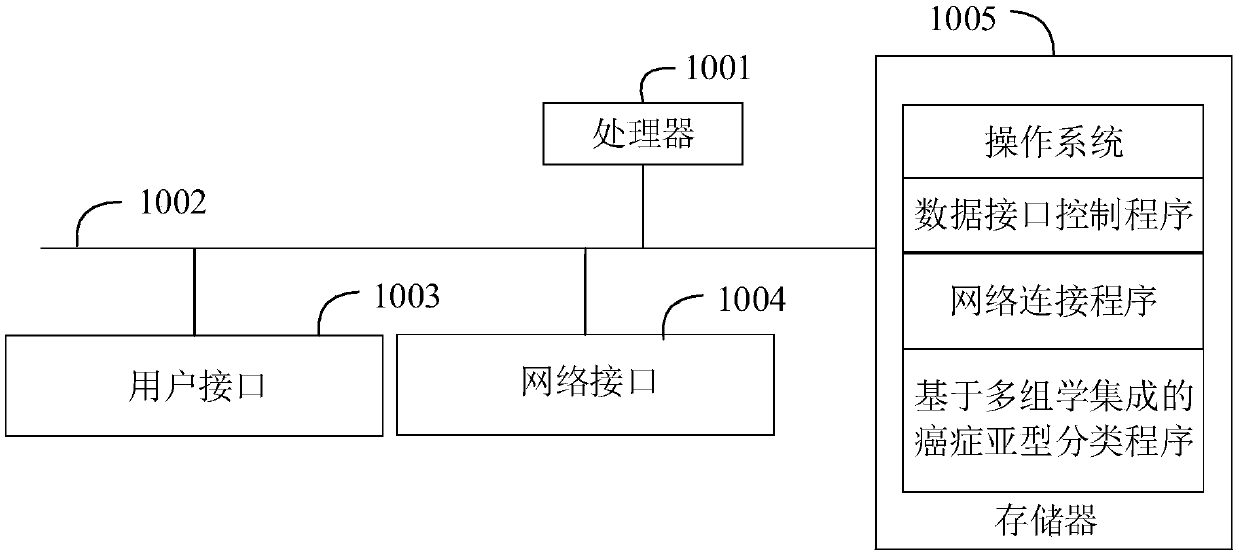
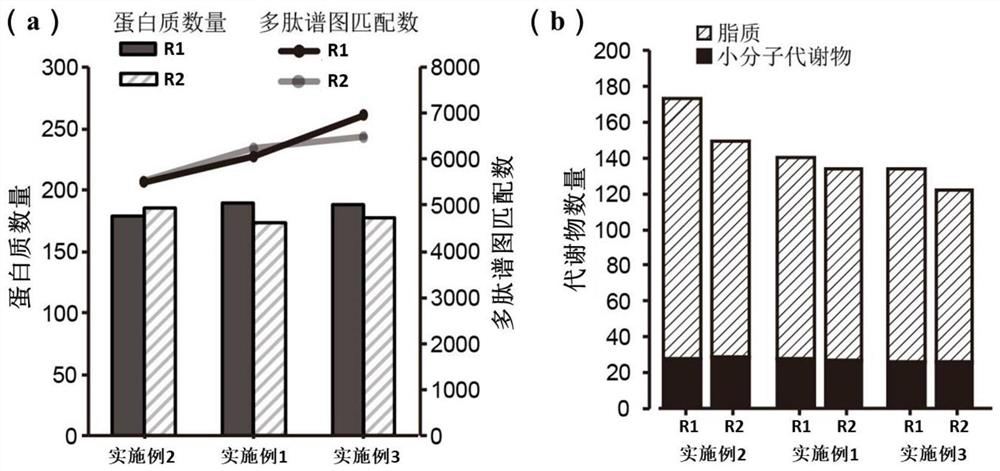
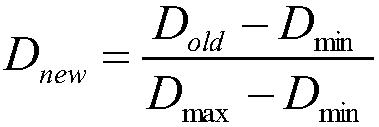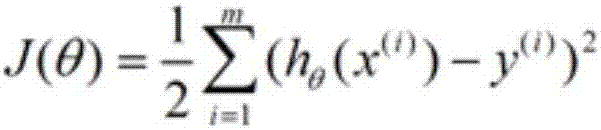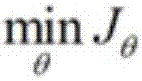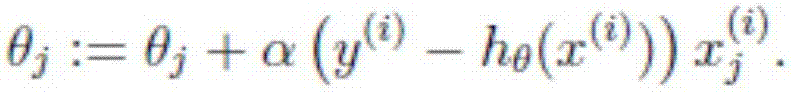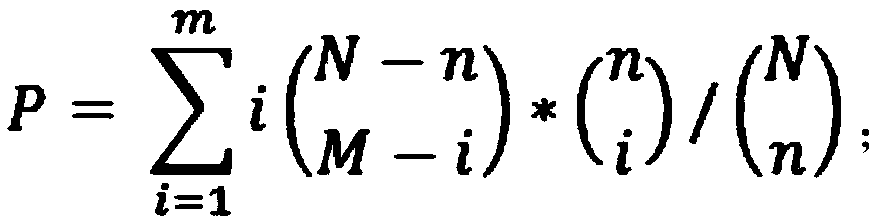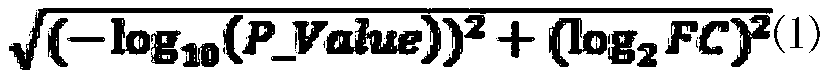Patents
Literature
70 results about "Multi omics" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Multiomics, multi-omics or integrative omics is a biological analysis approach in which the data sets are multiple " omes ", such as the genome, proteome, transcriptome, epigenome, and microbiome; in other words, the use of multiple omics technologies to study life in a concerted way.
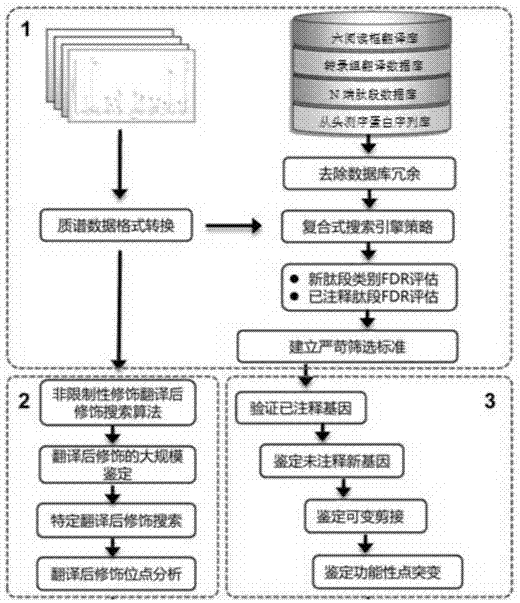
Bioinformatics method based on protein mass spectrum data annotation eukaryote genome
The invention discloses a bioinformatics method based on a protein mass spectrum data annotation eukaryote genome. The method specifically comprises the steps of 1, constructing a high-coverage eukaryote multi-omics sequence database; 2, removing eukaryotic protein sequence database redundancy; 3, conducting mass spectrum original data format conversion; 4, adopting a database searching engine with different algorithms, and retrieving mass spectrum data separately; 5, conducting peptide fragment spectrum matching and scoring on a retrieved and processed result; 6, screening result data after type FDR system evaluation; 7, verifying an annotated encoding gene; 8, authenticating a new gene which is not annotated; 9, authenticating alternative splicing; 10, authenticating functional point mutation; 11, aiming at protein posttranslational modification, conducting large-scale authentication; 12, conducting functional annotation of the new gene and the posttranslational modification. According to the bioinformatics method based on the protein mass spectrum data annotation eukaryote genome, the accuracy and sensitivity of protein mass spectrum data analysis are comprehensively improved, in-depth analysis and annotation for the eukaryote genome is achieved, and the method specifically has the advantages of being efficient, accurate and comprehensive.
Owner:湖北普罗金科技有限公司
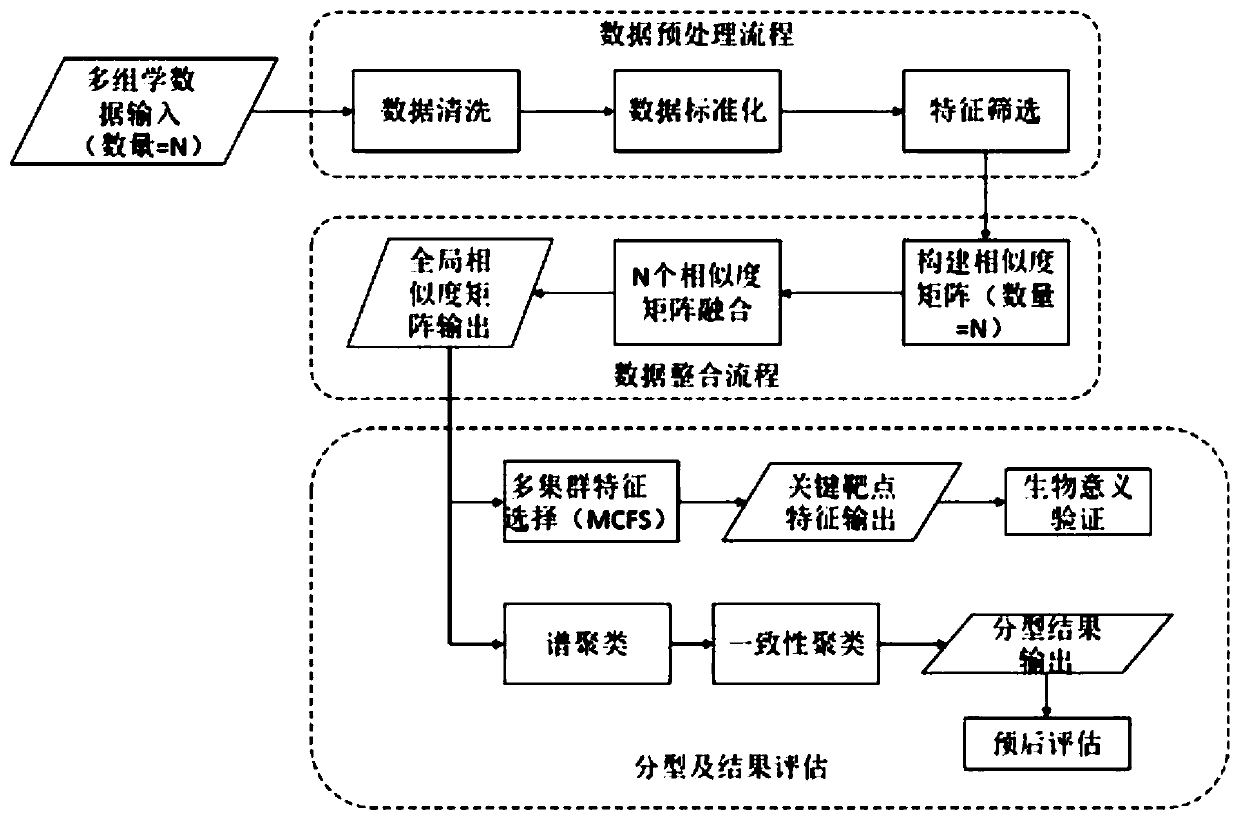
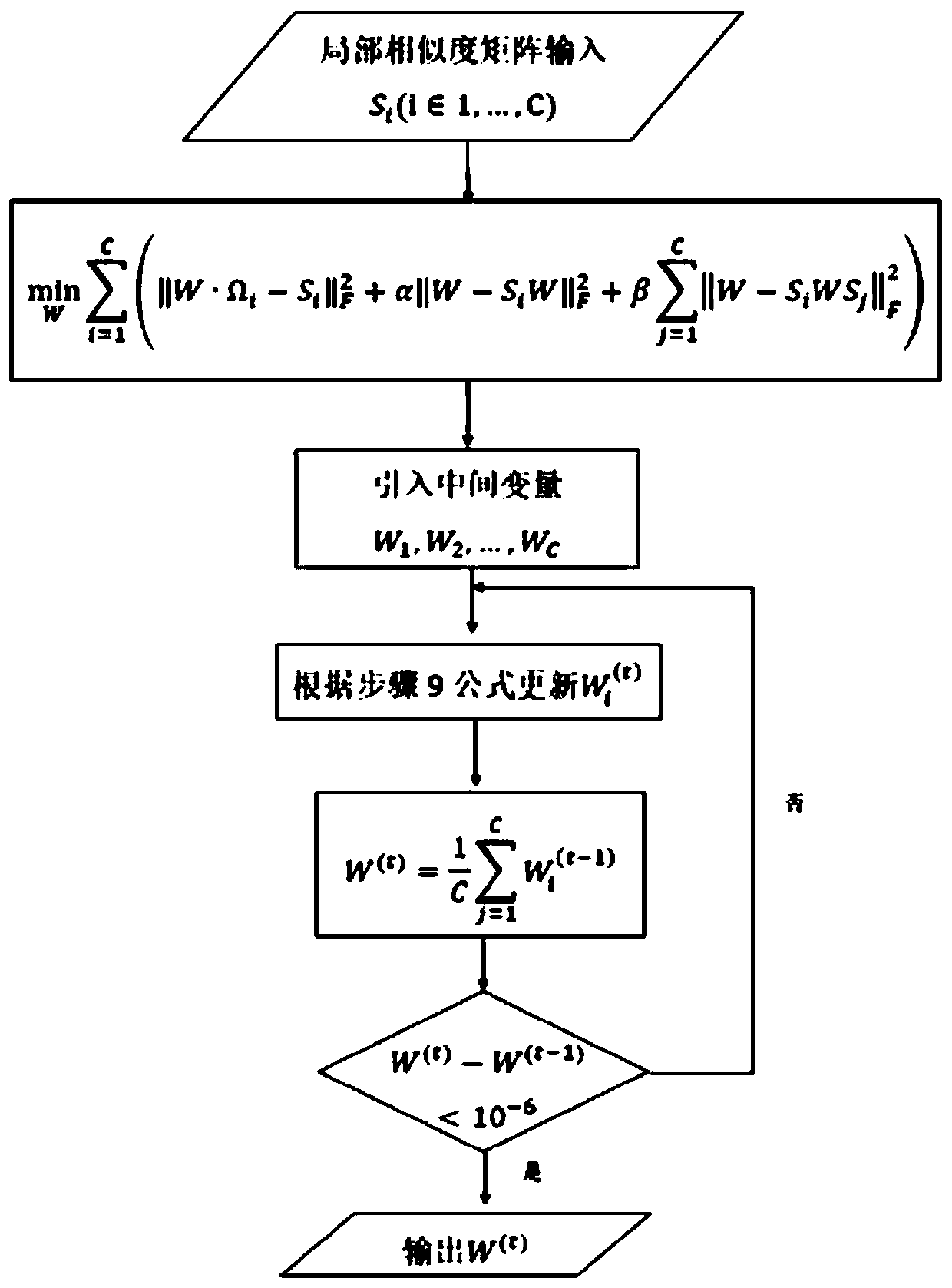
Multi-omics cancer data integrating and analyzing method based on similarity fusion
ActiveCN109994200AHigh precisionImprove stabilityMedical automated diagnosisProteomicsPattern recognitionMulti omics
The invention discloses a multi-omics cancer data integrating and analyzing method based on similarity fusion. The method comprises the steps of calculating a local similarity network, fusing multiplelocal similarity networks, typing according to a global similarity network, and backtracking the characteristic in an original data source according to the global similarity network. Compared with the prior art, the method is advantageous in that through modeling a similarity network connecting path which gradually advances, a fusion algorithm of multiple similarity networks is realized; a more complicated network structure can be described; and higher accuracy and higher stability are realized. Through a consistent alternating multiplier method, quick solving of a network fusion model is realized. The multi-omics cancer data integrating and analyzing method has advantages of utilizing the integrated global similarity network to typing of a cancer patient, obtaining types of the patientswith substantial prognosis difference, and combining with a multi-group characteristic selecting method for screening the key target characteristic. The selected characteristic has a potential of becoming a latent biological marker.
Owner:SOUTH CHINA UNIV OF TECH
An integrated method and system for identifying functional patient-specific somatic aberations using multi-omic cancer profiles
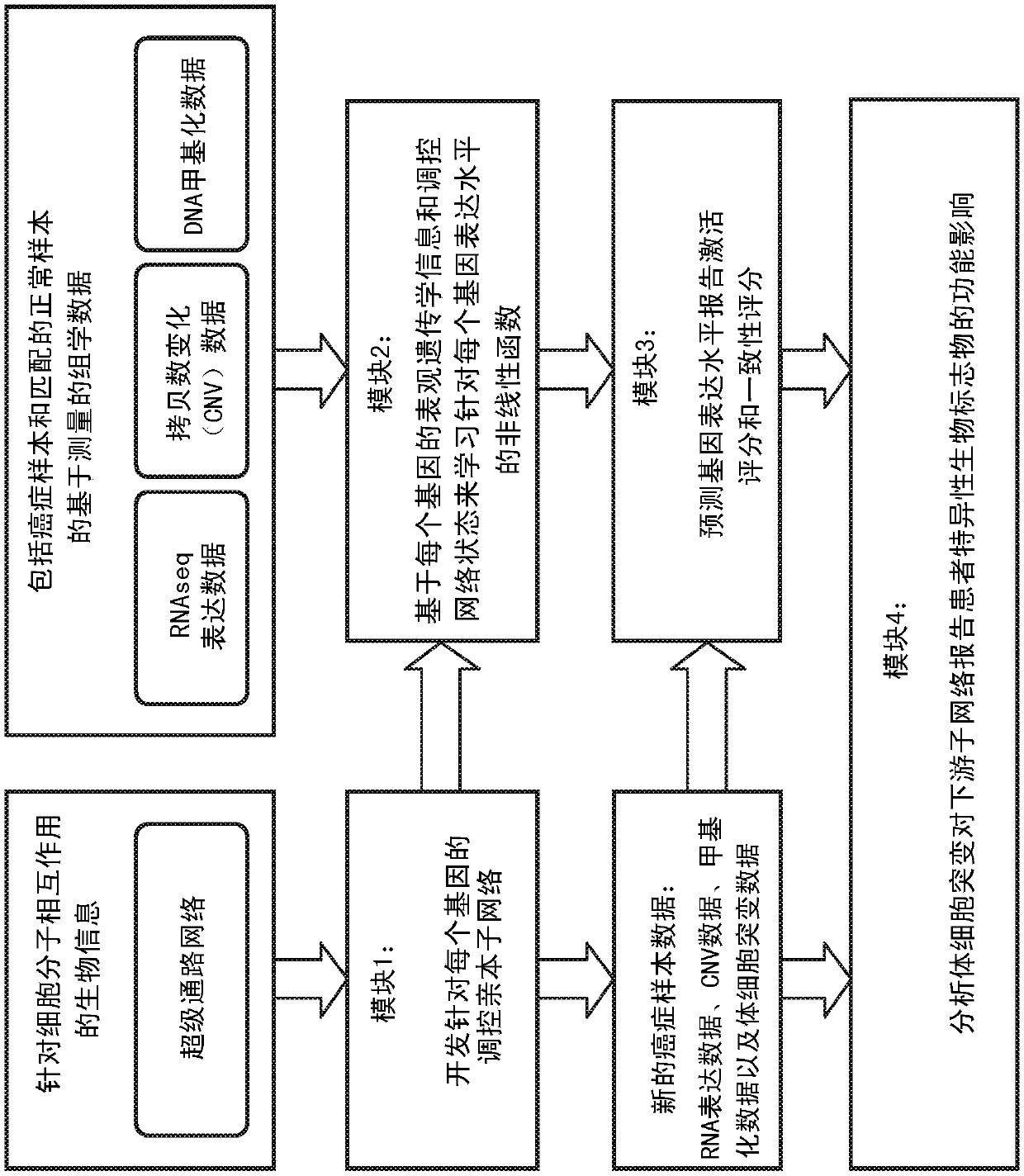
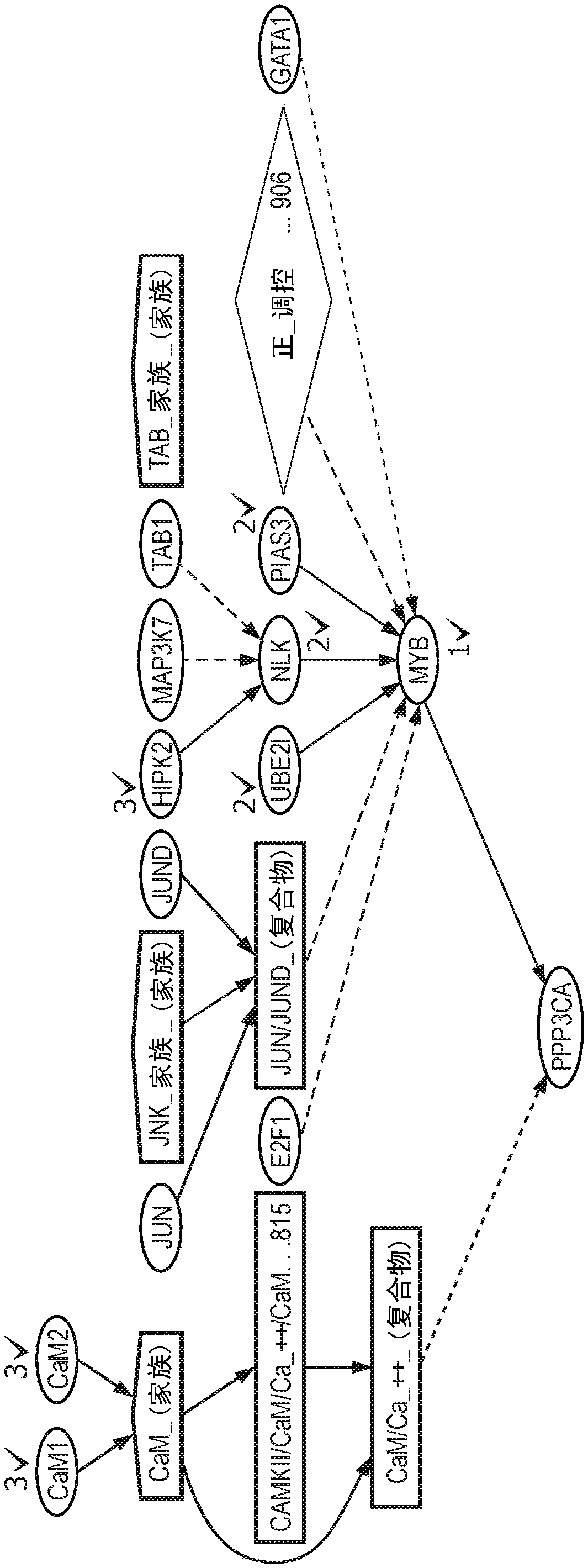
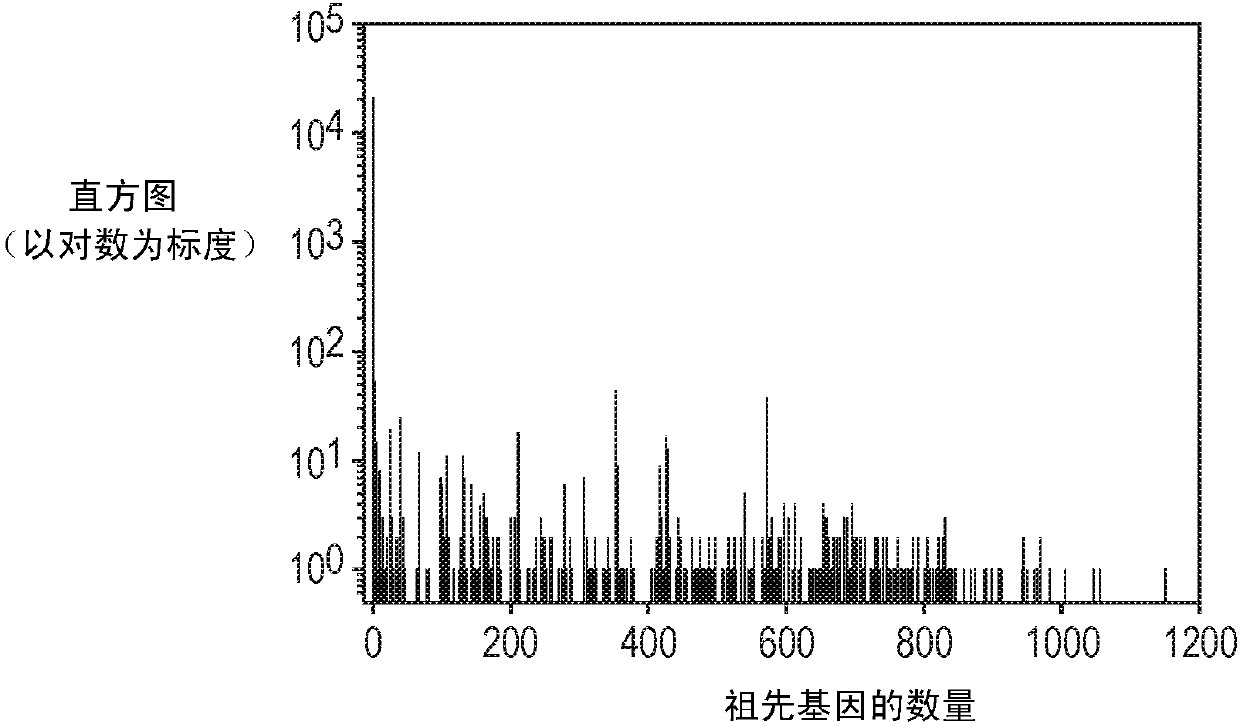
A system and method for determining the functional impact of somatic mutations and genomic aberrations on downstream cellular processes by integrating multi-omics measurements in cancer samples with community-curated biological pathways are disclosed. The method comprises the steps of extracting biological pathway information from well-curated biological pathway sources, using the pathway information to generate an upstream regulatory parent sub-network tree for each gene of interest, integrating measurement-based omic data for both cancer and normal samples to determine a nonlinear function for each gene expression level based on the gene's epigenetic information and regulatory network status, using the nonlinear function to predict gene expression levels and compare activation and consistency scores with inputted patient- specific gene expression data, and using the patient-specific gene expression predictions to identify significant deviations and inconsistencies in gene expressionlevels from expected levels in individual patient samples to identify potential biomarkers in providing predictive information in relation to cancer and cancer treatment.
