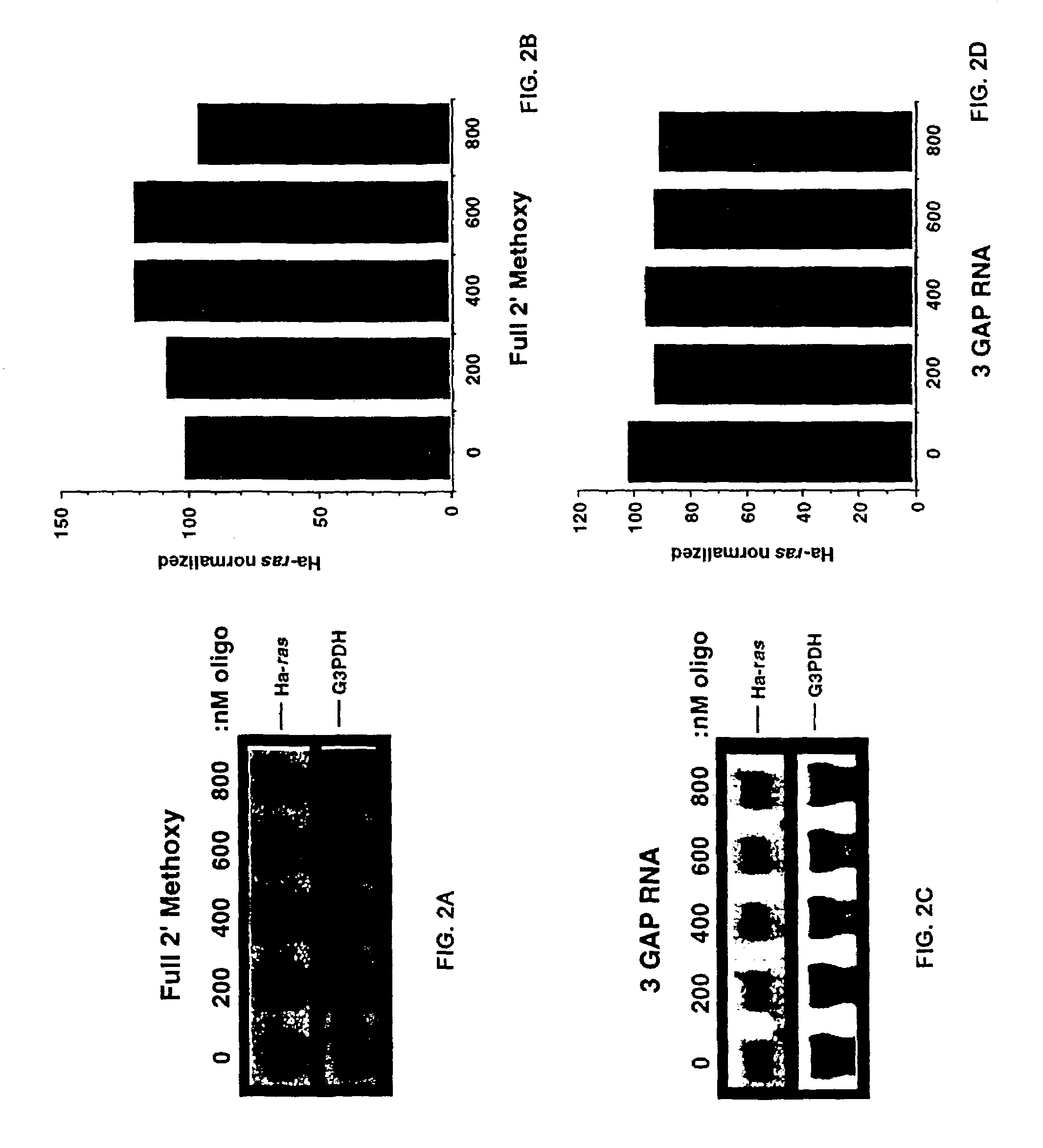
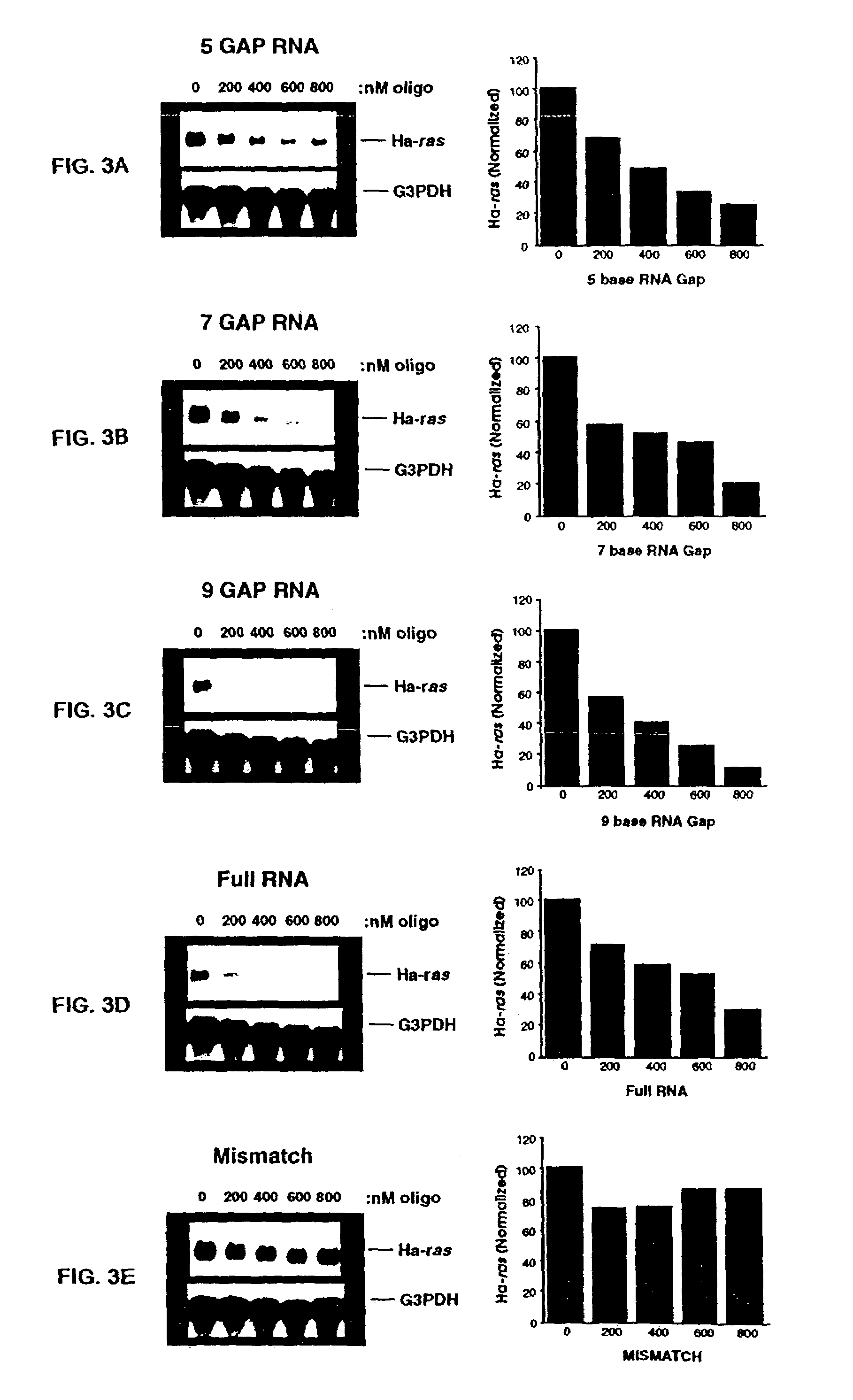
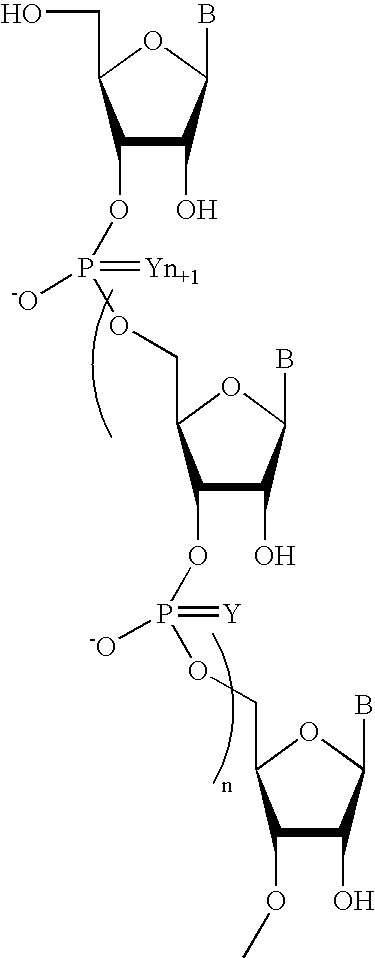
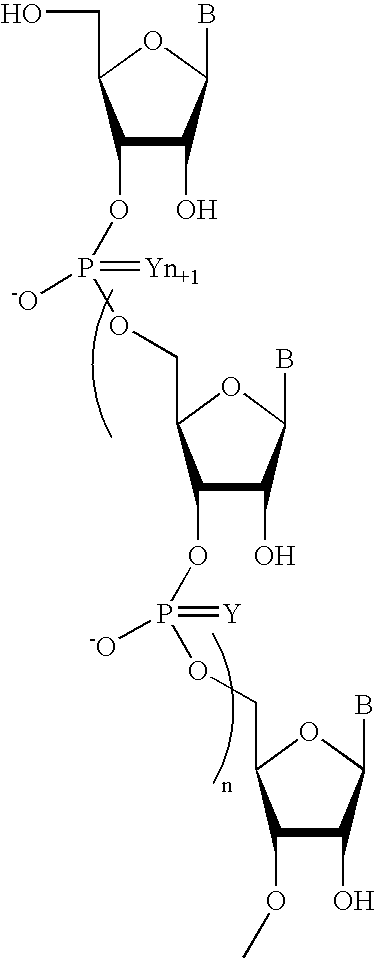
Patents
Literature
148 results about "Ribonucleoside" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A ribonucleoside is a type of nucleoside including ribose as a component. An example is cytidine.
