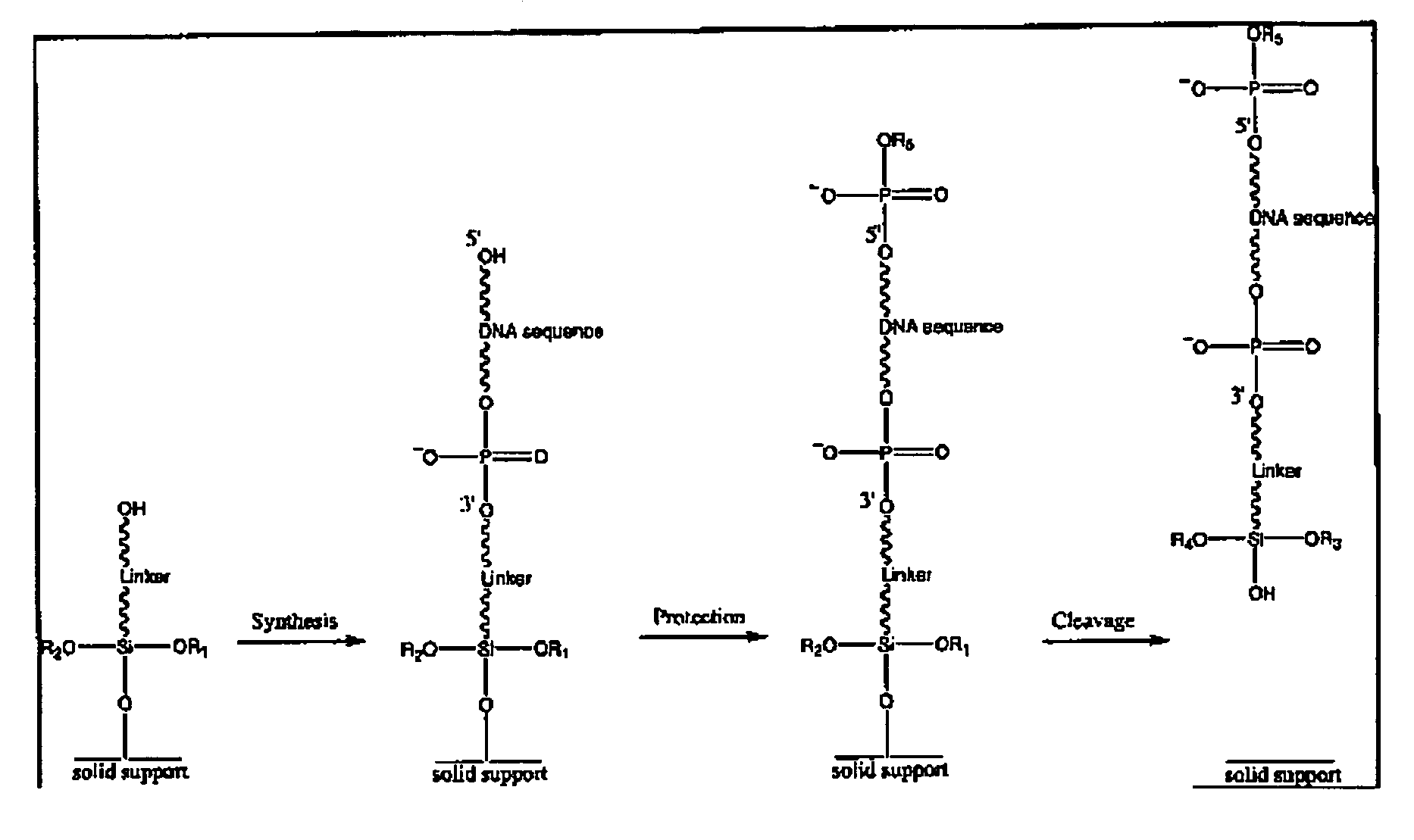
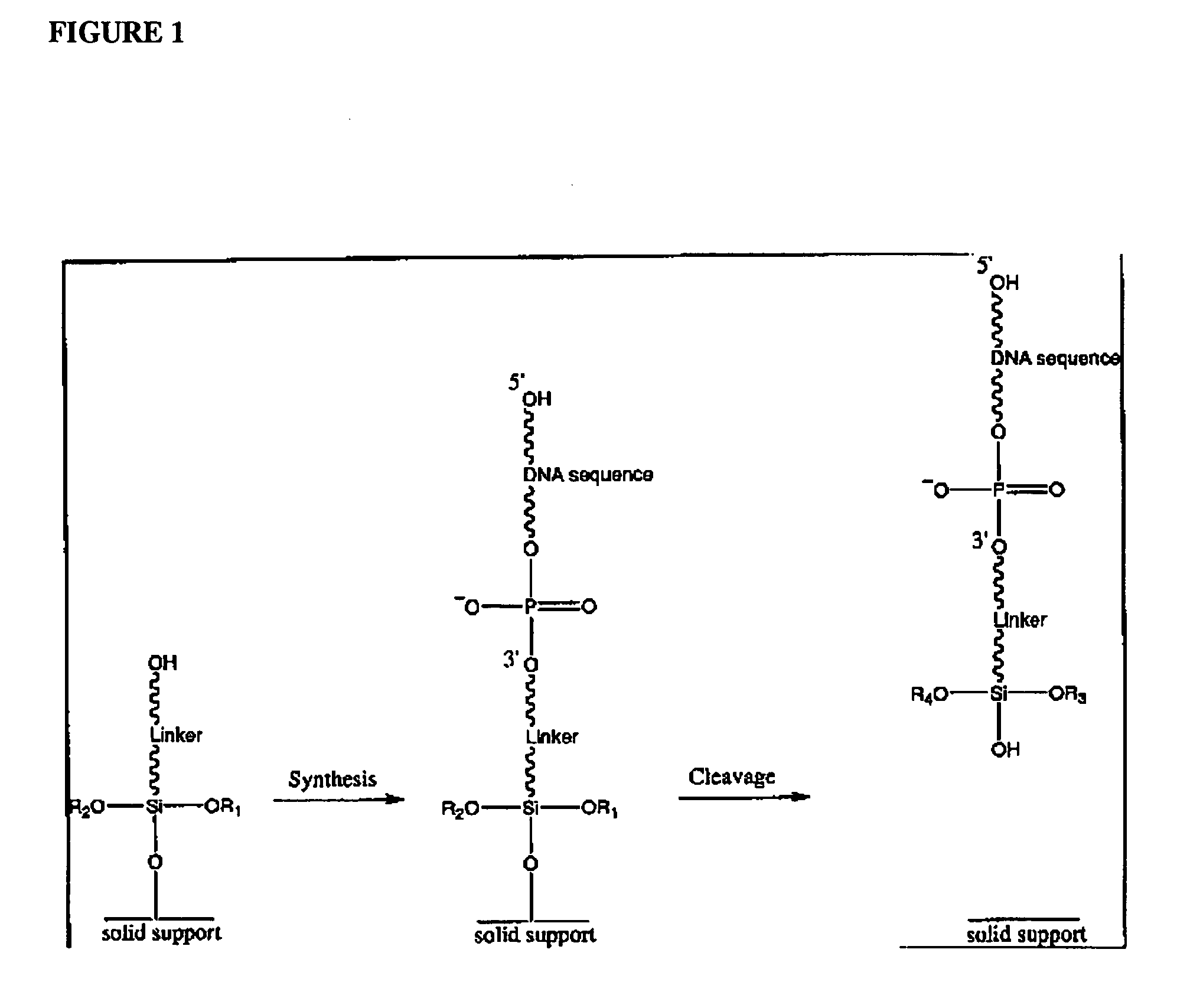
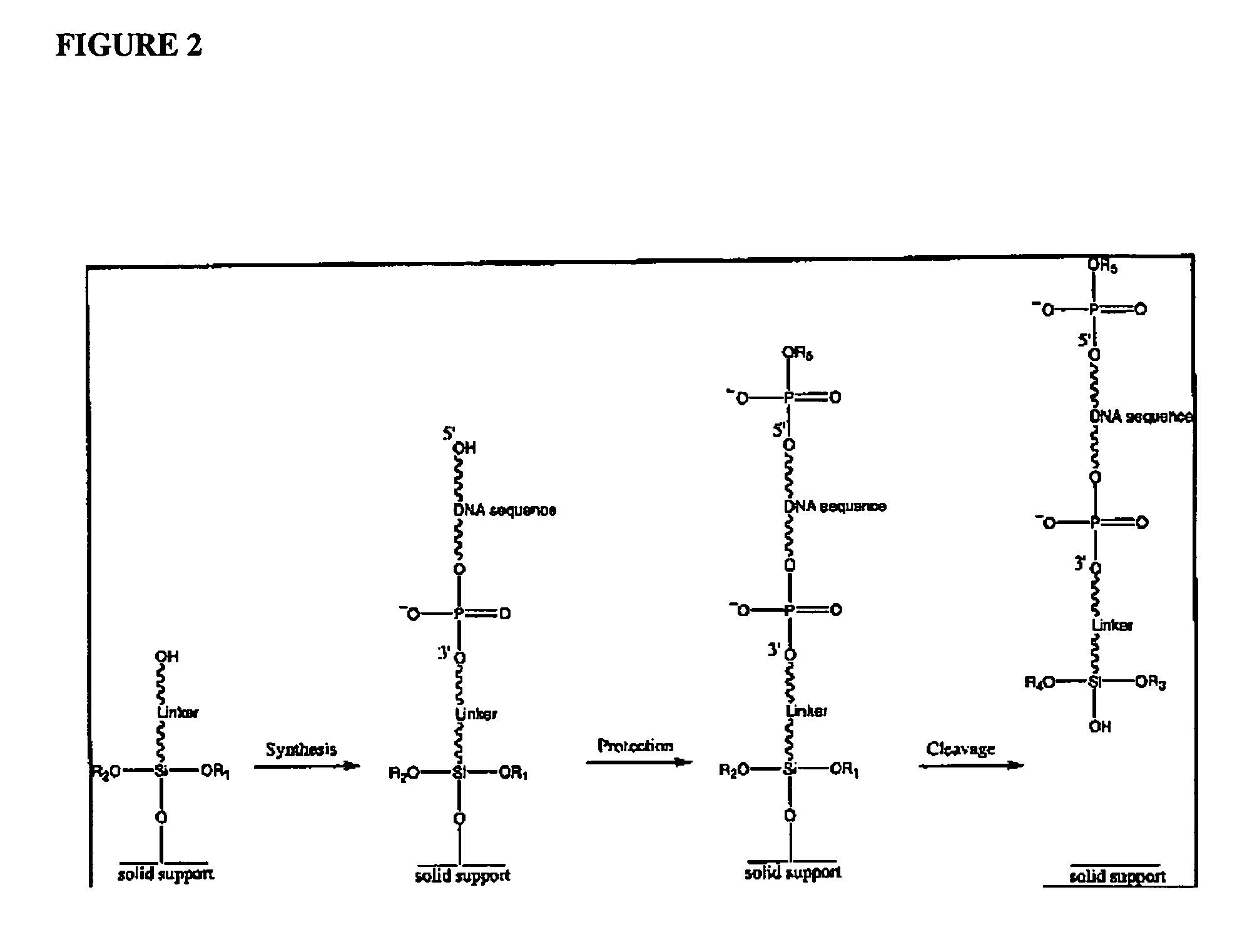
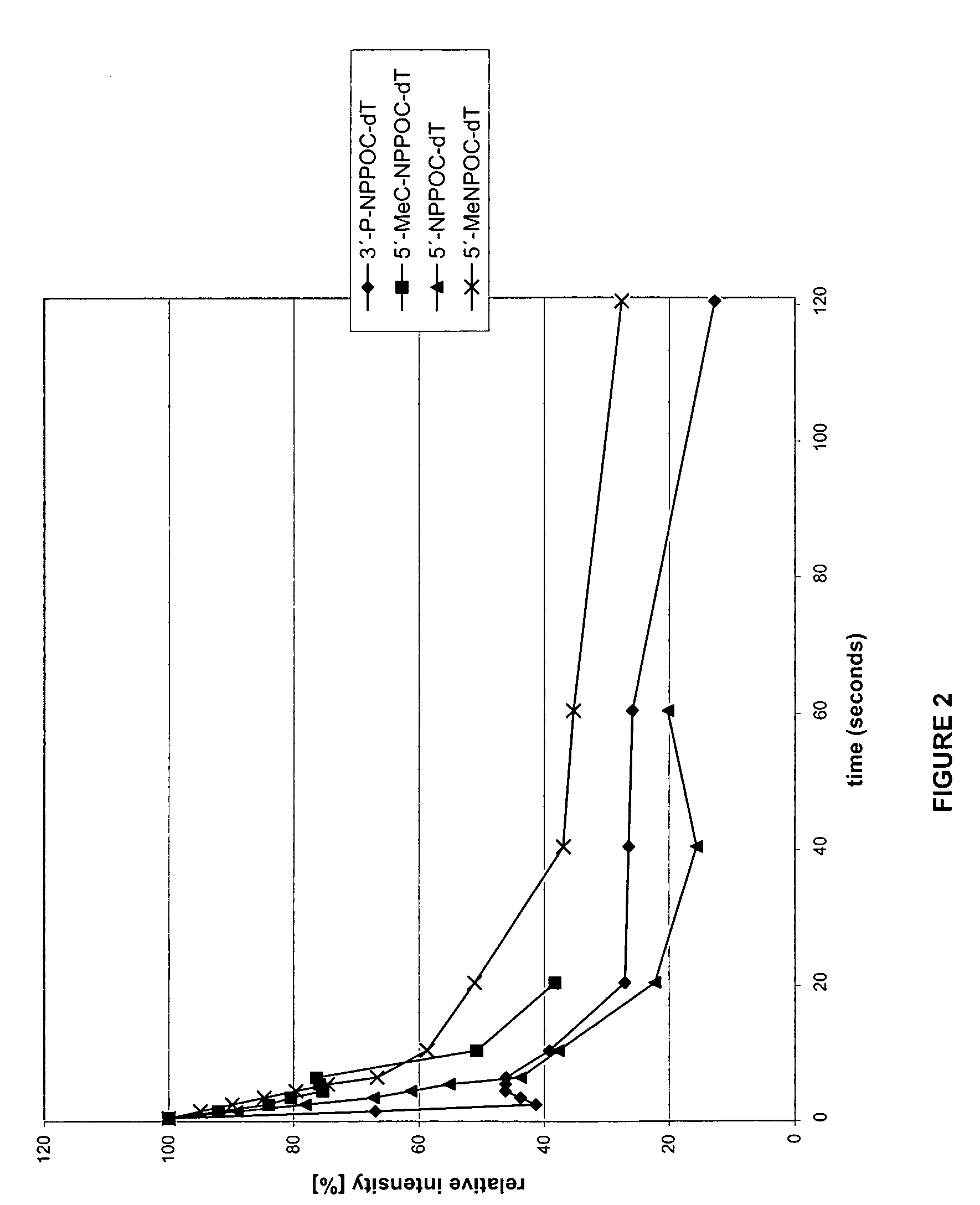
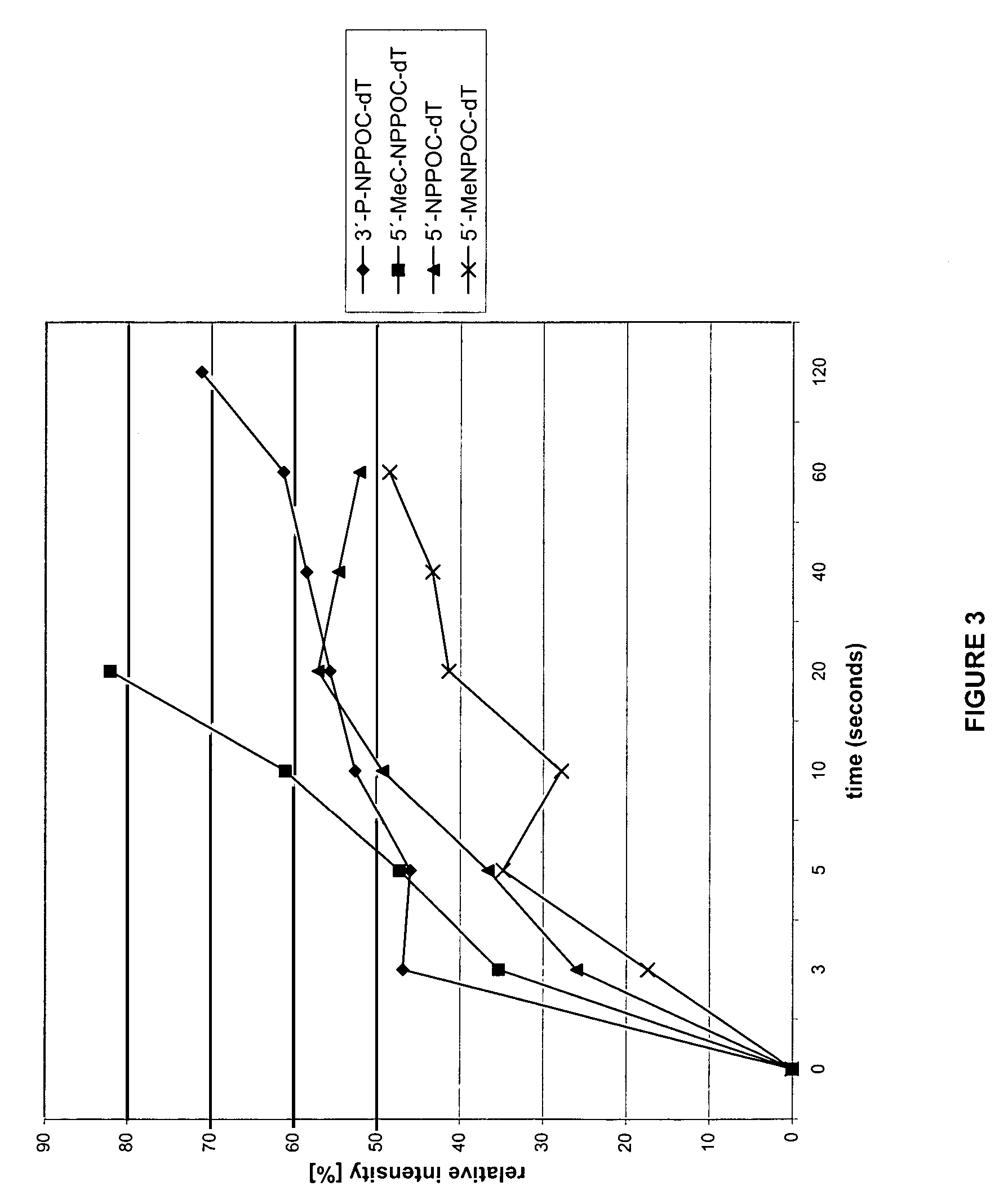
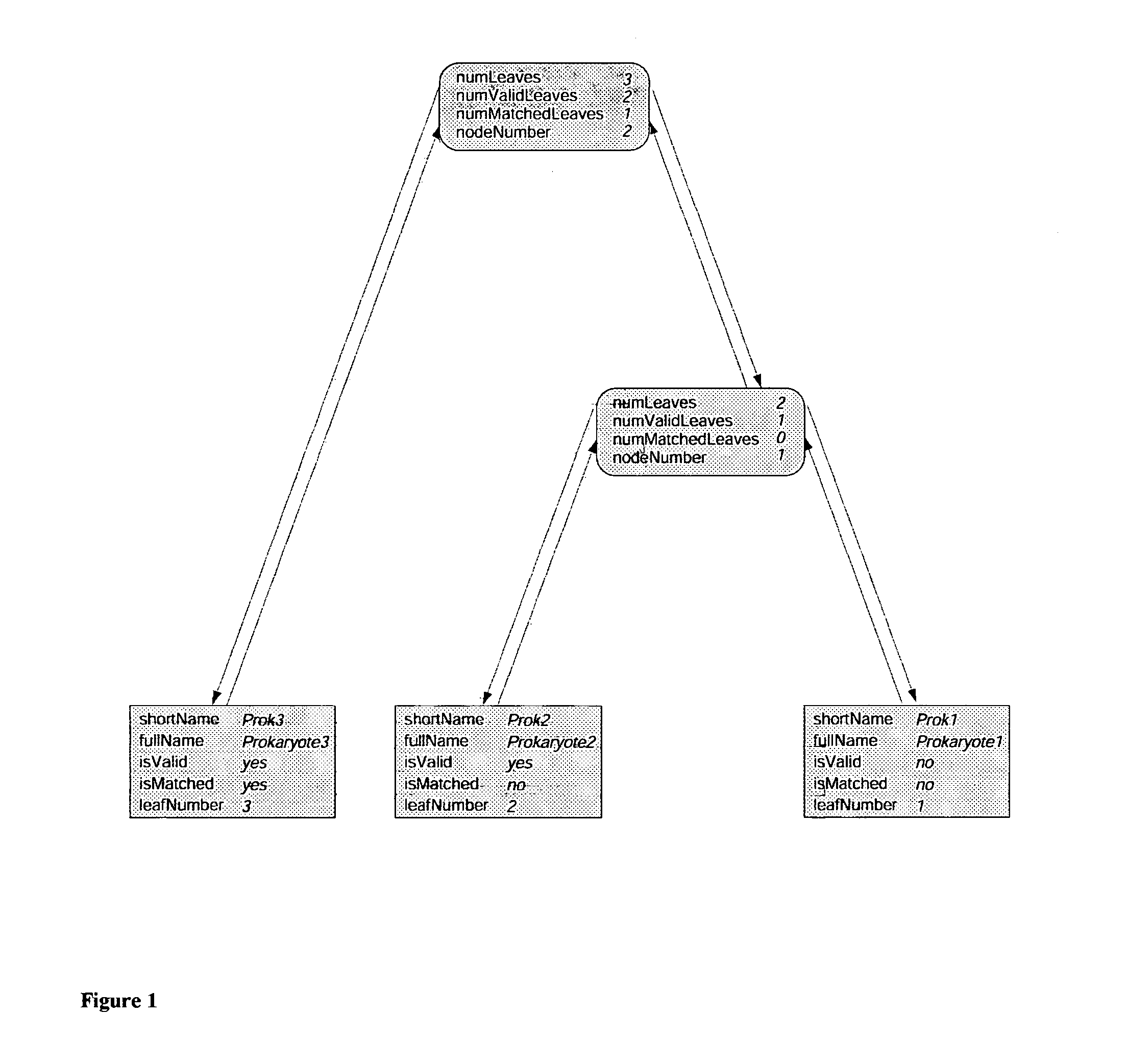
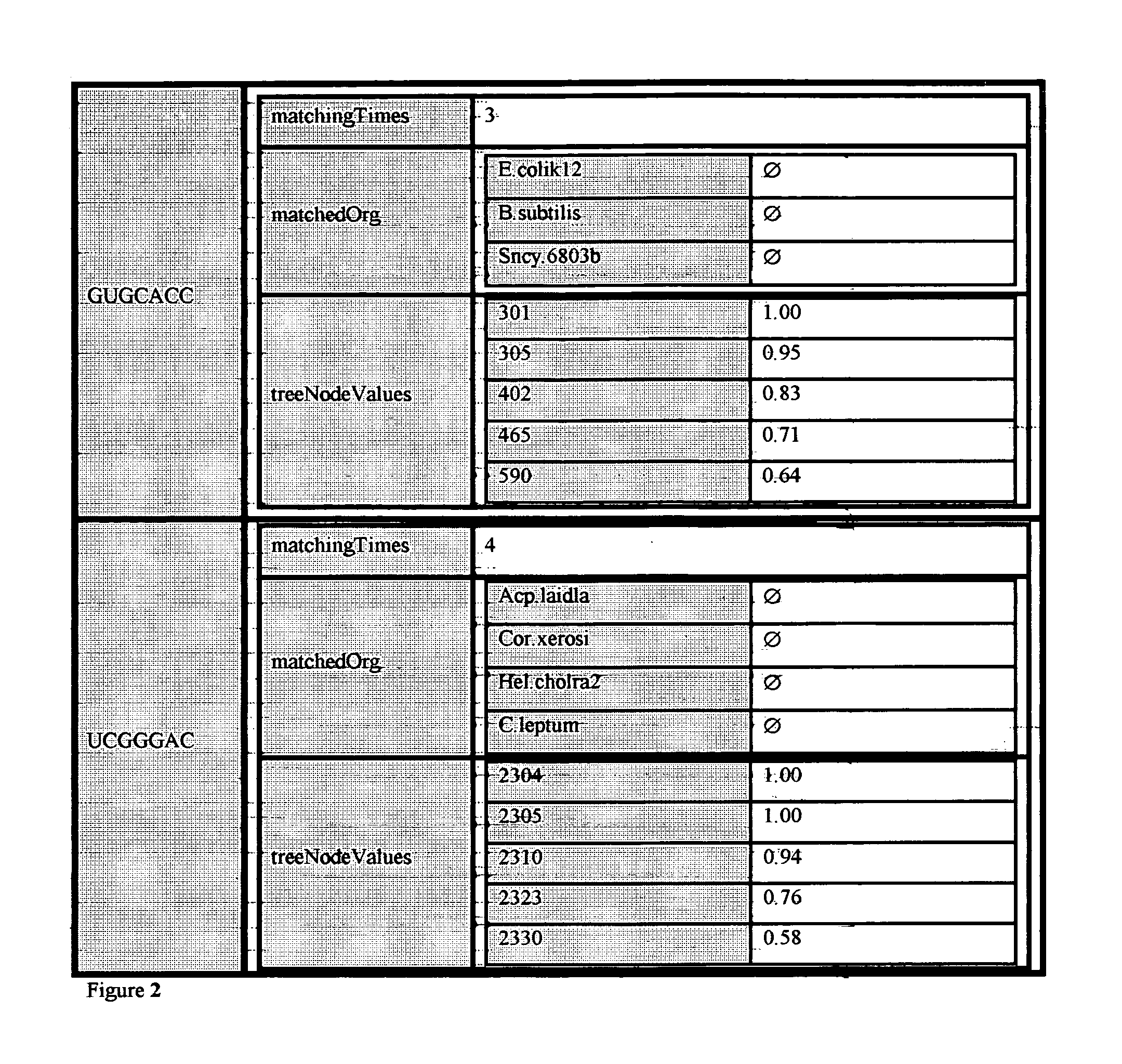
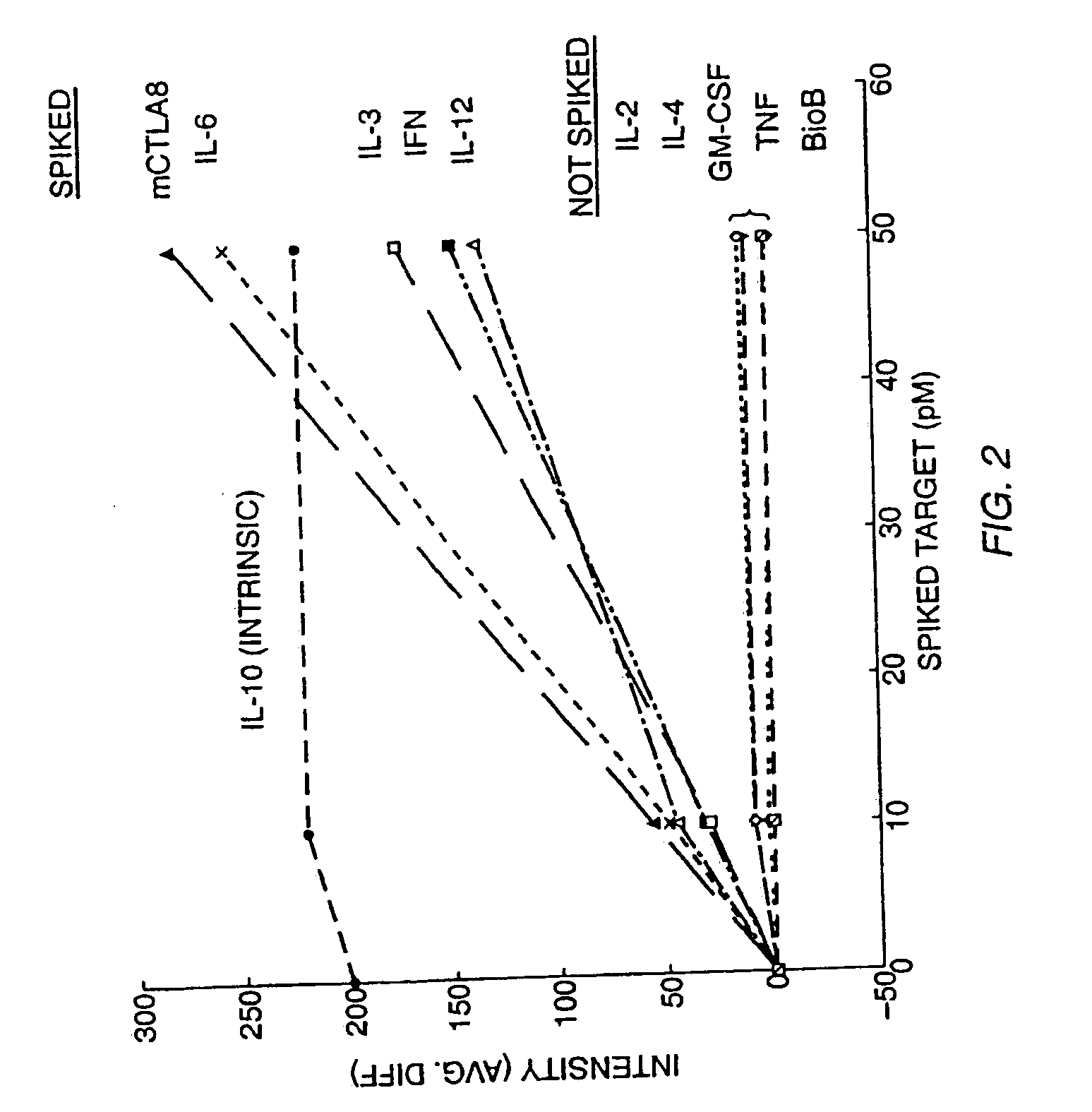
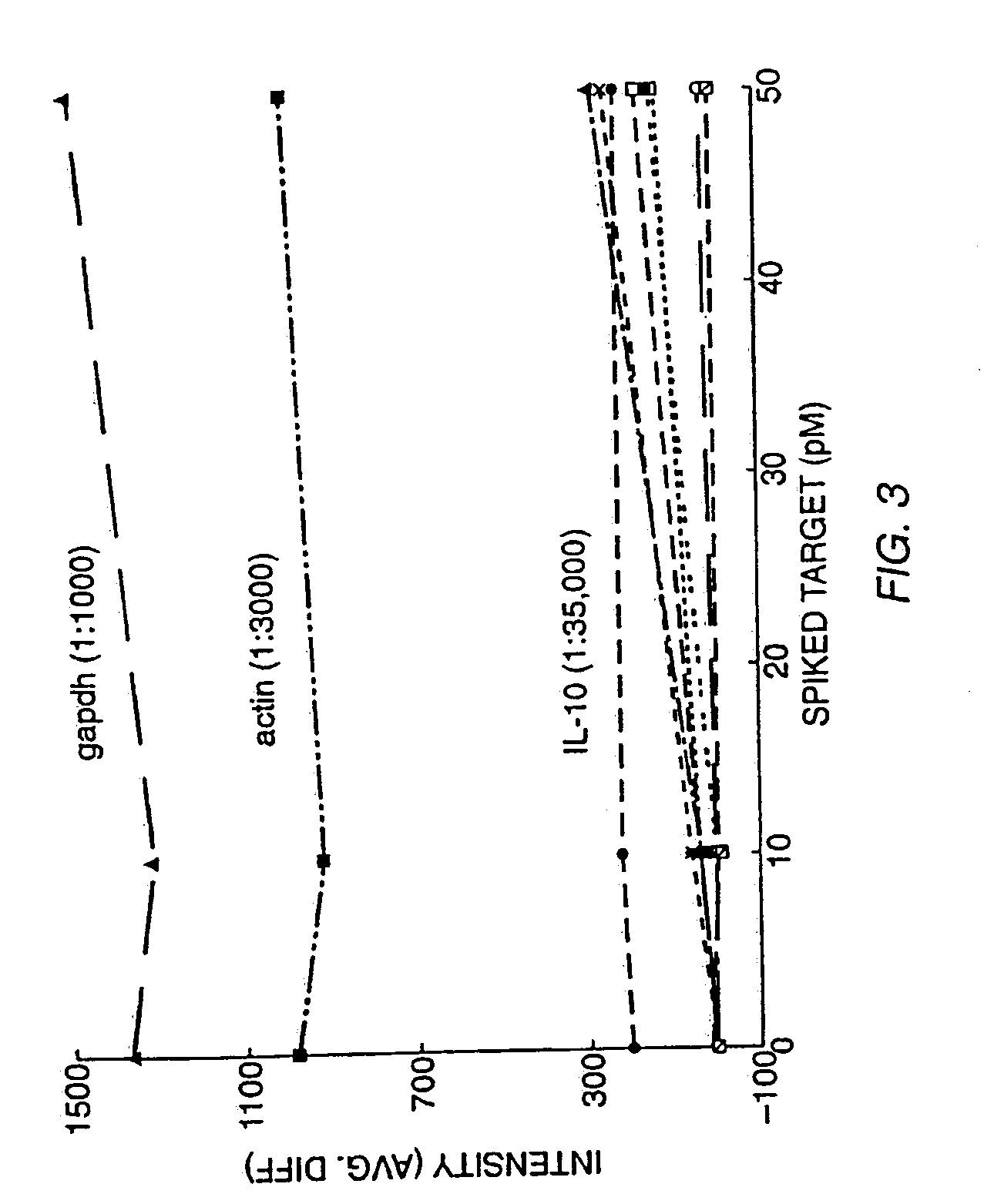
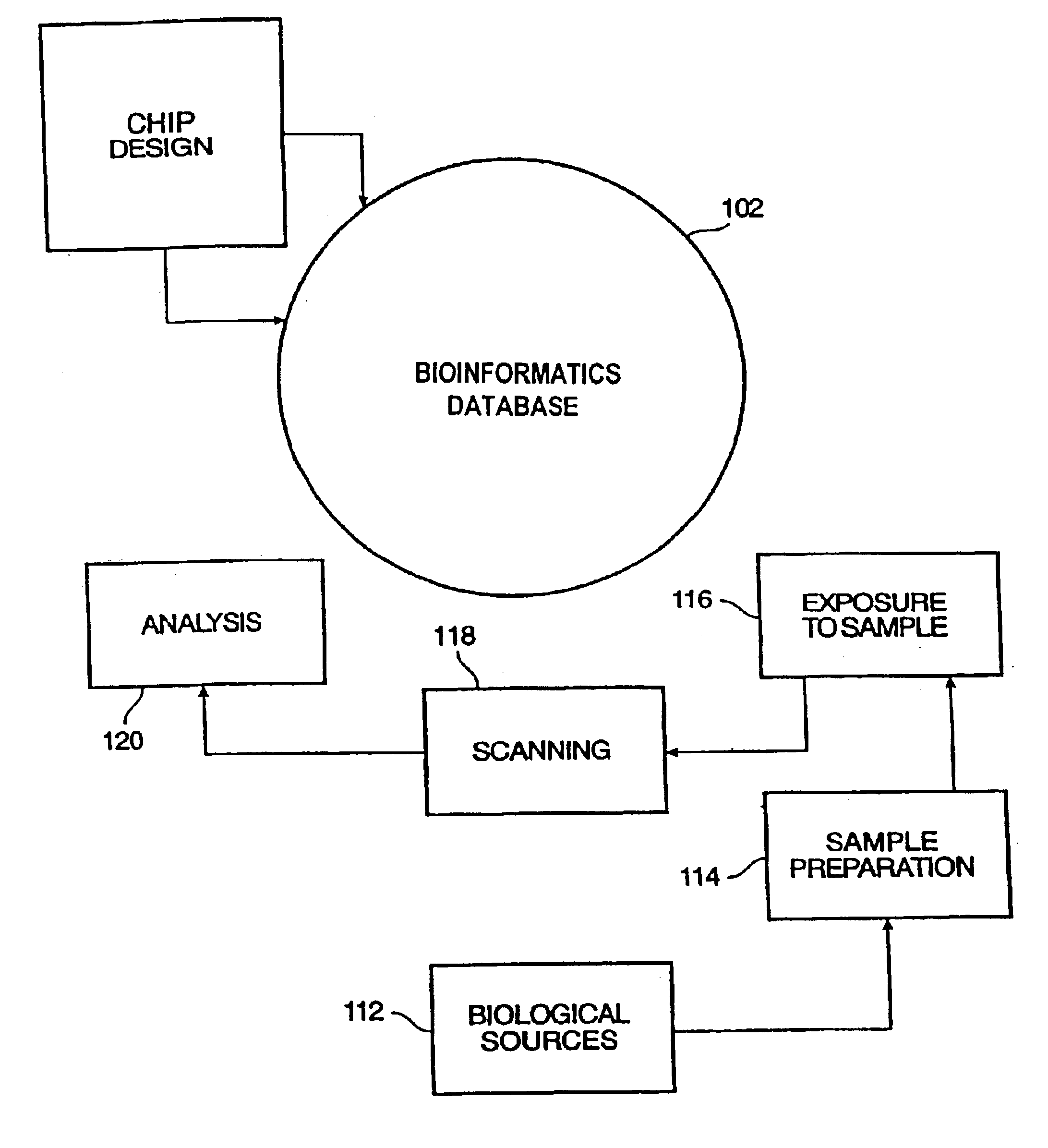
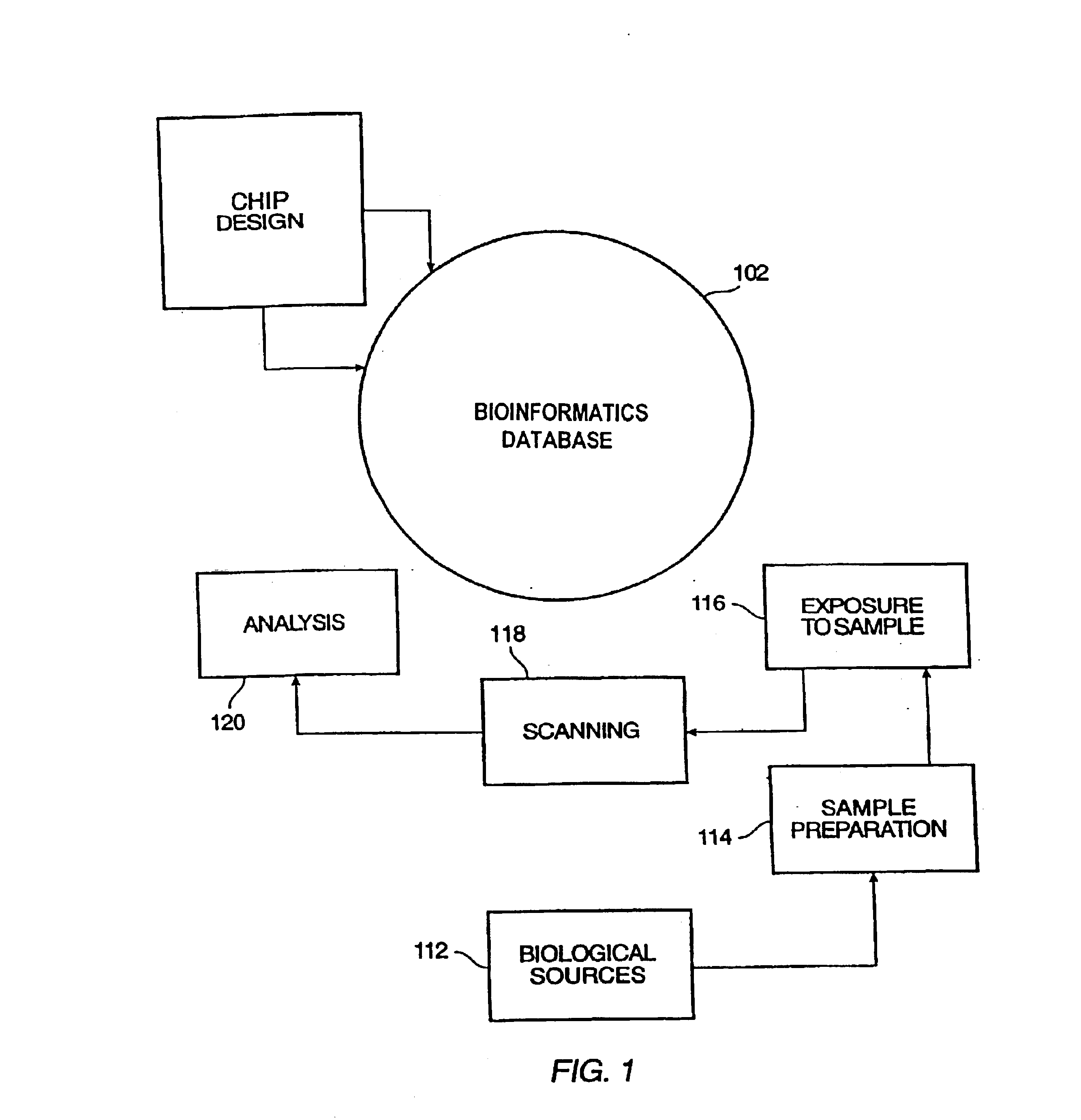
