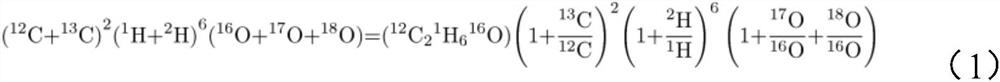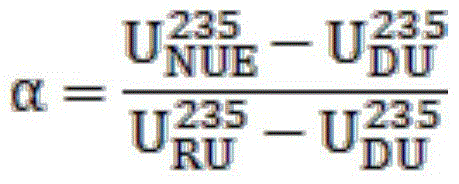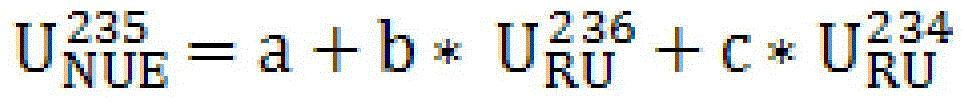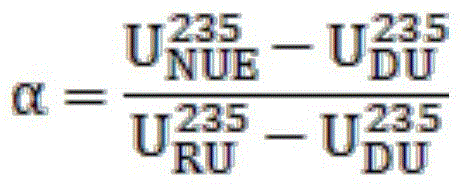Patents
Literature
52 results about "Isotope distribution" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
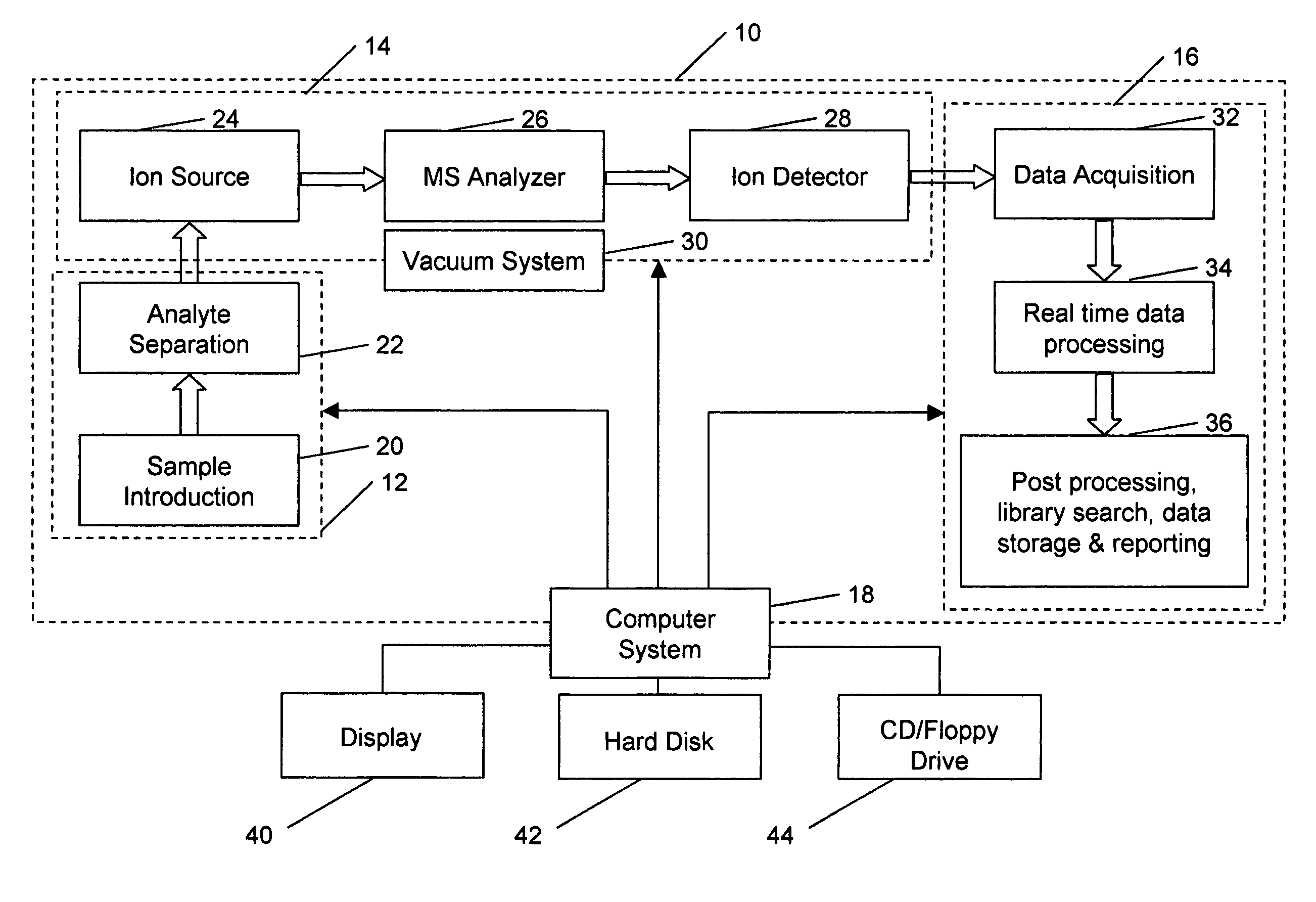
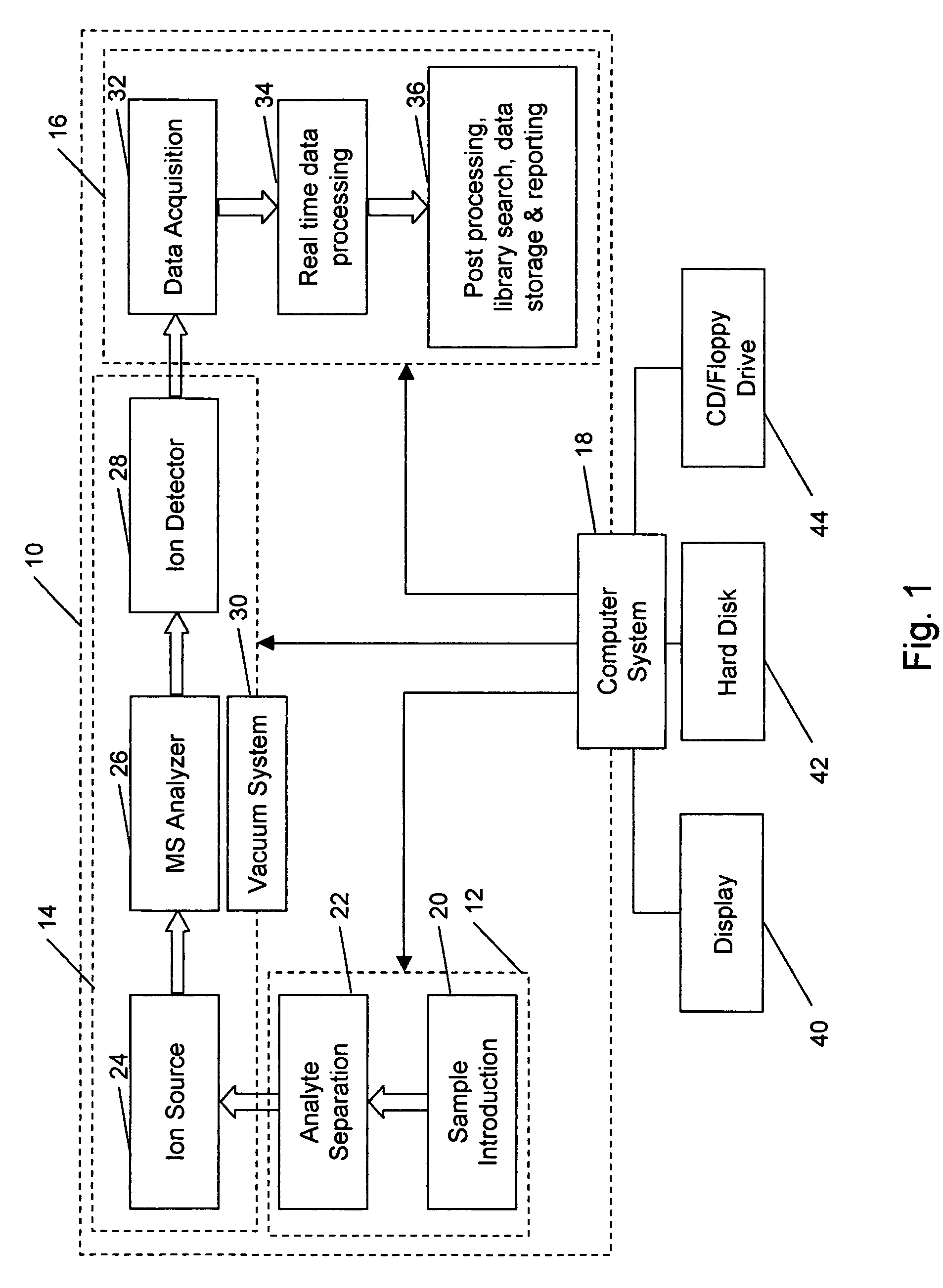
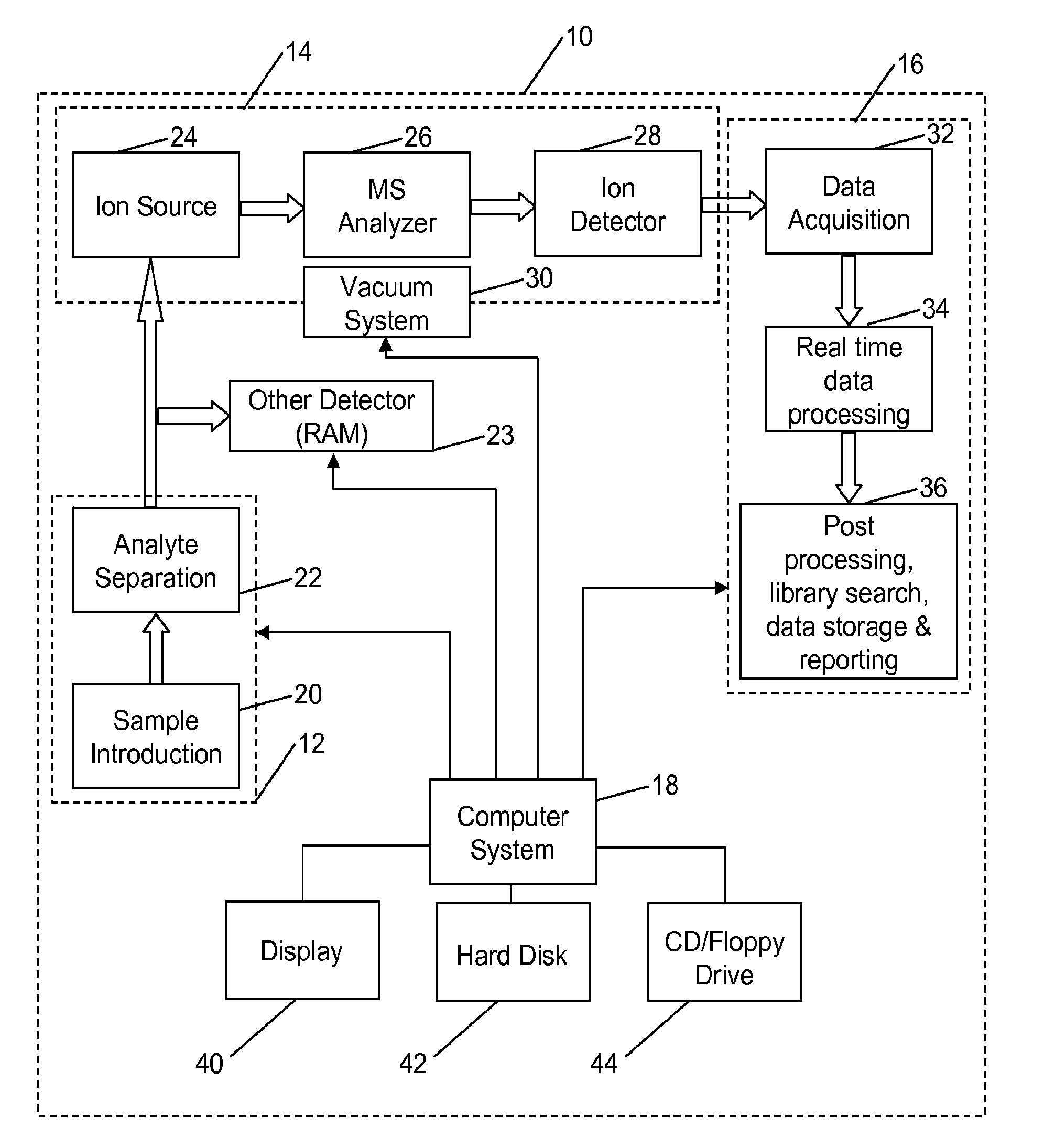
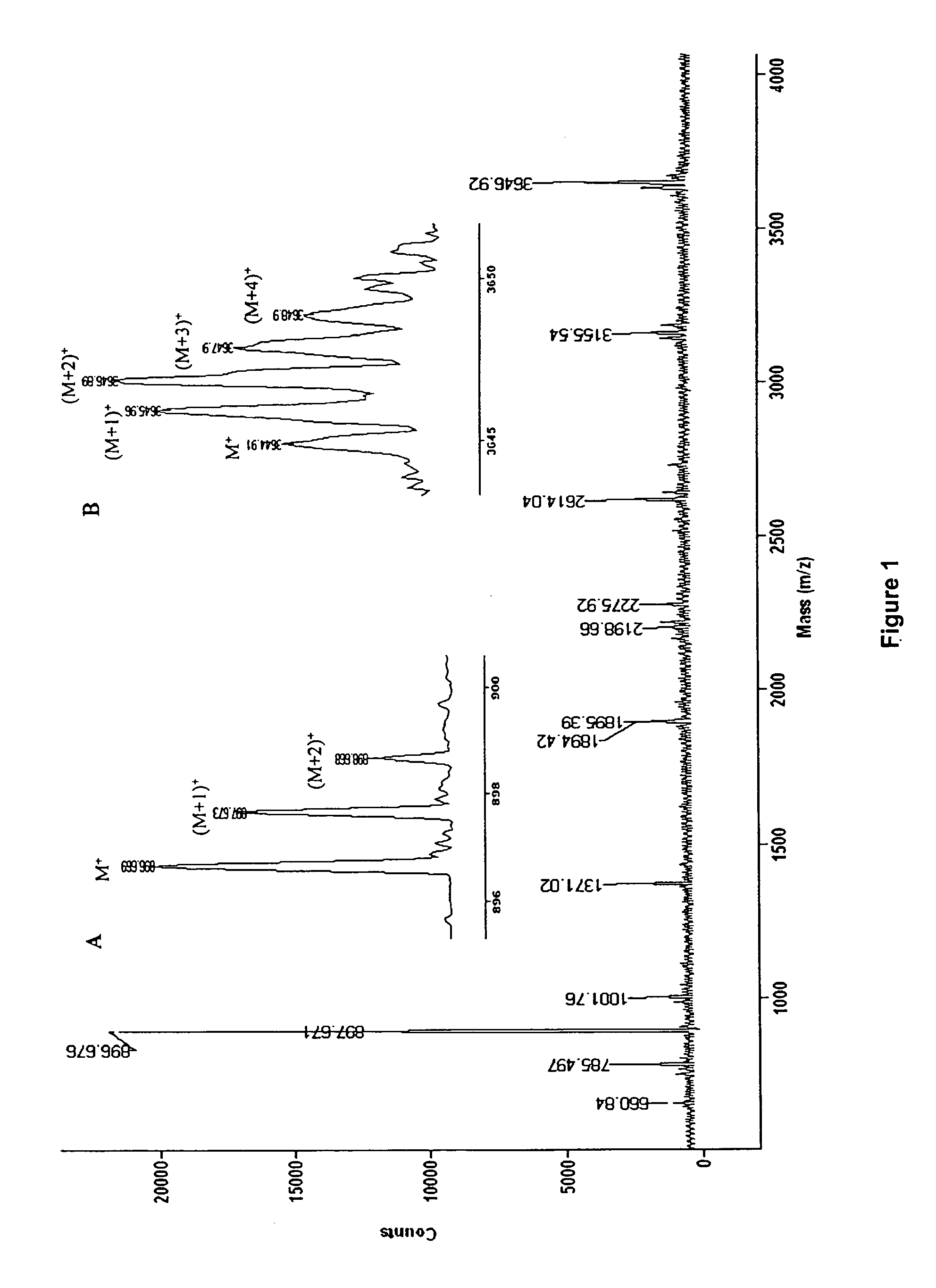
Aspects of mass spectral calibration
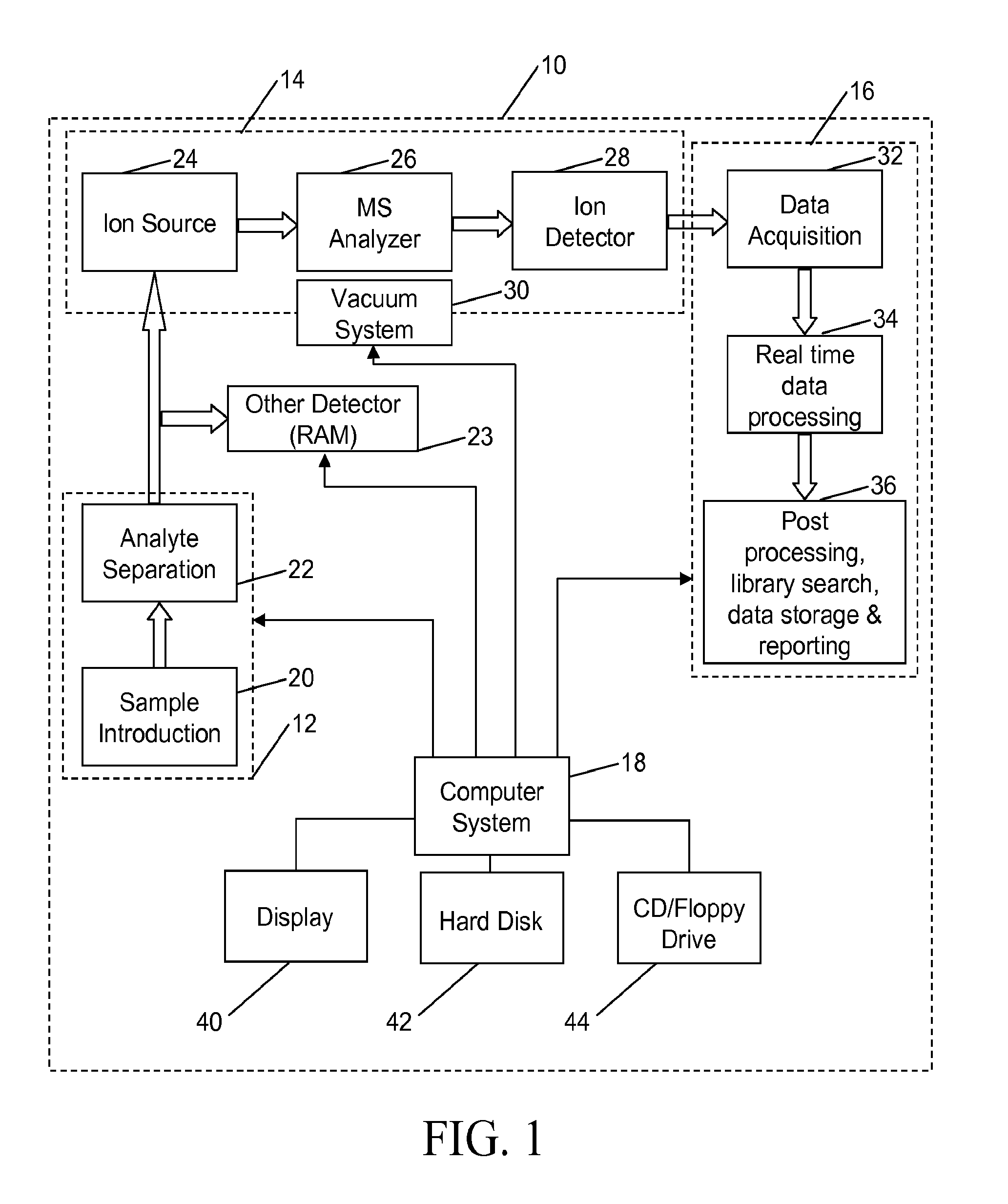
InactiveUS20060169883A1Improve accuracyHigh resolutionTime-of-flight spectrometersMaterial analysis by optical meansElemental compositionSpectral response
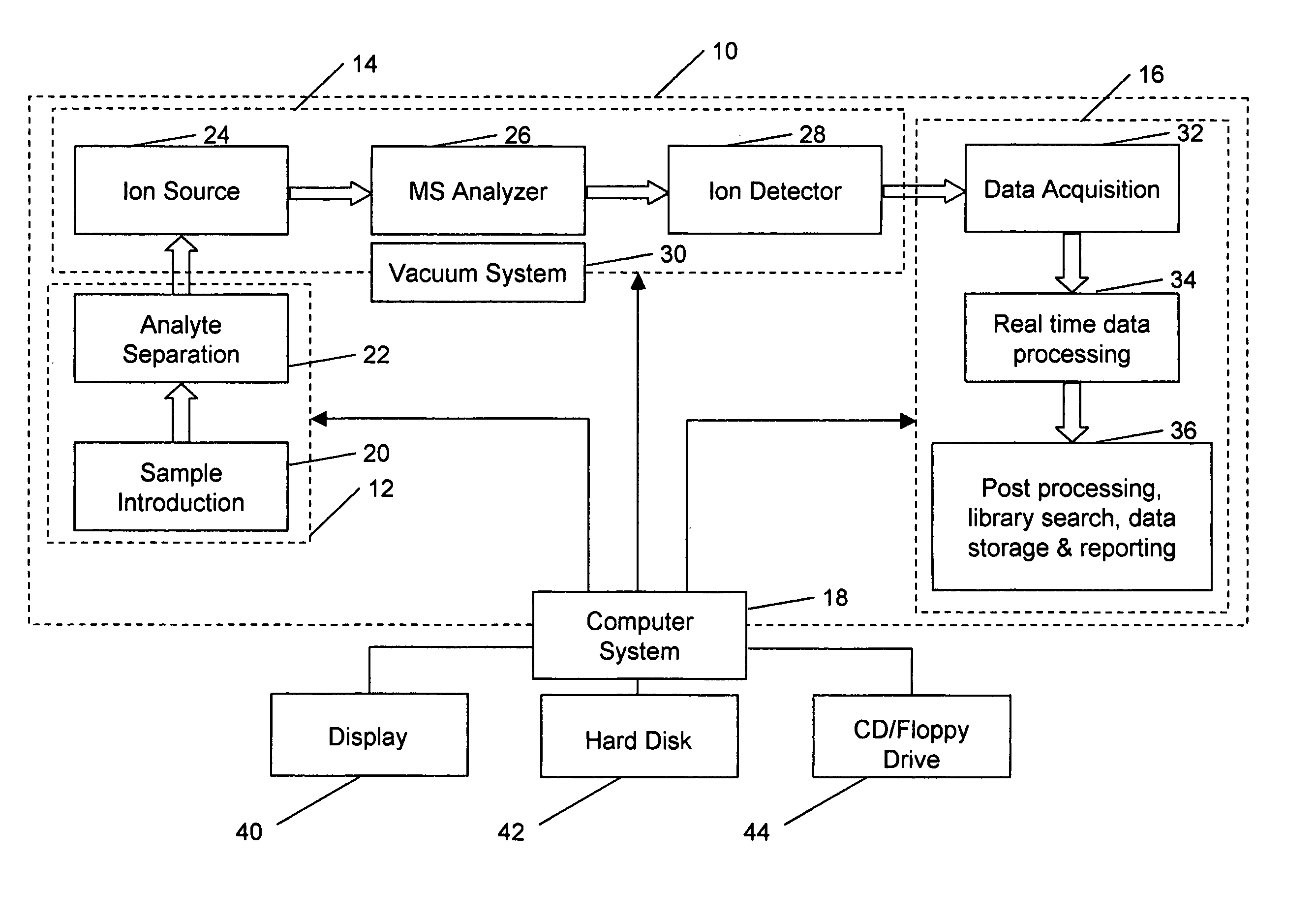
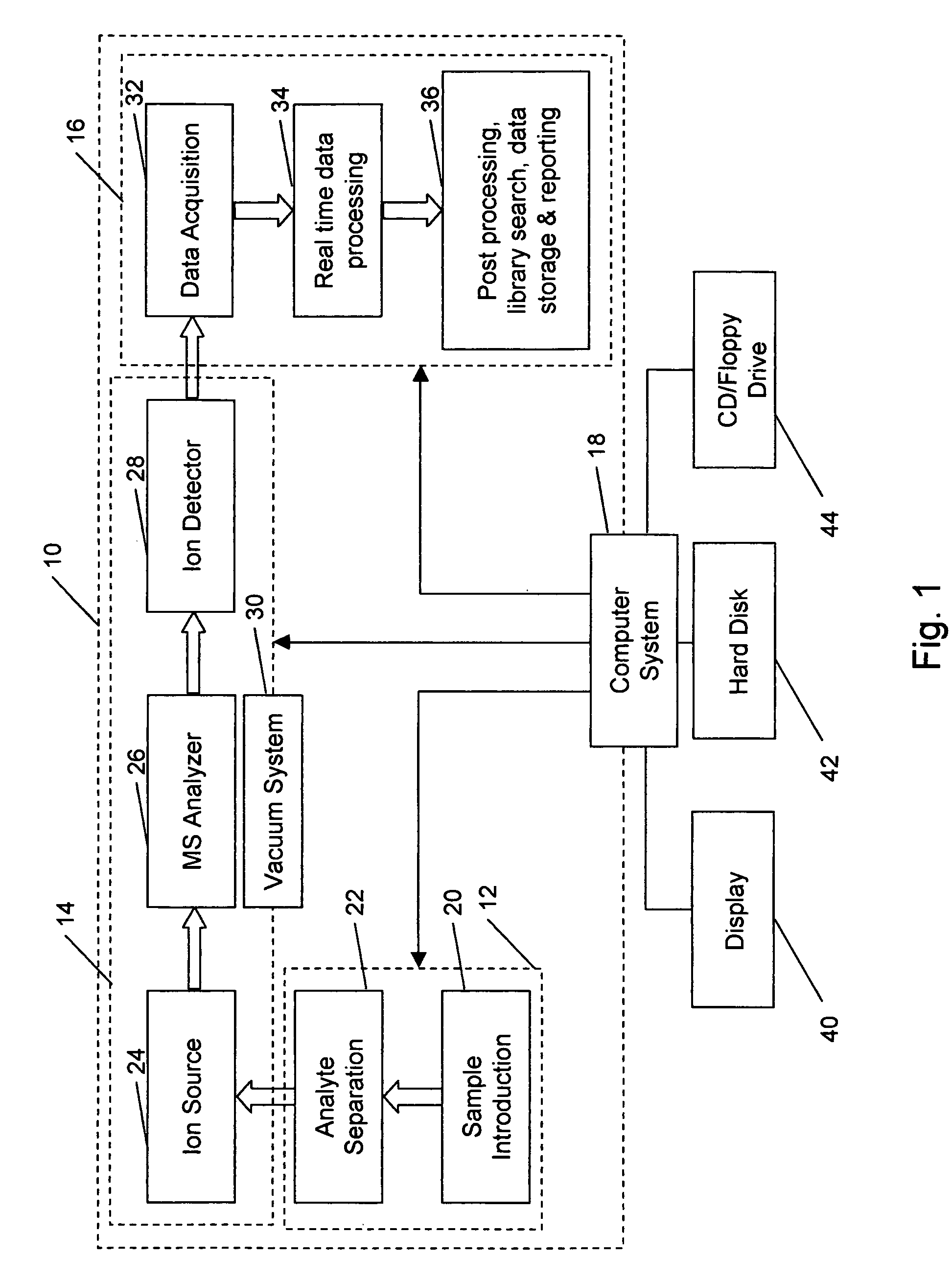
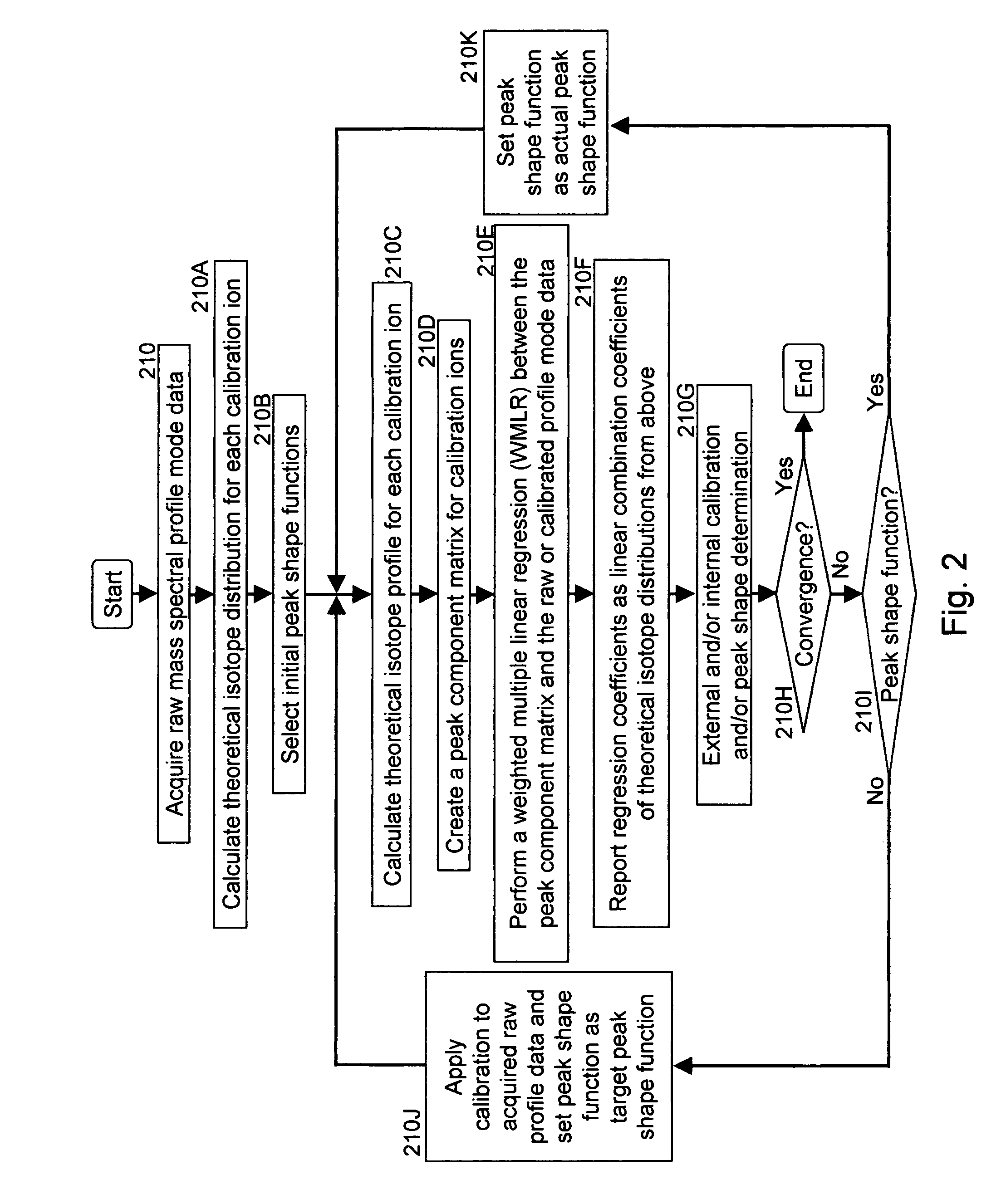
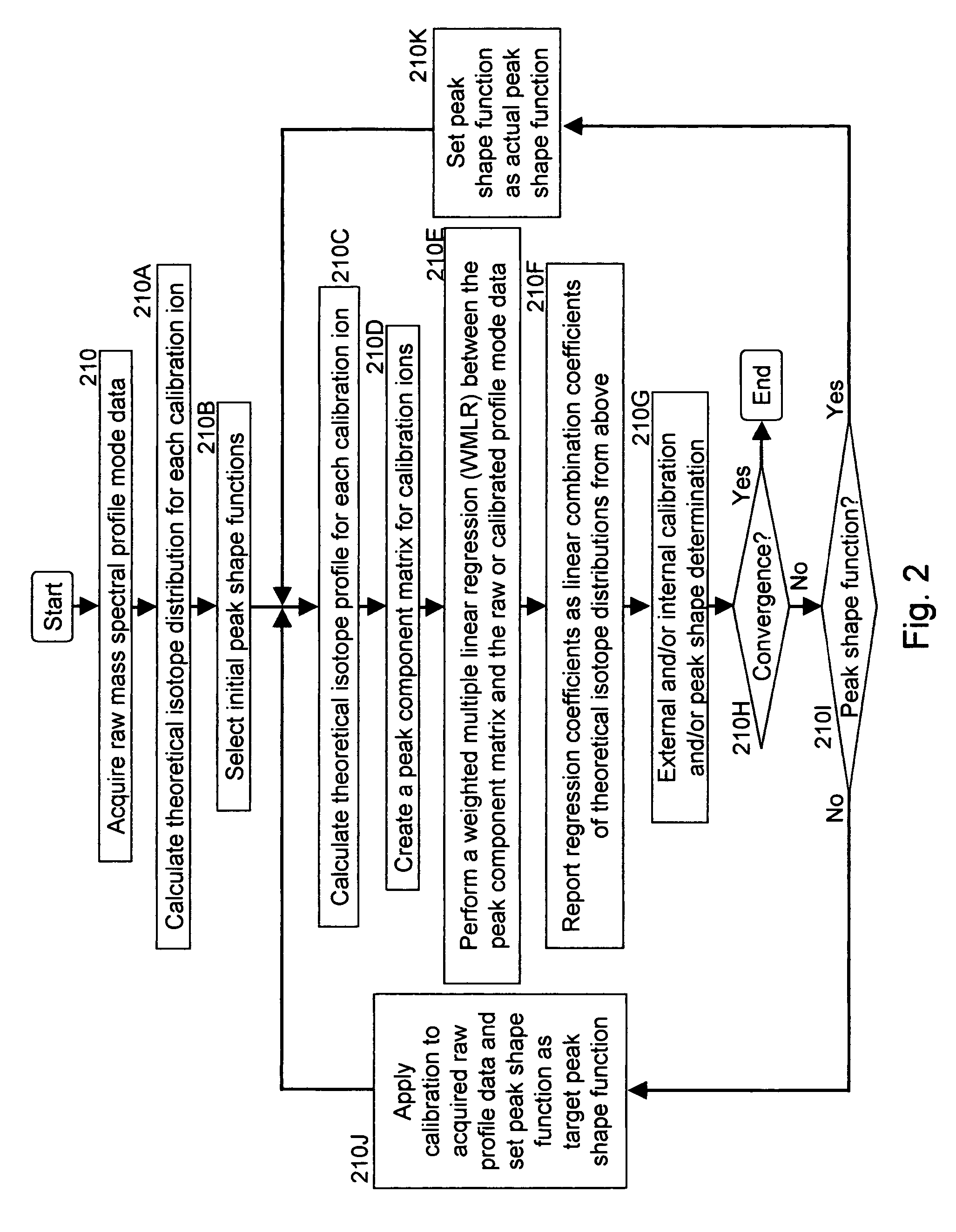
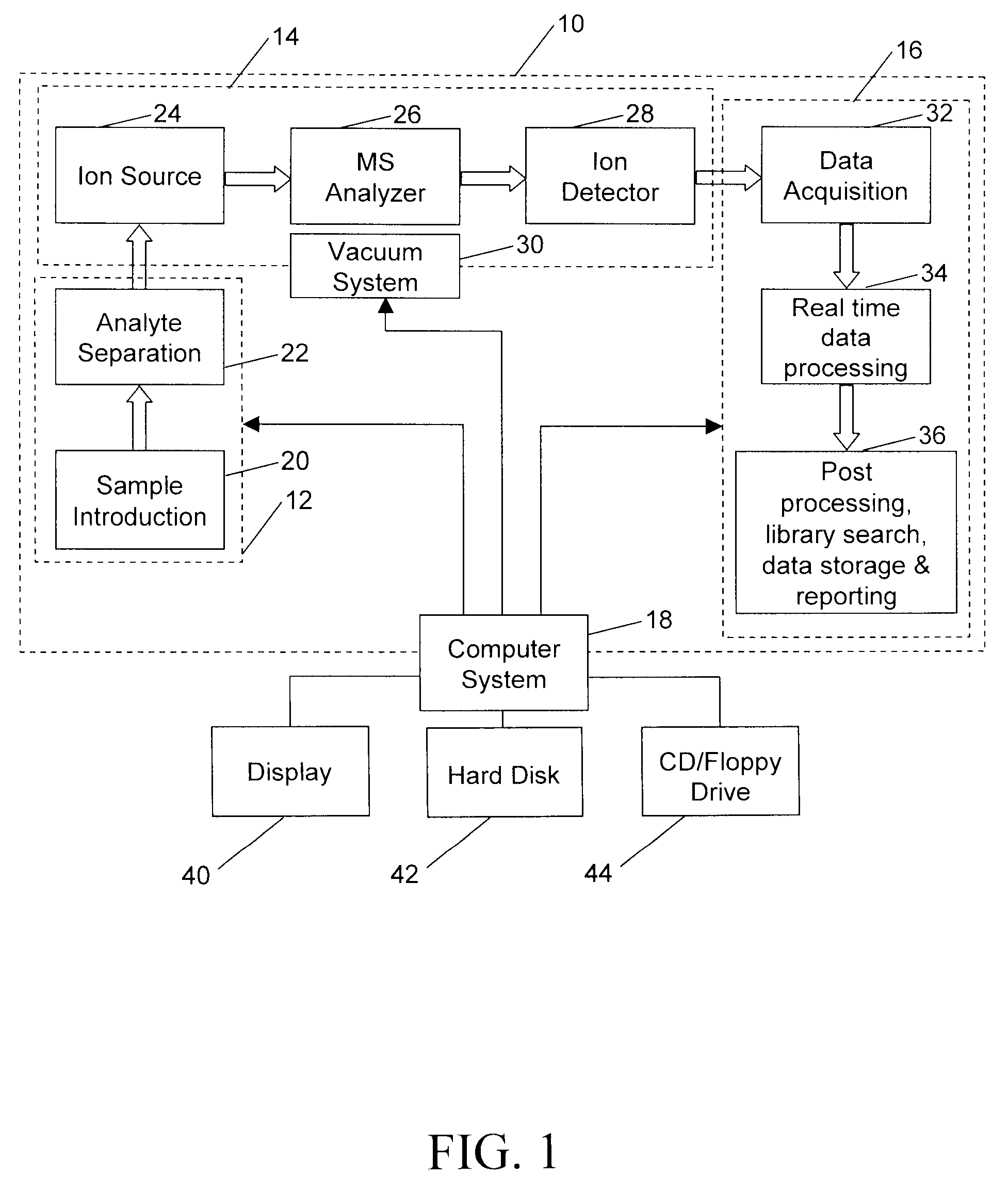
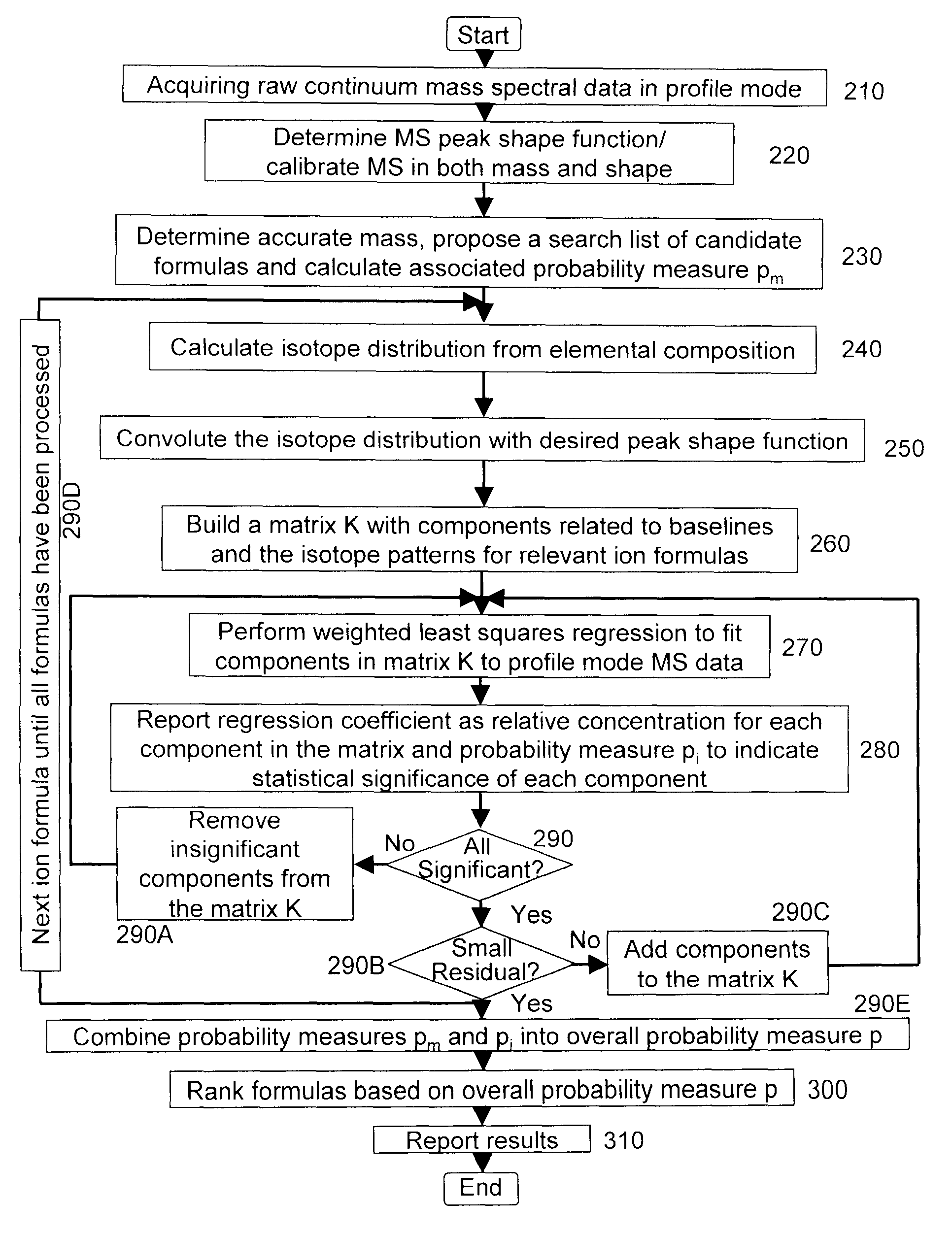
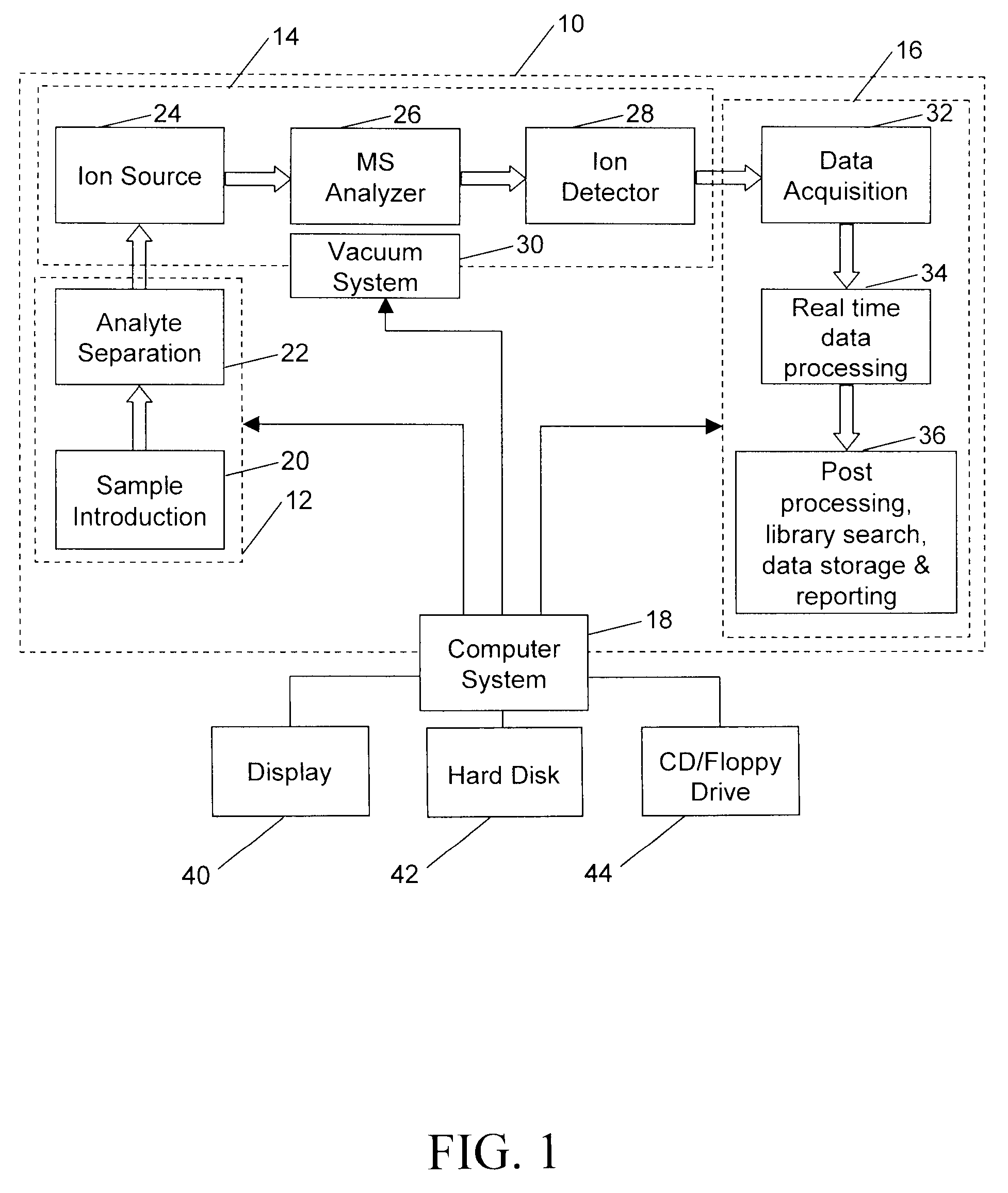
