Multi-omics data association relationship discovery method based on sparse matching
A technology of omics data and correlation, applied in the field of bioinformatics, can solve the problems of unutilized, too simple integrated research methods, insufficient sample size, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
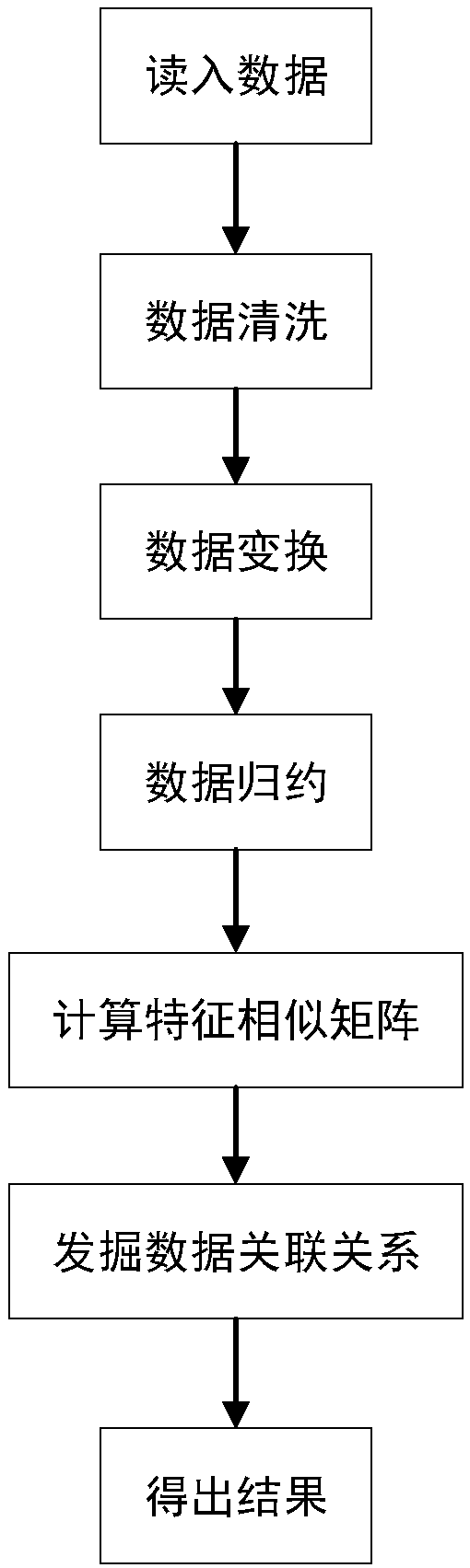
[0062] A multi-omics data association relationship discovery method based on sparse matching, the basic process is as follows figure 1 As shown, it includes: reading in data, data cleaning, data transformation, data reduction, calculating feature similarity matrix, mining data correlation, and obtaining results.
[0063] The scheme of multi-omics data association discovery based on sparse matching is as follows:
[0064] 1. Preprocess the input data, improve the quality of the data, and make the data better adapt to this method.
[0065] In the massive raw data, there are a lot of abnormal data and many problems such as too large dimensions, which seriously affect the execution efficiency of the model, and may even lead to deviations in the model results. Therefore, it is particularly important to perform data preprocessing. This method uses the following methods: Steps to preprocess the data:
[0066] 1.1 Data cleaning:
[0067] For the missing values in the data, the cu...
