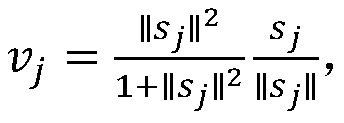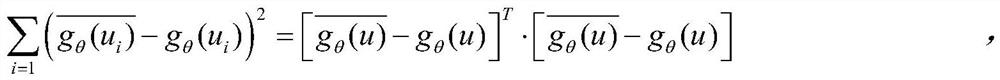Patents
Literature
70 results about "Protein recognition" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Recognition proteins are proteins that are embedded in the cellular membrane the allow cells to communicate with each other. receptor proteins are proteins which allow cells to attach to other cells to allow cell communication.
Gene overexpressed in cancer
InactiveUS20070037204A1High stringent conditionSuppress gene expressionTissue cultureFermentationAntigenAntigen Binding Fragment
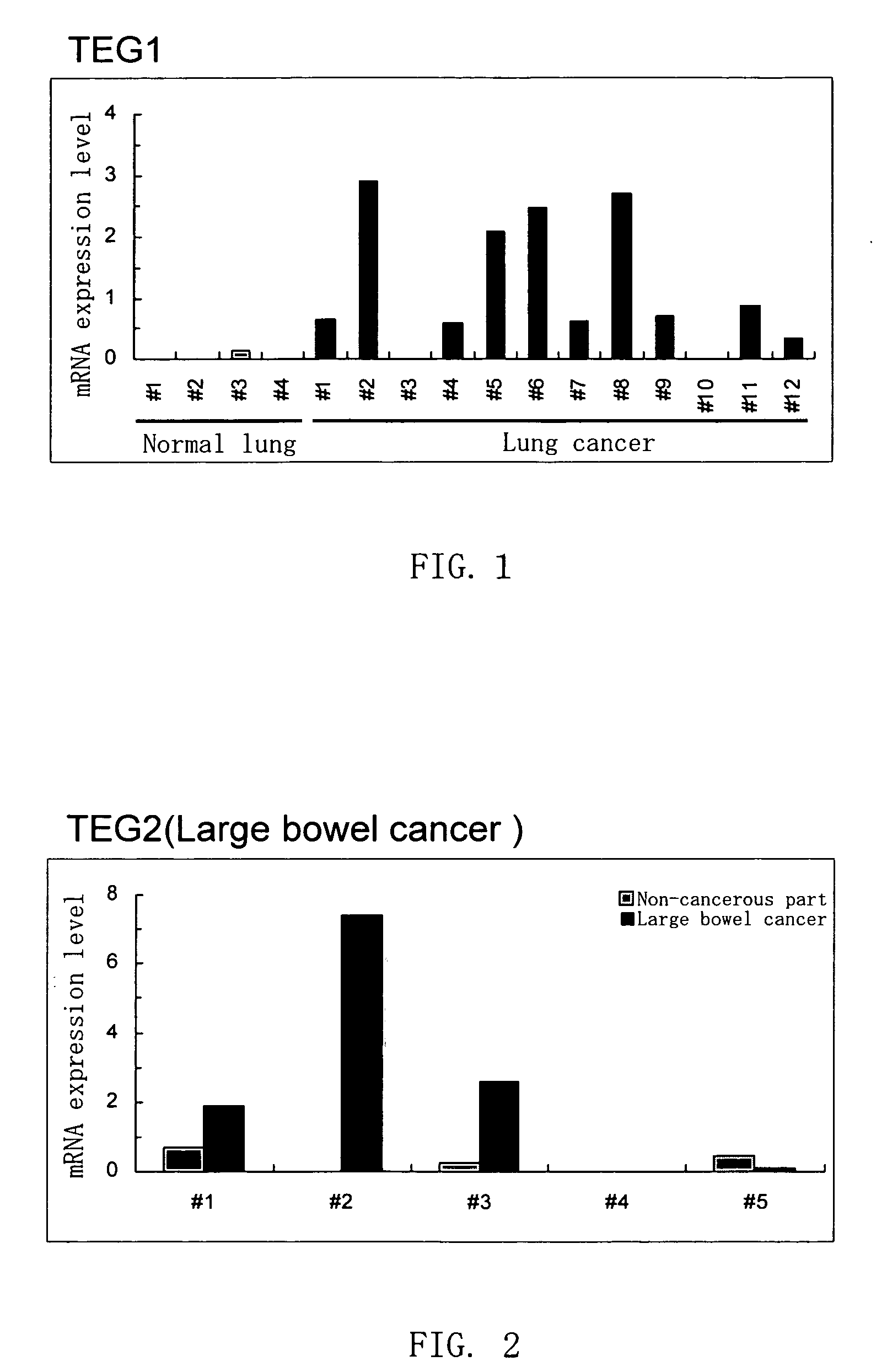
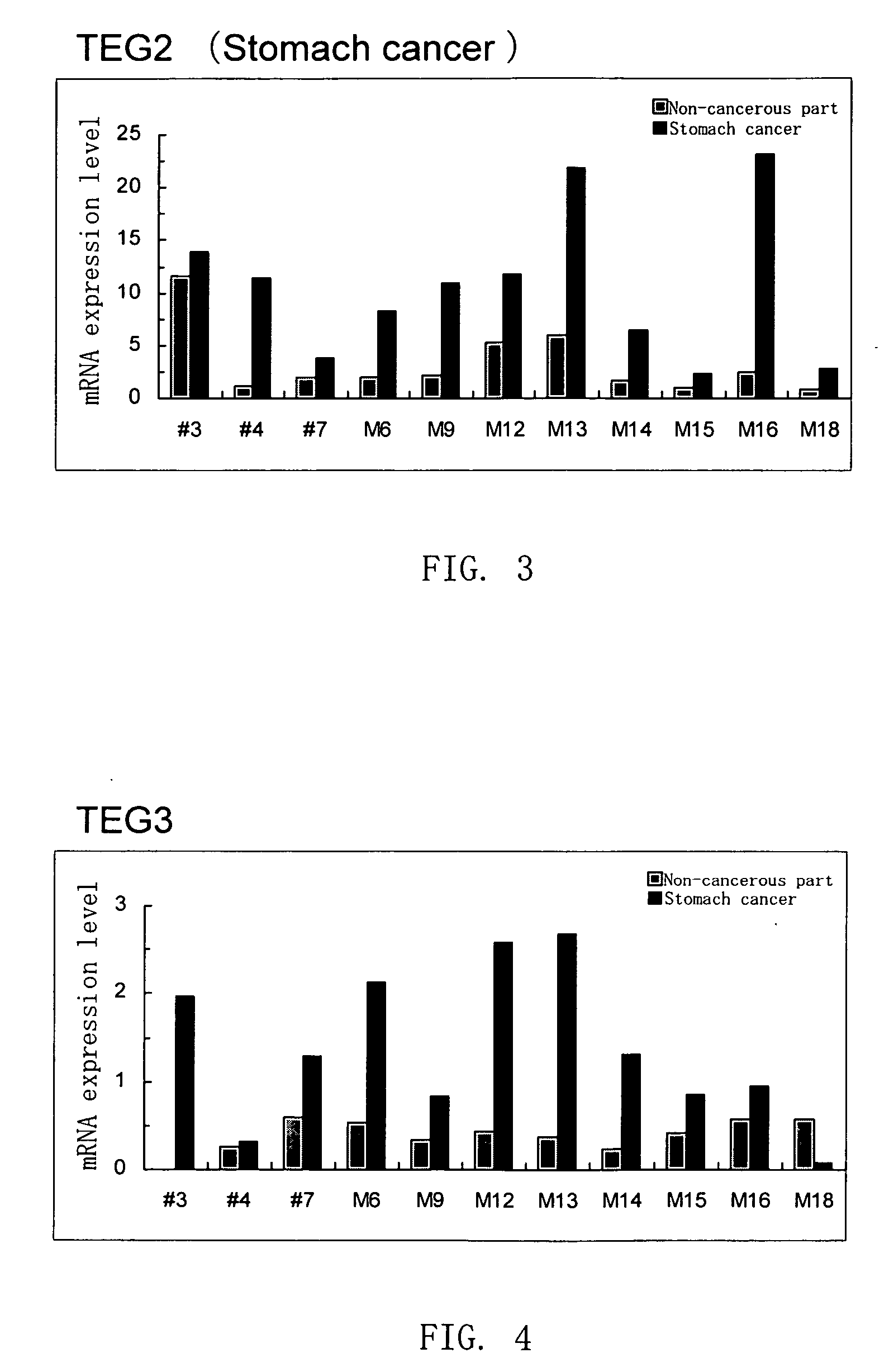
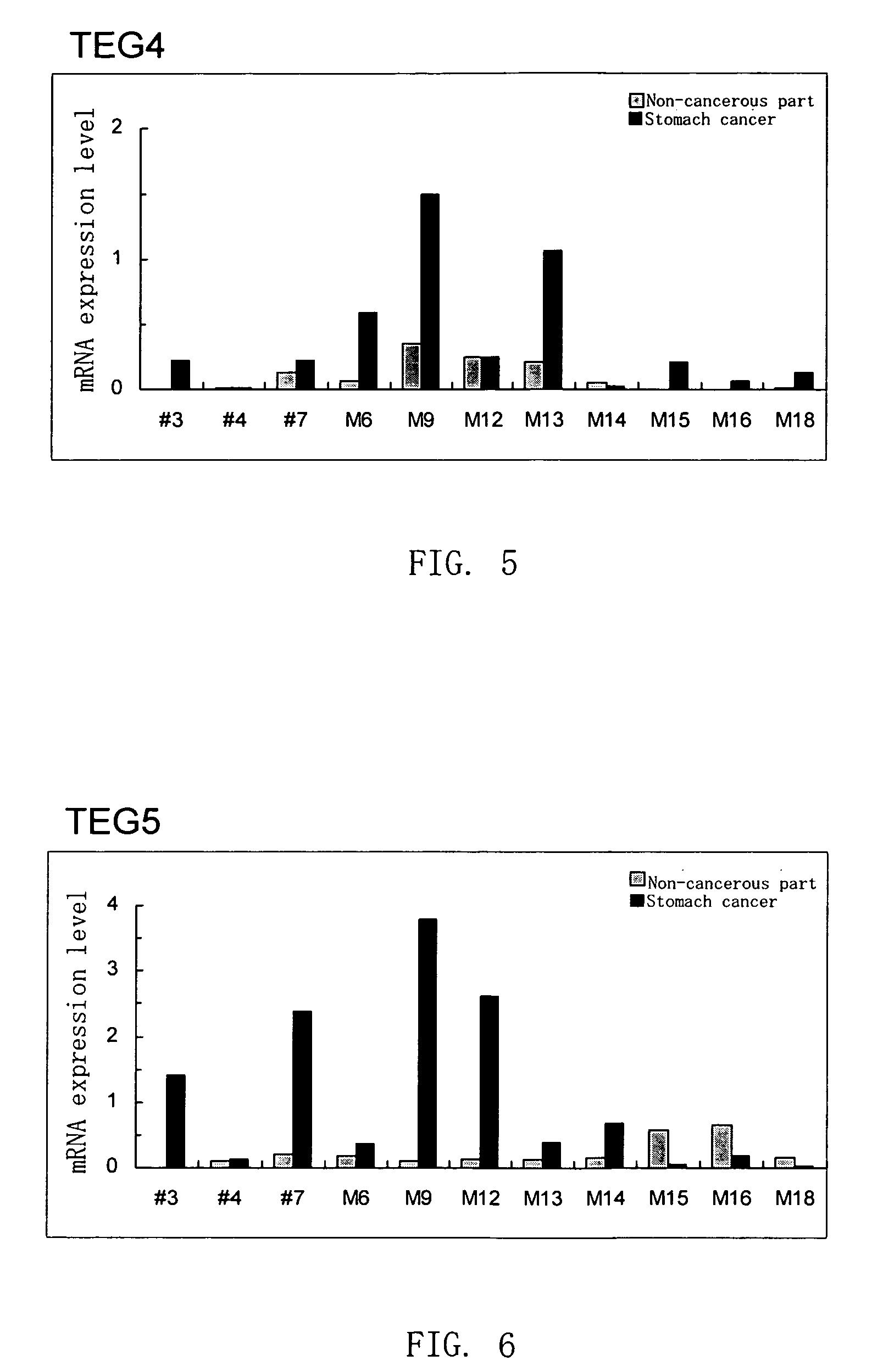
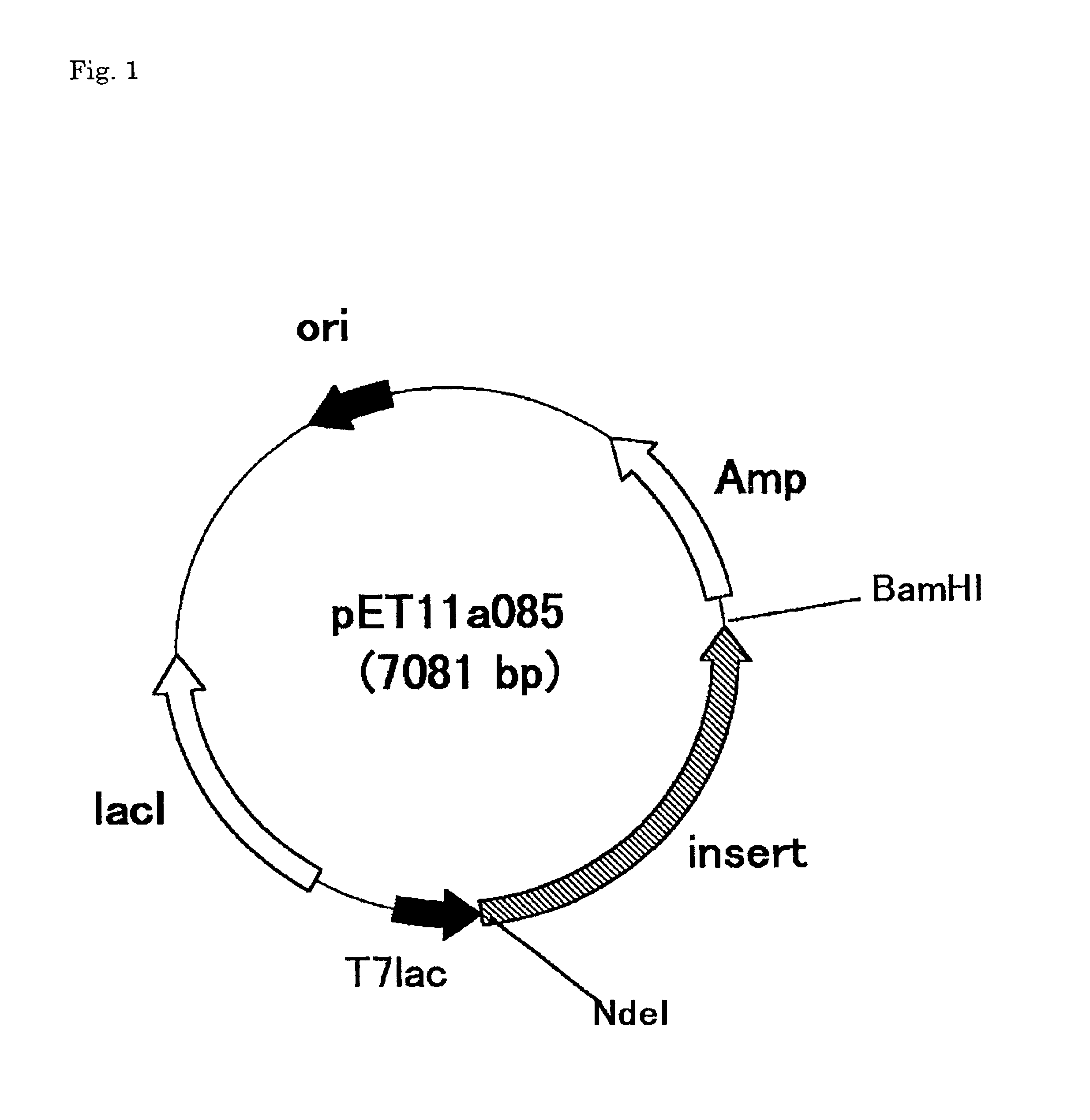
Disclosed are a protein encoded by a gene having a nucleotide sequence represented by any of SEQ ID NOs: 1 to 65 or a fragment thereof, an antibody recognizing the protein or antigen-binding fragment thereof, and a polynucleotide having a sequence comprising at least 12 consecutive nucleotides of a nucleotide sequence represented by any of SEQ ID NOs: 1 to 65 or a nucleotide sequence complementary thereto. The gene and the protein of the invention is useful for diagnosing and treating cancer.
Owner:ABURANTAI HIROYUKI +2
Protein molecular imprinting polyion liquid membrane electrochemical transducer
InactiveCN104142361ALarge specific surface areaGuaranteed stabilityMaterial analysis by electric/magnetic meansBiocompatibility TestingIonic liquid
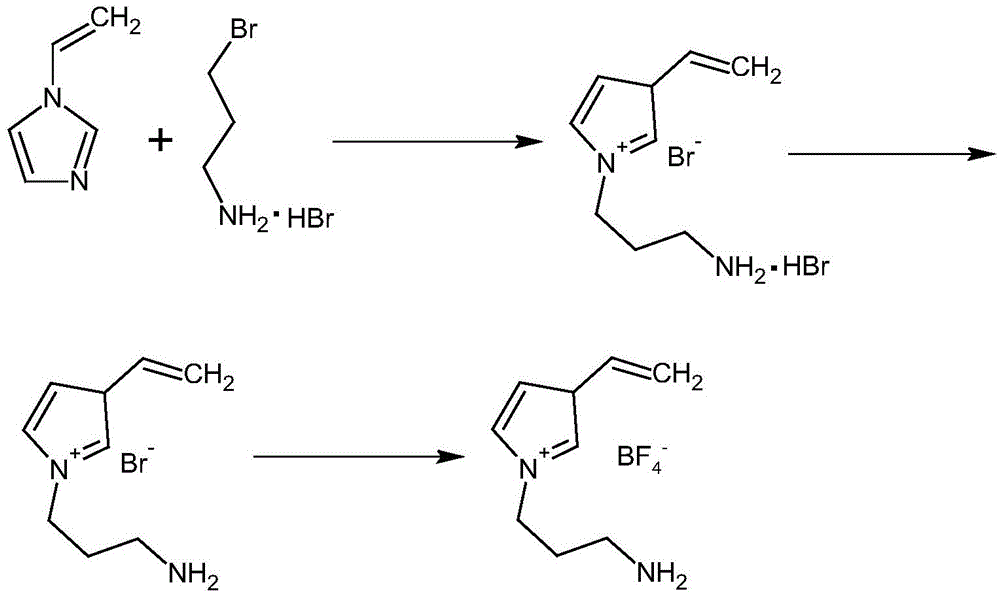
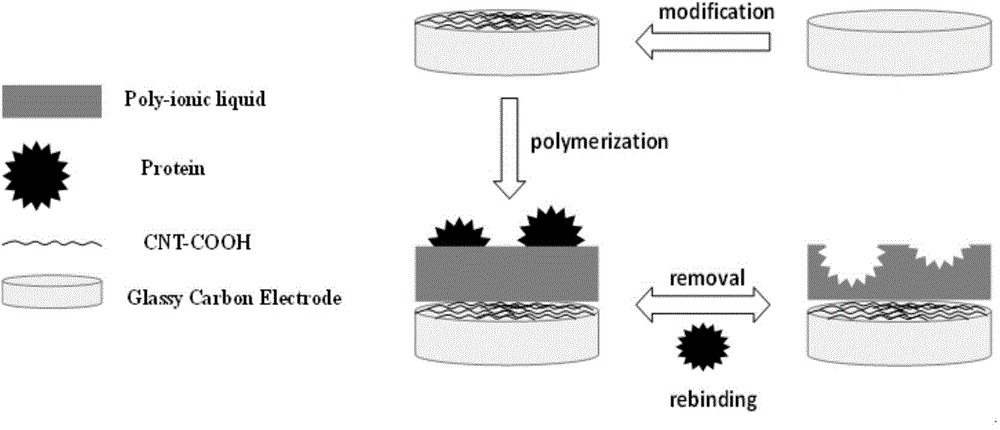
The invention relates to the technical field of electro-analytical chemistry and protein identification transducers, and discloses a synthesizing method of functionalized ionic liquid and a preparation method and application of a molecular imprinting electrochemical transducer composed of the functionalized ionic liquid, carboxylation multiwalled carbon nanotubes and glassy carbon electrodes. The preparation method of the molecular imprinting electrochemical transducer comprises the steps that the polymerizable amino-functionalized ionic liquid is used as a functional monomer, BSA is used as template protein, N, N'-methylene bisacrylamide is used as a cross-linking agent, an oxidation-reduction system composed of ammonium persulfate and TEMED is used as an initiator, after polymerization, a molecularly imprinted polymer film is formed on the surfaces of the glassy carbon electrodes decorating the carboxylation multiwalled carbon nanotubes, and then the template protein is eluted to obtain the molecular imprinting electrochemical transducer which can identify template protein in a specific mode. The molecular imprinting electrochemical transducer has the advantages of being simple in preparation, low in material cost, high in selectivity, good in biocompatibility, and capable of being used for identifying and detecting protein in aqueous solution.
Owner:SOUTH CENTRAL UNIVERSITY FOR NATIONALITIES
Diagnostics method
ActiveUS7572597B2Improve the level ofDetecting susceptibilityBacterial antigen ingredientsPeptide/protein ingredientsEpitopeProtein insertion
Method of diagnosing in an individual recent exposure to an agent which is a pathogen, vaccine or any other moiety which induces a cellular response, said method comprising determining in vitro whether the T cells of the individual recognise a protein from said agent having a length of at least 30 amino acids, to a greater extent than one or more peptide epitopes from the agent, a greater extent of recognition of the protein indicating that the individual has recently been exposed to the agent.
