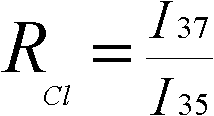Patents
Literature
177 results about "Mass number" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The mass number (symbol A, from the German word Atomgewicht (atomic weight)), also called atomic mass number or nucleon number, is the total number of protons and neutrons (together known as nucleons) in an atomic nucleus. It is approximately equal to the atomic (also known as isotopic) mass of the atom expressed in atomic mass units. Because protons and neutrons both are baryons, the mass number A is identical with the baryon number B as of the nucleus as of the whole atom or ion.
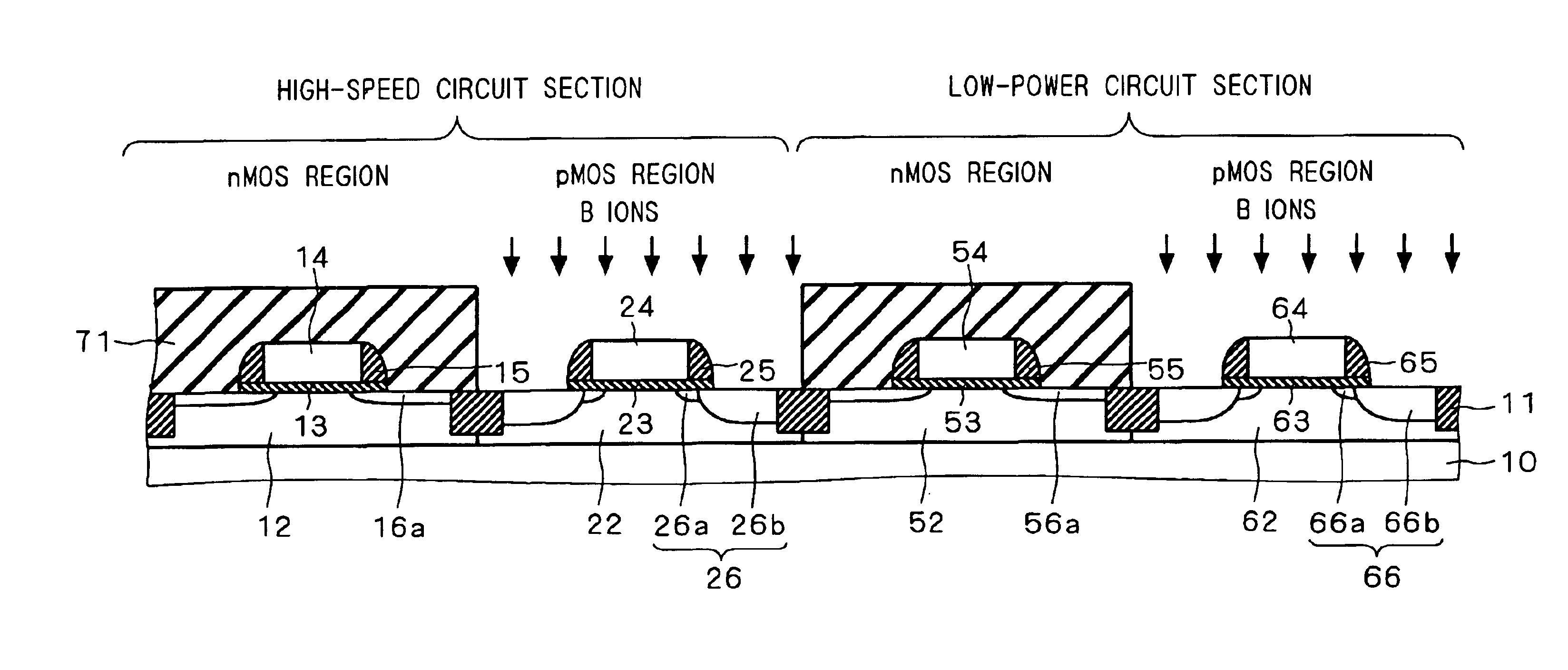
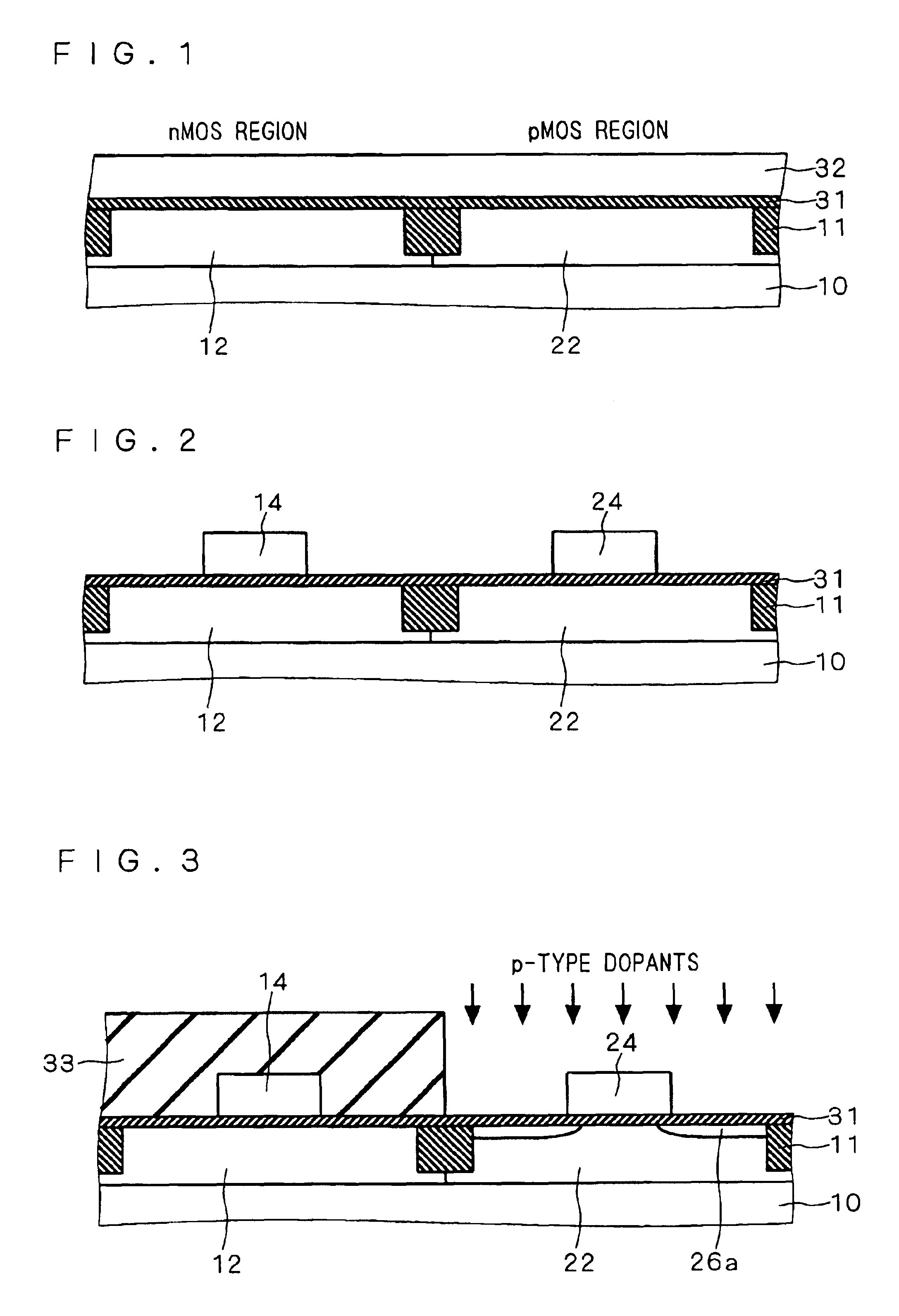
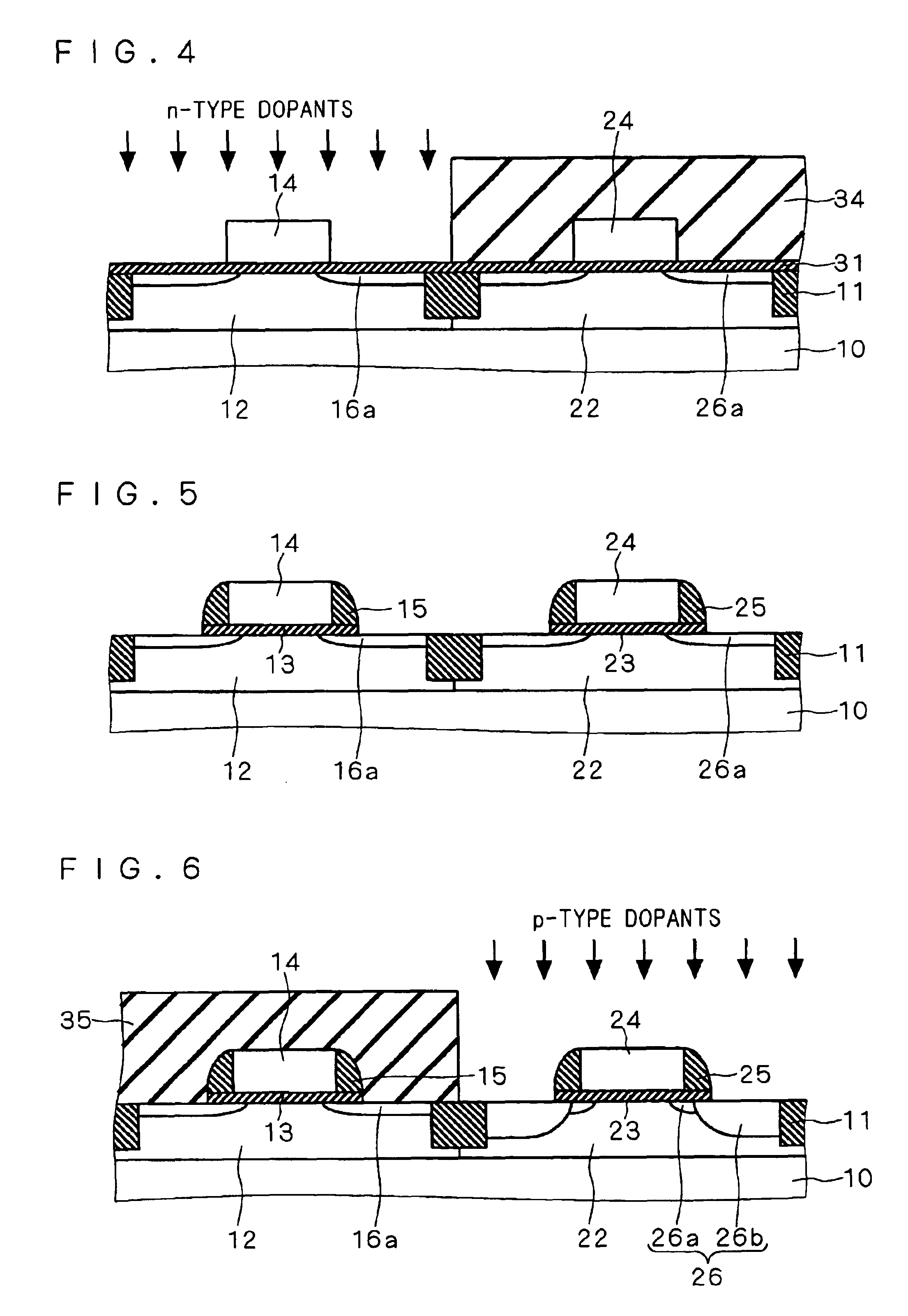
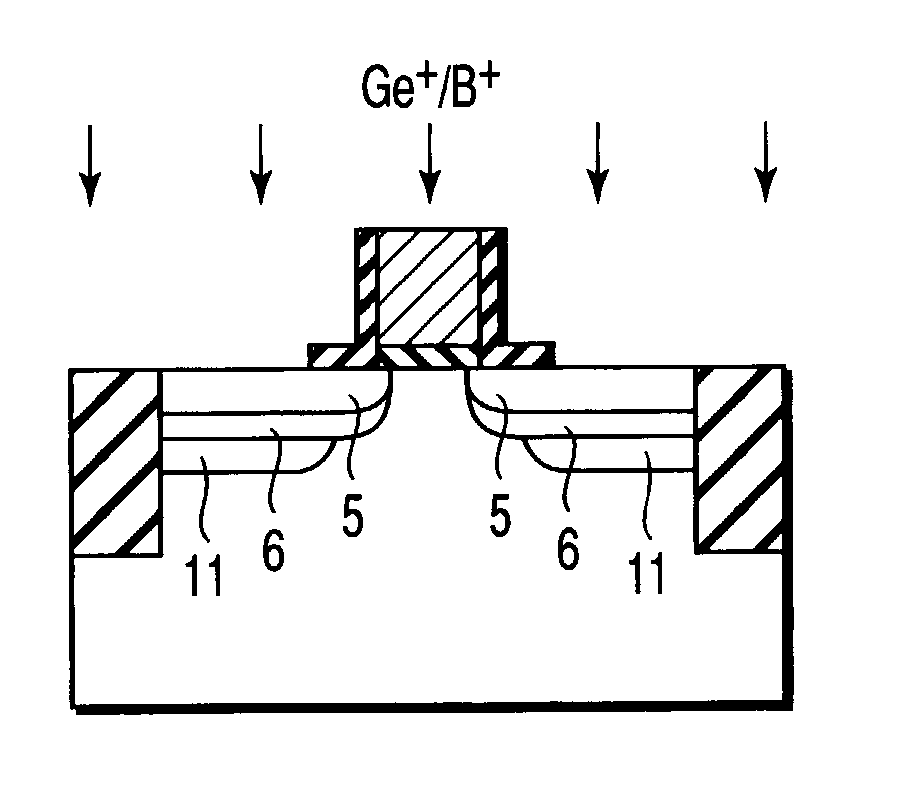
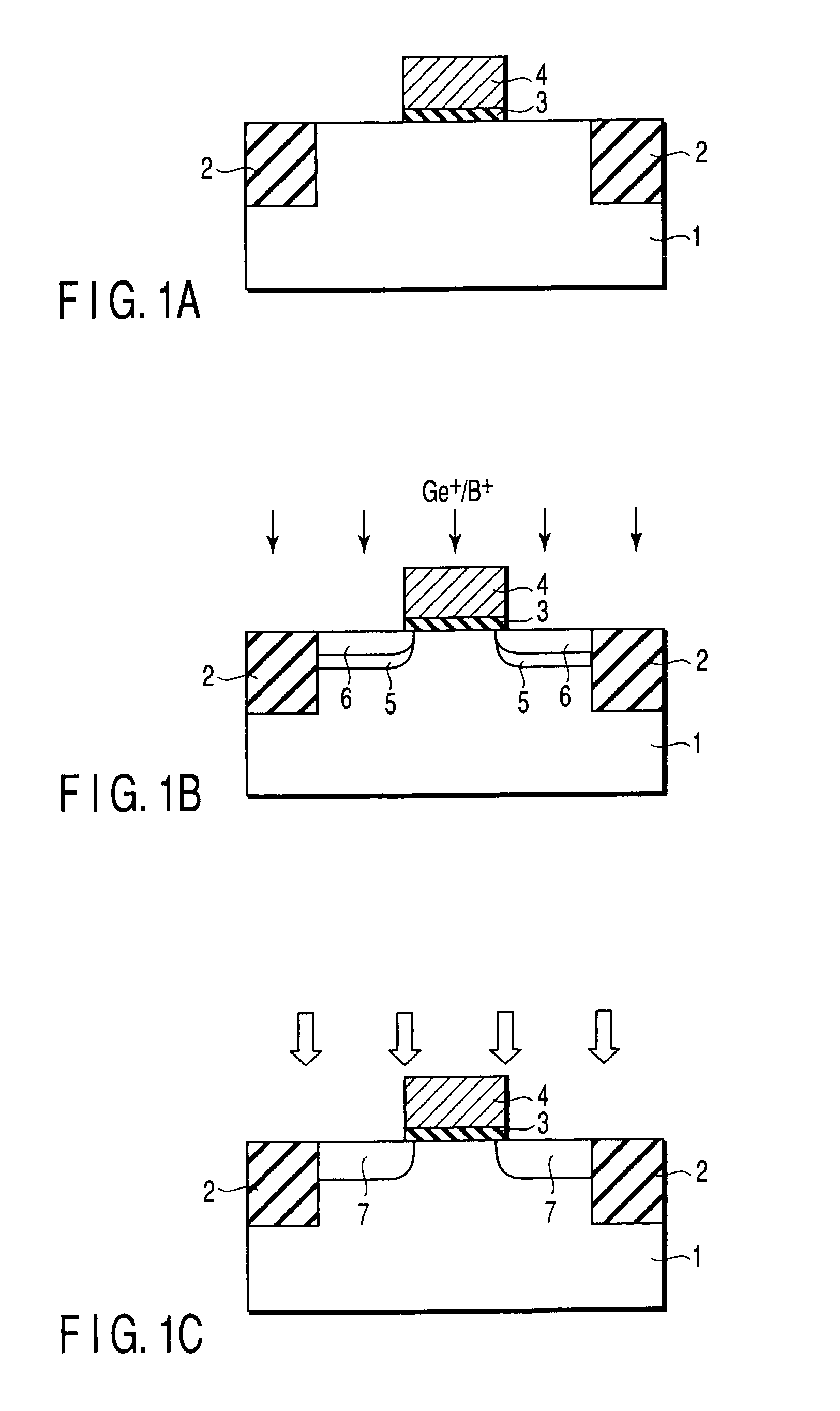
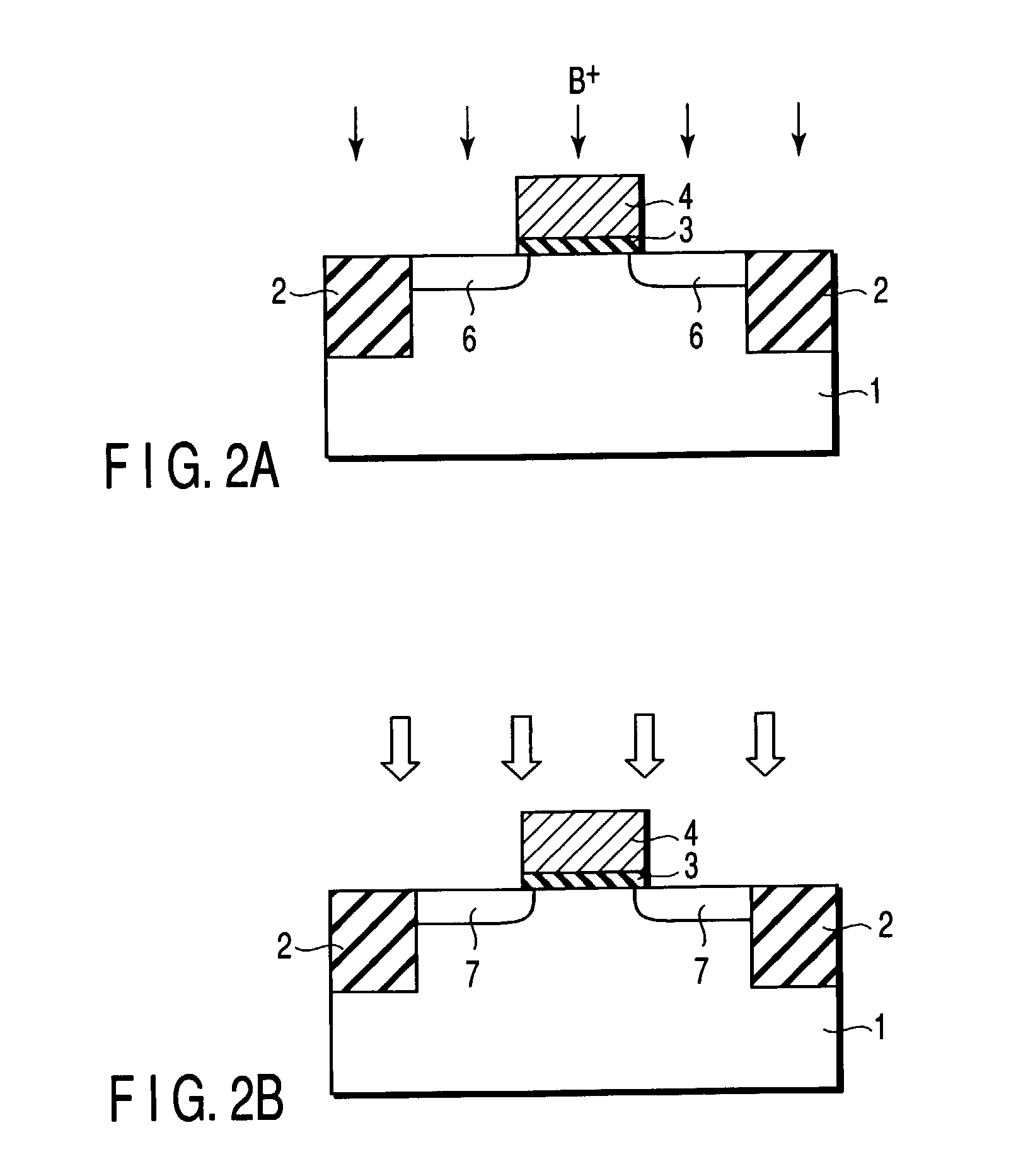
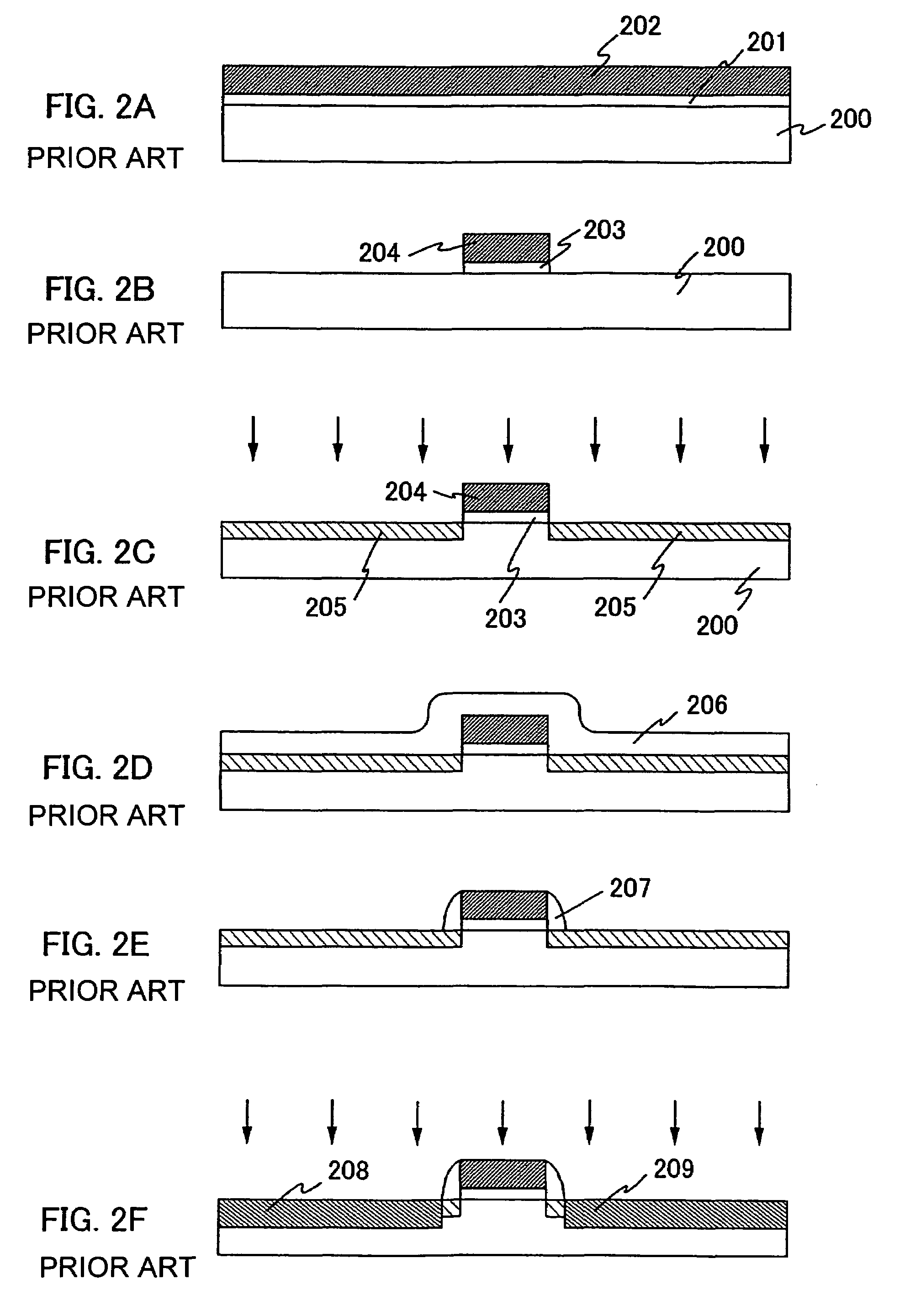
Semiconductor device including gate electrode for applying tensile stress to silicon substrate, and method of manufacturing the same
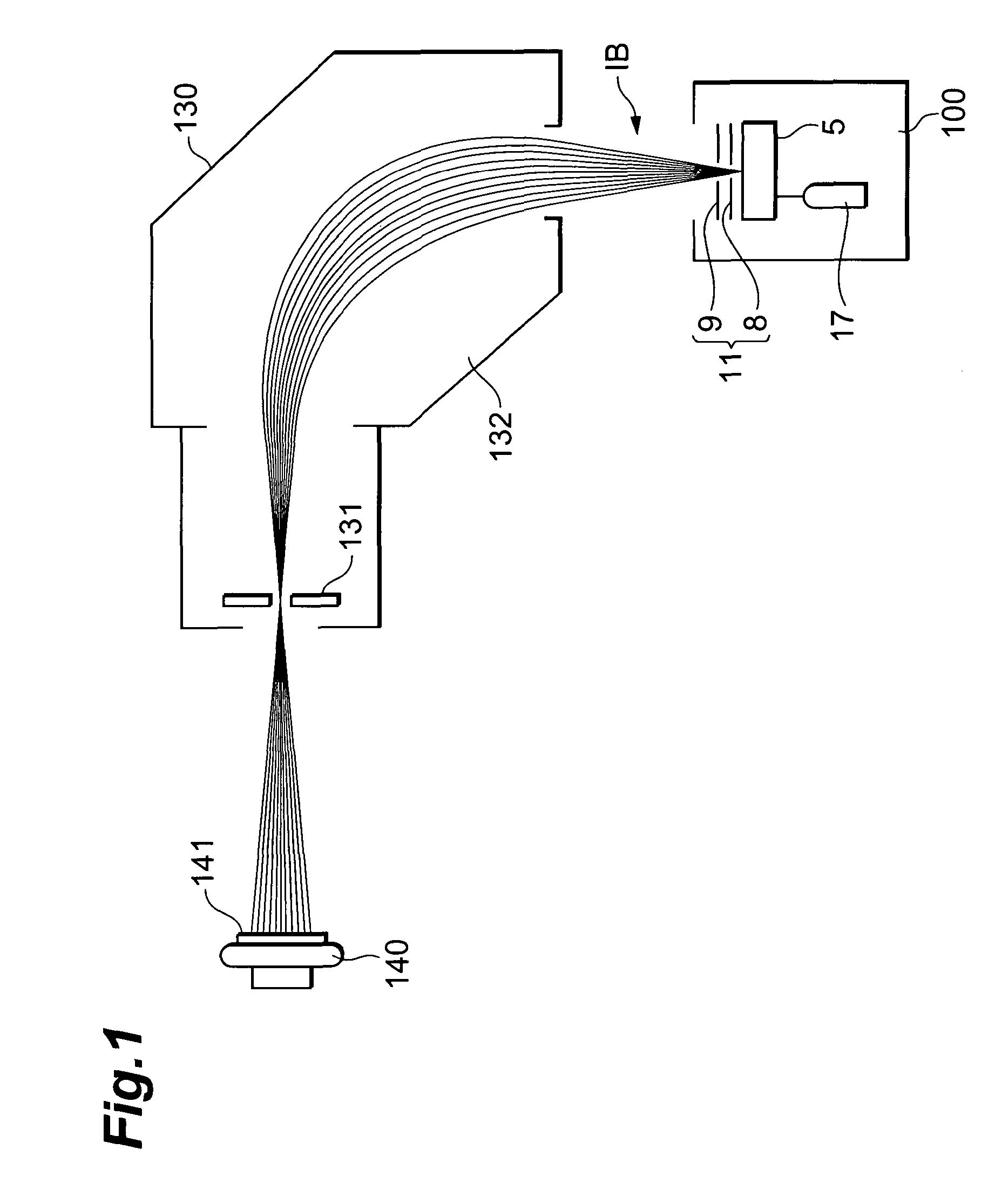
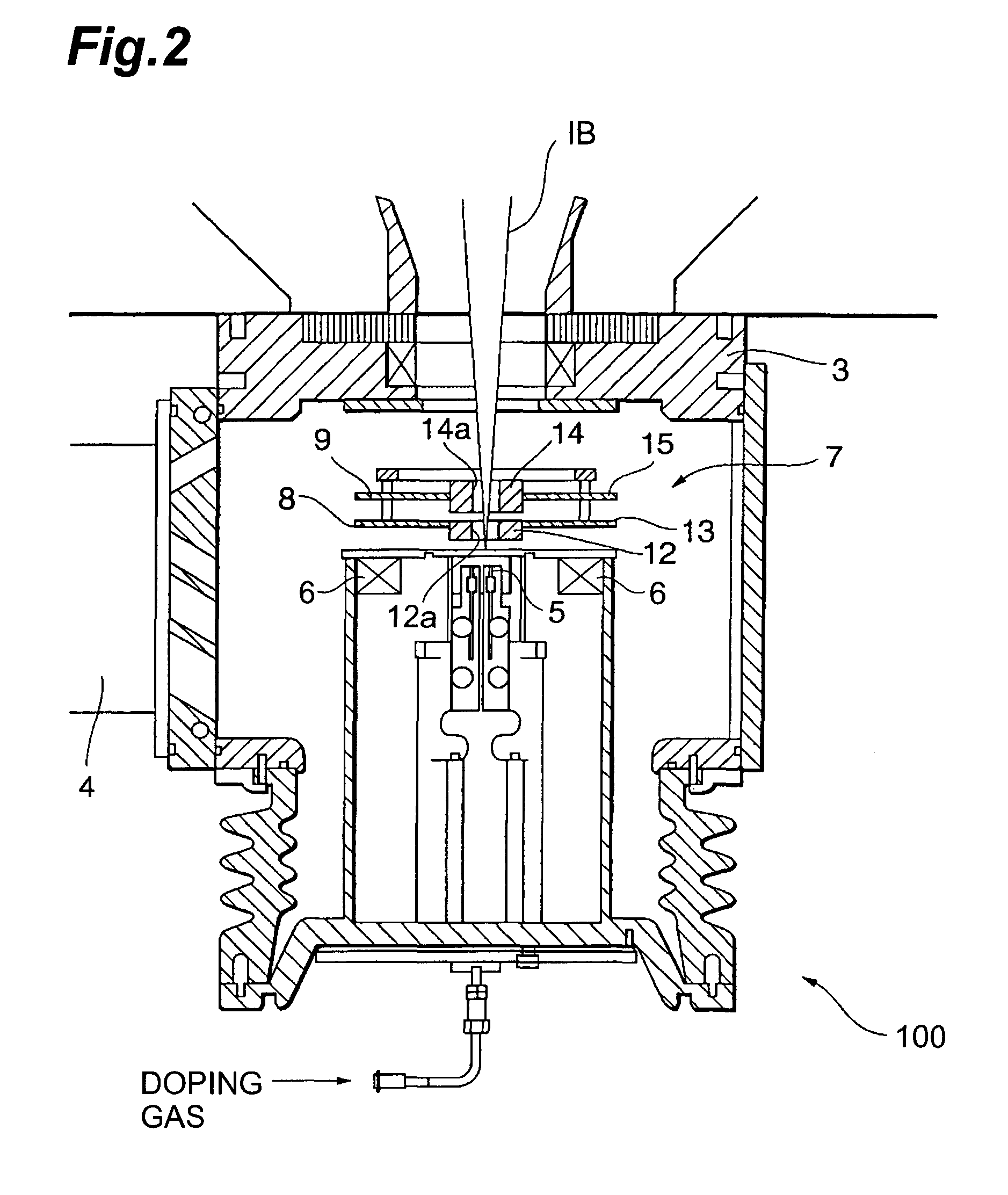
InactiveUS6906393B2High carrier mobilitySimplify manufacturing stepsTransistorSolid-state devicesMass numberCharge carrier mobility
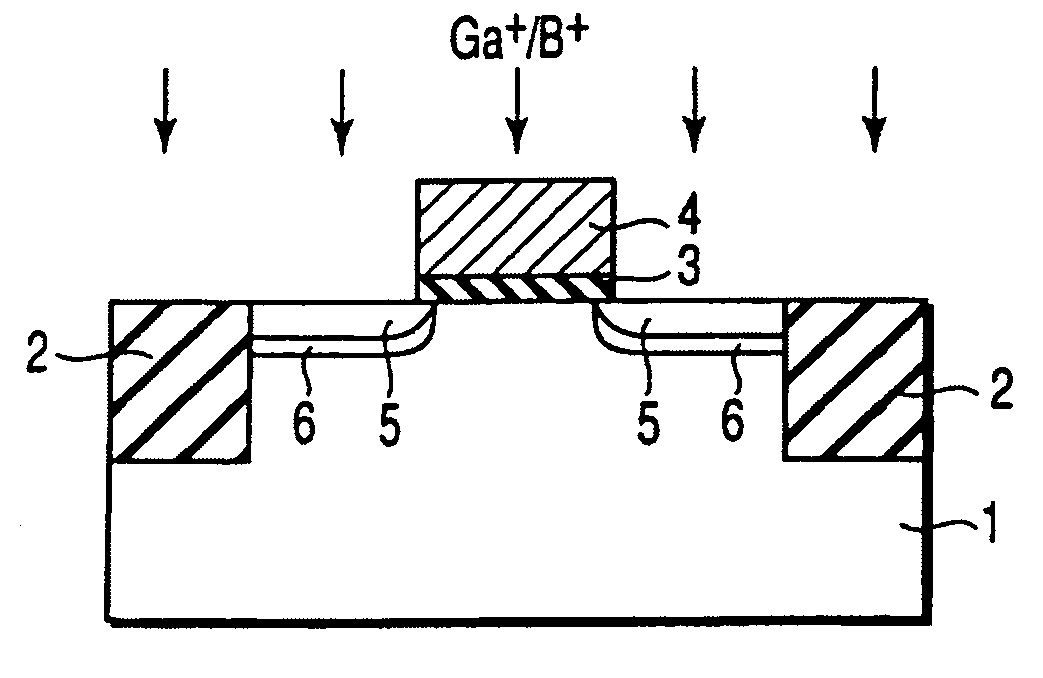
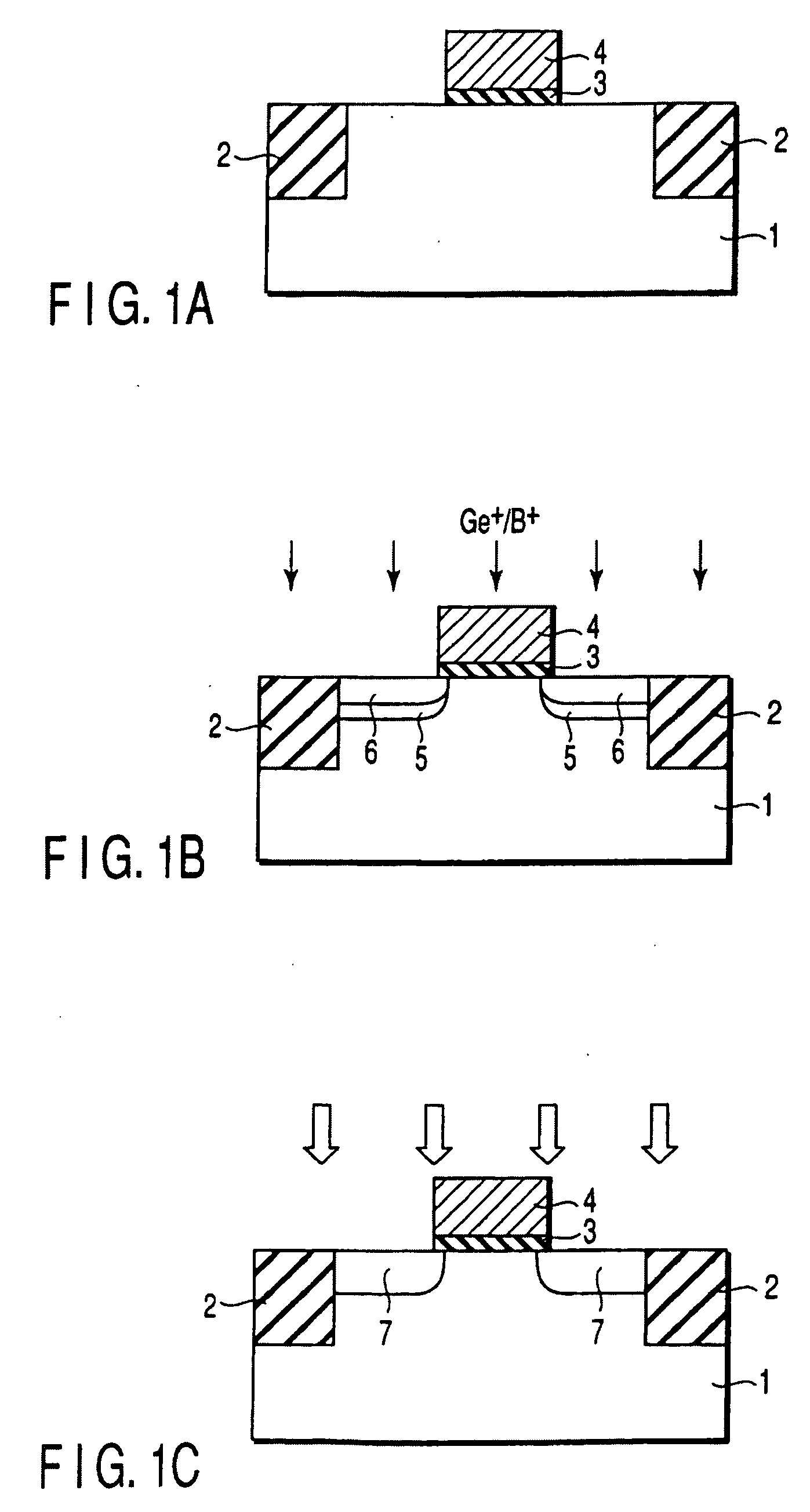
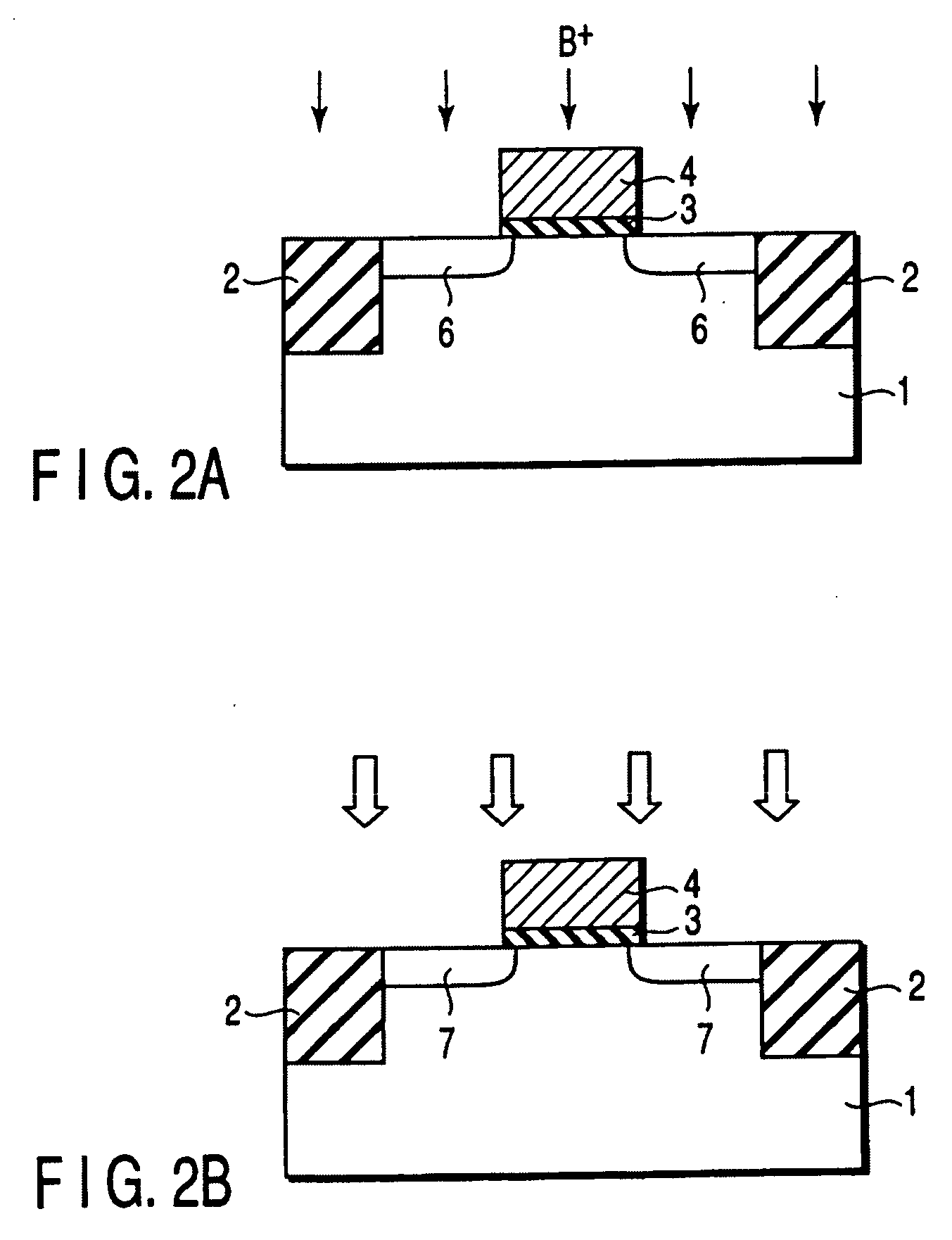
A gate insulating film (13) and a gate electrode (14) of non-single crystalline silicon for forming an nMOS transistor are provided on a silicon substrate (10). Using the gate electrode (14) as a mask, n-type dopants having a relatively large mass number (70 or more) such as As ions or Sb ions are implanted, to form a source / drain region of the nMOS transistor, whereby the gate electrode (14) is amorphized. Subsequently, a silicon oxide film (40) is provided to