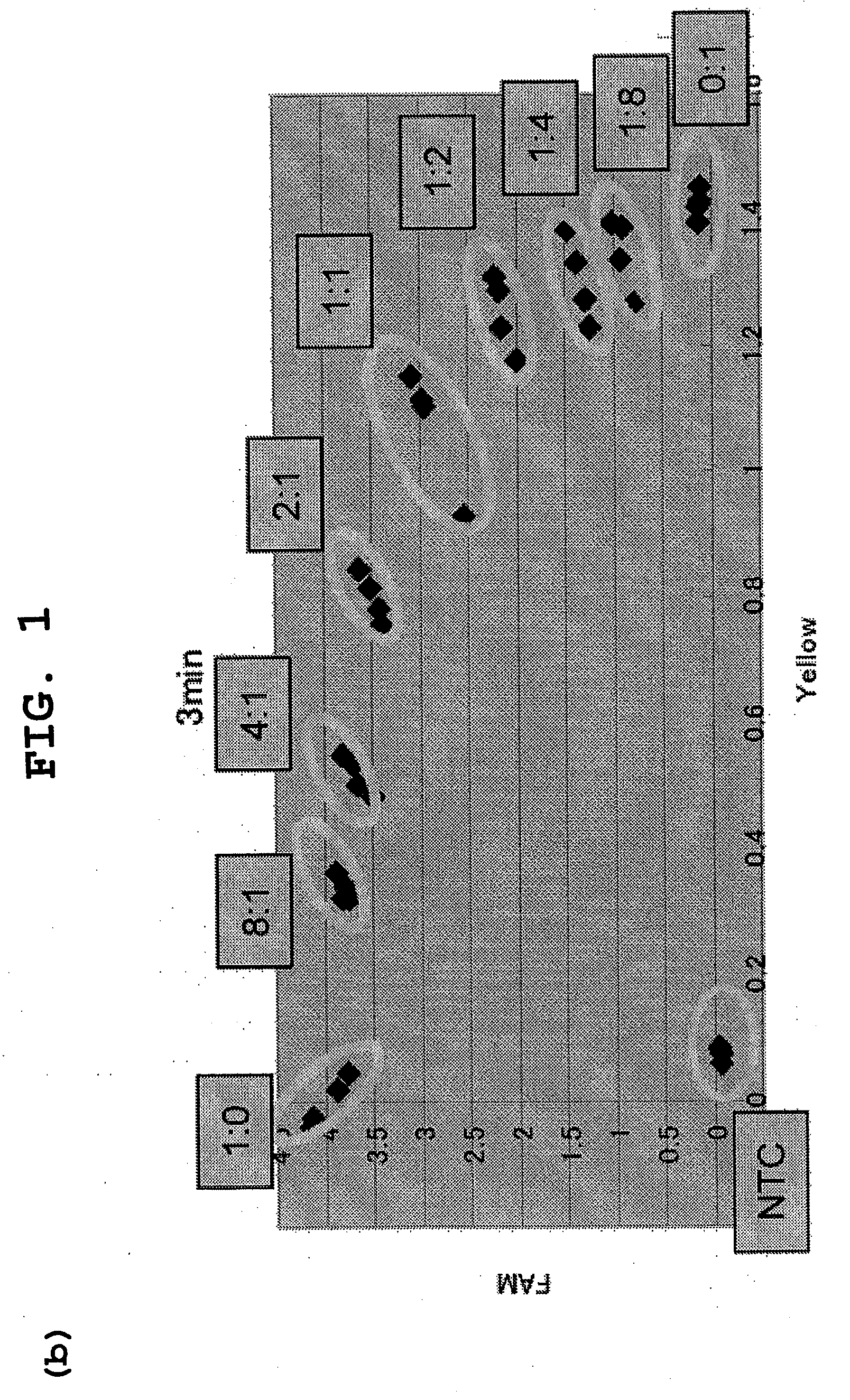
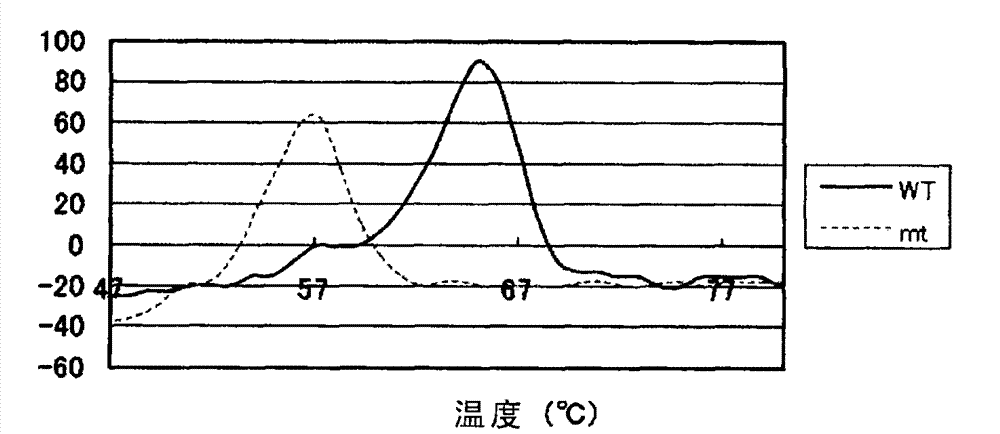
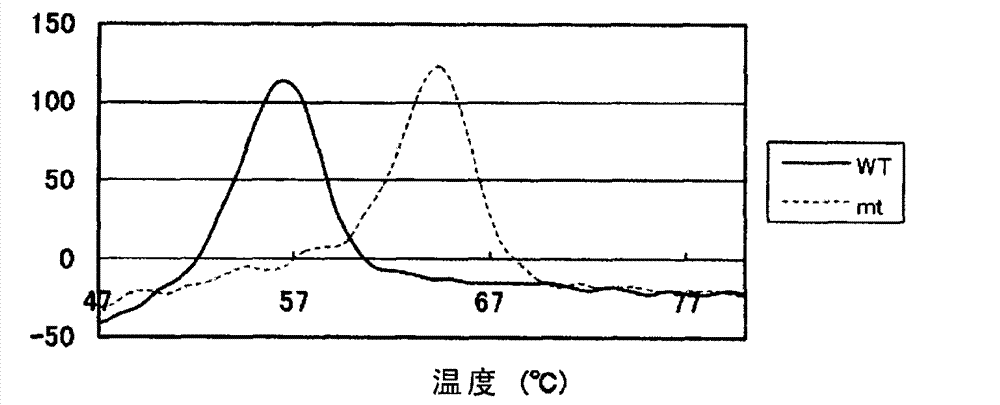
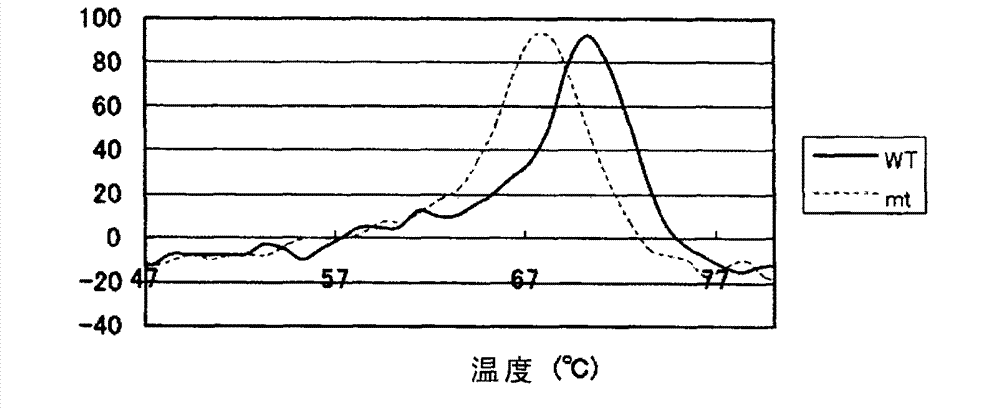
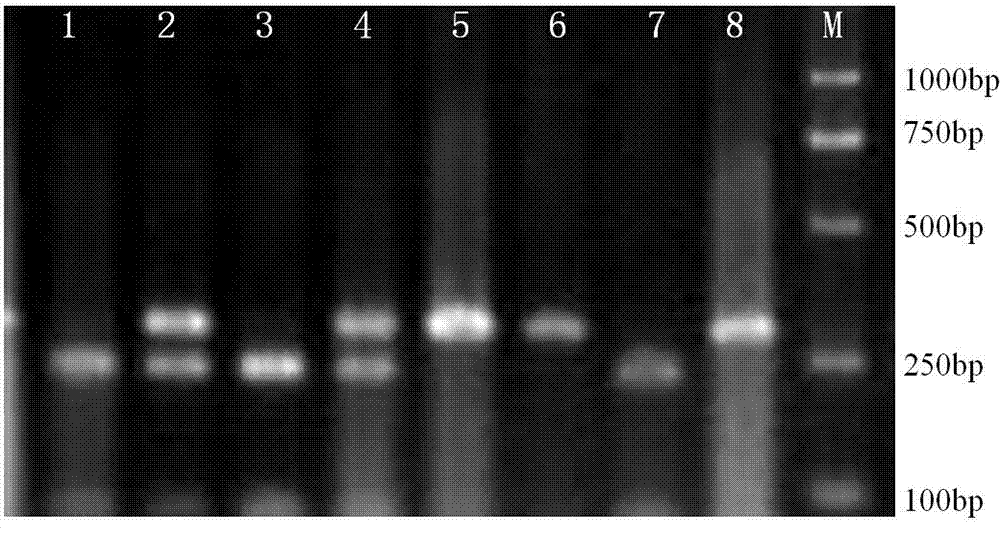
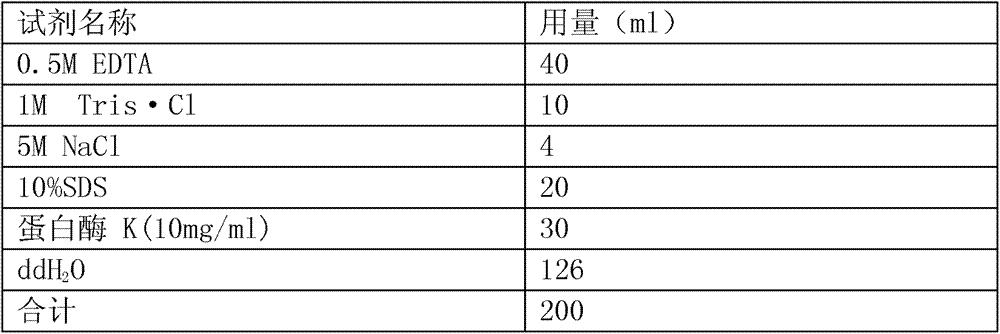
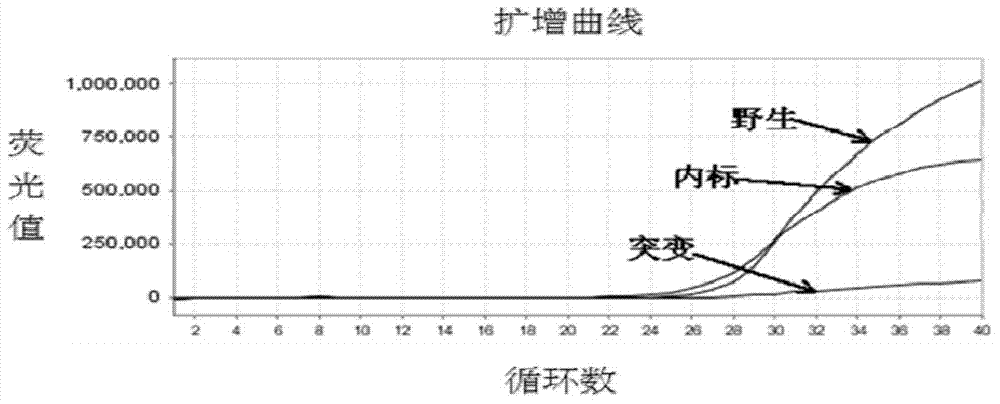
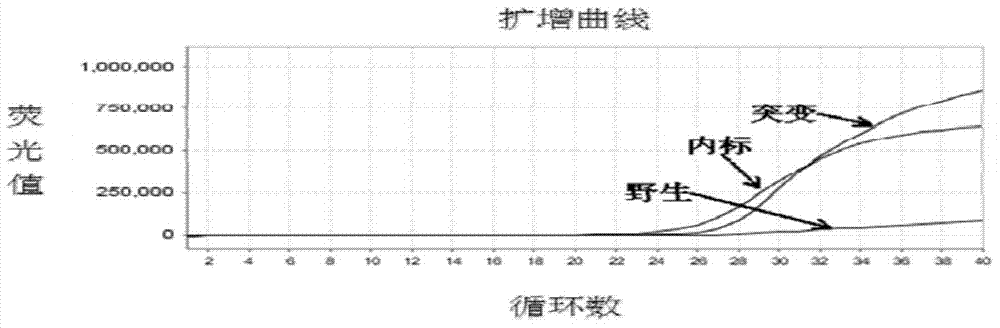
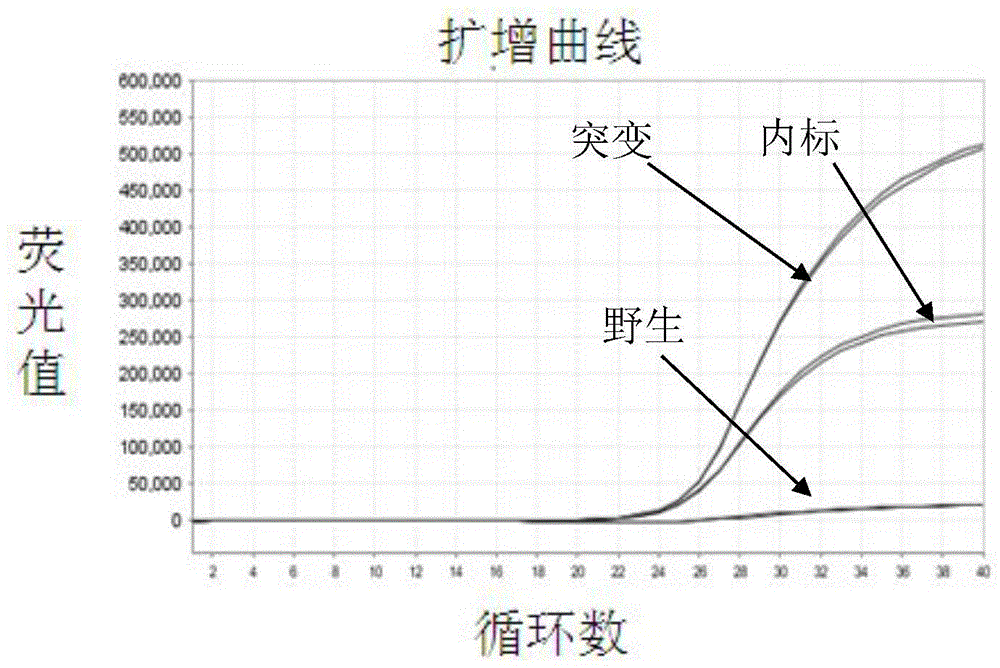
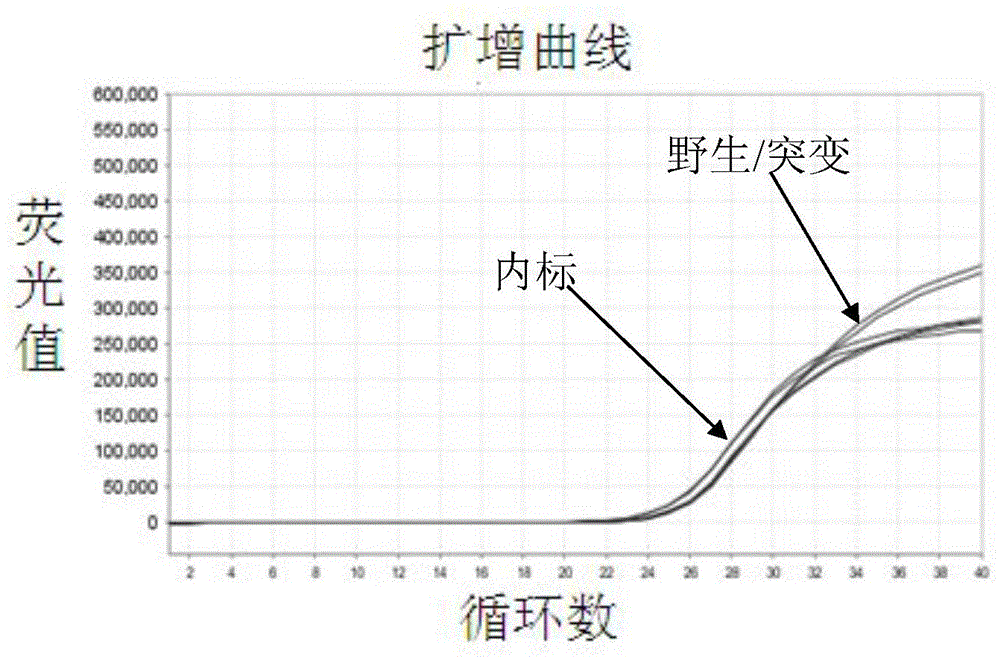
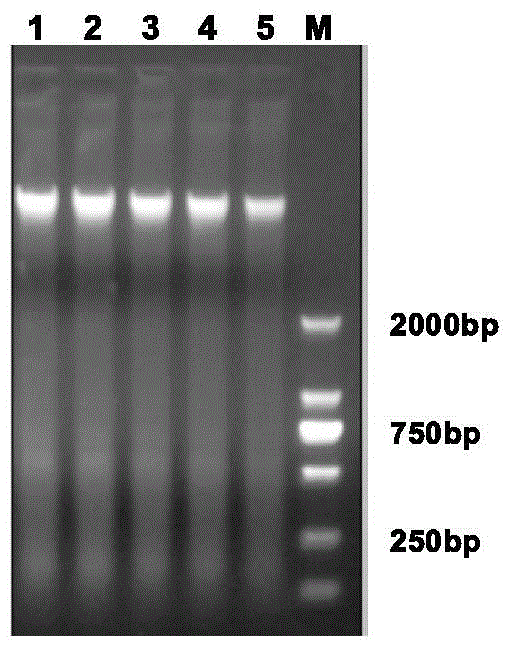
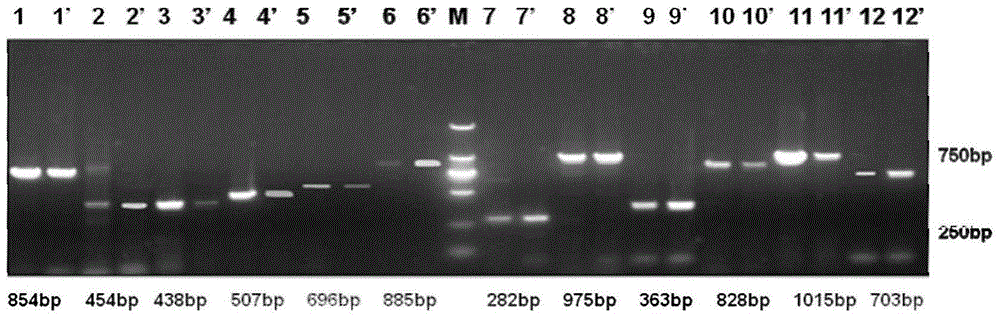
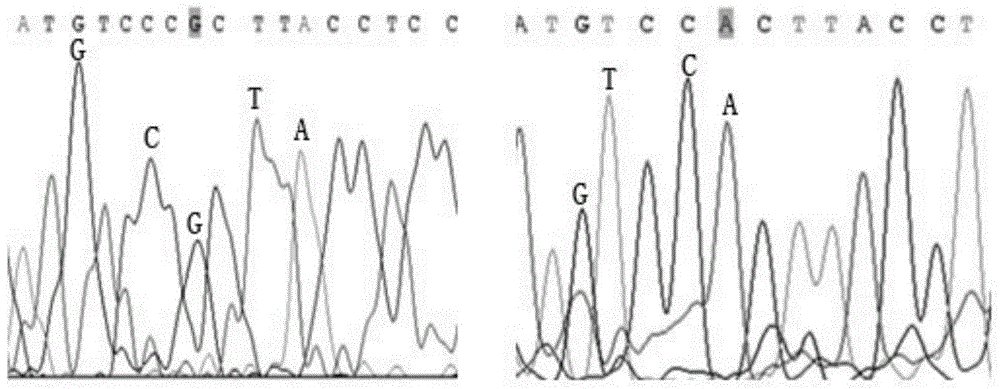
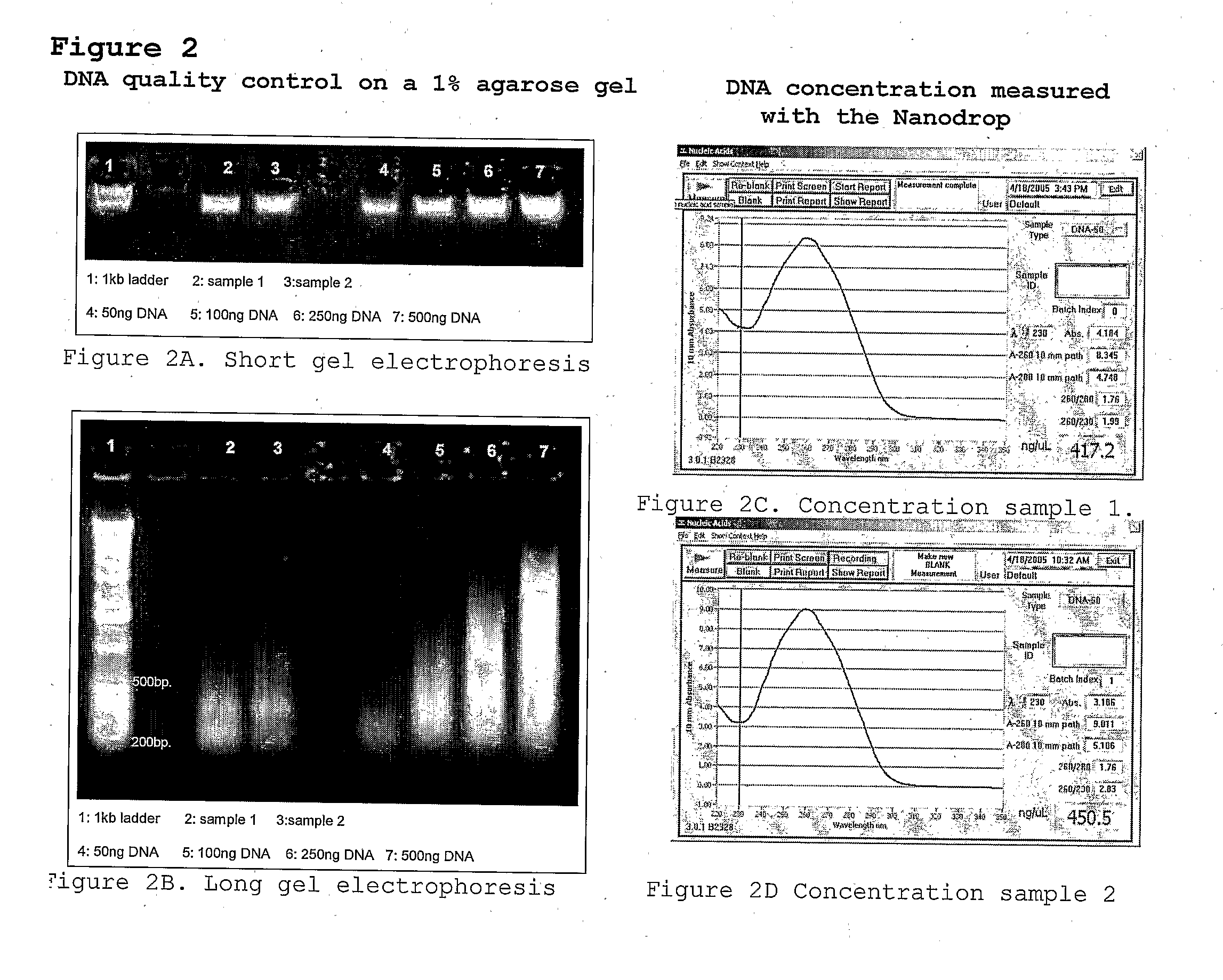
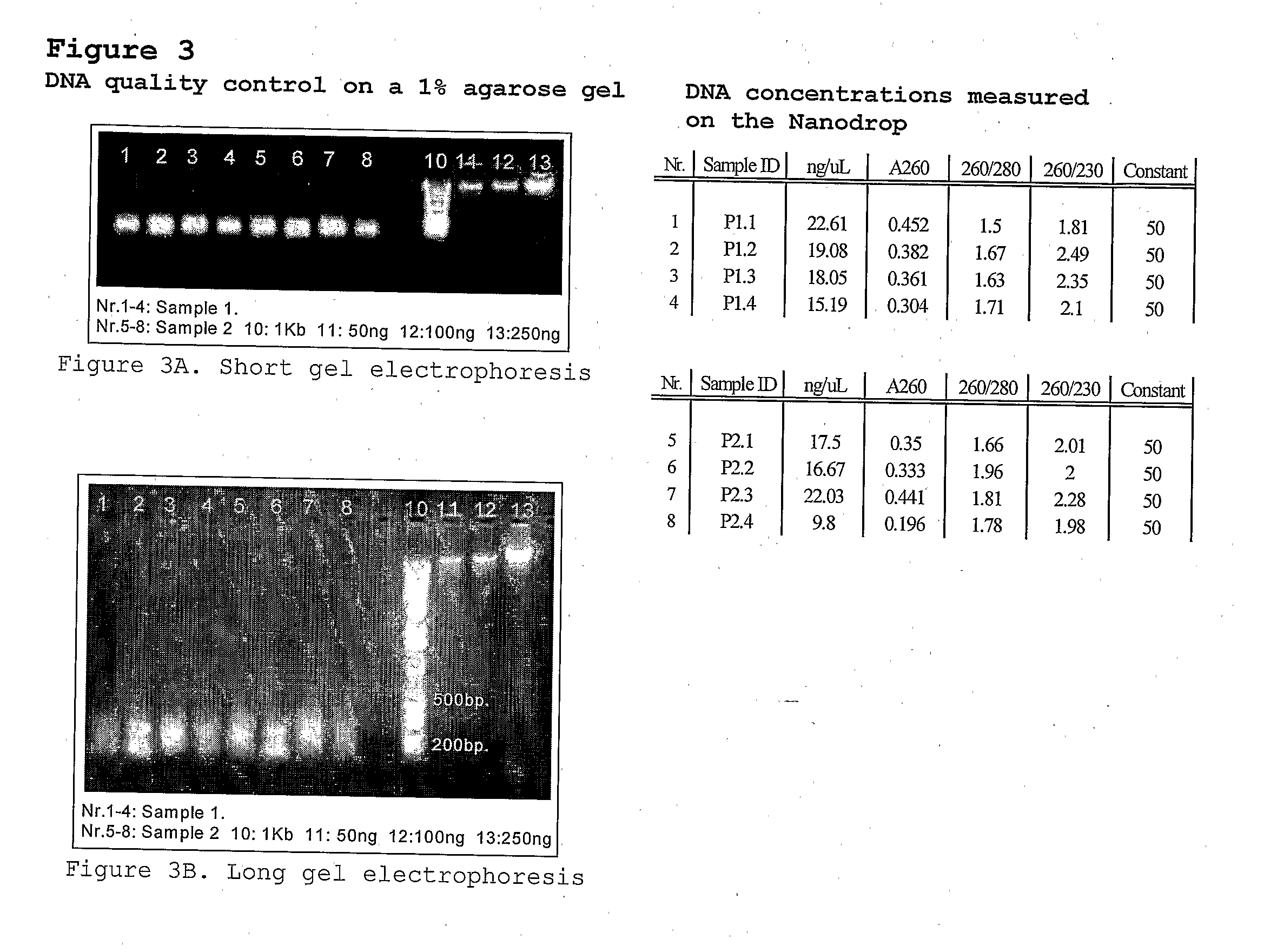
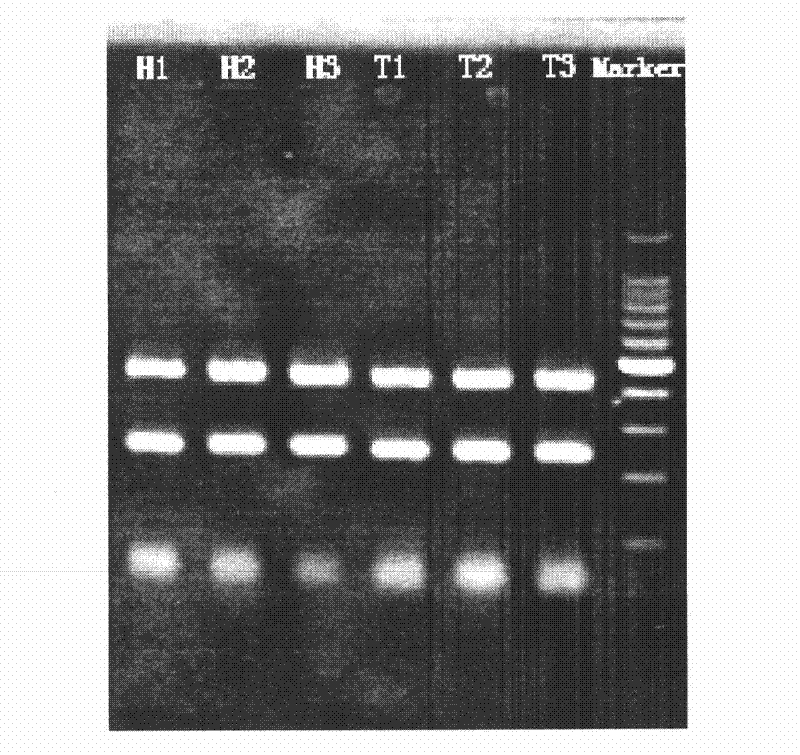
