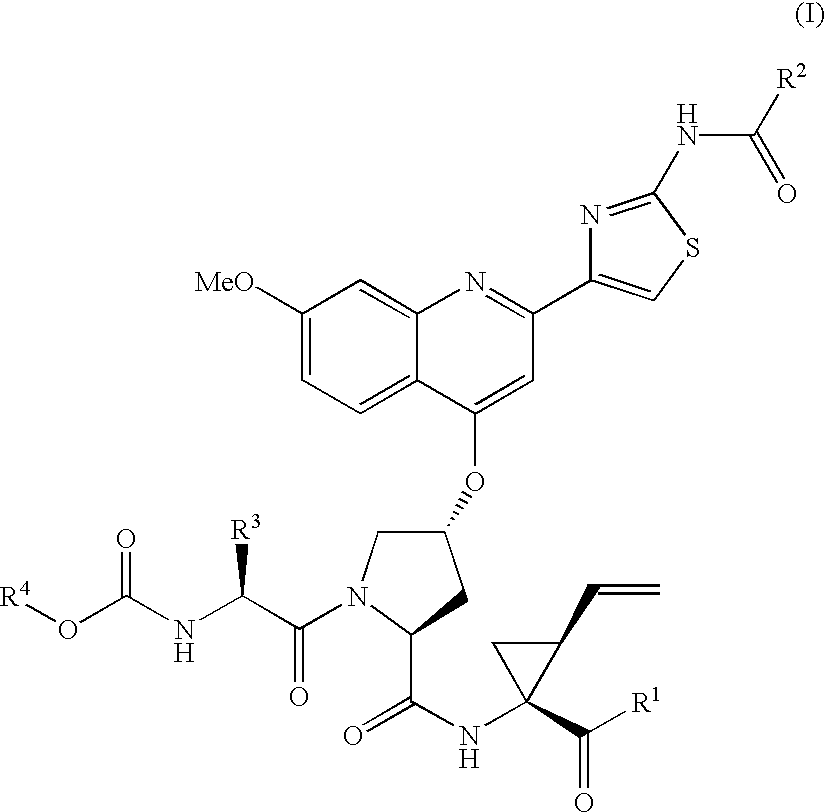
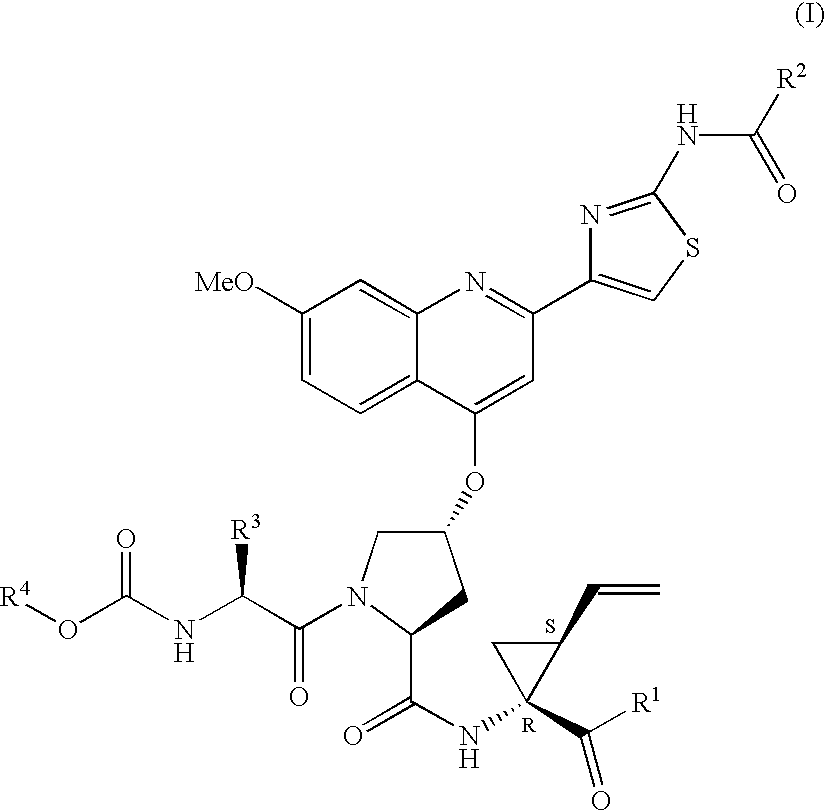
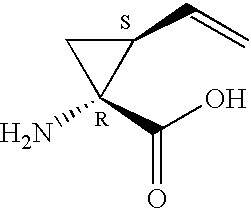
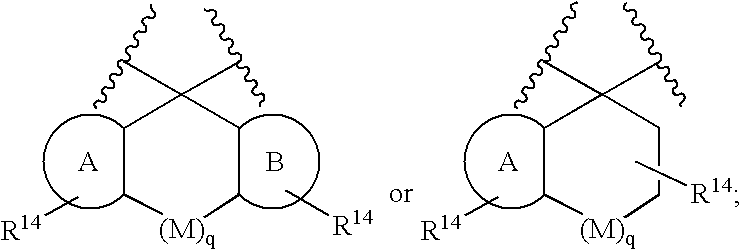
Patents
Literature
11394 results about "Protease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
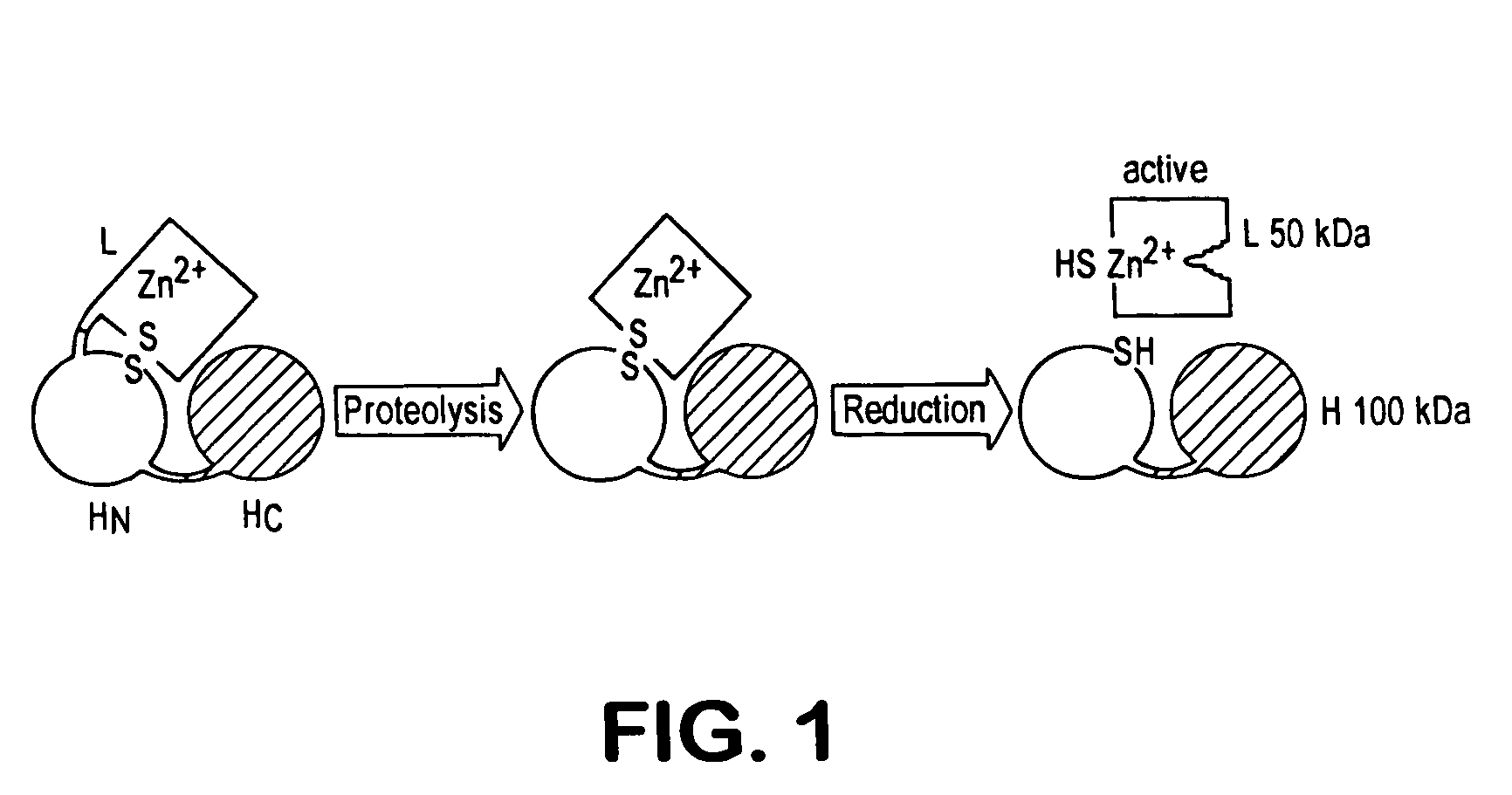
A protease (also called a peptidase or proteinase) is an enzyme that catalyzes (increases the rate of) proteolysis, the breakdown of proteins into smaller polypeptides or single amino acids. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of eaten proteins, protein catabolism (breakdown of old proteins), and cell signalling.
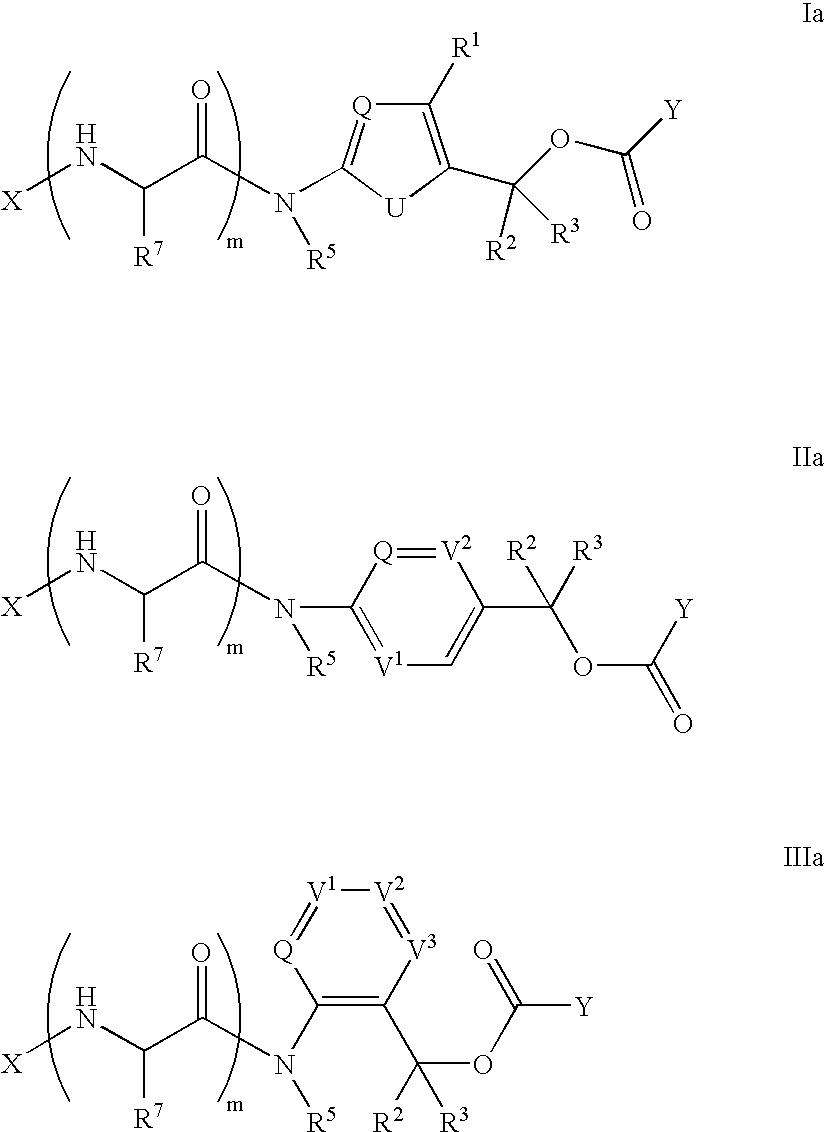
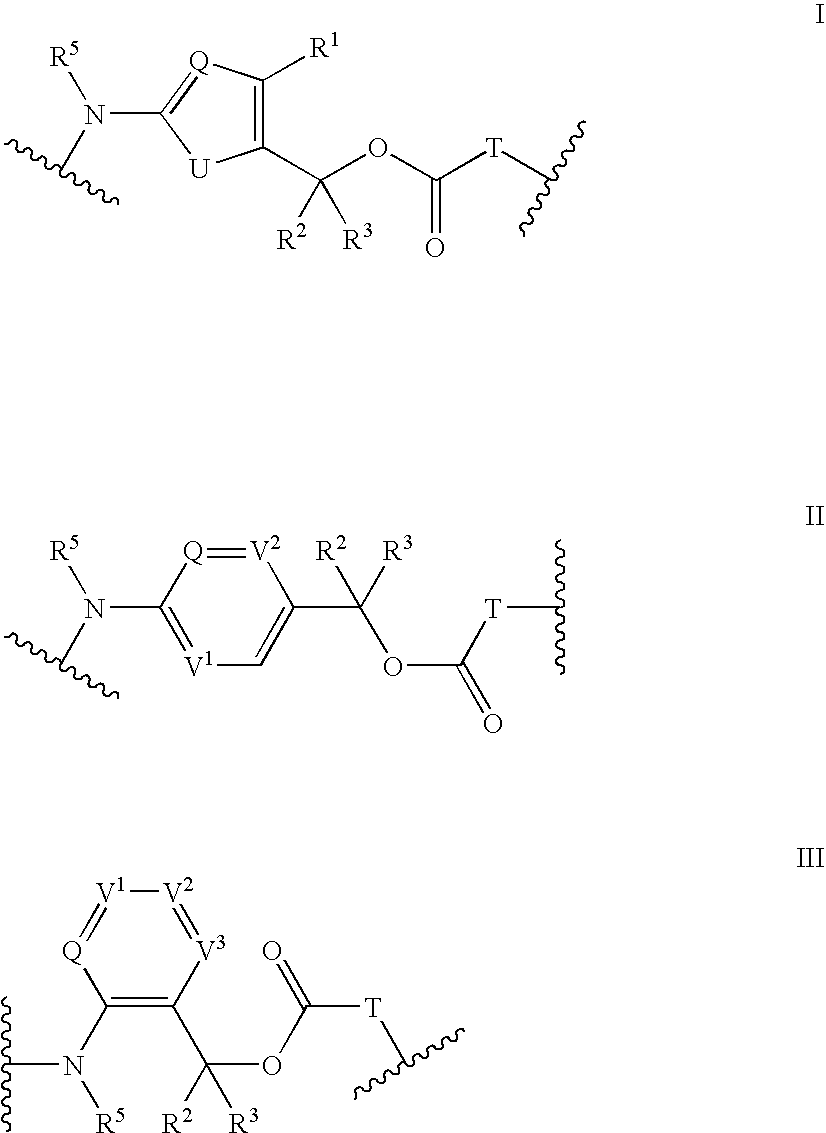
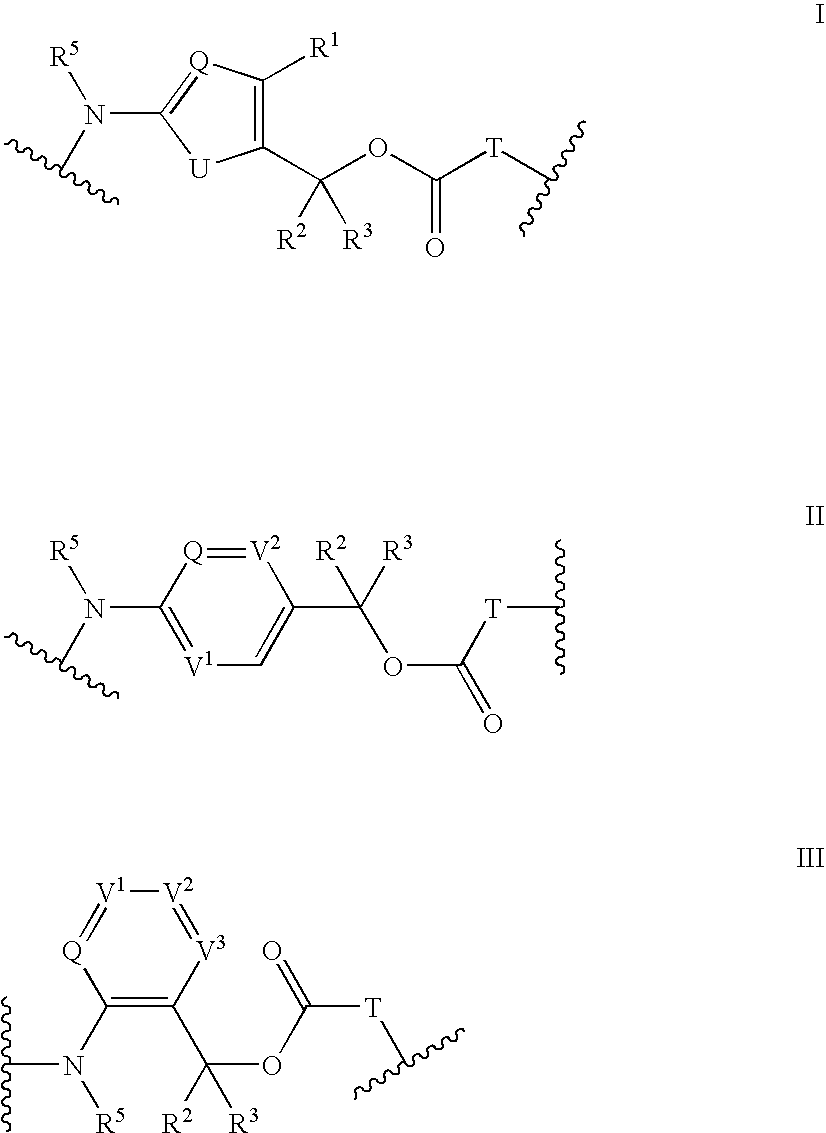
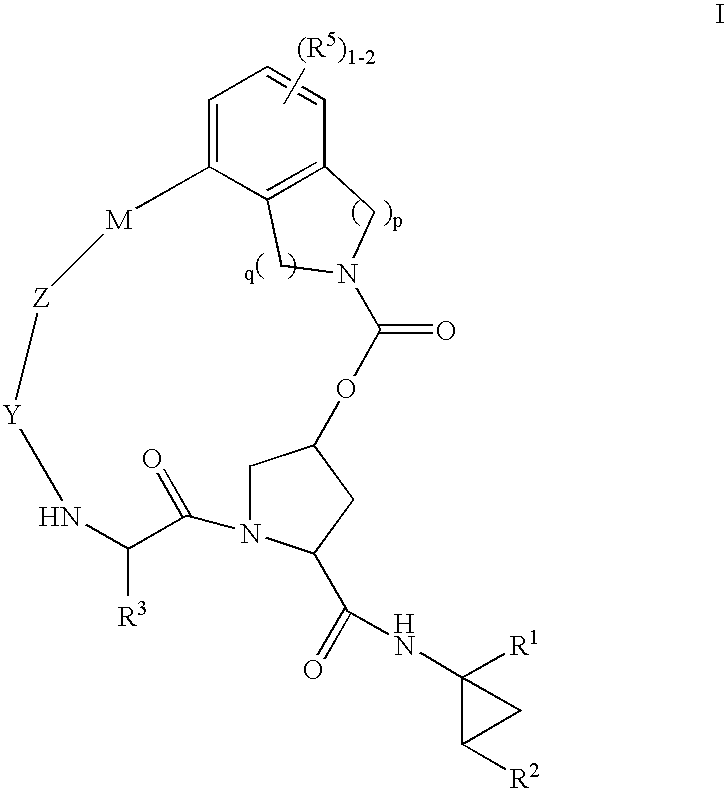
Heterocyclic self-immolative linkers and conjugates
The present invention provides heterocyclic linker compounds useful for linking drug moieties to ligands. The compounds also include drug-ligand conjugates comprising a ligand capable of targeting a selected cell population, and a drug connected to the ligand by a heterocyclic linker moiety. The linker moiety comprises a peptide sequence that is a substrate for an intracellular enzyme, for example a cathepsin, that cleaves the peptide at an amide bond. The peptide further contains a self-immolating moiety which connects the drug and the protein peptide sequence. Upon cleavage of the peptide sequence by an intracellular enzyme the self-immolating moiety cleaves itself from the drug moiety such that the drug moiety is in an underivatized and active form.