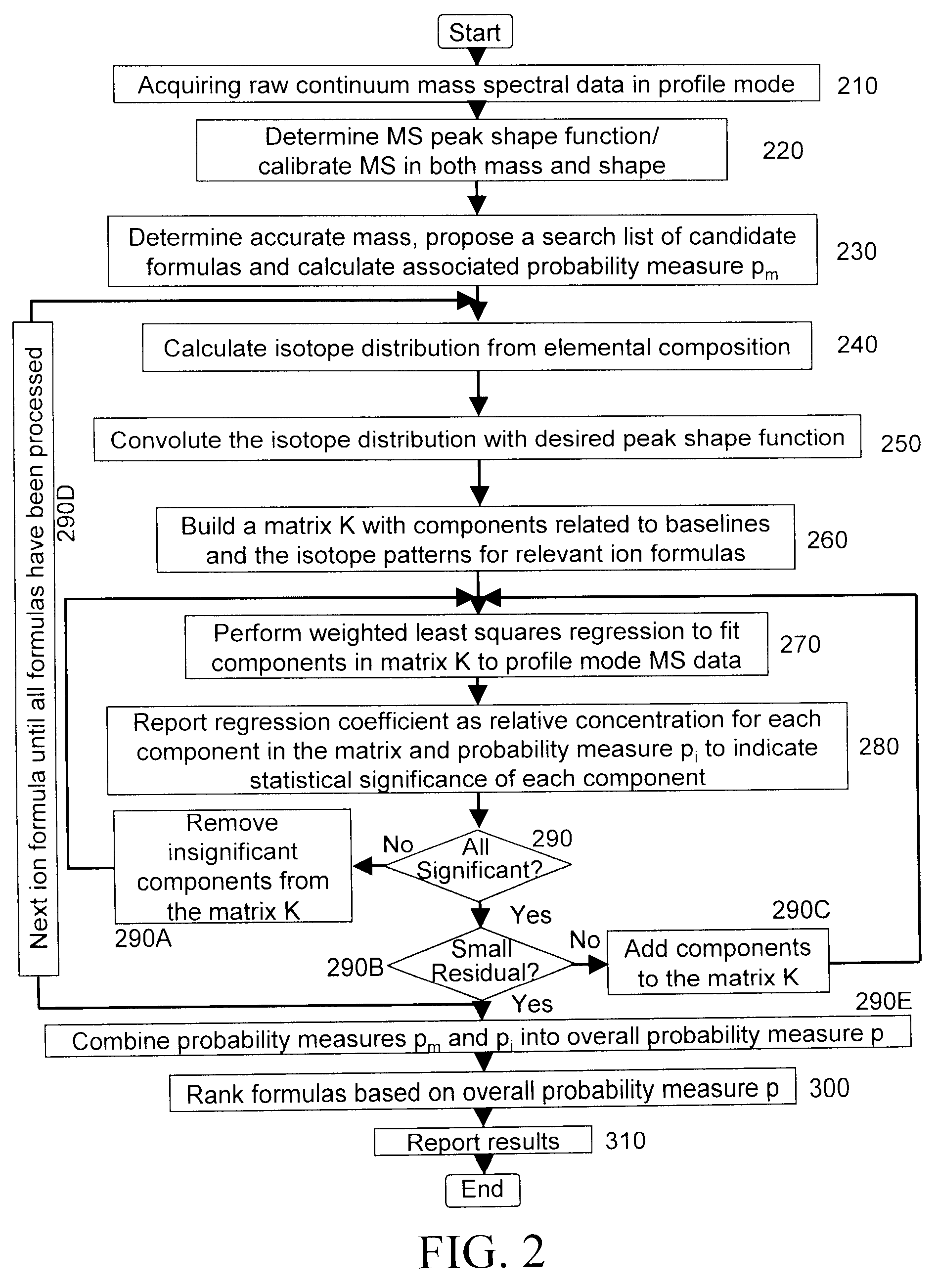
A method for calibrating and analyzing data from a mass spectrometer, comprising the steps of acquiring raw profile mode data containing mass spectral responses of ions with or without isotopes; calculating theoretical isotope distributions for each of at least one calibration ion based on elemental composition; convoluting the theoretical isotope distributions with an initial peak shape function to obtain theoretical isotope profiles for each ion; constructing a peak component matrix including the theoretical isotope profiles for calibration ions as peak components; performing a regression analysis between the raw profile mode mass spectral data and the peak component matrix; and reporting the regression coefficients as the relative concentrations for each of the components. A mass spectrometry system operated in accordance with the method and a computer readable medium having program code thereon for performing the method.
