Fast detection kit of donkey-origin component in food and application thereof
A donkey source and food technology, applied in the field of biological species identification, can solve the problems of inability to accurately quantitatively analyze samples, high homology of mitochondrial genes, and difficult qualitative detection, and achieve the effects of easy observation, good specificity, and easy quantification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Screening of donkey-sourced universal internal standard gene LOC106825524
[0041] By searching the gene information about donkeys in GenBank, the target genome was downloaded from NCBI and saved as ".FASTA" format. The whole genome information of the donkey was analyzed, and the homology analysis was carried out using BLAST and DNAMAN Version 4.0 software, and the LOC106825524 gene was screened out, which was located on the chromosome. 20 kinds of meat (respectively common chicken (LOC106825524lus LOC106825524lus), pheasant (Phasianuscolchicus), turkey (MeleagrisLOC106825524lopavo), black-bone chicken (LOC106825524lus domesticus brisson), pig (Sus scrofa), cattle (Bos taurus aries), sheep (Ovis ), duck (Anas platyrhynchos), goose (Goose calicivirus), dog (Canis lupus familiaris), rabbit (Oryctolagus cuniculus), yak (Bos mutus), yellow croaker (Pseudosciaena polyactis), horse (Equus caballus), donkey (Equus asinus) , mouse (Musmusculus), buffalo (Bubalus buba...
Embodiment 2
[0042] Example 2 Establishment of PCR detection method for donkey source components
[0043] Use primer premier5.0 software to design PCR primers for the LOC106825524 gene determined in Example 1, the 5' end of the upstream primer F is labeled with biotin (Biotin), and the 5' end of the downstream primer R is labeled with fluorescein (FITC).
[0044] See Table 1.
[0045] Table 1 PCR primer sequences
[0046]
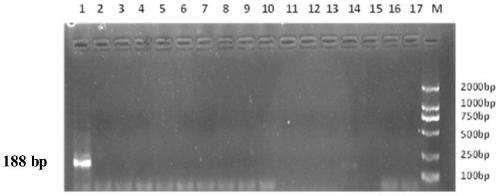
[0047] Donkey samples were quickly detected by PCR. The reaction system was 25 μL, including 10x reaction buffer, 0.4 mM dNTP, 0.2 μM primer F, 0.2 μM primer R, and 2U Taq PCR polymerase. The reaction program was 95°C for 5min; 95°C for 30s, 53°C for 30s, 72°C for 30s, a total of 30 cycles; 72°C for 5min. After the amplification, 2% agarose gel electrophoresis was used to determine the product, and the appearance of a specific band proved that the amplification was successful and contained the target gene. The result is as figure 2 Shown, donkey-specific interna...
Embodiment 3
[0048] Example 3 Establishment of detection method for donkey source component PCR product platinum palladium nanoparticle immunochromatographic test strip
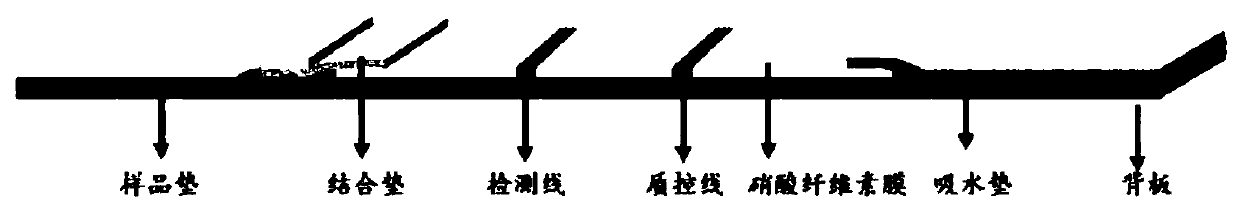
[0049] The platinum-palladium nanoparticle-labeled antibody was prepared by the sandwich method, and stored at 4°C for later use. Dilute the FITC antibody with buffer to the optimal concentration respectively. The distance between the test line (TL) and the quality control line (CL) is 4.5 mm, and sprayed on the NC membrane at 1.0 μL / cm respectively. The sprayed NC membrane was dried overnight at 37°C for later use. Test strips were cut to a width of 3.8 mm.
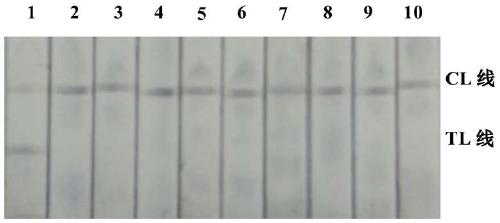
[0050] Fully mix the PCR reaction product with the buffer solution and drop it on the sample pad of the platinum-palladium nanoparticle immunochromatography test strip. At this time, the mixed solution passes through the binding pad and the NC membrane under the capillary power, and continues to move towards the water-absorbing pad. After 3 minutes, the test resu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com