Owner:KONINKLJIJKE PHILIPS NV +1
Multi-omic search engine for integrative analysis of cancer genomic and clinical data
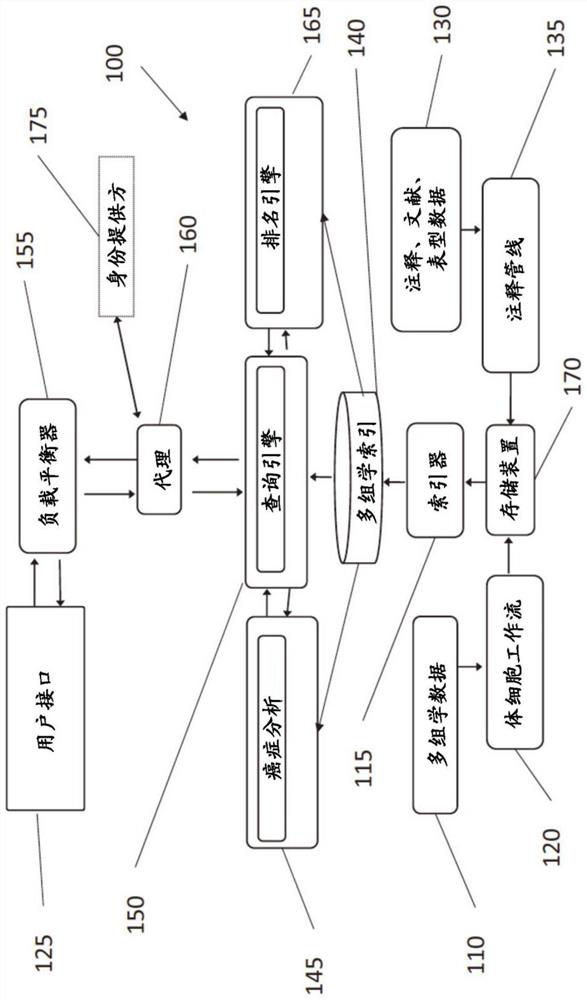
A method is provided for utilizing multi-omic data indices for tumor profiling. The method can comprise the steps of storing a plurality of multi-omic data indices, wherein each of the plurality of multi-omic data indices comprises cancer-specific tokenized data; ingesting additional multi-omic data and any annotations associated with the additional multi-omic data, the additional multi-omic data related to one or more indices; indexing the ingested additional multi-omic data and annotations while preserving gene names, gene variant names and multi-omic mapping between different data streams for the same patient in the specific index, to produce tokenized ingested additional multi-omic data; receiving a user query; selecting one or more relevant multi-omic data indices based on the user query; ranking the selected one or more multi-omic data indices based on at least one of clinical actionability, pathogenicity, feature weight, or frequency; and returning the ranked one or more multi-omic data indices to the user.
Owner:HUMAN LONGEVITY
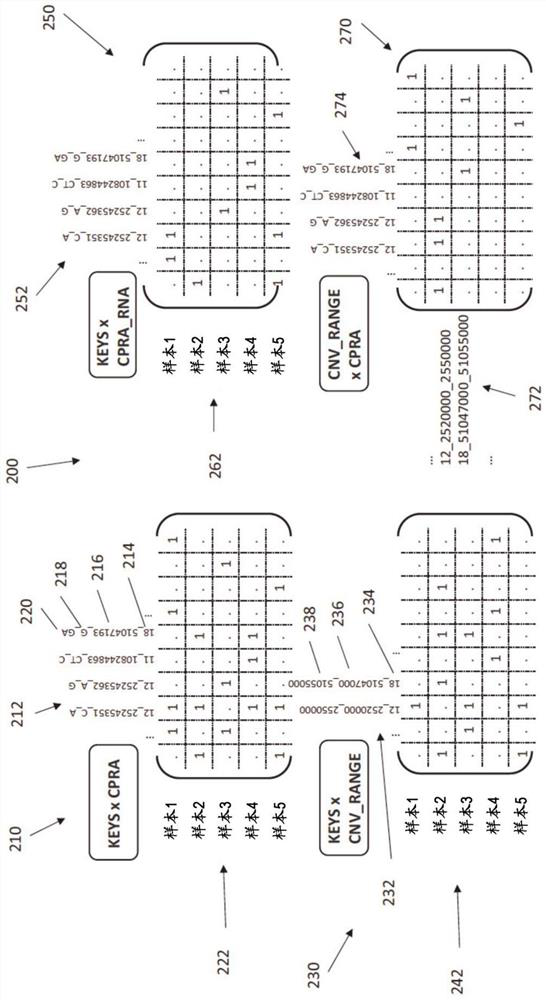
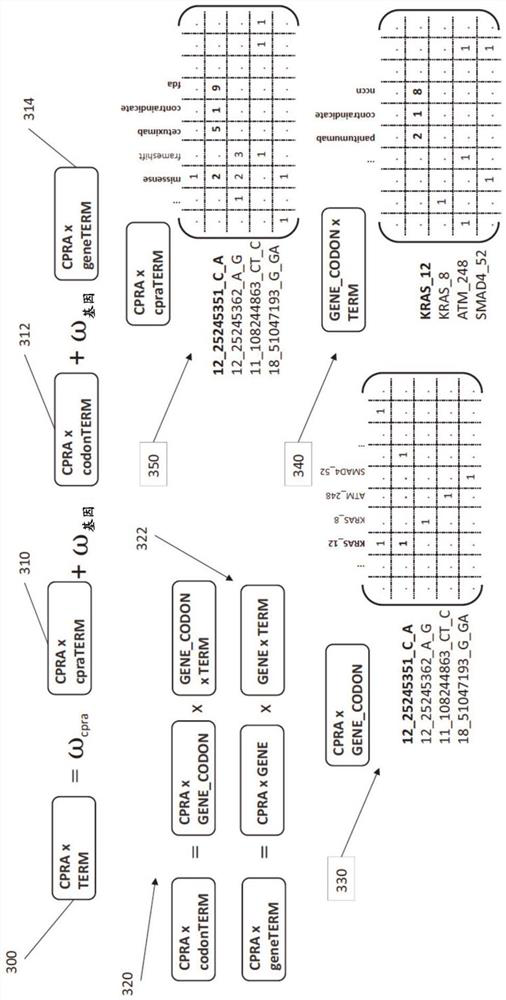
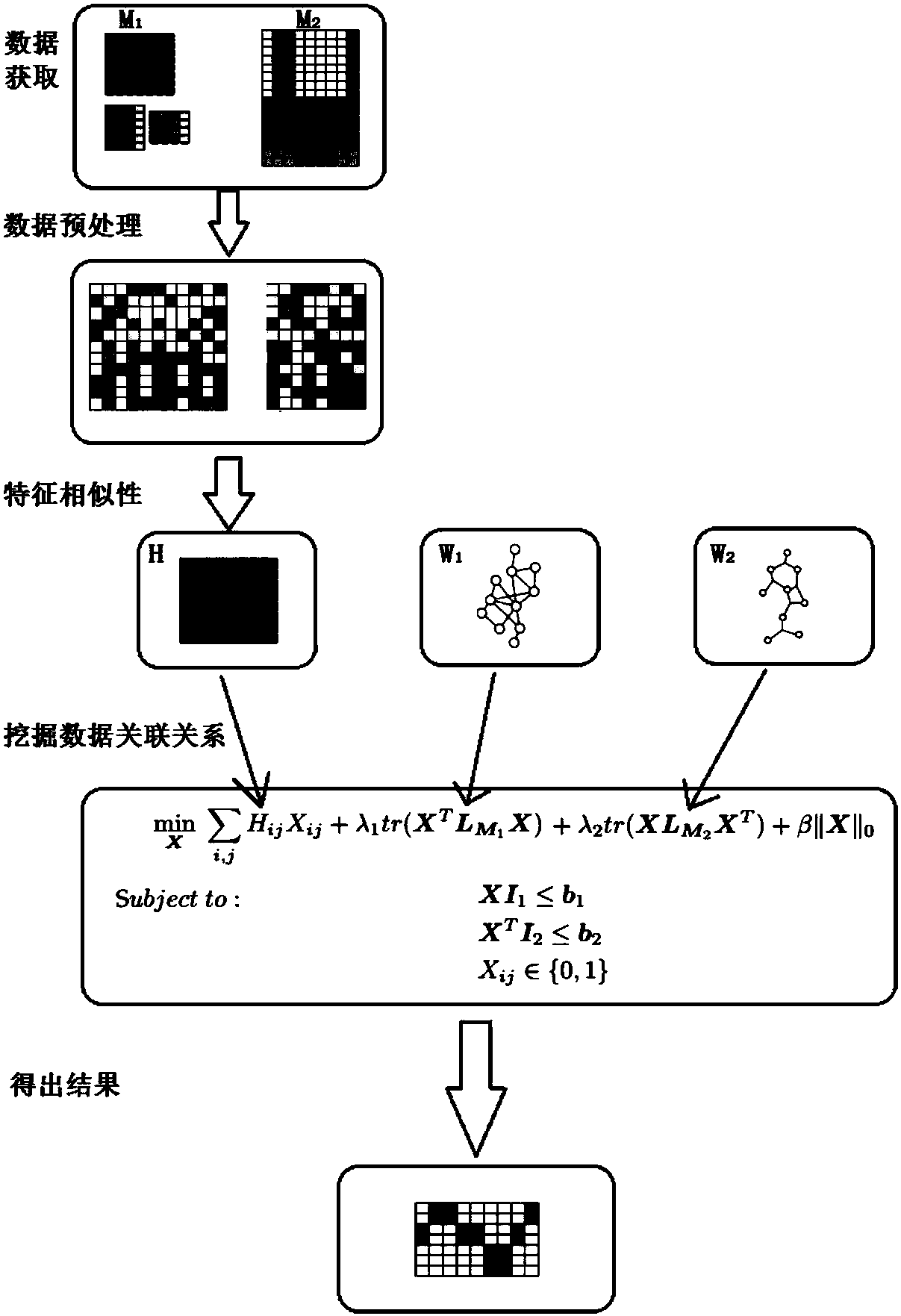
Multi-omics data association relationship discovery method based on sparse matching
ActiveCN108509771AHigh precisionImprove robustnessBiostatisticsSpecial data processing applicationsPrior informationMulti omics
The invention discloses a multi-omics data association relationship discovery method based on sparse matching. The method includes: preprocessing input data to improve quality of the data; selecting asuitable similarity measure according to data characteristics to calculate a similarity matrix among data features; and fusing prior information to mine potential association relationships among thedata features on the basis of a similarity network among the features. According to the method of the invention, the prior information of features of existing already-proven omics data can be fully utilized, impacts of noises on results can be reduced, uncertainty caused by data errors can be reduced, and accuracy and robustness of the results can be improved.
Owner:SOUTH CHINA UNIV OF TECH
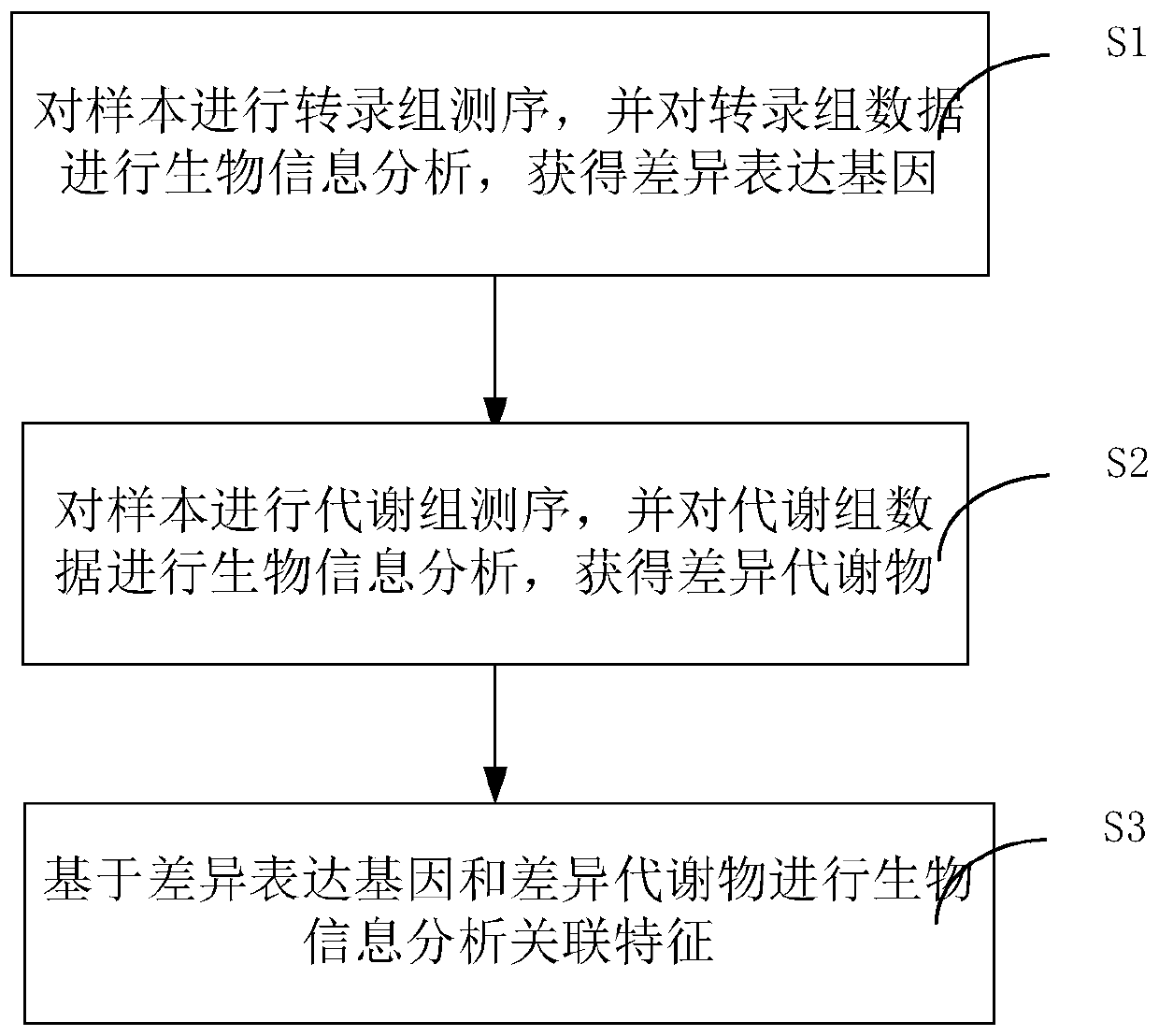
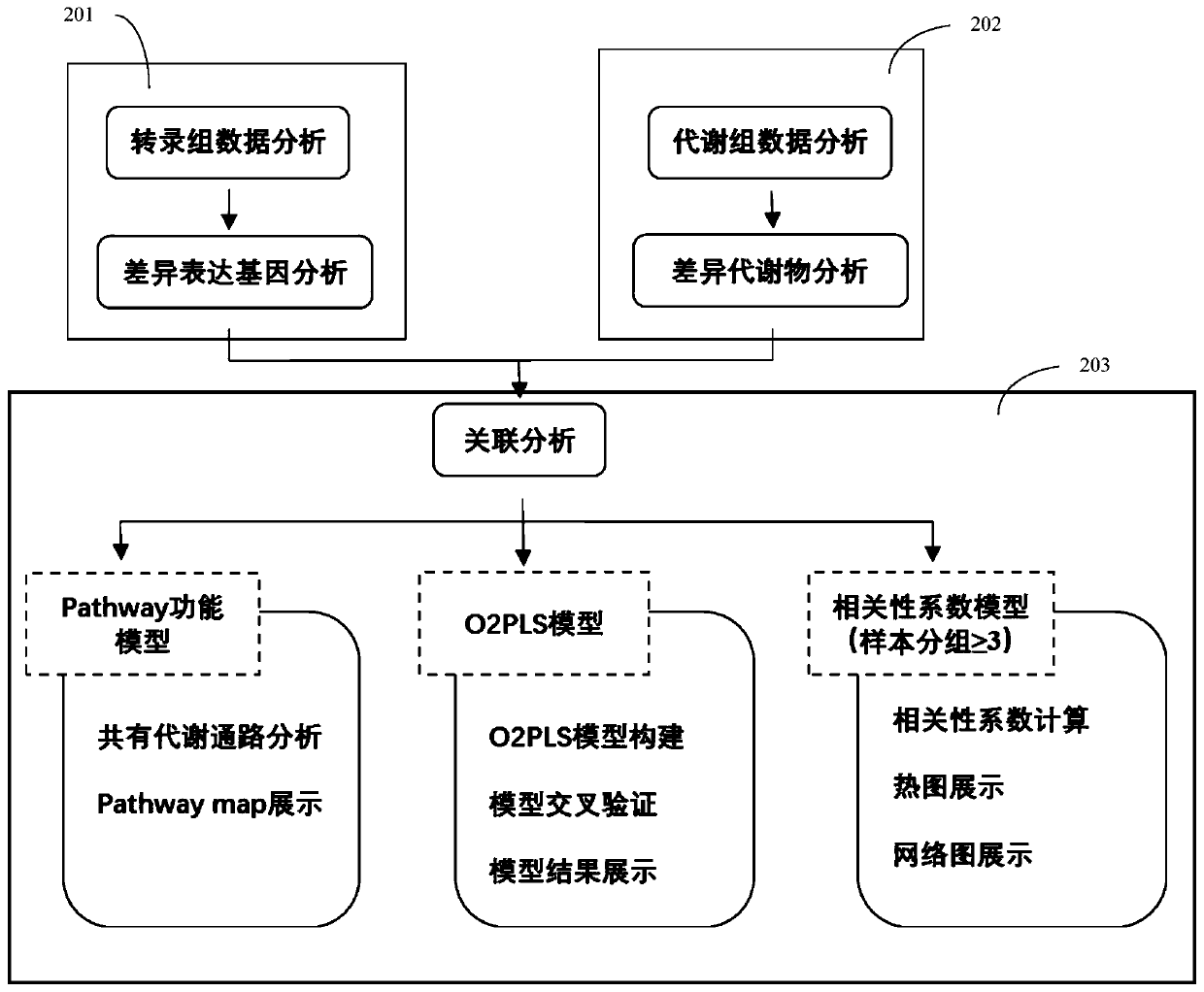
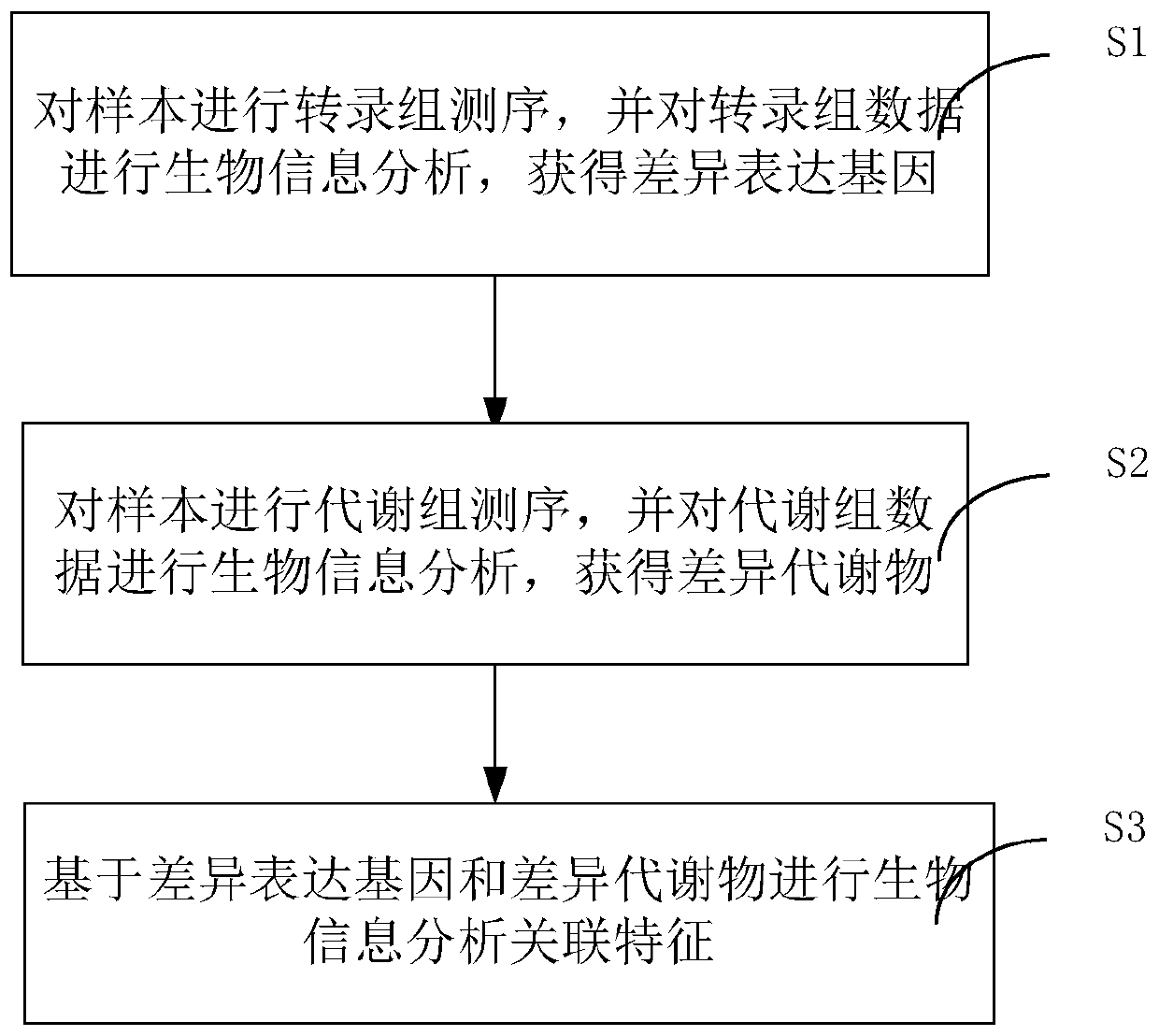
Method and system for association analysis of transcriptome and metabolome data
The invention discloses a method and a system for association analysis of transcriptome and metabolome data. The method comprises the following steps of S1, performing transcriptome sequencing on a sample, performing bioinformation analysis on the transcriptome data for obtaining a differential expression gene; S2, performing metabolome sequencing on the sample, and performing bioinformation analysis on the metabolome data for obtaining a differential metabolite; and S3, performing bioinformation analysis on the association characteristic based on the obtained differential expression gene andthe differential metabolite. The method and the system settle problems of one-sidedness of single omic sequencing data and low reliability of partial data in prior art. Furthermore, according to the method and the system, through information transmission, cooperation and coordination between biomolecules, a specific function is presented, thereby realizing more systematical multi-omics integratedanalysis and facilitating discloses of a complicated function mechanism.
