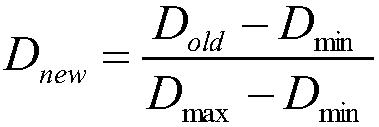Patents
Literature
290 results about "Omics" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics. Omics aims at the collective characterization and quantification of pools of biological molecules that translate into the structure, function, and dynamics of an organism or organisms.
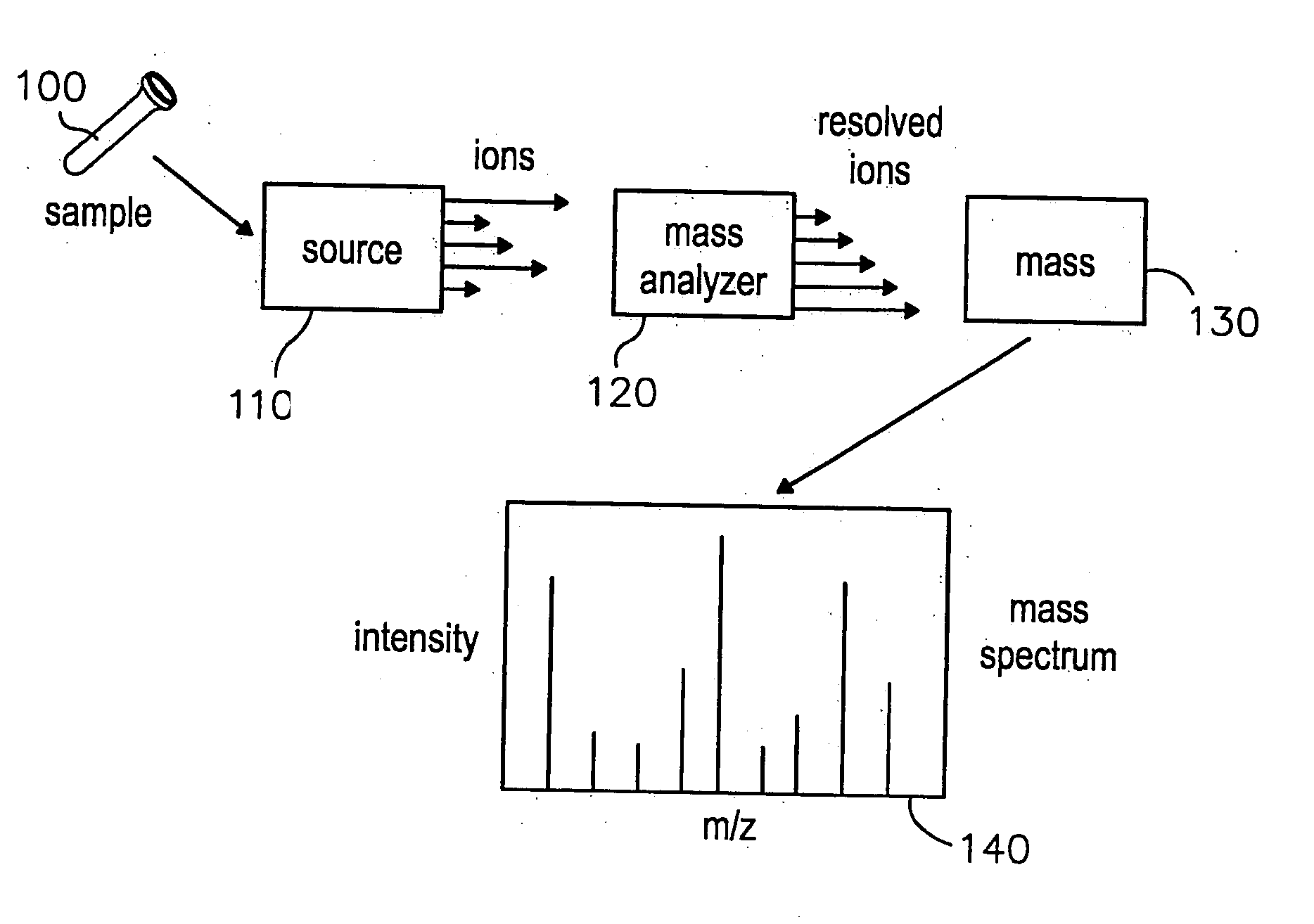
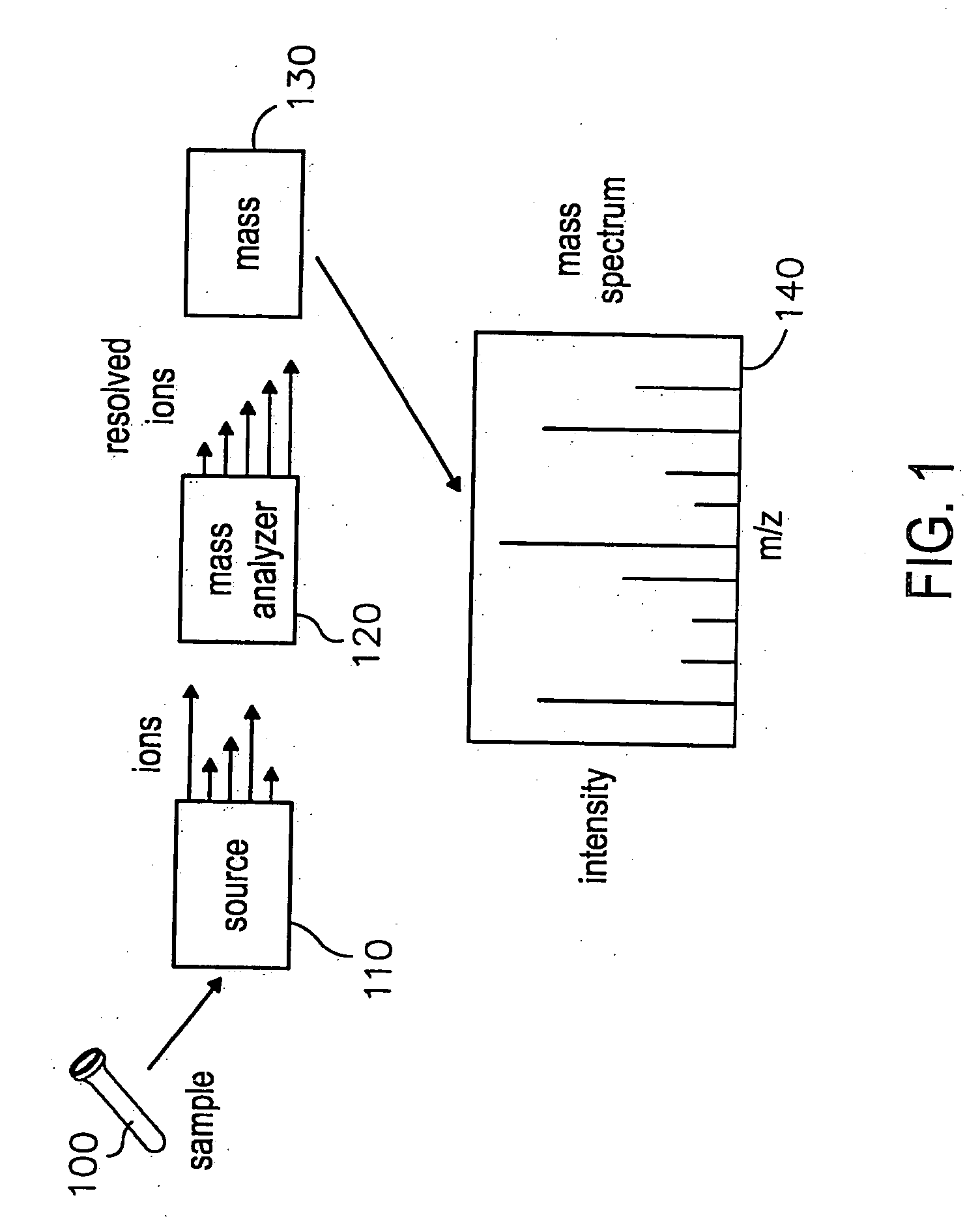
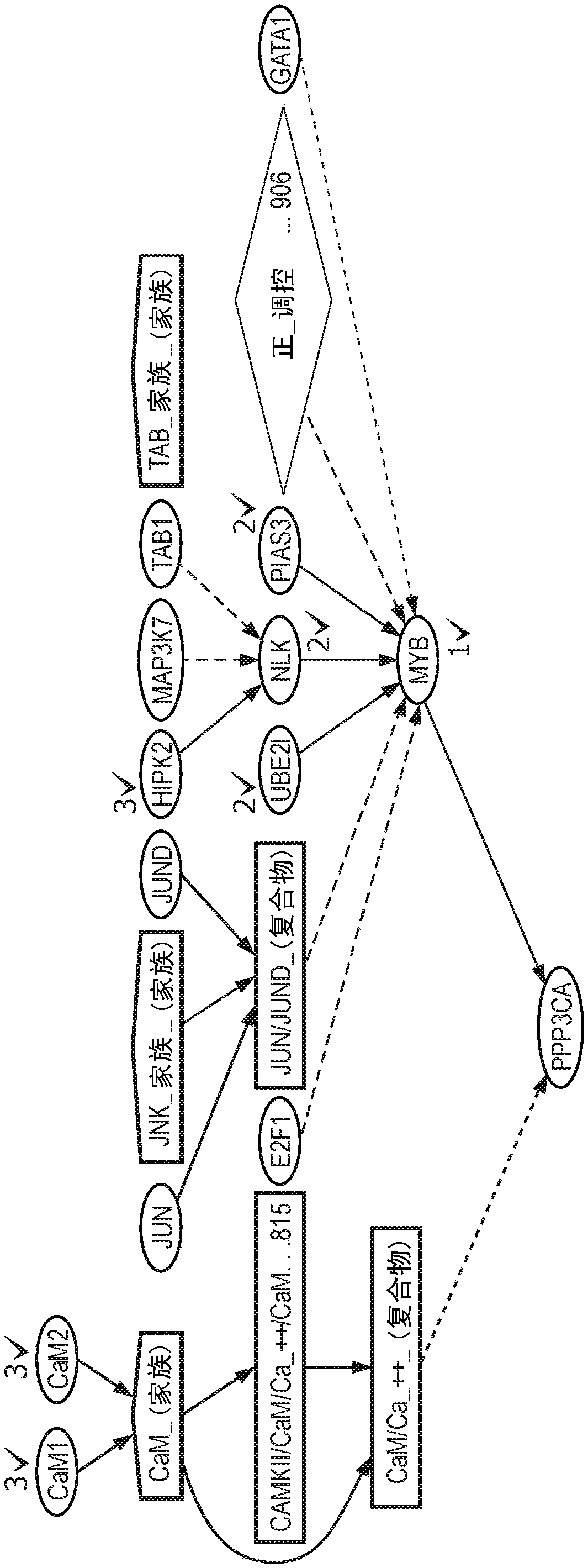
System and methods for navigating and visualizing multi-dimensional biological data
InactiveUS20060036425A1Easy to identifyFacilitate correlation2D-image generationData visualisationData setEeg data
Methods systems and computer readable media for manipulating and displaying sparse data contained within very large datasets. Particular applications includes those to proteomics and metabolomics. Navigation of the large data sets from a global perspective may be practically implemented, so that a user is not confined to a narrow slice of data at a time, thus providing context of the whole dataset while viewing data within the dataset.
Owner:AGILENT TECH INC
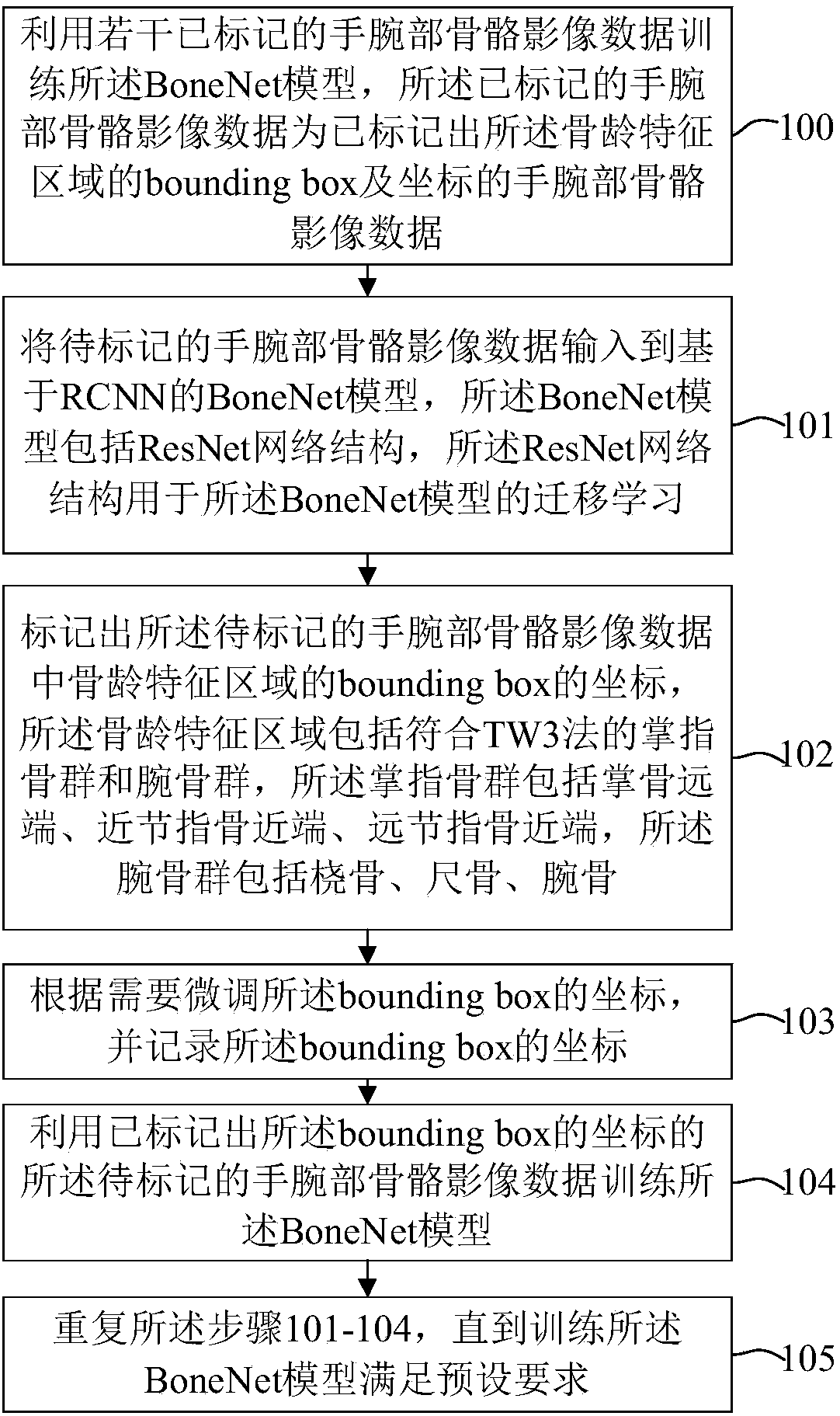
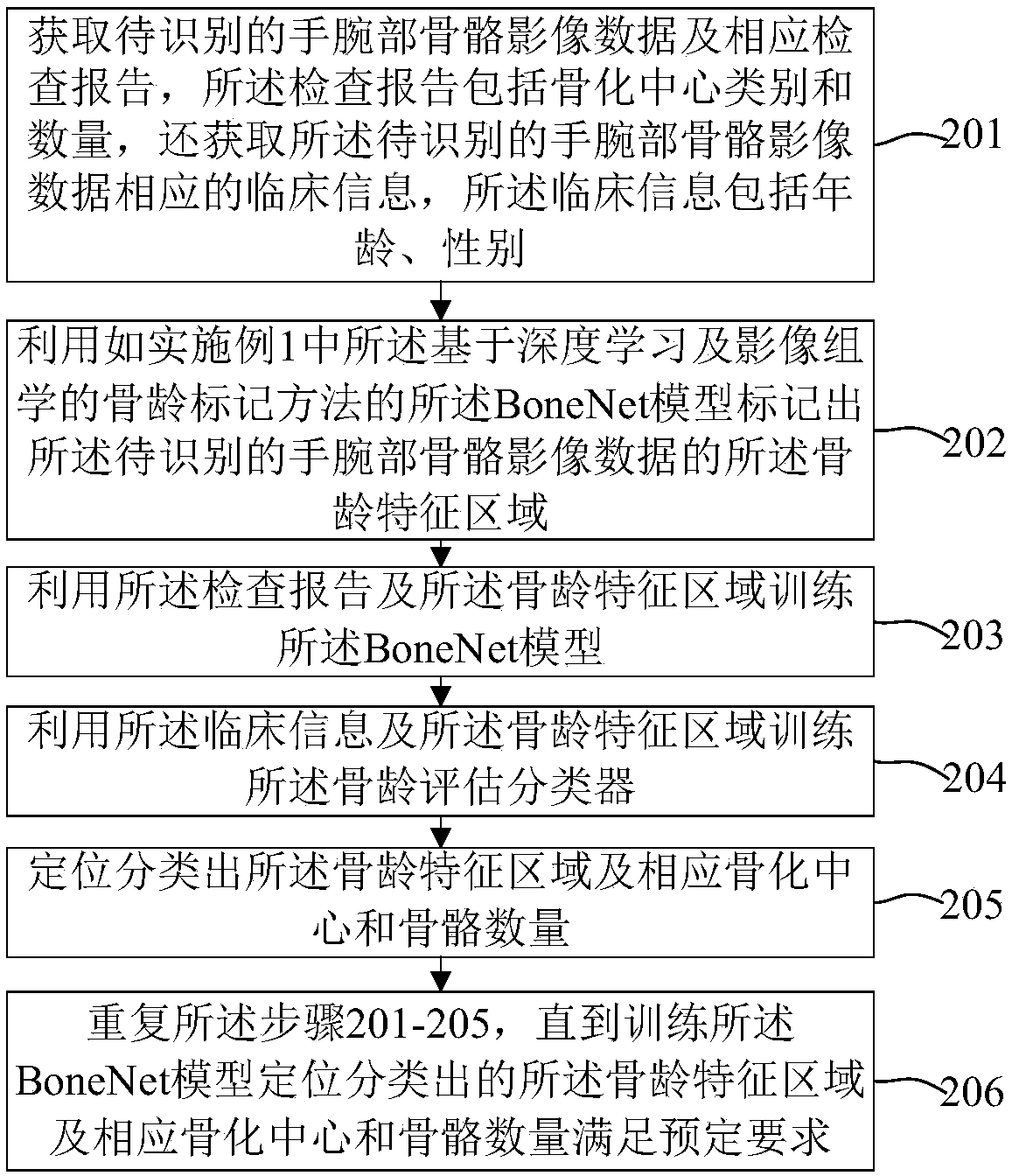
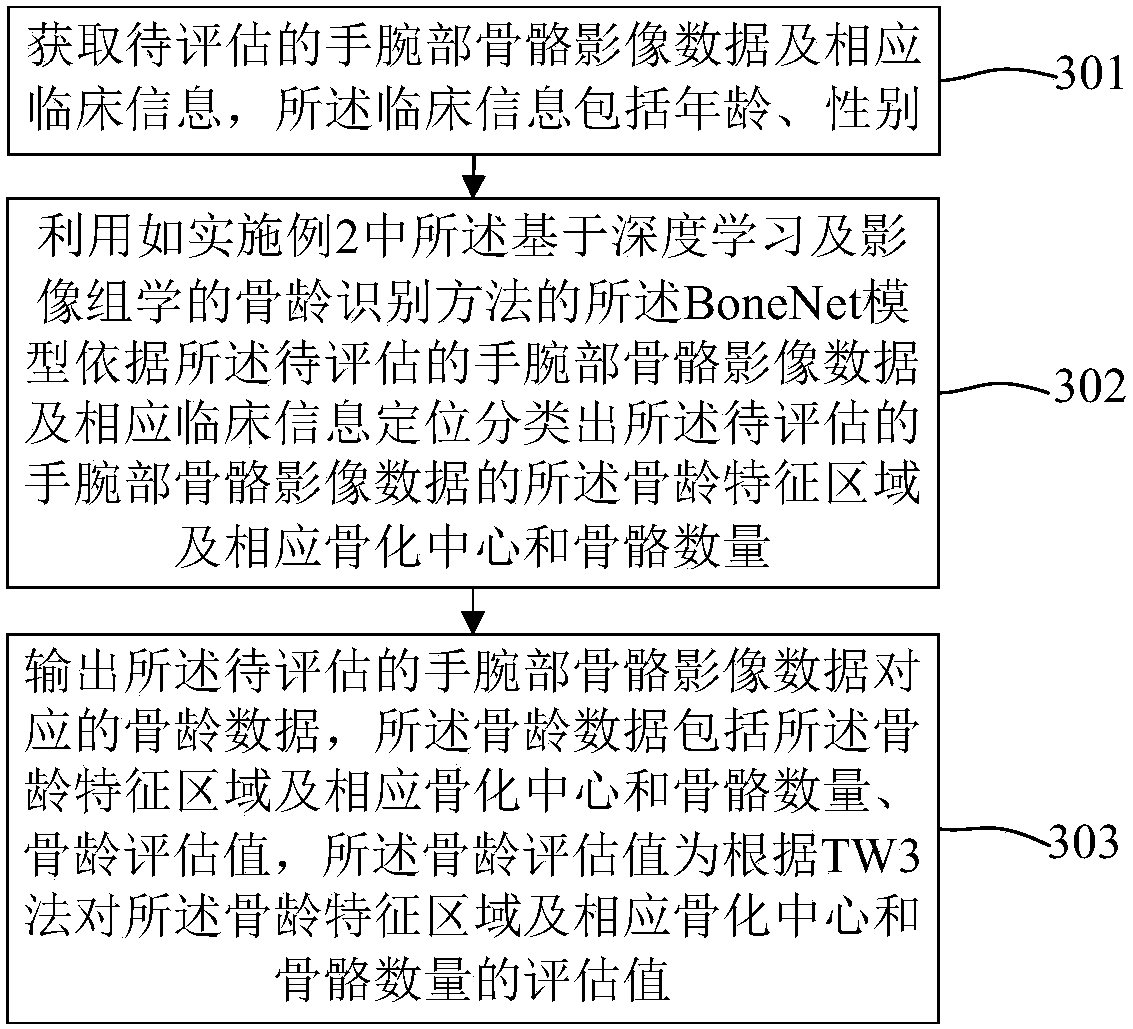
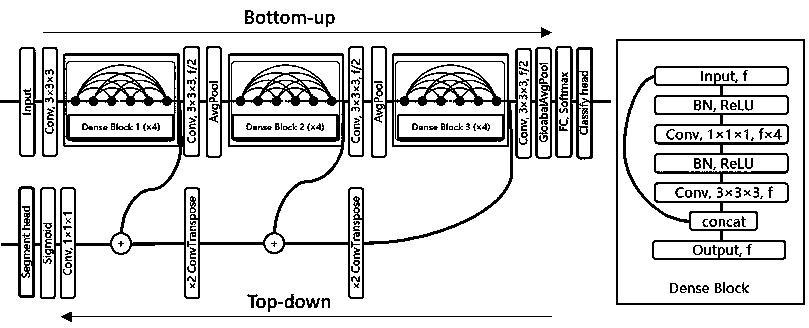
Bone age mark identification assessment method and system based on deep learning and image omics
ActiveCN107591200AImprove recognition accuracySolve the problem of insufficient training dataMedical automated diagnosisCharacter and pattern recognitionImaging dataPopulation statistics
The invention discloses a bone age mark identification assessment method and system based on deep learning and image omics. The bone age mark identification method includes the steps: performing preprocessing of window adjusting, alignment and standardization on the wrist bone image; using a bounding box to mark the bone age characteristic areas and mark the coordinates, wherein the bone age characteristic areas include a metacarphphalangeal group and a brachidium group according with a TW3 method; according to the requirement, performing augmentation processing, and inputting the wrist bone image data to a convolutional neural network of the area based on ResNet-101 to perform multi-task (positioning, classification and assessment) training at the same time; and based on the bone age characteristic areas, combining with the clinic information (demographic characteristics and inspection reports) to further train and improve the bone age assessment speed and accuracy. The bone age markidentification assessment method and system based on deep learning and image omics firstly utilize a small number of marked samples to perform preliminary training on the bone age model, and utilize the model with relatively higher positioning detection accuracy to automatically mark a large number of samples so as to realize automatic positioning, classification and bone age assessment of the bone age characteristic areas.
