Patents
Literature
49 results about "Protein expression profile" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Summary Protein expression profiling is defined in general as identifying the proteins expressed in a particular tissue, under a specified set of conditions and at a particular time, usually compared to expression in reference samples.
Protein expression profiling
InactiveUS6921642B2Easy to detectBioreactor/fermenter combinationsBiological substance pretreatmentsAnalyteProtein expression profile
Disclosed are compositions and methods for detecting small quantities of analytes such as proteins and peptides. The method involves associating a primer with an analyte and subsequently using the primer to mediate rolling circle replication of a circular DNA molecule. Amplification of the DNA circle is dependent on the presence of the primer. Thus, the disclosed method produces an amplified signal, via rolling circle amplification, from any analyte of interest. The amplified DNA remains associated with the analyte, via the primer, and so allows spatial detection of the analyte. The disclosed method can be used to detect and analyze proteins and peptides. Multiple proteins can be analyzed using microarrays to which the various proteins are immobilized. A rolling circle replication primer is then associated with the various proteins using a conjugate of the primer and a molecule that specifically binds the proteins to be detectable. Rolling circle replication from the primers results in production of a large amount of DNA at the sites in the array where the proteins are immobilized. The amplified DNA serves as a readily detectable signal for the proteins. The disclosed method can also be used to compare the proteins expressed in two or more different samples. The information generated is analogous to the type of information gathered in nucleic acid expression profiles. The disclosed method allows sensitive and accurate detection and quantitation of proteins expressed in any cell or tissue.
Owner:QIAGEN GMBH
Process for analyzing protein samples
InactiveUS7045296B2Reduce disulfide bondEasy labelingSamplingMicrobiological testing/measurementProtein expression profileUnit operation
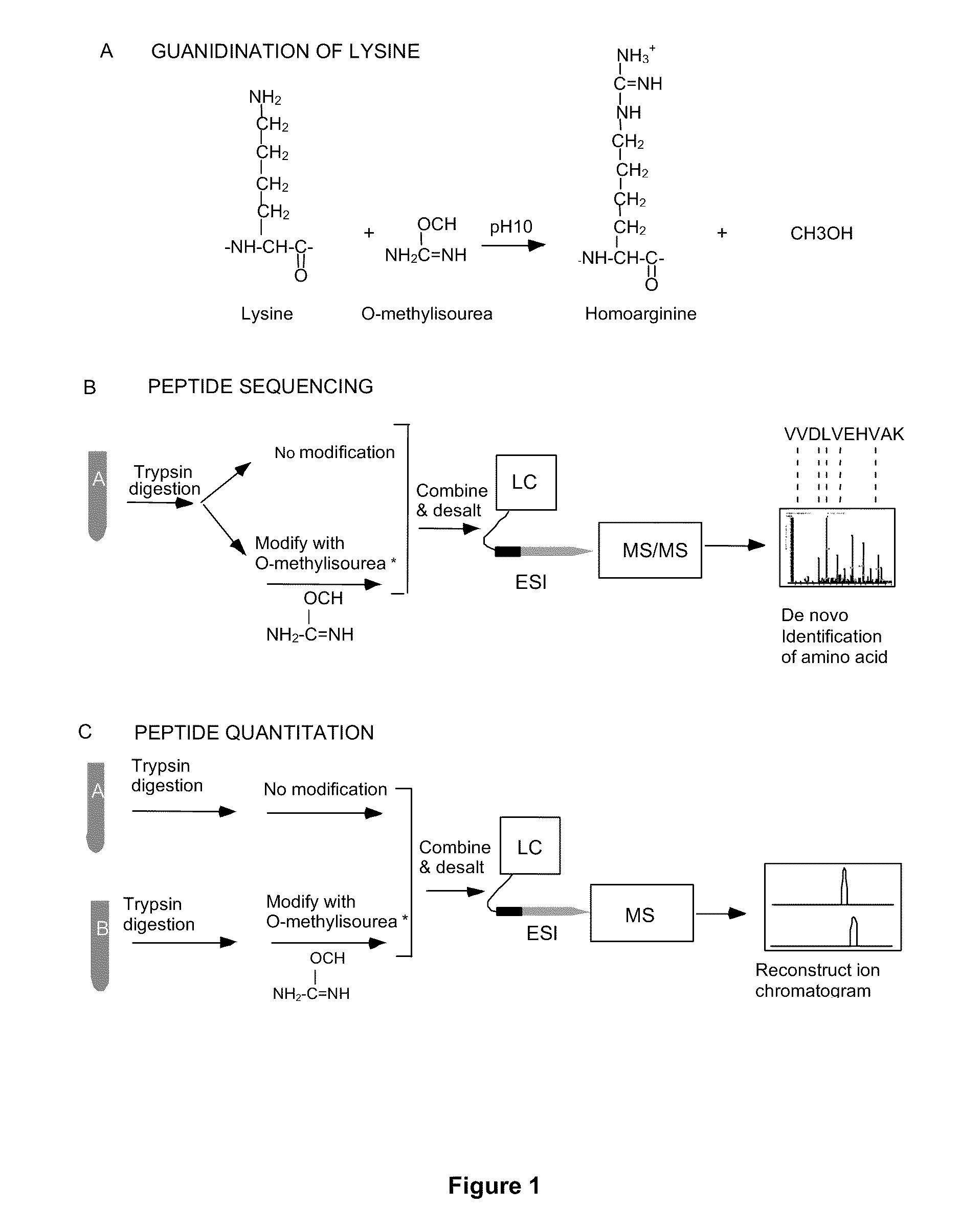
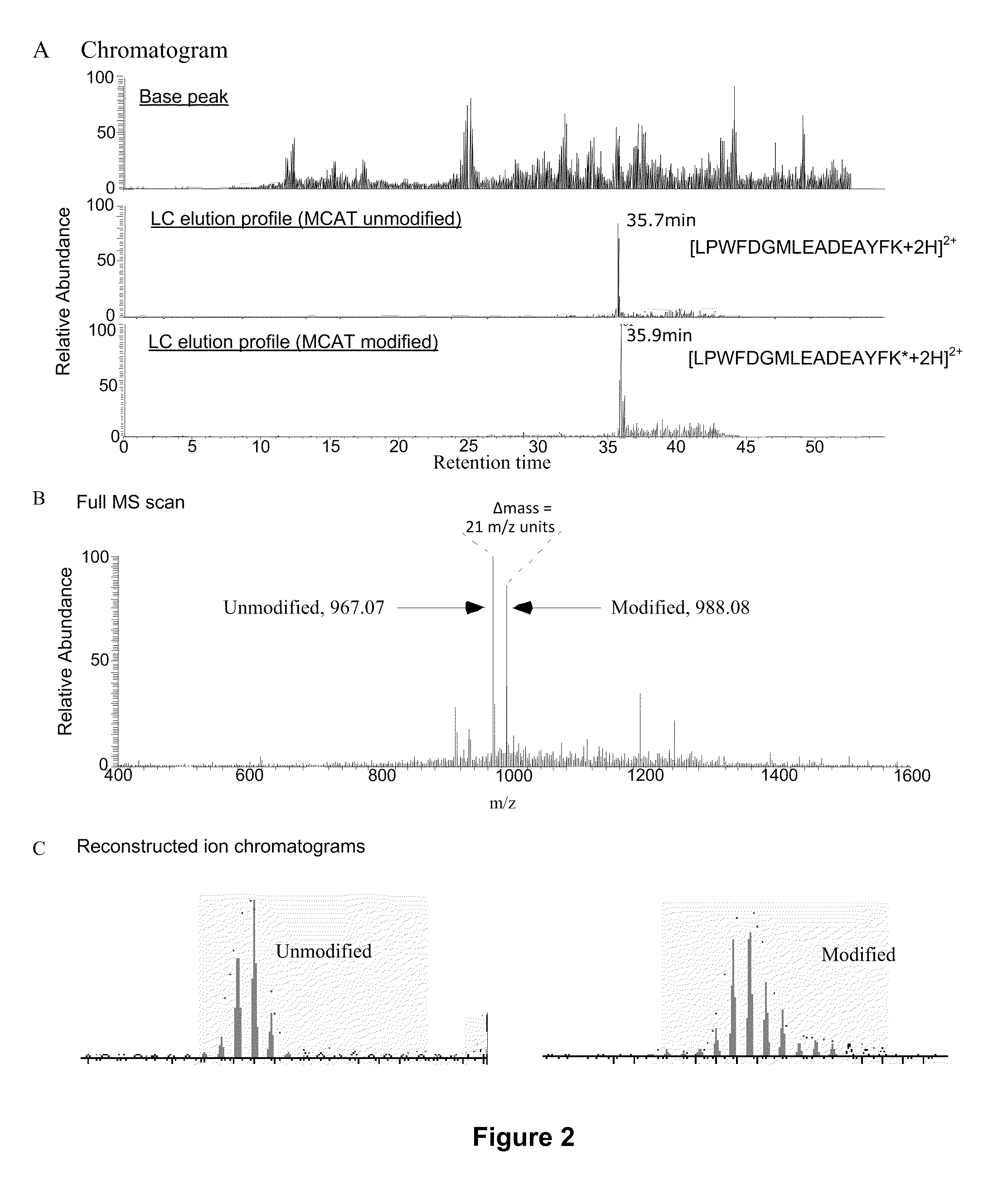
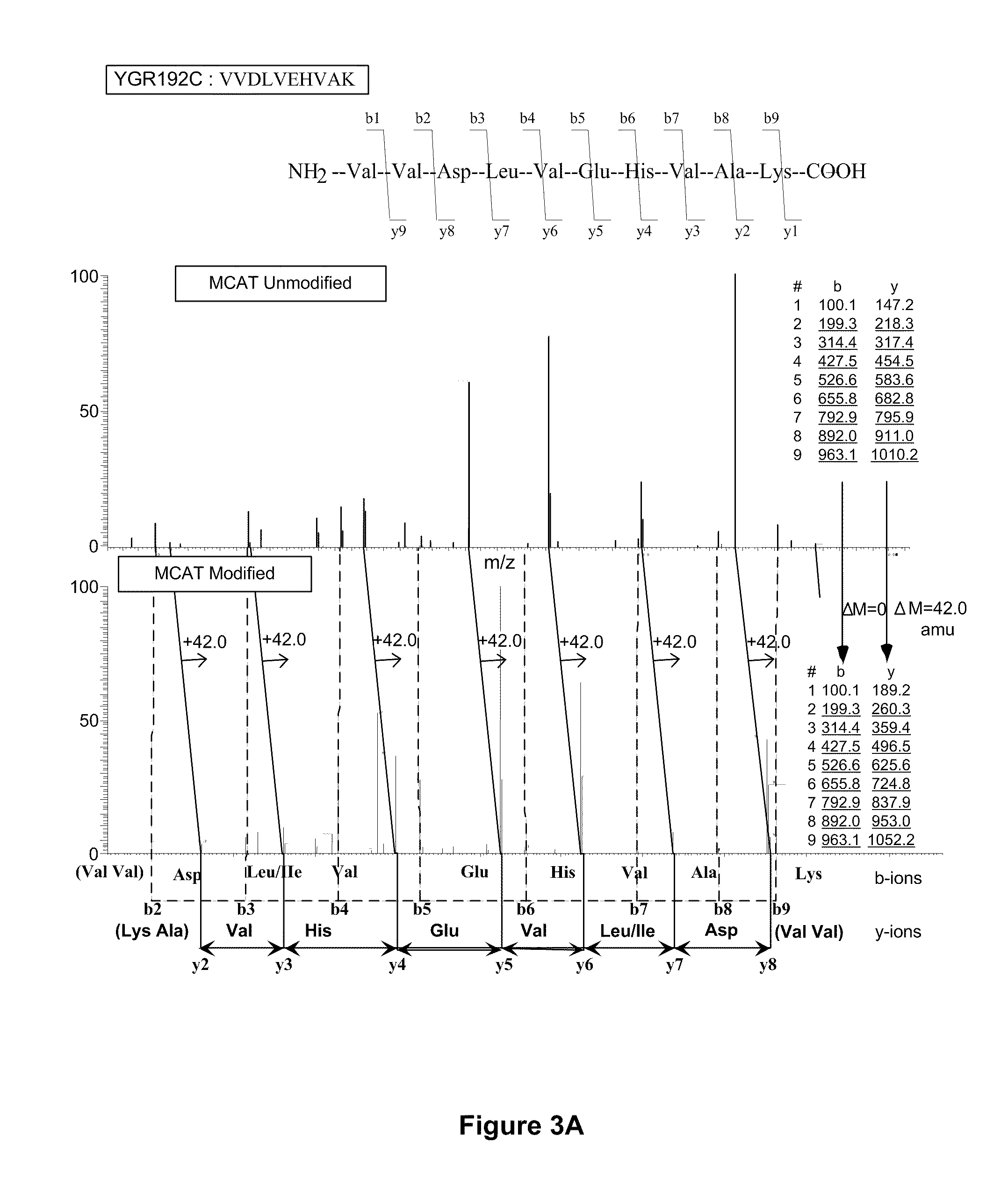
Methods using gel electrophoresis and mass spectrometry for the rapid, quantitative analysis of proteins or protein function in mixtures of proteins derived from two or more samples in one unit operation are disclosed. In one embodiment the method includes (a) preparing an extract of proteins from each of at least two different samples; (b) providing a set of substantially chemically identical and differentially isotopically labeled protein reagents; (c) reacting the extract of proteins from different samples of step (a) with a different isotopically labeled reagent from the set of step (b) to provide two or more sets of isotopically differentially labeled proteins; (d) mixing each of the two or more sets of isotopically labeled proteins to form a single mixture of isotopically differentially labeled proteins; (e) electrophoresing the mixture of step (d) by an electrophoresing method capable of separating proteins within the mixture; (f) digesting at least a portion of one or more separated proteins of step (e) and (g) detecting the difference in the expression levels of the proteins in the two samples by mass spectrometry based on one or more peptides in the sample of labeled peptides. The analytical method can be used for qualitative and particularly for quantitative analysis of global protein expression profiles in cells and tissues, i.e. the quantitative analysis of proteomes.