Owner:GUANGZHOU GENE DENOVO BIOTECH
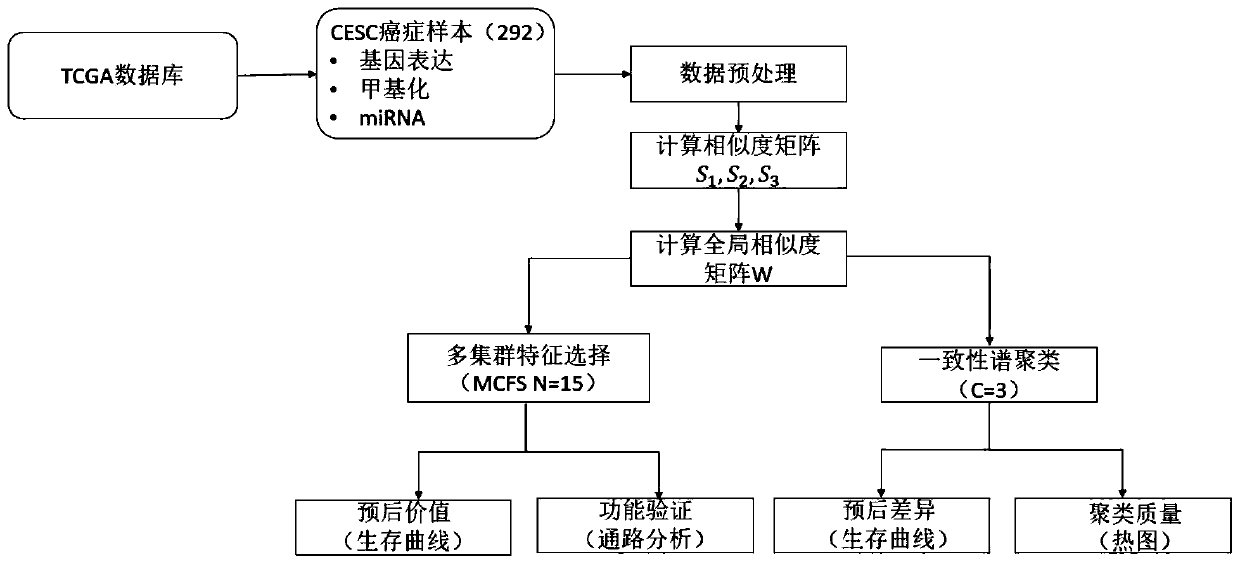
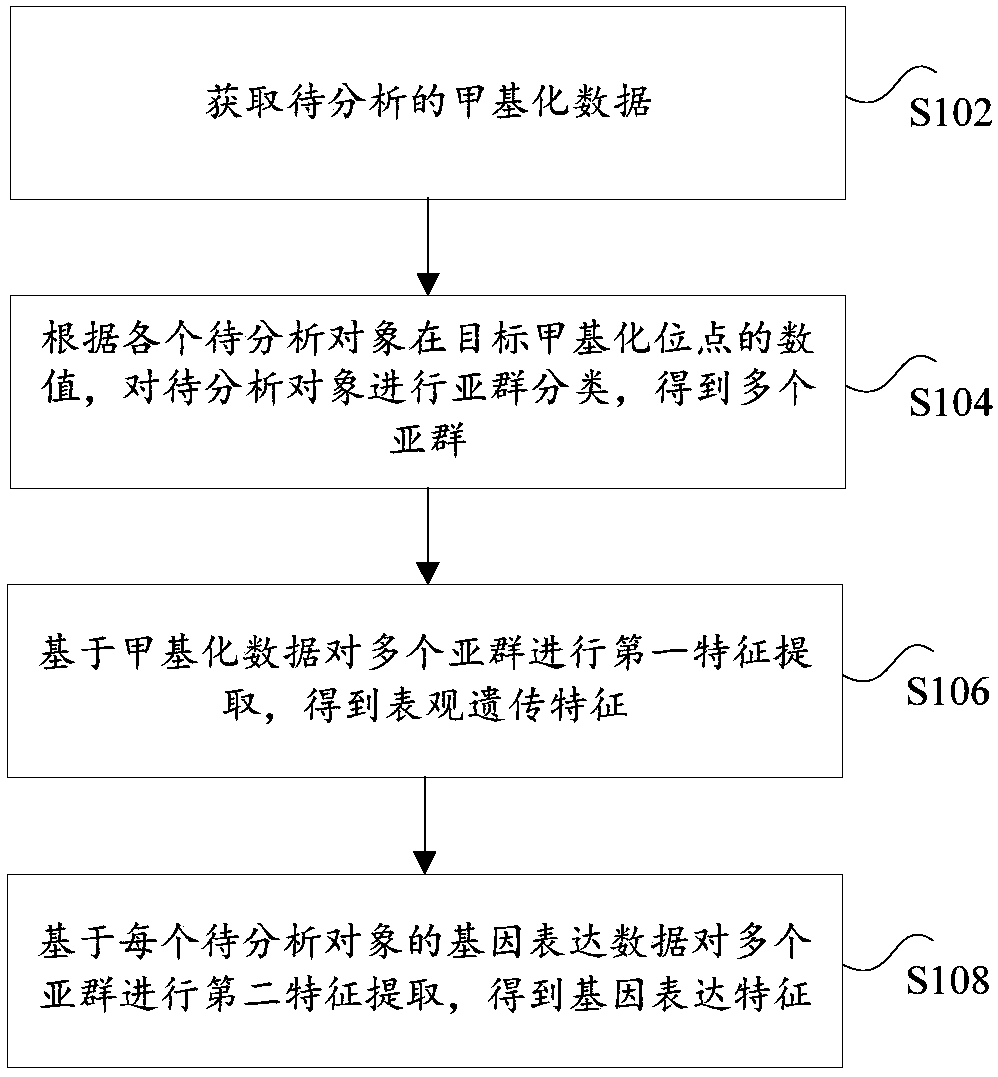
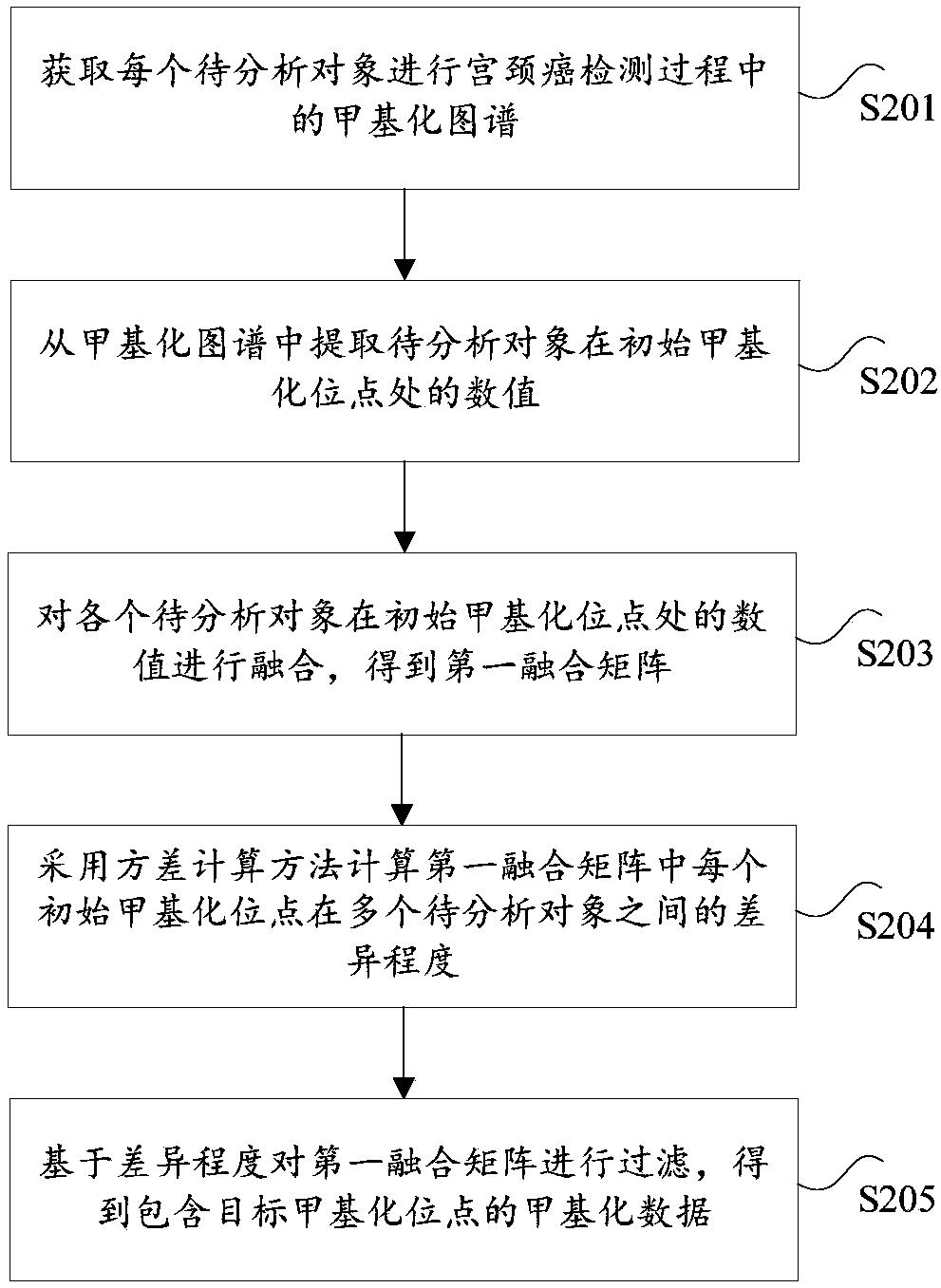
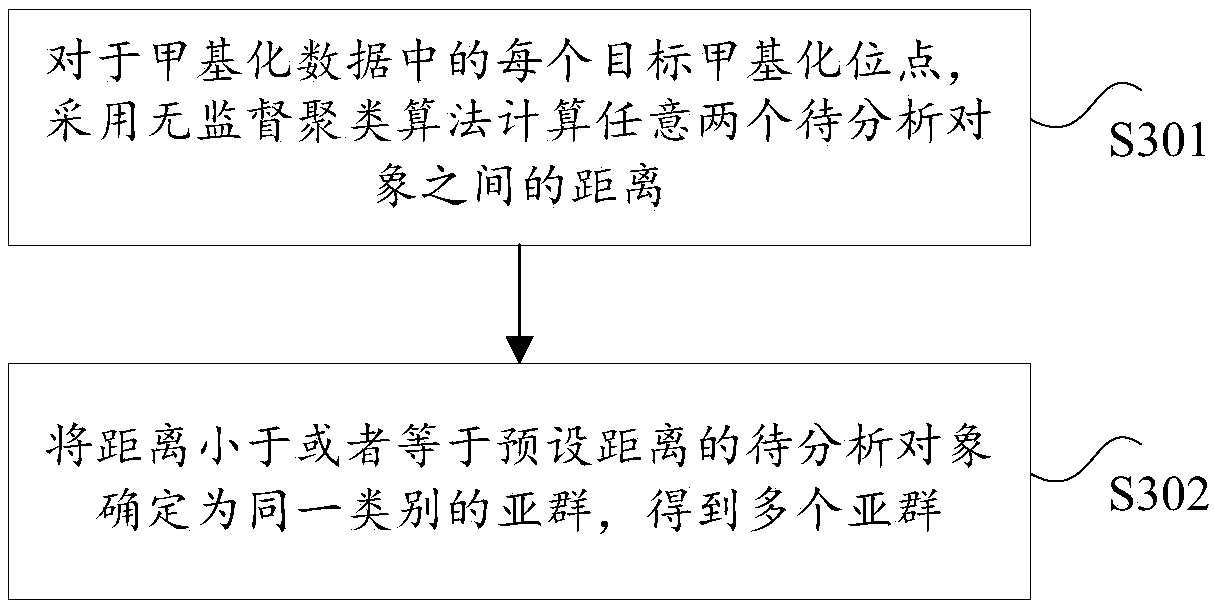
Multi-omics based cervical carcinoma characteristic obtaining method and system
PendingCN108949970AAccurate classificationMicrobiological testing/measurementSpecial data processing applicationsFeature extractionMulti omics
The invention provides a multi-omics based cervical carcinoma characteristic obtaining method and a system. The multi-omics based cervical carcinoma characteristic obtaining method comprises followingsteps: methylation data for analysis is obtained, wherein the methylation data is obtained in cervical carcinoma detection, and comprises target methylation site values of a plurality of objects foranalysis; based on the target methylation site values of the plurality of objects for analysis, the objects are subjected to subgroup classification so as to obtain a plurality of subgroups; the subgroups are subjected to first characteristic extraction based on the methylation data so as to obtain epigenetic characteristics; the subgroups are subjected to second characteristic extraction based onthe gene expression data of the objects for analysis so as to obtain gene expression characteristics. The transcription factor combination characteristics in epigenetic characteristics and the differential expression gene in gene expression characteristics can be used for determining target gene expression function characteristics, and solving problems in the prior art that the accuracy of classification of cervical carcinoma patients using a conventional method is poor, and characteristic extraction is not comprehensive.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
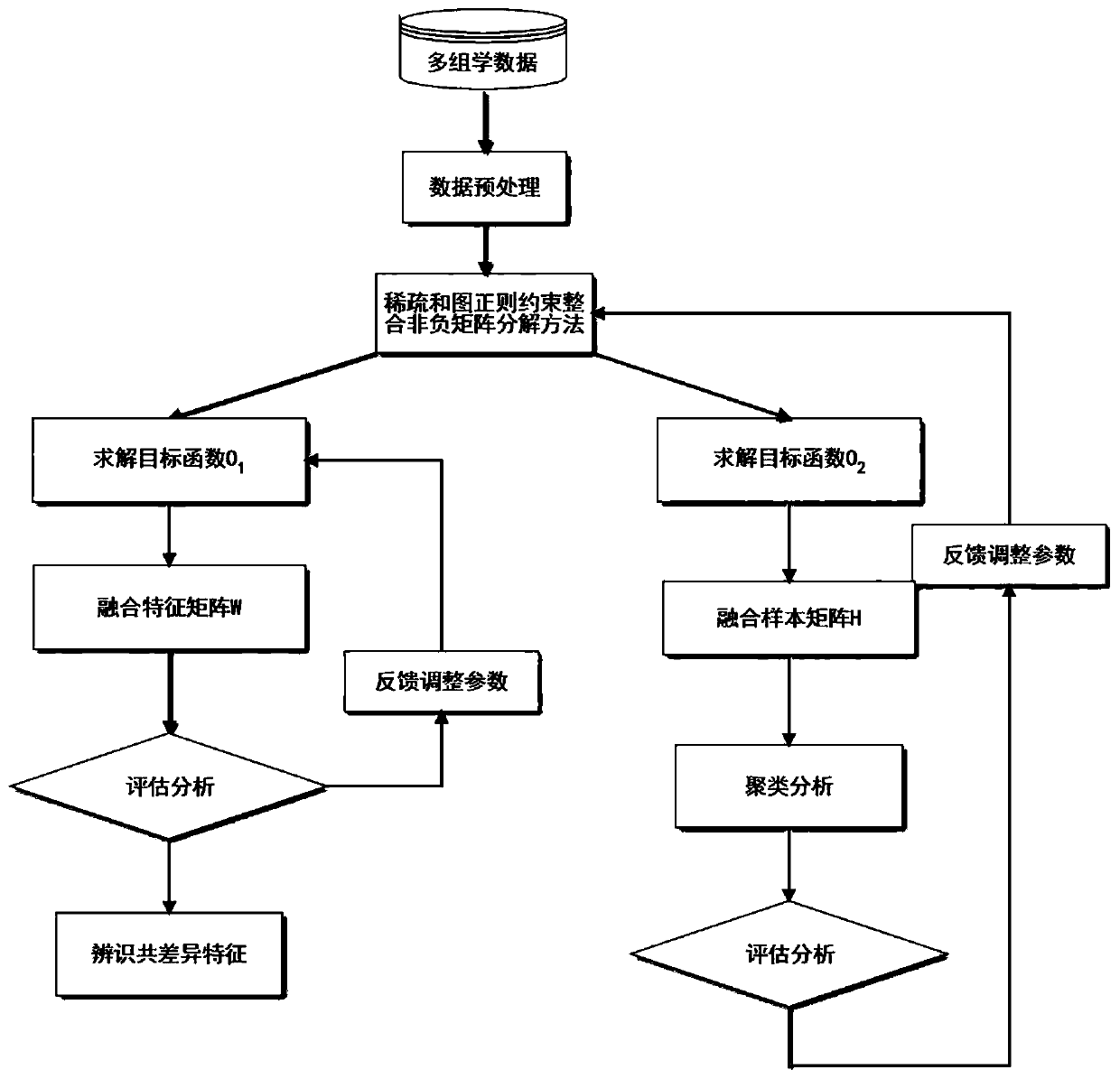
Sample clustering and feature recognition method based on integrated non-negative matrix factorization
ActiveCN110826635AImprove performanceImprove robustnessCharacter and pattern recognitionICT adaptationAlgorithmOmics
The invention discloses a sample clustering and feature recognition method based on integrated non-negative matrix factorization. The method comprises: 1, X = {X1, X2... XP} representing multi-view data composed of P different omics data matrixes of the same cancer; 2, constructing a diagonal matrix Q; 3, introducing graph regularization and sparse constraints into the integrated non-negative matrix factorization framework to obtain target functions O1 and O2; 4, solving the target function O1 to obtain a fusion feature matrix W and a coefficient matrix HI; solving the target function O2 to obtain a feature matrix WI and a fusion sample matrix H; 5, constructing an evaluation vector according to the fusion feature matrix W, and identifying common difference features according to the vector; 6, performing functional explanation on the identified common difference characteristics by using GeneCards; and 7, performing sample clustering analysis according to the fusion sample matrix. According to the method, the complementary and difference information of the multiple omics data can be fully utilized to identify the common difference characteristics, clustering analysis can be carriedout on the sample data provided by the multiple omics data, and a calculation method basis is provided for integrated research of different types of omics data.
Owner:QUFU NORMAL UNIV
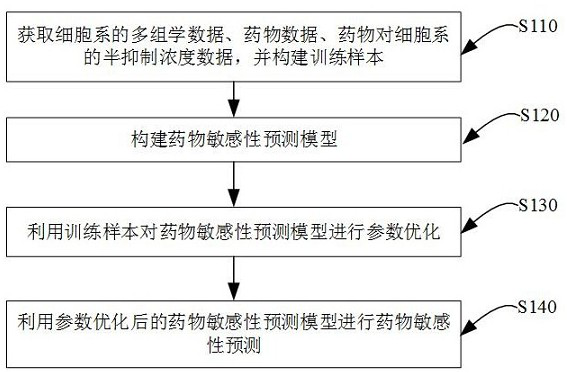
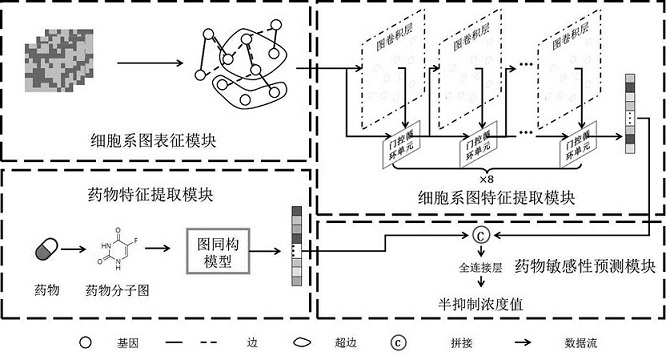
Drug sensitivity prediction method and device based on multi-omics data fusion
The invention discloses a drug sensitivity prediction method and device based on multi-omics data fusion, and belongs to the field of drug sensitivity detection. The method comprises the steps that three kinds of multi-omics information including genomics data, proteomics data and metabonomics data of individual cell lines are integrated through a cell line diagram characterization module, and a cell line multilateral diagram is obtained; according to the cell line multilateral graph, multi-omics information of a cell line and potential relation between expression products of genes in a multi-omics level are fully considered, then feature extraction is conducted on the cell line multilateral graph through a cell line graph feature extraction module, and node features and edge features in the cell line multilateral graph are fully extracted to serve as cell line features; and finally, a drug sensitivity prediction module is adopted to predict the semi-inhibitory concentration of the drug according to the cell line features and the drug features extracted based on the drug feature extraction module. According to the method, the prediction accuracy of the drug sensitivity is improved on the basis of comprehensively considering genomics data, proteomics data and metabonomics data.
Owner:ZHEJIANG UNIV
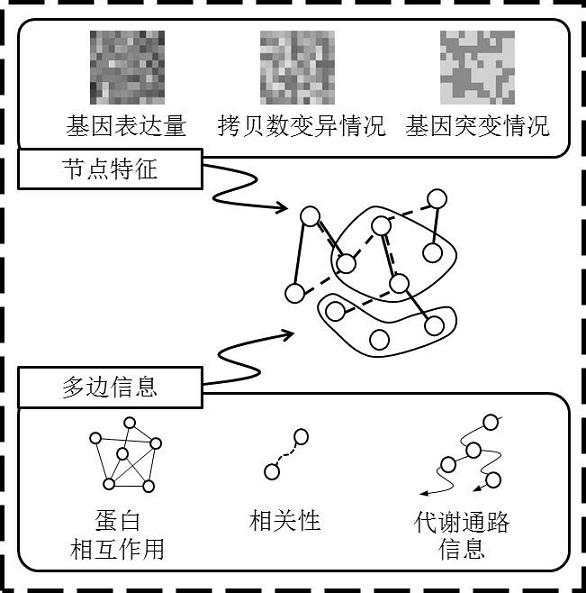
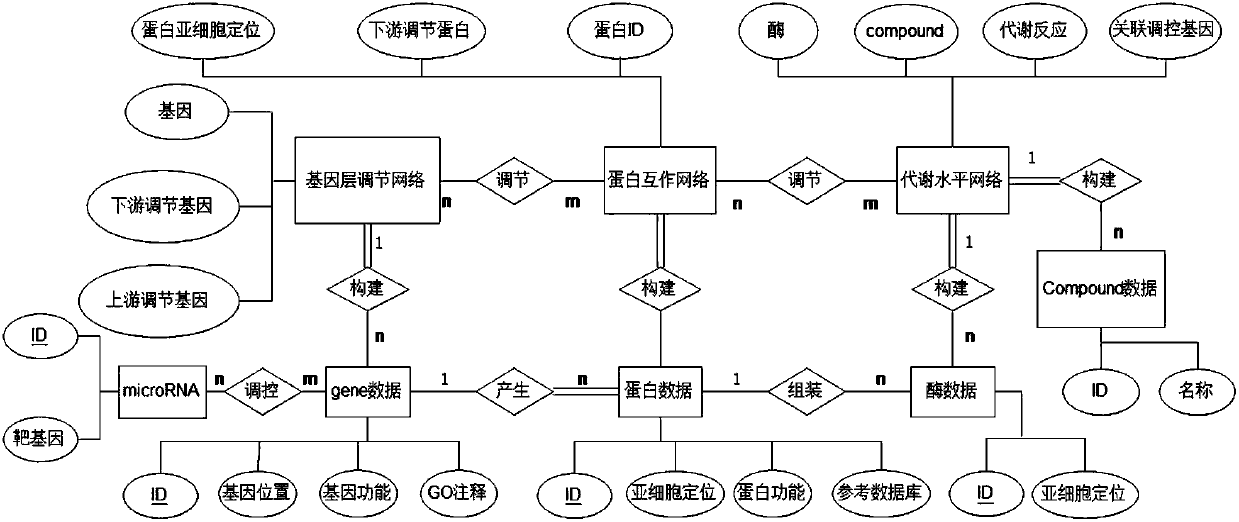
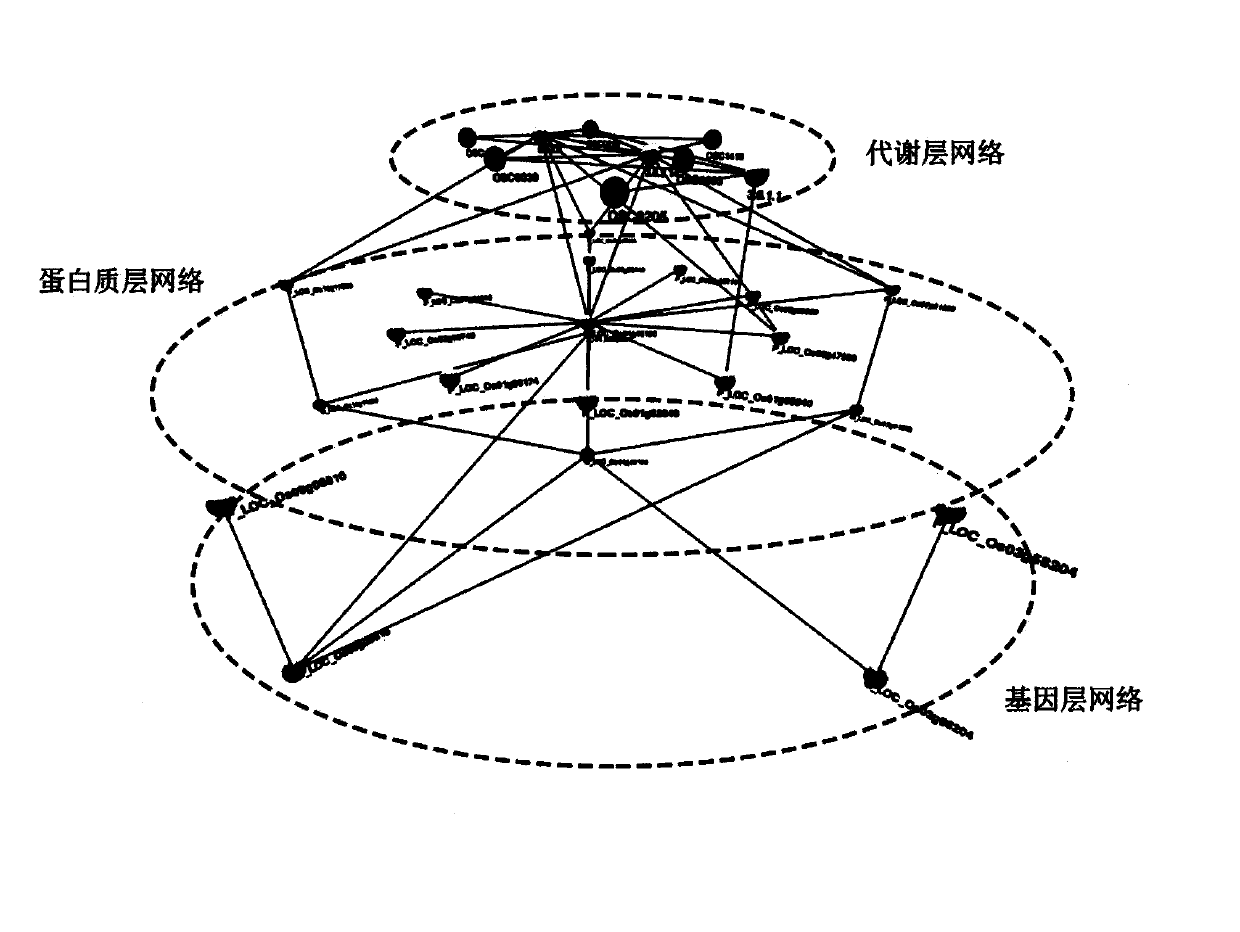
Plant whole-genome multilevel biological network reconstruction method based on multi-omics data integration
InactiveCN107862176ARealize 3D visualizationSpecial data processing applicationsReconstruction methodMetabolic network
The invention discloses a plant whole-genome multilevel biological network reconstruction method based on multi-omics data integration. The method carries out integration on a rice gene regulation andcontrol network, a protein interaction network and a metabolic network to finish the construction of a rice multilevel regulation and control network; and meanwhile, through the integration of expression profile data in different tissue growth processes of the rice, according to the constructed rice multilevel regulation and control network, a specific tissue multilevel gene regulation and control network is constructed so as to construct a first high-quality rice genome scale multilevel biological network of which the current data level is most integral. A new thought is provided for the construction of the plant multilevel biological network, and a constructed rice multilevel gene regulation and control network database realizes the 3D (Three-Dimensional) visualization of the multilevelregulation and control network.
Owner:ZHEJIANG UNIV
Method of analysis of regulatory relationship between genes or loci and disease based on multi-omics
The invention discloses a method of analysis of a regulatory relationship between genes or loci and a disease based on multi-omics, and belongs to the field of biological information and computer data analysis. The method includes the following steps that after a patient sample is collected, the pathogenesis is analyzed by transcriptome analysis, CHIP-Seq analysis, miRNA analysis and GWAS analysis respectively, and the mutant genes or loci and the confidence coefficients obtained are (g1, x1), (g2, x2), (g3, x3) and (g4, x4) respectively; the above analysis result is integrated by an estimation function htheta(x)=0.2+0.7*x1+0.6*x2+0.3*x3+0.9*x4, so that the regulatory relationship between the genes or the loci and the disease is analyzed. According to the method, the accuracy of analysis of the genes related to the disease by the estimation function reaches 87%.