Owner:CERNO BIOSCI
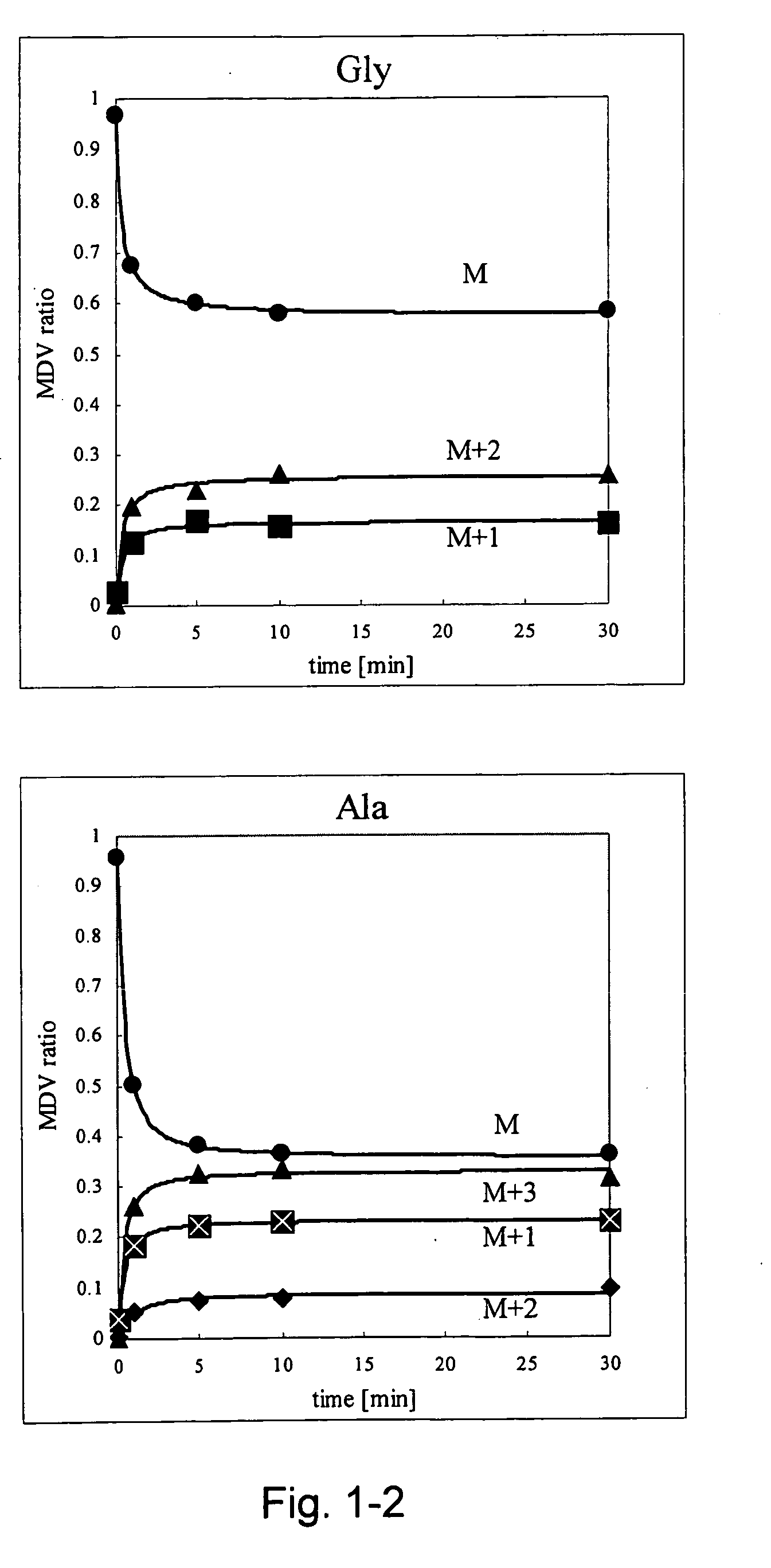
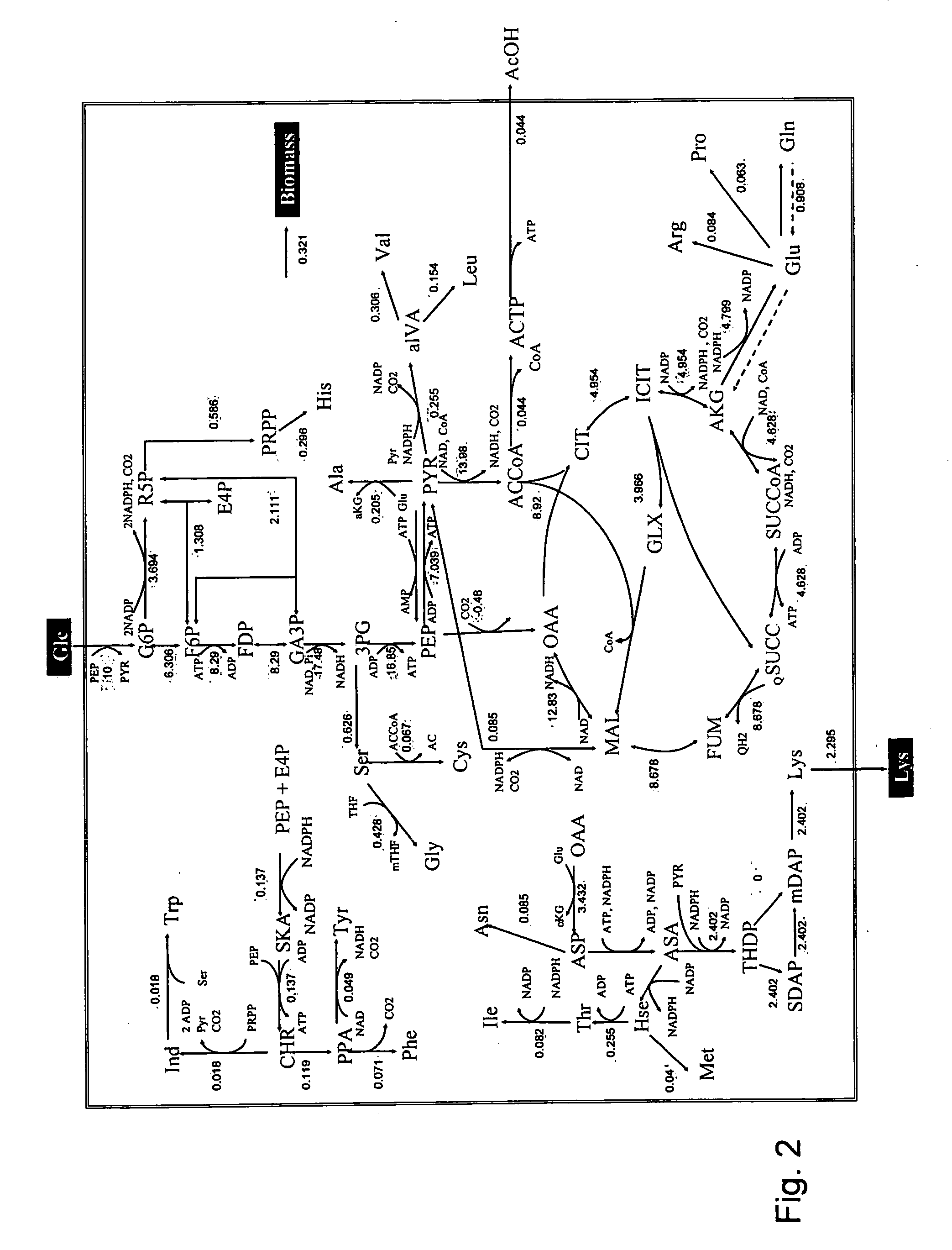
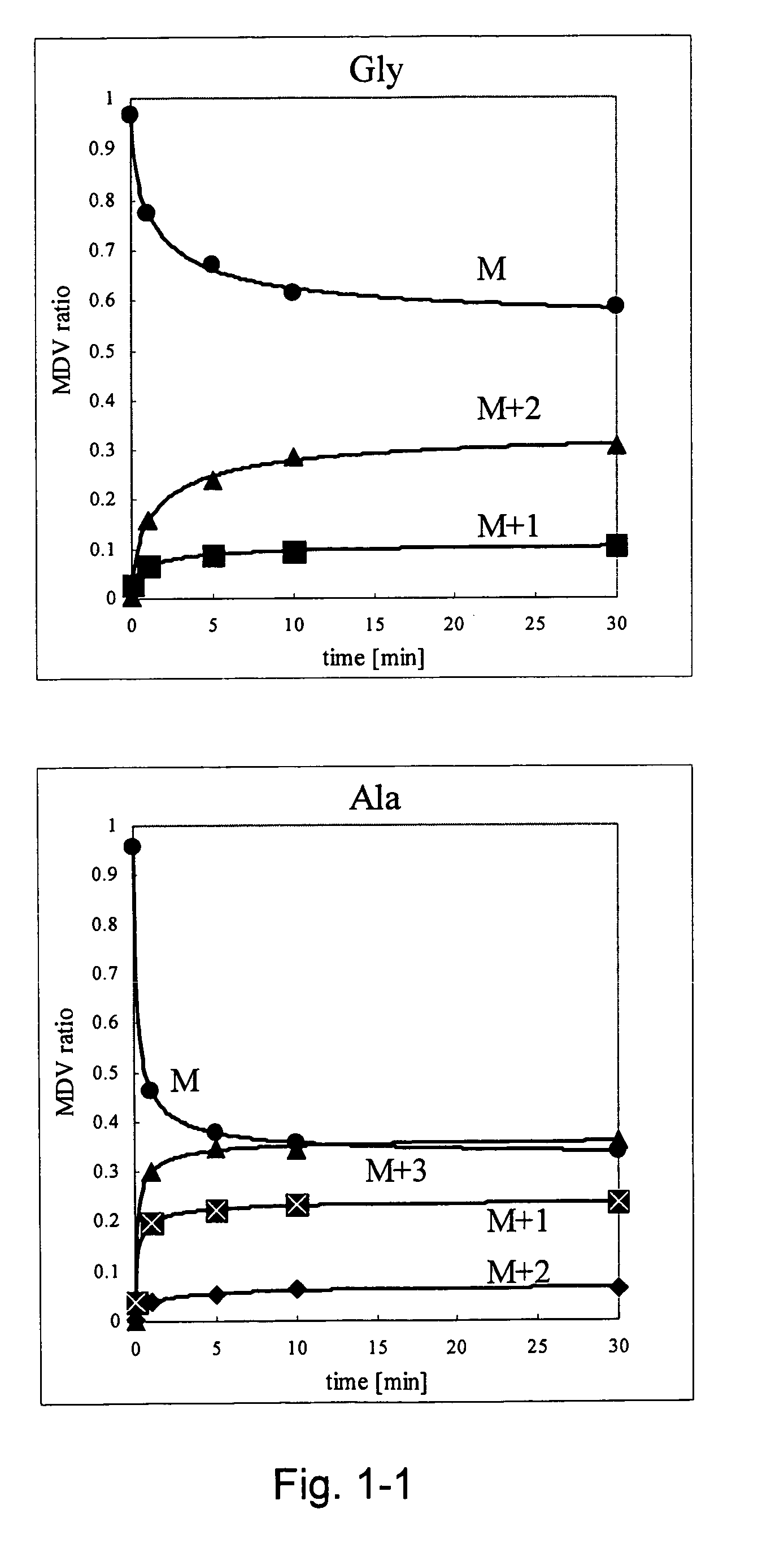
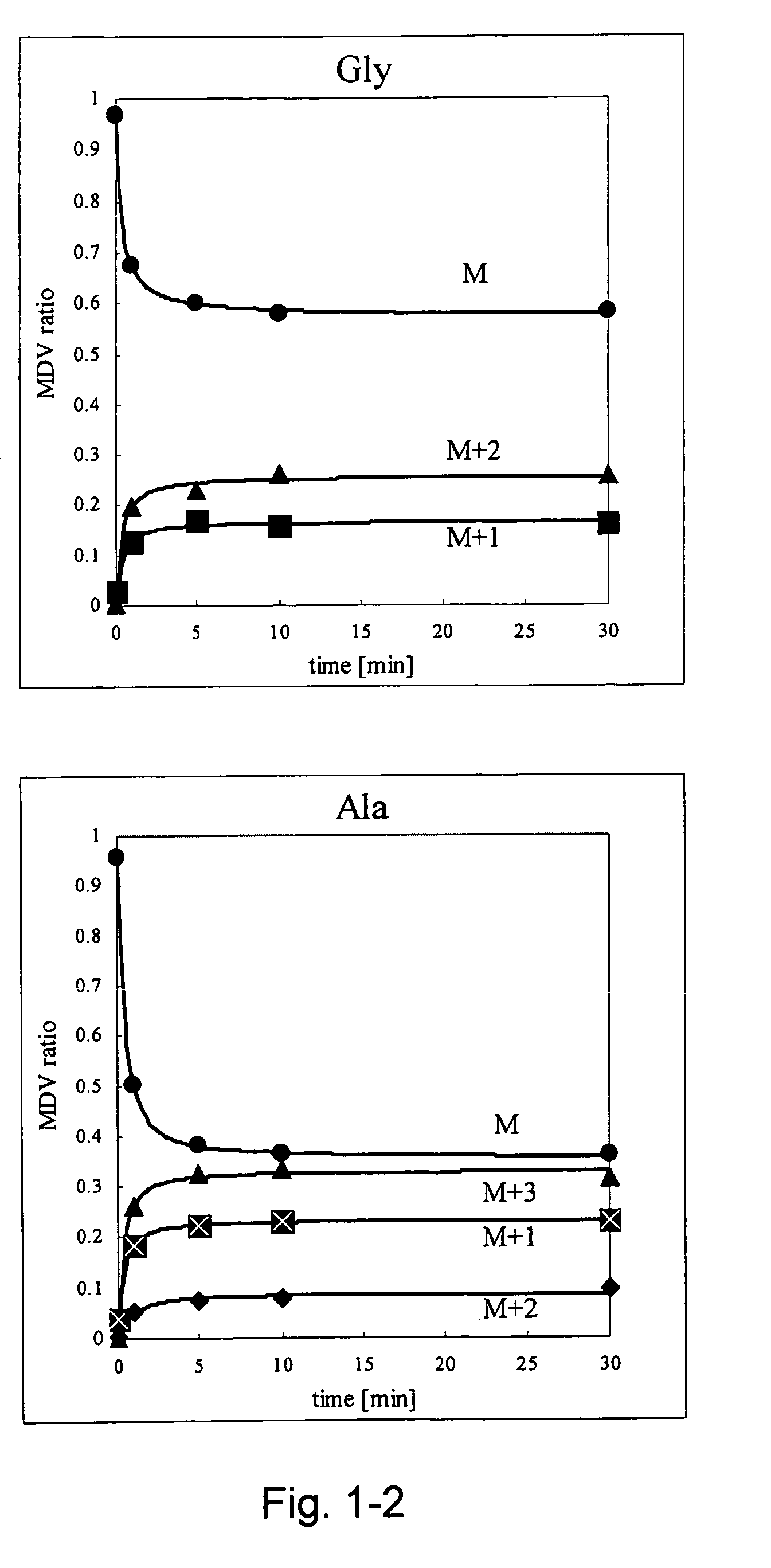
Intracellular metabolic flux analysis method using substrate labeled with isotope
InactiveUS20050175982A1Low costMicrobiological testing/measurementBiological testingDrug metabolismTime course
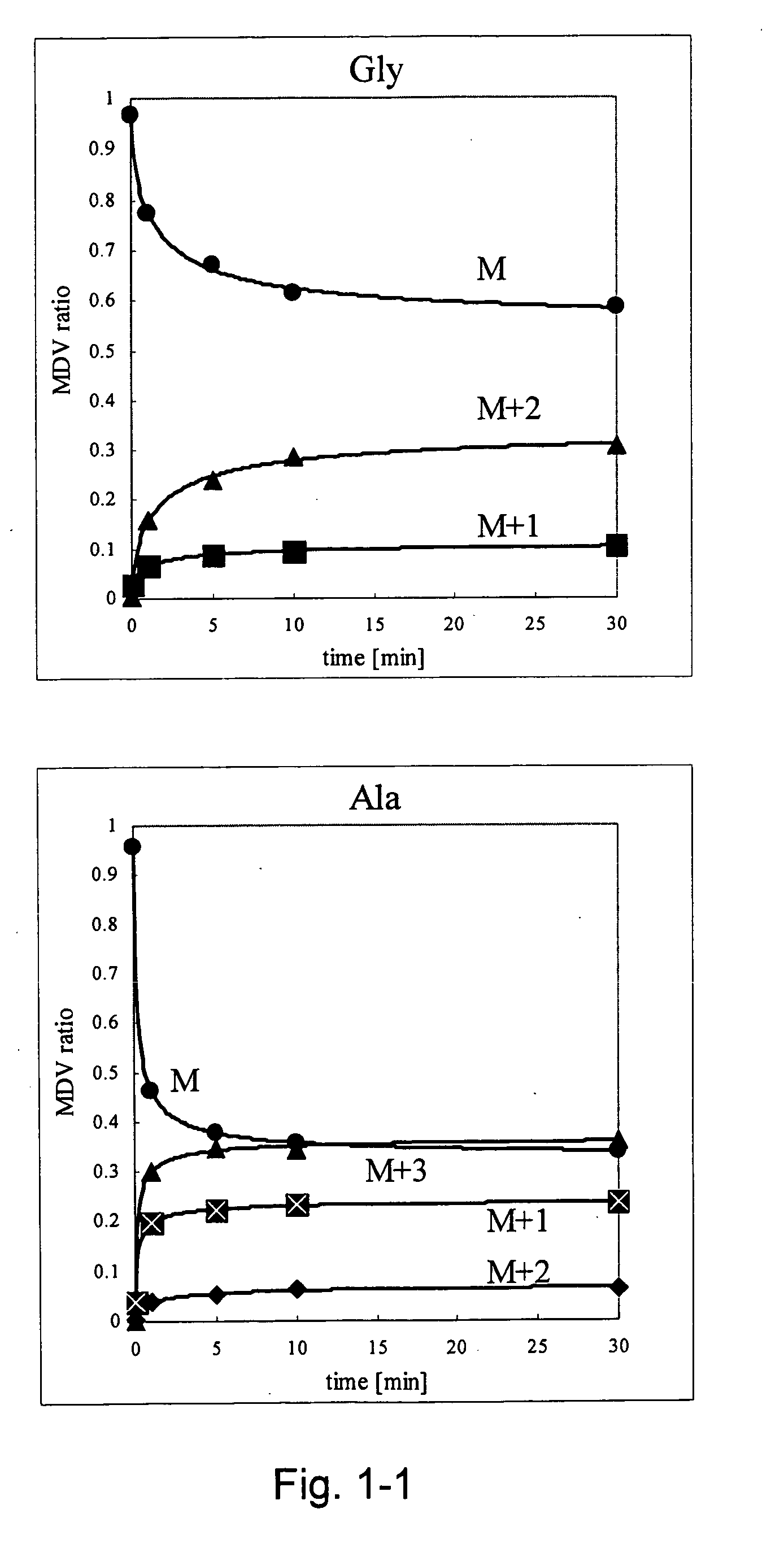
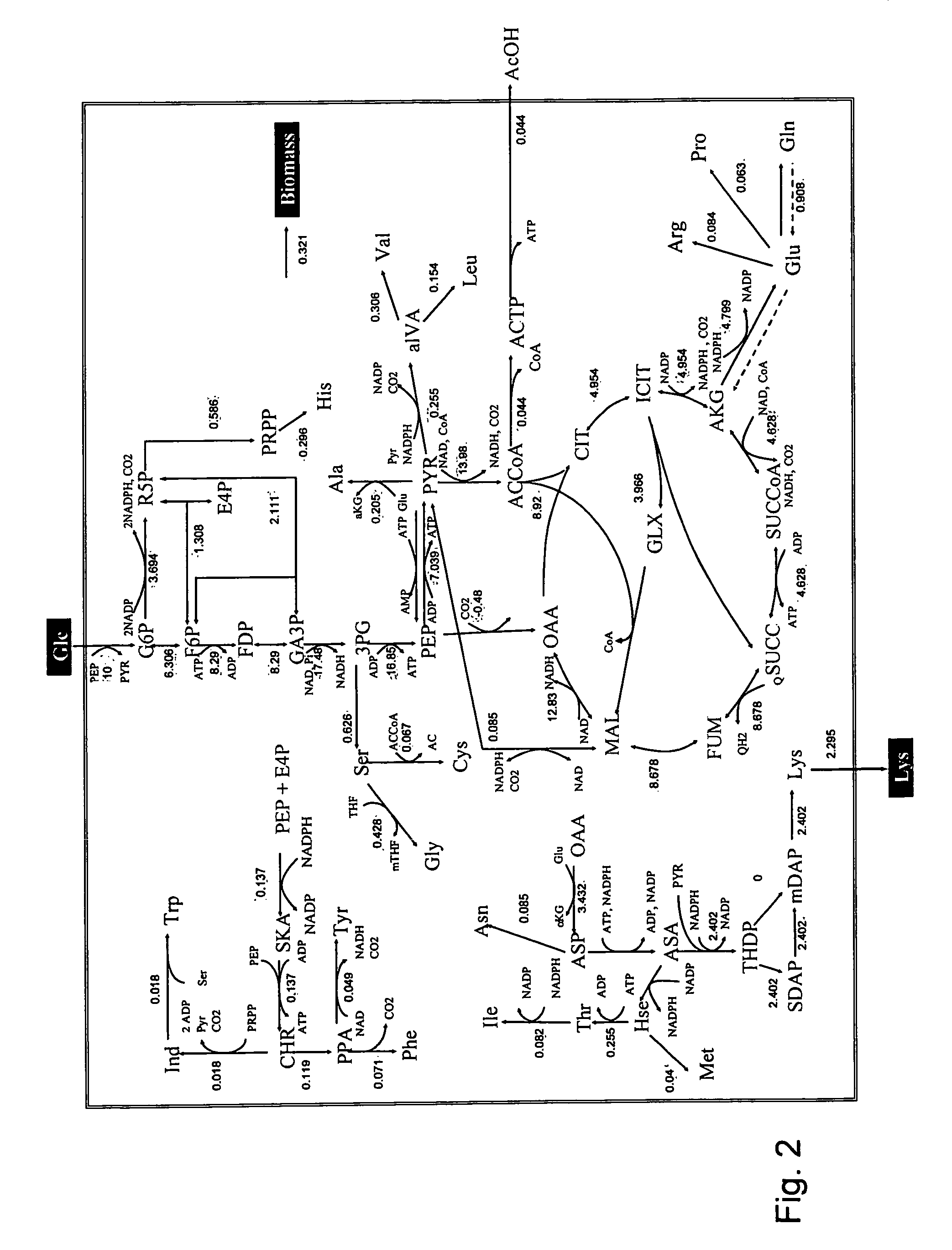
A method for intracellular metabolic flux analysis comprising (a) culturing cells in a medium not containing any isotope-labeled substrate to a target phase of the metabolic flux analysis, (b) adding an isotope-labeled substrate to the medium, and continuing culture and collecting samples from the medium in time course, (c) measuring isotope distribution in an intracellular metabolite contained in the samples collected in time course, (d) performing a regression analysis for measured data and calculating an isotope distribution ratio in a steady state, and (e) analyzing a metabolic flux of the cultured cells by using the calculated isotope distribution ratio.
Owner:AJINOMOTO CO INC
Aspects of mass spectral calibration
InactiveUS7348553B2Improve accuracyHigh resolutionIon-exchange process apparatusTime-of-flight spectrometersElemental compositionSpectral response
A method for calibrating and analyzing data from a mass spectrometer, comprising the steps of acquiring raw profile mode data containing mass spectral responses of ions with or without isotopes; calculating theoretical isotope distributions for each of at least one calibration ion based on elemental composition; convoluting the theoretical isotope distributions with an initial peak shape function to obtain theoretical isotope profiles for each ion; constructing a peak component matrix including the theoretical isotope profiles for calibration ions as peak components; performing a regression analysis between the raw profile mode mass spectral data and the peak component matrix; and reporting the regression coefficients as the relative concentrations for each of the components. A mass spectrometry system operated in accordance with the method and a computer readable medium having program code thereon for performing the method.
Owner:CERNO BIOSCI
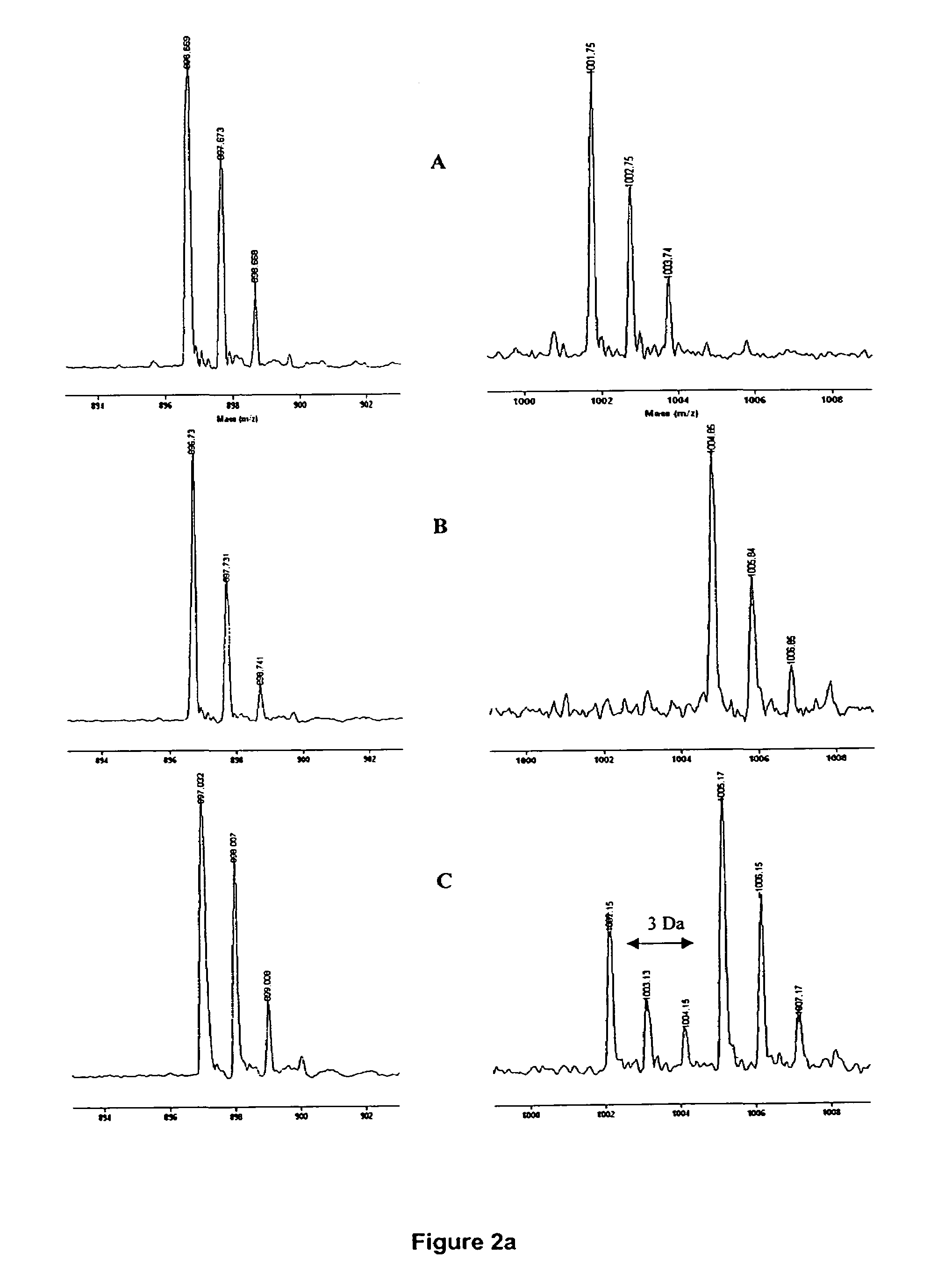
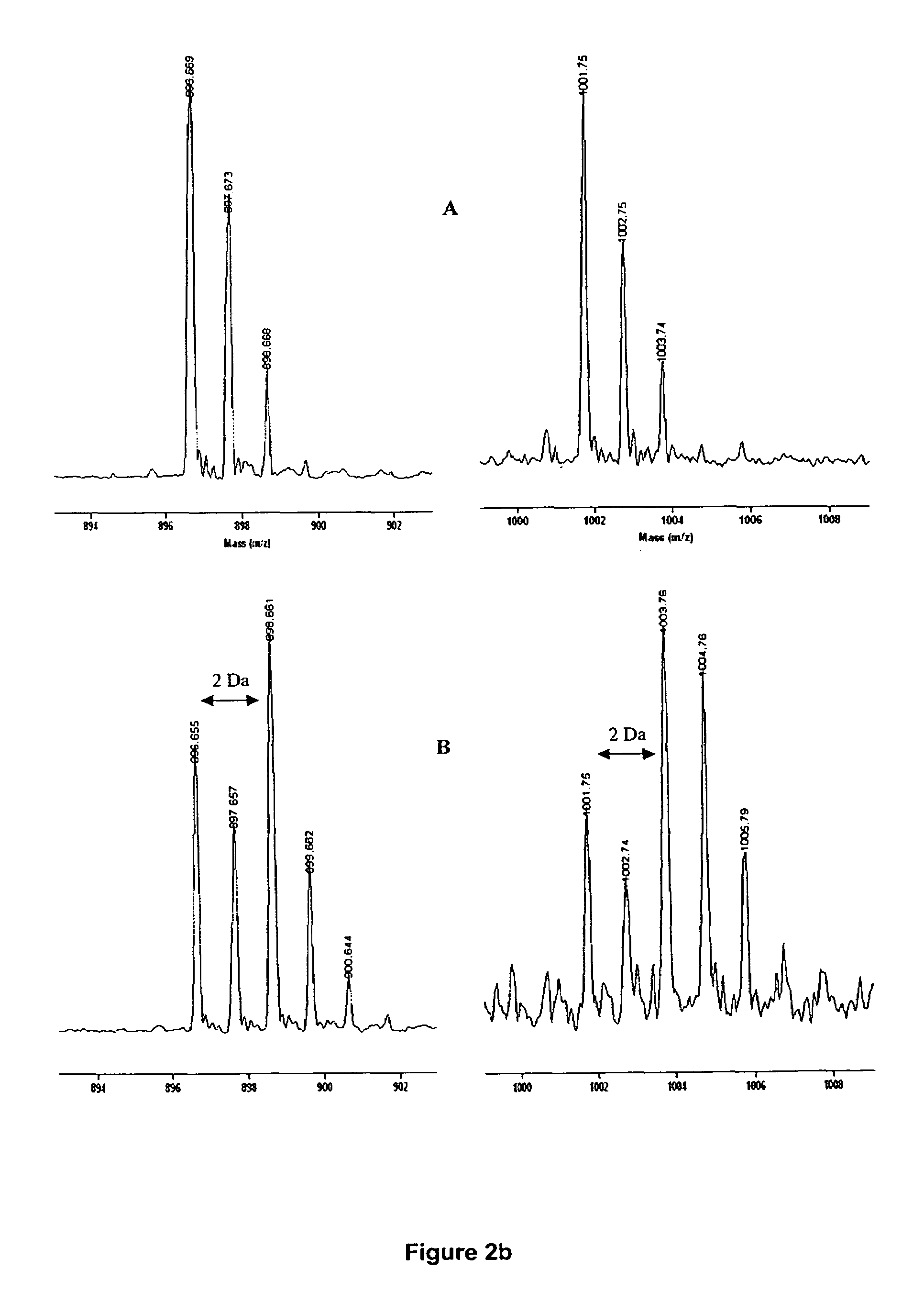
Determination of chemical composition and isotope distribution with mass spectrometry
InactiveUS20080052011A1Time-of-flight spectrometersComponent separationElemental compositionSpectral response
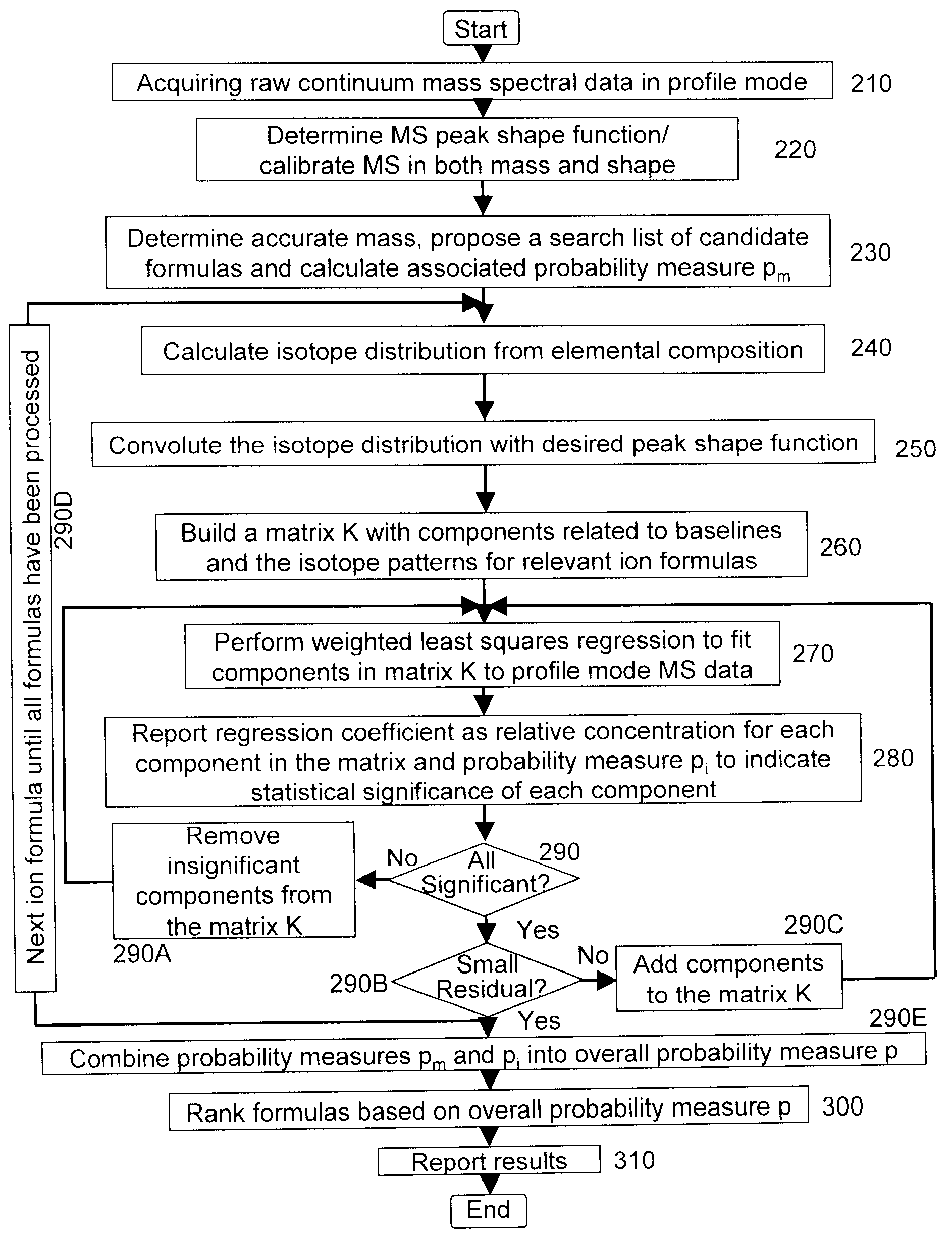
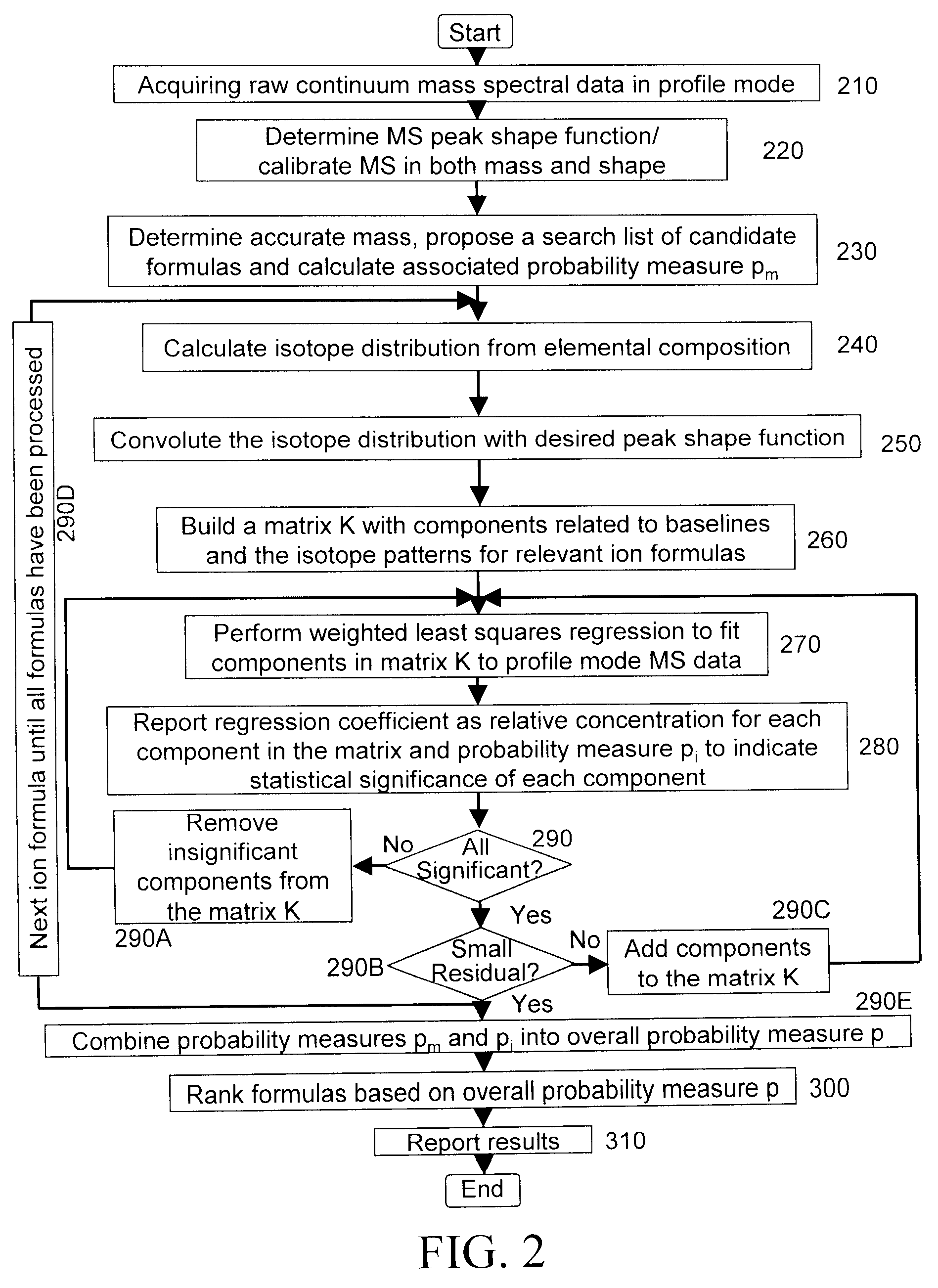
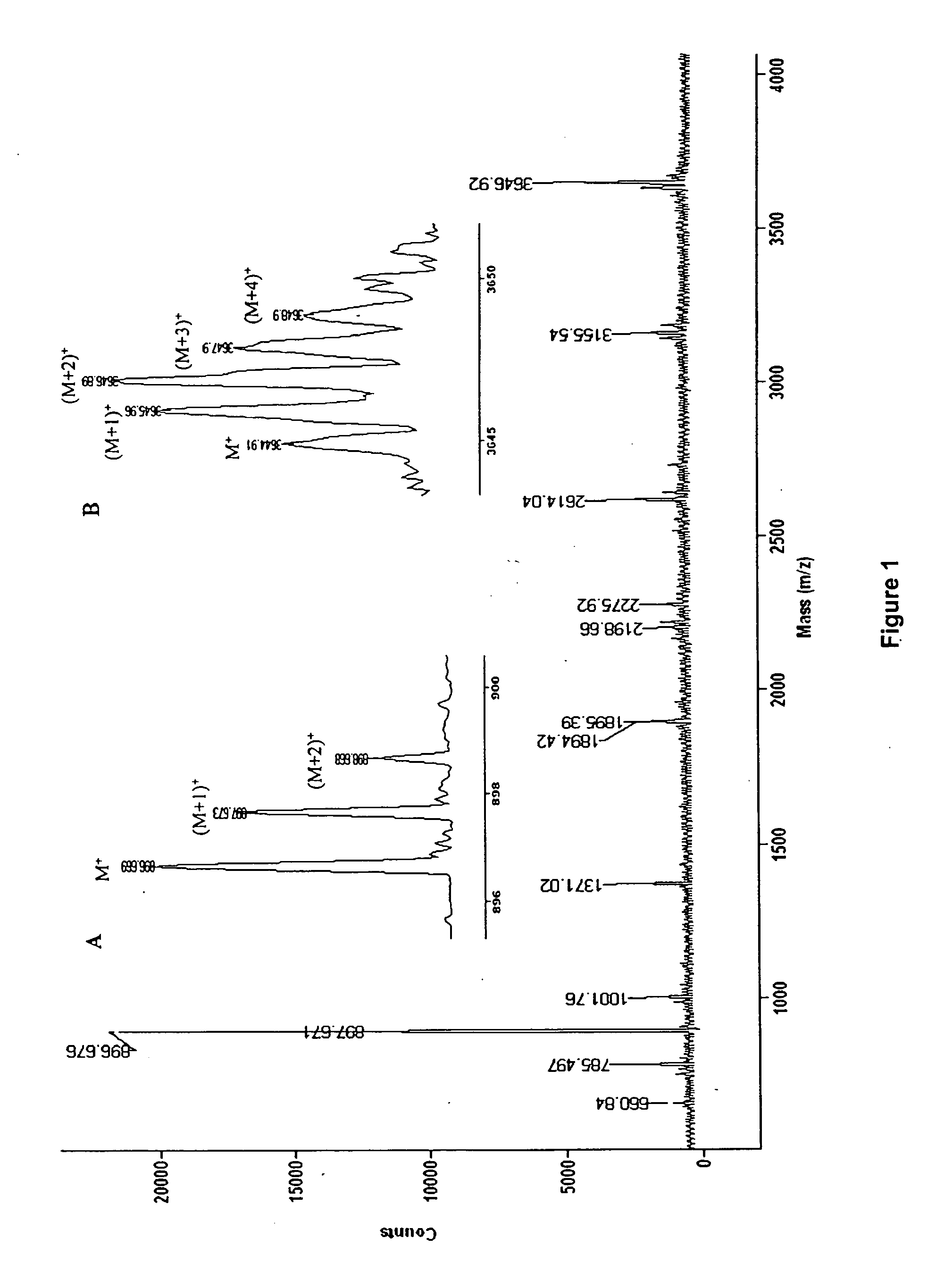
A method for determining elemental composition of ions from mass spectral data, comprising obtaining at least one mass measurement from mass spectral data; obtaining a search list of candidate elemental compositions whose exact masses fall within a given mass tolerance range from the accurate mass; reporting a probability measure based on a mass error; calculating an isotope pattern for each candidate elemental composition from the search list; constructing a peak component matrix including at least one of the isotope pattern and mass spectral data; performing a regression against at least one of isotope pattern, mass spectral data, and the peak component matrix; reporting a second probability measure for at least one candidate elemental composition based on the isotope pattern regression; and combining the two the probability measures into an overall probability measure. A method for determining elemental isotope ratios from mass spectral data, comprising obtaining measured mass spectral response; specifying the elemental composition of a given ion; specifying the initial isotope ratios for a given element in the ion; calculating the isotope pattern for the ion; constructing a peak component matrix including at least one of the isotope pattern and measured mass spectral response; performing a regression between measured mass spectral response and the peak component matrix; and reporting a regression residual and repeating the isotope pattern calculation, peak component construction, and regression process with updated isotope ratios to minimize this residual.
Owner:CERNO BIOSCI
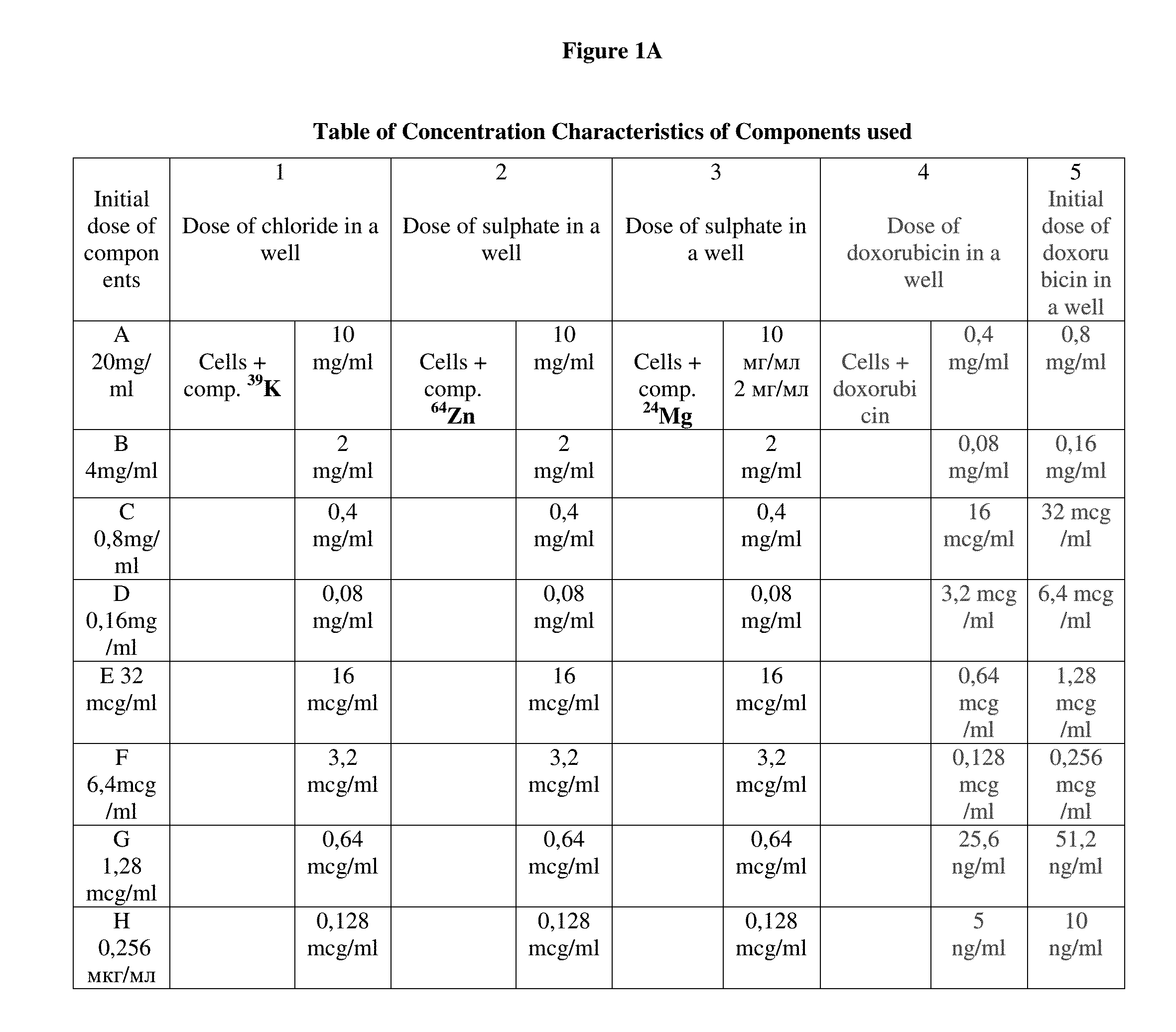
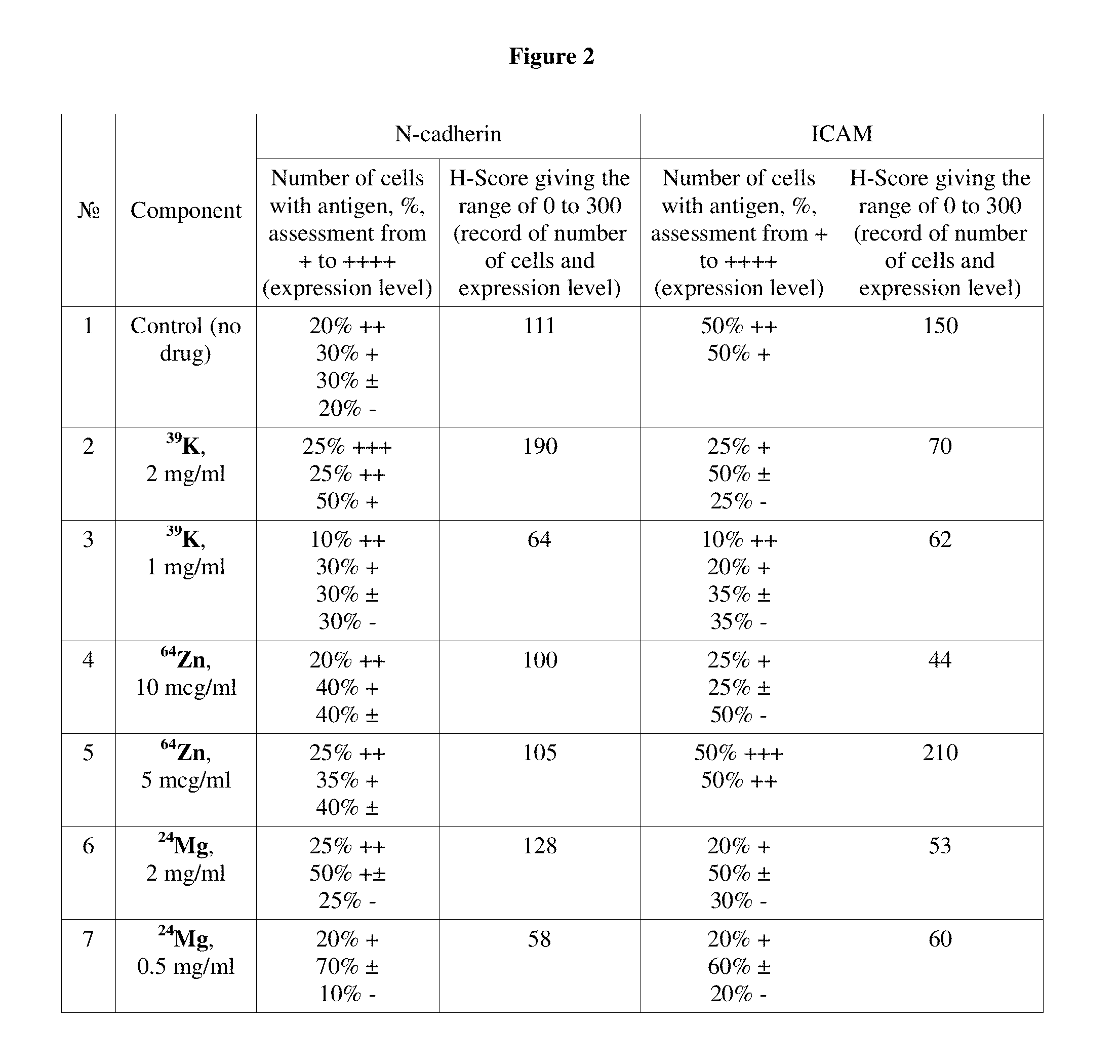
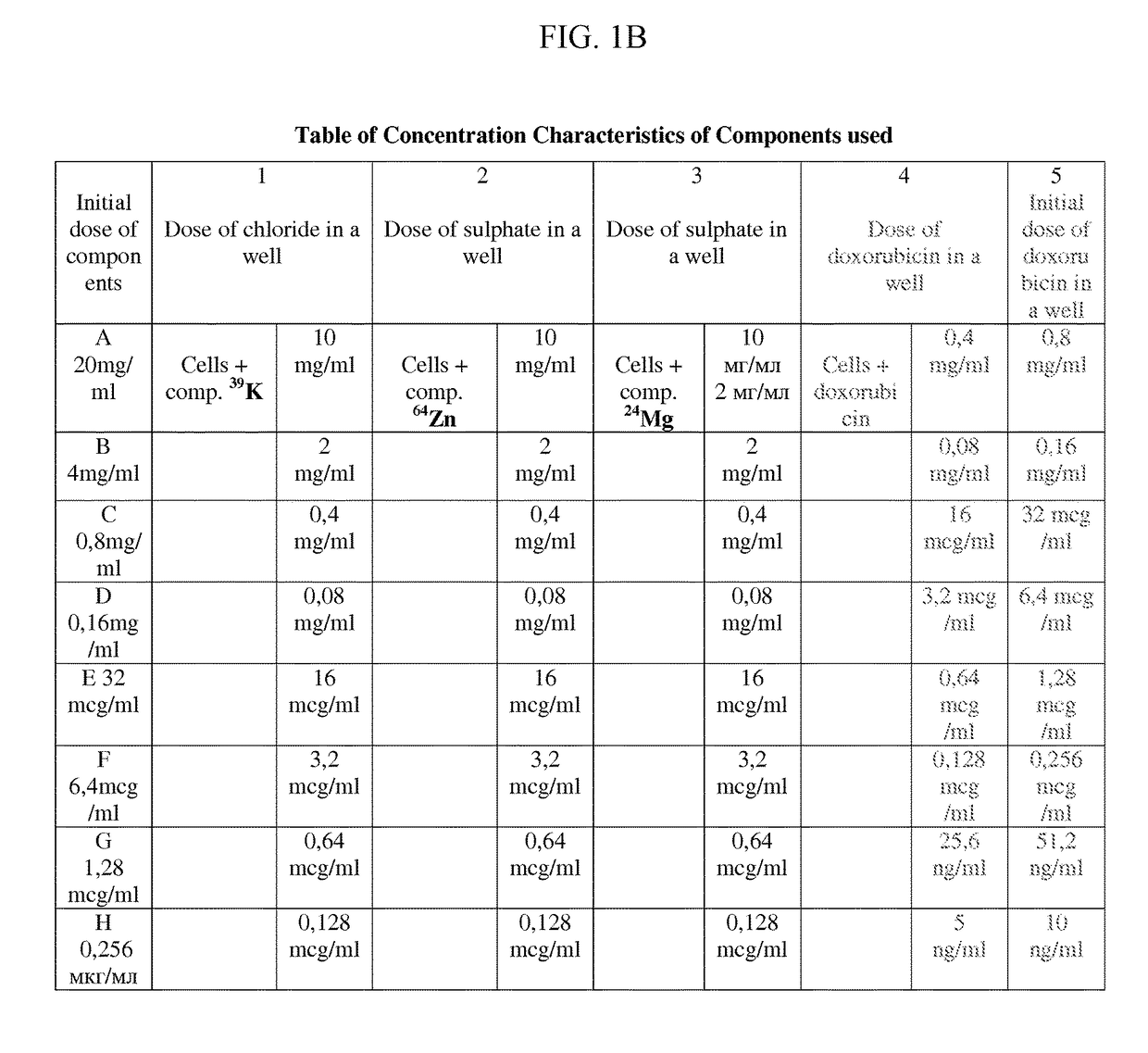
Pharmaceutical composition for improving health, cure abnormalities and degenerative disease, achieve Anti-aging effect of therapy and therapeutic effect on mammals and method thereof
