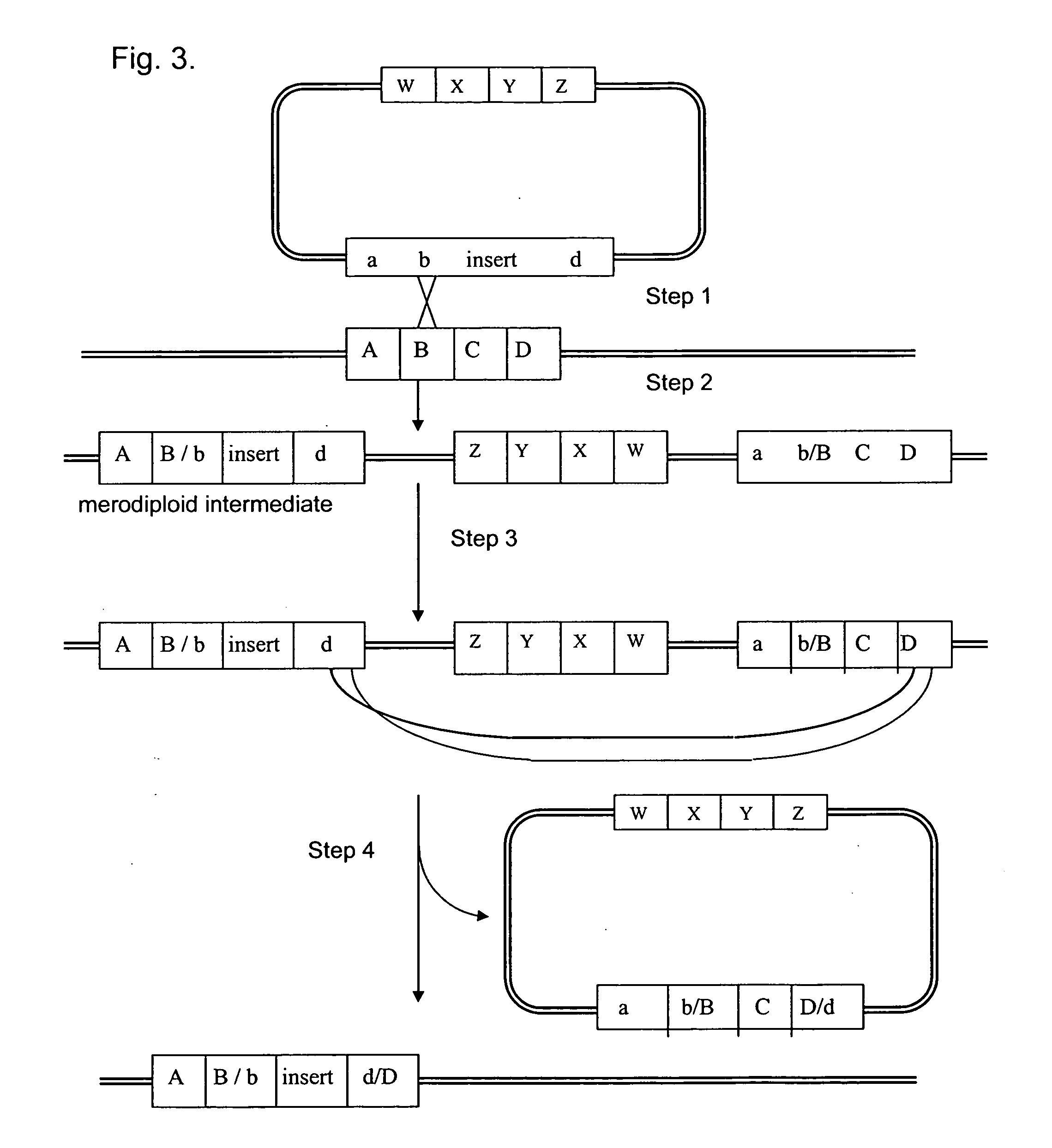
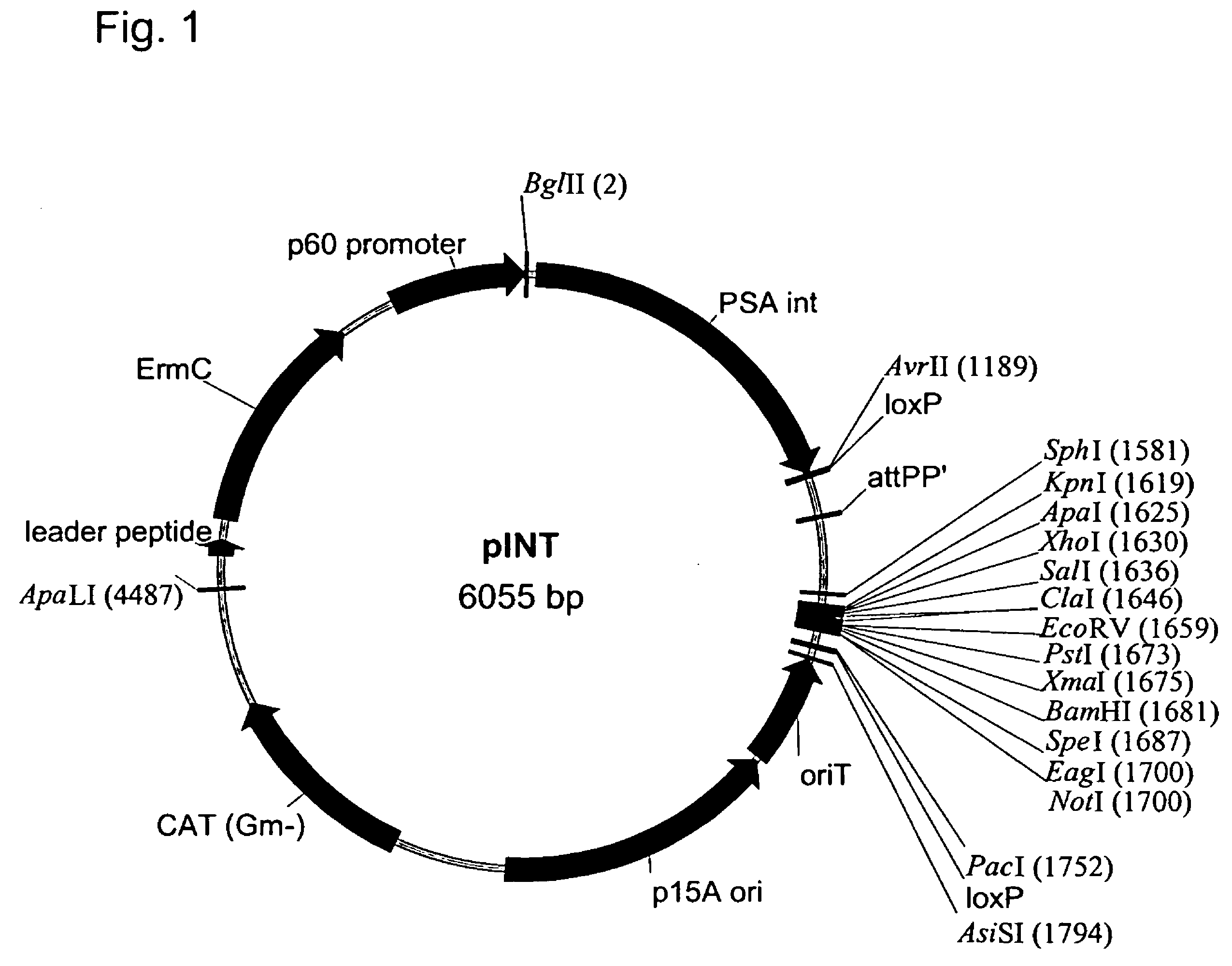
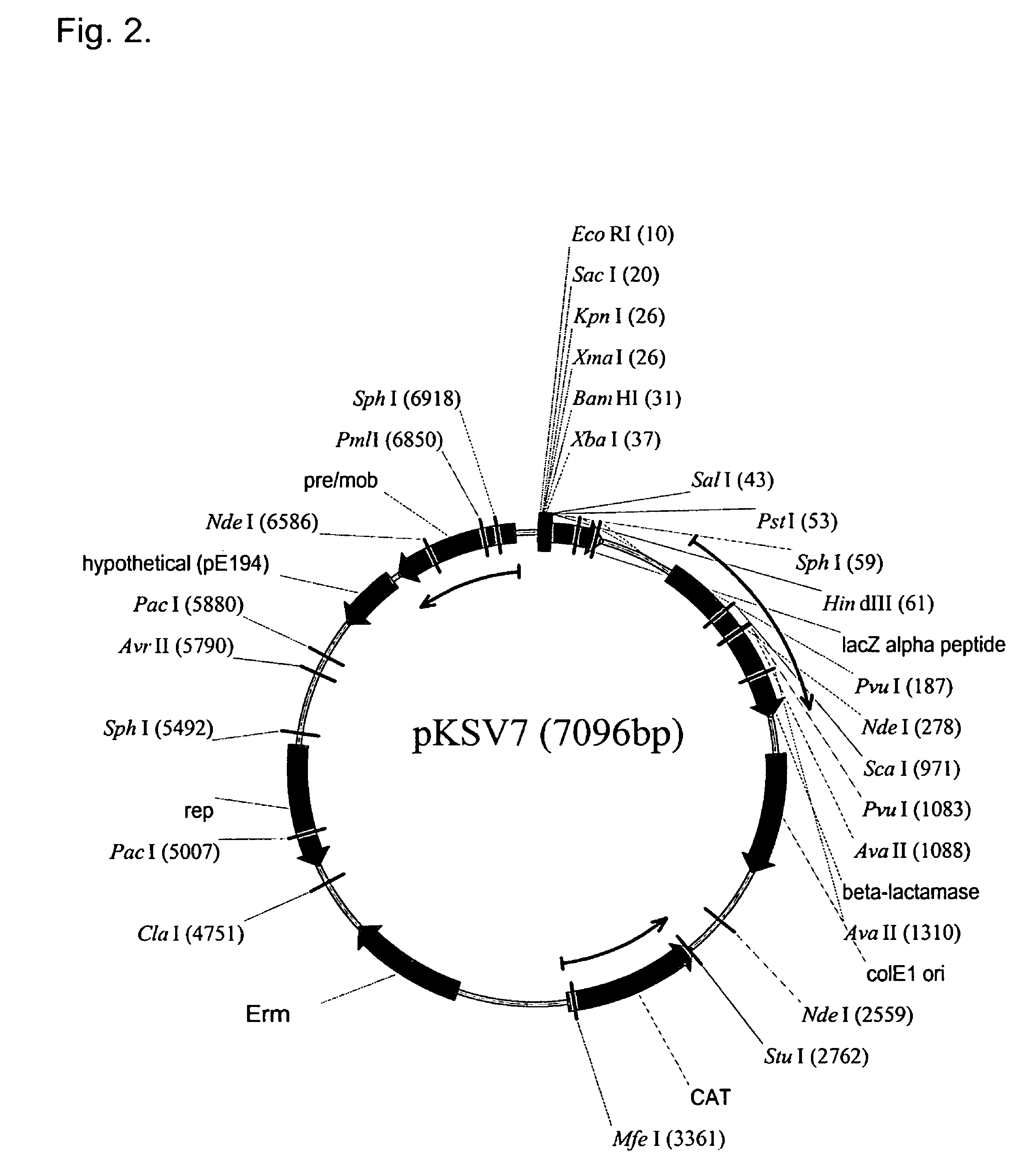
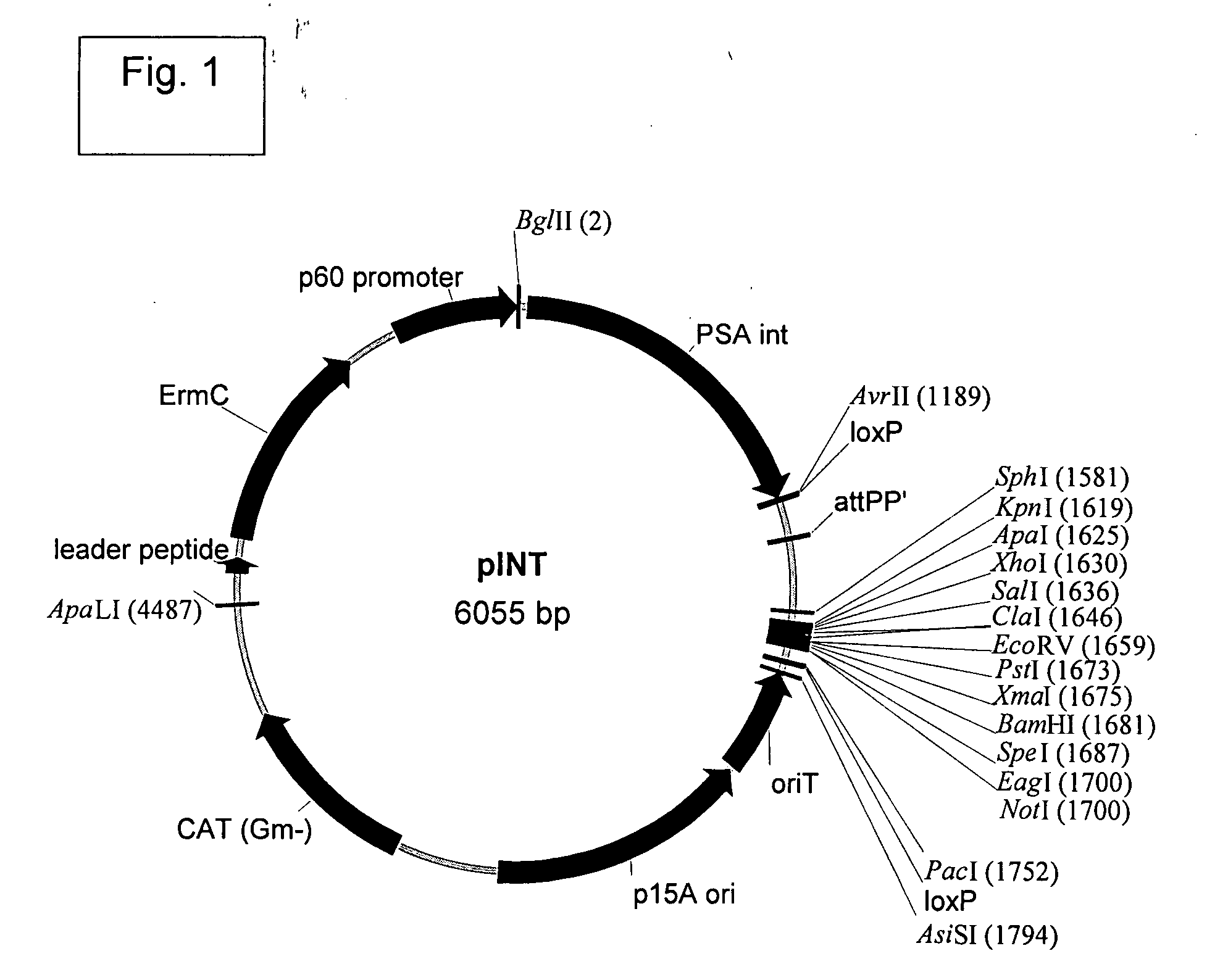
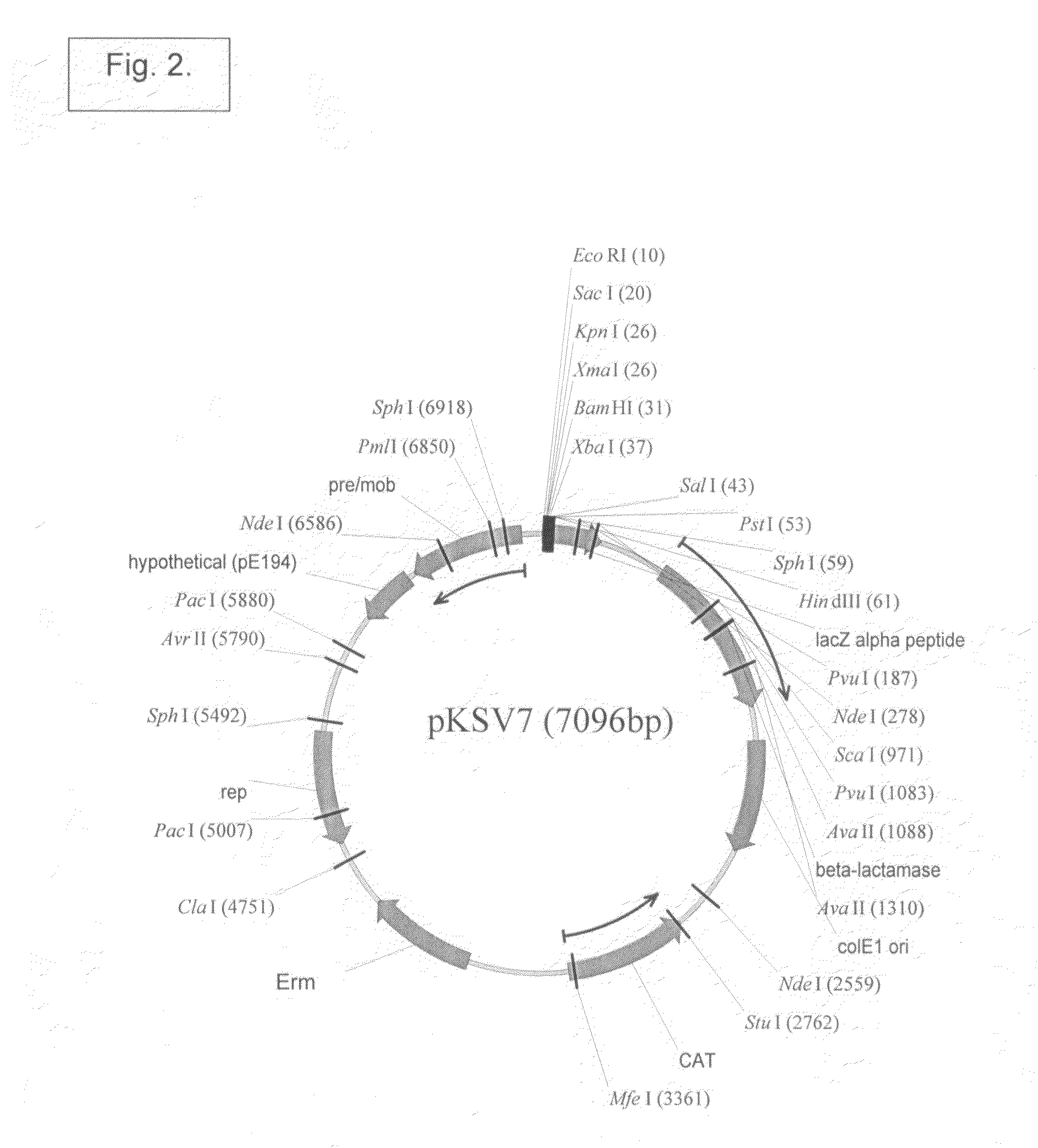
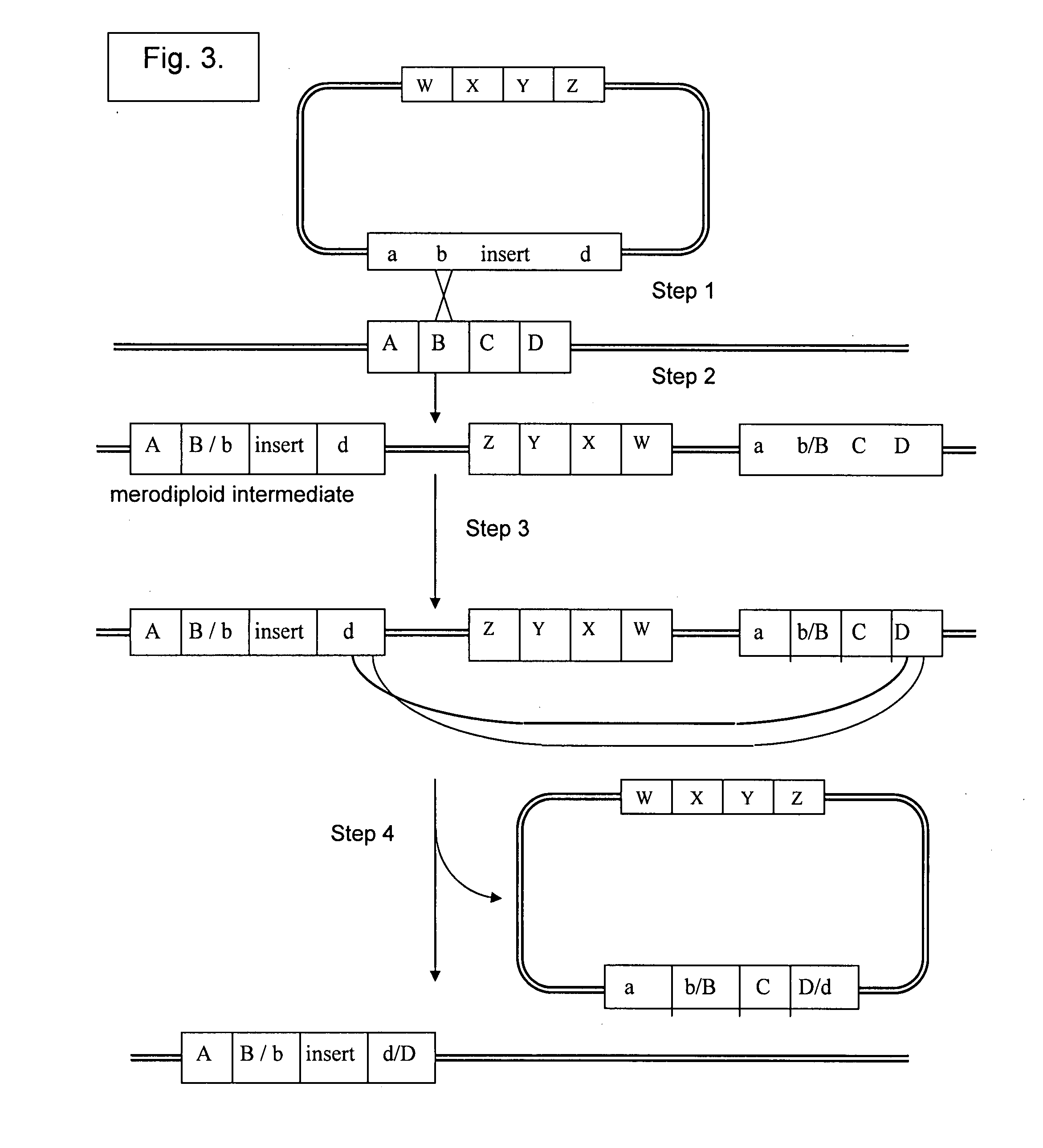
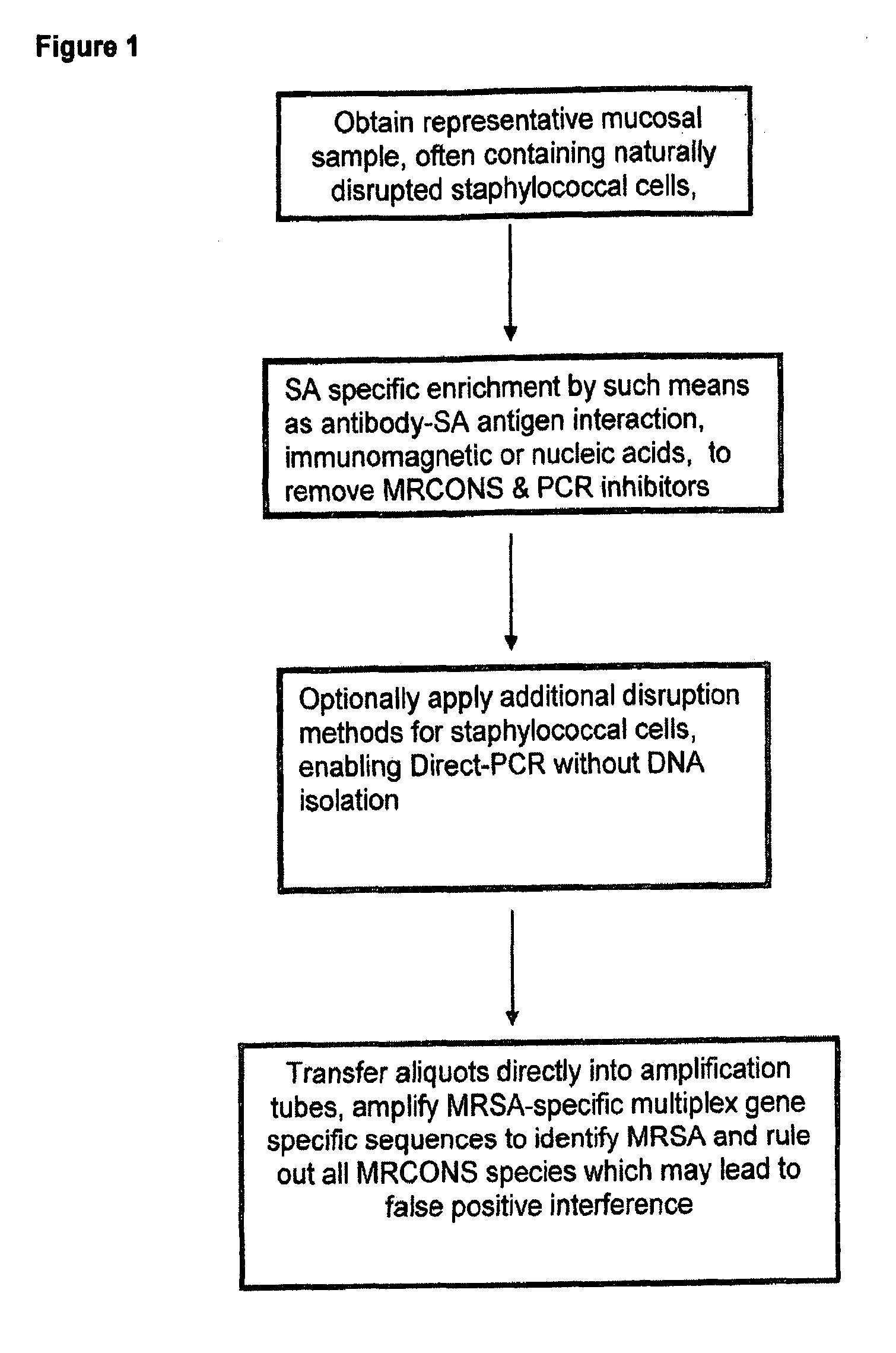
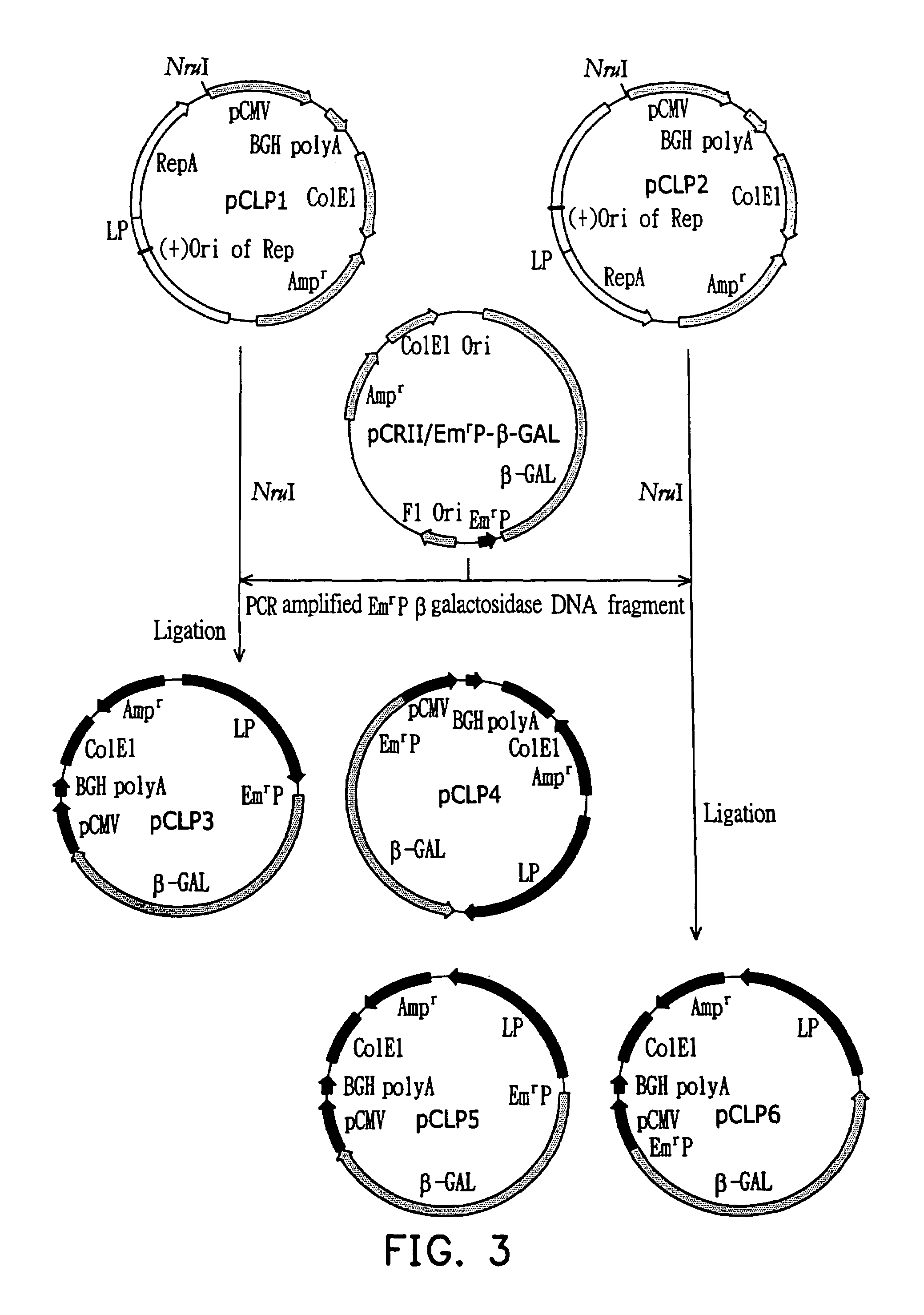
Patents
Literature
284 results about "Antibiotic resistance genes" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
One antibiotic resistance gene we found is called “blaNDM-1”. This gene confers resistance in bacteria to carbapenem antibiotics, one of our last resort remedies to infectious disease.
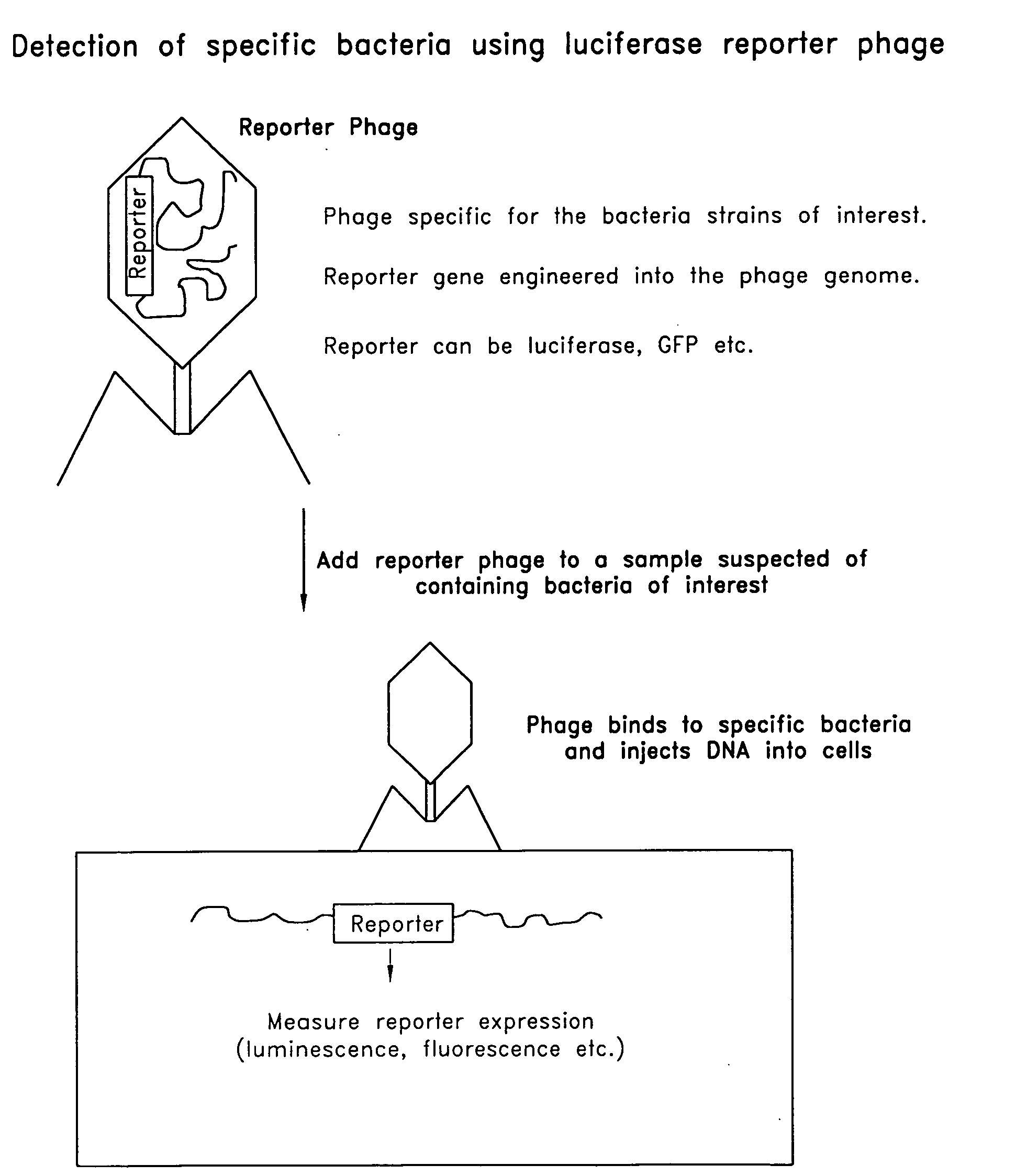
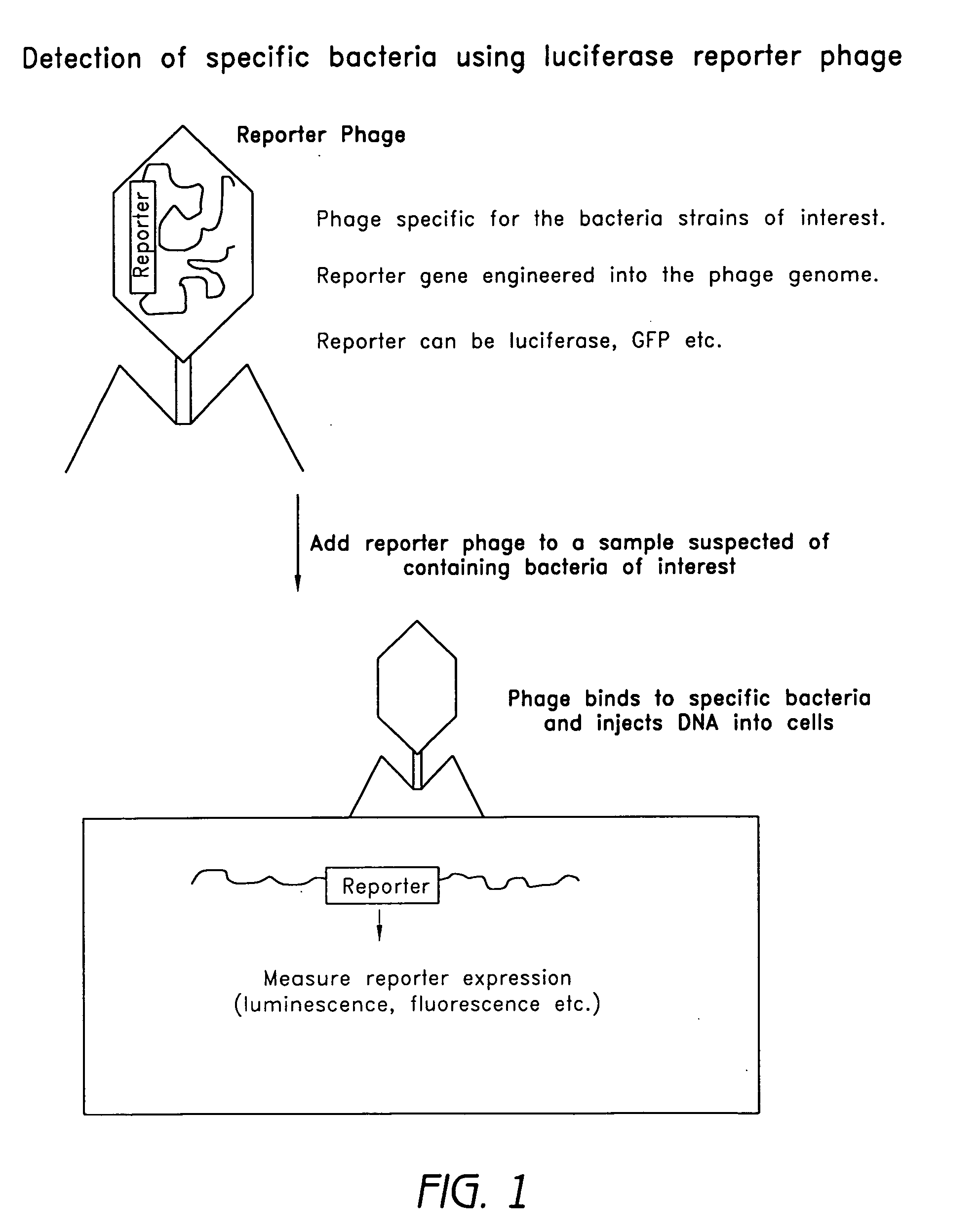
Reporter plasmid phage packaging system for detection of bacteria
InactiveUS20090155768A1Microbiological testing/measurementOther foreign material introduction processesBacteroidesOrigin of replication
The invention is related to a transducing particle that comprises a bacteriophage coat and a DNA core that comprises plasmid DNA comprising: a) a host-specific bacteriophage packaging site wherein the packaging site is substantially in isolation from sequences naturally occurring adjacent thereto in the bacteriophage genome, b) a reporter gene, c) a bacteria-specific promoter operably linked to said reporter gene, d) a bacteria-specific origin of replication, and optionally e) an antibiotic resistance gene. The invention includes phage transducing particles, methods of making transducing particles, and methods of using the transducing particles in bacterial detection.