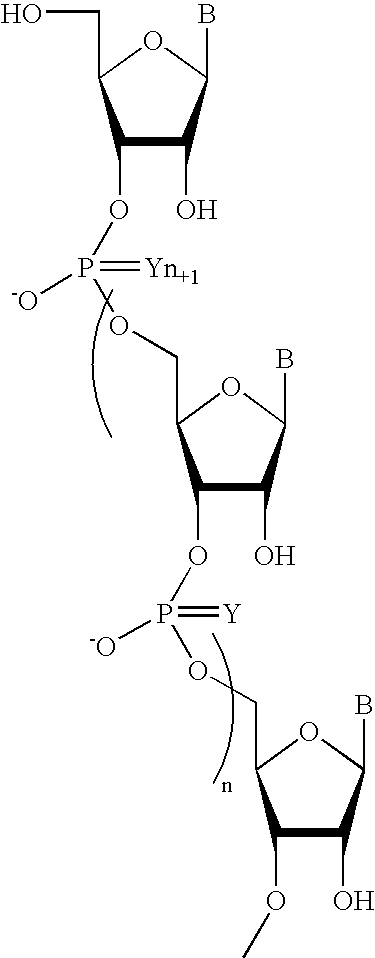
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432250B2High affinityStrong specificityHydrolasesPeptide/protein ingredientsOrganismResearch purpose
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432249B2High affinityStrong specificityPeptide/protein ingredientsHydrolasesADAMTS ProteinsOrganism
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
Oligonucleotides comprising a non-phosphate backbone linkage
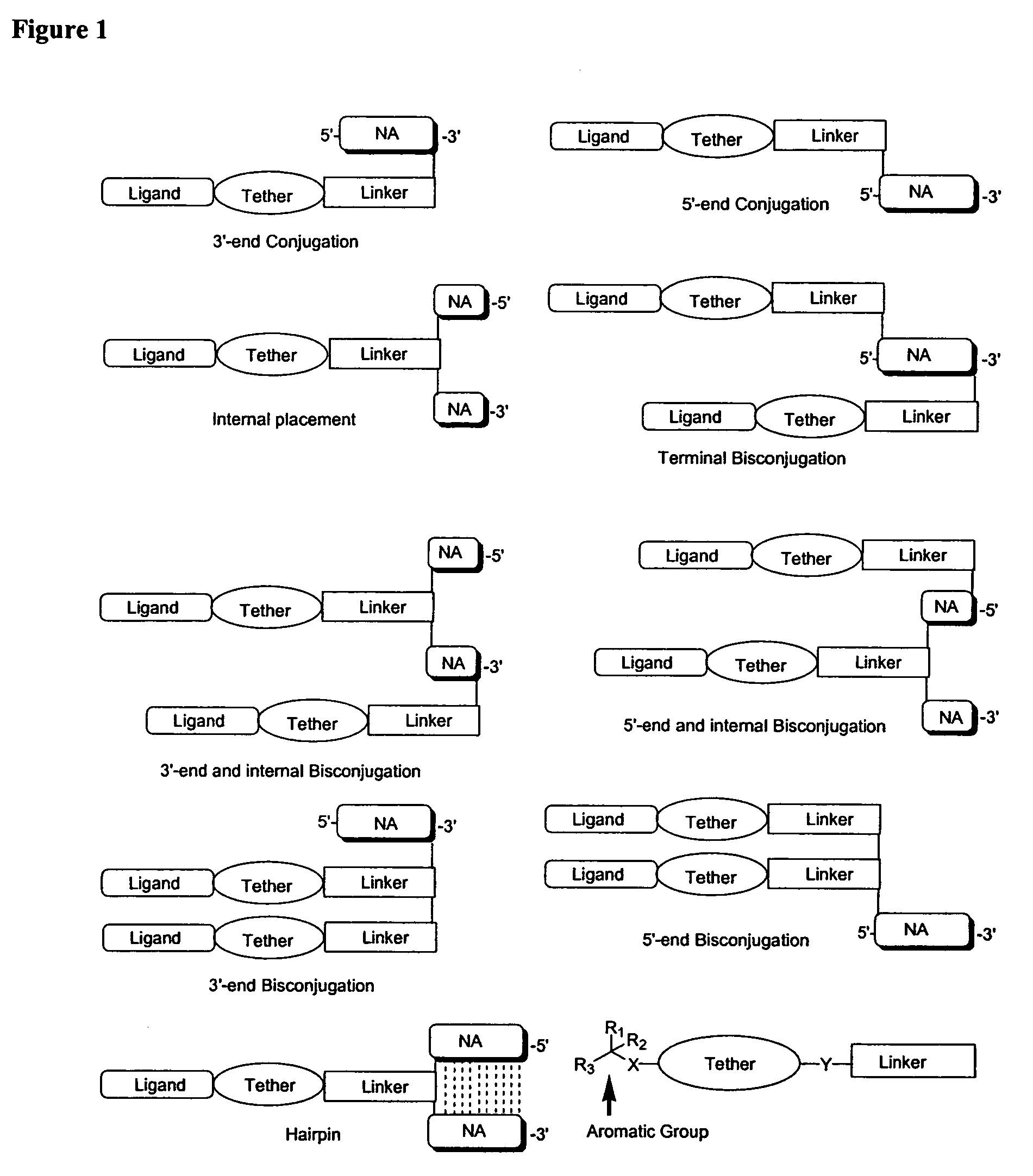
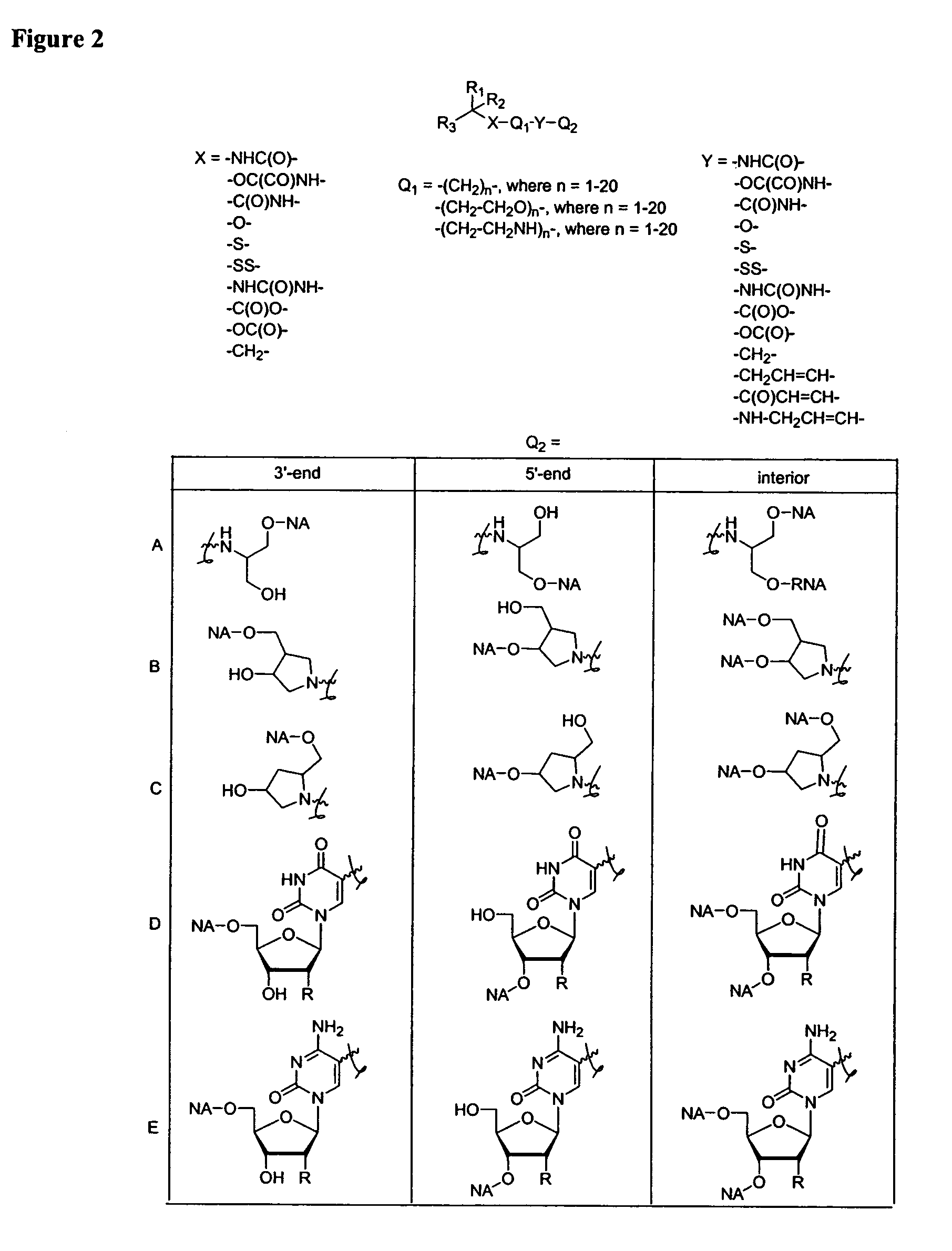
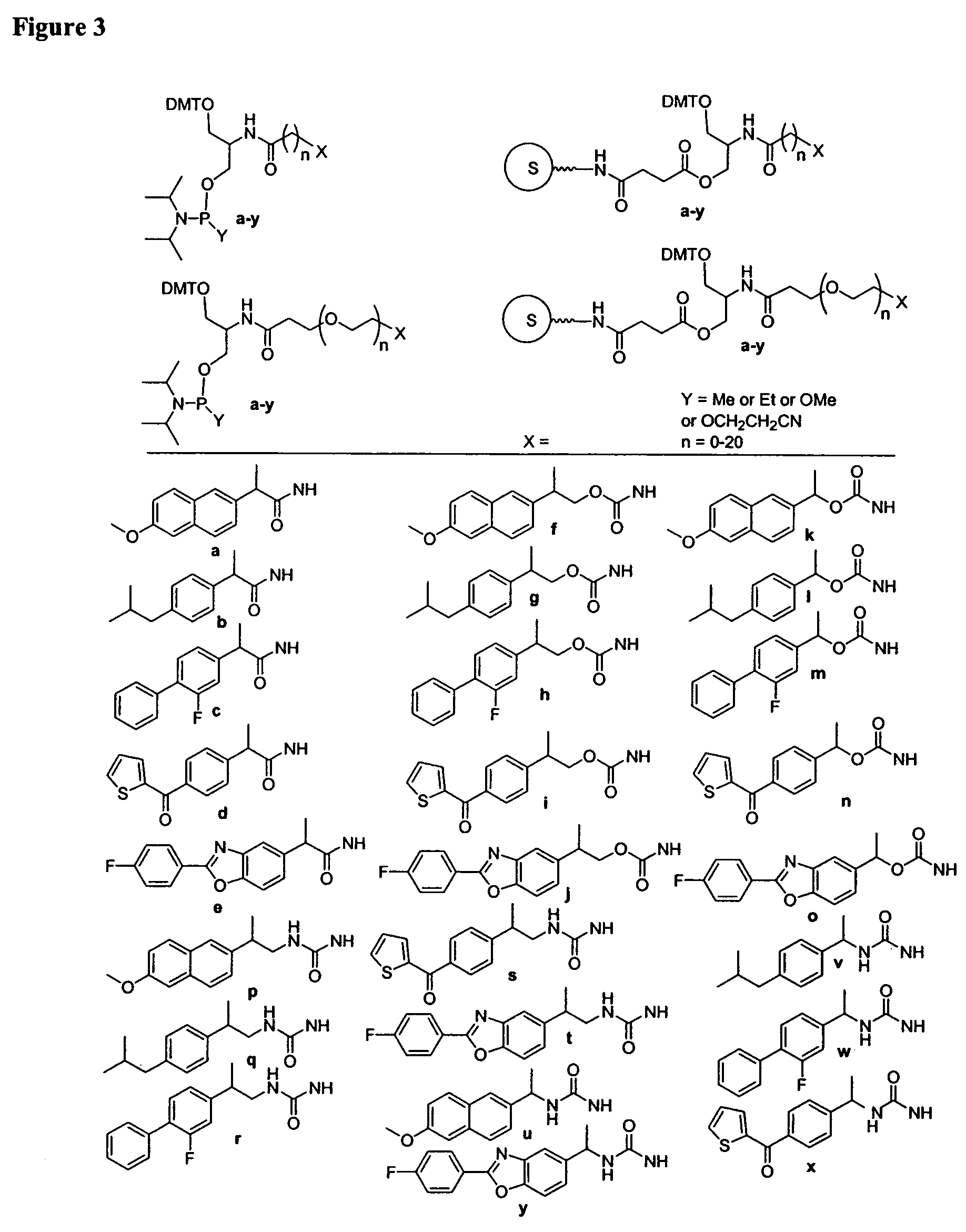
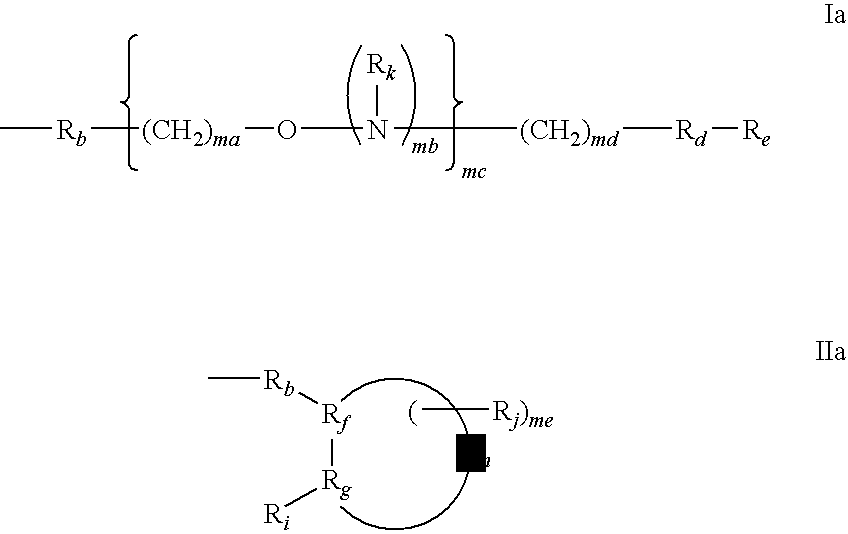
One aspect of the present invention relates to a ribonucleoside substituted with a phosphonamidite group at the 3′-position. In certain embodiments, the phosphonamidite is an alkyl phosphonamidite. Another aspect of the present invention relates to a double-stranded oligonucleotide comprising at least one non-phosphate linkage. Representative non-phosphate linkages include phosphonate, hydroxylamine, hydroxylhydrazinyl, amide, and carbamate linkages. In certain embodiments, the non-phosphate linkage is a phosphonate linkage. In certain embodiments, a non-phosphate linkage occurs in only one strand. In certain embodiments, a non-phosphate linkage occurs in both strands. In certain embodiments, a ligand is bound to one of the oligonucleotide strands comprising the double-stranded oligonucleotide. In certain embodiments, a ligand is bound to both of the oligonucleotide strands comprising the double-stranded oligonucleotide. In certain embodiments, the oligonucleotide strands comprise at least one modified sugar moiety. Another aspect of the present invention relates to a single-stranded oligonucleotide comprising at least one non-phosphate linkage. Representative non-phosphate linkages include phosphonate, hydroxylamine, hydroxylhydrazinyl, amide, and carbamate linkages. In certain embodiments, the non-phosphate linkage is a phosphonate linkage. In certain embodiments, a ligand is bound to the oligonucleotide strand. In certain embodiments, the oligonucleotide comprises at least one modified sugar moiety.
