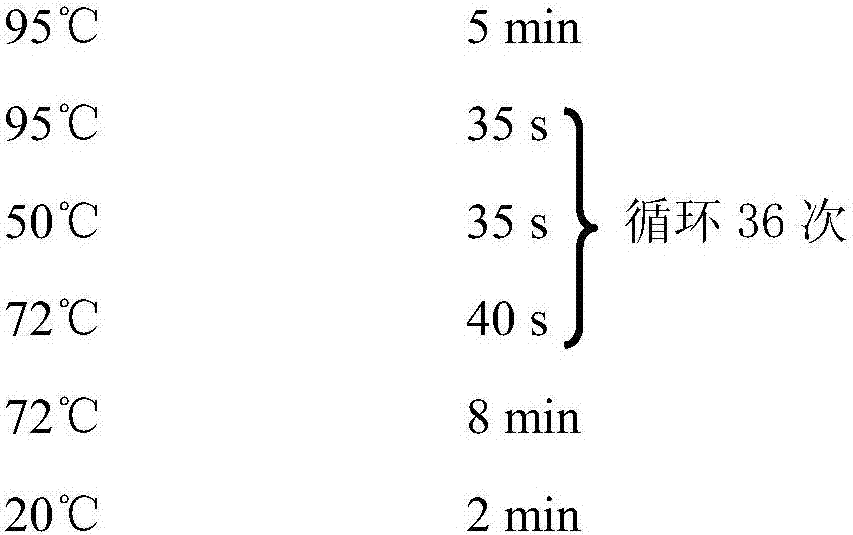
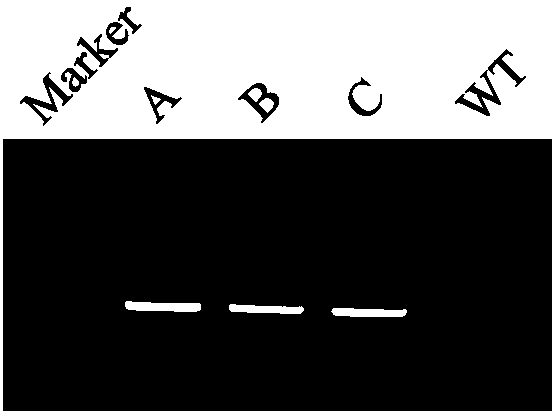
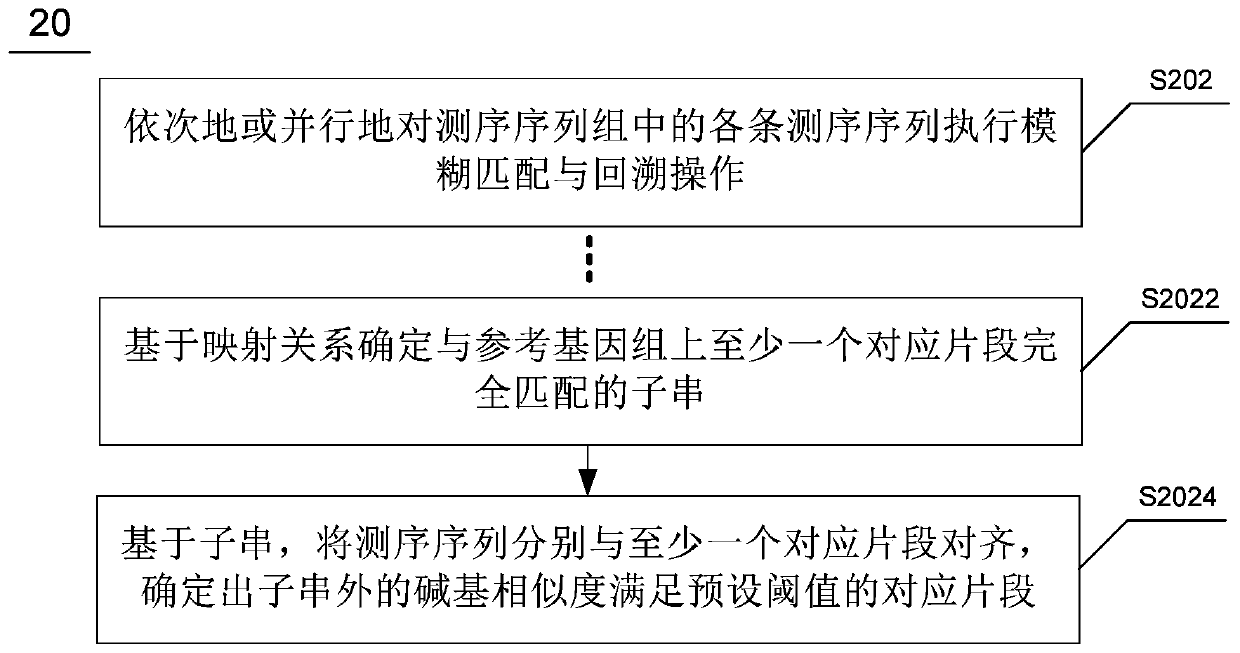
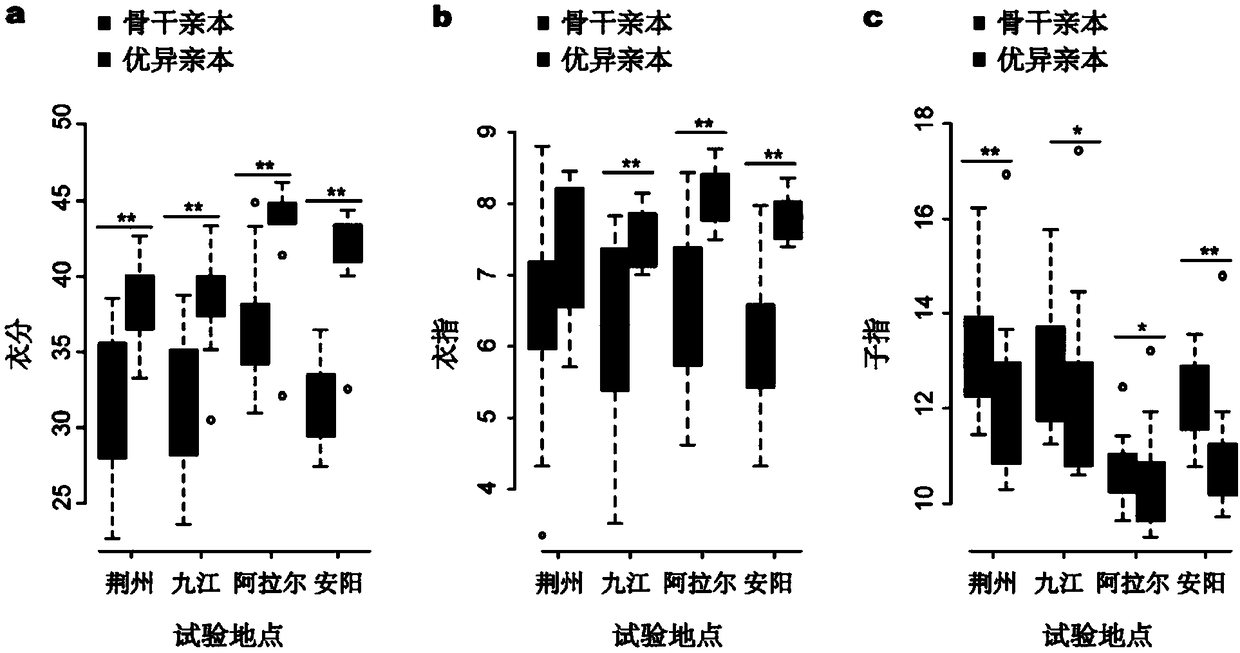
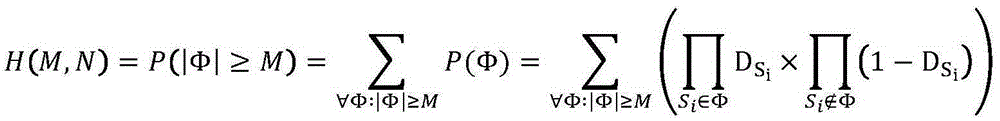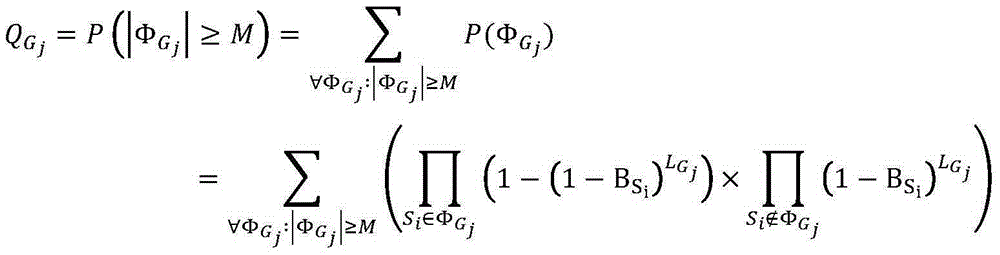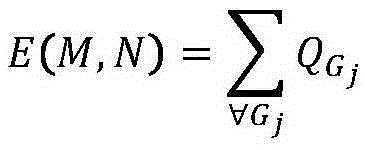Patents
Literature
123 results about "Genome resequencing" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
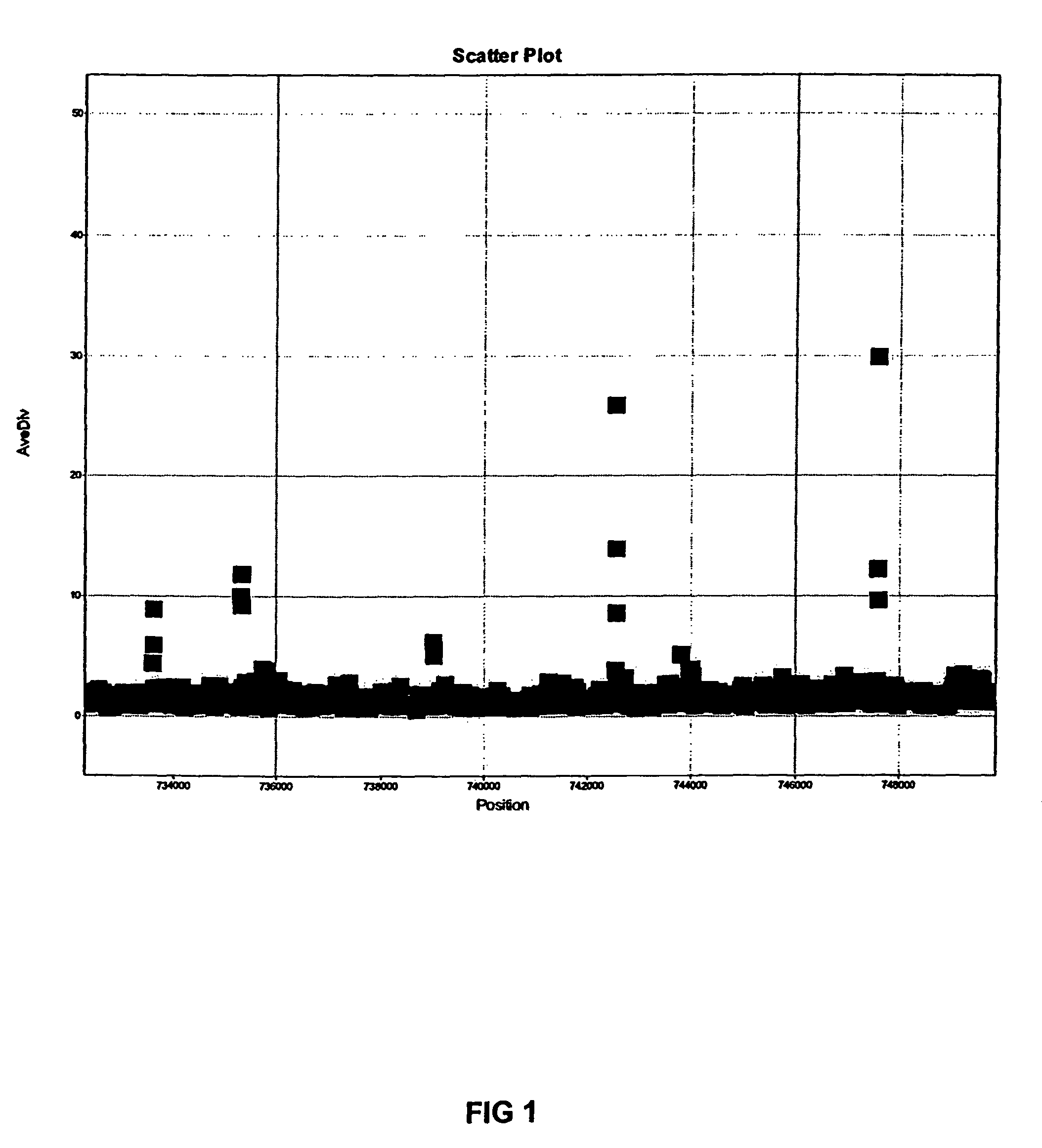
Whole genome sequencing (WGS) or genome resequencing are ideally suited for genome wide variant identifications and structural variation detection. These SNPs and insertion or deletions constitute the major disease biomarker for human and complex genomes.