Owner:UNITED STATES OF AMERICA
Engineered Listeria and methods of use thereof
InactiveUS20070207170A1Improve survivalIncreasing expression and secretionBiocideBacteriaHeterologous AntigensSite-specific recombination
The invention provides a bacterium containing a polynucleotide comprising a nucleic acid encoding a heterologous antigen, as well as fusion protein partners. Also provided are vectors for mediating site-specific recombination and vectors comprising removable antibiotic resistance genes.
Owner:ANZA THERAPEUTICS INC +1
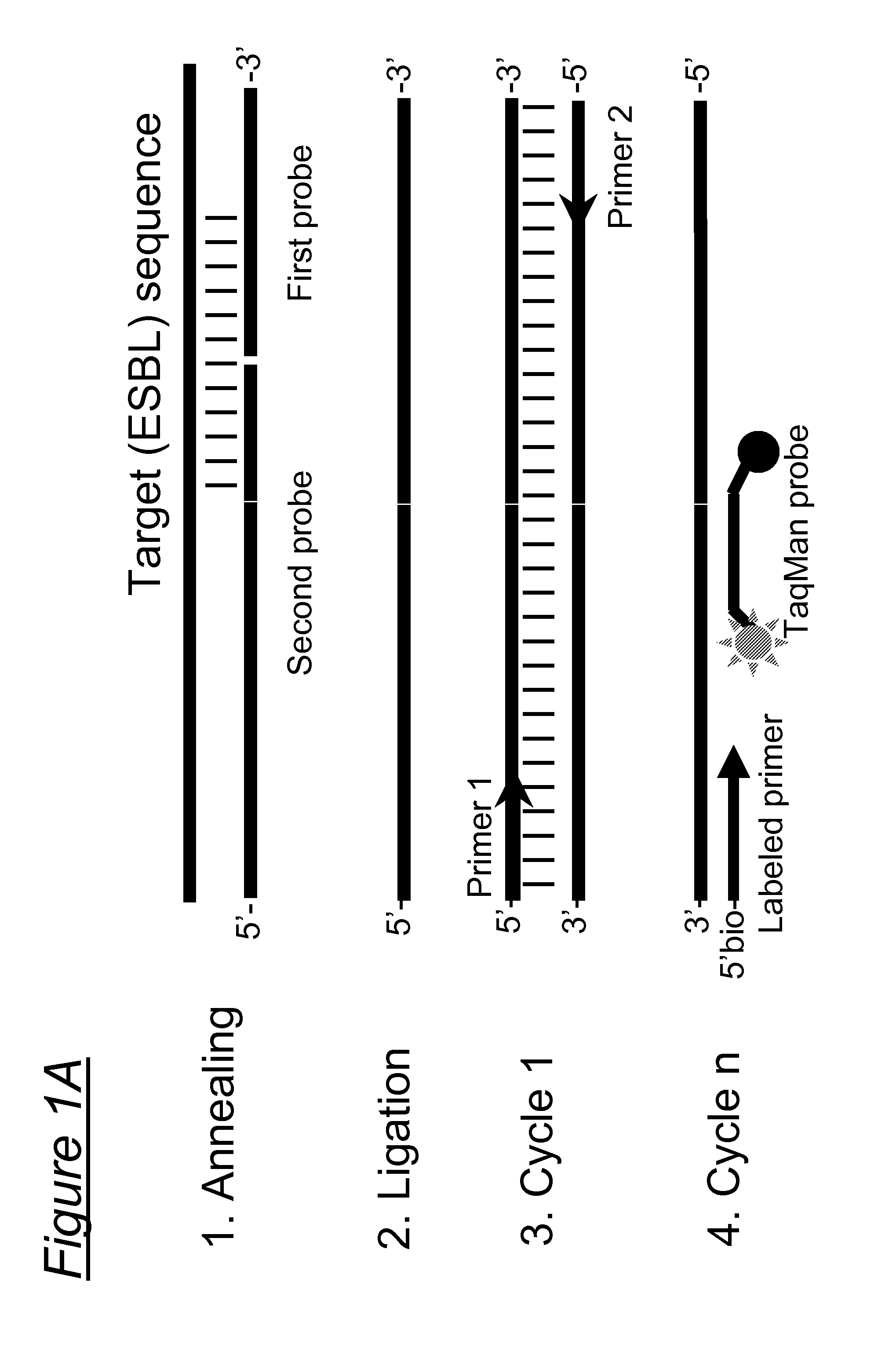
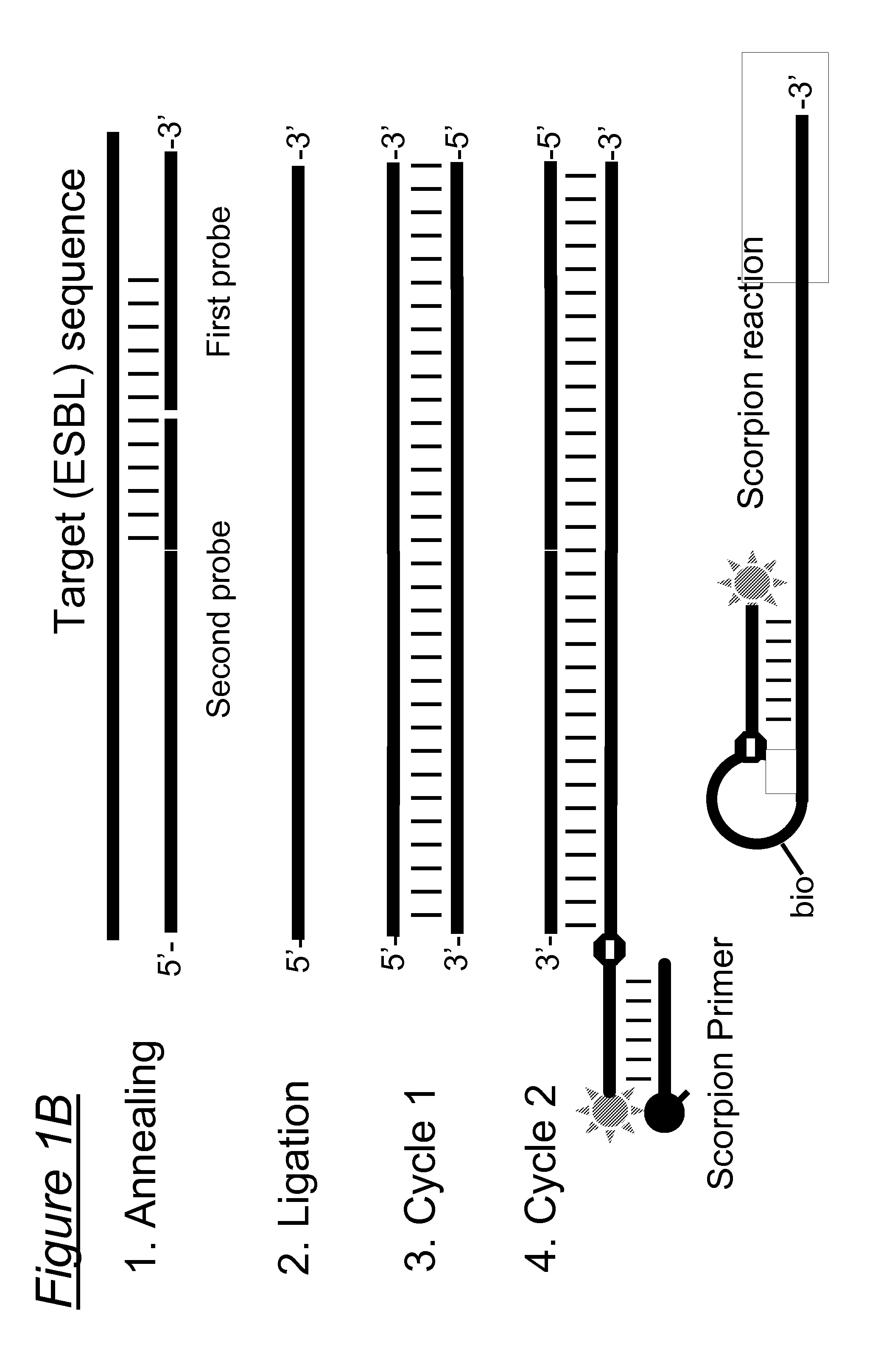
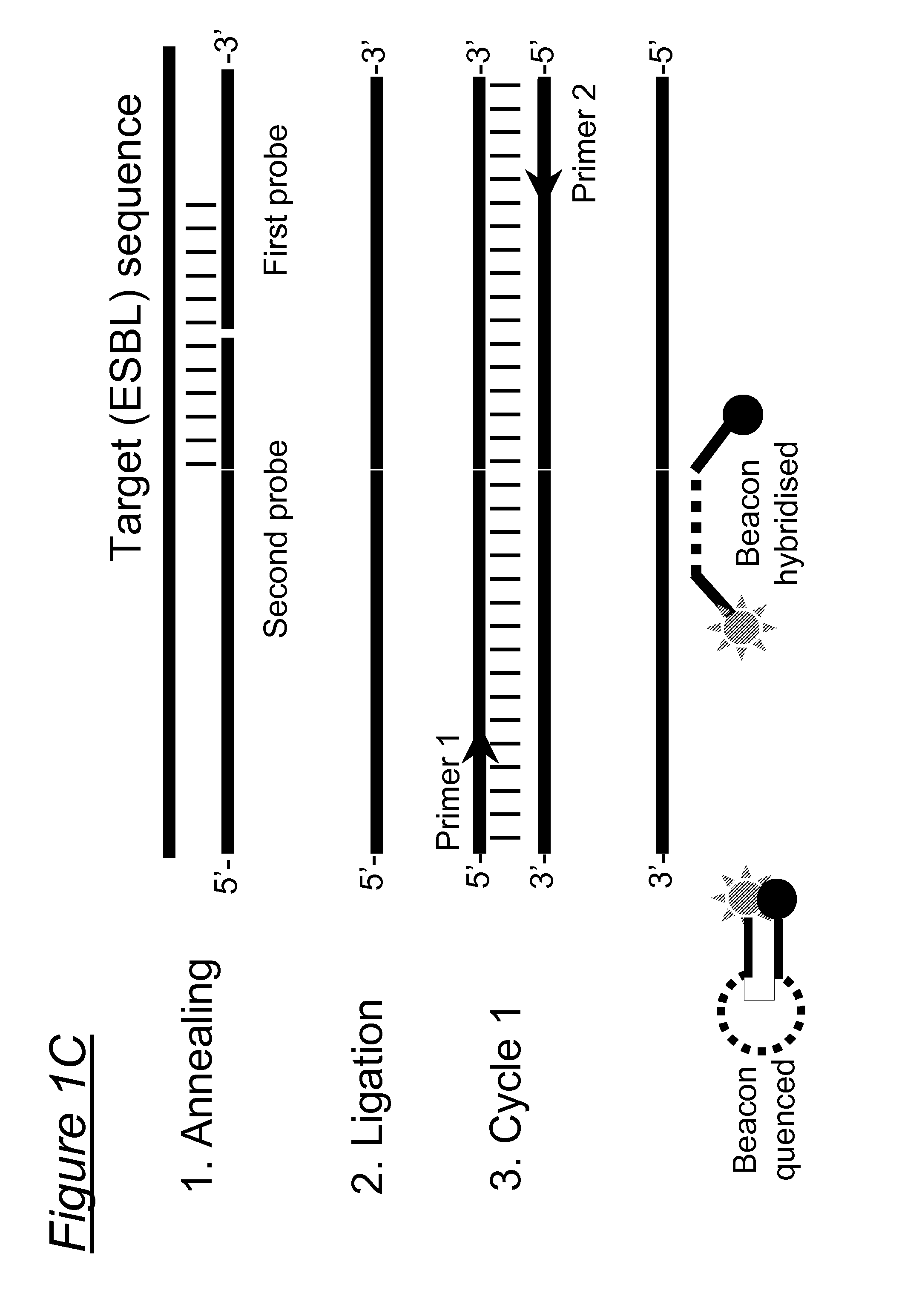
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20040185478A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
VCP-Based Vectors for Algal Cell Transformation
InactiveUS20090317904A1Improve efficiencyUnicellular algaeStable introduction of DNAChlorophyll aBiofuel
Provided herein are exemplary vectors for transforming algal cells. In exemplary embodiments, the vector comprises a Violaxanthin-chlorophyll a binding protein (Vcp) promoter driving expression of an antibiotic resistance gene in an algal cell. Embodiments of the invention may be used to introduce a gene (or genes) into the alga Nannochloropsis, such that the gene(s) are expressed and functional. This unprecedented ability to transform Nannochloropsis with high efficiency makes possible new developments in phycology, aquaculture and biofuels applications.
Owner:AURORA ALGAE
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20060263810A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsSugar derivativesMicrobiological testing/measurementNeisseria meningitidisListeria monocytogenes
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6′-IIa, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6′)-aph(2″), aad(6), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
Engineered Listeria and methods of use thereof
InactiveUS7935804B2Improve survivalIncreasing expression and secretionBiocideBacteriaHeterologousHeterologous Antigens
The invention provides a bacterium containing a polynucleotide comprising a nucleic acid encoding a heterologous antigen, as well as fusion protein partners. Also provided are vectors for mediating site-specific recombination and vectors comprising removable antibiotic resistance genes.
