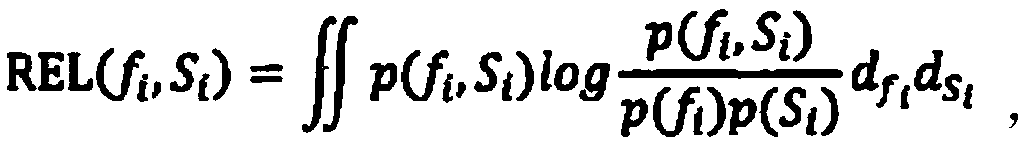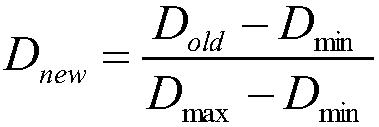Patents
Literature
132 results about "Omics data" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
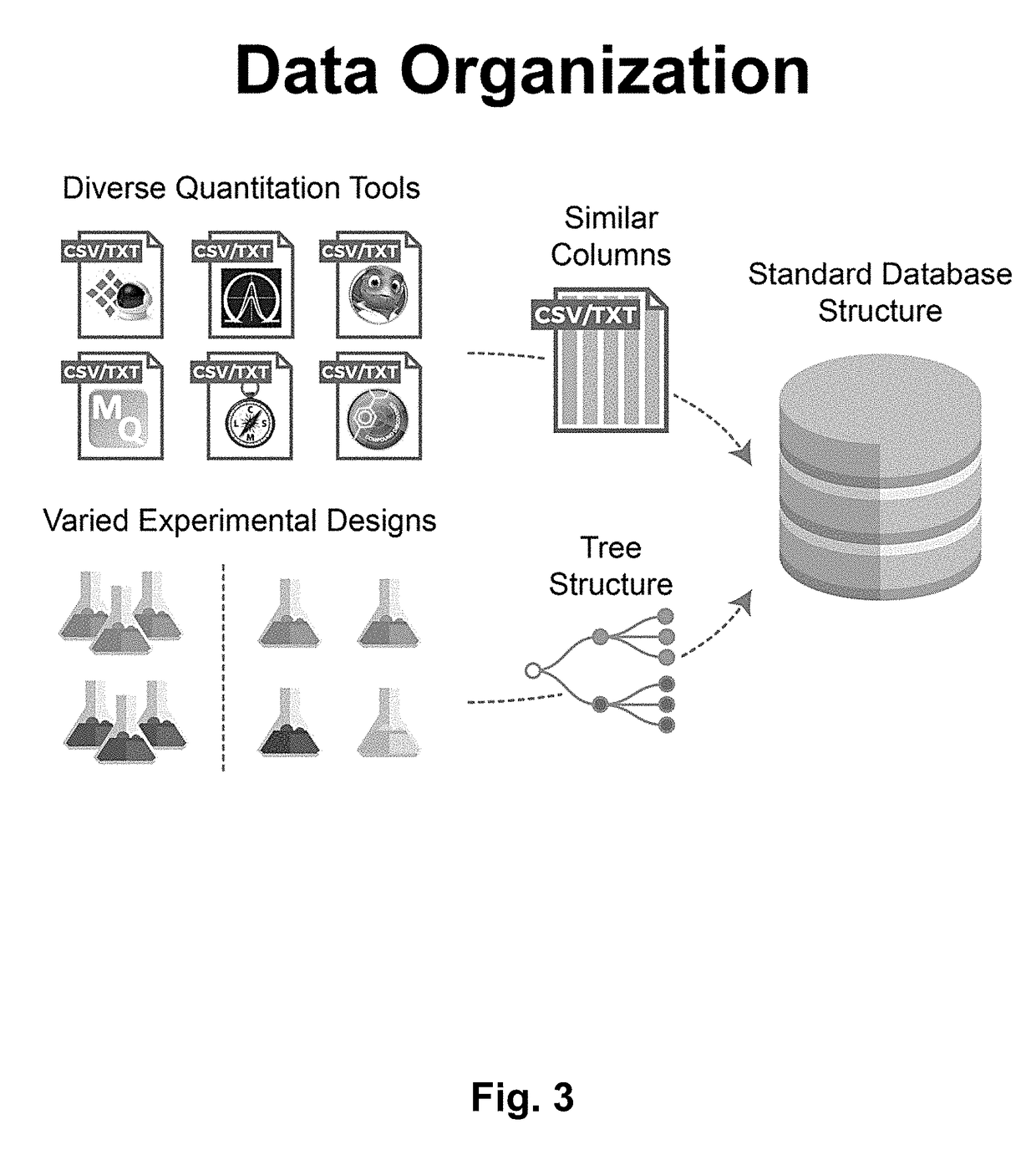
Omics Data Manager (ODM) enables data FAIRification, accelerating data-driven science in drug discovery, biomarker identification, agricultural crop development and the design of consumer good and personal healthcare products. ODM is built upon a modular architecture that can be deployed on-premise or in the cloud.
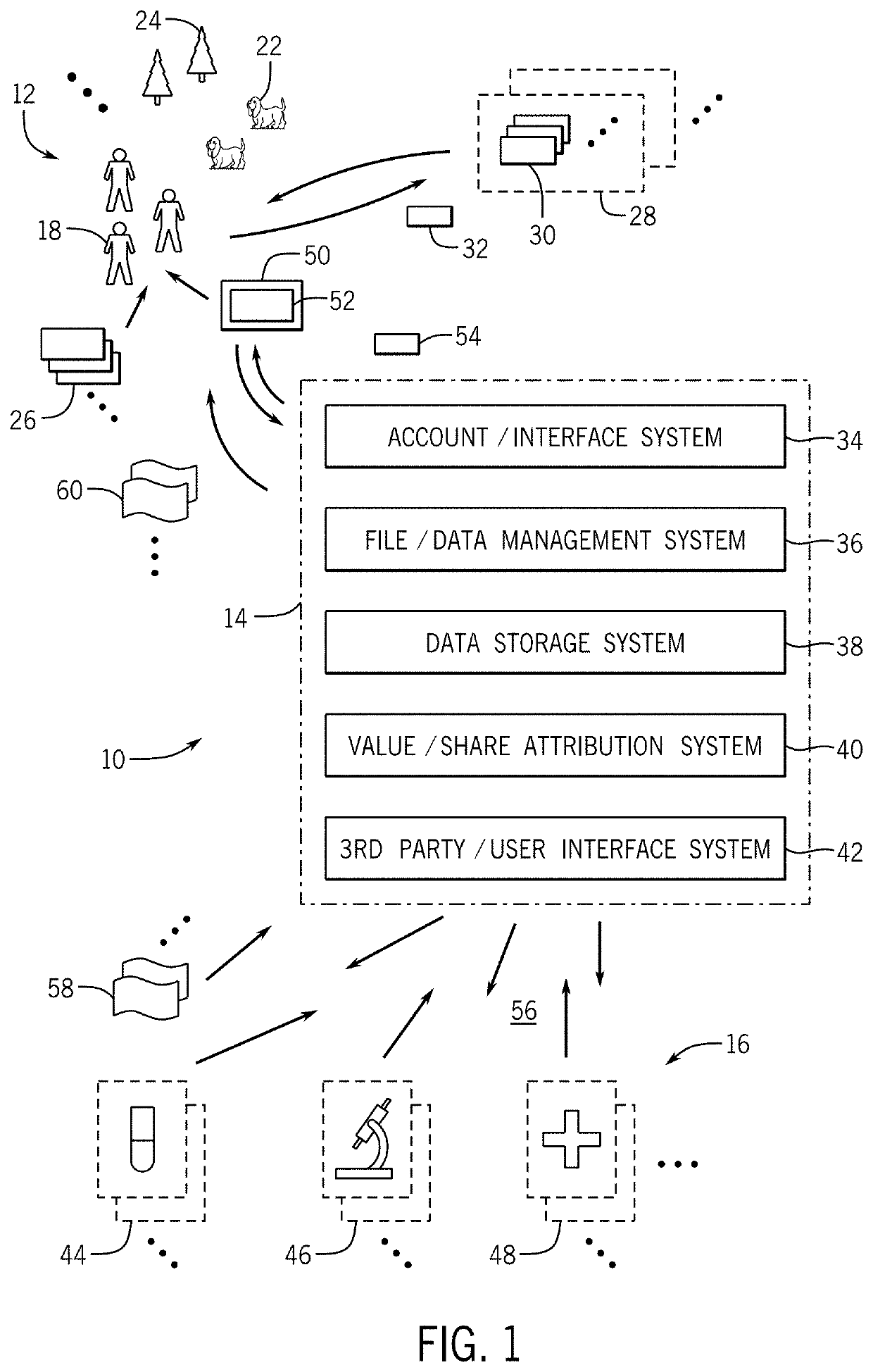
De-identification omic data aggregation platform with permitted third party access
InactiveUS20200035341A1Increase valueValue maximizationDatabase updatingFinancePersonalizationSystems management
A system and method are disclosed for the collection and aggregation of genomic, medical, and other data of interest for individuals and populations that may be of interest for analysis, research, pharmaceutical development, medical treatment, and so forth. Contributors become members of a community upon creation of an account and providing of data or files. The data is received and processed, such as to analyze, structure, perform quality control, and curate the data. Value or shares in one or more community databases are computed and attributed to each contributing member. The data is controlled to avoid identification or personalization. Third parties interested in the database information may contribute value (e.g., pay) for access and use. Value flows back to the members and to a system administrative entity.
Owner:LUNAPBC
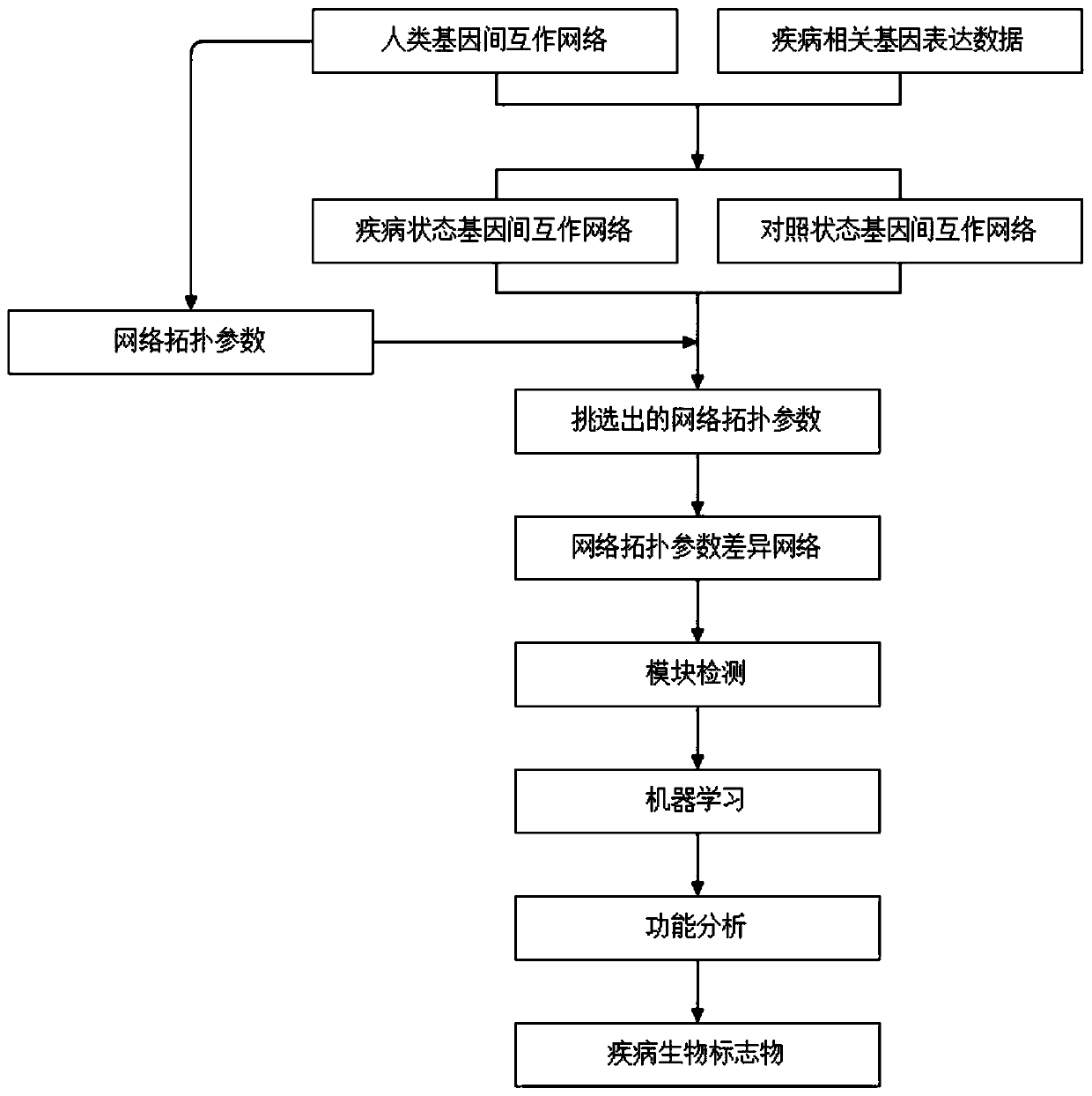
Cancer biomolecule marker screening method and system based on network topology parameters
ActiveCN110444248ACharacter and pattern recognitionProteomicsBiomarker identificationScreening method
The invention discloses a cancer biomolecule marker screening method and system based on network topology parameters. The method comprises the steps that a human gene interaction network and gene chipexpression data are acquired and integrated to obtain a gene interaction network based on gene expression data; a disease state and control state gene interaction network is constructed; network topology parameter difference genes of the disease state and control state gene interaction network are calculated, and a network topology parameter difference change network is obtained based on the network parameter difference genes; network module mining is performed on the network topology parameter difference network; feature selection is performed on an obtained difference network module to obtain genes capable of distinguishing normality and diseases in all modules; the classification effect of the genes selected from the modules on the diseases is detected, and the difference network module is screened according to the classification effect to serve as a biomolecule marker candidate. The invention provides a novel complex disease biomarker identification method based on omics data, andexperiments prove that the method has certain accuracy and effectiveness.
Owner:SHANDONG UNIV
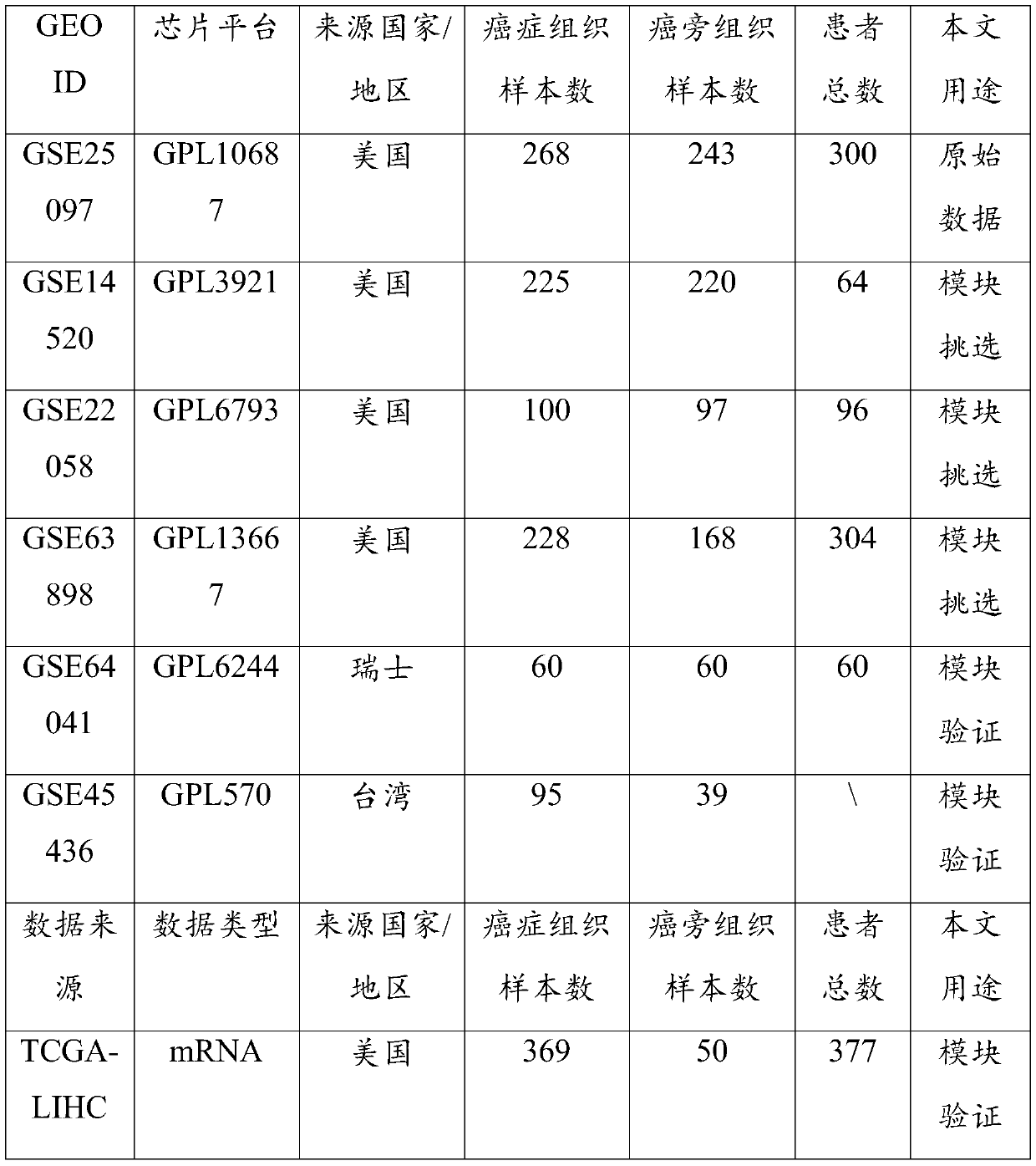
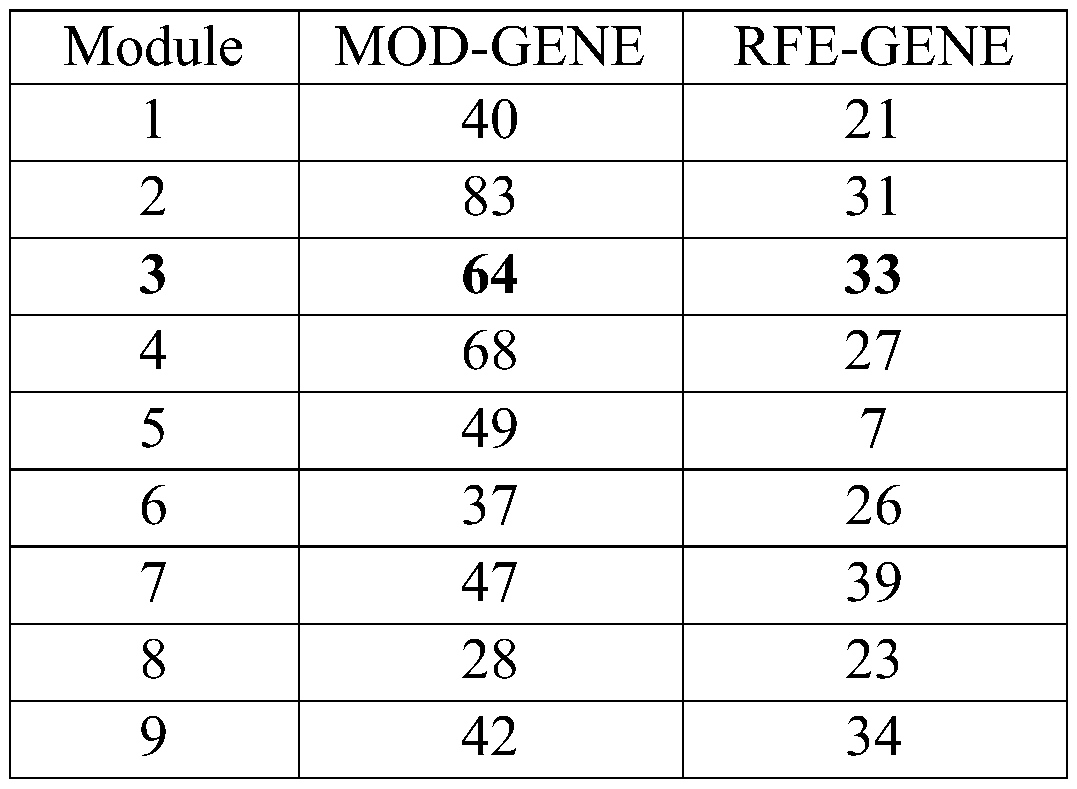
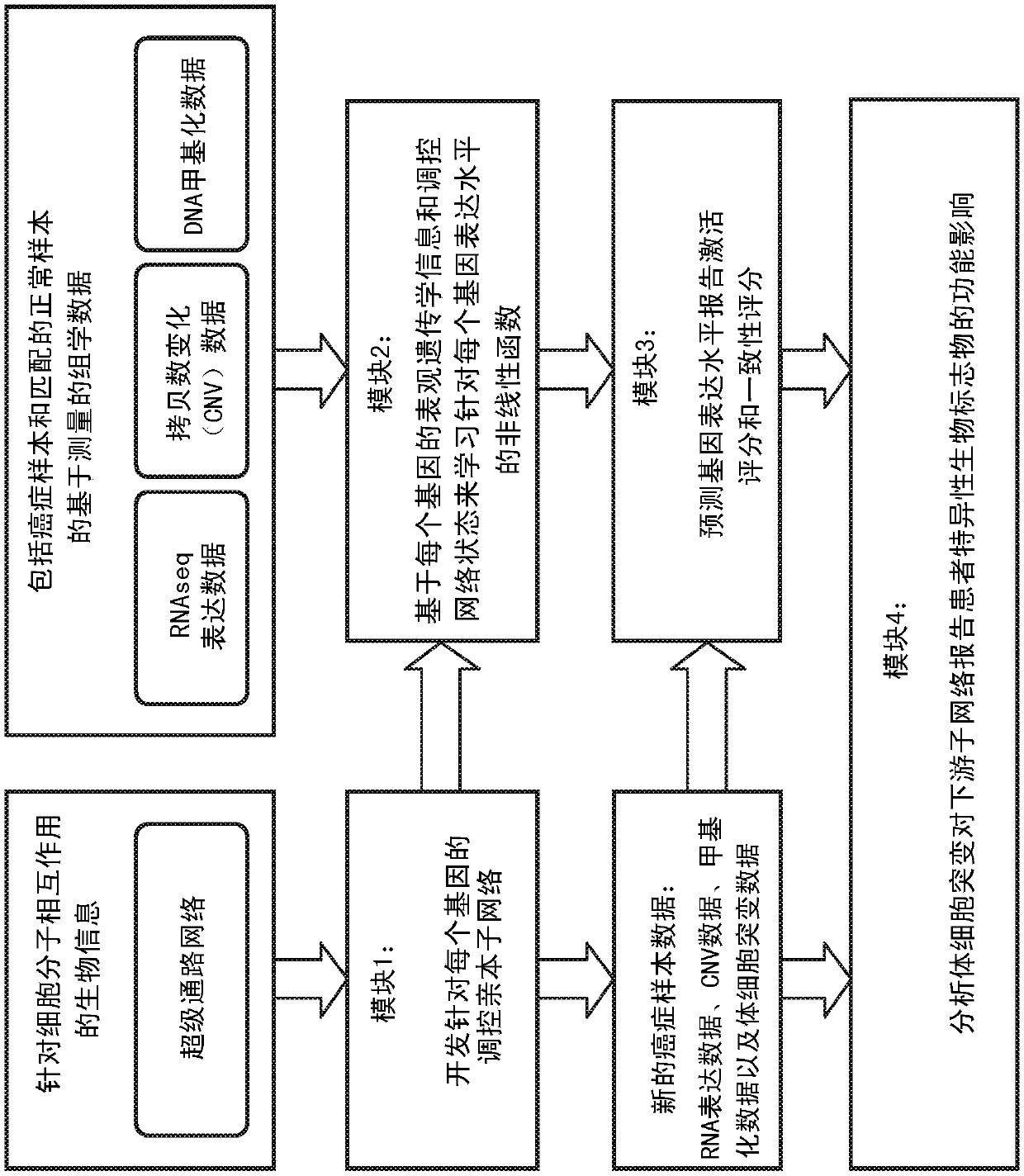
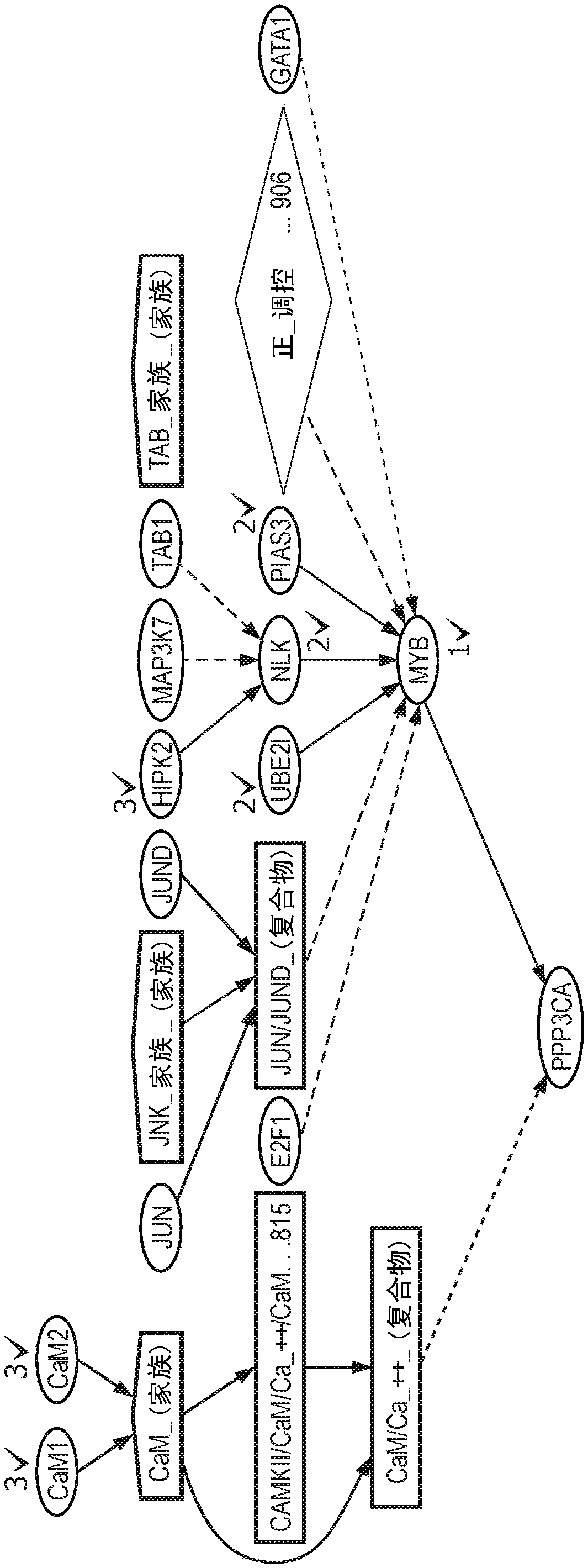
An integrated method and system for identifying functional patient-specific somatic aberations using multi-omic cancer profiles
A system and method for determining the functional impact of somatic mutations and genomic aberrations on downstream cellular processes by integrating multi-omics measurements in cancer samples with community-curated biological pathways are disclosed. The method comprises the steps of extracting biological pathway information from well-curated biological pathway sources, using the pathway information to generate an upstream regulatory parent sub-network tree for each gene of interest, integrating measurement-based omic data for both cancer and normal samples to determine a nonlinear function for each gene expression level based on the gene's epigenetic information and regulatory network status, using the nonlinear function to predict gene expression levels and compare activation and consistency scores with inputted patient- specific gene expression data, and using the patient-specific gene expression predictions to identify significant deviations and inconsistencies in gene expressionlevels from expected levels in individual patient samples to identify potential biomarkers in providing predictive information in relation to cancer and cancer treatment.