Patents
Literature
62 results about "Oligonucleotide Arrays" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
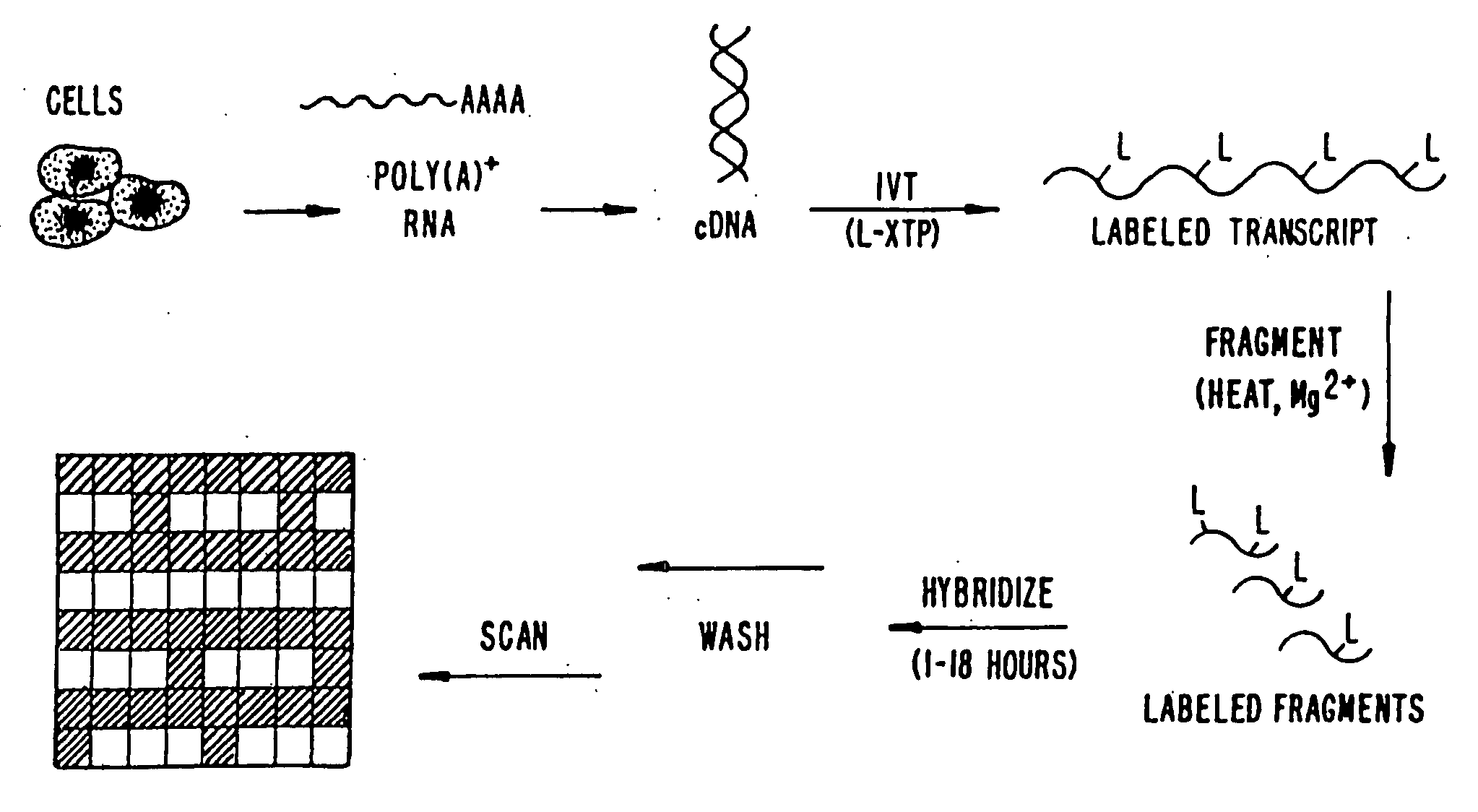
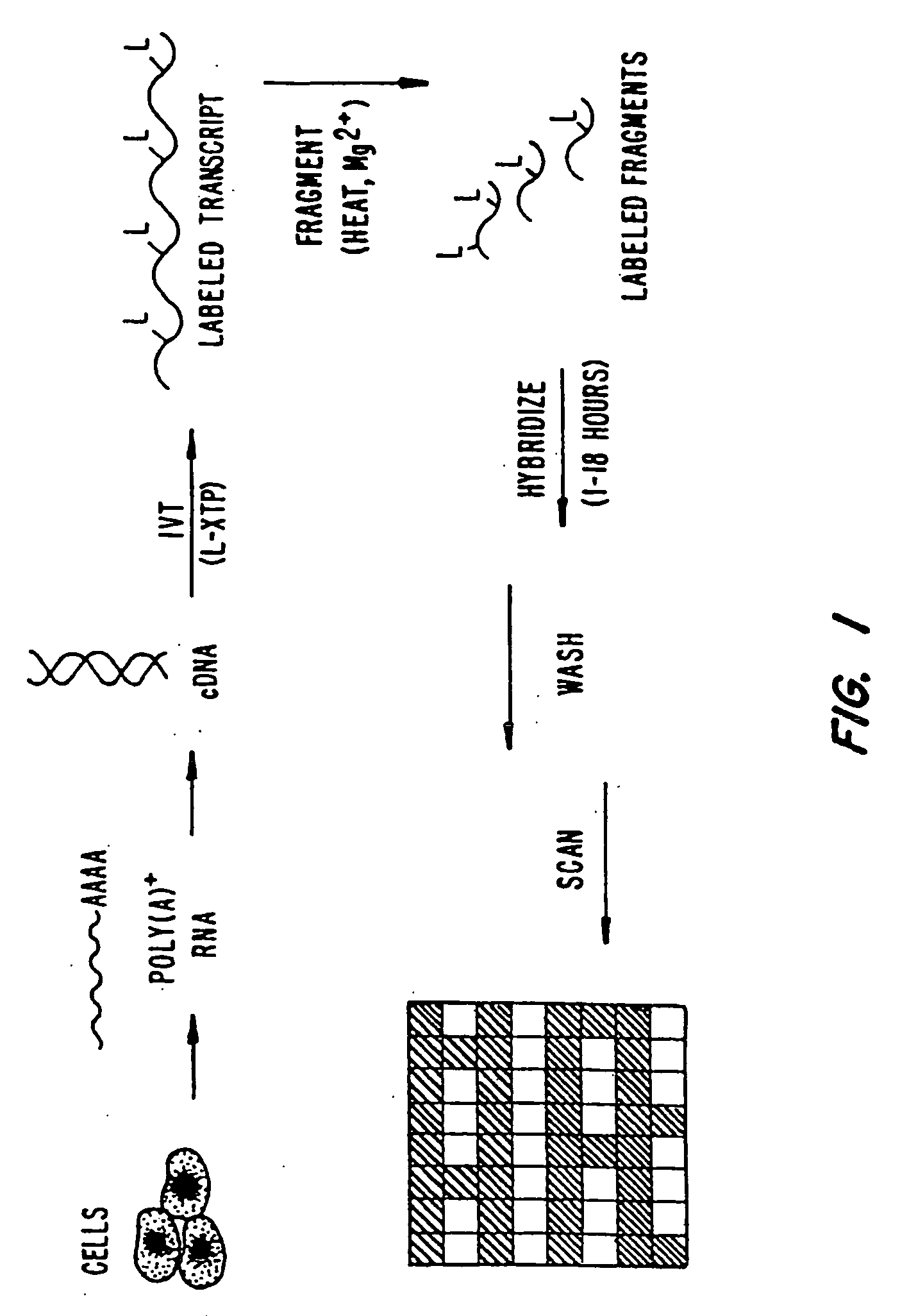
Oligonucleotide arrays and their use for sorting, isolating, sequencing, and manipulating nucleic acids
InactiveUS6322971B1Increase in hybridization specificityBioreactor/fermenter combinationsMaterial nanotechnologyHybridization ArrayBioinformatics
Ligation methods for manipulating nucleic acid stands and oligonucleotides utilizing hybridization arrays of immobilized oligonucleotides. The oligonucleotide arrays may be plain or sectioned, comprehensive or non-comprehensive. The immobilized oligonucleotides may in some cases be binary oligonucleotides having constant as well as variable segments. Some embodiments include amplification of ligated products.