Owner:ANZA THERAPEUTICS INC +1
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20030049636A1Low costRapid positioningMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:BERGERON MICHEL G +3
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20050042606A9Rapid bacterial identificationShorten the timeMicrobiological testing/measurementDepsipeptidesGenomic SegmentMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
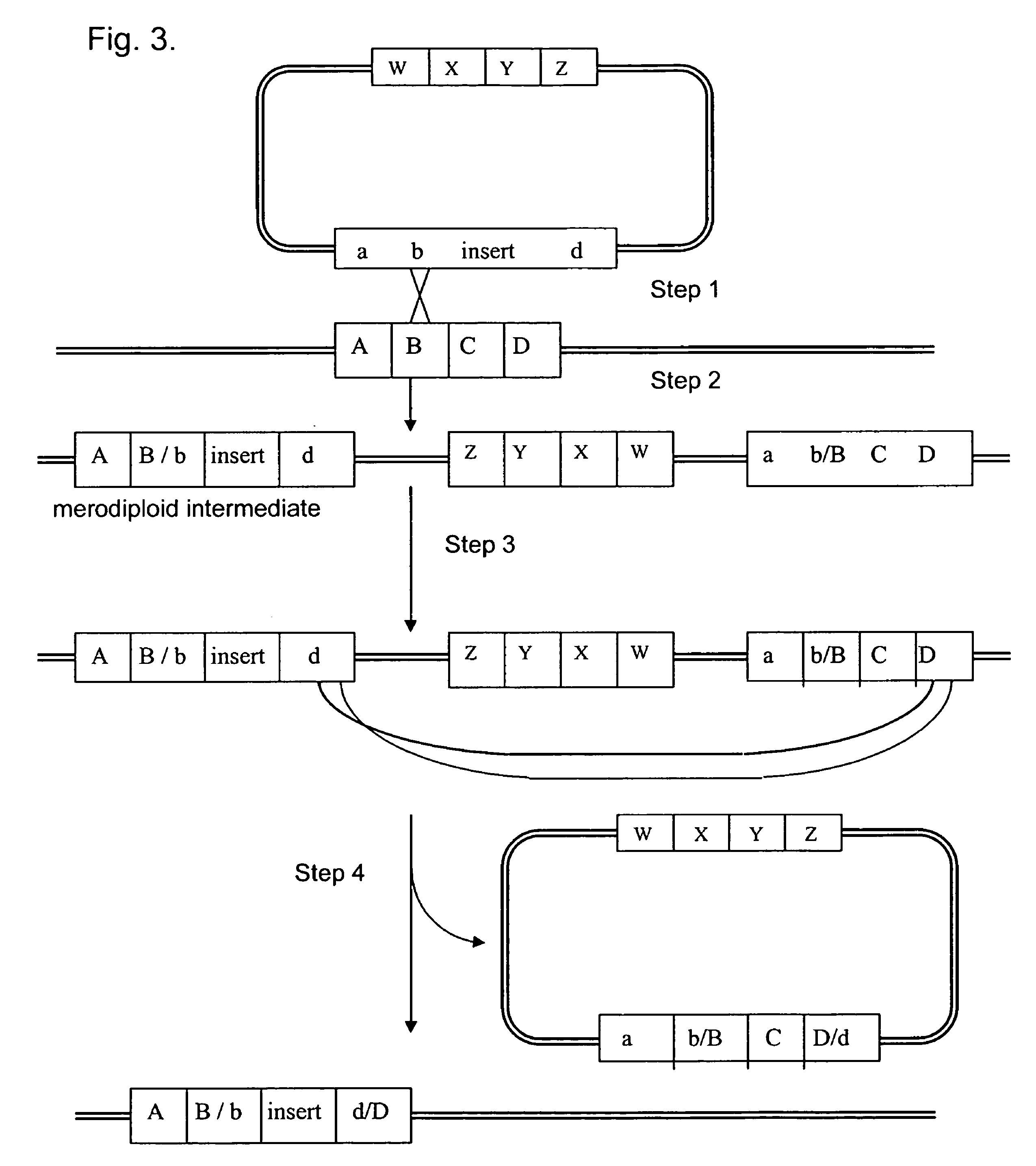
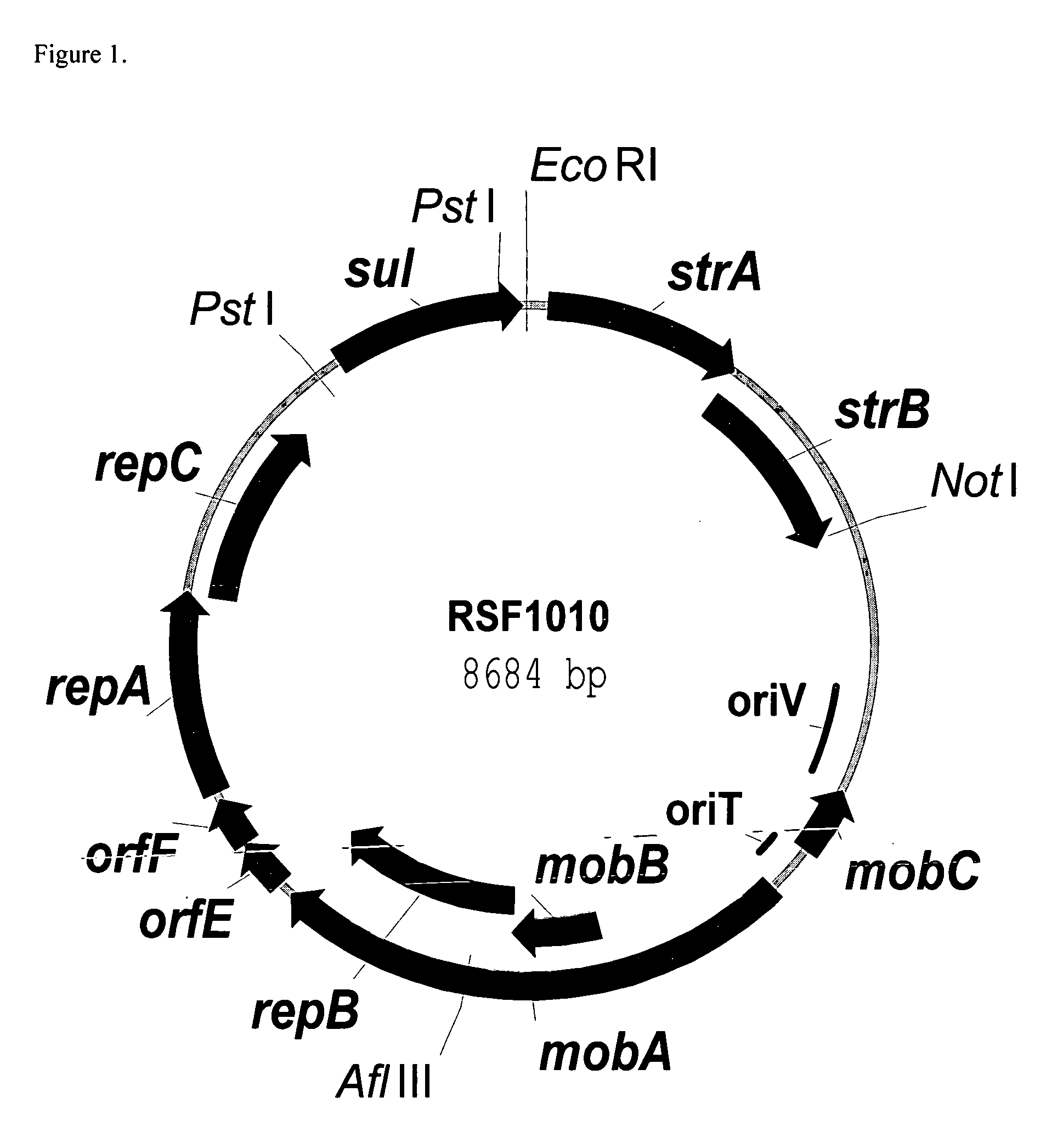
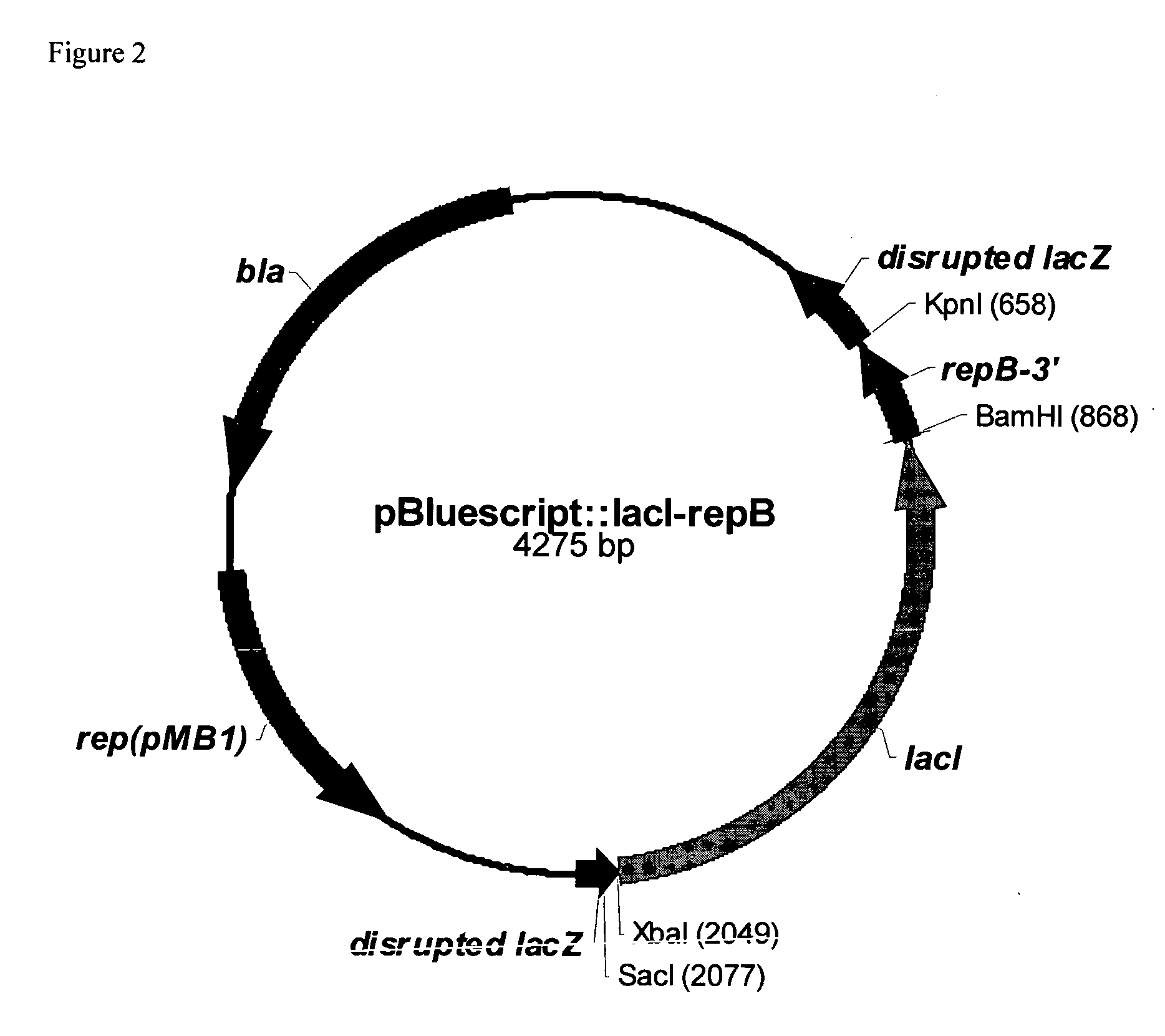
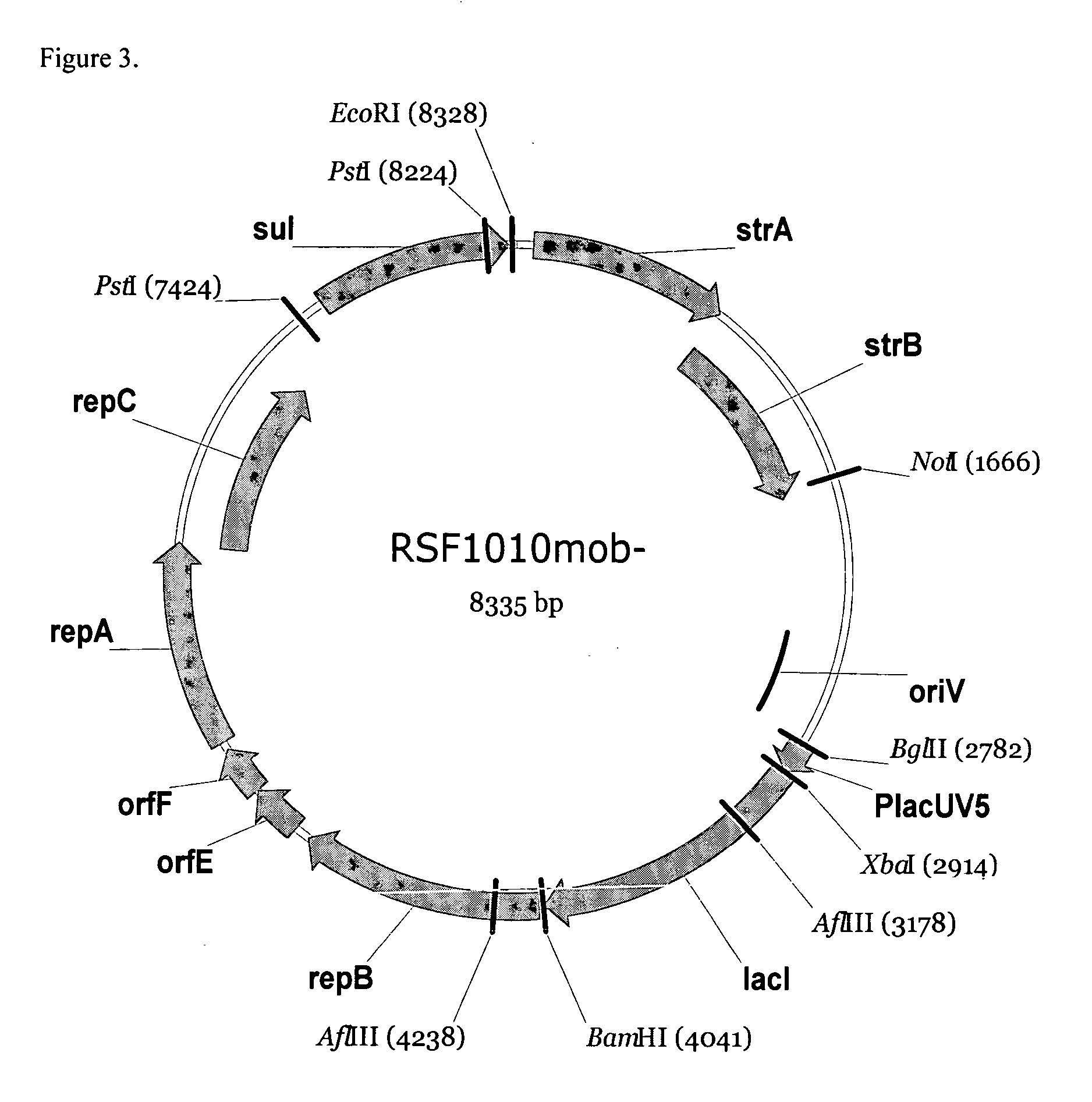
RSF1010 derivative Mob' plasmid containing no antibiotic resistance gene, bacterium comprising the plasmid and method for producing useful metabolites
A Mob− plasmid having a RSF1010 replicon, comprising a gene coding for Rep protein and said plasmid has been modified to inactivate gene related to mobilization ability. The present invention also describes a bacterium having an ability to produce useful metabolites, comprising the plasmid and said bacterium lack active thymidylate synthase coded by thyA gene and thymidine kinase coded by tdk gene, and a method for producing useful metabolites, such as native or recombinant proteins, enzymes, L-amino acids, nucleosides and nucleotides, vitamins, using the bacterium.
