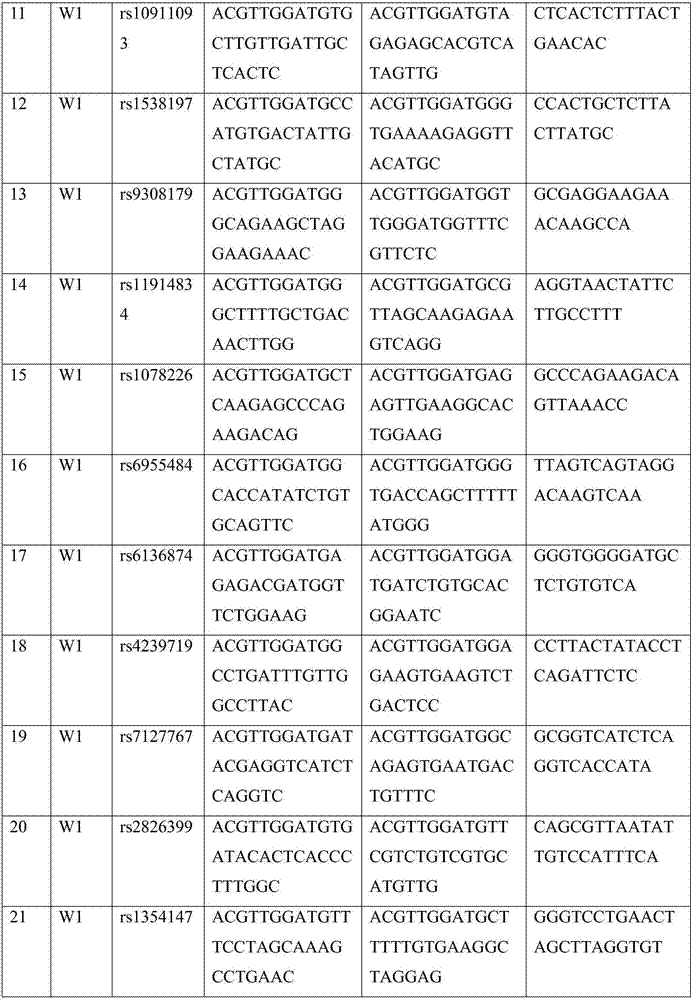
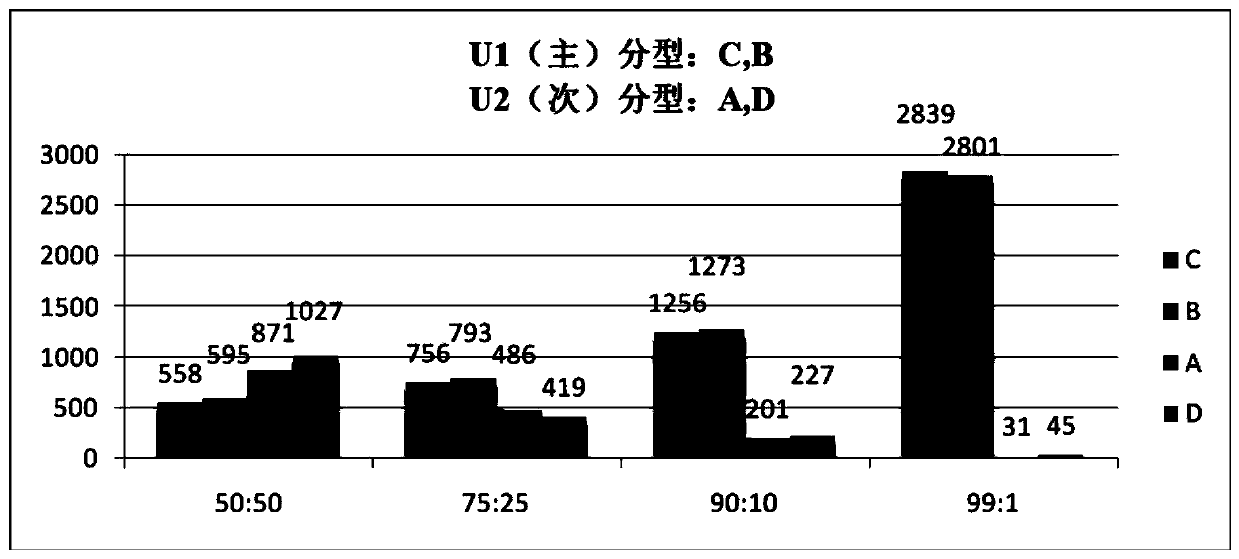
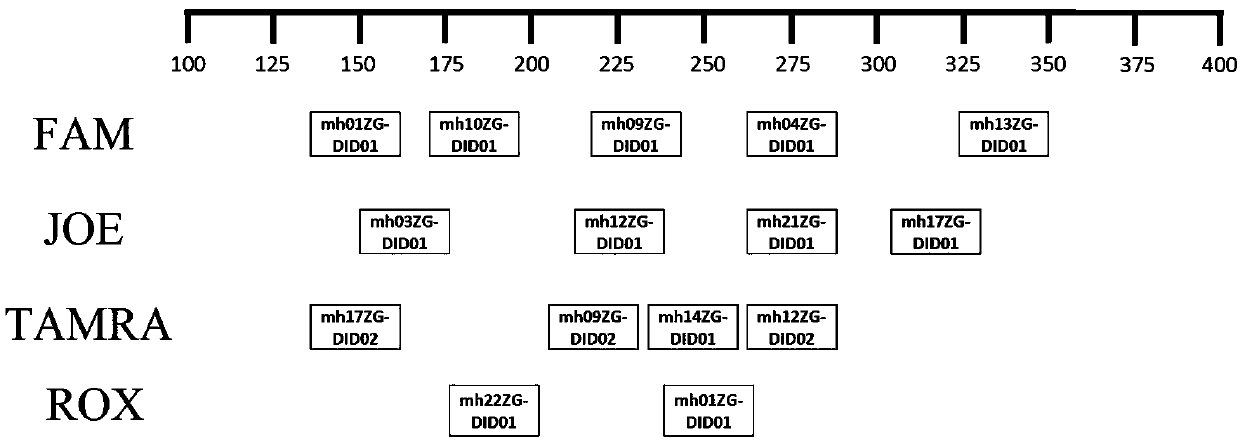
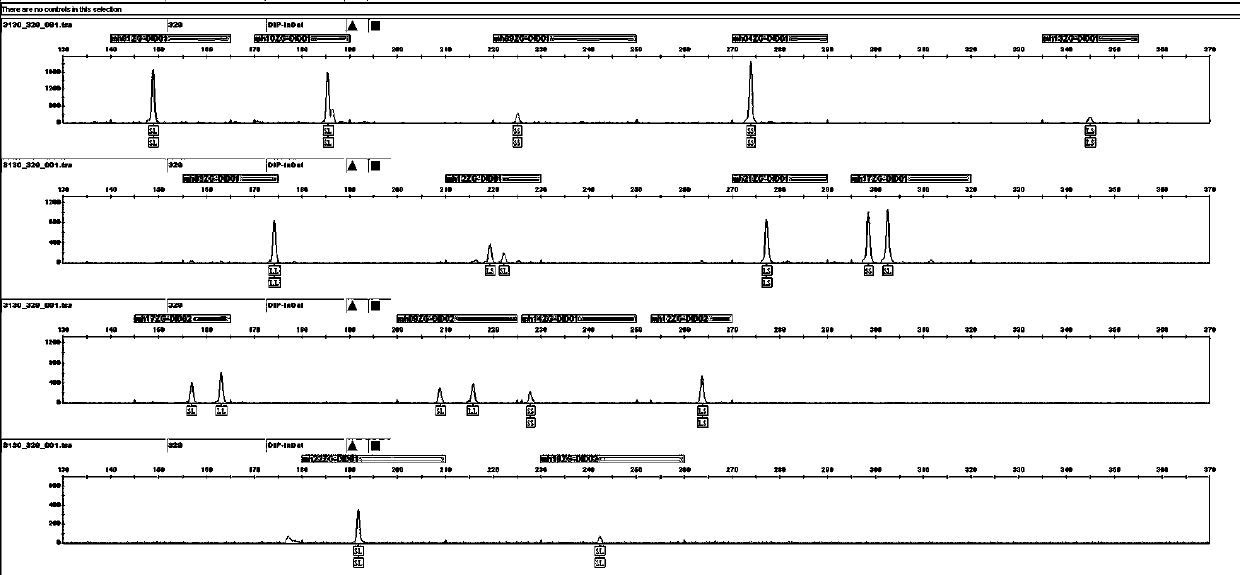
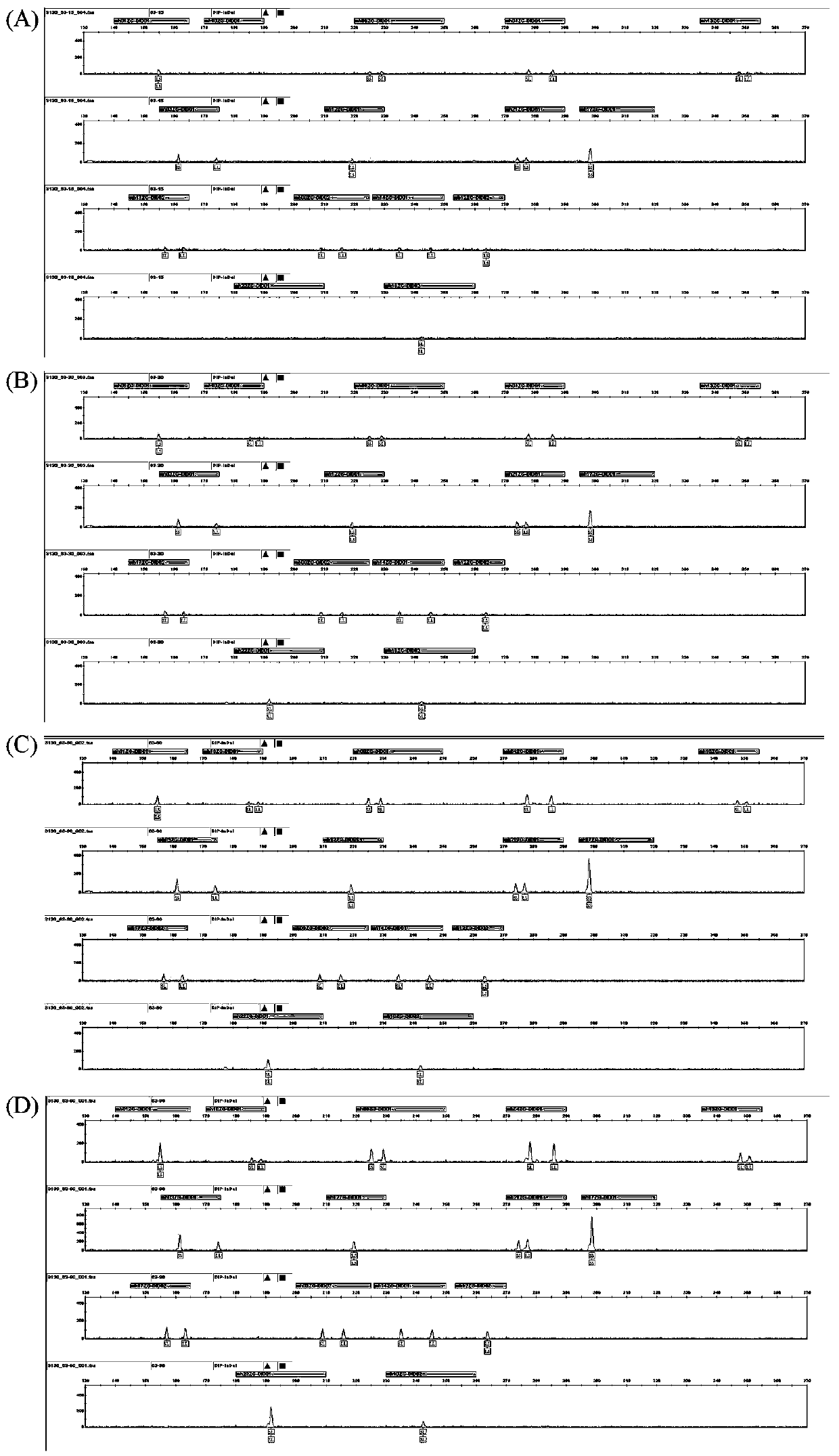
Patents
Literature
69results about How to "Low mutation rate" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Anti-aging methods and composition
InactiveUS20060204458A1Increase cell longevityFacilitate deliveryCosmetic preparationsToilet preparationsQuenchingOxidation resistant
Owner:HOLLOWAY WILLIAM D JR +1
Method for predicting membrane protein beta-barrel transmembrane area based on sparse coding and chain training
ActiveCN104615911AHigh precisionImprove predictive performanceSpecial data processing applicationsData setPosition-Specific Scoring Matrices
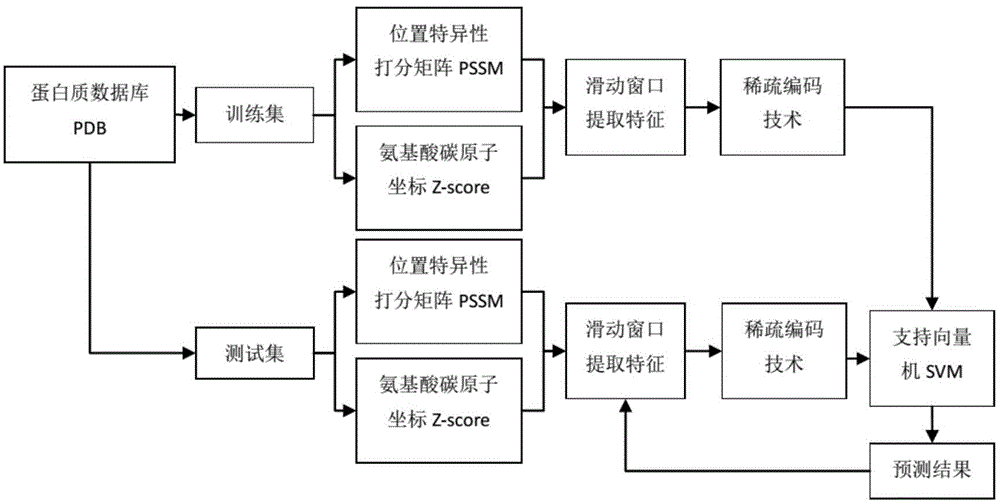
The invention provides a method for predicting a membrane protein beta-barrel transmembrane area based on sparse coding and chain training and relates to a sparse coding technology, a chain learning algorithm and a support vector machine. Structure prediction is conducted on the membrane protein beta-barrel transmembrane area through a computing method, and important information is provided for the research of the structures and functions of proteins. According to the method, the concept of digital image processing is introduced creatively, sparse coding is conducted on a protein feature matrix, and feature dimensionality reduction and denoising are achieved; a membrane protein beta-barrel data set is organized in a protein database PDB, a position-specific scoring matrix and a Z score are extracted and used as features, the position-specific scoring matrix represents amino acid evolution information, the Z score represents the position information of amino acid residues, a feature vector is extracted through a sliding window, multi-feature fusion is achieved, a chain learning algorithm training model based on a SVM classifier is provided, a predication effect is remarkably improved, and a Jakenife cross validation result shows that the precision can reach 92.5%.
Owner:SHANGHAI JIAO TONG UNIV
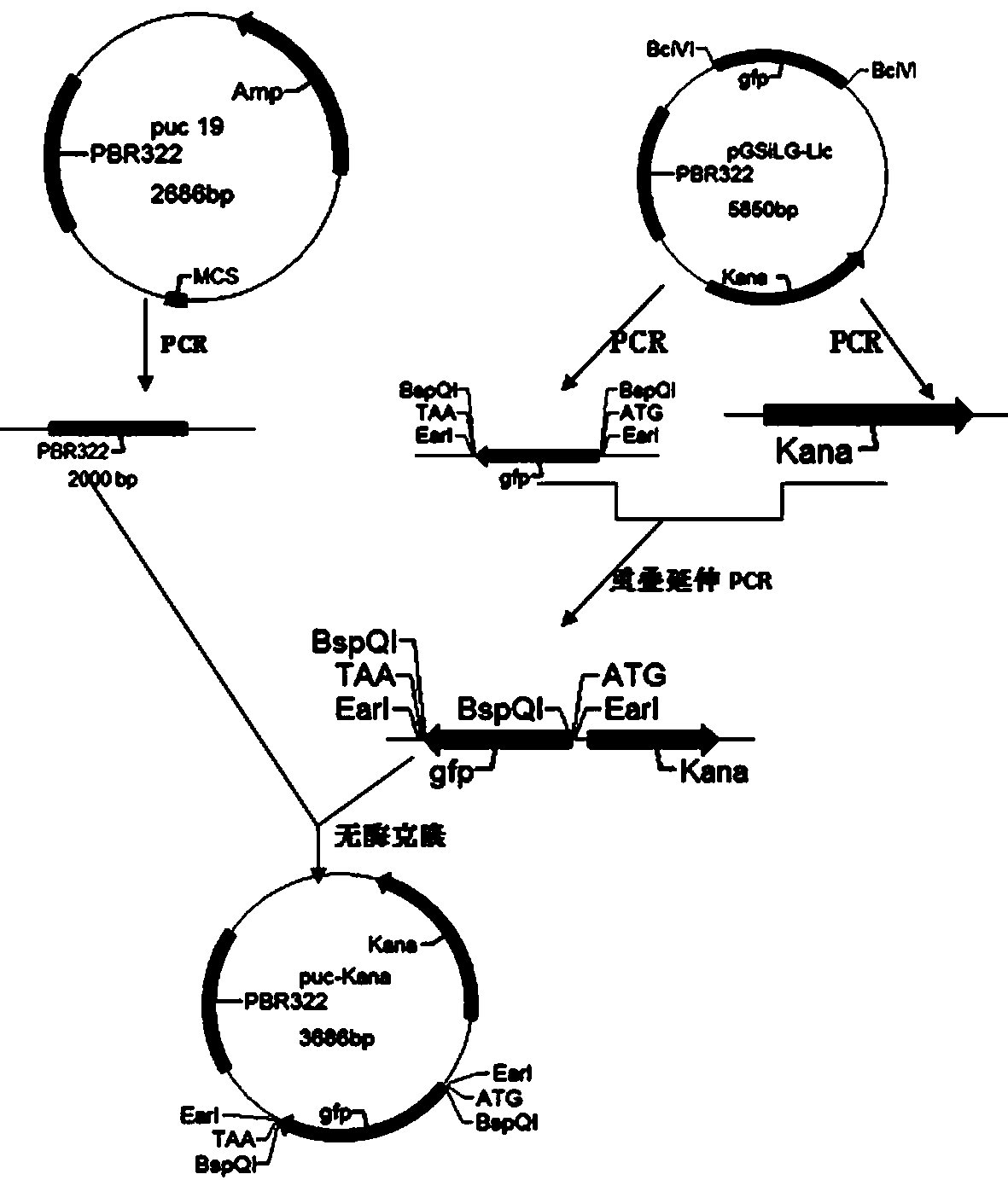
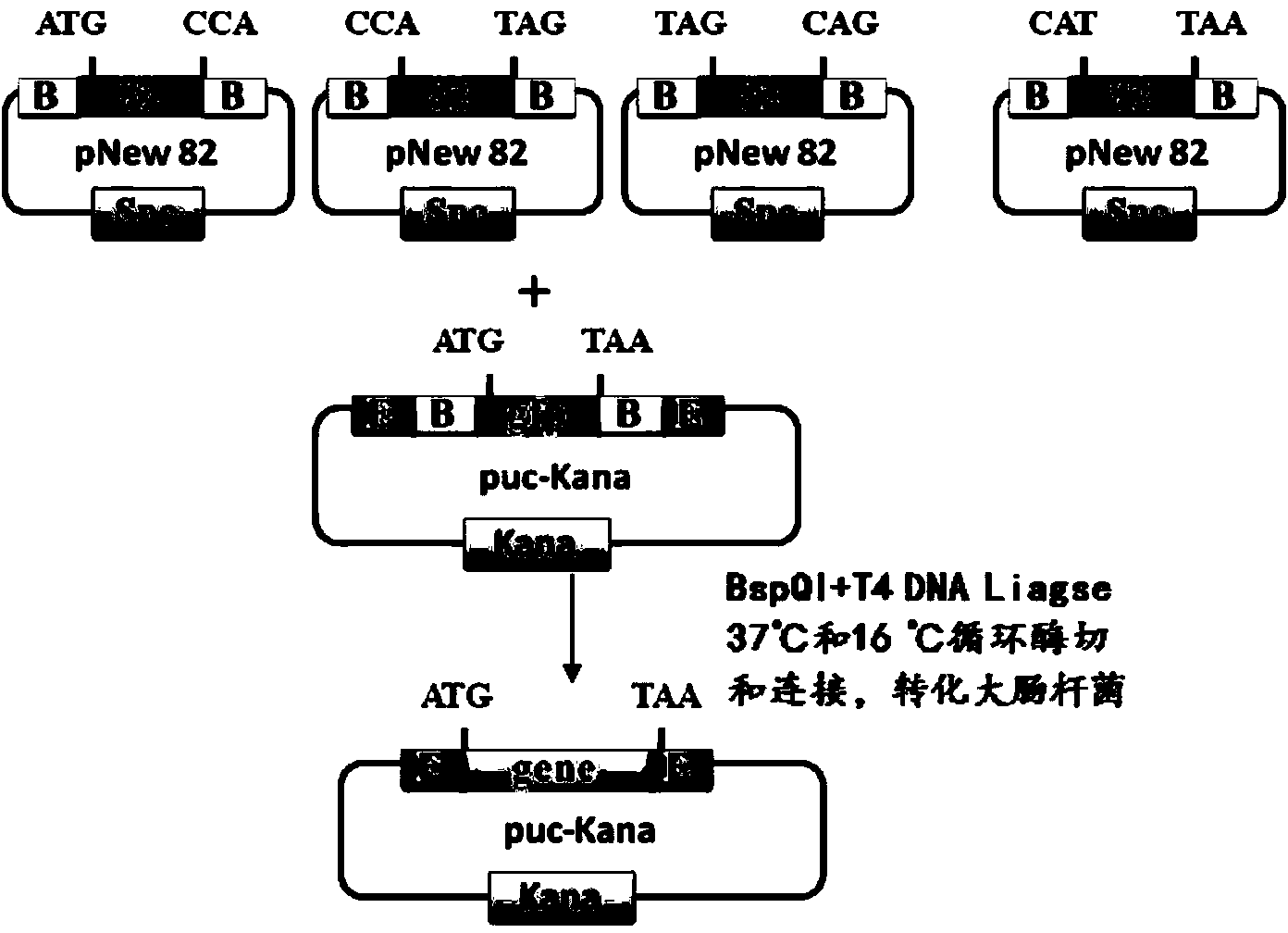
Method for synthesizing DNA fragments and assembling synthetic genes in Escherichia coli through one-step method
ActiveCN103725674AEasy to operateConditions for simple annealingVector-based foreign material introductionDNA preparationSynthesis methodsSmall fragment
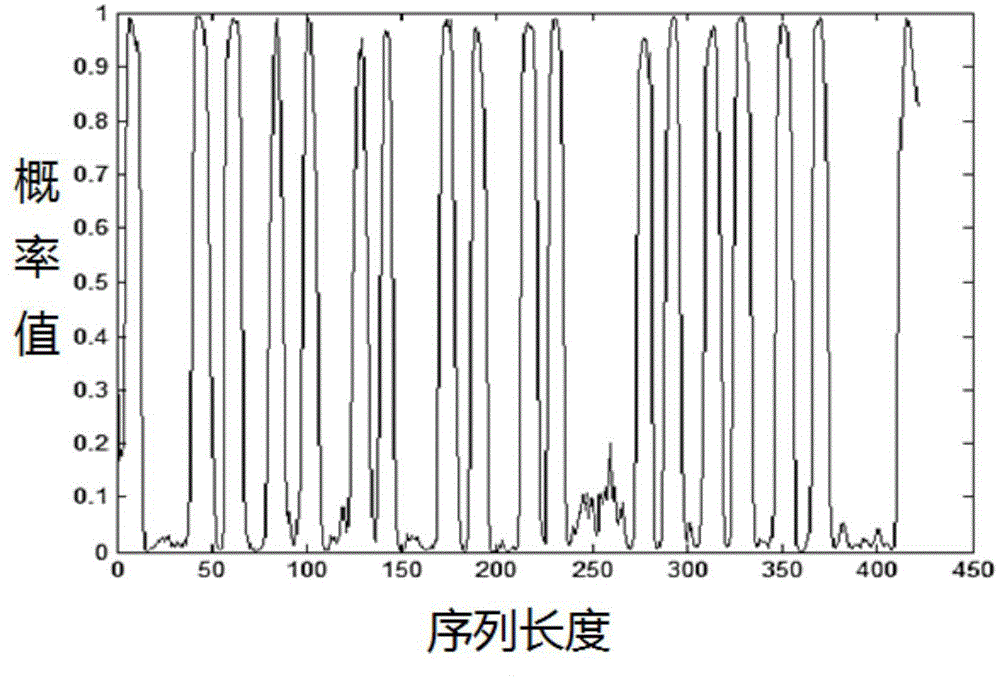
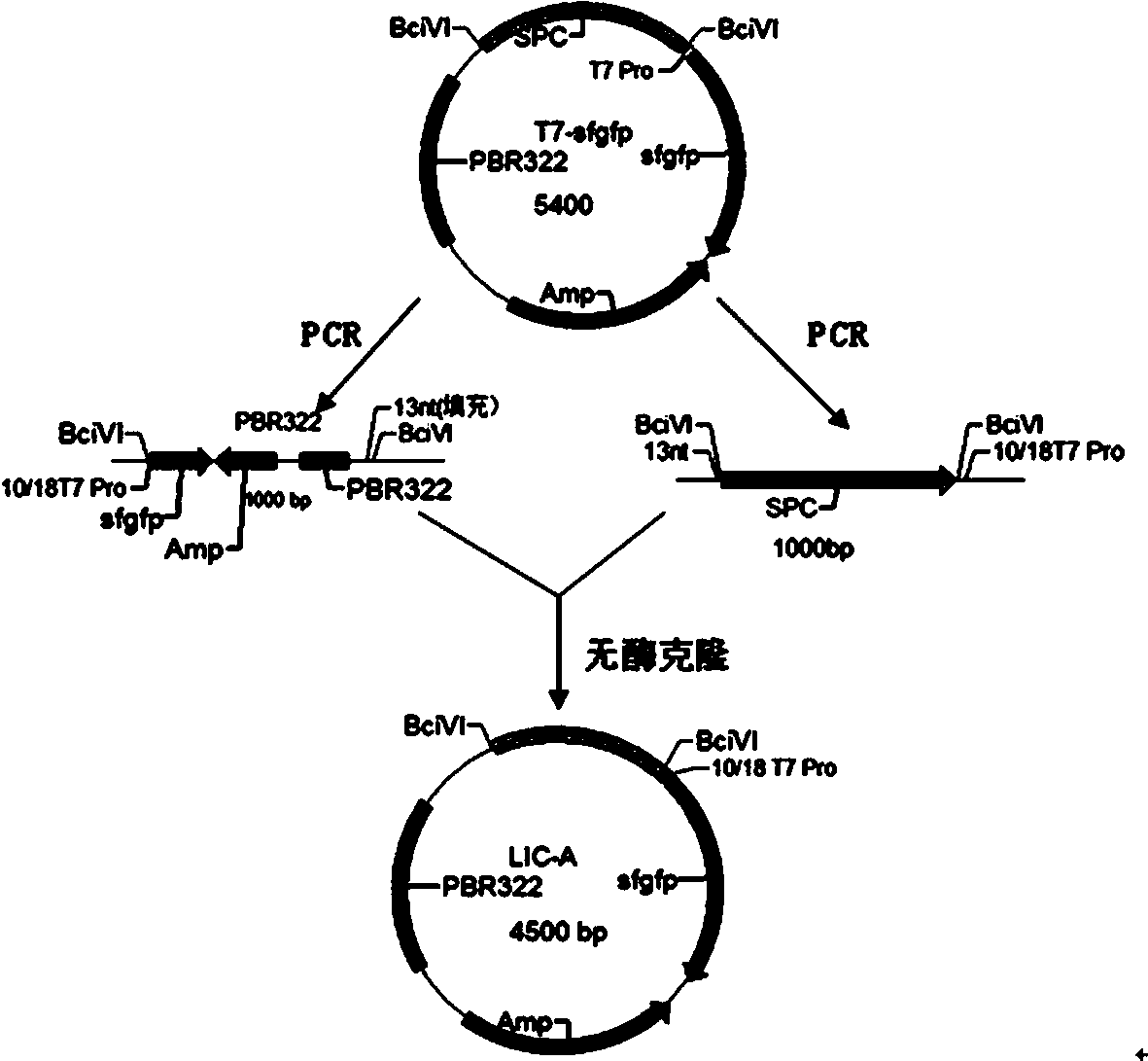