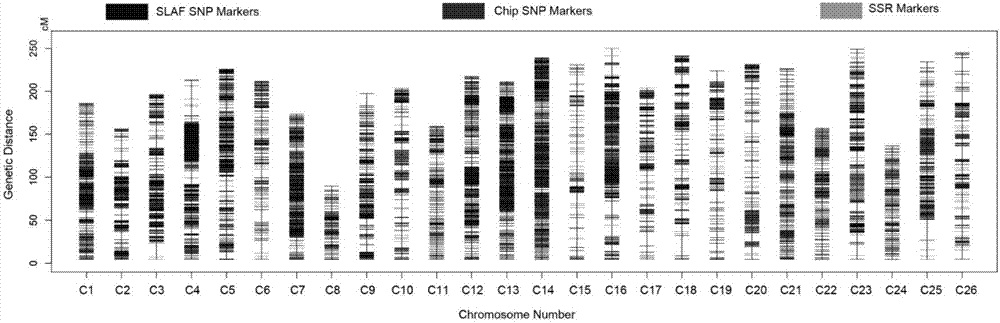
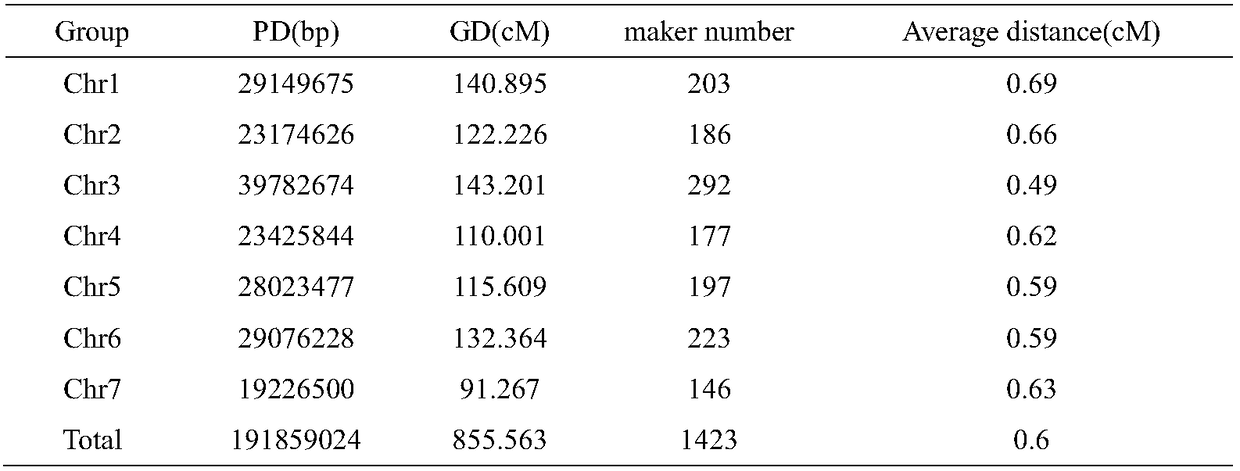
SNP molecular marker associated with chromosome 6 and fiber strength of upland cotton
ActiveCN106868131AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention belongs to the technical field of molecular breeding of cotton and discloses an SNP molecular marker associated with fiber strength of upland cotton as well as detection and application thereof. The SNP molecular marker is obtained by taking stable RIL group of cotton as the material through a genome resequencing method. When the SNP markers are used for carrying out molecular marker-assisted breeding selection, the breeding period can be greatly shortened; the breeding efficiency of the cotton can be improved; the cotton fiber strength can be improved.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Specific molecular marker for acquiring genders of bighead carps and screening method and application thereof
ActiveCN106947827AOvercome operabilityTo overcome the disadvantages such as time-consumingMicrobiological testing/measurementDNA/RNA fragmentationGenomic sequencingScreening method
The invention discloses a specific molecular marker for acquiring the genders of bighead carps and a screening method and application thereof. The screening method comprises the following steps: firstly, performing reduced-representation sequencing on 5 female bighead carps and 5 male bighead carps by using a 2b-RAD technology to identify a male specific sequence; then, performing genome resequencing and genome assembly on one of the male bighead carps, comparatively searching 2b-RAD specific sequences of the male bighead carps in a male bighead carp genome to obtain a long-fragment male bighead carp specific sequence, and designing a male specific primer. By adopting the screening method, a male specific sequence and a gender specific primer of the bighead carps are screened for the first time, and a PCR (Polymerase Chain Reaction) technology for generic gender determination is built. The method has the advantages of low cost, easiness, rapidness and accuracy in operation, and the like, and a reliable technical measure is provided for the development of a gender marker and gender determination of the bighead carps. The method can be popularized and applied in gender marker development and gender determination research of other fishes.
Owner:INST OF AQUATIC LIFE ACAD SINICA
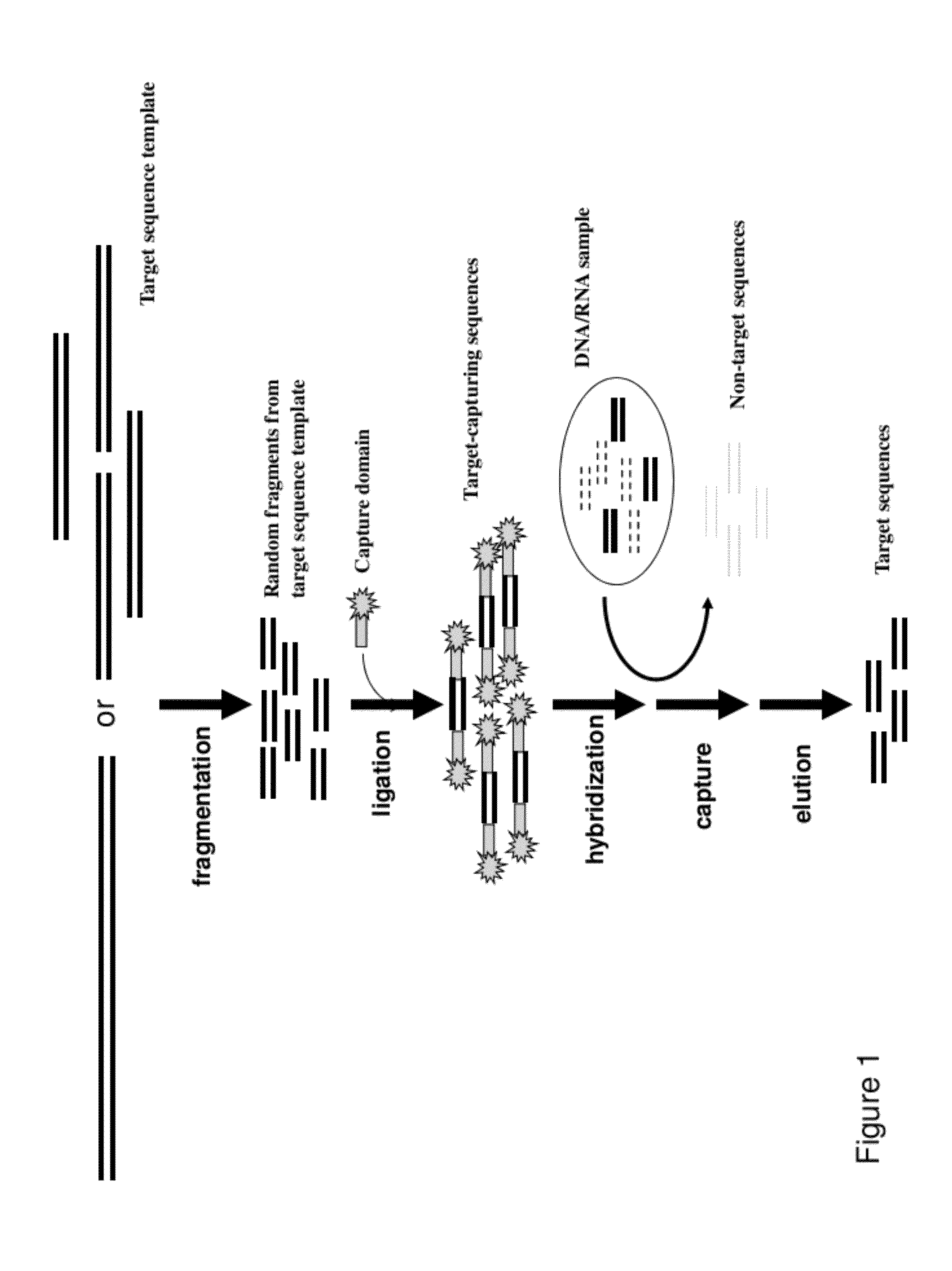
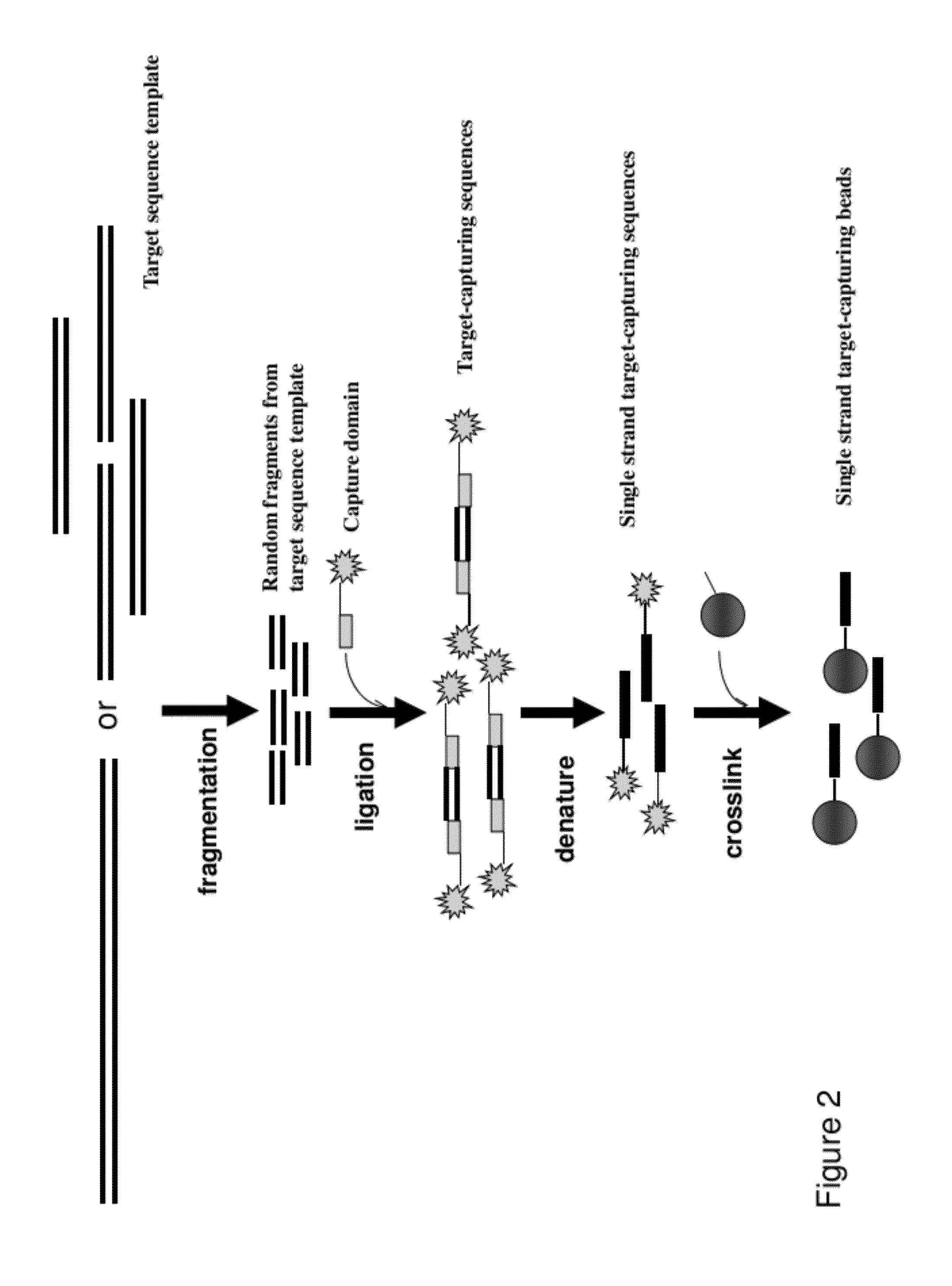
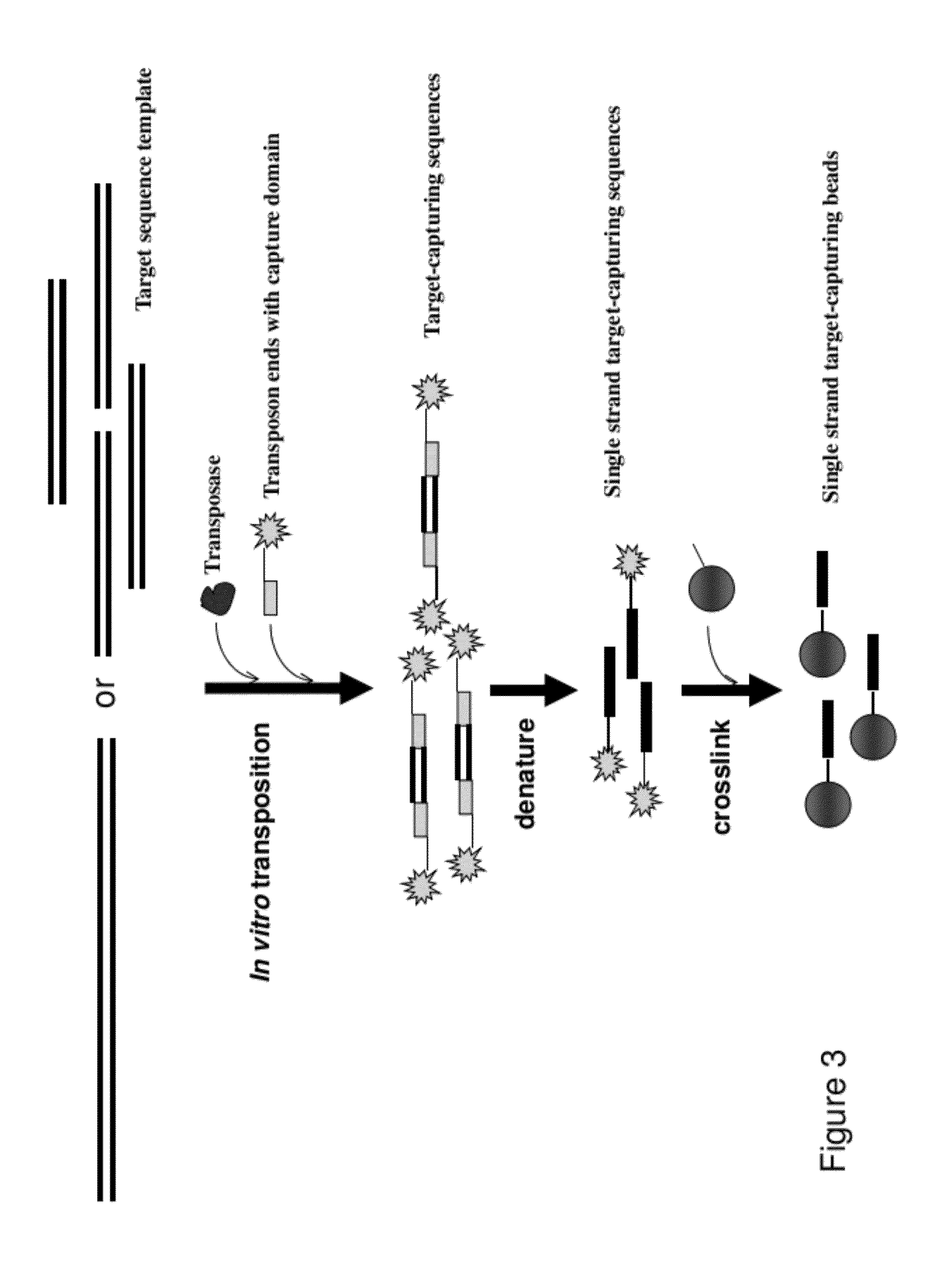
Methods, Compositions, and Kits for Making Targeted Nucleic Acid Libraries
InactiveUS20120258892A1Low costReduce designTransferasesLibrary creationTarget captureGenome resequencing
The present invention provides a method and a kit for selecting and enriching target sequences specific for a genomic region of interest or a subset of a transcriptome using a target-capturing sequence library. The target-capturing sequence library comprises random DNA fragments generated from a target sequence template encompassing all the target sequences. The present invention provides an efficient and cost-effective method of target selection for targeted genome resequencing.