Owner:ALNYLAM PHARM INC
Carba-nucleoside analogs for antiviral treatment
Owner:GILEAD SCI INC
Methods and means for enhancing RNA production
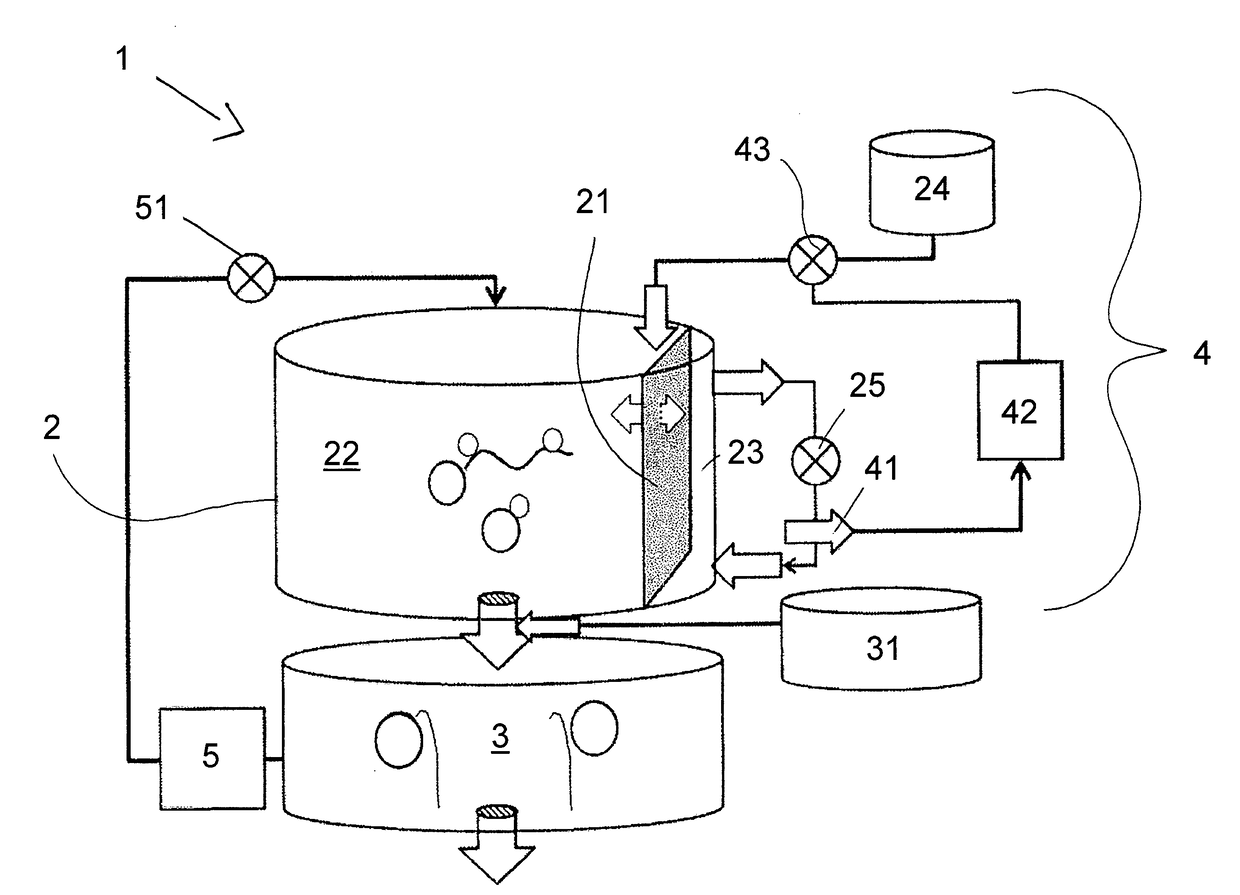
ActiveUS20170114378A1Improved and economical meanImproved and economical and methodBioreactor/fermenter combinationsBiological substance pretreatmentsRibonucleosideFiltration membrane
The present invention relates to a method for synthesizing an RNA molecule of a given sequence, comprising the step of determining the fraction (1) for each of the four nucleotides G, A, C and U in said RNA molecule, and the step of synthesizing said RNA molecule by in vitro transcription in a sequence-optimized reaction mix, wherein said sequence-optimized reaction mix comprises the four ribonucleoside triphosphates GTP, ATP, CTP and UTP, wherein the fraction (2) of each of the four ribonucleoside triphosphates in the sequence-optimized reaction mix corresponds to the fraction (1) of the respective nucleotide in said RNA molecule, a buffer, a DNA template, and an RNA polymerase. Further, the present invention relates to a bioreactor (1) for synthesizing RNA molecules of a given sequence, the bioreactor (1) having a reaction module (2) for carrying out in vitro RNA transcription reactions in a sequence-optimized reaction mix, a capture module (3) for temporarily capturing the transcribed RNA molecules, and a control module (4) for controlling the infeed of components of the sequence-optimized reaction mix into the reaction module (2), wherein the reaction module (2) comprises a filtration membrane (21) for separating nucleotides from the reaction mix, and the control of the infeed of components of the sequence-optimized reaction mix by the control module (4) is based on a measured concentration of separated nucleotides.
Owner:CUREVAC REAL ESTATE GMBH
Hybrid oligonucleotide phosphorothioates
The invention provides hybrid oligonucleotides having phosphorothioate or phosphorodithioate internucleotide linkages, and both deoxyribonucleosides and ribonucleosides or 2′-substituted ribonucleosides. Such hybrid oligonucleotides have superior properties of duplex formation with RNA, nuclease resistance, and RNase H activation.
Owner:UNIV OF MASSACHUSETTS WORCESTER
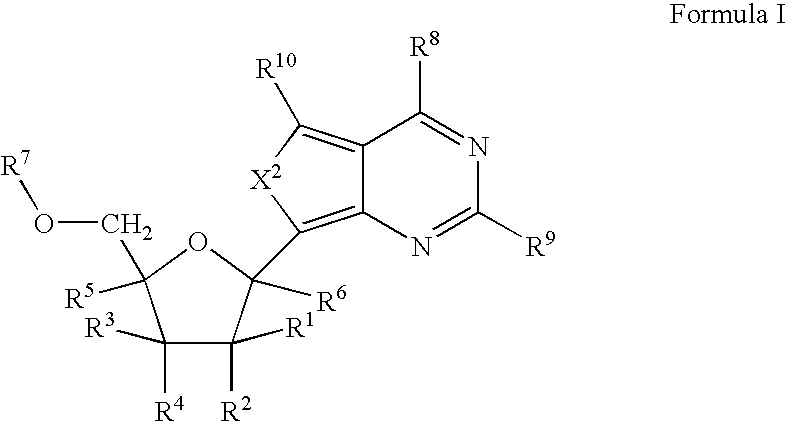
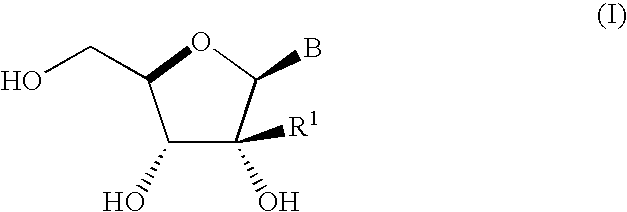
Ribonucleoside cyclic acetal derivatives for the treatment of RNA-dependent RNA viral infection
The present invention provides ribonucleoside 2′,3′-cyclic acetals of structural formula I which are precursors or prodrugs of inhibitors of RNA-dependent RNA viral polymerase. These compounds are precursors of inhibitors of RNA-dependent RNA viral replication and are useful for the treatment of RNA-dependent RNA viral infection. They are particularly useful as precursors or prodrugs of inhibitors of hepatitis C virus (HCV) NS5B polymerase, as precursors or prodrugs of inhibitors of HCV replication, and / or for the treatment of hepatitis C infection. The invention also describes pharmaceutical compositions containing such ribonucleoside 2′,3′-cyclic acetals alone or in combination with other agents active against RNA-dependent RNA viral infection, in particular HCV infection. Also disclosed are methods of inhibiting RNA-dependent RNA polymerase, inhibiting RNA-dependent RNA viral replication, and / or treating RNA-dependent RNA viral infection with the ribonucleoside 2′,3′-cyclic acetals of the present invention.
Owner:MERCK SHARP & DOHME CORP
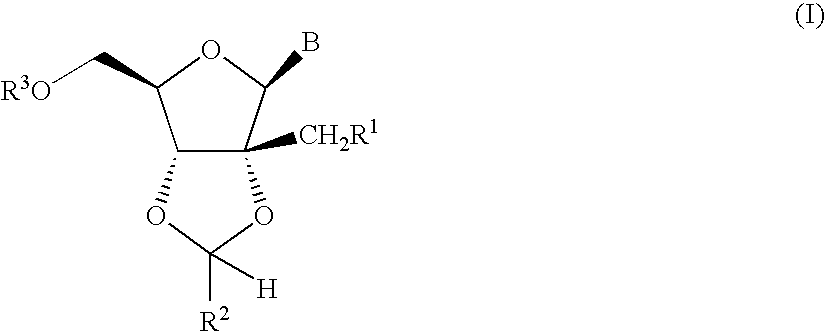
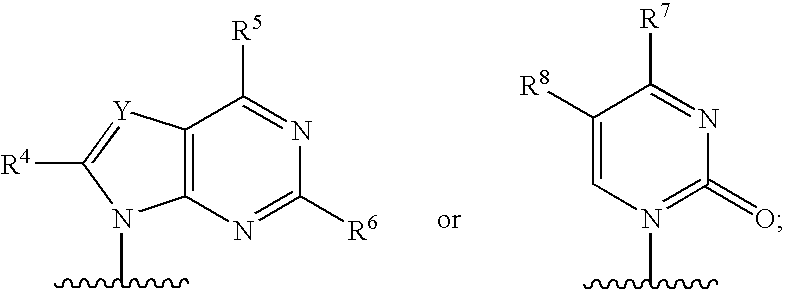
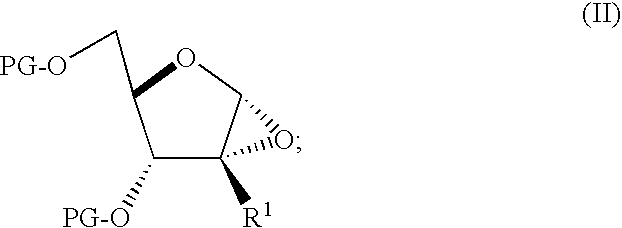
Process for preparing branched ribonucleosides from 1,2-anhydroribofuranose intermediates
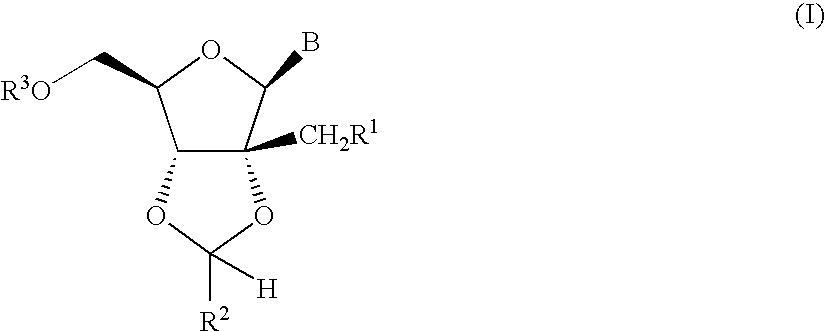
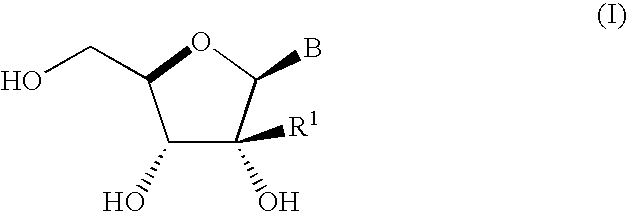
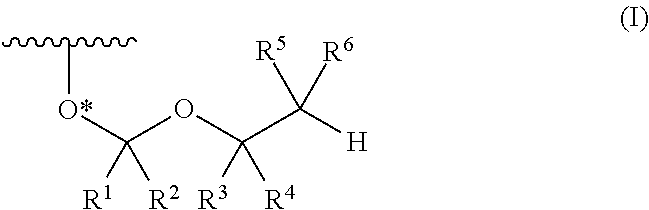
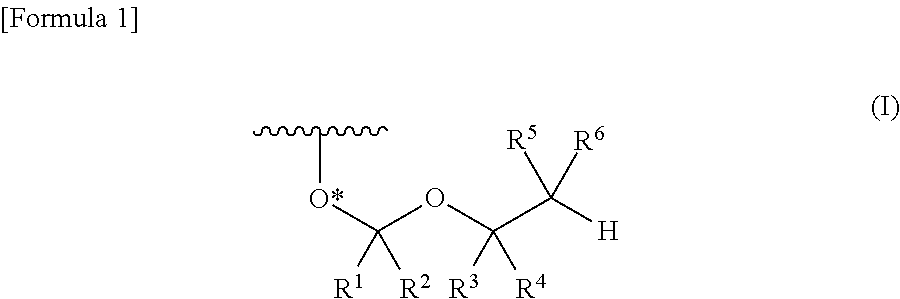
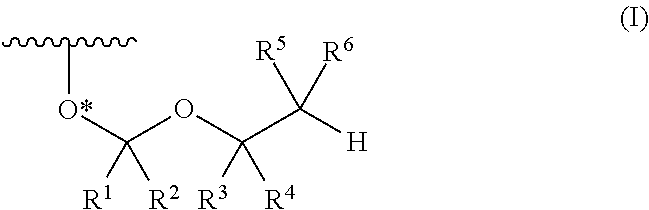
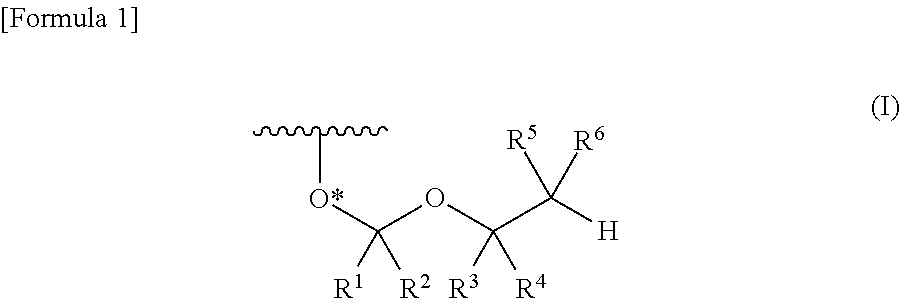
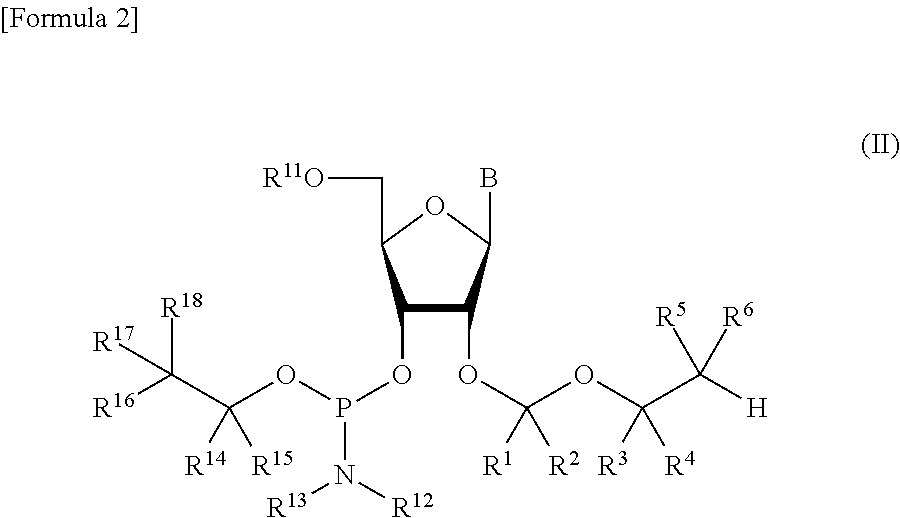
A process is provided for the preparation of branched-chain ribonucleosides of formula (I):from the 1,2-anhydroderivatives of formula (II).wherein PG is a hydroxyl protecting group, B is a purine or pyrimidine nucleobase, and R1 is C1-6 alkyl. The compounds of formula (I) are inhibitors of HCV polymerase useful in the treatment of HCV infection.