Owner:SEAGEN INC
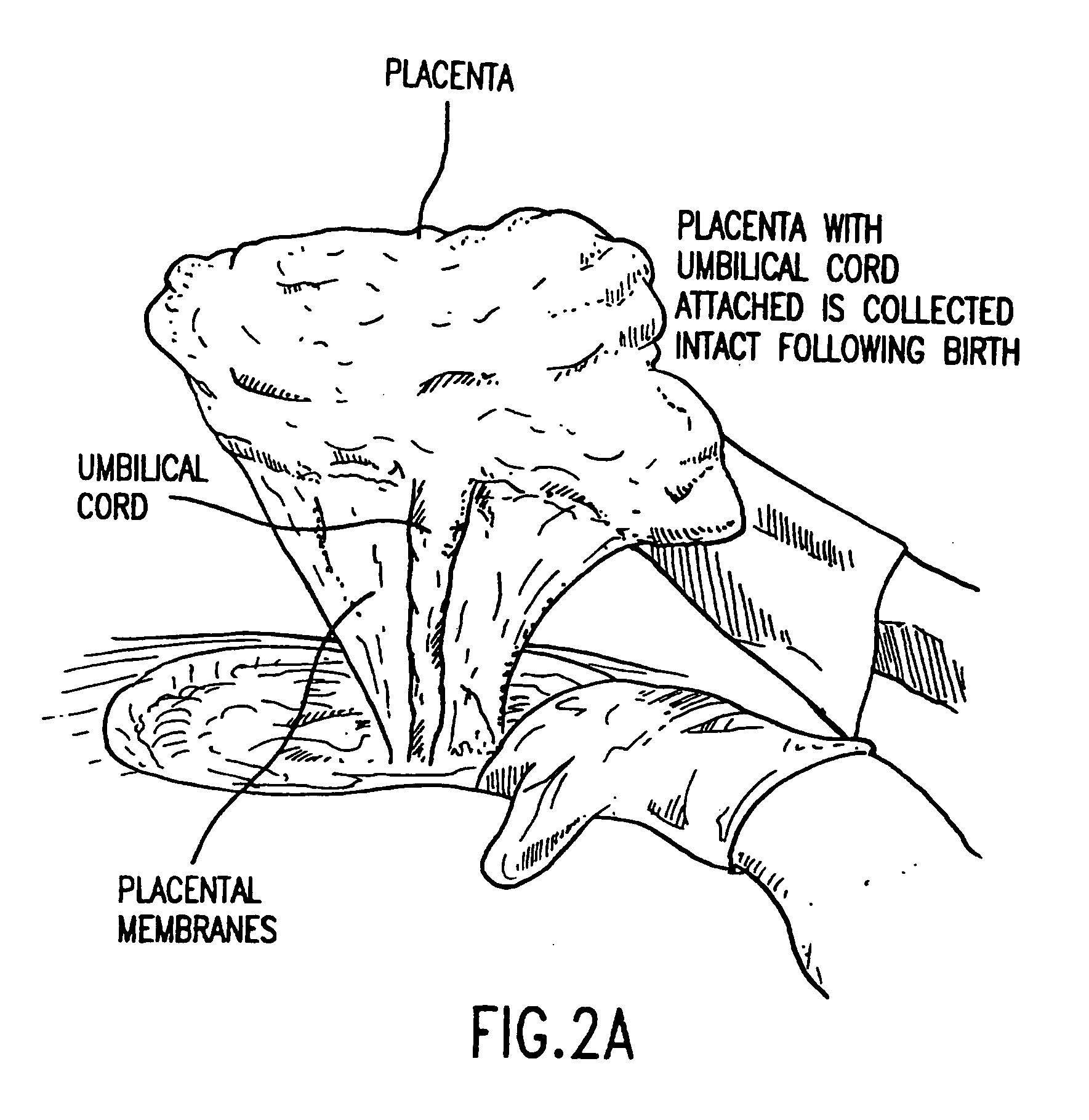
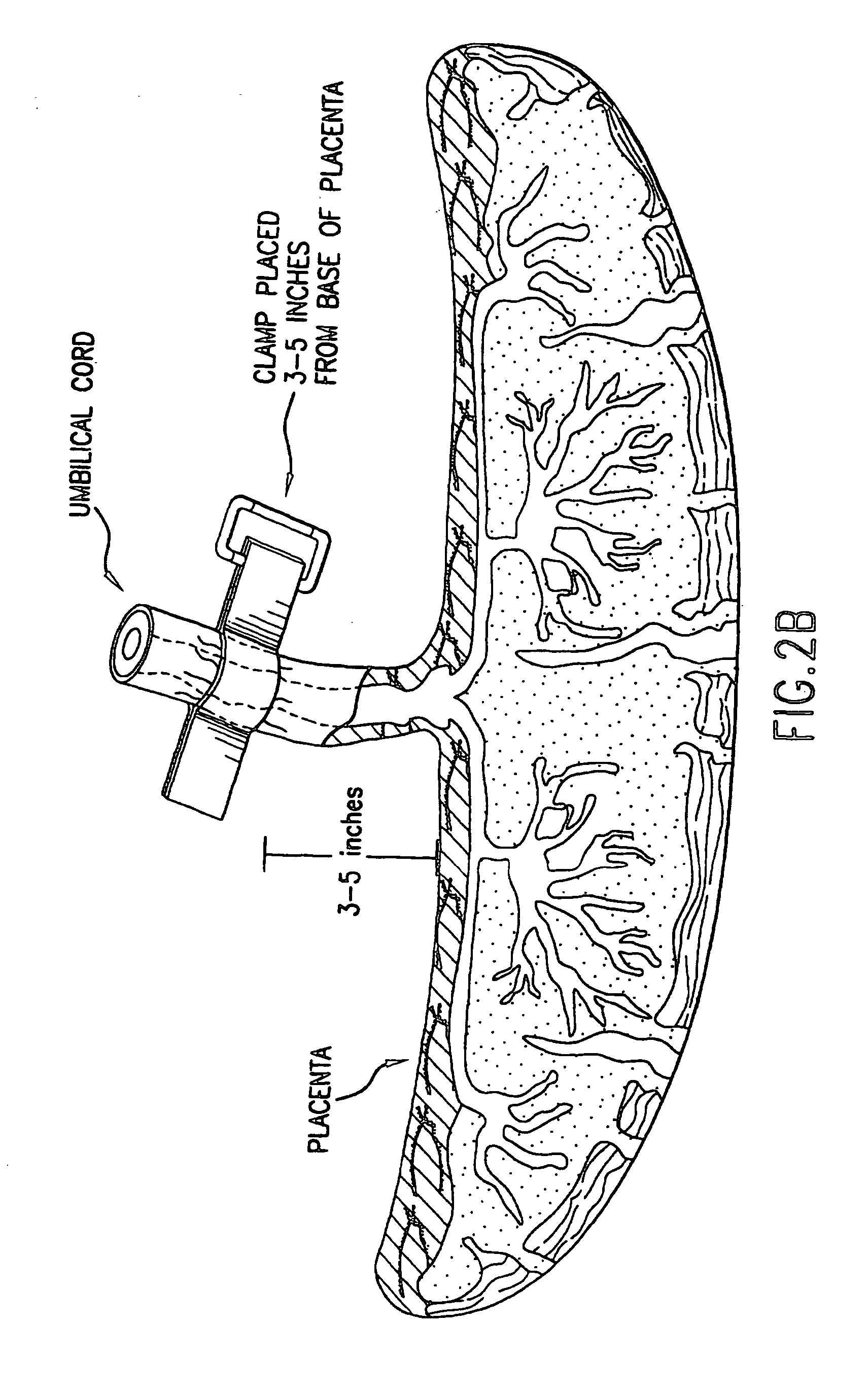
Composition for collecting and preserving placental stem cells and methods of using the composition
The present invention provides improved compositions and methods for the collection of stem cells from an organ, e.g., placenta. The invention provides a stem cell collection composition comprising an apoptosis inhibitor and, optionally, an enzyme such as a protease or mucolytic enzyme, vasodilator, necrosis inhibitor, oxygen-carrying perfluorocarbon, or an organ preserving compound. The invention provides methods of using the stem cell collection composition to collect stem cells and to preserve populations of stem cells.
Owner:CELULARITY INC
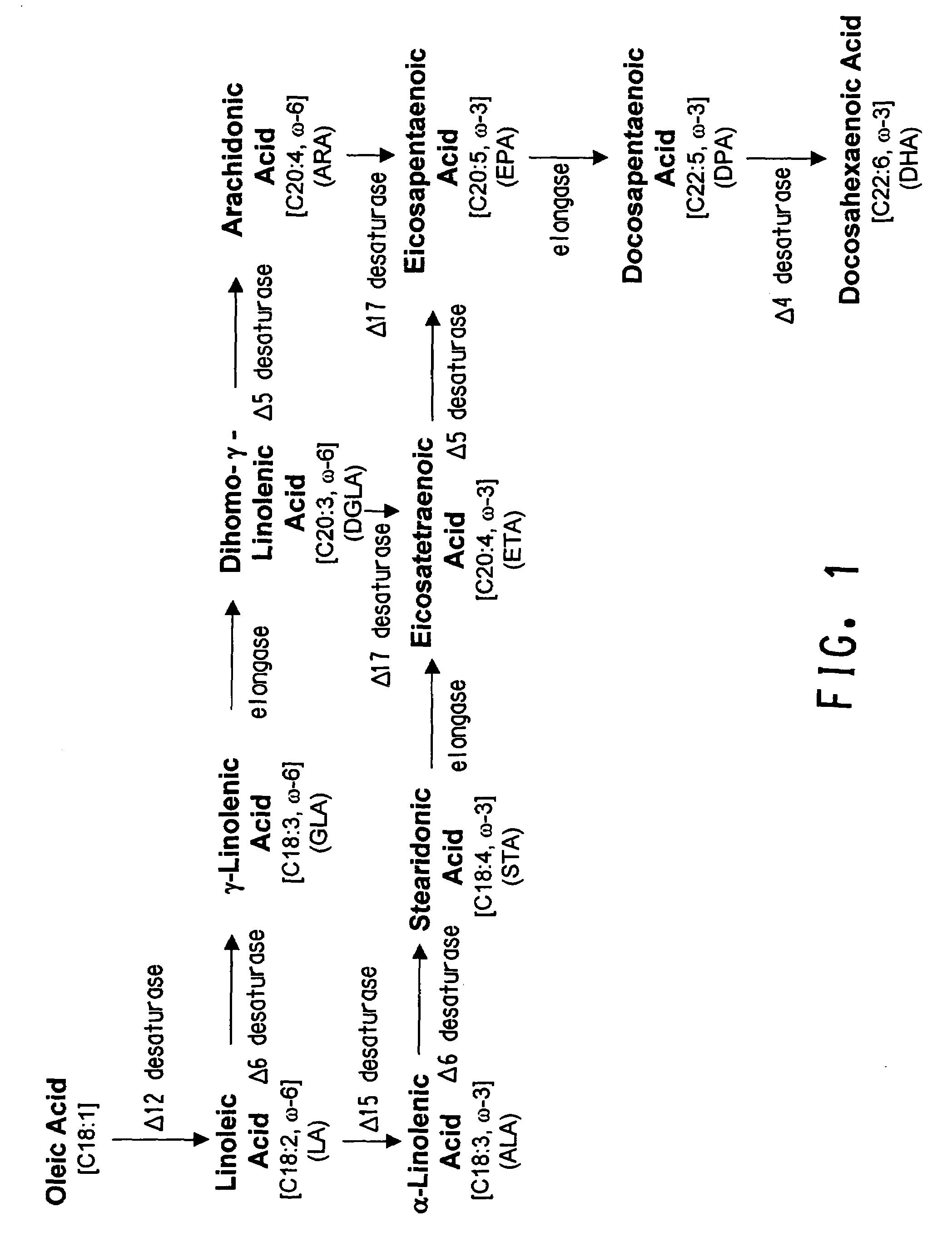
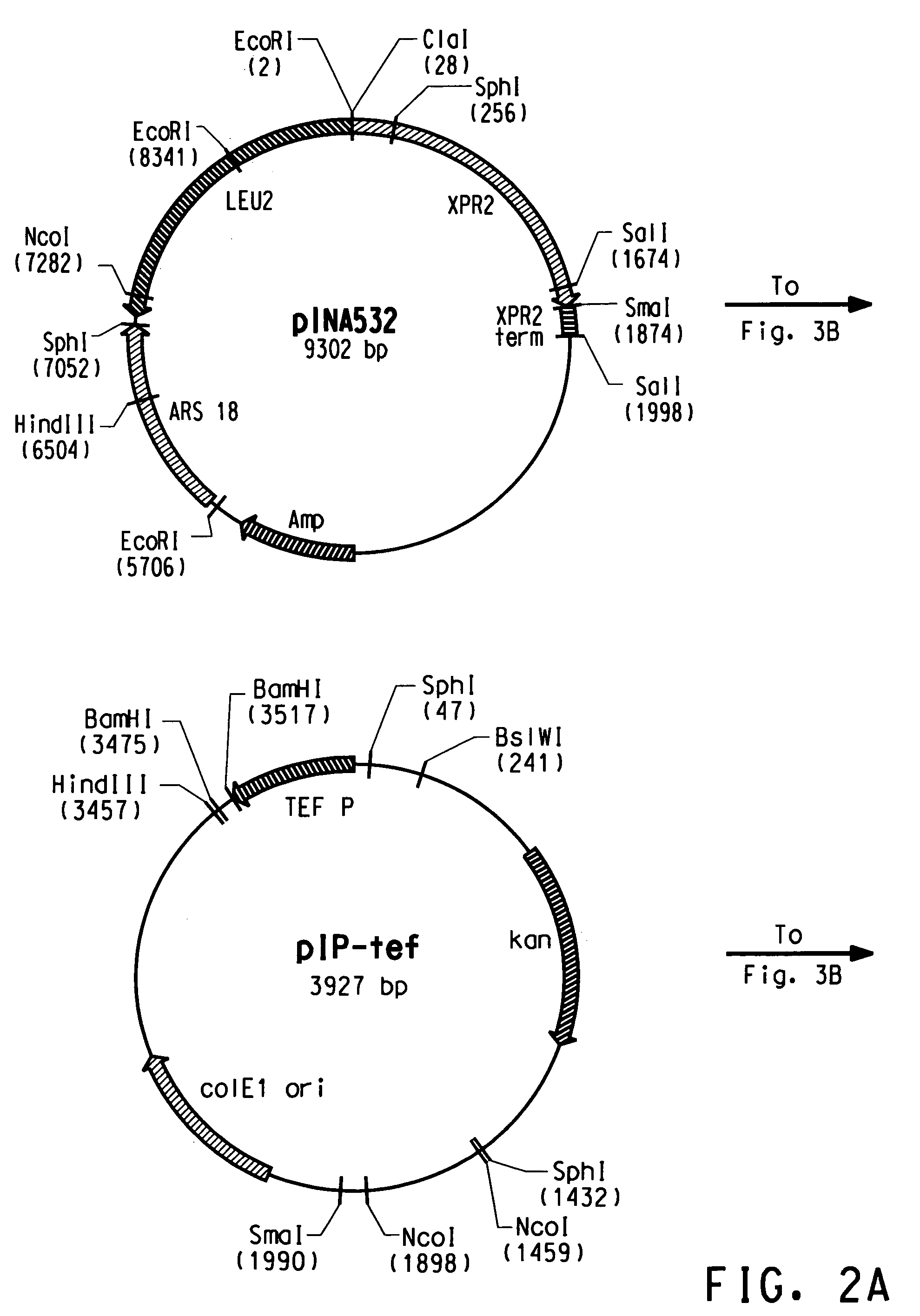
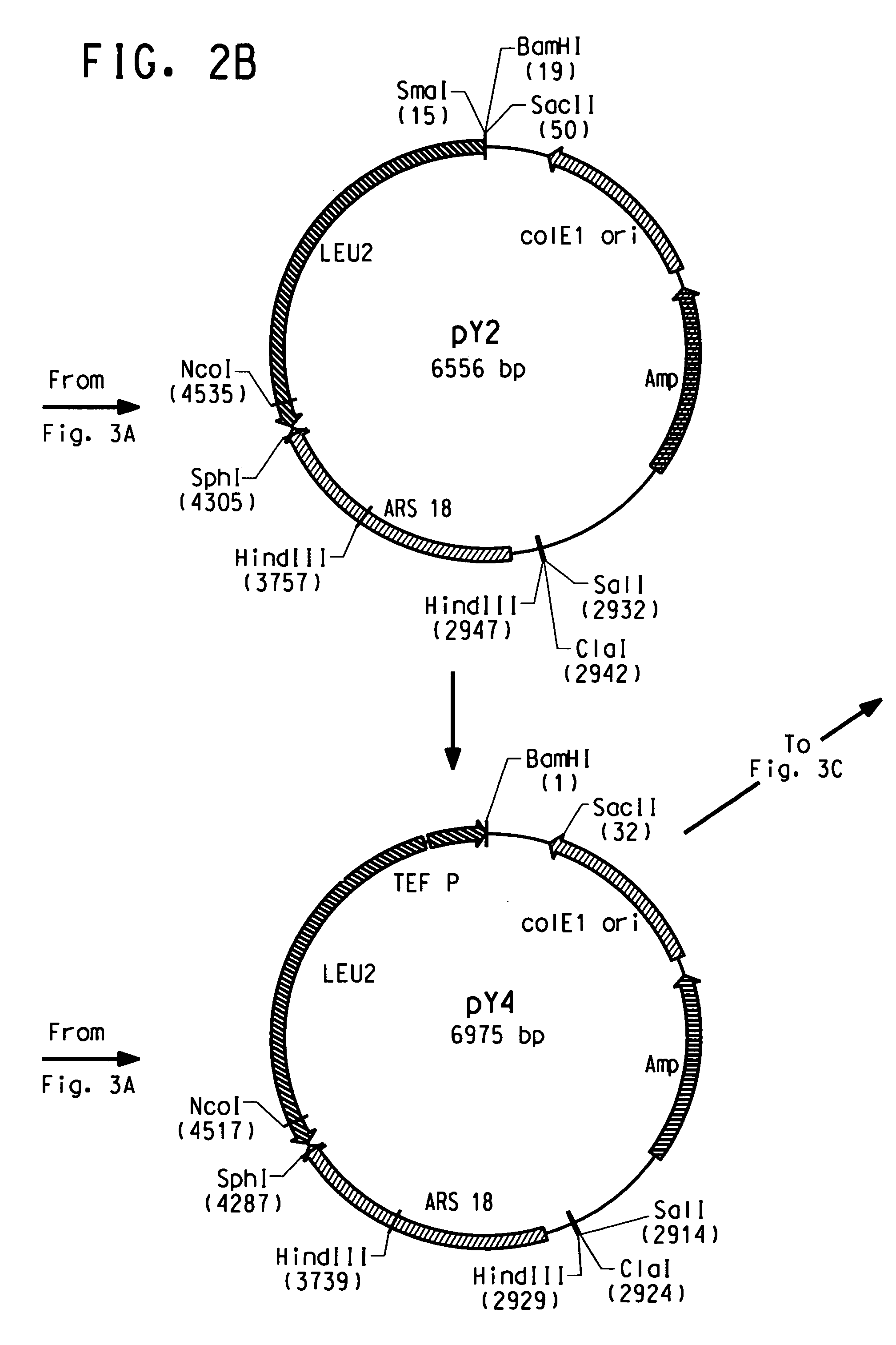
Codon-optimized genes for the production of polyunsaturated fatty acids in oleaginous yeasts
The present invention relates to fatty acid desaturases and elongases able to catalyze the conversion of linoleic acid (LA) to γ-linolenic acid (GLA); α-linoleic acid (ALA) to stearidonic acid (STA); GLA to dihomo-γ-linoleic acid (DGLA); STA to eicosatetraenoic acid (ETA); DGLA to ETA; eicosapentaenoic acid (EPA) to docosapentaenoic acid (DPA); and arachidonic acid (ARA) to EPA. Nucleic acid sequences encoding codon-optimized desaturases and elongases, nucleic acid sequences which hybridize thereto, DNA constructs comprising the codon-optimized desaturase or elongases, and recombinant host microorganisms expressing increased levels of desaturase or elongase are described.
