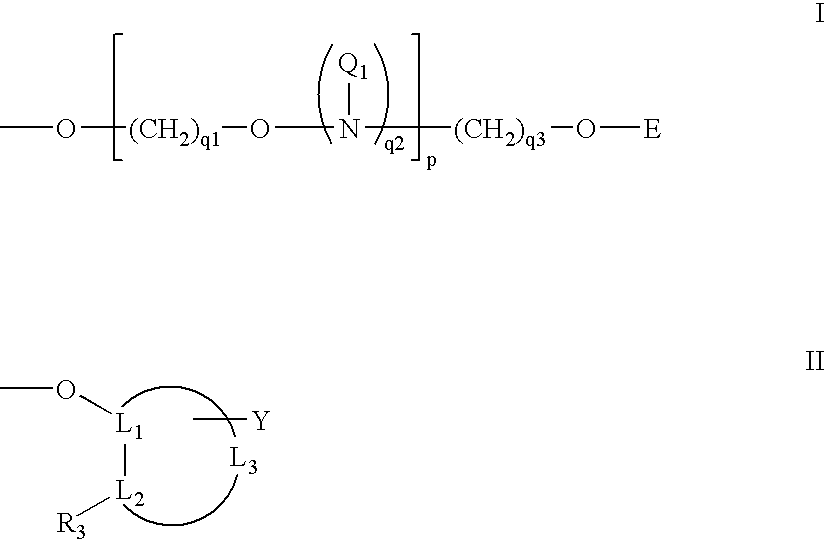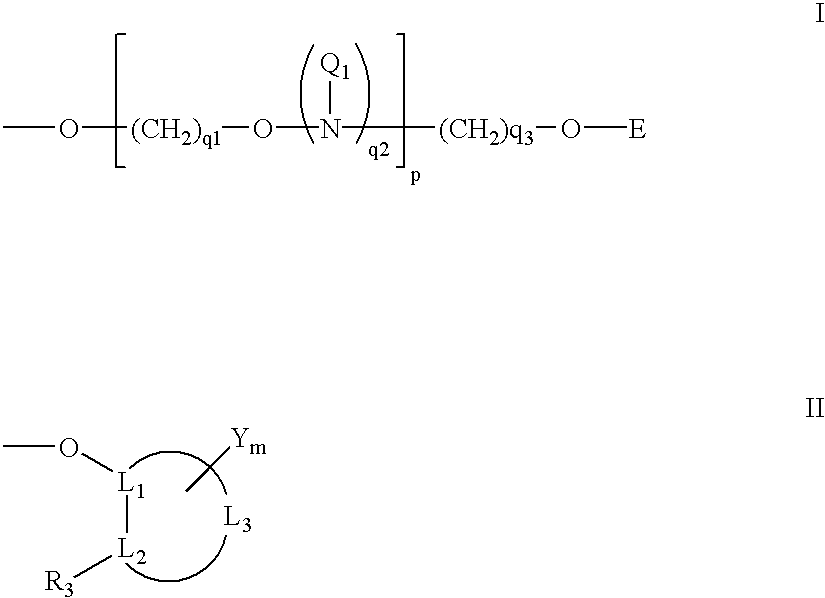Patents
Literature
203 results about "Nucleic Acid Strand" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
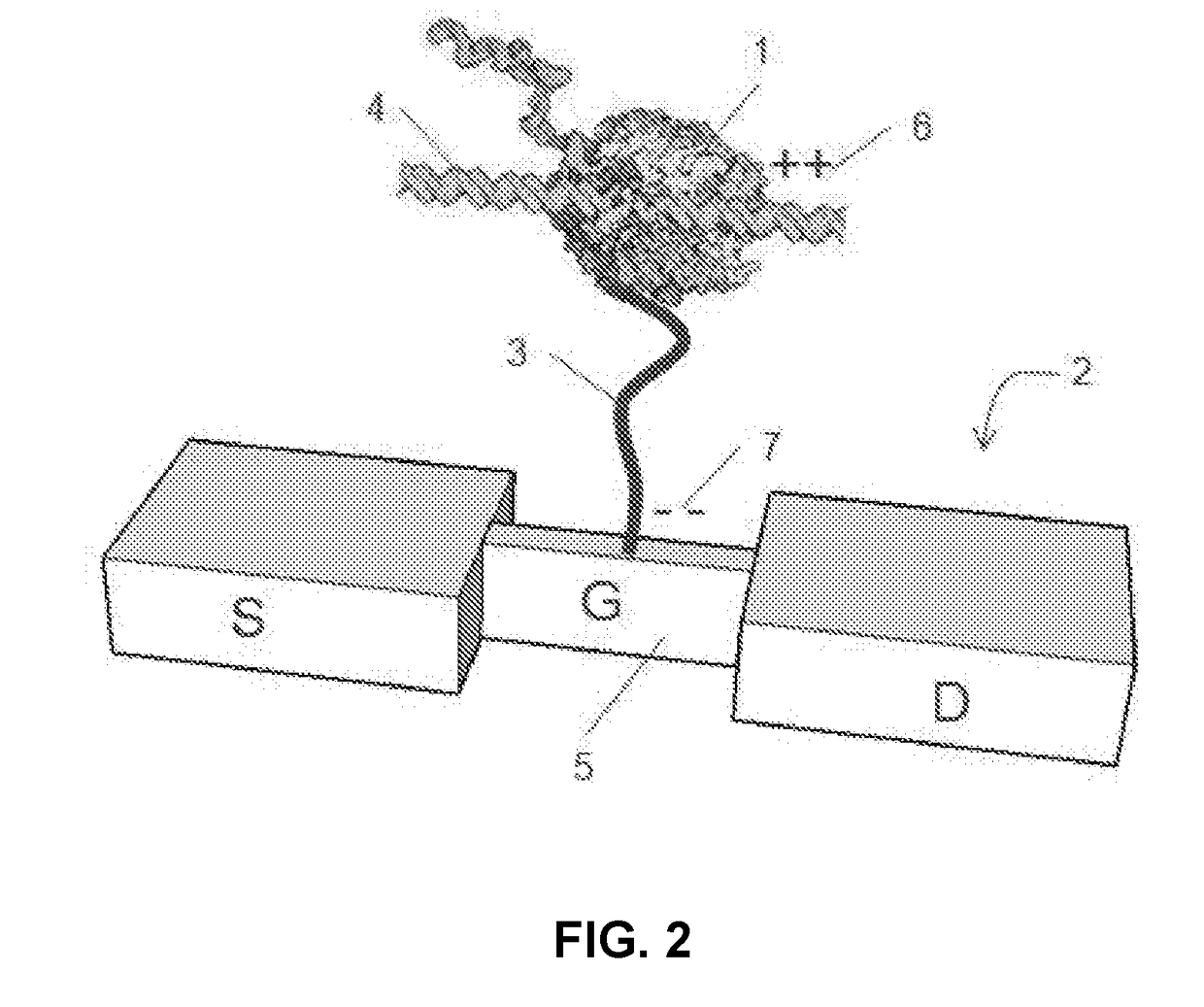
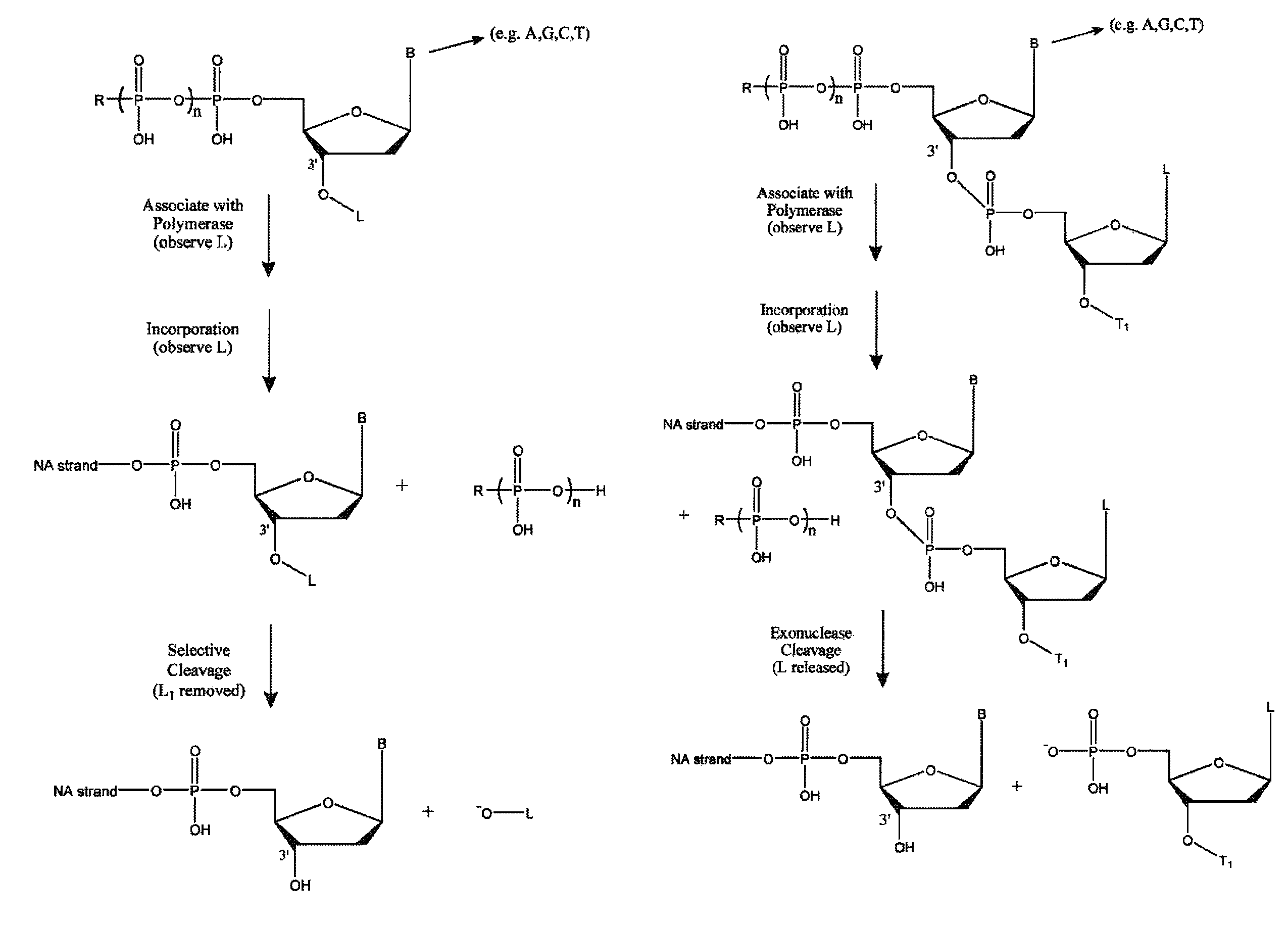
A single chain of nucleotides.
Ultra-fast nucleic acid sequencing device and a method for making and using the same
InactiveUS7001792B2Accurate identificationAccurately and effectively identifying bases of DNABioreactor/fermenter combinationsBiological substance pretreatmentsUltra fastNucleic acid sequencing
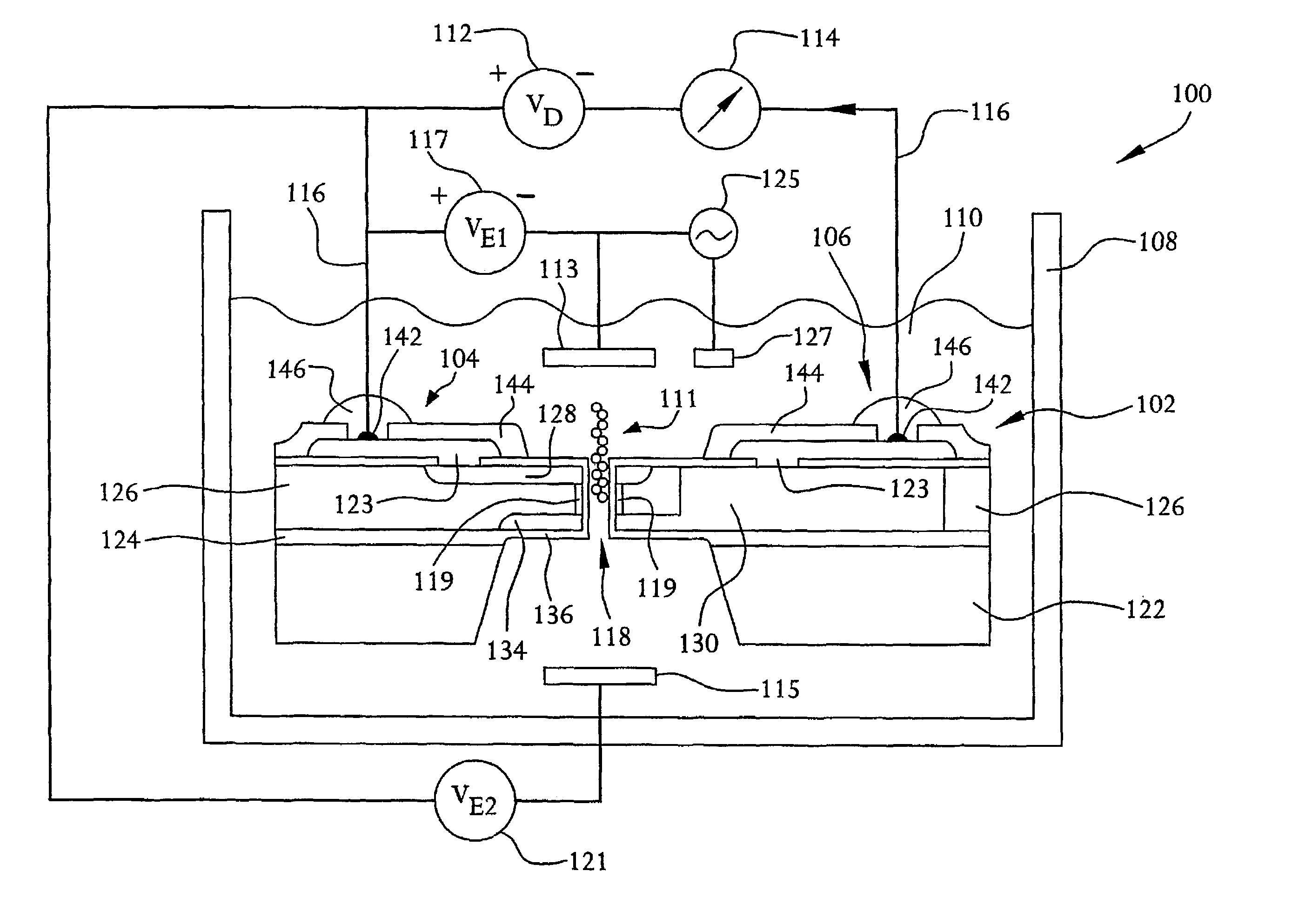
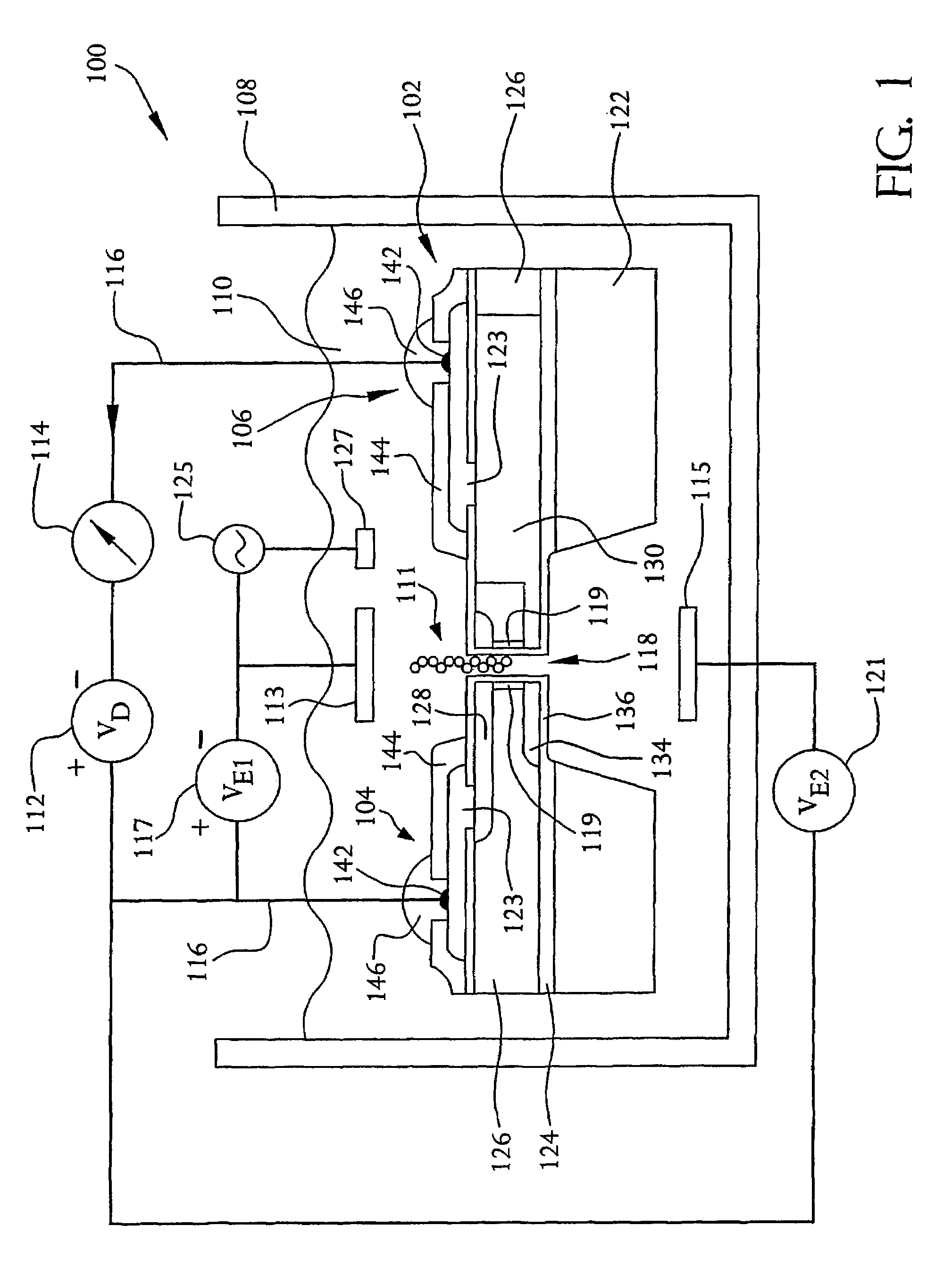
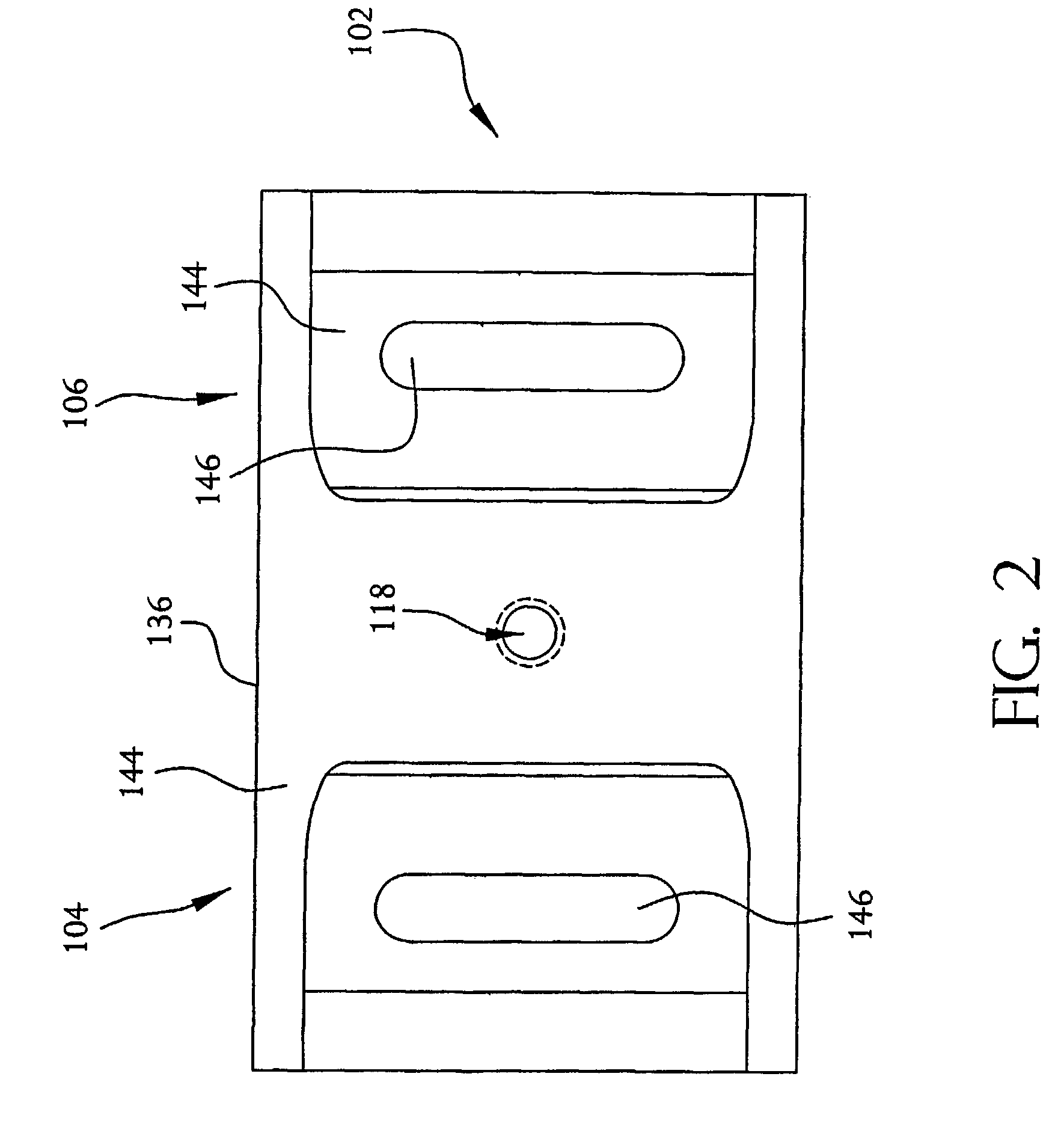
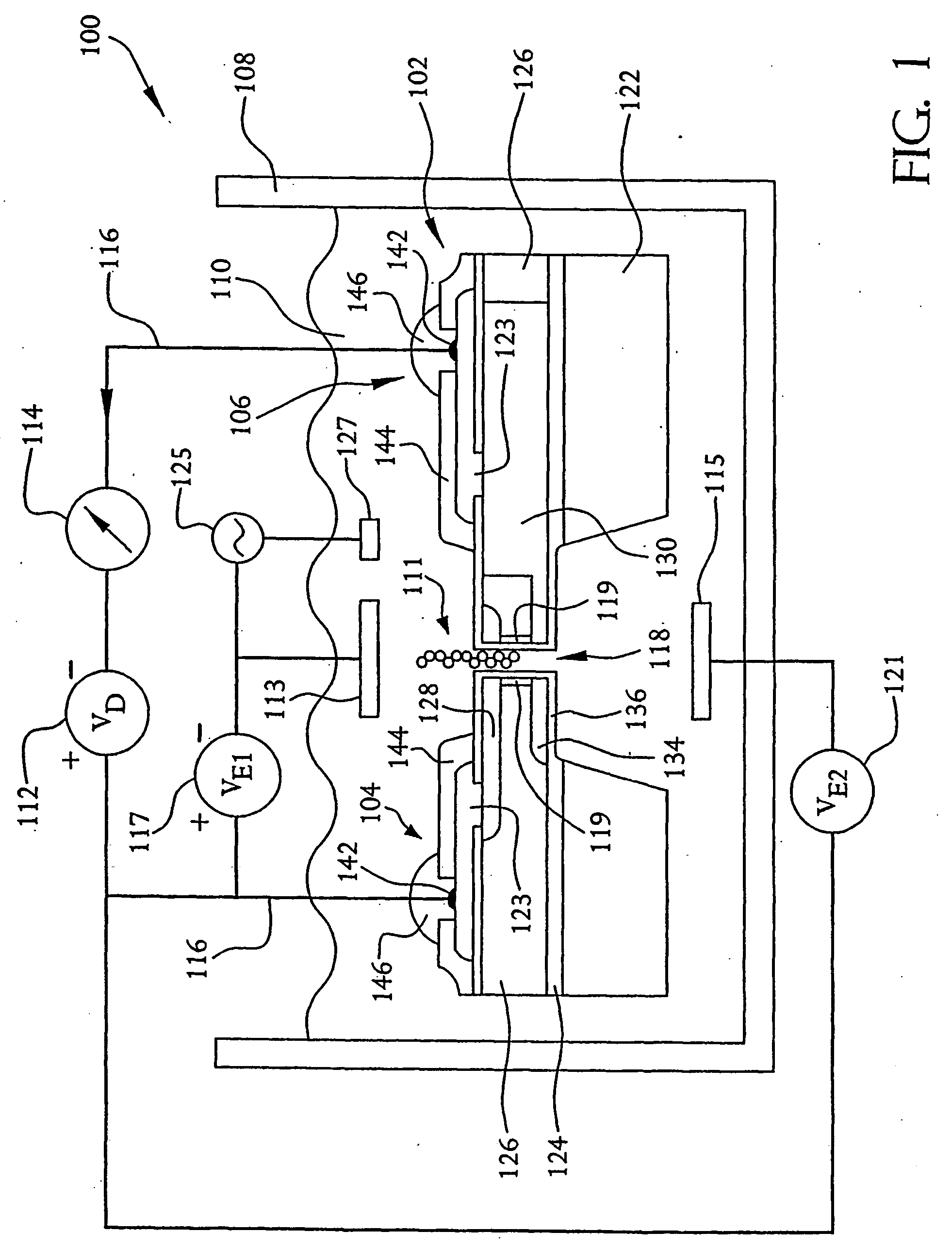
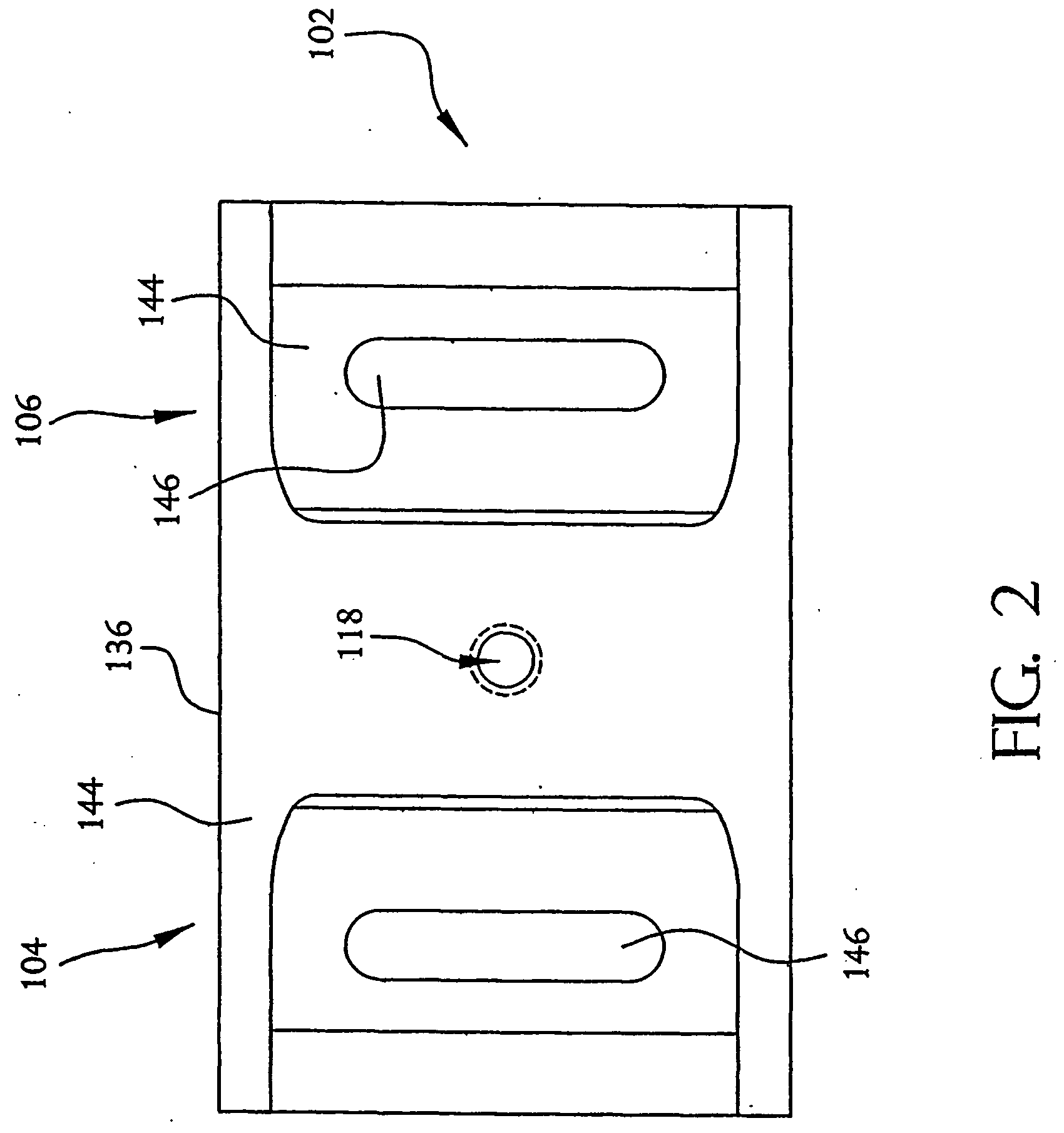
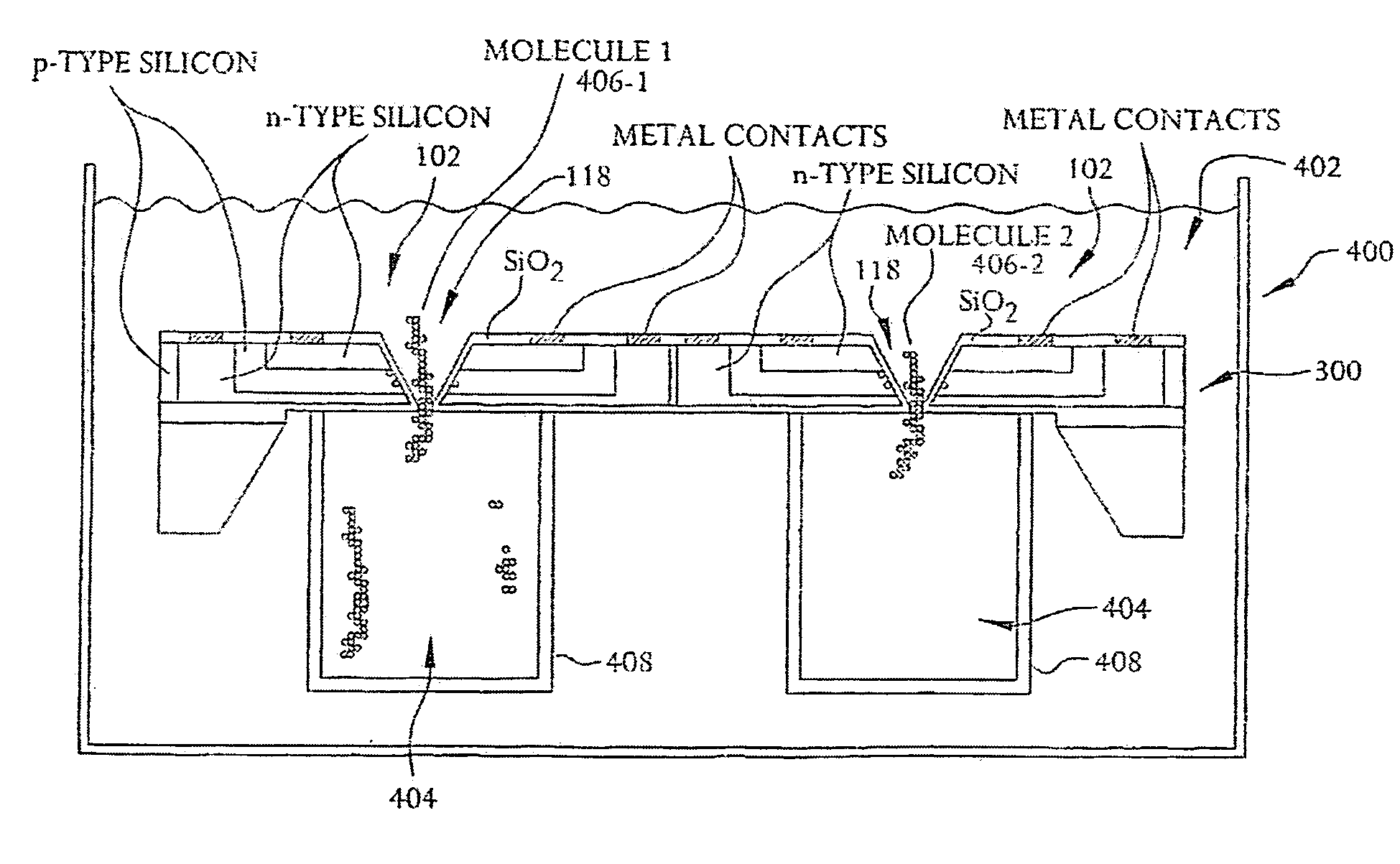
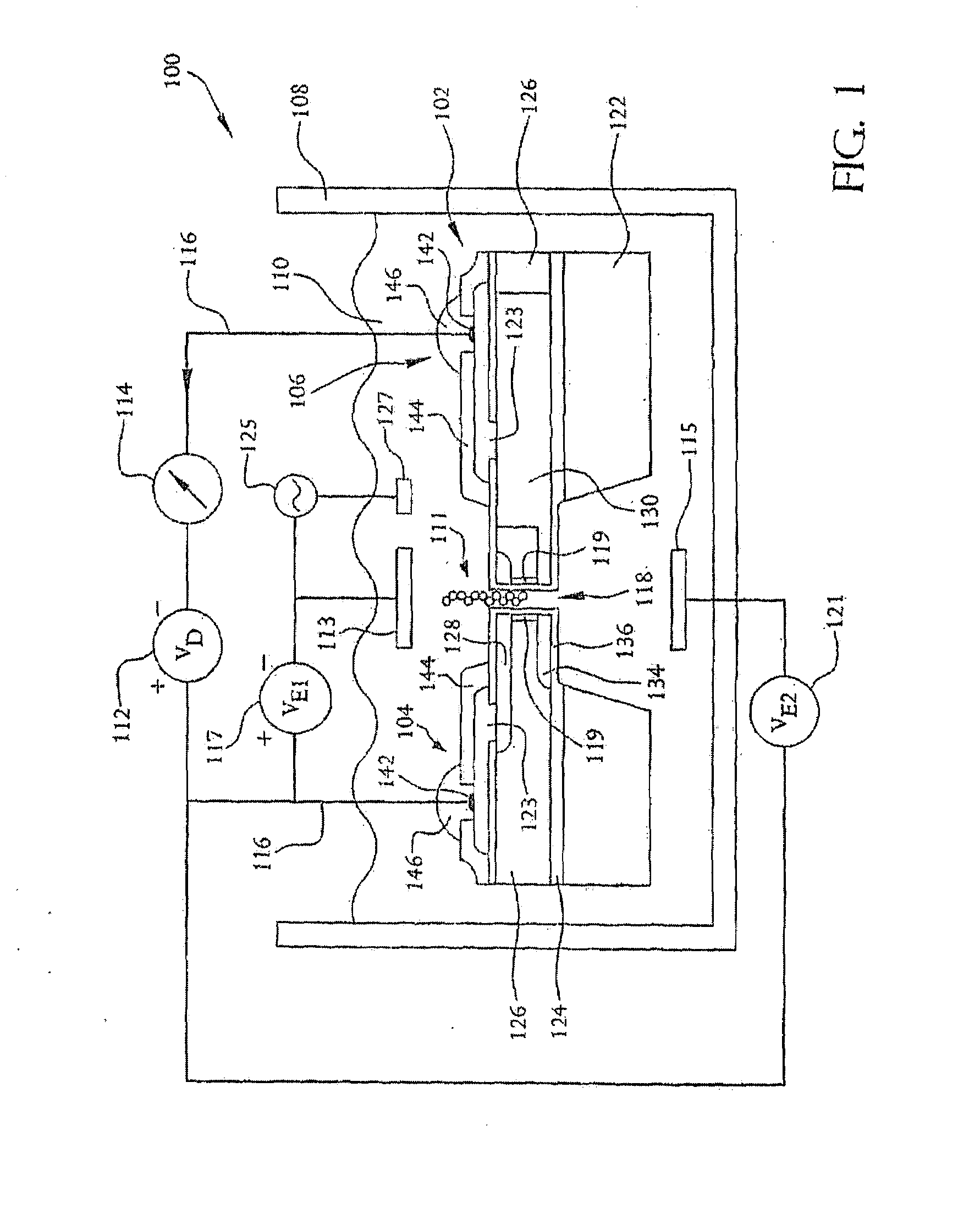
A system and method employing at least one semiconductor device, or an arrangement of insulating and metal layers, having at least one detecting region which can include, for example, a recess or opening therein, for detecting a charge representative of a component of a polymer, such as a nucleic acid strand, proximate to the detecting region, and a method for manufacturing such a semiconductor device. The system and method can thus be used for sequencing individual nucleotides or bases of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA). The semiconductor device includes at least two doped regions, such as two n-typed regions implanted in a p-typed semiconductor layer or two p-typed regions implanted in an n-typed semiconductor layer. The detecting region permits a current to pass between the two doped regions in response to the presence of the component of the polymer, such as a base of a DNA or RNA strand. The current has characteristics representative of the component of the polymer, such as characteristics representative of the detected base of the DNA or RNA strand.
Owner:LIFE TECH CORP
Oligonucleotide arrays and their use for sorting, isolating, sequencing, and manipulating nucleic acids
InactiveUS6322971B1Increase in hybridization specificityBioreactor/fermenter combinationsMaterial nanotechnologyHybridization ArrayBioinformatics
Ligation methods for manipulating nucleic acid stands and oligonucleotides utilizing hybridization arrays of immobilized oligonucleotides. The oligonucleotide arrays may be plain or sectioned, comprehensive or non-comprehensive. The immobilized oligonucleotides may in some cases be binary oligonucleotides having constant as well as variable segments. Some embodiments include amplification of ligated products.
Owner:UNIV OF MEDICINE & DENTISTRY OF NEW JERSEY
High speed parallel molecular nucleic acid sequencing
InactiveUS6982146B1Minimize shearImprove automationSugar derivativesMicrobiological testing/measurementNucleotidePolymerase L
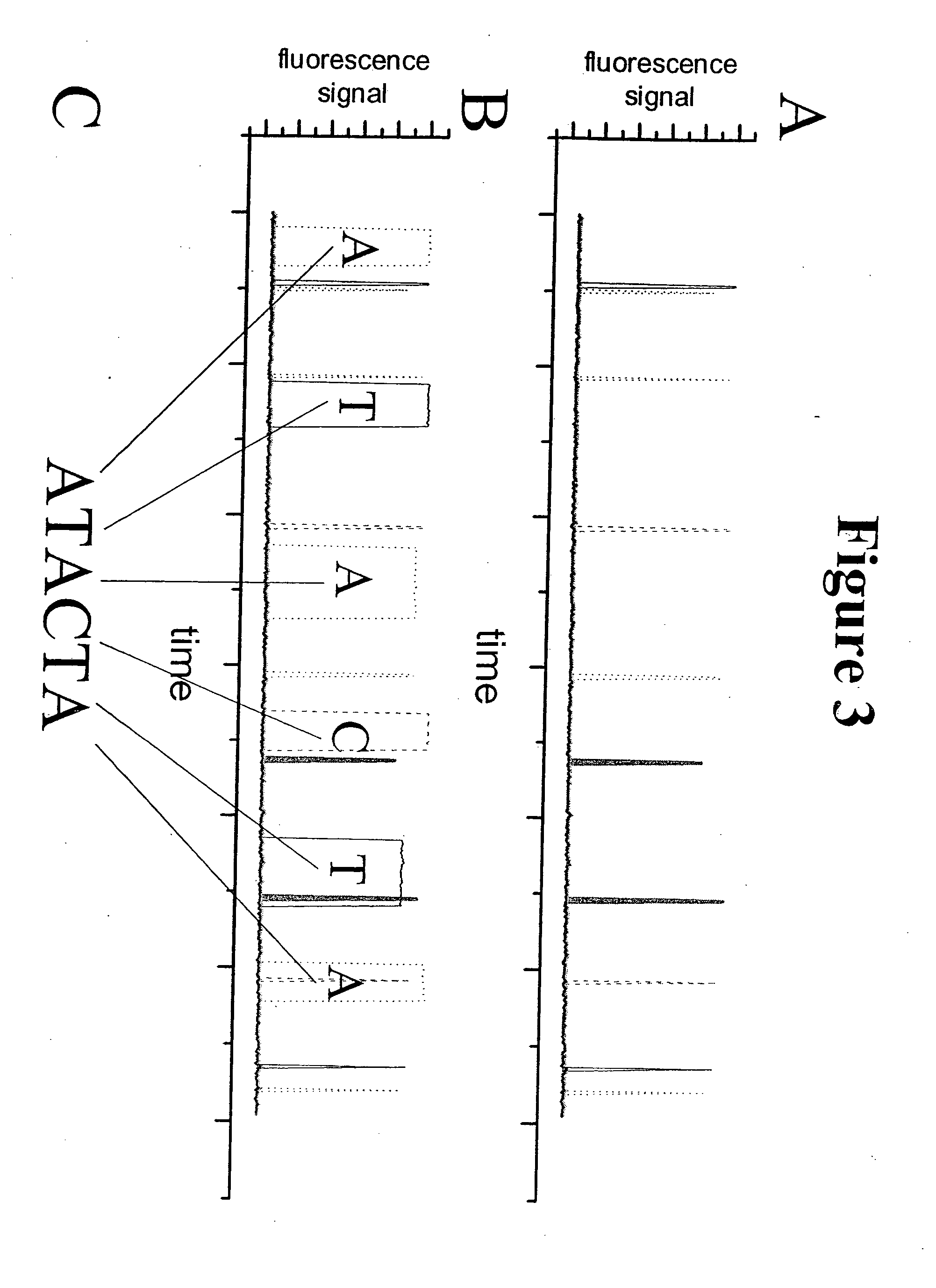