ActiveUS20160151415A1Good for healthAchieve therapeutic effectHeavy metal active ingredientsOrganic active ingredientsMedicineAdditive ingredient
A pharmaceutical composition of the present invention is used for improving health, cure abnormalities and degenerative disease; achieve anti-aging effect of therapy and therapeutic effect on mammals. The pharmaceutical composition includes a pharmaceutical carrier and an isotope selective ingredient including at least one of a chemical element and a chemical compound containing the chemical element whereby isotope distribution in the at least one of the chemical element and the chemical compound containing the chemical element is different from natural distribution of at least one of isotopes wherein the part of selected isotope of the chemical element ranges from 0 to 100%. A method of the present invention uses the inventive pharmaceutical composition to improve health, cure abnormalities and degenerative disease and achieve therapeutic effect on mammals.
Owner:VECTOR VITALE IP LLC
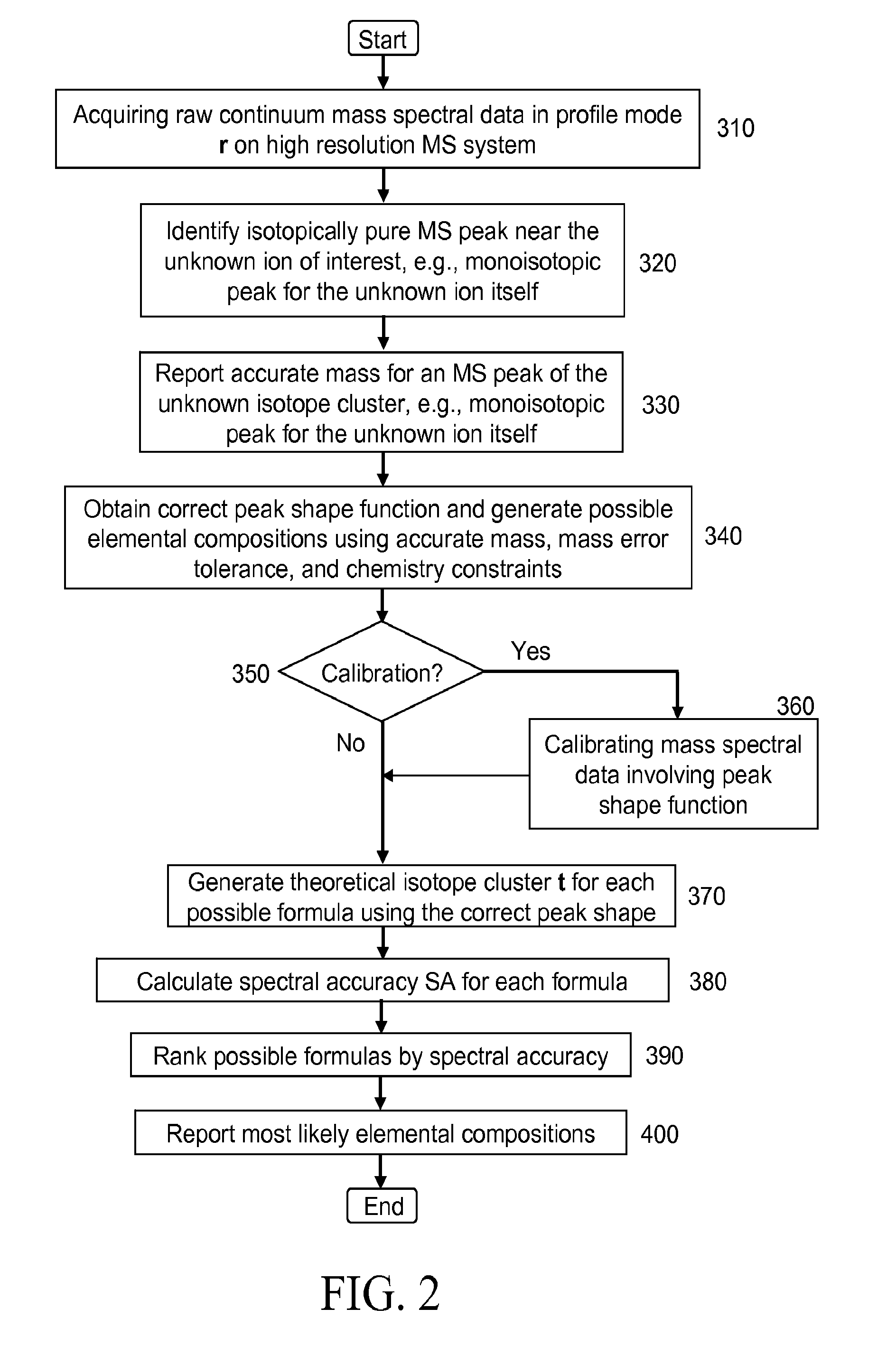
Self calibration approach for mass spectrometry
InactiveUS20100171032A1Stability-of-path spectrometersTime-of-flight spectrometersElemental compositionMass Spectrometry-Mass Spectrometry
Methods for analyzing mass spectral data, include acquiring profile mode mass spectral data containing at least on ion of interest whose elemental composition is determined; obtaining a correct peak shape function based on the actually measured peak shape of at least one of the isotypes of the same ion of interest; generating at least one possible elemental composition for the ion of interest; calculating a theoretical isotope cluster by applying correct peak shape function to the theoretical isotope distribution; comparing quantiatively the corresponding parts of the theoretical isotope cluster to that from acquired profile mode mass spectral data to obtain at least one of elemental composition determination, classification, or quantitation for the ion. A computer for and a computer readable medium having computer readable code thereon for performing the methods. A mass spectrometer having an associated computer for performing the methods.
Owner:CERNO BIOSCI
Pharmaceutical composition for improving health, cure abnormalities and degenerative disease, achieve anti-aging effect of therapy and therapeutic effect on mammals and method thereof
ActiveUS9861659B2Good for healthCure abnormalityOrganic active ingredientsInorganic active ingredientsMammalChemical compound
A pharmaceutical composition of the present invention is used for improving health, cure abnormalities and degenerative disease; achieve anti-aging effect of therapy and therapeutic effect on mammals. The pharmaceutical composition includes a pharmaceutical carrier and an isotope selective ingredient including at least one of a chemical element and a chemical compound containing the chemical element whereby isotope distribution in the at least one of the chemical element and the chemical compound containing the chemical element is different from natural distribution of at least one of isotopes wherein the part of selected isotope of the chemical element ranges from 0 to 100%. A method of the present invention uses the inventive pharmaceutical composition to improve health, cure abnormalities and degenerative disease and achieve therapeutic effect on mammals.
Owner:VECTOR VITALE IP LLC
Pharmaceutical composition for improving health, cure abnormalities and degenerative disease, achieve anti-aging effect of therapy and therapeutic effect on mammals and method thereof
ActiveUS10226484B2Good for healthCure abnormalityOrganic active ingredientsAluminium/calcium/magnesium active ingredientsMedicineAdditive ingredient
A pharmaceutical composition of the present invention is used for improving health, cure abnormalities and degenerative disease; achieve anti-aging effect of therapy and therapeutic effect on mammals. The pharmaceutical composition includes a pharmaceutical carrier and an isotope selective ingredient including at least one of a chemical element and a chemical compound containing the chemical element whereby isotope distribution in the at least one of the chemical element and the chemical compound containing the chemical element is different from natural distribution of at least one of isotopes wherein the part of selected isotope of the chemical element ranges from 0 to 100%. A method of the present invention uses the inventive pharmaceutical composition to improve health, cure abnormalities and degenerative disease and achieve therapeutic effect on mammals.