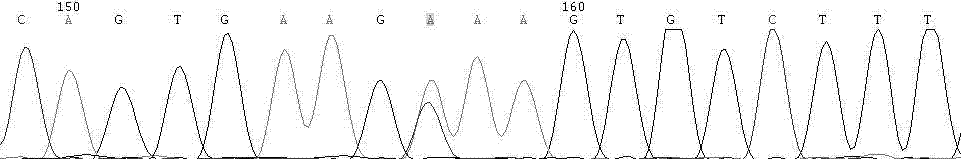
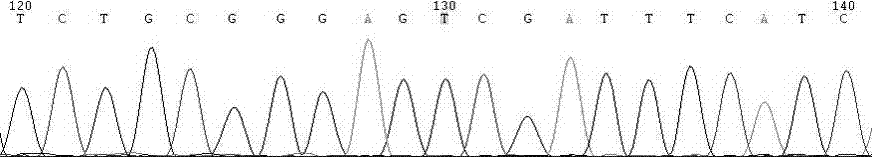
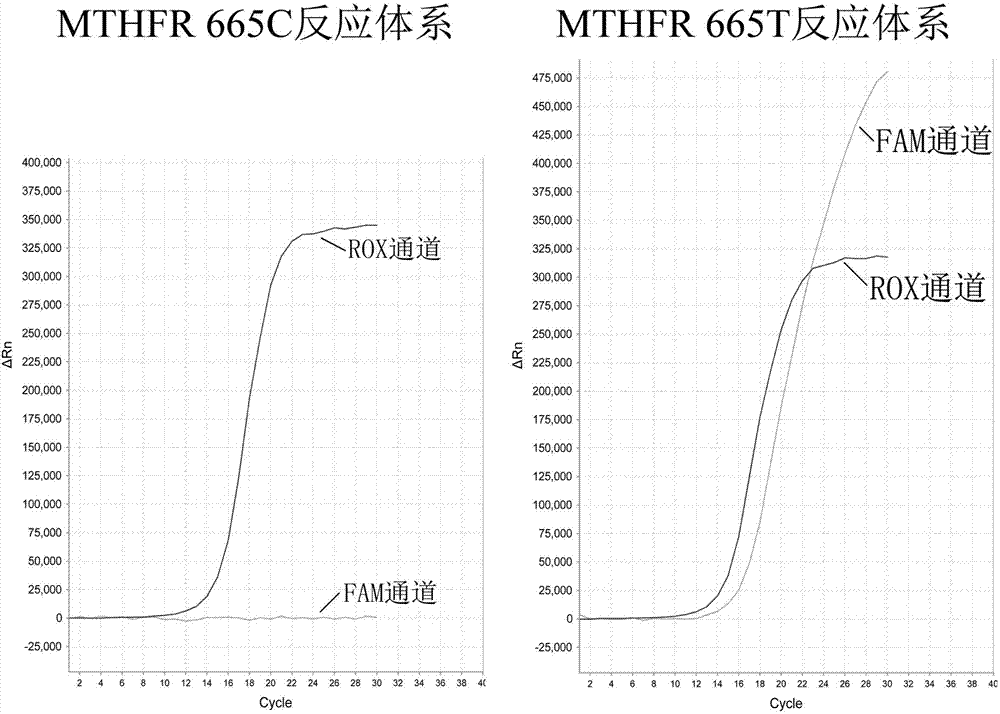
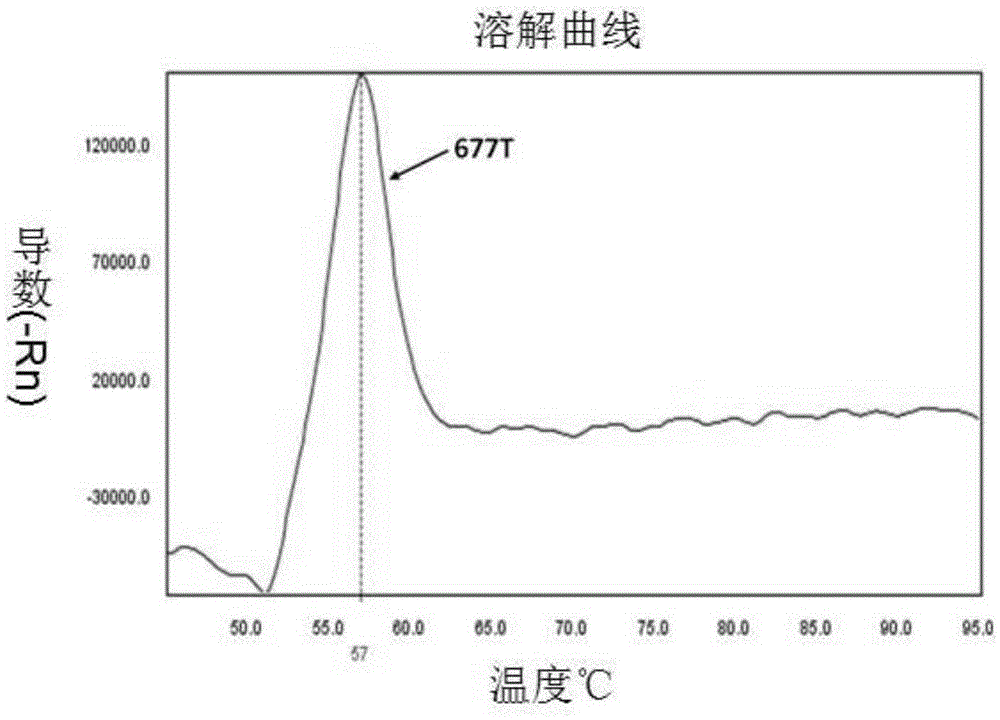
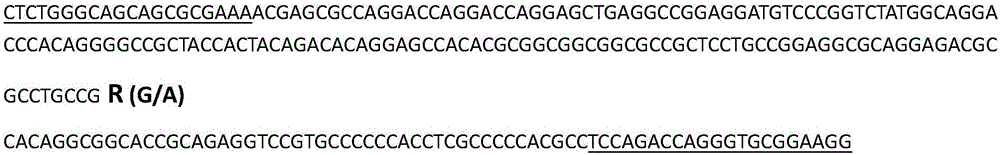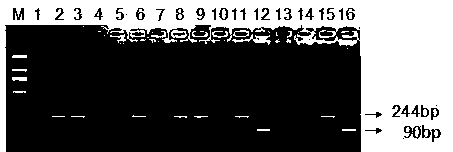Patents
Literature
507 results about "Polymorphism Detection" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Assay systems for genetic analysis
InactiveUS20130040375A1Improve throughputBioreactor/fermenter combinationsBiological substance pretreatmentsAssayPolymorphism Detection
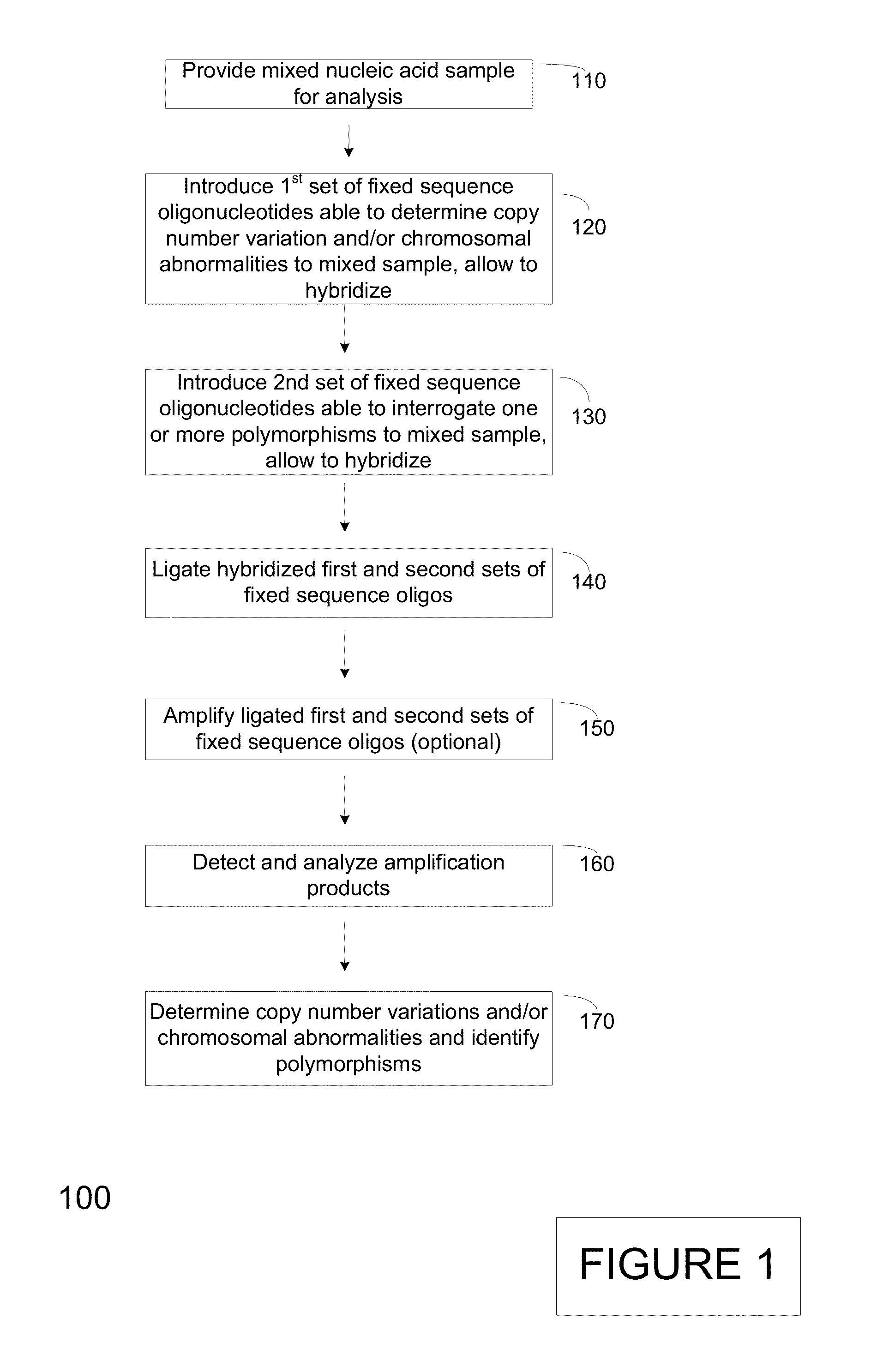
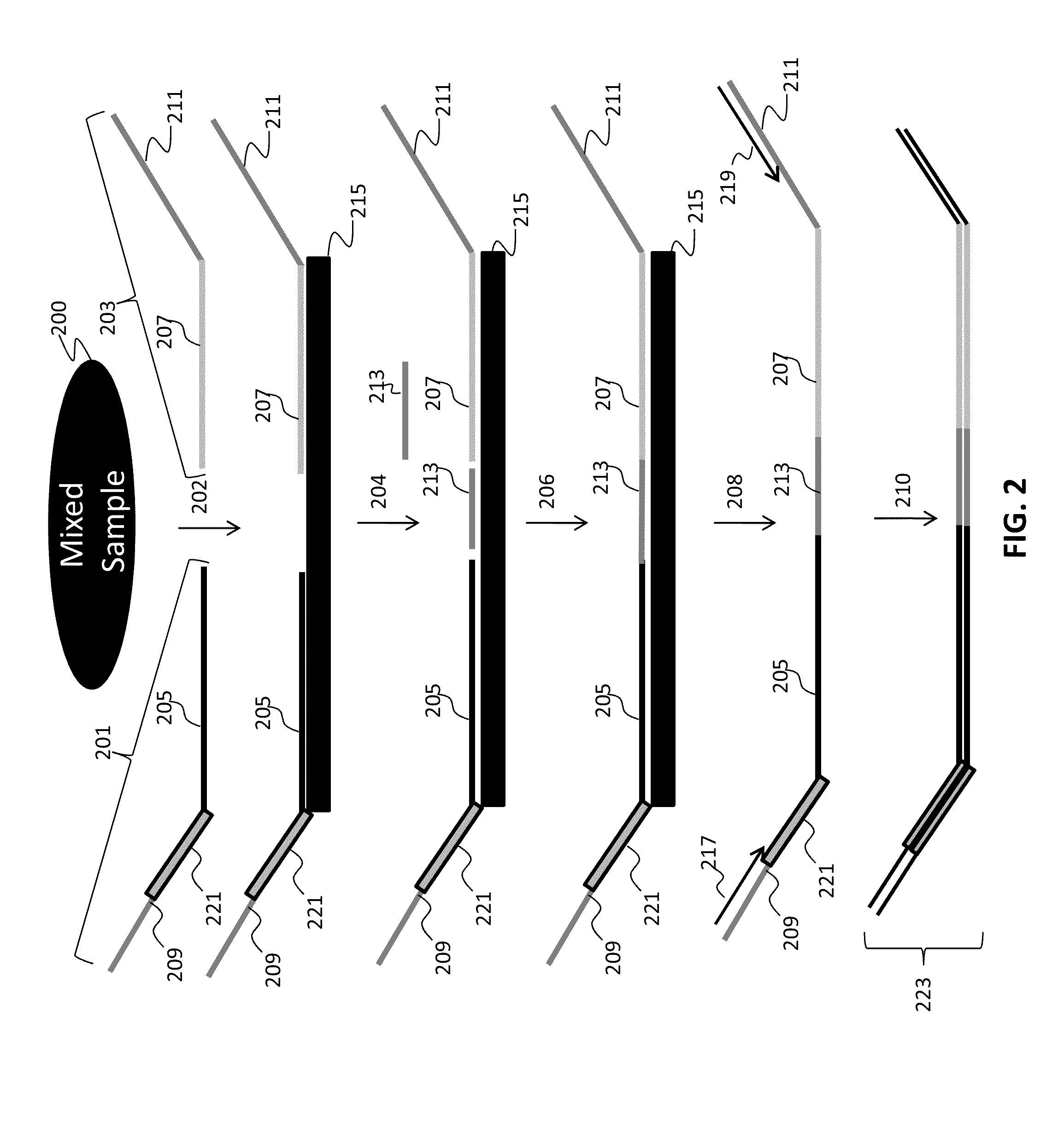
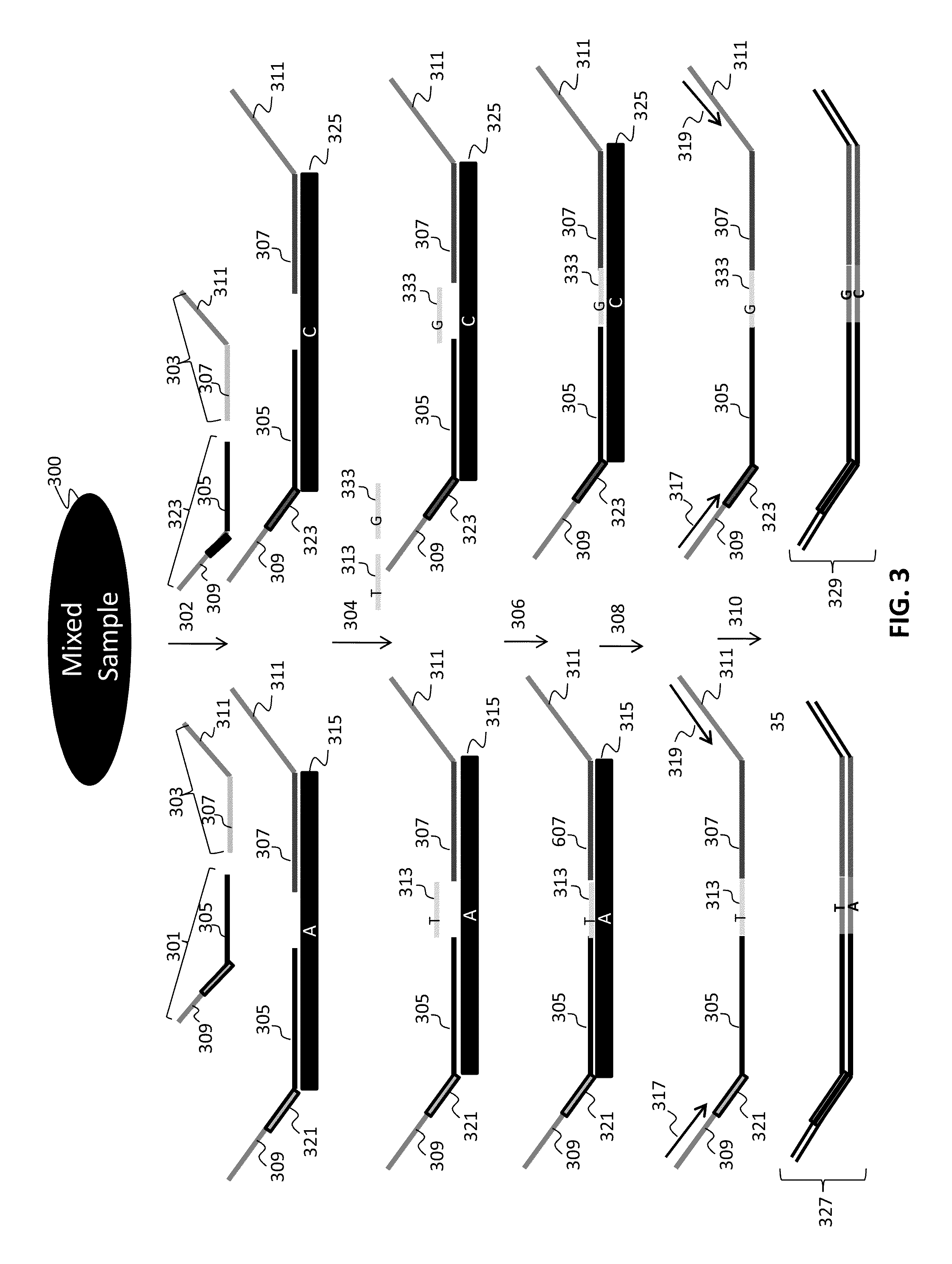
The present invention provides assay systems and methods for detection of copy number variation at one or more loci and polymorphism detection at one or more loci in a mixed sample from an individual.
Owner:TANDEM DIAGNOSTICS
Amplification based polymorphism detection
InactiveUS7250252B2Improve automationEasy to mergeSugar derivativesMicrobiological testing/measurementImproved methodPolymorphism Detection
An improved method of amplifying nucleic acids comprising the use of four discrete temperature steps in a thermocyclic amplification reaction, as well as, a method of detecting large nucleic acid insertions or deletions such as those that occur from gene duplication or deletion.
