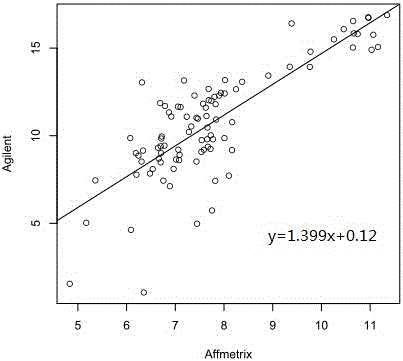
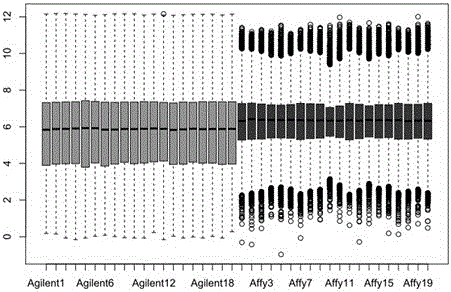
Patents
Literature
50 results about "Common gene" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
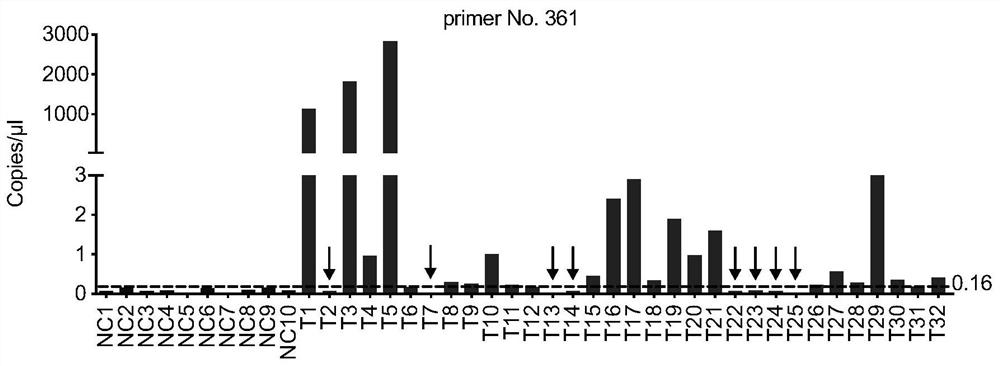
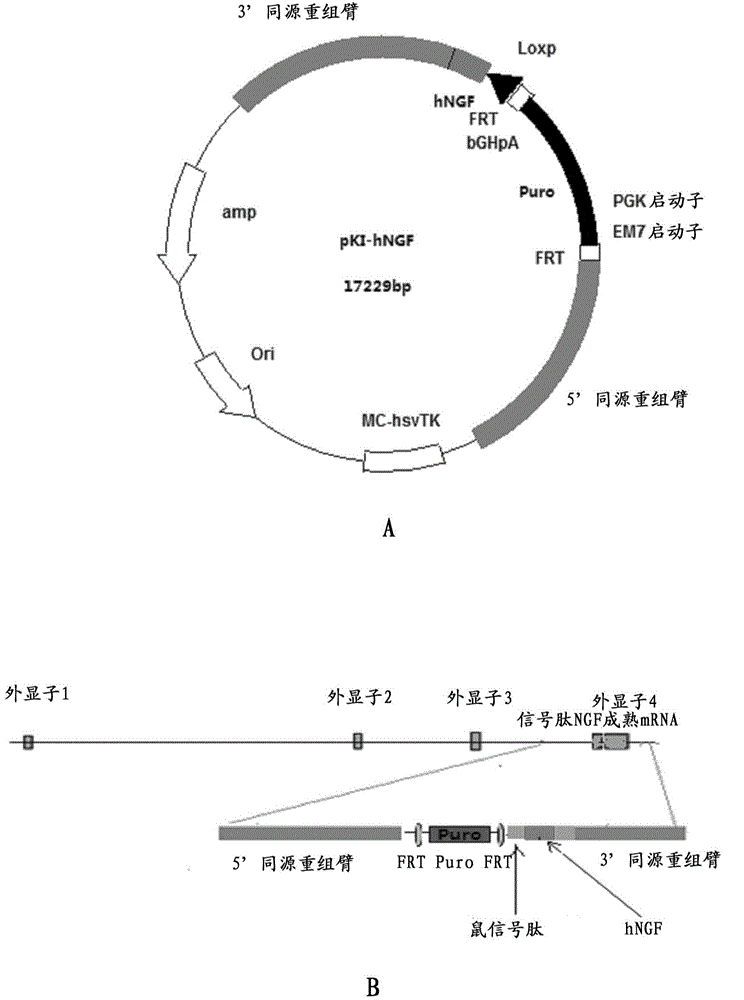
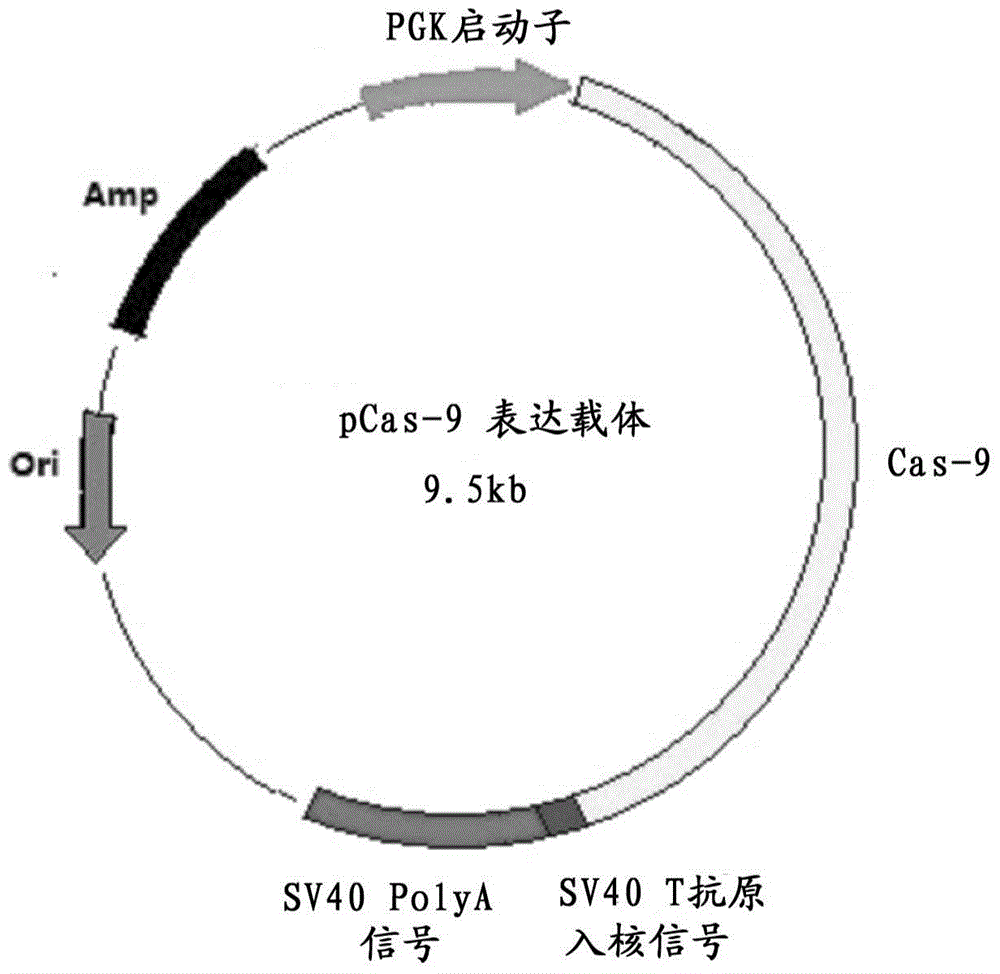
Preparation method for transgenic mice capable of producing human nerve growth factor
ActiveCN104561095AEasy to operateIncrease success rateAnimals/human peptidesVector-based foreign material introductionPlasmid dnaGenetically modified mouse
The invention discloses a method for producing transgenic mice through a homologous recombination technology. The method comprises the following steps: replacing mouse NGF genes on mouse chromosomes by human NGF genes through a Cas-9 / CRISPR gene knock-in technology, and obtaining the genes with homozygous human NGF genes through breeding and knocking the genes in mice, thus obtaining filial generation mice with salivary glands capable of secreting human NGF. Compared with the conventional targeting technology utilizing positive and negative double screening homologous recombination genes, the method disclosed by the invention is simple to operate, capable of being realized only by transfecting mouse embryonic stem cells by three plasmid DNAs or carrying out pronucleus injection on the zygotes of mice, and high in success rate which is up to 2-5% and remarkably higher than the mouse embryonic stem cell positive rate of 0.1% of the common gene targeting technology.
Owner:深圳市国创纳米抗体技术有限公司
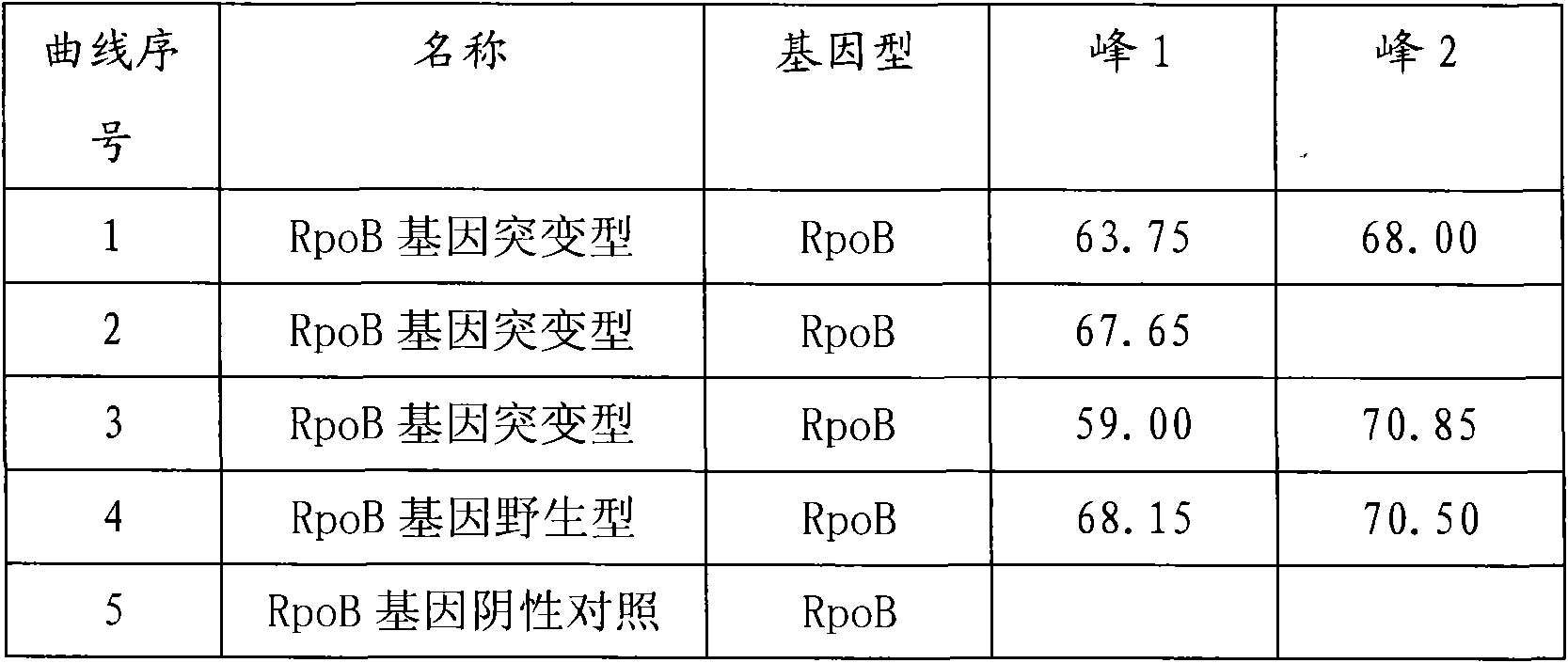
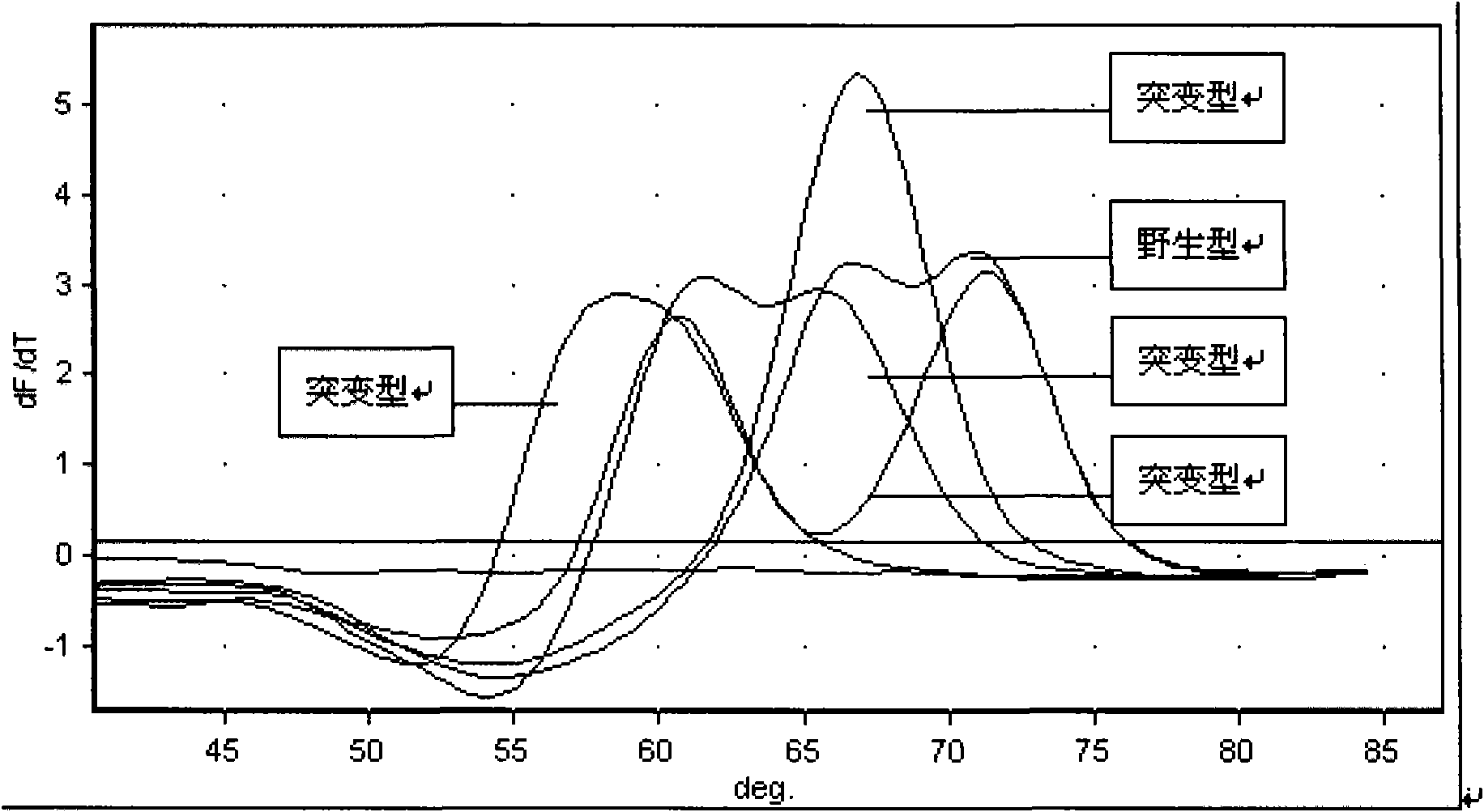
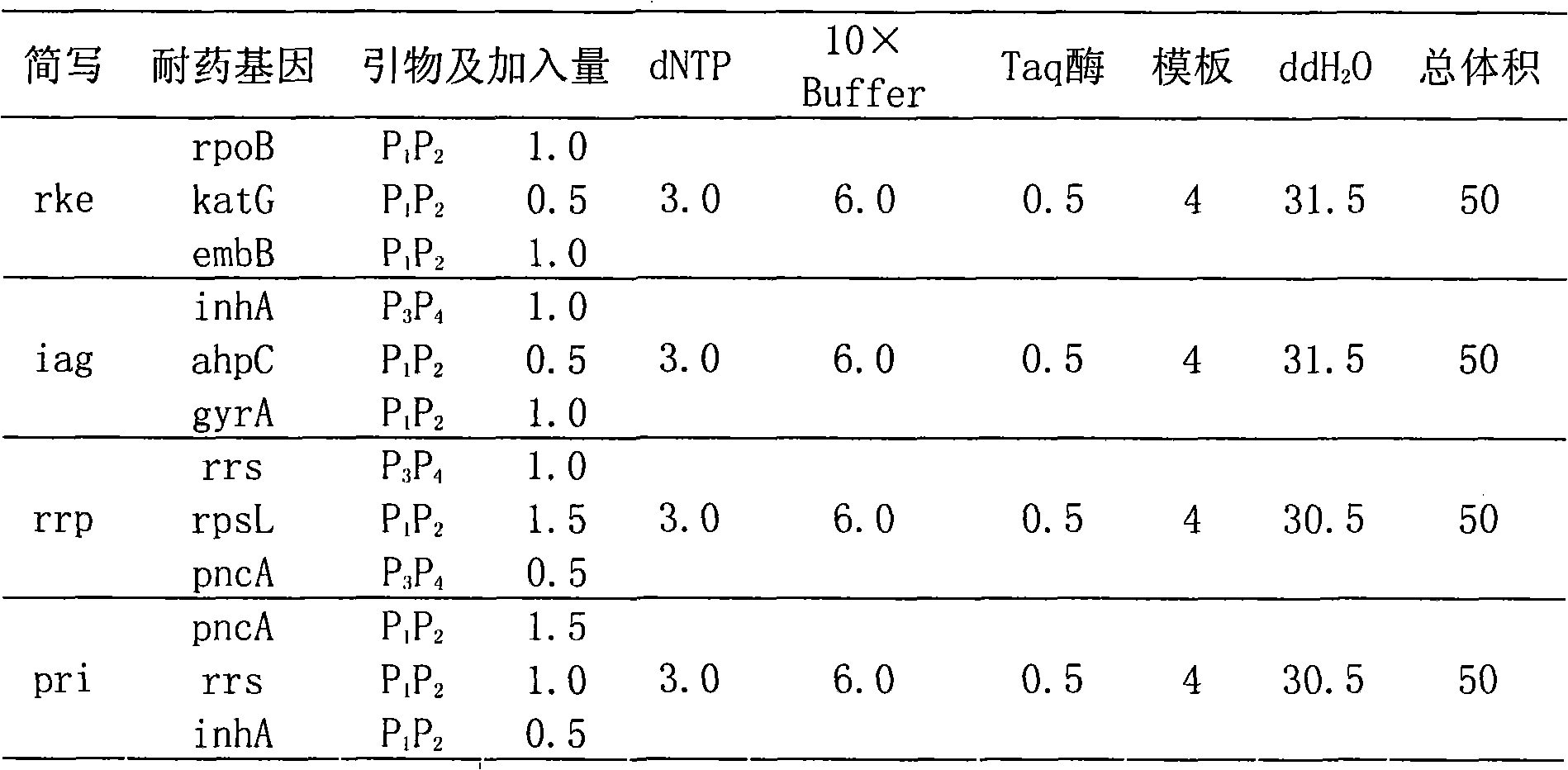
Method and kit for detecting multi-drug resistant mycobacterium tuberculosis (MDR-TB)
ActiveCN101845503AAccurate Diagnostic InformationShorten the timeMicrobiological testing/measurementFluorescence/phosphorescenceMulti-drug-resistant tuberculosisGenes mutation
The invention relates to a method adopting double-label probe detection and melting curve analysis for diagnosing the infection of multi-drug resistant mycobacterium tuberculosis and a kit which utilizes the method to detect multiple gene mutations related to drug resistant tuberculosis at the same time, and the invention belongs to the life science and biological technical field. The kit of the invention contains a primer designed for multiple gene mutations related to drug resistant, a double-label oligonucleotide probe capable of detecting multiple common gene mutation sites related to drug resistant tuberculosis and a DNA polymerases with heat stability and without 5' nuclease activity, and the kit can be used to detect at least 16 common gene mutation sites related to drug resistant tuberculosis under proper PCR reaction conditions. The detection method and kit of the invention can be used in the early diagnosis of multi-drug resistant tuberculosis and overcome the problems of the existing technology that the detection time is long, a great deal of manpower and large material resources are needed, the detection cost is high, etc.
