Bioinformatics method based on protein mass spectrum data annotation eukaryote genome
A technology of bioinformatics and eukaryotes, applied in the field of bioinformatics to annotate eukaryotic genomes based on protein mass spectrometry data, can solve the difficulty of obtaining full-length mRNA sequences, start codons and stop codons, and reduce Issues such as mass spectrum matching sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example
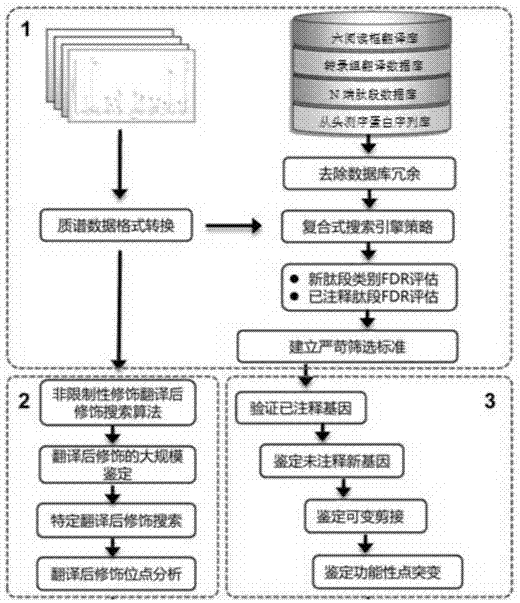
[0056] Aspergillus flavus mass spectrometry data 10G, establish six reading frame translation database, N-terminal peptide database, de novo predicted protein sequence database, transcriptome translation sequence database, integrated multi-omics database, construct high-coverage multi-omics sequence of eukaryotes database.
[0057] Download the non-coding RNA, pseudogene, non-coding gene sequence and EST sequence data of this species from the corresponding public database, and translate them into six different , A whole enzyme-cut peptide sequence with a length greater than 38.
[0058] (b) According to the integration strategy in step (e) of the first point, integrate the four types of databases created in the previous step into a redundant database.
[0059] (c) Filtering the constructed eukaryotic protein sequence database. If the same sequence appears in the eukaryotic protein sequence database and the deredundancy database, these sequences will be removed from the eukar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com