Patents
Literature
7766 results about "Mass spectrum analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
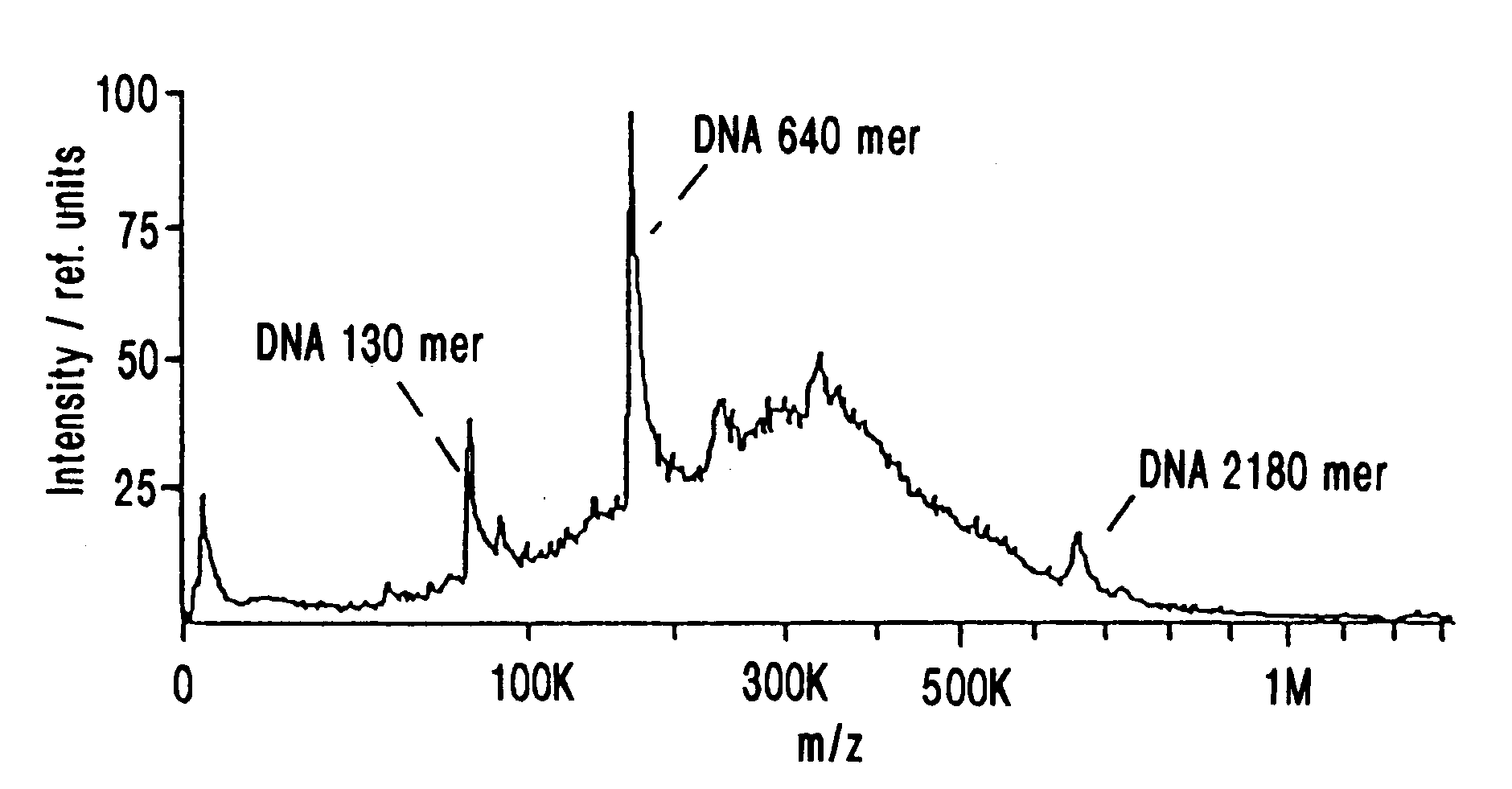