cover the gate electrode (14), at a temperature which is less than the one at which recrystallization of the gate electrode (14) occurs. Thereafter, thermal processing is performed at a temperature of about 1000° C., whereby high compressive residual stress is exerted on the gate electrode (14), and high tensile stress is applied to a channel region under the gate electrode (14). As a result, carrier mobility of the nMOS transistor is enhanced.
Owner:RENESAS ELECTRONICS CORP
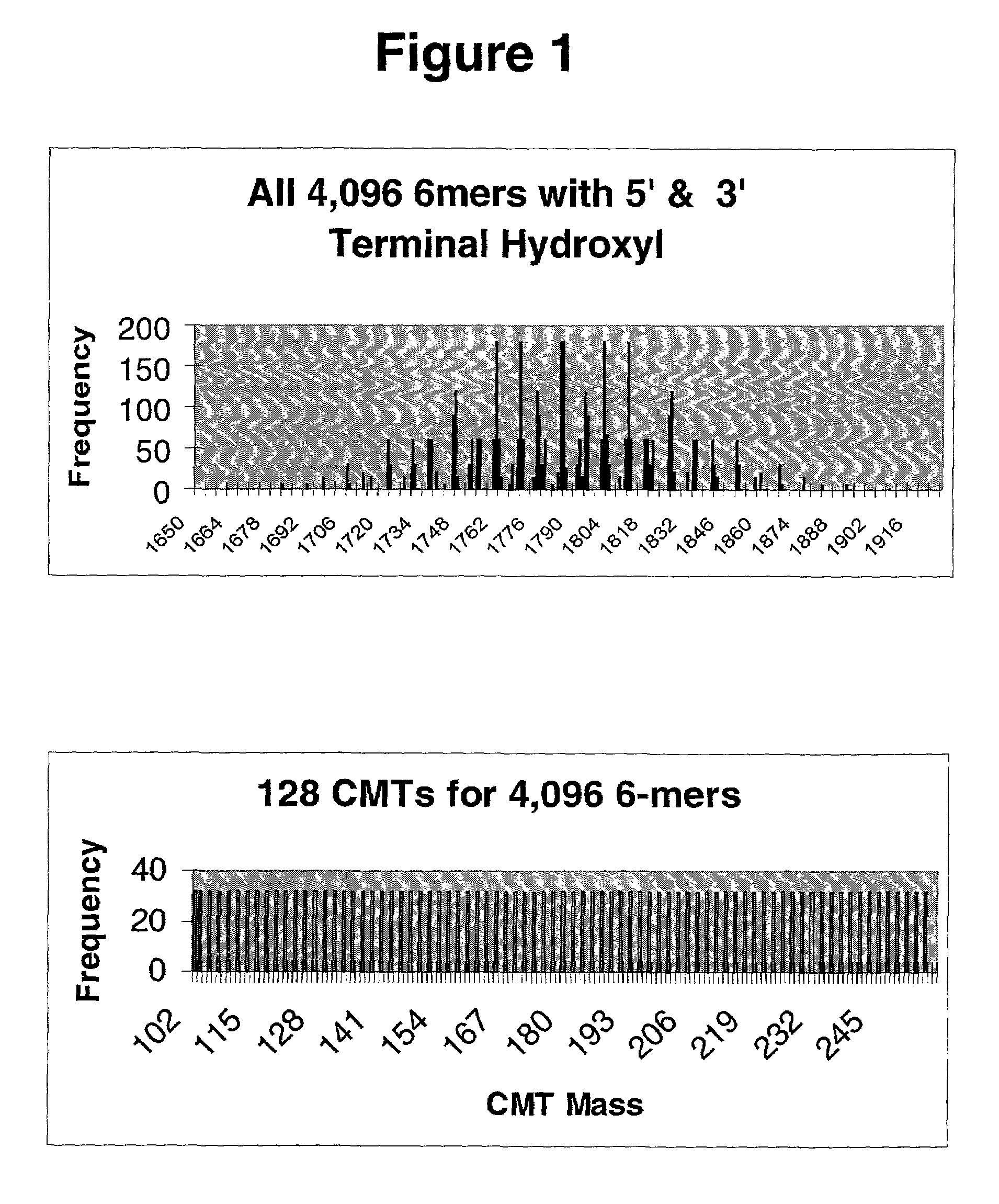
Method and reagents for analyzing the nucleotide sequence of nucleic acids
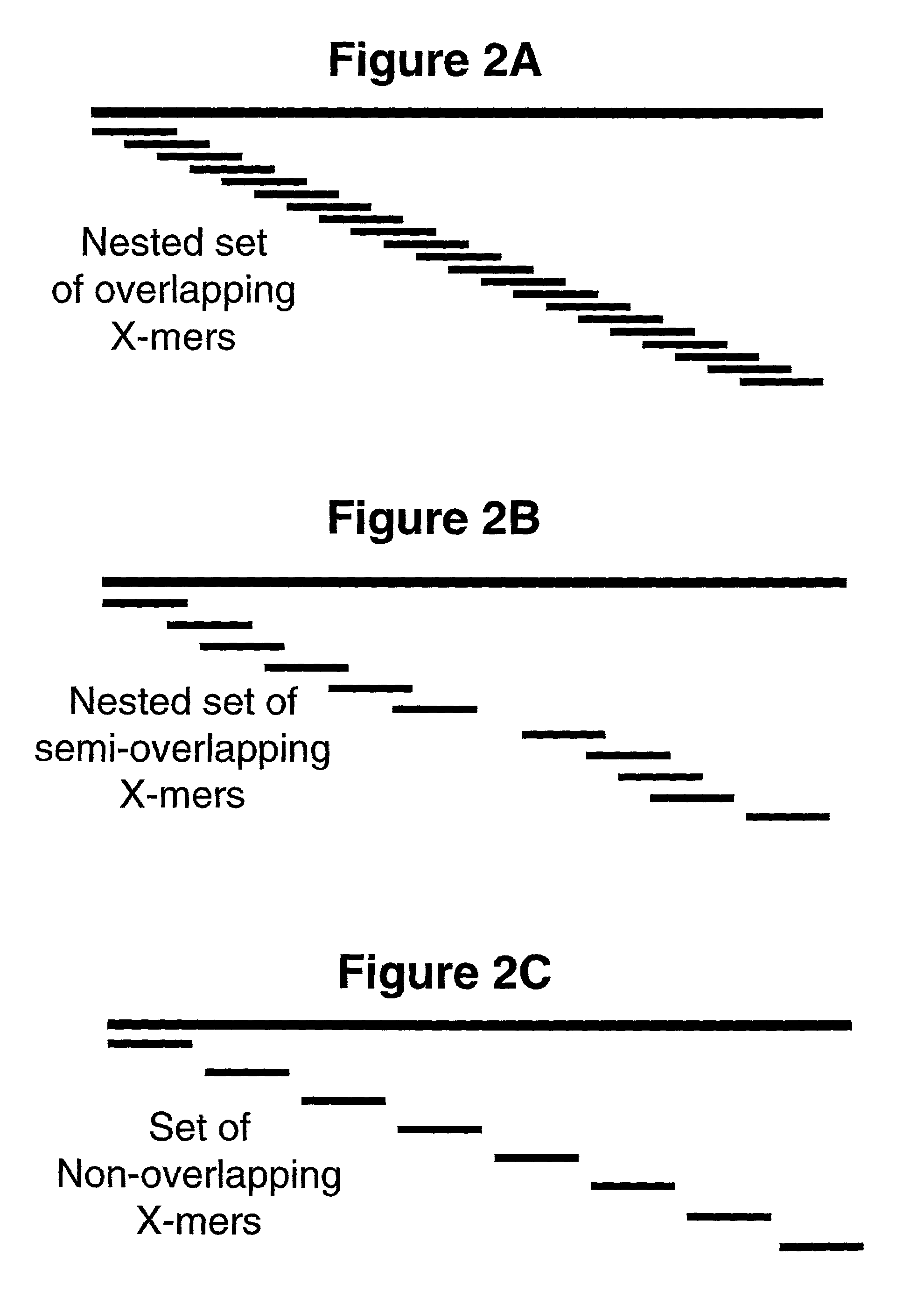
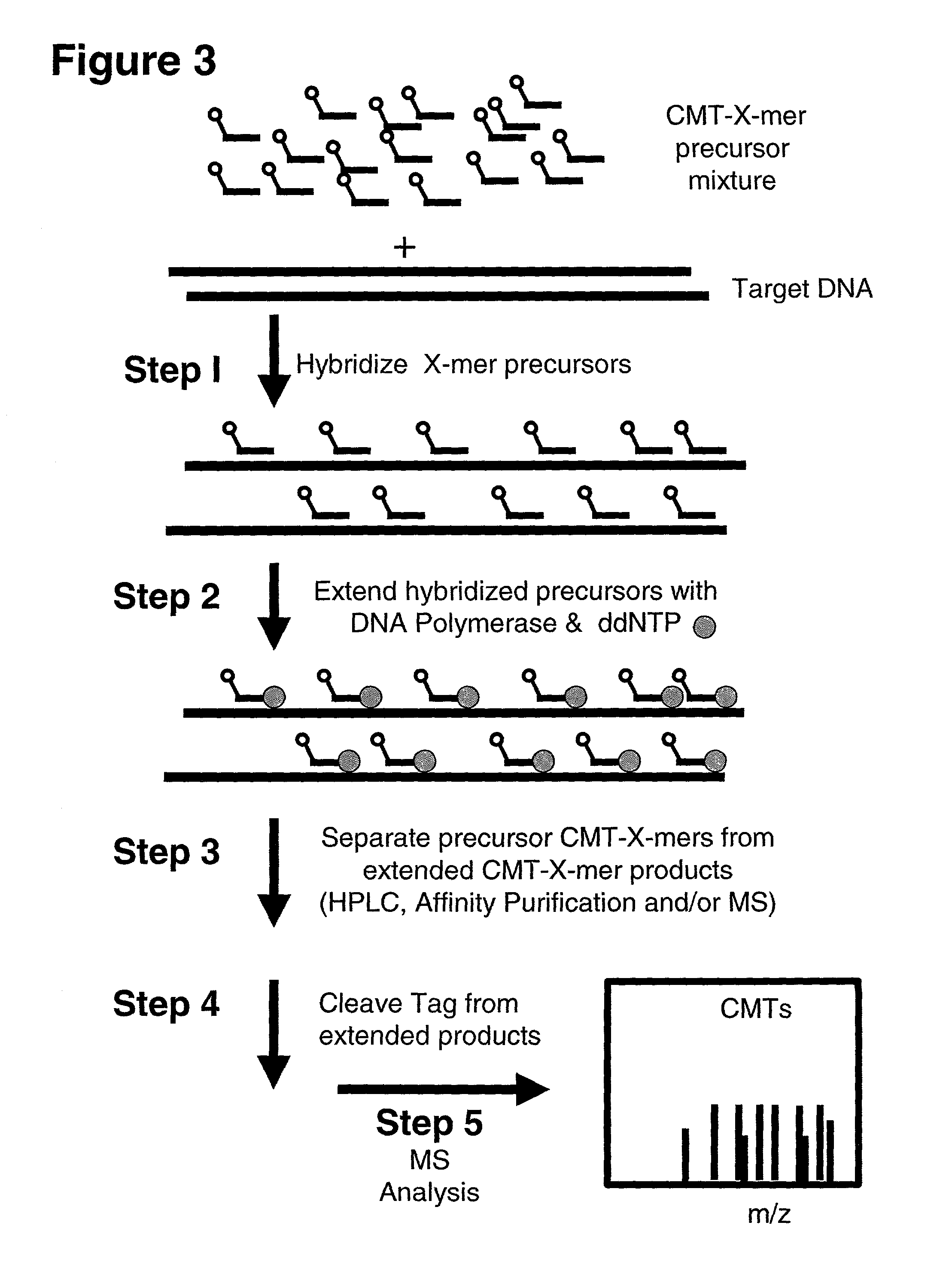
Methods and reagents are disclosed which provide for more sensitive, more accurate and higher through-put analyses of target nucleic acid sequences. The methods and reagents of the present invention may be generically applied to generally any target nucleic acid sequence and do not require a priori information about the presence, location or identity of mutations in the target nucleic acid sequence. The reagents of the invention are mixtures of oligonucleotide precursors having a high level of coverage and mass number complexity, and also having tags analyzable by mass spectrometry which are covalently linked to the precursors through cleavable bonds. A method is also disclosed for analyzing a target nucleic acid sequence employing the mixtures of oligonucleotide precursors having tags analyzable by mass spectrometry covalently linked to the oligonucleotide precursors through cleavable bonds, and chemical or enzymatic assays to alter the mass of the oligonucleotide precursors prior to mass spectral analysis. The enzymatic assay may be a polymerase extension assay or a ligation-based assay. The kits for carrying out the methods of the invention are also disclosed.
