Building method of long fragment nucleic acid library
A construction method and nucleic acid library technology, which is applied in the field of long-fragment nucleic acid library construction, can solve problems such as difficulty in assembly, and achieve the effects of improving assembly accuracy and shortening assembly time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1 Long fragment nucleic acid library construction method
[0071] (1) Preparation of large fragment library
[0072] ① Genomic DNA Fragmentation
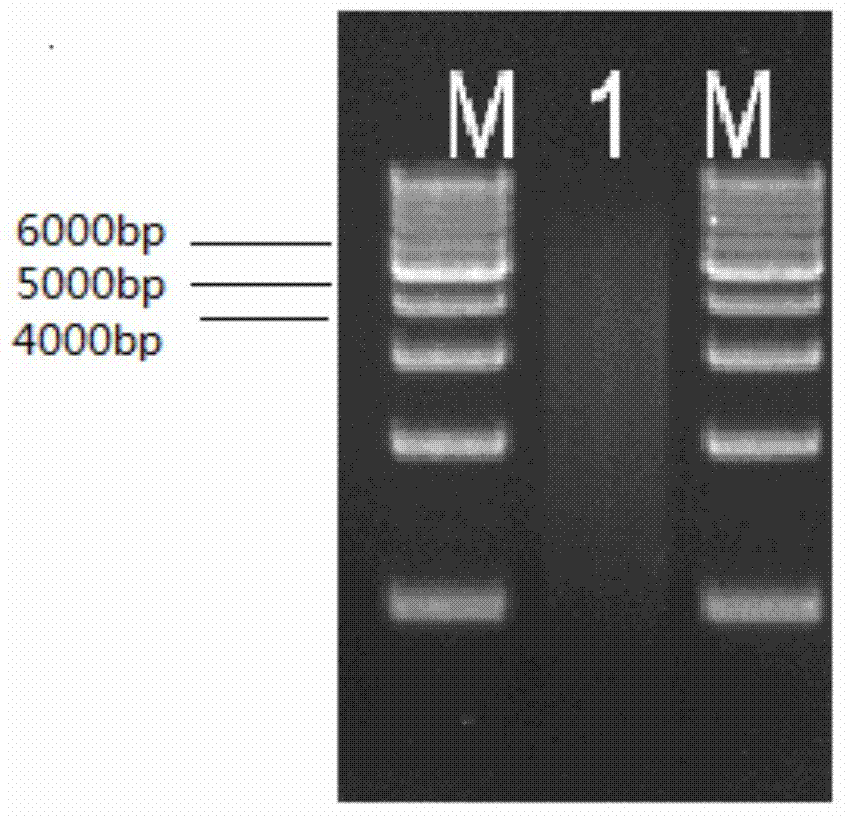
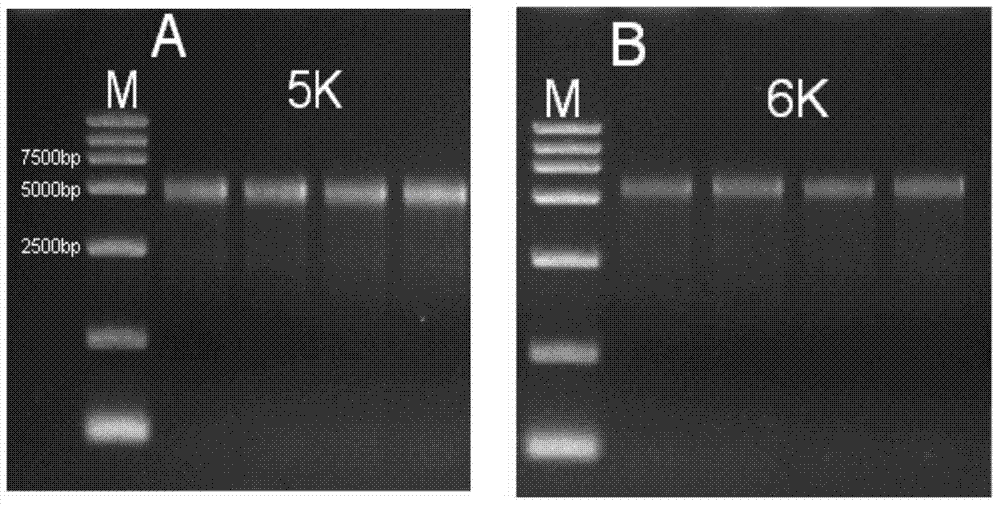
[0073] 20μg (Nanodrop quantitative) genomic DNA samples of rice Nipponbare were crushed by nebulization to obtain diffuse bands with concentrated bands of 5K and 6K, and the 5K and 6K fragments were recovered respectively. For electrophoresis detection, see figure 1 ;
[0074] ②End repair, adding A, adding joints
[0075] The purified large fragments were end-repaired with T4 DNA polymerase, T4PNK and Klenow enzyme and purified with a PCR product purification kit. After end repair and purification, the purified sample was treated and purified with A, and T4 DNA ligase was used to add a linker. The linker was MACCA (the NNN fixed position CCA in MANNN, to verify the linker connection efficiency), synthesized by Invitrogen, and the sequence was:
[0076] F: '-ACTTCCATCCCCATCCCCCATCCCCCATCCCGCTCTTCCGATCT;
[0077...
