Method and device for identifying glycopeptide segment
A glycosylation and peptide technology, which is applied in measuring devices, material analysis through electromagnetic means, instruments, etc., can solve the problems of many candidate results and low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
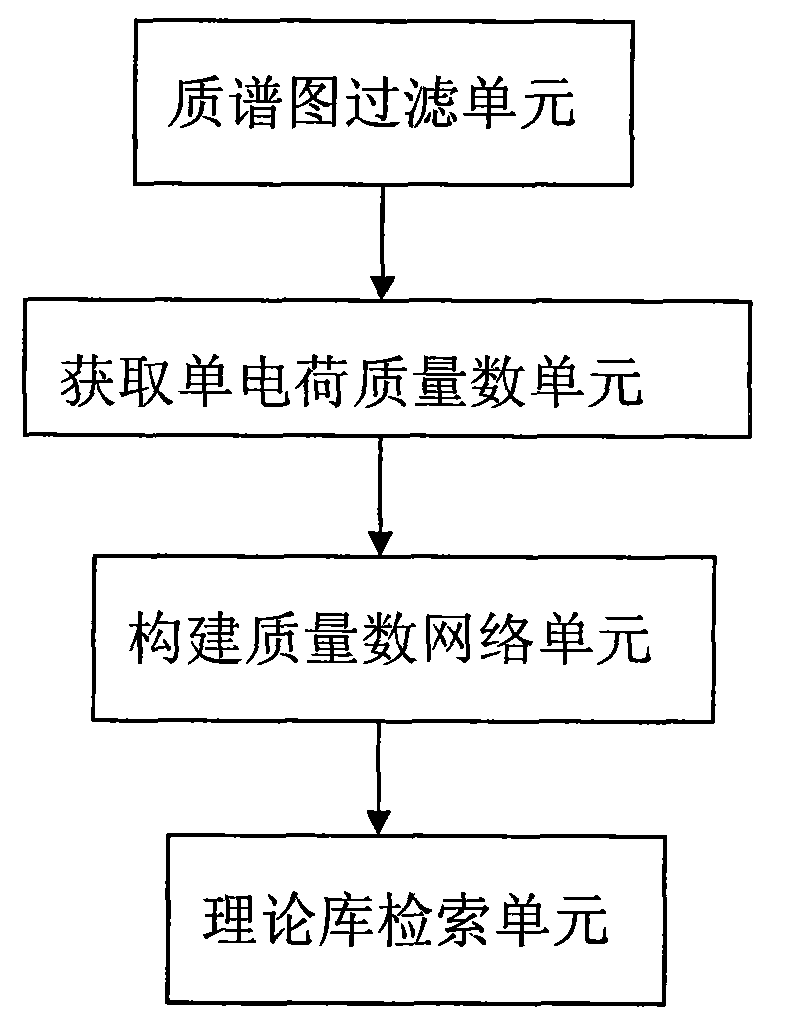
[0057] Example 1 Device for identifying glycosylated peptides
[0058] combined with image 3 , the device consists of:
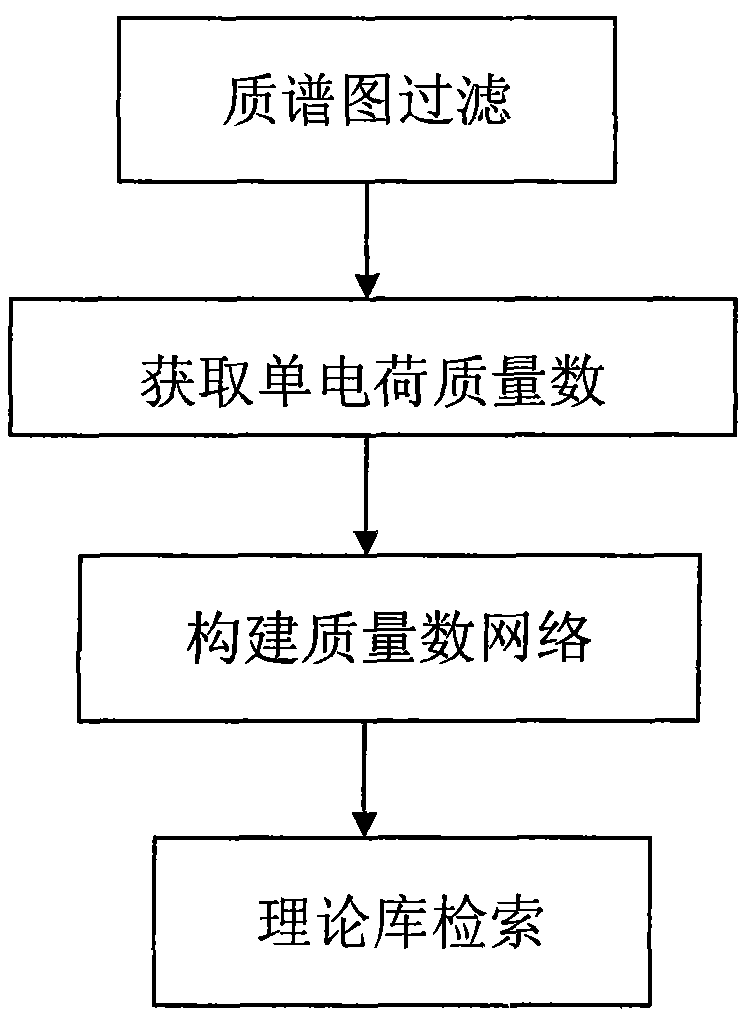
[0059] (1) a mass spectrum filtering unit, which selects a mass spectrum peak with a sufficiently high signal-to-noise ratio in the tandem mass spectrum mass spectrum of the glycosylated peptide sample;
[0060] (2) Obtain the single charge mass number unit: call the mass spectrum peak selected by the mass spectrum filter unit, if the source of the mass spectrum peak is a MALDI mass spectrum, then take the mass number of the mass spectrum peak as the single charge mass number, if the mass spectrum used for identification is an ESI mass spectrum, The mass number of the mass spectrum peak is deconvoluted to obtain its single charge mass number;