Owner:ABBOTT LAB INC
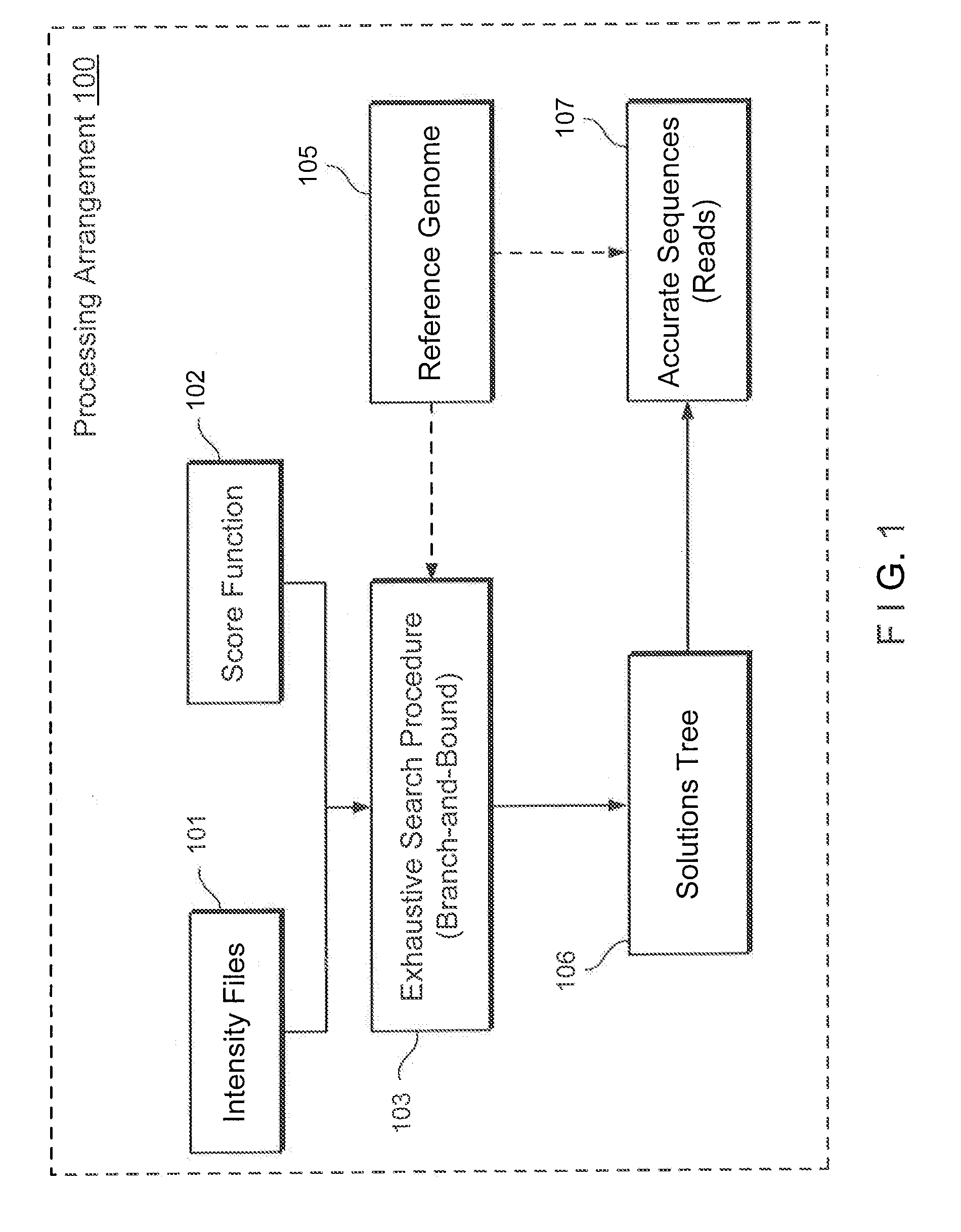
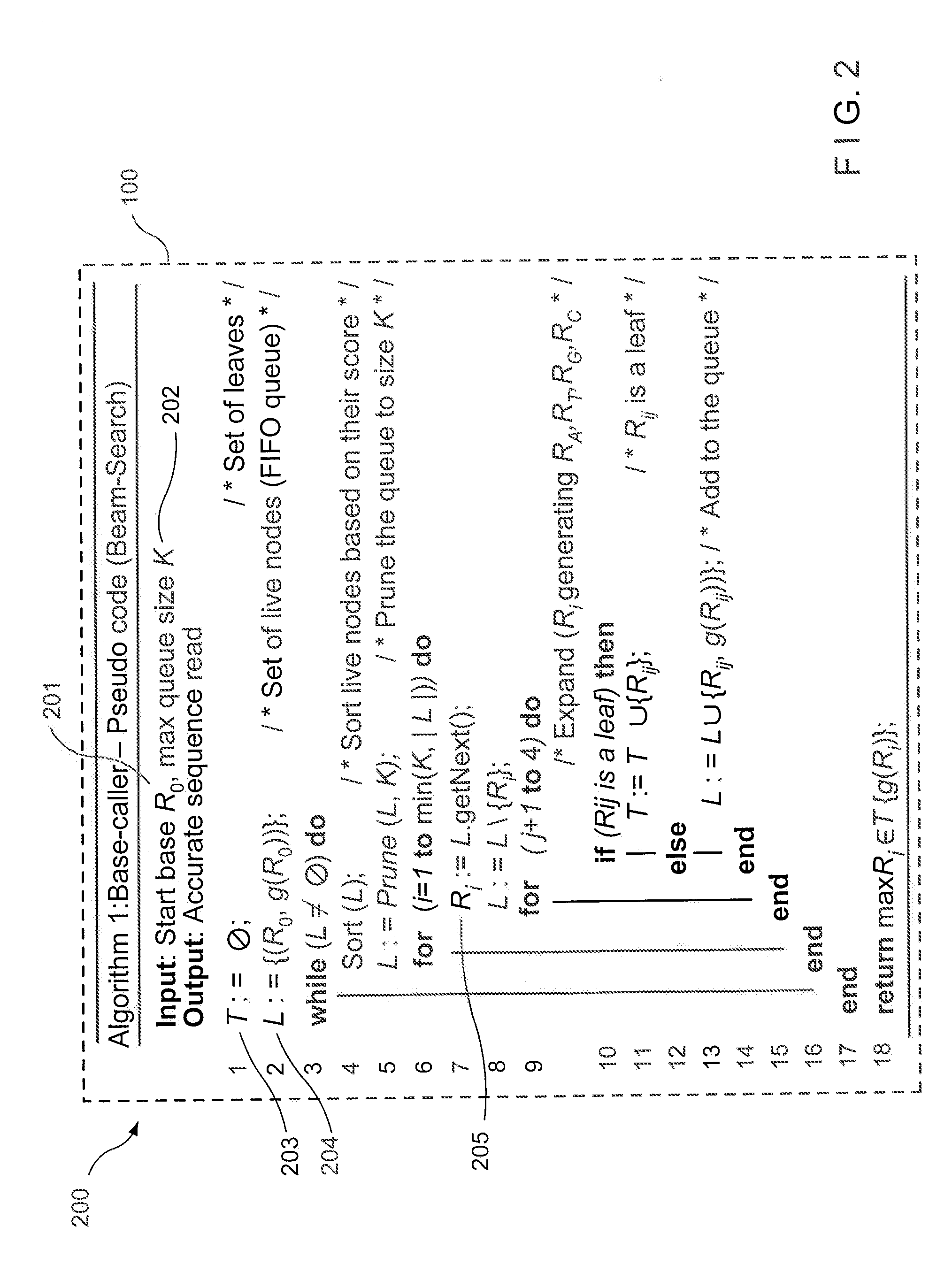
Method, computer-accessible medium and system for base-calling and alignment
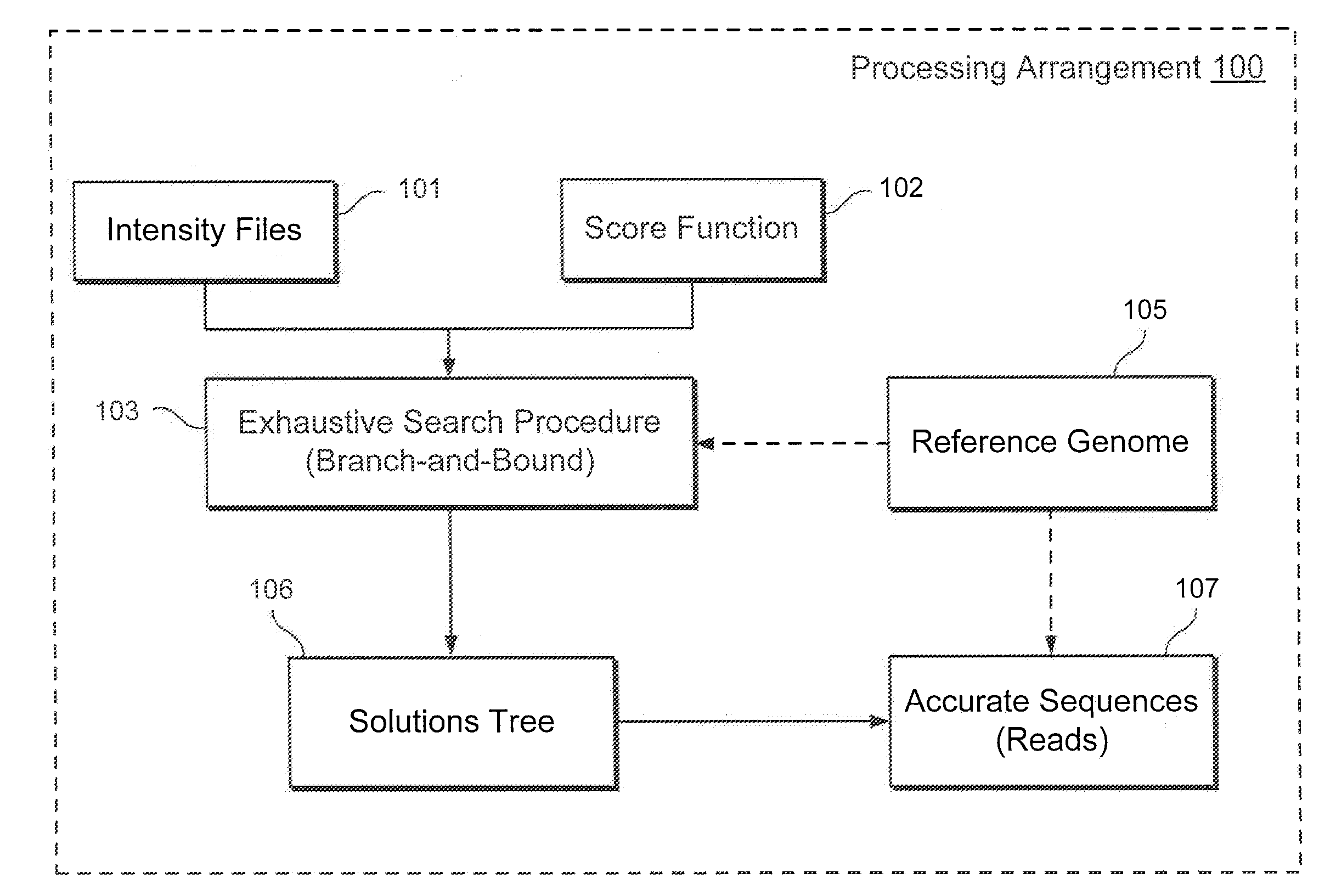
Exemplary methods, procedures, computer-accessible medium, and systems for base-calling, aligning and polymorphism detection and analysis using raw output from a sequencing platform can be provided. A set of raw outputs can be used to detect polymorphisms in an individual by obtaining a plurality of sequence read data from one or more technologies (e.g., using sequencing-by-synthesis, sequencing-by-ligation, sequencing-by-hybridization, Sanger sequencing, etc.). For example, provided herein are exemplary methods, procedures, computer-accessible medium and systems, which can include and / or be configured for obtaining raw output from a sequencing platform configured to be used for reading fragment(s) of genomes, obtaining reference sequences for the genomes obtained independently from the raw output, and generating a base-call interpretation and / or alignment using the raw output and the reference sequences. For example, a score function can be determined based on information associated with the sequencing platform that can be used to analyze polymorphisms based on the base-call interpretation and / or alignment.
Owner:NEW YORK UNIV
Novel method of detecting genetic polymorphism
InactiveUS20080286783A1Easy to detectRealized gainSugar derivativesMicrobiological testing/measurementPresent methodTyping
The present invention provides a novel polymorphism detecting method suitable for the detection and identification of copy number variation.Provided is a method of determining the genotype of a subject in a genomic region comprising an SNP site, comprising a step for performing typing of the SNP site by the invader assay with a DNA-containing sample comprising the genomic region from the subject as the template, wherein fluorescence is measured on a real time basis. The copy number ratio of both alleles is determined using the fluorescence intensity ratio of each allele at a time before saturation of fluorescence intensity. Preferably, the present method further comprises a step for amplifying the genomic region comprising an SNP site prior to the invader step. In this step of amplification, a plurality of regions comprising a plurality of SNP sites can be simultaneously amplified. Furthermore, the present method enables the determination of the copy number of each allele when combined with quantitative PCR.
Owner:RIKEN
Method for simultaneously detecting gene mutations of acetaldehyde dehydrogenase 2 and alcohol dehydrogenase 2
InactiveCN102758008ASimultaneous detection of genetic polymorphismsNo risk of contaminationMicrobiological testing/measurementFluorescence/phosphorescenceFluorescencePolymorphism Detection
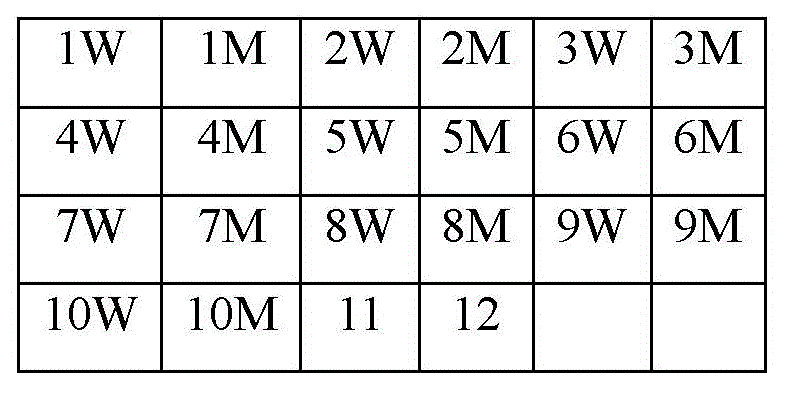
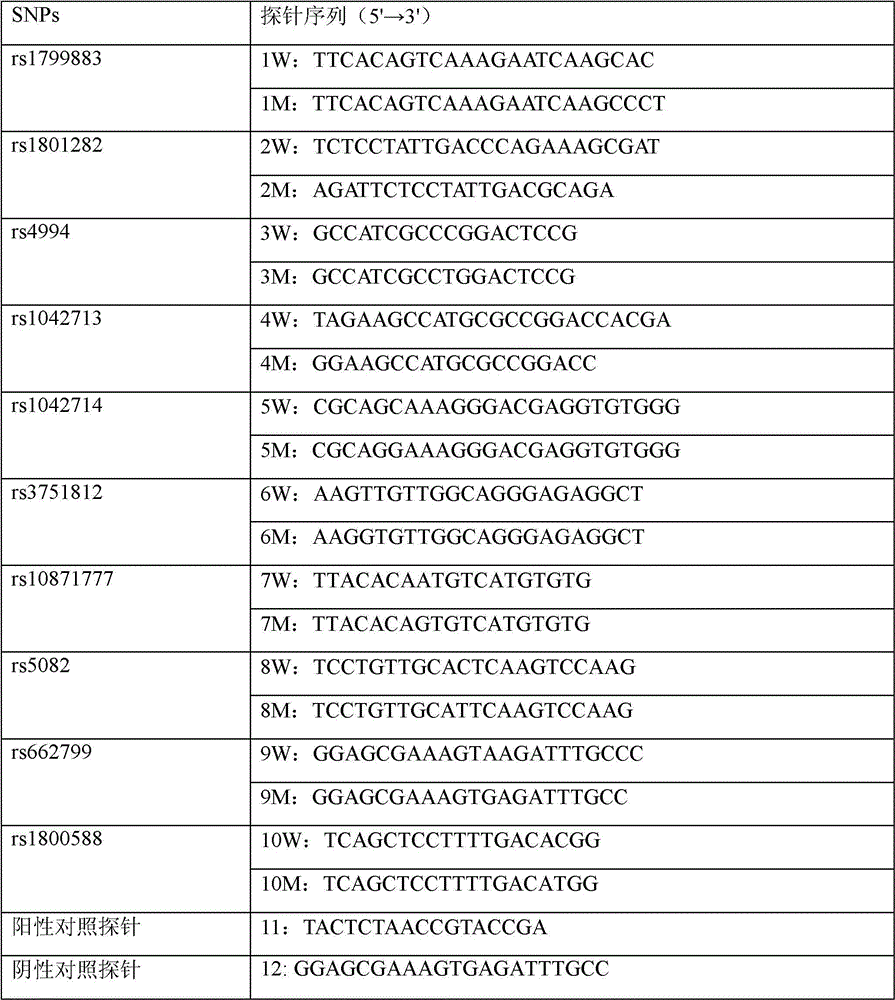
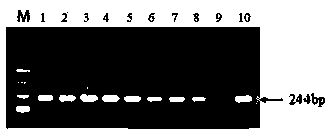
The invention relates to a method for simultaneously detecting gene mutations of acetaldehyde dehydrogenase 2 and alcohol dehydrogenase 2. Concretely, a probe effective for detection of genetic polymorphism of the ALDH2 gene rs671 and the ADH2 gene rs1229984, a method for simultaneously detecting gene mutations and a kit for the purpose are provided. Thus the invention provides a probe for polymorphism detection, namely a probe for detection of at at least one type of genetic polymorphism of the ALDH2 gene rs671 and the ADH2 gene rs1229984, characterized in that: the probe comprises at least 1 type of fluorescently labeled oligonucleotide selected from (P1) to (P3') defined in the description.
Owner:ARKRAY INC
Polymerase chain reaction-sequence based typing (PCR-SBT) method for ABO blood type genotyping and reagent
ActiveCN101921834ASolve the recombination phenomenonThe result is accurateMicrobiological testing/measurementSodium acetateHuman DNA sequencing
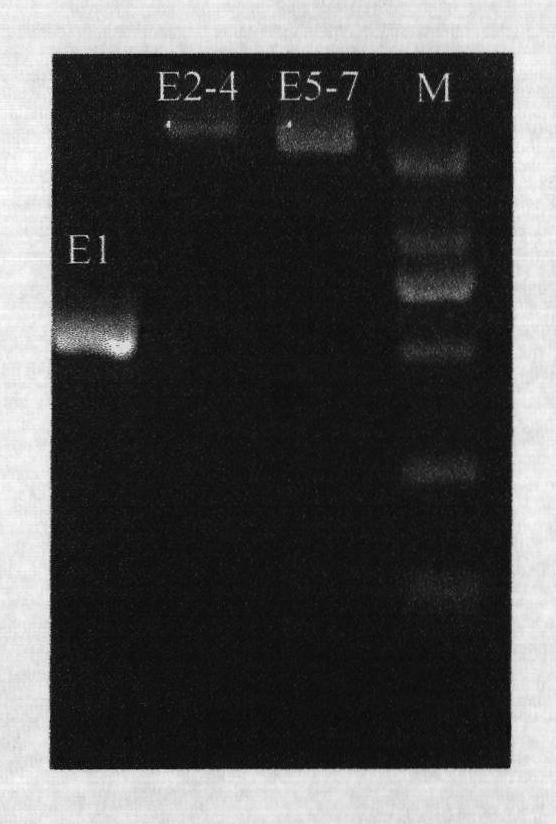
The invention provides a polymerase chain reaction-sequence based typing (PCR-SBT) method for ABO blood type genotyping. The method comprises the following steps of: preparing human genome DNA; amplifying segments of ABO gene exon 1, exons 2-4 and exons 5-7; performing double enzyme digestion purification on the obtained amplified products; performing a sequencing PCR reaction on the purified products; purifying the sequenced products by a sodium acetate-ethanol precipitation method and performing capillary electrophoresis sequencing; and analyzing the obtained sequences by using software to determine the genotype. The method has the advantages of solving the problems of identification of an ABO subtype, judgment of difficult blood types, discovery of a new mutational site, gene recombination among genes, genetic polymorphism detection and the like, exerting the characteristics of high flux and result accuracy of ABO genotyping operation by PCR-SBT, achieving great importance for the relative application in the fields of clinical transfusion medicinal research, genetics and the like and having important practical significance for medicinal research units, pharmic research and reagent development units.