Owner:WANG YAN
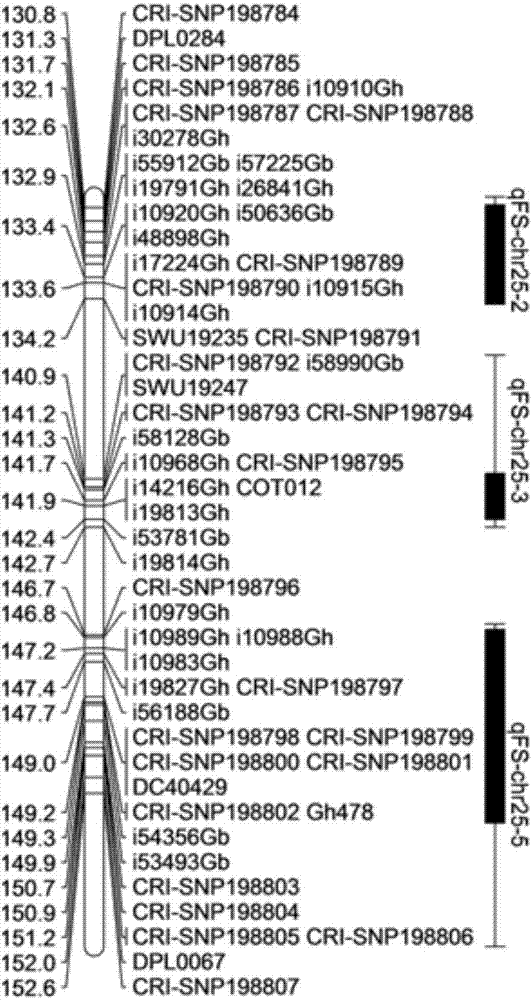
SNP molecular markers related to fiber strength of upland cotton 25# chromosome
ActiveCN107043813AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention relates to SNP molecular markers related to fiber strength of an upland cotton 25# chromosome. The invention belongs to the technical field of cotton molecular breeding and discloses the SNP molecular markers related to fiber strength of the upland cotton and detection and application thereof. The SNP molecular markers are obtained by taking a stable RIL group of cotton as a material by means of a genome re-sequencing method. The SNP markers disclosed by the invention are used for molecular marker-assisted selection, so that the breeding period can be greatly shortened, and the breeding efficiency of the cotton fiber strength can be increased.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Variable length probe selection
InactiveUS20050282209A1Improve accuracyImprove efficiencyBioreactor/fermenter combinationsBiological substance pretreatmentsGenome resequencingVariable length
The present invention provides novel method for increasing the efficiency and accuracy of high-throughput mutation mapping and genome resequencing by using a variable length probe selection algorithm to rationally select probes used in designing oligonucleotide arrays synthesized by Maskless Array Synthesis (MAS) technology. Also disclosed is a variable length probe selection algorithm used in designing such oligonucleotide arrays.
Owner:ROCHE NIMBLEGEN
Kiwi fruit InDel molecular marker and screening method and application thereof
ActiveCN106498075APrimer variant stabilitySpeed up the breeding processMicrobiological testing/measurementDNA/RNA fragmentationResearch ObjectActinidia
The invention discloses a kiwi fruit InDel molecular marker and a screening method and application thereof. Two different cold-proof Actinidia arguta genome DNAs are used as research subjects, resequencing of four variety genes is carried out, InDel primers are designed according to resequencing data, and PCR (polymerase chain reaction) amplification, agarose electrophoresis and non-denaturing polyacrylamide gel electrophoresis are carried out; polymorphism detection is carried in Actinidia arguta to screen out 14 pairs of InDel primers, the 14 pairs of InDel primers are applicable to the genetic diversity analysis for Actinidia chinensis, Actinidia deliciosa and the like and are also applicable to the researches, such as hybrid offspring authenticity identification, genetic map construction, and molecule-assisted breeding. The kiwi fruit InDel primers according to the embodiment of the invention has good mutation stability and low detection difficulty, allow InDel insertion / loss of large fragments, and allows agarose analysis, with steps that may be simplified.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Building method of long fragment nucleic acid library
InactiveCN104711250ARead longSolve assembly problemsMicrobiological testing/measurementLibrary creationGenome resequencingGenome
The invention provides a building method of a long fragment nucleic acid library. The method comprises the following steps: preparing a batch of Index label clusters, marking long fragment nucleic acid molecules in batch, using whole genome resequencing library building to build long fragment libraries suitable for illumine sequencing platforms, allowing all fragments to have connectors suitable for the illumine sequencing platforms under the action of transposase, carrying out PCR amplification, and screening the well build library. The method effectively solves the problem of influences of polyploid and repeated sequence in a genome de novo sequencing technology on the assembling accuracy, and can be used for assembling complex genomes and assisting de novo sequencing assembling to improve the assembling accuracy and shorten the assembling time.
Owner:BIOMARKER TECH
Breeding method of brassica napus radish cytoplasm sterile restorer line and application to brassica napus breeding
ActiveCN107347632AGenetic stabilityEarly removalMicrobiological testing/measurementPlant genotype modificationBrassicaBackcross population
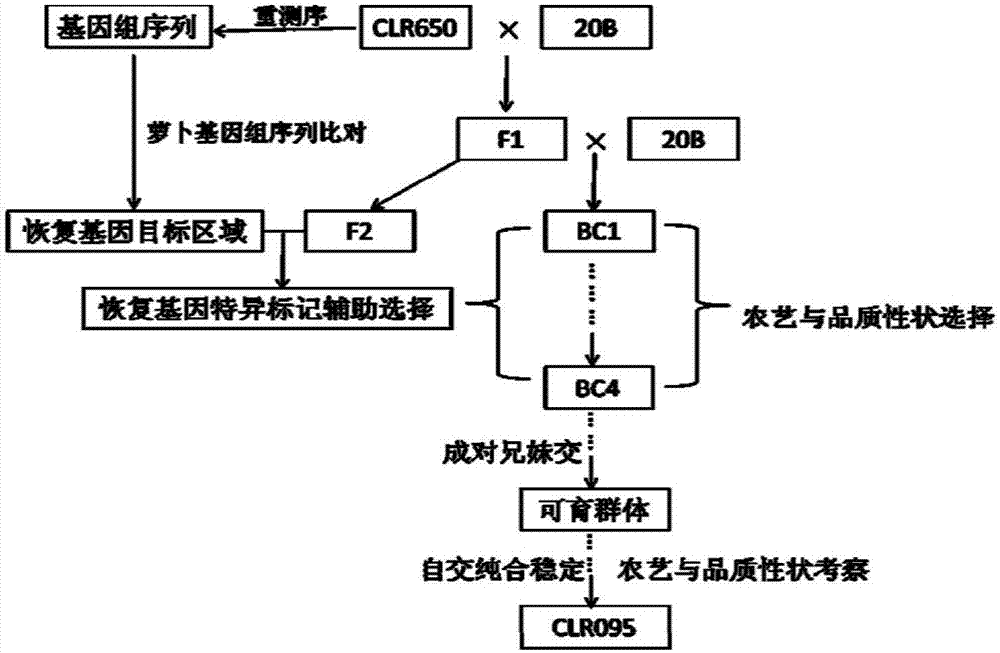
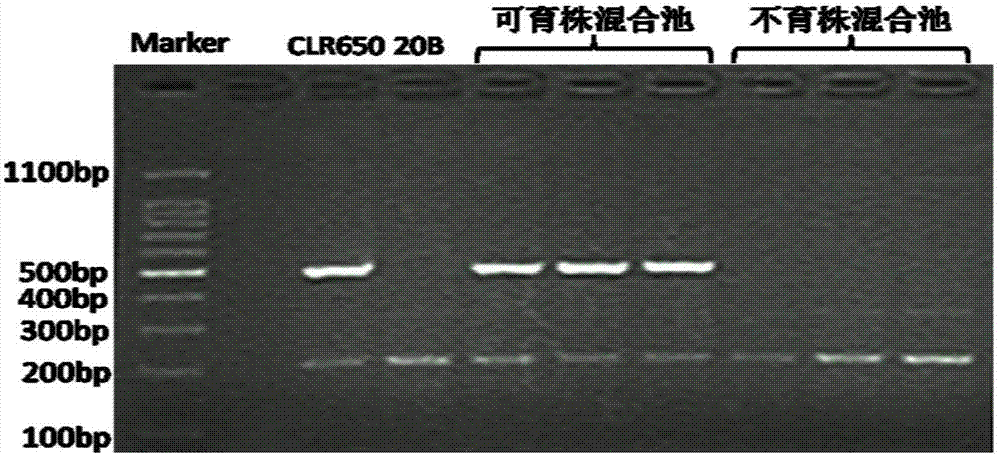
The invention relates to a breeding method of a brassica napus radish cytoplasm sterile restorer line. Through brassica napus radish cytoplasm sterile restorer material CLR650 whole genome resequencing data and radish genome sketch information comparison, specific molecular markers of linked exogenous radish fragments and restoring genes in CLR650 are developed; then, a target single strain of the brassica napus radish cytoplasm sterile restorer material CLR650 backcross population is bred in an auxiliary way through the specific molecular markers; a BC4 separation group is obtained; fertile plants in the BC4 group is subjected to multi-generation mixed selection by a mixed method until the mixed separation group with the fertile plant proportion exceeding 75 percent is obtained; finally, the homozygosis stabilization is performed through multi-generation selfing; the novel brassica napus radish cytoplasm sterile restorer line is obtained. The method provided by the invention overcomes the defects of too low restoring gene heritability and property homozygosis stabilization difficulty of the existing method.
Owner:湖南省作物研究所
Method for determining insertion site of transgenic line by re-sequencing technique
ActiveCN108034706AOvercome efficiencyOvercome operational complexityMicrobiological testing/measurementRe sequencingBiotechnology
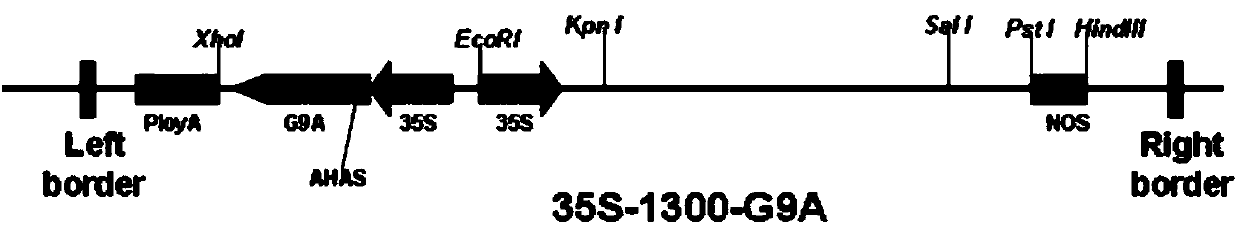
The invention discloses a method for determining an insertion site of a transgenic line by a re-sequencing technique. The method comprises the following steps of (1) extracting DNA (deoxyribonucleic acid) of a genome of the transgenic line, wherein the total amount is at least 2g; (2) re-sequencing the genome, wherein the data size is at least 12G; (3) using a T-DNA sequence of a transgenic carrier as a template, and comparing and analyzing the obtained sequencing data, so as to obtain information of the insertion site; (4) according to the information of the insertion site, designing a primer, and performing PCR (polymerase chain reaction) verification, so as to determine the insertion site of each transgenic line. The method for determining the insertion site of the transgenic line has the characteristics of maturity, simplicity and high success rate, and can be widely applied to determine the insertion site of the transgenic line.
Owner:ZHEJIANG UNIV
Method for rapidly discovering phenotype related gene based on probabilistic framework and resequencing technology
ActiveCN105404793AImprove research efficiencyMicrobiological testing/measurementSpecial data processing applicationsGenome resequencingForward genetics
The present invention discloses a method for rapidly discovering a phenotype related gene based on a probabilistic framework and a resequencing technology, and establishes a method based on the probabilistic framework. The method comprises: modeling a forward genetics study process based on a genome resequencing technology, estimating, by calculating four indexes, an effect of a design of each step in a study process on overall availability of the study, thereby guiding optimization of a current experimental scheme and analytical method, and achieving the purpose of rapidly discovering a gene possibly associated with a particular phenotype by using samples as few as possible. According to the method provided by the present invention, the estimation, according to four indexes, of the overall availability of the forward genetics study process based on the genome resequencing technology is firstly proposed, and a method success rate and a non-Mendelian phenotype significance are two indexes proposed creatively, and have a significant value on guiding the optimization of an overall study process.
