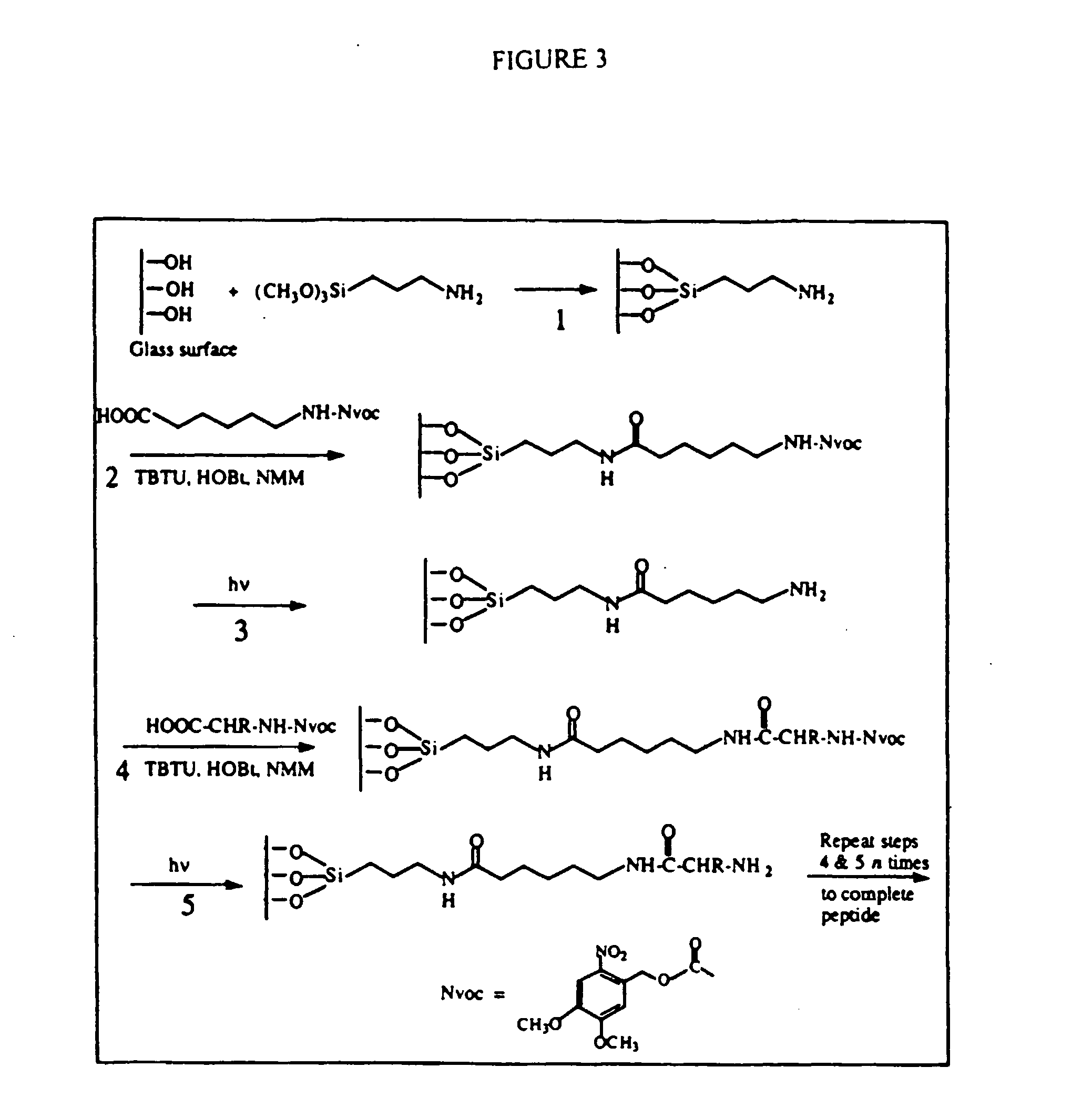
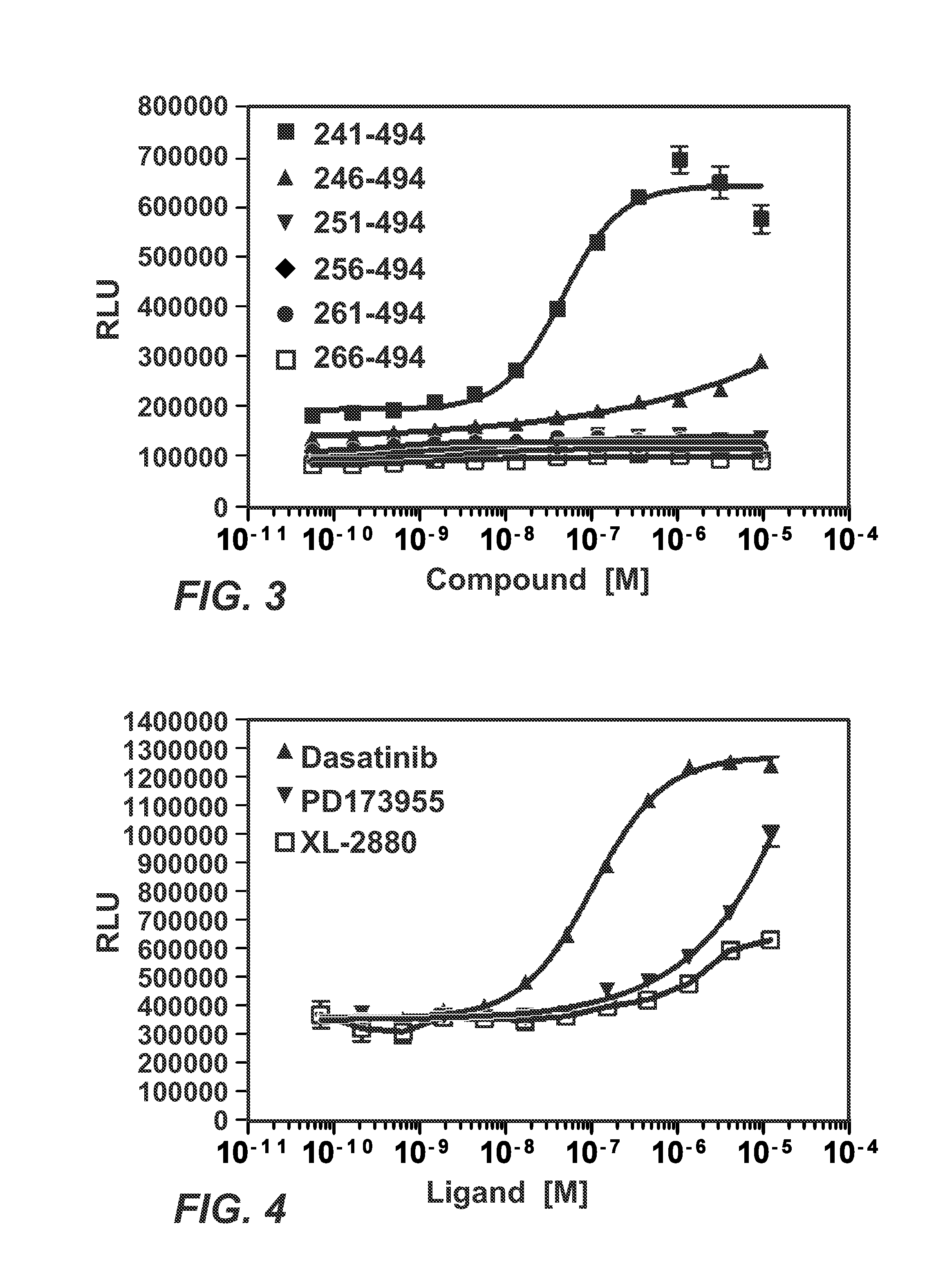
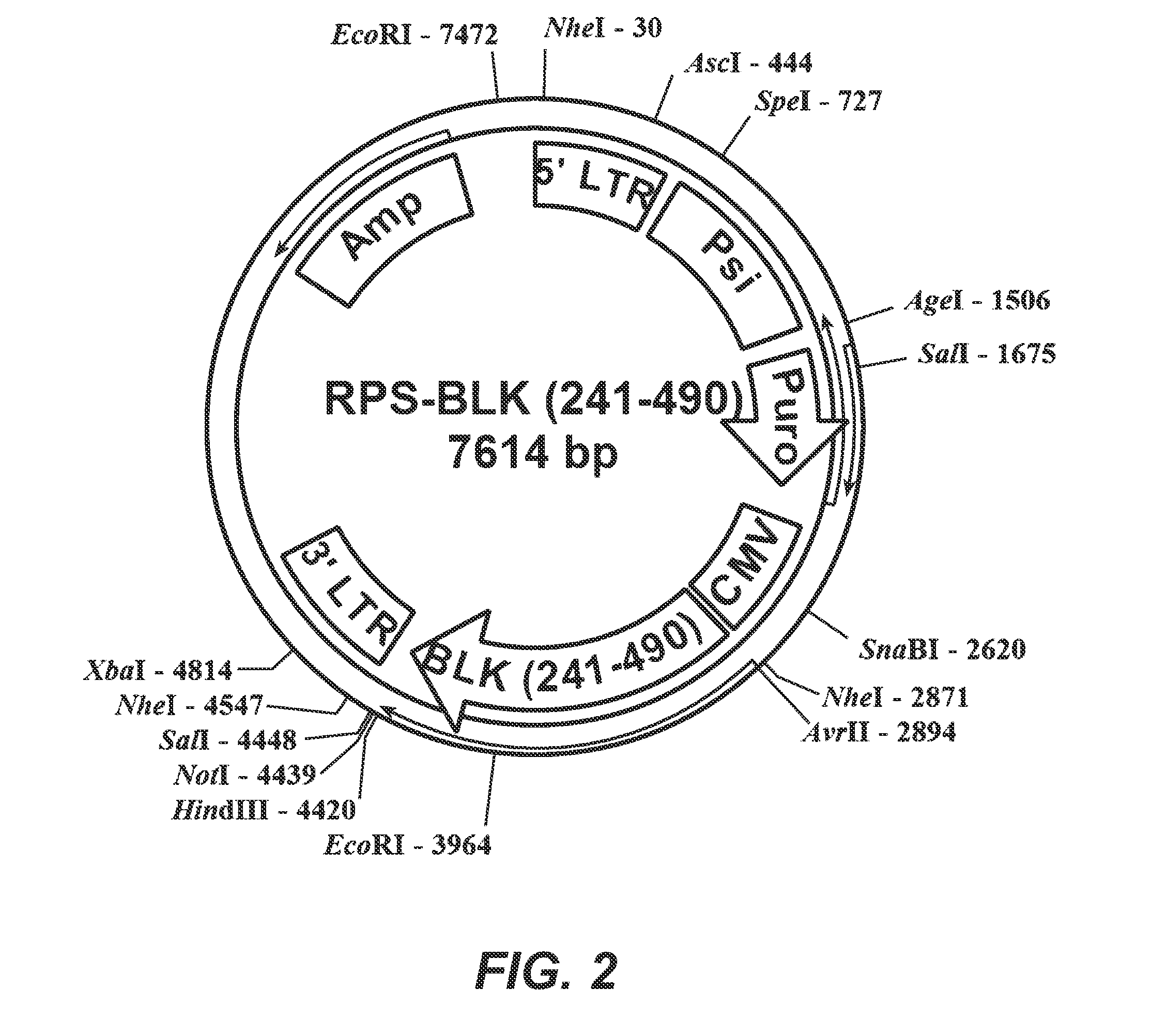
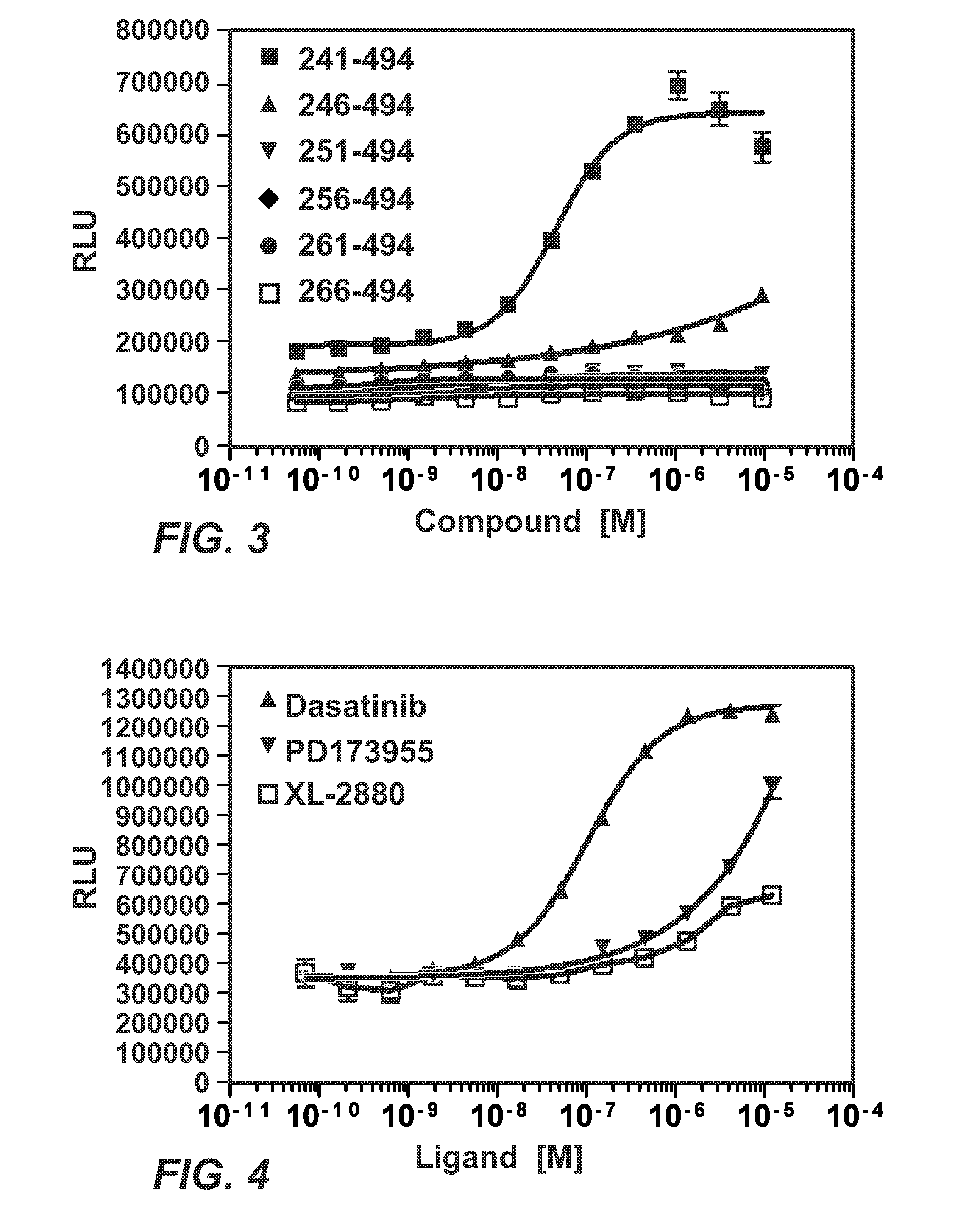
Patents
Literature
30 results about "Protein abundance" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Protein abundance is calculated from the sum of all unique normalised peptide ion abundances for a specific protein on each run. Alignment and co-detection of features means you have exactly the same number of quantified peptides identified on all runs so you can compare the sum of ion abundances between groups.
Methods for drug target screening
InactiveUS6165709AMicrobiological testing/measurementRecombinant DNA-technologyProtein targetProtein activity
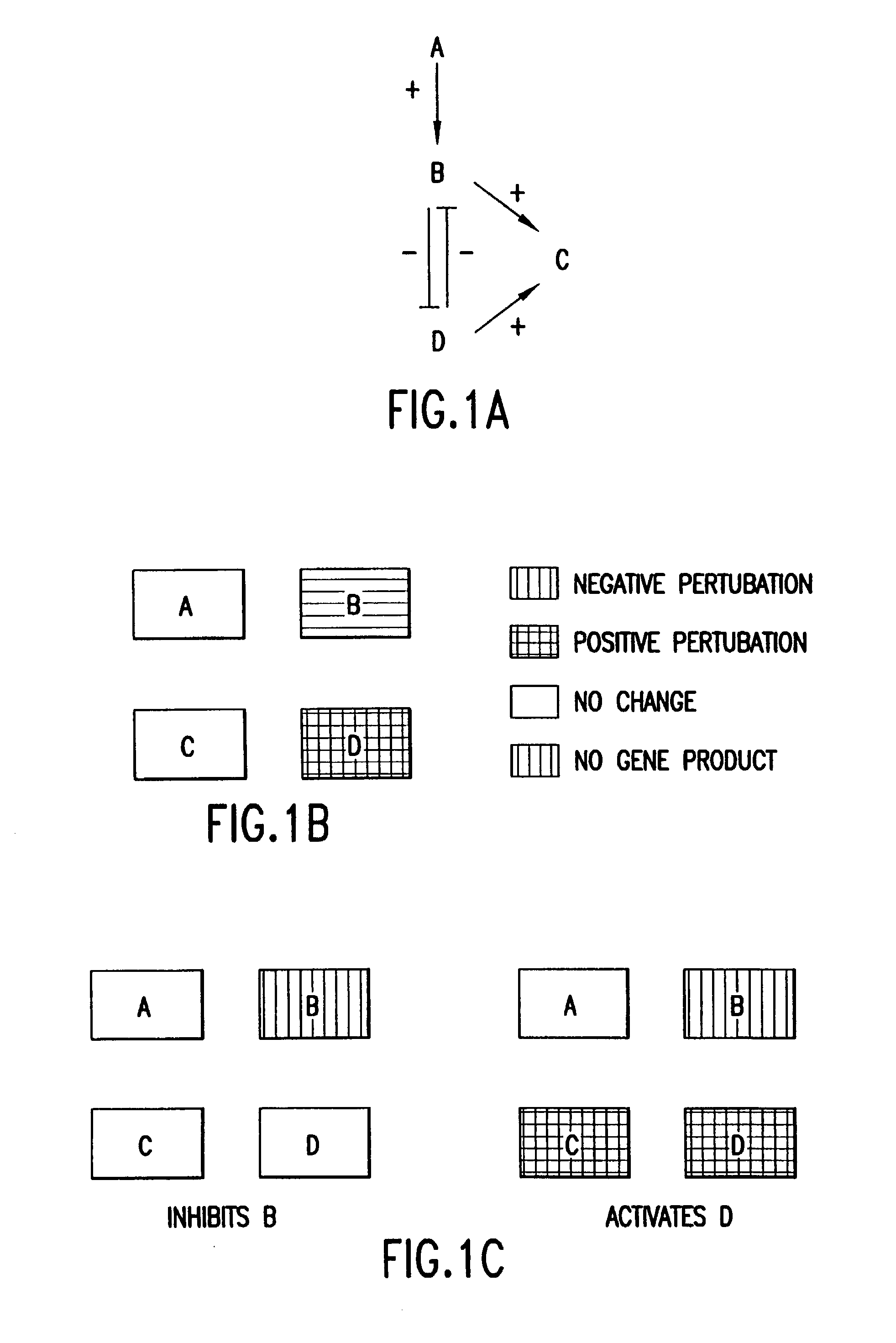
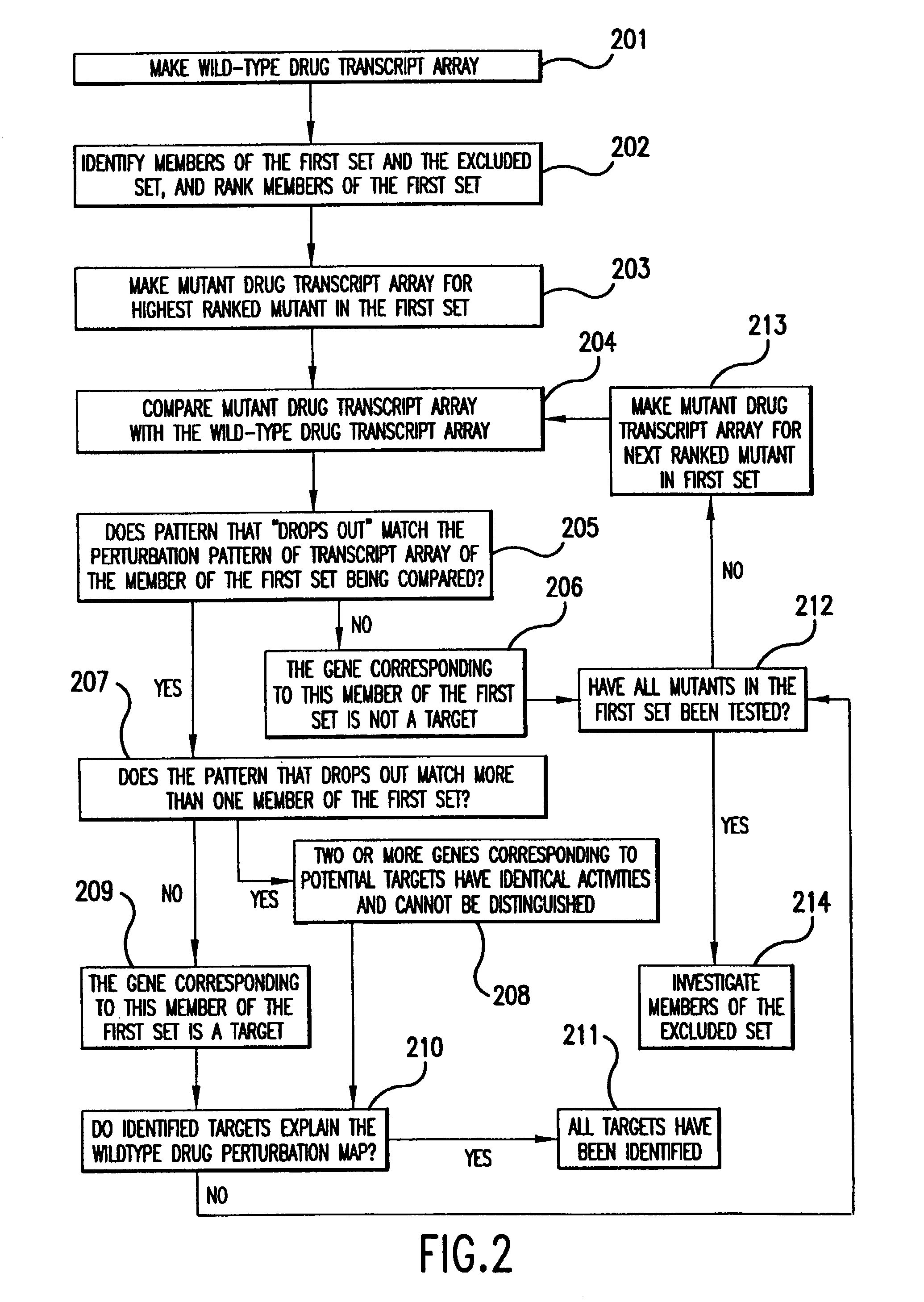
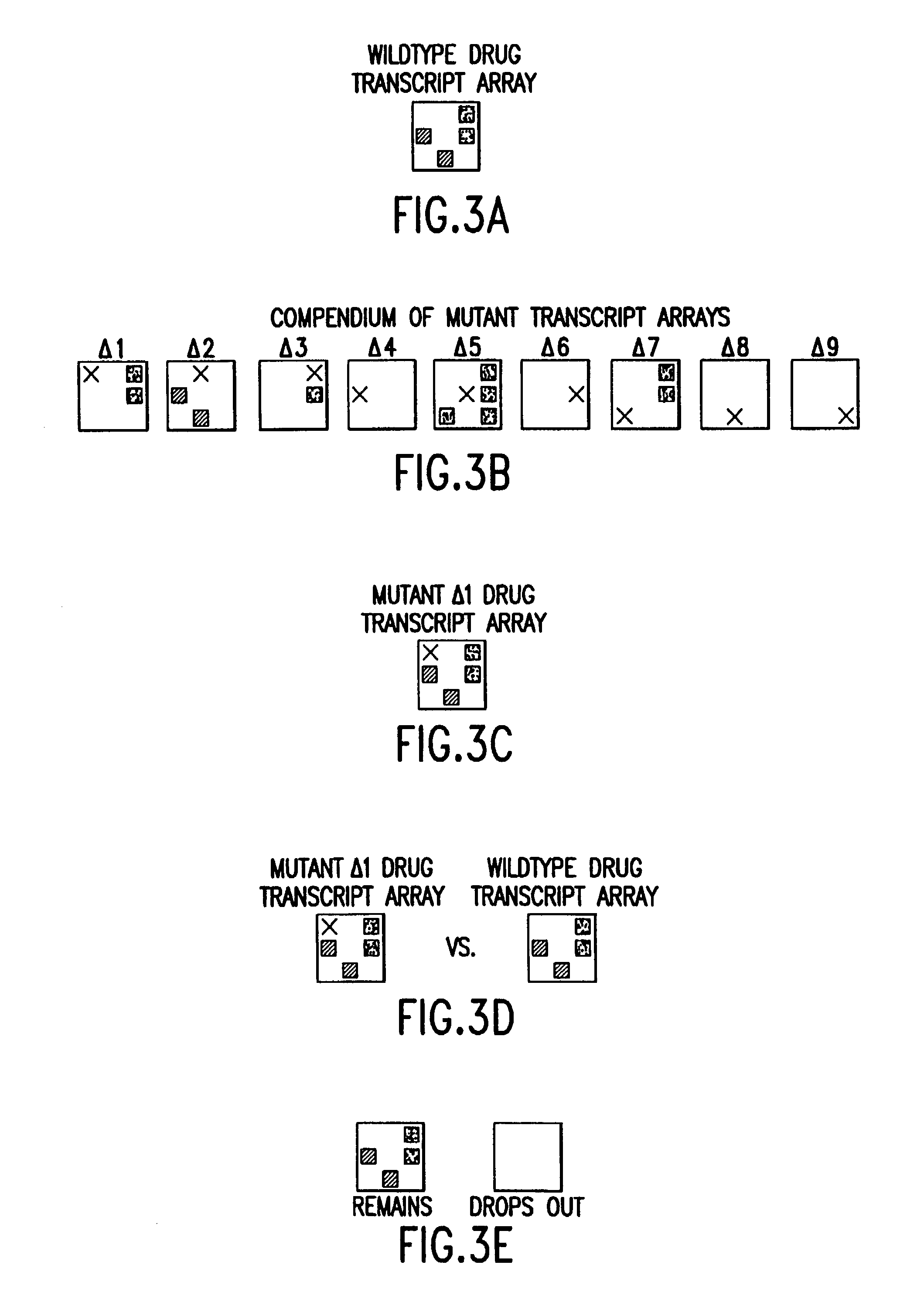
The present invention provides methods for identifying targets of a drug in a cell by comparing (i) the effects of the drug on a wild-type cell, (ii) the effects on a wild-type cell of modifications to a putative target of the drug, and (iii) the effects of the drug on a wild-type cell which has had the putative target modified of the drug. In various embodiments, the effects on the cell can be determined by measuring gene expression, protein abundances, protein activities, or a combination of such measurements. In various embodiments, modifications to a putative target in the cell can be made by modifications to the genes encoding the target, modification to abundances of RNAs encoding the target, modifications to abundances of target proteins, or modifications to activities of the target proteins. The present invention also provides methods for drug development based on the methods for identifying drug targets.