Owner:浙江省血液中心
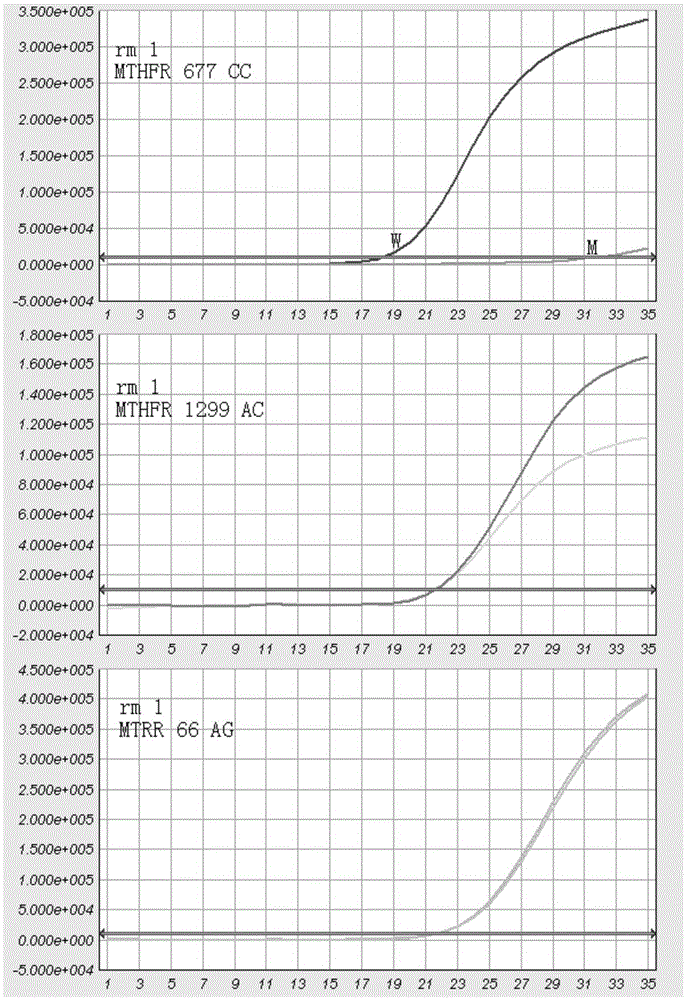
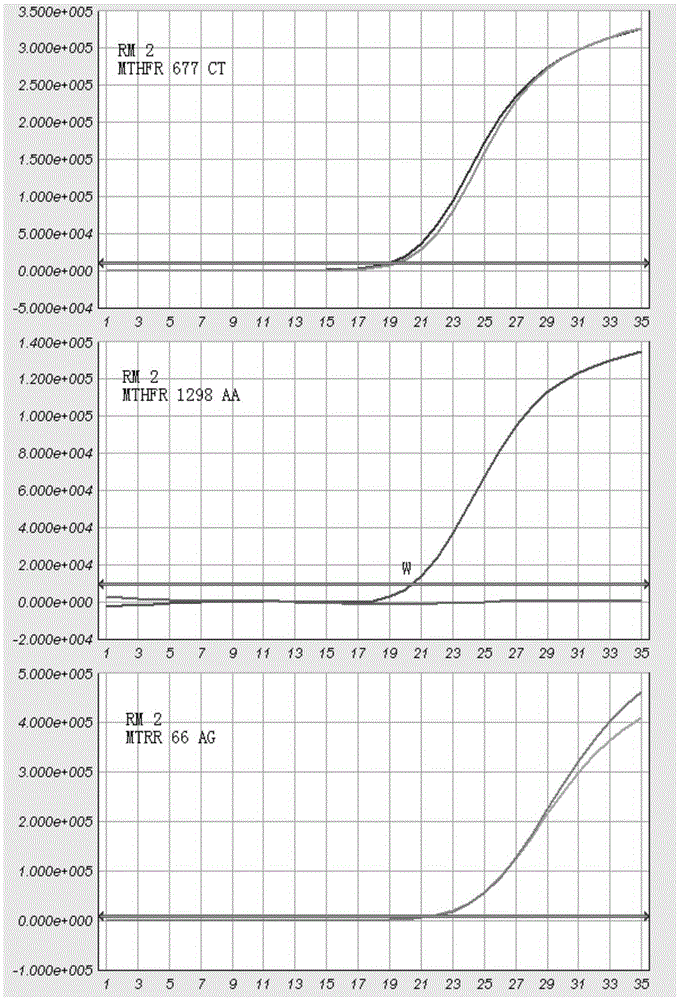
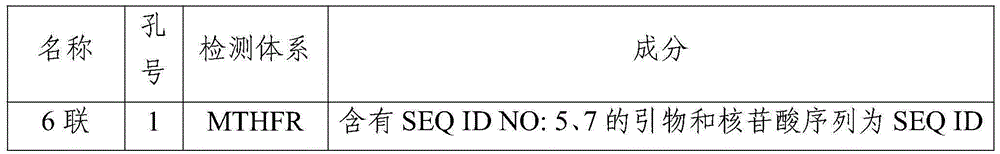
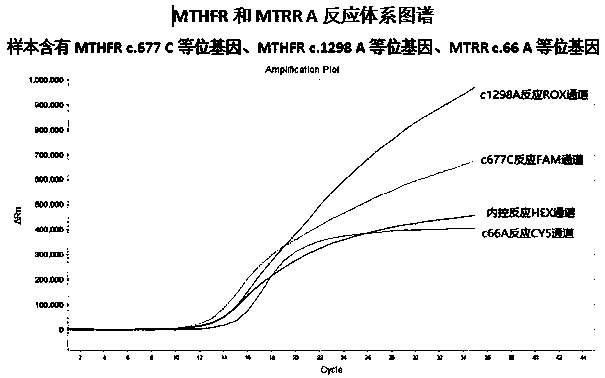
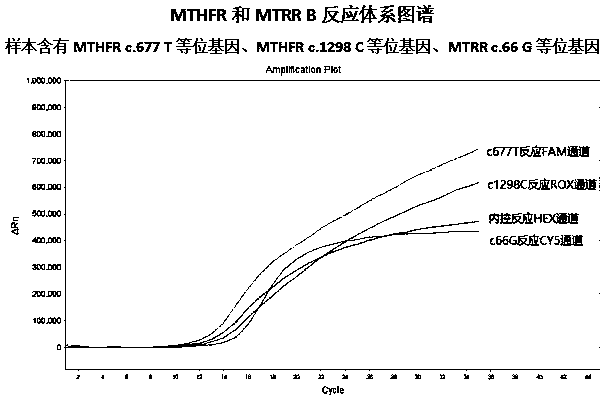
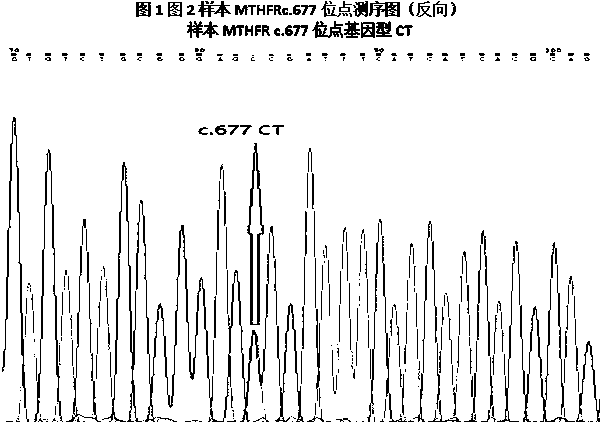
MTHFR and MTRR gene polymorphism detection primer group and kit
ActiveCN105256019AAccurate detectionHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationPolymorphism DetectionToxicity
The invention discloses an MTHFR and MTRR gene polymorphism detection primer group and a kit. Mutation primers and probes aiming at three genetic loci have the advantages of being high in specificity and sensitivity. The prepared kit can detect MTHFR677, MTHFR1298 and MTRR66 gene polymorphism conditions, operation is simple, the experimental period is short, and the primer group and the kit are safe, free of toxicity, low in cost and suitable for clinical large-scale use and popularization.
Owner:武汉海吉力生物科技有限公司
Molecule marking method for pig backfat thickness property
InactiveCN103497994ASolve the problems of high blindness and low selection accuracyShorten the generation intervalMicrobiological testing/measurementBOARAcyl coenzyme
The invention relates to a molecule marking method for pig backfat thickness property. The method comprises: extraction of pig genome DNA, design of a long-chain acyl-coenzyme Asynthetase 1 (ACSL1) gene seventh exon primer, amplification in vitro and genotype detection. For the first time, it is discovered that the polymorphism of the long-chain acyl-coenzyme Asynthetase 1 (ACSL1) gene seventeenth exon has significant correlation with the pig backfat thickness, and a method for detecting the restriction fragment polymorphism at mutation sites of the seventeenth exon is established. The molecule marking method is applicable to auxiliary selection on backfat thickness property during breeding of pigs, helps to realize early-stage breeding selection of boars, and even helps to accurately select breeding just when pigs are born, so that the generation interval is shortened, and the selection progress of the backfat thickness property is accelerated; and the method is simple in operation, condition requirements during a polymerase chain reaction are low, the length of amplified fragments is relatively short (292 bp), amplification is relatively easy, and the method helps to improve amplification efficiency and accuracy of genotype determining.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Human SLCO1B1 and ApoE (apolipoprotein E) gene polymorphism detection kit
InactiveCN104846085AEasy to detectAccurate Typing DetectionMicrobiological testing/measurementApolipoprotein e4SLCO1B1
The invention provides a human SLCO1B1 and ApoE (apolipoprotein E) gene polymorphism detection kit, comprising a PCR (polymerase chain reaction) buffer solution, dNTP (deoxy-ribonucleoside triphosphate), MgCl2, four groups of specific primers, four groups of specific probes, an internal standard system, HotStart Taq enzyme and UNG (uracil-N-glycosylase) enzyme. The detection kit has the advantages of high specificity, high sensitivity, ease and quickness of operation, high throughput, safety, objectiveness of result interpretation, and the like.
Owner:WUHAN YZY MEDICAL SCI & TECH
Sheep lambing number trait related molecular marker and application thereof
ActiveCN104099330AMicrobiological testing/measurementDNA/RNA fragmentationMarker-assisted selectionNucleotide
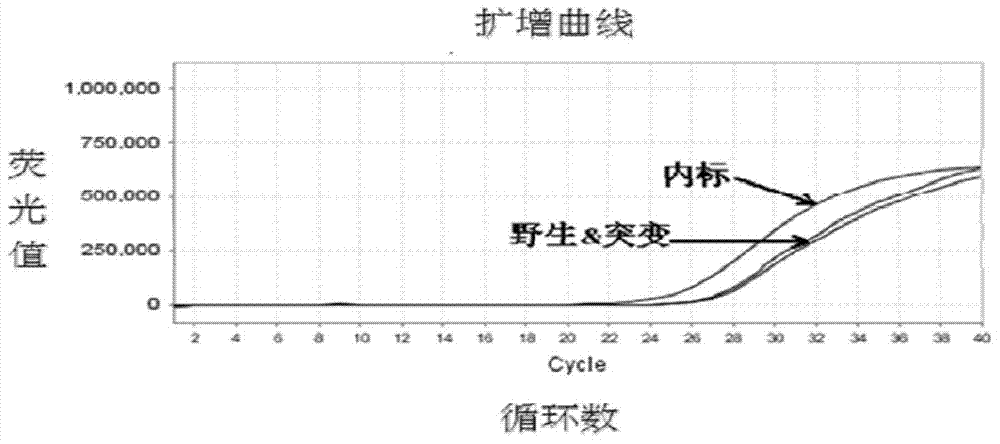
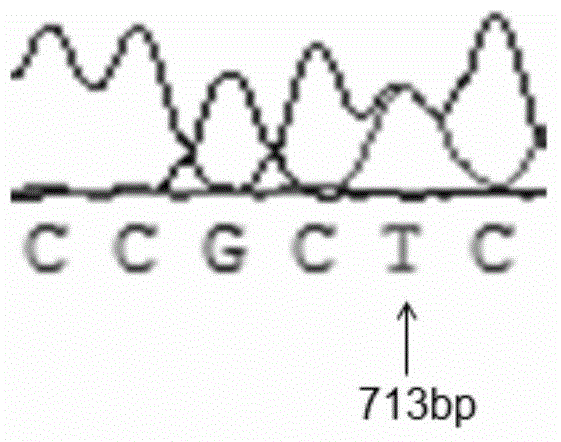
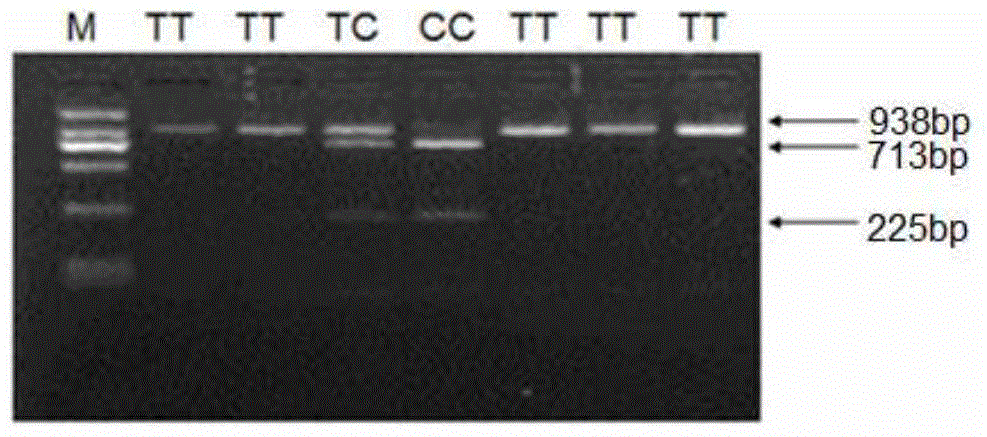
The invention belongs to the technical field of preparation of molecular markers for livestock, and particularly relates to a preparation method of a sheep lambing number trait related molecular marker serving as MAS (marker-assisted selection) as well as an application of the molecular marker. The molecular marker is obtained through LH (luteinizing hormone) beta gene cloning, and the nucleotide sequence is represented in SEQ ID NO.1. One C713-T713 base is substituted at 713 bp in the SEQ ID NO.1, so that BSrB I-RFLP (restricted fragment length polymorphisms) is caused. The invention further discloses a primer used in a DNA (deoxyribonucleic acid) sequence of an amplified LH beta gene part and a polymorphism detection method, and provides one novel molecular marker for MAS of the sheep lambing number trait.
Owner:甘肃润牧生物工程有限责任公司
Specific sequence specific primers-polymerase chain reaction primers and kit for human MTHFR and MTRR gene polymorphism detection
InactiveCN105002275AAccurate Typing DetectionStable Typing DetectionMicrobiological testing/measurementDNA/RNA fragmentationInternal standardPolymorphism Detection
The present invention relates to the fields of biotechnology and medicine, and provides specific sequence specific primers-polymerase chain reaction primer sequences and a kit for simultaneously detecting human MTHFR gene polymorphism and human MTRR gene polymorphism, wherein the kit contains the specific primers, specific probes, an internal standard system, Taq enzyme and UNG enzyme. According to the present invention, the primers and the kit have advantages of strong specificity, high sensitivity, simple and rapid operation, high-throughput, safety, objective result interpretation, and the like.