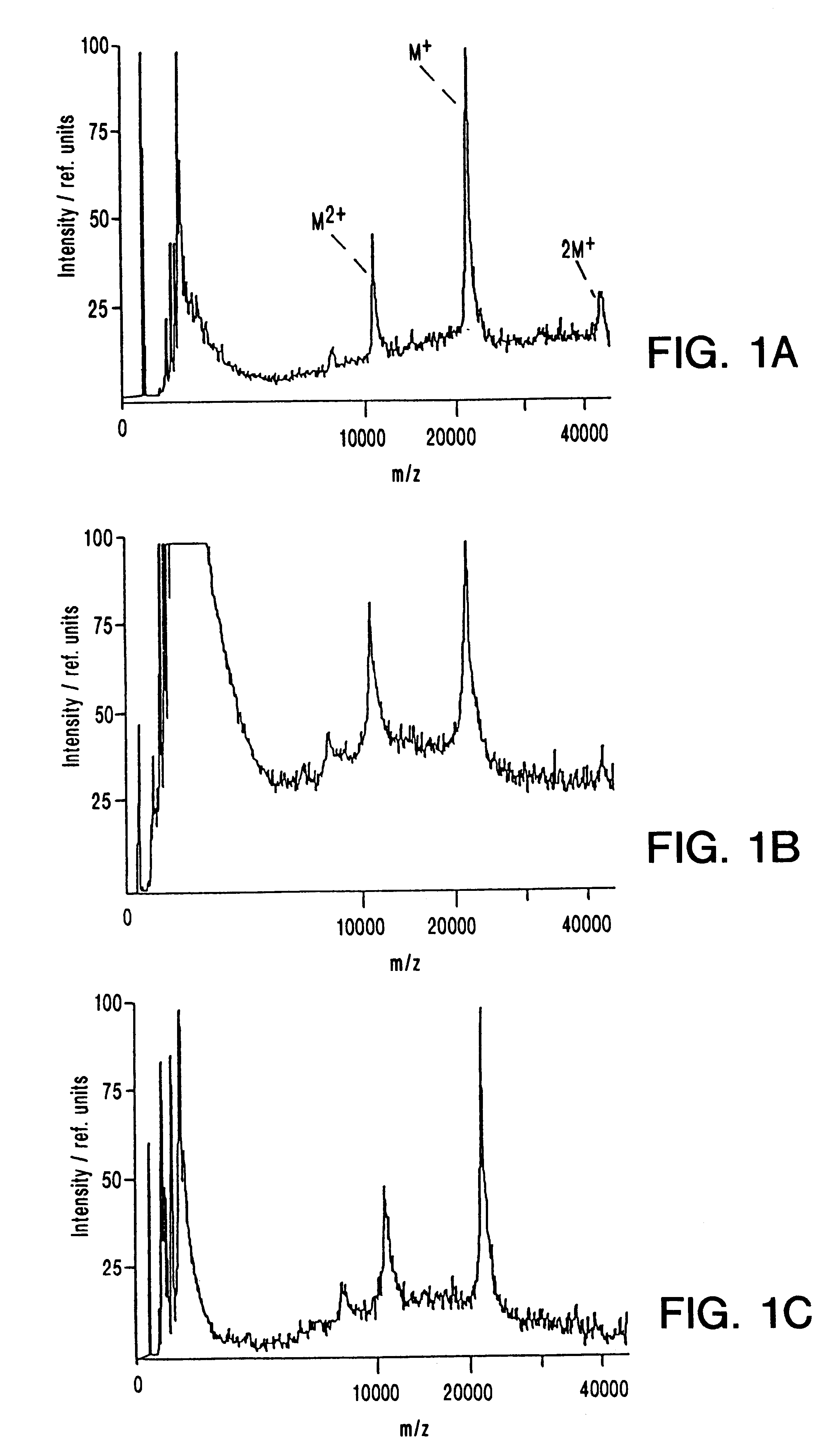
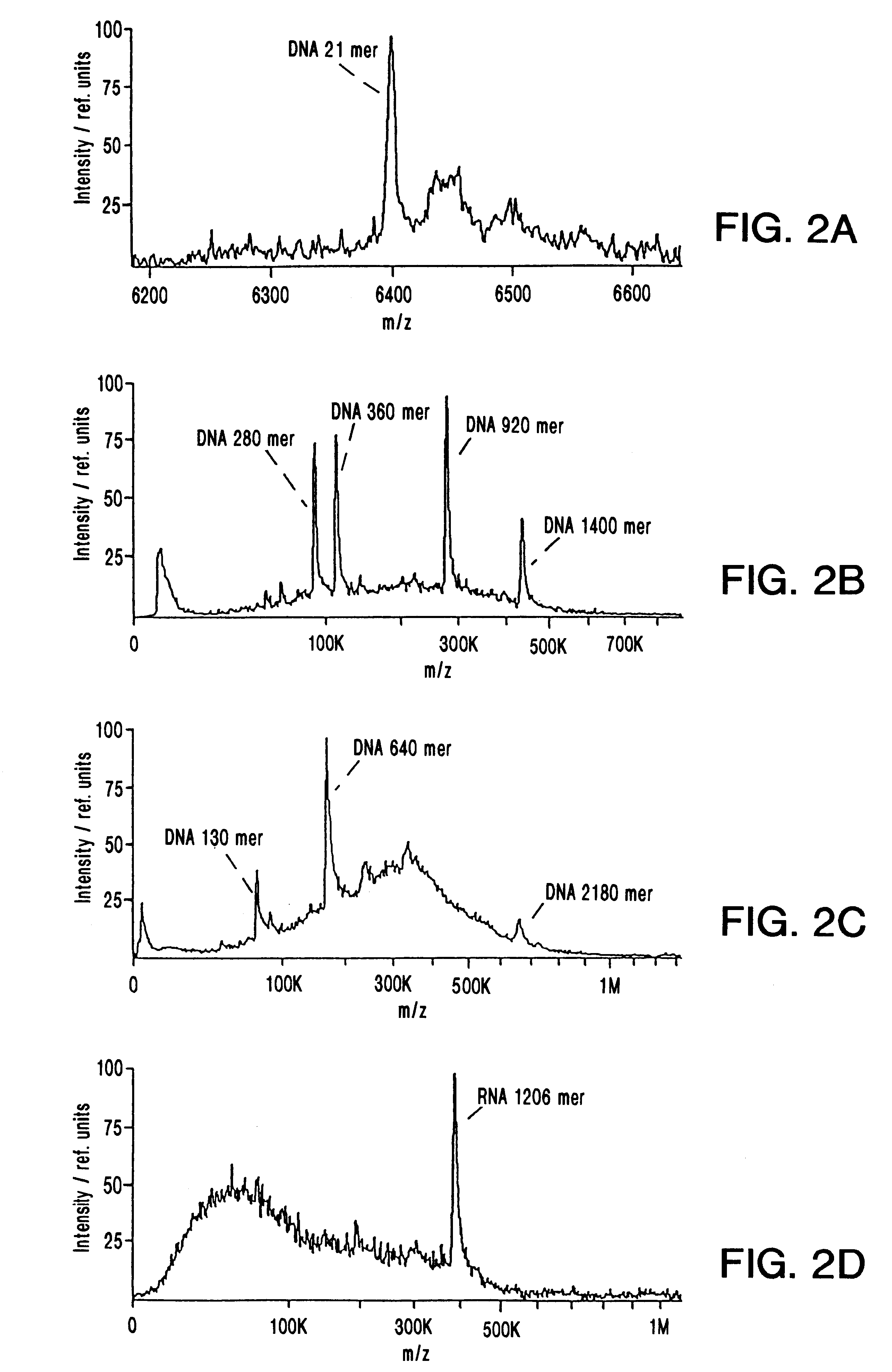
