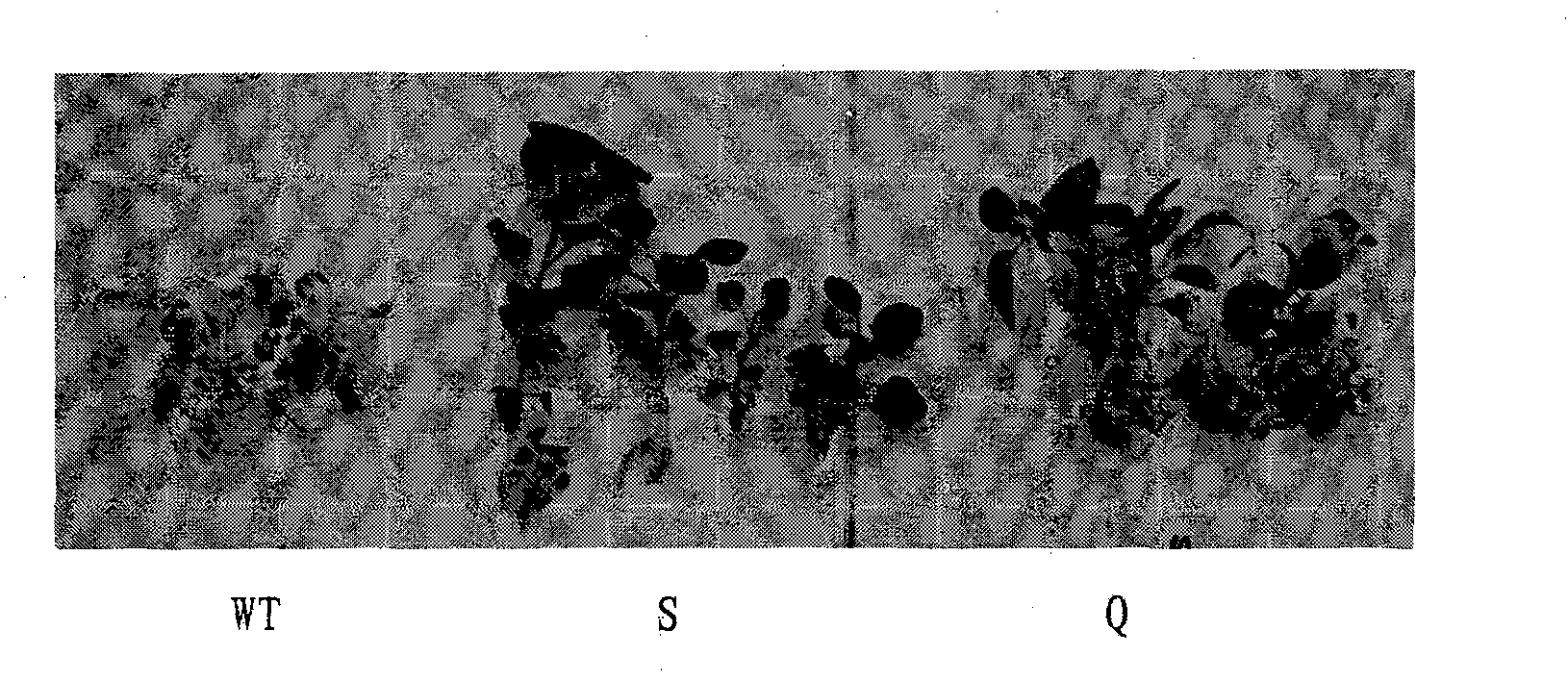
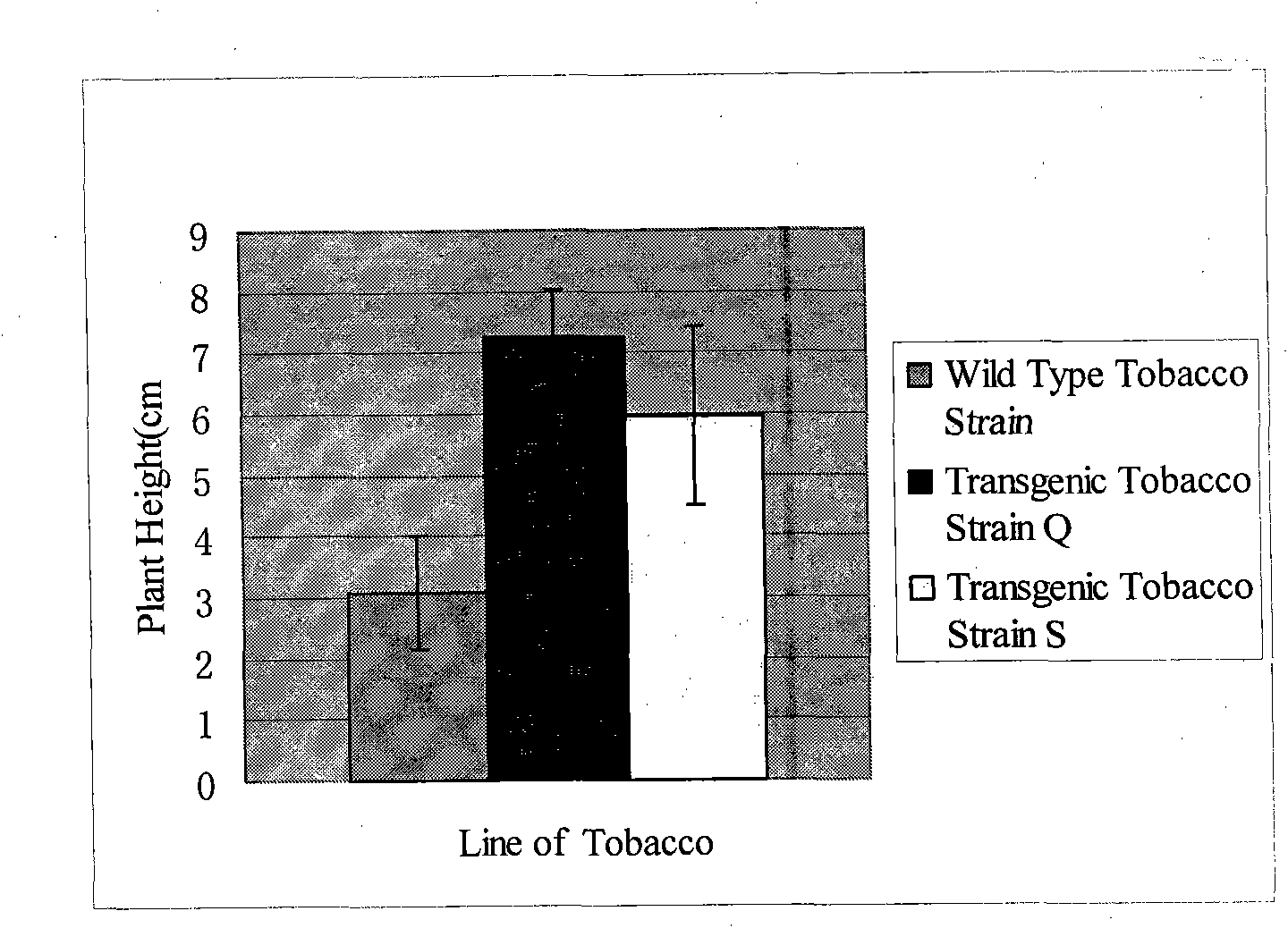
Patents
Literature
2343 results about "Resistant genes" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Resistant genes. Resistance genes are the genes in plant genomes that convey plant disease resistance against pathogens by producing resistance proteins. The main class of R-genes consist of a nucleotide binding domain and a leucine rich repeat domains .
Exogenous gene knocking-in and integrating system on basis of CRISPR/Cas9, method for establishing exogenous gene knocking-in and integrating system and application thereof
ActiveCN106191116ANucleic acid vectorVector-based foreign material introductionTarget geneIntegrated systems
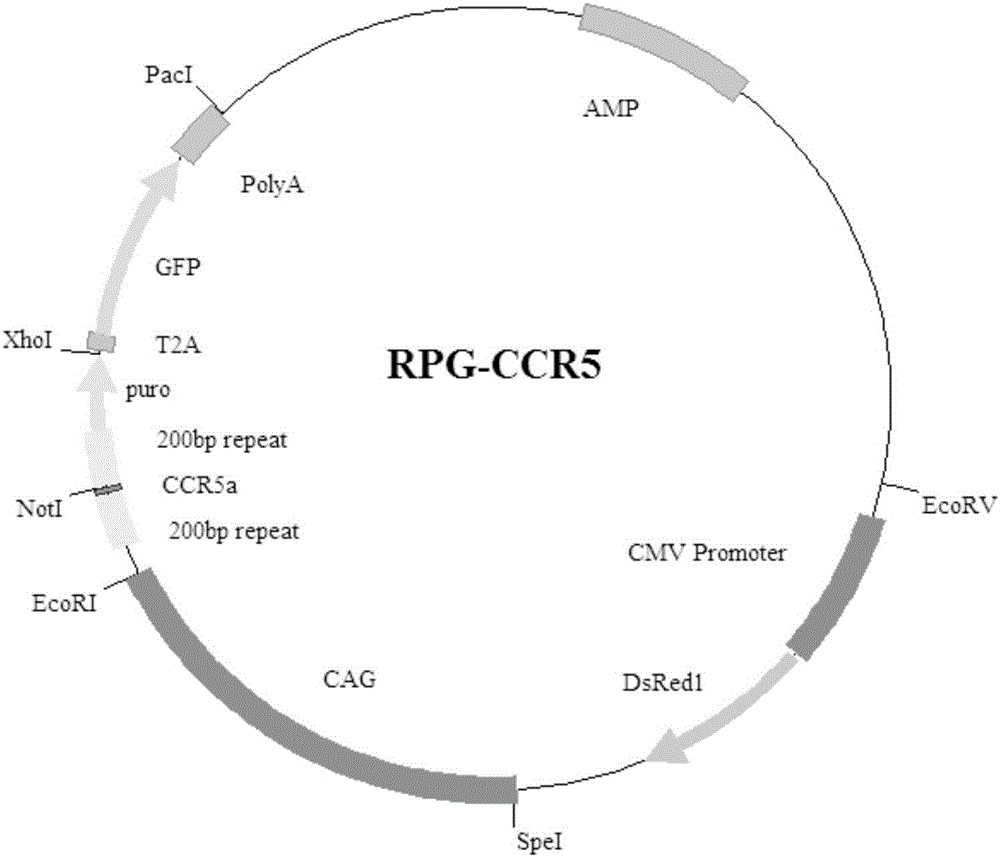
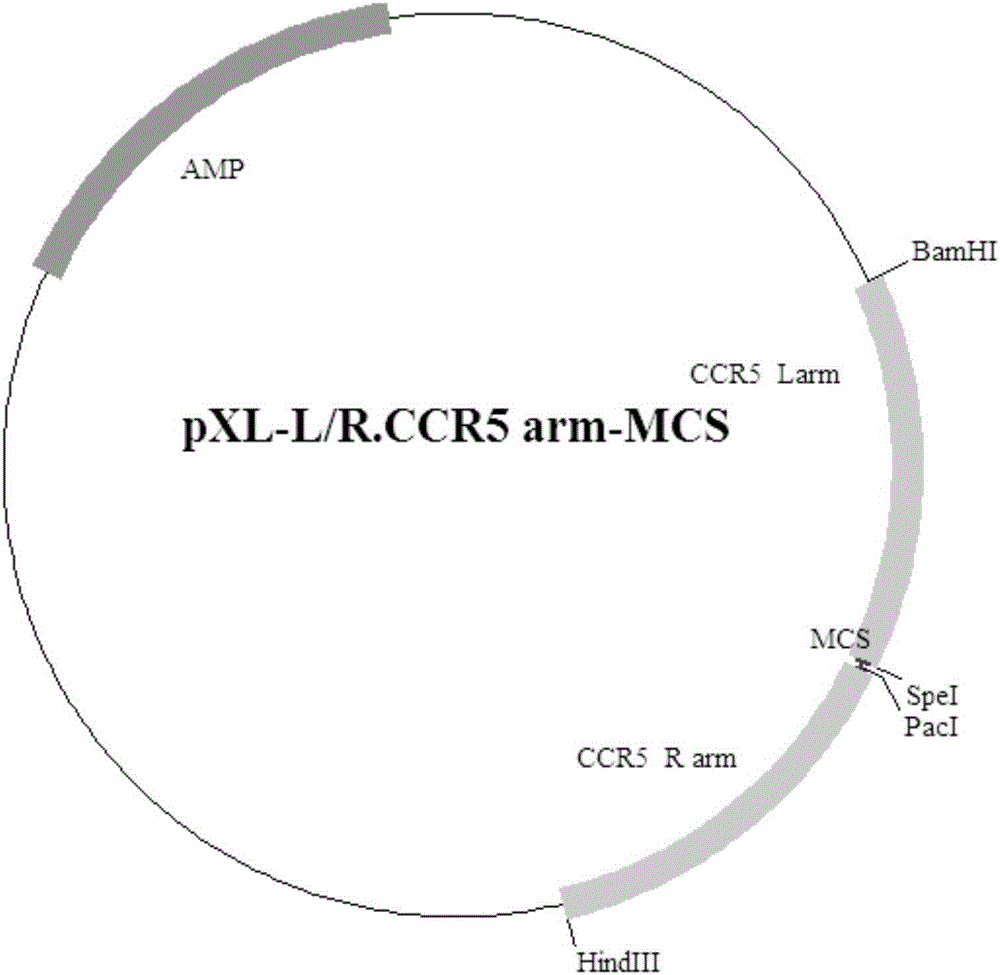
The invention provides an exogenous gene knocking-in and integrating system on the basis of CRISPR / Cas9, a method for establishing the exogenous gene knocking-in and integrating system and application thereof. The exogenous gene knocking-in and integrating system comprises vectors with report / donor functions and Cas9 expression vectors. Each report / donor vector comprises two target gent homologous arms and an exogenous sequence fragment positioned between the two target gene homologous arms; homologous sequences, which are positioned on a target gene, of the two target gene homologous arms of each report / donor vector are respectively positioned on two sides of a target sequence of the target gene and are connected with the target sequence of the target gene; the exogenous sequence fragments comprise promoters, resistant genes, shorn peptide sequences, report genes and polyA tails which are sequentially arrayed, two SSA repair homologous sequences of each resistant gene are inserted into the resistant gene, and the target sequence of each target gene is inserted in a space between the two corresponding SSA repair homologous sequences. The exogenous gene knocking-in and integrating system, the method and the application have the advantages that exogenous genes can be integrated with endogenous gene sequences in an efficient site-directed and accurately targeted manner, and double-chromosome allelic gene double-knocking-in can be efficiently carried out.