Owner:KONINKLJIJKE PHILIPS NV +1
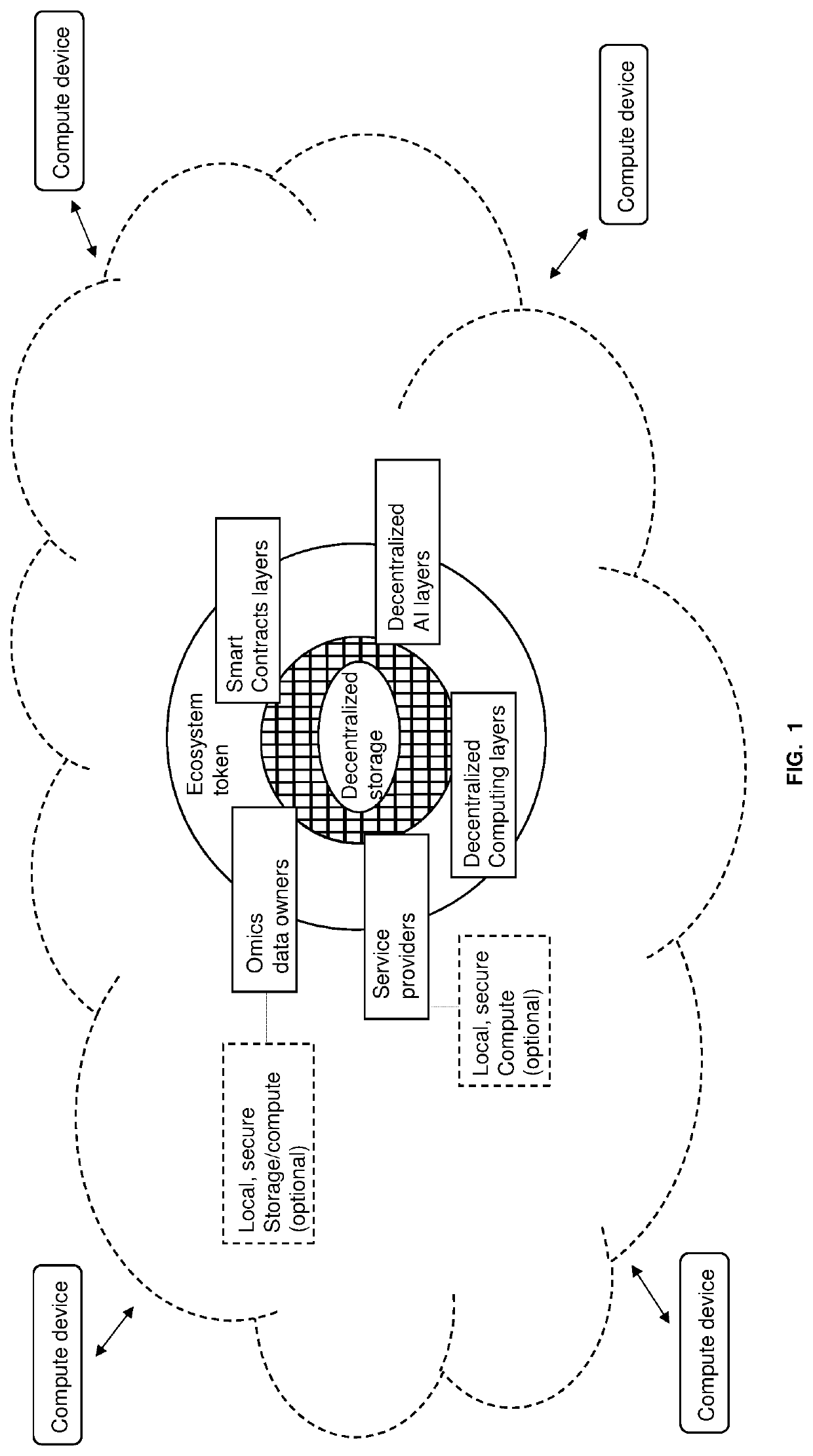
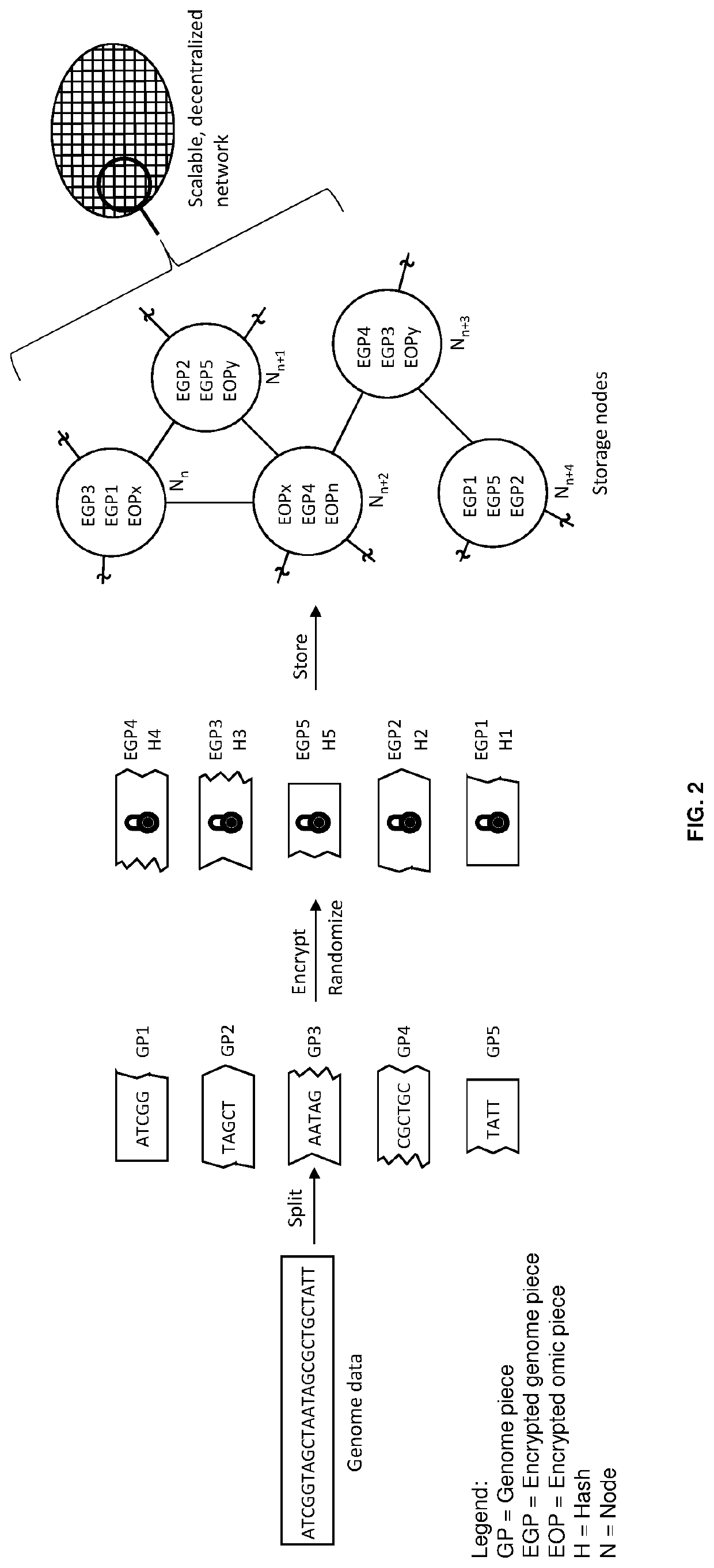
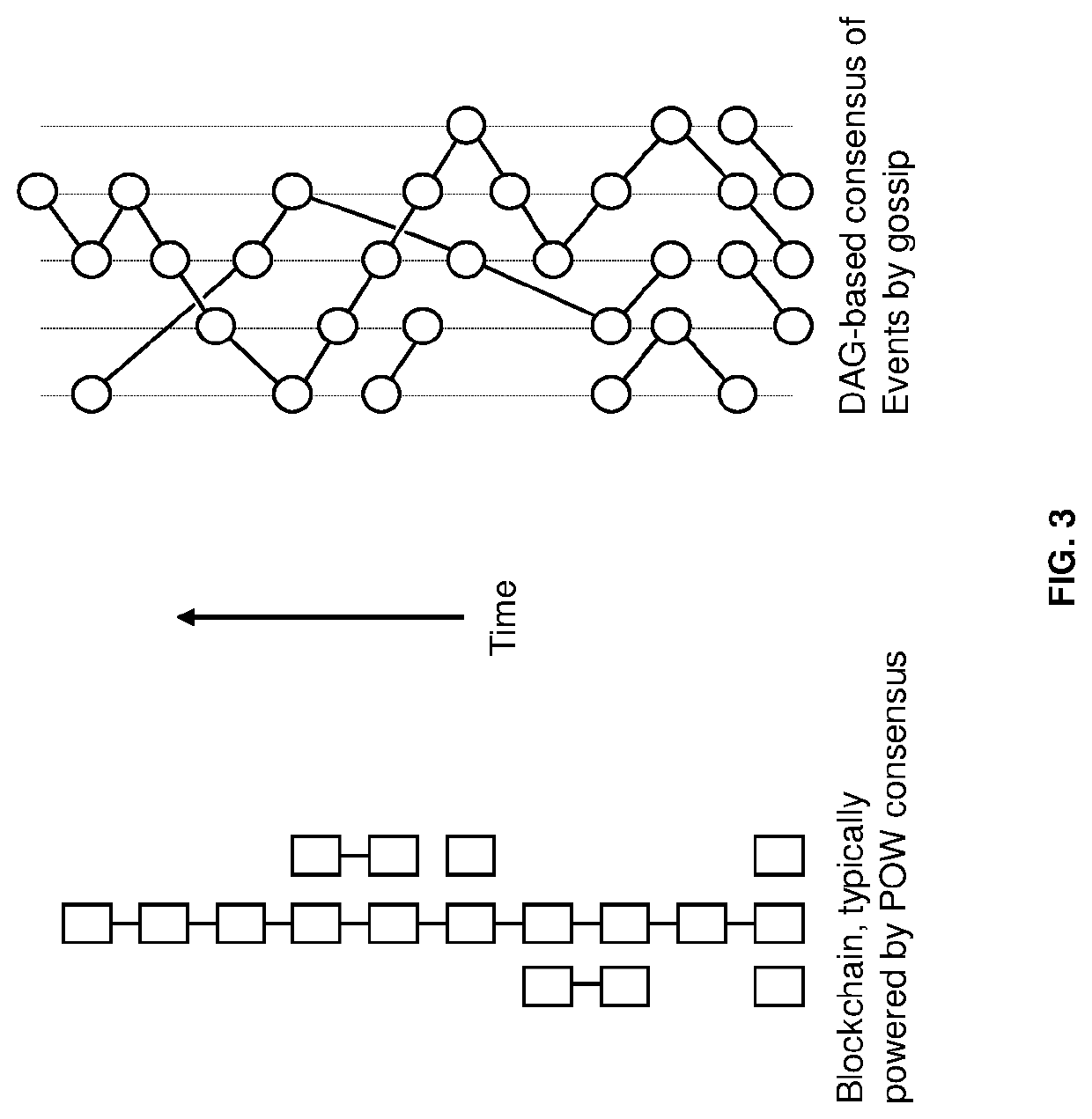
Methods for decentralized genome storage, distribution, marketing and analysis
PendingUS20200073560A1Easy to optimizeEasy to understandInput/output to record carriersData stream serial/continuous modificationData packBio molecules
Techniques for storing omics data that indicates long sequences of elements associated with a particular biological molecule include receiving digital omics data comprising over two kilobytes. The digital omics data is split into multiple partitions. The maximum partition size is much less than then a number of elements in a typical instance of the particular biological molecule. Each partition is encrypted. Each encrypted partition is inserted into a corresponding data packet that includes an owner field that uniquely indicates an owner of the omics data. Each data packet is uploaded into a non-centralized, peer-to-peer distributed storage network. Thus, genome data is encrypted and stored in a distributed, scalable, fully decentralized, fast, and highly secure network. The network further provides for decentralized computing, trustless validation of the genomes by way of oracles and trustless genome analysis by third party providers through smart contracts.
Owner:GENETIC INTELLIGENCE INC
Multi-omic search engine for integrative analysis of cancer genomic and clinical data
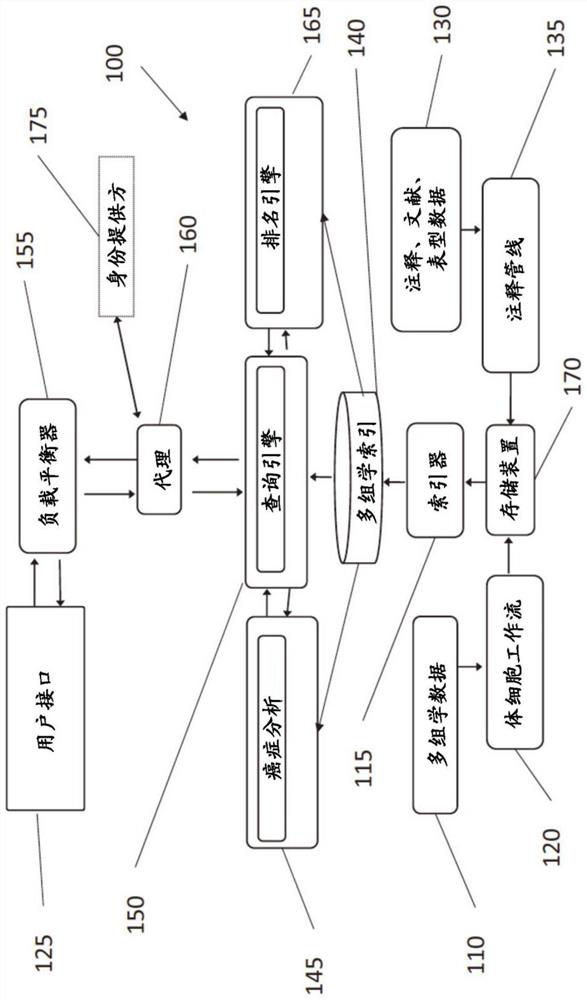
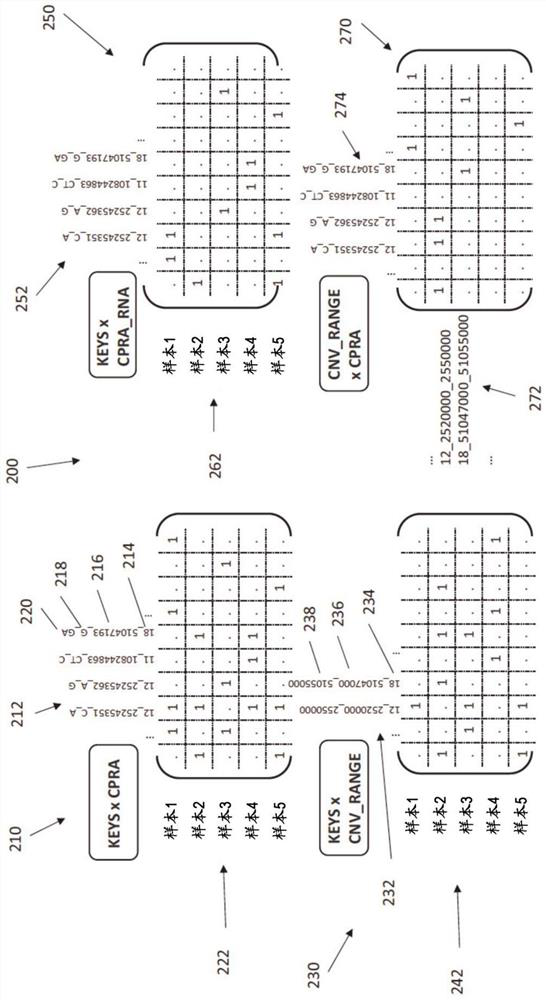
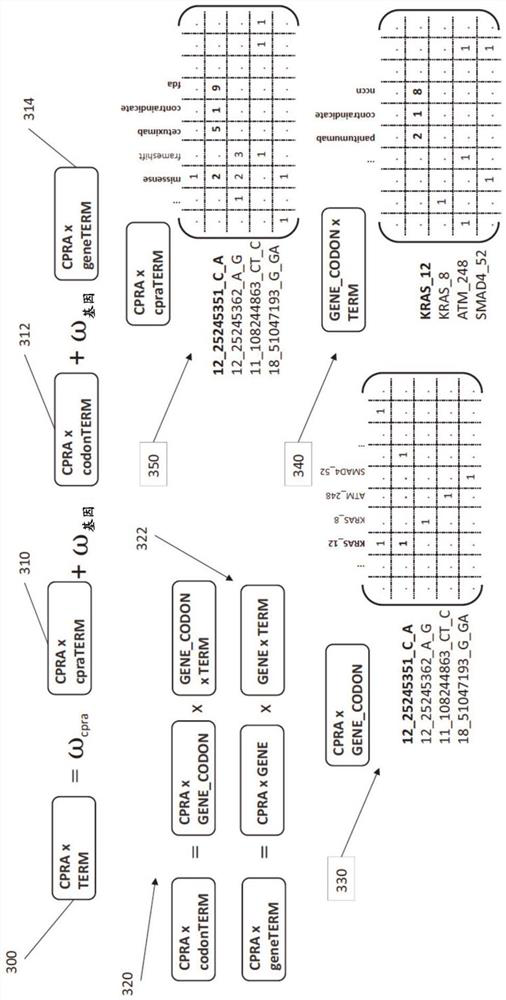
A method is provided for utilizing multi-omic data indices for tumor profiling. The method can comprise the steps of storing a plurality of multi-omic data indices, wherein each of the plurality of multi-omic data indices comprises cancer-specific tokenized data; ingesting additional multi-omic data and any annotations associated with the additional multi-omic data, the additional multi-omic data related to one or more indices; indexing the ingested additional multi-omic data and annotations while preserving gene names, gene variant names and multi-omic mapping between different data streams for the same patient in the specific index, to produce tokenized ingested additional multi-omic data; receiving a user query; selecting one or more relevant multi-omic data indices based on the user query; ranking the selected one or more multi-omic data indices based on at least one of clinical actionability, pathogenicity, feature weight, or frequency; and returning the ranked one or more multi-omic data indices to the user.
Owner:HUMAN LONGEVITY
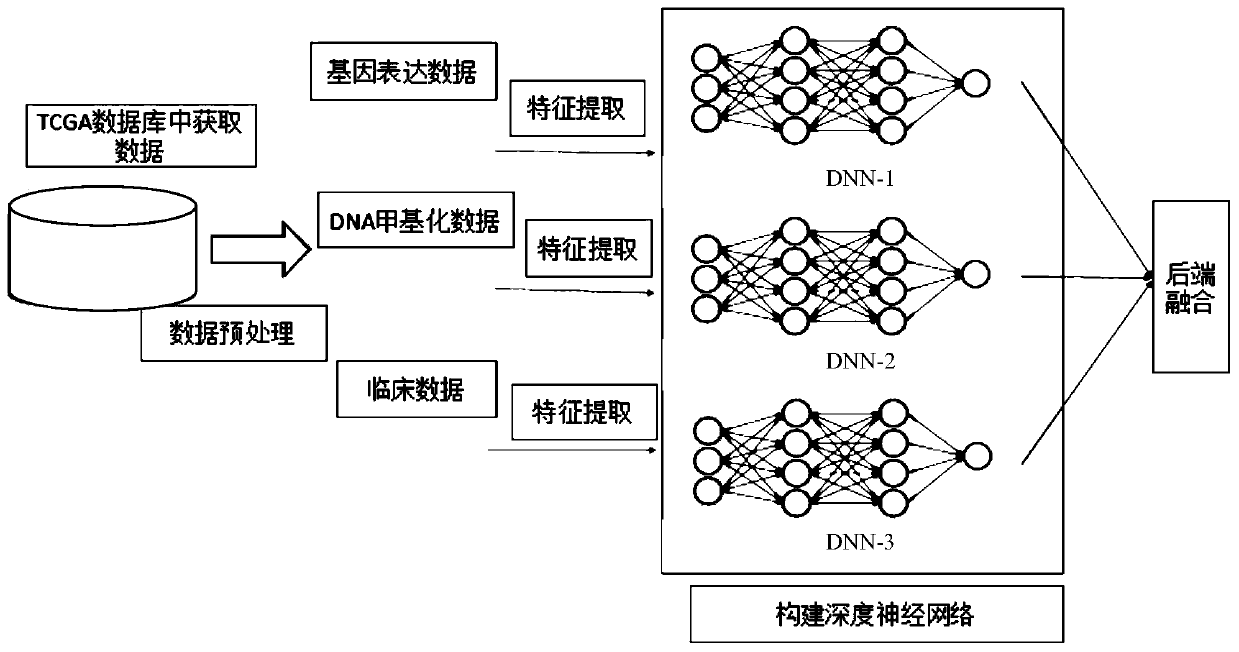
Prediction method for survival time of patient with breast cancer based on deep neural network
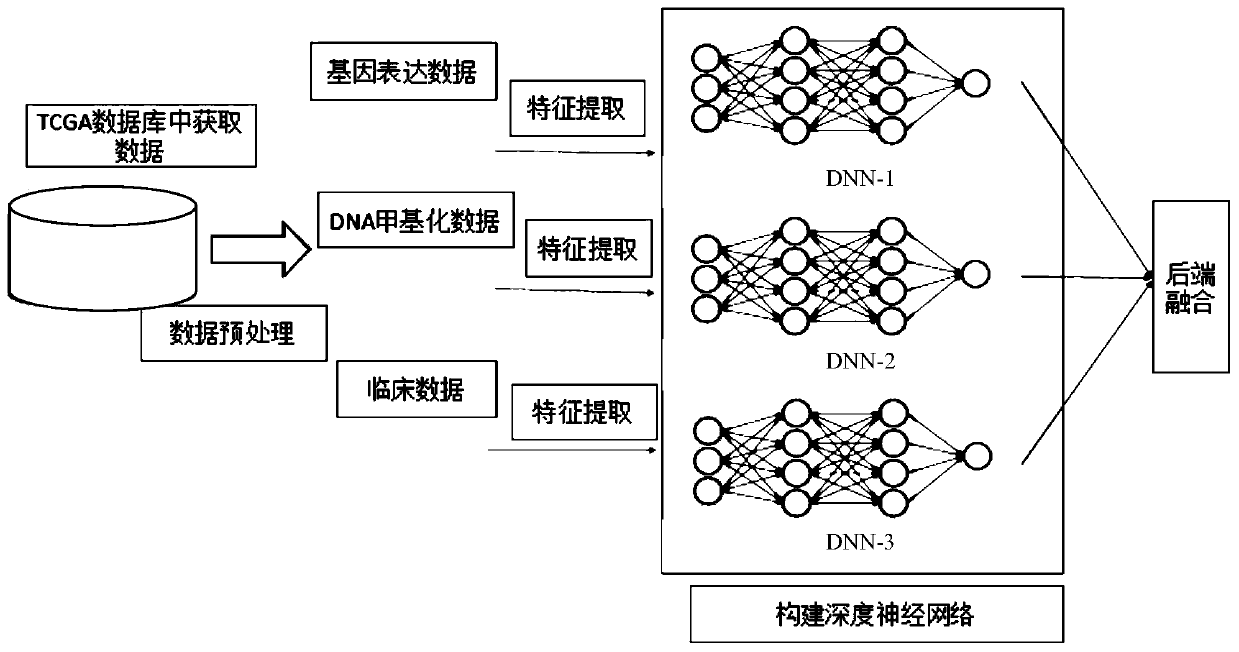
PendingCN111161882AImproving lifetime prediction performanceObvious effectMedical data miningNeural architecturesDNA methylationData set
The invention relates to a prediction method for the survival time of a patient with breast cancer based on a deep neural network. The method comprises the following steps: 1) acquiring data, whereinthe data is clinical data and omics data, and the omics data comprises gene expression data and DNA methylation data; 2) preprocessing the data; 3) extracting the features of a data set; and 4) constructing a deep neural network. According to the invention, survival time prediction is carried out by fusing the deep neural network with the multi-omics data of breast cancer; clinical data, gene expression data and DNA methylation data of breast cancer are obtained from a TCGA database, data features are extracted, deep neural network models are constructed respectively, and then back-end fusionis performed, so the performance of survival time prediction for breast cancer is improved, and a lifetime prediction model is obtained; and the method has a remarkable effect in prediction of the survival time of patients with breast cancer.