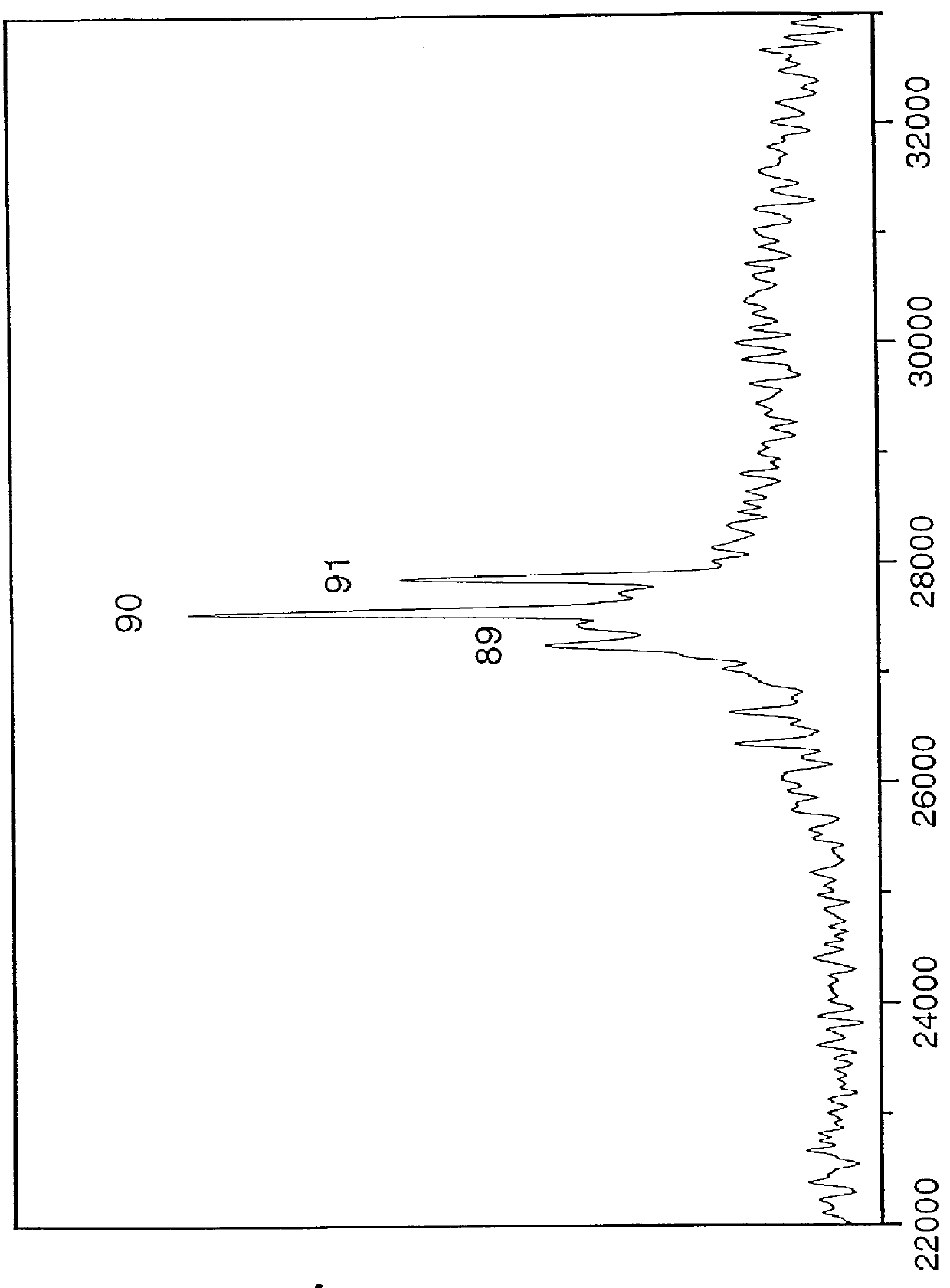