Owner:AGILENT TECH INC
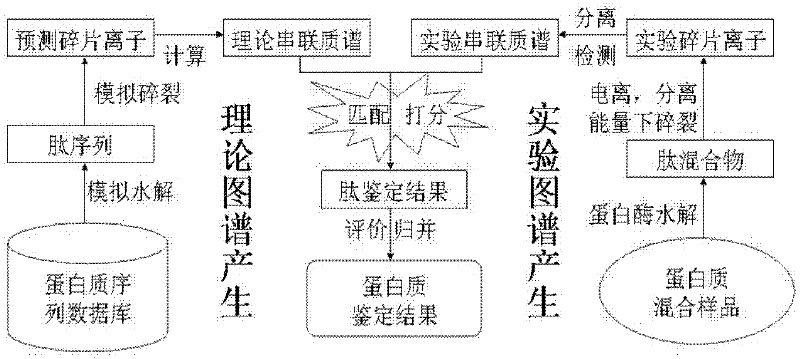
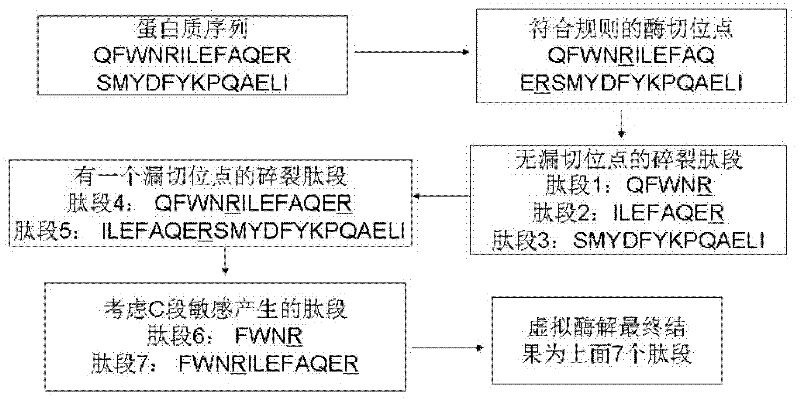
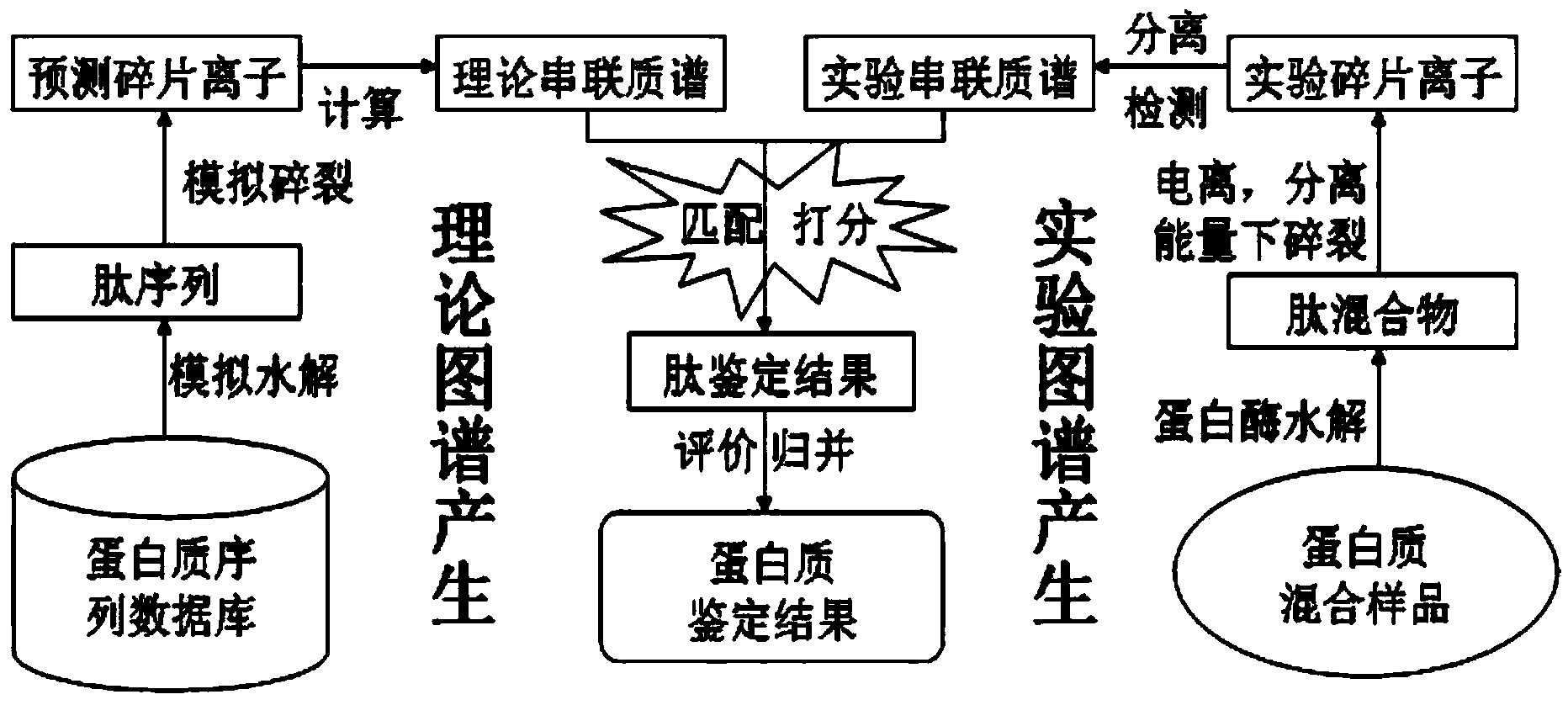
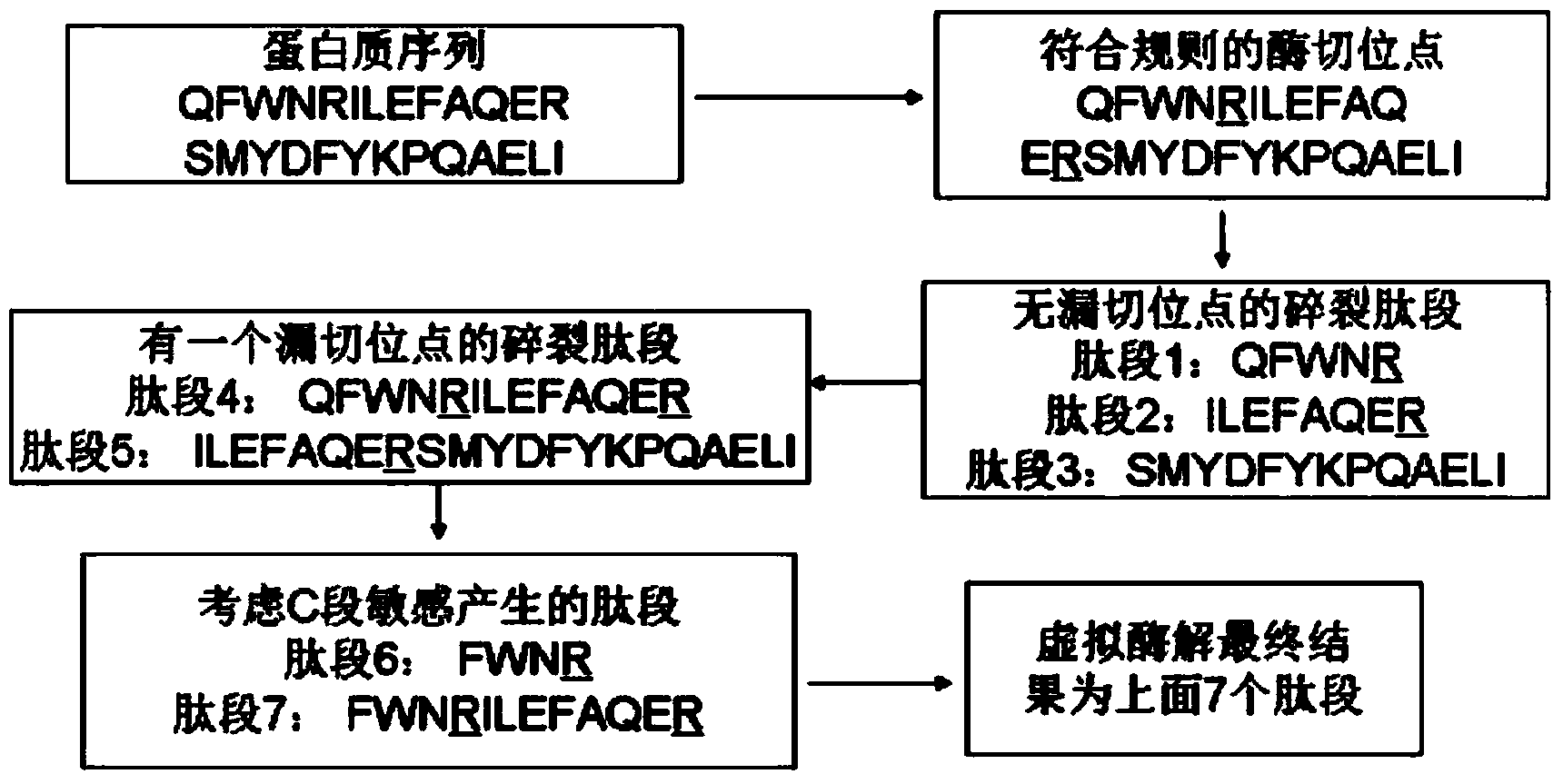
Protein secondary mass spectrometric identification method based on probability statistic model
InactiveCN102495127AMore identificationThe result of the identification method is excellentMaterial analysis by electric/magnetic meansProtein DatabasesMass number
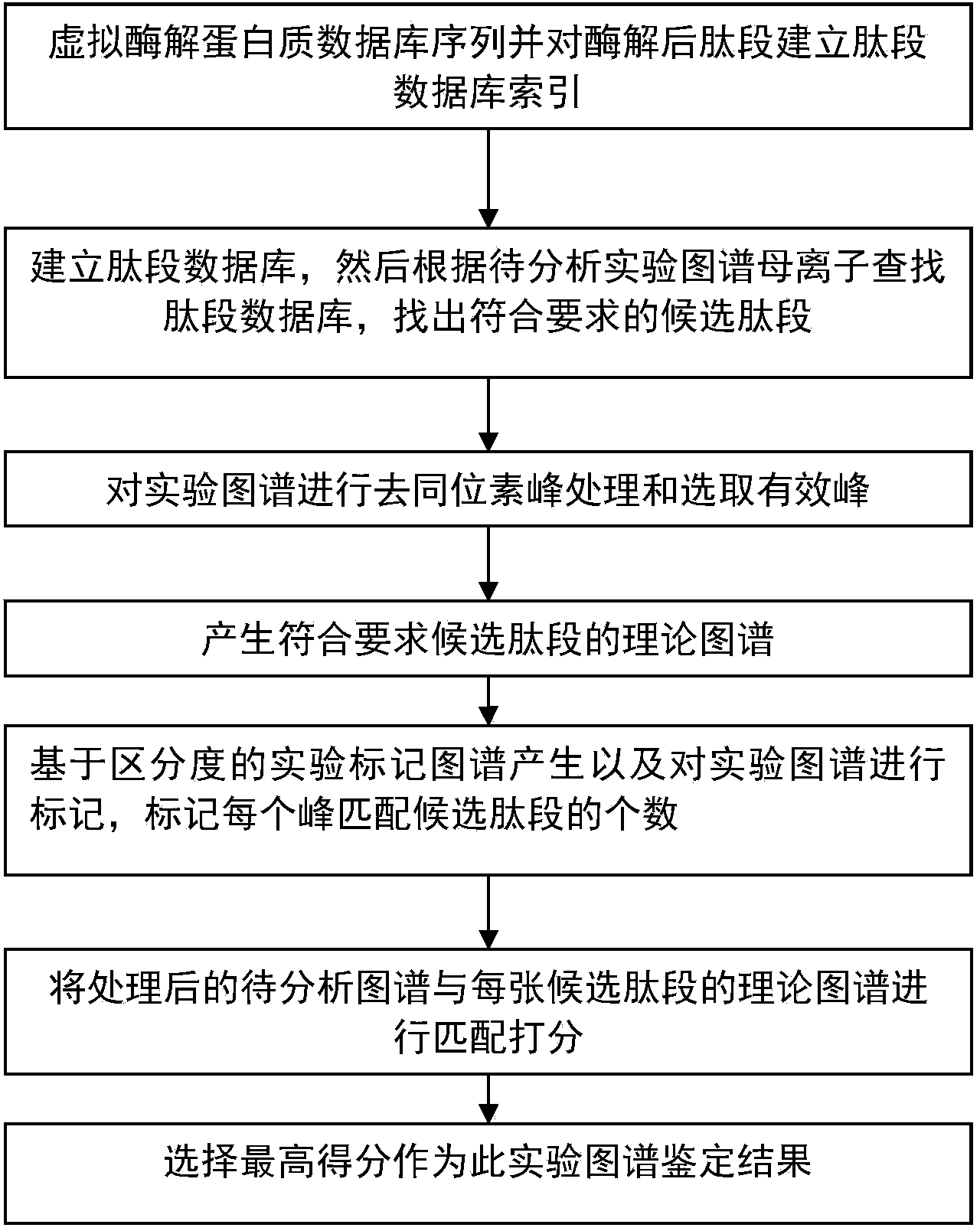
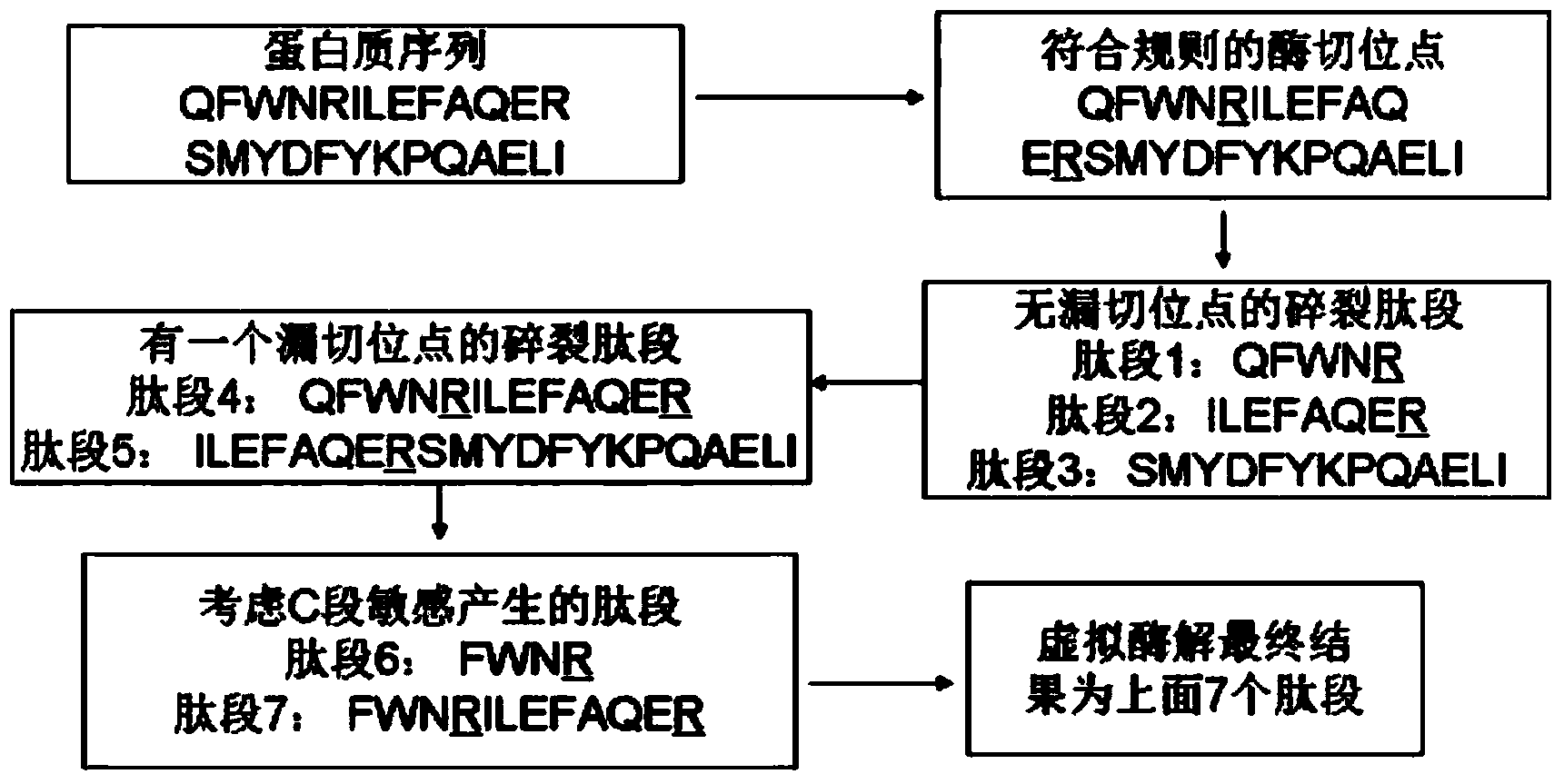
The invention discloses a protein secondary mass spectrometric identification method based on a probability statistic model. The method comprises the following steps of: firstly, virtualizing an enzymolysis protein database array, and establishing a peptide section database and a peptide section database index for peptide sections processed by the enzymolysis according to the mass number of the peptide sections; secondly, finding out standby peptide sections meeting the requirements from the peptide section database according to a nuclear-cytoplasmic ratio of parent ions in an experiment map to be analyzed, and generating a theoretical map meeting the requirements by all the standby peptide sections; thirdly, removing isotopes and noises from the experiment map to be analyzed; matching the processed experiment map to be analyzed and the theoretical map of each standby peptide section and grading, and selecting the standby peptide section with the highest score as an identification result of the experiment map; and finally, carrying out whole false positive control according to all the experiment map identification results. According to the invention, the quantity of effective massspectrums and the quantity of the protein peptide sections are higher than those of an existing algorithm; and the method has the advantages of capability of dynamically selecting peaks and fast operation speed.
Owner:JINAN UNIVERSITY
Semiconductor device and method of manufacturing the same
InactiveUS7091114B2Increase the number ofTransistorSemiconductor/solid-state device manufacturingMass numberLuminous intensity
Disclosed is a method for manufacturing a semiconductor device comprising implanting ions of an impurity element into a semiconductor region, implanting, into the semiconductor region, ions of a predetermined element which is a group IV element or an element having the same conductivity type as the impurity element and larger in mass number than the impurity element, and irradiating a region into which the impurity element and the predetermined element are implanted with light to anneal the region, the light having an emission intensity distribution, a maximum point of the distribution existing in a wavelength region of not more than 600 nm.
Owner:KK TOSHIBA
Ion implantation method, SOI wafer manufacturing method and ion implantation system
ActiveUS7064049B2Sufficient throughputIncrease currentVacuum evaporation coatingSputtering coatingMass numberIon implantation
The present invention provides an ion implantation method which can achieve sufficient throughput by increasing a beam current even in the case of ions with a small mass number or low-energy ions, an SOI wafer manufacturing method, and an ion implantation system. When ions are implanted by irradiating a semiconductor substrate with an ion beam, predetermined gas is excited in a pressure-reduced chamber to generate plasma containing predetermined ions, a magnetic field is formed by a solenoid coil or the like along an extraction direction when the ions are extracted to the outside of the chamber, and the ions are extracted from the chamber with predetermined extraction energy. The formation of the magnetic field promotes ion extraction, but this magnetic field has no influence on an advancing direction of the extracted ions. Therefore, the ion beam current can be kept at a high level-to contribute to the ion implantation.
Owner:APPLIED MATERIALS INC
Measuring method for quickly measuring contents of heavy metals in tobacco by using microwave digestion/ICP-MS method
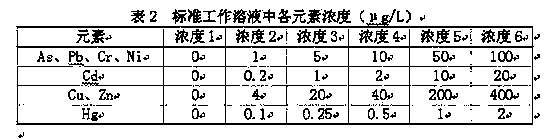
InactiveCN103412034AQuick breakdownReduce pollutionPreparing sample for investigationMaterial analysis by electric/magnetic meansMass numberPretreatment method
The invention discloses a measuring method for quickly measuring the contents of heavy metals in tobacco by using a microwave digestion / ICP-MS (Inductively Coupled Plasma Mass Spectrometry) method. According to the measuring method, a microwave digestion pretreatment method is adopted, an ICP-MS is adopted to measure the contents of arsenic, plumbum, cadmium, chromium, copper, zinc, nickel and mercury elements in a digestion liquid, 75As, 208Pb, 111Cd, 53Cr, 63Cu, 66Zn, 60Ni and 202Hg are selected as mass numbers of the to-be-measured elements to reduce mass spectrum interference, and an internal standard element Rh is added in an off-line manner to inhibit the matrix effect. For to-be-measured ions with a certain mass-to-charge ratio, mass spectrum signal response is directly proportional to the number of ions entering the ICP-MS, the concentration of the elements in a sample is measured by measuring the number of mass spectrum signals, and the contents of the elements are calculated by a standard curve method. The method is quick and simple, can efficiently and accurately measure the contents of heavy metals in tobacco and tobacco products, and provides an effective scheme for finding out the content level of heavy metals in cigarettes in China to reduce harm to human health.
Owner:江西省烟草公司抚州市公司 +2
Semiconductor device and manufacturing method thereof
InactiveUS20060121680A1Leakage currentShort-channel effectTransistorSemiconductor/solid-state device manufacturingMass numberDevice material
It is an object of the present invention to provide a semiconductor device superior in the decrease in leak current due to a short-channel effect and a manufacturing method thereof. In a process of forming a field-effect transistor over a single-crystal semiconductor substrate, an impurity is introduced to form an extension region and a single crystal lattice is broken to make the extension region amorphous. Alternatively, the impurity and an element having large mass number are introduced to break the single crystal lattice and make the extension region amorphous. Then, a laser beam with a pulse width of 1 fs to 10 ps and a wavelength of 370 to 640 nm is delivered to selectively activate the amorphous portion, so that the extension region is formed with a thickness of 20 nm or less.
Owner:SEMICON ENERGY LAB CO LTD
Nanometer all-silicon molecular sieve and its preparation method and use
InactiveCN102432032AHigh crystallinityRegular shapeLactams preparationMolecular sieve catalystsBeckmann rearrangementMass number
The invention provides a nanometer all-silicon molecular sieve. The nanometer all-silicon molecular sieve is prepared from tetrapropylammonium hydroxide, a silicon source, water and amino acids. A mole ratio of the silicon source to tetrapropylammonium hydroxide to water is 1: [0.05 to 0.50]: [15 to 65]. The mass of the used amino acids is 0.5 to 5% of that of the silicon source. A mole number and a mass number of silica represent a mole number and a mass number of the silicon source. The invention also provides a preparation method of the nanometer all-silicon molecular sieve, and a use of the nanometer all-silicon molecular sieve in a cyclohexanone-oxime gas phase beckmann rearrangement reaction. The nanometer all-silicon molecular sieve has a high degree of crystallization, regular morphology and adjustable particle sizes of 40 to 160 nanometers. In a cyclohexanone-oxime gas phase beckmann rearrangement reaction, the nanometer all-silicon molecular sieve shows excellent catalytic activity, selectivity and stability.