Owner:武汉古奥基因科技有限公司
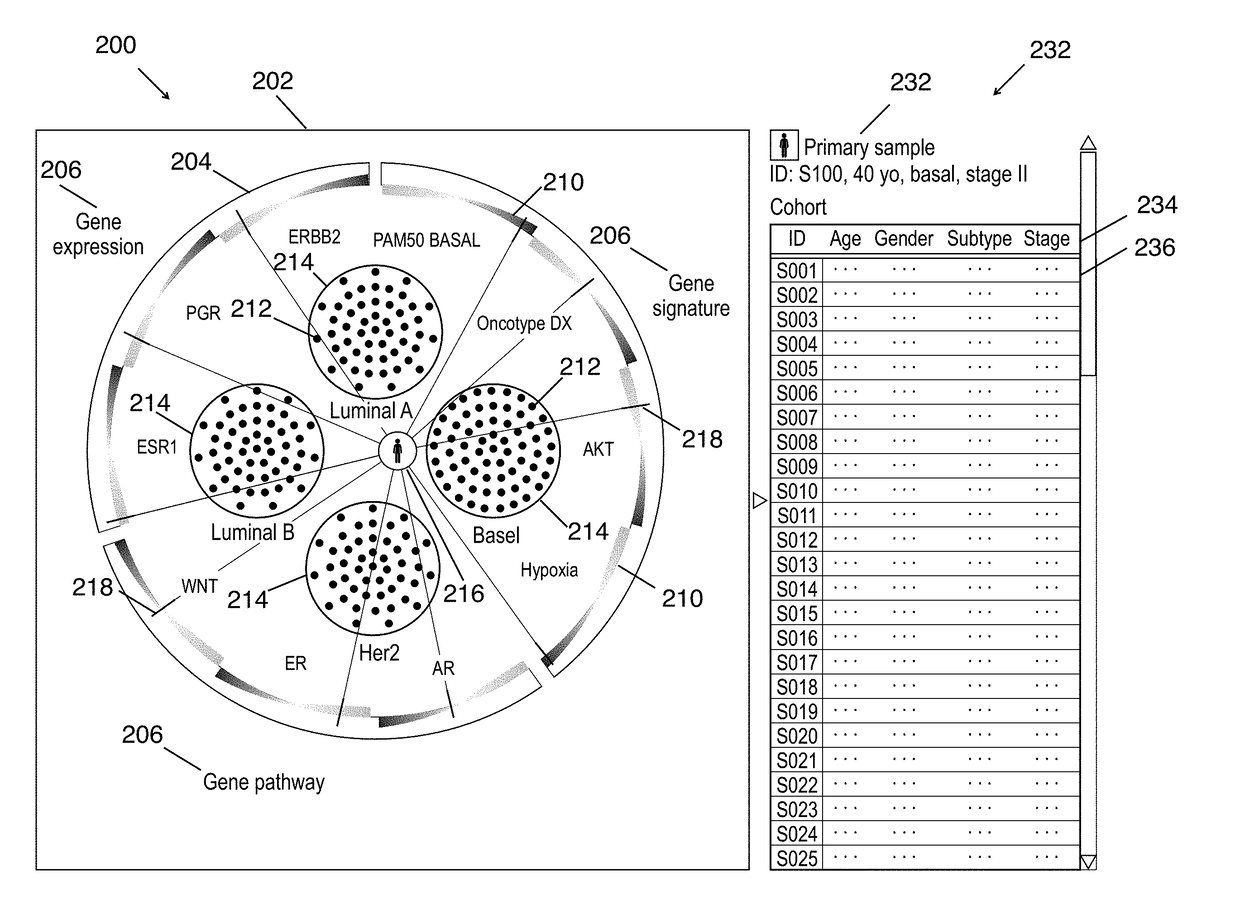
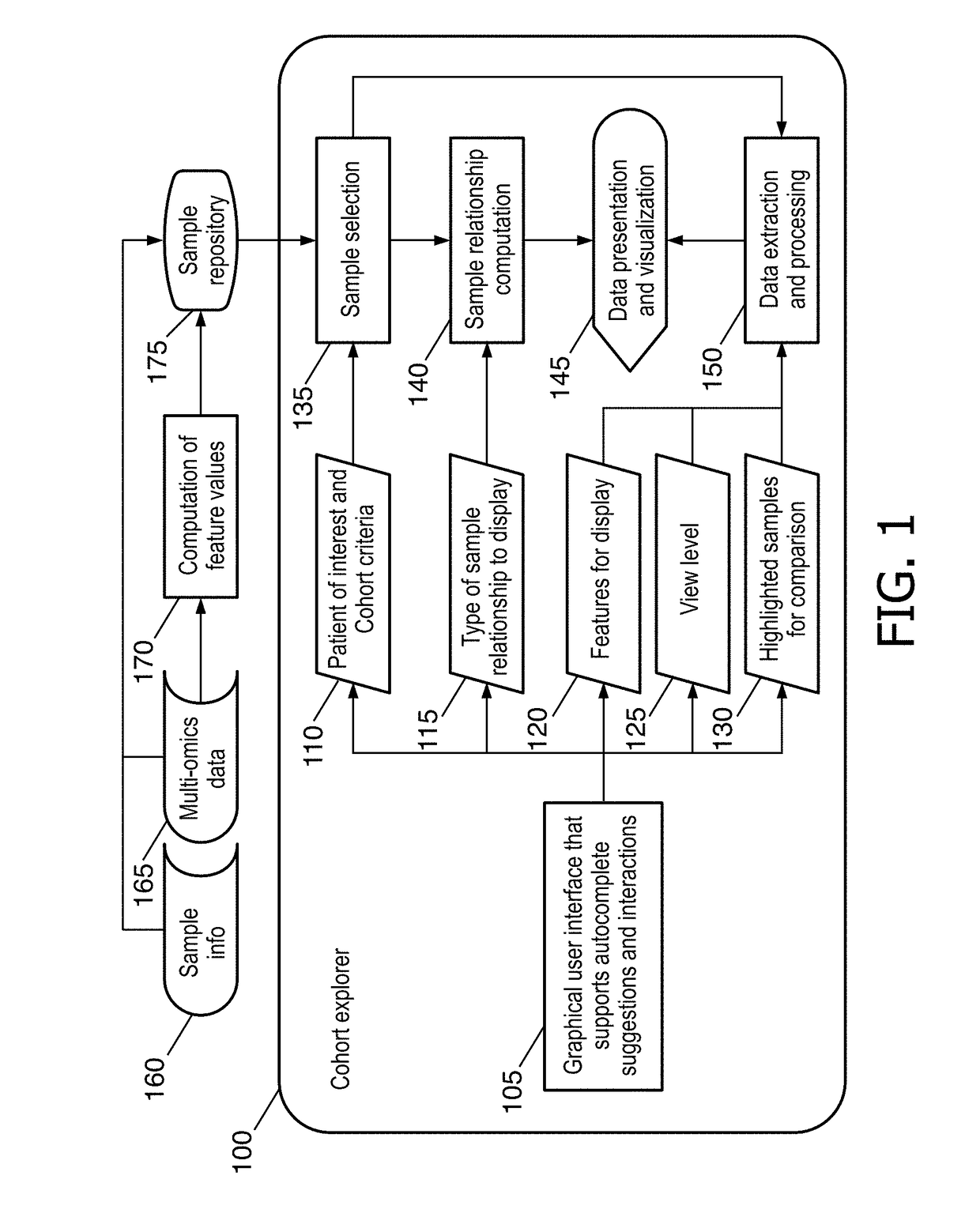
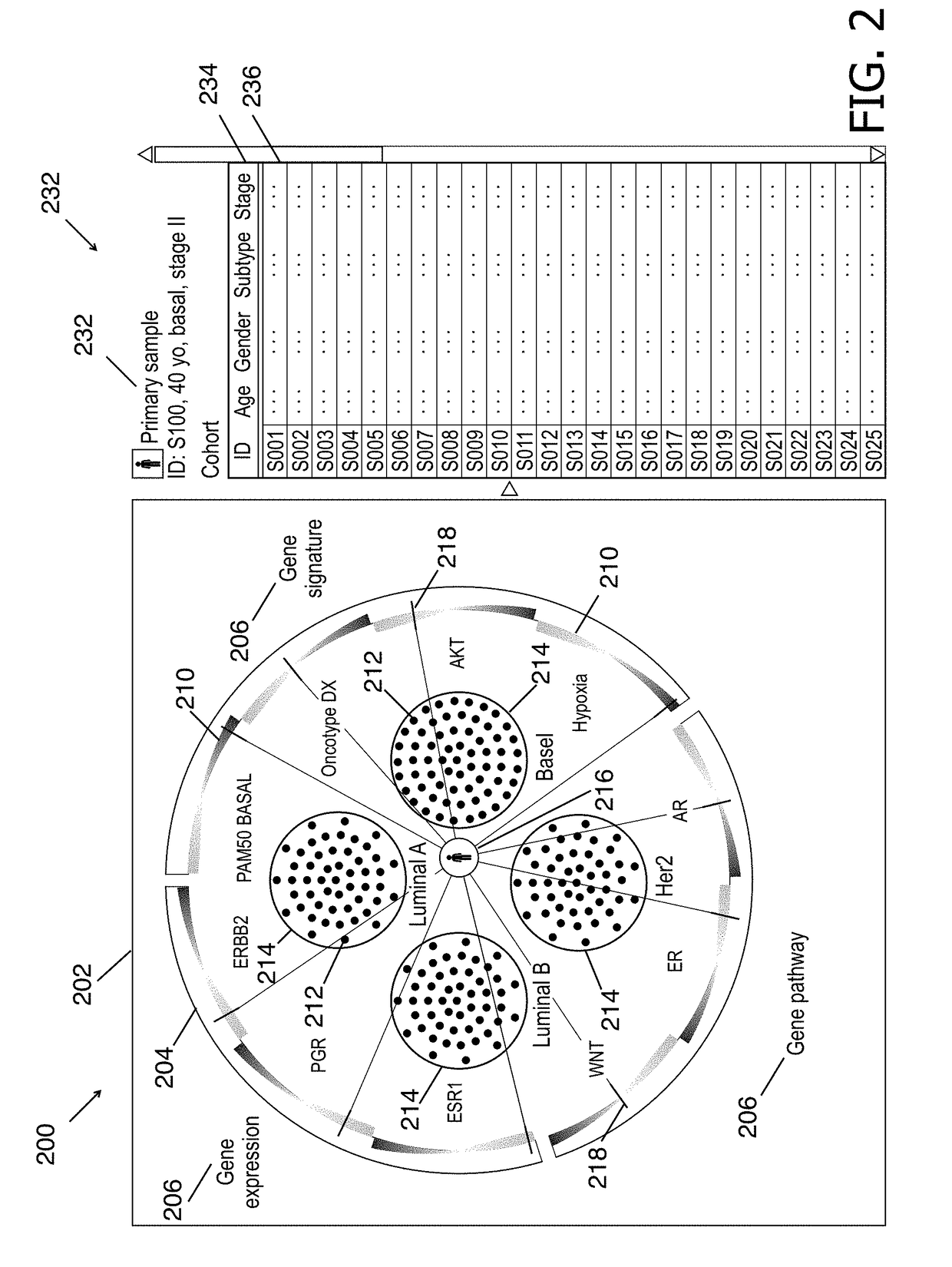
Cohort explorer for visualizing comprehensive sample relationships through multi-modal feature variations
InactiveUS20180330805A1Easy to demonstrateMedical data miningDrawing from basic elementsGenomicsMulti omics
A data-driven integrative visualization system and a method for visualization and exploration of the multi-modal features of a cohort of samples, is disclosed Specifically, a method for providing an interactive computation and visualization front-end of a genomics platform for presenting the complex multiparametric and high dimensional, multi-omic data of a patient with respect to a cohort of samples, that assists the user in understanding the similarities and differences across individual or groups of samples, identify correlation among different features and improve treatment planning and long term patient care, is described. The method may include obtaining and inputting multi-omic data of a patient and / or cohorts, identifying multi-modal feature variations and their relationships, and displaying this information in an interactive circular format on a GUI, from which the user can access further information. The system may provide an improved process of integrative analysis on a patient's multi-omic data for effective treatment planning.
Owner:KONINKLJIJKE PHILIPS NV
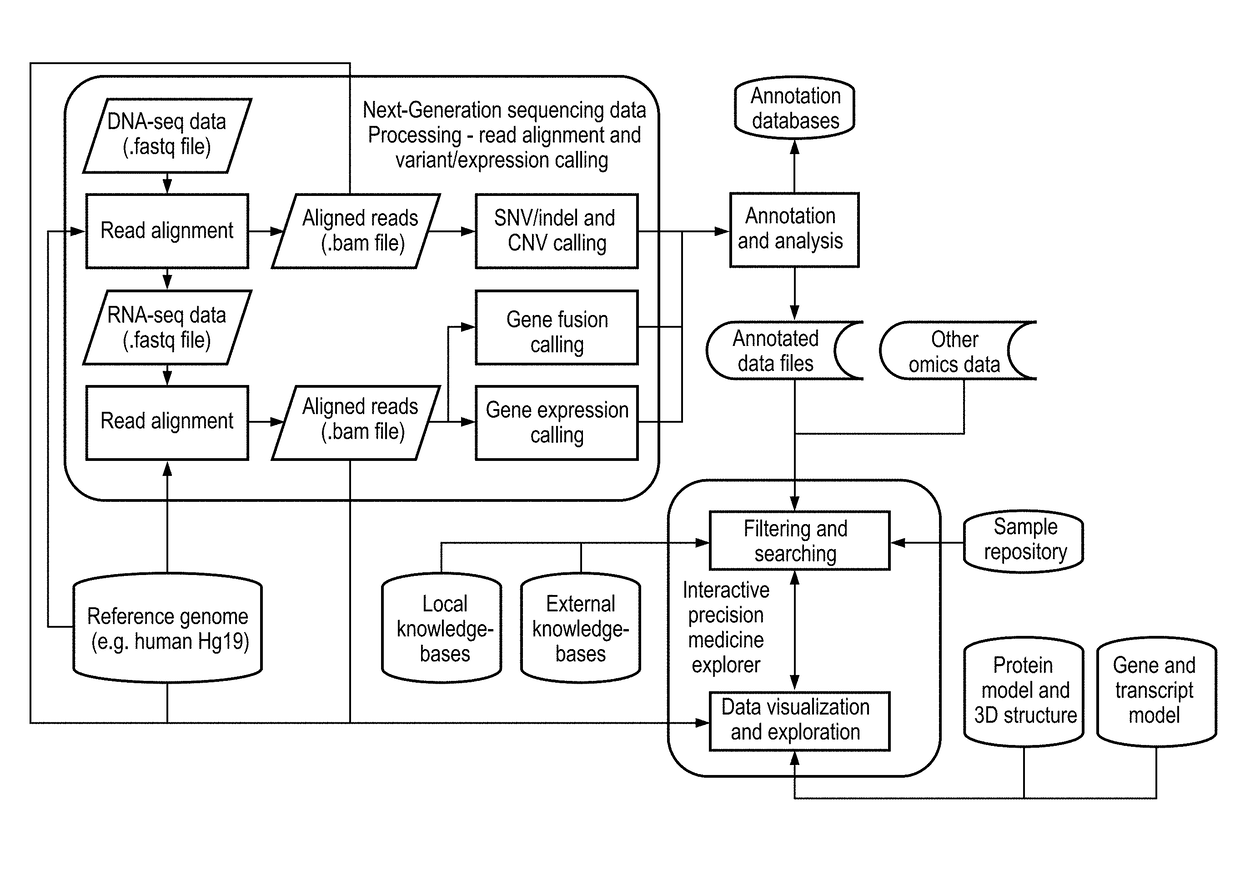
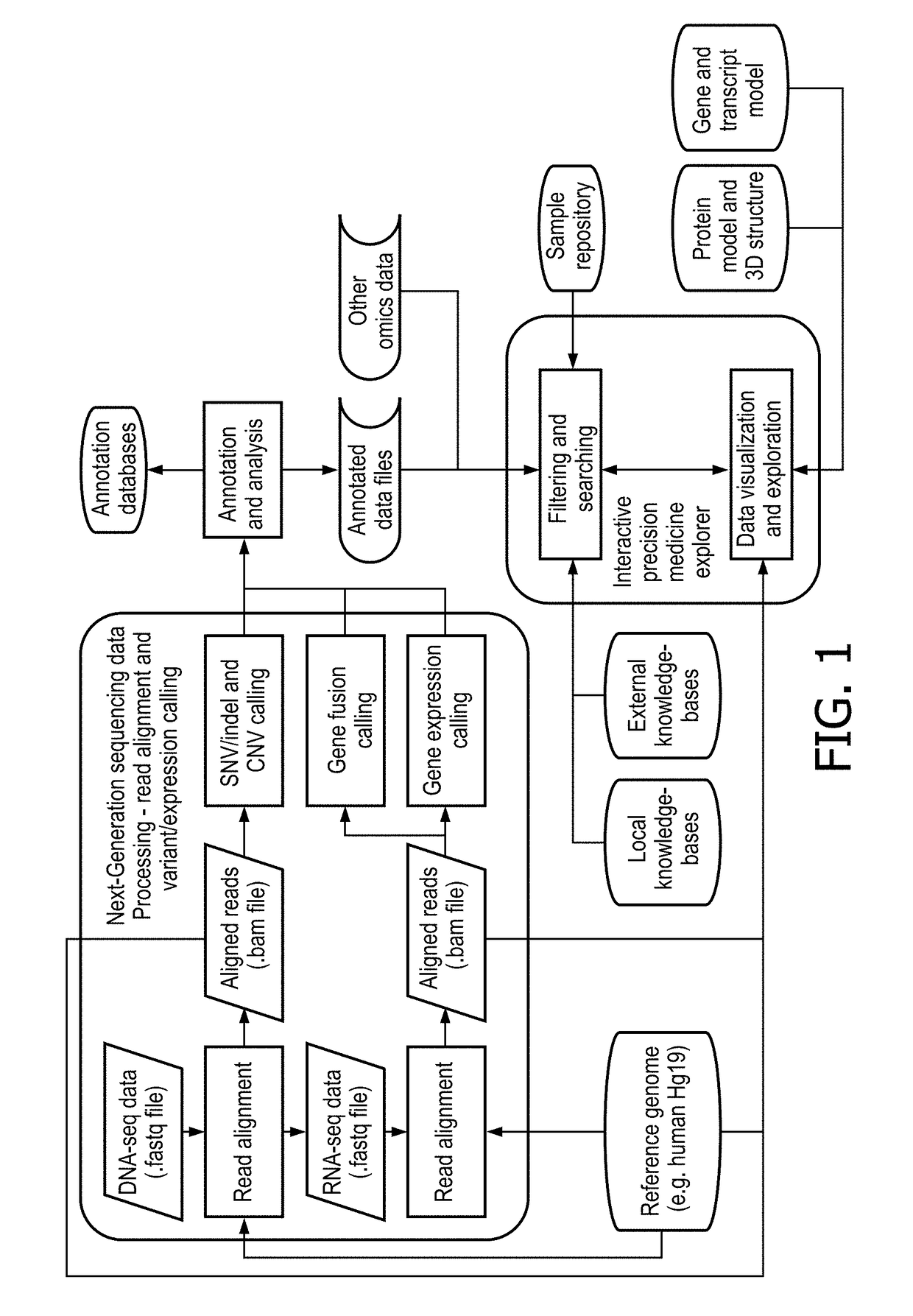
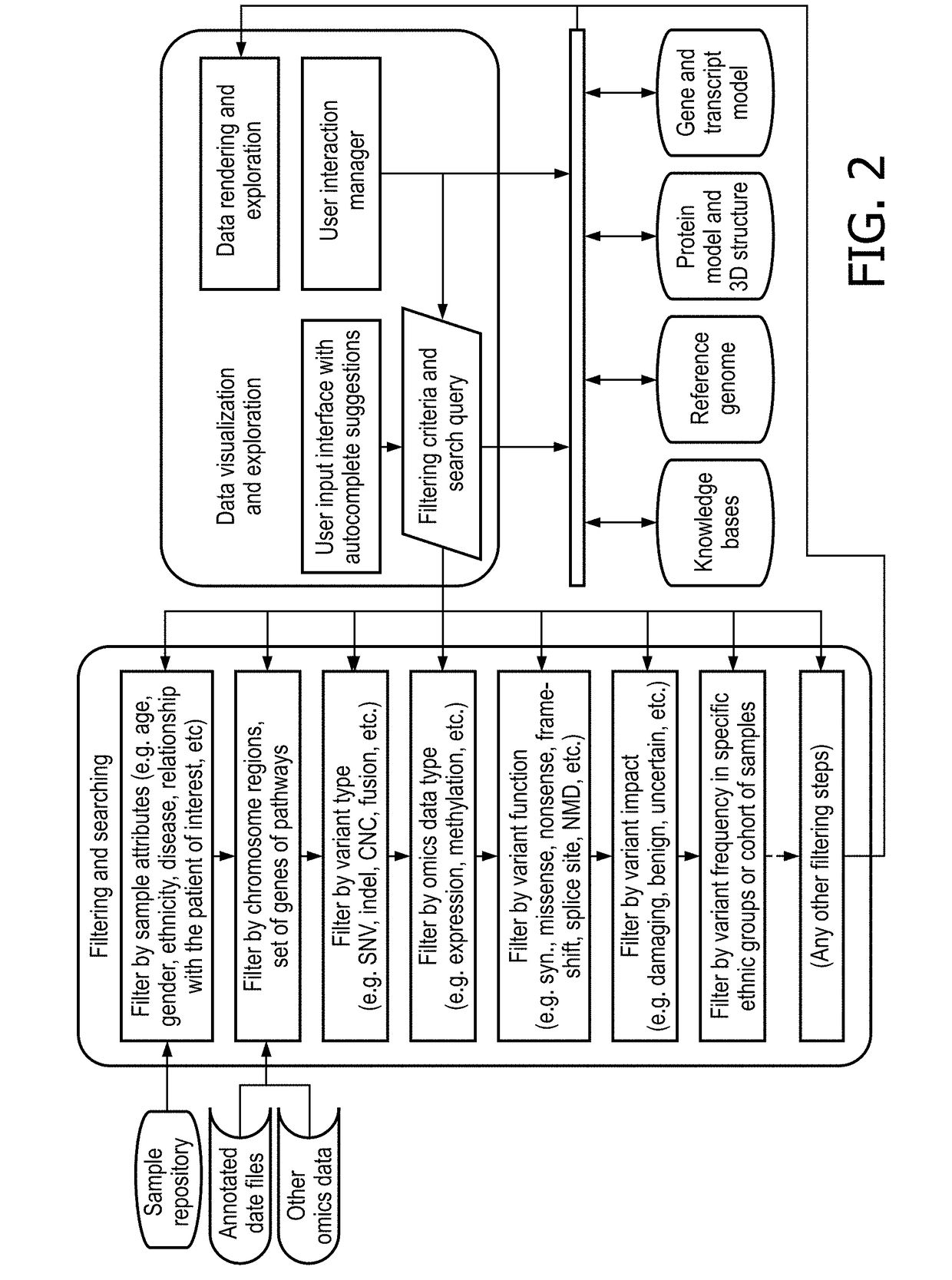
Interactive precision medicine explorer for genomic abberations and treatment options
PendingUS20180314795A1Improve viewing effectEasy to demonstrateMedical simulationData visualisationTreatment optionsMolecular level
A data-driven integrative visualization system and method for summarizing and presenting genomic aberrations, their drug responses and multi-omic data of a patient, is disclosed. Specifically, a method for displaying genomic aberrations and multi-omic data of a patient in an interactive tool which allows the medical practitioner to access underlying supporting biologic and scientific evidence from relevant knowledge bases through a set of graphical interactions, is described. The method comprises the steps of obtaining and inputting multi-omic data of a patient or cohorts, identifying genomic aberrations and their drug responses, and displaying this information in a first level interactive classical / circular ideogram in one or multiple layers on a GUI, from which the user can access and view further information on the gene and molecular levels. The system provides an improved process of integrative analysis on a patient's multi-omic data for effective treatment planning.