Owner:成都中科奥格生物科技有限公司
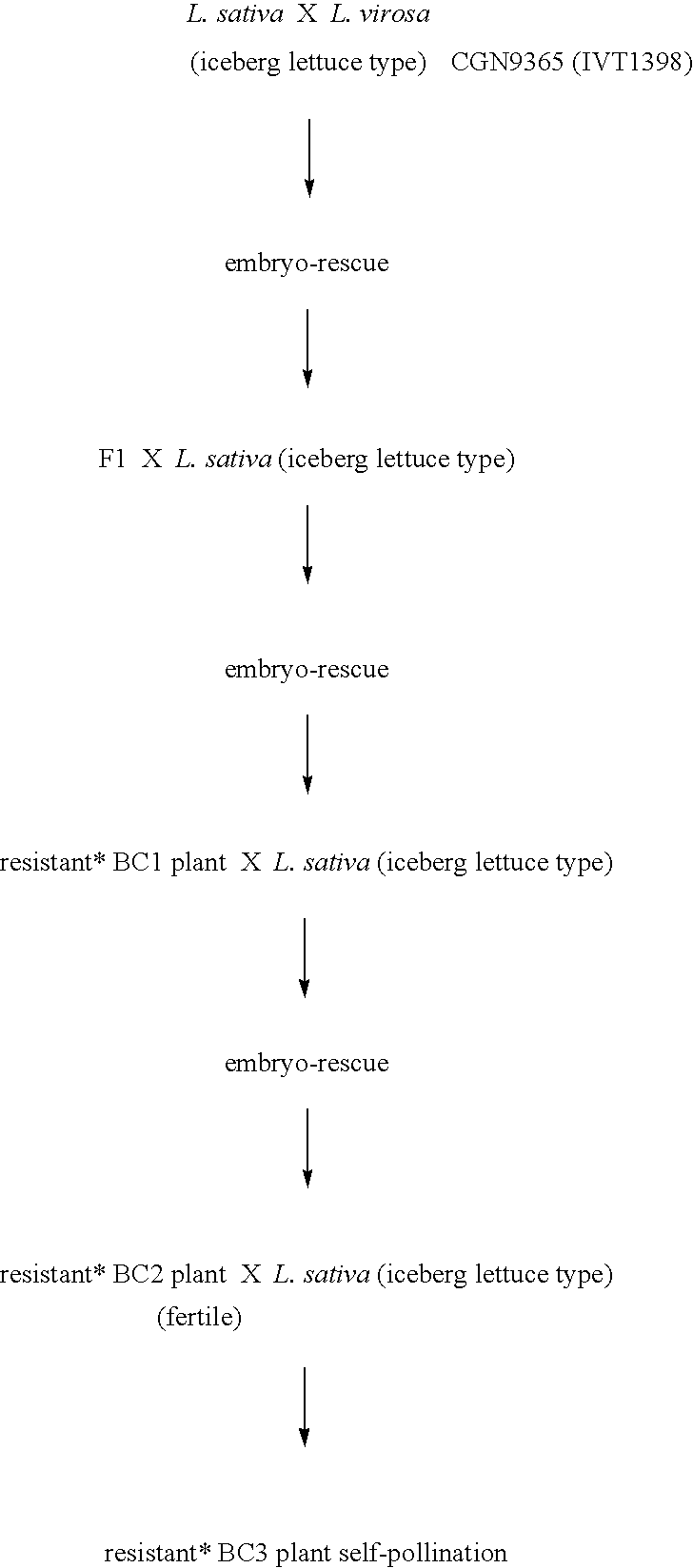
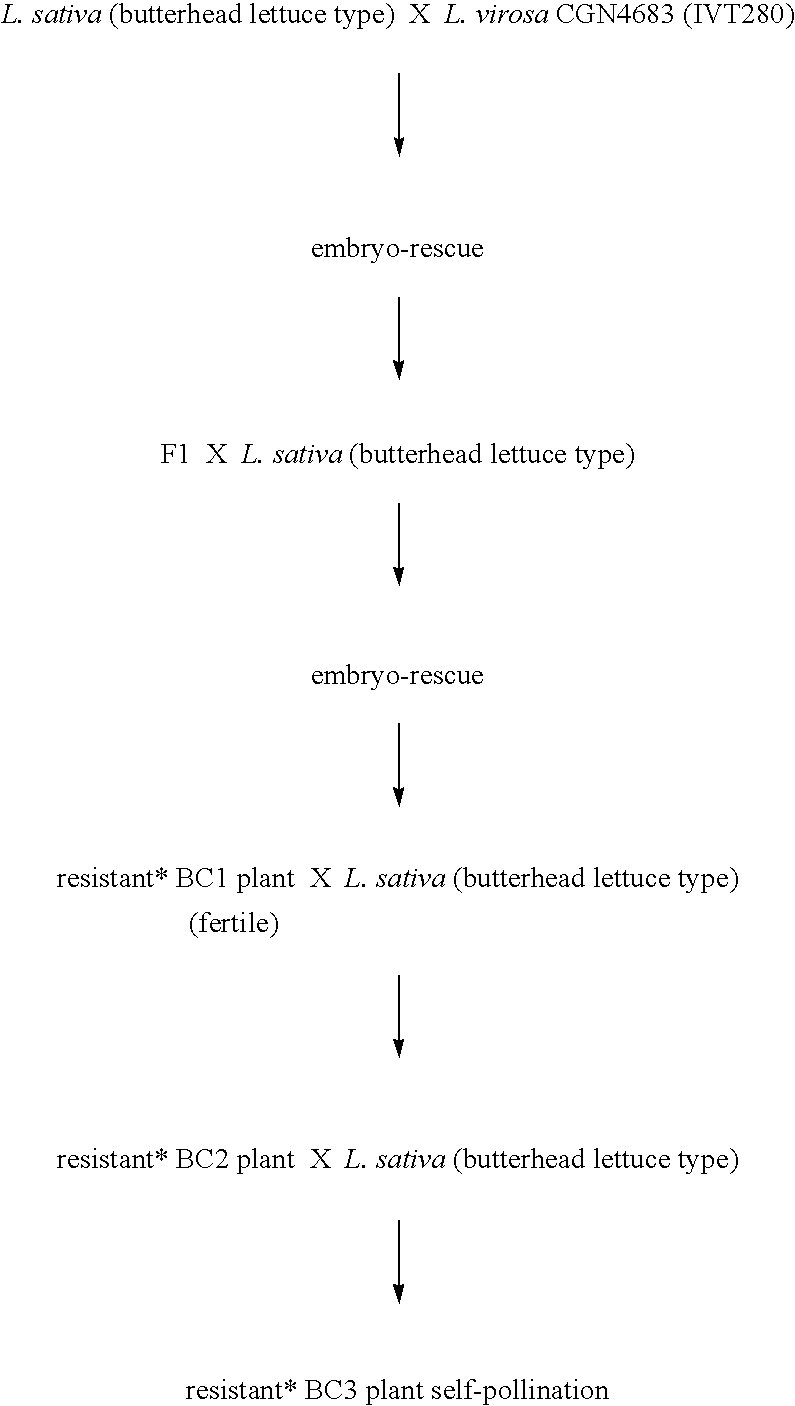
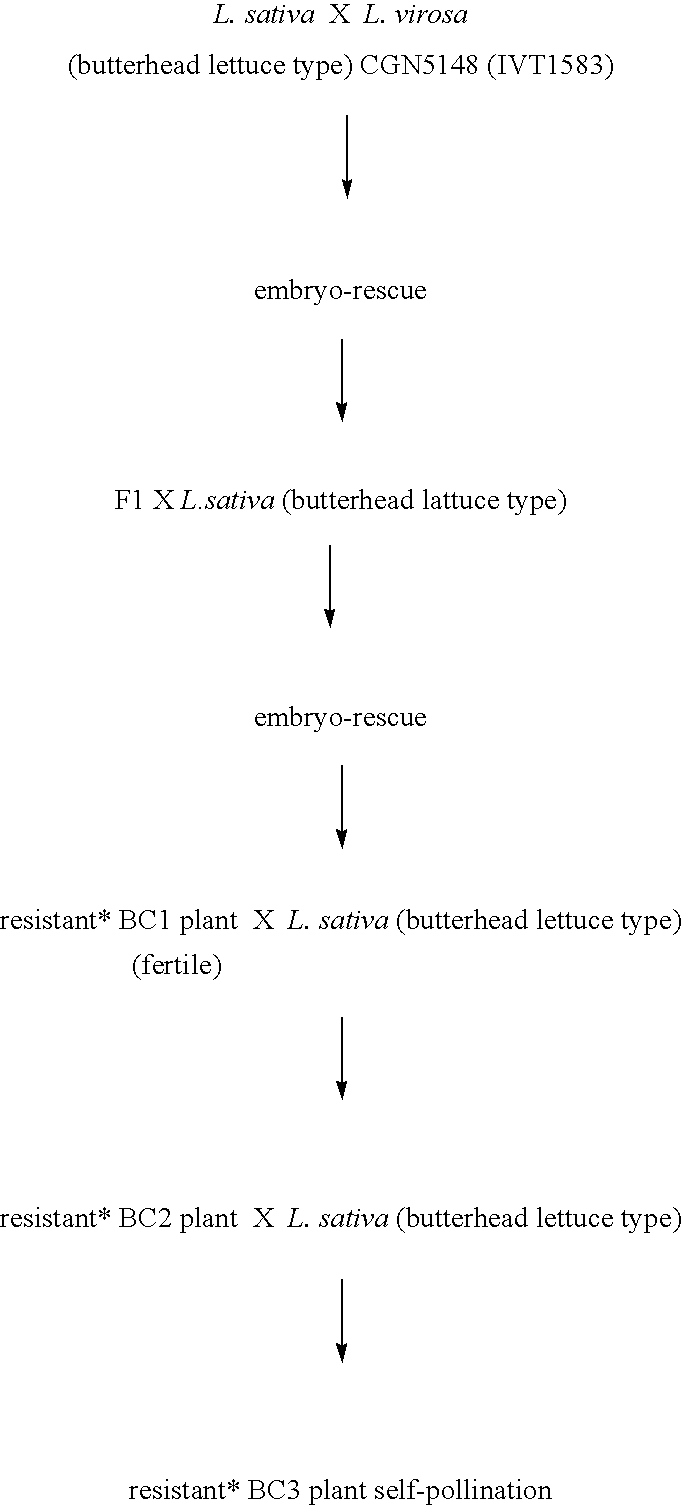
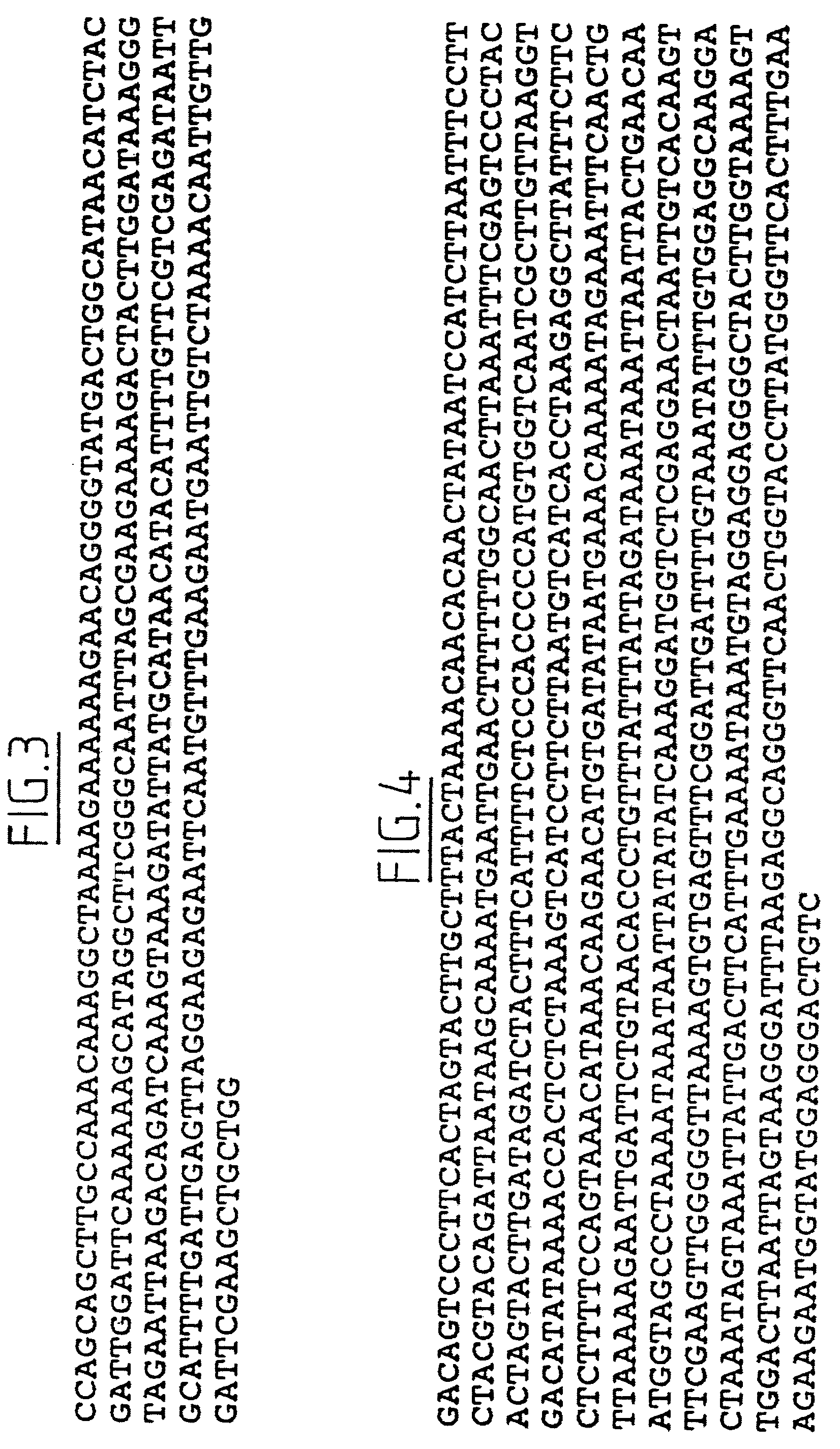
Lettuce plants having broad-spectrum Dm-resistance genes
InactiveUS6903249B2Lasting resistanceMicrobiological testing/measurementOther foreign material introduction processesBiotechnologyBremia lactucae
A method for obtaining a plant, in particular a cultivated lettuce plant (L. sativa), with a lasting resistance to a pathogen, in particular Bremia lactucae, comprising of providing one or more specific DNA markers linked to one or more resistance genes, determining the presence of one or more resistance genes in a plant using these DNA markers, subsequently crossing a first plant comprising one or more resistance genes with a second plant comprising one or more resistance genes, and selecting from the progeny a plant in which one or more resistance genes are present by using the DNA markers. The invention further relates to the plants obtained with this method, seeds and progeny of these plants, as well as progeny thereof.