A method and device is disclosed for high speed, automated sequencing of nucleic acid molecules. A nucleic acid molecule to be sequenced is exposed to a polymerase in the presence of nucleotides which are to be incorporated into a complementary nucleic acid strand. The polymerase carries a donor fluorophore, and each type of nucleotide (e.g. A, T / U, C and G) carries a distinguishable acceptor fluorophore characteristic of the particular type of nucleotide. As the polymerase incorporates individual nucleic acid molecules into a complementary strand, a laser continuously irradiates the donor fluorophore, at a wavelength that causes it to emit an emission signal (but the laser wavelength does not stimulate the acceptor fluorophore). In particular embodiments, no laser is needed if the donor fluorophore is a luminescent molecule or is stimulated by one. The emission signal from the polymerase is capable of stimulating any of the donor fluorophores (but not acceptor fluorophores), so that as a nucleotide is added by the polymerase, the acceptor fluorophore emits a signal associated with the type of nucleotide added to the complementary strand. The series of emission signals from the acceptor fluorophores is detected, and correlated with a sequence of nucleotides that correspond to the sequence of emission signals.
Owner:GOVERNMENT OF US SEC THE DEPT OF HEALTH & HUMAN SERVICES THE
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432250B2High affinityStrong specificityHydrolasesPeptide/protein ingredientsOrganismResearch purpose
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
Ultra-fast nucleic acid sequencing device and a method for making and using the same
InactiveUS20060154399A1Accurate identificationAccurately and effectively identifying bases of DNAMaterial nanotechnologySemiconductor/solid-state device testing/measurementDevice materialUltra fast
A system and method employing at least one semiconductor device, or an arrangement of insulating and metal layers, having at least one detecting region which can include, for example, a recess or opening therein, for detecting a charge representative of a component of a polymer, such as a nucleic acid strand, proximate to the detecting region, and a method for manufacturing such a semiconductor device. The system and method can thus be used for sequencing individual nucleotides or bases of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA). The semiconductor device includes at least two doped regions, such as two n-type regions implanted in a p-type semiconductor layer or two p-type regions implanted in an n-type semiconductor layer. The detecting region permits a current to pass between the two doped regions in response to the presence of the component of the polymer, such as a base of a DNA or RNA strand. The current has characteristics representative of the component of the polymer, such as characteristics representative of the detected base of the DNA or RNA strand.