Owner:MERCK SHARP & DOHME CORP
Double strand compositions comprising differentially modified strands for use in gene modulation
InactiveUS20070123484A1Suppress gene expressionOrganic active ingredientsBiocideRibonucleosideTRIM Motif
The present invention provides double stranded compositions wherein each strand is modified to have a motif defined by positioning of β-D-ribonucleosides and sugar modified nucleosides. More particularly, the present compositions comprise one strand having an alternating motif and another strand having a hemimer motif, a blockmer motif, a fully modified motif or a positionally modified motif. At least one of the strands has complementarity to a nucleic acid target. The compositions are useful for targeting selected nucleic acid molecules and modulating the expression of one or more genes. In preferred embodiments the compositions of the present invention hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA. The present invention also provides methods for modulating gene expression.
Owner:BHAT BALKRISHEN +4
Double strand compositions comprising differentially modified strands for use in gene modulation
The present invention provides double stranded compositions wherein each strand is modified to have a motif defined by positioning of β-D-ribonucleosides and sugar modified nucleosides. More particularly, the present compositions comprise one strand having an alternating motif and another strand having a hemimer motif, a blockmer motif, a fully modified motif or a positionally modified motif. At least one of the strands has complementarity to a nucleic acid target. The compositions are useful for targeting selected nucleic acid molecules and modulating the expression of one or more genes. In preferred embodiments the compositions of the present invention hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA. The present invention also provides methods for modulating gene expression.
Owner:BHAT BALKRISHEN +4
Method for preparing ribonucleoside phosphorothioate
ActiveUS8859755B2Efficient synthesisImprove efficiencySugar derivativesBulk chemical productionPhospholipinRibonucleoside
Owner:WAVE LIFE SCI LTD
Protective group for synthesis of RNA and derivative
ActiveUS8470987B2Small hindranceEfficient removalSugar derivativesGroup 5/15 element organic compoundsRibonucleosideRibose
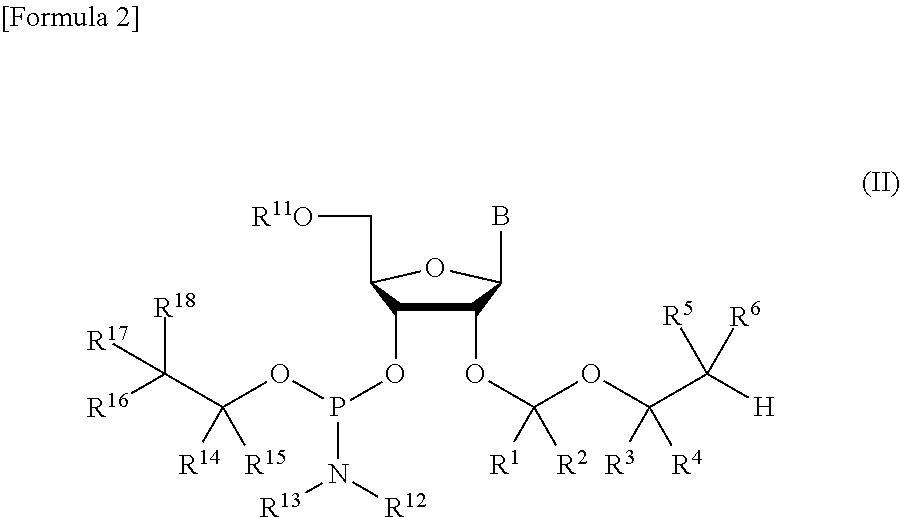
A protective group represented by the following general formula (I) (the oxygen atom attached with * represents oxygen atom of 2′-hydroxyl group of a ribonucleoside, a ribonucleotide or a derivative thereof; R1 and R2 both represent hydrogen atom, or represent a halogen atom, a C1-6 alkyl group, or a C1-6 halo-substituted alkyl group; R3 and R4 represent hydrogen atom, a halogen atom, a C1-6 alkyl group, or a C1-6 halo-substituted alkyl group; and R5 and R6 represent a halogen atom, a C1-6 halo-substituted alkyl group, cyano group, nitro group, or the like), which is stable under the reaction conditions of the nucleic acid synthetic cycles and has little steric hindrance, and can be removed under mild conditions using fluoride ions as a base.
Owner:WAVE LIFE SCI LTD
Induction of viral mutation by incorporation of miscoding ribonucleoside analogs into viral RNA
InactiveUS20050187180A1High mutation rateReduced viabilityOrganic active ingredientsSsRNA viruses positive-senseRibonucleosideViral replication
Owner:UNIV OF WASHINGTON
2'-O-modified RNA
ActiveUS8822671B2Improve abilitiesEasy to synthesizeSugar derivativesSugar derivatives preparationRibonucleosideHalogen
Owner:CHIRALGEN +1
Method for preparing ribonucleoside phosphorothioate
ActiveUS20130184450A1Improve efficiencyEfficient synthesisSugar derivativesBulk chemical productionRibonucleosideCombinatorial chemistry
Owner:WAVE LIFE SCI LTD
Oligonucleotides comprising a non-phosphate backbone linkage
Owner:ALNYLAM PHARMA INC
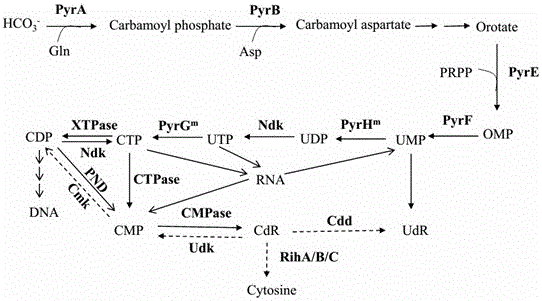
Recombinant microorganism for producing cytidine and method for producing cytidine
ActiveCN106754602ARealize large-scale industrial productionReduce manufacturing costBacteriaTransferasesRibonucleosideGenetic engineering
The invention provides a recombinant microorganism for producing cytidine and a method for producing the cytidine from the recombinant microorganism. A degradation and use gene of the cytidine is knocked off, and the gene encodes cytidine ammonialyase, ribonucleotide hydrolase, cytidine / uridine kinase and nucleoside transporter. Meanwhile key enzymes in biological synthesis process of the cytidine are over-expressed, including degrading cytidine triphosphoric acid to cytidine triphosphoric acid pyrophosphorylase of the cytidine monohosphoric acid, and catalyzing the cytidine monohosphoric acid to cytidine monohosphoric acid phosphorylase of the cytidine. In addition, pyrimidine nucleoside process is subjected to genetic engineering reform, and feedback inhibition of the synthesis process is relieved. By using a biological fermentation method, the cytidine yield greater than 20g / L can be achieved for a recombinant strain in a fermentation tank of 5L, industrial on-scale production can be achieved, and meanwhile the recombinant microorganism is low in cytidine production cost, small in pollution, green and environmental-friendly and relatively high in popularization and application value.
Owner:BIOSYNTHETICA INC
Novel protective group for synthesis of RNA and derivative
ActiveUS20110178284A1Small hindranceEfficient removalSugar derivativesOrganic compound preparationHydrogen atomRibonucleoside
A protective group represented by the following general formula (I) (the oxygen atom attached with * represents oxygen atom of 2′-hydroxyl group of a ribonucleoside, a ribonucleotide or a derivative thereof; R1 and R2 both represent hydrogen atom, or represent a halogen atom, a C1-6 alkyl group, or a C1-6 halo-substituted alkyl group; R3 and R4 represent hydrogen atom, a halogen atom, a C1-6 alkyl group, or a C1-6 halo-substituted alkyl group; and R5 and R6 represent a halogen atom, a C1-6 halo-substituted alkyl group, cyano group, nitro group, or the like), which is stable under the reaction conditions of the nucleic acid synthetic cycles and has little steric hindrance, and can be removed under mild conditions using fluoride ions as a base.