Owner:UNIV OF MEDICINE & DENTISTRY OF NEW JERSEY
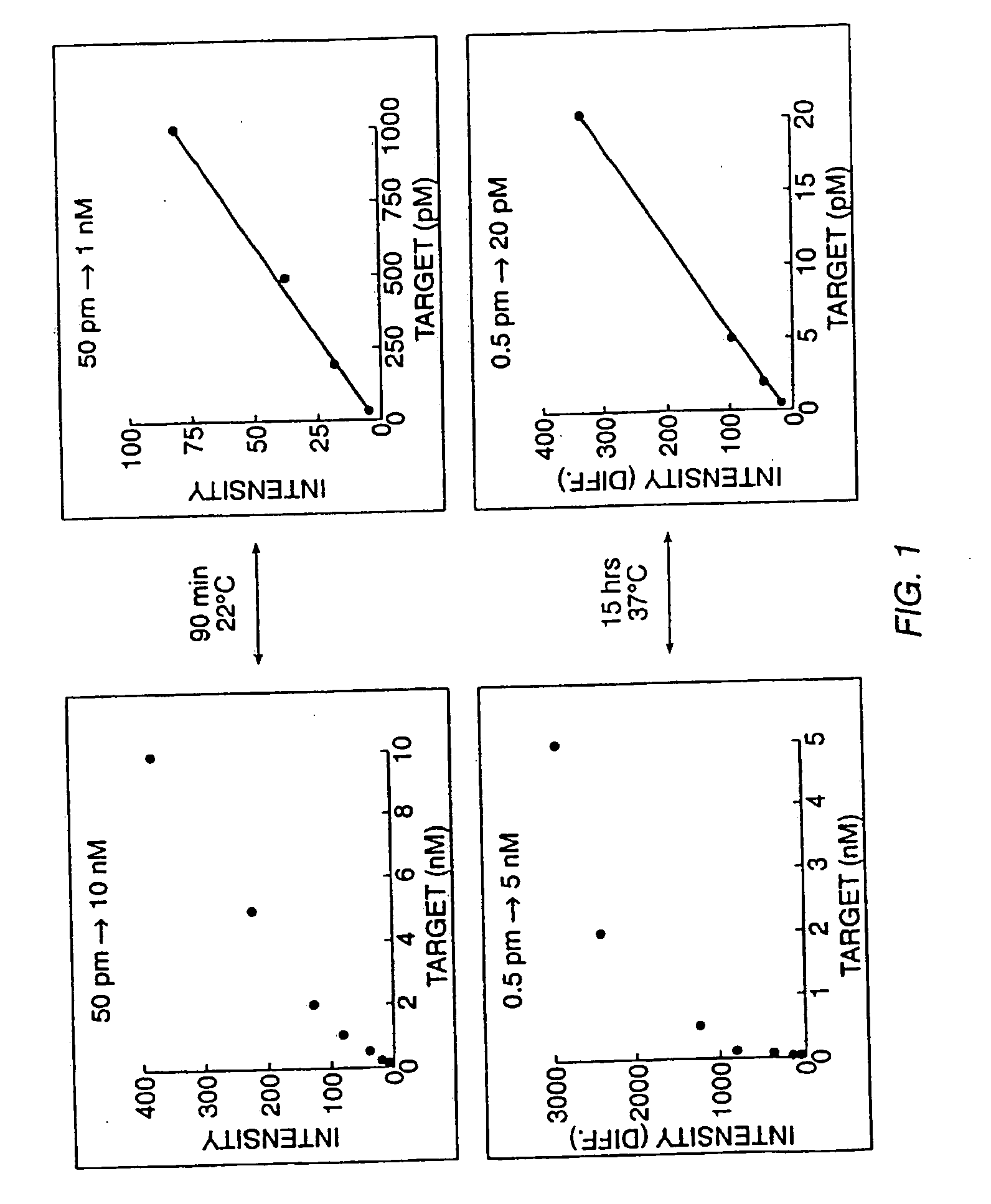
Detection of nucleic acid sequence differences using the ligase detection reaction with addressable arrays
InactiveUS7083917B2Avoid signalingFalse signalMaterial nanotechnologyBioreactor/fermenter combinationsNucleic acid sequencingNucleotide sequencing
The present invention describes a method for identifying one or more of a plurality of sequences differing by one or more single base changes, insertions, deletions, or translocations in a plurality of target nucleotide sequences. The method includes a ligation phase, a capture phase, and a detection phase. The ligation phase utilizes a ligation detection reaction between one oligonucleotide probe, which has a target sequence-specific portion and an addressable array-specific portion, and a second oligonucleotide probe, having a target sequence-specific portion and a detectable label. After the ligation phase, the capture phase is carried out by hybridizing the ligated oligonucleotide probes to a solid support with an array of immobilized capture oligonucleotides at least some of which are complementary to the addressable array-specific portion. Following completion of the capture phase, a detection phase is carried out to detect the labels of ligated oligonucleotide probes hybridized to the solid support. The ligation phase can be preceded by an amplification process. The present invention also relates to a kit for practicing this method, a method of forming arrays on solid supports, and the supports themselves.
Owner:CORNELL RES FOUNDATION INC
Expression monitoring by hybridization to high density oligonucleotide arrays
InactiveUS6927032B2Reduce complexityReduce background signalBioreactor/fermenter combinationsBiological substance pretreatmentsHigh densityOligonucleotide
This invention provides methods of monitoring the expression levels of a multiplicity of genes. The methods involve hybridizing a nucleic acid sample to a high density array of oligonucleotide probes where the high density array contains oligonucleotide probes complementary to subsequences of target nucleic acids in the nucleic acid sample. In one embodiment, the method involves providing a pool of target nucleic acids comprising RNA transcripts of one or more target genes, or nucleic acids derived from the RNA transcripts, hybridizing said pool of nucleic acids to an array of oligonucleotide probes immobilized on surface, where the array comprising more than 100 different oligonucleotides and each different oligonucleotide is localized in a predetermined region of the surface, the density of the different oligonucleotides is greater than about 60 different oligonucleotides per 1 cm2, and the olignucleotide probes are complementary to the RNA transcripts or nucleic acids derived from the RNA transcripts; and quantifying the hybridized nucleic acids in the array.
Owner:AFFYMETRIX INC
Sequencing by hybridization
InactiveUS6399364B1Improving the accuracy of an already very informationStable and reliableBioreactor/fermenter combinationsBiological substance pretreatmentsBioinformaticsOligonucleotide Arrays
A method of analysing a nucleic acid is disclosed which involves the use of a mixture of labelled oligonucleotides in solution and an array of immobilized oligonucleotides. The target nucleic acid is incubated with the mixture of labelled oligonucleotides. Those labelled oligonucleotides which hybridize are recovered and incubated with the array of immobilized oligonucleotides. Sequence information is obtained by observing the location of the label on the array. The method is particularly suitable for determining differences between nucleic acids.
Owner:AMERSHAM BIOSCIENCES UK LTD
Addressable oligonucleotide array of the rat genome
InactiveUS7314750B2Monitor gene expression levelBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid sequencingGenome
The invention provides nucleic acid sequences which are complementary, in one embodiment, to a wide variety of Rat genes. The invention provides the sequences in such a way as to make them available for a variety of analyses. In one embodiment the nucleic acid sequences provided are present as an array of probes that may be used to measure gene expression of at least 20,000 rat genes. As such, the invention relates to diverse fields impacted by the nature of molecular interaction, including chemistry, biology, medicine, and medical diagnostics.
Owner:AFFYMETRIX INC
Controlling use of oligonucleotide sequences released from arrays
InactiveUS7749701B2Bioreactor/fermenter combinationsSequential/parallel process reactionsEnzymeOligonucleotide
A method for controlling the use of oligonucleotide sequences released from arrays, comprises synthesizing a chemical array of oligonucleotides on a substrate under conditions for producing an array of cleavable oligonucleotides that are blocked from enzymatic reactions after cleavage. Methods may also include receiving a chemical array of cleavable oligonucleotides on a substrate, and cleaving the oligonucleotides from the array, wherein the oligonucleotides are blocked from enzymatic reactions after cleavage. Arrays, populations of oligonucleotides and kits are also provided to facilitate the methods.
Owner:AGILENT TECH INC
Long oligonucleotide arrays
InactiveUS7041445B2Improve binding efficiencyLow non-specific bindingBioreactor/fermenter combinationsSequential/parallel process reactionsBiologyOligonucleotide
Owner:TAKARA BIO USA INC
Detection of nucleic acid sequence differences using the ligase detection reaction with addressable arrays
InactiveUS20060183149A1Suitable for automationReduce in quantityMaterial nanotechnologySequential/parallel process reactionsNucleotideNucleic acid sequencing
The present invention describes a method for identifying one or more of a plurality of sequences differing by one or more single base changes, insertions, deletions, or translocations in a plurality of target nucleotide sequences. The method includes a ligation phase, a capture phase, and a detection phase. The ligation phase utilizes a ligation detection reaction between one oligonucleotide probe, which has a target sequence-specific portion and an addressable array-specific portion, and a second oligonucleotide probe, having a target sequence-specific portion and a detectable label. After the ligation phase, the capture phase is carried out by hybridizing the ligated oligonucleotide probes to a solid support with an array of immobilized capture oligonucleotides at least some of which are complementary to the addressable array-specific portion. Following completion of the capture phase, a detection phase is carried out to detect the labels of ligated oligonucleotide probes hybridized to the solid support. The ligation phase can be preceded by an amplification process. The present invention also relates to a kit for practicing this method, a method of forming arrays on solid supports, and the supports themselves.
Owner:CORNELL RES FOUNDATION INC
Method and apparatus for conducting an array of chemical reactions on a support surface
InactiveUS6921636B1Material nanotechnologyBioreactor/fermenter combinationsChemical reactionPeptide mimetic
The invention provides apparatus and methods for determining the nucleotide sequence of target nucleic acids using hybridization to arrays of oligonucleotides. The invention further provides apparatus and methods for identifying the amino acid sequence of peptides that bind to biologically active macromolecules, by specifically binding biologically active macromolecules to arrays of peptides or peptide mimetics.