Owner:WINNING HEALTH TECHNOLOGY GROUP CO LTD
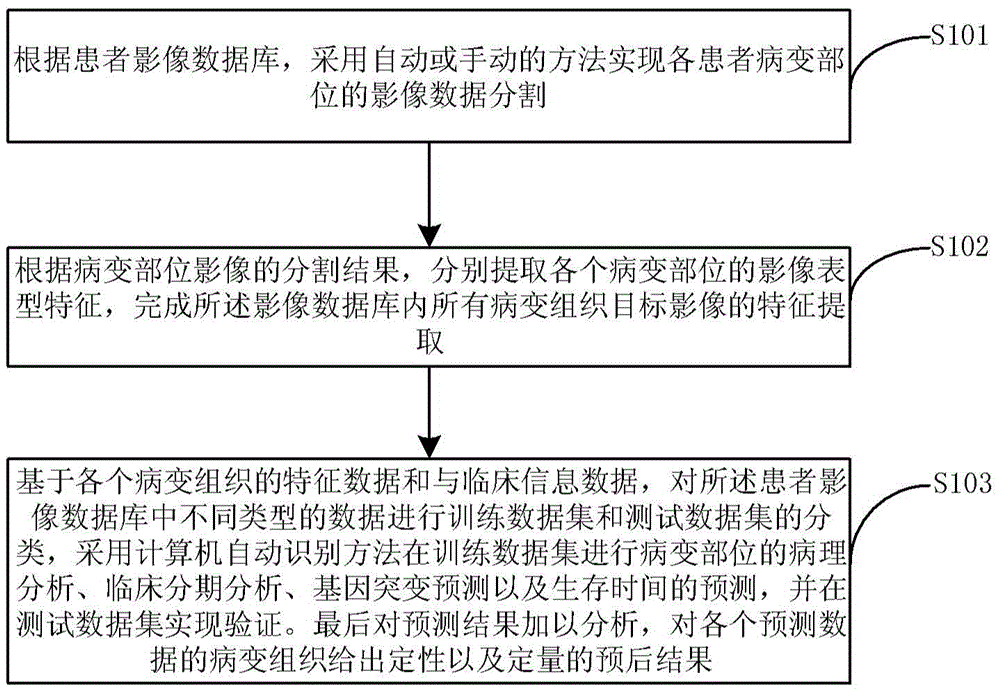
Image omics based lesion tissue auxiliary prognosis system and method
The invention discloses an image omics based lesion tissue auxiliary prognosis system and method. The method comprises the steps of extracting image data of lesion parts with an automatic or manual segmentation method from a big-data-volume patient image database; according to segmentation results of lesion part images, extracting image phenotypic characteristics of each lesion part, and finishing feature extraction of image data of all lesion parts in the patient image database; and based on characteristic data and clinical information data of each lesion part, performing training data set and test data set classification on the data in the patient image database, performing pathologic analysis, clinical stage analysis, gene mutation prediction and survival time prediction of the lesion parts in the training data set with a computer automatic identification method, and performing verification in the test data set. The method is capable of performing qualitative and quantitative prediction analysis on a specific individual and providing credible prediction and analysis results.
Owner:INST OF AUTOMATION CHINESE ACAD OF SCI
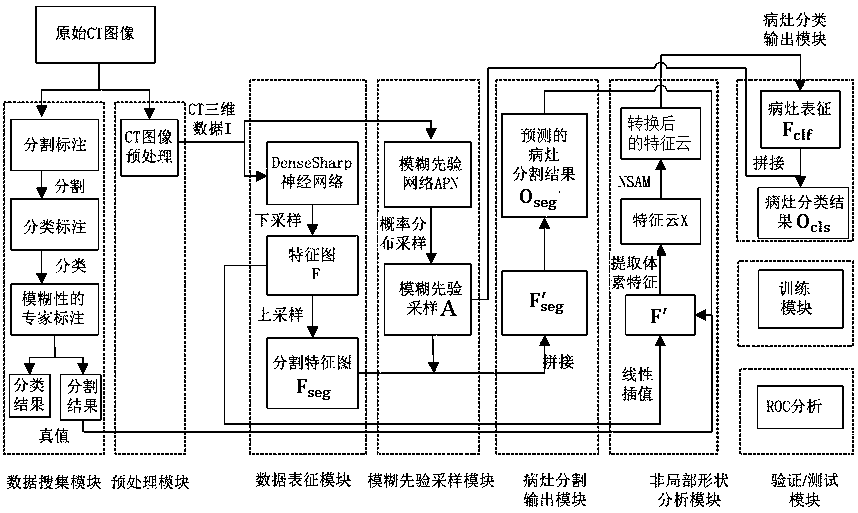
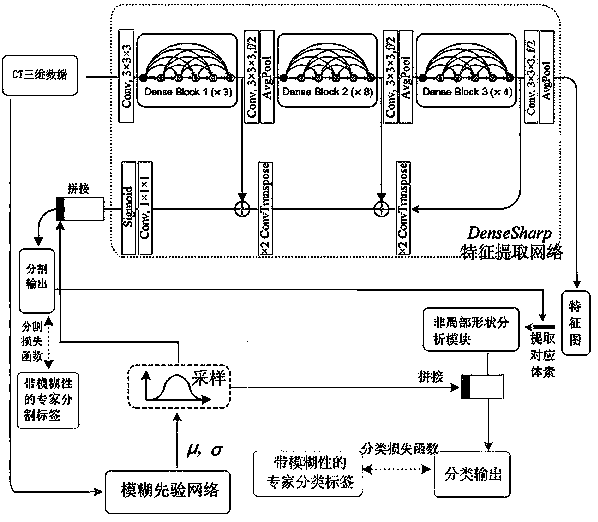
Focus classification system based on deep learning and probability imaging omics
ActiveCN110458249AShow ambiguityImprove robustnessImage enhancementImage analysisNerve networkAmbiguity
The invention relates to a focus classification system based on deep learning and probability imaging omics, and belongs to the technical field of medical image classification. The objective of the invention is to solve problems of ambiguity caused by classification ambiguity and low classification precision of an existing lesion classification system. According to the method, a deep convolutionalneural network is used as a main stem, a non-local shape analysis module is proposed to extract feature cloud of a focus on a medical image, interference of pixels around the focus on classificationjudgment is removed, and essential representation of the focus is obtained; meanwhile, the fuzziness of the label is captured; a fuzzy prior network is provided to simulate fuzzy distribution of different expert labels; ambiguity of expert annotation is displayed and modeled; the classification result of model training has better robustness, the fuzzy prior sample is combined with focus representation, a new focus classification system is constructed and can achieve controllability and probability; compared with a traditional convolutional neural network, a classification fuzziness problem isbetter solved, and higher classification precision can be acquired.
Owner:点内(上海)生物科技有限公司
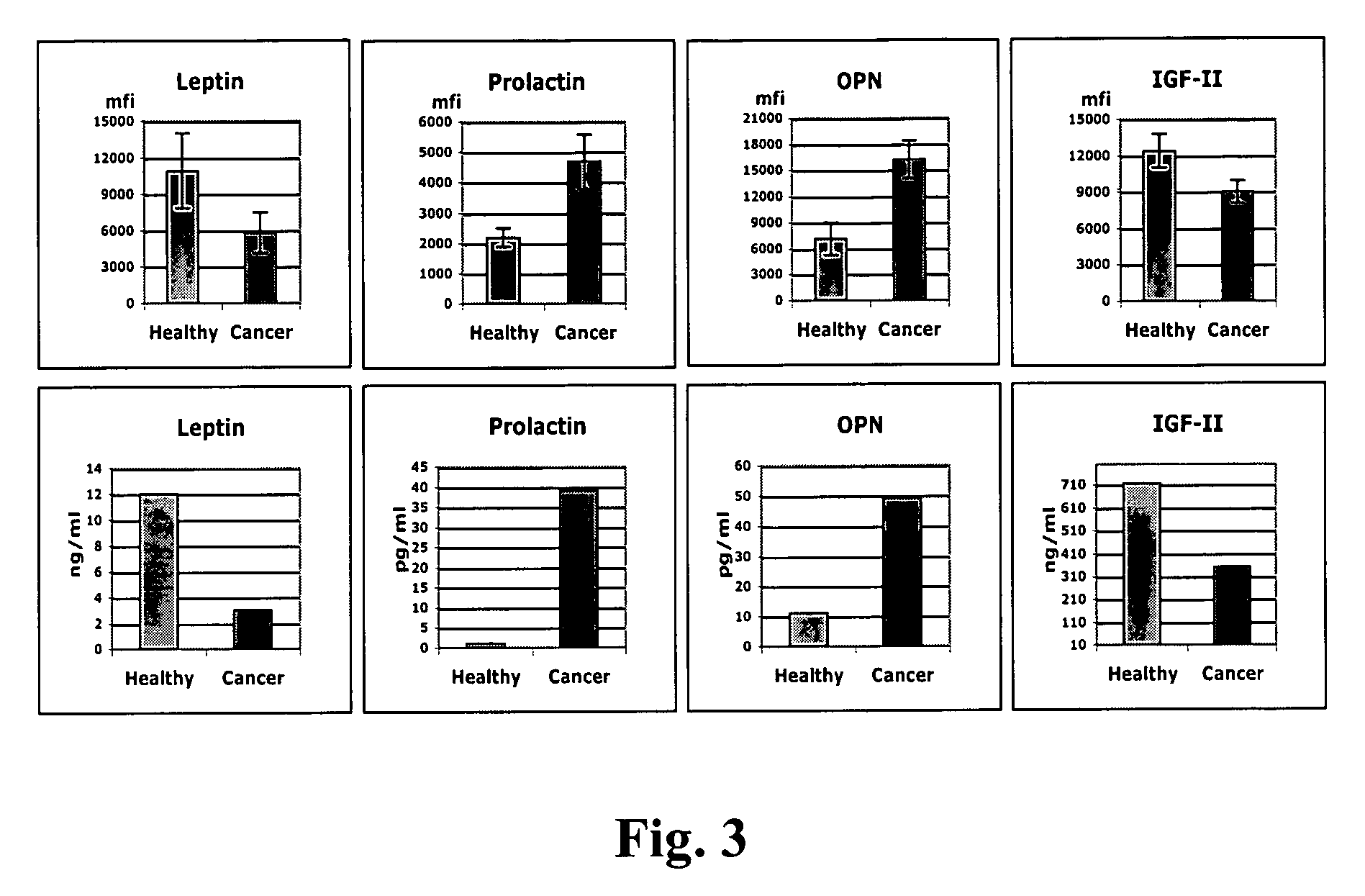
Identification of cancer protein biomarkers using proteomic techniques
The claimed invention describes methods to diagnose or aid in the diagnosis of cancer. The claimed methods are based on the identification of biomarkers which are particularly well suited to discriminate between cancer subjects and healthy subjects. These biomarkers were identified using a unique and novel screening method described herein. The biomarkers identified herein can also be used in the prognosis and monitoring of cancer. The invention comprises the use of leptin, prolactin, OPN and IGF-II for diagnosing, prognosis and monitoring of ovarian cancer.
Owner:YALE UNIV
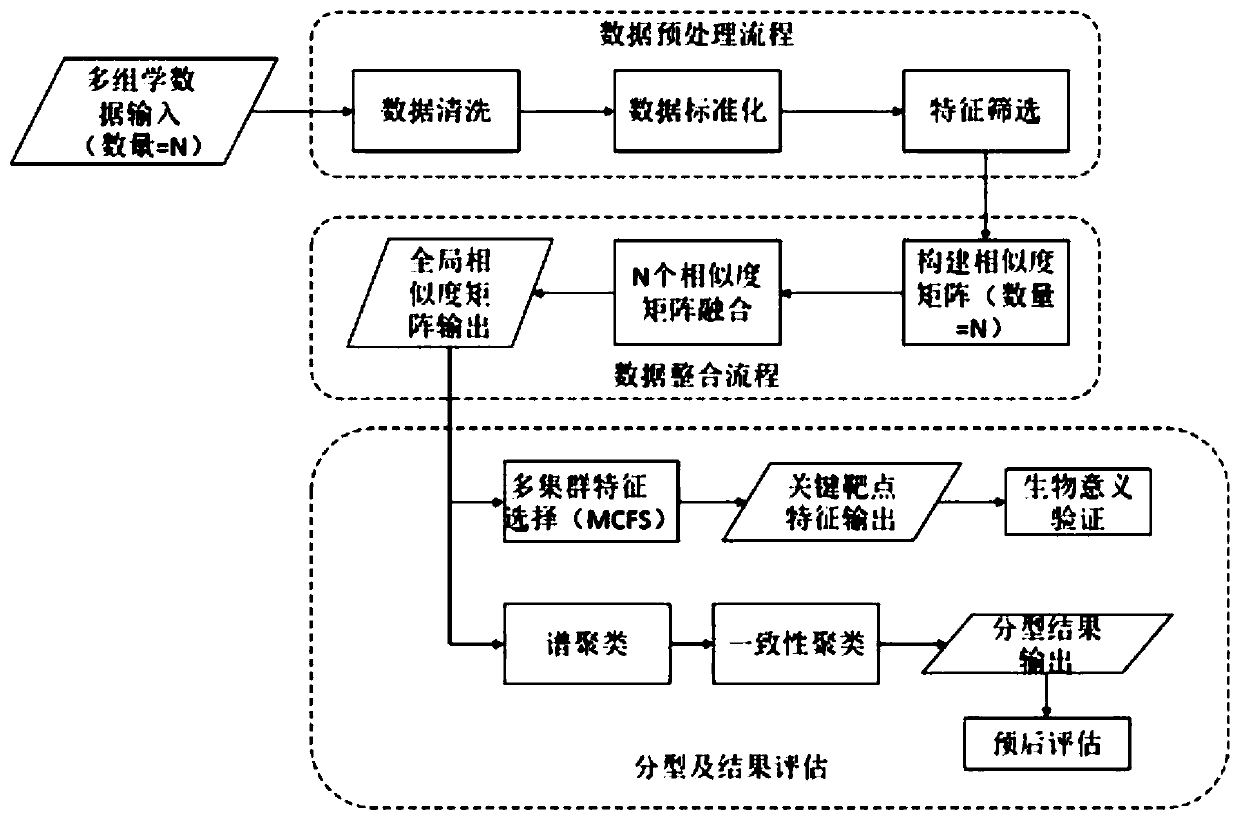
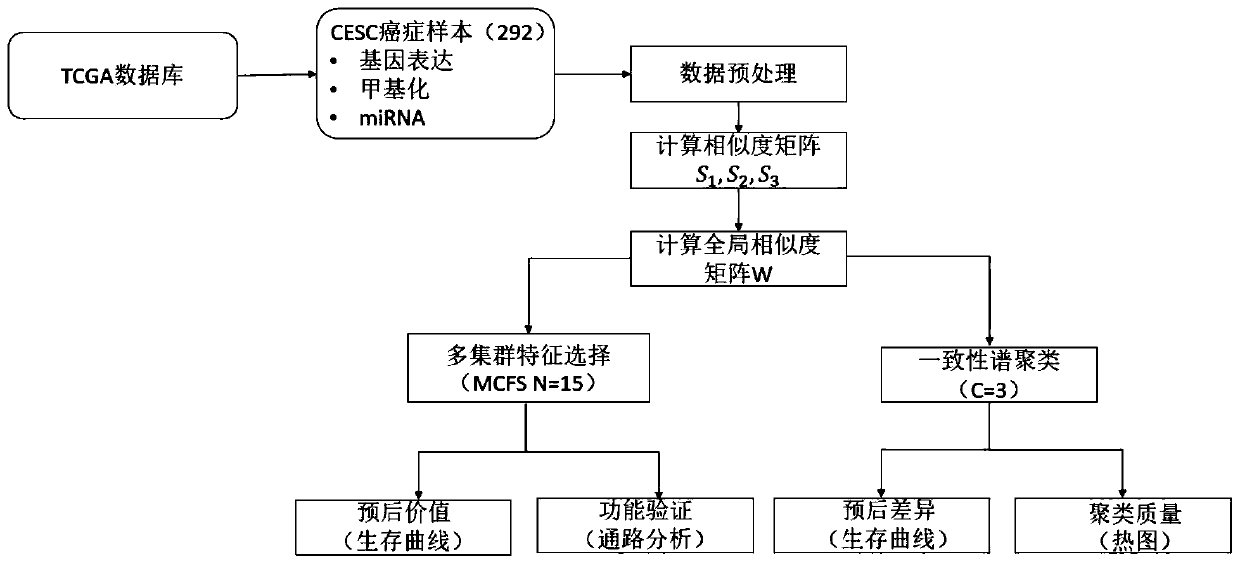
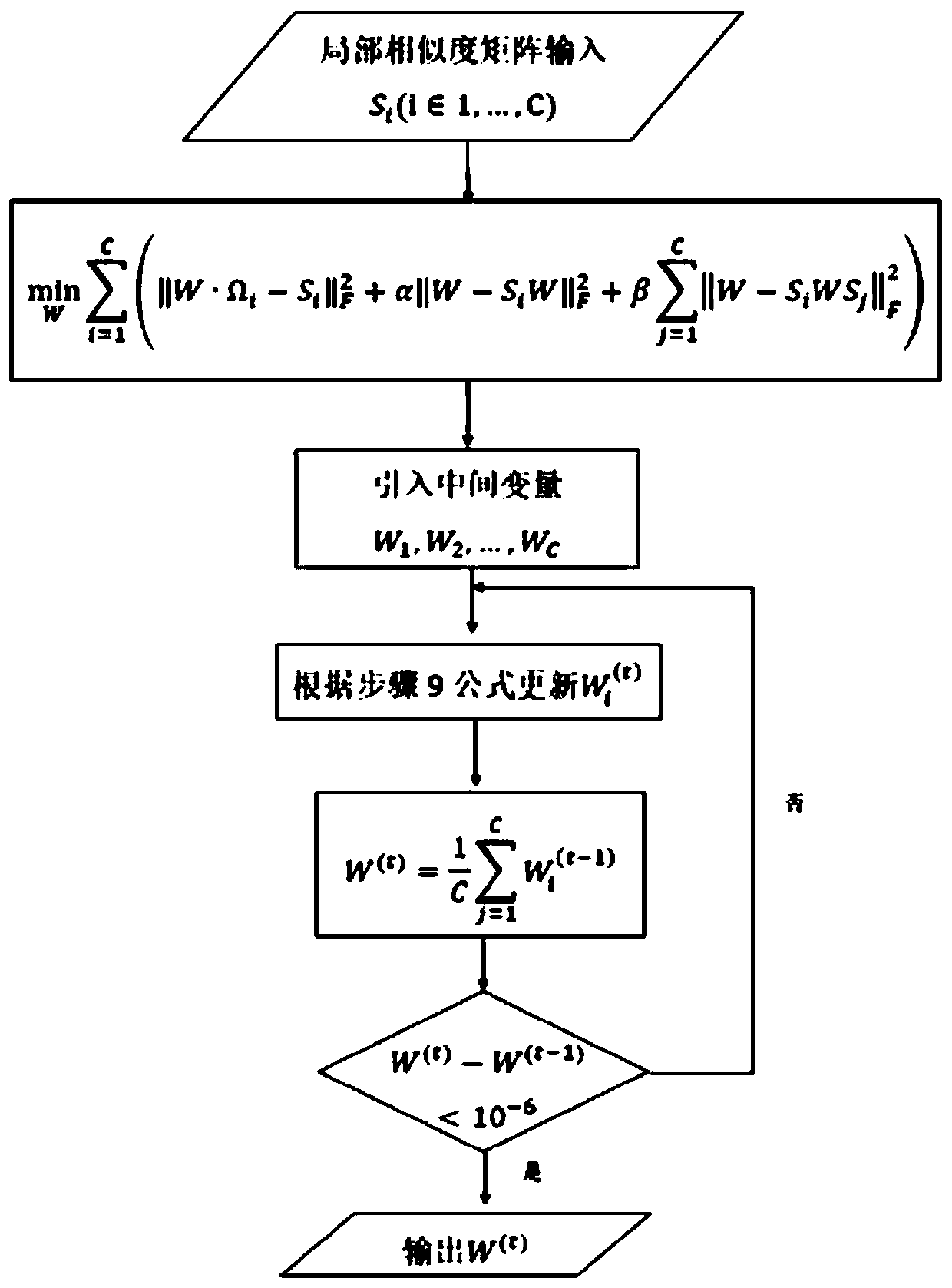
Multi-omics cancer data integrating and analyzing method based on similarity fusion
ActiveCN109994200AHigh precisionImprove stabilityMedical automated diagnosisProteomicsPattern recognitionMulti omics
The invention discloses a multi-omics cancer data integrating and analyzing method based on similarity fusion. The method comprises the steps of calculating a local similarity network, fusing multiplelocal similarity networks, typing according to a global similarity network, and backtracking the characteristic in an original data source according to the global similarity network. Compared with the prior art, the method is advantageous in that through modeling a similarity network connecting path which gradually advances, a fusion algorithm of multiple similarity networks is realized; a more complicated network structure can be described; and higher accuracy and higher stability are realized. Through a consistent alternating multiplier method, quick solving of a network fusion model is realized. The multi-omics cancer data integrating and analyzing method has advantages of utilizing the integrated global similarity network to typing of a cancer patient, obtaining types of the patientswith substantial prognosis difference, and combining with a multi-group characteristic selecting method for screening the key target characteristic. The selected characteristic has a potential of becoming a latent biological marker.
Owner:SOUTH CHINA UNIV OF TECH
Method for establishing prediction model of complex data
The invention provides a method for establishing a prediction model of complex data comprising following steps: A. obtaining high dimension omics data HDOD and determining a group of data objects having representativeness for the HDOD as examples; B. determining the similarity measurement between each data object in the HDOD and each example and accordingly establishing a similarity measurement matrix of the data objects and examples; C. selecting examples containing information from the examples through the penalized likelihood method by means of the similarity measurement matrix of the data objects and the examples; D. establishing a prediction model based on the selected examples. The method of the invention provides a natural quantification tool for discovering and verifying interaction among complex variables. The prediction model of the invention is suitable for large-scale database through searching based on similarity.