Owner:AJINOMOTO CO INC
Silver nanoparticles as Anti-microbial
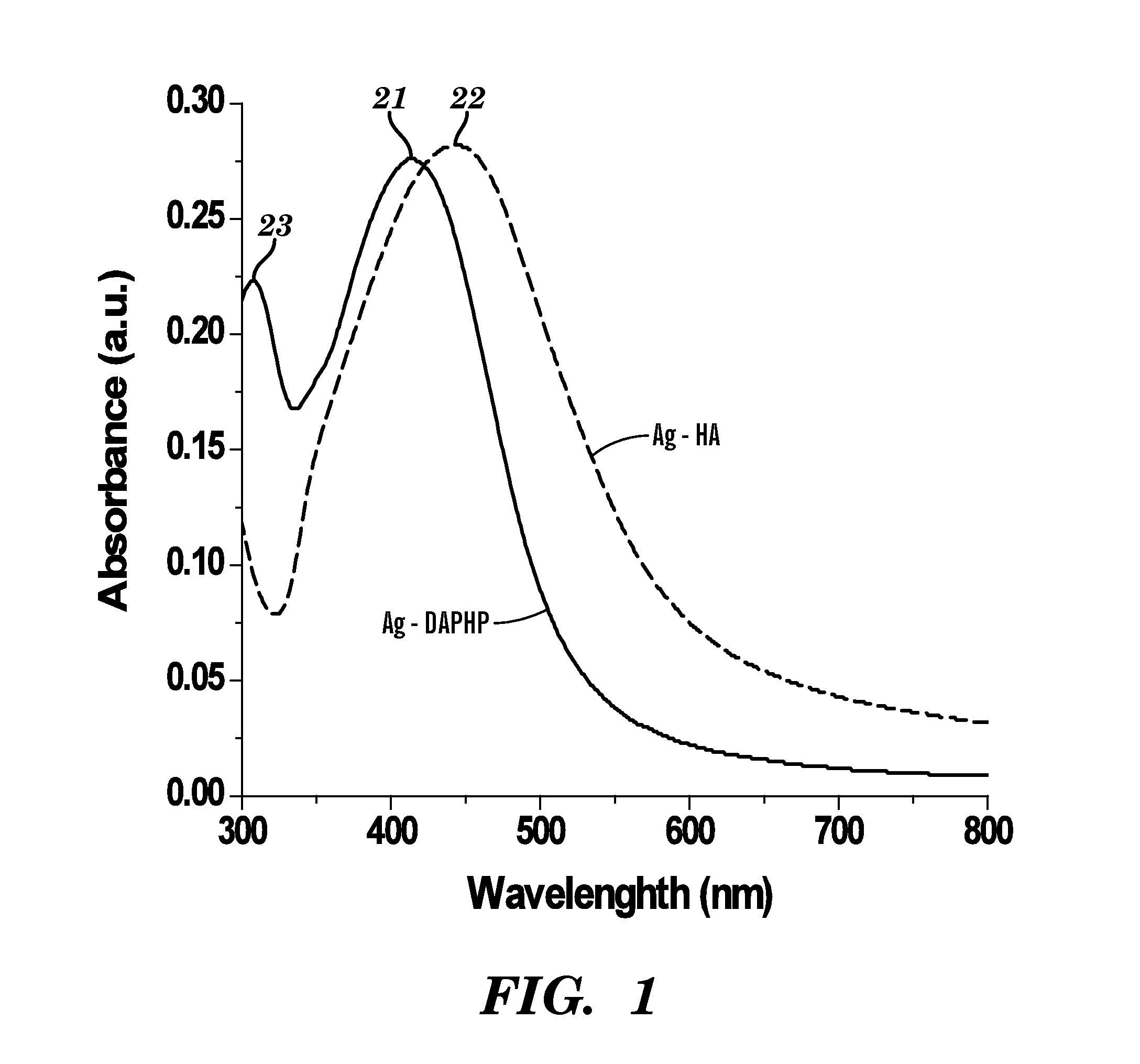
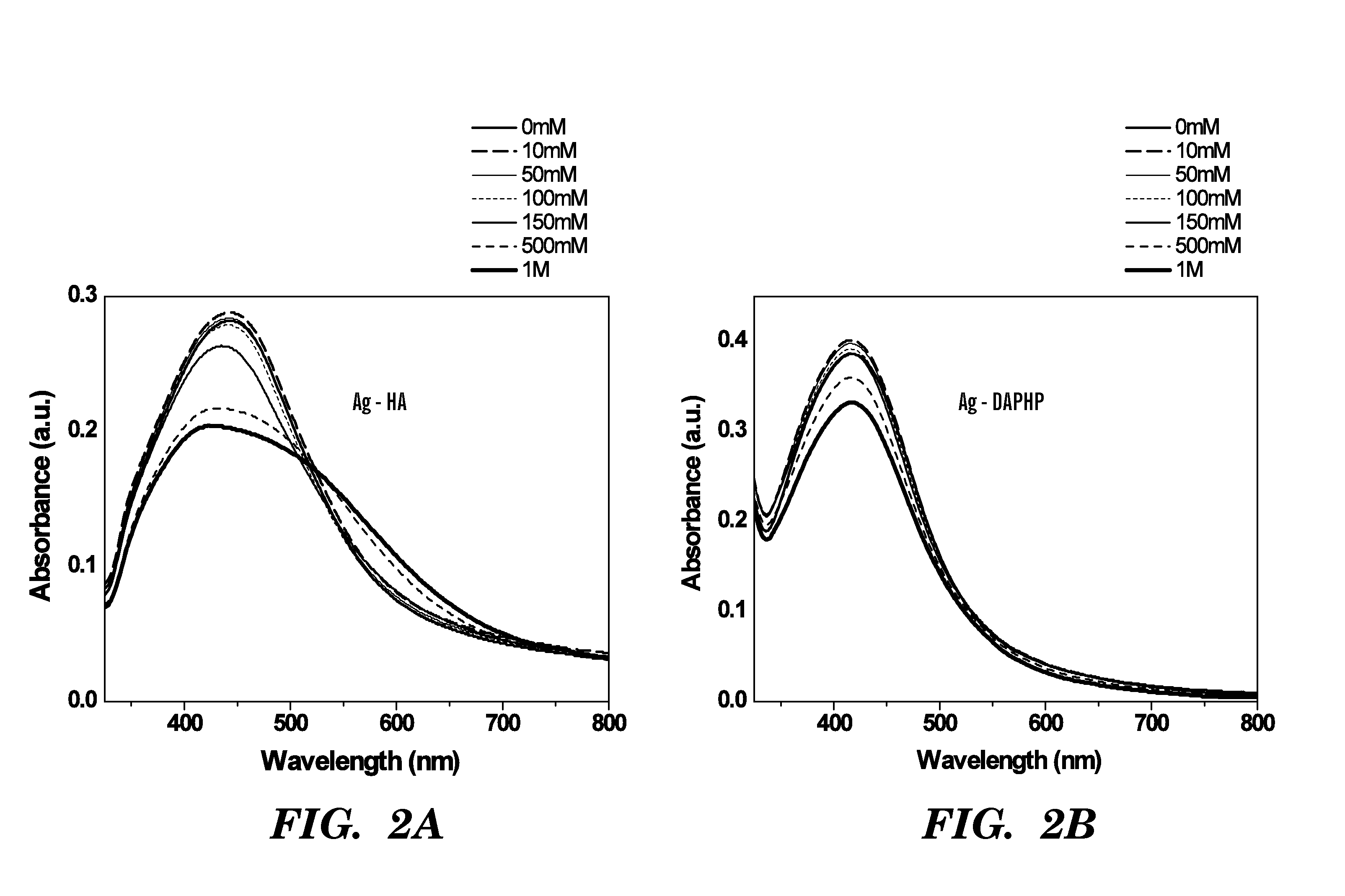
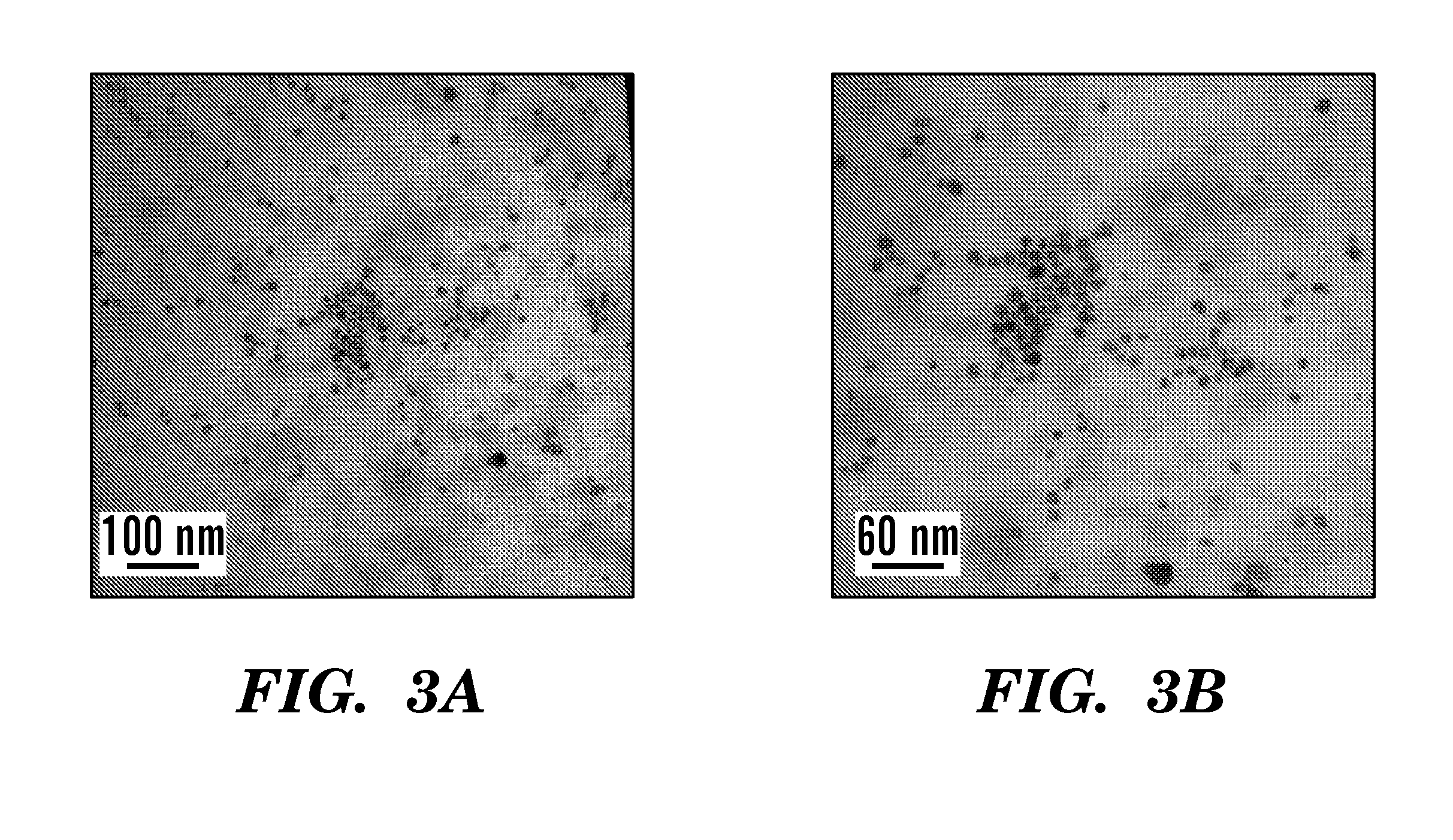
A silver nanocomposite, a formation method for forming the silver nanocomposite, and an application method utilizing the silver nanocomposite. The silver nanocomposite includes a silver nanoparticle conjugated to a glycosaminoglycan (GAG) or glucose. The formation method includes chemically reacting silver nitrate with a reducing agent to form a silver nanoparticle conjugated to the reducing agent of a GAG or glucose. The application method may include topically applying the silver nanocomposite to a wound or burn as an anti-microbial with respect to an antibiotic-resistant genotype in the wound or burn, wherein the silver nanocomposite topically applied includes the silver nanoparticle conjugated to the GAG of 2,6-diaminopyridinyl heparin (DAPHP) or hyaluronan (HA). The application method may include applying the silver nanocomposite as a coating to plastic, a catheter, or a surgical tool, wherein the silver nanocomposite applied as the coating includes the silver nanoparticle conjugated to the GAG of DAPHP.
Owner:VASCULAR VISION PHARMA
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20090053702A1Rapid and exponential in vitro replicationQuick identificationMicrobiological testing/measurementMicroorganism based processesBacteroidesStreptococcus pyogenes
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony.
Owner:GENEOHM SCI CANADA
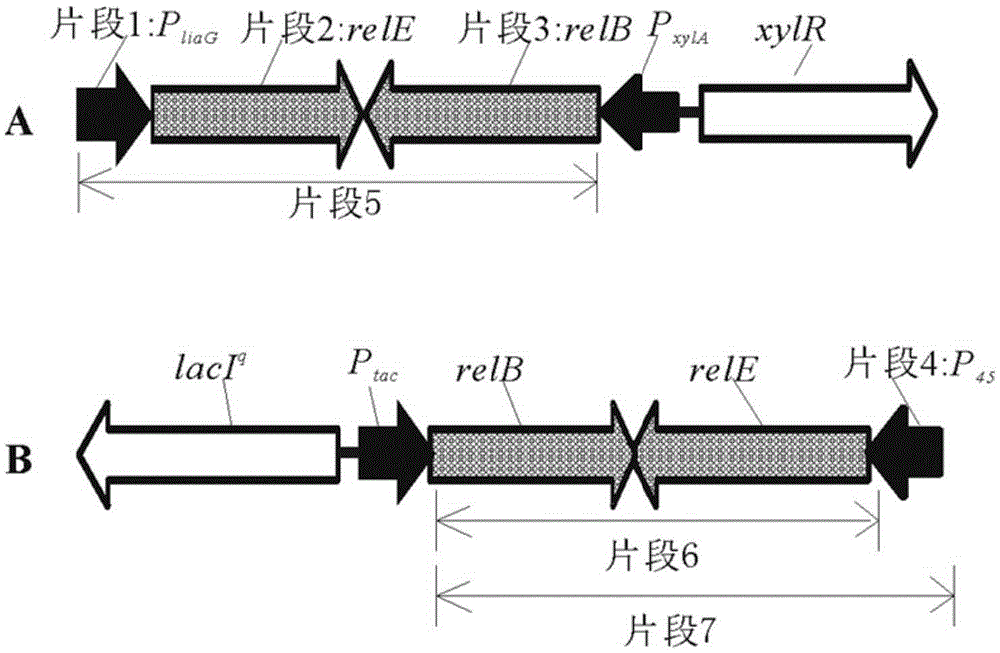
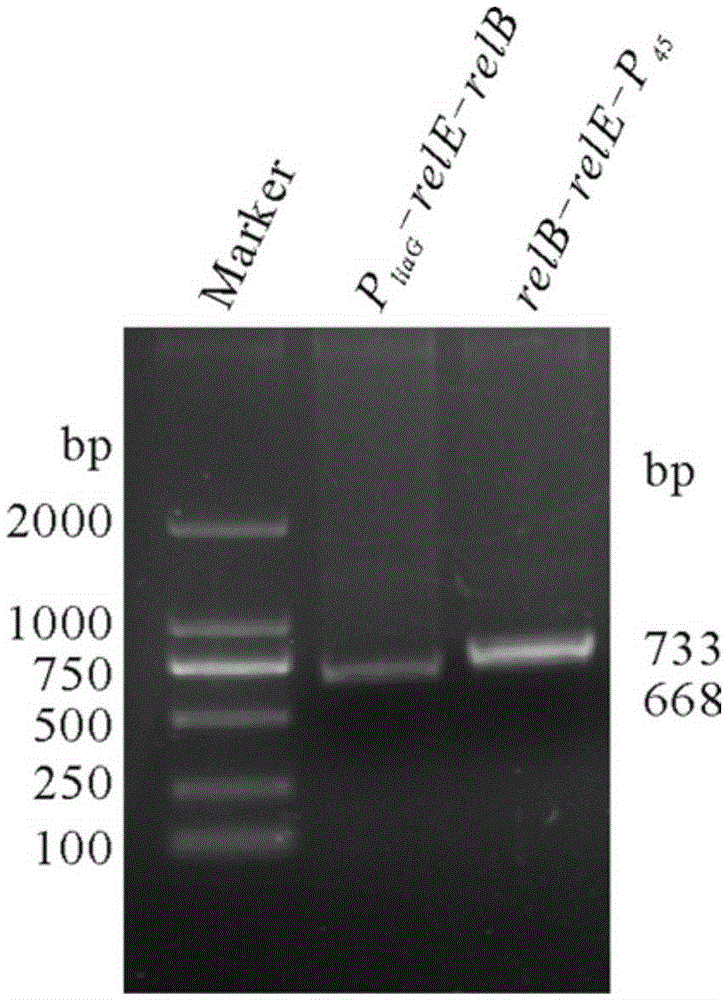
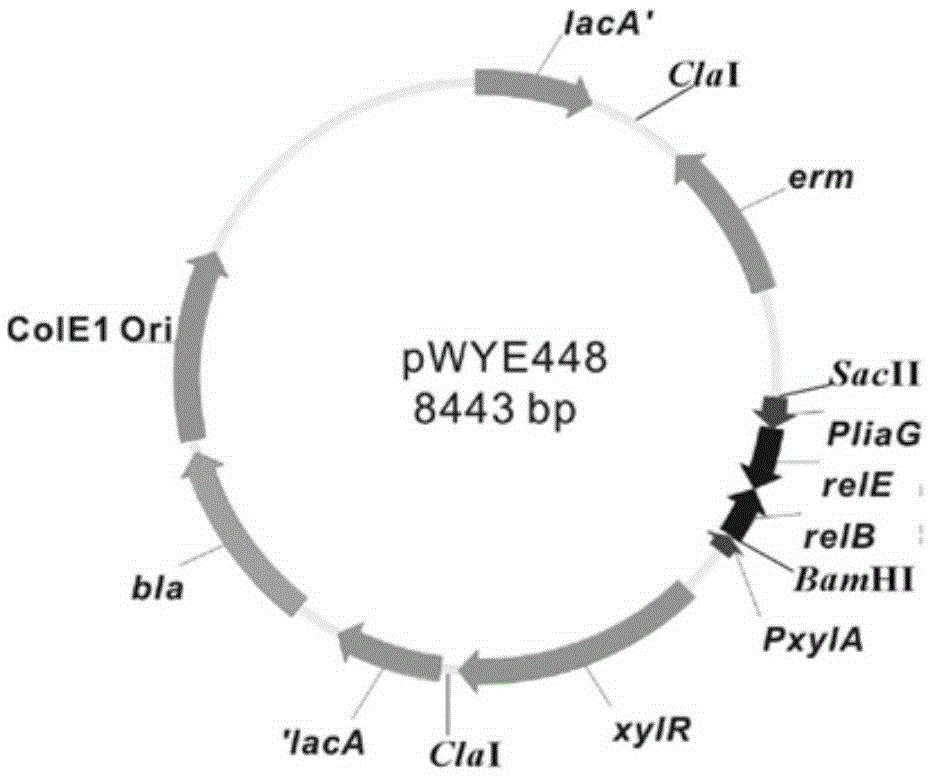
Bacterial traceless genetic manipulation vector and construction method and application thereof
ActiveCN106676119AAvoid toxicityImprove function and effectBacteriaVector-based foreign material introductionAntibiotic YToxin protein
The invention provides a bacterial traceless genetic manipulation vector. The vector contains a toxin inverse-screening element controlled by an antitoxin switch and an antibiotics resistance gene. The invention also provides a construction method of the vector and a method for application of the vector in bacterial traceless gene knock-out, knock-in, replacement, point mutation and insertion and deletion of large DNA fragments (larger than 194kb). By the utilization of a constitutive promoter for continuous expression of toxin protein and by the utilization of an inducible promoter for inducible expression of antitoxin protein, the toxin inverse-screening element (TCCRAS) controlled by the antitoxin switch is established, and the problem that specificity of existing genetic manipulation systems of gram-positive bacterium is not high, the systems are instable and screening is difficult is solved.
