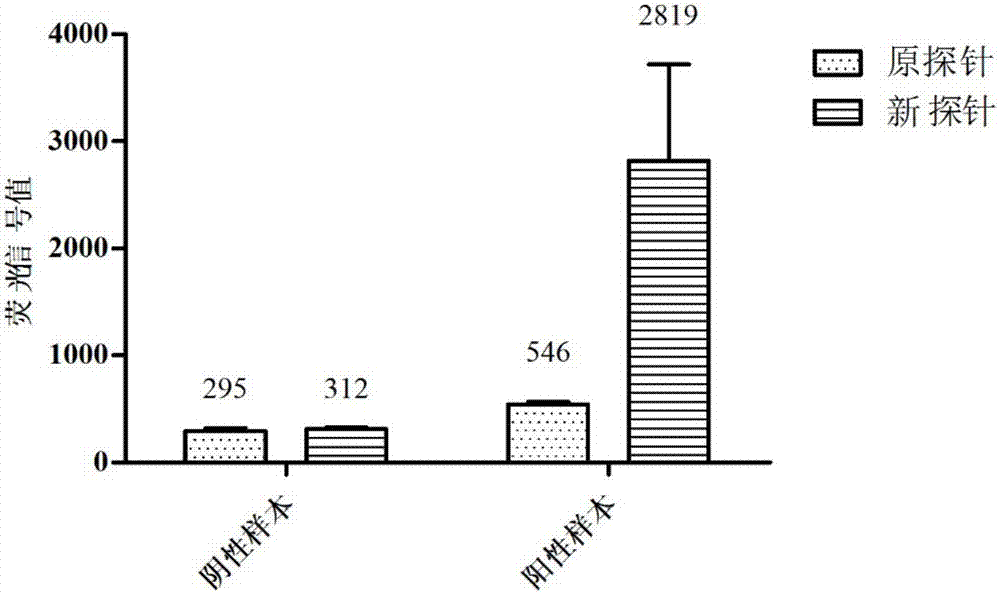
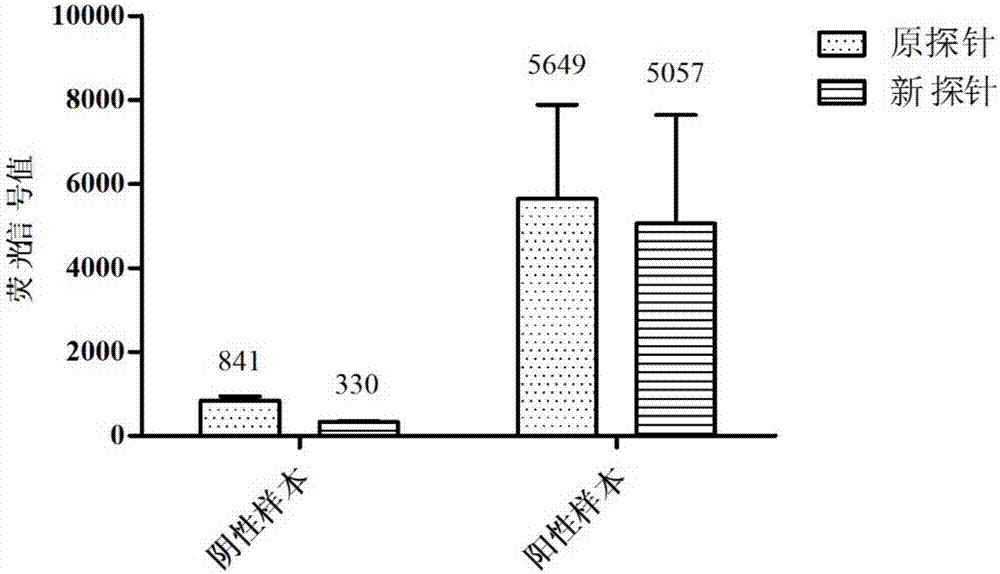
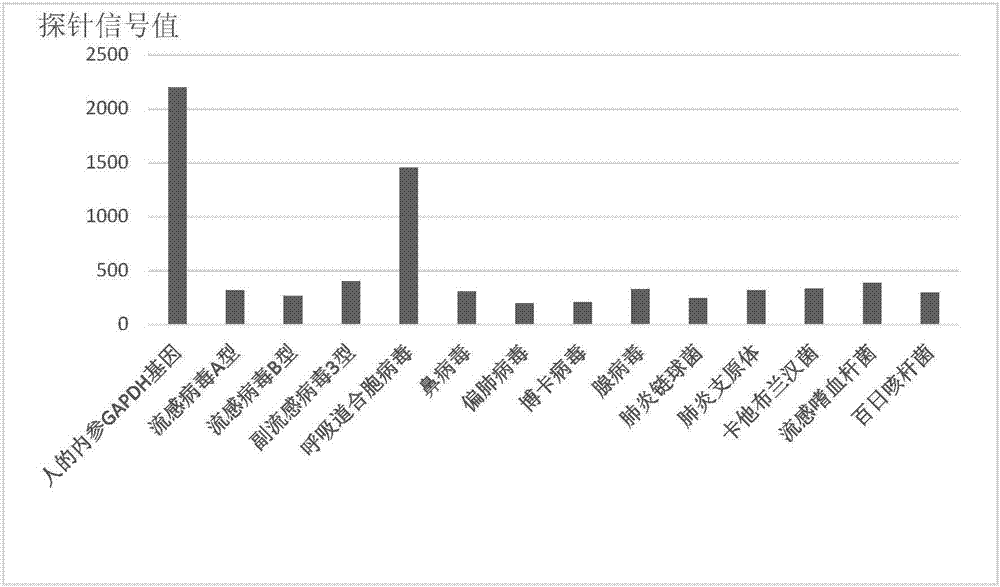
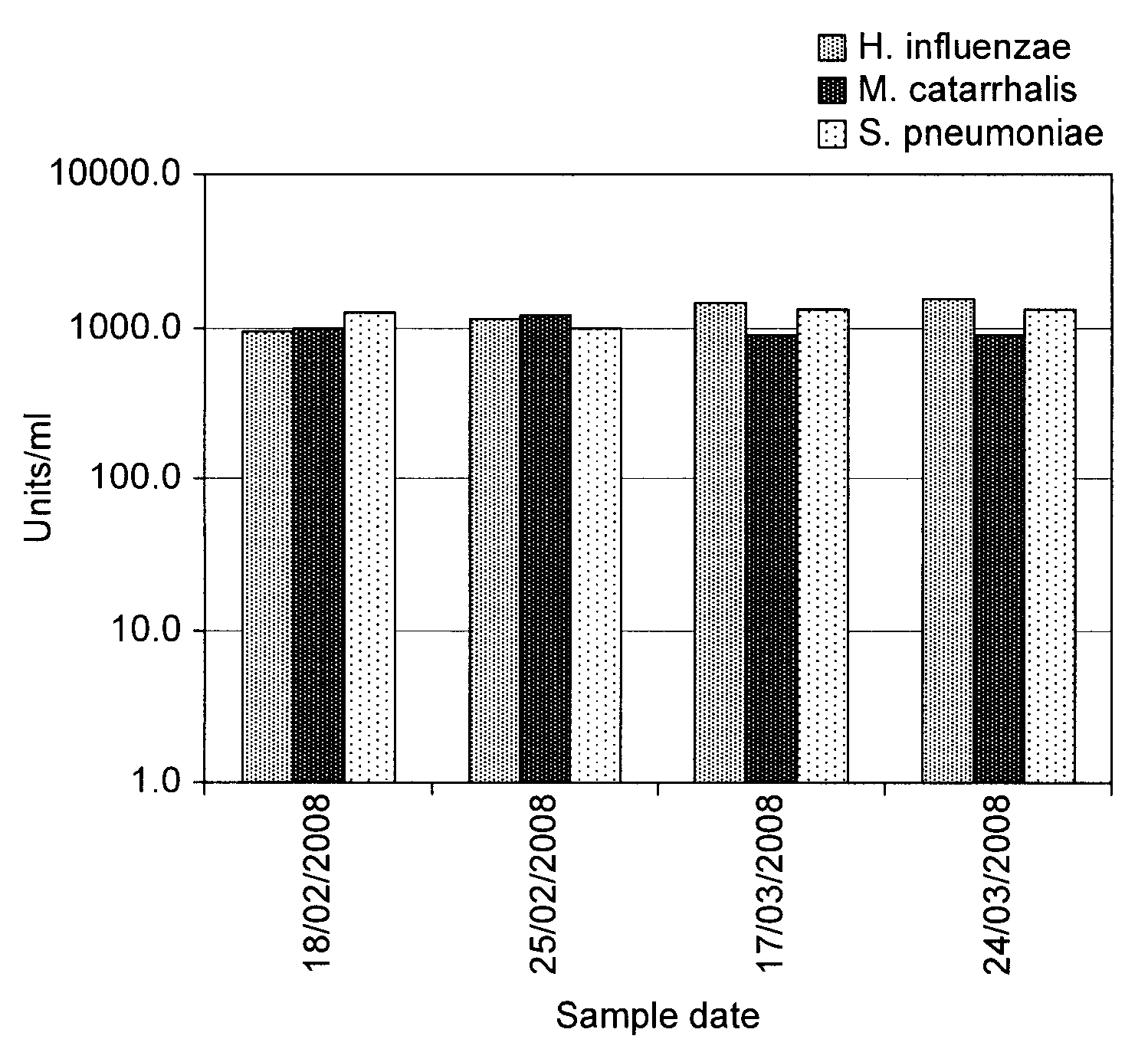
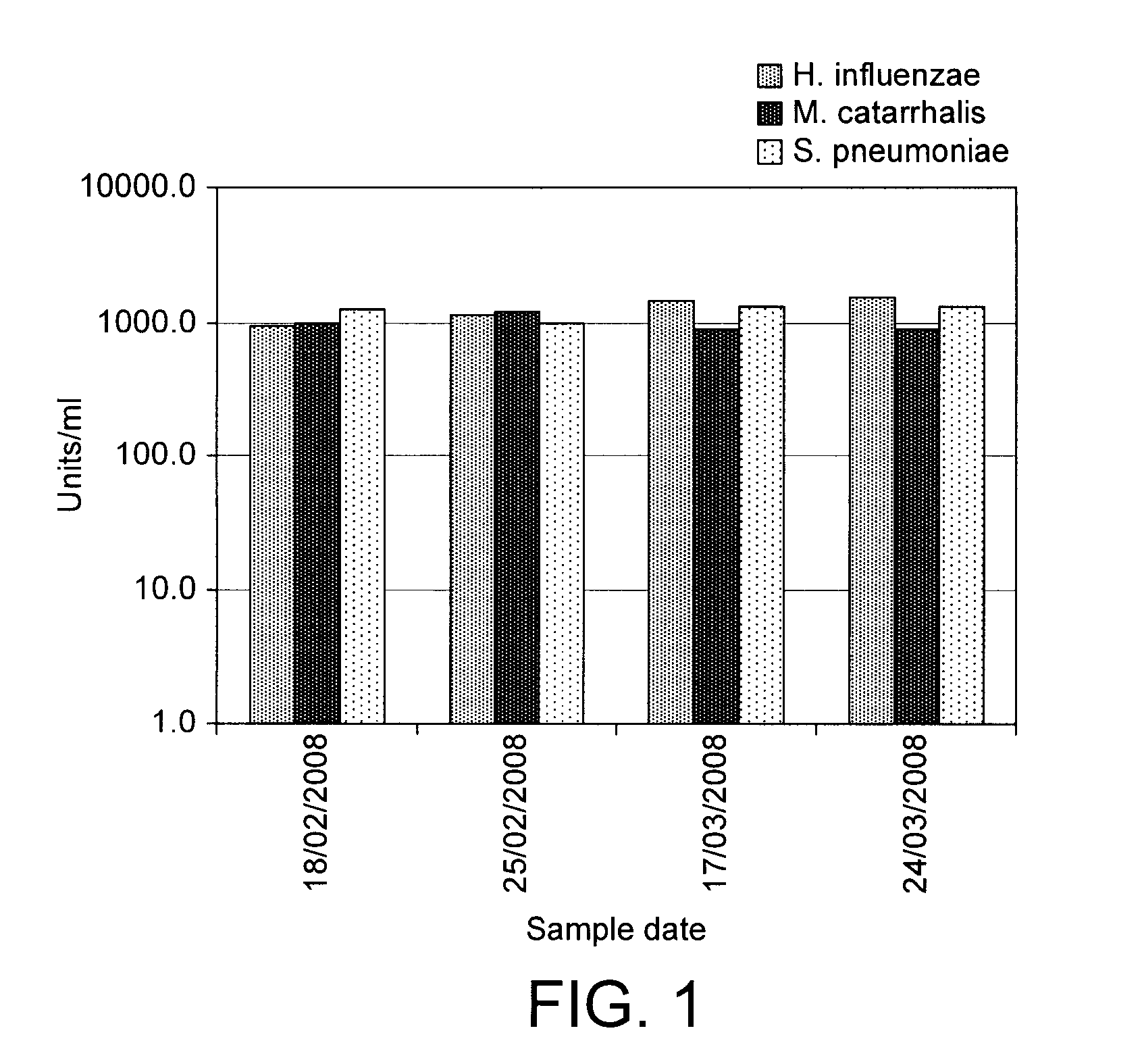
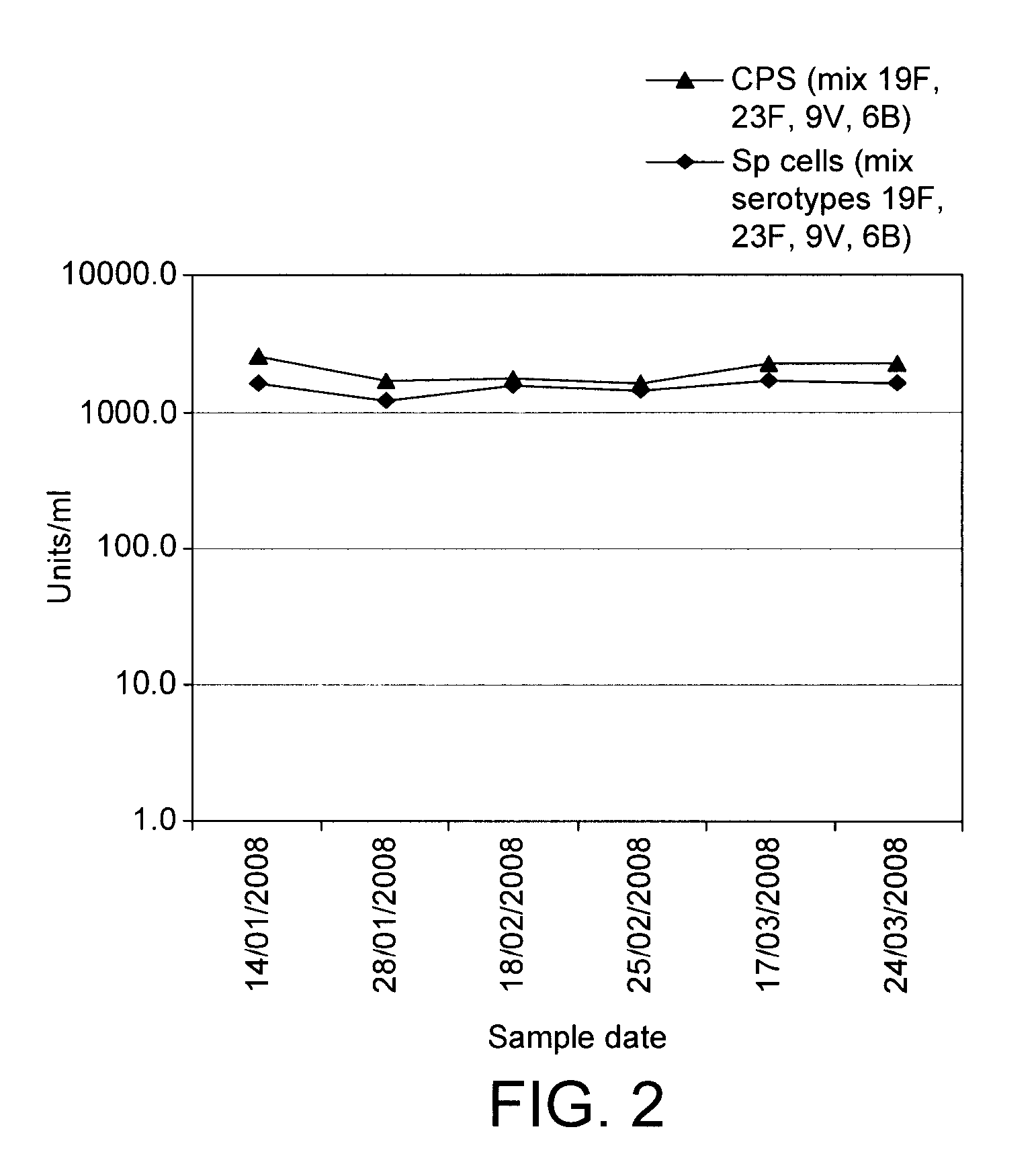
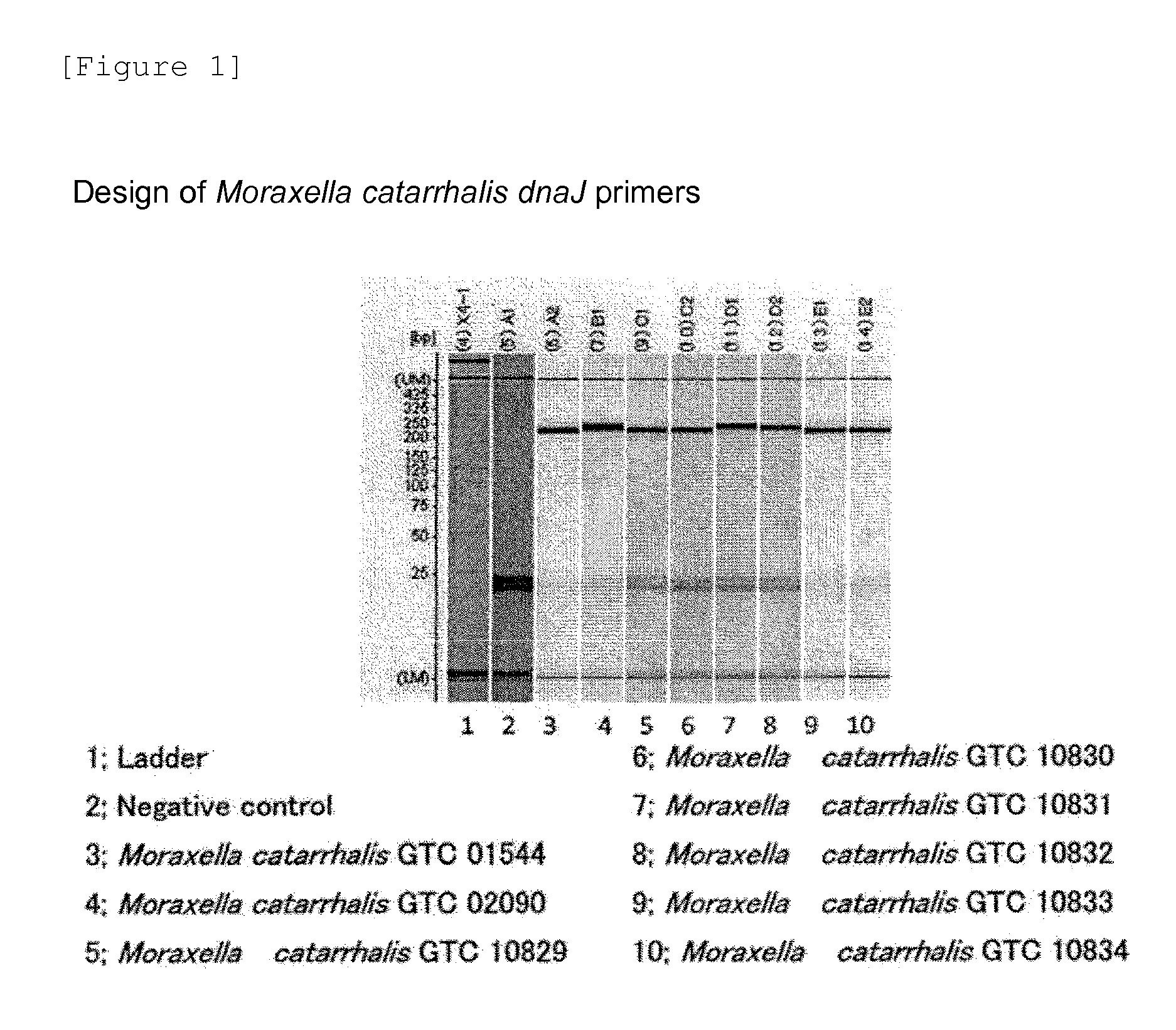
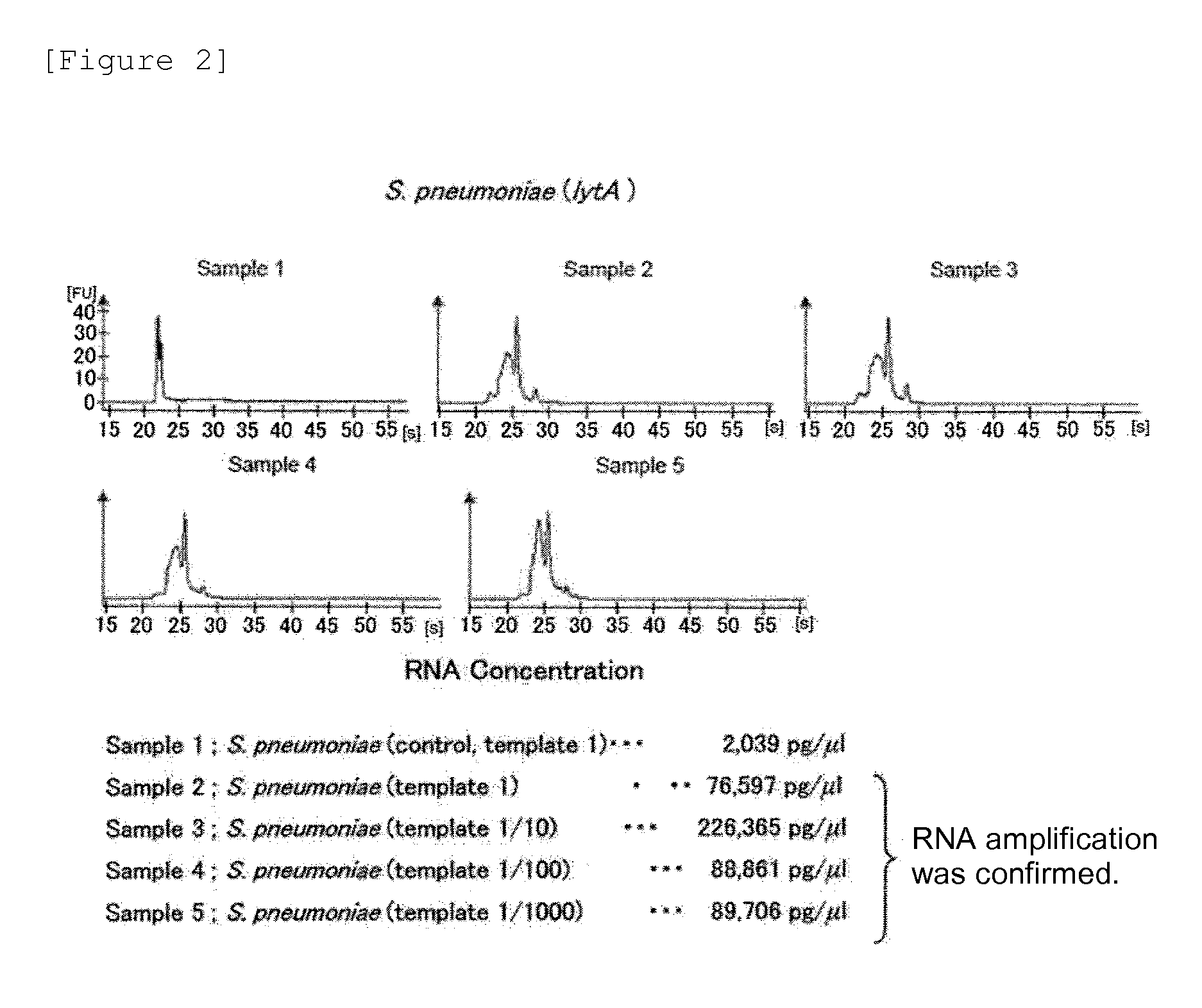
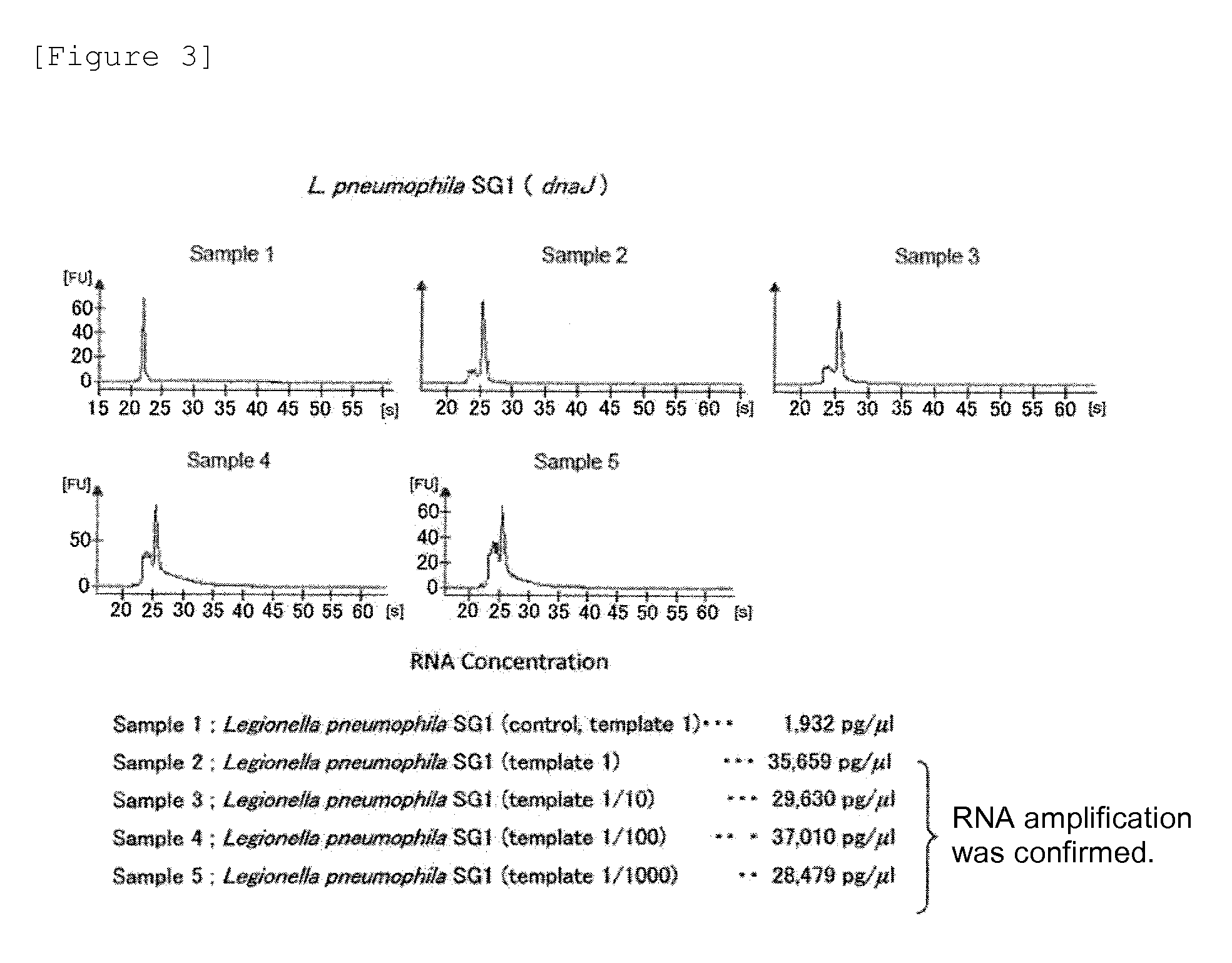
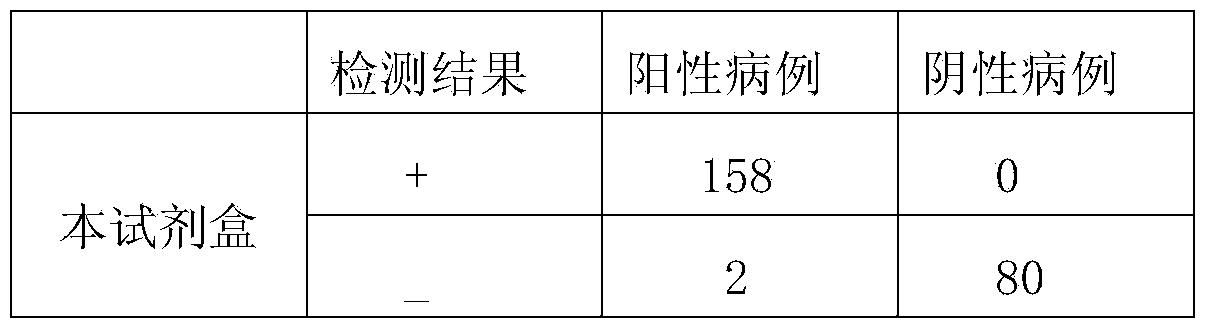
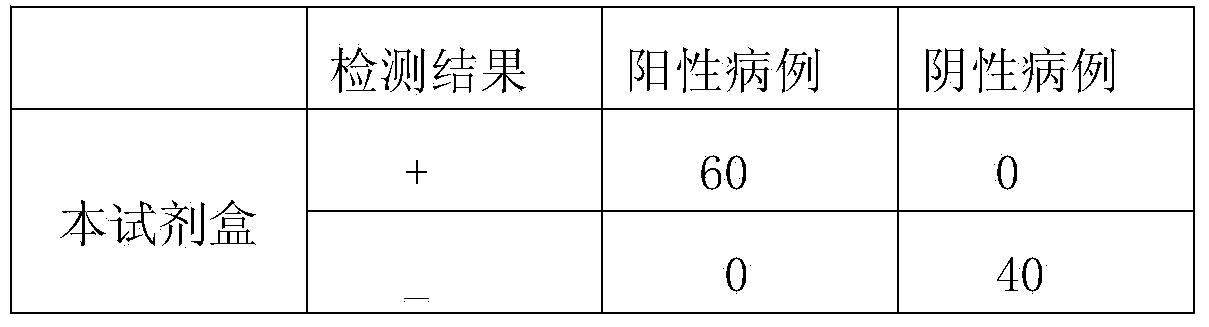
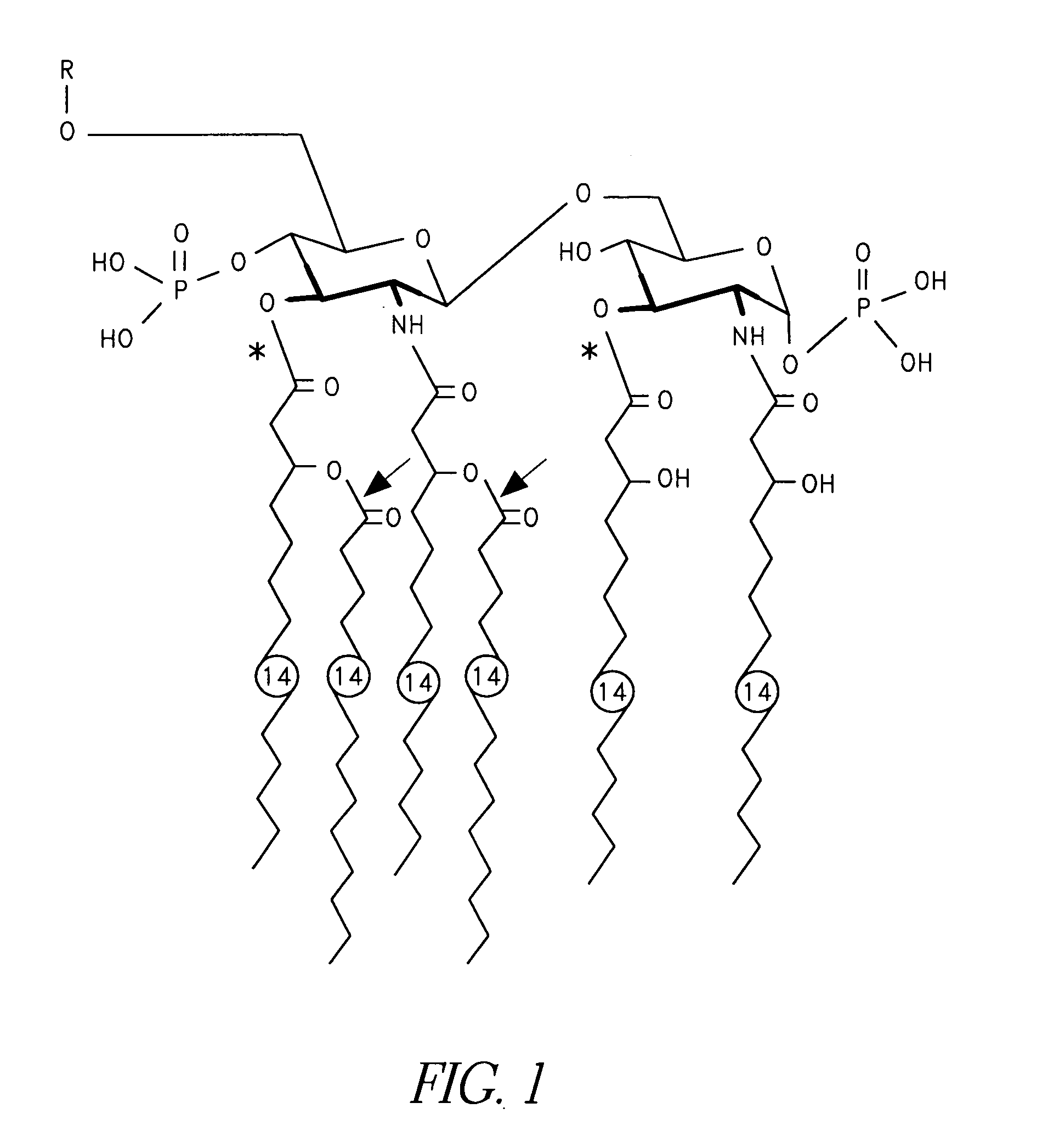
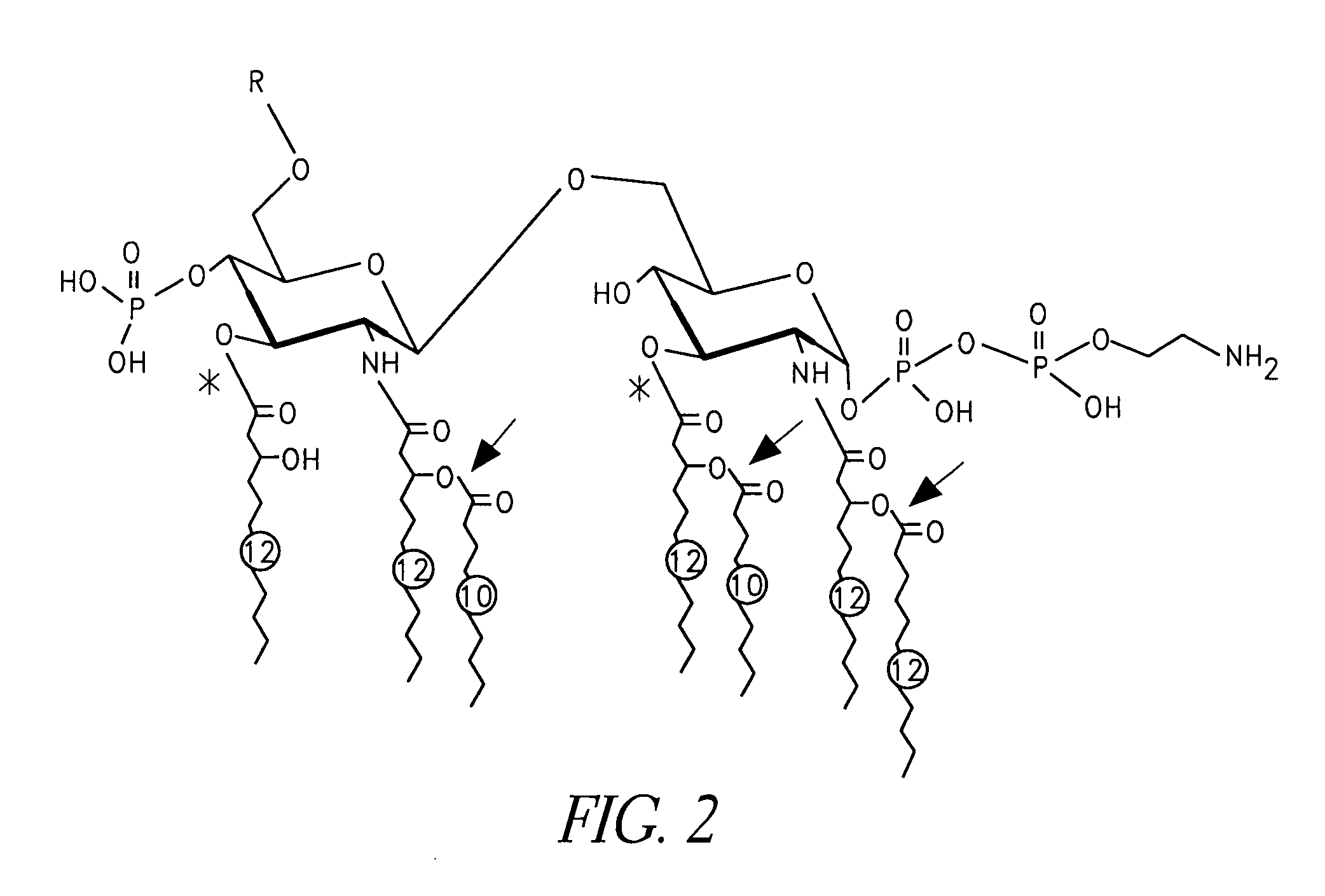
Patents
Literature
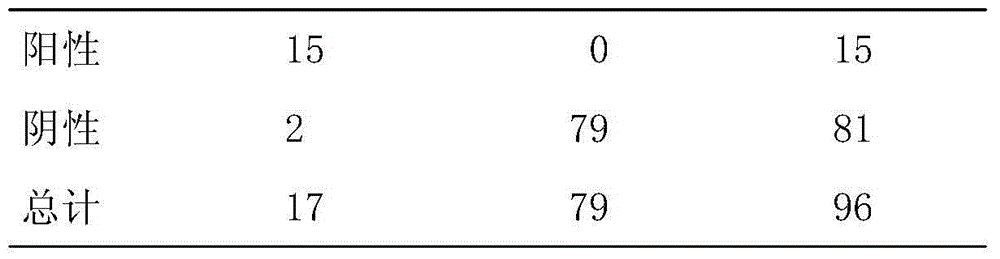
69 results about "Moraxella catarrhalis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
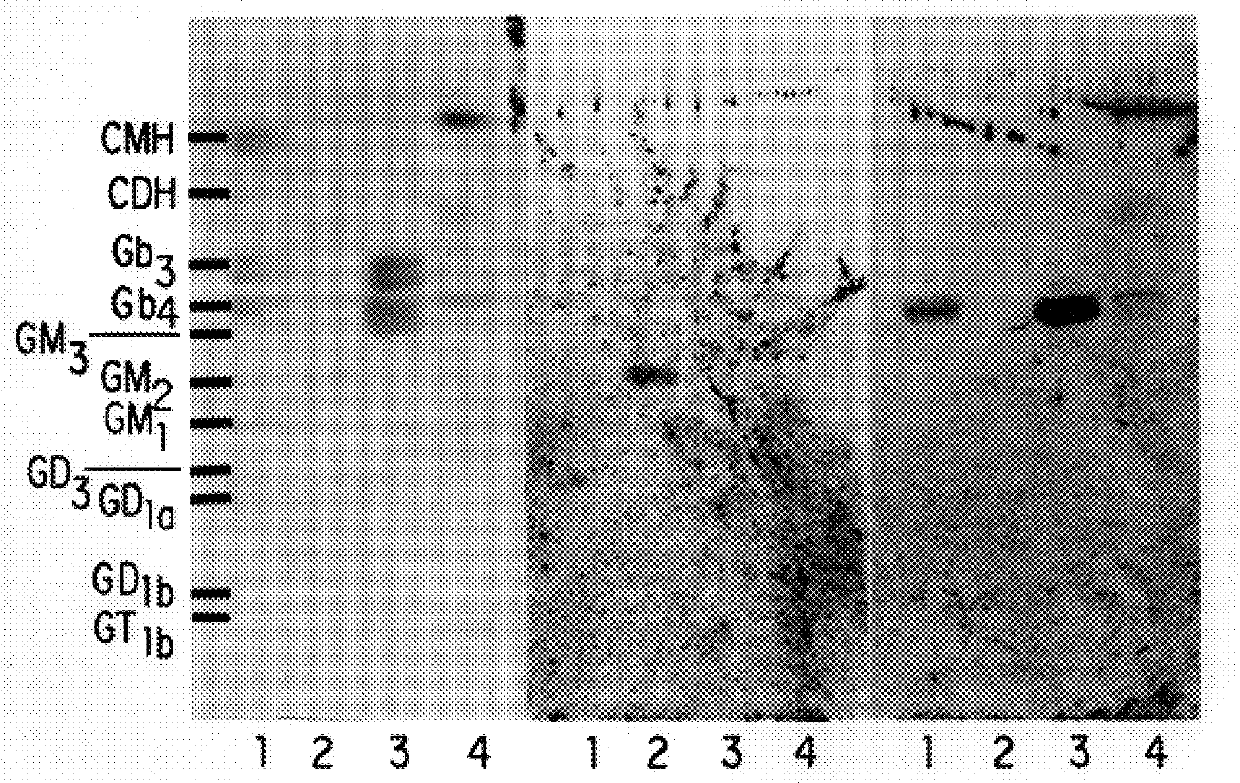
Moraxella catarrhalis is a fastidious, nonmotile, Gram-negative, aerobic, oxidase-positive diplococcus that can cause infections of the respiratory system, middle ear, eye, central