Owner:VECTOR VITALE IP LLC
Label-free protein quantification method based on mass spectrum
ActiveCN107328842AImprove detection coverageHigh throughput quantificationMaterial analysis by electric/magnetic meansDiseaseEnzyme digestion
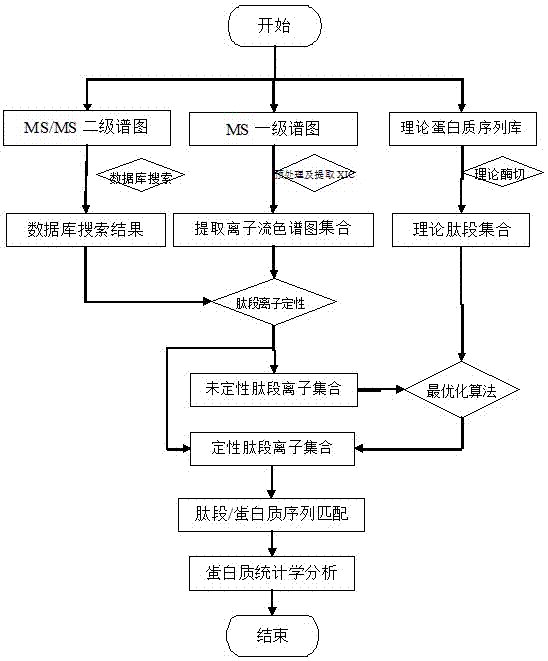
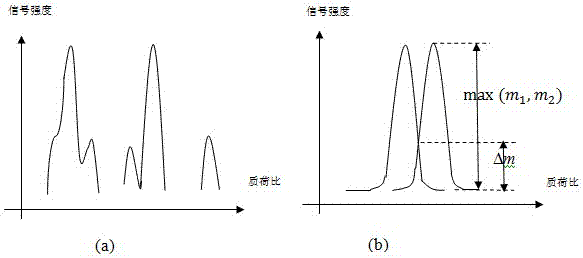
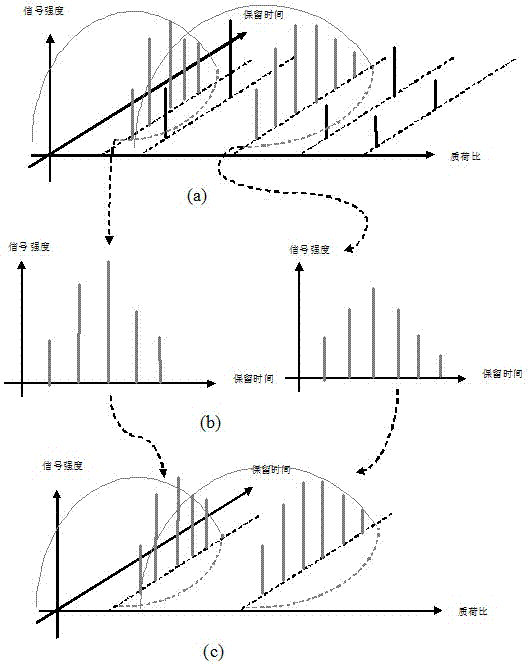
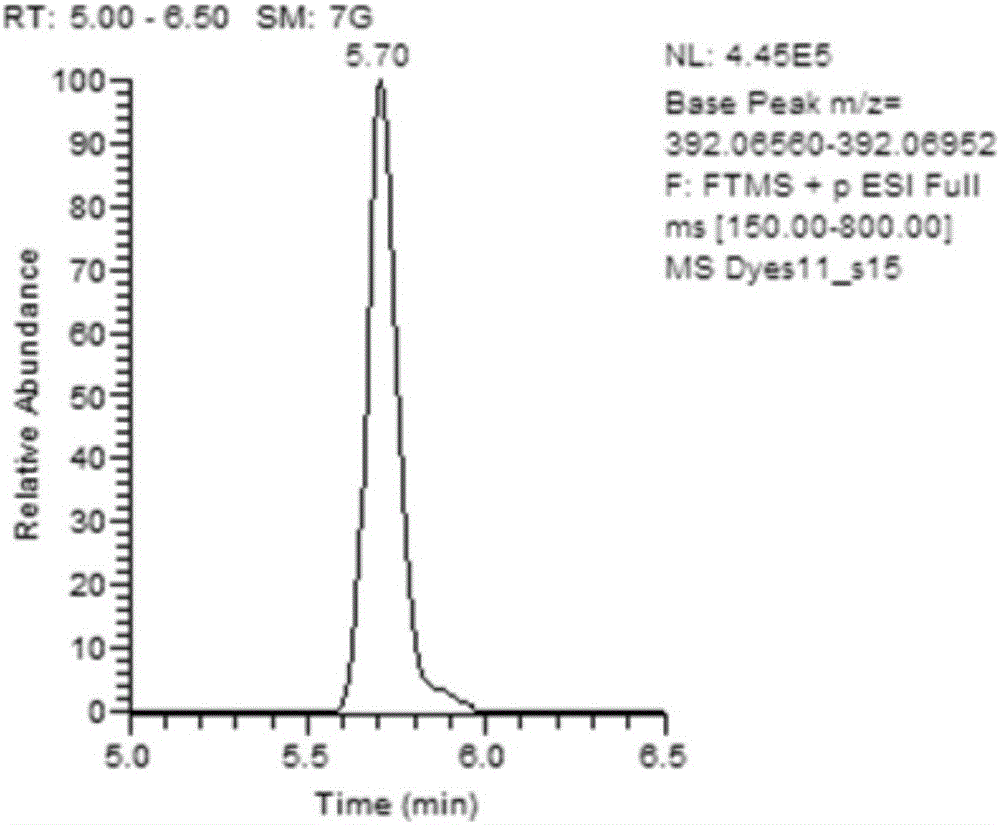
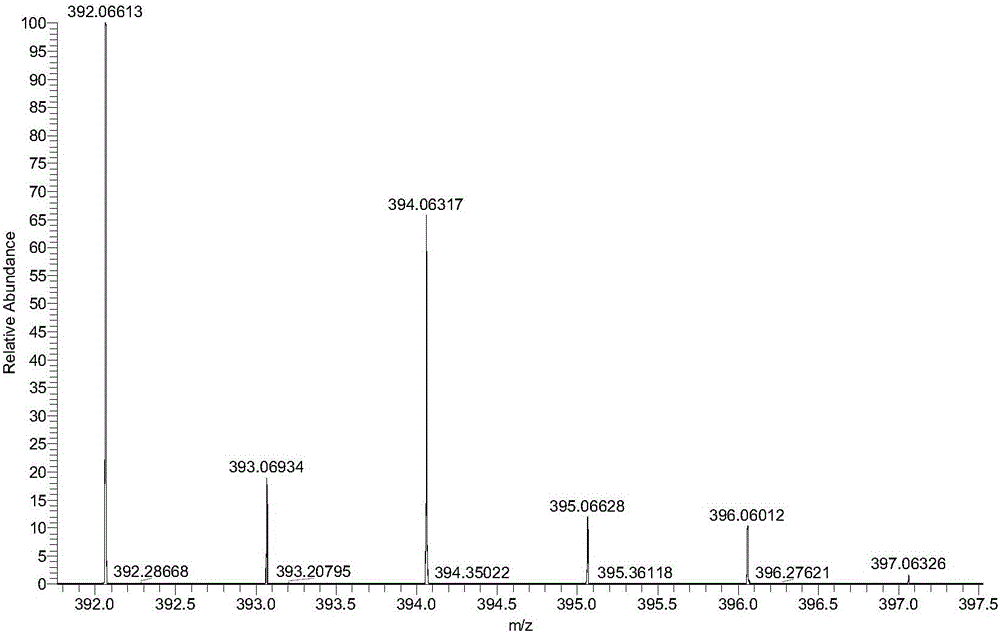
The invention discloses a label-free protein quantification method based on a mass spectrum. The label-free protein quantification method comprises the following steps: acquiring corresponding theoretical peptide fragment set by theoretical enzyme digestion of a theoretical protein sequence data bank; extracting extracted-ion-flow chromatograms (XIC) of all ions on a primary mass spectrum (MS) and calculating corresponding retention time (RS) and mass-to-charge ratios (M / Z) on the extracted-ion-flow chromatograms; establishing a multi-mapping relation table about isotopic distribution and retention time of the theoretical peptide fragment set and the extracted-ion-flow chromatograms. According to the method, a label-free protein quantification algorithm combined with an intelligent optimization algorithm is adopted, the optimal mapping relation between the theoretical peptide fragment set and the extracted-ion-flow chromatograms is obtained from the multi-mapping relation table by means of the intelligent optimization algorithm, and a quantification result is obtained. By means of efficient qualitative analysis of peptide fragments, higher protein detection coverage can be realized, so that high-throughput quantification of proteins is realized. Research proves that change of expressive abundance of the proteins in cells has great significance in early diagnosis of diseases.
Owner:EAST CHINA NORMAL UNIVERSITY
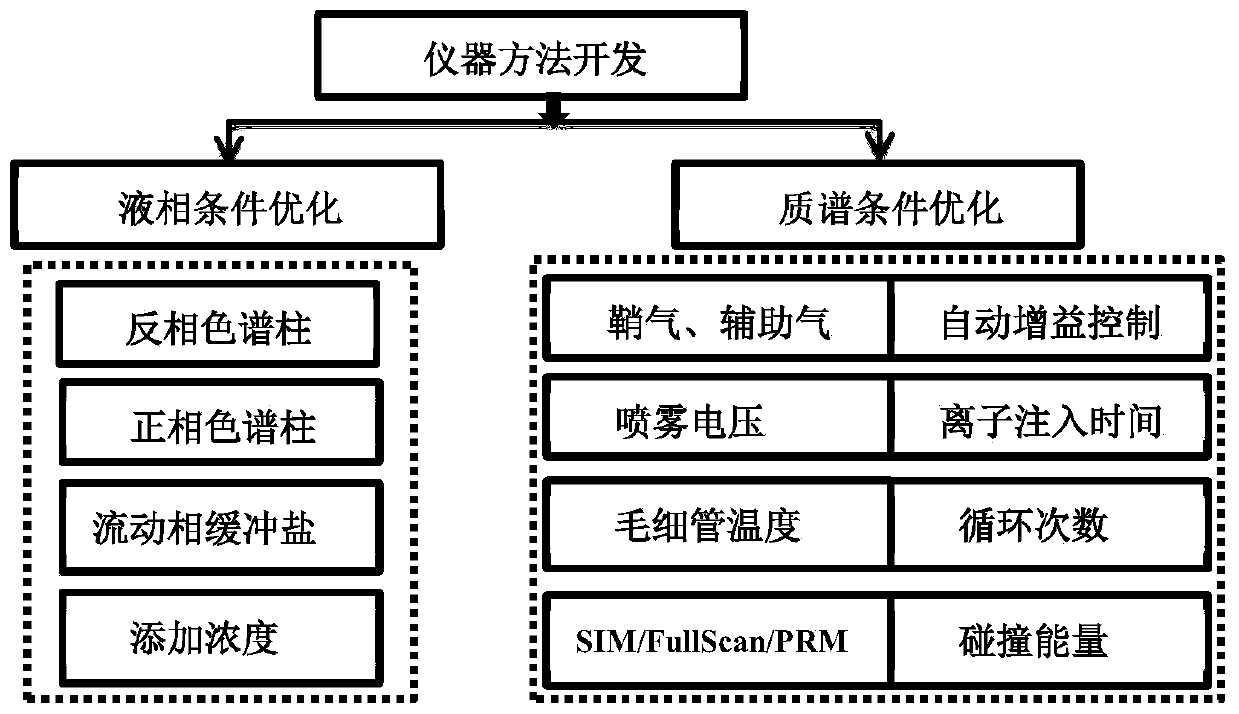
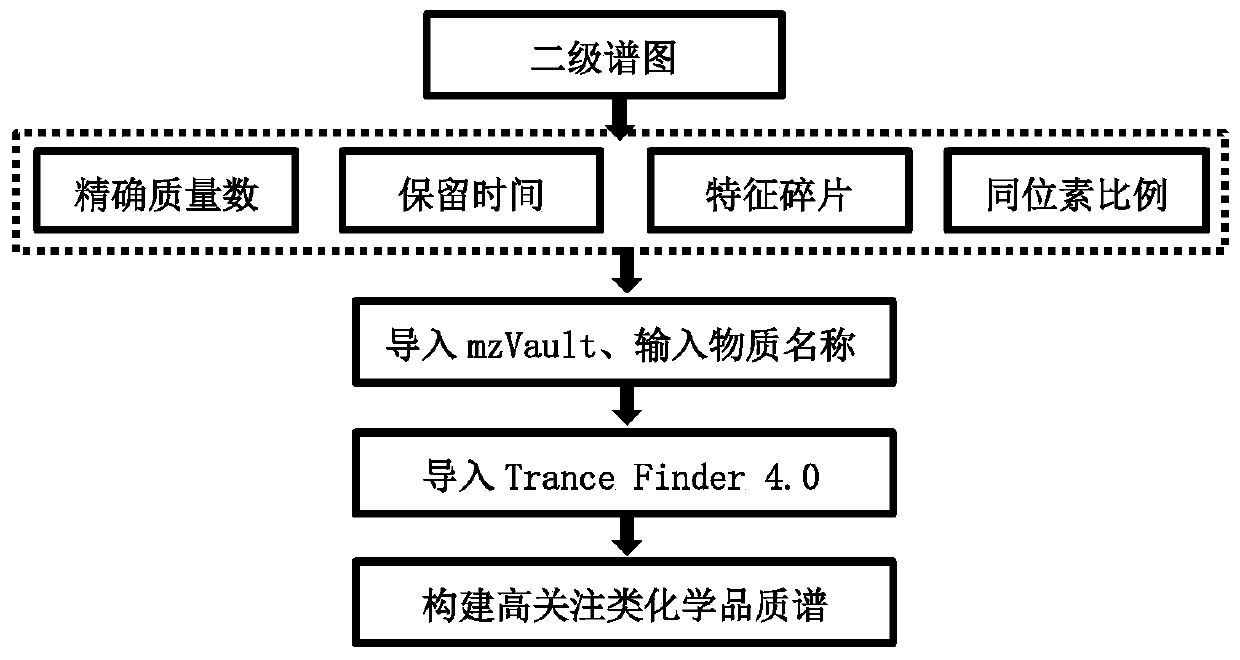
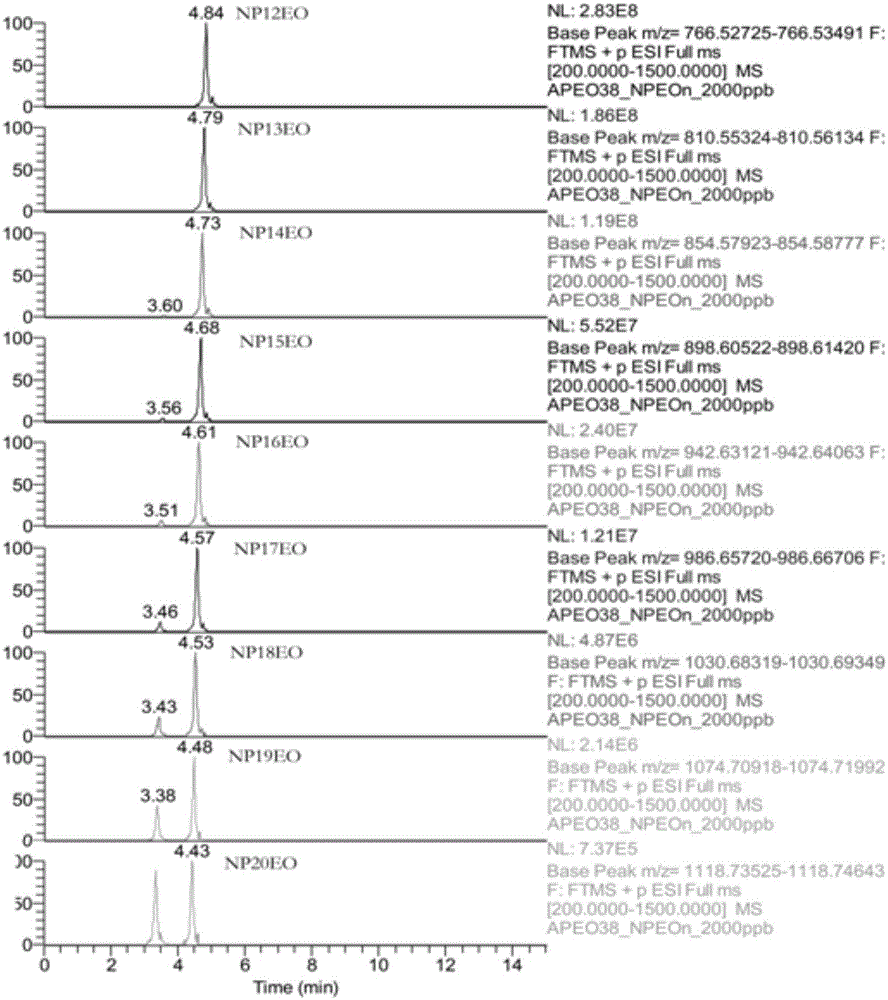
Method for rapidly screening highly-concerned chemicals in surface water and soil
PendingCN111380983ARich varietyAchieve accurate determinismComponent separationElectric vehicle charging technologySoil scienceFluid phase
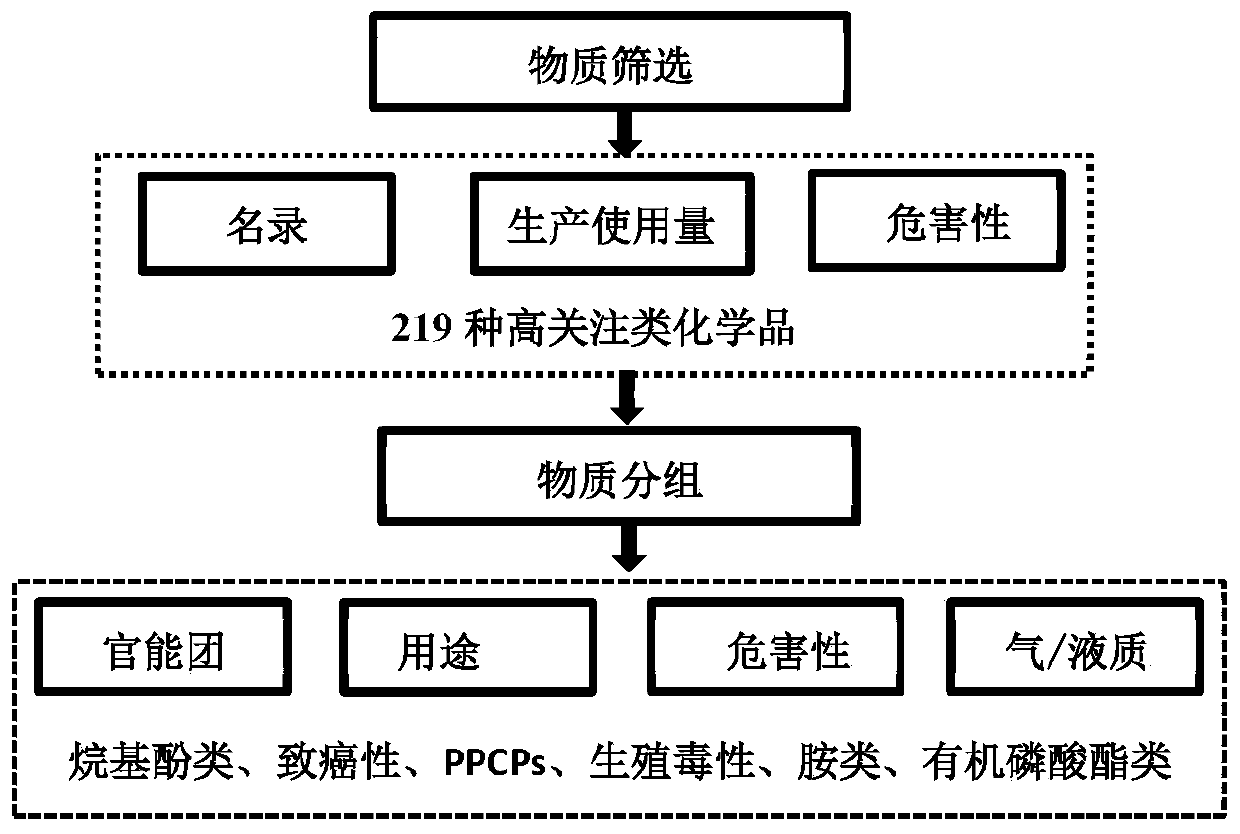
The invention discloses a method for rapidly screening highly-concerned chemicals in surface water and soil. The method comprises the steps of screening a highly-concerned chemical list, grouping substances, optimizing mass spectrum conditions and chromatographic conditions, constructing a mass spectrum library and rapidly screening the highly-concerned chemicals. According to the method, samplesin surface water and soil are rapidly screened by adopting ultra-high performance liquid chromatography-quadrupole rod / electrostatic field orbitrap high-resolution mass spectrum, and based on accuratemass number, retention time, characteristic fragments and isotope distribution proportion, the samples are searched and matched with a mass spectrum library of highly-concerned chemicals for rapid qualitative analysis. A screening result can be obtained within 70min, the sample detection and treatment time is greatly shortened, the detection efficiency is improved, and technical support is provided for risk assessment and risk management of highly-concerned chemicals.
Owner:NANJING INST OF ENVIRONMENTAL SCI MINIST OF ECOLOGY & ENVIRONMENT OF THE PEOPLES REPUBLIC OF CHINA
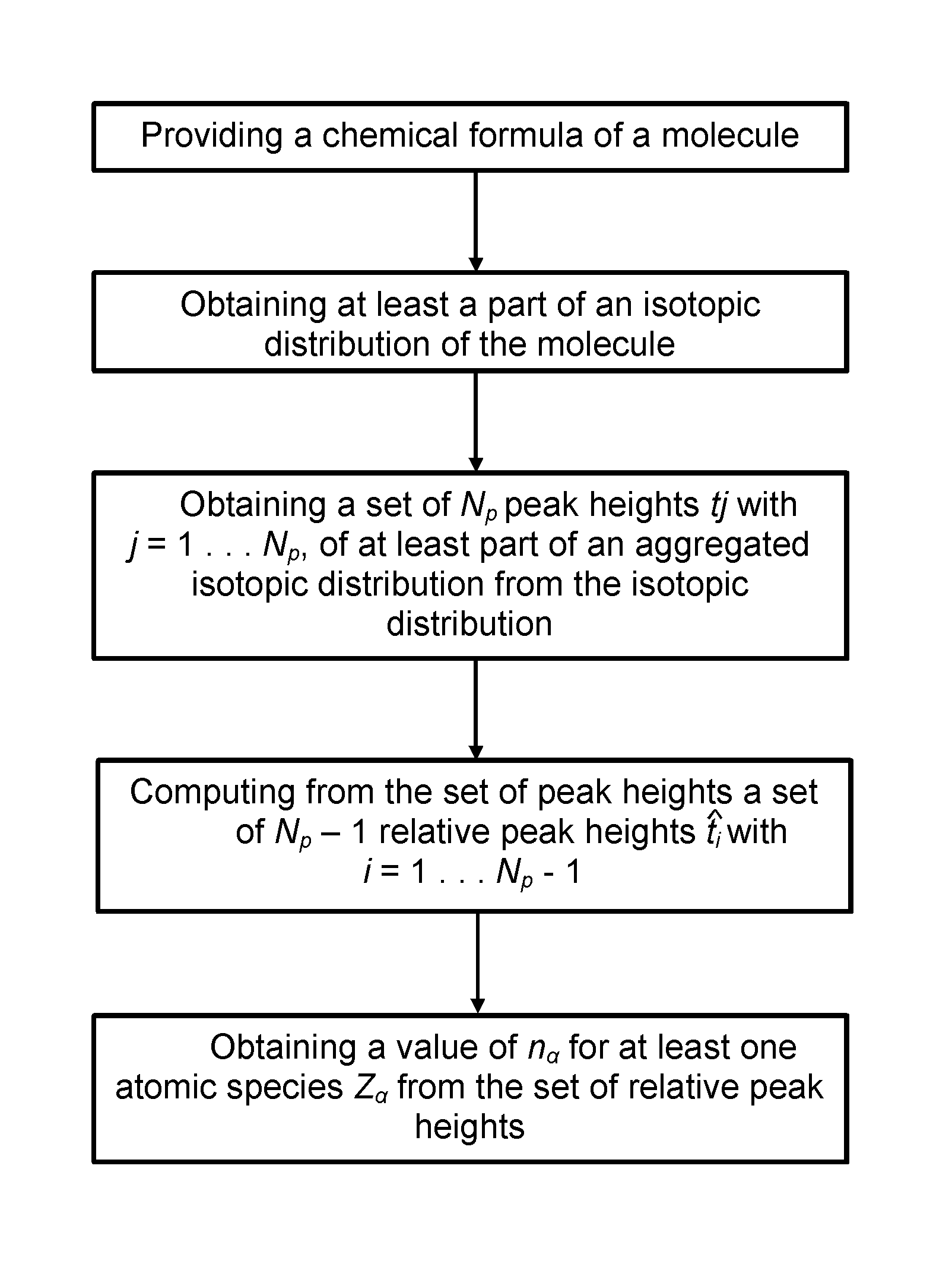
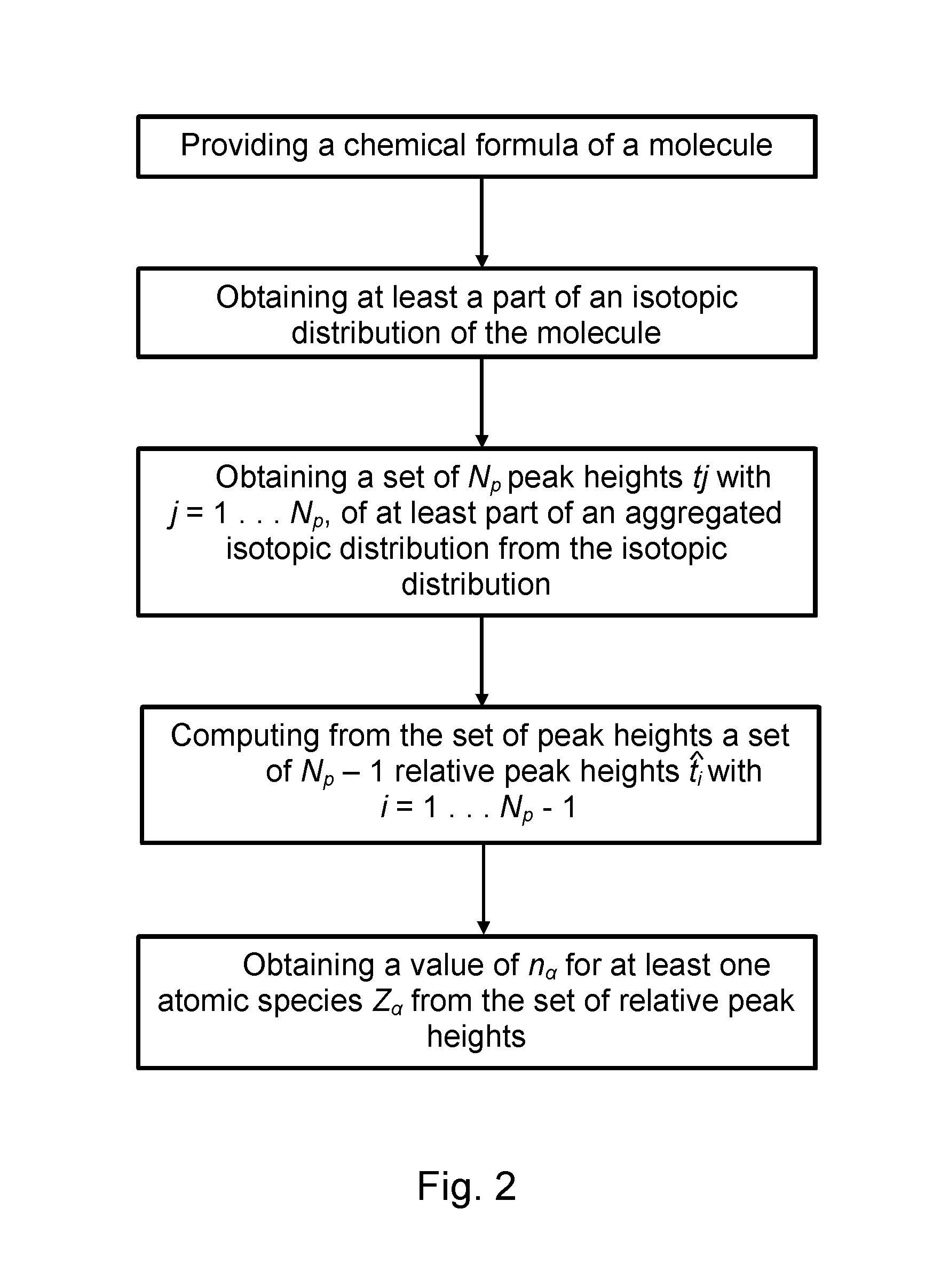
Method and device for computing molecular isotope distributions and for estimating the elemental composition of a molecule from an isotopic distribution
ActiveUS20130110412A1Molecular entity identificationComponent separationElemental compositionMass number
The current invention concerns a method for identifying the elemental composition of and / or quantifying the presence of mono-isotopic elements in a molecule in a sample by computing a solution of a system of linear equations ΣαEiαnα=Fi, whereby the set of numbers Fi comprises said set of relative peak heights from the aggregated isotopic distribution and the coefficients Eiα of said linear system comprise powers and / or power sums of roots rα,i<sub2>α< / sub2>. The present invention also provides a method for analysing at least part of an isotopic distribution of a sample, comprising obtaining data comprising at least one probability qj with which a j'th aggregated isotopic variant of said molecule with mass number Aj occurs within said aggregated isotopic distribution; and computing a probability qi with which an i'th aggregated isotopic variant occurs within said aggregated isotopic distribution, by taking a linear combination of said at least one probability qj.