Owner:WAVE LIFE SCI LTD
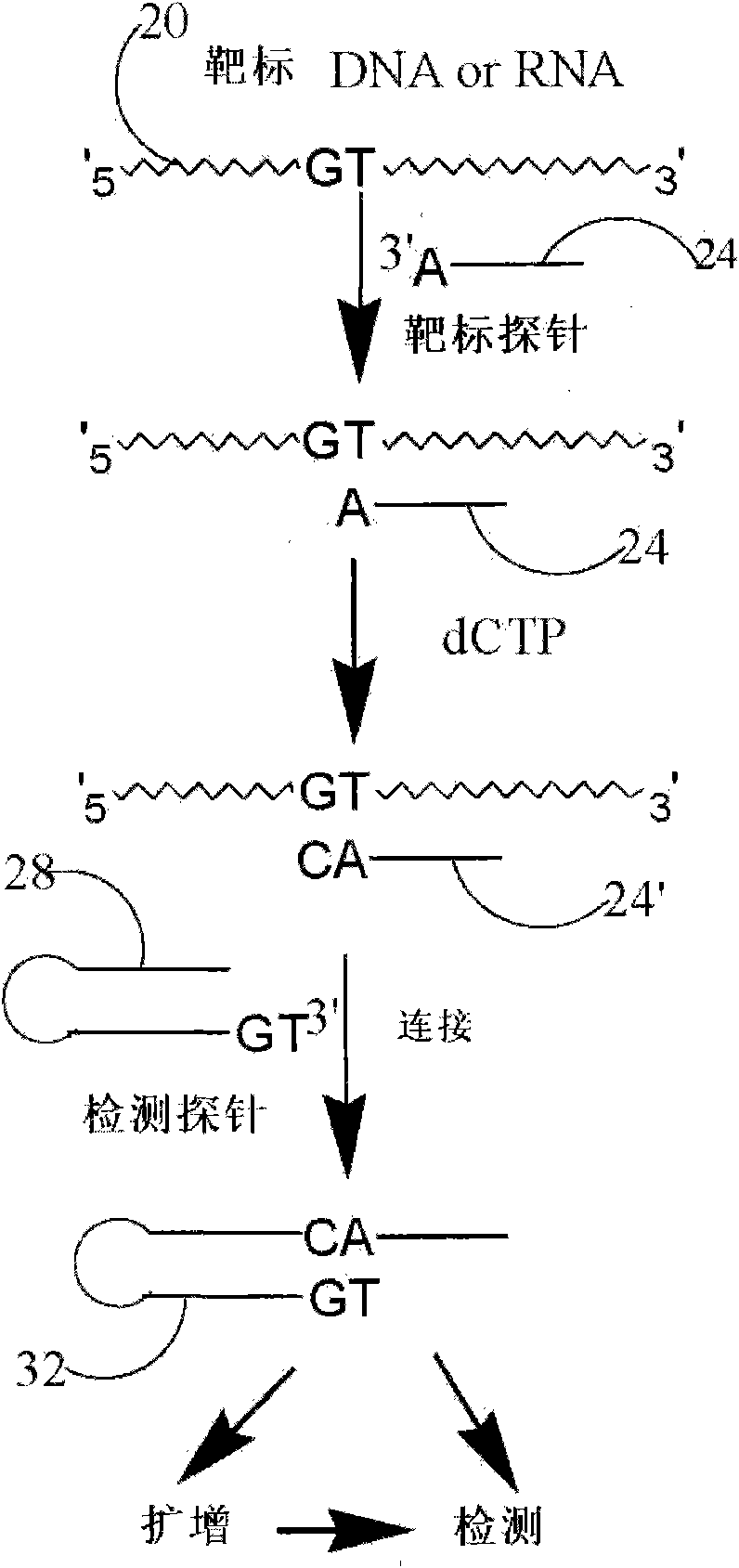
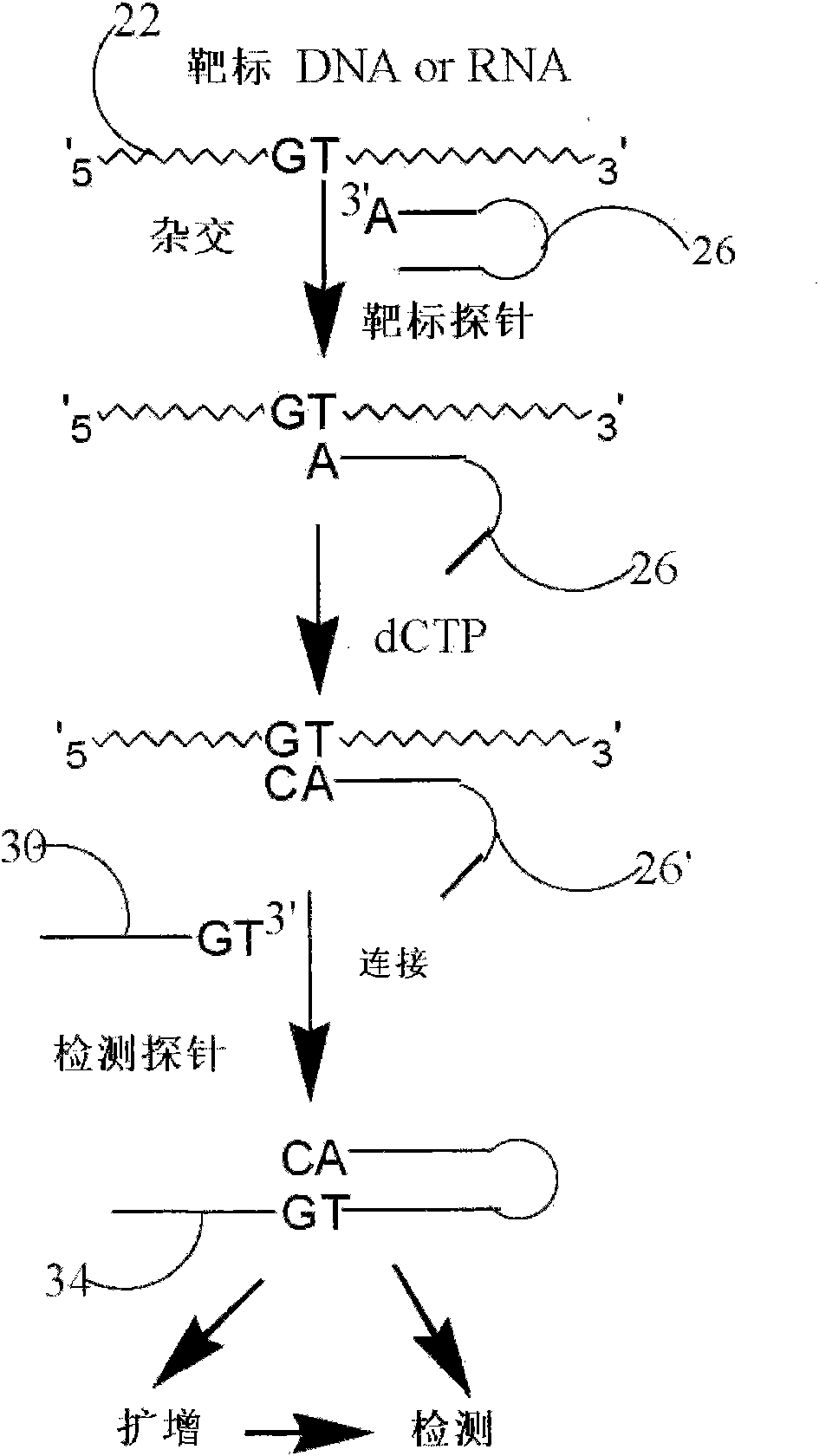
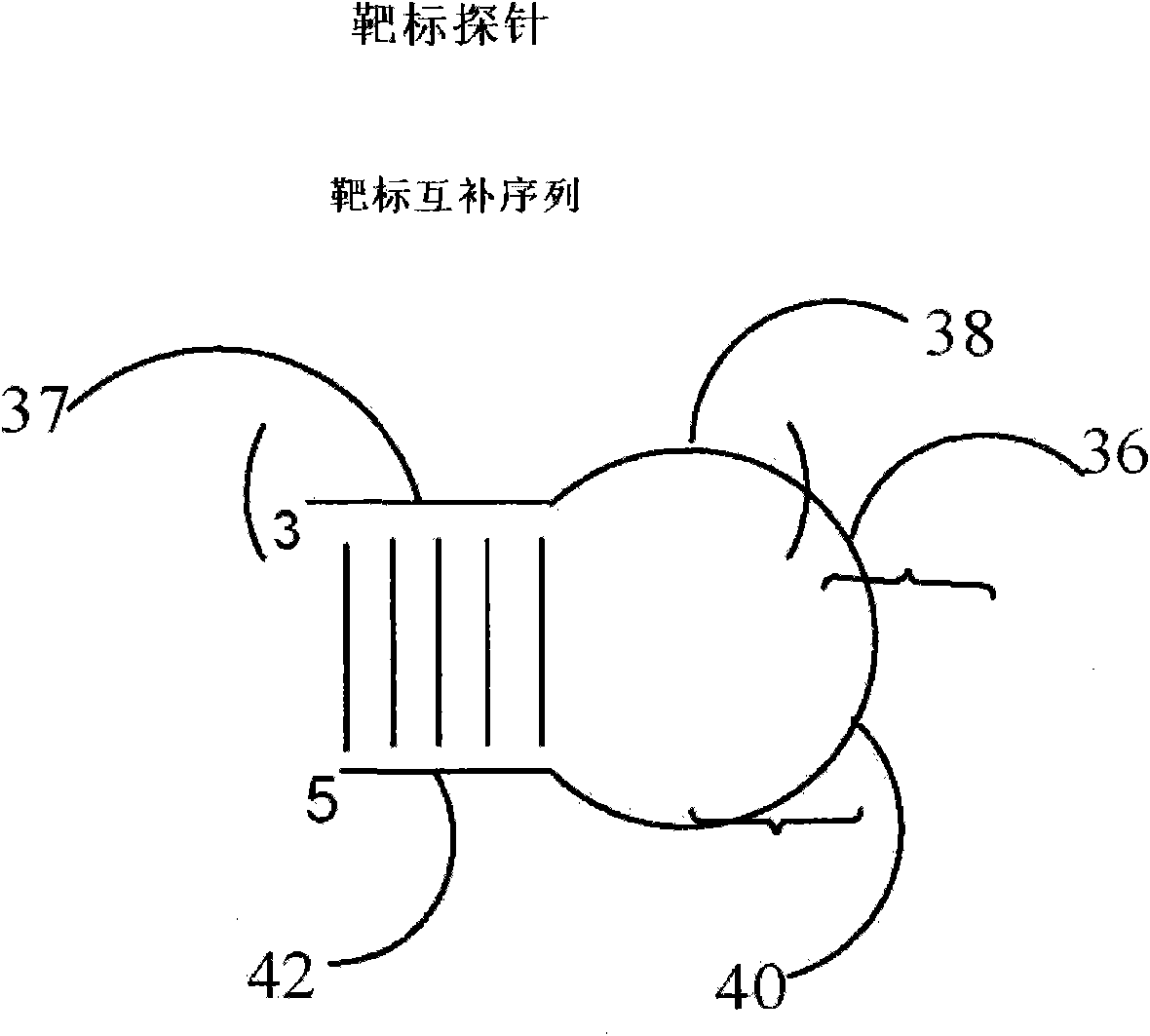
Method and kit for detecting nucleotide sequence
The invention relates to the technical field of biology, and discloses a method and a kit for detecting a target sequence in a polynucleotide analyte contained in a sample. The method comprises the following steps of: mixing the sample and a target probe under the condition of effectively forming a double-chain complex of a discovery substance and the probe, wherein the target probe has a sequence capable of being hybridized with the target sequence; reacting the probe in the complex in the presence of polymerase and one to three of four possible ribonucleoside triphosphotes to add one or more selected target-directed nucleotide bases at the 3' tail end of the probe so as to produce a modified probe; and hybridizing the modified probe and a detection probe, connecting the two probes to form a connecting product of the two probes, and detecting the existence condition of the connecting product.
Owner:中生方政生物技术股份有限公司
Isothermal nucleic acid amplification reaction reagent and isothermal nucleic acid amplification method
ActiveCN102816756AStrong specificityEfficient amplificationDNA preparationRibonucleosidePolyethylene glycol
The invention discloses an isothermal nucleic acid amplification reaction reagent which comprises Tris buffer solutions, potassium acetate, magnesium acetate, dithiothreitol, polyethylene glycol, adenosine triphosphate (ATP), deoxy-ribonucleoside triphosphate (dNTPs), phosphocreatine, creatine kinase, a primer group, single-strand binding (SSD) protein, colon bacillus helicase RecQ protein, UvsY protein and DNA polymerase. The invention further discloses an isothermal nucleic acid amplification method which includes steps of extracting DNA or performing inverse transcription after DNA extracting, and adding the isothermal nucleic acid amplification reaction reagent and reacting the mixture at a temperature in a range between 25 DEG C and 45 DEG C for 10 minutes to 60 minutes to complete the nucleic acid amplification. Compared with traditional polymerase chain reaction (PCR) technologies and according to the technology, nearly no professional instrument is needed, the operation is simple, technical requirements for operators are low, and the reaction time only needs about half an hour.
