Group of SNP markers for identifying donkey species, primer group of group of SNP markers and kit prepared by primer group
A primer set and labeling technology, applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve problems such as being easily affected by living environment and activities, spending a lot of time, and face recognition interference, etc. Achieve good generalization, easy operation, and short amplification detection fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1) Group data acquisition:
[0022] The whole genome resequencing data of 120 donkeys were obtained, and there was no biological relationship between 120 donkey individuals. 60 samples were used to screen SNP sites (training set), and the remaining 60 samples were used as a verification set.
[0023] 2) Reference genome acquisition:
[0024] The latest version of the donkey reference genome file was obtained from the UCSC database (http: / / genome.ucsc.edu / ).
[0025] 3) Raw data format conversion:
[0026] Use fastq-dump to convert the original .sra format to double-ended .fq format, and then use gzip to compress .fa to .fq.gz format.
[0027] 4) Data quality control:
[0028] FastQC (v0.11.5) was used for quality control of the raw data, and the sequencing quality was basically qualified and could be used for subsequent analysis.
[0029] 5) SNP loci typing
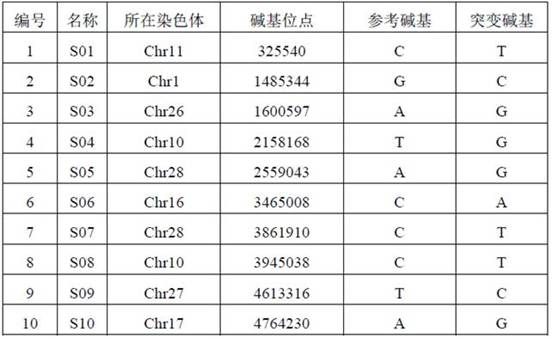
[0030] Use GATK to type the sample data to obtain a VCF file, including the location of the SNP site, etc. ...
Embodiment 2
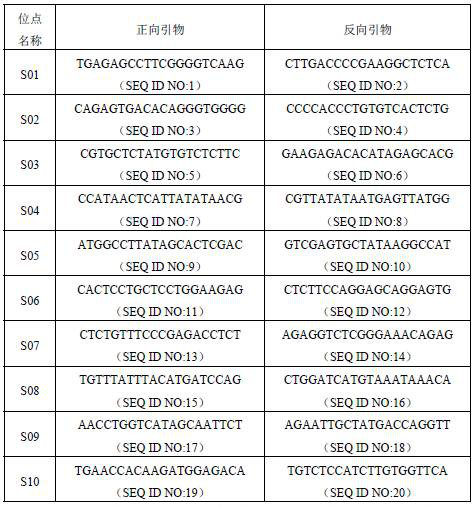
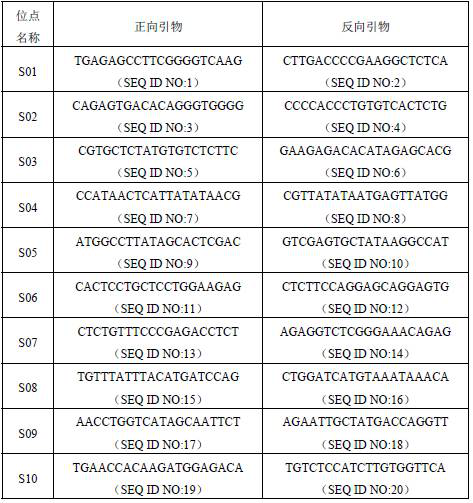
[0049] For each SNP site in Example 1, use the primer design software Primer Premier to design forward primers and reverse primers for PCR reactions in the conserved sequences within 250 bp of the core sequences of the above 10 SNP sites. Primers, as shown in Table 2.
[0050] Table 2. Forward and reverse reference primers for 10 SNP sites
[0051]
[0052] The primer set is designed according to the conserved sequence within 250 bp of both ends of the core sequence of at least one of the SNP sites described in the first aspect of the present invention. The primer set is designed using the conserved sequence within 250bp at both ends of the core sequence of each SNP site to achieve the amplification of each SNP site, thereby ensuring the accuracy of the amplification.
[0053] The primer set is selected from at least one pair of the following: sequences shown in SEQ ID NO:1~SEQ ID NO:20. When performing multiplex PCR reactions, the primer sequences should be chosen with g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com