Owner:KONINKLJIJKE PHILIPS NV
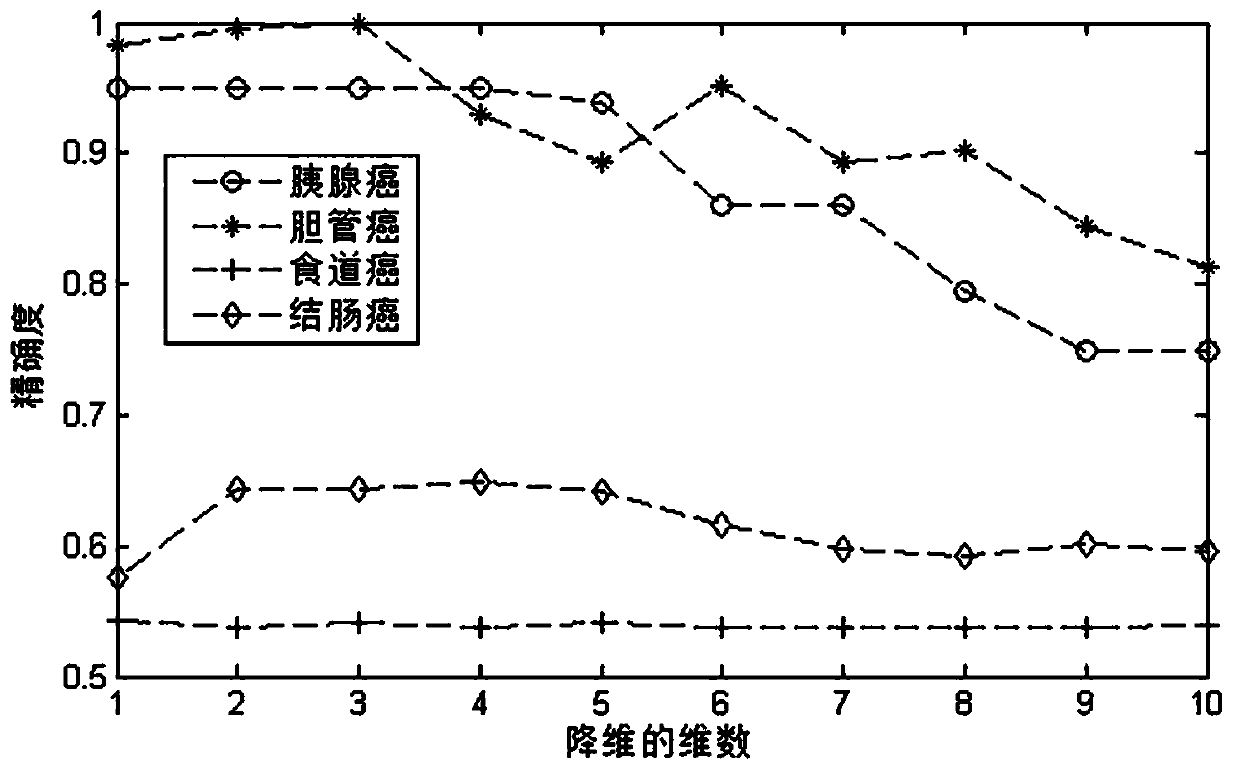
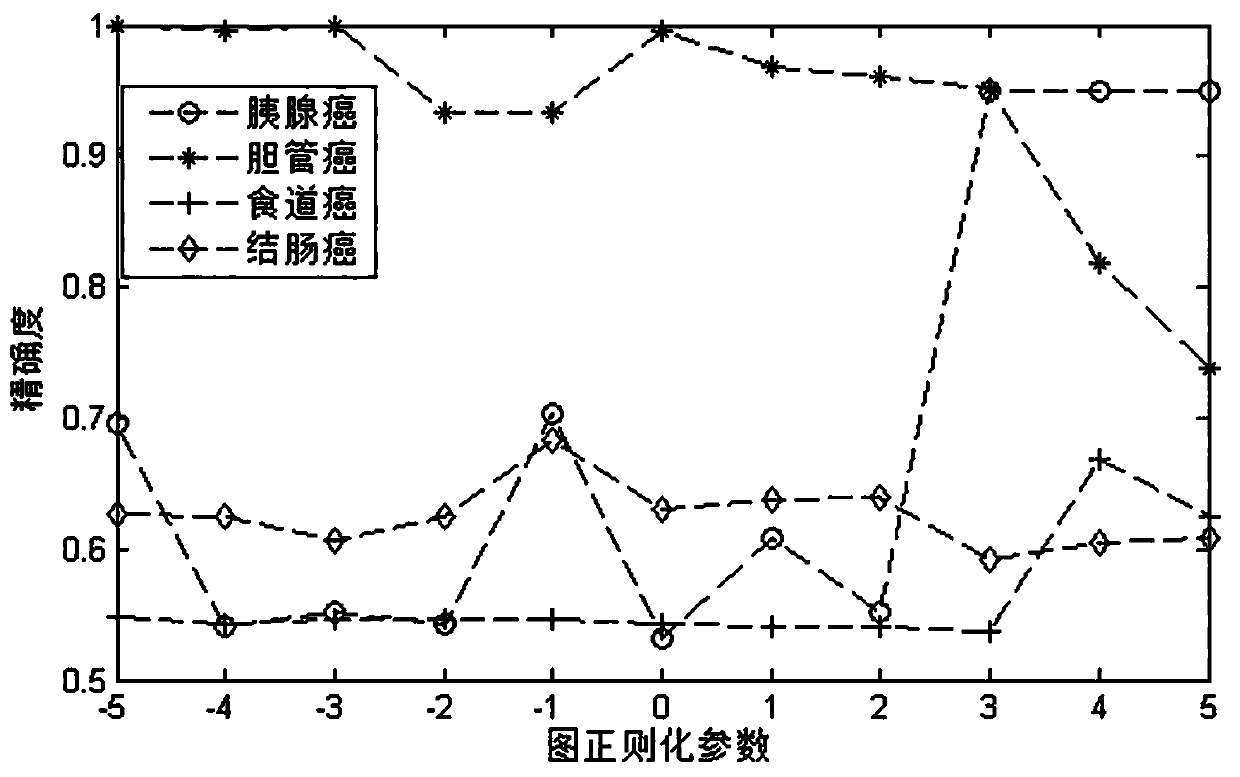
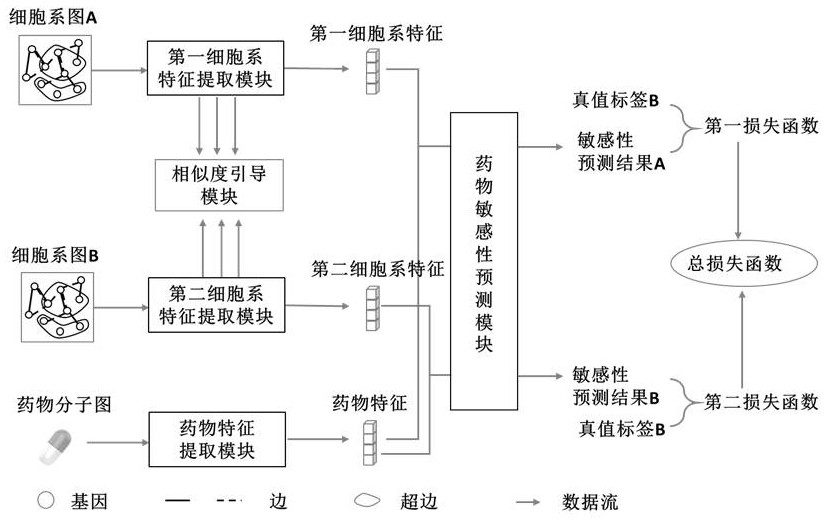
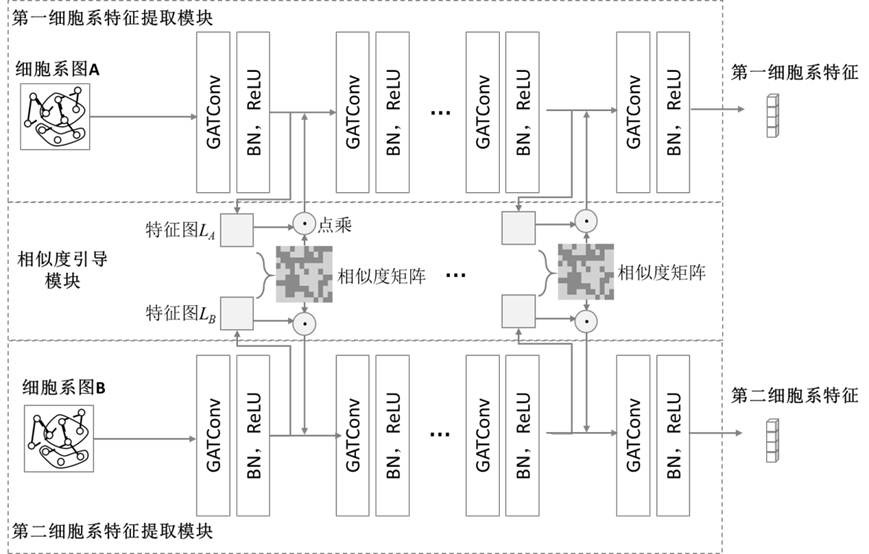
Drug sensitivity prediction method and device based on multi-omics similarity guidance
ActiveCN114255886AEfficient extractionGreat biological prior knowledgeBiostatisticsCharacter and pattern recognitionGenomicsMulti omics
The invention discloses a drug sensitivity prediction method and device based on multi-omics similarity guidance, and the method comprises the steps: a cell line diagram constructed based on multi-omics data of a cell line can fully integrate four types of multi-omics information of genomics data, transcriptomics data, proteomics data and metabonomics data of an individual cell line; compared with an existing cell line characterization mode, more kinds of omics information can be contained, and meanwhile the potential relation between products expressed by the cell line in the multi-omics level is fully considered; on the basis, a drug sensitivity prediction model for predicting the drug sensitivity based on the cell line diagram adopts a multi-omics similarity guiding mode, multi-omics similarity among individuals can be fully considered while multi-omics information of individual specificity is efficiently extracted, huge biological priori knowledge can be provided, and the drug sensitivity prediction model can be used for predicting the drug sensitivity based on the cell line diagram. Therefore, more accurate drug sensitivity prediction is realized.
Owner:ZHEJIANG UNIV
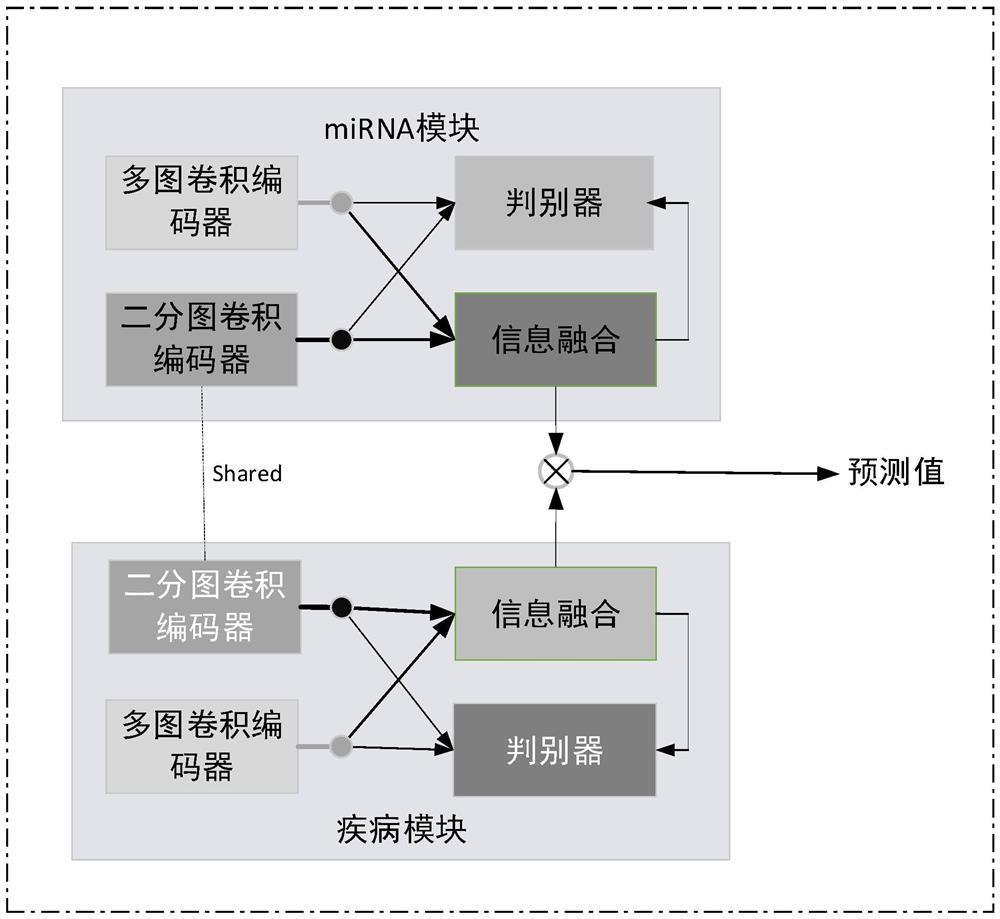
MiRNA-disease association prediction method and device based on graph neural network fusion multi-view information
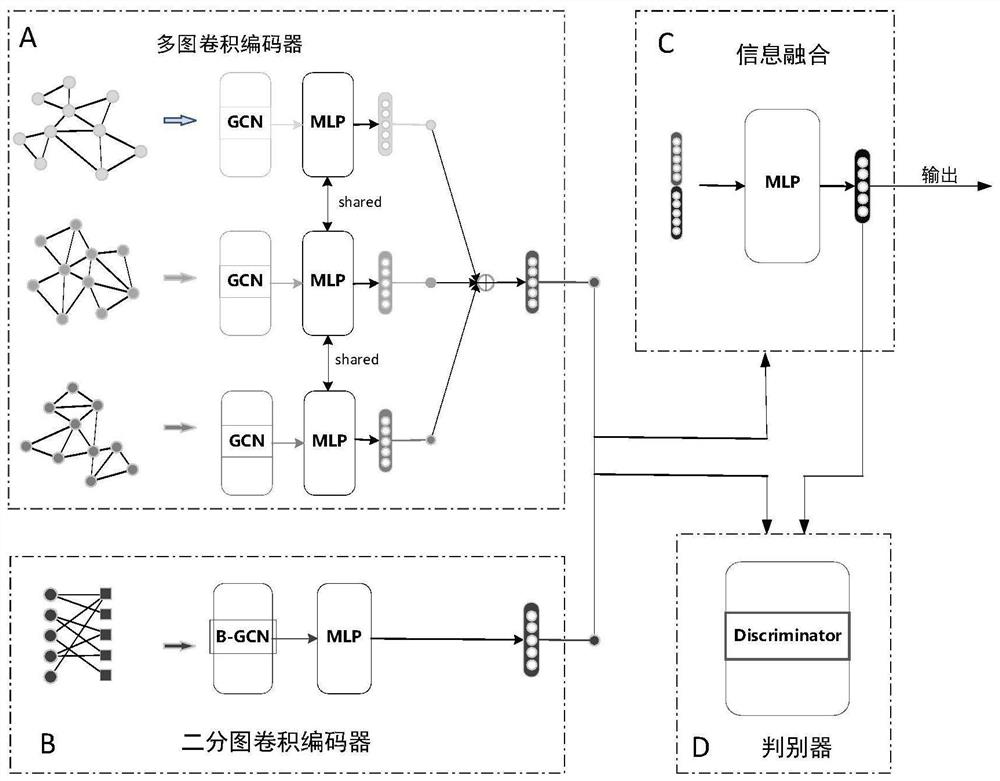
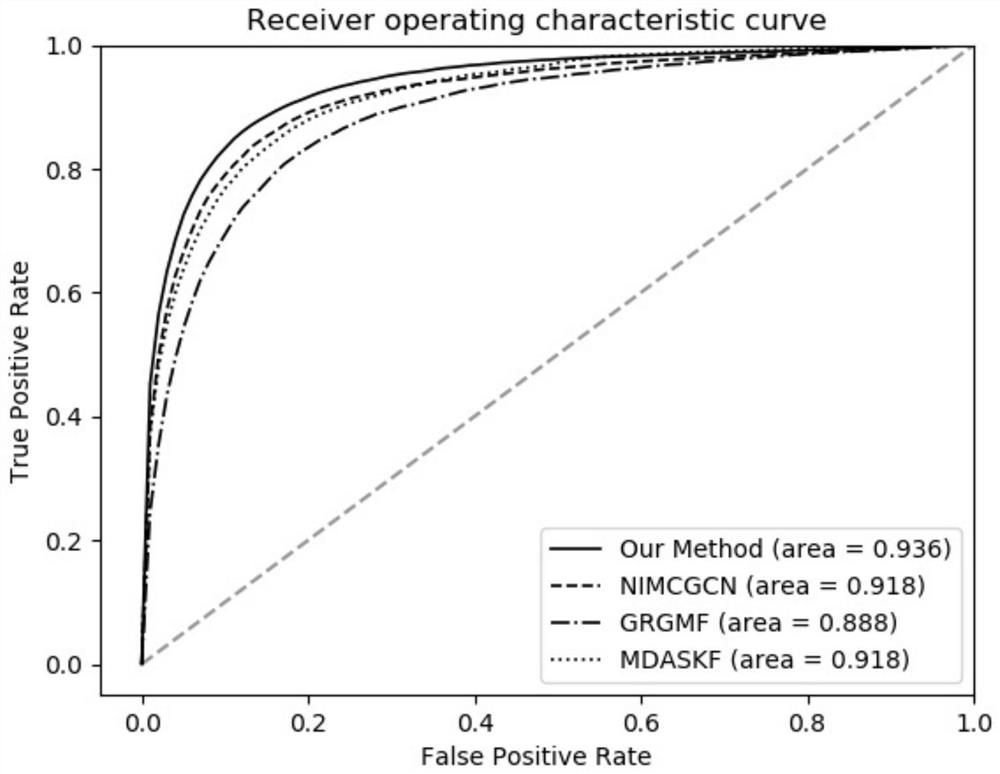
ActiveCN112784913AEasy to captureImprove forecast accuracyCharacter and pattern recognitionNeural architecturesMulti omicsPredictive methods
The invention discloses a miRNA-disease association prediction method and device based on graph neural network fusion multi-view information, and the method integrates miRNA-disease-related multi-omics data to construct a plurality of views, not only considers a plurality of homogeneous similarity networks, but also considers a heterogeneous bipartite network, extracts node features on all views in combination with a graph neural network and multi-view learning, captures dependency between global features and local features through a discriminator, and can better capture the complex nonlinear relation between miRNA and diseases.
Owner:HUNAN UNIV
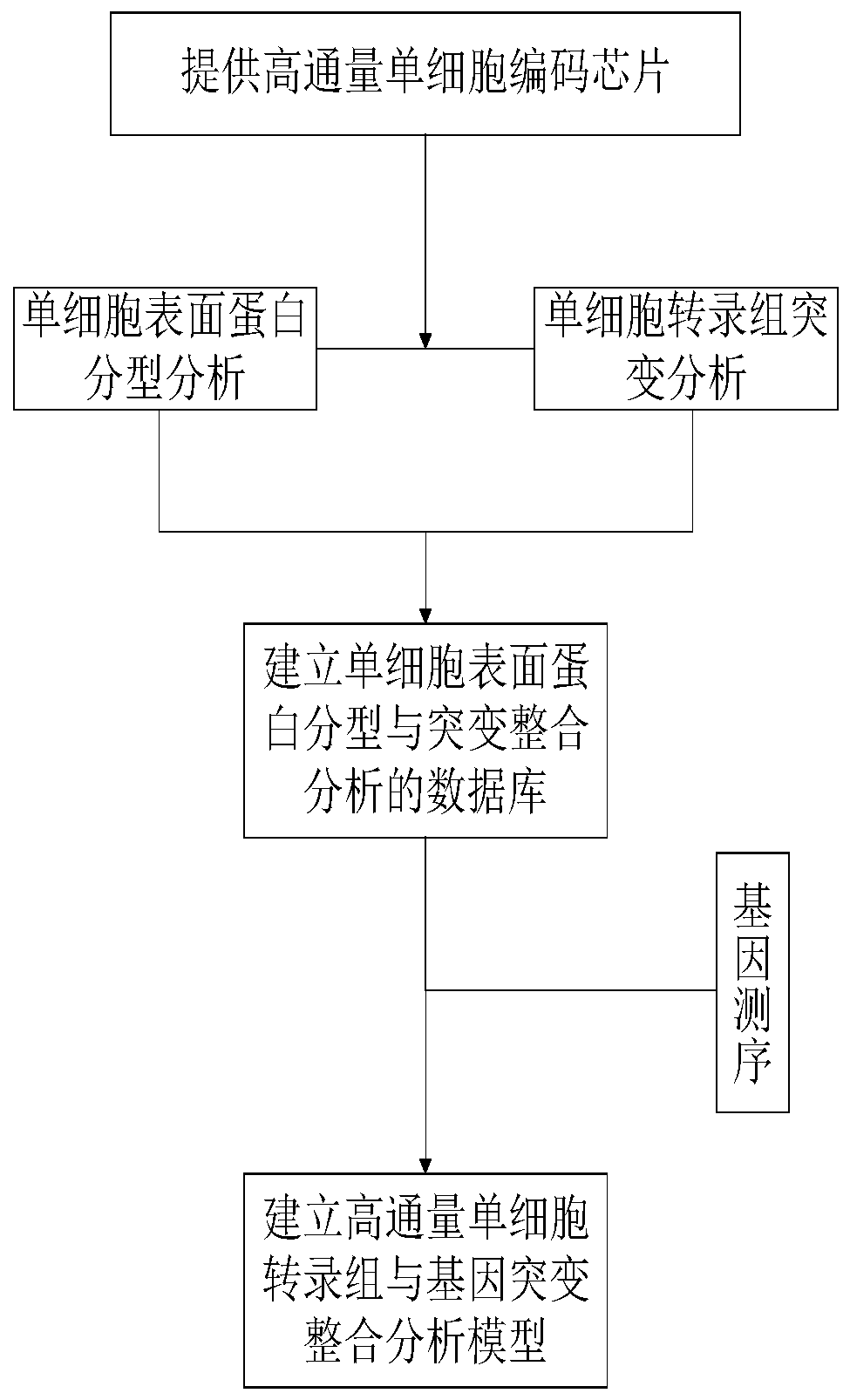
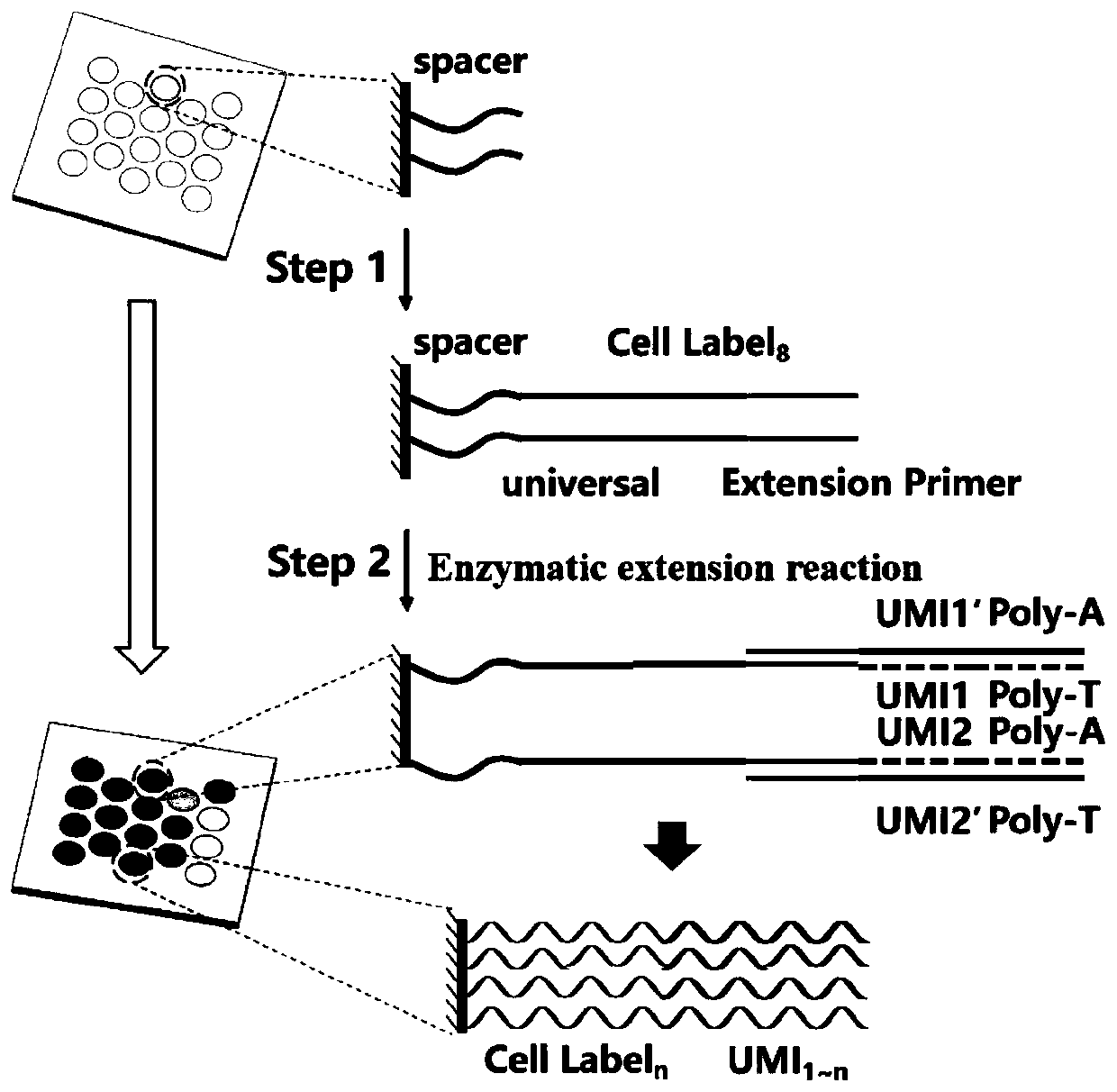
High-throughput single-cell transcriptome and gene mutation integration analysis method
InactiveCN110577983AAchieve a comprehensive understandingAchieve comprehensive characterizationMicrobiological testing/measurementSingle cell transcriptomeCell Surface Proteins
The invention discloses a high-throughput single-cell transcriptome and gene mutation integration analysis method. The high-throughput single-cell transcriptome and gene mutation integration analysismethod comprises the following steps that (1) a high-throughput single-cell encoding chip is provided; (2) single-cell surface protein parting analysis is conducted; (3) single-cell transcriptome mutation analysis is conducted; (4) a database for single-cell surface protein parting and mutation integration analysis is established; and (5) a high-throughput single-cell transcriptome and gene mutation integration analysis model is established. According to the high-throughput single-cell transcriptome and gene mutation integration analysis method, by designing the single-cell encoding chip withtriple encoding technologies of microporous spatial coordinates, cell nucleic acid labels and molecular nucleic acid labels and by combining the modes of single-cell surface protein parting, single-cell transcriptome mutation analysis and gene sequencing, gene mutation information, transcriptome information and protein expression information of single cells can correspond one by one, the completedatabase for high-throughput single-cell transcriptome and gene mutation integration analysis is formed, and the multi-omics integration analysis model is obtained.