Owner:FRED HUTCHINSON CANCER RES CENT
Apparatus, composition and method for proteome profiling
InactiveUS20050048566A1Sure easyPeptide librariesLibrary screeningProtein expression profileProtein abundance
The present invention is directed to a high throughput method for producing a large number of different antibodies, more specifically organized antibody microarrays. These antibodies and antibody microarrays can be used to rapidly assay protein abundance and identify types of proteins that are expressed in cells and tissues under a variety of conditions, or to compare protein expression profiles of different cells.
Owner:DELISI CHARLES +5
Methods and Compositions for Diagnosis of Ovarian Cancer
Methods and compositions are provided for diagnosing ovarian cancer in a mammalian subject, preferably in a serum or plasma sample of a human subject. The methods and compositions enable the detection or measurement in the sample or from a protein level profile generated from the sample, the protein level of one or more specified biomarkers. Comparing the protein level(s) of the biomarker(s) in the subject's sample or from protein abundance profile of multiple biomarkers, with the level of the same biomarker(s) or profile in a reference standard, permits the determination of a diagnosis of ovarian cancer, or the identification of a risk of developing ovarian cancer, or enables the monitoring of the status of progression or remission of ovarian cancer in the subject followed during a therapeutic protocol.
Owner:THE WISTAR INST OF ANATOMY & BIOLOGY
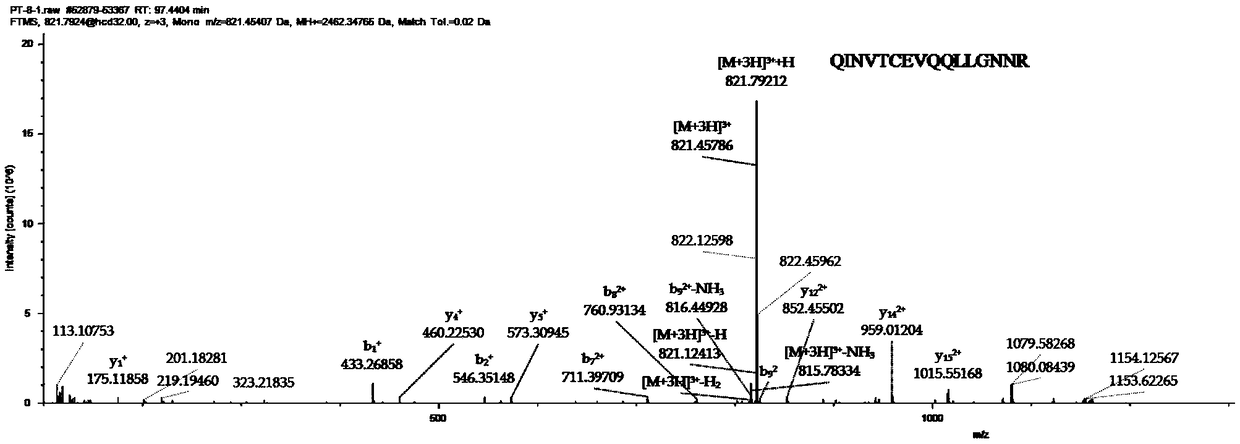
Absolute quantitation of protein contents based on exponentially modified protein abundance index by mass spectrometry
InactiveUS7910372B2Biological testingSpecial data processing applicationsCancer cellGene and protein expression
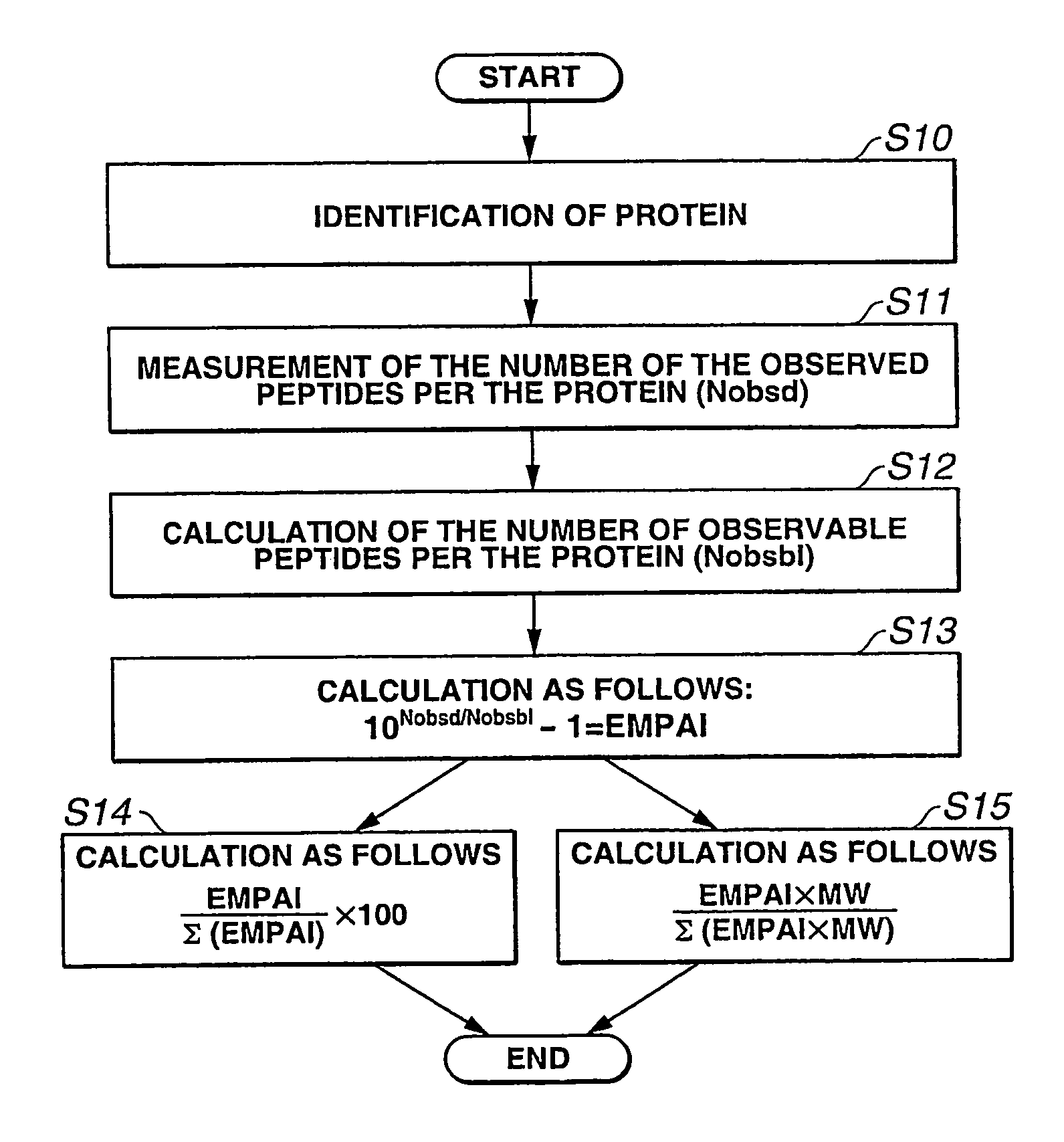
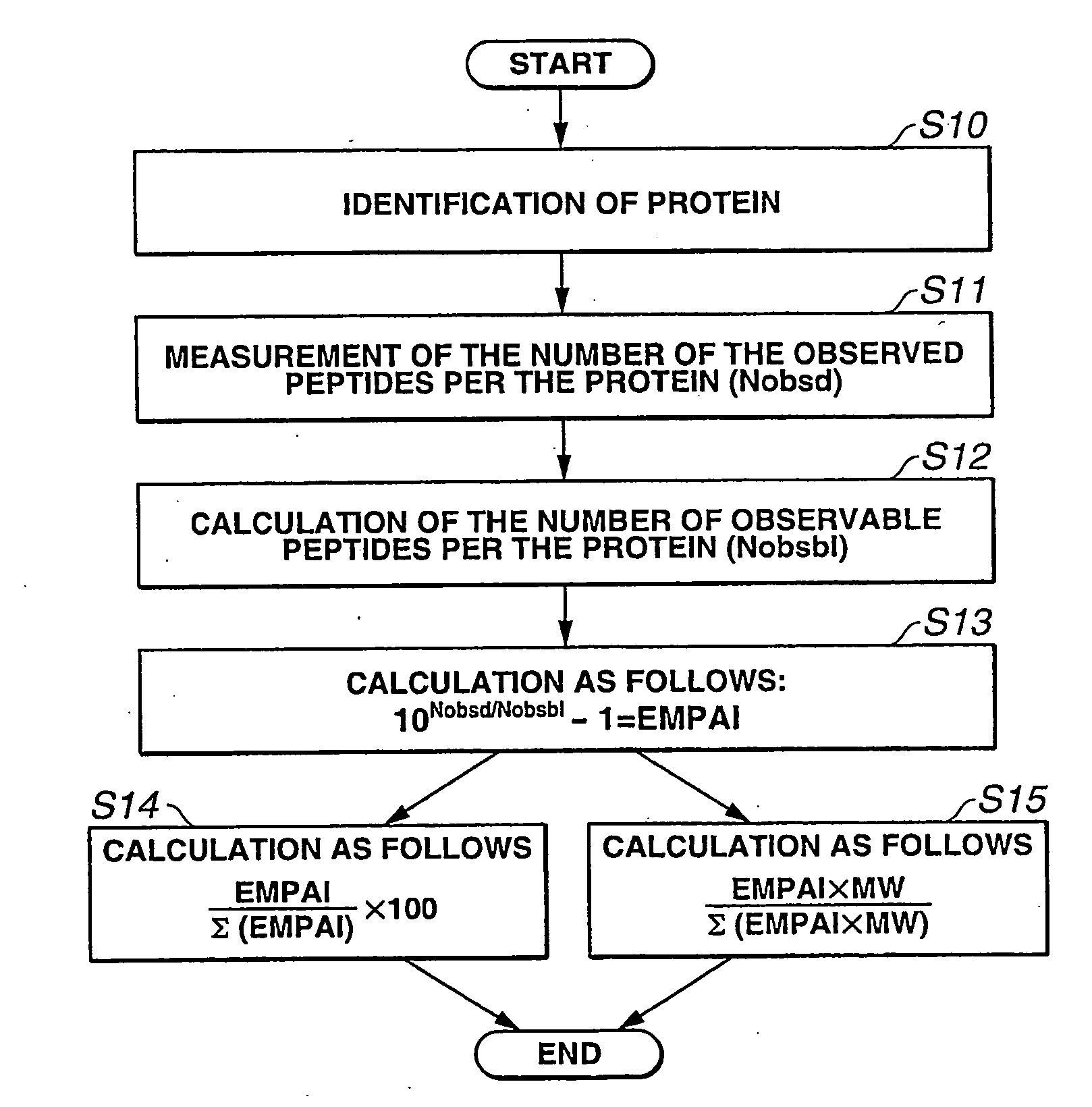
The present inventor has established protein abundance index (PAI, π) to determine the protein contents in a protein mixture solution using nanoLC-MSMS data. Digested peptides were analyzed by nanoLC-MS / MS and the obtained results were applied to a Mascot protein identification algorism based on tandem mass spectra. For absolute quantitation, PAI was converted to exponentially modified PAI (EMPAI, m π), which is proportional to protein contents in the protein mixture. EMPAI was successfully applied to comprehensive protein expression analysis and performed a comparison study between gene and protein expression in an HCT116 human cancer cells. Accordingly, the present invention provides a method and a computer program for quantifying the protein contents based on the protein abundance index.
Owner:EISIA R&D MANAGEMENT CO LTD
Absolute Quantitation of Protein Contents Based on Exponentially Modified Protein Abundance Index by Mass Spectrometry
InactiveUS20080319676A1Biological testingSpecial data processing applicationsCancer cellWhole cell lysate
The present inventor has established protein abundance index (PAI, π) to determine the protein contents in a protein mixture solution using nanoLC-MSMS data. Digested peptides were analyzed by nanoLC-MS / MS and the obtained results were applied to a Mascot protein identification algorism based on tandem mass spectra. PAI is defined as the number of observed peptides divided by the number of observable peptides per protein. PAI from different concentrations of serum albumin showed linear relationship to the logarithm of the protein concentration. This was also valid for 47 proteins in a mouse whole cell lysate analyzed by single run of nanoLC-MS / MS. On the other hand, Mascot protein scores as well as the number of identified peptides per protein were less correlated to the protein abundance. For absolute quantitation, PAI was converted to exponentially modified PAI (EMPAI, mπ), which is proportional to protein contents in the protein mixture. For the 47 proteins in the whole lysate, the deviation percentages of the EMPAI-based concentrations to the actual values were within 63% in average. EMPAI was successfully applied to comprehensive protein expression analysis and performed a comparison study between gene and protein expression in an HCT116 human cancer cells. Accordingly, the present invention provides a method and a computer program for quantifying the protein contents based on the protein abundance index.
Owner:EISIA R&D MANAGEMENT CO LTD
Plant milk
InactiveCN101480204AAdequate milk sourceClean milkFood preparationMilk substitutesHazardous substanceAdditive ingredient
The invention belongs to a largely improved milk-brewing technology in the food field by replacing milk produced in a physiological mechanism with milk produced by synthesizing milky juice obtained from green resources in a manual mechanism. The synthesized milk is prepared by using green marketable grain, beans, black beans, small red beans, sorghum and the like as main ingredients, adding natural auxiliary materials and condiments, fruits, and the like and processing the materials by a plurality of procedures. The material resources comprise the following seven categories: marketable grain, auxiliary materials, condiments, fruits, organic calcium bones, bird eggs and additives, thereby being healthy and natural plant milk with high nutrition, protein abundance, multiple elements and low fat. The synthesized milk can be drunk a long term safely and reliably by people without worry, has faint milk scent, suitable taste and scientific preparation and can be made into milk powder, liquid milk and various food compounds. Moreover, the invention is particularly suitable for infants, children, the middle-aged and the old to eat for a long time, does not contain melamine and any harmful substances so as not to damage the health of human beings and enables milking enterprises to make transition, remanufacture and regeneration without replacing the prior technology and equipment.
Owner:赵宇
Methods and Compositions for Diagnosis of Ovarian Cancer
InactiveUS20140274794A1Peptide librariesTumor rejection antigen precursorsPlasma samplesBlood plasma
Methods and compositions are provided for diagnosing ovarian cancer in a mammalian subject, preferably in a serum or plasma sample of a human subject. The methods and compositions enable the detection or measurement in the sample or from a protein level profile generated from the sample, the protein level of one or more specified biomarkers. Comparing the protein level(s) of the biomarker(s) in the subject's sample or from protein abundance profile of multiple biomarkers, with the level of the same biomarker(s) or profile in a reference standard, permits the determination of a diagnosis of ovarian cancer, or the identification of a risk of developing ovarian cancer, or enables the monitoring of the status of progression or remission of ovarian cancer in the subject followed during a therapeutic protocol.