Embodiment 2
[0121] Example 2 Long Fragment Nucleic Acid Library Construction Method and Sequence Assembly
[0122] (1) Preparation of large fragment library
[0123] ① Genomic DNA Fragmentation
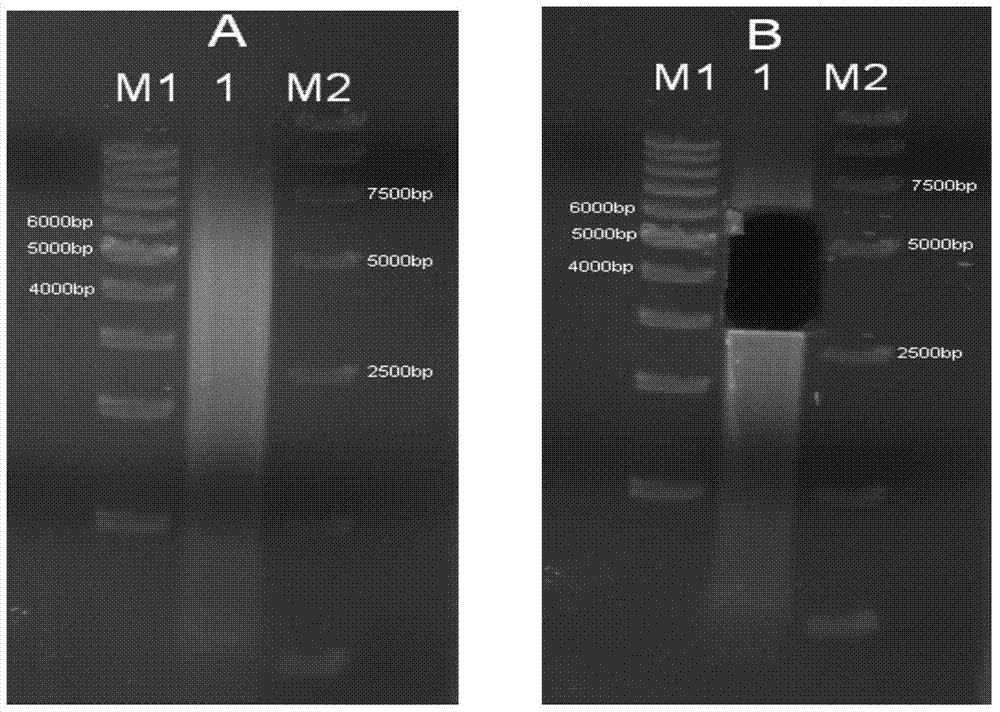
[0124] Take 20μg (Nanodrop quantification) of rice Nipponbare genomic DNA sample, sample number is C27, use the atomization method to break up the genomic DNA, obtain a diffuse band with a concentrated band of 10K, and recover the 10K fragments respectively. For the electrophoresis detection, see Figure 6 ;
[0125] ②End repair, adding A, adding joints
[0126] The purified large fragments were end-repaired with T4 DNA polymerase, T4PNK and Klenow enzyme and purified with a PCR product purification kit. After end repair and purification, the purified sample is treated and purified with A, and T4 DNA ligase is used to add a linker. The linker is MANNN, synthesized by Invitrogen, and the sequence is:
[0127] F:'-ACTTNNNTCCCNNNTCCNNNTCCNNNTCCCCGCTCTTCCGATC T;
[0128] R: 5'-GATCGGAAGAGCACACGT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com