Owner:无锡锐奇基因生物科技有限公司
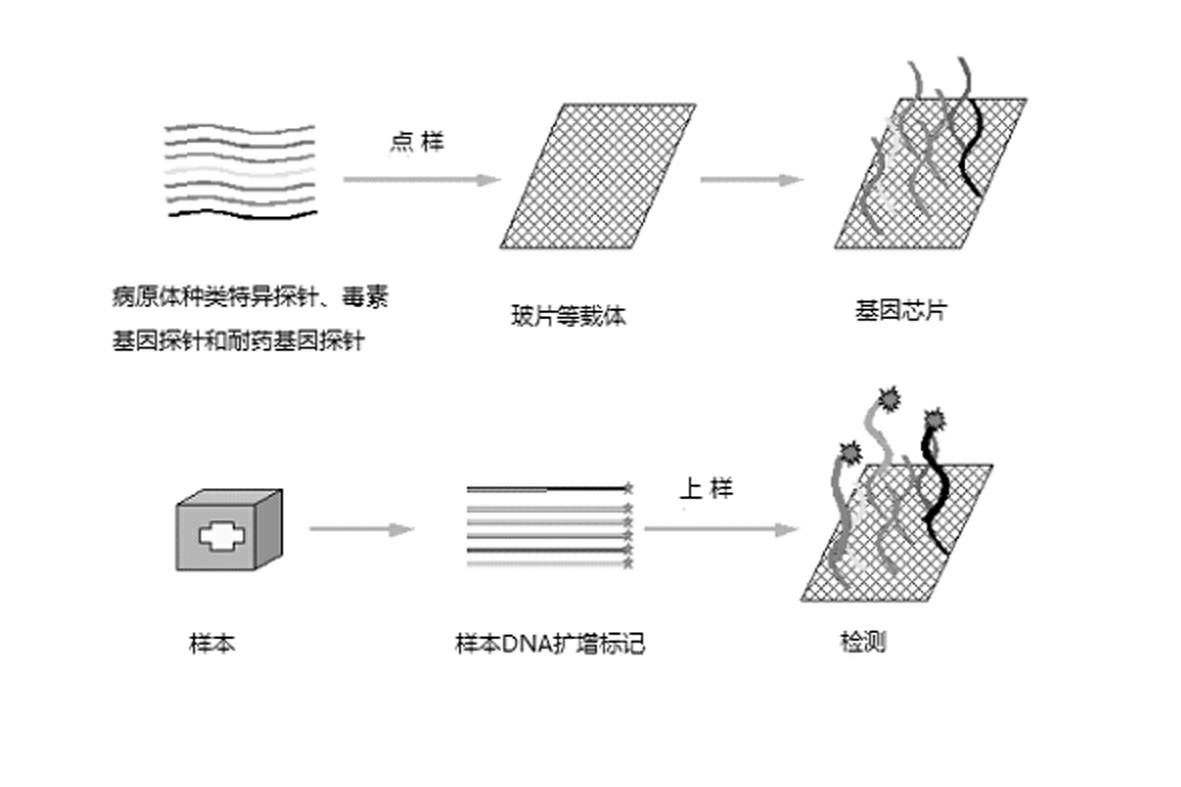
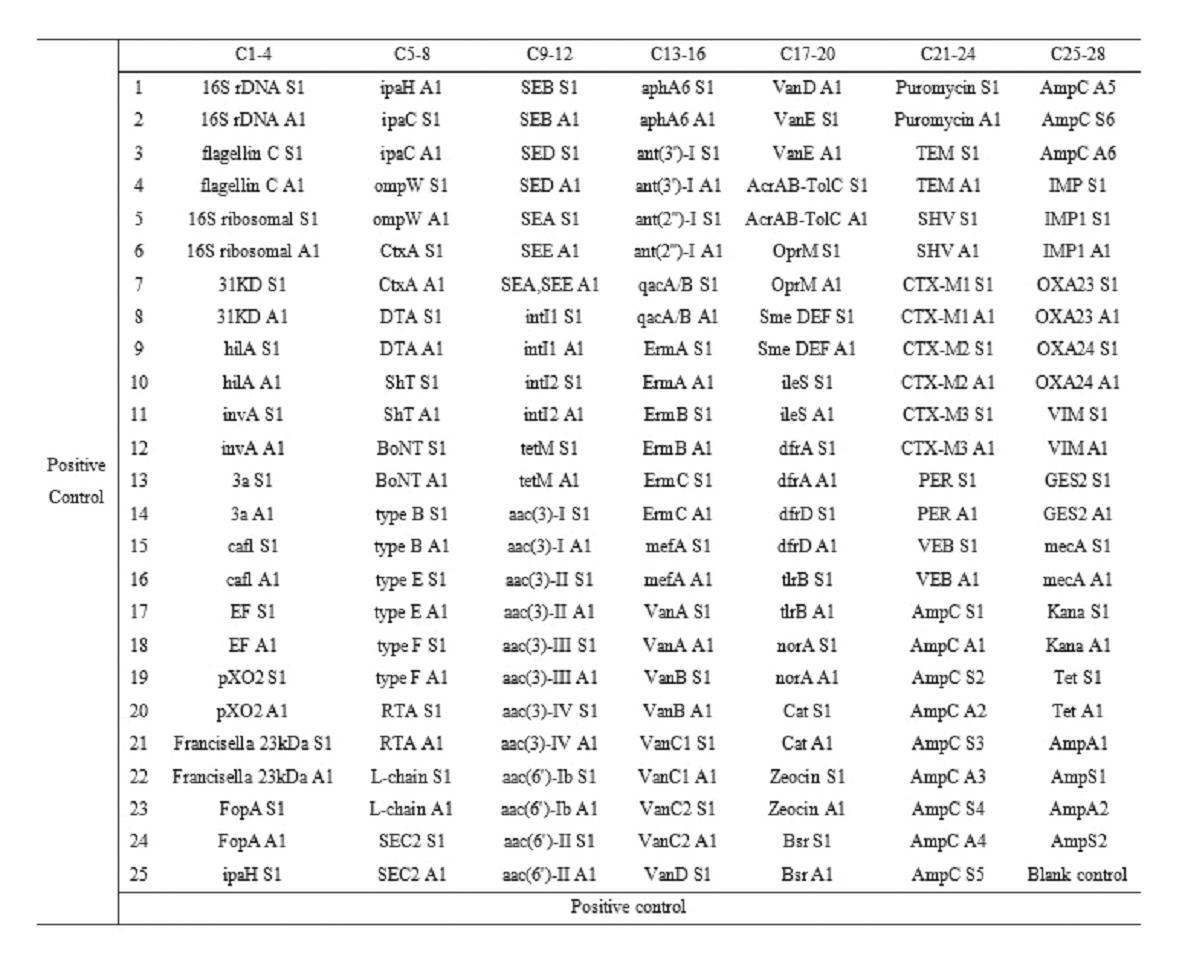
Gene chip for high-flux detection of pathogens and application thereof
InactiveCN102534013AStrong specificityDetermine the typeMicrobiological testing/measurementAgainst vector-borne diseasesYersinia pestisBrucella
The invention relates to a gene chip for high-flux detection of pathogens and application thereof. The gene comprises (1) a combination of 174 oligonucleotide probes of pathogen variety specific genes, toxin genes and drug-resistant genes; and (2) a probe array, which is formed by curing the oligonucleotide probes on a carrier material by arm molecules. The gene chip comprises 174 gene probes, namely 32 pathogen variety specific gene probes of the following 8 pathogens of Burkholderia mallei, Burkholderia pseudomallei, Brucella, salmonella, Yersinia pestis, Bacillus anthracis, comma bacillus and the like, 25 toxin gene probe of the following 7 toxins of diphtheria toxin, Shiga toxin, staphylococcus enterotoxin, choleratoxin and the like, and 117 drug-resistant gene probes of 17 drug-resistant genes of extended-spectrum beta-lactamase, cephalosporinase, carbapenemase, integrase gene, common gene engineering carrier drug-resistant gene and the like. The gene chip can be used to detect multiple pathogen variety specific genes, toxin genes and drug-resistant genes.
Owner:李越希
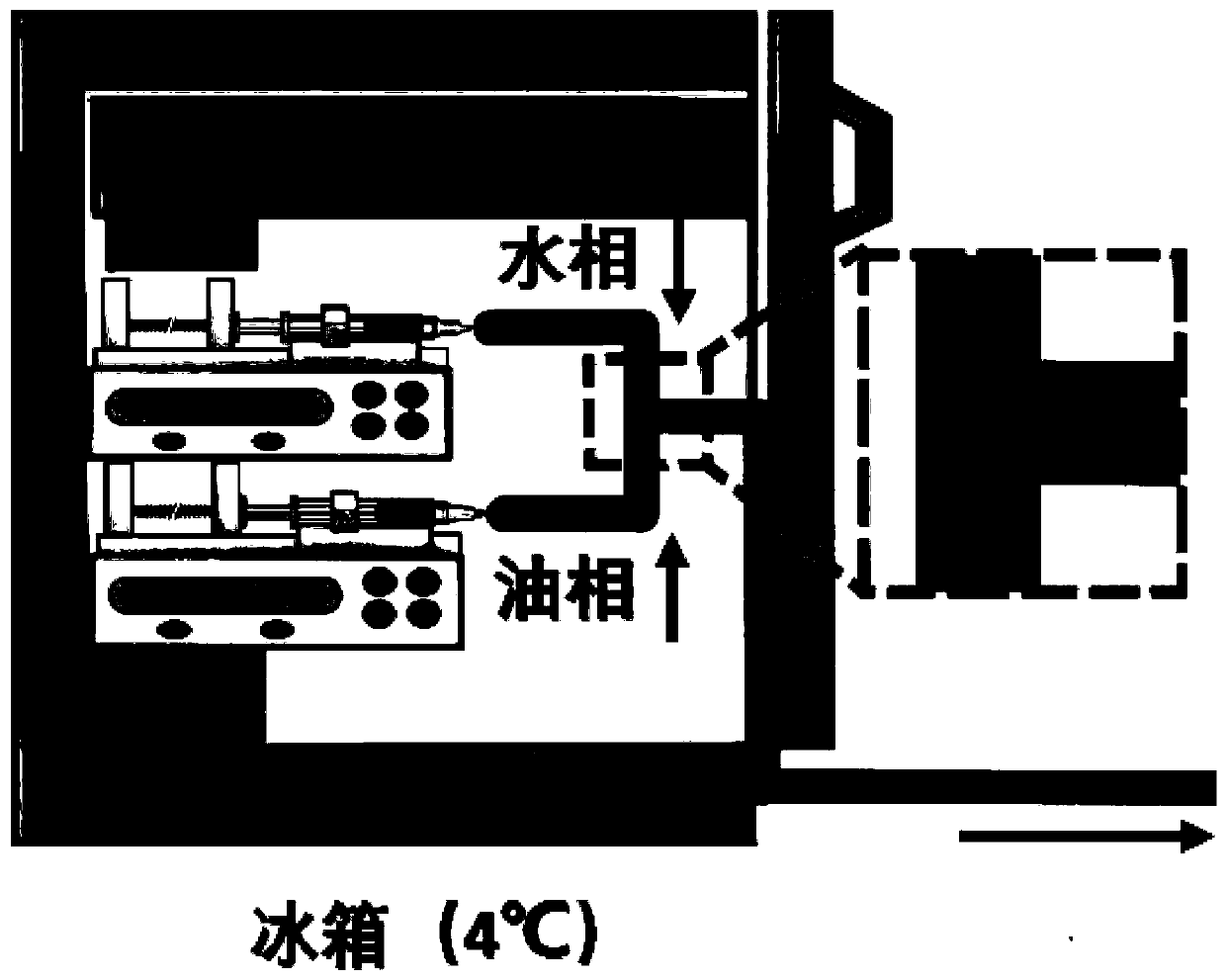
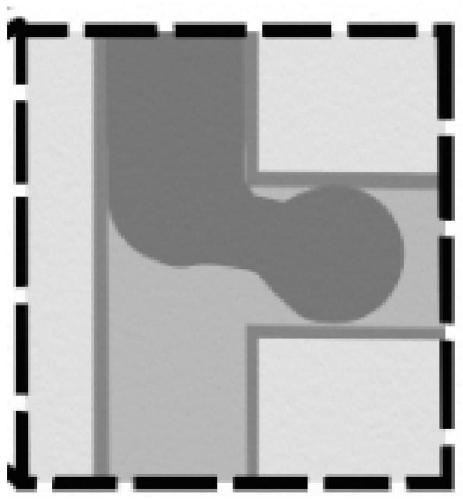
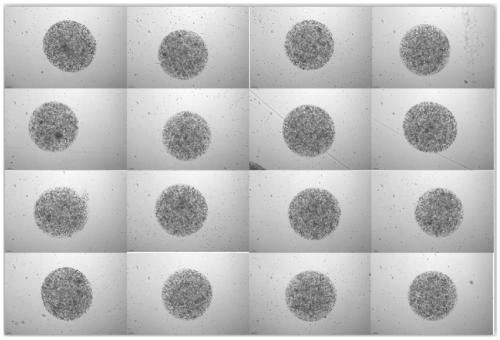
Preparation method of organoid spheres
ActiveCN110004111AThe size is easy to controlControllable and uniform in sizeArtificial cell constructsVertebrate cellsFluid controlMicrofluidics
The invention provides a preparation method of organoid spheres. The preparation method comprises the steps that matrigel containing cells and fluorocarbon oil are introduced into a three-way device to obtain cell spheres, and after culture is performed, the organoid spheres are formed; according to the preparation method of the organoid spheres, the micro-fluid control method is adopted for generating mono-disperse biomaterial organs, high-throughput production is achieved, the sizes, shapes and structures of organoids are controllable and uniform, and compared with a 2D tumor sensitivity predicting model, the organoids are combined with tumor micro-environment factors, and the prediction result is more accurate; compared with a PDX mouse tumor sensitivity predicting model, the modeling period is shorter, and the cost is lower; compared with common gene sequencing, the clinic predicting rate is higher; the preparation method of organoid spheres has the important significance and valuefor clinic application, and the important basis is supplied to detection of related results.
Owner:TSINGHUA BERKELEY SHENZHEN INST
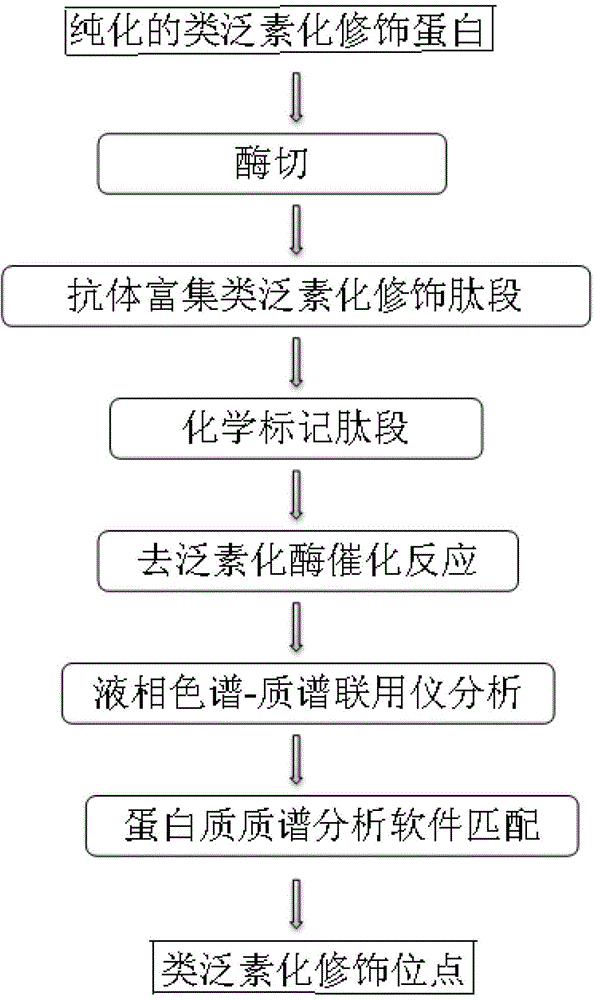
Method for identifying ubiquitin-like modification sites of proteins
The invention relates to a method for identifying the ubiquitin-like modification sites of proteins. The above specific enrichment labeling identification method comprises the following steps: carrying out enzymatic hydrolysis of purified ubiquitin-like modification proteins, enriching the ubiquitin-like modification enzyme digestion peptide fragments through a specific ubiquitin-like antibody, protecting the non-modification lysine amino residues in the enriched peptide fragments through utilizing chemical labeling, carrying out a deubiquinaning enzyme catalysis reaction to expose modified lysine on a substrate peptide fragment sequence, analyzing the peptide segment sequences through a liquid chromatograph-mass spectrometer, and matching through protein mass spectrum data analysis software to obtain the chemically modified peptide fragment sequence, wherein lysine sites containing free amino groups in the finally obtained sequence are the ubiquitin-like modification sites. Compared with common gene mutation methods for obtaining the sites at present, the method provided by the invention has an advantage that information about the ubiquitin-like modification sites of proteins can be rapidly, efficiently and accurately obtained without a tedious gene mutation technology.