Owner:DH TECH DEVMENT PTE +1
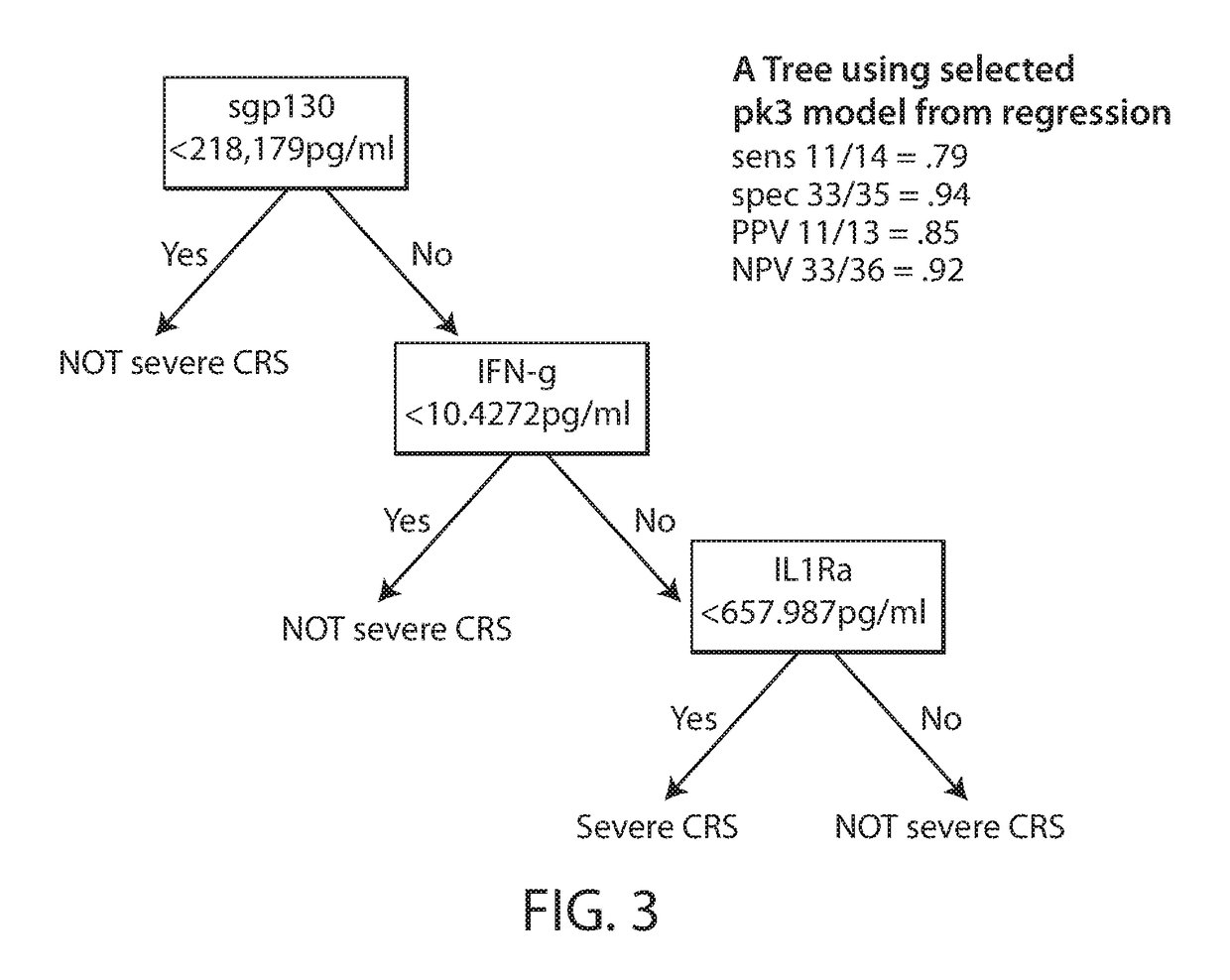
Biomarkers predictive of cytokine release syndrome
ActiveUS20180252727A1Reduce riskHigh activityOrganic active ingredientsAntipyreticAnalyteProtein expression profile
Owner:NOVARTIS AG +1
Protein expression profile database
InactiveUS20050048564A1Peptide librariesLibrary screeningSequence databaseProtein expression profile
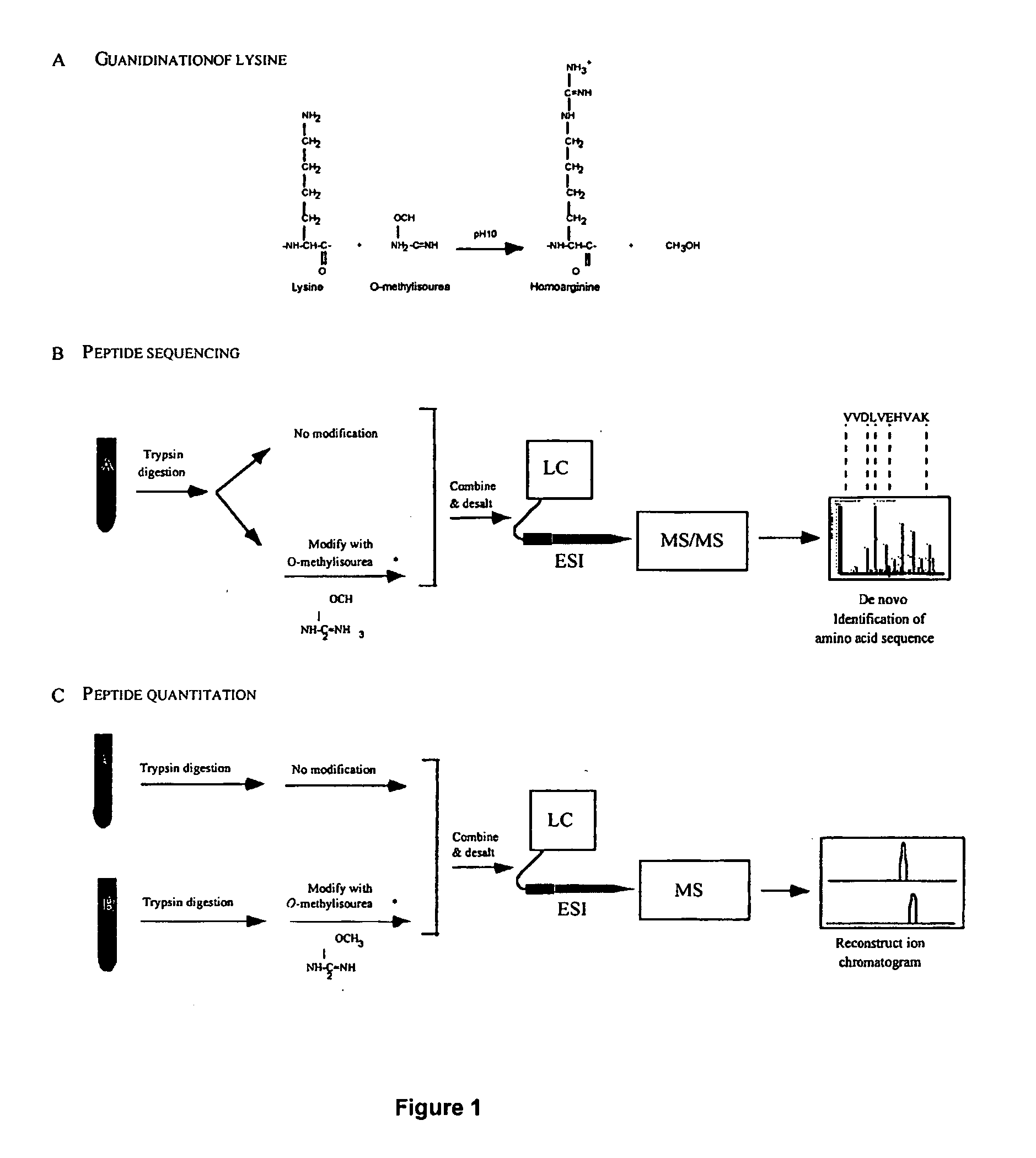
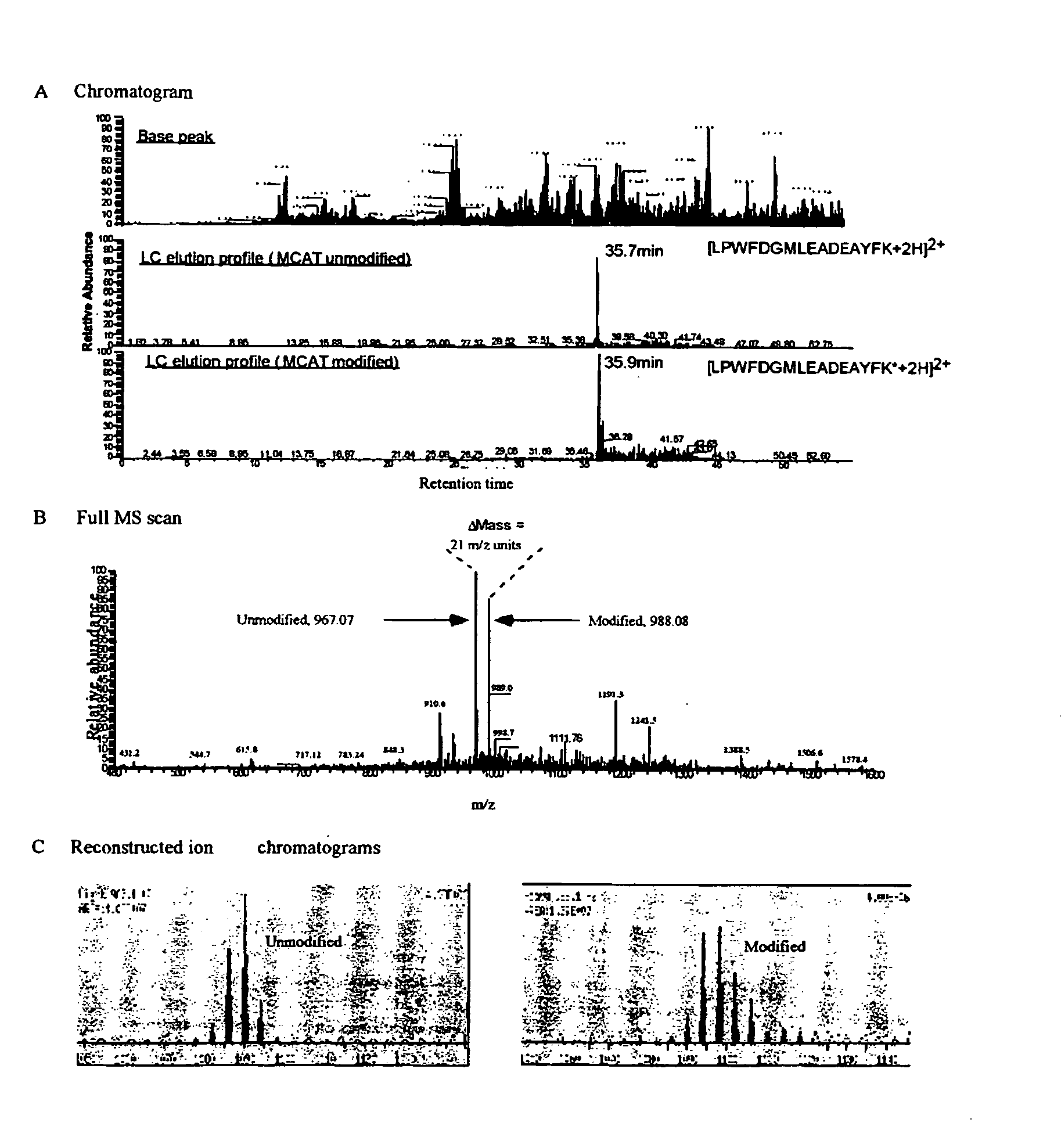
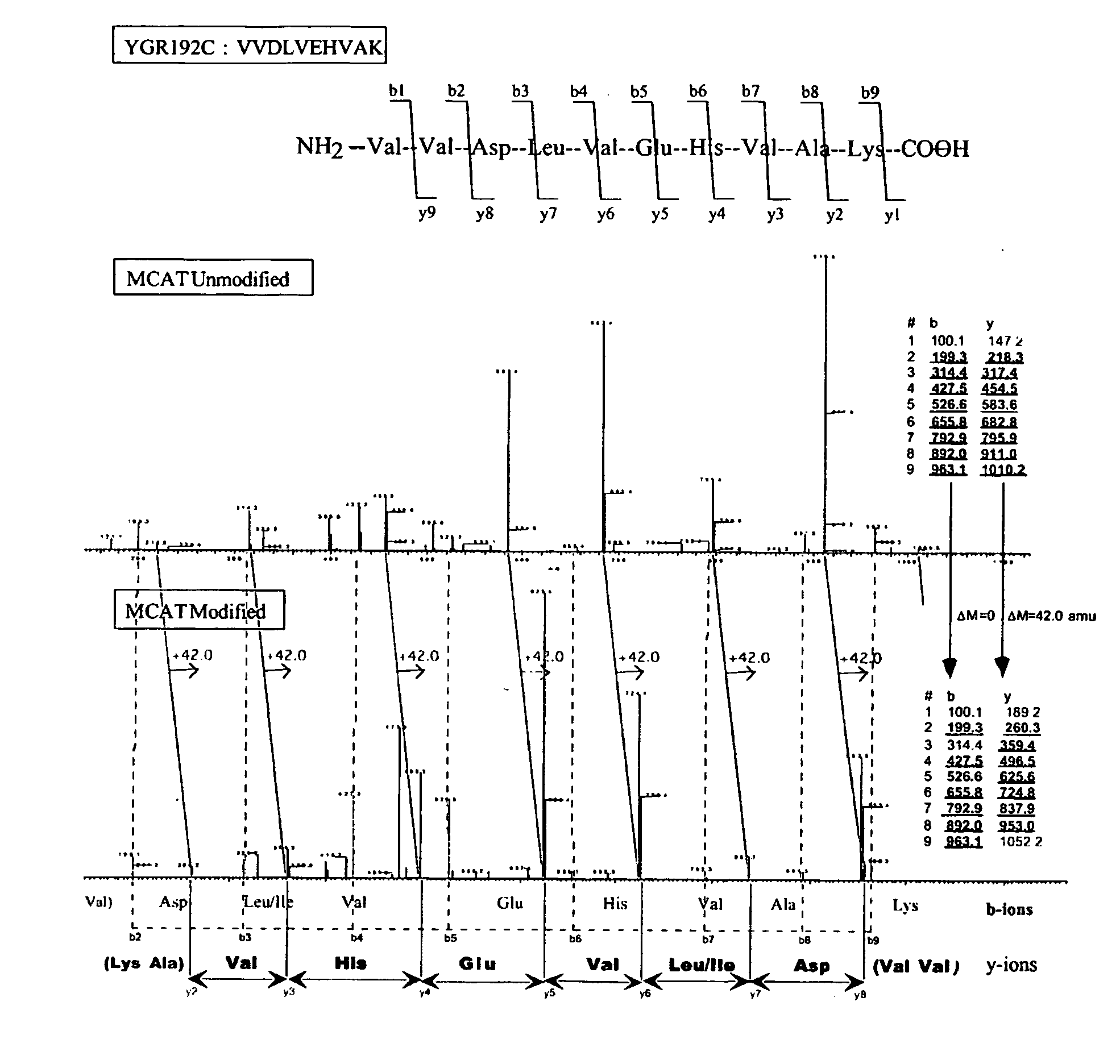
This invention describes the use of peptide profiling to identify, characterize, and classify biological samples. In complex samples, many thousands of different peptides will be present at varying concentrations. The invention uses liquid chromatography and similar methods to separate peptides, which are then identified and quantified using mass spectrometry. By identification it is meant that the correct sequence of the peptide is established through comparisons with genome sequence databases, since the majority of peptides and proteins are unannotated and have no ascribed name or function. Quantification means an estimate of the absolute or relative abundance of the peptide species using mass spectrometry and related techniques including, but not limited to, pre- or post-experimental stable or unstable isotope incorporation, molecular mass tagging, differential mass tagging, and amino acid analysis.
Owner:EMILI ANDREW +1
Rapid quantitative analysis of proteins or protein function in complex mixtures
InactiveUS7544518B2Facilitates quantitative determinationFacilitates quantitative determination of the absolute amountsComponent separationMaterial analysis by electric/magnetic meansIsotopic labelingProtein expression profile
Analytical reagents and mass spectrometry-based methods using these reagents for the rapid, and quantitative analysis of proteins or protein function in mixtures of proteins. The methods employ affinity labeled protein reactive reagents having three portions: an affinity label (A) covalently linked to a protein reactive group (PRG) through a linker group (L). The linker may be differentially isotopically labeled, e.g., by substitution of one or more atoms in the linker with a stable isotope thereof. These reagents allow for the selective isolation of peptide fragments or the products of reaction with a given protein (e.g., products of enzymatic reaction) from complex mixtures. The isolated peptide fragments or reaction products are characteristic of the presence of a protein or the presence of a protein function in those mixtures. Isolated peptides or reaction products are characterized by mass spectrometric (MS) techniques. The reagents also provide for differential isotopic labeling of the isolated peptides or reaction products which facilitates quantitative determination by mass spectrometry of the relative amounts of proteins in different samples. The methods of this invention can be used for qualitative and quantitative analysis of global protein expression profiles in cells and tissues, to screen for and identify proteins whose expression level in cells, tissue or biological fluids is affected by a stimulus or by a change in condition or state of the cell, tissue or organism from which the sample originated.
Owner:UNIV OF WASHINGTON
Rapid quantitative analysis of proteins or protein function in complex mixtures
InactiveUS20050233399A1Facilitates quantitative determinationFacilitates quantitative determination of the absolute amountsComponent separationMaterial analysis by electric/magnetic meansChemistryProtein expression profile
Analytical reagents and mass spectrometry-based methods using these reagents for the rapid, and quantitative analysis of proteins or protein function in mixtures of proteins. The methods employ affinity labeled protein reactive reagents having three portions: an affinity label (A) covalently linked to a protein reactive group (PRG) through a linker group (L). The linker may be differentially isotopically labeled, e.g., by substitution of one or more atoms in the linker with a stable isotope thereof. These reagents allow for the selective isolation of peptide fragments or the products of reaction with a given protein (e.g., products of enzymatic reaction) from complex mixtures. The isolated peptide fragments or reaction products are characteristic of the presence of a protein or the presence of a protein function in those mixtures. Isolated peptides or reaction products are characterized by mass spectrometric (MS) techniques. The reagents also provide for differential isotopic labeling of the isolated peptides or reaction products which facilitates quantitative determination by mass spectrometry of the relative amounts of proteins in different samples. The methods of this invention can be used for qualitative and quantitative analysis of global protein expression profiles in cells and tissues, to screen for and identify proteins whose expression level in cells, tissue or biological fluids is affected by a stimulus or by a change in condition or state of the cell, tissue or organism from which the sample originated.
Owner:UNIV OF WASHINGTON
Differential protein expression patterns related to disease states
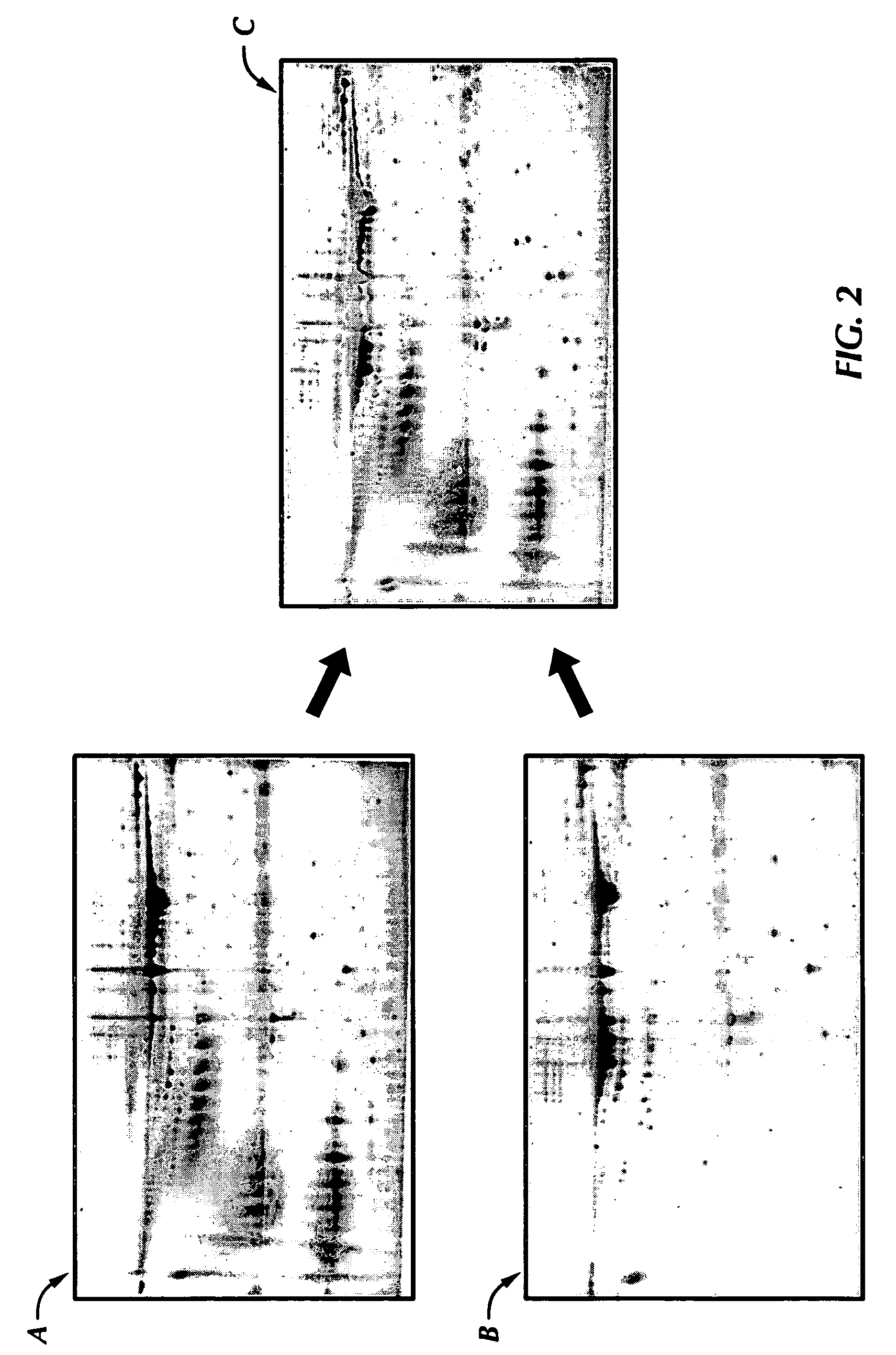
InactiveUS20060068452A1Electrophoretic profilingCharacter and pattern recognitionBiologyProtein expression profile
The present invention is a method for determining protein expression profiles in disease or an altered biological state. The method is based on the use of two-dimensional (2D) gel electrophoresis where the gel images are assigned two different colors. The gels are then compared by an overlay procedure that allows for identification and quantification of unique proteins by determining which colors are detected in the superimposed images and the density of those colors.