Owner:OXFORD IMMUNOTEC
Method for analyzing the amount of intraabdominal adipose tissue
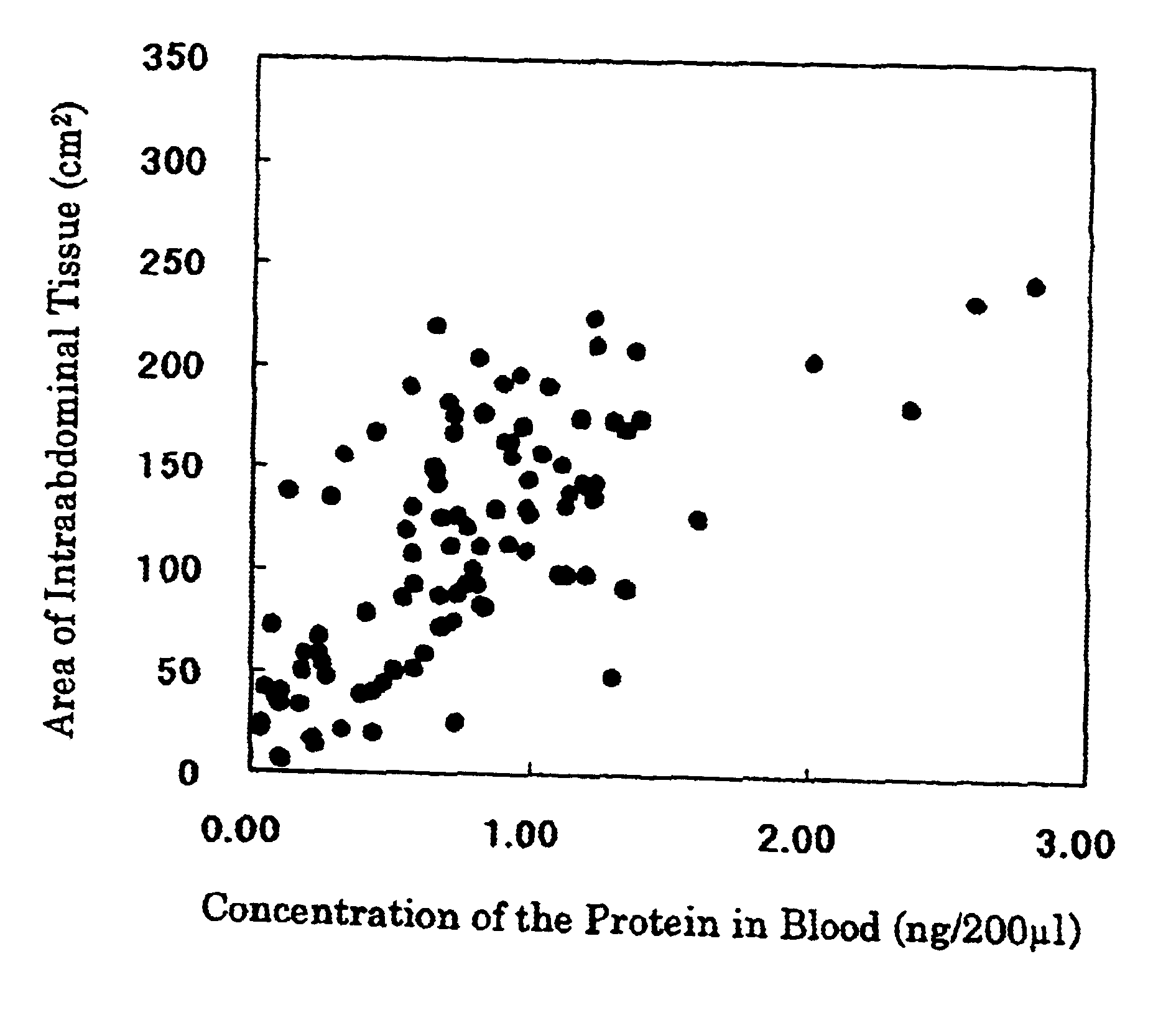
The present invention provides a method for analyzing the amount of intraabdominal adipose tissue comprising:[0002]determining the amount of intraabdominal tissue in a test-animal from the concentration of a protein in a body fluid, tissue or cell of said test-animal or in a prepared test solution from the body fluid, tissue or cell of said test-animal, based on the positive relationship between the amount of the intraabdominal adipose tissue in an animal and the concentration of a protein in a body fluid, tissue or cell of said animal or in a prepared solution from the body fluid, tissue or cell of said animal,[0003]wherein the protein is (1) encompassed by the amino acid sequence shown in SEQ ID NO:1 or is (2) recognized by an antibody against a protein encompassed by the amino acid sequence shown in SEQ ID NO:1.
Owner:SUMITOMO CHEM CO LTD +1
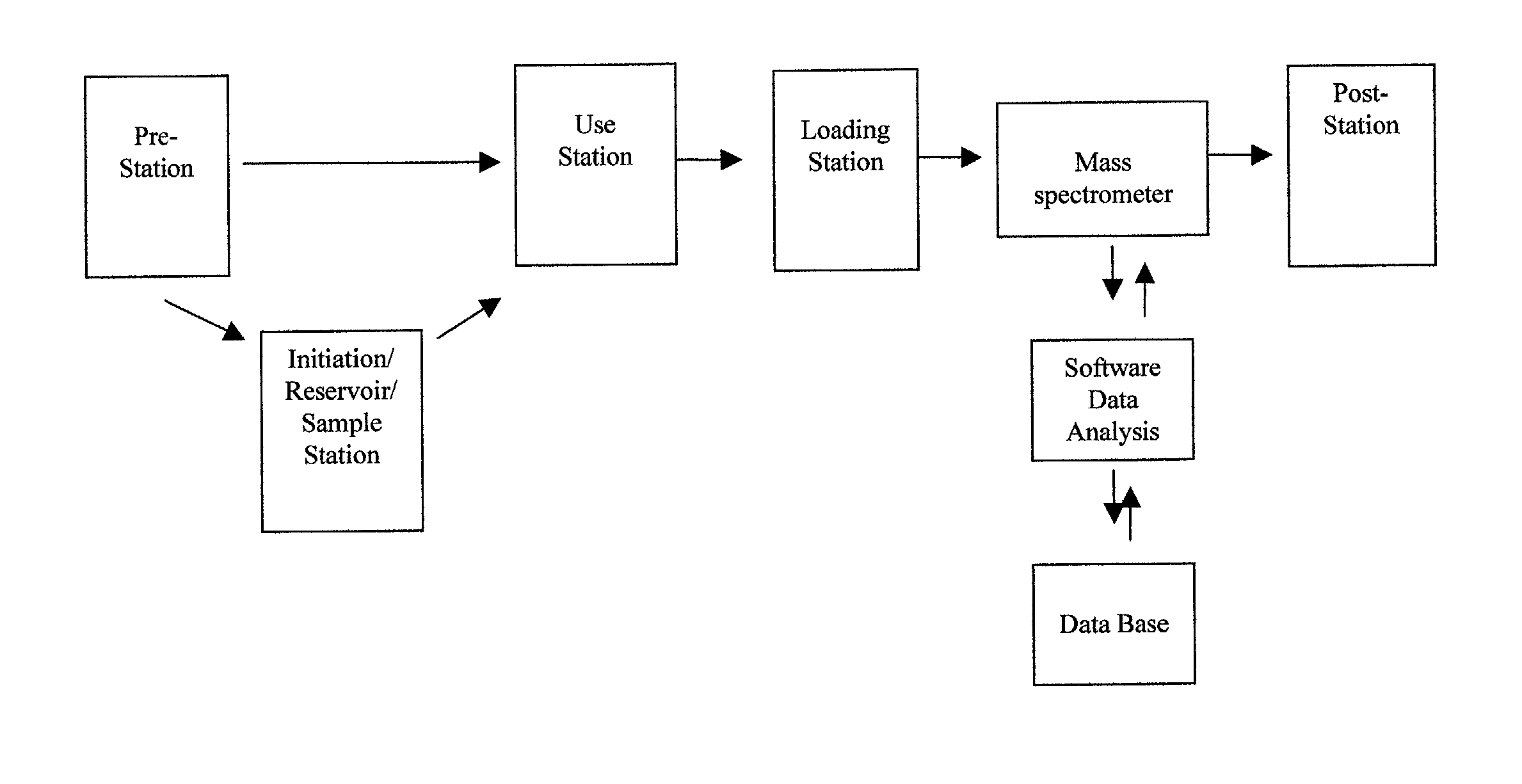
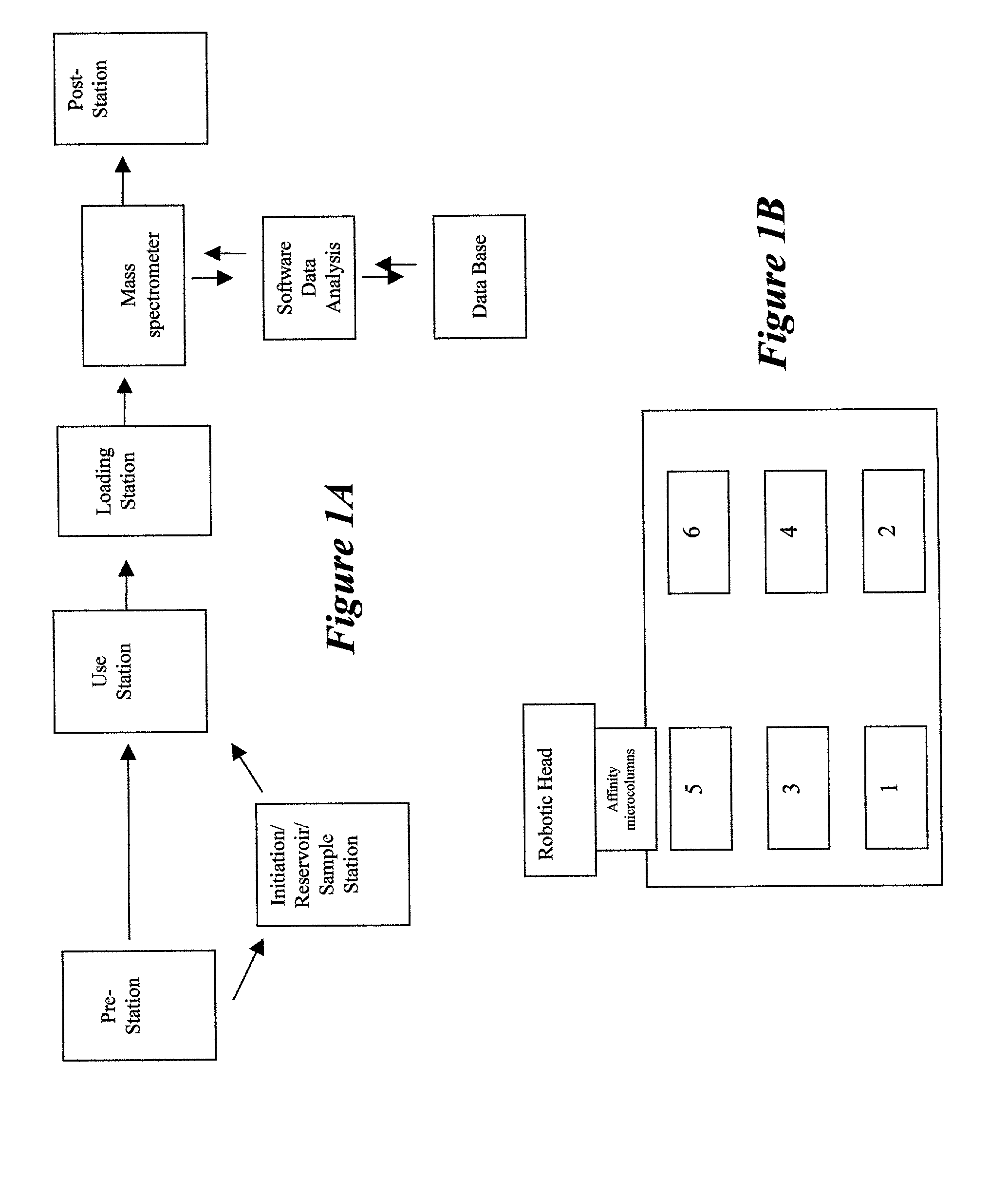
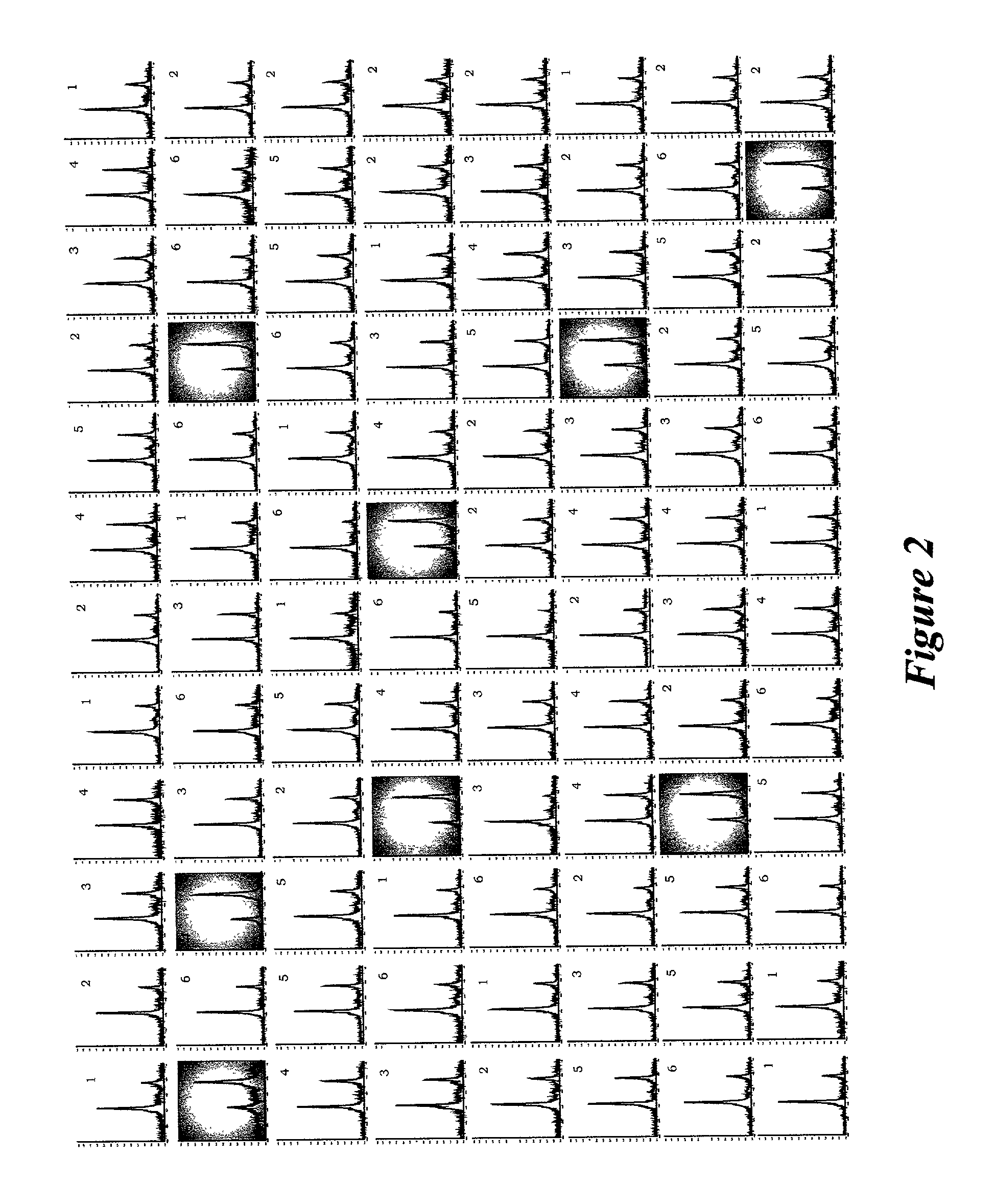
Integrated system for analysis of biomolecules
InactiveUS20020110904A1Material nanotechnologyBioreactor/fermenter combinationsProtein targetMass analyzer
Owner:INTRINSIC BIOPROBES
Temperature-sensitive protein molecular engram monolithic column and preparation method and application thereof
InactiveCN101816927ASensitive to temperatureReduce manufacturing costOther chemical processesPeptide preparation methodsCross-linkFunctional monomer
The invention discloses a temperature-sensitive protein molecular engram monolithic column and a preparation method and application thereof, belonging to high molecular material preparation technology and application thereof in the field of protein separation and analysis, and particularly relating to a novel protein molecular engram monolithic column. A simple and moderate method is adopted to combine an inorganic silica gel monolithic column skeleton, protein serves as a template, and a temperature-sensitive functional monomer and cross-linking agent ingraft protein engram material on the surface of the inorganic silica gel so as to obtain the temperature-sensitive protein molecular engram monolithic column which has stronger template protein recognition capability. The temperature-sensitive protein molecular engram monolithic column prepared with the method of the invention combines the single identification characteristic of molecular engram with the characteristic of on-line separation of the monolithic column, has simple operation, moderate condition and low cost and provides a new method for removing high-abundance protein for the Journal of Proteome Research.
Owner:NANKAI UNIV
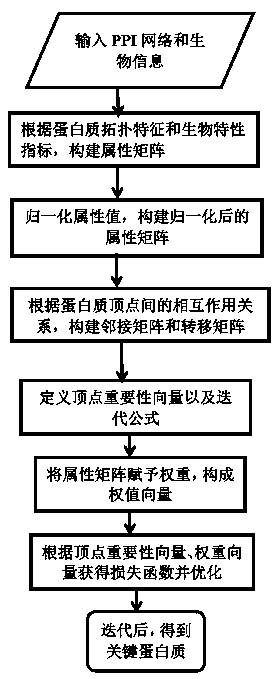
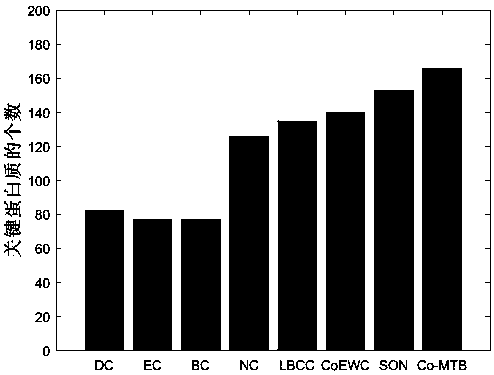
Key-protein recognition method based on fusion of biological and topological features
ActiveCN108733976AImprove accuracyImprove recognition efficiencySpecial data processing applicationsProtein Interaction NetworksEuclidean vector
The invention belongs to the field of bioinformatics technology, and particularly relates to a key-protein recognition method based on fusion of biological and topological features. The method includes: assigning a score, which indicates an importance degree of each protein vertex, to the protein vertex, wherein the scores of all the vertices constitute a vector of n columns; giving initial valuesof the scores, constituting the attribute values of the protein vertices according to biological information and values of topological characteristics, and constituting an attribute matrix; and finally, carrying out descending arrangement according to the scores, and outputting k proteins corresponding to the scores, namely final results. Combination of the topological characteristics of a protein interaction network and the protein biological-attributes facilitates identification of key proteins, and improves efficiency of key-protein recognition.
Owner:YANGZHOU UNIV
Diagnostics method
ActiveUS20050208594A1Improve the level ofPrevent considerable morbidityBacterial antigen ingredientsPeptide/protein ingredientsInducer CellsT cell
Method of diagnosing in an individual recent exposure to an agent which is a pathogen, vaccine or any other moiety which induces a cellular response, said method comprising determining in vitro whether the T cells of the individual recognise a protein from said agent having a length of at least 30 amino acids, to a greater extent than one or more peptide epitopes from the agent, a greater extent of recognition of the protein indicating that the individual has recently been exposed to the agent.