nervous system, and joints of humans. It causes the infection of the host cell by sticking to the host cell using trimeric autotransporter adhesins.
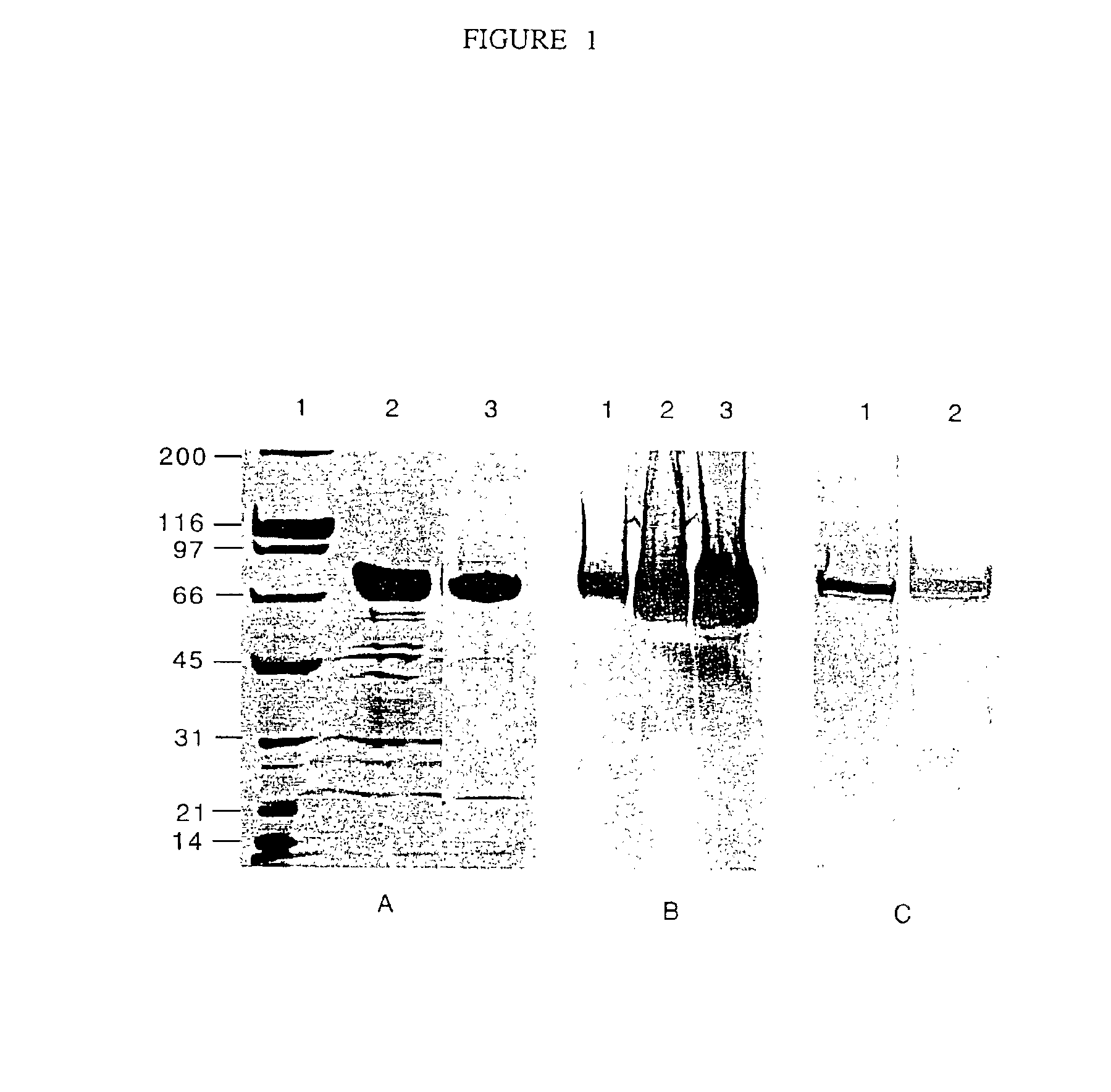
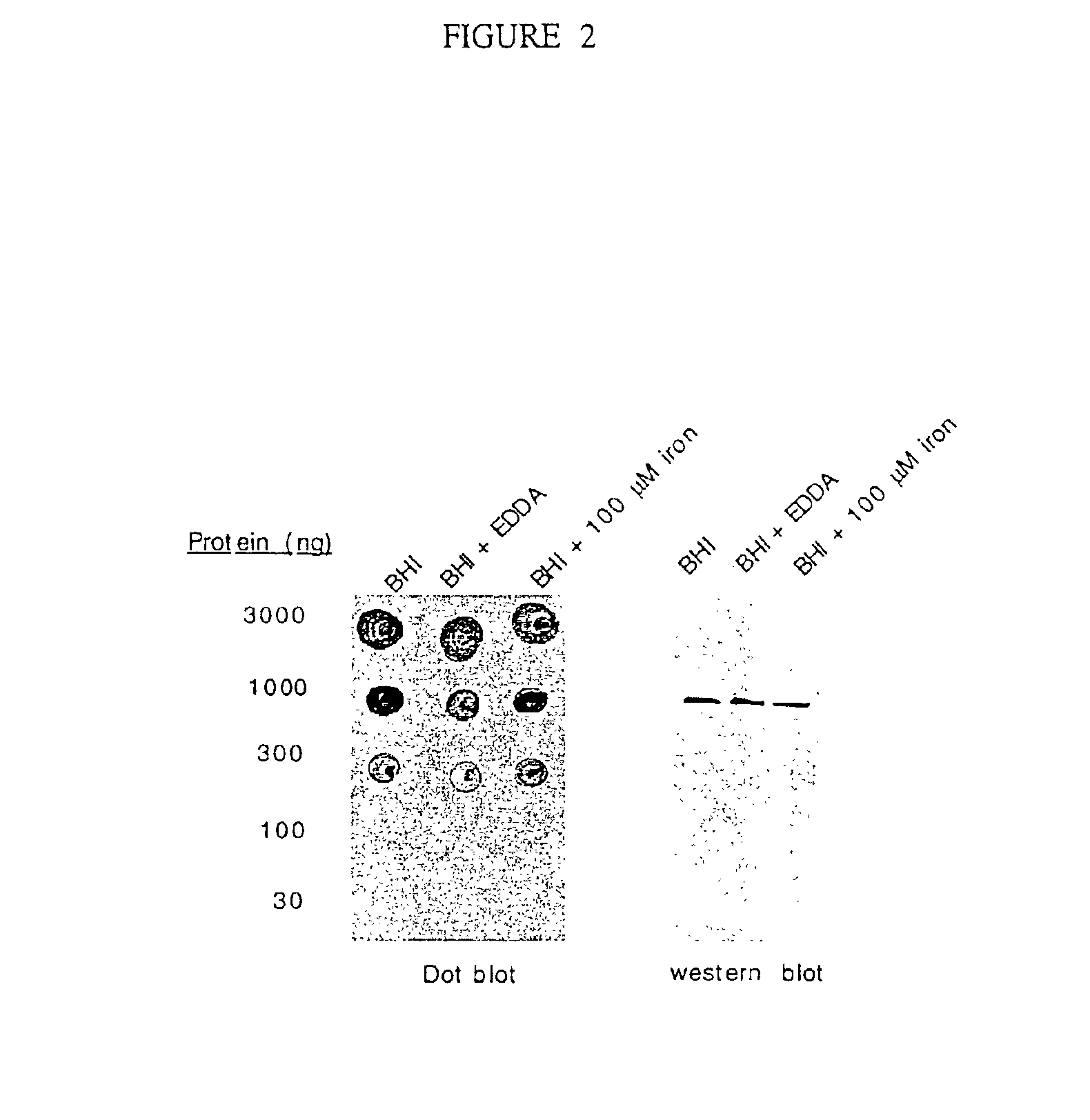
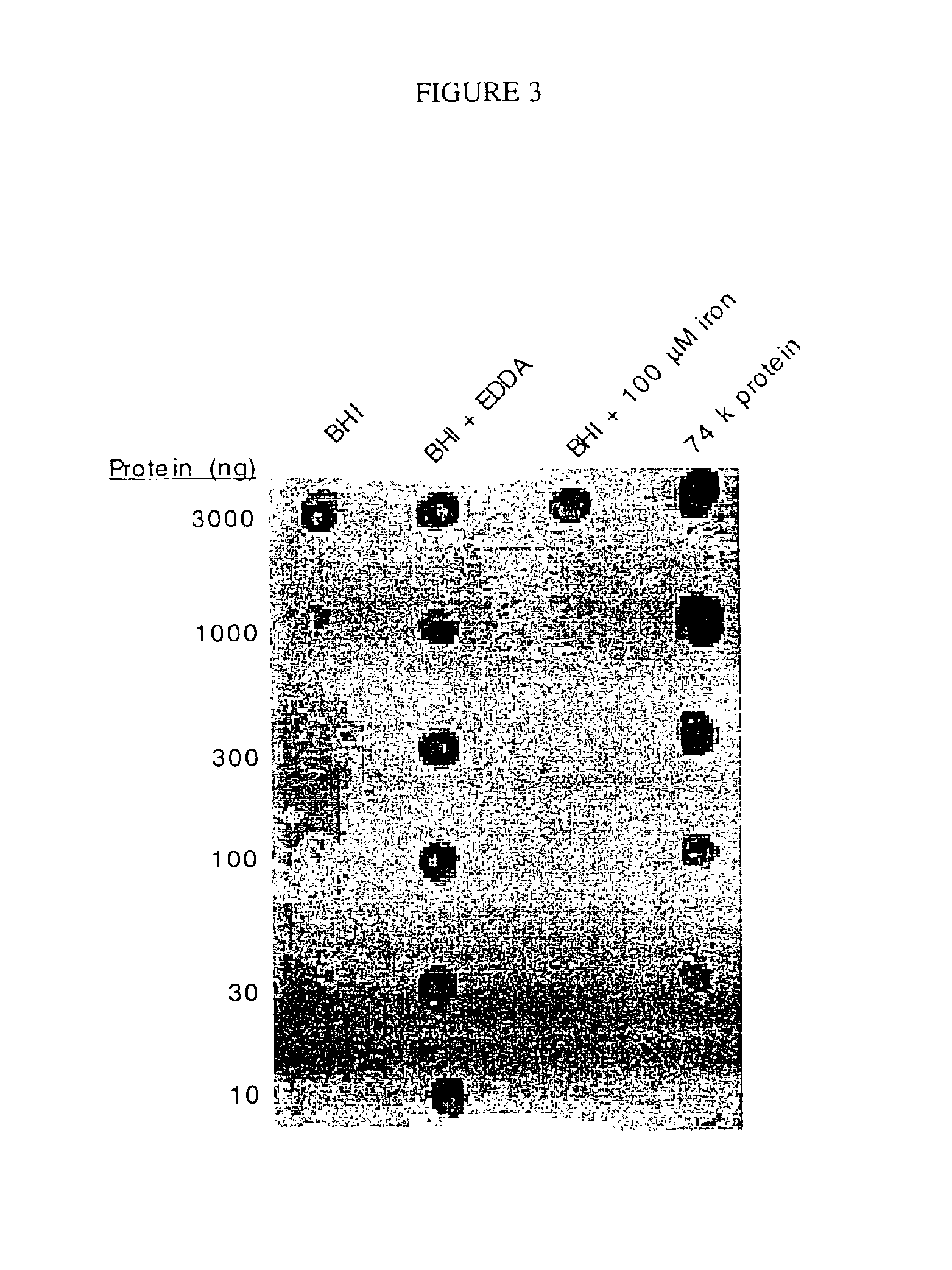
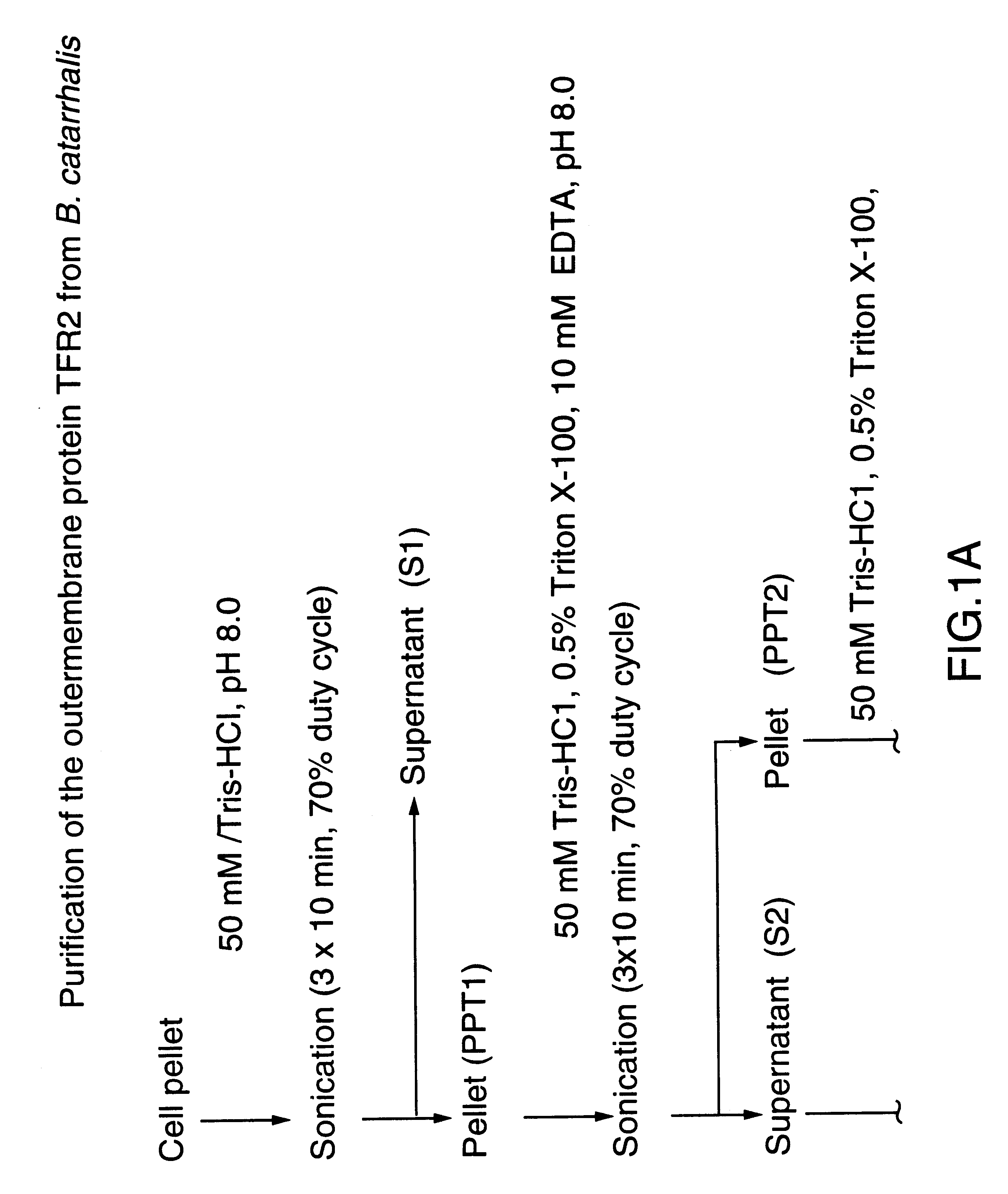
74-kilodalton outer membrane protein from Moraxella catarrhalis
InactiveUS6899885B1Enhance antibody responseLower titerBacterial antigen ingredientsBacteriaDiseaseKilodalton
A protein from M. catarrhalis, designated the 74 kD protein, is isolated and purified. The 74 kD protein has an amino-terminal amino acid sequence which is conserved among various strains of M. catarrhalis. The protein has a molecular weight of approximately 74.9 kD as measured on a 10% SDS-PAGE gel, while its molecular weight as measured by mass spectrometry is approximately 74 kD. The 74 kD protein is used