Owner:NEOGENOMICS INC
Apparatus, composition and method for proteome profiling
InactiveUS20050048566A1Sure easyPeptide librariesLibrary screeningProtein expression profileProtein abundance
The present invention is directed to a high throughput method for producing a large number of different antibodies, more specifically organized antibody microarrays. These antibodies and antibody microarrays can be used to rapidly assay protein abundance and identify types of proteins that are expressed in cells and tissues under a variety of conditions, or to compare protein expression profiles of different cells.
Owner:DELISI CHARLES +5
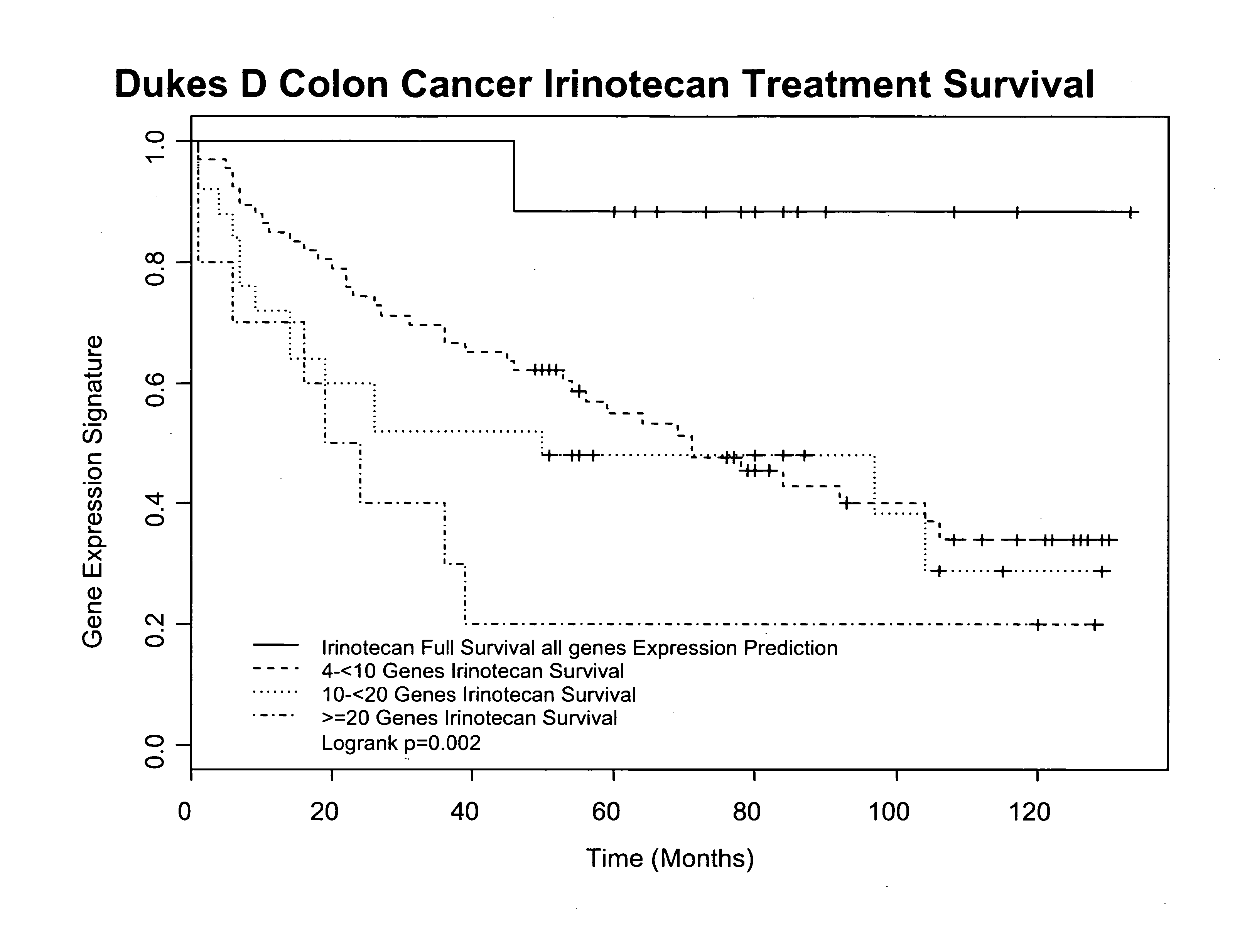
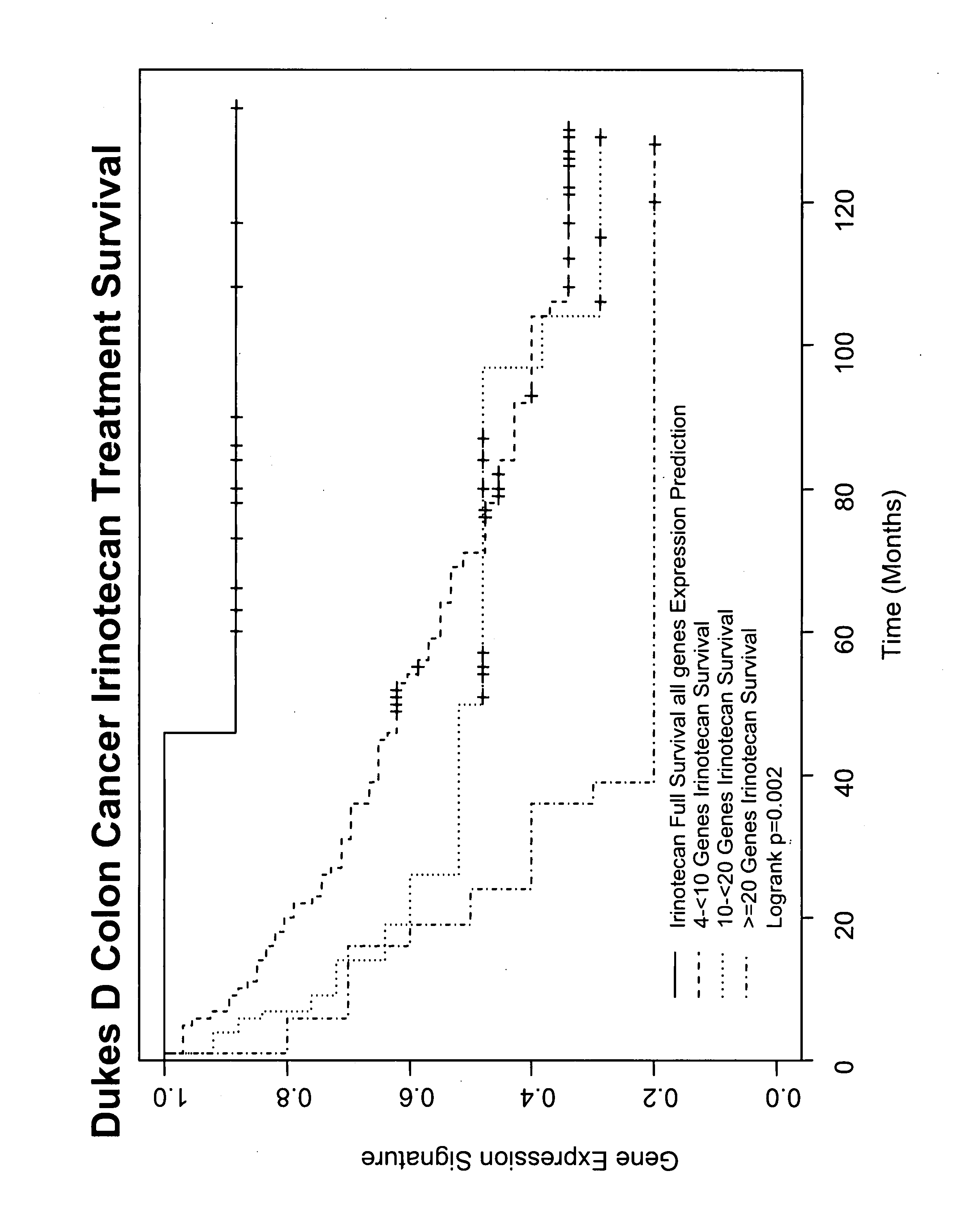
Gene and protein expression profiles associated with the therapeutic efficacy of irinotecan
InactiveUS20080076134A1Microbiological testing/measurementDisease diagnosisProtein expression profileIrinotecan
The present invention includes gene and protein expression profiles indicative of whether a cancer patient is likely to respond to treatment with irinotecan. By identifying such responsiveness, a treatment provider may determine in advance those patients who would benefit from such treatment, as well as identify alternative therapies for non-responders. The present invention further provide methods of using the gene and / or protein expression profiles and assays for identifying the presence of a gene and / or protein expression profile in a patient sample.
Owner:NUCLEA BIOMARKERS
Gene and protein expression profiles associated with the therapeutic efficacy of egfr-tk inhibitors
InactiveUS20090263819A1Microbiological testing/measurementBiological material analysisProtein expression profileNon responders
The present invention provides protein and gene expression profiles indicative of whether a patient afflicted with non-small cell lung cancer is likely to be responsive to treatment with a therapeutic compound that is a EGFR-TK inhibitor. By identifying such responsiveness, a treatment provider may determine in advance those patients who would benefit from such treatment, as well as identify alternative therapies for non-responders. The present invention further provide methods of using the gene and protein expression profiles, and assays for identifying the presence of a gene or protein expression profile in a patient sample.