Owner:METRIGEN
Method for processing a nucleic acid sample by swinging a segment of a cartridge wall, a system and a cartridge for performing such a method
InactiveUS6872566B2Good reproducibilitySimple meansBioreactor/fermenter combinationsBiological substance pretreatmentsEffective surfaceEngineering
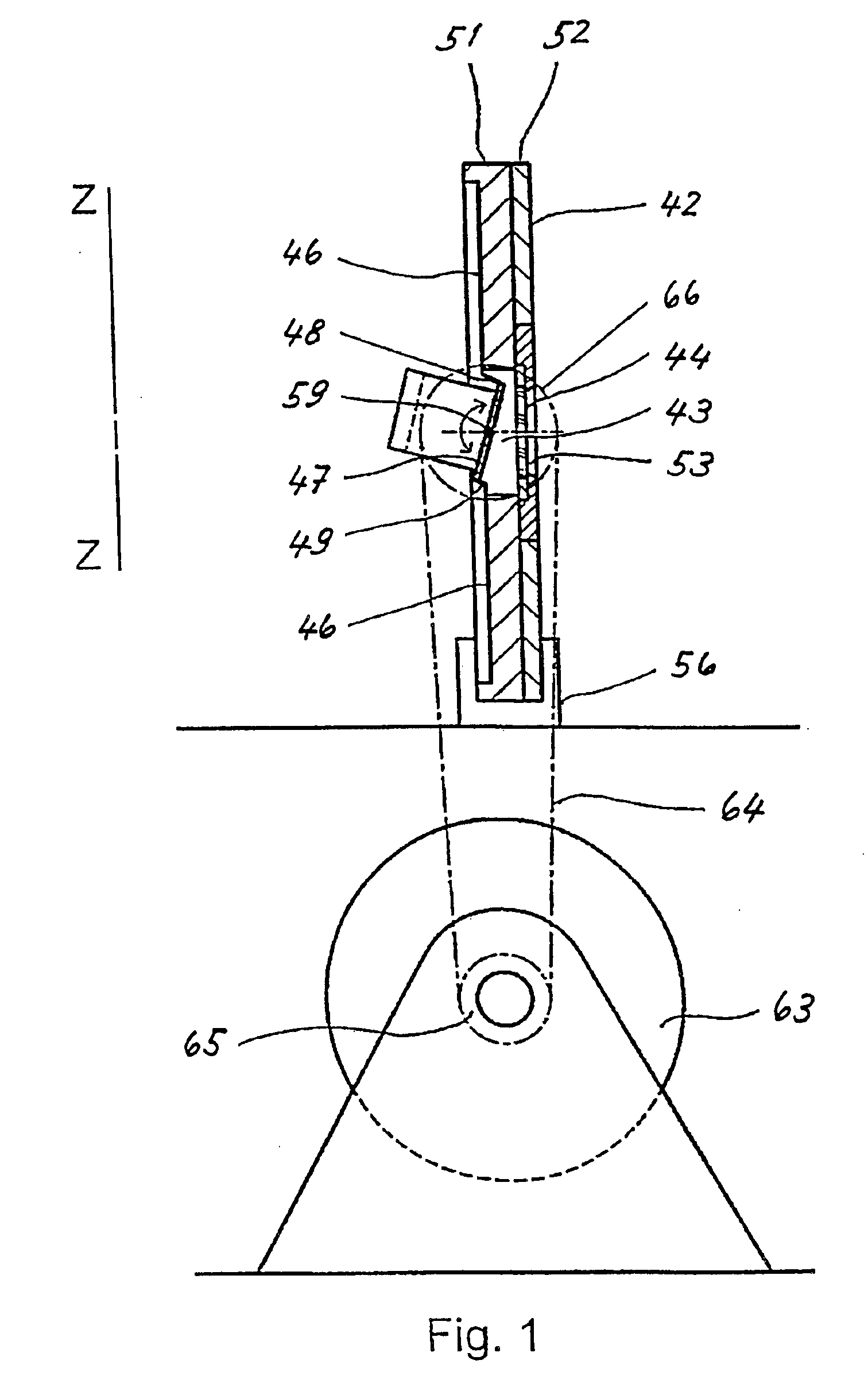
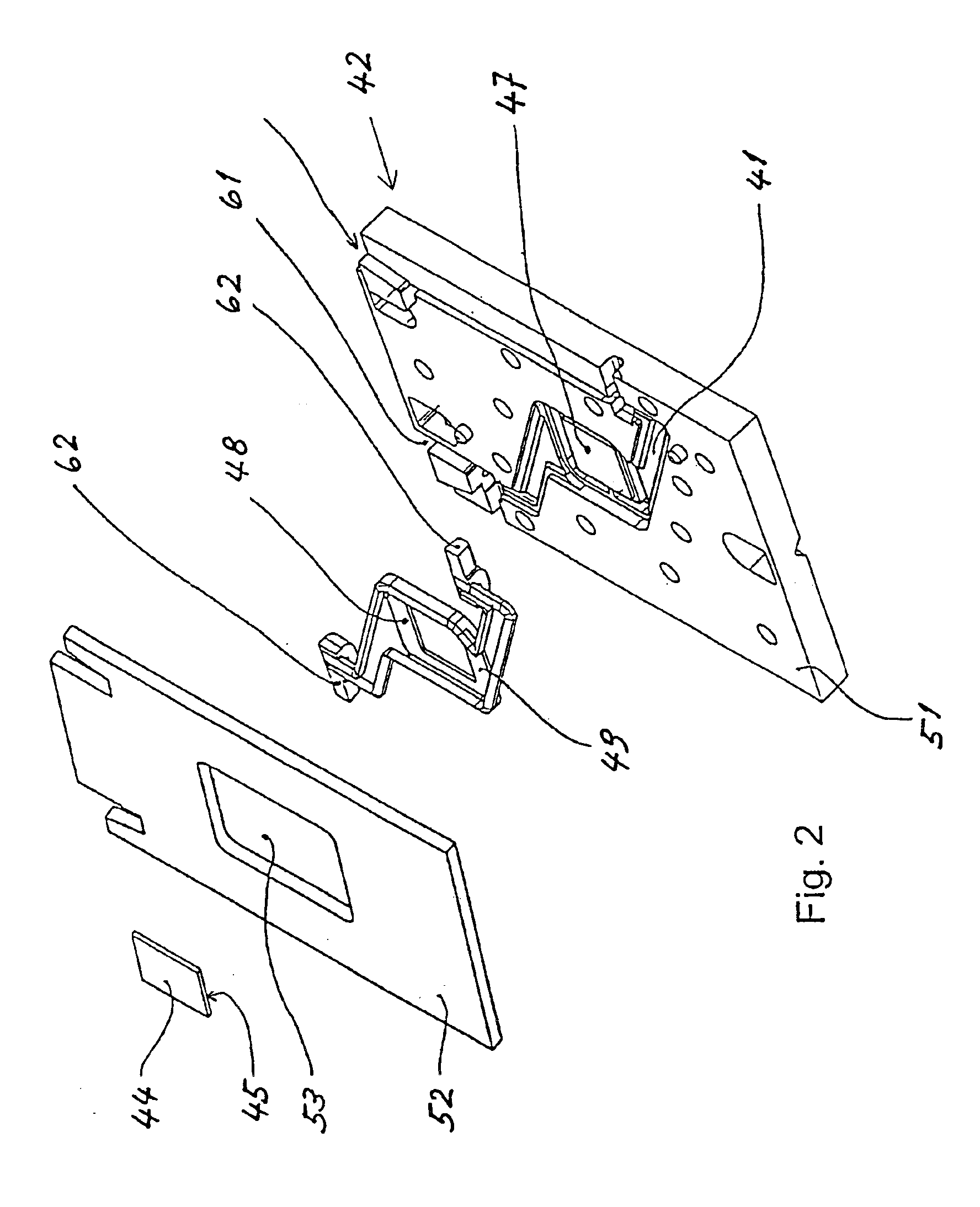
A method for processing a nucleic acid sample contained in a liquid comprises: (a) introducing said liquid into a chamber (41) of a cartridge (42) which contains a chip shaped carrier (44) having an active surface (45) which carries an array of oligonucleotides, said active surface (45) facing the inner surface of a wall (46) of said cartridge,said chamber (41) having a narrow interior and including a channel (43), a portion of said channel lying between said active surface (45) of said chip shaped carrier (44) and the inner surface of said wall (46),a rigid segment (47) of said wall (46) being adapted to be swung of a predetermined angle back and forth about a torsion bar (59), swinging of that segment (47) in one sense moving one end thereof towards said active surface (45), and swinging of that segment (47) in the opposite sense moving said one end of that segment (47) away from said active surface (45),(b) positioning said cartridge (42) into a cartridge holder (56) which holds said cartridge, said positioning being effected before or after introduction of said liquid containing a sample into said chamber (41), and(c) swinging said rigid segment (47) of said wall (46) of said predetermined angle back and forth about said torsion bar (59) in order to cause relative motion of the liquid contained in said channel (43) with respect to said active surface (45) of said chip shaped carrier (44).
Owner:ROCHE MOLECULAR SYST INC
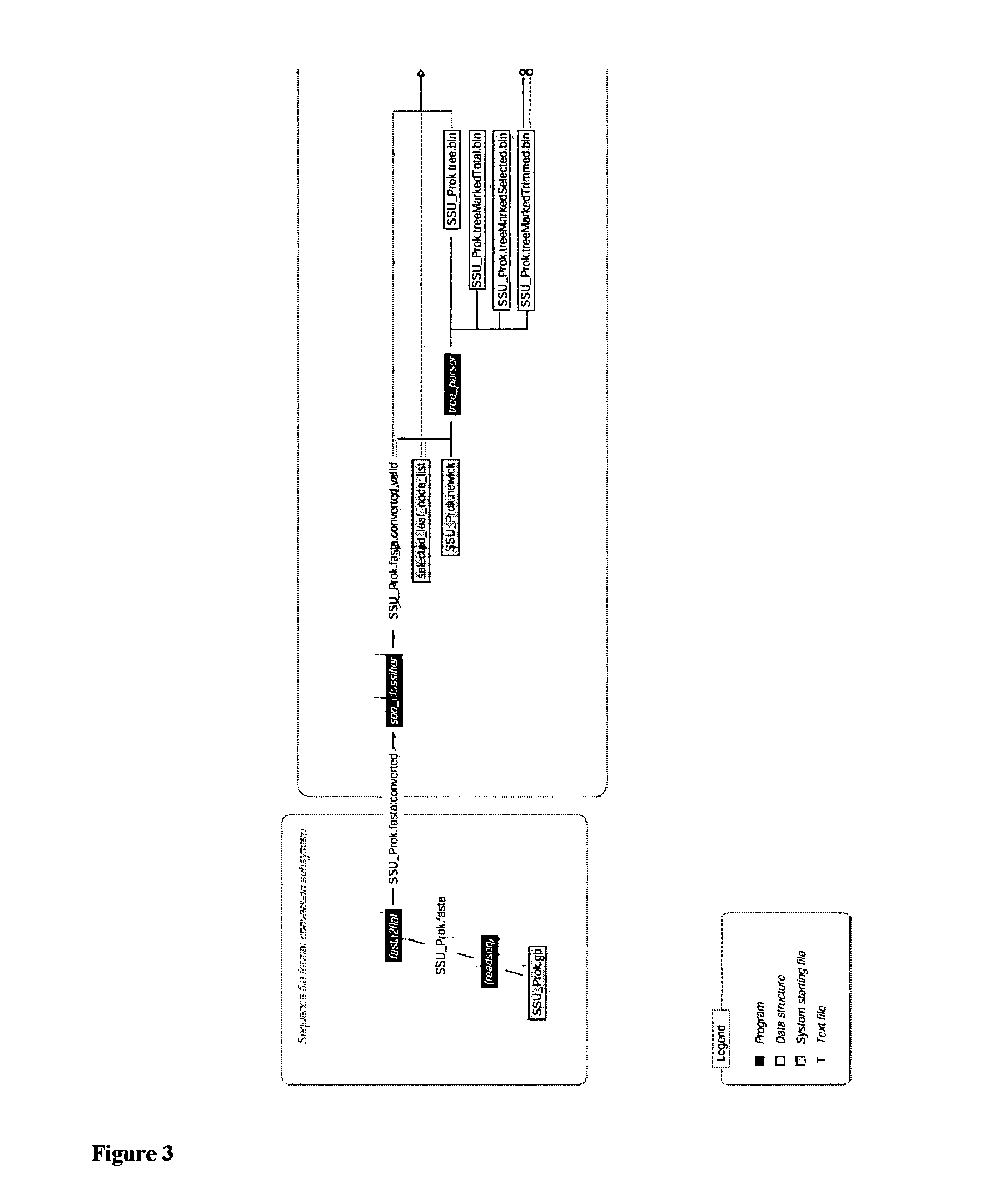
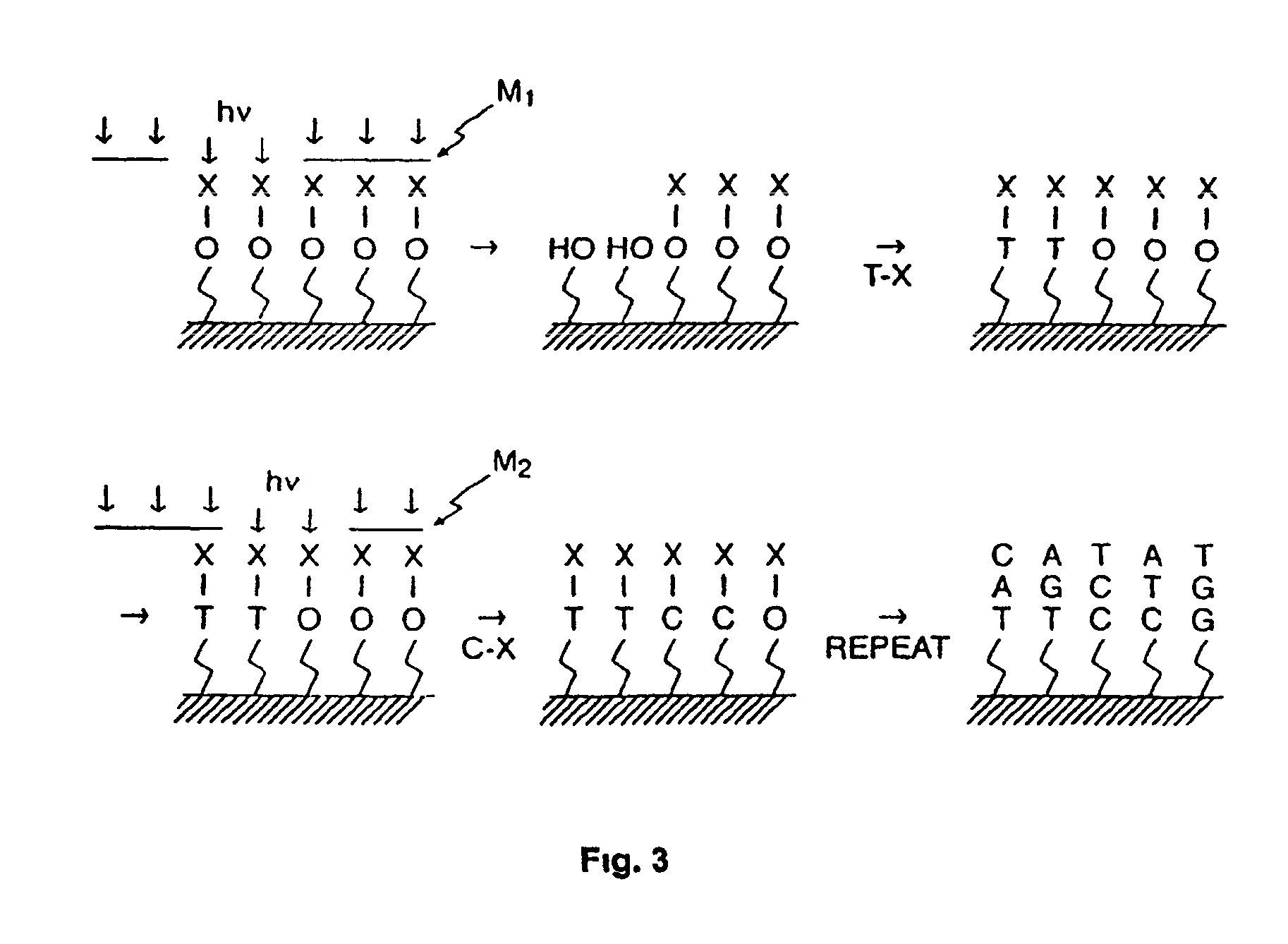
Materials and Methods for the Photodirected Synthesis of Oligonucleotide Arrays
ActiveUS20090270279A1Increasing intrafilm diffusion coefficientWeak molecular weightMaterial nanotechnologyPeptide librariesPhotoacid generatorArray element
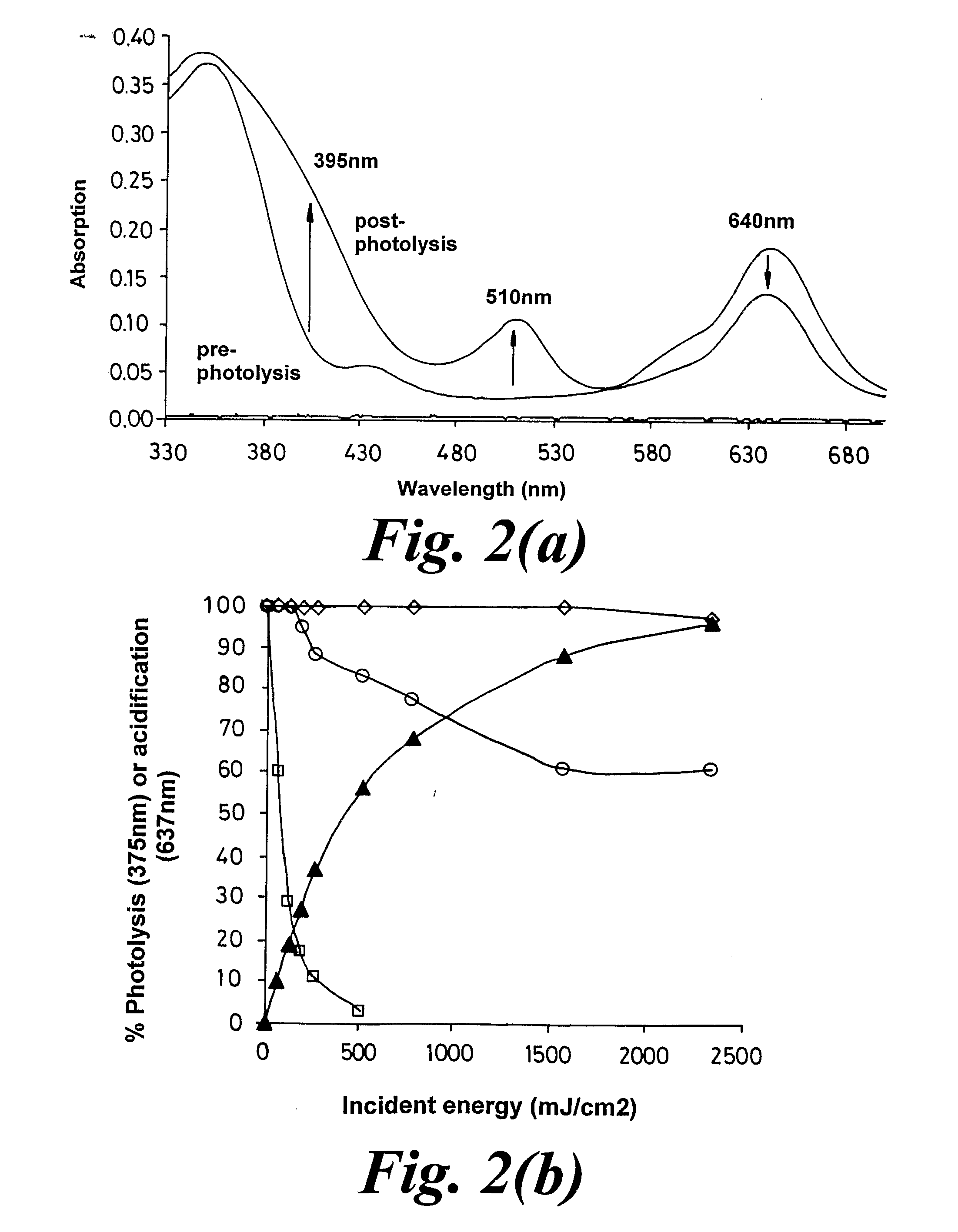
Materials and Methods for photodirected synthesis of oligonucleotide arrays on a solid substrate by photodirected synthesis are disclosed which employ a film formed from (i) a photoacid generator that on photolysis generates acid that is capable of directly removing the protecting group of the linker molecules or oligonucleotides and (ii) a polymer substantially lacking electronegative heteroatoms that are capable of hydrogen bonding with photogenerated acid. Methods of synthesizing an oligomer arrays are also described that use a film that restricts diffusion of reactants and products on the substrate during synthesis of the array and which includes a precursor of a deprotecting reagent. The method involves removing one or more of the non-required products of the deprotection reaction from the reactive array elements at which they are produced during the reaction, in order to displace the deprotection reaction towards completion.