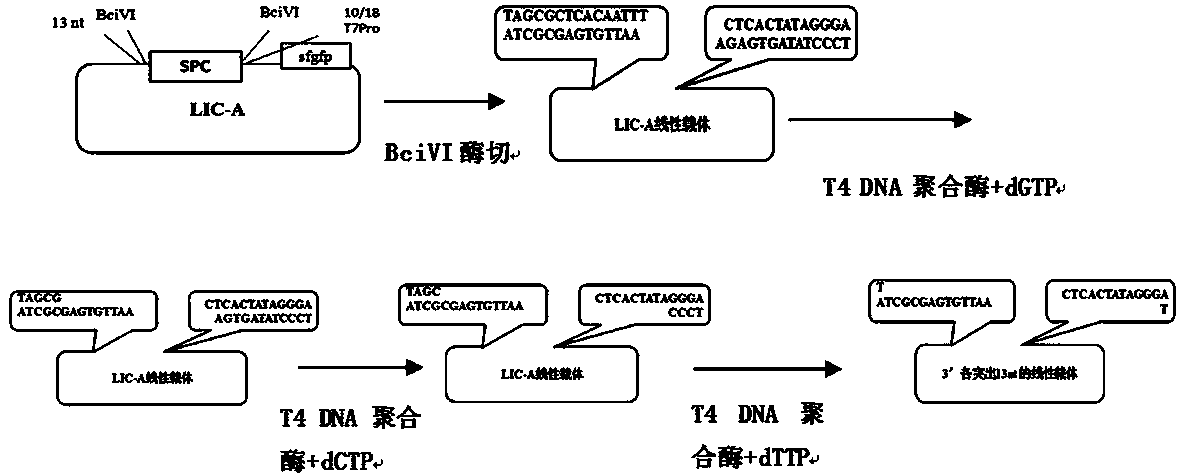
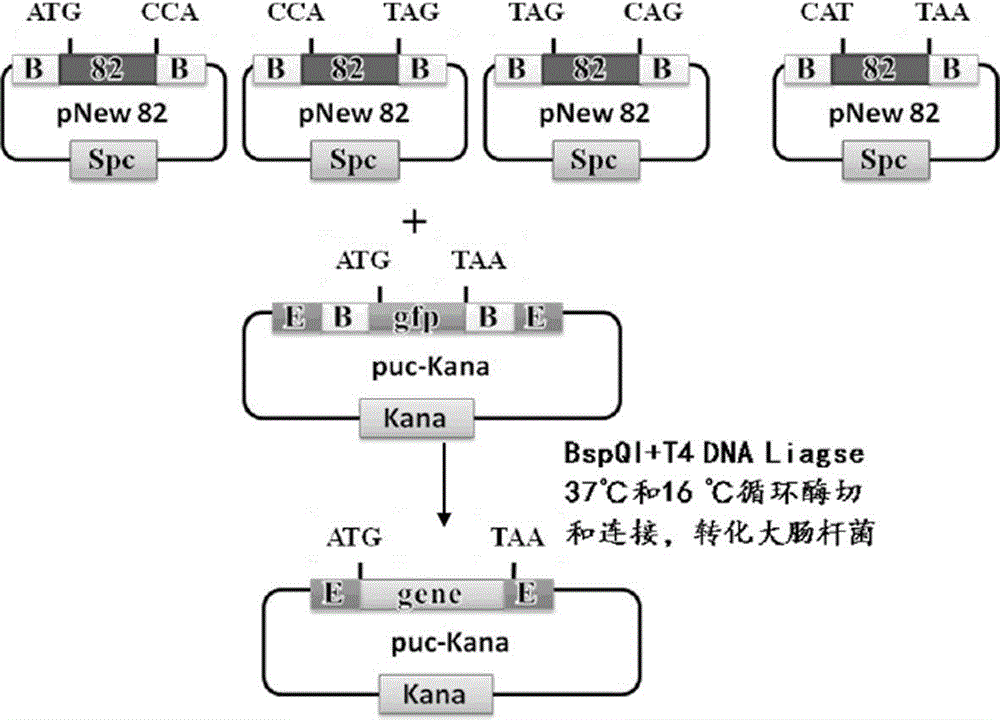
The invention provides a method for synthesizing DNA fragments and assembling synthetic genes in Escherichia coli through a one-step method. The method mainly comprises the following steps: constructing LIC vector plasmid LIC-A; preparing a LIC-A linear vector; constructing a receptor vector puc-Kana for assembling the DNA fragments; selecting target genes, and dividing small fragments of about 300bp for synthesizing; and assembling multiple small-fragment DNA into large-fragment genes by utilizing a Jinmen cloning reaction. The method is simple in operation, the operations of cloning and sequencing are not needed to be repeatedly performed, direct annealing is realized, the cloning efficiency is high, luminous report genes such as sfgfp and gfp are used, the screening method is intuitive, the error rate of the synthesis method is extremely low, the period of the synthetic genes is short, the cost is greatly saved, and the time is saved. Moreover, the method is suitable for synthesizing various genes and is particularly effective for large-gene synthesis.
Owner:WUHAN GENECREATE BIOLOGICAL ENG CO LTD
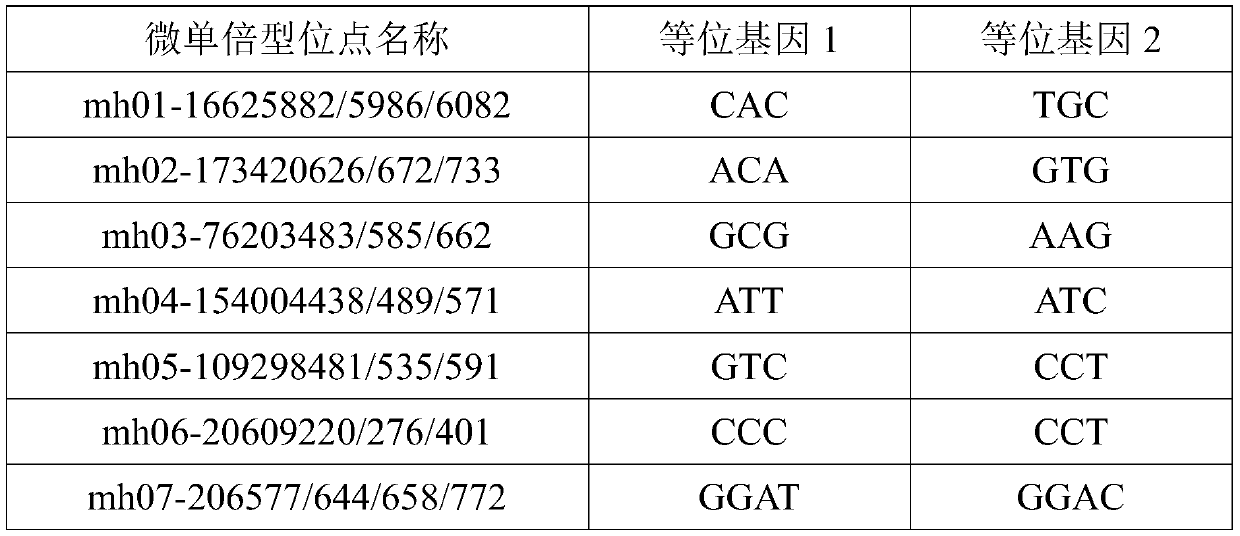
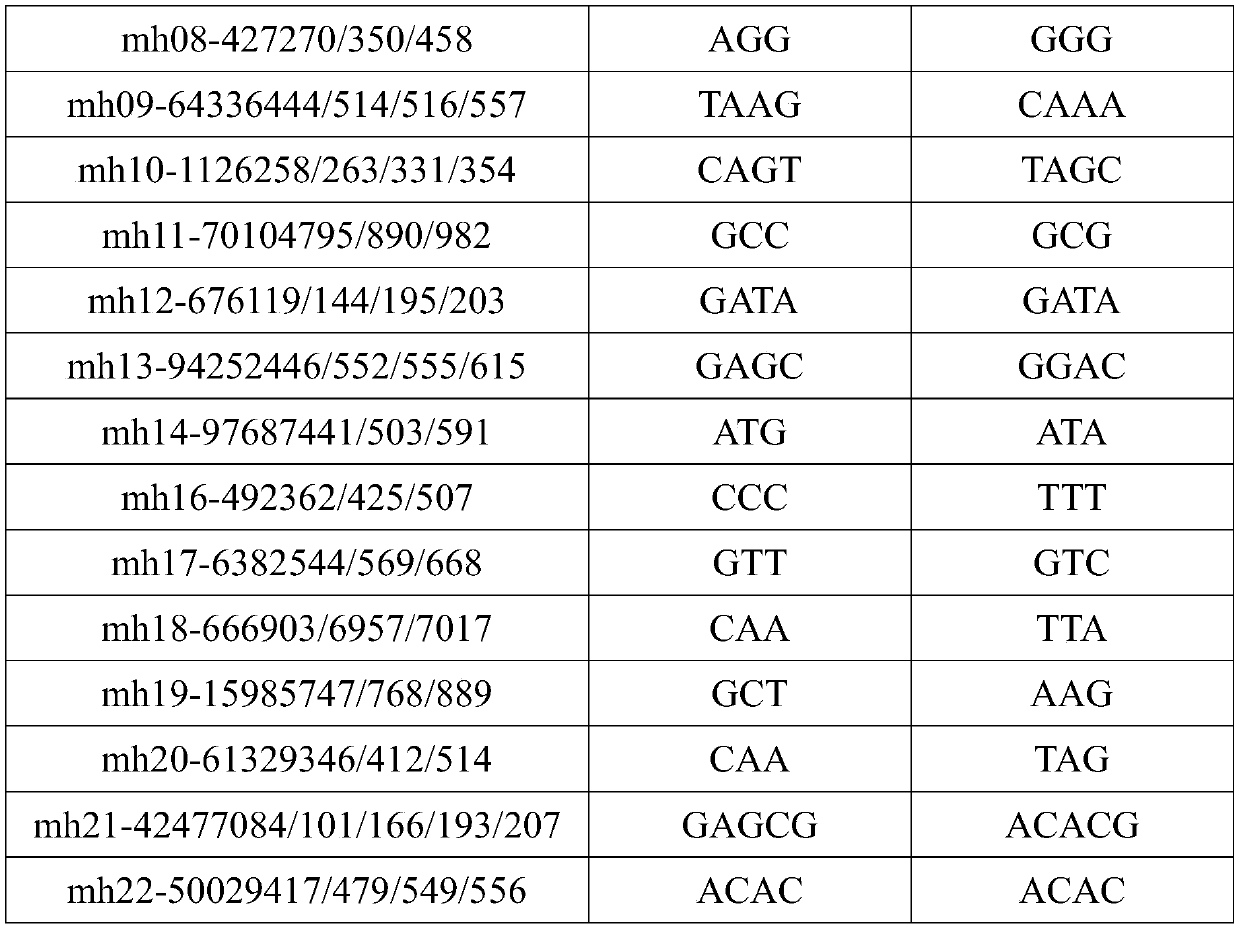
Multiplex amplification system, and next generation sequencing typing kit and typing method for 21 micro-haplotype sites
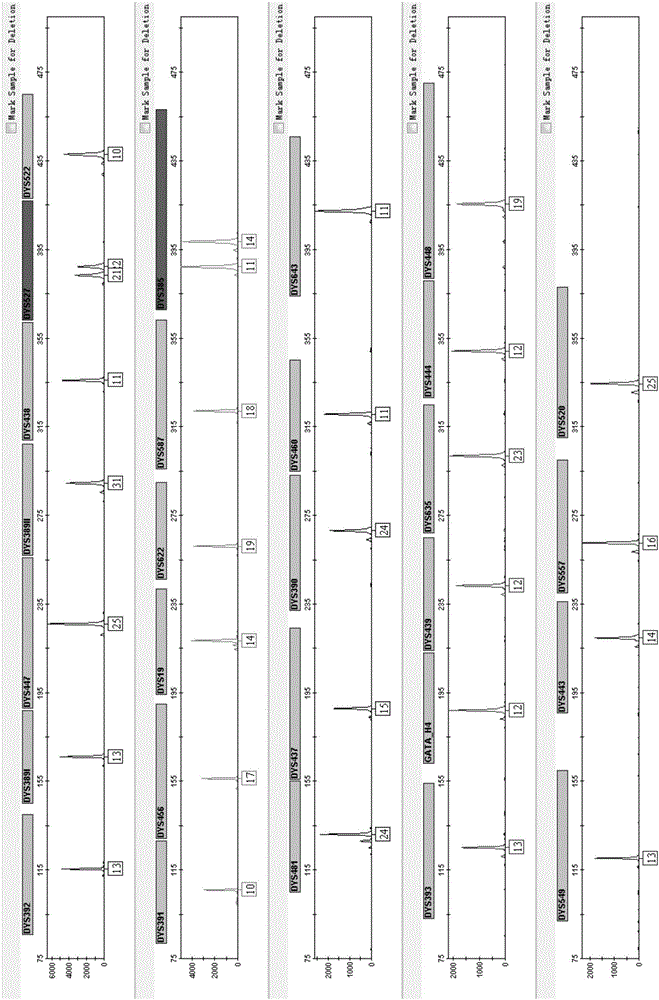
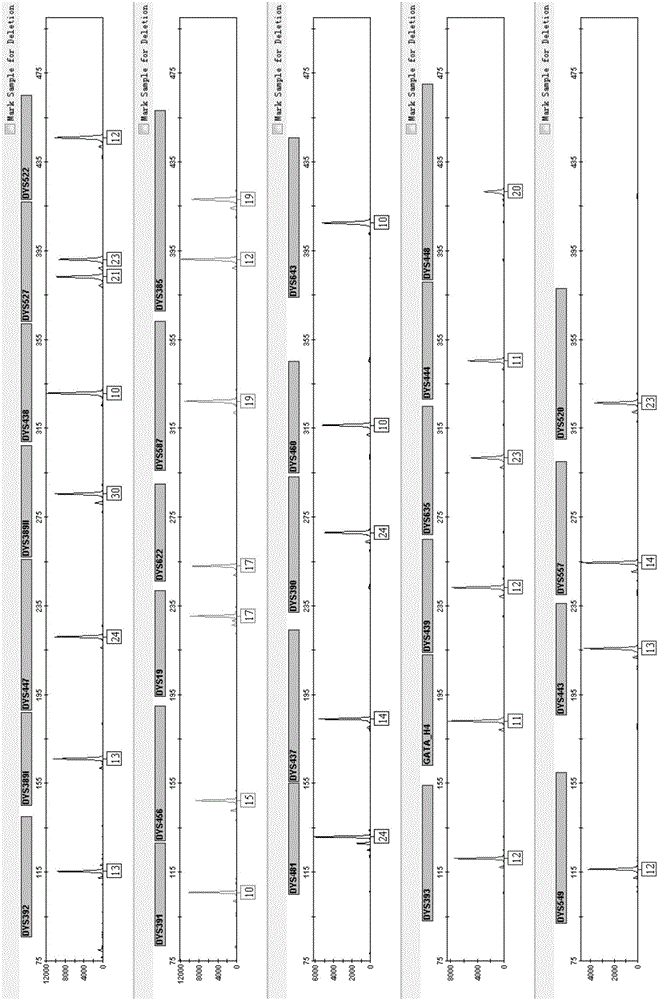
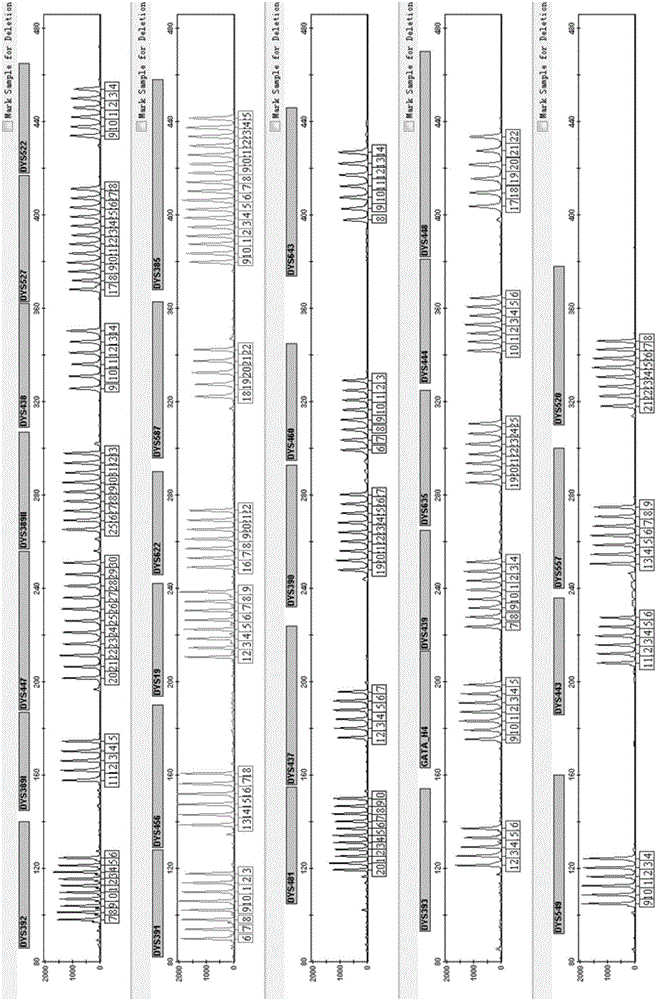
ActiveCN110218781AShorten the lengthLow mutation rateMicrobiological testing/measurementSequence analysisStr typingAllele frequency
The invention relates to the technical field of molecular biology, and especially relates to a multiplex amplification system, and a next generation sequencing typing kit and typing method for 21 micro-haplotype sites. The 21 micro-haplotype sites in the multiplex amplification system are derived from 21 autosomes, are balanced in allele frequency, are mutually independent between the sites, and have the advantages of polymorphism and low mutation rate. The kit contains 53 PCR-amplified single-end specific primers shown in SEQ ID NO. 1-SEQ ID NO.53, and can detect the 21 microhaplotype sites in a same reaction system. The multiplex amplification system, the kit and the typing method provided by the invention can solve the technical problem that a current STR typing technology cannot obtainan ideal typing result, and allele loss may occur due to excessive number of SNP sites.