Owner:RUIJIN HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Drug-resistance gene film chip for detecting mycobacterium tuberculosis
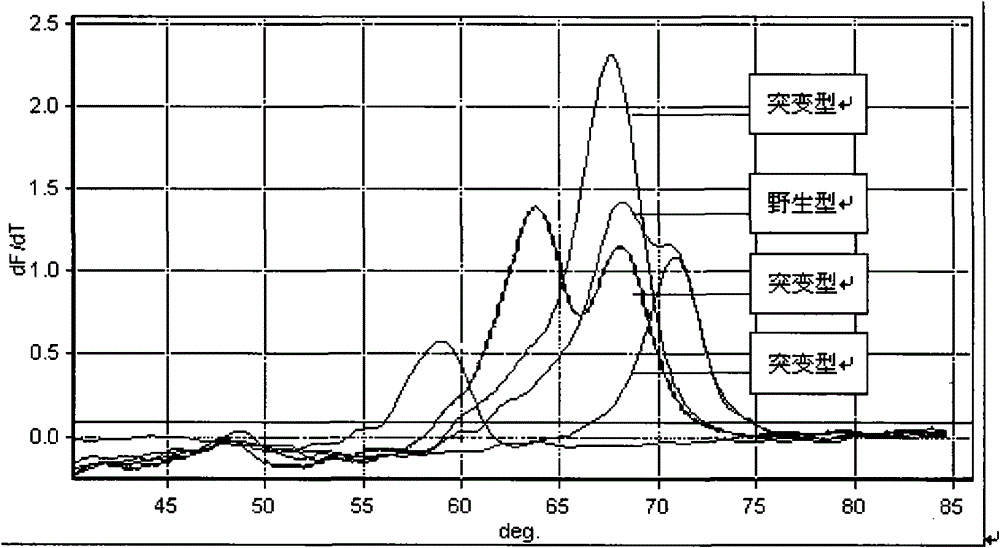
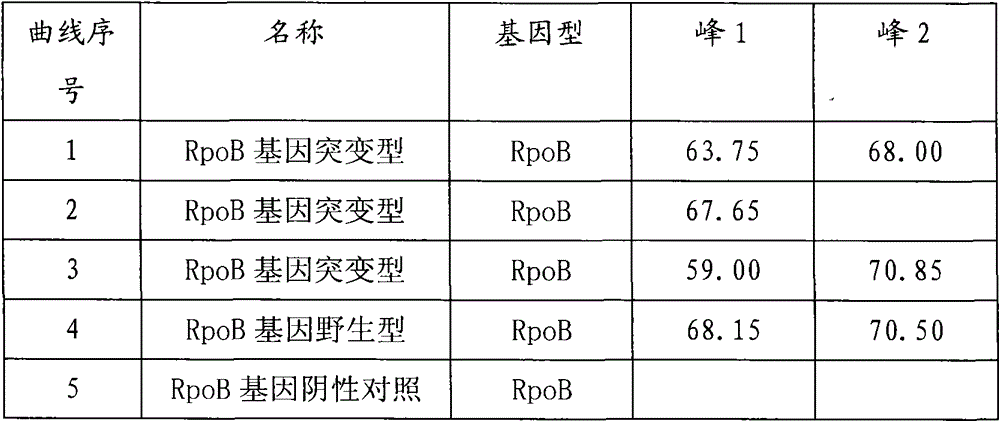
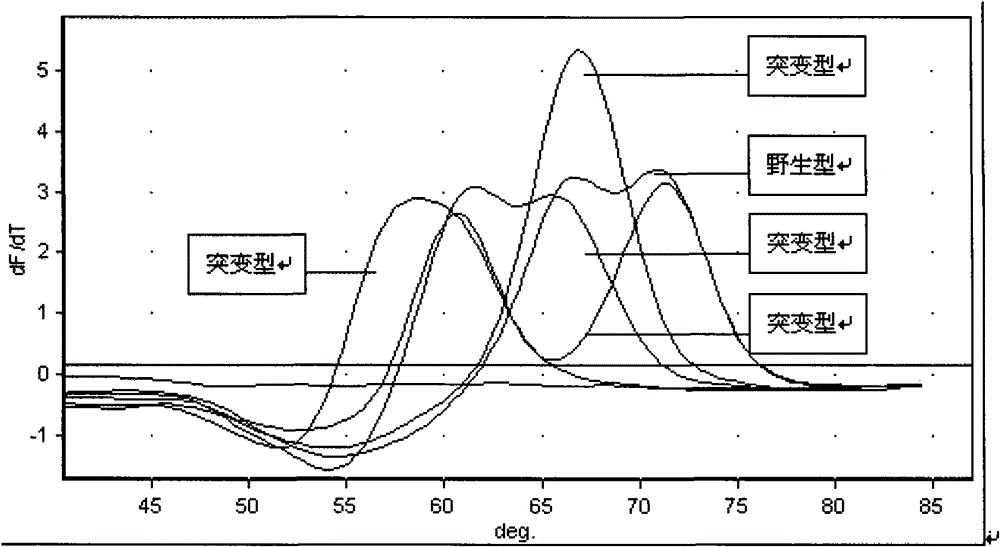
InactiveCN101580879AHigh sensitivityTo achieve the purpose of detectionNucleotide librariesMicrobiological testing/measurementRpoBBiotin
A drug-resistance gene film chip for detecting mycobacterium tuberculosis is prepared by designing 54 probe spotting on a nylon film by aiming at the mutant sites of mycobacterium tuberculosis rpoB, kagG, embB, inhA, ahpC, gyrA, rrs, rpsL and pncA gene; 12 pairs of specific primers with biotin-labeled at 5' end are utilized; a sample DNA is subjected to triple PCR and amplified to form a large amount of gene fragment products with biotin; the amplified products and the probes on the film chip carry out specific hybridization; and then film washing, enzyme-linking and color reaction are carried out, thus preparing the chip. The gene film chip and the detection method thereof can detect the common gene mutations of the mycobacterium tuberculosis on the drug resistance of drugs such as isoniazid, rifampicin, streptomycin, ethambutol, pyrazinamide, quinolone and the like at one step, and are applicable to extracorporeal detection sputum sample, clinical isolation strains and mycobacterium tuberculosis multi-drug resistant gene in the organization sample.
Owner:GUANGXI MEDICAL UNIVERSITY
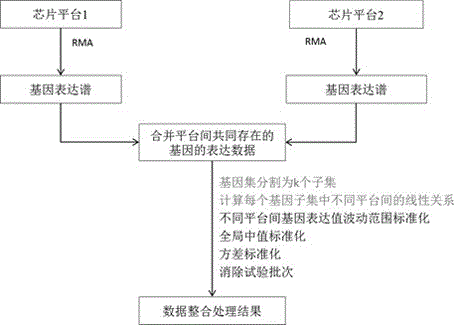
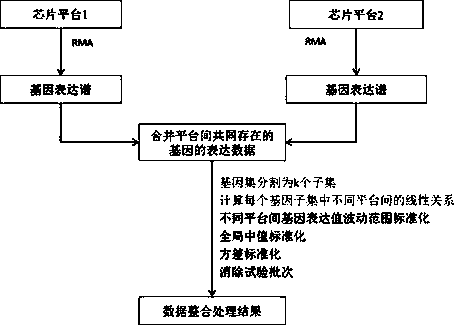
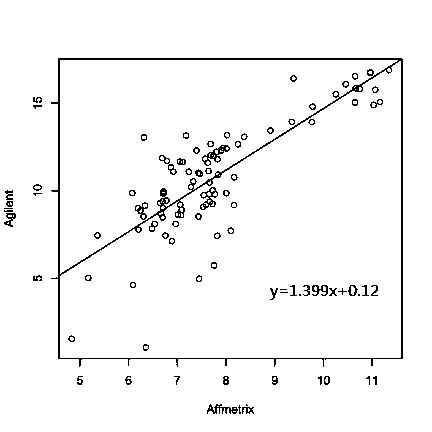
Integration method for gene expression data by crossing chip platforms
ActiveCN103745137AEliminate experimental batch effectsImprove performanceSpecial data processing applicationsGene expression matrixExpression gene
The invention belongs to the technical field of biological information. The invention provides a method for integrating gene expression data by crossing a plurality of different chip platforms; the method comprises the following steps: the standard preprocessing is implemented for the gene expression profile of the plurality of chip platforms; the common gene expression data in the different chip platforms is merged; genes are divided into k subsets according to the expression similarity of the genes on the plurality of chip platforms; the expression linear relation exps1=as*exps2+bs of the different chip platforms in every gene subset can be calculated by the least square method; the gene expression values of the different chip platforms are standardized into the same change range by using the formula exp1=X*A.*exp2+X*B to obtain standard gene expression matrixes, wherein the implications of symbols are defined as the specification.
Owner:艾吉泰康(嘉兴)生物科技有限公司
LAMP method for detecting enterobacteriaceae food-borne pathogenic bacteria, nucleic acid and primer pairs
ActiveCN105803064AThe detection process is fastReliable resultsMicrobiological testing/measurementAgainst vector-borne diseasesFood bornePathogenic bacteria
The invention relates to an LAMP method for detecting enterobacteriaceae food-borne pathogenic bacteria, nucleic acid and primer pairs and belongs to the technical field of food safety detection.The LAMP method includes the following steps that a common gene sequence of the enterobacteriaceae pathogenic bacteria is screened out through bioinformatics and comparative genomics, and the specific amplification primer pairs are designed according to the sequence; an LAMP detection system is established by optimizing reaction conditions.The invention further relates to nucleic acid with the base sequence shown in SEQ ID NO:1 and a group (three pairs) of primers.The base sequences of the primers are shown in SEQ ID NO:2, SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO:5, SEQ ID NO:6 and SEQ ID NO:7.Compared with the prior art, the detection method is used for detecting enterobacteriaceae, detection time is short, cost is low, higher practicality is achieved, the detection result is specific, and result judgment is simple.
Owner:杭州海关技术中心
Method and system for comparative genomics
A method and system for representing a similarity between at least two genomes that includes detecting gene clusters which are common to the at least two genomes and representing the common gene clusters in a PQ tree. The PQ tree includes a first internal node (P node), that allows permutation of the children thereof, and a second internal node (Q node), that maintains unidirectional order of the children thereof.
Owner:IBM CORP
Liver echinococcus gene segment screening method, amplification primer and kit
PendingCN113355431AImprove accuracyImprove clinical outcomesMicrobiological testing/measurementSequence analysisHuman DNA sequencingEchinococcus multilocularis
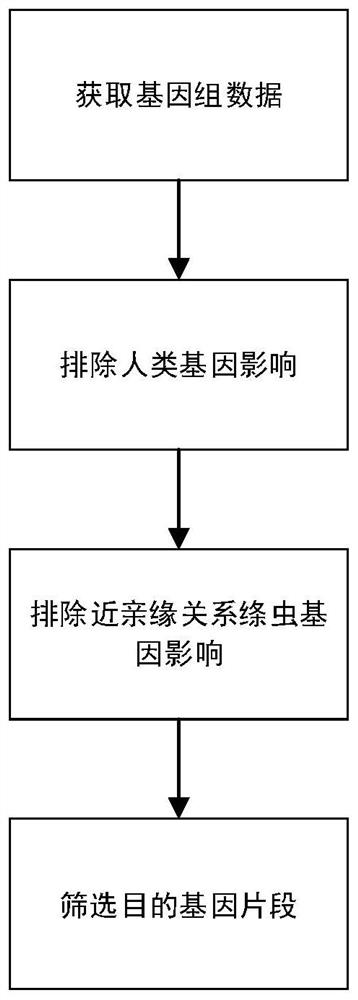
The invention discloses a liver echinococcus gene segment screening method, an amplification primer and a kit.The screening method comprises the following steps: eliminating an influence of a human genome and a close genetic relationship tapeworm group genome from whole genomes of echinococcus granulosus and echinococcus multilocularis; and screening to obtain a third echinococcus granulosus gene segment, a third echinococcus multilocularis gene segment and a common gene segment, and designing three types of amplification primers by using three types of the gene segments respectively. A primer pair group for detecting echinococcosis of human tissues is obtained by further screening and a kit and a use method of the kit are provided based on the primer pair group. False positive results caused by human genes or close genetic relationship tapeworm genes existing in to-be-detected tissue DNA is avoided from the source, the to-be-detected DNA aiming at the primer during design is a human tissue sample, the false negative results in clinical detection are remarkably reduced, specific primers have higher accuracy and higher specificity, and clinical use effects of the primer pair and the kit are obviously enhanced.
Owner:WEST CHINA HOSPITAL SICHUAN UNIV
Detection method of germs in traditional zymotic soybean paste
InactiveCN101619355AAvoid churnDo not change the contentMicrobiological testing/measurementMaterial analysis by electric/magnetic meansMicroorganismMicrobiology
The invention relates to a detection method of germs in soybean paste, in particular to a detection method of germs in traditional zymotic soybean paste, solving the problem that the prior PCR-DGGE technology can not accurately detect the germs in the traditional zymotic soybean paste. The detection method comprises the following steps: (1) the DNA extraction of a germ common gene group of a traditional zymotic soybean paste sample; (2) a V3 section of 16S rDNA of PCR amplified germs; and (3) the PCR-DGGE of the germs. The method of the invention thoroughly clear up other contents of the traditional zymotic soybean paste except microbial cells while avoiding the loss of the species and the amount of microorganism whenever possible, and the whole detection process does not change the DNA content, reduces the final analysis bias and ensures the precision of a detection result.
Owner:HEILONGJIANG UNIV
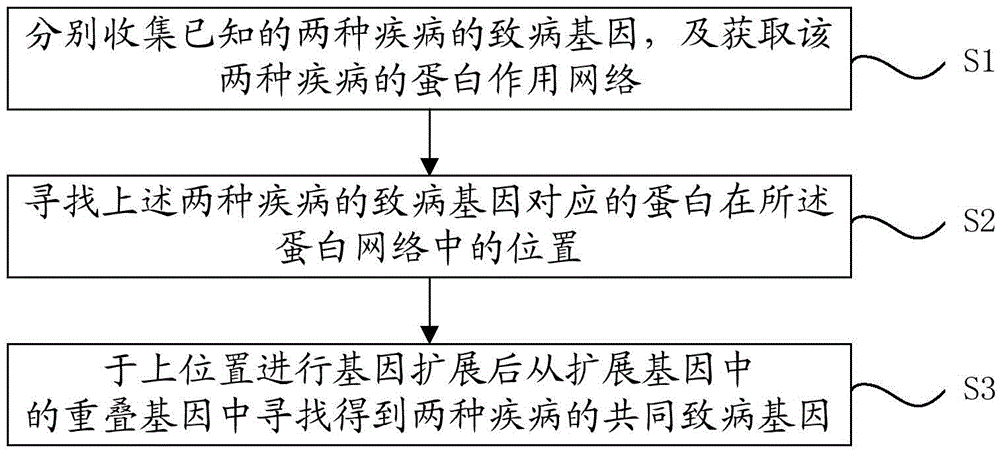
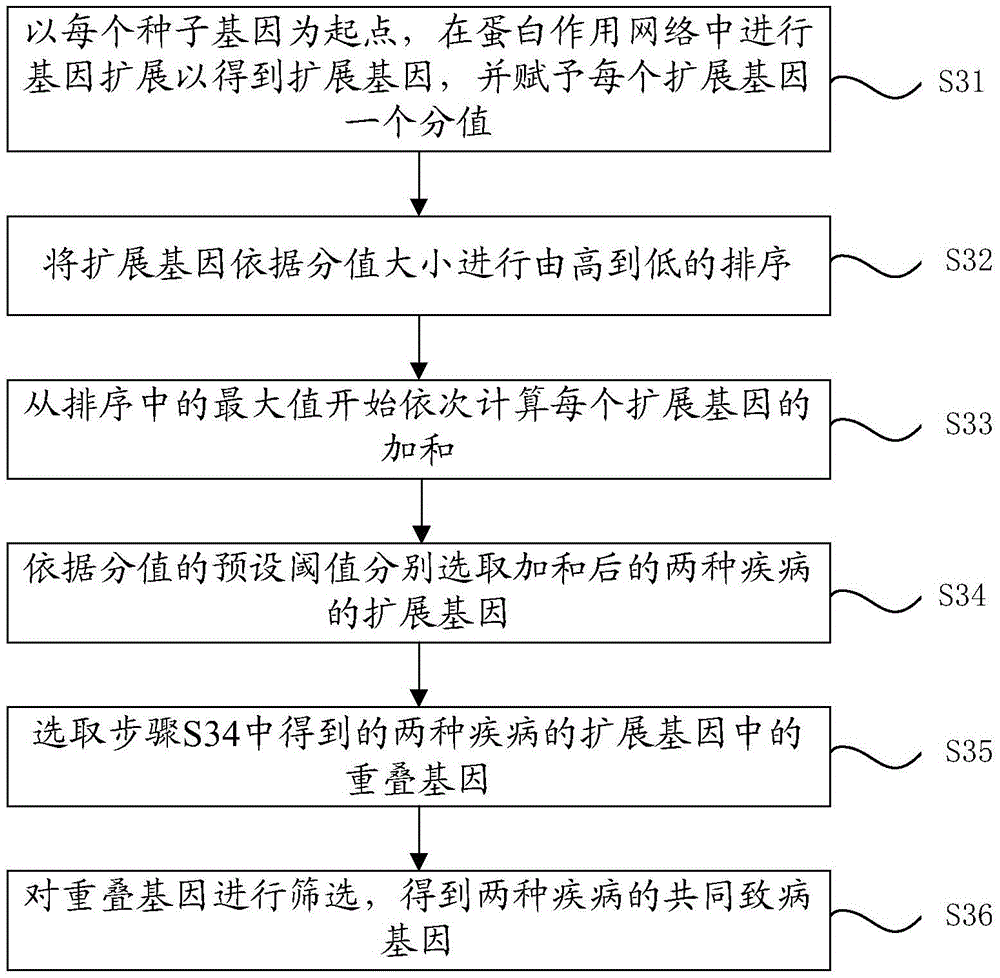
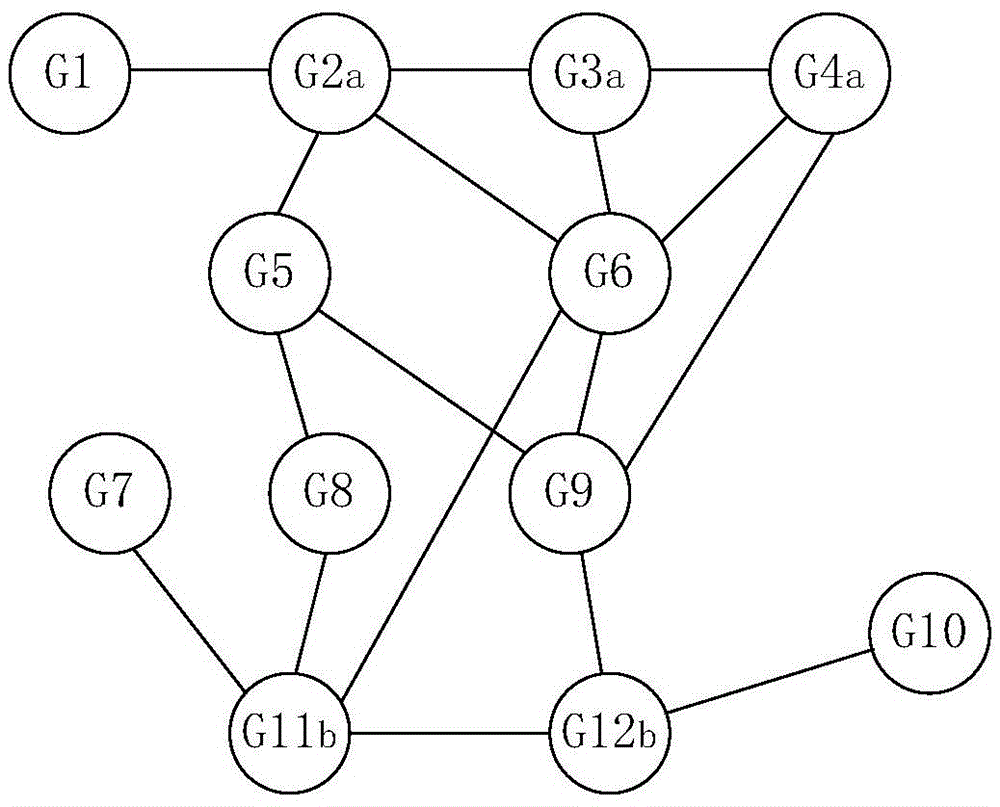
Method for predicting common disease-causing genes of two diseases
InactiveCN105631244AChoose accuratelyPrecision medicineProteomicsGenomicsAnalysis methodGene expansion
The invention provides a method for predicting common disease-causing genes of two diseases. The method includes the following steps that known disease-causing genes of the two diseases are collected respectively, and a protein function network of the two diseases is obtained; positions of protein corresponding to the disease-causing genes of the two diseases in the protein network are searched for; after gene expansion is conducted based on the positions, the common disease-causing genes of the two diseases are searched for and obtained from overlapped genes in the expansion genes. According to the method, gene expansion is conducted in a protein function network diagram corresponding to the genes of the two diseases, the common disease-causing genes of the two diseases is selected from common genes in the expansion genes, a random walking method, a gene set enrichment and analysis method and a hypergeometric inspection method are applied in the predicting process, and thus the common disease-causing genes can be selected more accurately. By means of the method, the key drive genes possibly influencing the two diseases can be predicted, and precise medical treatment can be achieved.
Owner:SHANGHAI JIAO TONG UNIV
Visual LAMP detection kit for streptococcic mastitis pathogenic bacteria
InactiveCN103789410AVisualization of test resultsThe detection method is simpleMicrobiological testing/measurementMicroorganism based processesWater bathsUltraviolet
The invention discloses a visual LAMP (Loop-Mediated Isothermal Amplification) detection kit for streptococcic mastitis pathogenic bacteria, wherein the inventor designs two specific inner primers and two specific outer primers according to the common gene conservative area of the streptococcic mastitis pathogenic bacteria by virtue of adopting the loop-mediated isothermal amplification technique, so that the visual LAMP detection kit is obtained. A method for detecting the streptococcic mastitis pathogenic bacteria by using the LAMP detection kit only needs a common water bath kettle in stead of expensive PCR (Polymerase Chain Reaction) instrument, and the method is simple to operate and low in cost, but higher in sensitivity than PCR detection; the detection results are visual and can be obtained through judgment by naked eyes, and the results do not need to be observed through gel electrophoresis ultraviolet analysis and development. The visual LAMP detection kit can be used for overcoming the defects of long time taken, high workload, easy cross contamination, complex operation and the like of the existing detection technology, and is more specific and sensitive than the conventional detection method, and thus applicable to quick detection in basic-level veterinary stations and dairy farms; besides, the visual LAMP detection kit has excellent application prospect.