Owner:EI DU PONT DE NEMOURS & CO
Bacterial expression of protease inhibitors and variants thereof
ActiveUS20050202535A1Reduce disulfide bondBacteriaPeptide/protein ingredientsProteinase activityProtease
Owner:GENENCOR INT INC
Dipeptidyl peptidase inhibitors
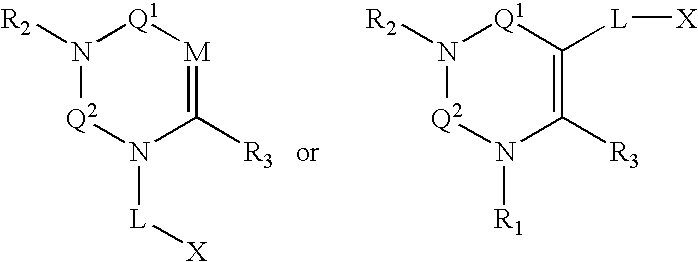
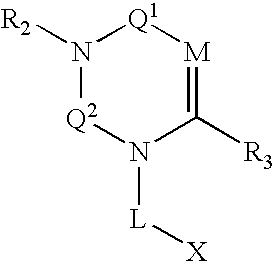
Compounds, pharmaceuticals, kits and methods are provided for use with DPP-IV and other S9 proteases that comprise a compound comprising: wherein M is N or CR4; Q1 and Q2 are each independently selected from the group consisting of CO, SO, SO2, and C═NR9; and each L, X, R1, R2, and R3 are as defined herein.
Owner:TAKEDA PHARMA CO LTD
Nucleic acid arrays for detecting gene expression associated with human osteoarthritis and human proteases
InactiveUS20050118625A1Help studyIncrease heightBioreactor/fermenter combinationsMaterial nanotechnologyProteinase activityPolynucleotide
The present invention provides nucleic acid arrays and methods of using the same for expression profiling of human protease and / or osteoarthritis genes. The nucleic acid arrays of the present invention include one or more substrate supports. A substantial portion of all polynucleotide probes that are stably attached to the substrate support(s) can hybridize under stringent or nucleic acid array hybridization conditions to human protease or osteoarthritis genes. In one embodiment, the nucleic acid arrays of the present invention include a plurality of probe sets, each of which can hybridize under stringent or nucleic acid array hybridization conditions to a different respective tiling sequence selected from Attachment C, or the complement thereof.
Owner:WYETH LLC
Hepatitis C inhibitor tri-peptides
ActiveUS7091184B2Better pharmacokinetic profileNot significant inhibitory activityBiocideDipeptide ingredientsHcv ns3 proteaseHepatitis C
Compounds of formula (I):wherein R1 is hydroxyl or sulfonamide derivative; R2 is t-butyl or —CH2—C(CH3)3 or —CH2-cyclopentyl; R3 is t-butyl or cyclohexyl and R4 is cyclobutyl, cyclopentyl or cyclohexyl; or a pharmaceutically acceptable salt thereof, are described as useful as inhibitor of the HCV NS3 protease.
Owner:BOEHRINGER INGELHEIM INT GMBH
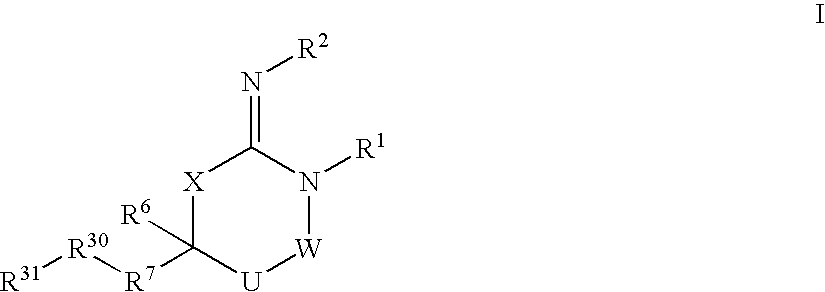
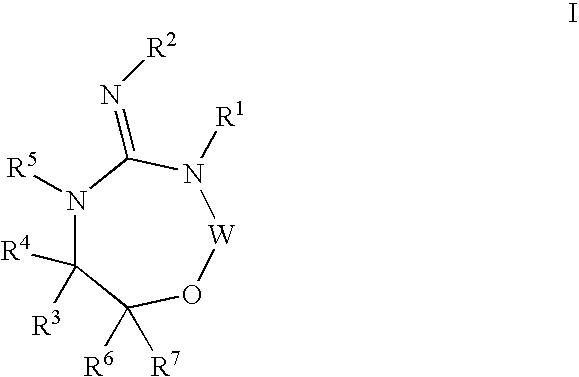
Heterocyclic aspartyl protease inhibitors
Disclosed are compounds of the formula I or a stereoisomer, tautomer, or pharmaceutically acceptable salt or solvate thereof, wherein W is a bond, —C(═S)—, —S(O)—, —S(O)2—, —C(═O)—, —O—, —C(R6)(R7)—, —N(R5)— or —C(═N(R5))—; X is —O—, —N(R5)— or —C(R6)(R7)—; provided that when X is —O—, U is not —O—, —S(O)—, —S(O)2—, —C(═O)— or —C(═NR5)—; U is a bond, —S(O)—, —S(O)2—, —C(O)—, —O—, —P(O)(OR15)—, —C(═NR5)—, —(C(R6)(R7))b— or —N(R5)—; wherein b is 1 or 2; provided that when W is —S(O)—, —S(O)2—, —O—, or —N(R5)—, U is not —S(O)—, —S(O)2—, —O—, or —N(R5)—; provided that when X is —N(R5)— and W is —S(O)—, —S(O)2—, —O—, or —N(R5)—, then U is not a bond; and R1, R2, R3, R4, R5, R6, and R7 are as defined in the specification; and pharmaceutical compositions comprising the compounds of formula I. Also disclosed is the method of inhibiting aspartyl protease, and in particular, the methods of treating cardiovascular diseases, cognitive and neurodegenerative diseases, and the methods of inhibiting of Human Immunodeficiency Virus, plasmepins, cathepsin D and protozoal enzymes. Also disclosed are methods of treating cognitive or neurodegenerative diseases using the compounds of formula I in combination with a cholinesterase inhibitor or a muscarinic m1 agonist or m2 antagonist.