Owner:NUCLEA BIOMARKERS
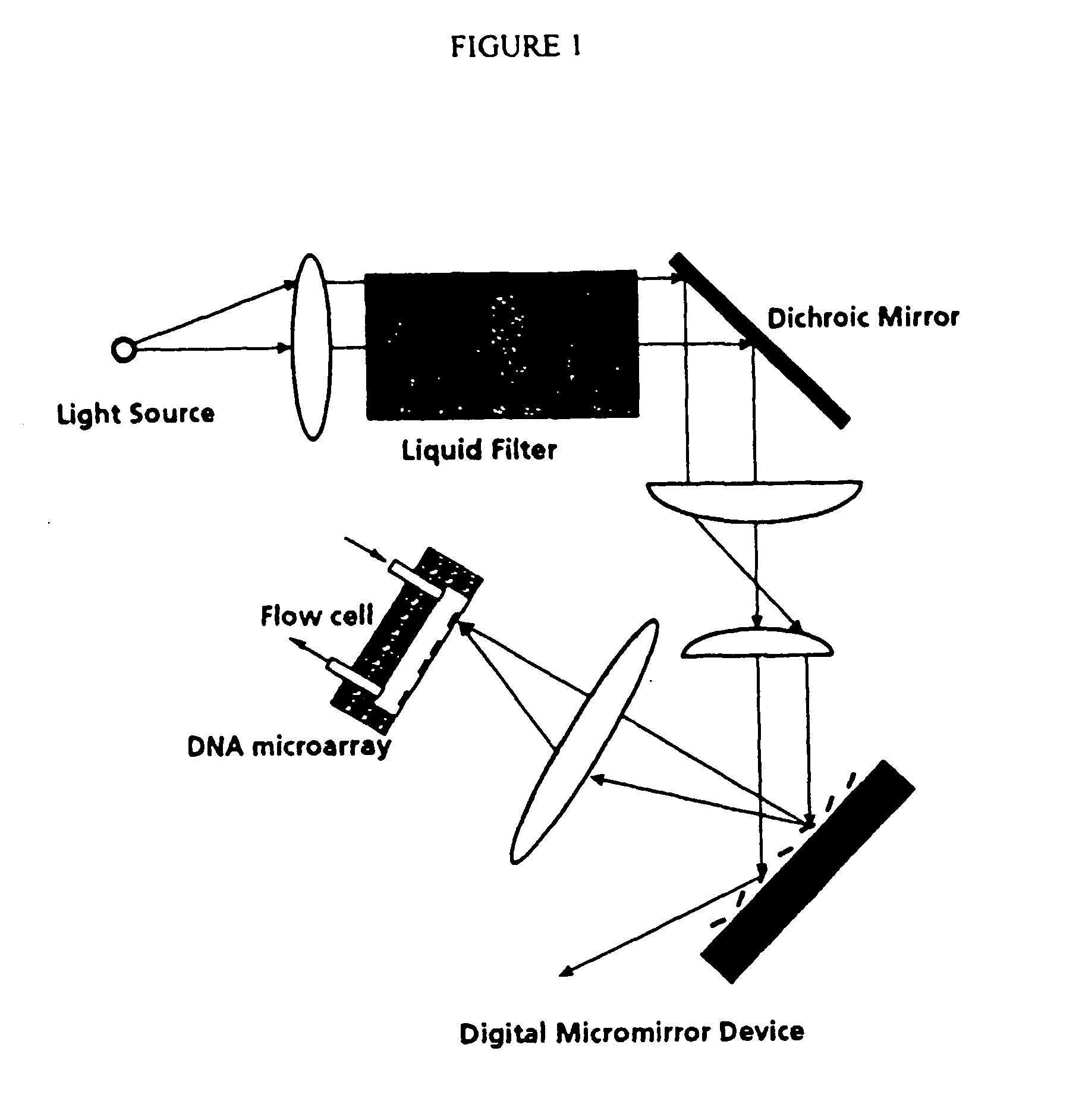
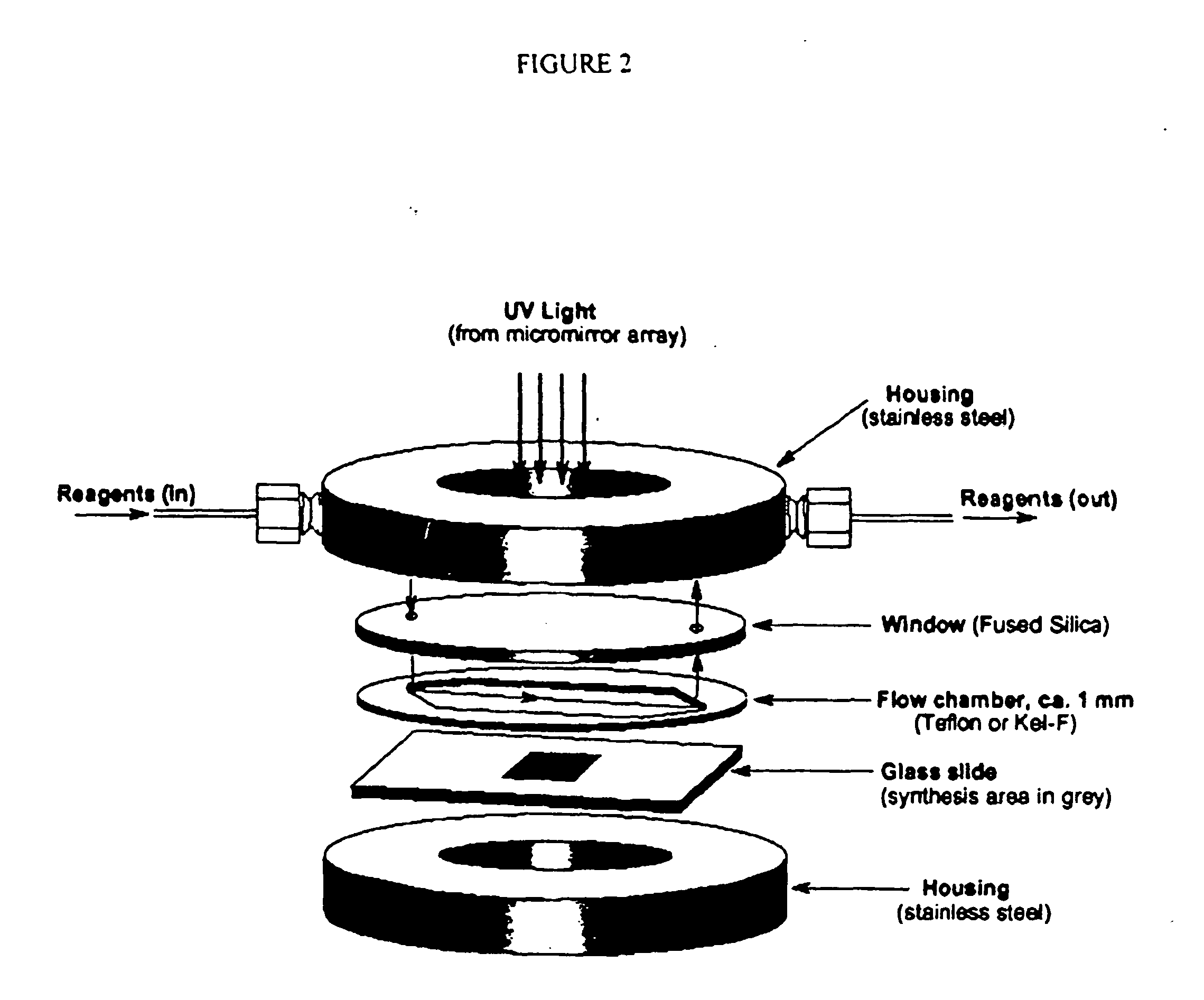
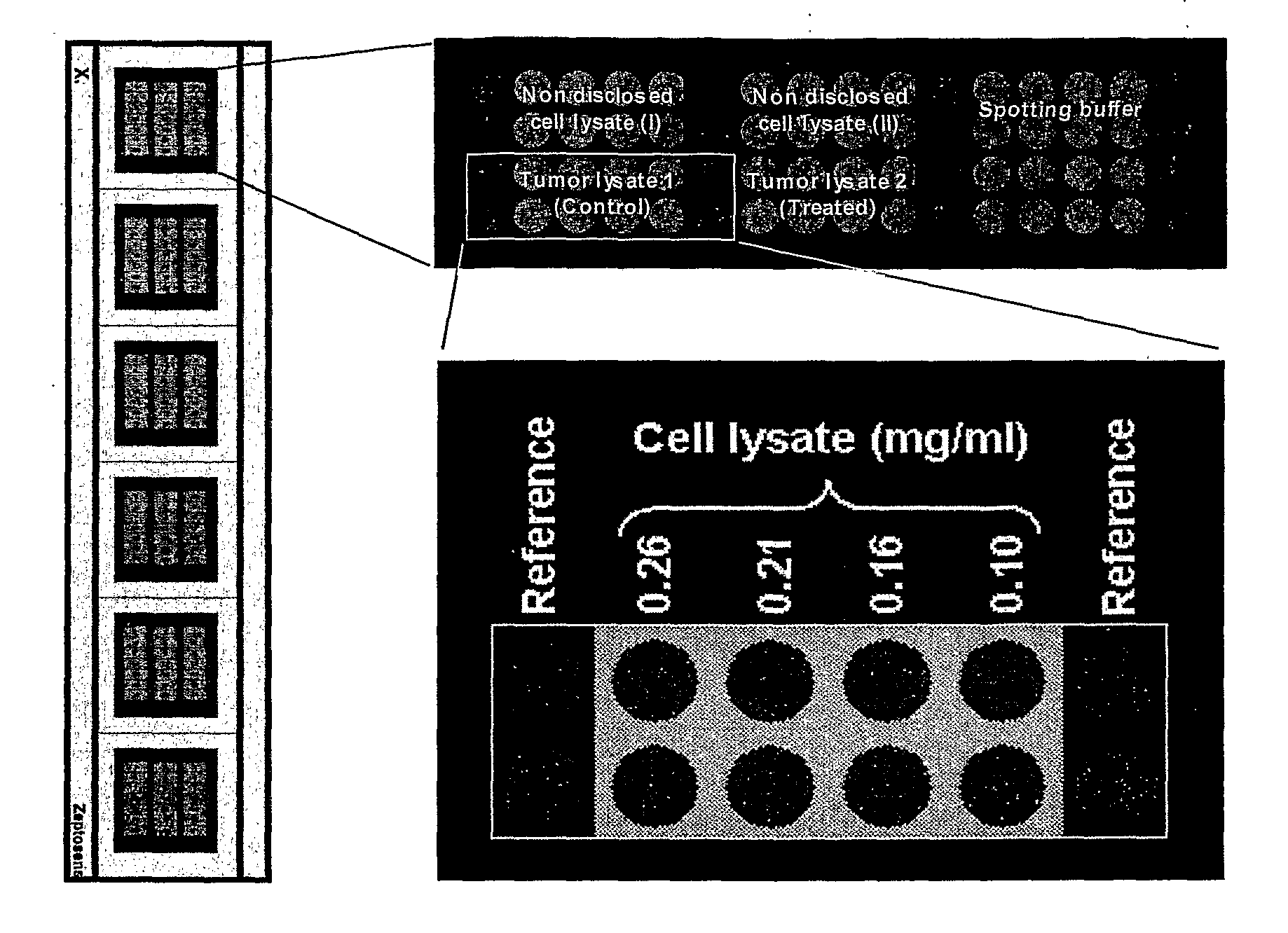
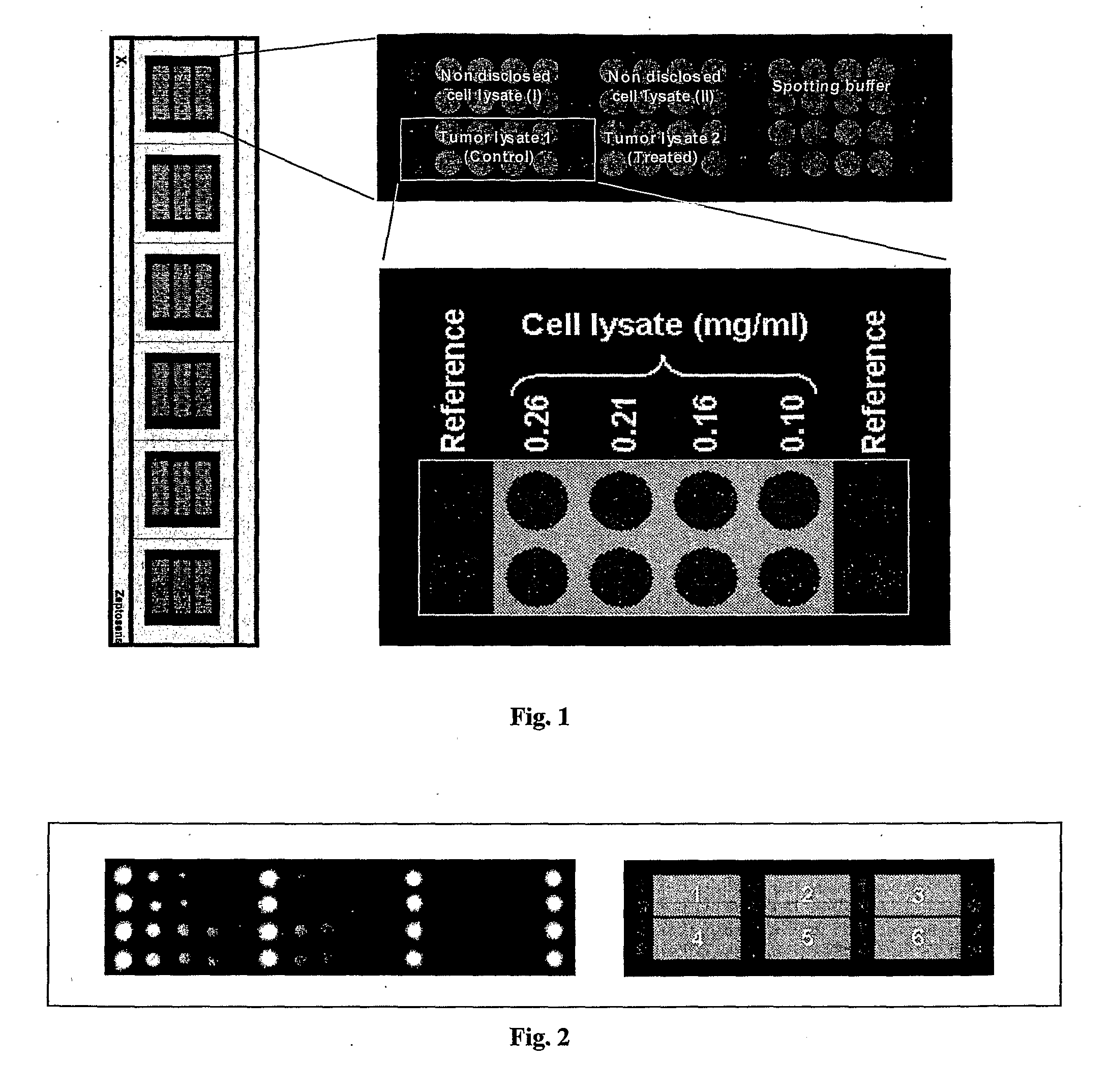
Analytical Platform and Method for Generating Protein Expression Profiles of Cell Populations
InactiveUS20080020409A1Bioreactor/fermenter combinationsBiological substance pretreatmentsProtein targetProtein expression profile
The present invention is related to analytical platforms and methods performed therewith for generating qualitative and / or quantitative protein expression profiles, in particular differential protein expression profiles, of cell populations comprising: generating lysates of one or more populations of cells, the lysates comprising a plurality of proteins expressed by the respective cell populations, providing an essentially planar solid support, depositing at discrete sites small quantities of the cell lysates, in diluted or undiluted form directly on said solid support or on an adhesion-promoting layer applied on said solid support, thereby creating one or more one- or two-dimensional arrays of discrete measurement areas on said solid support, applying a number of binding reagents as specific binding partners for the proteins contained in cell lysates in discrete measurement areas and to be detected and, if adequate, one or more detection reagents on said one or more arrays of measurement areas, the binding reagents and the detection reagents being applied sequentially or in a single addition-step, after binding of the detection reagents to the binding reagents, to the one or more arrays of discrete measurement areas for e.g. global analysis of signaling pathways or screening antibody sets / libraries against protein targets for best specificity, selectivity and affinity, and measuring and recording optical signals emanating from said one or more arrays of discrete measurement areas in a locally resolved manner, wherein said essentially planar solid support is non-porous and an optionally applied adhesion-promoting layer has a thickness of less than 1 μm.
Owner:BAYER TECH SERVICES GMBH
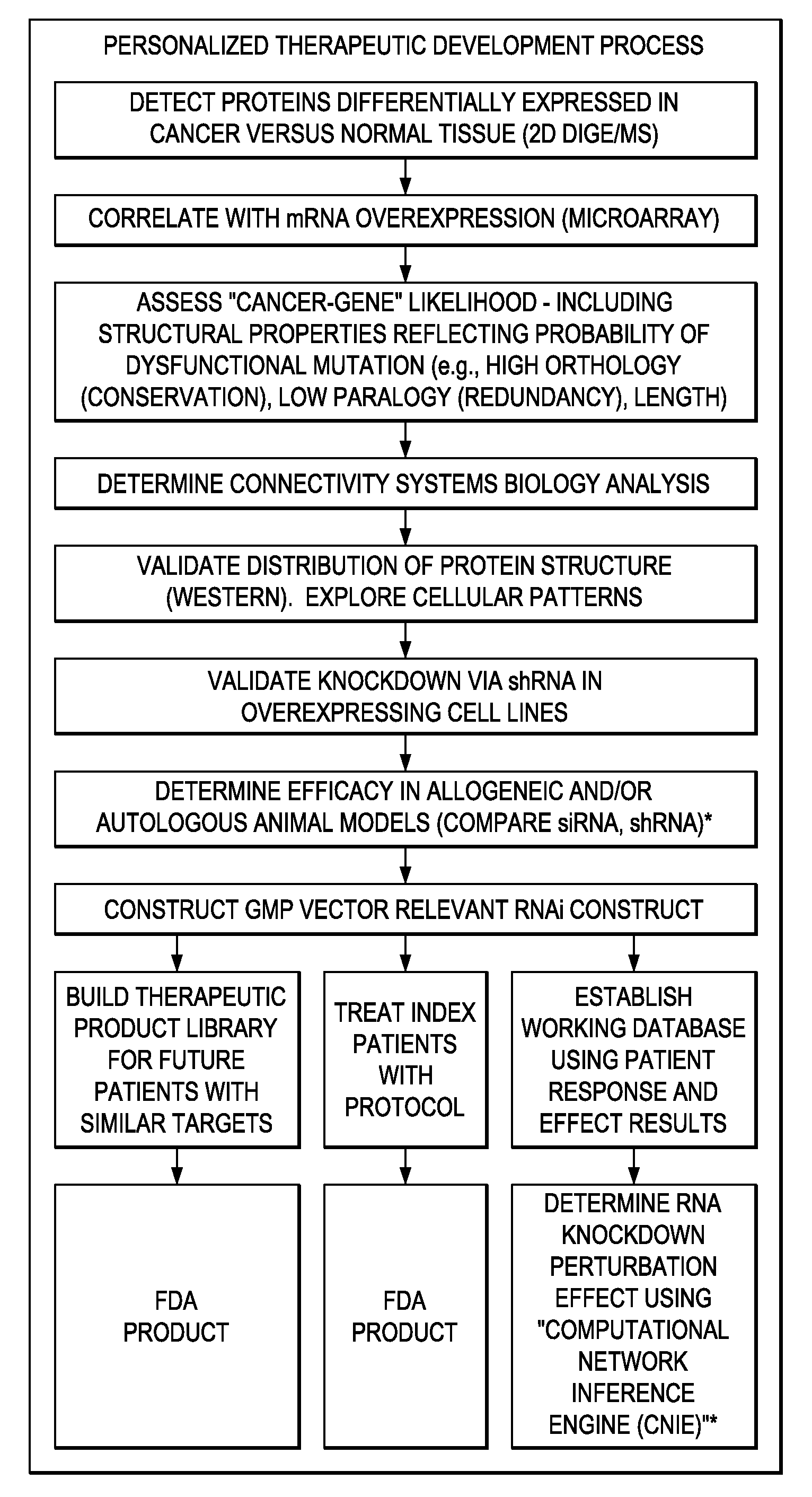
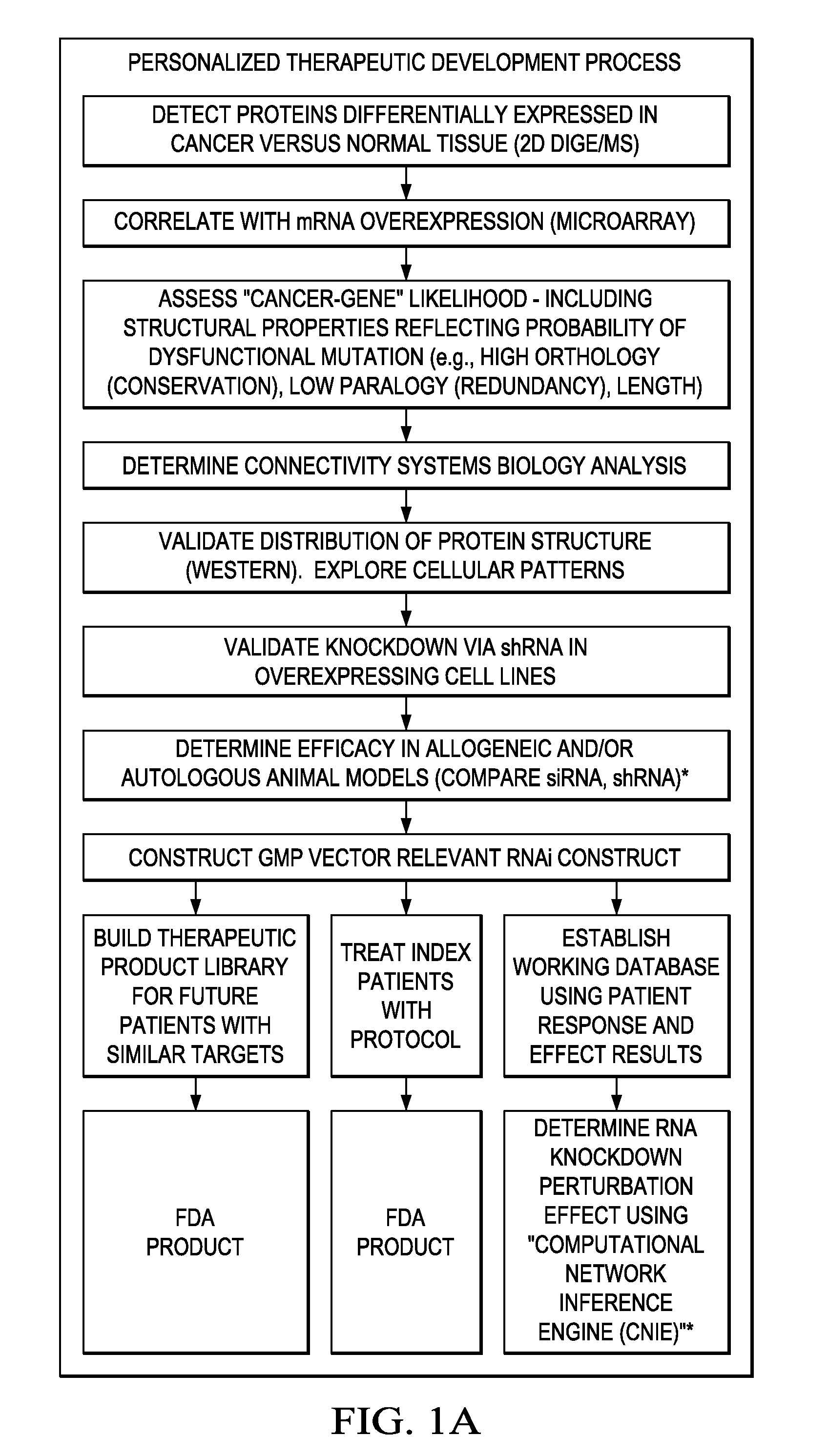