Owner:THE WISTAR INST OF ANATOMY & BIOLOGY
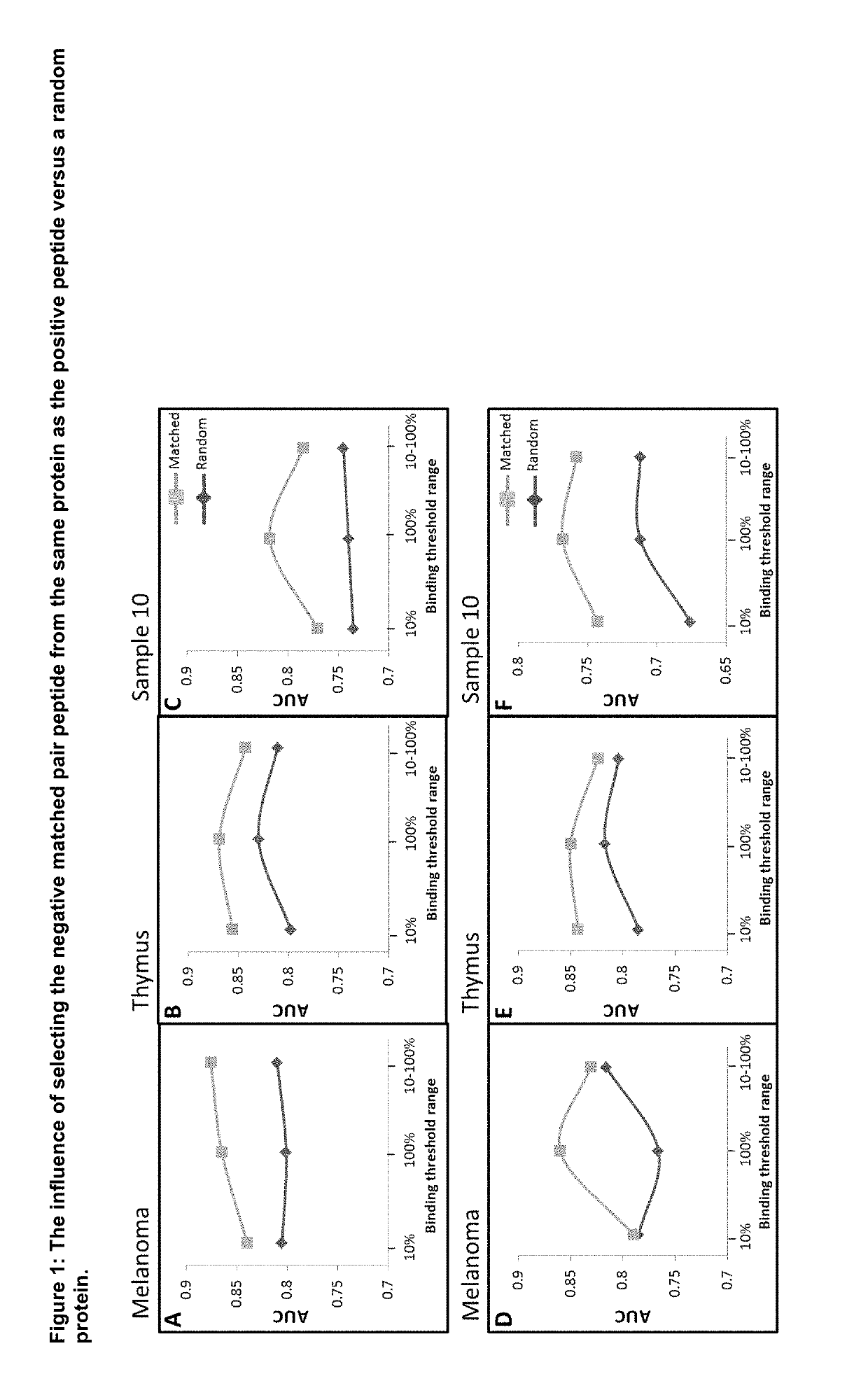
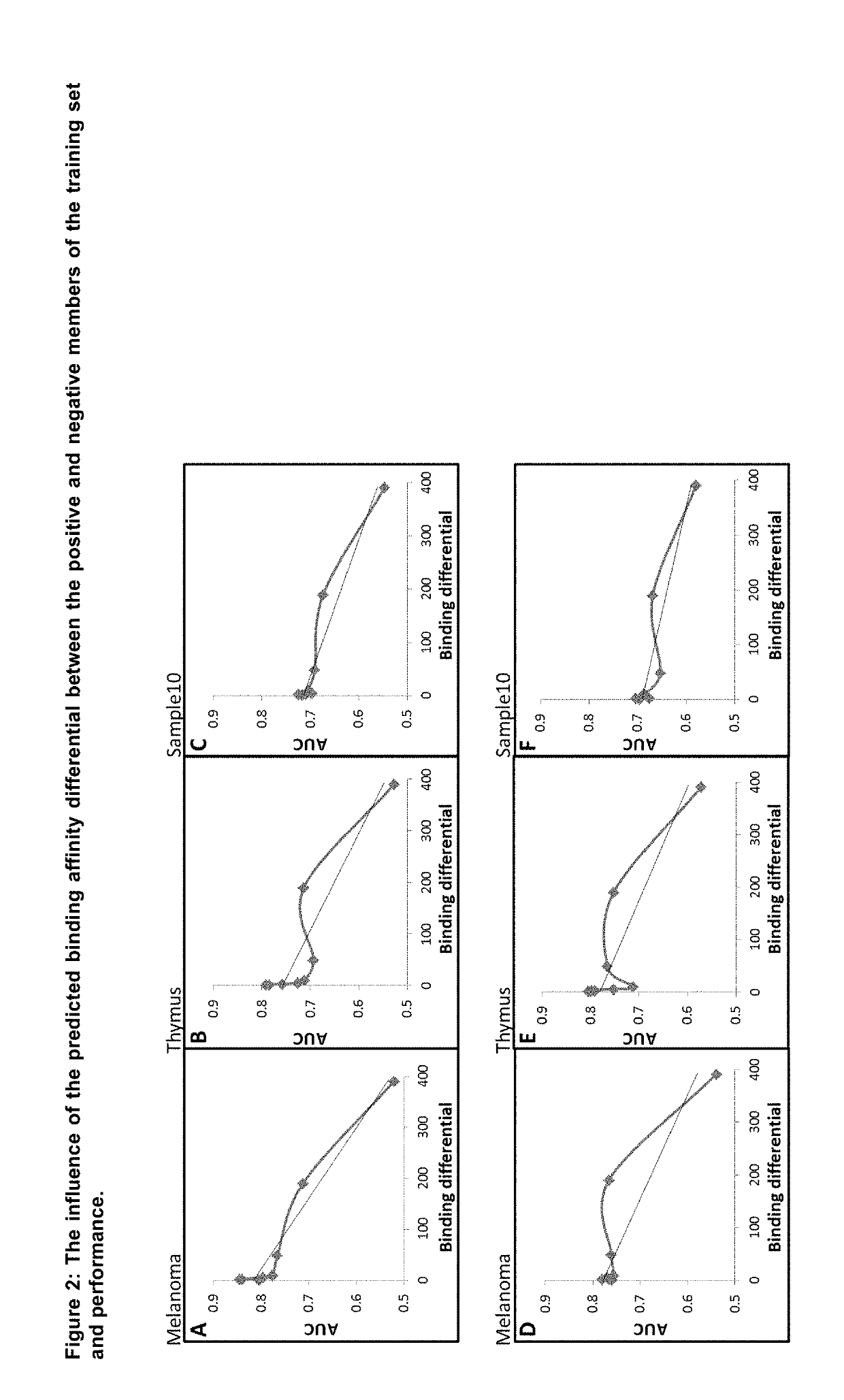
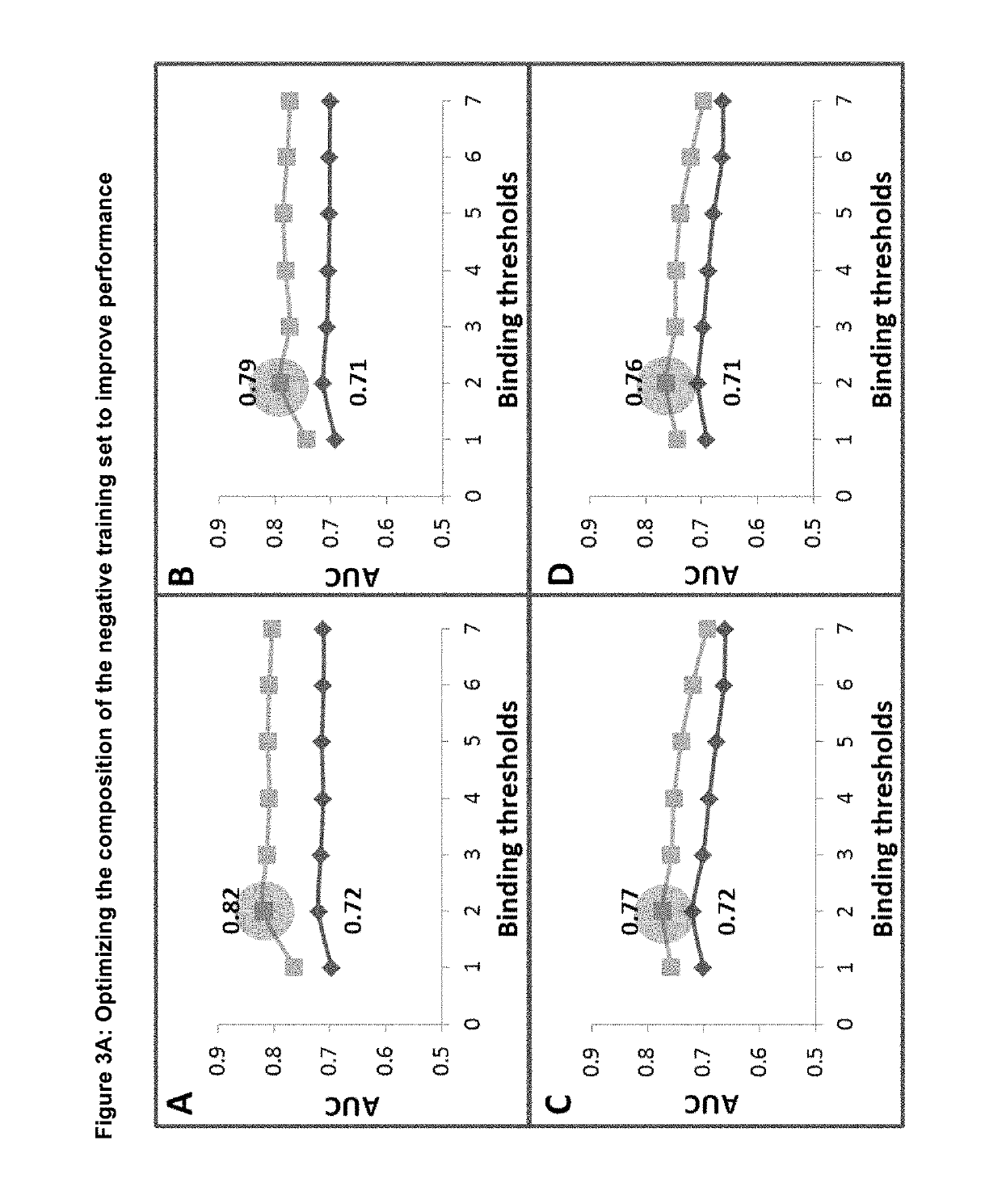
Machine Learning Algorithm for Identifying Peptides that Contain Features Positively Associated with Natural Endogenous or Exogenous Cellular Processing, Transportation and Histocompatibility Complex (MHC) Presentation
PendingUS20190311781A1Reduce riskGood processing characteristicsMathematical modelsKernel methodsMHC restrictionData set
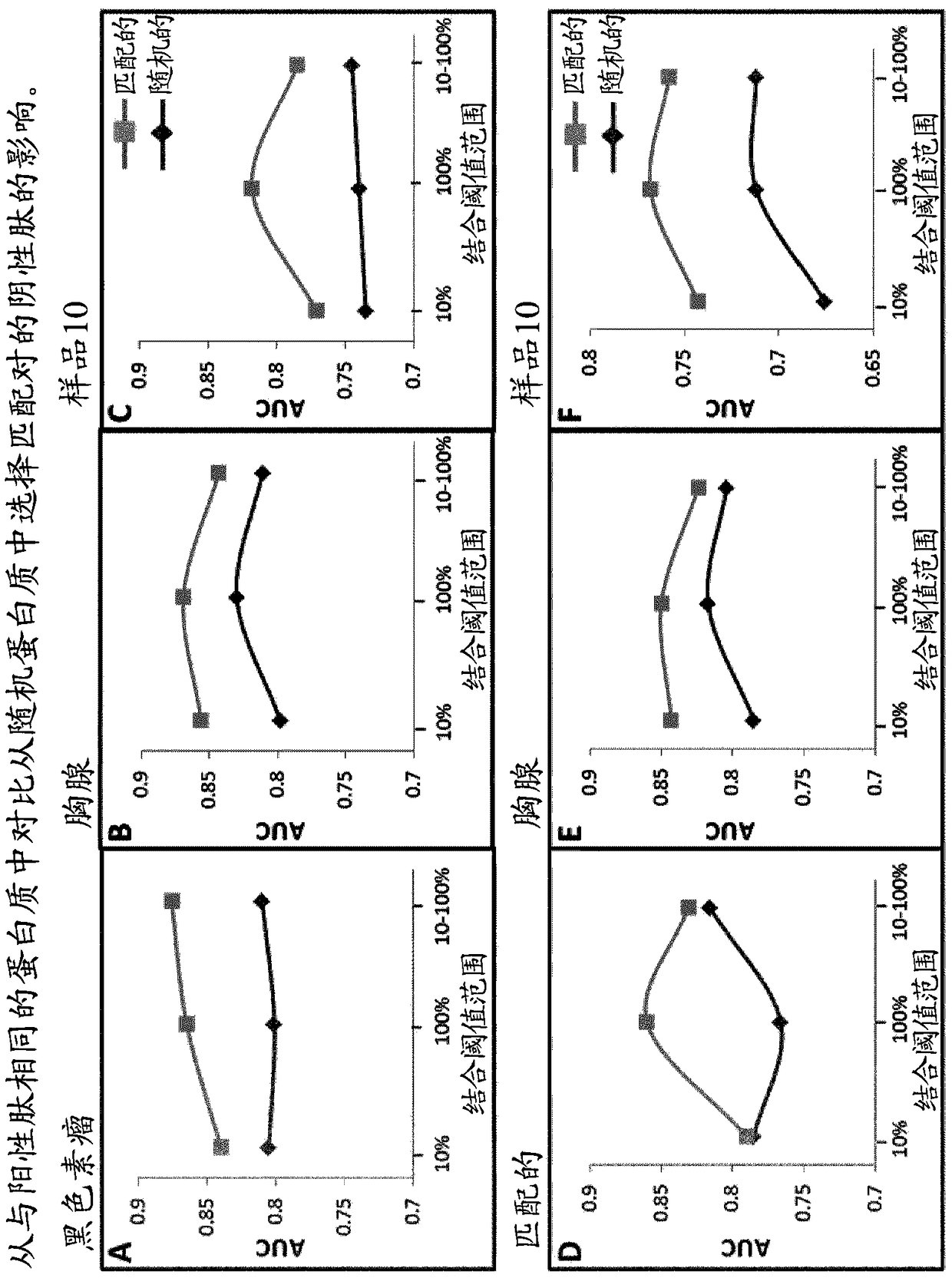
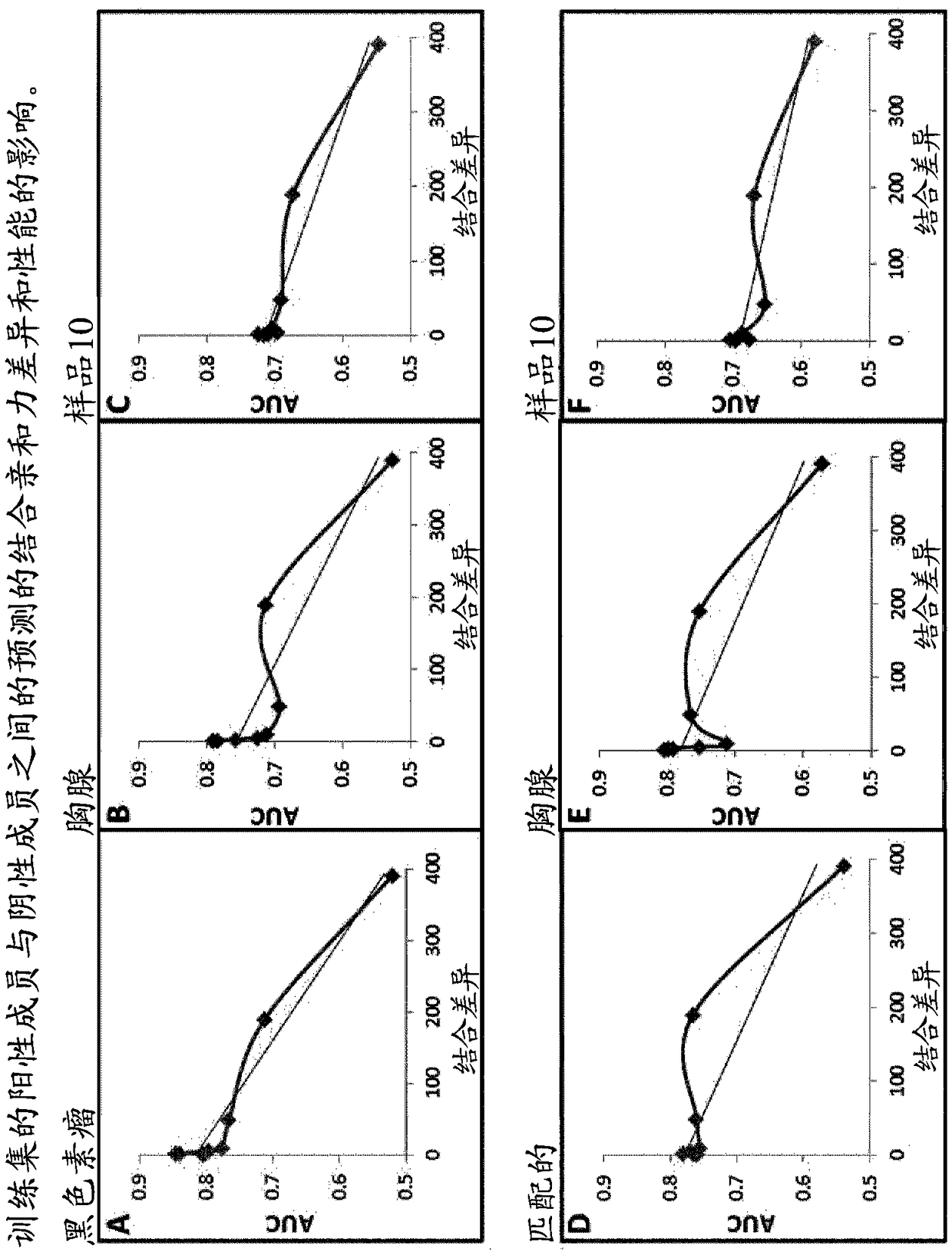
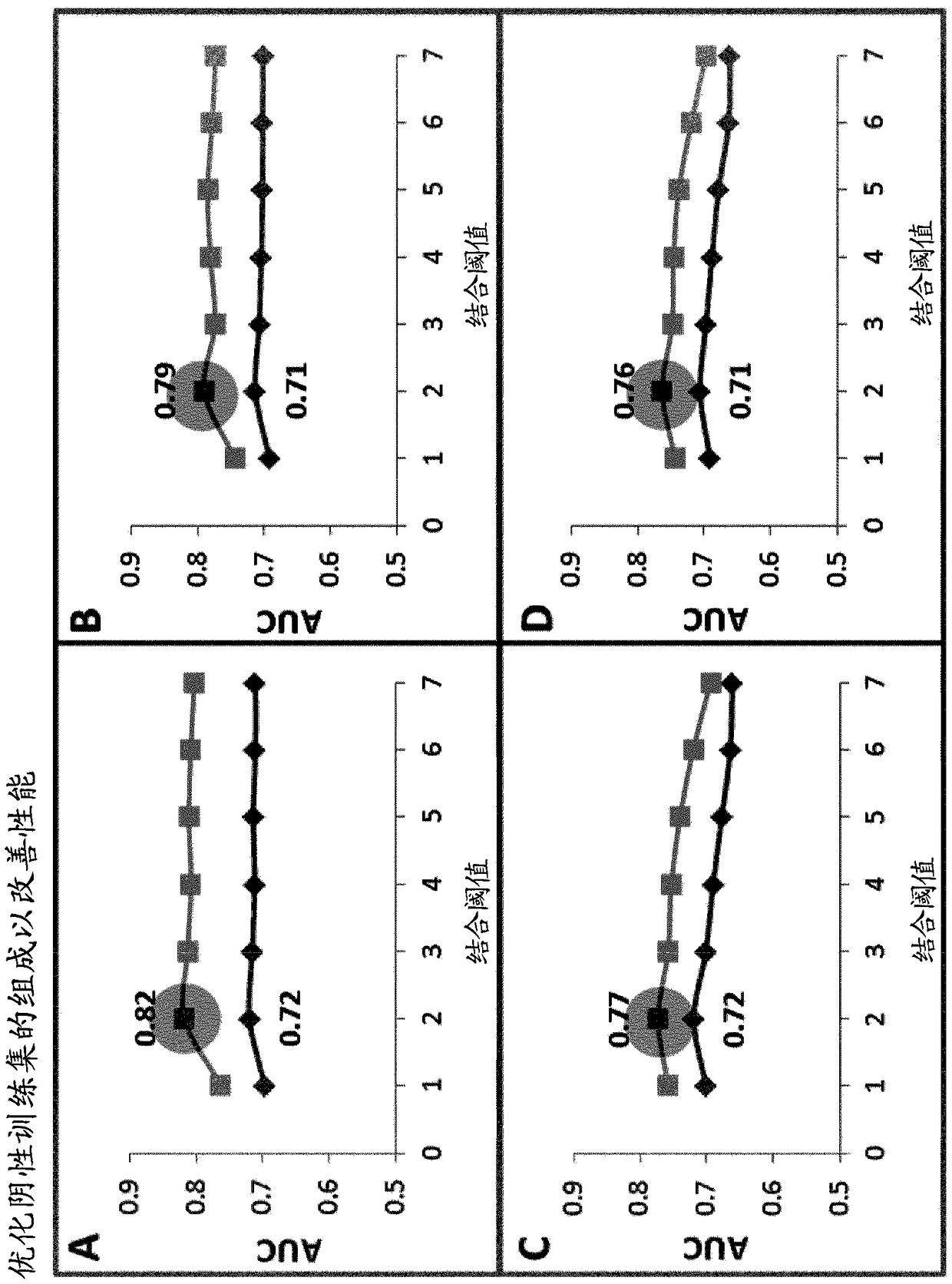
The present invention provides a method for identifying peptides that contain features positively associated with natural endogenous or exogenous cellular processing, transportation and major histocompatibility complex (MHC) presentation. In particular, the invention / method controls for the influence of protein abundance, stability and HLA / MHC binding on processing and presentation, enabling a machine-learning algorithm or statistical inference model trained using the method to be applied to any test peptide regardless of its HLA / MHC restriction i.e. the algorithm operates in a HLA / MHC-agnostic manner. This is attained through the building of positive and negative data sets of peptide sequences (peptides identified or inferred from surface bound or secreted MHC / peptide complexes in the literature, and those which are not). Specifically, the positive and negative data sets comprise a multiplicity of pairings between individual entries, in which both sequences of a pair are of equal or similar length, and are derived from the same source protein, and / or have similar binding affinities, with respect to the HLA / MHC molecule from which the peptide of the positive peptide is restricted.