Owner:SUZHOU INST OF BIOMEDICAL ENG & TECH CHINESE ACADEMY OF SCI
Cancer survival analysis system based on multiple tasks and multiple modes
PendingCN112687327AImprove multimodal fusionImprove the ability of multi-task learningMedical data miningBiostatisticsMulti modal dataData pre-processing
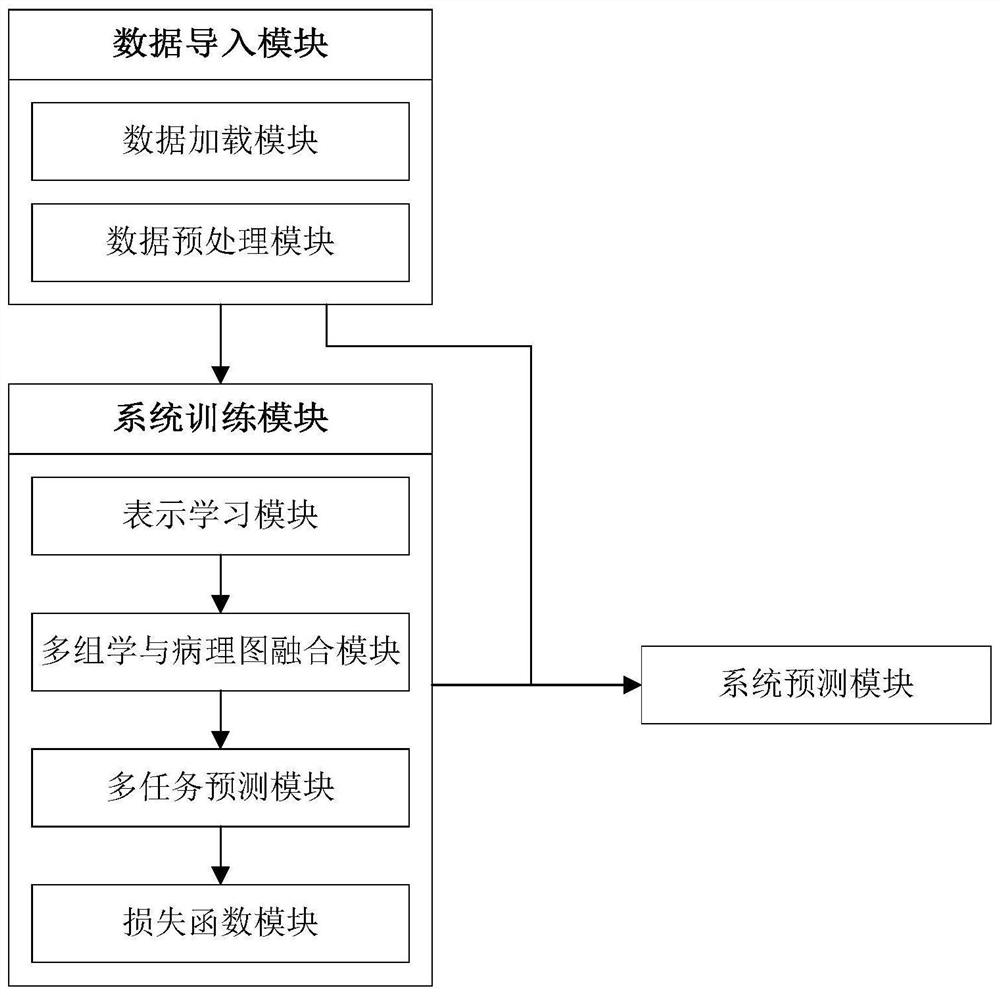
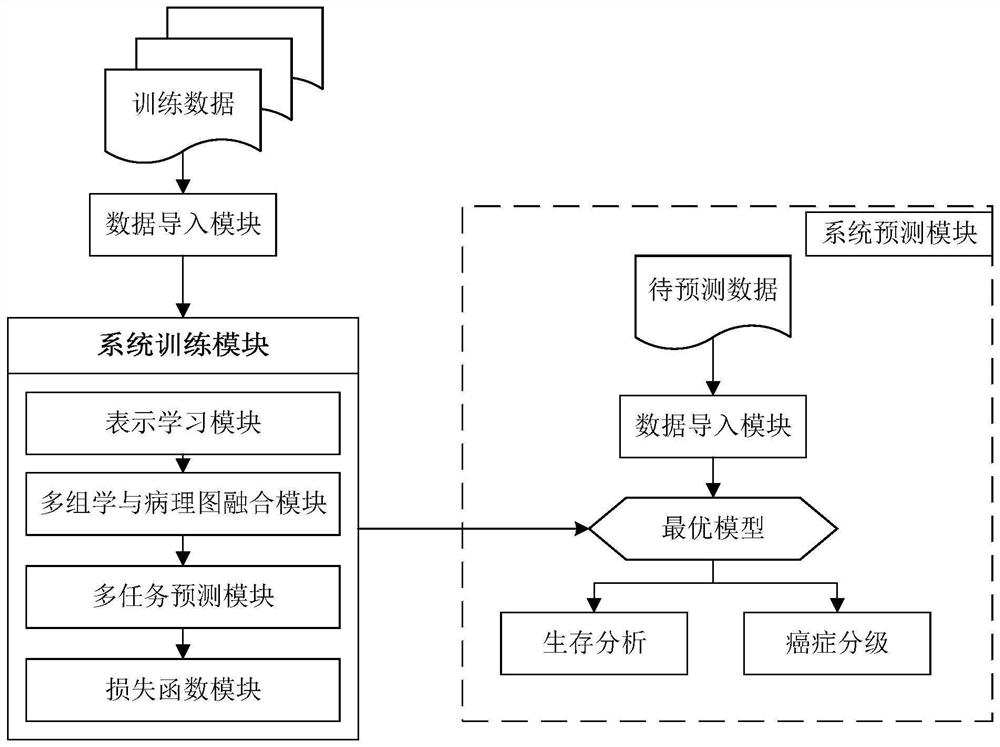
The invention discloses a cancer survival analysis system based on multiple tasks and multiple modes, and the system comprises a data importing module which comprises a data loading module and a data preprocessing module; a system training module, comprising a representation learning module, a multi-omics and pathological map fusion module, a multi-task prediction module and a loss function module; and a system prediction module, used for carrying out survival analysis and grading according to the multi-omics data and the pathological map of the cancer patient. According to the invention, the deep learning technology and the multi-task learning technology are combined with the multi-omics and pathological map data, so that the complementary characteristics among the multi-modal data can be captured, and the sharing relevance among multiple tasks can also be captured, thereby forming a cancer survival analysis system based on multiple tasks and multiple modalities, and providing automatic survival analysis and cancer grading results.
Owner:中山依数科技有限公司
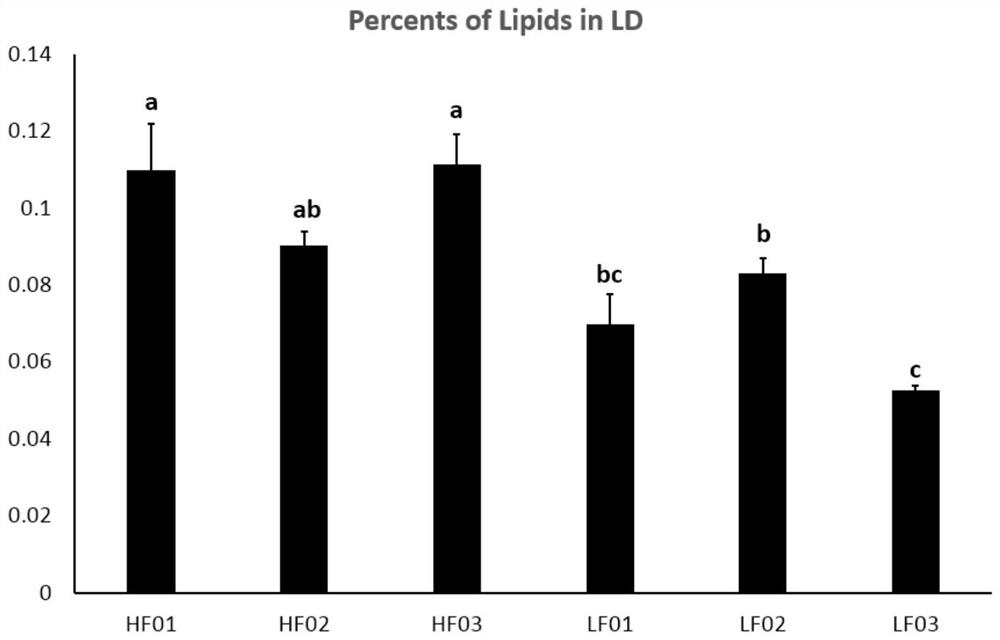
Multi-omics-based mining method for key gene of fat deposition trait of Nanyang black pigs
The invention discloses a multi-omics-based mining method for a key gene of a fat deposition trait of Nanyang black pigs. According to the method, in order to solve the problem of the existing methodsthat the accuracy of mining and identifying of swine fat deposition trait genes is low, transcriptomes of musculi longissimus dorsi of six pairs of full-sib or half-sib Nanyang black pig individualsof 6 months old with extreme back fat thickness and intramuscular fat content are sequenced by using a high-flux sequencing technology, important fat deposition candidate genes are sought and identified on the basis of phenotype determination and under the assistance of bioinformatics analysis, meanwhile, two groups of Nanyang black pigs are subjected to genome resequencing to find out important fat deposition candidate difference genes, between-group remarkable-difference-expression genes are screened through integrating transcriptome data, re-sequenced data and QTL database information and using bioinformatics and functional genomics methods, and gene functions and signal channels of the between-group remarkable-difference-expression genes are predicted.
Owner:NANYANG NORMAL UNIV
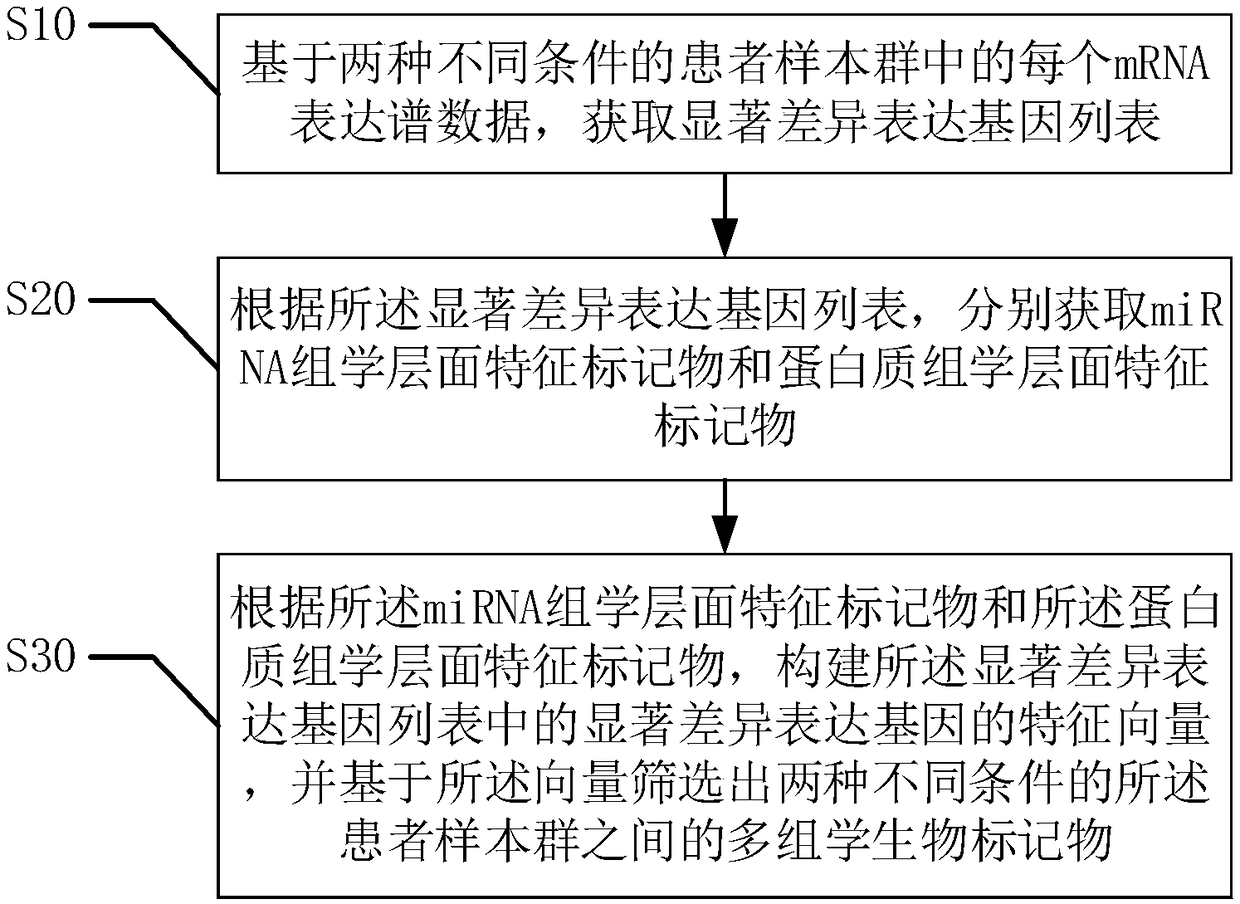
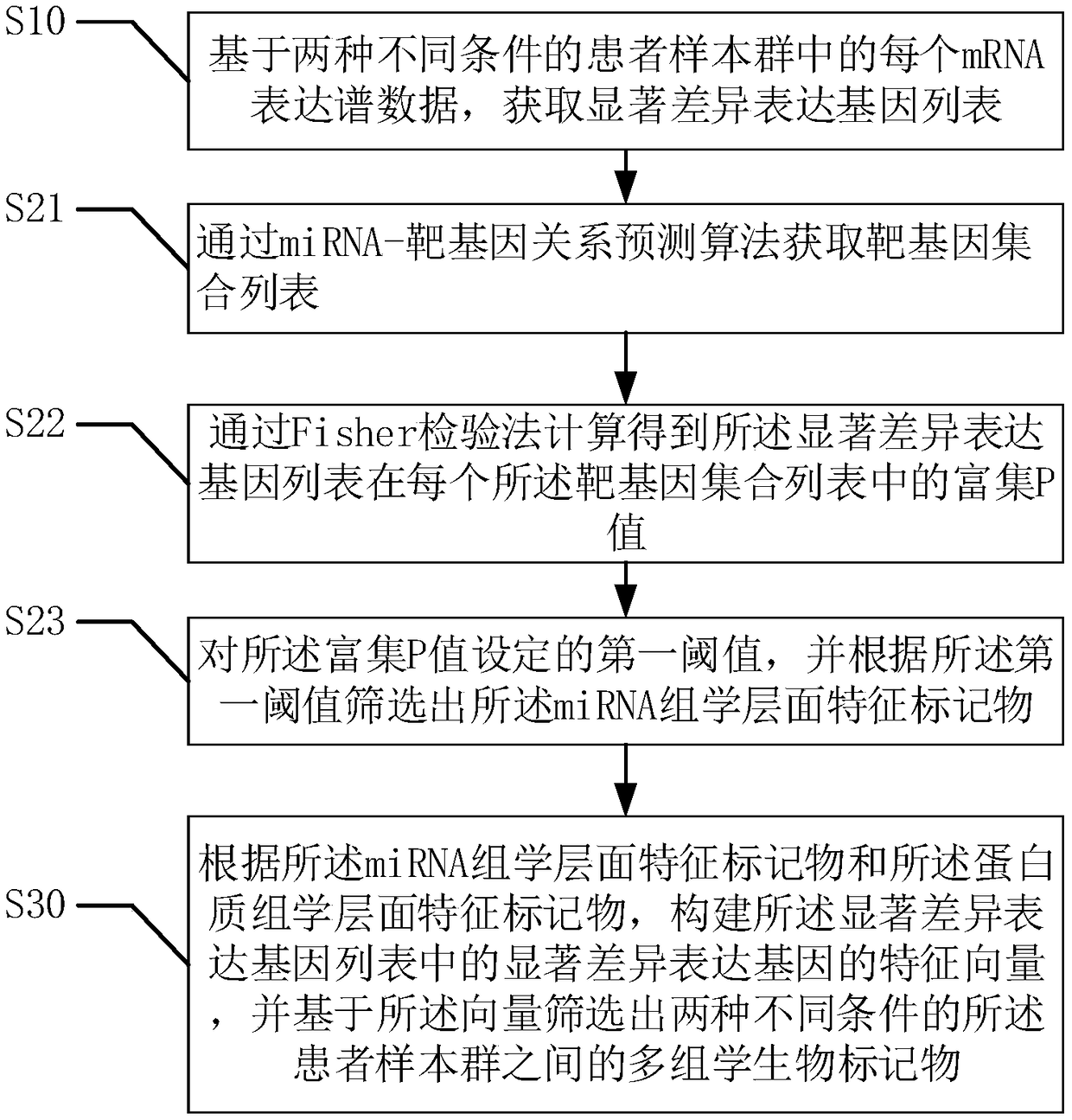
Biomarker system screening method based on multi-omics
ActiveCN108830045AImprove accuracySpecial data processing applicationsProteomic ProfileScreening method
The invention provides a biomarker system screening method based on multi-omics. The method includes according to expression profile data of each mRNA in patient sample clusters under two different conditions, acquiring a significant difference expression gene list; according to the significant difference expression gene list, acquiring miRNA omics-level characteristic markers and proteomics-levelcharacteristic markers; creating feature vectors of significant difference expression genes in the significant difference expression gene list, and screening multi-omics biomarkers among the patientsample clusters under two different conditions according to the vectors. The biomarker system screening method has the advantages that the biomarkers are screened according to transcriptomics-level data, miRNA omics-level data and proteomics-level data, so that the accuracy of the screening method is high; result coverage is comprehensive for multi-factor multi-level measurement, and great convenience is provided to researchers in the life science field in screening and further exploration of the biomarkers.
Owner:SHENZHEN INST OF ADVANCED TECH
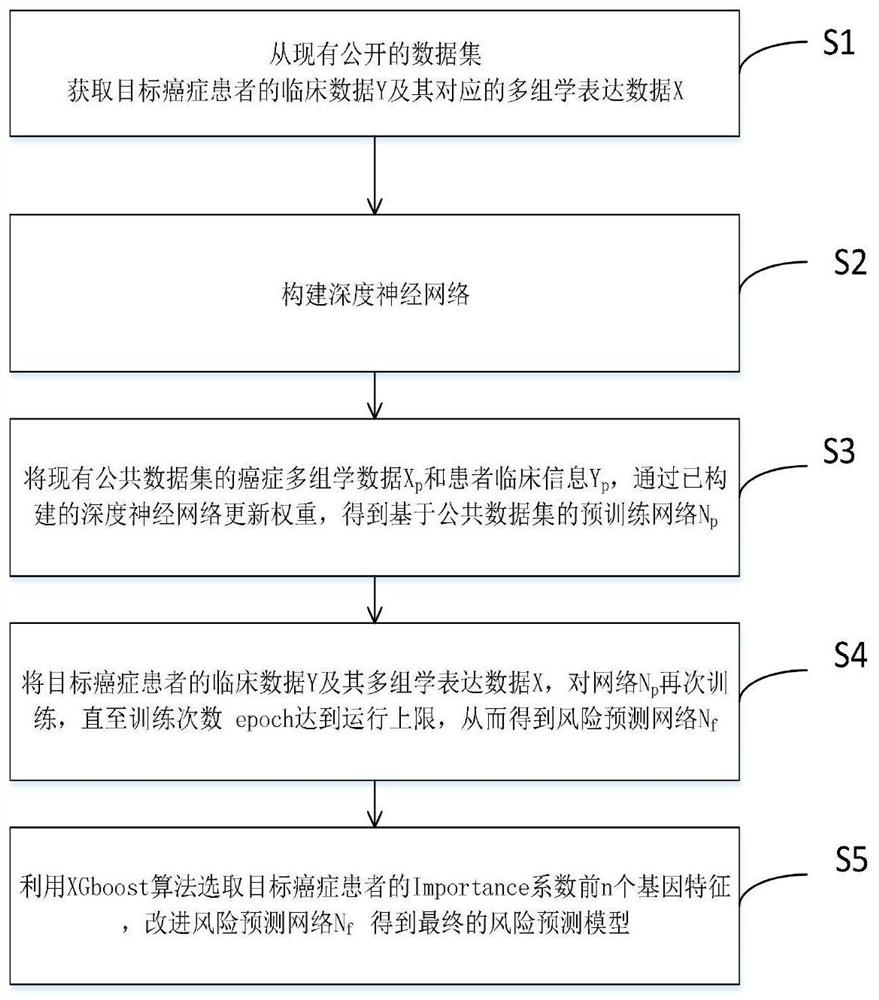
Deep learning method for predicting prognosis risk of cancer patient based on multi-omics data
PendingCN112820403AImprove robustnessEpidemiological alert systemsMedical automated diagnosisData setGene Feature
The invention discloses a deep learning method for predicting the prognosis risk of a cancer patient based on multi-omics data, which is used for predicting the prognosis risk of the cancer patient and comprises the following steps: S1, acquiring clinical data Y of a target cancer patient and corresponding multi-omics expression data X from an existing public data set; S2, constructing a deep neural network; S3, updating the weight theta of the cancer multi-omics data Xp and the clinical information Yp of the patient of the existing common data set through the constructed deep neural network to obtain a pre-training network Np based on the common data set; S4, training the network Np again until the training frequency epoch reaches the operation upper limit, thereby obtaining a risk prediction network Nf; and S5, selecting the first n gene features of the Importance coefficient of the target cancer patient by using an XGboost algorithm, and improving the risk prediction network Nf to obtain a final risk prediction model. According to the method, the robustness of the prediction model is improved, and the prognosis risk of the cancer patient is predicted more accurately by using multi-omics data.
Owner:SUN YAT SEN UNIV
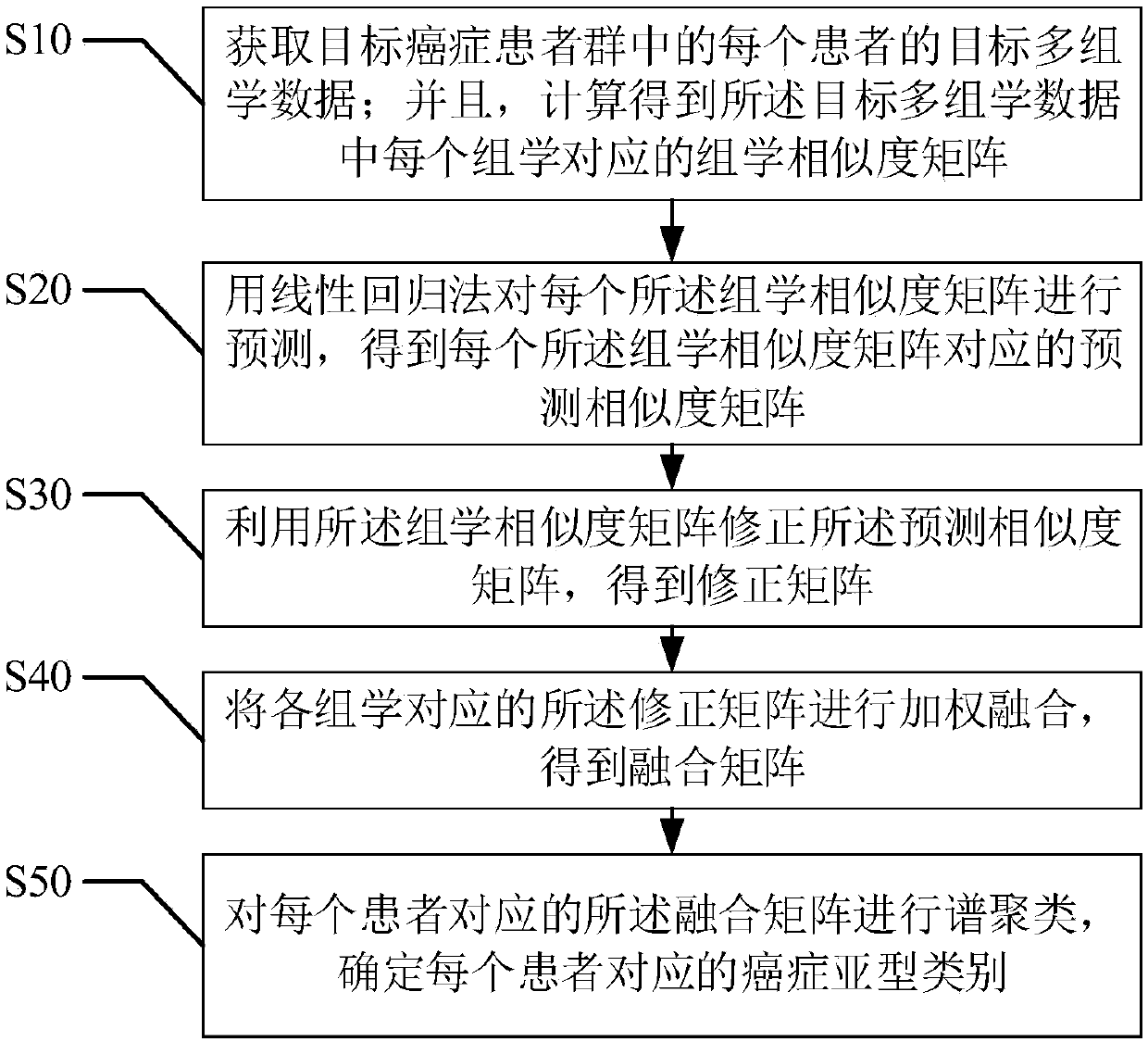
Cancer subtype classification method based on multi-omics integration
ActiveCN111291777AImprove accuracyConvenient researchBiostatisticsCharacter and pattern recognitionMedicineMulti omics
The invention provides a cancer subtype classification method based on multi-omics integration. The method comprises the steps of obtaining target multi-omics data of each patient in a target cancer patient group; calculating to obtain an omics similarity matrix; predicting each omics similarity matrix to obtain a predicted similarity matrix; correcting the prediction similarity matrix by utilizing the omics similarity matrix to obtain a correction matrix; performing weighted fusion to obtain a fusion matrix; and performing spectral clustering on the fusion matrix, and establishing a cancer subtype category label corresponding to the fusion matrix of each patient. The accuracy of classification and evaluation of the cancer subtypes is improved, the patients are classified through a more flexible integration method, the data analysis efficiency is improved, and convenience is provided for research on the cancer subtypes.
Owner:SHENZHEN INST OF ADVANCED TECH
Multi-omics data combined analysis method
The invention discloses a multi-omics data combined analysis method. The multi-omics data combined analysis method comprises the following steps of (A), performing coexpression network analysis on each single-omics index data in to-be-analyzed multi-omics data, and finding a respective expression model; and (b), according to the overlapping relation between respective expression modules of different omics data, screening interaction modules which are remarkably correlated in the to-be-analyzed multi-omics data. The multi-omics data combined analysis method according to the invention is not restricted by the number of omics data, and random multiple groups can be utilized. Furthermore the method does not depend on a data source. The index data (such as gene expression magnitude, apparent methylation degree, SNP mutation rate) which can measure the corresponding omics can be used as input data.
Owner:BGI TECH SOLUTIONS
System and method of analyzing association change pattern from multi-omics data
InactiveCN109300502ABig progressImprove compatibilityInstrumentsMolecular structuresData setMutual correlation
The invention discloses a system and a method of analyzing an association change pattern from multi-omics data. The system comprises an omics data set containing multiple omics data, a binary algorithm unit for preprocessing the omics data set and an association rule Apriori algorithm unit for mining the mutual association pattern of each molecule change. Different omics data can be effectively integrated, and the analysis association change pattern in the multi-omics data can be well mined.