Owner:PHARMACOPEIA DRUG DISCOVERY +1
Method and system for rapid biomolecular recognition of amino acids and protein sequencing
InactiveUS6846638B2Low costHigh sensitivityBioreactor/fermenter combinationsBiological substance pretreatmentsProtein Sequence DeterminationDigestion
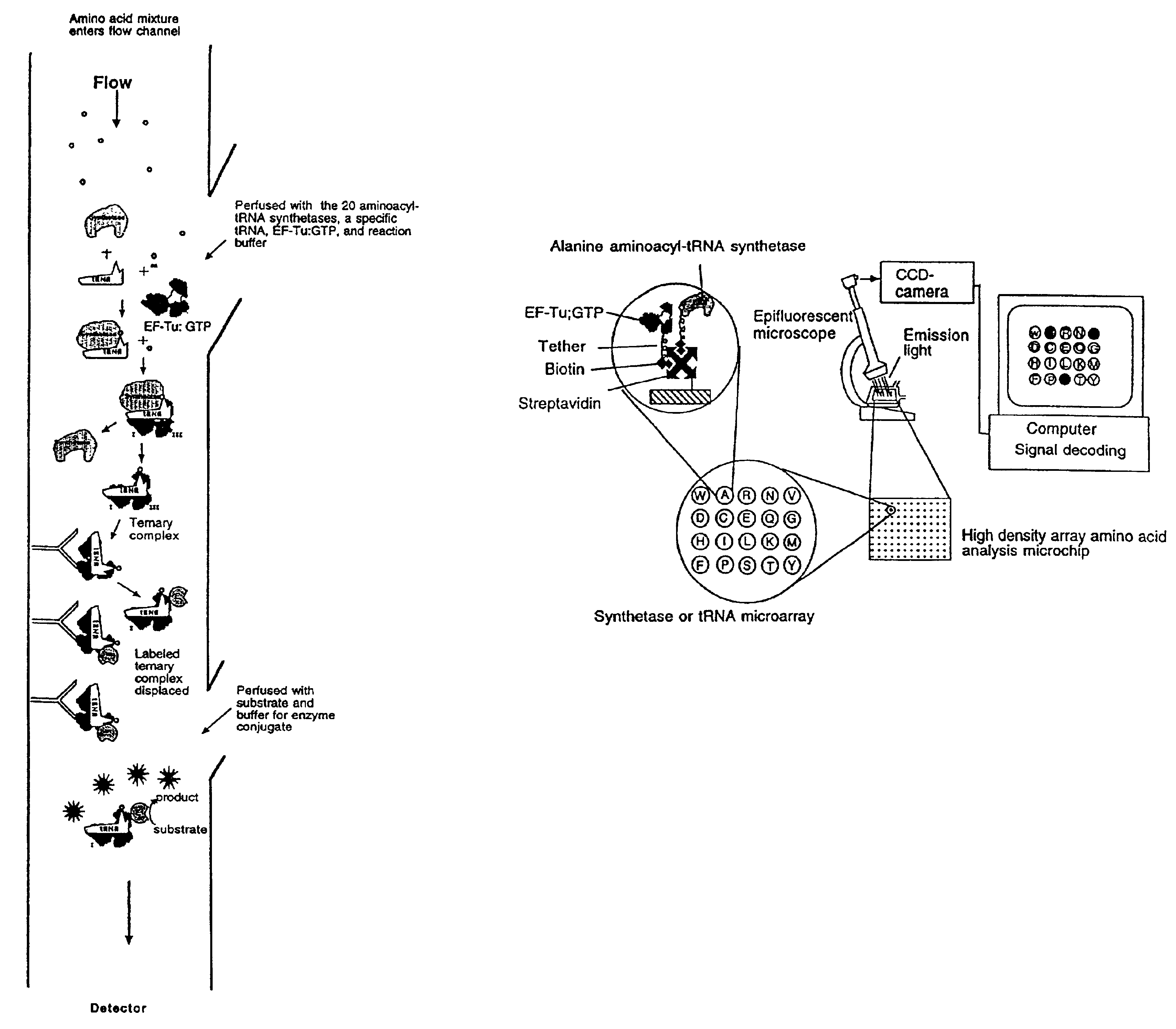
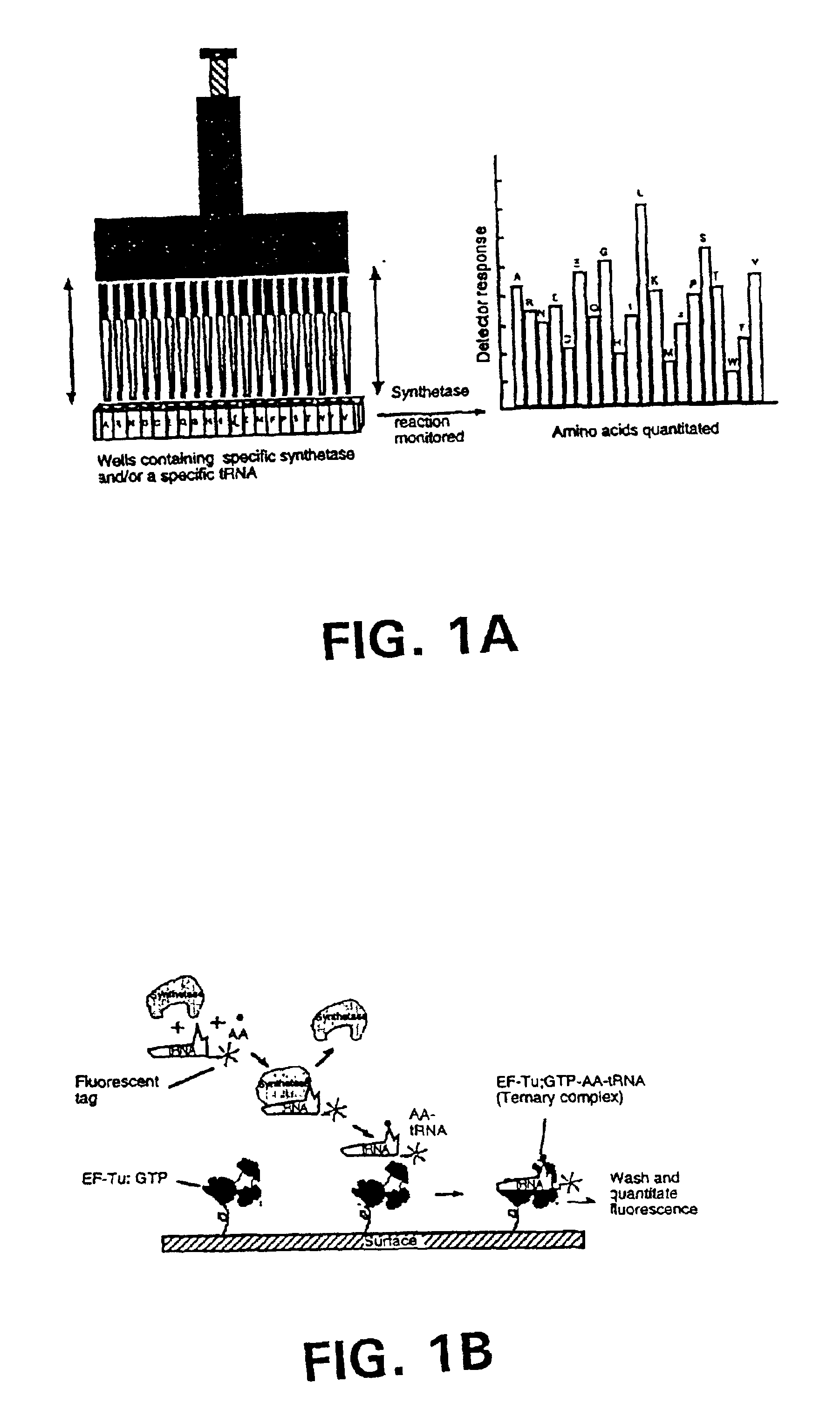
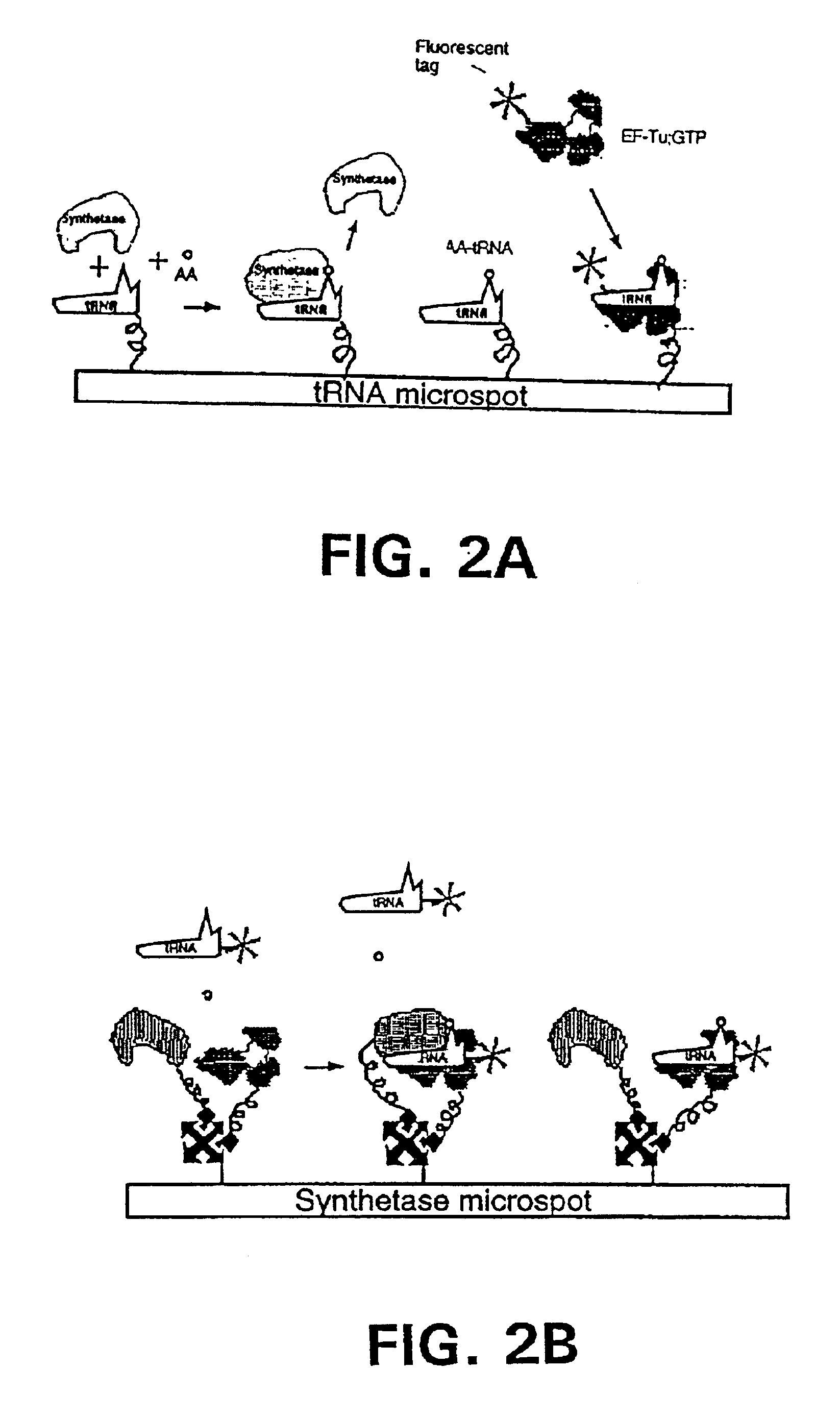
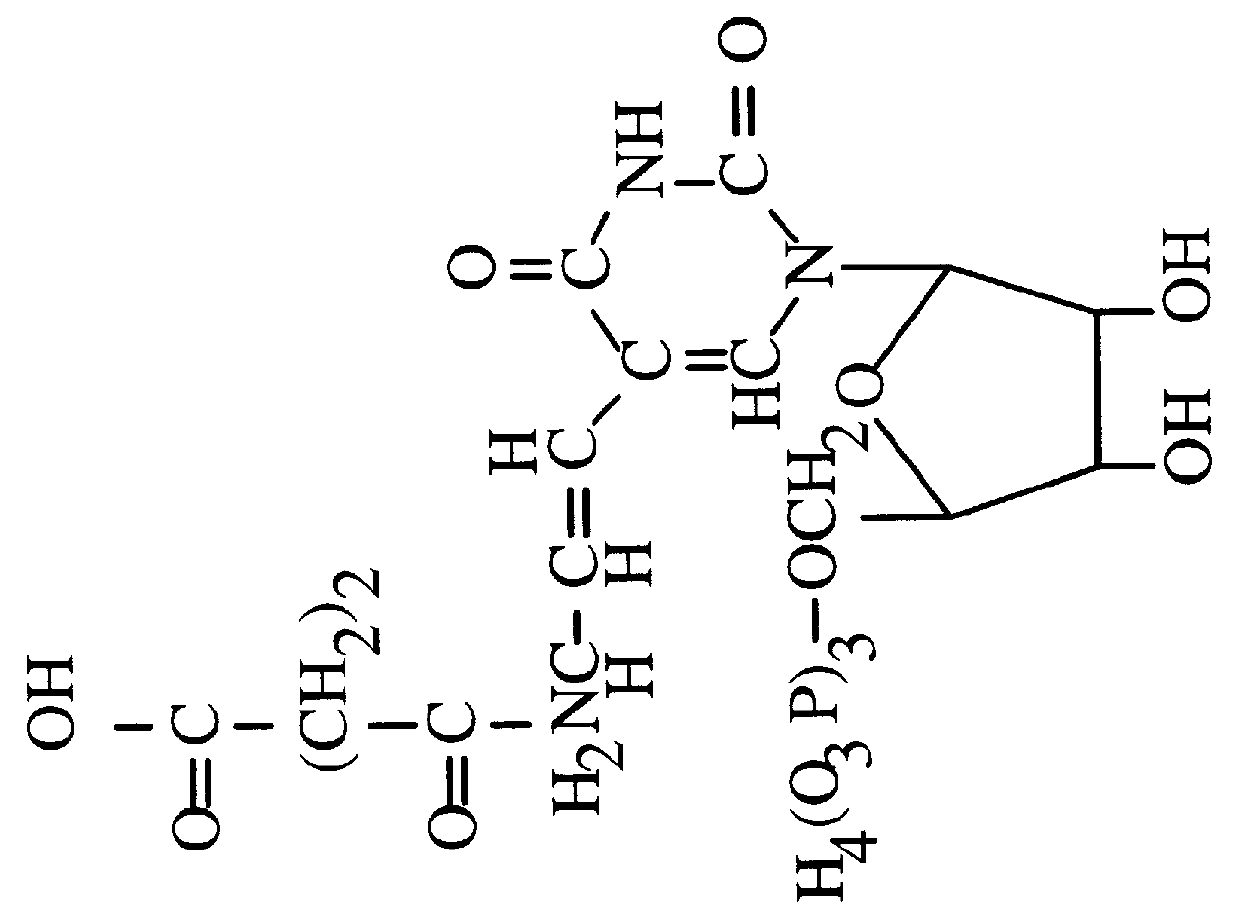
Methods, compositions, kits, and apparatus are provided wherein the aminoacyl-tRNA synthetase system is used to analyze amino acids. The method allows very small devices for quantitative or semi-quantitative analysis of the amino acids in samples or in sequential or complete proteolytic digestions. The methods can be readily applied to the detection and / or quantitation of one or more primary amino acids by using cognate aminoacyl-tRNA synthetase and cognate tRNA. The basis of the method is that each of the 20 synthetases and / or a tRNA specific for a different amino acid is separated spatially or differentially labeled. The reactions catalyzed by all 20 synthetases may be monitored simultaneously, or nearly simultaneously, or in parallel. Each separately positioned synthetase or tRNA will signal its cognate amino acid. The synthetase reactions can be monitored using continuous spectroscopic assays. Alternatively, since elongation factor Tu:GTP (EF-Tu:GTP) specifically binds all AA-tRNAs, the aminoacylation reactions catalyzed by the synthetases can be monitored using ligand assays. Microarrays and microsensors for amino acid analysis are provided. Additionally, amino acid analysis devices are integrated with protease digestions to produce miniaturized enzymatic sequenators capable of generating either N- or C-terminal sequence and composition data for a protein or peptide. The possibility of parallel processing of many samples in an automated manner is discussed.
Owner:NANOBIODYNAMICS
Systematic evolution of ligands exponential enrichment: blended selex
InactiveUS6083696AHigh affinityDesired effectSugar derivativesMicrobiological testing/measurementAla-Ala-Pro-ValKetone
A method is described for generating blended nucleic acid ligands containing non-nucleic acid functional units. Specifically, a SELEX identified RNA ligand to the integrin gpIIbIIIa is conjugated to the peptide Gly-Arg-Gly-Asp-Thr-Pro (SEQ ID NO:1). This blended RNA ligand inhibits the biological activity of gpIIbIIIa with high specificity. Also described is a single-stranded DNA ligand to elastase coupled to N-methoxysuccinyl-Ala-Ala-Pro-Val-chloromethyl (SEQ ID NO:3) ketone. This elastase blended nucleic acid ligand inhibits the biological activity of elastase.
Owner:GILEAD SCI INC
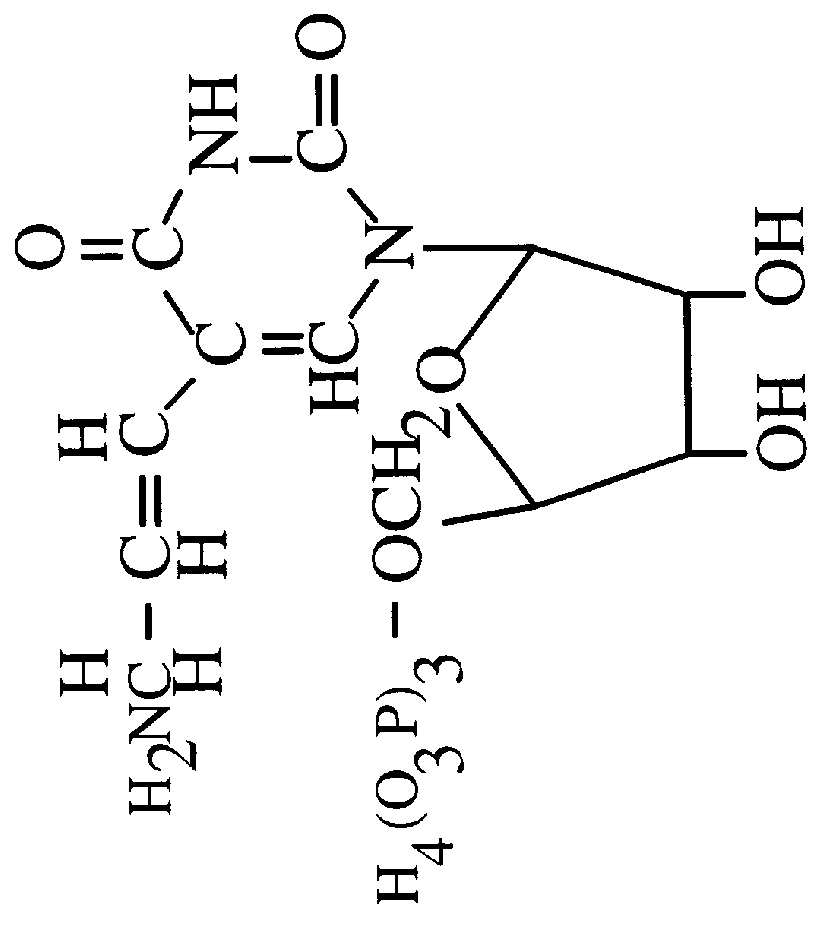
Thermostable protease
InactiveUS6358726B1Improve efficiencyImprove expression levelPolypeptide with localisation/targeting motifHydrolasesProteinase activityProtease
A hyperthermostable protease having the amino acid sequence represented by the SEQ ID NO:1 of the Sequence Listing or a sequence derived therefrom by deletion, substitution, insertion or addition of one to several amino acid residues, a gene encoding the hyperthermostable protease, and a process for preparing the protease, aiming at providing by genetic engineering techniques a hyperthermophile protease which is advantageous for industrial use.