Owner:LIFE TECH CORP
Methods and compositions for isolating nucleic acid sequence variants
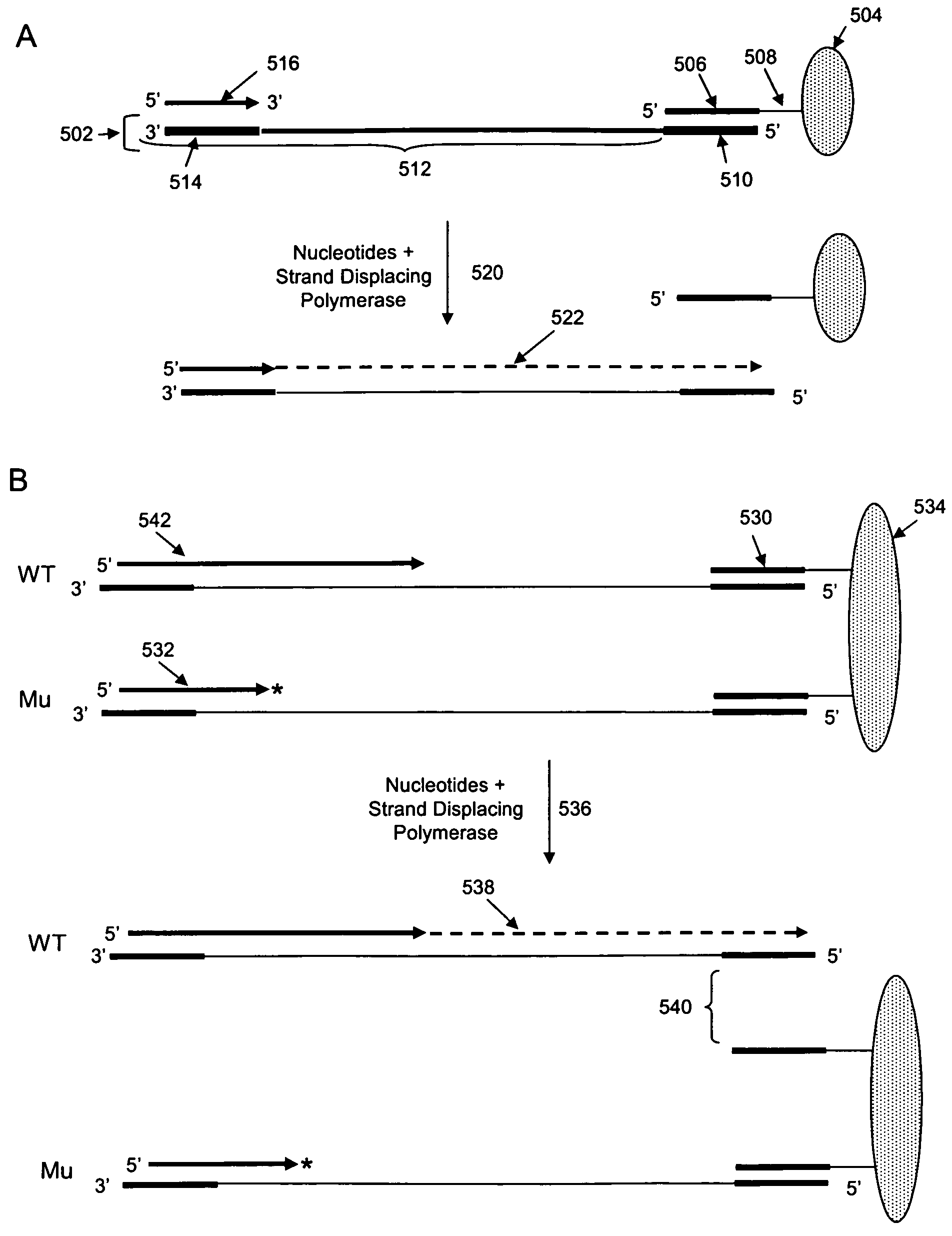
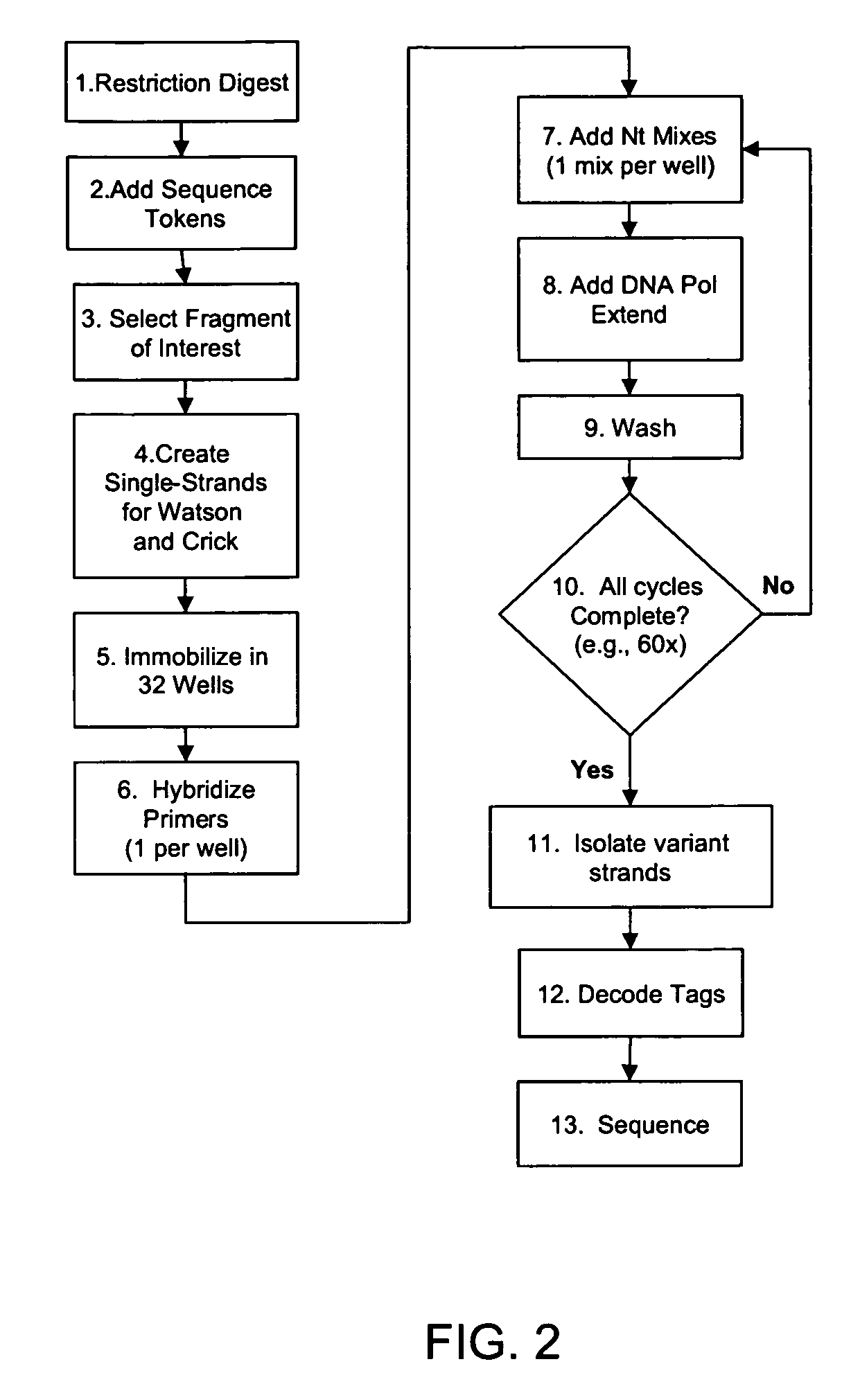
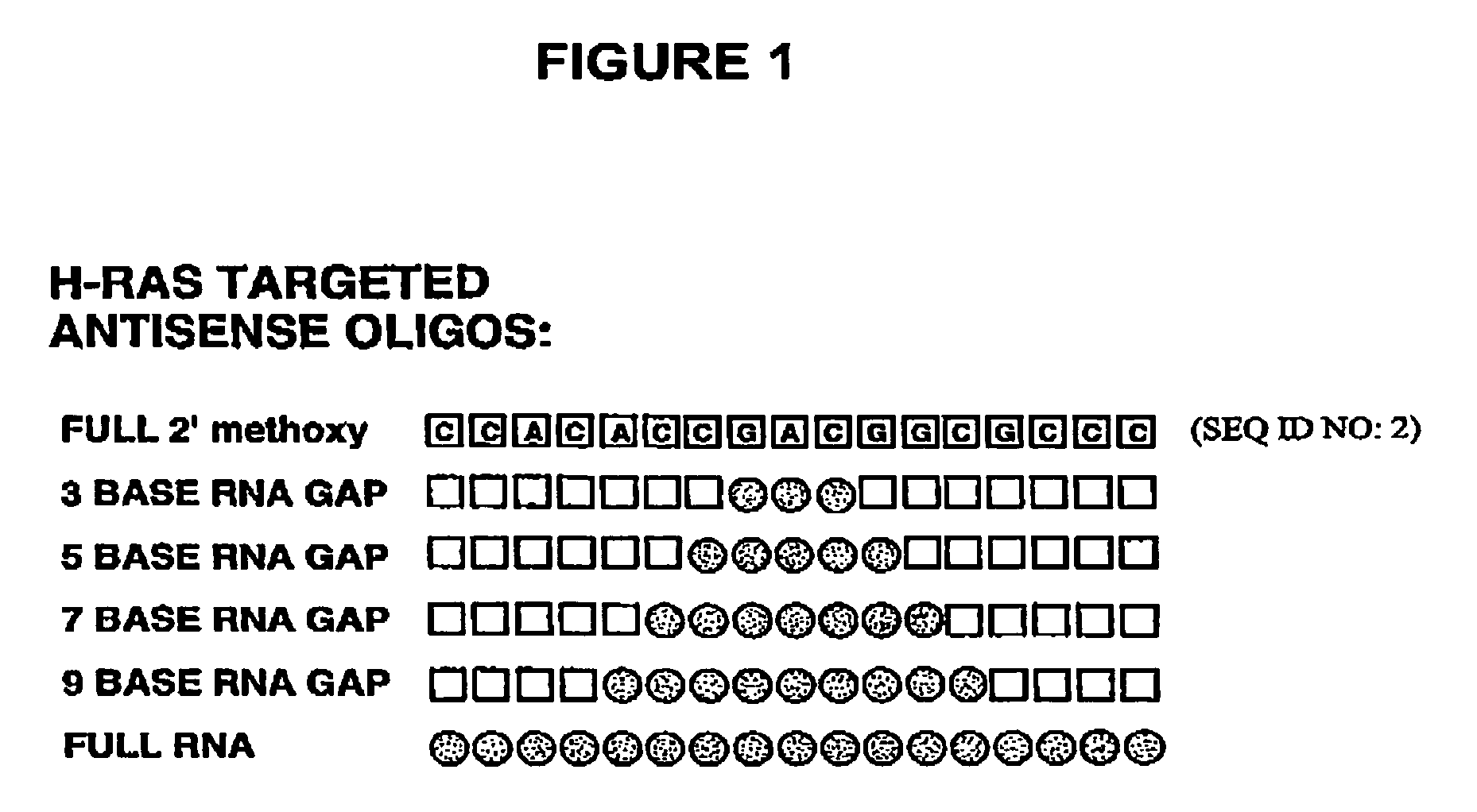
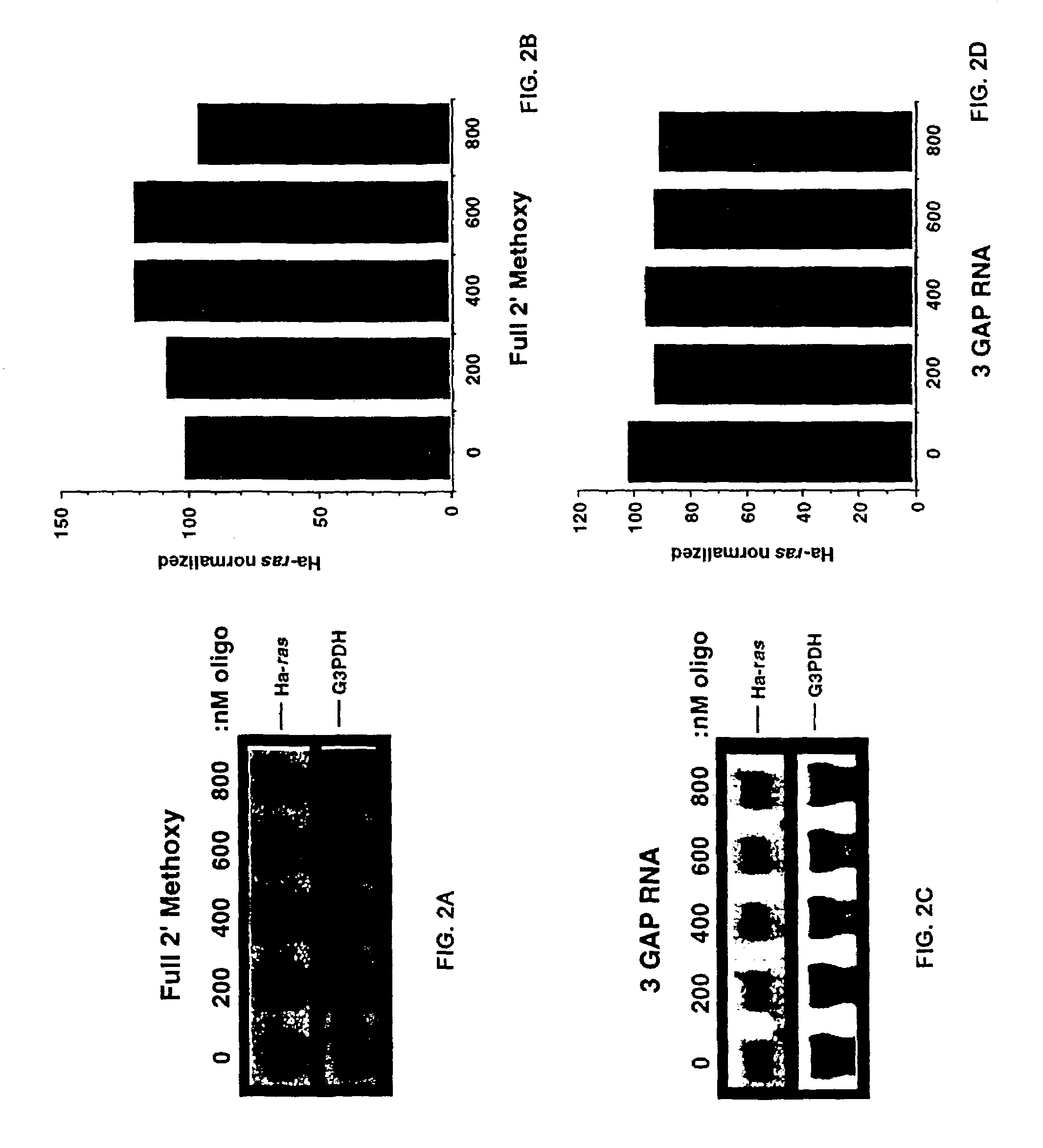
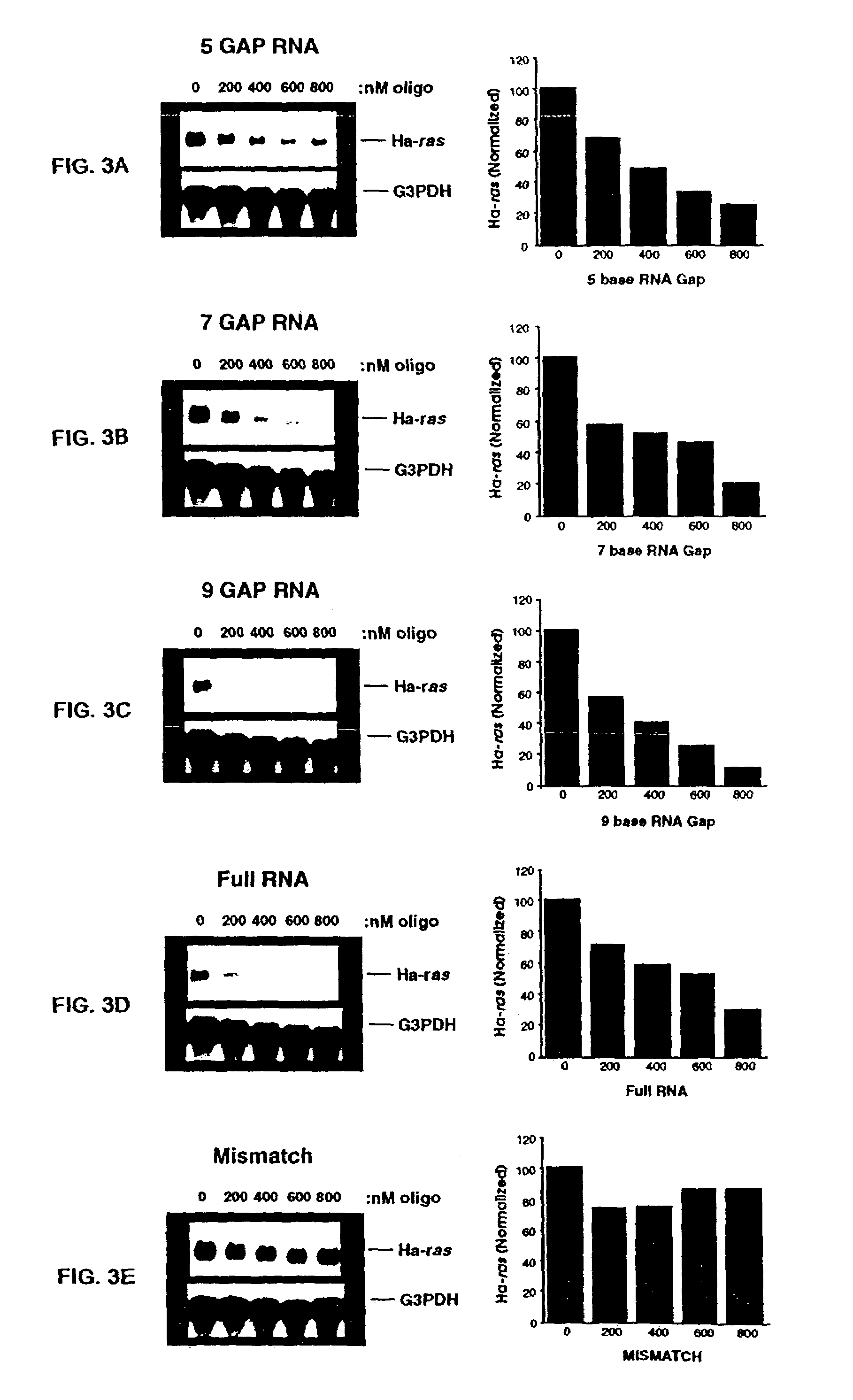
The invention is drawn to isolating sequence variants of a genetic locus of interest using a modified iterative primer extension method. The nucleic acids analyzed are generally single stranded and have a reference sequence which is used as a basis for performing iterative single nucleotide extension reactions from a hybridized polymerization primer. The iterative polymerization reactions are configured such that polymerization of the strand will continue if the sequence of the nucleic acid being analyzed matches the reference sequence, whereas polymerization will be terminated if the nucleic acid being analyzed does not match the reference sequence. Nucleic acid strands that have mutations can be isolated using a variety of methods and sequenced to determine the precise identity of the mutation / polymorphism. By performing the method on both strands of the nucleic acid being analyzed, virtually all possible mutations can be identified.
Owner:AGENCY FOR SCI TECH & RES
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7432249B2High affinityStrong specificityPeptide/protein ingredientsHydrolasesADAMTS ProteinsOrganism
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
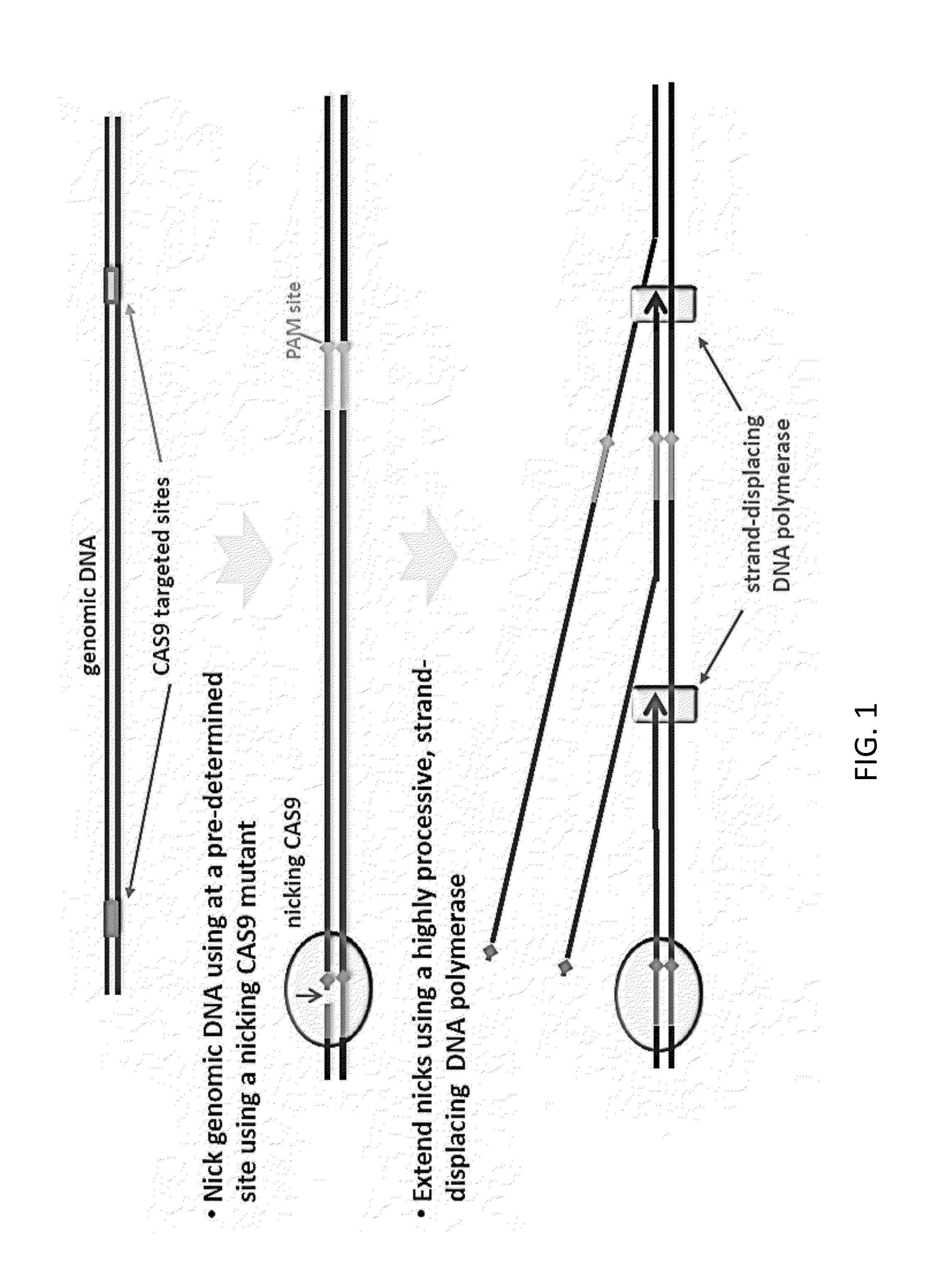
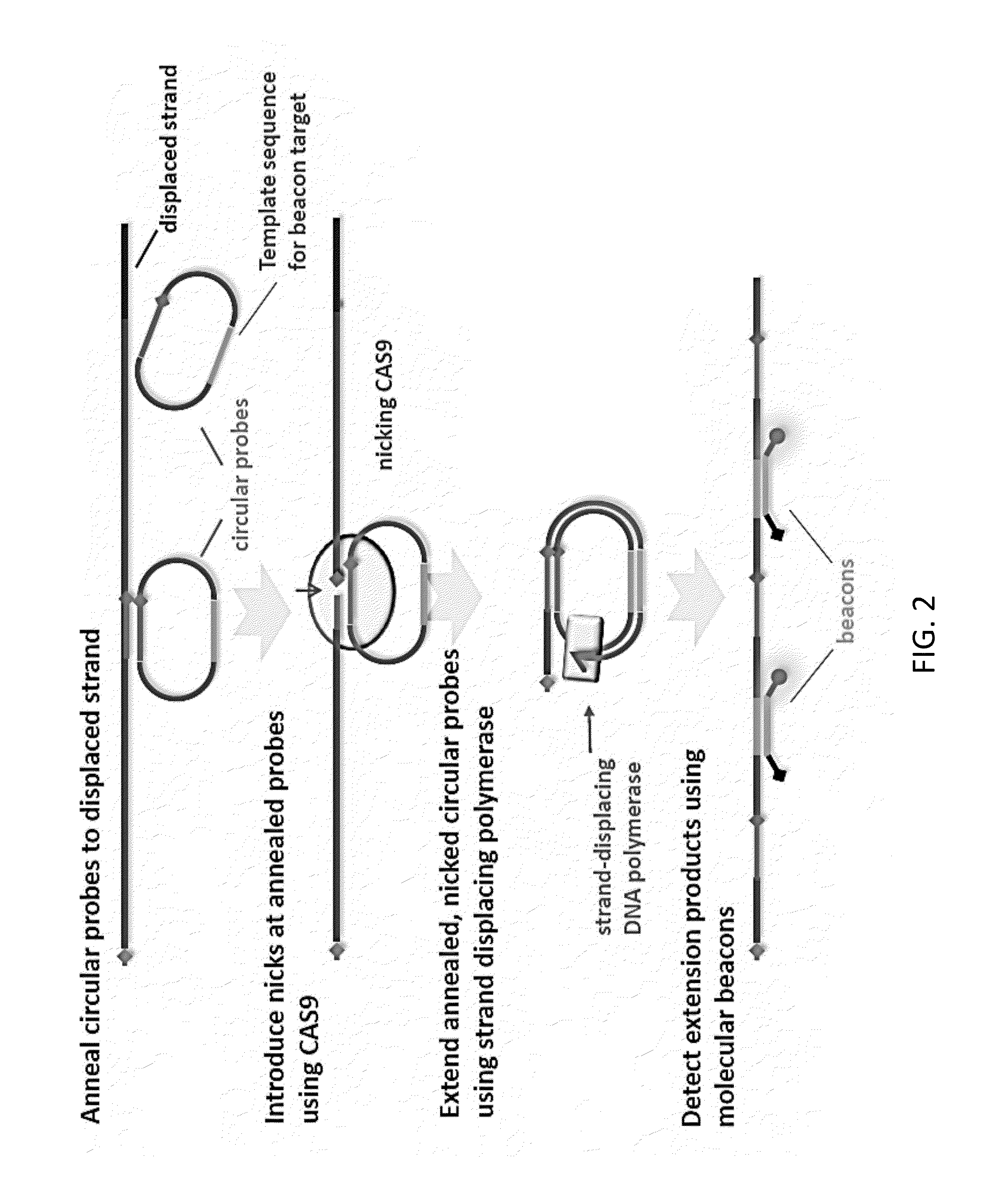
CAS9-based Isothermal Method of Detection of Specific DNA Sequence
ActiveUS20150211058A1Microbiological testing/measurementVector-based foreign material introductionDNANucleic acid
The present invention relates to an isothermal method for detecting in a sample a target nucleic acid strand.