Owner:INST OF MICROBIOLOGY - CHINESE ACAD OF SCI
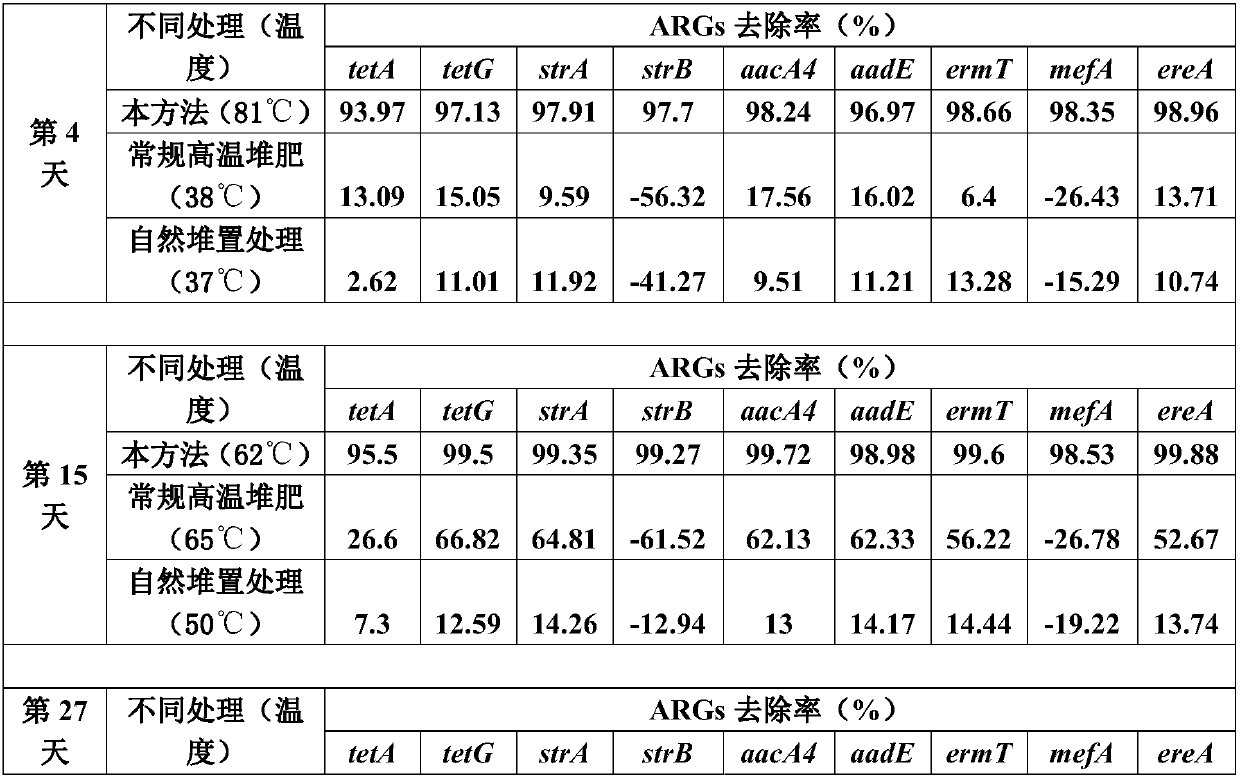
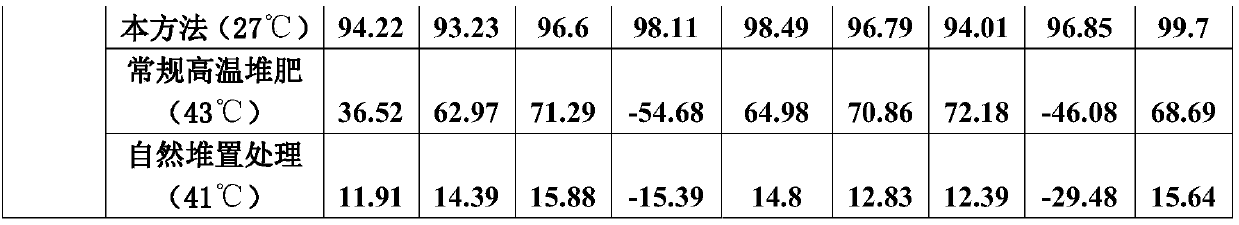
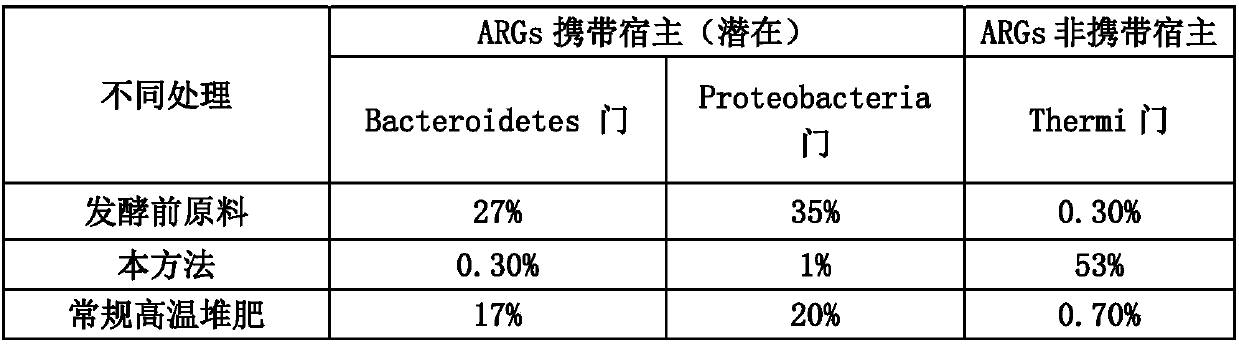
Method for quickly reducing antibiotics and resistance genes in organic solid waste
ActiveCN108033817AReduce the risk of gene transferNo reboundBio-organic fraction processingBacteriaGene transferBacteroidetes
The invention discloses a method for quickly reducing antibiotics and resistance genes in organic solid waste, and belongs to the field of organic solid waste treatment. The method has the advantagesthat ultrahigh-temperature aerobic fermentation is carried out on the organic solid waste by the aid of aerobic zymocyte capable of tolerating the temperatures of at least 80 DEG C under the conditionthat pile temperatures are not lower than 80 DEG C under the control for at least 5-7 days, and accordingly effects of quickly and stably reducing the antibiotics and the resistance genes in the organic solid waste can be realized; the antibiotics and the ARGs (antibiotic resistance genes) can be quickly degraded by means of ultrahigh-temperature aerobic fermentation, integral microbial communitystructures in piles further can be changed, 90% of microorganisms (mainly including Proteobacteria and Bacteroidetes) which carry the ARGs can be killed, risks of gene transfer of the ARGs can be reduced, diffusion of the ARGs can be controlled from the source, and the ARGs can be guaranteed against rebounding; the organic solid waste can be treated, and double effects of efficiently removing antibiotic residues and resistance gene pollution can be realized; exogenous heating can be omitted, energy can be produced only by the aid of metabolism of thermophilic microorganisms to reach the highfermentation temperatures, and accordingly the method is low in energy consumption and is environmentally friendly.
Owner:FUJIAN AGRI & FORESTRY UNIV
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20090047671A1Rapid bacterial identificationShorten the timeMicrobiological testing/measurementMicroorganism based processesBacteroidesMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
Assays, compositions and methods for detecting drug resistant micro-organisms
InactiveUS20130065790A1Microbiological testing/measurementLibrary screeningMicroorganismResistant genes
The present invention relates to assays, compositions and methods for the detection and discrimination of specific gene sequences encoding antibiotic resistance in a sample. The nucleotide sequences encoding antibiotic resistance genes are uniquely identified. In particular, these sequences are hybridized to capture probes, enabling real-time PCR. For this purpose, the capture probes may also be covalently linked to amplification primers. Alternatively, detection of amplified products with hybridization to capture probes may be performed after amplification by hybridization to capture probes bound to a solid support such as microarrays and microspheres (beads). Finally, detection of amplification products during amplification in real-time using capture probes may be combined with detection of such amplification products after amplification using the same and / or different capture probes.
Owner:CHECK POINTS HLDG
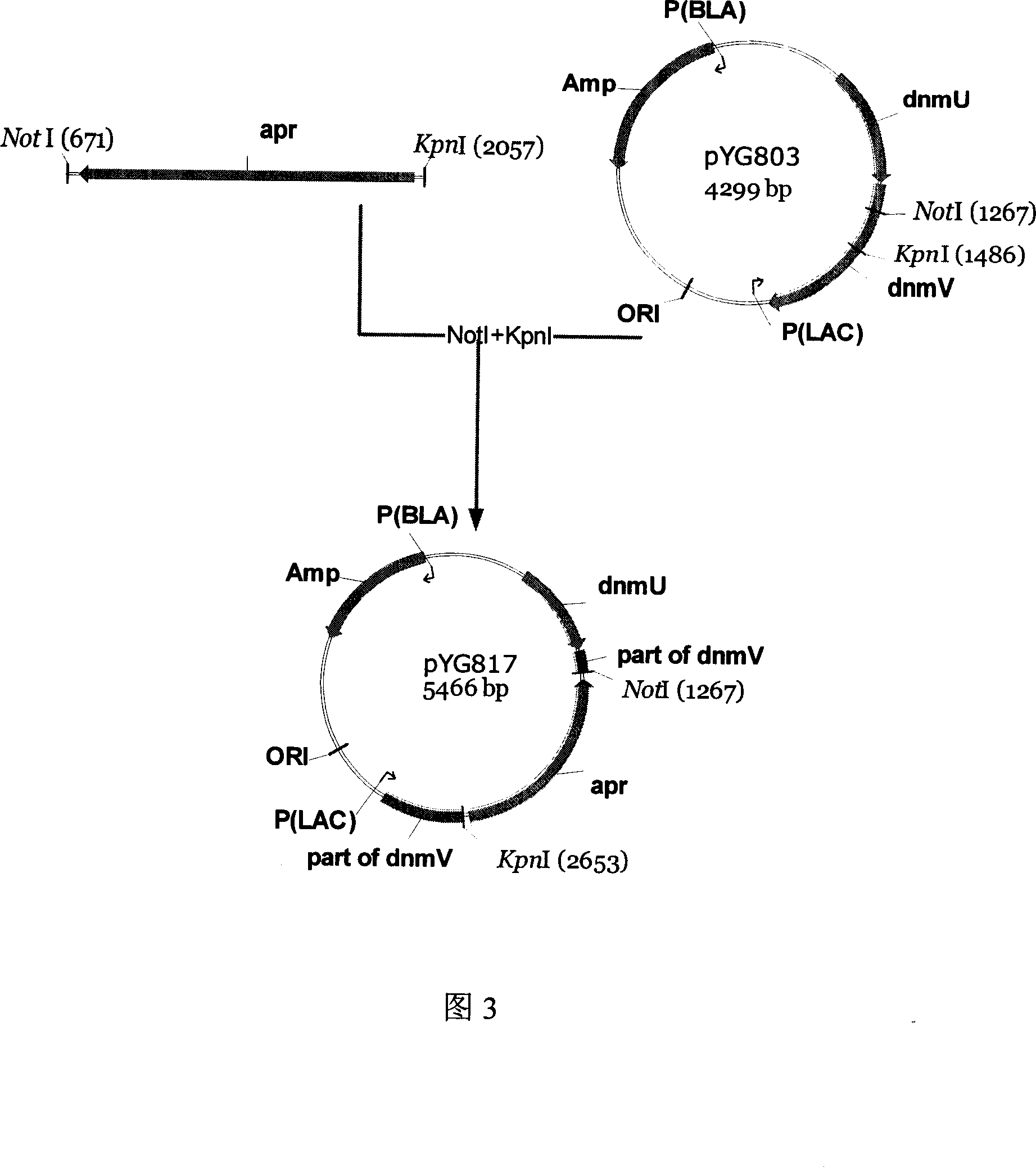
Engineering bacterium capable of producing anthracene ring antibiotics and application of the same
The invention discloses an engineering bacterium through anthracene nucleus antibiotic and appliance, which is characterized by the following: inserting or replacing restriction enzymes site of dnmV gene nucleic acid order of sky blue rufus streptomycete with antibiotic against property gene and surface cerubidin synthetic gene; blocking-up engineering bacterium of dnmV gene; or transforming expressing carrier of anthracene nucleus antibiotic synthetic gene to lead streptomycin (S.lividans)TK24; extracting expressing plasmid from S.lividans TK24; leading-in sky blue rufus streptomycete gene only with antibiotic against property gene to block dnmV gene; blocking-up abrupt change bacterium. This invention can produce cerubidin and / or analogue, such as anthracene nucleus antibiotic.
Owner:ZHEJIANG HISUN PHARMA CO LTD +1
Method for abating antibiotics resistance gene pollution in water
InactiveCN103159357AEliminate pollutionMature technologyEnergy based wastewater treatmentMultistage water/sewage treatmentParticulatesSuspended particles
The invention discloses a method for abating antibiotics resistance gene pollution in water and belongs to the technical field of water treatment. The method disclosed by the invention aims at solving the problem of the water pollution caused by antibiotics resistance gene. The method comprises the following steps of: firstly removing the content of suspended particles in water through coagulating sedimentation of an ultraviolet ray irradiation method to obtain a further ultraviolet projection distance in subsequent processing; then, through the ultraviolet radiation, generating strong oxidizing substances such as hydroxyl radical on one hand and reacting the substances with the chemical substances composing the antibiotics resistance gene for changing the chemical natures thereof so that the genetic information contained in the antibiotics resistance gene can not be sent, destroying the molecular structure of the antibiotics resistance gene through the ultraviolet physical radiation on the other hand so that the molecular structure is broken into free base segments and the integrity of the genetic information contained therein is lost, thereby achieving the aim of abating antibiotics resistance gene pollution.
Owner:INST OF URBAN ENVIRONMENT CHINESE ACAD OF SCI
Construction method for domestic silkworm silk glandulae biological factory and pharmacy use
InactiveCN101270367AClear efficacyHigh biosecurityAnthropod material medical ingredientsFermentationBiotechnologyFluorescence
The invention discloses a method for building a domestic silkworm silk gland biologic plant. A recombinant transgene vector which is provided with domestic silkworm silk protein specificity promoter controlled extraneous gene expression cassettes, an enhancer activity component, antibiotic resistant genes and fluorescent protein reporter genes and based on a piggyBAC transposon is established through gene engineering operations. The transgene vector is introduced into primarily laid eggs, special antibiotics are fed to hatched newly exuviated silkworms continuously until the fourth instar. In this way, silkworms which can resist the specific antibiotics and are provided with fluorescent protein reporter genes are obtained; target genes are detected; and transgene silkworms are manufactured by breeding. By adopting the silk gland tissues of the transgene silkworms, protein drugs can be obtained. With the method disclosed by the invention, transgene domestic silkworm silk gland plants can prepare protein drugs at low manufacture cost without potentially hazardous rhabdovirus. The method is safe and is practically valuable.
Owner:SUZHOU UNIV
Artificial wetland capable of strongly removing antibiotic and resistance gene from culture waste water
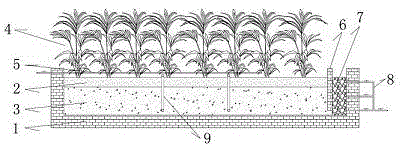
InactiveCN103332791ARich denitrification pathwayGood removal effectSustainable biological treatmentBiological water/sewage treatmentConstructed wetlandBrick
The invention discloses an artificial wetland system capable of strongly removing an antibiotic and an antibiotic resistance gene from culture waste water, and relates to waste water treatment equipment which is an artificial wetland ecological treatment device capable of efficiently removing nitrogen and phosphorus elements, high-residual veterinary antibiotic contents and the antibiotics resistance gene from the livestock culture waste water. The system comprises a vertical subsurface flow artificial wetland device, adopts a filling manner of large size grading difference 'positive and negative particle size mixture' filling and paving, and takes red soil, waste bricks, and zeolite as main wetland fillings; the red soil, the waste bricks and the zeolite have large specific surface areas, good adsorptive capacity to nitrogen, phosphorus, the antibiotic and the antibiotic resistance gene, and high biological holding; and continuous, high-efficiency, and steady operation of the artificial wetland ecological system is ensured. The system has the advantages of low cost, low energy consumption, low possibility of blocking, high removal efficiency of the antibiotic and the antibiotic resistance gene, high efficiency of denitrification and dephosphorization, small occupied area, simple structure, convenient operation management and the like.
Owner:INST OF URBAN ENVIRONMENT CHINESE ACAD OF SCI
Simple method for efficiently extracting DNA (deoxyribonucleic acid) from soil samples
ActiveCN103993007APracticalHigh degree of reliabilityMicroorganism based processesDNA preparationFreeze thawingHigh concentration
The invention provides a simple method for efficiently extracting DNA (deoxyribonucleic acid) from soil samples. According to the simple method, the DNA in environmental samples is fully released and exposed by using combination of methods such as glass bead grinding, liquid nitrogen repetitive freeze-thawing, SDS (sodium-dodecyl sulphate), lysozyme and a protease-K method for the different soil samples, the DNA purification is carried out through multiple phenol imitation extraction processes, and the determination of the DNA extraction efficiency is carried out by adding an internal standard bacterial strain Xcc8004. The simple method has simple requirements on sample pretreatment; the quantity of the required environmental samples is small; the obtained DNA has relatively high concentration and purity; the extraction efficiency is relatively high; the analysis and detection of antibiotic resistance gene pollution in the complex environmental samples can be met; and the method is simple and reliable and has practical application value.
Owner:NANKAI UNIV
Single-resistance escherichia coli-bacillus subtilis shuttle expression vector and application thereof
InactiveCN104232675AEasy genetic manipulationEase of plasmid transformationBacteriaHydrolasesEscherichia coliAgricultural science
The invention discloses a single-resistance escherichia coli-bacillus subtilis shuttle expression vector and application thereof. The bacillus subtilis shuttle expression vector provided by the invention comprises the following elements: a bacillus subtilis formed strong promoter P43, a sequence which encodes signal peptide, multiple cloning sites, an antibiotics resistance gene (kalamycin or chloramphenicol), a bifunctional promoter sequence which can start resistance gene expression of kalamycin or chloramphenicol in escherichia coli and bacillus subtilis, and copy homing sequences of bacillus subtilis and escherichia coli. The expression vector disclosed by the invention not only can be stably copied in escherichia coli, but also can be stably copied in bacillus subtilis, and expresses homologous and heterologous proteins. All vectors just contain one resistance gene, the plasmid of which is less than 400bp, so that gene operation and plasmid conversion are easy to carry out, and the conversion ratio is high. Besides the expression vector of an exogenous gene, the vector can be further used for screening promoters and constructing a gene knock-out vector.
Owner:WUHAN RUIHENGDA BIOLOGICAL ENG
Engineered listeria and methods of use thereof
InactiveUS20120121643A1Improve survivalIncreasing expression and secretionBacteriaAntibody mimetics/scaffoldsHeterologous AntigensNucleotide
The invention provides a bacterium containing a polynucleotide comprising a nucleic acid encoding a heterologous antigen, as well as fusion protein partners. Also provided are vectors for mediating site-specific recombination and vectors comprising removable antibiotic resistance genes.