Owner:THE INST OF CANCER RES ROYAL CANCER HOSPITAL
Expression monitoring by hybridization to high density oligonucleotide arrays
InactiveUS20080227653A1Material nanotechnologySequential/parallel process reactionsHigh densityNucleic acid sequencing
The present invention provides methods for comparing and identifying differences in nucleic acid sequences using a plurality of sequence specific recognition reagents (i.e., probes comprising a nucleic acid complementary to a nucleic acid sequence in collections to be compared) bound to a solid surface.
Owner:AFFYMETRIX INC
Variable length probe selection
InactiveUS20050282209A1Improve accuracyImprove efficiencyBioreactor/fermenter combinationsBiological substance pretreatmentsGenome resequencingVariable length
The present invention provides novel method for increasing the efficiency and accuracy of high-throughput mutation mapping and genome resequencing by using a variable length probe selection algorithm to rationally select probes used in designing oligonucleotide arrays synthesized by Maskless Array Synthesis (MAS) technology. Also disclosed is a variable length probe selection algorithm used in designing such oligonucleotide arrays.
Owner:ROCHE NIMBLEGEN
Methods of enzymatic discrimination enhancement and surface-bound double-stranded DNA
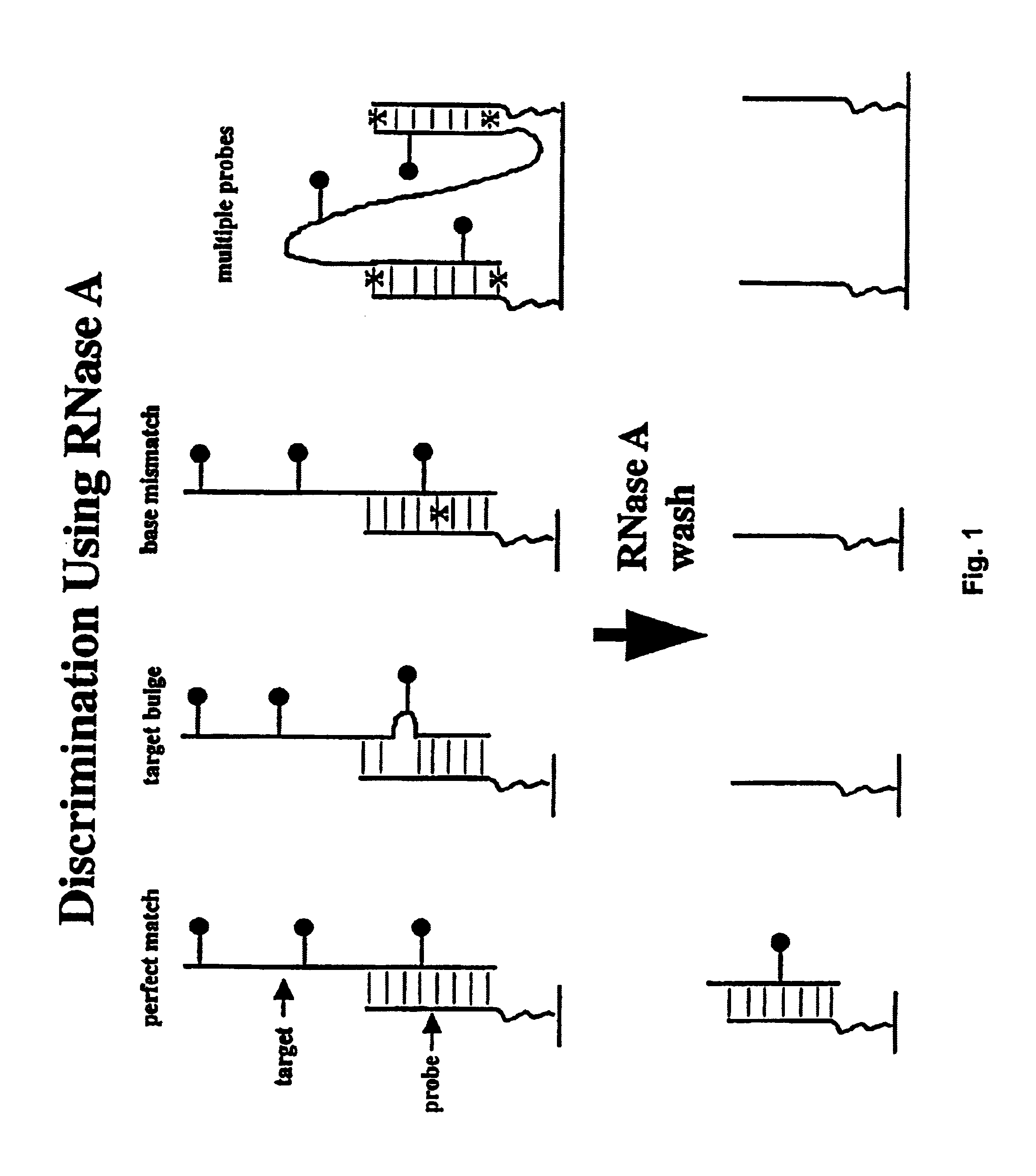
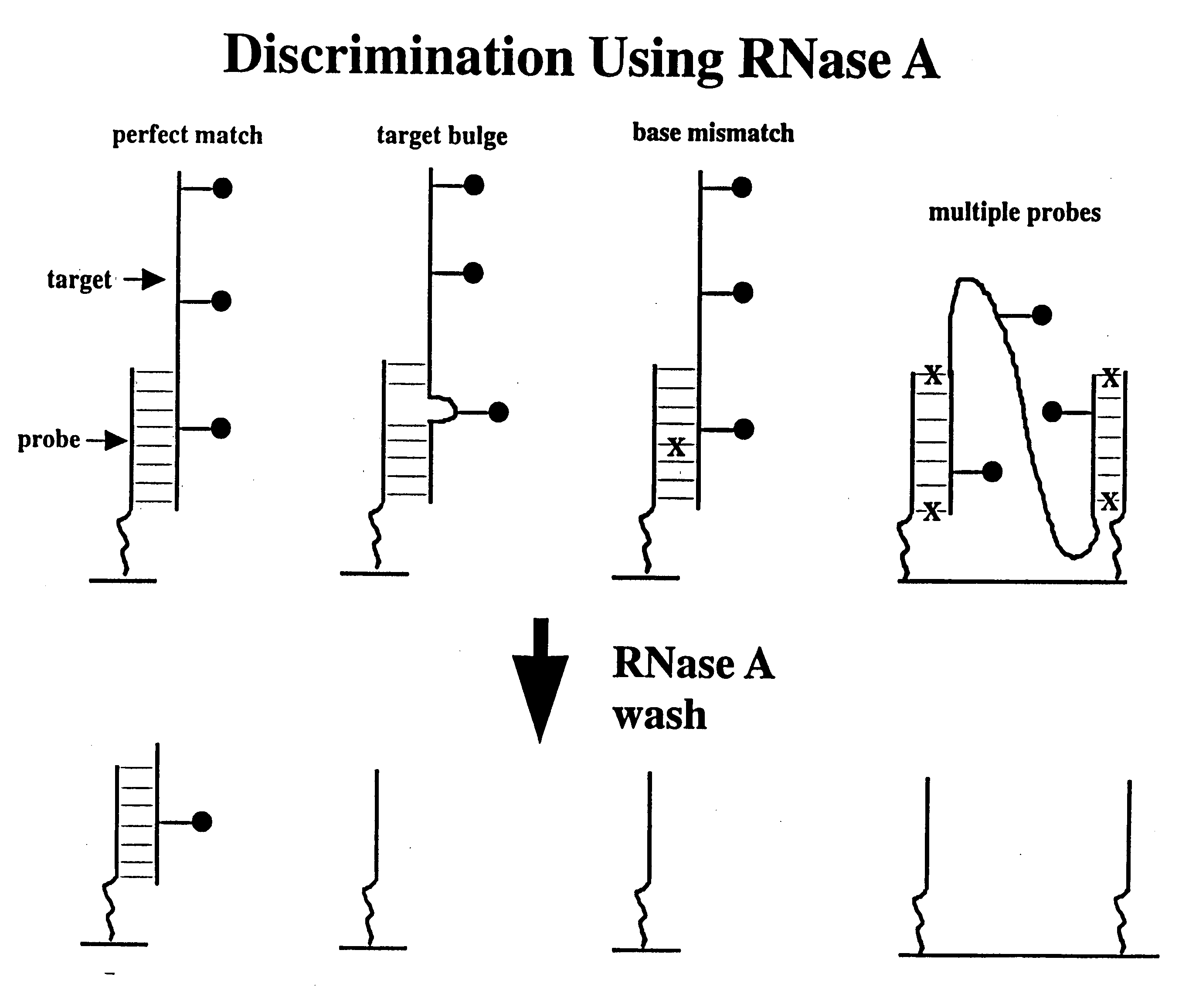
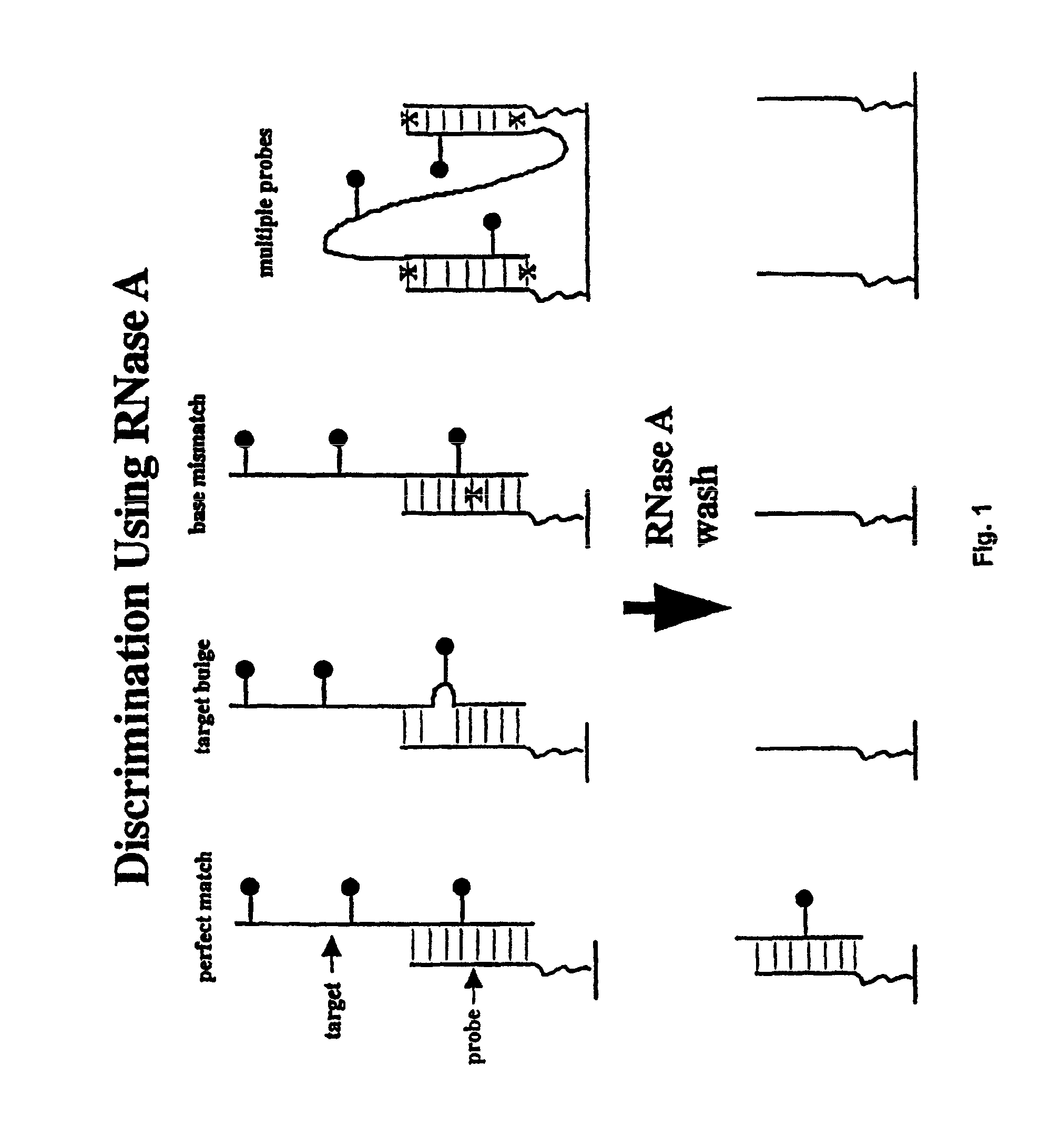
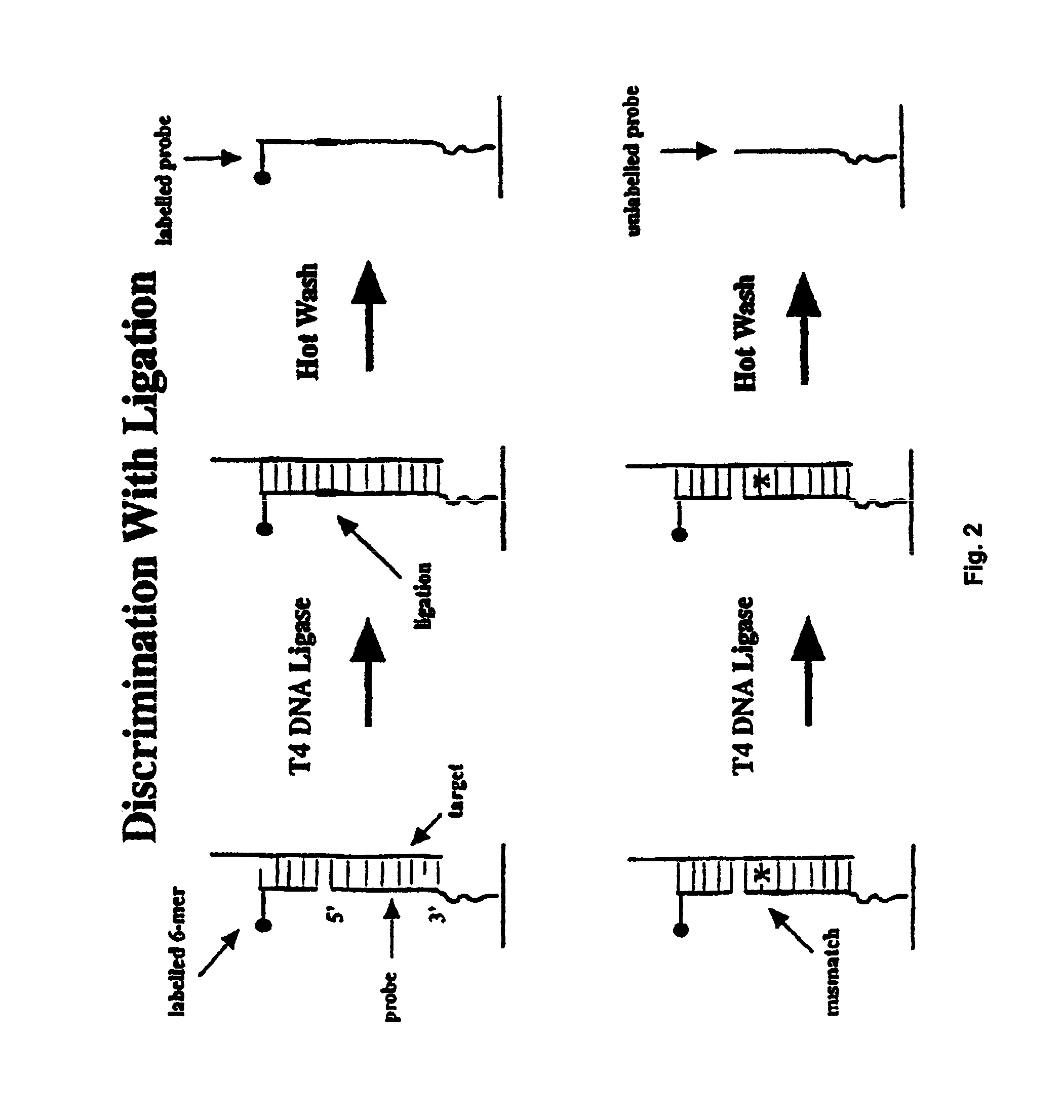
InactiveUS6974666B1Quality improvementSure easySequential/parallel process reactionsSugar derivativesScreening proceduresSignal on
Methods for discriminating between fully complementary hybrids and those that differ by one or more base pairs and libraries of unimolecular, double-stranded oligonucleotides on a solid support. In one embodiment, the present invention provides methods of using nuclease treatment to improve the quality of hybridization signals on high density oligonucleotide arrays. In another embodiment, the present invention provides methods of using ligation reactions to improve the quality of hybridization signals on high density oligonucleotide arrays. In yet another embodiment, the present invention provides libraries of unimolecular or intermolecular, double-stranded oligonucleotides on a solid support. These libraries are useful in pharmaceutical discovery for the screening of numerous biological samples for specific interactions between the double-stranded oligonucleotides, and peptides, proteins, drugs and RNA. In a related aspect, the present invention provides libraries of conformationally restricted probes on a solid support. The probes are restricted in their movement and flexibility using double-stranded oligonucleotides as scaffolding. The probes are also useful in various screening procedures associated with drug discovery and diagnosis. The present invention further provides methods for the preparation and screening of the above libraries.
