Method and element for screening different-intensity ribosome binding sites and promoters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
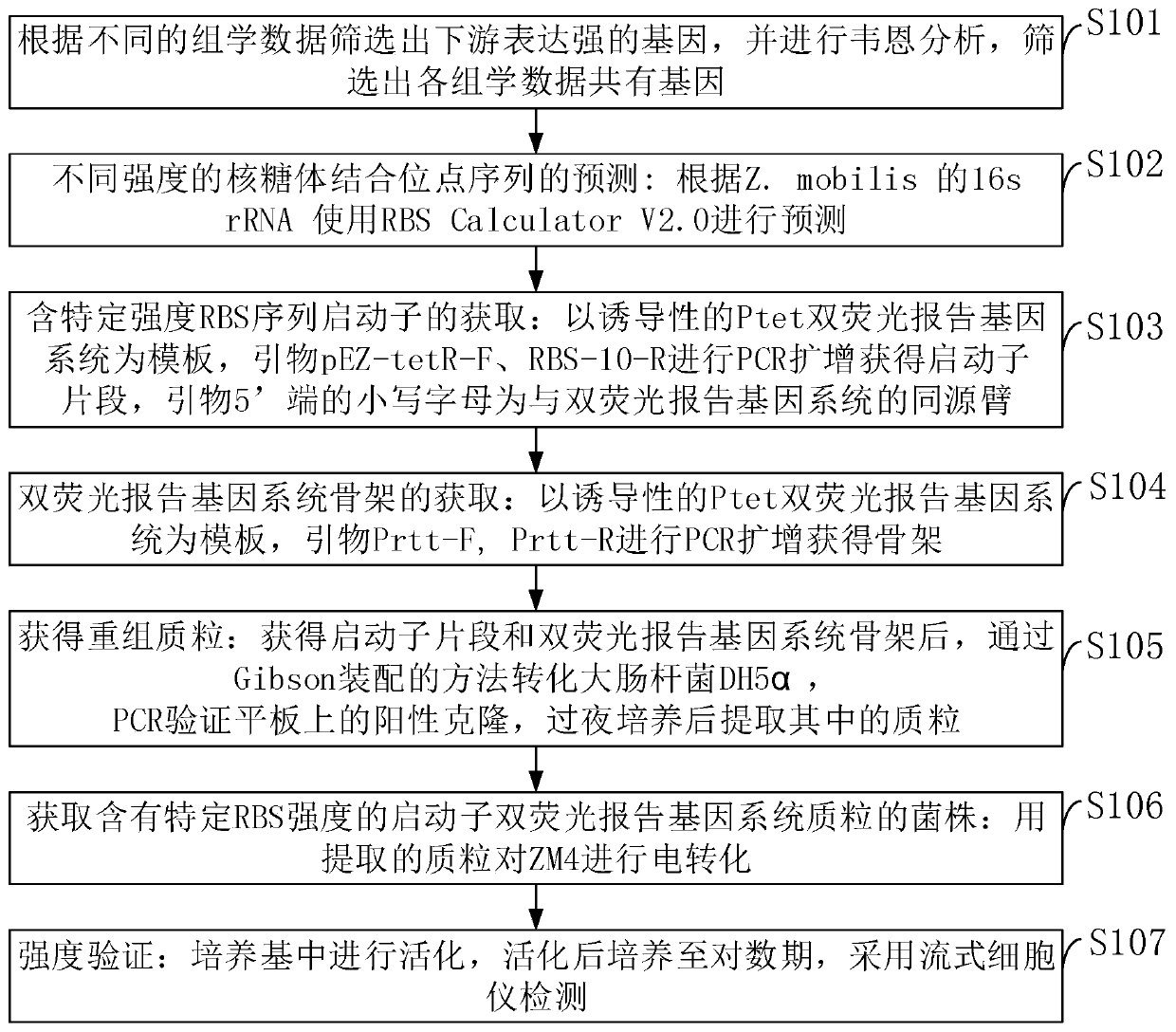
[0089] The different-strength ribosome binding sites and promoter screening methods of Zymomonas mobilis provided by the embodiments of the present invention include:
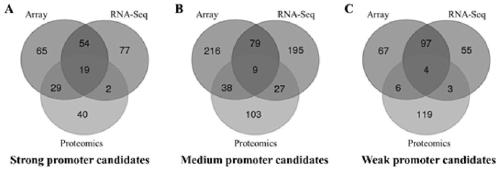
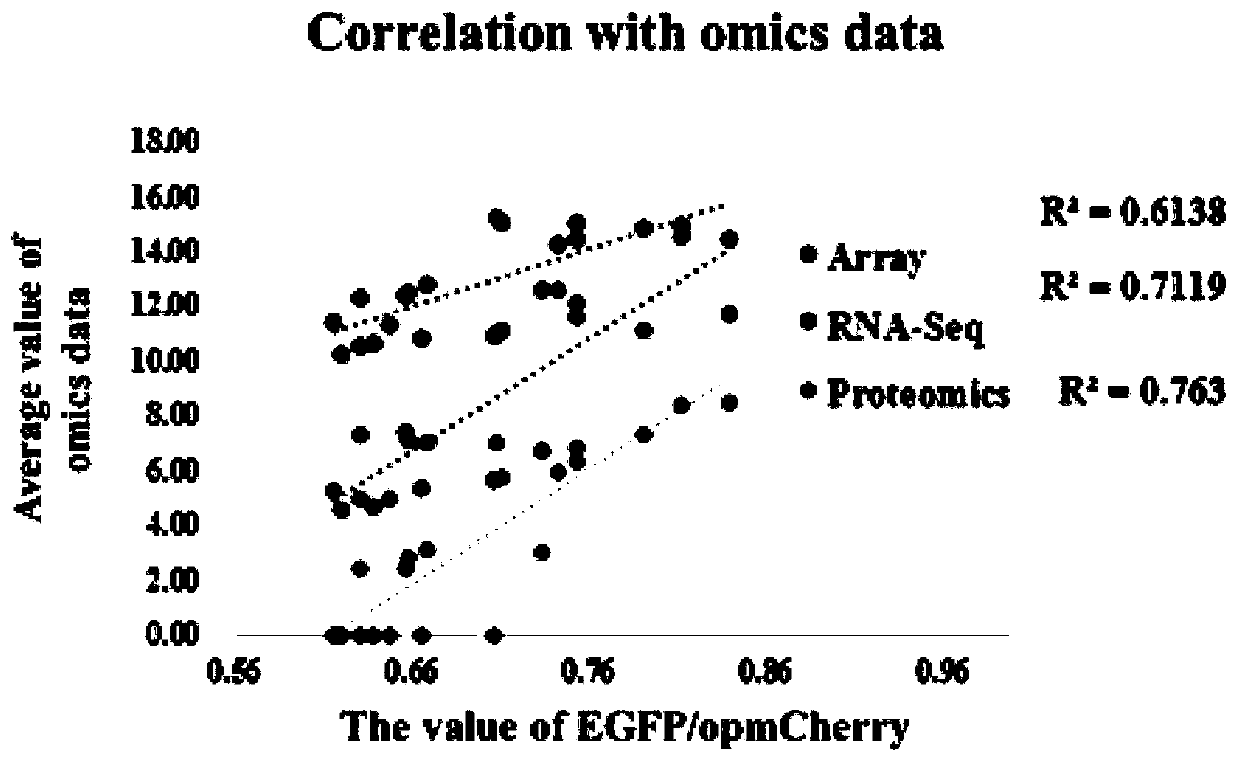
[0090] 1) Firstly, genes with strong downstream expression are screened out according to different omics data, and Venn analysis is performed to screen out genes shared by all omics data. In different omics data, each gene is ranked according to the average value under all conditions, and the promoters ranked above 90% are defined as strong promoters, those ranked 40-60% are moderately strong promoters, and ranked 10 Below % are weak promoters. 19 strong promoters, 9 medium-strength promoters and 10 weak promoters were screened.
[0091] like figure 2 As shown, the Venn analysis provided in the embodiment of the present invention screens promoters with different strengths.
[0092] 2) Prediction of ribosome binding site sequences (RBS) with different strengths: Prediction is performed using RBS Calculator V...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com