Owner:ONCOIMMUNITY AS
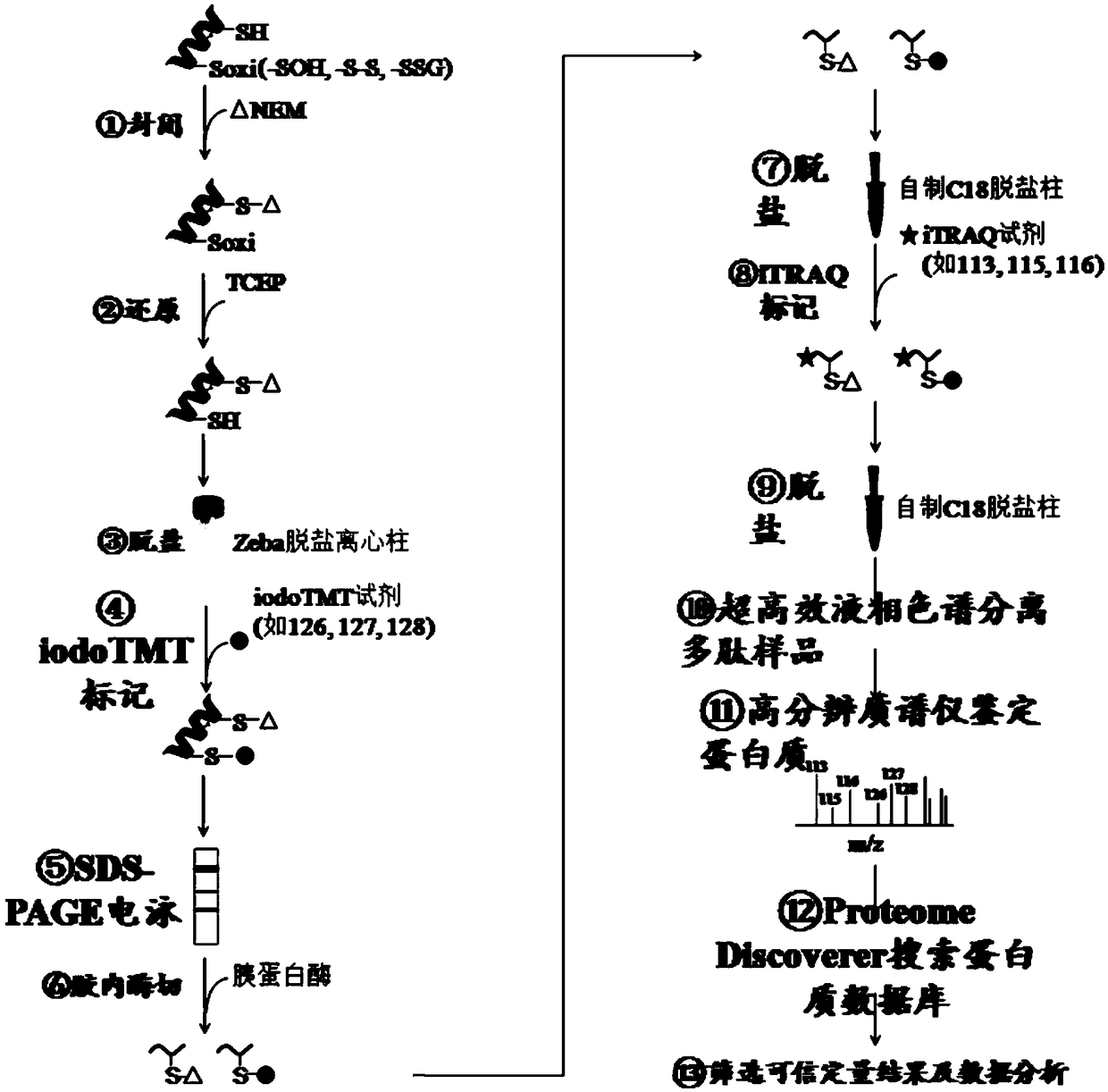
Method for simultaneous quantification of protein abundance and the cysteine oxidation level and application of method
InactiveCN109142561AAccurate judgment of redox levelImprove efficiencyComponent separationCysteine thiolateItraq reagent
Owner:SHANGHAI NORMAL UNIVERSITY
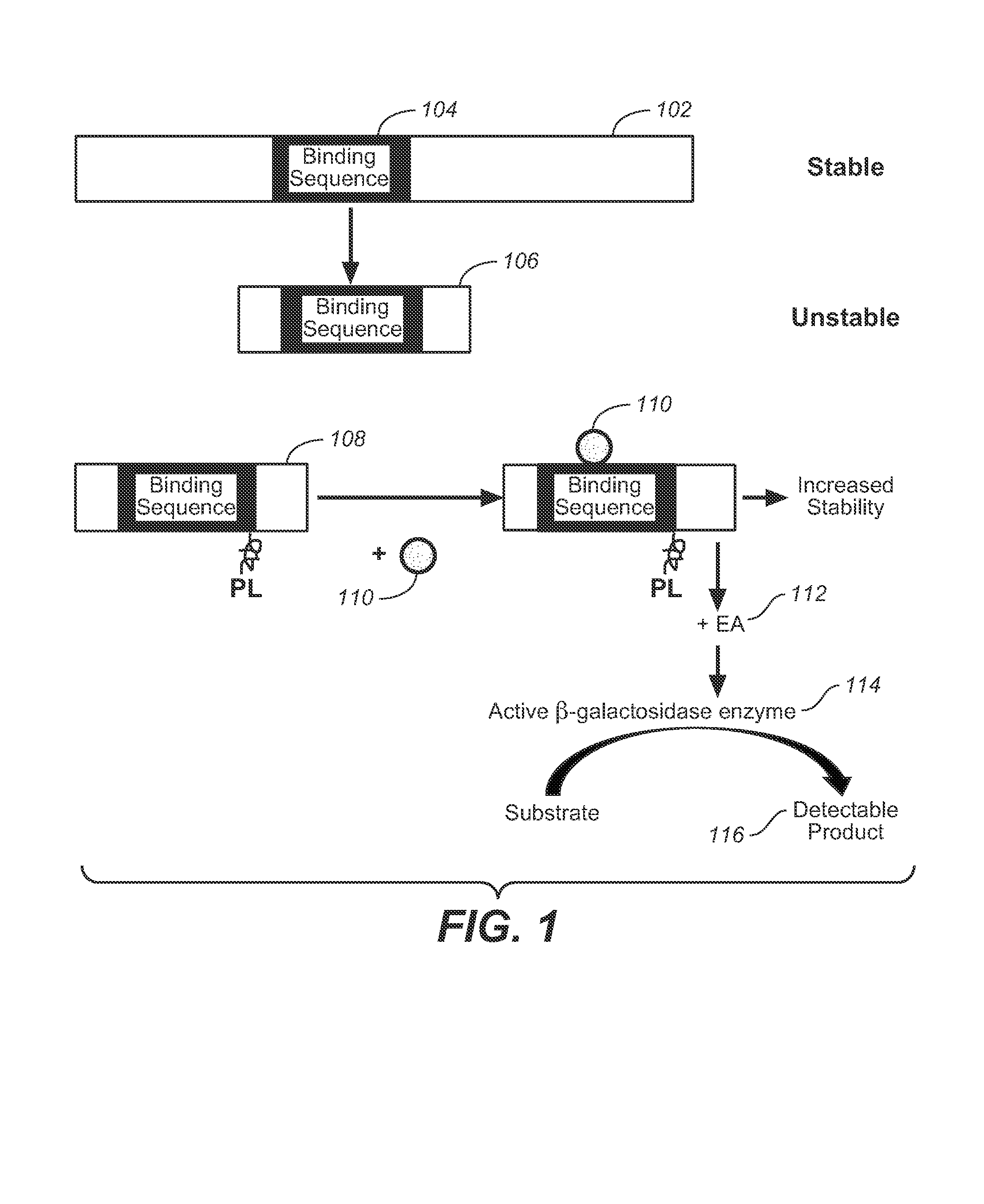
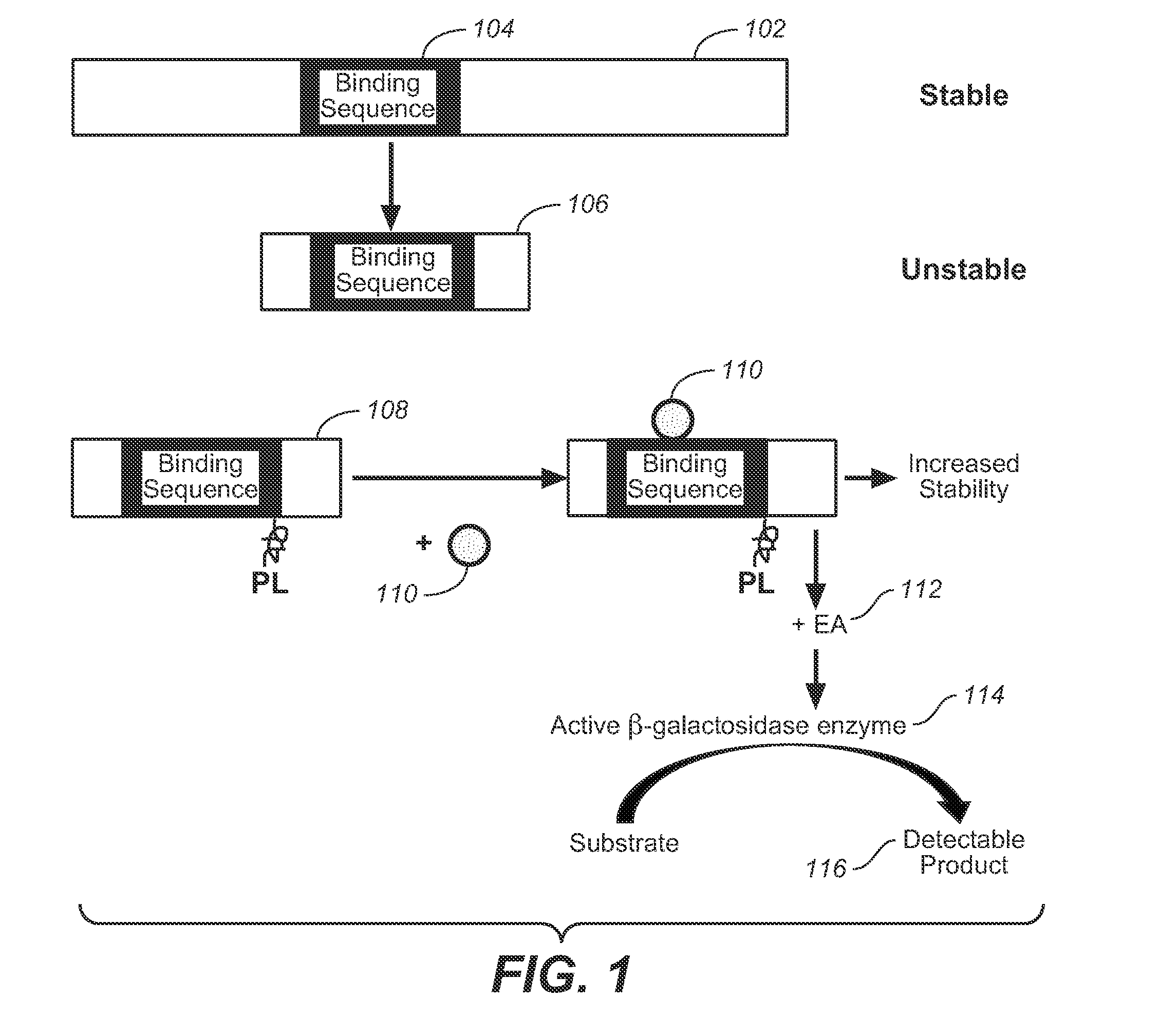
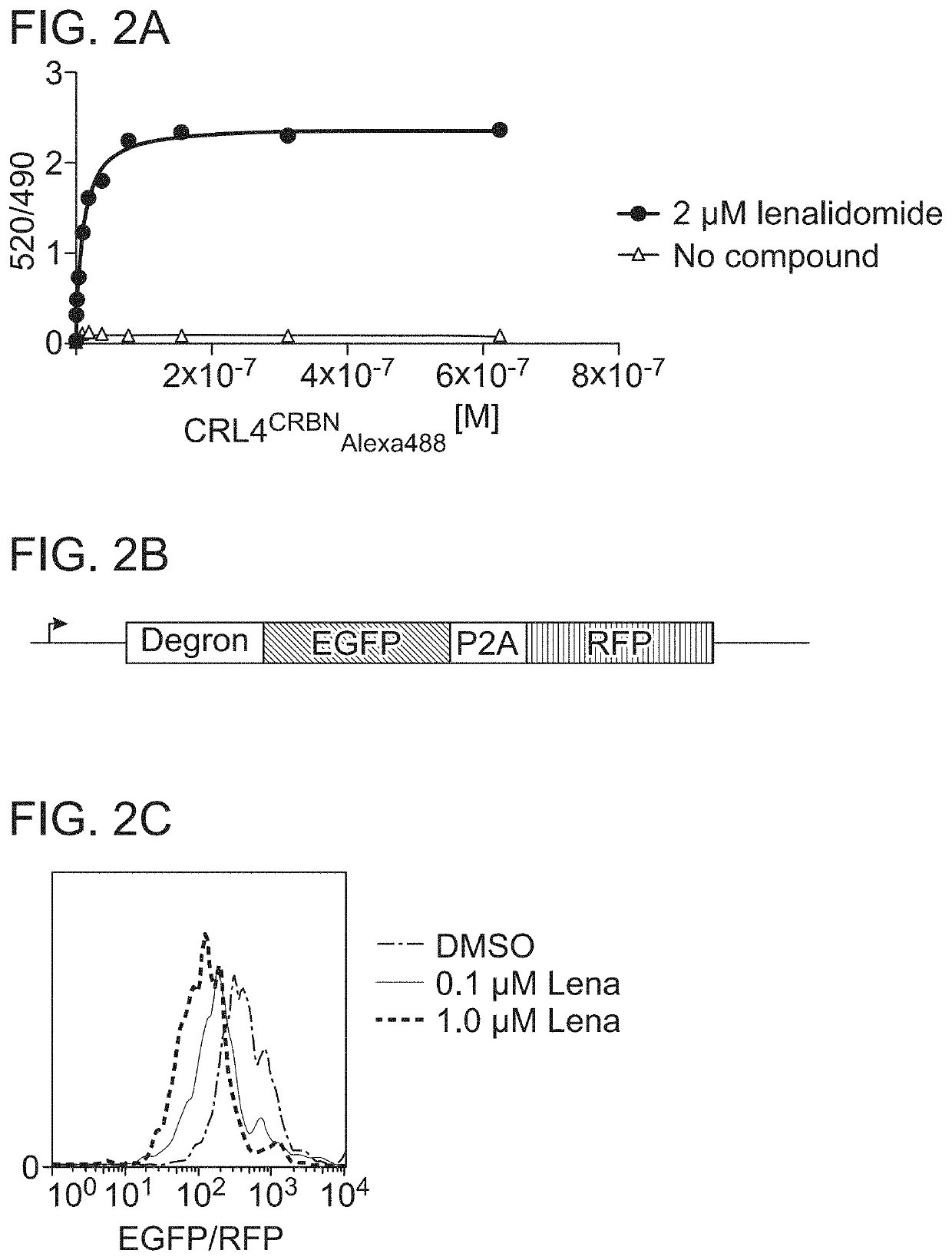
Detection of intracellular binding events by measuring protein abundance
Methods and compositions are provided to measure the binding of a test compound to a target peptide by measuring the effect of the compound on the abundance of the target peptide inside a cell. The target peptide may bind the test compound at an active site or an allosteric site, and it has been found that such binding may stabilize the target peptide against cellular degradation. The target peptide will preferably comprise a destabilizing mutation which shortens the half life of the target peptide within the cell, typically a mammalian cell. Test compounds, including small molecules, have been found to stabilize target peptides. Also provided are systems and kits for use in practicing the methods.
Owner:DISCOVERYX CORP
Method used for large-scale screening of protein biomarkers
ActiveCN108956791AReduce the amount requiredQuantitative restrictionsComponent separationReference sampleScreening method
The invention discloses a method used for large-scale screening of protein biomarkers. The method comprises following steps: 1, a biological tissue check sample and a reference sample (a pathologicalsample) are subjected to homogenization and digestion; 2, an obtained lysate is subjected to enzymatic hydrolysis and desalting; 3, LC-MS / MS liquid chromatography-mass spectrometry is adopted for protein identification; 4, mass spectrometry frequency or mass spectrum peak intensity are compared for analysis of sample protein abundance change, MS1 is taken as a base, the integration of the signal intensity of each peptide segment on the LC-MS is calculated, MS2 identification results are taken as quantification base, and large scale correction is carried out; and 5, analysis results of corrected data are adopted for screening of obviously up-regulated or down-regulated protein biomarkers. In operation, no expensive marking reagent is needed; the cost is relatively low; screening amount of each time is large; and screening results are accurate.
Owner:湖北普罗金科技有限公司
X-linked lymphoproliferative syndrome diagnostic kit and application thereof
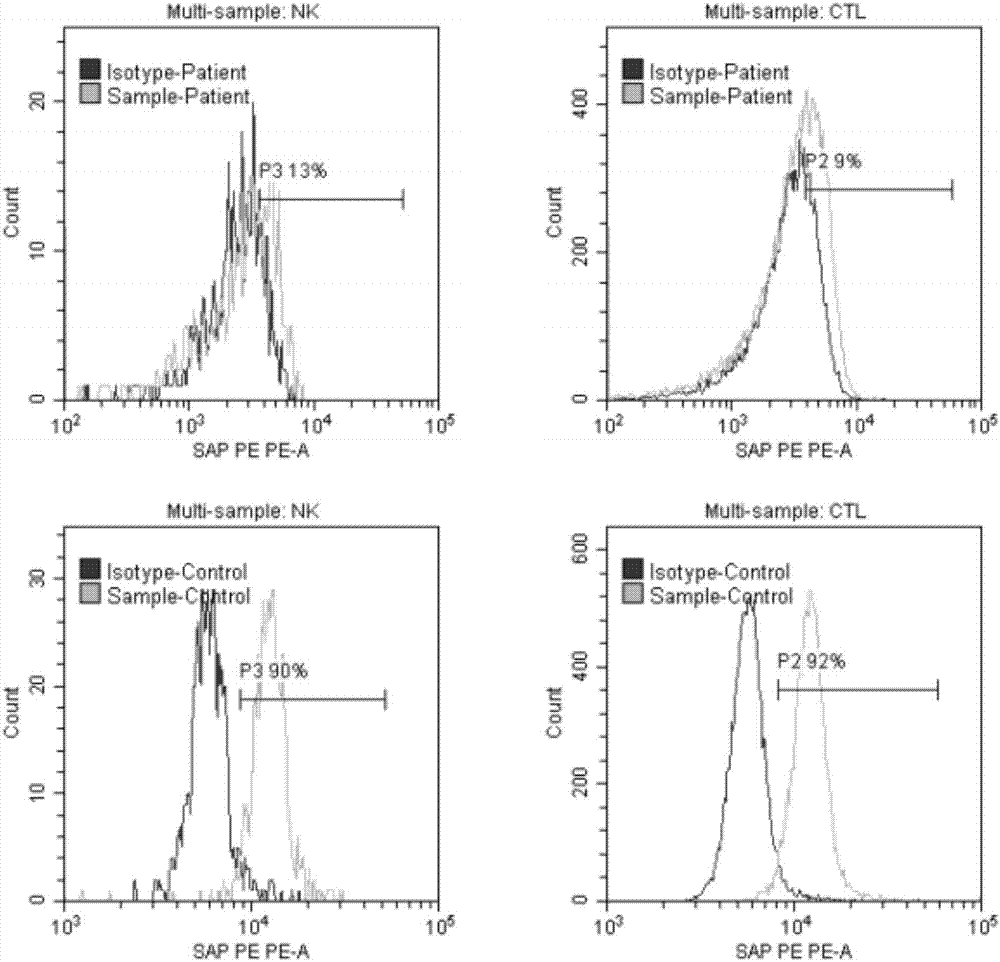
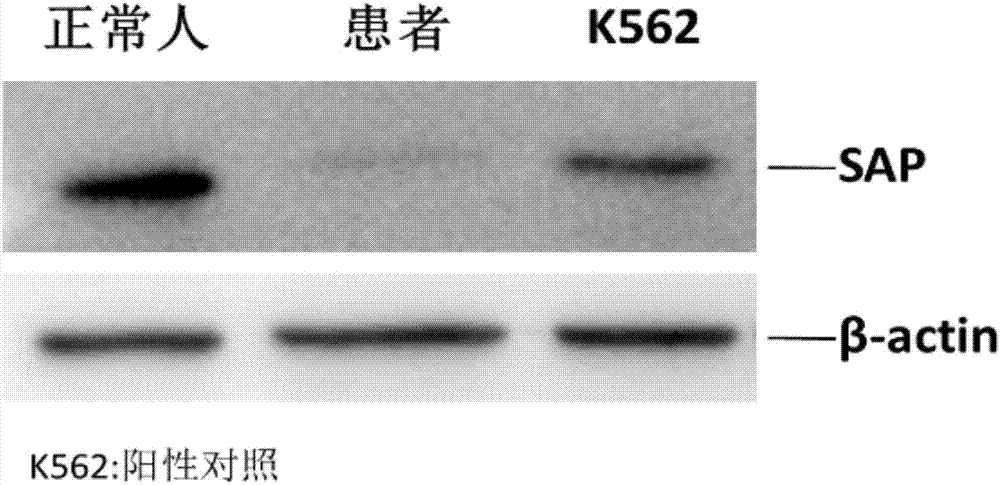
InactiveCN107037219AQuick checkImprove throughputIndividual particle analysisBiological testingX-Linked Lymphoproliferative SyndromeFluorescence
The invention relates to the technical field of medical diagnosis, and specifically relates to an X-linked lymphoproliferative syndrome diagnostic kit and application thereof. The kit comprises anti-human CD 3, CD56, CD8a and SAP flow fluorescent antibodies, an isotype control fluorescent antibody of the anti-human SAP flow fluorescent antibody, an anti-human SAP Western Blot antibody, and a secondary antibody corresponding to the anti-human SAP Western Blot antibody. A method for detecting SAP protein abundance in CTL and NK cells based on the diagnostic kit is quick to detect, high in flux, high in accuracy, and small in human factor influence.
Owner:BEIJING FRIENDSHIP HOSPITAL CAPITAL MEDICAL UNIV
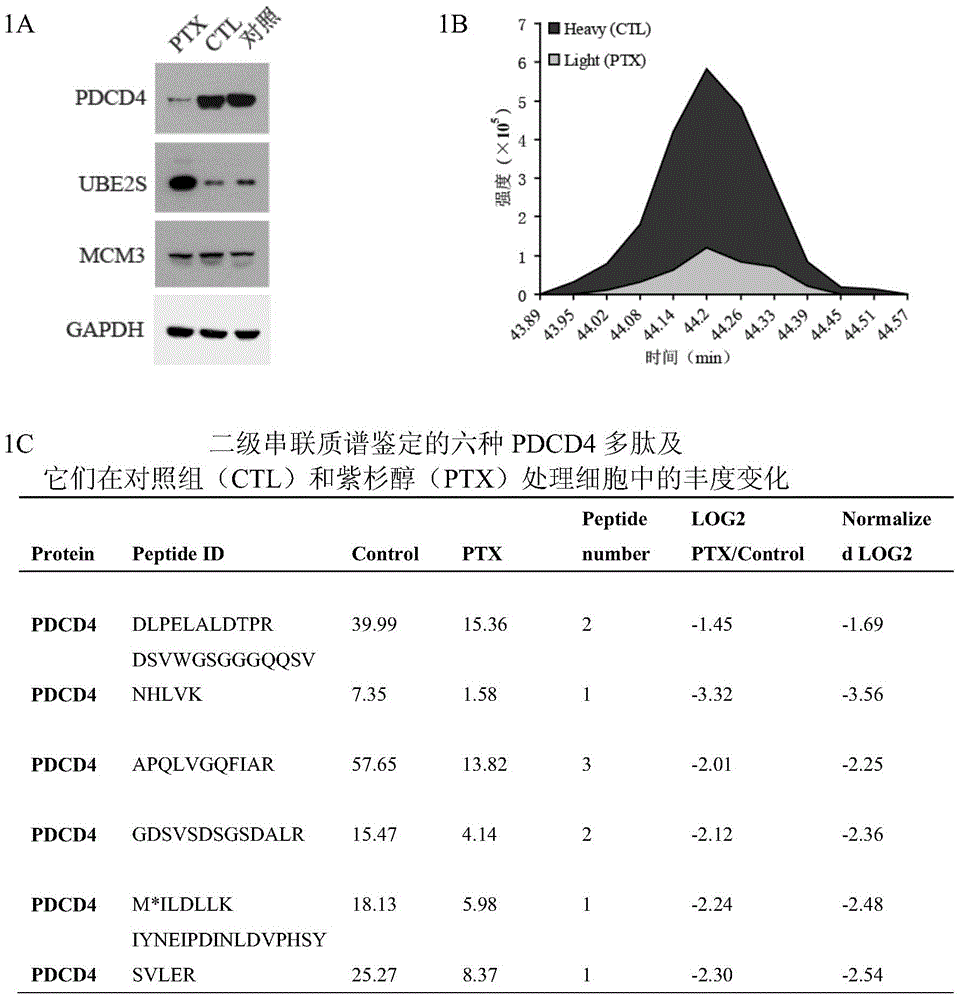
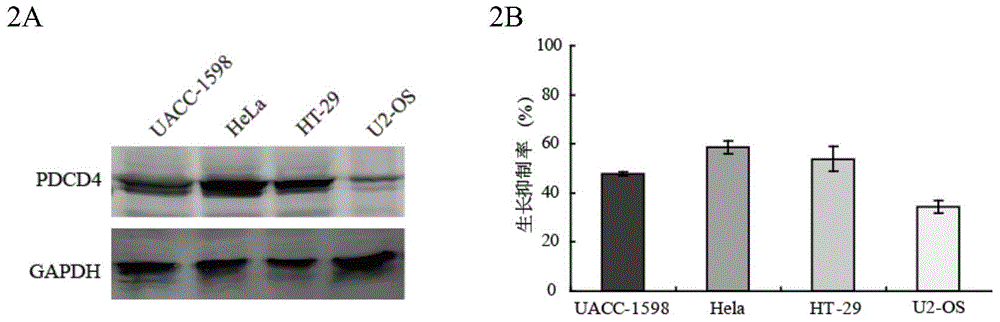
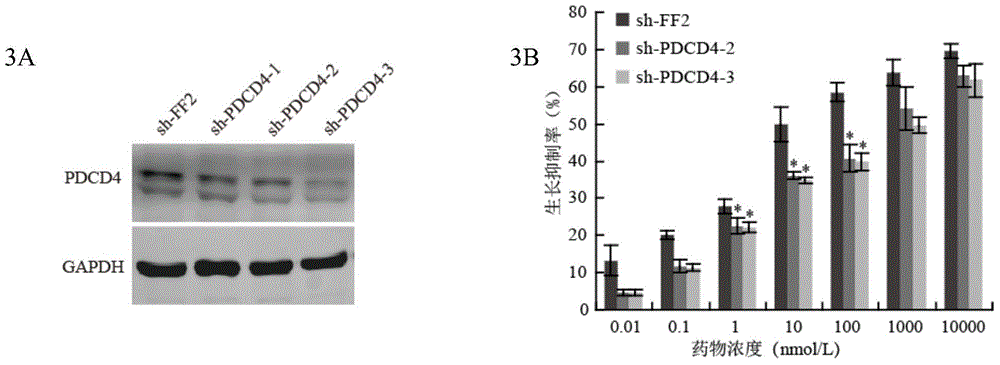
Application of anti-PDCD4 (Programmed Cell Death 4) antibody in preparation of detection reagent for predicting personalized medicine sensitivity of paclitaxel or derivative drug of paclitaxel
InactiveCN104155446AThe detection method is simpleMature technologyDisease diagnosisDocetaxelGene silencing
The invention discloses application of an anti-PDCD4 (Programmed Cell Death 4) antibody in preparation of a detection reagent for predicting personalized medicine sensitivity of paclitaxel or a derivative drug of paclitaxel. The variation of protein abundance of whole cells in tumor cells treated by paclitaxel is analyzed by using a quantitative proteomics technology, the results shows that paclitaxel reduces the expression level of PDCD4 and the sensitivity of the tumor cells to paclitaxel is in positive correlation with the expression level of PDCD4 in cells. PDCD4 in the tumor cells is subject to gene silencing or overexpression and the result further proves that the sensitivity of the cells to paclitaxel is in positive correlation with the expression level of PDCD4 in the cells. Meanwhile, researches on survival prognosis of clinical lung cancer patients shows that under the same paclitaxel combined adjuvant chemotherapy, the survival prognosis of patients with low expression level of PDCD4 is poor and the survival time of patients with high expression level of PDCD4 is long. Based on the research results, the invention provides a reference index and a detection and application method for guiding personalized medicine of paclitaxel or the derivative drug (such as docetaxel) of the paclitaxel.