Owner:OXFORD IMMUNOTEC
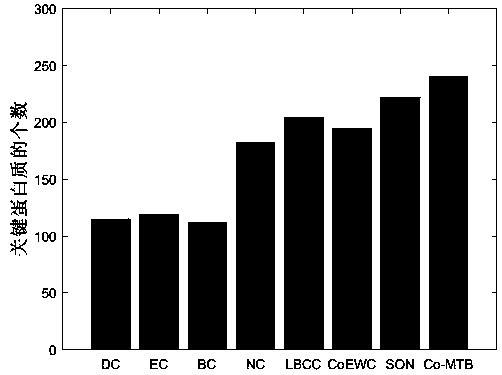
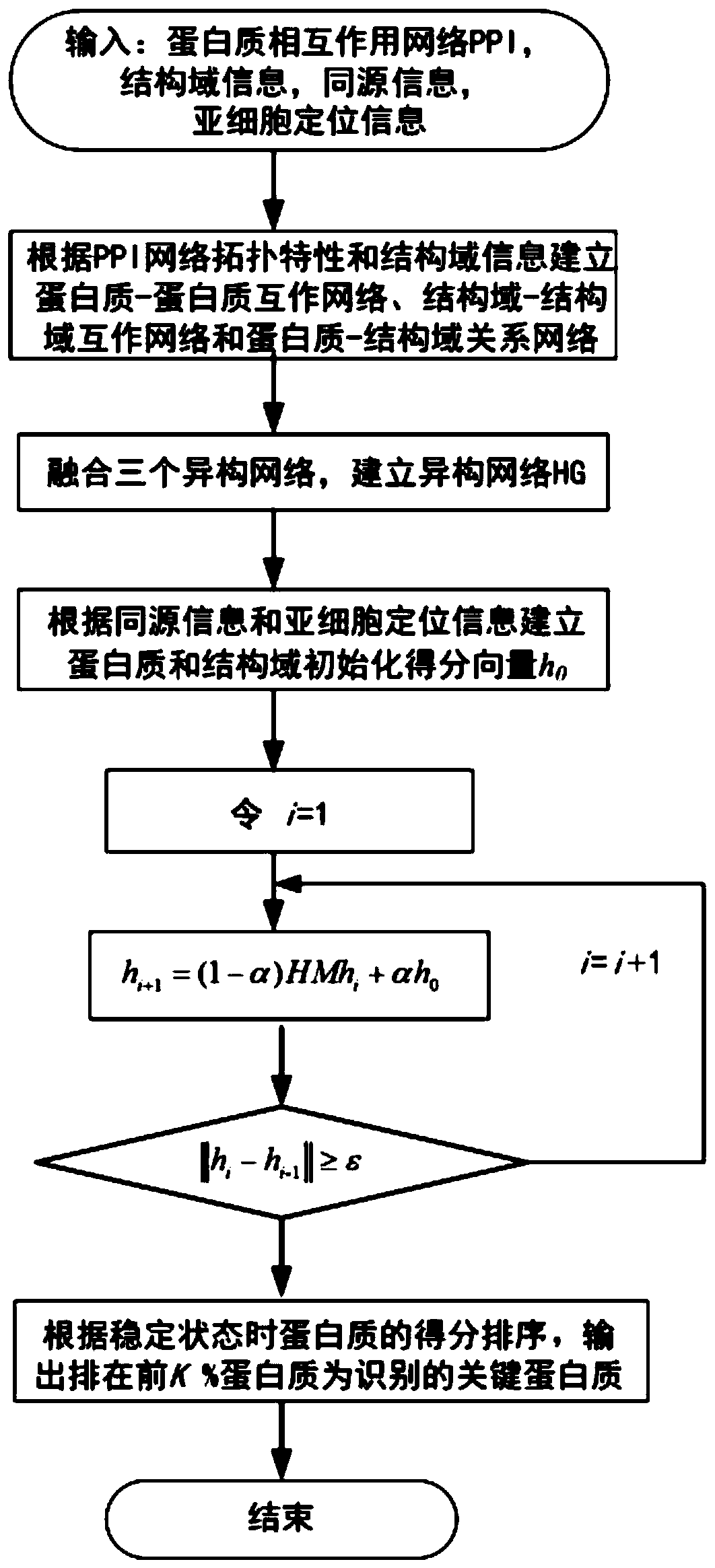
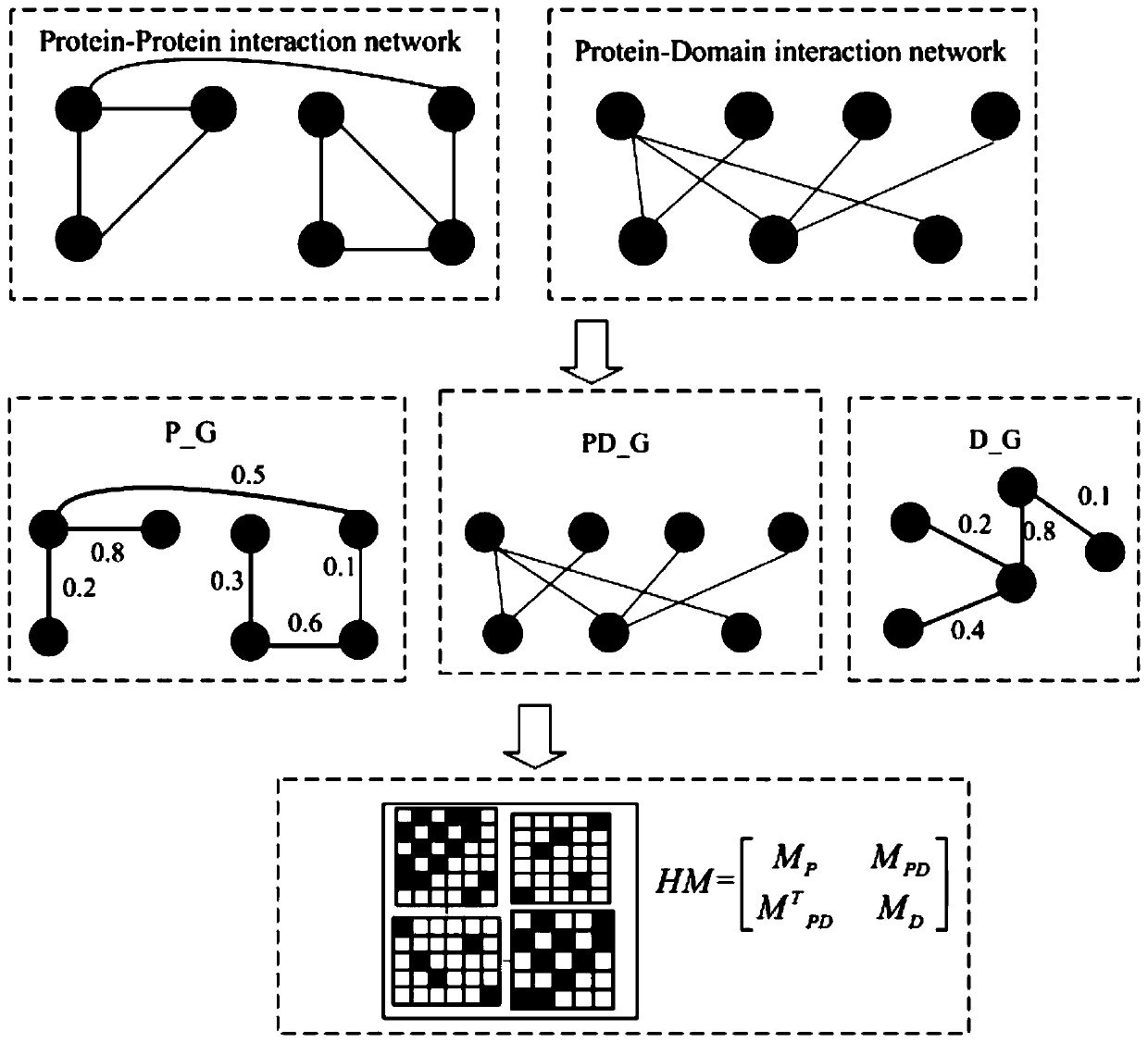
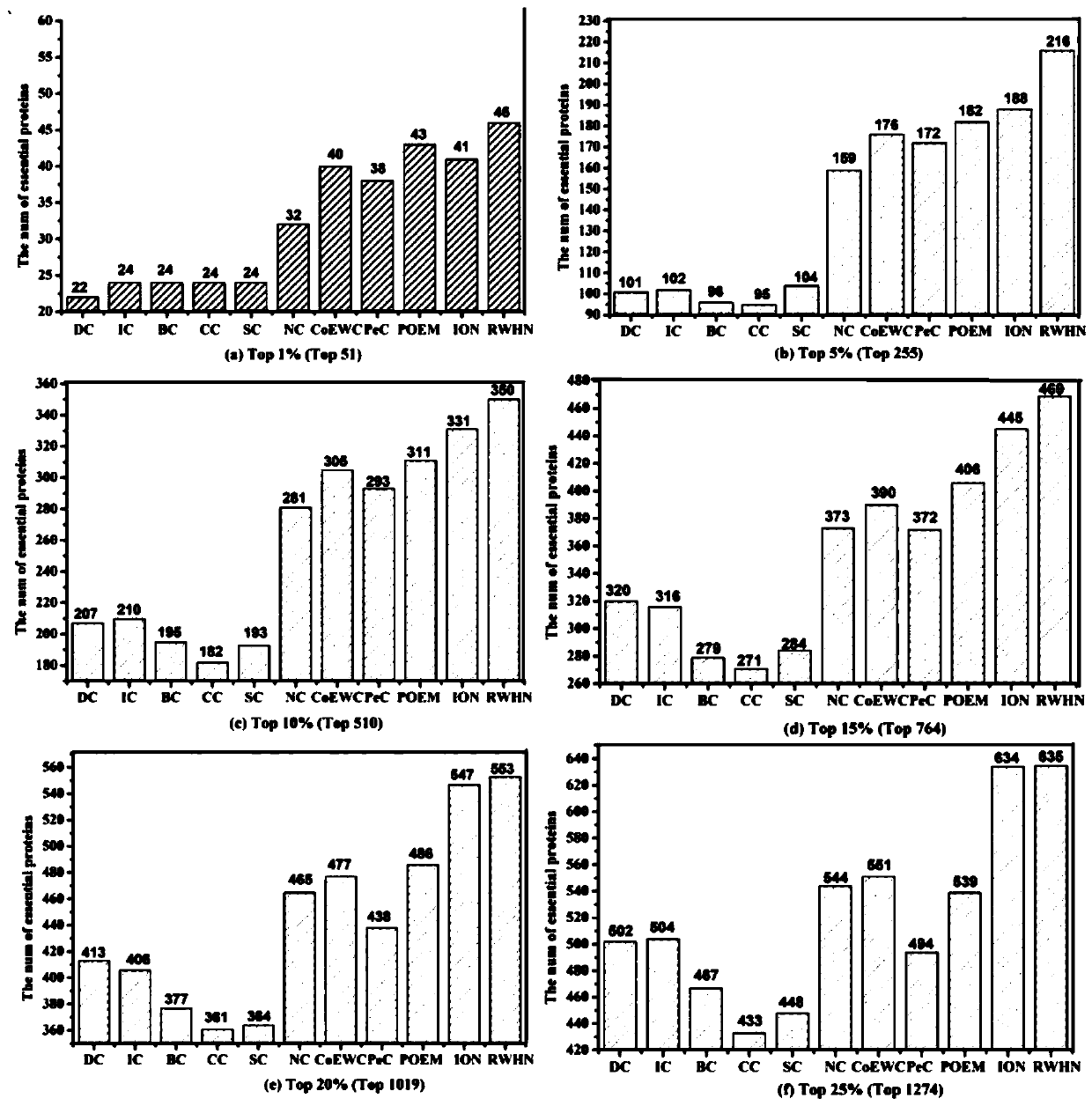
Key protein recognition method based on heterogeneous biological network fusion
ActiveCN109801674AImprove recognition accuracyBiostatisticsInstrumentsYeast ProteinsHeterogeneous network
The invention discloses a key protein recognition method based on heterogeneous biological network fusion. The key protein recognition method includes the steps: obtaining a yeast protein interactionnetwork topology, protein domain information, protein homology information, and protein subcellular localization information; respectively establishing a protein-protein interaction network P_G, a structural domain-structural domain interaction network D_G and a protein-structural domain network PD_G; fusing the three networks PD_G three networks, and establishing a heterogeneous network HG; establishing an initialized score vector h0 of the protein and structural domain; iteratively calculating the score vector h(i)t( / i) of the protein and structural domain based on a random walk model untila steady state is achieved; and sorting according to the score of the protein in the steady state, and identifying the key protein. The key protein recognition method based on heterogeneous biologicalnetwork fusion improves the fusion mode of multi-source biological data in the research of key protein recognition methods, and greatly improves the recognition accuracy of key proteins.
Owner:CHANGSHA UNIVERSITY
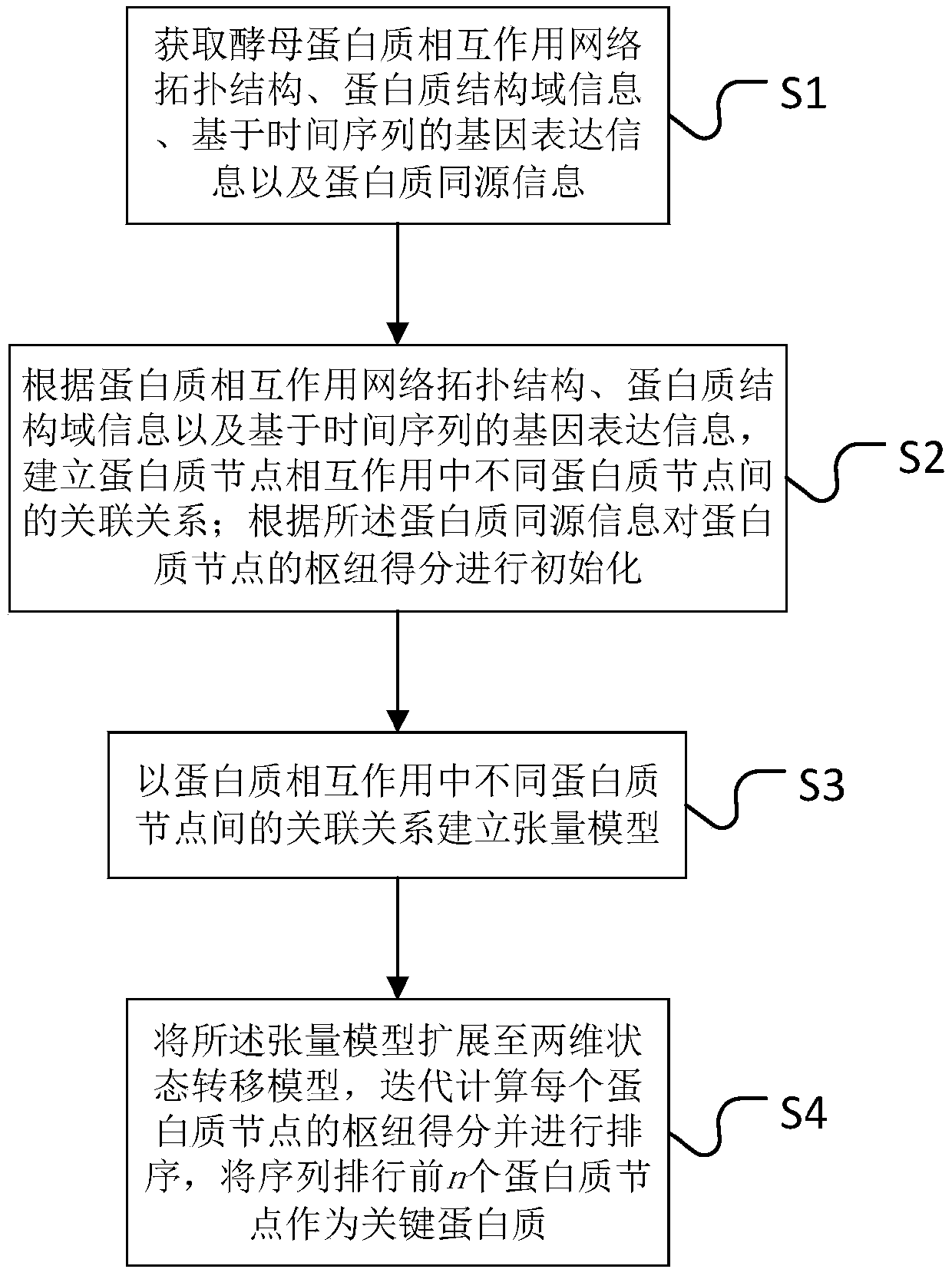
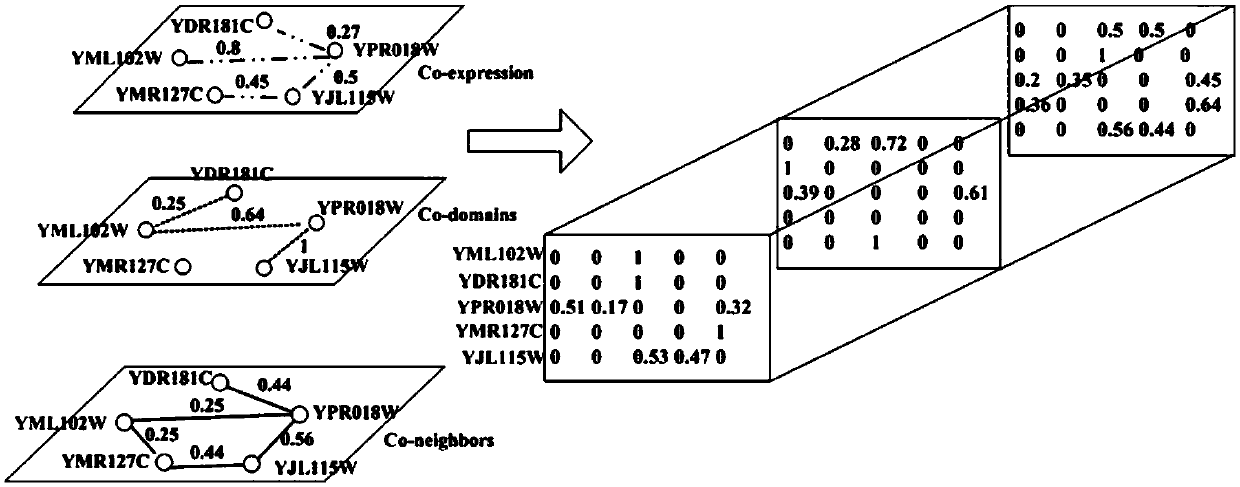
Key protein recognition method based on tensor random walking
The invention discloses a key protein recognition method based on tensor random walking. The method comprises the steps that a protein mutual impacting network topological structure, protein structural domain information, genetic expression information based on time sequence and protein homologous information are obtained; a correlation relationship of different protein nodes in the mutual impacting of protein nodes is constructed according to the above information; hub scores of protein nodes are initialized according to the protein homologous information; a tensor model is established according to the correlation relationship of different protein nodes in the protein mutual impacting; the hub scores of each protein nodes which are obtained by iterative computation based on the tensor model are sorted, and the top n protein nodes are taken as key proteins. The method has the advantages that the method is simple and effective, and compared with other methods, tests on multiple data sets show that the method has better prediction performance on key protein recognition.
Owner:CHANGSHA UNIVERSITY
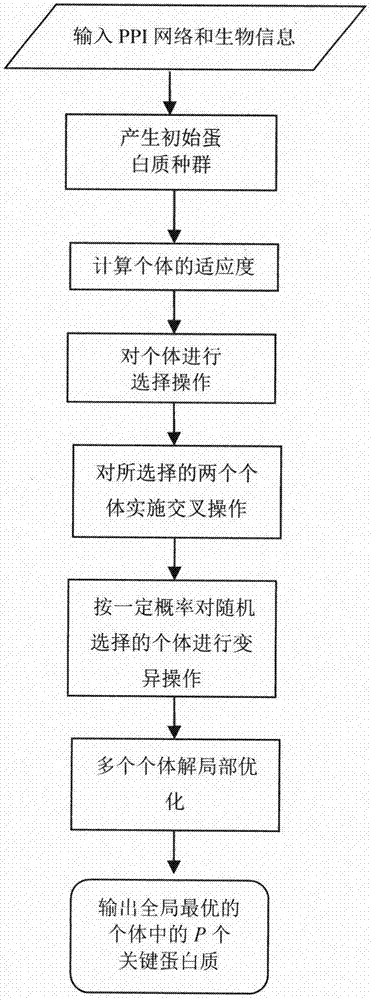
Method for identifying key protein based on genetic algorithm in PPI network
ActiveCN107092812APrediction is accurateImprove reliabilityProteomicsGenomicsAlgorithmMethod selection
The invention relates to an algorithm for identifying a key protein based on a genetic algorithm in a PPI network. The algorithm comprises the steps of generating an initial population in a protein interaction network; calculating the fitness of individuals; performing selection operation by a roulette wheel method; performing crossing operation and mutation operation among the randomly selected individuals; and performing local optimization on multiple individual solutions. According to the algorithm, the defects of existing methods are overcome; indexes are optimized and bio-information is fused, so that the reliability is higher and lots of unnecessary calculations are reduced; and the predicted key protein can be locally optimized, so that the efficiency of key protein identification is improved, and the application range and practicality of the technology in the field of the bio-information are expanded and improved.
Owner:YANGZHOU UNIV
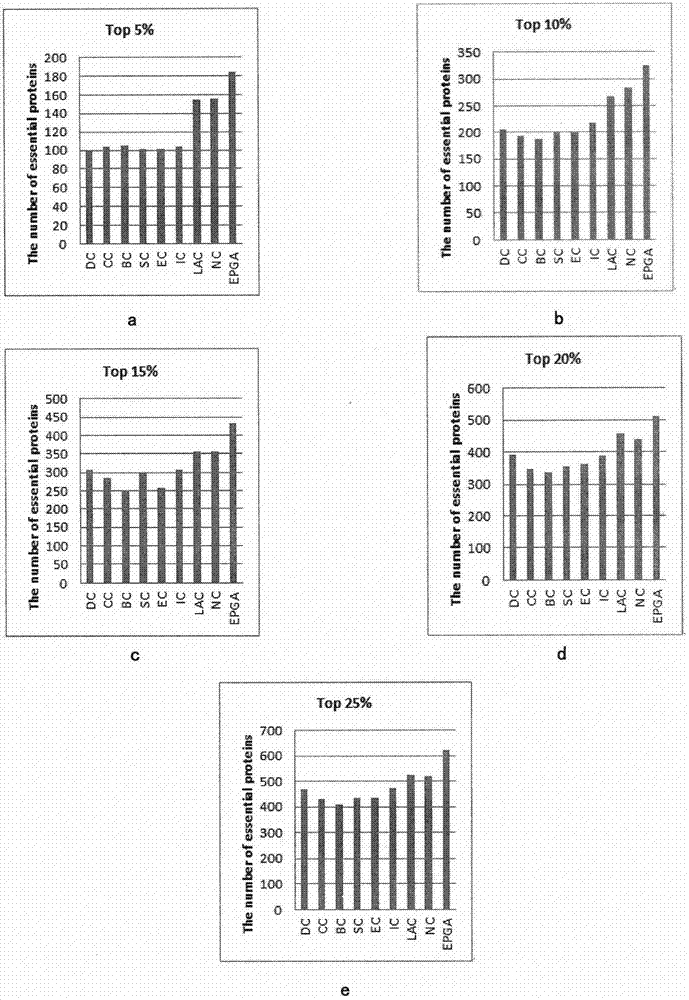
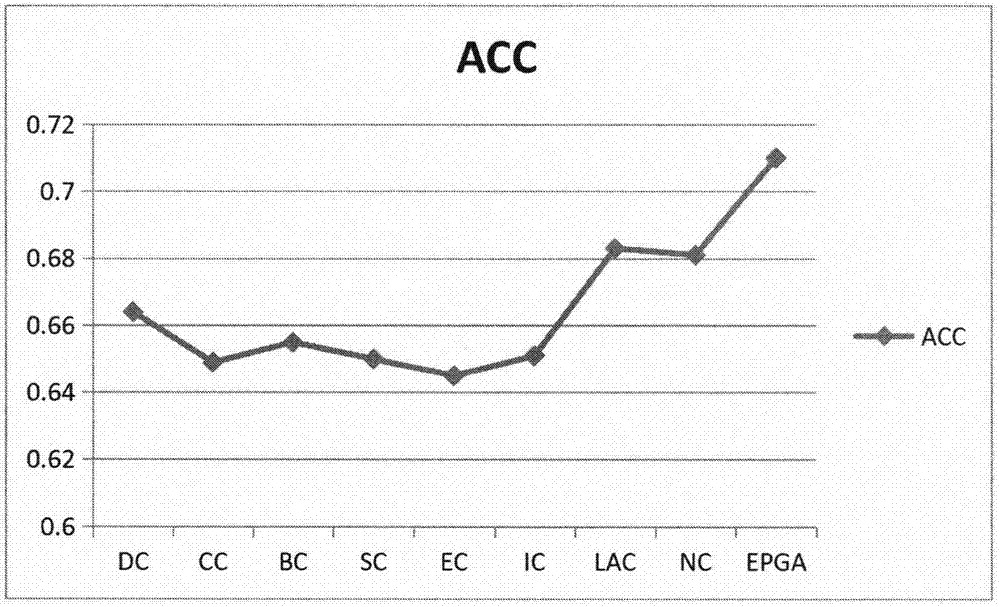
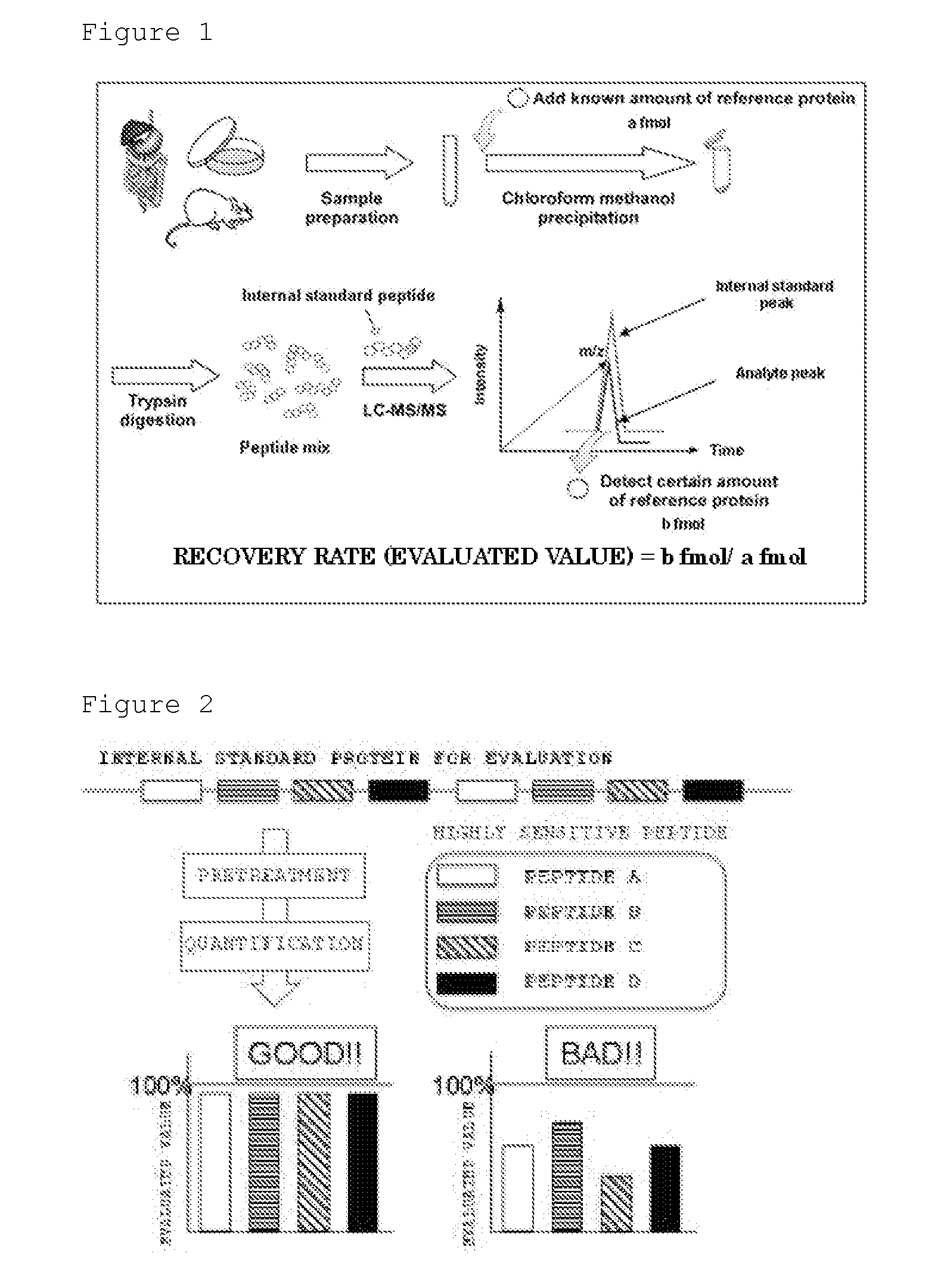
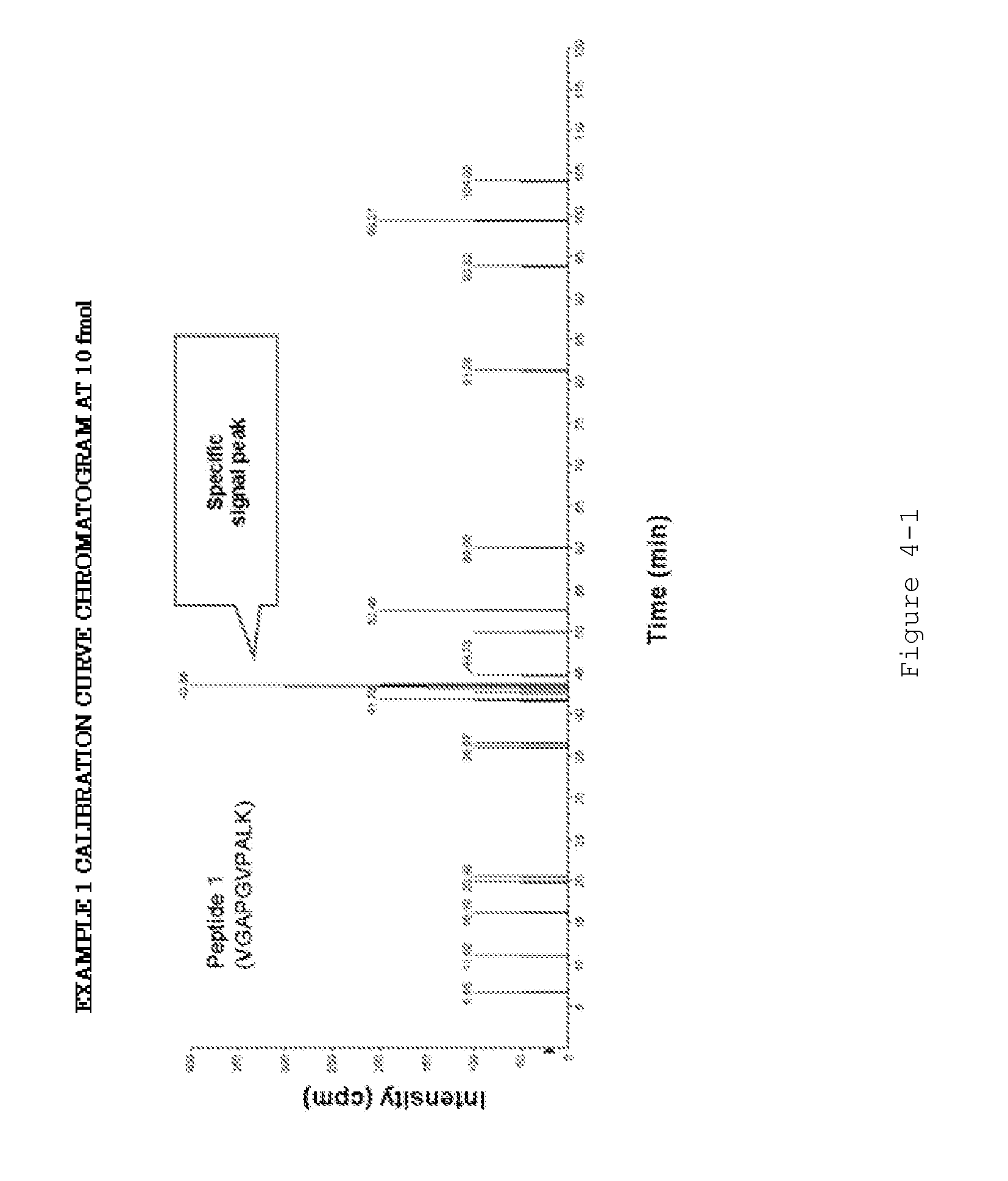
Evaluation Peptide For Use In Quantification Of Protein Using Mass Spectrometer, Artificial Standard Protein, And Method For Quantifying Protein
InactiveUS20120021446A1High general versatilityAddressing Insufficient SensitivityPeptide/protein ingredientsMicrobiological testing/measurementMass spectrometryEnzyme
Disclosed is an evaluation peptide for evaluating the efficiency of a pretreatment in the quantification of a protein using a mass spectrometer, having high reliability and high general versatility. Also disclosed is an artificial standard protein comprising the evaluation peptide. Further disclosed is a method for quantifying a protein utilizing the artificial standard protein. Specifically disclosed is a method for selecting a peptide which consists of an amino acid sequence not agreeing with that in a naturally occurring protein and a variant thereof and capable of being detected by mass spectrometry and which has an amino acid that can be recognized by a protein-digesting enzyme, and using the peptide as an evaluation peptide for use in the quantification of a protein by a mass spectrometer.