to prepare a vaccine composition which elicits a protective immune response in a mammalian host, to protect the host against disease caused by M. catarrhalis.
Owner:WYETH HOLDINGS CORP
Moraxella catarrhalis outer membrane protein-106 polypeptide, gene sequence and uses thereof
The invention discloses the Moraxella catarrhalis outer membrane protein-106 (OMP106) polypeptide, polypeptides derived therefrom (OMP106-derived polypeptides), nucleotide sequences encoding said polypeptides, and antibodies that specifically bind the OMP106 polypeptide and / or OMP106-derived polypeptides. Also disclosed are immunogenic, prophylactic or therapeutic compositions, including vaccines, comprising OMP106 polypeptide and / or OMP106-derived polypeptides. The invention additionally discloses methods of inducing immune responses to M. catarrhalis and M. catarrhalis OMP106 polypeptides and OMP106-derived polypeptides in animals.
Owner:EMERGENT PROD DEV GAITHERSBURG INC
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20050042606A9Rapid bacterial identificationShorten the timeMicrobiological testing/measurementDepsipeptidesGenomic SegmentMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
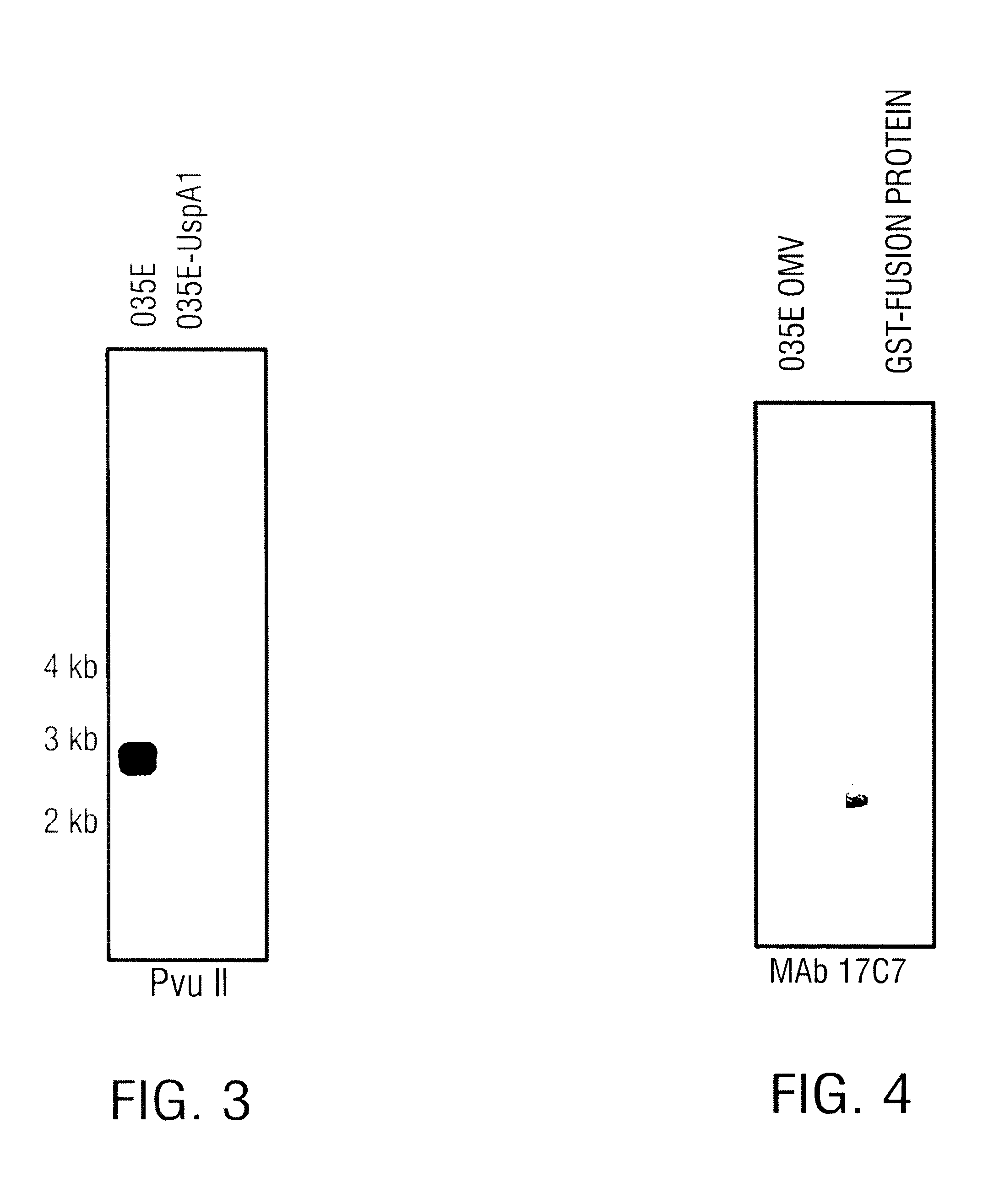
USPA1 and USPA2 antigens of Moraxella catarrhalis
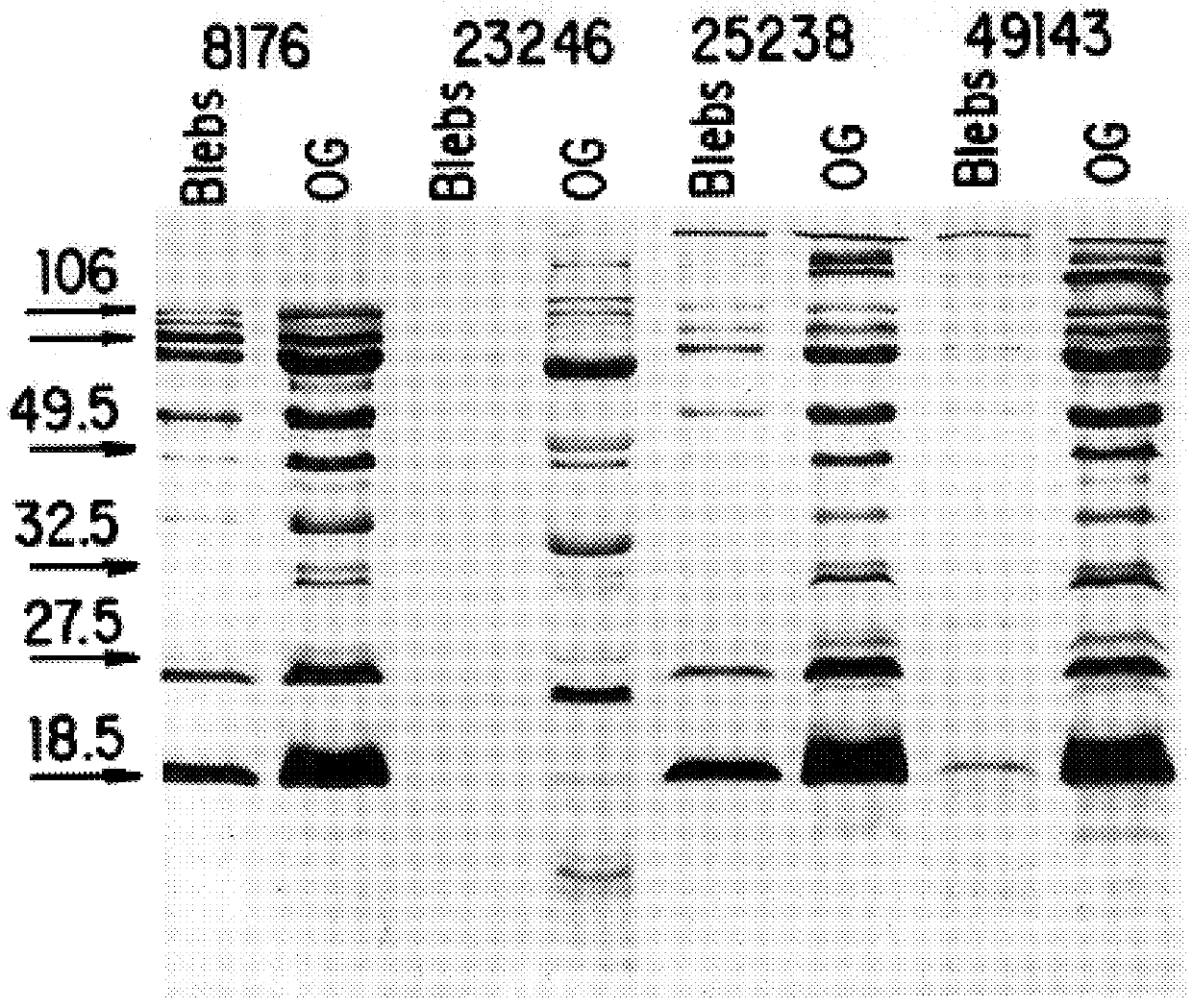
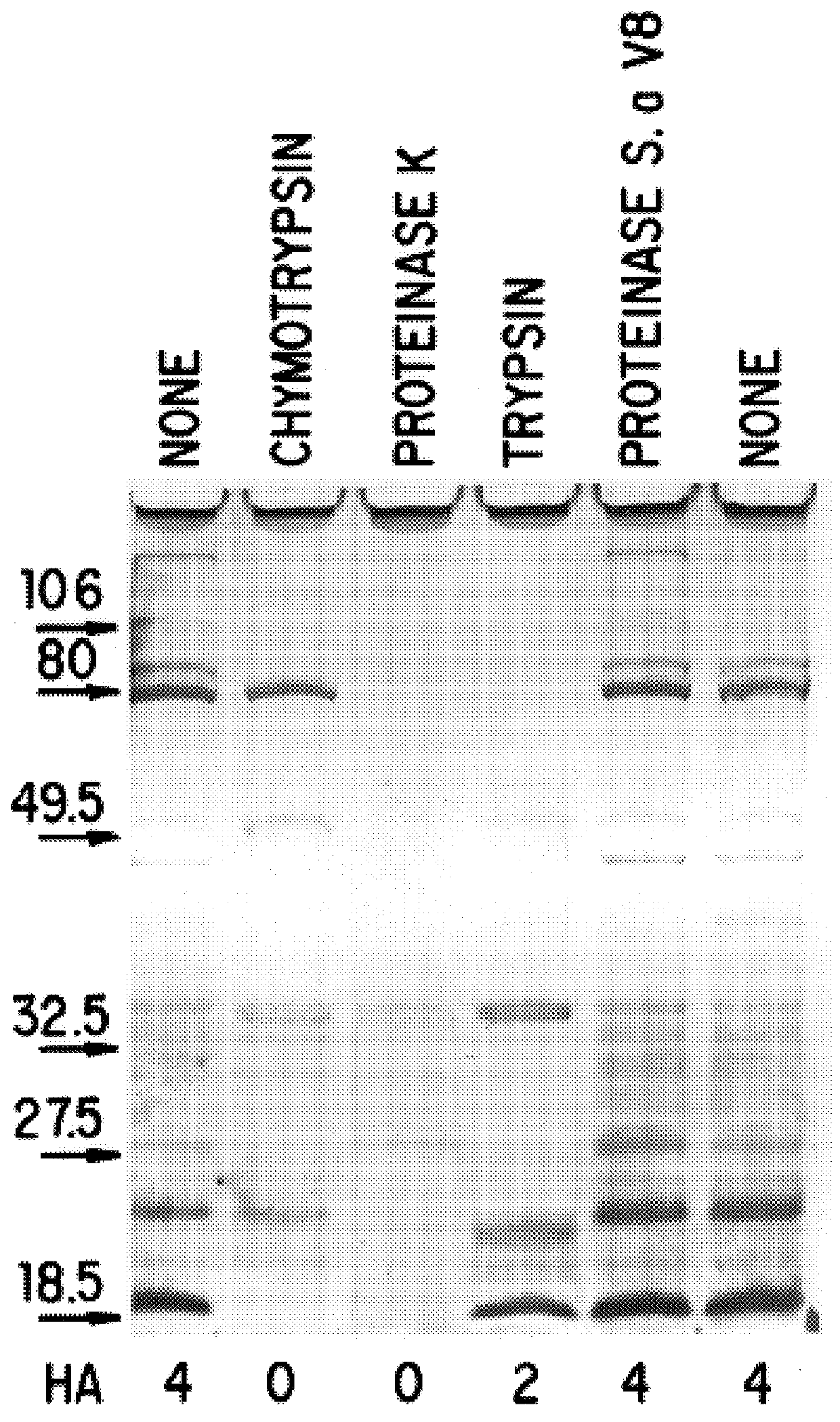
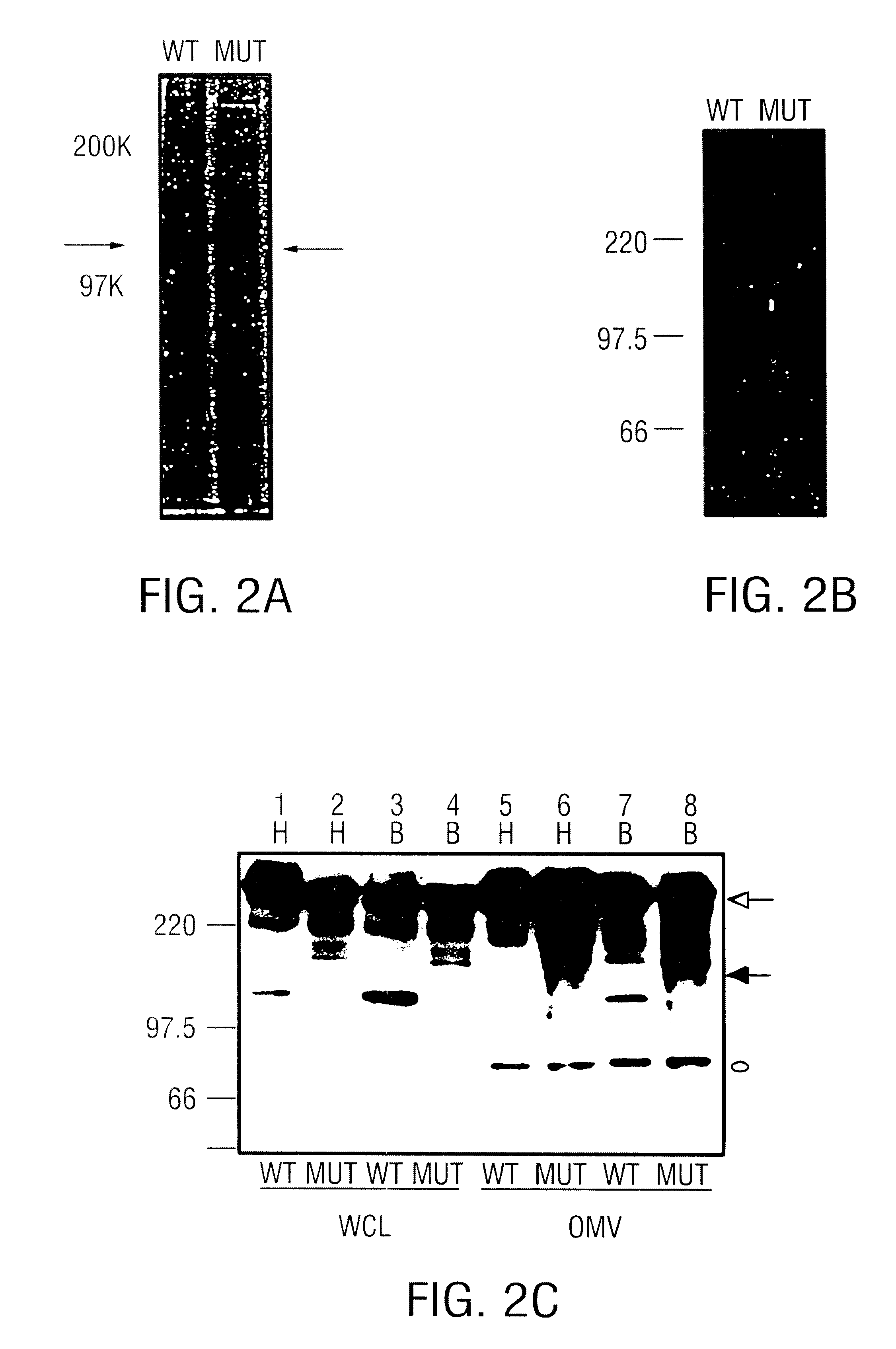
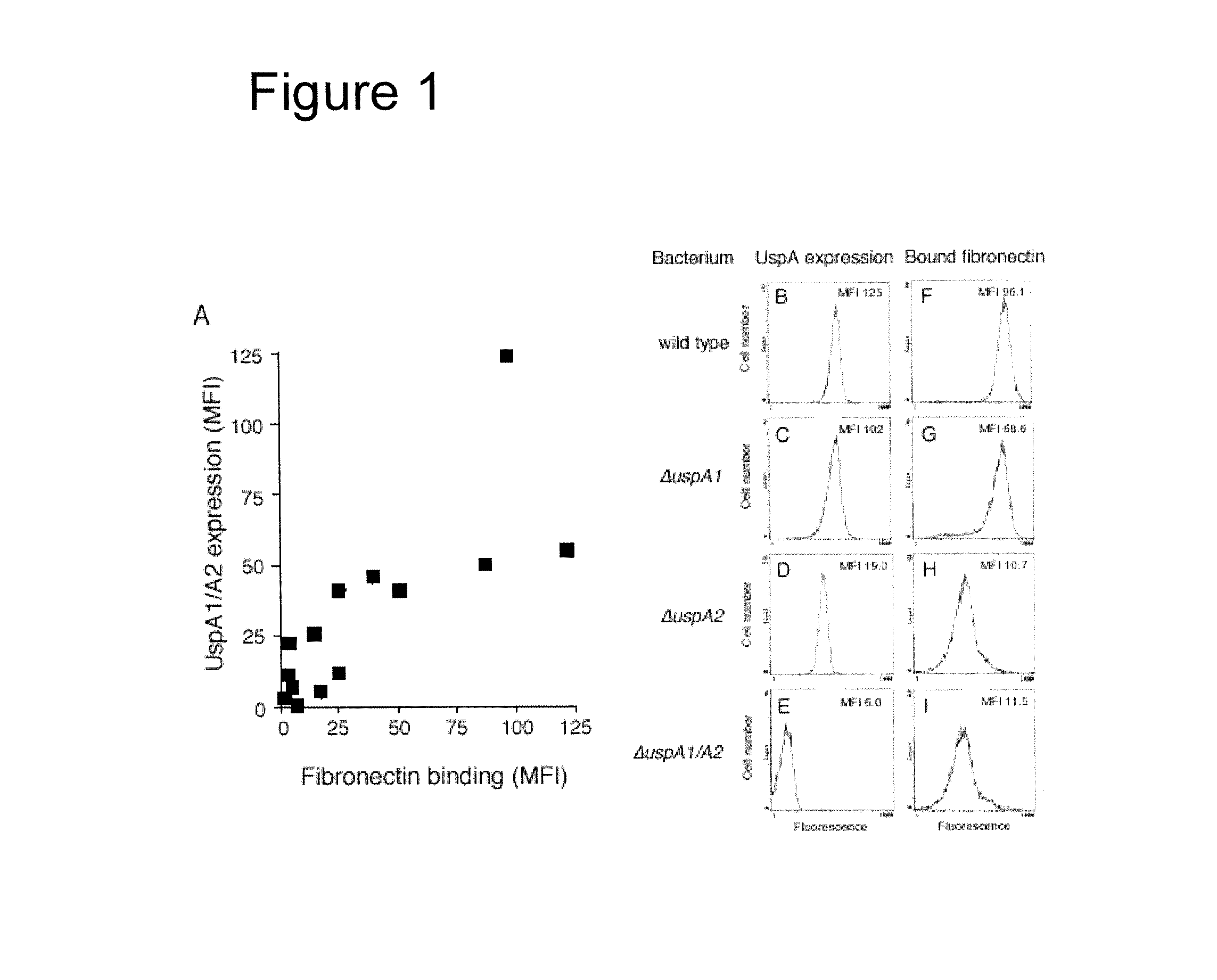
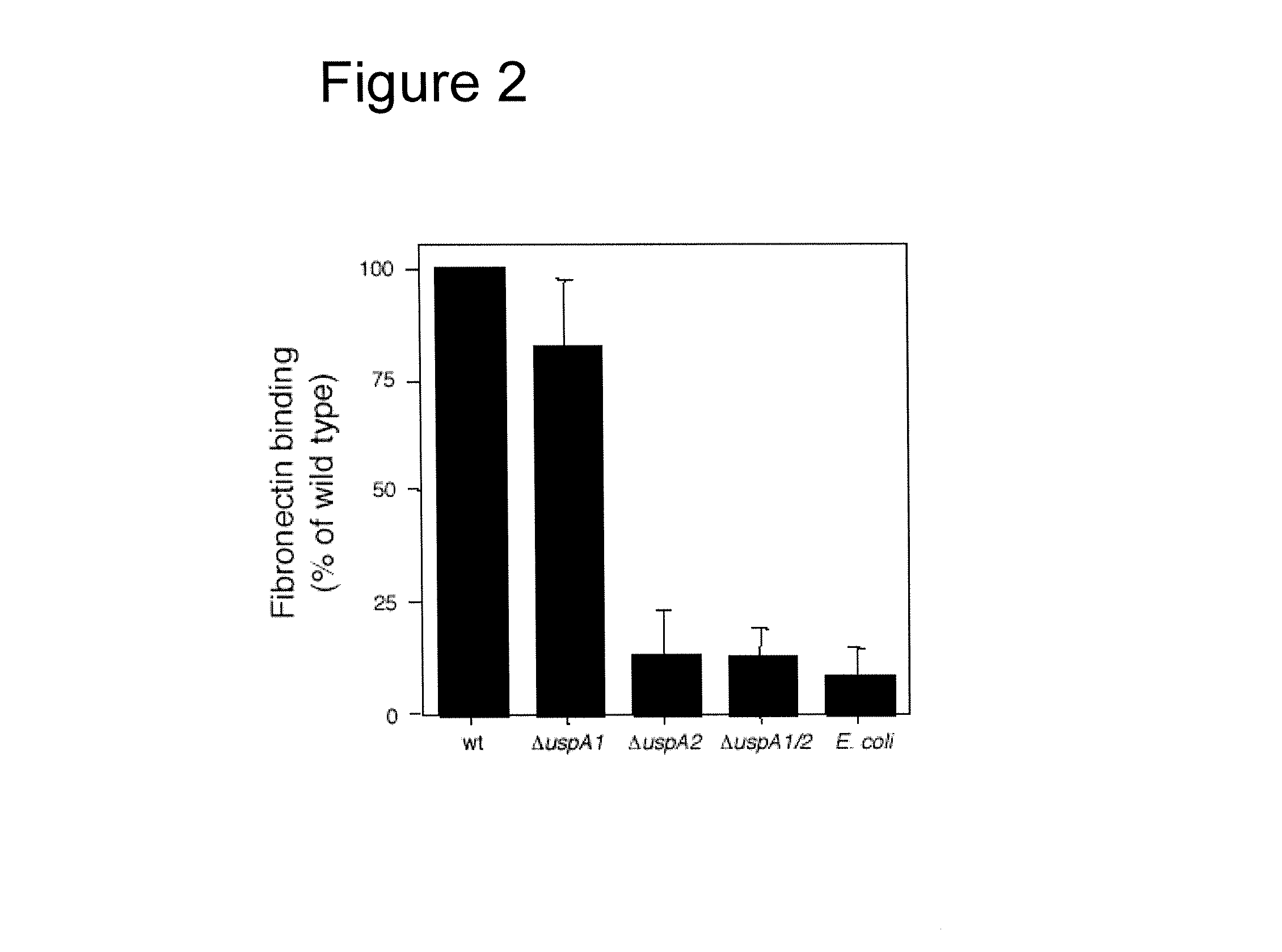
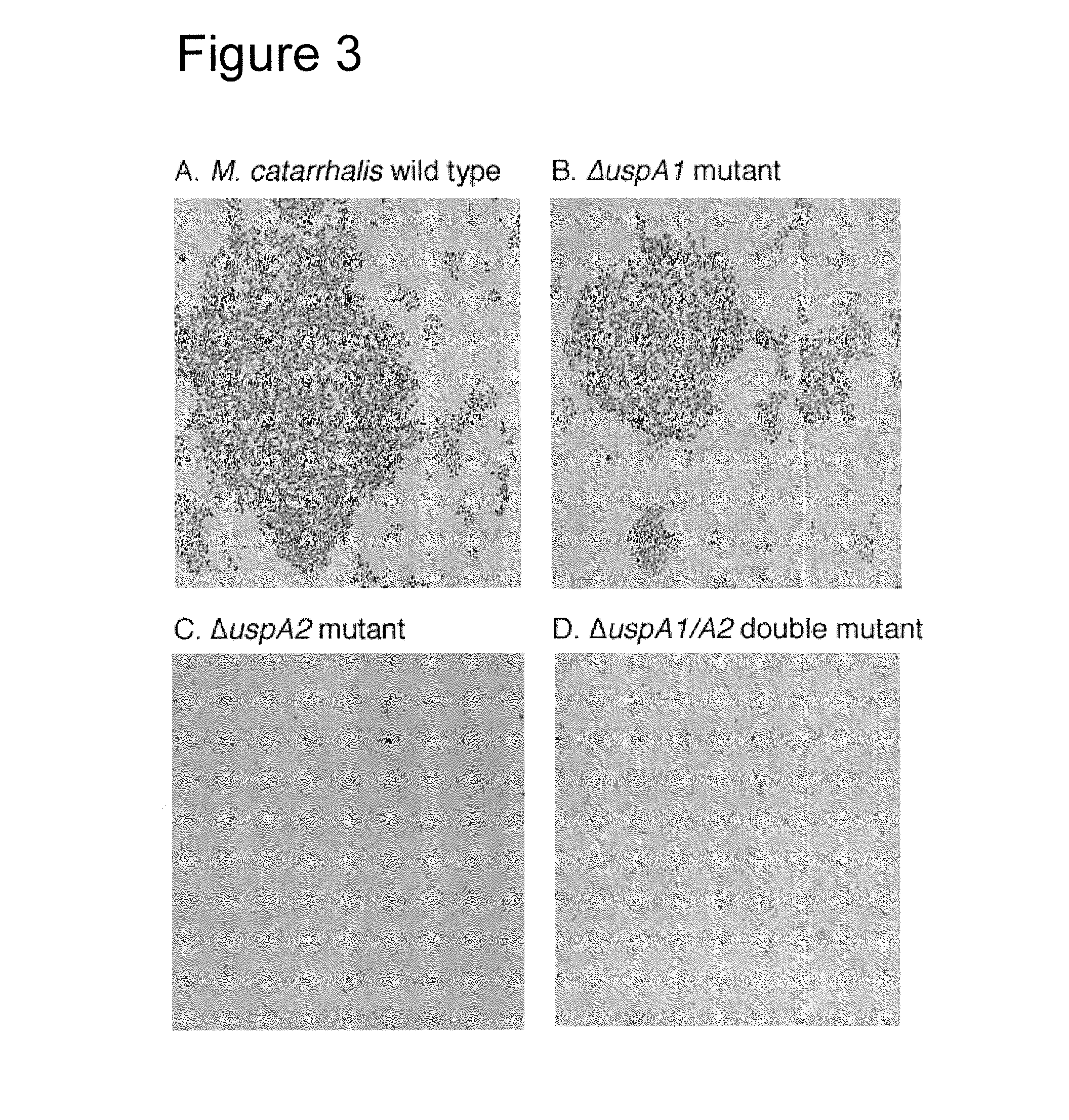
The present invention discloses the existence of two novel proteins UspA1 and UspA2, and their respective genes uspA1 and uspA2. Each protein encompasses a region that is conserved between the two proteins and comprises an epitope that is recognized by the MAb 17C7. One or more than one of these species may aggregate to form the very high molecular weight form (i.e. greater than 200 kDa) of the UspA antigen. Compositions and both diagnostic and therapeutic methods for the treatment and study of M. catarrhalis are disclosed.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST +1
Removal of bacterial endotoxin in a protein solution by immobilized metal affinity chromatography
InactiveUS6942802B2Ion-exchange process apparatusIon-exchanger regenerationProtein solutionBacteroides
The present invention relates to the purification of polypeptides and the removal of endotoxin via immobilized metal affinity chromatography (IMAC). More specifically, the invention relates to methods for removing bacterial endotoxin in a protein solution. In specific embodiments, the invention relates to the elimination of endotoxin from Moraxella catarrhalis outer membrane proteins.