Owner:HARBIN MEDICAL UNIVERSITY
Detection of intracellular binding events by measuring protein abundance
Methods and compositions are provided to measure the binding of a test compound to a target peptide by measuring the effect of the compound on the abundance of the target peptide inside a cell. The target peptide may bind the test compound at an active site or an allosteric site, and it has been found that such binding may stabilize the target peptide against cellular degradation. The target peptide will preferably comprise a destabilizing mutation which shortens the half life of the target peptide within the cell, typically a mammalian cell. Test compounds, including small molecules, have been found to stabilize target peptides. Also provided are systems and kits for use in practicing the methods.
Owner:DISCOVERYX CORP
Pharmaceutical composition for use in improving quality of scalp or skin, wound healing, or improving quality of hair
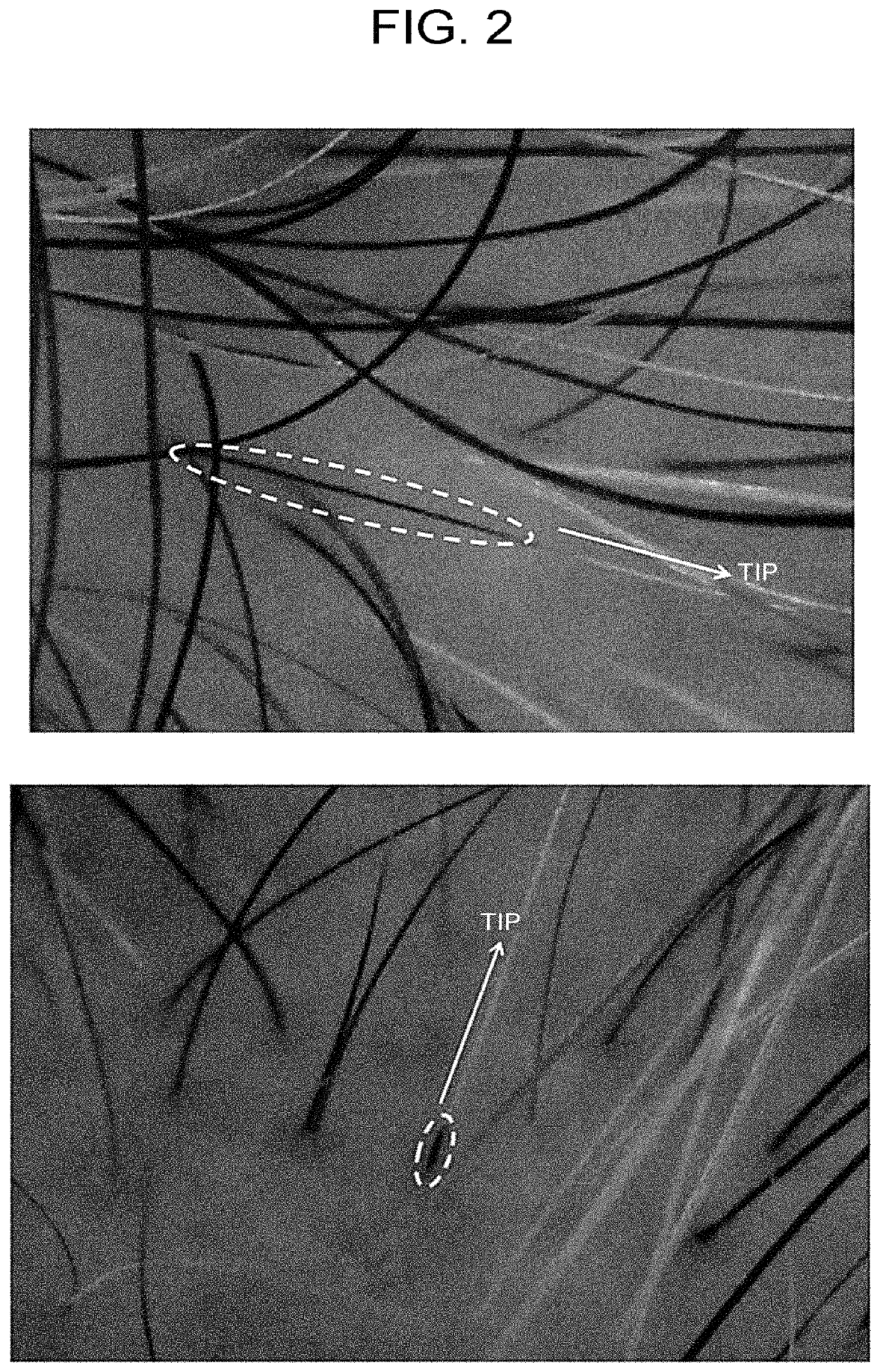
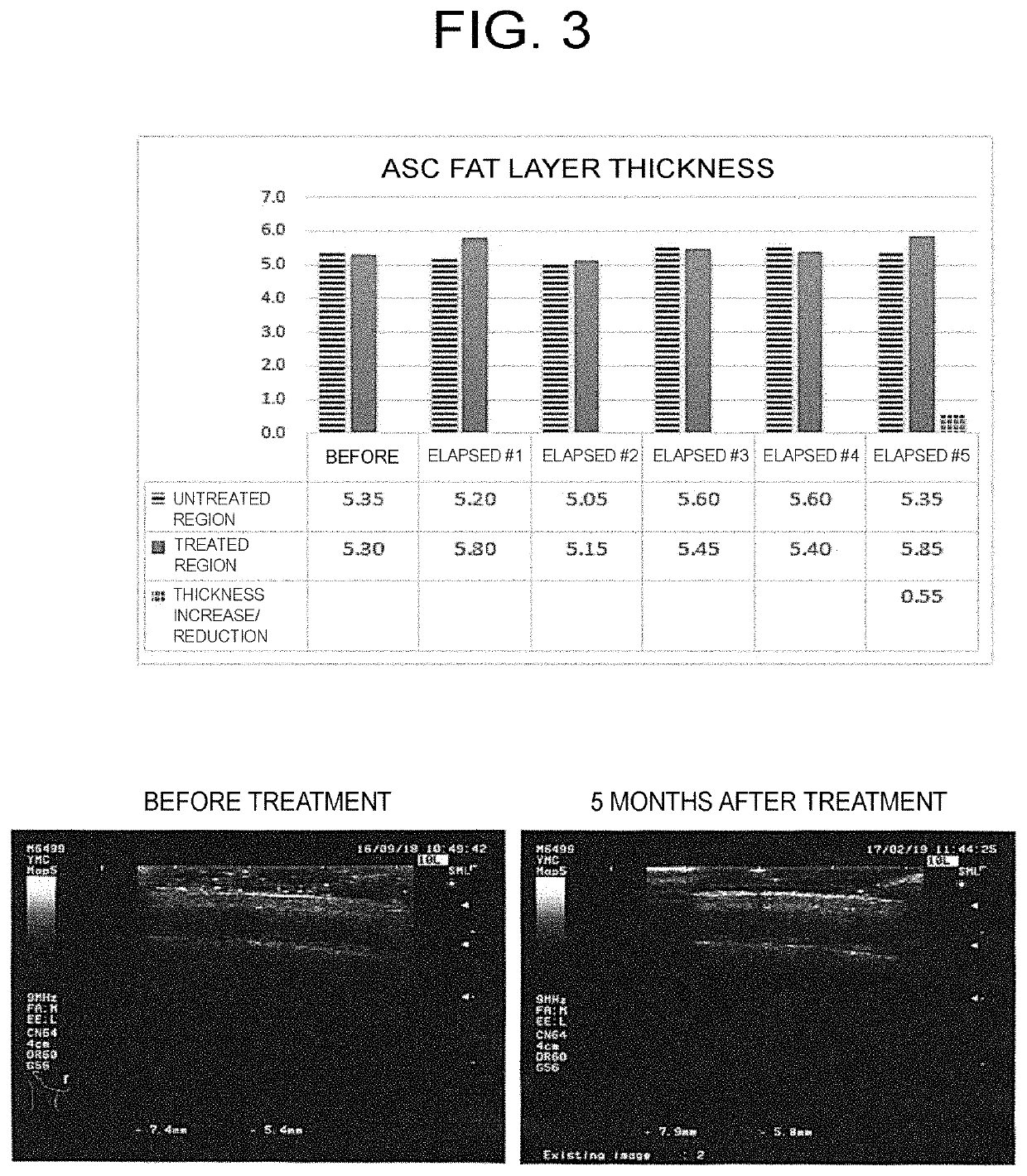
ActiveUS20200101114A1Shorten the timeEasily propagatedCosmetic preparationsHair cosmeticsWound healingBULK ACTIVE INGREDIENT
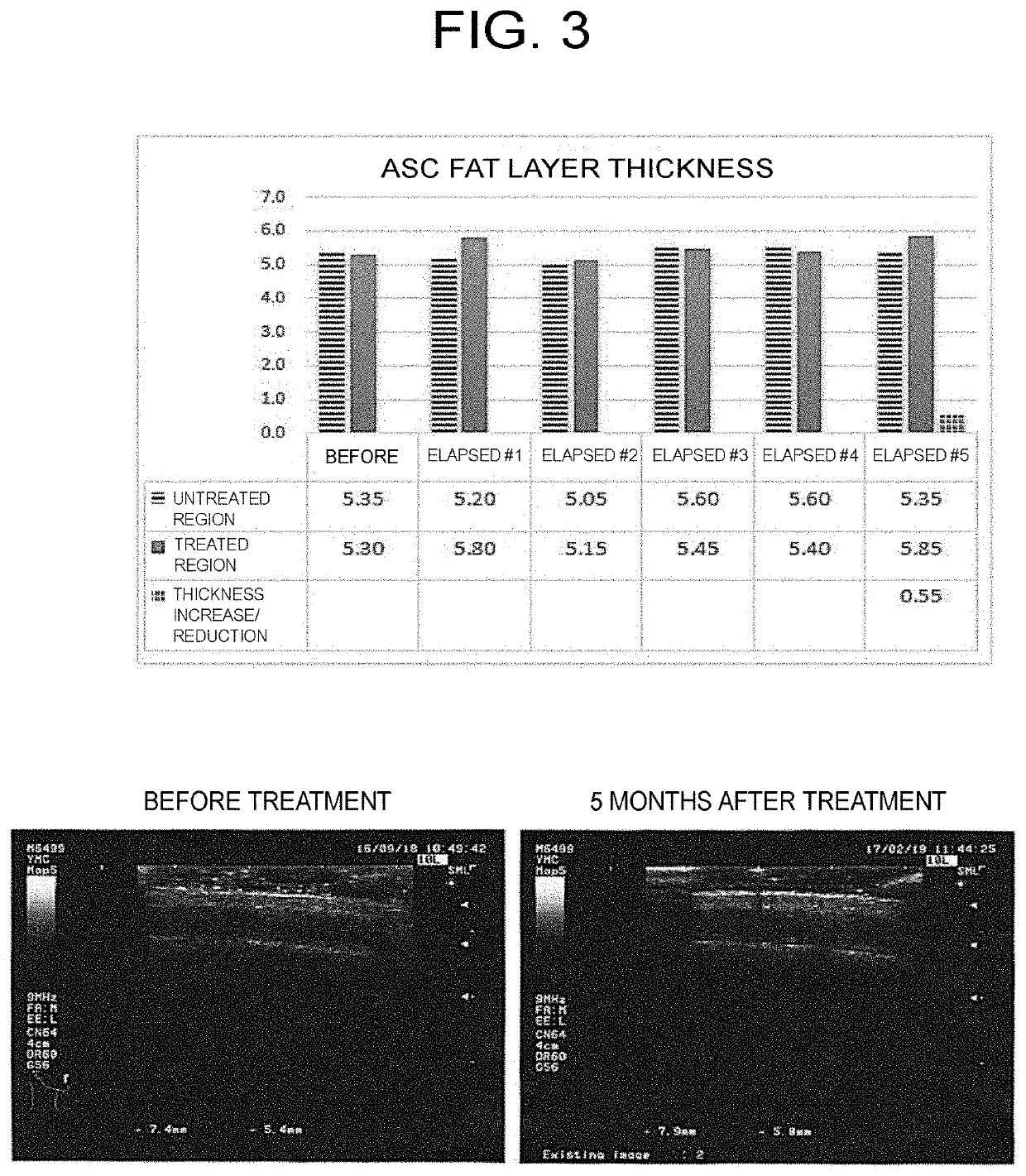
The present invention provides a pharmaceutical composition for use in modifying scalp or skin, promoting wound healing, or modifying hair, comprising a secretion from adipose stem cells as an active ingredient, wherein 0.3 to 0.6 μg of the secretion in terms of protein abundance per site in scalp or skin is administered.
Owner:FUKUOKA HIROTARO
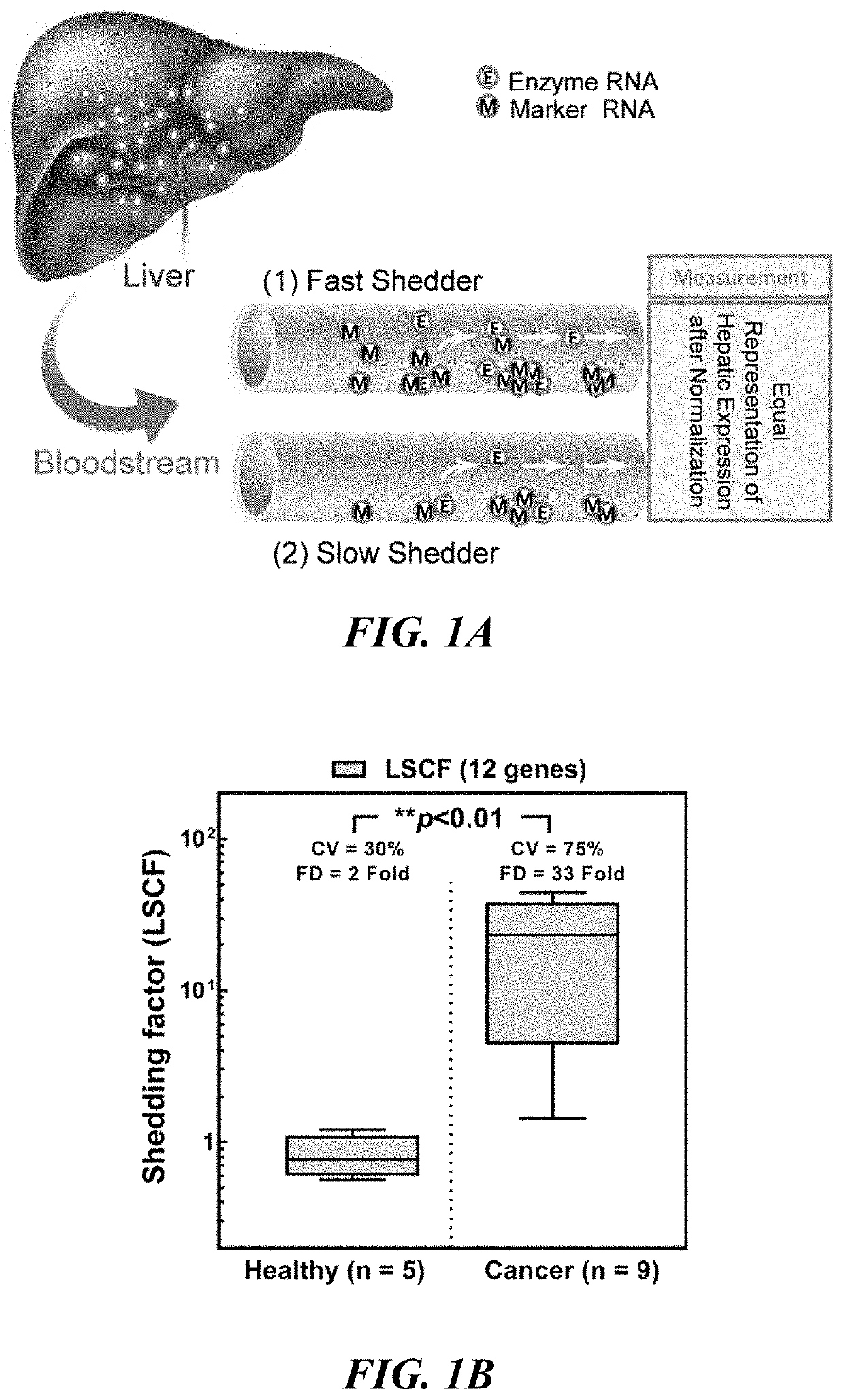
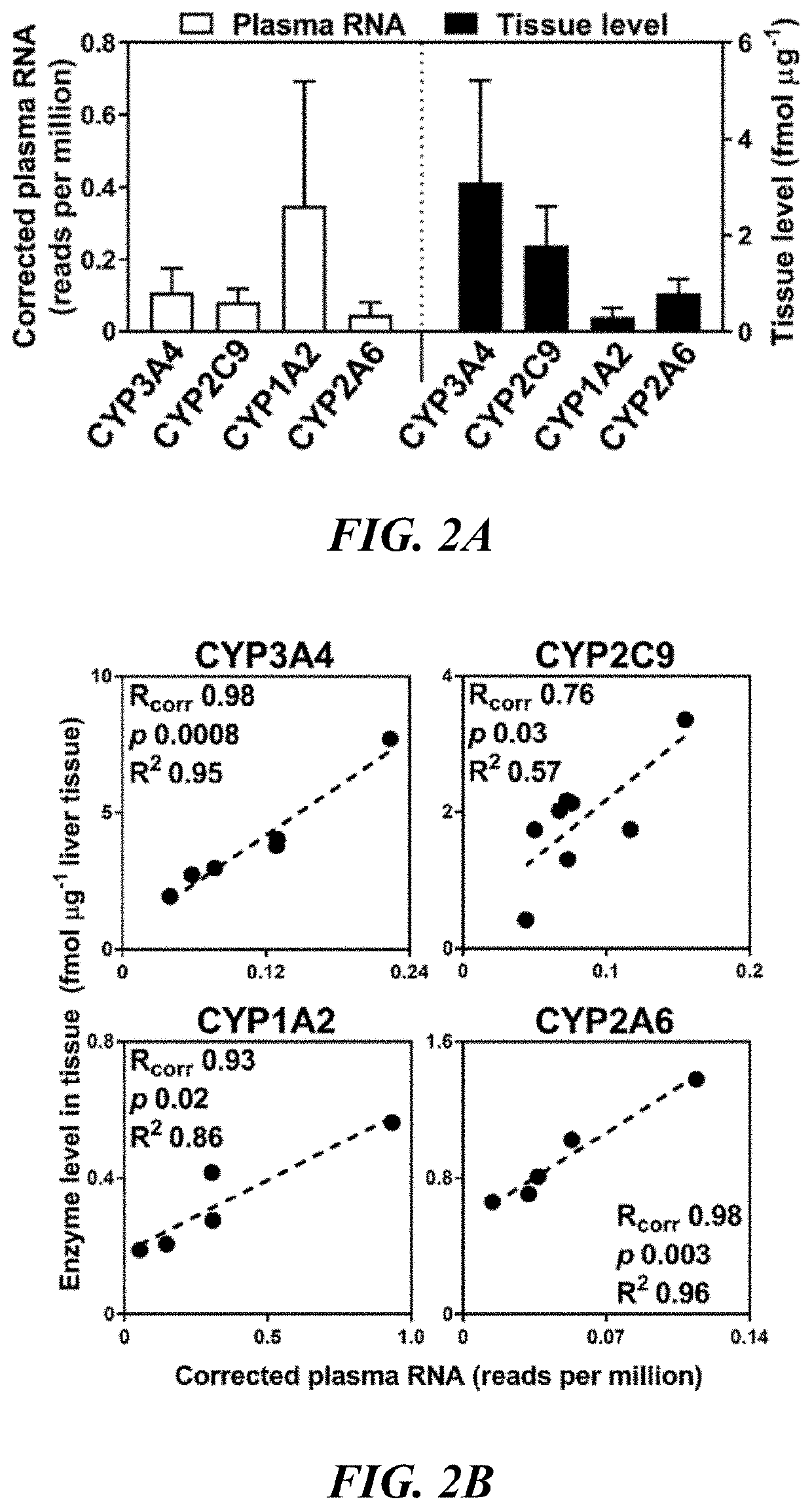
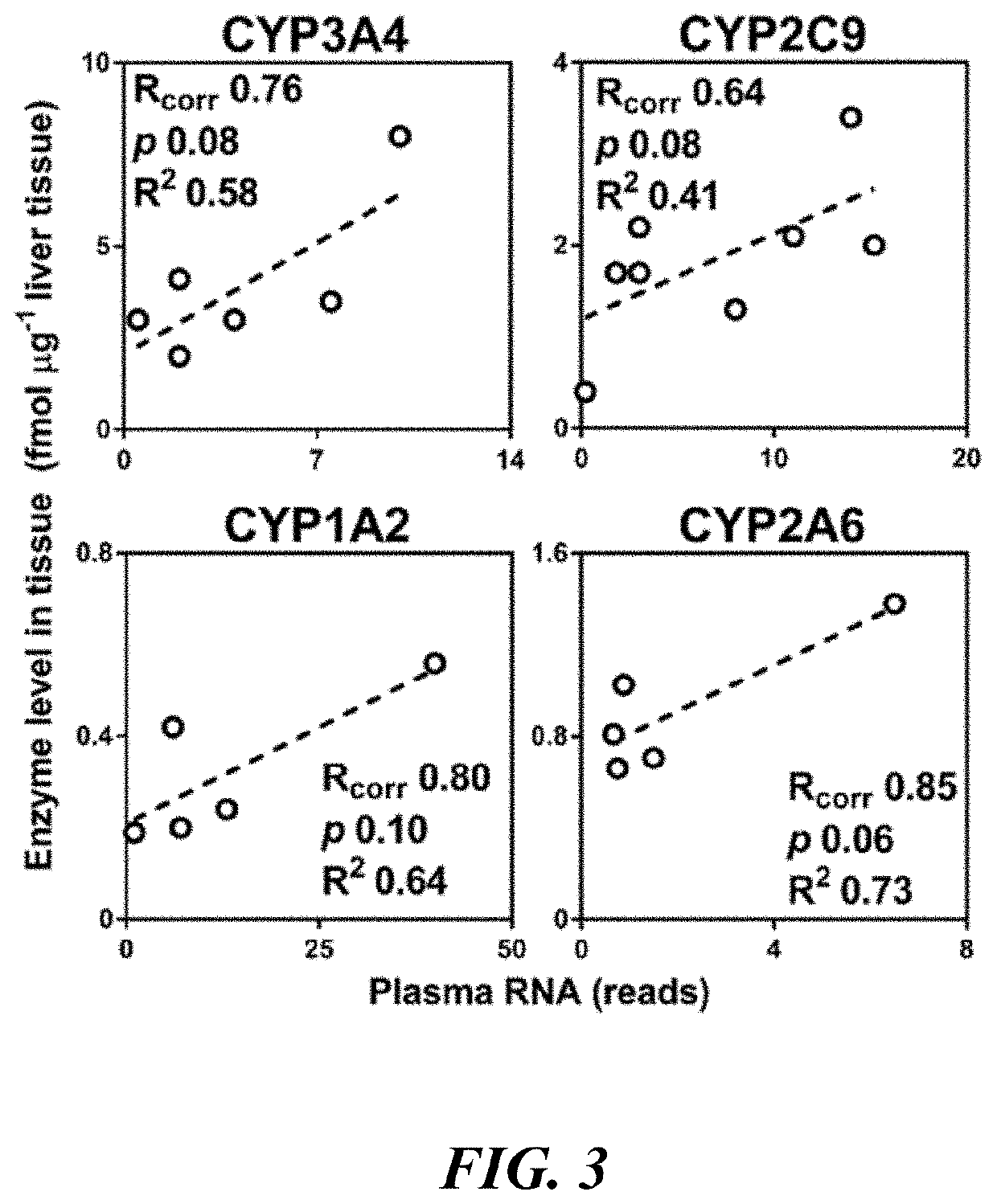
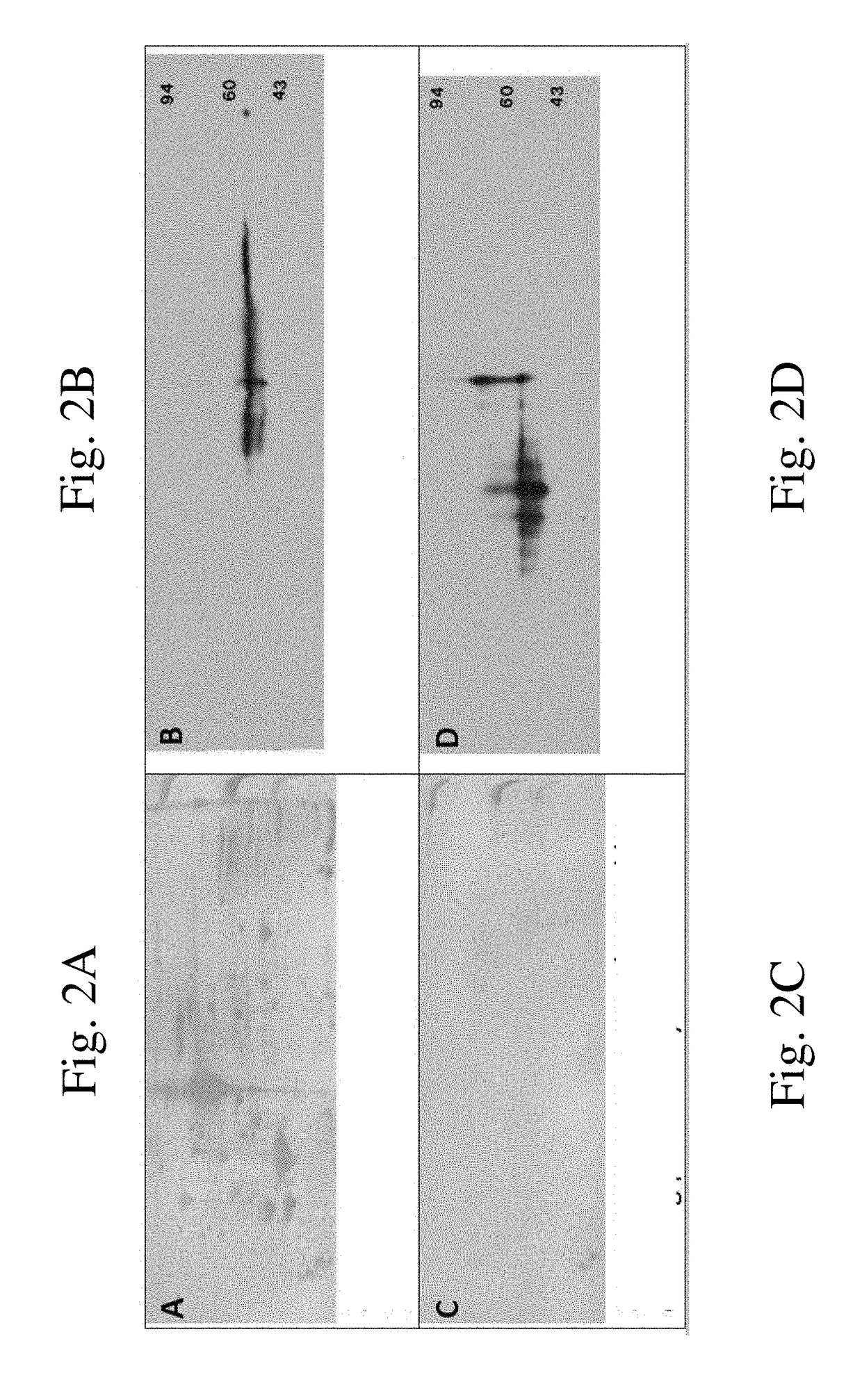
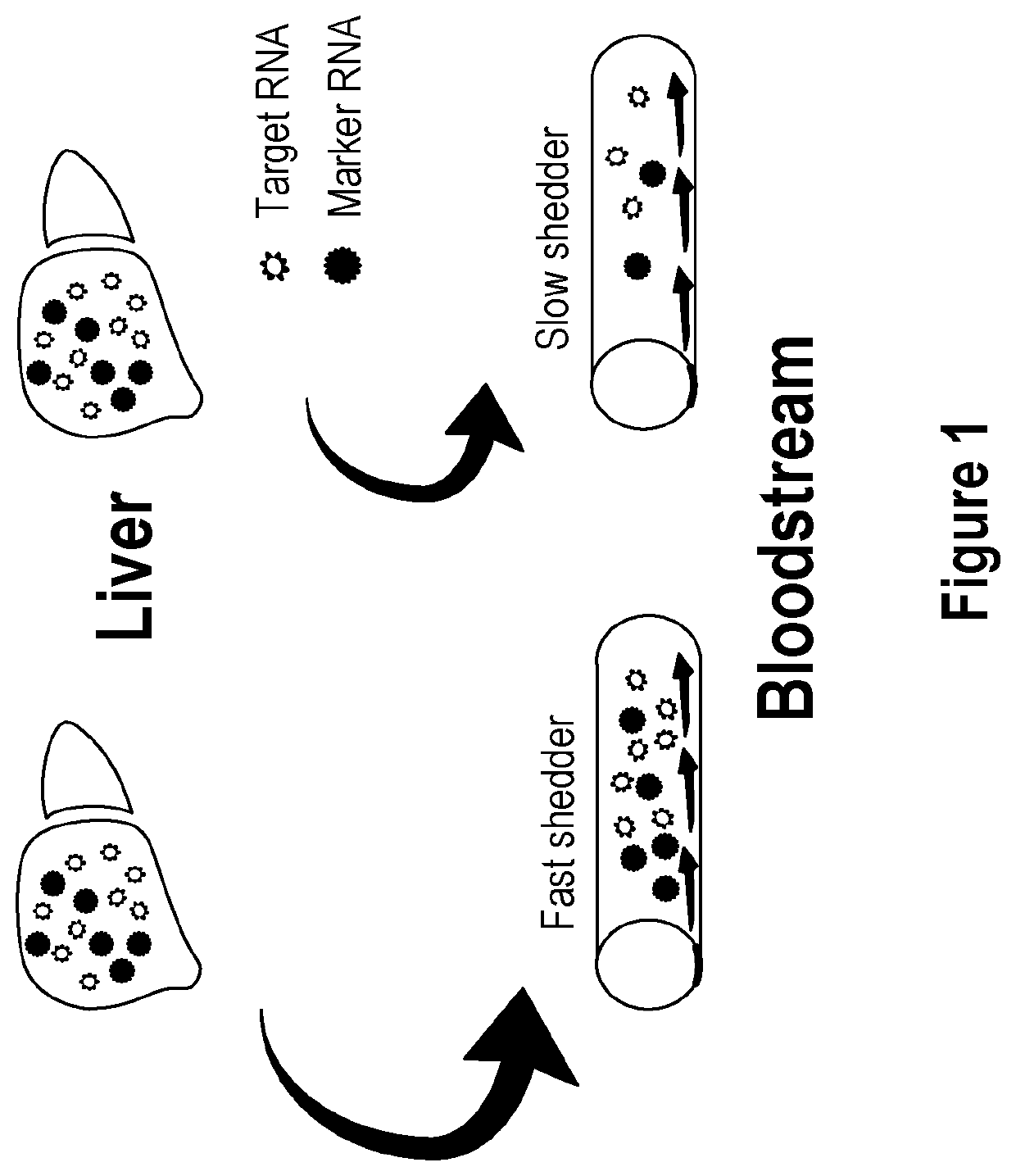
Methods and apparatus for quantifying protein abundance in tissues via cell free ribonucleic acids in liquid biopsy
Methods, systems and apparatus are provided for quantifying the amount of at least a first cell free RNA (cfRNA) present in a liquid biopsy obtained from an individual subject. The first cfRNA may encode a protein that functions in the clearance of xenobiotic compounds from the body of the subject. Quantification of the amount of the first cfRNA is normalised to the individual and permits the construction of more accurate virtual models that facilitate improved personalised medicine, dosage regimens and clinical trials.
Owner:CERTARA USA INC
Machine learning algorithm for identifying peptides that contain features positively associated with natural endogenous or exogenous cellular processing, transportation and major histocompatibility complex (MHC) presentation
The present invention provides a method for identifying peptides that contain features positively associated with natural endogenous or exogenous cellular processing, transportation and major histocompatibility complex (MHC) presentation. In particular, the invention / method controls for the influence of protein abundance, stability and HLA / MHC binding on processing and presentation, enabling a machine-learning algorithm or statistical inference model trained using the method to be applied to any test peptide regardless of its HLA / MHC restriction i.e. the algorithm operates in a HLA / MHC-agnostic manner. This is attained through the building of positive and negative data sets of peptide sequences (peptides identified or inferred from surface bound or secreted MHC / peptide complexes in the literature, and those which are not). Specifically, the positive and negative data sets comprise a multiplicity of pairings between individual entries, in which both sequences of a pair are of equal or similar length, and are derived from the same source protein, and / or have similar binding affinities, with respect to the HLA / MHC molecule from which the peptide of the positive peptide is restricted.