Owner:AGILENT TECH INC
Oligoribonucleotides and ribonucleases for cleaving RNA
InactiveUS7629321B2High affinityStrong specificityHydrolasesPeptide/protein ingredientsOrganismResearch purpose
Oligomeric compounds including oligoribonucleotides and oligoribonucleosides are provided that have subsequences of 2′-pentoribofuranosyl nucleosides that activate dsRNase. The oligoribonucleotides and oligoribonucleosides can include substituent groups for increasing binding affinity to complementary nucleic acid strand as well as substituent groups for increasing nuclease resistance. The oligomeric compounds are useful for diagnostics and other research purposes, for modulating the expression of a protein in organisms, and for the diagnosis, detection and treatment of other conditions susceptible to oligonucleotide therapeutics. Also included in the invention are mammalian ribonucleases, i.e., enzymes that degrade RNA, and substrates for such ribonucleases. Such a ribonuclease is referred to herein as a dsRNase, wherein “ds” indicates the RNase's specificity for certain double-stranded RNA substrates. The artificial substrates for the dsRNases described herein are useful in preparing affinity matrices for purifying mammalian ribonuclease as well as non-degradative RNA-binding proteins.
Owner:IONIS PHARMA INC
High speed parallel molecular nucleic acid sequencing
InactiveUS20060292583A1Minimize shearImprove automationBioreactor/fermenter combinationsBiological substance pretreatmentsPolymerase LFluorophore
A method and device is disclosed for high speed, automated sequencing of nucleic acid molecules. A nucleic acid molecule to be sequenced is exposed to a polymerase in the presence of nucleotides which are to be incorporated into a complementary nucleic acid strand. The polymerase carries a donor fluorophore, and each type of nucleotide (e.g. A, T / U, C and G) carries a distinguishable acceptor fluorophore characteristic of the particular type of nucleotide. As the polymerase incorporates individual nucleic acid molecules into a complementary strand, a laser continuously irradiates the donor fluorophore, at a wavelength that causes it to emit an emission signal (but the laser wavelength does not stimulate the acceptor fluorophore). In particular embodiments, no laser is needed if the donor fluorophore is a luminescent molecule or is stimulated by one. The emission signal from the polymerase is capable of stimulating any of the donor fluorophores (but not acceptor fluorophores), so that as a nucleotide is added by the polymerase, the acceptor fluorophore emits a signal associated with the type of nucleotide added to the complementary strand. The series of emission signals from the acceptor fluorophores is detected, and correlated with a sequence of nucleotides that correspond to the sequence of emission signals.
Owner:THE GOVERNMENT OF US REPRESENTED BY THE SEC OF THE DEPT OF HEALTH & HUMAN SERVICES
Method for nucleotide detection
ActiveUS20120196758A1Prevent degradationMicrobiological testing/measurementLibrary screeningGallic acid esterNucleic acid detection
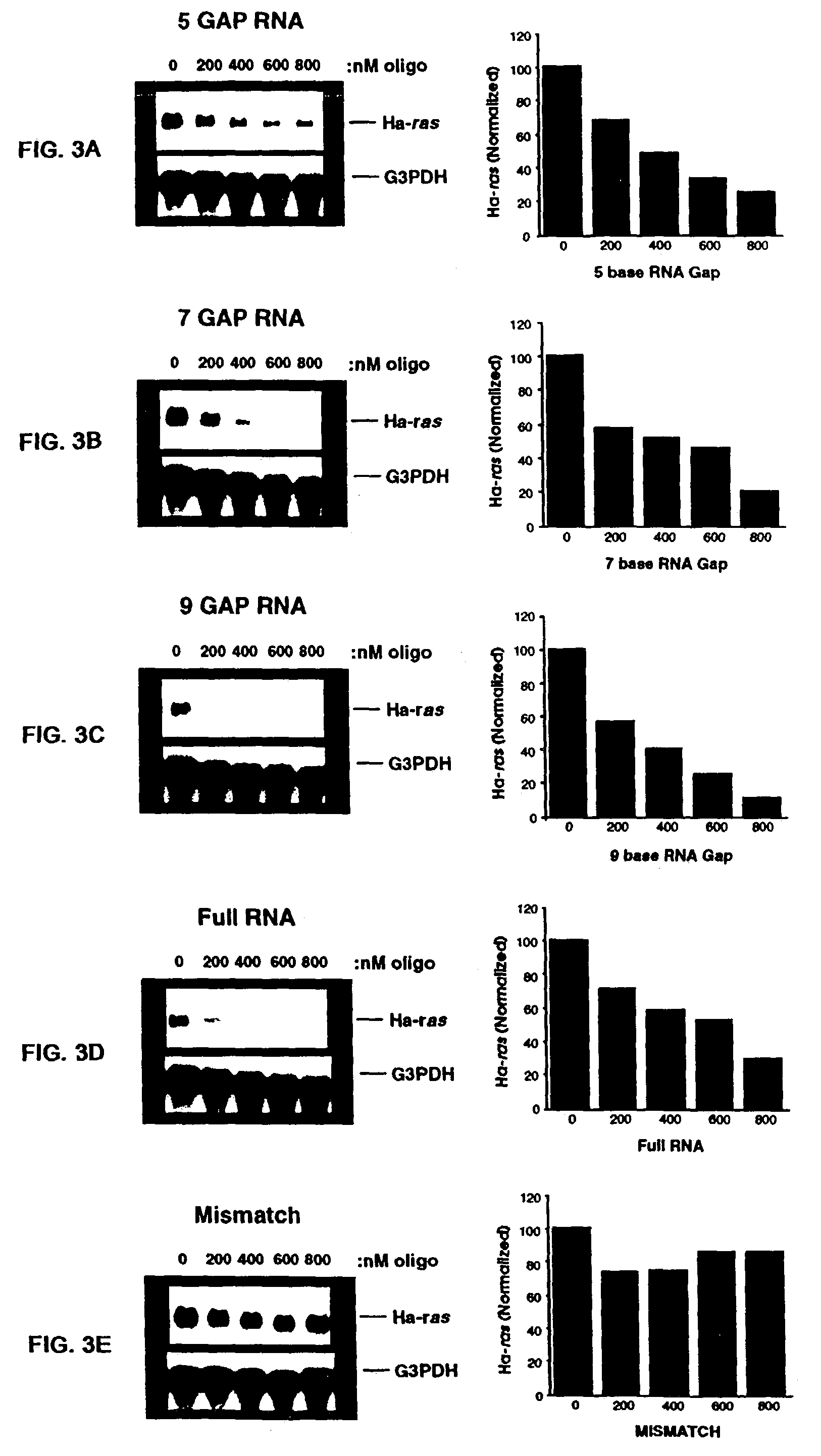
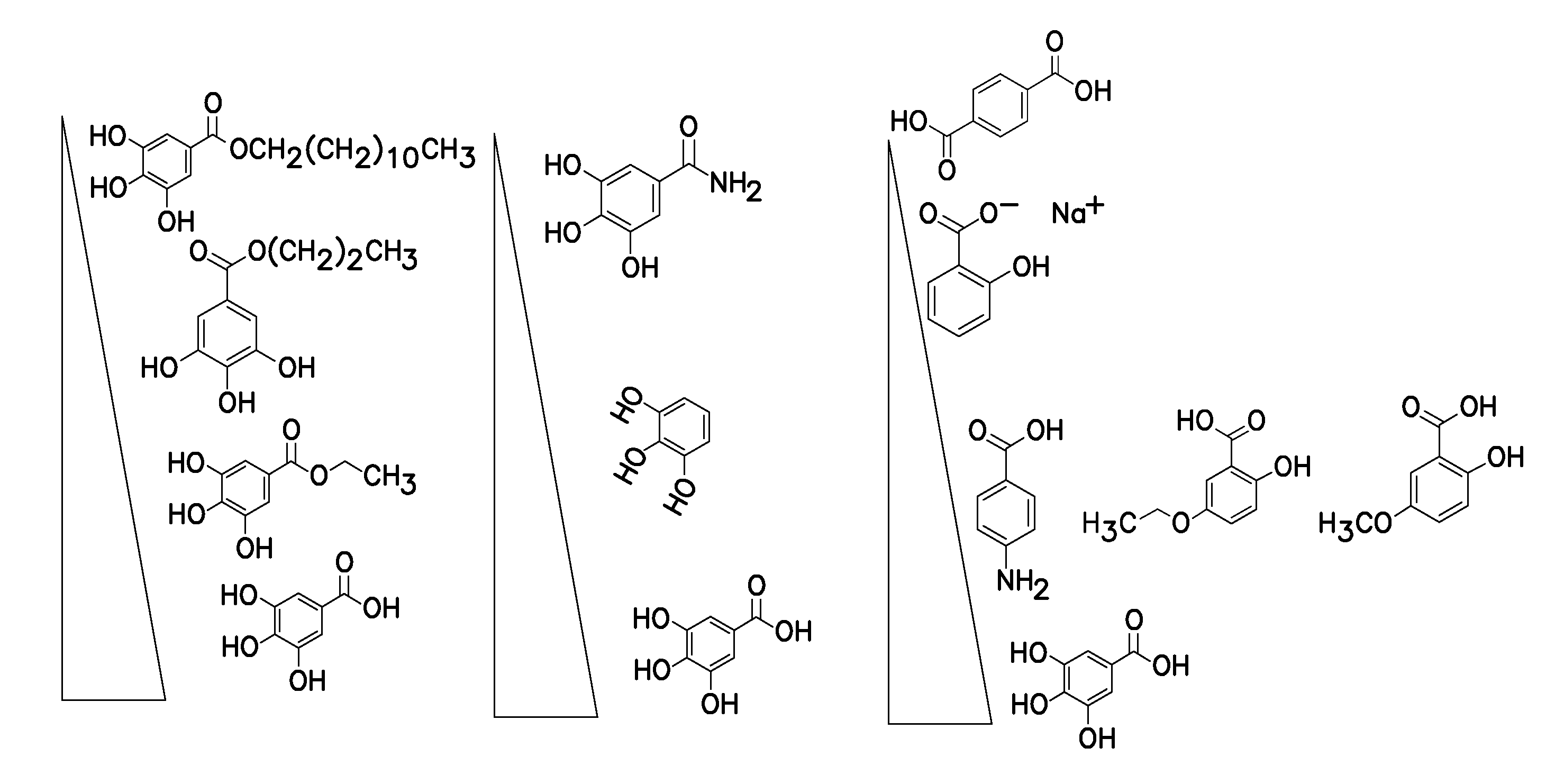
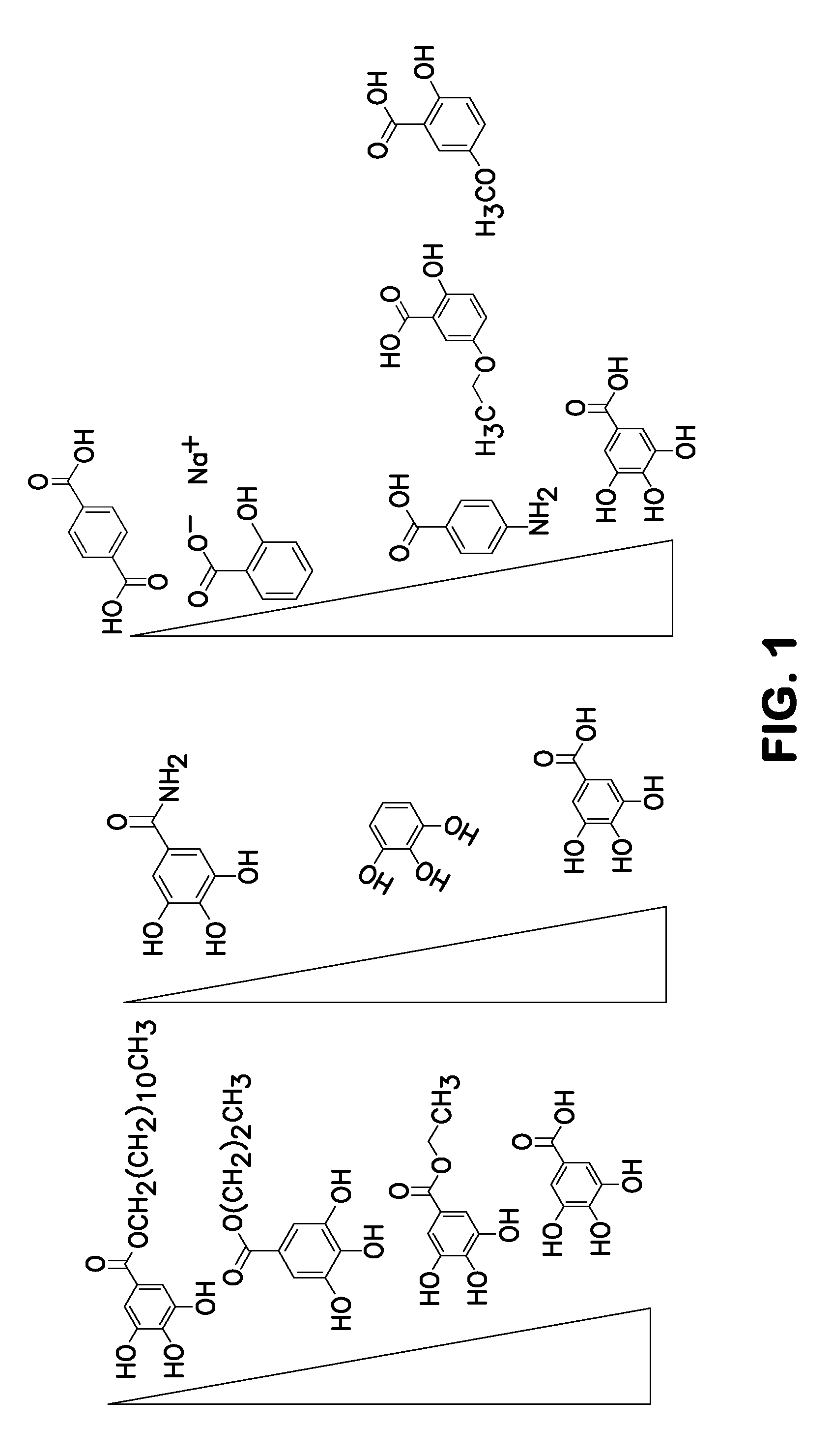
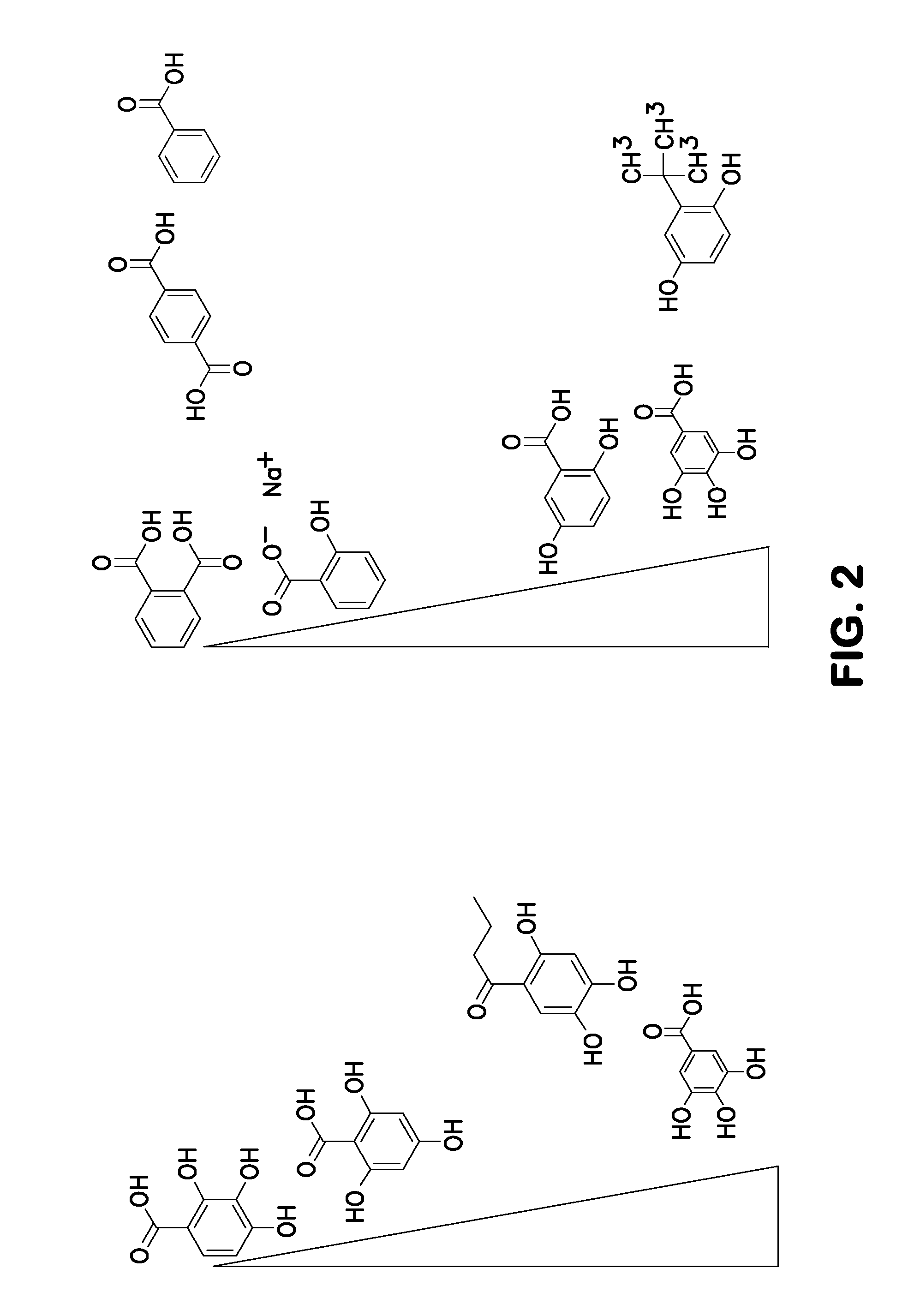
A method of inhibiting light-induced degradation of nucleic acids includes irradiating a portion of the nucleic acids in the presence of a detection solution comprising a polyphenolic compound. A method of detecting a nucleic acid having a fluorescent tag includes irradiating at least a portion of the nucleic acid with light of a suitable wavelength to induce a fluorescence emission and detecting the fluorescence emission. Optionally, the polyphenolic compound is gallic acid, a lower alkyl ester thereof, or mixtures thereof. A kit includes one or more nucleotides, an enzyme capable of catalyzing incorporation of the nucleotides into a nucleic acid strand and a polyphenolic compound suitable for preparing a detection solution.
Owner:ILLUMINA INC
Compositions and methods to prevent AAV vector aggregation
ActiveUS7704721B2Increase ionic strengthImprove efficiencyRecovery/purificationSsDNA virusesFreeze thawingIonic strength
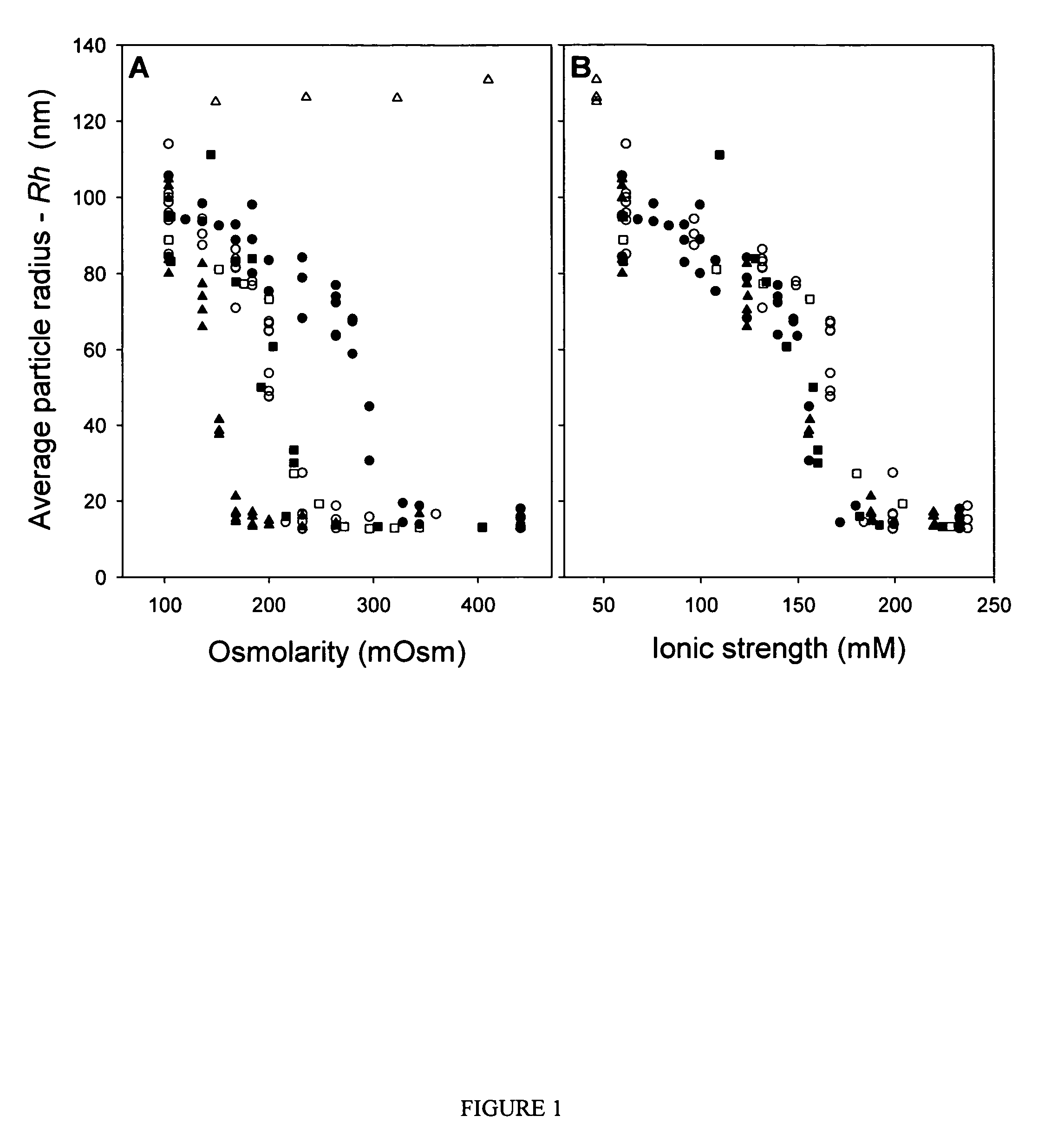
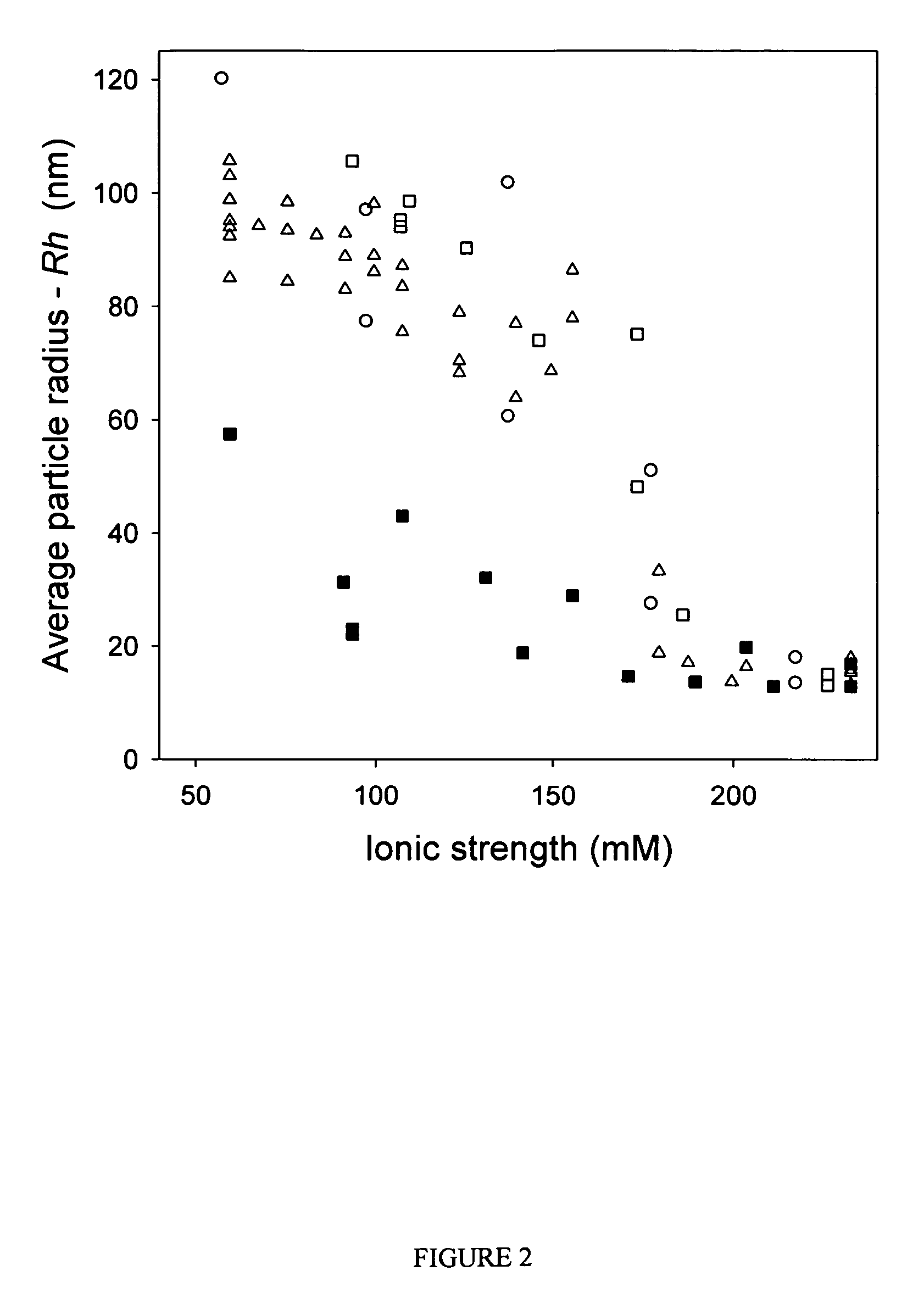
Compositions and methods are provided for preparation of concentrated stock solutions of AAV virions without aggregation. Formulations for AAV preparation and storage are high ionic strength solutions (e.g. μ˜500 mM) that are nonetheless isotonic with the intended target tissue. This combination of high ionic strength and modest osmolarity is achieved using salts of high valency, such as sodium citrate. AAV stock solutions up to 6.4×1013 vg / mL are possible using the formulations of the invention, with no aggregation being observed even after ten freeze-thaw cycles. The surfactant Pluronic® F68 may be added at 0.001% to prevent losses of virions to surfaces during handling. Virion preparations can also be treated with nucleases to eliminate small nucleic acid strands on virions surfaces that exacerbate aggregation.