Owner:WYETH HOLDINGS CORP
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20070009947A1Rapid and exponential in vitro replicationQuick identificationMicrobiological testing/measurementDepsipeptidesBacteroidesMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20090053702A1Rapid and exponential in vitro replicationQuick identificationMicrobiological testing/measurementMicroorganism based processesBacteroidesStreptococcus pyogenes
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony.
Owner:GENEOHM SCI CANADA
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20090047671A1Rapid bacterial identificationShorten the timeMicrobiological testing/measurementMicroorganism based processesBacteroidesMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
Nucleic acids encoding 3-ketoacyl-ACP reductase from Moraxella catarrahalis
InactiveUS6632636B1Extended half-lifeIncrease intracellular stabilitySugar derivativesBacteriaPDAT enzymeOpen reading frame
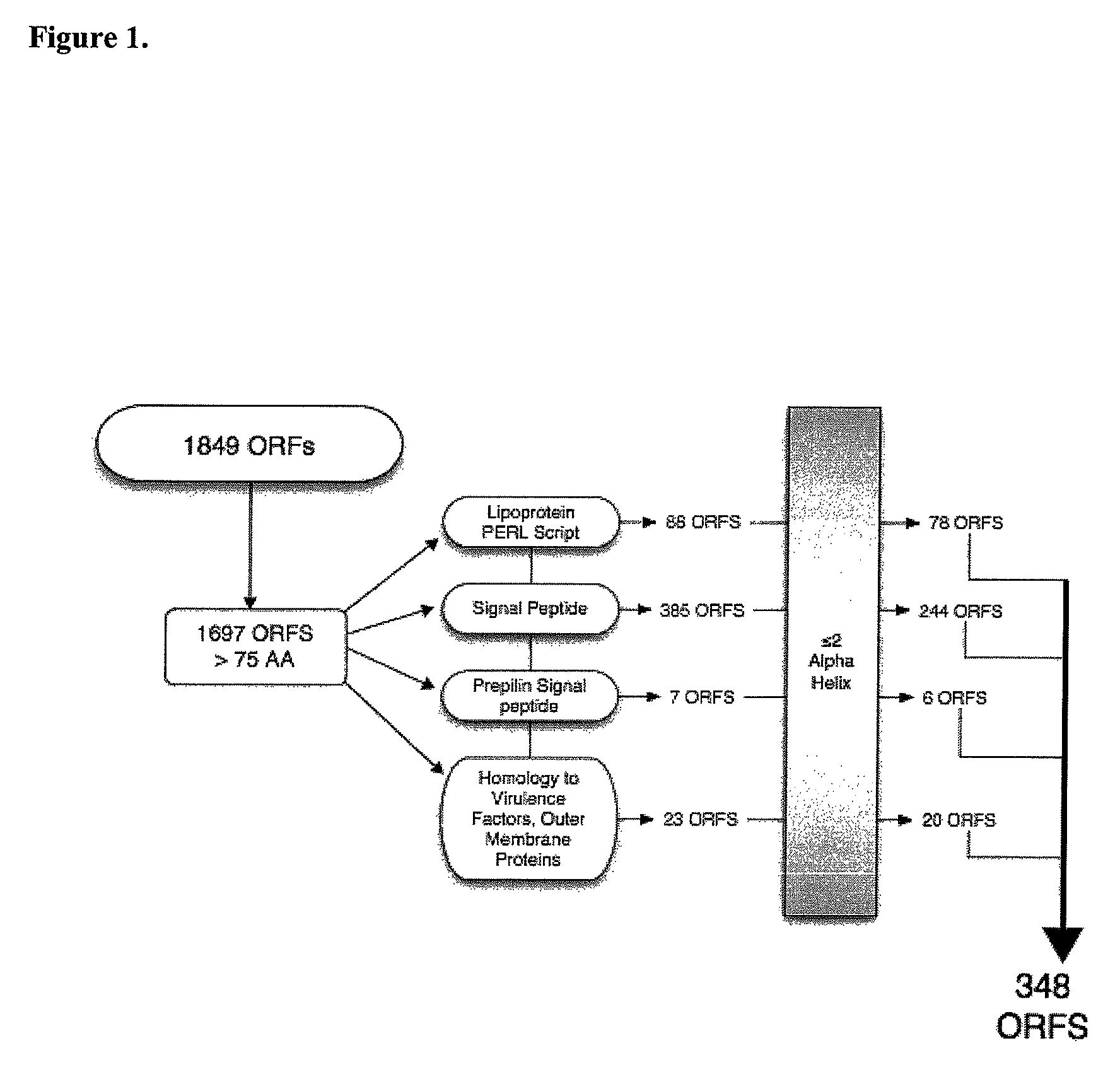
The present invention provides the genomic sequences of a library of purified nucleic acid molecules, or their complements, comprising the genome of <HIL><PDAT>Moraxella catarrhalis< / ITALIC><PDAT>. The invention also provides the identification of open reading frames contained within the nucleic acid molecules of the library. The present invention further provides for the use of the nucleic acid molecules, their complements or fragments, and proteins or portions thereof for identifying ligands and useful diagnostic and therapeutic compositions. In addition the invention provides for vectors, host cells and methods for producing <HIL><PDAT>M. catarrhalis < / ITALIC><PDAT>proteins or portions thereof.< / PTEXT>
Owner:MERCK & CO INC
Gene chip for detecting pathogens of lower respiratory tract and reagent kit
ActiveCN101748193AImprove accuracyGood repeatabilityMicrobiological testing/measurementAgainst vector-borne diseasesK pneumoniaeStaphylococcus aureus
The invention provides a gene chip for detecting pathogens of lower respiratory tract, which comprises a solid phase carrier and an oligonucleotide probe fixed on the solid phase carrier, wherein the oligonucleotide probe comprises a sequence selected from one or more DNA sequences of staphylococcus aureus, 16s-23s rDNA intergenic spacer region of Klebsiella pneumoniae or Acinetobacter baumannii, 16S gene of pseudomonas aeruginosa or haemophilus influenzae, gyrB gene of streptococcus pneumoniae or legionella pneumophila and copB gene of moraxella catarrhalis or the complementary DNA or RNA sequence. The invention further provides a reagent kit containing the gene chip. The utilization of the gene chip and the reagent kit can detect the pathogens of the lower respiratory tract; furthermore, the operation is simple, the accuracy is high and the repeatability is strong.
Owner:TIANJIN BIOCHIP TECH CO LTD
Prevention and Treatment of Otitis Media with Non-Pathogenic Bacterial Strains
InactiveUS20090214594A1Effective preventionEffective treatmentSenses disorderBacterial antigen ingredientsBacteroidesBacterial strain
This invention relates to a composition comprising a bacterial strain capable of boosting the systemic immune response, a bacterial strain capable of exerting bacteriostatic effects on pathogens associated with development of otitis media such as Streptococcus pneumoniae, untypeable Haemophilus influenzae and Moraxella catarrhalis and a bacterial strain capable of exerting bacteriocidal effects on such pathogens. The invention further extends to the use of such a composition in the prevention or treatment of otitis media.
Owner:NESTEC SA
Method and kit for fast detection of moraxella catarrhalis based on magnetic resolution and quantum dot labelling
ActiveCN105203754AFast separationIt has the effect of synergistic amplification of multiple signalsFluorescence/phosphorescenceAntigenMagnetic bead
The invention provides a method for detection of moraxella catarrhalis based on magnetic resolution and quantum dot labelling. The method comprises the following steps: (1) preparing anti-moraxella catarrhalis nano immunomagnetic beads; (2) preparing quantum dot labelled anti-moraxella catarrhalis nanoprobes; (3) dissolving a sample to be detected with a PBST buffer solution, adding the anti-moraxella catarrhalis immune nano magnetic beads into the dissolved solution, carrying out magnetic separation after sufficient mixing and reaction, washing with the PBST buffer solution, adding the quantum dot labelled anti-moraxella catarrhalis nanoprobes into the obtained precipitate, carrying out magnetic separation after a reaction, washing with the PBST buffer solution, and detecting the fluorescence value with a fluorescence micro-plate reader. The method is accurate, fast, and high in sensitivity, and has very high practical value in the aspects of clinical diagnosis, etiological identification, epidemiological surveys and the like of moraxella catarrhalis.
Owner:湖北诺美华抗体药物技术有限公司
Administration of negamycin or deoxynegamycin for the treatment of bacterial infections
The invention provides a method for treating bacterial infections. In one aspect, the invention comprises orally administering a pharmaceutical composition to an animal, wherein the composition comprises a pharmaceutically acceptable excipient and an antibacterial effective amount of negamycin, or a pharmaceutically acceptable salt, prodrug or isomer thereof. An aspect of the invention also relates to a method of treating a bacterial infection, wherein the method comprises intravenously administering a pharmaceutical composition to an animal, and wherein the composition comprises a pharmaceutically acceptable excipient and an antibacterial effective amount of deoxynegamycin, or a pharmaceutically acceptable salt, prodrug or isomer thereof. An aspect of the invention also relates to a method of treating a bacterial infection, wherein the method comprises administering to an animal an antibacterial effective amount of negamycin or deoxynegamycin, or a pharmaceutically acceptable salt, prodrug or isomer thereof, and wherein the infecting bacteria are selected from a group of bacteria consisting of the following: Acinetobacter baumanii, Citrobacter freundii, Enterobacter aerogenes, haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus MRSA, Staphylococcus aureus GISA, Staphylococcus epidermis, Streptococcus pneumoniae PenR, Streptococcus pneumoniae PenS and Streptococcus pyogenes.
