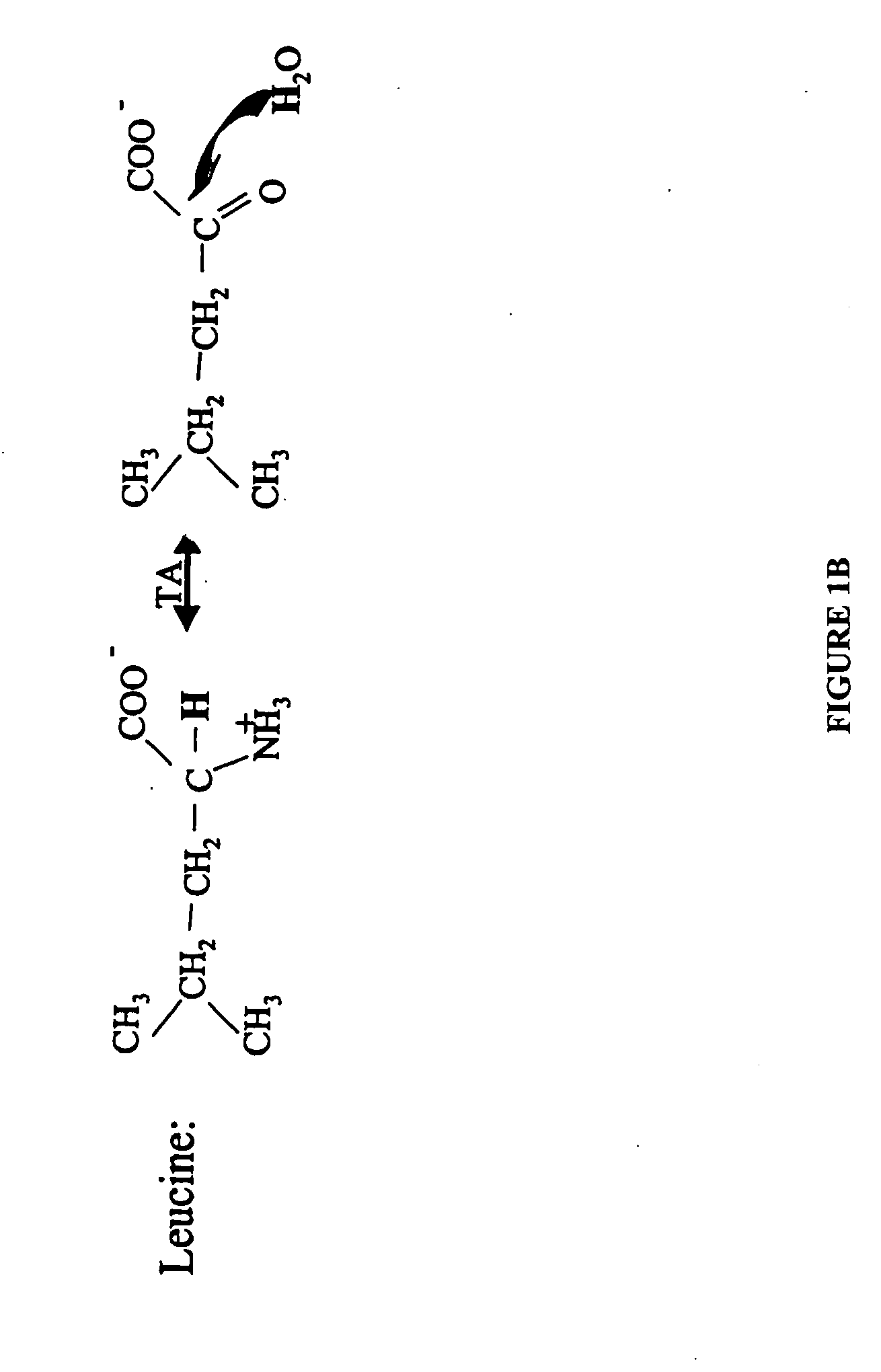
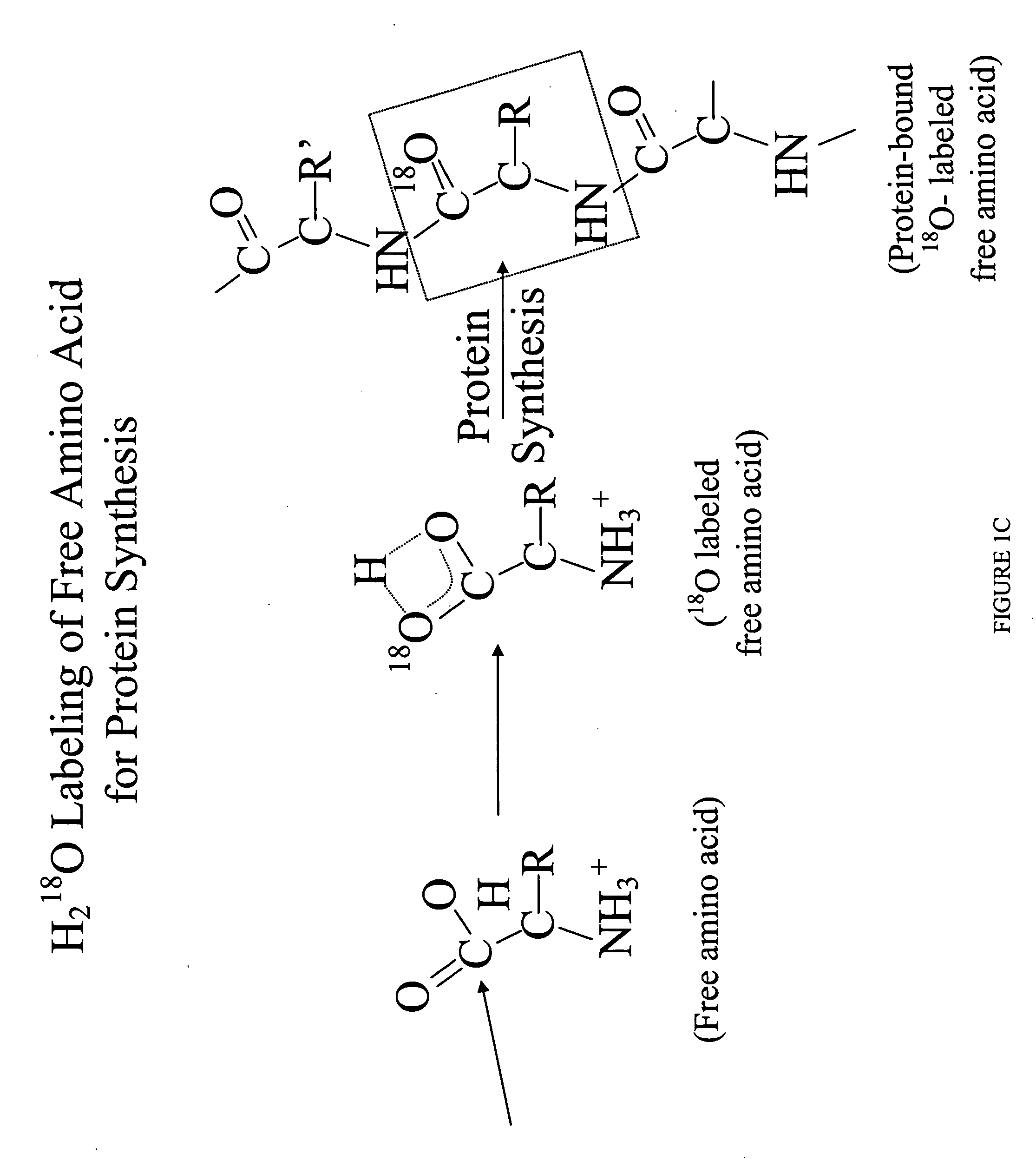
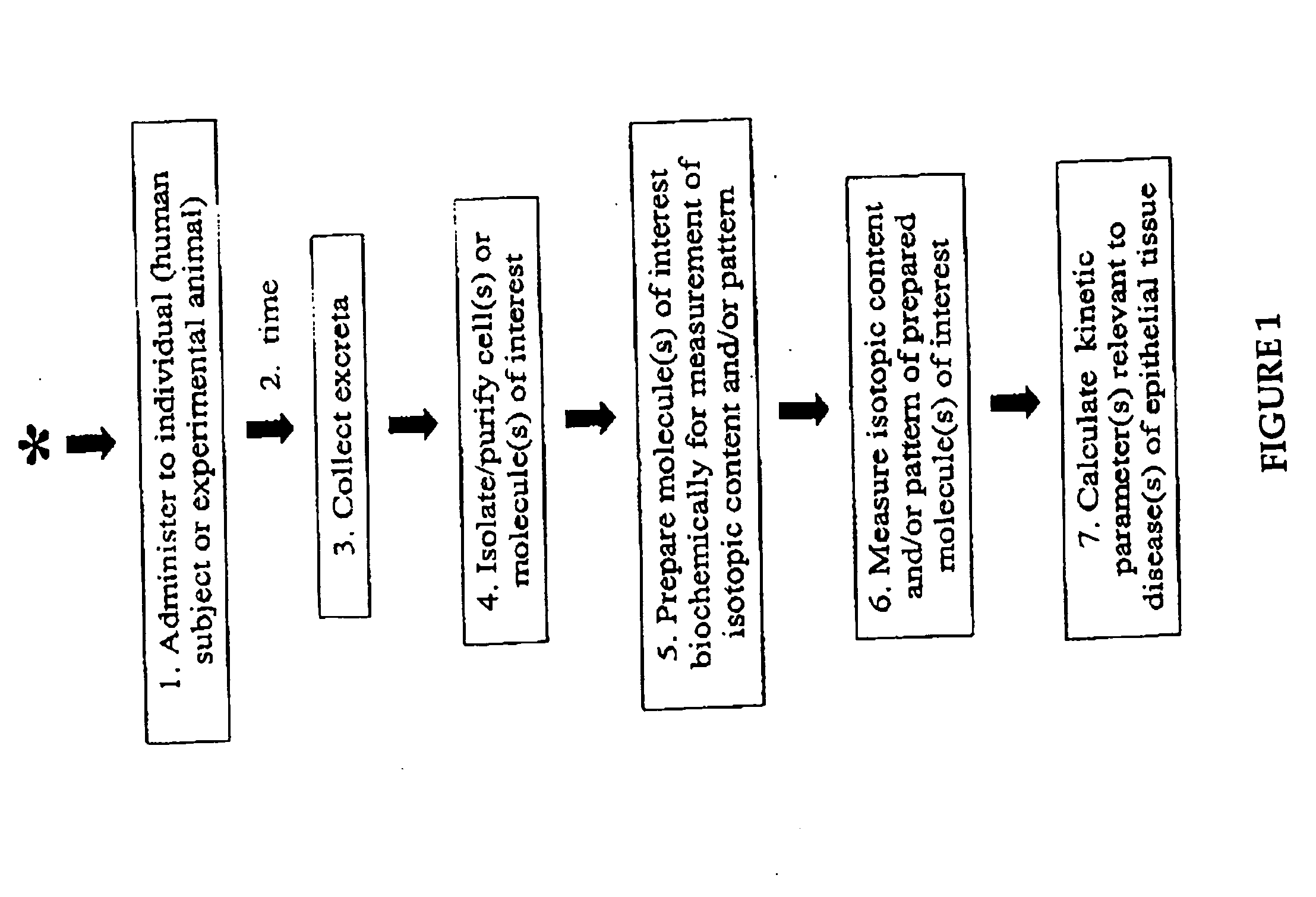
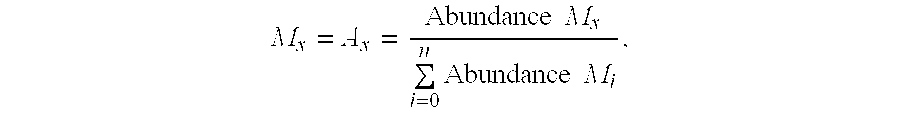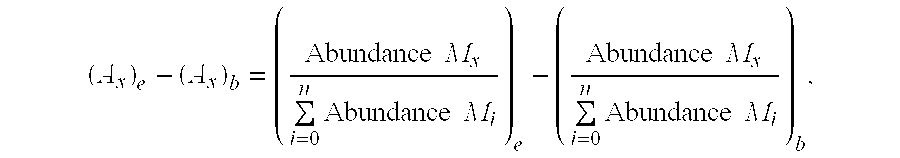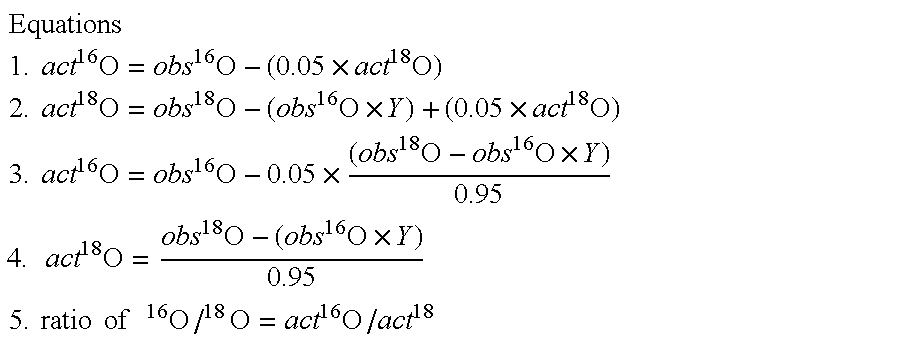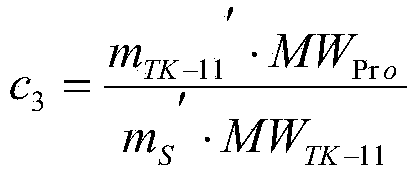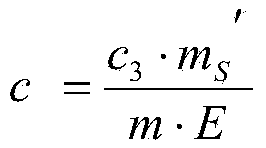Patents
Literature
322 results about "Isotopic labeling" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
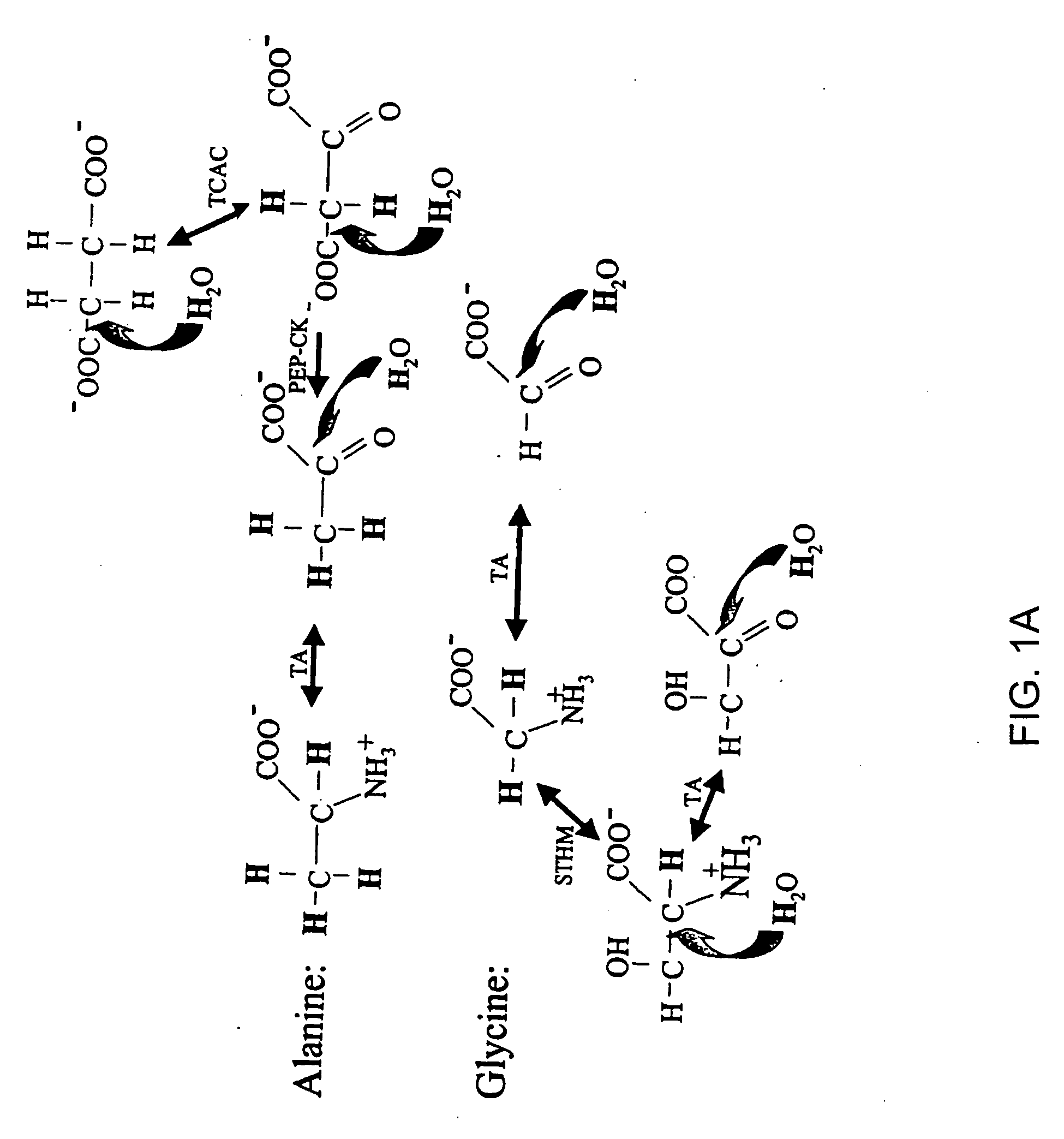
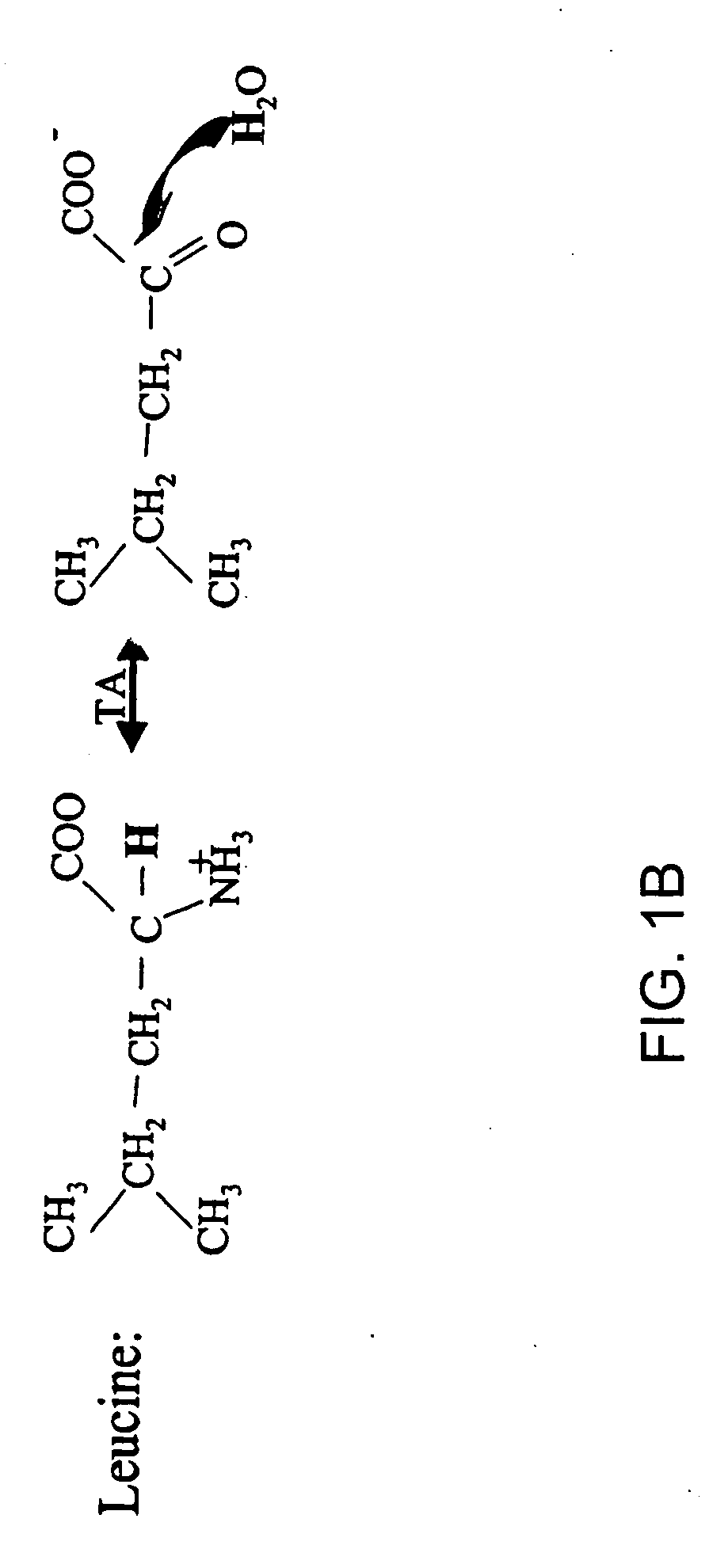
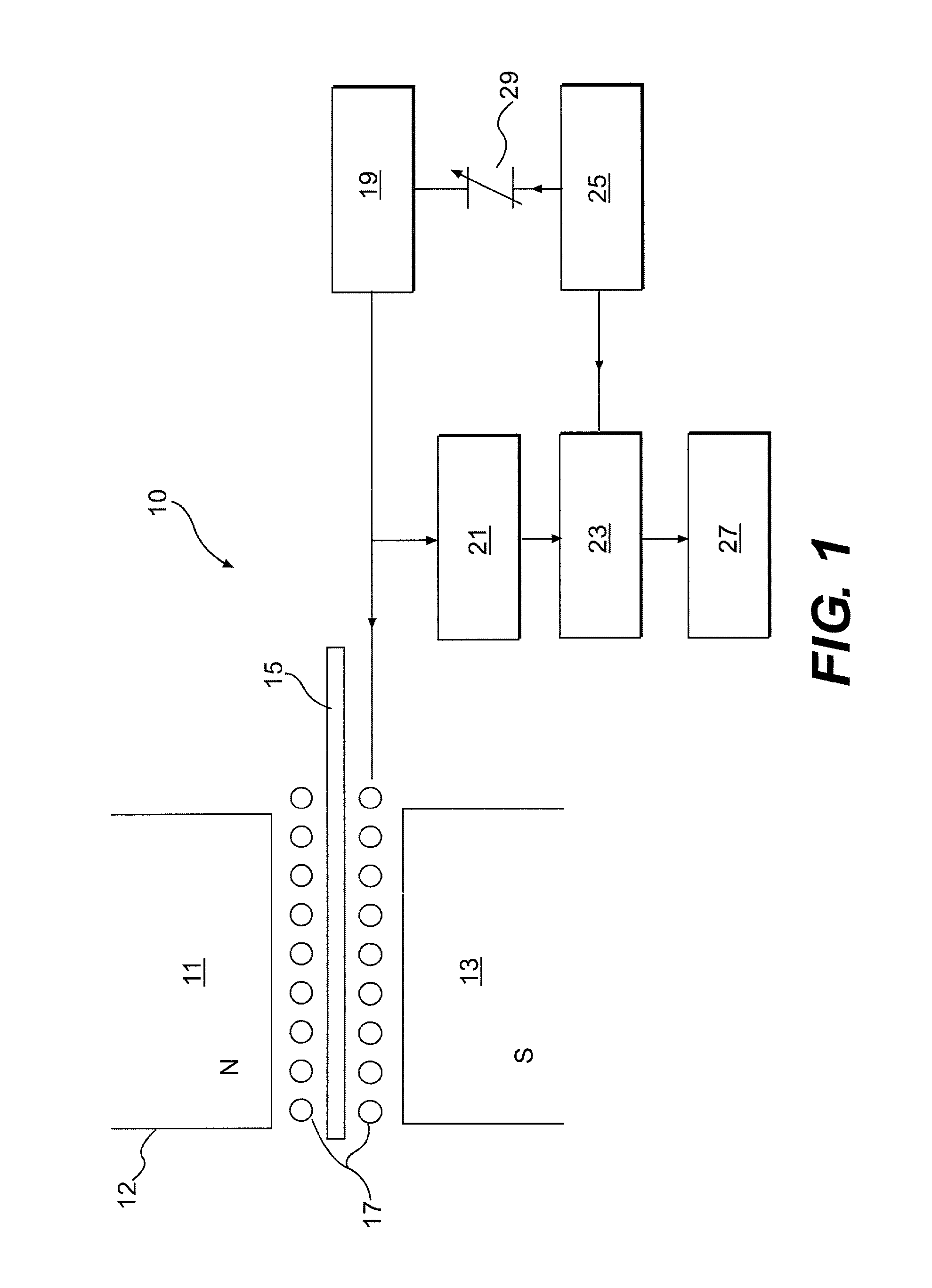
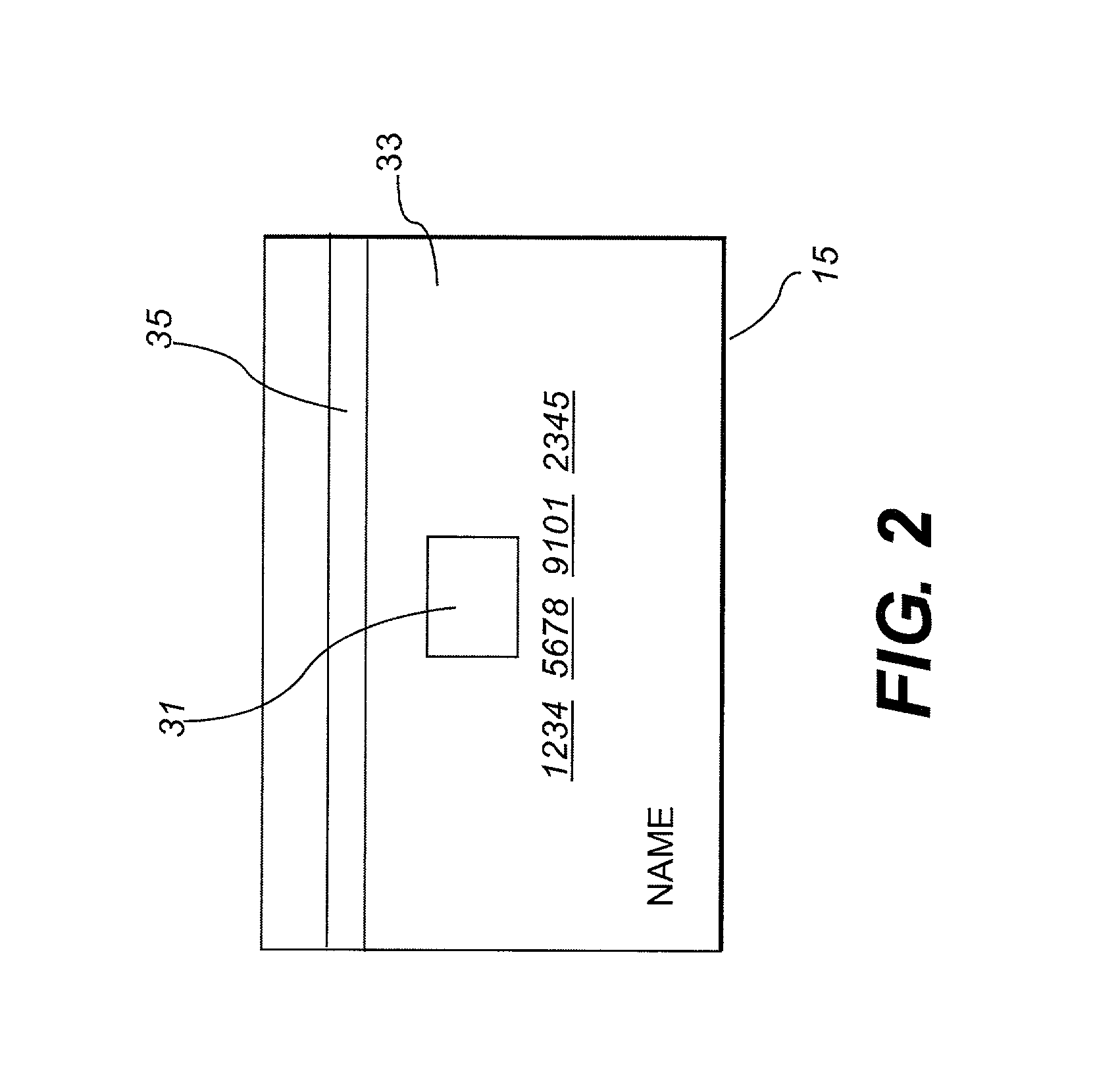
Isotopic labeling (or isotopic labelling) is a technique used to track the passage of an isotope (an atom with a detectable variation in neutron count) through a reaction, metabolic pathway, or cell. The reactant is 'labeled' by replacing specific atoms by their isotope. The reactant is then allowed to undergo the reaction. The position of the isotopes in the products is measured to determine the sequence the isotopic atom followed in the reaction or the cell's metabolic pathway. The nuclides used in isotopic labeling may be stable nuclides or radionuclides. In the latter case, the labeling is called radiolabeling.