Owner:GENZYME CORP
Ultra-Fast Nucleic Acid Sequencing Device and a Method for Making and Using the Same
InactiveUS20080119366A1Accurate identificationAccurately and effectively identifying bases of DNAMaterial nanotechnologyMicrobiological testing/measurementA-DNAPolymer
A system and method employing at least one semiconductor device, or an arrangement of insulating and metal layers, having at least one detecting region which can include, for example, a recess or opening therein, for detecting a charge representative of a component of a polymer, such as a nucleic acid strand proximate to the detecting region, and a method for manufacturing such a semiconductor device. The system and method can thus be used for sequencing individual nucleotides or bases of ribonucleic acid (RNA) or deoxyribonucleic acid (DNA). The semiconductor device includes at least two doped regions, such as two n-typed regions implanted in a p-typed semiconductor layer or two p-typed regions implanted in an n-typed semiconductor layer. The detecting region permits a current to pass between the two doped regions in response to the presence of the component of the polymer, such as a base of a DNA or RNA strand. The current has characteristics representative of the component of the polymer, such as characteristics representative of the detected base of the DNA or RNA strand.
Owner:LIFE TECH CORP
Enzymes for the detection of nucleic acid sequences
InactiveUS20030134349A1Simple structureLow detection sensitivitySugar derivativesHydrolasesA-siteA-DNA
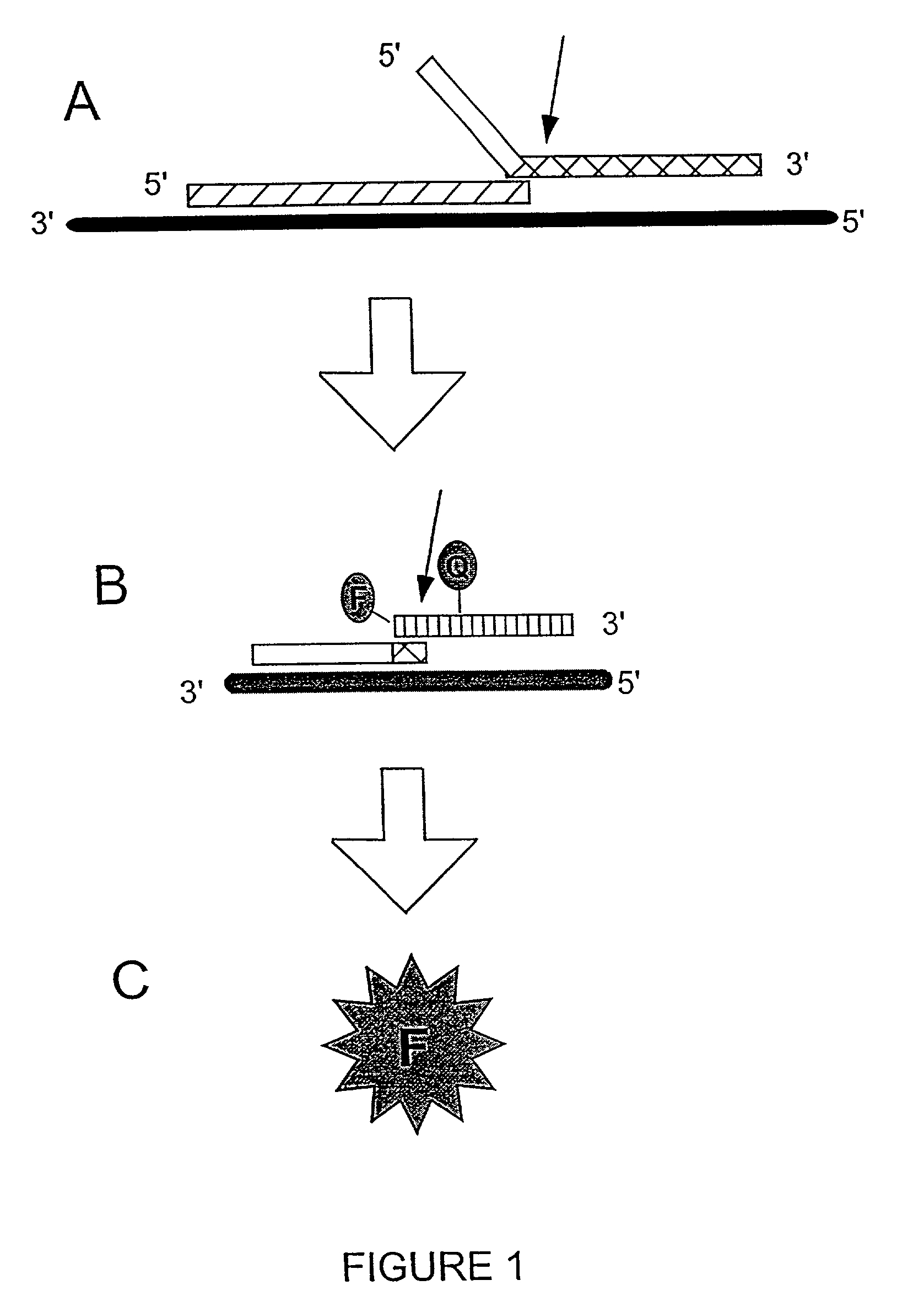
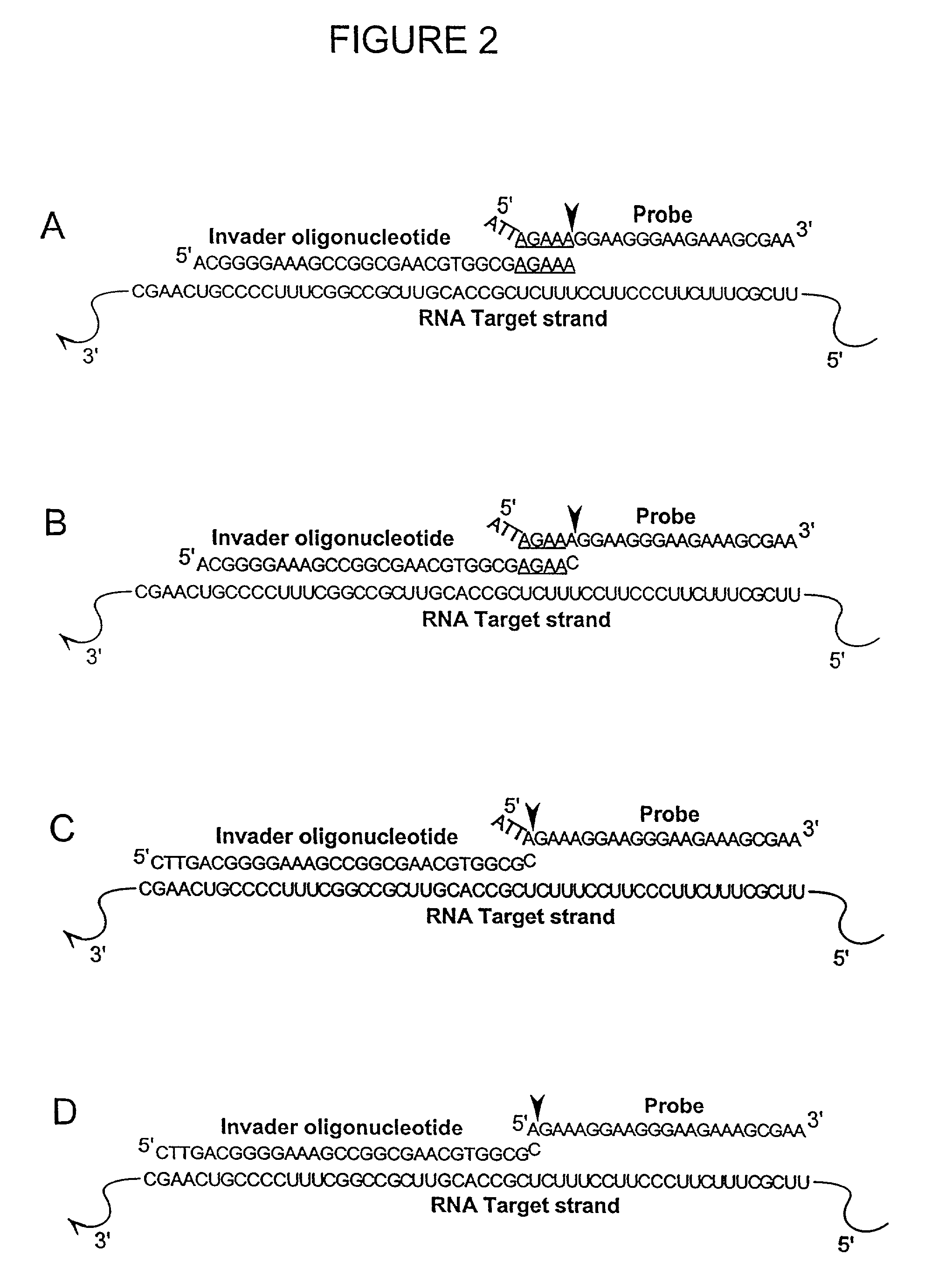
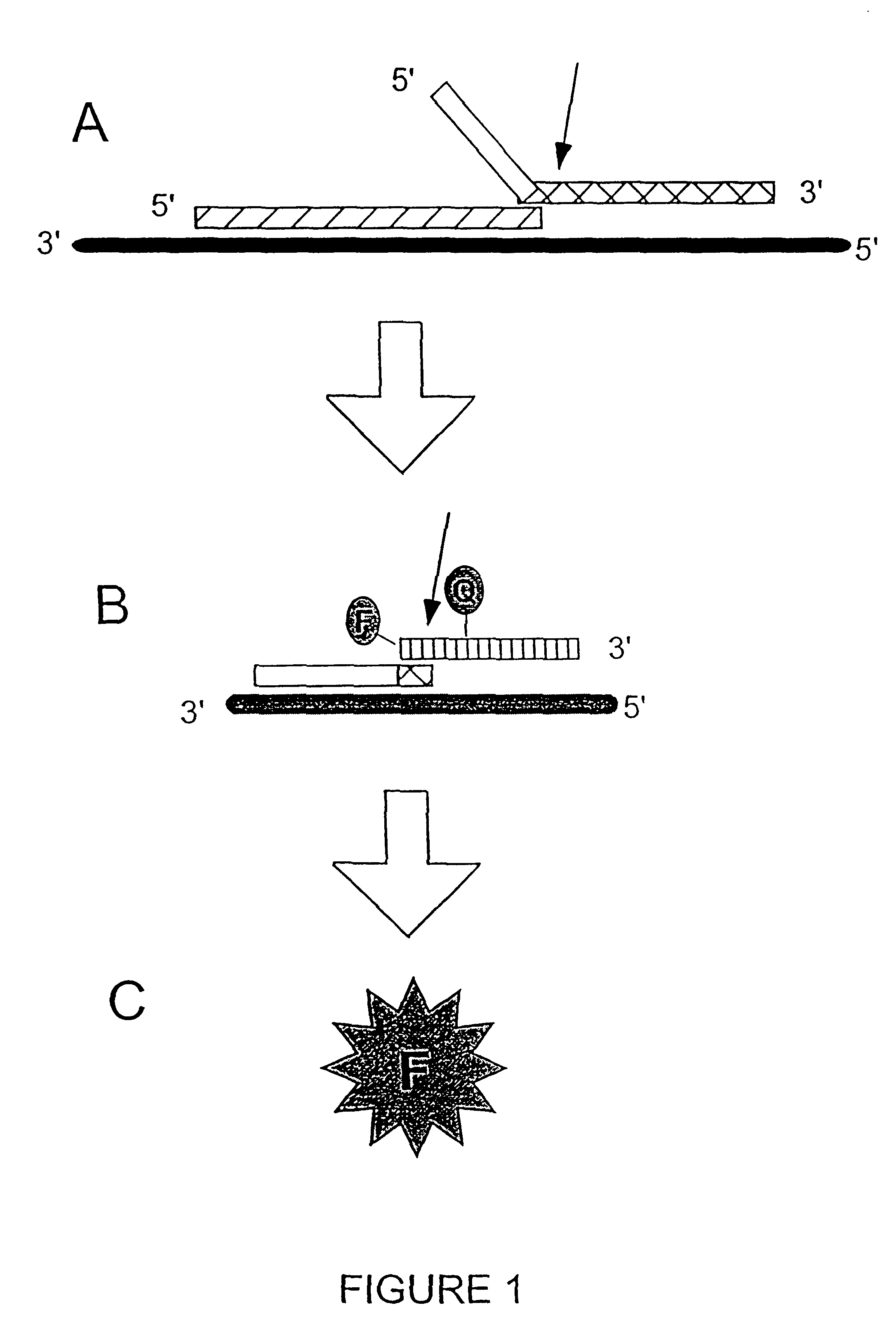
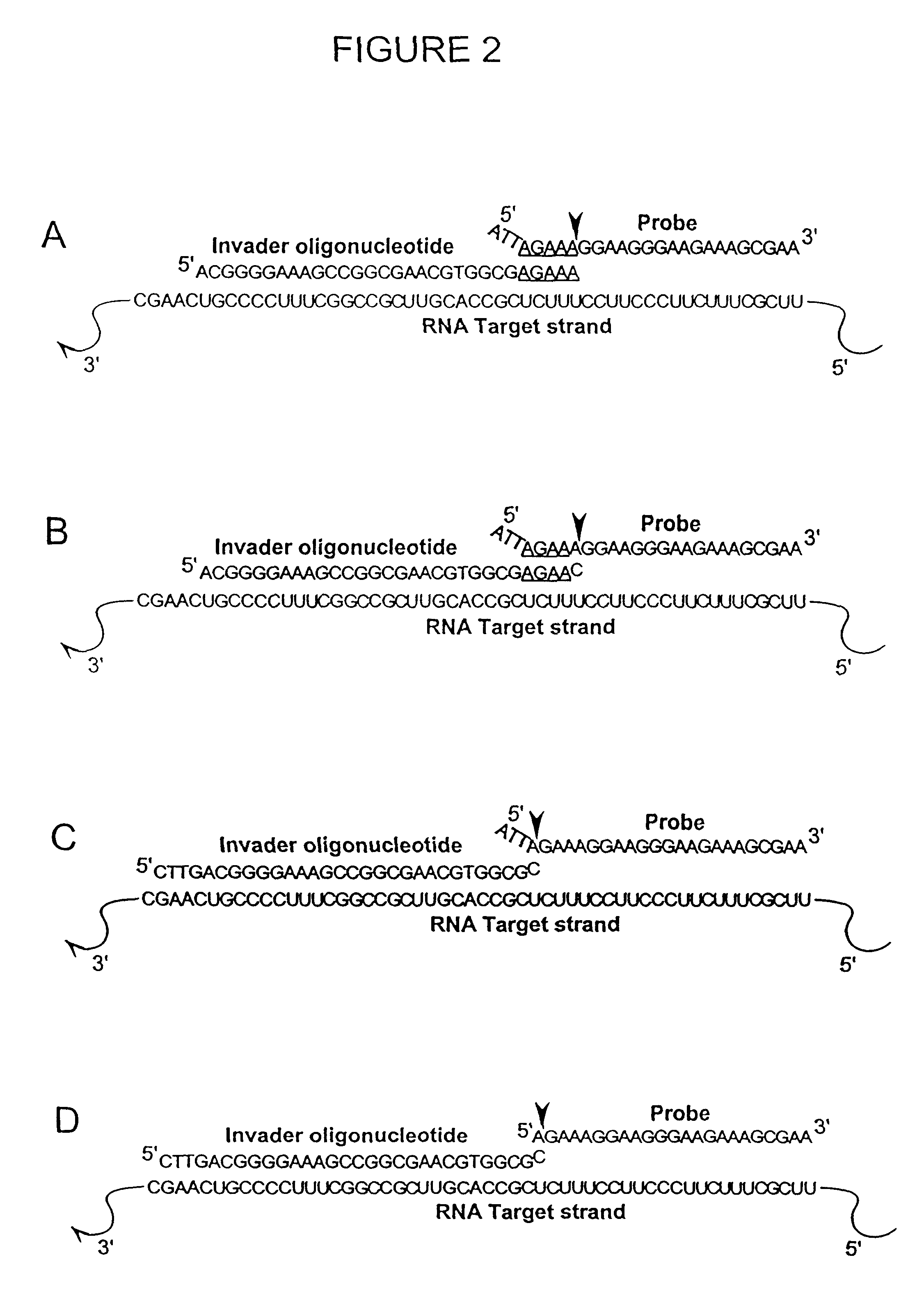
The present invention relates to novel enzymes designed for direct detection, characterization and quantitation of nucleic acids, particularly RNA. The present invention provides enzymes that recognize specific nucleic acid cleavage structures formed on a target RNA sequence and that cleave the nucleic acid cleavage structure in a site-specific manner to produce non-target cleavage products. The present invention provides enzymes having an improved ability to specifically cleave a DNA member of a complex comprising DNA and RNA nucleic acid strands.
Owner:GEN PROBE INC
Enzymes for the detection of nucleic acid sequences
The present invention relates to novel enzymes designed for direct detection, characterization and quantitation of nucleic acids, particularly RNA. The present invention provides enzymes that recognize specific nucleic acid cleavage structures formed on a target RNA sequence and that cleave the nucleic acid cleavage structure in a site-specific manner to produce non-target cleavage products. The present invention provides enzymes having an improved ability to specifically cleave a DNA member of a complex comprising DNA and RNA nucleic acid strands.
Owner:GEN PROBE INC
Nucleic acids coding for adhesion factor of group B streptococcus, adhesion factors of group B streptococcus and further uses thereof
InactiveUS7485710B2Assess the effectiveness of the potential antagonistAntibacterial agentsSugar derivativesMicrobiologyTGE VACCINE
The present invention is related to nucleic acids coding for adhesion factors of group B streptococcus, adhesion factors of group B streptococcus and uses thereof. More particularly, the present invention is related to a polypeptide being such adhesion factors and comprising an amino acid sequence, whereby the amino acid sequence is selected from the group comprising SEQ ID NO 11 to SEQ ID NO 20, and the use of such polypeptide for the manufacture of a vaccine.