Owner:AFFYMETRIX INC
Novel photolabile protective groups for improved processes to prepare oligonucleotide arrays
ActiveUS20100292458A1Easy to handleEasy to storeEsterified saccharide compoundsSugar derivativesArylNucleotide
Owner:NIGU CHEM
Ratio-based oligonucleotide probe selection
InactiveUS7047141B2Accurately indicatedBioreactor/fermenter combinationsBiological substance pretreatmentsMethod selectionNucleic acid sequencing
Disclosed herein are methods of selecting probes to target nucleic acid sequences, methods of making oligonucleotide arrays comprising such probes, and methods of using such arrays. Also, described herein are oligonucleotide arrays comprising probes selected by a method of the invention.
Owner:GE HEALTHCARE BIO SCI CORP
Oligonucleotide arrays to monitor gene expression and methods for making and using same
InactiveUS20060010513A1Microbiological testing/measurementOther foreign material introduction processesAntigenCell phenotype
The present invention provides an oligonucleotide array capable of identifying genes and related pathways involved with the induction of a particular phenotype by a cell line, e.g., the genes and related pathways involved with the induction of transgene expression by the cell line. The invention is particularly useful when there is little or no information about the genome of the cell line being studied, because it provides methods for identifying consensus sequences for known and previously undiscovered genes, and for designing oligonucleotide probes to the identified consensus sequences. Additionally, when the array is to be used to determine optimal conditions for expression of a transgene by the cell line, the invention teaches methods of including oligonucleotide probes to transgene sequences in the array. The invention also provides methods of using the array to identify genes and related pathways involved with the induction of a particular cell line phenotype. The invention also provides novel polynucleotides of undiscovered genes (i.e., a gene that had not been sequenced and / or shown to be expressed by CHO cells) and novel polynucleotides involved with the induction of a particular cell phenotype, e.g., increased survival when grown under stressful culture conditions, increased transgene expression, decreased production of an antigen, etc. These novel polynucleotides are termed novel CHO sequences and differential CHO sequences, respectively. The invention also provides genetically engineered expression vectors, host cells, and transgenic animals comprising the novel nucleic acid molecules of the invention. The invention additionally provides antisense and RNAi molecules to the nucleic acid molecules of the invention. The invention further provides methods of using the polynucleotides of the invention.
Owner:WYETH LLC
Mucosal gene signatures
InactiveUS20110059445A1Inhibit expressionInhibitory activityMicrobiological testing/measurementImmunoglobulinsMucosal BiopsyUlcerative colitis
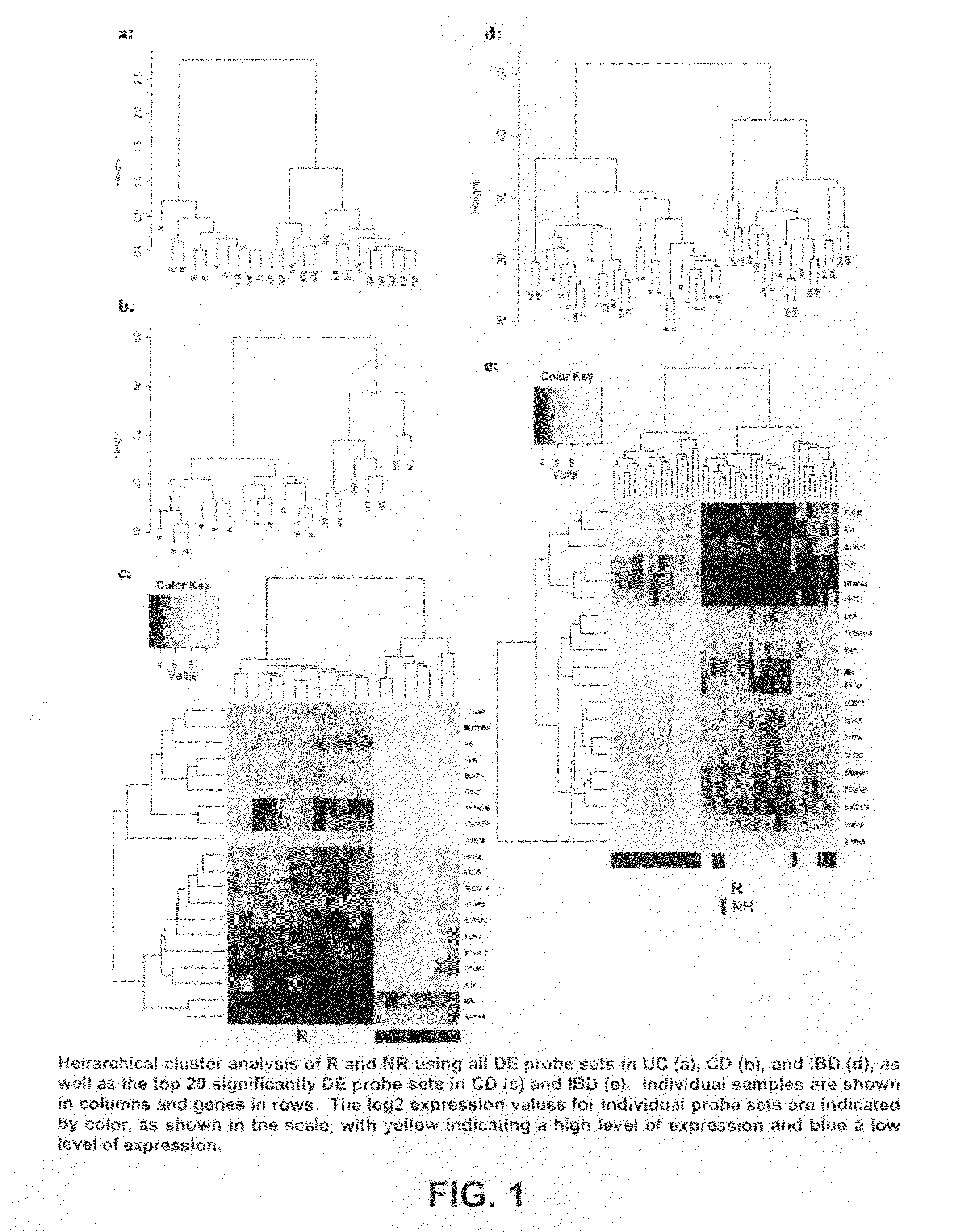
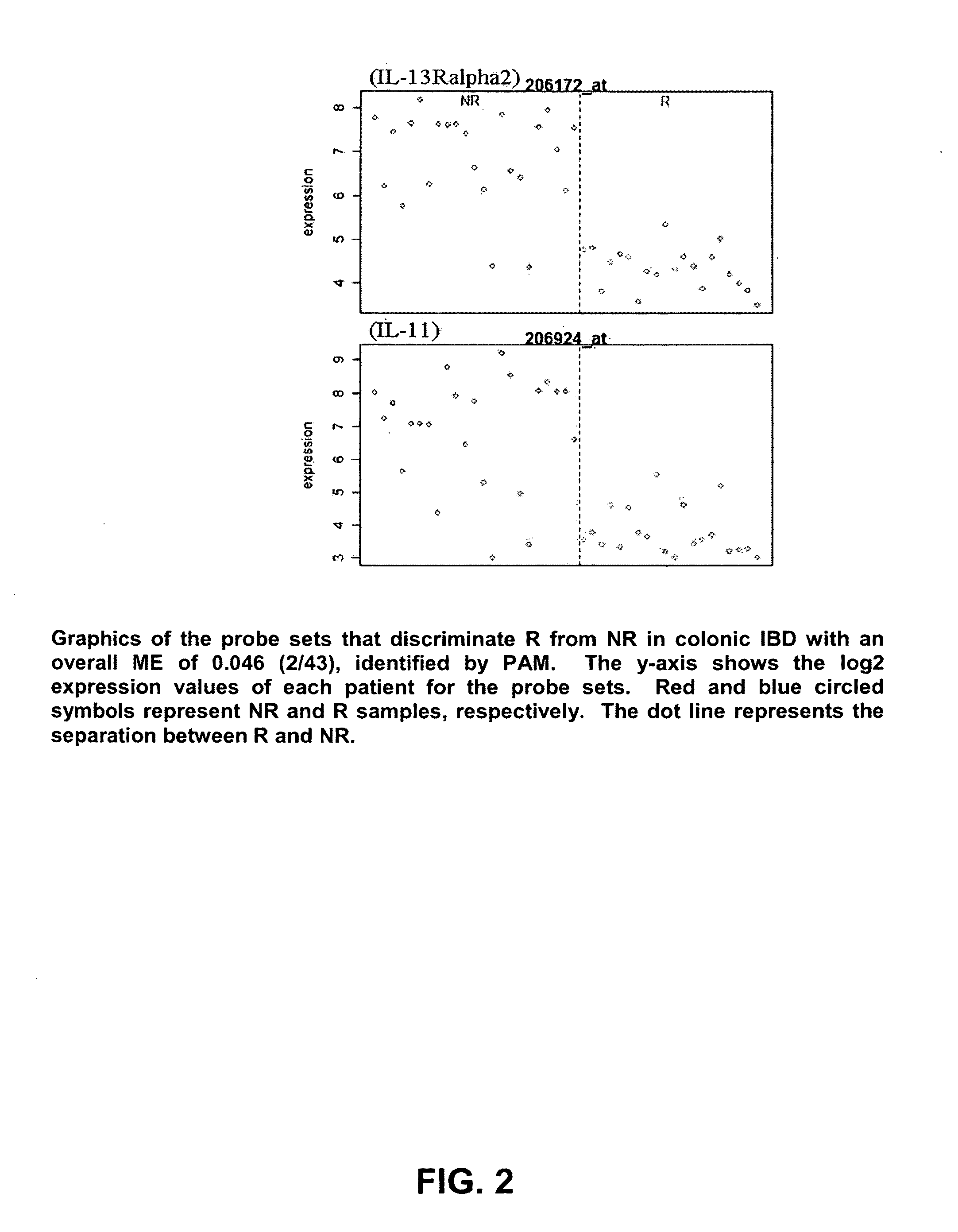
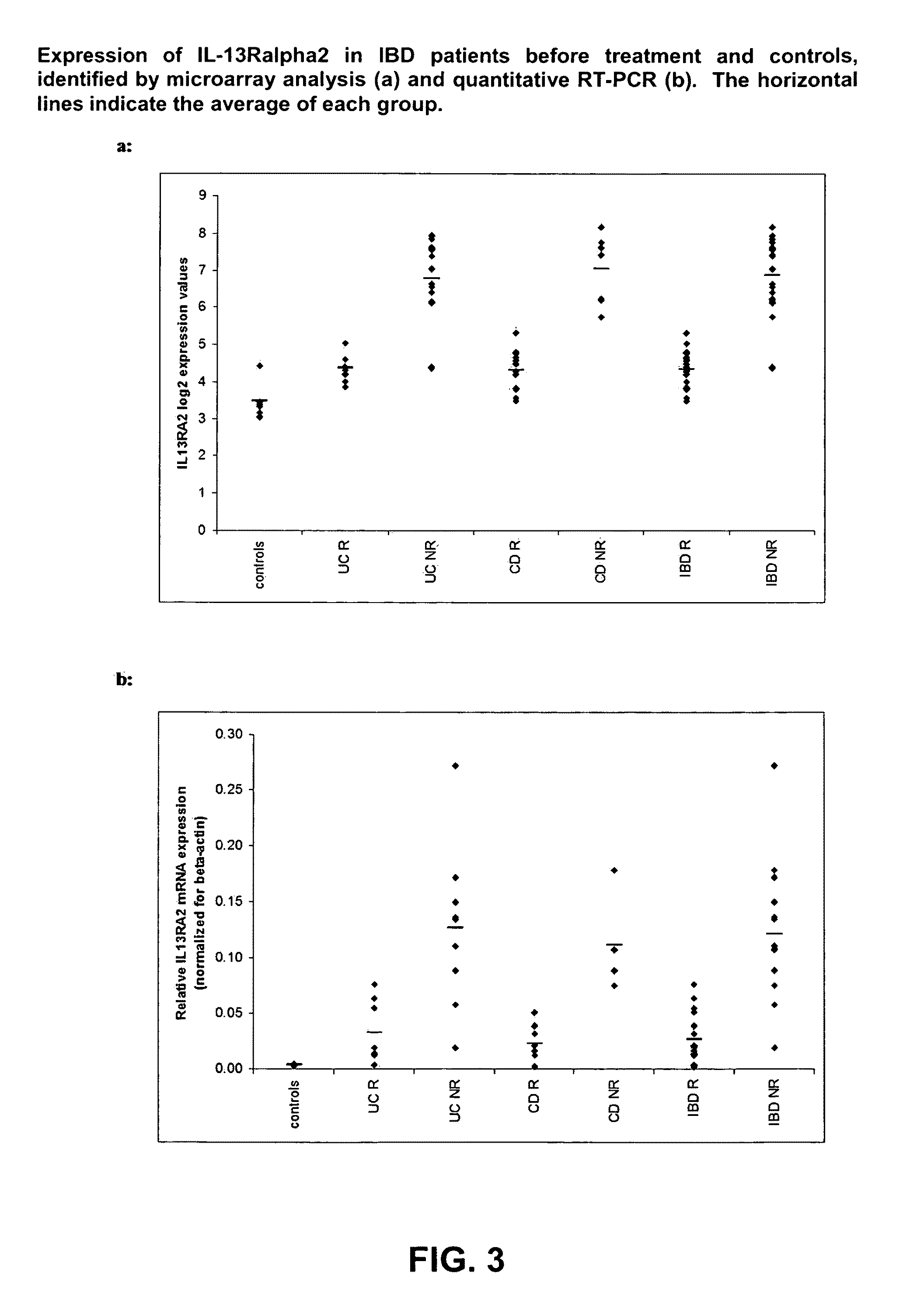
Infliximab (IFX) is an effective treatment for Crohn's disease (CD) and ulcerative colitis (UC) not responding to standard therapy. Thirty percent to forty percent of patients however do not improve and the response is often incomplete. We identified mucosal gene signatures predictive of response to EFX using high-density oligonucleotide arrays. Eight UC patients and twelve CD patients showed healing. In UC, only one probe set was differentially expressed in responders compared with non-responders, i.e., IL-13R(alpha)2. At PAM analysis, two probe sets, representing IL-13Ralpha2 and IL-I 1, separated IBD responders from non-responders with an overall misclassification error rate of 0.046 (2 / 43), with 100% sensitivity and 91.3% specificity. The IL-13R(alpha)2 probe set was a top-ranked probe set in all our analyses using both LIMMA and PAM strategies. Our gene array studies of mucosal biopsies identified IL-13R(alpha)2 in IBD as a predictor of response or non-response to IFX.