Owner:SHANTOU UNIV MEDICAL COLLEGE
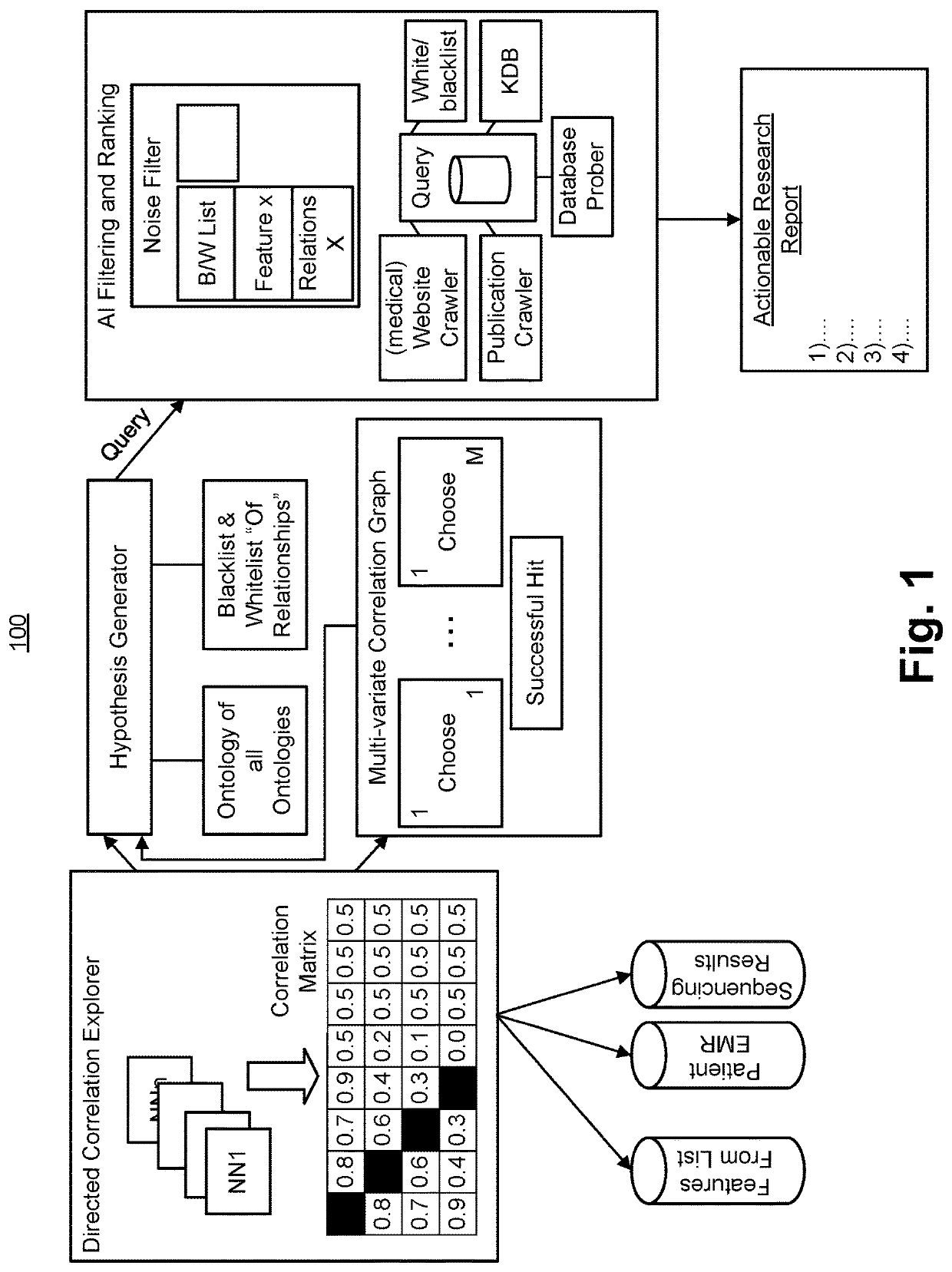
Artificial intelligence engine for directed hypothesis generation and ranking
PendingUS20220261668A1Quality improvementRelational databasesBiological modelsDiseaseSearch analytics
An artificial intelligence engine for directed hypothesis generation and ranking uses multiple heterogeneous knowledge graphs integrating disease-specific multi-omic data specific to a patient or cohort of patients. The engine also uses a knowledge graph representation of ‘what the world knows’ in the relevant bio-medical subspace. The engine applies a hypothesis generation module, a semantic search analysis component to allow fast acquiring and construction of cohorts, as well as aggregating, summarizing, visualizing and returning ranked multi-omic alterations in terms of clinical actionability and degree of surprise for individual samples and cohorts. The engine also applies a moderator module that ranks and filters hypotheses, where the most promising hypothesis can be presented to domain experts (e.g., physicians, oncologists, pathologists, radiologists and researchers) for feedback. The engine also uses a continuous integration module that iteratively refines and updates entities and relationships and their representations to yield higher quality of hypothesis generation over time.
Owner:TEMPUS LABS INC
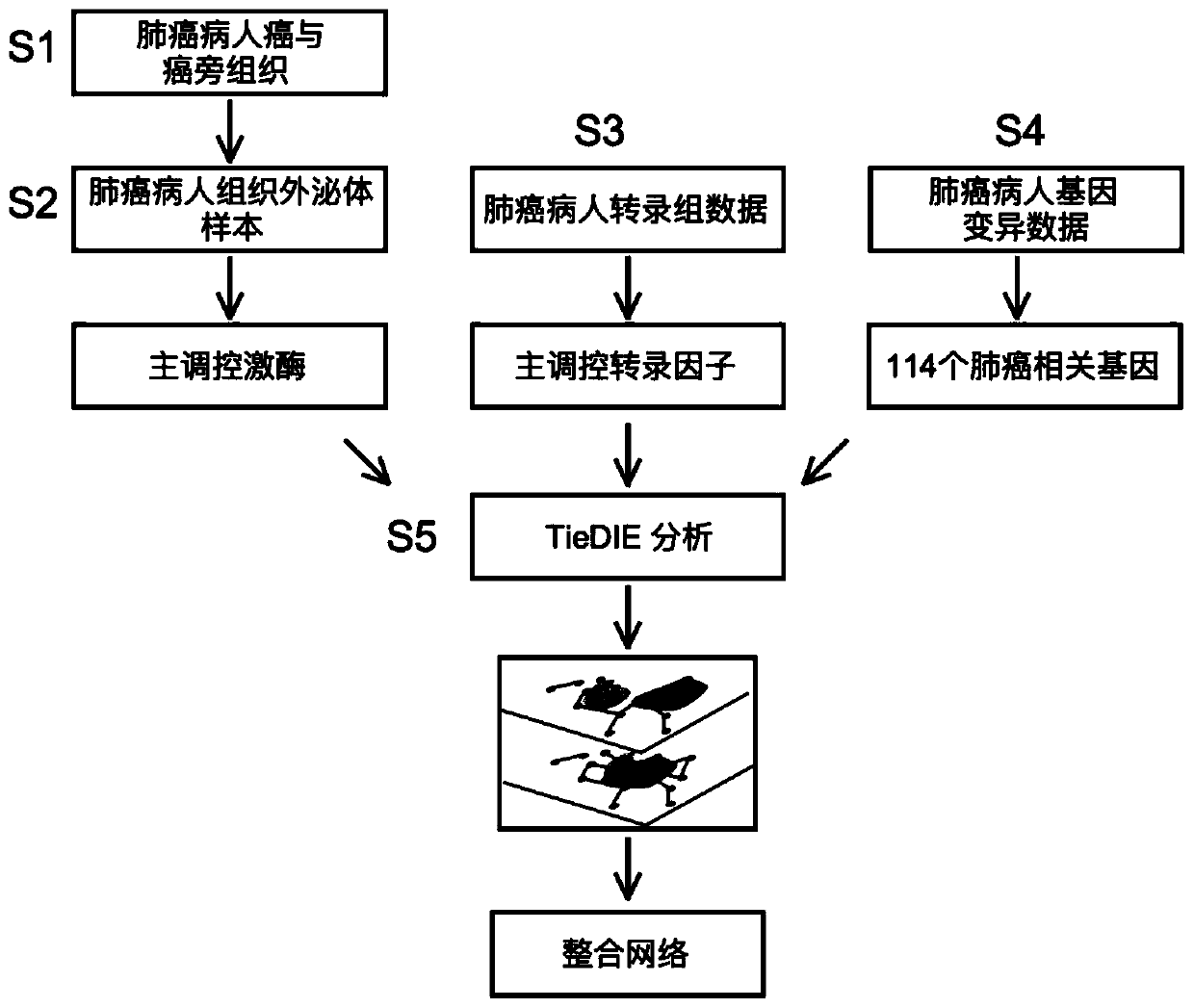
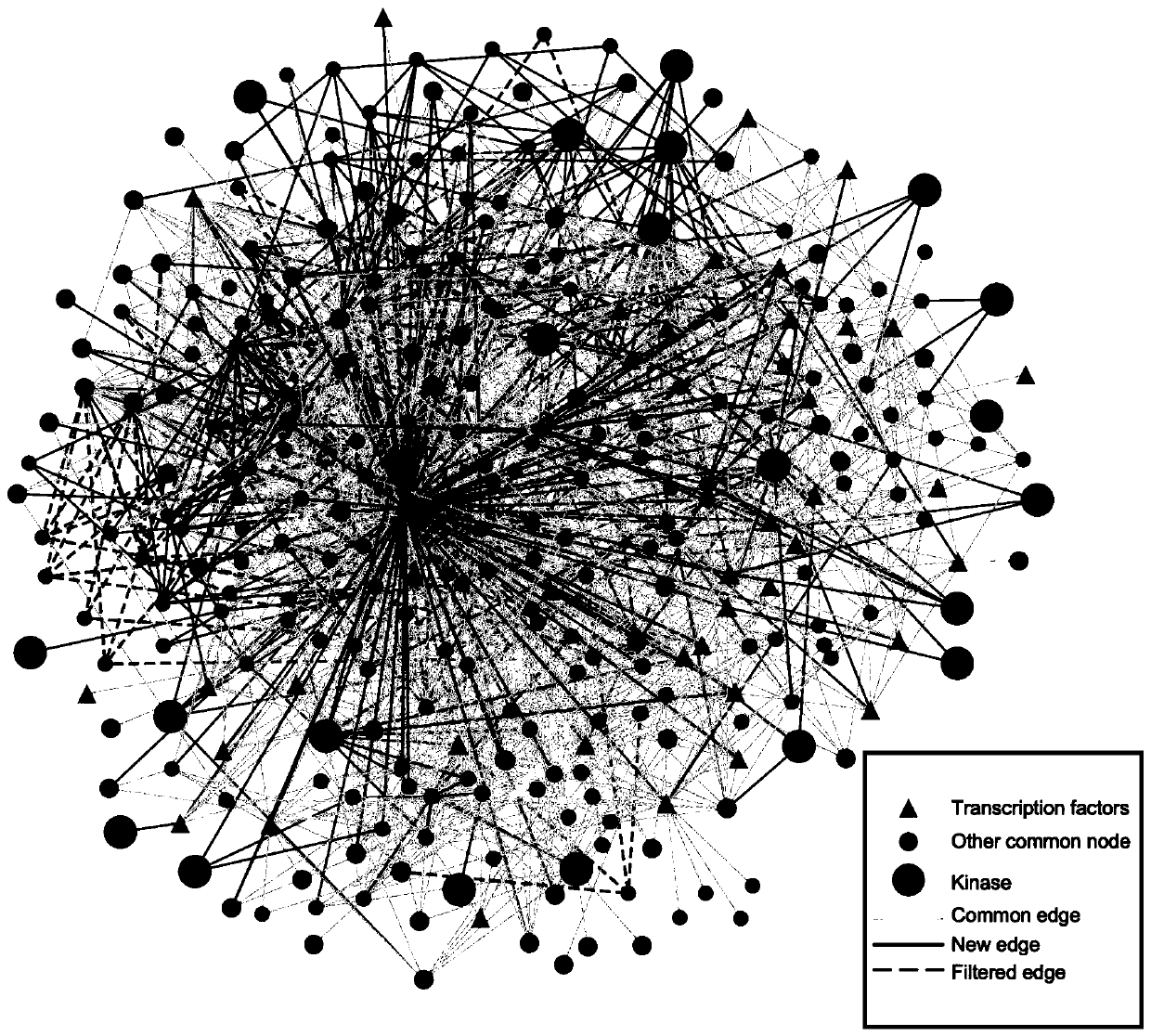
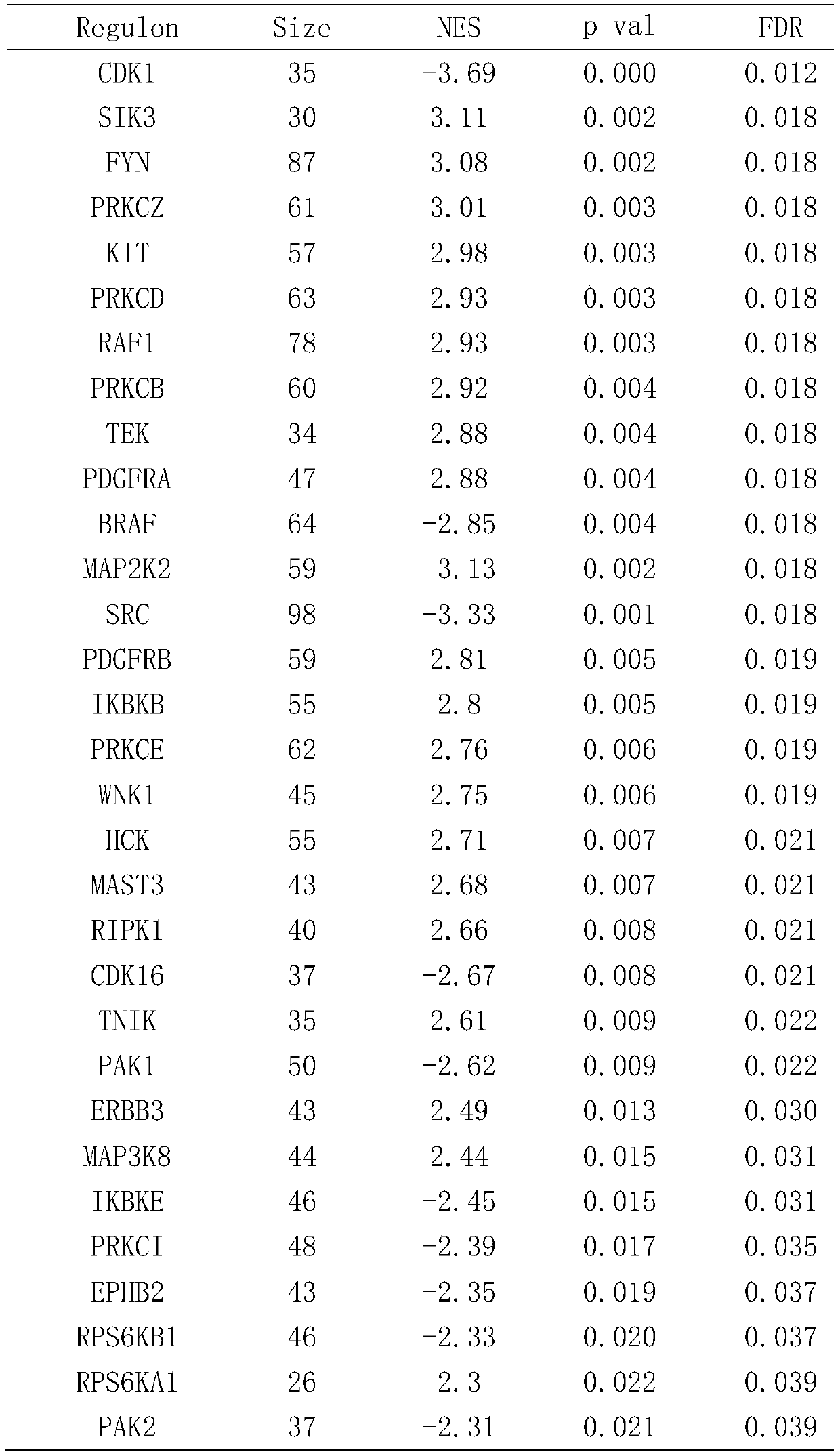
Tissue exosome phosphorylated proteome-based multi-omics analysis method
ActiveCN110957007ALarge amount of informationUniversalMicrobiological testing/measurementProteomicsSequence analysisDisease
The invention provides a tissue exosome phosphorylated proteome-based multi-omics analysis method. The method comprises the following steps of: S1, separating exosomes of a tissue sample and specifically enriching phosphorylated proteins; S2, carrying out sequencing analysis on the enriched phosphorylated protein to obtain main regulation kinase; S3, analyzing transcriptome data of the tissue sample to obtain a main regulation transcription factor; S4, analyzing the genome variation data of the tissue sample to obtain a tumor-related gene; and S5, performing strategy analysis on the analysis data obtained in the steps S2, S3 and S4 to obtain a cancer-related main regulation kinase network and a drug action target. The method is suitable for the joint analysis technology of phosphorylated proteome sequencing data, transcriptome data and gene mutation data of any disease exosome sample, and a foundation is laid for exploring markers related to diseases and selecting more effective and accurate disease treatment targets.
Owner:SHANGHAI JIAO TONG UNIV
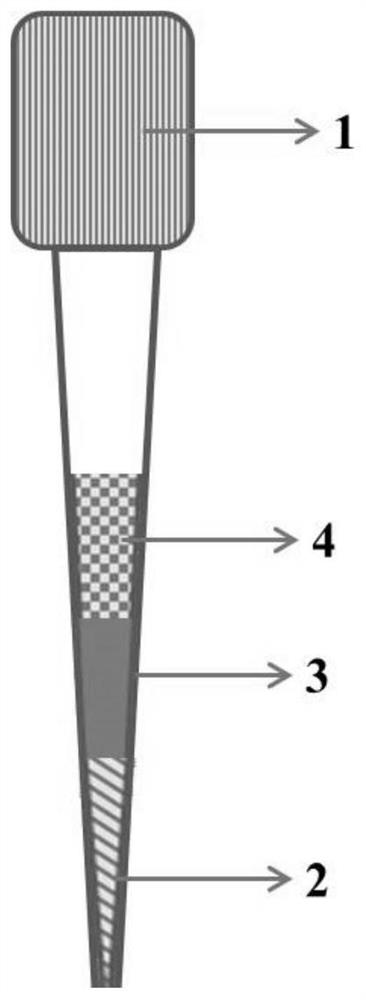
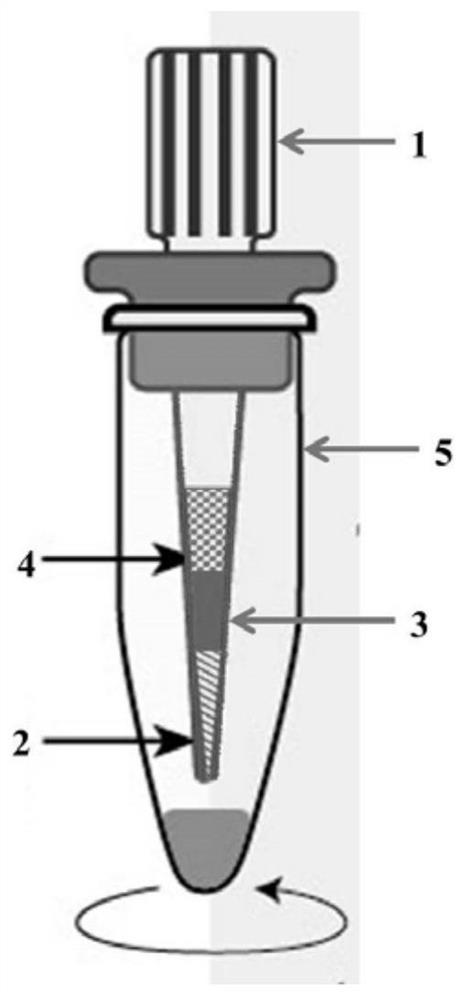
Sample pretreatment method for multi-omics analysis, and application thereof
ActiveCN111766325AEasy to operateAchieve enrichmentComponent separationBlood collectionPretreatment method
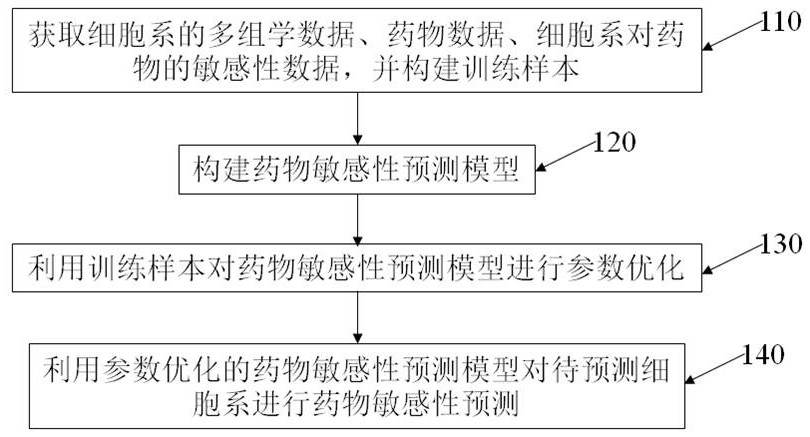
The invention provides a sample pretreatment method for multi-omics analysis, and application thereof. The sample pretreatment method comprises the following steps: (1) adding a to-be-detected sampleto a sample pretreatment device, centrifuging, and loading the obtained sample to obtain a loaded sample; (2) eluting the sample to obtain an eluate and an eluted sample, and collecting the eluate toobtain a metabonomics analysis sample; and (3) carrying out enzymolysis on the eluted sample, transferring the sample to a solid-phase extraction membrane, eluting, and collecting the obtained eluateto obtain a proteomics analysis sample. According to the sample pretreatment method, single-drop blood collection, dry sample transportation, enrichment of a to-be-detected object and preparation of the multi-omics analysis sample can be integrally carried out, and the same liquid phase mass spectrometry device is adopted to carry out mass spectrometry on a plurality of omics samples from a singledrop of plasma, such that the process from sample pretreatment to analysis result obtaining can be controlled within 6 h so as to achieve rapid, high-sensitivity and high-accuracy analysis detection.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA
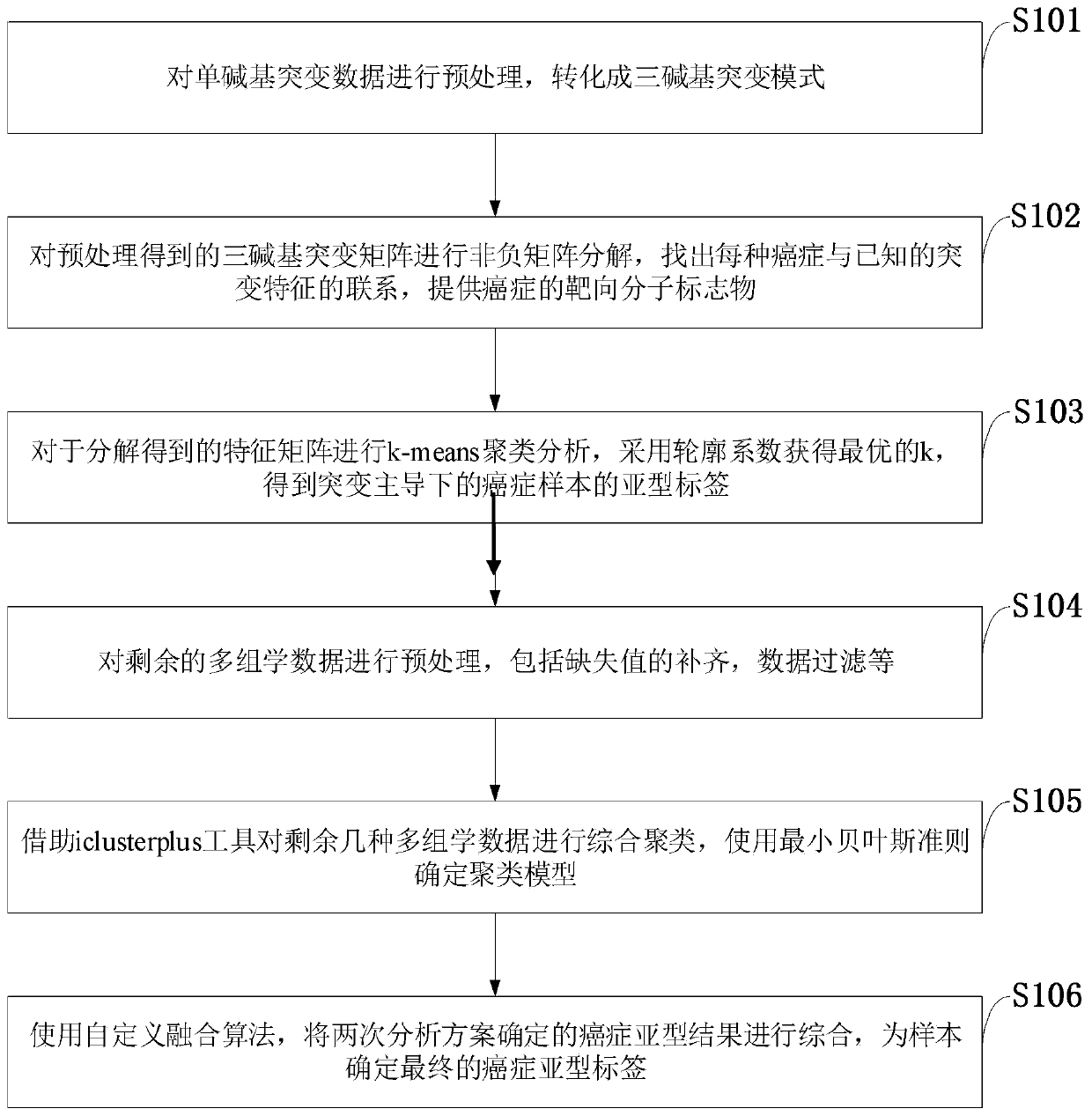
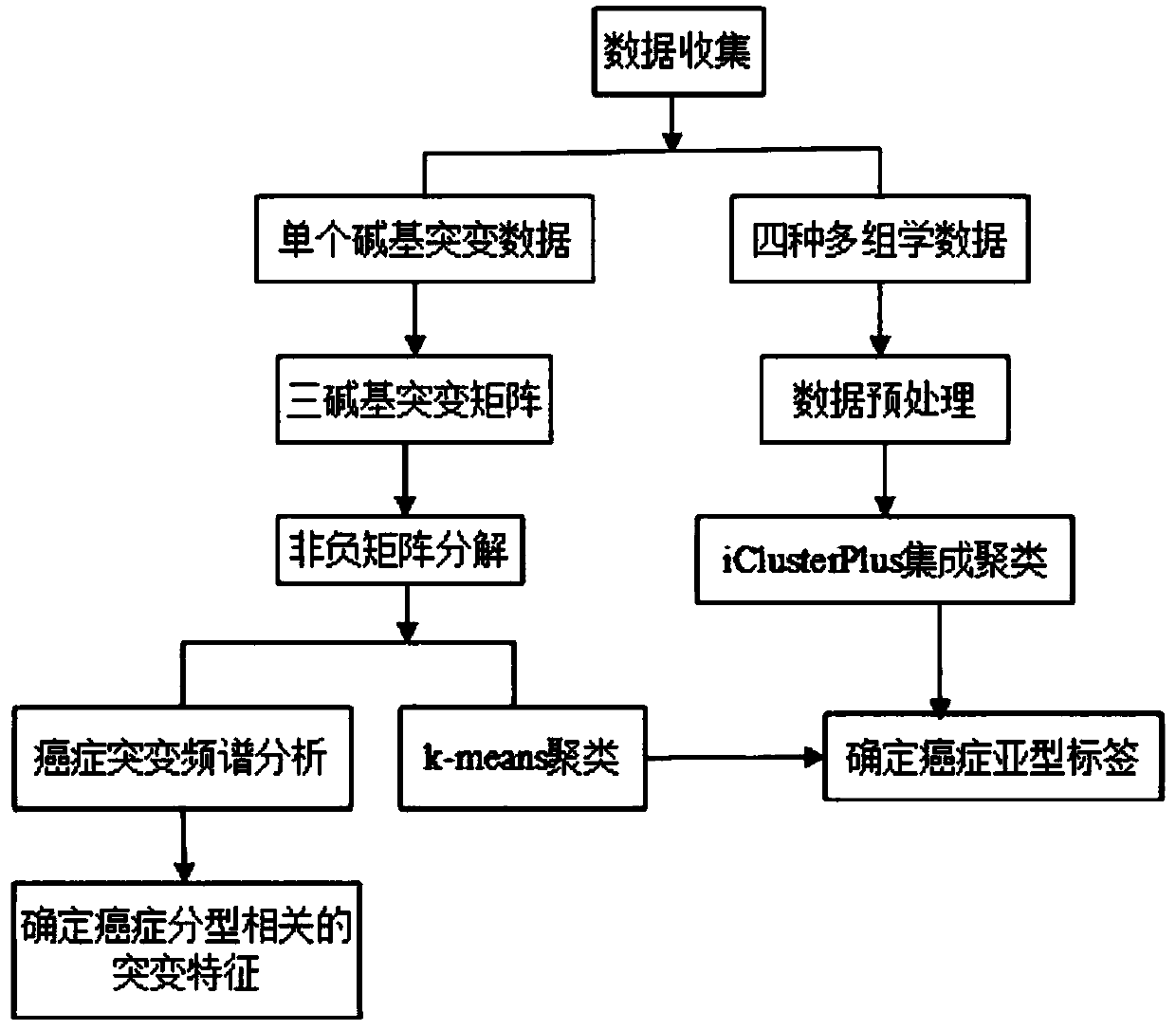
Cancer typing information processing method based on multi-omic data
ActiveCN110379460AGet goodEasy to operateSequence analysisInstrumentsInformation processingCancer type
The invention belongs to the field of biology and medical gene technology, and discloses a cancer typing information processing method based on multi-omic data. The method comprises the steps of preprocessing single-base mutation data of the multi-omic data for converting to a three-base mutation mode; performing non-negative matrix decomposition on the three-base mutation matrix which is obtainedthrough preprocessing, performing k-means cluster analysis on the decomposed characteristic matrix, selecting an optimal k by means of a contour coefficient, and obtaining a subtype label of a cancersample in guidance of mutation; afterwards preprocessing the residual omic data, performing integrated cluster analysis by means of an iclusterplus tool, determining a cluster model according to a minimum Bayes criterion, performing subtype definition on the sample again, then combining the results in two times of typing by means of a customized algorithm, and determining a final cancer subtype label of the sample. The cancer typing information processing method has advantages of convenient data acquisition, convenient tool operation and relatively high result reliability.