Owner:WUHAN YZY MEDICAL SCI & TECH
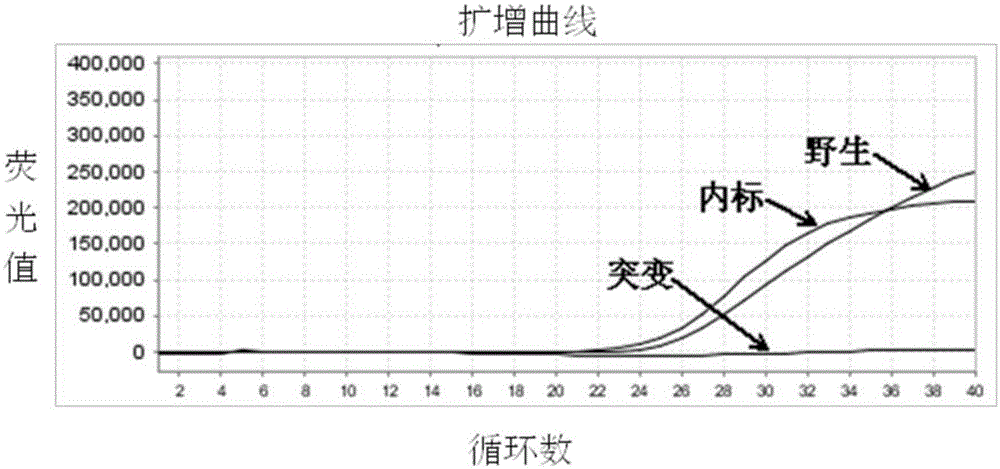
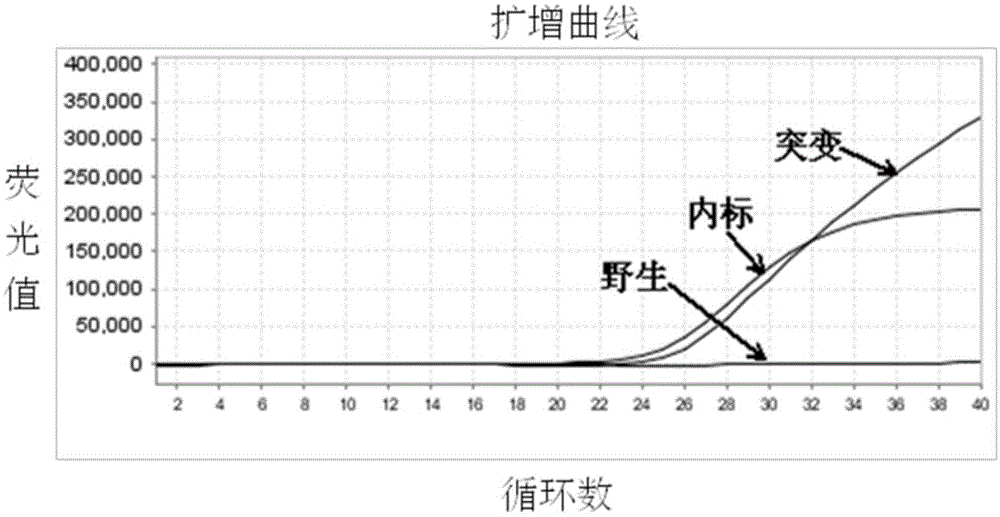
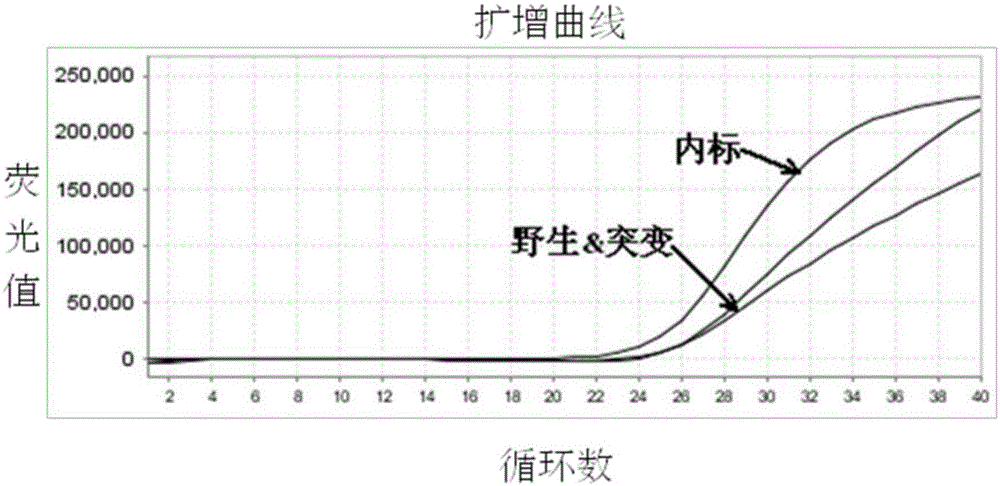
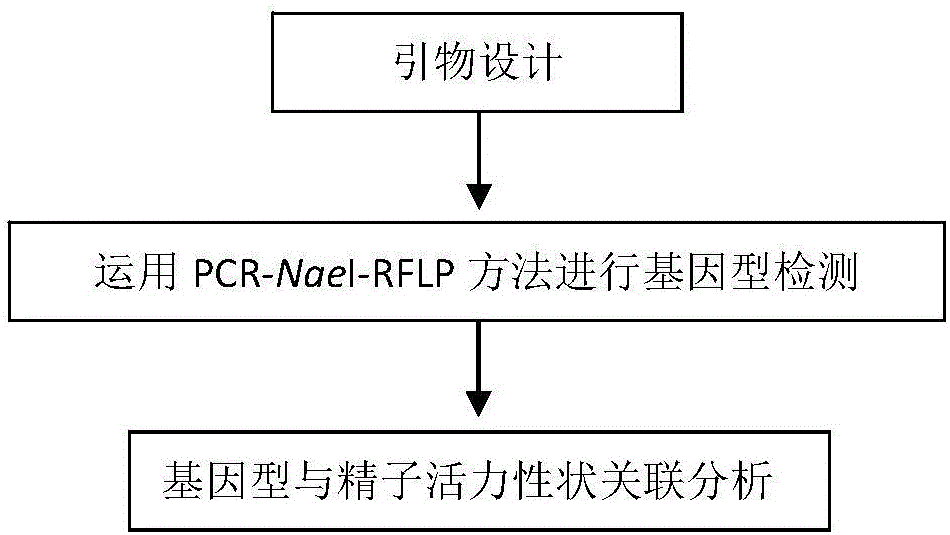
Molecular marker related to sperm activity character of boar and application
The invention belongs to the technical field of livestock molecular marker preparation, and concretely relates to a molecular marker related to a sperm activity character of a boar and application. The molecular marker is screened from a PRM2 gene; a nucleotide sequence of the molecular marker is as shown in SEQ ID NO: 1. An allelic mutation R exists in nucleotide in position 169 of a sequence table, R is G or A, and the mutation causes the polymorphism of NaeI-RFLP. The invention further discloses a primer pair for amplifying PRM2 gene SNP (Single Nucleotide Polymorphism), and the primer pair is also the primer pair for detecting the screened molecular marker provided by the invention. The invention builds a method for detecting the polymorphisms of the sperm activity character of the boar. Association analytic application shows that the molecular marker provided by the invention can be used as the marker for assistant selection of the sperm activity character of the boar.
Owner:HUAZHONG AGRI UNIV
Molecular marking method for pig litter size
InactiveCN101768642AShorten the generation intervalSpeed up the selection processMicrobiological testing/measurementEnzyme digestionAnimal science
The present invention relates to a molecular marking method for pig litter size. The method comprises the steps of pig genome DNA extraction, exon 6 primer design of sex hormone binding globulin (SHBG) genes, amplification in vitro and genotype detection. The present invention first discovers pig SHBG genes, discovers the obvious relationship between the polymorphism of exon 6 and pig litter size, and develops a restriction fragment enzyme digestion polymorphism detection method for detecting exon 6 mutational sites. The method can be used for the auxiliary selection of litter size during pig breeding, can realize the early selection of parent pigs and even can accurately select parent pigs when the pigs are born. Generation interval is largely shortened, and selection progress is accelerated. The method has the advantages of simple operation, low condition requirements in polymerase chain reaction processes, short amplification fragment length (217bp), easy amplification, high amplification efficiency and accurate genotype judgement.
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY MEDICINE ANHUI ACAD OF AGRI SCI
Method for detecting polymorphism of flora of prawn culture water body
InactiveCN101724690AEffective qualitativeEffective quantitative analysisMicrobiological testing/measurementFluorescence/phosphorescencePrawnFluorescence
The invention discloses a method for detecting polymorphism of a flora of a prawn culture water body. The method comprises the following steps: (1) extracting total genomic DNA of mixed microbes in a prawn culture water body sample; (2) designing a specificity T-RFLP-PCR universal primer and performing PCR circulating reaction; (3) purifying a product and performing enzyme cutting on DNA by a specific restriction enzyme Hae III; (4) performing ionophortic separation on DNA segments by 1 percent agarose gel, performing fluorescent scanning on the DNA segments; (5) analyzing a polymorphism structure of a microbe flora; and (6) performing quantitative detection on predominant bacteria of the flora through fluorescence in situ hybridization technology. The method improves detection technology combining T-RFLP-PCR with FISH, uses fluorescence to mark a specific primer; and compared with the modern technology, the method has the characteristics of high repeatability, sensitivity, rapidness, accuracy, stability and the like, and can qualitatively and quantitatively analyze the ecological diversity of the microbes in the prawn culture water body along with time change and the dynamic change of a composite structure of the predominant flora.
Owner:SOUTH CHINA UNIV OF TECH
Human HLA-B*5801 gene polymorphism detection kit
InactiveCN106929591AAccurate detectionHigh sensitivityMicrobiological testing/measurementInternal standardHLA-B
The invention relates to a human HLA-B*5801 gene polymorphism detection kit. The human HLA-B*5801 gene polymorphism detection kit comprises PCR damping liquid, a specific primer, a specific probe, an interior label system, a Taq enzyme, a UNG enzyme, a weakly-positive control group and a blank control group, and also comprises a blood treating agent; a blood sample is simply treated and can be directly subjected to PCR amplification, the DNA extracting process is omitted, and operating time is saved. According to the human HLA-B*5801 gene polymorphism detection kit, the SNP probe is used in cooperation with the technology of the ARMS primer, and it is achieved that two different gene types are detected in one pipe; meanwhile, the interior label system is designed and used for monitoring the quality of the sample, and the weakly-positive control group and the blank control group are designed and used for monitoring the quality of the kit. The human HLA-B*5801 gene polymorphism detection kit for detecting HLA-B*5801 alleles has the advantages of being high in specificity and sensitivity, rapid and easy to operate, safe, objective in result interpretation and the like when being used for detecting HLA-B*5801 alleles.
Owner:WUHAN YZY MEDICAL SCI & TECH
Human CYP2C19 gene polymorphism detection specific primer and kit
InactiveCN104988149AAccurate Typing DetectionStable Typing DetectionMicrobiological testing/measurementDNA/RNA fragmentationHigh fluxNucleotide
The invention belongs to the field of biotechnologies and medicine, and provides a specific primer used for human CYP2C19 gene polymorphism detection. The nucleotide sequences of the primer are shown in SEQ ID NO:1-9. The invention further provides a kit for human CYP2C19 gene polymorphism detection. The primer and the kit are used for human CYP2C19 gene polymorphism detection and have the advantages of being high in specificity and sensitivity, easy and fast to operate and safe and having high flux, and result interpretation is objective.
Owner:WUHAN YZY MEDICAL SCI & TECH
Sudden cardiac death mutant gene detection kit
ActiveCN104561310AChange bad habitsAchieve the purpose of preventionMicrobiological testing/measurementHigh risk populationsCvd risk
The invention relates to the field of molecular biology and medical science, and in particularly relates to a sudden cardiac death mutant gene detection kit which is high in accuracy and good in predictability. The kit is used for performing polymorphism detection on 12 SNP sites of 8 major genes related to sudden cardiac death; three forward and reverse specific primers are respectively adopted for each SNP site by combining the characteristics of specific allelic gene PCR (polymerase chain reaction) and temperature gradient descent PCR; and the mutation of the 12 SNP sites can be simultaneously determined by virtue of program amplification of the temperature gradient descent PCR and agarose gel electrophoretic analysis. The detection kit provided by the invention is strong in specificity, high in detection rate, high in efficiency and low in cost, and can be used for screening high-risk population of sudden cardiac death, evaluating the risk degree of having the sudden cardiac death for a detected patient, and making clear pathogenic factors of patient sudden death from a gene level, thereby providing a new way of preventing, diagnosing and treating clinical sudden death.
Owner:谢怡
Method for high-throughput aflp-based polymorphism detection
ActiveUS20090269749A1Efficient screeningImprove signal-to-noise ratioSugar derivativesMicrobiological testing/measurementGenotypingPolymorphism Detection
The invention relates to a method for the high throughput discovery, detection and genotyping of one or more genetic markers in one or more samples, comprising the steps of restriction endonuclease digest of DNA, adaptor-ligation, optional pre-amplification, selective amplification, pooling of the amplified products, sequencing the libraries with sufficient redundancy, clustering followed by identification of the genetic markers within the library and / or between libraries and determination of (co-) dominant genotypes of the genetic markers.
Owner:KEYGENE NV
Kit for detecting genotyping and IL28-site polymorphism of hepatitis C virus (HCV)
The invention relates to a kit for detecting the genotype and IL28-site polymorphism of a hepatitis C virus (HCV), in particular to a kit for detecting the genotype and IL28-site polymorphism of an HCV, which is prepared by using a nucleic acid reverse dot hybridization technology. The kit comprises an amplification reagent, a low-density chip using a nylon membrane as a carrier and a hybridization reagent, and can simultaneously detect the genotype of the HCV and the IL28B gene polymorphism closely related to the HCV treatment, thereby providing a more reliable basis for the individual hepatitis C treatment.
Owner:DAAN GENE CO LTD
Genetic marker related to growth trait of goat and application thereof
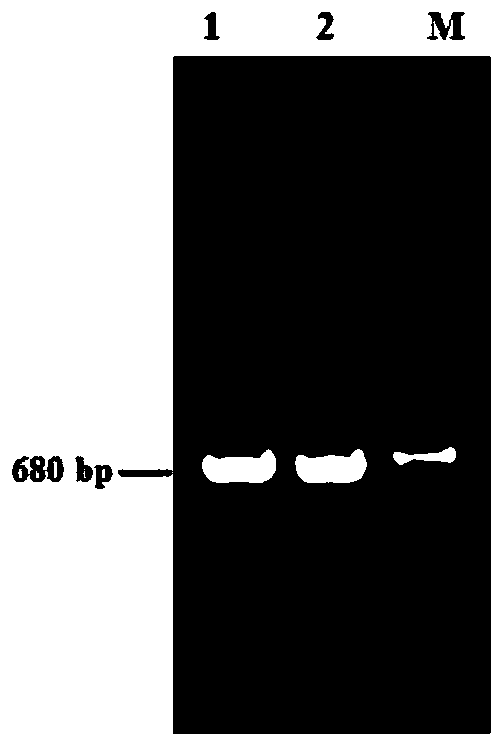
The invention belongs to the technical field of preparation of molecular markers and particularly relates to a genetic marker related to growth trait of a goat and application thereof. The genetic marker is a gene fragment which is cloned from an SKIP gene of the goat, shown in a sequence table SEQID NO:1 and related to the growth trait of the goat, wherein the sequence length is 680bp. An allele mutant (base substitution) is arranged at 413bp of the sequence as shown in the sequence table SEQID NO:1, and the mutant leads to Bg1I-RFLP polymorphism. The invention further discloses a preparation method of the genetic marker and application of the genetic marker in detecting polymorphism of the growth trait of the goat. The genetic marker provided by the invention can be applied to marker assisted selection of the goat.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
Kiwi fruit InDel molecular marker and screening method and application thereof
ActiveCN106498075APrimer variant stabilitySpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationResearch ObjectActinidia
The invention discloses a kiwi fruit InDel molecular marker and a screening method and application thereof. Two different cold-proof Actinidia arguta genome DNAs are used as research subjects, resequencing of four variety genes is carried out, InDel primers are designed according to resequencing data, and PCR (polymerase chain reaction) amplification, agarose electrophoresis and non-denaturing polyacrylamide gel electrophoresis are carried out; polymorphism detection is carried in Actinidia arguta to screen out 14 pairs of InDel primers, the 14 pairs of InDel primers are applicable to the genetic diversity analysis for Actinidia chinensis, Actinidia deliciosa and the like and are also applicable to the researches, such as hybrid offspring authenticity identification, genetic map construction, and molecule-assisted breeding. The kiwi fruit InDel primers according to the embodiment of the invention has good mutation stability and low detection difficulty, allow InDel insertion / loss of large fragments, and allows agarose analysis, with steps that may be simplified.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Primer, probe, fluorescent PCR kit and method for detecting polymorphism of human MTHFR (Methylene Tetrahydrofolate Reductase) gene
ActiveCN102808026AEasy to readHigh specificity of PCR detectionMicrobiological testing/measurementFluorescence/phosphorescenceNucleotideFluorescent pcr
The invention provides a primer, probe, fluorescent PCR kit and method for detecting nucleotide polymorphisms of two sites of c.665 and c.1286 in an MTHFR (Methylene Tetrahydrofolate Reductase) gene. The nucleotide types of the two polymorphic sites of c.665 and c.1286 in the MTHFR gene are detected on a real-time fluorescent quantitative PCR (Polymerase Chain Reaction) technology platform according to the principle of 'allelic specific PCR'.