Individualized cancer therapy
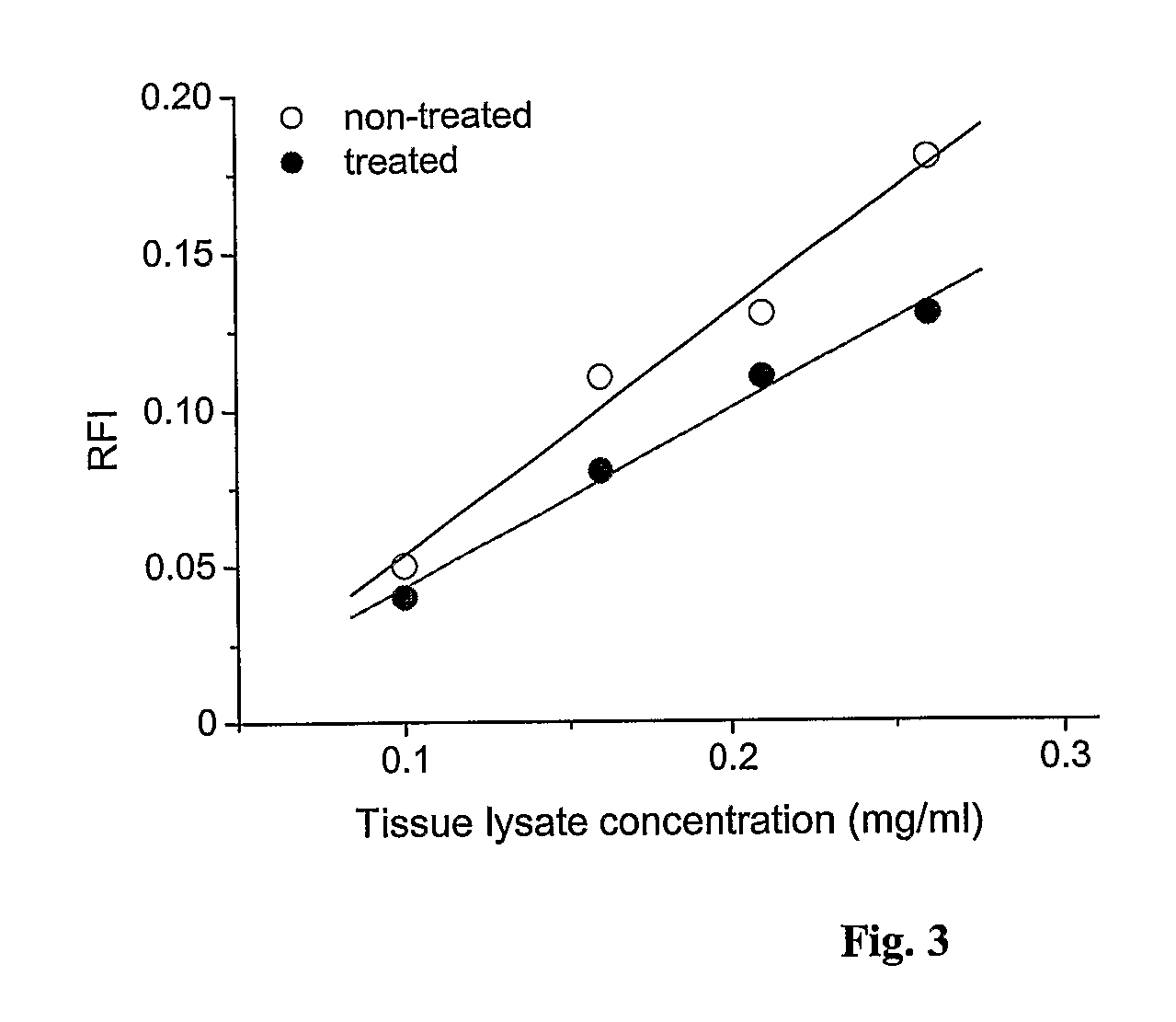
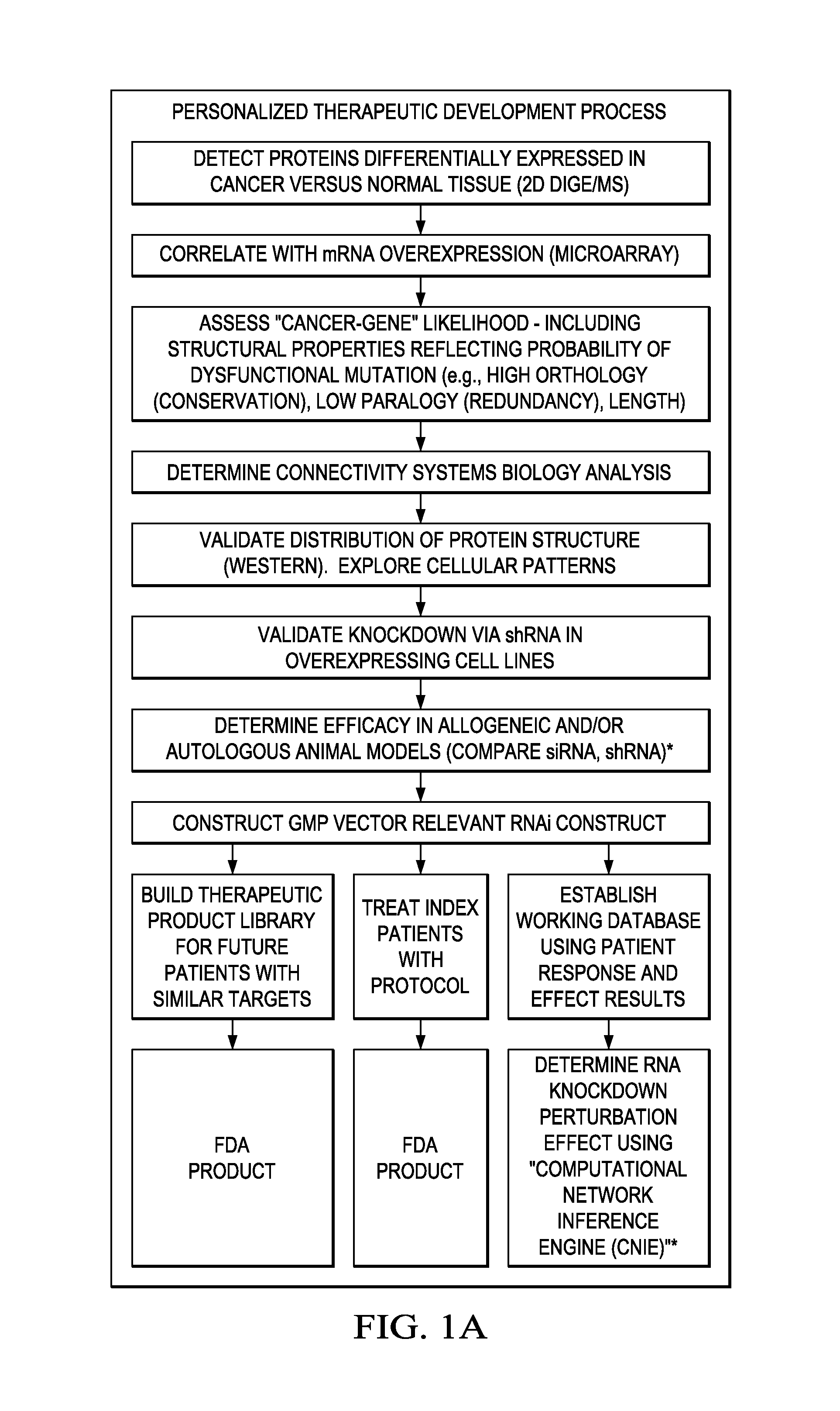
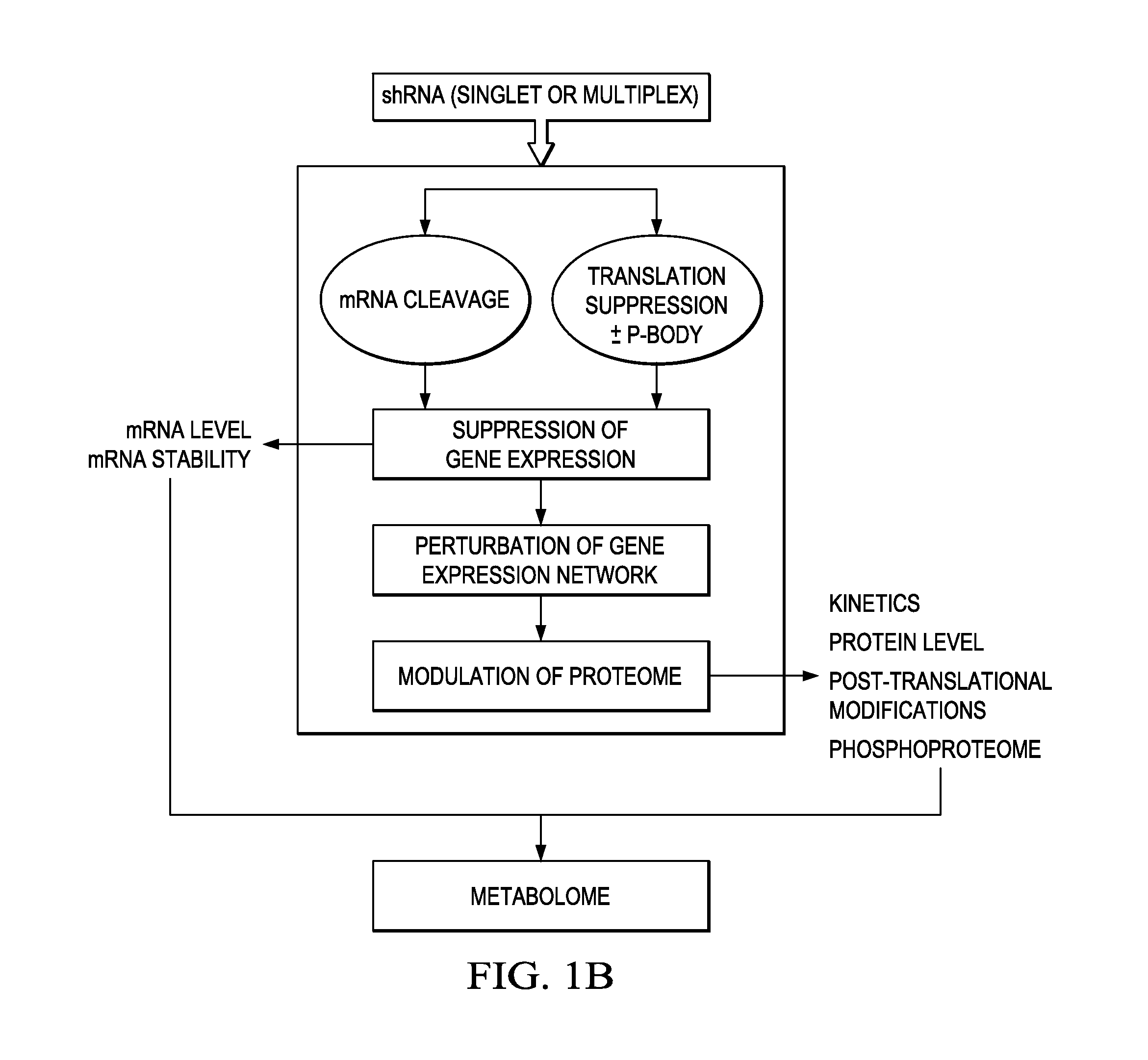
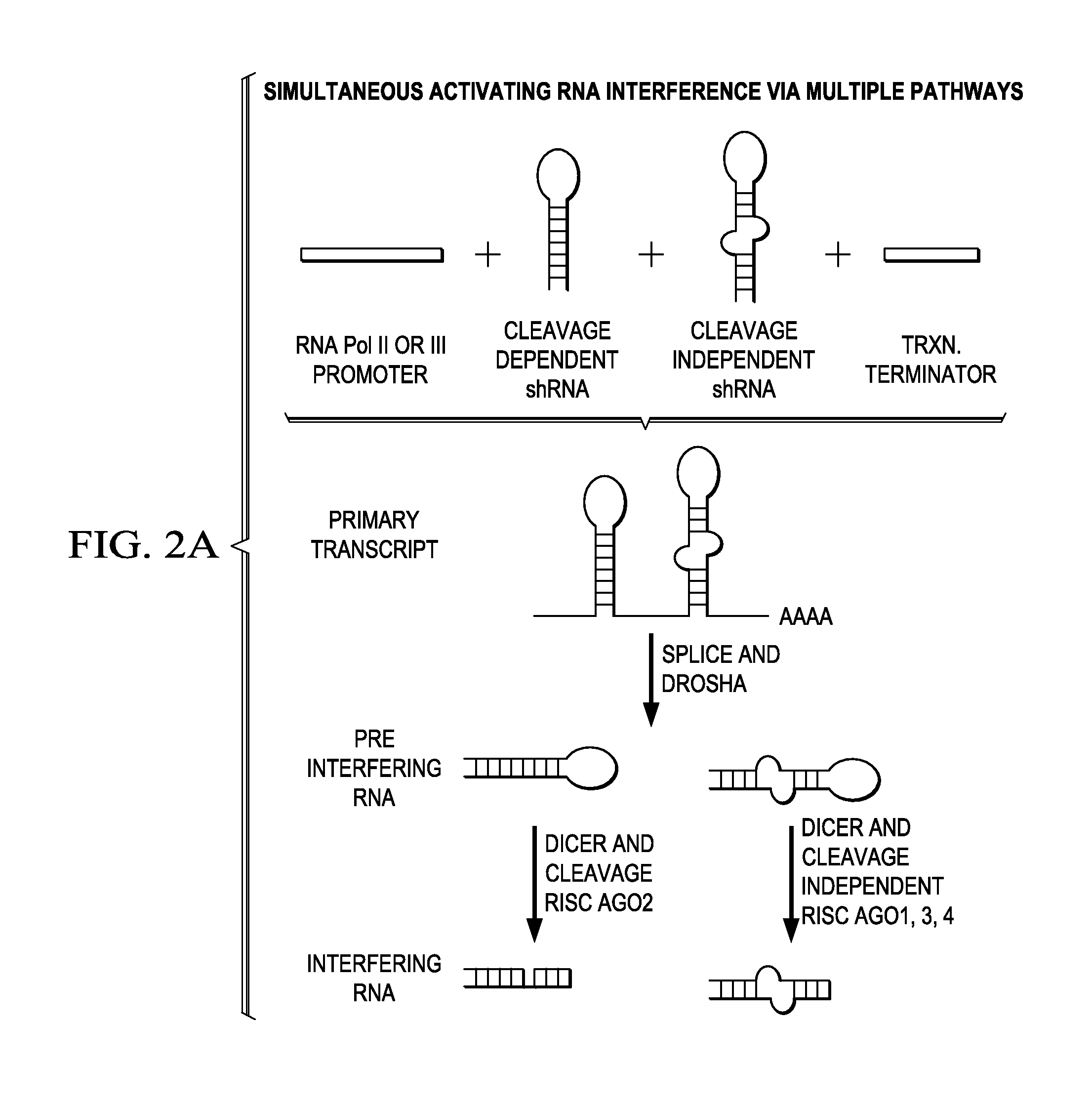
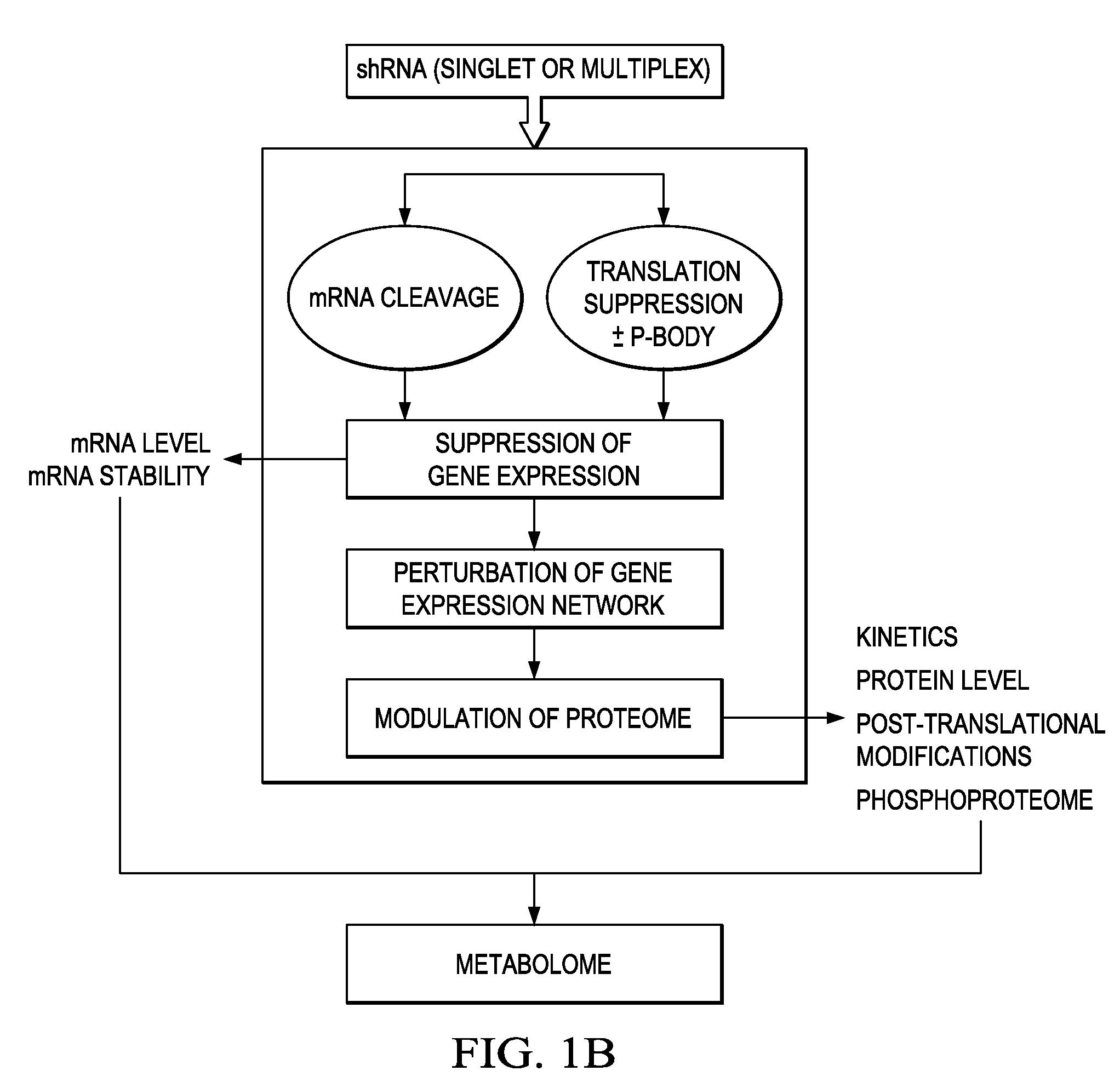
In certain embodiments, the invention provides methods for treating cancer, comprising: (a) obtaining a specimen of cancer tissue and normal tissue from a patient; (b) extracting total protein and RNA from the cancer tissue and normal tissue; (c) obtaining a protein expression profile of the cancer tissue and normal tissue; (d) identifying over-expressed proteins in the cancer tissue; (e) comparing the protein expression profile to a gene expression profile; (f) identifying at least one prioritized protein target by assessing connectivity of each said over-expressed protein to other cancer-related or stimulatory proteins; (g) designing a first RNA interference expression cassette to modulate the expression of at least one gene encoding the prioritized target protein; (h): designing a first RNA interference expression cassette to modulate the expression of at least one gene encoding a protein of higher priority in the signaling pathway in which the first protein is a component; (i) incorporating the first cassette into a first delivery vehicle; (j) providing a patient with an effective amount of the first delivery vehicle; (k) extracting total protein and RNA from the treated cancer tissue; (l) identifying over-expressed proteins in the treated cancer tissue; (m) designing a second RNA interference expression cassette to modulate the expression of a second prioritized protein in the treated tissue; (n) incorporating the second cassette into a second delivery vehicle; (o) providing the previously treated patient with an effective amount of the second delivery vehicle; (p) identifying a novel protein signal following prior treatment with protein specific knockdown; (q) identifying a gene mutation provided by gene sequencing / microarray on assessment of other protein signals; and (r) identifying of a novel protein signal as a result of determination of the gene mutation and assessment of other protein signals to, directly or indirectly, modify the expression (i.e., production) of such proteins.