Owner:SHENZHEN INST OF ADVANCED TECH
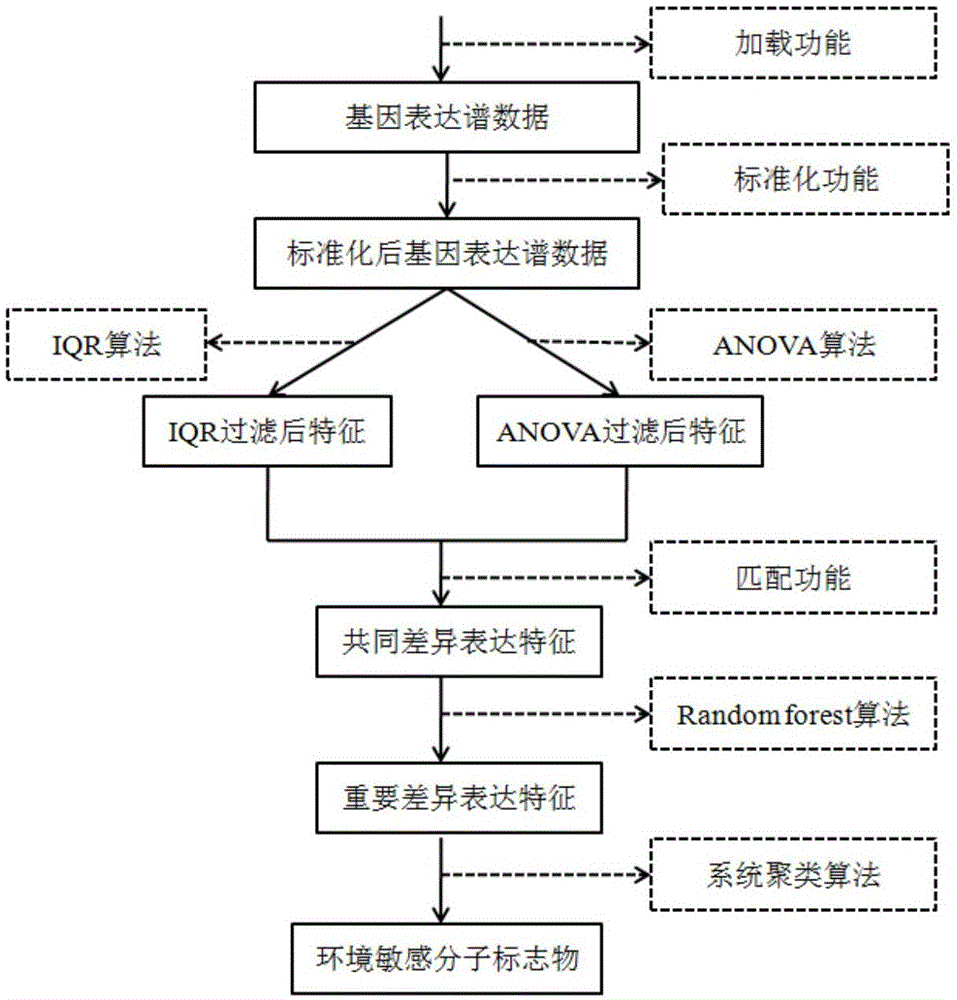
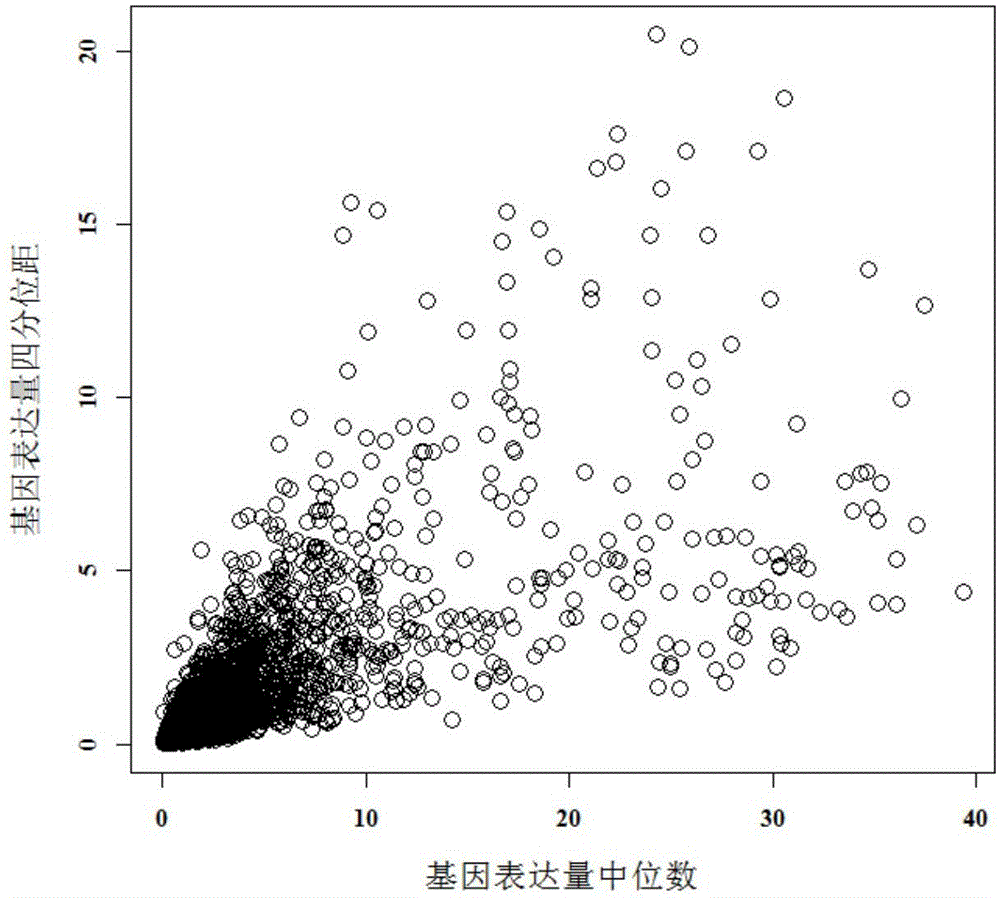
Method for screening environmentally sensitive biomolecules
ActiveCN105117617AEasy to filterPrecise screeningSpecial data processing applicationsBio moleculesData set
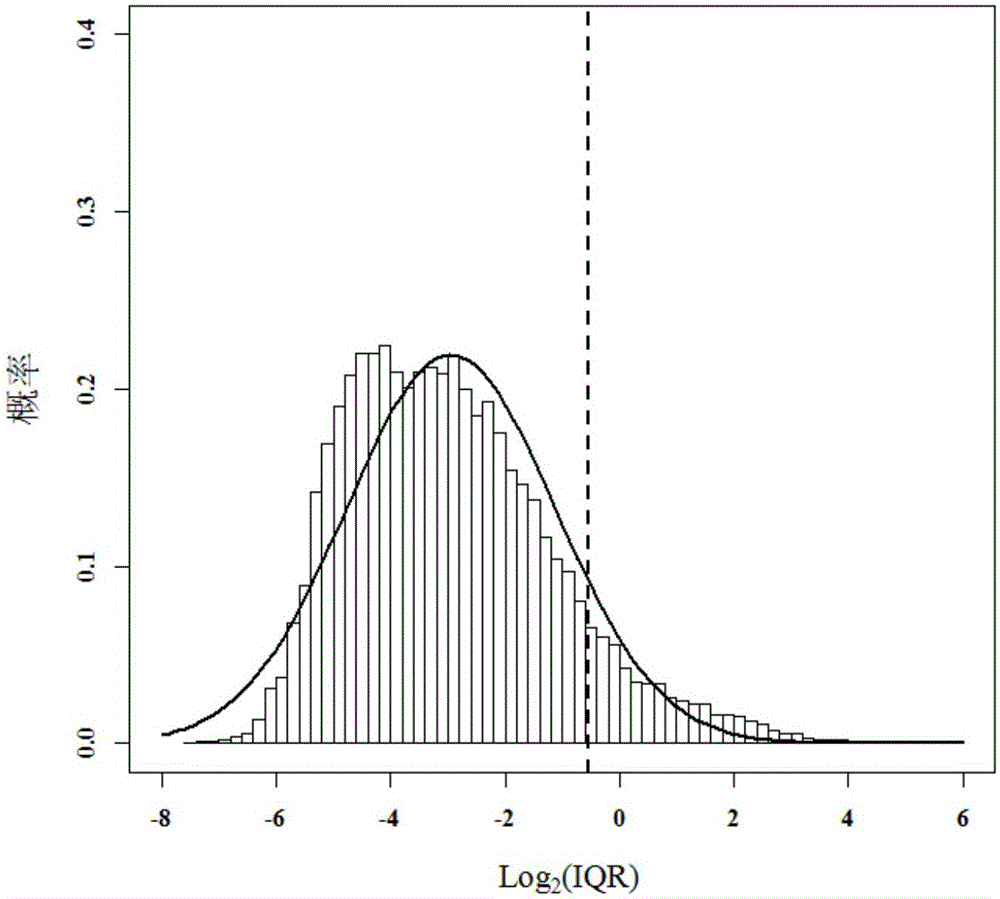
The present invention discloses a method for screening environmentally sensitive biomolecules. The method comprises the following steps of: performing loading and standardization processing on omics data; calculating an interquartile range and a set threshold value of a feature; using the threshold value for performing filtering with an interquartile range algorithm to obtain a differential expression feature; and then using a variance analysis algorithm for filtering a standardized data set to obtain a differential expression feature. In combination with the two algorithms, features of common differential expression changes are matched and sequenced with a random forests algorithm so as to obtain an important differential expression feature. On this basis, by cluster analysis, environmentally sensitive molecular markers are determined. The combination algorithm for screening the environmentally sensitive molecule markers, provided by the present invention, shortens calculation time, improves accuracy, gives a feature importance sequence, rapidly locates an environmentally sensitive target molecule for a biologist, reveals a biological response mechanism for early warning and prevention, and provides an efficient and convenient data processing tool.
Owner:DALIAN MARITIME UNIVERSITY
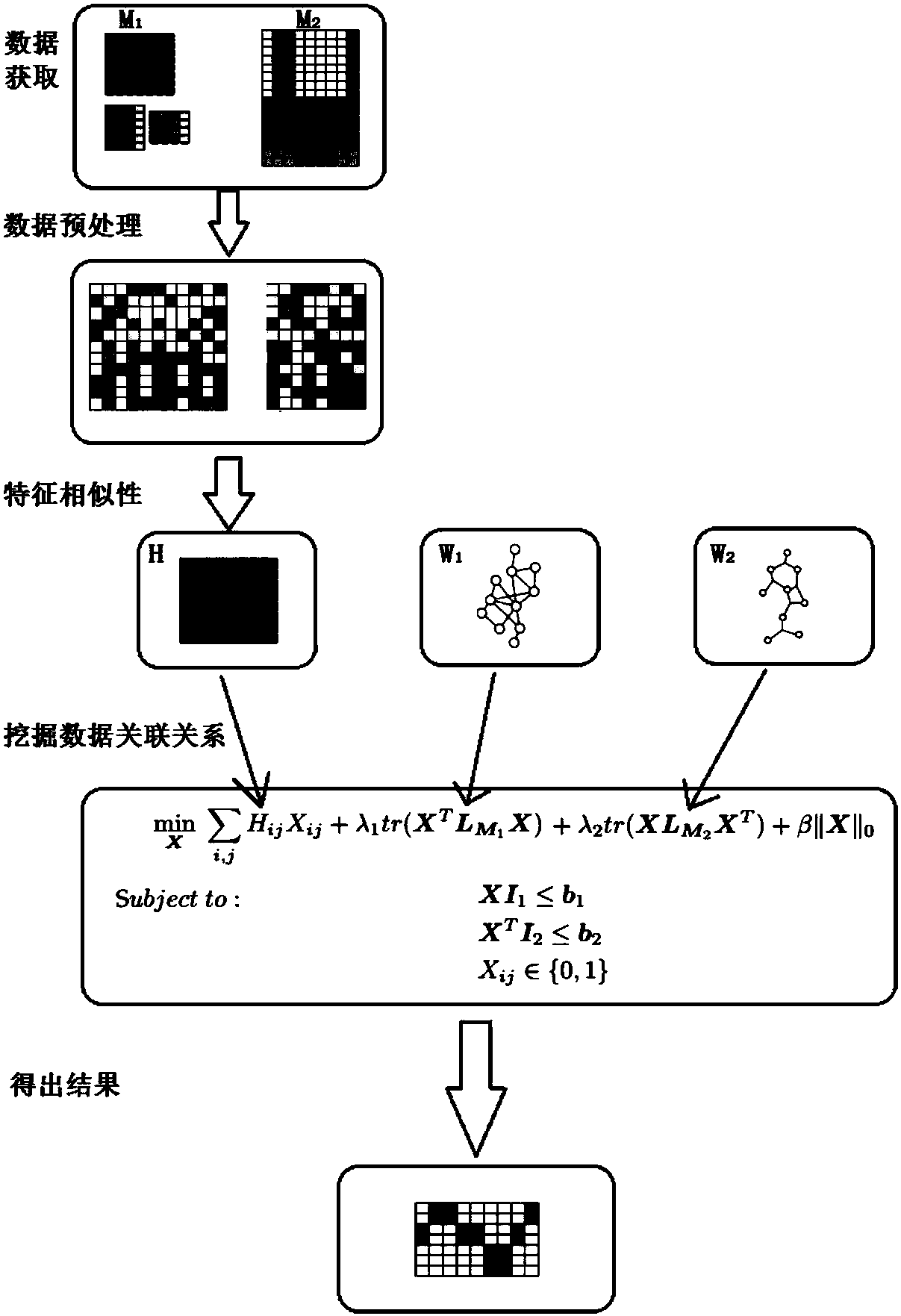
Multi-omics data association relationship discovery method based on sparse matching
ActiveCN108509771AHigh precisionImprove robustnessBiostatisticsSpecial data processing applicationsPrior informationMulti omics
The invention discloses a multi-omics data association relationship discovery method based on sparse matching. The method includes: preprocessing input data to improve quality of the data; selecting asuitable similarity measure according to data characteristics to calculate a similarity matrix among data features; and fusing prior information to mine potential association relationships among thedata features on the basis of a similarity network among the features. According to the method of the invention, the prior information of features of existing already-proven omics data can be fully utilized, impacts of noises on results can be reduced, uncertainty caused by data errors can be reduced, and accuracy and robustness of the results can be improved.
Owner:SOUTH CHINA UNIV OF TECH
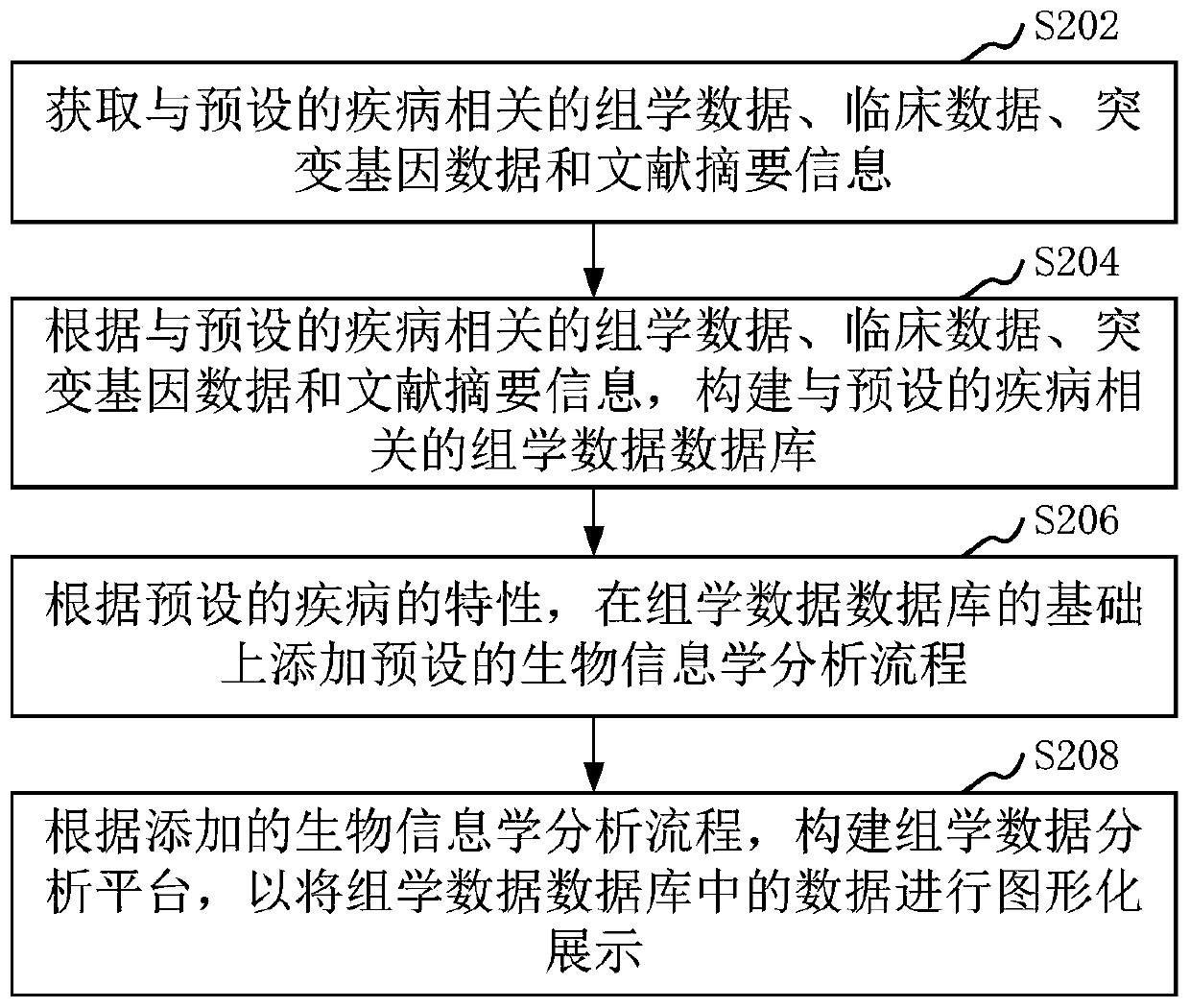
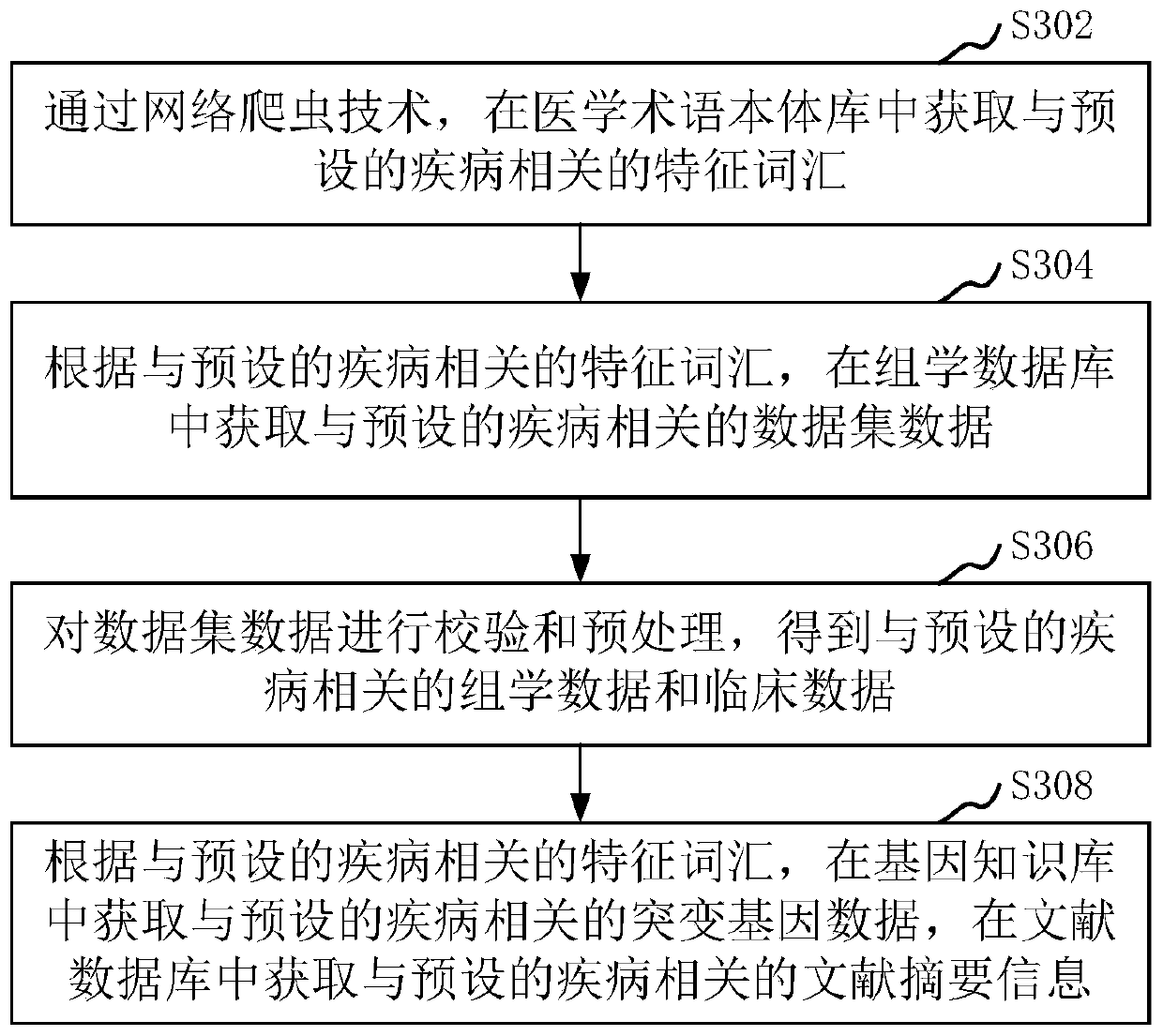
Method and device for constructing omics data analysis platform, and computer device
ActiveCN110570905AImprove system performanceQuality improvementMedical data miningProteomicsDiseaseCandidate Gene Association Study
The invention relates to a method and device for constructing an omics data analysis platform, a computer device and a storage medium. The method includes: obtaining omics data, clinical data, mutation gene data, and literature summary information related to a preset disease; according to the omics data, the clinical data, the mutation gene data, and the literature summary information related to the preset disease, building a omics data database related to the preset disease; according to the characteristics of the preset disease, adding a preset bioinformatics analysis process based on the omics data database; according to the added bioinformatics analysis process, constructing an omics data analysis platform to graphically display the data in the omics data database. The method can construct a systematic and high-quality omics data analysis platform by which a user can group, standard analyze and functionally annotate candidate gene samples based on the clinical characteristics of acertain type of disease intuitively and conveniently.
Owner:GENERAL HOSPITAL OF PLA
Liver cancer image omics data processing method, system and device and storage medium
ActiveCN110175978AStable evaluation effectOvercoming the disadvantage of not considering tumor heterogeneityImage enhancementImage analysisUpper abdomenOmics data
The invention discloses a liver cancer image omics data processing method, system and device and a storage medium. The method comprises the steps of obtaining an upper abdomen CT image of a liver cancer patient, calculating image omics data corresponding to the upper abdomen CT image, calculating a linear combination of the plurality of image omics characteristic values, and dividing the liver cancer patient into a hepatic artery chemoembolization reaction group or a hepatic artery chemoembolization reaction-free group according to a magnitude relation between the predicted value and a presetthreshold value. The used imaging omics technology can extract the tumor feature information contained in the CT image from the three-dimensional perspective in an omnibearing mode, and the defect that tumor heterogeneity is not considered in an existing method is overcome; and patients are divided into a reaction group and a non-reaction group according to scores of the imaging characteristics ofpatients, so that the clinical guiding significance is good, and a better decision reference is provided for clinical workers. The method is widely applied to the technical field of data processing.