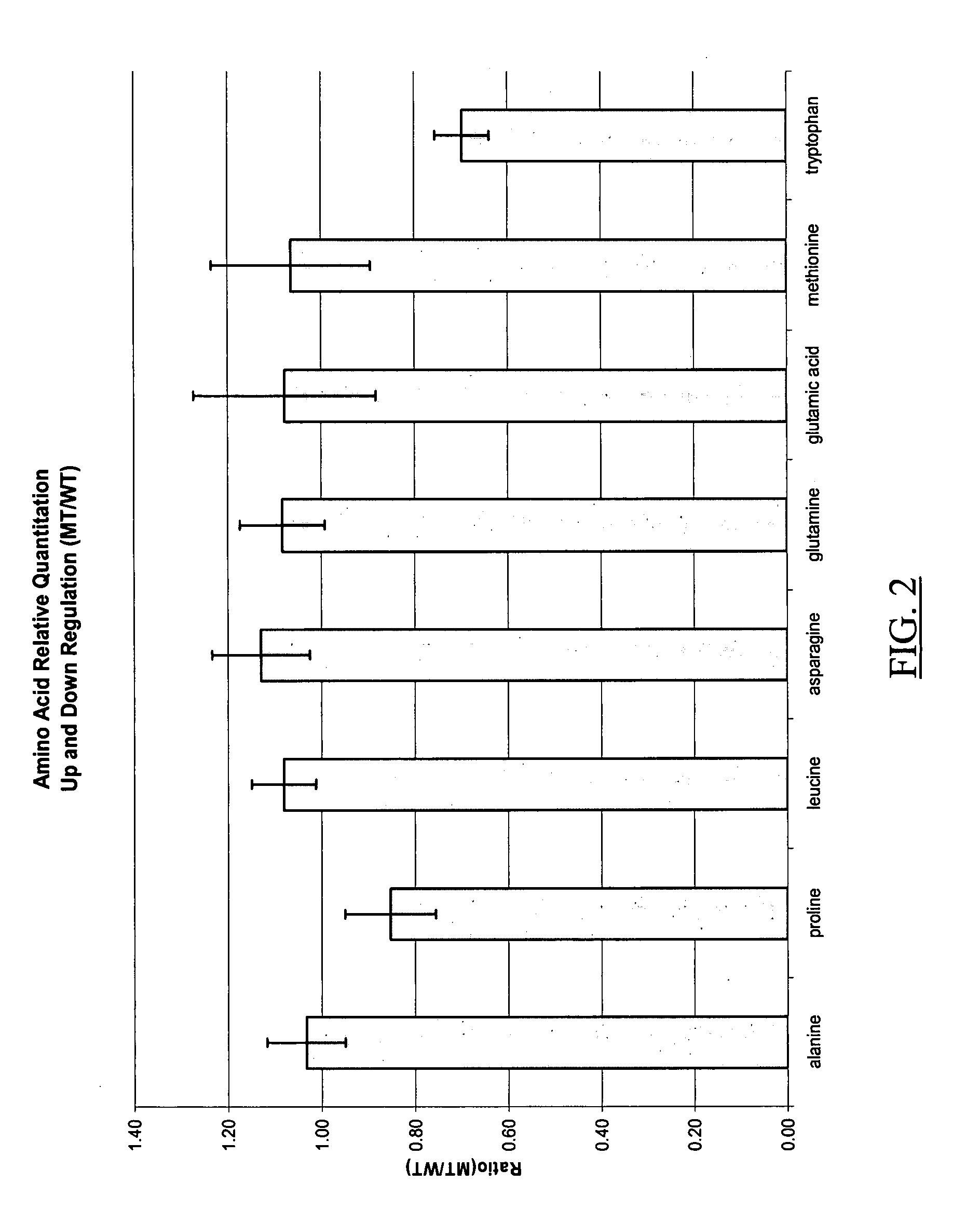
Molecular flux rates through critical pathways measured by stable isotope labeling in vivo, as biomarkers of drug action and disease activity
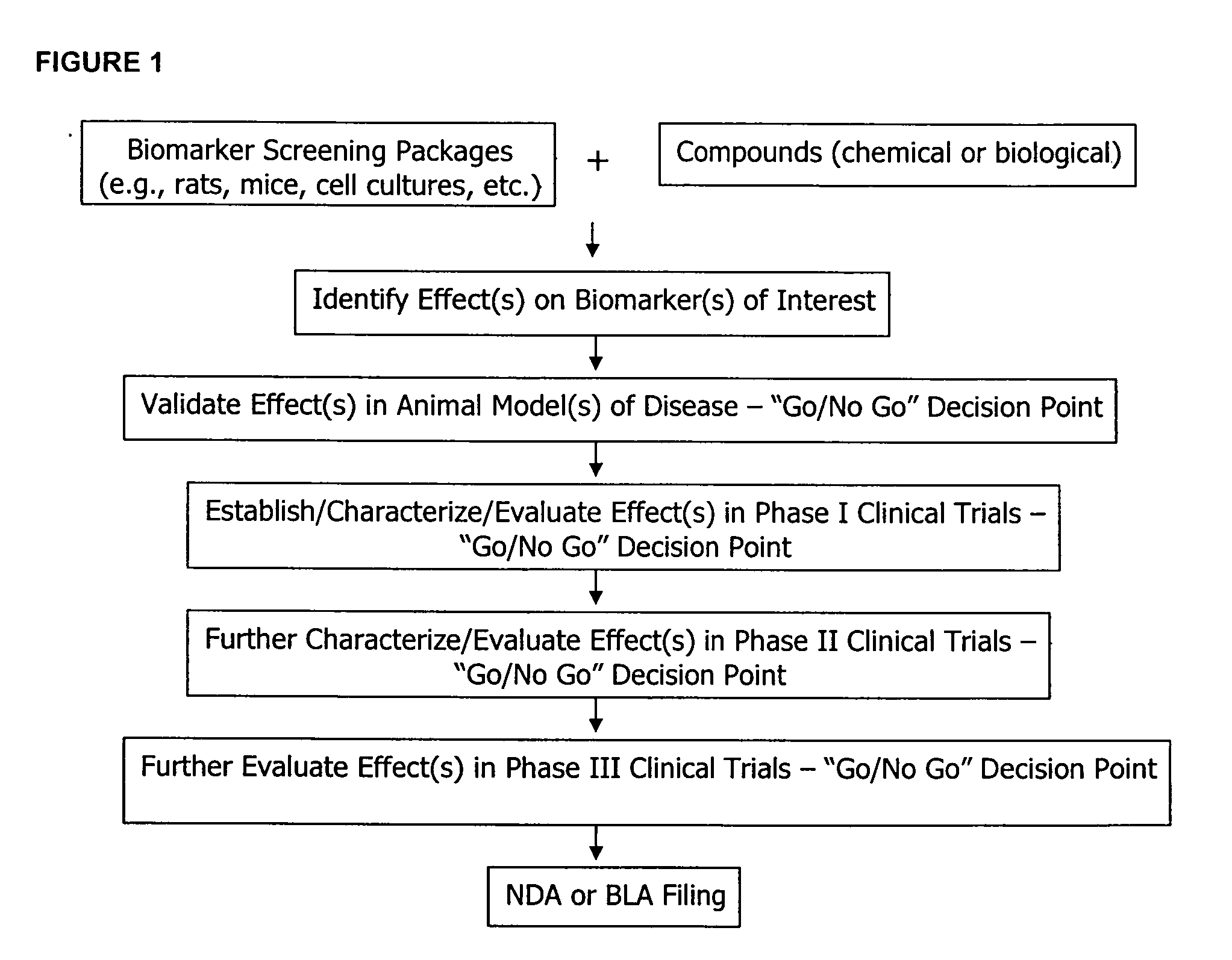
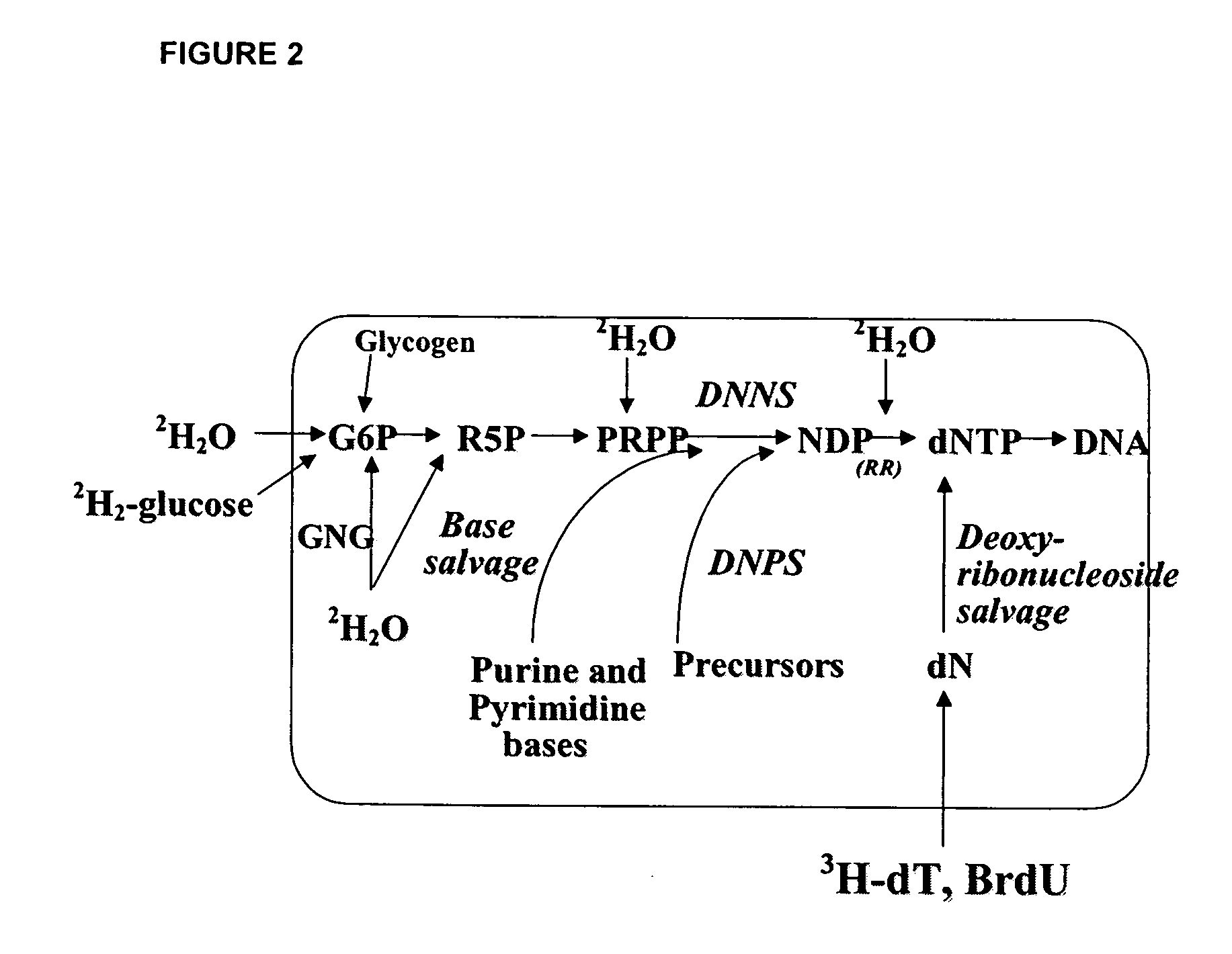
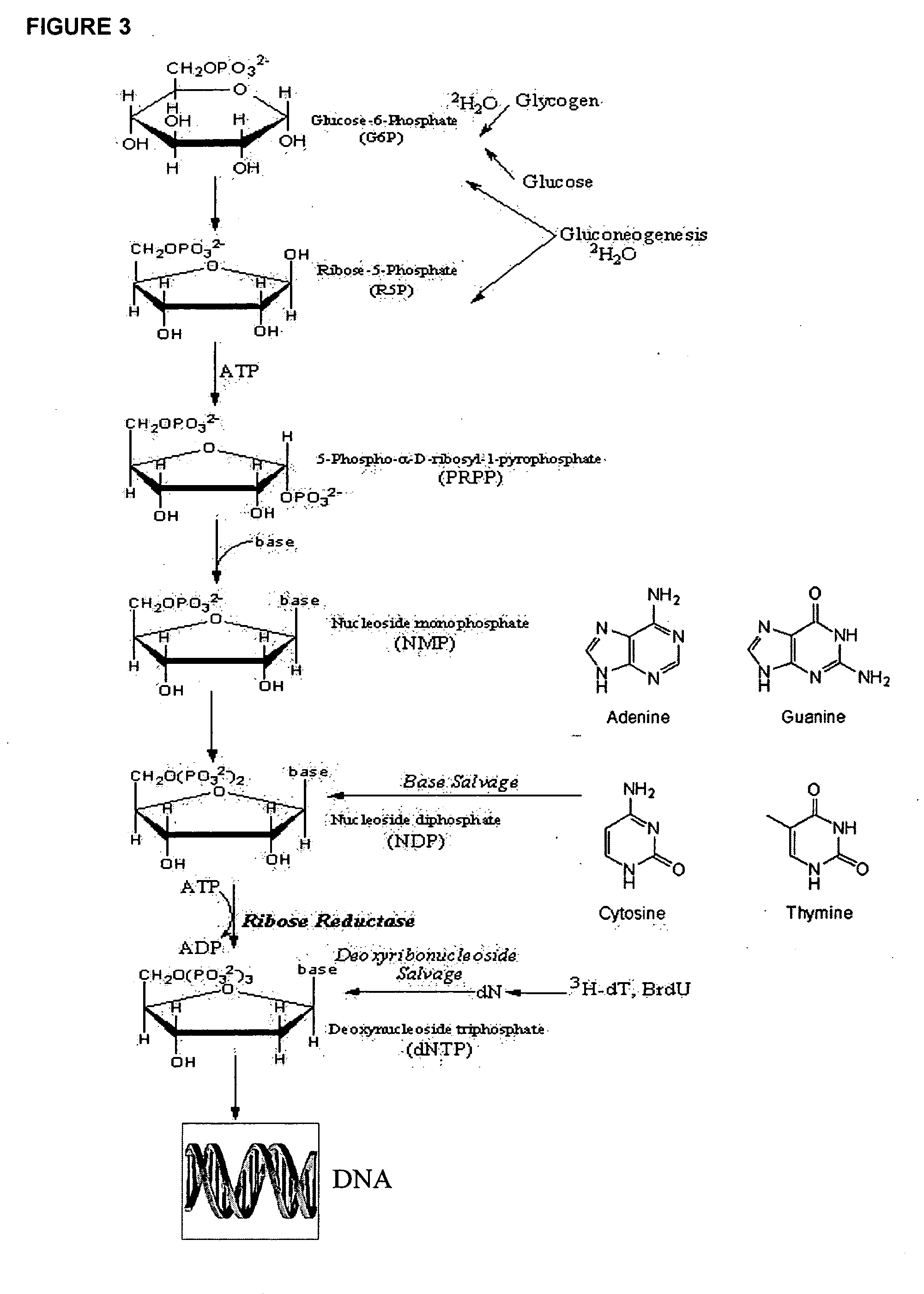
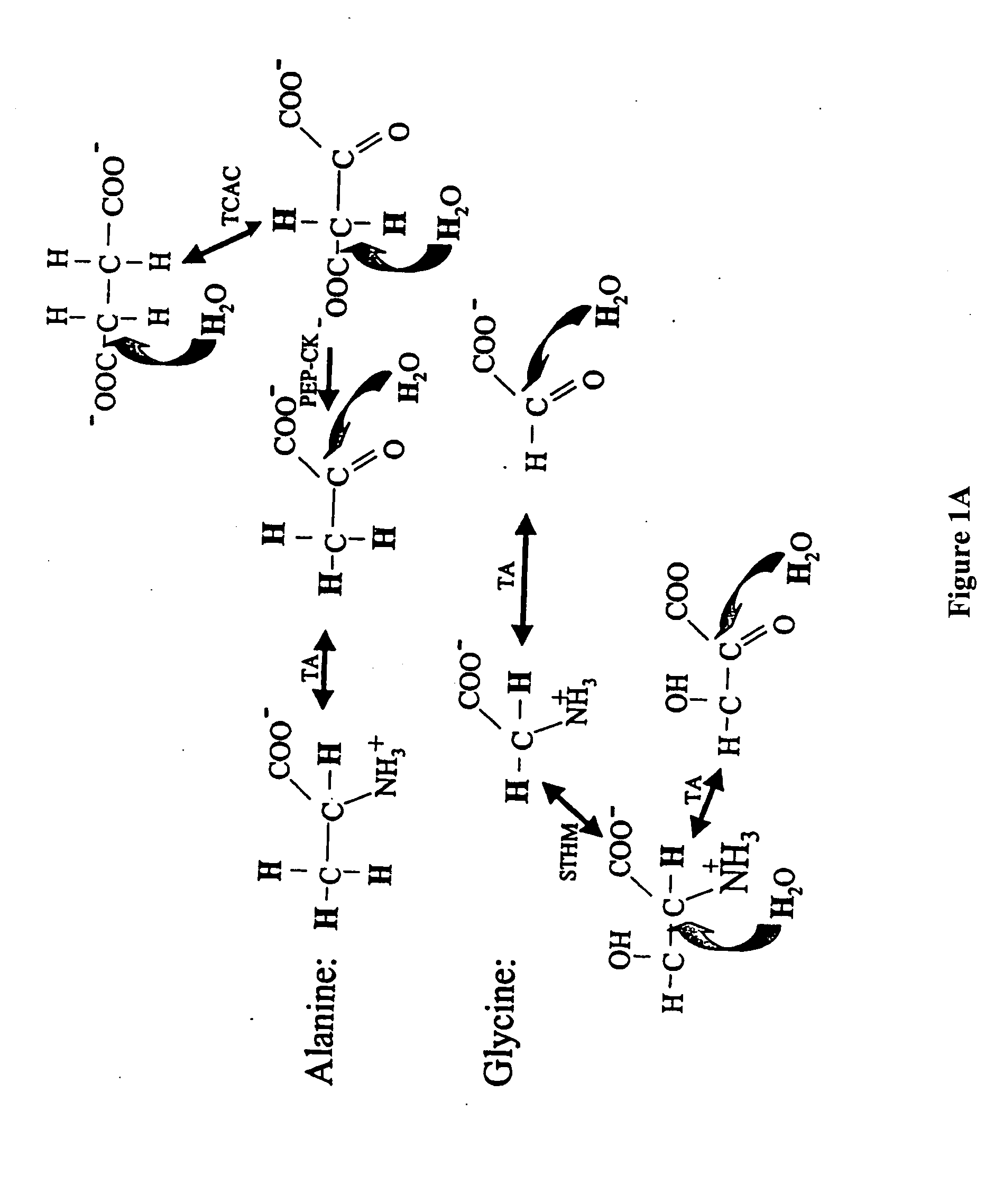
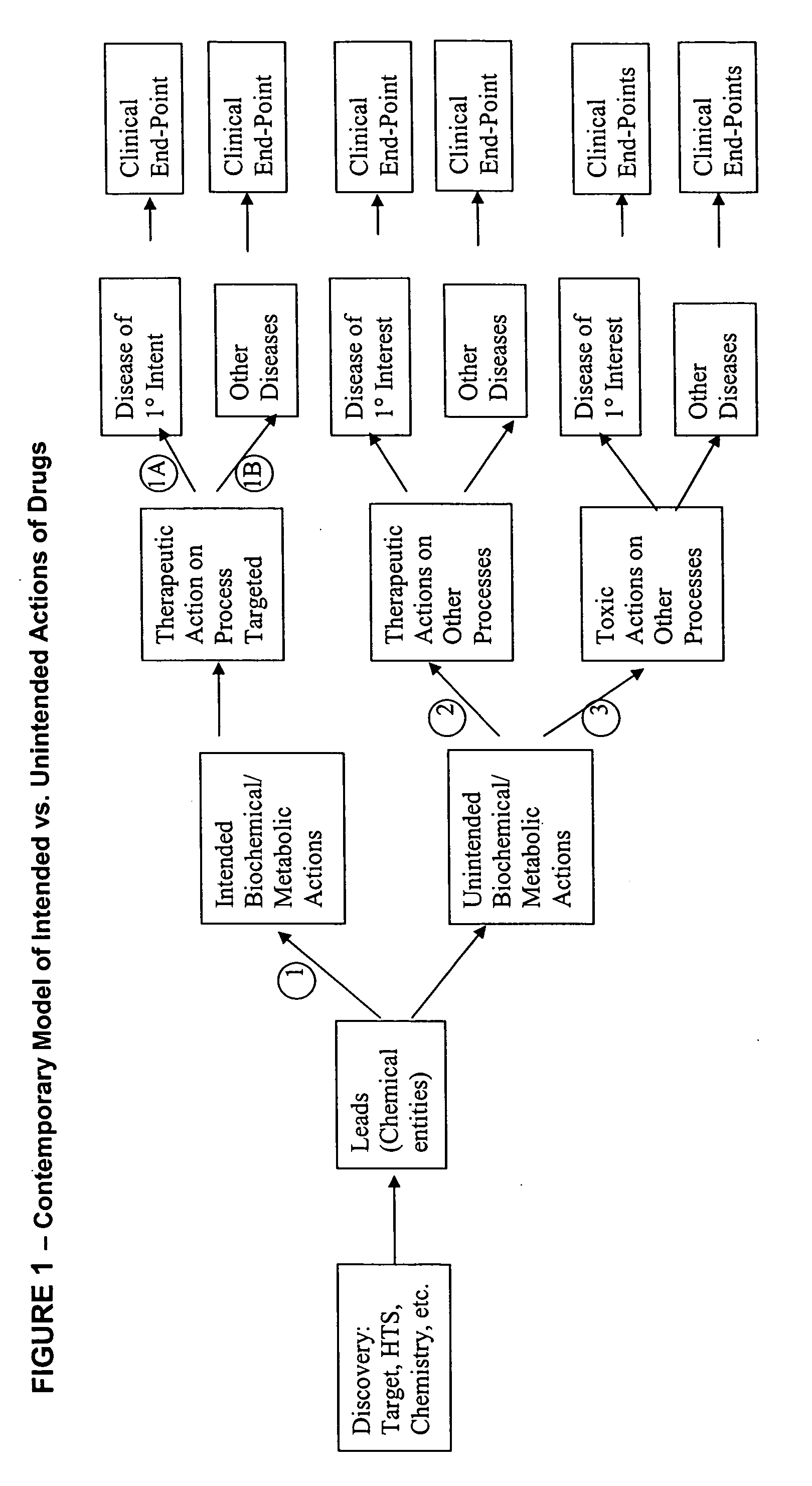
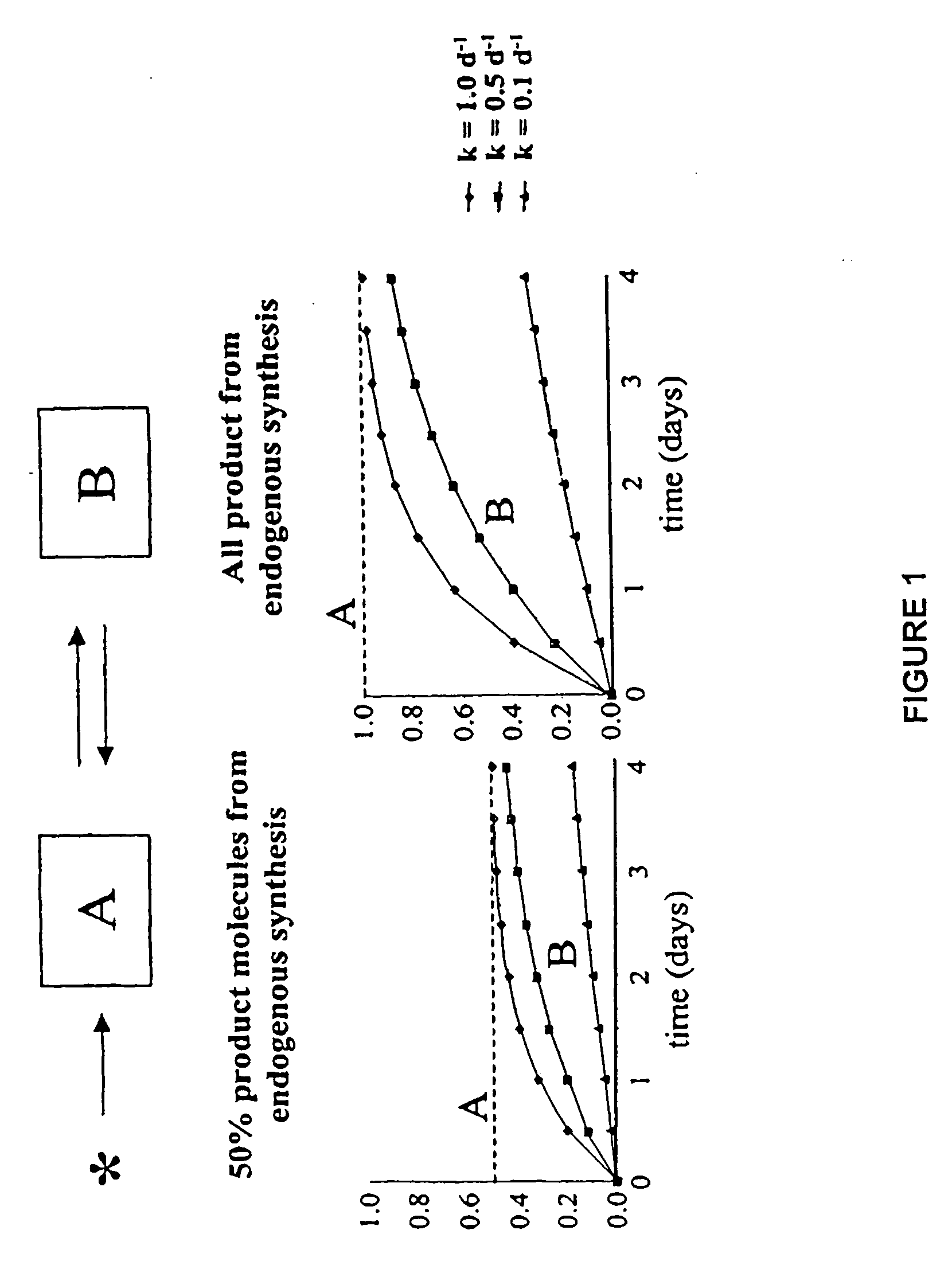
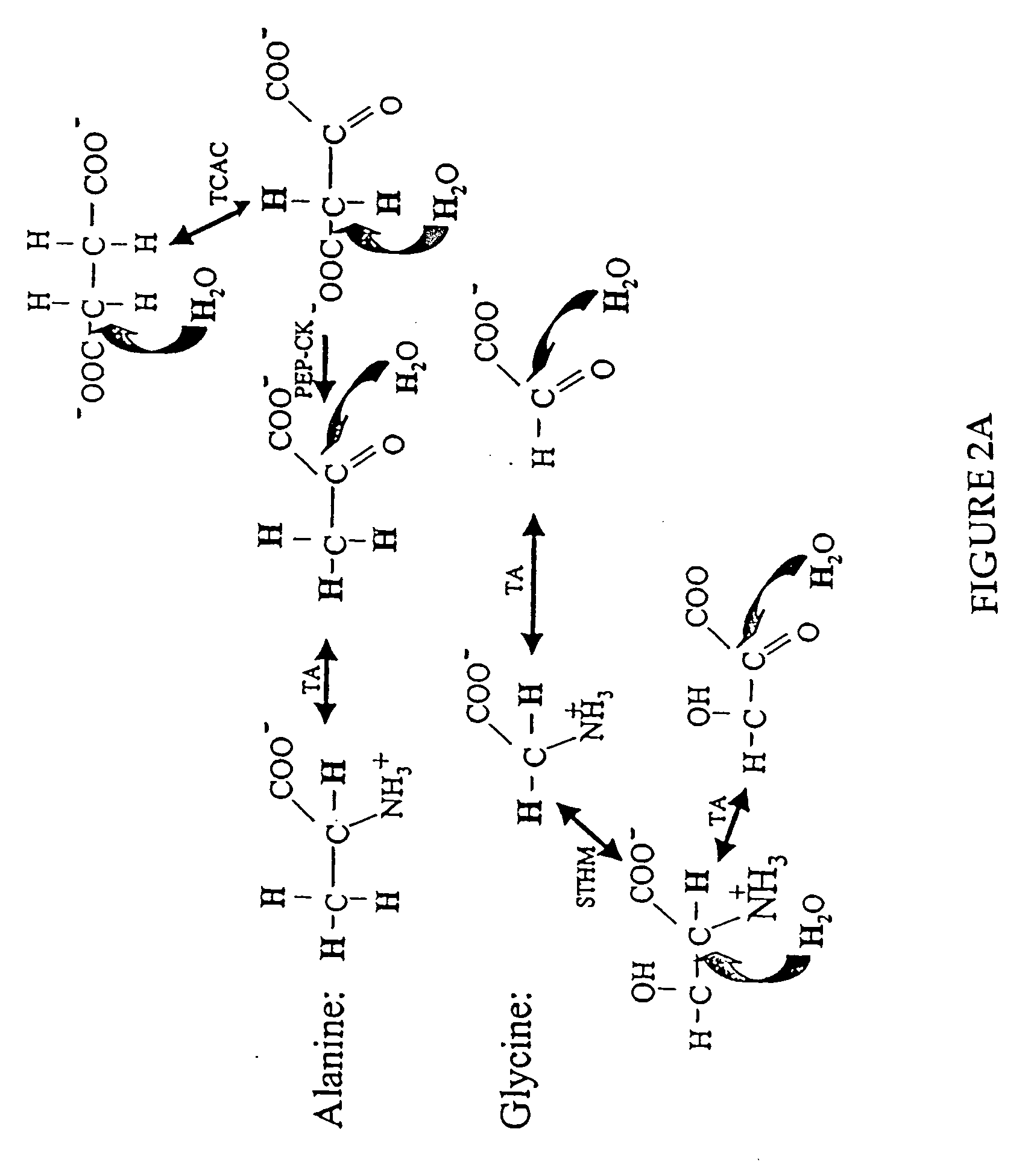
The methods described herein enable the evaluation of compounds on subjects to assess their therapeutic efficacy or toxic effects. The target of analysis is the underlying biochemical process or processes (i.e., metabolic process) thought to be involved in disease pathogenesis. Molecular flux rates within the one or more biochemical processes serve as biomarkers and are quantitated and compared with the molecular flux rates (i.e., biomarker) from control subjects (i.e., subjects not exposed to the compounds). Any change in the biomarker in the subject relative to the biomarker in the control subject provides the necessary information to evaluate therapeutic efficacy of an administered drug or a toxic effect and to develop the compound further if desired. In one aspect of the invention, stable isotope-labeled substrate molecules are administered to a subject and the label is incorporated into targeted molecules in a manner that reveals molecular flux rates through one or more metabolic pathways of interest. By this method, a comparison between subjects and control subjects reveals the effects of the chemical entity or entities on the biomarkers. This, in turn, allows for the identification of potential therapeutic uses or toxicities of the compound. Combinations of compounds can also be systematically evaluated for complementary, synergistic, or antagonistic actions on the metabolic pathways of interest, using the methods of the present invention as a strategy for identifying and confirming novel therapeutic or toxic combinations of compounds.
Owner:RGT UNIV OF CALIFORNIA
Methods for comparing relative flux rates of two or more biological molecules in vivo through a single protocol
ActiveUS20060204439A1Ease of of half-lifeEase of of levelsIn-vivo radioactive preparationsSugar derivativesDiseaseBio molecules
The invention relates to techniques for measuring and comparing relative molecular flux rates of different biological molecules by administering isotope-labeled water to one or more tissues or individuals and comparing the molecular flux rates of two or more biological molecules, including biological molecules in different chemical classes. The methods find use in several applications including diagnosing, prognosing, or monitoring a disease, disorder, or condition, the in vivo high-throughput screening of chemical entities and biological factors for therapeutic effects in various disease models, and the in vivo high-throughput screening of chemical entities and biological factors for toxic effects.
Owner:RGT UNIV OF CALIFORNIA
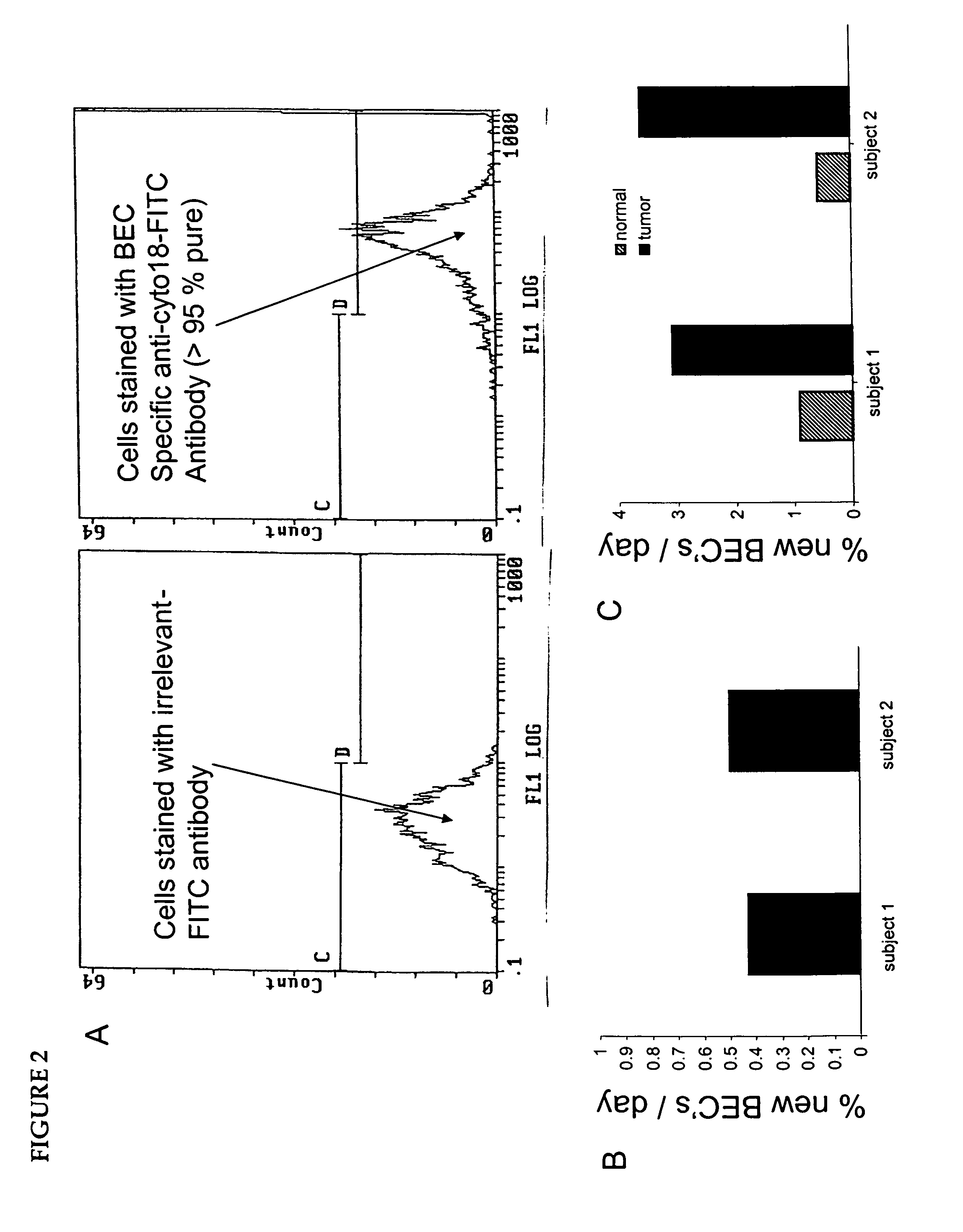
Isolation of epithelial cells or their biochemical contents from excreta after in vivo isotopic labeling