Owner:ENZA ZADEN DE ENKHUIZER ZAADHANDEL
Brown planthopper resistant rice gene Bph9 as well as molecular marker and application thereof
ActiveCN103667309AClear functionGood effectBacteriaMicrobiological testing/measurementBinding siteProtein
The invention provides a brown planthopper resistant rice gene Bph9 as well as a molecular marker and application thereof. The brown planthopper resistant rice gene Bph9 has a nucleotide sequence as shown in SEQ ID No.1 and a cDNA (complementary Desoxvribose Nucleic Acid) sequence as shown in SEQ ID No.2. The Bph9 gene is located between molecular markers InD2 and RM28466. The molecular markers closely linked with the gene also includes one of RM28438, InD28450, InD28453, InD14, InD28432, RM28481 and RM28486 which can be used for sieving the rice containing the brown planthopper resistant gene Bph9. The Bph9 gene belongs to the NBS-LRR (Nucleotide Binding Site-Leucine Rich Repeat) gene family; the encoded protein is related to the plant disease resistance; the Bph9 gene is transferred into common rice by virtue of genetic transformation and hybridization, and thus the resistance of rice to brown planthopper can be improved, so that the damage of brown planthopper is decreased, and the purposes of yield increase and yield stabilization are realized.
Owner:WUHAN UNIV
Method for obtaining a plant with a lasting resistance to a pathogen
InactiveUS7501555B2Sugar derivativesMicrobiological testing/measurementResistant genesBremia lactucae
A method for obtaining a plant, in particular a cultivated lettuce plant (L. sativa), with a lasting resistance to a pathogen, in particular Bremia lactucae, comprised of providing one or more specific DNA markers linked to one or more resistance genes, determining the presence of one or more resistance genes in a plant using these DNA markers, subsequently crossing a first plant comprising one or more resistance genes with a second plant comprising one or more resistance genes, and selecting from the progeny a plant in which one or more resistance genes are present by using the DNA markers. The invention further relates to the plants obtained with this method, seeds and progeny of these plants, as well as progeny thereof.
Owner:ENZA ZADEN BEHEER BV
Polynucleotide Encoding a Maize Herbicide Resistance Gene and Methods for Use
Owner:EI DU PONT DE NEMOURS & CO
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20040185478A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
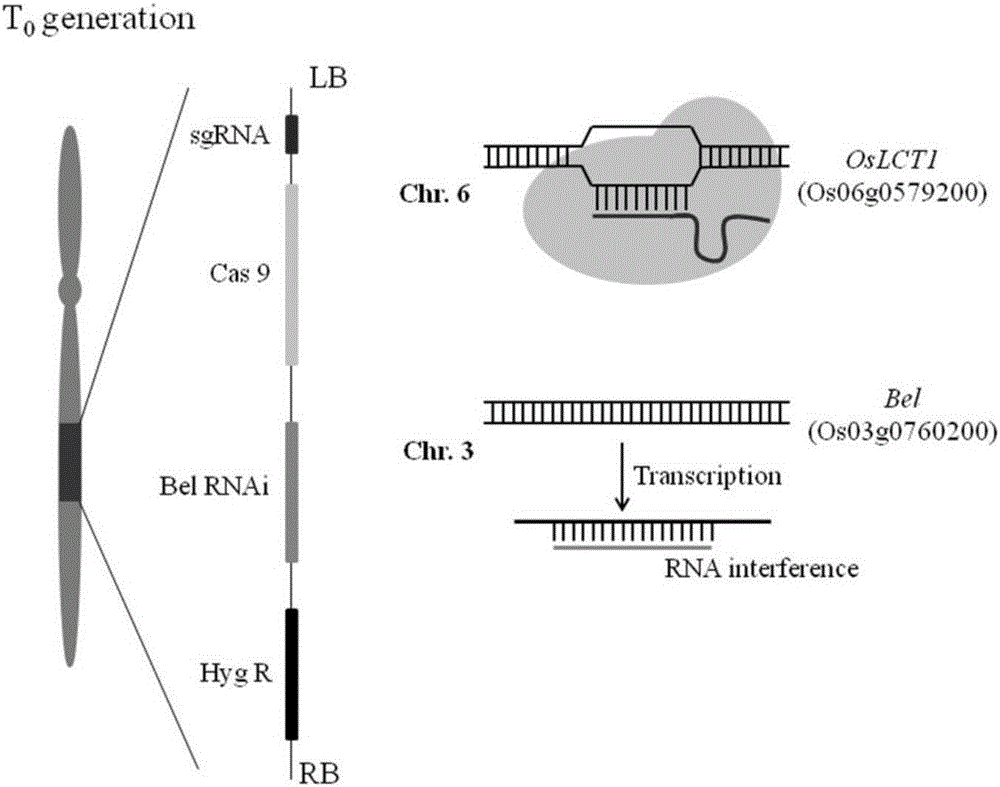
Recombinant vector and non-GMO gene editing plant screening method
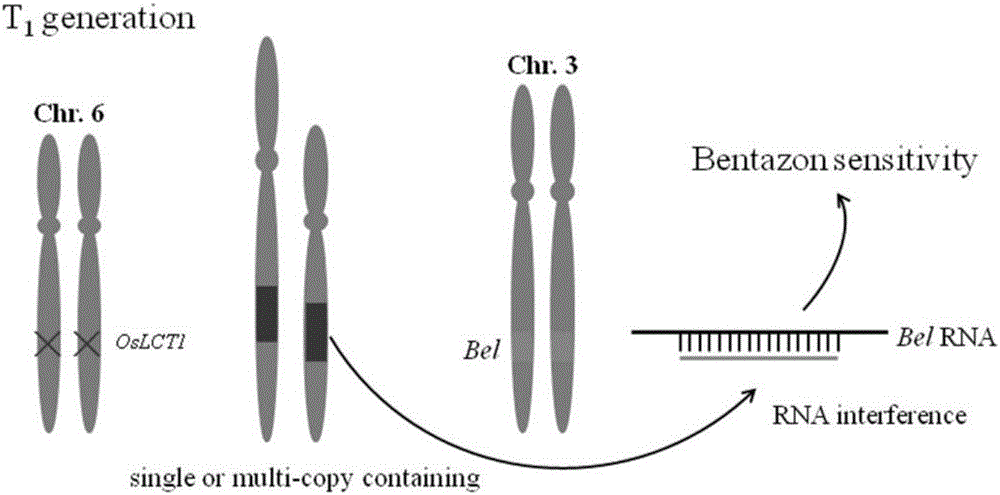
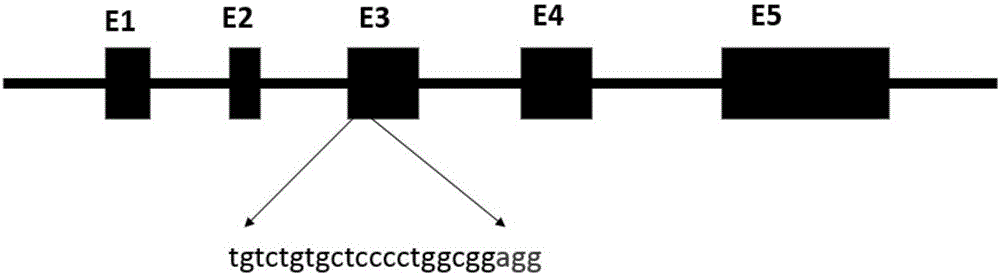
ActiveCN106222193AInexpensive screening methodSimple screening methodVector-based foreign material introductionAngiosperms/flowering plantsResistant genesHerbicide screen
The invention discloses a recombinant vector and a non-GMO gene editing plant screening method. An initial carrier in the recombinant vector is a CRISPR / Cas9 carrier which contains sgRNA gene, Cas9 gene and screening label gene and is used for plant gene editing, the recombinant vector carries a BelRNAi expression element, and a hairpin RNA interference fragment interfering with bentazone resistance gene is generated through transcription of the BelRNAi expression element. By introducing the BelRNAi expression element to the initial carrier and conducting bentazone herbicide screening on future generation plants, offspring with target gene mutated are reserved, and it is also ensured that the mutated offspring do not contain transgenic fragments; screening method is cheaper, simpler and more effective.
Owner:ZHEJIANG UNIV +1
Production method for Fbxo40 gene knockout pigs
InactiveCN105821049AConvenient researchLow costNucleic acid vectorFermentationDiseaseAgricultural science
The invention provides a production method for Fbxo40 gene knockout pigs. The production method comprises the steps that a CRISPR-Cas9 targeting vector and a PGK-Neo resistant gene are jointly transfected into a porcine embryonic fibroblast to obtain a G418-resistant positive monoclonal cell, an Fbxo40 gene in the positive monoclonal cell generates insertion or deletion mutation, a reading frame generates frame shift and stops in advance, then the obtained cell is cloned to serve as a donor cell for nuclear transfer, and an oocyte serves as a receptor oocyte for nuclear transfer; cloned embryos are obtained through a somatic cell nuclear transfer technique, the high-quality cloned embryos are transferred into the fallopian tube of an oestrous sow, and the Fbxo40 gene knockout pigs are obtained through whole-period development. According to the production method, the Fbxo40 knockout pigs are efficiently obtained through a CRISPR-Cas9 gene editing technique at the low cost, and animal models are supplied to research of muscle development and diseases related to the muscles.