Owner:NANFANG HOSPITAL OF SOUTHERN MEDICAL UNIV
Calculation method for calculating lung cancer genotyping by using PET/CT image
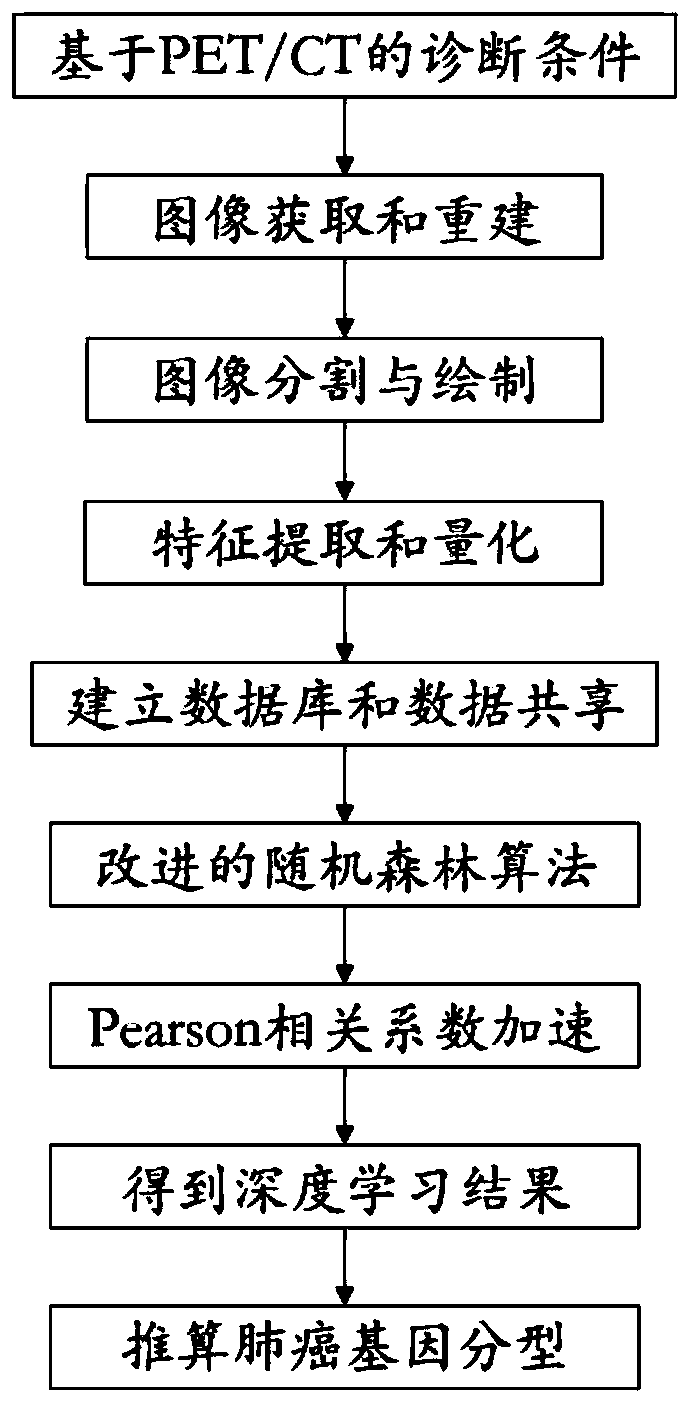
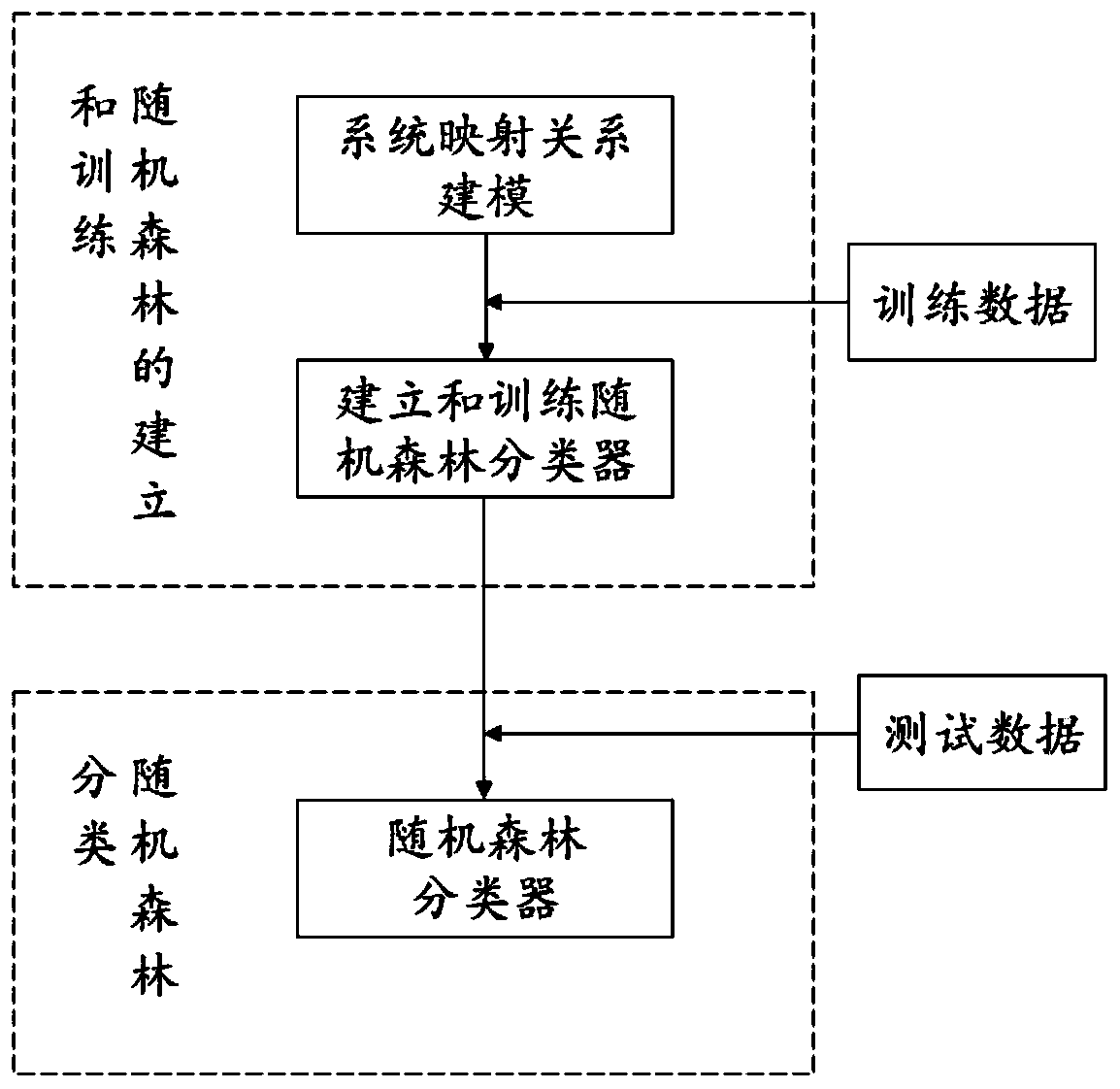
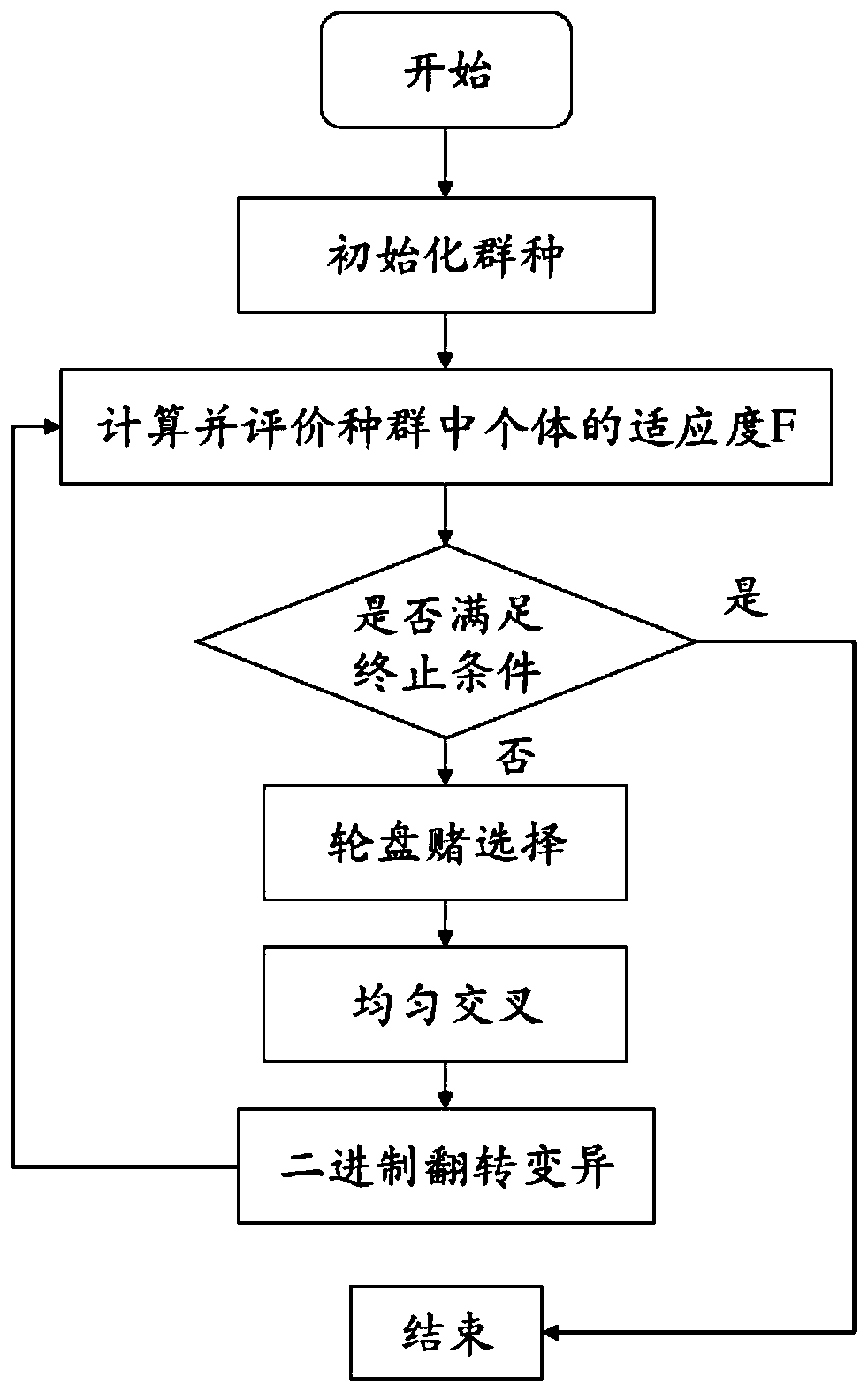
ActiveCN111445946APuncture injuryImprove diagnostic efficiencyMedical automated diagnosisCharacter and pattern recognitionGenomicsCorrelation coefficient
The invention relates to a calculation method for calculating lung cancer genotyping by using a PET / CT image. Outlines of lesions are sketched layer by layer, three-dimensional volume recombination iscarried out on a two-dimensional tumor region to generate a three-dimensional interested volume, and characteristic data is extracted from the three-dimensional interested volume; once the lesion (tumor) area is determined, image features can be extracted; image omics data is directly extracted and analyzed, and a radiogenomics data model is established; a function or mathematical model is established according to the extracted characteristics of each classification of the radiogenomics; an improved random forest algorithm is established; model prediction is improved by using a Pearson correlation coefficient; and lung cancer genotyping is calculated according to deep learning, thereby improving the diagnosis efficiency of the lung cancer.
Owner:SHANDONG RES INST OF TUMOUR PREVENTION TREATMENT +2
Precise intelligent diagnosis and treatment big data system
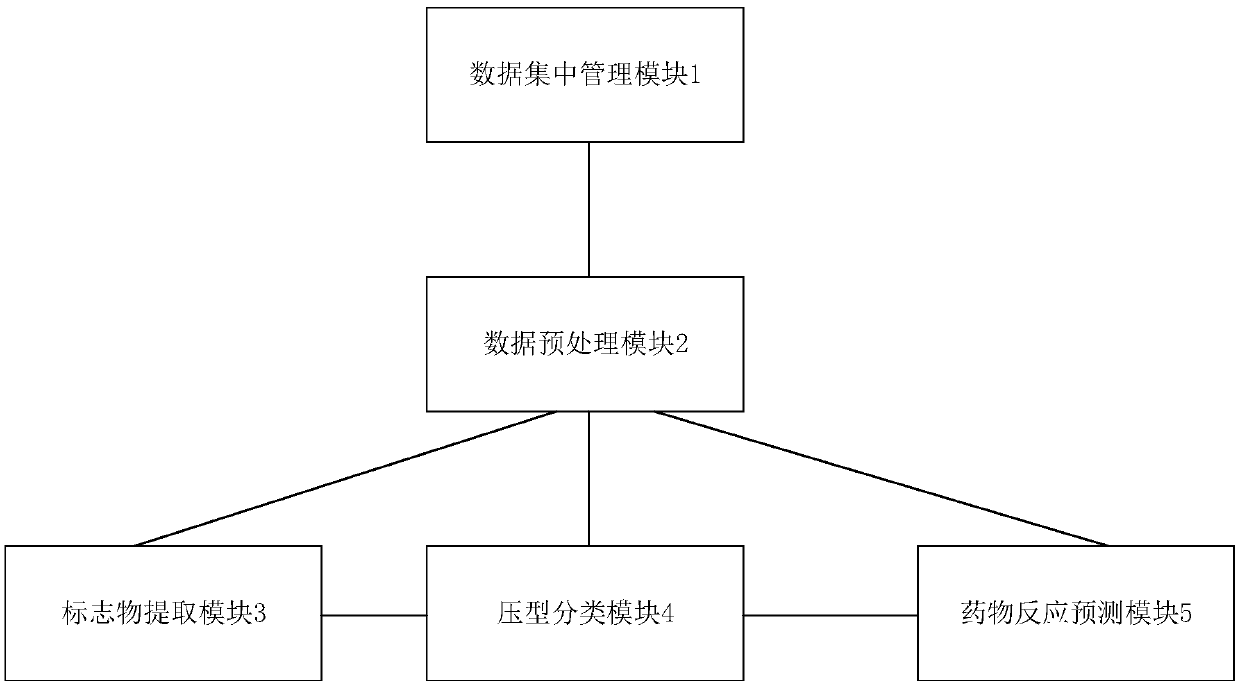
ActiveCN109599157AAchieve sharingRealize big data managementMedical data miningDrug and medicationsMedical recordData system
The invention relates to a precise intelligent diagnosis and treatment big data system which comprises a centralized data management module, a data preprocessing module, a marker extraction module, asubtype classification module and a medicine reaction prediction module, wherein the centralized data management module is used for centrally managing clinical electronic medical history data and omics data of multiple medical institutes; the data preprocessing module is used for preprocessing the centrally managed data and used for establishing a relationship dependency net on the basis of biomedicine characteristics; the marker extraction module is used for extracting characteristics genes of patients to obtain marker sets on the basis of preprocessed data; the subtype classification moduleis used for classifying subtypes of the patients and used for confirming corresponding groups of the patients; the medicine reaction prediction module is used for establishing medicine reaction prediction models and used for predicting reactions of the patients upon different medicines according to the medicine reaction prediction models. Compared with the prior art, the system is capable of achieving effective management on medical data and medicine reaction prediction, and intelligentization can be achieved.
Owner:TONGJI UNIV
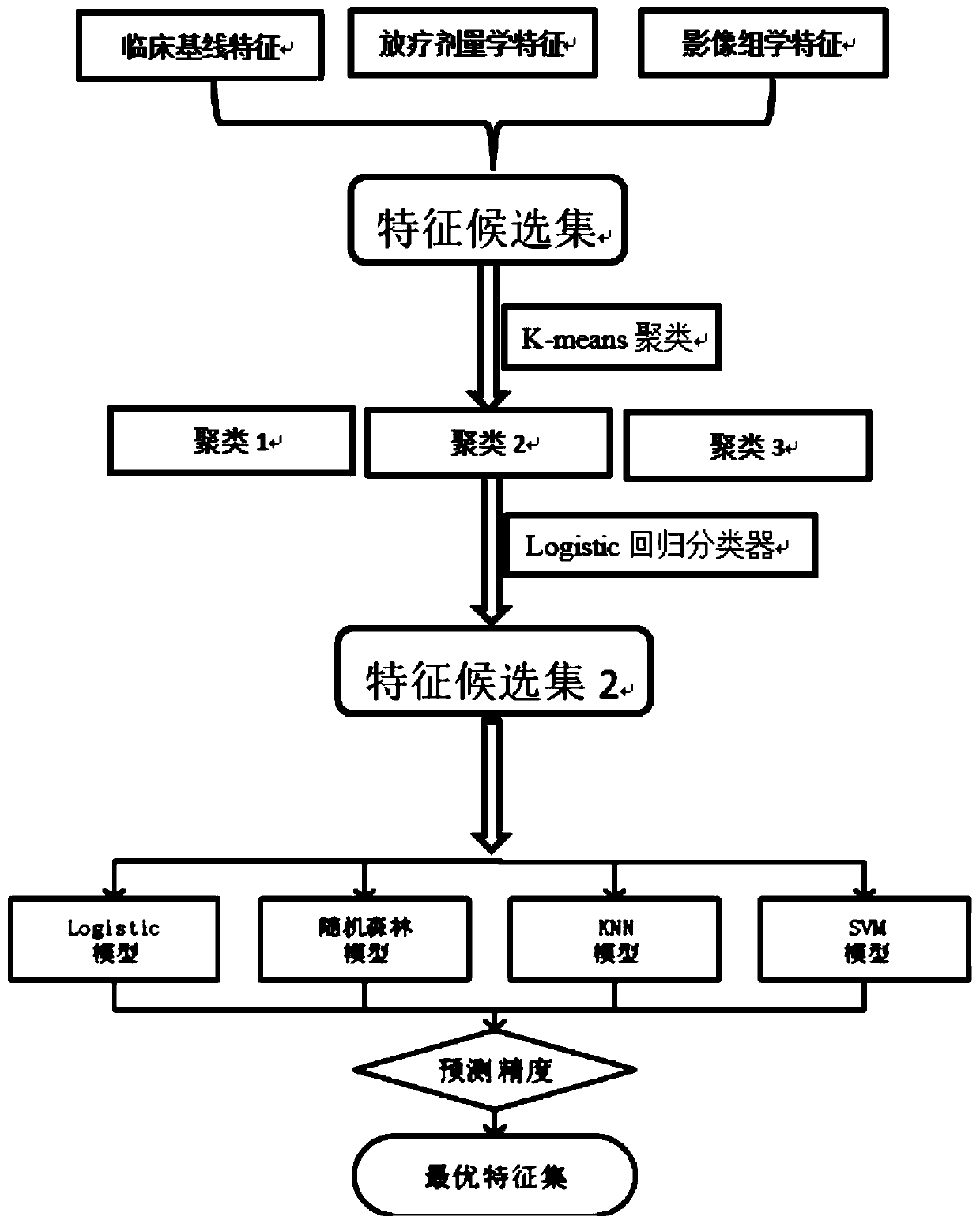
Method for predicting complications of normal tissue organs after tumor radiotherapy
ActiveCN110604550AGood treatment effectQuality improvementDiagnostic recording/measuringSensorsTumor targetLife quality
The invention relates to a technology for predicting clinical complications after radiotherapy of tumor patients, in particular to a method for predicting complications of normal tissue organs after tumor radiotherapy based on the multimodal image omics characteristics and the radiotherapy dosimetry characteristics. The method mainly comprises the steps: S1, a multimodal image database is established; S2, image data of endangered organs near a tumor target area are extracted; S3, the image omics characteristics of the endangered organs are extracted, and the characteristics of normal organ image data are extracted; S4, parameters of the image phenotypic characteristics of the endangered organs are extracted according to image segmentation results; S5, the image omics characteristics are analyzed; S6, parameters of the exposure doses of the endangered organs are extracted; and S7, the characteristics are collected and extracted. The complications after radiotherapy and chemotherapy of the tumor patients are predicted through the image omics data, effective treatment and intervention can be provided for the patients in time through the reliable, safe and high-precision prediction method, the complications are reduced, and thus the treatment effect on the patients and the later life quality of the patients are improved.
Owner:CANCER CENT OF GUANGZHOU MEDICAL UNIV
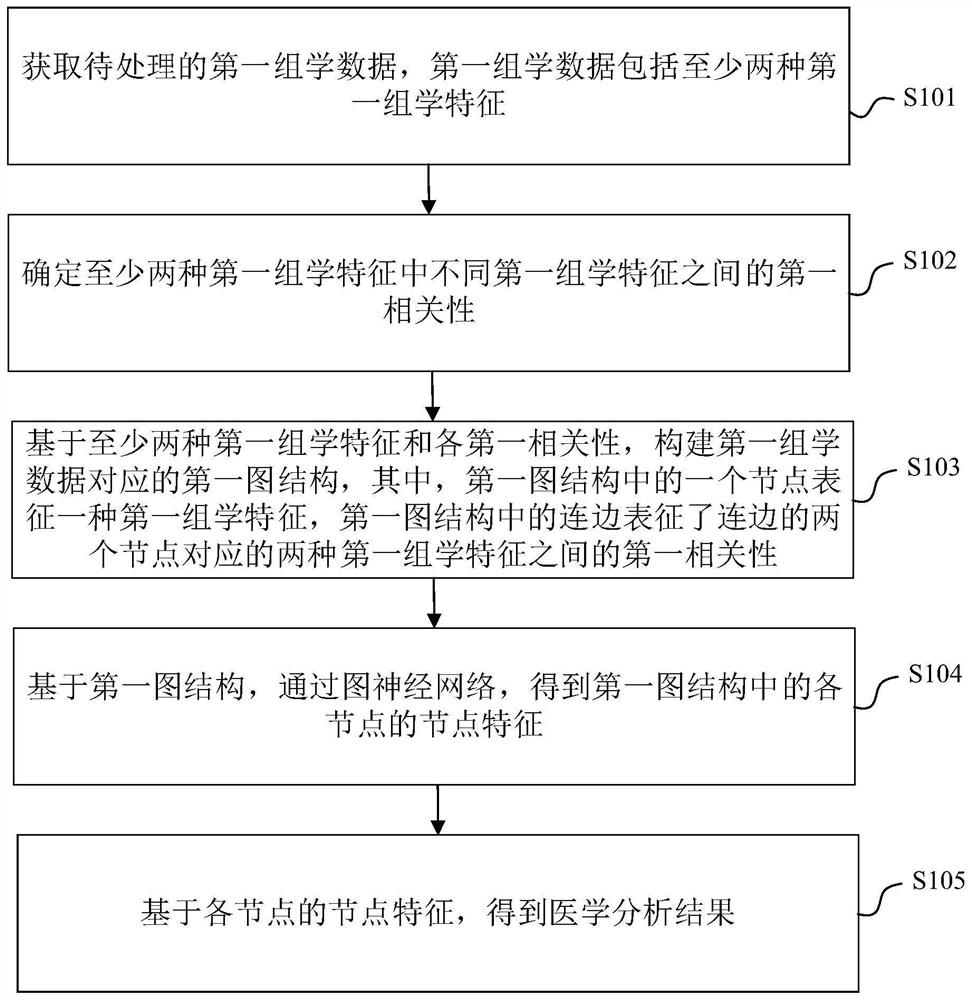
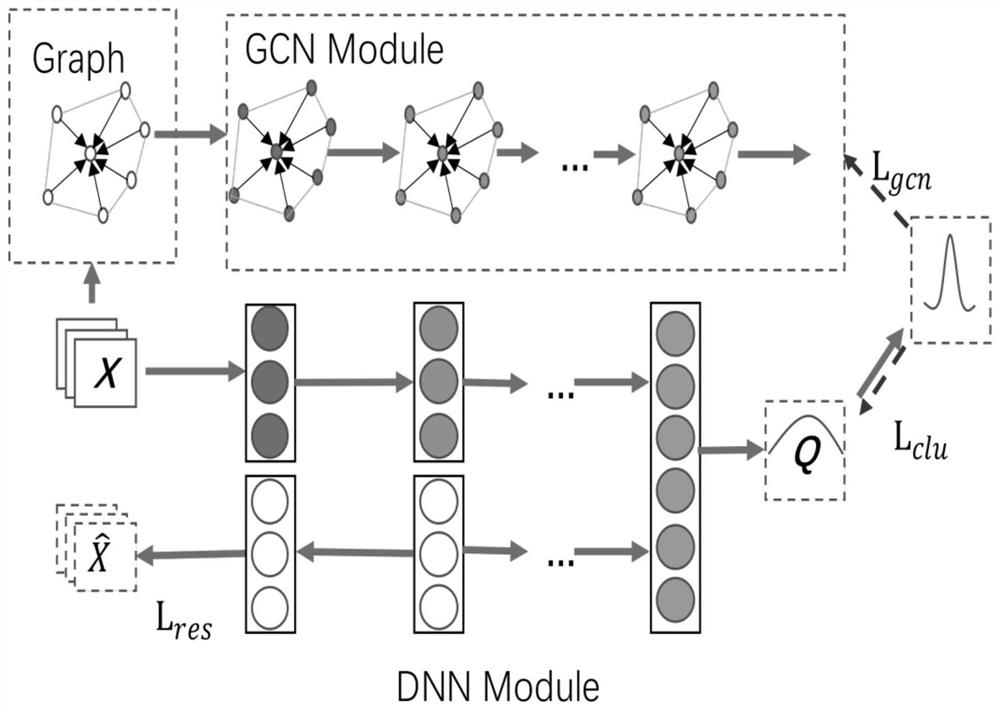
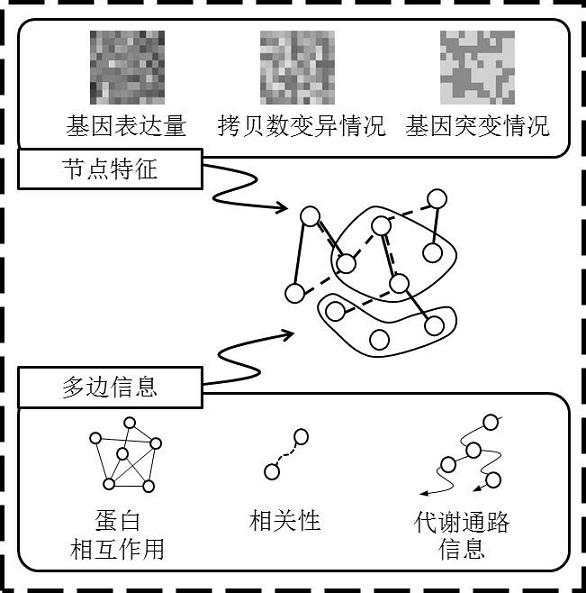
Omics data processing method and device based on a graph neural network, equipment and medium
ActiveCN112364880AIncrease contentAccurate results of medical analysisMedical simulationHealth-index calculationEngineeringCloud data
The embodiment of the invention provides an omics data processing method and device based on a graph neural network, equipment and a medium, and relates to the technical fields of medical treatment, artificial intelligence, cloud data and the like. The method comprises the following steps: acquiring first omics data to be processed, wherein the first omics data comprises at least two first omics characteristics; determining a first correlation between different omics features in the at least two first omics features; constructing a first graph structure corresponding to the first omics data based on the at least two first omics features and each first correlation, one node in the first graph structure representing one first omics feature, and a connecting edge in the first graph structurerepresenting the first correlation corresponding to two nodes of the connecting edge; based on the first graph structure, obtaining node features of each node in the first graph structure through a graph neural network; and obtaining a medical analysis result based on the node features of each node. According to the invention, the node features can reflect the correlation among the features, and the obtained result is more accurate.