The methods of the present invention allow for the non-invasive or minimally invasive isolation of epithelial cells or their biochemical contents from excreta after in vivo isotopic labeling. Once isolated, the labeled epithelial cells or their labeled biochemical contents find use as kinetic biomarkers of diseases of epithelial tissues including epithelial cancers and epithelial proliferative disorders such as psoriasis and benign prostate hyperplasia. The biomarkers are useful in diagnosing a disease of epithelial tissue origin, monitoring therapeutic efficacy of compounds or combinations of compounds used to treat diseases of epithelial tissue origin, screen new chemical entities or biological factors or combinations of chemical entities or biological factors, or mixtures thereof, for therapeutic activity in animal models of diseases of epithelial tissue origin or in human clinical trials, or test for toxicity on epithelial tissues from exposure to therapeutic compounds or new chemical entities or environmental chemicals such as industrial or occupational chemicals, environmental pollutants, food additives, and cosmetics.
Owner:RGT UNIV OF CALIFORNIA
Methods for comparing relative flux rates of two or more biological molecules in vivo through a single protocol
ActiveUS20050019251A1Ease of of half-lifeEase of of levelsIn-vivo radioactive preparationsSugar derivativesHigh-Throughput Screening MethodsIsotopic labeling
The invention relates to techniques for measuring and comparing relative molecular flux rates of different biological molecules by administering isotope-labeled water to one or more tissues or individuals and comparing the molecular flux rates of two or more biological molecules, including biological molecules in different chemical classes. The methods find use in several applications including diagnosing, prognosing, or monitoring a disease, disorder, or condition, the in vivo high-throughput screening of chemical entities and biological factors for therapeutic effects in various disease models, and the in vivo high-throughput screening of chemical entities and biological factors for toxic effects.
Owner:RGT UNIV OF CALIFORNIA
Imaging Agents
InactiveUS20070082879A1High yieldExcessive reactionBiocideIn-vivo radioactive preparationsRheniumAbnormal tissue growth
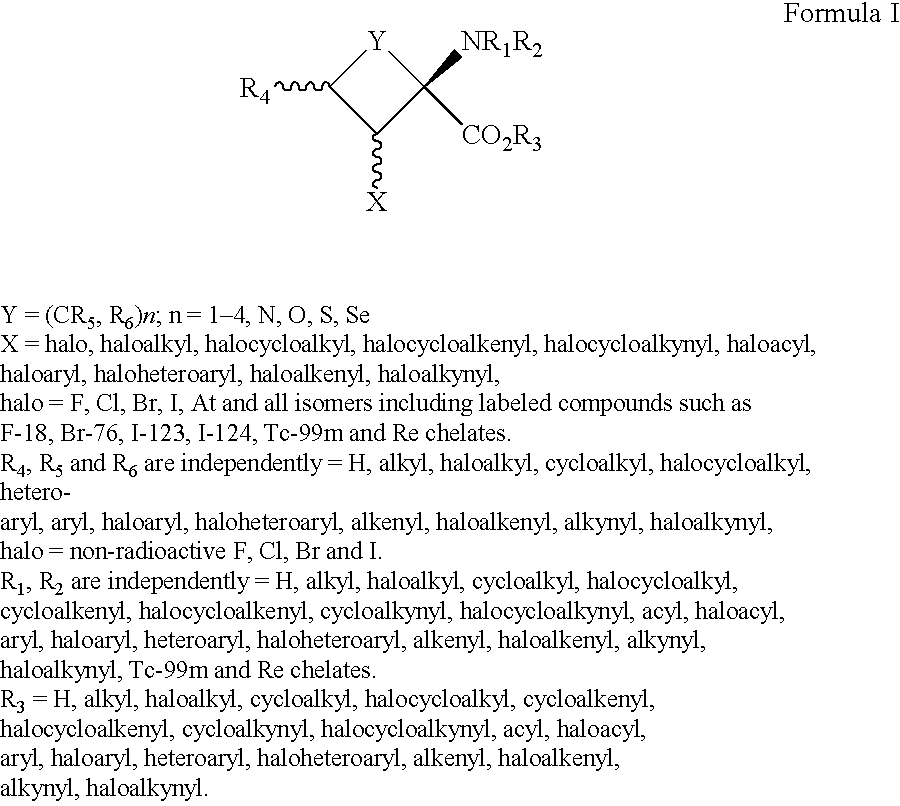
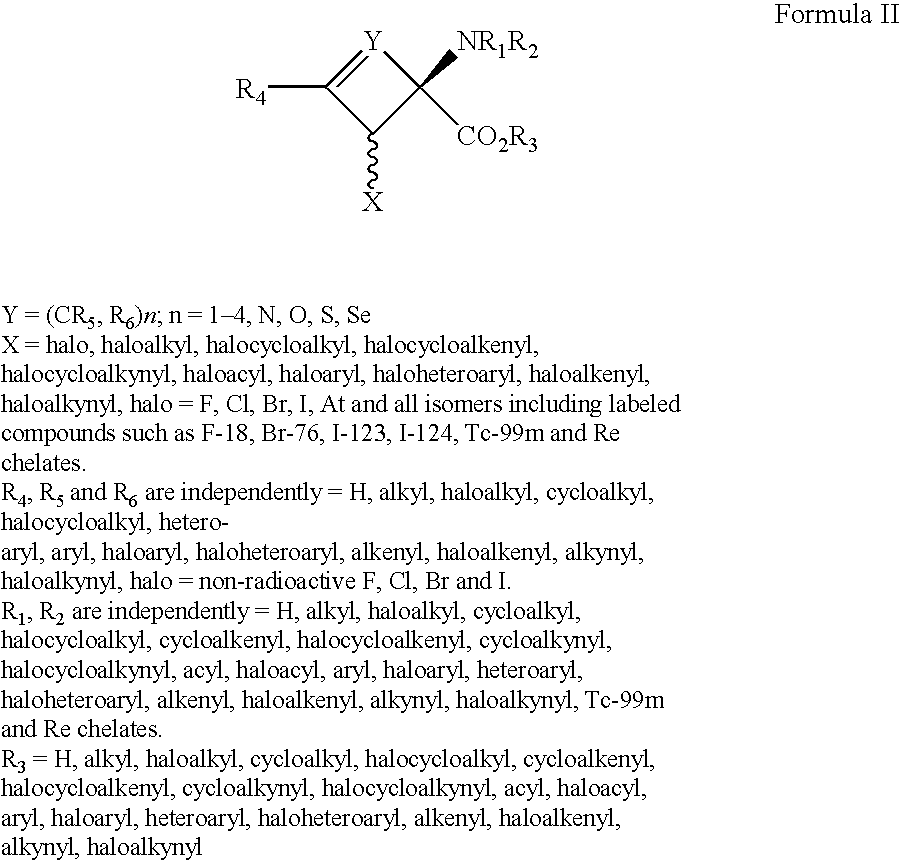
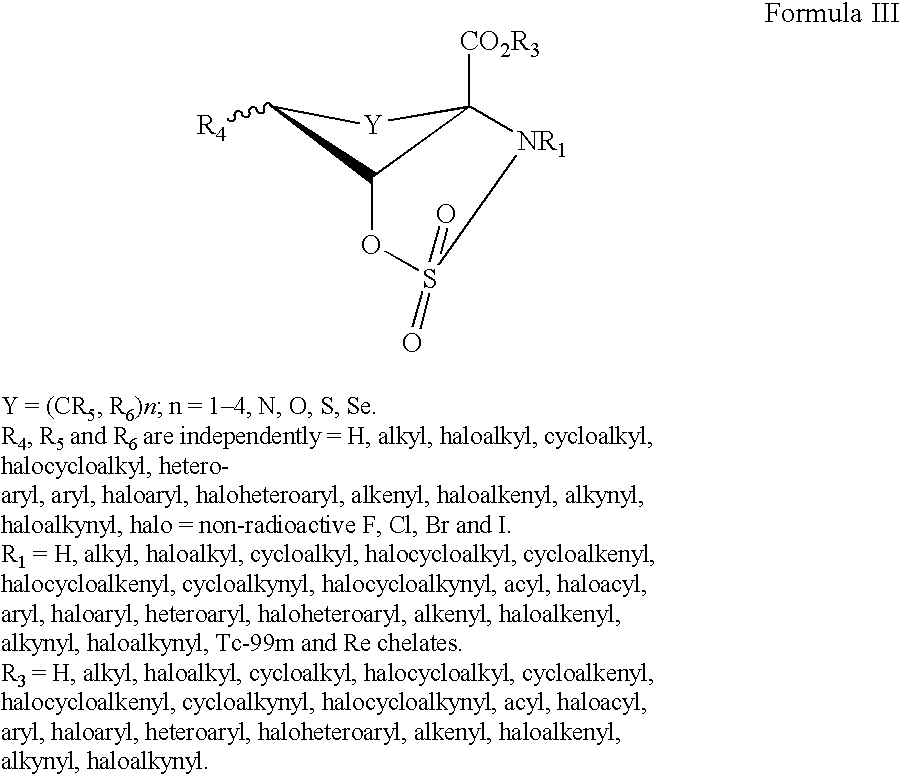
The present invention provides novel amino acid compounds useful in detecting and evaluating brain and body tumors. These compounds have the advantageous properties of rapid uptake and prolonged retention in tumors and can be labeled with halogen isotopes such as fluorine-18, iodine-123, iodine-124, iodine-125, iodine-131, bromine-75, bromine-76, bromine-77, bromine-82, astatine-210, astatine-211, and other astatine isotopes. These compounds can also be labeled with technetium and rhenium isotopes using known chelation complexes. The compounds disclosed herein bind tumor tissues in vivo with high specificity and selectivity when administered to a subject. Preferred compounds show a target to non-target ratio of at least 2:1, are stable in vivo and substantially localized to target within 1 hour after administration. Preferred compounds include 1-amino-2-[18F]fluorocyclobutyl-1-carboxylic acid (2-[18F]FACBC) and 1-amino-2-[18F]fluoromethylcyclobutyl-1-carboxylic acid (2-[18F]FMACBC). The labeled amino acid compounds of the invention are useful as imaging agents in detecting and / or monitoring tumors in a subject by PET or SPECT.