Owner:赵乐平
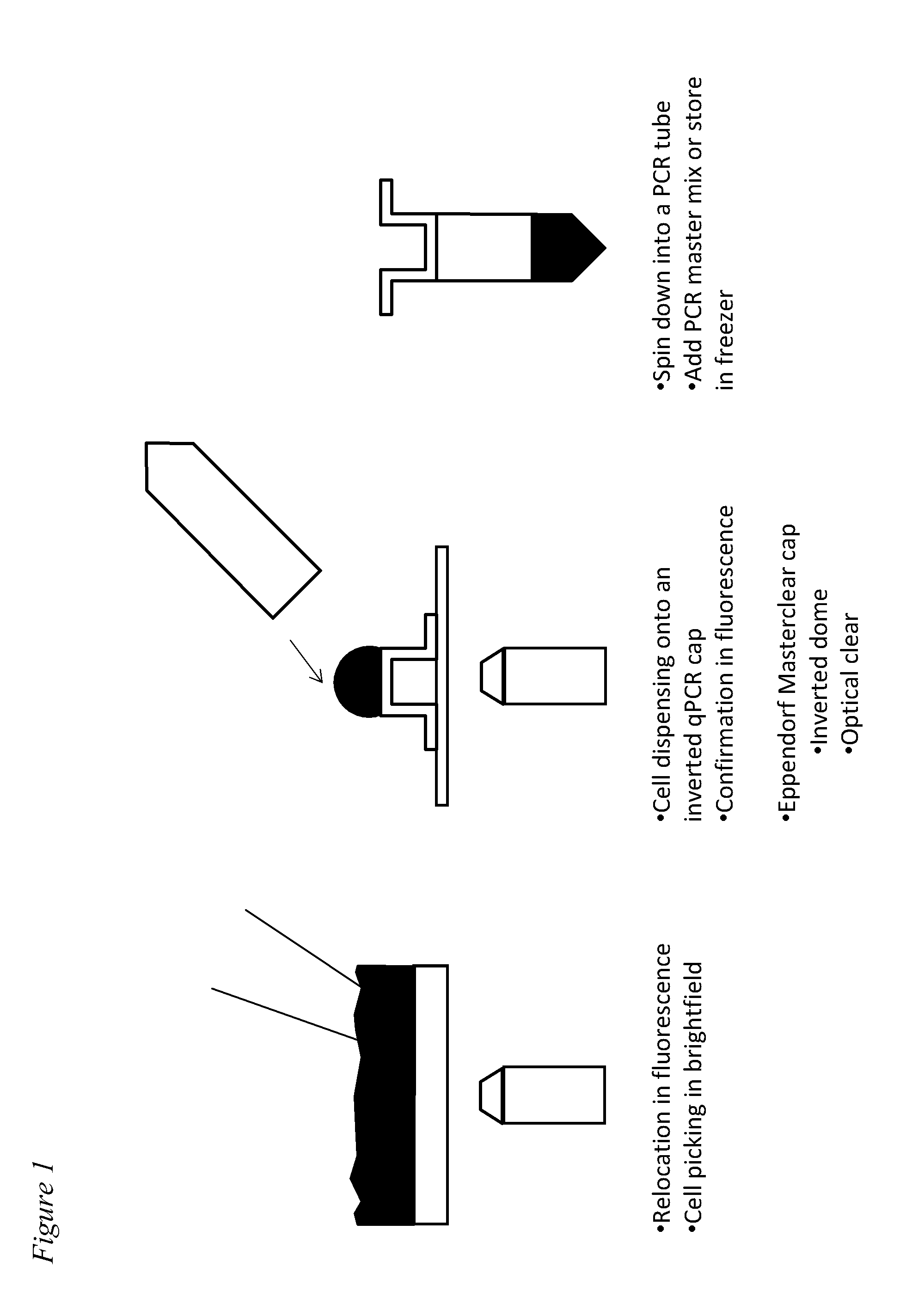
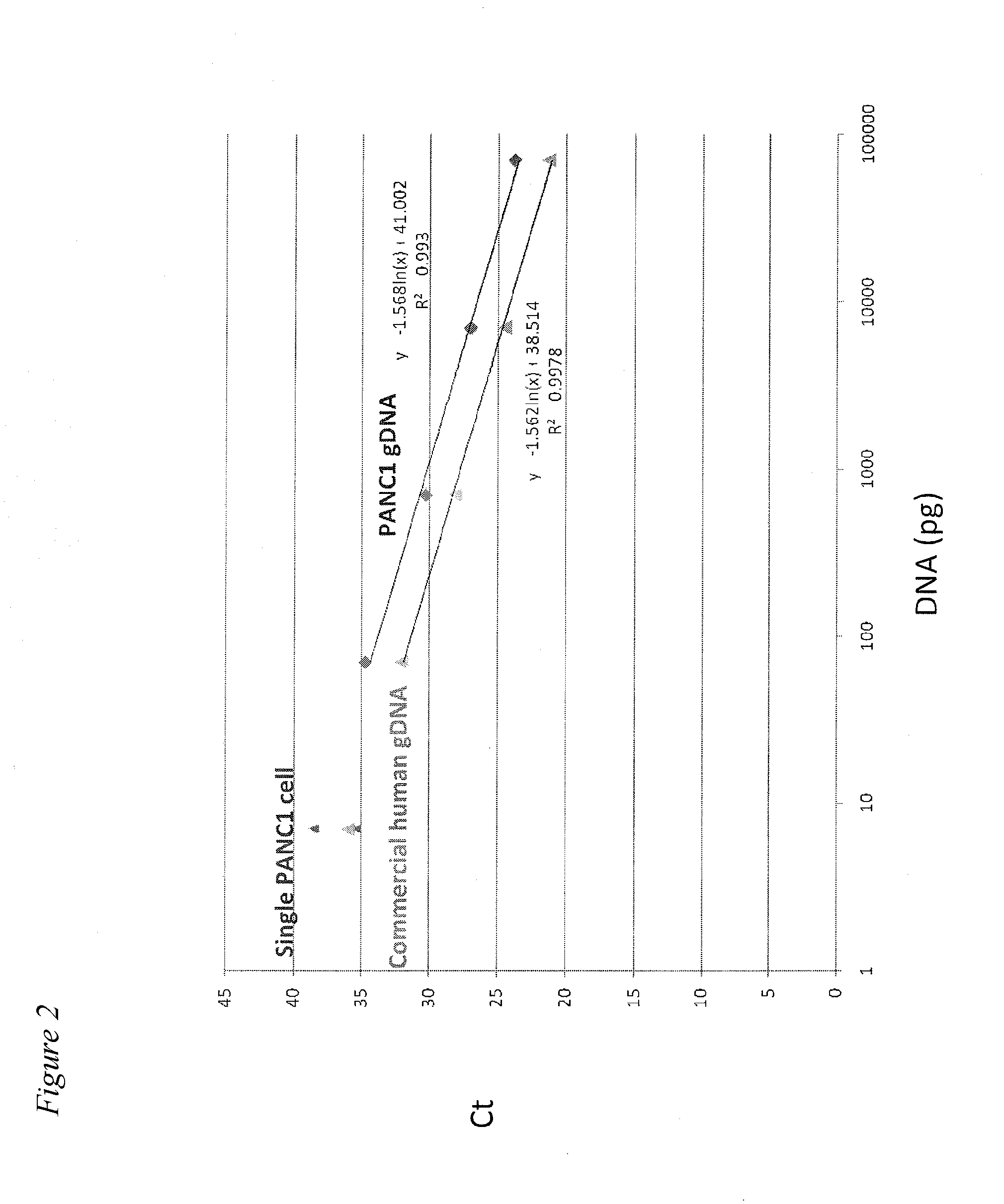
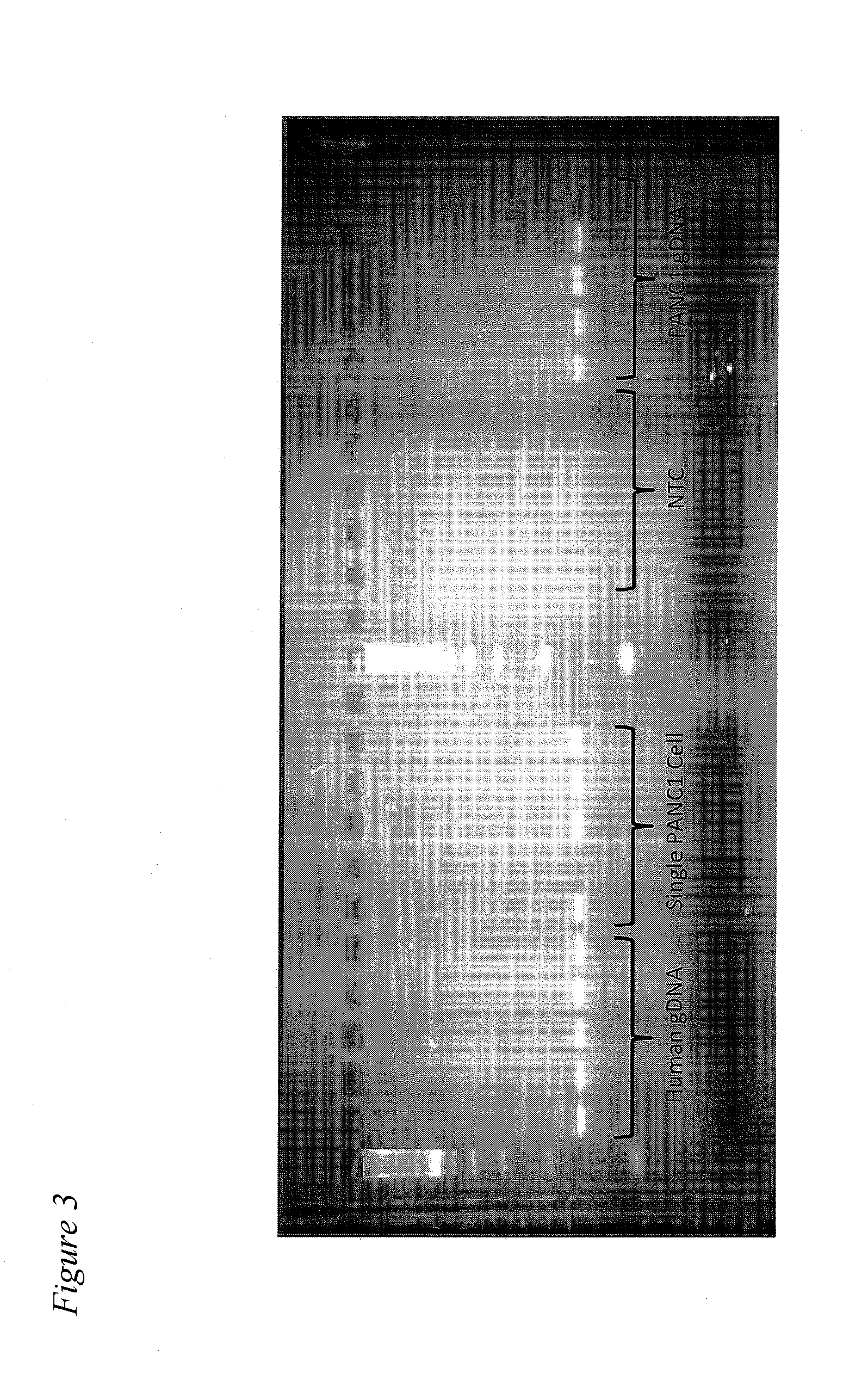
Methods for obtaining single cells and applications of single cell omics
InactiveUS20140308669A1Microbiological testing/measurementMaterial analysisCirculating cancer cellTumor cells
The present application provides methods for obtaining single cells from a sample. Methods for isolating and analyzing molecular features obtained from a single cell are also disclosed herein. For example, individual circulating tumor cells (CTCs) from a sample such as a patient's blood sample can be identified and obtained using methods disclosed herein, and picked for further analysis.
Owner:THE SCRIPPS RES INST
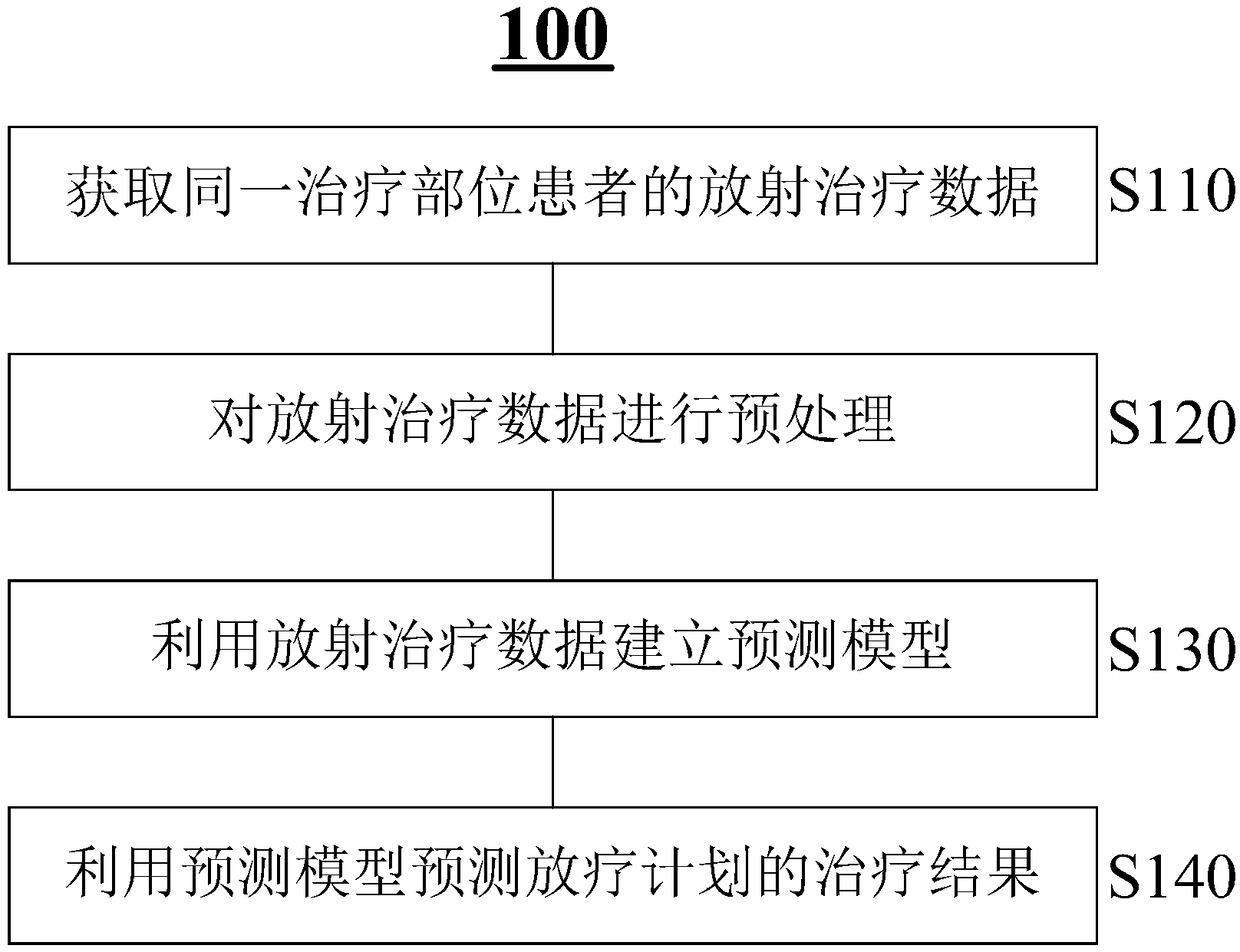
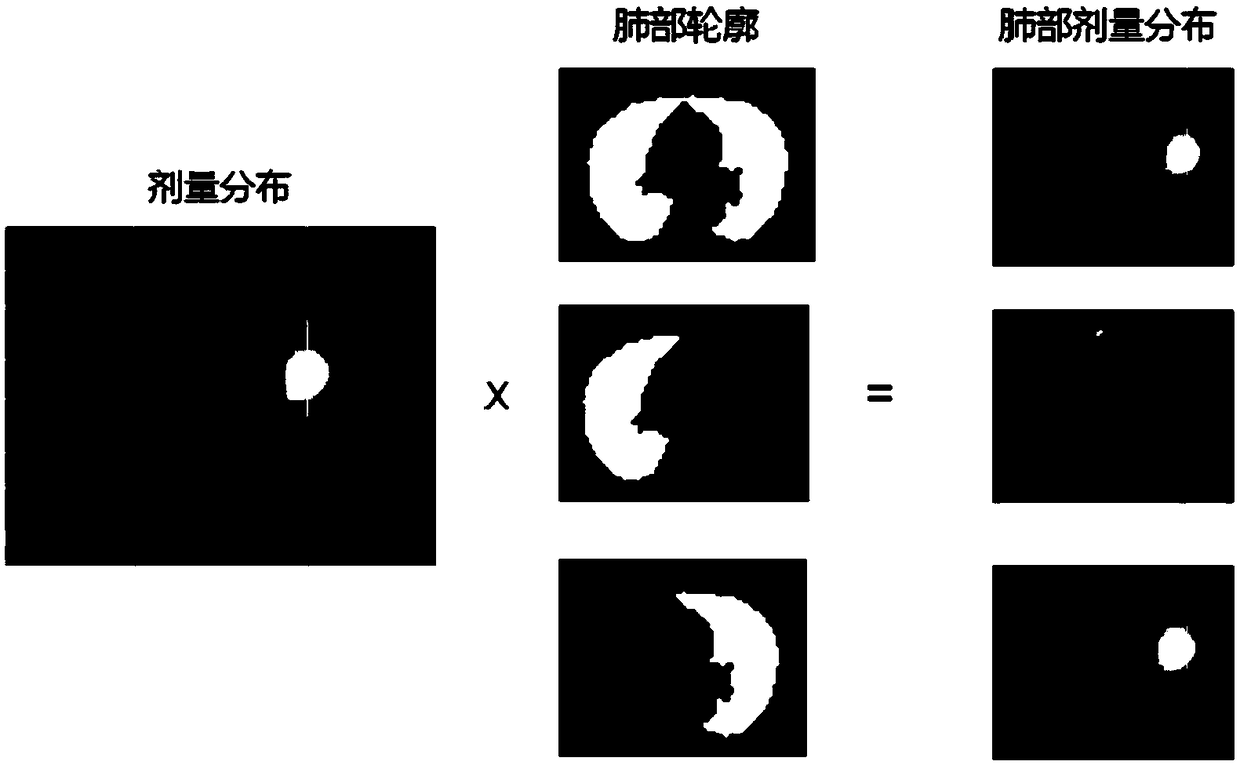
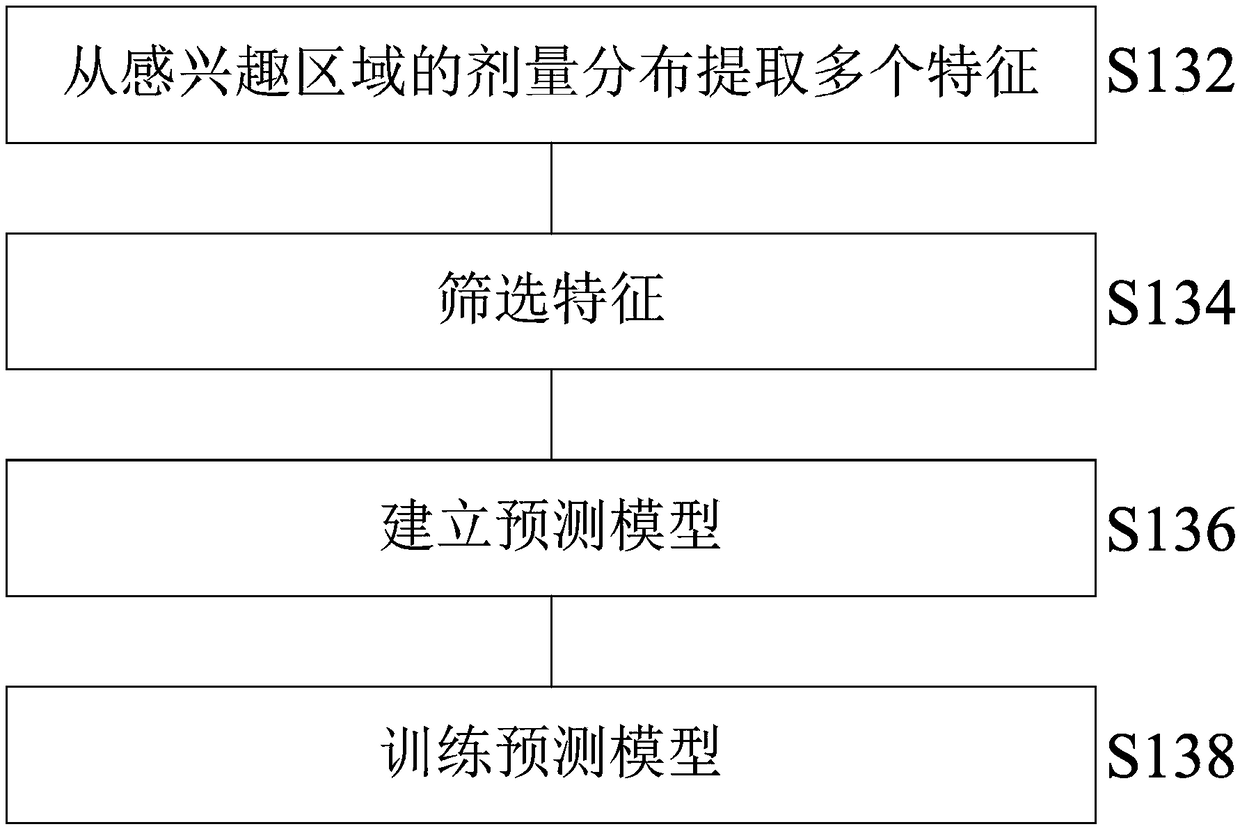
Method and system for predicting radiotherapy results based on dose omics
InactiveCN108766563ARealize personalized customizationImprove accuracyMechanical/radiation/invasive therapiesMedical automated diagnosisTreatment effectProgram planning
The present invention relates to a method and a system for predicting radiotherapy results based on dose omics. According to an embodiment, a method for predicting radiotherapy results may include: acquiring radiotherapy data of a patient at the same treatment site, wherein the radiotherapy data includes historical radiotherapy plan data and treatment result data; establishing a prediction model using the radiotherapy data; and using the prediction model to predict treatment results of a radiotherapy plan. The method of the invention can more effectively improve the prediction accuracy, not only can more accurately predict the tumor treatment effect, but also can more accurately predict the radiation damage of normal tissues, and thus can be widely applied to various radiotherapy situations.
Owner:戴建荣 +1
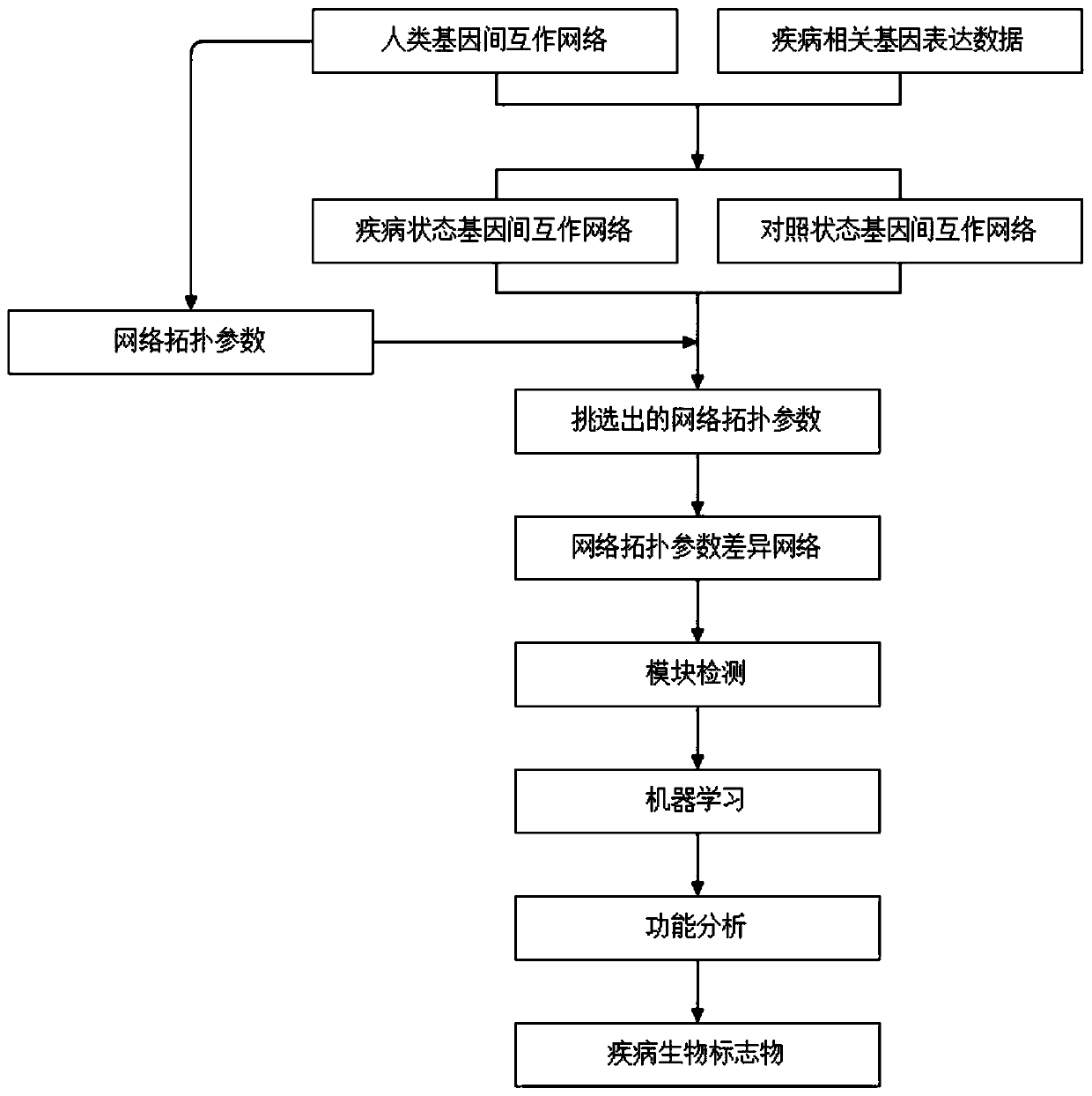
Cancer biomolecule marker screening method and system based on network topology parameters
ActiveCN110444248ACharacter and pattern recognitionProteomicsBiomarker identificationScreening method
The invention discloses a cancer biomolecule marker screening method and system based on network topology parameters. The method comprises the steps that a human gene interaction network and gene chipexpression data are acquired and integrated to obtain a gene interaction network based on gene expression data; a disease state and control state gene interaction network is constructed; network topology parameter difference genes of the disease state and control state gene interaction network are calculated, and a network topology parameter difference change network is obtained based on the network parameter difference genes; network module mining is performed on the network topology parameter difference network; feature selection is performed on an obtained difference network module to obtain genes capable of distinguishing normality and diseases in all modules; the classification effect of the genes selected from the modules on the diseases is detected, and the difference network module is screened according to the classification effect to serve as a biomolecule marker candidate. The invention provides a novel complex disease biomarker identification method based on omics data, andexperiments prove that the method has certain accuracy and effectiveness.