Owner:VLAAMSE INSTELLING VOOR TECHNOLOGISCH ONDERZOEK NV VITO
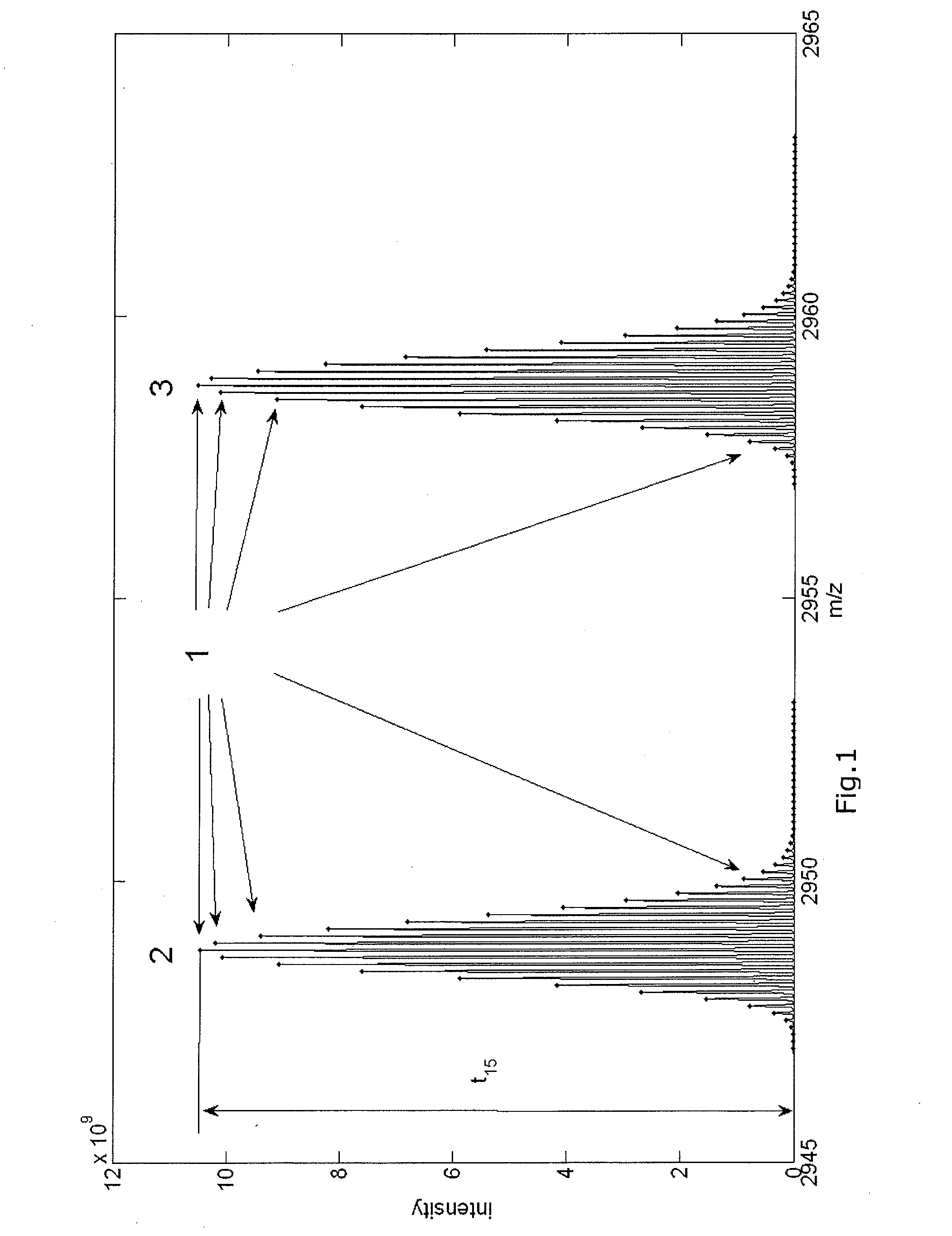
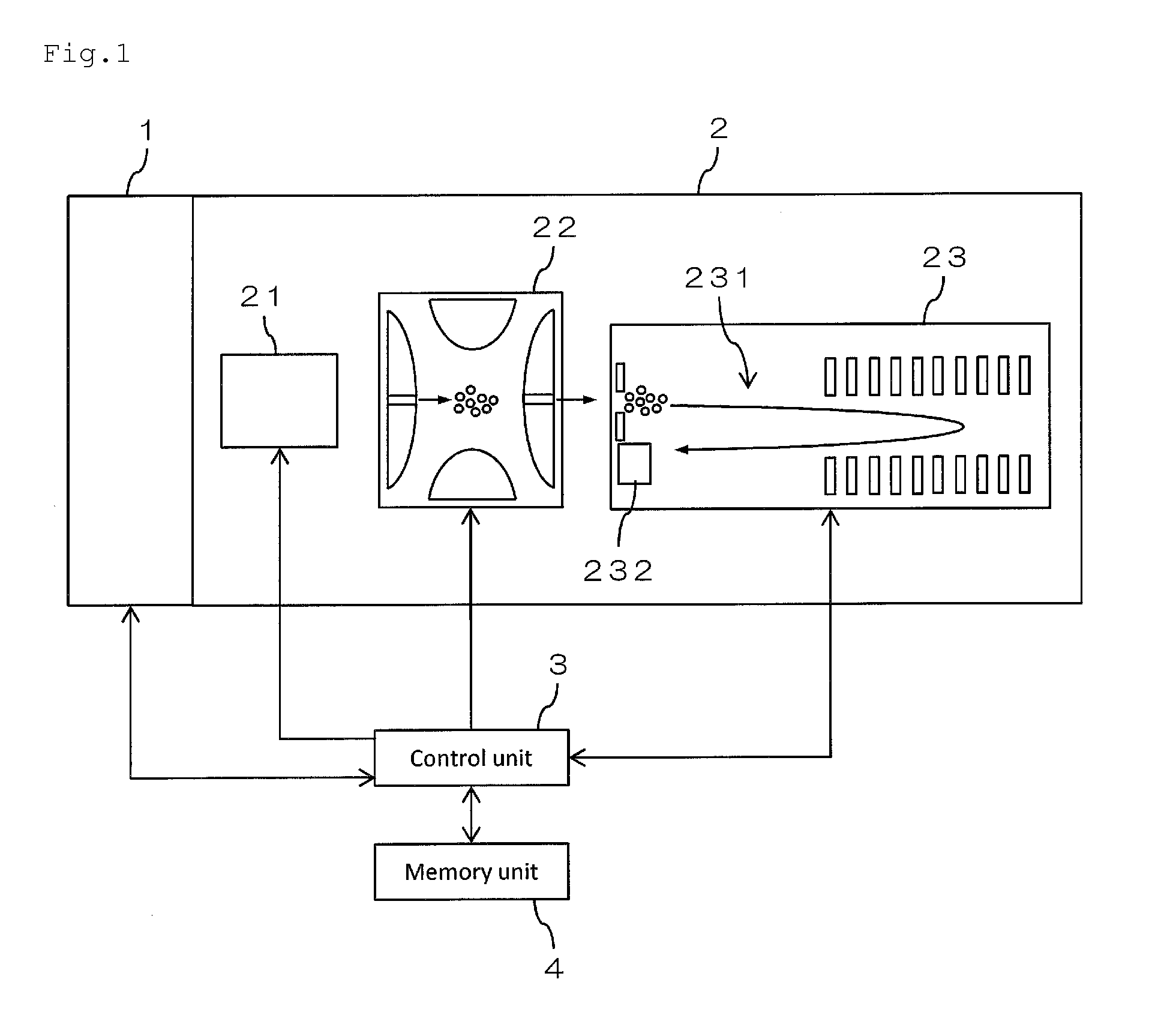
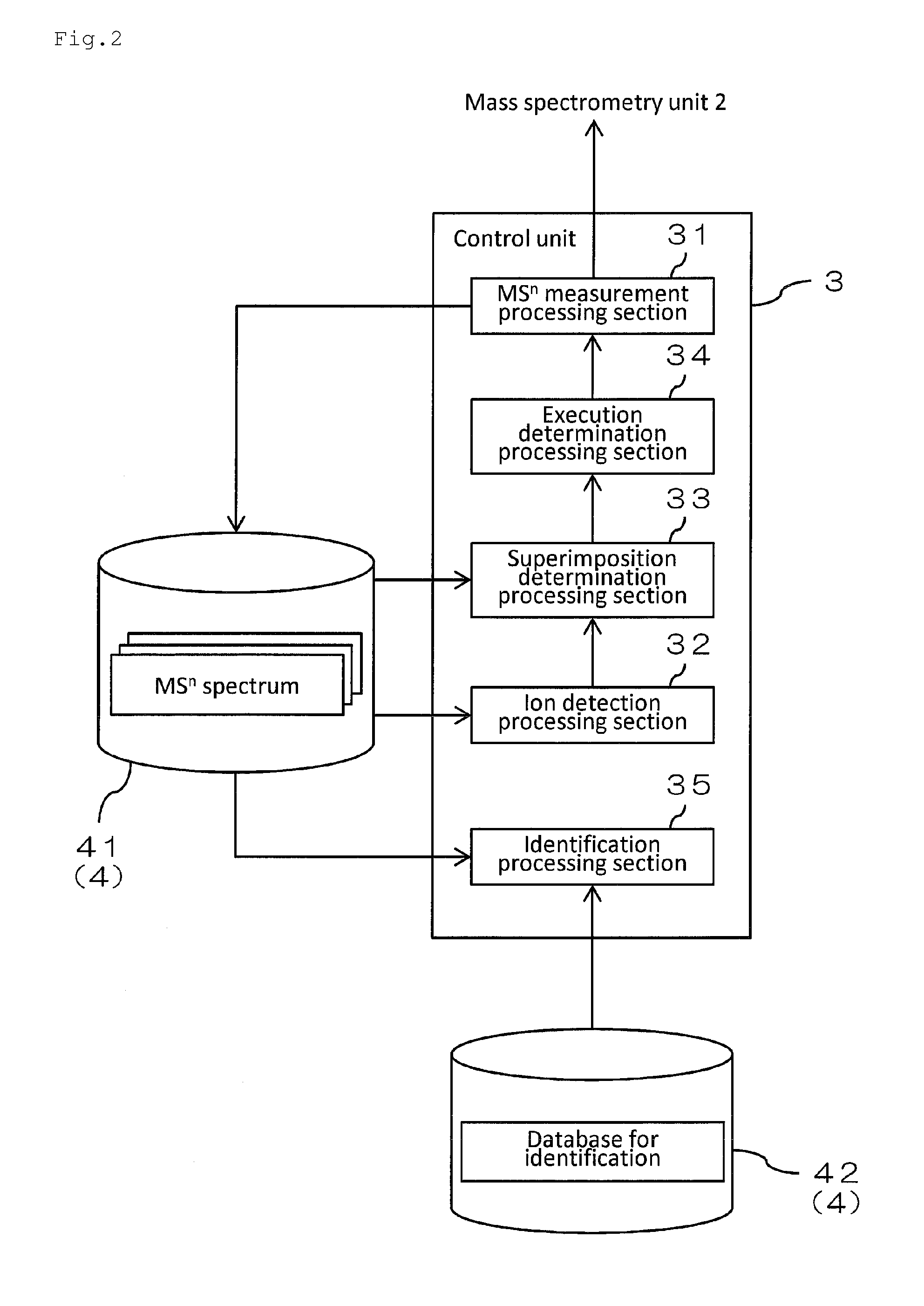
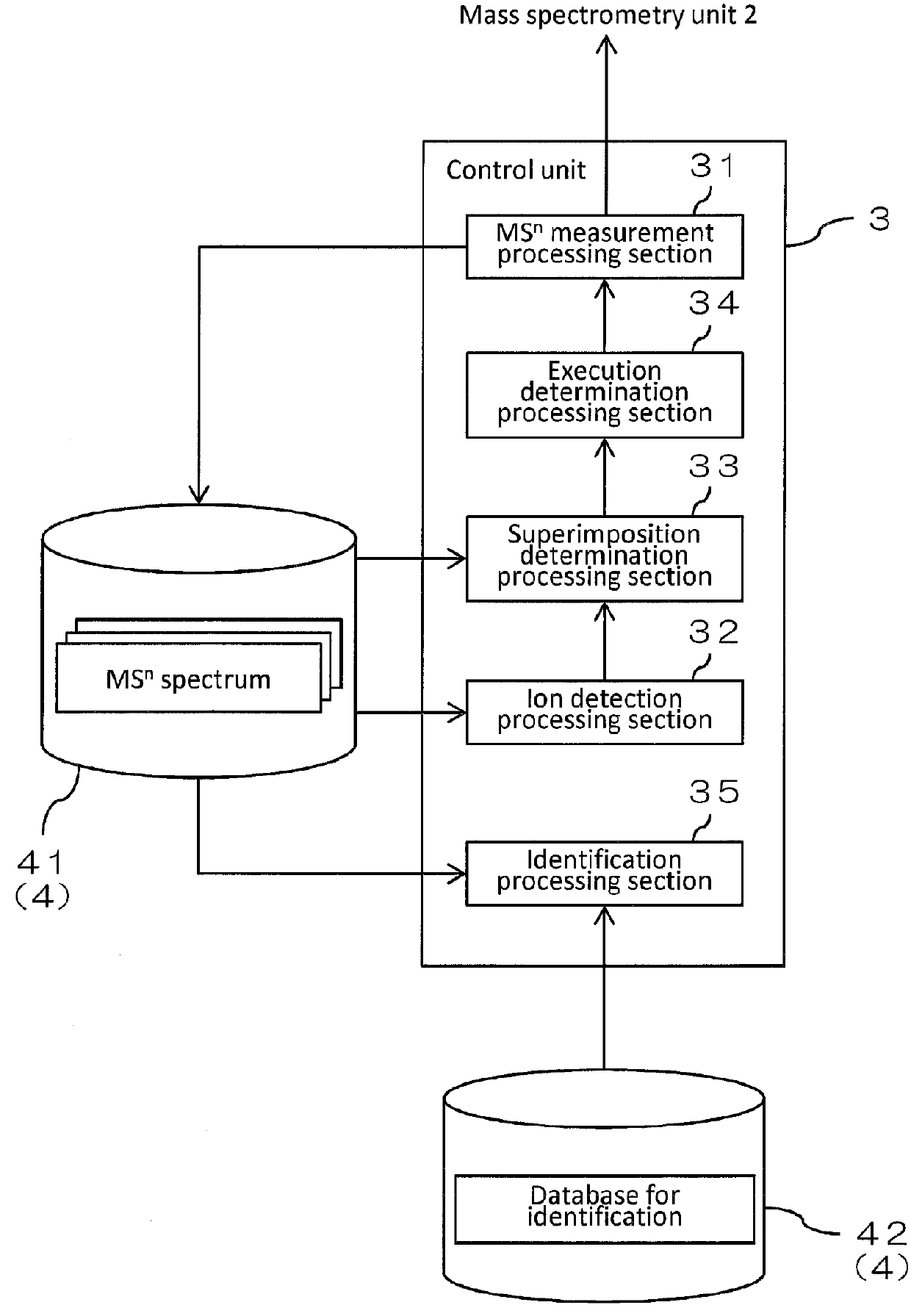
Mass spectrometer and mass spectrometry method
InactiveUS20150219606A1Reduced measurement timeDecrease sample consumptionComponent separationParticle spectrometer methodsRetention timeMass Spectrometry-Mass Spectrometry
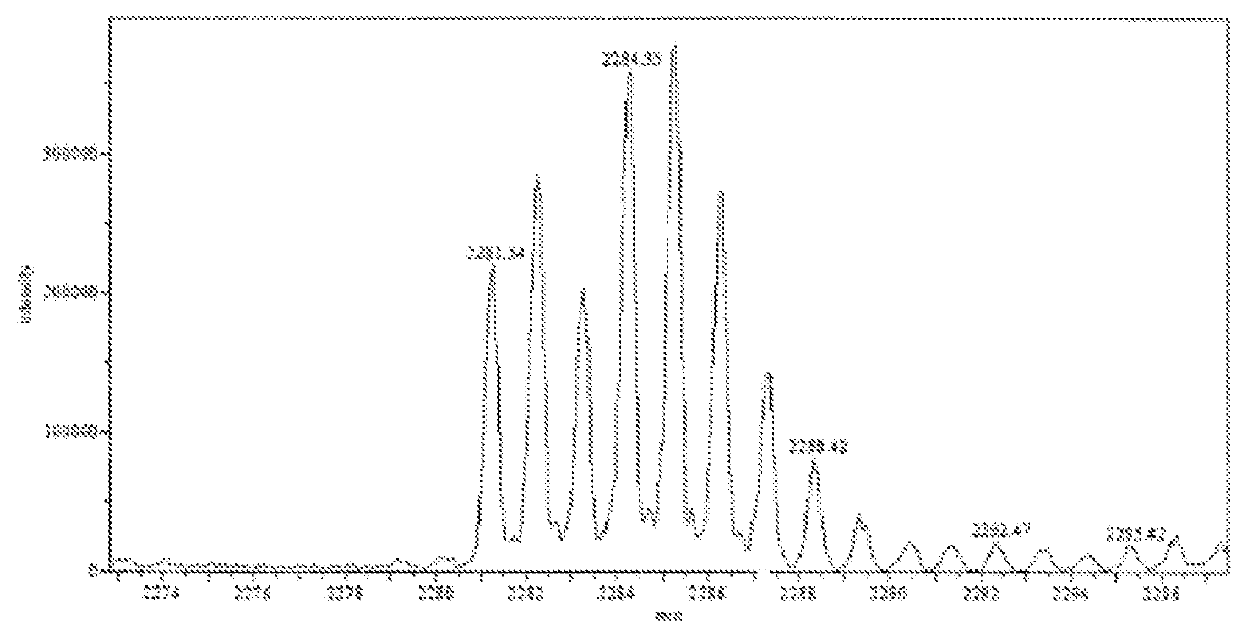
There are provided a mass spectrometer and a mass spectrometry method which can realize shortening of the measurement time and reduction of the consumption of a sample. Ions, in which the intensity distribution forms a peak waveform at both of each retention time and each mass-to-charge ratio (peaks P11, P21, P22 and P32) are detected as MS / MS precursor ions based on three-dimensional information of a retention time, a mass-to-charge ratio and an intensity. Whether or not MS3 analysis is performed for each ion is determined beforehand based on whether or not the isotopic distributions of a plurality of ions are superimposed at each retention time rt1 to rt3. Ions (peaks P21 and P22) for which MS3 analysis is performed and ions (peaks P11 and P32) for which MS3 analysis is not performed can be hereby determined at the time the MS spectrum is measured to detect MS / MS precursor ions.
Owner:SHIMADZU CORP
Intracellular metabolic flux analysis method using substrate labeled with isotope
InactiveUS8510054B2Low costMicrobiological testing/measurementBiological testingTime courseCulture cell
Owner:AJINOMOTO CO INC
Method for simultaneously screening and analyzing forbidden and restricted dye in textiles
InactiveCN105891316AImprove detection efficiencyReduce testing costsComponent separationMaterial analysis by electric/magnetic meansMass numberPositive sample
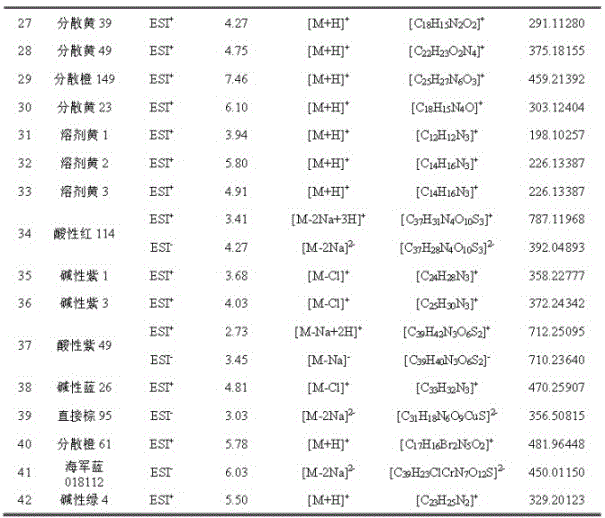
The invention discloses a method for simultaneously screening and analyzing forbidden and restricted dye in textiles. The method includes steps of (1), stripping colors of samples and extracting the samples to obtain extract liquid; (2), separating the samples by the aid of reversed phase liquid chromatograms, measuring the samples by the aid of high-resolution mass spectra and acquiring data of chromatogram peak retention time, accurate mass numbers of parent ions, isotope distribution of the parent ions and the like; (3), screening and analyzing the samples, to be more specific, determining that the samples are positive samples if high-resolution mass spectrum data of the samples simultaneously meet conditions, or determining that the samples are negative samples if the high-resolution mass spectrum data of the samples do not simultaneously meet the conditions, quantitatively analyzing the samples, to be more specific, extracting the accurate mass numbers of the parent ions of the samples, computing chromatogram peak areas, comparing the chromatogram peak areas to chromatogram peak areas of standard solution and quantitatively analyzing the samples. The conditions include that the allowable deviation of the retention time is + / -0.2 min, the allowable deviation of the accurate mass numbers of the parent ions is + / -5 ppm, and matching threshold values of the isotope distribution of the parent ions are larger than 70%. The method has the advantages that the samples are treated in such detection modes that the samples are treated at one step and are injected at one step and multiple compound analysis results are obtained at one step, accordingly, the detection efficiency can be greatly improved, and the detection cost can be reduced; the method is simple, convenient and speedy and is high in recycling rate and operability.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Determination of chemical composition and isotope distribution with mass spectrometry
InactiveUS7904253B2Time-of-flight spectrometersComponent separationElemental compositionChemical composition
A method for determining elemental composition of ions from mass spectral data, comprising obtaining at least one mass measurement from mass spectral data; obtaining a search list of candidate elemental compositions whose exact masses fall within a given mass tolerance range from the accurate mass; reporting a probability measure based on a mass error; calculating an isotope pattern for each candidate elemental composition from the search list; constructing a peak component matrix including at least one of the isotope pattern and mass spectral data; performing a regression against at least one of isotope pattern, mass spectral data, and the peak component matrix; reporting a second probability measure for at least one candidate elemental composition based on the isotope pattern regression; and combining the two the probability measures into an overall probability measure. A method for determining elemental isotope ratios from mass spectral data.
Owner:CERNO BIOSCI
Stable isotope, site-specific mass tagging for protein identification
InactiveUS20050124014A1Improve accuracyImprove efficiencyBacteriaSamples introduction/extractionPresent methodProtein-protein complex
Proteolytic peptide mass mapping as measured by mass spectrometry provides an important method for the identification of proteins, which are usually identified by matching the measured and calculated m / z values of the proteolytic peptides. A unique identification is, however, heavily dependent upon the mass accuracy and sequence coverage of the fragment ions generated by peptide ionization. The present invention describes a method for increasing the specificity, accuracy and efficiency of the assignments of particular proteolytic peptides and consequent protein identification, by the incorporation of selected amino acid residue(s) enriched with stable isotope(s) into the protein sequence without the need for ultrahigh instrumental accuracy. Selected amino acid(s) are labeled with 13C / 15N / 2H and incorporated into proteins in a sequence-specific manner during cell culturing. Each of these labeled amino acids carries a defined mass change encoded in its monoisotopic distribution pattern. Through their characteristic patterns, the peptides with mass tag(s) can then be readily distinguished from other peptides in mass spectra. The present method of identifying unique proteins can also be extended to protein complexes and will significantly increase data search specificity, efficiency and accuracy for protein identifications.
Owner:LOS ALAMOS NATIONAL SECURITY
Method for determining oil sources by using sulfur isotopes in monomer sulfur-containing compounds of crude oil
ActiveCN105510561AAchieve separationSolve difficult problemsComponent separationMaterial testing goodsAlkaneEthane Dichloride
The invention provides a method for determining oil sources by using sulfur isotopes in monomer sulfur-containing compounds of crude oil. The method comprises the following steps: crude oil is dissolved in ethanol, dichloromethane or dichloroethane, an alkylating reagent is added, and filtrate is obtained after a reaction; ethanol, dichloromethane or dichloroethane are removed from the filtrate, and oily substances are obtained; the oily substances and an alkane solvent of C5-C7 are mixed, and separation is carried out for deposition; the deposition is dissolved in dichloromethane, trichloromethane or acetonitrile, a first dealkylating reagent is added, and a solution A is obtained; extraction is carried out, and a thiophene compound and a solution B are obtained; a second dealkylating reagent is added into the solution B for carrying out a second dealkylation reaction, and a solution C is obtained; extraction is carried out, and a thioether compound is obtained; sulfur isotopic distribution of monomers in the thiophene compound and the thioether compound as well as sulfur isotopic distribution of sulfate in an oil-generating rock stratum system are determined; two sulfur isotopic distributions are compared, and the stratum whose isotopic distribution is most close to is the oil source layer.
Owner:PETROCHINA CO LTD
Method for quickly screening and accurately confirming alkylphenol ethoxylates in textiles
ActiveCN107436334AEffectively respond to health and safety emergenciesEliminate distractionsComponent separationChromatographic separationMass number
The invention discloses a method for quickly screening and accurately confirming alkylphenol ethoxylates in textiles. The method includes preparing alkylphenol ethoxylates extract liquor; carrying out high-performance liquid phase chromatographic separation and high-resolution mass spectrometric determination; carrying out screening and confirming analysis, determining that samples are positive samples if high-performance liquid phase chromatographic / high-resolution mass spectrometric data of the samples simultaneously meet conditions that the allowable deviation of chromatographic peak retention time is + / -0.2 min, the allowable deviation of accurate mass numbers of parent ions is + / -5 ppm, matching thresholds of isotopic distribution of the parent ions are higher than 60% and the allowable deviation of accurate mass numbers of fragment ions is + / -5 ppm, or determining that the samples are negative samples if the high-performance liquid phase chromatographic / high-resolution mass spectrometric data of the samples do not simultaneously meet the conditions; extracting the accurate mass numbers of the parent ions in the extract liquor, computing peak areas of chromatographic peaks, comparing the extract liquor to standard solution and carrying out quantitative analysis. The method has the advantages that the method is in high-throughput detection modes for acquiring analysis results for diversified compounds in the short-chain and long-chain alkylphenol ethoxylates at one step by means of one-step treatment and one-step sample injection, accordingly, the detection efficiency can be improved, and the detection cost can be reduced.
Owner:INSPECTION & QUARANTINE TECH CENT SHANDONG ENTRY EXIT INSPECTION & QUARANTINE BUREAU
Arc discharge with improved isotopic mixture of mercury
InactiveUS8339043B1Reduction in resonance trappingImprove lighting efficiencyElectroluminescent light sourcesVessels or leading-in conductors manufactureResonance radiationIsotope distribution
In a mercury-containing arc discharge device for converting electrical energy into resonance radiation energy, the isotopic distribution of the mercury in the device is altered from that of natural mercury so as to reduce imprisonment time of resonance radiation and thereby increase the efficiency of conversion of electrical energy into resonance radiation. The mercury-199 isotope content of the mercury is lower than that in natural mercury and the mercury-201 and / or mercury-204 isotopes are greater than those in natural mercury.