Owner:ANZA THERAPEUTICS INC
Methods and Compositions Including Diagnostic Kits For The Detection In Samples Of Methicillin-Resistant Staphylococcus Aureus
InactiveUS20120077684A1Cost-effective management and controlMicrobiological testing/measurementLibrary screeningSCCmecStaphylococcus saprophyticus
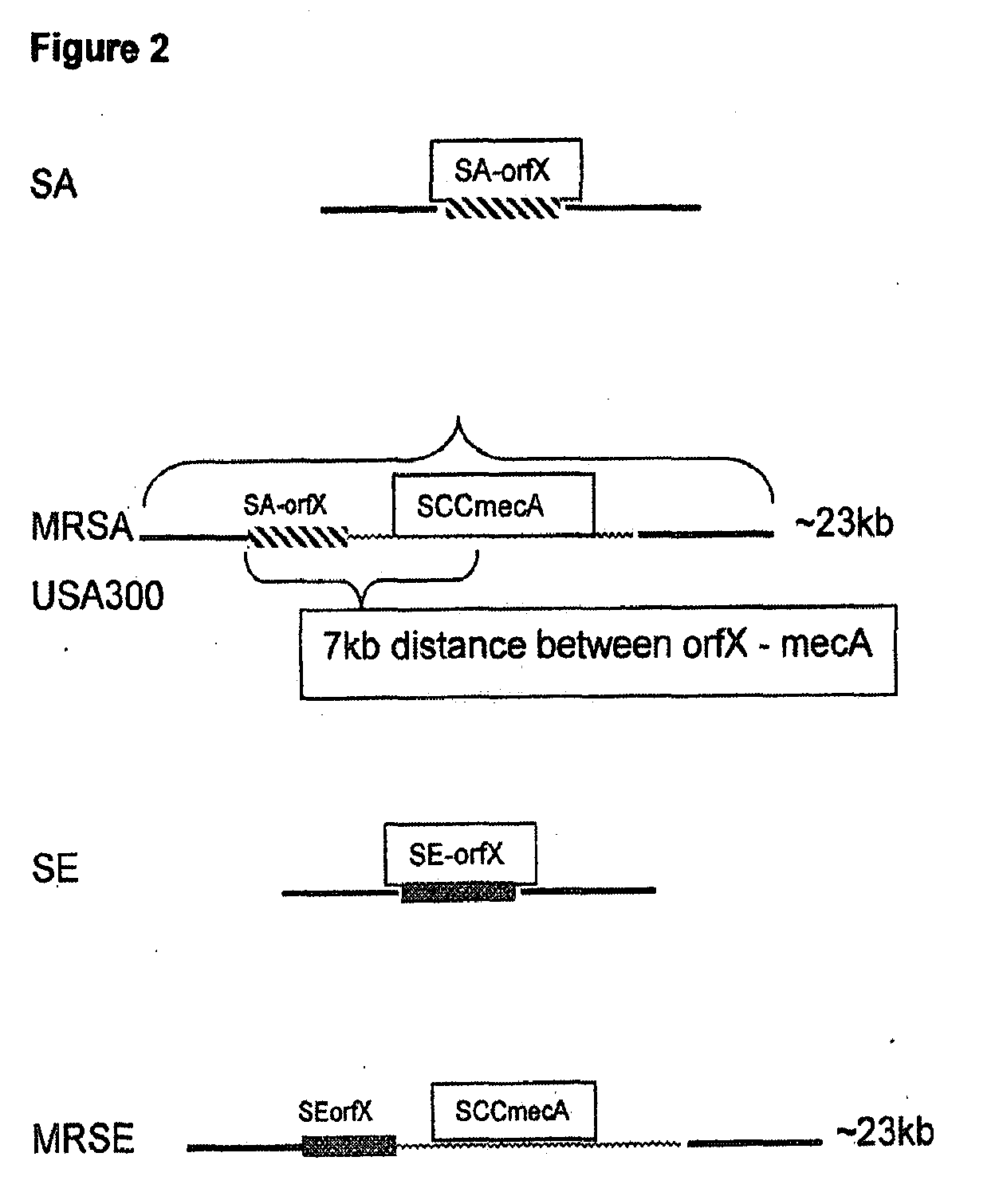
The present invention provides methods, compositions and diagnostic kits for the detection of Staphylococcus Aureus (SA) and antibiotic resistant forms and variants thereof, such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Staphylococcus aureus (VRSA), mupirocin-resistant Staphylococcus aureus (mupSA), and the like, in a sample population. The invention preferably involves the improvements of bacterial sampling by means of SA enrichment, followed by SA cell disruption and amplification procedures incorporating the use of multiplex assays for SA specific genes, such as mecA and coagulase negative Staphylococci (CONS) specific genes such as tufA, for SA identification and identification of its known species. This provides means for controlling for the thirty or more known CONS species in assessing SA samples, especially those CONS species that may carry antibiotic resistance genes, such as SCCmec.
Owner:ZEUS SCI
High-grade oxidization method for removing antibiotic resistance genes in sewage
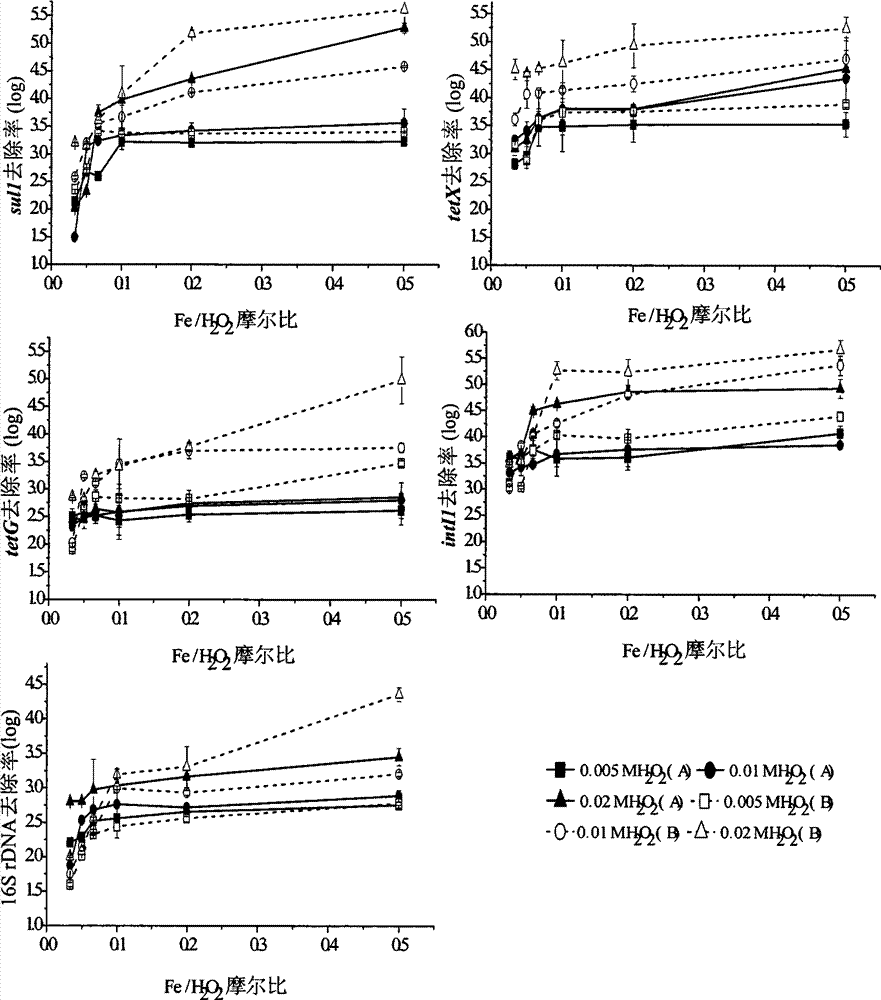
ActiveCN104773903ASimple and fast operationLow costMultistage water/sewage treatmentBiotechnologyFenton oxidation
The invention discloses a high-grade oxidization method for removing antibiotic resistance genes in sewage. In the provided method, the antibiotic resistance genes in sewage are removed by the following steps: filtering the sewage by grating, performing precipitation separation, carrying out Fenton oxidation, performing UV / H2O2 oxidation, disinfecting, and discharging. In the Fenton oxidation method, the influences of Fe2+ / H2O2 mole ratio, H2O2 concentration, initial pH value and reaction time are all taken into account, and in the UV / H2O2 oxidation method, the influences of the illumination time, the addition amount of H2O2, pH value, and the like are considered so as to find the optimal operation parameters. The provided method can effectively remove the antibiotic resistance gene in sewage, and provides a novel technology for cutting off the propagation of antibiotic resistance genes in sewage.
Owner:NANJING UNIV
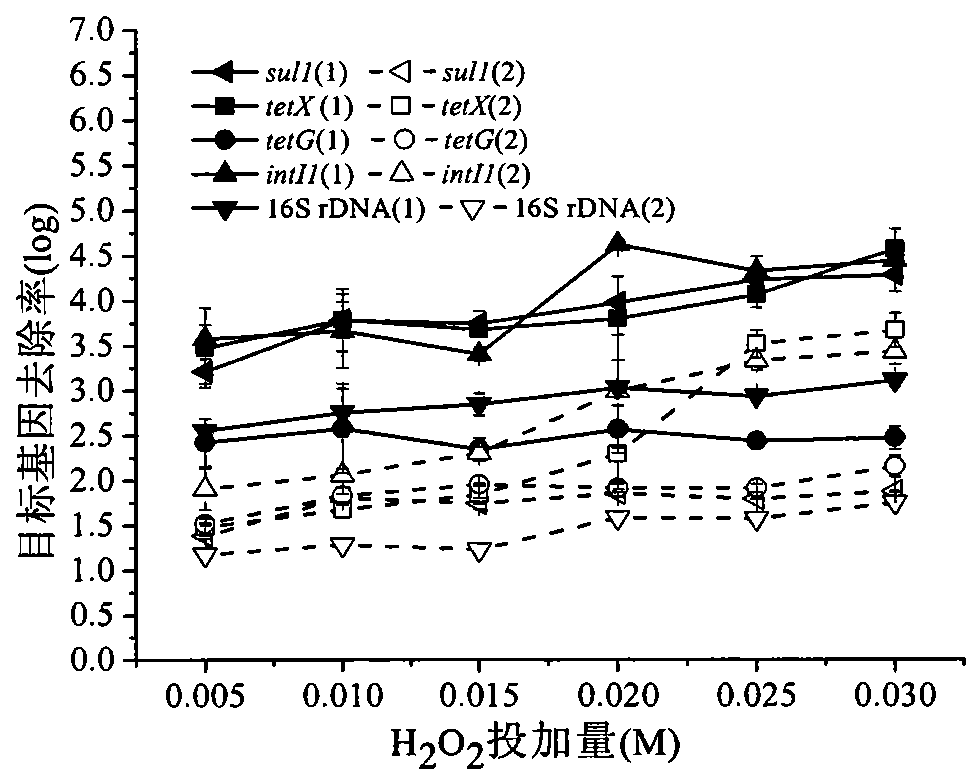
Method for eliminating antibiotics resistance gene pollution in water by hydrogen peroxide coupled ultrasonic method
InactiveCN103159314AStrong physical damageEliminate pollutionWater/sewage treatment by irradiationNature of treatment waterChemical reactionUltrasonic radiation
The invention discloses a method for eliminating antibiotics resistance gene pollution in water by a hydrogen peroxide coupled ultrasonic method and belongs to the technical field of water treatment. The method provided by the invention is used for solving the problem that the water is polluted by the antibiotics resistance gene. The method is characterized in that the method of feeding hydrogen peroxide into water in combination with ultrasonic radiation is employed so that the antibiotics resistance gene is broken from the point of chemical reaction and basic groups, riboses and phosphoric acids in the antibiotics resistance gene are oxidized by means of strong oxidizing property of the hydrogen peroxide, and consequently, the chemical nature of the antibiotics resistance gene is changed and has no ability of storing and transferring genetic information, on one hand, and on the other hand, many cavitation bubbles are formed in water by means of ultrasonic. When the cavitation bubbles are broken, strong physical destruction effect is generated on the antibiotics resistance gene in water so that the double helix structure of the gene is decomposed and the nucleotide chain of the gen is broken; as a result, the genetic information contained in the nucleotide chain cannot be transferred; therefore, the purpose of eliminating the antibiotics resistance gene pollution is achieved.
Owner:INST OF URBAN ENVIRONMENT CHINESE ACAD OF SCI
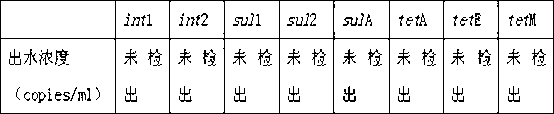
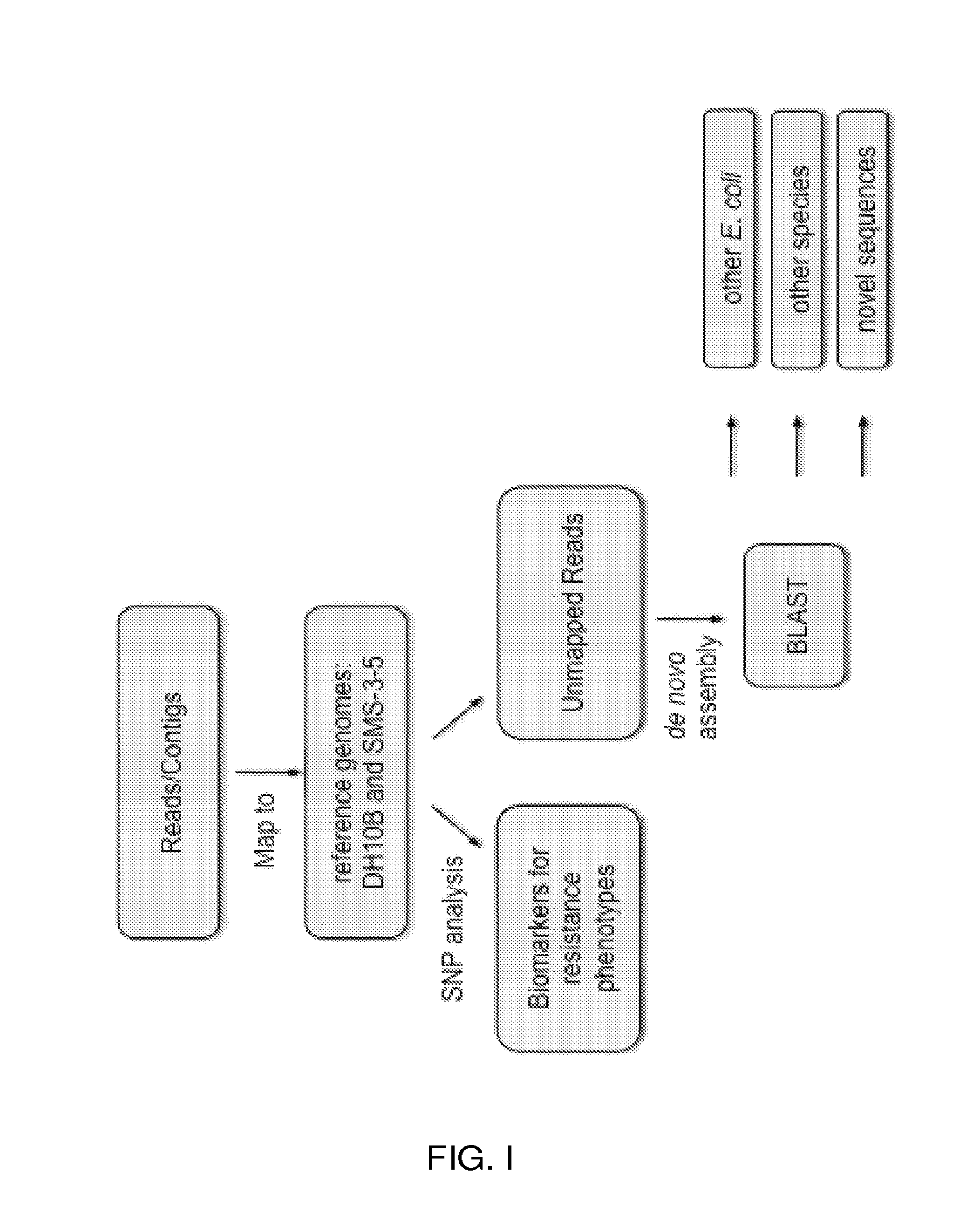
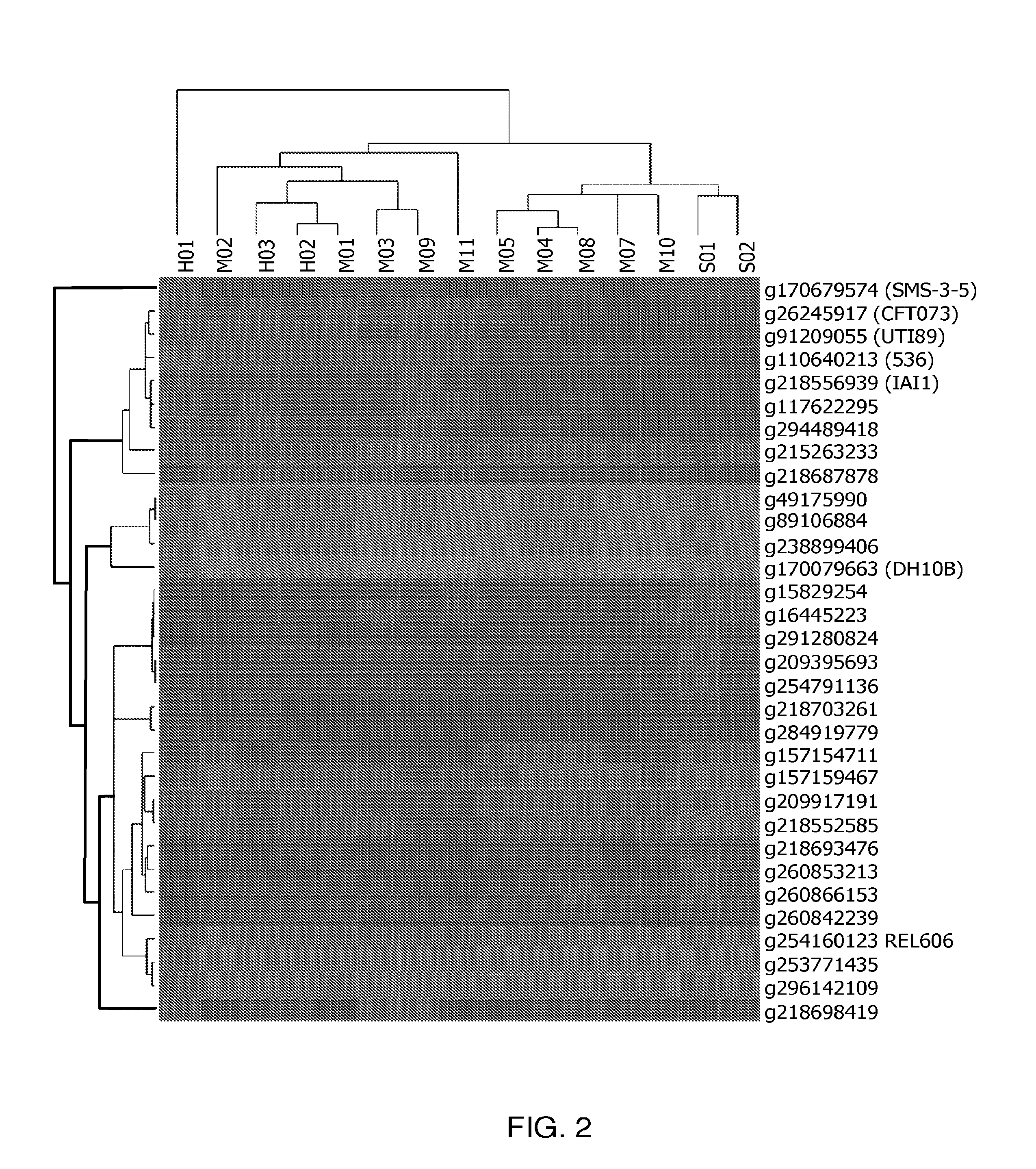
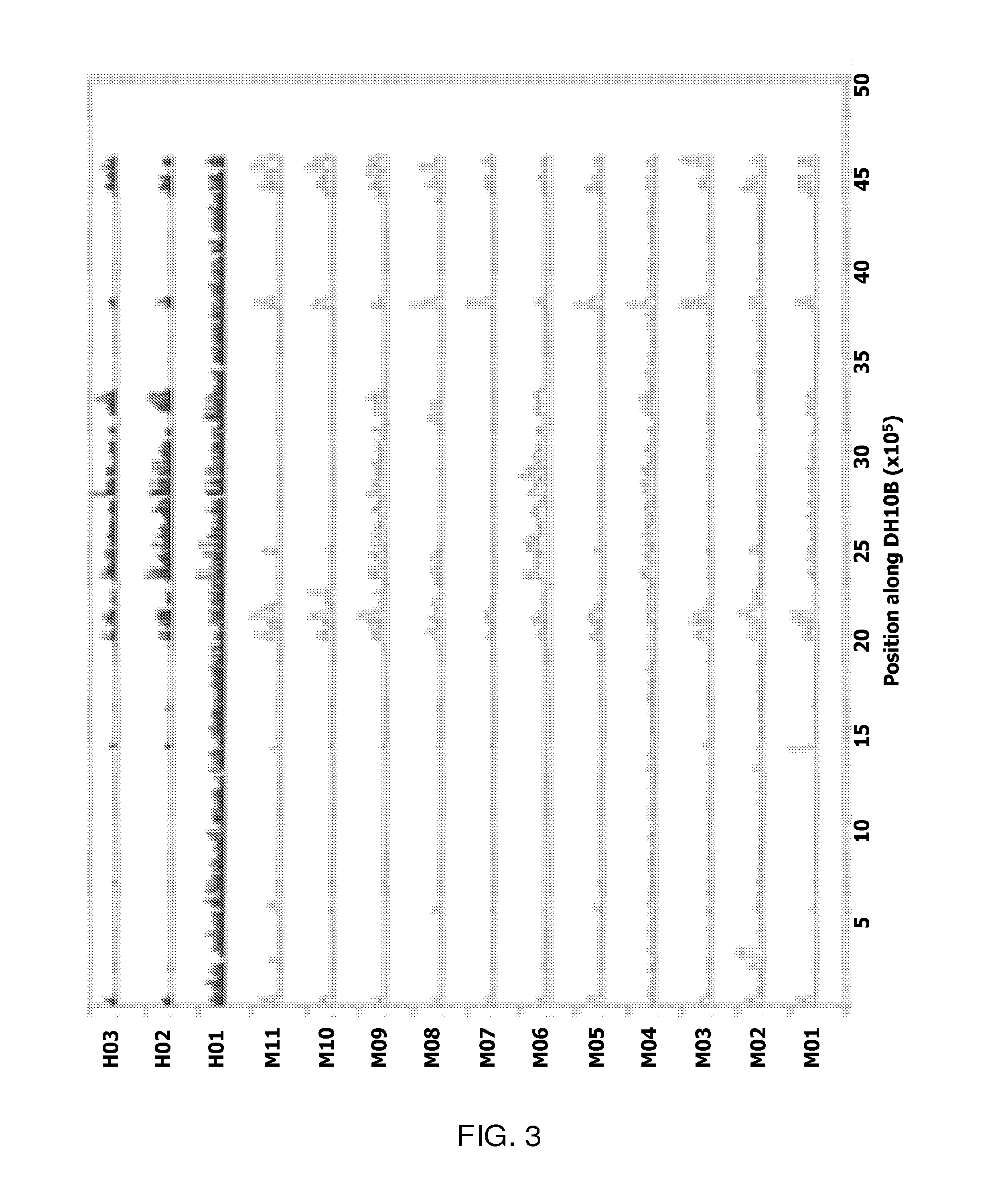
Genomic approach to the identification of biomarkers for antibiotic resistance and susceptibility in clinical isolates of bacterial pathogens
InactiveUS20140030712A1Faster and easy and accurate diagnosisEliminate empirical prescribingMicrobiological testing/measurementProteomicsBacteroidesAntibiotic Y
In certain embodiments, the present invention concerns genotypic identification of bacteria that are resistant to a bacteria and subsequent determination of an appropriate therapy. In specific embodiments, a high-throughput genotypic detection method for biomarkers for antibiotic resistance and susceptibility allows efficient prescription practice and increases the likelihood of a successful therapeutic outcome. In certain embodiments, the information from the genotypic detection method is utilized for determining antibiotics that should be avoided or, alternatively, employed.