Owner:SHANDONG UNIV
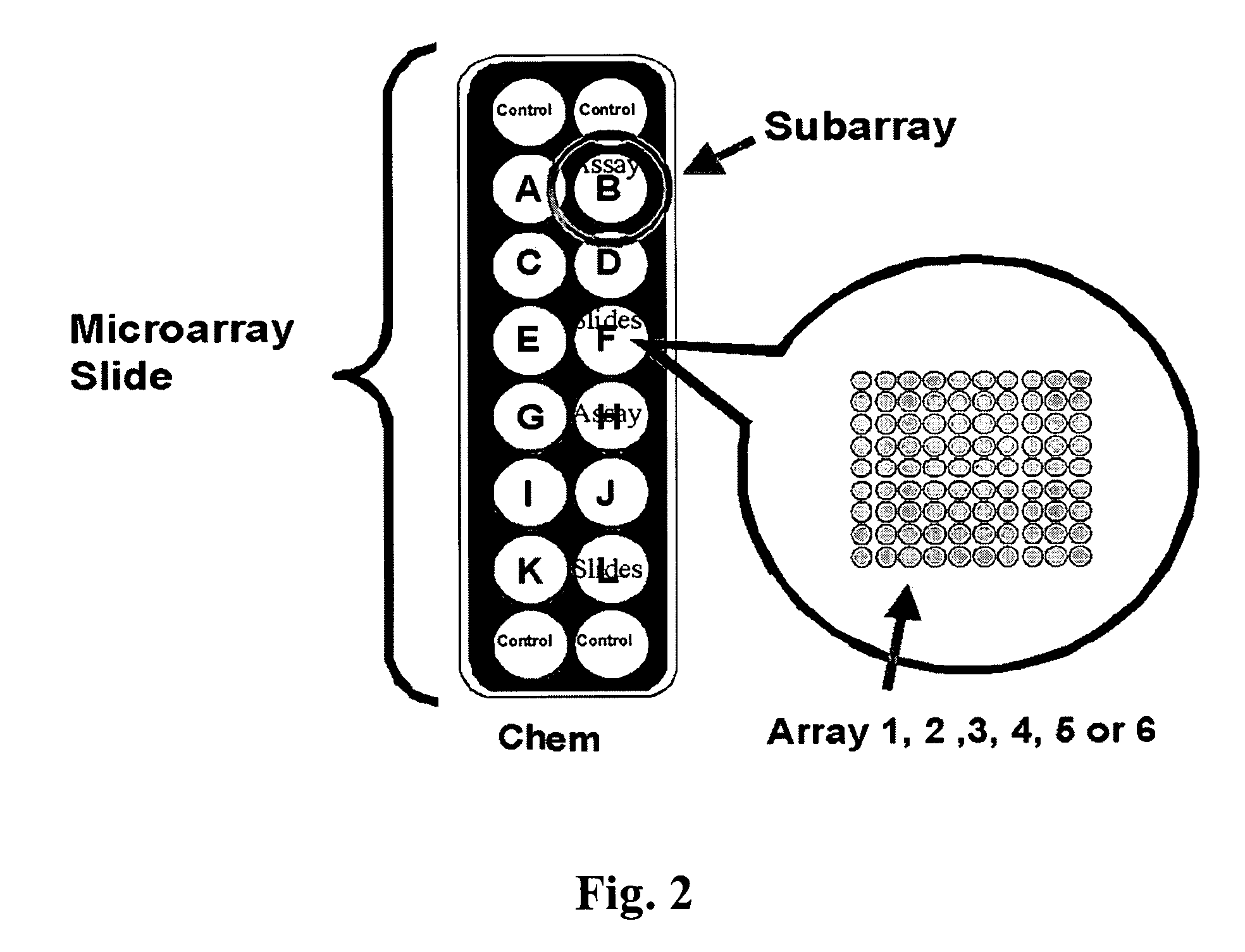
Programmable Arrays
Biomolecule arrays on a substrate are described which contain a plurality of biomolecules, such as coding nucleic acids and / or isolated polypeptides, at a plurality of discrete, isolated, locations. The arrays can be used, for example, in high throughput genomics and proteomics for specific uses including, but not limited molecular diagnostics for early detection, diagnosis, treatment, prognosis, monitoring clinical response, and protein crystallography.
Owner:ARIZONA STATE UNIVERSITY +1
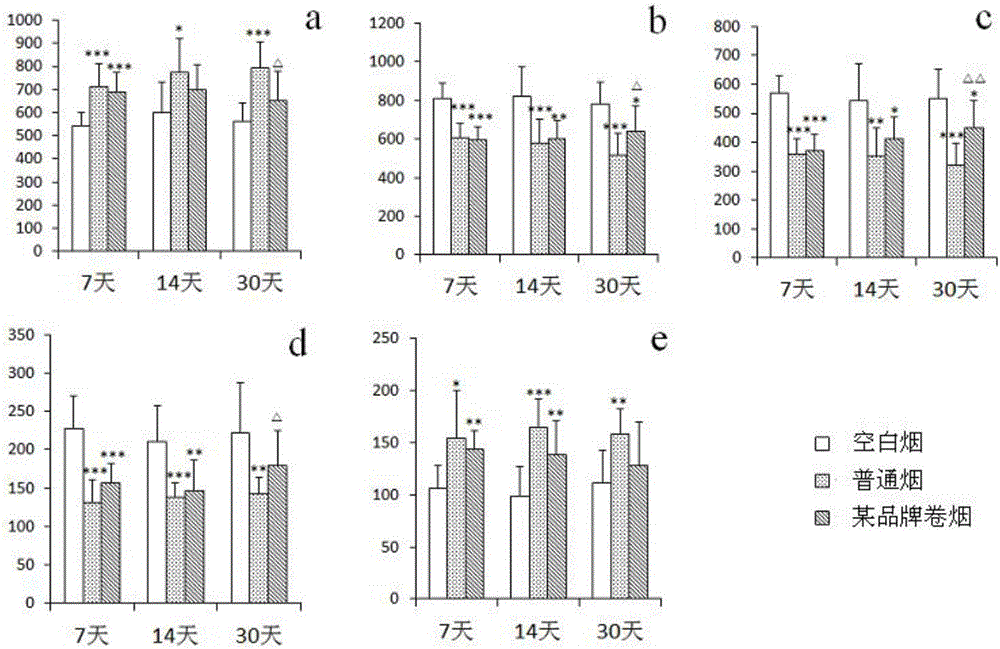
Test method for characterizing smoke exposure biological effect based on metabonomics
InactiveCN106814164ASimple and fast operationEase of evaluationComponent separationAnalysing gaseous mixturesEndogenous metabolismMetabolite
The invention discloses a test method for characterizing the biological effects of smoke exposure based on metabolomics. The test method adopts the metabolomics analysis method, and the biomarker groups related to the biological effects of smoke exposure are tested in different drug administration groups. The changes between groups are used to characterize the biological responses of experimental animals placed in the smoke exposure environment. The present invention is easy to operate, and evaluates the influence of mainstream smoke on endogenous metabolites in rats, and compares the similarities and differences between the effects of natural herbal added tobacco and ordinary cigarettes on endogenous metabolites, in order to evaluate the harm reduction effect of cigarettes and its associated mechanisms provide a reliable tool.
Owner:CHINA TOBACCO JIANGXI IND CO LTD
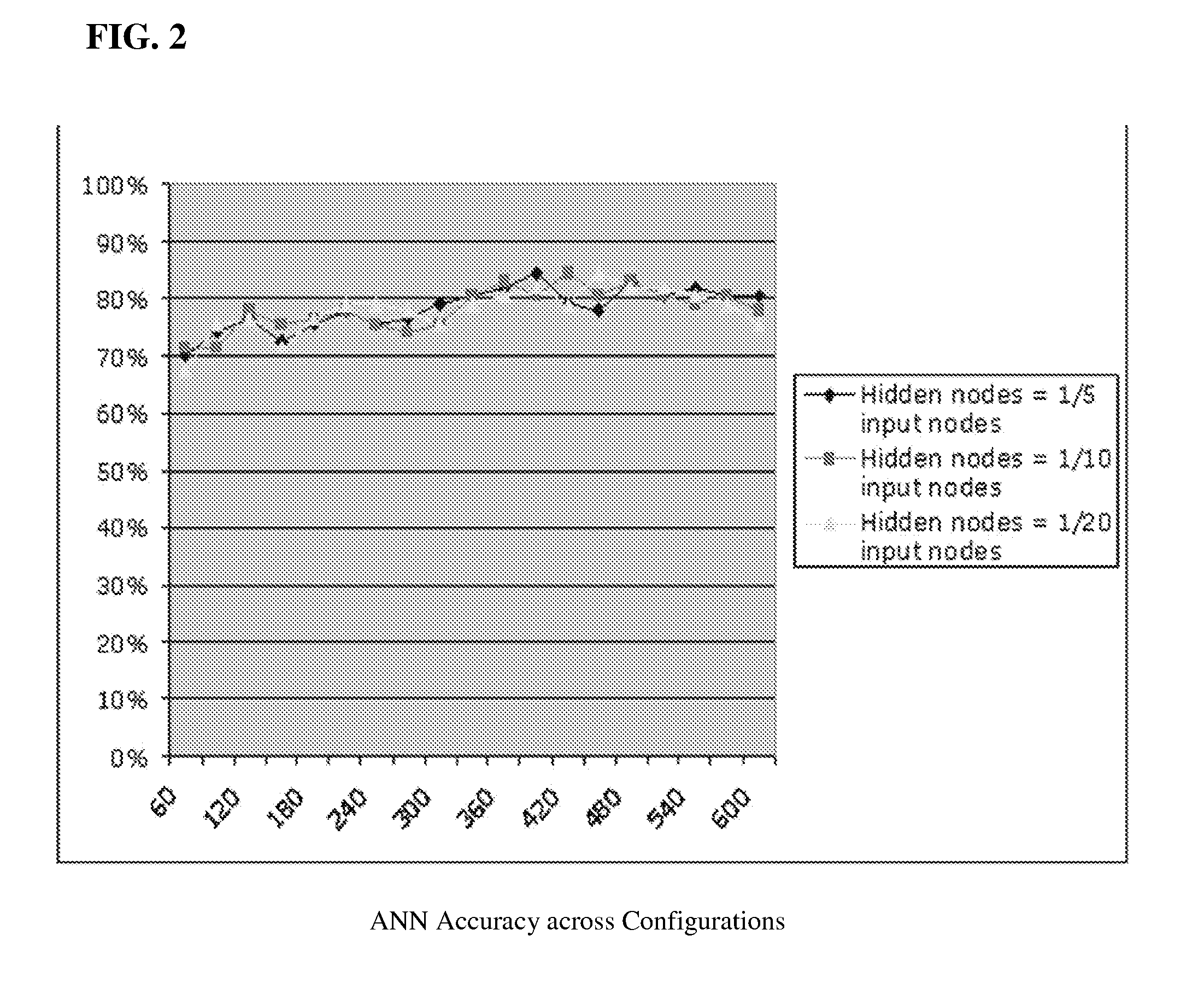
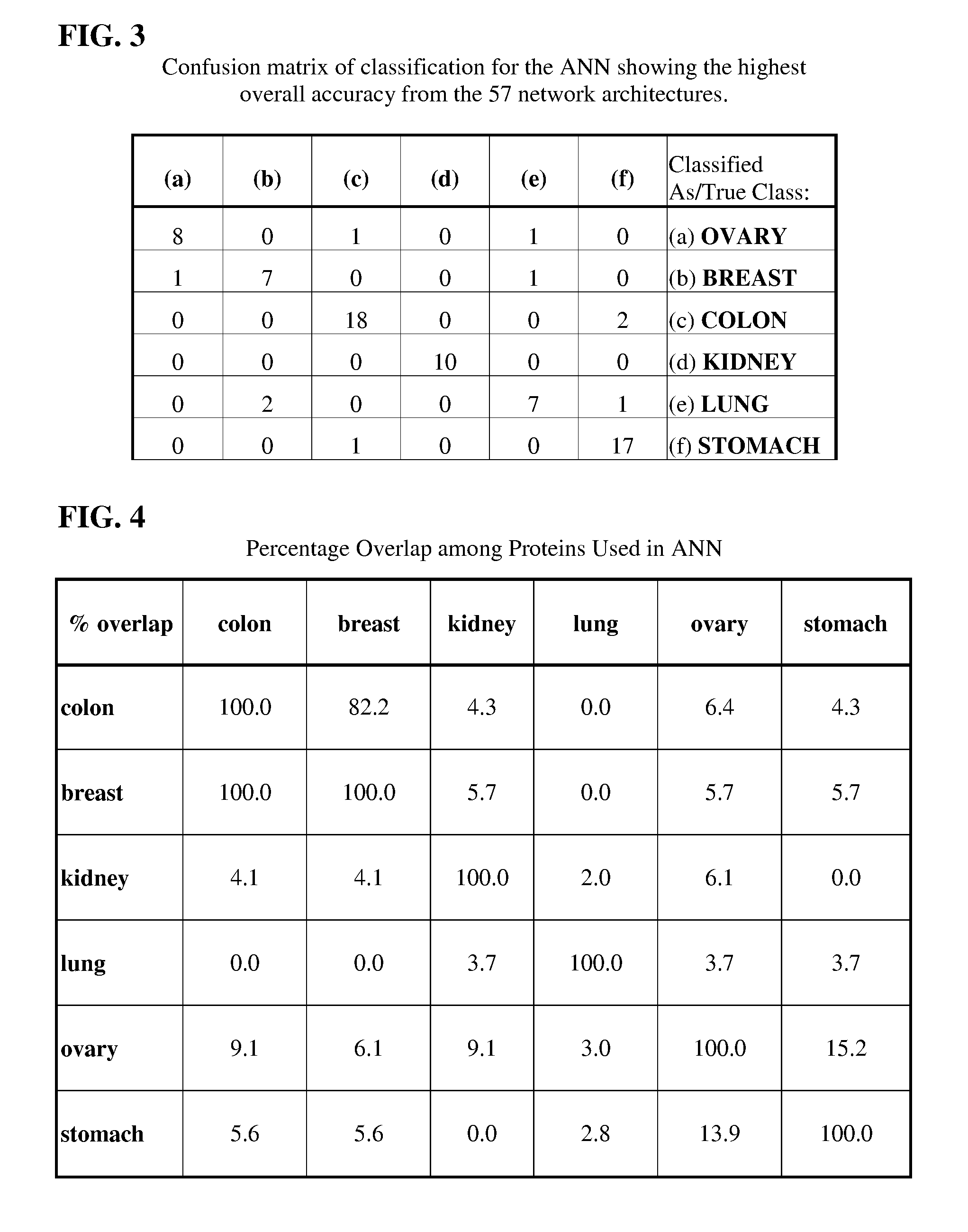
Artificial neural network proteomic tumor classification
Here the inventors describe a tumor classifier based on protein expression. Also disclosed is the use of proteomics to construct a highly accurate artificial neural network (ANN)-based classifier for the detection of an individual tumor type, as well as distinguishing between six common tumor types in an unknown primary diagnosis setting. Discriminating sets of proteins are also identified and are used as biomarkers for six carcinomas. A leave-one-out cross validation (LOOCV) method was used to test the ability of the constructed network to predict the single held out sample from each iteration with a maximum predictive accuracy of 87% and an average predictive accuracy of 82% over the range of proteins chosen for its construction.
Owner:UNIV OF SOUTH FLORIDA +1
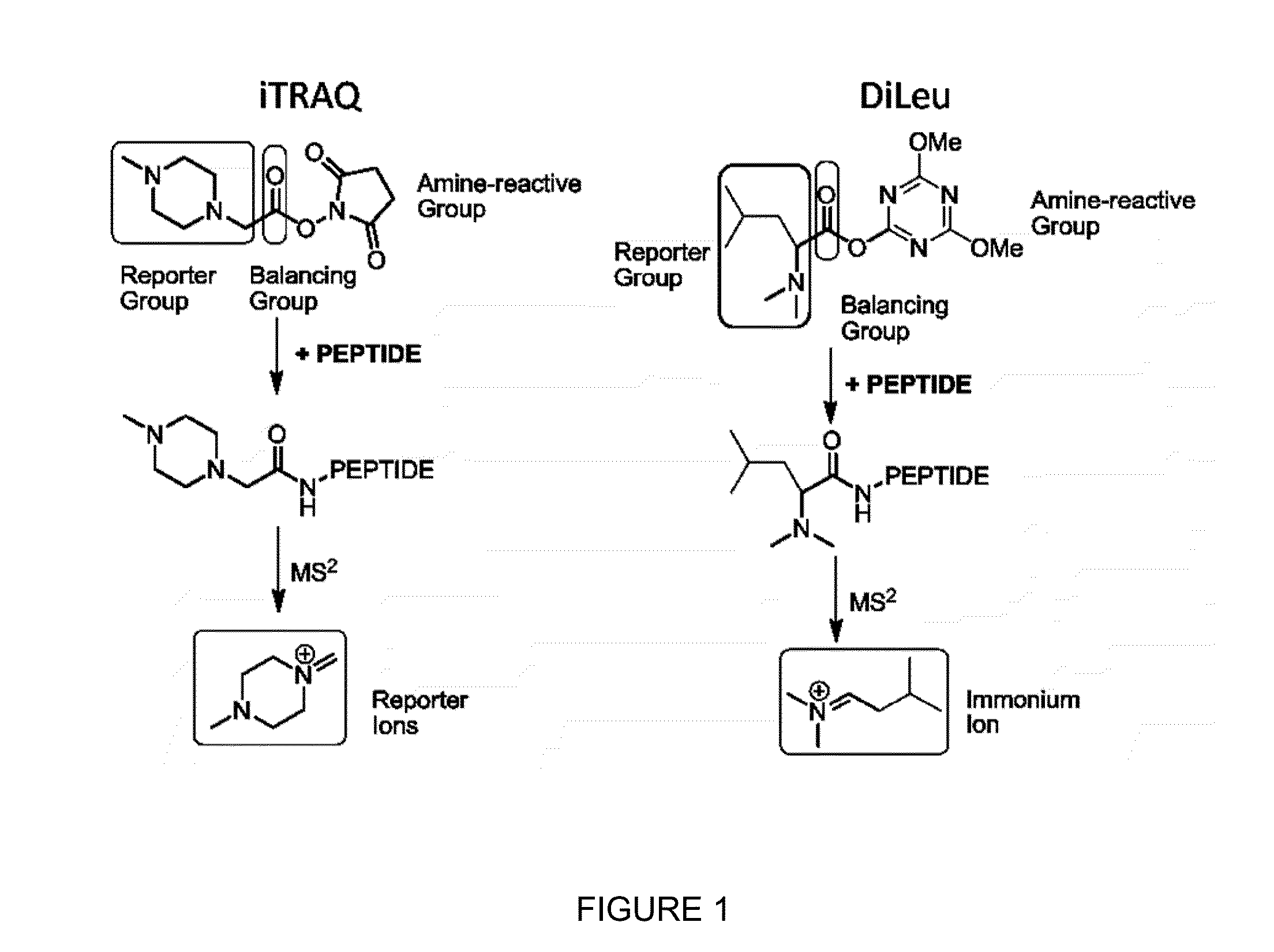
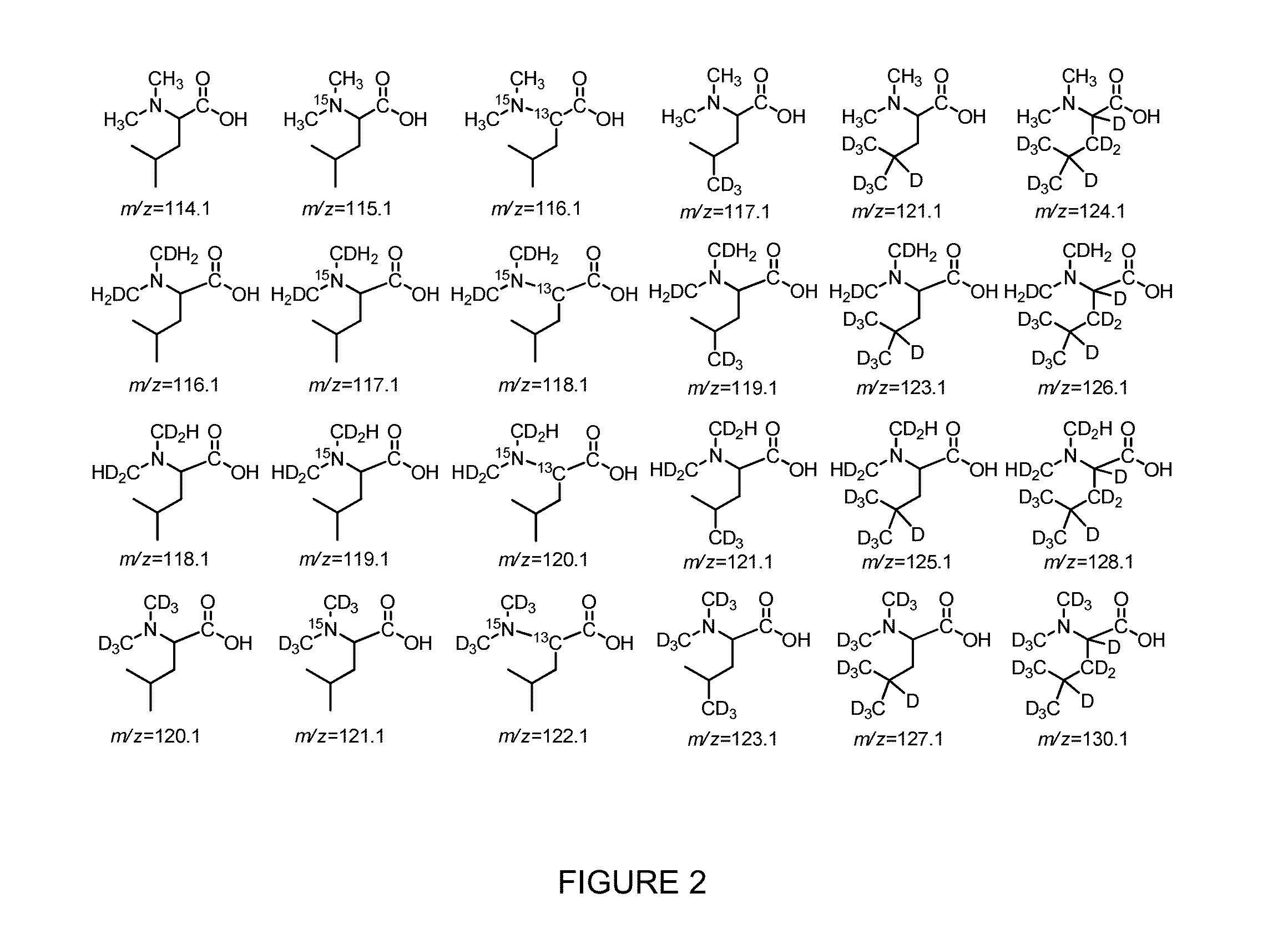
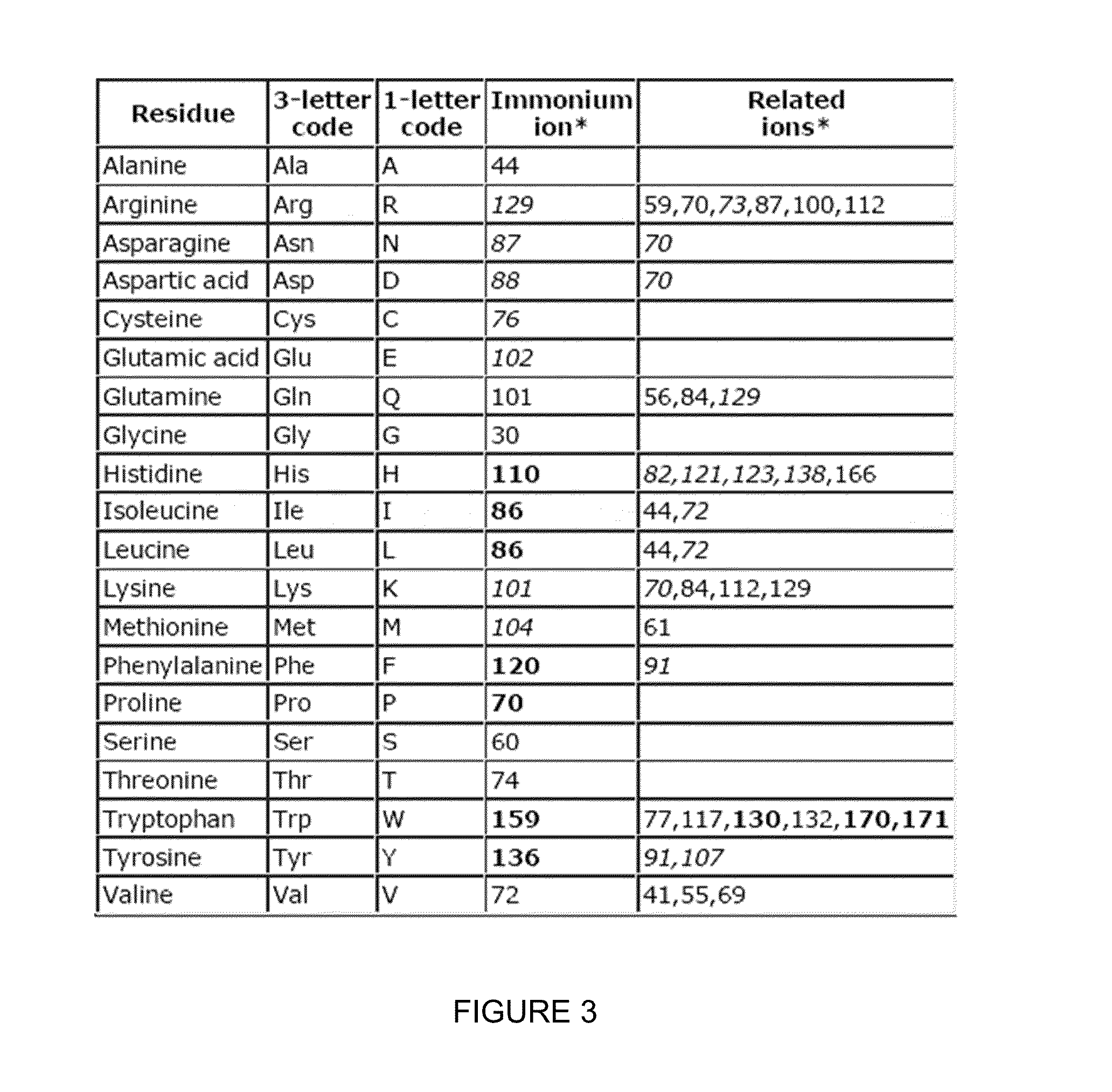
Novel Isobaric Tandem Mass Tags for Quantitative Proteomics and Peptidomics
ActiveUS20130078728A1High quantitation efficacyLow costOrganic chemistryBiological testingCombinatorial chemistryIsotope
Compositions and methods of tagging peptides and other molecules using novel isobaric tandem mass tagging reagents, including novel N,N-dimethylated amino acid 8-plex and 16-plex isobaric tandem mass tagging reagents. The tagging reagents comprise: a) a reporter group having at least one atom that is optionally isotopically labeled; b) a balancing group, also having at least one atom that is optionally isotopically labeled, and c) an amine reactive group. The tagging reagents disclosed herein serve as attractive alternatives for isobaric tag for relative and absolute quantitation (iTRAQ) and tandem mass tags (TMTs) due to their synthetic simplicity, labeling efficiency and improved fragmentation efficiency.