Owner:JIANGSU QITIAN GENE BIOTECHNOLOGY CO LTD
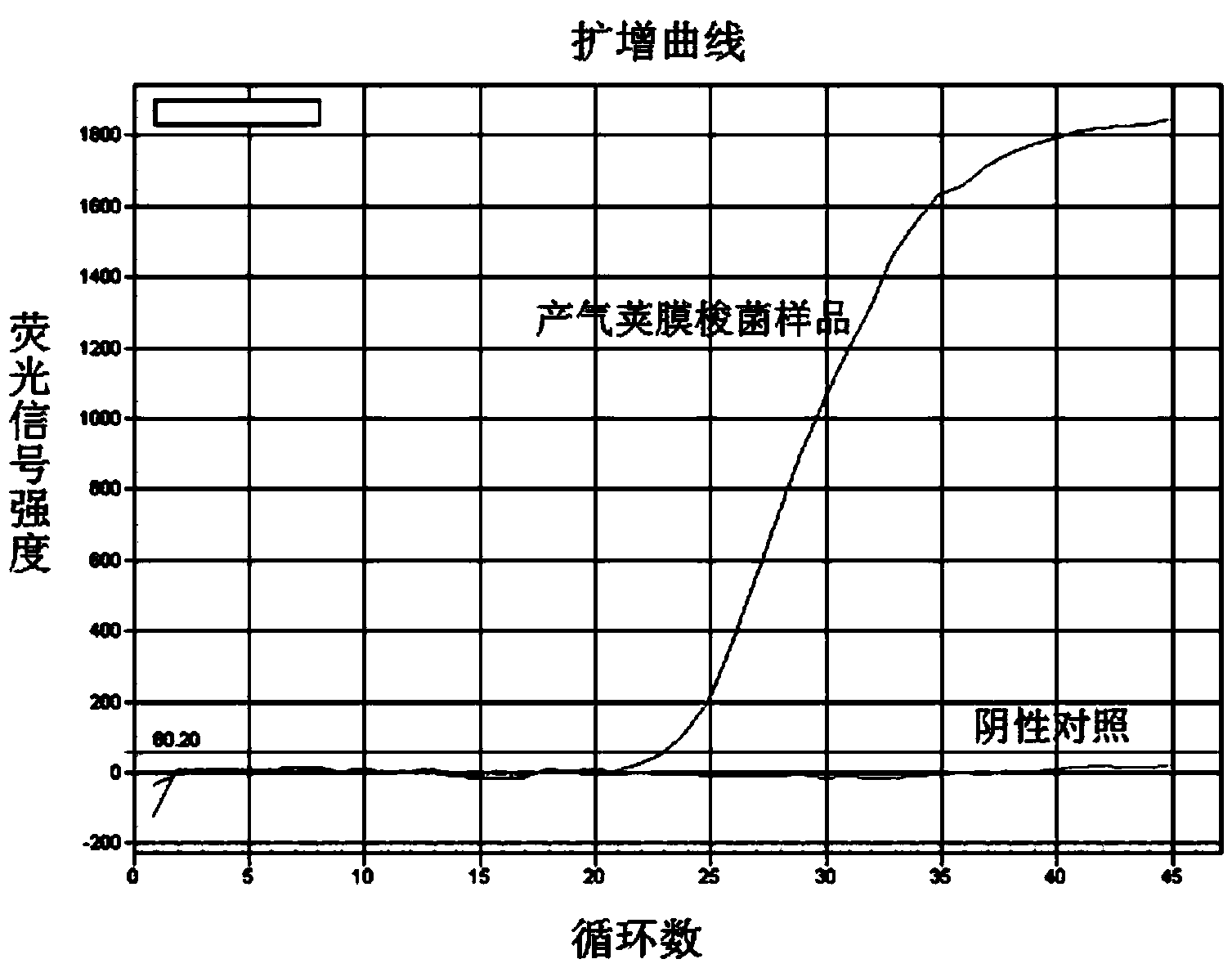
Quadruple fluorescent PCR (Polymerase Chain Reaction) detection kit for common chicken food-borne bacteria and application method thereof
InactiveCN103436626AQuick checkHigh detection sensitivityMicrobiological testing/measurementMicroorganism based processesDiseaseBacteroides
The invention discloses a quadruple fluorescent PCR (Polymerase Chain Reaction) detection kit for common chicken food-borne bacteria and an application method thereof. The kit contains a fluorescent PCR solution and standards of Clostridium perfringens, Salmonella enteritidis, Escherichia coli O157: H7 and Campylobacter jejuni, wherein the fluorescent PCR solution contains Taq DNA (deoxyribonucleic acid) polymerase, a PCR buffer solution, Mg<2+>, dNTPs (deoxy-ribonucleoside triphosphates), four pairs of bacterial specific primers and four kinds of corresponding fluorescent probes. According to the detection kit and the application method thereof, four kinds of chicken-carried main food-borne pathogenic bacteria, namely Clostridium perfringens, Salmonella enteritidis, Escherichia coli O157: H7 and Campylobacter jejuni, can be rapidly and accurately detected by using quadruple fluorescent PCR, and the detection kit can be applied to the assay of bacteria, the diagnosis of diseases and epidemiological investigation.
Owner:INST OF ANIMAL SCI & VETERINARY HUBEI ACADEMY OF AGRI SCI
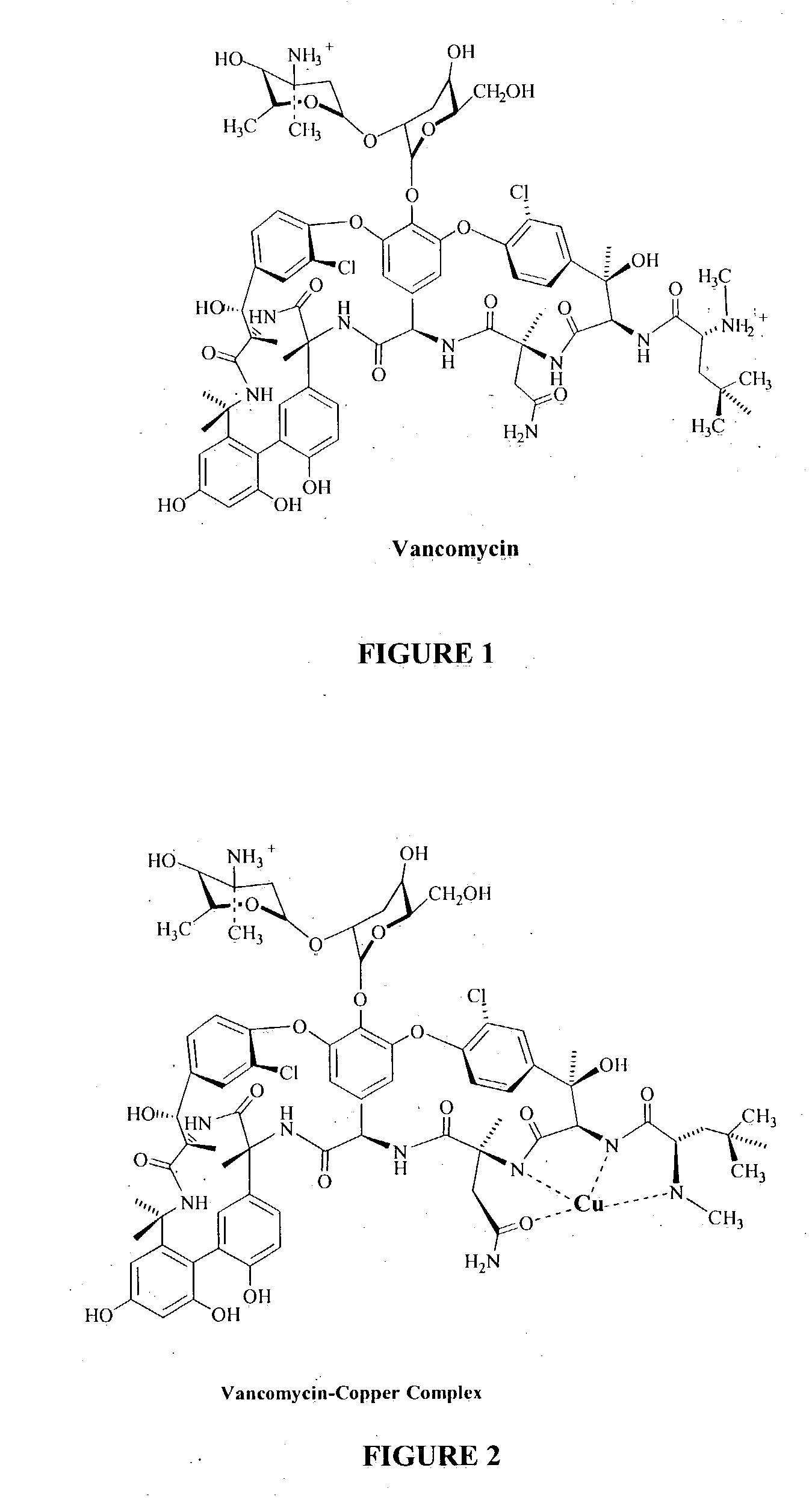
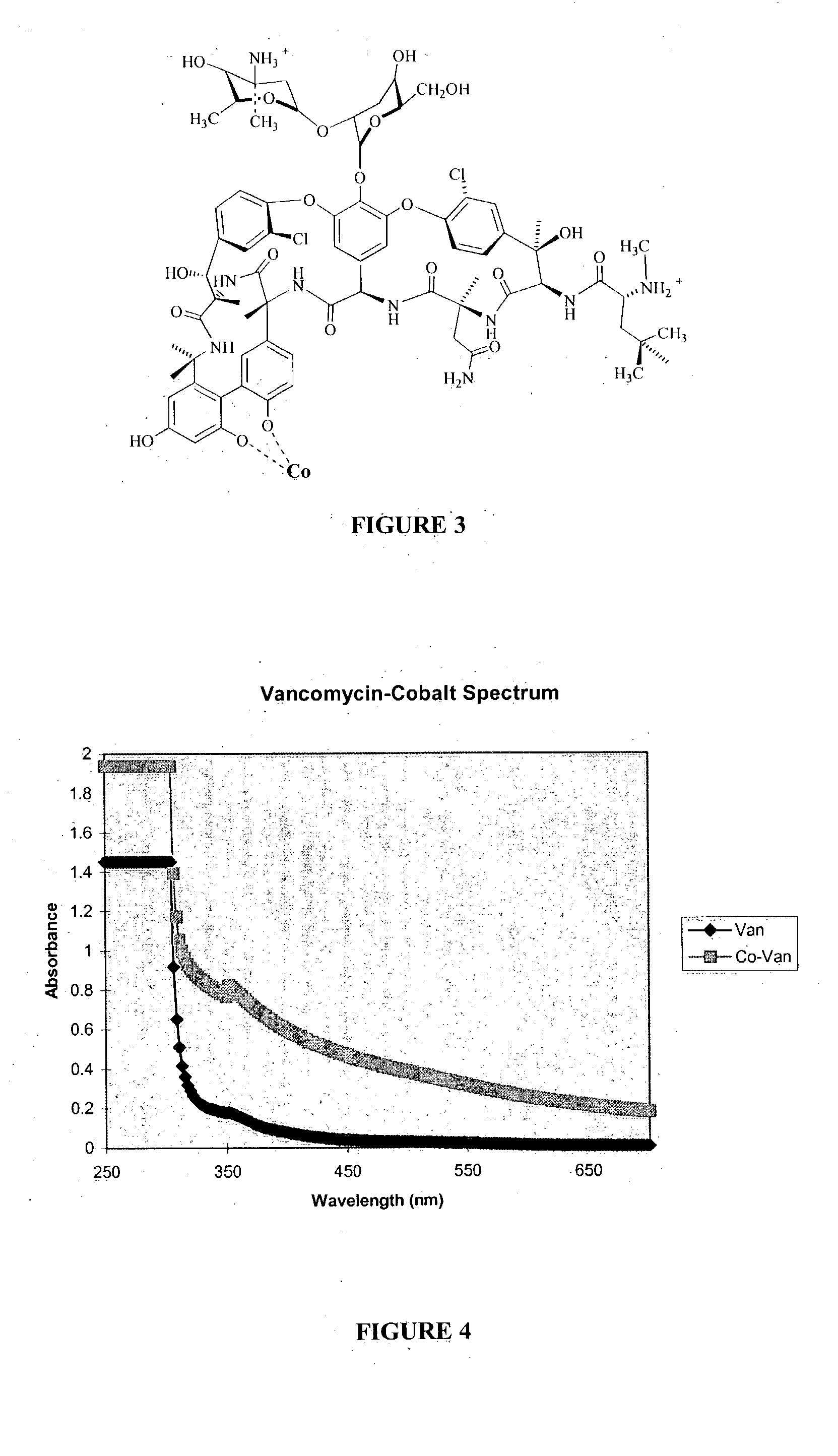
Antibiotic-metal complexes in the detection of gram-positive bacteria and other biological analytes
Complexes of antibiotics and metals are provided that are useful in detecting microorganisms, including gram-positive bacteria, Mycobacteria, permeabilized gram-negative bacteria, protozoans and other biological analytes, and are particularly useful in detecting gram-positive bacteria. The complexes are preferably chelated complexes wherein the antibiotic is a glycopeptide antibiotic, quinolone antibiotic, ribonucleoside antibiotic, mixtures thereof, and analogs and derivatives thereof, and (b) a detectable label comprising a transition or lanthanide metal. The complexes provide for chemiluminescent, fluorescent or magnetic detection of the analyte. Methods of synthesizing and using the complexes are also provided.