Owner:CHINA AGRI UNIV
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20060263810A1Reduce usageDetermine rapidly the bacterial resistance to antibioticsSugar derivativesMicrobiological testing/measurementNeisseria meningitidisListeria monocytogenes
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6′-IIa, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6′)-aph(2″), aad(6), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:GENEOHM SCI CANADA
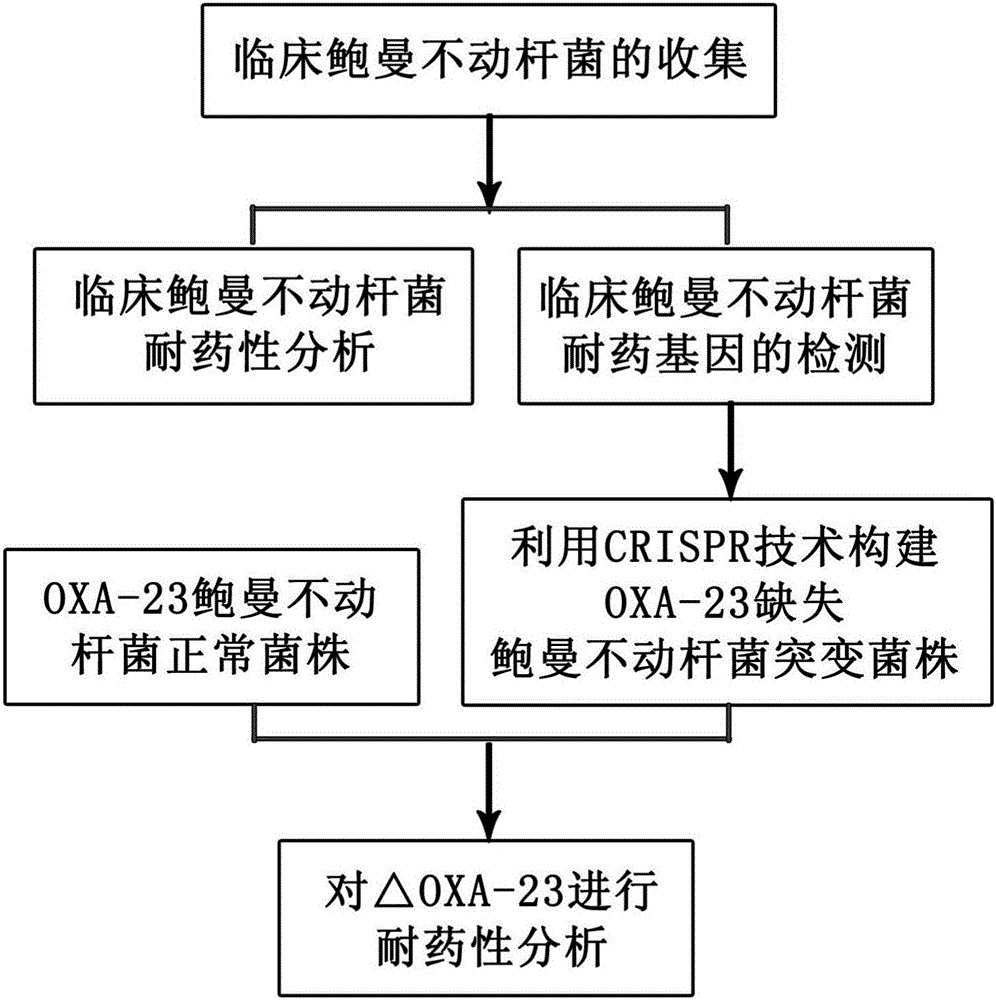
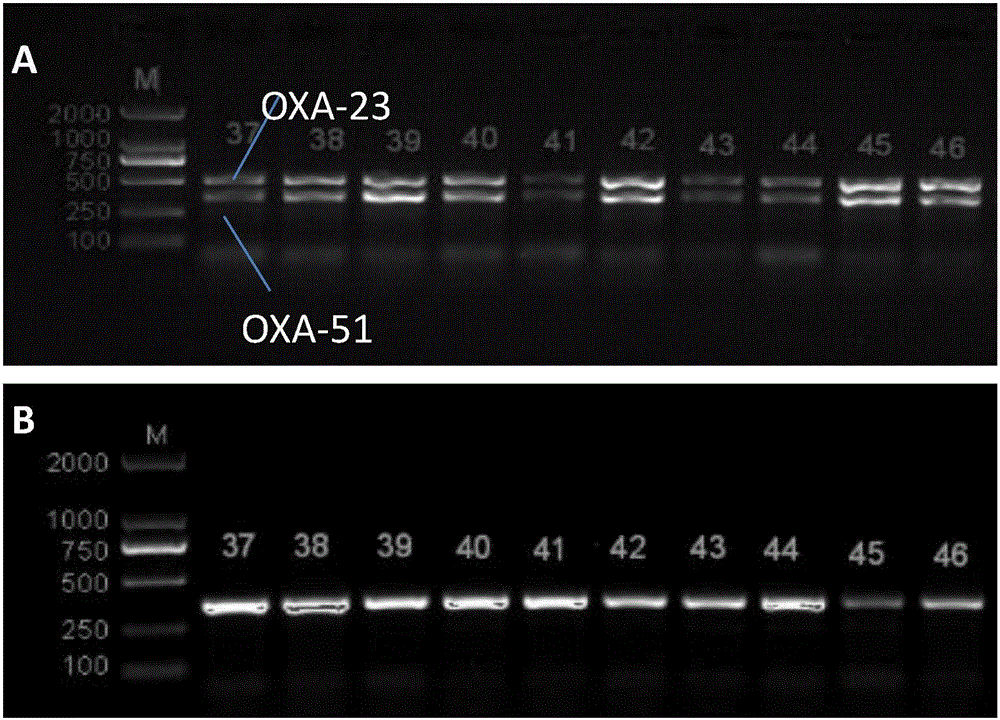
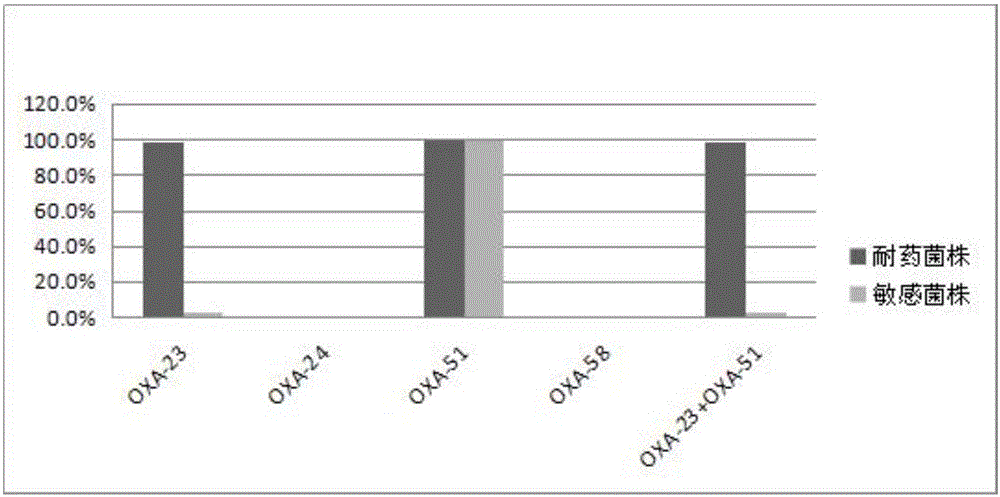
Method for deleting drug resistant genes of acinetobacter baumannii (AB) through CRISPR-Cas9
InactiveCN106544353ASensitivity reversalEasy to operateVectorsMicroorganism based processesBacteroidesMulti drug resistant
The invention discloses a method for deleting drug resistant genes of acinetobacter baumannii (AB) through CRISPR-Cas9. The method comprises the steps that firstly, AB is collected clinically, and drug resistance analysis and statistics are conducted after the AB is subjected to drug resistance measurement through a drug sensitive slip method; secondly, the multi-drug resistant AB is subjected to DNA extraction through a lysis-boiling method, and then amplification analysis is conducted by mean of well designed drug resistant gene primers of the AB; and thirdly, according to drug resistant gene detection in the former step, the AB containing an OXA-23 gene is selected, CRISPR / Cas9 and sgRNA plasmids are established and transferred into the AB containing the OXA-23 gene, an OXA-23 gene deletion AB mutant strain is established, and drug resistance analysis is conducted on the OXA-23 gene deletion AB mutant strain. The method is easy to operate and high in knocking-out efficiency, and a novel method and a novel concept are provided for preventing spreading of the drug resistant genes and treating drug resistant bacteria.
Owner:GENERAL HOSPITAL OF NINGXIA MEDICAL UNIV
Analysis method and system of metagenome data
ActiveCN108334750AImprove accuracyReduce processingSequence analysisSpecial data processing applicationsEndocarditisResistant genes
The invention relates to an analysis method and a system of metagenome data. According to the invention, a preliminary species identification result of a sample is obtained on the basis of a k-Mer algorithm, a part or all of supporting sequences are extracted on the basis of the preliminary species identification result, and the preliminary species identification result is verified by using a blast algorithm to judge whether the preliminary species identification result is a reported detected species or not. The method and system disclosed by the invention can lower false positivity, quickly and accurately obtain the reported detected species of the sample in a short time, and are compatible with various mainstream sequencing platforms, thereby being suitable for second-generation sequencing technologies and third sequencing technologies; the method and system of the invention can also accurately identify drug-resistant genes and drug-resistant mutation sites of the sample and map thedrug-resistant genes and the drug-resistant mutation sites of the sample to the reported detected species. Furthermore, the system disclosed by the invention can be used for identifying pathogenic microorganisms, especially endocarditis pathogens to overcome the defect that the endocarditis pathogens are difficultly cultured.
Owner:SIMCERE DIAGNOSTICS CO LTD +2
Species-specific, genus-specific and universal DNA probes and amplification primers to rapidly detect and identify common bacterial and fungal pathogens and associated antibiotic resistance genes from clinical specimens for diagnosis in microbiology laboratories
InactiveUS20030049636A1Low costRapid positioningMicrobiological testing/measurementFermentationBacteroidesNeisseria meningitidis
DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample DNA from (i) any bacterium, (ii) the species Streptococcus agalactiae, Staphylococcus saprophyticus, Enterococcus faecium, Neisseria meningitidis, Listeria monocytogenes and Candida albicans, and (iii) any species of the genera Streptococcus, Staphylococcus, Enterococcus, Neisseria and Candida are disclosed. DNA-based methods employing amplification primers or probes for detecting, identifying, and quantifying in a test sample antibiotic resistance genes selected from the group consisting of blatem, blarob, blashv, blaoxa, blaZ, aadB, aacC1, aacC2, aacC3, aacA4, aac6'-lla, ermA, ermB, ermC, mecA, vanA, vanB, vanC, satA, aac(6')-aph(2''), aad(6'), vat, vga, msrA, sul and int are also disclosed. The above microbial species, genera and resistance genes are all clinically relevant and commonly encountered in a variety of clinical specimens. These DNA-based assays are rapid, accurate and can be used in clinical microbiology laboratories for routine diagnosis. These novel diagnostic tools should be useful to improve the speed and accuracy of diagnosis of microbial infections, thereby allowing more effective treatments. Diagnostic kits for (i) the universal detection and quantification of bacteria, and / or (ii) the detection, identification and quantification of the above-mentioned bacterial and fungal species and / or genera, and / or (iii) the detection, identification and quantification of the above-mentioned antibiotic resistance genes are also claimed.
Owner:BERGERON MICHEL G +3
Specific and universal probes and amplification primers to rapidly detect and identify common bacterial pathogens and antibiotic resistance genes from clinical specimens for routine diagnosis in microbiology laboratories
InactiveUS20050042606A9Rapid bacterial identificationShorten the timeMicrobiological testing/measurementDepsipeptidesGenomic SegmentMoraxella catarrhalis
The present invention relates to DNA-based methods for universal bacterial detection, for specific detection of the common bacterial pathogens Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Proteus mirabilis, Streptococcus pneumoniae, Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Staphylococcus saprophyticus, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis as well as for specific detection of commonly encountered and clinically relevant bacterial antibiotic resistance genes directly from clinical specimens or, alternatively, from a bacterial colony. The above bacterial species can account for as much as 80% of bacterial pathogens isolated in routine microbiology laboratories. The core of this invention consists primarily of the DNA sequences from all species-specific genomic DNA fragments selected by hybridization from genomic libraries or, alternatively, selected from data banks as well as any oligonucleotide sequences derived from these sequences which can be used as probes or amplification primers for PCR or any other nucleic acid amplification methods. This invention also includes DNA sequences from the selected clinically relevant antibiotic resistance genes. With these methods, bacteria can be detected (universal primers and / or probes) and identified (species-specific primers and / or probes) directly from the clinical specimens or from an isolated bacterial colony. Bacteria are further evaluated for their putative susceptibility to antibiotics by resistance gene detection (antibiotic resistance gene specific primers and / or probes). Diagnostic kits for the detection of the presence, for the bacterial identification of the above-mentioned bacterial species and for the detection of antibiotic resistance genes are also claimed. These kits for the rapid (one hour or less) and accurate diagnosis of bacterial infections and antibiotic resistance will gradually replace conventional methods currently used in clinical microbiology laboratories for routine diagnosis. They should provide tools to clinicians to help prescribe promptly optimal treatments when necessary. Consequently, these tests should contribute to saving human lives, rationalizing treatment, reducing the development of antibiotic resistance and avoid unnecessary hospitalizations.
Owner:GENEOHM SCI CANADA
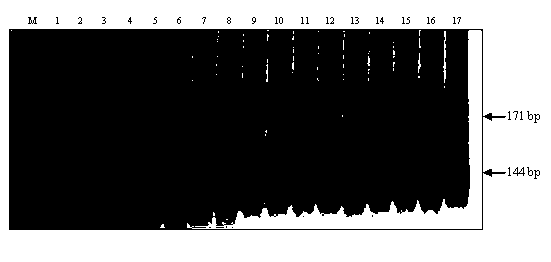
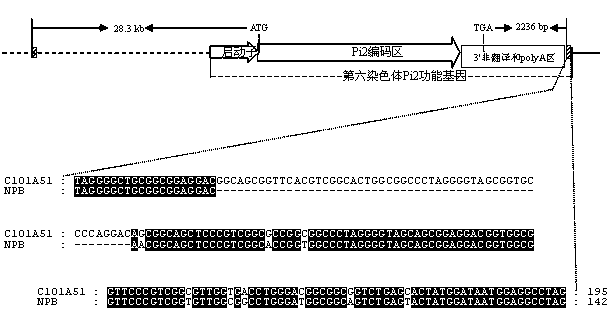
Gene-specific molecular marker Pi2SNP of rice blast-resistant gene Pi2 as well as preparation method and application thereof
ActiveCN103320437AStrong specificityHigh sequence identityMicrobiological testing/measurementDNA preparationAgricultural scienceEnzyme digestion
The invention discloses a gene-specific molecular marker Pi2SNP of a rice blast-resistant gene Pi2 as well as a preparation method and application thereof. The molecular marker Pi2SNP is a nucleotide fragment II in a specific banding pattern with the rice blast-resistant gene Pi2 obtained by amplifying F1 and R1 from the total DNA (Deoxyribose Nucleic Acid) of the rice blast-resistant variety carrying rice blast-resistant gene Pi2 by use of a primer to obtain a nucleotide fragment I and then performing enzyme digestion of the nucleotide fragment I by use of restriction endonuclease Pst I or Hinf I. The molecular marker is the first Pi2 gene-specific SNP marker developed for the sequence in the Pi2 gene, and has the advantages of high specificity and low cost and high flux in practical application, and can be widely applied to the colonies with different inheritance backgrounds. By adopting the molecular marker, the utilization efficiency of the gene in molecular marker-assisted selective breeding, gene pyramiding breeding and transgenic breeding can be improved.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
Herbicide resistance gene and application thereof
InactiveCN101892247AExpand the spectrum of weed controlImprove the ability to resist adversity stress such as saltBacteriaMicroorganism based processesPlant cellEukaryotic plasmids
The invention discloses a herbicide resistance gene and an application thereof. The herbicide resistance gene comprises a base sequence in SEQ ID NO:1. The invention also discloses a herbicide resistance protein and a plant expression plasmid, wherein the herbicide resistance protein contains an amino acid sequence in SEQ ID NO:2, and the plant expression plasmid contains the herbicide resistancegene. The herbicide resistance gene is a resistance gene of an HPPD inhibitor type herbicide and is led into plant cells through the gene engineering technology to prepare a transgenic plant which has natural resistance to the HPPD inhibitor type herbicide and simultaneously has high stress resistance such as salt resistance and the like.
Owner:HEBEI UNIVERSITY
Method and kit for detecting mycobacterium tuberculosis and drug-resistant gene mutation thereof
ActiveCN101144099AImprove diffusivityIncrease concentrationMicrobiological testing/measurementFermentationResistant genesMicrobiology
The present invention relates to a method and a reagent kit which are used for detecting the existence of the mycobacterium tuberculosis in the clinical biological sample and the mutations of the drug-resistant genes, in particular to a method and a reagent kit which are used for quickly detecting the existence of the mycobacterium tuberculosis in the biological samples such as the clinical sputamentum, the bronchoalveolar lavage fluid, the blood, the ascites, and the cerebrospinal fluid, etc. with the reverse dot-blot hybridization technology.