Owner:WISCONSIN ALUMNI RES FOUND
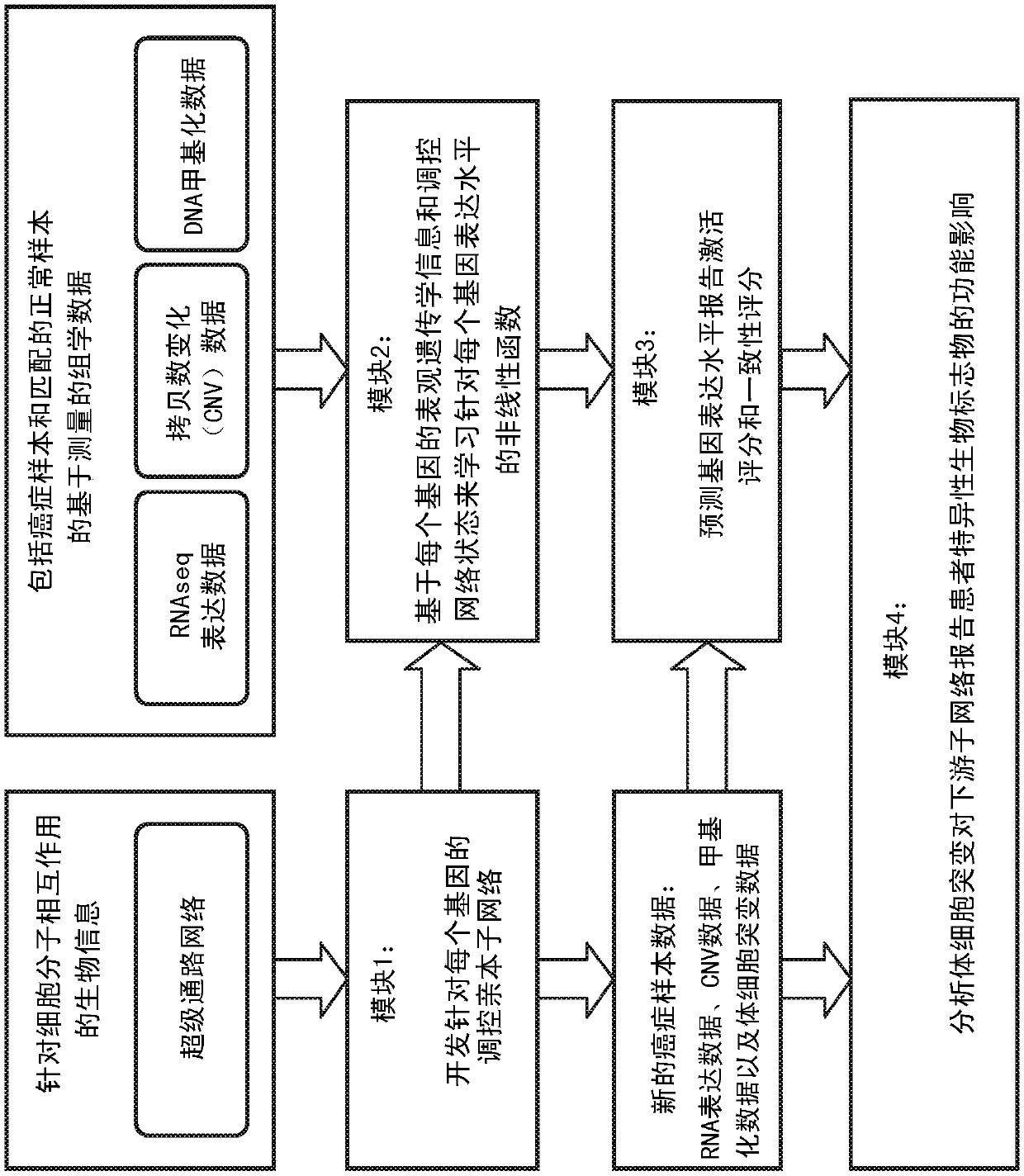
An integrated method and system for identifying functional patient-specific somatic aberations using multi-omic cancer profiles
A system and method for determining the functional impact of somatic mutations and genomic aberrations on downstream cellular processes by integrating multi-omics measurements in cancer samples with community-curated biological pathways are disclosed. The method comprises the steps of extracting biological pathway information from well-curated biological pathway sources, using the pathway information to generate an upstream regulatory parent sub-network tree for each gene of interest, integrating measurement-based omic data for both cancer and normal samples to determine a nonlinear function for each gene expression level based on the gene's epigenetic information and regulatory network status, using the nonlinear function to predict gene expression levels and compare activation and consistency scores with inputted patient- specific gene expression data, and using the patient-specific gene expression predictions to identify significant deviations and inconsistencies in gene expressionlevels from expected levels in individual patient samples to identify potential biomarkers in providing predictive information in relation to cancer and cancer treatment.
Owner:KONINKLJIJKE PHILIPS NV +1
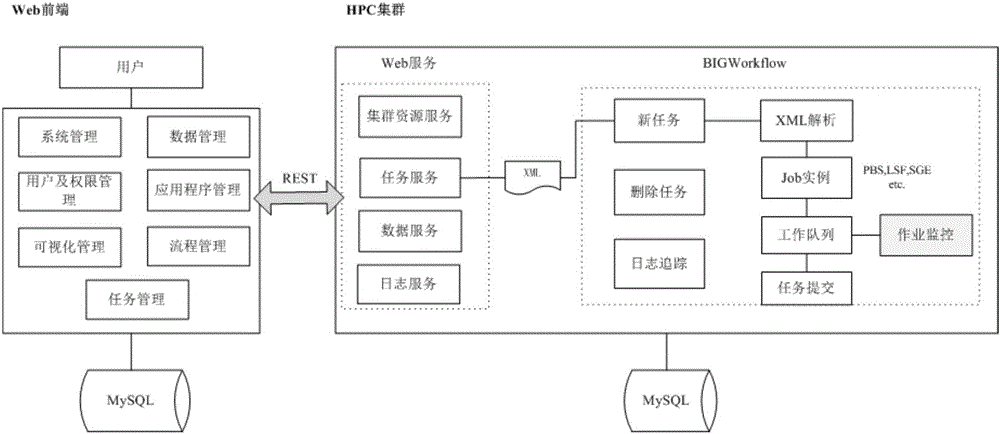
Cloud platform system and method oriented to biological omics big data calculation
InactiveCN106022007AImprove securityEasy to deployTransmissionBioinformaticsSystems managementCluster systems
The invention discloses a cloud platform system and method oriented to biological omics big data calculation, and relates to the technical field of maintenance or management devices. The system comprises a system management module, a data management module, an application management module, a process management module, a task management module, a data visualized operation module and a user and authority management module. The cloud platform system is seamlessly connected with a high-performance calculation cluster system through a distributed type calculation and management mode of the high-performance calculation cluster system, the WEB technology and the computer remote calling, remote controlling and cloud calculating and other technological means, the management and utilization of big data are achieved, and the deep mining, analysis and utilization of biological omics big data by means of online, visual and free customization processes and tools are achieved. By means of the system, the application of the high-performance calculation cluster system in the field of biological omics big data can be promoted, and the deep mining, analysis and industrial application of biological omics big data can also be promoted.
Owner:BEIJING INST OF GENOMICS CHINESE ACAD OF SCI CHINA NAT CENT FOR BIOINFORMATION
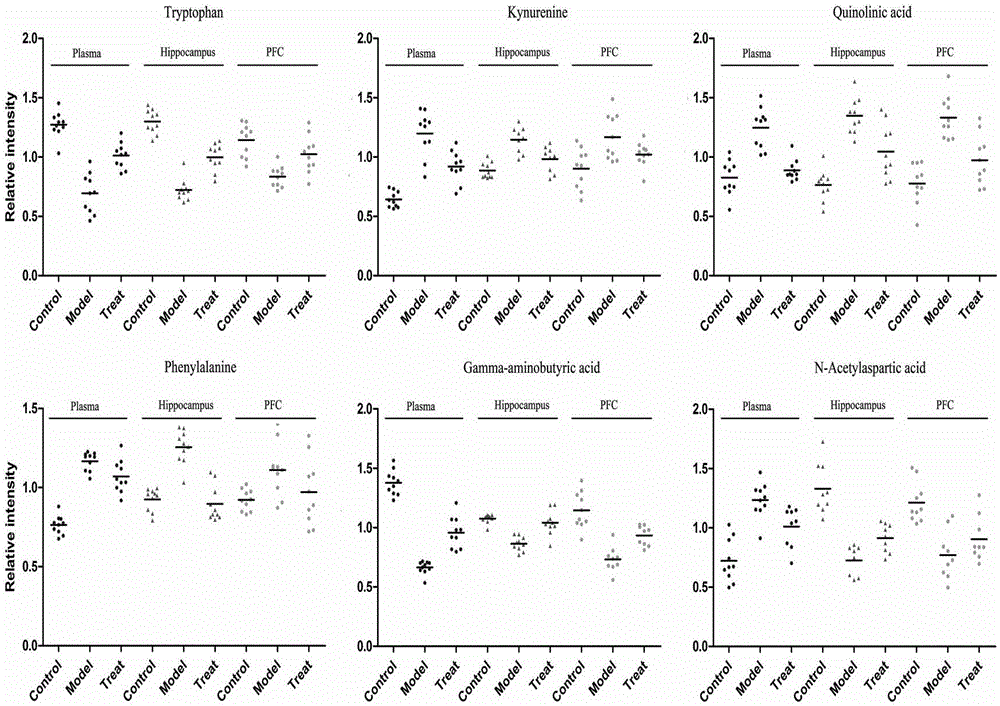
Experimental method for animal model depression degree evaluation based on metabonomics
InactiveCN106770857AAccurate measurementComprehensive reflection of metabolic changesComponent separationMass spectrum analysisBehavioral experiment
The invention discloses an experimental method for animal model depression degree evaluation based on metabonomics. The experimental method comprises the following steps: comprehensively and accurately determining endogenous small molecule metabolites in a biological sample by utilizing a metabonomics platform based on high-resolution mass spectrum, screening different metabolites among different samples and different groups according to the obtained metabolic spectrum data by combining a single-variable multi-variable statistical method, utilizing a common different metabolite among three samples, comparing correlation of compounds in a blood plasma sample and a brain tissue sample, and taking a comparison result as a basis of reflecting change of metabolome in brain tissues by utilizing a blood plasma marker; meanwhile, taking a model group and a control group as a training set to construct a model and an administration group as predication capability of a validation set test model, which is an index for evaluating the depression degree by utilizing the change of the blood plasma metabolome. The experimental method disclosed by the invention can be used for solving the problems that the traditional behavioral experiment has many uncontrollable factors, preparation is relatively complex and animal subjectivity is stronger.
Owner:NANJING MEDICAL UNIV
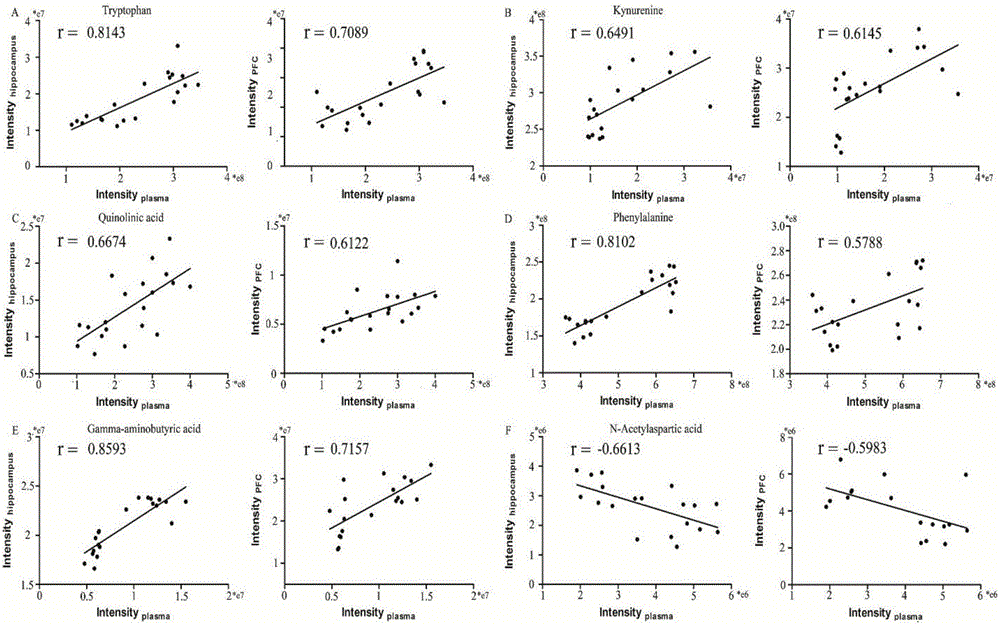
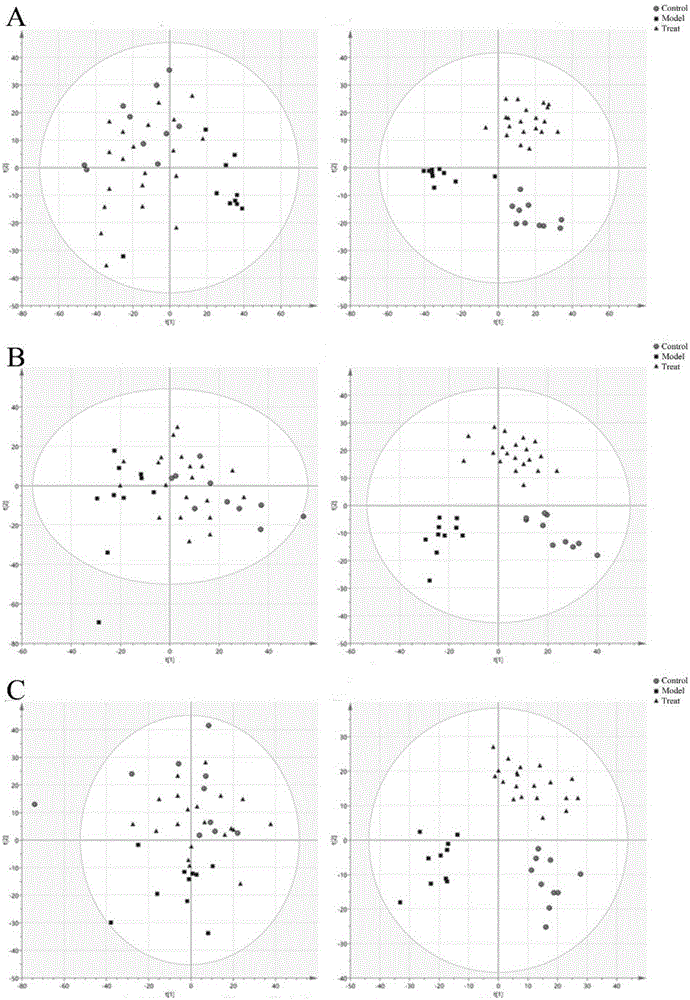
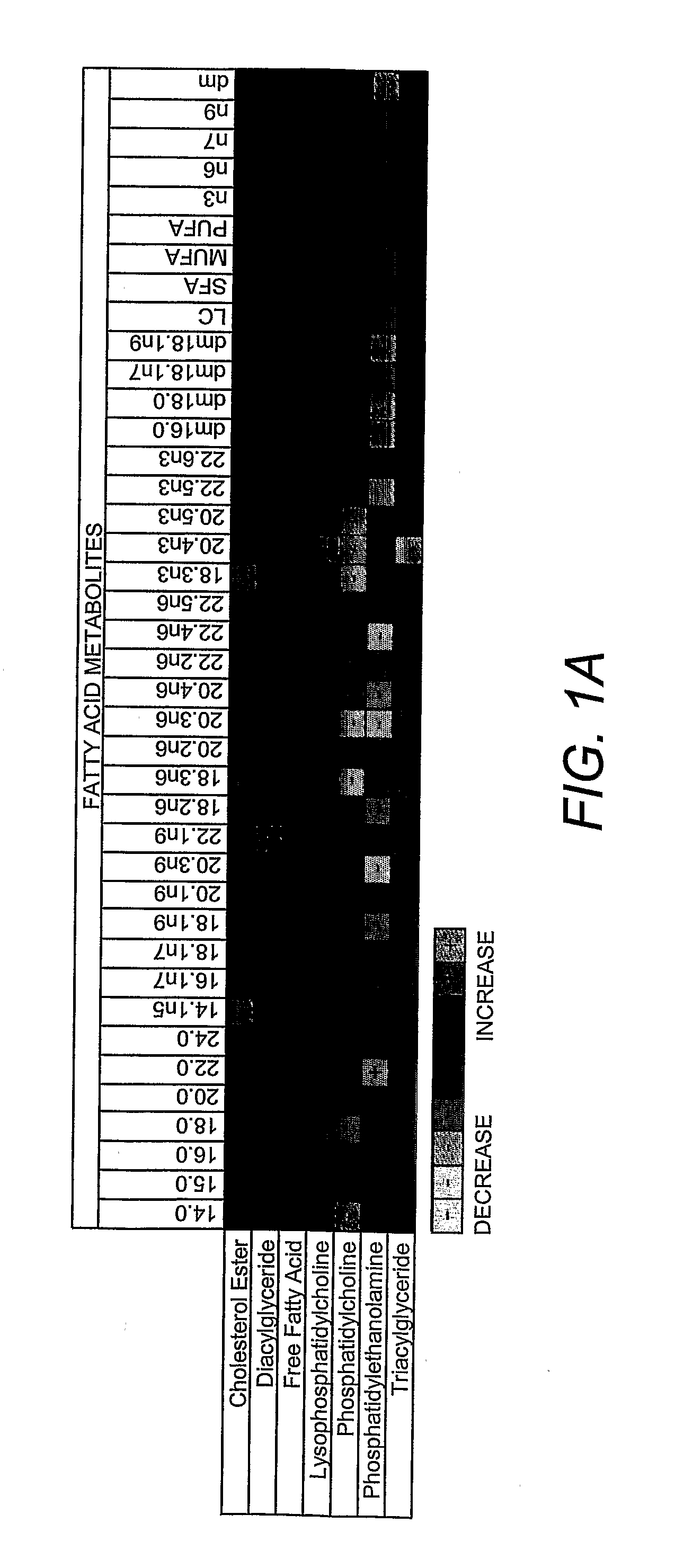
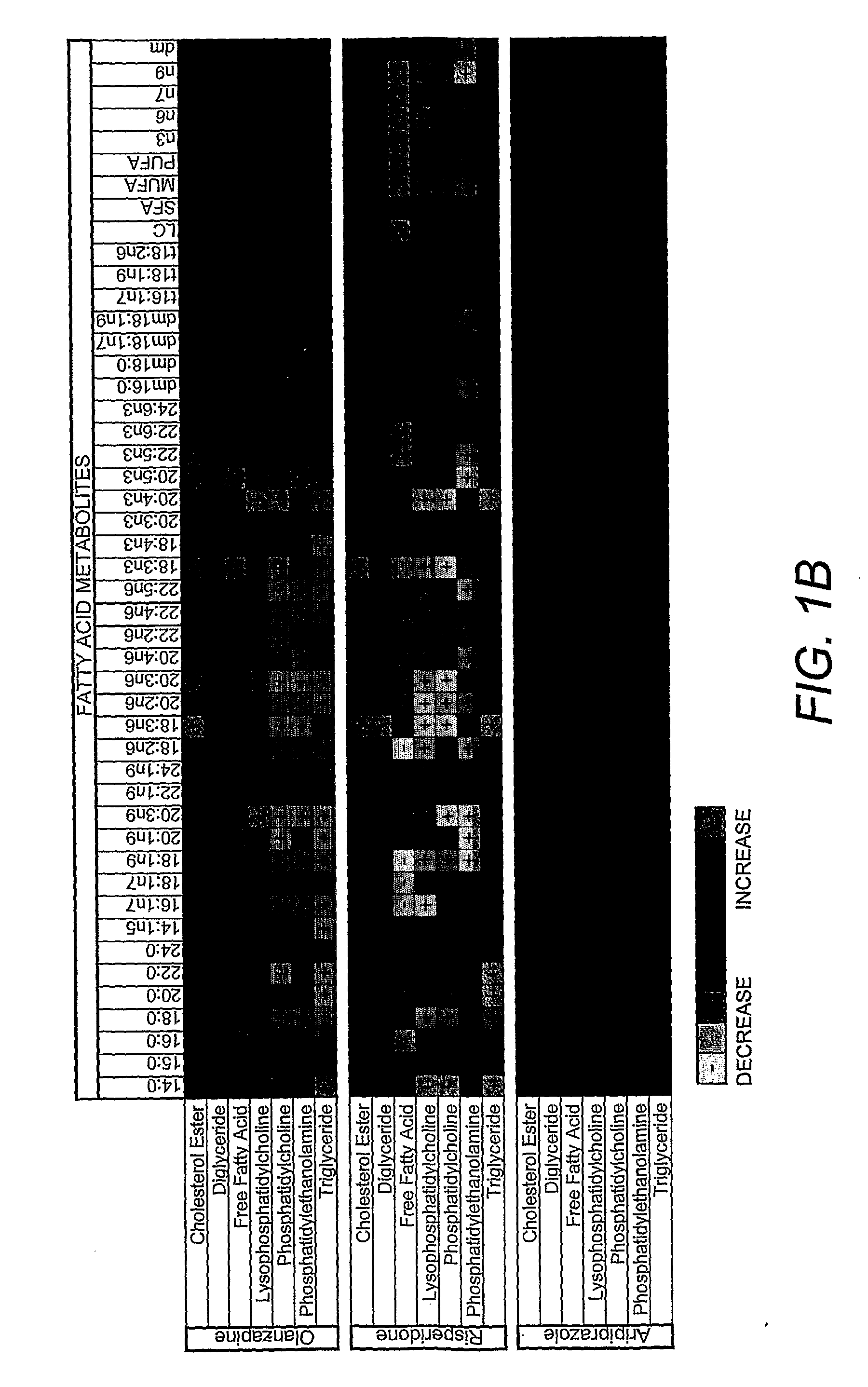
Lipidomics approaches for central nervous system disorders
InactiveUS20090305323A1Less metabolic side effectEasy to determineMicrobiological testing/measurementDisease diagnosisDiseaseLipid formation
The present invention has utilized the power of lipidomics to profile lipid metabolites and to characterize changes in lipid metabolism as they relate to CNS disorders. Lipidomic signatures can guide the development of diagnostic, prognostic and surrogate markers for CNS disorders; identification of new targets for drug design based on highlighted perturbed pathways; stratify patients with CNS disorders as to which pathways are impaired, and facilitate the determination of which patients with CNS disorders are candidates for a particular therapy, i.e. provide the tools for a personalized approach to therapy; identify which patients are responding or are developing side effects to a treatment; design of modified antipsychotics that have less metabolic side effects and enhanced activity; overcome the lag phase in response to some treatments; and find better combination therapies for CNS disorders that target the pathways that are impaired (e.g., impairments in lipid and / or carbohydrate metabolism).