Owner:XIDIAN UNIV
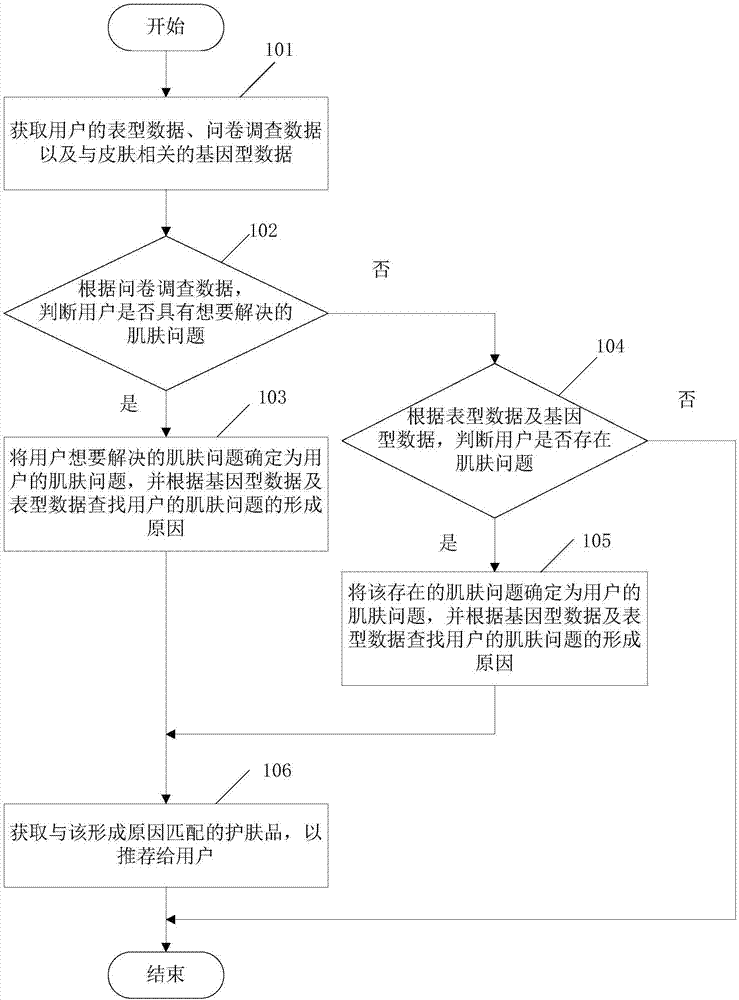
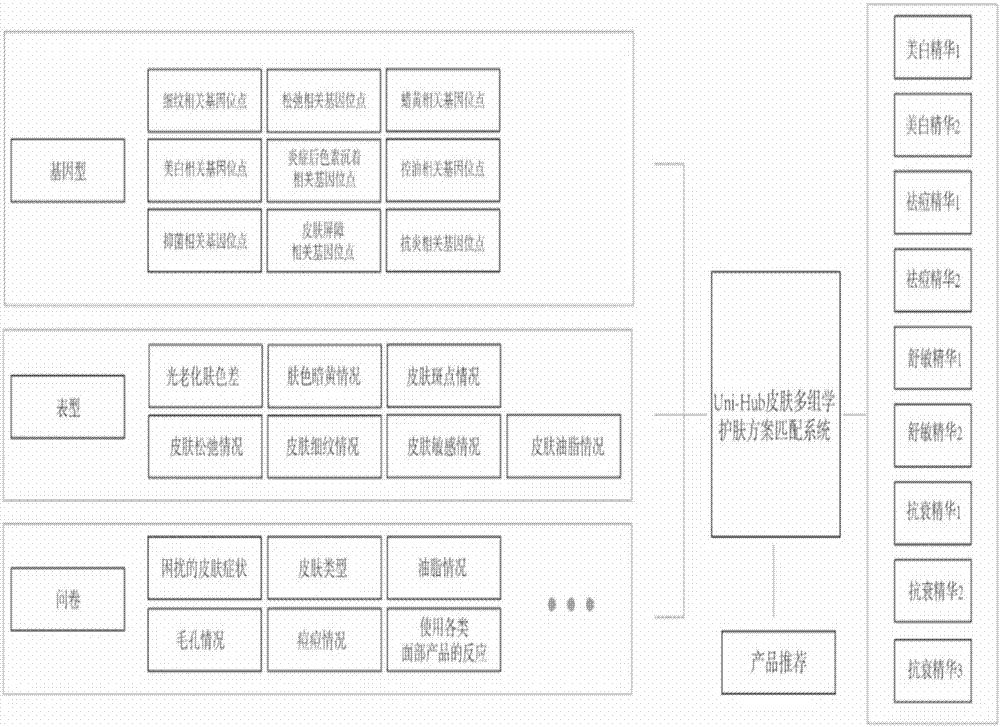
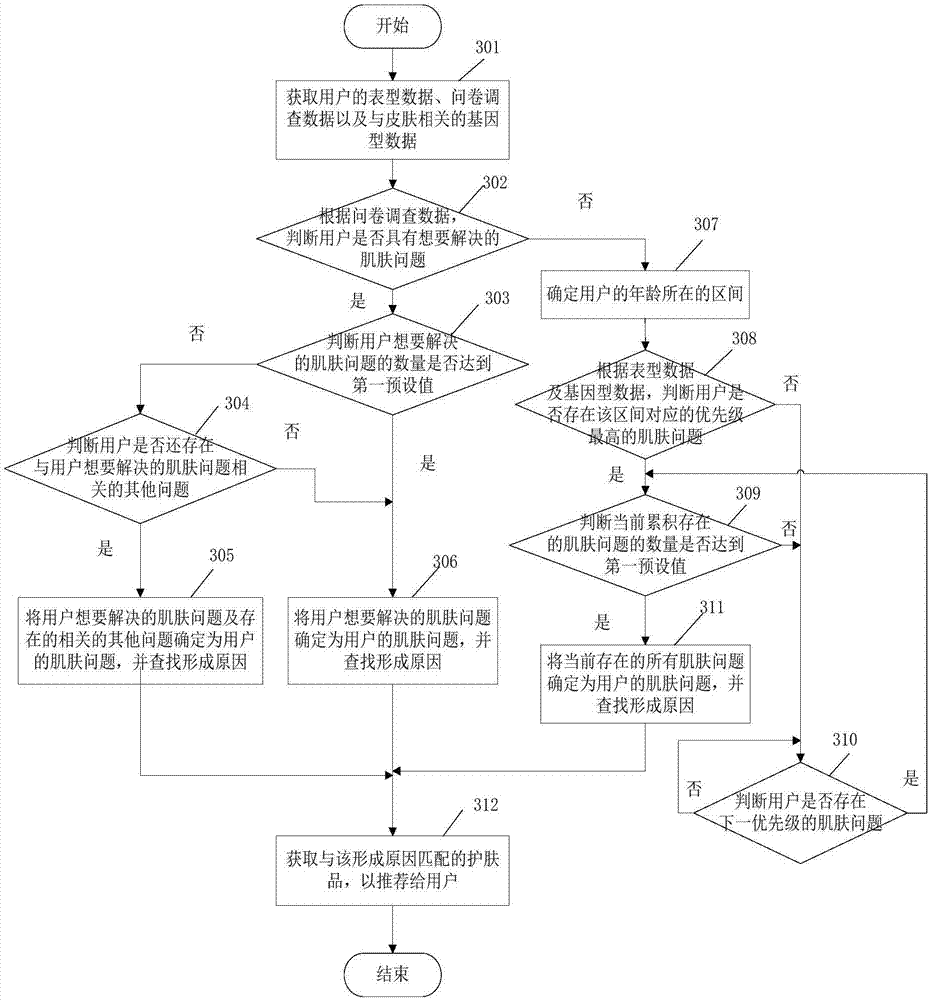
Skincare matching method based on skin multi-omics and terminal
The embodiment of the invention relates to the field of biotechnology, and discloses a skincare matching method based on skin multi-omics and a terminal. The method includes: acquiring phenotype data,questionnaire survey data and skin-related genotype data of a user; determining a skin problem of the user according to the genotype data, the phenotype data and the questionnaire survey data, and finding a formation cause of the skin problem of the user; and acquiring a skincare product, which matches the formation cause, to recommend the same to the user. When the embodiment of the invention solves the skin problem of the user, the phenotype data and the questionnaire survey data of the user are considered, the skin-related genotype data are also referred to, finding of the fundamental cause of the skin problem is facilitated, and the user is helped in finding more potential skin problems. In addition, the embodiment recommends the skincare product to the user according to the formationcause of the skin problem, and also facilitates improvement of precision of the recommended skincare product.
Owner:INERTIA SHANGHAI BIOTECHNOLOGY CO LTD +1
Multi-omics integration accurate pig breeding method
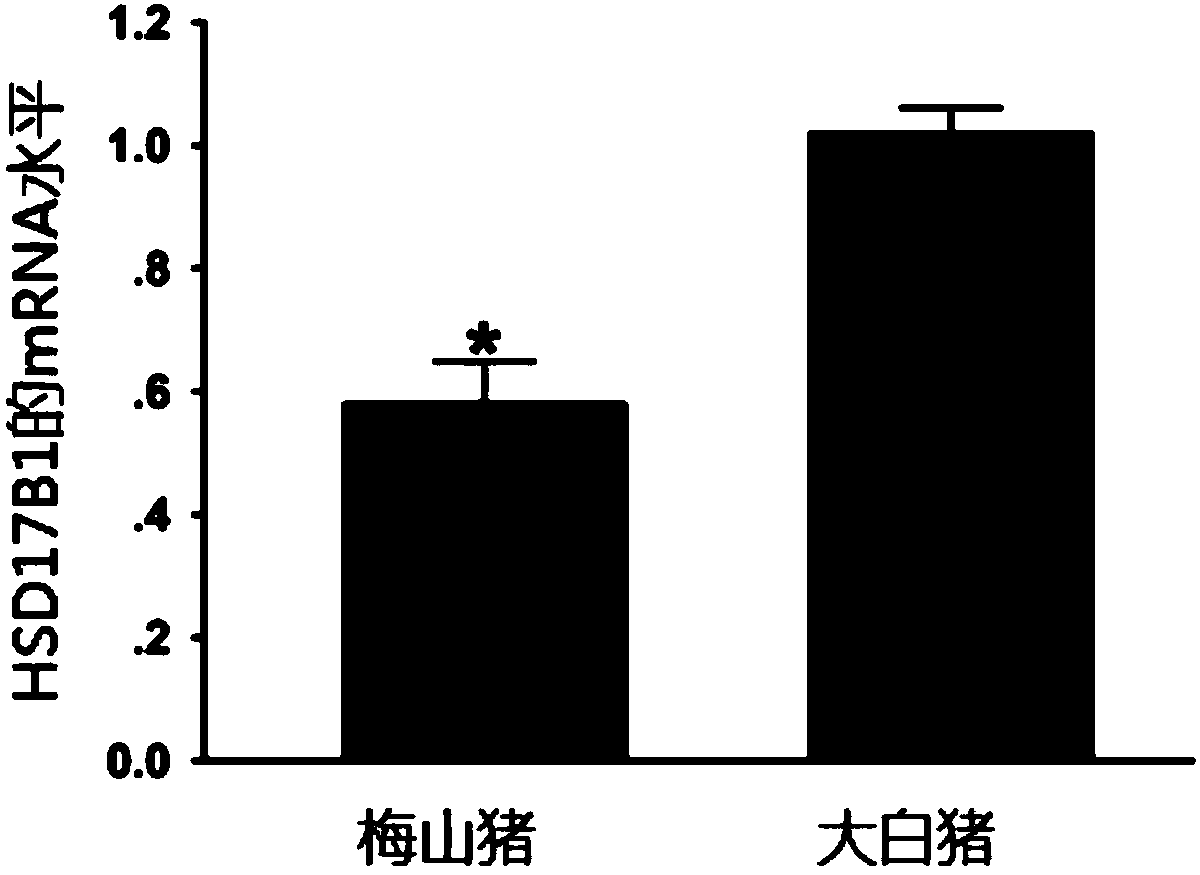
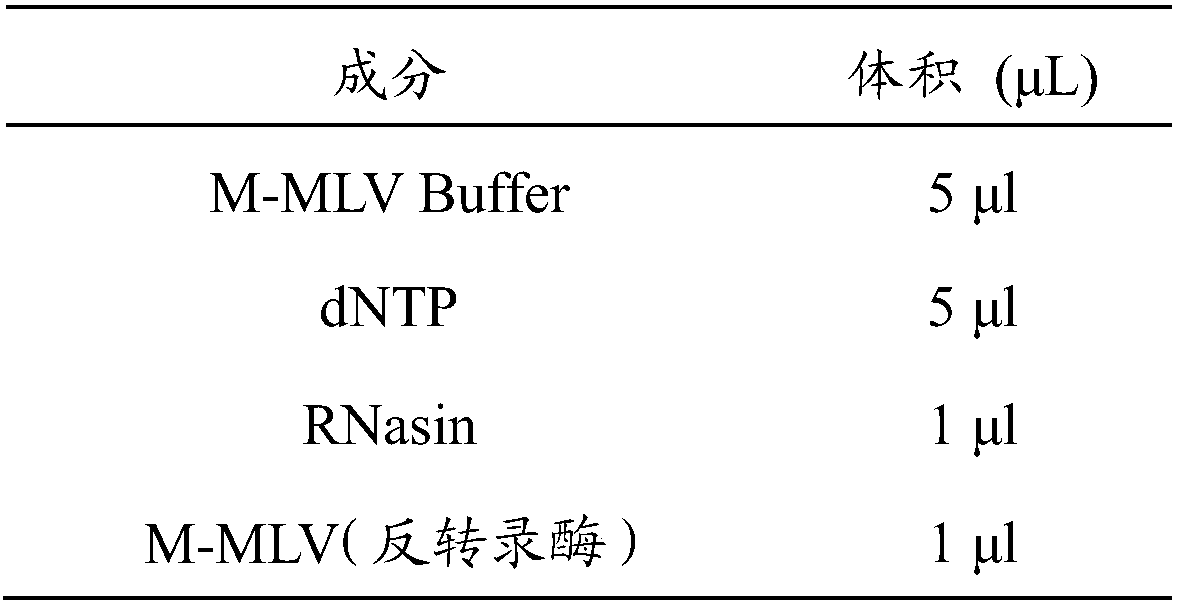
InactiveCN109486957AFacilitating Breeding SelectionEasy to breedMicrobiological testing/measurementAnimal scienceGenome editing
The invention discloses a method for identifying or auxiliary identifying of pig farrowing characters and relates to a multi-omics integration accurate pig breeding method. The pig farrowing characters are determined through determination of mRNA expression quality of a pig HSD17B1 gene. Particularly, by acquisition of total RNA of pig ovary tissue cells, reverse transcription and quantitative PCR, the mRNA expression quality of the pig HSD17B1 gene is determined, and pigs in low mRNA expression quality are high in litter size and / or litter weight. In addition, by adoption of the identification method and artificial selection, high-litter-size pig breeding can be implemented. The identification method and application thereof to breeding are independent of genome editing means, acceptance of people is improved, and new breakthrough in breeding pig breeding is realized.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
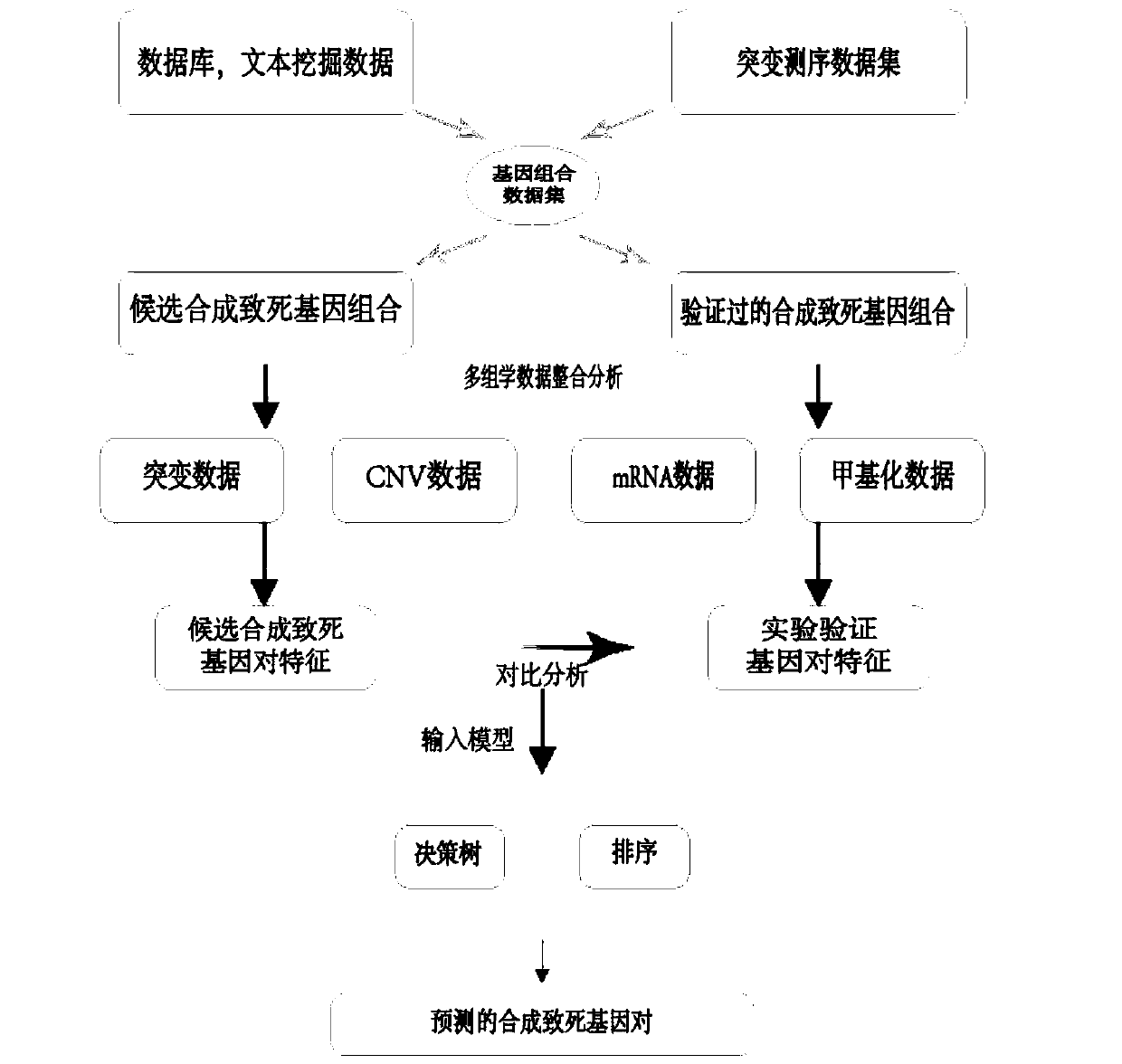
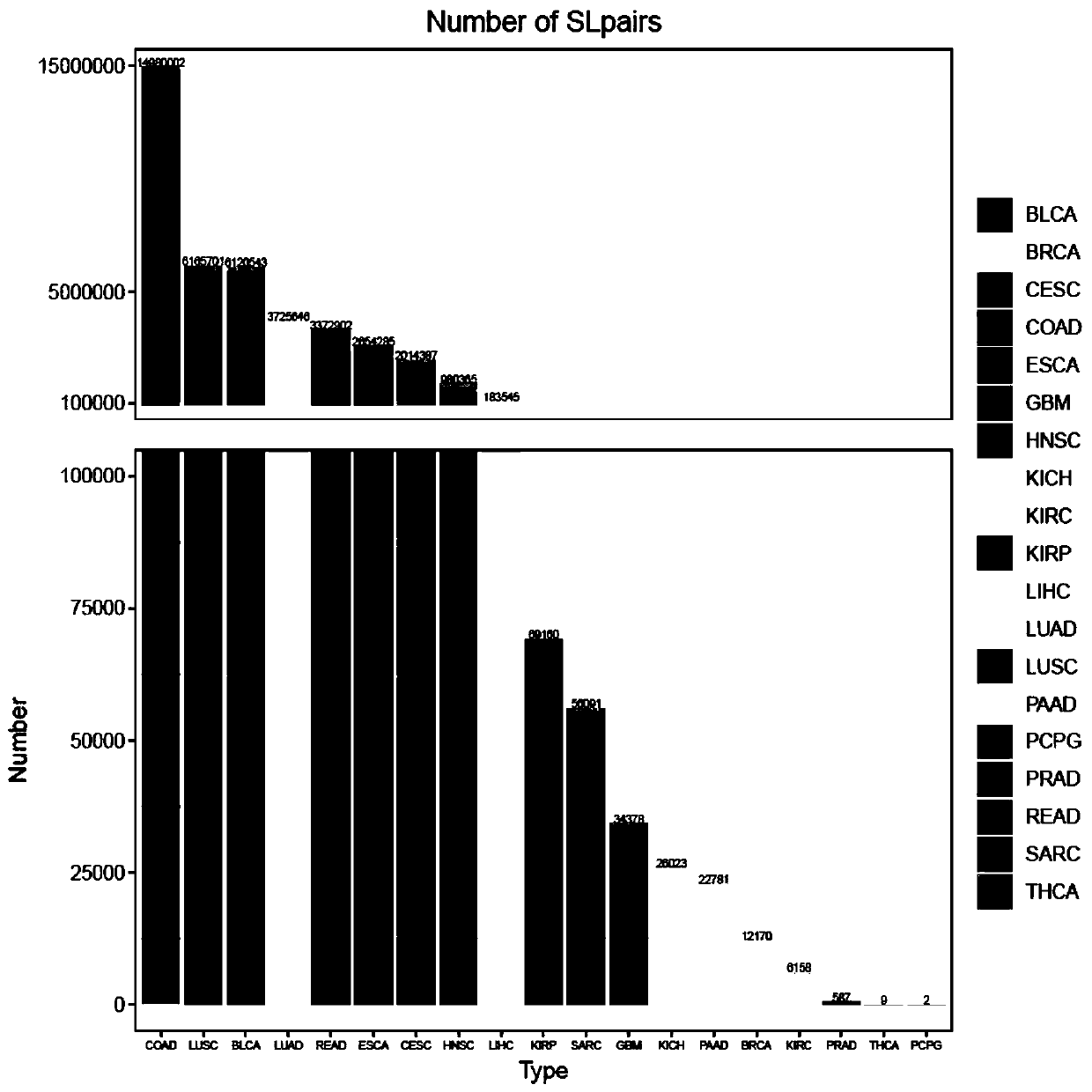
Method for predicting cancer synthesis lethal gene pairs based on decision tree and linear regression model
The invention relates to the field of gene prediction, and discloses a cancer synthesis lethal gene pair prediction method based on a decision tree model and a linear regression model. The method is mainly divided into a data training stage and a synthetic lethal gene pair testing stage. The method sequentially comprises the steps: firstly, extracting the data, which contains all mutant gene paircoverage rates, DNA methylation, mRNA expression profiles and copy number variation, from multi-omics data to serve as model feature values to be clustered, removing the false positive, performing normalization processing, and training a decision tree model and a linear regression model; secondly, respectively predicting synthetic lethal gene pairs possibly existing in various cancers through utilizing the decision tree model and the linear regression model to obtain distribution maps of the synthetic lethal gene pairs in different cancers; and finally, comparing the two models to obtain 508 pairs of synthetic lethal genes existing in the pan-cancer. According to the method, the synthetic lethal gene pairs possibly existing in various cancers can be accurately predicted, so a basis is provided for accurate treatment of the cancers.
Owner:NANJING UNIV OF POSTS & TELECOMM
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com