Owner:HEBEI MEDICAL UNIVERSITY
Genetic marker for human individual recognition and/or paternity identification and detection method thereof and kit
ActiveCN107541554ABroad multi-racial adaptabilityThe test result is accurateMicrobiological testing/measurementDNA/RNA fragmentationAllele frequencyCommercial kit
The invention discloses a genetic marker combination for human individual recognition and / or paternity identification and a detection method thereof. By conducting whole-genome SNPs unbiased scanningon multiple races, a SNPs site combination widely applicable to the multiple races is screened, and 116 autosome SNPs sites which come from 37 groups in different regions in the whole world, have thehypermorph allel frequency and low difference and achieve independent inheritance and 12 X chromosome SNPs sites which come from 37 groups in different regions in the whole world, have the hypermorphallel frequency and low difference and achieve independent inheritance are screened from 25,580,678 SNPs sites in a genome-wide scale. Compared with an existing commercial kit, the SNPs site combination has higher and wider multiracial adaptation, the cumulative individual recognition probability and the cumulative probability of exclusion are both significantly superior to those of the commercialkit, the detection result is more accurate, and a great application prospect is achieved.
Owner:SUN YAT SEN UNIV
Methods and systems for thermodynamic evolution
ActiveUS8909580B2Maximize dissipation of energyLow mutation rateCAD circuit designKnowledge representationControl systemSelf adaptive
Methods and systems for thermodynamic evolution. Adaptive control systems are constructed based on the property of volatile matter to self-organize to maximize the dissipation of energy. The logical state of sensory nodes in a node circuit are set and projected into a network. Then, the system evaluates logical state of processing nodes by summing input currents of processing nodes and project processing node's state into network. The strength of processing node is increased such that logical state of sensory node matches with logical states of processing node by utilizing plasticity rule. The system is configured to maximize energy dissipation by creating weight structures to stabilize nodes with logical state. The internal positive feedback of node circuit forces competition between nodes such that one node is driven to high logical state and other nodes to low logical state.
Owner:KNOWM TECH
Detection method of low requency mutated DNA fragment and library building method
ActiveCN108192955AEasy to useImprove detection limitMicrobiological testing/measurementLibrary creationMutation detectionEngineering
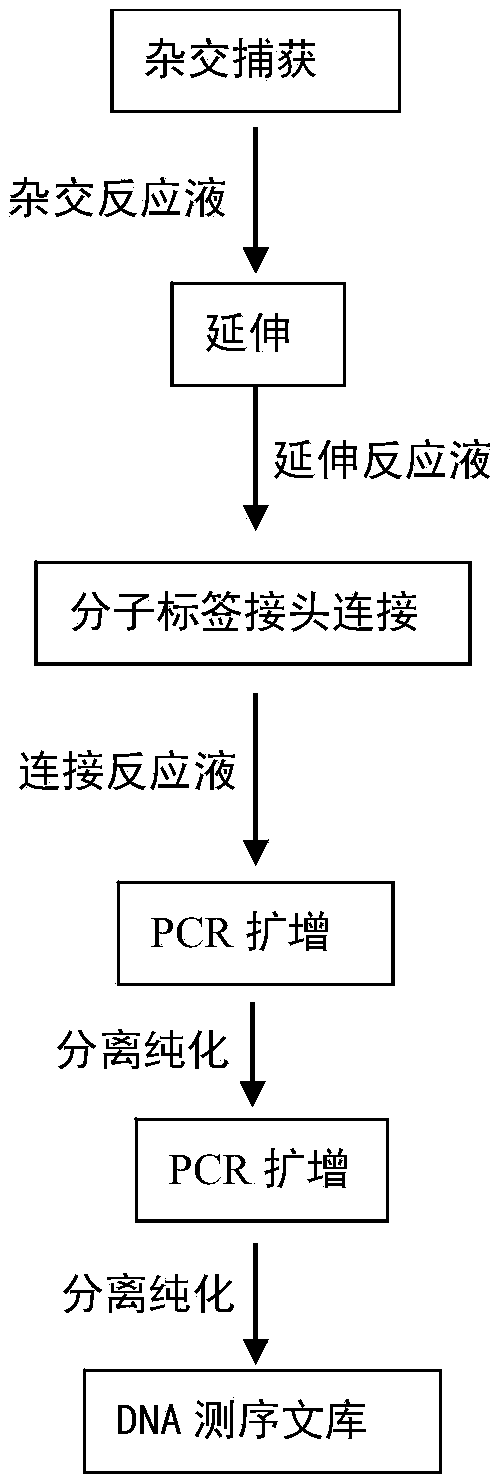
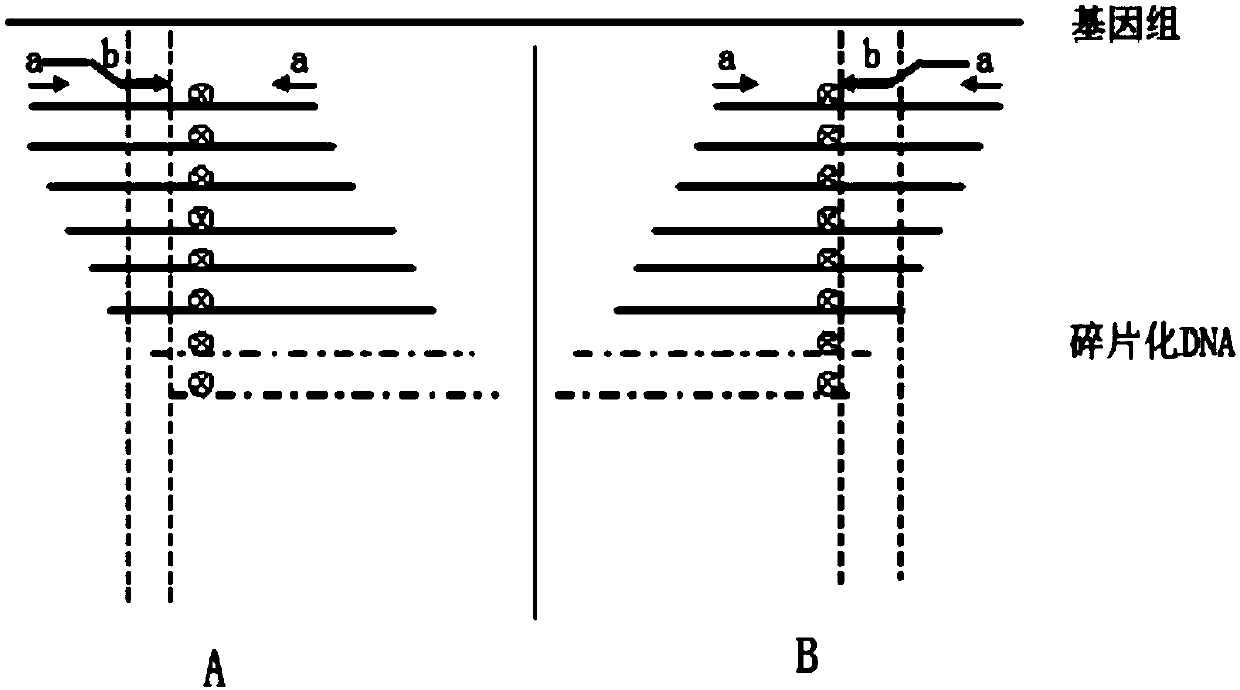
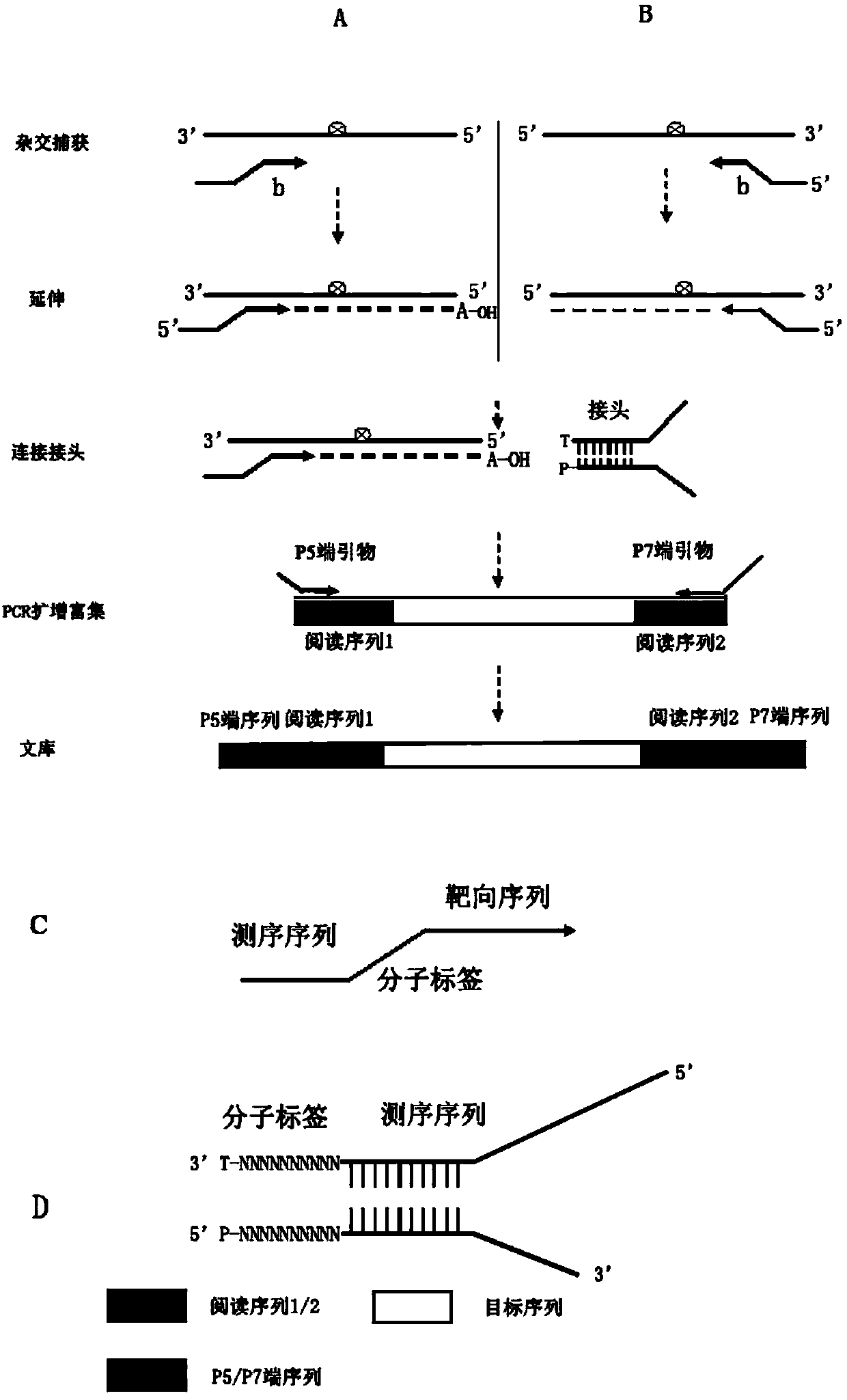
The invention belongs to the technical field of gene engineering, and particularly relates to a detection method of a low frequency mutated DNA fragment and a library building method. The technical problems that in the existing detection technique, the DNA fragment capturing efficiency is low, wrong mutation is introduced in the library building process, the detection limit is low, and the sensitivity is low are solved by adopting the hybrid capturing and molecular tagging technique and meanwhile combining with a special library building mode, the library specificity is improved, the false positive rate is lowered, and the template utilization rate is improved.
Owner:HUNAN YEARTH BIOTECHNOLOGICAL CO LTD
Compound amplification detection kit containing forty InDel genetic polymorphic sites of human X chromosome
ActiveCN110578009ALow mutation rateHigh sensitivityMicrobiological testing/measurementInsertion deletionX chromosome
The invention discloses a compound amplification detection kit containing forty InDel genetic polymorphic sites of human X chromosome. The forty InDel sites of the X chromosome and an individual identification location point are amplified simultaneously by a primer group. Compared with a traditional STR genetic marker, an insertion deletion genetic marker used in the compound amplification detection kit has the advantages of low mutation rate and high sensitivity; and the polymorphism detection sites involved in the compound amplification detection kit combine the advantages of an InDel genetic marker and the special genetic characteristics of the X chromosome, and effective supplementary means can be provided for identification of difficult or special cases.
Owner:GUANGDONG HUAMEI ZHONGYUAN BIOLOGICAL SCI & TECH +2
Quick detection kit for chicken-origin ingredient in food and feed and application of quick detection kit
InactiveCN106434959AEasy to quantifyCopy number stabilityMicrobiological testing/measurementDNA/RNA fragmentationAdditive ingredientNucleotide
The invention provides a quick detection kit for a chicken-origin ingredient in food and feed and relates to the technical field of biological species identification. A general chicken-origin internal standard gene Actb is screened first; a nucleotide sequence of the gene Actb is as shown as SEQ ID NO.1 (Sequence Identifier Number 1); the gene Actb is located on a chromosome; single copy is easy to quantify; and the gene Actb has a constant copy number in chicken species, without allelic variation and can serve as a target gene for chicken-origin identification. LAMP amplification primers are designed by taking the gene as a target sequence; nucleotide sequences of the LAMP amplification primers are as shown as SEQ ID NO. 2-5; the LAMP amplification primers and an LAMP reaction solution constitute the detection kit; the detection kit can detect the chicken-origin ingredient in the food and the feed quickly and sensitively, and the sensitivity reaches 10pg. The kit is simple in use method, low in cost, high in specificity and very applicable to field real-time detection and has a wide application prospect in animal-origin food surveillance and meat product adulteration defense, and a reaction result is easy to observe.
Owner:CHINA AGRI UNIV
Primer-free gene synthesis method
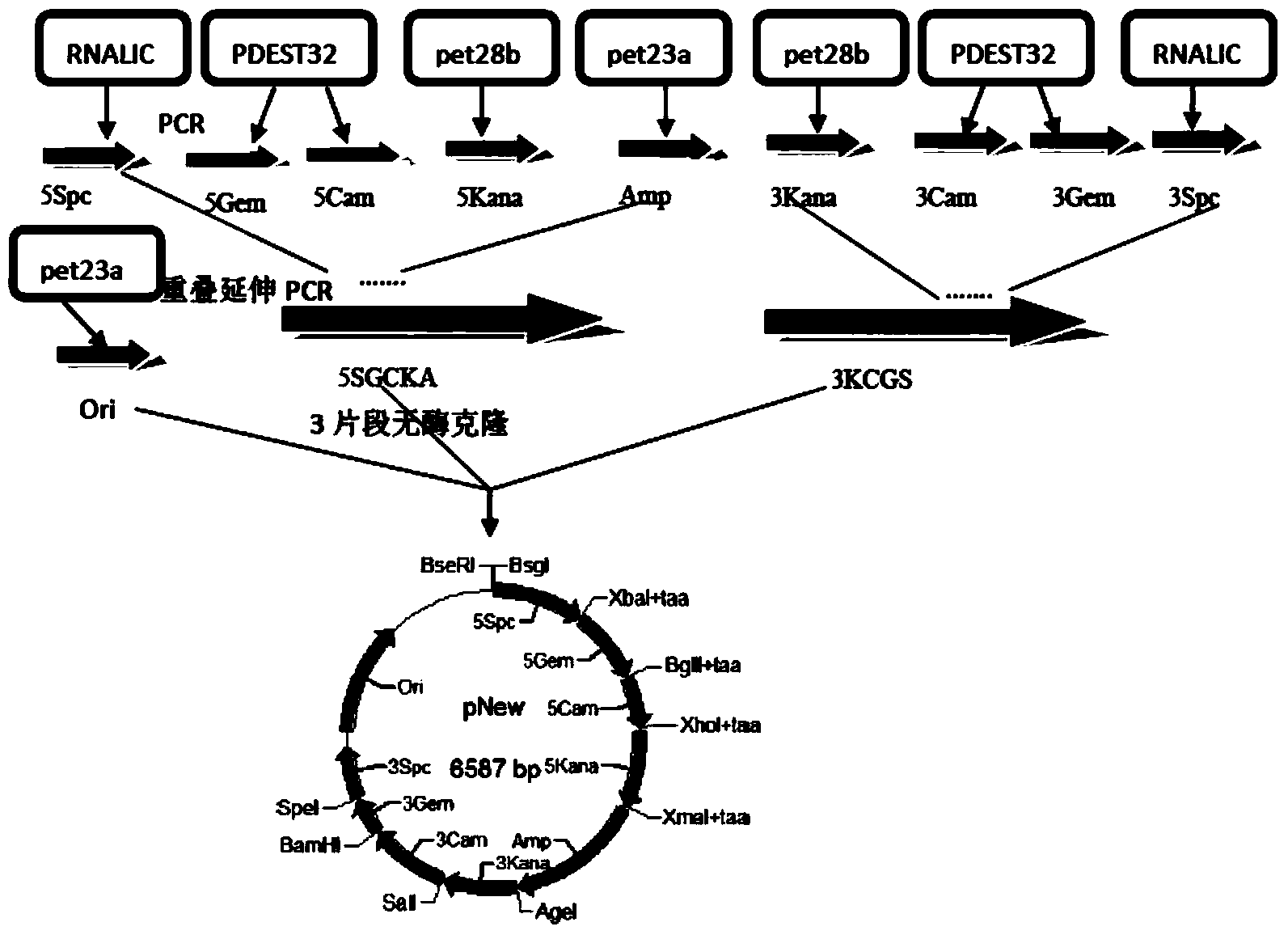
InactiveCN104109683AWork simplification and efficiencyWill not mutateNucleotide librariesMicroorganism librariesEnzyme digestionNucleotide
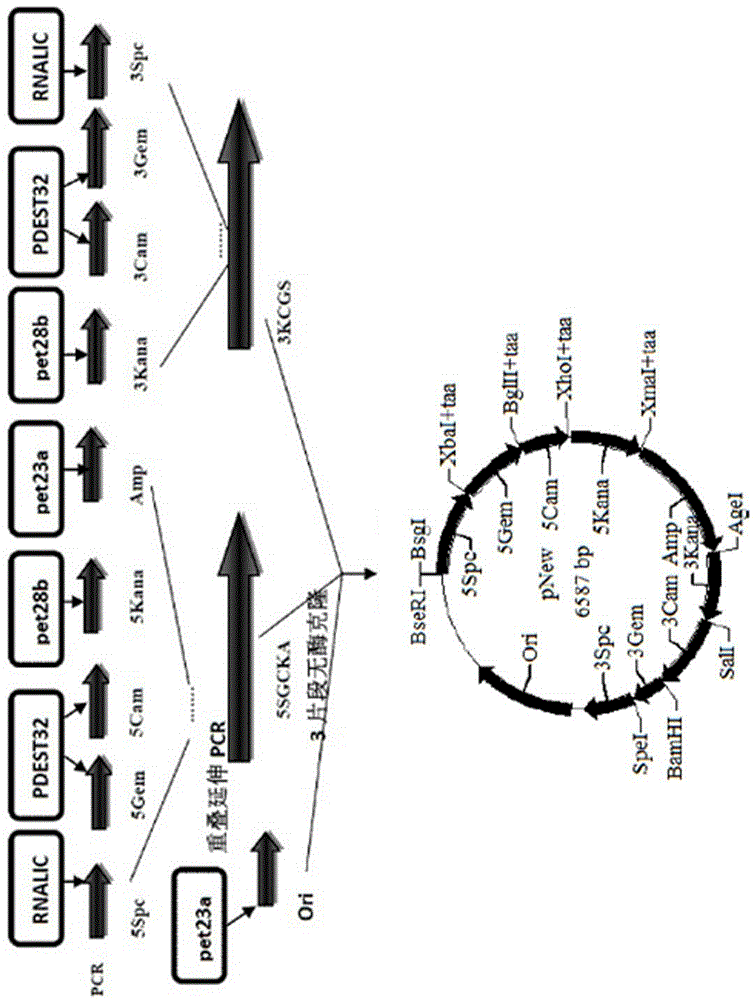
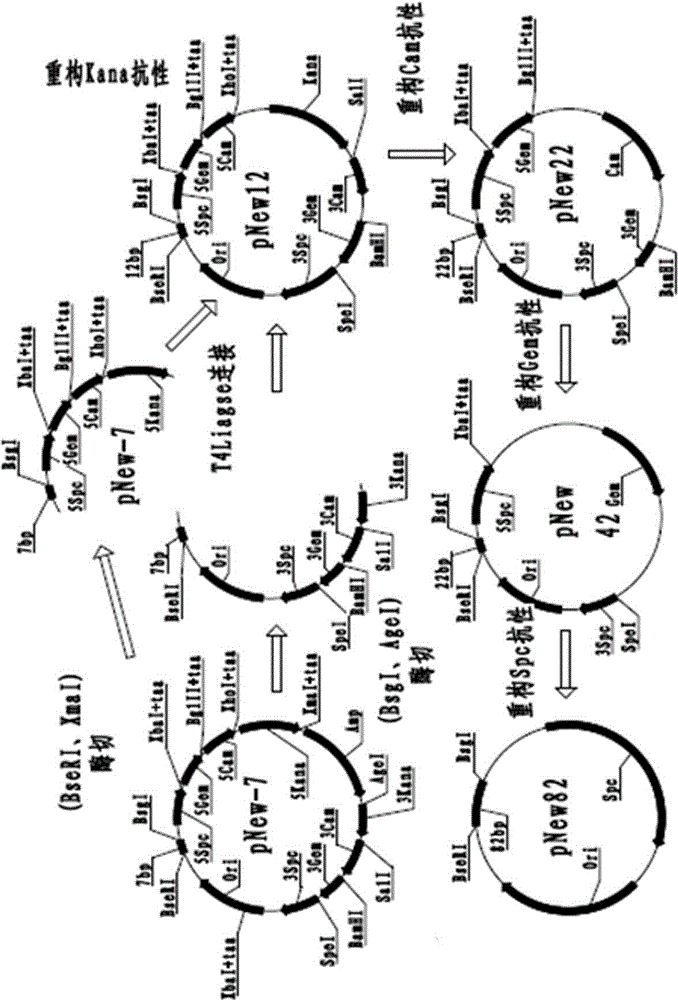
The invention discloses a primer-free gene synthesis method and belongs to the field of artificial synthesis of genes. The invention discloses a pNew carrier plasmid, and the pNew carrier plasmid has a nucleotide sequence shown in the formula of SEQ ID NO.33. The invention also discloses a carrier bank which is constructed from pNew carrier plasmids and contains 16384(47) plasmids. The invention further discloses a primer-free gene synthesis method. The primer-free gene synthesis method comprises the following steps of 1, carrying out grouping on targeted gene DNA sequences to be synthesized according to a ratio of 82bp per fragment, 2, finding plasmids corresponding to the grouped fragments in the constructed plasmid bank, 3, carrying out enzyme digestion on the corresponding plasmids, carrying out screening by antibiotic and carrying out reconstruction to obtain a plasmid containing 82bp of the targeted gene sequences, and 4, splicing the gene fragments by a golden-gate cloning reaction to obtain a complete targeted gene. The primer-free gene synthesis method is free of primers, has a low synthesis cost, a very low mutation rate and high accuracy, can synthesize special gene sequences such as a highly repetitive sequence and Poly A, has simple processes and can realize automatic operation.
