Integrated system for analysis of biomolecules
a biomolecule and integrated system technology, applied in the field of integrated systems for biomolecule analysis, can solve the problems of overlapping the scope of investigation using the identification approach, the relative high degree of variability, and the equally subject to complications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
An Integrated Mass Spectrometric Immunoassay System
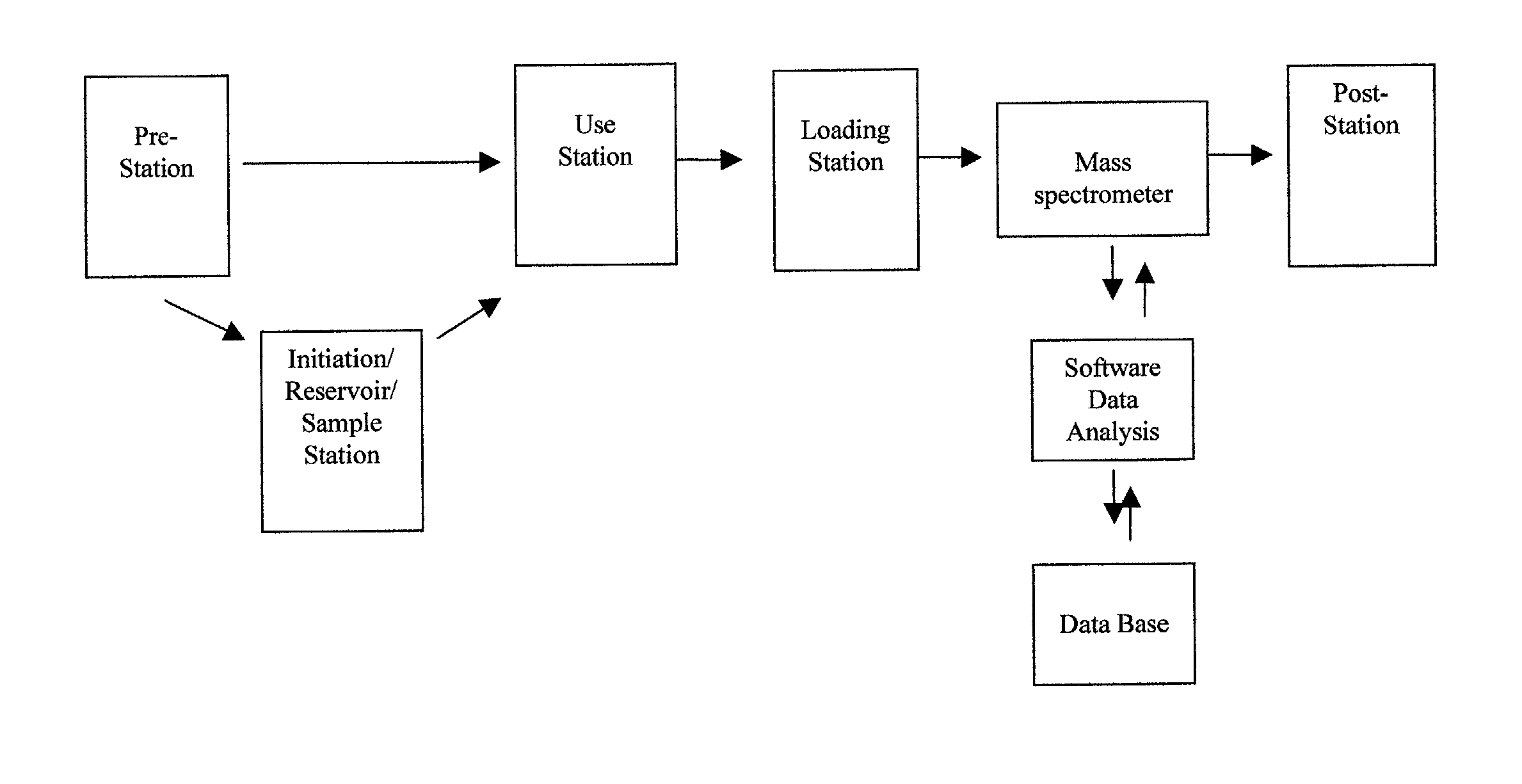
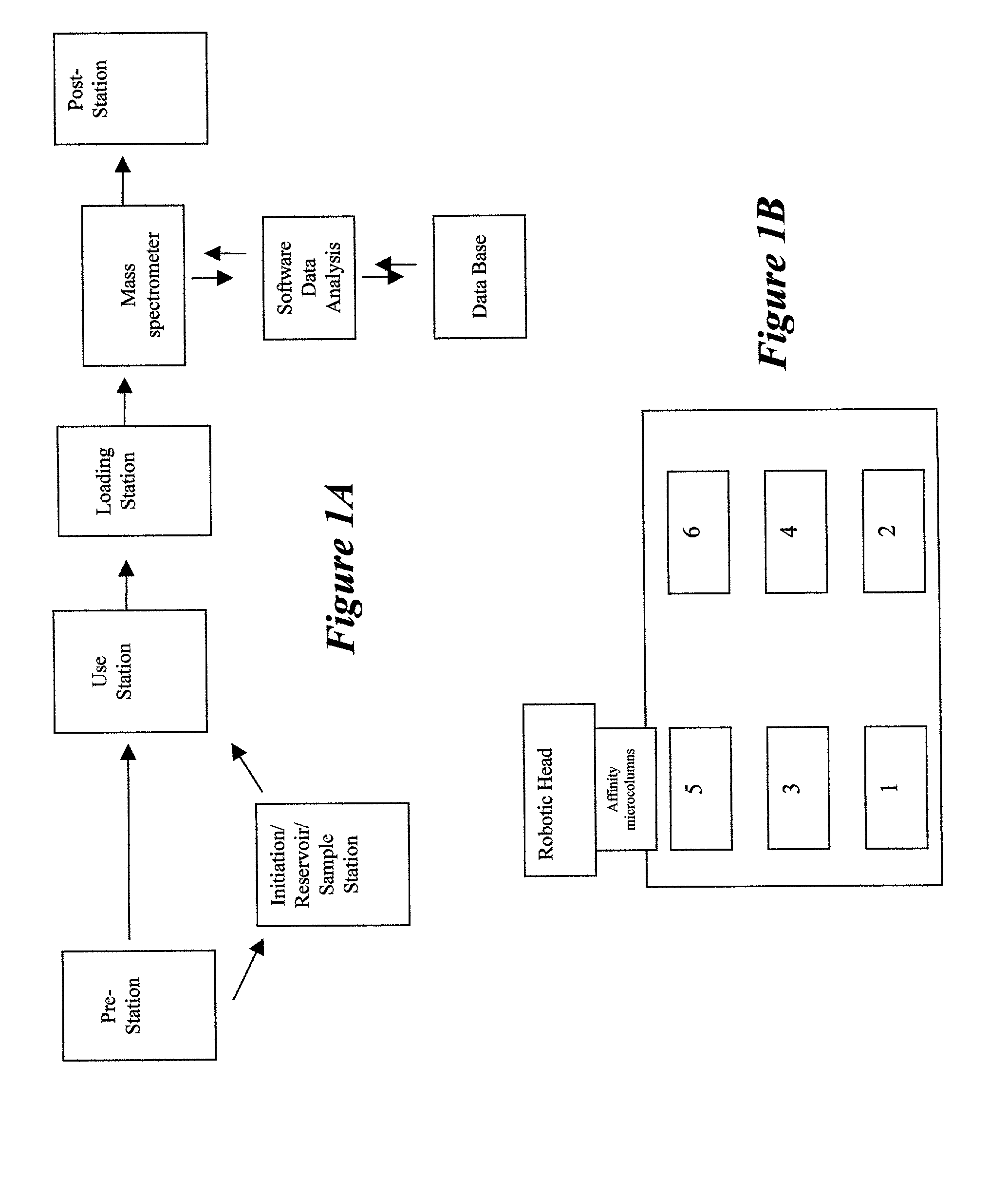
[0031] The present invention provides an integrated system capable of selectively retrieving and concentrating specific biomolecules from biological media for subsequent high-performance analyses, quantifying targeted proteins, recognizing variants of targeted biomolecules (e.g., splice variants, point mutations and post-translational modifications) and elucidating their nature, analyzing for, and identifying, ligands interacting with targeted biomolecules, and high-throughput screening of large populations of samples using a single, unified, economical, multiplexed and parallel processing platform.
[0032] The preferred embodiment of the integrated system comprises molecular traps, such as affinity microcolumns, derivatized mass spectrometer targets, mass spectrometers capable of multi-sample input and robotics with processing / data analysis interactive database. The present invention also includes methods and processes for use of the...
