Method for simultaneous quantification of protein abundance and the cysteine oxidation level and application of method
A cysteine and protein technology, which is applied in the field of simultaneous quantification of protein abundance and cysteine oxidation level, can solve the problems of inability to accurately determine changes in protein redox levels, and inability to simultaneously quantify protein abundance, etc. Conducive to detection and quantification, improved efficiency, and rapid identification by mass spectrometry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1. Redox Proteomic Analysis of Spinach Leaf High Temperature Response
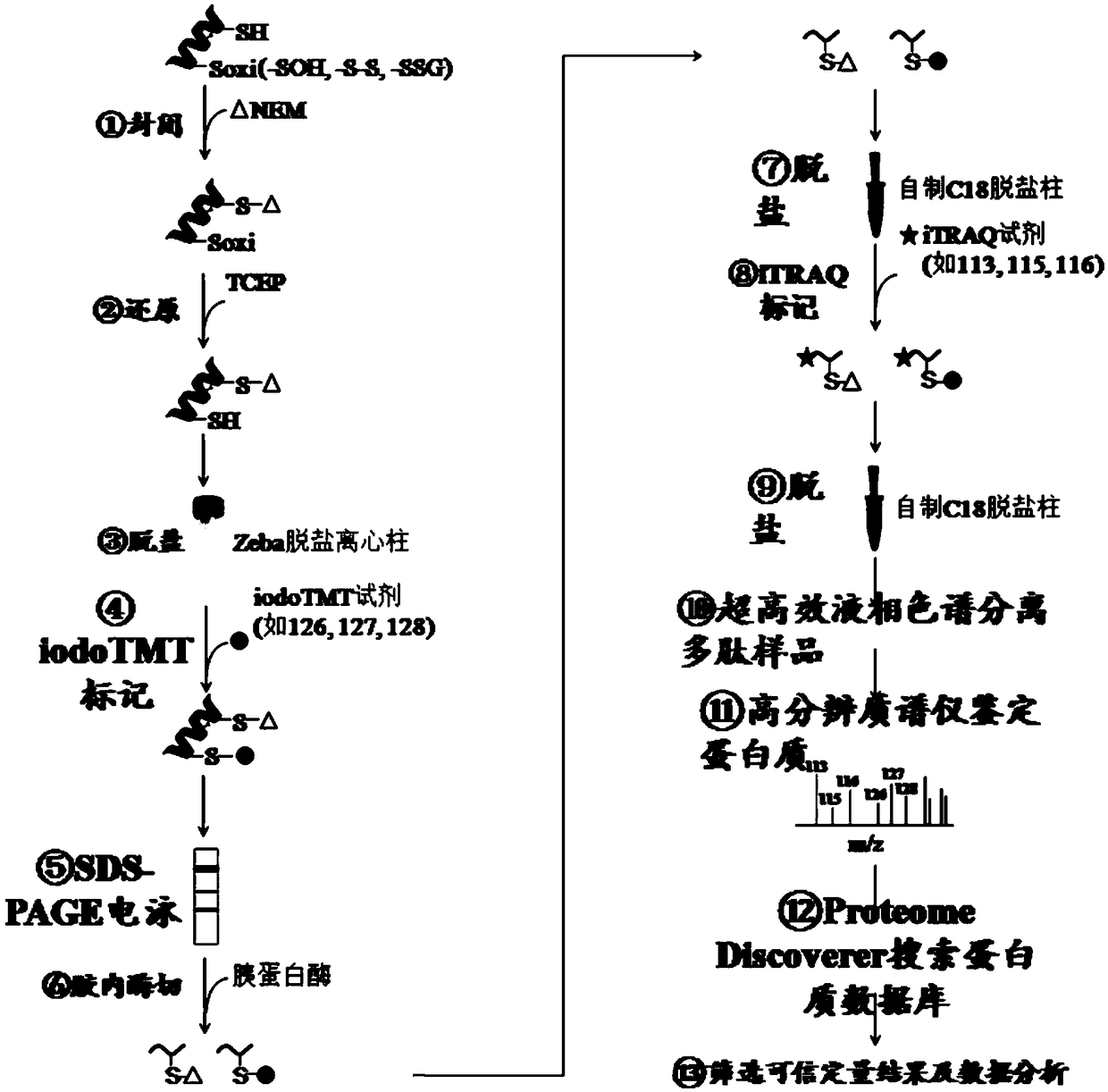
[0047] attached figure 1 An experimental flow diagram of the method for simultaneous quantification of protein abundance and cysteine oxidation levels is shown. Include the following steps:
[0048] 1. Apply the extract that has been added with NEM to extract the spinach (Spinacia oleracea L.) leaf protein treated at 37°C for 0h, 2h and 4h, then use acetone to precipitate the protein, and then apply HES buffer (50mM HEPES, 1mM EDTA, 0.1%SDS, pH 8.0) to reconstitute the protein.