Owner:TAKARA HOLDINGS
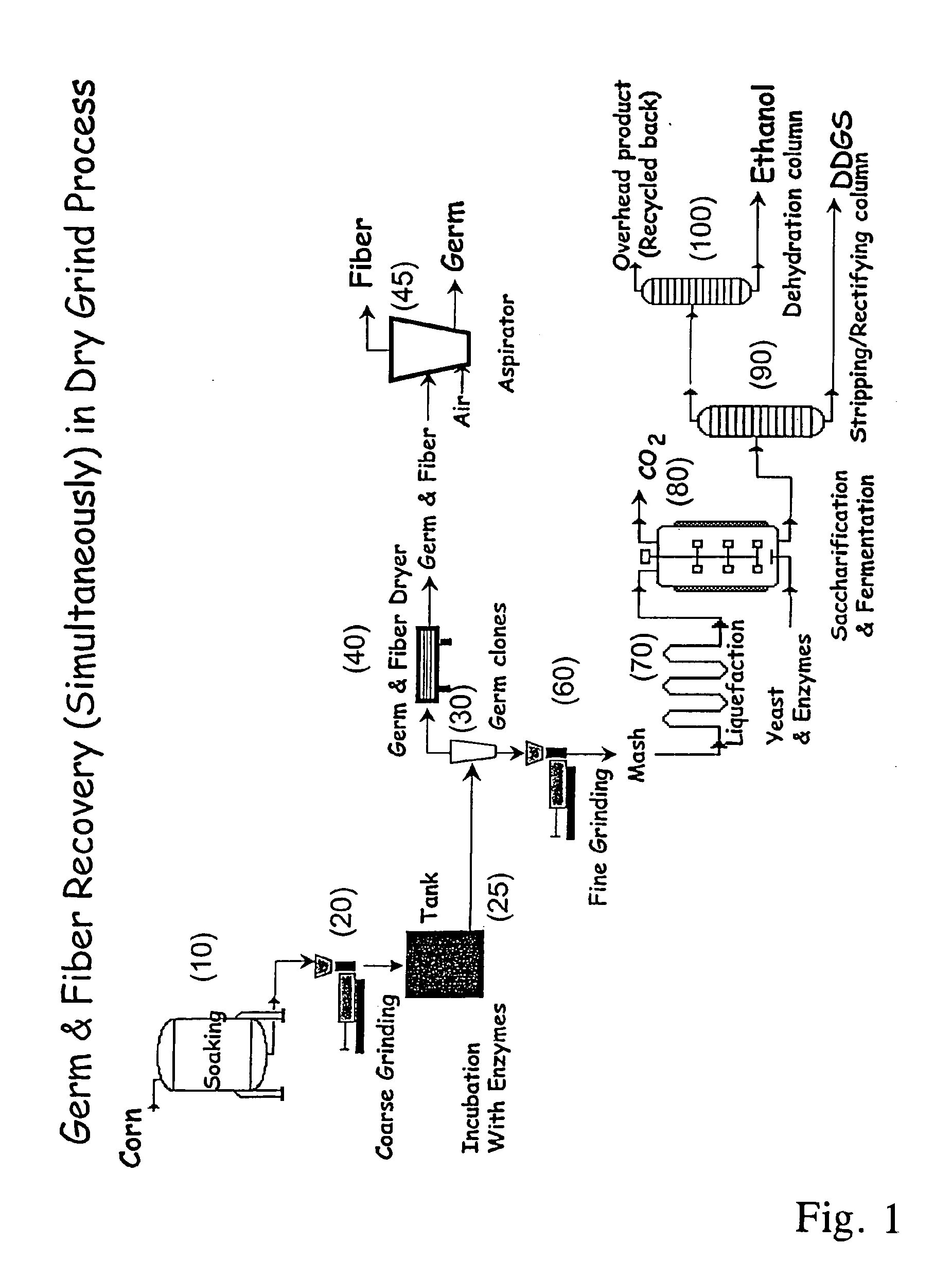
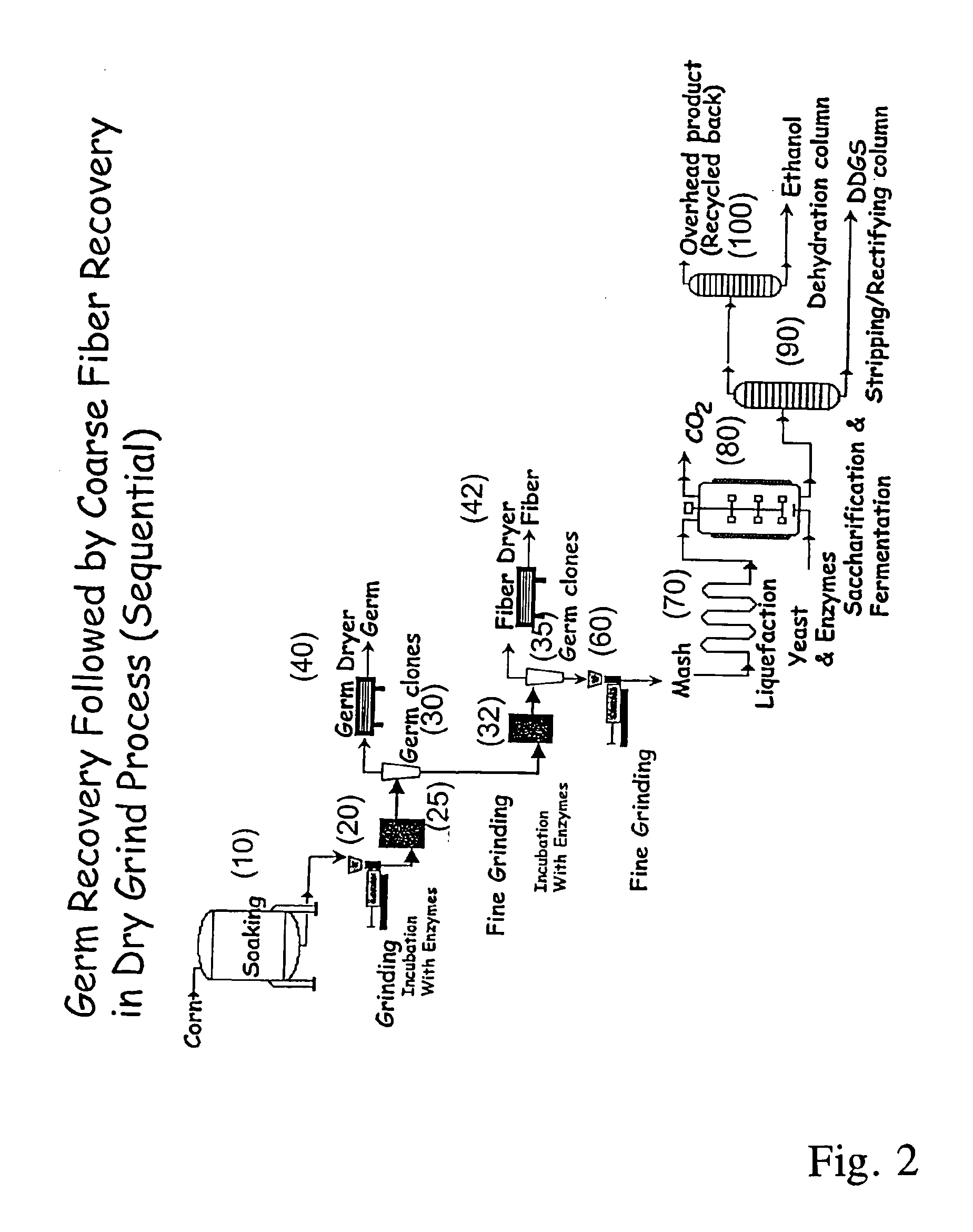
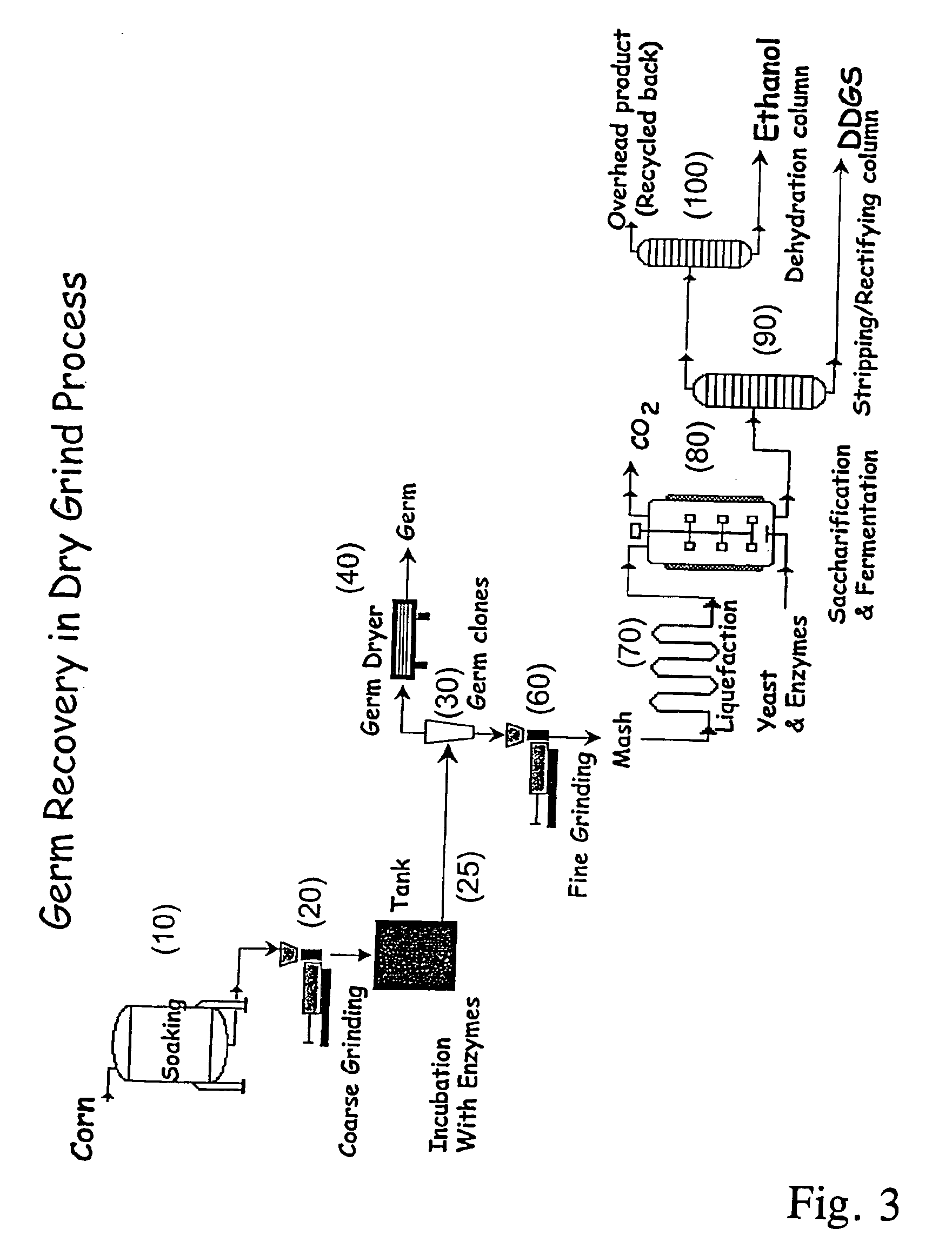
Processes for recovery of corn germ and optionally corn coarse fiber (pericarp)
A process for recovering corn germ and corn coarse fiber from corn in a dry grind process, involving soaking corn kernels in water to produce soaked corn kernels, grinding the soaked corn kernels to produce a ground corn slurry, and incubating the ground corn slurry with at least one enzyme (amylase(s), protease(s), cell wall degrading enzyme(s), or mixtures thereof, and optionally other enzyme(s)) to increase the specific gravity of the slurry to about 10-about 16 Baume so that the corn germ and corn coarse fiber floats to the top of the slurry, recovering the corn germ and the corn coarse fiber, and optionally producing ethanol from the slurry no longer containing the corn germ and corn coarse fiber. The process does not involve the addition of starch, a salt, a sugar syrup, or mixtures thereof to the slurry.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
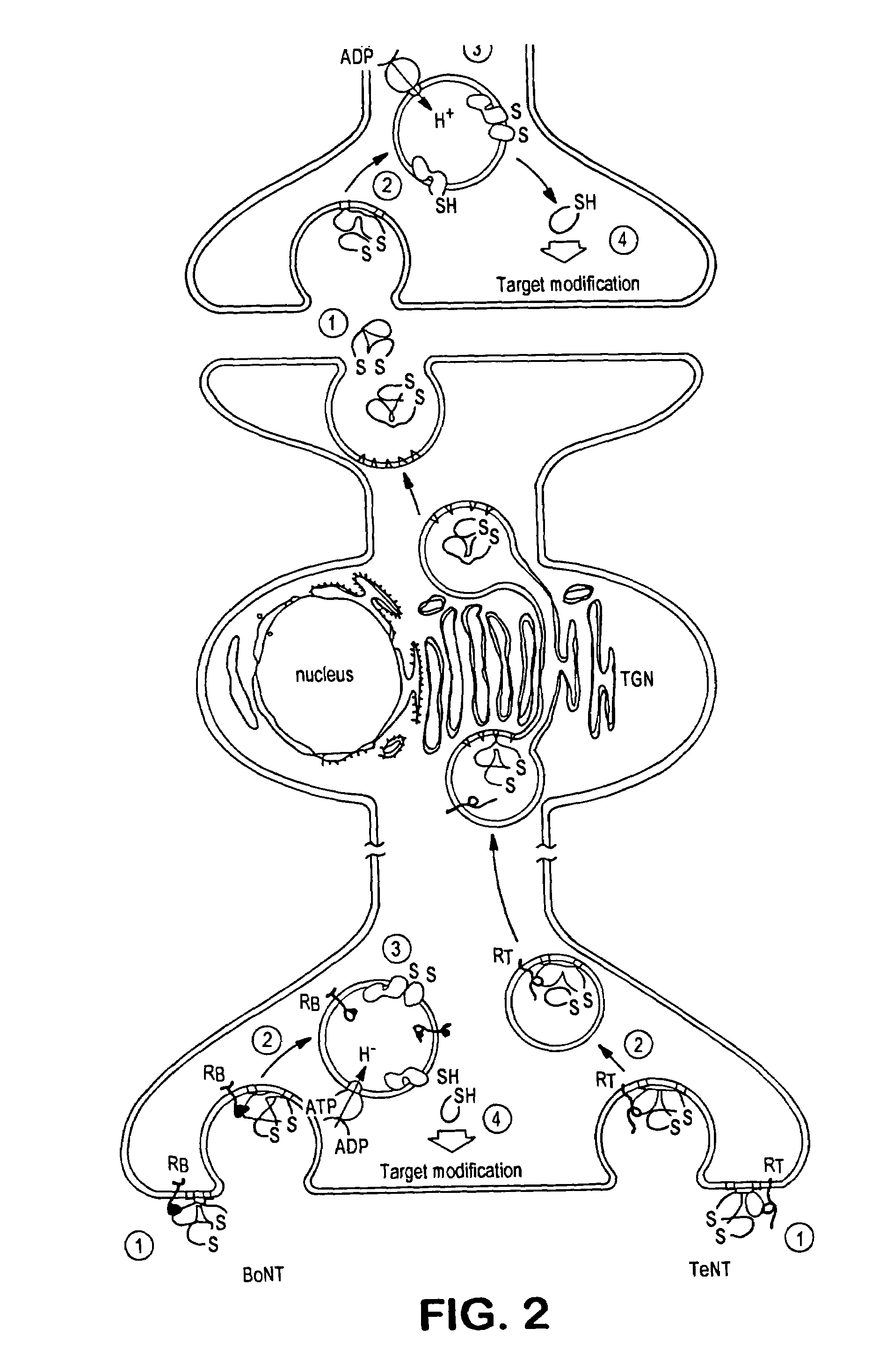
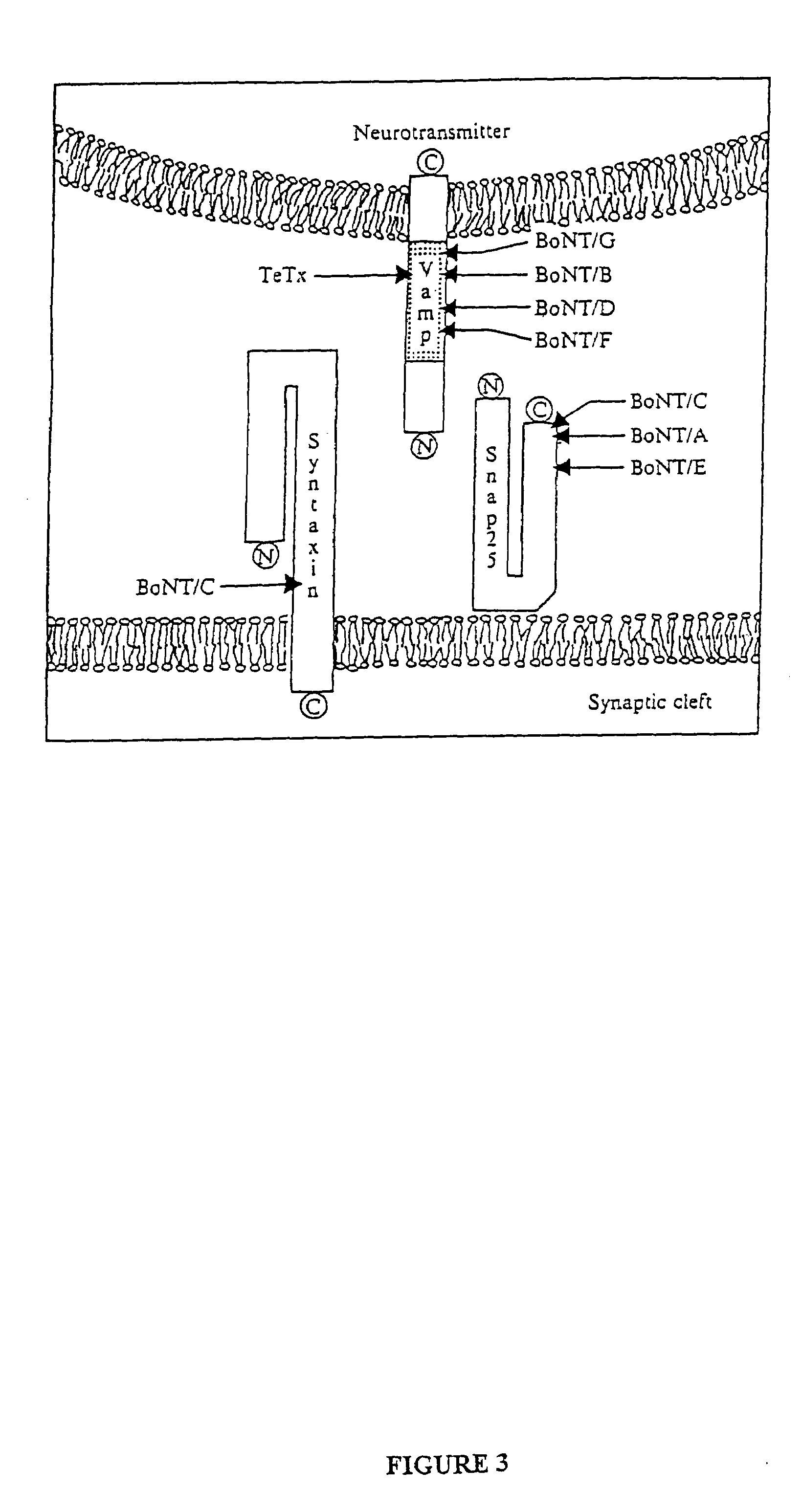
Fret protease assays for botulinum serotype A/E toxins
InactiveUS7208285B2Used to determineDecreased acceptor fluorescence intensityBacteriaSugar derivativesFluorophoreReceptor for activated C kinase 1
The present invention provides clostridial toxin substrates useful in assaying for the protease activity of any clostridial toxin, including botulinum toxins of all serotypes as well as tetanus toxins. A clostridial toxin substrate of the invention contains a donor fluorophore; an acceptor having an absorbance spectrum overlapping the emission spectrum of the donor fluorophore; and a clostridial toxin recognition sequence that includes a cleavage site, where the cleavage site intervenes between the donor fluorophore and the acceptor and where, under the appropriate conditions, resonance energy transfer is exhibited between the donor fluorophore and the acceptor.