Owner:GUANGXI UNIV
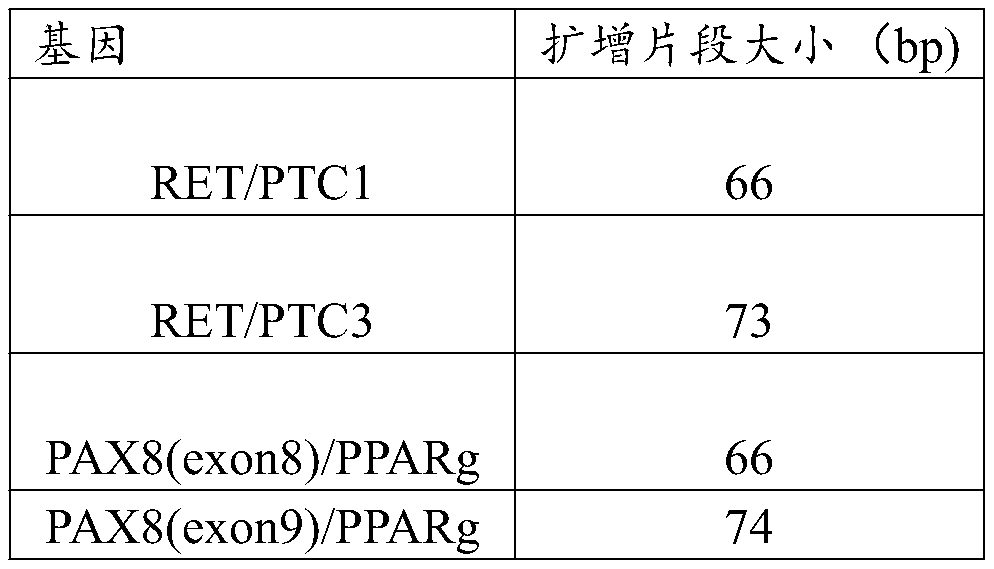
Detection method for thyroid cancer related gene fusion mutation and kit
PendingCN111349694AEasy to detectQuick checkMicrobiological testing/measurementElectrophoresesTotal rna
The invention provides a detection method for thyroid cancer related gene fusion mutation and a kit and relates to the technical field of gene engineering. The detection method comprises the followingsteps: performing detection, namely extracting total RNA (ribonucleic acid) or miRNA (micro ribonucleic acid) from a detection sample; performing reverse transcription on the total RNA or mRNA of thesample so as to obtain cDNA (complementary deoxyribonucleic acid), and performing fusion gene target specific PCR (polymerase chain reaction) amplification by using fusion gene target specific PCR amplification primers; performing electrophoresis detection on specific PCR amplification products so as to obtain types and sizes of fragments of the amplification products; and according to electrophoresis results, judging whether sample genes have thyroid cancer related gene fusion mutation or not. By adopting the method provided by the invention, three types of common gene fusion in thyroid cancer can be simply, conveniently and rapidly detected at an RNA level, and the method is high in sensitivity and applicable to clinical use.
Owner:深圳市新合生物医疗科技有限公司
Kit for detecting pathogenic bacteria of mucor lungs as well as use method and application of kit
ActiveCN112725508ARealize detectionMicrobiological testing/measurementMicroorganism based processesPullulanRhizopus oryzae
The invention discloses a kit for detecting pathogenic bacteria of mucor lungs as well as a use method and an application of the kit. The kit of the present invention comprises a universal primer capable of being present in the form of a mixture, wherein the universal primer is capable of binding to a common gene conserved region of rhizopus oryzae, rhizomucor micranthum, aureobasidium pullulans, rhizopus stolonifer, and Cunninghamia lanceolata. The kit provided by the invention can realize one-time detection of various mucor fungi, the detection time does not exceed 2 hours, and the specific type of mucor can be further determined in one-time detection.
Owner:上海捷诺生物科技有限公司
Method and element for screening different-intensity ribosome binding sites and promoters
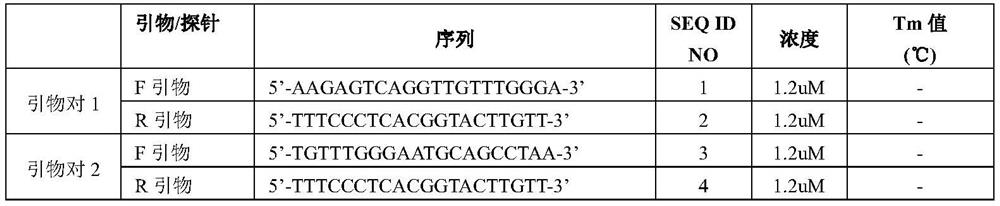
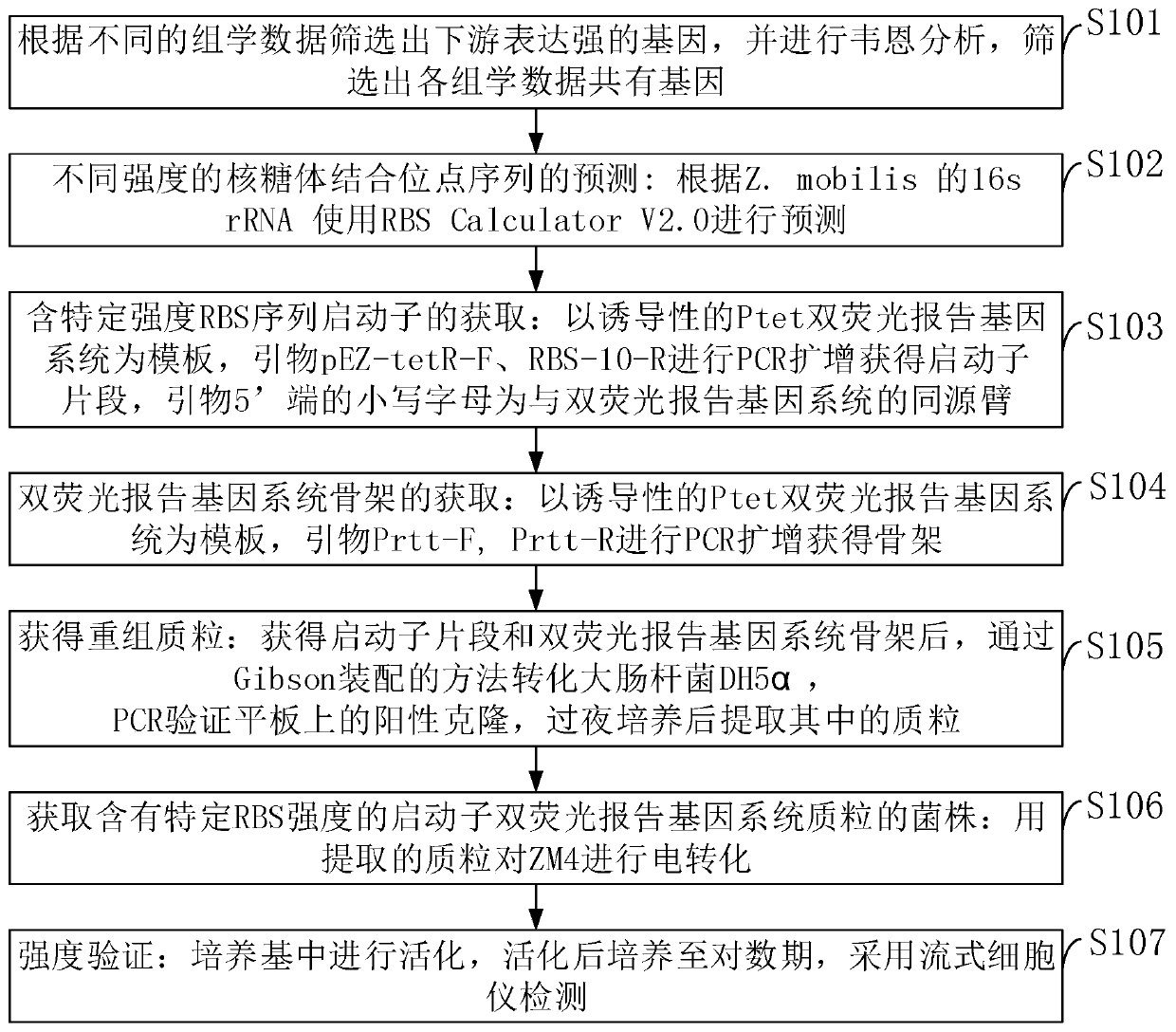
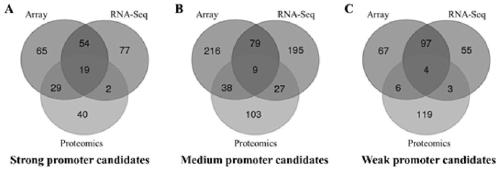
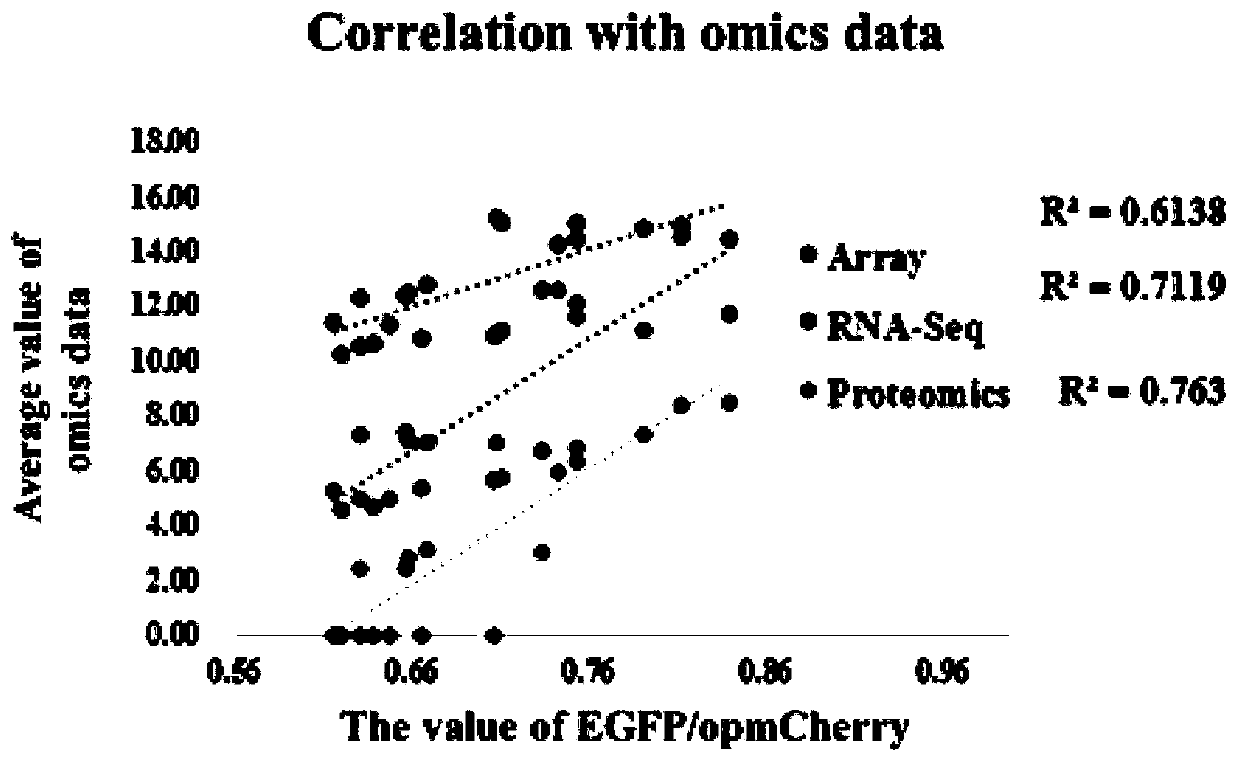
InactiveCN109852631AEfficient screeningGood correlationMicrobiological testing/measurementVector-based foreign material introductionGenomicsFluorescence
The invention belongs to the technical field of zymomonas mobilis, and discloses a method and an element for screening different-intensity ribosome binding sites and promoters. The method comprises the following steps: firstly, screening genes with strong downstream expression according to different genomics data, carrying out Wayne analysis, and screening common gene of the genomics data; predicting the binding site sequence of ribosome with different intensities; acquiring an RBS sequence promoter with specific intensity; acquiring a double-fluorescence reporter gene system framework; acquiring recombinant plasmid; acquiring a strain of the double-fluorescence reporter gene system plasmid containing the promoter with specific RBS intensity; and performing intensity verification. The method can integrate existing systematic biological data, and quickly and efficiently screen promoters with specific intensity. The intensities of the RBS and the promoter which are predicted and screenedby the method have good correlation with experimental data (respectively is R2)0.9 and R2)0.7).
Owner:HUBEI UNIV
Standard substance for extensive oncogene detection and preparation method and application thereof
ActiveCN111334505AFit closelyMicrobiological testing/measurementDNA/RNA fragmentationInsertion deletionCancer genome
The invention provides a standard substance for extensive oncogene detection. The standard substance includes 13 mutation sites verified by Droplet Digital PCR (ddPCR) and 500X high-throughput whole exome sequencing verified site information, has more than 700 variation sites of over 330 genes, also includes common structural variations of cancer genomes, such as common gene SNV, base insertion deletion and the like, has corresponding mutation site frequencies within a range of 1% to 100%, and is wide in application scene and platform. The standard substance can be used for evaluating stability, specificity and sensitivity of the working flow from sample extraction to biological information analysis, evaluating each sample treatment method and detecting performance differences among platforms. The standard substance belongs to a major innovation in the industry, and has wide application prospect and great industrial application value.
Owner:菁良科技(深圳)有限公司
Kit for detecting human natural killer cell immunoglobulin-like receptor KIR genotyping
ActiveCN112442525AHigh detection throughputMeet testing needsMicrobiological testing/measurementDNA/RNA fragmentationMultiplexReceptor
The invention provides 16 common genes for detecting a human natural killer cell immunoglobulin-like receptor KIR, wherein the 16 common genes comprise 14 KIR functional genes (2DL1, 2DL2, 2DL3, 2DL4,2DL5A / B, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3 and 3DS1) and 2 pseudo genes (2DP1 and 3DP1), and a combination of a PCR amplification primer group and a Taqman probe of the common variant ofthe 2DS4; and the combination comprises six primer groups. The invention also provides a kit containing the combination of the PCR amplification primer group and the Taqman probe. The invention has the following technical effects that: an ARMS analysis method is combined with a Taqman multiplex fluorescence PCR technology to carry out qualitative typing detection on the KIR gene; furthermore, improvement is carried out on the basis of a traditional ARMS; various defects of an existing detection method are overcome; and the method disclosed by the invention has wide application prospects and clinical reference value.
Owner:JIANGSU WEIHE BIOTECH
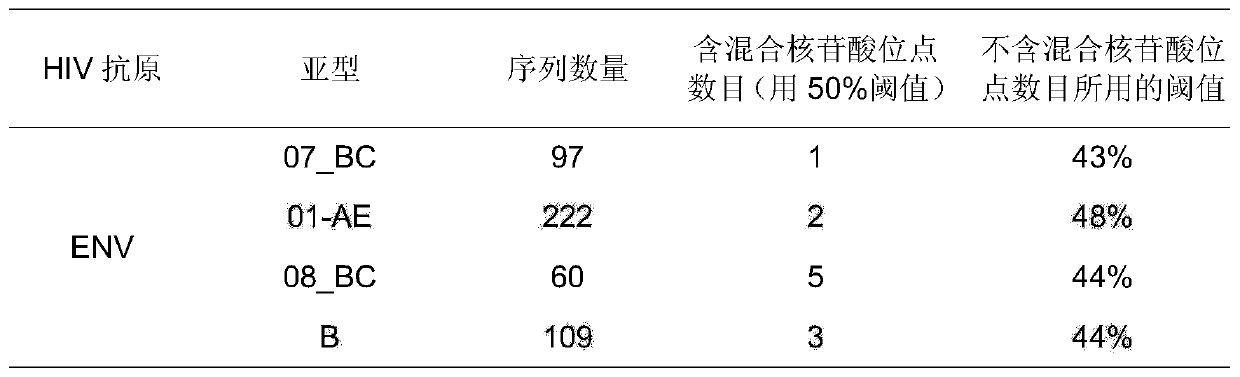
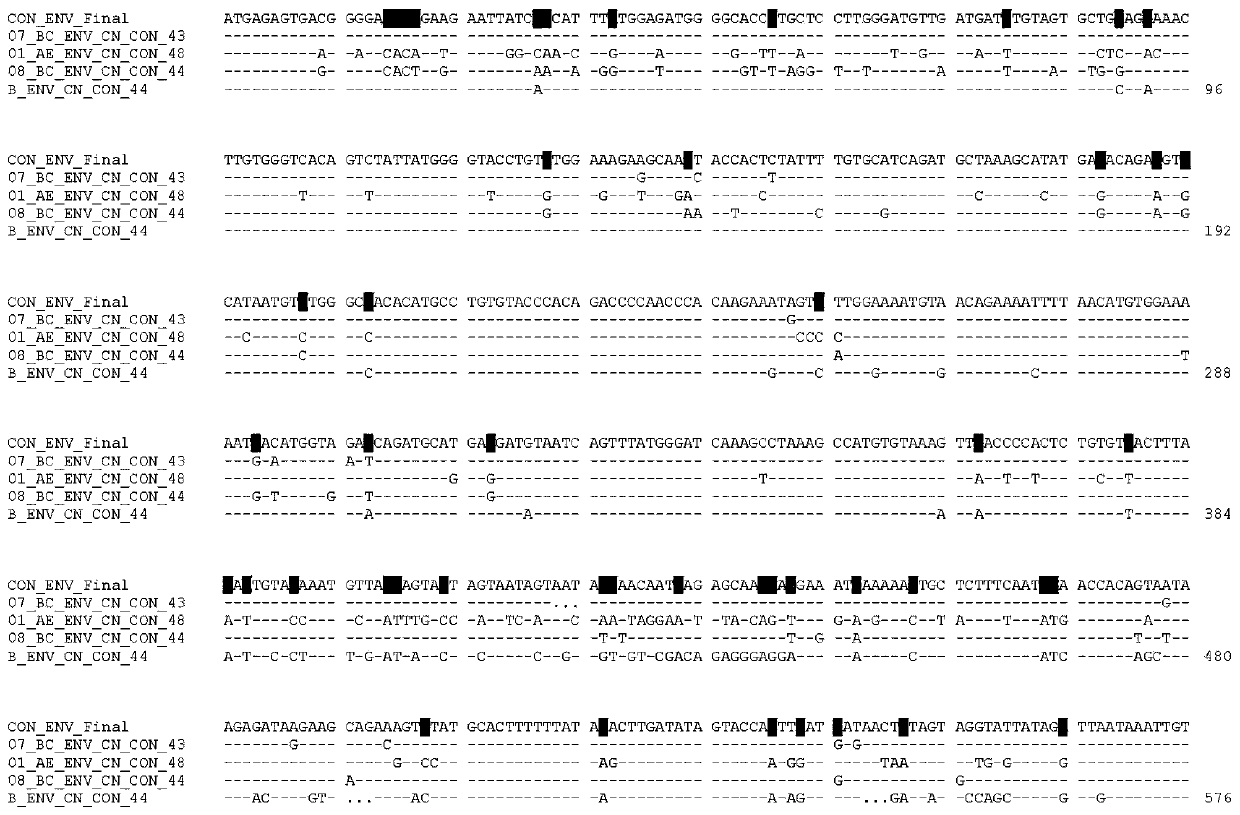
Envelope protein trimer immunogen capable of inducing HIV-1 broad spectrum and neutralizing antibody and application thereof
ActiveCN110627911AGood immune resultsStrong broad spectrumAntibody mimetics/scaffoldsViral antigen ingredientsProtein trimerSequence design
The invention relates to the field of biomedicine, relates to an envelope protein trimer immunogen capable of inducing HIV-1 broad spectrum and neutralizing antibody and application thereof, in particular to an envelope protein Env common gene sequence designed based on HIV-1 Chinese epidemic strain and a stable envelope protein gp120 trimer immunogen obtained based on the sequence. The inventionfurther relates to the application of the HIV-1 envelope protein gp120 trimer immunogen in preparation of AIDS vaccines.