Owner:ZHEJIANG UNIV
Comparative genomic resequencing
The present invention is an improved method of resequencing DNA using microarrays to rapidly map and identify SNPs, deletions and amplification events present in the genome of an organism. The method is performed by hybridizing a reference and a test genome to two separate arrays with each array exhibiting a specific intensity pattern. The intensity differences between the reference and the test genome arrays are used to produce a mutation map. The mapped differences are resequenced on a set of resequencing arrays to identify specific genetic mutations.
Owner:ROCHE NIMBLEGEN
Molecular marks closely linked to glomerella leaf spot resistant genetic loci of apple and application
ActiveCN106520762AMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention provides molecular marks closely linked to glomerella leaf spot resistant genetic loci of an apple and an application of the molecular marks. The molecular marks include 6 SNP marks and 5 InDel marks, which are SNP3955, SNP4236, SNP4257, SNP4299, SNP4336, SNP4432, InDel4199, InDel4227, InDel4254, InDel4305 and InDel4334 respectively. In addition, the invention provides a mark screening method and the application of the marks. SNP and InDel marks on two sides of an Rgls gene are selected out in combination with a preliminary locating result of the Rgls gene (located between SSR marks S0304673 and S0405127) by utilizing SNP and InDel marks developed through a whole genome re-sequencing technology; 18 SNP loci and 30 InDel loci are analyzed through an HRM curve analysis technology; and 6 SNP marks and 5 InDel marks are screened out to be closely linked to the loci of the Rgls gene.
Owner:QINGDAO AGRI UNIV
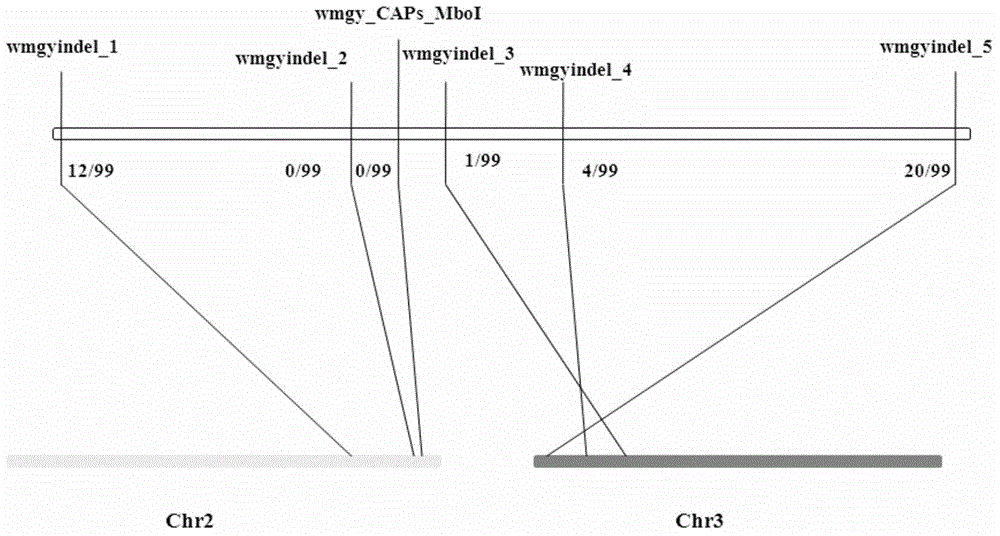
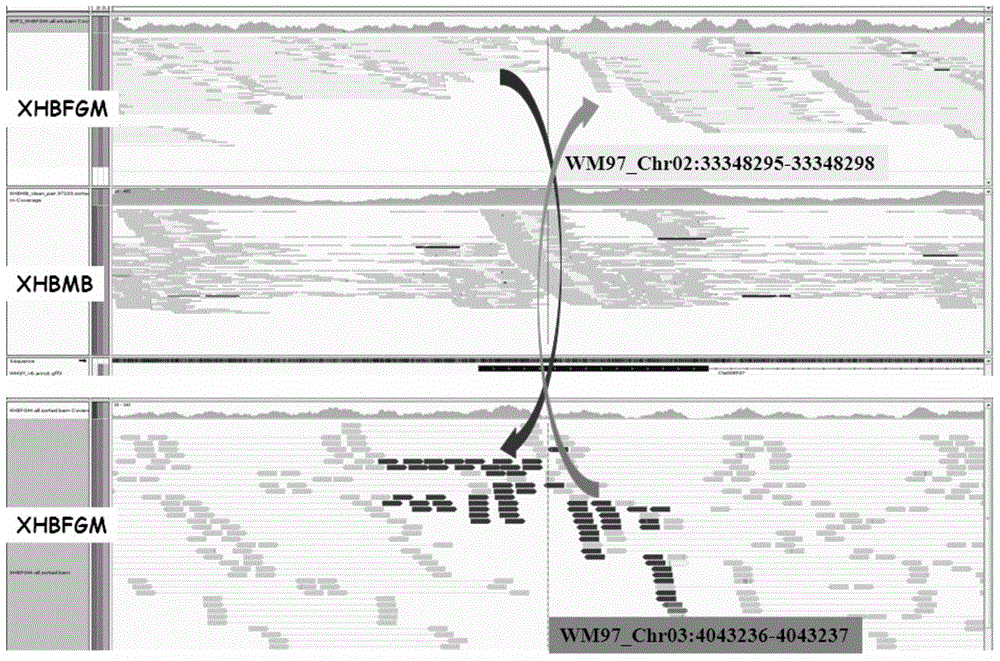
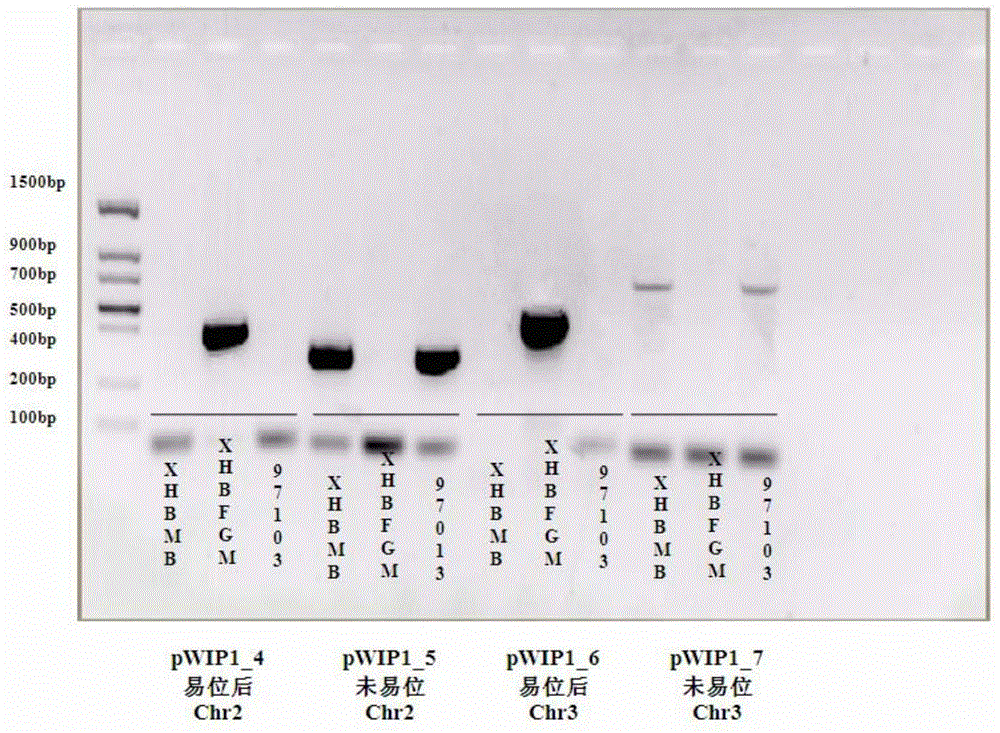
Watermelon female lines gene C1WIP1 and chromosome translocation and linkage marker
ActiveCN103937785AFast and accurate cloningReduce the cost of seed productionMicrobiological testing/measurementFermentationGenome resequencingA-DNA
The invention belongs to the molecular biology field, and more specifically relates to watermelon female lines gene(i) C1WIP1( / i) and chromosome translocation and linkage marker, which comprises a DNA sequence of watermelon female lines gene(i) gy( / i) gene(i) C1WIP1( / i), its acquisition method, a molecular marker linked with the DNA sequence, a chromosome translocation section due to appearance of the watermelon female lines and its acquisition method. According to the invention, a classic gene positioning method and a whole genome re-sequencing technology are organically combined, and watermelon female lines gene(i) C1WIP1( / i) is rapidly and accurately cloned; a PCR technology is used for confirming that a watermelon spontaneous mutant material XHBFGM is appeared by female lines phenotype due to chromosome translocation, and a preliminary research is carried out on the effect mechanism of (i) C1WIP1( / i); By aiming at the existence of the chromosome translocation, the molecular marker linked with watermelon female lines gene(i) C1WIP1( / i) is designed, the watermelon female lines phenotype which takes XHBFGB as a transfer material can be rapidly detected, the molecular marker is used for assistant breeding, seed production cost of watermelon is greatly reduced, and the seed purity is increased.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Kit for guiding human mental disease medication and detection method thereof
ActiveCN109777870AMicrobiological testing/measurementDNA/RNA fragmentationDiseaseHuman DNA sequencing
The invention provides a kit for guiding human mental disease medication and a detection method thereof. The kit can simultaneously type 16 gene loci through one reaction, and comprises 16 pairs of amplification primers for amplifying 16 gene segments; the sequences of 16 pairs of the amplification primers are specifically SEQ ID No.1-SEQ ID No.32; the kit also comprises 16 extension primers, andthe sequences of the 16 extension primers are specifically SEQ ID No.33-SEQ ID No.48. According to the kit and the detection method thereof, through typing based on multiple PCRs and a mass spectrum sequencing technology, human genome mutation related to mental disease medication can be accurately detected; relative to whole genome re-sequencing and whole-exome sequencing, and through the kit andthe detection method of the kit, the experiment cost is also greatly reduced, the period is shortened, and therefore the kit and the detection method of the kit have the all-important practical application value.
Owner:SHANGHAI CONLIGHT MEDICAL LTD
SNP locus closely related to orientation of capsicum fruit and its universal molecular marker, acquisition method and application thereof
ActiveCN110305978AAdvantages and Notable ImprovementsReduce planting sizeMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGermplasm
The present invention provides a mononucleotide polymorphism (SNP) loci highly associated with fruit orientation obtained by using genomic resequencing data and genome-wide association analysis (GWAS). A F2 segregating population with fruit orientation traits is used, by constructing two extreme mixing pools facing the phenotype, RNA sequencing (BSR) is carried out on the mixed pools, the SNP locus closely linked to the orientation of the capsicum fruit is obtained, the molecular markers obtained by the two strategies are used to finely position the orientation traits, and a tightly linked enzyme-cleaved polymorphic sequence (CAPS) marker CaUP12 is developed. CaUP12 is used to identify capsicum germplasm resources, and the accuracy rate reached 95.6%. The F2 segregating population is identified by CaUP12, and the accuracy rate reached 100%. The present invention provides the closely related molecular marker for the cloning of capsicum fruit control genes and capsicum breeding, which has important application value.
Owner:HUAZHONG AGRI UNIV
Method for simultaneously locating two character-related genes
ActiveCN110675915AImprove positioning efficiencyReduce R&D investment costsSequence analysisHybridisationMolecular breedingGenome resequencing
The invention belongs to the technical field of molecular genetics and molecular breeding, and discloses a method for simultaneously locating two character-related gene loci by using recombination phenotype grouping analysis. The method, based on the analysis of a genome re-sequenced or transcriptome sequenced recombination phenotype individual group, simultaneously locates two independent character-related gene loci, specifically comprises: constructing a separate population by using two parents with different characters; selecting, from the population, two recombination phenotype individualswith character phenotypes of different parental types; classifying the two recombination phenotype individuals into two types: one of which has a character A of a maternal type and a character B of apaternal type, and the other one of which has a character A of a paternal type and a character B of a paternal type; re-sequencing or transcriptome sequencing a mixed pool of parents and the two individual groups or the genomes of the individuals to calculate a difference of the SNP frequencies of a parental type corresponding to the SNPs having different parents in the individual group, and simultaneously locating two character-related gene loci.