Owner:KATHOLIEKE UNIV LEUVEN
Method for detecting cytosine methylations
InactiveUS8241855B2Easy to detectMicrobiological testing/measurementMaterial analysis by electric/magnetic meansGenomic DNAPolymerase L
A method is described for the detection of 5-methylcytosine in genomic DNA samples. First, a genomic DNA from a DNA sample is chemically converted with a reagent, whereby 5-methylcytosine and cytosine react differently. Then the pretreated DNA is amplified with the use of a polymerase with primers of different sequence. In the next step, the amplified genomic DNA is hybridized to an oligonucleotide array and PCR products are obtained, which must be provided with a label. Alternatively, the PCR products can be extended in a primer extension reaction, wherein the extension products are also provided with a label. In the last step, the extended oligonucleotides are investigated for the presence of the label.
Owner:QIAGEN GMBH
Oligonucleotide arrays for high resolution HLA typing
Owner:FRED HUTCHINSON CANCER RES CENT +1
Controlling use of oligonucleotide sequences released from arrays
InactiveUS20070037175A1Bioreactor/fermenter combinationsSequential/parallel process reactionsNucleotideEnzyme
A method for controlling the use of oligonucleotide sequences released from arrays, comprises synthesizing a chemical array of oligonucleotides on a substrate under conditions for producing an array of cleavable oligonucleotides that are blocked from enzymatic reactions after cleavage. Methods may also include receiving a chemical array of cleavable oligonucleotides on a substrate, and cleaving the oligonucleotides from the array, wherein the oligonucleotides are blocked from enzymatic reactions after cleavage. Arrays, populations of oligonucleotides and kits are also provided to facilitate the methods.
Owner:AGILENT TECH INC
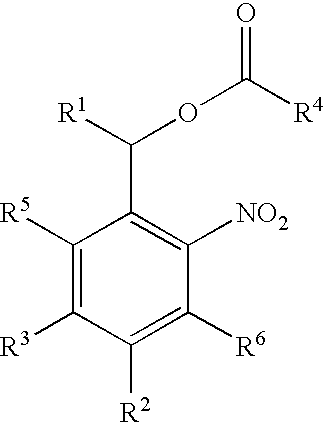
Photolabile protective groups for improved processes to prepare oligonucleotide arrays
InactiveUS7759513B2Reduced deprotection timeImprove cycle efficiencyEsterified saccharide compoundsSugar derivativesArylCombinatorial chemistry
The present invention discloses novel and improved nucleosidic and nucleotidic compounds that are useful in the light-directed synthesis of oligonucleotides, as well as, methods and reagents for their preparation. These compounds are characterized by novel photolabile protective groups that are attached to either the 5′- or the 3′-hydroxyl group of a nucleoside moiety. The photolabile protective group is comprised of a 2-(2-nitrophenyl)-ethyoxycarbonyl skeleton with at least one substituent on the aromatic ring that is either an aryl, an aroyl, a heteroaryl or an alkoxycarbonyl group. The present invention includes the use of the aforementioned compounds in light-directed oligonucleotide synthesis, the respective assembly of nucleic acid microarrays and their application.
Owner:NIGU CHEM
Methods for determining the genetic affinity of microorganisms and viruses
InactiveUS8214153B1Particle separator tubesMicrobiological testing/measurementBacteroidesIsotopic labeling
Selecting which sub-sequences in a database of nucleic acid such as 16S rRNA are highly characteristic of particular groupings of bacteria, microorganisms, fungi, etc. on a substantially phylogenetic tree. Also applicable to viruses comprising viral genomic RNA or DNA. A catalogue of highly characteristic sequences identified by this method is assembled to establish the genetic identity of an unknown organism. The characteristic sequences are used to design nucleic acid hybridization probes that include the characteristic sequence or its complement, or are derived from one or more characteristic sequences. A plurality of these characteristic sequences is used in hybridization to determine the phylogenetic tree position of the organism(s) in a sample. Those target organisms represented in the original sequence database and sufficient characteristic sequences can identify to the species or subspecies level. Oligonucleotide arrays of many probes are especially preferred. A hybridization signal can comprise fluorescence, chemiluminescence, or isotopic labeling, etc.; or sequences in a sample can be detected by direct means, e.g. mass spectrometry. The method's characteristic sequences can also be used to design specific PCR primers. The method uniquely identifies the phylogenetic affinity of an unknown organism without requiring prior knowledge of what is present in the sample. Even if the organism has not been previously encountered, the method still provides useful information about which phylogenetic tree bifurcation nodes encompass the organism.
Owner:TECH LICENSING CO LLC
Photolabile esters and their uses
InactiveUS7301049B2Satisfactory coefficientEfficient photolysisMaterial nanotechnologyBiocideNucleotideMedicinal chemistry
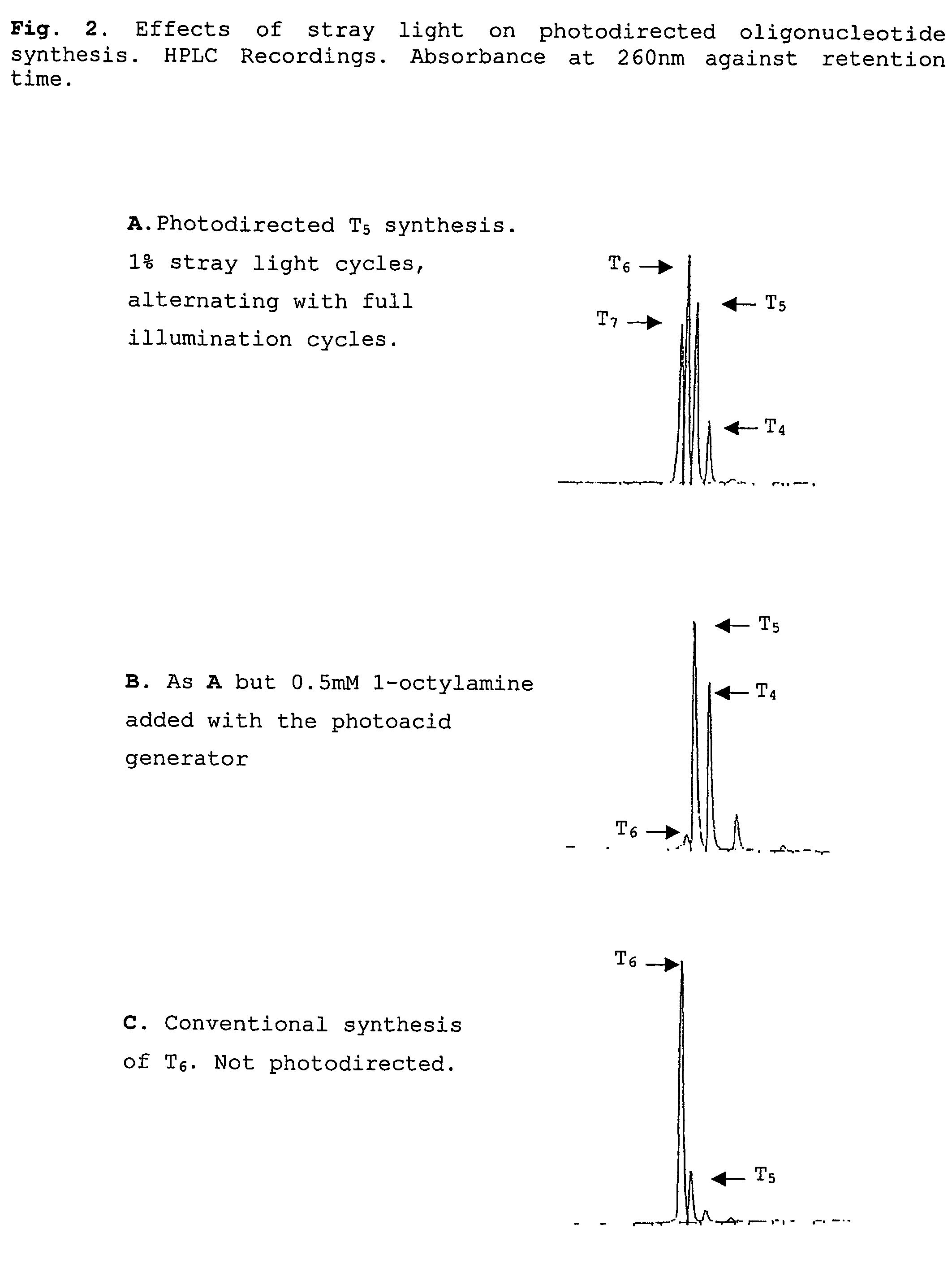
Compounds which are capable of generating acid on photolysis are disclosed, and the uses of these compounds, especially for deprotecting the termini of nucleic acid molecules or peptides during synthesis of arrays. The compounds described herein may be employed in the detritylation of 5′-O-dimethoxytrityl (DMT) protected nucleotides by photolysing the compounds to generate an acid capable of removing the DMT group allowing oligonucleotide arrays to be synthesised using readily available 5′-O-DMT-nucleoside-3′-O-phosphoramidite monomers conventionally used in solid phase nucleic acid synthesis. A method of avoiding the effects of stray light in projection lithography techniques is also disclosed.
Owner:THE INST OF CANCER RES
Methods of enzymatic discrimination enhancement and surface bound double-stranded DNA
InactiveUS20060292579A1Quality improvementReadily determined and verifiedSequential/parallel process reactionsSugar derivativesScreening proceduresSignal on
Methods for discriminating between fully complementary hybrids and those that differ by one or more base pairs and libraries of unimolecular, double-stranded oligonucleotides on a solid support. In one embodiment, the present invention provides methods of using nuclease treatment to improve the quality of hybridization signals on high density oligonucleotide arrays. In another embodiment, the present invention provides methods of using ligation reactions to improve the quality of hybridization signals on high density oligonucleotide arrays. In yet another embodiment, the present invention provides libraries of unimolecular or intermolecular, double-stranded oligonucleotides on a solid support. These libraries are useful in pharmaceutical discovery for the screening of numerous biological samples for specific interactions between the double-stranded oligonucleotides, and peptides, proteins, drugs and RNA. In a related aspect, the present invention provides libraries of conformationally restricted probes on a solid support. The probes are restricted in their movement and flexibility using double-stranded oligonucleotides as scaffolding. The probes are also useful in various screening procedures associated with drug discovery and diagnosis. The present invention further provides methods for the preparation and screening of the above libraries.
Owner:AFFYMETRIX INC
Oligonucleotide arrays for high resolution HLA typing
Arrays of HLA Class I oligonucleotide probes on a solid support are provided, wherein the probes are sufficient to represent at least 80% of the known polymorphisms in exons 2 and 3 of the HLA Class I locus.
Owner:FRED HUTCHINSON CANCER RES CENT +1
Genetic markers for tumors
InactiveUS7105293B2Raise the possibilityBioreactor/fermenter combinationsBiological substance pretreatmentsGeneticsOligonucleotide Arrays
Sets of genetic markers for specific tumor classes are described, as well as methods of identifying a biological sample based on these markers. Also described are diagnostic, prognostic, and therapeutic screening uses for these markers, as well as oligonucleotide arrays comprising these markers.
Owner:WHITEHEAD INST FOR BIOMEDICAL RES +1
Methods of enzymatic discrimination enhancement and surface-bound double-stranded DNA
InactiveUS8236493B2Quality improvementReadily determined and verifiedMicrobiological testing/measurementLibrary screeningScreening proceduresSignal on
Methods for discriminating between fully complementary hybrids and those that differ by one or more base pairs and libraries of unimolecular, double-stranded oligonucleotides on a solid support. In one embodiment, the present invention provides methods of using nuclease treatment to improve the quality of hybridization signals on high density oligonucleotide arrays. In another embodiment, the present invention provides methods of using ligation reactions to improve the quality of hybridization signals on high density oligonucleotide arrays. In yet another embodiment, the present invention provides libraries of unimolecular or intermolecular, double-stranded oligonucleotides on a solid support. These libraries are useful in pharmaceutical discovery for the screening of numerous biological samples for specific interactions between the double-stranded oligonucleotides, and peptides, proteins, drugs and RNA. In a related aspect, the present invention provides libraries of conformationally restricted probes on a solid support. The probes are restricted in their movement and flexibility using double-stranded oligonucleotides as scaffolding. The probes are also useful in various screening procedures associated with drug discovery and diagnosis. The present invention further provides methods for the preparation and screening of the above libraries.
Owner:AFFYMETRIX INC
Expression monitoring to high density oligonucleotide arrays
InactiveUS20050202500A1Reduce complexityReduce background signalMicrobiological testing/measurementRecombinant DNA-technologyNucleotideGenetics
This invention provides methods of monitoring the expression levels of a multiplicity of genes. The methods involve hybridizing a nucleic acid sample to a high density array of oligonucleotide probes where the high density array contains oligonucleotide probes complementary to subsequences of target nucleic acids in the nucleic acid sample. In one embodiment, the method involves providing a pool of target nucleic acids comprising RNA transcripts of one or more target genes, or nucleic acids derived from the RNA transcripts, hybridizing said pool of nucleic acids to an array of oligonucleotide probes immobilized on surface, where the array comprising more than 100 different oligonucleotides and each different oligonucleotide is localized in a predetermined region of the surface, the density of the different oligonucleotides is greater than about 60 different oligonucleotides per 1 cm2, and the olignucleotide probes are complementary to the RNA transcripts or nucleic acids derived from the RNA transcripts; and quantifying the hybridized nucleic acids in the array.
Owner:AFFYMETRIX INC
Method and apparatus for providing a bioinformatics database
InactiveUS6882742B2Improve translationCordless telephonesBioreactor/fermenter combinationsBioinformatics databasesComputer science
System and method for organizing information relating to polymer probe array chips including oligonucleotide array chips. A database model is provided which organizes information relating to sample preparation, chip layout, application of samples to chips, scanning of chips, expression analysis of chip results, etc. The model is readily translatable into database languages such as SQL. The database model scales to permit mass processing of probe array chips.
Owner:AFFYMETRIX INC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com