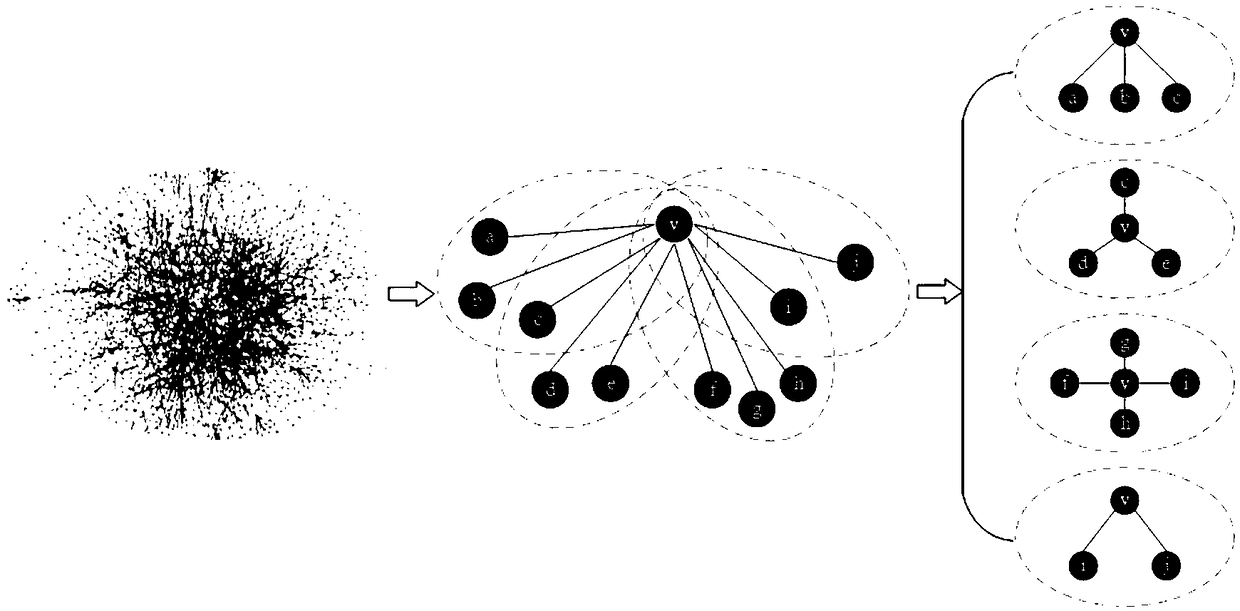
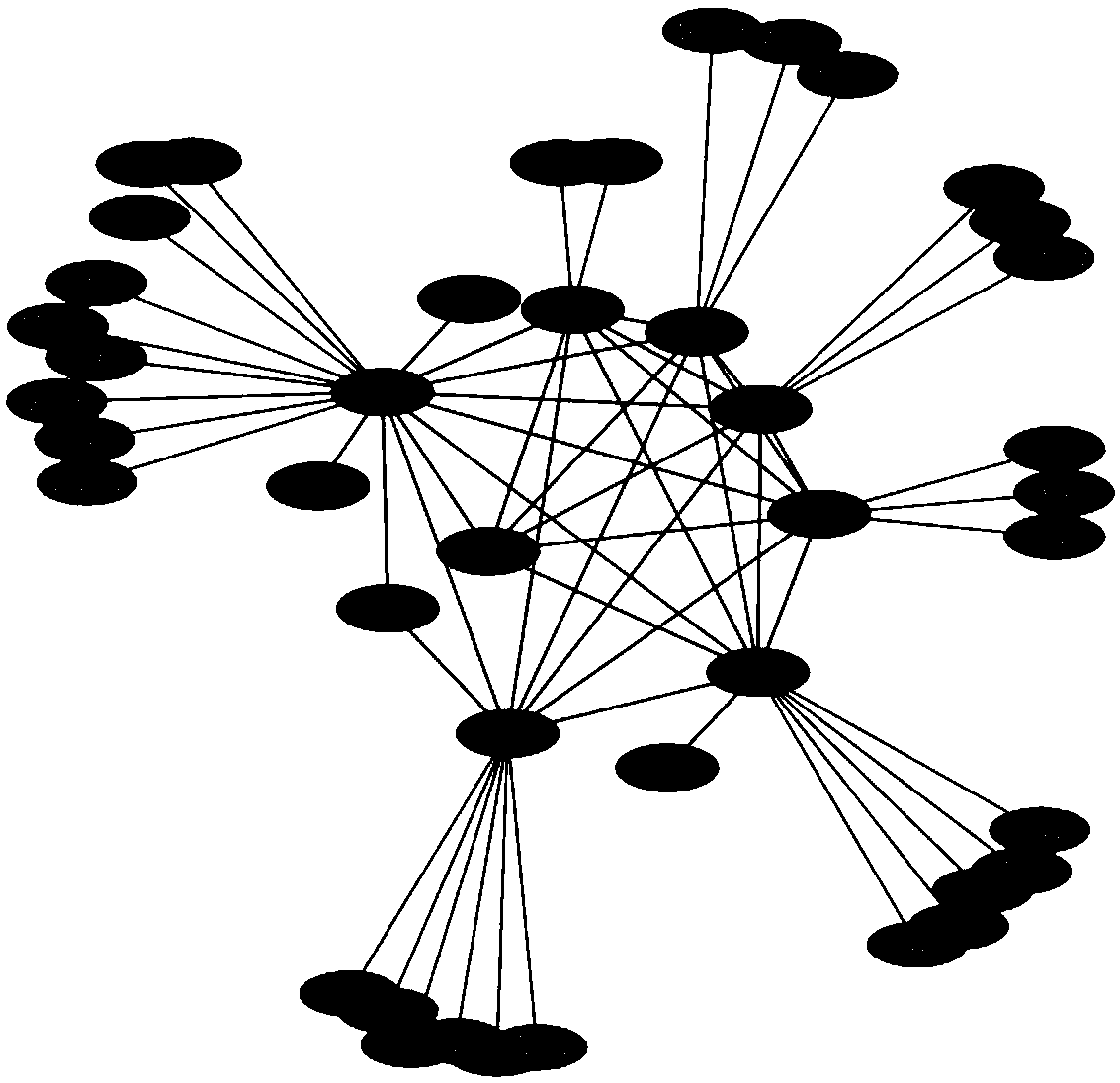
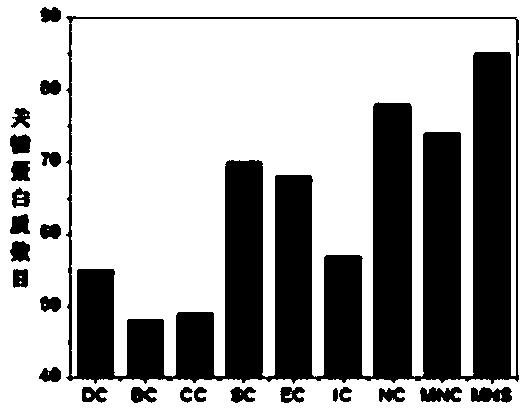
Key protein recognition method based on largest neighbor sub-network
An identification method and protein technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of mixed false positive and false negative data, inability to efficiently predict key proteins, and incomplete interaction data. achieve the effect of improving the accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] Biological data sets used in the present invention: Yeast is the most widely used species in current research, and there are a large number of key proteins of yeast species determined experimentally in existing public databases. The yeast protein interaction data set used in the present invention is downloaded from the DIP database, and self-interactions and repeated interactions in the data set are removed to obtain the original protein interaction network. Due to deficiencies in experimental techniques and other aspects, some false positive and false negative data inevitably exist in protein interaction data. In order to reduce the impact of these erroneous data on the identification of key proteins, the present invention uses yeast protein subsets in the COMPARTMENTS database. Cell localization information partitions the original network. The database integrates relevant subcellular localization information from databases such as UniProtKB, MGI, SGD, FlyBase, and Wor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com