Owner:WUHAN GENECREATE BIOLOGICAL ENG CO LTD +1
Selection methods
A rational method for obtaining a novel molecule capable of a desired interaction with a substrate of interest comprising selecting hosts or replicators which encode said novel molecules based upon cell or replicator growth caused by the desired interaction of the novel molecule and a selection molecule expressed by said host.
Owner:WOHLSTADTER JACOB NATHANIEL
Multi-color fluorescence multiplex amplification kit for amplifying STR gene locuses of human Y chromosome and application of kit
InactiveCN106520976AStrong material adaptabilityHigh polymorphismMicrobiological testing/measurementFluorescenceGene
The invention discloses a multi-color fluorescence multiplex amplification kit for amplifying STRgene locuses of a human Y chromosome and application of the kit. Thirty STR gene locuses can be detected in a single amplification system. Compared with the prior art, the kit has the advantages that (1) the gene locuses included in the kit can guarantee the identity of the same family parting and the difference of different family partings due to the advantages high polymorphism, low mutation rate, reasonable frequency distribution and high individual identification; and (2) the kit is extremely high in detection material applicability and can detect various samples.
Owner:AGCU SCIENTECH +1
Method for inducing somatic embryos of corn
InactiveCN102172217AReduce consumptionReduce adverse effectsHorticulture methodsPlant tissue cultureBio engineeringSomatic cell
The invention provides a method for inducing somatic embryos of corn and belongs to the technical field of biological engineering. The method comprises the following steps of: inducing corn explants in a callus inducing culture medium; transferring the explants undergoing inducing culture to a subculture medium; propagating the somatic embryos in the subculture medium; and putting the propagated somatic embryos into differentiation culture medium for differentiation and seedling. By the method, a plurality of somatic embryos of corn can be induced with high efficiency, and the induced somaticembryos of corn have the characteristics of realizing the differentiation of both poles at the same time by long-time subculture infinite propagation and without the control of exogenous hormones, along with high differentiation rate and seedling rate; and because influence of exogenous hormones is avoided in the differentiation process, the mutation rate of somatic cells is relatively lowered atthe seedling stage of corn. The invention can provide an excellent and practical acceptor system for various trans-genetic methods (such as an agrobacterium-mediated method and a gene gun method).
Owner:JILIN UNIV
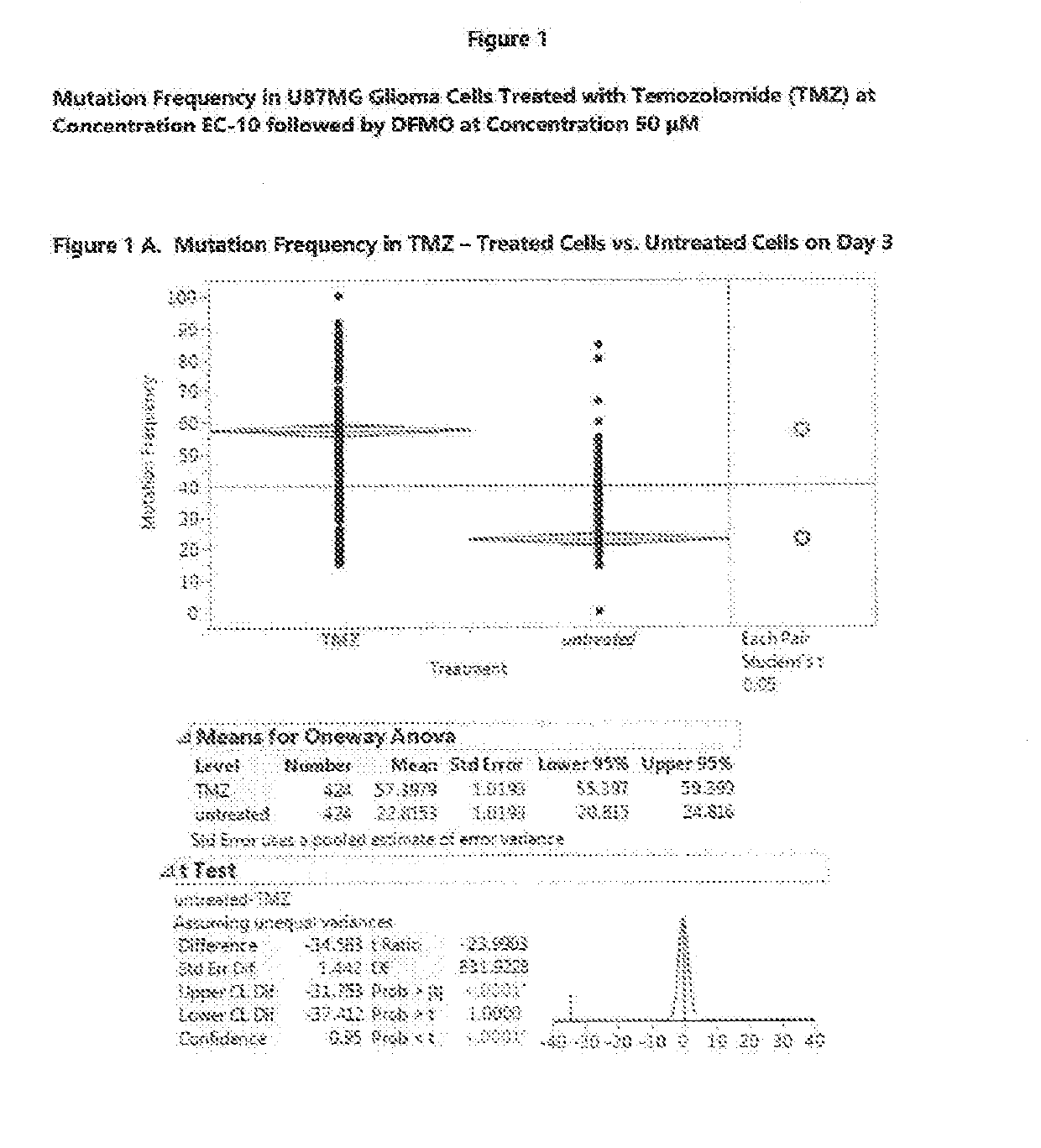
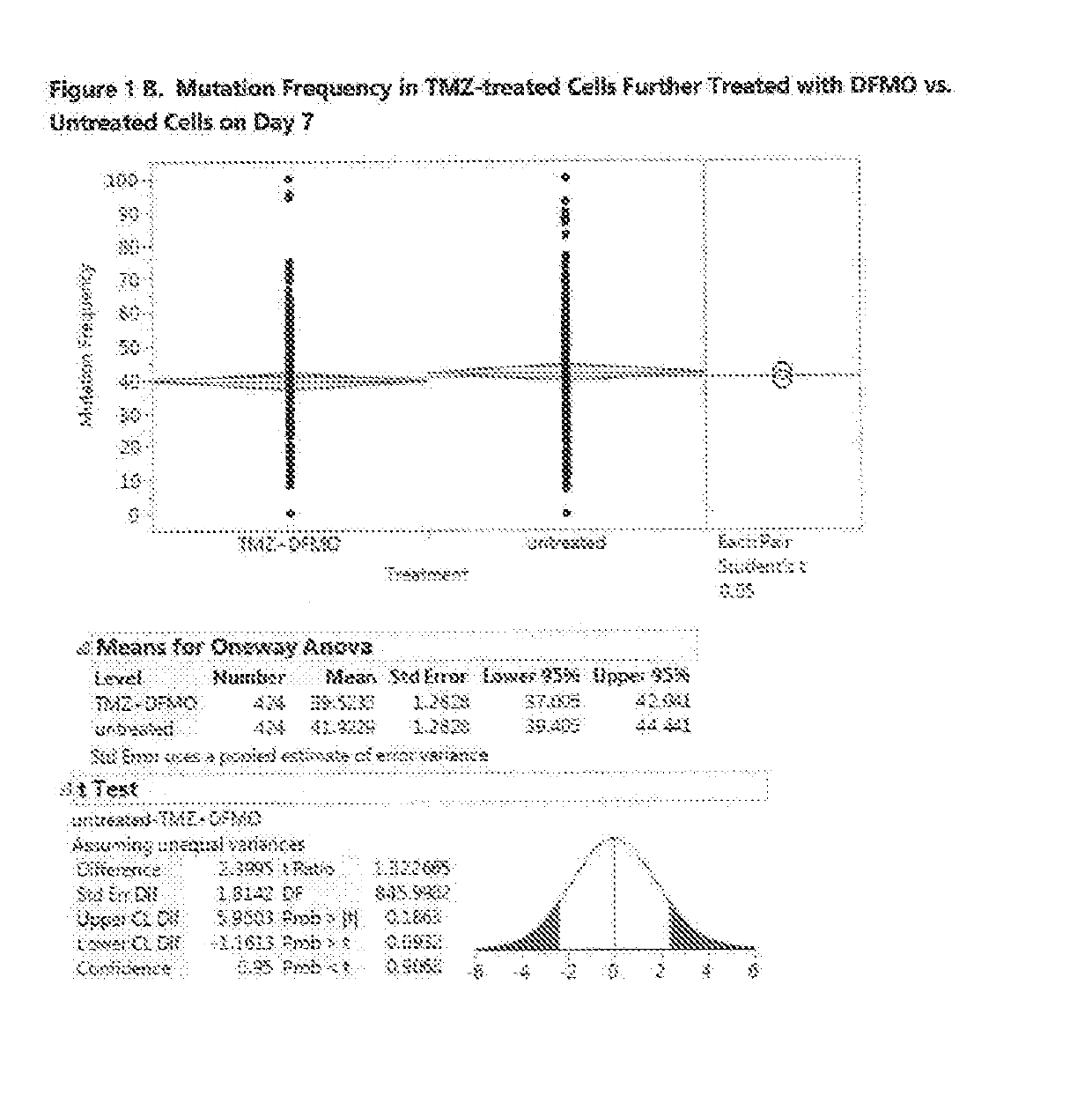
Compositions and methods for use of eflornithine and derivatives and analogs thereof to treat cancers, including gliomas
InactiveUS20190133985A1Low mutation rateShorten the progressOrganic active ingredientsPharmaceutical delivery mechanismAdenosineMalignant CNS Tumor
Eflornithine is an agent that can be used to treat glioma, especially glioma of WHO Grade II or Grade III such as anaplastic glioma. Eflornithine can suppress or prevent mutations in glioma which can cause the glioma to progress to a higher grade. Compositions and methods can include eflornithine or a derivative or analog of eflornithine, together with other agents such as conventional anti-neoplastic agents for treatment of glioma, inhibitors of polyamine transport, polyamine analogs, or S-adenosylmethionine decarboxylase inhibitors. Eflornithine or derivatives or analogs thereof can also be used to treat a range of non-glioma malignancies, including both malignancies of the central nervous system and other malignancies.