Owner:TENCENT TECH (SHENZHEN) CO LTD
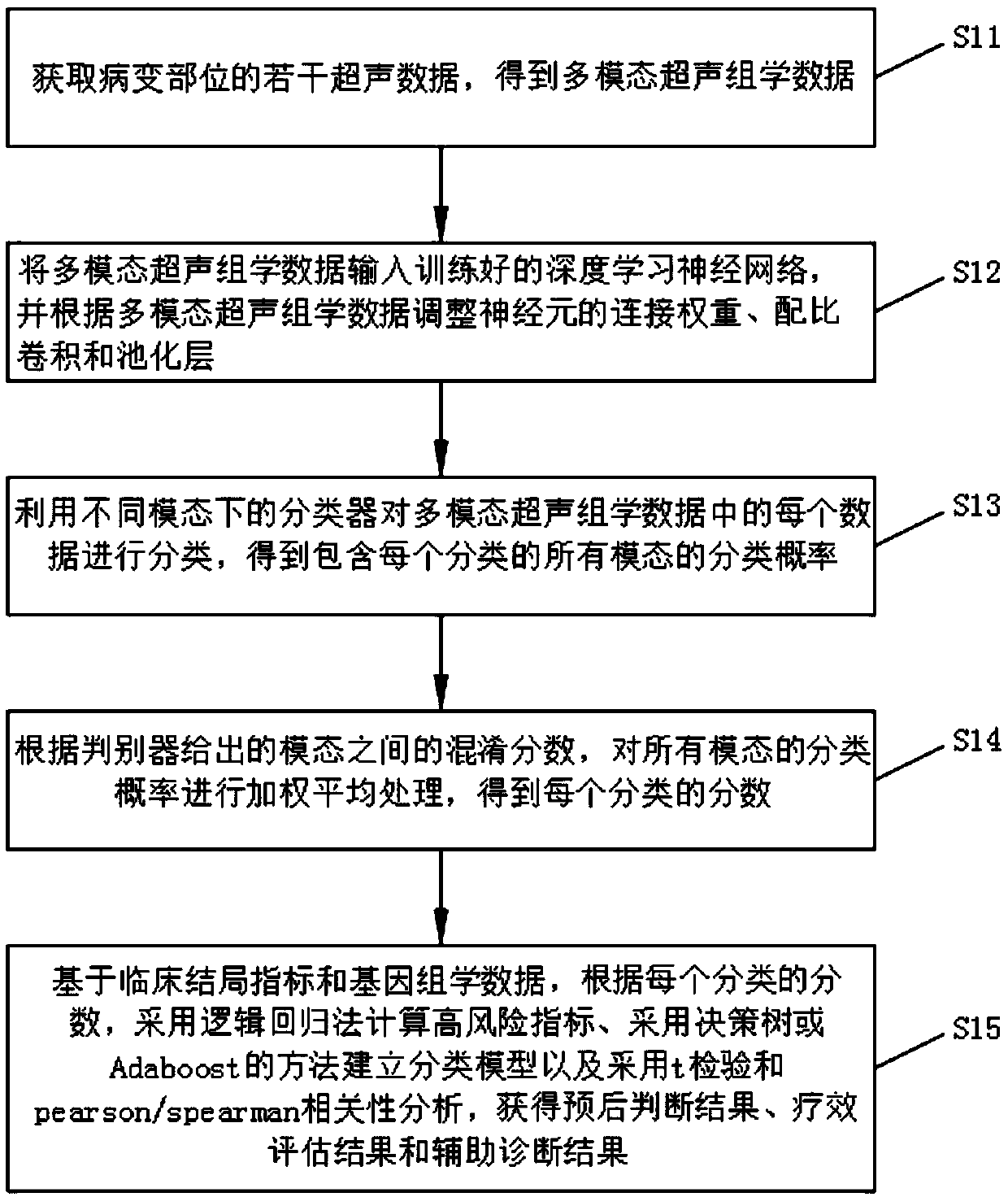
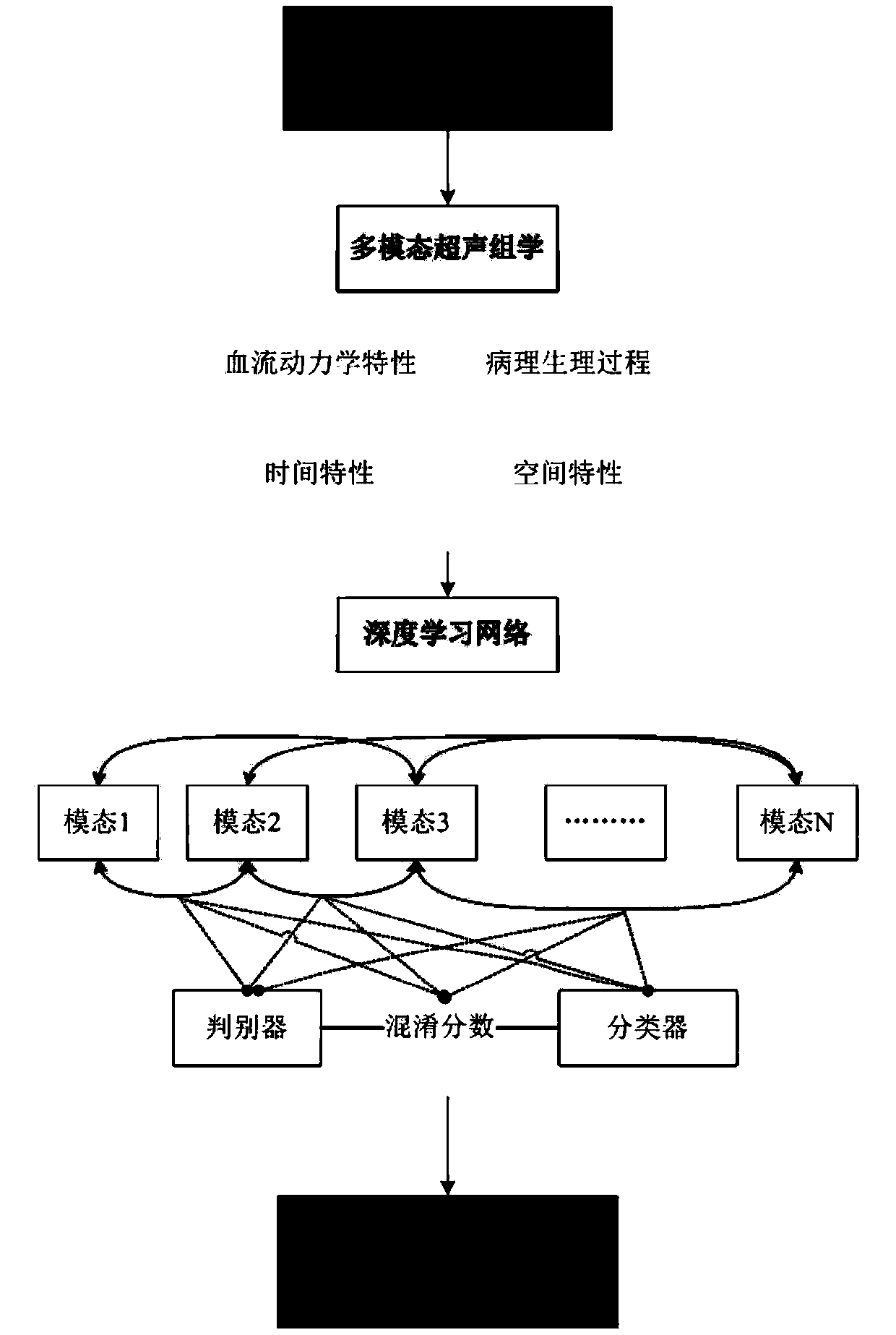
A disease intelligent analysis method and system based on ultrasound omics and deep learning
PendingCN109558896AImprove accuracyReduce complexityMedical automated diagnosisCharacter and pattern recognitionDiscriminatorPattern recognition
The invention discloses an intelligent disease analysis method and system based on ultrasonic omics and deep learning, and the method comprises the steps of obtaining a plurality of pieces of ultrasonic data of a lesion site, and obtaining multi-modal ultrasonic omics data; inputting the multi-modal ultrasound omics data into a deep learning neural network, and adjusting the connection weight, theproportion convolution and the pooling layer of neurons according to the multi-modal ultrasound omics data to obtain adjusted multi-modal ultrasound omics data; and classifying the adjusted multi-modal ultrasonic omics data by using classifiers in different modes, obtaining a score of each classification through a discriminator, and obtaining prognosis judgment, curative effect evaluation and anauxiliary diagnosis result according to the score of each classification. Compared with an existing method for intelligently analyzing diseases by using single-mode ultrasonic data, the technical scheme of the invention optimizes the deep learning network from the aspects of data and model design according to the characteristics of multi-mode ultrasonic omics data, and improves the accuracy and prediction value of intelligent analysis of diseases.
Owner:THE FIRST AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
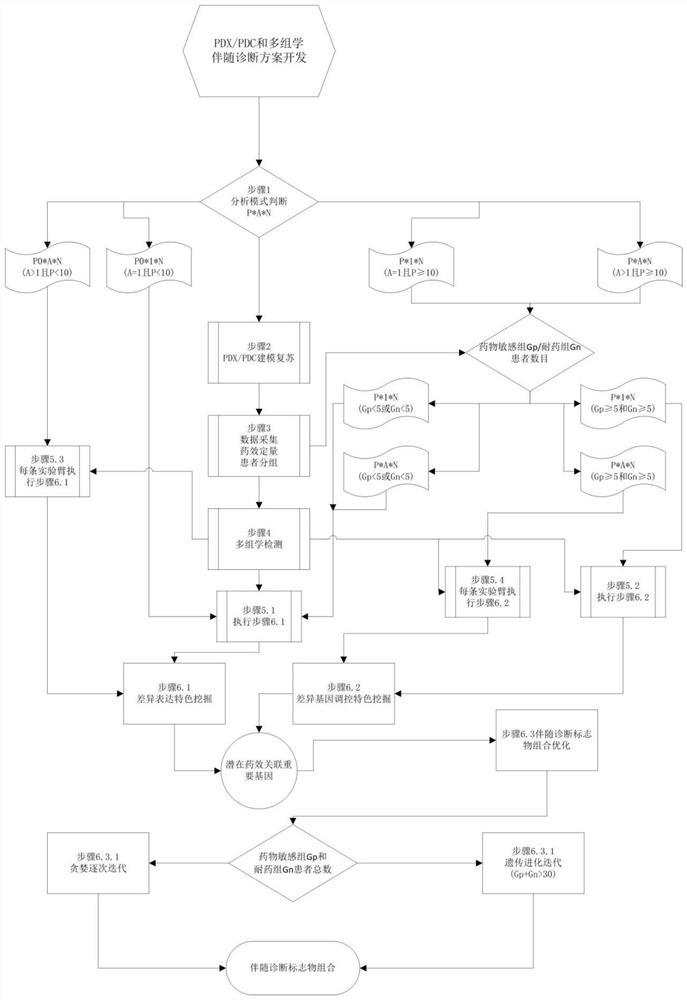
Accompanying diagnosis model based on a PDX or PDC model drug sensitivity experiment and multi-omics detection analysis and application
Owner:上海朴岱生物科技合伙企业(有限合伙)
Multi-omics intelligent diagnosis system based on deep learning
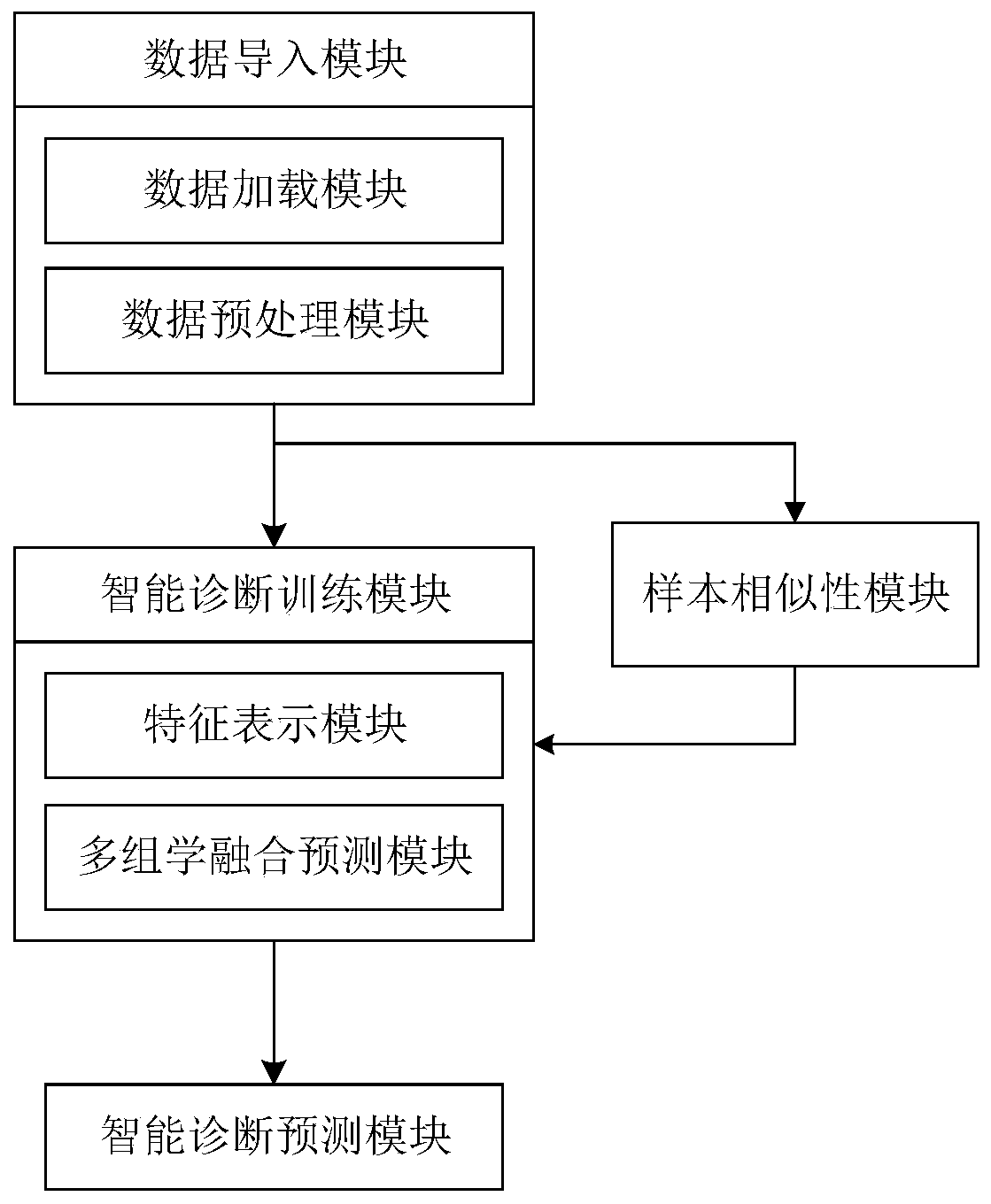
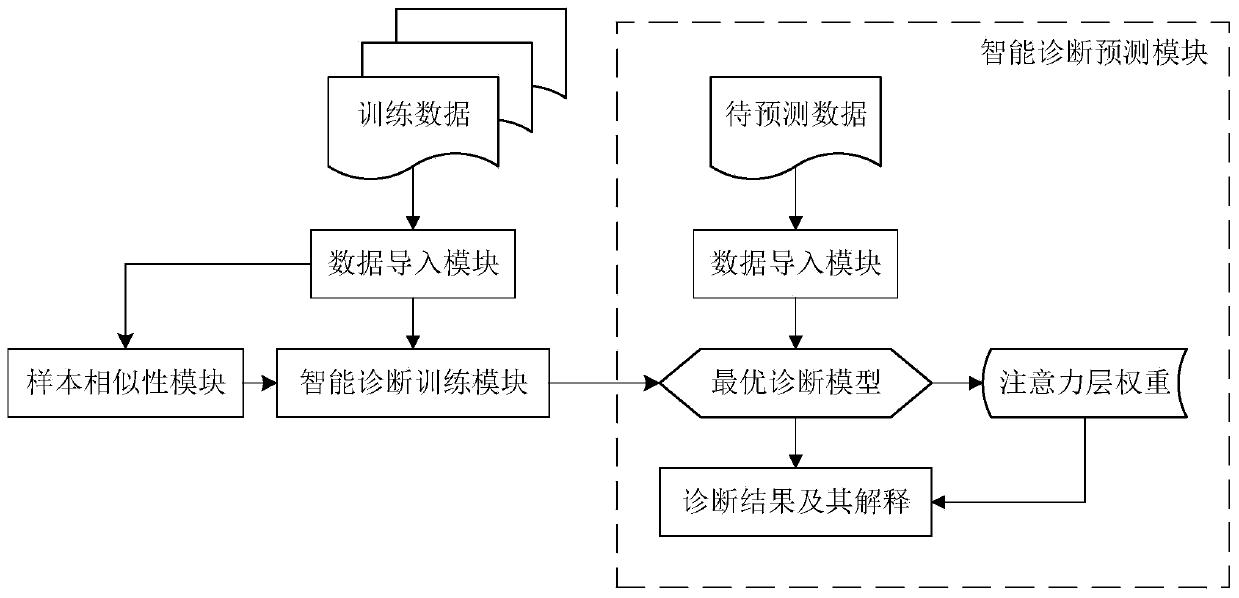
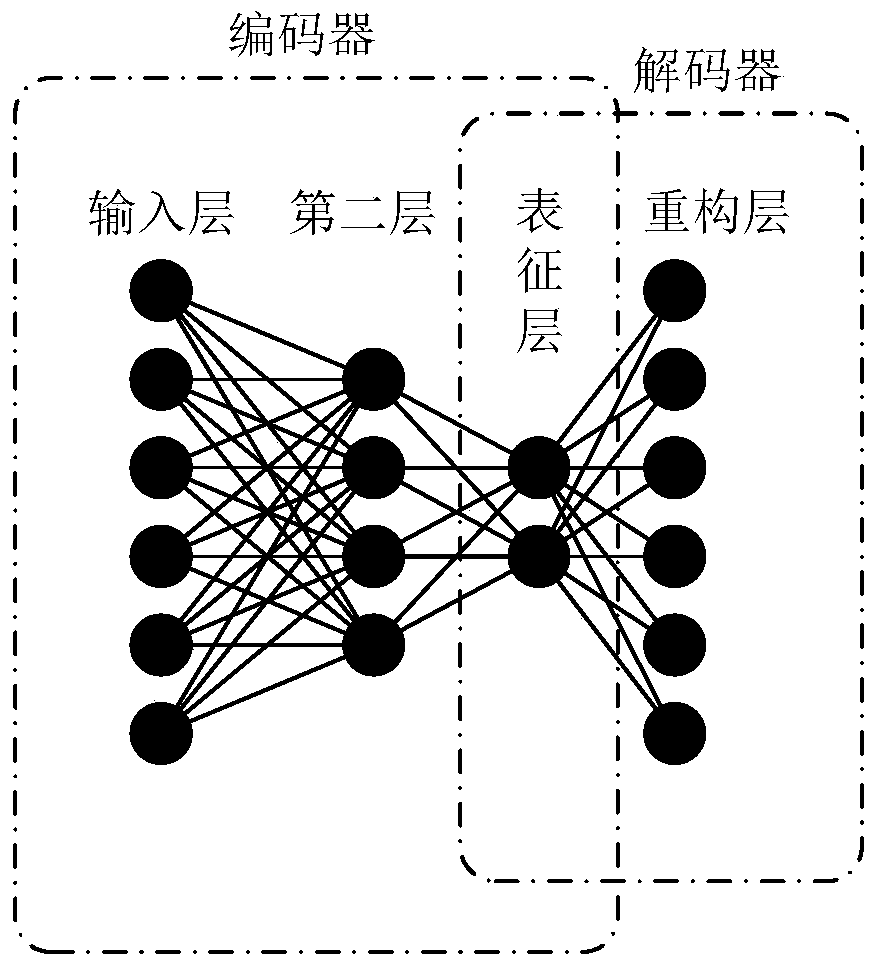
ActiveCN111028939AImprove accuracyUnderstand the formation principleMedical data miningMedical automated diagnosisDiseaseOmics data
The invention discloses a multi-omics intelligent diagnosis system based on deep learning, and the system comprises a data importing module which is used for loading multi-omics data and clinical data, and preprocesses the data; a sample similarity module which is used for constructing a multi-omics sample similarity matrix; an intelligent diagnosis training module which is used for carrying out feature representation by utilizing an automatic encoder, carrying out multi-omics feature fusion by utilizing a multi-view attention mechanism neural network and integrating a sample similarity moduleresult into a training process to finally obtain an optimal diagnosis model; and an intelligent diagnosis and prediction module which is used for carrying out intelligent diagnosis according to the multiple omics data and providing result explanation. According to the invention, the deep learning technology is combined with the multi-omics data, and the diagnosis result and interpretability of the disease are provided, so that the multi-omics intelligent diagnosis system based on deep learning is formed, the disease diagnosis capability is improved, and the interpretability of the diagnosis result is provided.
Owner:SOUTH CHINA UNIV OF TECH
Clustering analysis method and system for multi-omics data
PendingCN113392894AHigh utility valueCharacter and pattern recognitionNeural learning methodsCluster algorithmData set
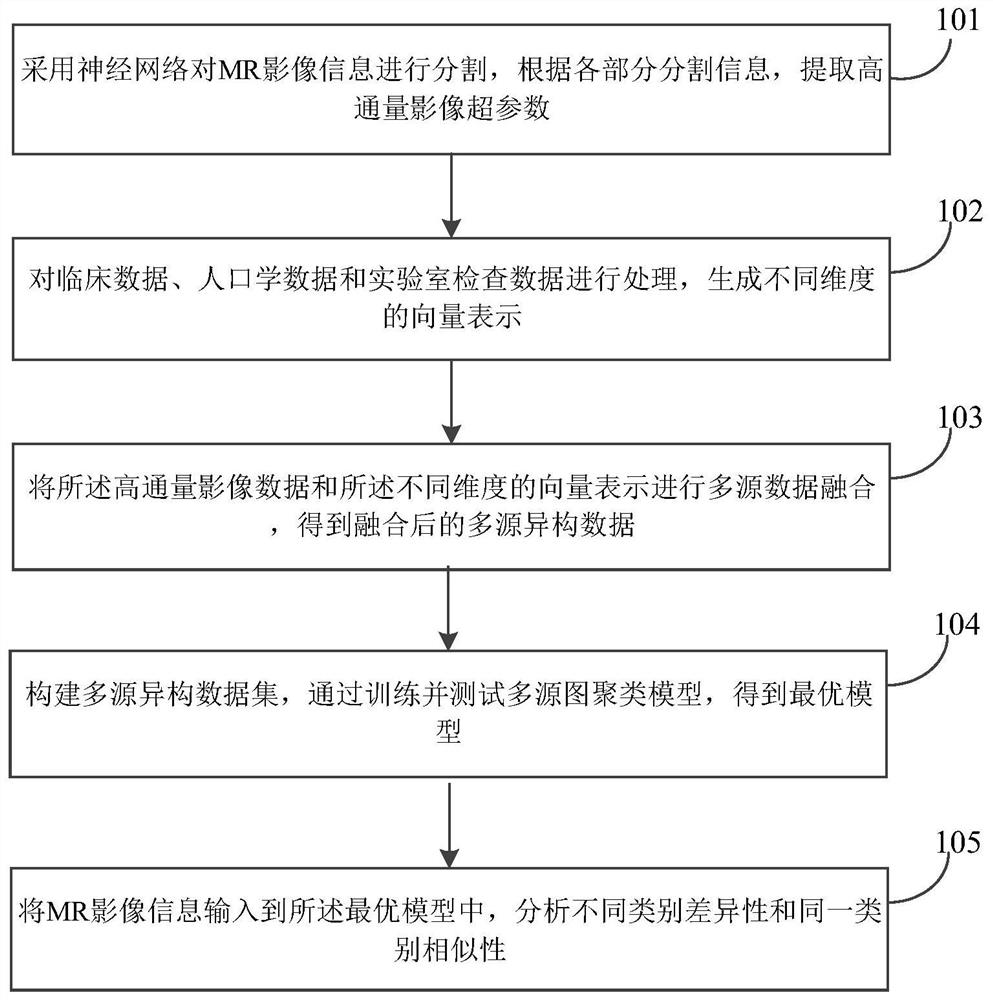
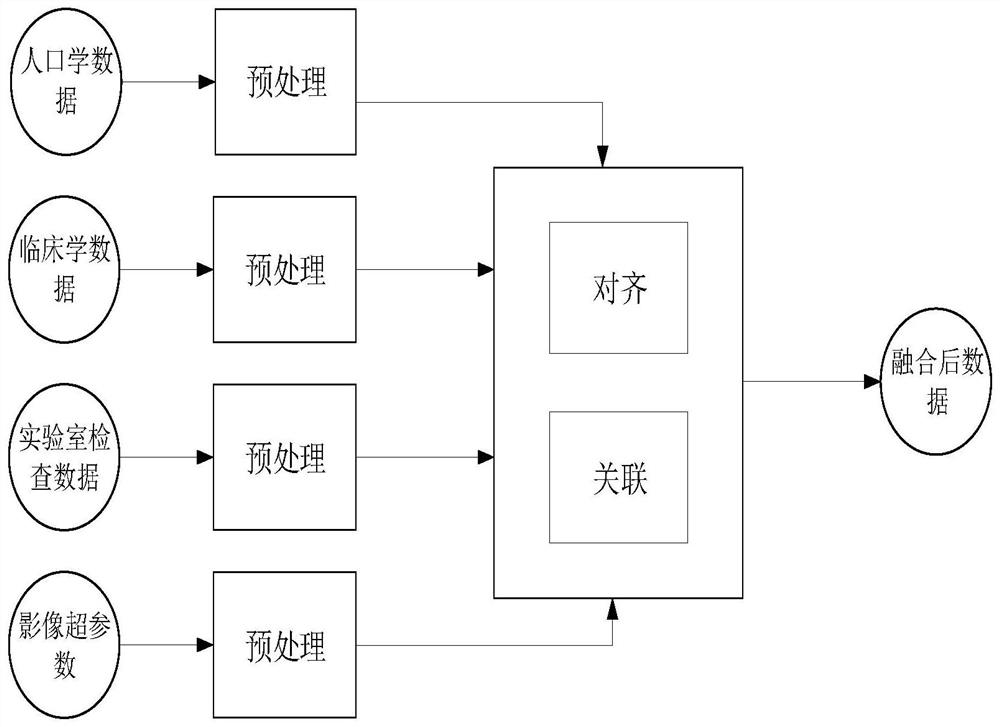
The embodiment of the invention discloses a clustering analysis method and system for multi-omics data, and the method comprises the steps: segmenting MR image information through employing a neural network, and extracting a high-throughput image hyper-parameter according to the segmentation information of each part; processing the clinical data, the demographic data and the laboratory inspection data to generate vector representations of different dimensions; performing multi-source data fusion on the high-throughput image data and the vector representations of different dimensions to obtain fused multi-source heterogeneous data; constructing a multi-source heterogeneous data set, and training and testing a multi-source graph clustering model to obtain an optimal model; inputting MR image information into the optimal model, and analyzing differences of different categories and similarities of the same category. A graph structure mode is adopted, the correlation condition between data is visually expressed, different features are captured, the model is more robust, the efficient clustering algorithm based on the graph neural network model is achieved, and the practical value is very high.