Owner:GRADALIS
Method for correlating gene expression profiles with protein expression profiles
InactiveUS7299134B2Microbiological testing/measurementMaterial analysis by electric/magnetic meansProtein expression profileGene and protein expression
Owner:CIPHERGEN BIOSYSTEMS INC
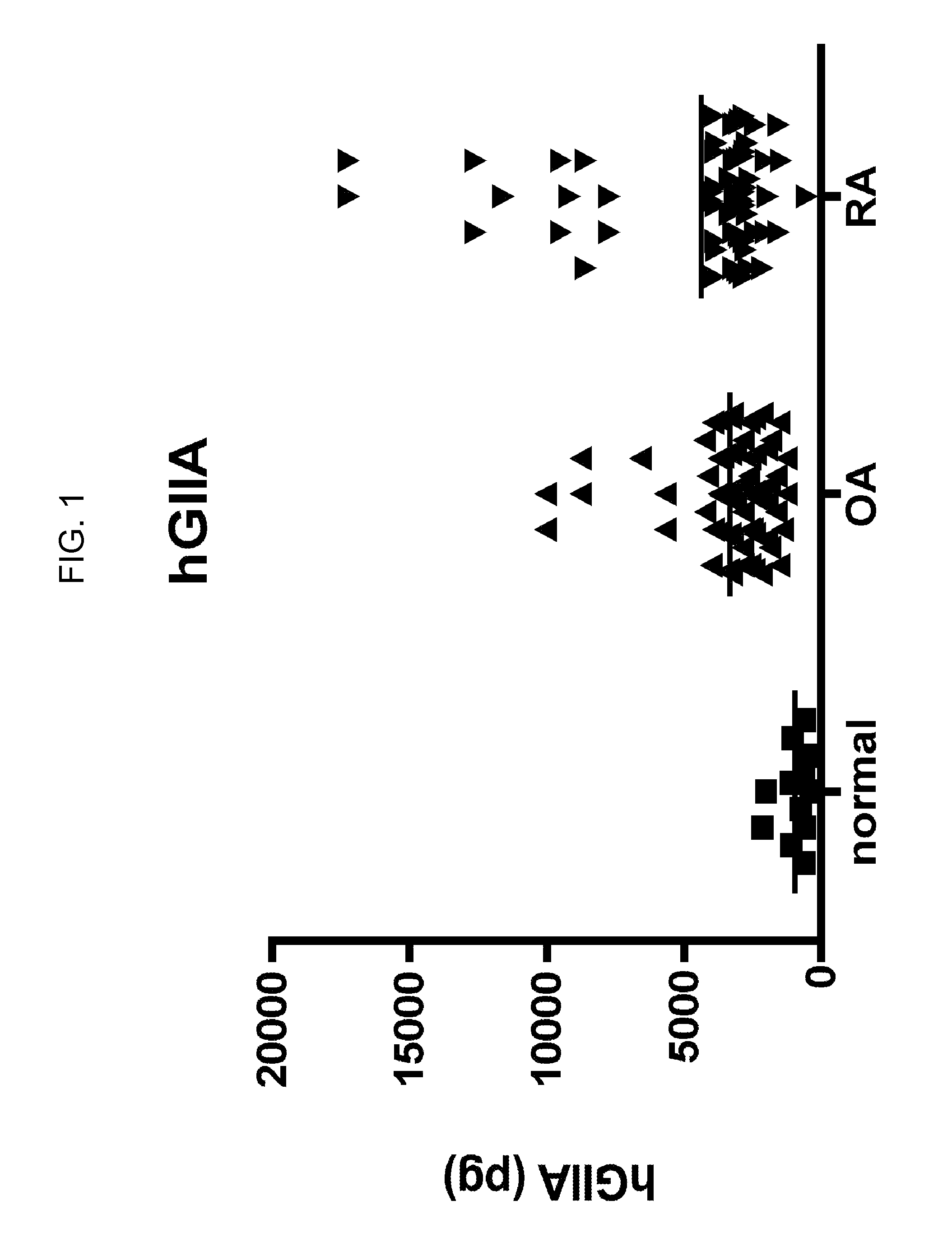
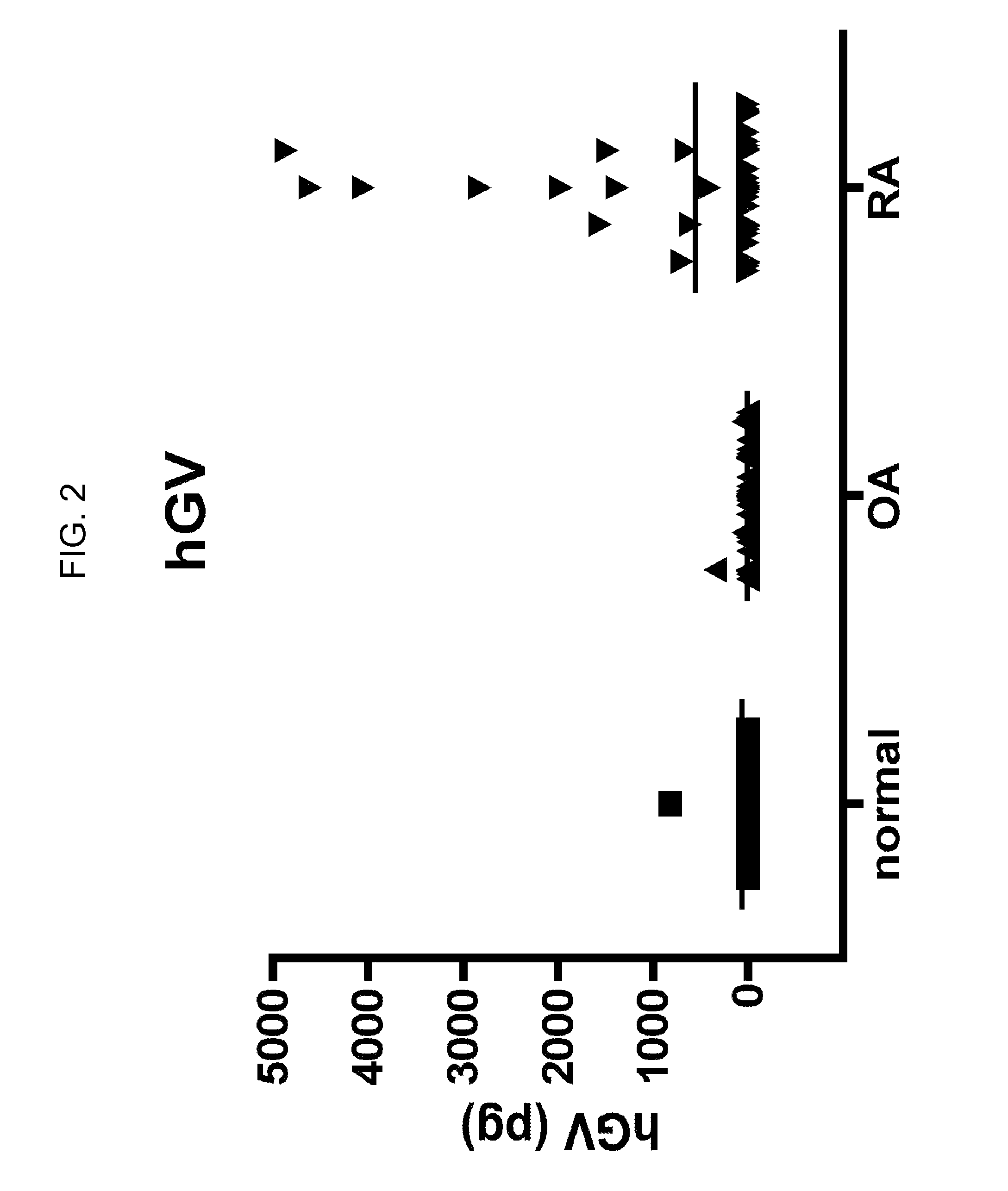
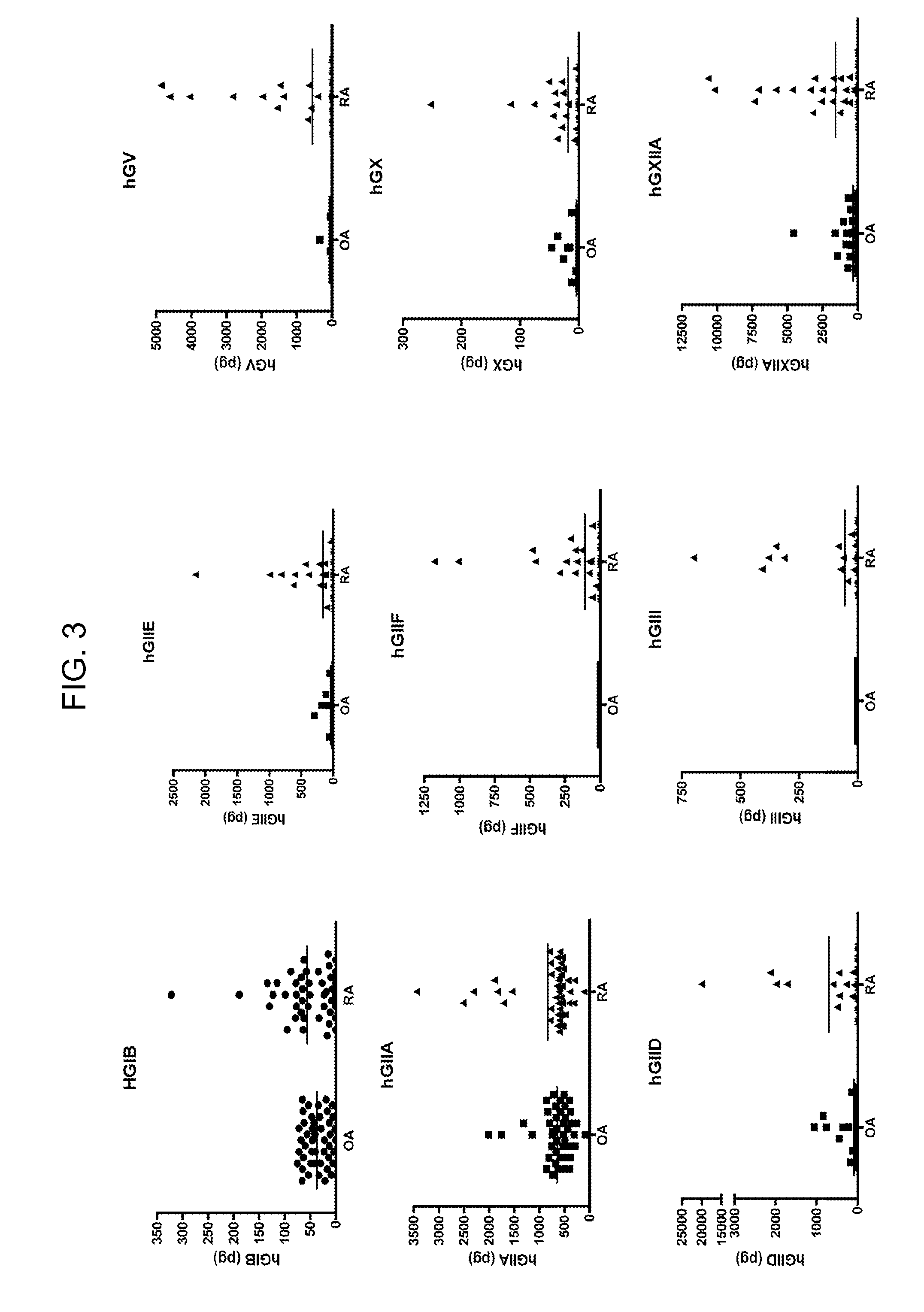
Protein profile for osteoarthritis
InactiveUS20100292154A1High expressionReduce expressionPeptide librariesSugar derivativesProtein profilingProtein expression profile
The present invention relates to the identification and use of protein expression profiles with clinical relevance to osteoarthritis (OA). In particular, the invention provides the identity of marker proteins whose expression is correlated with OA and OA progression. Methods and kits are described for using these protein expression profiles in the study and / or diagnosis of OA, in the determination of the degree of advancement of OA, and in the selection and / or monitoring of treatment regimens. The invention also relates to the screening of drugs that modulate expression of these proteins or nucleic acid molecules encoding these proteins, in particular for the development of disease-modifying OA agents.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC
Protein profile for osteoarthritis
ActiveUS20070207480A1Solid foundationCompound screeningApoptosis detectionProtein profilingProtein expression profile
The present invention relates to the identification and use of protein expression profiles with clinical relevance to osteoarthritis (OA). In particular, the invention provides the identity of marker proteins whose expressions are correlated with OA, OA subtype, and / or OA progression. Methods and kits are described for using these protein expression profiles in the study and / or diagnosis of OA, in the determination of the degree of advancement of OA, and in the selection and / or monitoring of treatment regimens. The invention also relates to the screening of drugs that modulate expression of these proteins or nucleic acid molecules encoding these proteins, in particular for the development of disease-modifying OA agents.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC
Lung cancer marker SBP-1 and its application
The invention relates to the molecular biology and tumor medicine fields, and relates to an application of a lung cancer marker SBP-1 in lung cancer diagnosis, and a method for detecting lung cancer by taking SBP-1 as the marker. According to the invention, a comparative proteomics method is employed for comparative analysis of protein expression profile of a lung cancer cell line / tissue and normal control cell / tissue, and a mass spectrum identifies the partial difference protein points. SBP-1 protein is a difference point, and enables high expression in cancer cells by comparing with that of the normal cells, and the difference is obvious. The invention employs RT-PCR and an immunohistochemical technology to respectively detect mRNA of SBP-1 and expression level of protein expression in normal tissue, the result displays that the expression of SBP-1 in the lung cancer tissue is obviously higher than that of its paired normal tissue, so that SBP-1 can be taken as a diagnosis marker of lung cancer. The invention also provides a lung cancer diagnostic kit and a detection method used for lung cancer.
Owner:SHANGHAI PUBLIC HEALTH CLINICAL CENT
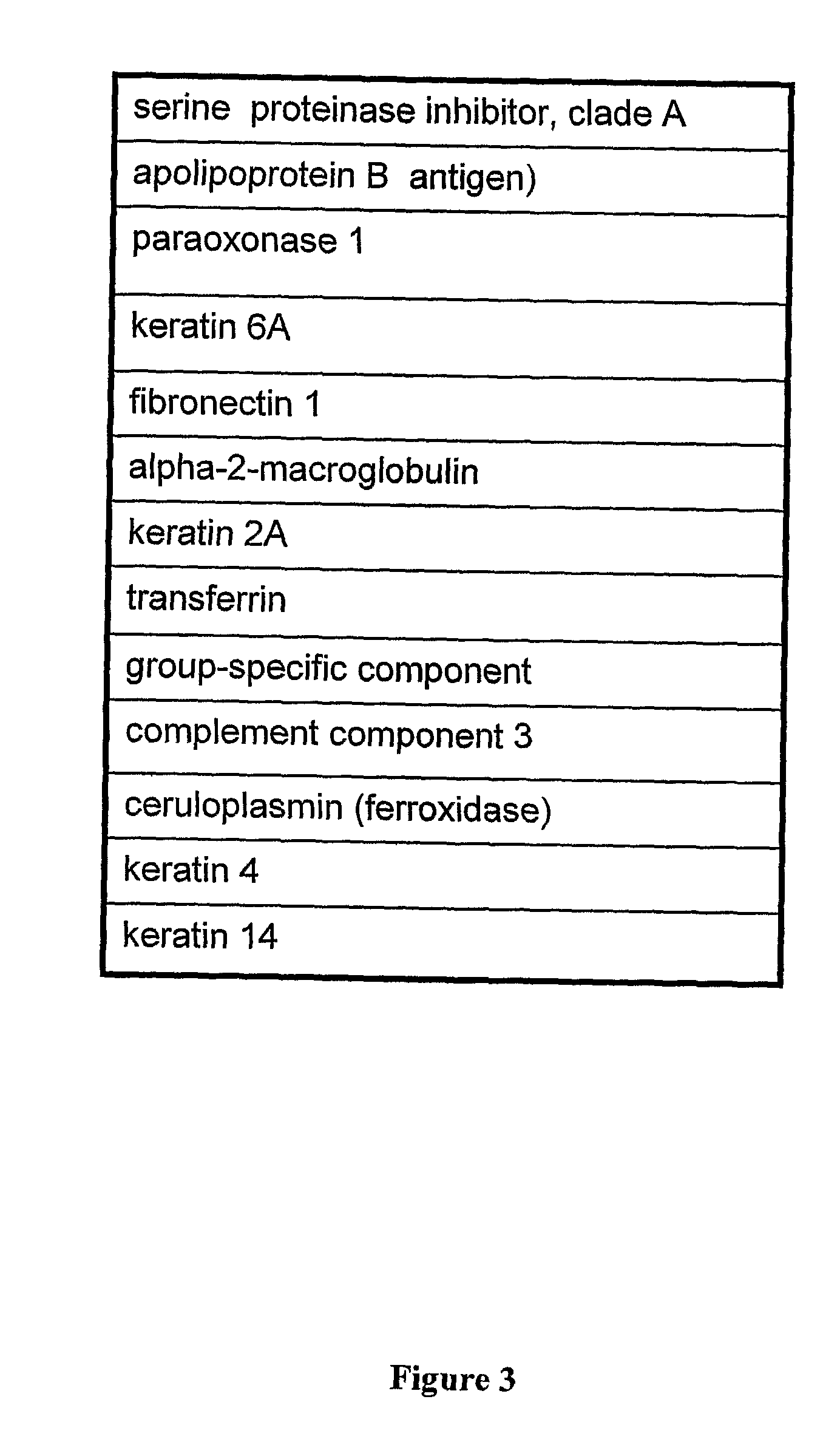
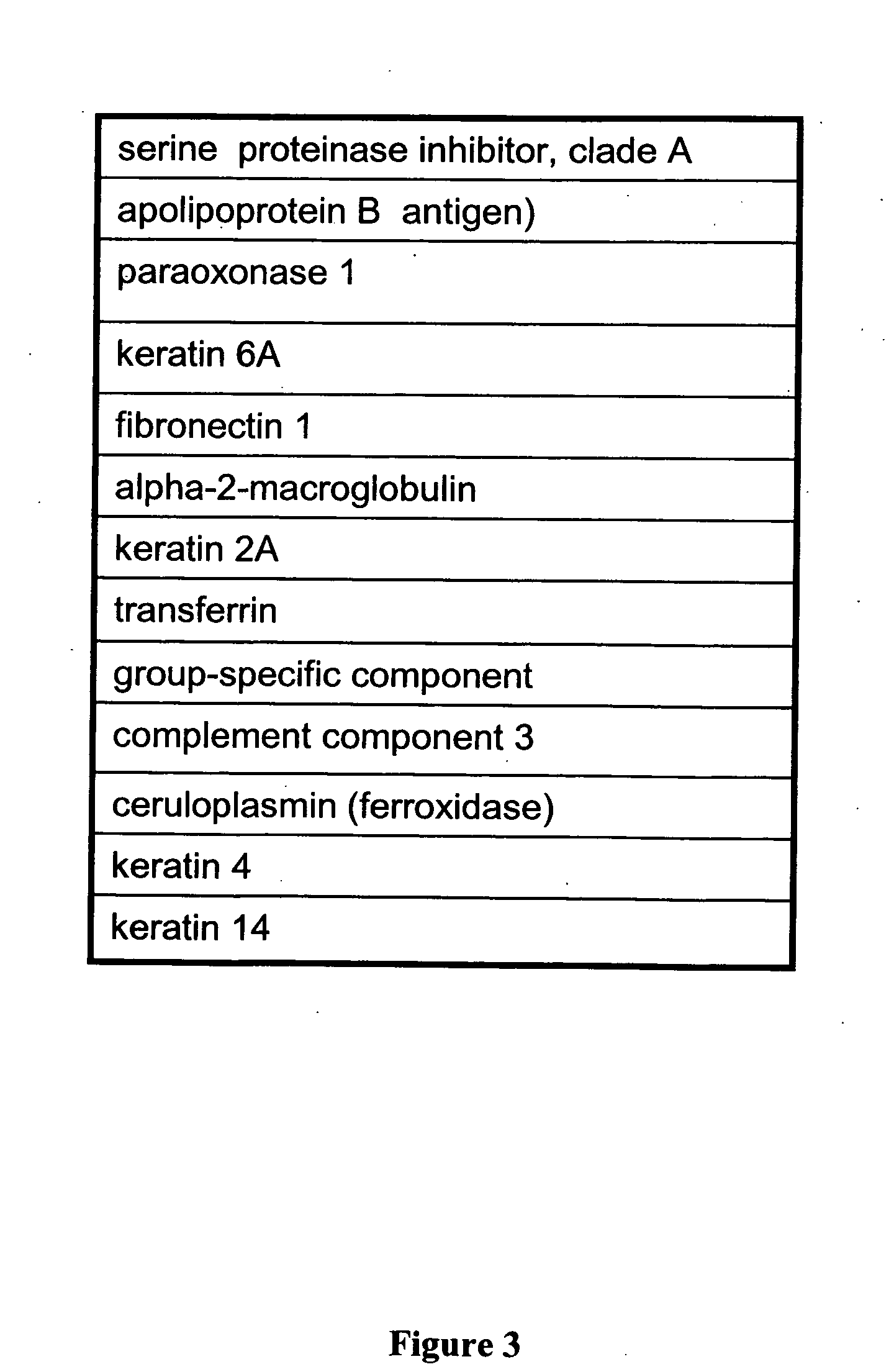
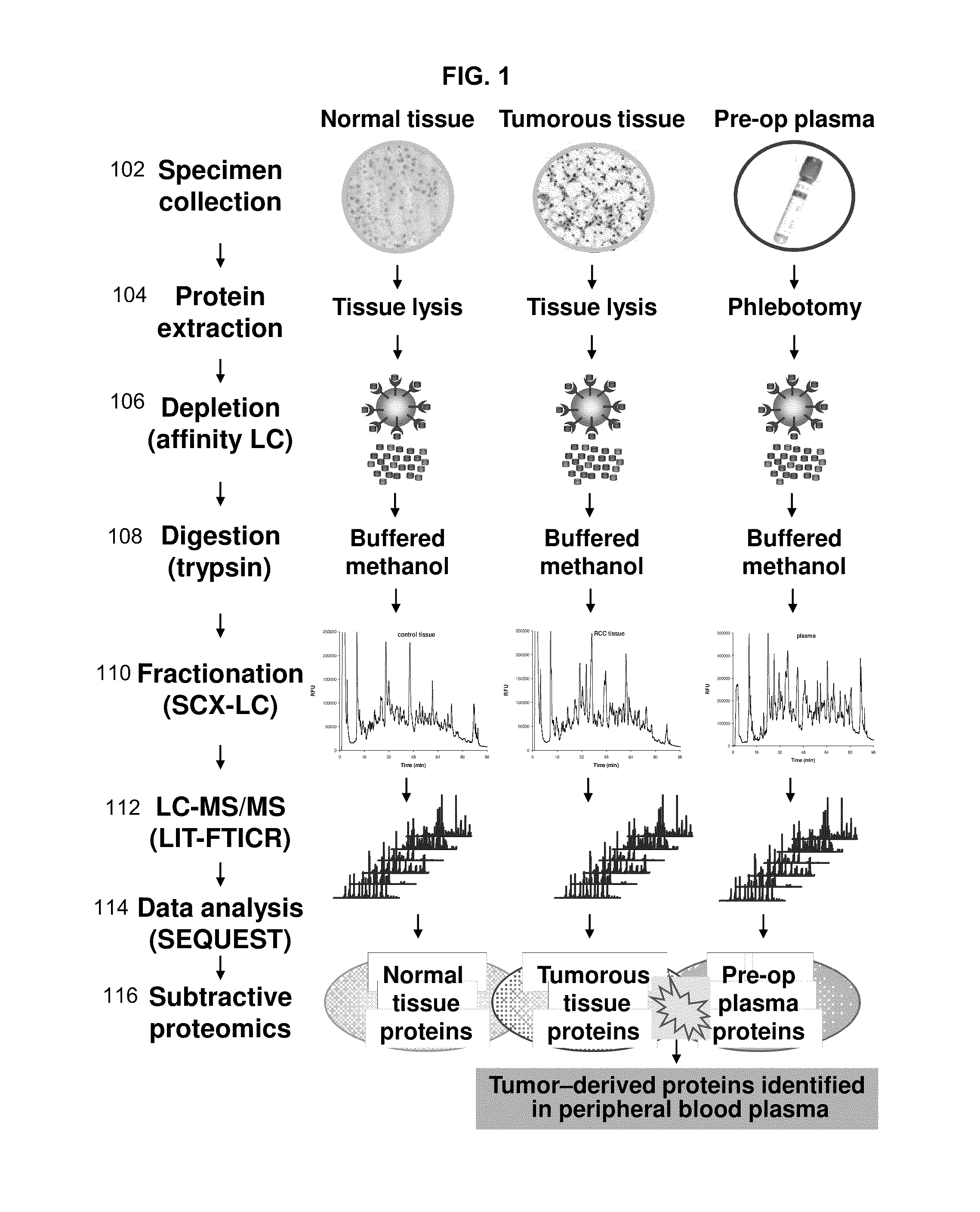
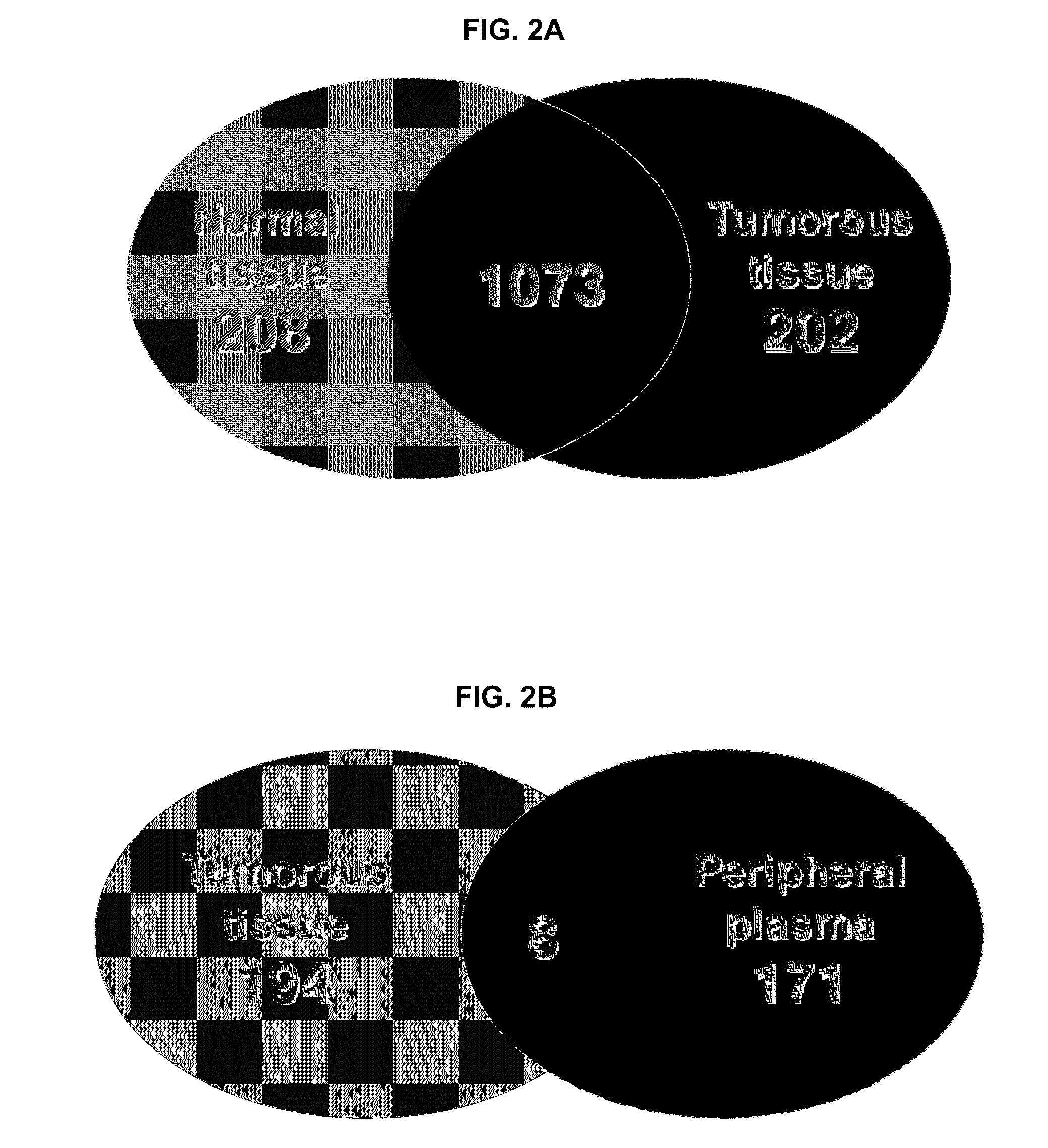
Renal cell carcinoma biomarkers
InactiveUS20120009201A1Easy to findOrganic active ingredientsCompound screeningTumor BiomarkersProtein expression profile
Disclosed herein is a method of identifying a tumor biomarker. In one example, a tumor biomarker is identified by obtaining a peripheral biological fluid sample from a subject with a tumor as well as a tumor sample and an adjacent non-tumor sample from such subject. A protein expression profile is detected in the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample. The protein expression profiles of the peripheral biological fluid sample, tumor sample and adjacent non-tumor sample are then compared, wherein an increase in expression of a specific protein in the tumor sample and peripheral biological fluid sample but not in the adjacent non-tumor sample indicates that the specific protein is a biomarker of the tumor. Also disclosed herein is a gene profiling signature that can be used to diagnosis a subject with renal cell carcinoma (RCC) or to identify agents with therapeutic potential to treat RCC. Thus, methods of diagnosing a subject with RCC are disclosed. Methods are also provided for identifying agents that alter an activity of a RCC biomarker.
Owner:UNITED STATES OF AMERICA
Protein biomarkers and therapeutic targets for osteoarthritis
InactiveUS20120190042A1Solid foundationMicrobiological testing/measurementDisease diagnosisProtein expression profileBiology
The present invention relates to the identification and use of protein expression profiles with clinical relevance to osteoarthritis. In particular, the invention provides the identity of marker proteins whose expressions are correlated with OA, and / or OA progression. Methods and kits are described for using these protein expression profiles in the study and / or diagnosis of OA, in the determination of the degree of advancement of OA, and in the selection and / or monitoring of treatment regimens. The invention also relates to the screening of drugs that modulate expression of these proteins or nucleic acid molecules encoding these proteins, in particular for the development of disease-modifying OA agents.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +1