Owner:VALNEVA AUSTRIA GMBH
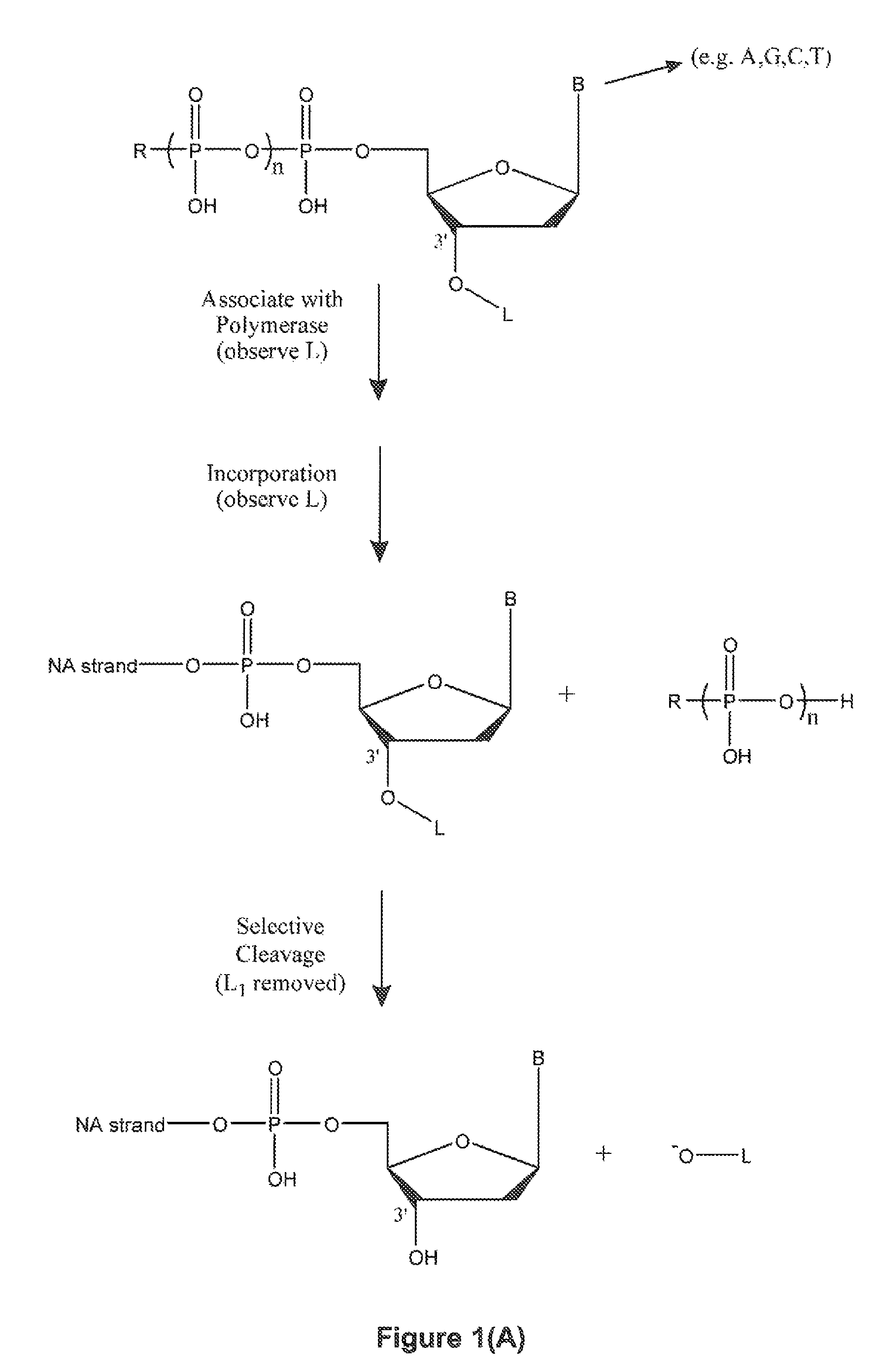
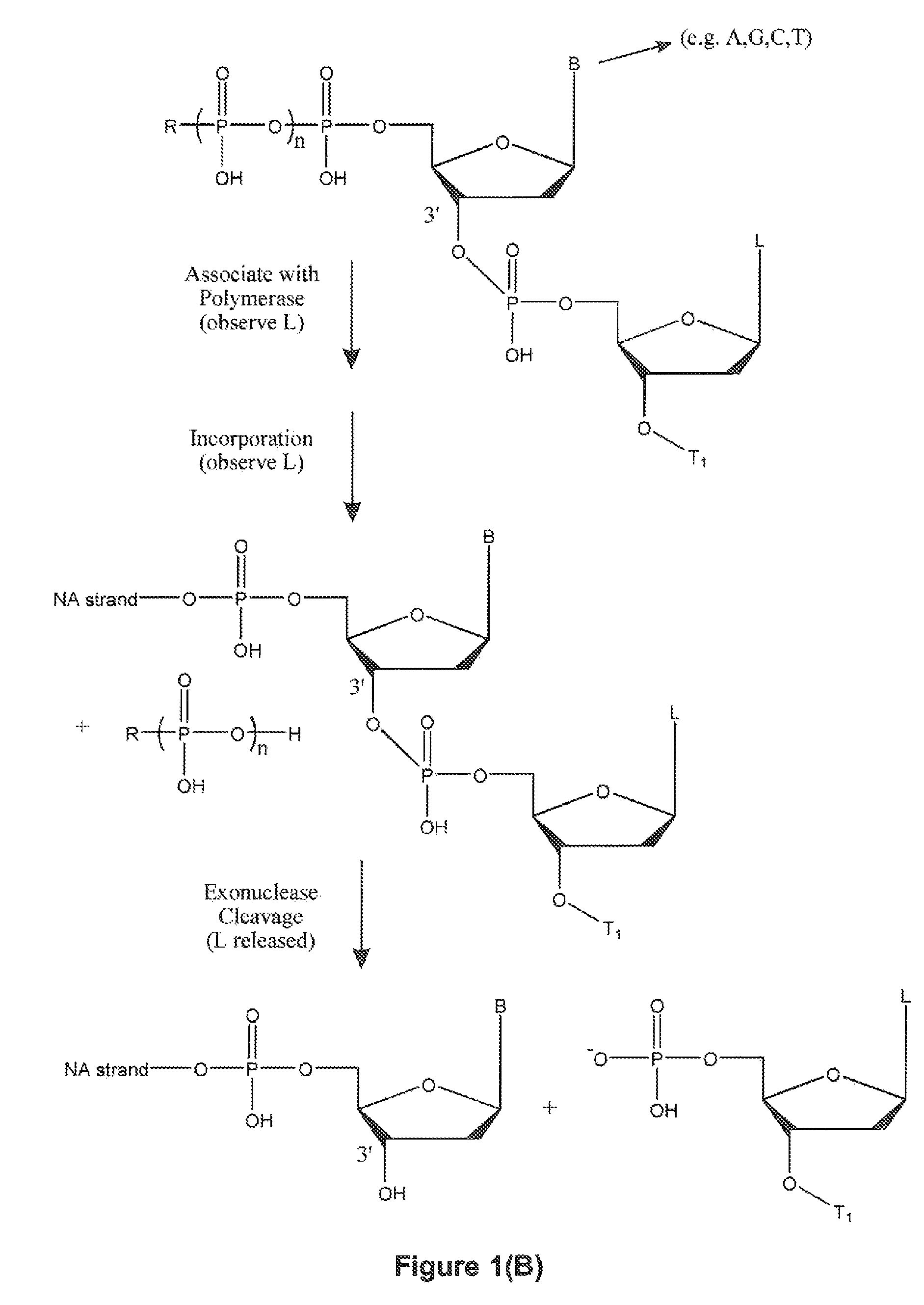
Method for sequencing nucleic acid molecules
InactiveUS20050164255A1Eliminate needExemption stepsBioreactor/fermenter combinationsBiological substance pretreatmentsOligonucleotide primersPolymerase L
The present invention is directed to a method of sequencing a target nucleic acid molecule having a plurality of bases. In its principle, the temporal order of base additions during the polymerization reaction is measured on a molecule of nucleic acid, i.e. the activity of a nucleic acid polymerizing enzyme on the template nucleic acid molecule to be sequenced is followed in real time. The sequence is deduced by identifying which base is being incorporated into the growing complementary strand of the target nucleic acid by the catalytic activity of the nucleic acid polymerizing enzyme at each step in the sequence of base additions. A polymerase on the target nucleic acid molecule complex is provided in a position suitable to move along the target nucleic acid molecule and extend the oligonucleotide primer at an active site. A plurality of labelled types of nucleotide analogs are provided proximate to the active site, with each distinguishable type of nucleotide analog being complementary to a different nucleotide in the target nucleic acid sequence. The growing nucleic acid strand is extended by using the polymerase to add a nucleotide analog to the nucleic acid strand at the active site, where the nucleotide analog being added is complementary to the nucleotide of the target nucleic acid at the active site. The nucleotide analog added to the oligonucleotide primer as a result of the polymerizing step is identified. The steps of providing labelled nucleotide analogs, polymerizing the growing nucleic acid strand, and identifying the added nucleotide analog are repeated so that the nucleic acid strand is further extended and the sequence of the target nucleic acid is determined.
Owner:CORNELL RES FOUNDATION INC
Monoazole ligand platinum analogs
InactiveUS7759488B2Easily transported into tumor cellStrong hydrogen bondingOrganic active ingredientsPlatinum organic compoundsPurinePt element
Disclosed herein are novel platinum-based analogs with a single substituted azole ligand: RN═NR7, wherein the RN═NR7 functional group is covalently bonded to the platinum through nitrogen of NR7. The analogs also have nitrogen donor ligands capable of forming hydrogen bonds with the bases in DNA or RNA, and one or more leaving groups which can be displaced by water, hydroxide ions or other nucleophiles, which is thought to form active species in vivo, and then, form cross-linked complexes between nucleic acid strands, principally between purines in DNA (or RNA), i.e., at the Guanine or Adenine bases, thereof. These platinum analogs may also be more easily transported into tumor cells, due to their increased lipophilicity and are likely to be useful as anti-neoplastic agents, and in modulating or interfering with the synthesis or replication or transcription of DNA or translation or function of RNA in vitro or in vivo, as they are potentially capable of forming a platinum coordinate complex with an intact or nascent DNA or RNA and thereby interfering with cellular synthesis, transcription or replication of nucleic acid polynucleotides.
Owner:BIONUMERIK PHARMA INC
Active-electrode integrated biosensor array and methods for use thereof
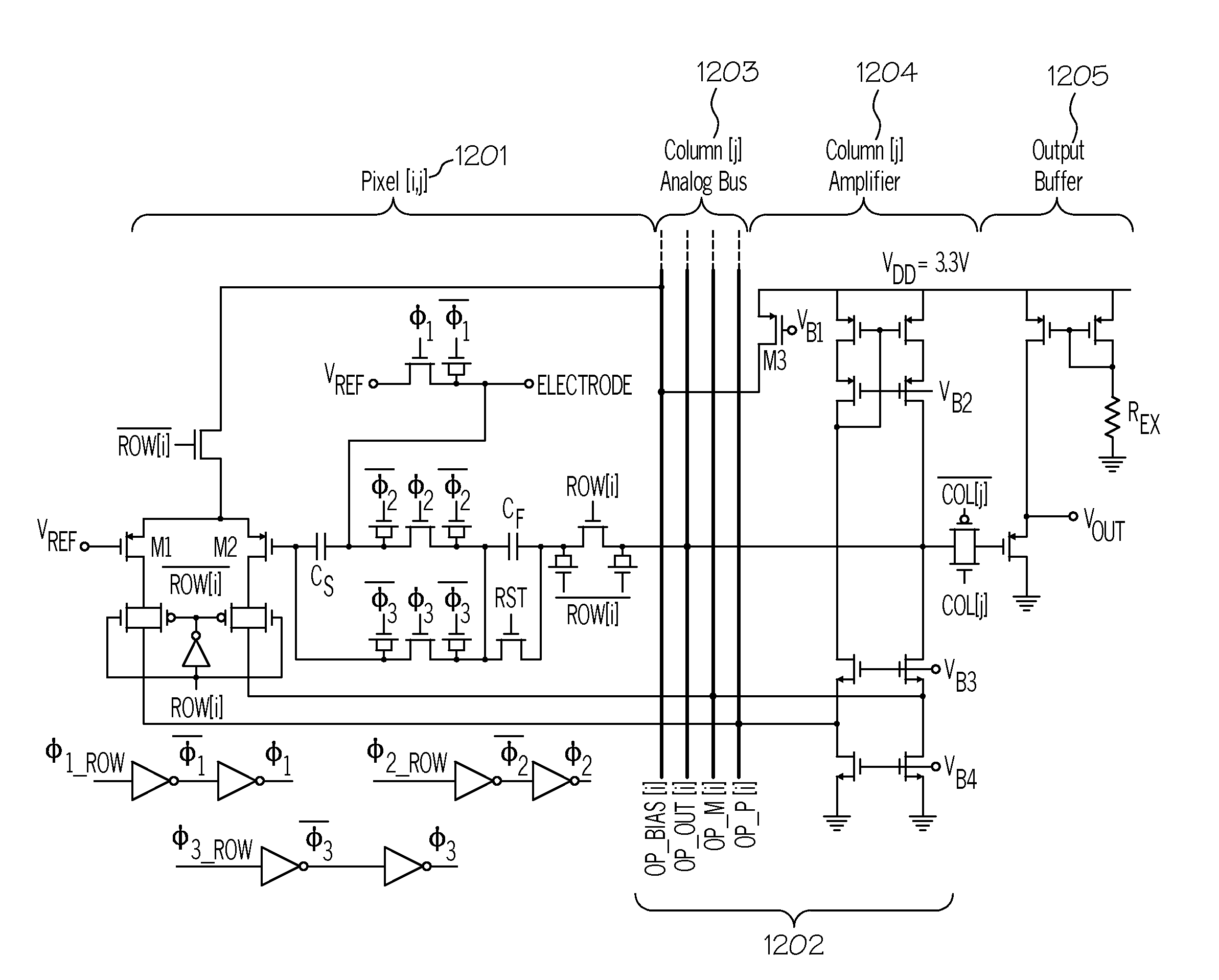
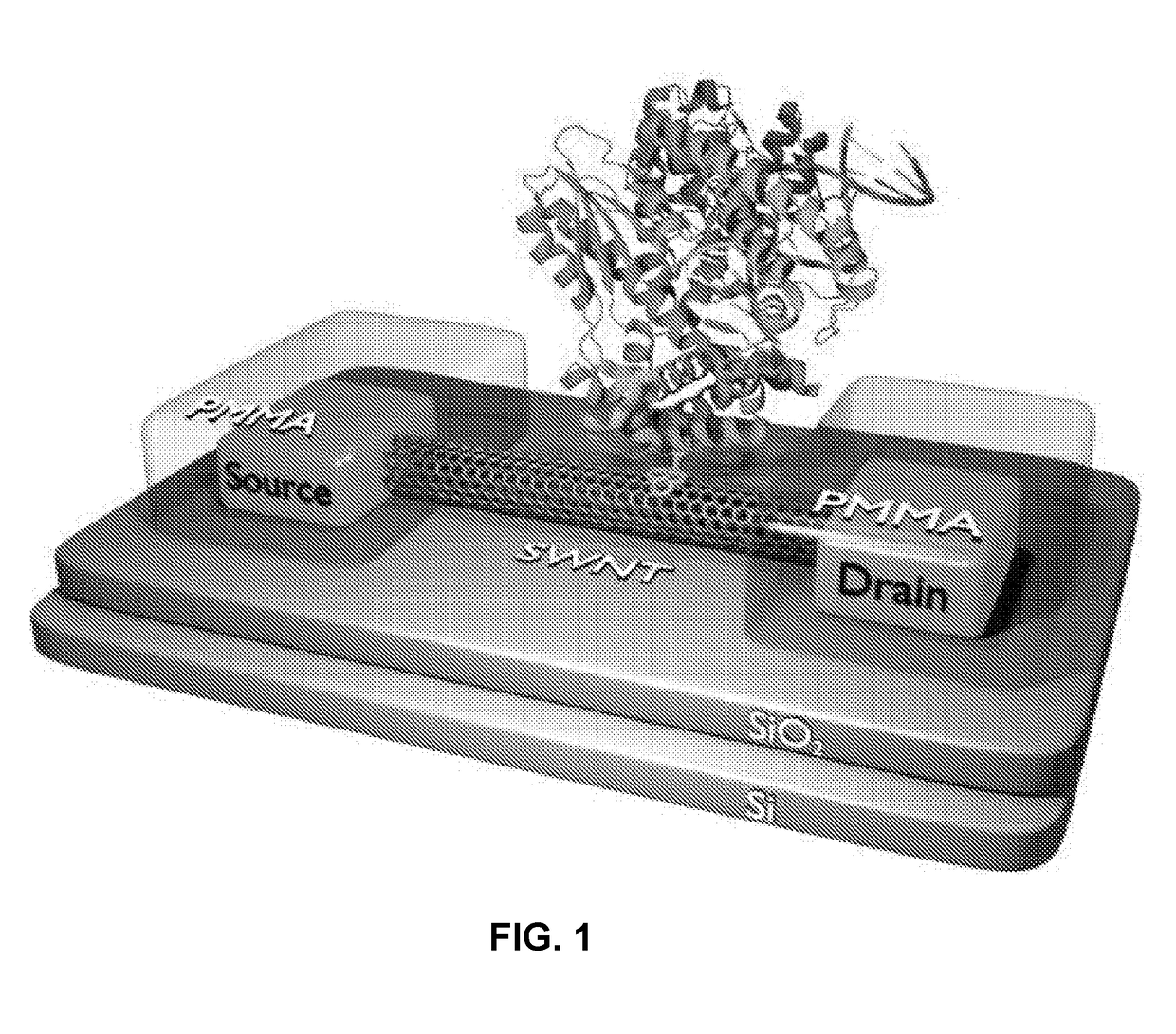
ActiveUS20130345065A1Not compatibleImmobilised enzymesBioreactor/fermenter combinationsCMOSBiosensor array
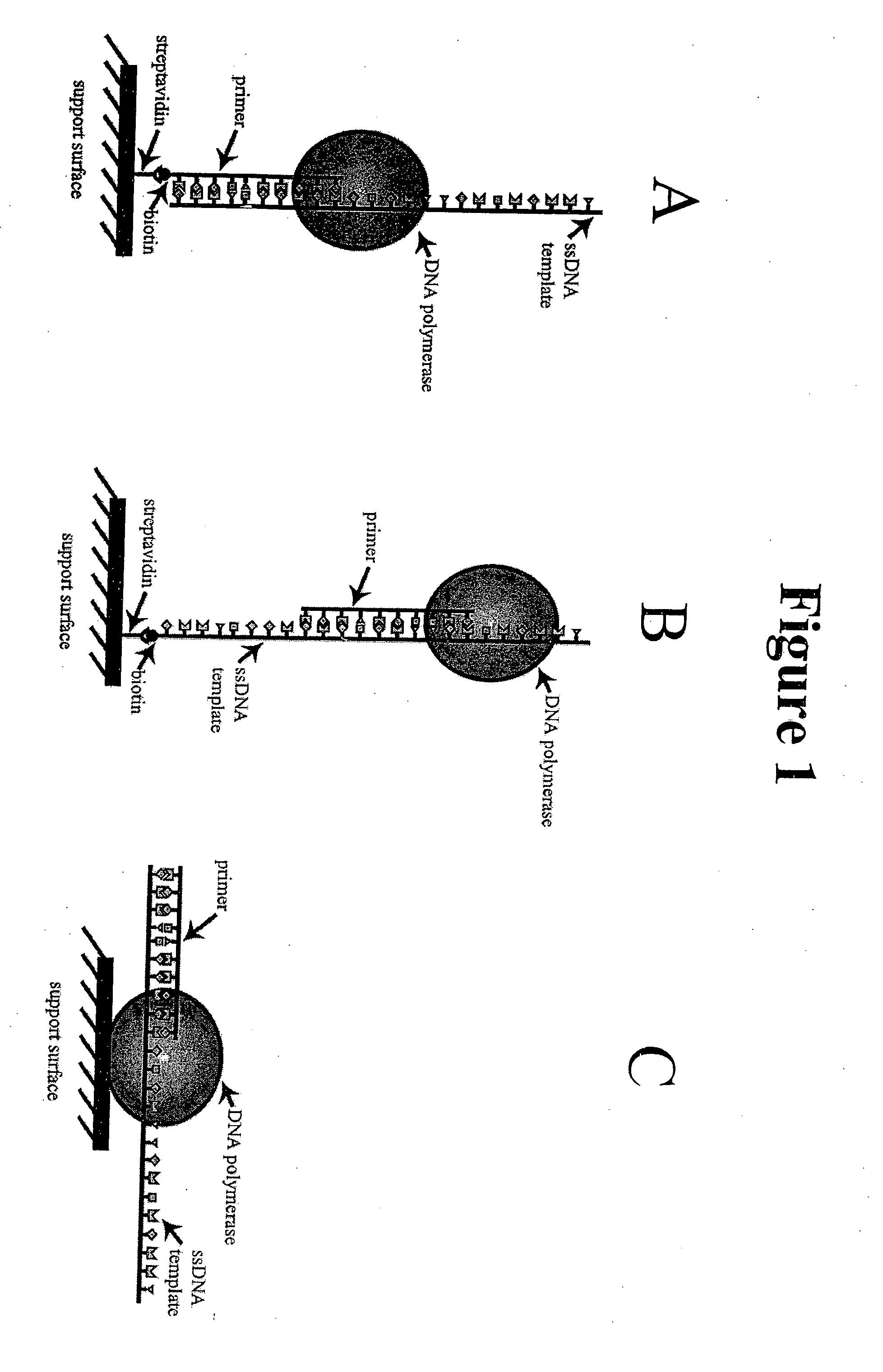
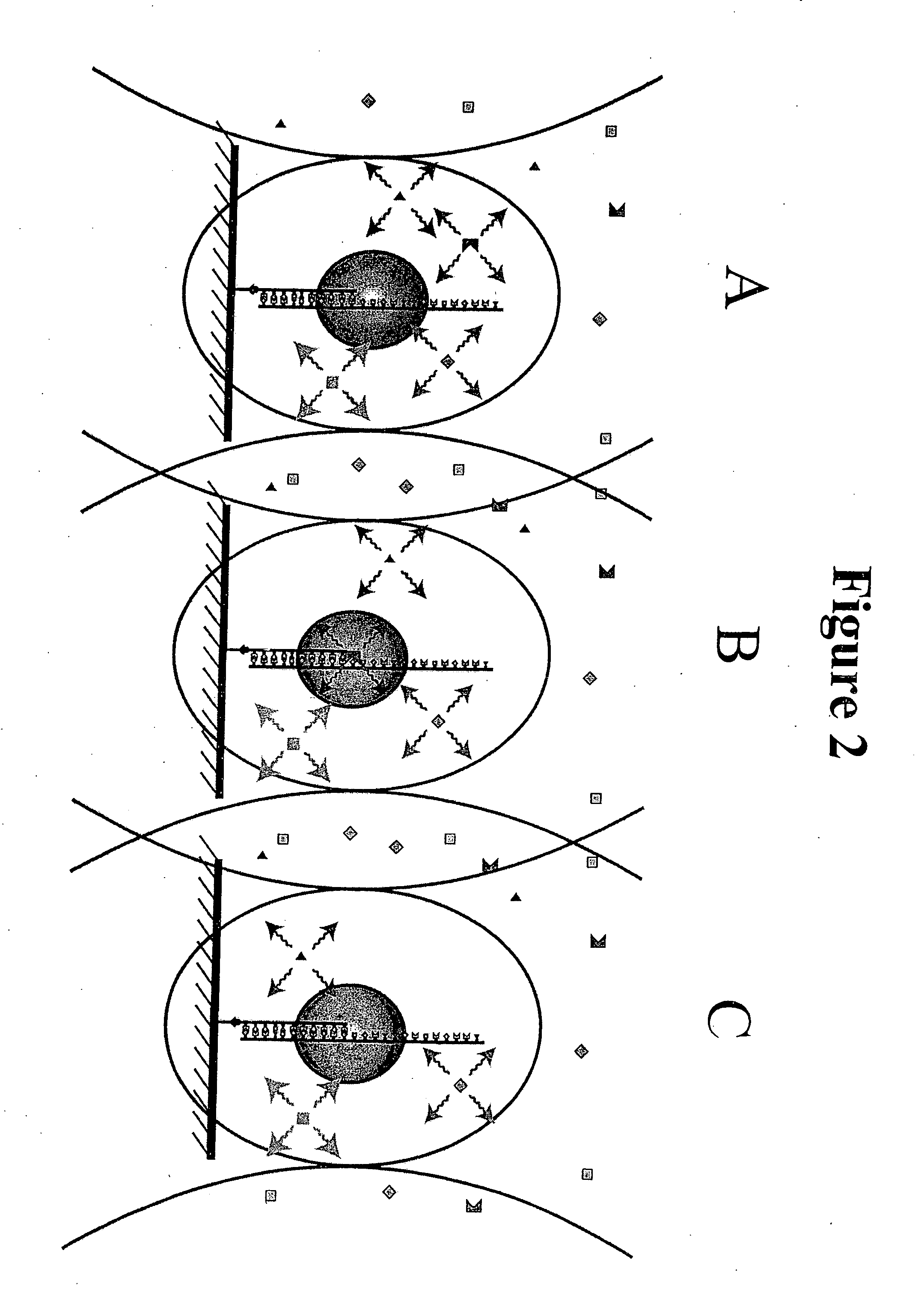
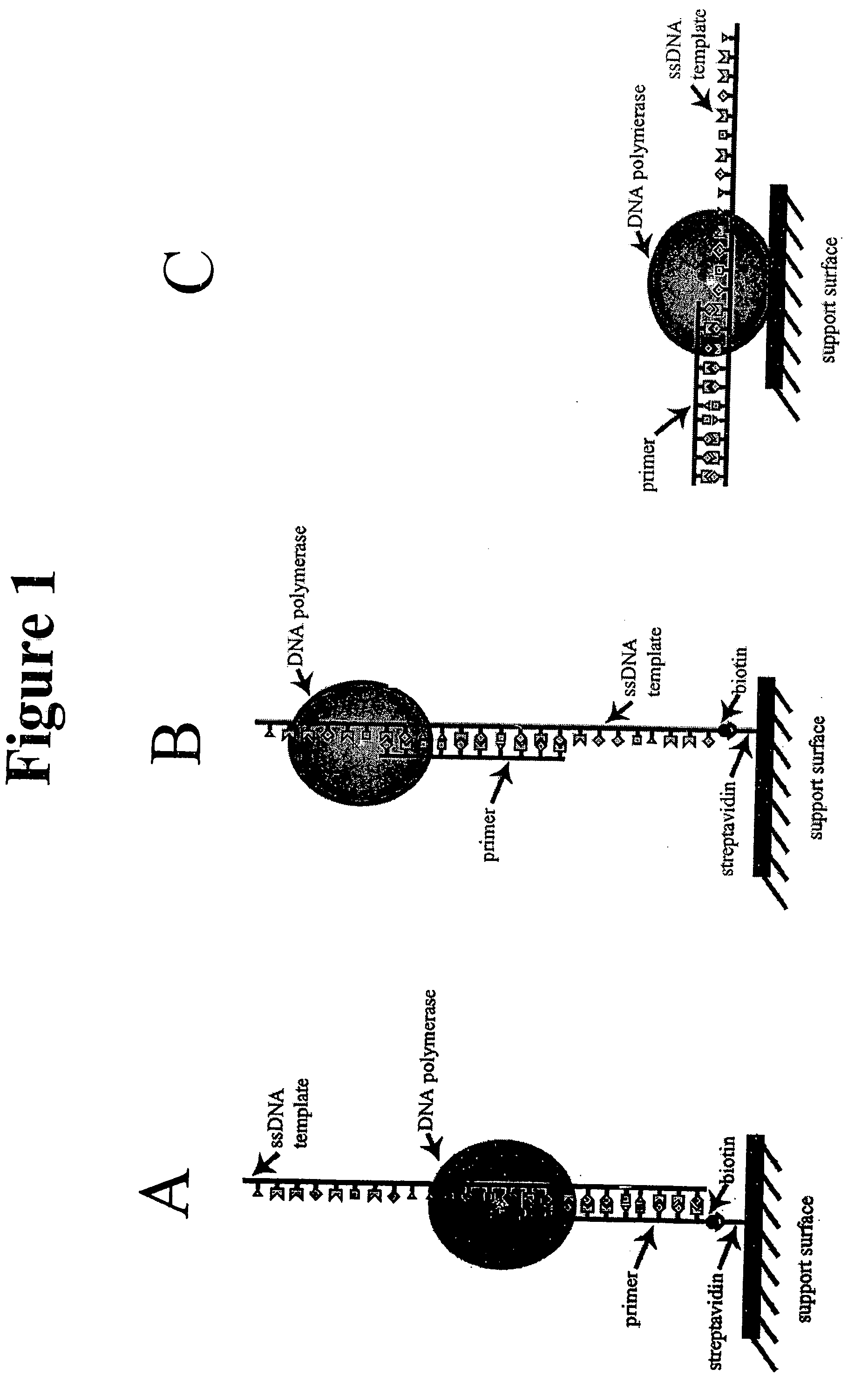
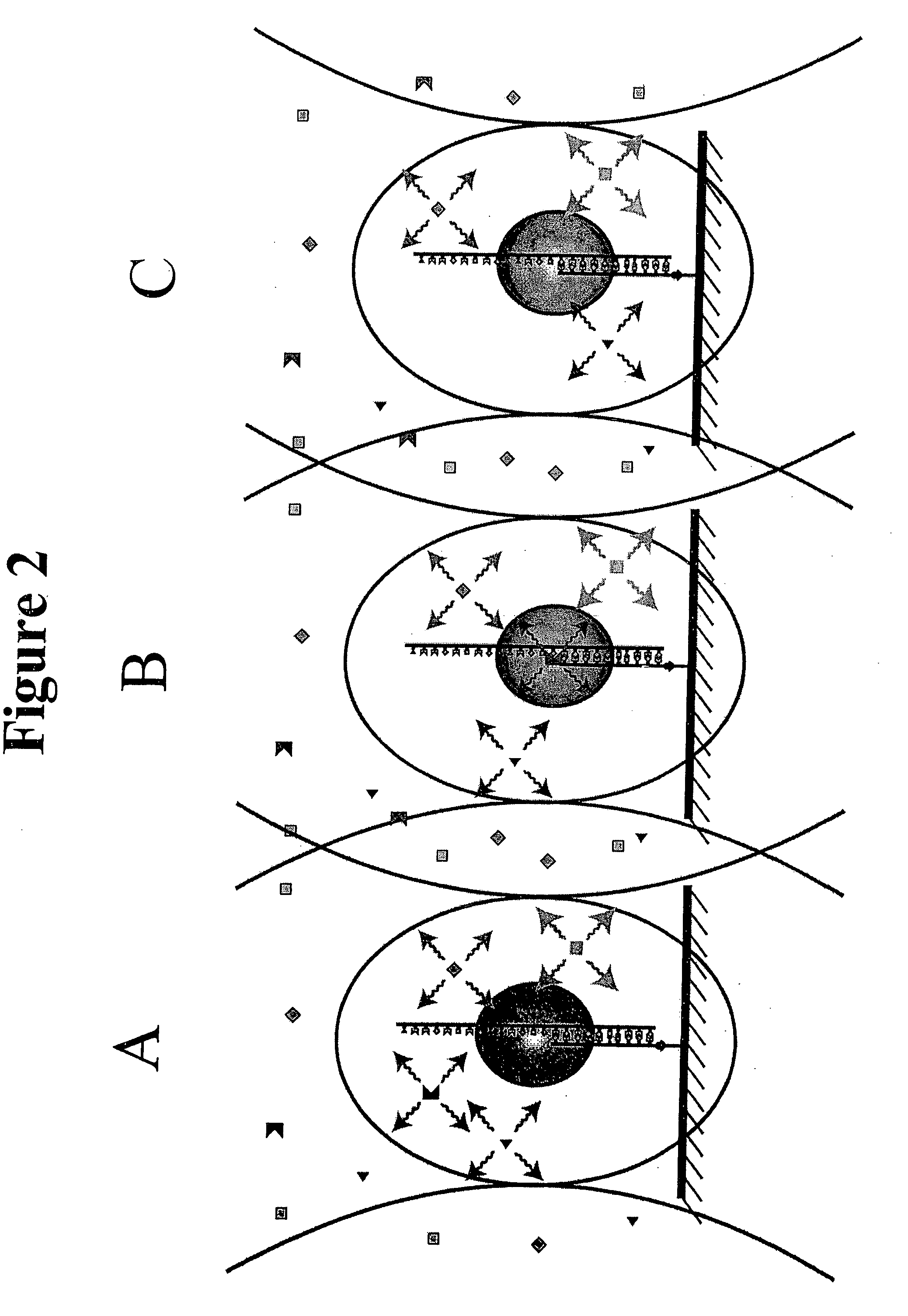
A method and device for performing DNA sequencing and extracting structural information from unknown nucleic acid strands. The device includes a microwell structure, where identical DNA strands are immobilized within the microwell structure on a surface of a micro-bead, an active electrode or a porous polymer. The device further includes a CMOS-integrated semiconductor integrated circuit, where the CMOS-integrated semiconductor integrated circuit includes metal layers on a silicon substrate, where the metal layers form an active electrode biosensor. In addition, a sensing electrode is formed by creating openings in a passivation layer of the CMOS-integrated semiconductor integrated circuit to hold a single bead, on which the DNA strands are immobilized.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Nucleic acids coding for adhesion factor of group b streptococcus, adhesion factors of group b streptococcus and further uses thereof
InactiveUS20060115479A1Assess the effectiveness of the potential antagonistAntibacterial agentsHydrolasesMicrobiologyTGE VACCINE
The present invention is related to nucleic acids coding for adhesion factors of group B streptococcus, adhesion factors of group B streptococcus and uses thereof. More particularly, the present invention is related to a polypeptide being such adhesion factors and comprising an amino acid sequence, whereby the amino acid sequence is selected from the group comprising SEQ ID NO 11 to SEQ ID NO 20, and the use of such polypeptide for the manufacture of a vaccine.
Owner:VALNEVA AUSTRIA GMBH
Oligonucleotides having A-DNA form and B-DNA form conformational geometry
InactiveUS6737520B2High affinityImproved nuclease resistanceSugar derivativesGenetic material ingredientsNucleotideA-DNA
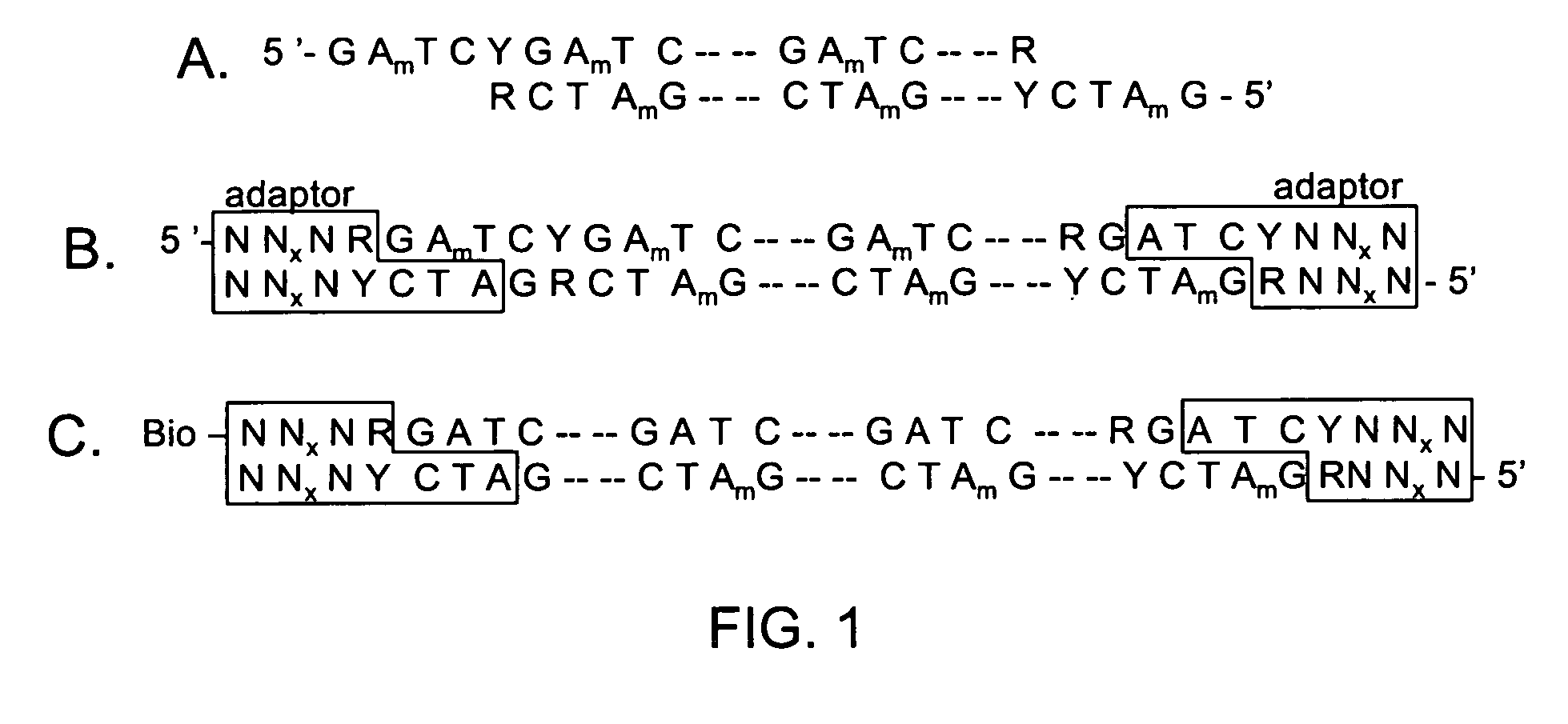
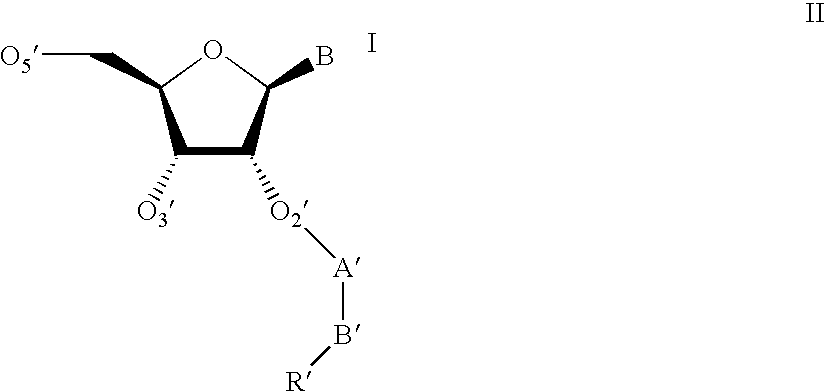
Modified oligonucleotides containing both A-form conformation geometry and B-from conformation geometry nucleotides are disclosed. The B-form geometry allows the oligonucleotide to serve as substrates for RNase H when bound to a target nucleic acid strand. The A-form geometry imparts properties to the oligonucleotide that modulate binding affinity and nuclease resistance. By utilizing C2' endo sugars or O4' endo sugars, the B-form characteristics are imparted to a portion of the oligonucleotide. The A-form characteristics are imparted via use of either 2'-O-modified nucleotides that have 3' endo geometries or use of end caps having particular nuclease stability or by use of both of these in conjunction with each other.
Owner:IONIS PHARMA INC
Device for the amplification of dna, comprising a microwave energy source
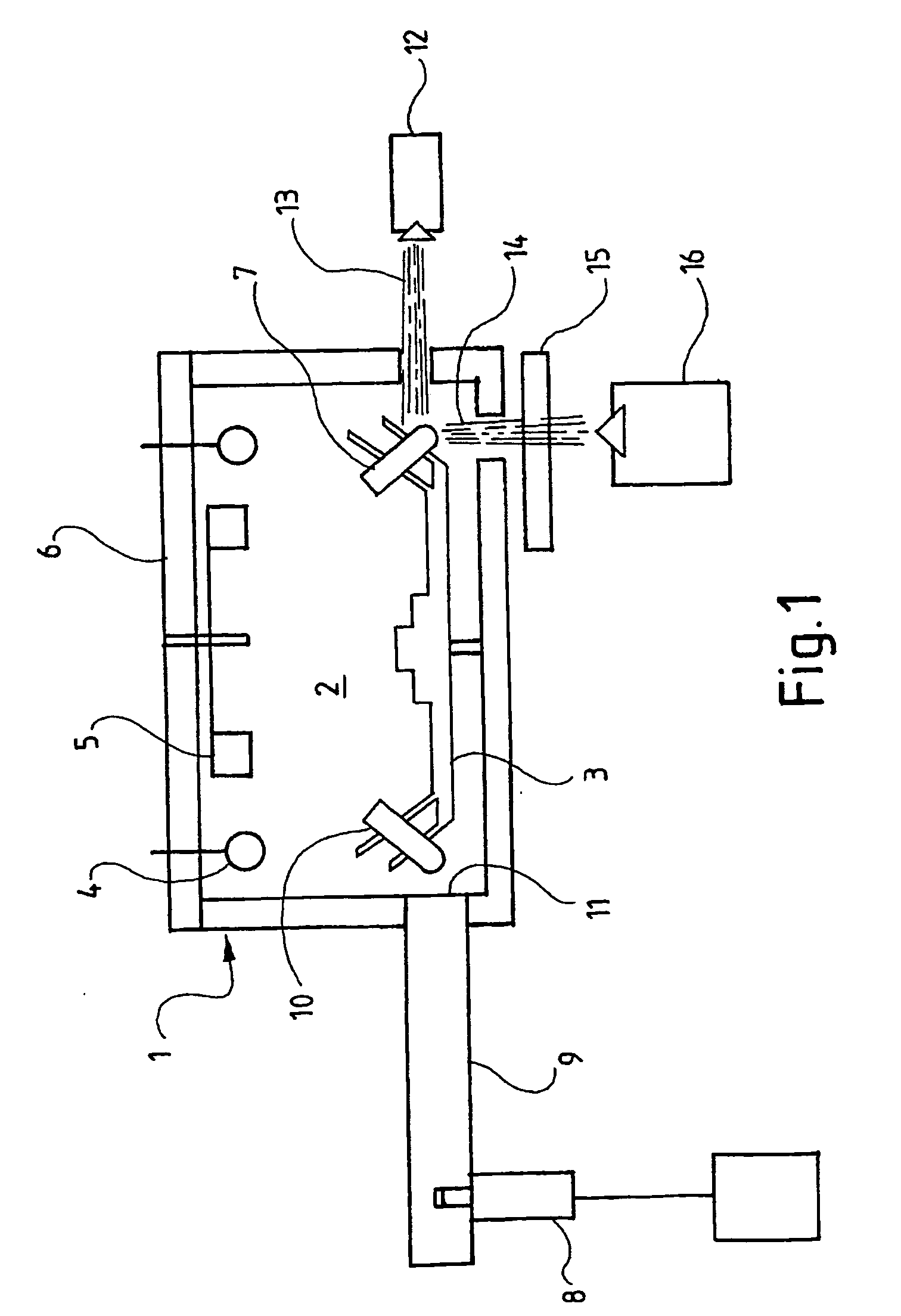
InactiveUS20050233324A1Shorten cycle timeFaster cycle timeBioreactor/fermenter combinationsBiological substance pretreatmentsMicrowaveAmplification dna