Owner:VERSICOR
Interaction of Moraxella catarrhalis with epithelial cells, extracellular matrix proteins and the complement system
ActiveUS8092811B2Improve abilitiesEnhanced decrease in <i>MAntibacterial agentsBiocideCell-Extracellular MatrixComplement system
Owner:ARNE FORSGREN AB
Method for stimulating immune response against Moraxella catarrhalis
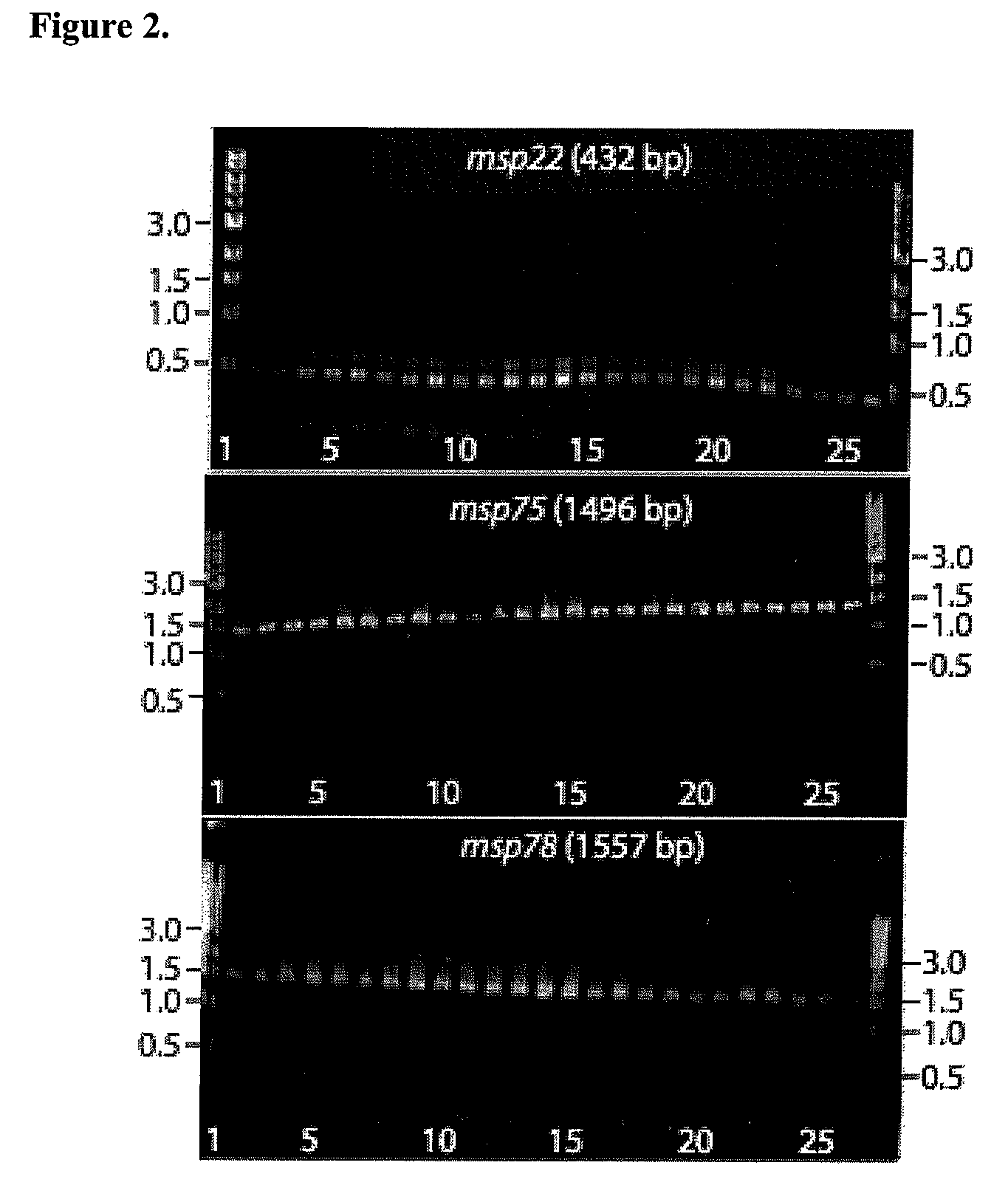
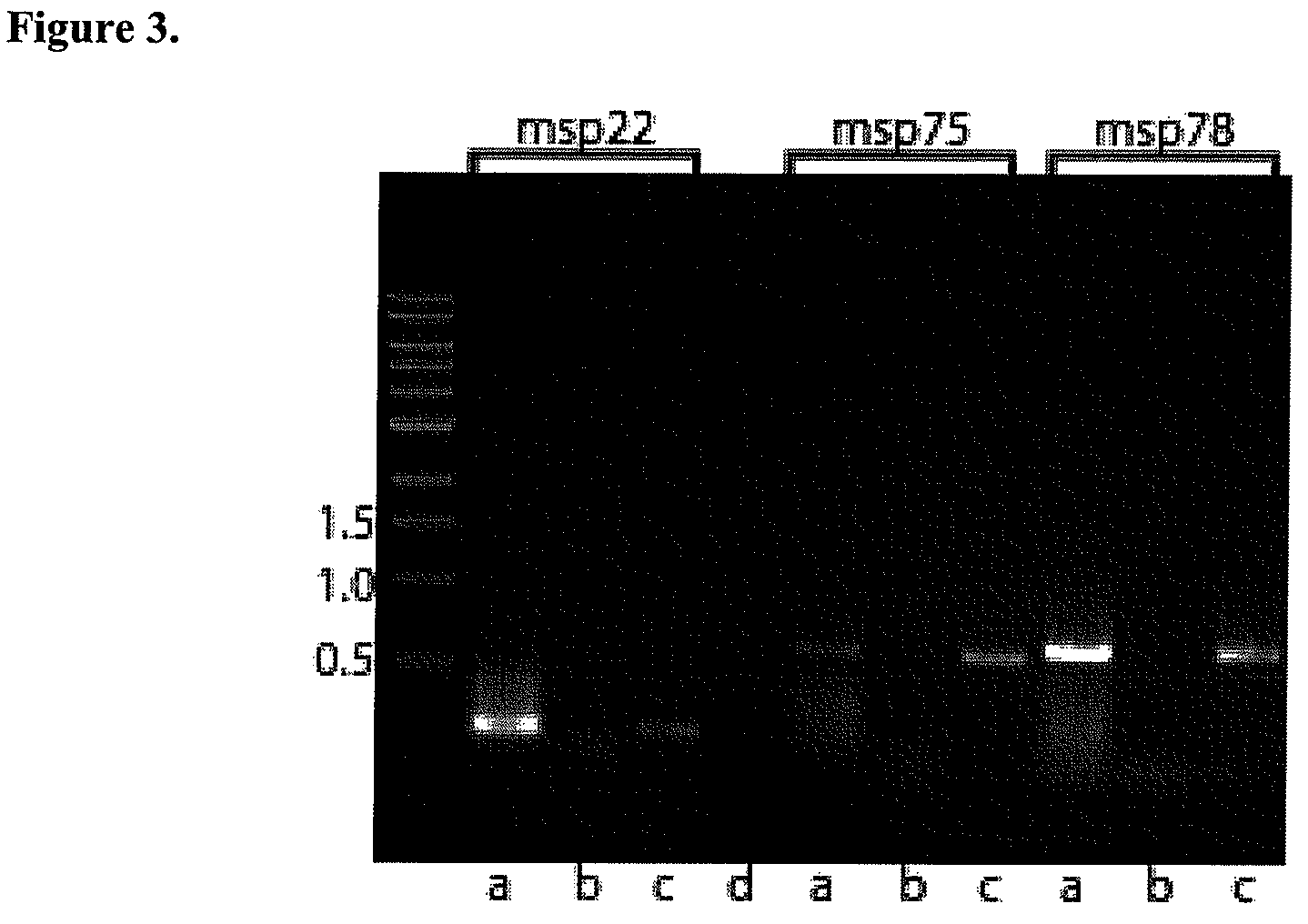
ActiveUS7811589B2Increase ratingsBacterial antigen ingredientsPeptide/protein ingredientsMoraxella catarrhalisMoraxella
Provided is a method for stimulating in an individual an immune response against M. catarrhalis. The method is performed by administering to an individual a composition that contains at least one isolated M. catarrhalis protein in an amount effective to stimulate an immune response against M. catarrhalis in the individual. The M. catarrhalis proteins used in the method of the invention are M. catarrhalis proteins Msp22, Msp75, Msp78, Protein 28, Protein 99, Protein 238, and combinations thereof.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Multi-component vaccine to protect against disease caused by Haemophilus influenzae and Moraxella catarrhalis
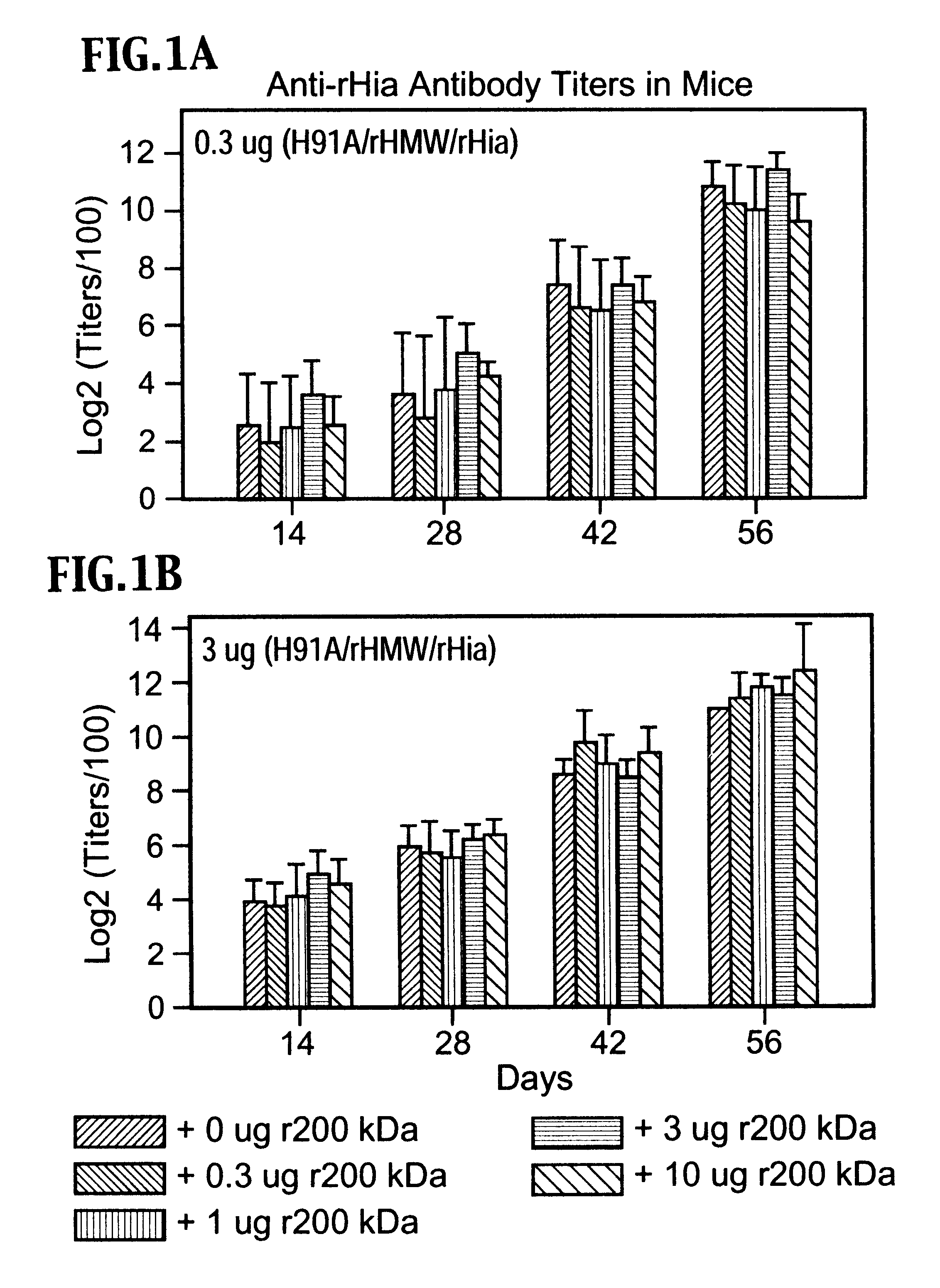
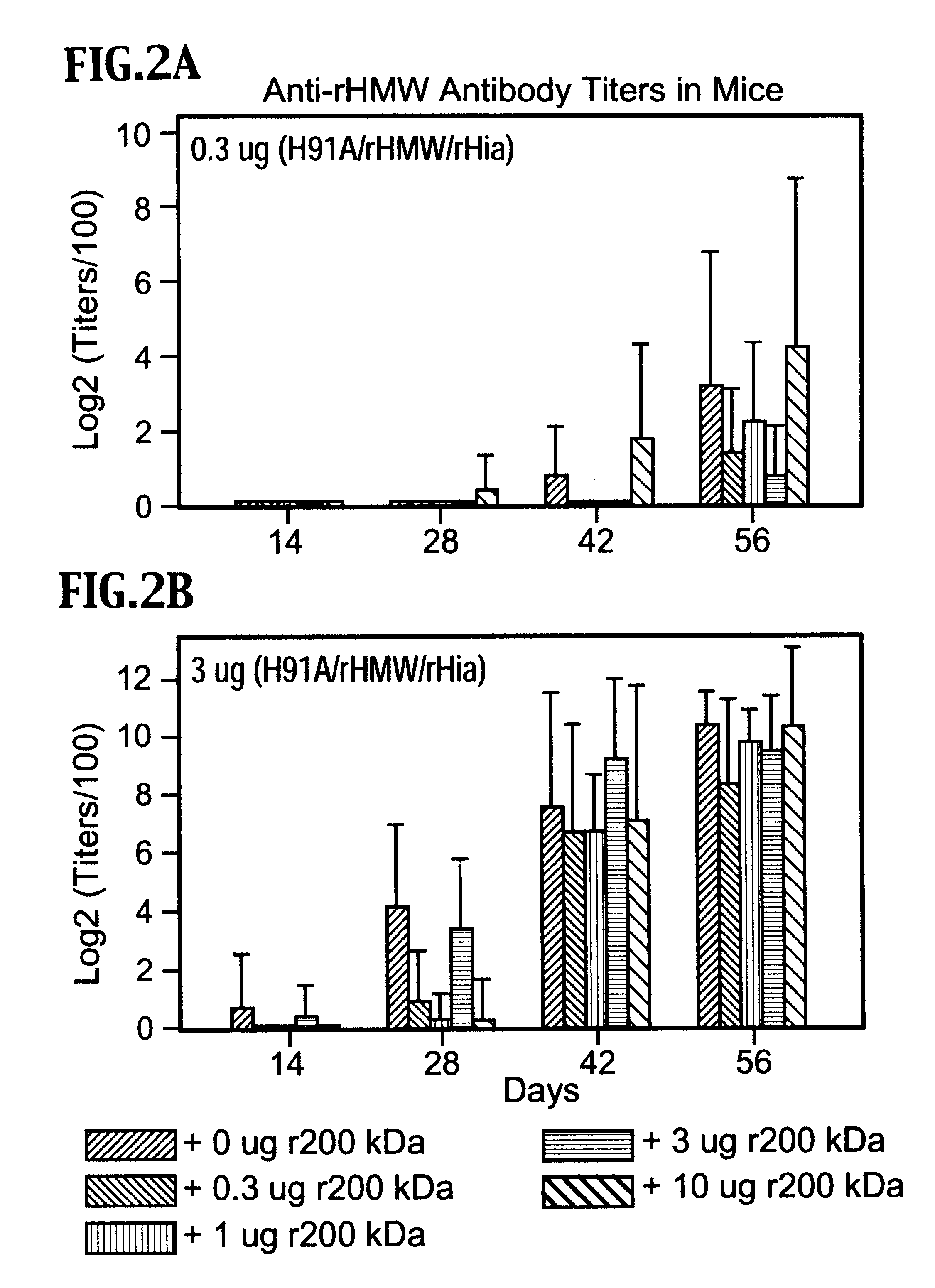
InactiveUS6391313B1Enhanced primary anti-H91A Hin4 responseLower immune responseAntibacterial agentsBacterial antigen ingredientsDiseaseHaemophilus
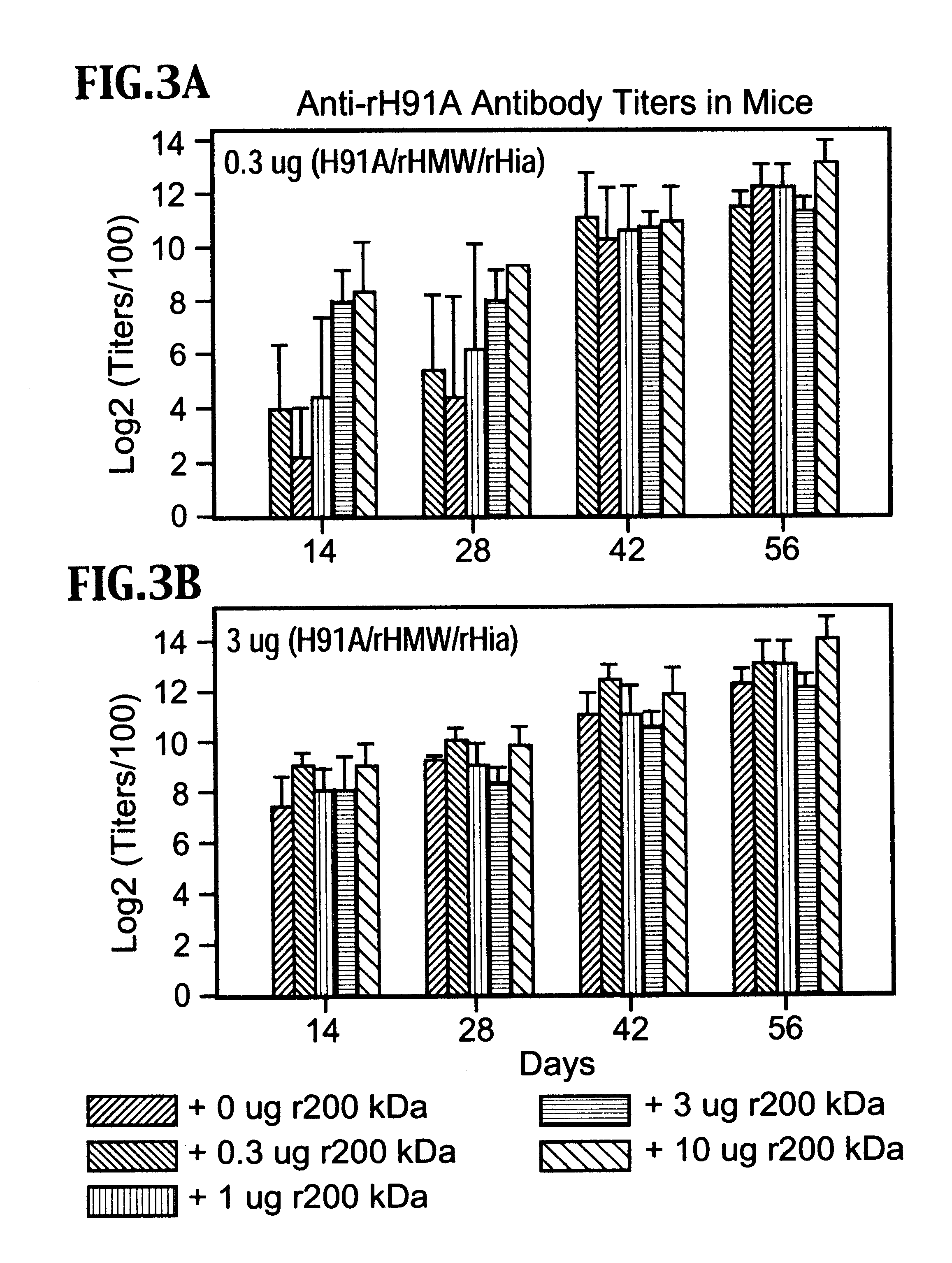
A multi-valent immunogenic composition confers protection on an immunized host against infection caused by both Haemophilus influenzae and Moraxella catarrhalis. Such composition comprises at least four antigens comprising at least one antigen from Haemophilus influenzae, and at least one antigen from Moraxella catarrhalis. Three of the antigens are adhesins. High molecular weight (HMW) proteins and Haemophilus influenzae adhesin (Hia) proteins of non-typeable Haemophilus and a 200 kDa outer membrane protein of Moraxella catarrhalis comprise the adhesin components while the other antigen is a non-proteolytic analog of Hin47 protein. Each component does not impair the immunogenicity of the others. The multi-valent immunogenic composition may be combined with DTP component vaccines, which may also include non-virulent poliovirus and PRP-T, to provide a component vaccine without impairment of the immunogenic properties of the other antigens.