example 2
High Throughput Protein Quantification in the Presence of Variants
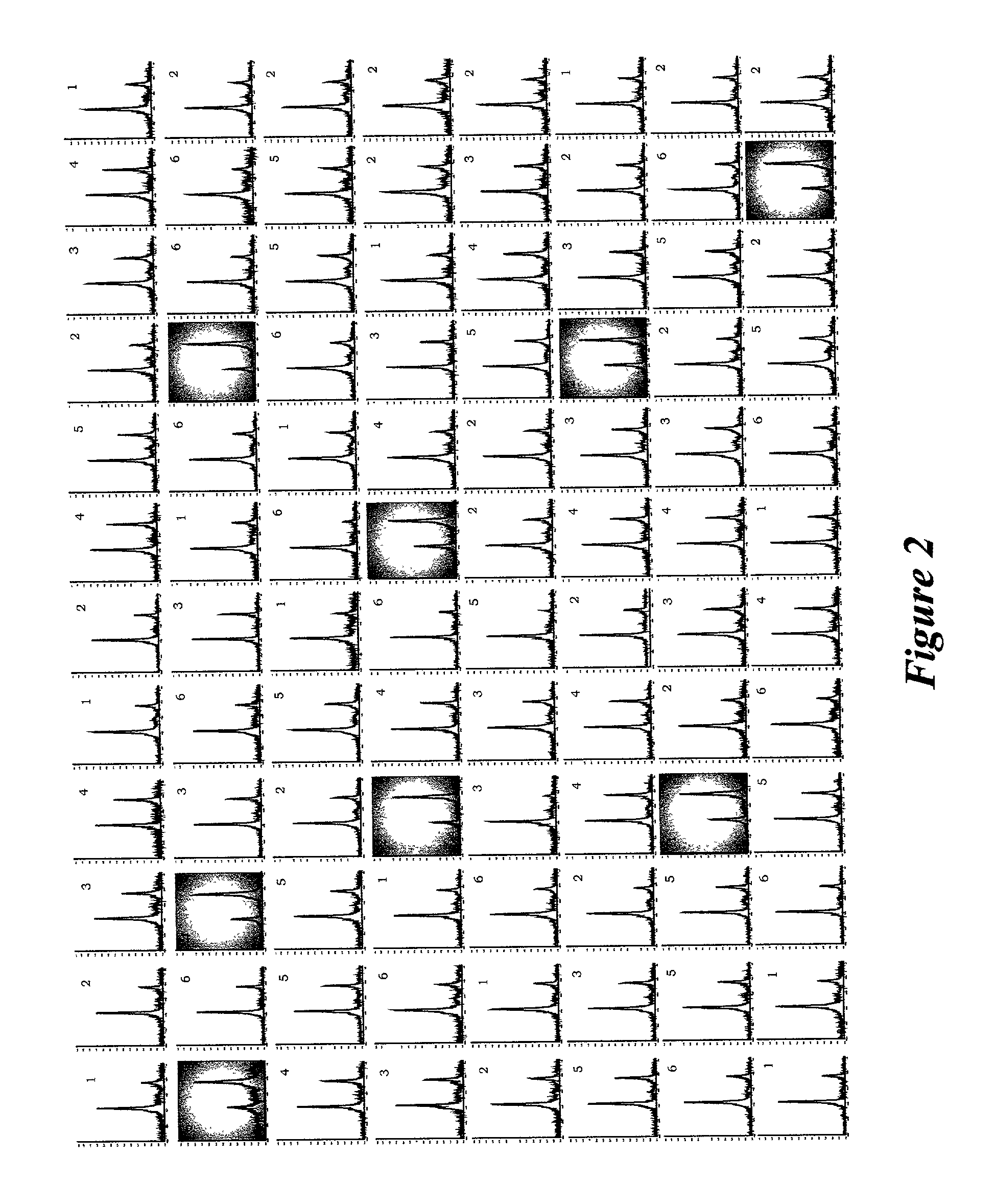
[0046] With reference to FIG. 4, a high-throughput quantitative analysis of .beta..sub.2m from human plasma samples was performed using the integrated system and methods described herein. The samples from six individuals were prepared as described in FIG. 2. Eighty-eight wells of the 96-well sample plate received 15 .mu.L plasma aliquots (the samples from the six individuals were randomized on the 96-well plate), 7.5 .mu.L of equine plasma (undiluted) and 128 .mu.L of HBS buffer. A series of dilutions of a 7.6.times.10.sup.-4 mg / mL standard solution of purified human .beta..sub.2m were prepared (spanning a concentration range of 7.6.times.10.sup.-4 to 1.14.times.10.sup.-4 mg / mL) and used as samples (15 .mu.L of each) in the last column (8 wells) on the 96-well plate. Parallel sampling processing and MALDI-TOF MS analysis was performed as described for FIG. 2, using the polyclonal anti-.beta..sub.2m microcolumns. The m...
example 3
High Throughput Screening for Genetic and Posttranslational Variants
[0049] With reference to FIG. 7, a qualitative high-throughput screening of transthyretin (TTR) for posttranslational modification (PTM) and point mutations (PM) was performed using the integrated system and methods described herein. Aliquots of diluted (5-fold) human plasma samples collected from six individuals were prepared for parallel screening on a 96-well plate. Each well received a 15 .mu.L plasma aliquot (the samples from the six individuals were randomized on the 96-well plate), and 135 .mu.L of HBS buffer. Parallel sampling processing entailed simultaneous incubation / capture of the 96 samples on 96 anti-TTR derivatized microcolumns. The polyclonal anti-TTR microcolumns were made via glutaraldehyde-mediated coupling of the antibodies to amino-coated / modified microcolumns. Captured proteins were eluted from the microcolumn array with a small volume of MALDI matrix (saturated ACCA solution) and stamped onto ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentrations | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| multiple processing | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com