Owner:SUZHOU KUANGYUAN MOLECULAR BIOTECH
Primer pair, fluorescence probe and kit for detecting polymorphism of MTHFR gene
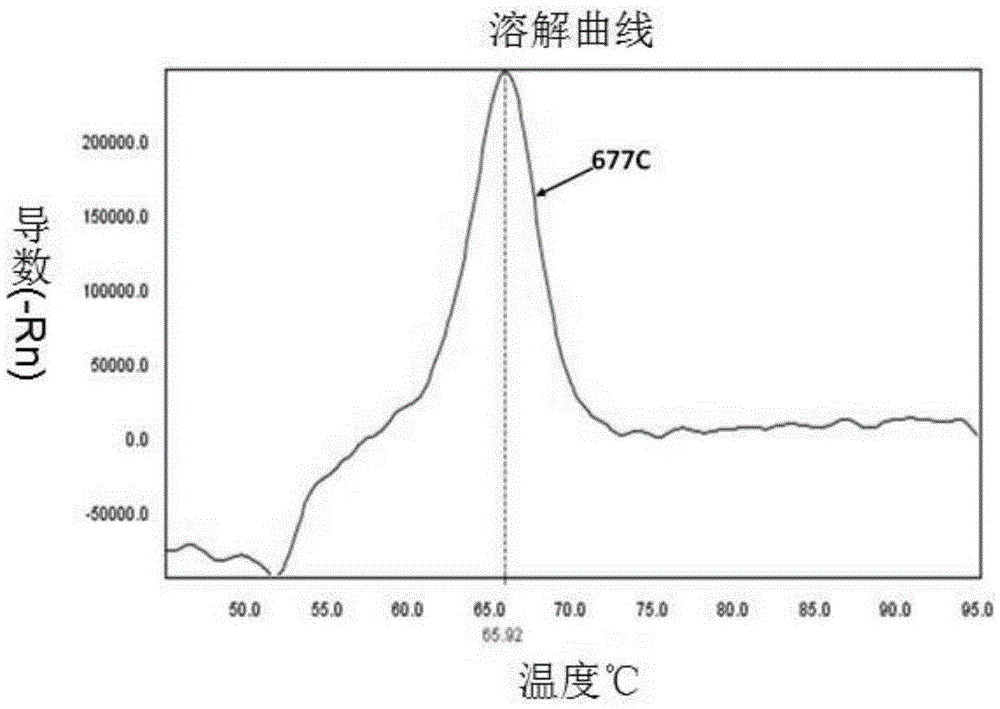
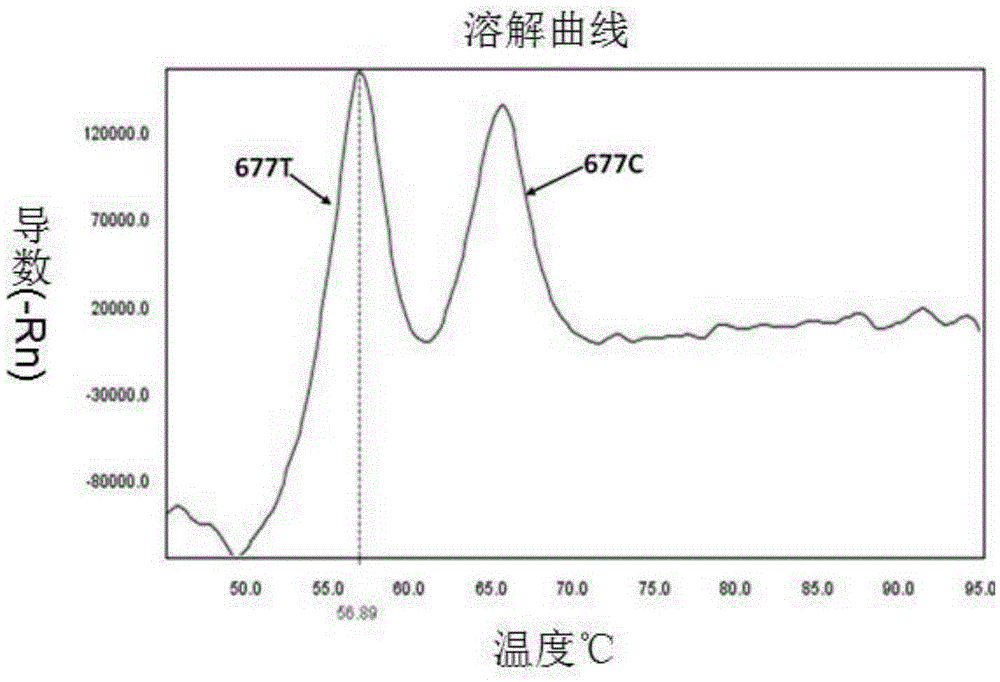
InactiveCN105296621AHigh detection specificityEasy to readMicrobiological testing/measurementDNA/RNA fragmentationFluorescencePcr ctpp
The invention provides a primer pair, specific oligonucleotide fluorescence probe and kit for detecting the nucleotide polymorphism of a C677T locus of an MTHFR gene. The new specific primer pair and the corresponding fluorescence probe are designed, PCR amplification is performed by adopting an asymmetric PCR technology, melting curve analysis is performed after PCR amplification is finished, and a genetype is judged through the melting peak at specific temperature. According to the kit, the MTHFR gene can be amplified in a highly specific mode, detection of three kinds of genetypes of the C677T locus in a single-pipe PCR system can be completed, the detection specificity is high, the result is easy to judge and read, the operating steps are simple, the detection cost is low, the cycle is short, and the efficiency is high.
Owner:智海生物工程(北京)股份有限公司
Hypertension gene polymorphism fluorescent PCR (polymerase chain reaction) solubility curve detecting kit and application thereof
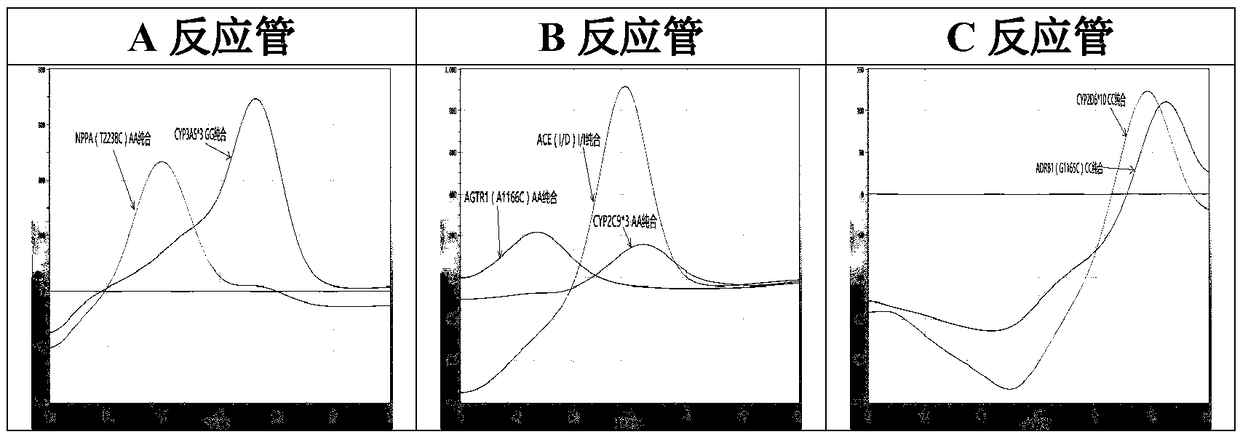
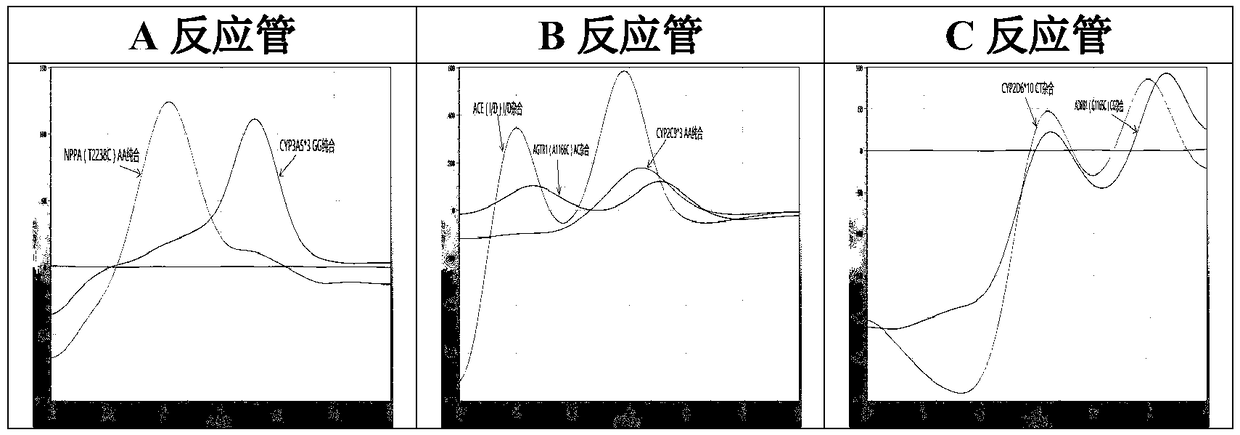
PendingCN109457024AEasy to operateShorten the timeMicrobiological testing/measurementDNA/RNA fragmentationSolubilityRibonucleoside
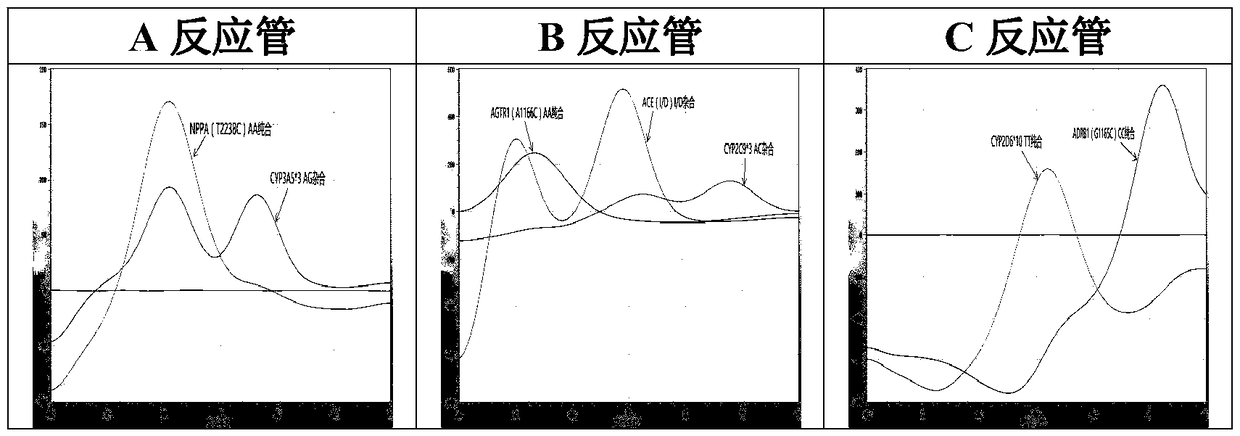
The invention relates to a hypertension gene polymorphism fluorescent PCR solubility curve detecting kit and application thereof, and a hypertension medication gene multiple-PCR solubility curve detecting method. The hypertension gene polymorphism fluorescent PCR solubility curve detecting kit mainly comprises a primer combination, a probe combination, and PCR reaction agents such thermally activated polymerases, PCR buffer solution and dNTPs (deoxy-ribonucleoside triphosphates) and involves polymorphism detection of 7 hypertension-related genes including CYP2D6*10, CYP2C9*3, ADRB1(1165G)C), AGTR1(1166A)C), CYP3A5*3, NPPA(T2338C) and ACE(I / D); the three reaction agents are applied to perform PCR amplification and solubility analysis to determine the related polymorphism of the 7 genes to guide clinical customized application of 5 majors types of hypertension drugs. The hypertension gene polymorphism fluorescent PCR solubility curve detecting kit can easily complete detection through 3reaction tubes, and meanwhile, save a nucleic acid extraction process, simplify operation steps and achieve a simple interpretation method.
Owner:众福健康科技(杭州)有限公司
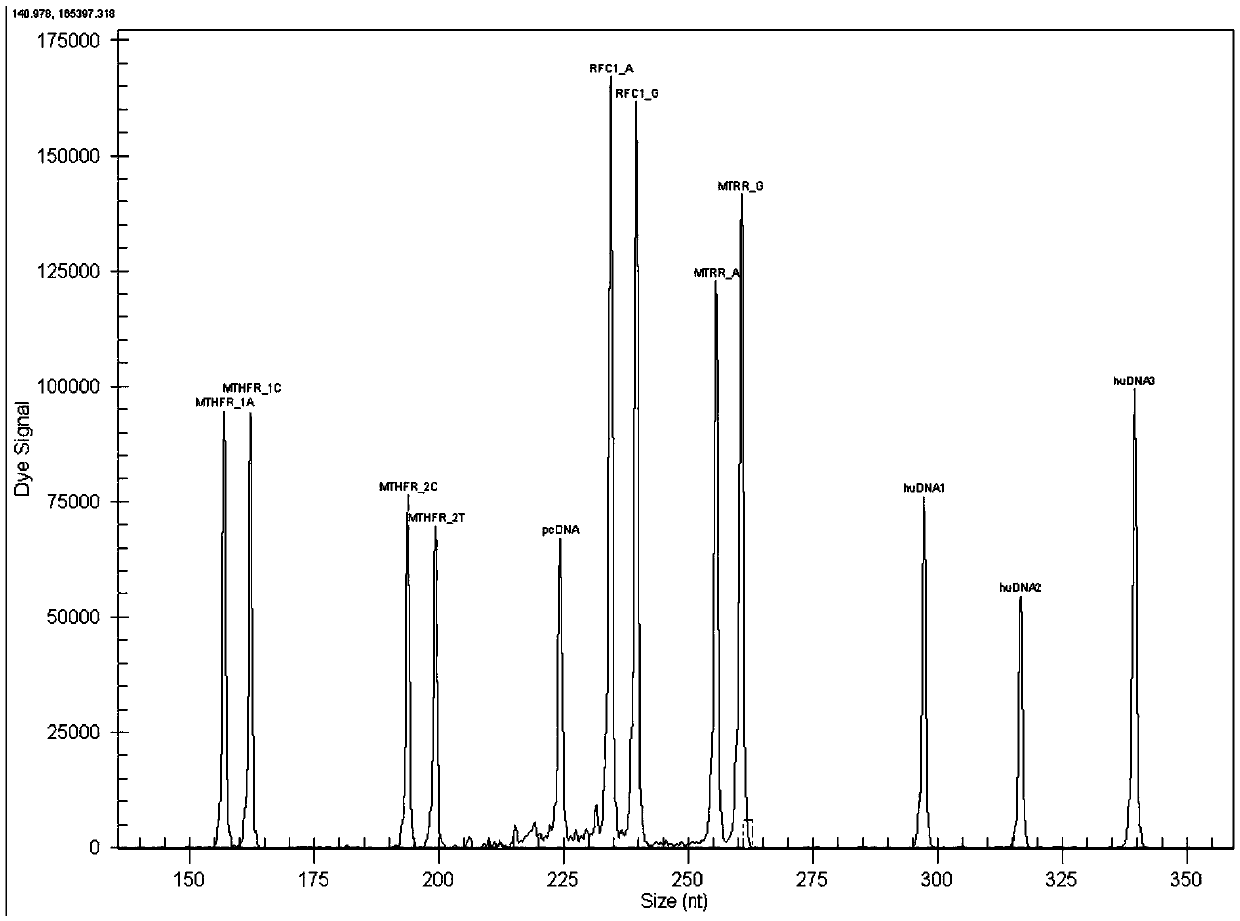
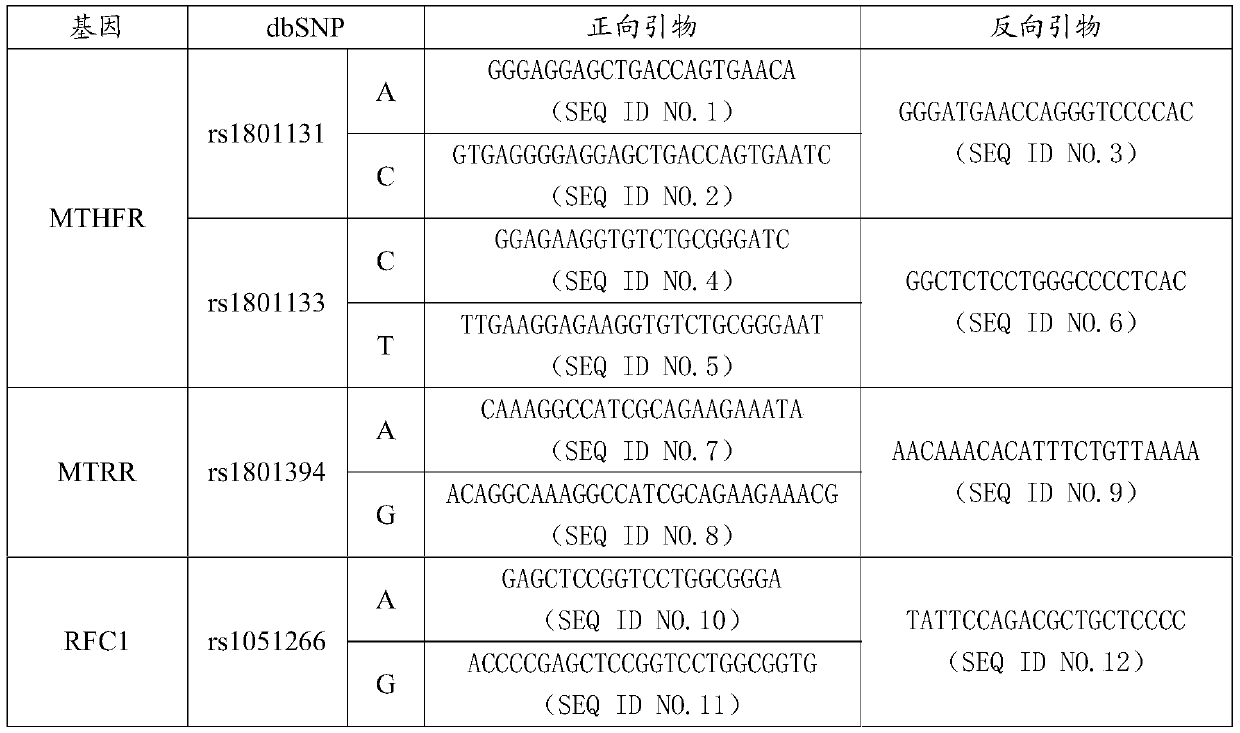
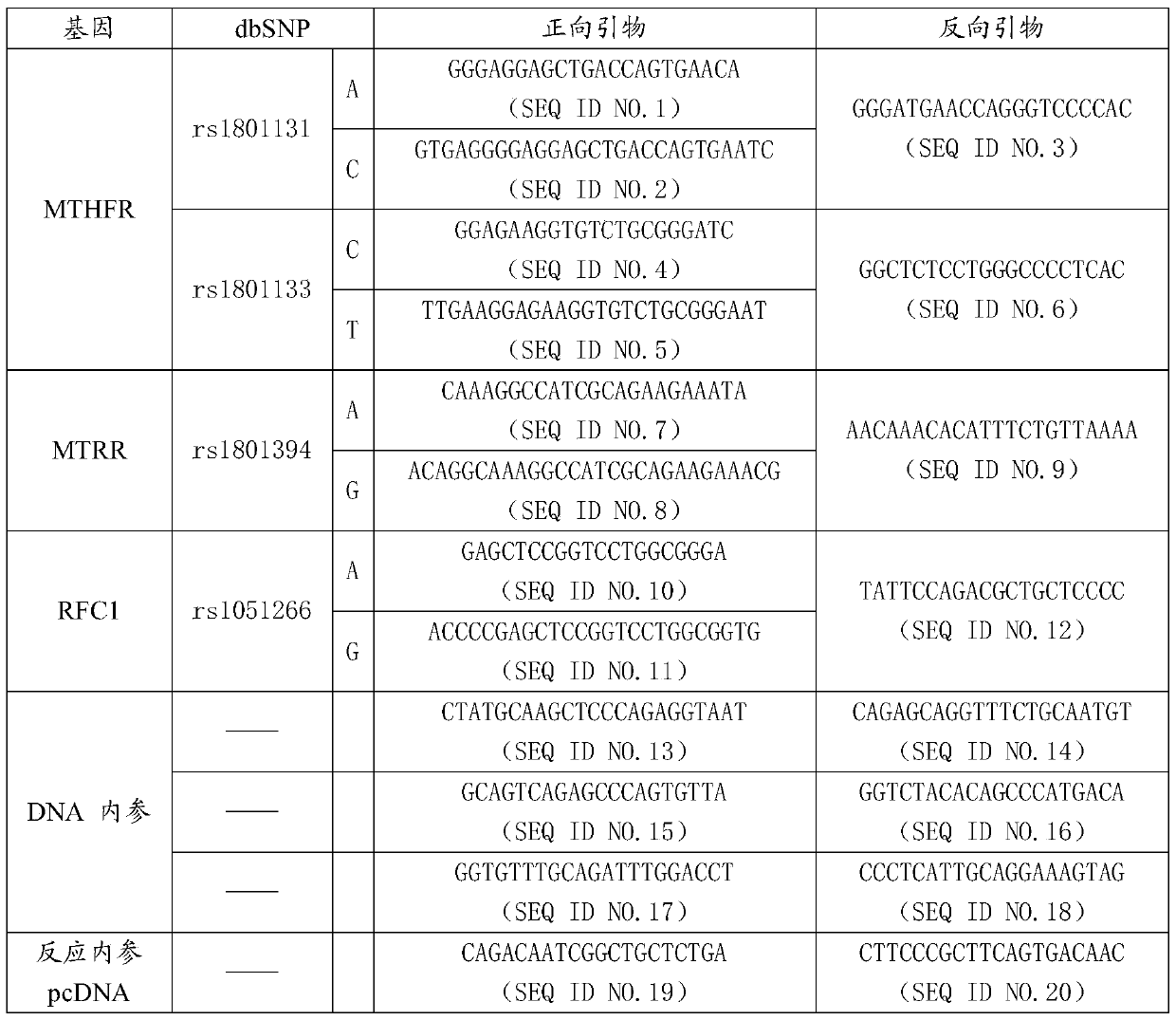
MTHFR, MTRR and RFC1 gene polymorphism detection primer combination and kit and application of MTHFR, MTRR and RFC1 gene polymorphism detection kit
ActiveCN105463122ASave production costSave testing costMicrobiological testing/measurementDNA/RNA fragmentationPositive controlRFC1
The invention discloses an MTHFR, MTRR and RFC1 gene polymorphism detection primer combination and kit and application of the MTHFR, MTRR and RFC1 gene polymorphism detection kit. The detection kit comprises ultrapure water, an X solution, a 10*PCR buffering solution, a PCR primer, a 25 mM magnesium chloride solution, DNA polymerase and a positive control product, wherein the PCR primer comprises a forward and reverse amplification primer, a DNA internal reference forward and reverse amplification primer and a reaction internal reference forward and reverse amplification primer of different gene types, the three primers are located at four SNP sites on a gene related to folic acid metabolism, and the gene sequence of the PCR primer is shown in SEQ ID NO.1-NO.20. The detection kit has the advantages of being high in specificity, accuracy, flux and reliability, low in cost and free of a false-negative result.