Owner:PARADIGM DIAGNOSTICS
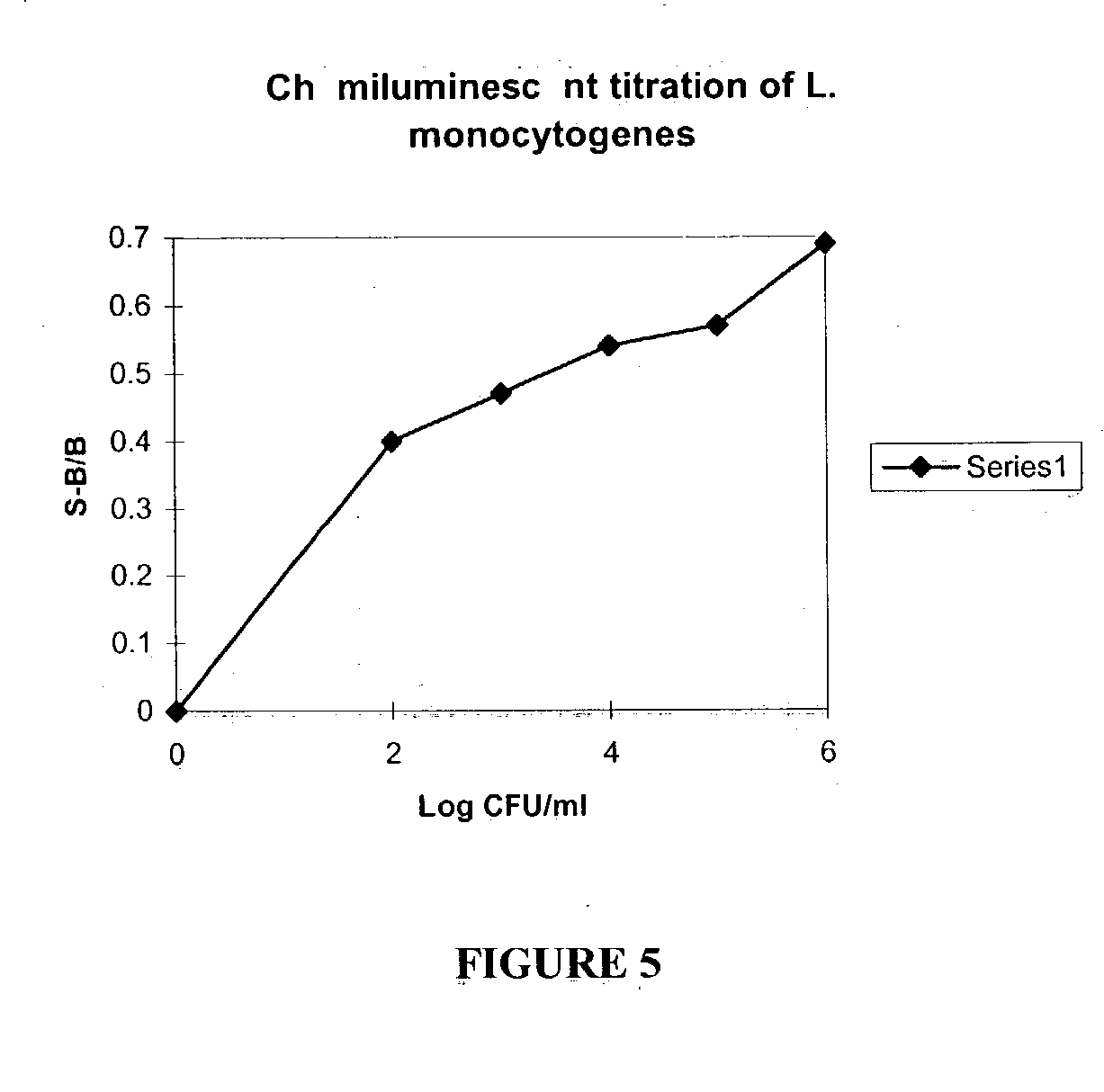
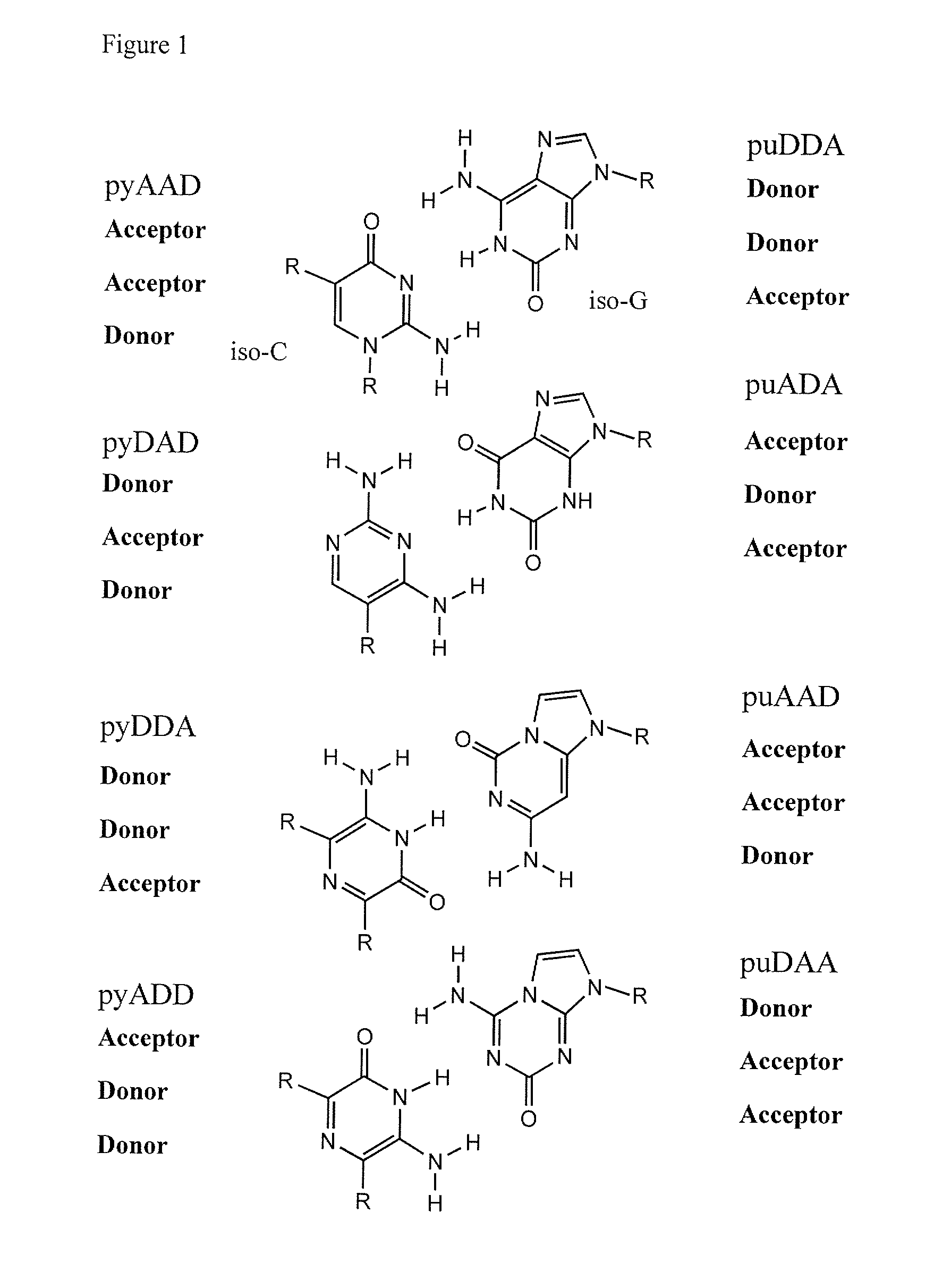
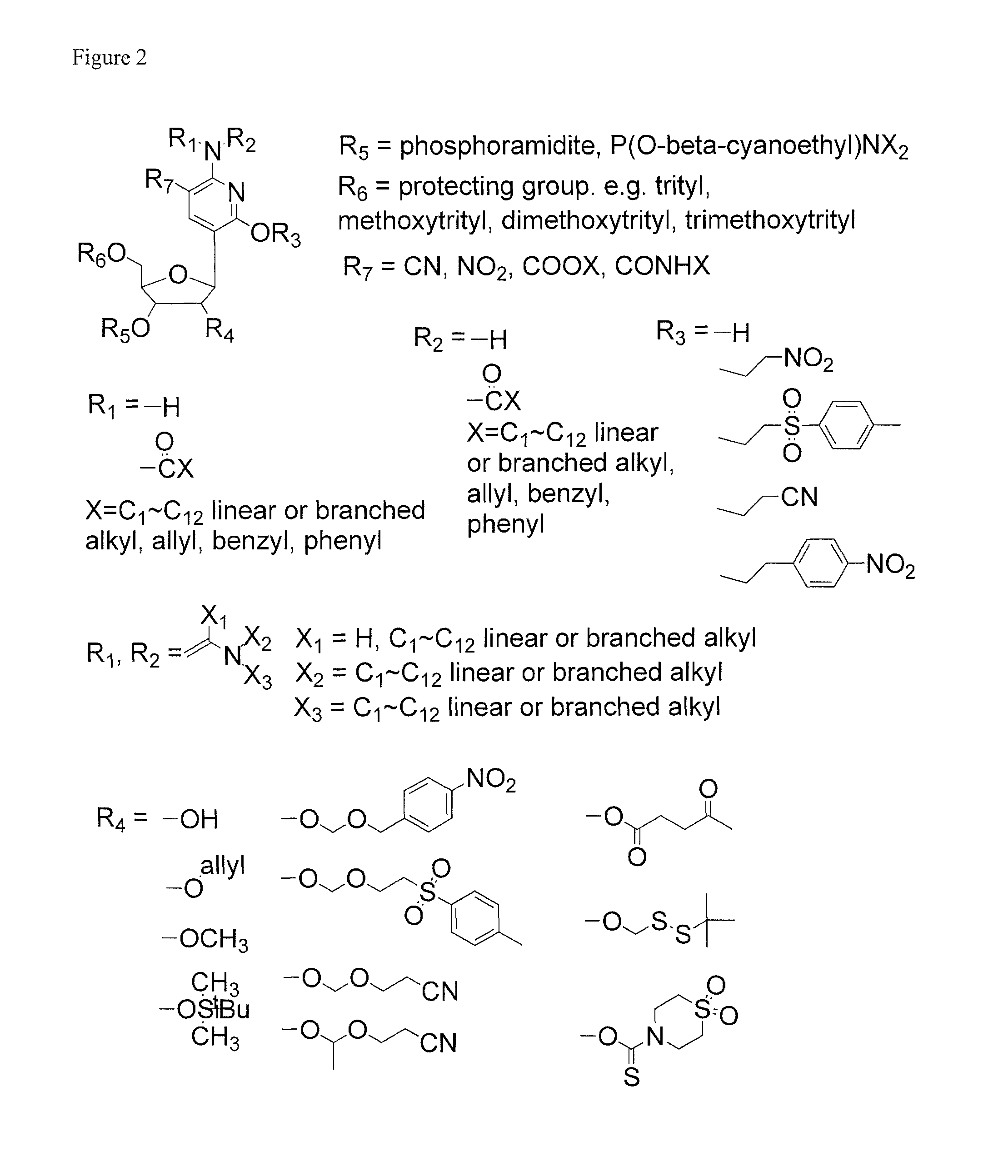
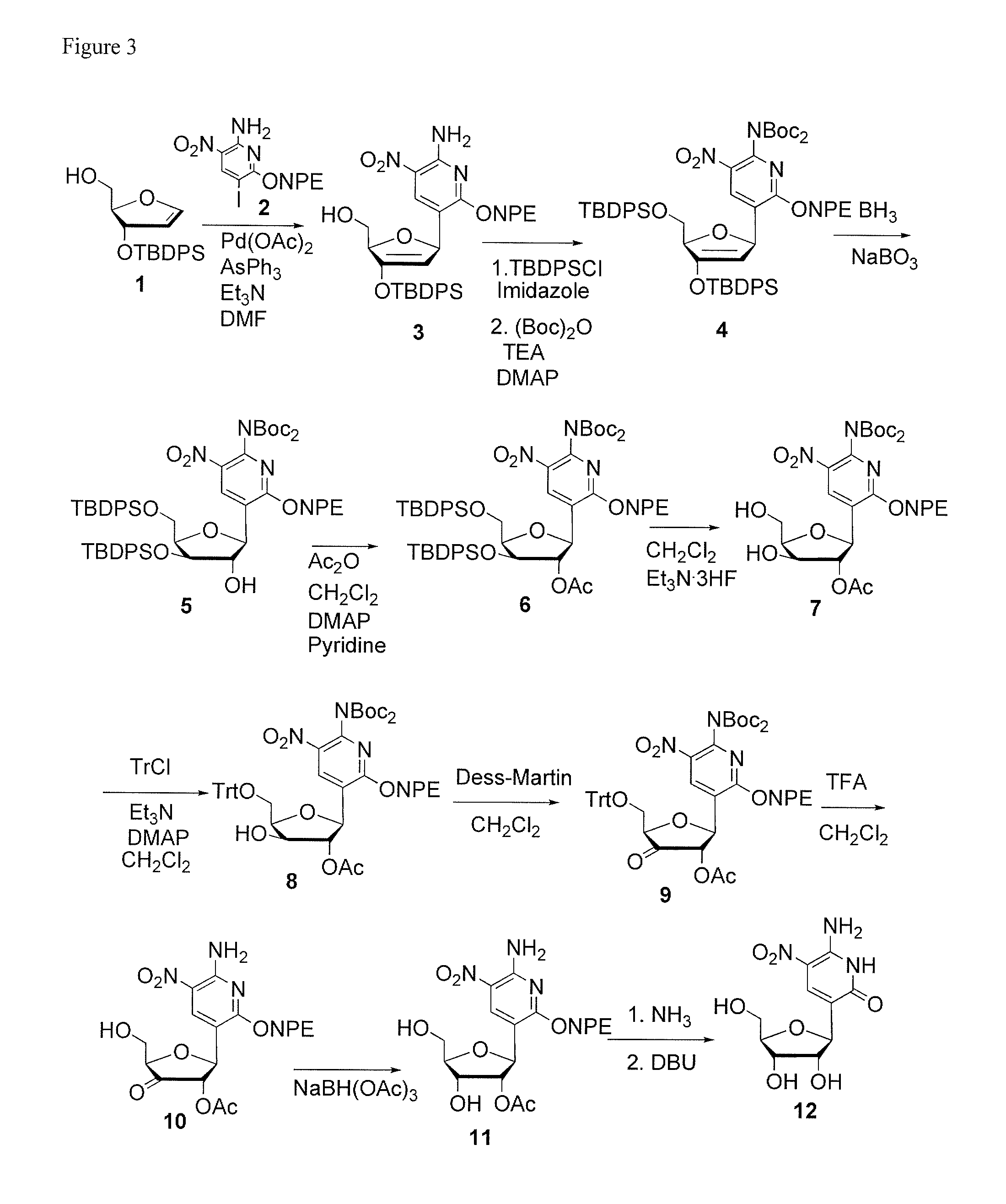
Ribonucleoside analogs with novel hydrogen bonding patterns
This invention relates to nucleoside, nucleotide, and oligonucleotide analogs that incorporate non-standard nucleobase analogs, defined to be those that present a pattern of hydrogen bonds to a paired nucleobase analog in a complementary strand that is different from the pattern presented by adenine, guanine, cytosine, and thymine. The invention is specifically concerned with nucleotide analogs that present the donor-donor-acceptor, hydrogen bonding patterns on pyrimidine analogs, and especially those that are analogs of ribonucleotides, including protected ribonucleotides suitable for phosphoramidite-based synthesis of RNA. The heterocycles on these nucleoside analogs are aminopyridones that have electron withdrawing groups attached to the position analogous to the 5-position of the ring in standard pyrimidines, including nitro, cyano, and carboxylic acid derivatives.