Owner:瓴域影诺北京科技有限公司
Sample clustering and feature recognition method based on integrated non-negative matrix factorization
ActiveCN110826635AImprove performanceImprove robustnessCharacter and pattern recognitionICT adaptationAlgorithmOmics
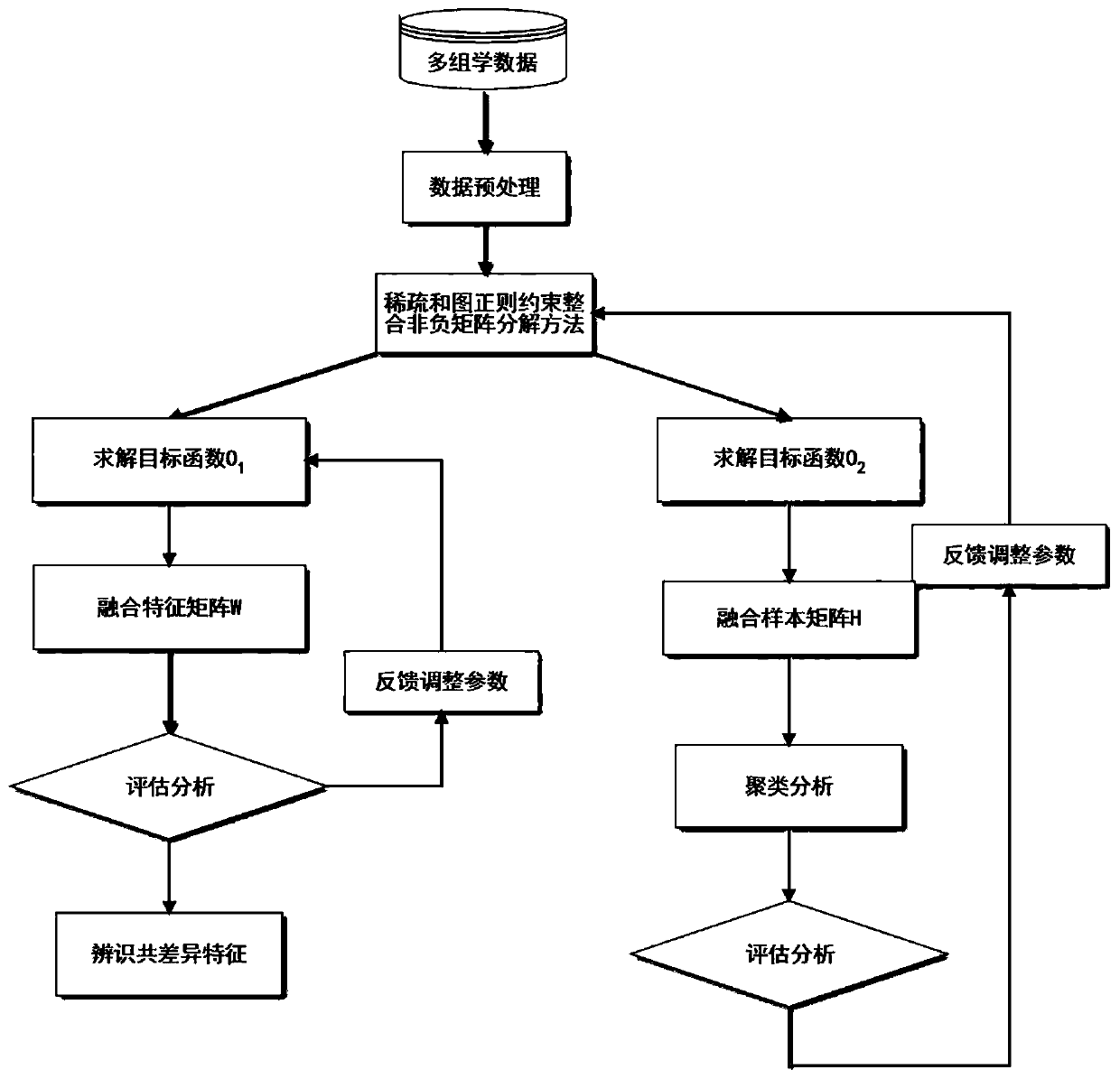
The invention discloses a sample clustering and feature recognition method based on integrated non-negative matrix factorization. The method comprises: 1, X = {X1, X2... XP} representing multi-view data composed of P different omics data matrixes of the same cancer; 2, constructing a diagonal matrix Q; 3, introducing graph regularization and sparse constraints into the integrated non-negative matrix factorization framework to obtain target functions O1 and O2; 4, solving the target function O1 to obtain a fusion feature matrix W and a coefficient matrix HI; solving the target function O2 to obtain a feature matrix WI and a fusion sample matrix H; 5, constructing an evaluation vector according to the fusion feature matrix W, and identifying common difference features according to the vector; 6, performing functional explanation on the identified common difference characteristics by using GeneCards; and 7, performing sample clustering analysis according to the fusion sample matrix. According to the method, the complementary and difference information of the multiple omics data can be fully utilized to identify the common difference characteristics, clustering analysis can be carriedout on the sample data provided by the multiple omics data, and a calculation method basis is provided for integrated research of different types of omics data.
Owner:QUFU NORMAL UNIV
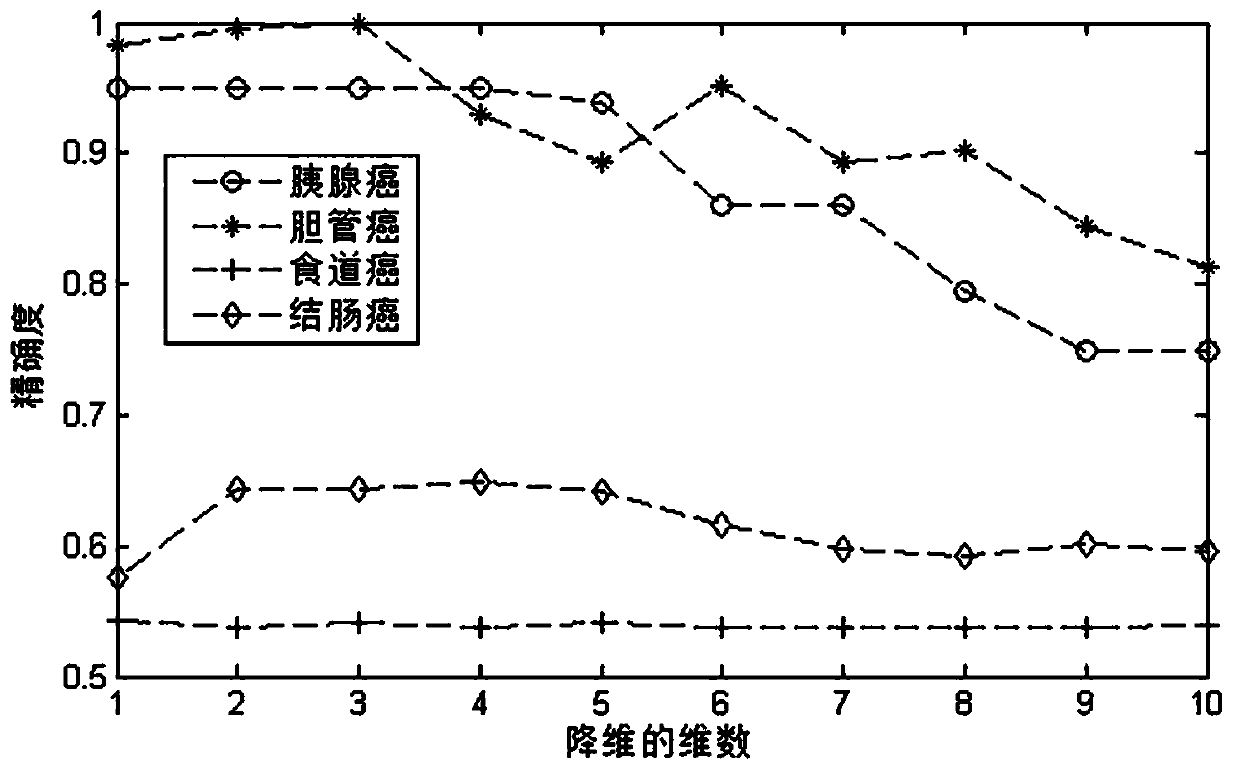
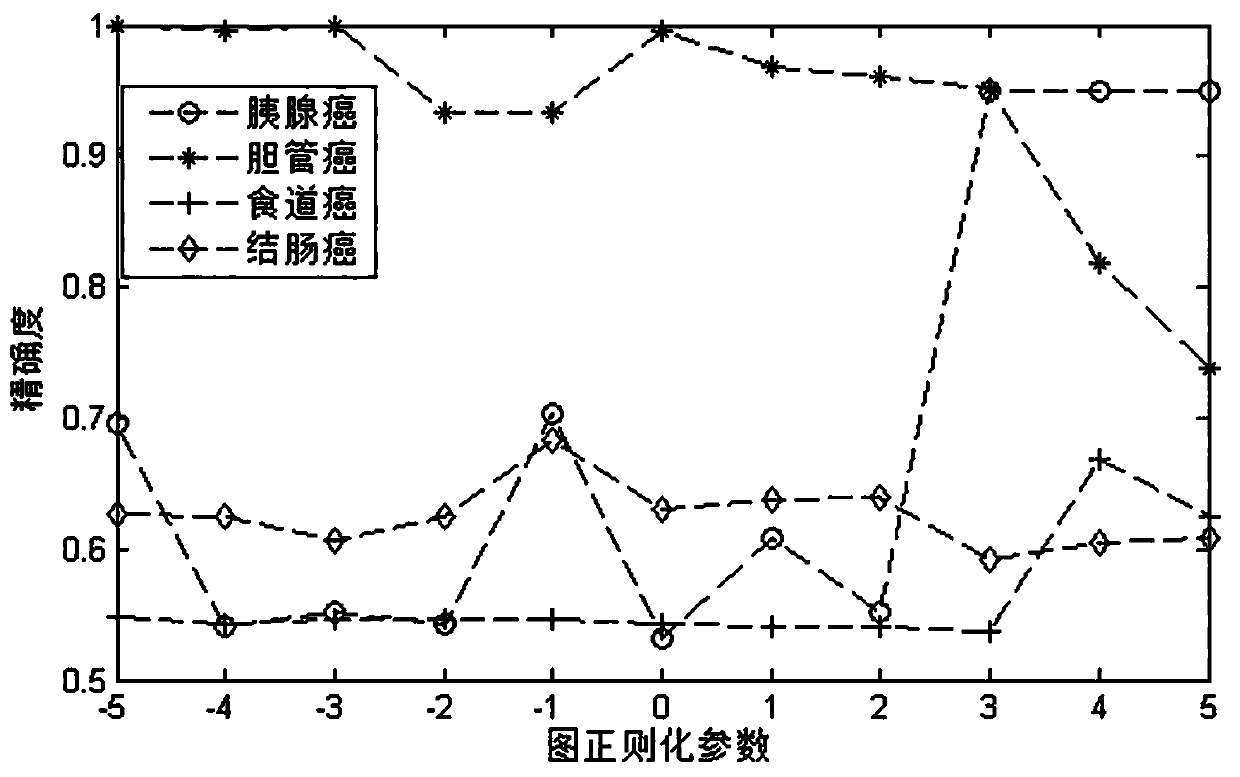
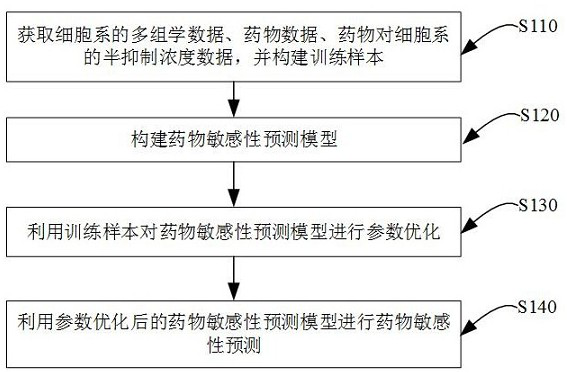
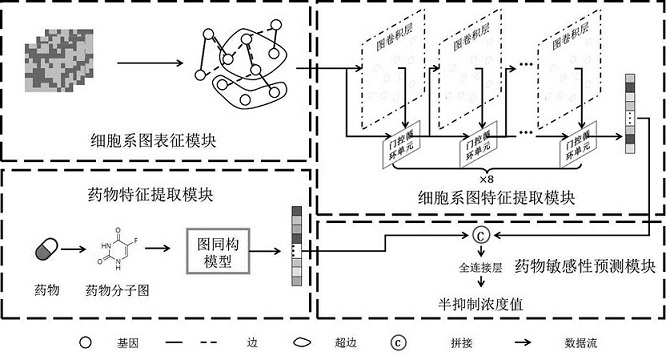
Drug sensitivity prediction method and device based on multi-omics data fusion
The invention discloses a drug sensitivity prediction method and device based on multi-omics data fusion, and belongs to the field of drug sensitivity detection. The method comprises the steps that three kinds of multi-omics information including genomics data, proteomics data and metabonomics data of individual cell lines are integrated through a cell line diagram characterization module, and a cell line multilateral diagram is obtained; according to the cell line multilateral graph, multi-omics information of a cell line and potential relation between expression products of genes in a multi-omics level are fully considered, then feature extraction is conducted on the cell line multilateral graph through a cell line graph feature extraction module, and node features and edge features in the cell line multilateral graph are fully extracted to serve as cell line features; and finally, a drug sensitivity prediction module is adopted to predict the semi-inhibitory concentration of the drug according to the cell line features and the drug features extracted based on the drug feature extraction module. According to the method, the prediction accuracy of the drug sensitivity is improved on the basis of comprehensively considering genomics data, proteomics data and metabonomics data.
Owner:ZHEJIANG UNIV
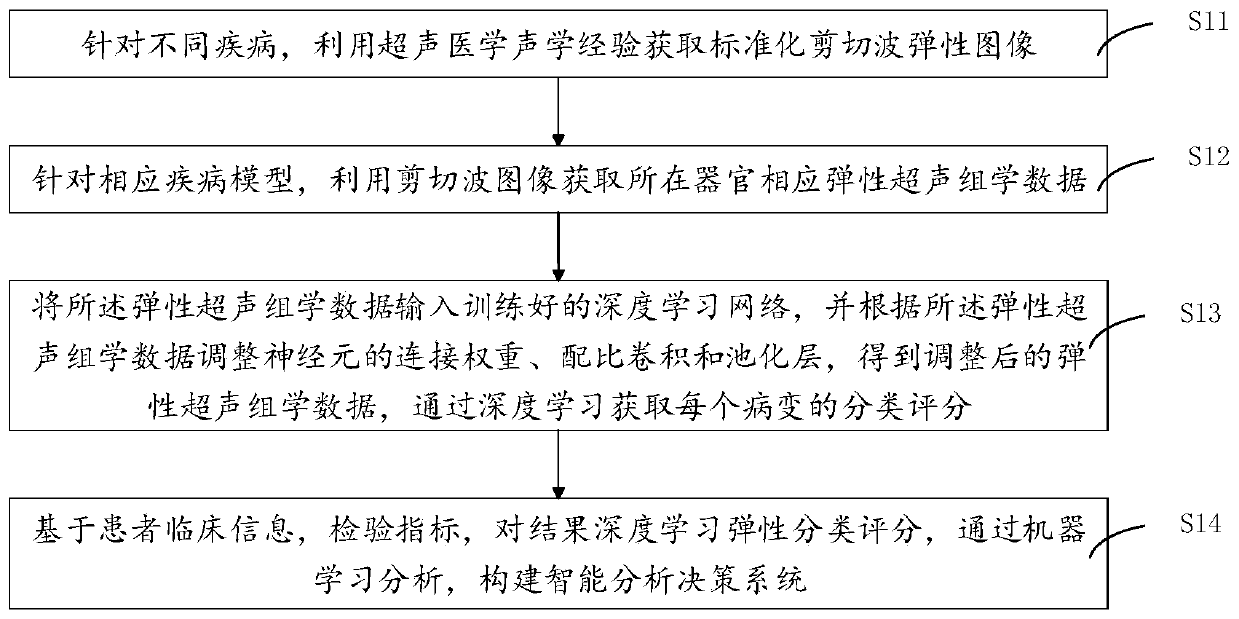
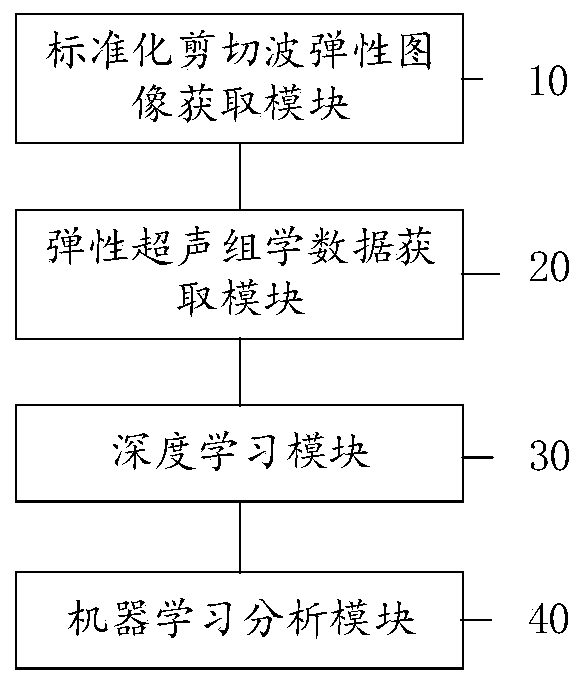
Ultrasonic omics depth analysis method and system based on shear wave elastography
ActiveCN111275706AGood repeatabilityImprove accuracyImage enhancementImage analysisDiseaseData acquisition
The invention discloses an ultrasonic omics depth analysis method and system based on shear wave elastic imaging, and the method comprises the steps: obtaining a standardized shear wave elastic imagethrough the ultrasonic medical acoustic experience for different diseases; aiming at the corresponding disease model, utilizing the shear wave image to obtain corresponding elastic ultrasonic omics data of the organ; inputting the elastic ultrasonic omics data into a trained deep learning network, adjusting the connection weight, proportion convolution and pooling layer of neurons according to theelastic ultrasonic omics data to obtain adjusted elastic ultrasonic omics data, and obtaining the classification score of each lesion through deep learning; based on patient clinical information andexamination indexes, results are subjected to deep learning elastic classification scoring, and a deep analysis decision system is constructed through machine learning analysis. According to the invention, the repeatability of boundary data acquisition and the adaptability of image analysis can be improved, and a deep analysis decision system is constructed to improve the accuracy of an auxiliaryanalysis result.
Owner:THE FIRST AFFILIATED HOSPITAL OF SUN YAT SEN UNIV
Method for identifying litchi varieties by using wide targeted metabonomics technology
PendingCN111721857AEasy to distinguishComponent separationMultivariate statisticalStatistical analysis
The invention discloses a method for identifying litchi varieties by using a wide targeted metabonomics technology. The method comprises the following steps: (1) obtaining a known litchi variety endogenous small molecule metabolite peak area database through litchi sample preparation, litchi sample detection and analysis, litchi sample qualitative and quantitative analysis and metabonomics data processing and analysis, screening to obtain 69 common difference significant metabolites, and generating a heatmap; and (2) obtaining the peak area of the endogenous small molecule metabolite of the unknown variety, combining the peak area with the peak area database of the known variety, carrying out multivariate statistical analysis, carrying out clustering analysis on the screened common differential metabolite, and judging the known variety which is firstly gathered with the variety to be identified as the variety to be identified or the similar variety. According to the method, the endogenous small molecule metabolites are obtained by utilizing a wide targeted metabonomics analysis technology, common difference significance metabolites of litchis are obtained through multivariate statistical analysis, and a visual map is generated and can be used for litchi variety identification.