Owner:NEC奥克尔姆内特公司
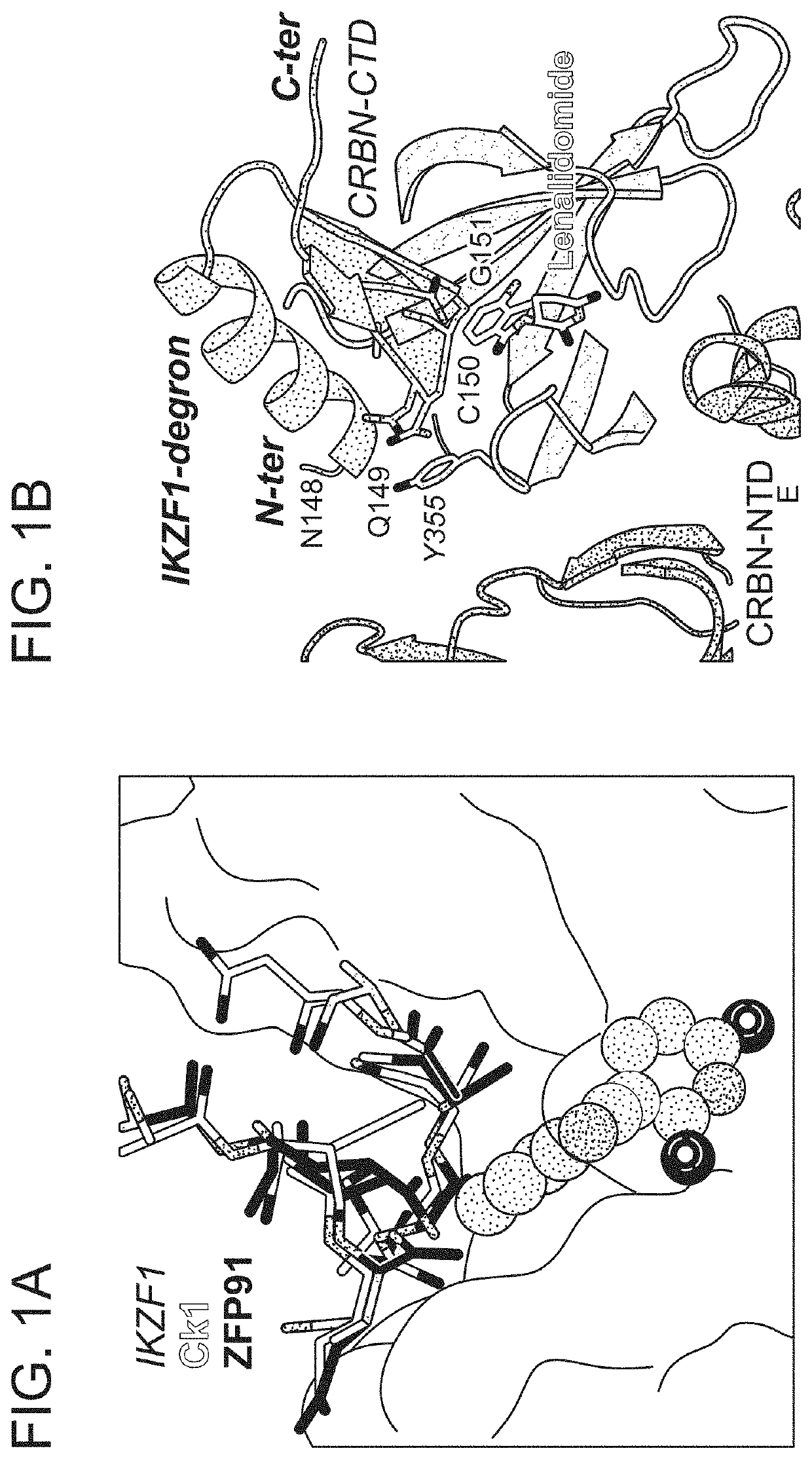
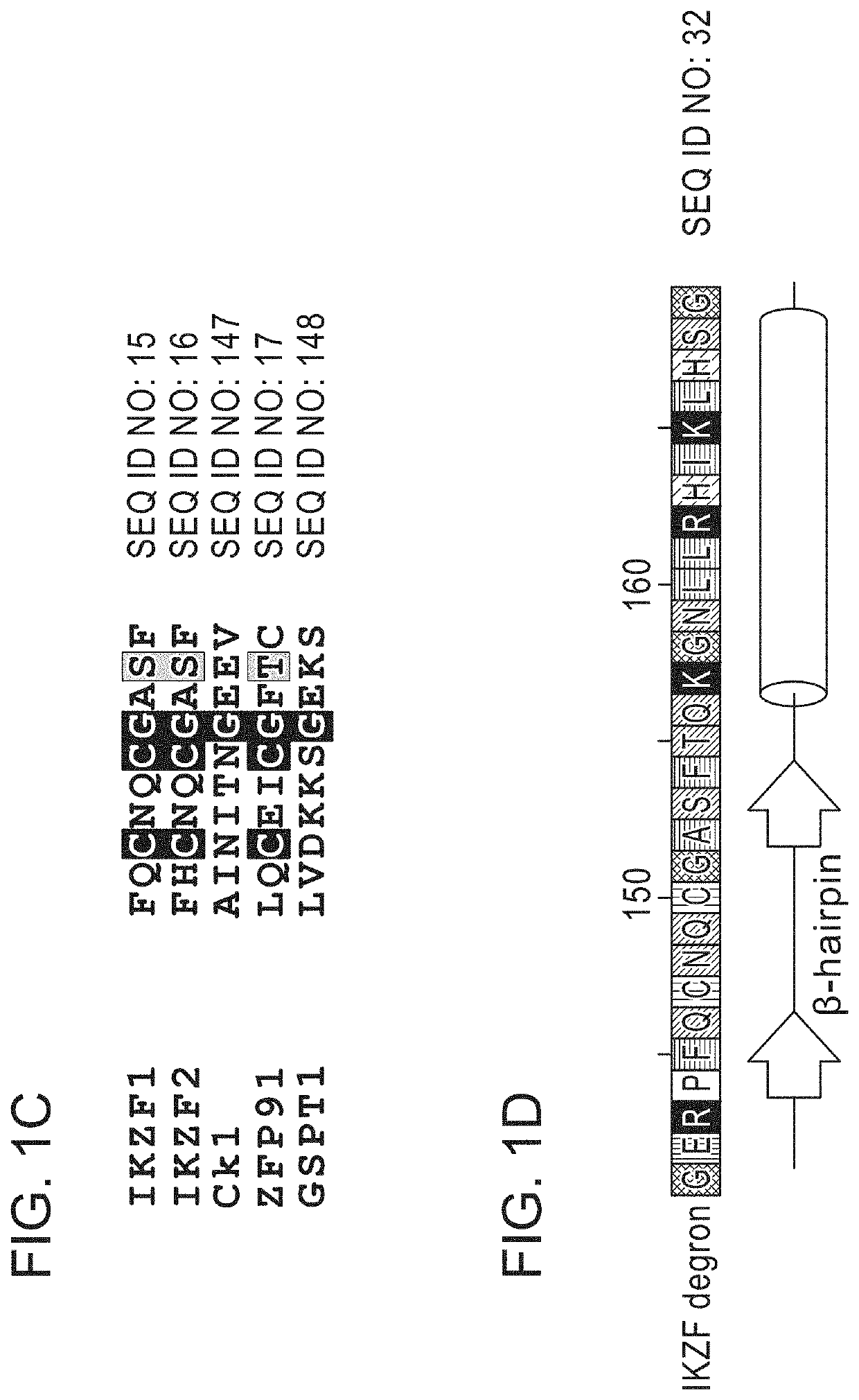
Peptide tags for ligand induced degradation of fusion proteins
PendingUS20220098251A1Minimal modificationUniversal applicabilityOrganic active ingredientsPeptide/protein ingredientsProtein abundanceCell biology
Described herein are compositions and methods for modulating protein abundance in a target-specific manner via degron tags.
Owner:DANA FARBER CANCER INST INC +1
Protein standard compositions and methods of making and using the same
ActiveUS20180127802A1Microbiological testing/measurementTransferasesPhosphorylationProtein abundance
The invention generally relates to protein standard compositions and methods of making the same. Also contemplated are kits including the protein standard compositions or kits for making the protein standard compositions and methods of using the protein standard compositions to quantify the abundance of a phosphorylated protein in a sample.
Owner:KENDRICK LAB INC
Methods and apparatus for generating a virtual model of xenobiotic exposure using transcriptomics analysis of liquid biopsy samples
PendingUS20220215898A1Improve the level ofReduce variationChemical property predictionMicrobiological testing/measurementCell freeProtein
Processes are provided for establishing a virtual physiologically based pharmacokinetic (PBPK) model in a population comprised of a plurality of individual subjects that has been or may be exposed to a xenobiotic molecule. The processes are derived from the identification of an abundance of a protein that is involved in absorption; distribution; localization; biotransformation; and excretion of the xenobiotic molecule from a liquid biopsy of corresponding cell free RNA. Personalised PBPK models for precision dosing, as well as methods of treatment are also provided.
Owner:CERTARA USA INC
Identification method of individual differential expression protein
PendingCN114220484AReduce the impactGuaranteed stabilityBiostatisticsProteomicsDecreased protein SGenetics
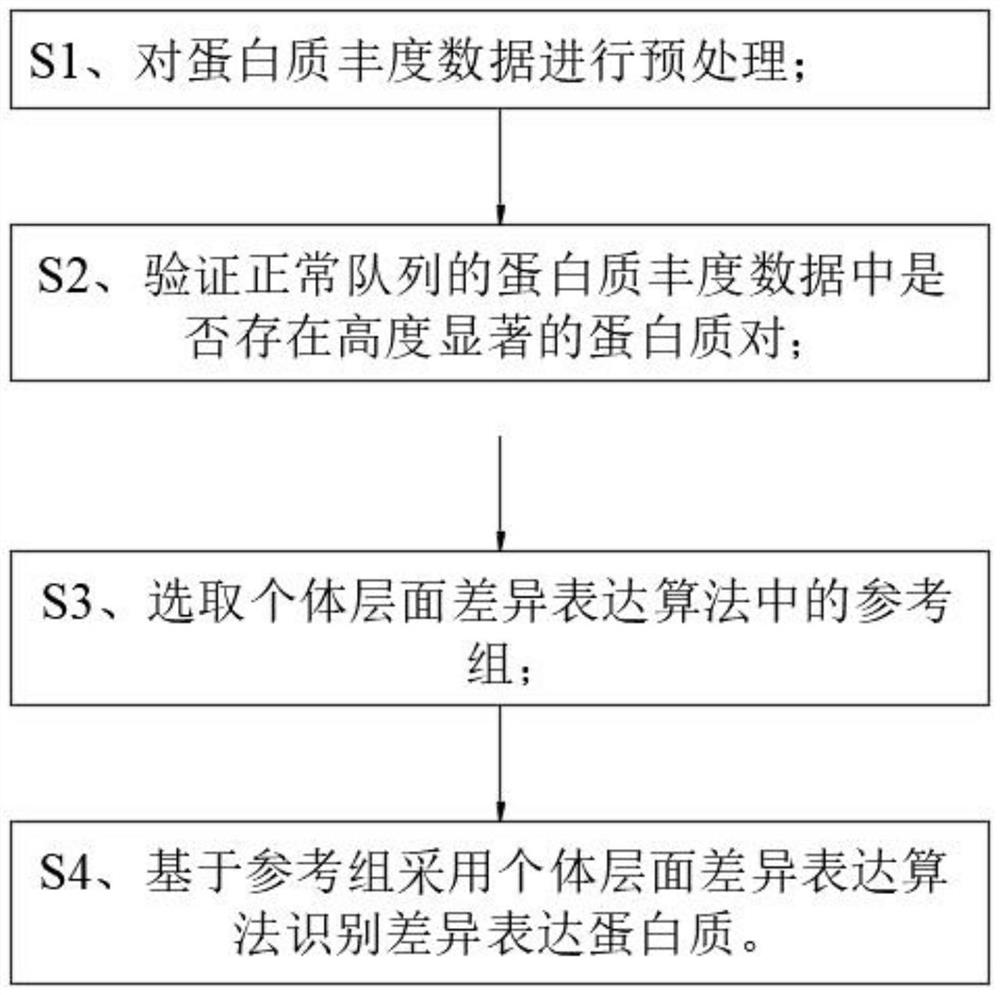
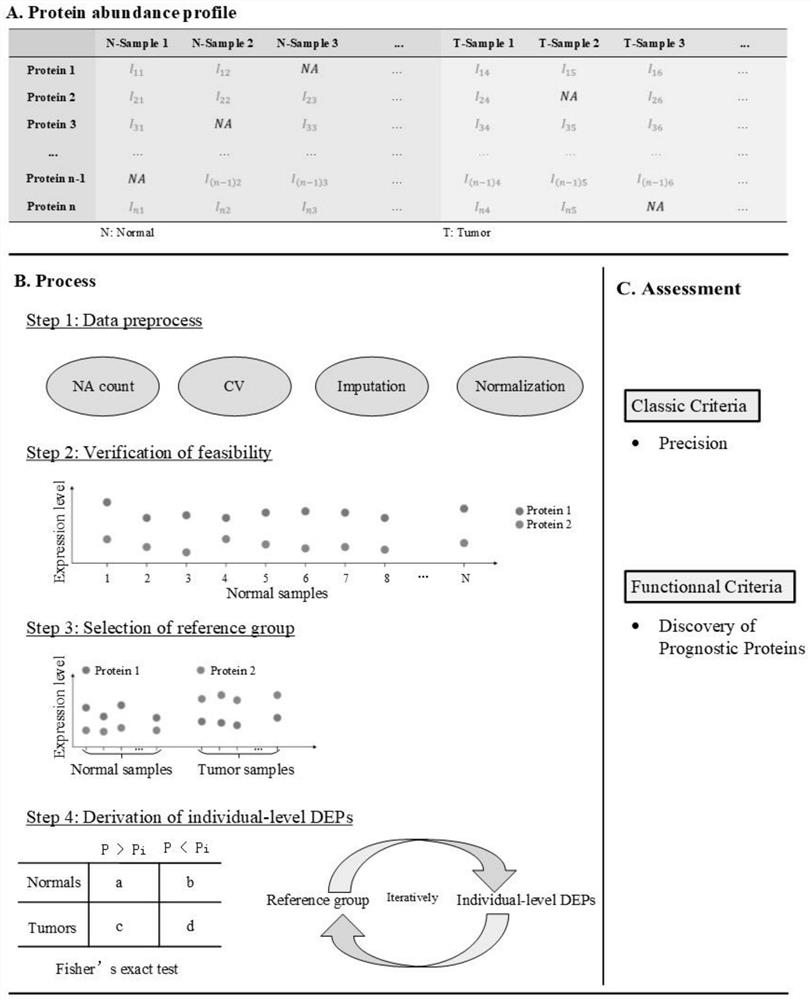
The invention discloses a method for identifying individual differential expression proteins. The method comprises the following steps: S1, preprocessing protein abundance data; s2, verifying whether highly significant protein pairs exist in the protein abundance data of the normal queue; s3, selecting a reference group in an individual level differential expression algorithm; s4, identifying differential expression proteins by adopting an individual level differential expression algorithm based on the reference group; the method comprises the following steps: firstly, based on the characteristics of protein abundance data, preprocessing the protein abundance data, removing proteins with high variation coefficients and high missing value proportions, and reducing the influence of protein quantitative errors on individual differential protein results; and secondly, a reference group is selected based on the protein stability pair, and proteins with differential expression in the reference group are continuously optimized by using an iteration method, so that the stability of the reference group is ensured, the proteins with differential expression can be effectively identified on the individual level, the identification precision is high, and the application prospect is good.