[0061] (3) Construct the mass number network unit: call the single-charge mass number obtained from the single-charge mass number unit, perform pairwise subtraction to obtain the mass difference number, and then us...
Embodiment 2
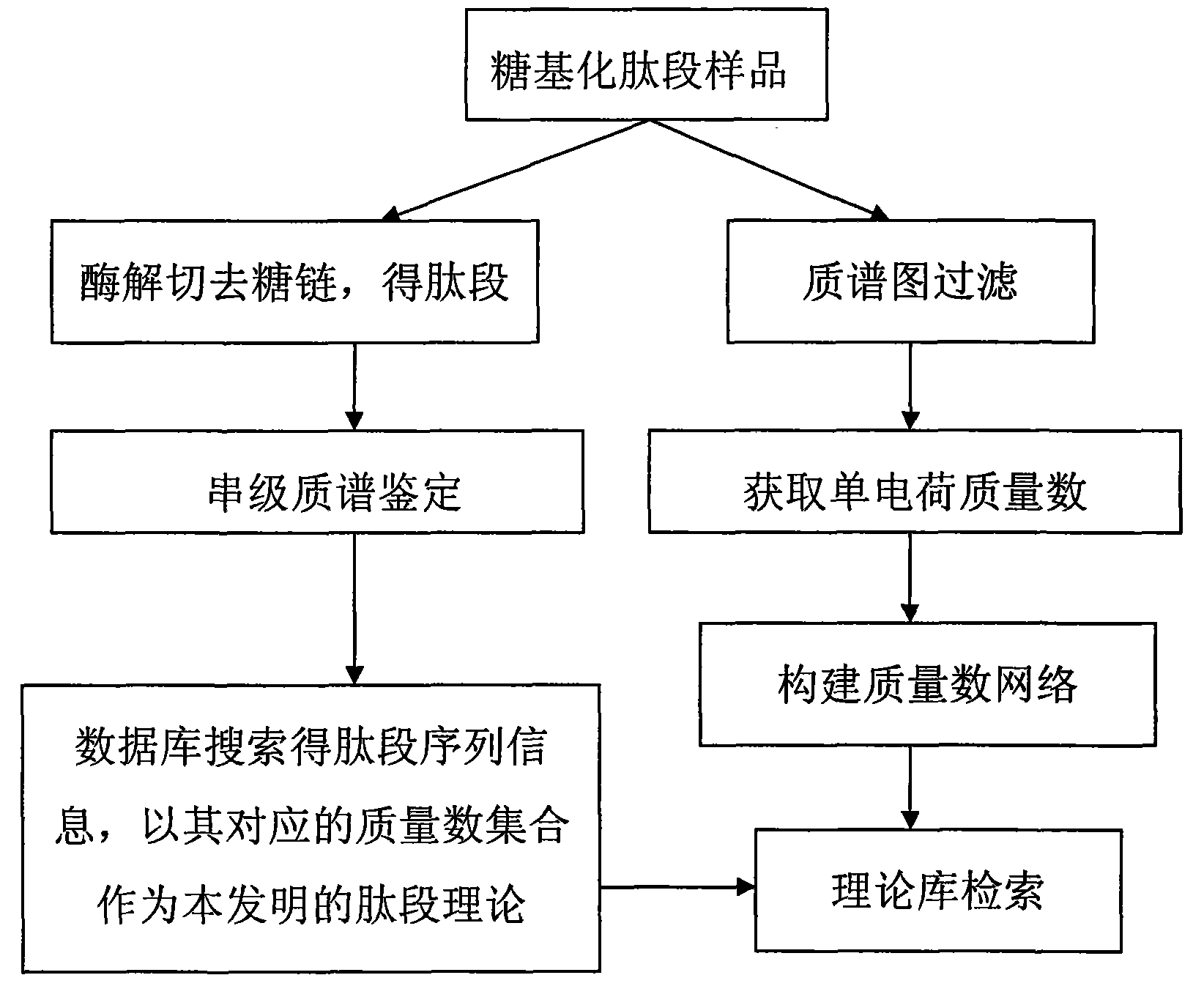
[0064] Example 2 Identification of glycosylated peptides of asialofetuin (Asialofetuin)
[0065] The glycosylated peptide of the standard glycoprotein asialofetuin (Asialofetuin) is used as an unknown sample for identification to test the feasibility and reliability of the method of the present invention.
[0066] experimental method:
[0067] 1. Sample preparation: dissolve the unknown glycoprotein sample in 25mM NH 4 HCO 3 Medium, heat denatured. After the solution was cooled, trypsin was added (according to the ratio of trypsin: glycoprotein sample mass ratio of 1:50), and the enzymolysis was carried out overnight at 37°C. The next day, the enzymatic hydrolysis solution was vacuum-centrifuged and dried for use in subsequent experiments.
[0068] 2. Mass spectrometry identification: AXIMA-QITTM MALDI TOF MS was used for tandem mass spectrometry identification. 2,5-dihydroxybenzoic acid (DHB) was dissolved in 30 vt% acetonitrile aqueous solution containing 0.1 vt% triflu...
Embodiment 3
[0094] Example 3 Identification of glycosylated peptides of horseradish peroxidase (HRP)
[0095] The glycosylated peptide of the standard glycoprotein horseradish peroxidase (Horseradish Peroxidase, HRP) is used as an unknown sample for identification to test the feasibility and reliability of the method of the present invention.
[0096] Mass spectrometer: LTQ-Orbitrap mass spectrometer
[0097] experimental method:
[0098] 1. Sample preparation: dissolve the unknown glycoprotein sample in 25mM NH 4 HCO 3 Medium, heat denatured. After the solution was cooled, trypsin was added (according to the ratio of trypsin: glycoprotein sample mass ratio of 1:50), and the enzymolysis was carried out overnight at 37°C. The next day, the enzymatic hydrolysis solution was vacuum-centrifuged and dried for use in subsequent experiments.
[0099] 2. Mass spectrometry identification: ESI-LTQ-Orbitrap XL Systems was used for tandem mass spectrometry identification, and the proteolytic pep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com