Owner:TOHOKU UNIV
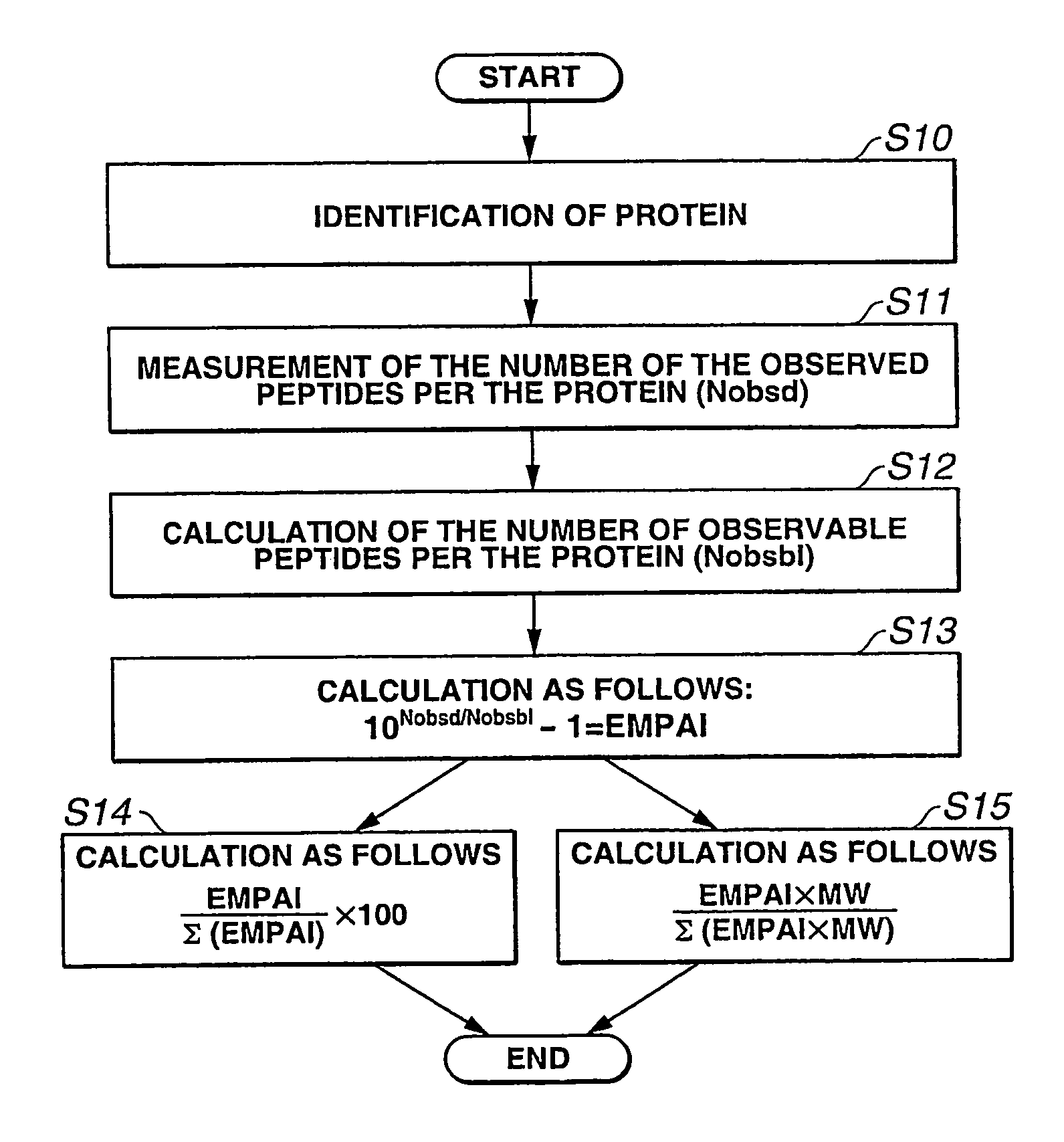
Absolute quantitation of protein contents based on exponentially modified protein abundance index by mass spectrometry
InactiveUS7910372B2Biological testingSpecial data processing applicationsCancer cellGene and protein expression
The present inventor has established protein abundance index (PAI, π) to determine the protein contents in a protein mixture solution using nanoLC-MSMS data. Digested peptides were analyzed by nanoLC-MS / MS and the obtained results were applied to a Mascot protein identification algorism based on tandem mass spectra. For absolute quantitation, PAI was converted to exponentially modified PAI (EMPAI, m π), which is proportional to protein contents in the protein mixture. EMPAI was successfully applied to comprehensive protein expression analysis and performed a comparison study between gene and protein expression in an HCT116 human cancer cells. Accordingly, the present invention provides a method and a computer program for quantifying the protein contents based on the protein abundance index.
Owner:EISIA R&D MANAGEMENT CO LTD
Method for identifying key protein by adopting improved flower pollination algorithm
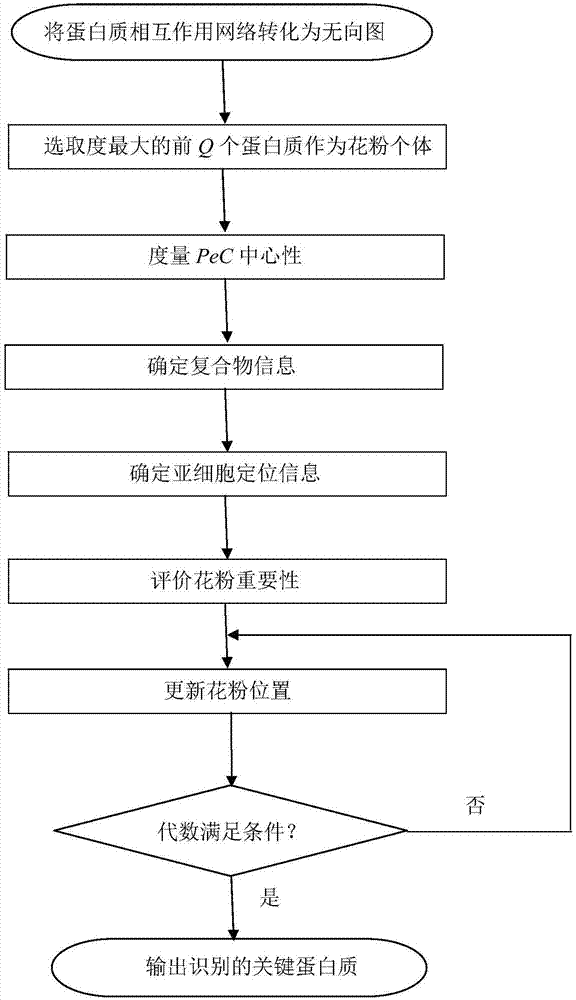
ActiveCN107885971AImprove accuracyUnderstanding Growth Regulatory ProcessesSystems biologySpecial data processing applicationsUndirected graphPollen
The invention discloses a method for identifying a key protein by adopting an improved flower pollination algorithm. A protein-protein interaction network is converted into an undirected graph, previous Q proteins having the largest choice degree serve as pollen individuals, the PeC centrality is measured, compound information is determined, subcellular localization information is determined, pollen significance is evaluated, a pollen position is updated, and an identified key protein is output. The topological attributes of the protein network are considered and the biological characteristicsof the protein network are fused during evaluation of the pollen significance, and the key protein can be accurately identified. A simulation experiment result shows that the index properties such asaccuracy, specificity and sensitivity are more excellent. Compared with other key protein identifying methods, the key protein identifying process is achieved by combining with the topological attributes and biological characteristics of the protein network, and the key protein identifying accuracy is improved.
Owner:SHAANXI NORMAL UNIV
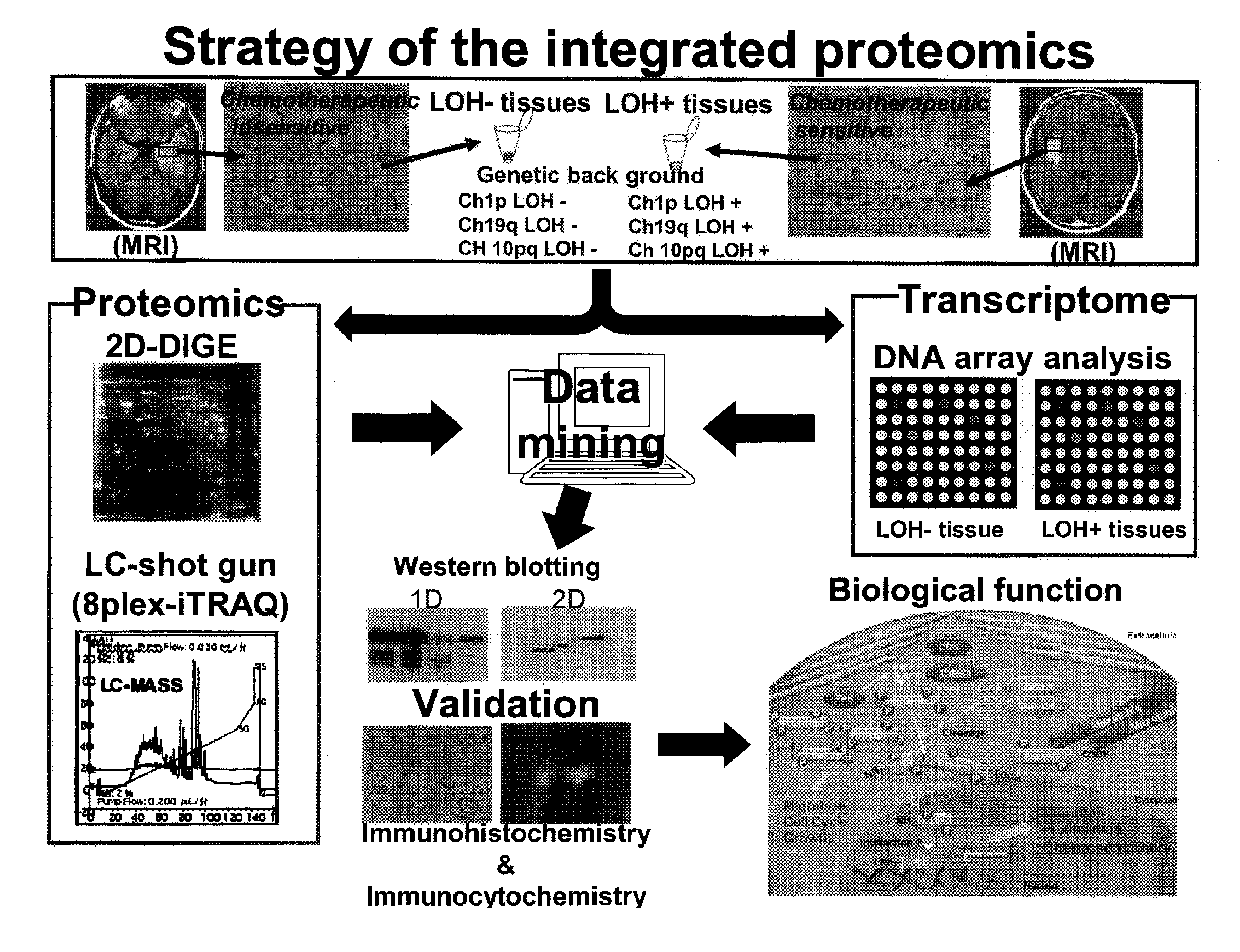
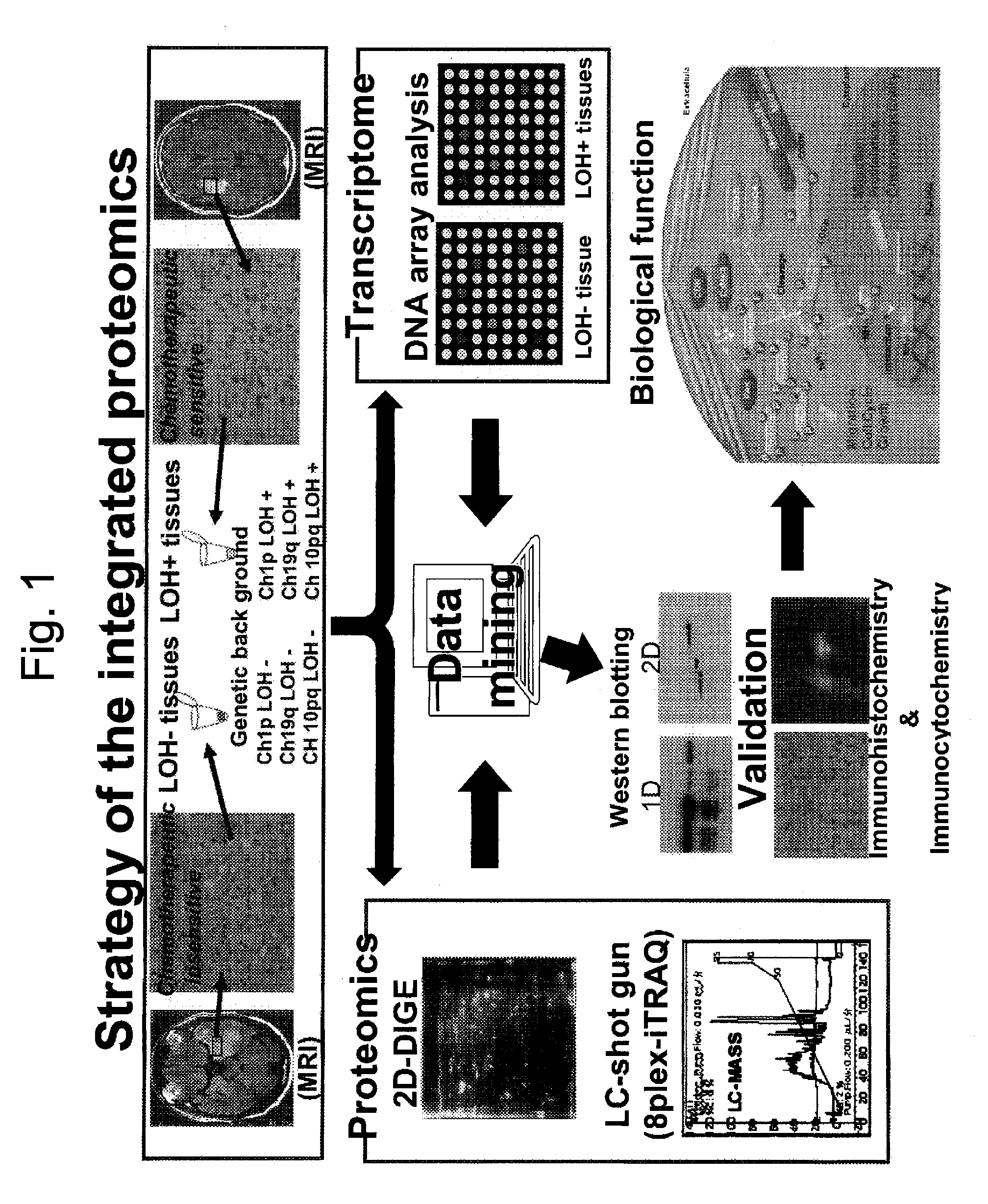
Method for generating data set for integrated proteomics, integrated proteomics method using data set for integrated proteomics that is generated by the generation method, and method for identifying causative substance using same
InactiveUS20130023574A1Easy to analyzeEffective applicationBiocideOrganic active ingredientsProteomics methodsSubstance use
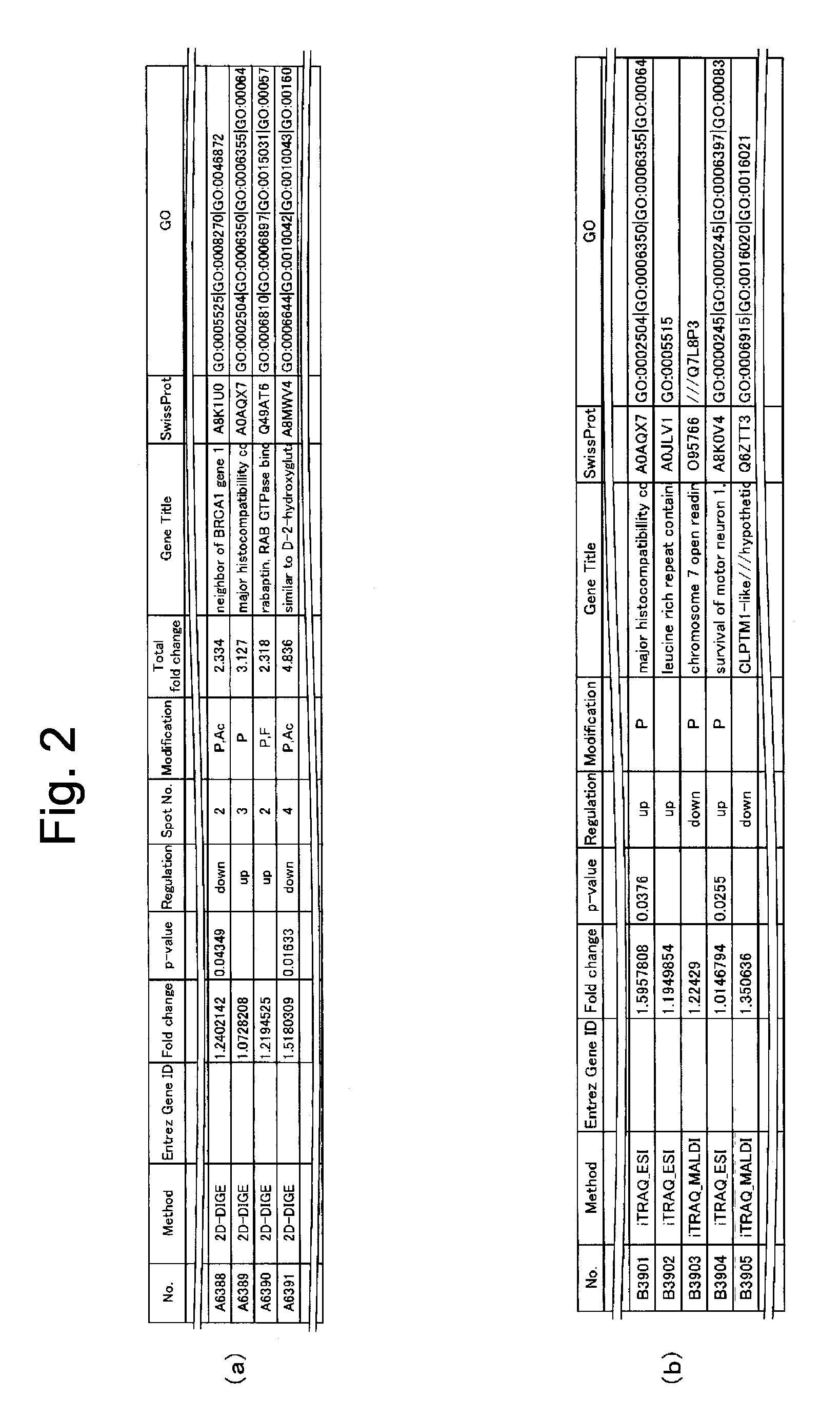
Provided are a method for generating a data set for integrated proteomics analysis, whereby expression level variations of both of proteins and genes can be integrally united together and, moreover, highly accurate and appropriate analysis results can be obtained compared with the existing cases where the expression variation amount of proteins or genes is singly analyzed, an integrated proteomics analysis method, a method for identifying a protein causative of a disease or the like using these methods, and a method of using the same???. The aforesaid method for generating a data set for integrated proteomics analyses comprises: a protein identity number-assigning step for assigning common identity numbers to the expression variation amount data of individual proteins; a gene identity number-assigning step for assigning common identity numbers to the expression variation amount data of individual genes; a data-binding step for binding together the set of the expression variation amount data; a data-rejecting step for rejecting, among the individual expression variation amount data constituting the thus bound data set, data showing a p-value equal to or greater than a specific level; and a data-selecting step for selecting one data, from a set of data having the same common identity number assigned thereto, on the basis of a definite requirement to thereby generate the data set to be subjected to integrated proteomics analyses. Further, the data set for integrated proteomics analyses thus generated is subjected to GO analysis and network analysis to thereby identify a protein causative of a disease, a pathological condition or the like. Furthermore, the causative protein thus identified is usable, for example, a tumor marker or a clinical target.
