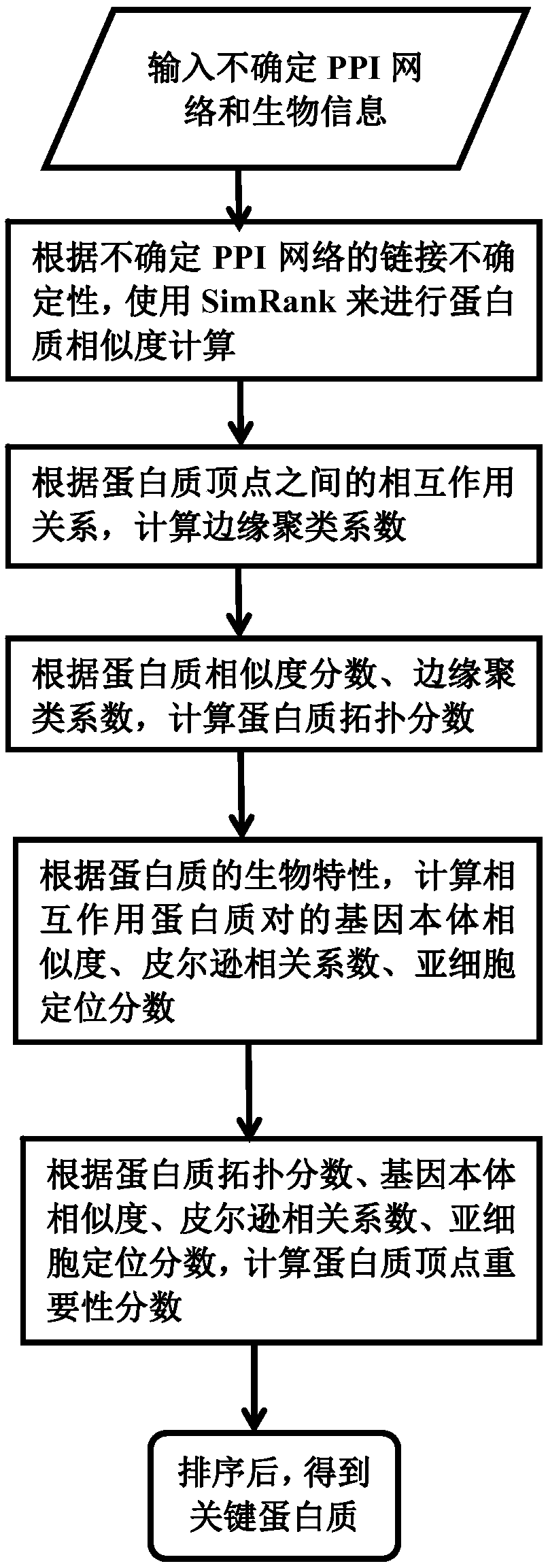
A key protein identification method in an uncertain protein interaction network
A recognition method, protein technology, applied in proteomics, character and pattern recognition, genomics, etc., can solve the problems of ignoring the uncertainty of protein interaction network, false positives in protein interaction data, noise in PPI network, etc. , to achieve the effect of expanding the scope of application and practicability, overcoming negative effects, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