Owner:JILIN UNIV
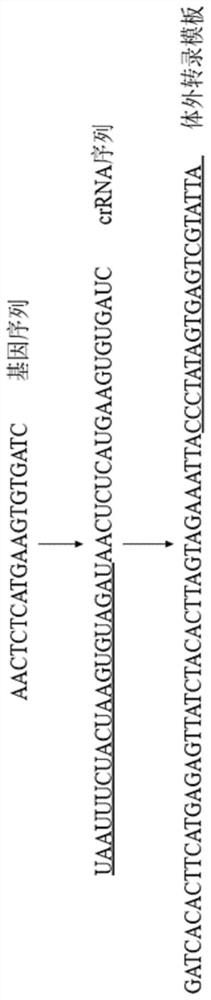
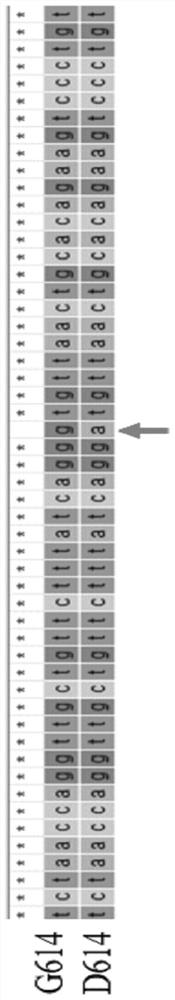
Method for identifying D614G mutation in SARS-CoV-2 based on CRISPR-Cas12a
PendingCN113584223AEasy to masterEfficient use ofMicrobiological testing/measurementDNA/RNA fragmentationRecombinaseVirus
The invention provides a crRNA molecule and a method for detecting D614G mutation in SARS-CoV-2 by using CRISPR-Cas12a. The method for detecting D614G mutation in SARS-CoV-2 by using the CRISPR-Cas12a technology through the crRNA is simple, convenient, easy to implement, rapid and specific; when a recombinase polymerase nucleic acid amplification technology is combined, the sensitivity is 100 copies / [mu]L, the sensitivity is extremely high, compared with a common gene sequencing method, the method can achieve the identification of a sample with lower virus content, and is suitable for large-scale screening.
Owner:中国人民解放军疾病预防控制中心
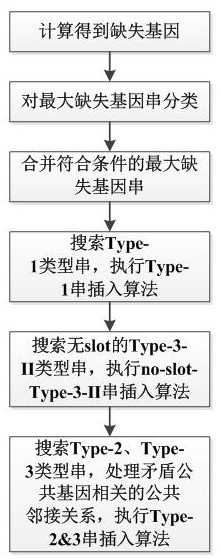
Double-sided genome segment filling method and device based on segment contiguous group
PendingCN112634989AImprove search speedImprove accuracyHybridisationInstrumentsGenomic SegmentContig
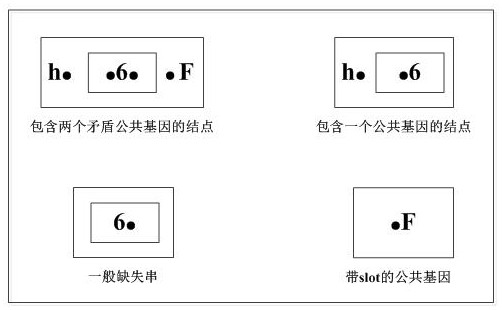
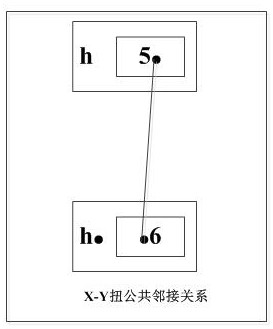
The invention discloses a genome segment filling method and device based on a segment contiguous group. The method comprises the following steps: calculating to obtain a deletion gene; classifying the maximum deletion gene string; merging the maximum deletion gene strings meeting the conditions; searching a Type-1 type string, and executing a Type-1 string insertion algorithm; searching for a Type-3-II type string without slot, and executing a no-slot-Type-3-II string insertion algorithm; searching Type-2 and Type-3 type strings, processing a common adjacency relation related to the contradictory common gene, and executing Type 2& 3-string insertion algorithm. Herein, the calculation is carried out based on the fragment contigs, the form is more general, and application is wider; the filling method is high in search speed and high in filling efficiency, can reduce the time and space complexity of genome segment filling based on the segment contiguous group, and improves the sensitivity and specificity of filling.
Owner:SHANDONG JIANZHU UNIV
Method for identifying ubiquitin-like modification sites of proteins