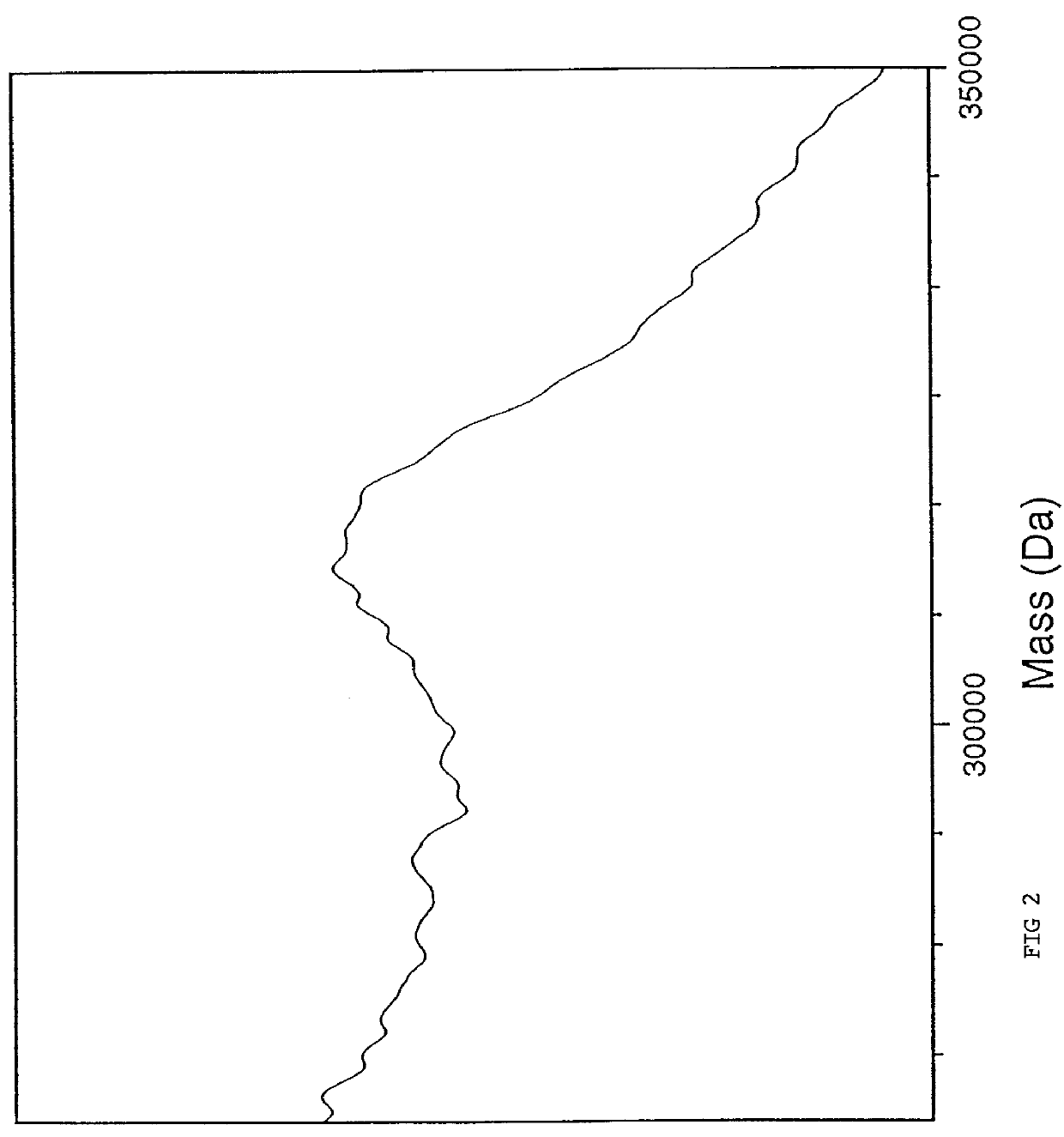
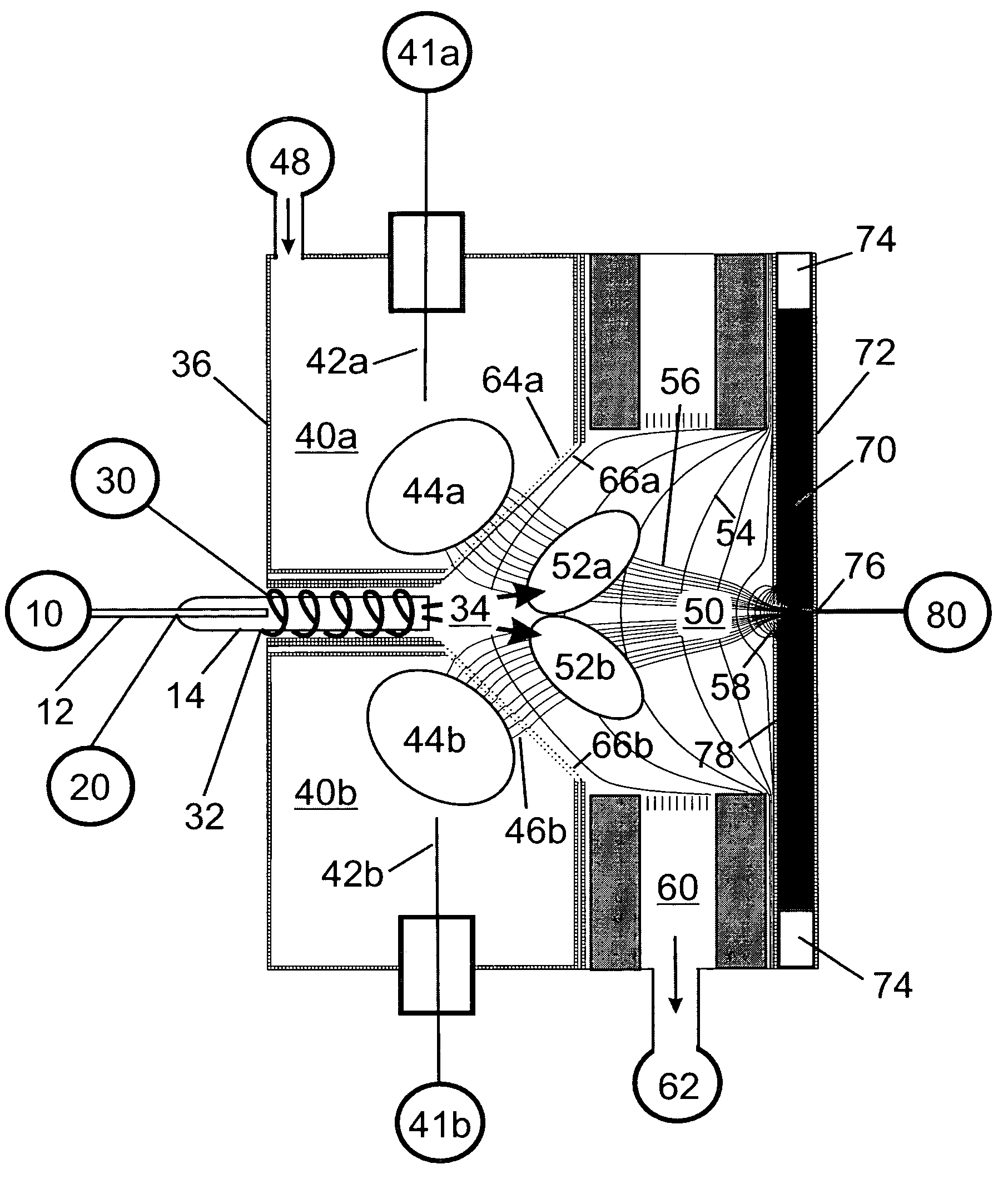