Owner:QINGDAO AGRI UNIV
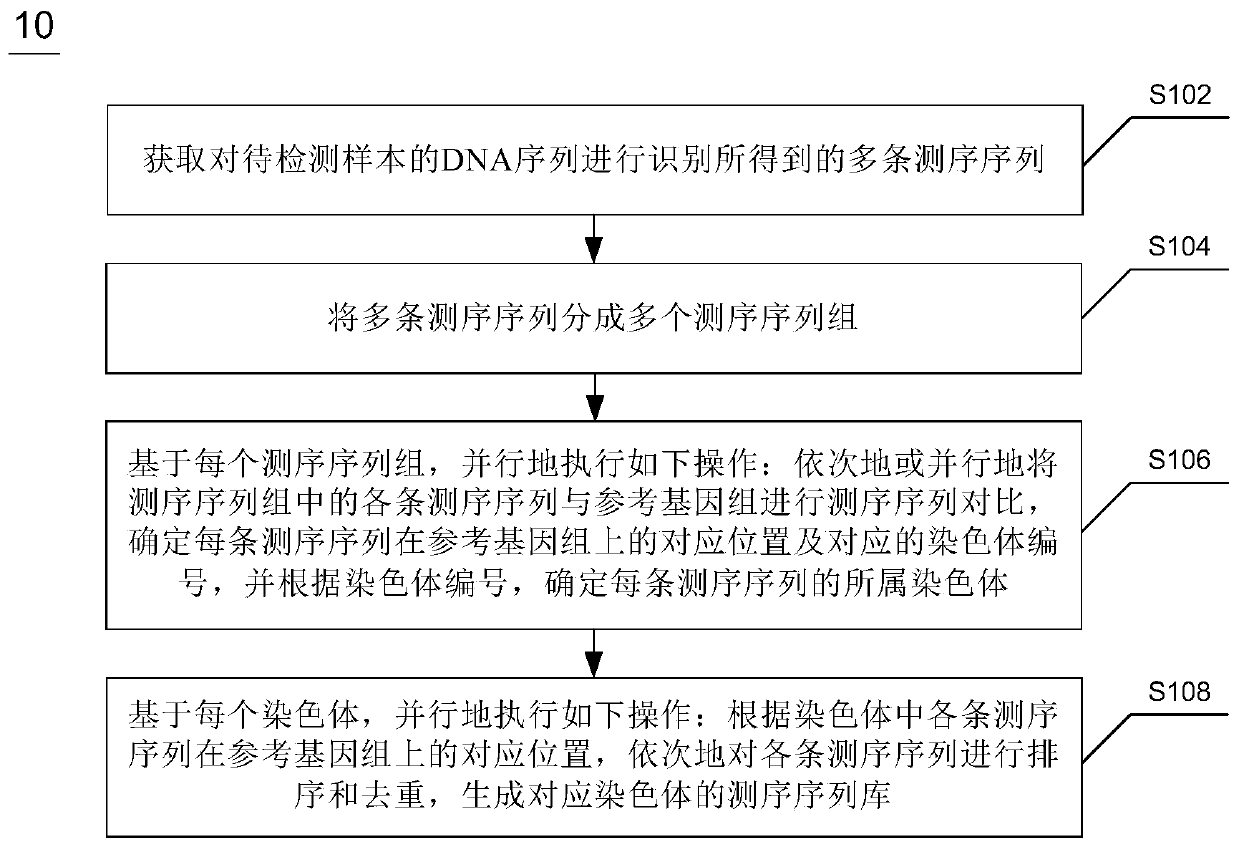
Whole genome re-sequencing analysis and method for whole genome re-sequencing analysis
ActiveCN110797088ATake advantage ofShorten the timeBiostatisticsProteomicsSequence analysisGenome resequencing
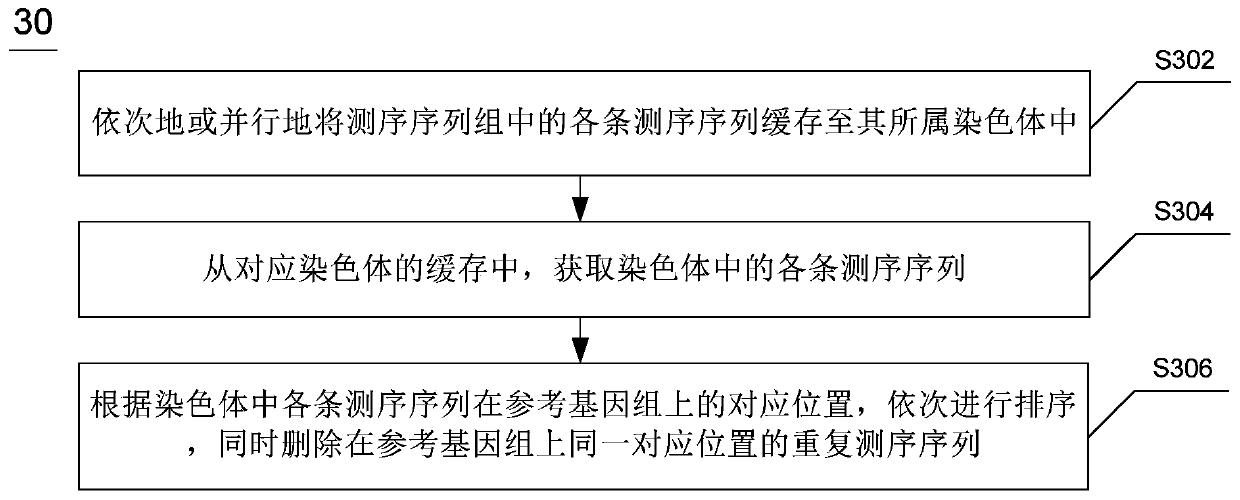
The invention discloses whole genome re-sequencing analysis and a method for the whole genome re-sequencing analysis. The method for whole genome re-sequencing analysis comprises the following steps of: acquiring a plurality of sequencing sequences obtained by identifying a DNA sequence of a sample to be detected, dividing the plurality of sequencing sequences into a plurality of sequencing sequence groups, based on each sequencing sequence group, executing the following operations in parallel: sequentially or parallelly comparing each sequencing sequence in the sequencing sequence group witha reference genome, and determining a corresponding position of each sequencing sequence on the reference genome and a corresponding chromosome number, and according to the corresponding position of each sequencing sequence on the reference genome and the corresponding chromosome number, conducting sequencing and duplicate removal on each sequencing sequence, and generating a sequencing sequence library corresponding to each chromosome.
Owner:南京医基云医疗数据研究院有限公司
Method for rapidly identifying transgenic or gene editing material and insertion site thereof by using whole genome resequencing data
ActiveCN110556165AAccurate insertionReduce sensitivityMicrobiological testing/measurementProteomicsGenome resequencingInsertion site
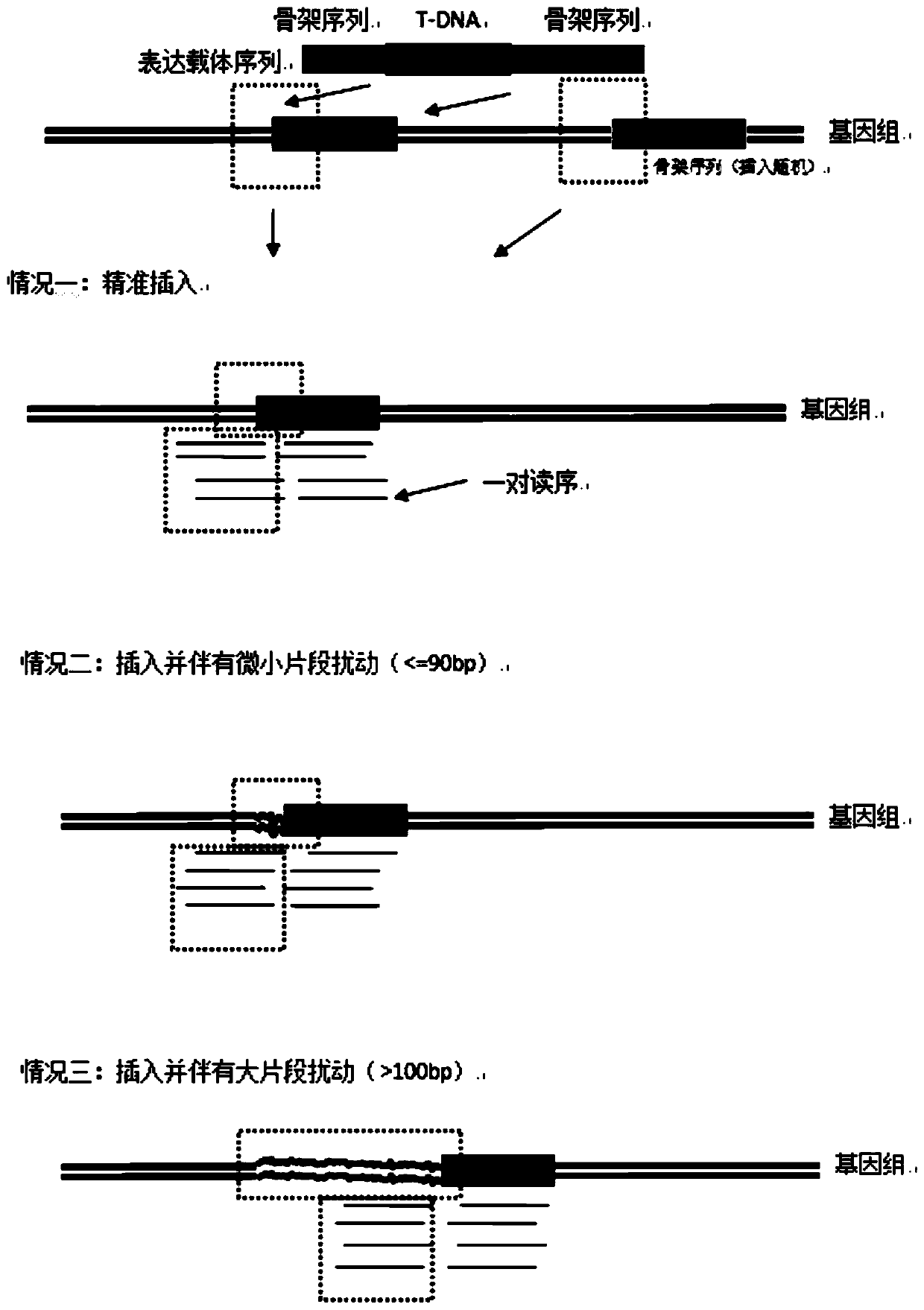
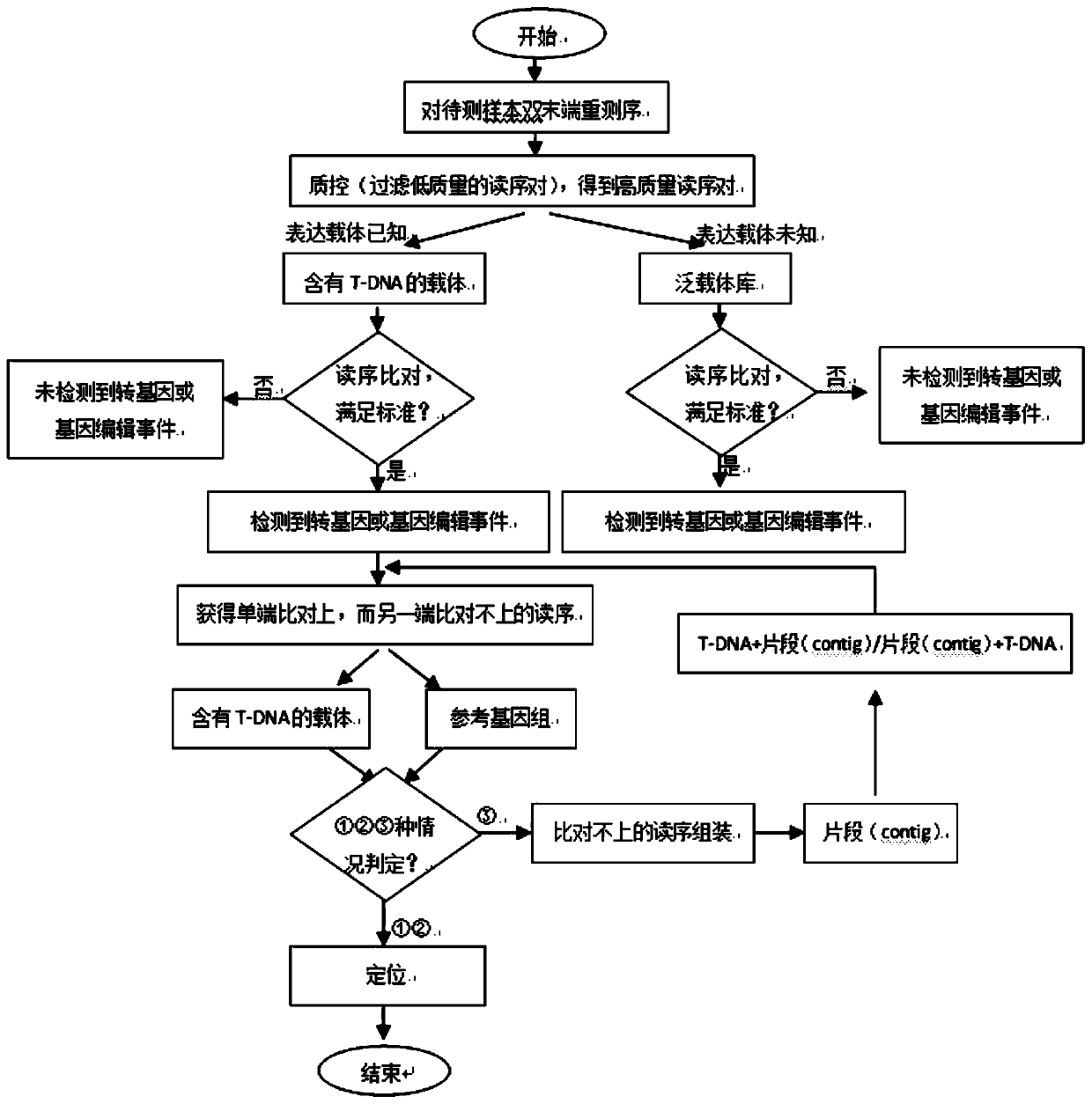
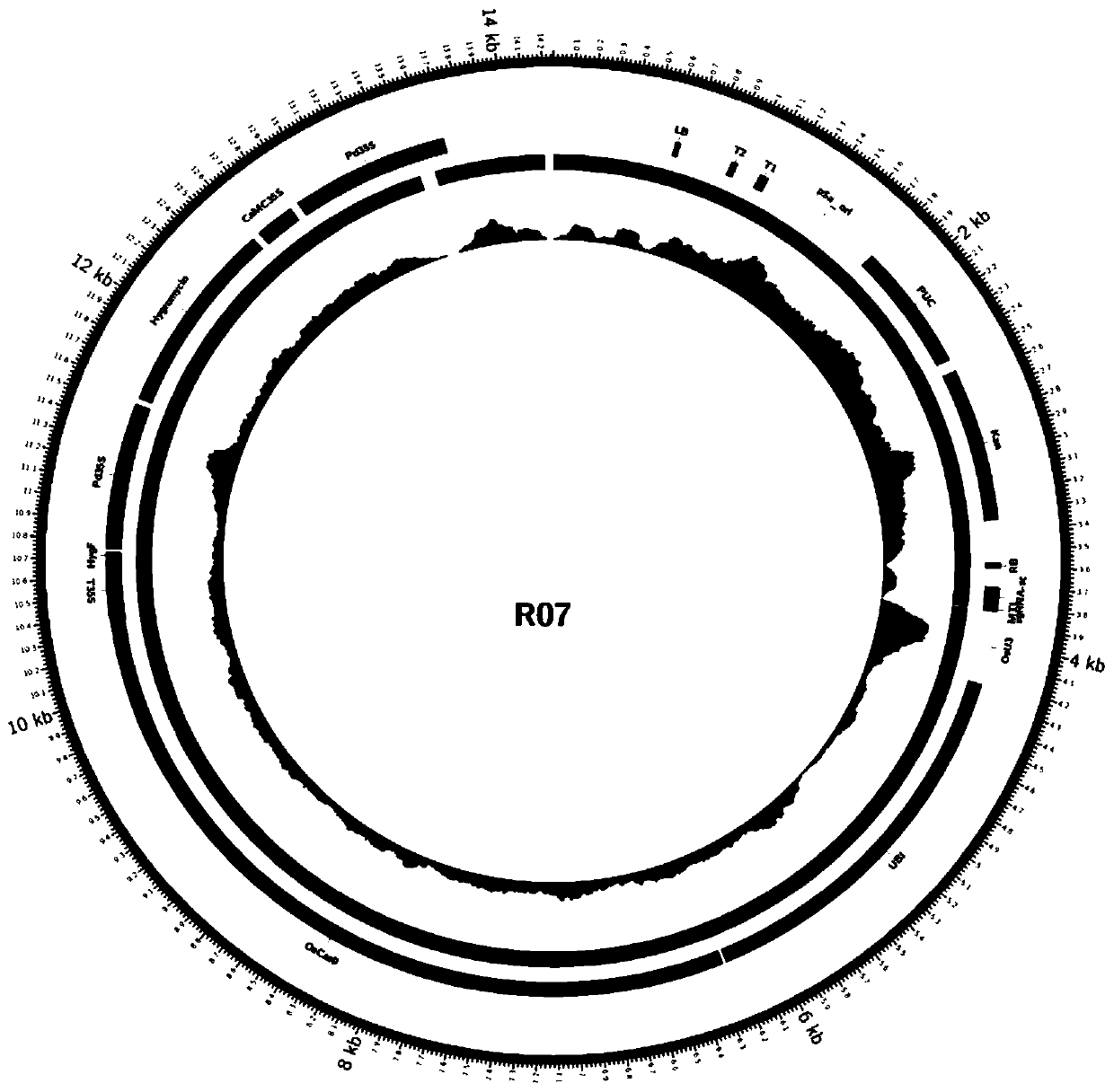
The invention discloses a method for rapidly identifying a transgenic or gene editing material and an insertion site thereof by using whole genome resequencing data. The method comprises the followingsteps: extracting a genome DNA; obtaining the double-end sequencing data of the whole genome; judging whether an expression vector sequence inserted into the plant to be detected and containing the T-DNA sequence is known or not; judging whether the to-be-detected plant has a transgenic event or a gene editing event or not, and whether a skeleton sequence transfer event occurs or not; and determining an insertion site of the T-DNA sequence. According to the invention, the method combines with a bioinformatics analysis means; under the condition that an expression vector is known or unknown, whether a transgenosis or gene editing event occurs or not is identified; under the condition that an expression vector is known, the accurate positioning, direction, copy number and flanking sequenceinformation of a target sequence inserted into a genome can be rapidly and accurately given; whether a skeleton sequence, namely a sequence except the target sequence, is inserted into the genome or not can also be identified, and the same positioning is given.