Owner:NINGBO HEALTH GENE TECHNOLOGIES CO LTD
Molecular identification method of cytoplasm male sterile restoring line
InactiveCN104342434AClear resultThe resulting band is stableMicrobiological testing/measurementDNA/RNA fragmentationMolecular identificationMicrobiology
The invention belongs to the technical filed of biology, and relates to a molecular identification method of a cytoplasm male sterile restoring line, and concretely provides a CAPS-labeled primer used for identifying the cytoplasm male sterile restoring line, and a nucleotide sequence is shown as SEQ ID NO. 2 and SEQ ID NO.3. The method employs the CAPS label for a polymorphism detection of molecule level of a cytoplasm male sterile restoring line material, The method has the advantages of fast identification speed, high accuracy and simple operation, and is very suitable for indoor identification of purity of the cytoplasm male sterile restoring line material and molecular marker-assisted selection of a new restoring line material.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
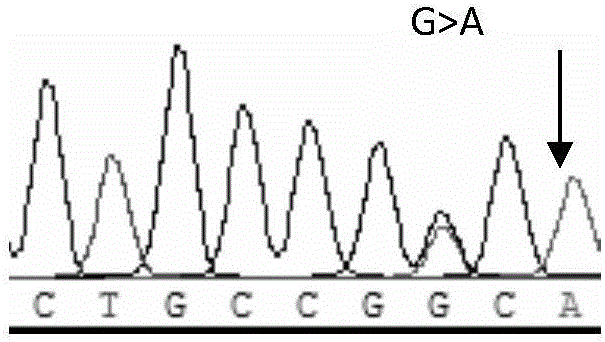
Preparation method of electrochemical sensor for FGFR3 (fibroblast growth factor receptor 3)-1138G>A gene polymorphism detection
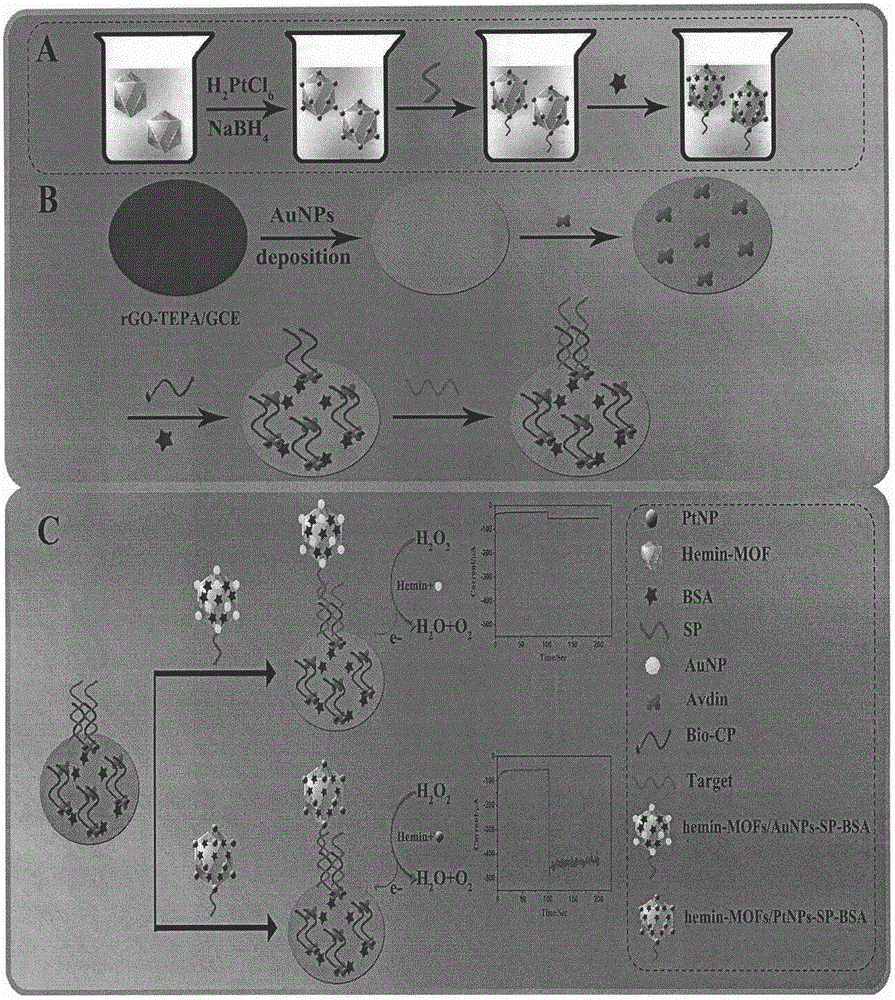
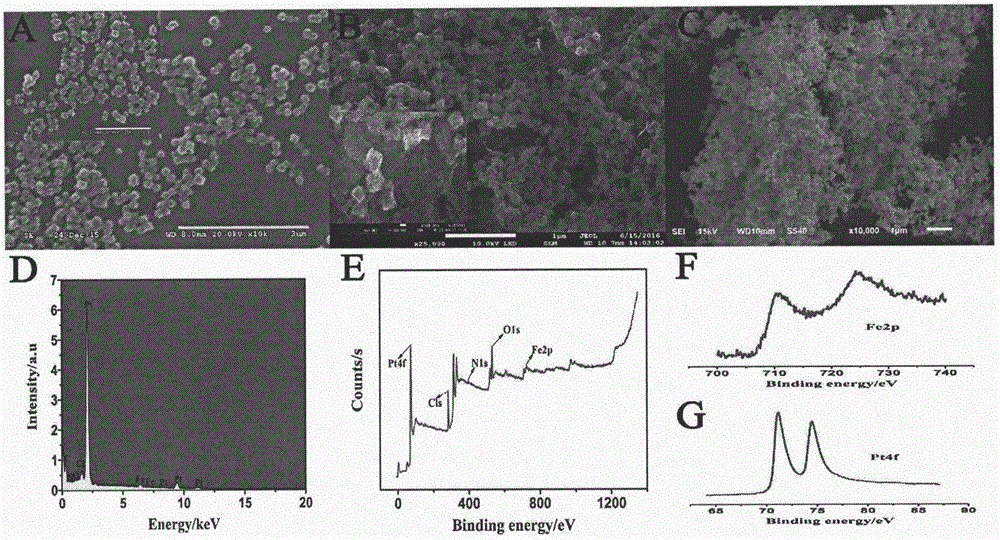
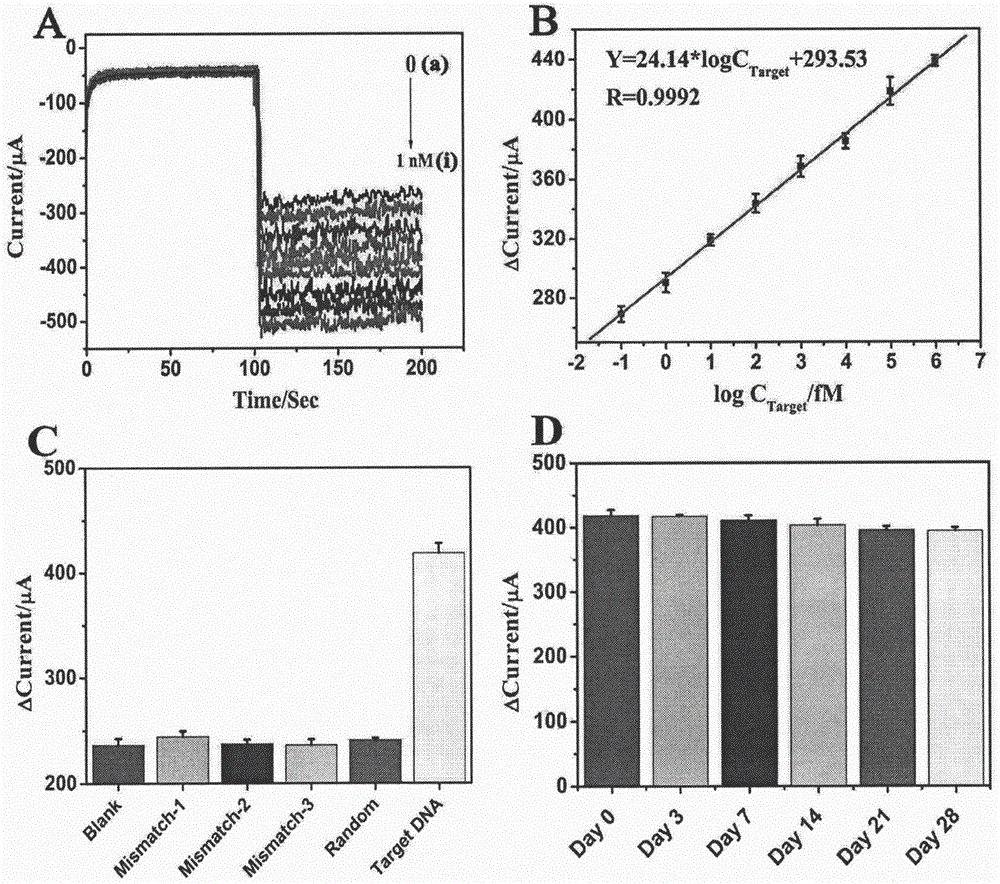
ActiveCN106483176AImprove catalytic performanceHigh sensitivityMaterial analysis by electric/magnetic meansPrenatal diagnosisHemin
The invention relates to a preparation method and application of an electrochemical sensor for achondroplasia prenatal diagnosis gene-fibroblast growth factor receptor 3 (FGFR3) gene polymorphism detection, belonging to the technical field of electrochemical detection. The preparation method is characterized by comprising the following steps: synthesizing a Hemin-MOFs composite material, reducing platinum nanoparticles onto the Hemin-MOFs composite material, and mixing a single-strand DNA signal probe with the composite material to obtain a biological signal probe; and carrying out layer-by-layer self-assembly by using reducible graphene oxide tetraethylenepentamine, nano gold and avidin to fix the biotinylated DNA capture probe, thereby preparing the electrochemical sensor for FGFR3-1138G>A gene polymorphism detection. The sensor is successfully used for detecting FGFR3-gene single base mutation. The sensor has the advantages of high sensitivity and high specificity, and is quick and convenient for detection. The invention provides a new detection method for prenatal noninvasive diagnosis of achondroplasia.
Owner:CHONGQING MEDICAL UNIVERSITY
Gene chip for weight control, preparation method, using method and kit of gene chip
InactiveCN102912452AQuick checkAccurate detectionNucleotide librariesMicrobiological testing/measurementNucleotideNucleotide sequencing
The invention relates to a gene chip for weight control, a preparation method, a using method and a kit of the gene chip. The gene chip comprises a solid phase carrier, and a specific oligonucleotide probe for gene mononucleotide polymorphic site detection related to fat fixed on the solid phase carrier, and a nucleotide sequence of the oligonucleotide probe is shown in SEQ ID No.1-20. The invention also discloses the preparation method of the gene chip, the using method, and the kit comprising the gene chip. Each related gene polymorphism detection system related to fat is provided. SNPs (Single Nucleotide Polymorphisms) related to a fat gene in a sample to be detected can be rapidly and accurately detected. The related gene polymorphism detection system has high sensitivity, and significance on scientific weight loss to control weight, and can be widely applied to people who want to lose weight. The gene locus selects specific SNPs of Chinese people, and has pertinence to the Chinese people, and a PCR (polymerase chain reaction) primer has good specificity and can better ensure a success rate of an experiment.
Owner:湖北维达健基因技术有限公司
Molecular marker related to lambing number character of sheep and application thereof
ActiveCN104046626AAuxiliary selection is activeEfficient technical meansMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMarker-assisted selection
The invention belongs to the technical field of preparation of molecular markers for livestock, and particularly relates to a preparation method and application of a molecular marker which is related to lambing number character of sheep and used for sheep marker assisted selection. The molecular marker is obtained by FSHR gene cloning, and the nucleotide sequence of the molecular marker is as shown in a sequence table SEQ ID NO:1. One T88 - C88 base substitution is generated at the 88 bp of the sequence table SEQ ID NO:1, thus resulting in BsiEI-RFLP digestion polymorphism. The invention also discloses a primer for amplifying a part of DNA (deoxyribonucleic acid) sequence of an FSHR gene and a method used for polymorphism detection, and provides a new molecular marker for selection assisted by the marker related to lambing number character of sheep.
Owner:甘肃兰天同和农业有限公司
Full premix MTHFR and MTRR multiple PCR gene polymorphism detection kit and method thereof
InactiveCN107841537AActivity is not affectedStability is not affectedMicrobiological testing/measurementDNA/RNA fragmentationNucleotidePcr ctpp
The invention provides a complete premixed multiple primer-specific PCR detection kit and method for detecting nucleotide polymorphisms at three sites of c.677 and c.1298 of the folic acid metabolic pathway and c.1298 of the MTRR gene. "Allele-specific PCR" principle, design multiple primer-specific PCR, realize two-tube reaction on the real-time fluorescent quantitative PCR technology platform, and simultaneously detect three folic acid metabolic pathway MTHFR genes c.677 and c.1298 and MTRR gene c.66 The nucleotide type of the site. In addition, modified Taq DNA polymerase and disaccharide PCR protective agent are added to the reaction system to increase the stability of the reaction system and realize the long-term stability of the pre-mixed detection system.
Owner:SUZHOU KUANGYUAN MOLECULAR BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com