Individualized cancer therapy
In certain embodiments, the invention provides methods for treating cancer, comprising: (a) obtaining a specimen of cancer tissue and normal tissue from a patient; (b) extracting total protein and RNA from the cancer tissue and normal tissue; (c) obtaining a protein expression profile of the cancer tissue and normal tissue; (d) identifying over-expressed proteins in the cancer tissue; (e) comparing the protein expression profile to a gene expression profile; (f) identifying at least one prioritized protein target by assessing connectivity of each said over-expressed protein to other cancer-related or stimulatory proteins; (g) designing a first RNA interference expression cassette to modulate the expression of at least one gene encoding the prioritized target protein; (h): designing a first RNA interference expression cassette to modulate the expression of at least one gene encoding a protein of higher priority in the signaling pathway in which the first protein is a component; (i) incorporating the first cassette into a first delivery vehicle; (j) providing a patient with an effective amount of the first delivery vehicle; (k) extracting total protein and RNA from the treated cancer tissue; (l) identifying over-expressed proteins in the treated cancer tissue; (m) designing a second RNA interference expression cassette to modulate the expression of a second prioritized protein in the treated tissue; (n) incorporating the second cassette into a second delivery vehicle; (o) providing the previously treated patient with an effective amount of the second delivery vehicle; (p) identifying a novel protein signal following prior treatment with protein specific knockdown; (q) identifying a gene mutation provided by gene sequencing / microarray on assessment of other protein signals; and (r) identifying of a novel protein signal as a result of determination of the gene mutation and assessment of other protein signals to, directly or indirectly, modify the expression (i.e., production) of such proteins.
Owner:GRADALIS
Methods and compositions for elucidating protein expression profiles in cells
InactiveUS20060134629A1VirusesMicrobiological testing/measurementProtein expression profileTissues types
The present invention relates generally to methods and compositions for the identification of differential protein expression patterns and concomitantly the active genetic regions that are directly or indirectly involved in different tissue types, disease states, or other cellular differences desirable for diagnosis or for targets for drug therapy.
Owner:LINK CHARLES +9
Method of diagnosing neoplasms
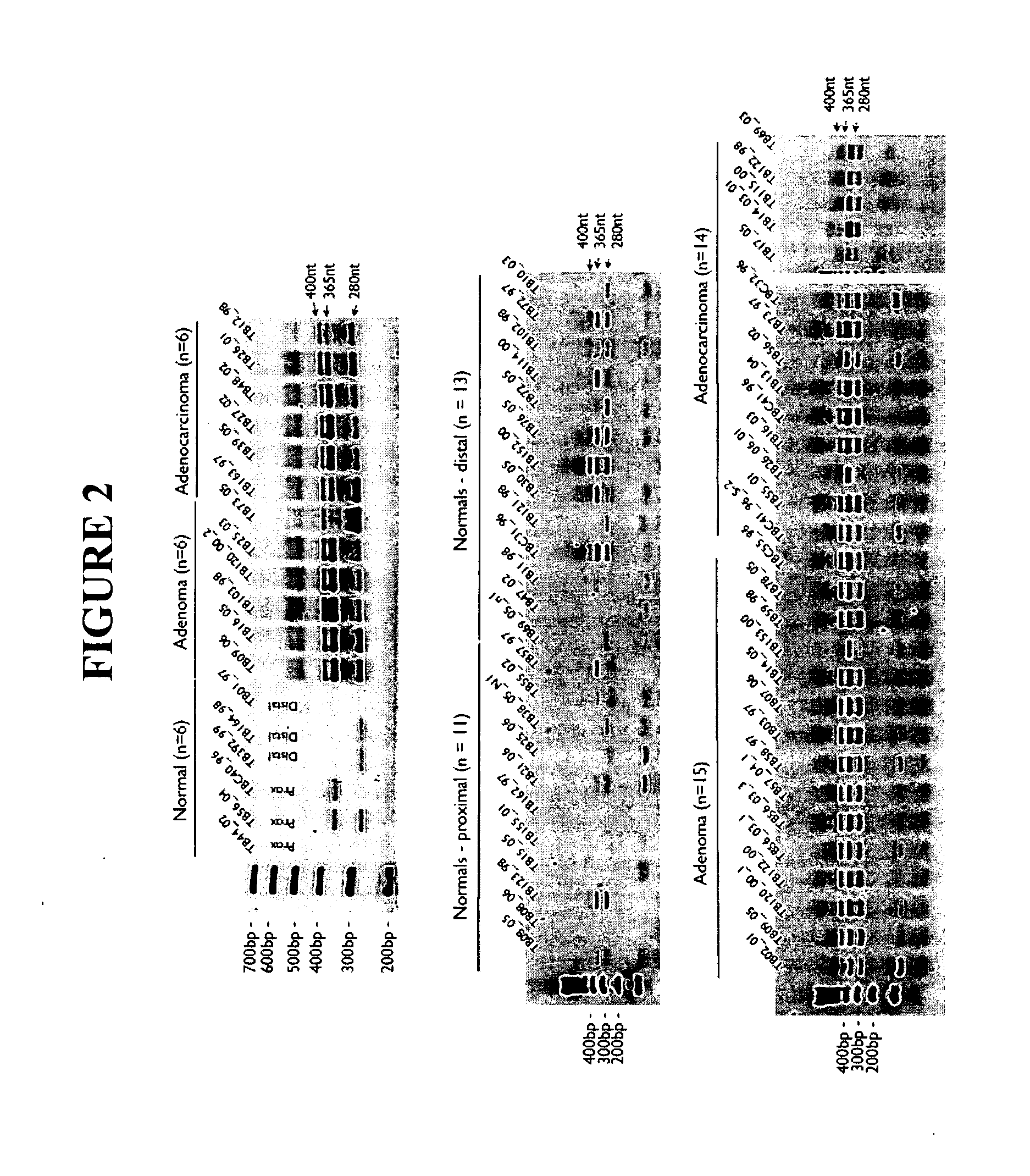
The present invention relates generally to a nucleic acid molecule, the RNA and protein expression profiles of which are indicative of the onset, predisposition to the onset and / or progression of a large intestine neoplasm. More particularly, the present invention is directed to a nucleic acid molecule, the expression profiles of which are indicative of the onset and / or progression of a colorectal neoplasm, such as an adenoma or an adenocarcinoma. The expression profiles of the present invention are useful in a range of applications including, but not limited to, those relating to the diagnosis and / or monitoring of colorectal neoplasms, such as colorectal adenomas and adenocarcinomas. Accordingly, in a related aspect the present invention is directed to a method of screening a subject for the onset, predisposition to the onset and / or progression of a large intestine neoplasm by screening for modulation in the expression profile of said nucleic acid molecule markers.
Owner:CLINICAL GENOMICS PTY LTD +1
Antibody-based system for detection of differential protein expression patterns
InactiveUS20060014301A1Character and pattern recognitionMaterial analysisDiseaseProtein expression profile
The present invention is a kit and method for identifying the presence or absence of a protein expression profile that is known to be associated with a particular disease or an altered biological state. The method is based on the combination of a known protein expression pattern biomarker with the use of an antibody-based detection system. The images of two antibody-based detection systems are compared by an overlay procedure to determine protein expression patterns in biological samples.
Owner:NEOGENOMICS INC
Biomarkers for prediction of breast cancer
InactiveUS20120149594A1Clinical decisionMicrobiological testing/measurementImmunoglobulins against animals/humansEarly breast cancerCalcification
The invention provides gene expression profiles (GEPs), protein expression profiles (PEPs) as well as gene / protein expression profiles (GPEPs) and methods for using them to identify those patients who are likely to progress to breast cancer after detection of suspicious calcifications and / or fibrocystic disease by standard imaging techniques, e.g., mammography, MRI or ultrasound. The present invention further allows a treatment provider to identify those patients who are most likely to develop breast cancer to initiate and / or adjust treatment options for such patients accordingly.