Owner:EMORY UNIVERSITY
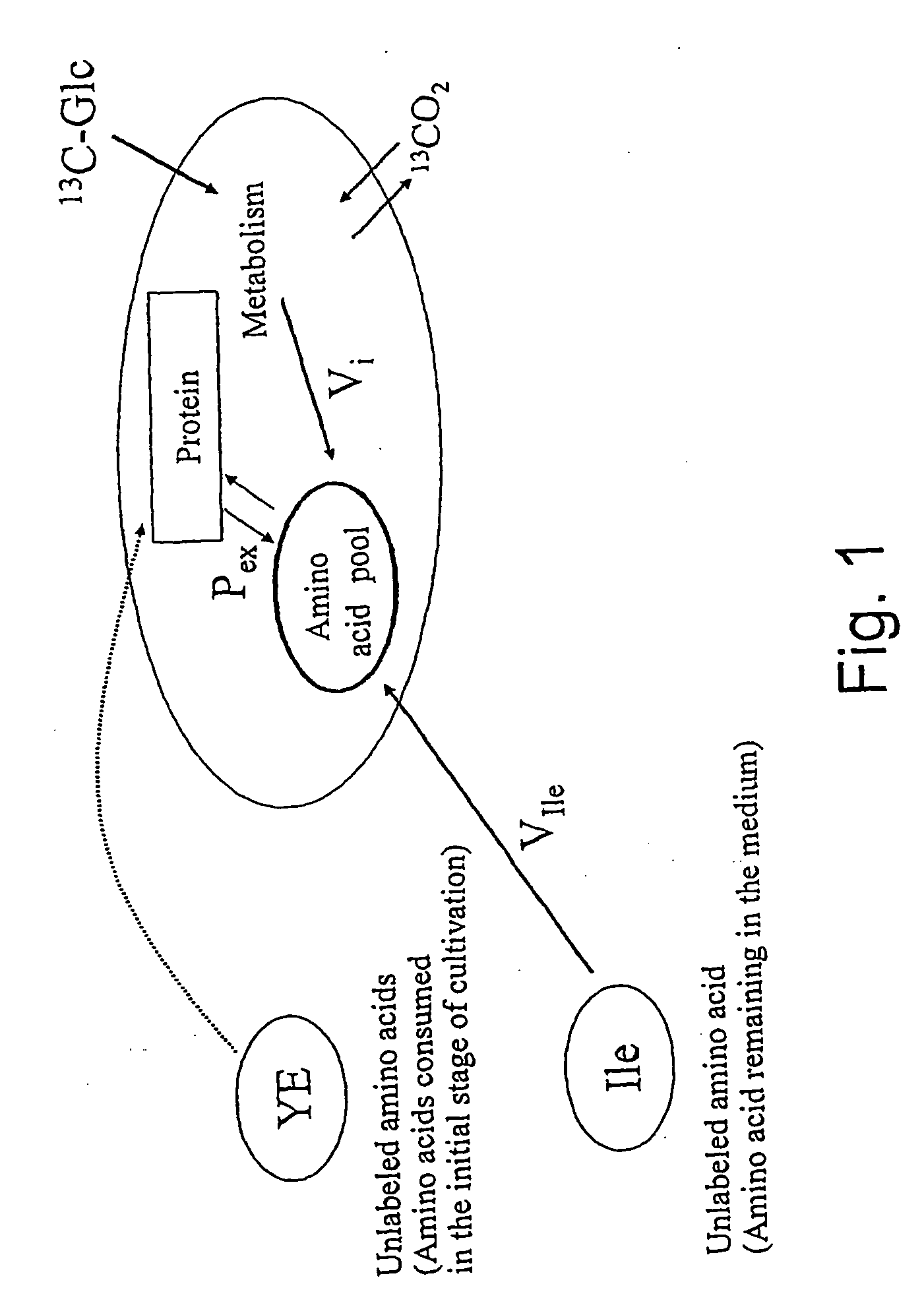
Intracellular metabolic flux analysis method using substrate labeled with isotope
InactiveUS20050175982A1Low costMicrobiological testing/measurementBiological testingDrug metabolismTime course
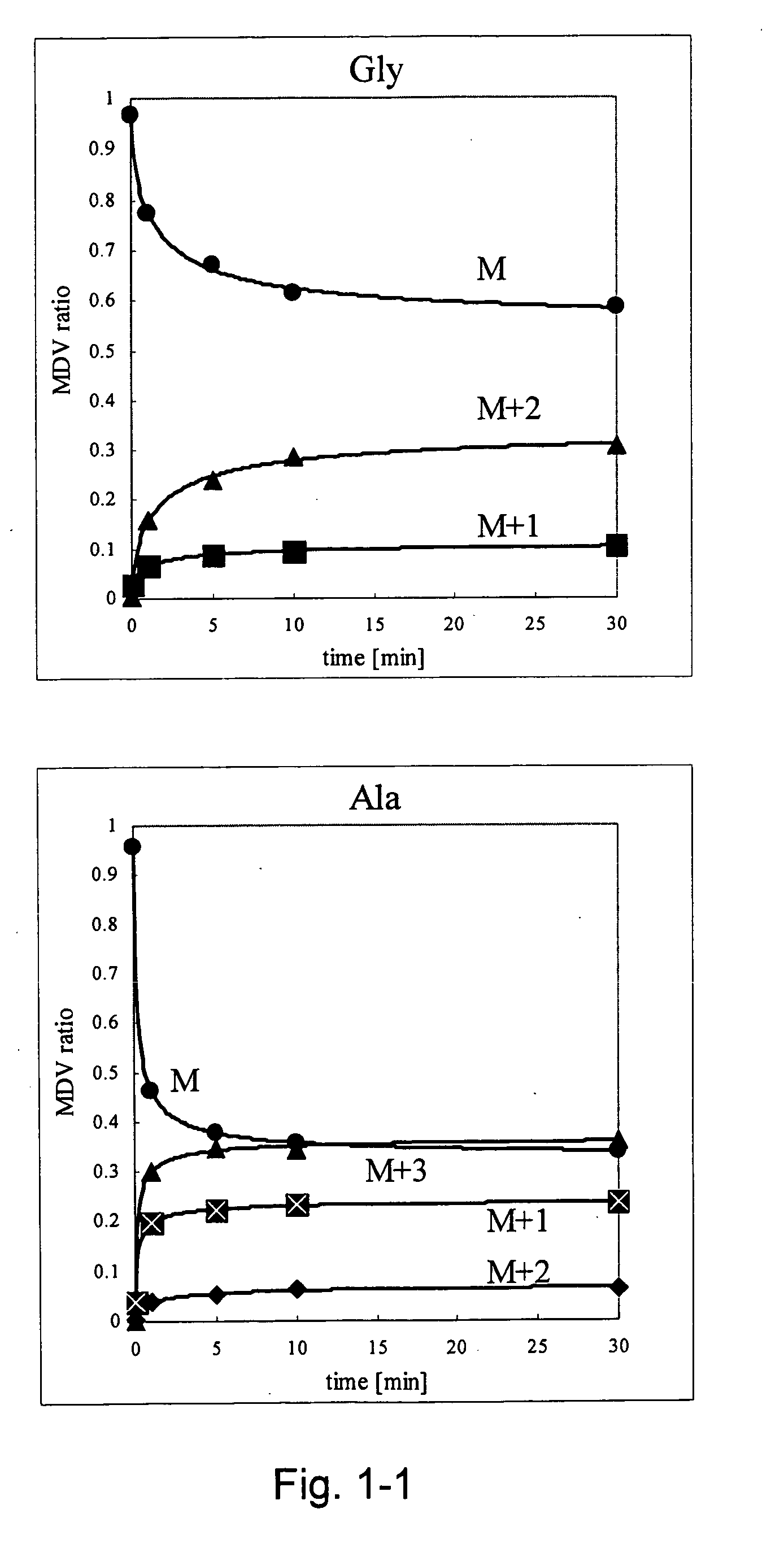
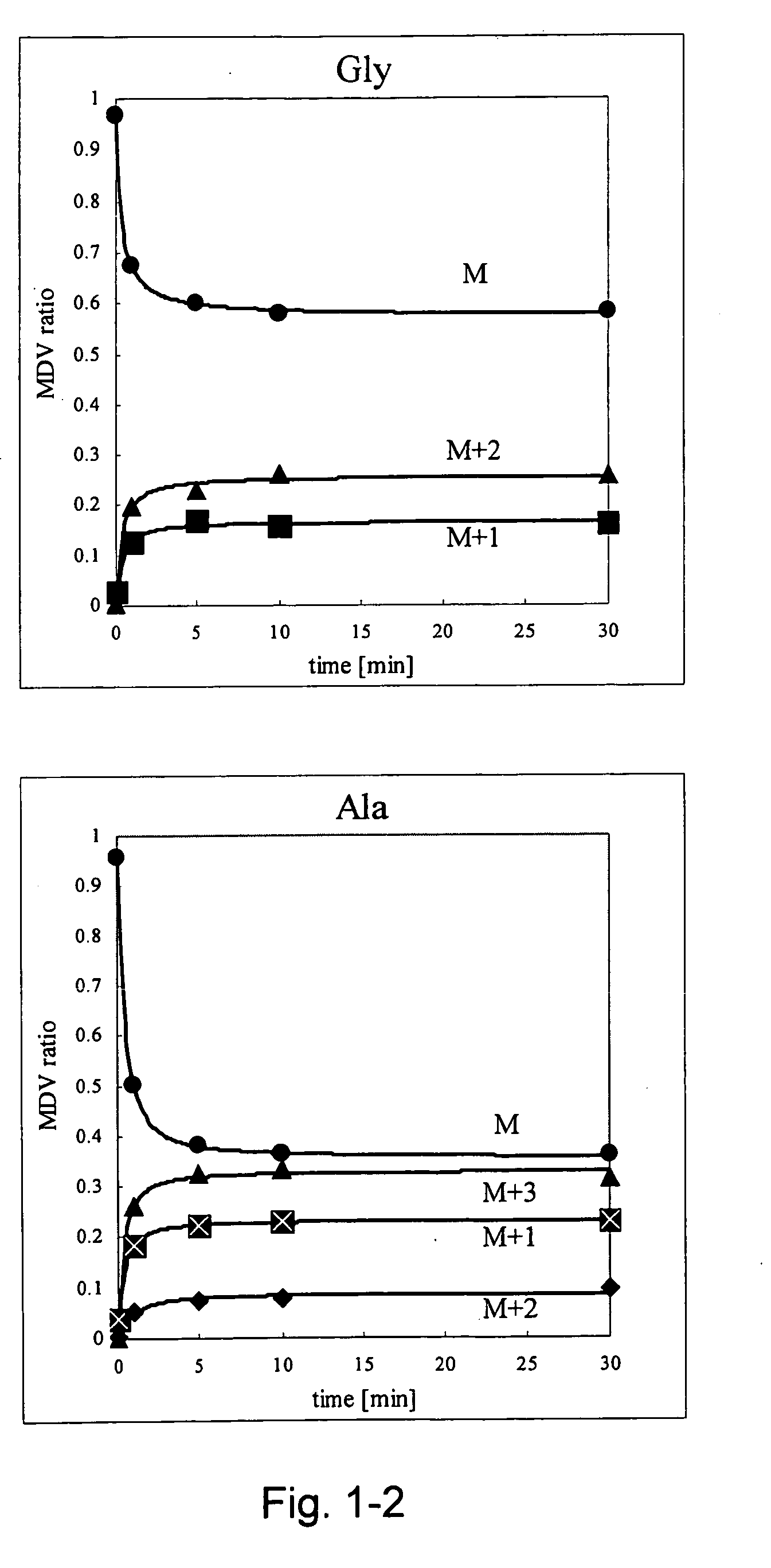
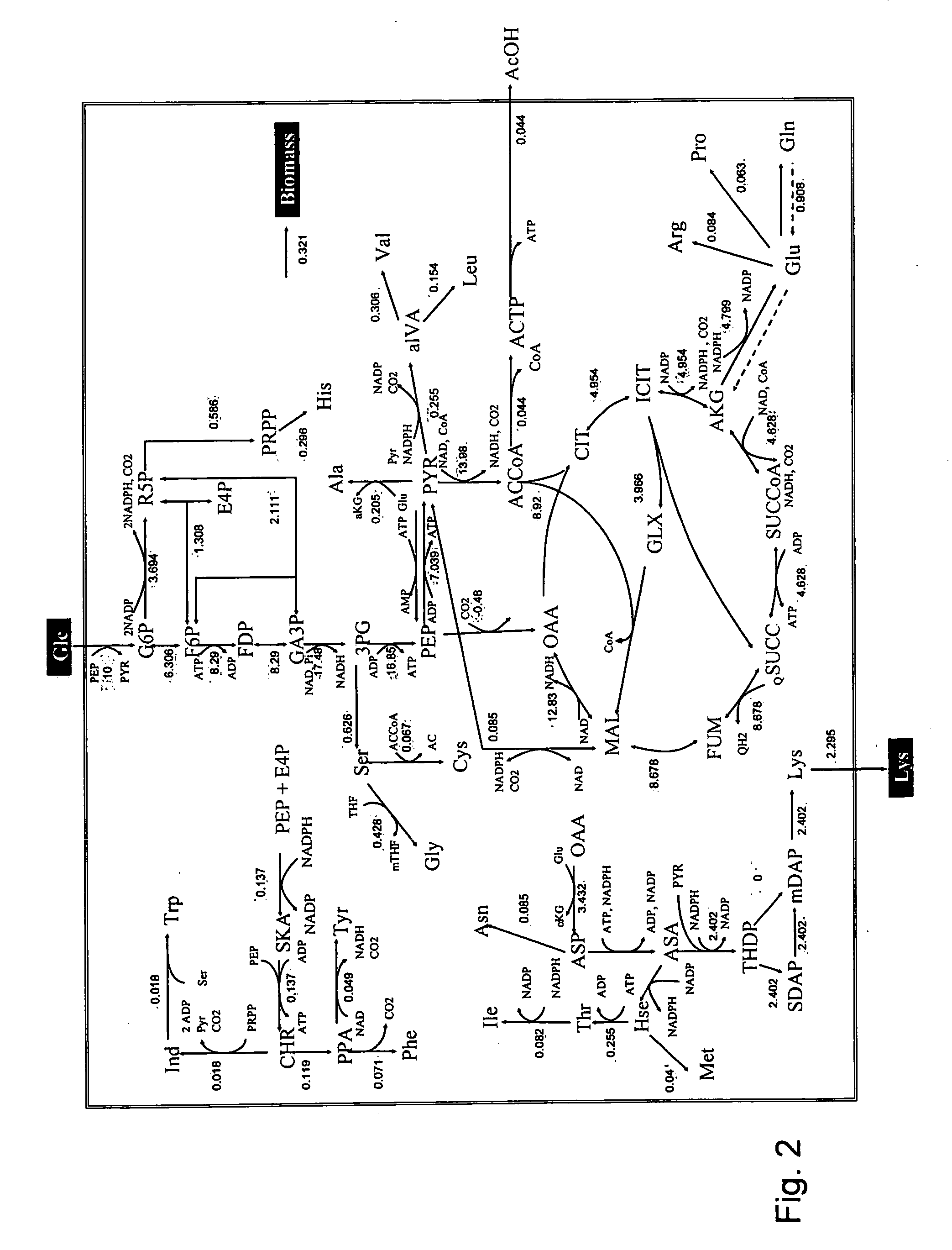
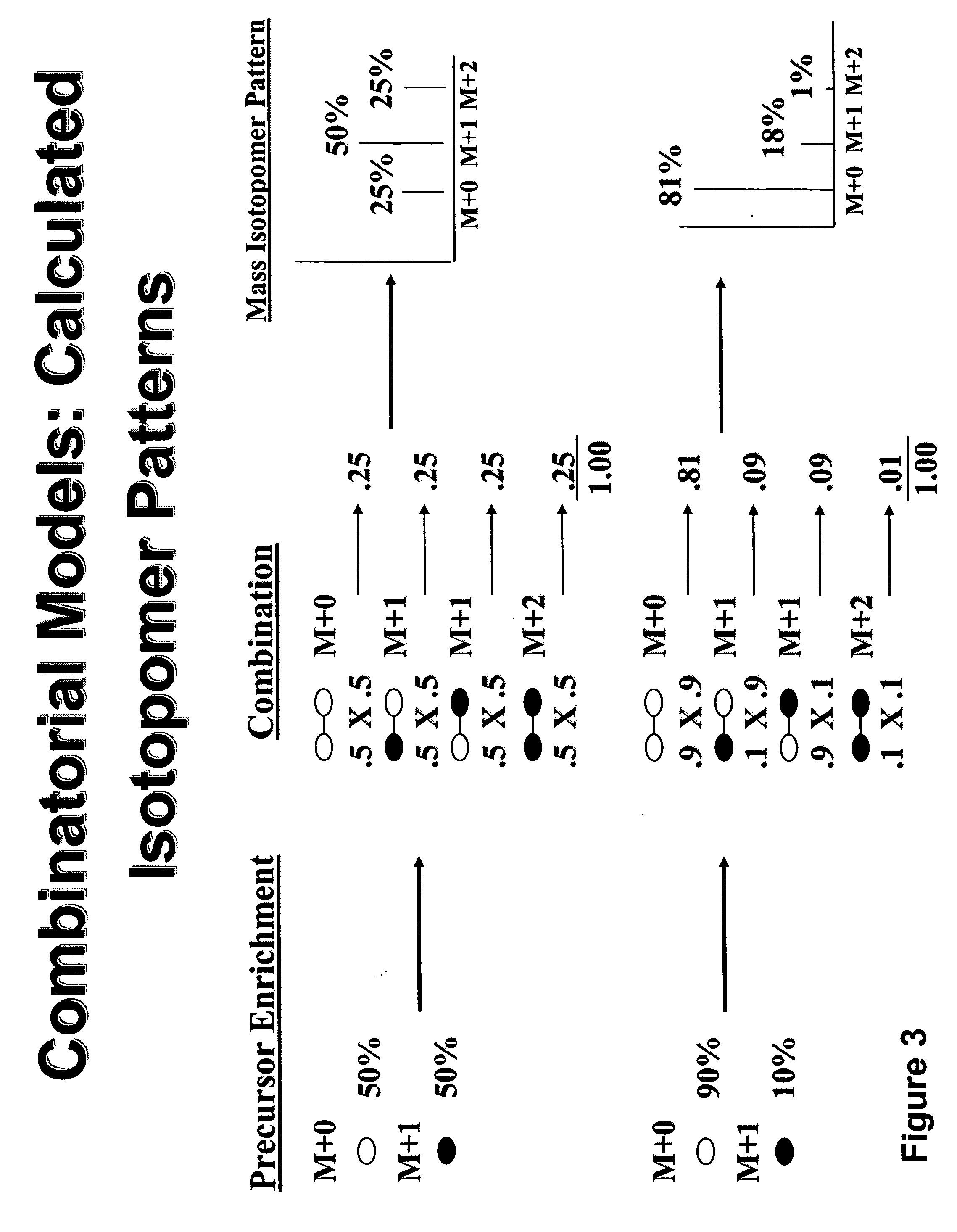
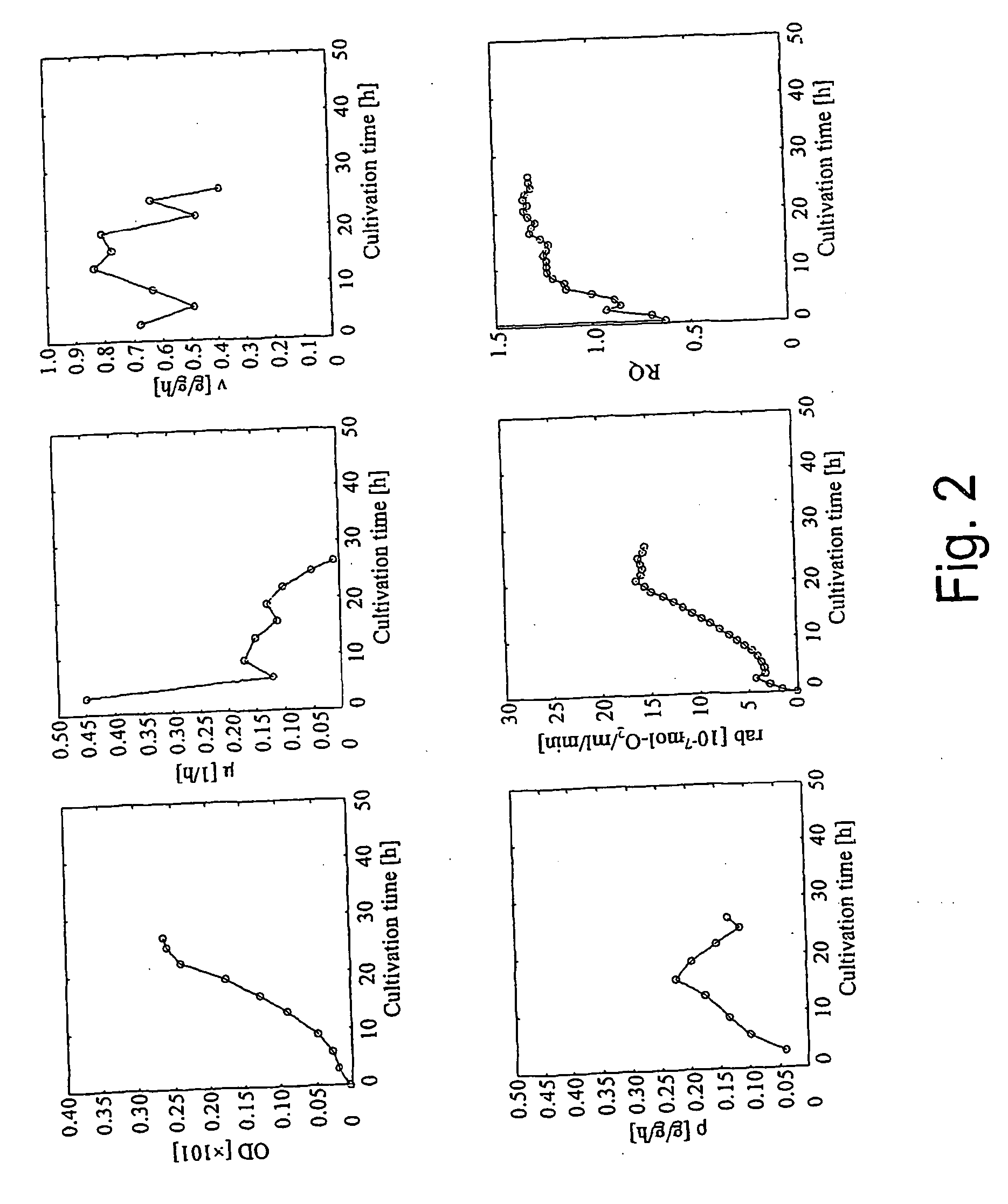
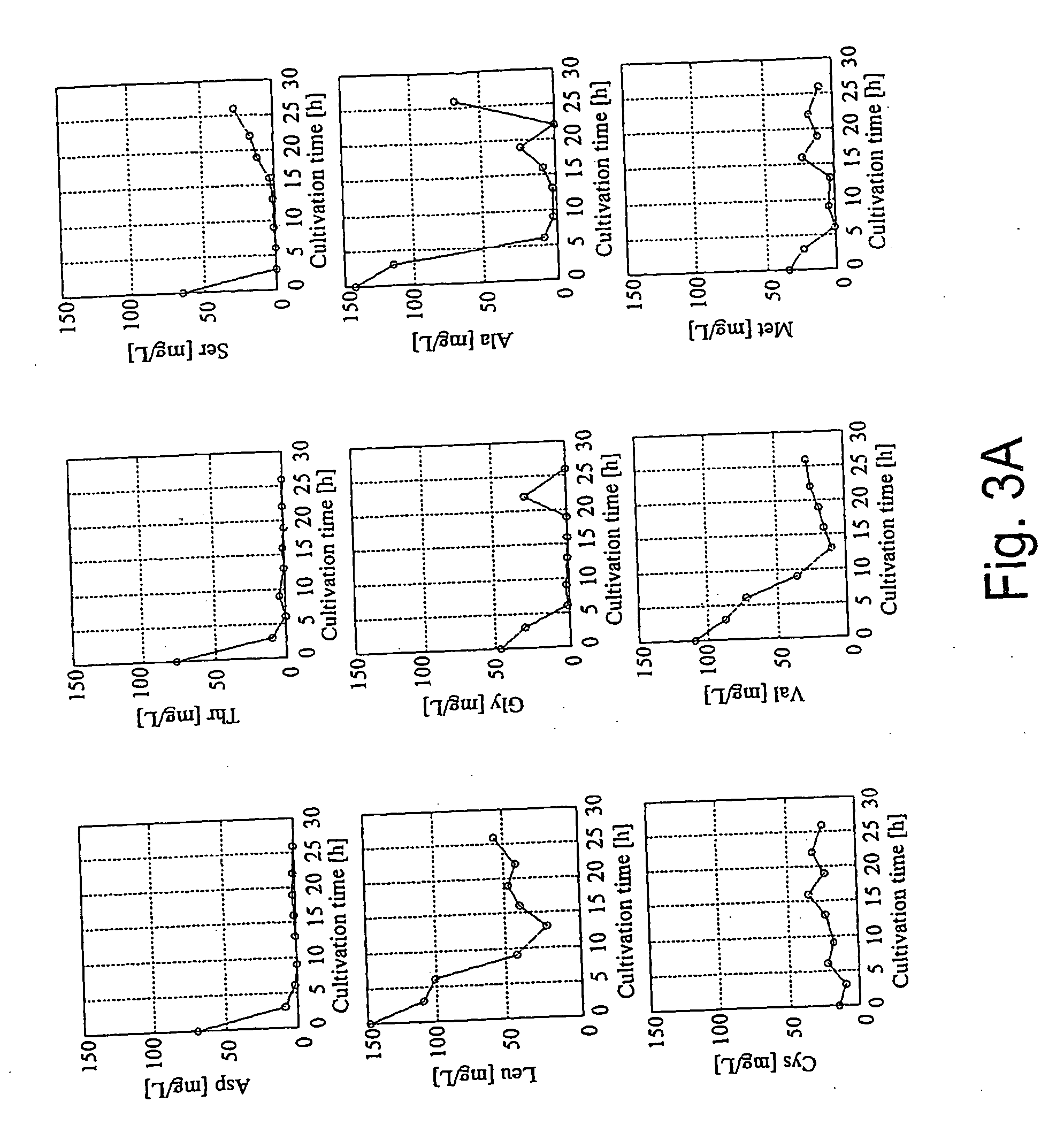
A method for intracellular metabolic flux analysis comprising (a) culturing cells in a medium not containing any isotope-labeled substrate to a target phase of the metabolic flux analysis, (b) adding an isotope-labeled substrate to the medium, and continuing culture and collecting samples from the medium in time course, (c) measuring isotope distribution in an intracellular metabolite contained in the samples collected in time course, (d) performing a regression analysis for measured data and calculating an isotope distribution ratio in a steady state, and (e) analyzing a metabolic flux of the cultured cells by using the calculated isotope distribution ratio.