Owner:ZHEJIANG UNIV
Method for identifying authenticity of brassica rapa variety and special SSR (simple sequence repeat) primer combination of method
ActiveCN111235302AGuarantee authenticityProtection of rights and interestsMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention belongs to the field of molecular markers and detection thereof and in particular relates to a method for identifying authenticity of a brassica rapa variety and a special SSR (simple sequence repeat) primer combination of the method. An SSR locus for identifying authenticity of a brassica rapa variety is obtained through data digging according to a reference genome Brassica_rapa V1.5 of brassica rapa and a reference genome Brassica olerecea V2.1 of brassica oleracea together with whole genome resequencing data of 40 genetic resources; the SSR primer combination is selected froma first SSR primer pair to a twenty-third SSR primer pair which are respectively applied to PCR (polymerase chain reaction) amplification on a first SSR locus to a twenty-third SSR locus, and are respectively optimally selected as nucleotide sequences which have 85-100% of homology with nucleotide sequences of SEQ ID NO:1-46 in a sequence table. The method can be applied to authenticity identification on the brassica rapa variety for a whole life cycle from seeds, and technical support is provided for protection on genetic resources and new varieties of brassica rapa.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Mutation site of XI-type osteogenesis imperfecta pathogenic gene FKBP10 and application of mutation site
ActiveCN106755395AEasy to identifyGuaranteed FeaturesMicrobiological testing/measurementDNA preparationDiagnosis methodsGenome resequencing
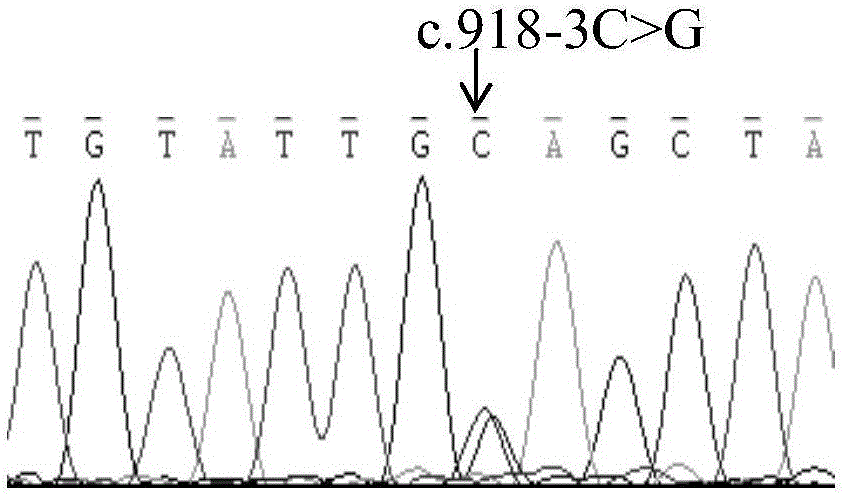
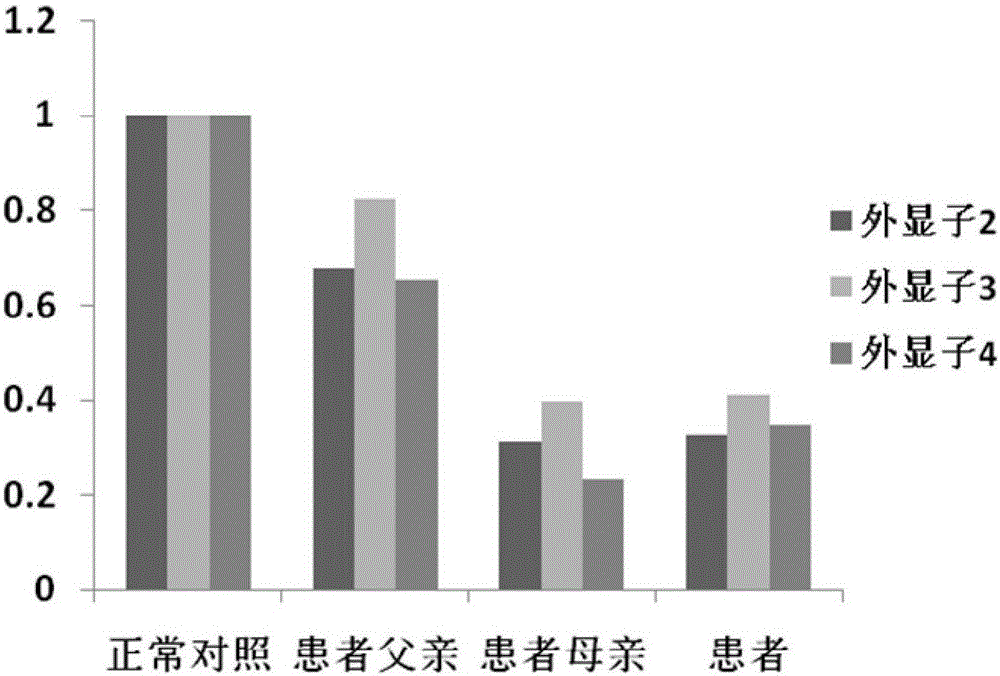
The invention relates to a mutation site of an XI-type osteogenesis imperfecta pathogenic gene FKBP10 and application of the mutation site. A whole genome resequencing technology is combined with comprehensive technologies of digital PCR, Sanger sequencing and the like; whole genome resequencing is carried out for clinic suspected osteogenesis imperfect patients; the condition that copy number variation exists between an exon 2 to an exon 4 of the FKBP10 gene and meanwhile, a heterozygous shear site mutation that c.918-3C is greater than G is discovered by combining with bioinformatics. Copy number variation and heterozygous shear mutation between the exon 2 to the exon 4 of the FKBP10 gene are further validated through digital PCR and a traditional PCB-based Sanger sequencing technology separately. Through discovery of the copy number variation and the heterozygous shear mutation, basis and reference are provided for discussing pathogenesis of XI-type osteogenesis imperfect and enriching and developing a diagnosis method for clinical diagnosis and treatment, and a basic basis is provided for early pathogenic gene screening and treatment.
Owner:SHANDONG FIRST MEDICAL UNIV & SHANDONG ACADEMY OF MEDICAL SCI
Chicken whole-genome SNP chip and use thereof
ActiveUS20210139962A1Improve accuracyEffectivenessMicrobiological testing/measurementAnimal husbandryBiotechnologyGenome resequencing
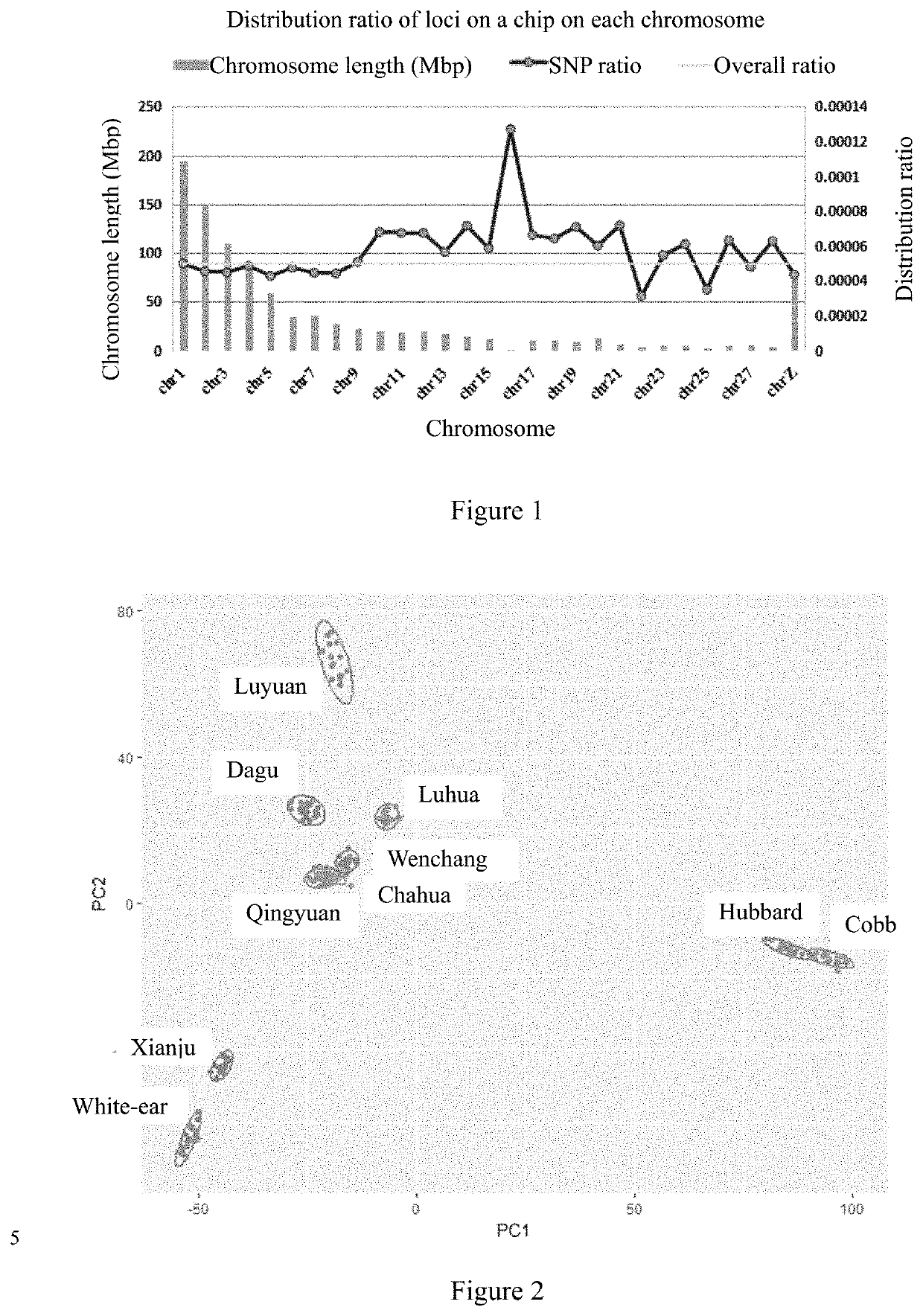
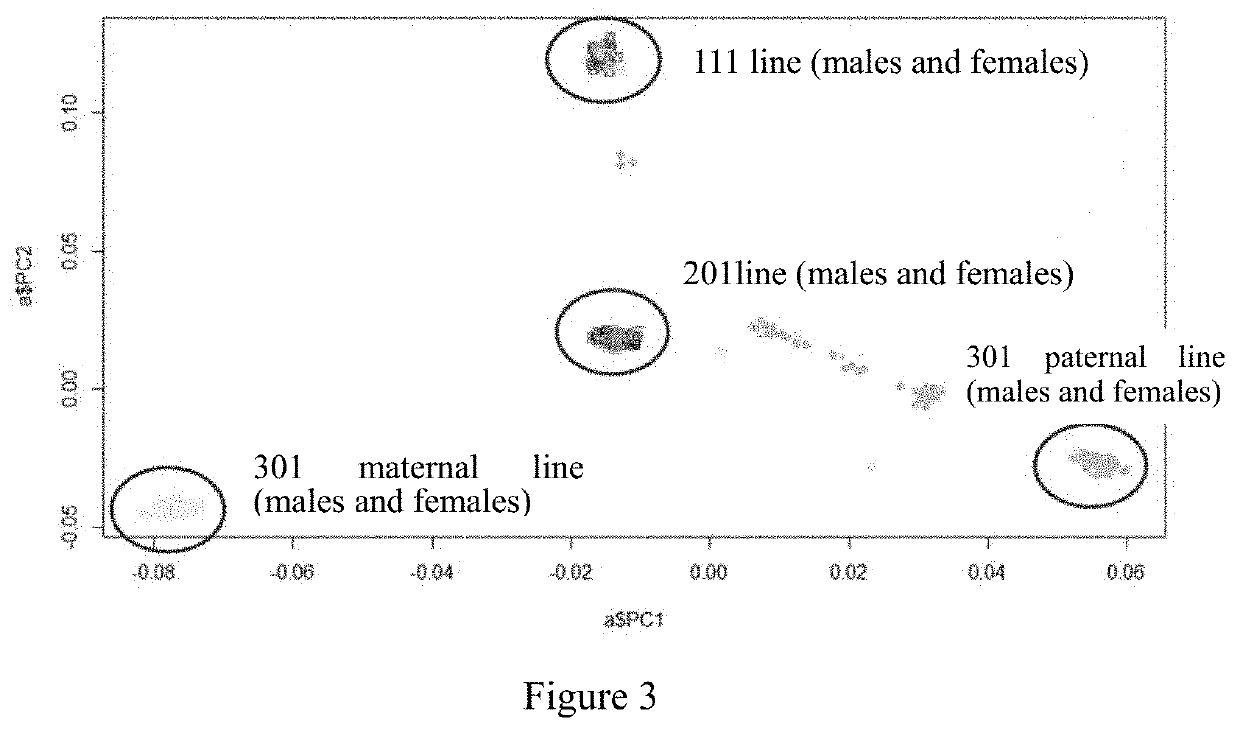
Provided in the present invention is a chicken whole-genome SNP chip and application thereof. There are a total of 50,000 SNP loci on the chip: including 19,600 SNP loci for white-feather broilers, yellow-feather and partridge chickens having a MAF value greater than 0.05 and uniformly distributed across the genome which were derived from the data of the whole-genome resequencing of main indigenous chicken breeds in China and introduced chicken breeds; 14,000 SNP loci associated with economic traits, and 16,400 SNP loci for making up for the genomic regions that are not covered by the first two types of probes. The 50,000 SNP loci on the chicken whole-genome SNP chip of the present invention have DNA sequences represented by SEQ ID NOs. 1 to 50,000. The SNP loci on the chip are uniformly distributed across the whole genome, and associated with traits such as feed efficiency, meat production rate, lipid metabolism, meat quality, general resistance to diseases, reproduction and the like, and the chip has moderate through-put and low cost, and could be used universally for chicken breeds at indigenous and abroad.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
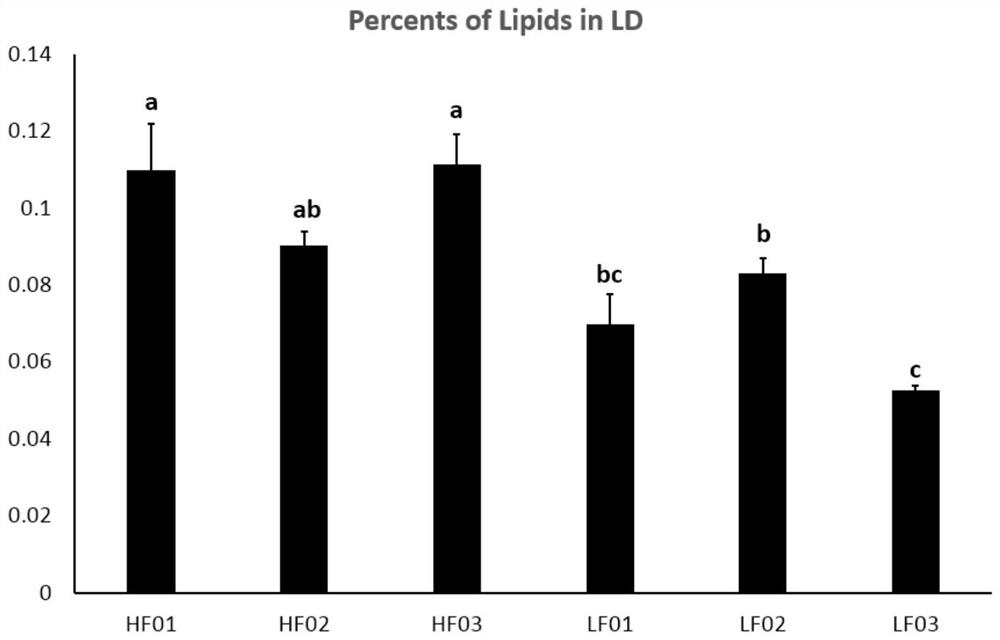
Multi-omics-based mining method for key gene of fat deposition trait of Nanyang black pigs