Owner:NUCLEA BIOMARKERS
Aptamer barcoding
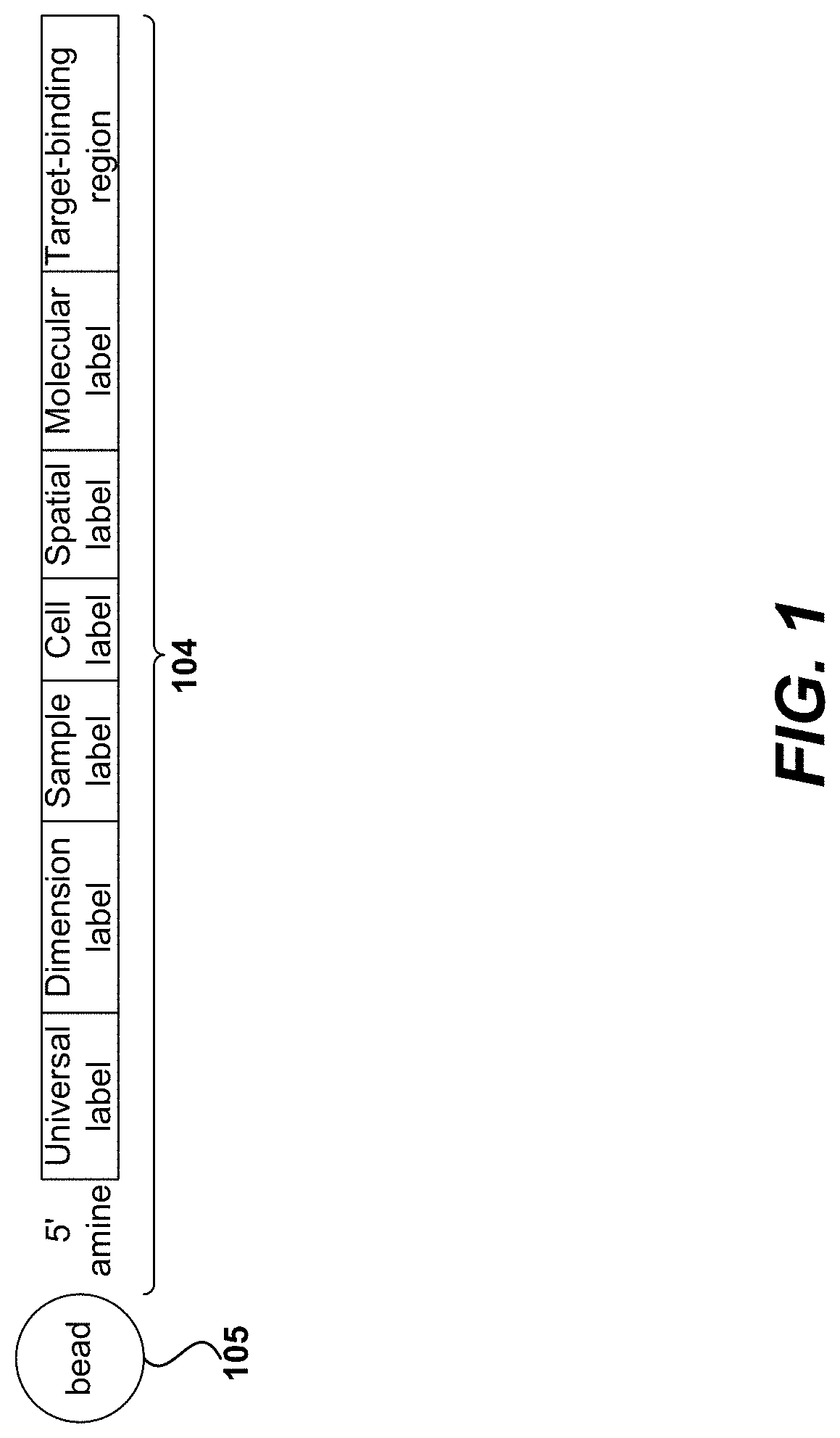
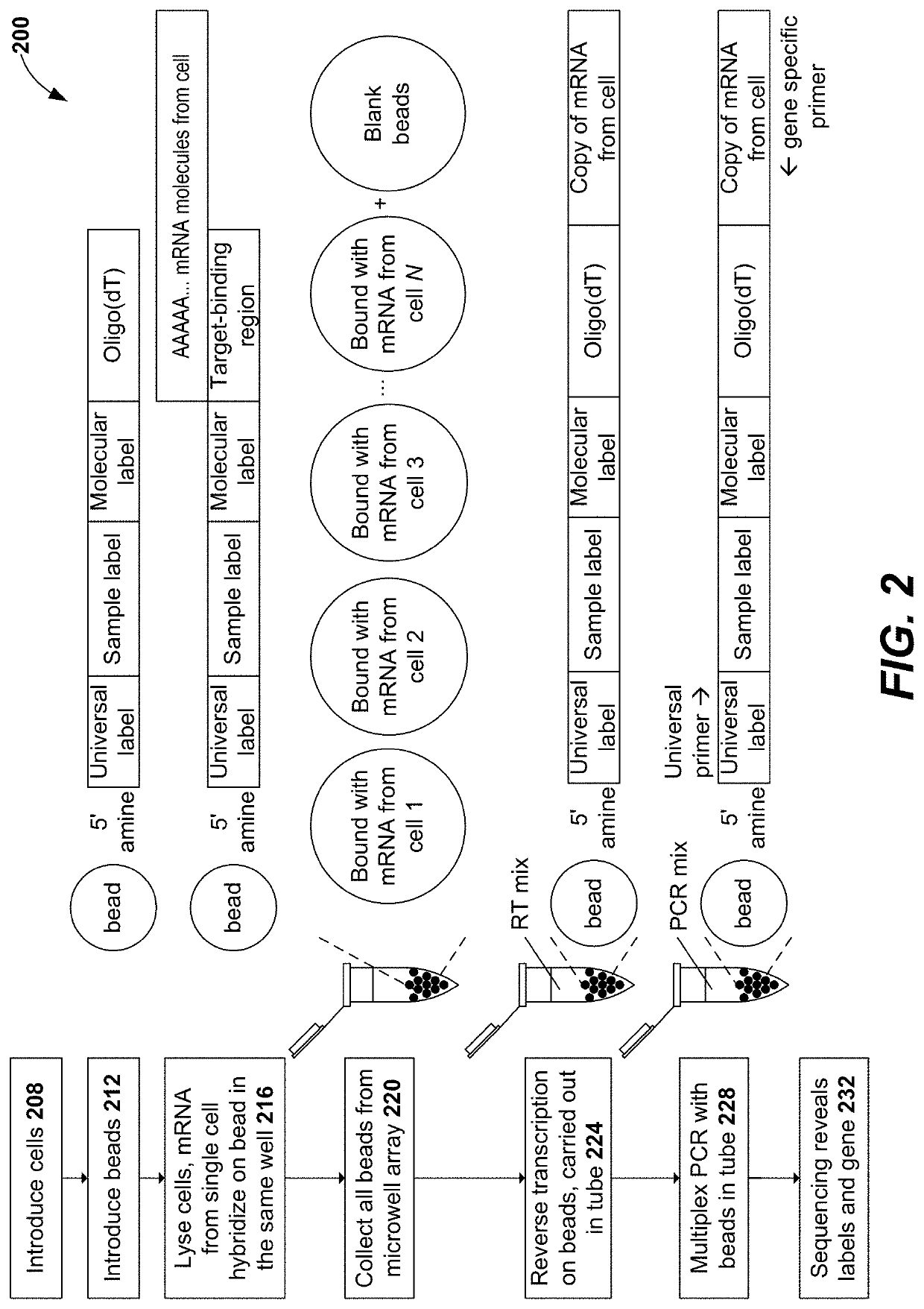
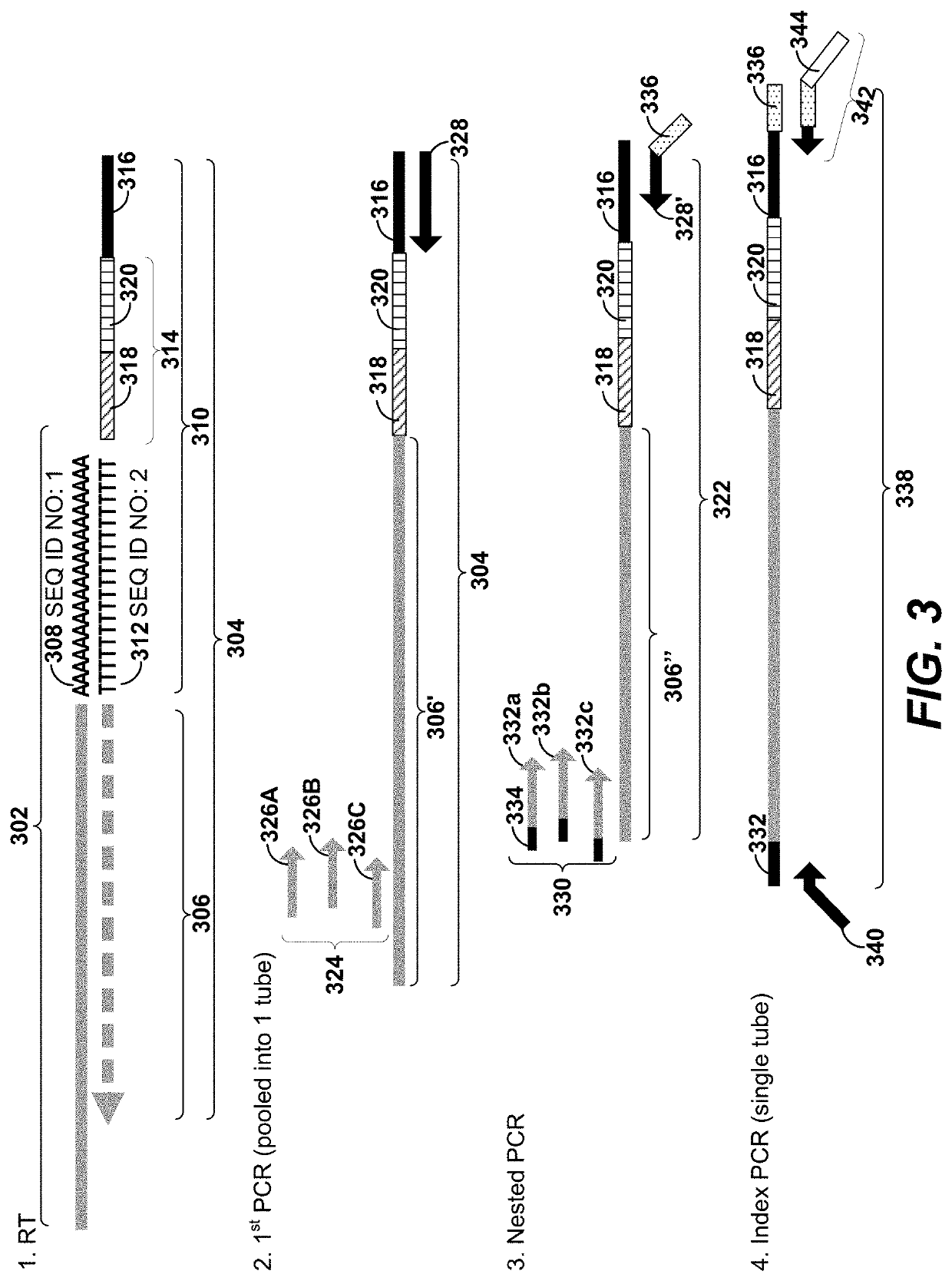
Disclosed herein include systems, methods, compositions, and kits for sample identification and protein expression profiling. A sample indexing composition, or a composition for protein expression profiling, can comprise, for example, a protein binding aptamer associated with an oligonucleotide, such as a sample indexing oligonucleotide. Different oligonucleotides can have different sequences. Sample origin of cells, or protein expression profiles of cells, can be determined based on the sequences of the oligonucleotides by, for example, barcoding the oligonucleotides.
Owner:BECTON DICKINSON & CO
Systems and methods for multiplexed biomarker quantitation using single cell segmentation on sequentially stained tissue
Improved systems and methods for the analysis of digital images are provided. More particularly, the present disclosure provides for improved systems and methods for the analysis of digital images of biological tissue samples. Exemplary embodiments provide for: i) segmenting, ii) grouping, and iii) quantifying molecular protein profiles of individual cells in terms of sub cellular compartments (nuclei, membrane, and cytoplasm). The systems and methods of the present disclosure advantageously perform tissue segmentation at the sub-cellular level to facilitate analyzing, grouping and quantifying protein expression profiles of tissue in tissue sections globally and / or locally. Performing local-global tissue analysis and protein quantification advantageously enables correlation of spatial and molecular configuration of cells with molecular information of different types of cancer.
Owner:莱卡微系统 CMS +1
Markers and diagnostic methods for metastasis
InactiveUS8735329B2Peptide/protein ingredientsLibrary screeningLymphatic SpreadProtein expression profile
The present invention provides methods for the prediction, prognosis and / or diagnosis of metastasis. The present invention also provides proteins (or the related nucleic acid sequences) or protein expression profiles which are predictive and / or prognostic for metastasis. The invention thus relates to the use of said proteins and the corresponding amino acid or nucleic acid sequences for the prediction, prognosis or diagnosis of metastasis.
Owner:KATHOLIEKE UNIV LEUVEN
Protein Expression Profile Database
InactiveUS20100137151A1Library screeningBiological testingSequence databaseProtein expression profile
This invention describes the use of peptide profiling to identify, characterize, and classify biological samples. In complex samples, many thousands of different peptides will be present at varying concentrations. The invention uses liquid chromatography and similar methods to separate peptides, which are then identified and quantified using mass spectrometry. By identification it is meant that the correct sequence of the peptide is established through comparisons with genome sequence databases, since the majority of peptides and proteins are unannotated and have no ascribed name or function. Quantification means an estimate of the absolute or relative abundance of the peptide species using mass spectrometry and related techniques including, but not limited to, pre- or post-experimental stable or unstable isotope incorporation, molecular mass tagging, is differential mass tagging, and amino acid analysis.
Owner:EMILI ANDREW +1
Methods and compositions for typing molecular subgroups of medulloblastoma
ActiveUS20140155400A1Quick and accurate identificationOrganic active ingredientsDisease diagnosisPositive controlBeta-catenin
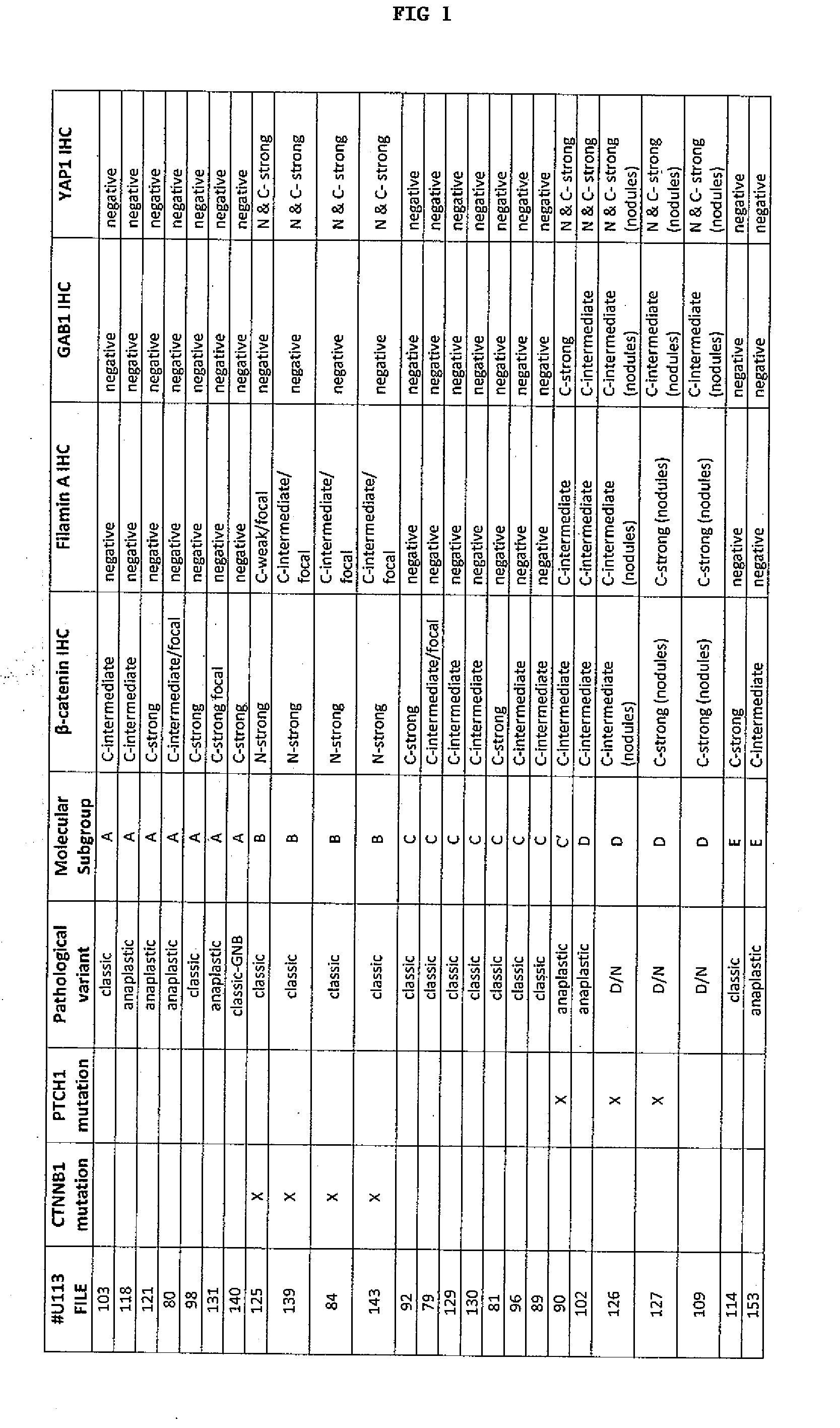
Immunohistochemical methods and compositions for the typing of molecular subgroups of medulloblastomas are provided. The methods comprise determining a protein expression profile for a sample obtained from a medulloblastoma by detecting expression of GAB 1, filamin A, or at least two biomarker proteins selected from the group consisting of β-catenin, YAP1, GAB1, and filamin A, and typing the medulloblastoma as a WNT pathway tumor, a SHH pathway tumor, or a non-WNT / non-SHH tumor based on this protein expression profile. Kits for typing a medulloblastoma according to these three molecular subgroups are provided. The kits comprise at least two antibodies, wherein each of said antibodies specifically binds to a distinct biomarker protein selected from the group consisting of β-catenin, YAP1, GAB1, and filamin A, and can optionally comprise one or more of instructions for use, reagents for detecting antibody binding to one or more of said biomarker proteins, and one or more positive control samples.
Owner:ST JUDE CHILDRENS RES HOSPITAL INC
Method for correlating gene expression profiles with protein expression profiles
InactiveCN1636068AMicrobiological testing/measurementMaterial analysis by electric/magnetic meansProtein insertionProtein expression profile
Owner:CIPHERGEN BIOSYSTEMS INC
Secreted phospholipase a2 biomarkers for arthritis
ActiveUS20120122117A1Accurate diagnosisDiagnosing determiningPeptide/protein ingredientsMammal material medical ingredientsSecreted Phospholipases A2Protein expression profile
The present invention relates to the use of protein expression profiles of sPLA2 isoforms with clinical relevance to osteoarthritis (OA). In particular, the invention provides methods for diagnosing OA or determining risk factors for development of OA based on expression of sPLA2-IIA.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC +2
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com