Owner:HUNAN UNIV
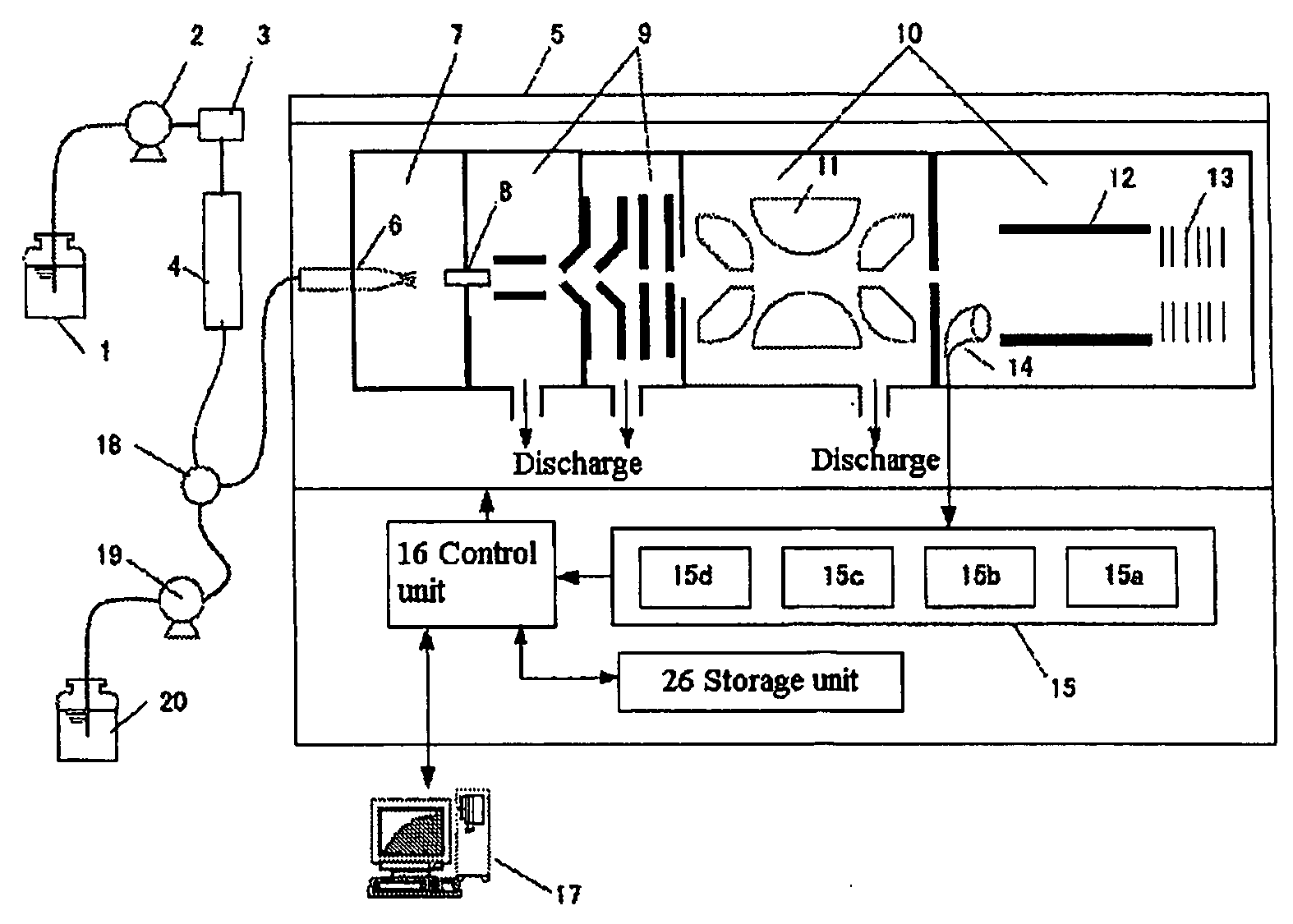
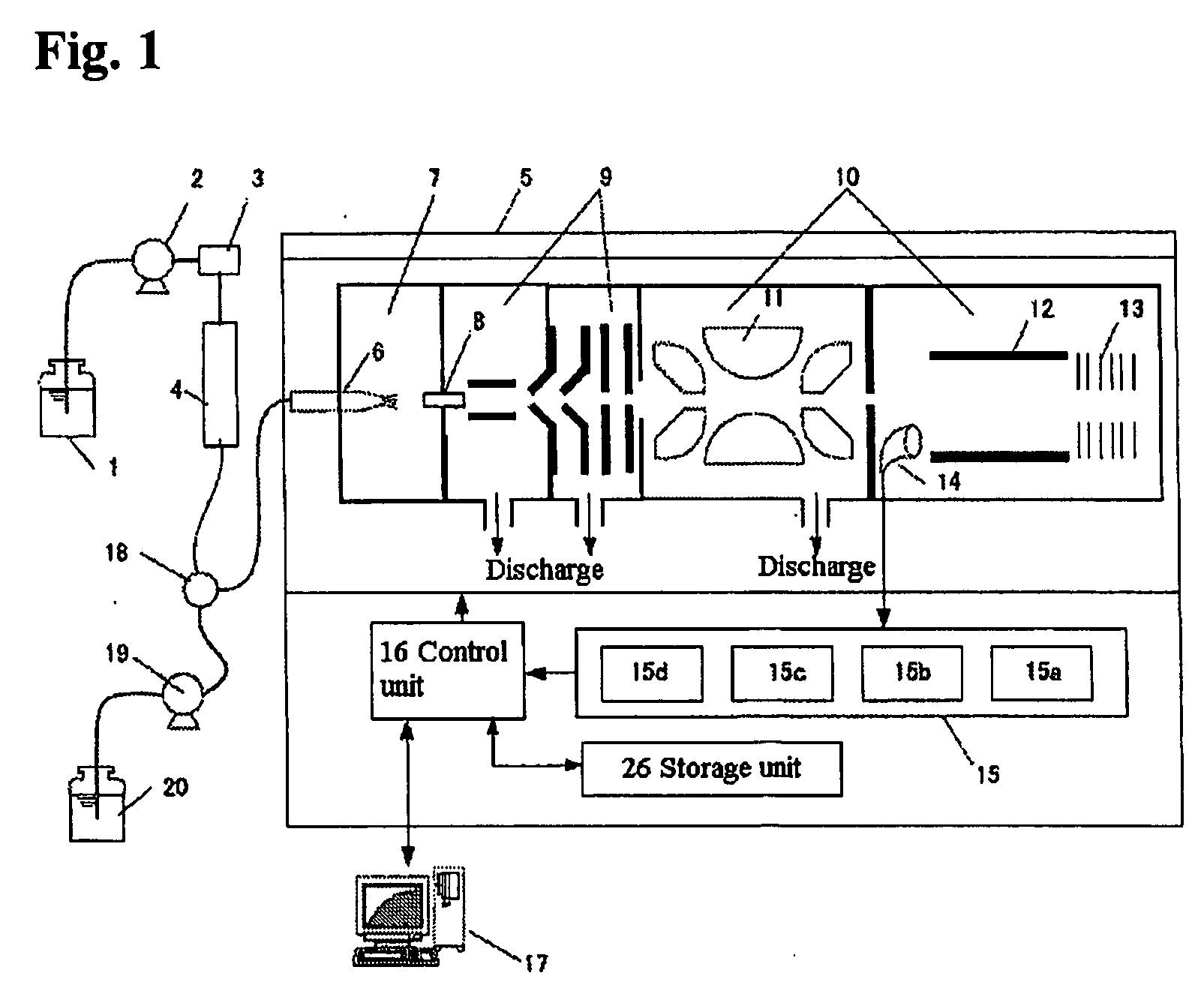
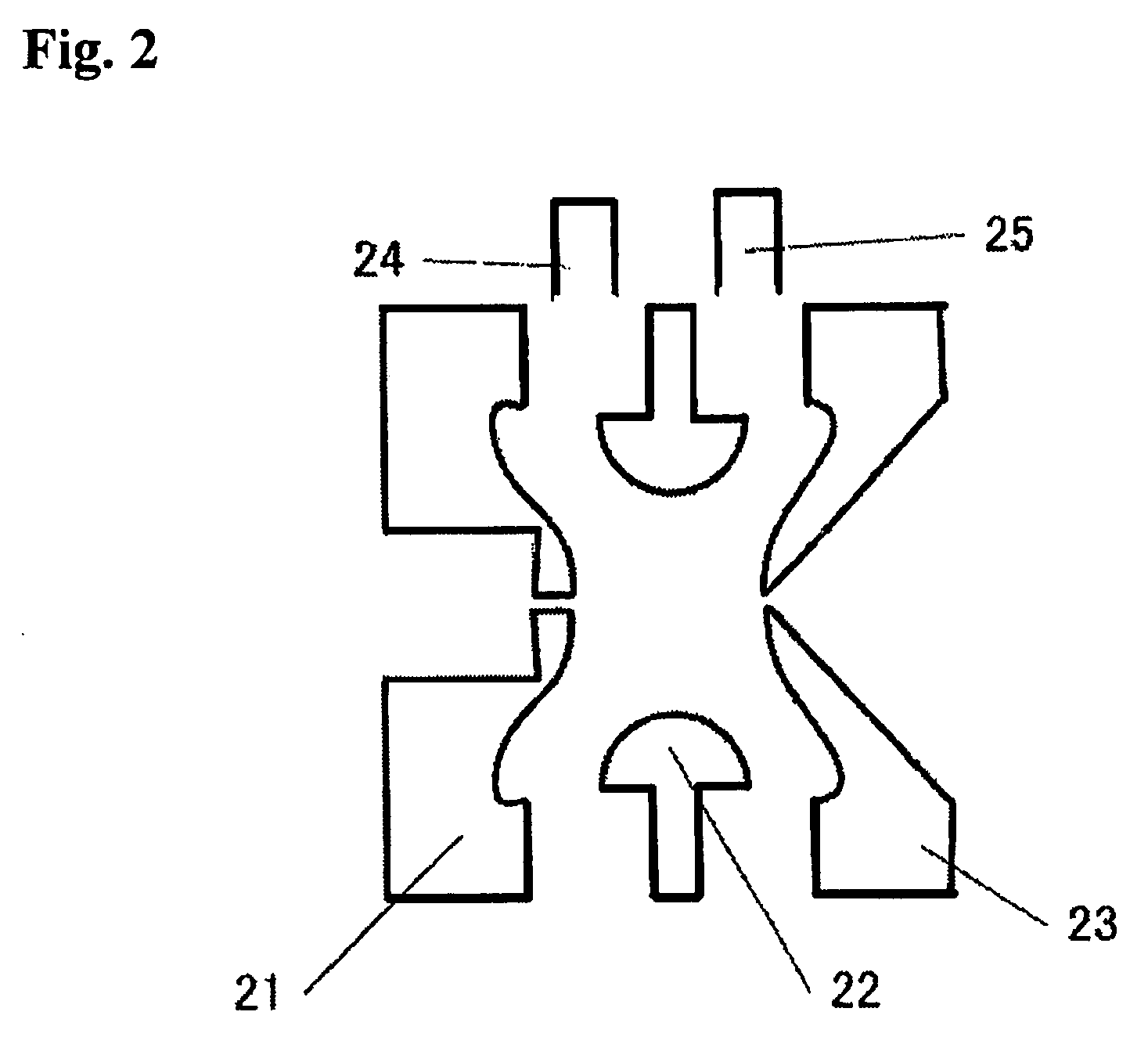
Time of flight mass spectrometer
InactiveUS7355168B2Improve accuracyPreferable mass spectrumTime-of-flight spectrometersIsotope separationMass numberMass analyzer
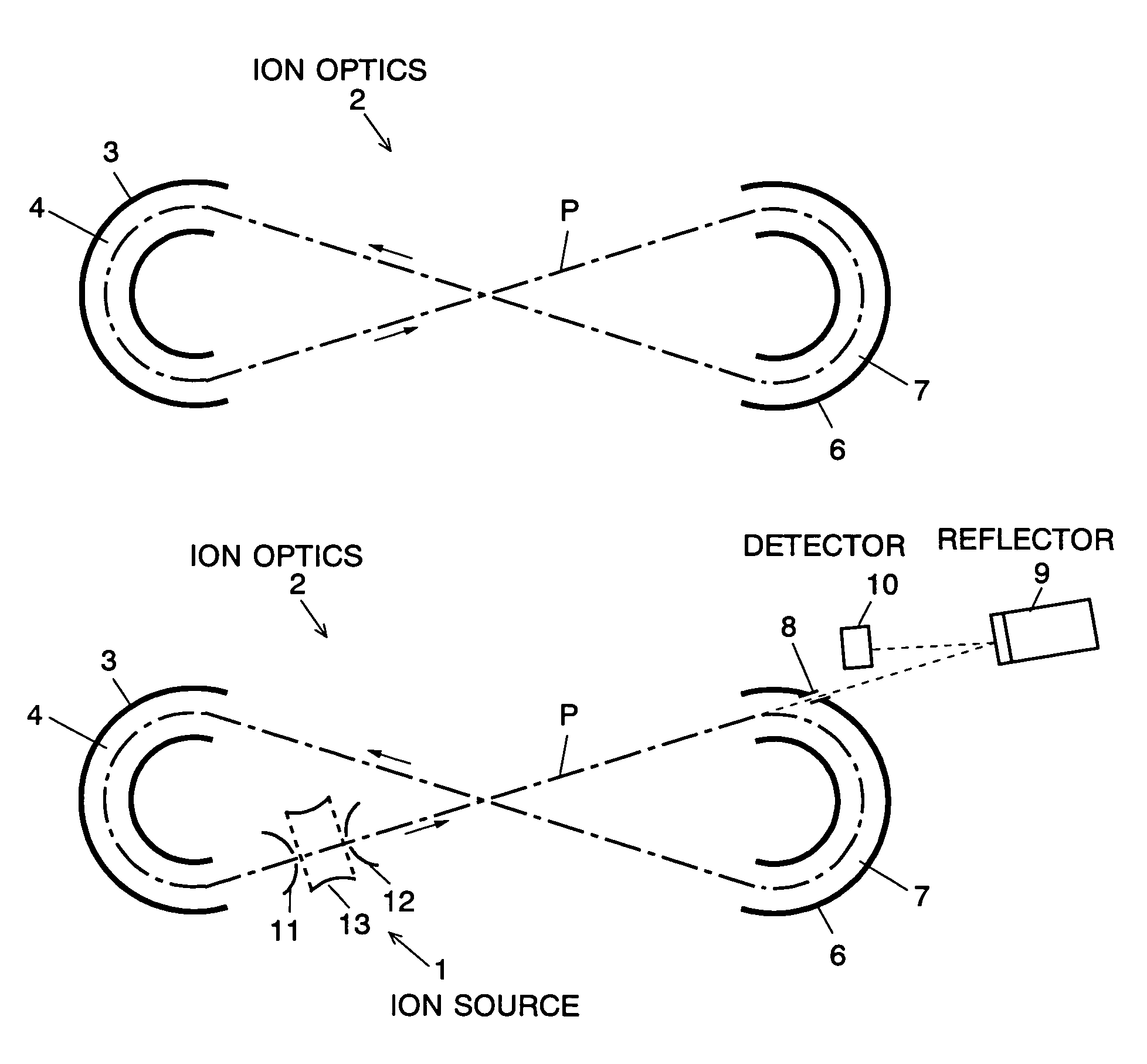
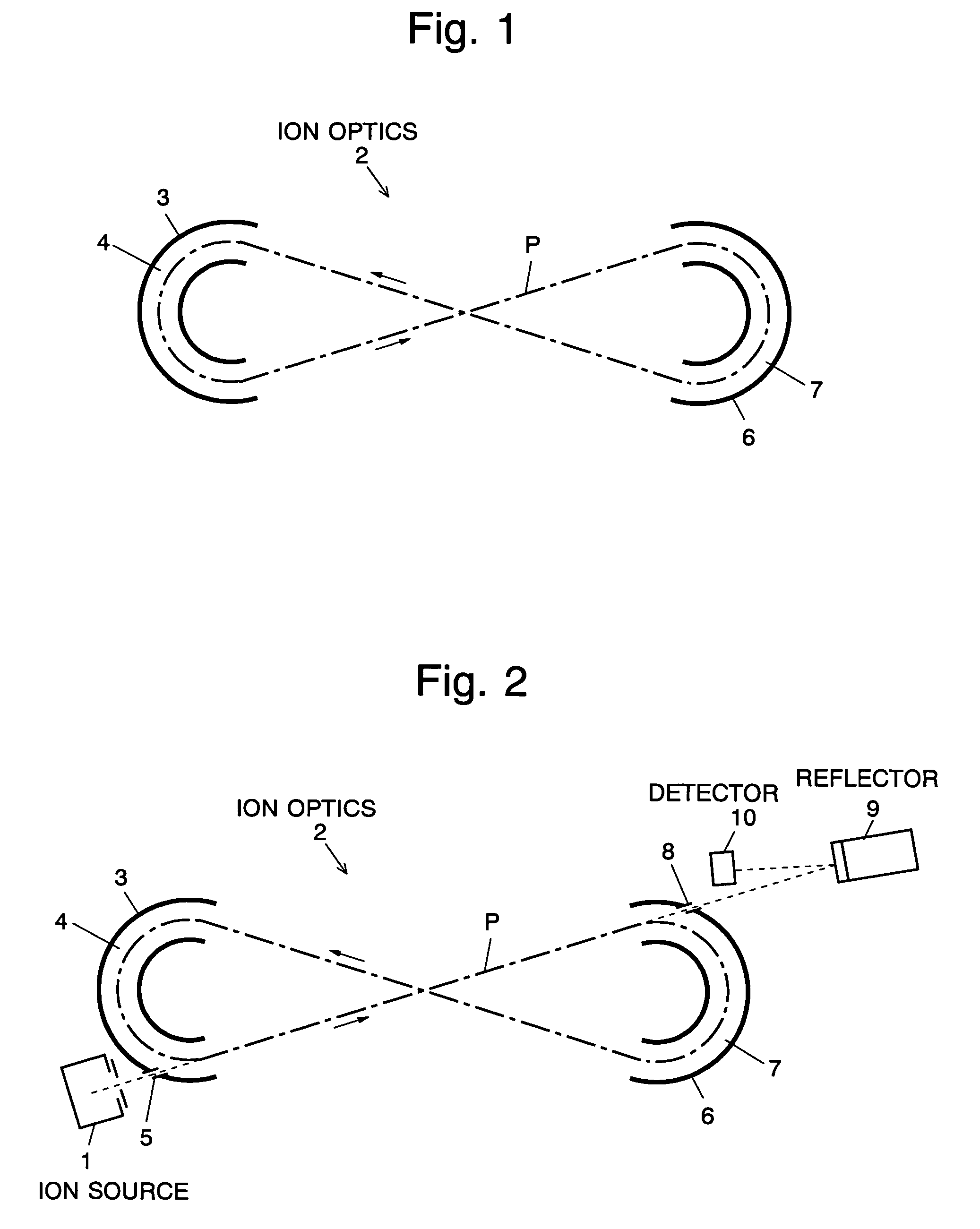
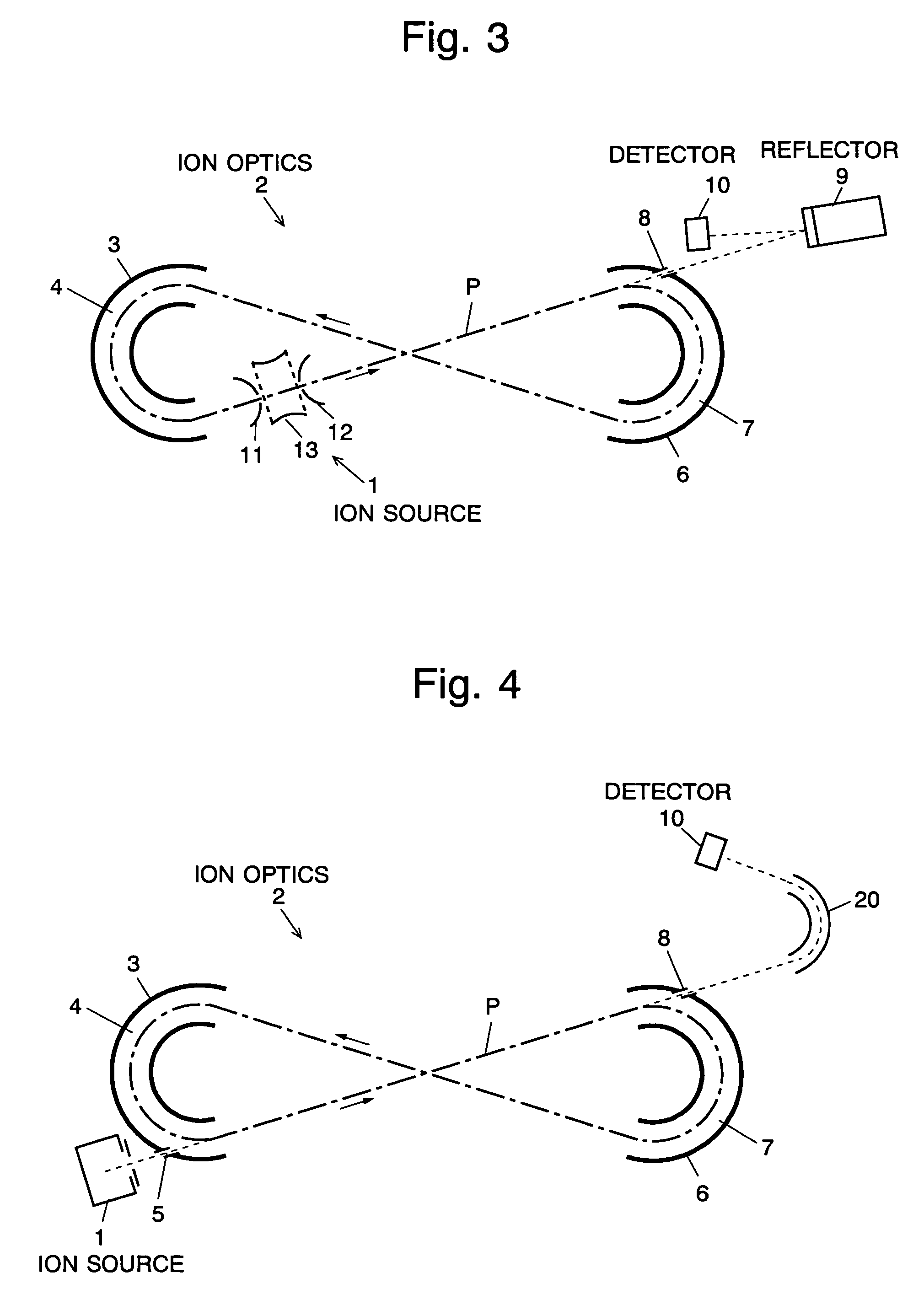
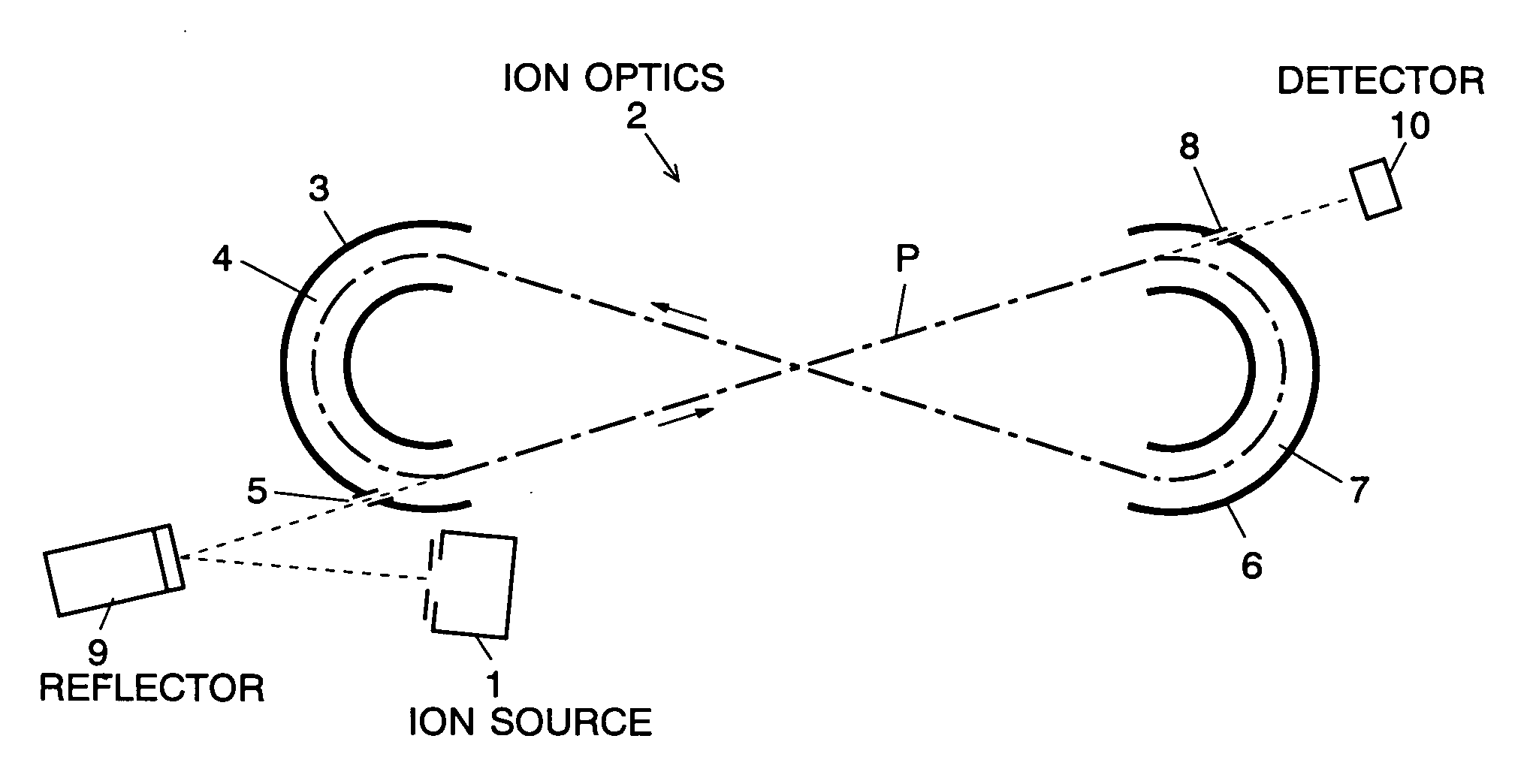
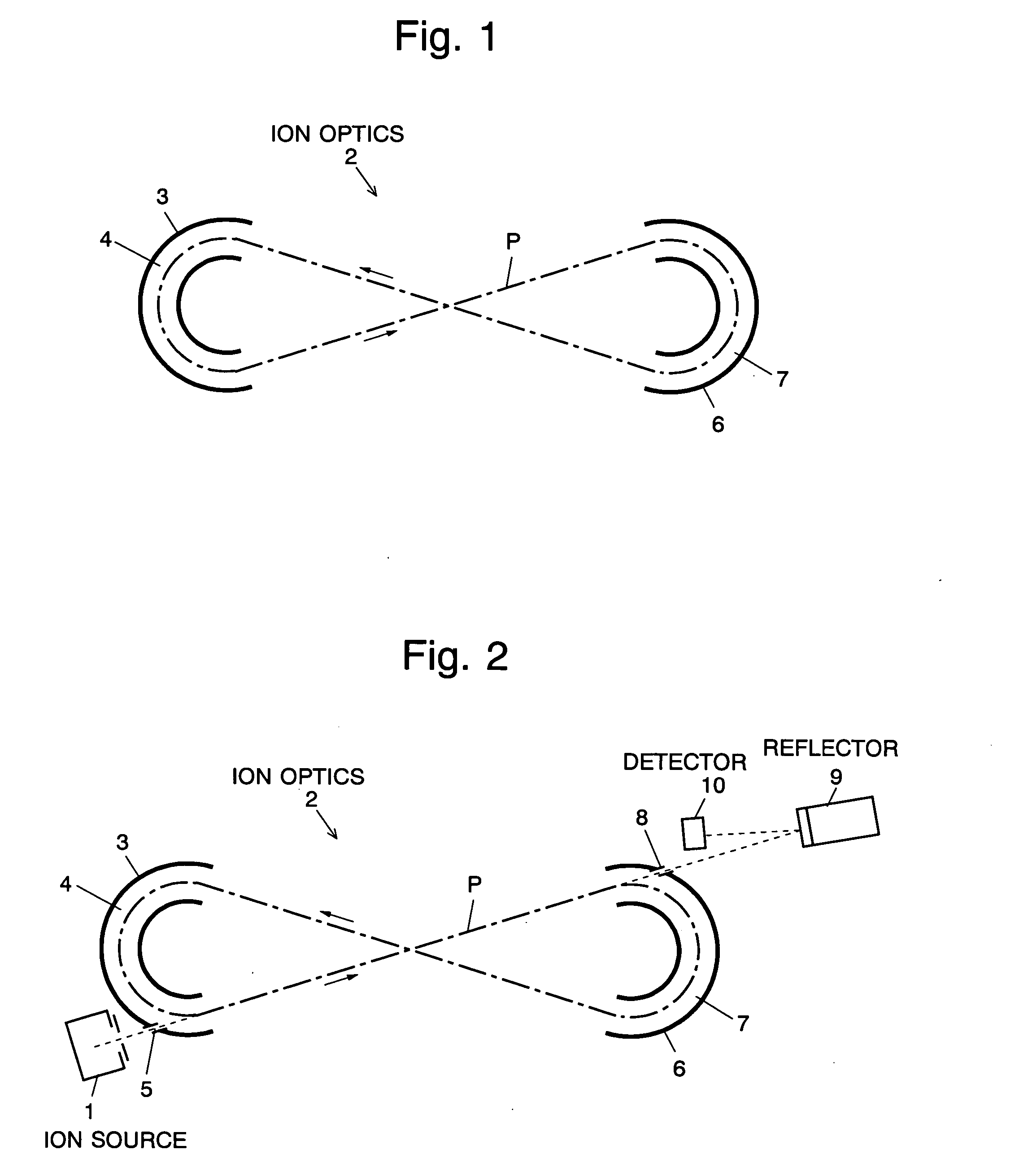
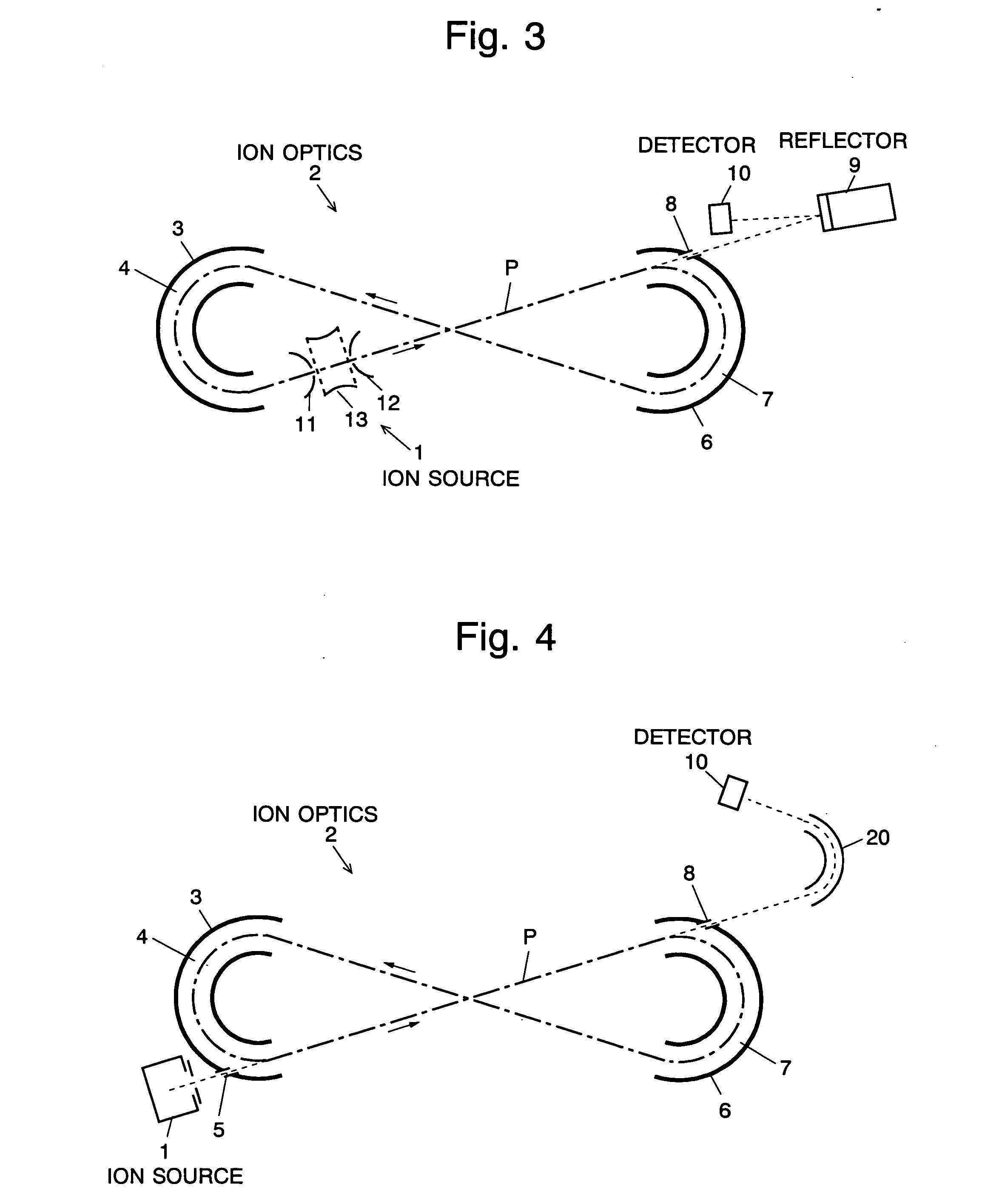
The present invention provides a time of flight mass spectrometer having an ion optics forming a multi-turn track, which is capable of time-focusing the ions while allowing the multi-turn track to be configured in an unlimited and highly variable manner. In a specific form of the invention, a reflector 9 is provided on the flight path between the position where the ions leave the loop orbit P and the ion detector 10 located outside the loop orbit P, and the condition of the electric field generated by the reflector 9 is appropriately determined. Thus, even if the ions cannot be well time-focused by the ion optics 2 creating the sector-shaped electric fields 4 and 7, it is possible to compensate the time-focusing performance with the reflector 9 to achieve a good performance of time-focusing of the ion throughout the overall system wherein the ions leave the ion source 1 and finally reach the ion detector 10. Thereby, the ions can reach the ion detector 10 at approximately the same time even if the ions having the same mass number have different levels of energy at the moment they leave the ion source 1.