Owner:GUANGZHOU DARUI BIOTECH
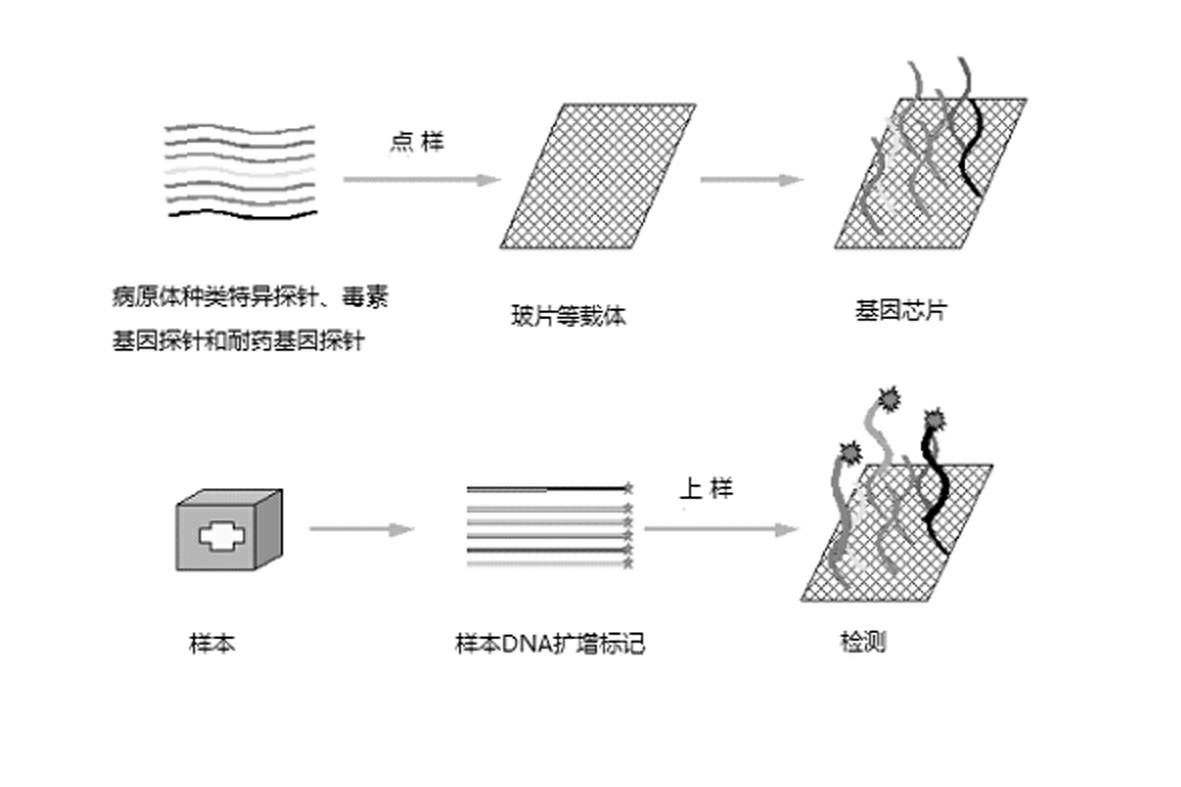
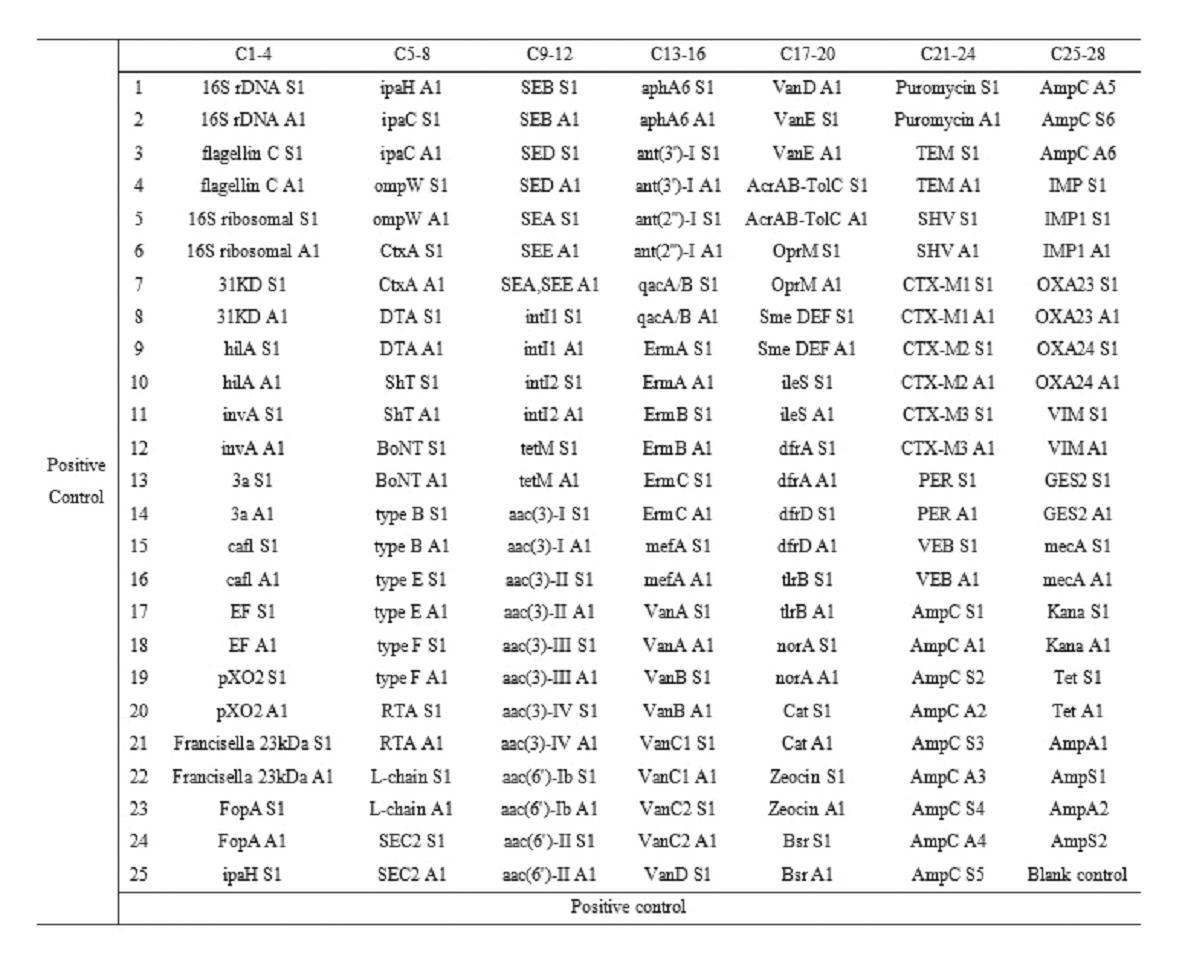
Gene chip for high-flux detection of pathogens and application thereof
InactiveCN102534013AStrong specificityDetermine the typeMicrobiological testing/measurementAgainst vector-borne diseasesYersinia pestisBrucella
The invention relates to a gene chip for high-flux detection of pathogens and application thereof. The gene comprises (1) a combination of 174 oligonucleotide probes of pathogen variety specific genes, toxin genes and drug-resistant genes; and (2) a probe array, which is formed by curing the oligonucleotide probes on a carrier material by arm molecules. The gene chip comprises 174 gene probes, namely 32 pathogen variety specific gene probes of the following 8 pathogens of Burkholderia mallei, Burkholderia pseudomallei, Brucella, salmonella, Yersinia pestis, Bacillus anthracis, comma bacillus and the like, 25 toxin gene probe of the following 7 toxins of diphtheria toxin, Shiga toxin, staphylococcus enterotoxin, choleratoxin and the like, and 117 drug-resistant gene probes of 17 drug-resistant genes of extended-spectrum beta-lactamase, cephalosporinase, carbapenemase, integrase gene, common gene engineering carrier drug-resistant gene and the like. The gene chip can be used to detect multiple pathogen variety specific genes, toxin genes and drug-resistant genes.
Owner:李越希
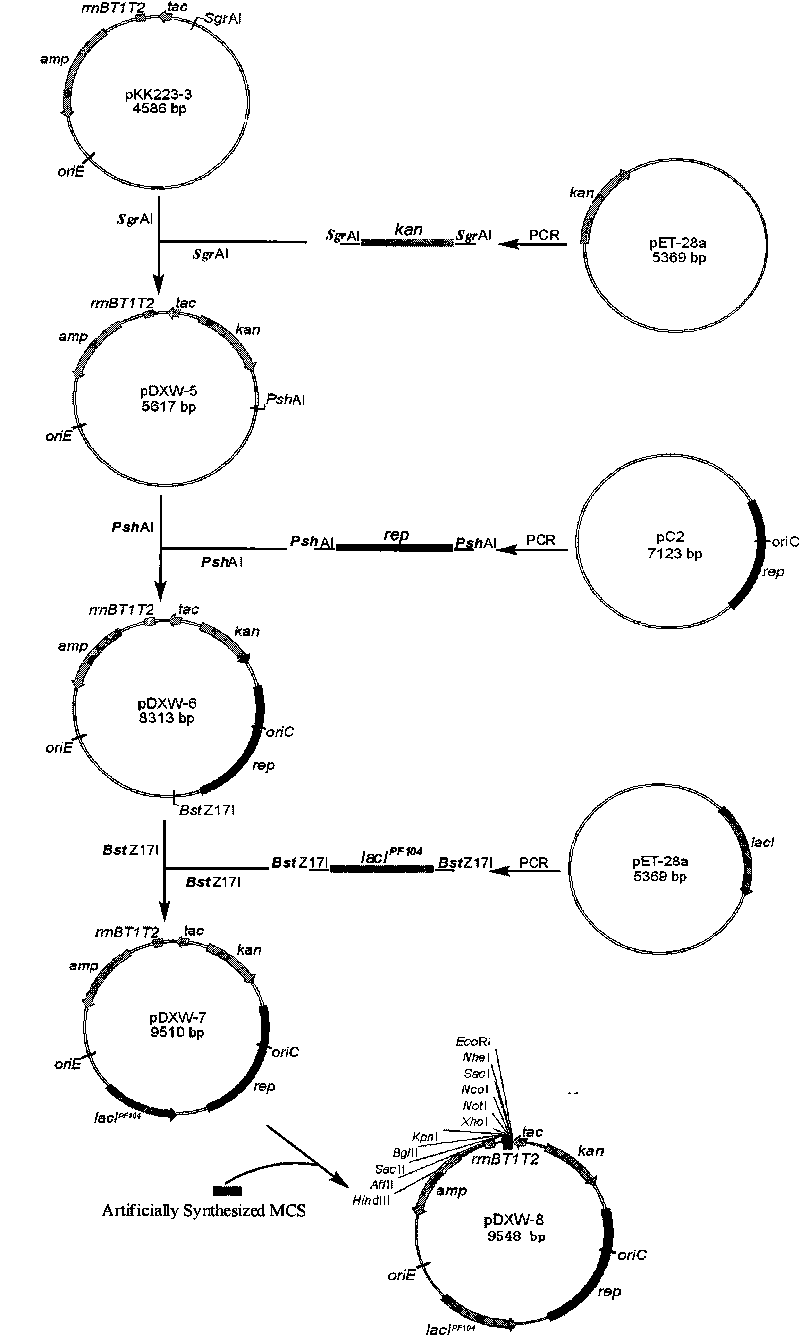
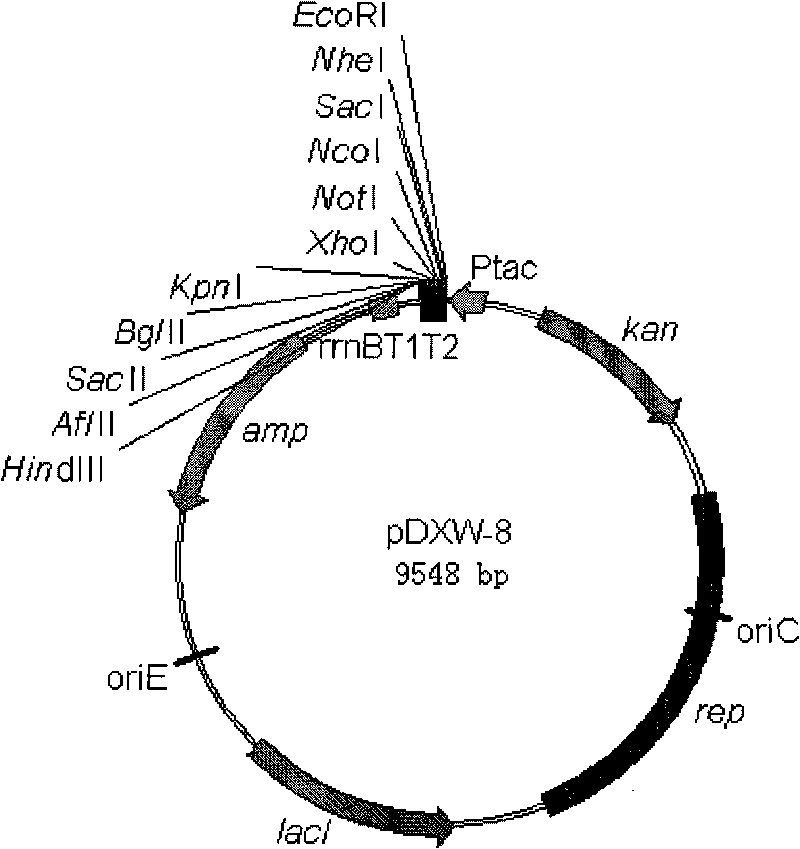
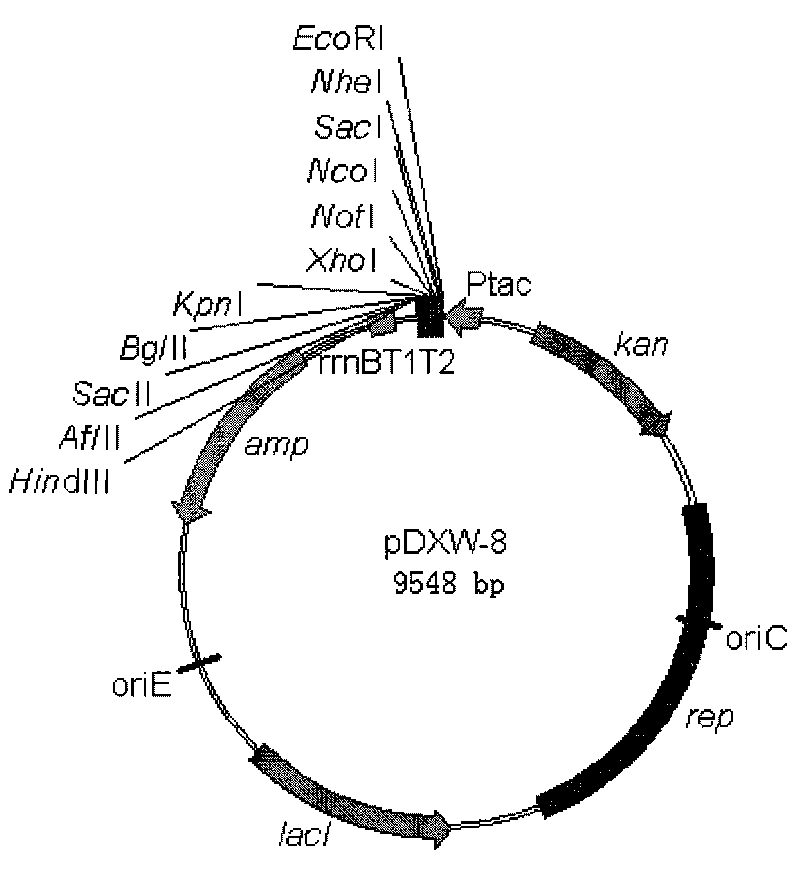
Colibacillus-corynebacterium inducible expression carrier pDXW-8 and building method thereof
ActiveCN101693901AEffective cloningReduced activityMicroorganism based processesVector-based foreign material introductionCorynebacterium efficiensEscherichia coli
A colibacillus-corynebacterium inducible expression carrier pDXW-8 and a building method thereof belong to the technical field of genetic engineering. The inducible expression carrier pDXW-8 can be copied and stably exists in colibacillus and corynebacterium and comprises a tac promoter, a terminator rrnBT1T2, a resistance marker card kanamycin mark resistance gene, a negative regulation gene laclPE104 and a multi-cloning site, wherein the tac promoter plays a transcript promoting function in the colibacillus and the corynebacterium, the terminator rrnBT1T2 has a transcript stopping function, the resistance marker card kanamycin mark resistance gene is used for selecting transformants of the corynebacterium, the negative regulation gene is used for severely controlling expression of the tac promoter, and the multi-cloning site includes 11 signal limited incision enzyme sites. In the corynebacterium, the carrier is moderate in foreign protein expression, and is particularly adaptable to study on corynebacterium metabolic engineering.