Embodiment 2
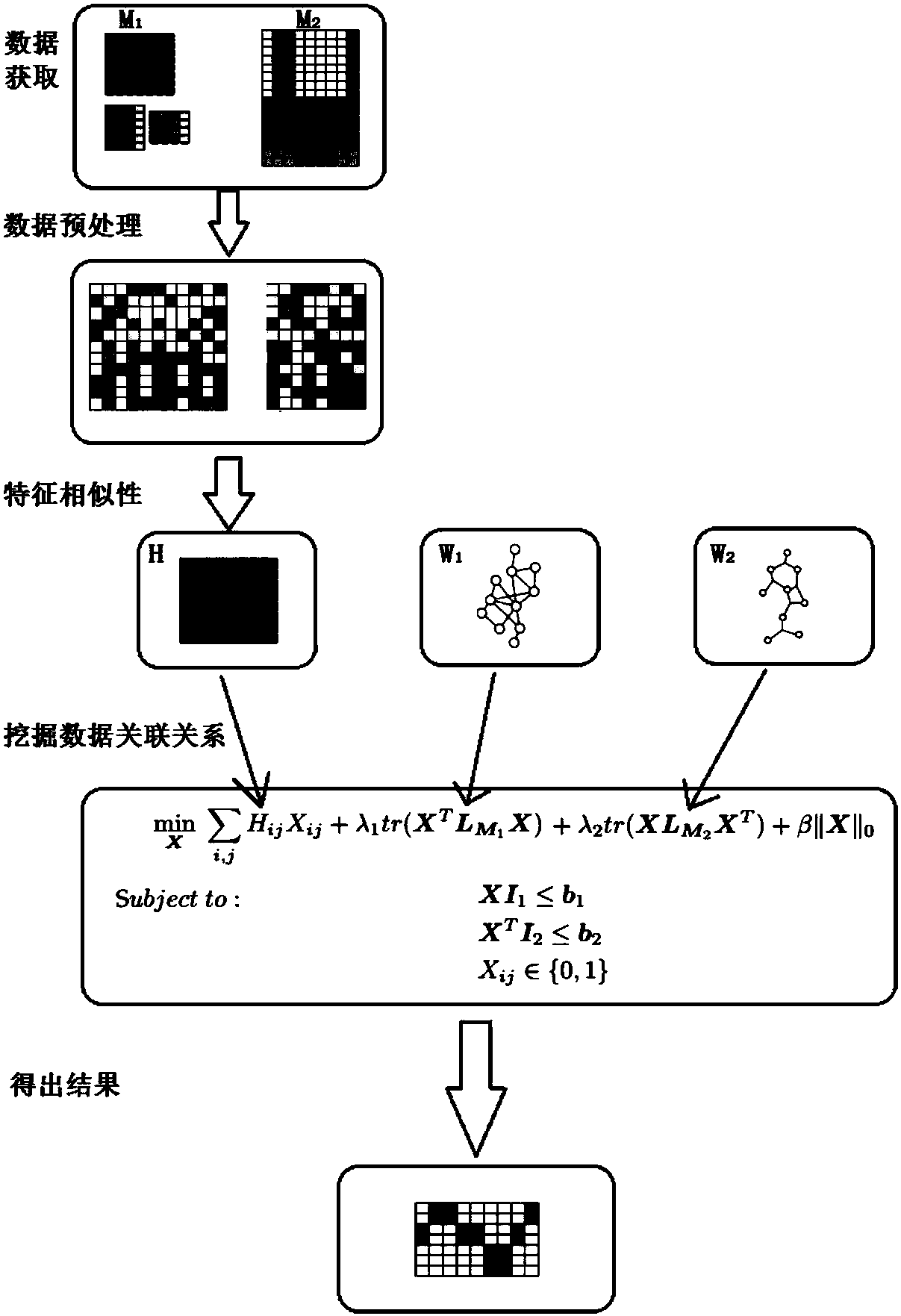
[0120] A multi-omics data association discovery method based on sparse matching, such as figure 2 Shown is a specific schematic diagram of the method, including:
[0121] Step 1: Acquire gene expression data M 1 and drug response data M 2 , gene-gene association network W 1 and the drug-drug association network W 2 ;
[0122] Step 2: Separate gene expression data M 1 and drug response data M 2 Perform data preprocessing.
[0123] Step 3: Utilize gene expression data M 1 and drug response data M 2 Calculate the gene-drug similarity measure matrix H.
[0124]Step 4: The gene-drug similarity measure matrix H, gene-gene association network W 1 and the drug-drug association network W 2 , as the model input to mine the association relationship between genes and drugs.
[0125] Step 2 specifically includes:
[0126] Step 2.1: Clean the data. For the missing values in the data, the cubic spline interpolation method is used to interpolate them; for the outliers in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com