Owner:SHIMADZU CORP
A nucleus fuel with carborundum as inertial base and its making method
ActiveCN101188147AImprove thermal conductivityEasy to prepareNuclear energy generationReactors manufactureMass numberEngineering
The invention provides a nuclear fuel and a preparation method thereof, wherein, the nuclear fuel takes carborundum as an inert basal body. The carborundum inert basal body nuclear fuel equably disperses fuel globules of zirconia fuel to the carborundum basal body, and the mass number dispersed in the carborundum basal body is 5-35 percent. The nuclear fuel is a column-shaped or ring-shaped ceramic-ceramic complex fuel core block. The nuclear fuel has good thermal conduction performance and irradiation-resistant performance, and the chemical durability is high. The preparation method equably disperses fuel globules of zirconia fuel to the carborundum basal body, and then ceramic complex carborundum inert basal body nuclear fuel of the zirconia fuel globules equably dispersed in the carborundum basal body are formed through rapid agglomeration of electric sparks. The carborundum inert basal body nuclear fuel is the safe and environment-friendly nuclear fuel. The preparation method is simple and reliable.
Owner:NUCLEAR POWER INSTITUTE OF CHINA
Time of flight mass spectrometer
InactiveUS20060192110A1The degree of freedom becomes largerSimple configurationParticle separator tubesIsotope separationTime-of-flight mass spectrometryMass number
The present invention provides a time of flight mass spectrometer having an ion optics forming a multi-turn track, which is capable of time-focusing the ions while allowing the multi-turn track to be configured in an unlimited and highly variable manner. In a specific form of the invention, a reflector 9 is provided on the flight path between the position where the ions leave the loop orbit P and the ion detector 10 located outside the loop orbit P, and the condition of the electric field generated by the reflector 9 is appropriately determined. Thus, even if the ions cannot be well time-focused by the ion optics 2 creating the sector-shaped electric fields 4 and 7, it is possible to compensate the time-focusing performance with the reflector 9 to achieve a good performance of time-focusing of the ion throughout the overall system wherein the ions leave the ion source 1 and finally reach the ion detector 10. Thereby, the ions can reach the ion detector 10 at approximately the same time even if the ions having the same mass number have different levels of energy at the moment they leave the ion source 1.
Owner:SHIMADZU CORP
Mass spectrometer
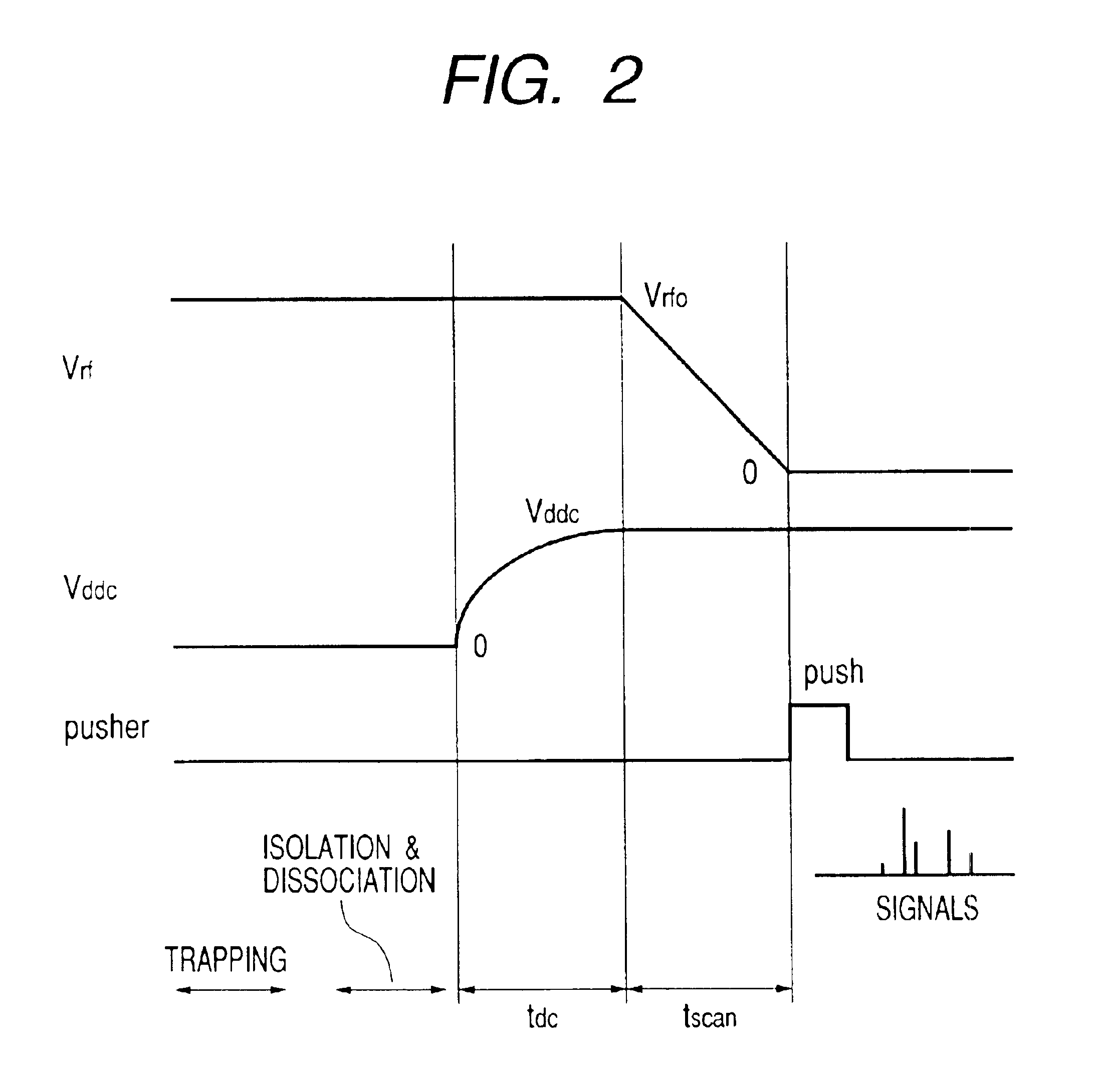
InactiveUS6852972B2Stability-of-path spectrometersTime-of-flight spectrometersMass numberIon trap mass spectrometry
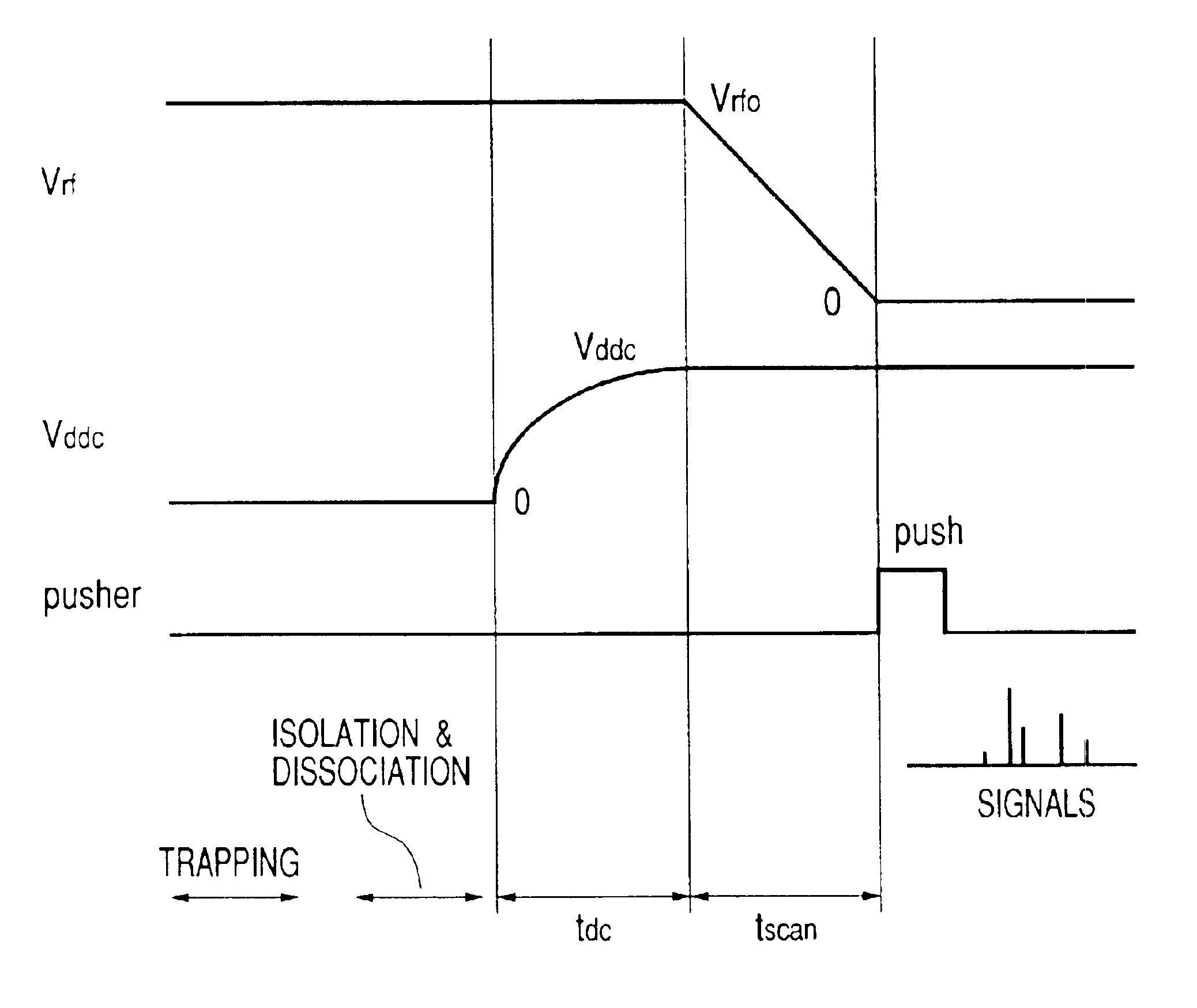
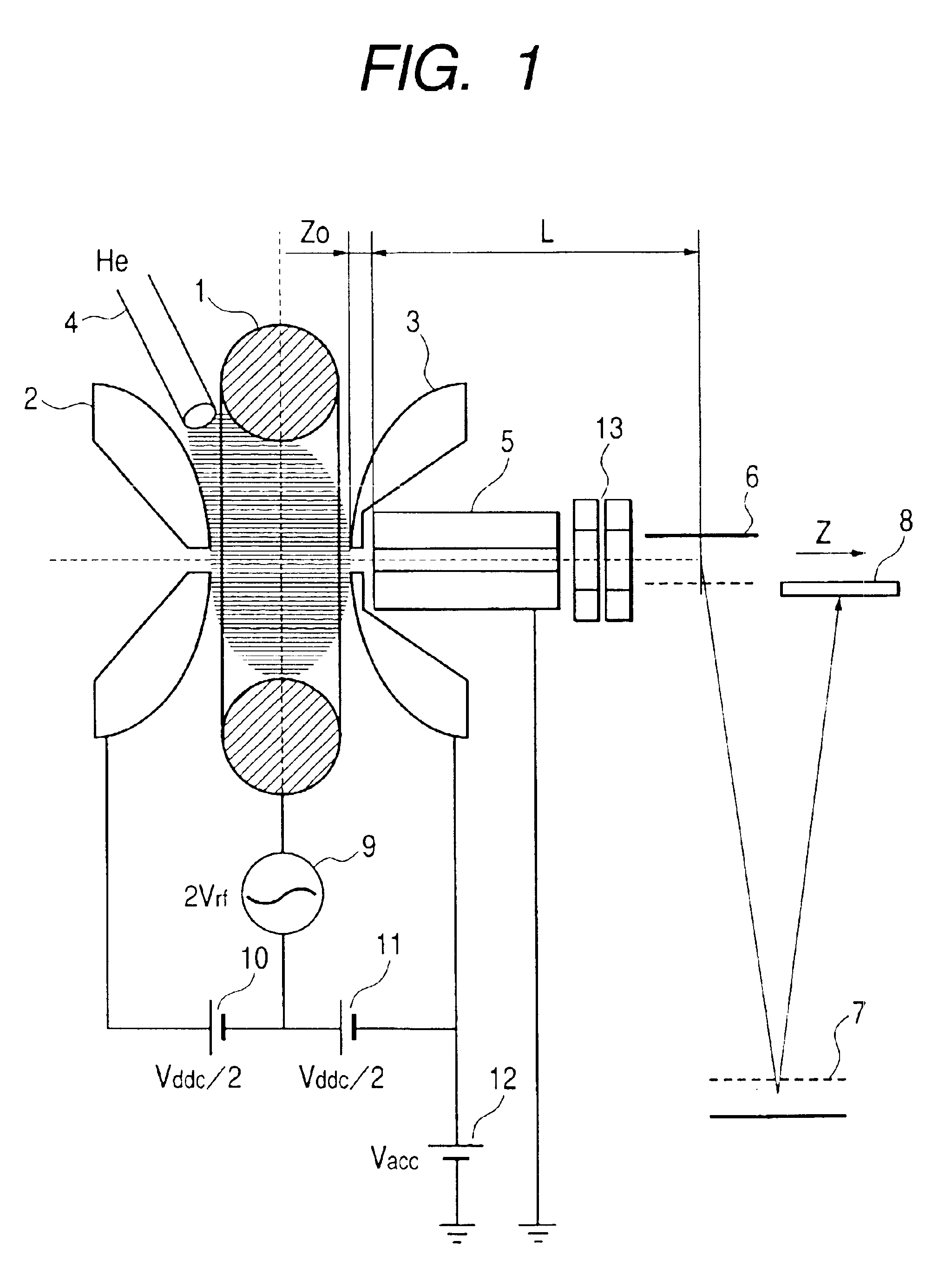
Heavy ions are ejected earlier than light ions sequentially at almost zero energy and they are accelerated at a fixed voltage so as to be guided to a pusher of a TOF spectrometer. After ions are ejected in a procedure of giving an electric field gradient to an ion trap and linearly decreasing its RF voltage, a condition of spatially focusing ions having all mass numbers of a single point on the pusher is found. The focused ions are vertically accelerated using the pusher to perform the TOF mass spectrometry.
Owner:HITACHI HIGH-TECH CORP
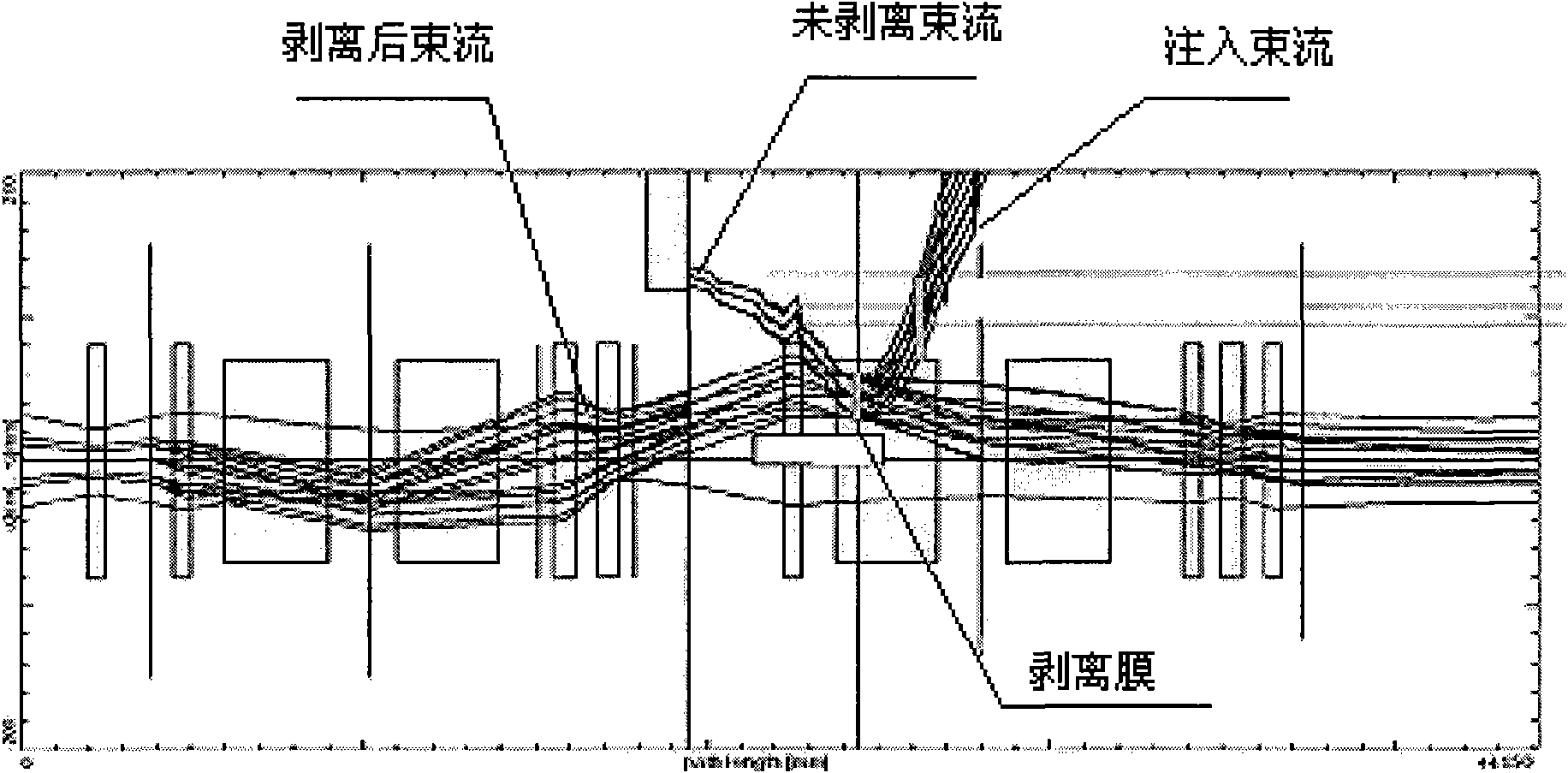
Method and device for implanting heavy ion beams into synchrotron
ActiveCN101631419ASimple process requirementsSolution to short lifeMagnetic resonance acceleratorsMass numberHeavy ion beam
The invention mainly relates to a method and a device for implanting non-full stripping heavy ion beams which are provided by a cyclotron and have mass numbers less than or equal to 40 into a synchrotron. The method comprises the following steps: 1. accelerating the heavy ion beams which are not fully stripped and have mass numbers less than or equal to 40 until the energy is 5.0-10.0MeV / u and the intensity of the heavy ion beams is 2.0-6.0mu A; 2. deflecting the heavy ion beams with mass numbers less than or equal to 40 in the step 1 to a stripping foil and embossing the central track of the synchrotron to the position of the stripping foil in the position of an implanting point; 3. reducing the embossment of the central track in the step 2, implanting the stripped heavy ion beams to fill in the space of the synchrotron and enabling the heavy ion beams to carry out cyclotron in the synchrotron. The invention also provides the device for implanting the heavy ion beams into the synchrotron. The device has the advantages of simple device, convenient operation and high implantation efficiency and can realize multiple choices of the charge states of the heavy ion beams by adjusting the thickness of the stripping foil.