Owner:ALLERGAN INC
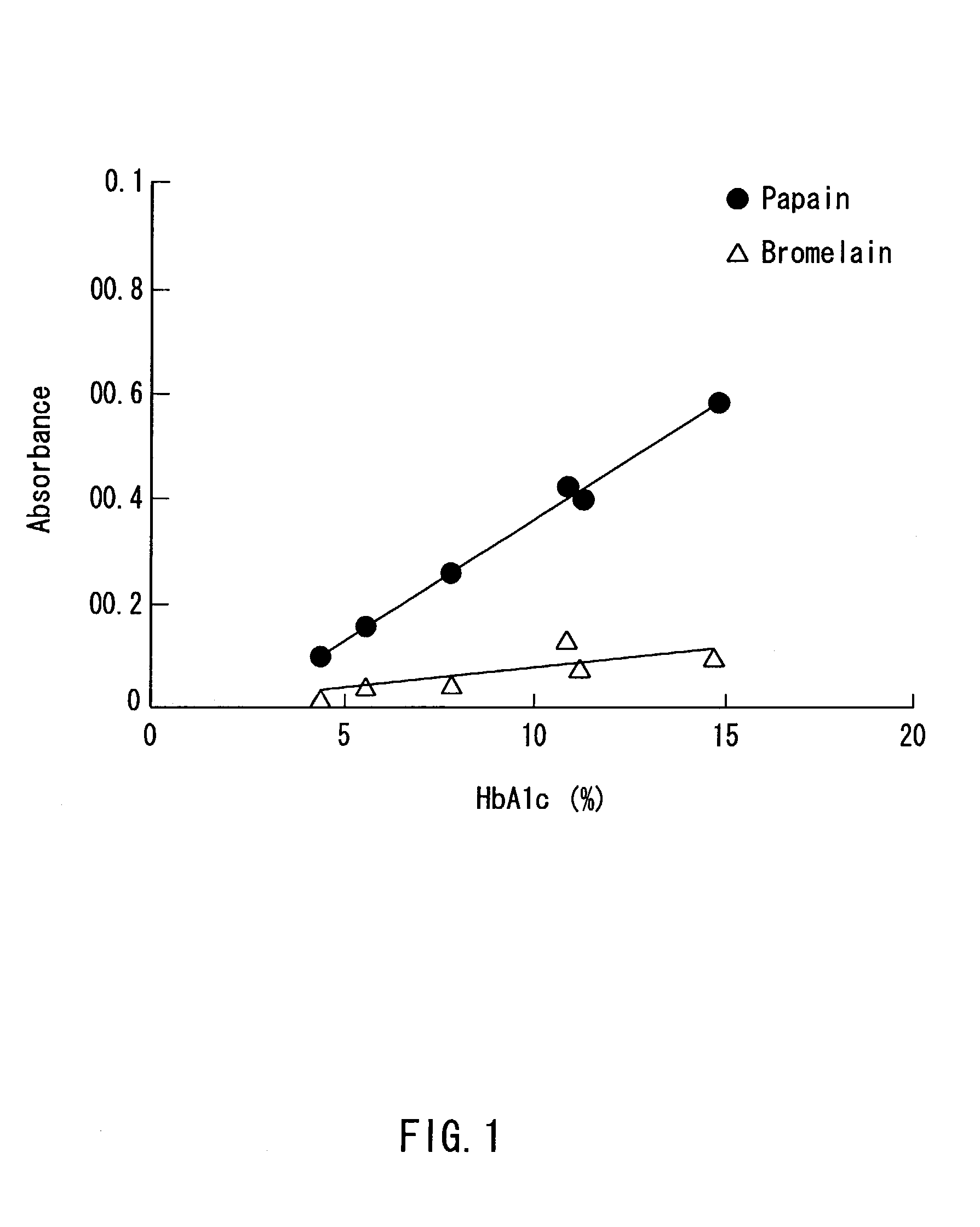
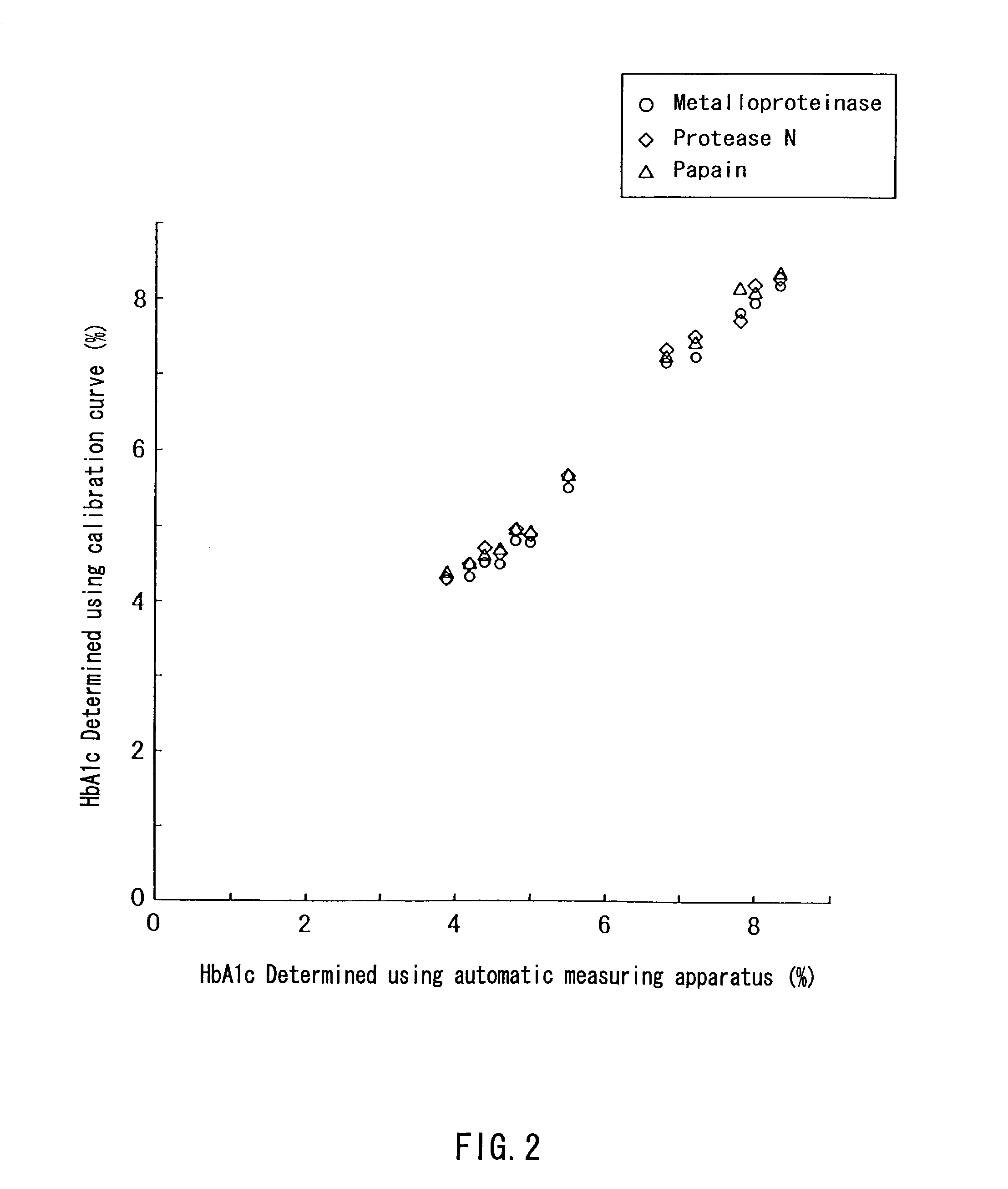
Method of selectively determining glycated hemoglobin
InactiveUS7235378B2Easily and accurately determinedEliminate the effects ofMicrobiological testing/measurementBiological testingProteinase activityHemoglobin F
Owner:ARKRAY INC
Supplemented and unsupplemented tissue sealants, methods of their production and use
ActiveUS7189410B1Low antigenicityDecreasing thrombogenicityAntibacterial agentsOrganic active ingredientsTissue sealantVascular dilatation
This invention provides a fibrin sealant bandage, wherein said fibrin sealant may be supplemented with at least one composition selected from, for example, one or more regulatory compounds, antibody, antimicrobial compositions, analgesics, anticoagulants, antiproliferatives, anti-inflammatory compounds, cytokines, cytotoxins, drugs, growth factors, interferons, hormones, lipids, demineralized bone or bone morphogenetic proteins, cartilage inducing factors, oligonucleotides polymers, polysaccharides, polypeptides, protease inhibitors, vasoconstrictors or vasodilators, vitamins, minerals, stabilizers and the like. Also disclosed are methods of preparing and / or using the unsupplemented or supplemented fibrin sealant bandage.
Owner:AMERICAN NAT RED CROSS
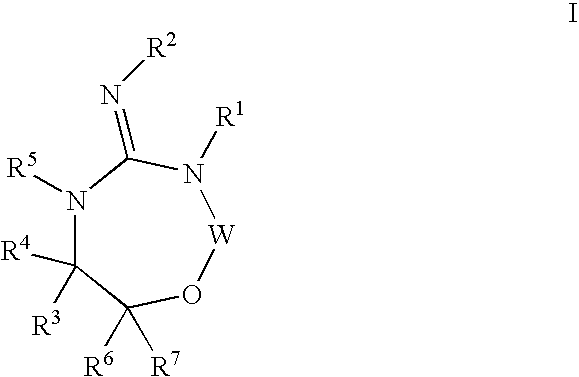
Aspartyl protease inhibitors
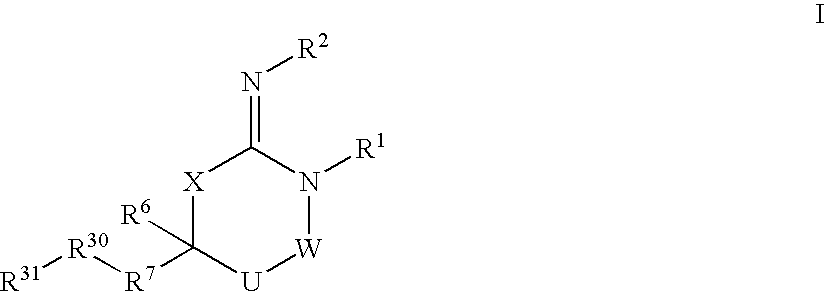
Disclosed are compounds of formula I or a stereoisomer, tautomer, or pharmaceutically acceptable salt or solvate thereof, U, W, X, R1, R2, R6, R7, R30 and R31 are as described above in the specification. Also disclosed is the method of inhibiting aspartyl protease, and in particular, the methods of treating cardiovascular diseases, cognitive and neurodegenerative diseases. Also disclosed are methods of treating cognitive or neurodegenerative diseases using the compounds of formula I in combination with a cholinesterase inhibitor or a muscarinic m1 agonist or m2 antagonist.
Owner:MERCK SHARP & DOHME LLC
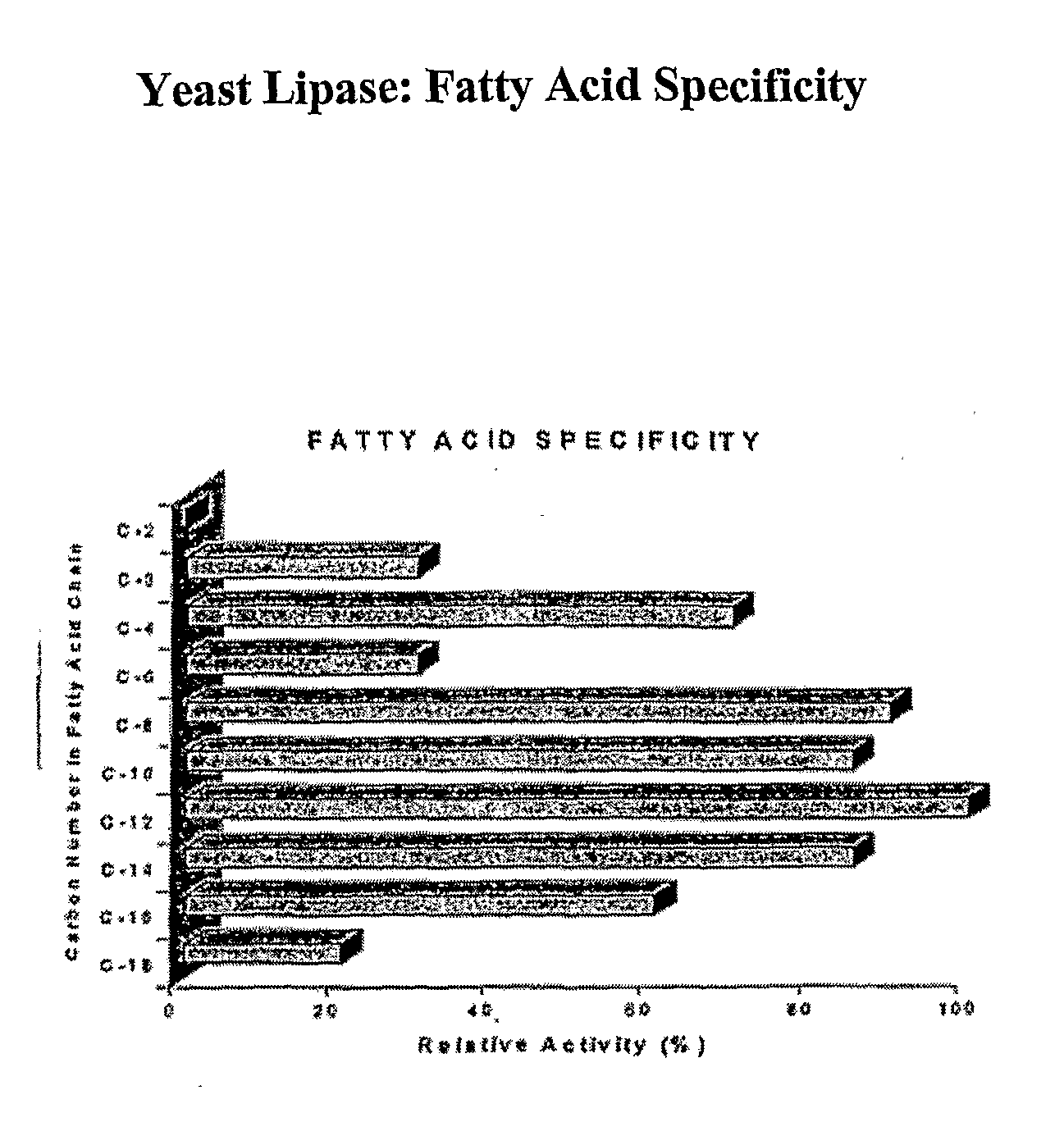
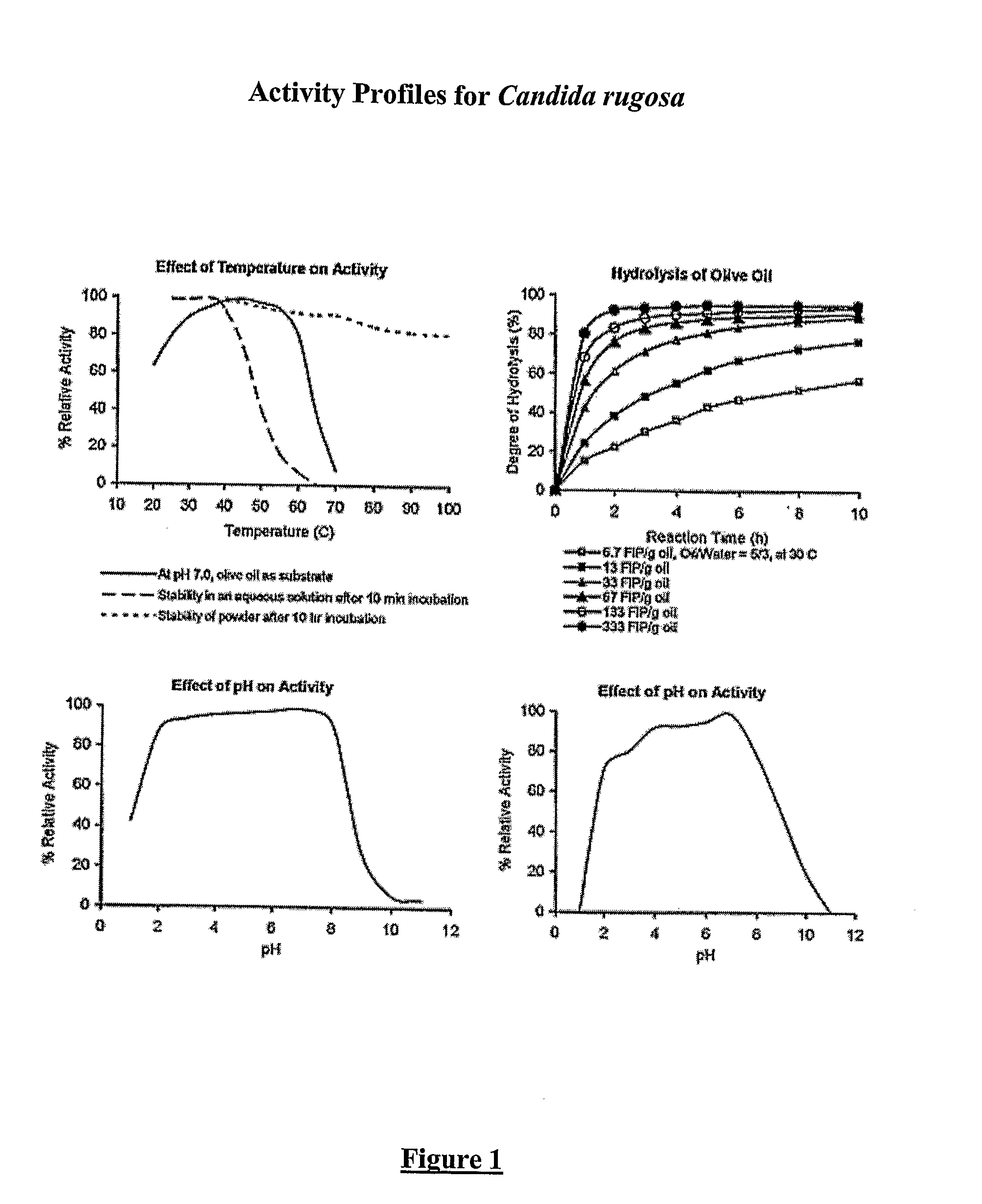
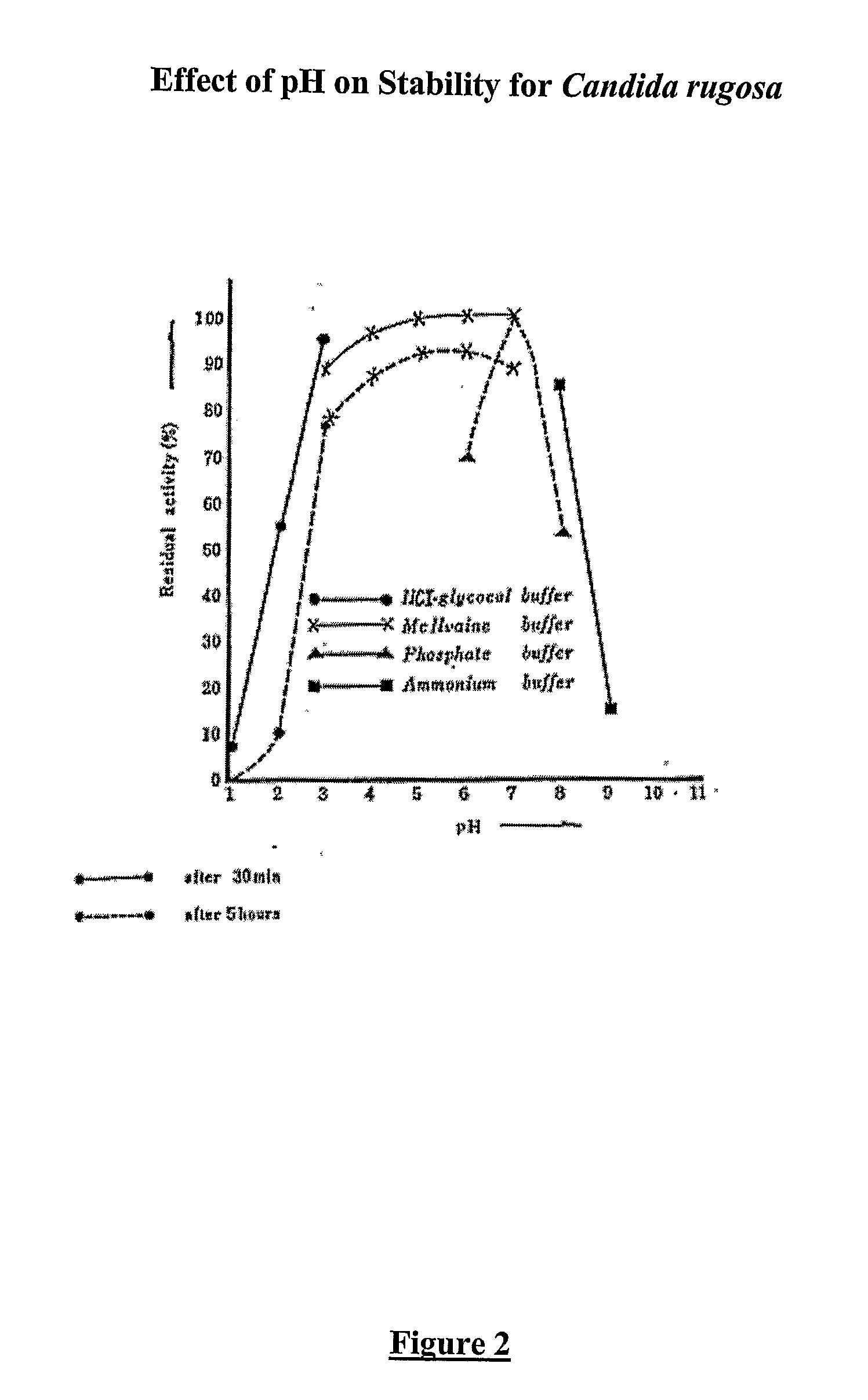
Composition With a Fungal (Yeast) Lipase and Method For Treating Lipid Malabsorption in Cystic Fibrous as Well as People Suffering From Pancreatic Lipase Insufficiency
The invention provides compositions and methods for treating pancreatic enzyme insufficiency, such as the pancreatic enzyme insufficiency associated with cystic fibrosis. The invention also provides compositions comprising lipase from Candida cylindracea, alone or in combination with amylase or amyloglucosidase, protease and / or lactase. Furthermore, the invention discloses methods for treating pancreatic enzyme insufficiency comprising administering compositions to patients in need thereof.
