Five-color fluorescence STR parting method for synchronously detecting 23 gene loci and special kit of method
A gene locus and locus technology, applied in the biological field, can solve the problem of waste of DNA database data resources, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1, the preparation of the compound amplification system based on 23 gene loci
[0043] 1. Screening of 23 gene loci
[0044] In order to meet the requirements of the amount of information, individual recognition and compatibility of the detection of STR gene loci, the inventors of the present invention adopted five-color fluorescent labels to increase the number of accommodated loci, and at the same time increased the compatibility of the kit’s loci, making it compatible with white The basic standard allelic loci of the race, yellow race, black race and brown race. The inventors of the present invention finally screened a total of 23 gene loci including 21 autosomal STR gene loci, 1 sex identification locus AMEL and 1 indel locus Yindel. The 21 autosomal STR loci are D3S1358, D13S317, D7S820, D16S539, D8S1179, Penta D, D19S433, D5S818, D21S11, TPOX, D1S1656, D6S1043, D2S441, D12S391, D2F8, D13PO38CS, E5S01, Penta and FGA.
[0045] 2. Screening of primers ...
Embodiment 2
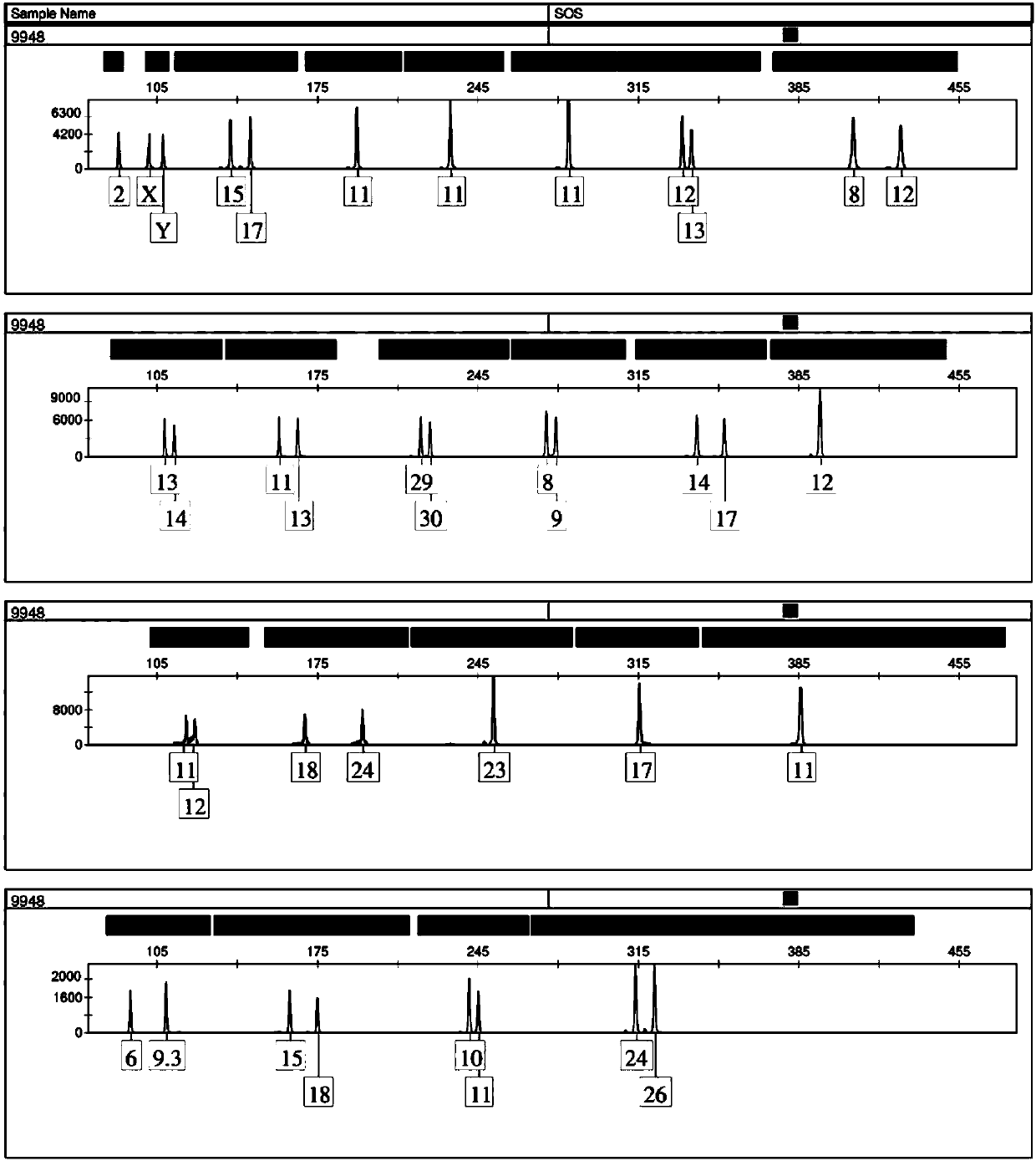
[0099] The type detection of the DNA standard 9948 based on the multiple amplification system of 23 gene loci prepared in Example 2 and Example 1
[0100] DNA standard 9948 (concentration: 10ng / μL) is a product of Xinhai Biotechnology Co., Ltd.