Owner:JIANGNAN UNIV
Rice blast resistance gene Pi7 and application thereof
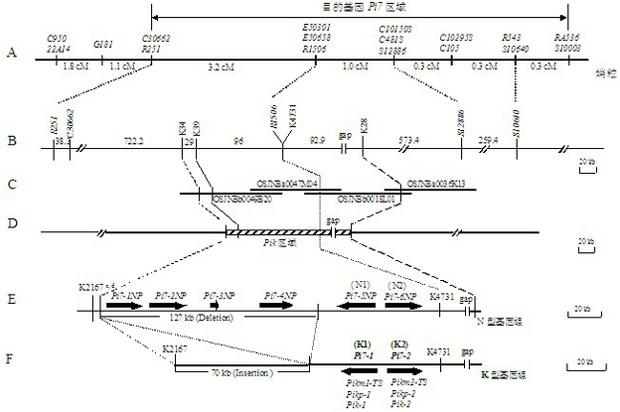
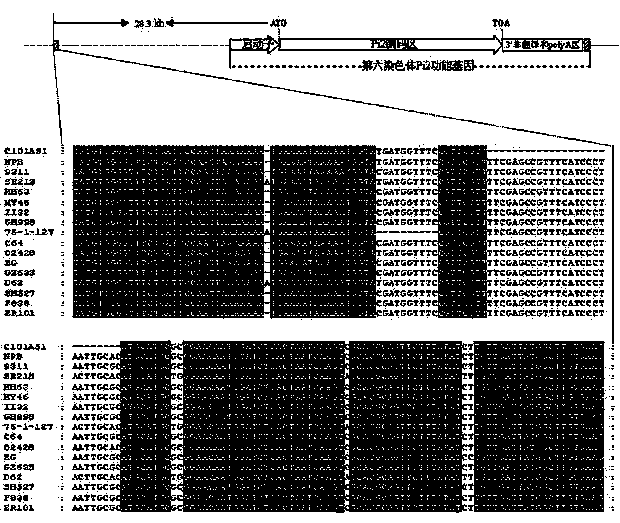
InactiveCN102094027AObvious advantagesObvious effectPlant peptidesFermentationBiotechnologyBinding site
The invention discloses a rice blast resistance gene Pi7 and application thereof, and provides a nucleotide sequence of the rice blast resistance gene Pi7 and an amino acid polypeptide sequence coded thereby. The nucleotide sequence comprises two genes which belong to members of a coiled-coil nucleotide binding site leucine-rich repeat (CC-NBS-LRR) resistance gene family and are constitutive expression genes. The invention also provides application of the gene to the aspect of transforming rice or breeding disease-resistant varieties of other plants and application of a molecular marker produced according to the gene sequence to breeding.
Owner:SOUTH CHINA AGRI UNIV
Brown planthopper resistant gene in rice and use thereof
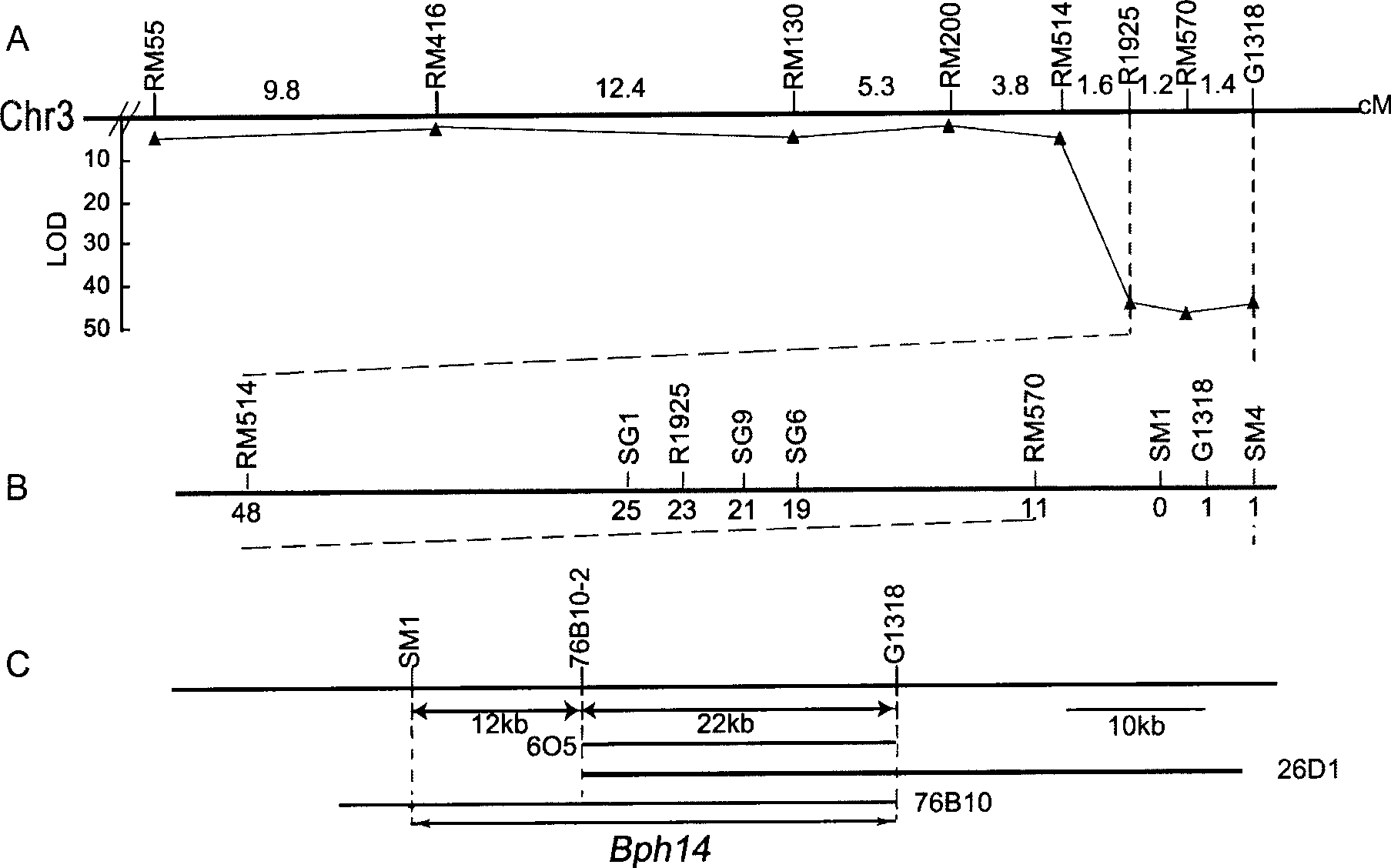
The invention provides a brown planthopper resistance gene Bph14 in rice. The gene has a nucleotide sequence shown in SEQ ID No. 1 and a cDNA sequence thereof is shown in SEQ ID No.2. The gene Bph14 of the invention belongs to CC-NBS-LRR gene family, and coded protein is related to plant disease resistance. The gene Bph14 has function of resisting the brown planthopper. Through genetic transformation and hybridization, the gene Bph14 is transferred into normal rice varieties, which can improve the resistance of the rice to the brown planthopper, thus reducing the damage caused by the brown planthopper and achieving the purposes of increasing and stabilizing the production of rice.
Owner:WUHAN UNIV
Molecular marker of rice-blast-resistant gene Pi2 and application of molecular marker
InactiveCN104073487AChoose a clear goalImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyNucleotide
The invention belongs to the field of crop biotechnology and in particular relates to a molecular marker of a rice-blast-resistant gene Pi2 and application of the molecular marker. The upstream and downstream molecular markers are based on specific insertion / deletion loci polymorphism of nucleotide sequences shown in SEQ ID NO.1 and SEQ ID NO.2. The two molecular markers are co-dominant molecular markers developed for close linkage regions on two sides of the functional gene Pi2, and have the advantages of being high in accuracy and capable of being widely applied to different breeding parents. By detecting the two molecular markers, whether a detected rice material has the rice-blast-resistant gene Pi2 or not can be accurately judged. By virtue of a molecular marker combination, whether a hybrid transferred progeny plant genome of a parent of the functional gene Pi2 and other parents has the functional gene Pi2 or not and the homozygous state of the functional gene Pi2 can be accurately detected, and the breeding efficiency of a rice-blast-resistant rice variety with the functional gene Pi2 is improved.