Owner:TRUE HEALTH IP LLC +2
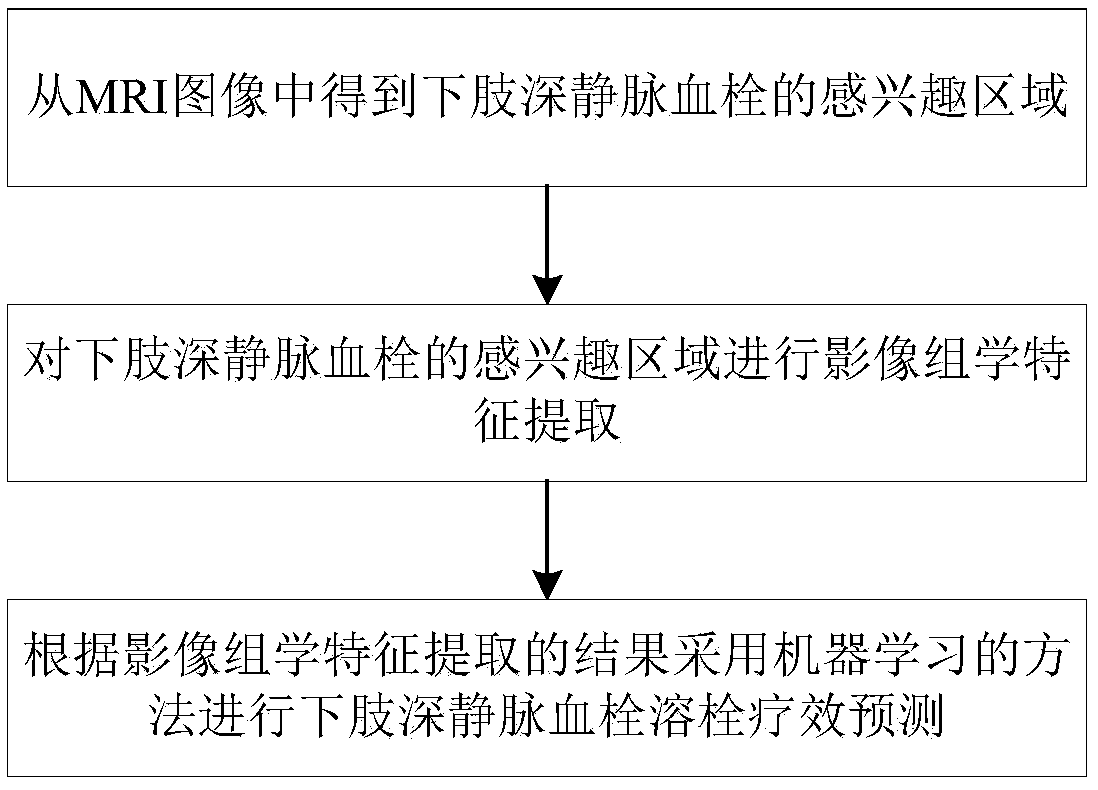
A lower limb deep venous thrombosis thrombolysis curative effect prediction method and system based on machine learning
InactiveCN109598266AThrombolytic efficacy evaluation results are accurateImprove efficiencyMedical referencesRecognition of medical/anatomical patternsPredictive methodsThrombus
The invention discloses a lower limb deep venous thrombosis thrombolysis curative effect prediction method and system based on machine learning. The method comprises the steps: obtaining an area of interest of lower limb deep venous thrombosis from an MRI image; Performing image omics feature extraction on the region of interest of the lower limb deep venous thrombosis; And predicting the curative effect of lower limb deep venous thrombosis thrombolysis by adopting a machine learning method according to an image omics feature extraction result. The invention discloses a lower limb deep venousthrombosis thrombolysis curative effect prediction method and system based on machine learning. lower limb deep venous thrombolysis curative effect prediction is performed through image omics featureextraction and a machine learning method; The MRI imaging omics method and the machine learning technology are combined to predict the thrombolysis curative effect of the deep venous thrombosis of the lower limbs, the curative effect evaluation work can be completed through prediction before thrombolysis treatment, the method does not depend on experience of doctors any more, and the thrombolysiscurative effect prediction result is more accurate and higher in efficiency. The method can be widely applied to the field of medical image processing.
Owner:SHENZHEN UNIV +1
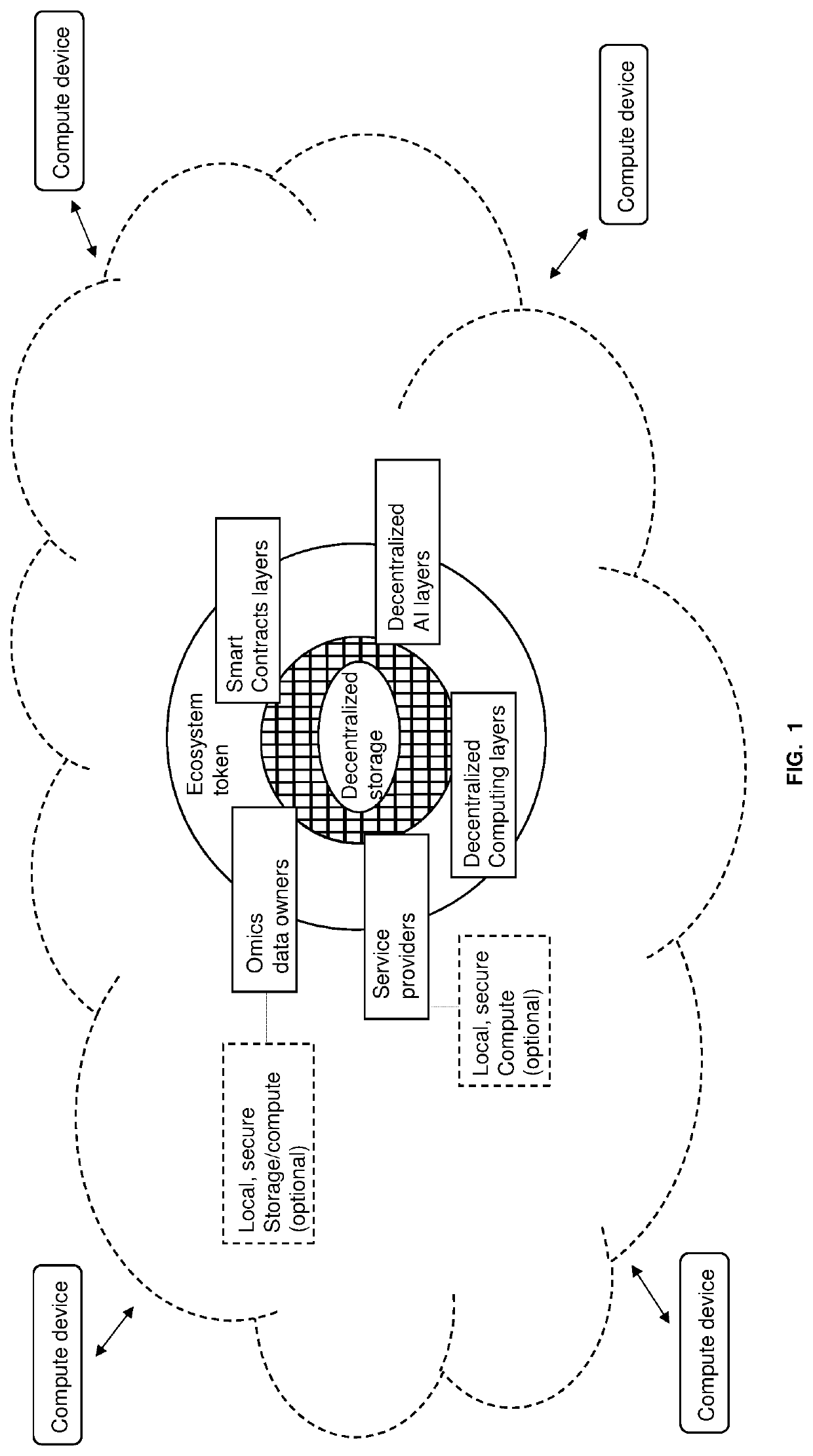
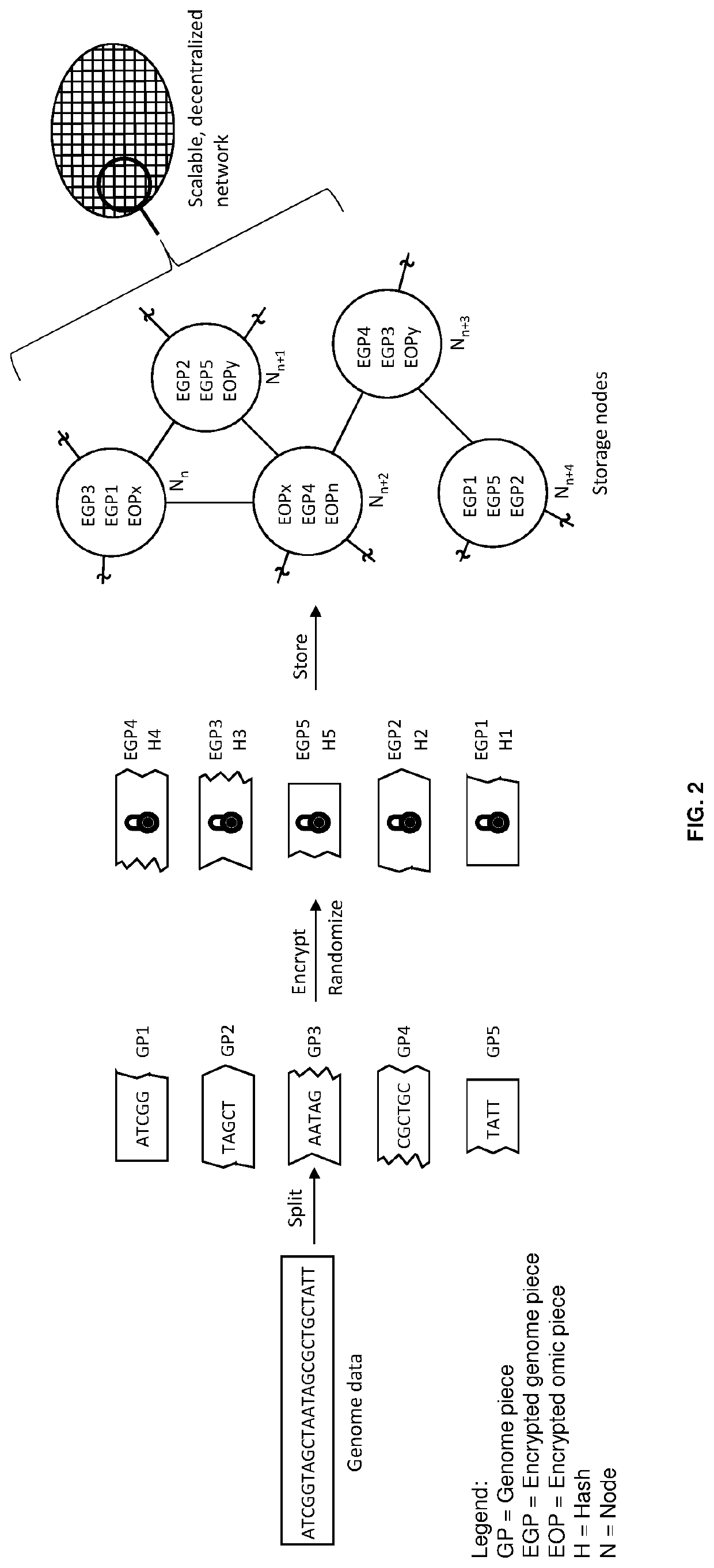
Methods for decentralized genome storage, distribution, marketing and analysis
PendingUS20200073560A1Easy to optimizeEasy to understandInput/output to record carriersData stream serial/continuous modificationData packBio molecules
Techniques for storing omics data that indicates long sequences of elements associated with a particular biological molecule include receiving digital omics data comprising over two kilobytes. The digital omics data is split into multiple partitions. The maximum partition size is much less than then a number of elements in a typical instance of the particular biological molecule. Each partition is encrypted. Each encrypted partition is inserted into a corresponding data packet that includes an owner field that uniquely indicates an owner of the omics data. Each data packet is uploaded into a non-centralized, peer-to-peer distributed storage network. Thus, genome data is encrypted and stored in a distributed, scalable, fully decentralized, fast, and highly secure network. The network further provides for decentralized computing, trustless validation of the genomes by way of oracles and trustless genome analysis by third party providers through smart contracts.
Owner:GENETIC INTELLIGENCE INC
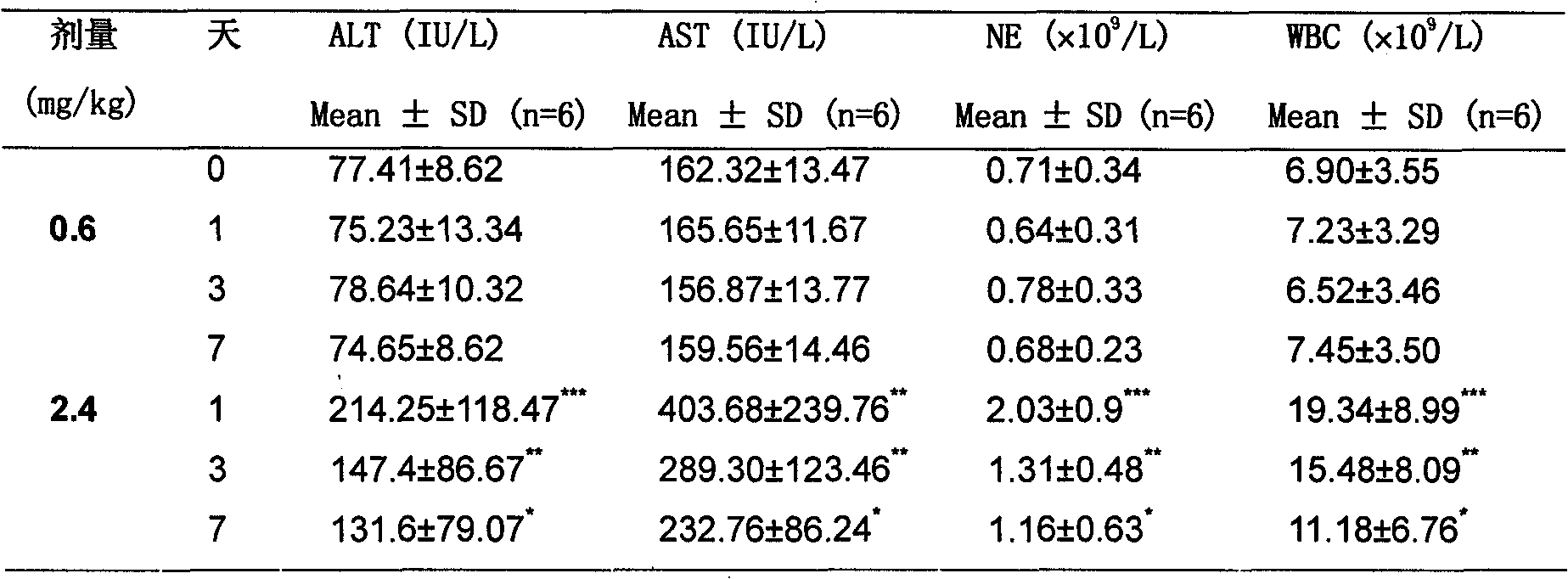
Method for quantitatively evaluating medicament toxicity by using metabonomic technology
InactiveCN101813680AReduce deviationImprove objectivityComponent separationNMR - Nuclear magnetic resonanceMathematical model
The invention discloses a method for quantitatively evaluating medicament toxicity by using metabonomic technology, which is characterized by comprising the following steps: comprehensively and quantitatively measuring small molecule compounds in a biological sample by using the measuring technology based on mass spectrum, nuclear magnetic resonance and the like, then establishing a multi-dimensional spatial mathematical model by adopting a multi-variable data processing method, calculating a relative distance between an administration group and a control group and taking the relative distance as a quantitative index for evaluating the medicament toxicity so as to solve the difficult problem that the medicament toxicity evaluation lacks the quantitative evaluating index. Compared with the conventional toxicity evaluating method, the method has the advantages of wide application, sensitivity, simple and convenient sampling, no harm to the body, and capability of reflecting the toxicity function more comprehensively, and providing a comprehensive and reliable quantitative evaluating method for toxicity evaluation in new medicament research and development and pharmacologic research.
Owner:CHINA PHARM UNIV
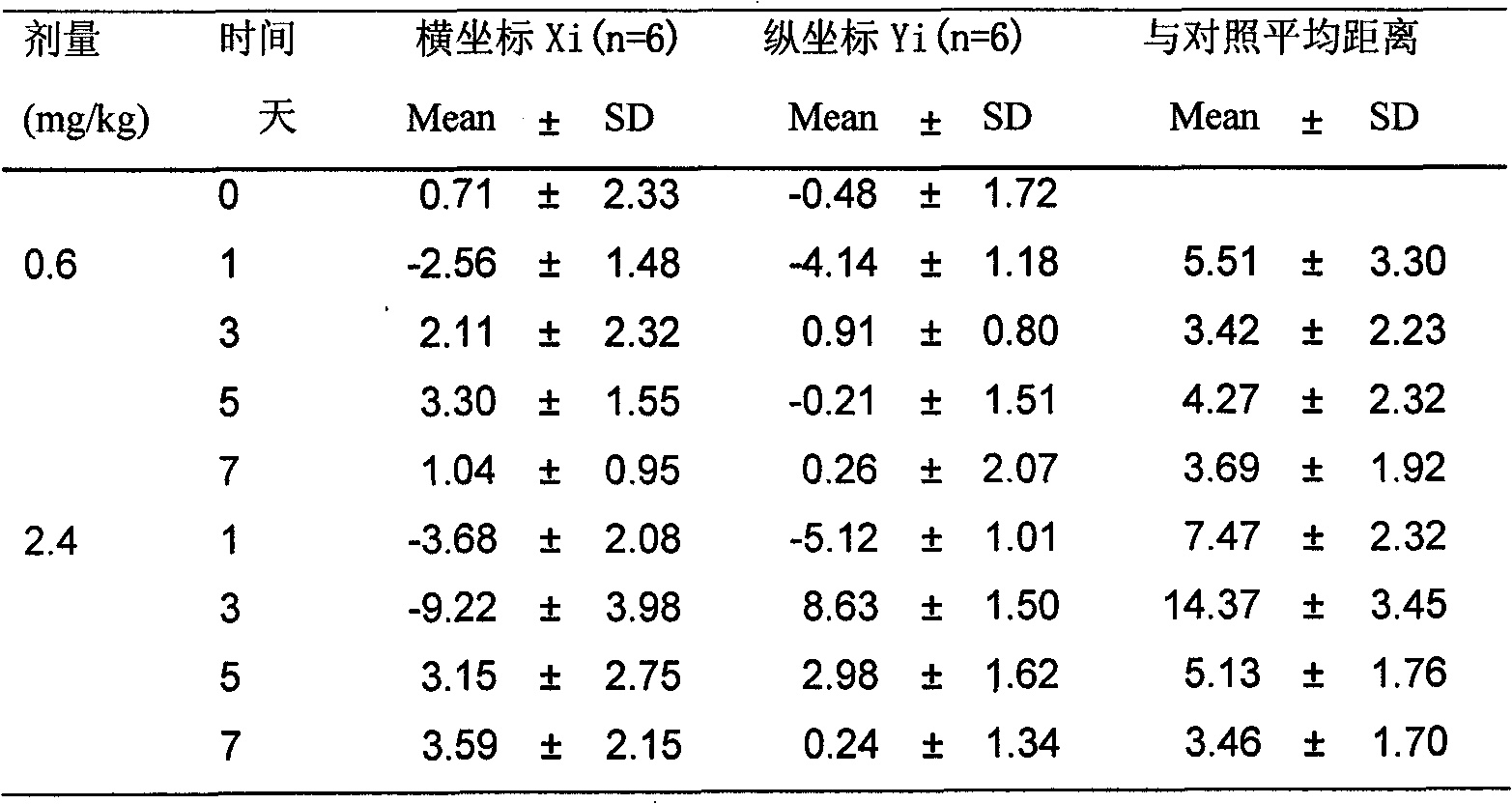
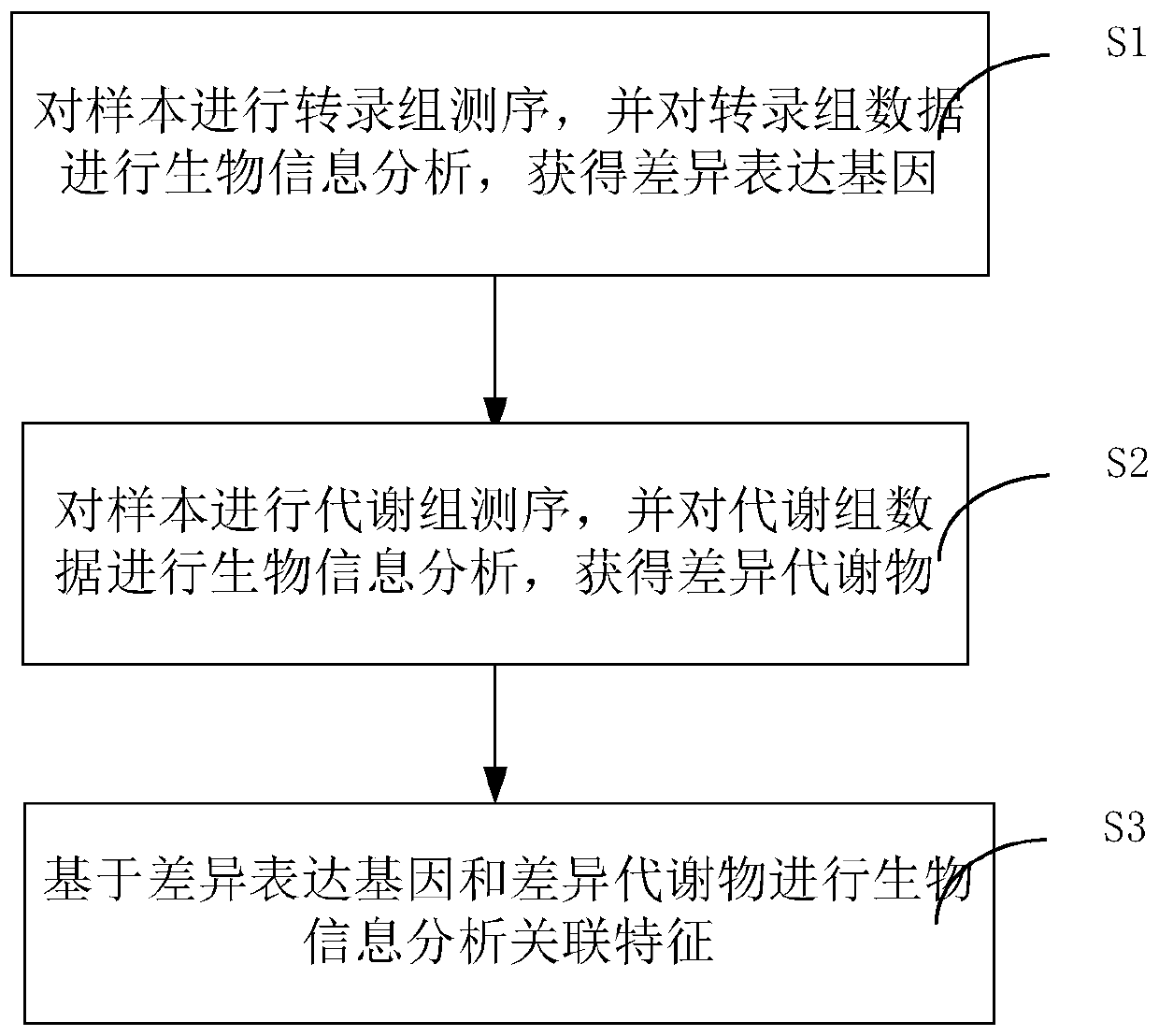
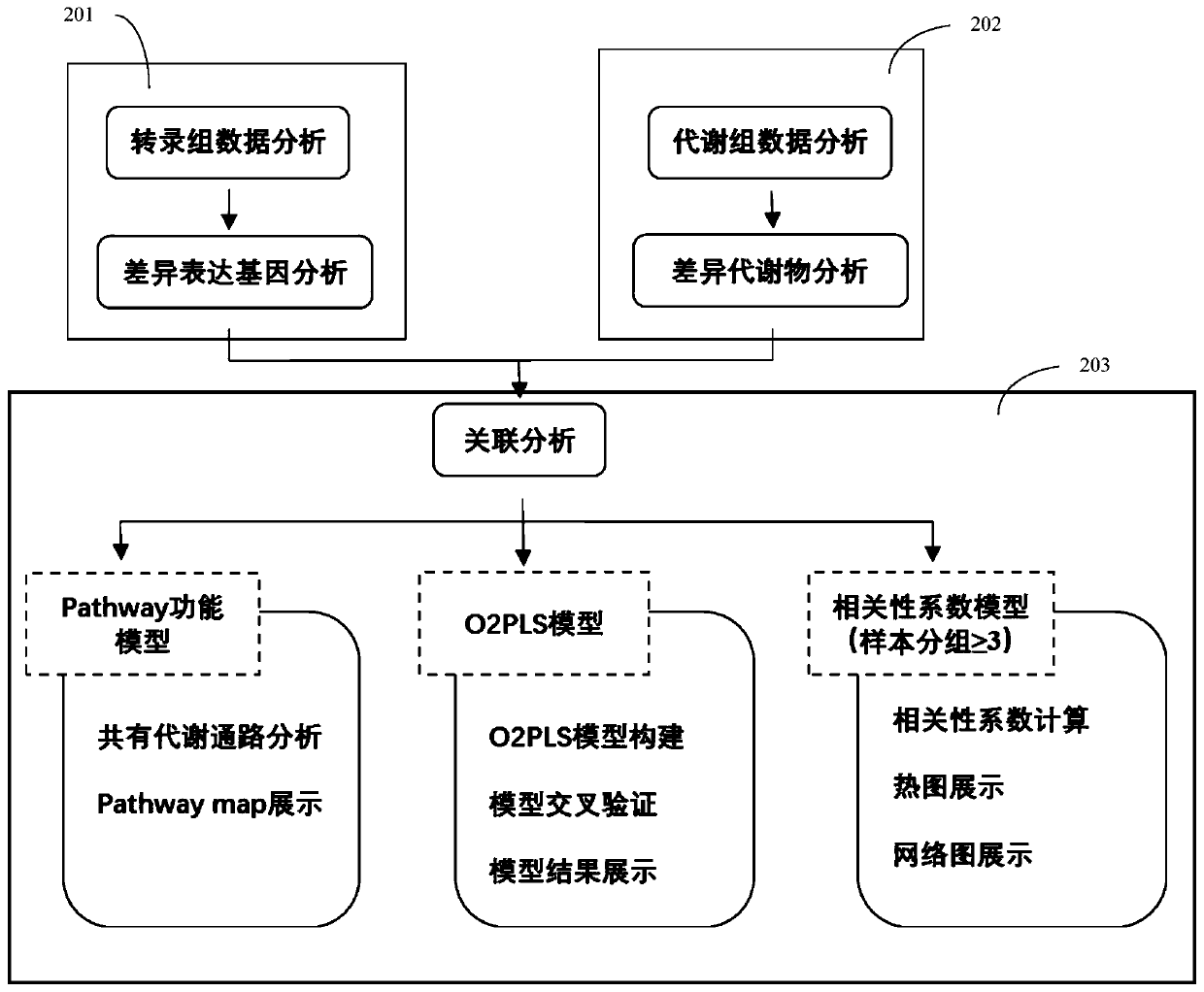
