Method for identifying biological essential protein
A protein and biological technology, applied in the field of life sciences, can solve problems such as troublesome and complex verification of model correctness, and achieve the effect of improving accuracy, reducing training time and
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
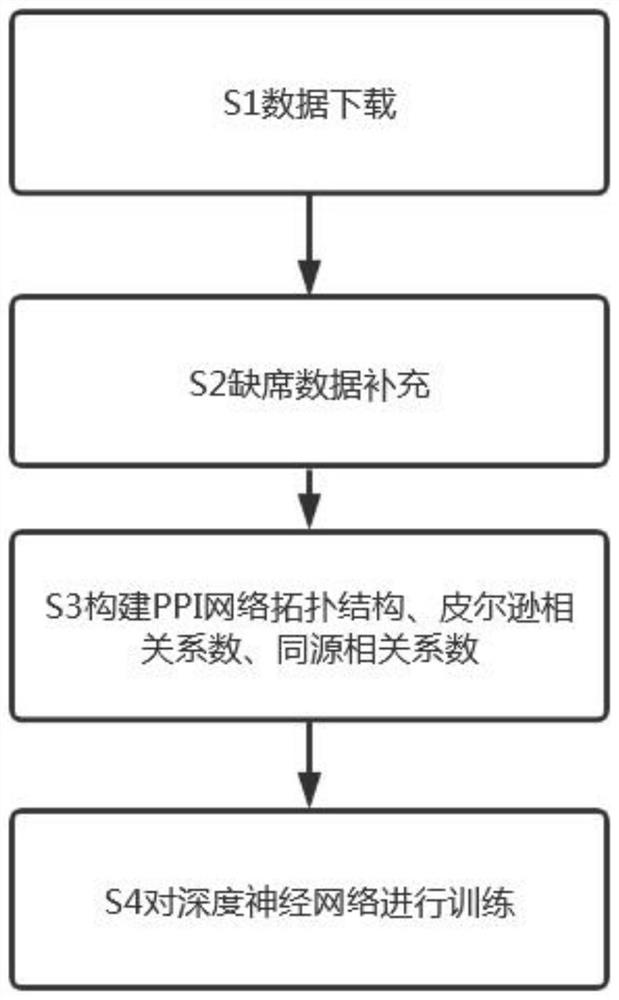
[0071] The object of the present invention is to provide a method for identifying biologically essential proteins, comprising the following steps,
[0072] S1 data download:
[0073] The present invention obtains yeast protein information from DIP and GAVIN data sets respectively. After removing self-interactions and repeated interactions, the DIP dataset resulted in 5093 proteins, 24743 pairs of interactions and 1167 essential proteins. The GAVIN dataset provides 1855 proteins, 7669 pairs of interactions, and 714 basic proteins.
[0074] In addition, information on homologous proteins was downloaded from the InParanoid database (Version 7), which contains 100 genome-wide pairwise comparisons. Additionally, gene expression data for yeast were downloaded from the dataset provided by Tu BP.
[0075] Finally, the present invention will further download a data set containing 1285 Saccharomyces cerevisiae essential genes from four databases of MIPS, SGDP, DEG and SGD as a benchm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com