Owner:ORBUS THERAPEUTICS INC
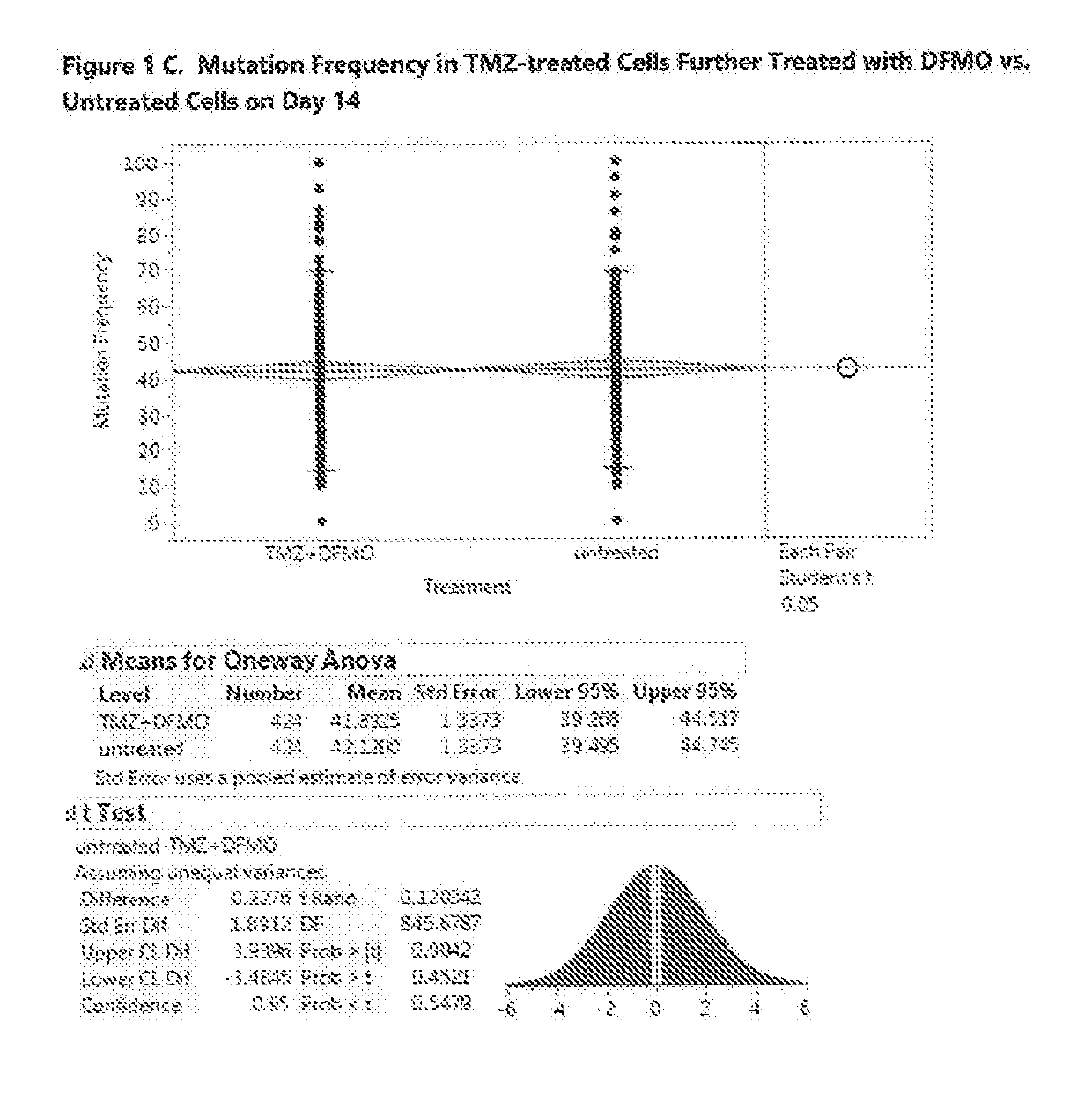
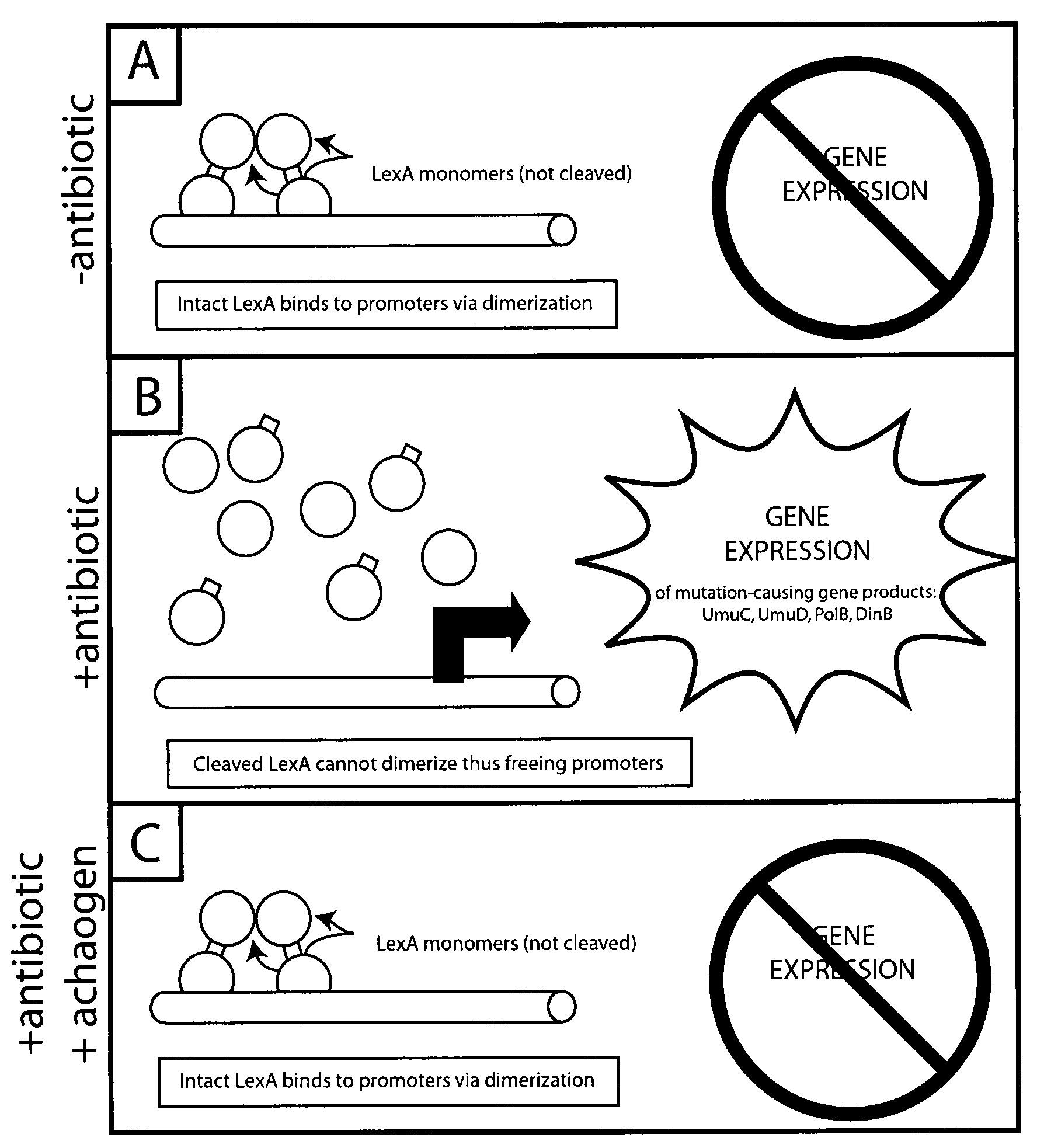
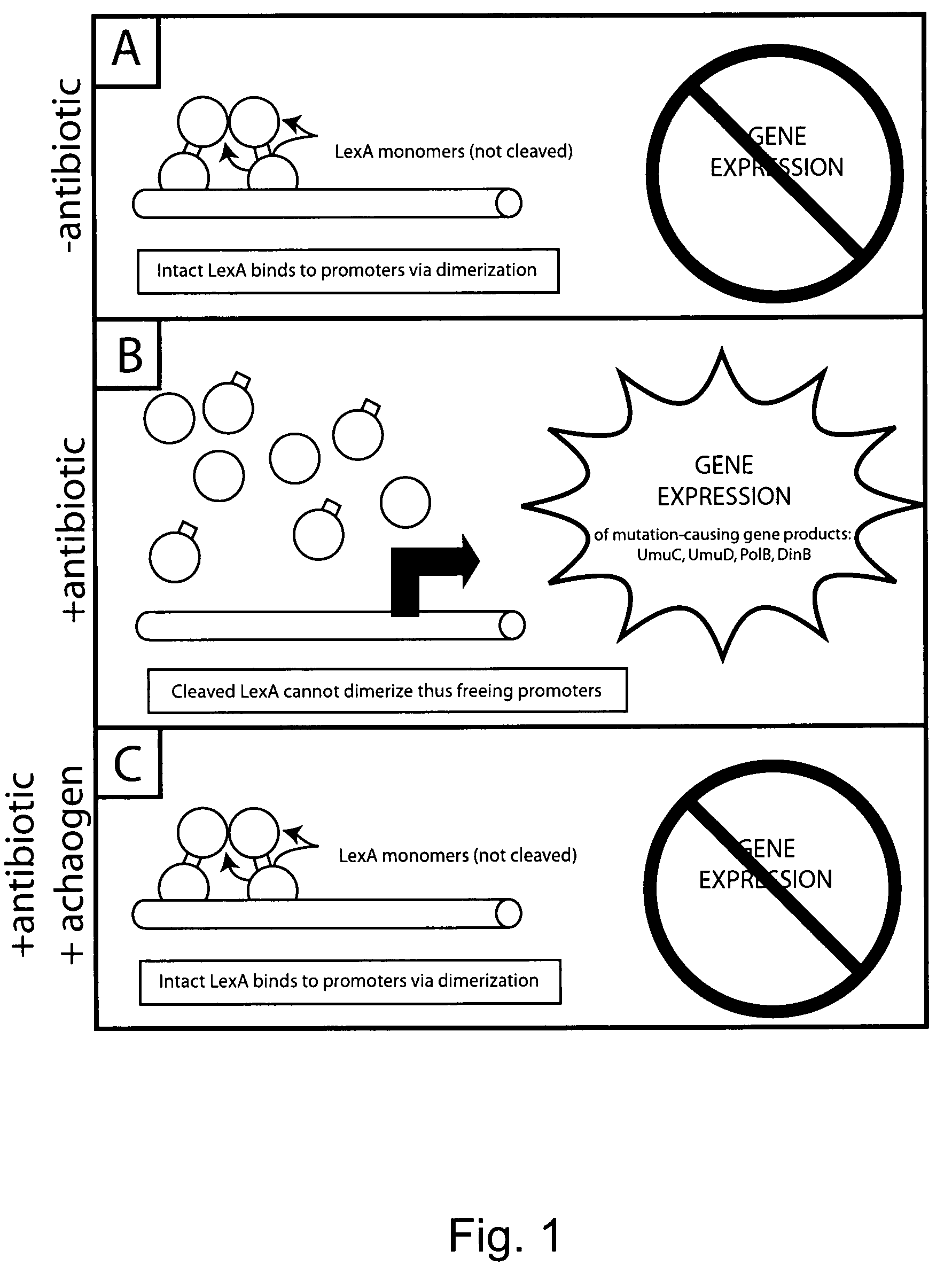
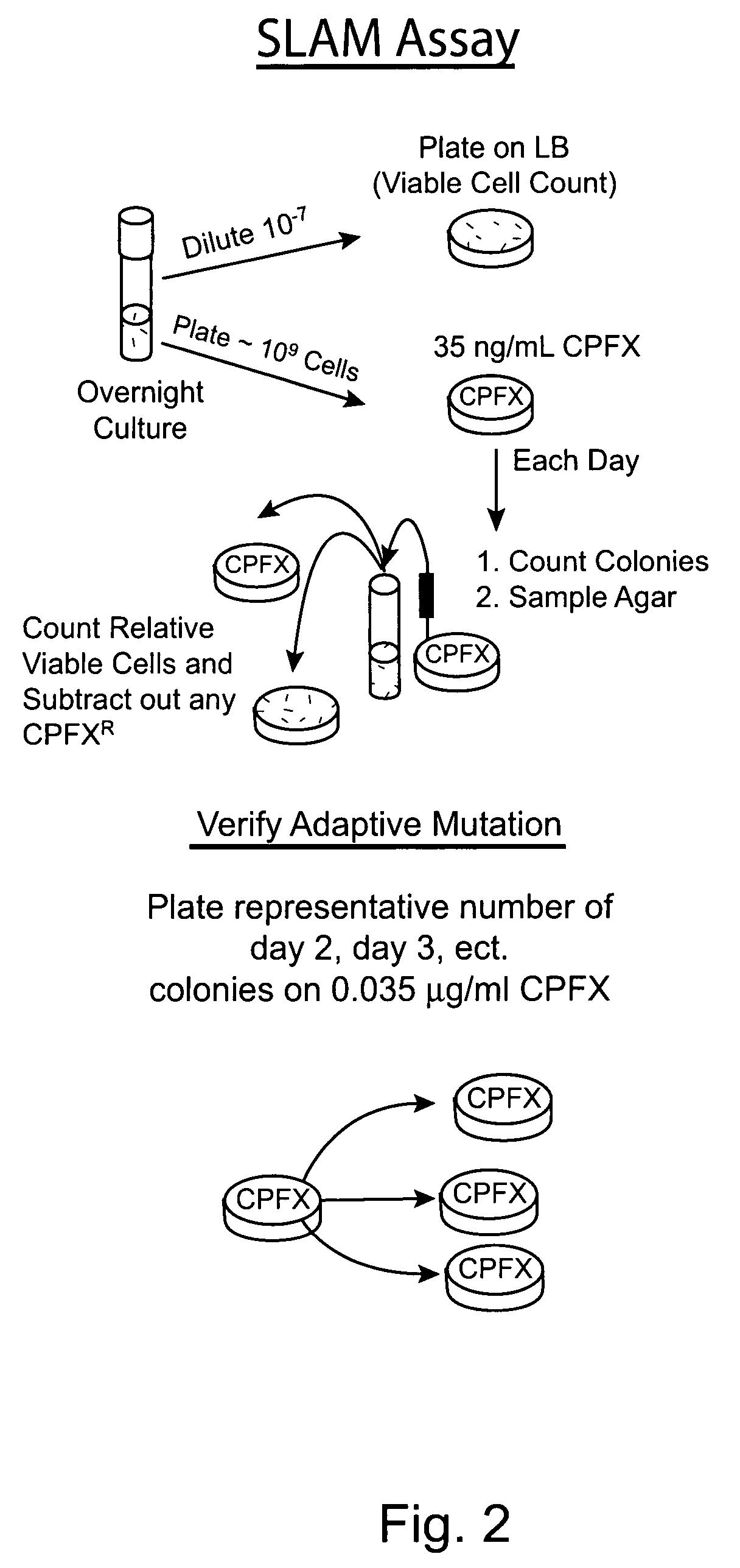
Compositions and methods to reduce mutagenesis
InactiveUS7455840B2Reduce probabilityLow mutation rateAntibacterial agentsBiocideAntibiotic resistanceDrug resistance
The present invention provides methods and compositions for said inhibition of drug resistance. In one embodiment, said invention provides methods and compositions for said inhibition of antibiotic resistance. The invention generally involves said administration of achaogens, agents that inhibit said mutational process, to inhibit said evolution of drug resistance. Also, described herein are compositions that are suitable for use as achaogens.
Owner:THE SCRIPPS RES INST +1
Kit for 35 insertion/deletion sites in forensic individual identification
InactiveCN108546762ALow mutation rateHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationCapillary electrophoresisFluorescence
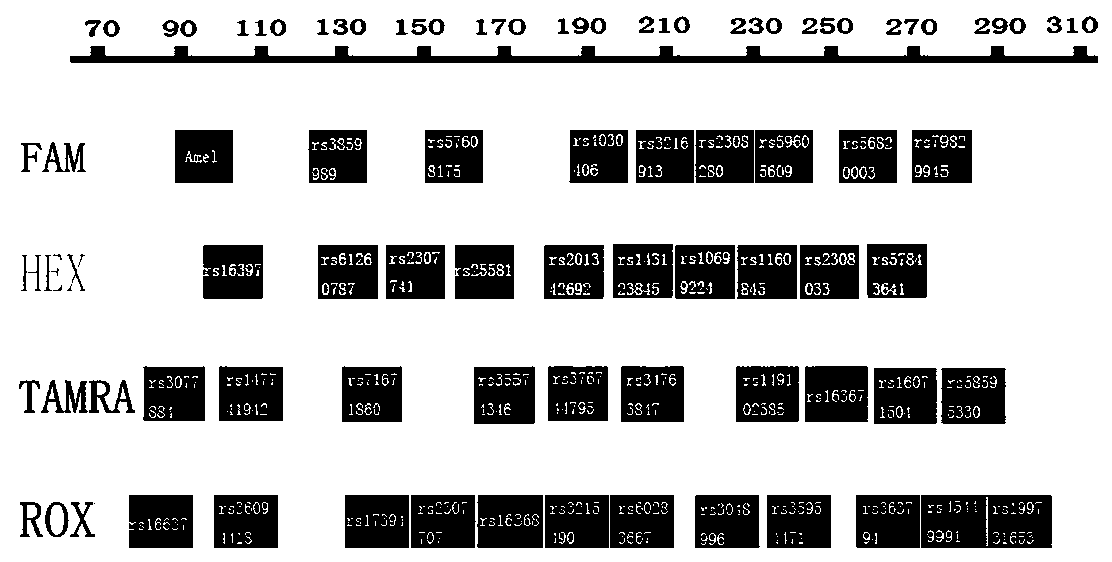
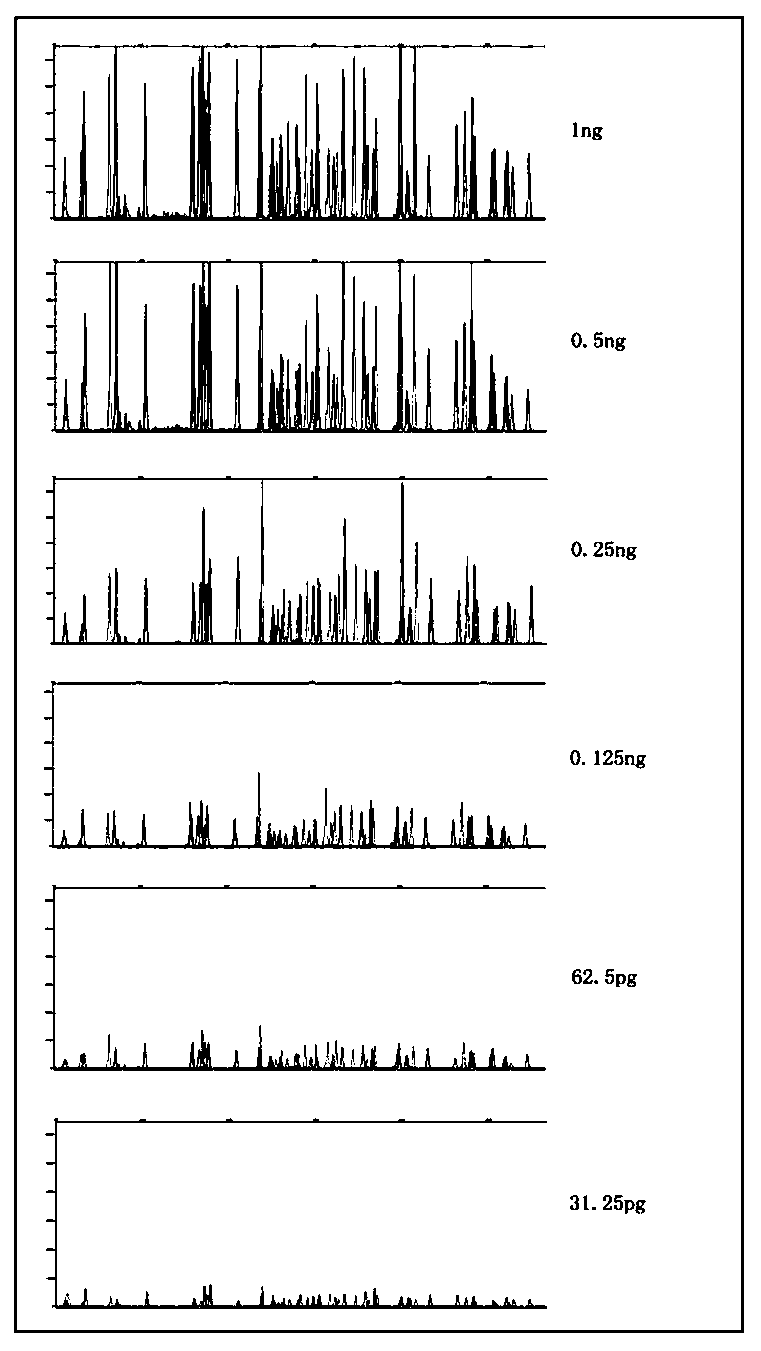
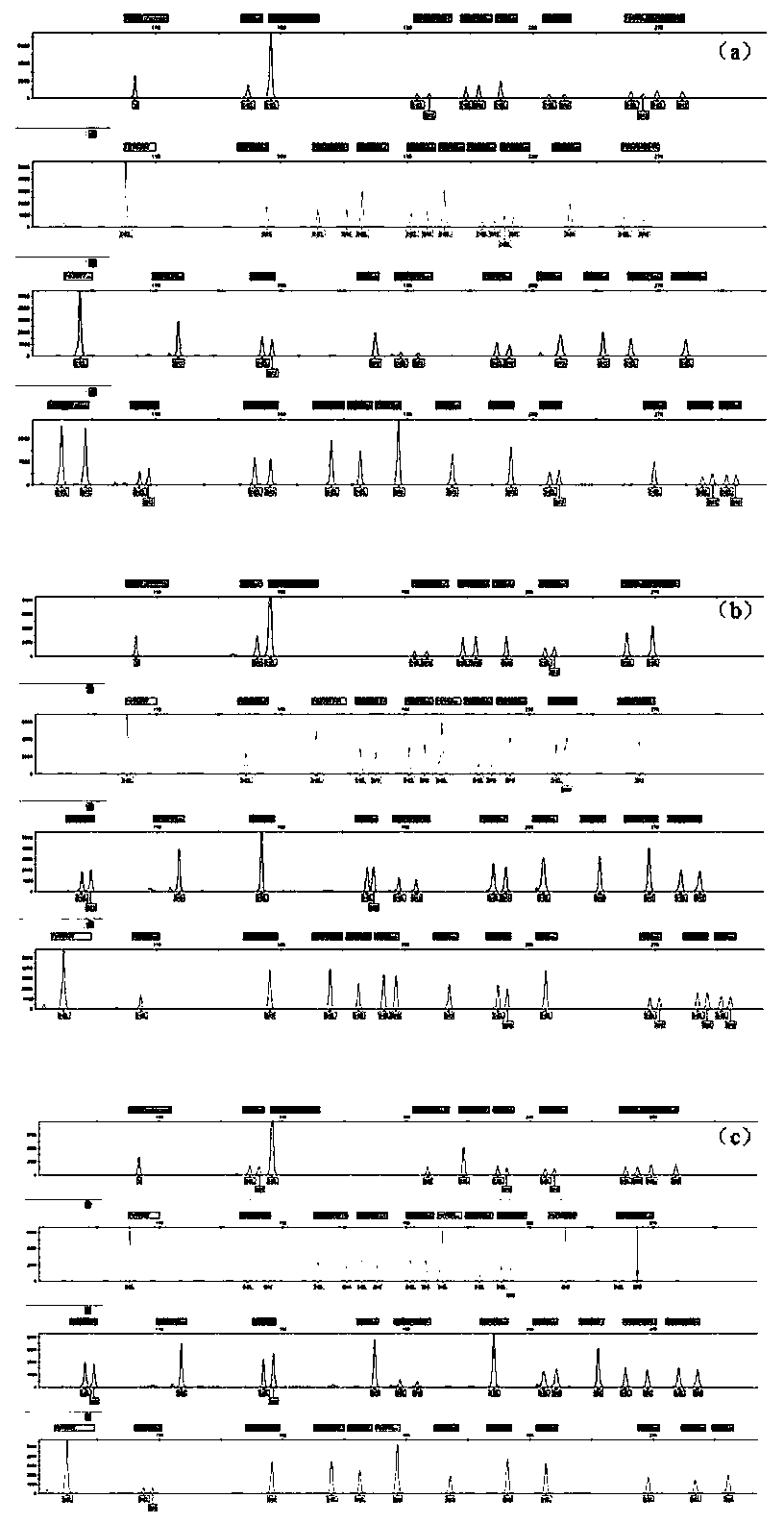
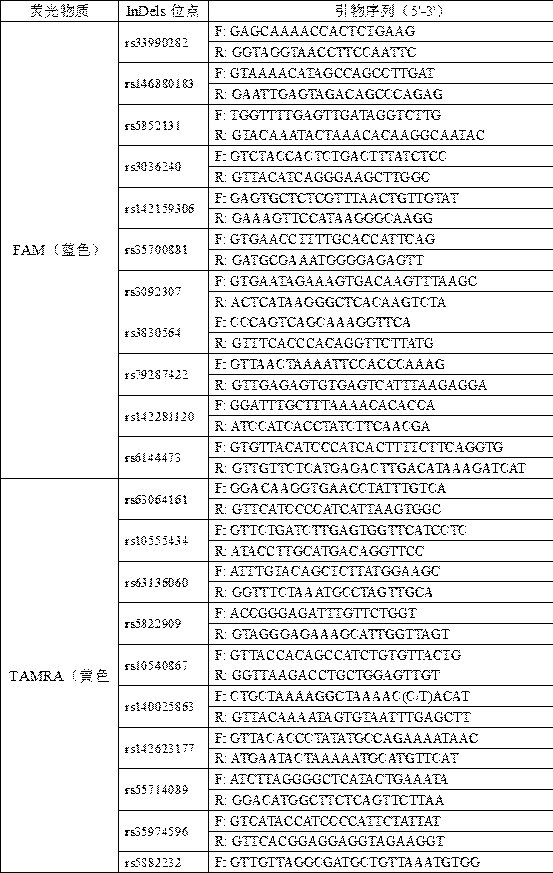
The invention belongs to the field of new research of forensic medicine, and particularly relates to a composite amplification typing detection system kit and a detection method for 35 new insertion / deletion (InDel) sites in forensic individual identification research. According to the present invention, the primers of InDel sites are respectively subjected to fluorescent labeling with four fluorescent substances (FAM, HEX, TAMRA and ROX), such that the 35 sites are subjected to composite amplification in the same amplification system, and are detected synchronously; the InDel sites are the length polymorphism molecular genetic markers, can be typed by using the capillary electrophoresis method, and are easily applied and promoted in the basic forensic department; the kit is compatible with various biological samples, and has advantages of high sensitivity, strong specificity and simple and quick operation; and the 35 InDel sites have high cumulative individual recognition rate in unrelated healthy individuals, can meet the needs of forensic individual identification, and further can provide new and effective detection reagents for forensic practice.