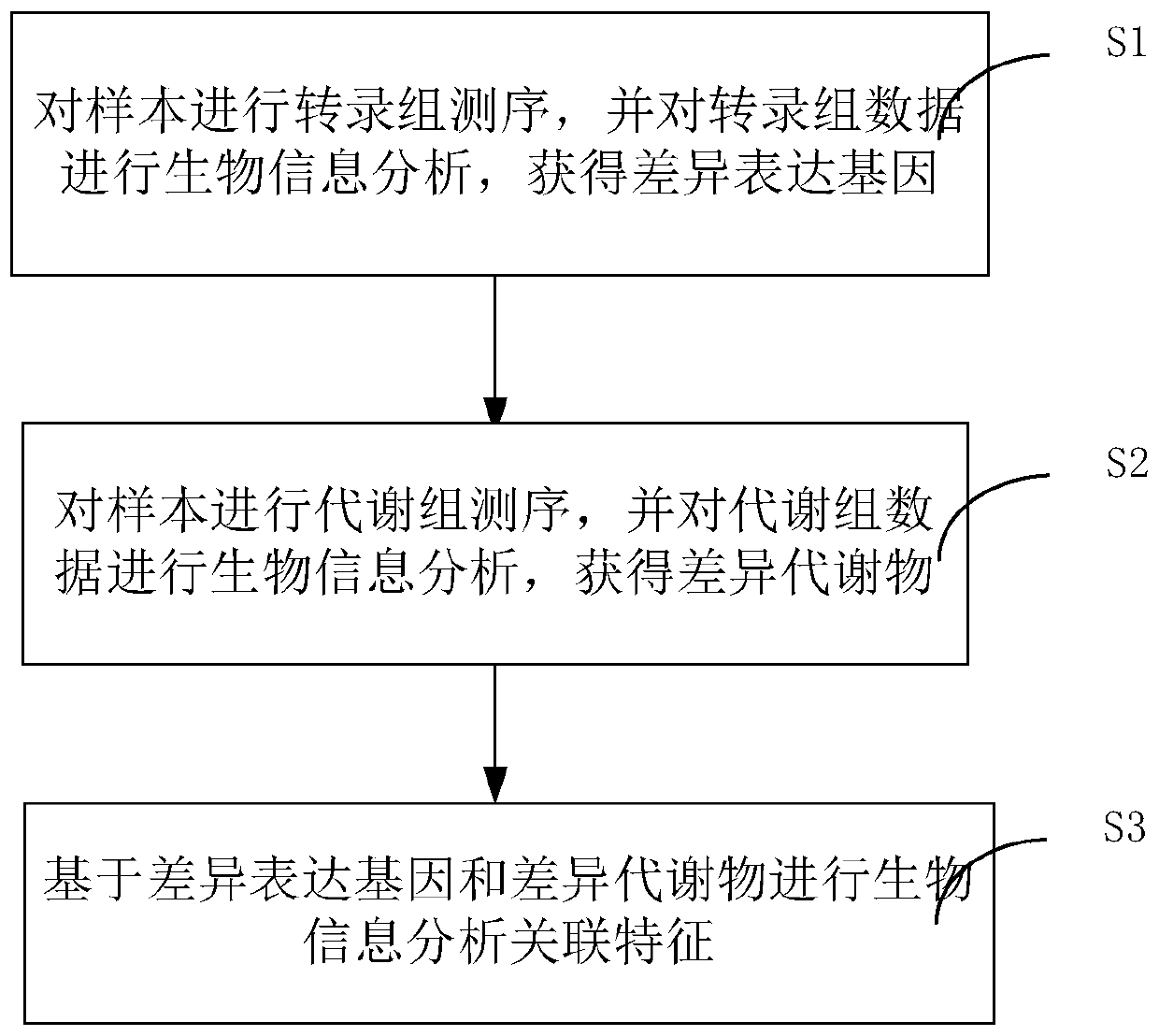
Method and system for association analysis of transcriptome and metabolome data
The invention discloses a method and a system for association analysis of transcriptome and metabolome data. The method comprises the following steps of S1, performing transcriptome sequencing on a sample, performing bioinformation analysis on the transcriptome data for obtaining a differential expression gene; S2, performing metabolome sequencing on the sample, and performing bioinformation analysis on the metabolome data for obtaining a differential metabolite; and S3, performing bioinformation analysis on the association characteristic based on the obtained differential expression gene andthe differential metabolite. The method and the system settle problems of one-sidedness of single omic sequencing data and low reliability of partial data in prior art. Furthermore, according to the method and the system, through information transmission, cooperation and coordination between biomolecules, a specific function is presented, thereby realizing more systematical multi-omics integratedanalysis and facilitating discloses of a complicated function mechanism.
Owner:GUANGZHOU GENE DENOVO BIOTECH
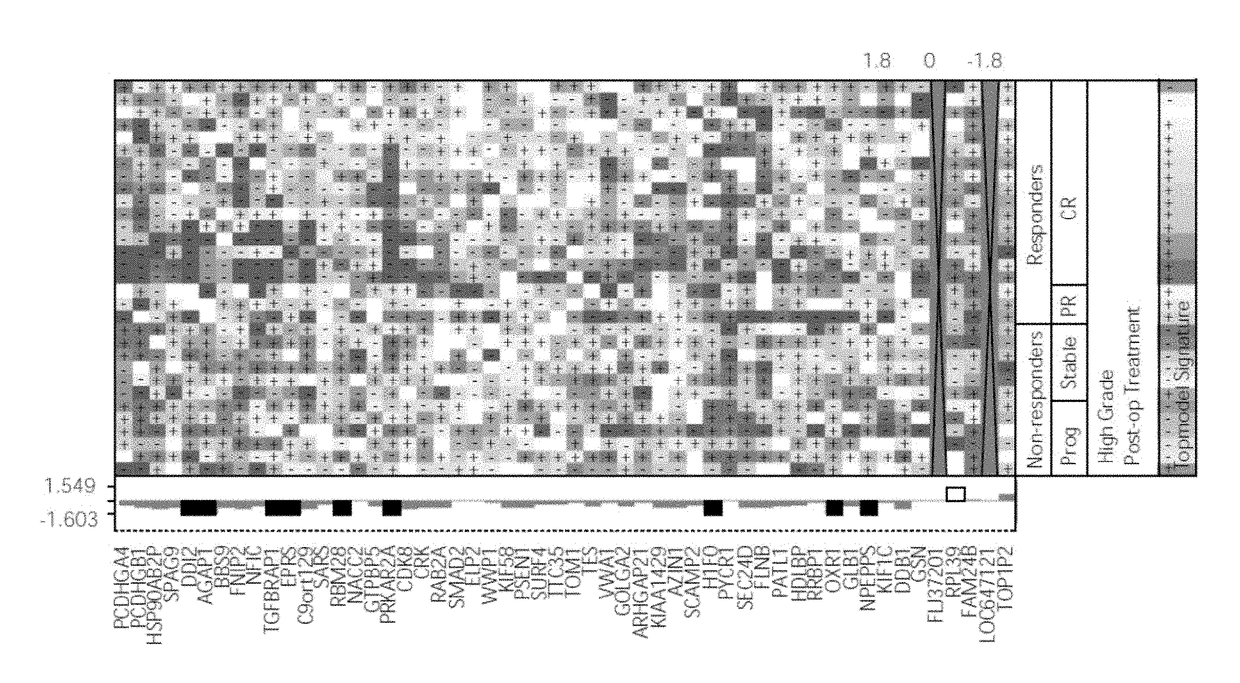
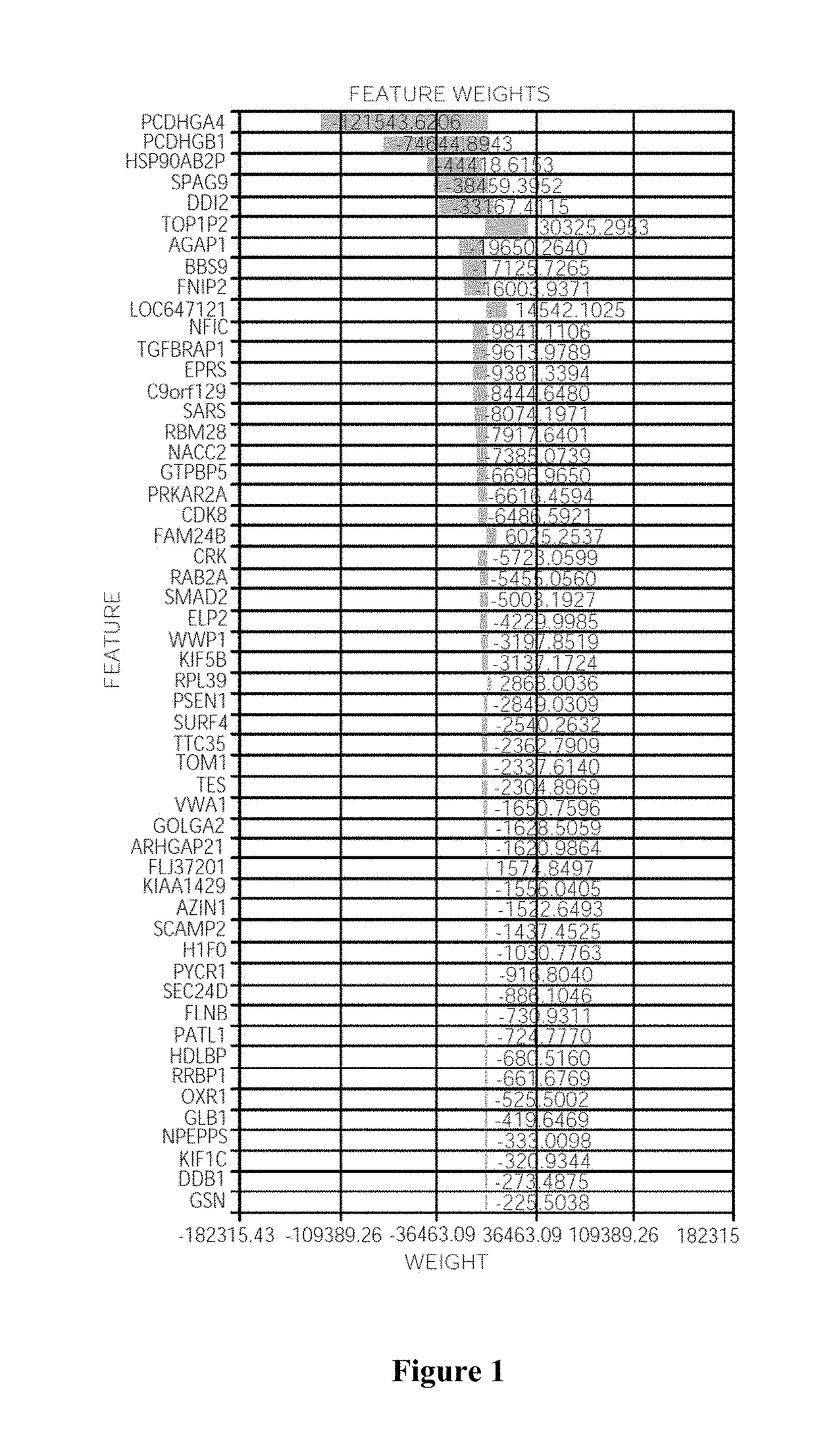
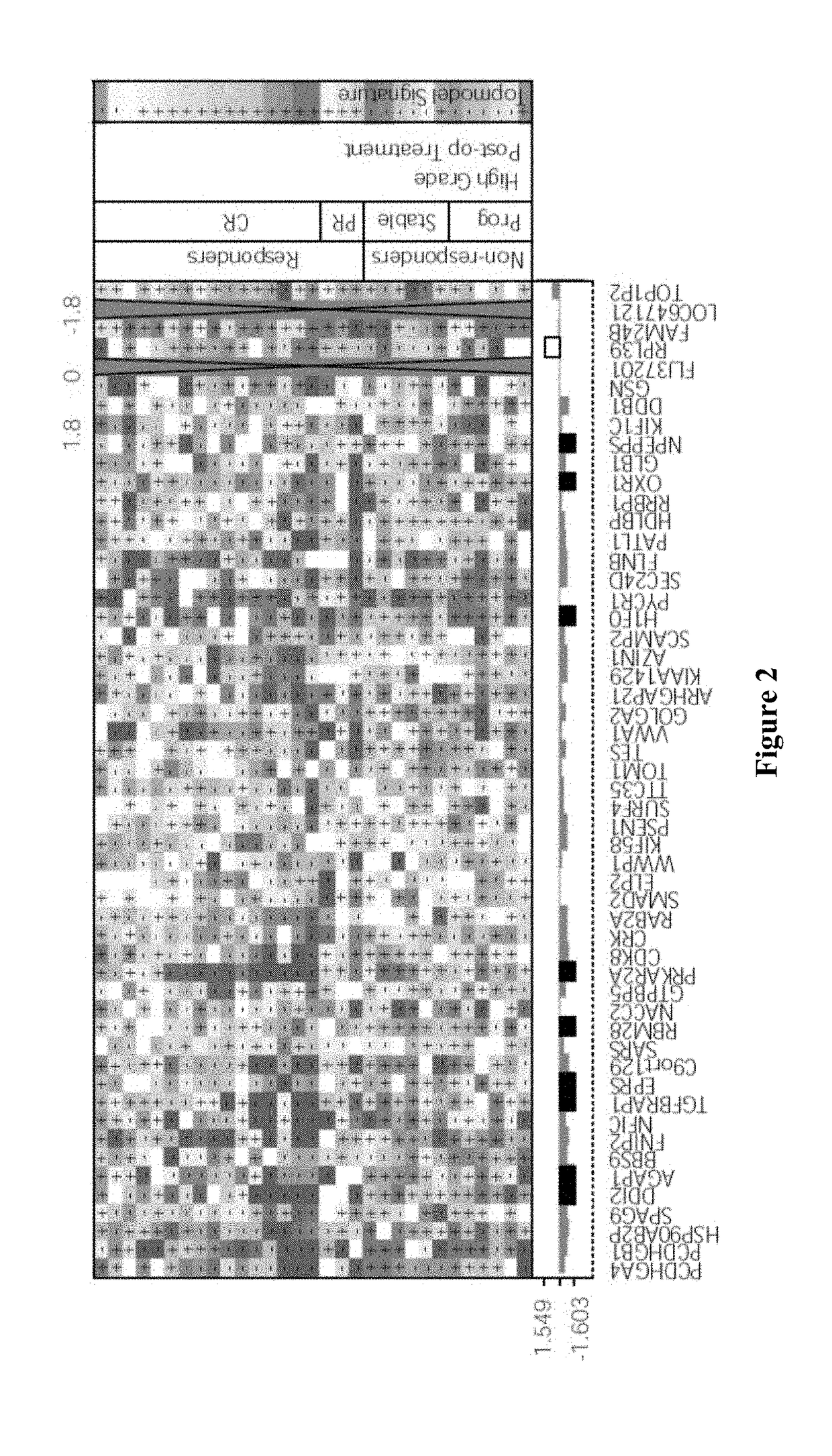
Systems and Methods for Response Prediction to Chemotherapy in High Grade Bladder Cancer
ActiveUS20180004905A1Improve forecast accuracyImprove accuracyMedical simulationEnsemble learningBladder cancerPredictive systems
Contemplated systems and methods allow for prediction of chemotherapy outcome for patients diagnosed with high-grade bladder cancer. In particularly preferred aspects, the prediction is performed using a model based on machine learning wherein the model has a minimum predetermined accuracy gain and wherein a thusly identified model provides the identity and weight factors for omics data used in the outcome prediction.
Owner:NANTOMICS LLC
Reflector TOF with high resolution and mass accuracy for peptides and small molecules
Many applications in the study of metabolics and proteomics require measurements on peptides and small molecules with high resolving power and mass accuracy. These are often present in complex mixtures and sensitivity over a relatively broad mass range, speed of analysis, reliability, and ease of use are very important. The present invention is a time-of-flight mass spectrometer providing optimum performance for these and similar applications.
Owner:VIRGIN INSTR CORP
Boron-containing diacylhydrazines
The present disclosure provides boron-containing diacylhydrazines having Formula I:and the pharmaceutically acceptable salts and solvates thereof, wherein R1, R2, R3, R4, and R5 are defined as set forth in the specification. The present disclosure also provides the use of boron-containing diacylhydrazines is ecdysone receptor-based inducible gene expression systems. Thus, the present disclosure is useful for applications such as gene therapy, treatment of disease, large scale production of proteins and antibodies, cell-based screening assays, functional genomics, proteomics, metabolomics, and regulation of traits in transgenic organisms, where control of gene expression levels is desirable.
Owner:PRECIGEN INC
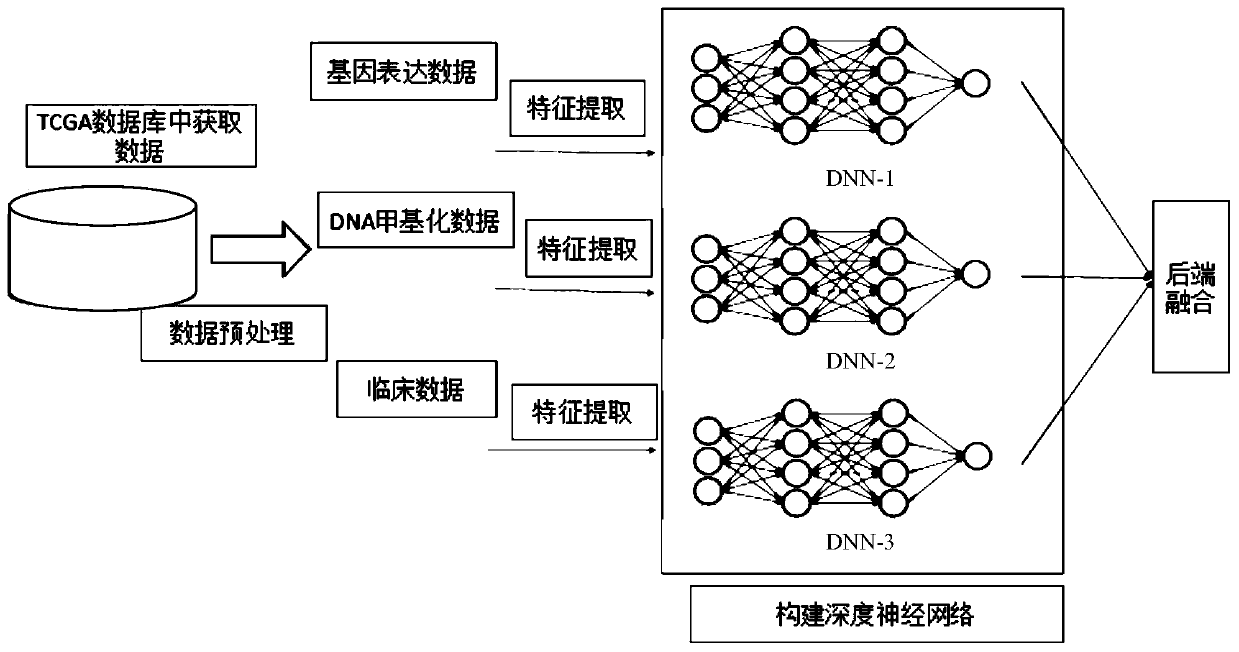
Prediction method for survival time of patient with breast cancer based on deep neural network
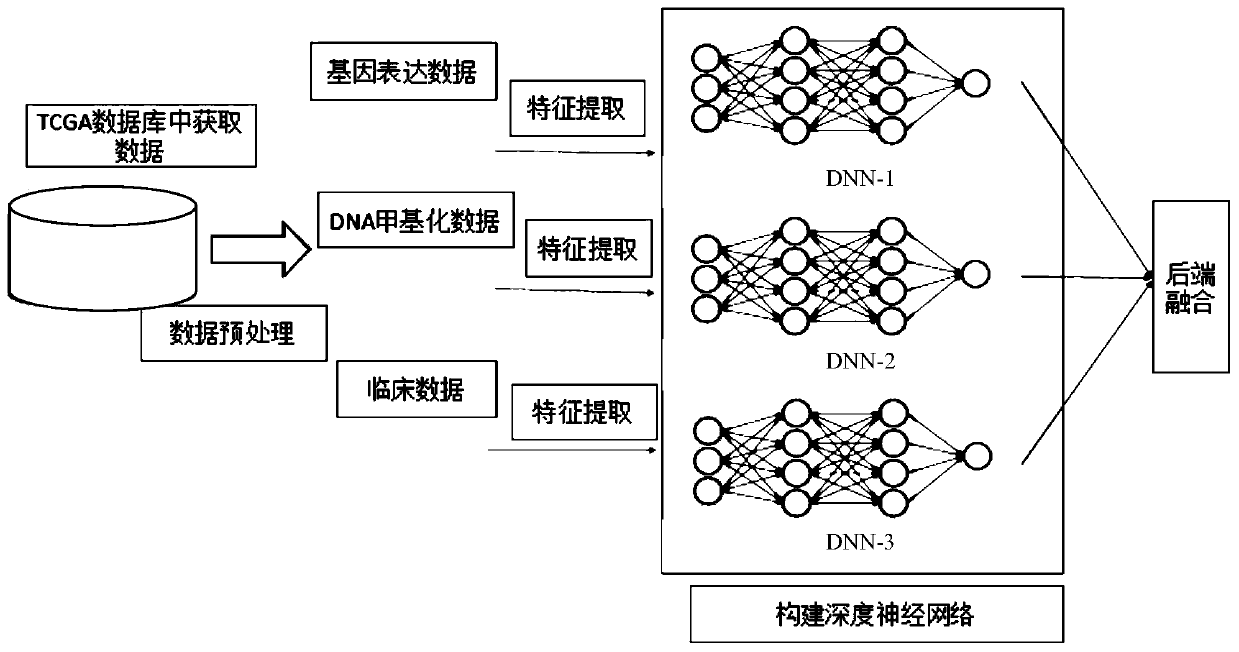
PendingCN111161882AImproving lifetime prediction performanceObvious effectMedical data miningNeural architecturesDNA methylationData set
The invention relates to a prediction method for the survival time of a patient with breast cancer based on a deep neural network. The method comprises the following steps: 1) acquiring data, whereinthe data is clinical data and omics data, and the omics data comprises gene expression data and DNA methylation data; 2) preprocessing the data; 3) extracting the features of a data set; and 4) constructing a deep neural network. According to the invention, survival time prediction is carried out by fusing the deep neural network with the multi-omics data of breast cancer; clinical data, gene expression data and DNA methylation data of breast cancer are obtained from a TCGA database, data features are extracted, deep neural network models are constructed respectively, and then back-end fusionis performed, so the performance of survival time prediction for breast cancer is improved, and a lifetime prediction model is obtained; and the method has a remarkable effect in prediction of the survival time of patients with breast cancer.