Owner:BIO CAT
Novel nutritional food products for improved digestion and intestinal absorption
InactiveUS20100239559A1Sustained stabilityAvoiding unstable breakdown of the enzyme and large overdosingHydrolasesPeptide/protein ingredientsDigestionNovel food
The present invention is directed to novel food products, e.g., nutritional food products and infant formula, which contain one ore more enzymes selected from lipase, protease, and amylase that have been formulated / stabilized to have sustained stability in an aqueous medium. Such formulations are intended to provide a greater degree of compliance based on their ability to be incorporated into aqueous media while avoiding unstable breakdown of the enzyme and large overdosing due to expected breakdown when exposed to an aqueous environment, including saliva. Further described in the invention are additives packaged with instructions for combination with an aqueous medium, and instructions for the administration of the resulting mixture to a subject. In certain embodiments, enzyme insufficient patients, e.g., infants and elderly persons, would find particular benefit from the food products described herein.
Owner:BETH ISRAEL DEACONESS MEDICAL CENT INC
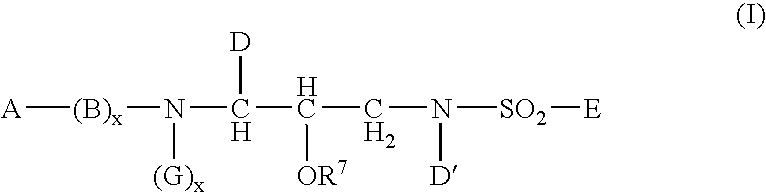
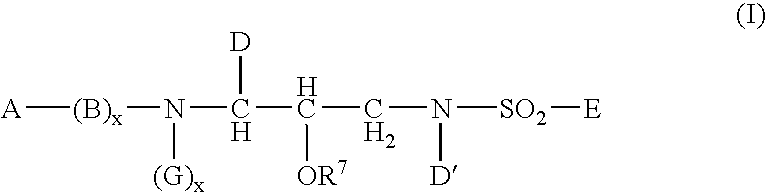
Sulphonamide derivatives as prodrugs of aspartyl protease inhibitors
InactiveUS20050148548A1Good water solubilityImprove bioavailabilityBiocideSugar derivativesSulfur drugPatient compliance
The present invention relates to prodrugs of a class of sulfonamides which are aspartyl protease inhibitors. In one embodiment, this invention relates to a novel class of prodrugs of HIV aspartyl protease inhibitors characterized by favorable aqueous solubility, high oral bioavailability and facile in vivo generation of the active ingredient. This invention also relates to pharmaceutical compositions comprising these prodrugs. The prodrugs and pharmaceutical compositions of this invention are particularly well suited for decreasing the pill burden and increasing patient compliance. This invention also relates to methods of treating mammals with these prodrugs and pharmaceutical compositions.
Owner:VERTEX PHARMA INC
Compositions and methods for treating pancreatic insufficiency
InactiveUS20060121017A1Stable enzyme componentEffective low dose treatment regimenPeptide/protein ingredientsHydrolasesProteinase activityPancreas
The present invention relates to compositions for the treatment of conditions, including pancreatic insufficiency. The compositions of the present invention comprise lipase, protease and amylase in a particular ratio that provides beneficial results in patients, such as those afflicted with pancreatic insufficiency. This invention also relates to methods using such compositions for the treatment of pancreatic insufficiency.
Owner:ELI LILLY & CO
Aspartyl protease inhibitors
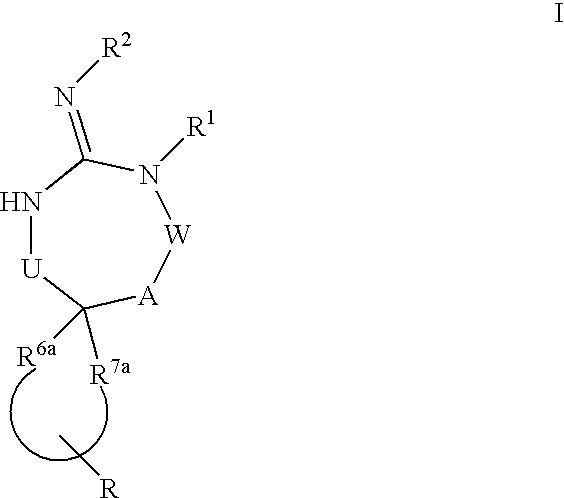
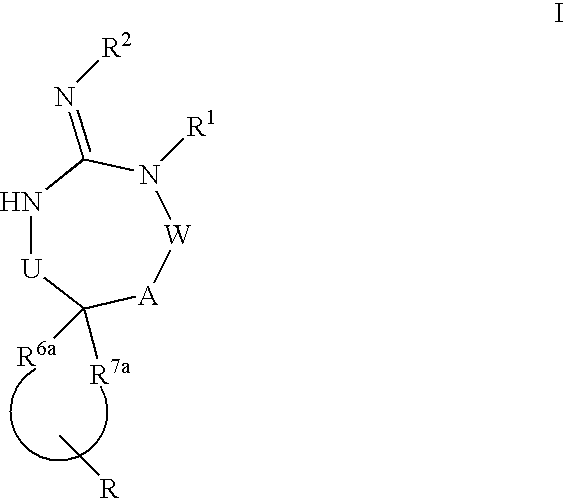
Disclosed are compounds of the formula I or a stereoisomer, tautomer, or pharmaceutically acceptable salt or solvate thereof, wherein U, W, A, R, R1, R2, R6a and R7, are as defined in the specification; and pharmaceutical compositions comprising the compounds of formula I. Also disclosed is the method of inhibiting aspartyl protease, and in particular, the methods of treating cardiovascular diseases, cognitive and neurodegenerative diseases. Also disclosed are methods of treating cognitive or neurodegenerative diseases using the compounds of formula I in combination with a cholinesterase inhibitor or a muscarinic m1 agonist or m2 antagonist.
Owner:MERCK SHARP & DOHME LLC +1
Baby Care Skin Protectant Compositions for Diaper Rash
The present invention provides a comprehensive solution to skin problems of infants and incontinent adults related to diaper rash, also known as diaper dermatitis. This is based on certain novel divalent metal and quaternary ammonium complexes (ion-pairs) of zeolites (that are made by an in-situ process), which in synergistic combination with certain other compositions, provide a comprehensive treatment for diaper rash that encompasses the following aspects: (1) deactivation of lipase and protease enzymes on skin surface, (2) the controlled-release delivery of skin protectant compositions, such as divalent metal zinc cation, (3) trapping of acidic and alkaline chemicals deposited on skin from body exudates and enzyme activity, (4) controlled-release delivery of anti-inflammatory agents, and COX and LOX enzyme inhibitors, (5) controlled-release delivery of antibacterial and antifungal compositions, and (6) absorption of excess moisture in the diaper zone.
Owner:BIODERM RES
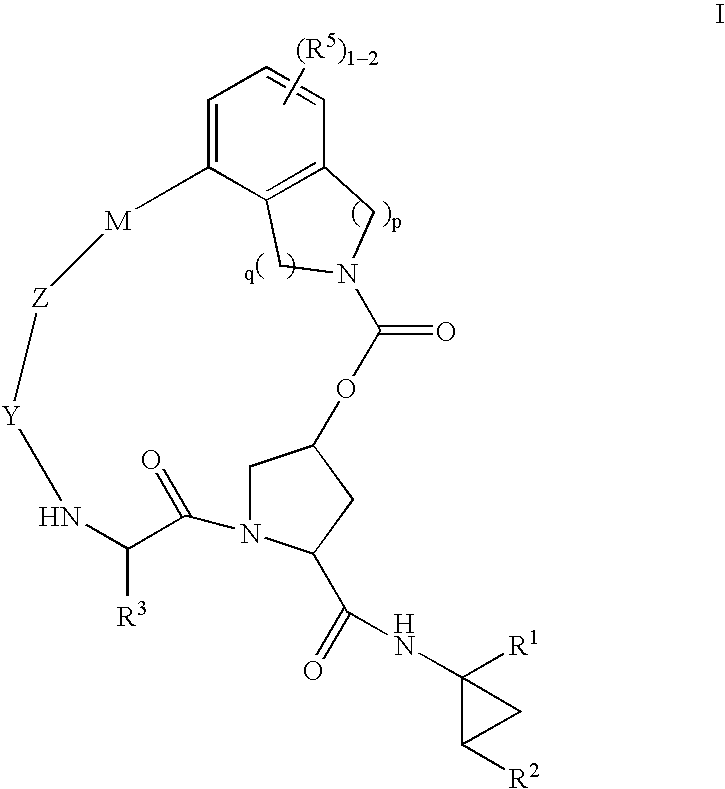
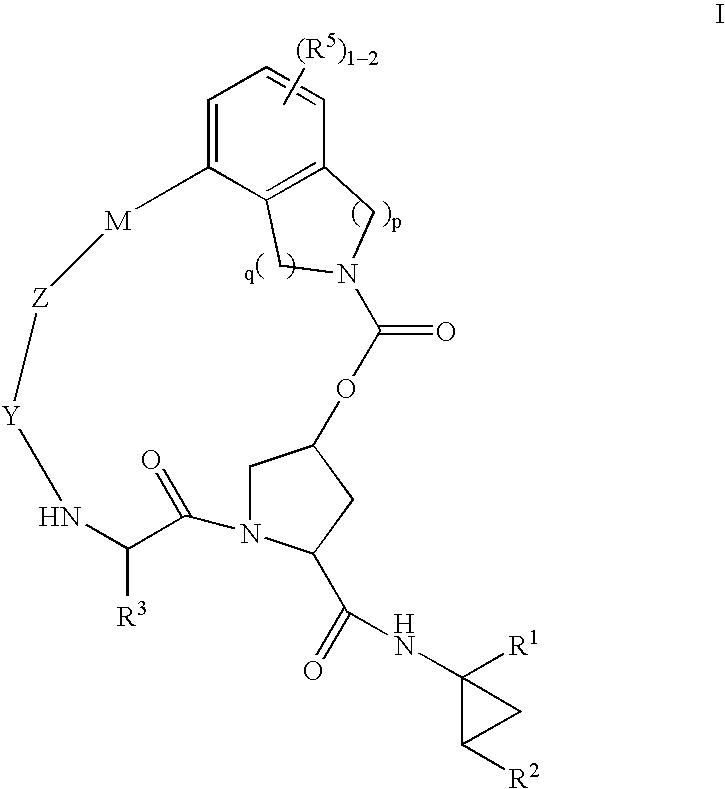
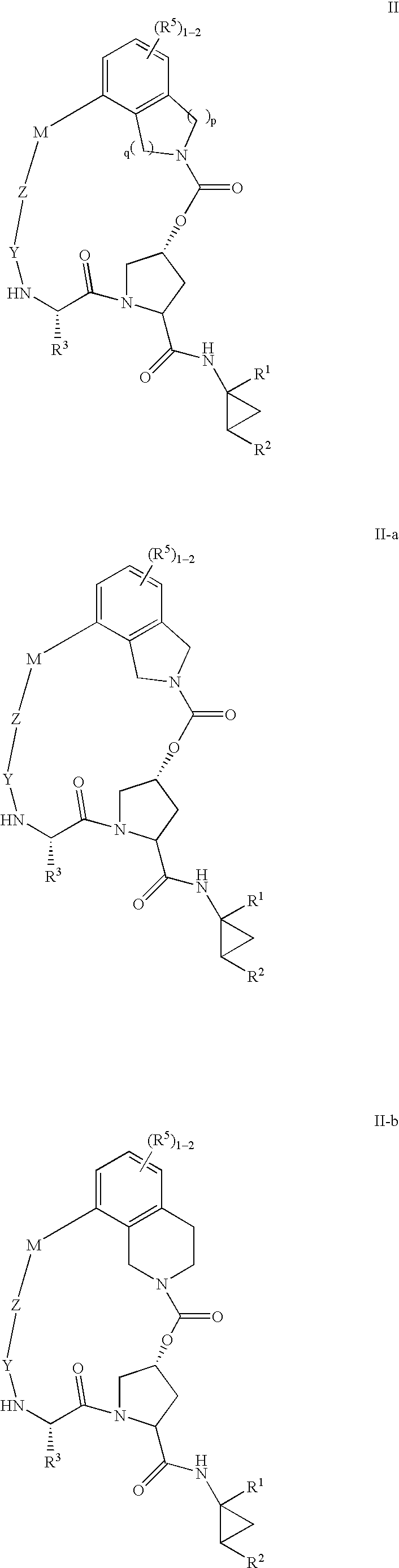
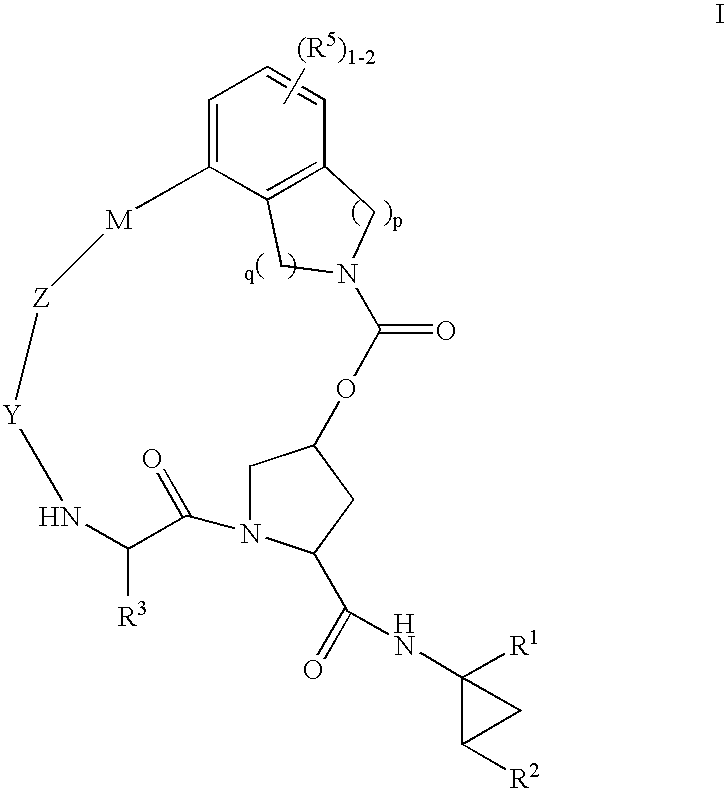
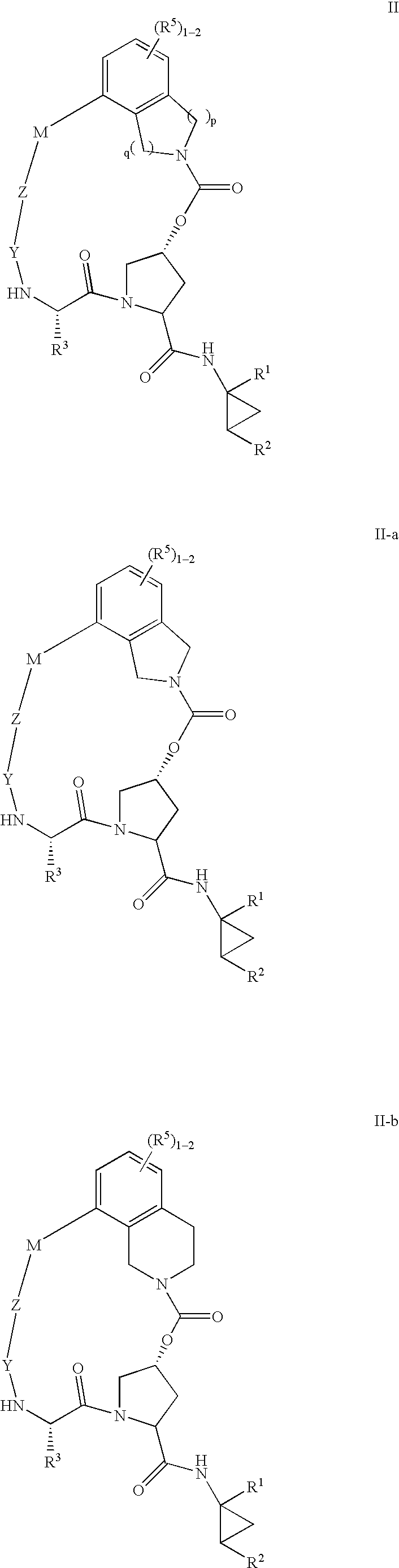
Macrocyclic heterocyclic aspartyl protease inhibitors
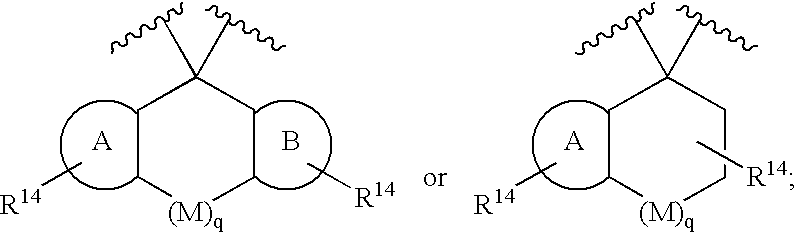
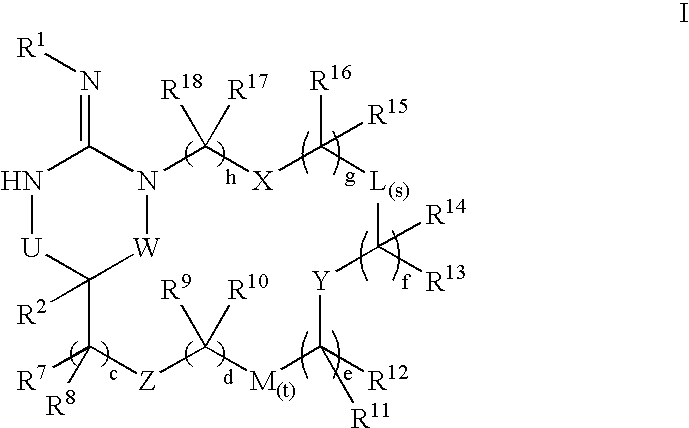
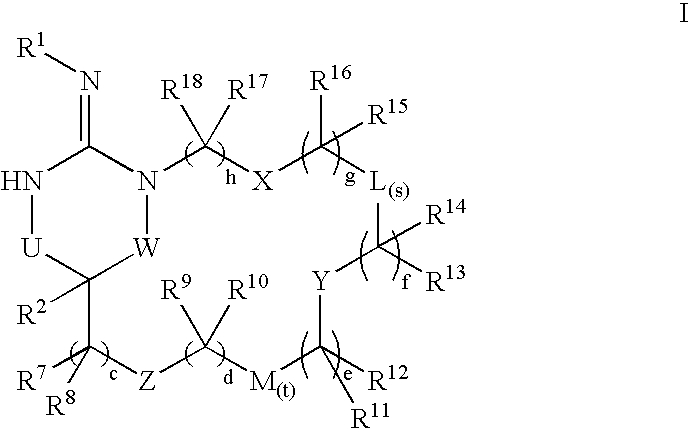
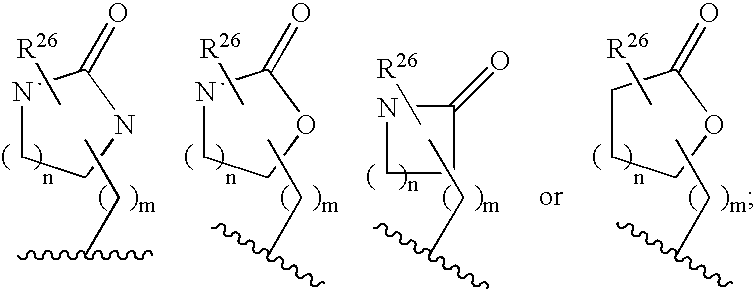
Disclosed are compounds of the formula I or a stereoisomer, tautomer, or pharmaceutically acceptable salt, solvate or ester thereof, wherein U, W, X, L, Y, M, Z, c, d, e, f, g, h, s, t, R1, R2, R7, R8, R9, R10, R11, R12, R13, R14, R15, R16, R17 and R18 are as defined in the specification; and pharmaceutical compositions comprising the compounds of formula I. Also disclosed is the method of inhibiting aspartyl protease, and in particular, the methods of treating cardiovascular diseases, cognitive and neurodegenerative diseases. Also disclosed are methods of treating cognitive or neurodegenerative diseases using the compounds of formula I in combination with a cholinesterase inhibitor or a muscarinic m1 agonist or m2 antagonist.