Owner:ANDERSON JAMES BERNHARD
Stable isotope, site-specific mass tagging for protein identification
InactiveUS7125685B2Improve accuracyImprove efficiencyBacteriaSamples introduction/extractionPresent methodProtein-protein complex
Proteolytic peptide mass mapping as measured by mass spectrometry provides an important method for the identification of proteins, which are usually identified by matching the measured and calculated m / z values of the proteolytic peptides. A unique identification is, however, heavily dependent upon the mass accuracy and sequence coverage of the fragment ions generated by peptide ionization. The present invention describes a method for increasing the specificity, accuracy and efficiency of the assignments of particular proteolytic peptides and consequent protein identification, by the incorporation of selected amino acid residue(s) enriched with stable isotope(s) into the protein sequence without the need for ultrahigh instrumental accuracy. Selected amino acid(s) are labeled with 13C / 15N / 2H and incorporated into proteins in a sequence-specific manner during cell culturing. Each of these labeled amino acids carries a defined mass change encoded in its monoisotopic distribution pattern. Through their characteristic patterns, the peptides with mass tag(s) can then be readily distinguished from other peptides in mass spectra. The present method of identifying unique proteins can also be extended to protein complexes and will significantly increase data search specificity, efficiency and accuracy for protein identifications.
Owner:LOS ALAMOS NATIONAL SECURITY
Method for identification of the monoisotopic mass of species of molecules
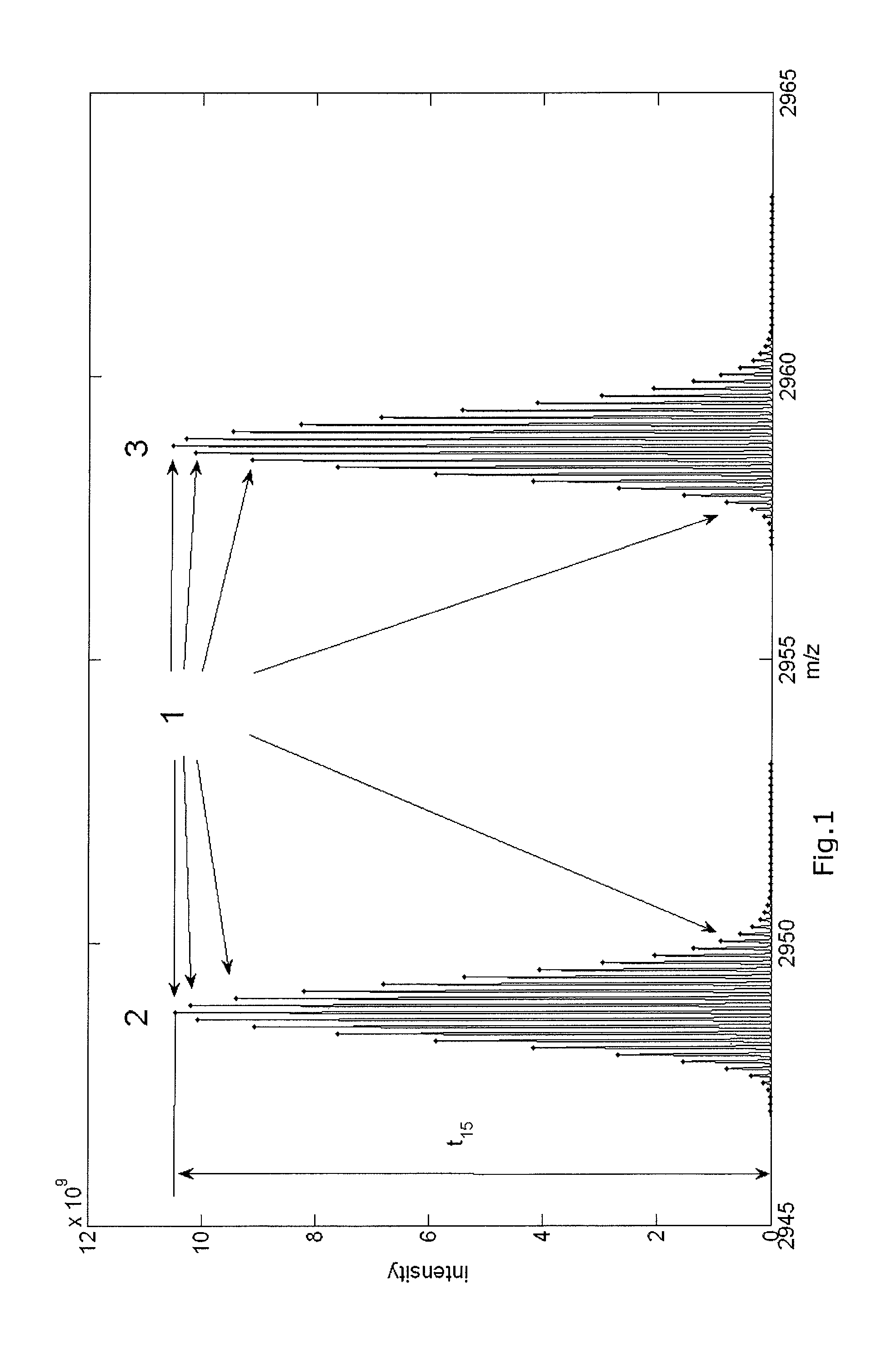
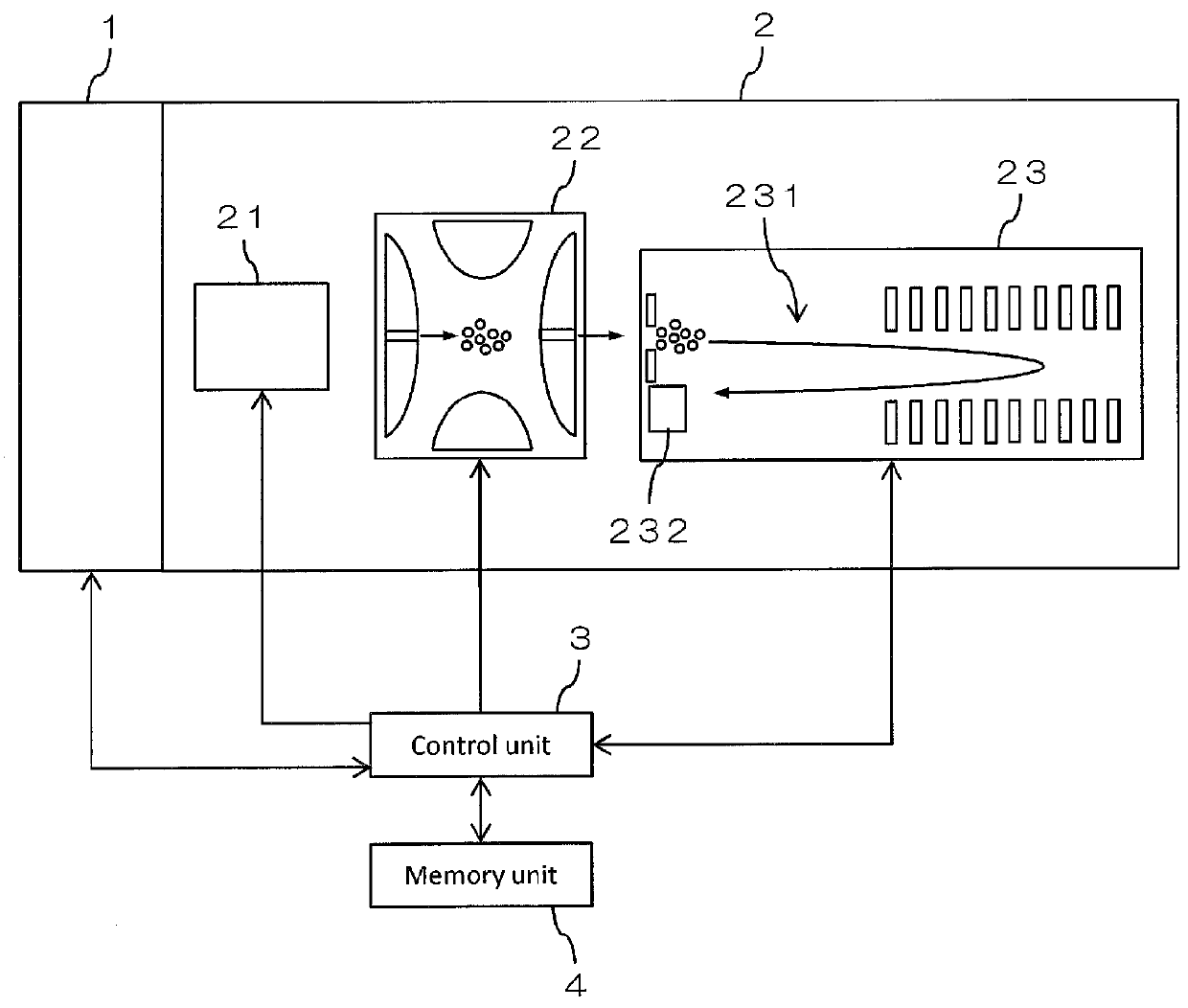
ActiveUS10593530B2Improve recognition accuracyImprove performanceParticle separator tubesComponent separationPhysical chemistryMass analyzer
A method for identification of the monoisotopic mass or a parameter correlated to the mass of the isotopes of the isotope distribution of at least one species of molecules contained in a sample and / or originated from a sample by at least an ionization process includes measuring a mass spectrum of the sample with a mass spectrometer, dividing at least one range of measured m / z values of the mass spectrum into fractions, assigning at least some of the fractions to one processor of several provided processors, deducing for each of the at least one species of molecules an isotope distribution of their ions having a specific charge z, deducing from at least one deduced isotope distribution the monoisotopic mass or a parameter correlated to the mass of the isotopes of the isotope distribution of the species of molecules.
Owner:THERMO FISHER SCI BREMEN
Method and device for determining the molecular weight of compound
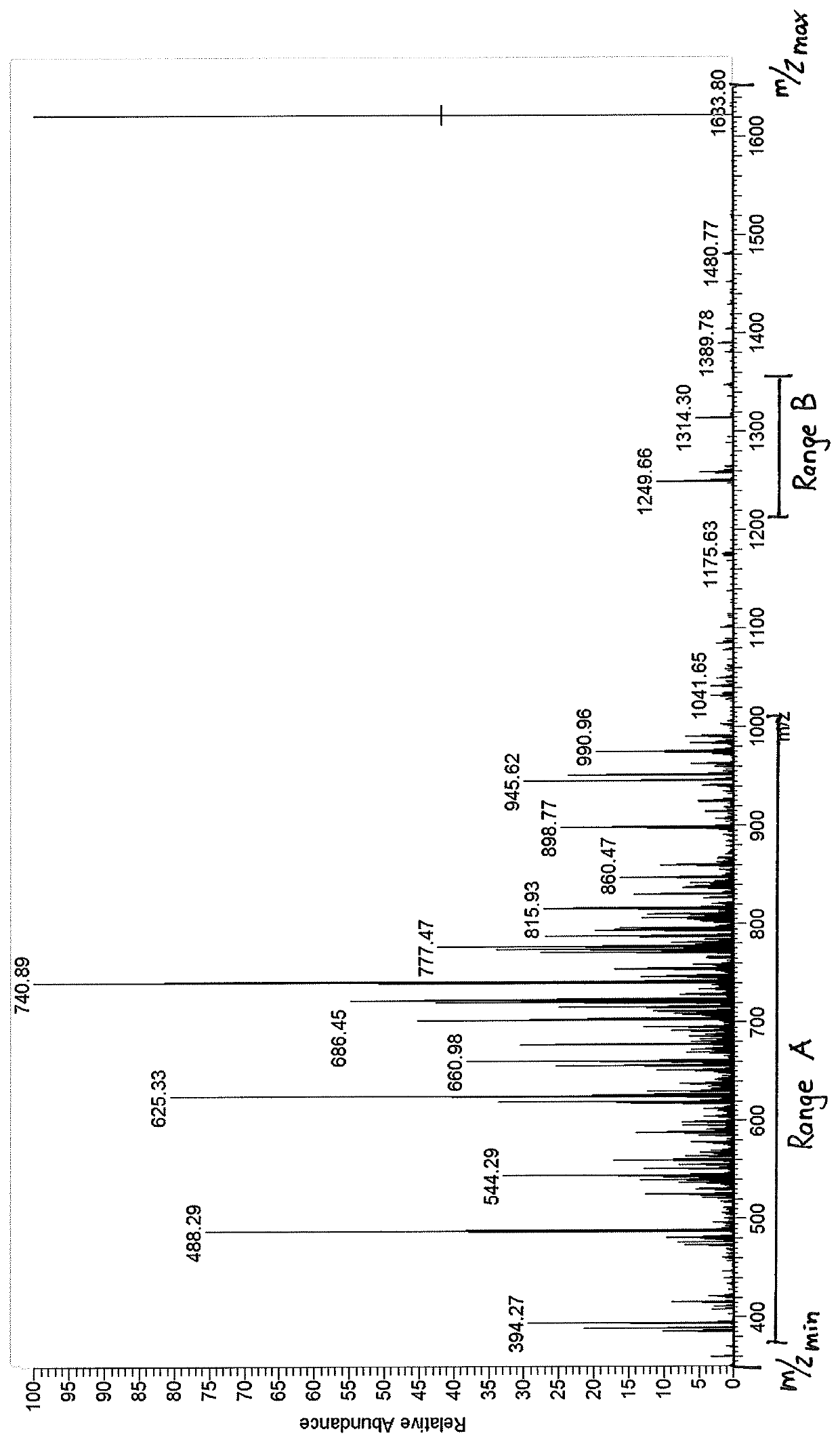
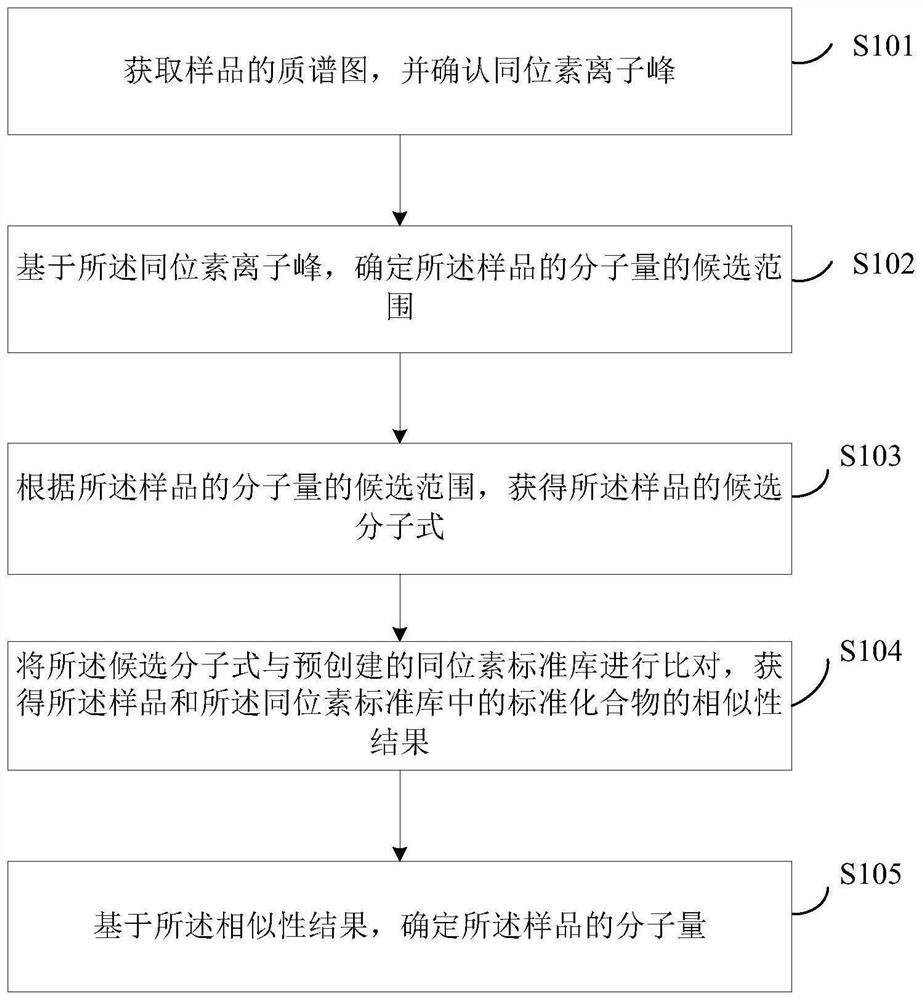
PendingCN112289386ALow limit of quantitationLow detection limitMolecular entity identificationMaterial analysis by electric/magnetic meansChemical compoundPhysical chemistry
The invention discloses a method and a device for determining the molecular weight of a compound. The method comprises the following steps: acquiring a mass spectrogram of a sample and determining anisotope ion peak; determining a candidate range of the molecular weight of the sample based on the isotope ion peak; obtaining a candidate molecular formula of the sample according to the candidate range of the molecular weight of the sample; comparing the candidate molecular formulas with a pre-created isotope standard library to obtain a similarity result of the sample and a standard compound inthe isotope standard library, the isotope standard library comprising molecular formulas screened based on an isotope principle; and determining the molecular weight of the sample based on the similarity result. Similarity comparison is carried out on the organic matter to be detected and isotope distribution to determine the molecular weight of the organic matter, the detection sensitivity is improved, the compound quantification limit and detection limit are reduced, and the compound molecular weight detection accuracy is improved.
Owner:CHINA AGRI UNIV
Method and device for computing molecular isotope distributing and for estimating the elemental composition of a molecule from an isotopic distribution
ActiveUS9411941B2Molecular entity identificationComponent separationElemental compositionMass number
The current invention concerns a method for identifying the elemental composition of and / or quantifying the presence of mono-isotopic elements in a molecule in a sample by computing a solution of a system of linear equations ΣαEiαnα=Fi, whereby the set of numbers Fi comprises said set of relative peak heights from the aggregated isotopic distribution and the coefficients Eiα of said linear system comprise powers and / or power sums of roots rα,i<sub2>α< / sub2>. The present invention also provides a method for analyzing at least part of an isotopic distribution of a sample, comprising obtaining data comprising at least one probability qj with which a j'th aggregated isotopic variant of said molecule with mass number Aj occurs within said aggregated isotopic distribution; and computing a probability qi with which an i'th aggregated isotopic variant occurs within said aggregated isotopic distribution, by taking a linear combination of said at least one probability qj.
Owner:VLAAMSE INSTELLING VOOR TECHNOLOGISCH ONDERZOEK NV VITO
Mass spectrometer and mass spectrometry method
InactiveUS9230784B2Reduced measurement timeReduce consumptionDynamic spectrometersComponent separationRetention timeMass Spectrometry-Mass Spectrometry
There are provided a mass spectrometer and a mass spectrometry method which can realize shortening of the measurement time and reduction of the consumption of a sample. Ions, in which the intensity distribution forms a peak waveform at both of each retention time and each mass-to-charge ratio (peaks P11, P21, P22 and P32) are detected as MS / MS precursor ions based on three-dimensional information of a retention time, a mass-to-charge ratio and an intensity. Whether or not MS3 analysis is performed for each ion is determined beforehand based on whether or not the isotopic distributions of a plurality of ions are superimposed at each retention time rt1 to rt3. Ions (peaks P21 and P22) for which MS3 analysis is performed and ions (peaks P11 and P32) for which MS3 analysis is not performed can be hereby determined at the time the MS spectrum is measured to detect MS / MS precursor ions.
Owner:SHIMADZU CORP
Method for identifying abnormal energy metabolism pathway of depression based on stable isotope tracing metabonomics
The invention belongs to the technical field of stable isotope tracing metabonomics, and provides a method for identifying an abnormal energy metabolism pathway of depression based on stable isotope tracing. The method comprises the steps of carrying out stable isotope labeling on an energy metabolism precursor substance glucose, carrying out comprehensive characterization on an energy metabolismcompound by utilizing a normal-phase chromatographic column and a reversed-phase chromatographic column, and determining an exact metabolic pathway of energy metabolism abnormality of depression according to isotope distribution characteristics of glucose downstream metabolites. The method provided by the invention has important scientific value for knowing the pathogenesis of depression and researching and developing novel antidepressant drugs, and provides a new method for exploring the action mechanism of diseases, discovering drug targets and researching and developing new drugs.
Owner:SHANXI UNIV
Photosynthetic plant tracing box
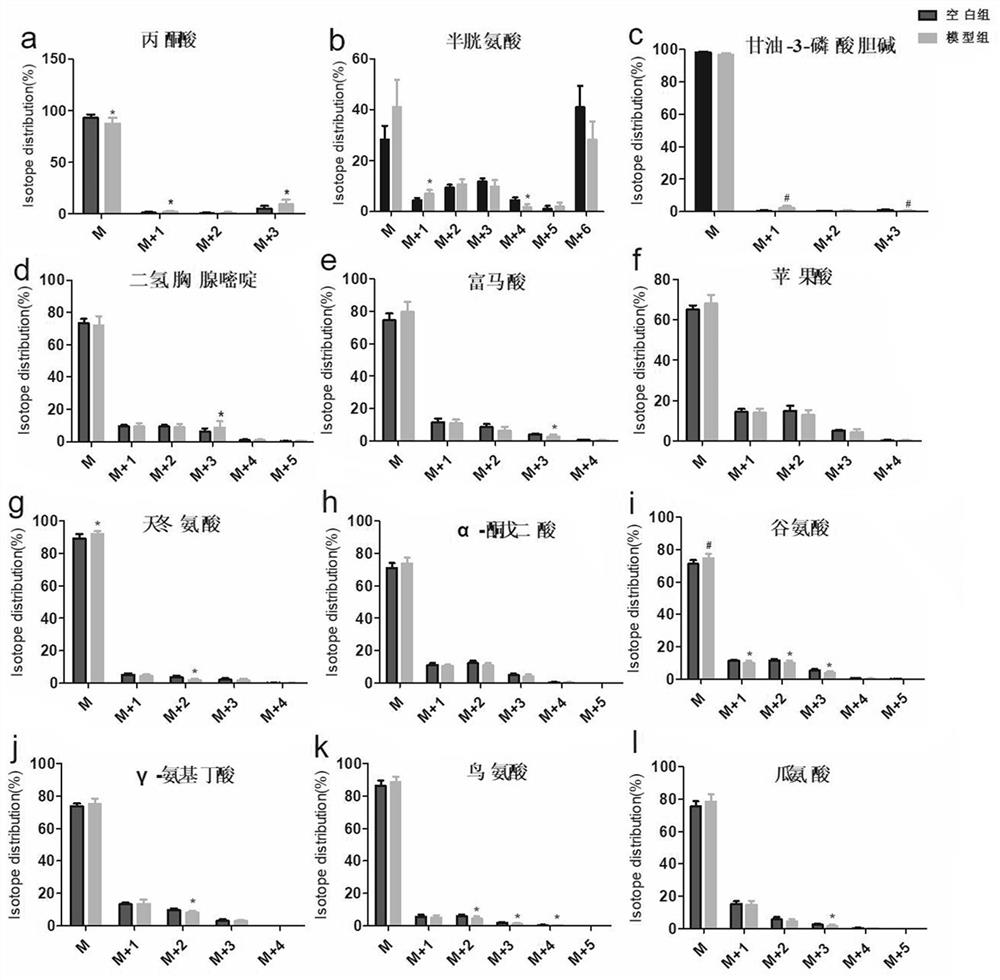
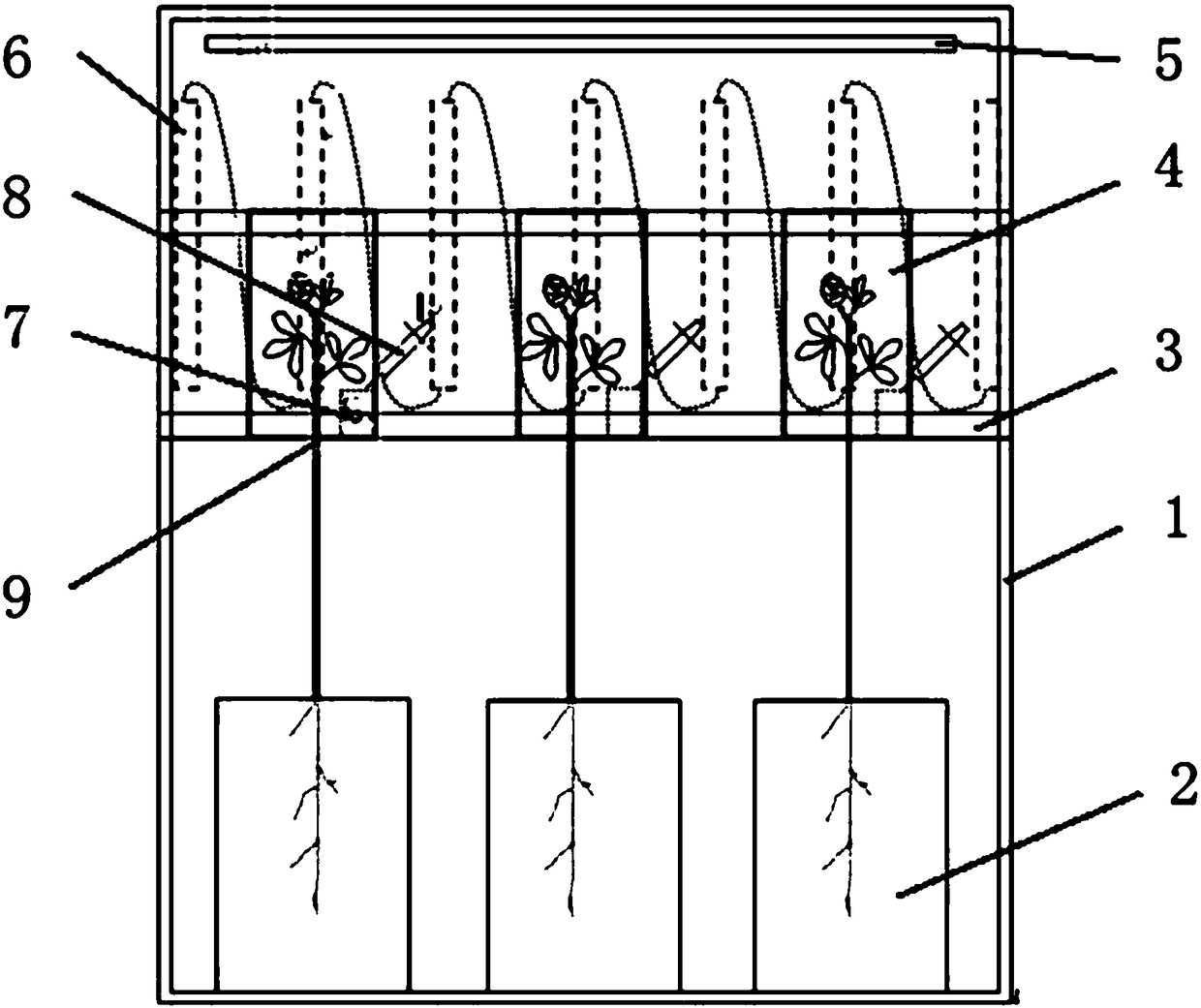
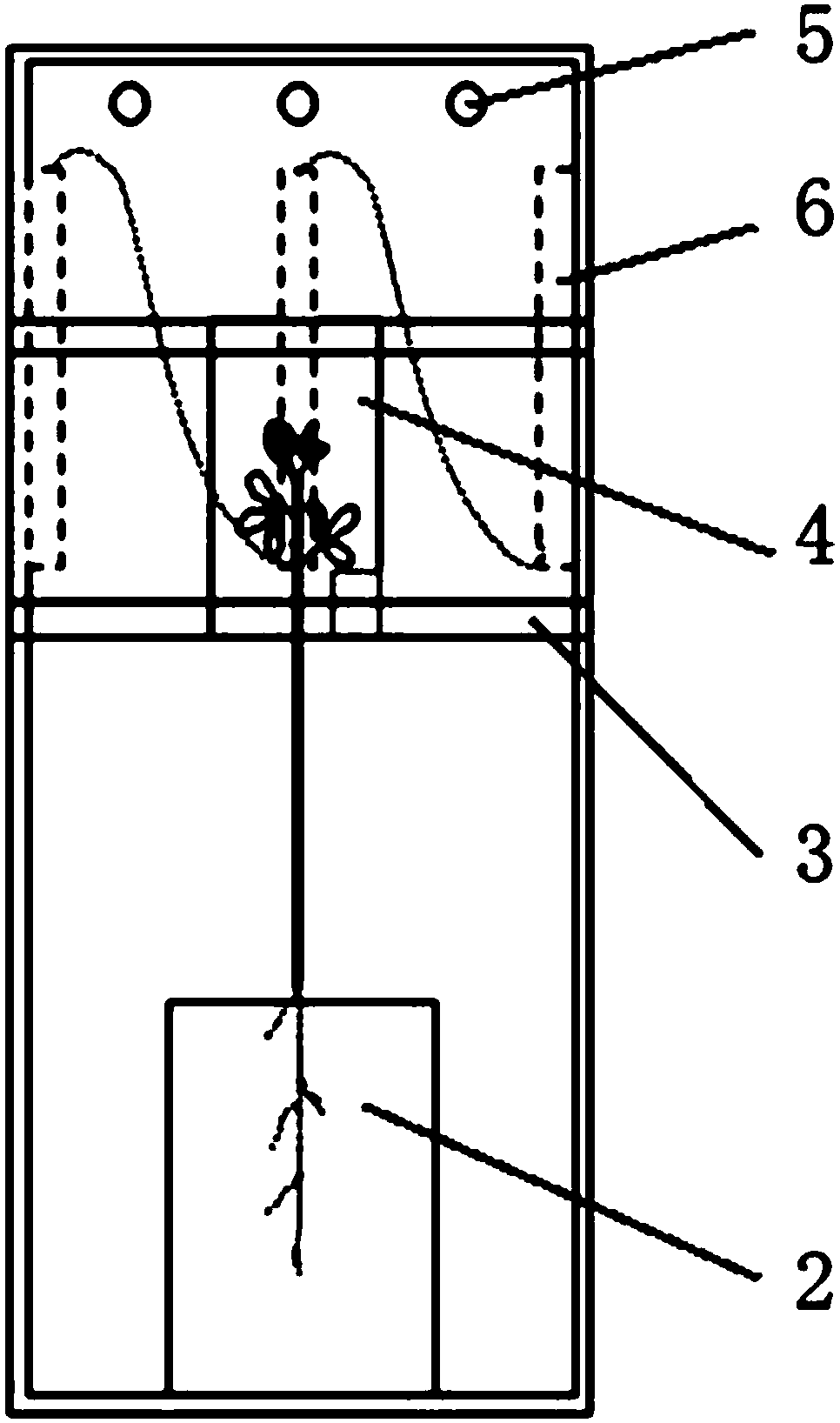
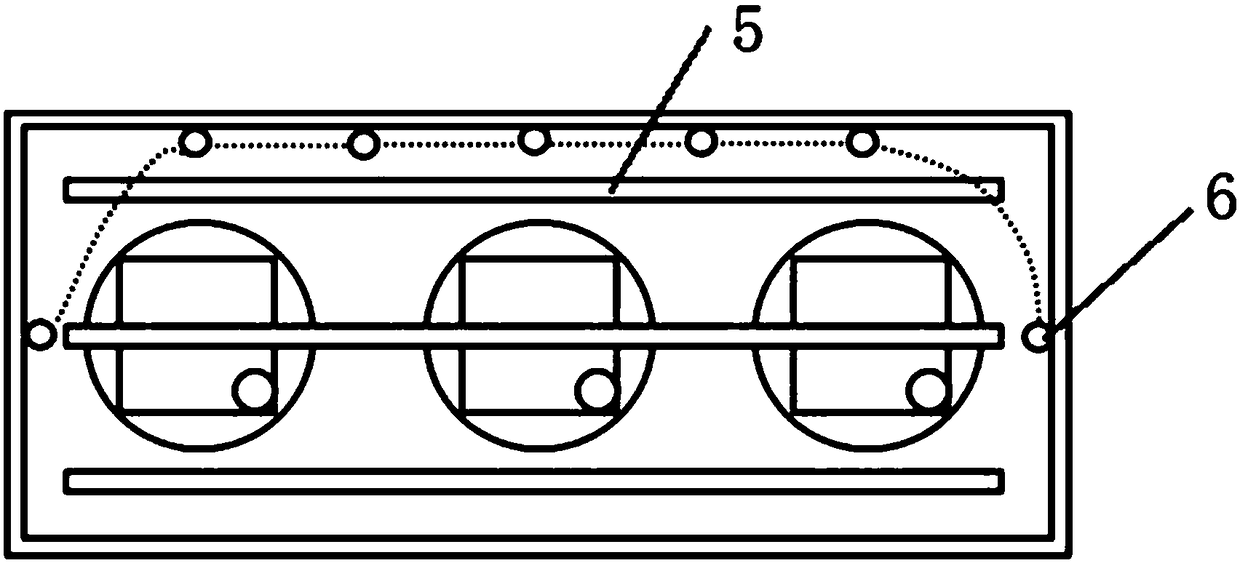
PendingCN108093930AAvoid interferenceImprove test accuracyClimate change adaptationGreenhouse cultivationIsotopes of carbonIsotope distribution
The invention relates to the technical field of plant test, in particular to a photosynthetic plant tracing box suitable for research of isotope distribution and transport of carbon in photosynthesisof plants. The tracing box includes a box, root culture dishes, a hanging plate, plant leaf sealing inner shells, a lamp strip and condensing tubes. The root culture dishes are arranged at the bottomof the box, the plant leaf sealing inner shells are arranged on the hanging plate, connection holes are formed in the bottoms of the plant leaf sealing inner shells, plant leaves are placed in the plant leaf sealing inner shells, and roots are embedded in the root culture dishes through the connection holes. The plant leaves are sealed through the inner shells to avoid outside interference and improve the test accuracy. The plant leaf sealing inner shells are arranged on the hanging plate to achieve a transport experiment of various isotopes of carbon element.
