Method for detecting new resistance genes in environmental sample based on Nanopore metagenome sequencing
A detection method and resistance gene technology, applied in the field of detection of new resistance genes in environmental samples based on Nanopore metagenomic sequencing, can solve problems such as the spread of drug-resistant bacteria and drug-resistant genes, health threats, etc., and achieve high comparison accuracy , Assembly accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1. DNA extraction and purification
[0033] The DNA extracted using the kit DNeasy PowerSoil Kit (QIAGEN, USA) has high DNA length and purity (the average length of the DNA fragment is about 15kb, A 260 / 280≈1.8, A 260 / 230≈2.0), and the DNA quality can be Meet the requirements of the Nanopore third-generation sequencing library construction.
[0034] Purification of DNA by 1% agarose gel electrophoresis and gel cutting recovery can increase the average length of DNA samples (that is, remove short fragments and some impurities), thereby improving the quality of library construction.
[0035] The Monarch DNA Gel Extraction Kit (NEB, USA) was used for gel recovery, and the concentration of purified DNA was determined using NanoDrop (ND-One, Thermo Fisher Scientific, USA). For the gel recovery method, refer to the kit instructions.
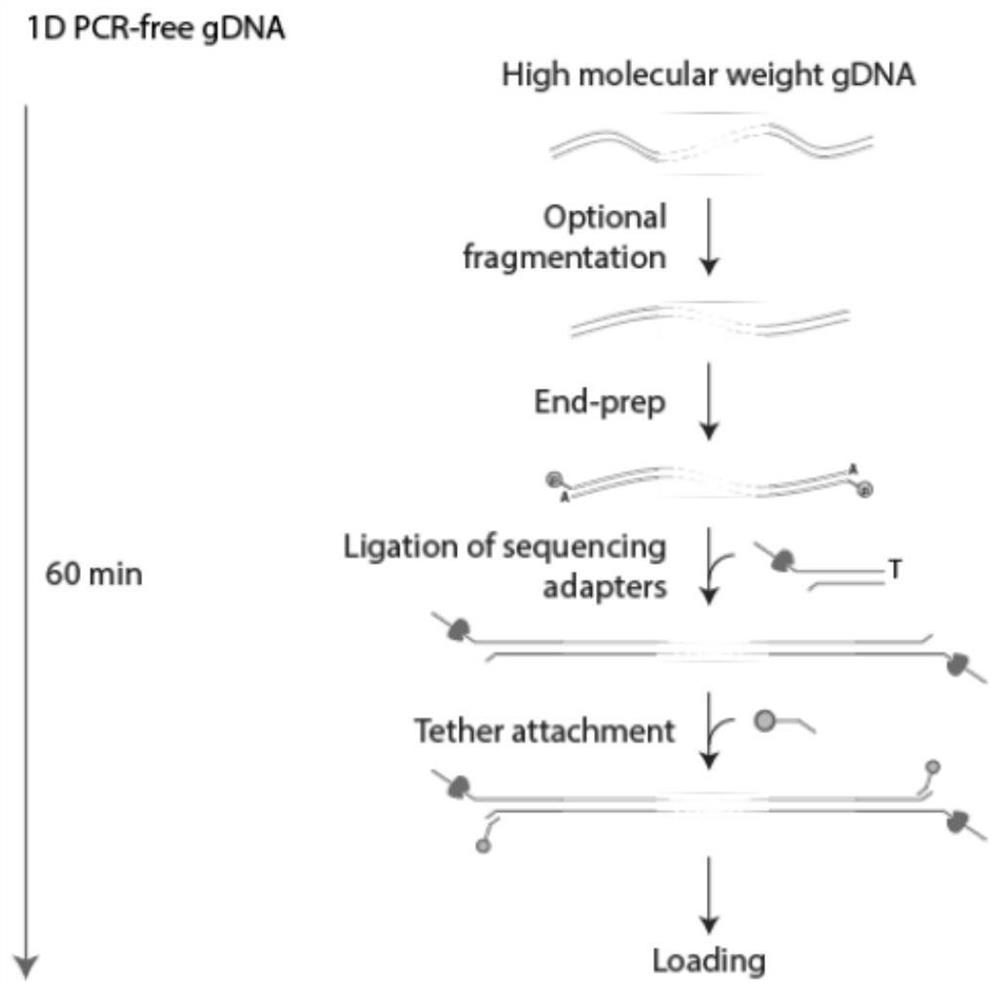
[0036] 2. Oxford Nanopore Technologies (ONT) third-generation metagenomic sequencing
[0037] Use the Ligation Sequencing Kit 1D (SQK-LSK108)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com