Owner:朱波峰 +3
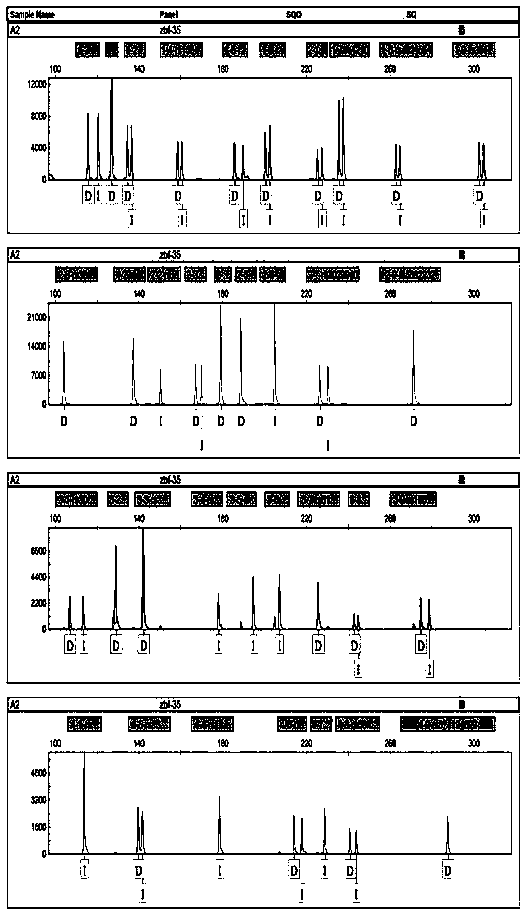
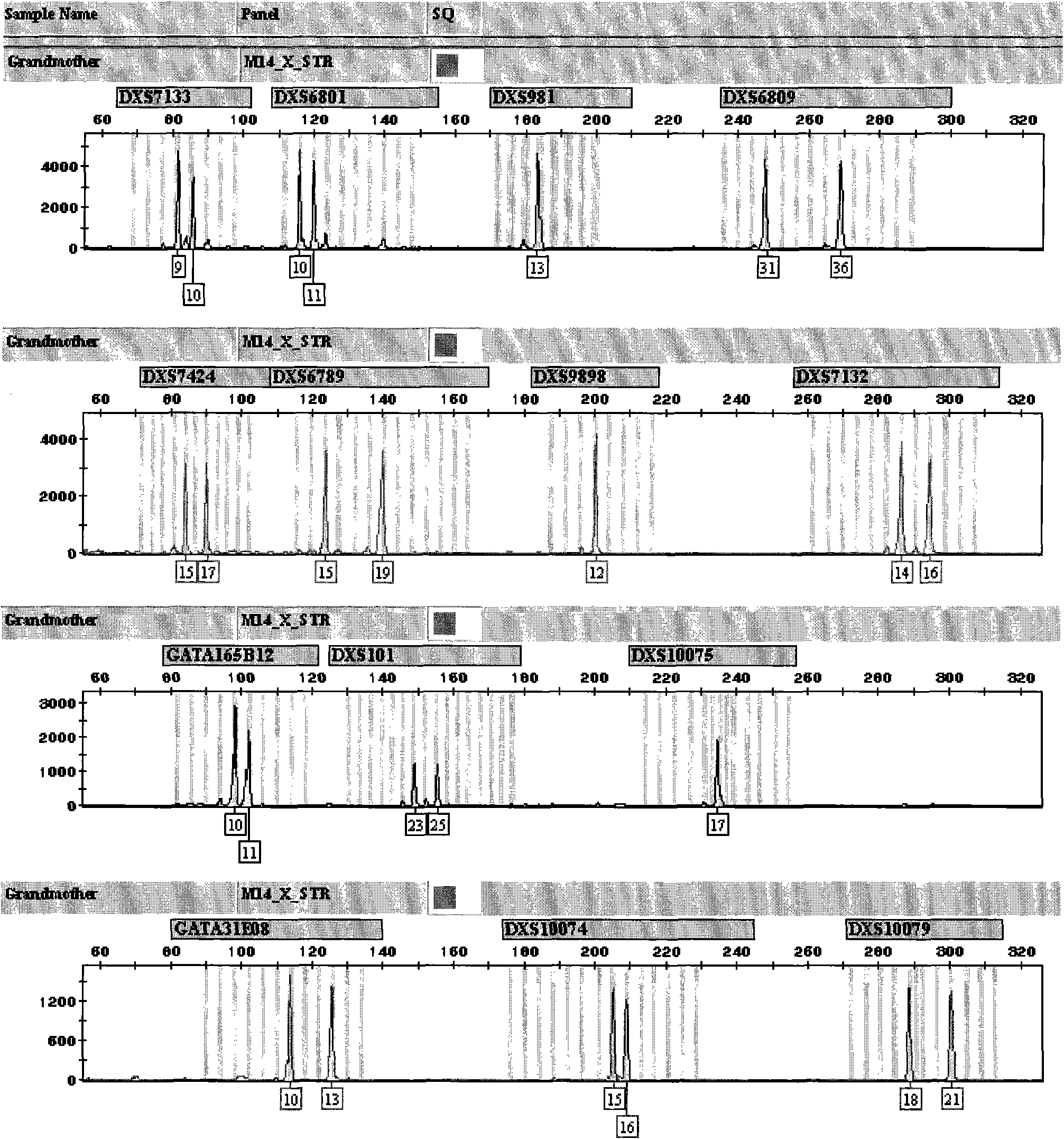
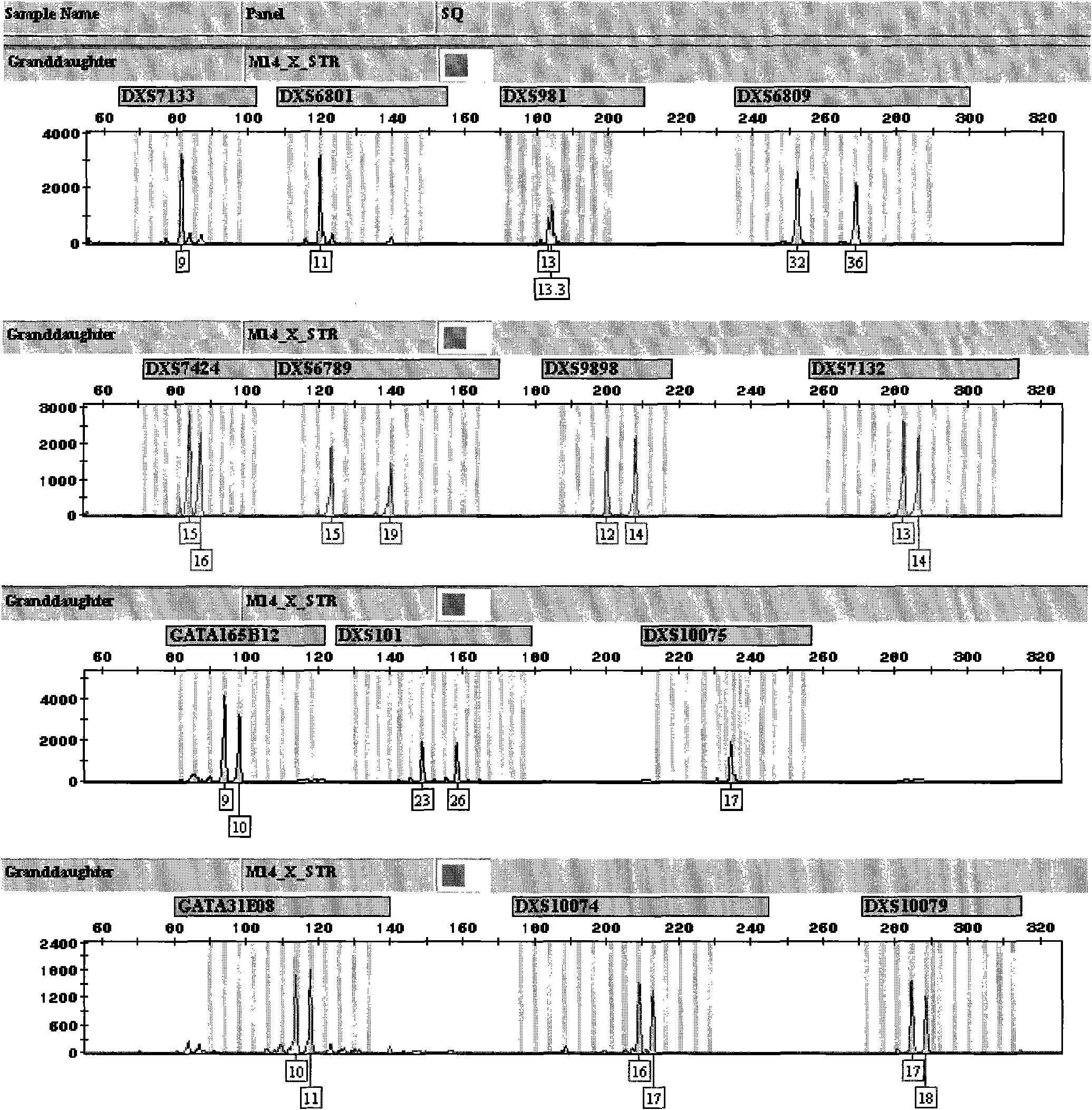
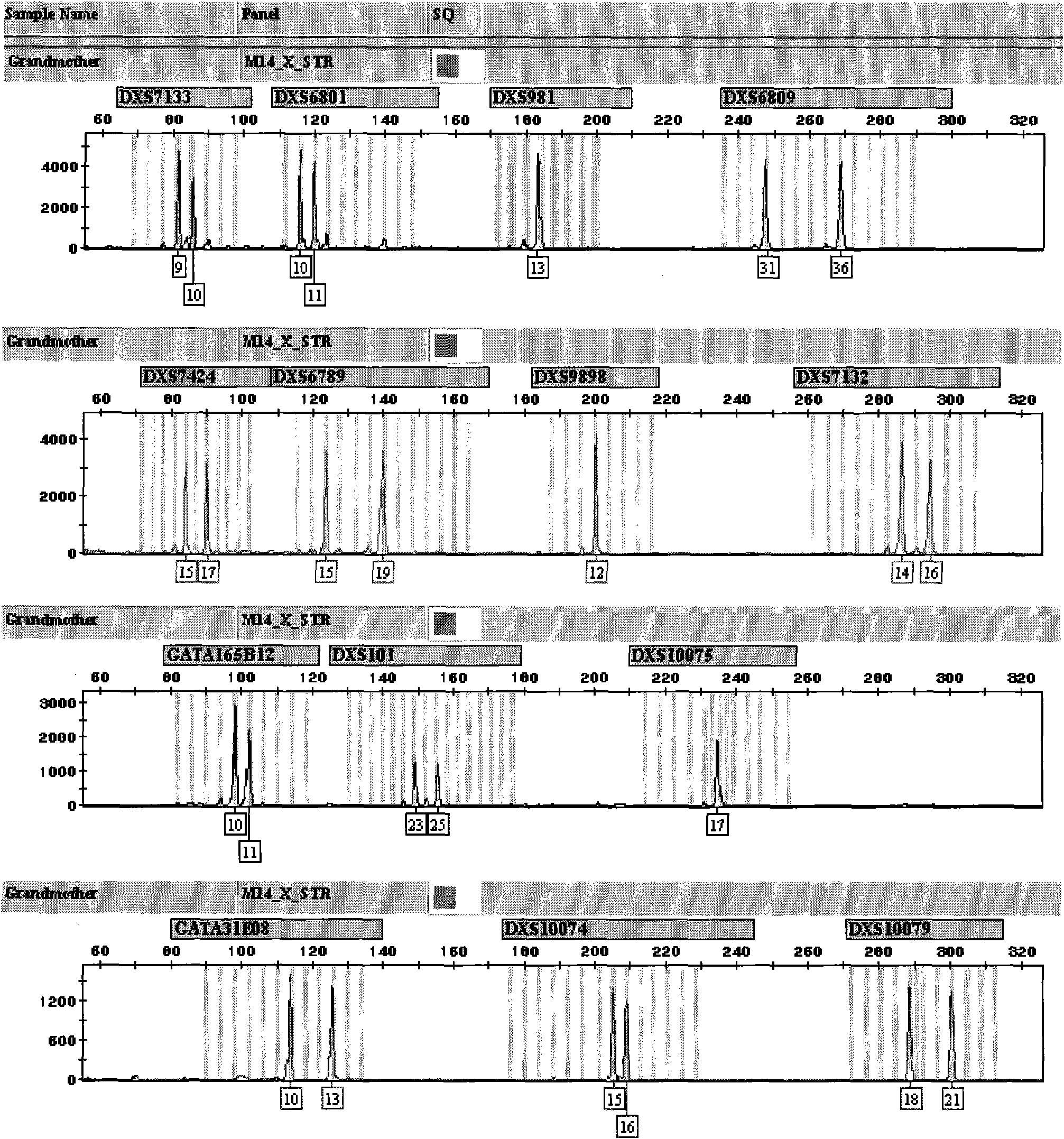
X-STR locus fluorescence labeling composite amplification system and application thereof
InactiveCN101787396AHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceElectrophoresisFluorescence
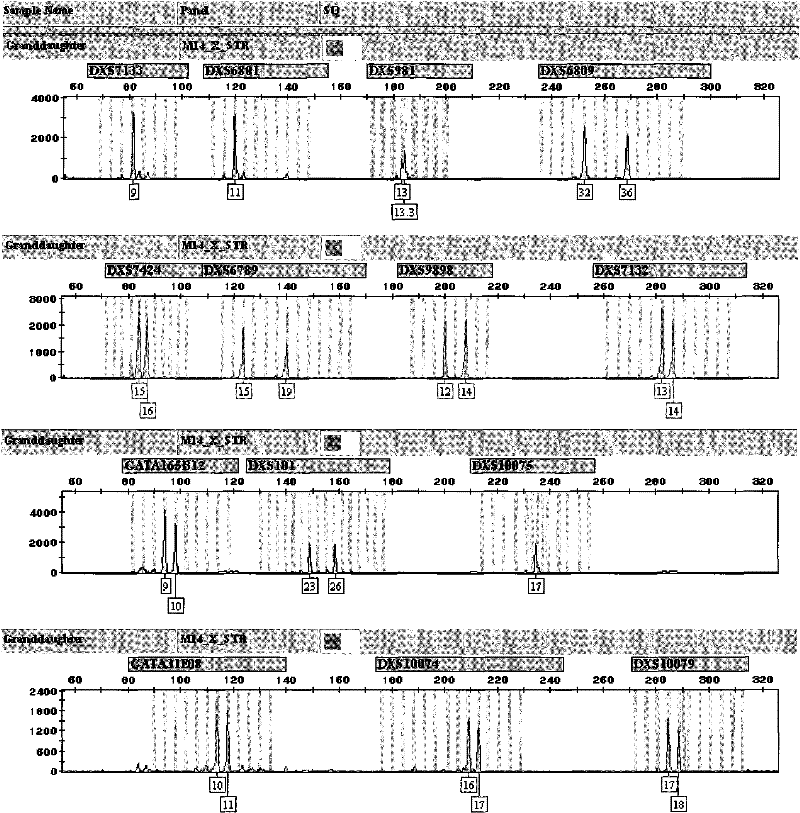
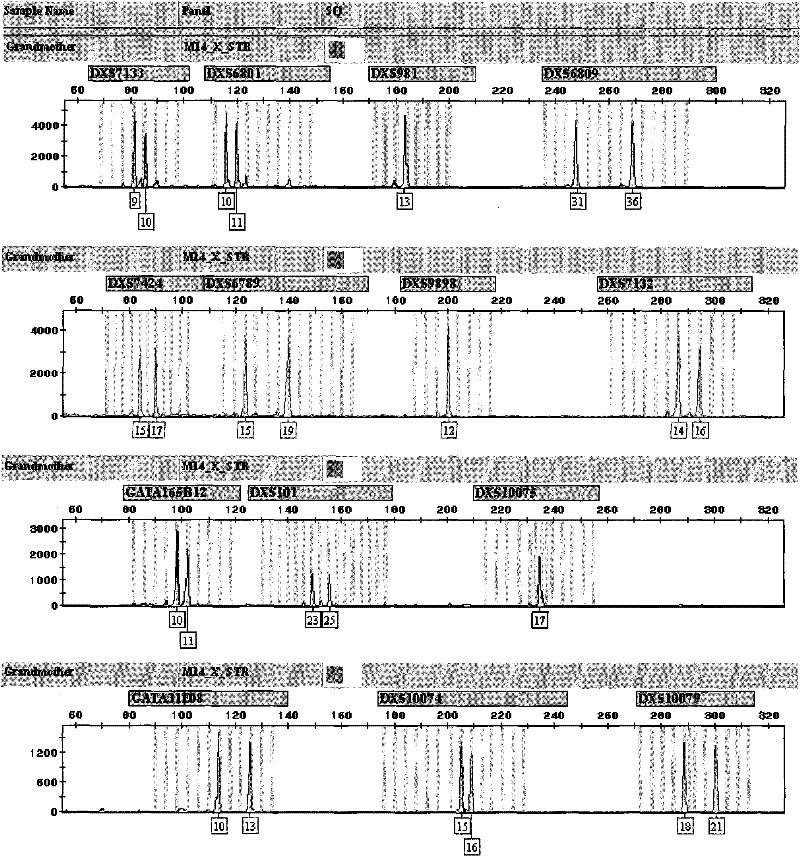
The invention discloses an X-STR locus fluorescence labeling composite amplification system and an application thereof. The composite amplification system comprises 14 loca in total: DXS7133, DXS6801, DXS981, DXS6809, DXS7424, DXS6789, DXS9898, DXS7132, GATA165B12, DXS101, DXS10075, GATA31E08, DXS10074 and DXS10079. The 14 X-STR loca are amplified simultaneously in a composite amplification system in a fast and convenient manner; the PCR product can be standardized and internationalized and ensure relative accuracy of different lab data with genetic analyzer electrophoresis and automatic result analysis.
Owner:SUN YAT SEN UNIV
Primer-free gene synthesis method based on plasmid library
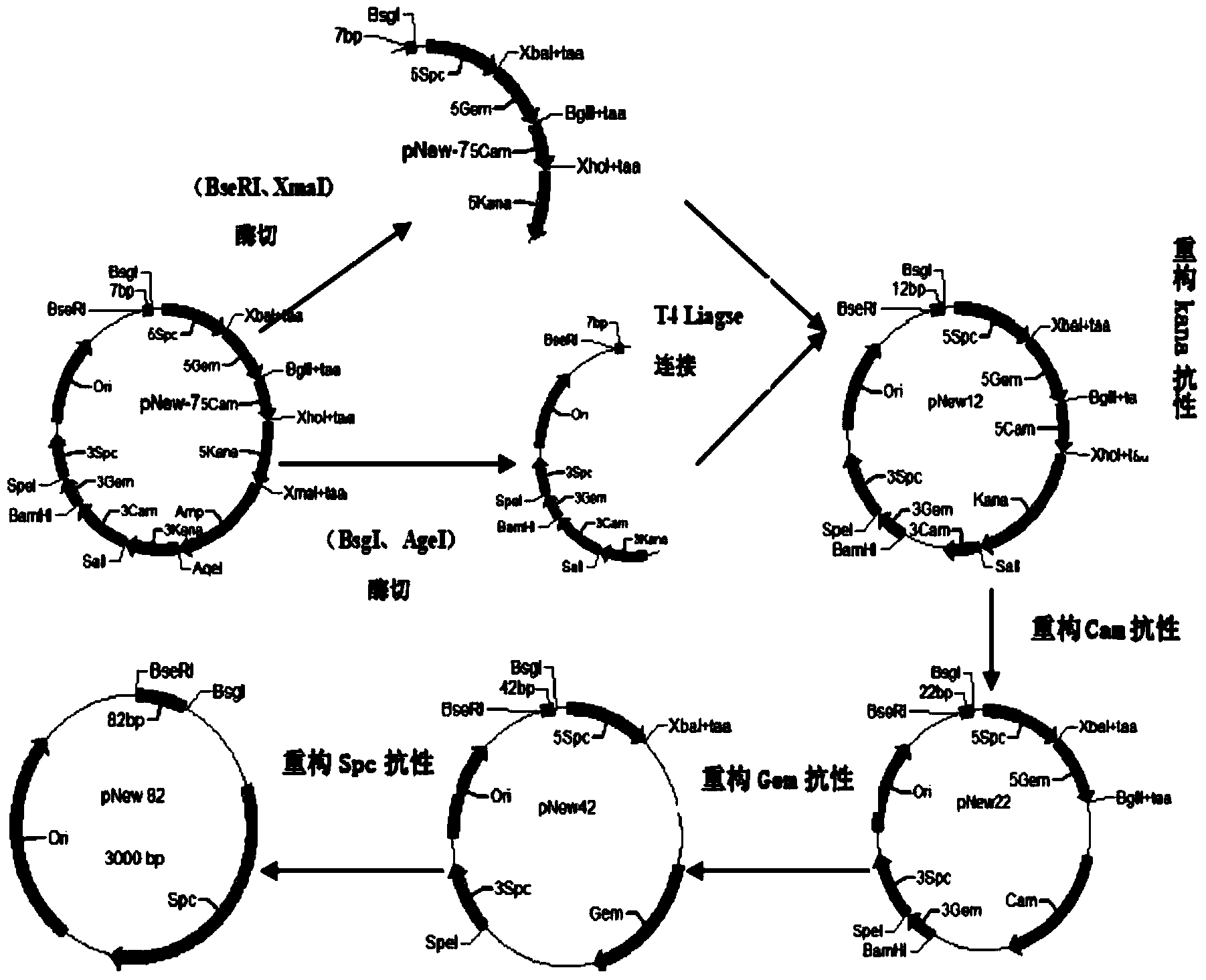
ActiveCN103952396ALow costImprove accuracyMicroorganism librariesLibrary creationEnzyme digestionSynthesis methods
The invention provides a primer-free gene synthesis method based on a plasmid library, and the method is as follows: constructing a plasmid library containing 16384 plasmids (any combination sequence of four basic groups of 47 and 7bp), starting from a constructed plasmid containing 7 bp, dividing a selected target gene according to each 82bp, successively reconstructing four resistance genes Kana, cam, gem and spc by enzyme digestion and enzyme connection operation to obtain a plurality of 82bp plasmids, and assembling the 82bp plasmids into long fragment genes. According to the method, after construction of the plasmid library containing the 16384 plasmids (any combination sequence of four basic groups of 47 and 7bp), various target genes can be synthesized without any primer. The synthesis method has the advantages of simple operation, extremely low mutation rate, no need of sequencing, low cost and less amount of screening, is conductive to constructing a mutant library, can realize solution operation, and is easy to realize automation.