Owner:AJINOMOTO CO INC
Method for high-throughput screening of compounds and combinations of compounds for discovery and quantification of actions, particularly unanticipated therapeutic or toxic actions, in biological systems
InactiveUS20050202406A1Compounds screening/testingMicrobiological testing/measurementLiving systemsIsotopic labeling
The invention enables high-throughput screening of compounds in living systems to detect unanticipated or unintended biological actions. The invention also allows for screening, detection, and confirmation of new indications for approved drugs. Screening and detection of toxic effects of compounds also can be achieved by using the methods of the invention. The methods comprise administering isotope-labeled substrates to a living system so that the label is incorporated into molecules in a manner that reveals flux rates through metabolic pathways thought to be involved in a disease. Comparisons between living systems exposed to compounds and living systems not so exposed reveals the effects of the compounds on the flux rates through the metabolic pathways. Combinations or mixtures of compounds can be systematically screened to detect unanticipated or unintended biological actions, including synergistic actions, in the same manner.
Owner:RGT UNIV OF CALIFORNIA
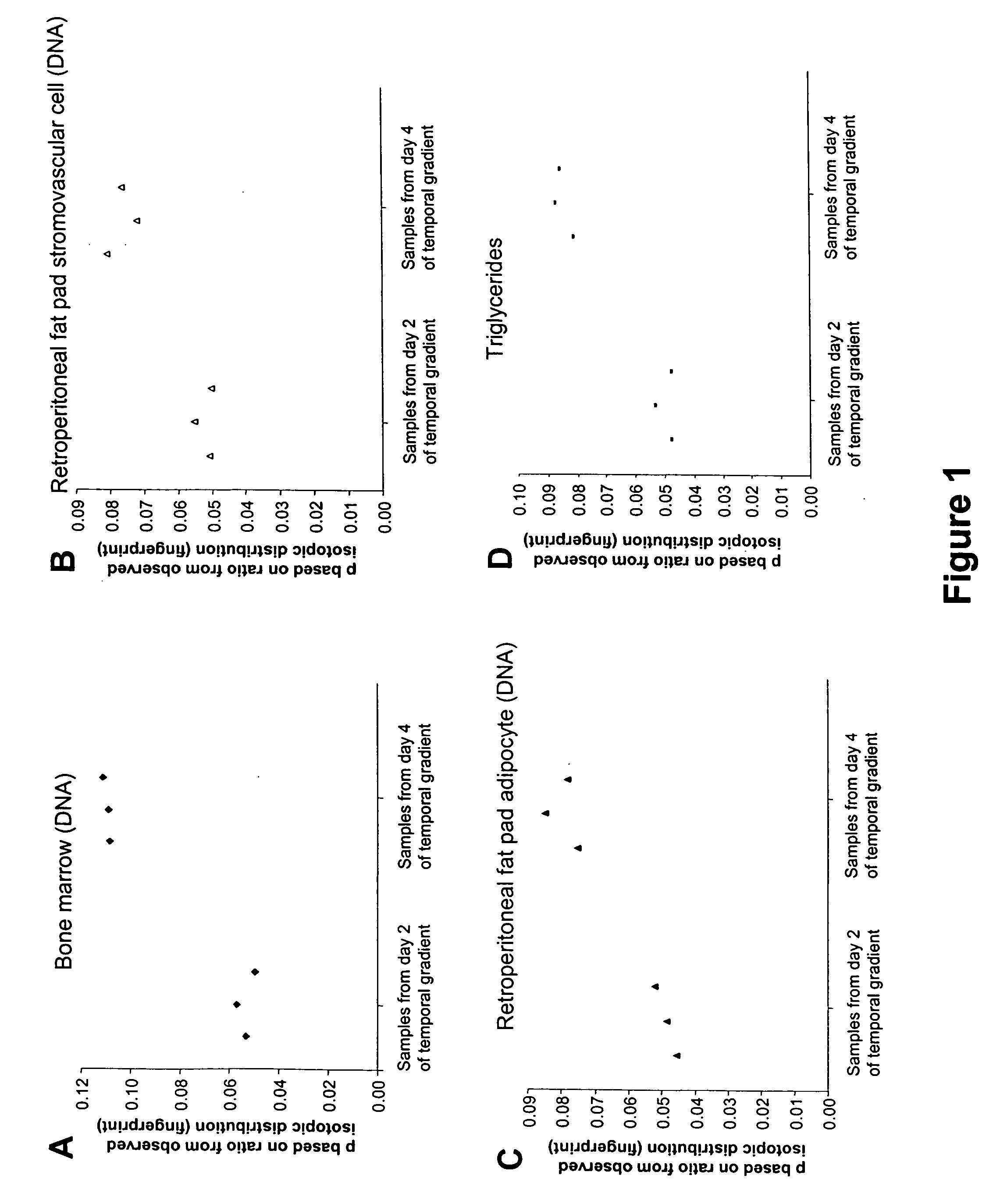
Temporal or spatial characterization of biosynthetic events in living organisms by isotopic fingerprinting under conditions of imposed isotopic gradients
InactiveUS20050201937A1Sugar derivativesMicrobiological testing/measurementBiological bodyIsotopic labeling
The invention provides methods useful for establishing timing or spatial location of a biosynthetic event in a living organism, without disrupting the event of interest and without disrupting the living organism. A temporal or spatial gradient of an isotopically labeled biochemical precursor is created, which serves to isotopically fingerprint (i.e., definitively mark) when or where biosynthesis occurs. The methods of the invention are broadly applicable to a variety of medical, public health, and diagnostic applications, as well as for establishing sequences of biochemical events that occur within a living organism.
Owner:RGT UNIV OF CALIFORNIA
Intracellular metabolic flux analysis method using substrate labeled with isotope
InactiveUS20060105322A1Reduce analysis errorReduce errorsMicrobiological testing/measurementBiological testingIsotopic labelingIsotope
A method for analyzing an intracellular metabolic flux comprising determining the intracellular metabolic flux from analytical values of cells cultured in a medium containing an isotope-labeled substrate as a carbon source on the basis of an intracellular metabolic flux model constructed for the intracellular metabolic flux to be analyzed, wherein (a) influence of an exchange reaction between an intracellular metabolite and a cell component produced by integration of the intracellular metabolite is considered, (b) uptake of a compound in a medium into cells, which compound is identical to an intracellular metabolite and unlabeled with an isotope, is considered, or (c) carbon dioxide used in a fixation reaction is assumed as carbon dioxide produced in a production reaction.
Owner:AJINOMOTO CO INC
Methods for measuring the rates of replication and death of microbial infectious agents in an infected
InactiveUS20060105339A1Inhibit growthAvoid maintenanceMicrobiological testing/measurementDisease diagnosisMicroorganismBiological body
The present invention provides methods and kits useful for determining rates of replication and destruction of an infectious agent within an infected host organism. In the methods of the invention, an isotopically-labeled precursor molecule is administered to an infected host, and is given sufficient time to pass into the host's metabolic pools into a biochemical component of the infectious agent. The isotopic content and / or pattern or the rate of change of the isotopic content and / or pattern of the biochemical component is then measured to determine the rate of replication (growth) of the infectious organism while in the host. Alternatively, isotopic decay of labeled molecular components of the infectious agent is measured over time after discontinuing administration of the isotopically labeled precursor molecule to determine the rate of destruction (death) of the infectious agent while in the host. Thus, using methods of the invention, in vivo sensitivity of infectious agents to drug agents may be determined, in order to optimize therapy of the infected host.
Owner:RGT UNIV OF CALIFORNIA
Method for single oxygen atom incorporation into digested peptides using peptidases
InactiveUS20060105415A1Highly accurate quantification methodAccurately quantify different protein populationsSamplingMicrobiological testing/measurementIsotopic labelingProteomics
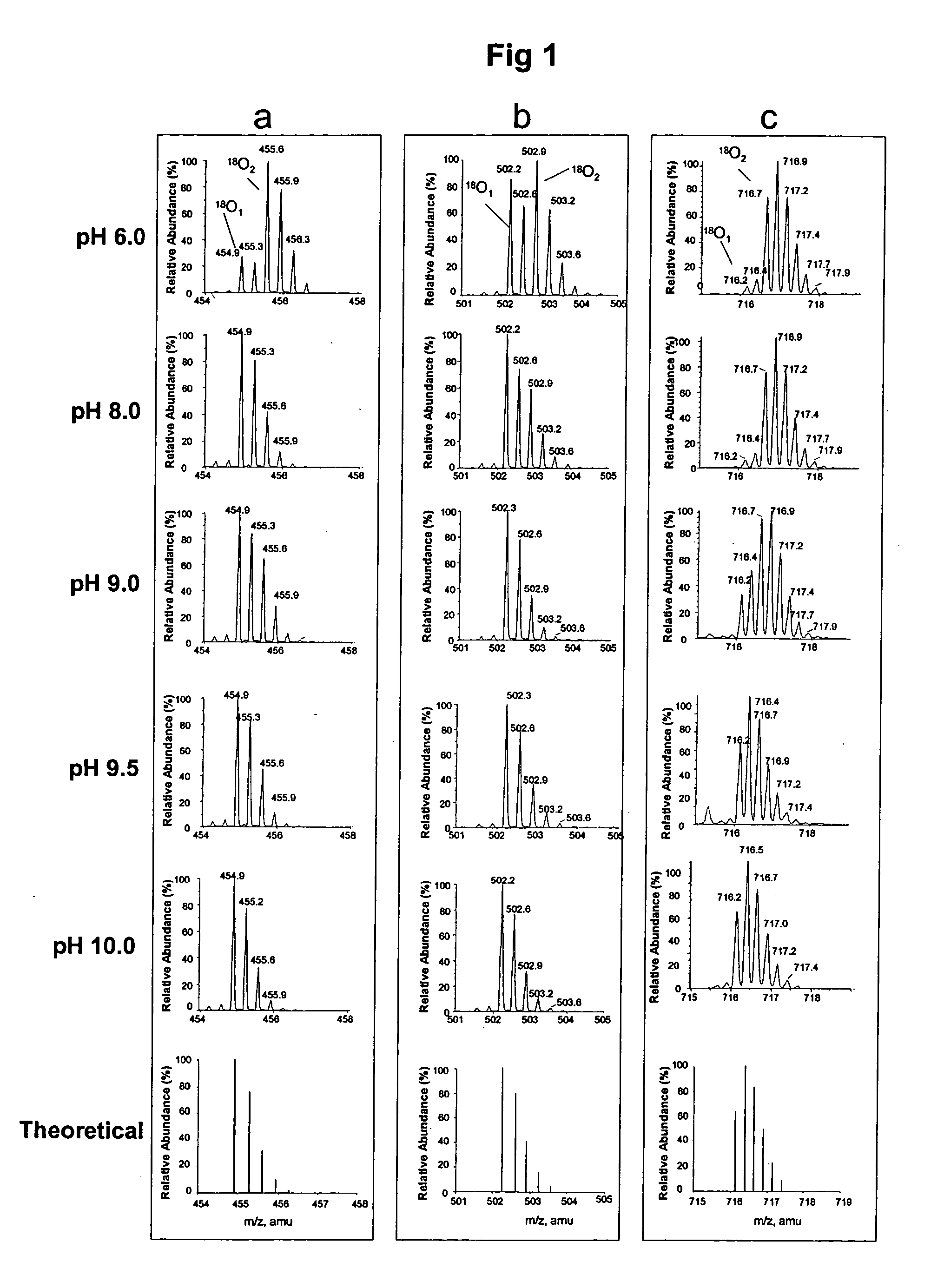
Optimized enzymatic conditions incorporate a single oxygen atom into digested peptides using a peptidase. The incorporation of a single oxygen atom is especially useful for proteolytic 18O labeling in comparative proteomics. The optimized proteolytic 18O labeling minimizes the generation of a mixture of isotopic isoforms of the peptides resulting from incorporation of either one or two 18O atoms. The outcome is accurate quantification of isotopically labeled peptides.
Owner:UNIVERSITY OF NORTH DAKOTA +1
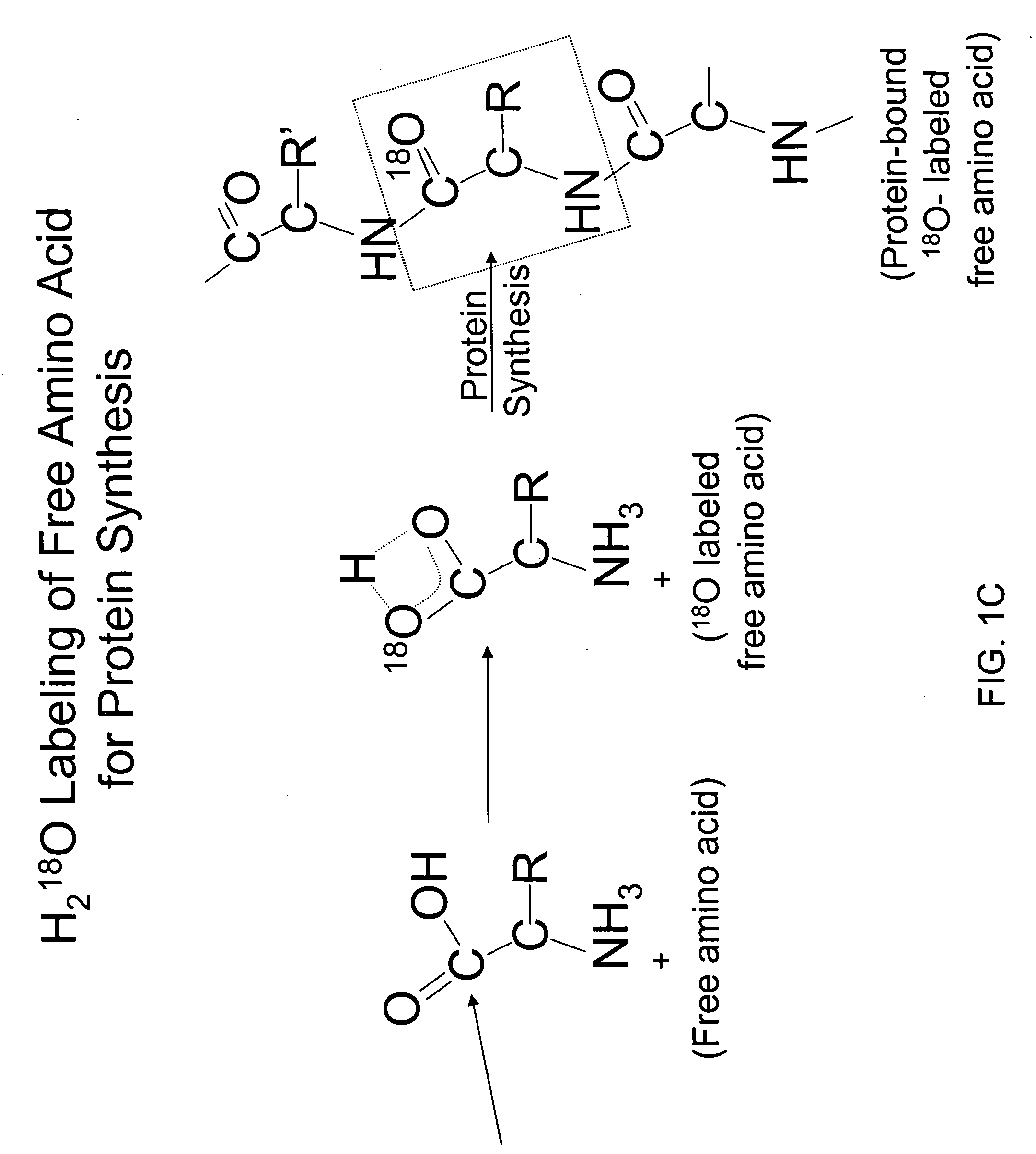
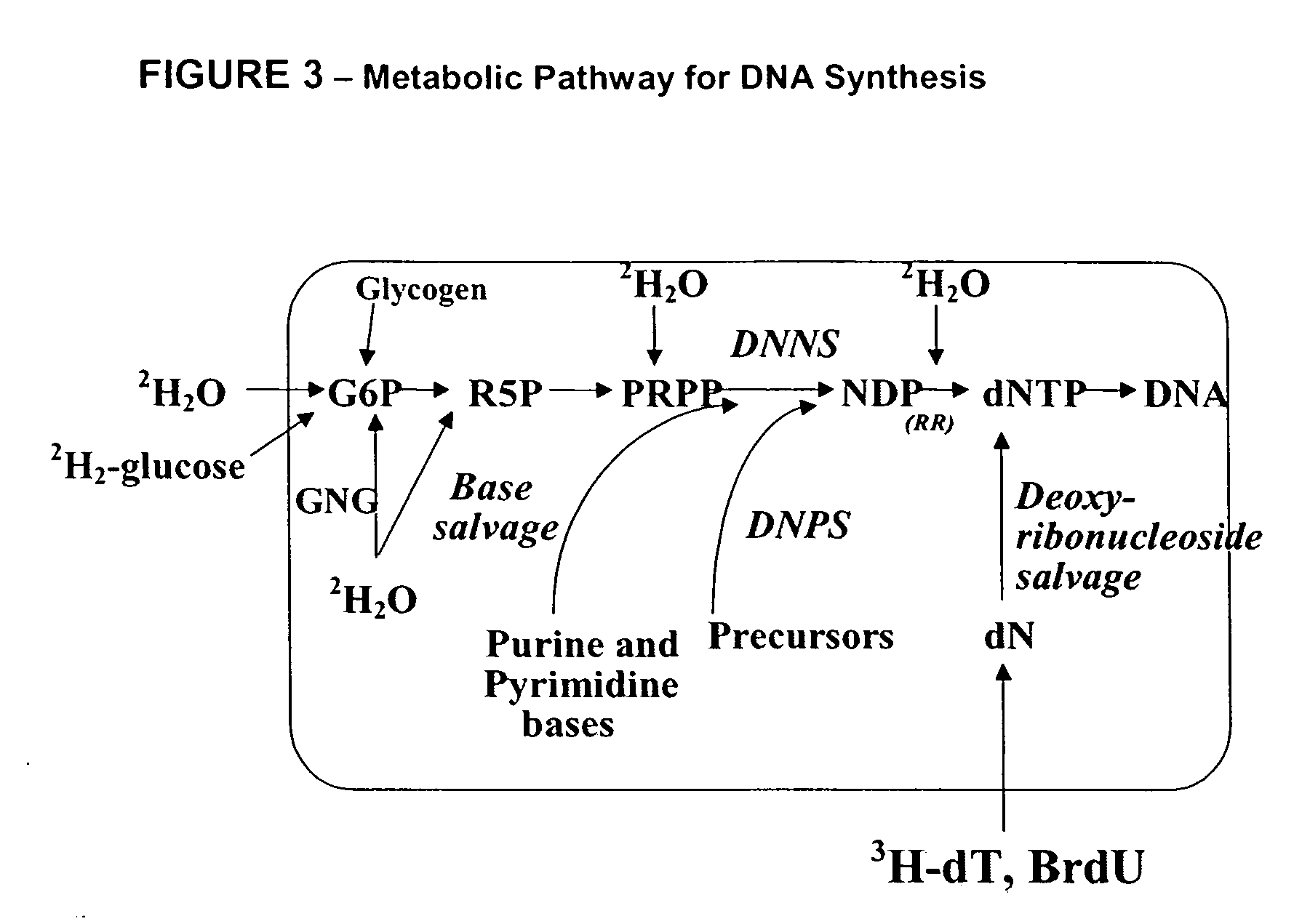
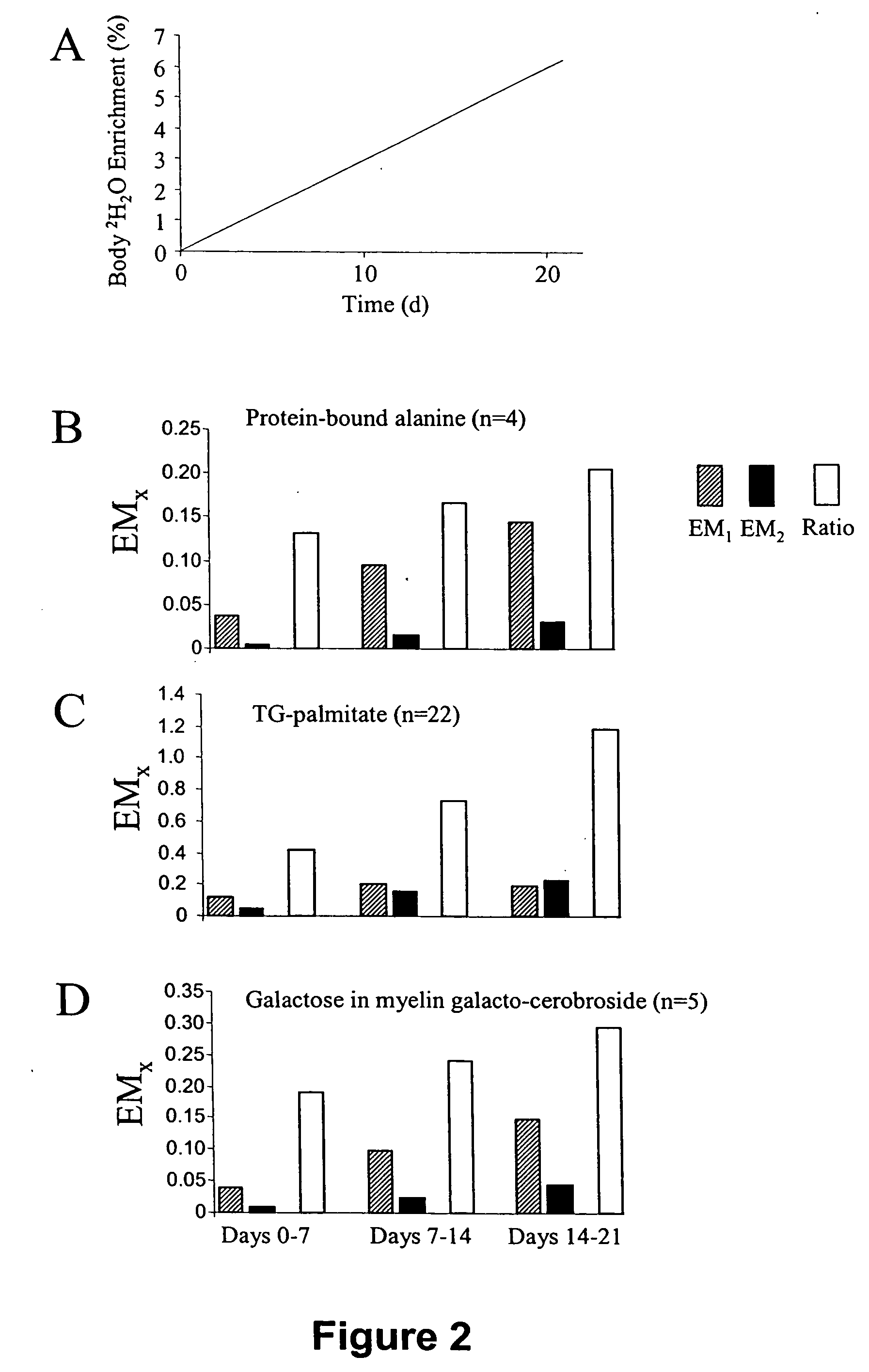
Measurement of protein synthesis rates in humans and experimental systems by use of isotopically labeled water
InactiveUS20060029549A1Biological testingIn-vivo testing preparationsIsotopic labelingProtein biosynthesis
Provided herein are methods for measuring protein biosynthesis by using 2H2O or radioactive 3H2O and applicable uses thereof.
Owner:RGT UNIV OF CALIFORNIA
Screening for enzyme stereoselectivity utilizing mass spectrometry
InactiveUS20020127609A1Improved screening sensitivityImprove detection limitParticle separator tubesMicrobiological testing/measurementEnzymeBiology
Methods of screening for enzyme stereoselectivity that include detecting isotopically labeled products by mass spectrometry are provided. The methods are particularly suitable for screening enzyme libraries for stereoselectivity.
Owner:MAXYGEN
Rapid quantitative analysis of proteins or protein function in complex mixtures
InactiveUS7544518B2Facilitates quantitative determinationFacilitates quantitative determination of the absolute amountsComponent separationMaterial analysis by electric/magnetic meansIsotopic labelingProtein expression profile
Analytical reagents and mass spectrometry-based methods using these reagents for the rapid, and quantitative analysis of proteins or protein function in mixtures of proteins. The methods employ affinity labeled protein reactive reagents having three portions: an affinity label (A) covalently linked to a protein reactive group (PRG) through a linker group (L). The linker may be differentially isotopically labeled, e.g., by substitution of one or more atoms in the linker with a stable isotope thereof. These reagents allow for the selective isolation of peptide fragments or the products of reaction with a given protein (e.g., products of enzymatic reaction) from complex mixtures. The isolated peptide fragments or reaction products are characteristic of the presence of a protein or the presence of a protein function in those mixtures. Isolated peptides or reaction products are characterized by mass spectrometric (MS) techniques. The reagents also provide for differential isotopic labeling of the isolated peptides or reaction products which facilitates quantitative determination by mass spectrometry of the relative amounts of proteins in different samples. The methods of this invention can be used for qualitative and quantitative analysis of global protein expression profiles in cells and tissues, to screen for and identify proteins whose expression level in cells, tissue or biological fluids is affected by a stimulus or by a change in condition or state of the cell, tissue or organism from which the sample originated.
Owner:UNIV OF WASHINGTON
Measurement of protein synthesis rates in humans and experimental systems by use of isotopically labeled water
InactiveUS20050147558A1In-vivo radioactive preparationsPeptide/protein ingredientsIsotopic labelingProtein biosynthesis
Provided herein are methods for measuring protein biosynthesis by using 2H2O or radioactive 3H2O and applicable uses thereof.