Owner:INST OF MODERN PHYSICS CHINESE ACADEMY OF SCI
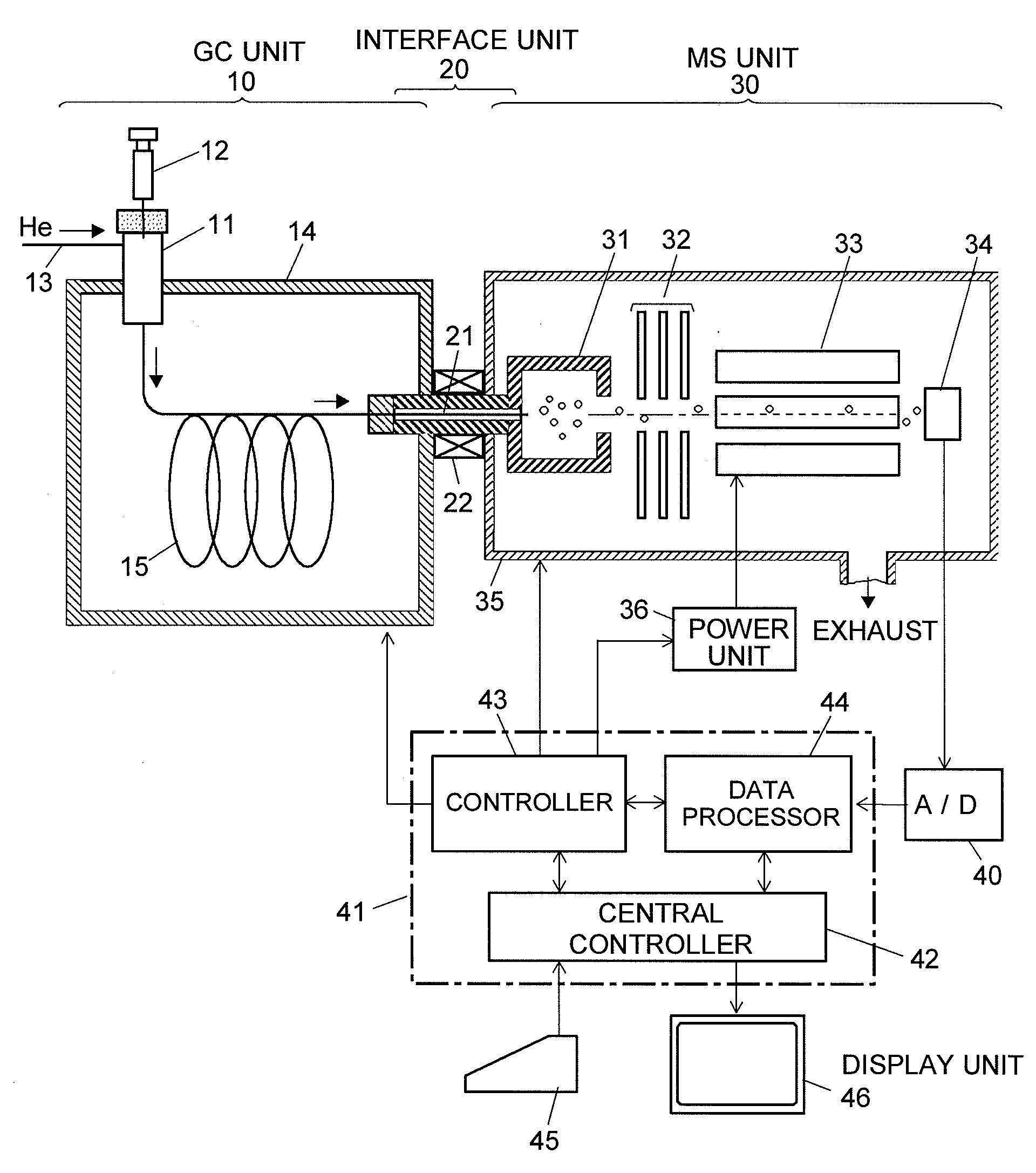
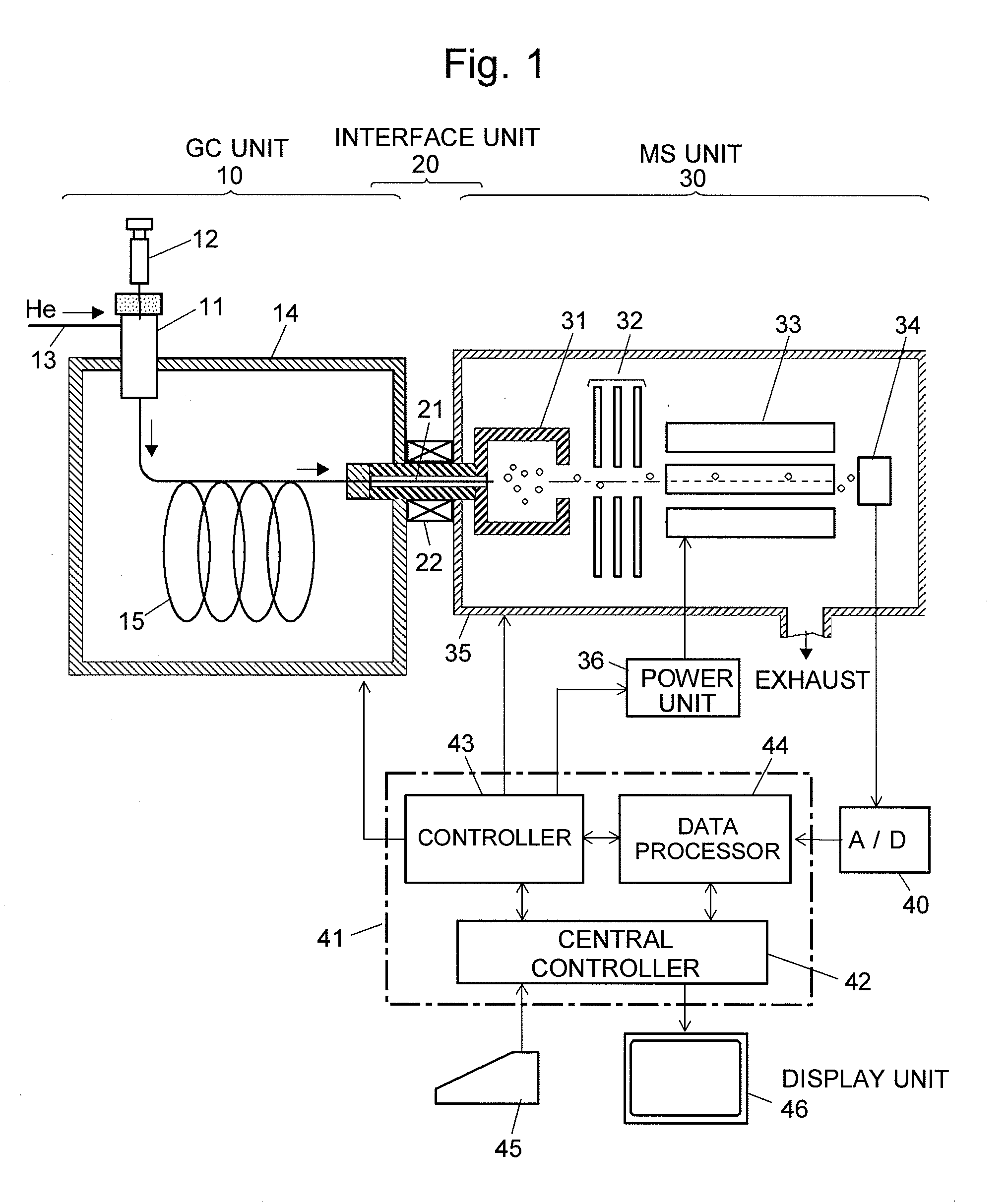
Chromatograph mass spectrometer
InactiveUS20070284520A1Easy to measurePrecise shiftDynamic spectrometersComponent separationMass numberComputational physics
An exact centroid spectrum with a mass number corrected is determined from a profile spectrum adjacent to a plurality of peaks. Regarding a profile spectrum determined by a mass spectrometer, overlapping with adjacent peaks occurs, and compounds having a plurality of peaks with different overlapping degrees is measured, a correction function is created from a relationship between an overlapping degrees with respect to the plurality of peaks and a shift of the mass number, and a centroid peak is corrected by the correction function when the profile spectrum is converted into the centroid spectrum.
Owner:SHIMADZU CORP
Protein second-level mass spectrum identification method based on peak intensity recognition capability
ActiveCN104076115AThe result of the identification method is excellentImprove identification efficiencyComponent separationSpecial data processing applicationsProtein DatabasesMass number
The invention discloses a protein second-level mass spectrum identification method based on peak intensity recognition capability. The method comprises the following steps: firstly, virtualizing enzymatically hydrolyzed protein database sequence, establishing a peptide fragment database and a peptide fragment database index for peptide fragments subjected to enzymatic hydrolysis according to the mass number of the peptide fragments; then, finding out candidate peptide fragments conforming to the requirement from the established peptide fragment database according to the mass number of parent ions without charges in a to-be-analyzed experiment spectrum; then removing an isotopic peak and selecting an effective peak from the to-be-analyzed experiment spectrum so as to generate a theory spectrum of the candidate peptide fragments conforming to the requirement, counting peak intensity information of different ions, calculating the peak intensity recognition capability of different types of ions at different intervals, marking each candidate peptide fragment based on the peak intensity recognition capability, and selecting the peptide fragment with the highest mark as the authentication result of the experiment spectrum; and finally, performing quality control on the authentication result. The number of valid mass spectra and the number of valid protein peptide fragments, which are authenticated by the method, are both higher than those obtained by an existing algorithm; peaks can be selected dynamically; the running speed is high.
Owner:广州辉骏生物科技股份有限公司
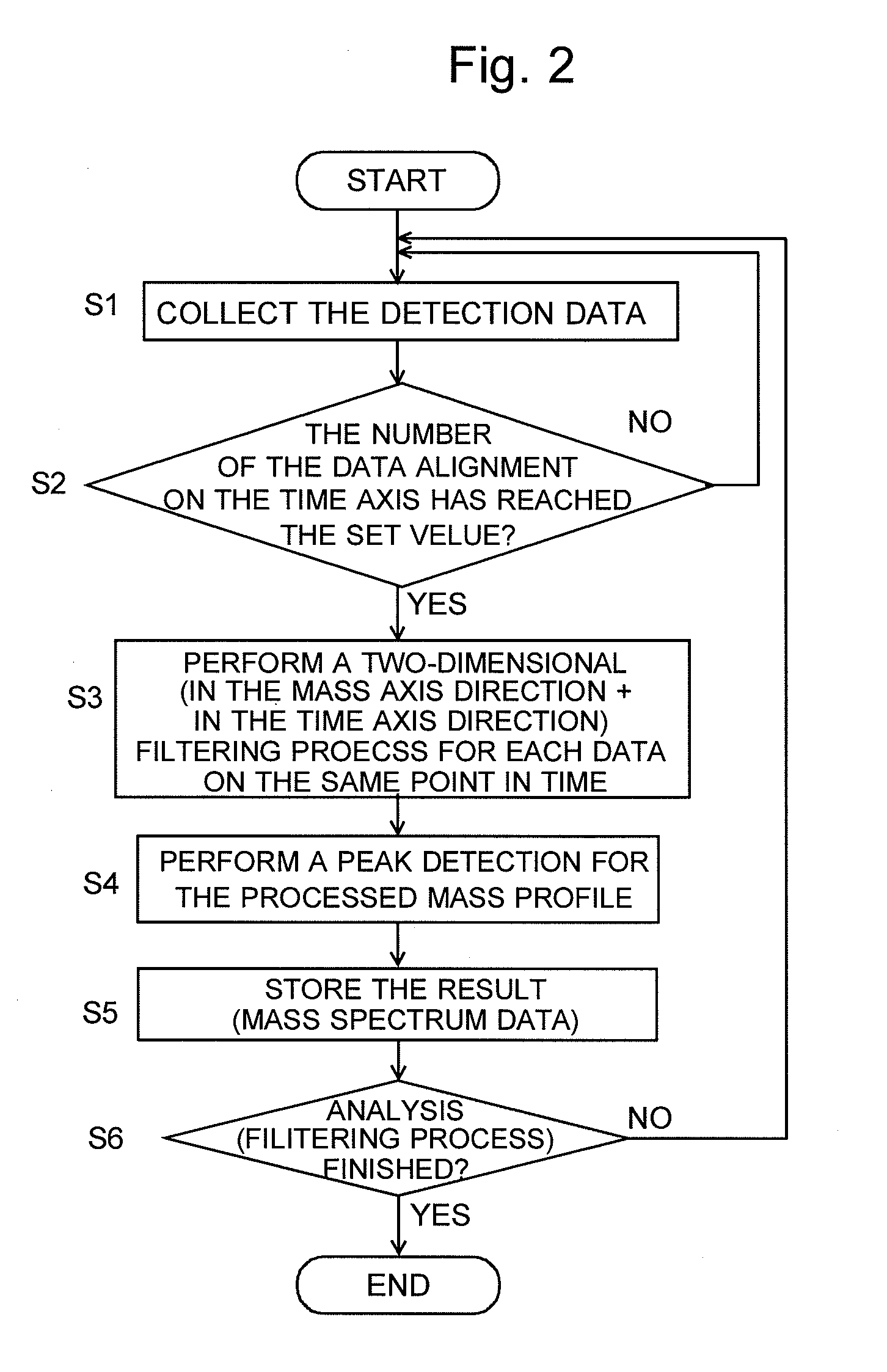
Chromatograph mass analysis data processing apparatus
InactiveUS20090199620A1Improve quality resolutionImprove detection accuracyParticle separator tubesComponent separationMass numberImage resolution
In the case where a given mass range is repeatedly scanned to sequentially create mass spectra and create a mass chromatogram or the like, when a number of data, which are arranged on the mass axis and constitute a mass profile of the given mass range, are collected for a predetermined number of the alignment (S1 and S2), a two-dimensional filtering process is performed by correcting the target data, for each data aligned in the mass axis direction, using the data contiguous before and after in the mass axis direction and the data contiguous before and after in the time axis direction (S3). For the mass profile constituted of the data thus processed, a peak detection is performed (S4) and the peak's mass number is determined to create a mass spectrum (S5). Consequently, the mass numbers' fluctuation in the mass spectrum is moderated, the mass resolution is increased, and the accuracy of the mass chromatogram is also increased.
Owner:SHIMADZU CORP
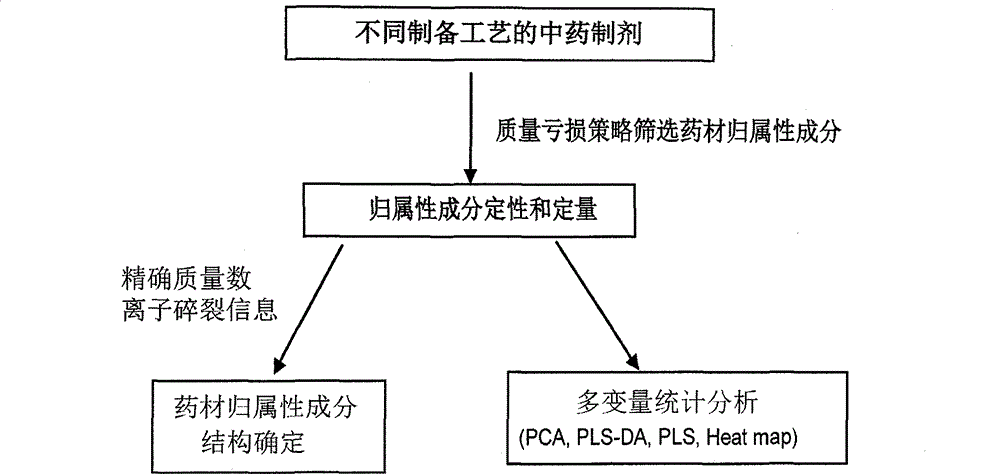
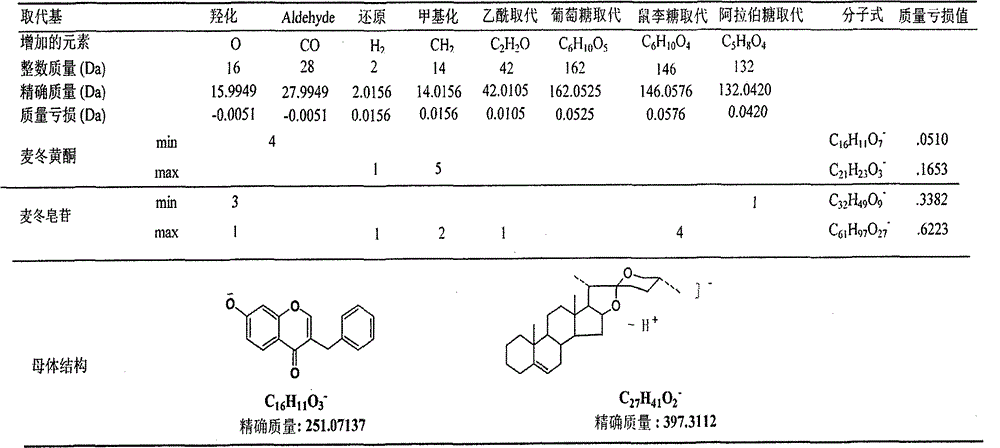
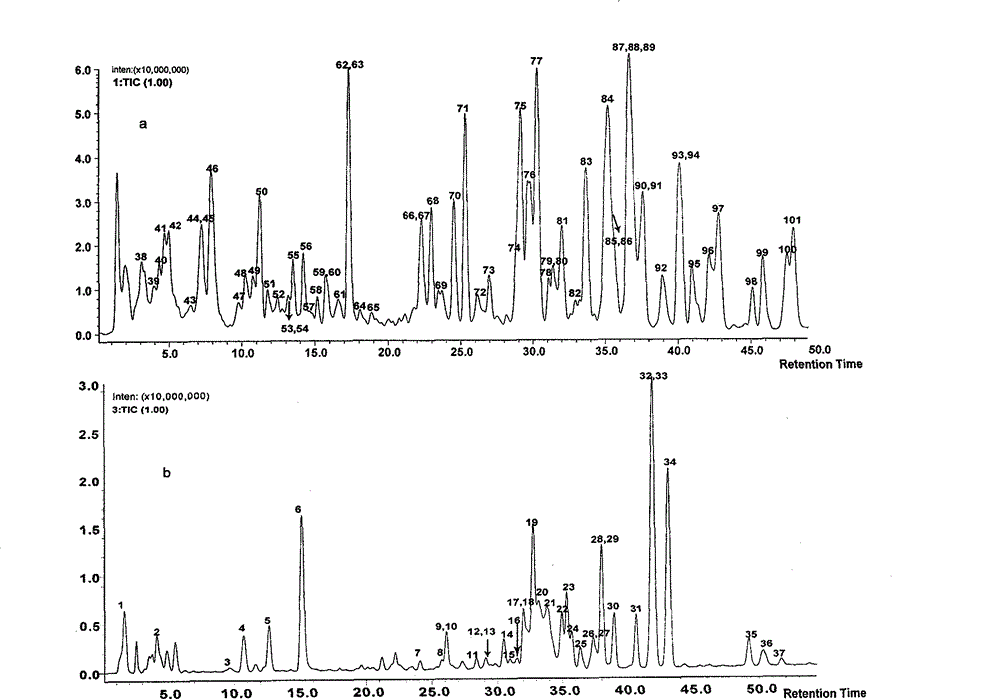
Method for evaluating traditional Chinese medicine preparation making technology based on metabonomics technology
InactiveCN102749409AAvoid processing powerAvoid generalizationComponent separationMass numberMedicine
The invention discloses a method for evaluating a traditional Chinese medicine preparation making technology based on a metabonomics technology, which comprises the following steps: 1, an orthogonal experiment design or a parallel contrast experiment design are used for acquiring traditional Chinese medicine preparation products with different preparation technologies; 2, the prepared products are subjected to sample pre-treatment; 3, a liquid phase series high resolution mass spectrometer is used for acquiring a chromatogram of the traditional Chinese medicine preparation product; 4, the belongingness components of the traditional Chinese medicine are found in the chromatogram according to mass loss strategy; 5, each component structure can be deducted according to fragment information obtained by determined accurate mass number and collision induced dissociation; 6, all belongingness components content in each product are subjected to quantitative analysis; and 7) the influence of the preparation technology on the preparation can be evaluated through multivariable data statistics analysis method. The method of the invention has the advantages of accuracy and sensitivity, and is not depend on standard compounds, and the change of belongingness components of the traditional Chinese medicine in the preparation product can be integrally traced.
Owner:CHINA PHARM UNIV
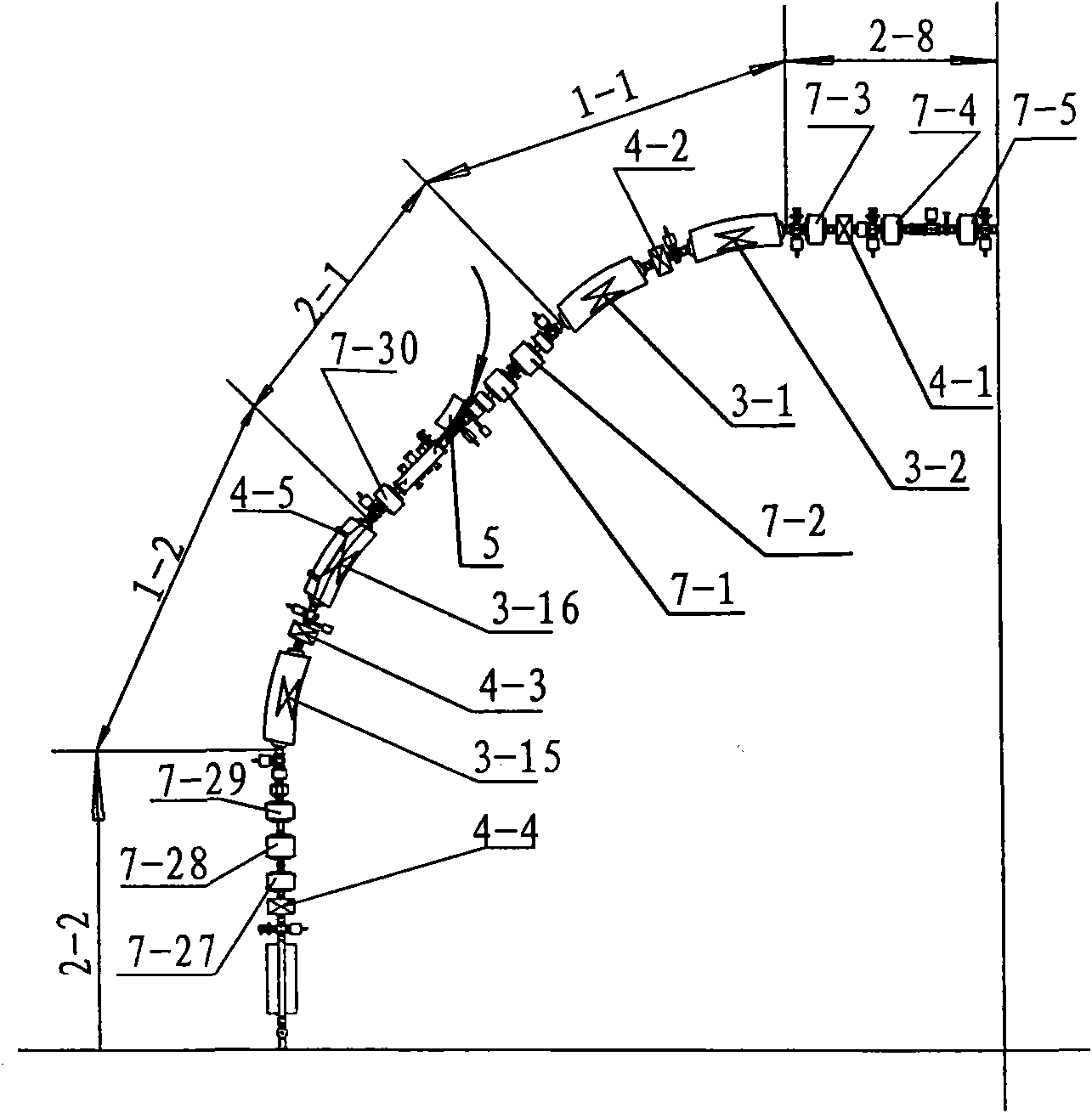
Accelerator used for cancer therapy with protons-heavy ion beams
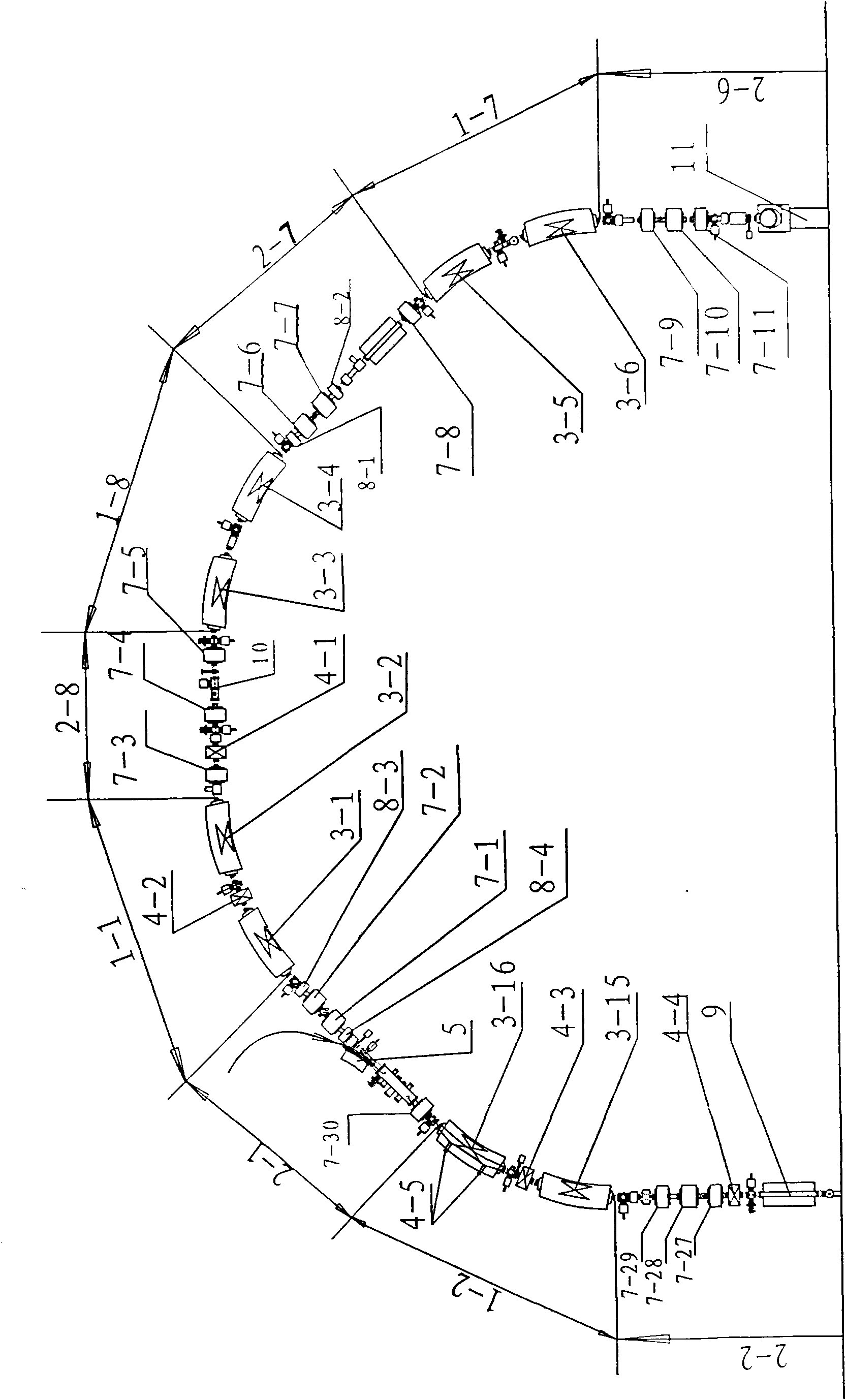
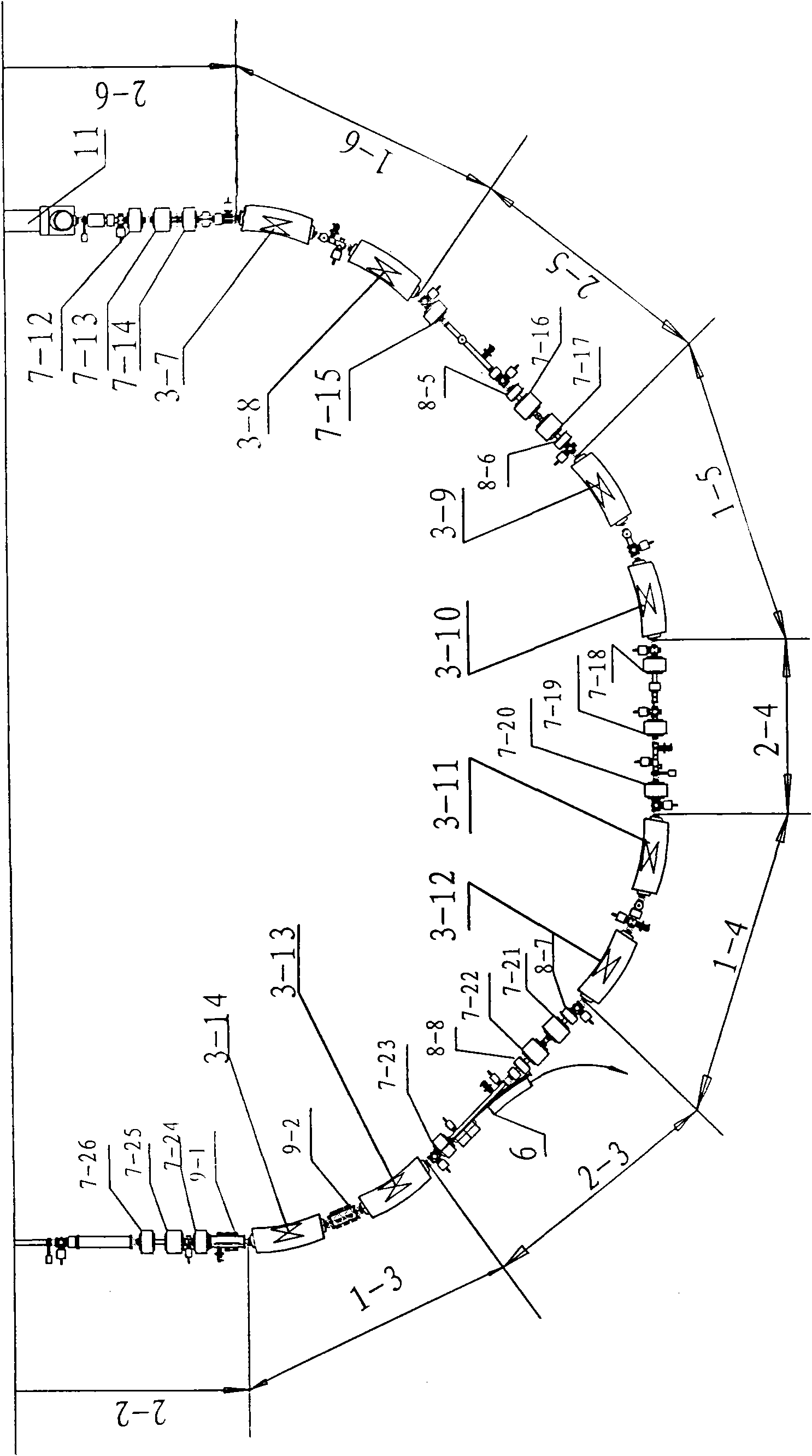
ActiveCN101631420AFulfillment requirementsTo achieve the requirements of the beam current parametersMagnetic resonance acceleratorsRadiation therapyMass numberSynchrotron
The invention relates to an accelerator which combines a cyclotron and a synchrotron and is used for cancer therapy with protons-heavy ion beams, comprising an ion source E and the synchrotron C. The accelerator is characterized in that the synchrotron C comprises a septum magnet implanting device (5), a septum magnet extracting device (6), a radio-frequency accelerating device (9), eight bending sections of the synchrotron (1-1 to 1-8) and eight straight sections of the synchrotron (2-1 to 2-8), which are connected by an ion beam transmission line F; the ion beam implantation end of the synchrotron C is provided with a sector focusing cyclotron A or / and a separated sector focusing cyclotron B. By adopting the method of combining the cyclotron and the synchrotron, a set of accelerator devices required by cancer therapy with protons and heavy ion beams is set up to realize the requirements of clinical therapy on beam parameters; the mass numbers of the protons and the heavy ions are less than or equal to 40; the protons and heavy ion beams have wide energy range from 10MeV / u to 400MeV / u and high flux (the ion number provided by each pulse is up to 10<7-10> magnitude).