Owner:NAT UNIV CORP KUMAMOTO UNIV
Fabrication method for protein electrochemical imprint sensor of modified magnetic electrode by one-step deposition method
ActiveCN109085225AEasy to makeReduce consumptionMaterial analysis by electric/magnetic meansProtein targetProtein molecules
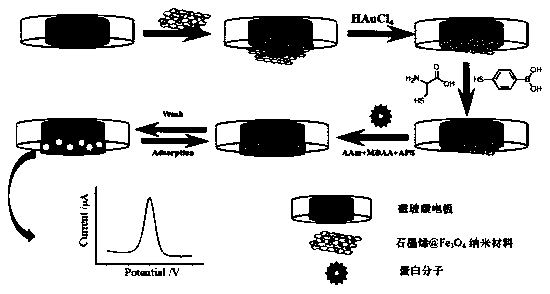
The invention discloses a fabrication method for a protein electrochemical imprint sensor of a modified magnetic electrode by a one-step deposition method, and relates to the technical field of electrical analysis chemistry and protein recognition sensors. The fabrication method comprises the steps of 1) preparing a graphene@Fe3O4 nanometer composite material; 2) fabricating a graphene@Fe3O4@Au modified magnetic electrode by the one-step electrodeposition method; 3) introducing boric acid, amino and carboxyl active group into a surface of the modified magnetic electrode so that a plurality ofrecognition sites are provided for protein imprint; 4) pre-assembling protein; 5) performing electrochemical polymerization on a protein imprint membrane; and 6) washing template protein with an eluent so as to build the protein molecular imprint electrochemical sensor. The magnetic modified electrode fabricated by the method is beneficial for introducing the active group, increasing the recognition sites, improving selectivity, expanding surface area and improving conductivity of electrode surface, the built protein imprint electrochemical sensor is high in sensitivity and can be used repeatedly, and target protein can be specially recognized.
Owner:MINJIANG UNIV
Method for knocking out fungus genes
InactiveCN105331628AImprove stabilityAvoiding SCRaMbLE EffectsMicroorganism based processesVector-based foreign material introductionBiotechnologyFungal genome
The invention belongs to the technical field of gene engineering and discloses a method for knocking out fungus genes. The method includes: respectively introducing nucleotide sequences identified by Vika recombinase into the upstream and downstream of target genes in fungus genomes; identifying protein with Vika recombinase activity, shearing the nucleotide sequences identified by the Vika recombinase, and knocking out the target genes to obtain fungi with the target genes being knocked out. The method has the advantages that a Vika-vox recombinase system is successfully applied to fungus gene knocking out, knocking out of the target genes is achieved, a new method is provided for fungus gene knocking out, and function researches and performance optimization of fungus genes are benefited.
Owner:TIANJIN UNIV
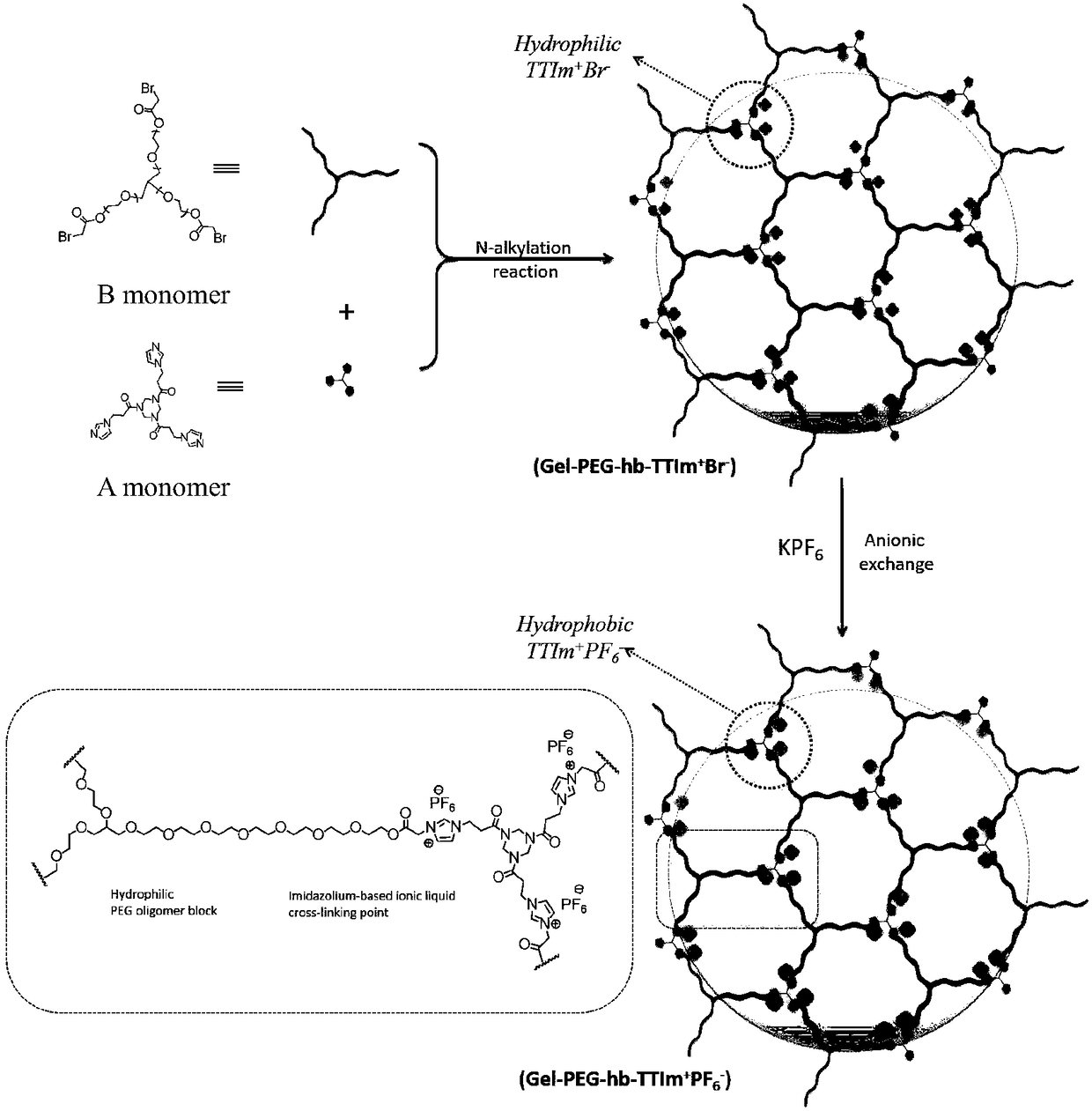
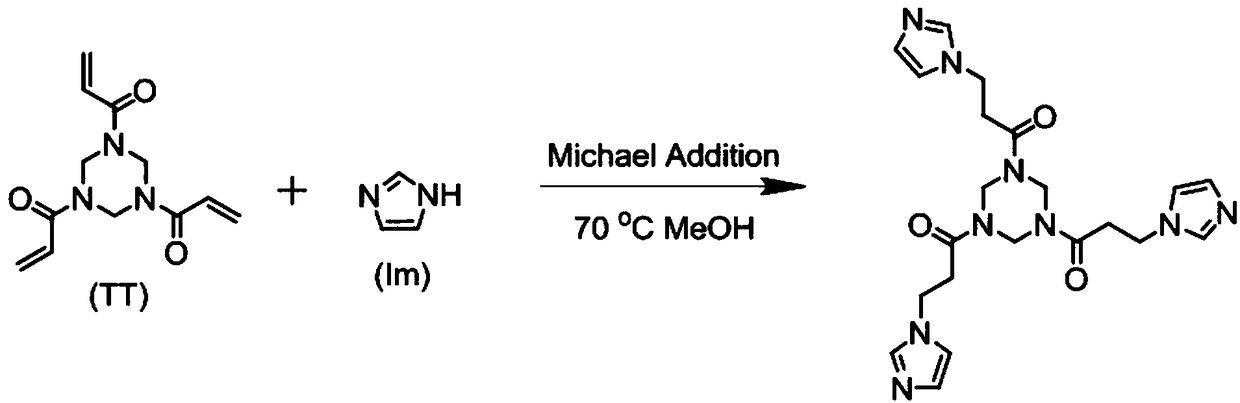
Polyionic liquid gel with nanopores and method thereof
InactiveCN108484900ASolve the problem of no network cross-linked structure and poor adsorption capacityIncrease contactOrganic chemistryOther chemical processesCrystallographyMolecular imprinting
The invention discloses a polyionic liquid gel with nanopores and a method thereof. The polyionic liquid gel has a monomer structure shown as a formula (1), wherein in the formula (1), R is an N heterocyclic ring and comprises one of FORMULAE, X comprises Br-, Cl- or FORMULA and n is 6-30. The polyionic liquid gel has nanopores, comprises a three-dimensional hyperbranched topological structure, shows excellent adsorption capacity, and has a huge application potential in the field of dyes, heavy metal ion adsorption, controlled medicine release, molecular imprinting, protein recognition and thelike.
Owner:XIJING UNIV
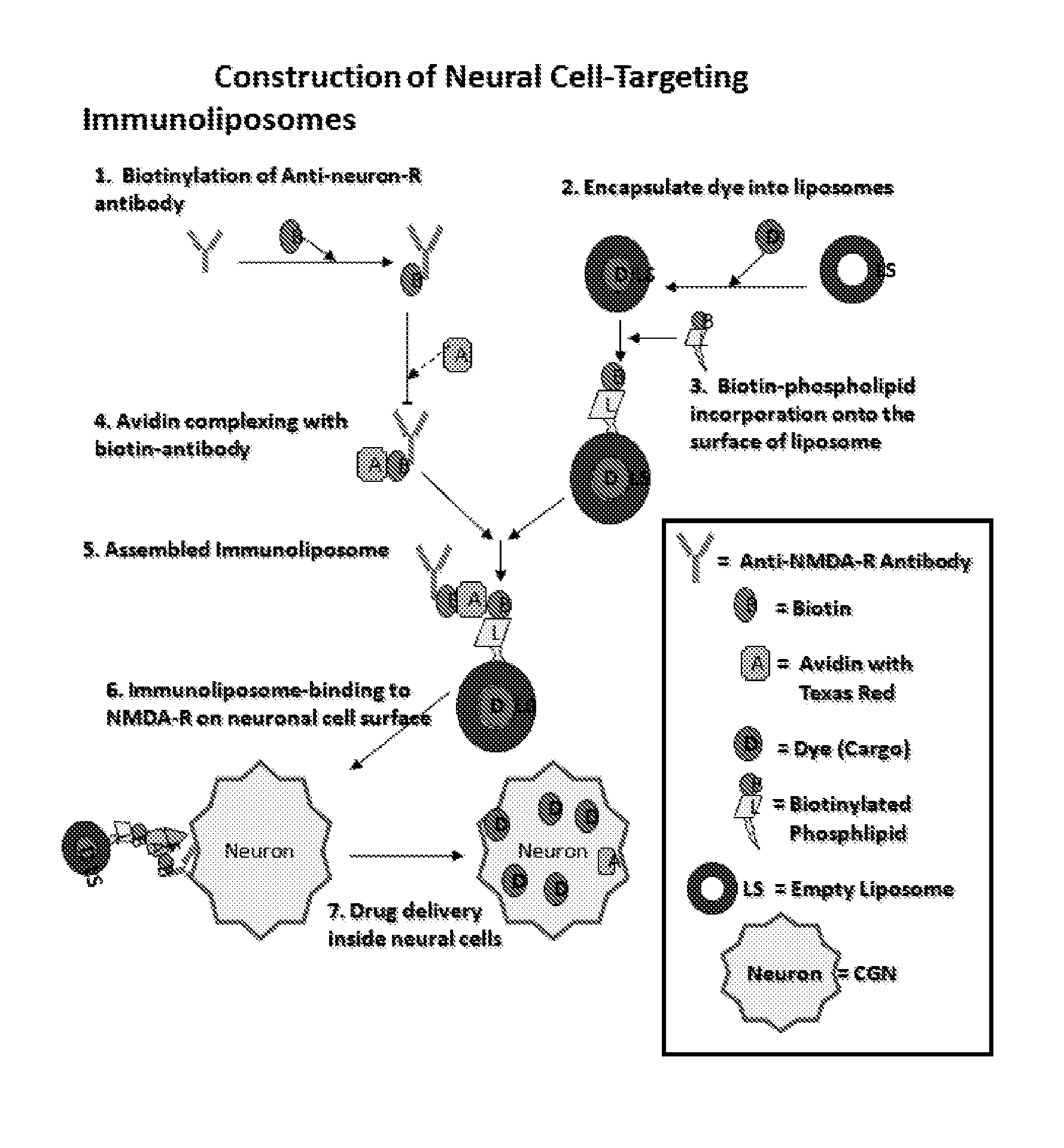
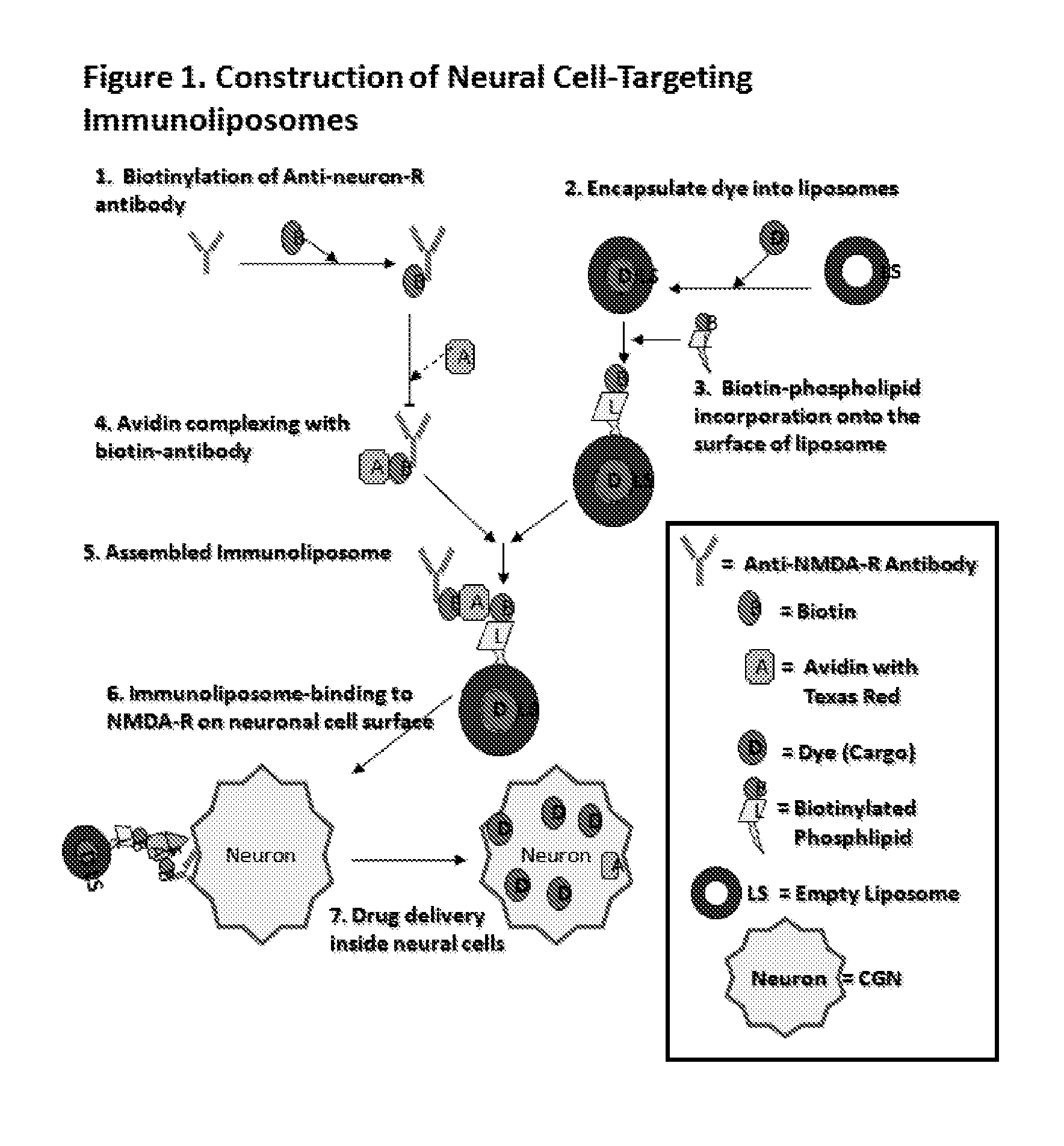
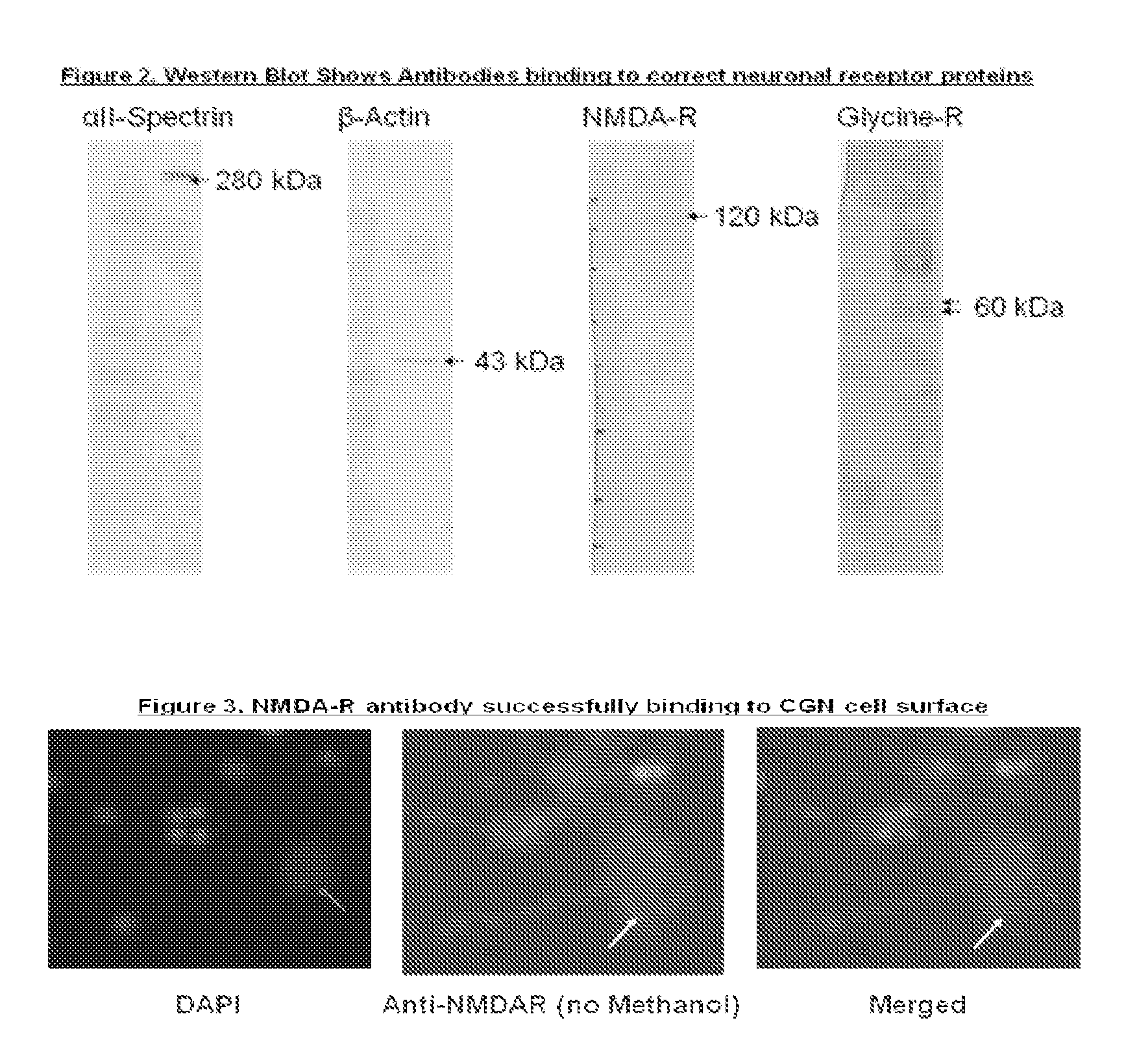
Antibody bound synthetic vesicle containing molecules for deliver to central and peripheral nervous system cells
A process is provided of delivering at least one active agent cargo molecule into an neuronal cell whereby a cargo molecule is placed within a synthetic vesicle such as a liposome and a biotinylated protein such as an antibody is bound to the synthetic vesicle to form a protein bound synthetic vesicle whereby the protein recognizes a receptor expressed on the surface of a neuronal cell, and exposing the protein bound synthetic vesicle to the cell until the cargo molecule is delivered into the neuronal cell. Numerous cargo molecules are delivered by the inventive synthetic vesicle including a calpain inhibitor and a caspase inhibitor. The protein illustratively targets a cellular receptor for a ligand such as glutamate, glycine, dopamine, nicotine, muscarine, acetylcholine, or serotonin, and the like.