The invention relates to a method for identifying the ubiquitin-like modification sites of proteins. The above specific enrichment labeling identification method comprises the following steps: carrying out enzymatic hydrolysis of purified ubiquitin-like modification proteins, enriching the ubiquitin-like modification enzyme digestion peptide fragments through a specific ubiquitin-like antibody, protecting the non-modification lysine amino residues in the enriched peptide fragments through utilizing chemical labeling, carrying out a deubiquinaning enzyme catalysis reaction to expose modified lysine on a substrate peptide fragment sequence, analyzing the peptide segment sequences through a liquid chromatograph-mass spectrometer, and matching through protein mass spectrum data analysis software to obtain the chemically modified peptide fragment sequence, wherein lysine sites containing free amino groups in the finally obtained sequence are the ubiquitin-like modification sites. Compared with common gene mutation methods for obtaining the sites at present, the method provided by the invention has an advantage that information about the ubiquitin-like modification sites of proteins can be rapidly, efficiently and accurately obtained without a tedious gene mutation technology.
Owner:RUIJIN HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Group of nucleotide sequences for detecting gene mutants of hepatitis B virus (HBV) and use thereof
InactiveCN102146486AImprove detection efficiencyReduce testing costsMicrobiological testing/measurementDNA/RNA fragmentationNucleotideNucleotide sequencing
The invention relates to a group of nucleotide sequences for detecting the gene mutants of HBV, which comprise at least one of the nucleotide sequences from SEQ ID No.1 to SEQ ID No.15. The invention also provides the use of the sequences in detection of the gene mutants of HBV. When the nucleotide sequences are used, 10 common gene mutation loci of HBV can be detected. When used for detecting the gene mutants of the HBV, the nucleotide sequences have the advantages of high throughput, high sensitivity and high specificity, are low in detection cost and have a practical clinic promotion and application value.
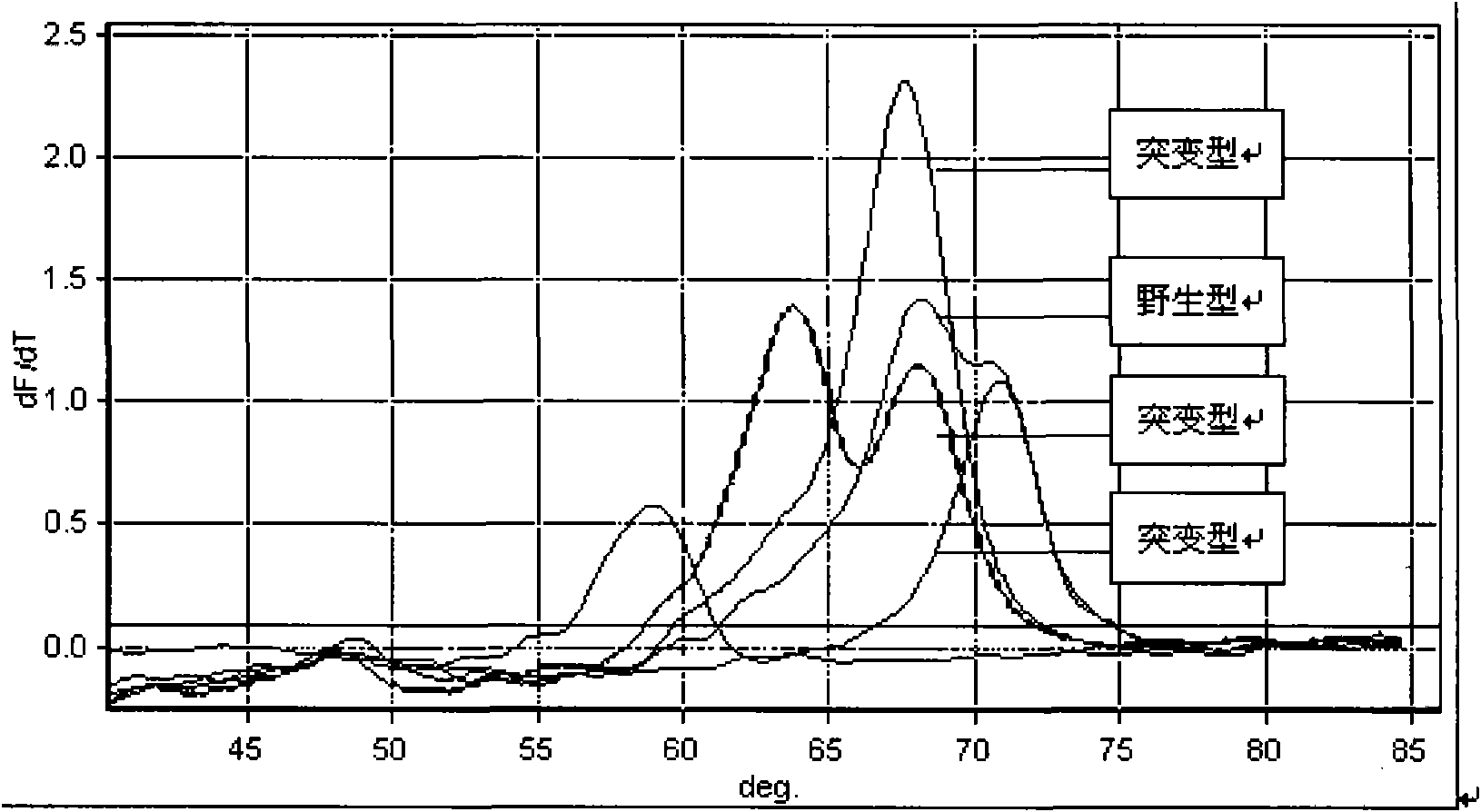
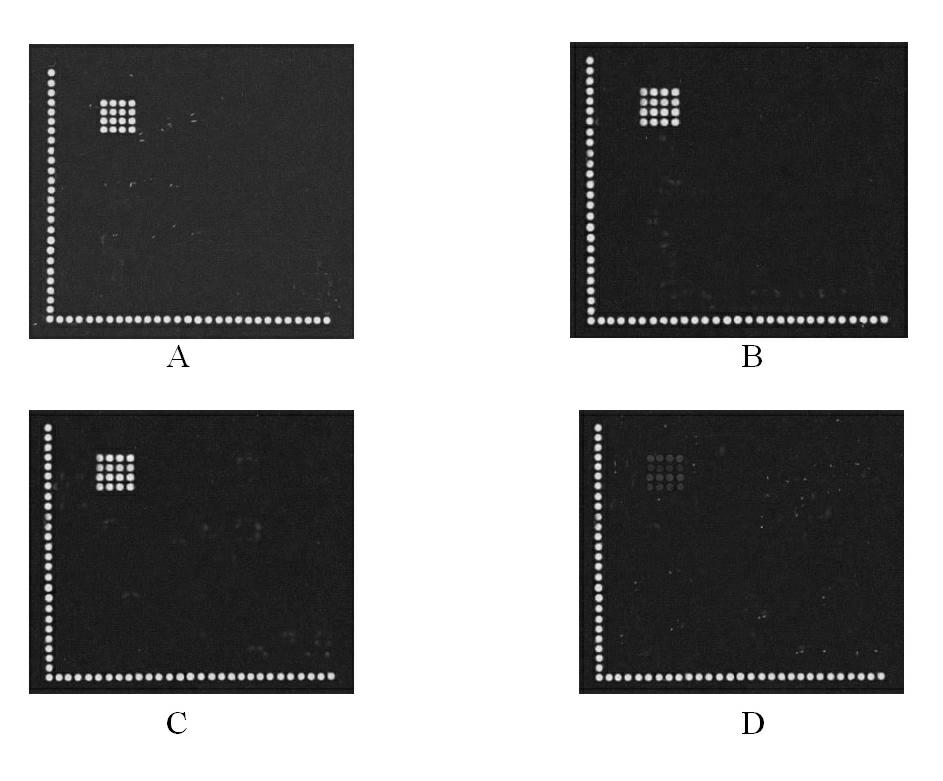
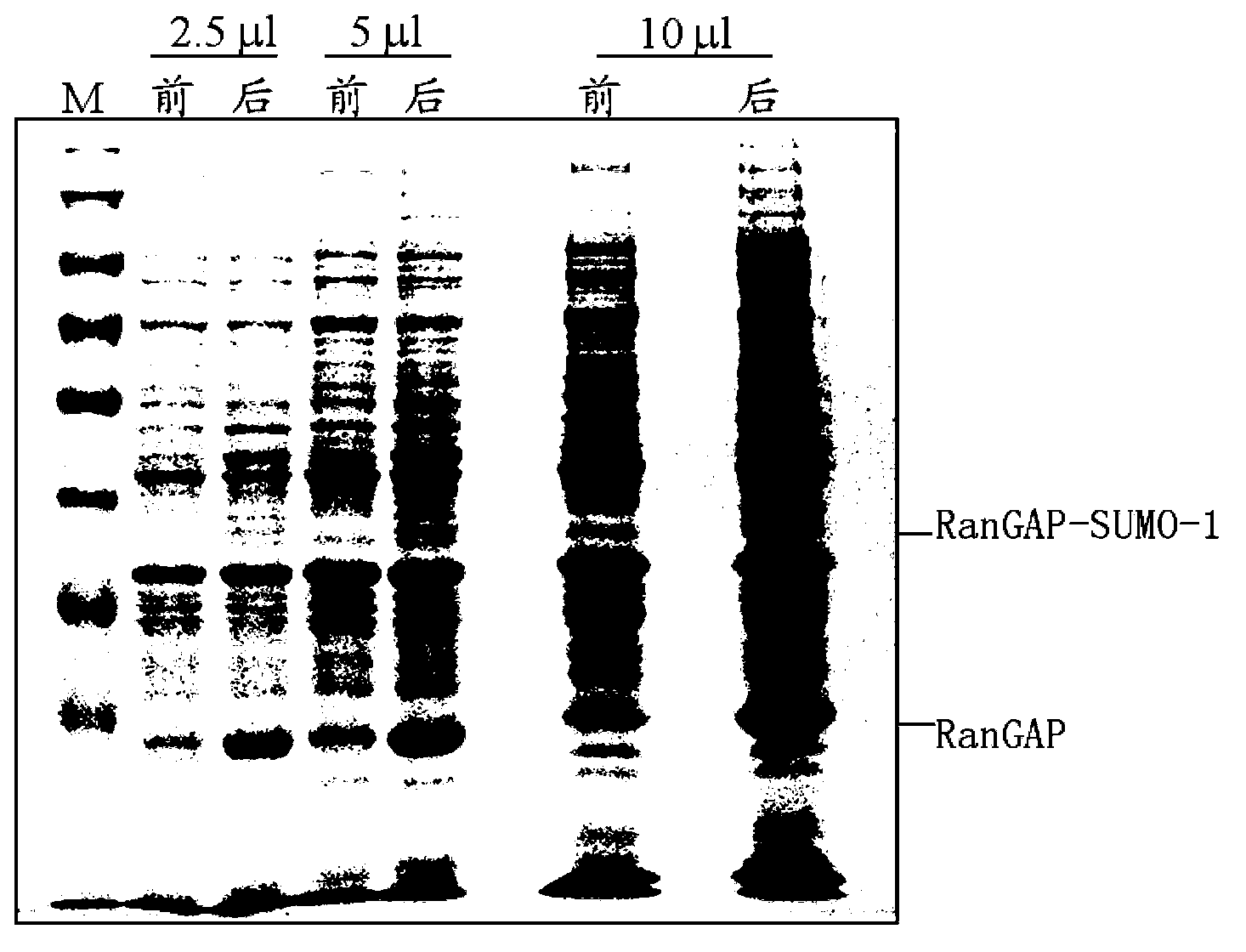
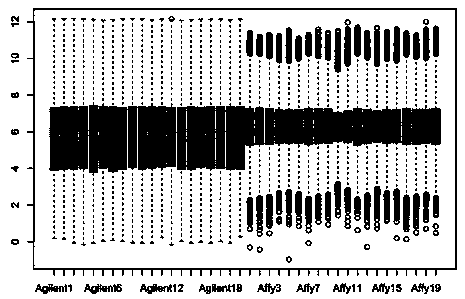
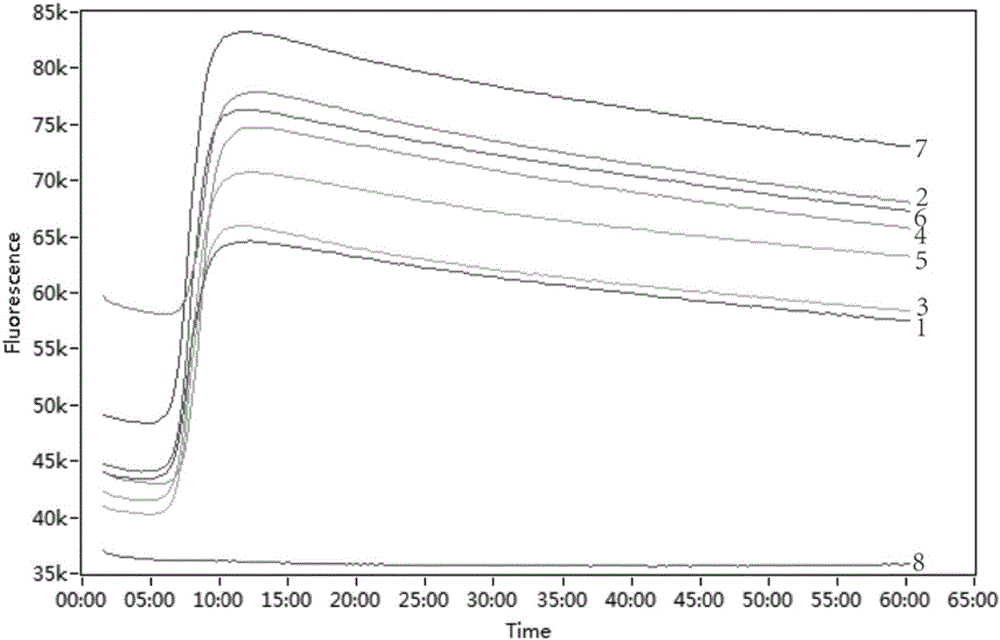
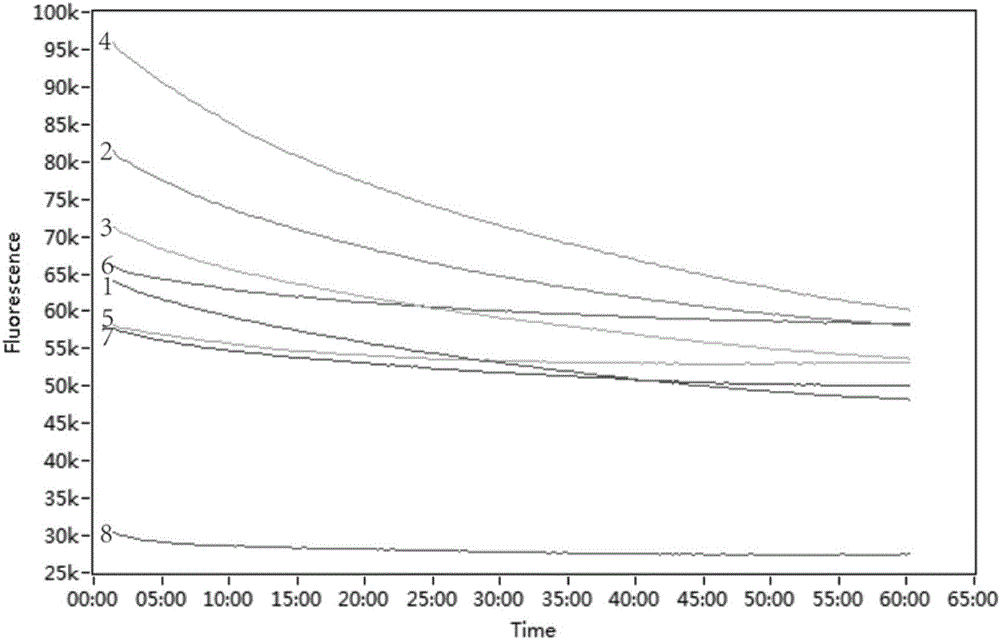
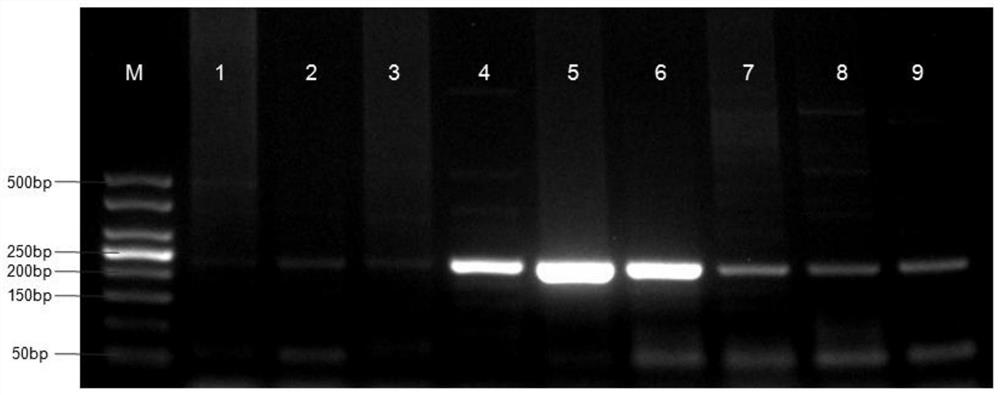
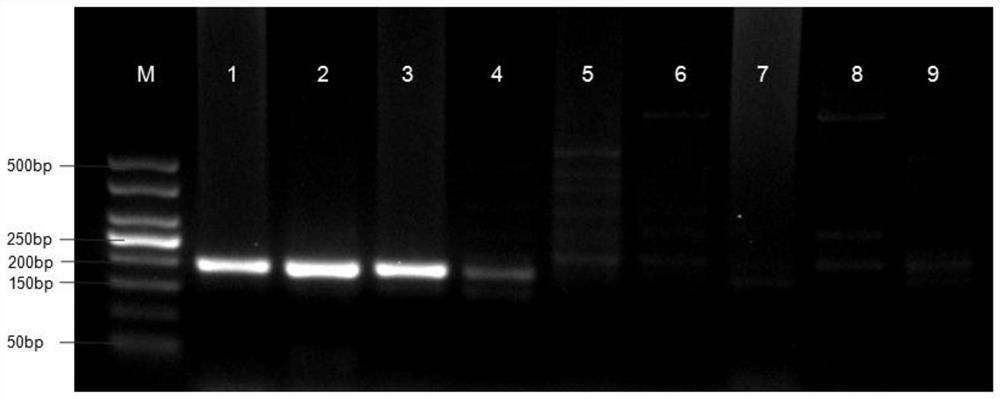
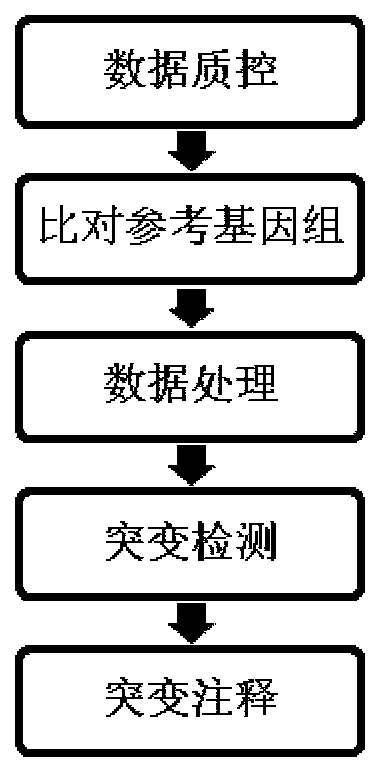
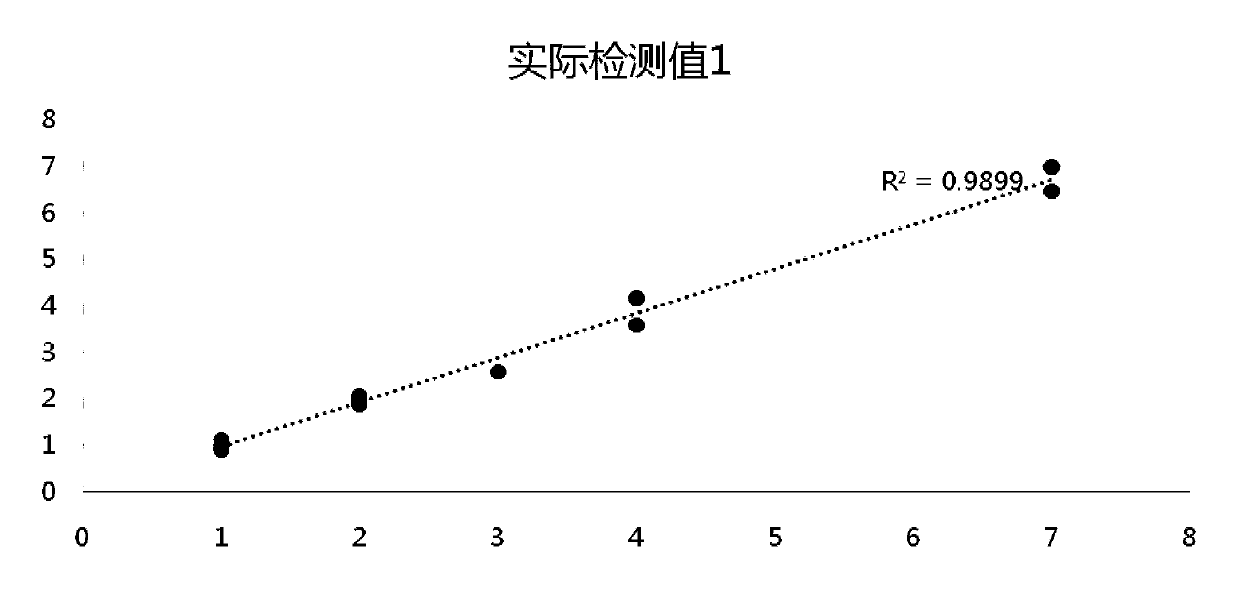
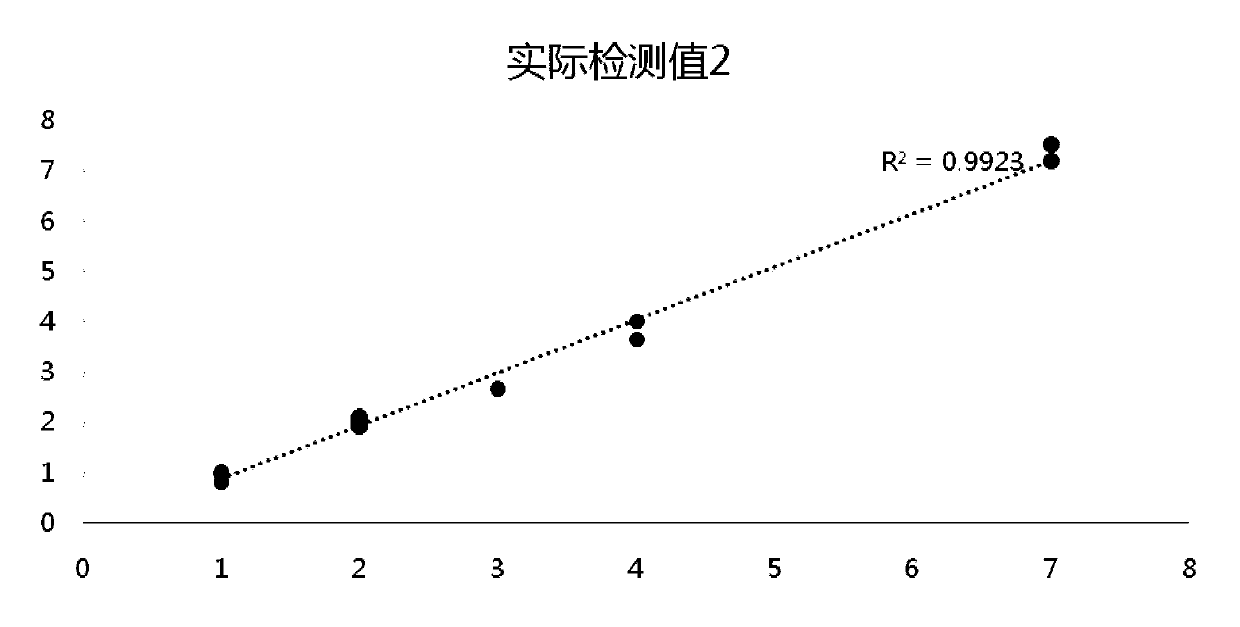
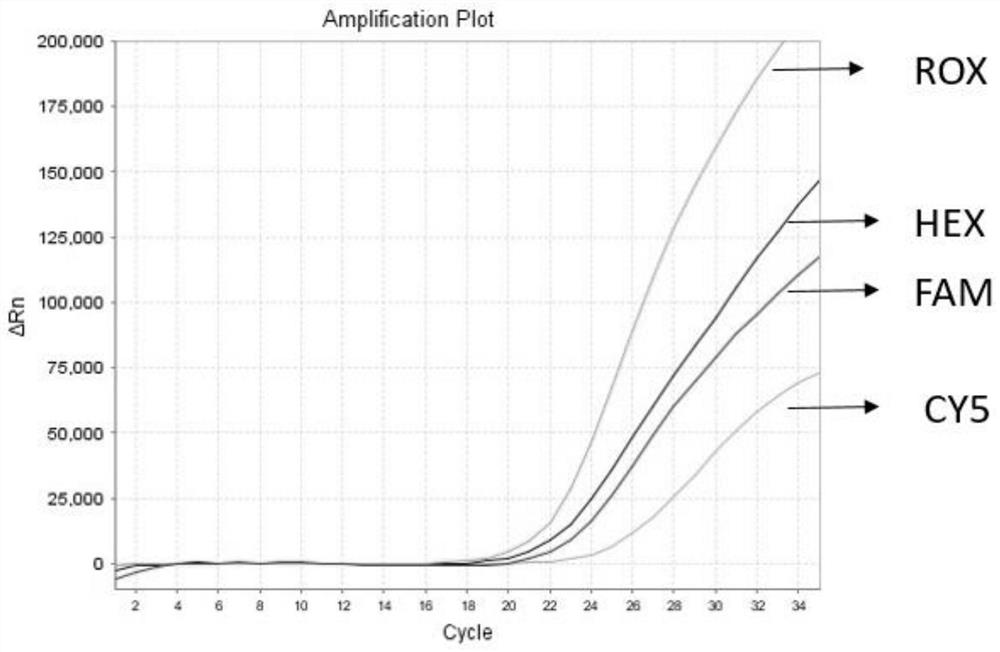
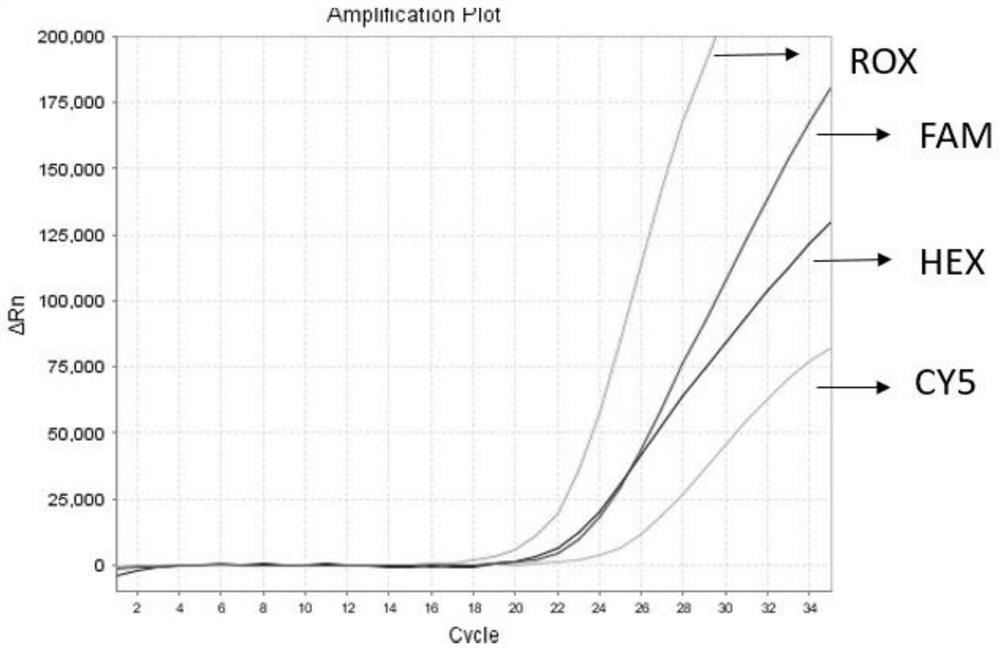
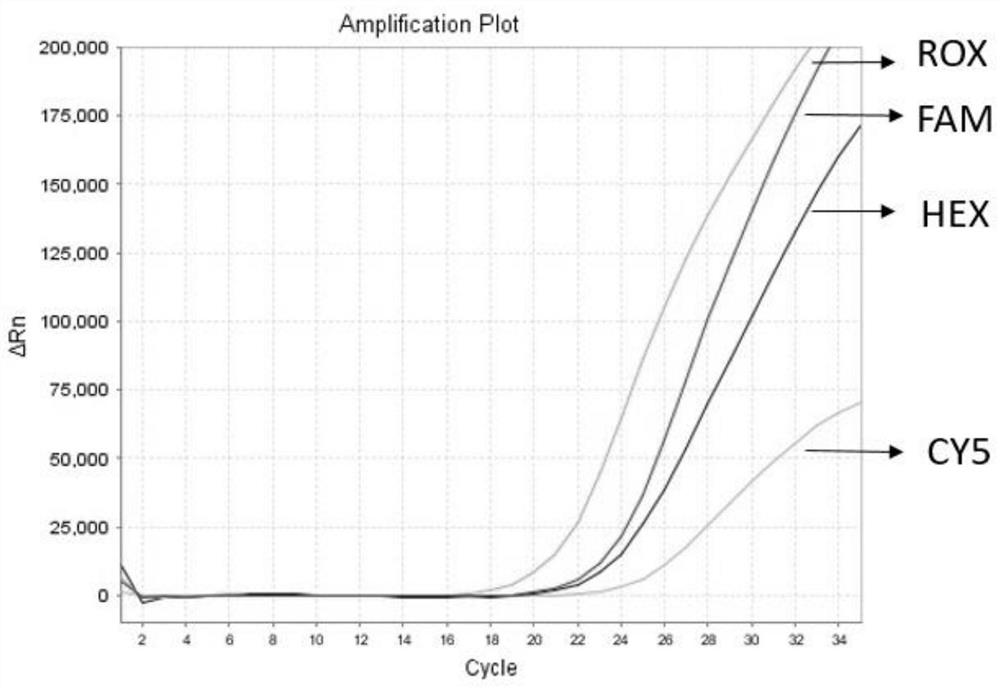
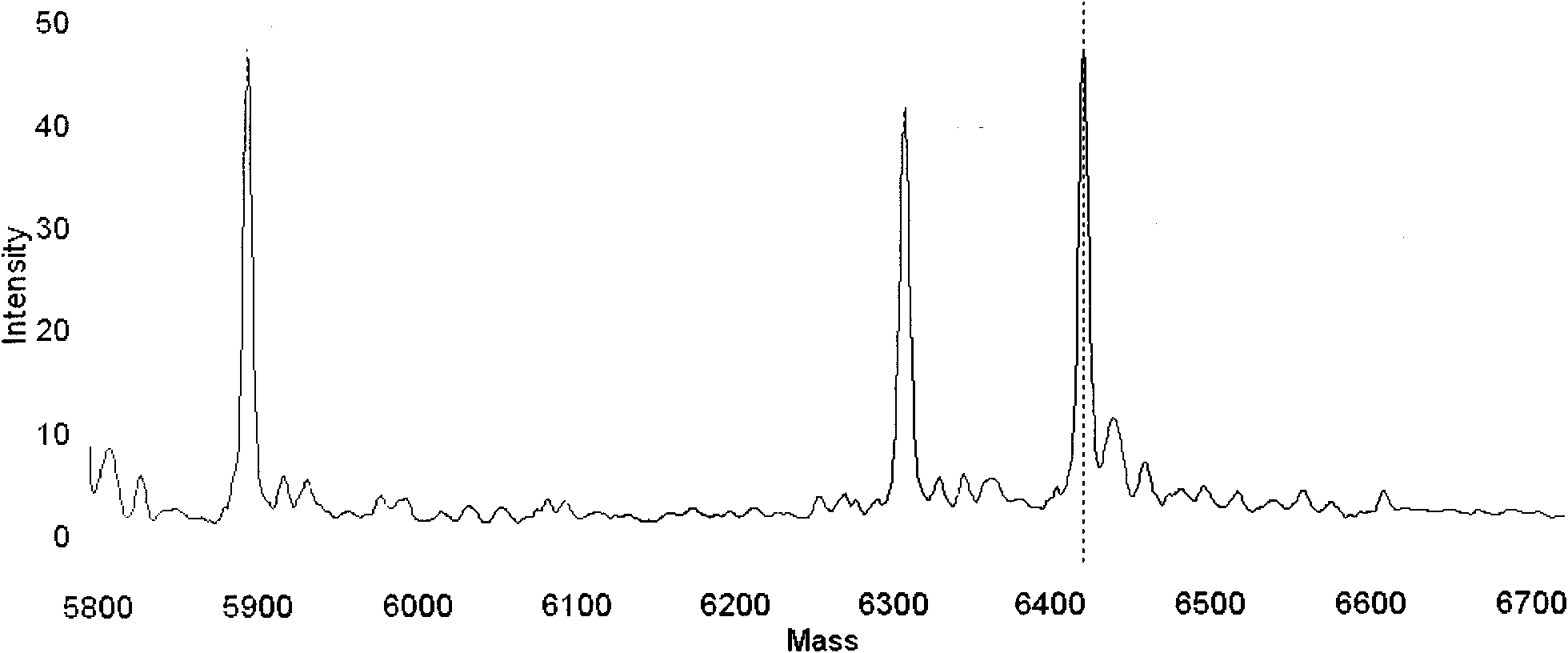
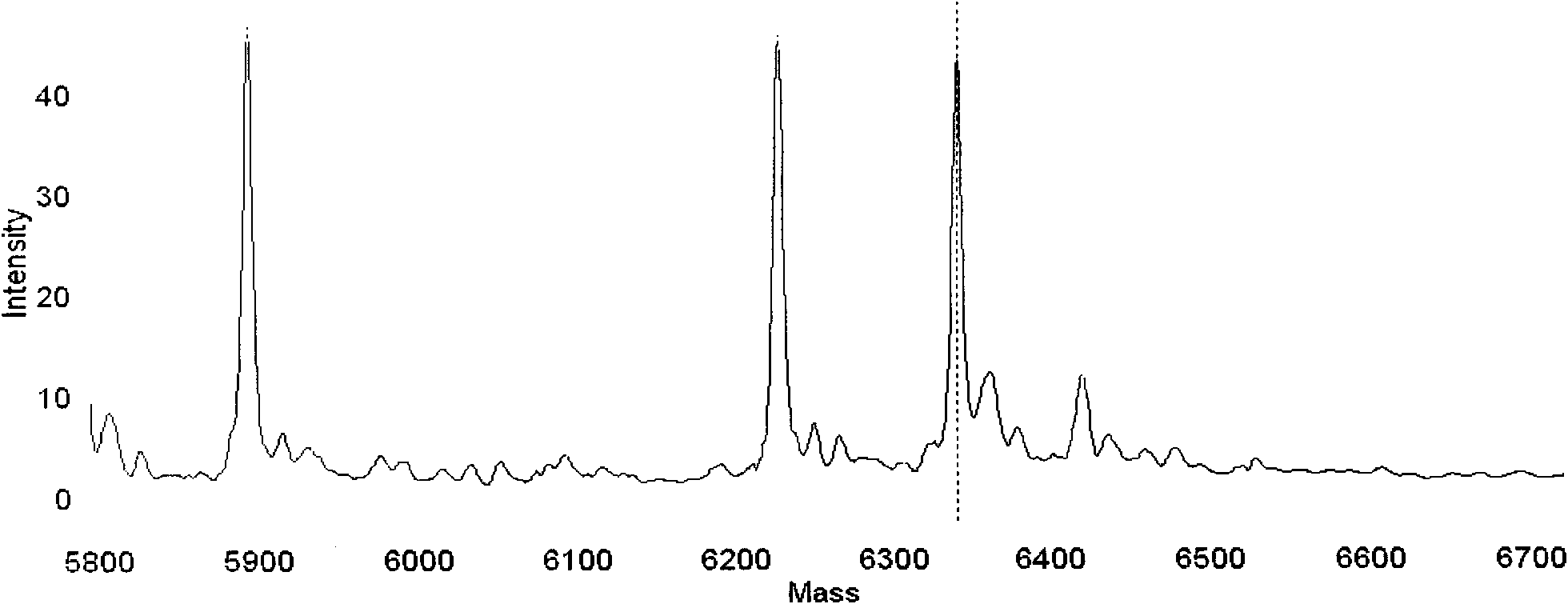
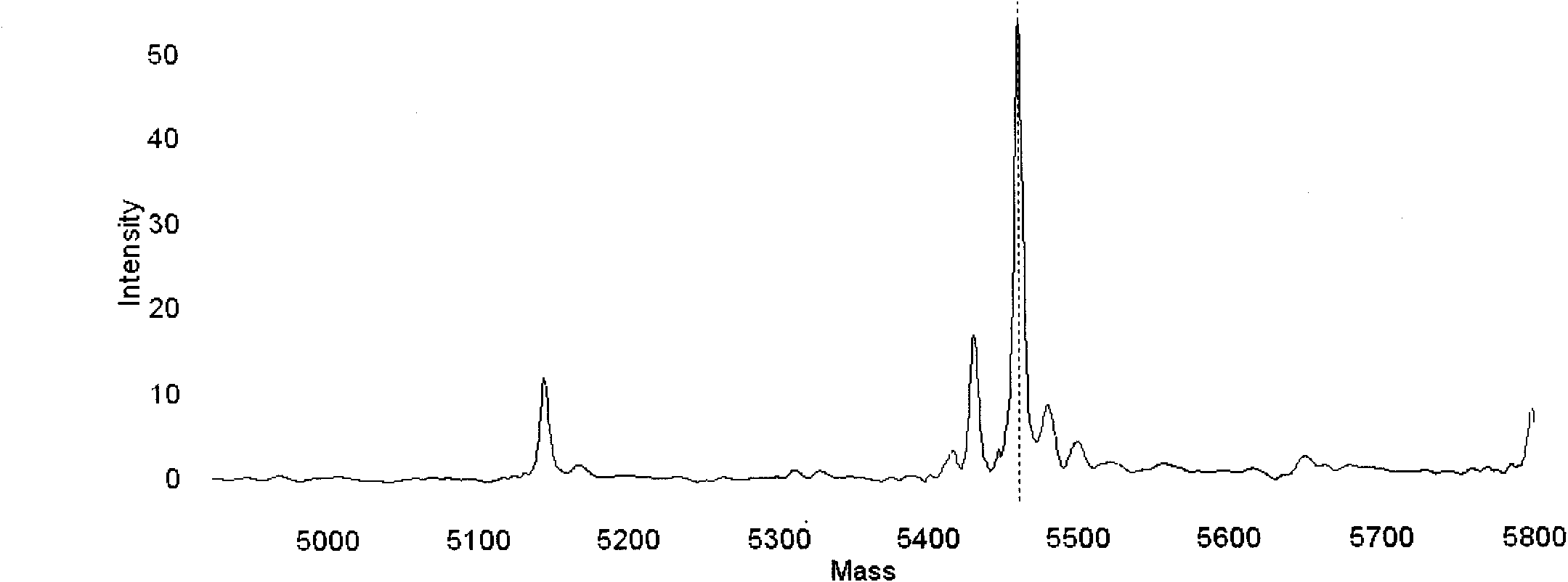