The invention relates to a device for the amplification of DNA in a reaction mixture, the device (1) comprising a heated chamber (2) including a rotor (3) for holding a plurality of reaction vessels for reaction mixtures, a drive means for the rotor, a microwave energy source (8) with means for controlled delivery of said energy to the reaction mixtures, and a system (14-16) for determining denaturation of double-stranded DNA. The invention also provides a method for the amplification of a nucleic acid strand. In the first step of the method, a reaction mixture is formed comprising the target nucleic acid strand, nucleotides, a primer, a thermostable nucleic acid polymerase, and, if necessary, a reagent for the detection of denaturation of double-stranded DNA. In the second step, the mixture is incubated at a temperature which allows synthesis of a nucleic acid strand complementary to the target nucleic acid strand. The third step comprises denaturing double-stranded DNA formed in the second step by microwave energisation of the reaction mixture with monitoring of the mixture to determine the denaturation end point. The reaction mixture is allowed to cool to a temperature at which primer anneals to the target nucleic acid strand in the fourth step. The second to fourth steps are repeated until a desired level of amplification is achieved.
Owner:CORBETT LIFE SCI
Field-effect apparatus and methods for sequencing nucleic acids
The present disclosure provides a method for sequencing nucleic acids. The method can include polymerase catalyzed incorporation of nucleotides into a nascent nucleic acid strand against a nucleic acid template, wherein the polymerase is attached to a charge sensor that detects nucleotide incorporation events. One or more non-natural nucleotide types that each produce a unique signatures at the charge sensor can be used to uniquely identify different nucleotides in the template nucleic acid.
Owner:RGT UNIV OF CALIFORNIA +1
Method for sequencing nucleic acids by observing the uptake of nucleotides modified with bulky groups
InactiveUS20070059733A1Bioreactor/fermenter combinationsMaterial nanotechnologySurface stressPresent method
The present methods and apparatus concern nucleic acid sequencing by incorporation of nucleotides into nucleic acid strands. The incorporation of nucleotides is detected by changes in the mass and / or surface stress of the structure. In some embodiments of the invention, the structure comprises one or more nanoscale or microscale cantilevers. In certain embodiments of the invention, each different type of nucleotide is distinguishably labeled with a bulky group and each incorporated nucleotide is identified by the changes in mass and / or surface stress of the structure upon incorporation of the nucleotide. In alternative embodiments of the invention only one type of nucleotide is exposed at a time to the nucleic acids. Changes in the properties of the structure may be detected by a variety of methods, such as piezoelectric detection, shifts in resonant frequency of the structure, and / or position sensitive photodetection.
Owner:INTEL CORP
Single molecule sequencing with two distinct chemistry steps
Owner:PACIFIC BIOSCIENCES
Oligonucleotides having A-DNA form and B-DNA form conformational geometry
InactiveUS7119184B2High affinityImproved nuclease resistanceSugar derivativesPhosphorus organic compoundsNucleotideA-DNA
Modified oligonucleotides containing both A-form conformation geometry and B-from conformation geometry nucleotides are disclosed. The B-form geometry allows the oligonucleotide to serve as substrates for RNase H when bound to a target nucleic acid strand. The A-form geometry imparts properties to the oligonucleotide that modulate binding affinity and nuclease resistance. By utilizing C2′ endo sugars or O4′ endo sugars, the B-form characteristics are imparted to a portion of the oligonucleotide. The A-form characteristics are imparted via use of either 2′-O-modified nucleotides that have 3′ endo geometries or use of end caps having particular nuclease stability or by use of both of these in conjunction with each other.
Owner:IONIS PHARMA INC
Method for suppressing nonspecific hybridization in primer extension method
InactiveUS6033851ASuppress hybridizationEasily caughtSugar derivativesMicrobiological testing/measurementNon specificNucleic acid
PCT No. PCT / JP95 / 02535 Sec. 371 Date May 30, 1997 Sec. 102(e) Date May 30, 1997 PCT Filed Dec. 11, 1995 PCT Pub. No. WO96 / 17932 PCT Pub. Date Jun. 13, 1996An object of the present invention is to suppress nonspecific extension reaction of the primer in the primer extension method. The primer extension reaction to form a nucleic acid strand complementary to a nucleic acid template strand with the use of a primer according to the present invention is characterized in that the reaction between the primer and the template strand is carried out in the presence of a nucleic acid or a derivative thereof which is complementary to said primer and has an affinity for said primer is equivalent to or less than that of said primer for the nucleic acid template strand.
Owner:WAKUNAGA SEIYAKU KK
Method for sequencing nucleic acid molecules
InactiveUS20050158761A1Small sizeEliminate needBioreactor/fermenter combinationsBiological substance pretreatmentsOligonucleotide primersPolymerase L
The present invention is directed to a method of sequencing a target nucleic acid molecule having a plurality of bases. In its principle, the temporal order of base additions during the polymerization reaction is measured on a molecule of nucleic acid, i.e. the activity of a nucleic acid polymerizing enzyme on the template nucleic acid molecule to be sequenced is followed in real time. The sequence is deduced by identifying which base is being incorporated into the growing complementary strand of the target nucleic acid by the catalytic activity of the nucleic acid polymerizing enzyme at each step in the sequence of base additions. A polymerase on the target nucleic acid molecule complex is provided in a position suitable to move along the target nucleic acid molecule and extend the oligonucleotide primer at an active site. A plurality of labelled types of nucleotide analogs are provided proximate to the active site, with each distinguishable type of nucleotide analog being complementary to a different nucleotide in the target nucleic acid sequence. The growing nucleic acid strand is extended by using the polymerase to add a nucleotide analog to the nucleic acid strand at the active site, where the nucleotide analog being added is complementary to the nucleotide of the target nucleic acid at the active site. The nucleotide analog added to the oligonucleotide primer as a result of the polymerizing step is identified. The steps of providing labelled nucleotide analogs, polymerizing the growing nucleic acid strand, and identifying the added nucleotide analog are repeated so that the nucleic acid strand is further extended and the sequence of the target nucleic acid is determined.