Owner:INST OF MODERN PHYSICS CHINESE ACADEMY OF SCI
Method for online-analyzing organic monomer chlorine or bromine isotope and based on chemical ionization
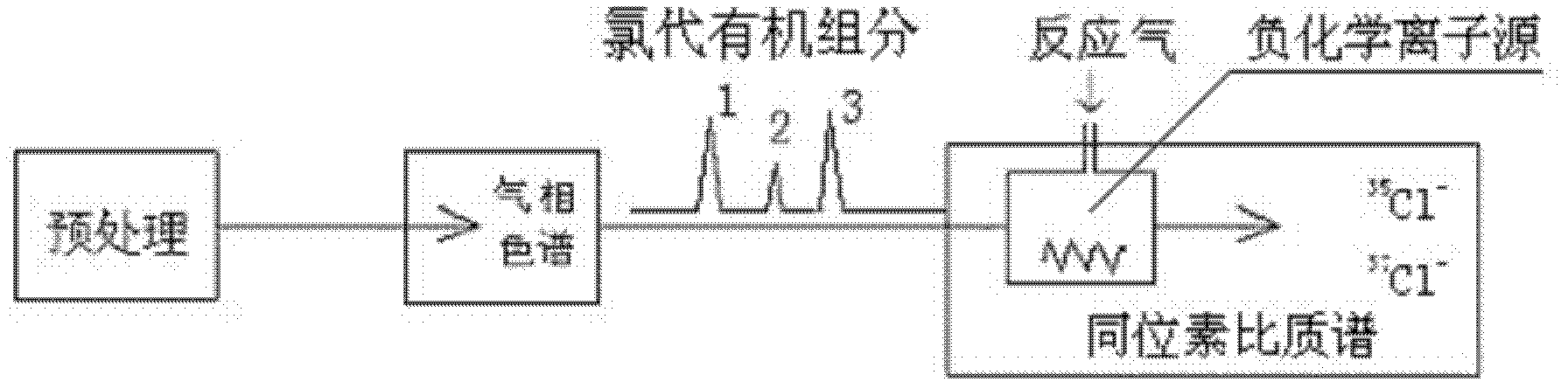
InactiveCN102590379AAvoid separationSimplify the process of online analysisComponent separationMass numberGas phase
The invention aims at providing a method for online-analyzing organic monomer chlorine or bromine isotope, which adopts a direct negative chemical ionization to replace an original complex organic monomer conversion process. The method is characterized by including the steps: A. gathering chlorine or bromine substitute; B. separating gas chromatography: placing the gathered chlorine or bromine substitute into the gas chromatography to be separated; C. conducting negative chemical ionization: components analyzed through chromatography directly flow into a negative chemical ion source through acapillary column, the negative chemical ion source replaces an original electron bombardment source configured on an isotope ratio mass spectrum to conduct the negative chemical ionization; D. mass spectrometric detecting: on a gas isotope ratio mass spectrum, selecting a faraday receiving cup corresponding to mass number of chlorine or bromine substitute directly, and receiving electronegative isotope fragment ions generated by the negative chemical ionization. The process is conducted under high vacuum state, and degree of vacuum is higher than 1.0*10<-3> Pa. The method greatly simplifies aprocess of online-analyzing organic monomer chlorine or bromine isotope and can form various special devices for detecting organic monomer isotope ratio.
Owner:INST OF HYDROGEOLOGY & ENVIRONMENTAL GEOLOGY CHINESE ACAD OF GEOLOGICAL SCI
Method for non-targeted quick screening of unknown residual pesticides in imported grains by FaPEx-UPLC-Q-TOF
ActiveCN106896176AAvoid False Positive ResultsImprove accuracyComponent separationMass numberAcetic acid
The invention relates to the field of pesticide detection and discloses a method for non-targeted quick screening of unknown residual pesticides in imported grains by FaPEx-UPLC-Q-TOF. The method for the non-targeted quick screening of the unknown residual pesticides in the imported grains is established by using a FaPEx-UPLC-Q-TOF principle. The method comprises the following steps: extracting the unknown residual pesticides in the imported grains by adopting an acetic acid and acetonitrile solution; carrying out purification and concentration by using a FaPEx solid-phase extraction column; detecting by UPLC-Q-TOF; carrying out database matching by using an exact mass number of characteristic ions of a target compound, isotope matching, secondary fragment information and retention time, and screening suspected unknown pesticides. The result shows that by using the method, the residual pesticides in the imported grains can be quickly screened without a reference standard substance. The method has the advantages of quickness, accuracy and high analysis throughput, and can provide an important method and basis for quick screening and quality control of the residual pesticides in the imported grains.
Owner:舟山出入境检验检疫局综合技术服务中心
Method for rapidly screening perfluorinated compounds and precusor substances thereof in fish meat
ActiveCN106124678AEasy to handleImplement synchronous extractionComponent separationMass numberPhospholipid
The invention discloses a method for rapidly screening perfluorinated compounds and precusor substances thereof in fish meat, and belongs to the technical field of aquatic product detection. According to the method, a pass-type Oasis PRiME HLB solid-phase extraction and purification strategy is adopted, a sample pretreatment process is simplified, impurity interference components such as fat and phospholipids in a sample matrix are removed, and simultaneous extraction of multiple perfluorinated compounds and precusor substances thereof is achieved; meanwhile, the advantages of a liquid chromatography-quadrupole rod / electrostatic field orbitrap high resolution mass spectrum are utilized, the qualitative capacity for a complex matrix is enhanced by obtaining the accurate mass number and rich fragment ion information, the quantifying effect of the high resolution mass spectrum is improved, high-throughput qualitative screening for simultaneously determining 18 PFCs and 21 precusor substances through single injected sampling is achieved, and a high-sensitive quantitative result is obtained.
Owner:YELLOW SEA FISHERIES RES INST CHINESE ACAD OF FISHERIES SCI
Secondary protein mass spectrum identification method based on mass-to-charge ratio error recognition capability
InactiveCN104034792AThe identification result is excellentImprove identification efficiencyMaterial analysis by electric/magnetic meansMass numberDatabase index
The invention discloses a secondary protein mass spectrum identification method based on mass-to-charge ratio error recognition capability. The method comprises the following steps: virtualizing an enzymolysis protein databank sequence, establishing a peptide fragment databank and a peptide fragment databank index for each peptide fragment after being subjected to enzymolysis according to the mass number of the peptide fragment, finding out each candidate peptide fragment which meets the requirement in the established peptide fragment databank according to the mass number after charges of mother ions are removed in an experiment spectrum to be analyzed, performing isotopic peak removal and selecting an effective peak for processing, generating an experiment mark spectrum which meets a candidate peptide fragment theoretic spectrum, counting mass error information of different ions, calculating the mass-to-charge ratio error recognition capabilities of the different ions in different areas, scoring the candidate peptide fragments based on the mass-to-charge ratio error recognition capabilities, selecting the peptide fragment with the highest score as the identification result of the experiment spectrum, and performing the overall mass control on the identification result. The numbers of effective spectrums and peptide fragments identified by using the method are both greater than that of a conventional algorithm, the peak can be dynamically selected, and the operation speed is high.
Owner:YUNNAN MINZU UNIV
Semiconductor device and method of manufacturing the same
InactiveUS20060273392A1TransistorSemiconductor/solid-state device manufacturingMass numberDevice material
Disclosed is a method for manufacturing a semiconductor device comprising implanting ions of an impurity element into a semiconductor region, implanting, into the semiconductor region, ions of a predetermined element which is a group IV element or an element having the same conductivity type as the impurity element and larger in mass number than the impurity element, and irradiating a region into which the impurity element and the predetermined element are implanted with light to anneal the region, the light having an emission intensity distribution, a maximum point of the distribution existing in a wavelength region of not more than 600 nm.
Owner:KK TOSHIBA
Mass spectrometer and method of analyzing isomers
ActiveUS20050258355A1Shorten the timeSave amountStability-of-path spectrometersComponent separationMass numberIonic strength
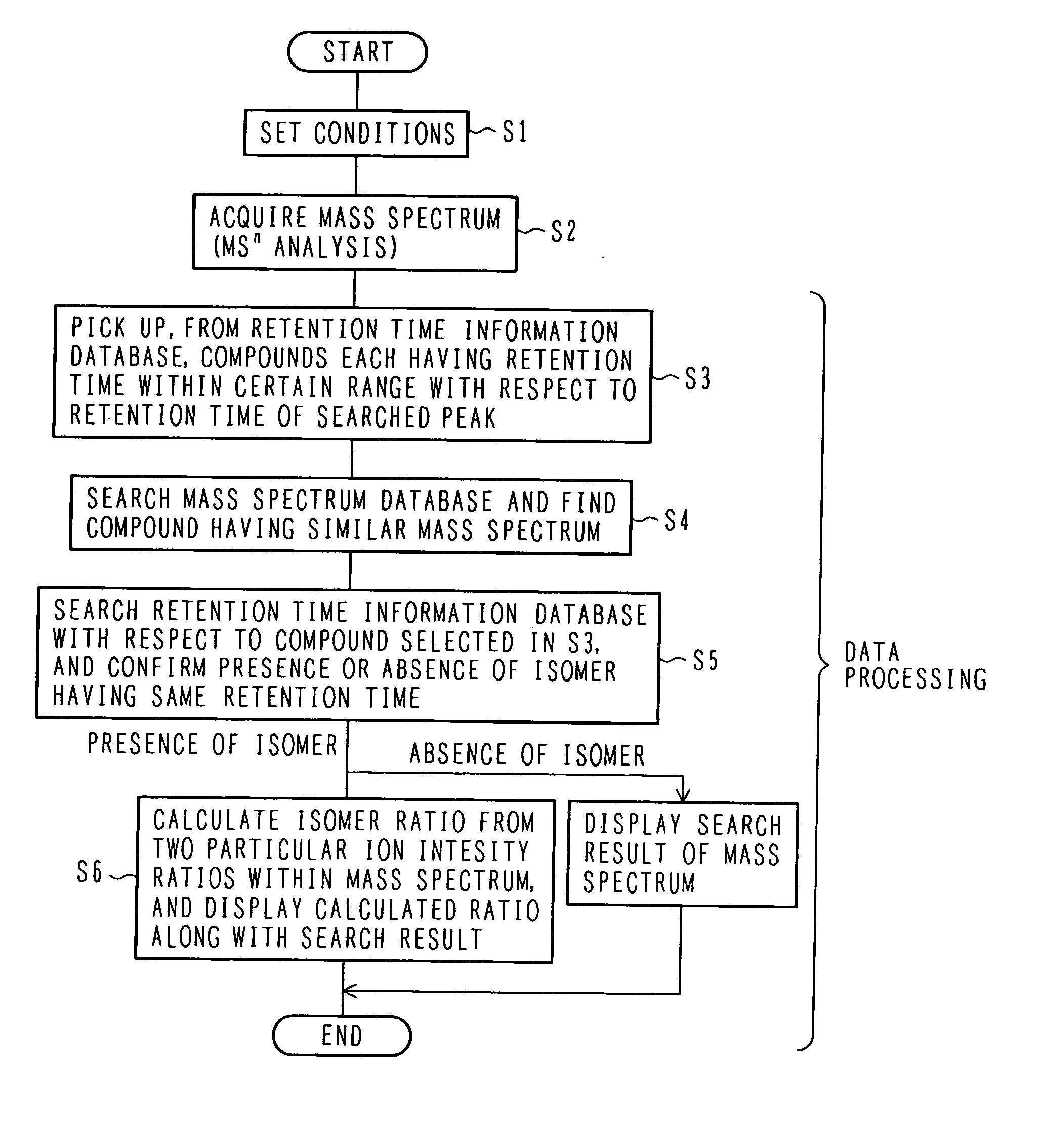
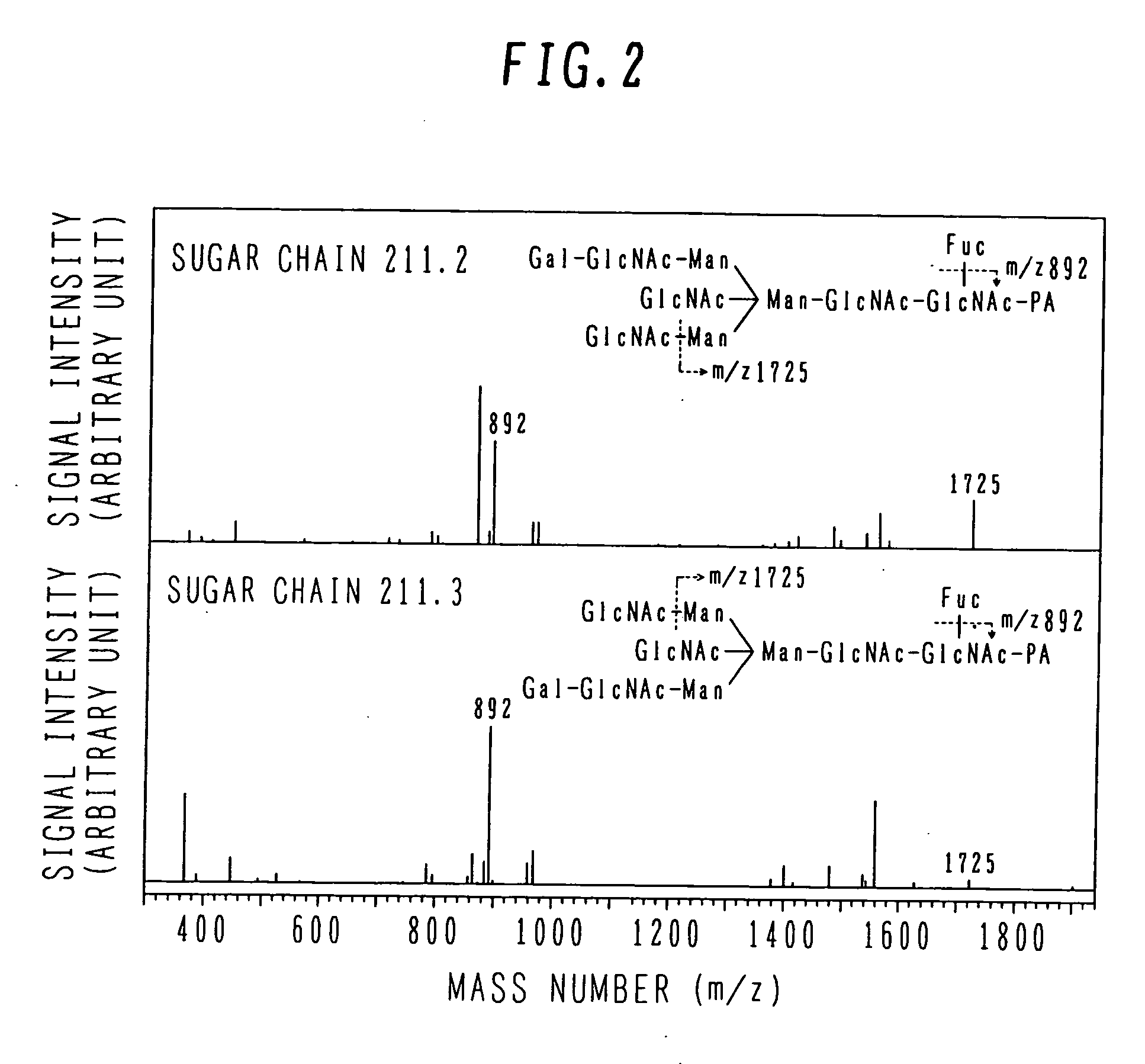
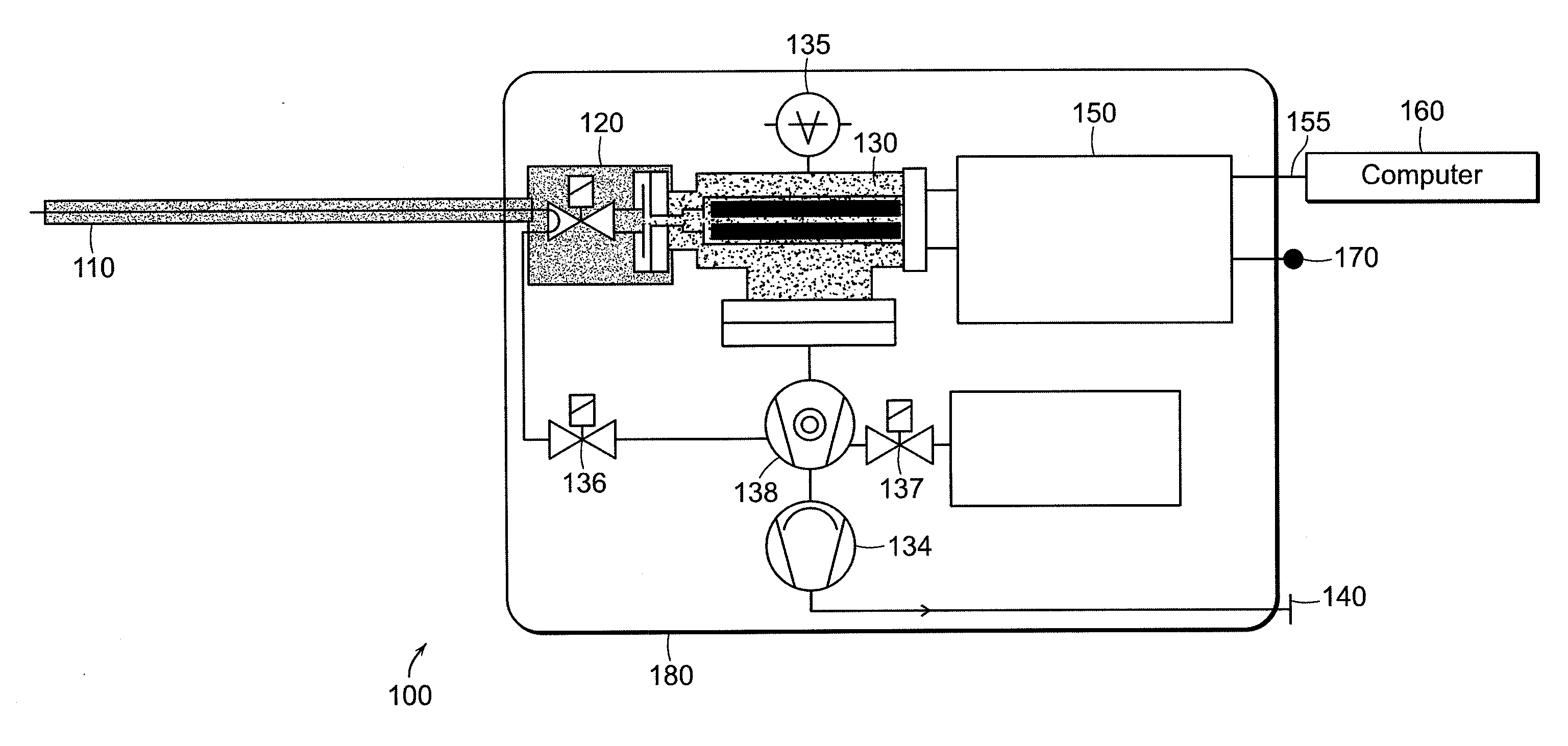
The invention intends to determine a presence ratio between mixed compounds which are difficult to separate from each other by a separation unit and cannot be discriminated by MS. In a mass spectrometer with an MSn analysis function for ionizing a sample eluted from a separation unit for separating the sample into individual components, fragmenting an ion of a desired mass number, and performing a mass analysis of fragment ions, the mass spectrometer includes a database storing correlation information between an isomer presence ratio and a particular ion intensity ratio in a mass spectrum per isomer. An isomer ratio can be clarified even for a mixed sample of isomers, such as enantiomers, which are difficult to separate from each other by the separation unit and cannot be discriminated by MS.
Owner:HITACHI HIGH-TECH CORP
Detection of ethanol emission from a spark ignition engine operating on gasohols
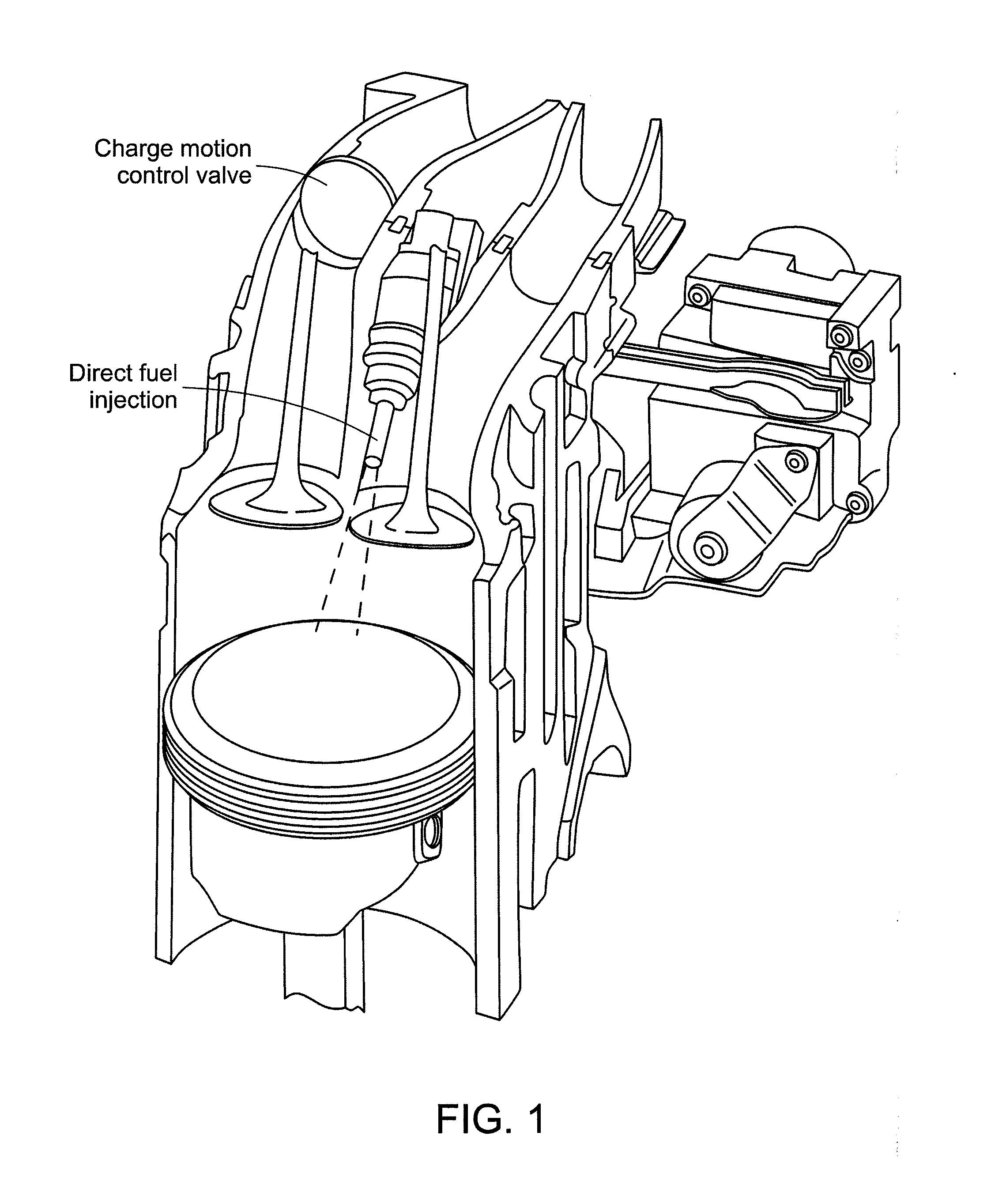
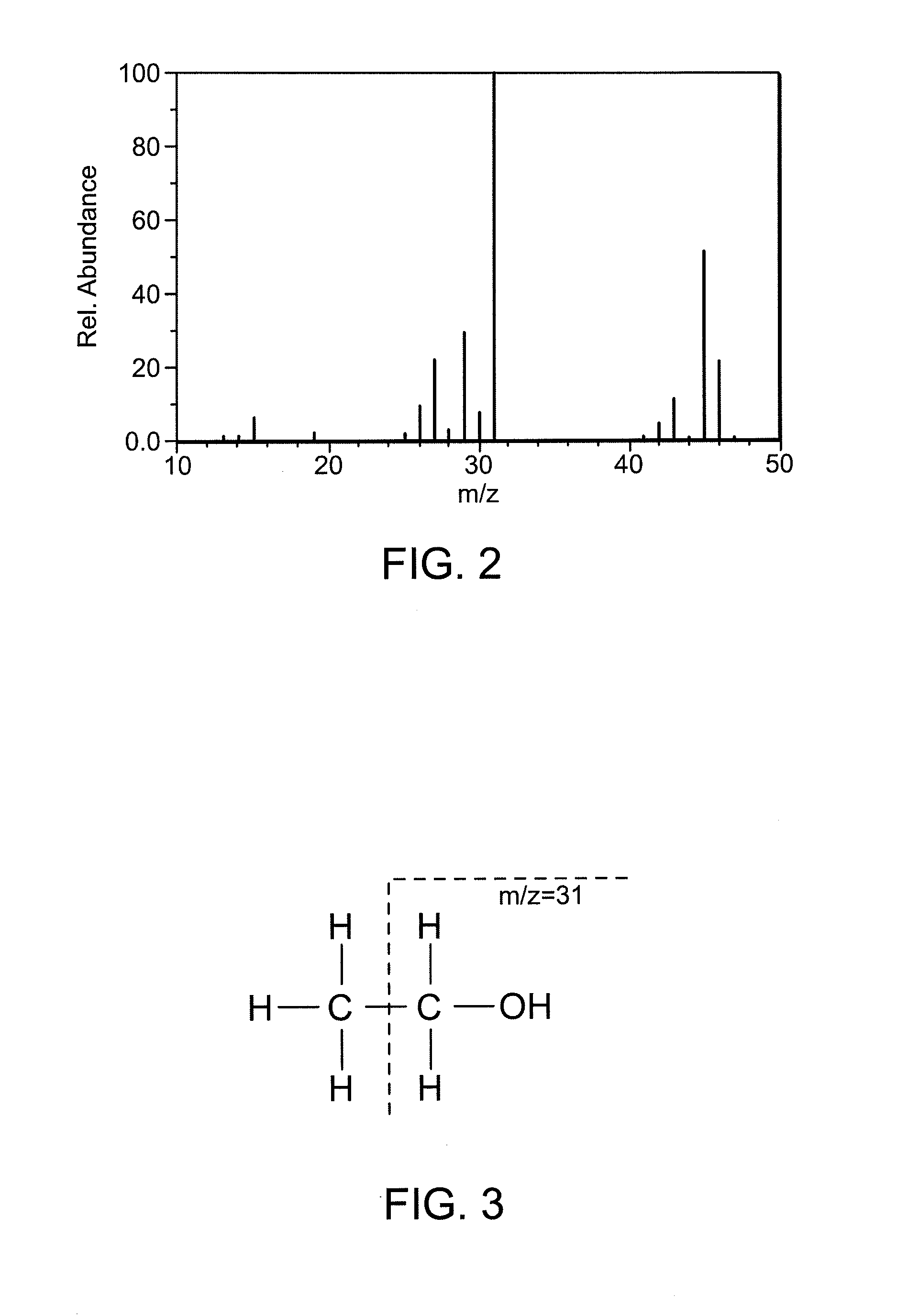
InactiveUS20120089343A1Minimize impactWeaken energyInternal combustion piston enginesComponent separationMass numberGasoline
Ethanol emissions from a direct ignition spark ignition are measured using mass spectrometry. The method exploits specific fragment ions from ethanol. Ethanol contributes ions of mass number 31, and no other gas species produces ions at this mass number. The method and a device for implementing the method can be used for online detection of ethanol in emissions from engines burning E85 or other ethanol / gasoline mixtures.