[0101] 1. Acquisition of DNA Standard Diluent
[0102] Take 1 μL of DNA standard 9948 and add 9 μL of TE buffer to obtain a dilution of DNA standard 9948 (concentration: 1 ng / μL).
[0103] 2. Take 15 μL of the complex amplification system based on 23 gene loci prepared in step 4 of Example 1, add 0.4 μL hot start Taq enzyme (including 2U), 1 μL DNA standard 9948 dilution and 8.6 μL nuclease-free water , to get the reaction system.
[0104] 3. Put the reaction system obtained in step 2 into a Bioer G-1000 thermal cycler for PCR amplification to obtain PCR amplification products. See Table 6 for the temperature cycle conditions of the Bioer G-1000 thermal cycler.
[0105] Table 6. Temperature Cycling Conditions
[0106]
[0107]...
Embodiment 3
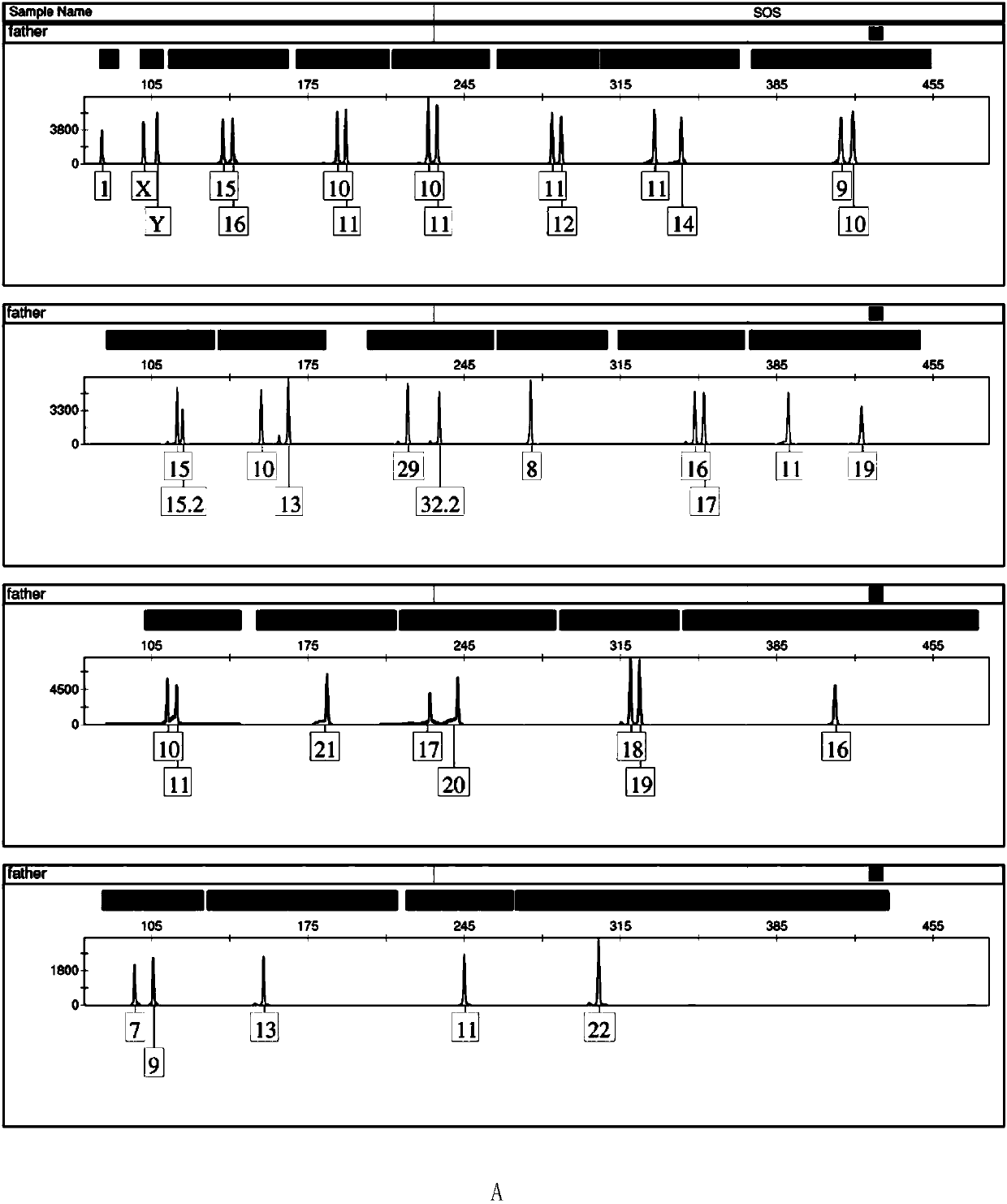
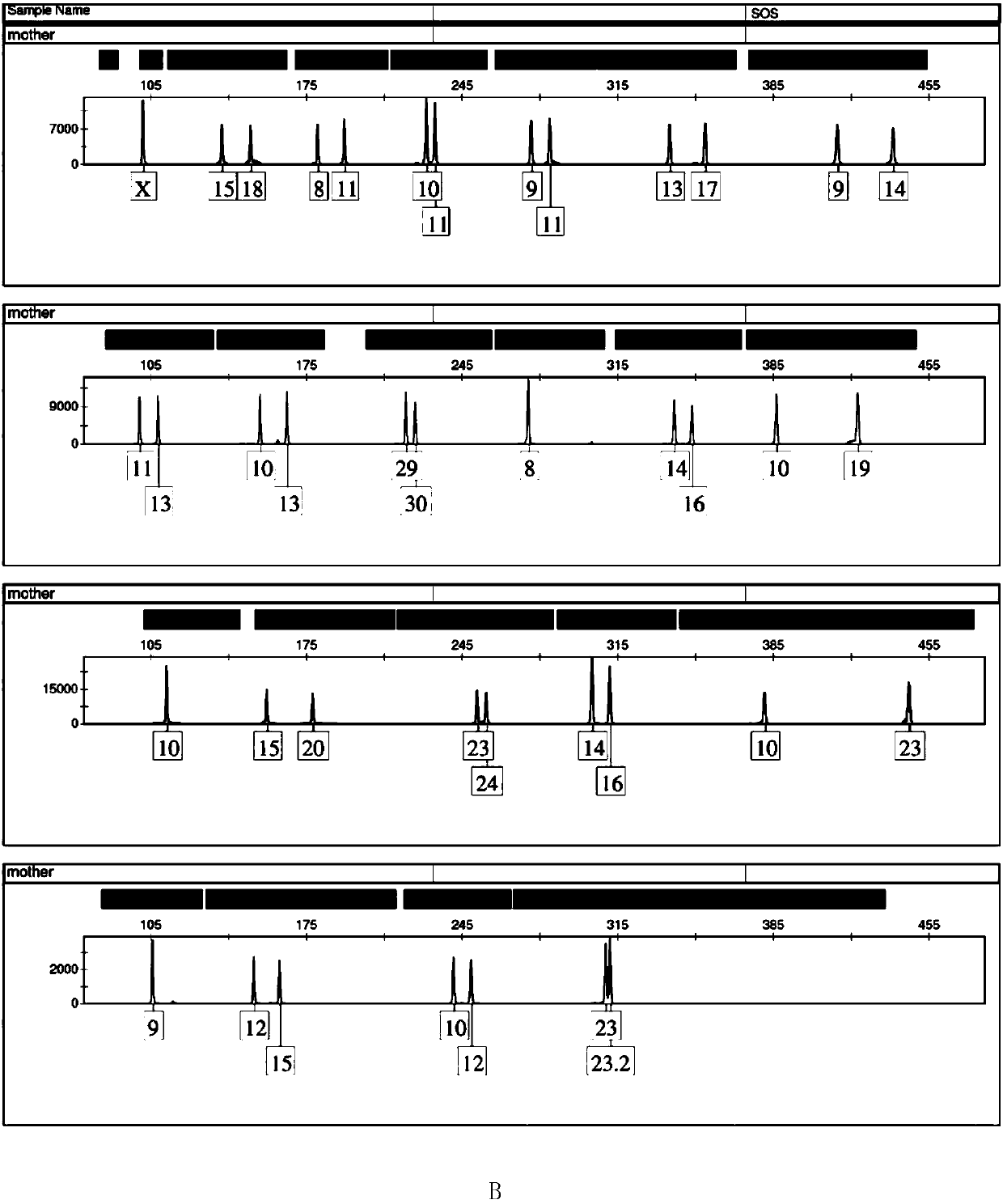
[0117] Example 3, the typing detection of a triplet family based on the multiple amplification system of 23 gene loci prepared in Example 1
[0118] The blood samples of the triplet family (the identification result is identified) were provided by Guangdong Huada Forensic Evidence Identification Institute.
[0119] 1. Obtaining genomic DNA from family blood samples
[0120] Genomic DNA was extracted from the blood samples of triplet families by chelex-100 method.
[0121] 2. Take 15 μL of the complex amplification system based on 23 gene loci prepared in step 4 of Example 1, add 0.4 μL of hot-start Taq enzyme (including 2 U), 1 μL of genomic DNA from the family blood sample and 8.6 μL of nuclease-free water , to get the reaction system.
[0122] 3. Same as step 3 of embodiment 2.
[0123] 4. Same as step 4 of embodiment 2.
[0124] 5. Same as step 5 of embodiment 2.
[0125] 6. Same as step 6 of embodiment 2.
[0126] 7. Same as Step 7 of Example 2.
[0127] The typing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com