Owner:CORNELL RES FOUNDATION INC
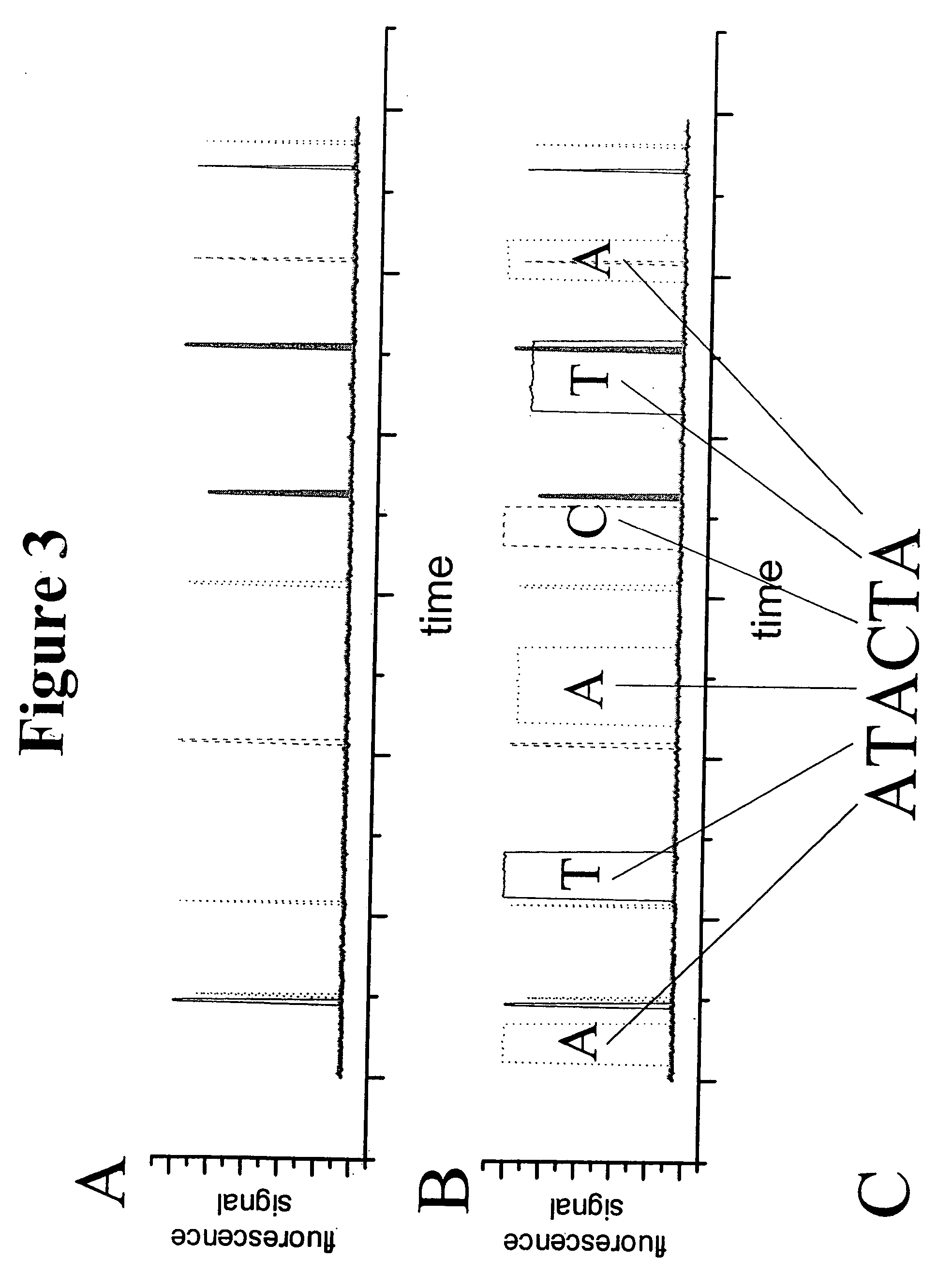
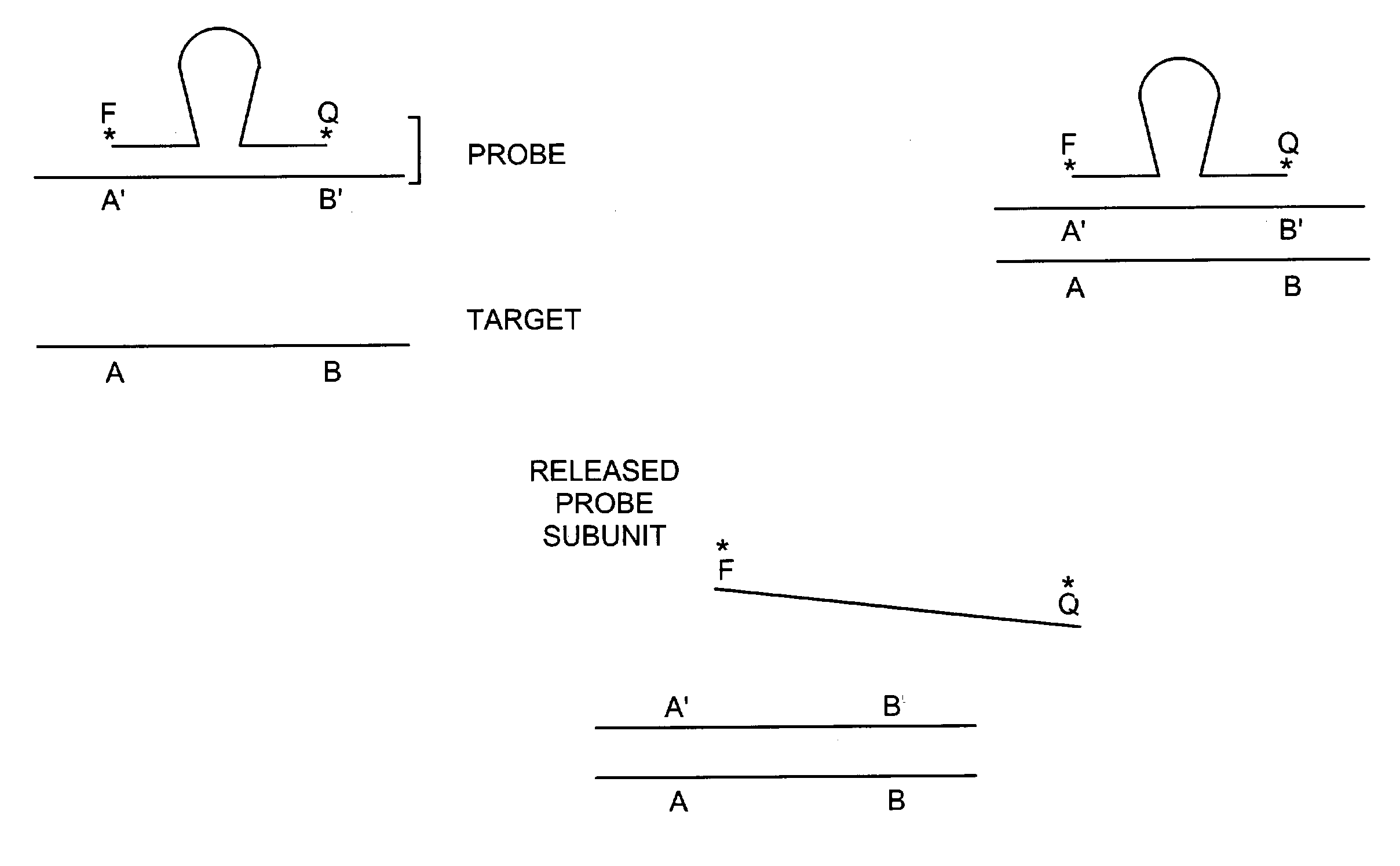
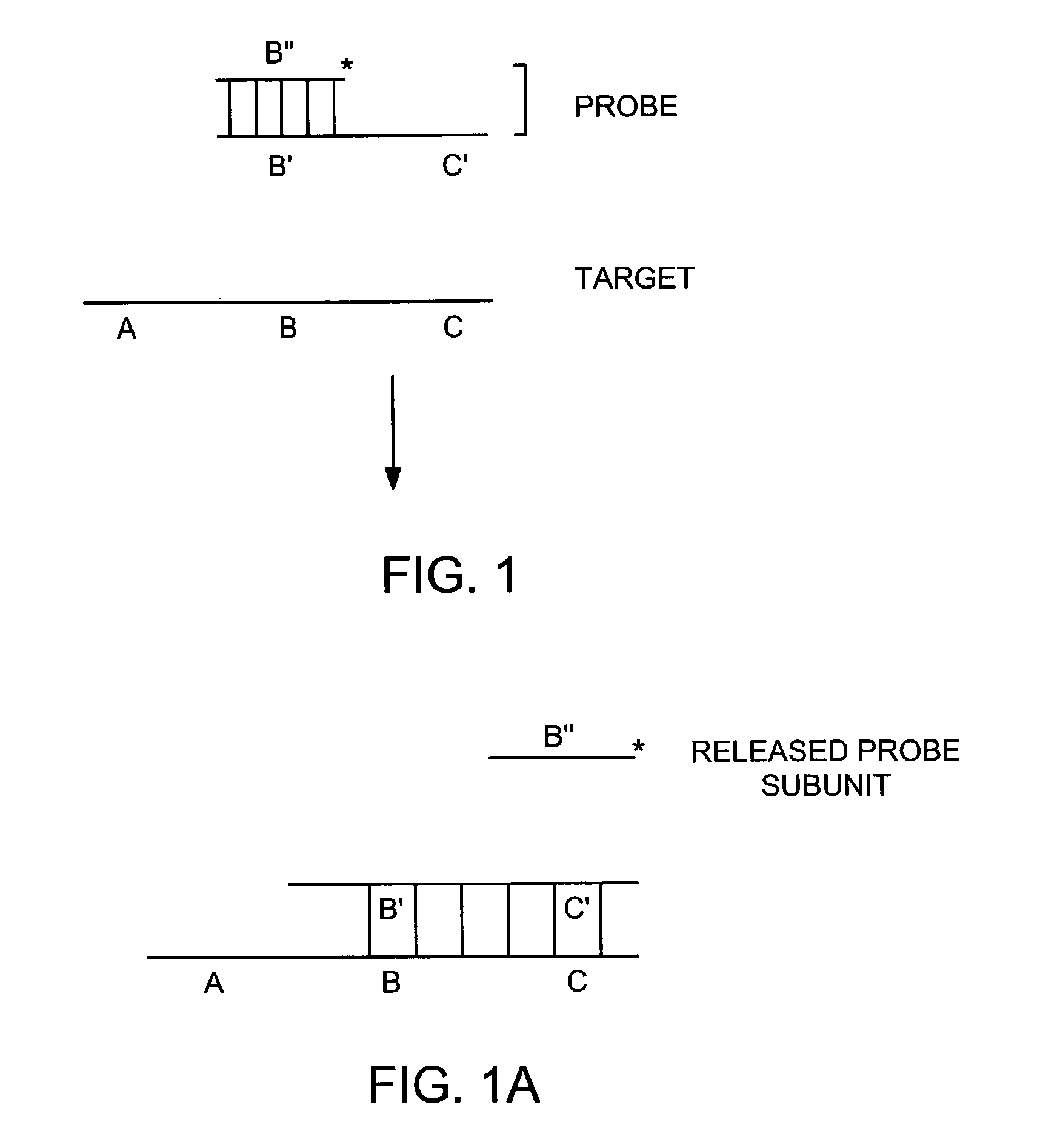
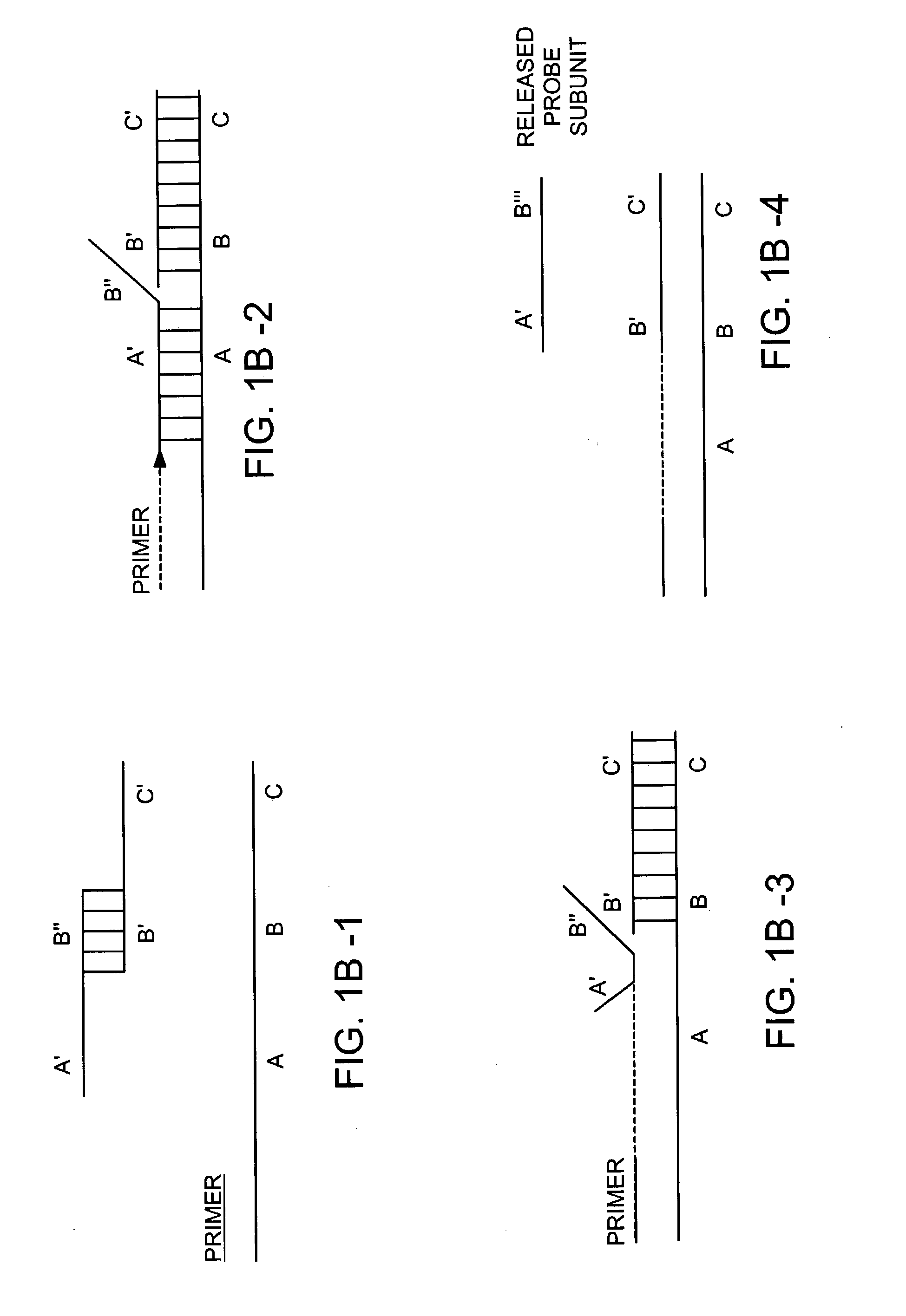
Methods for detection of a target nucleic acid using multi-subunit probes
InactiveUS7183052B2Bind less effectivelyEfficient measurementSugar derivativesMicrobiological testing/measurementPolymerase LNucleic acid sequencing
The invention relates to a method of generating a signal indicative of the presence of a target nucleic acid sequence in a sample, comprising forming a complex by incubating a sample comprising a target nucleic acid sequence with a probe comprising a first and second subunit, and / or an upstream primer, and binding the probe to the target nucleic acid such that the first and second subunits dissociate to release the first subunit and generate a signal. In certain embodiments, the upstream primer is extended with a nucleic acid polymerase to displace at least a portion of the first subunit of the probe from the target nucleic acid strand and dissociate the first and second subunits to release the first subunit of the probe and generate a signal.
Owner:STRATAGENE INC US
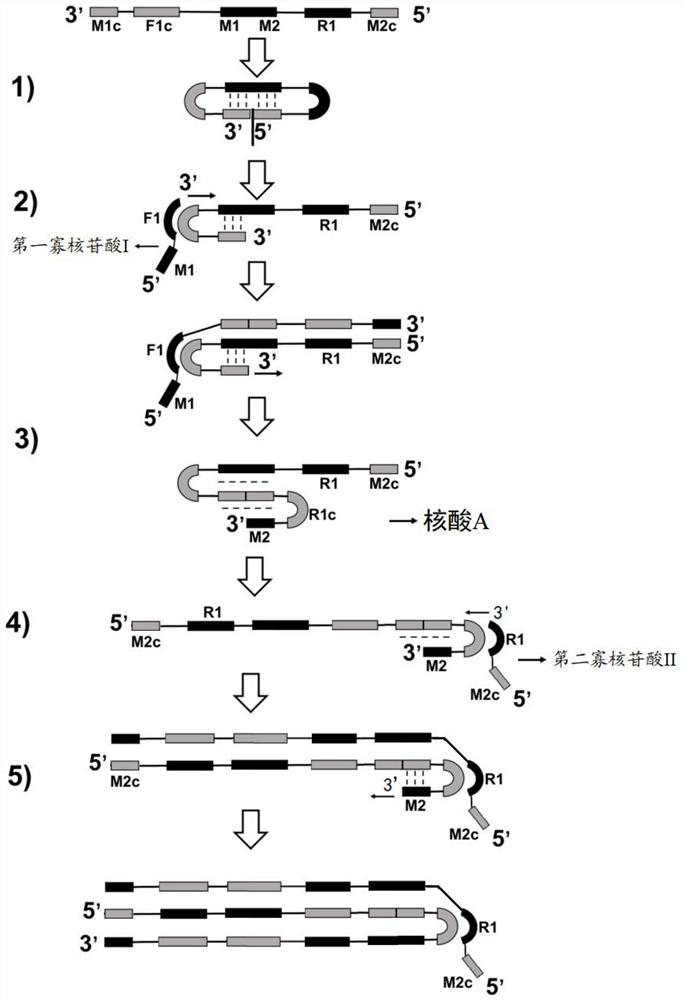
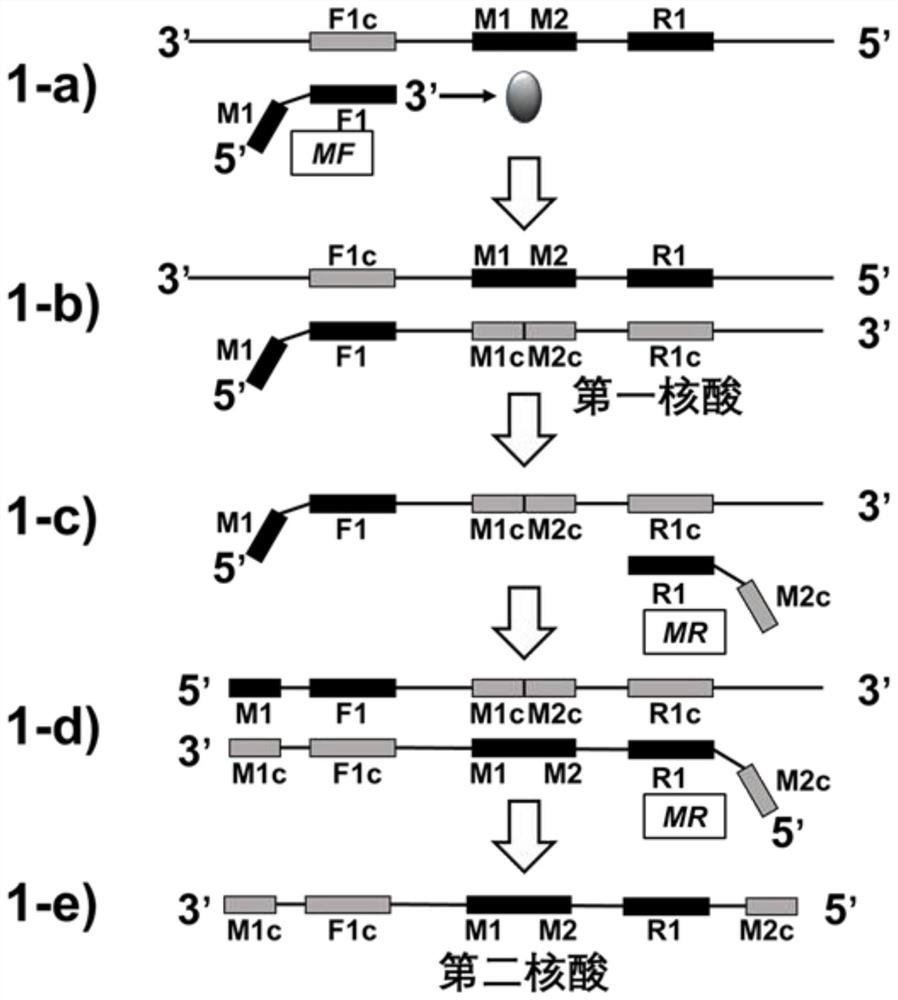
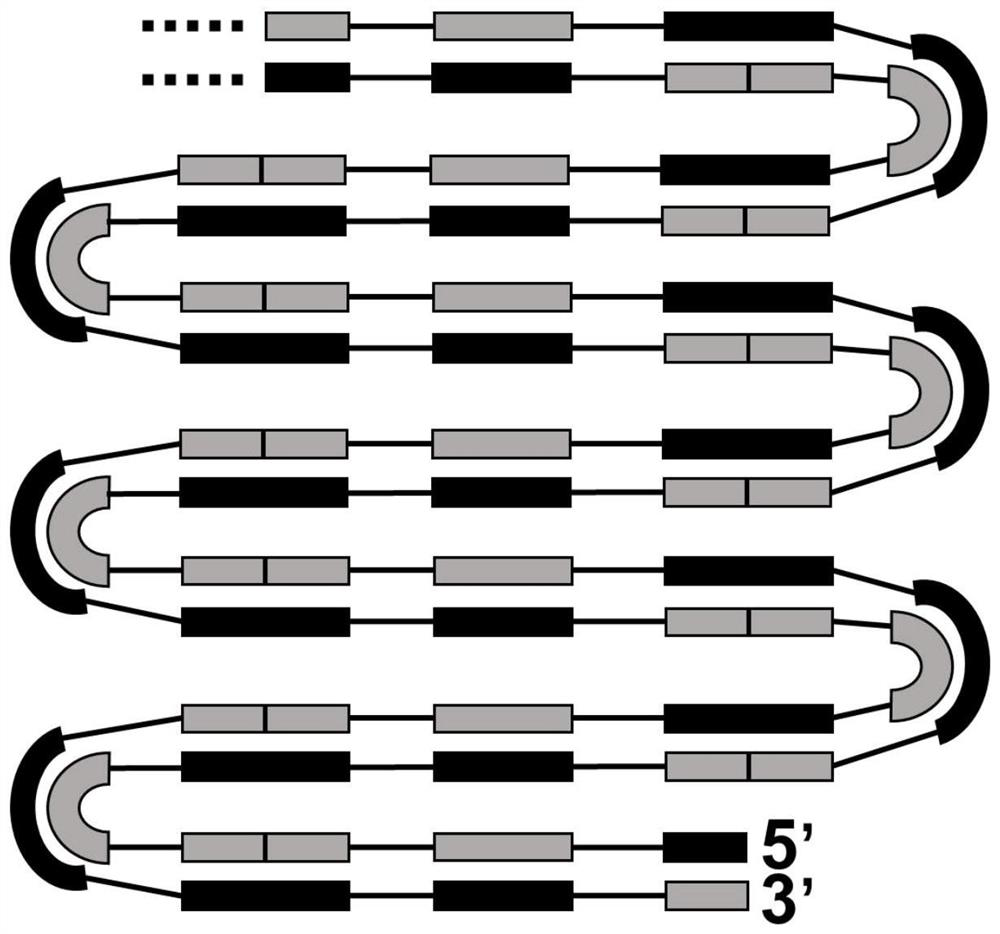
Method for synthesizing nucleic acid under constant temperature condition, kit and application
The invention discloses a method for synthesizing nucleic acid under a constant temperature condition, a kit and application. The method comprises the steps that a nucleic acid is provided, the nucleic acid is sequentially composed of an M1c region, an F1c region, an M region, an R1 region and an M2c region in the 3'to 5 'direction, the M region comprises an M1 region and an M2 region, and annealing of the M2c region at the 5'end and the M1c region at the 3' end of the nucleic acid and the M region on the same chain can form a closed ring structure; a first oligonucleotide I and a second oligonucleotide II of a primer are respectively annealed with an F1c region and an R1c region of the nucleic acid, and the nucleic acid is used as a template to react so that a nucleic acid chain continuously extends. According to the nucleic acid synthesis method disclosed by the invention, the length of the core primer is about 30bp and is shortened by 10bp compared with the core primer which is about 40bp and is used by LAMP (Loop-Mediated Isothermal Amplification) and CAMP (Cyclic Amplified Polymorphism), so that about 1 / 4 of primer cost can be saved, and meanwhile, the reaction rate of nucleic acid synthesis is also improved.
Owner:国科宁波生命与健康产业研究院
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com