Owner:MASSACHUSETTS INST OF TECH
Novel nuclear pore membrane
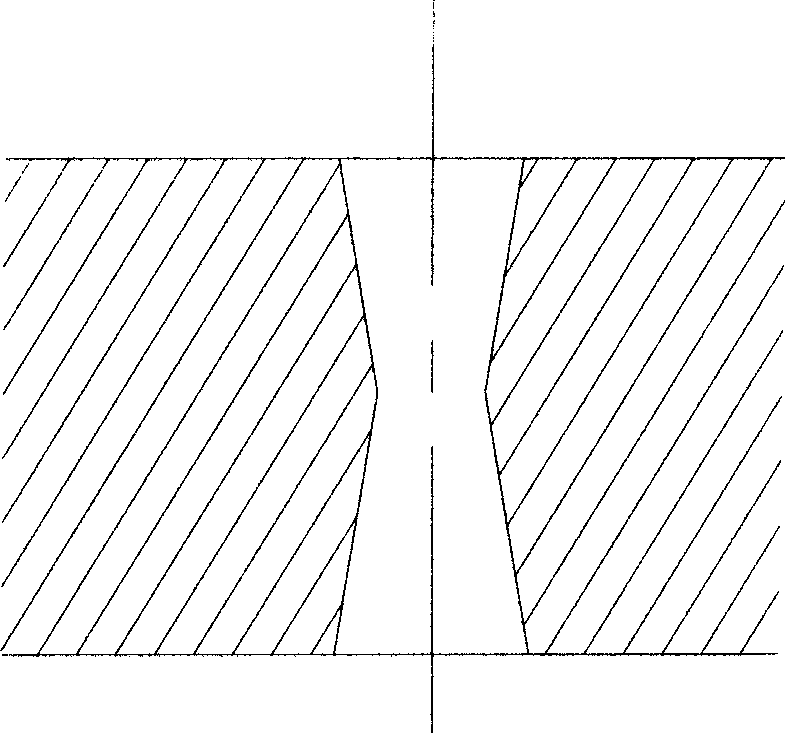
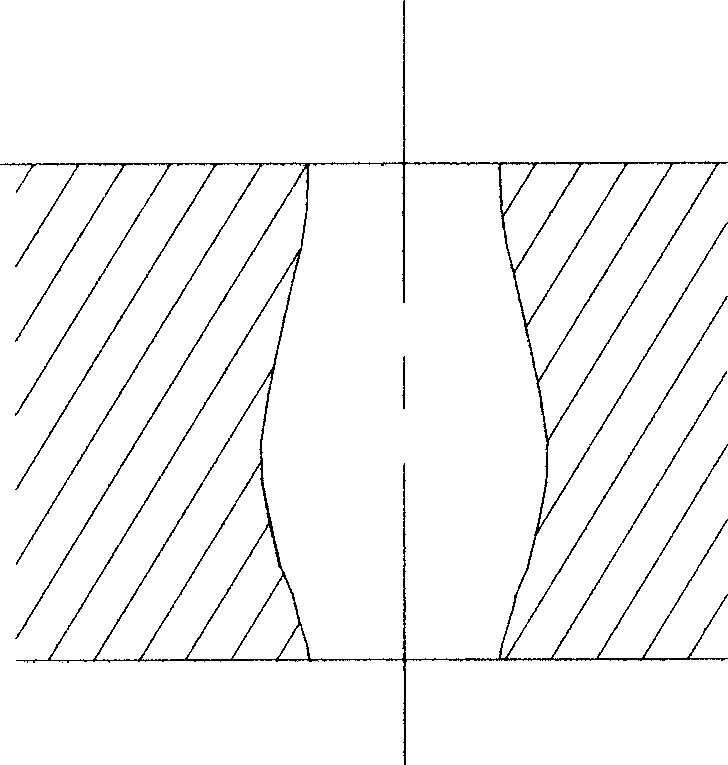
InactiveCN101380547AIncreased etch sensitivityReduce taperSemi-permeable membranesMass numberMembrane surface
The invention relates to a novel nuclear pore membrane (or nuclear track membrane), generally the nuclear track always has conicity after etching, the millipore of the nuclear pore membrane is single-cone or double-cone, the center aperture of the millipore of the double cone is less than the aperture on both surfaces of the membrane. Particles are blocked in the millipore easily when filtering, and the membrane is blocked easily. Meanwhile, the ions with lower mass number are utilized for irradiating the electrolyte membranes, for the nuclear track etching sensitivity is lower, conicity is larger, the nuclear pore membrane with small aperture is made hard. The center aperture of the millipore of the nuclear pore membrane made by the invention is larger than the diameters of both surfaces of the membrane or is of column form capillary tube shape. The shape of the millipore causes the particles larger than the membrane surface aperture to be trapped on the membrane surface, and the nuclear pore membrane is not blocked easily. Meanwhile, a new method is provided for making nuclear pore membranes by utilizing the ions with lower mass number.
Owner:毕明光
Method and device for identifying glycopeptide segment
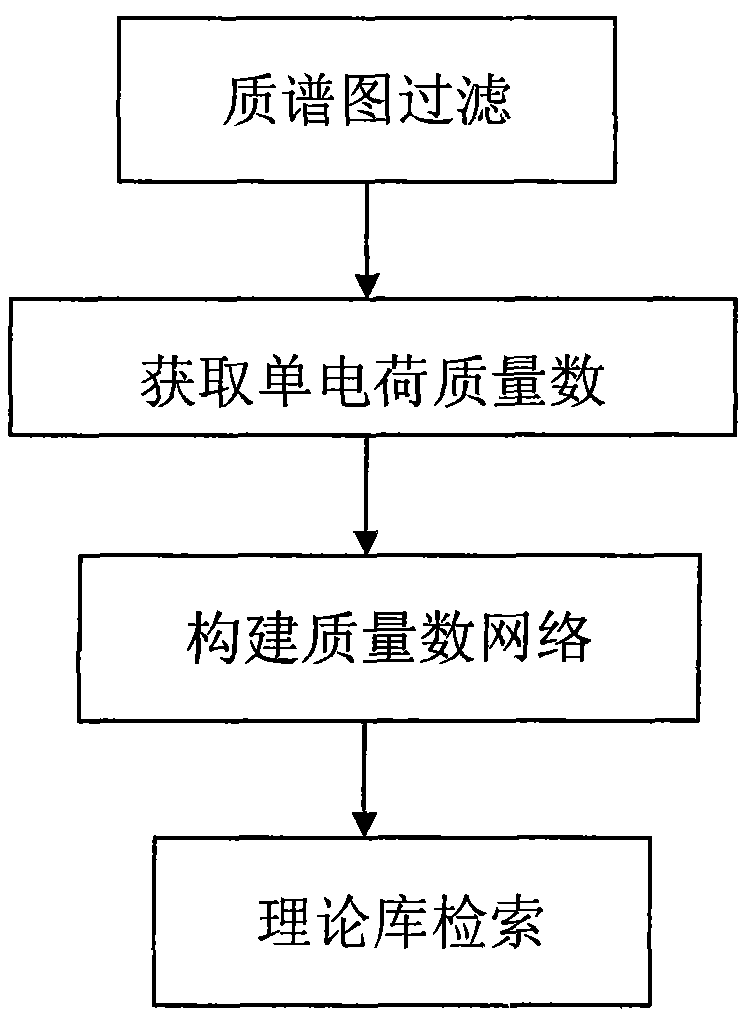
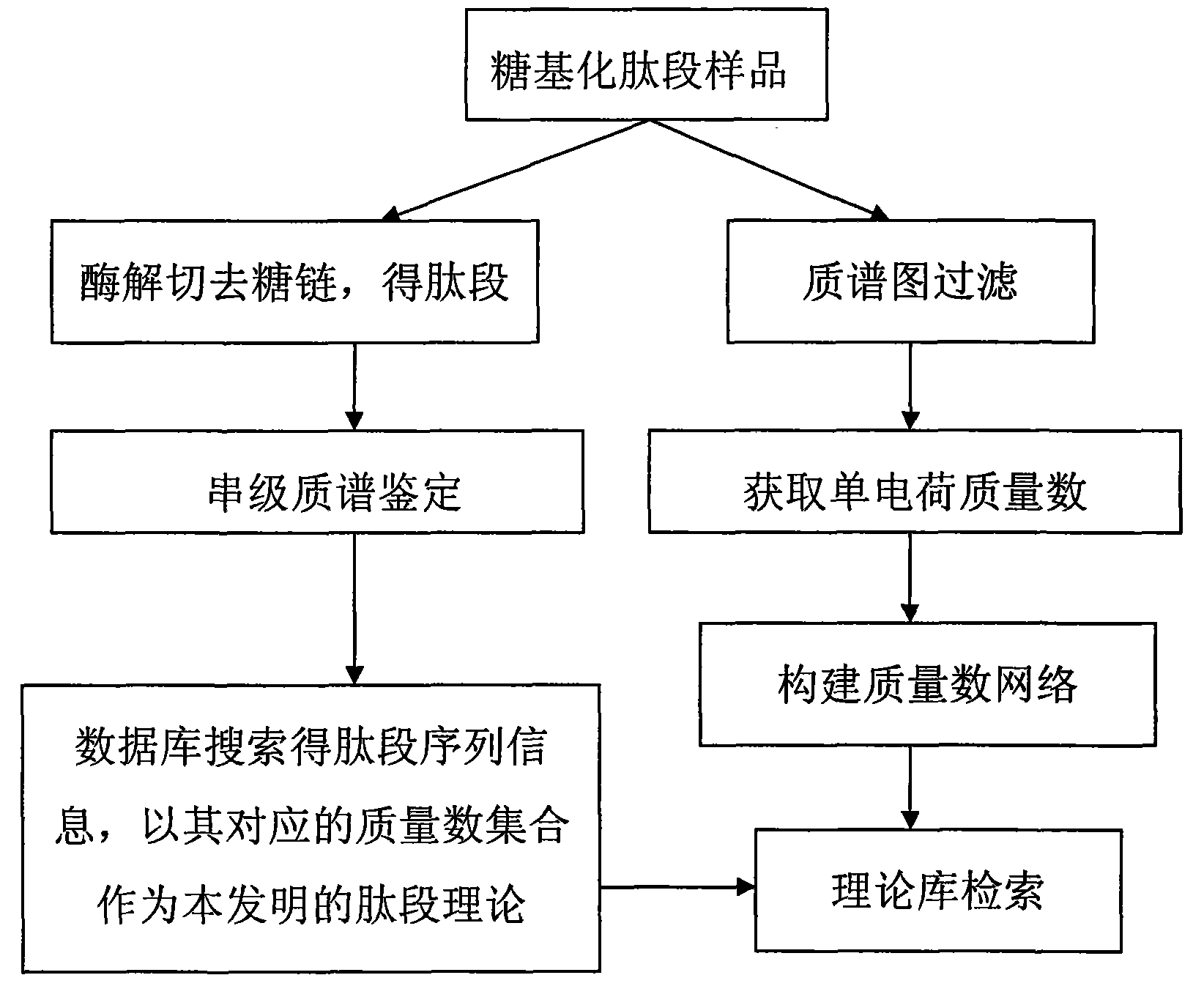
InactiveCN102072932AImprove accuracyReduce false positive resultsMaterial analysis by electric/magnetic meansMass numberIdentification device
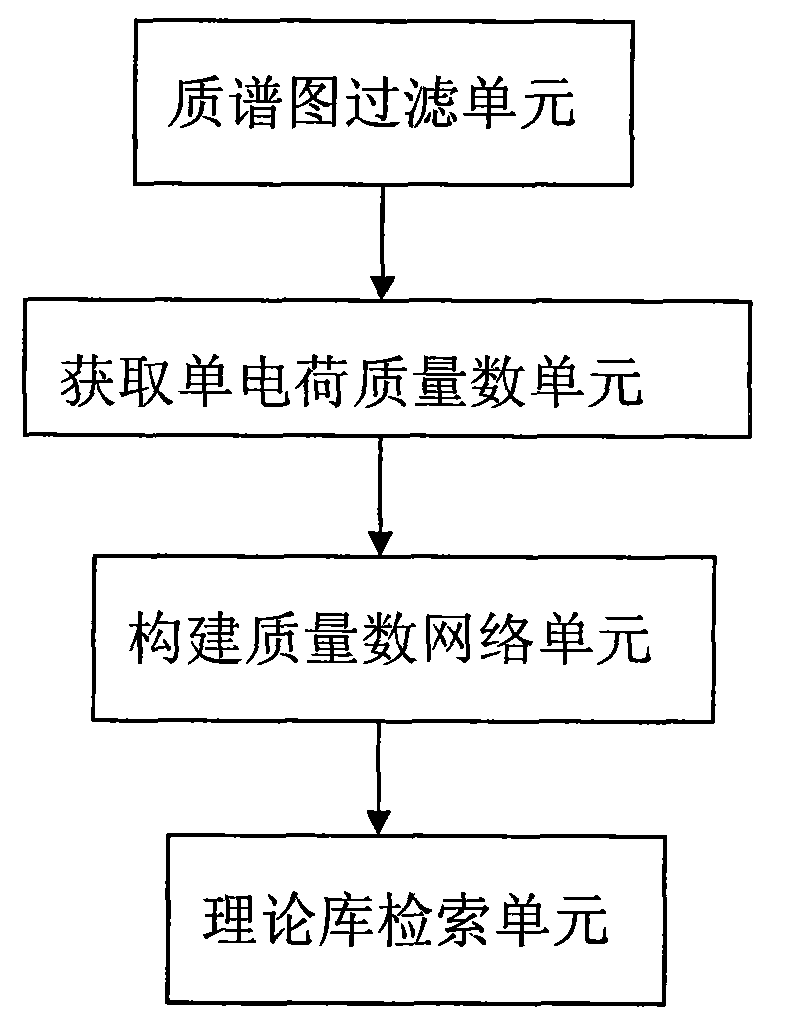
The invention discloses a method for identifying a glycopeptide segment, which comprises the following steps: filtering a mass spectrogram; acquiring a single-charge mass number; constructing a mass number network; and retrieving a theory library. The invention also discloses a device for identifying the glycopeptide segment, which comprises a mass spectrogram filtering unit, a single-charge massnumber acquisition unit, a mass number network construction unit and a theory library retrieving unit. Compared with the conventional method for identifying the glycopeptide segment, the identification method and the identification device can efficiently acquire more accurate identification results in which false positive results are obviously reduced.
Owner:FUDAN UNIV
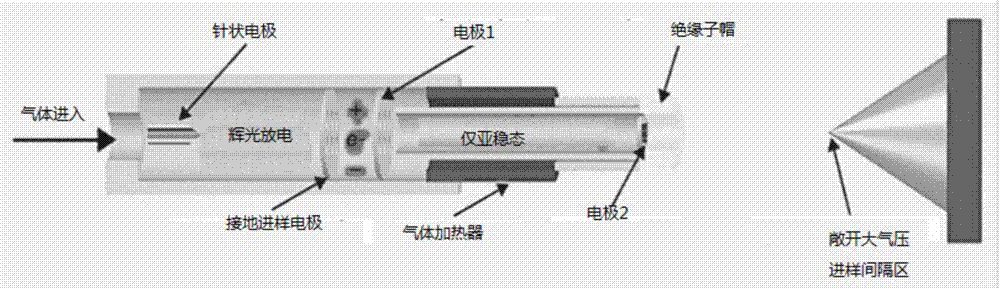
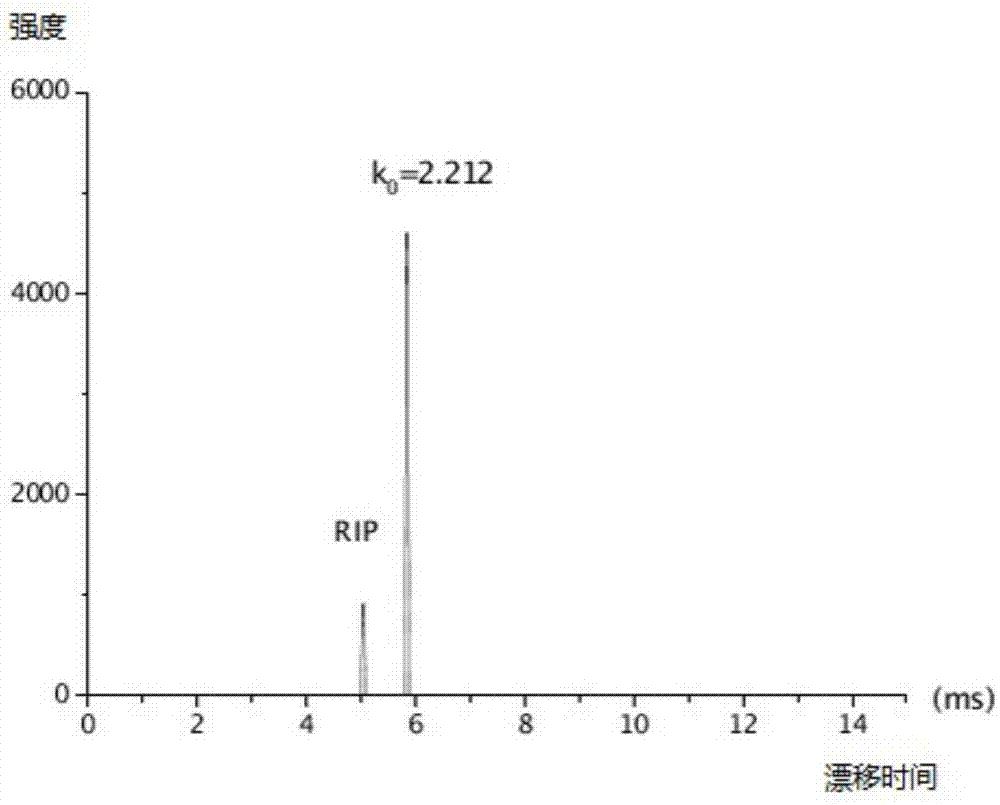
Method for establishing a reduced mobility - mass spectra database for quickly screening drugs and application
InactiveCN107462625ASimple operating systemQuick responseMaterial analysis by electric/magnetic meansChromatographic separationMass number
Provided are a method for establishing a reduced mobility - mass spectra database for quickly screening drugs and application. An ion mobility spectroscopy is combined with real-time direct analysis of ion source time-of-flight mass spectrometry, and a database is established and includes reduced mobility (k0) obtained by the ion mobility spectroscopy and also includes a mass number of a quasi-molecular ion peak generated by real-time direct analysis of the ion source time-of-flight mass spectrometry, as well as a mass number of a fragment ion peak generated by a collision induced ionization mode, the obtained database is utilized to perform high-throughput drug screening, sample processing is simple, no chromatographic separation is required, test results are accurate, reliable and easy to operate, and fake positive results are avoided. The established rapid screening method can be used for the rapid detection of multiple drugs with the detection limit range of 0.02-2 micrograms / mL.
Owner:SHANGHAI CRIMINAL SCI TECH RES INST
Semiconductor device and manufacturing method thereof
InactiveUS7521326B2Suppress DiffuseSignificant timeTransistorSemiconductor/solid-state device manufacturingMass numberDevice material
It is an object of the present invention to provide a semiconductor device superior in the decrease in leak current due to a short-channel effect and a manufacturing method thereof. In a process of forming a field-effect transistor over a single-crystal semiconductor substrate, an impurity is introduced to form an extension region and a single crystal lattice is broken to make the extension region amorphous. Alternatively, the impurity and an element having large mass number are introduced to break the single crystal lattice and make the extension region amorphous. Then, a laser beam with a pulse width of 1 fs to 10 ps and a wavelength of 370 to 640 nm is delivered to selectively activate the amorphous portion, so that the extension region is formed with a thickness of 20 nm or less.
Owner:SEMICON ENERGY LAB CO LTD
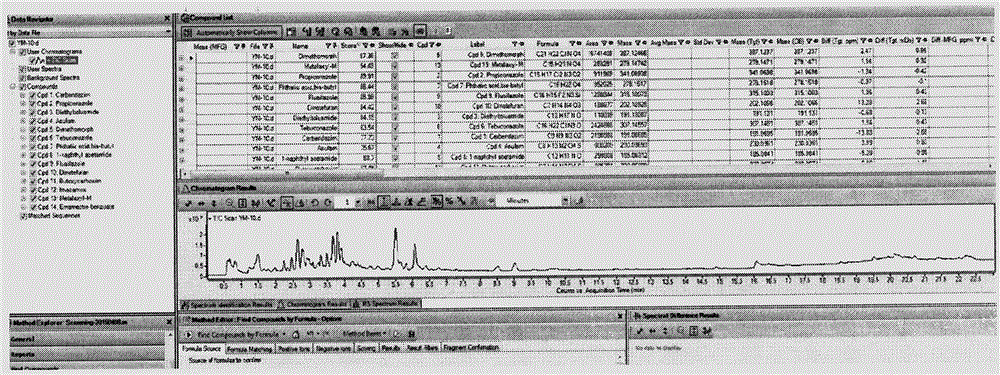
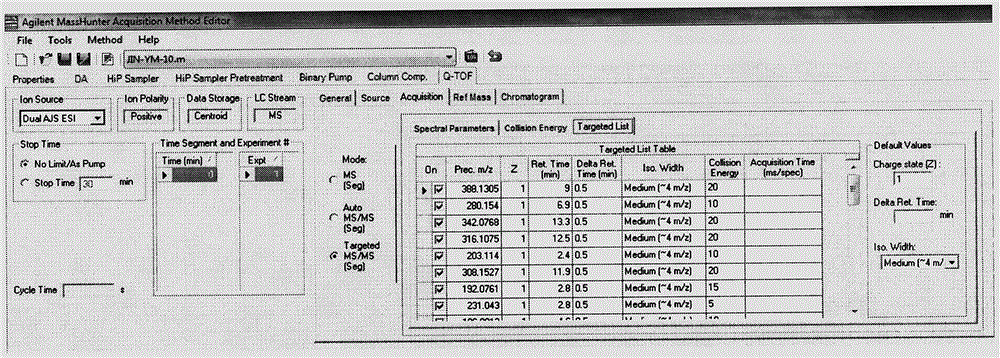
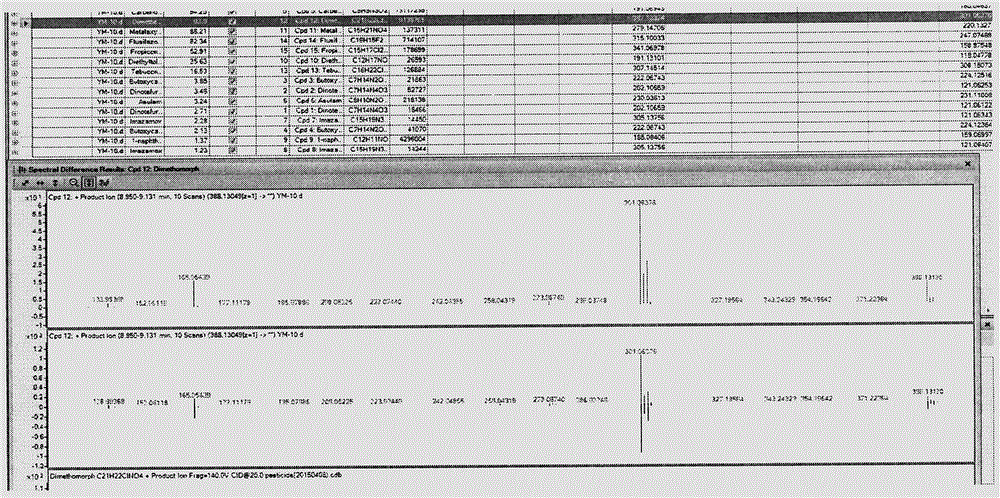
LC-Q-TOF/MS detection technology for 544 pesticide residues in kernel fruit
The invention provides an LC-Q-TOF / MS detection technology for 544 pesticide residues in kernel fruit. In a TOF / MS mode, the LC-Q-TOF / MS detection technology separately determines the retention time of each pesticide standard under designated gas chromatography-mass spectrography conditions, determines the ionization form and chemical formula of a compound under the condition of an ESI source and acquires the accurate mass number of the parent ion of each compound so as to form a TOF / MS database. In a Q-TOF / MS mode, the LC-Q-TOF / MS detection technology separately acquires the fragment ion mass spectrum of each pesticide standard under the condition of three to five different collision energies and introduces acquired information into PCDL software so as to form a Q-TOF / MS database. Through comparison of the retention time, mass spectrometry information and tandem mass spectrometry information of a kernel fruit sample, whether the sample contains pesticide residues or not can be determined; and compounds with high primary scores are subjected to secondary confirmation, and if secondary scores are high, it is confirmed that related pesticide residues are detected. The LC-Q-TOF / MS detection technology provided by the invention has the advantages of fastness, high throughput, high accuracy, high reliability and the like, and can accurately screen pesticides in the kernel fruit.
Owner:CHINESE ACAD OF INSPECTION & QUARANTINE
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com