Method for determining insertion site of transgenic line by re-sequencing technique
A technology of insertion sites and transgenes, applied in the field of plant genetic engineering, can solve the problems of cumbersome steps, high operation requirements, and high technical requirements, and achieve the effect of simple operation process, low method threshold and wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
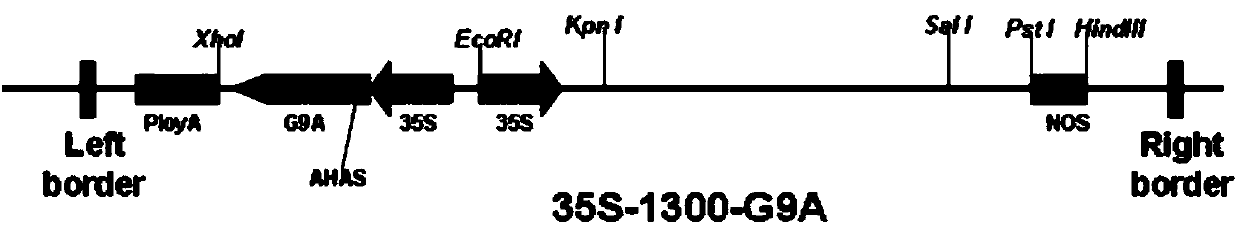
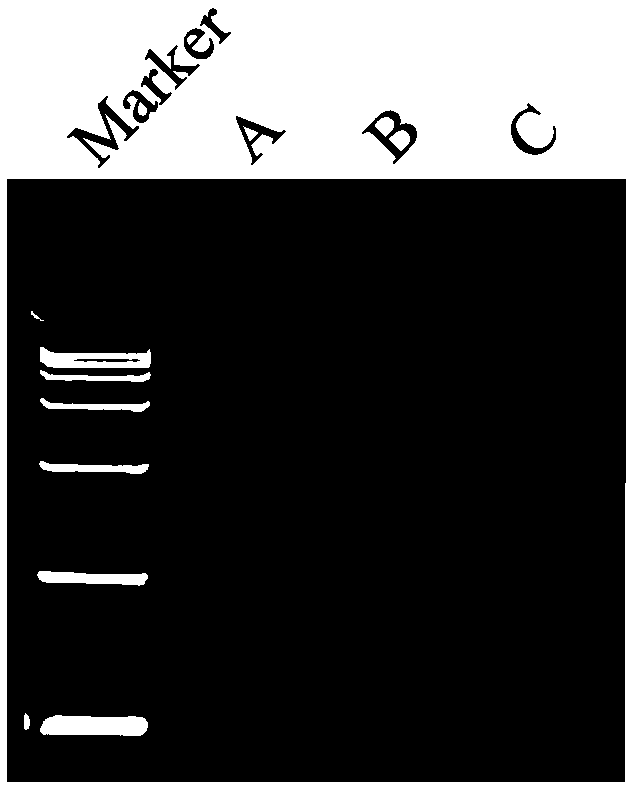
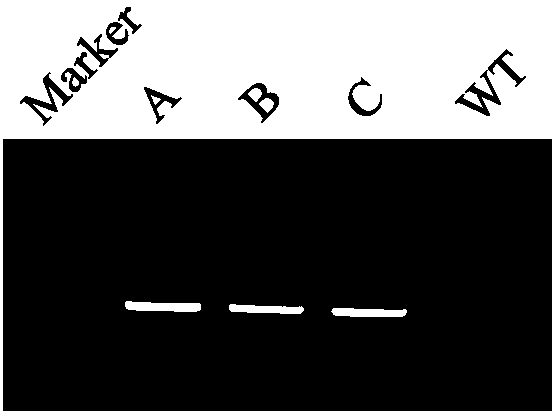
[0045]1) Randomly take 3 transgenic strains A, B, and C (all are transgenic strains obtained by transgenic Agrobacterium using the transgenic carrier 35S-1300-G9A), after 30 days of normal culture, each take 100 mg of leaves, and put them under liquid nitrogen After grinding, add 1000 μL 80°C 1.5×CTAB (15g CTAB; 75ml 1M Tris-HCl, pH 8.0; 30ml 0.5M EDTA, pH 8.0; 61.4gNaCl; constant volume 1L, stirring and dissolving with a stir bar for 2h); 65°C water bath for 30min; Add an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1 volume ratio) and slowly invert it up and down for about 15 minutes until the lower liquid phase turns dark green; centrifuge at 3000r / min for 15 minutes at room temperature; take the supernatant in a new 10ml Add an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1 volume ratio) to the centrifuge tube and slowly invert it up and down for about 15 minutes; centrifuge at 3000r / min for 15 minutes at room temperature; take the supernatant and ad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com