Owner:WUHAN GENECREATE BIOLOGICAL ENG CO LTD
X-STR locus fluorescence labeling composite amplification system and application thereof
InactiveCN101787396BHigh sensitivityHigh individual recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceFluorescenceElectrophoresis
Owner:SUN YAT SEN UNIV
Cultivation method of dendrobium officinale group
InactiveCN107509633AImprove germination rateLow mutation ratePlant tissue cultureHorticulture methodsDiseaseEconomic benefits
The invention discloses a cultivation method of dendrobium officinale group. The cultivation method of dendrobium officinale group includes the following steps in order: 1) breeding: a. protocorm induction, b. protocorm differentiation, c. bud proliferation, d. seedling strengthening and rooting, 2) seedling selection, 3) removal out of a bottle, 4) cultivation, and 5) field management prevention and control of diseases and pests. The cultivation method provided by the invention has the advantages of high seed germination rate, low mutation rate, high seedling survival rate, good growth vigor, low lesion rate, and high economic benefits.
Owner:安徽徽生源生物科技股份有限公司
Method for identifying human-derived genetic relationship through two-nucleotide repeated microsatellite
ActiveCN106521017AEliminate distractionsAvoid interferenceMicrobiological testing/measurementCapillary electrophoresisElectrophoresis
Owner:INST OF MEDICAL BIOLOGY CHINESE ACAD OF MEDICAL SCI
Pharmaceutical composition for treating tumors or cancers and application thereof
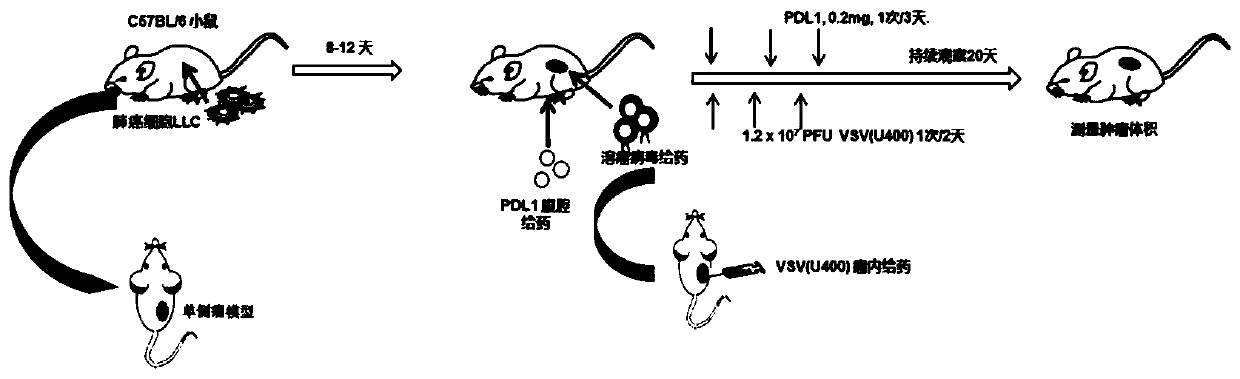
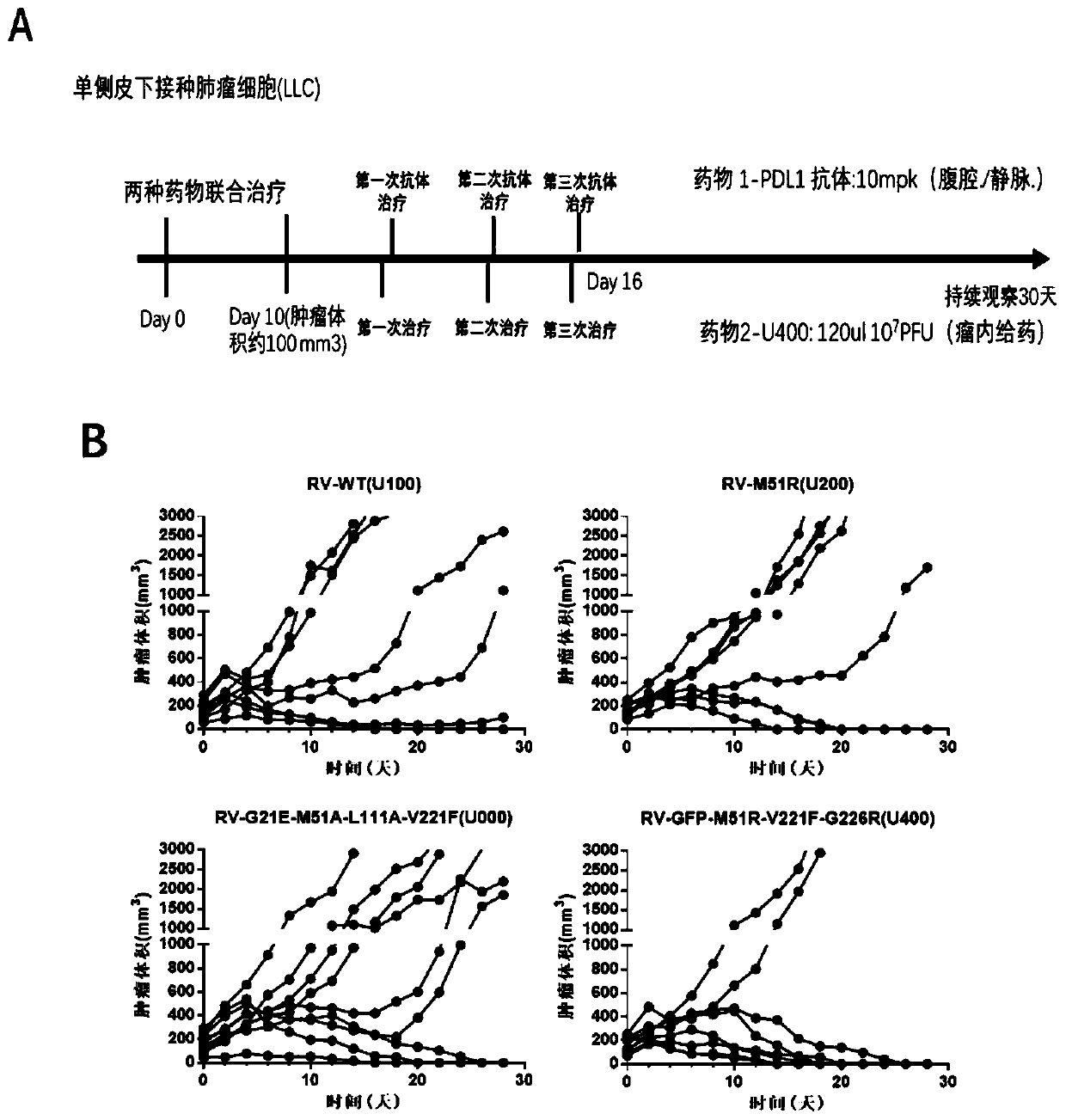
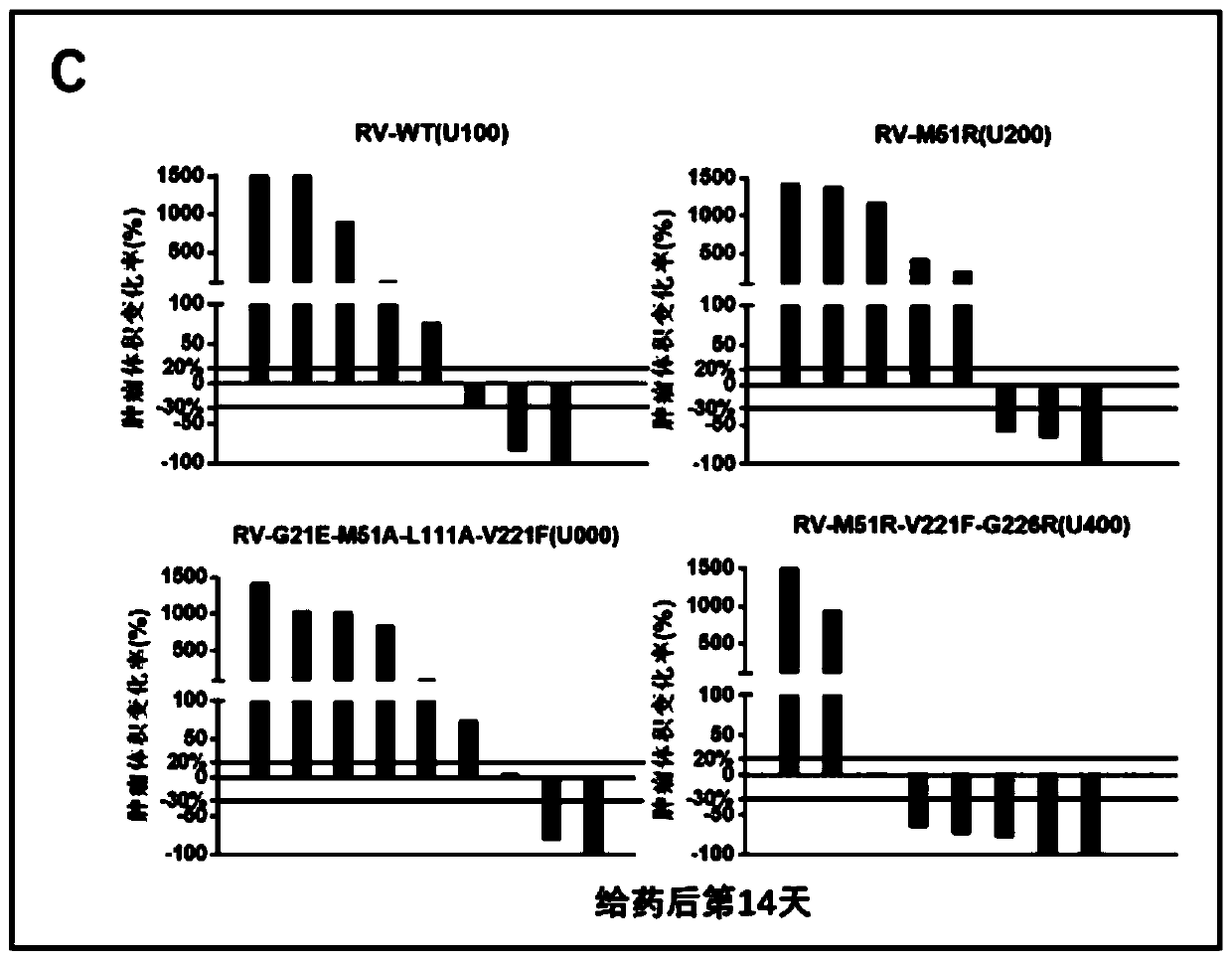
InactiveCN111494432ABreak inhibitionPromote specific killingUnknown materialsAntibody ingredientsGenes mutationAntiendomysial antibodies
The invention relates to a pharmaceutical composition for treating tumors or cancers and application thereof, in particular to a pharmaceutical composition which comprises vesicular stomatitis virusesdelivered through direct injection or systemic administration or intratumoral delivery and is combined with PDL1 antibody intravenous administration to generate a synergistic effect in treatment of metastatic lung cancer. Besides, the oncolytic virus after gene mutation and the PDL1 antibody have the characteristic of recognizing tumor cells, normal cells are not damaged, and the oncolytic virusand the PDL1 antibody are sequentially combined for use to generate a synergistic effect in curative effect.
Owner:FANTASIA BIOPHARMA ZHEJIANG CO LTD
44 InDels locus composite amplification detection kit for forensic individual identification of degradation samples
PendingCN109762909ALow mutation rateHigh sensitivityMicrobiological testing/measurementFluorescenceIdentification rate
The invention belongs to the field of forensic new technology research, and particularly relates to a composite amplification typing detection system kit and detection method for suitable for forensicdegradation sample individual identification and sex confirmation application research of 43 insertion / deletion and 1 amelogenin loci. The four different colors of fluorescent substances (FAM, HEX, TAMRA and ROX) are used for carrying out fluorescence labeling grouping on the primers of 44 loci, so that the 44 loci can be simultaneously subjected to composite amplification in the same amplification system. The InDels loci provided by the invention can be used for typing detection of degradation samples due to the fact that amplified fragments are short. The 44 loci contained in the system kithave high accumulated individual identification rate in irrelevant healthy individuals, and can meet the needs of individual identification of forensic degradation biological samples.
Owner:朱波峰 +3
Fast detection kit of donkey-origin component in food and application thereof
ActiveCN109811069AHigh copy numberEasy to quantifyMicrobiological testing/measurementDNA/RNA fragmentationReference genesNucleotide
The invention provides a fast detection kit of a donkey-origin component in food, and relates to the technical field of biological species identification. According to the fast detection kit of the donkey-origin component in the food, firstly, a donkey-origin universal endogenous reference gene LOC106825524 is screened out, a nucleotide sequence of the donkey-origin universal endogenous referencegene LOC106825524 is as shown in SEQ ID NO.1, and a copy number of the donkey-origin universal endogenous reference gene LOC106825524 in the donkey species is constant so that the donkey-origin universal endogenous reference gene LOC106825524 can be used as a target gene for identifying a donkey origin. By using the gene as a target sequence, PCR amplification primers are designed, the PCR amplification primers and PCR reaction liquid amplify together, and a PCR reaction product is used for platinum-palladium nanoparticle immunochromatographic test strip detection. PCR reaction in combinationwith platinum-palladium nanoparticle-based immunochromatographic test strip detection forms the fast detection kit, the donkey-origin component in the food can be detected fast and sensitively, and the detection sensitivity can reach 0.8%. The kit is simple in use method and low in cost, a reaction result is easy to observe, the specificity is good, and the kit is suitable for on-site real-time detection, and has wide application prospects on the aspects of animal-origin food surveillance and meat product adulteration resistance.
Owner:CHINA AGRI UNIV
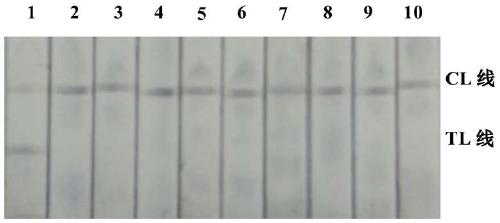
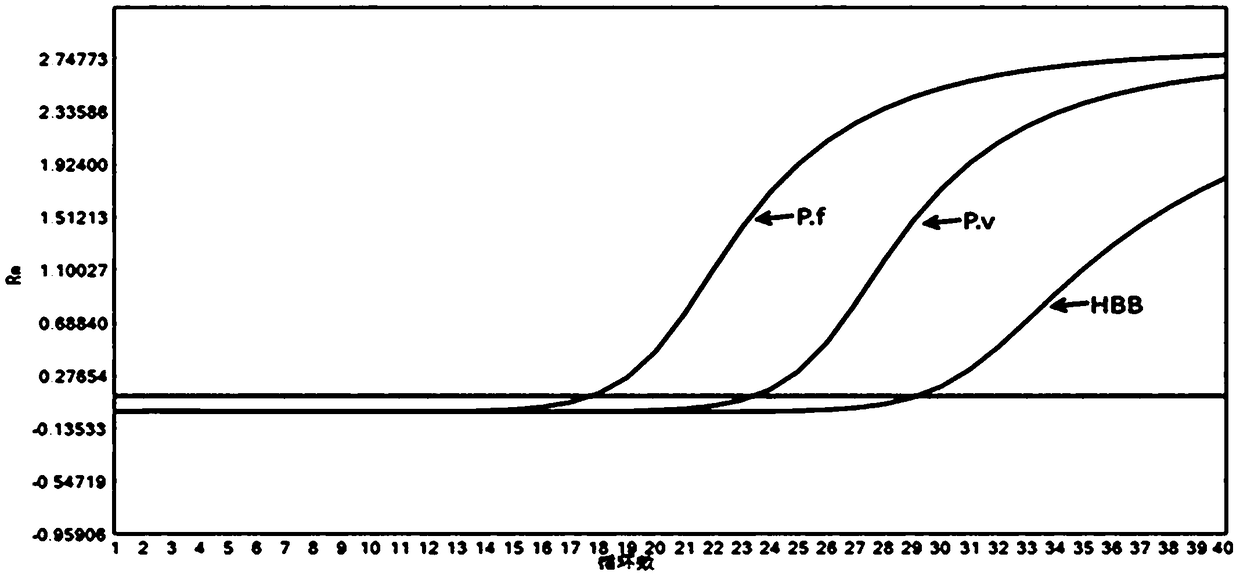
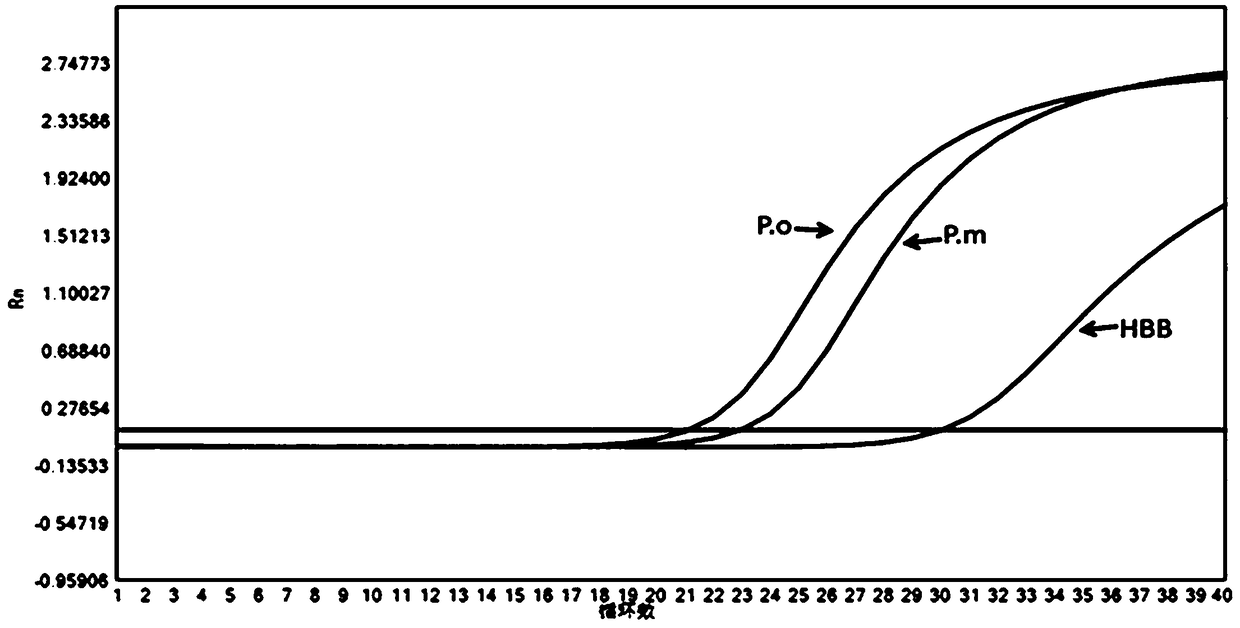
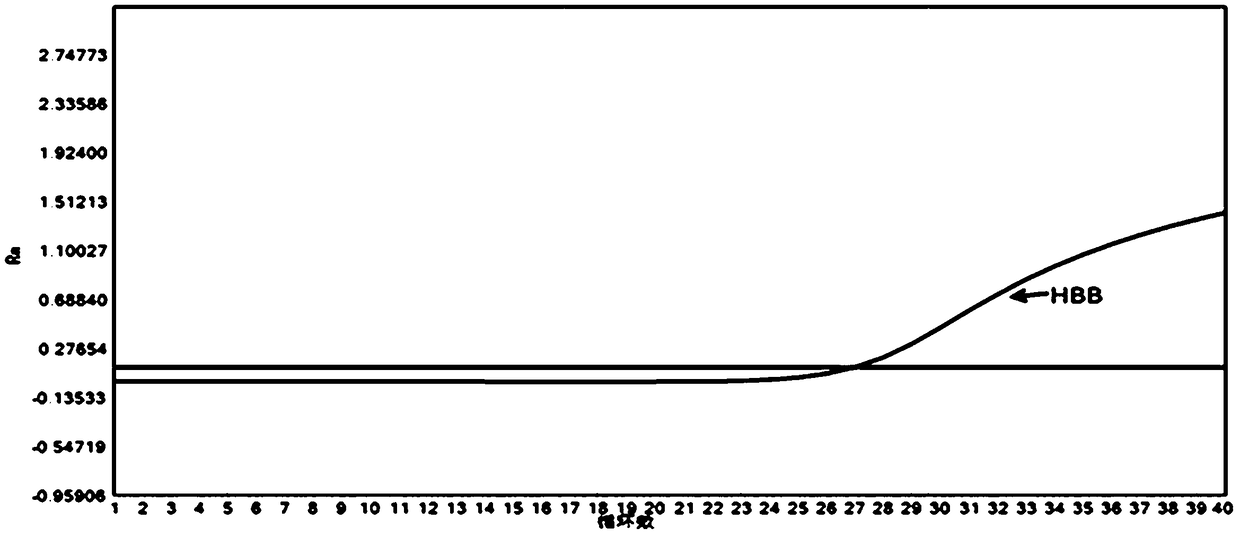
Plasmodium nucleate type detection kit and application method thereof
InactiveCN109486962ALess immune stressLow mutation rateMicrobiological testing/measurementAgainst vector-borne diseasesTypingEnzyme
The invention discloses a plasmodium nucleate type detection kit and an application method thereof. The kit comprises a PCR reaction solution I comprising a mixed primer, a PCR reaction solution II comprising a typing probe, a Taq / UNG enzyme mixed solution, and a positive reference and negative reference comprising recombinant plasmids of P.v, P.f, P.o and P.m mitochondria cox gene specific area sequence; the mixed primer comprises a primer sequence used for amplifying a P.v, P.f, P.o and P.m mitochondria cox gene specific areas, and a primer sequence for amplifying an HBB gene; and the typingprobe comprises a sequence for typing identifying the P.v, P.f, P.o and P.m mitochondria cox 3 gene pecific areas and a probe sequence for identifying the HBB gene. By adopting the design of the invention, the plasmonium in a sample can be detected, and four plasmonium can be accurately typed; and the plasmonium mitochondria cox3 gene is adopted as a target gene to perform the detection, so thatthe specificity and the sensitivity are higher.