Owner:SHENZHEN INST OF ADVANCED TECH
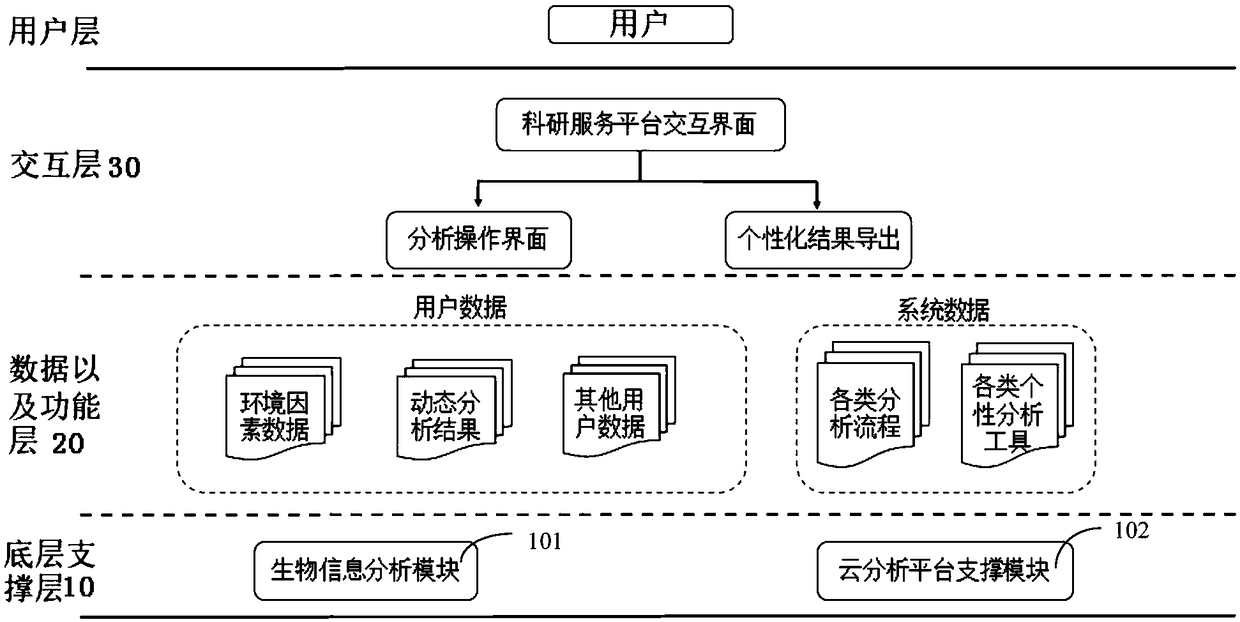
Microbial omics online analysis platform framework based on genomics and bioinformatics
ActiveCN109448788AEasy to analyzeReal-time analysisData visualisationBiostatisticsGenomicsInteraction layer
The invention discloses a microbial omics online analysis platform framework based on genomics and bioinformatics. The framework comprise a bottom layer supporting layer, a data and function layer andan interaction layer, wherein the bottom layer supporting layer comprises a biological information analysis module and a cloud platform supporting module, and the biological information analysis module is used for personalized analysis of 16S, ITS, 18S and microbial metagenome sequencing through the cloud platform supporting module, interacting with the data and function layer and the interactionlayer and finally conducting displaying to users through an interaction interface; the cloud platform supporting module comprises software and hardware conditions required by the cloud platform; thedata and the function layer is used for providing user data and system data to the bottom layer supporting layer; the interaction layer is used for presenting analysis results of the biological information analysis module to the users through the interaction interface. The defects of a traditional high-throughput sequencing scientific research service mode can be overcome so that scientific research workers without biological information bases can select parameters independently according to needs, one-key analysis result generation can be achieved, graphs are completely dynamic, and real-timeanalysis is facilitated.
Owner:GUANGZHOU GENE DENOVO BIOTECH
Systems medicine platform for personalized oncology
ActiveUS20140330583A1Easy to optimizeEasy to identifyData processing applicationsData visualisationMetabolitePatient characteristics
Owner:GEORGETOWN UNIV
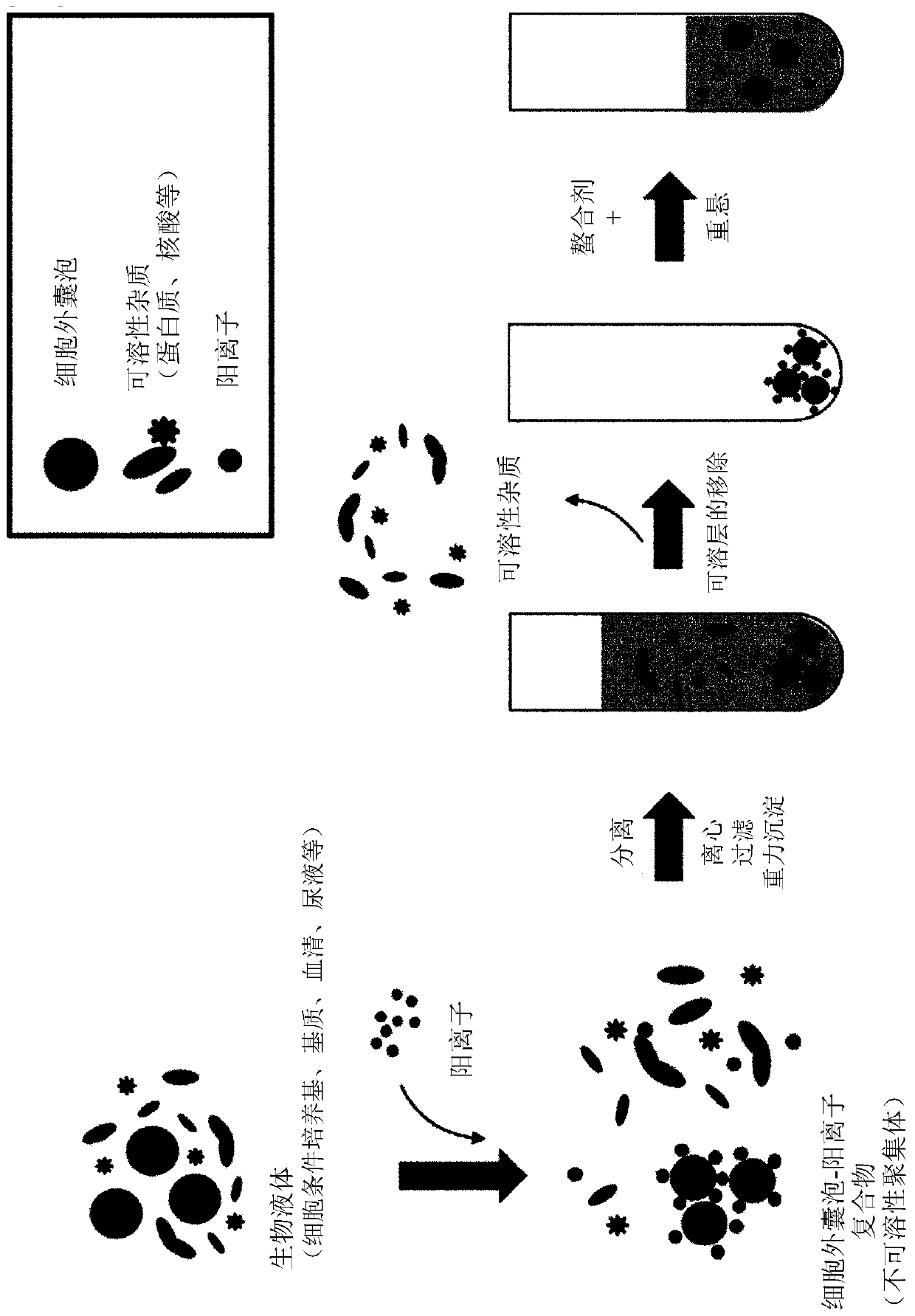
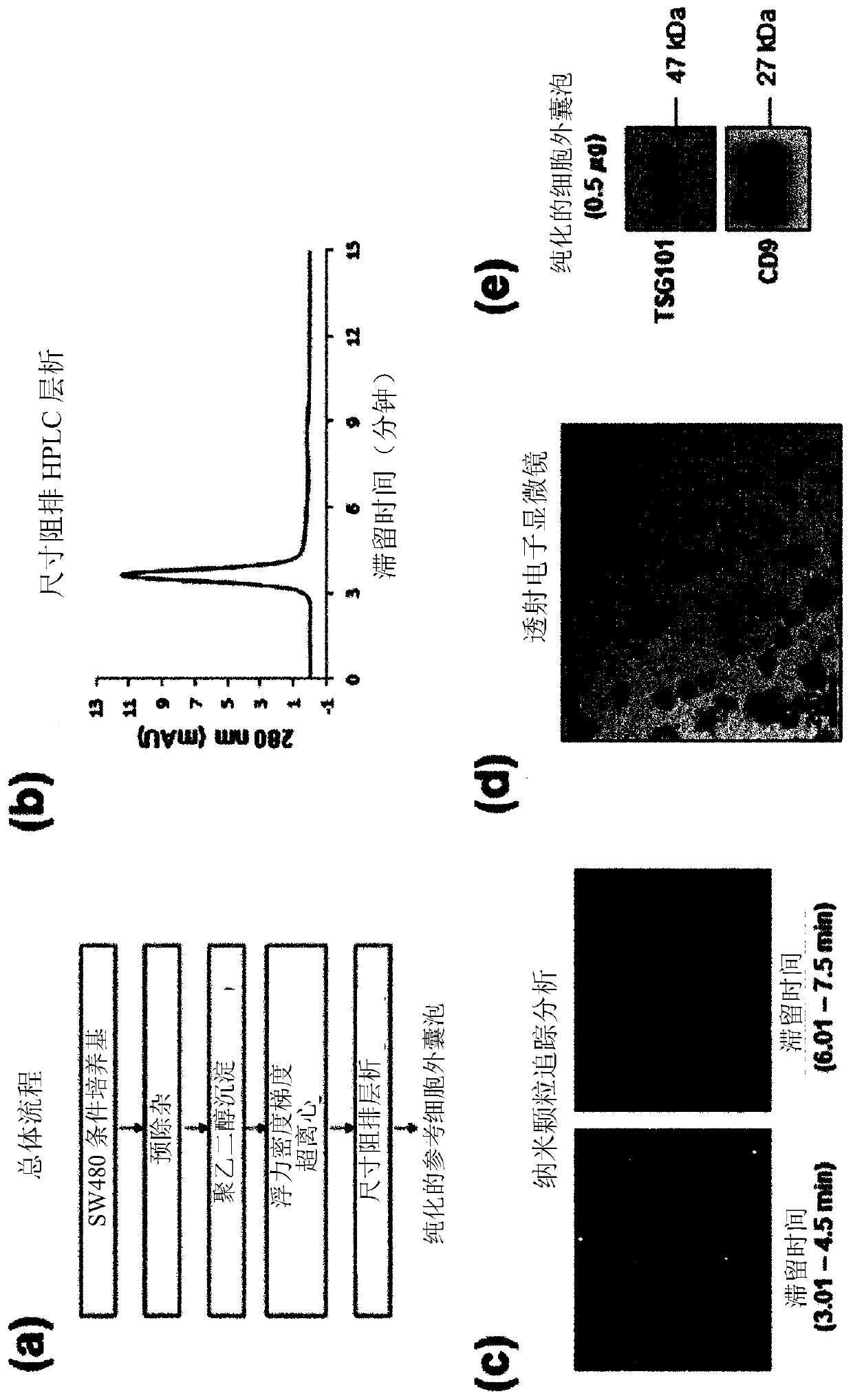
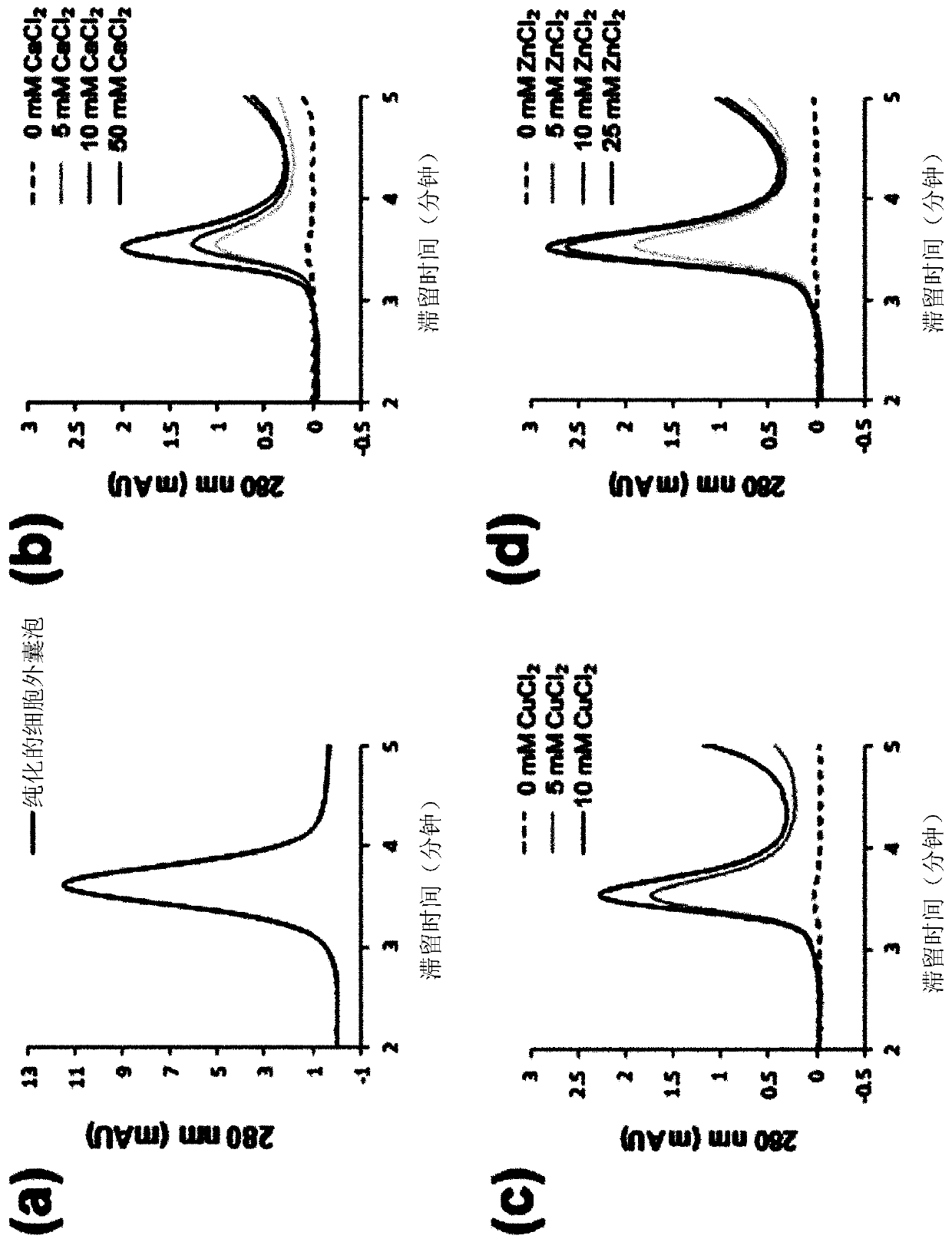
Method for isolating extracellular vesicles using cations
PendingCN111148828AEfficient separationKeep shapeCell dissociation methodsCation exchanger materialsExtracellular vesicleDisease
The present invention relates to a method for isolating extracellular vesicles using cations, and more particularly, to a method for isolating extracellular vesicles from various samples by using theaffinity between the extracellular vesicles and cations. A method for isolating extracellular vesicles according to the present invention does not require expensive equipment, can be applied irrespective of sample amount, and has the advantage of being capable of efficiently isolating the extracellular vesicles while preserving the shape or characteristics thereof. Moreover, the method according to the present invention can be combined with existing isolation methods to maximize extracellular vesicle isolation efficiency, and can be applied to disease diagnosis, disease treatment, and multi-omics research using isolated extracellular vesicles, as well as to research on the properties of extracellular vesicles.
Owner:ROSETTA EXOSOME CO LTD
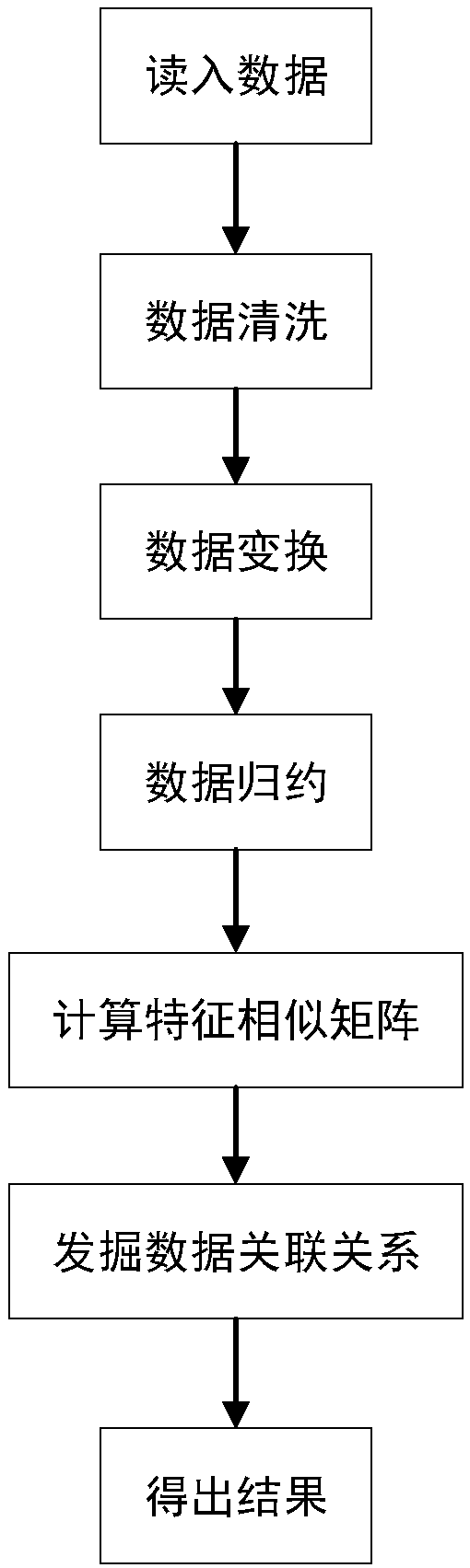
Multi-omics data association relationship discovery method based on sparse matching
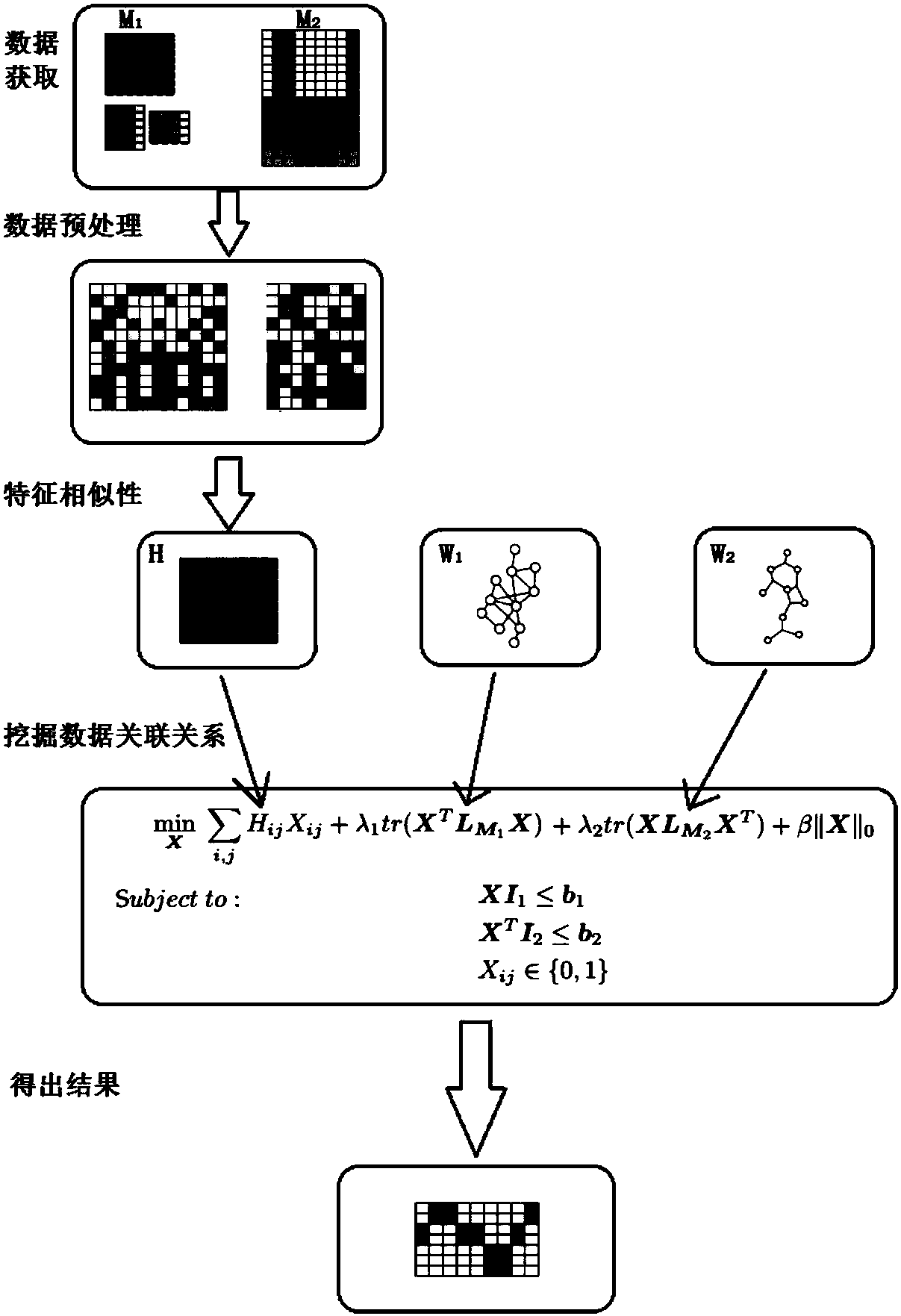
ActiveCN108509771AHigh precisionImprove robustnessBiostatisticsSpecial data processing applicationsPrior informationMulti omics
The invention discloses a multi-omics data association relationship discovery method based on sparse matching. The method includes: preprocessing input data to improve quality of the data; selecting asuitable similarity measure according to data characteristics to calculate a similarity matrix among data features; and fusing prior information to mine potential association relationships among thedata features on the basis of a similarity network among the features. According to the method of the invention, the prior information of features of existing already-proven omics data can be fully utilized, impacts of noises on results can be reduced, uncertainty caused by data errors can be reduced, and accuracy and robustness of the results can be improved.
Owner:SOUTH CHINA UNIV OF TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com