Owner:BIOLOGICAL TECH INST OF FUJIAN ACADEMY OF AGRI SCI
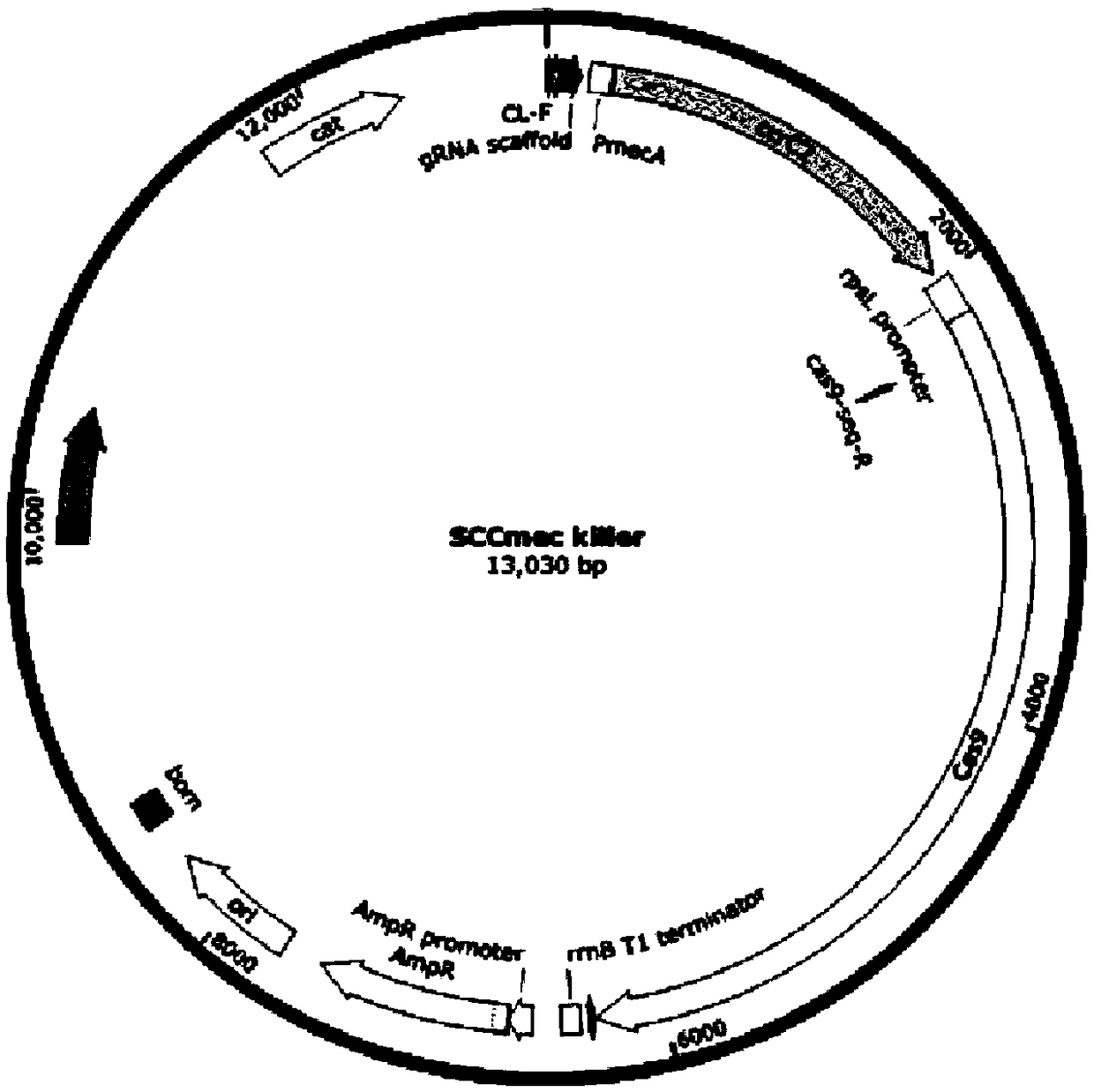
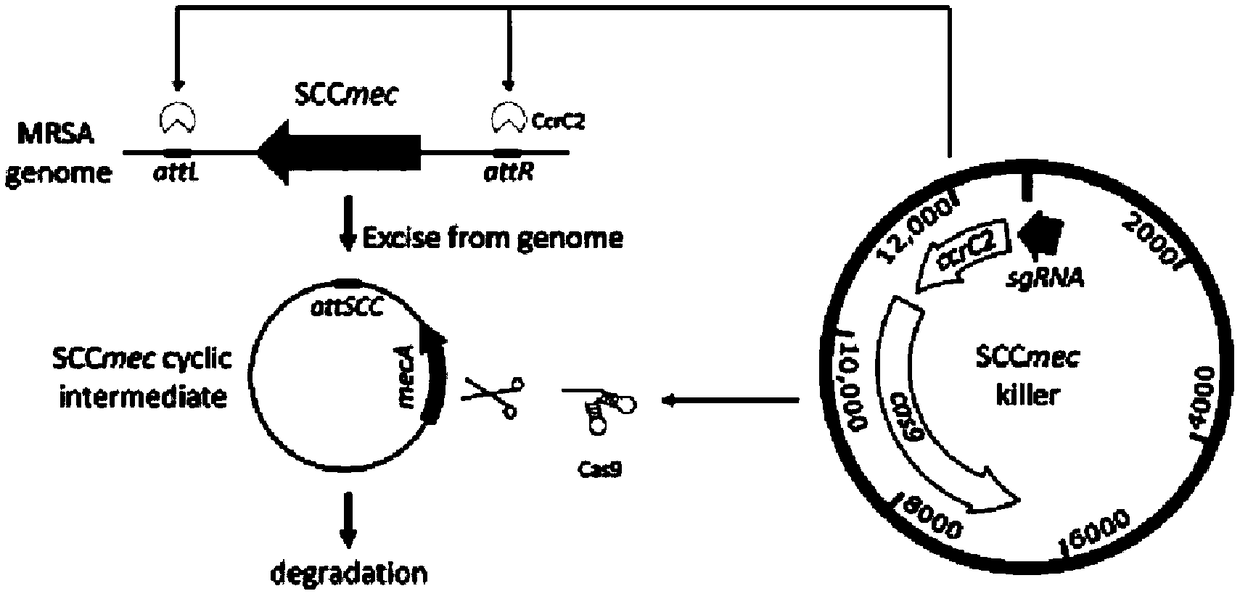
Gene drive carrier and construction method thereof
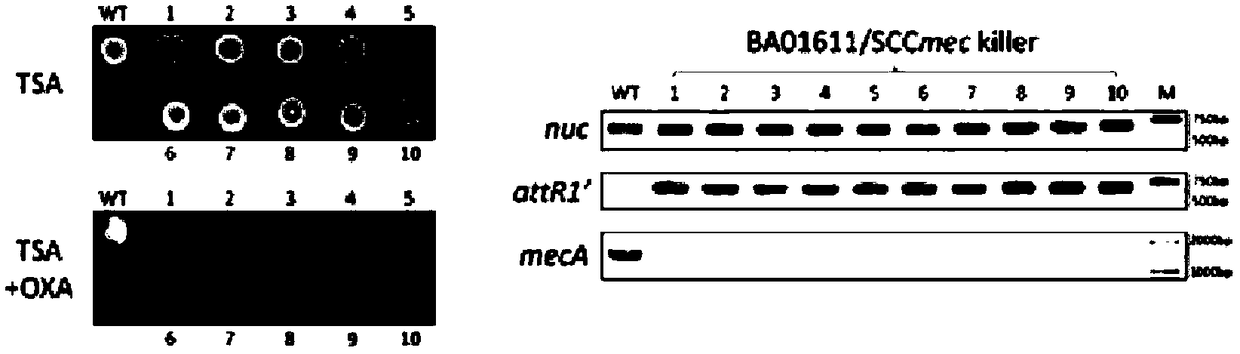
InactiveCN108707620AAvoid getting againElimination of the methicillin resistance geneVector-based foreign material introductionResistant genesCarrier system
The invention belongs to the field of bioengineering and particularly relates to a Gene drive carrier and a construction method thereof. The construction method includes the steps of 1), artificiallysynthesizing a cap5A promoter and an sgRNA fragment; 2), amplifying a cas9 gene; 3), amplifying a plasmid skeleton; 4), amplifying an rpsL promoter; 5), assembling an empty carrier; 6), inserting a spacer targeted at the formula described; 7), constructing the carrier. In the method, a novel chromosome box recombinase CcrC2 and CRISPR-Cas9 technology are combined in an attempting manner, an SCCmeckiller carrier system, namely the Gene driver carrier is constructed, the Gene drive acts on methicillin-resistant staphylococcus aureus MRSA, staphylococcal chromosome box SCCmec is targeted to remove from the methicillin-resistant staphylococcus aureus MRSA, and methicillin-resistant genes are thereby eliminated.
Owner:NORTHWEST A & F UNIV
Asymmetric magnetic mesoporous silica rod supporting chemotherapeutic and gene drugs and application thereof to tumor diagnosis and treatment
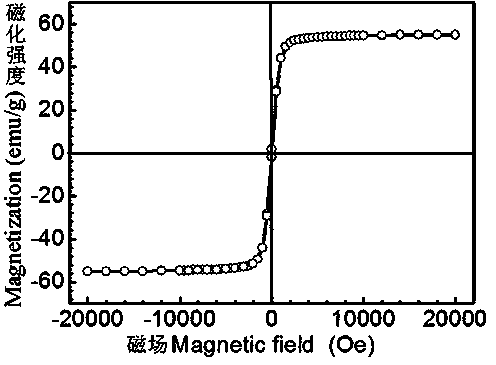
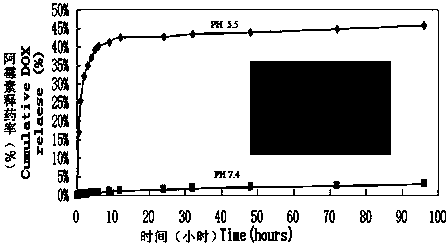
InactiveCN104225599AGood treatment effectReal-time monitoring of treatment effectGenetic material ingredientsInorganic non-active ingredientsBiocompatibility TestingMesoporous silica
The invention relates to the field of nanometer drug carriers, and concretely relates to an asymmetric magnetic mesoporous silica rod supporting chemotherapeutic and gene drugs and application thereof to tumor diagnosis and treatment. The asymmetric magnetic mesoporous silica rod is prepared by employing spherical magnetic ferrite nanoparticles and ethyl orthosilicate through a sol-gel method, and the asymmetric magnetic mesoporous silica rod is subjected to surface functionalization modification, and is successively loaded with a chemotherapeutic drug, coated by a positive high-molecular polymer and loaded with a gene drug, so that a target product is obtained. The chemotherapeutic drug is connected with the silica rod through functionalization of the mesoporous surface, and the silica rod is endowed with the pH-responsive drug release characteristic, also the biocompatibility of the composite material is increased and the in-vivo cycling time is prolonged, and gene is supported in an electrostatic adsorption mode. The composite material is injected into a living body via an intravenous route, the characteristics of nanoparticle in-vivo passive targeting, gene guiding and pH-responsive drug release of the composite material are utilized, also the cooperativity of the multidrug resistant gene and the chemotherapeutic drug is utilized, and in-vitro magnetic targeting, NMR imaging and other technologies are applied to diagnosis and treatment of malignant tumors.
Owner:JILIN UNIV
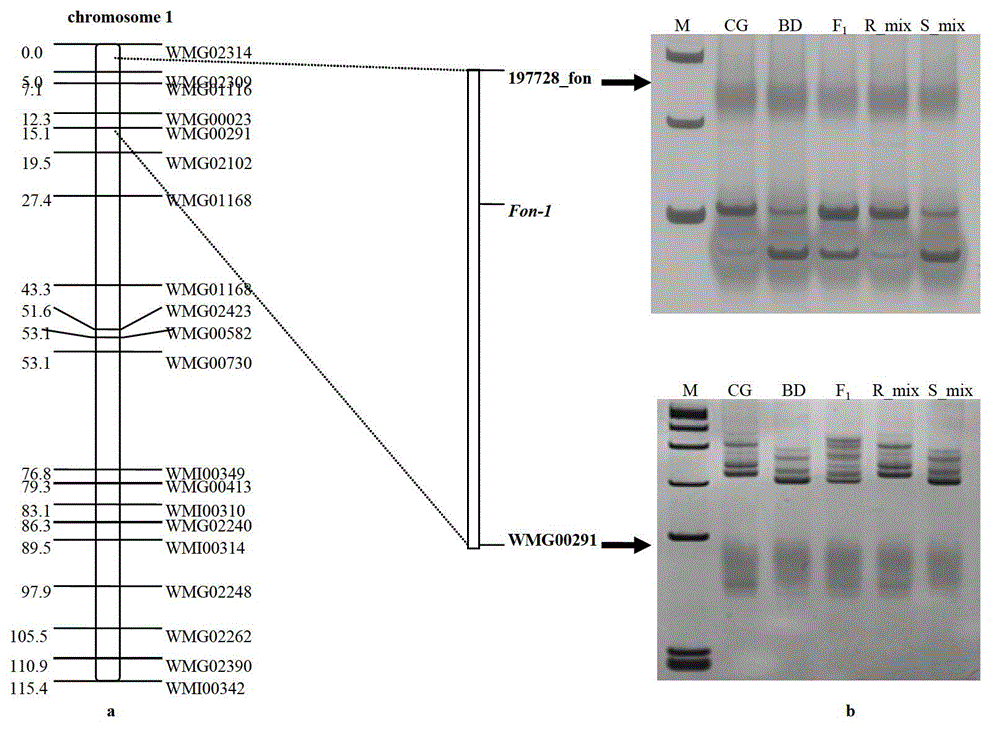
SNP loci linked with blight resistant gene Fon-1 in watermelon, and markers thereof
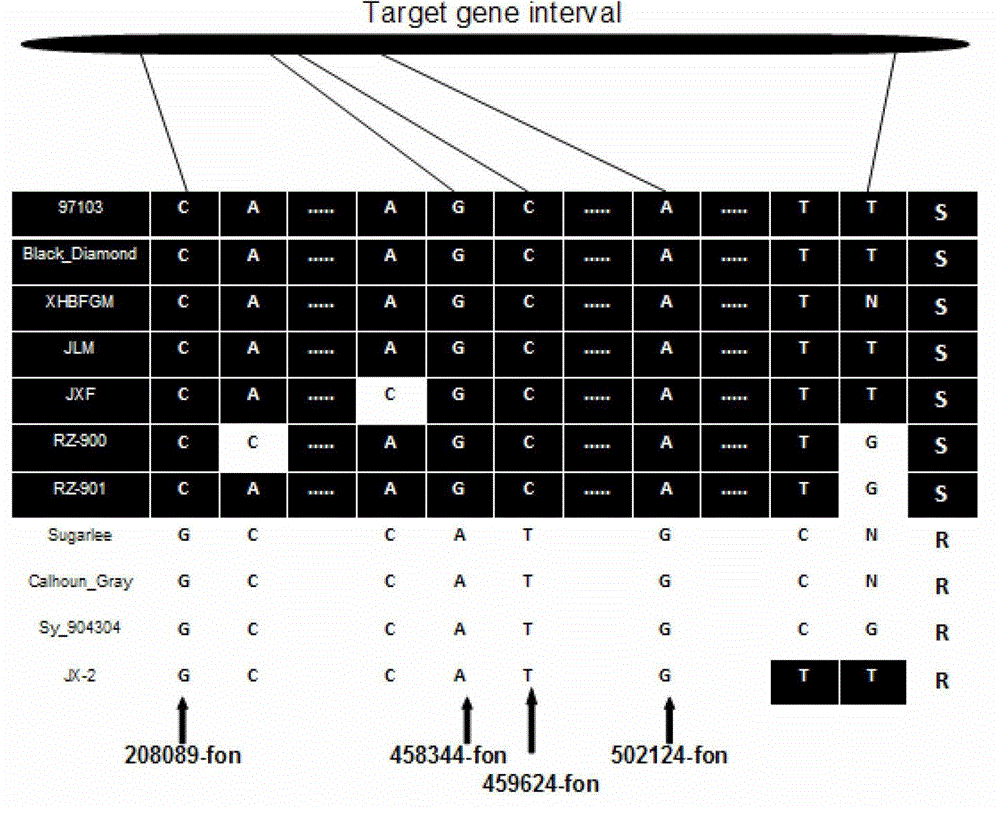
ActiveCN103146691AShorten the timeTo achieve the purpose of assisted breedingDNA preparationDNA/RNA fragmentationResistant genesTest material
The invention relates to the fields of gene sequences and acquiring methods thereof, and concretely relates to SNP loci linked with a blight resistant gene Fon-1 in a watermelon, markers thereof, and acquiring methods of the loci and the markers. The SNP loci linked with a blight resistant gene Fon-1 in a watermelon and the markers thereof have nucleotide base sequences represented by SEQ ID NO:1-8 in a sequence table. The acquiring method of the loci comprises the following steps: 1, selecting a tested material; 2, carrying out preliminary positioning of the blight resistant gene Fon-1 in the watermelon; 3, acquiring candidate SNP; and 4, acquiring the SNP loci closely linked with the blight resistant gene Fon-1 in the watermelon, and verifying the candidate SNP loci. According to the invention, the phenotype identification and the molecular detection of F2 generation separation populations and natural populations are carried out through utilizing a series of markers to confirm the close linkage degree of the series of the SNP loci with target properties; and the SNP loci are utilized to design the markers respectively, and the markers can be utilized to carry out the initial-stage screening of the kind in order to reach a molecule assisted breeding purpose, so the breeding period is substantially shortened, and an important scientific research meaning is possessed.