Owner:BANYAN BIOMARKERS INC
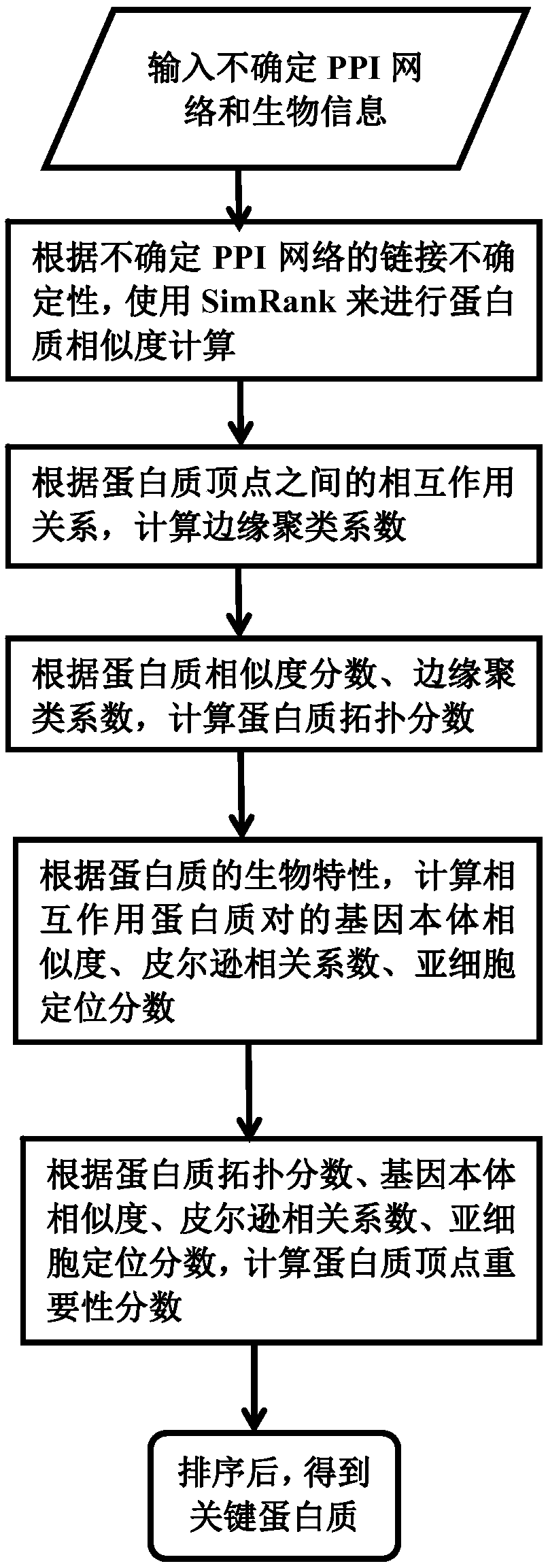
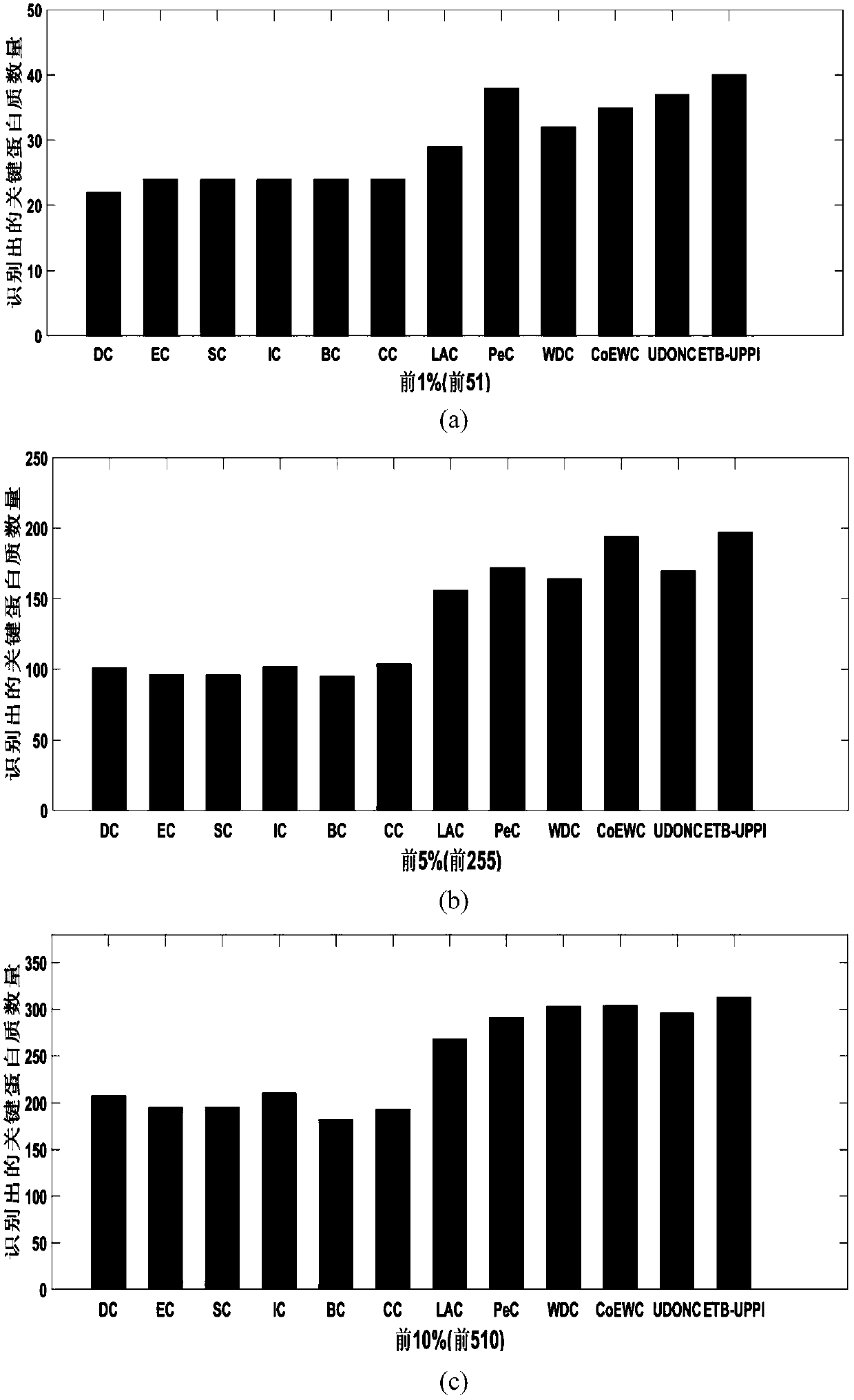
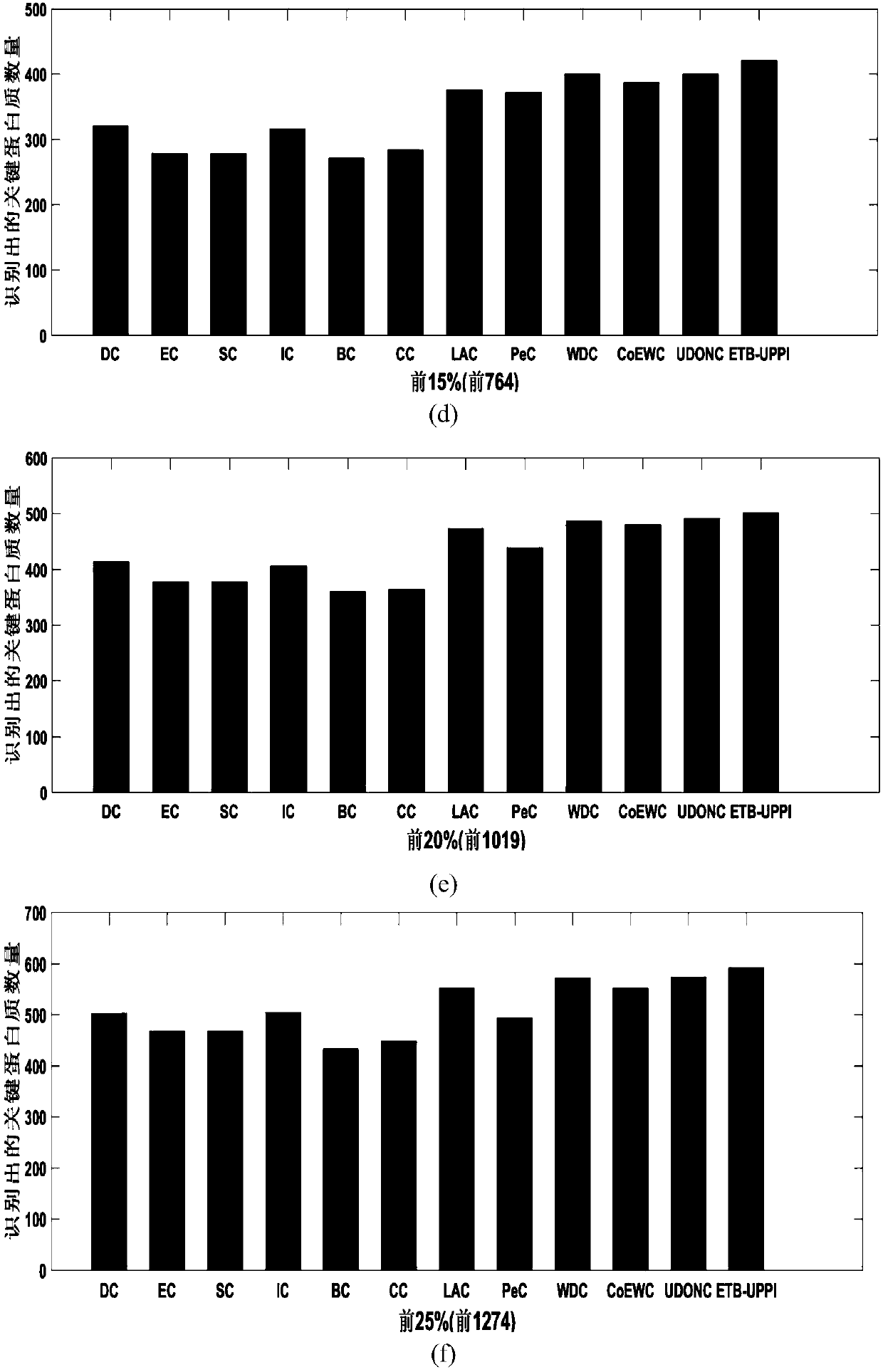
A key protein identification method in an uncertain protein interaction network
PendingCN109686403AImprove accuracyAvoid negative effectsCharacter and pattern recognitionProteomicsProtein recognitionProtein identification
SimRank is used for carrying out protein similarity calculation based on a key protein identification method in an uncertain protein interaction network. A SimRank calculation problem in an uncertainnetwork is converted into SimRank calculation in a deterministic network; and then, considering the topological characteristics of the protein interaction network and the biological characteristics ofthe protein, and calculating an edge clustering coefficient, gene ontology similarity, a Pearson's correlation coefficient and a subcellular localization score to obtain an importance score. And finally, ranking the scores from large to small, and outputting the first k proteins corresponding to the scores as final results. On the basis of an uncertain interaction network, the accuracy of key protein recognition is improved by fusing biological attributes and topological characteristics, meanwhile, the prediction result is more accurate, the prediction efficiency is improved, the applicationrange of the technology in the field of biological information is widened, and the practicability of the technology in the field of biological information is improved.
Owner:YANGZHOU UNIV
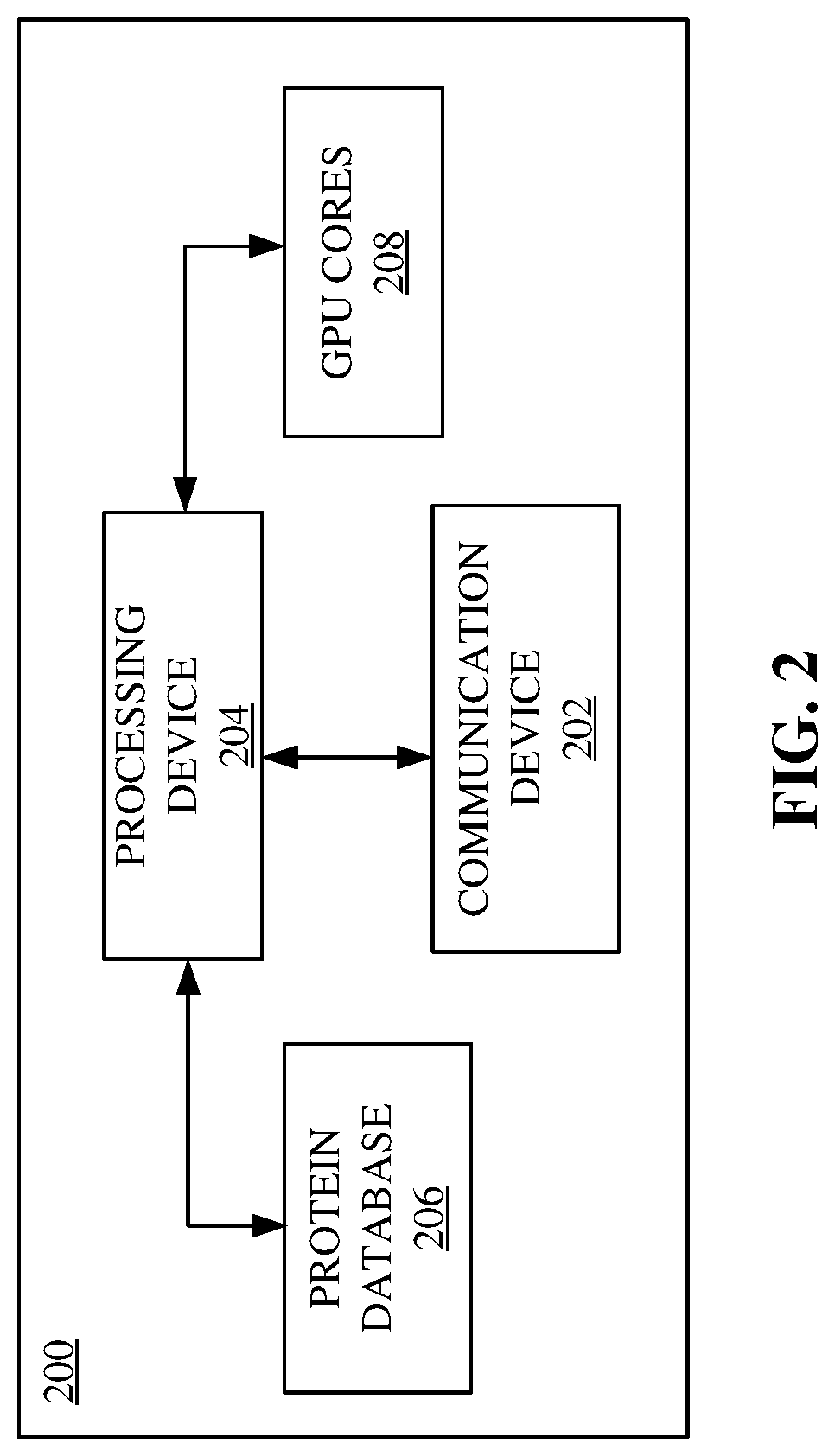
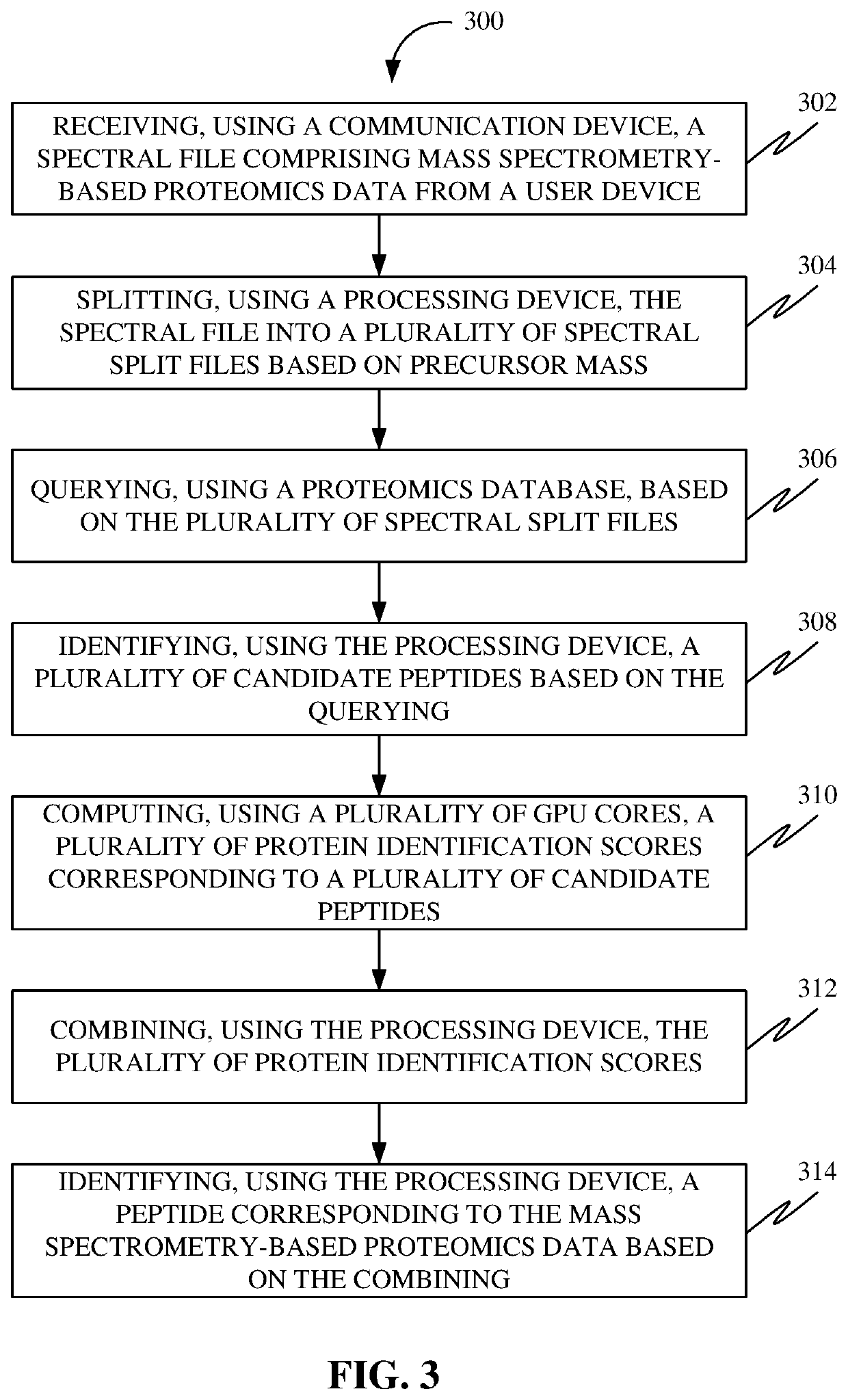
Methods, systems, apparatuses and devices for accelerating execution of a search query for peptide identification
A system for accelerating execution of a search query for peptide identification is disclosed. The system may include a communication device configured for receiving a spectral file including mass spectrometry-based proteomics data from a user device. Further, the system may include a processing device configured for splitting the spectral file into spectral split files based on precursor mass, identifying candidate peptides based on querying, combining protein identification scores, and identifying a peptide corresponding to the mass spectrometry-based proteomics data based on the combining. Further, the system may include a protein database configured for querying based on the plurality of spectral split files. Further, the system may include a plurality of GPU cores communicatively coupled to the processing device configured for computing the plurality of protein identification scores corresponding to candidate peptides.
Owner:BRUKER SCI LLC
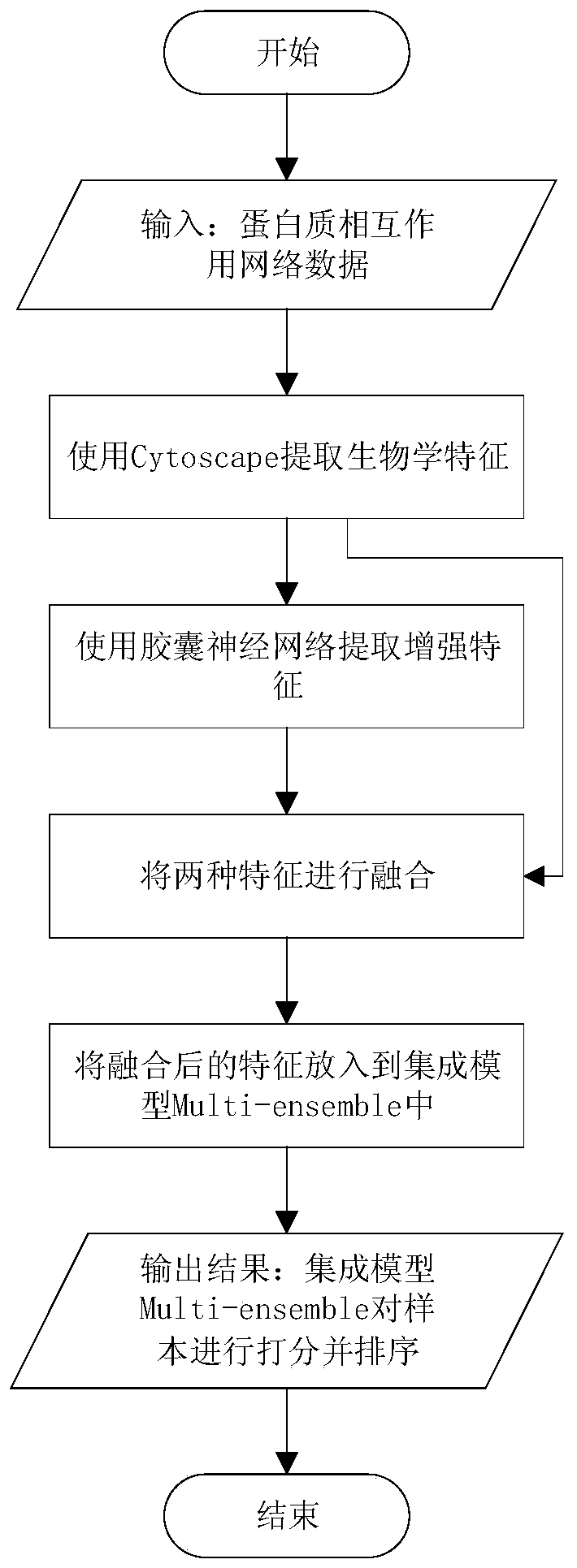
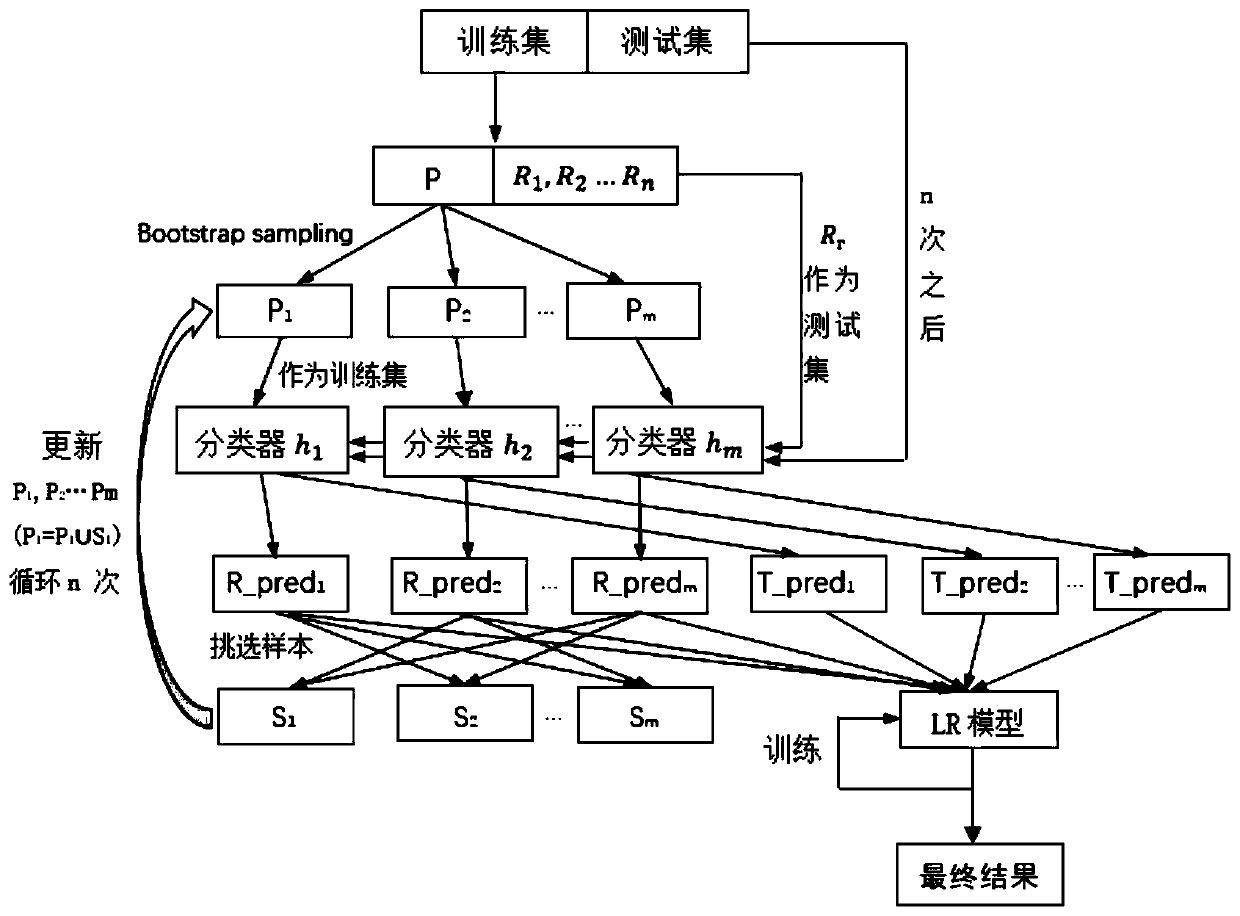
Key protein identification method based on capsule neural network and ensemble learning
ActiveCN111584010AImprove accuracyEnsemble learningBiostatisticsProtein Interaction NetworksEnsemble learning
The invention discloses a key protein identification method based on a capsule neural network and ensemble learning. The key protein identification method comprises the following steps: 1, extractingeight biological characteristics of protein in a protein interaction network by utilizing a Cytoscape tool; 2, extracting deep enhanced features of the eight biological characteristics by using a capsule neural network; 3, connecting the biological characteristics with the protein enhancement characteristics; 4, putting the connected characteristics obtained in the step 3 into an integrated modelMulti-enseable, training the model, and predicting a new key protein by using the trained integrated model; 5, outputting a result. Compared with the initial biological characteristics, the enhanced characteristics extracted through the capsule neural network can improve the accuracy of predicting the key protein by some machine learning models, and the accuracy of predicting the key protein by the machine learning model can be further improved by fusing the initial biological characteristics and the enhanced characteristics.