Owner:AVENTIS PASTUER LTD
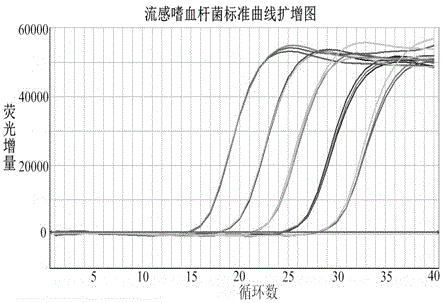
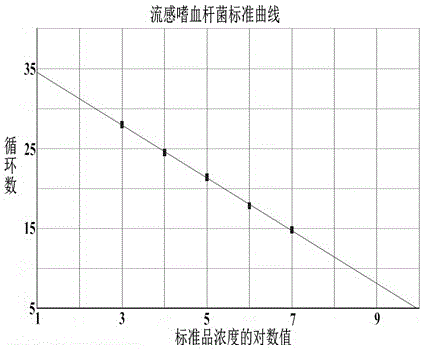
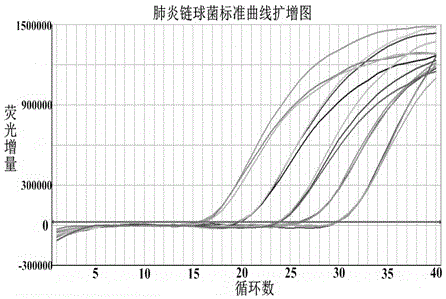
Multiple quantitative PCR (polymerase chain reaction) kit for quick combined detection of four bacteria difficult to cultivate and identify
ActiveCN105063218ARapid identificationIdentification is simple and effectiveMicrobiological testing/measurementMicroorganism based processesMoraxella catarrhalisBacilli
The invention provides a multiple quantitative PCR (polymerase chain reaction) kit for quick combined detection of four bacteria difficult to cultivate and identify. The multiple quantitative PCR kit comprises four PCR reaction systems, wherein the four PCR reaction systems comprise AllGlo fluorescence probes and forward and reverse primers aimed at the following four pathogens which cause child bacterial pneumonia and are clinically difficult to cultivate and identify: haemophilus influenza, streptococcus pneumonia, moraxella catarrhalis and legionella pneumophila. According to the multiple quantitative PCR kit, the design is reasonable, the infection of the four pathogens, clinically difficult to cultivate and identify, of child bacterial pneumonia can be easily, conveniently, quickly and parallelly detected in a reaction tube at the same time, the situation that four bacteria are detected at the same time through single-tube PCR is achieved, quantitative detection is achieved, the kit is easy and quick to operate, high in sensitivity, good in specificity and repeatability, accurate and reliable in result, early specific diagnosis, prevention and treatment can be provided for patients suffering from infantile pneumonia according to the bacterial infection titer, and the kit has great clinical practicability for interdicting an infection source, reducing infection or mixed infection of the four bacteria and monitoring the clinical curative effect.
Owner:HANGZHOU FIRST PEOPLES HOSPITAL
Removal of bacterial endotoxin in a protein solution by immobilized metal affinity chromatography
InactiveUS20050186218A1Reduce concentrationProtein recovery is greatIon-exchange process apparatusBacterial antigen ingredientsProtein solutionEndotoxin removal
The present invention relates to the purification of polypeptides and the removal of endotoxin via immobilized metal affinity chromatography (IMAC). More specifically, the invention relates to methods for removing bacterial endotoxin in a protein solution. In specific embodiments, the invention relates to the elimination of endotoxin from Moraxella catarrhalis outer membrane proteins.
Owner:WYETH HOLDINGS CORP
Suspension microbead array system for detecting common pathogens of respiratory tract infection
ActiveCN107254554AConducive to targeted treatmentLow costMicrobiological testing/measurementMicroorganism based processesBacteroidesMoraxella catarrhalis
The invention relates to a suspension microbead array system for detecting common pathogens of the respiratory tract infection. The system detects influenza A virus, influenza B virus, parainfluenza virus, human metapneumovirus, rhinovirus, bocavirus, adenovirus, respiratory syncytial virus, mycoplasma pneumoniae, moraxella catarrhalis, haemophilus influenza, streptococcus pneumoniae and bordetella pertussis in one experiment. The suspension microbead array system finishes the detection of 8 viruses, 4 bacteria and 1 mycoplasma in one experiment, prevents leak detection to the greatest extent, and is beneficial for the targeted clinical treatment; the cost of reagents, if divided on the detection of each index, is greatly lowered, so that the medical cost is reduced.
Owner:广东辉锦创兴生物医学科技有限公司
PREVENTION AND TREATMENT OF OTITIS MEDIA USING IgA ENRICHED MILK
InactiveUS20100303830A1Deterioration levelSenses disorderMilk immunoglobulinsPhysiologyMoraxella nonliquefaciens
This invention relates to a composition suitable for use in the prevention or treatment of otitis media comprising IgA derived from mature bovine milk and having specificity for at least one of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis. The invention further extends to the use of such a composition in the prevention or treatment of otitis media.
Owner:NESTEC SA
Method for detecting pneumonia causative bacteria using nucleic acid chromatography
InactiveUS20130023443A1Quick and accurate identificationNucleotide librariesMicrobiological testing/measurementBacteroidesStaphylococcus aureus
Provided are a method and a kit for accurately and rapidly detecting ten types of targeting pneumonia bacteria: Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, Klebsiella pneumoniae, Pseudomonas aeruginosa, Moraxella catarrhalis, methicillin-resistant Staphylococcus aureus (MRSA), and Staphylococcus aureus. A set of primer pairs directed to their respective target regions contained in the DnaJ gene, etc., of the ten types of pneumonia causative bacteria is designed for the ten bacterial strains and used to amplify gene products. A set of bacterial strain-specific probe pairs is further designed for the ten bacterial strains such that the probe pairs hybridize with the amplification products via sequences in the respective target regions differing from the sequences hybridized by the set of primer pairs. A first probe-bound labeled high molecular carrier in which plural types of first probes for the pneumonia bacteria are bound to a labeled high molecular carrier and a solid-phase second probe-carrying developing support are used as the set of probe pairs to perform nucleic acid chromatography.
Owner:YAMAGUCHI TECH LICENSING ORG
Transferrin receptor protein of moraxella
InactiveUS6190668B1Improving immunogenicityImprove responseAntibacterial agentsSenses disorderDiseaseCell mass
An isolated and purified non-denatured transferrin receptor protein of a Moraxella strain, particularly M. catarrhalis, has an apparent molecular mass of about 80 to about 90 kDa, as determined by SDS-PAGE. The transferrin receptor protein or a fragment analog thereof is useful in diagnostic applications and immunogenic compositions, particularly for in vivo administration to a host to confer protection against disease caused by a strain of Moraxella. The transferrin receptor protein is isolated from strains of Moraxella catarrhalis by a procedure including extraction of agent soluble proteins of a cell mass produced by cultivating the strain under iron-starved conditions. The transferrin receptor protein is selectively solubilized from the extracted cell mass and purified.
Owner:AVENTIS PASTUER LTD
Method and kit for performing quick co-detection on anti-Mc (Moraxella catarrhalis) IgM (Immunoglobulin M) and IgG (Immunoglobulin G) antibodies based on magnetic separation and multi-color quantum dot labeling
ActiveCN104181306AFast separationIt has the effect of synergistic amplification of multiple signalsBiological material analysisBiological testingSerum igeCo detection
The invention discloses a method and a kit for performing quick co-detection on anti-Mc (Moraxella catarrhalis) IgM (Immunoglobulin M) and IgG (Immunoglobulin G) antibodies based on magnetic separation and multi-color quantum dot labeling. The kit consists of anti-Mc antibody capturing nano magnetic beads with an anti-Mc IgM and IgG antibody gathering function, anti-human IgM and IgG antibody nano probes labeled by multi-color quantum dots, quality control substances and a PBST buffering solution, wherein the quality control substances comprise a positive quality control substance and a negative quality control substance; the positive quality control substance is serum, in which anti-Mc IgM and IgG antibodies of Mc infected people are respectively positive; the negative quality control substance is serum, in which anti-Mc IgM and IgG are respectively negative. The kit and the method have the advantages of simplicity, quickness and high sensitivity, and can be used for carrying out synchronous detection on the anti-Mc IgM and IgG antibodies.
Owner:HUBEI UNIV OF TECH +1
Intranasal immunization with detoxified lipooligosaccharide from nontypeable Haemophilus influenzae or Moraxella catarrhalis
The invention relates to intranasal immunization with detoxified lipooligosaccharide from nontypeable Haemophilus influenzae or Moraxella catarrhalis.
Owner:US DEPT OF HEALTH & HUMAN SERVICES
Nucleotide sequences of moraxella catarrhalis genome
InactiveUS20040067554A1Retains and enhances biological activity and stability and lifespanHigh expressionDepsipeptidesGenetic engineeringOpen reading frameNucleotide
Owner:MERCK & CO INC
Kit for simultaneously detecting streptococcus pneumoniae, legionella pneumophila and Moraxella catarrhalis
PendingCN112481401AStrong specificityFast detection methodMicrobiological testing/measurementMicroorganism based processesDisease monitoringMoraxella catarrhalis
Owner:AUTOBIO DIAGNOSTICS CO LTD
BASB019 proteins and genes from moraxella catarrhalis, antigens, antibodies, and uses
Owner:GLAXOSMITHKLINE BIOLOGICALS SA
Method for stimulating immune response against moraxella catarrhalis
ActiveUS20090169577A1Increase ratingsBacterial antigen ingredientsPeptide/protein ingredientsMoraxella catarrhalisMoraxella
Provided is a method for stimulating in an individual an immune response against M. catarrhalis. The method is performed by administering to an individual a composition that contains at least one isolated M. catarrhalis protein in an amount effective to stimulate an immune response against M. catarrhalis in the individual. The M. catarrhalis proteins used in the method of the invention are M. catarrhalis proteins Msp22, Msp75, Msp78, Protein 28, Protein 99, Protein 238, and combinations thereof.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
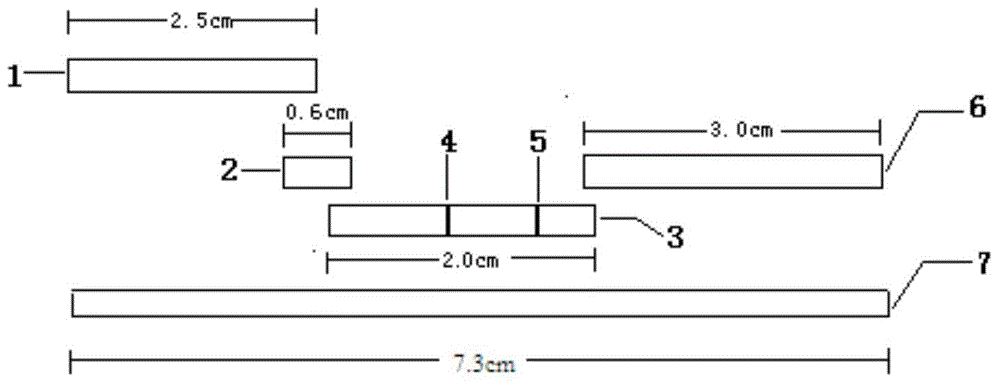
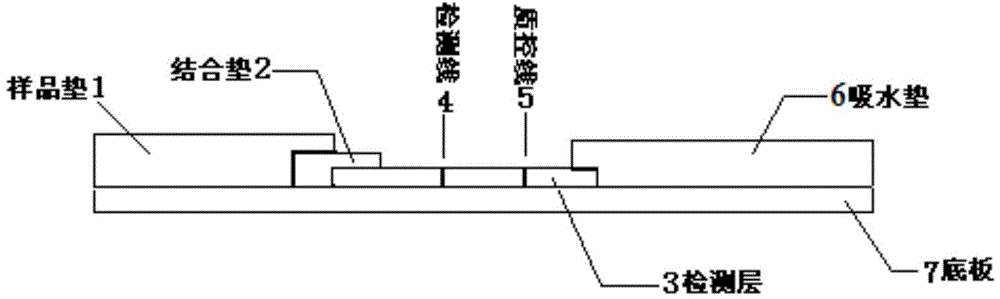
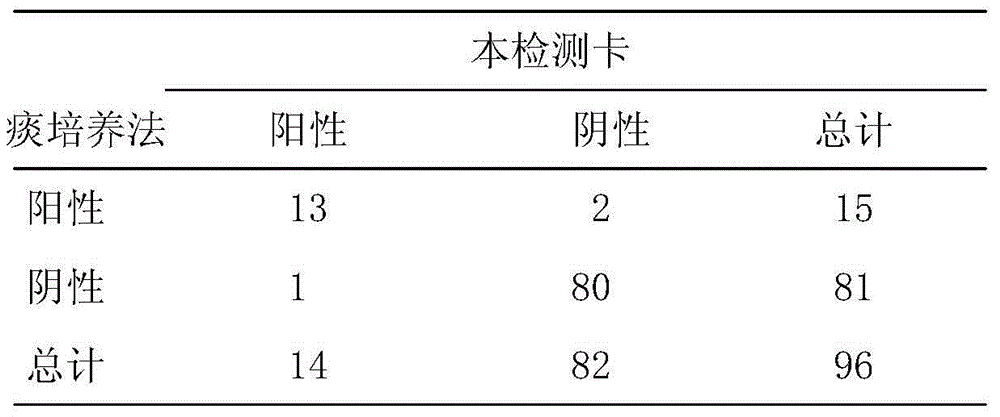
Moraxella catarrhalis quantum dot-immunochromatography detection card and preparation method and application thereof
The invention provides a moraxella catarrhalis (Mc) quantum dot-immunochromatography detection card and a preparation method and an application thereof. The detection card comprises a bottom plate, a sample pad, a water absorption pad, a conjugate pad and a detection layer, wherein the conjugate pad is coated by a quantum dot labelled anti-Mc nano-probe; the detection layer is formed by a solid-phase nitrocellulose membrane with a detection line and a quality control line; the detection line is coated by a mouse anti-Mc UspA1 protein polyclonal antibody; the quality control line is coated by anti-rabbit IgG (immunoglobulin); the detection layer is stuck to the bottom plate; the conjugate pad and the water absorption pad are respectively arranged above the two ends of the detection layer and are respectively stuck to the detection layer and the bottom plate after partially overlapping with the detection layer; the sample pad is arranged above the conjugate pad and is stuck to the conjugate pad and the bottom plate respectively after partially overlapping with the conjugate pad. The detection card has the advantages of simplicity and convenience in operation, high detection speed, capability of quantifying, high sensitivity and the like.
Owner:湖北诺美华抗体药物技术有限公司
Prevention and treatment of otitis media with non-pathogenic bacterial strains
InactiveCN101460181AHinder group buildingIncrease the number ofMilk preparationSenses disorderBacteroidesBacterial strain
This invention relates to a composition comprising a bacterial strain capable of boosting the systemic immune response, a bacterial strain capable of exerting bacteriostatic effects on pathogens associated with development of otitis media such as Streptococcus pneumoniae, untypeable Haemophilus influenzae and Moraxella catarrhalis and a bacterial strain capable of exerting bacteriocidal effects on such pathogens. The invention further extends to the use of such a composition in the prevention or treatment of otitis media.
Owner:NESTEC SA
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com