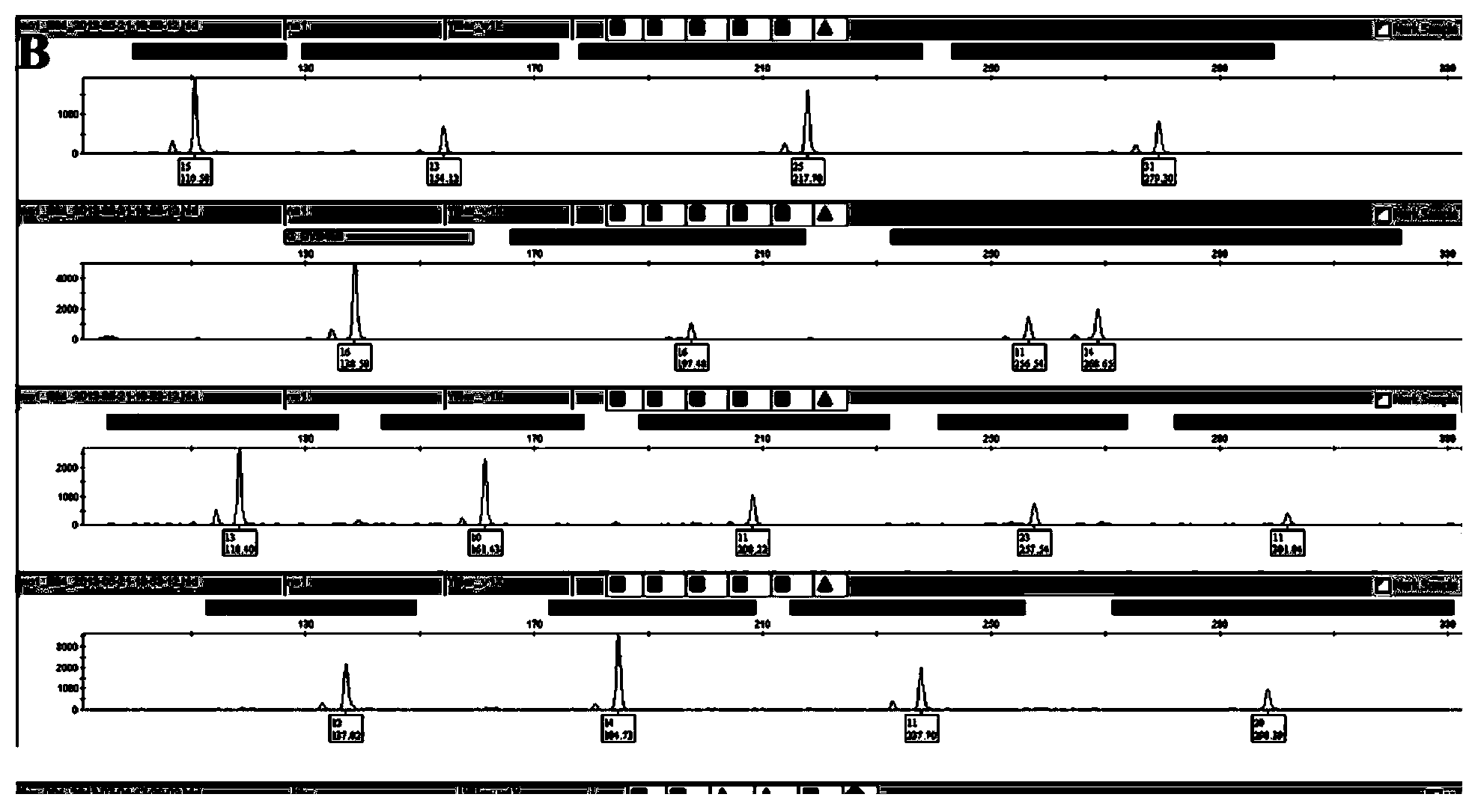
Positive reference for STR typing
The invention discloses a positive reference for STR typing. The positive reference for STR typing is prepared according to STR gene locus sites discovered in forensic molecular genetics. Experimental results show that the positive reference for STR typing can totally replace human genome DNA of 9948 for STR typing detection. Thus, the positive reference for STR typing can be used as a standard substance for a plurality of commercial kits and has important application value.
Owner:INST OF FORENSIC SCI OF MIN OF PUBLIC SECURITY
Popular searches
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com