Owner:POMOLOGY RES INST GUANGDONG ACADEMY OF AGRI SCI
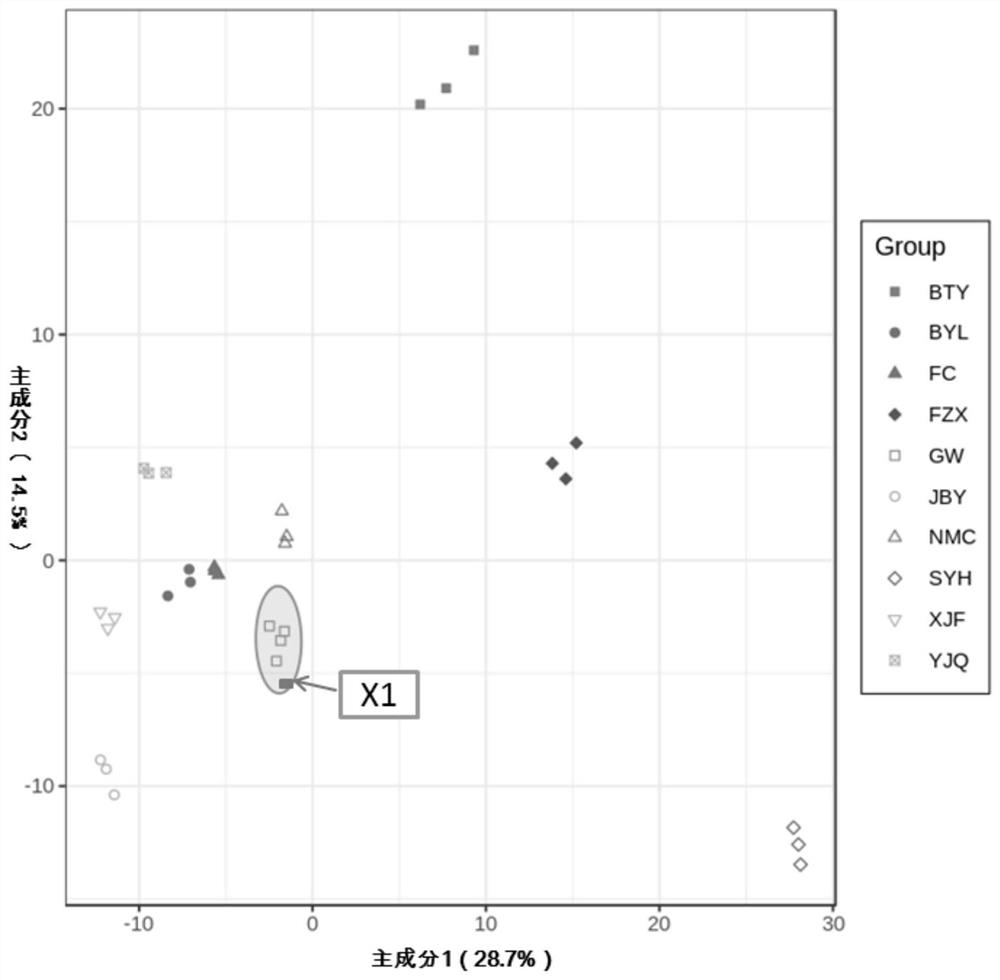
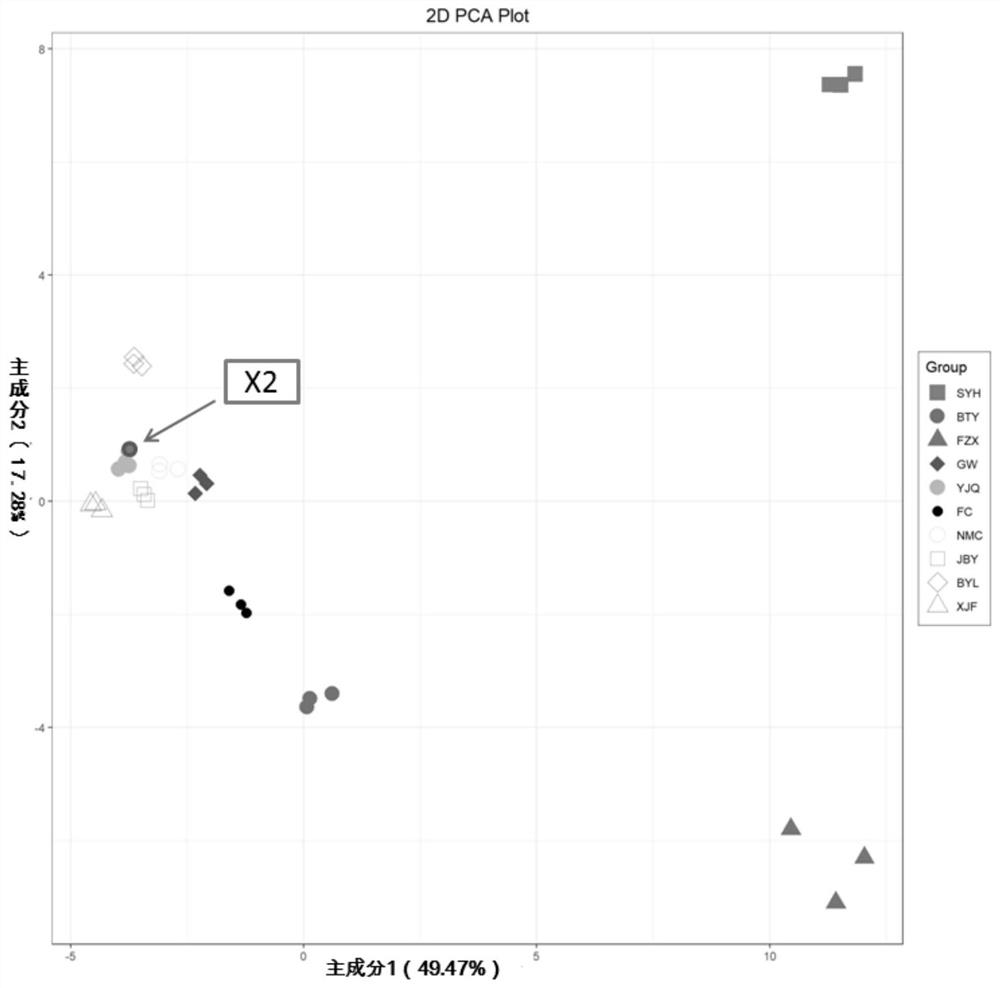
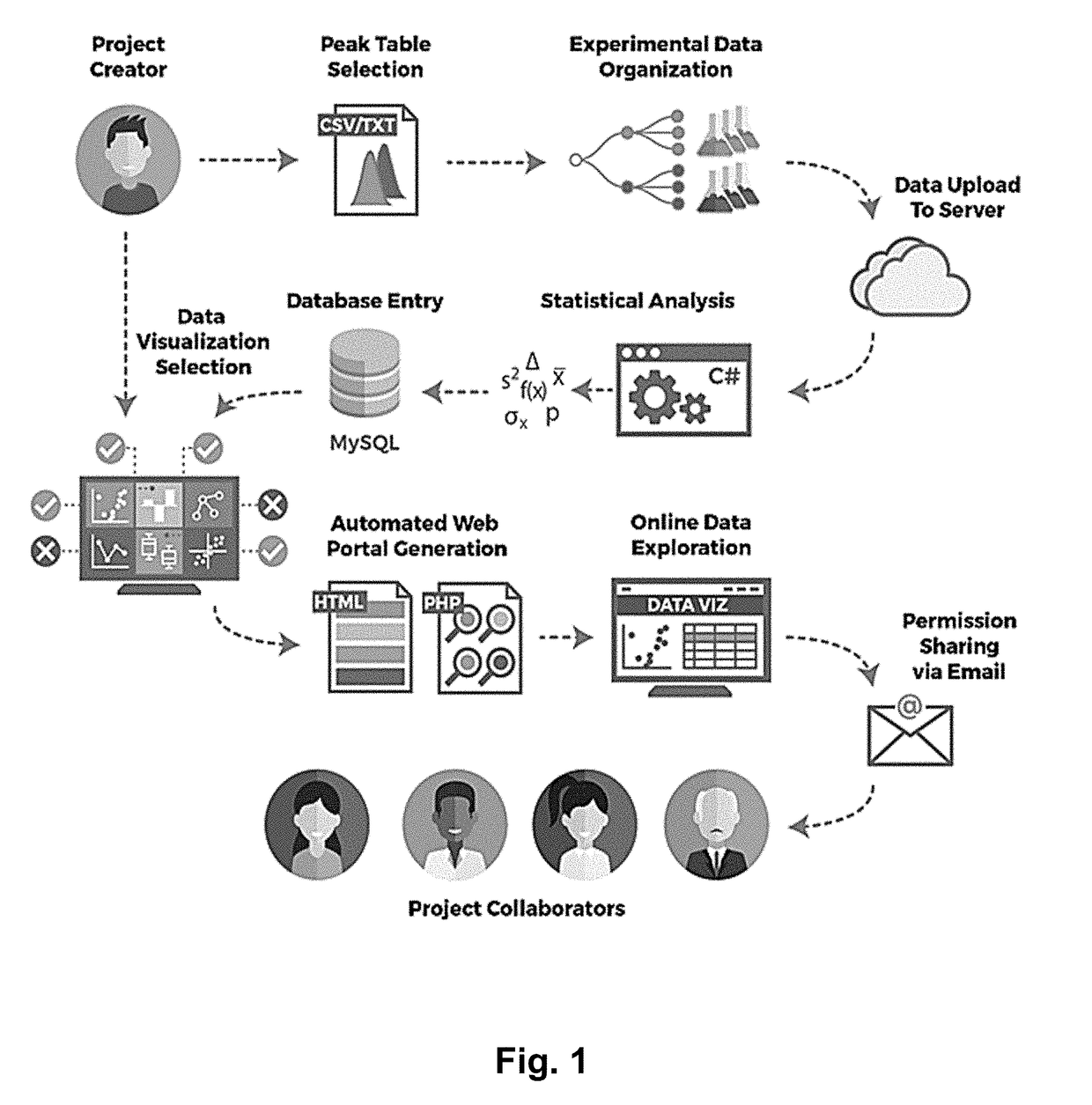
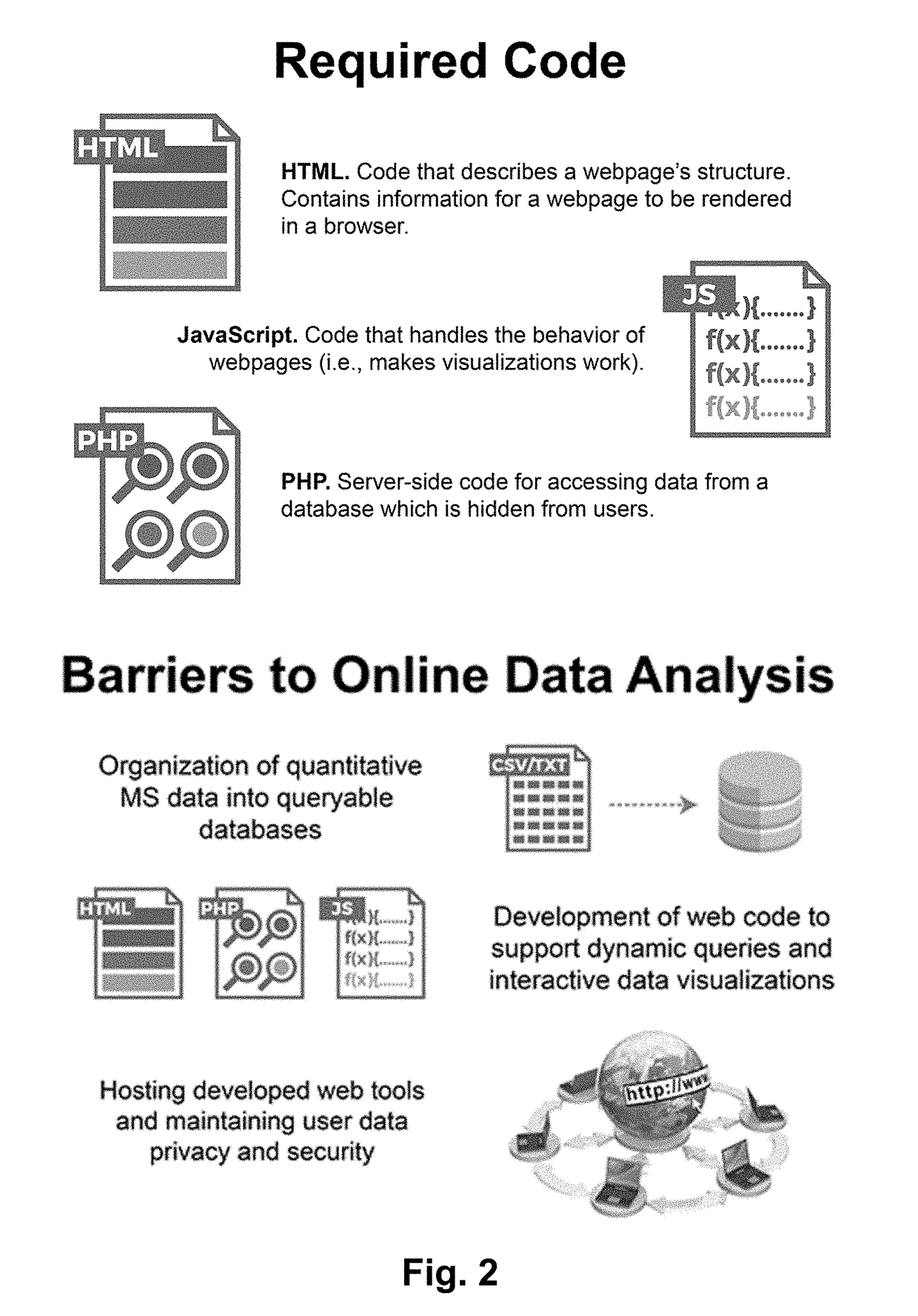
Web-Based Data Upload and Visualization Platform Enabling Creation of Code-Free Exploration of MS-Based Omics Data
InactiveUS20190034047A1Easy to uploadEasy to compareParticle separator tubes2D-image generationElectronic formMass Spectrometry-Mass Spectrometry
The present invention provides a platform that enables codeless generation of online mass spectrometry data exploration portals. The platform facilitates upload of generic spreadsheets containing processed mass spectrometry results (e.g., peak tables) and enables on-the-fly hierarchical organization of data. Following data upload, platform users can select individual visualizations to add to their custom web portal from a menu of options. Based on these selections, a complete webpage is constructed with all associated functionality embedded. These custom web portals can then be shared with collaborators and other researchers at the discretion of the creator via a developed user permissions sharing scheme.
Owner:WISCONSIN ALUMNI RES FOUND
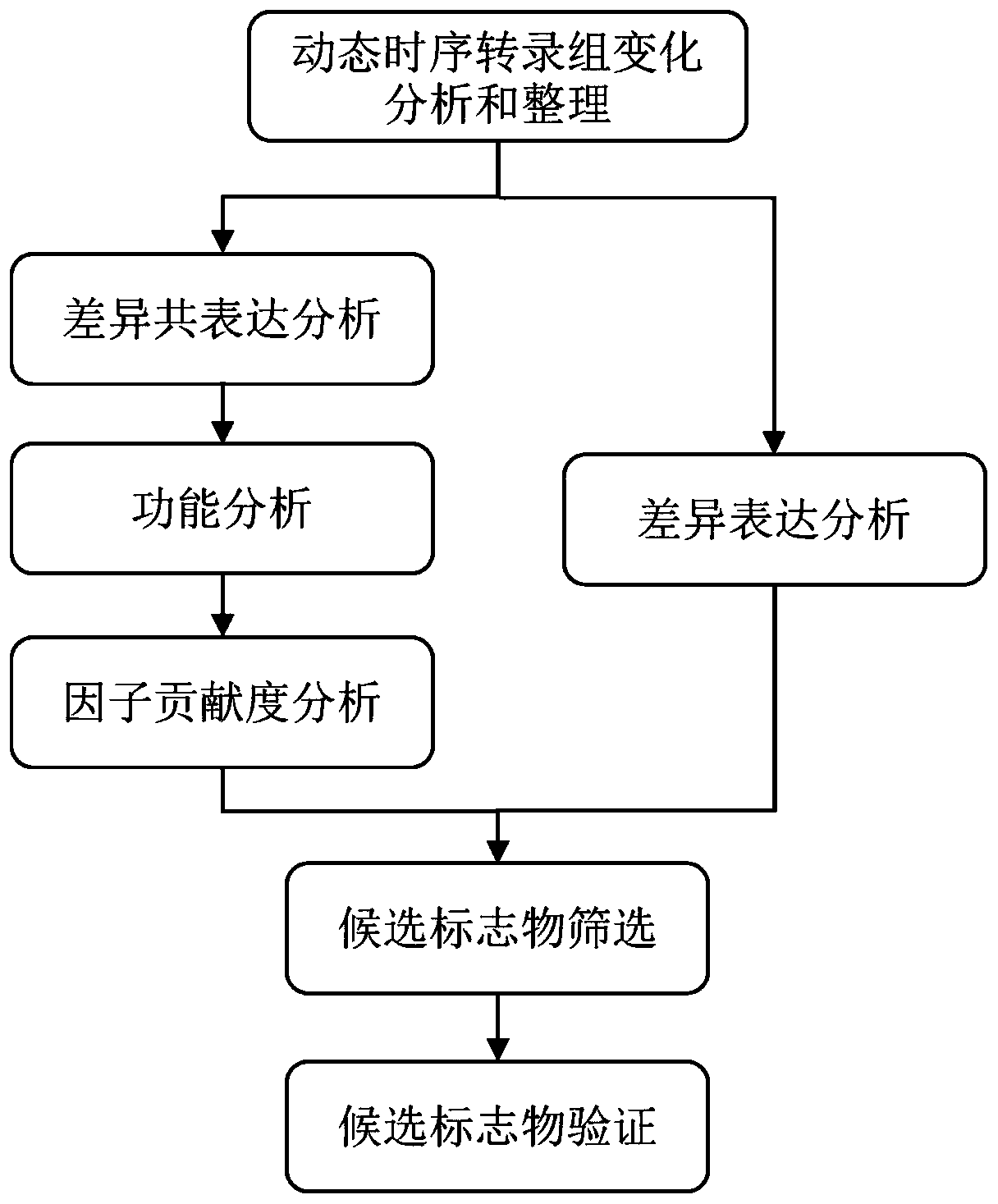
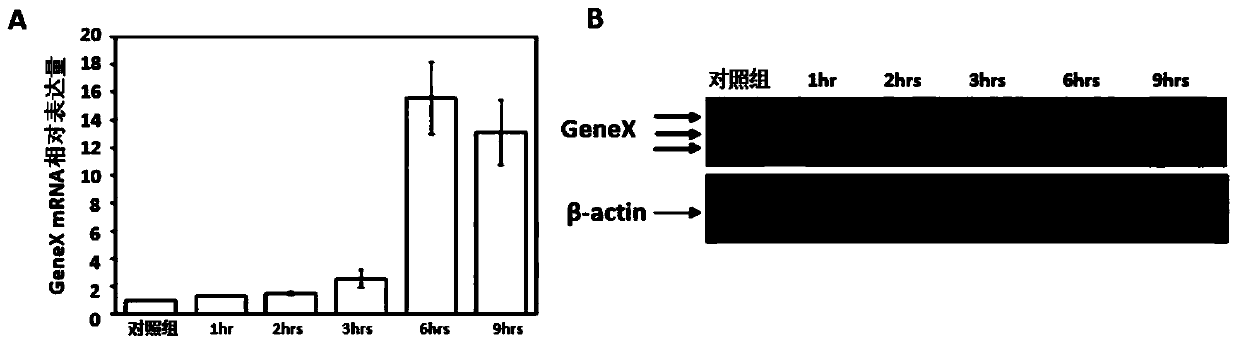
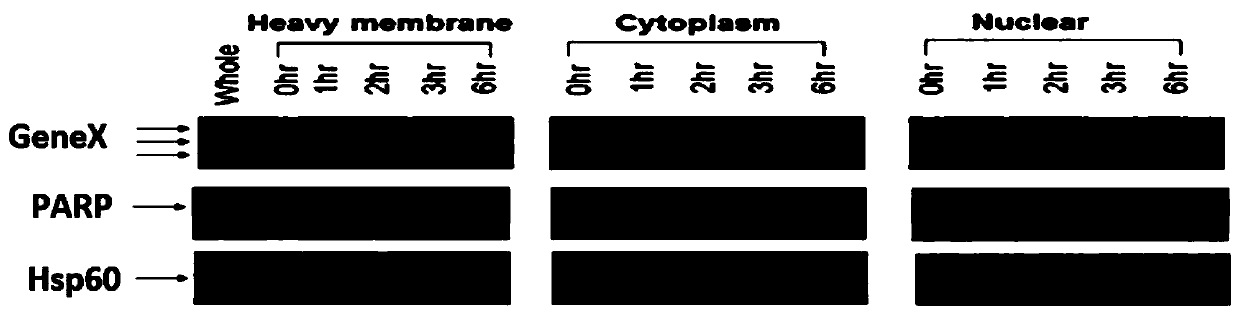
Method and system for finding molecular marker based on transcriptome time sequence dynamic change and gene function association
ActiveCN110379459AOrientationHigh verification success rateProteomicsGenomicsProcessed GenesHigh flux
The invention discloses a method and a system for finding a molecular marker based on transcriptome time sequence dynamic change and gene function association. The method comprises the steps of acquiring a target pathophysiological process and a dynamic time sequence transcriptome change which accords with the pathophysiological process through multi-time-sequence sampling and high-flux transcriptome sequencing technology on a sample; through gene function association analysis, evaluating an association strength between a different coexpression set and the target pathophysiological process gene set, thereby finding the robust reliable marker with pathophysiological meaning in omics data. According to the method, the internal association between time sequence association and omics indexes is sufficiently used; the marker finding process is supported and verified by different types of information, thereby realizing high robustness. Therefore relatively low requirement for data measuringprecision and method parameter optimizing requirement is realized. The personnel without abundant practice experience and analysis experience can effectively use the method and the system.
Owner:杭州新范式生物医药科技有限公司
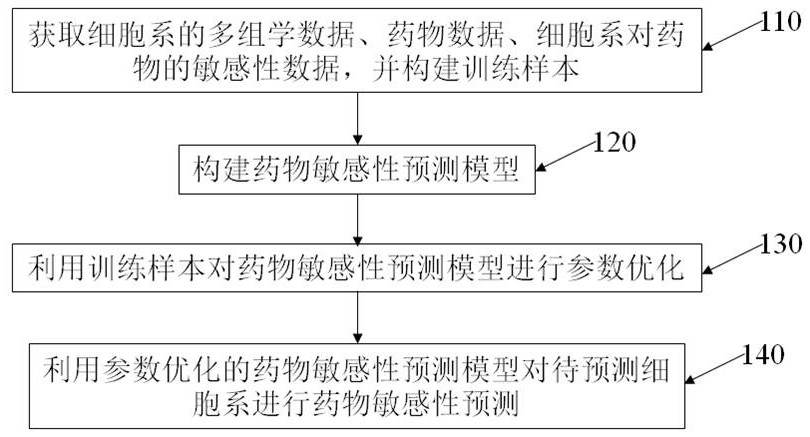
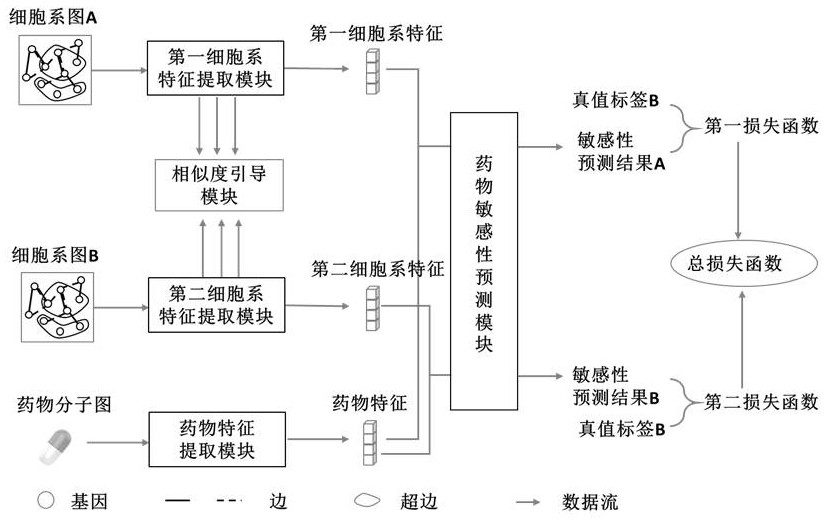
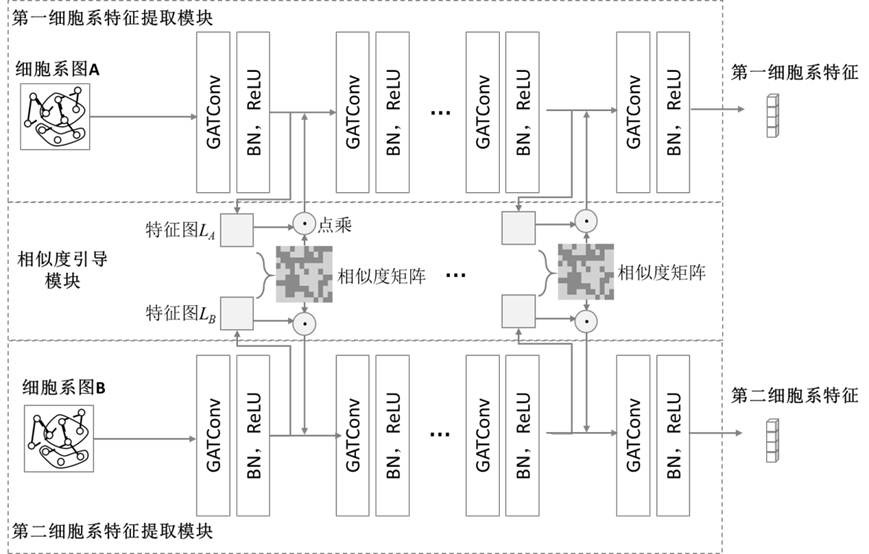
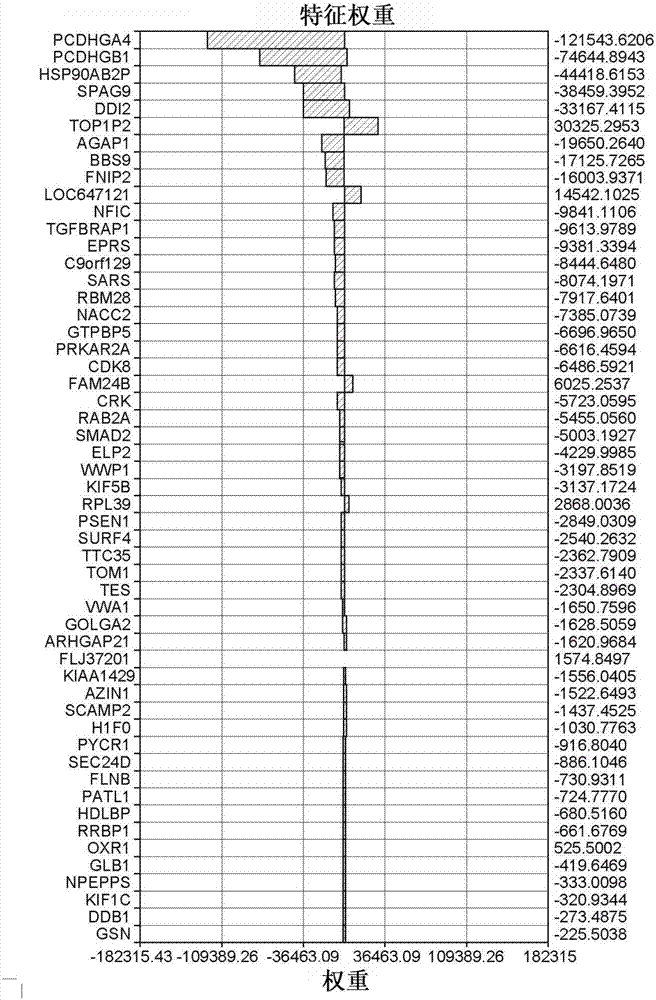
Drug sensitivity prediction method and device based on multi-omics similarity guidance
ActiveCN114255886AEfficient extractionGreat biological prior knowledgeBiostatisticsCharacter and pattern recognitionGenomicsMulti omics
The invention discloses a drug sensitivity prediction method and device based on multi-omics similarity guidance, and the method comprises the steps: a cell line diagram constructed based on multi-omics data of a cell line can fully integrate four types of multi-omics information of genomics data, transcriptomics data, proteomics data and metabonomics data of an individual cell line; compared with an existing cell line characterization mode, more kinds of omics information can be contained, and meanwhile the potential relation between products expressed by the cell line in the multi-omics level is fully considered; on the basis, a drug sensitivity prediction model for predicting the drug sensitivity based on the cell line diagram adopts a multi-omics similarity guiding mode, multi-omics similarity among individuals can be fully considered while multi-omics information of individual specificity is efficiently extracted, huge biological priori knowledge can be provided, and the drug sensitivity prediction model can be used for predicting the drug sensitivity based on the cell line diagram. Therefore, more accurate drug sensitivity prediction is realized.