Owner:NORTHWEST NORMAL UNIVERSITY
Method for applying uranium recycled from pressurized water reactor to heavy water reactor
ActiveCN105825904AAvoid the effects of high radioactivityCost-effective useNuclear energy generationReactors manufactureProduction lineNatural uranium
The invention provides a method for applying uranium recycled from a pressurized water reactor to a heavy water reactor. The method replaces natural uranium with natural uranium equivalent to produce a fuel for the heavy water reactor on a heavy water reactor fuel production line. The natural uranium equivalent is prepared through the following steps: (1) measuring the metal uranium content, uranium isotope composition and / or uranium isotope distribution of recycled uranium and depleted uranium by using a mass spectrometer; (2) according to the uranium isotope composition of the recycled uranium and depleted uranium, calculating the mixing ratios alpha and 1-alpha of the recycled uranium and depleted uranium and target value of natural uranium equivalent according to a formula or mixing computation procedure so as to achieve the effect equivalent to the effect of natural uranium; 3) accurately measuring the recycled uranium and depleted uranium and then carrying out physical mixing so as to form natural uranium equivalent; (4) carrying out sampling analysis on the natural uranium equivalent formed through mixing; and (5) replacing natural uranium with the natural uranium equivalent to produce the fuel for the heavy water reactor. The method provided by the invention avoids the influence of high radioactivity of the recycled uranium, realizes economic high-efficiency utilization of the recycled uranium and reduces cost of the fuel of the heavy water reactor.
Owner:THIRD QINSHAN NUCLEAR POWER
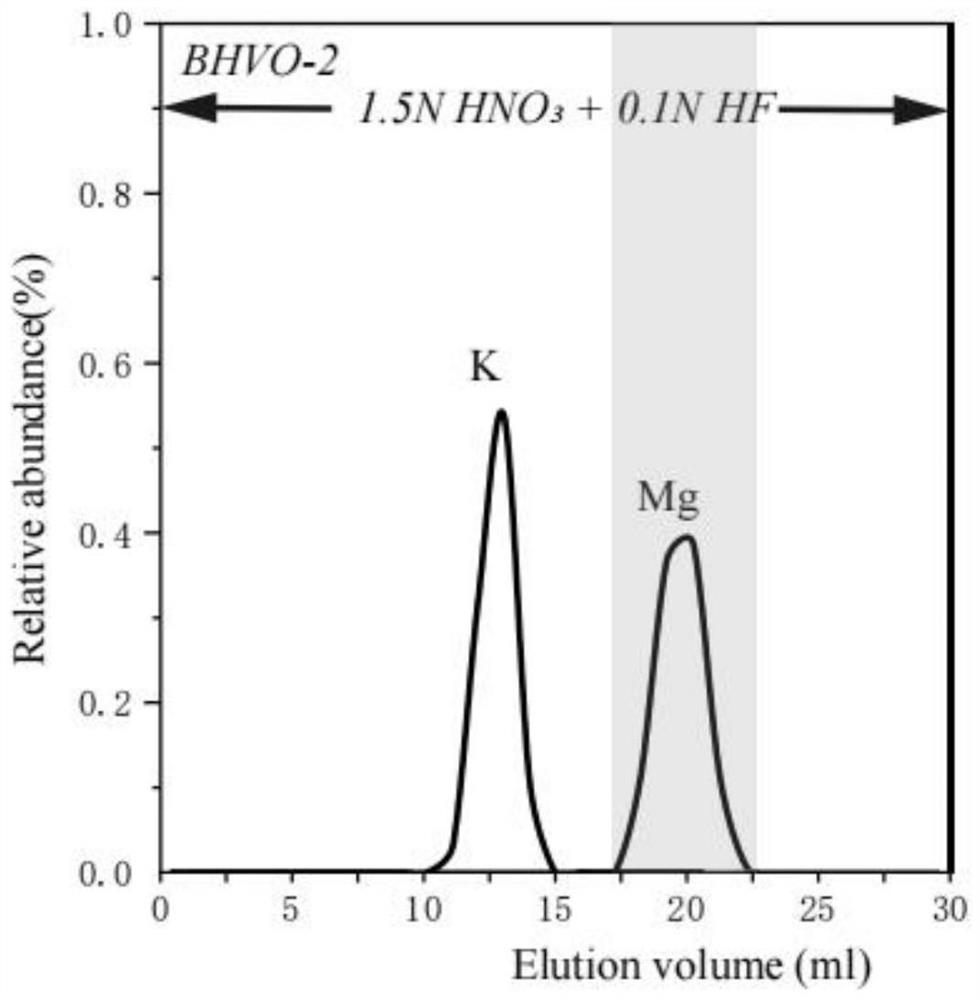
Analysis method for magnesium isotope composition
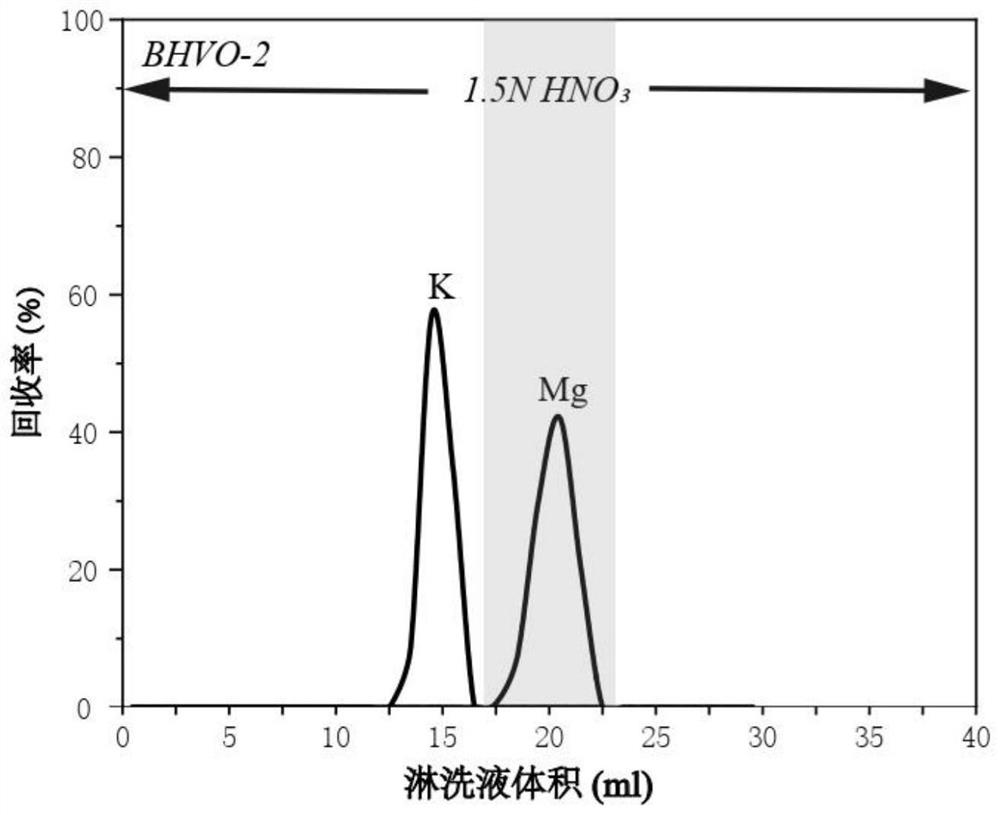
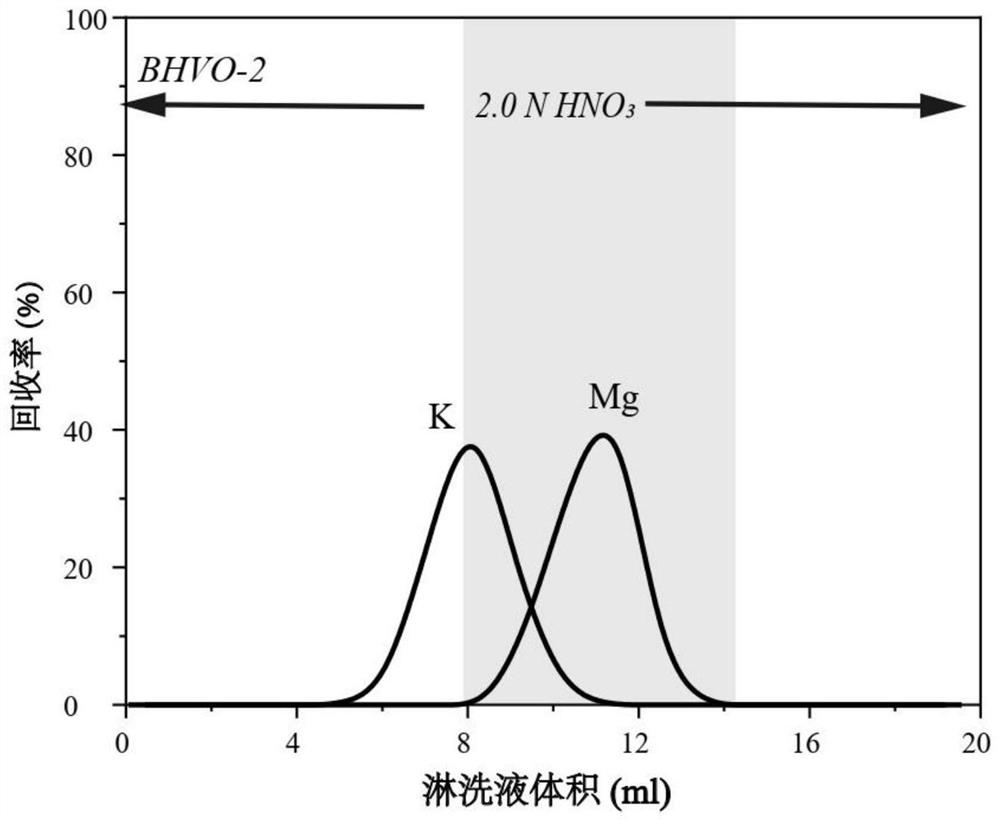
PendingCN114295762AReduce crossoverEfficient separationComponent separationIsotopic compositionPhysical chemistry
The invention provides an analysis method for magnesium isotope composition, and belongs to the field of geochemistry. Comprising the following steps: digesting a sample to be detected to obtain a digested substance; the digested substance is mixed with mixed acid, a digested sample solution is obtained, and the mixed acid comprises nitric acid and hydrofluoric acid; carrying out cation exchange chromatographic column separation and purification on the digested sample solution to obtain an Mg isotope solution sample; and carrying out multi-receiving plasma mass spectrometry detection on the Mg isotope solution sample to obtain an Mg isotope distribution ratio, and calculating according to a formula 1 to obtain the Mg isotope composition. The method is applicable to separation of magnesium elements in samples with different magnesium contents, and separation of magnesium elements in low-magnesium samples can be completed simply, conveniently and efficiently; the recovery rate of the Mg element in various standard samples is close to 100%, the recovery rate is high, and the Mg isotope composition in the samples with different magnesium contents can be accurately measured.
Owner:CHINA UNIV OF GEOSCIENCES (BEIJING) +1
Method for automatically parsing stable isotope labeled sugar chain quantification mass spectrum data
ActiveCN109738532AImprove Quantitative AccuracyImprove robustnessComponent separationBiological testingStable Isotope LabelingSugar
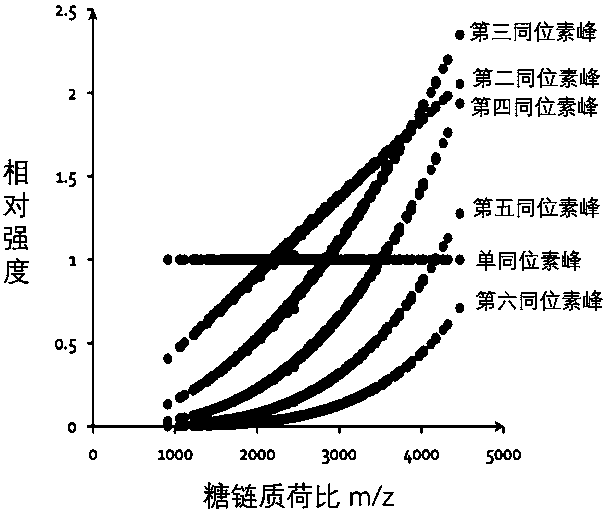
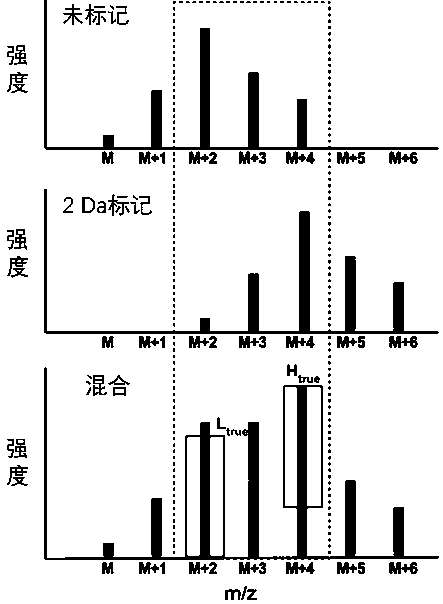
The invention belongs to the technical field of protein analysis, and particularly relates to a method for automatically parsing stable isotope labeled sugar chain quantification mass spectrum data. According to the invention, a tool gQuant is utilized to correct mass of an isotope labeled sugar chain produced by an MALDI-MS so as to carry out accurate relative quantification; the tool sufficiently considers the characteristics of isotope labeled relative quantification data; isotopes are alternatively and accurately corrected; moreover, particularly, an algorithm for carrying out quantification on the basis of the highest theoretical isotope intensity peak is set by utilizing a theoretical isotope distribution mode and a mass spectrum fracturing property of the sugar chain; and the methodhas higher quantification accuracy and robustness for a high-molecular-weight and low-abundance sugar chain component.
Owner:FUDAN UNIV
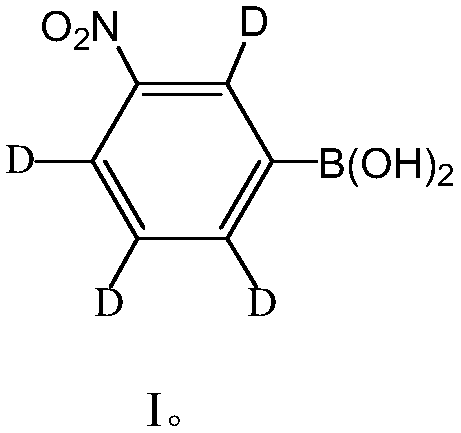
Deuteration-3-nitrobenzene boric acid and preparation method and application thereof
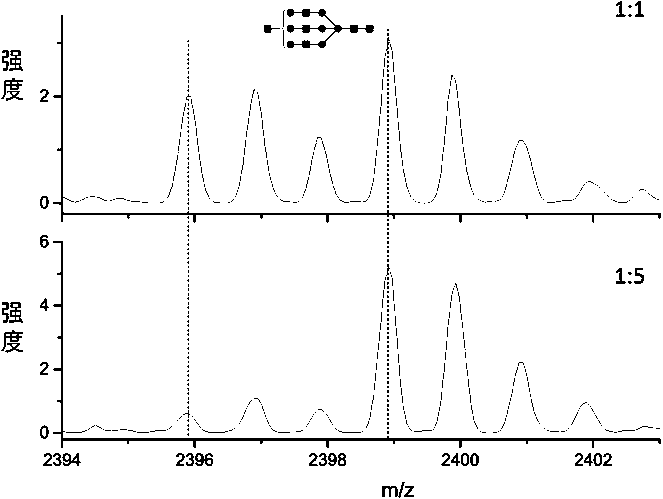
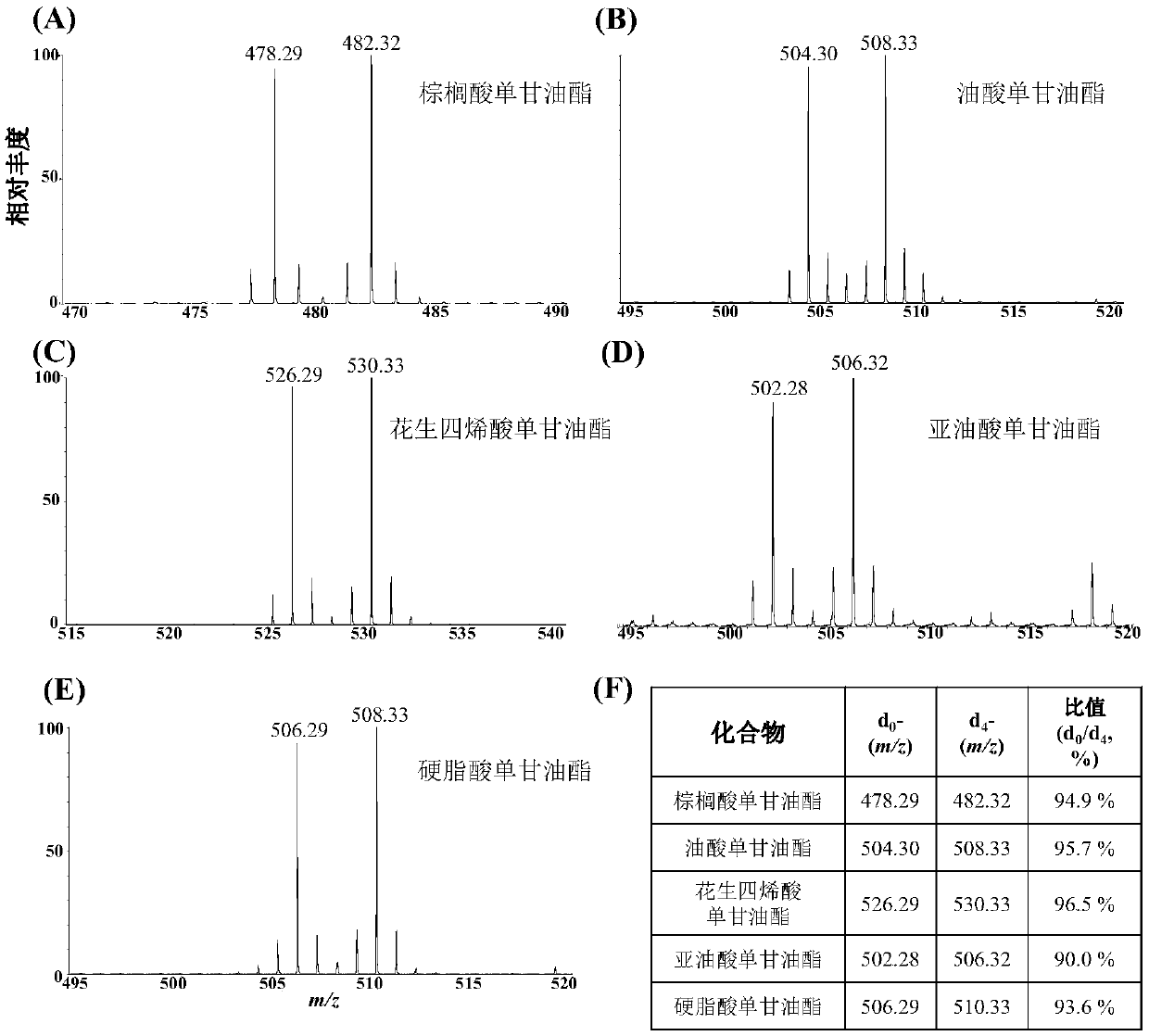
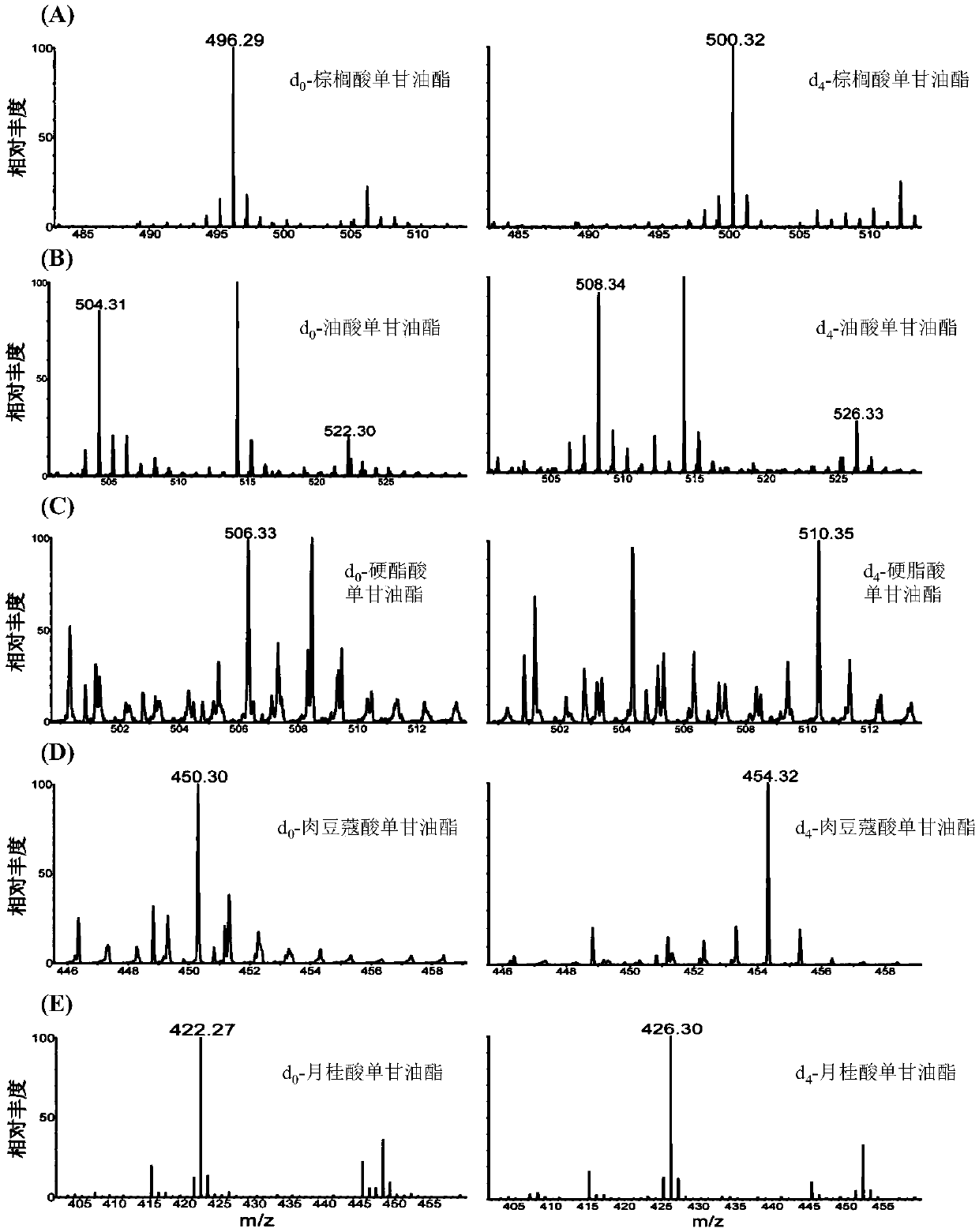
ActiveCN109096316AImprove featuresImprove accuracyComponent separationGroup 3/13 element organic compoundsMonoglycerideNitrobenzene
The invention disclose deuteration-3-nitrobenzene boric acid and a preparation method and application thereof. The compound is capable of improving the mass spectrum response of monoglyceride after being subjected to derivatization with monoglyceride; the special isotope distribution property of boron atoms is utilized to improve the specificity of monoglyceride detection and the identification accuracy; the deuteration-3-nitrobenzene boric can be used in match with a light marking agent in detection and relative quantitative analysis of monoglyceride in a complex biospecimen.
Owner:CHINA PHARM UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com