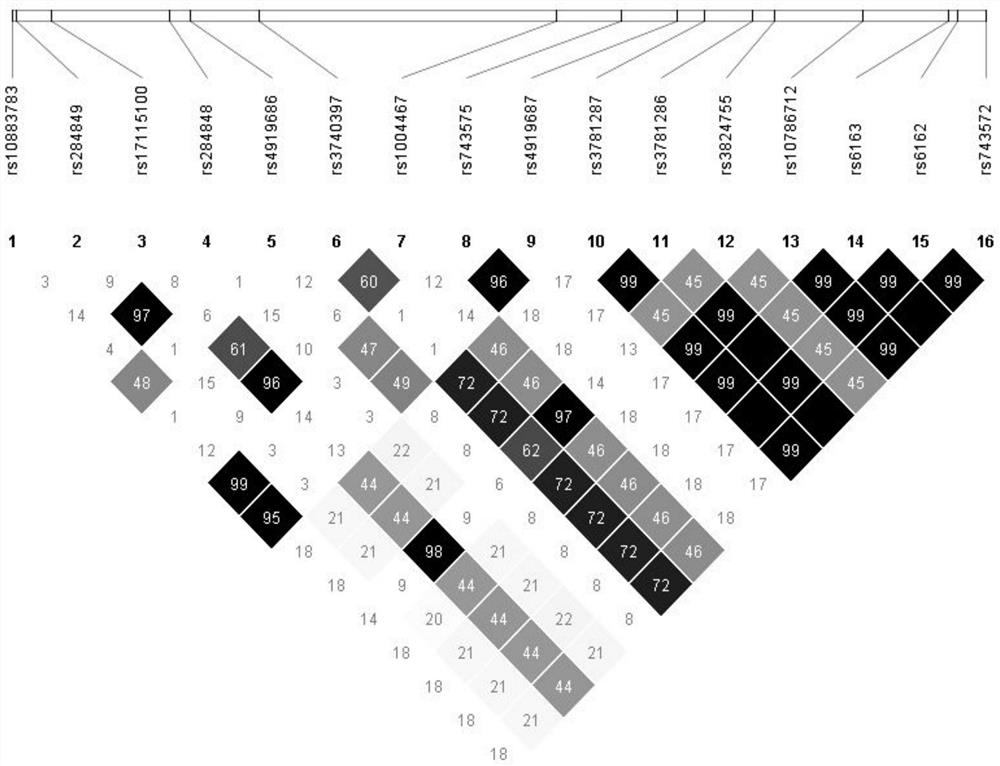
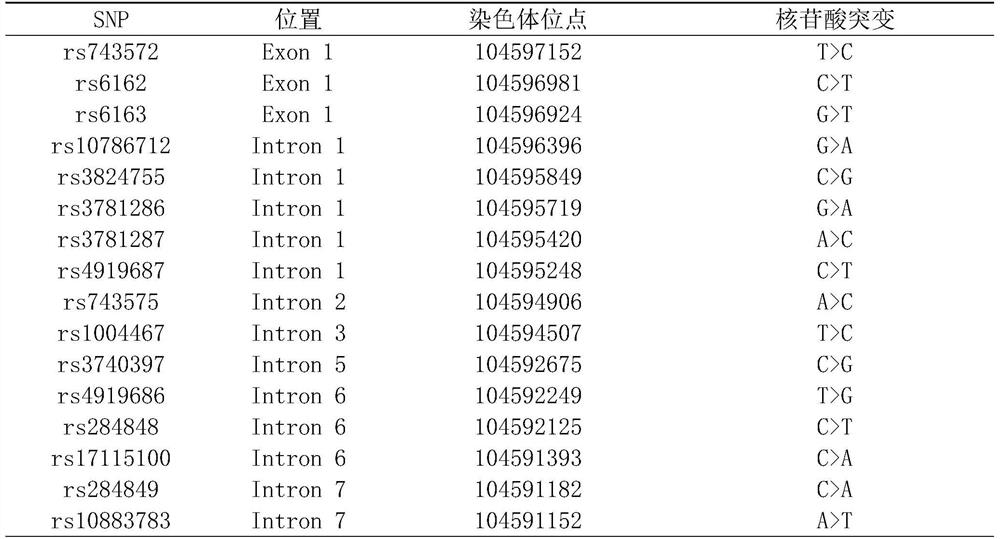
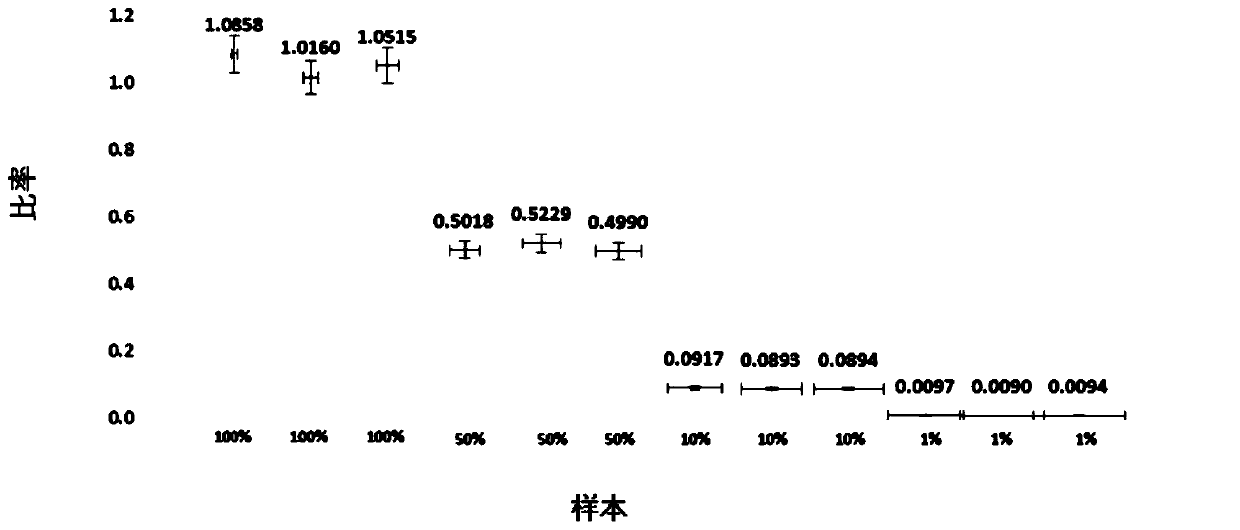
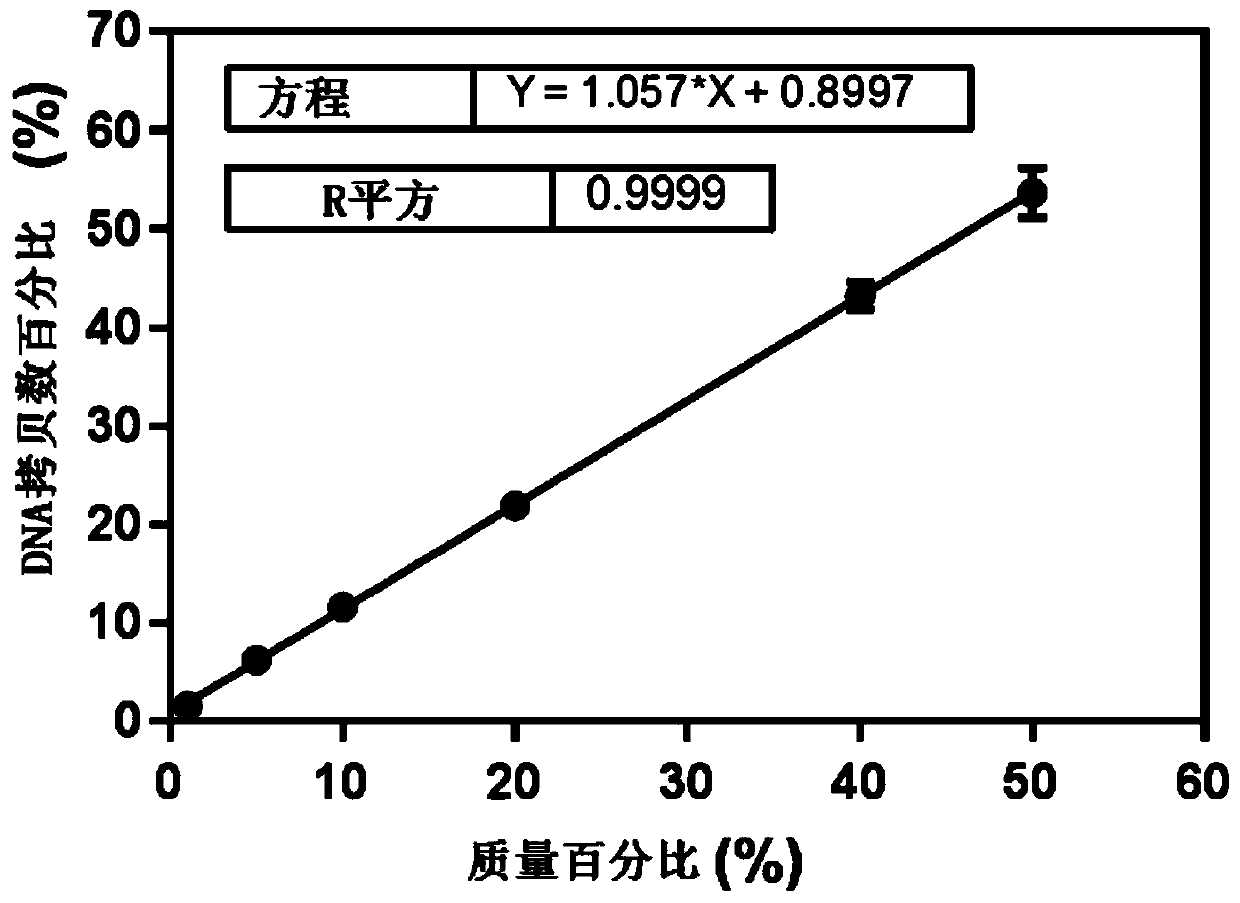
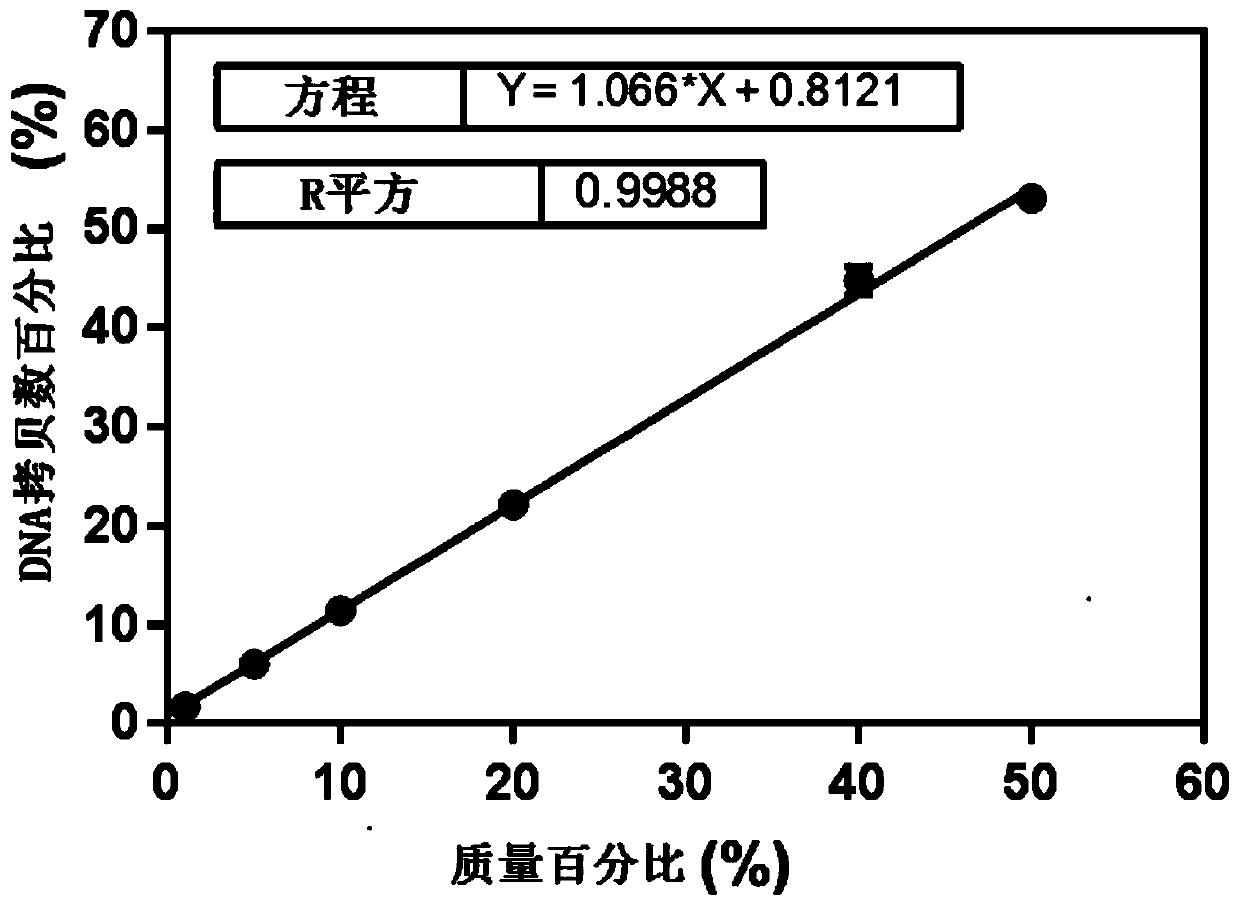
Detection method for common gene mutations in 17α-hydroxylase deficiency applicable to the Chinese population
ActiveCN106884041BMake up for deficienciesHas practical valueMicrobiological testing/measurementGenes mutationMedicine
The invention relates to a method for detecting common gene mutations of 17α hydroxylase deficiency applicable to the Chinese population, which is characterized in that it comprises the following steps: Step (1): selecting multiple SNPs on the genes to be detected in patients and carriers site; step (2): clarify the genotype of the SNP site; step (3): use software to perform haplotype analysis on multiple SNP sites; step (4): find out the gene mutation The haplotype of the haplotype is obtained to obtain the result of the haplotype analysis; step (5): the result is verified in the normal population. The invention is very suitable for Chinese people, can make up for the deficiencies and deficiencies in this field, and has very practical value.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Method for quantitatively detecting components of vigna umbellata and vigna angularis in food by double digital PCR
ActiveCN110408682AAccurate quantitative detectionEffective quantitative detectionMicrobiological testing/measurementVigna umbellataFluorescence
The invention provides a method for quantitatively detecting the components of vigna umbellata and vigna angularis in food by double digital PCR. The method comprises the steps that a dual-channel detection method is adopted, a digital PCR system is used for simultaneously detecting two kinds of fluorescent signals, probes for detecting a vigna angularis species-specific gene sequence and a vignaumbellata and vigna angularis common gene sequence are labeled as FAM and VIC separately, through the copy number concentration of the vigna angularis species-specific gene sequence and the vigna umbellata and vigna angularis common gene sequence which is measured in a same PCR reaction system, the percentage of the DNA copy number of vigna angularis in the vigna umbellata and vigna angularis is calculated, and the mass percentage of the vigna angularis components in the vigna umbellata and vigna angularis components is converted. According to the method, the mass percentage content of the vigna angularis components in a mixture of the vigna umbellata and the vigna angularis can be accurately and quickly detected.
