Identification method of individual differential expression protein
A technology of differential expression and identification method, which is applied in proteomics, instrumentation, genomics, etc., can solve problems such as performance improvement, and achieve the effect of ensuring stability, good application prospects, and high recognition accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
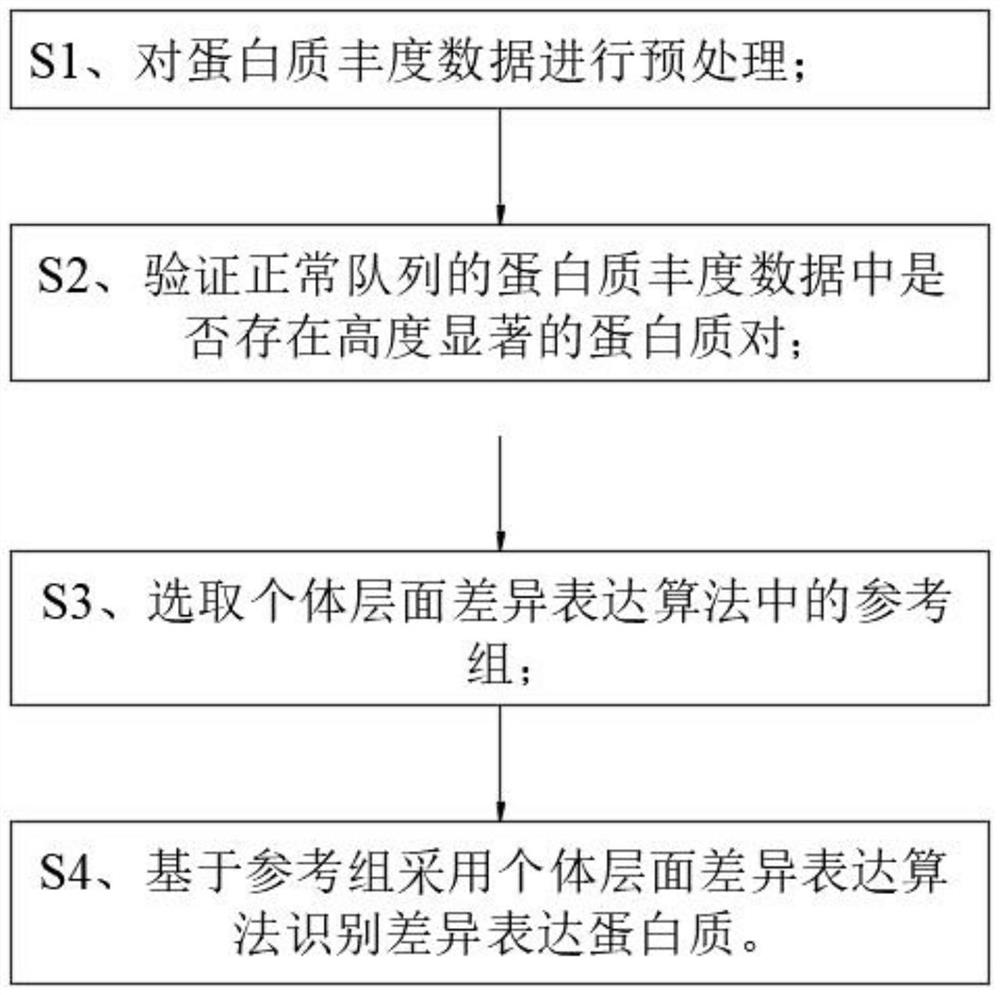
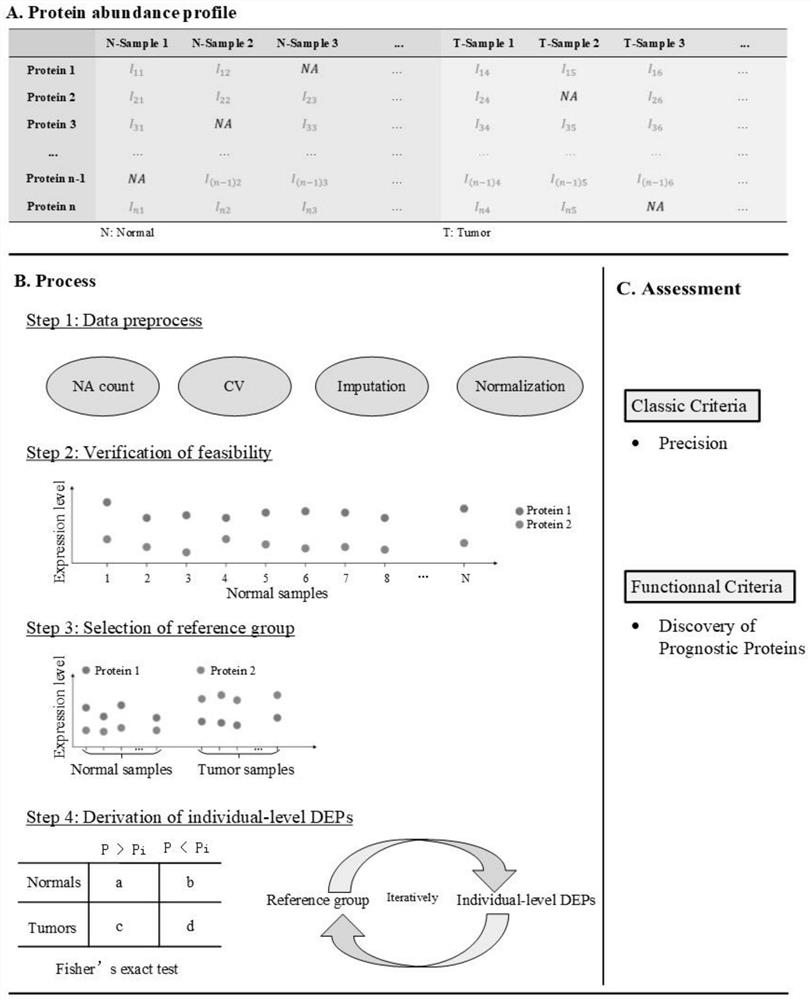
[0042] Such as Figure 1 to Figure 4 As shown, the present invention discloses a method for identifying individual differentially expressed proteins, the method comprising the following steps:
[0043] S1. Preprocessing the protein abundance data;
[0044] The specific process of step S1 is:
[0045] S11. Perform statistics on the ratio of missing values for each protein in all samples in the protein abundance data, filter proteins by setting the upper limit of the missing value ratio, and remove proteins whose missing value ratio exceeds the threshold in the protein abundance data;
[0046]S12. By calculating the coefficient of variation of proteins in the same cohort, it is judged whether the difference in protein abundance among different individuals is due to heterogeneity among individuals or quantitative errors;
[0047] Proteins with a high coefficient of variation in the same cohort in step S12 are identified as caused by quantitative errors, and are screened out; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com