Owner:RGT UNIV OF CALIFORNIA
Ionizable isotopic labeling reagents for relative quantification by mass spectrometry
ActiveUS20080050833A1Improve ETD analysisConvenient charging statusOrganic compound preparationComponent separationMetaboliteIsotopic labeling
Relative quantification of metabolites by Electrospray Ionization Mass Spectrometry (ESI-MS) requiring a mechanism for simultaneous analysis of multiple analytes in two or more samples. Labeling reagents that are reactive to particular compound classes and differ only in their isotopic kit facilitating relative quantification and providing tangible evidence for the existence of specific functional groups. Heavy and light isotopic forms of methylacetimidate were synthesized and used as labeling reagents for quantification of amine-containing molecules, such as biological samples. Heavy and light isotopic forms of formaldehyde and cholamine were also synthesized and used independently as labeling reagents for quantification of amine-containing and carboxylic acid-containing molecules, such as found in biological samples. Advantageously, the labeled end-products are positively charged under normal acidic conditions involving conventional Liquid Chromatography Mass Spectrometry (LC / MS) applications. Labeled primary and secondary amine and carboxylic acid end-products also generated higher signals concerning mass-spectra than pre-cursor molecules and improved sensitivity. Improved accuracy concerning relative quantification was achieved by mixing heavy and light labeled Arabidopsis extracts in different ratios. Labeling strategy was further employed to ascertain differences in the amounts of amine-containing metabolites for two strains of Arabidopsis seeds.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
Reagents and methods for labeling terminal olefins
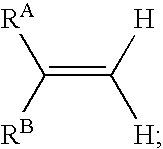
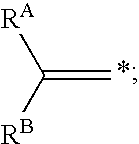
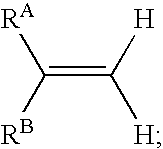
InactiveUS20060045846A1In-vivo radioactive preparationsGroup 3/13 element organic compoundsArylIsotopic labeling
In one aspect, the present invention provides a method for labeling a terminal olefin, the method comprising a step of treating a terminal olefin substrate having the structure: with a labeled ethylene reagent in the presence of a suitable catalyst under suitable olefin metathesis reaction conditions to form a labeled terminal olefin having the structure: wherein RA and RB are independently hydrogen, or an aliphatic, alicyclic, heteroaliphatic, heterocyclic, aryl or heteroaryl moiety, with the proviso that RA and RB are not each hydrogen, or RA and RB taken together with the carbon atom to which they are attached form an alicyclic or heterocyclic moiety; and * denotes the presence of an isotopic label on the terminal carbon atom.
Owner:EISIA R&D MANAGEMENT CO LTD
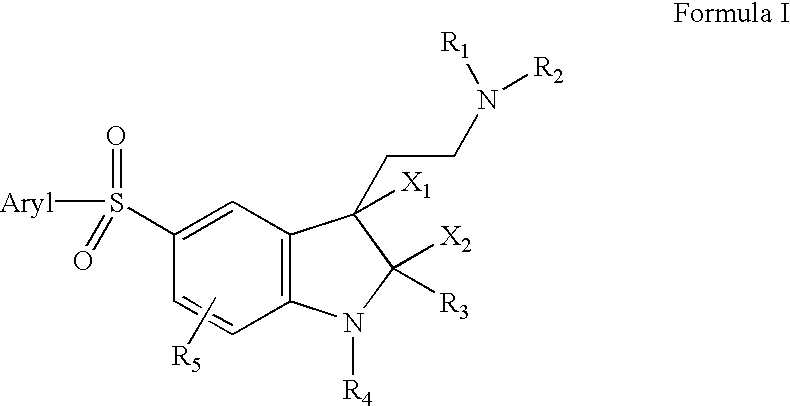
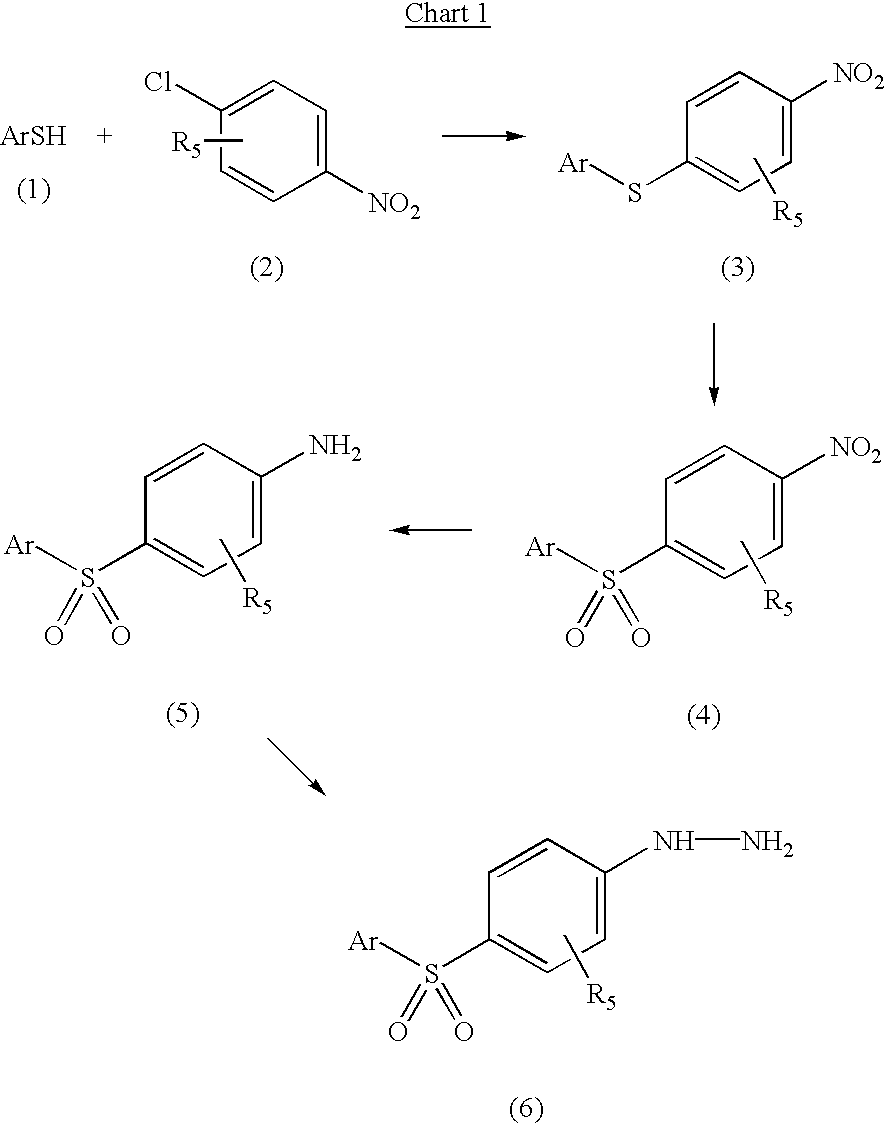
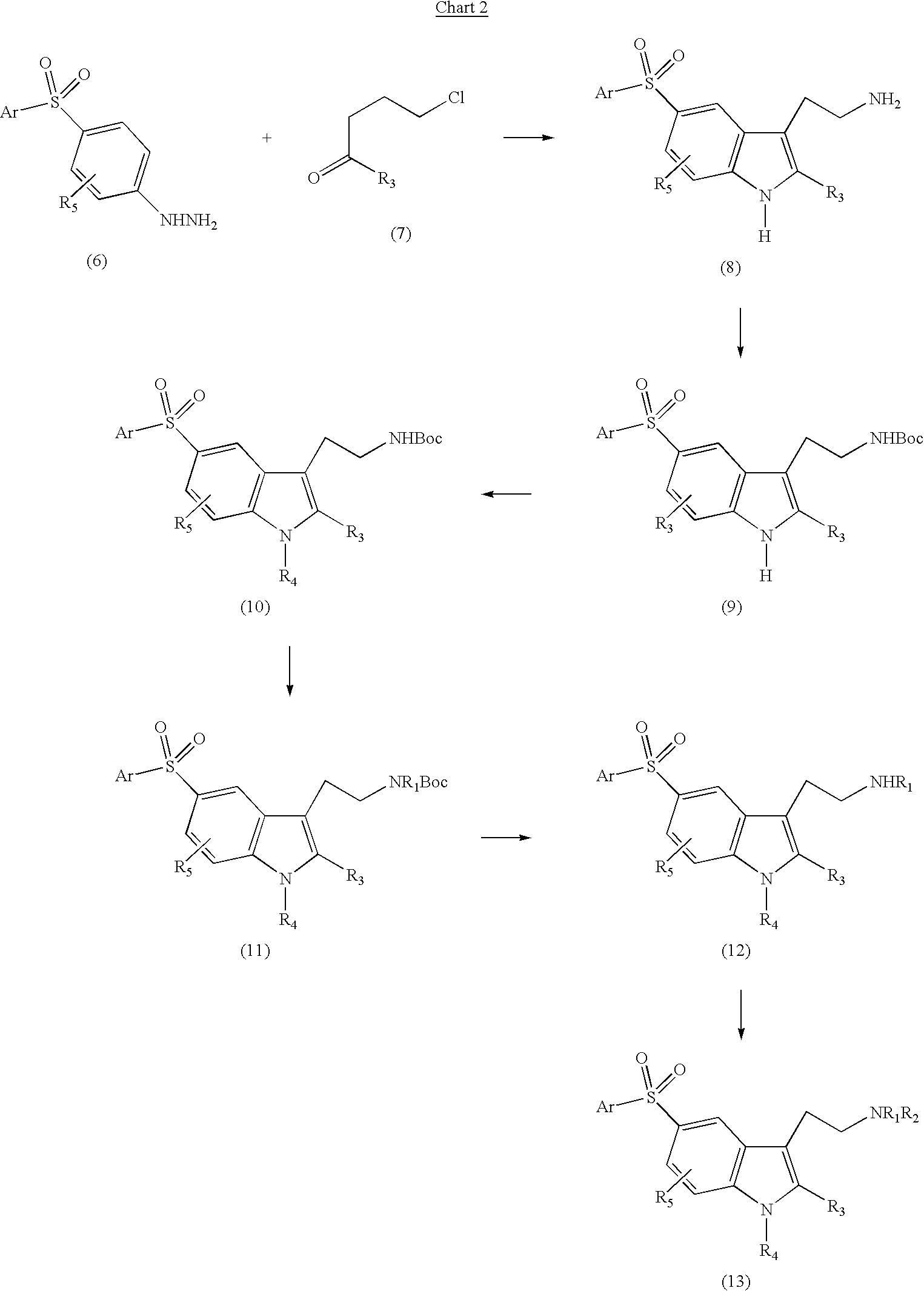
5-arylsulfonyl indoles useful for treating disease
InactiveUS20030060498A1Good metabolic stabilityProlong half-life in vivoBiocideNervous disorderDiseaseNMR - Nuclear magnetic resonance
The invention provides derivatives of 5-arylsulfonyl indole and 5-arylsulfonyl indoline compounds which may be in the form of pharmaceutical acceptable salts or compositions that are useful in treating central nervous system diseases such as anxiety and depression. The invention also includes intermediates and processes to make the compounds, isotopically-labeled forms of the compounds and the use of the isotopically labeled forms of the compounds to perform nuclear magnetic resonance imaging and positron emission tomography.
Owner:PHARMACIA & UPJOHN CO
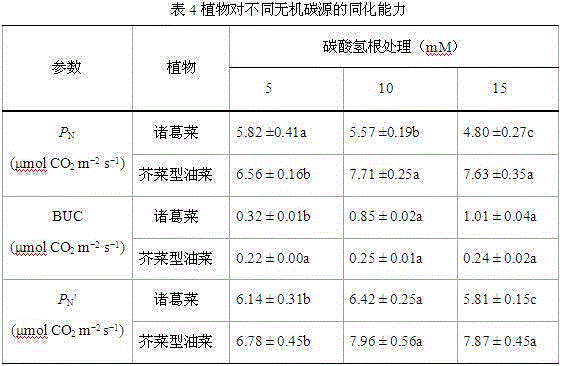
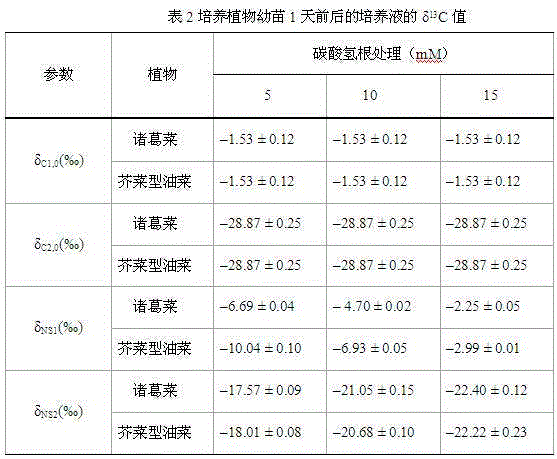
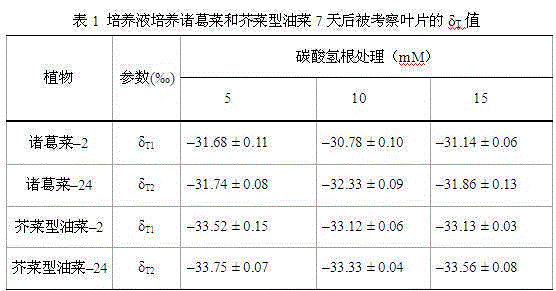
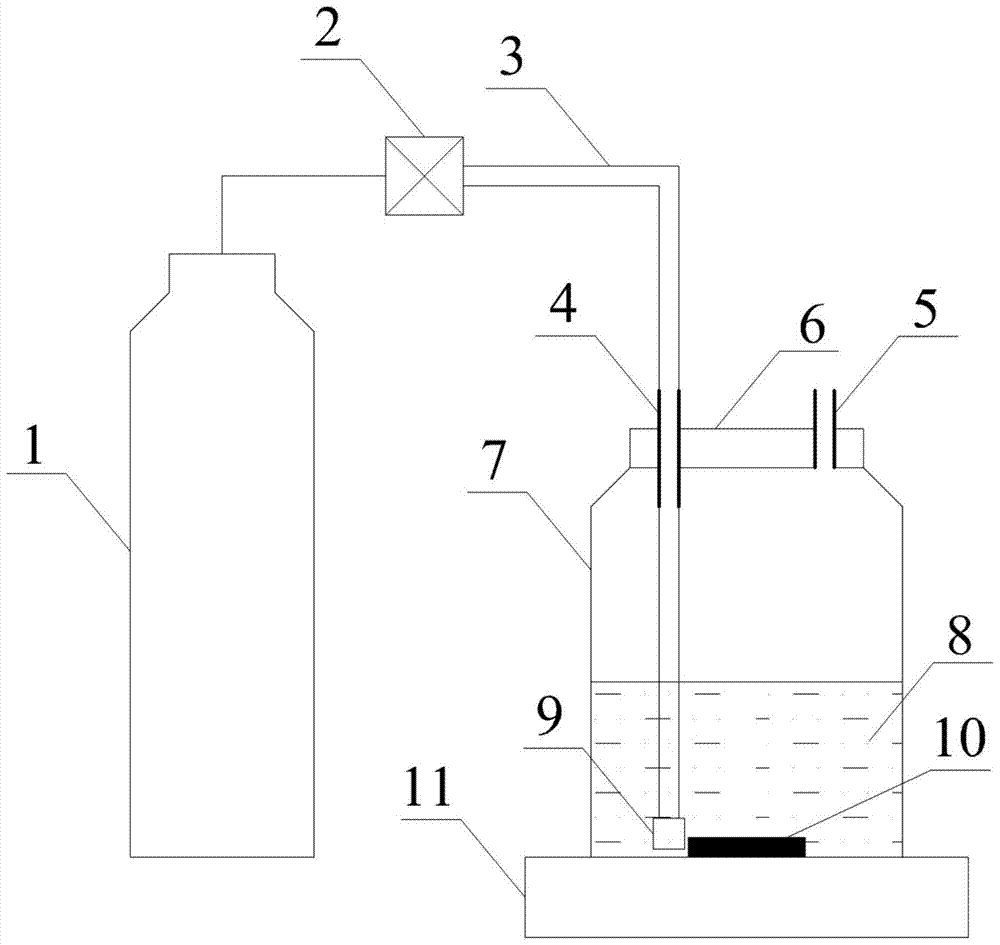
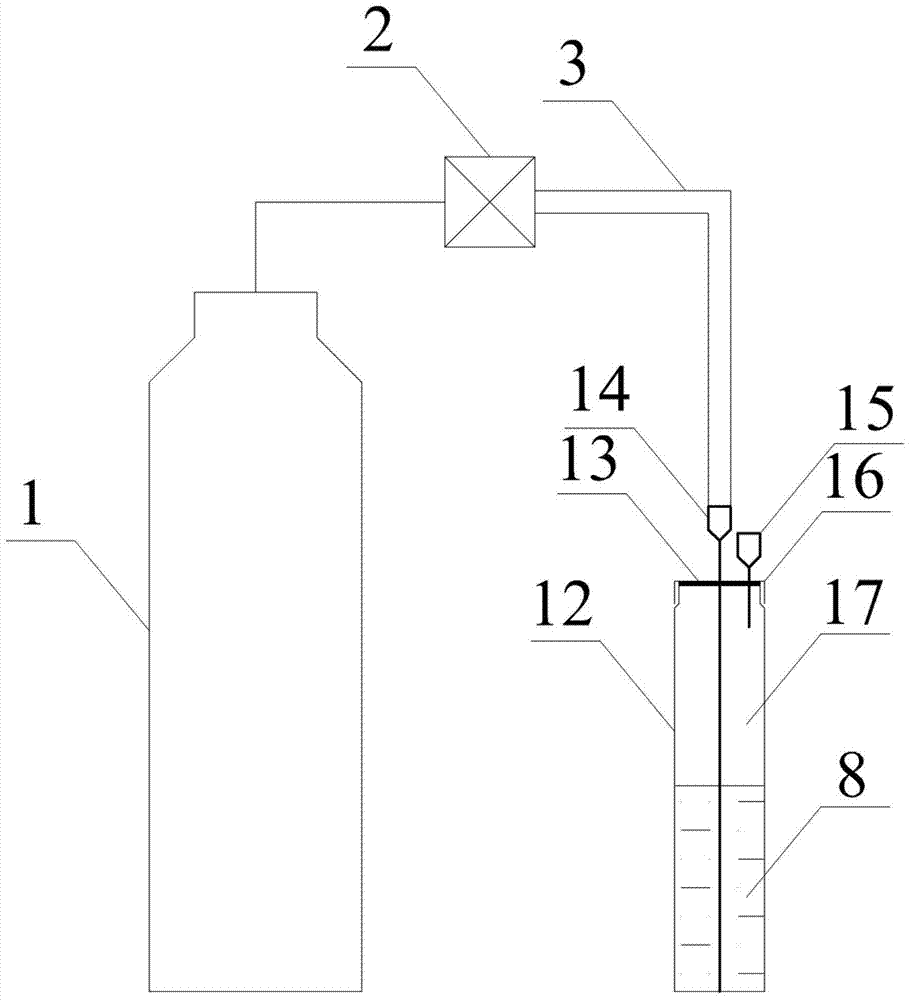
Method used for determining plant total photosynthesis carbon assimilation capacity
ActiveCN105067772AOvercome the lack of ability to exploitEasy accessMaterial analysisCarbon assimilationIsotopic labeling
The invention discloses a method used for determining plant total photosynthesis carbon assimilation capacity. According to the method, double-way isotope labeling and a double source isotope mixed module are adopted to determine using ratio of plant on added bicarbonate ions and using ratio of plant on bicarbonate ions come from atmospheric carbon dioxide dissolving; a photosynthesis device is adopted to determine photosynthesis assimilation capacity of plant on atmospheric carbon dioxide; and plant total photosynthesis carbon assimilation capacity is obtained finally. The method is used for measuring using capacity of plant on added bicarbonate ions, and measuring using capacity of plant on bicarbonate ions come from atmospheric carbon dioxide dissolving; and influences of old carbon on determination results are avoided; so that obtained plant total photosynthesis carbon assimilation capacity data is reliable, and result accuracy is high.
Owner:INST OF GEOCHEM CHINESE ACADEMY OF SCI
Method for determining N-DAMO (nitrite-dependent anaerobic methane oxidation) rate
The invention discloses a method for determining an N-DAMO (nitrite-dependent anaerobic methane oxidation) rate, belongs to the field of environmental research, and discloses a method for determining an N-DAMO rate of soil, sediments or activated sludge by using an isotope tracer technique. The method disclosed by the invention comprises a sample mixing and subpackaging device, an anaerobic environment creation device and an anaerobic environment creation method, a method for pretreating samples before being determined, and a sample determining and calculating method. Under the conditions of high-purity helium protection, samples are uniformly mixed and subpackaged, and then an anaerobic environment is created by using a high-purity helium to blow the sample mixture. After residual NO2<->, NO3<-> and O2 in a reaction system are consumed by pre-culturing, an electron acceptor and a labeled substrate <13>C-CH4 are added into a reactant, and by measuring the contents of <45>CO2 generated by reacting at different time nodes, an N-DAMO rate is calculated. According to the method, equipment is simple and the operation is easy, and a strict anaerobic environment can be created without being operated in an anaerobic tank. An N-DAMO rate is calculated by labeling a reaction substrate by using an isotope and determining a labeled product, and the calculation of the N-DAMO rate is not affected by other physical and chemical changes.