Owner:XIAMEN UNIV
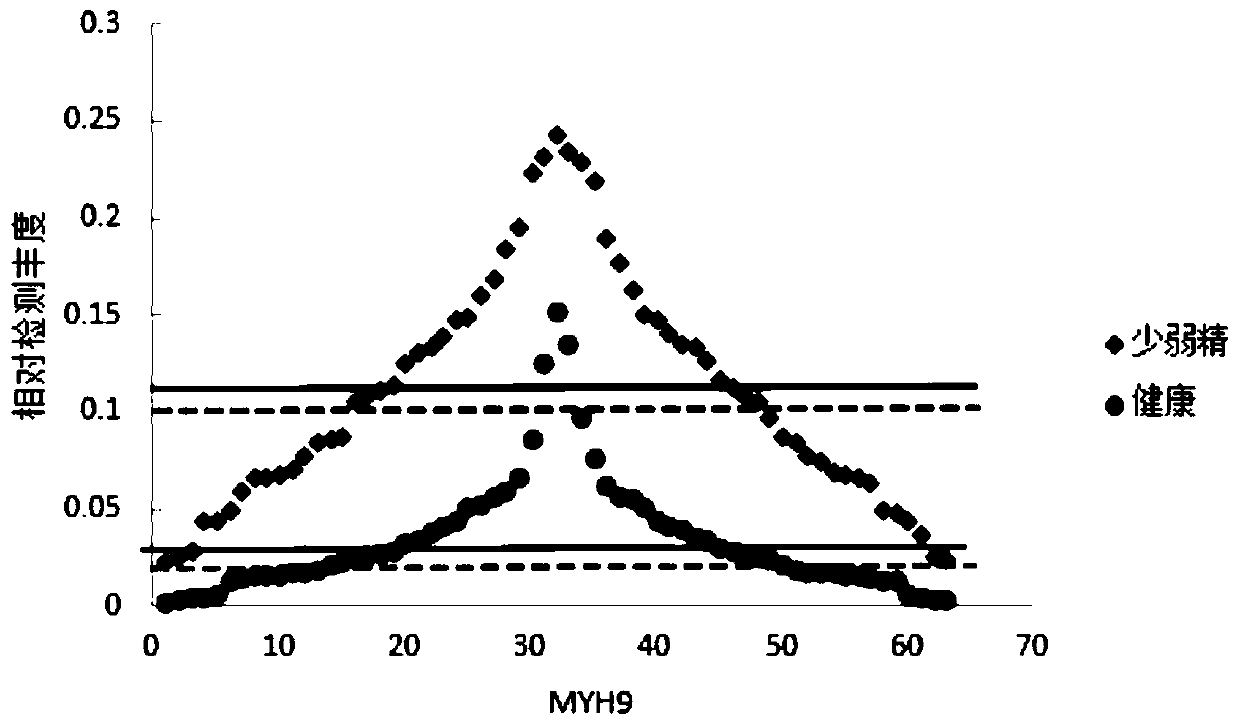
Application of MYH9 in preparation of diagnostic reagent for severe oligoasthenozoospermia and asthenozoospermia
PendingCN111537749AGood correlationDisease diagnosisBiological testingProteomics methodsBiochemistry
Owner:山东立菲生物产业有限公司
Method for detecting interaction and affinity between ligand and protein
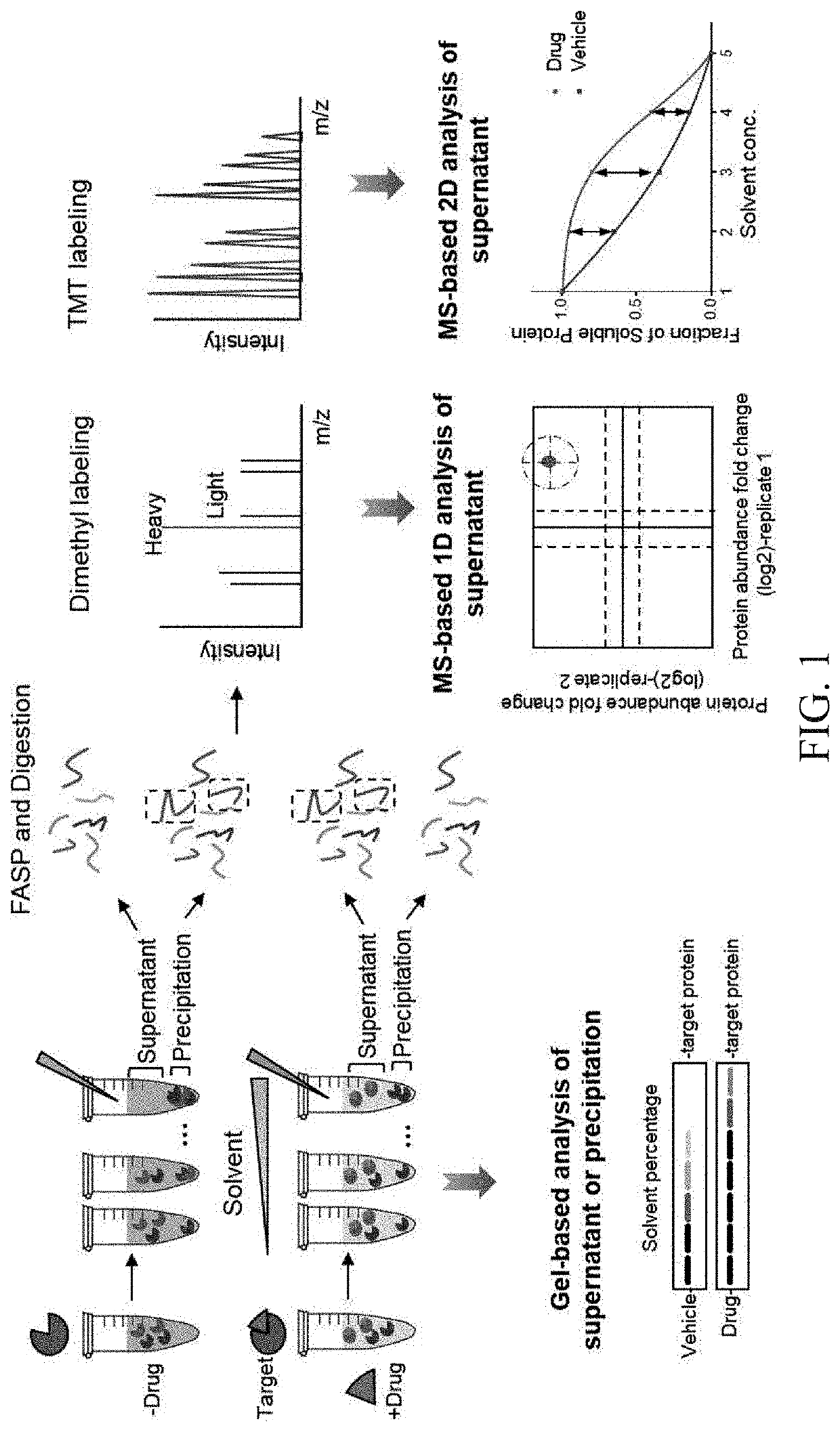
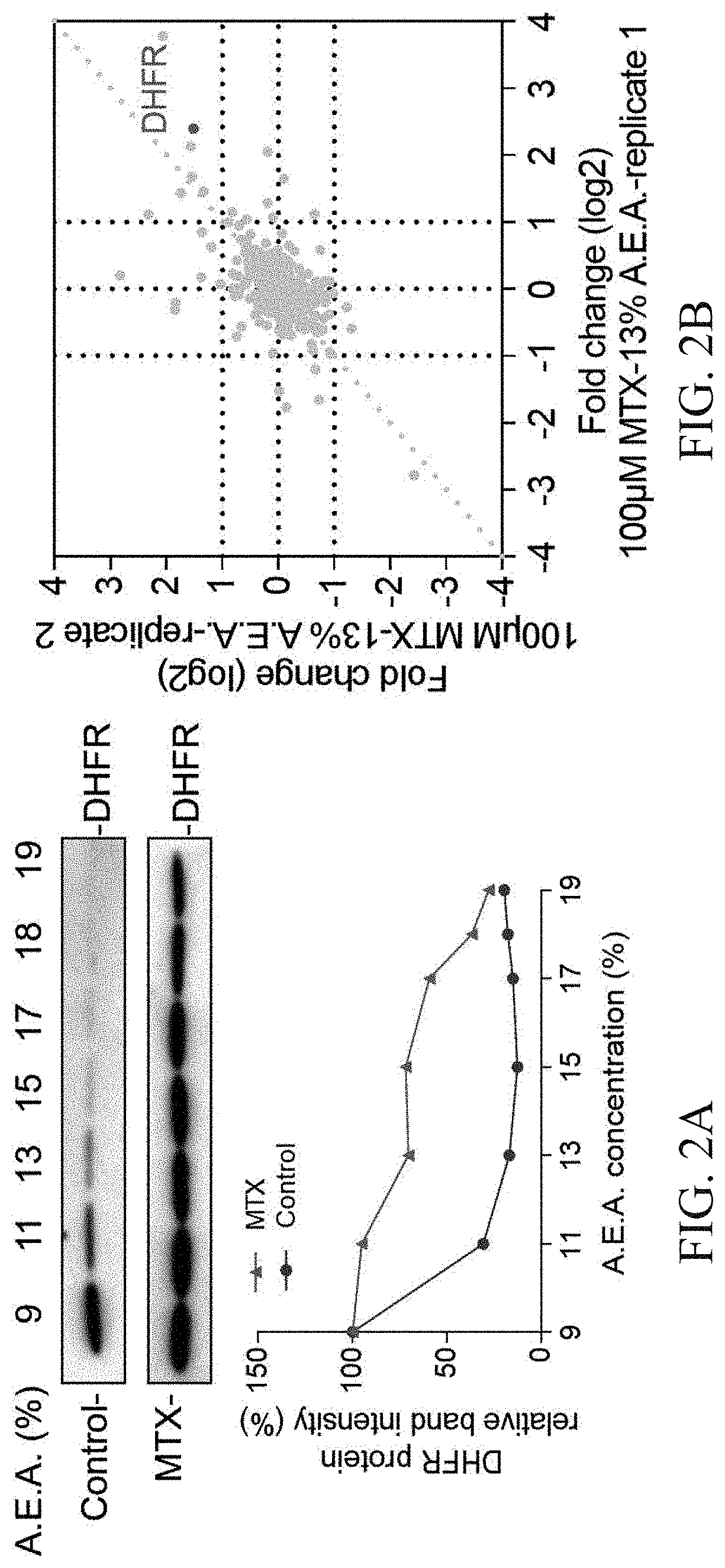
A method of solvent-induced protein precipitation (SIP) for detecting the interaction of ligands with proteins in a complex protein sample. After the equal amount of solvent is added to the protein samples with and without a ligand to denature and precipitate the proteins, the protein abundances in supernatant and / or precipitate in the ligand group and the control group are measured by quantitative technology. The target protein(s) of a ligand is / are determined by comparing the differences of protein abundances in the ligand group and the control group. The affinity between a ligand and its targets can be evaluated by dose dependent experiments. This method does not require the chemical modification of the ligand and has the feature of high specificity. Furthermore, in certain embodiments, the targets identified by SIP method are complementary to those identified by thermal proteome profiling (TPP) method.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
Method for identifying main components for treating mastitis in coronet aqueous extract
The invention discloses a method for identifying main components for treating mastitis in a coronet aqueous extract, and belongs to the technical field of traditional Chinese medicine identification. The method comprises the following steps: screening component peaks with a remarkable effect on treating mastitis in a coronet aqueous extract, identifying a single-component peak with an optimal effect on treating mastitis, and carrying out liquid chromatography-mass spectrometry combined analysis on the single-component peak with the optimal effect from the protein level, so as to obtain a single-component peak with an optimal effect on treating mastitis. According to the score, the protein abundance, the polypeptide modification site and the secondary mass spectrum, identifying the main component for treating mastitis in the single-component peak with the optimal effect; by screening and identifying active ingredients with anti-inflammatory action in a single-component peak of the antler plate, a powerful experimental reference is provided for deep development and clinical application of the antler plate; the screening and verification problems of antler plates and even animal traditional Chinese medicine complex components are solved, and the method has great significance in narrowing the range of traditional Chinese medicine effective components and screening and determining the traditional Chinese medicine effective components.
Owner:长春科技学院
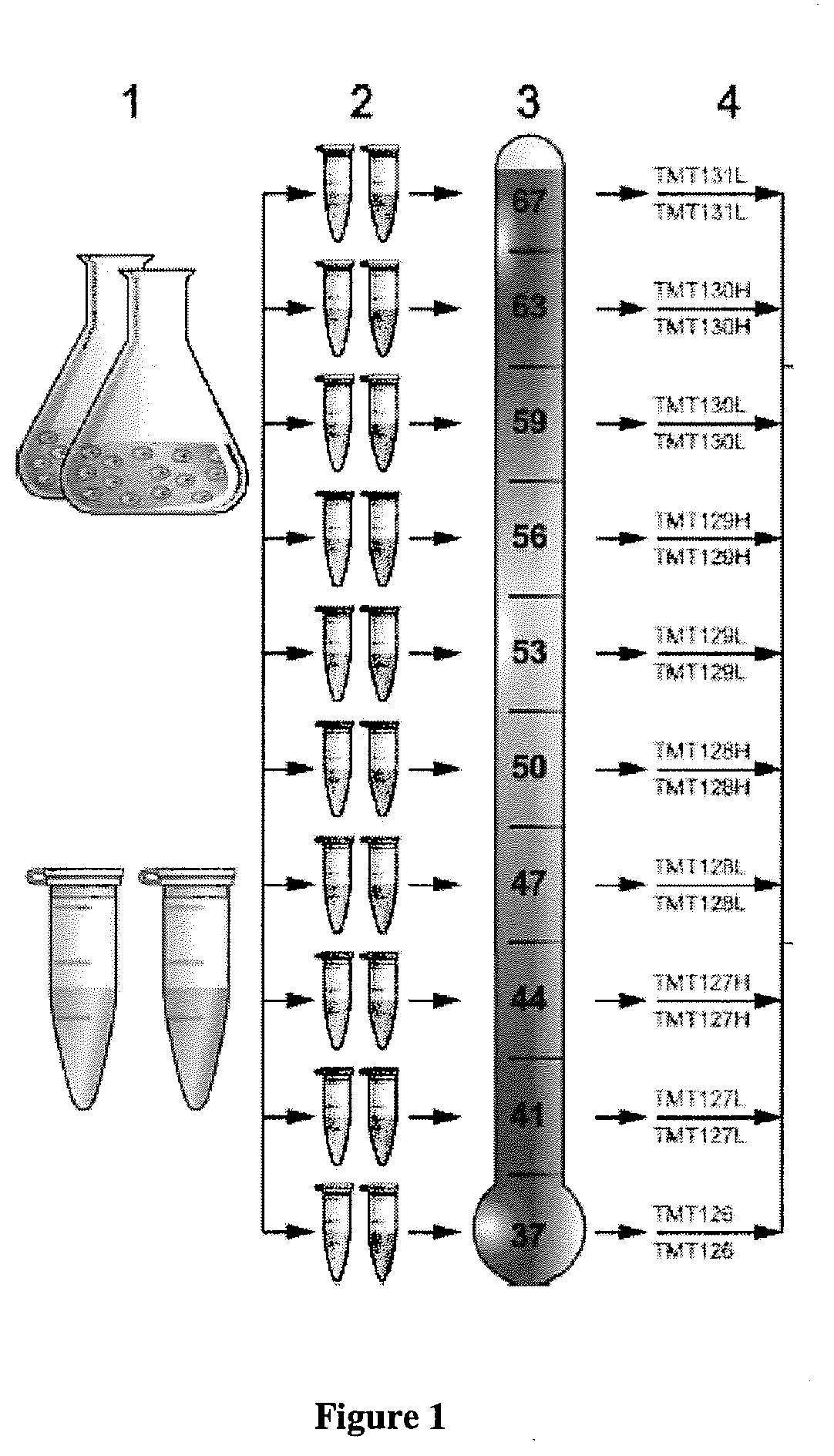
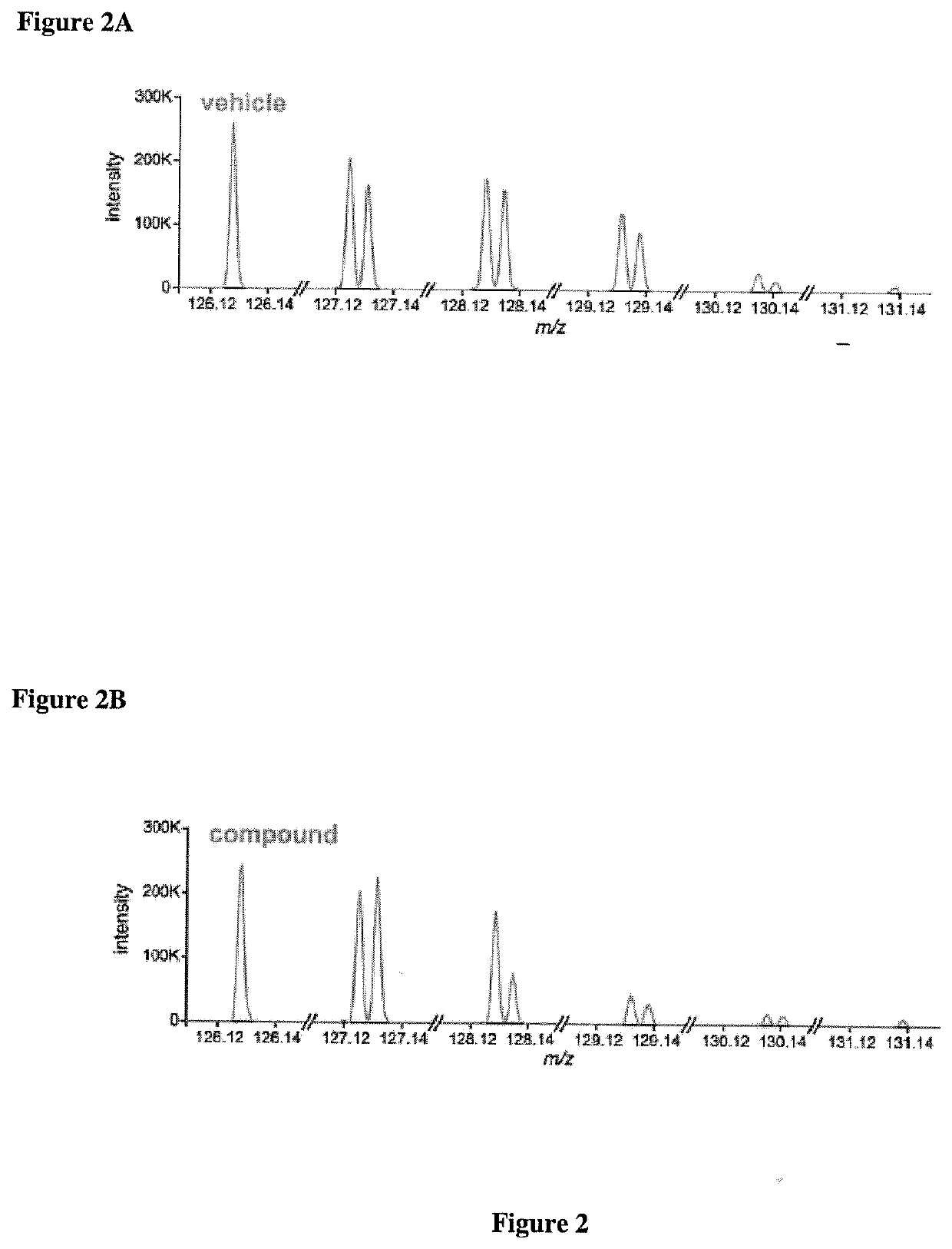
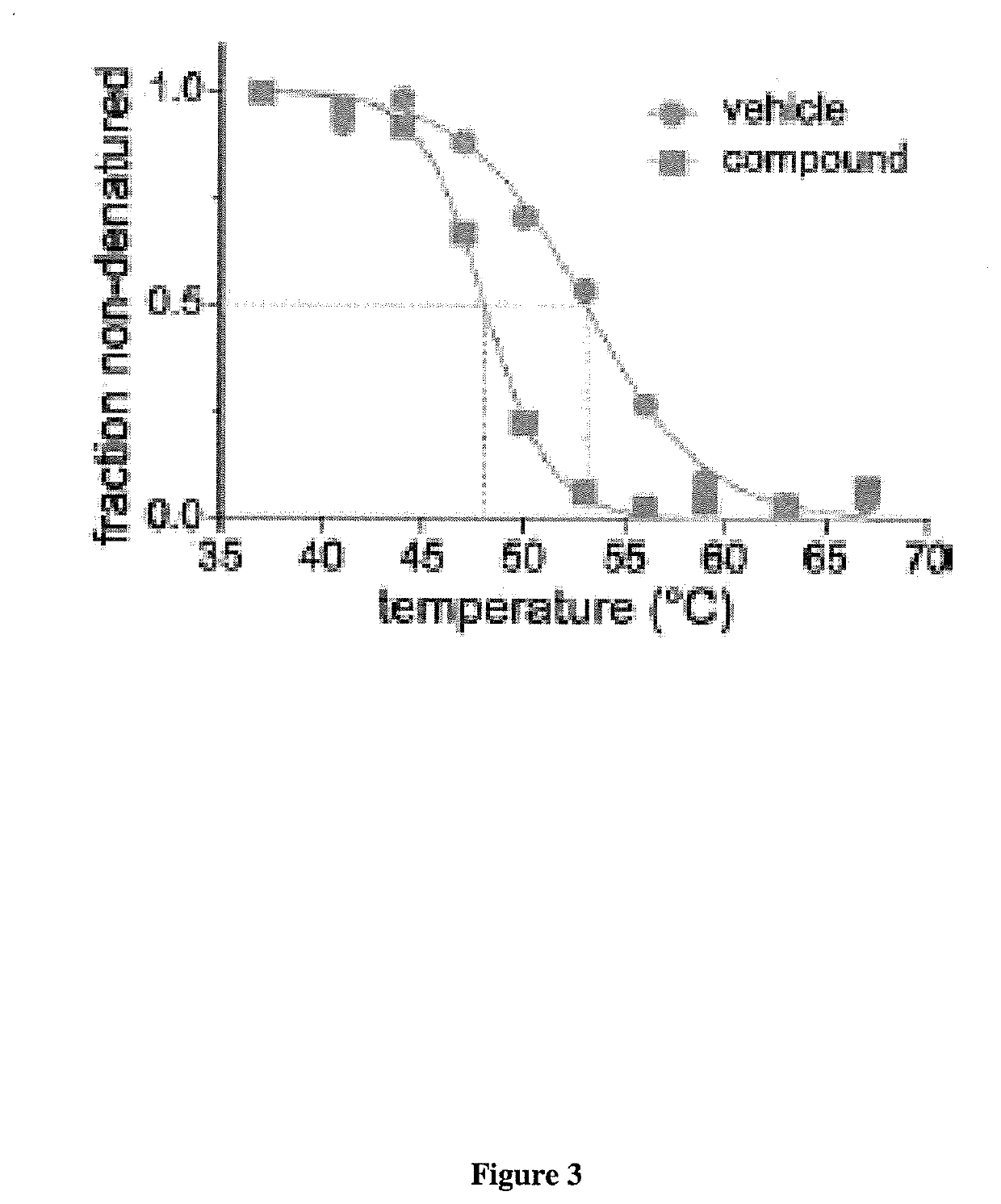
Microfluidic Devices And Methods For Cellular Thermal Shift Assays
InactiveUS20190358630A1Increases effective diffusivityIncrease ratingsCompound screeningApoptosis detectionMechanism of actionDrug target
The invention relates to thermal shift assays implemented to uncover ligand protein binding interactions in whole cells or cell extracts. In particular embodiments, the invention provides improved methods and devices for performing thermal shift assays for determining both drug targets and drug mechanism of action. For example, the invention performs thermal shift assays in microfluidic devices and determines ligand-protein binding by relative protein abundance measurements.