Owner:KUNMING UNIV OF SCI & TECH
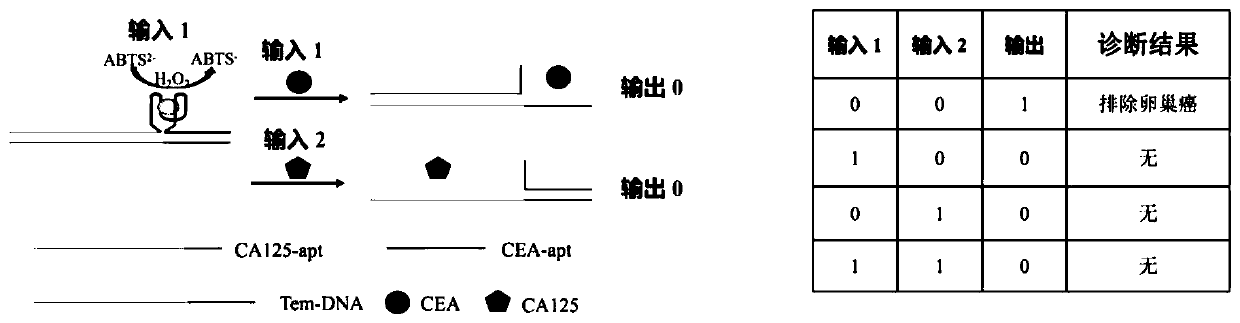
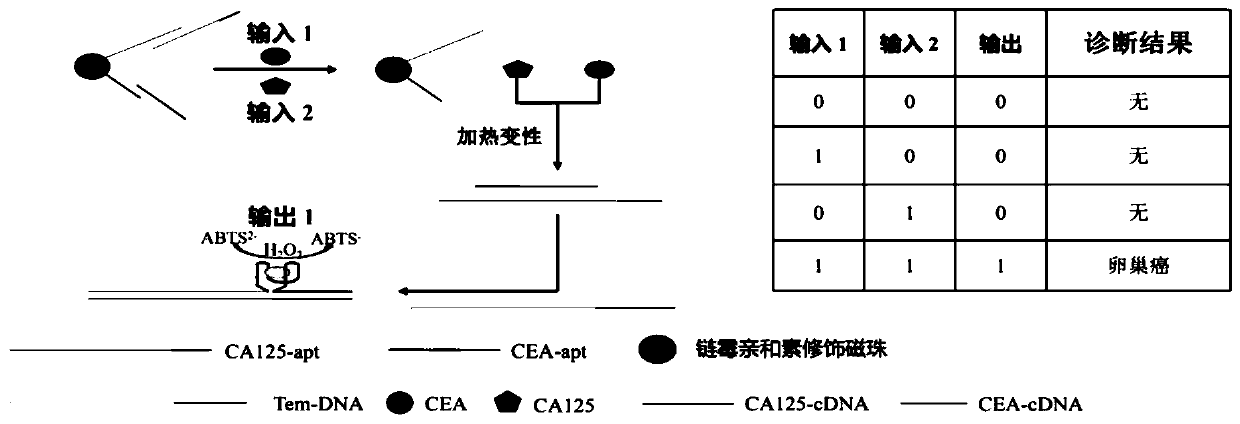
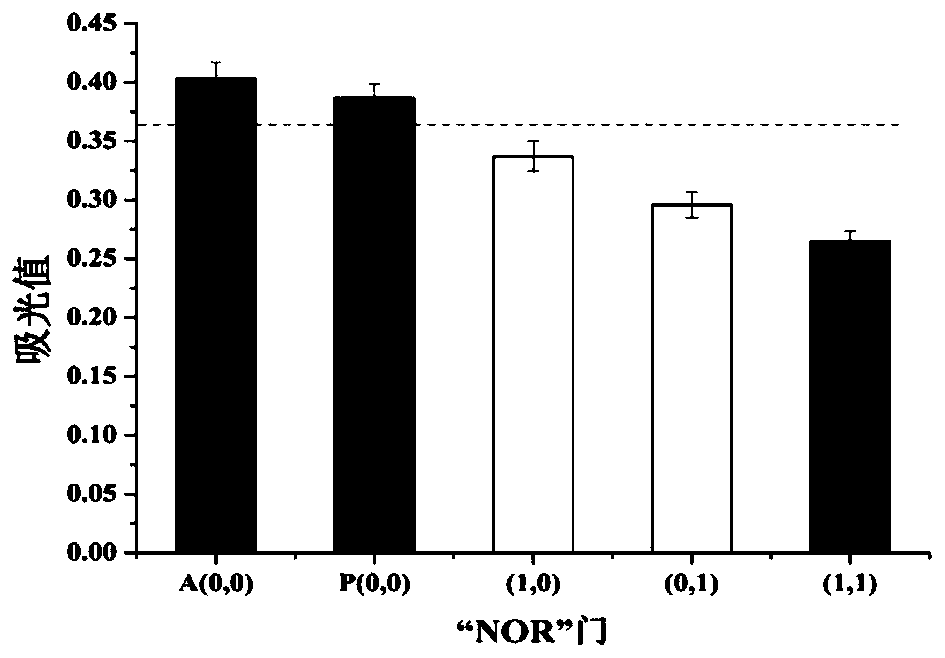
Method for ovarian cancer screening based on ovarian cancer marker and logic gate operation
ActiveCN110426519AHigh sensitivityStrong specificityColor/spectral properties measurementsAptamerObject based
The invention discloses a method for ovarian cancer screening based on an ovarian cancer marker and a logic gate operation, and belongs to the technical field of biomedicine. The protein-nucleic acidrecognition technology, the signal amplification technology and a logic gate are combined, aptamers of CEA and CA125 are used for target recognition, signal transformation and amplification are performed through a G-quadruplex-heme complex, and a method for simultaneous detection of tumor markers is established. According to the method, an optical biosensor with high sensitivity and strong specificity is constructed for achieving the joint detection of the CEA and the CA125, and an ovarian cancer screening result is provided based on the logic gate; through the combination of optical detectionand the signal amplification technology, the detection sensitivity is improved; the universality of the detection principle makes the method also suitable for detection of different objects based ondifferent protein recognition elements.
Owner:JIANGNAN UNIV
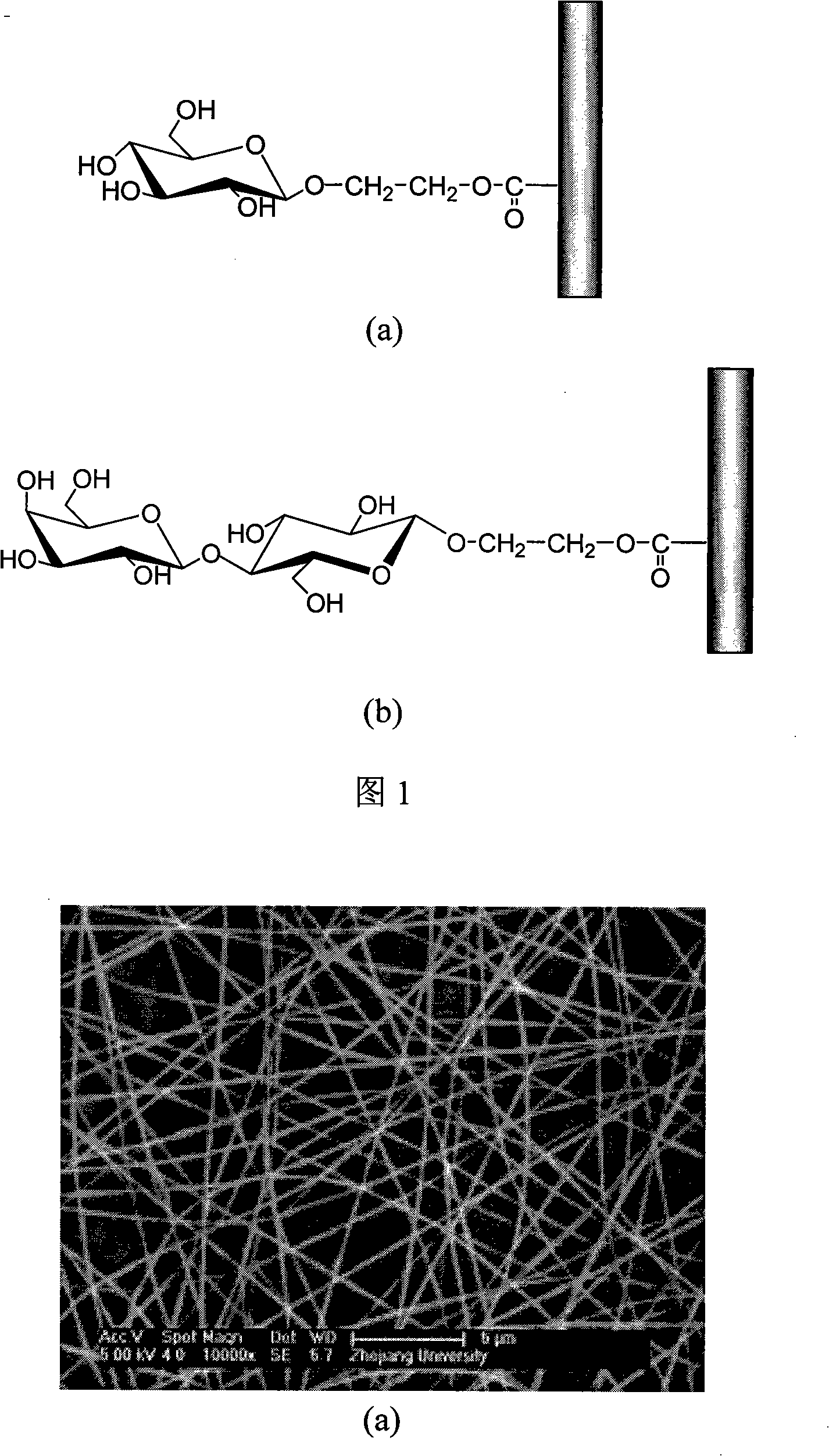
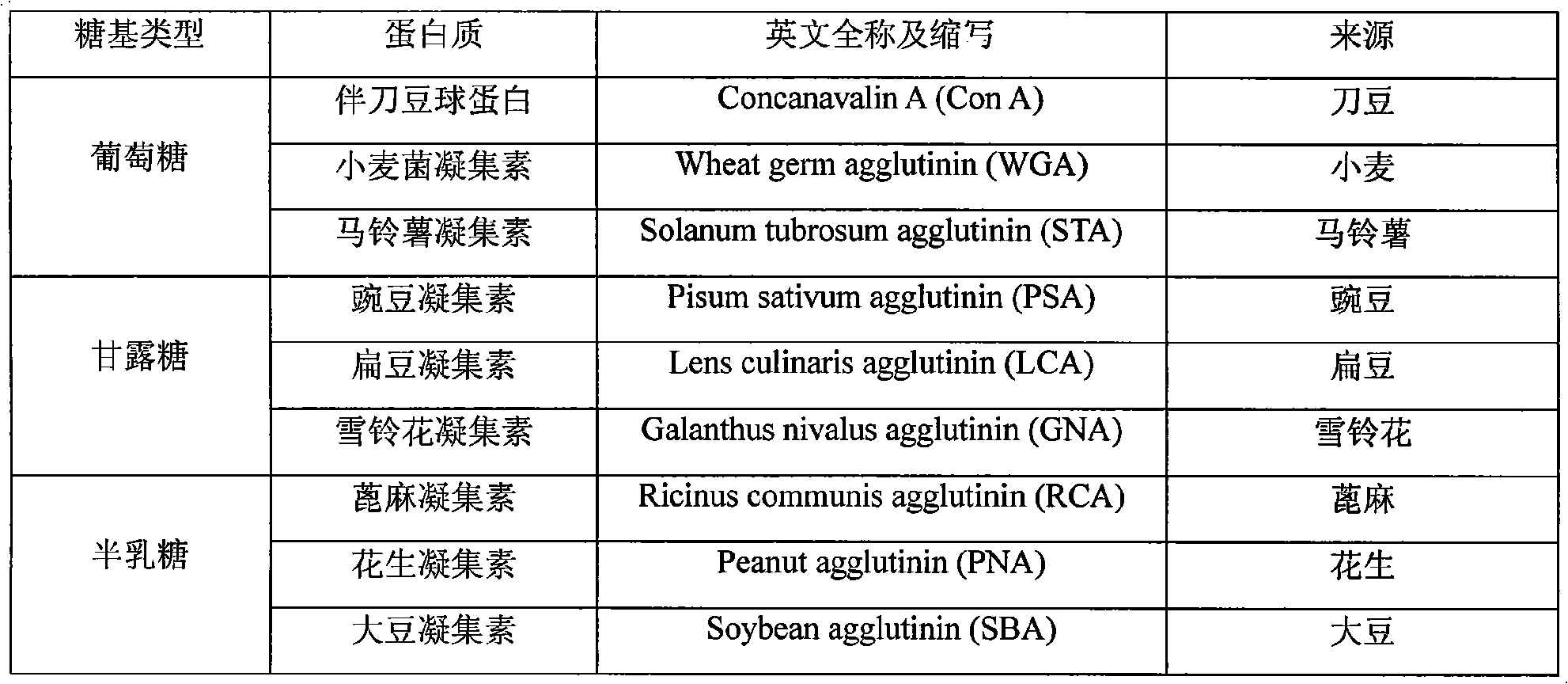
Preparation method of glycosylated nano-fibre for identifying proteinicsubstance and applications
InactiveCN101285221AEasy to makeHigh surface areaFilament/thread formingPeptide preparation methodsProtein solutionSodium methoxide
The invention discloses a method for preparing glycosylation nanometer fiber for protein recognition and applying the same. The method comprises the following steps that: acrylonitrile group copolymer is used as a raw material; acrylonitrile group copolymer nanometer fiber with controllable diameter is prepared by means of electrostatic spinning; sugar monomer which is protected by acetyl is fixed on the surface of nanometer fiber under the catalysis of boron trifluoride etherate; acetyl group is stripped down with methanol solution of sodium methoxide; and affinity nanometer fiber for glycosylation is obtained. With high specific surface area of the nanometer fiber, high porosity and a special biological function of glycosyl, the glycosylation nanometer fiber with a high immobilization rate can be immersed directly into protein solution, or the glycosylation nanometer fiber is superposed in multilayer to arrange in a membrane exchange filter, which is used for separation and purification of the protein. The glycosylation nanometer fiber for protein recognition has the advantages of simple preparation method and, repeatable use for many times and is favorable for industrialization production and so on.
Owner:ZHEJIANG UNIV
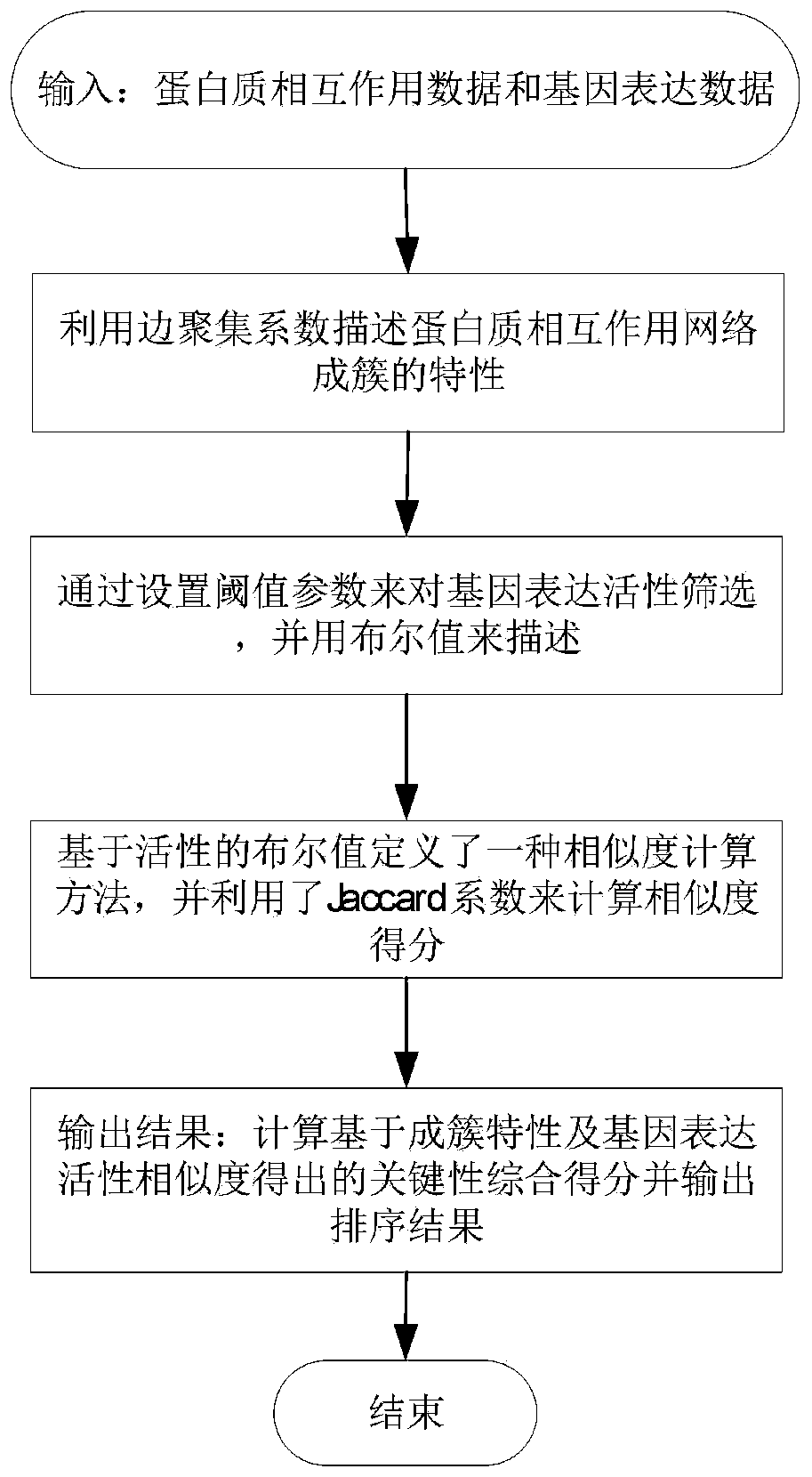
Key protein identification method based on protein clustering characteristics and activity co-expression
ActiveCN111128292AImprove accuracyExcellent featuresSystems biologyInstrumentsProtein Interaction NetworksProtein cluster
The invention relates to a key protein identification method based on protein clustering characteristics and activity co-expression. The method comprises the following specific steps: describing the clustering characteristics of a protein interaction network by utilizing an edge aggregation coefficient; setting gene activity expression by setting a threshold parameter, and describing by adopting aBoolean value; defining a calculation method based on the Boolean value of gene activity expression, and calculating the score of activity co-expression by utilizing a Jaccard coefficient; finally obtaining the key comprehensive scores based on the protein clustering characteristics and the activity co-expression, outputting a sorting result, and using the protein with the high key comprehensivescore after top sorting (top N is taken as a threshold value) as a key protein. According to the key protein identification method, the influence of gene expression data noise is eliminated, and the key protein identification method is superior to a centrality measurement method and a key protein prediction method with the same input data set in the aspects of identification accuracy, specificity,sensitivity and the like.
