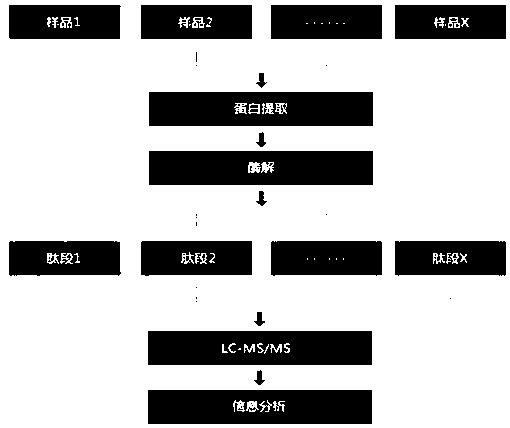
Method used for large-scale screening of protein biomarkers
A biomarker and protein technology, applied in the field of molecular biology, can solve the problems of high cost, small dynamic range, and poor consistency, and achieve the effect of low cost, large number of screenings, and small total amount of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Take 4 g of mouse liver tissue control samples and 4 g of pathological samples, cut into pieces, and transfer them to a homogenizer. Add PMSF / isopropanol stock solution (2 mL) to pre-cooled lysis buffer. Quickly add 2mL of pre-cooled lysis buffer into the homogenizer, and grind thoroughly under ice bath conditions. Transfer the tissue grinding solution to a 1.5mL centrifuge tube and centrifuge at 4°C (14000rpm, 10min). After centrifugation, draw the supernatant and transfer it to a new 1.5mL centrifuge tube, which is the crude tissue protein extract. The obtained protein extract can be stored at -20°C for subsequent experiments.
[0027] The above protein extracts were sampled with a spotting instrument and fixed in 96-well plates for 1 hour of reaction. Block the plate with a solution containing skim milk to prevent non-affinity substances from sticking to the chip. Take out the material in the orifice plate, use a glass rod to crush the gel, dissolve it with buffe...
Embodiment 2
[0054] Take 4g of mouse spleen tissue control sample and 4g of pathological tissue and put them in 4ml of protein lysate A (10mmol / LMgCl2, 1mmol / LKCl, 1mmol / L pyrenetetraylsulfonyl fluoride, 1mmol / L dithiothreitol), and mix well. Slurry until the particles are invisible to the naked eye, then add a few drops of 3mmol / L sucrose and mix well, centrifuge at 10000r / min at 4°C for 30min, and discard the supernatant. Dissolve the precipitate in 3-5 times protein lysate (0.5mol / L KCl) and mix well, centrifuge at 10000r / min at 4°C for 30min, take the supernatant and store it at -80°C for future use.
[0055] The above protein extracts were sampled with a spotting instrument and fixed in 96-well plates for 1 hour of reaction. Block the plate with a solution containing skim milk to prevent non-affinity substances from sticking to the chip. Take out the material in the orifice plate, use a glass rod to crush the gel, dissolve it with buffer solution (0.3% bromophenol blue solution: 0.3%...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com