Owner:MERCK SHARP & DOHME LLC
Aspartyl protease inhibitors
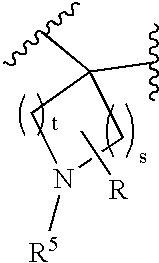
Disclosed are compounds of the formula I or a stereoisomer, tautomer, or pharmaceutically acceptable salt or solvate thereof, wherein W, R1, R2, R3, R4, R5, R6, and R7 are as defined in the specification; and pharmaceutical compositions comprising the compounds of formula I. Also disclosed is the method of inhibiting aspartyl protease, and in particular, the methods of treating cardiovascular diseases, cognitive and neurodegenerative diseases, and the methods of inhibiting of Human Immunodeficiency Virus, plasmepins, cathepsin D and protozoal enzymes. Also disclosed are methods of treating cognitive or neurodegenerative diseases using the compounds of formula I in combination with a cholinesterase inhibitor or a muscarinic antagonist.
Owner:SCHERING CORP
Method of removing a biofilm
The present invention relates to a method of removing a biofilm, which comprises at least the following steps, carried out simultaneously or consecutively: a) a solution comprising an enzyme mixture containing at least one enzyme chosen from the group of proteases, at least one enzyme chosen from the group of esterases and an amylase is prepared; b) a solution comprising a detergent with an alkaline pH is prepared; and c) said solutions are applied, by washing or by circulation, to the surface to be treated. It also relates to a kit intended for removing a biofilm, which comprises the solutions defined above and the compositions comprising the said solutions.
Owner:KARINE MARION +1
Bone matrix compositions and methods
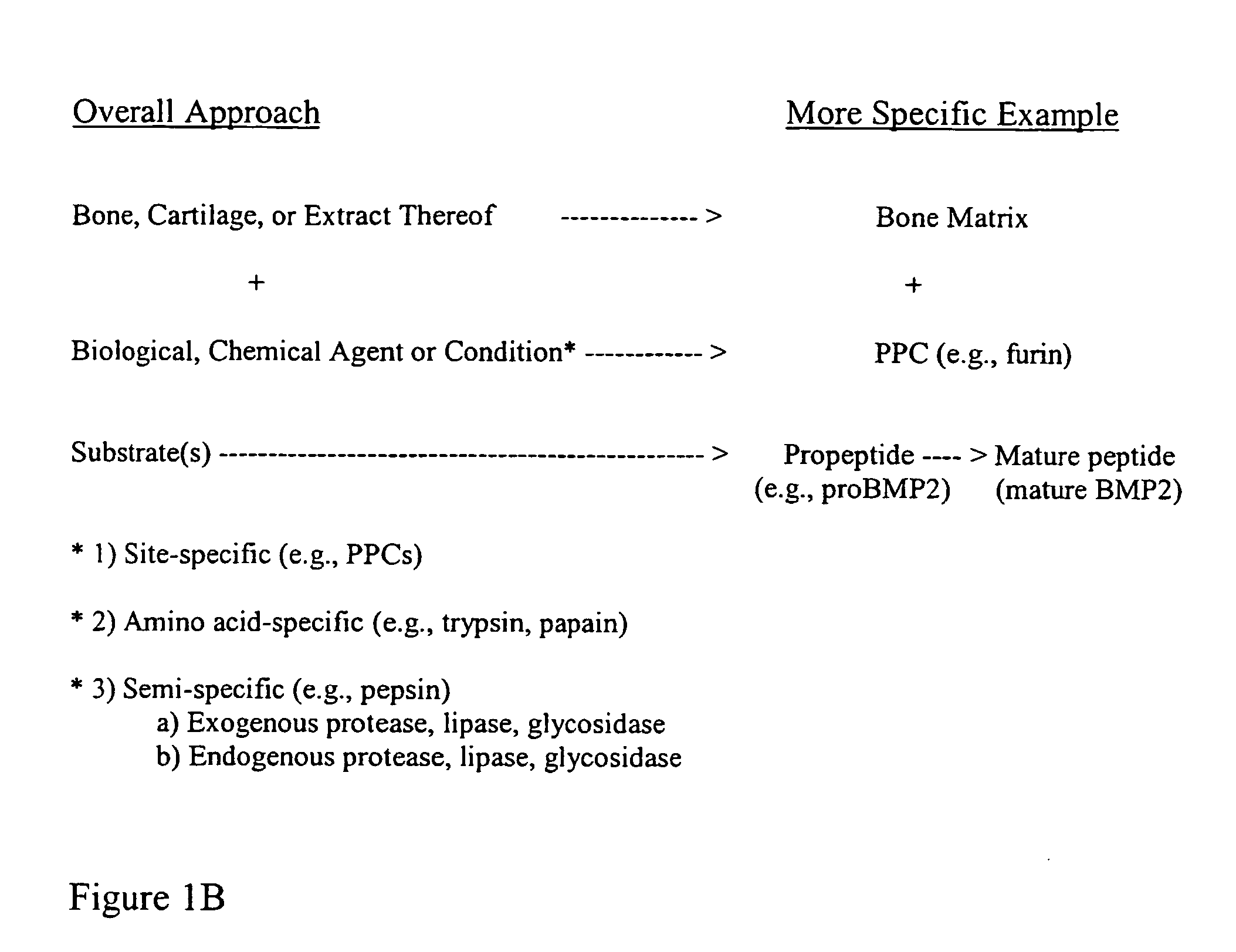
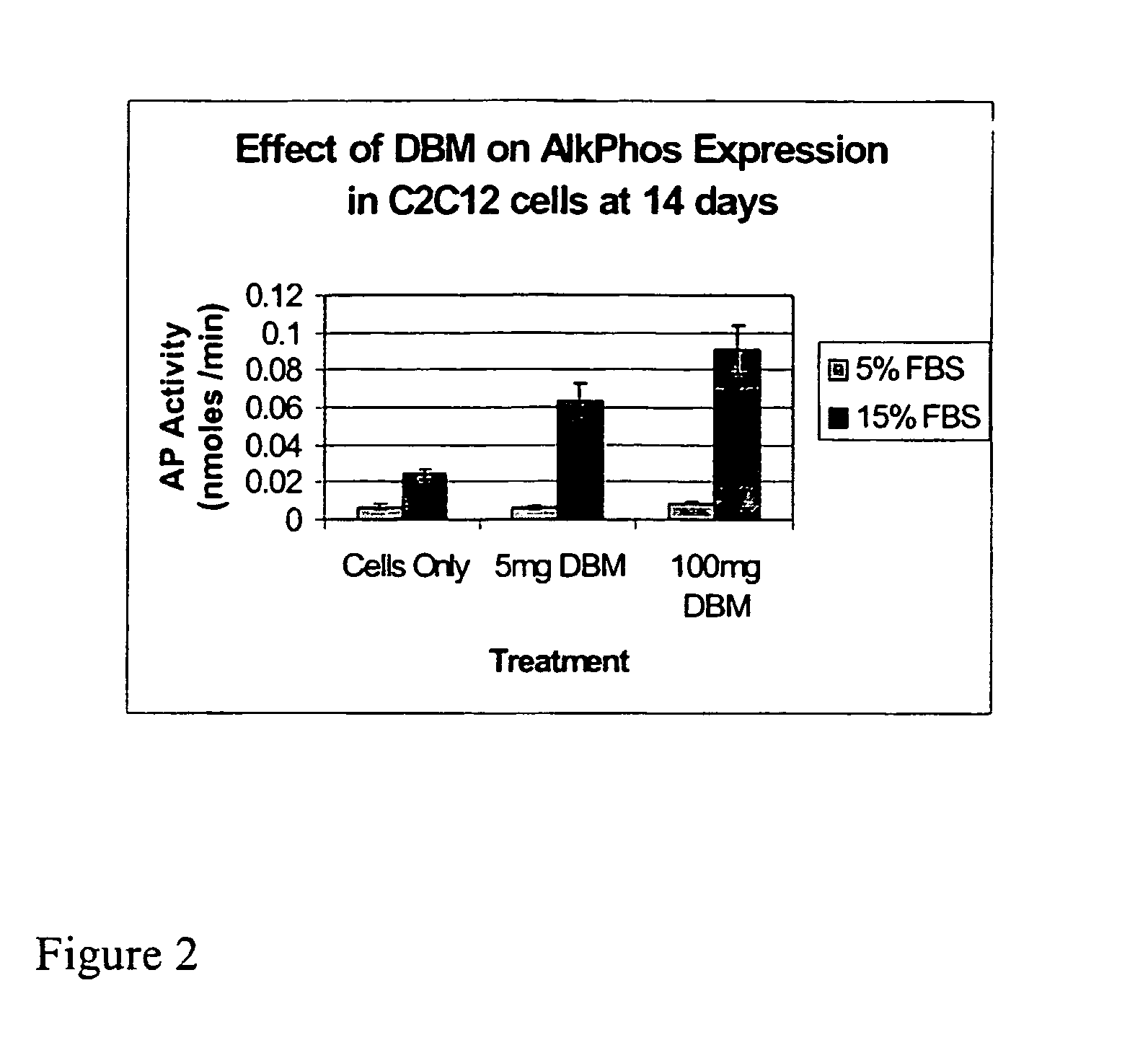
ActiveUS20070154563A1Good osteoinductivityHigh activityHydrolysed protein ingredientsBone implantOsteoblastLine of therapy
The present invention provides methods of improving the osteogenic and / or chondrogenic activity of a bone matrix, e.g., a dermineralized bone matrix (DBM), by exposing the bone matrix to one or more treatments or conditions. In preferred embodiments the bone matrix is derived from human bone. The treatment or condition may alter the structure of the bone matrix and / or cleave one or more specific proteins. Cleavage may generate peptides or protein fragments that have osteoinductive, osteogenic, or chondrogenic activity. Preferred treatments include collagenase and various other proteases. The invention further provides improved bone and cartilage matrix compositions that have been prepared according to the inventive methods and methods of treatment using the compositions. The invention further provides methods of preparing, testing, and using the improved bone matrix compositions. Ona assay comprises exposing relatively undifferentiated mesenchymal cells to a bone matrix composition and measuring expression of a marker characteristic of osteoblast or chondrocyte lineage(s). Increased expression of the marker relative to the level of the marker in cells that have been exposed to a control matrix (e.g., an inactivated or untreated matrix) indicates that the treatment or condition increased the osteogenic and / or chondrogenic activity of the bone matrix. Suitable cells include C2C12 cells. A suitable marker is alkaline phosphatase. The inventive methods increase the osteogenic and / or chondrogenic activity of human DBM when tested using this assay system.
Owner:WARSAW ORTHOPEDIC INC
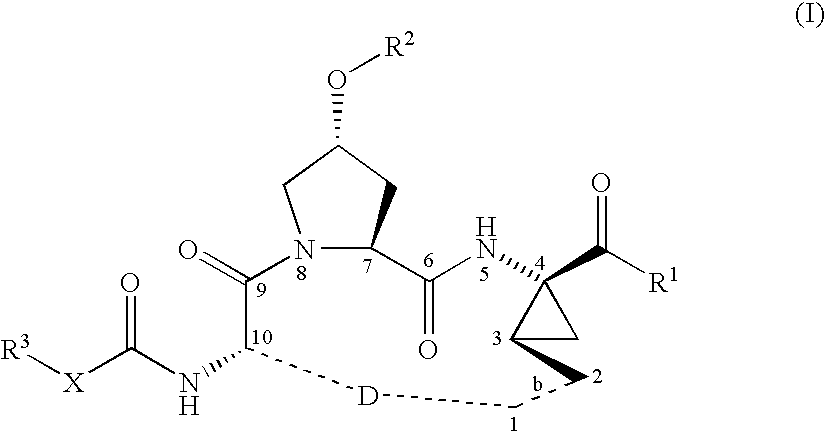
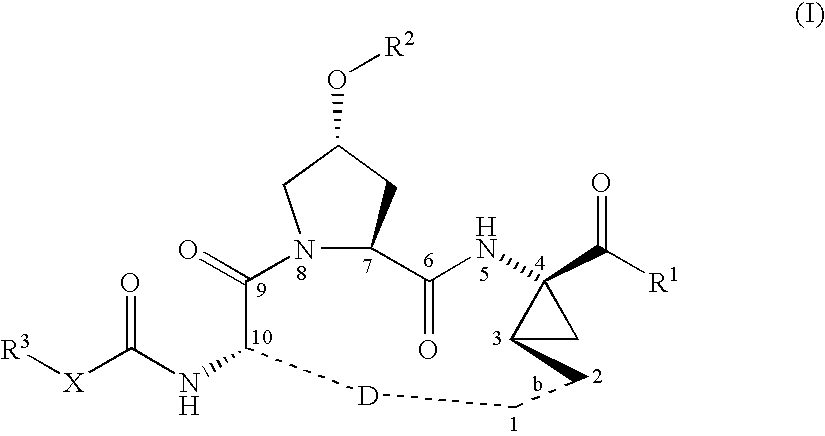
Macrocyclic peptides active against the hepatitis C virus
ActiveUS20050192212A1Inhibition of replicationEffective treatmentBiocideAntiviralsMacrocyclic peptideHcv ns3 protease
Compounds of formula (I): wherein R1, R2, X, R3, D, and the dotted line b are as defined herein; or a pharmaceutically acceptable salt or ester thereof, are useful as inhibitors of the HCV NS3 protease.
Owner:BOEHRINGER INGELHEIM INT GMBH
Supplemented and unsupplemented tissue sealants, methods of their production and use
InactiveUSRE39321E1Decreasing thrombogenicityLow antigenicityAntibacterial agentsOrganic active ingredientsTissue sealantVascular dilatation
This invention provides a fibrin sealant dressing, wherein said fibrin sealant may be supplemented with at least one composition selected from, for example, one or more regulatory compounds, antibody, antimicrobial compositions, analgesics, anticoagulants, antiproliferatives, antiinflammatory compounds, cytokines, cytotoxins, drugs, growth factors, interferons, hormones, lipids, demineralized bone or bone morphogenetic proteins, cartilage inducing factors, oligonucleotides polymers, polysaccharides, polypeptides, protease inhibitors, vasoconstrictors or vasodilators, vitamins, minerals, stabilizers and the like. Also disclosed are methods of preparing and / or using the unsupplemented or supplemented fibrin sealant dressing.
Owner:AMERICAN NAT RED CROSS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com