The invention discloses a multi-omics-based mining method for a key gene of a fat deposition trait of Nanyang black pigs. According to the method, in order to solve the problem of the existing methodsthat the accuracy of mining and identifying of swine fat deposition trait genes is low, transcriptomes of musculi longissimus dorsi of six pairs of full-sib or half-sib Nanyang black pig individualsof 6 months old with extreme back fat thickness and intramuscular fat content are sequenced by using a high-flux sequencing technology, important fat deposition candidate genes are sought and identified on the basis of phenotype determination and under the assistance of bioinformatics analysis, meanwhile, two groups of Nanyang black pigs are subjected to genome resequencing to find out important fat deposition candidate difference genes, between-group remarkable-difference-expression genes are screened through integrating transcriptome data, re-sequenced data and QTL database information and using bioinformatics and functional genomics methods, and gene functions and signal channels of the between-group remarkable-difference-expression genes are predicted.
Owner:NANYANG NORMAL UNIV
Method for identifying authenticity of pepper varieties and special SSR primer combination thereof
ActiveCN111270004AGuarantee authenticityImprove representationMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetics
The invention belongs to the field of molecular markers and detecting thereof, and particularly relates to a method for identifying the authenticity of pepper varieties and a special SSR primer combination thereof. SSR loci for identifying the authenticity of the pepper variety are obtained by performing data mining according to a reference genome Capsicum.annum.L of pepper and 40 parts of germplasm resource whole genome re-sequencing data; the SSR primer combination is selected from the group consisting of the first SSR primer pair to the twenty-second SSR primer pair which are used for PCR amplification of the first SSR site to the twenty-second SSR site respectively, and preferably, the first SSR primer pair to the twenty-second SSR primer pair are nucleotide sequences with the homologyof 85%-100% with the nucleotide sequences shown as SEQ ID NO: 1-44 in the sequence table respectively. The method can be used for identifying the authenticity of the pepper variety in the full life cycle from seeds, and technical support is provided for pepper germplasm resource and new variety protection.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
Method for identifying quantitative trait loci (QTL) and genes related to tender skin colors of cucumbers
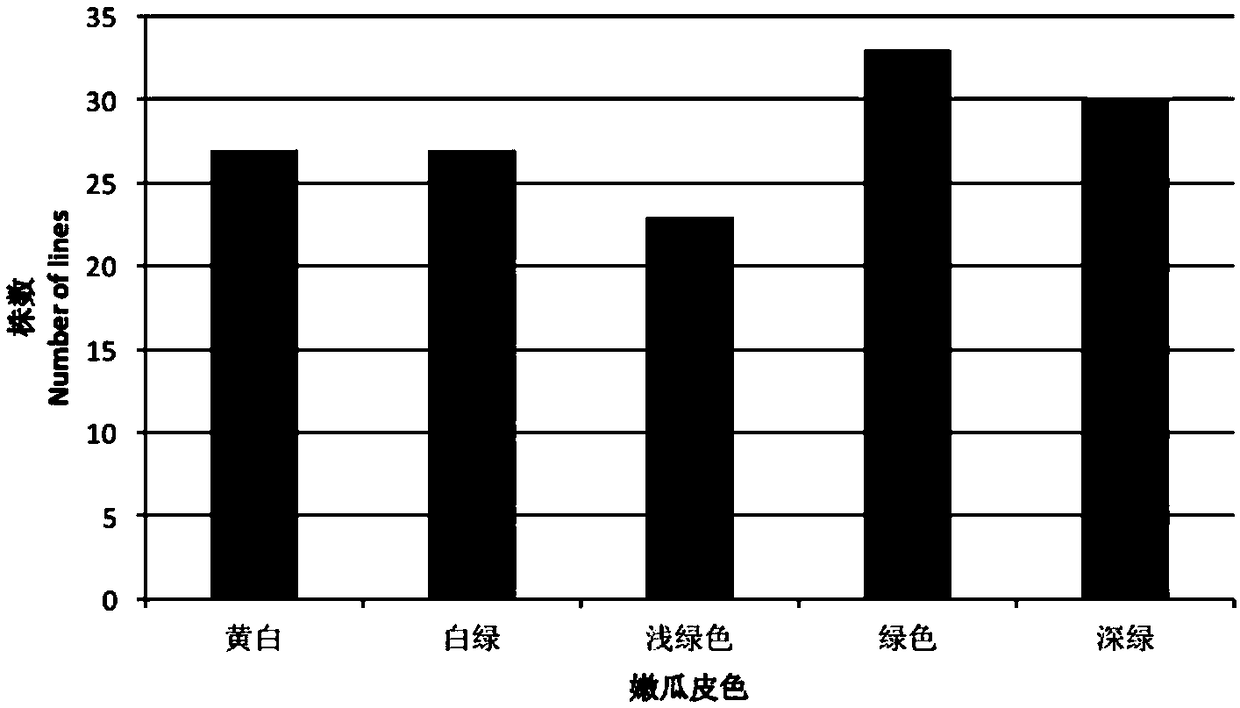
ActiveCN109321673AIdentify the genetic mechanismMicrobiological testing/measurementDNA/RNA fragmentationStatistical analysisGenotype Analysis
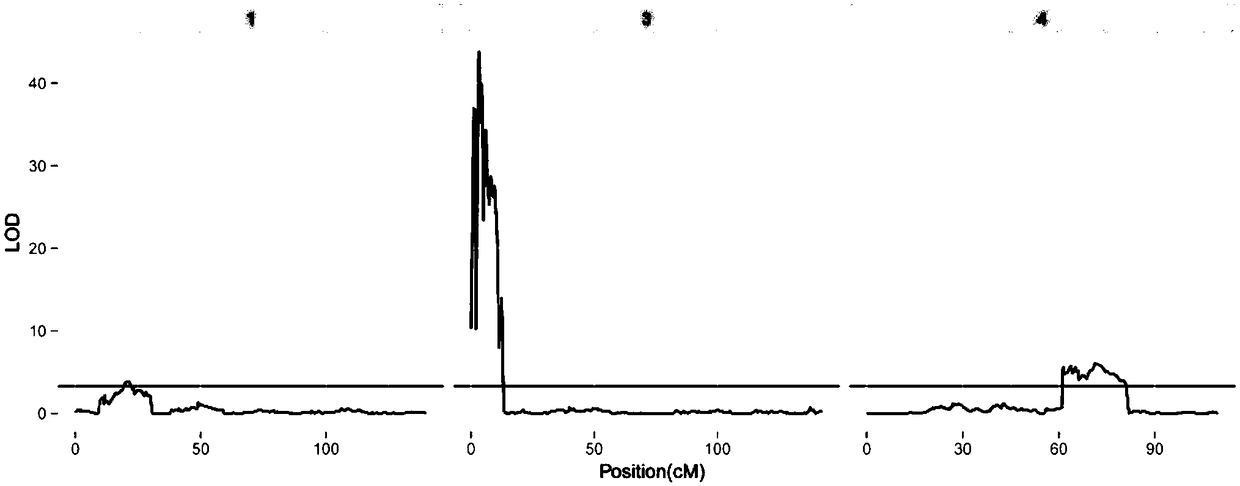
The invention discloses a method for identifying quantitative trait loci (QTL) and genes related to the tender skin colors of cucumbers. The method comprises the steps of (1) carrying out hybridization on a female parent and a male parent to obtain F1 for constructing recombinant inbred lines (RILs); (2) identifying the tender skin colors of all plants and carrying out statistical analysis of data; (3) extracting genome DNAs of all the RIL plants and the parents thereof; (4) carrying out genome-wide resequencing on the genome DNAs of the parents and the RIL plants, and carrying out single nucleotide polymorphism (SNP) mutation detection and genotyping analysis; (5) determining the LOD threshold of all phenotypes and QTL segments corresponding to same; (6) comparing the SNP detection results in the QTL segments with a cucumber reference genome, searching for related genes, and performing gene function annotation. The method provided by the invention has the advantages that the method adopts a high-throughput sequencing method to perform genotype identification on self-created high generation RILs to obtain a molecular marker number and a high-density linkage map which can notcannotbe reached by the traditional method, so that a support is provided for efficient and accurate identification of the genes related to the tender skin colors of the cucumbers.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Variable length probe selection
InactiveUS7636637B2Improve accuracyImprove efficiencyBioreactor/fermenter combinationsBiological substance pretreatmentsGenome resequencingVariable length
The present invention provides novel method for increasing the efficiency and accuracy of high-throughput mutation mapping and genome resequencing by using a variable length probe selection algorithm to rationally select probes used in designing oligonucleotide arrays synthesized by Maskless Array Synthesis (MAS) technology. Also disclosed is a variable length probe selection algorithm used in designing such oligonucleotide arrays.