Owner:UNIV OF COLORADO THE REGENTS OF
Pharmaceutical composition for use in improving quality of scalp or skin, wound healing, or improving quality of hair
ActiveUS11406668B2Shorten the timeEasily propagatedCosmetic preparationsHair cosmeticsWound healingBULK ACTIVE INGREDIENT
The present invention provides a pharmaceutical composition for use in modifying scalp or skin, promoting wound healing, or modifying hair, comprising a secretion from adipose stem cells as an active ingredient, wherein 0.3 to 0.6 μg of the secretion in terms of protein abundance per site in scalp or skin is administered.
Owner:FUKUOKA HIROTARO
Methods for drug target screening
InactiveUS7122312B1Microbiological testing/measurementOther foreign material introduction processesProtein targetScreening method
The present invention provides methods for identifying targets of a drug in a cell by comparing (i) the effects of the drug on a wild-type cell, (ii) the effects on a wild-type cell of modifications to a putative target of the drug, and (iii) the effects of the drug on a wild-type cell which has had the putative target modified of the drug. In various embodiments, the effects on the cell can be determined by measuring gene expression, protein abundances, protein activities, or a combination of such measurements. In various embodiments, modifications to a putative target in the cell can be made by modifications to the genes encoding the target, modification to abundances of RNAs encoding the target, modifications to abundances of target proteins, or modifications to activities of the target proteins. The present invention also provides methods for drug development based on the methods for identifying drug targets.
Owner:FRED HUTCHINSON CANCER RES CENT
A dynamic regulation system controlled by combination of promoters
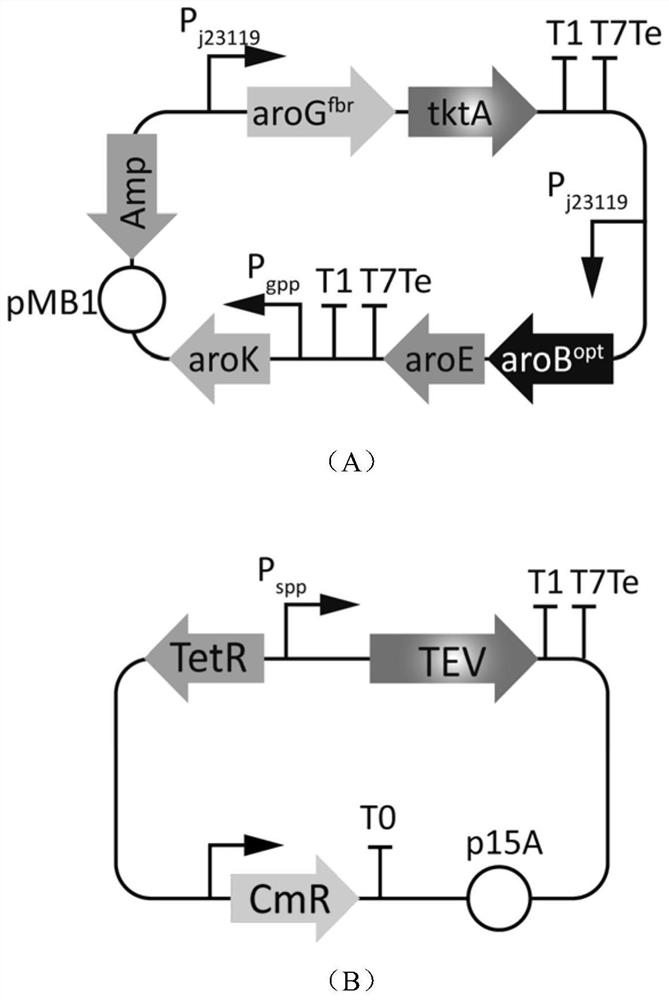
ActiveCN109576199BReduce growth loadSimple designBacteriaTransferasesShikimate kinaseEscherichia coli
The invention discloses a promoter combination controlling dynamic regulation and control system and discloses the application of the promoter combination controlling dynamic regulation and control system and application thereof to the production of shikimic acid, and belongs to the technical field of bioengineering. Two types of promoters with completely different transcription characteristics are screened through a molecular biology method, and comprise a growth period correlated promoter and a stable period promoter. According to the property of specifically identifying cut specific short peptide of proteinase, a protein abundance regulation and control system, which does not need manual control and exogenous addition of an inducing agent, is designed and constructed. A dynamic regulation and control gene path targeting shikimate kinase is introduced into engineering escherichia coli, so that the capability of producing shikimic acid without exogenously adding aromatic amino acid and the inducing agent in an inorganic salt culture medium is realized. According to the dynamic regulation and control system, the yield of the produced shikimic acid can reach 21.2 g / L and the conversion rate reaches 0.24 g / g glucose.
Owner:JIANGNAN UNIV
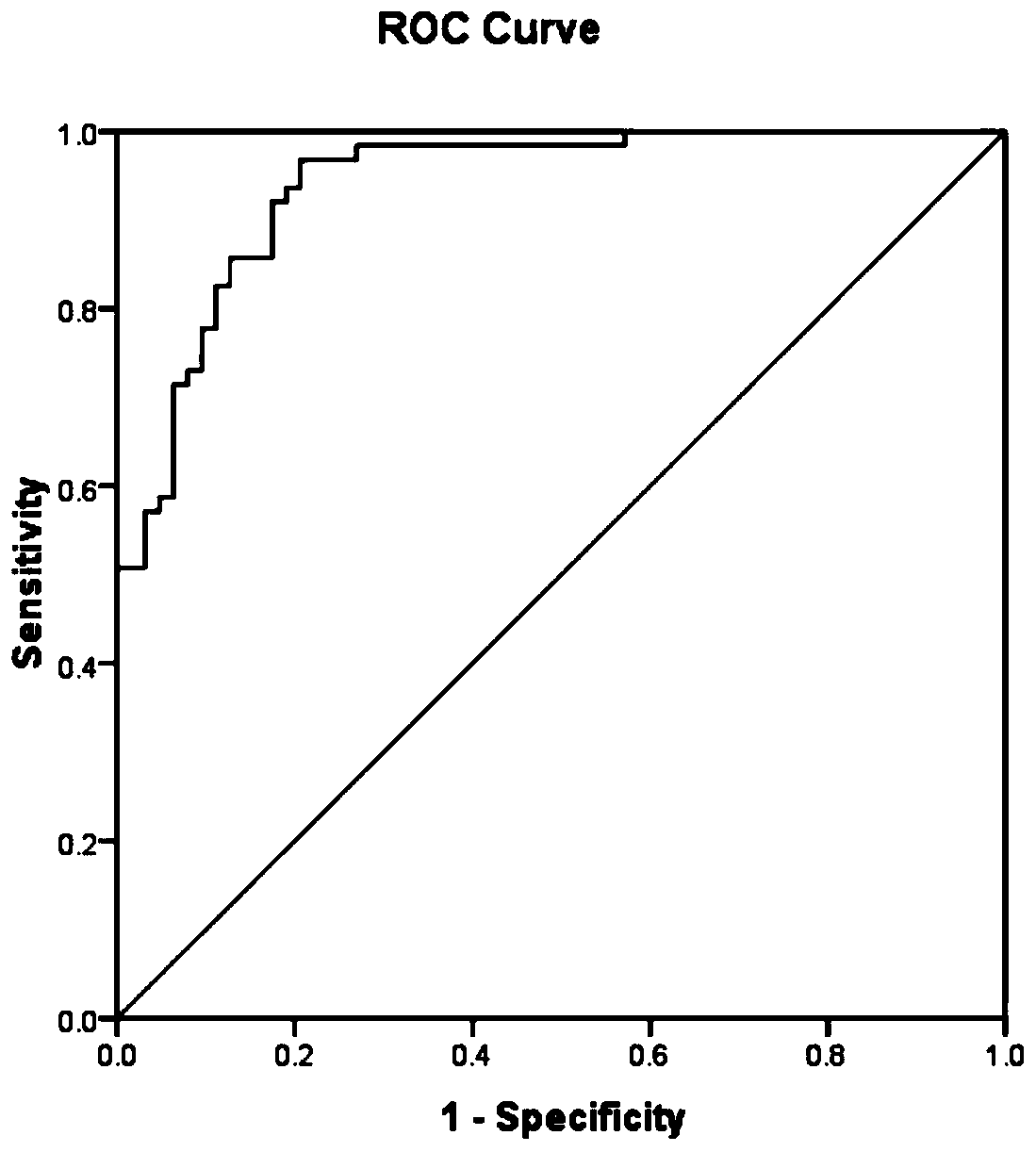
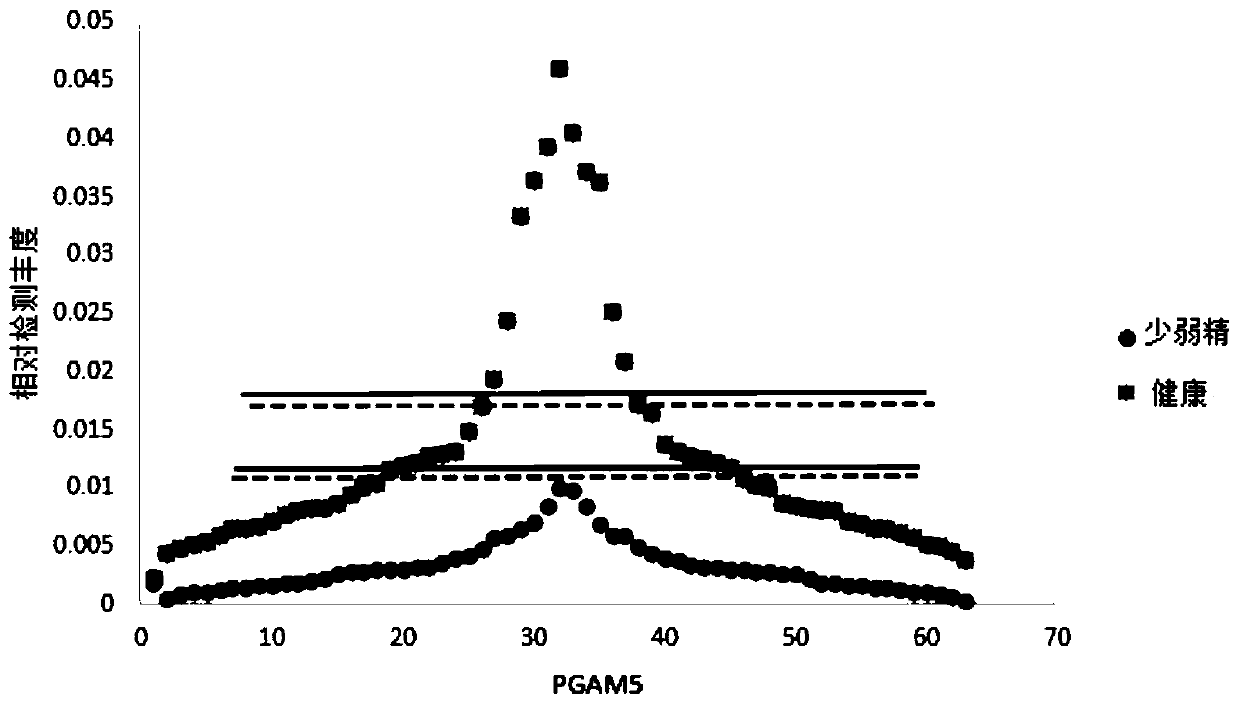
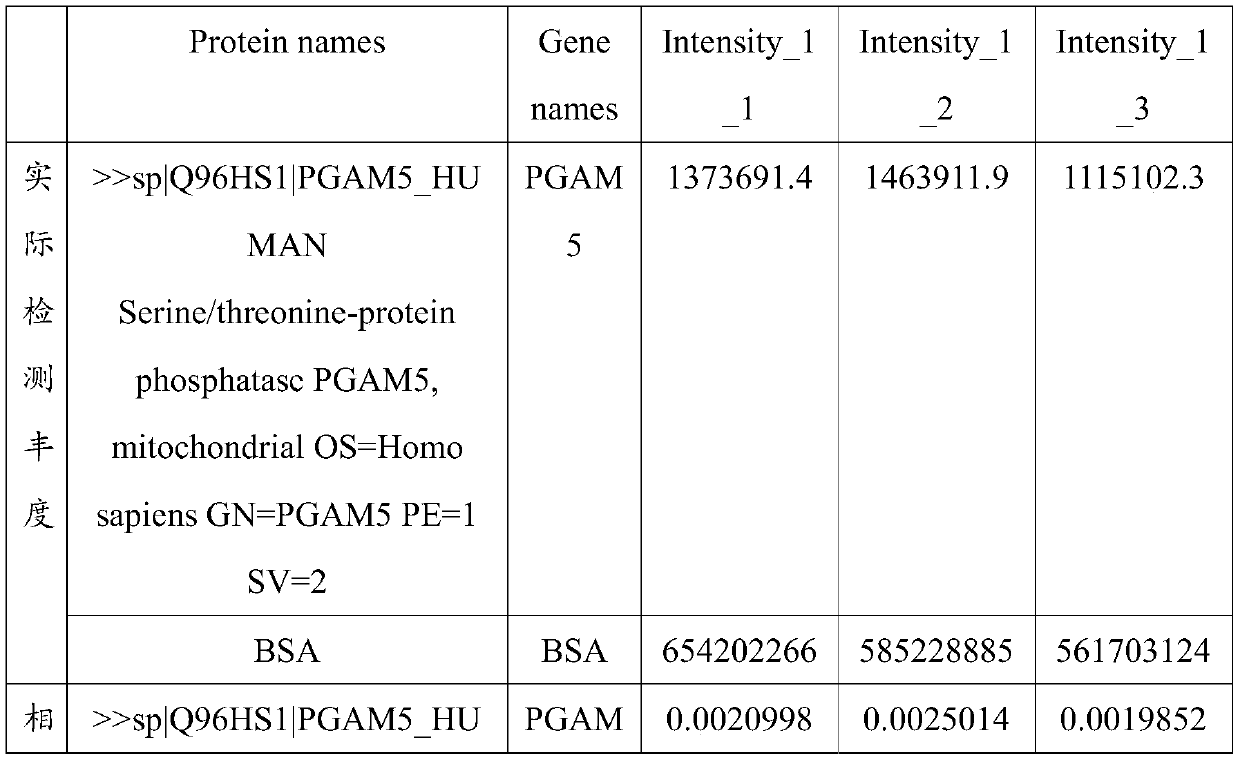
Application of PGAM5 as diagnostic marker and therapeutic target for oligospermia and asthenozoospermia
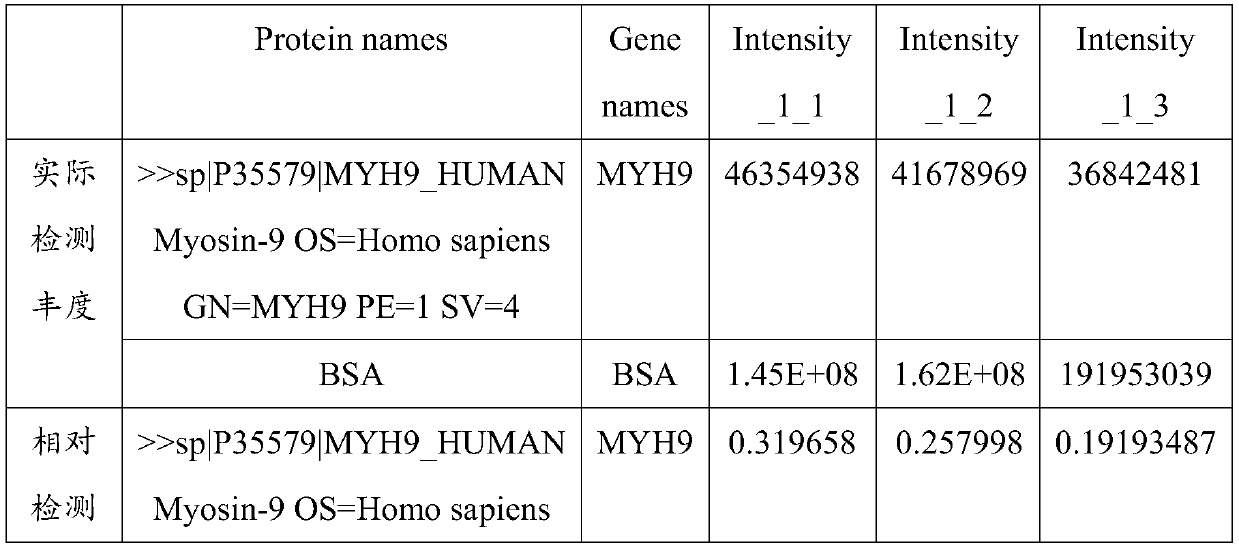
PendingCN111518868AImprove detection valueImprove accuracyMicrobiological testing/measurementDisease diagnosisProteomics methodsDisease patient
The invention belongs to the technical field of diagnosis for oligospermia and asthenozoospermia, and particularly relates to application of PGAM5 as a diagnostic marker and a therapeutic target for oligospermia and asthenozoospermia. On the basis of a proteomics method, protein data of normal patient samples and patient samples with oligospermia and asthenozoospermia are analyzed, BSA protein signal strength is taken as the standard to detect protein abundance of the samples, and differential protein of the normal patient samples and the diseased patient samples is obtained. By means of detection with the method, remarkable down-regulation of the PGAM5 in the samples with severe oligospermia and asthenozoospermia is determined, the PGAM5 has good credibility when serving as the diagnosticbiomarker, and the PGAM5 is applied to development of related detection reagents and kits. Meanwhile, the research result is conducive to clarifying the pathogenesis of oligospermia and asthenozoospermia, contributes to development of target drugs for oligospermia and asthenozoospermia and has great clinical significance.
Owner:山东立菲生物产业有限公司
Method for quantifying protein abundance by taking metal cluster as artificial antibody
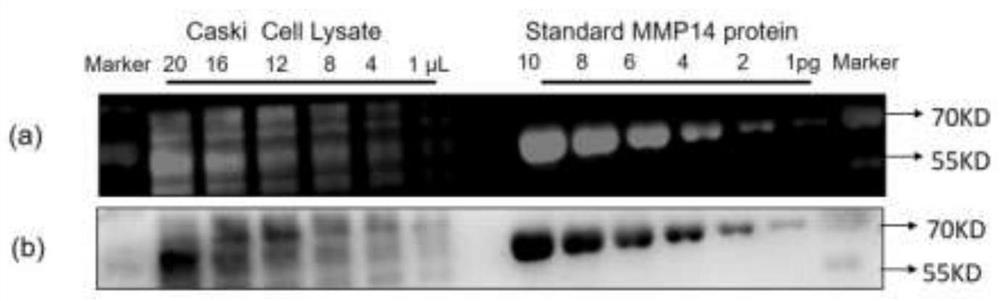
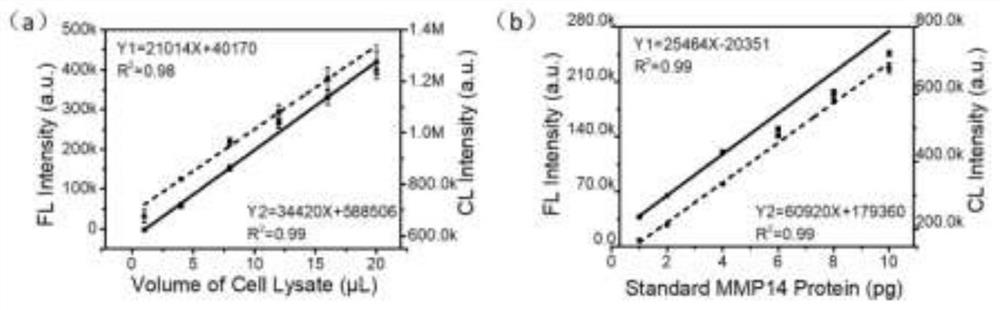
PendingCN112485452AQuick checkAccurate detectionChemiluminescene/bioluminescenceBiological testingAntiendomysial antibodiesProtein target
The invention discloses a method for quantifying protein abundance by taking a metal cluster as an artificial antibody, and relates to a method for detecting and quantifying low-abundance protein in cells, tissue extracts or serum. The method comprises the following steps: specifically identifying target protein separated by polyacrylamide gel electrophoresis in cells, tissue lysate or serum by the artificial antibody, and quantitatively detecting the abundance of the target protein by analyzing an intrinsic fluorescence signal of the artificial antibody and a chemiluminescence signal generated by a catalytic substrate of the artificial antibody. The artificial antibody is composed of a metal cluster core and a targeting peptide of a targeting target protein modified on the metal cluster core. By using the method provided by the invention, the expression quantity of the protein in the cell, tissue or serum biological sample can be quickly and accurately analyzed quantitatively.
Owner:BEIJING UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com