Owner:BENNER STEVEN A +1
Preparation method of oxidation coenzyme I
ActiveCN102605026AMild reaction conditionsEfficient reaction conditions under mild conditionsFermentationRibonucleosideAdenosine
The invention relates to a preparation method of an oxidation coenzyme I. According to the method, nicotinamide ribonucleoside (NR) and adenosine disodium triphosphate (ATP-Na2) react with each other to obtain the oxidation coenzyme I (NAD+, Nicotinamide Adenine Dinucleotide+) in a buffer solution with the pH of 5.0 to 8.0 at a temperature of 30 DEG C to 40 DEG C under the catalytic action of nicotinamide ribonucleoside kinase (NRK) under the condition of the existence of divalent metal ions. According to the invention, a biocatalysis method is adopted, the oxidation coenzyme is prepared by utilizing an NRK one pot process, the reaction system is simple, the conditions are mild, and the preparation method has wide industrialization application prospect.
Owner:ENZYMEWORKS
LAMP (loop-mediated isothermal amplification) rapid detection kit and detection method for salmonella
InactiveCN102199665AResolution cycleResolve SensitivityMicrobiological testing/measurementAgainst vector-borne diseasesRibonucleosideFluorescence
The invention belongs to the field of biological products, and relates to a detection kit and a detection method for rapidly detecting salmonella by utilizing an LAMP (loop-mediated isothermal amplification) technique. The kit is composed of a salmonella genome extracting reagent and an LAMP reaction reagent, wherein the salmonella genome extracting reagent comprises the following components: SDS (sodium dodecyl sulfate), protease K, CTAB (cetyltrimethyl ammonium bromide) / NaCl solution and NaAC; and the LAMP reaction reagent is composed of a reaction liquid A and a reaction liquid B, the reaction liquid A comprises the following components: 10*Lampbuffer, Bst DNA (deoxyribonucleic acid) polymerase, dNTP (deoxy-ribonucleoside triphosphate), an interprimer 1, an interprimer 2, an outer primer 1, an outer primer 2, lycine and ultrapure water, and the reaction liquid B is a fluorescent dye. The invention can be used to carry out LAMP detection on salmonella, has the characteristics of rapid detection, high accuracy, high sensitivity and convenient field application, and can be widely applied in the fields of veterinarian, food and exit-entry quarantine, etc.
Owner:镇江出入境检验检疫局检验检疫综合技术中心
Hypertension gene polymorphism fluorescent PCR (polymerase chain reaction) solubility curve detecting kit and application thereof
PendingCN109457024AEasy to operateShorten the timeMicrobiological testing/measurementDNA/RNA fragmentationSolubilityRibonucleoside
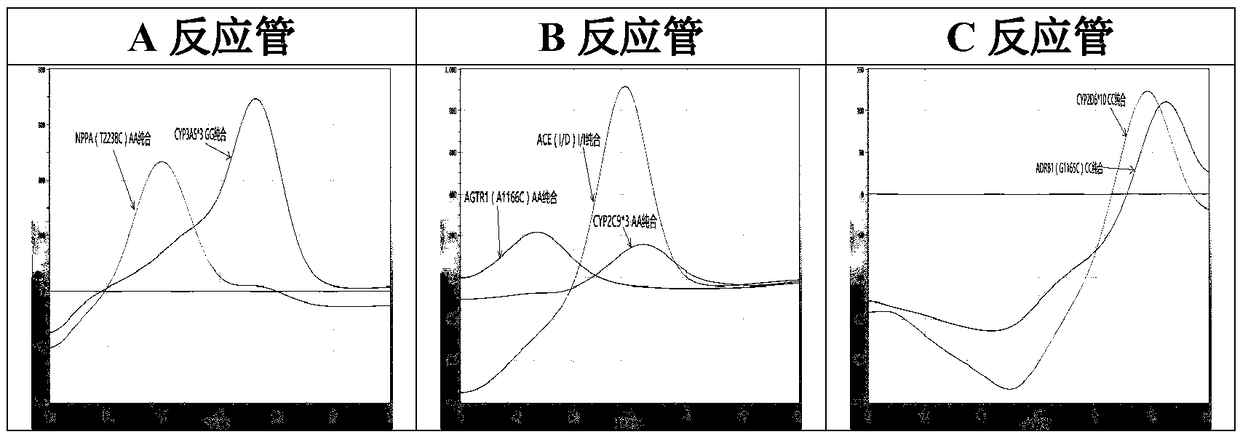
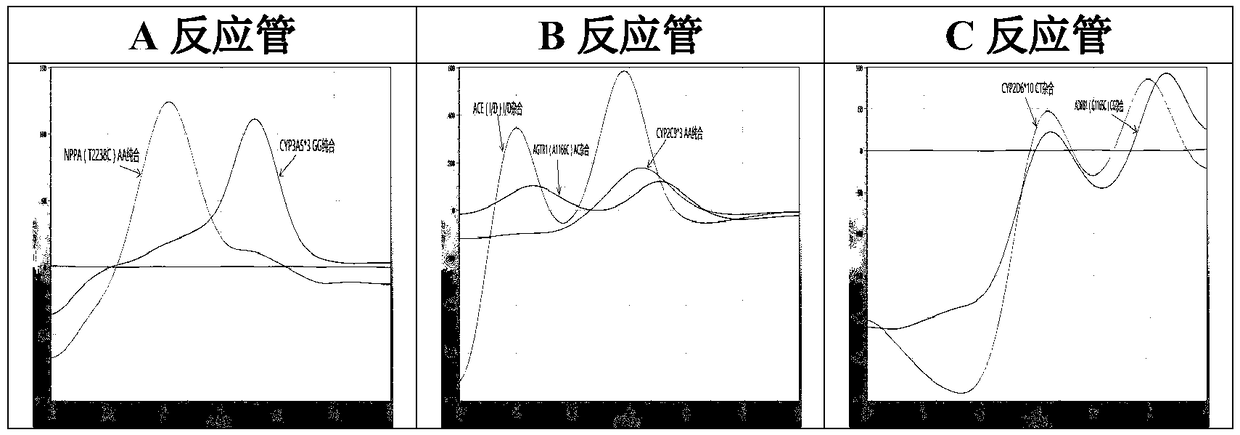
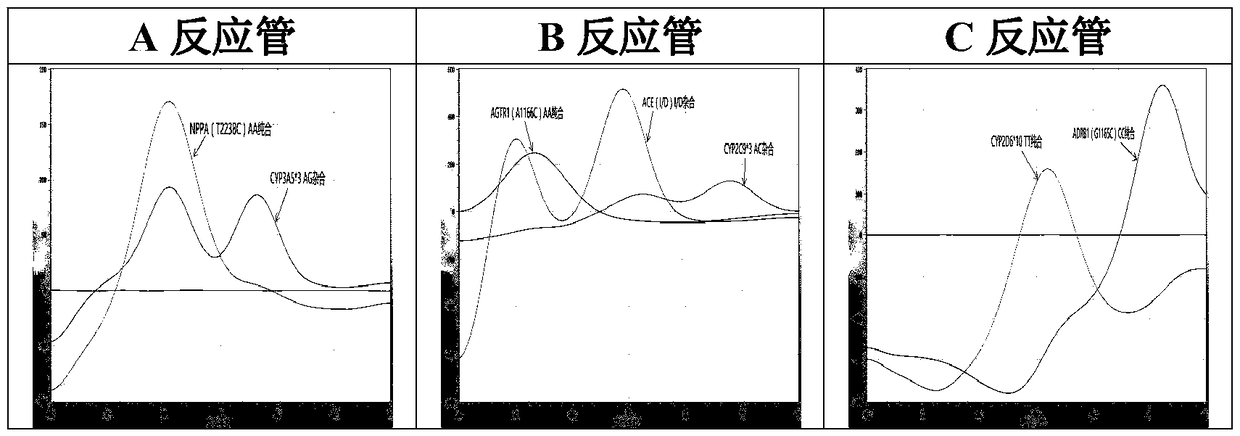
The invention relates to a hypertension gene polymorphism fluorescent PCR solubility curve detecting kit and application thereof, and a hypertension medication gene multiple-PCR solubility curve detecting method. The hypertension gene polymorphism fluorescent PCR solubility curve detecting kit mainly comprises a primer combination, a probe combination, and PCR reaction agents such thermally activated polymerases, PCR buffer solution and dNTPs (deoxy-ribonucleoside triphosphates) and involves polymorphism detection of 7 hypertension-related genes including CYP2D6*10, CYP2C9*3, ADRB1(1165G)C), AGTR1(1166A)C), CYP3A5*3, NPPA(T2338C) and ACE(I / D); the three reaction agents are applied to perform PCR amplification and solubility analysis to determine the related polymorphism of the 7 genes to guide clinical customized application of 5 majors types of hypertension drugs. The hypertension gene polymorphism fluorescent PCR solubility curve detecting kit can easily complete detection through 3reaction tubes, and meanwhile, save a nucleic acid extraction process, simplify operation steps and achieve a simple interpretation method.
Owner:众福健康科技(杭州)有限公司
Conjugated double strand compositions for use in gene modulation
InactiveUS20090306178A1Increased nuclease resistanceIncrease resistanceOrganic active ingredientsDrug compositionsRegulator geneDouble strand
The present invention provides conjugated double stranded compositions wherein each strand is modified to have a motif defined by positioning of β-D-ribonucleosides and / or sugar modified nucleosides. More particularly, the present compositions comprise a linked conjugate group on one strand and a non hybridizing region of 2′-modified nucleosides on the other strand. Each strand further comprises one or more phosphorothioate internucleoside linkage. The compositions are useful for targeting selected nucleic acid molecules and modulating the expression of one or more genes. In preferred embodiments the compositions of the present invention hybridize to a portion of a target RNA resulting in loss of normal function of the target RNA. The present invention also provides methods for modulating gene expression.
Owner:IONIS PHARMA INC
Prevention of and countermeasures against mitochondrial disease
InactiveUS20050256186A1Reduce adverse effectsGood effectBiocideNervous disorderDiseaseRibonucleoside
The present invention relates to an agent containing L-arginine, 0.2 to 20 parts by weight of L-ascorbic acid per 1 part by weight of L-arginine, and, if desired, at least one selected from the group consisting of ribonucleic acids, ribonucleotides and ribonucleosides. The agent can treat mitochondrial disease which shows a variety of symptoms caused by dysfunction of mitochondria in cells. L-arginine contained in the agent for treating mitochondrial disease of the present invention increases an NO radical level to thereby dilate the arteries. L-ascorbic acid serves to mitigate a harsh taste and acrid feeling accompanied with the intake of L-arginine and eliminate excessive NO radicals.
Owner:MORISHIGE FUMIE
Method for determining DNA nucleotide sequence
PCT No. PCT / JP95 / 02254 Sec. 371 Date May 5, 1997 Sec. 102(e) Date May 5, 1997 PCT Filed Nov. 6, 1995 PCT Pub. No. WO96 / 14434 PCT Pub. Date May 17, 1996Disclosed is a method for determining a nucleotide sequence of DNA product amplified by polymerase chain reaction not requiring removal of primers and / or 2'-deoxyribonucleoside-5'-triphosphates and / or derivatives thereof, which comprises reacting ribonucleoside-5'-triphosphates comprising ATP, GTP, CTP, UTP and derivatives thereof and one or more of 3'-deoxyribonucleotide-5'-triphosphates comprising 3'-dATP, 3'-dGTP, 3'-dCTP, 3'-dUTP and derivatives thereof in the presence of an RNA polymerase and a DNA product which has been amplified by polymerase chain reaction and contains a promoter sequence for the RNA polymerase to afford a nucleic acid transcription product, separating the obtained nucleic acid transcription product and determining a nucleic acid sequence from the resluting separated fractions.
Owner:RIKEN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com