Owner:ROCHE NIMBLEGEN
Method utilizing liquid phase capture to carry out gene typing of large yellow croaker genome
InactiveCN106191246AStrong specificityImprove capture efficiencyMicrobiological testing/measurementRe sequencingTyping
The invention discloses a method utilizing liquid phase capture to carry out gene typing of large yellow croaker genome. The method comprises the following steps: subjecting a large yellow croaker to whole genome re-sequencing, at the same time, carrying out large yellow croaker whole genome SNP / InDel mining to provide high quality SNP labeled sites; screening and utilizing SNP labels obtained by large yellow croaker whole genome re-sequencing to design probes; utilizing the large yellow croaker whole genome to establish a large yellow croaker genome library; utilizing a liquid phase to capture the large yellow croaker genome sections; and subjecting the captured large yellow croaker genome sections to high throughput sequencing and SNP / InDel gene typing. The provided method effectively solves the problem that a whole gene simplified genome typing technology may cause unevenness of label density and most labels are originated from non-coding regions. According to the method, four probes are designed to effectively improve the specificity of hybrid of sequences where each SNP stays, the efficiency of section capture is improved; and the method has an important application in the large scale typing of large yellow croakers with special SNP labels.
Owner:JIMEI UNIV
Identifying and breeding method of SNP marker for increasing cotton fiber yield and high-yield cotton
ActiveCN109055593AShorten the cultivation periodImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationFiberRe sequencing
The invention relates to the field of cotton SNP markers and specifically relates to an identifying and breeding method of SNP marker for increasing cotton fiber yield and high-yield cotton. In the invention, an SNP marker remarkably improving the fiber yield character is identified by analyzing the genome re-sequencing data of an excellent parent with obviously improved fiber yield character andthe pedigree material of the excellent parent, a high-fiber yield material can be selected by selecting beneficial allelic variation (GG genotype) of the marker site, thus the selection efficiency andaccuracy are noticeably enhanced, and the cultivation years of the high-fiber yield material are obviously shortened.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
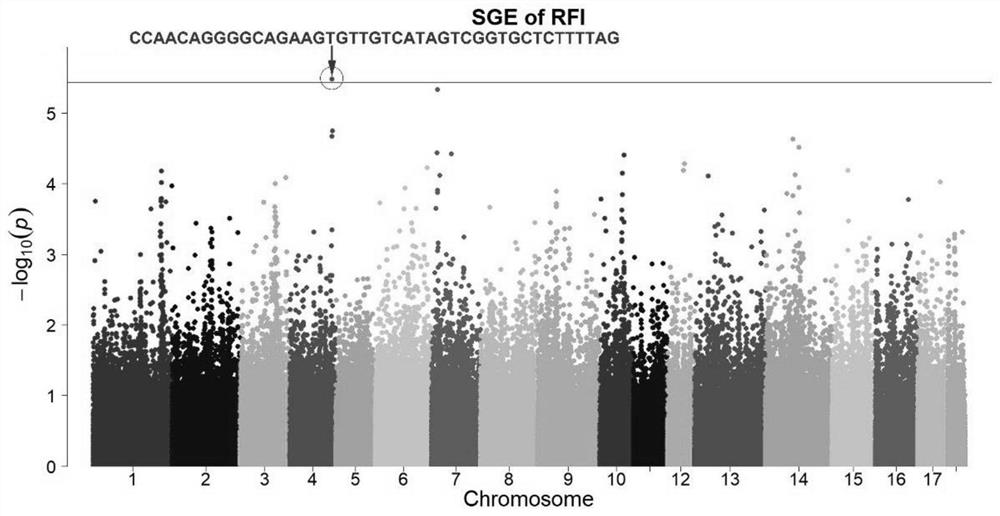
Method for positioning social genetic haplotype site of pig residual feed intake
PendingCN112029867AAccurate identificationImprove reliabilityMicrobiological testing/measurementProteomicsBiotechnologyMedicine
The invention discloses a method for positioning a social genetic haplotype site of pig residual feed intake, which is used for identifying a key haplotype of a social genetic effect of the pig residual feed intake by utilizing chip data, whole genome resequencing data, genotype filling, whole genome haplotype construction and a haplotype-based GWAS means. The method has the advantages of accurateidentification and high reliability.
Owner:SICHUAN AGRI UNIV
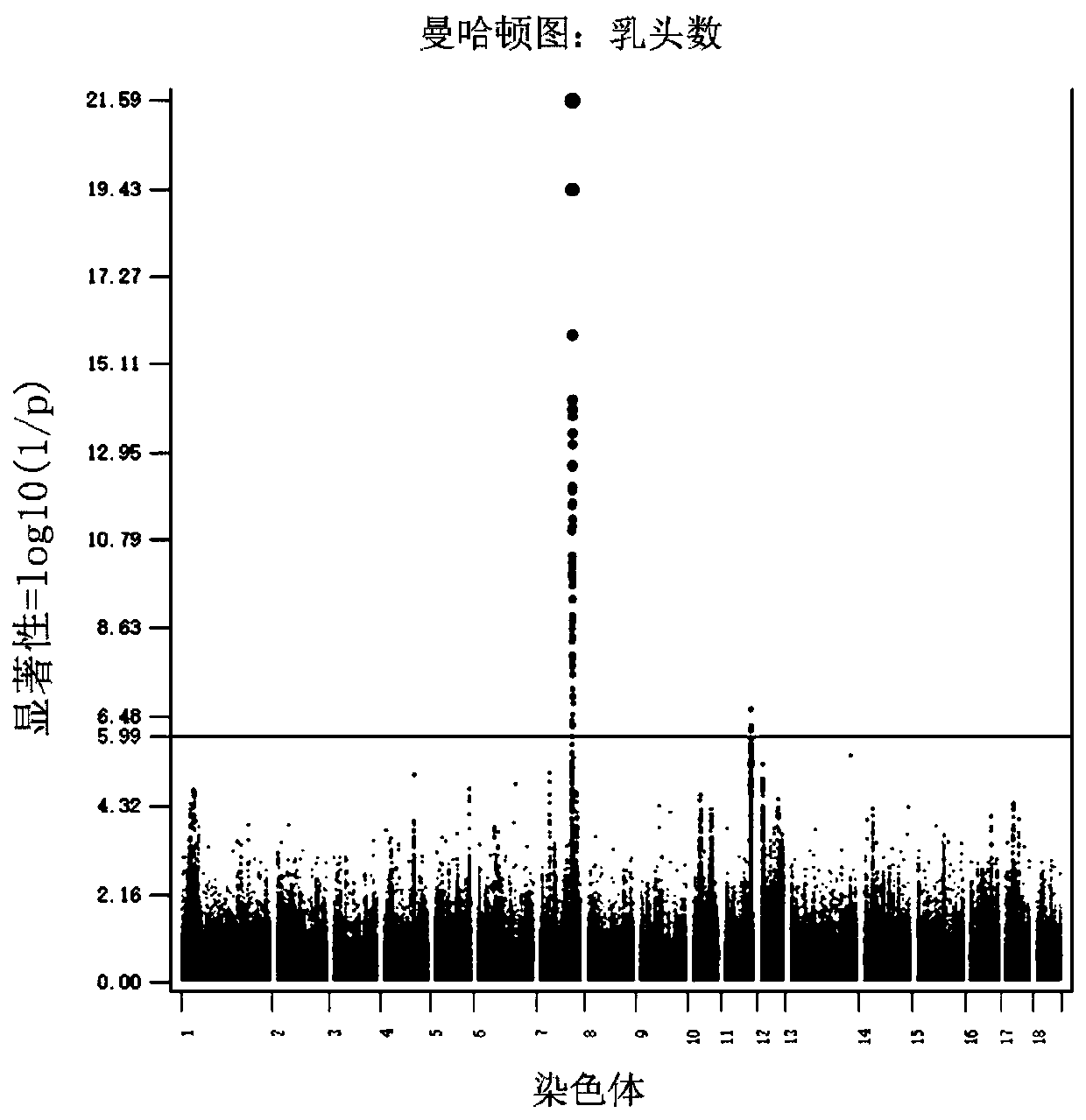
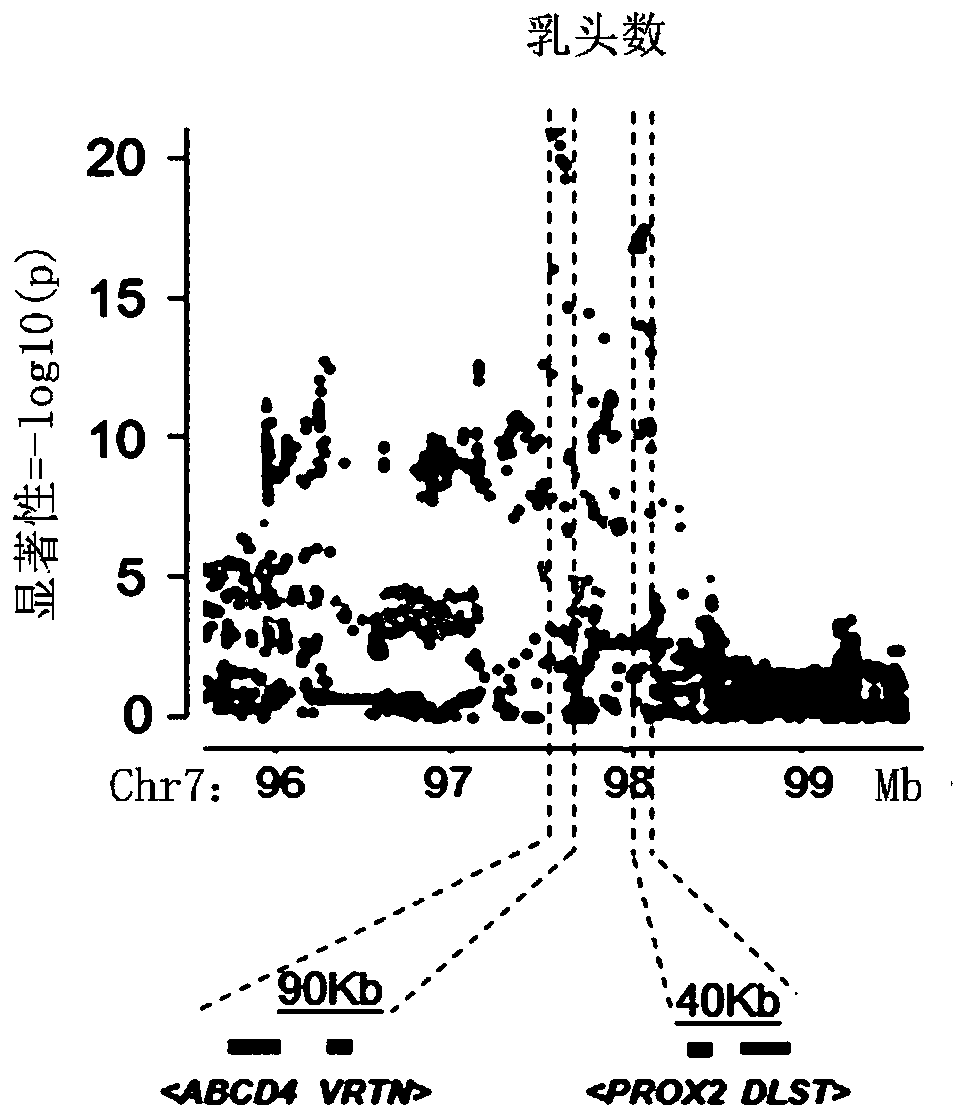
SNP sites related to number of pig nipples and detection method and application thereof
ActiveCN110714082AQuality improvementEarly predictionMicrobiological testing/measurementProteomicsBiotechnologyTn5 transposase
The invention relates to SNP sites related to the number of pig nipples and a detection method and application thereof. The SNP sites are located at a chr7:97570528 of a genomic version Ensembl Sscrofa 11.1, and alleles of the sites are A and G. According to the SNP sites related to the number of the pig nipples and the detection method and application thereof, Tn5 transposase is used for establishing a library, low-depth sequencing is carried out on a core population of 3,616 Duroc pigs with important economic performance records, and high-quality whole-genome resequencing data are obtained through padding. Through GWAS analysis of the characteristics of the number of the pig nipples, seven SNP sites which significantly affect the number of the pig nipples are detected, and a newly discovered SNP site SSC7: 97570528 with the most significant effect is included. The SNP sites can be used as molecular markers for the breeding of dominant populations of the number of the pig nipples.
Owner:CHINA AGRI UNIV
Lotus root InDel molecular marker and development method and application thereof
ActiveCN106544446AIncrease success rateFlexible detection of InDel markersMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityMethod development
The invention discloses a set of lotus root InDel molecular markers and a development method and application thereof. According to the development method, genome resequencing data of a lotus variety, namely Qitou, and a seed lotus variety, namely space No.36, are compared with a Chinese ancient lotus seed reference genome; InDel loci different from one another among the lotus variety, namely Qitou, the seed lotus variety, namely space No.36, and the Chinese ancient lotus seed reference genome are screened; PCR primers are designed to conduct detection; and a set of 46 pairs of primers is selected according to the positions of the primers on lotus root chromosomes. The development method can be applied to the studies such as construction of a genetic map, QTL location, molecular assisted breeding and genetic diversity analysis of a lotus root. The InDel polymorphic markers developed by the development method have the advantages of being high in success rate, flexible in detection, simple in operation, low in cost and the like.
Owner:SHANDONG RICE RES INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com