Owner:京研益农(北京)种业科技有限公司
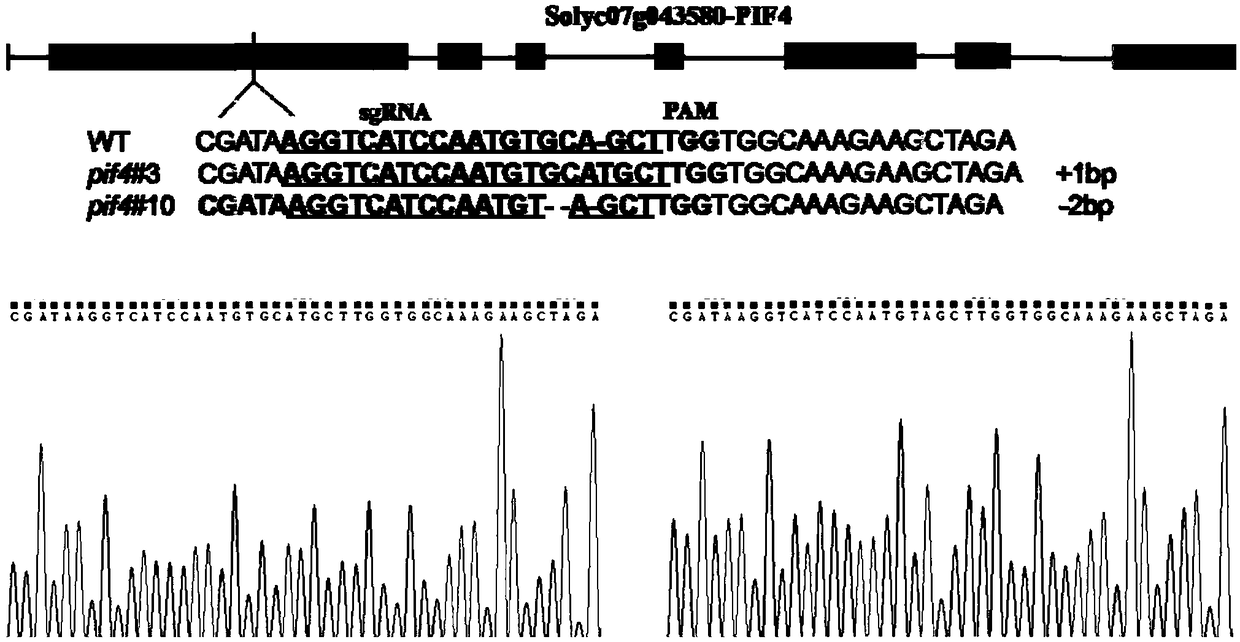
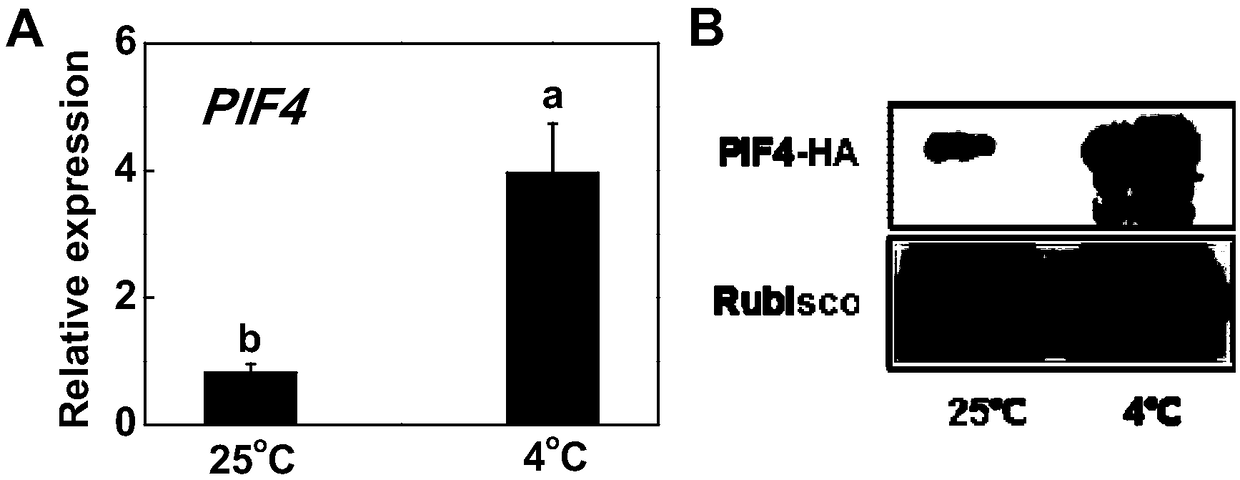
Tomato Sl1PIF4 gene, protein and application thereof to improving low temperature resistance of plant
ActiveCN109456394AImprove low temperature resistancePromote accumulationPlant peptidesFermentationBiotechnologyPlant hormone
The invention discloses a tomato S1lPIF4 gene, protein and application thereof to improving the low temperature resistance of plant. The nucleotide sequence of the S1lPIF4 gene is SEQ ID NO.1, and thecorresponding amino acid sequence is as shown in SEQ ID NO. 2. A tomato SlIPIF4 overexpression plant or a gene knockout plant through the gene means to regulate the expression level of the S1lPIF4 gene in order to do research on the regulatory mechanism of the S1lPIF4 gene on the low temperature resistance of tomato. According to the result, overexpression of S1lPIF4 at the lower temperature canpromote accumulation of plant hormone abscisic acid (ABA) and jasmonic acid (JA), inhibits accumulation of gibberellin (GA), further induces expression of the low temperature resistant gene of the tomato, and finally improves the low temperature resistance of the tomato. Therefore, the S1lPIF4 gene of the tomato enhances the low temperature resistance by inducing formation of hormones ABA and JA in the body. The tomato S1lPIF4 gene provides the gene resource for breeding new varieties of low temperature resistant tomato, has the good potential application value, and lays theoretical basis fordoing research on tomato plant distress response signal mechanism and molecular mechanism of being tolerant to adverse environment.
Owner:ZHEJIANG UNIV
Unit cell fungus, engineering fungus prepared through motion ferment and application
InactiveCN1600850ALess quantityIncreased conversion efficiency to ethanolBacteriaMicrobiological testing/measurementResistance mutationBiology
This invention relates to motion fermentation monocell engineering bacterium, its prepn. method and its application in fermentation for producing ethanol. In the prepn. of said engineering bacterium, the resistance gene of tetracycline is inserted into glucose-fructose oxide-erductase gene of motion fermentation monocell baterium XW101, endo zyme being used for digesting said recombination plasmid to form linear plasmid, after the transformation of motion fermentation monocell bacterium XW101, screening the strain having tetracycline resistance mutation. Carbon source is used as substrate, with this inventive engineering bacterium to produce ethanol with high ethanol prodn. yield and less by-produt-sorbicolan.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
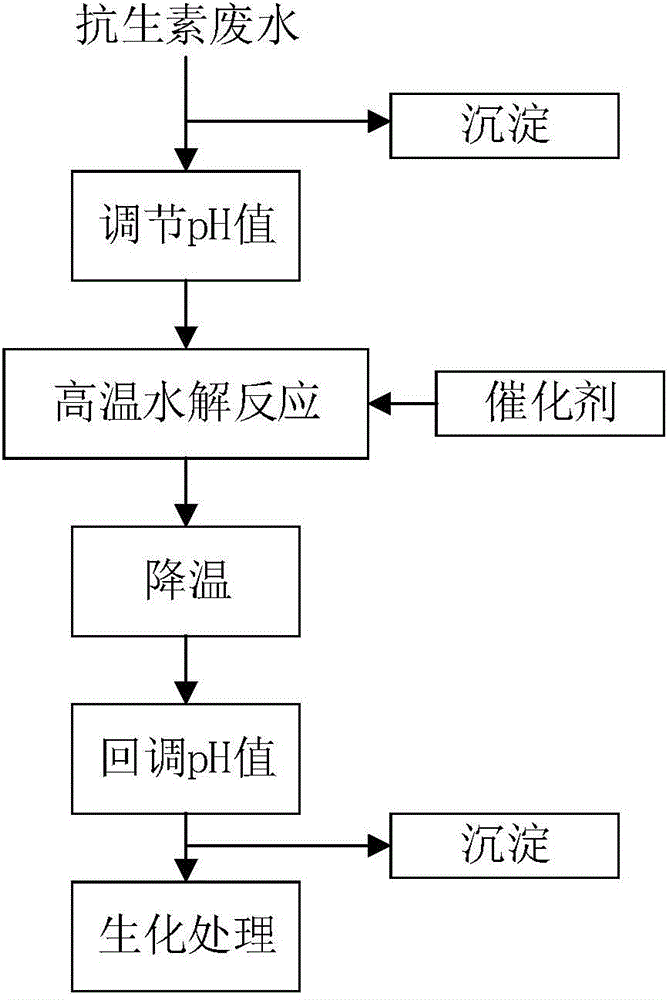
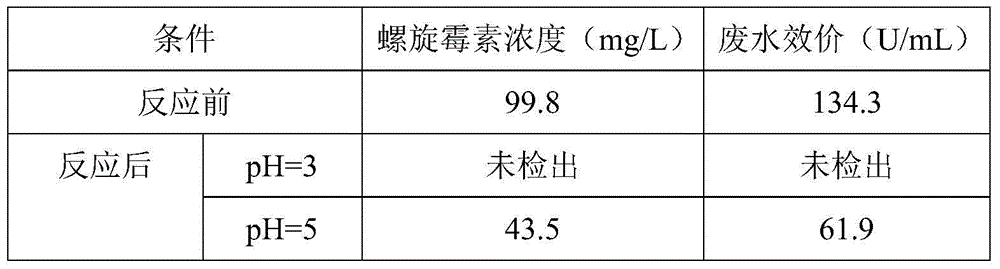
Pretreatment method for removing antibiotics in ferment antibiotic pharmaceutical wastewater
ActiveCN105084442ALower titerEfficient decompositionMultistage water/sewage treatmentWater/sewage treatment by heatingMicroorganismResistant bacteria
The invention provides a pretreatment method for removing antibiotics in ferment antibiotic pharmaceutical wastewater. The method removes antibiotics in the ferment antibiotic pharmaceutical wastewater mainly through adjustment of the pH value of the wastewater and high-temperature catalytic hydrolysis, and the treated pharmaceutical wastewater without antibiotics can enter a subsequent biochemical treatment process for treatment. The method provided by the invention can basically remove antibiotics in the pharmaceutical wastewater, reduce inhibition effect of high-concentration antibiotics on microbes, lower down difficulty in treatment of the wastewater by using the subsequent biochemical process and decrease generation of drug-resistant bacteria and drug-resistant genes in subsequent biochemical treatment.
Owner:RES CENT FOR ECO ENVIRONMENTAL SCI THE CHINESE ACAD OF SCI
Biochip for detecting drug resistant genes of helicobacter pylori clarithromycin and preparation method and application thereof
InactiveCN101665824ADetermining drug resistanceSimple methodNucleotide librariesMicrobiological testing/measurementResistant genesClarithromycin
The invention belongs to the fields of genetic engineering and medical inspection, and relates to a biochip for detecting drug resistant genes of drug resistant genes and a preparation method and an application thereof. The biochip is prepared by the following steps: carrying out aldehyde processing for a chip base; connecting a 5' end of a probe with a molecule arm of poly (dT)10; then carrying out amino modification for the 5' end of the probe; aiming at the mutants at the following positions of helicobacter pylori 23S rRNA genes by the probe: A2115G, A2142G, A2142C, A2143G, A2143C, T2182C and G2224A; and fixing the probe on the chip base. The biochip has high sensitivity, good specificity and low detection cost, can distinguish the difference of monobasic bases, and is suitable for being popularized and used in establishment units.
Owner:周玉贵
Beet stress-resistant yield and sugar increasing series composite growth regulator and systematic chemical regulation method
InactiveCN101946803AIncrease productionIncrease the sugar contentBiocidePlant growth regulatorsDiseaseAlkali soil
The invention relates to beet stress-resistant yield and sugar increasing series composite growth regulator and a systematic chemical regulation method. The series composite growth regulator comprises chemical control I composite growth regulator, chemical control II composite growth regulator, chemical control III composite growth regulator and chemical IV composite growth regulator; the beet stress-resistant yield and sugar increasing systematic chemical control method sprays the four regulators on the surface of leaves respectively according to the four growth stages of the beet: seedling stage, leafage forming stage, root and sugar increasing stage and sugar accumulation stage, induces the expression of the beet stress-resistant genes, sufficiently explores the potential capability of resisting saline and alkali, drought and diseases of the beet in the full growth stage, and increases the yield and sugar content. In the saline-alkali soil with average salt content of 0.51% and Ph value of 8.4 in west Jilin where the annual precipitation is less than 300 mm, the average beet yield is 53.33 tons per hectare, an increase of 27.26%, and the sugar content is 17.6%, an increase of 2.1%.
Owner:JILIN ACAD OF AGRI SCI
Clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers, primers and selection method of clubroot-resistant plant
InactiveCN103275972AImprove breeding efficiencyShorten the breeding processMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyResistant genes
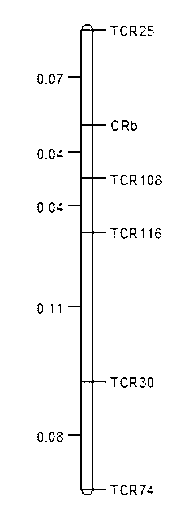
The invention provides 5 clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers, primers and a selection method of a clubroot CRb-resistant plant. The 5 clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers comprise TCR25, TCR108, TCR116, TCR30 and TCR74. Distances of the gene CRb respectively to the TCR25, the TCR108, the TCR116, the TCR30 and the TCR74 are 0.07cM, 0.04cM, 0.08cM, 0.19cM and 0.27cM. The 5 clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers can be used for molecular marker-assistant selection of clubroot-resistant genes of brassica such as Chinese cabbage and Shanghai Bak choi. In assistant selection, the 5 clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers are used simultaneously so that theoretical selection accuracy is 100%. The 5 clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers have good repeatability, high reliability and a low detection cost and saves time and labor.
Owner:SHENYANG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com