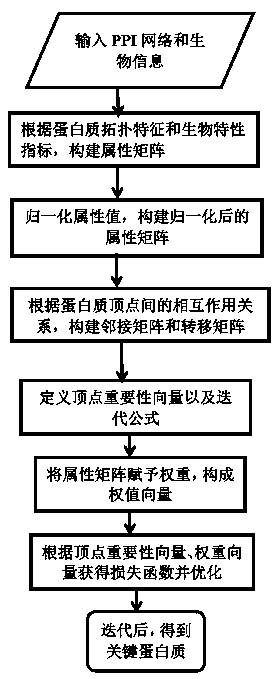
Key-protein recognition method based on fusion of biological and topological features
A technology of topological features and identification methods, applied in the field of biological information, can solve the problems of lack of key protein biological characteristics, low accuracy of key proteins, dependence on the accuracy of protein interaction networks, etc., to expand the scope of application and practicability, and improve efficiency. , the effect of improving the accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
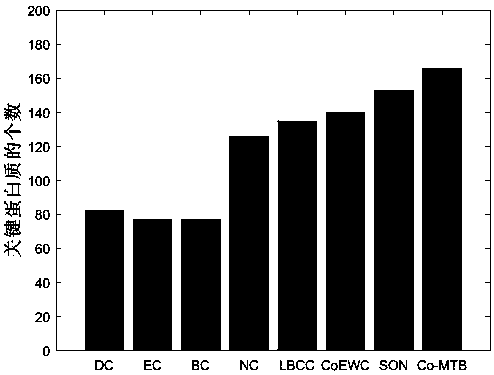
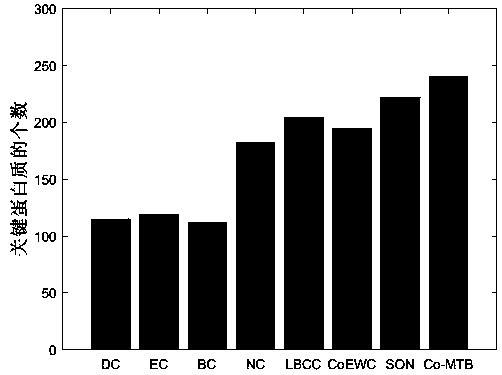
[0077] The method proposed in this paper (Co-MTB) will be compared with the existing methods of DC, EC, BC, NC, LBCC, SON, COEWC in DIP dataset and MIPS dataset. For each method, we select the top 100 to 600 protein results as the candidate set.
[0078] The prediction results of the DIP data set are as follows Figure 2a , Figure 2b , Figure 2c , Figure 2d , Figure 2e , Figure 2f shown. The method Co-MTB proposed in this paper can achieve better results than other methods in identifying key proteins. When taking the top 100-600 proteins as candidate proteomes, the prediction accuracy of Co-MTB is 25%, 19%, 23%, 24%, 24%, 28% higher than CoEWC, respectively. However, Co-MTB improves the prediction accuracy by 11%, 8%, 8%, 6%, 7% and 5% at six levels compared with the recently developed method SON.
[0079] The prediction results of the MIPS data set are as follows Figure 3a , Figure 3b , Figure 3c , Figure 3d , Figure 3e , Figure 3f shown. The algorit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com