Owner:HUNAN NORMAL UNIVERSITY
A method for predicting association relationship between metabolites and diseases based on a KATZ model
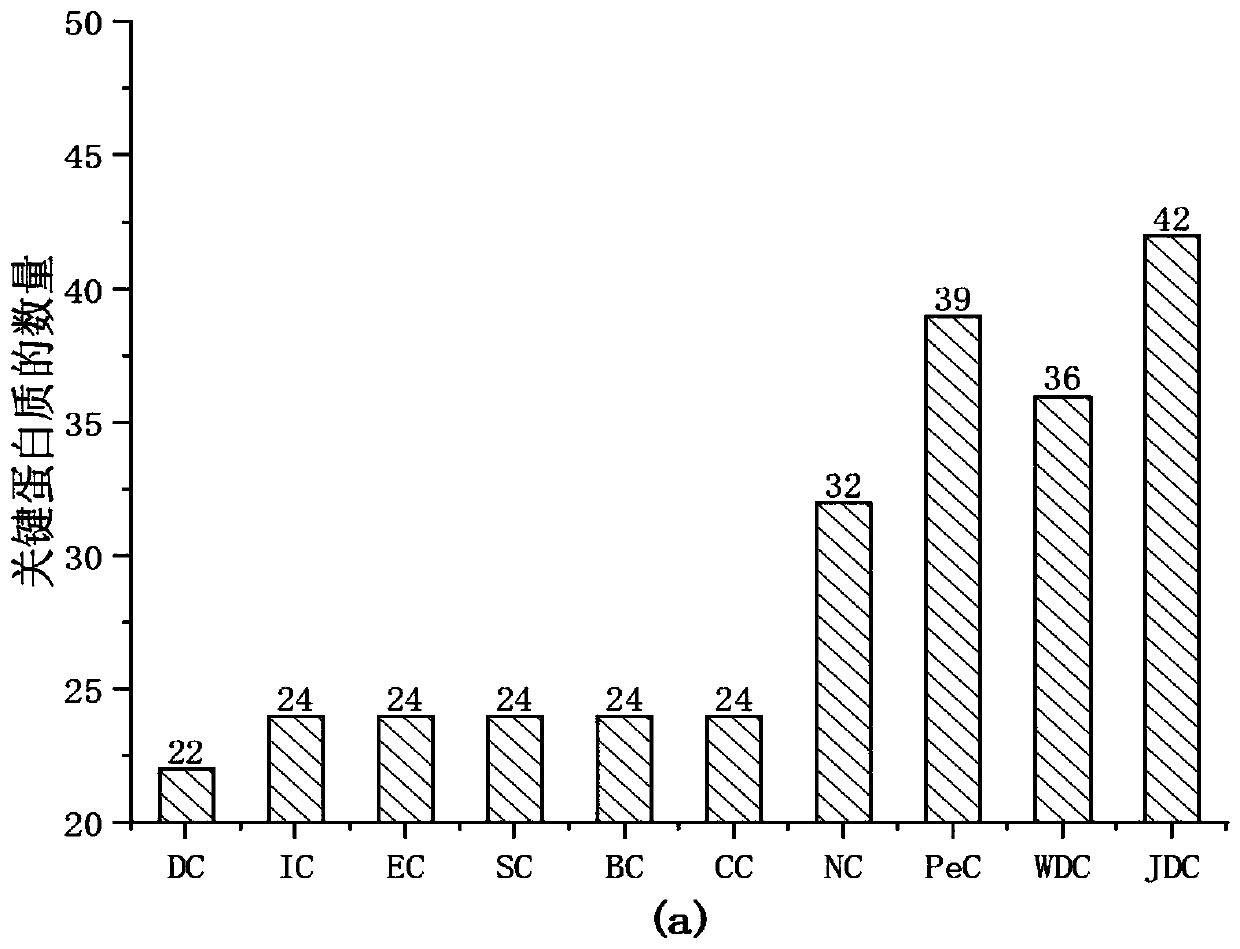
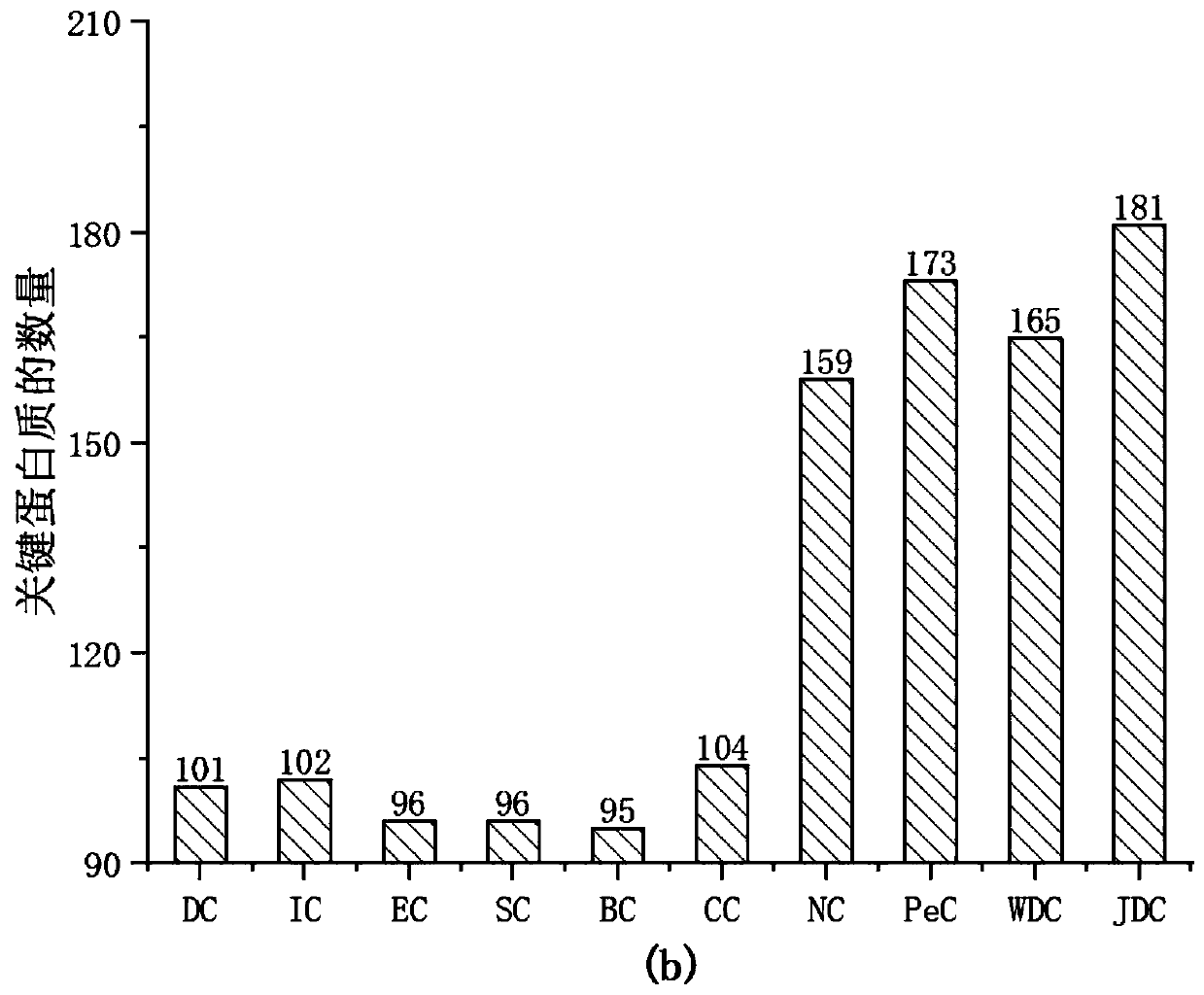
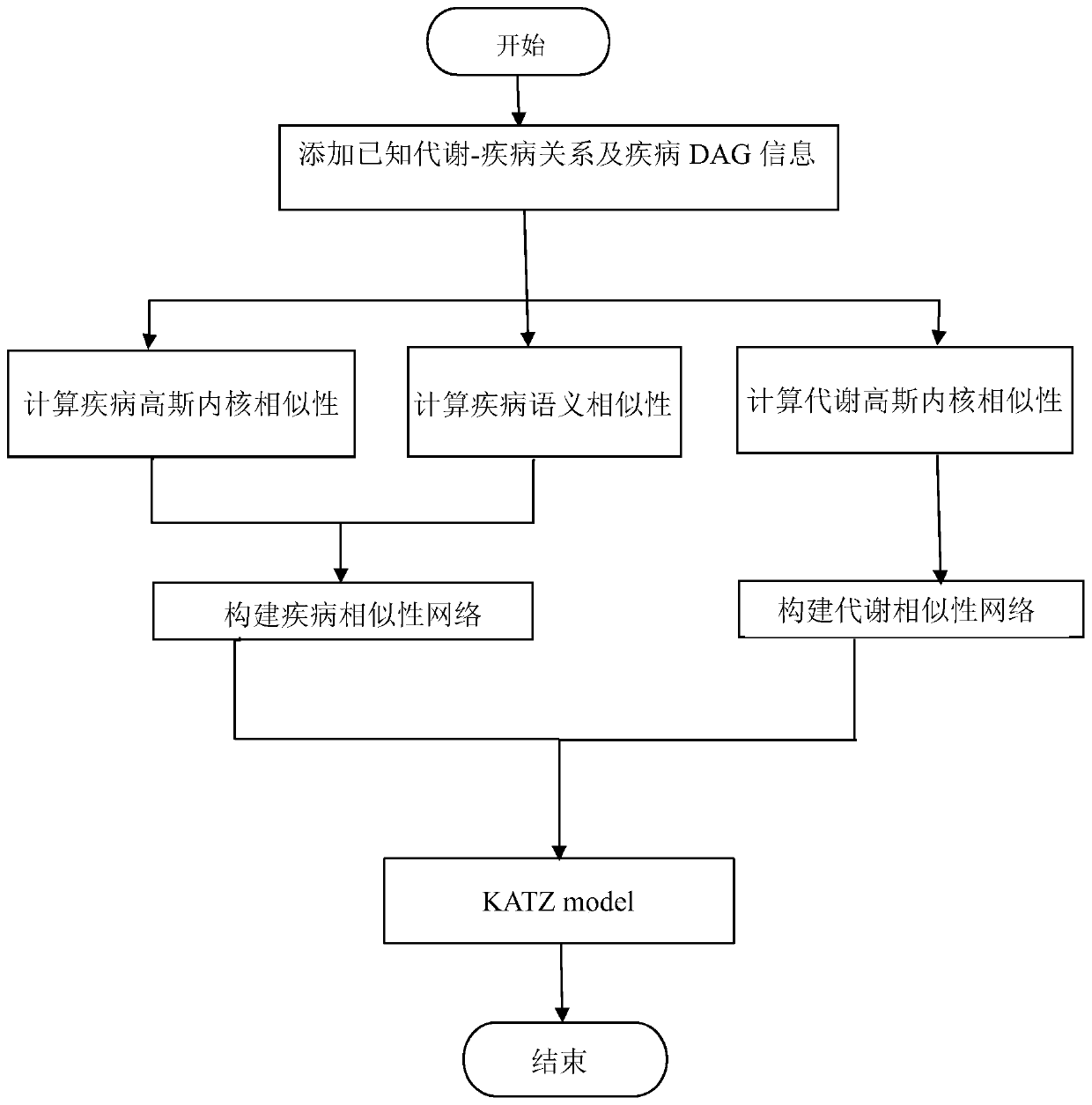
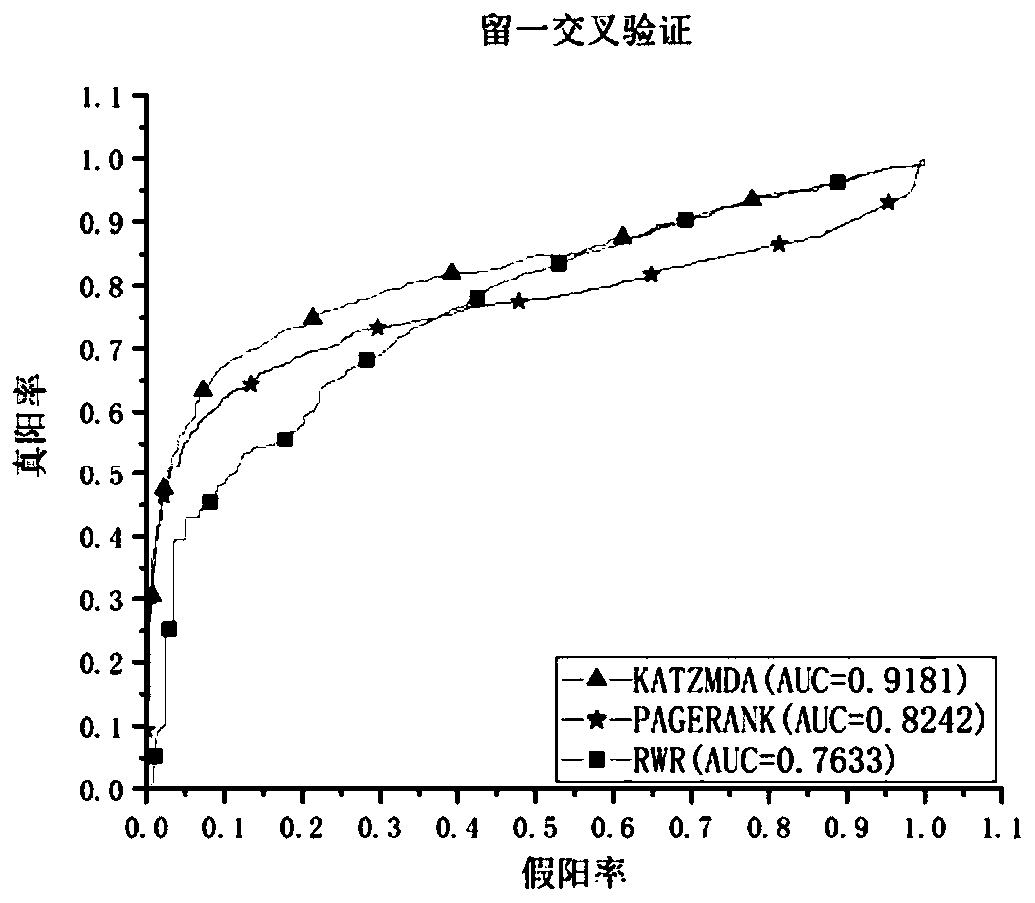
PendingCN110610763AImprove accuracyImprove predictive performanceMedical data miningMedical automated diagnosisDiseaseMetabolite
The invention discloses a method for predicting association relationship between metabolites and diseases based on a KATZ model. The method is characterized by converting known metabolite and diseaserelationship into a relationship network, calculating corresponding disease and metabolite similarity, constructing disease and metabolite similarity networks, predicting the relationship between metabolites and diseases through the KATZ model, verifying the accuracy of the predicted relationship through several cross validation methods, and further verifying the feasibility of the method throughcase analysis. The method can predict a new relationship between the metabolites and diseases, and a part of the relationship is verified by literatures and is not recorded by a database temporarily;verification results show that AUC index performance is excellent; and compared with other key protein recognition methods, the method improves the accuracy in mining potential relations of the prediction method by fusing the biological characteristics (semantic similarity) and topological characteristics (Gaussian kernel similarity) of the diseases.
Owner:SHAANXI NORMAL UNIV
Method for identifying biological essential protein
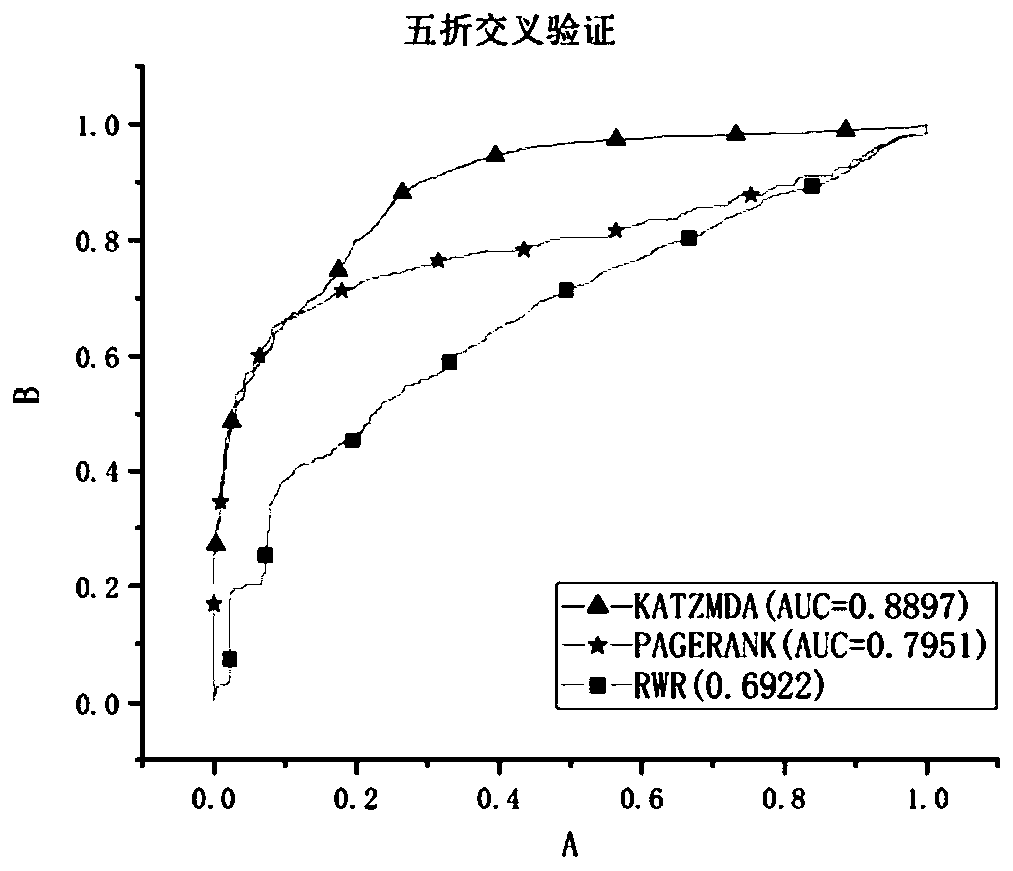
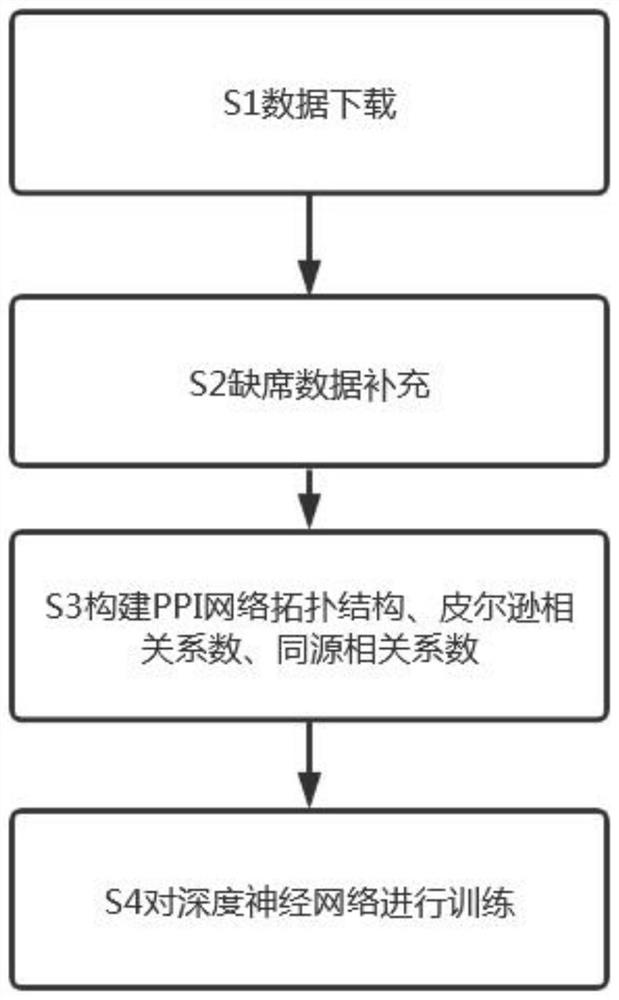
The invention provides a method for constructing necessary protein recognition between multi-dimensional biological attribute information and PPI topological characteristics by using a deep neural network, and in order to solve the problem that gene expression data is not comprehensive, absent data is supplemented so as to improve robustness. And the convergence speed of the deep neural network is reduced by respectively constructing a PPI network topology structure, a Pearson's correlation coefficient and a homologous correlation coefficient. And finally, searching an optimal incidence relation of three features of the degree of the node, the Pearson's correlation coefficient and the homologous correlation coefficient through the deep neural network, thereby improving the recognition precision of the essential protein.
Owner:GUIZHOU UNIV +1
Machine learning for protein identification
Methods for identifying a peptide by analyzing a linear readout representative of at least a portion of at least two amino acids along the peptide using a machine learning model, wherein the machine learning model is trained on linear readouts representative of a set of peptides of known sequence are provided. Methods of training a machine learning model on linear readouts representative of a set of known peptides, and systems for performing the methods of the invention are also provided.
Owner:TECHNION RES & DEV FOUND LTD
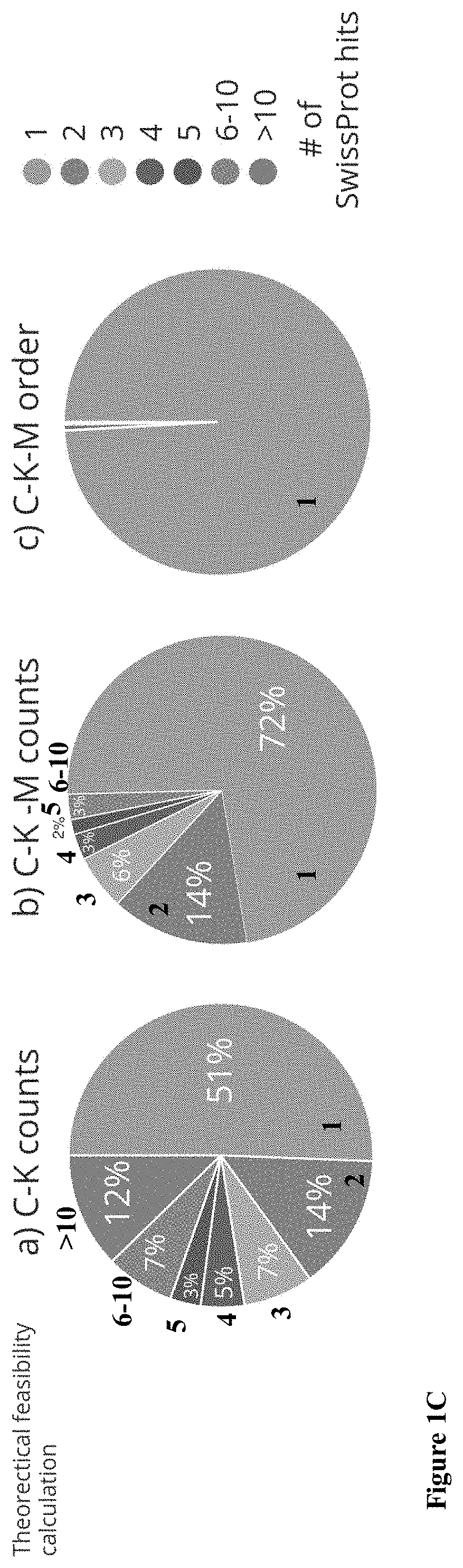
Screening Method for Specific Protein in Proteome Comprehensive Analysis
InactiveUS20090132171A1Efficiently narrowedGood repeatabilityMaterial analysis by electric/magnetic meansBiological testingSAA proteinMass Spectrometry-Mass Spectrometry
A screening method for a specific protein in a proteome analysis comprises: (a) obtaining samples containing a protein or protein digest from a cell or tissue in a specific group and a control group; (b) analyzing the samples obtained in the step (a) with a mass spectrometer, thereby obtaining mass spectrometry data; (c) analyzing the mass spectrometry data obtained in the step (b) using an arbitrary database searching software, thereby acquiring a protein list containing items for specifying proteins and indexes for identifying the proteins, for each of the samples; (d) averaging values of the indexes for each of the items in all of the protein lists acquired in the step (c), and acquiring protein list models of the specific group and the control group, containing the average values of the indexes; (e) calculating a difference between the average values for each of the items, between the protein list models of the specific group and the control group obtained in the step (d), and acquiring one protein list in which the items are rearranged in the order of the difference between the average values; and (f) selecting a protein with a large difference between the average values, from the protein list acquired in the step (e).
Owner:JCL BIOASSAY CORP
Key protein recognition method based on largest neighbor sub-network
ActiveCN108804871AImprove accuracySpecial data processing applicationsSAA proteinProtein Interaction Networks
The invention discloses a key protein recognition method based on a largest neighbor sub-network. The method comprises the steps of firstly performing sub-network division on an obtained protein interaction network according to sub-cell positioning information, and dividing proteins located in the same sub-cell position in the same sub-network, thereby obtaining a plurality of protein sub-networkswith an interaction relation in the same sub-cell interval; and then, performing topological feature analysis on the largest sub-network corresponding to each protein and where each protein is located, and calculating a critical comprehensive score value based on common neighbors, wherein the higher score value indicates that the protein tends more to be critical, thereby predicting potential critical proteins by means of score sorting. On the basis of simplicity and practicability, the accuracy of critical protein recognition can be well improved, and important reference values and practicalvalues are provided for researchers to perform experimental analysis and deeper research of the practical proteins.
Owner:CENT SOUTH UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com