Owner:RES CENT FOR ECO ENVIRONMENTAL SCI THE CHINESE ACAD OF SCI
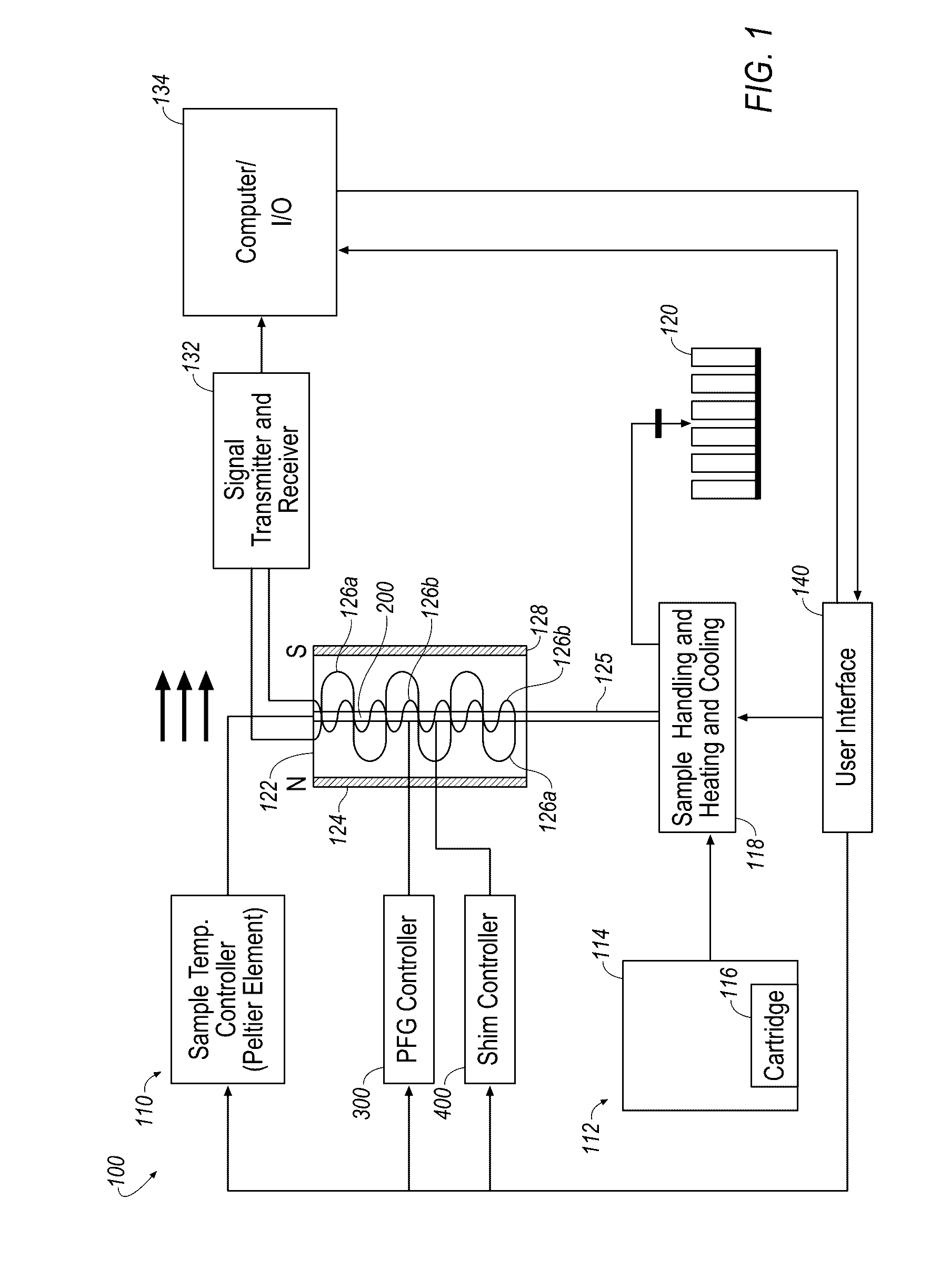
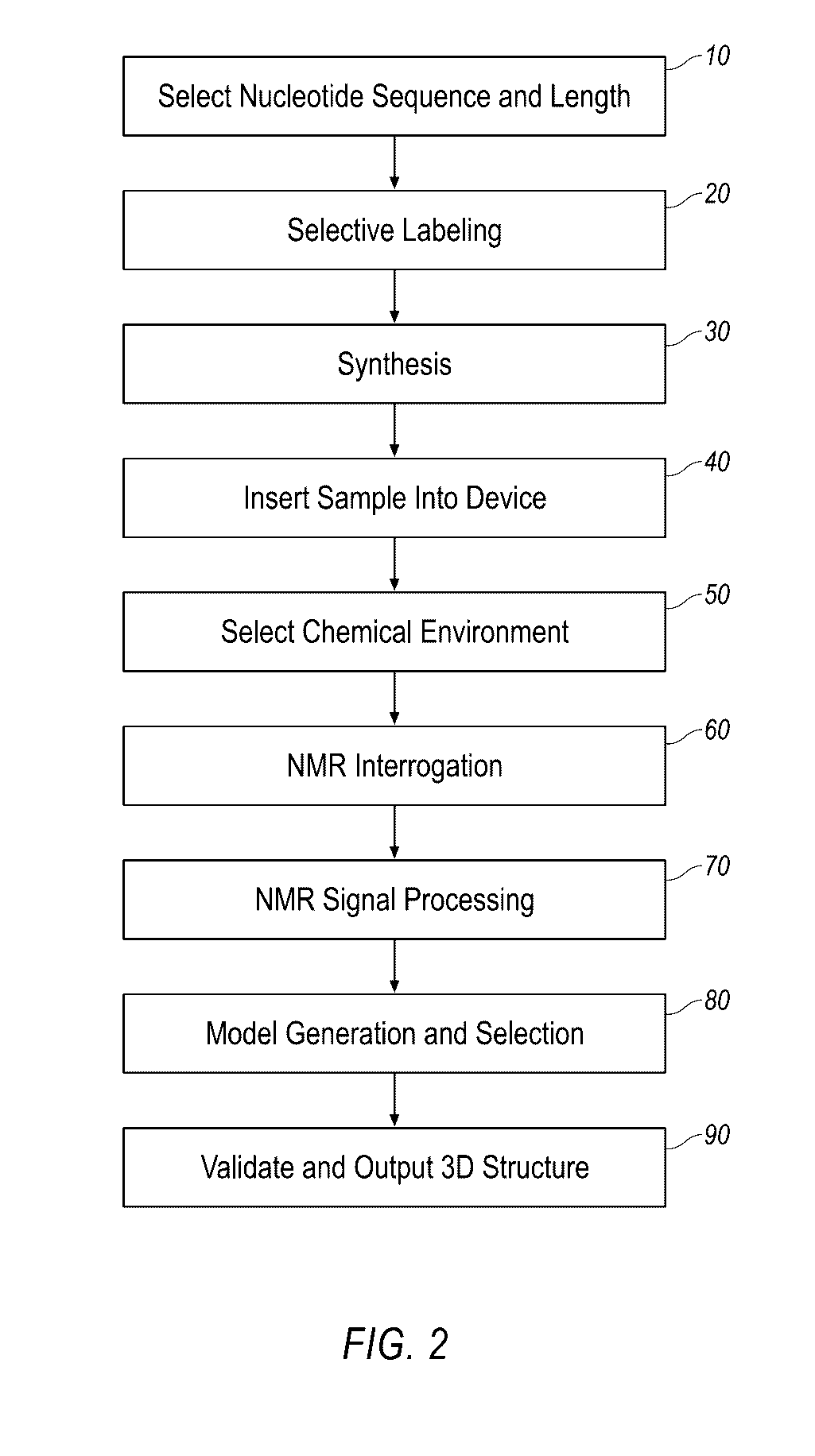
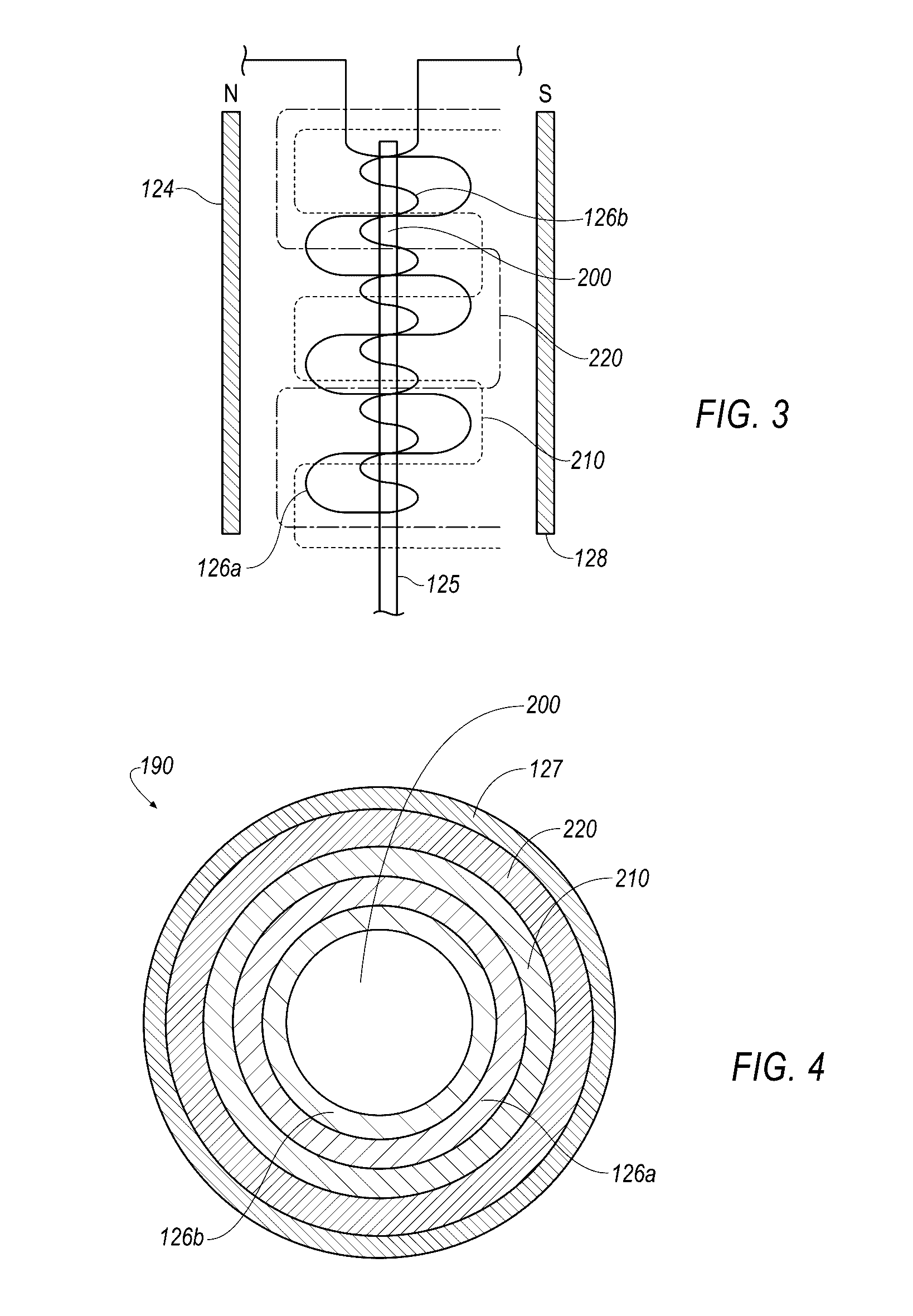
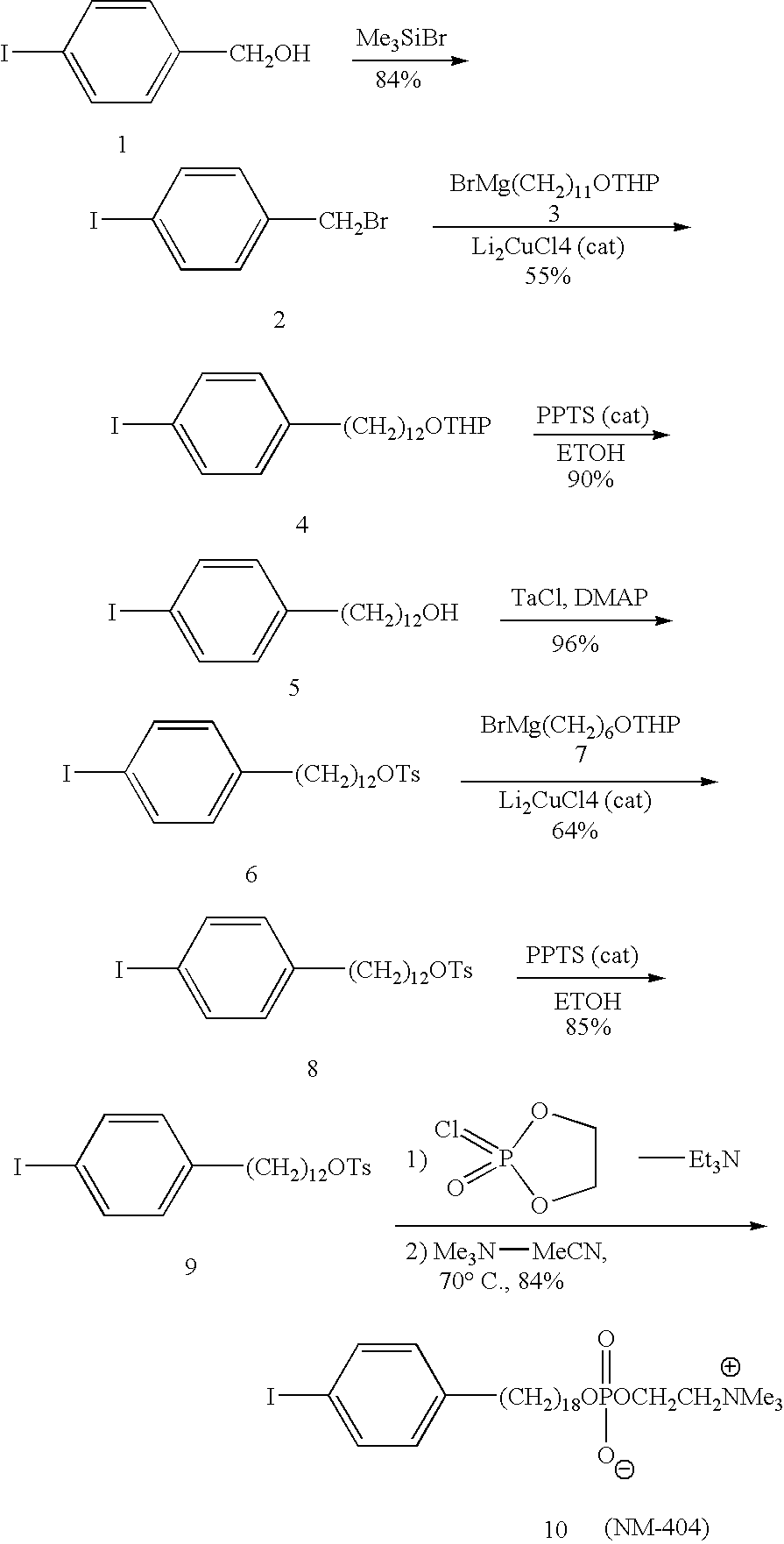
Small molecule binding pockets in nucleic acids
Described herein is technology for determining the 2-D or 3-D atomic resolution structure of a polynucleotide bound to and / or interacting with another molecule, for example a small molecule. In some aspects of the technology, NMR and isotopic labeling strategies are used. The technology described herein is useful for a plurality of applications including but not limited to drug discovery and chemical biology probe discovery.
Owner:NYMIRUM
Virtual colonoscopy with radiolabeled phospholipid ether analogs
ActiveUS20060013767A1Radioactive preparation carriersX-ray constrast preparationsAbnormal tissue growthIsotopic labeling
The present invention provides agents and methods for dual modality virtual colonoscopy that gives both anatomical and functional information using hybrid CT / PET scanning. In preferred embodiments, the present invention provides radiolabeled tumor-specific agents and methods for distinguishing benign polyps from malignant tumors. In further embodiments, the present invention provides compositions and methods useful for distinguishing morphological and functional subregions of a selected region of tissue based on relative levels of phospholipid metabolism. Preferred radiolabeled tumor-specific agents are phospholipid ether analogs labeled with a halogen radiosiotope. In certain preferred embodiments, the compositions including radiolabeled phospholipid ether analogs have therapeutic actions in addition to functionally identifying malignant tissue.
Owner:CELLECTAR
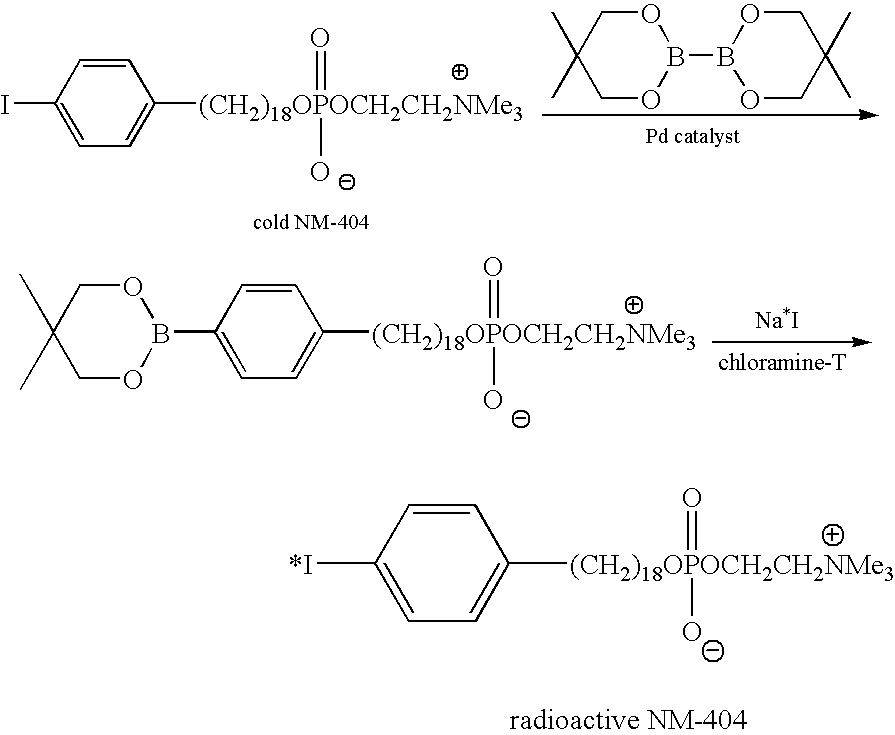
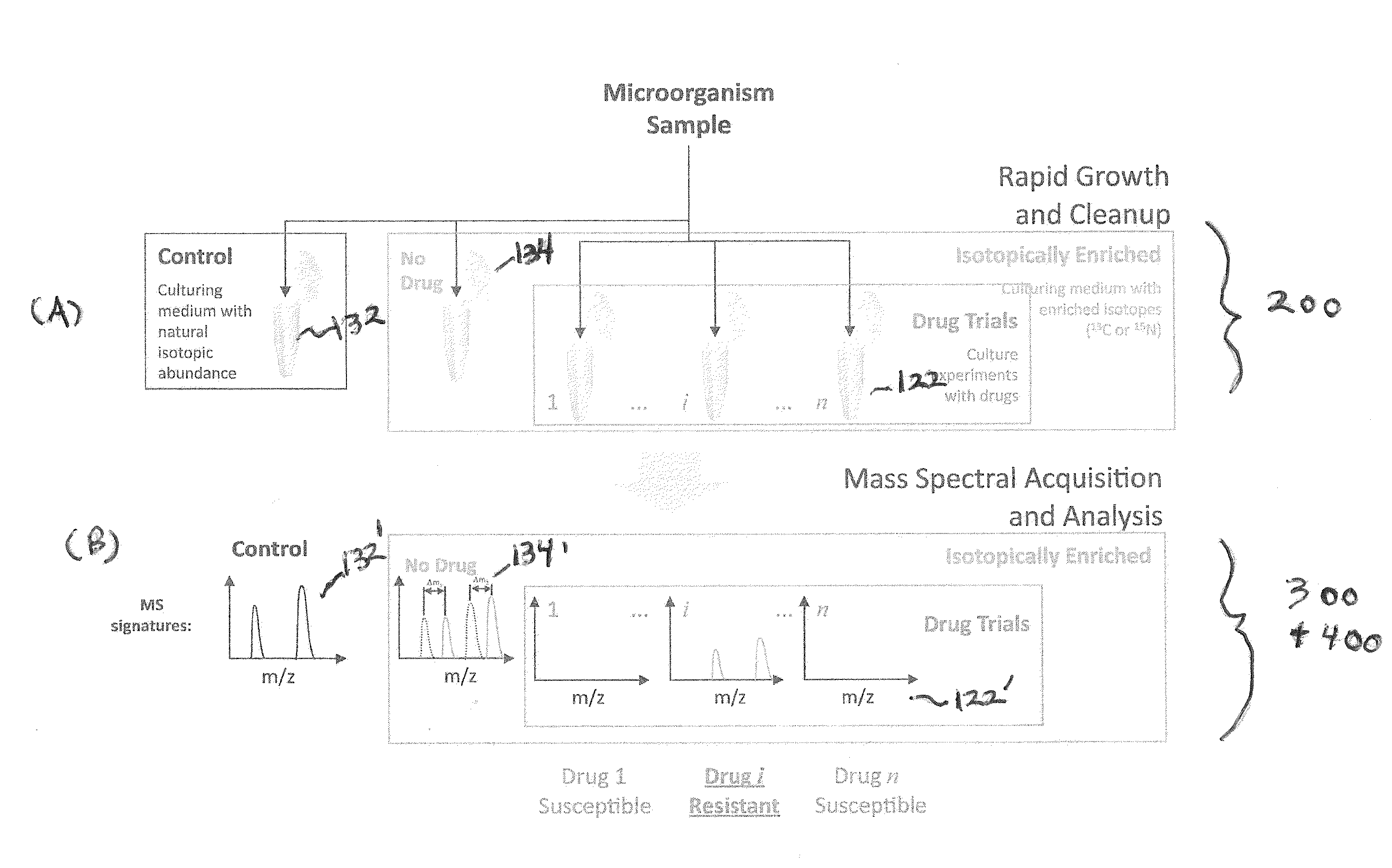
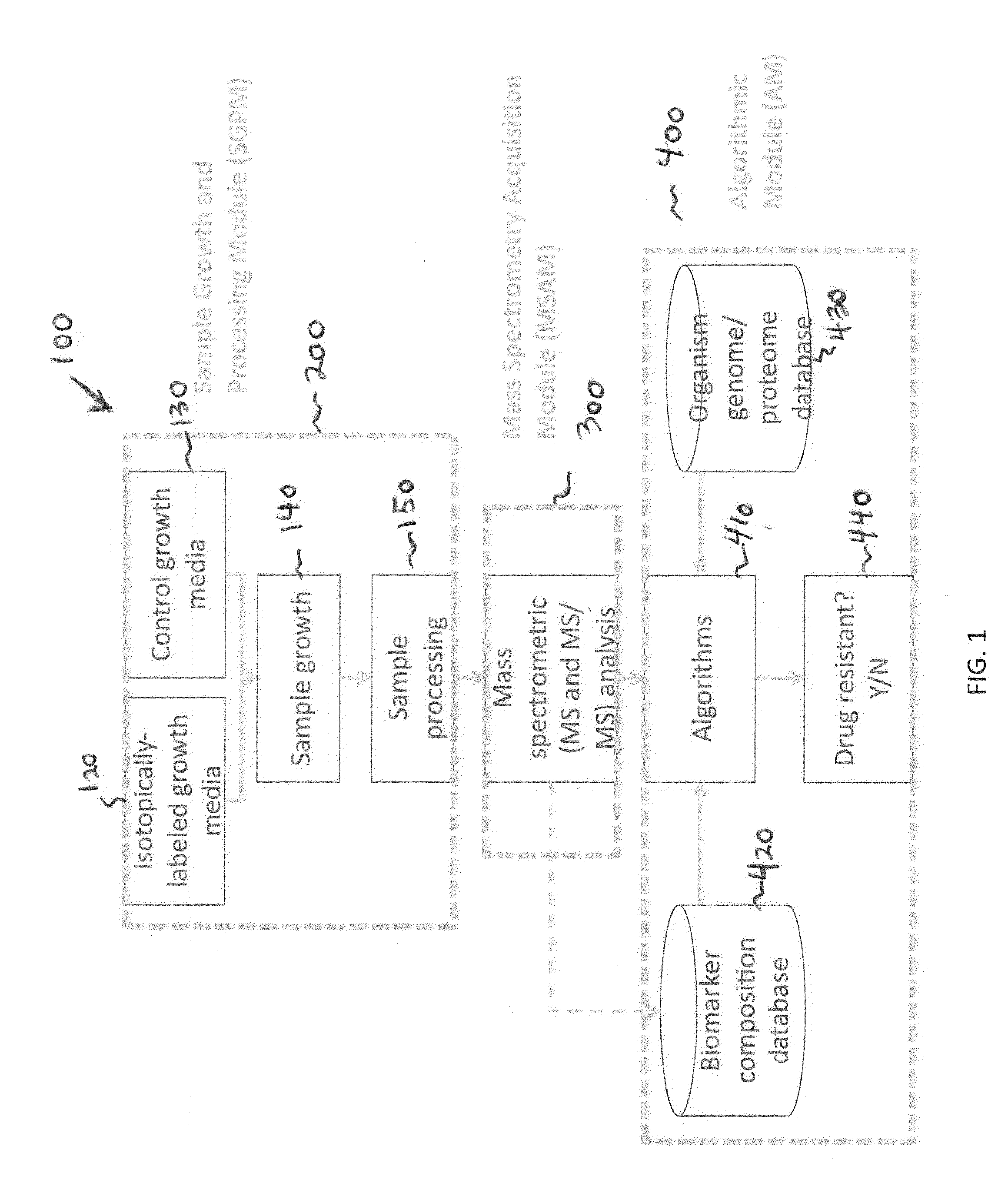
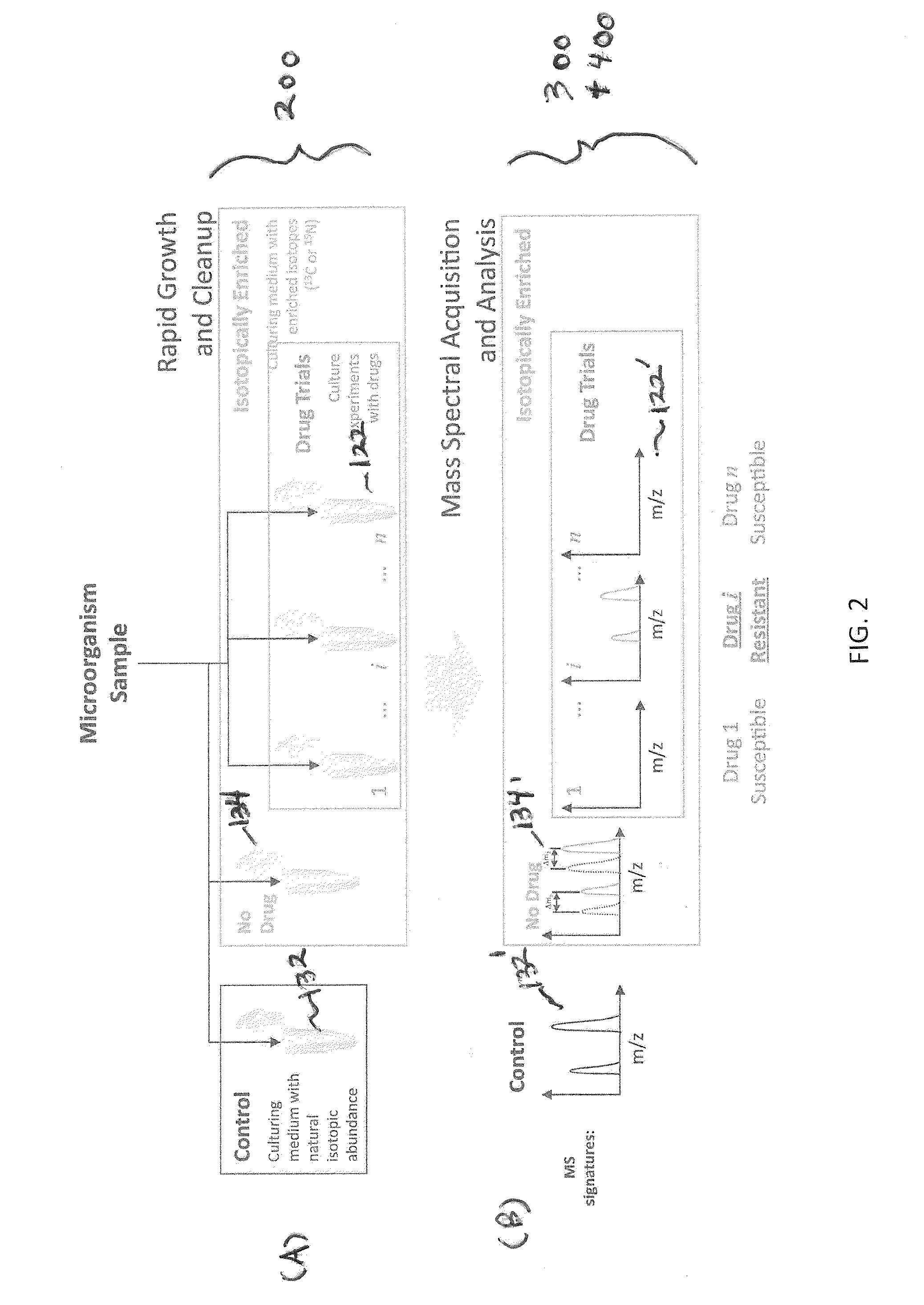
Systems and methods for determining drug resistance in microorganisms
ActiveUS8481281B2Accurate resistanceSludge treatmentVolume/mass flow measurementMicroorganismIsotopic labeling
The present invention is based on the discovery that drug resistance in microorganisms can be rapidly and accurately determined using mass spectrometry. A mass spectrum of an intact microorganism or one or more isolated biomarkers from the microorganism grown in drug containing, isotopically-labeled media is compared with a mass spectrum of the intact microorganism or one or more isolated biomarkers from the microorganism grown in non-labeled media without the drug present. Drug resistance is determined by predicting and detecting a characteristic mass shift of one or more biomarkers using algorithms. The characteristic mass shift is indicative that the microorganism is growing in the presence of the drug and incorporating the isotopic label into the one or more biomarkers, resulting in change in mass.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
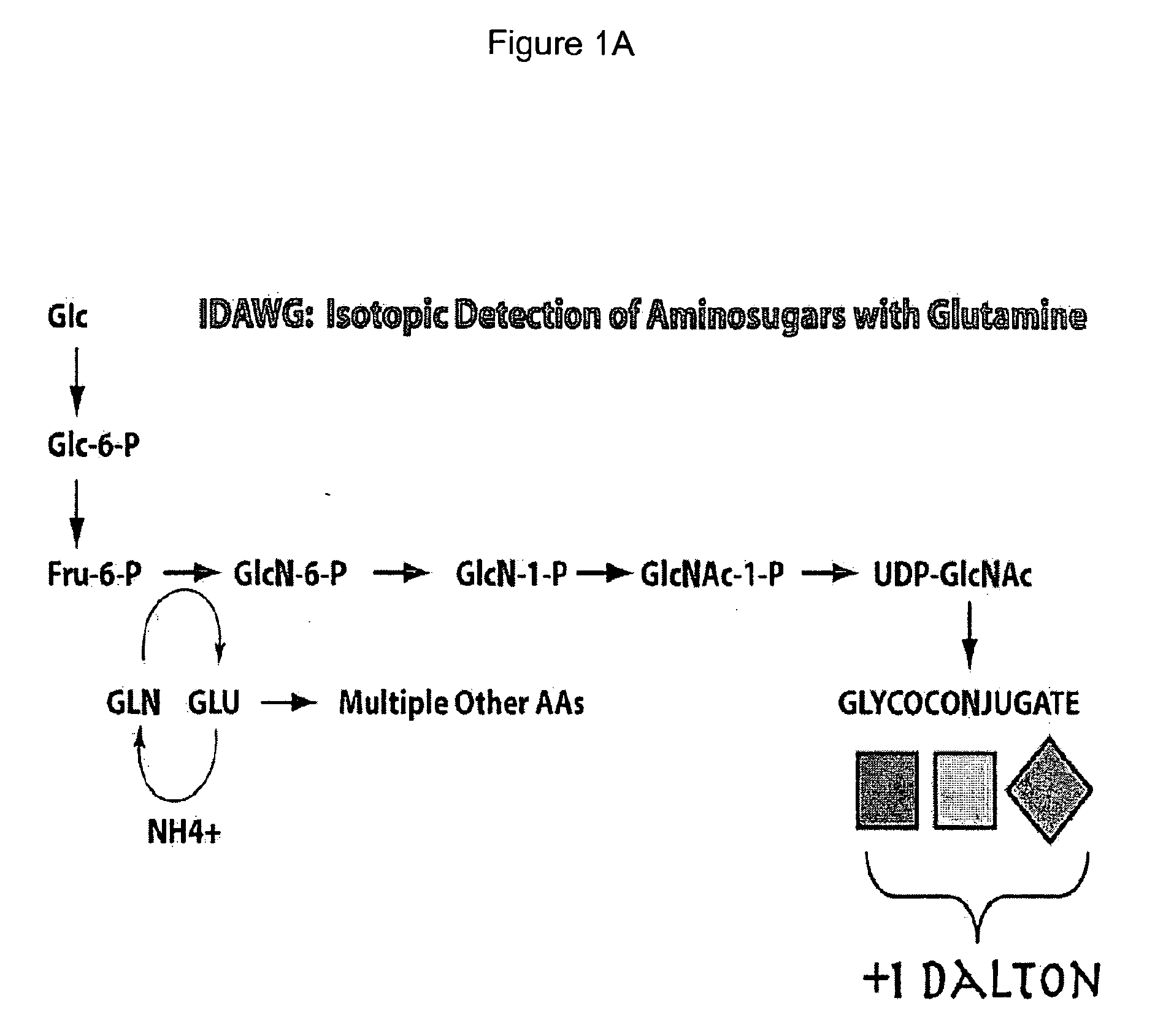
Vivo isotopic labeling method for quantitative glycomics
InactiveUS20100297609A1High analysisImprove throughputMicrobiological testing/measurementAssay labelsBiological cellIsotopic labeling
The present invention relates to a method of isotopically labeling glycans and in facilitating high throughput quantitative / comparative analysis of glycomic compositions of biological cells. The method is applicable inter alia for identifying differentiated cells and their glycomic characteristics, differentiation conditions, disease and / or therapeutic progression, diagnosing disease states, determining drug activity, establishing manufacturing efficiencies and for determining the half-life of glycans in cells.