[0049] 2. Add 5mM TCEP reduced protein to the protein solution and incubate at 50°C for 70min.
[0050] 3. Desalt the protein sample by referring to the Zeba desalting spin column desalting instructions for protein samples.
[0051] 4. Apply iodoTMT tags 126, 127 and 128 to label leaf protein samples treated at 37°C for 0h, 2h and 4h, respectively. After the reaction was completed, 20 mM DTT was added to term...
Embodiment 2
[0072] Example 2. Redox proteomics analysis of chloroplast oxidative stress response of star grass
[0073] 1. The analysis process is the same as in Example 1, except that the sample analyzed is that Puccinellia tenuiflora responds to 0mM, 5mM and 10mM H 2 o 2 Stressed chloroplast protein samples. 0 mM, 5 mM and 10 mM H were labeled with iodoTMT tags 126 and 129, 127 and 130, and 128 and 131, respectively 2 o 2 Stressed chloroplast protein samples; 0mM, 5mM and 10mM H 2 o 2 Stressed chloroplast protein samples.
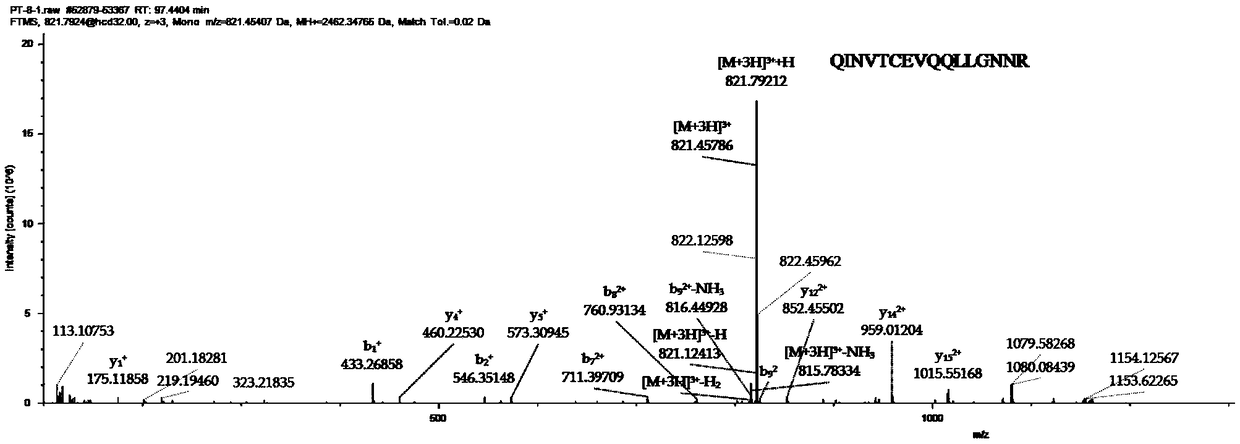
[0074] 2. When using Waters nanoAcquity UPLC and Thermal Orbitrap Fusion high-resolution mass spectrometer to identify polypeptide samples, except that the setting parameters of HCD collision energy are inconsistent with those in Example 1, other parameters are the same as in Example 1. image 3 Example of a secondary spectrum for the peptide "QINVTCEVQQQLLGNNR" with HCD collision energy set to 32%, Figure 4 Example of a secondary spectrum for the peptide "QI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com