Owner:ZHEJIANG UNIV
Systems and methods for response prediction to chemotherapy in high grade bladder cancer
Contemplated systems and methods allow for prediction of chemotherapy outcome for patients diagnosed with high-grade bladder cancer. In particularly preferred aspects, the prediction is performed using a model based on machine learning wherein the model has a minimum predetermined accuracy gain and wherein a thusly identified model provides the identity and weight factors for omics data used in the outcome prediction.
Owner:NANTOMICS LLC
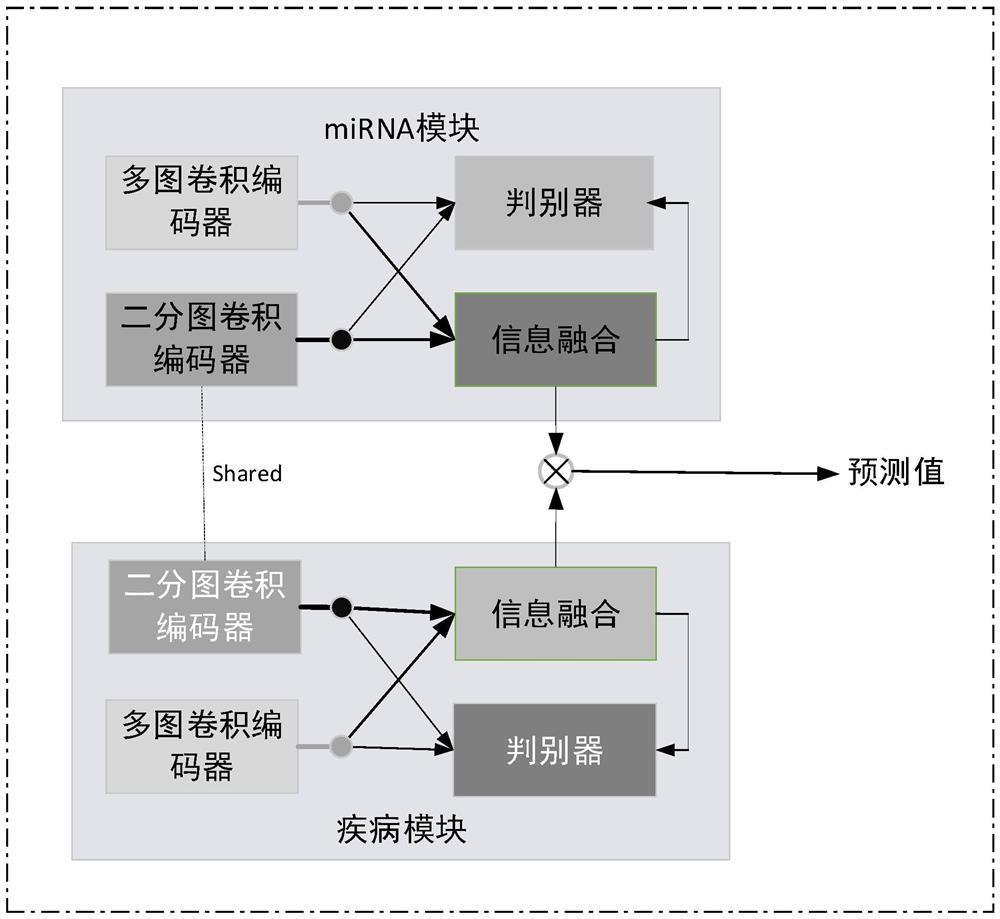
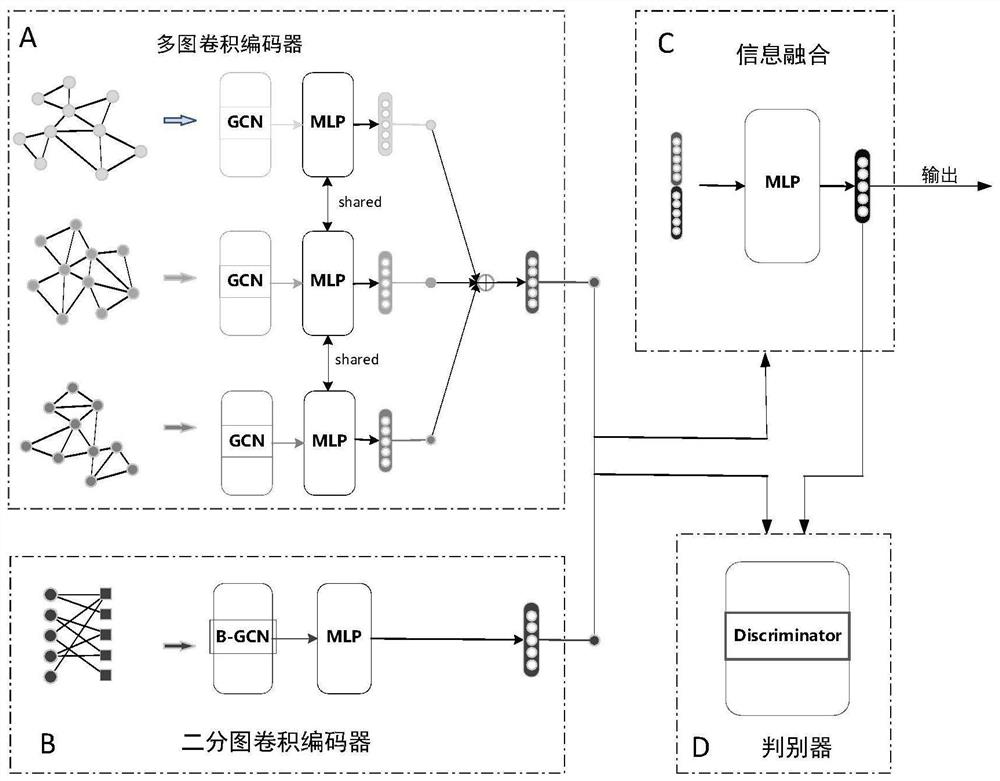
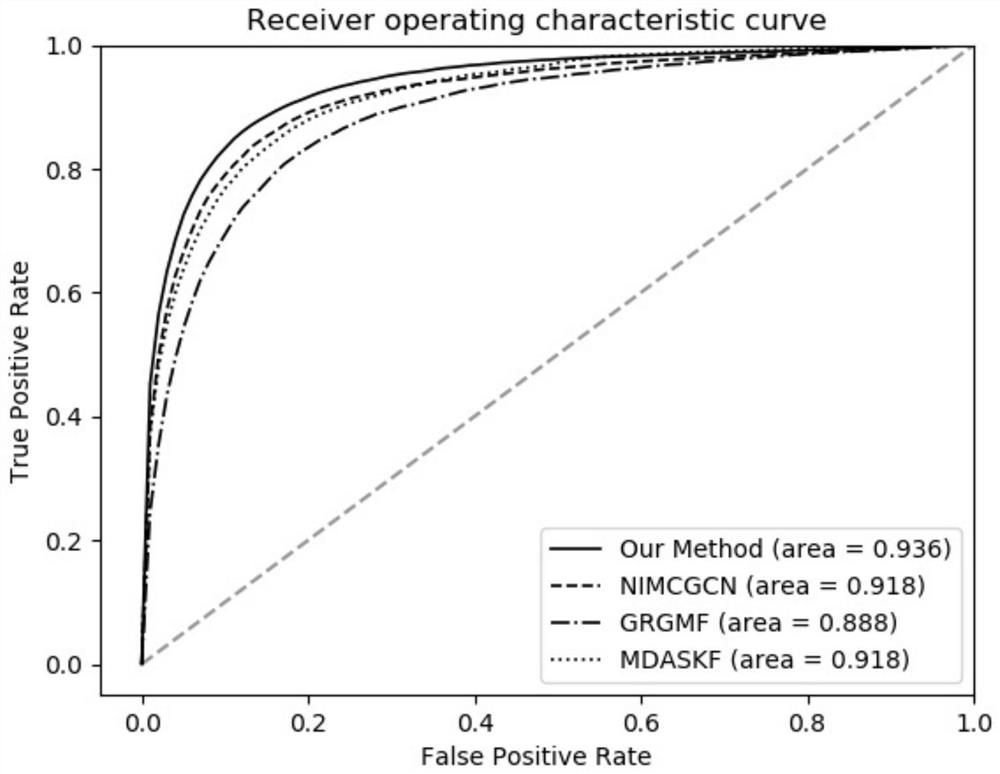
MiRNA-disease association prediction method and device based on graph neural network fusion multi-view information
ActiveCN112784913AEasy to captureImprove forecast accuracyCharacter and pattern recognitionNeural architecturesMulti omicsPredictive methods
The invention discloses a miRNA-disease association prediction method and device based on graph neural network fusion multi-view information, and the method integrates miRNA-disease-related multi-omics data to construct a plurality of views, not only considers a plurality of homogeneous similarity networks, but also considers a heterogeneous bipartite network, extracts node features on all views in combination with a graph neural network and multi-view learning, captures dependency between global features and local features through a discriminator, and can better capture the complex nonlinear relation between miRNA and diseases.
Owner:HUNAN UNIV
Cancer survival analysis system based on multiple tasks and multiple modes
PendingCN112687327AImprove multimodal fusionImprove the ability of multi-task learningMedical data miningBiostatisticsMulti modal dataData pre-processing
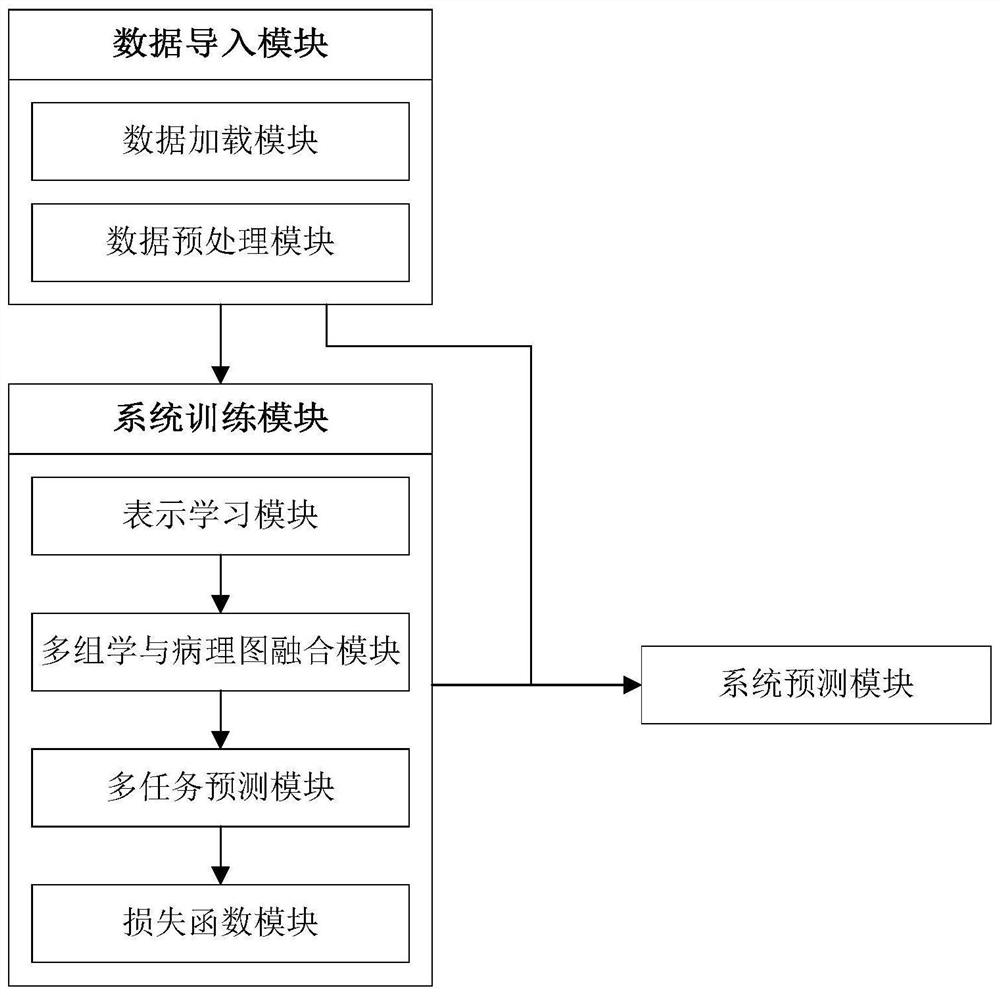
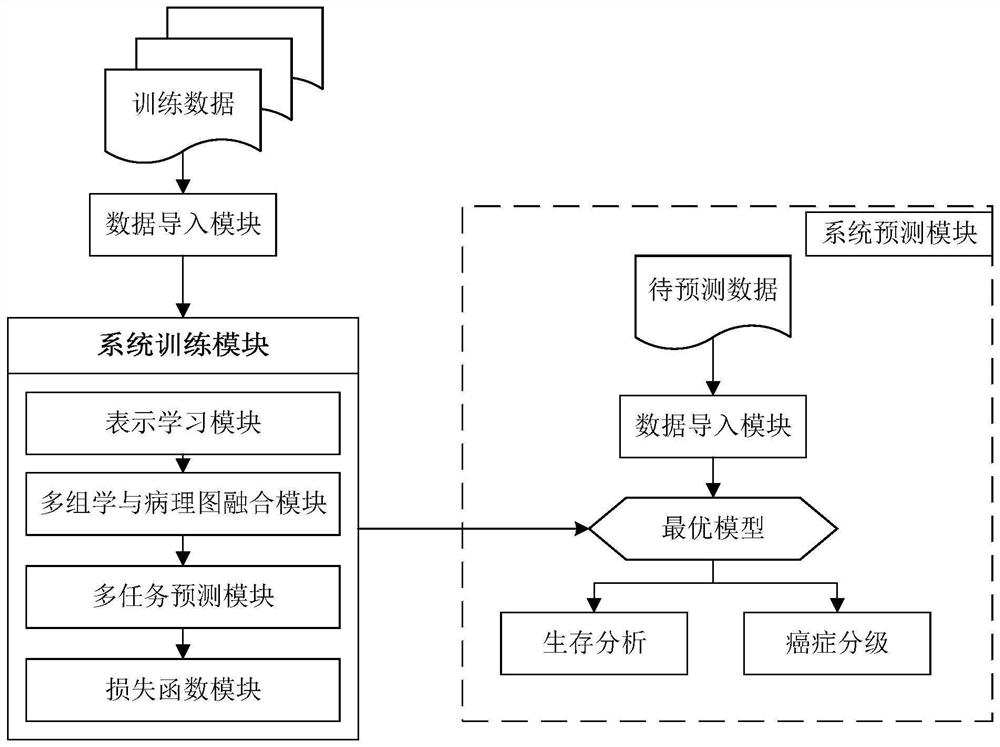
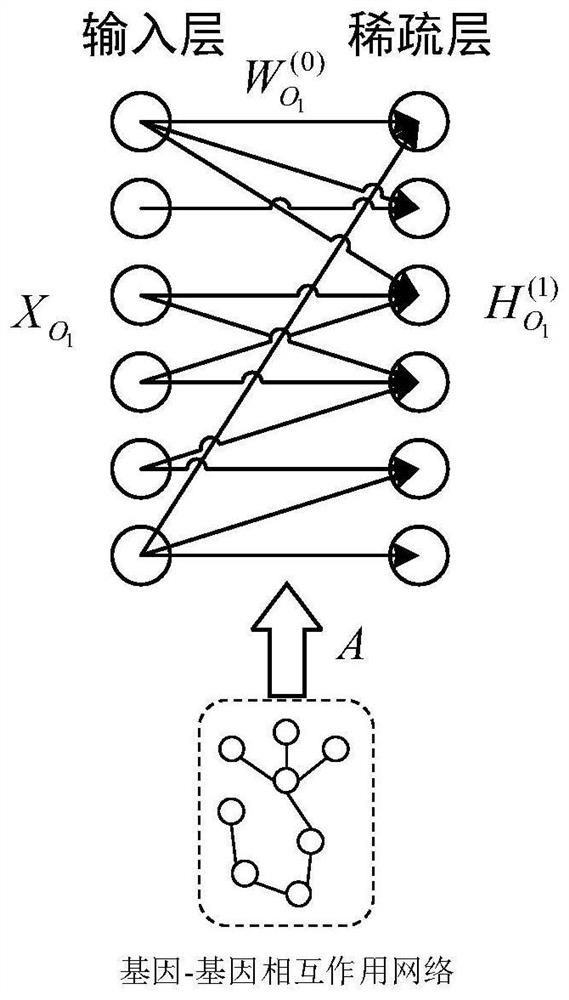
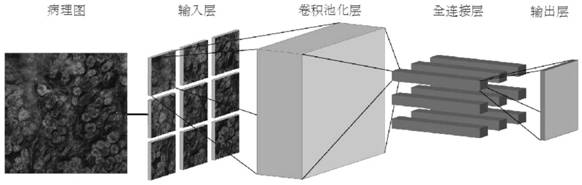
The invention discloses a cancer survival analysis system based on multiple tasks and multiple modes, and the system comprises a data importing module which comprises a data loading module and a data preprocessing module; a system training module, comprising a representation learning module, a multi-omics and pathological map fusion module, a multi-task prediction module and a loss function module; and a system prediction module, used for carrying out survival analysis and grading according to the multi-omics data and the pathological map of the cancer patient. According to the invention, the deep learning technology and the multi-task learning technology are combined with the multi-omics and pathological map data, so that the complementary characteristics among the multi-modal data can be captured, and the sharing relevance among multiple tasks can also be captured, thereby forming a cancer survival analysis system based on multiple tasks and multiple modalities, and providing automatic survival analysis and cancer grading results.
Owner:中山依数科技有限公司
Endogastric biopsy Raman image auxiliary diagnosis method and system based on artificial intelligence
PendingCN113539476AVerification of identification abilityVerify availabilityImage enhancementImage analysisPattern recognitionG i endoscopy
The invention belongs to the technical field of medical equipment, and particularly relates to an endogastric biopsy Raman image auxiliary diagnosis method and system based on artificial intelligence. The artificial intelligence technology is applied to gastroscope stimulated Raman scattering endoscopic biopsy tissue image auxiliary diagnosis for the first time, and the method comprises the step: after histopathology image information is obtained by means of the stimulated Raman scattering microimaging technology, adopting the image classification and image omics data analysis based on a deep learning neural network and machine learning, and constructing an endogastric biopsy Raman image auxiliary diagnosis system. The system comprises a stomach tissue Raman image data preprocessing module, a neural network model, an algorithm module for training the neural network model, a neural network fine tuning module and a test module; compared with an existing traditional endoscope diagnosis and treatment system, the system has the advantages that real-time, rapid and intelligent diagnosis support in the endoscope examination process is achieved, and pathologists do not need to conduct explanation.
Owner:FUDAN UNIV
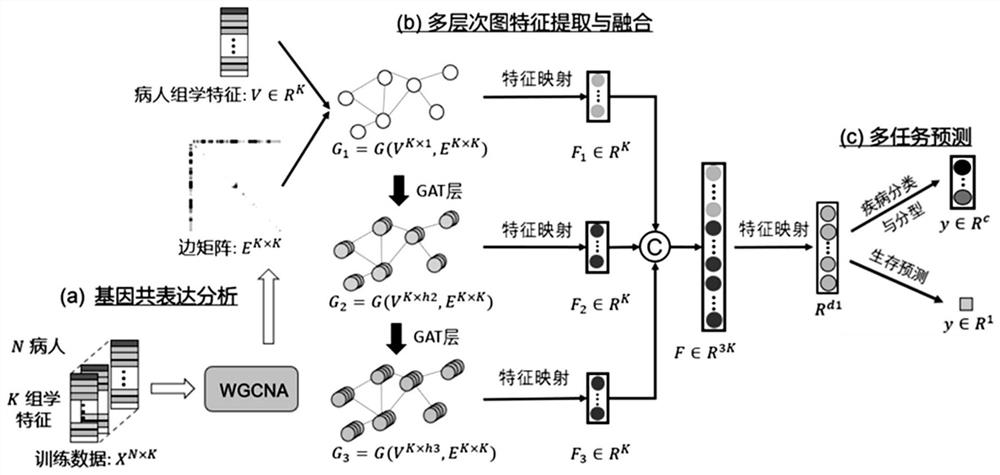
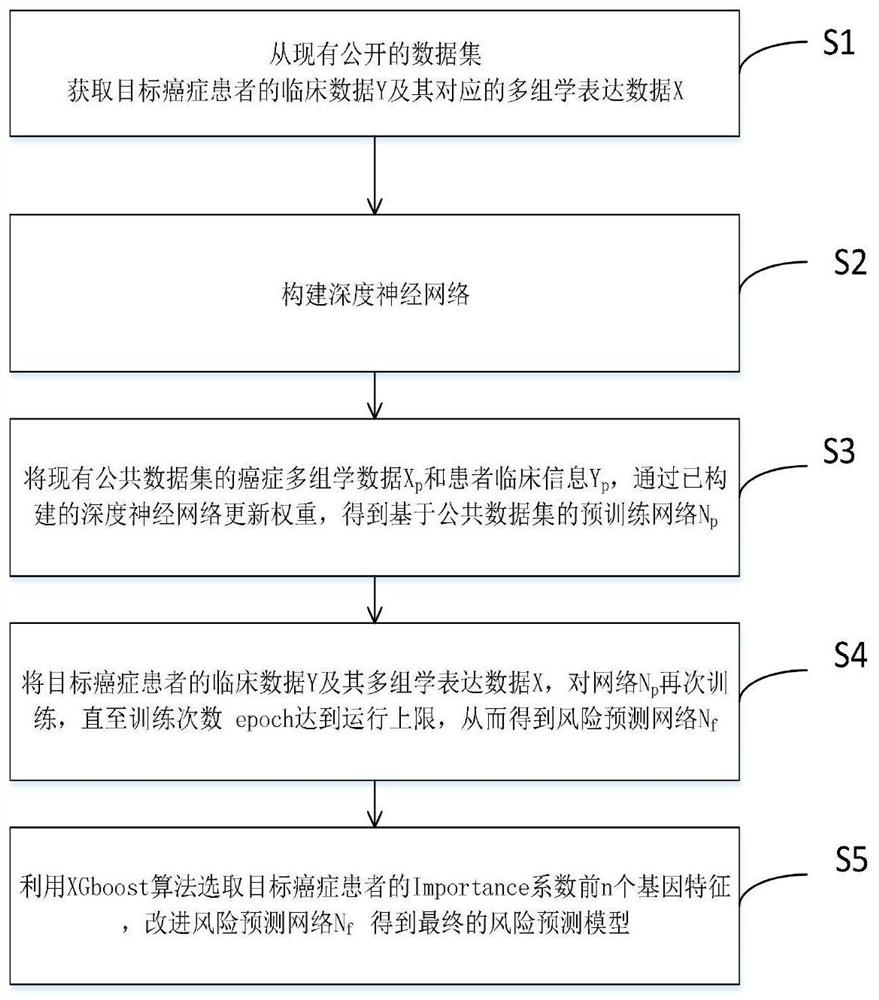
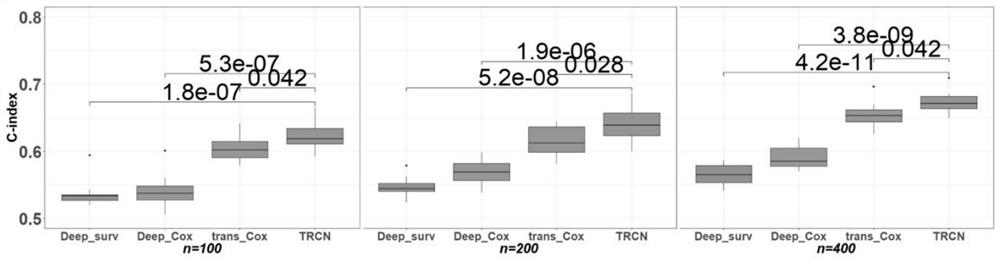
Deep learning method for predicting prognosis risk of cancer patient based on multi-omics data
PendingCN112820403AImprove robustnessEpidemiological alert systemsMedical automated diagnosisData setGene Feature
The invention discloses a deep learning method for predicting the prognosis risk of a cancer patient based on multi-omics data, which is used for predicting the prognosis risk of the cancer patient and comprises the following steps: S1, acquiring clinical data Y of a target cancer patient and corresponding multi-omics expression data X from an existing public data set; S2, constructing a deep neural network; S3, updating the weight theta of the cancer multi-omics data Xp and the clinical information Yp of the patient of the existing common data set through the constructed deep neural network to obtain a pre-training network Np based on the common data set; S4, training the network Np again until the training frequency epoch reaches the operation upper limit, thereby obtaining a risk prediction network Nf; and S5, selecting the first n gene features of the Importance coefficient of the target cancer patient by using an XGboost algorithm, and improving the risk prediction network Nf to obtain a final risk prediction model. According to the method, the robustness of the prediction model is improved, and the prognosis risk of the cancer patient is predicted more accurately by using multi-omics data.
Owner:SUN YAT SEN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com