Owner:合肥欧创基因生物科技有限公司
Crispr-based treatment of friedreich ataxia
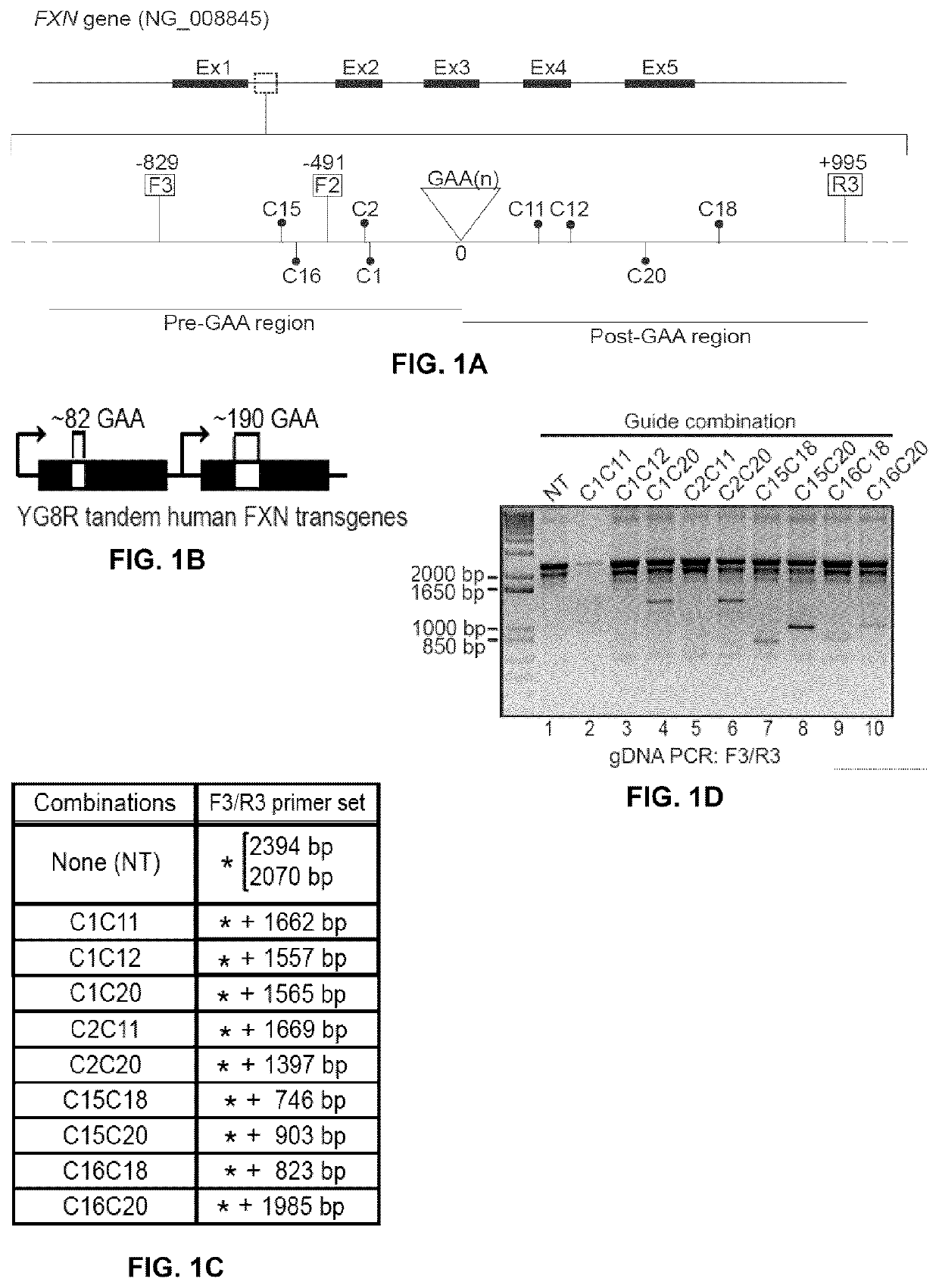
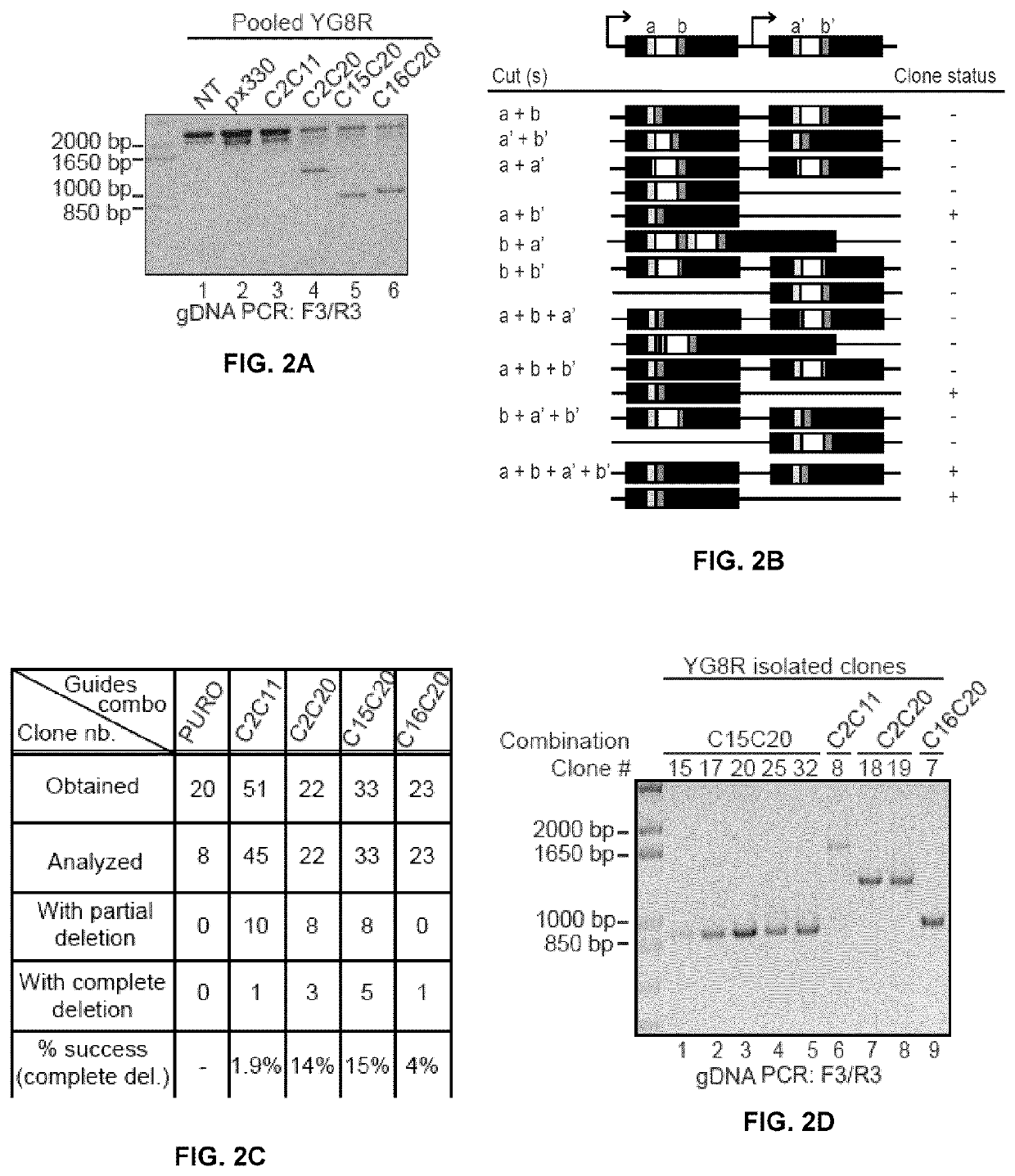
InactiveUS20200056206A1Low mutation rateIncreasing baseline levelOrganic active ingredientsNervous disorderNucleotideMedicine
Methods of modifying a frataxin gene are disclosed, comprising removing some or all of endogenous GAA trinucleotide repeats within the frataxin gene, e.g., within an intron (e.g., intron 1) of the frataxin gene. The removal may be effected using a CRISPR / CAS nuclease system. Such modification may be used to increase frataxin expression in the cell, and also to treat a subject suffering from Friedreich ataxia. Reagents, kits and uses of the method are also disclosed, for example to modify a frataxin gene and to treat a subject suffering from Friedreich ataxia.
Owner:UNIV LAVAL
System for subtyping 59 micro-haplotypes genetic markers for individual identification in forensic medicine and application thereof
ActiveCN111118169AHigh polymorphismReduce false positive typingMicrobiological testing/measurementDNA/RNA fragmentationMedicineForensic Pharmacy
Owner:SUN YAT SEN UNIV
Ancestral polymorphism prediction method based on big data artificial intelligence algorithm
PendingCN112233724AHighly polymorphicGood predictive analyticsBiostatisticsProteomicsStructure analysisAlgorithm
The invention discloses an ancestral polymorphism prediction method based on a big data artificial intelligence algorithm. The method comprises the following steps: S1, selecting thousand-person genome data; S2, screening a plurality of microhaplotype sites from a part of different crowds, and carrying out size classification according to the average effective allele number of the different crowds; S3, carrying out group structure analysis on part of different crowds, and carrying out source classification on the different crowds; and S4, selecting a plurality of microhaplotype sites accordingto the information degree value and the average effective allele value. According to the invention, the microhaplotype is used for detecting and classifying the genes of the crowds, so that the method has the advantages of good high polymorphism, low mutation rate and length, can well predict and analyze the genes and sources of the crowds, and is high in accuracy; a plurality of diallelic SNP loci, a plurality of triallelic SNP loci and a plurality of microhaplotype loci are selected according to indexes such as information degree, fixed indexes and effective allele number, so that the accuracy of ancestral sources is respectively high.
Owner:深圳市盛景基因生物科技有限公司
Human DIP-InDel locus fluorescent labeling kit and detection method thereof
ActiveCN110628921ADirect lockProtection securityMicrobiological testing/measurementFluorescenceNon invasive
The invention discloses a human DIP-InDel locus fluorescent labeling kit and a detection method thereof. The kit comprises 45 specific multiplex PCR primers with nucleotide sequences shown in SEQ NO.1-45 and 4 pairs of fluorescent labeled universal primers with nucleotide sequences shown in SEQ NO.46-53. By amplifying and comparing 15 DIP-InDel loci, trace components in double-human unbalanced mixed sample are detected and excluded, an individual is confirmed, and the kit can be applied to aspects such as double-human mixed stain medicolegal expertise and non-invasive fetal paternity test.
Owner:SHANXI MEDICAL UNIV
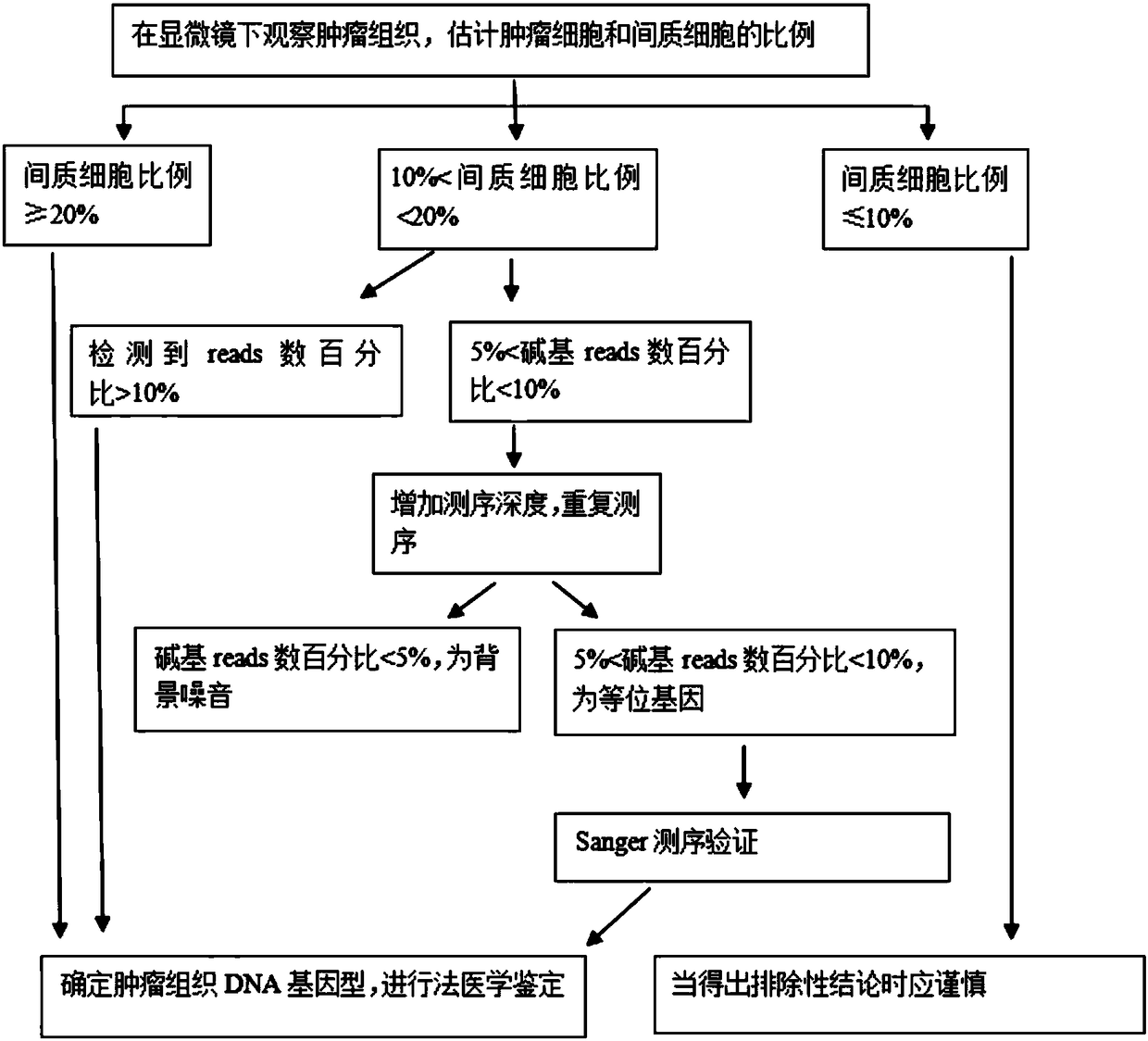
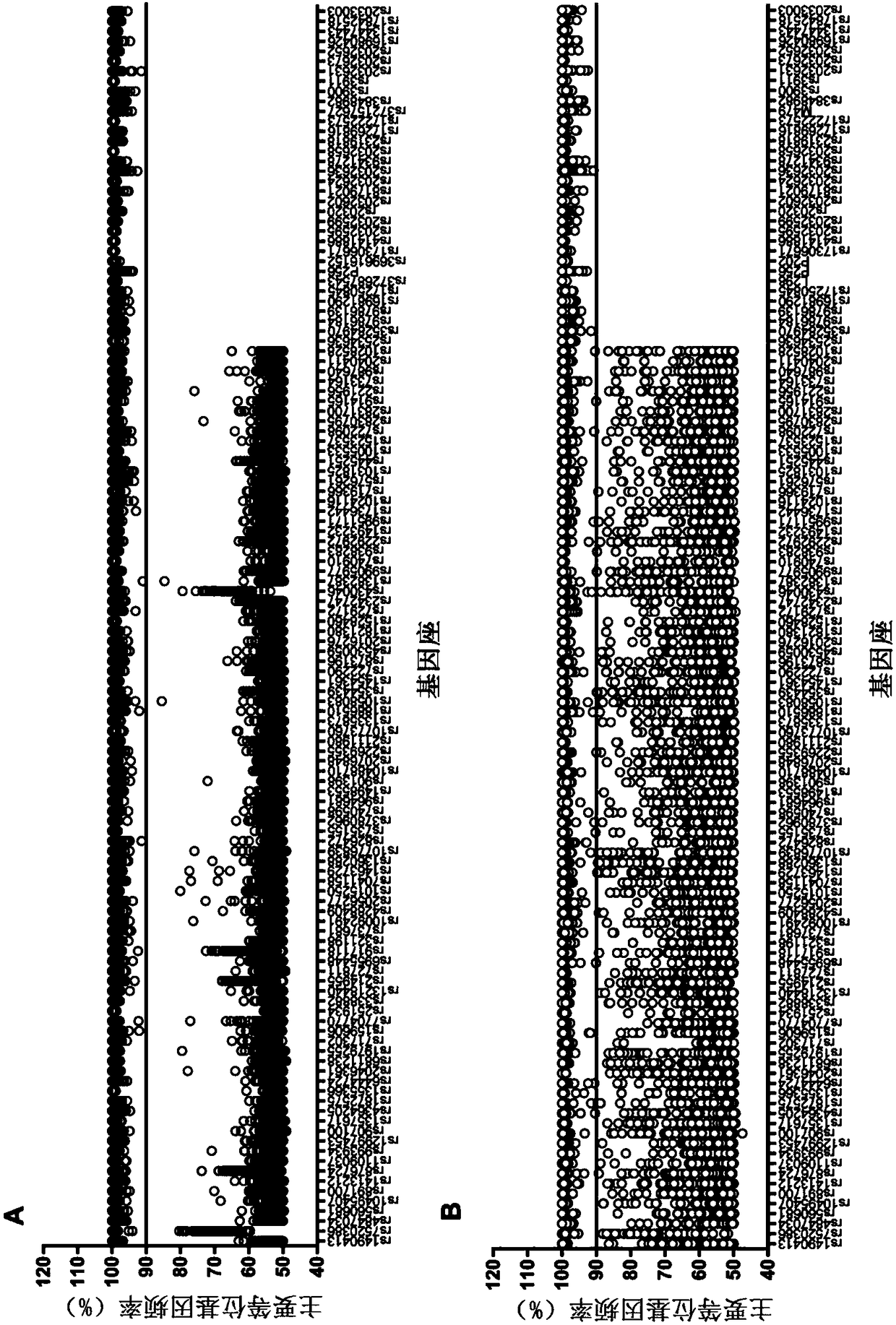
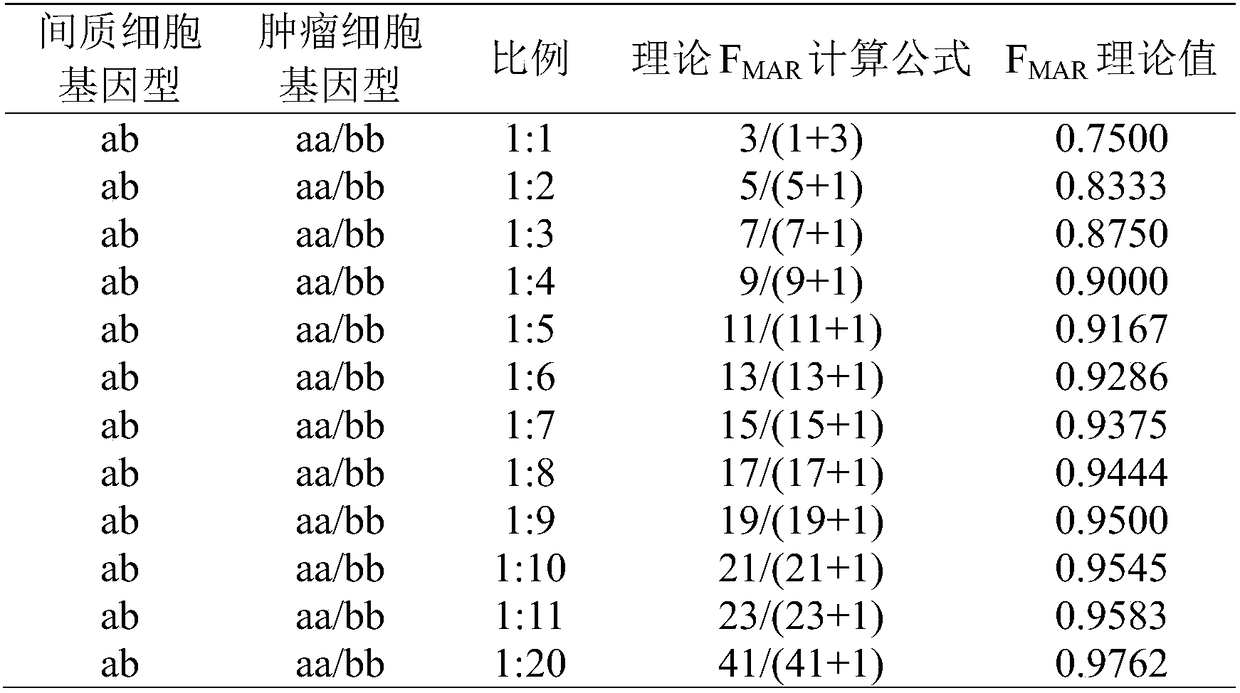
Method for judging specific individual attribution of malignant tumor tissues, and application of method
ActiveCN108504734ASimple and fast operationJudgment of affiliationMicrobiological testing/measurementGenomic sequencingGenotyping
The invention relates to a method for judging specific individual attribution of malignant tumor tissues. The method comprises the following operating steps: respectively extracting cell DNA of the malignant tumor tissues to be tested and cell DNA of the normal tissues of a specific individual; quantifying and diluting the DNA, and then performing genome sequencing by using a next-generation sequencing technology to obtain original genomic data; performing judgment of single nucleotide polymorphism according to the original genomic data; judging the attribution of the malignant tumor tissues and the specific individual based on the result of the judgment. The method has the characteristics of being high in accuracy, strong in expansibility, rapid and flexible, and the like, and can more accurately detect the genotyping of the malignant tumor tissues, thereby being applied to forensic identification of the malignant tumor tissues; the method can be used for judging whether the malignanttumor tissues to be tested are derived from a specific individual or not.
Owner:HEBEI MEDICAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com