Owner:TECH CENT OF GUANGZHOU CUSTOMS
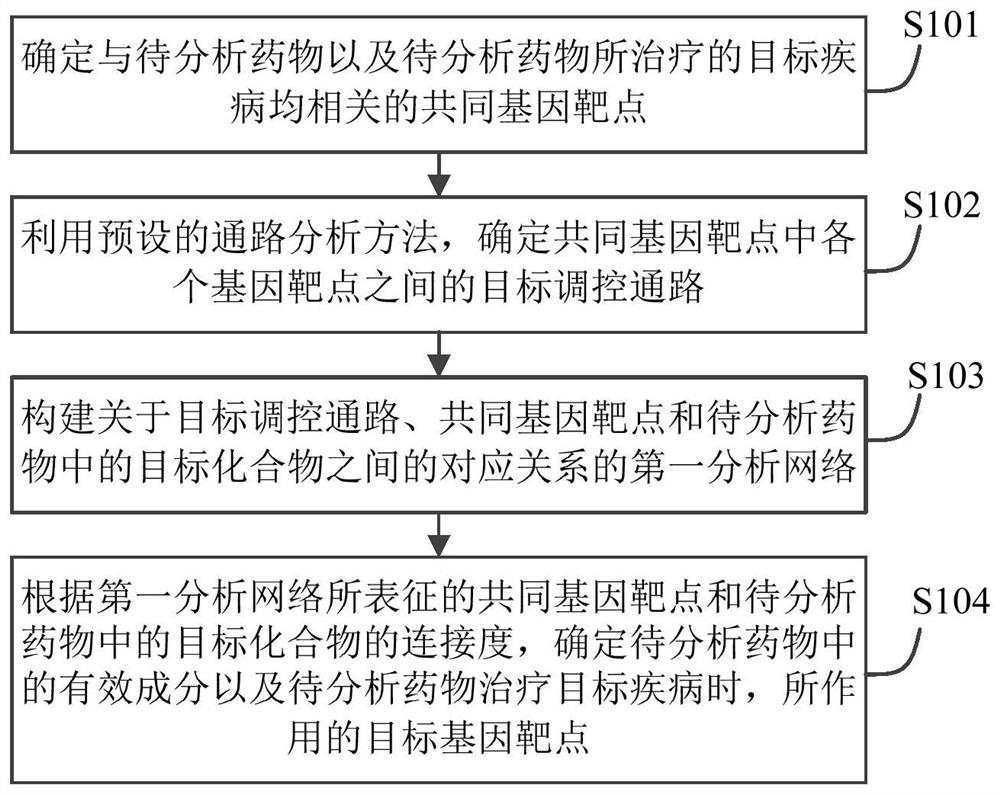
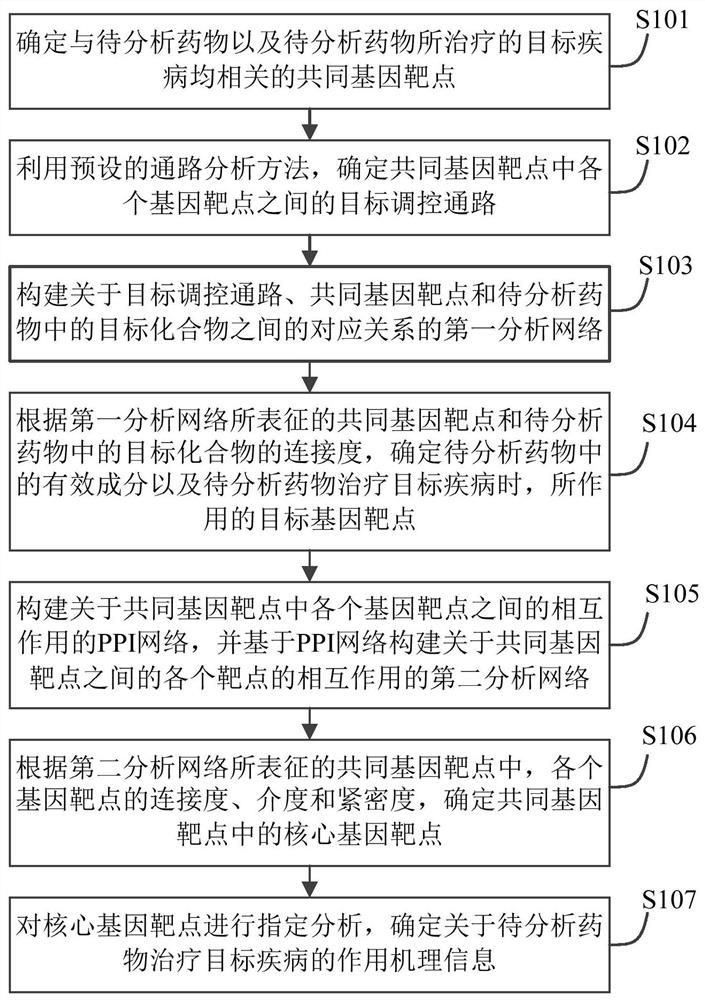
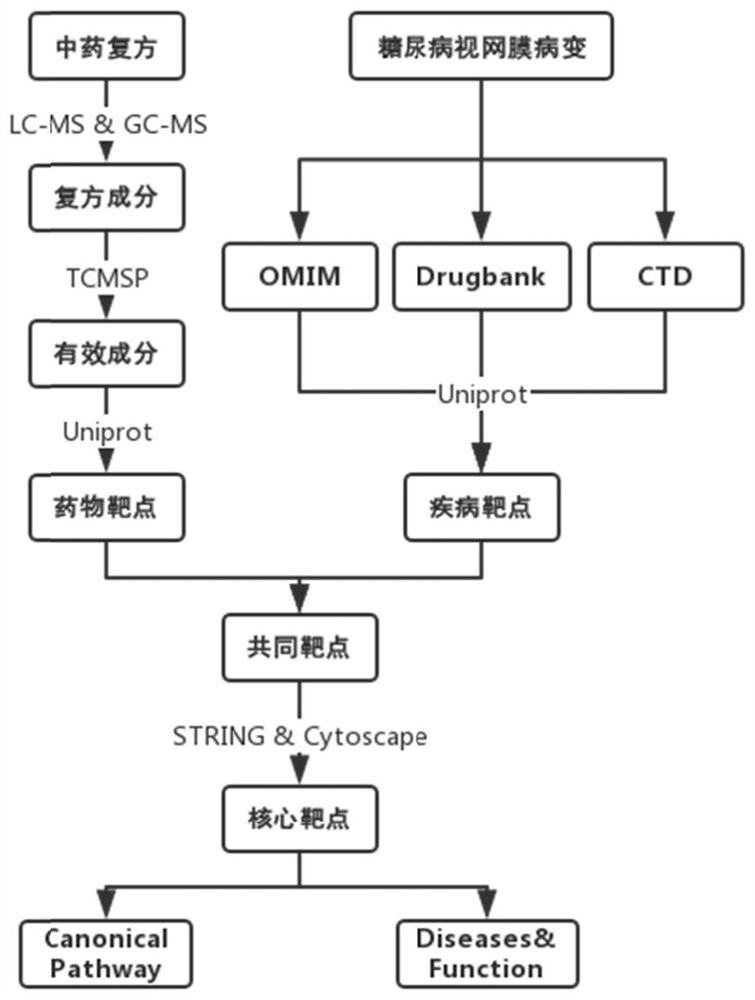
Drug action mechanism analysis method and device and electronic equipment
PendingCN113643751AProduce therapeutic effectAlternative medicinesDrug referencesDiseaseGene targeting
The embodiment of the invention provides a drug action mechanism analysis method and device and electronic equipment, and relates to the technical field of computers. The method comprises the following steps: determining common gene targets related to a to-be-analyzed drug and a target disease treated by the to-be-analyzed drug; determining a target regulatory pathway between the gene targets in the common gene targets; constructing a first analysis network about the corresponding relationship among the target regulatory pathway, the common gene target and the target compound in the to-be-analyzed drug; and according to the common gene target represented by the first analysis network and the connectivity of the target compound in the to-be-analyzed drug, determining the effective components in the to-be-analyzed drug and the target gene target acted by the to-be-analyzed drug when the to-be-analyzed drug treats the target disease. Compared with the prior art, by applying the scheme provided by the embodiment of the invention, the action mechanism of the traditional Chinese medicine compound can be efficiently and systematically analyzed so as to comprehensively screen the effective components in the traditional Chinese medicine compound and the gene targets on which the effective components act.
Owner:TIANJIN UNIV OF TRADITIONAL CHINESE MEDICINE
Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population
ActiveCN106884041AMake up for deficienciesHas practical valueMicrobiological testing/measurementHaplotypeNormal population
The invention relates to a detection method of of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population. The detection method is characterized by comprising the following steps: (1) selecting a plurality of SNP sites from to-be-detected genes of a patient and a carrier; (2) determining genotype of the SNP sites; (3) conducting haplotype analysis on the plurality of SNP sites by virtue of software; (4) finding out haplotype of gene mutation, so as to obtain a haplotype analysis result; and (5) verifying the result in a normal population. The detection method provided by the invention is quite applicable to Chinese population and is capable of overcoming shortcomings and deficiencies in the field; therefore, the detection method has a high practical value.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Kit for detecting drug resistant mycobacterium tuberculosis (MDR-TB)
ActiveCN101845503BAccurate Diagnostic InformationShorten the timeMicrobiological testing/measurementFluorescence/phosphorescenceMulti-drug-resistant tuberculosisMycobacterium Infections
The invention relates to a method adopting double-label probe detection and melting curve analysis for diagnosing the infection of multi-drug resistant mycobacterium tuberculosis and a kit which utilizes the method to detect multiple gene mutations related to drug resistant tuberculosis at the same time, and the invention belongs to the life science and biological technical field. The kit of the invention contains a primer designed for multiple gene mutations related to drug resistant, a double-label oligonucleotide probe capable of detecting multiple common gene mutation sites related to drug resistant tuberculosis and a DNA polymerases with heat stability and without 5' nuclease activity, and the kit can be used to detect at least 16 common gene mutation sites related to drug resistant tuberculosis under proper PCR reaction conditions. The detection method and kit of the invention can be used in the early diagnosis of multi-drug resistant tuberculosis and overcome the problems of the existing technology that the detection time is long, a great deal of manpower and large material resources are needed, the detection cost is high, etc.
Owner:无锡锐奇基因生物科技有限公司
A cross-chip platform gene expression data integration method
ActiveCN103745137BEliminate experimental batch effectsImprove performanceSpecial data processing applicationsGene expression matrixLeast squares
The invention belongs to the technical field of biological information. The invention provides a method for integrating gene expression data by crossing a plurality of different chip platforms; the method comprises the following steps: the standard preprocessing is implemented for the gene expression profile of the plurality of chip platforms; the common gene expression data in the different chip platforms is merged; genes are divided into k subsets according to the expression similarity of the genes on the plurality of chip platforms; the expression linear relation exps1=as*exps2+bs of the different chip platforms in every gene subset can be calculated by the least square method; the gene expression values of the different chip platforms are standardized into the same change range by using the formula exp1=X*A.*exp2+X*B to obtain standard gene expression matrixes, wherein the implications of symbols are defined as the specification.
Owner:艾吉泰康(嘉兴)生物科技有限公司
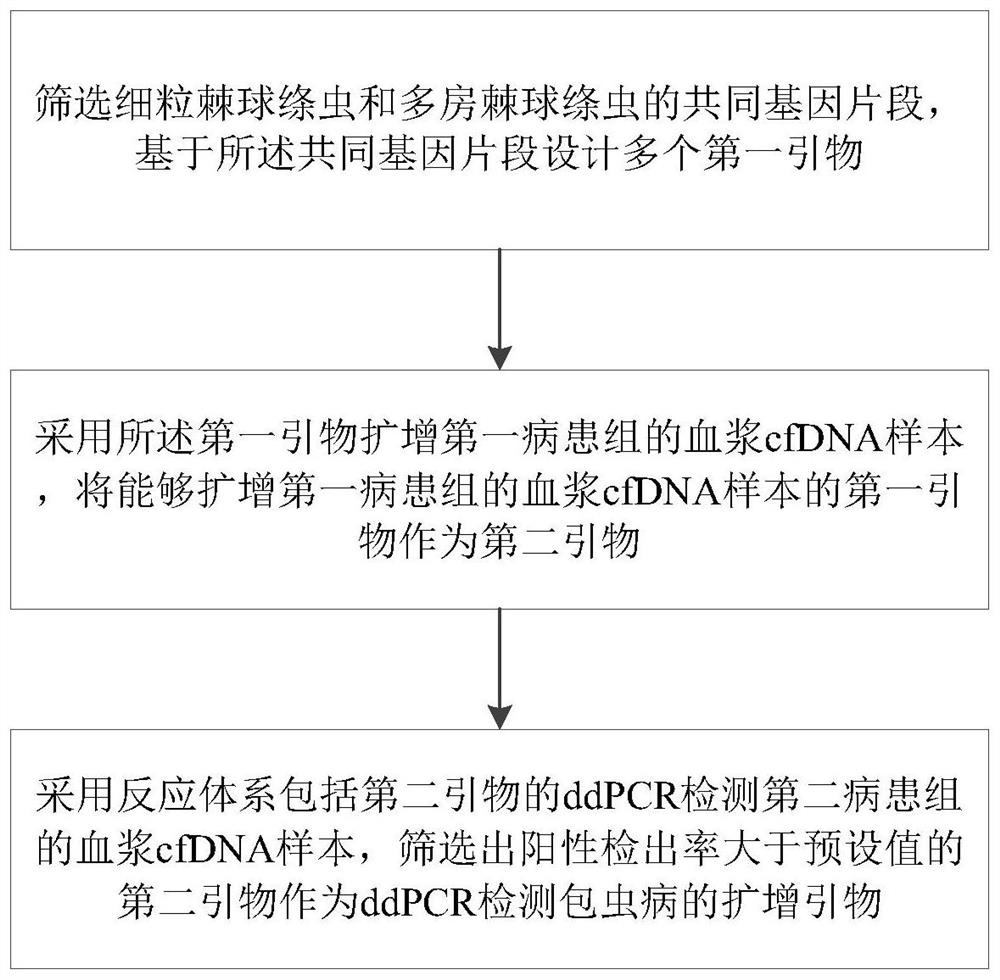
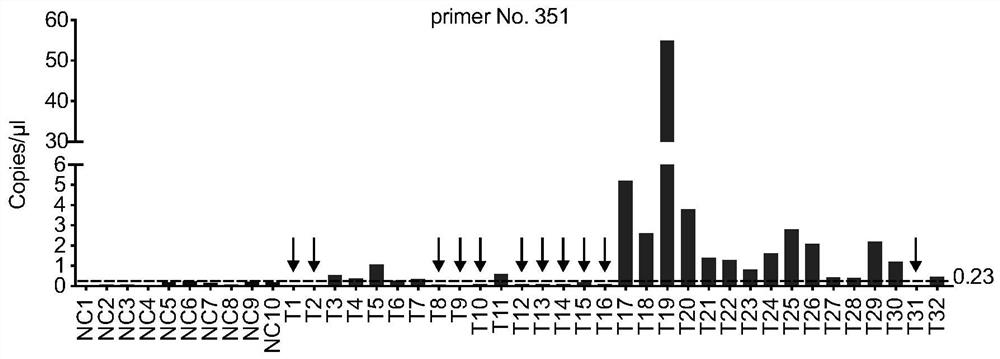
Amplification primer for detecting echinococcosis through ddPCR and construction method and application of amplification primer
PendingCN113637774ARealize ultra-early diagnosisShorten the judgment periodMicrobiological testing/measurementDNA/RNA fragmentationPatient groupBlood plasma
The invention discloses a primer construction method for detecting hepatic echinococcosis through ddPCR, an amplification primer and application of the amplification primer. The primer construction comprises the following steps that common gene segments of echinococcus granulosus and echinococcus multilocularis are screened, and a plurality of first primers are designed based on the common gene segments; a plasma cfDNA sample of a first patient group is amplified by adopting the first primers, and the first primers capable of amplifying the plasma cfDNA sample of the first patient group are taken as second primers; and a plasma cfDNA sample of a second patient group is detected by adopting ddPCR of a reaction system comprising the second primers, and the second primers with the positive detection rate greater than a preset value are screened out as a target amplification primer. The amplification primer can amplify DNA fragments of echinococcus which are released into peripheral blood and exist in the form of plasma free DNA, by combining with a ddPCR method, whether human plasma is infected with echinococcosis can be diagnosed by detecting the human plasma, so that the ultra-early diagnosis of echinococcus granulosus and echinococcus multilocularis is realized, and a detection tool which is sensitive enough is provided for research and development of an echinococcosis precise therapy.
Owner:WEST CHINA HOSPITAL SICHUAN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com

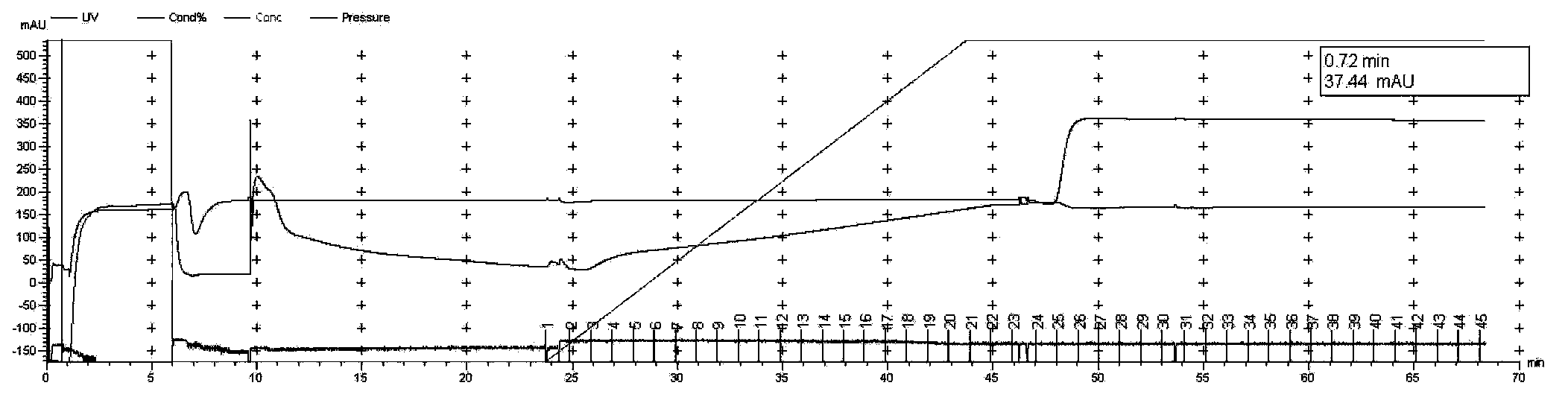







![Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population](https://images-eureka.patsnap.com/patent_img/17e323f1-487a-4706-9f23-69e558d90032/HDA0000880693730000011.png)
![Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population](https://images-eureka.patsnap.com/patent_img/17e323f1-487a-4706-9f23-69e558d90032/BDA0000880693720000031.png)
![Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population Detection method of 17[alpha] hydroxylase deficiency common gene mutation applicable to Chinese population](https://images-eureka.patsnap.com/patent_img/17e323f1-487a-4706-9f23-69e558d90032/BDA0000880693720000032.png)