Owner:UNIV OF GEORGIA RES FOUND INC
Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics
ActiveUS20130078728A1High quantitation efficacyLow costOrganic chemistryBiological testingCombinatorial chemistryIsotope
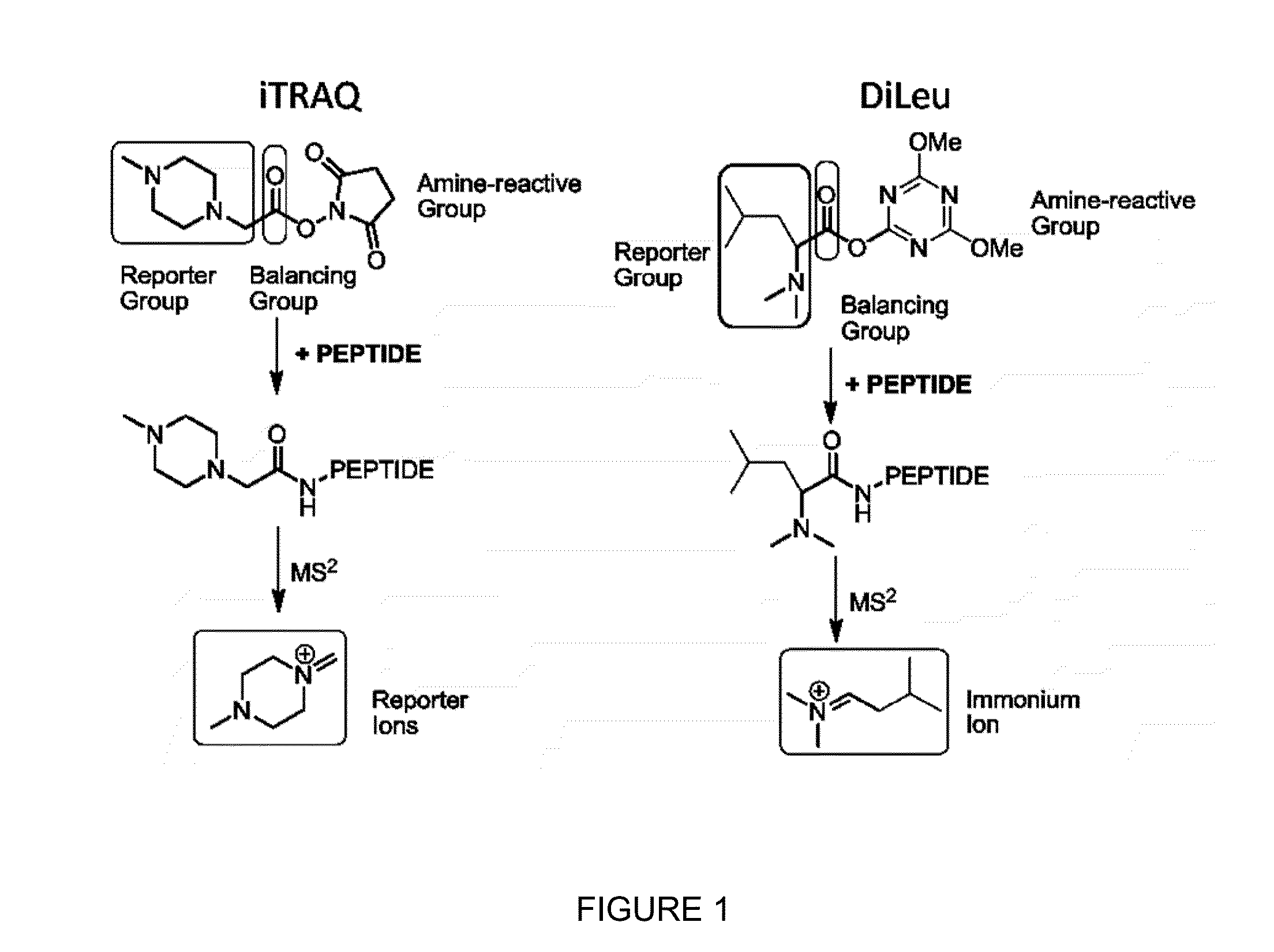
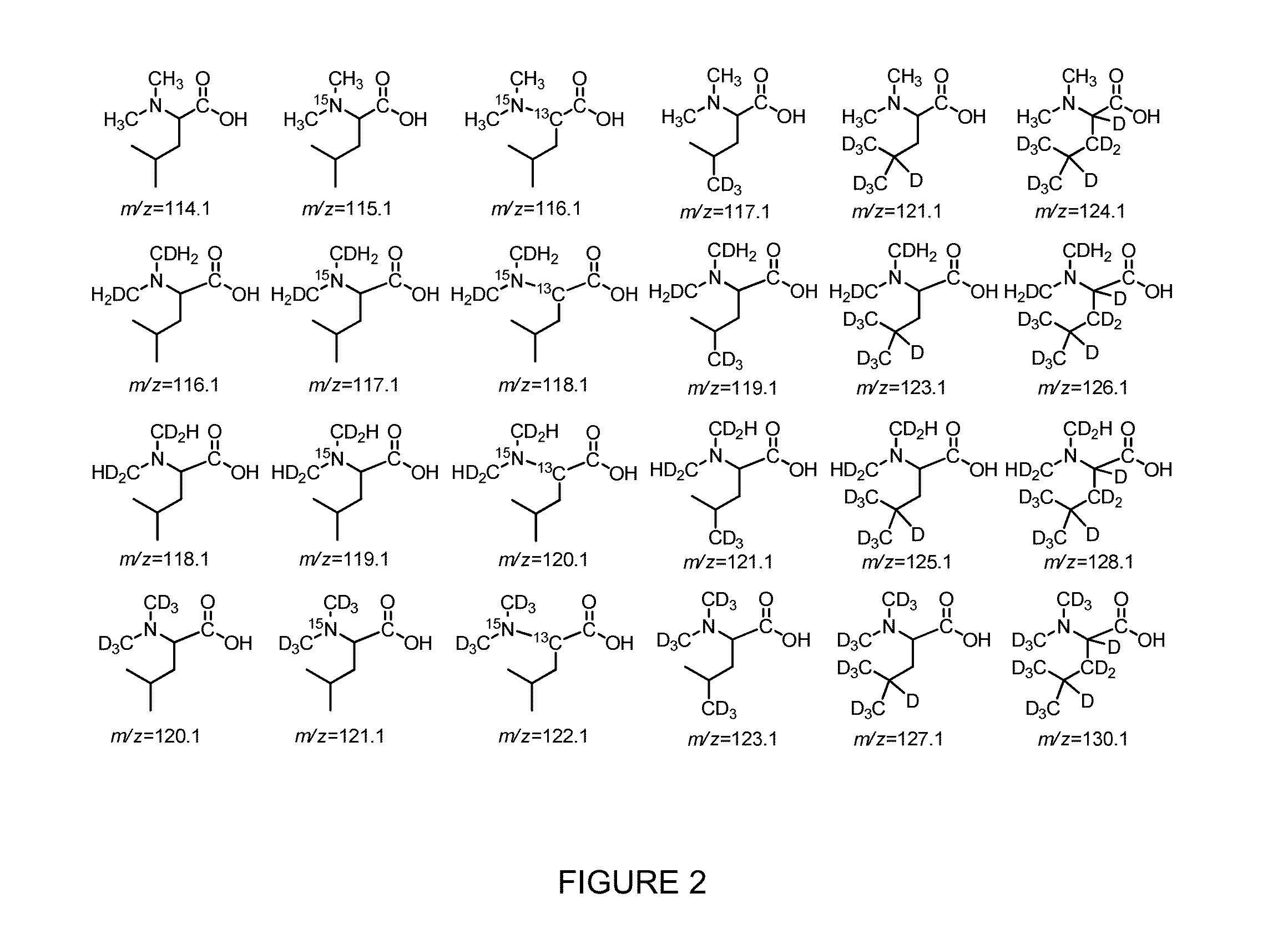
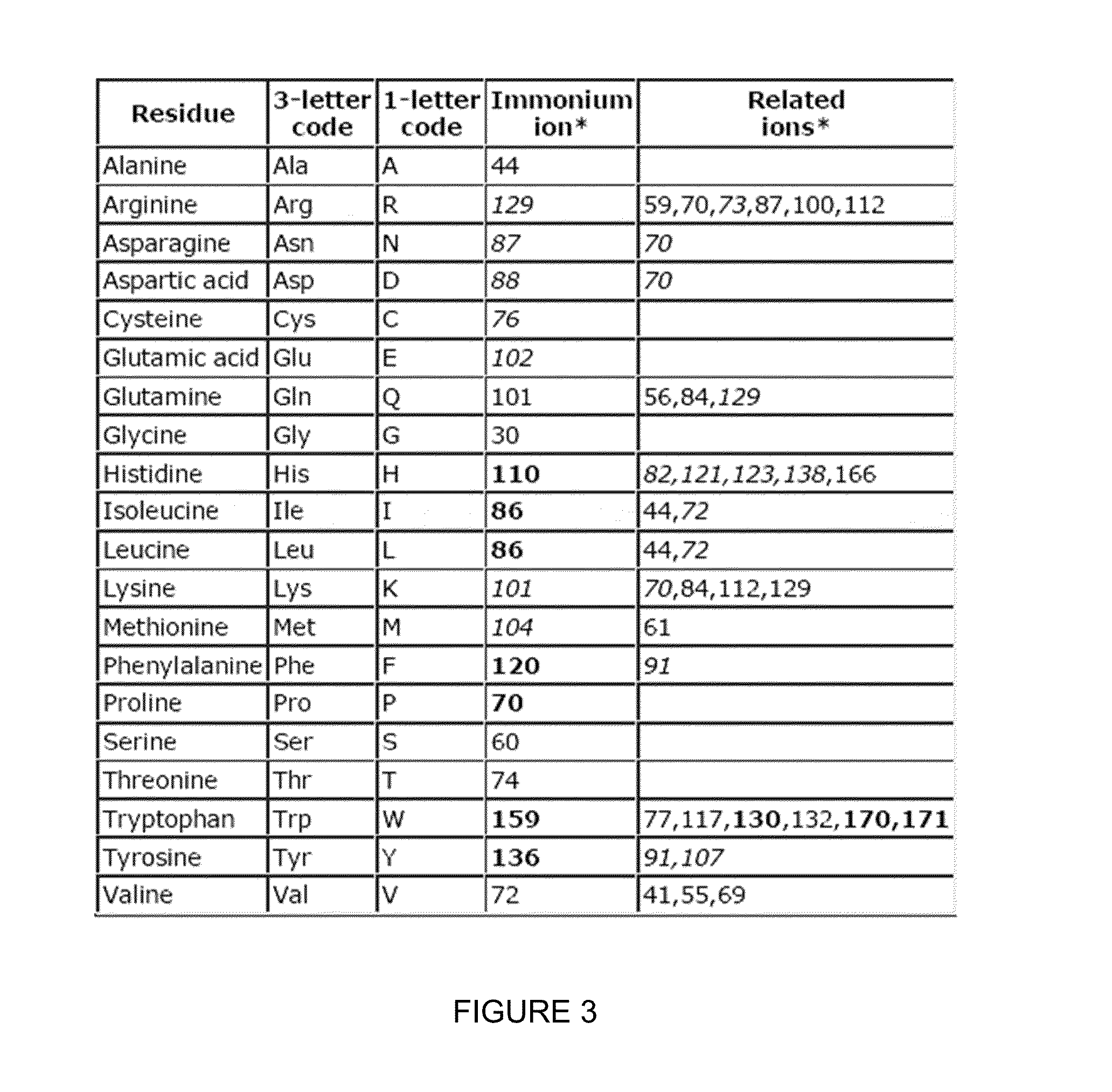
Compositions and methods of tagging peptides and other molecules using novel isobaric tandem mass tagging reagents, including novel N,N-dimethylated amino acid 8-plex and 16-plex isobaric tandem mass tagging reagents. The tagging reagents comprise: a) a reporter group having at least one atom that is optionally isotopically labeled; b) a balancing group, also having at least one atom that is optionally isotopically labeled, and c) an amine reactive group. The tagging reagents disclosed herein serve as attractive alternatives for isobaric tag for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMTs) due to their synthetic simplicity, labeling efficiency and improved fragmentation efficiency.
Owner:WISCONSIN ALUMNI RES FOUND
Method for determining content of beta-lactoglobulin in milk powder
The invention discloses a method for determining the content of beta-lactoglobulin in milk powder. The method comprises the following steps: (1) selecting TK-11 as a specific peptide fragment and conducting beta-lactoglobulin quantification; (2) synthesizing the peptide fragment, and conducting isotope labeling on the peptide fragment; (3) accurately quantifying the synthesized standard peptide fragment by adopting an isotope dilution mass spectrography; (4) conducting sample treatment and enzyme digestion; (5) hygroplasm analysis, namely filtering the solution after enzyme digestion through a filter film, and then carrying out selective ion scanning analysis on the filtrate by adopting mass spectrography; and (6) calculating according to a formula, so as to complete the determination on the content of beta-lactoglobulin in milk powder. By adopting the method, determination results are accurate and reasonable.
Owner:NAT INST OF METROLOGY CHINA +1
Use of isotopes to provide information to an article
InactiveUS20040106205A1Material analysis using wave/particle radiationPaper-money testing devicesIsotopic labelingSpectroscopy
The present invention relates to a method for associating information with an article, an isotopic labeling composition to label an article with information using at least two different isotopes of an element, and an article labeled with the labeling composition. In the labeling composition, the isotopes of the labeling composition have an abundance ratio that is detectably different from the natural abundance ratio thereof. By detecting the abundance ratio(s) of the isotopes in the labeling composition, via laser ablation / magnetic spectroscopy, an NQR spectrometer, an NMR spectrometer, an IR spectrometer, or a microwave spectrometer, information about the article can be determined. The invention may be used to identify, authenticate or determine the source or origin of an article or to provide detailed information about an article.
Owner:STEVENSON NIGEL R +3
Nuclear magnetic resonance measuring method
ActiveUS20100225314A1Reduce impactImprove signal-to-noise ratioElectric/magnetic detectionMeasurements using NMRNMR - Nuclear magnetic resonanceIsotopic labeling
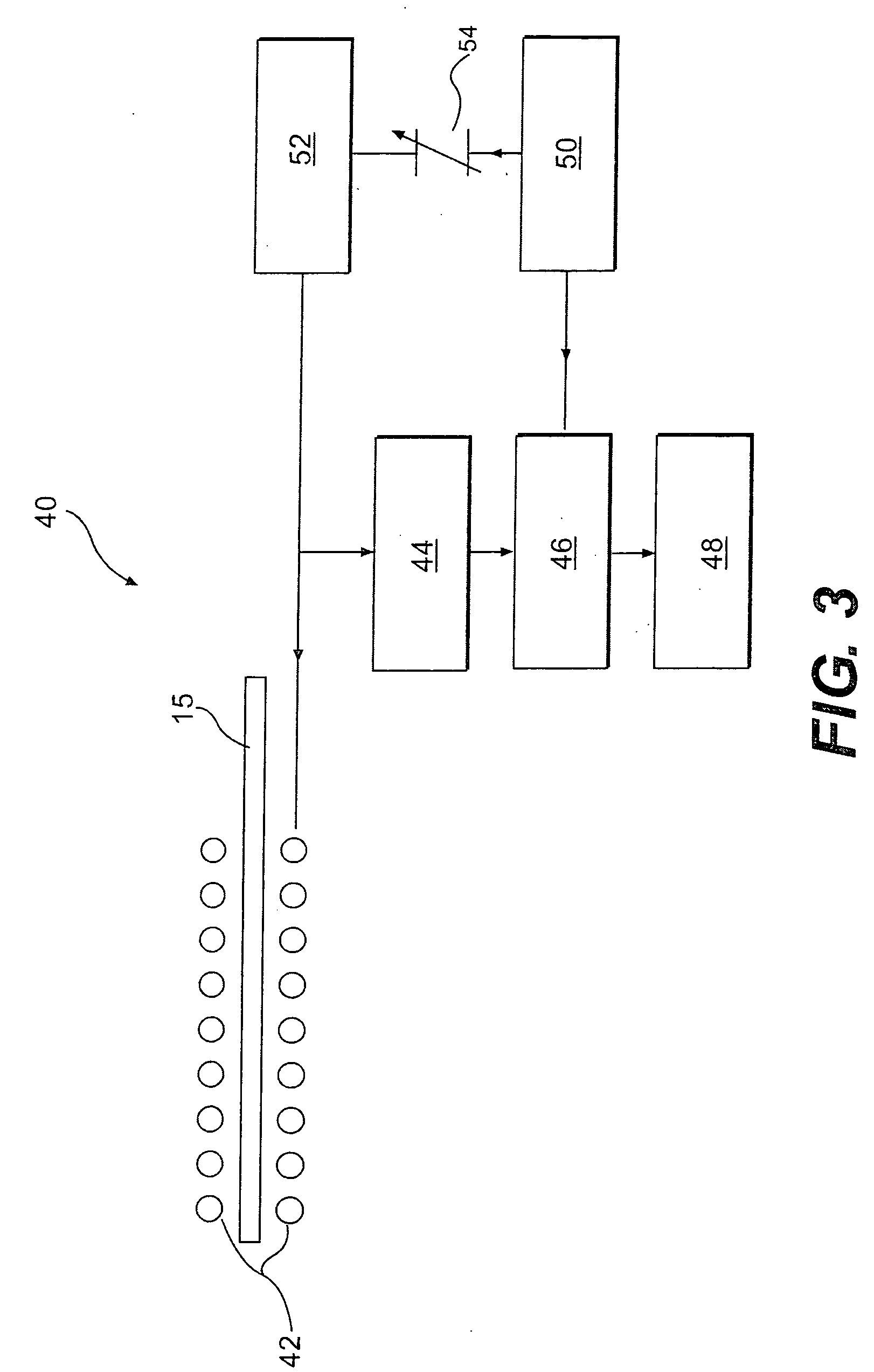
The present invention provides a method for measuring nuclear magnetic resonance that employs a compound in which a plurality of nuclei is labeled with isotopes as a probe agent, highly selectively and highly sensitively obtains a nuclear magnetic resonance signal of the above described probe agent, and can attach a spatial positional information to the above described nuclear magnetic resonance signal, and an apparatus therefore.
Owner:CANON KK
Acid-labile isotope-coded extractant (ALICE) and its use in quantitative mass spectrometric analysis of protein mixtures
InactiveUS6902936B2Improve purification effectEasy to analyze and useAnalysis using chemical indicatorsMaterial analysis using wave/particle radiationDual columnBiopolymer
The method of the invention provides novel compounds, termed acid-labile isotope-coded extractants (ALICE), for quantitative mass spectrometric analysis of protein mixtures. The compounds contain a thiol-reactive group that is used to capture cysteine-containing peptides from all peptide mixtures, an acid-labile linker, and a non-biological polymer. One of the two acid-labile linkers is isotopically labeled and therefore enables the direct quantitation of peptides / proteins through mass spectrometric analysis. Because no functional proteins are required to capture peptides, a higher percentage of organic solvent can be used to solubilize the peptides, particularly hydrophobic peptides, through the binding, washing and eluting steps, thus permitting much better recovery of peptides. Moreover, since the peptides are covalently linked to the non-biological polymer (ALICE), more stringent washing is allowed in order to completely remove non-specifically bound species. Finally, peptides captured by ALICE are readily eluted from the polymer support under mild acidic condition with high yield and permit the direct down stream mass spectrometric analysis without any further sample manipulation. In combination with our novel dual column two dimensional liquid chromatography-mass spectrometry (2D-LC-MS / MS) design, the ALICE procedure proves to a general approach for quantitative mass spectrometric analysis of protein mixtures with better dynamic range and sensitivity.
Owner:GENETICS INST INC
Use of isotopes to provide information to an article
InactiveUS20030030558A1Digital data processing detailsBurglar alarm mechanical actuationIsotopic labelingSpectroscopy
The present invention relates to a method for associating information with an article, an isotopic labeling composition to label an article with information using at least two different isotopes of an element, and an article labeled with the labeling composition. In the labeling composition, the isotopes of the labeling composition have an abundance ratio that is detectably different from the natural abundance ratio thereof. By detecting the abundance ratio(s) of the isotopes in the labeling composition, via laser ablation / magnetic spectroscopy, an NQR spectrometer, an NMR spectrometer, an IR spectrometer, or a microwave spectrometer, information about the article can be determined. The invention may be used to identify, authenticate or determine the source or origin of an article or to provide detailed information about an article.
Owner:THERAGENICS CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com