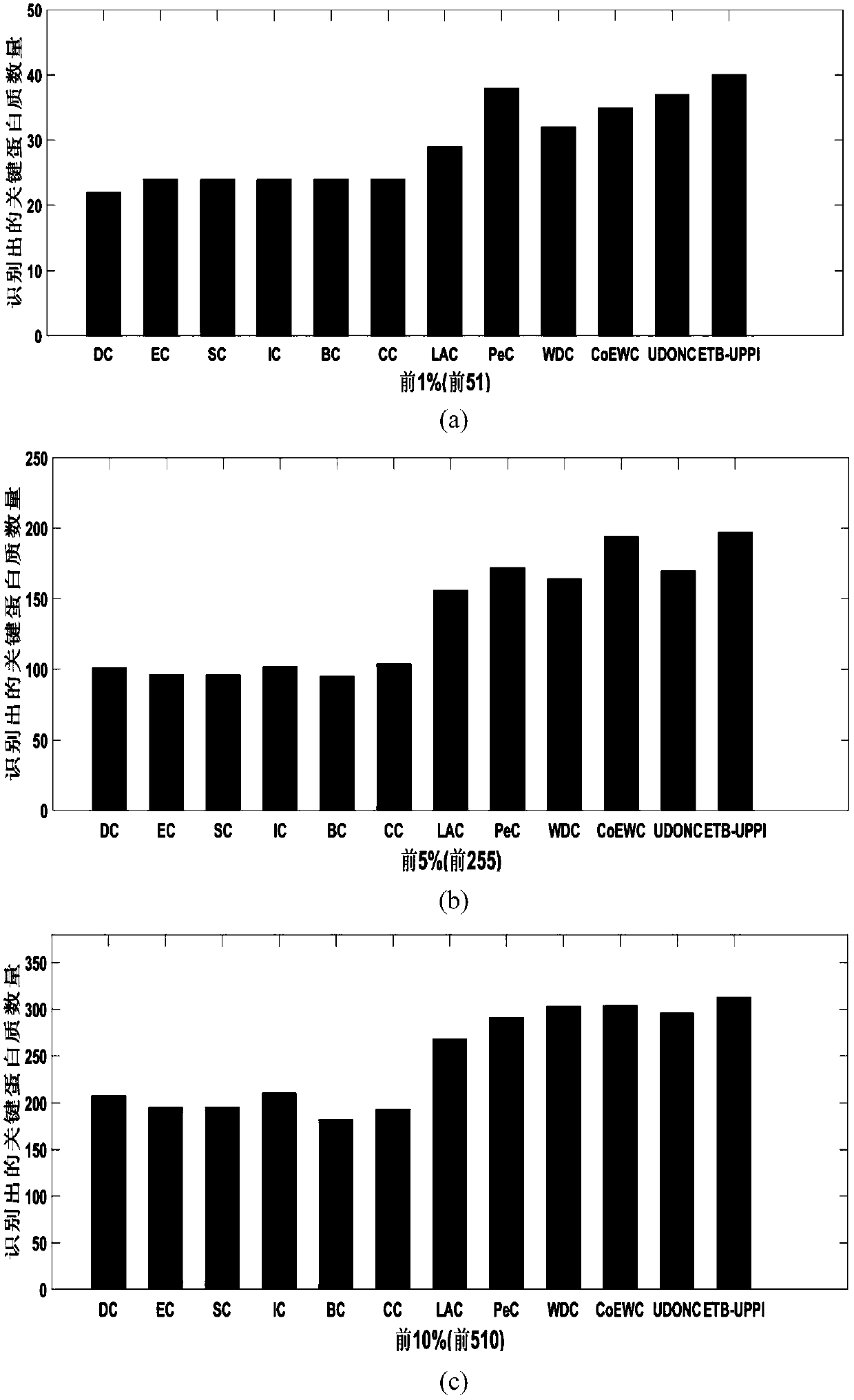
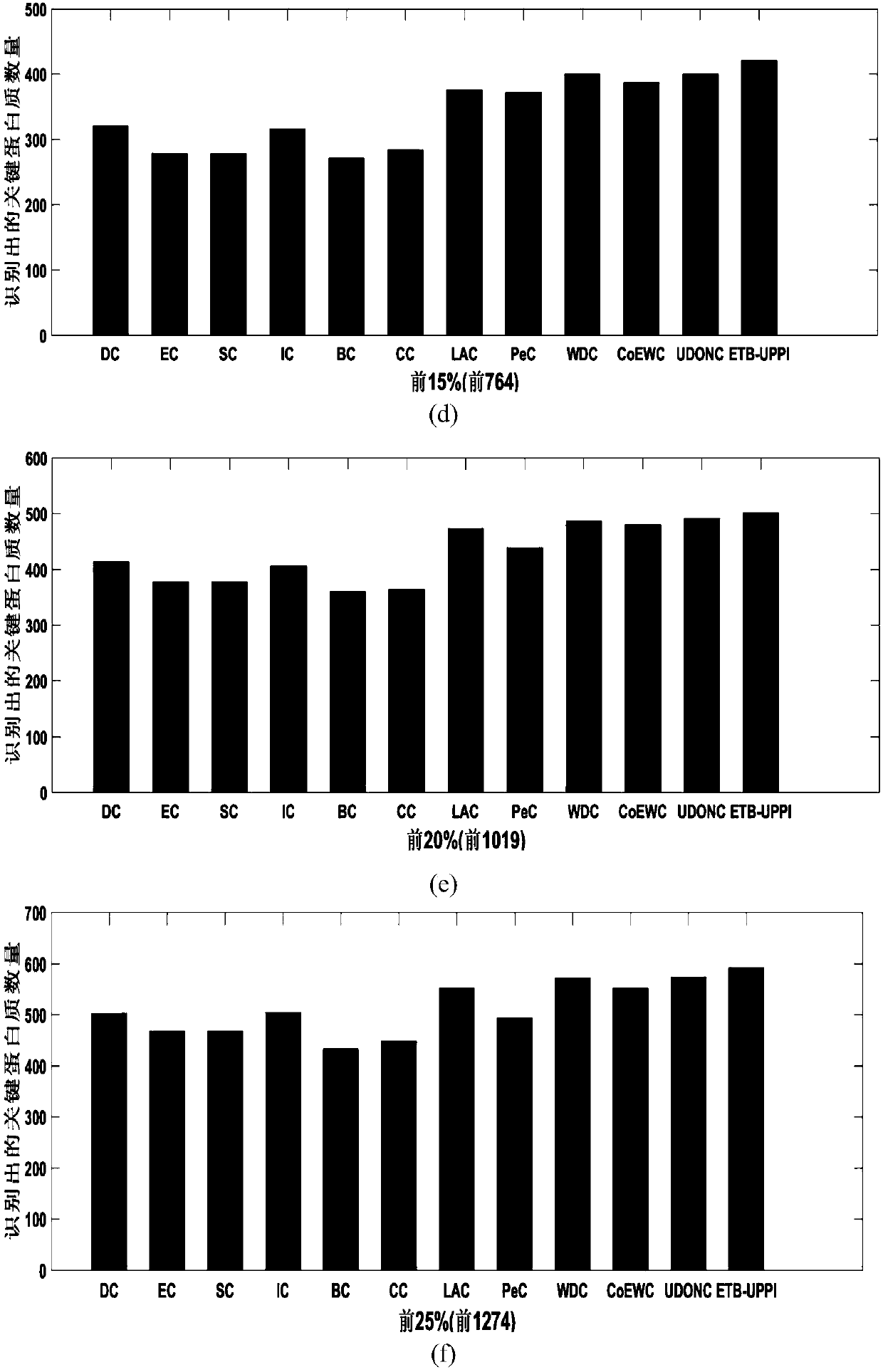
[0081] In order to verify the performance of the method (ETB-UPPI) proposed by the present invention, the present invention compares the quantity of key proteins identified with other various methods (DC, EC, SC, IC, NC, BC, CC, LAC, PeC, WDC , CoEWC, UDONC) were compared in four datasets (DIP, MIPS, Gavin, Krogan). For each method, the present invention selects the protein identification results of the top 1%, top 5%, top 10%, top 15%, top 20% and top 25% as candidate sets, and the proteins in each candidate set are compared with the key of the standard The protein sets are intersected to obtain the number of real key proteins in the candidate set. The experimental results are as follows figure 2 shown.
[0082] The prediction results of the DIP data set are as follows figure 2 shown. The method ETB-UPPI proposed by the present invention can obtain better results than other methods in identifying key proteins. When the top 1%, top 5%, top 10%, top 15%, top 20% and top 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com