Owner:BAYLOR COLLEGE OF MEDICINE
Method for removing antibiotic resistance genes through ionizing irradiation
The invention discloses a method for removing antibiotic resistance genes through ionizing irradiation. Antibiotic mushroom dregs are treated with an ionizing irradiation technology (including gamma-rays and high-power electron beams generated by an electronic accelerator) to damage DNA of microbial cells, so that the resistance genes are effectively removed, and residual antibiotics can be decomposed simultaneously. The method is efficient and wide in application range; radiation can be performed at the normal temperature, no chemical reagents or only a few chemical reagents are required to be added, and no secondary pollution can be caused. The method can be applied to removal of the resistance genes in the antibiotic mushroom dregs, can be applied to removal of the resistance genes in water, soil and sludge and has wide application prospects in the environmental field.
Owner:TSINGHUA UNIV
Method for removing antibiotic resistance gene from water source water
The invention provides a method for removing antibiotic resistance gene from water source water. According to the method, the antibiotic resistance gene is removed through an entire set of processes comprising coagulating sedimentation, sand filtration, O3 contact tower processing, biological active carbon filtering tank processing, high-pressure CO2 disinfection, coagulating sedimentation, reverse osmosis filtering, and chlorination and water discharging. With the method, spreading of resistance gene in the environment can be effectively controlled, such that a problem of the harm of resistance gene spreading in water source water upon human health is solved, and a scientific and reliable technology is provided for human health and environment protection.
Owner:NANJING UNIV
Lac shuttle vectors
The invention discloses a Lac shuttle vector, comprising at least (a) a region which regulates a plasmid copy number; (b) an eukaryotic gene expression cassette, which comprises at least an eukaryotic gene transcriptional promoter sequence, a multiple cloning site and a transcriptional terminator sequence; (c) a lactic acid bacteria plasmid sequence, which comprises a plus origin of replication, and a nucleic acid sequence encoding for a protein which relates to the lactic acid bacteria plasmid replication; and (d) a non-antibiotic resistance selection gene and the promoter sequence thereof. The Lac shuttle vector features a non-antibiotic resistance gene as a selection marker, which is useful in pharmaceuticals and foods.
Owner:ANAWRAHTA BIOTECH
Construction method of gene engineering strain without plasmid and antibiotic resistance screening marker
InactiveCN102604983AWon't cause floodingSuitable for industrial productionBacteriaMicroorganism based processesBiotechnologyEscherichia coli
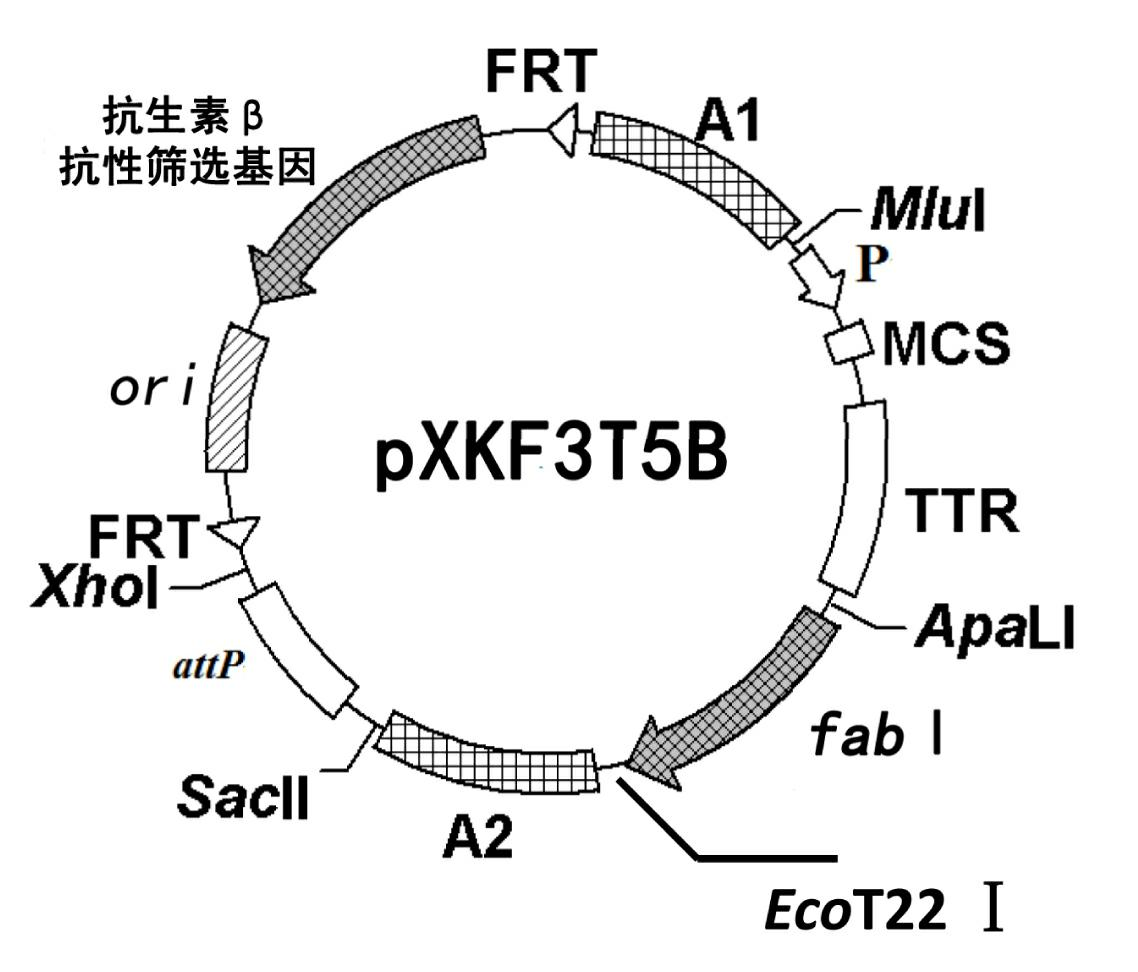
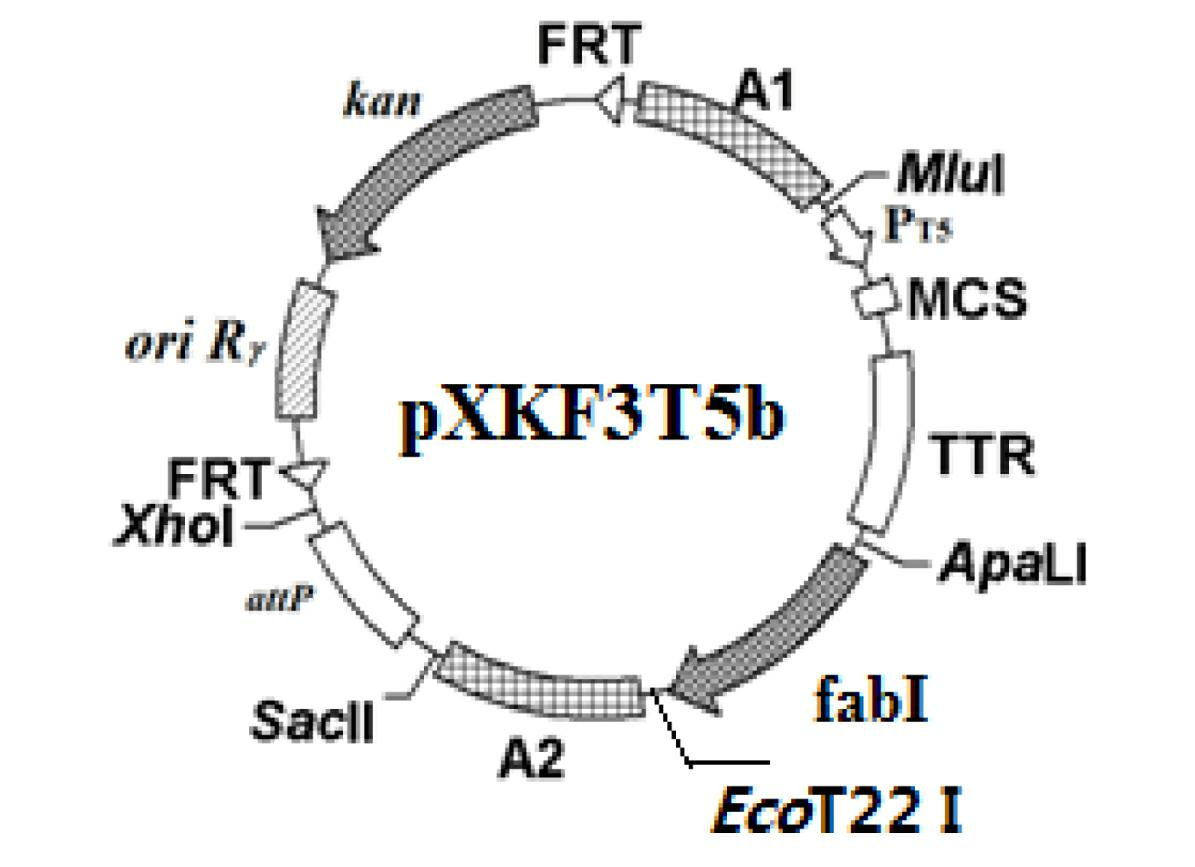
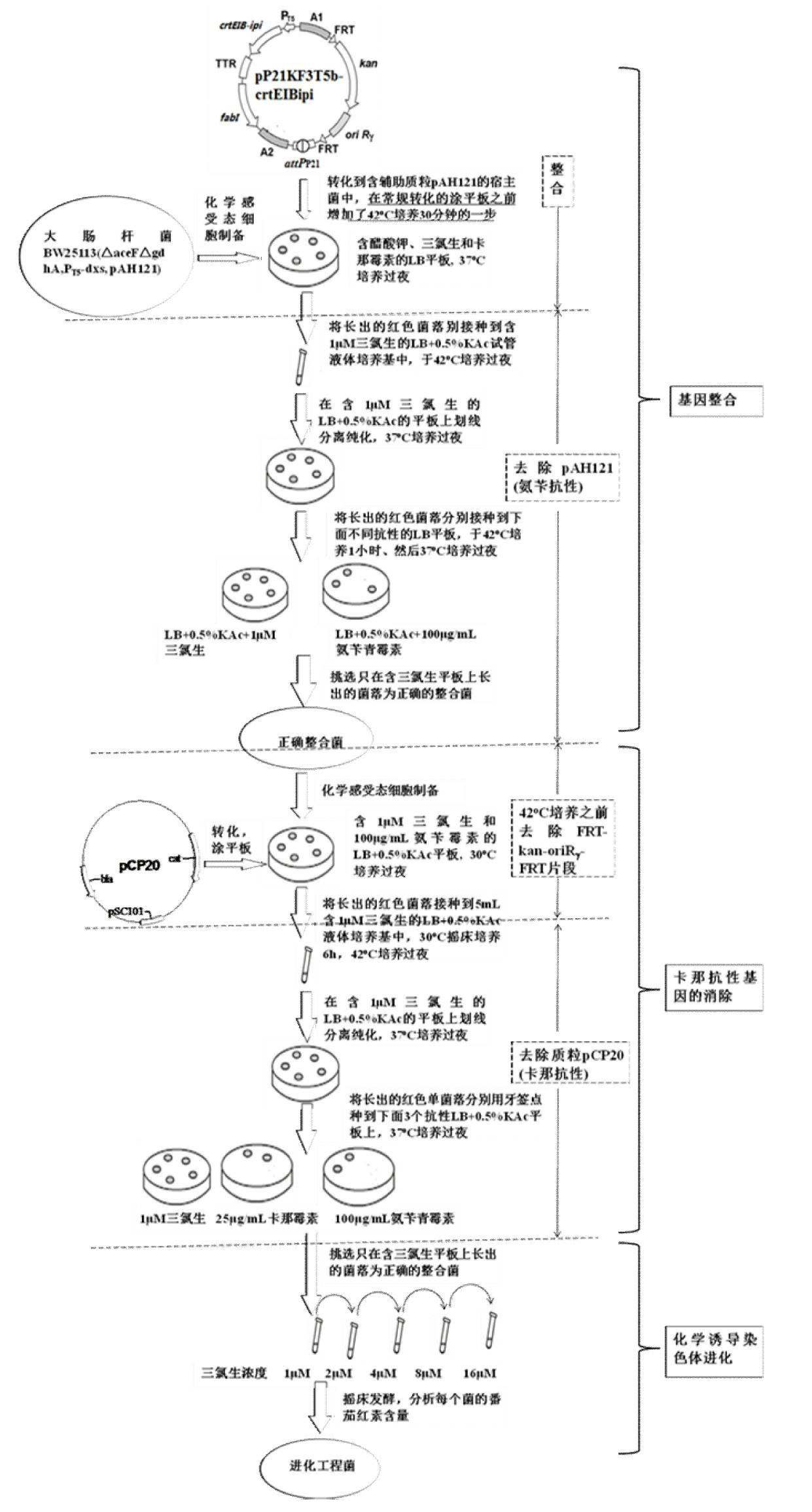
The invention discloses a construction method of a gene engineering strain without plasmid and antibiotic resistance screening marker. A pXKF3T5b plasmid is used as the carrier to transform a target gene into a host cell, and then an auxiliary plasmid for expressing the integrase is removed, a kanamycin resistance gene in the integrated carrier is removed, the chromosome is induced by the triclosan to evolve and the evolved strain is fermented and screened, so as to obtain the gene engineering strain without plasmid but with high gene copy number; moreover, a screen marker of the pXKF3T5b plasmid is the fabI gene (for coding enoyl-acetyl carrier protein reductase) in Escherichia coli and the fabI gene is the triclosan resistance gene, thus the obtained gene engineering strain does not have the antibiotic resistance screening marker, furthermore can not cause the spread of the antibiotics drug-resistant bacteria in the environment and can be suitable for the industrialized production. In addition, the target gene of the gene engineering strain is integrated into the chromosome, thus the gene engineering strain can not be lost during sub cultures, the genetic stability is high, and the production performance is stable.
Owner:刘紫琦
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com