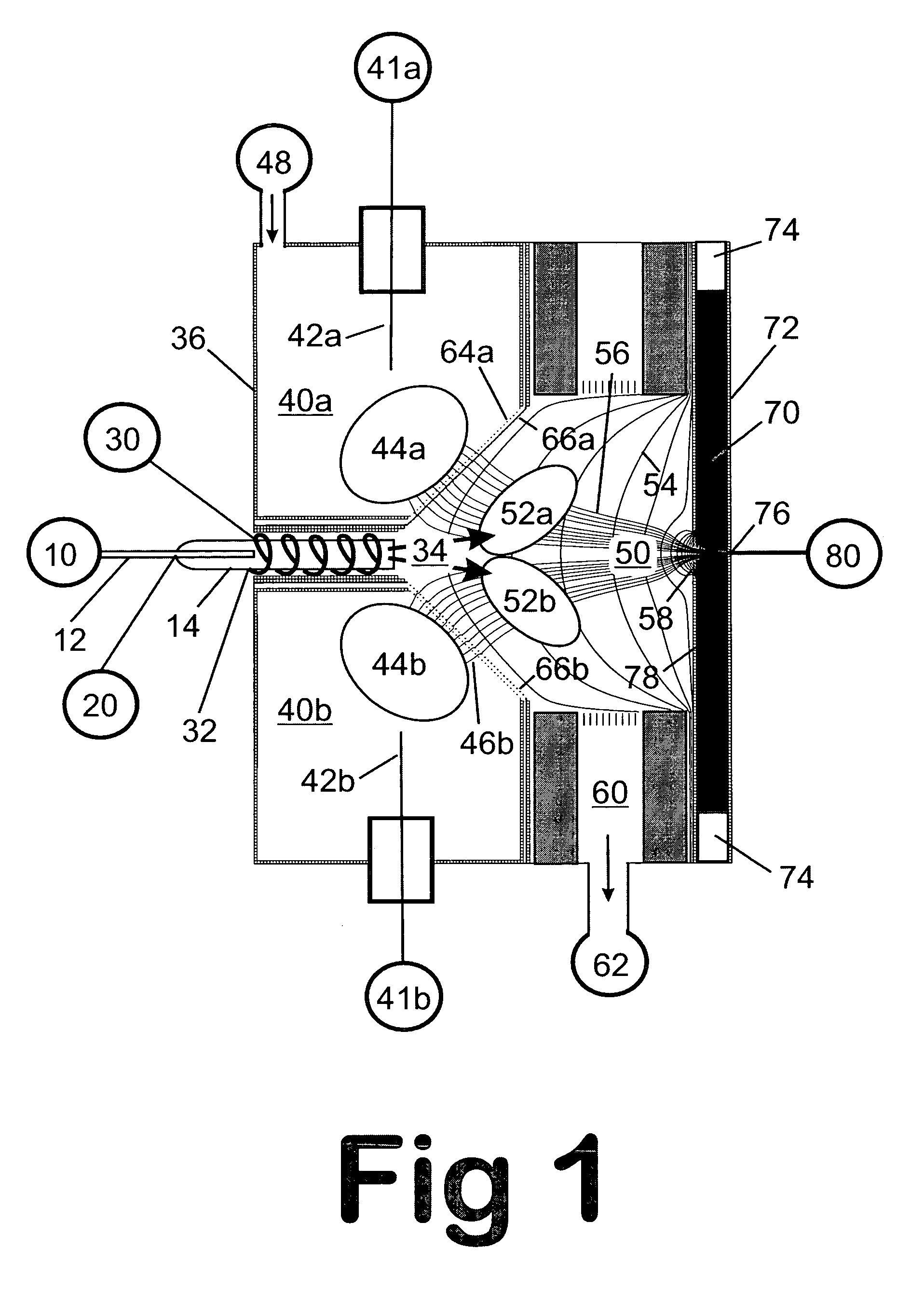
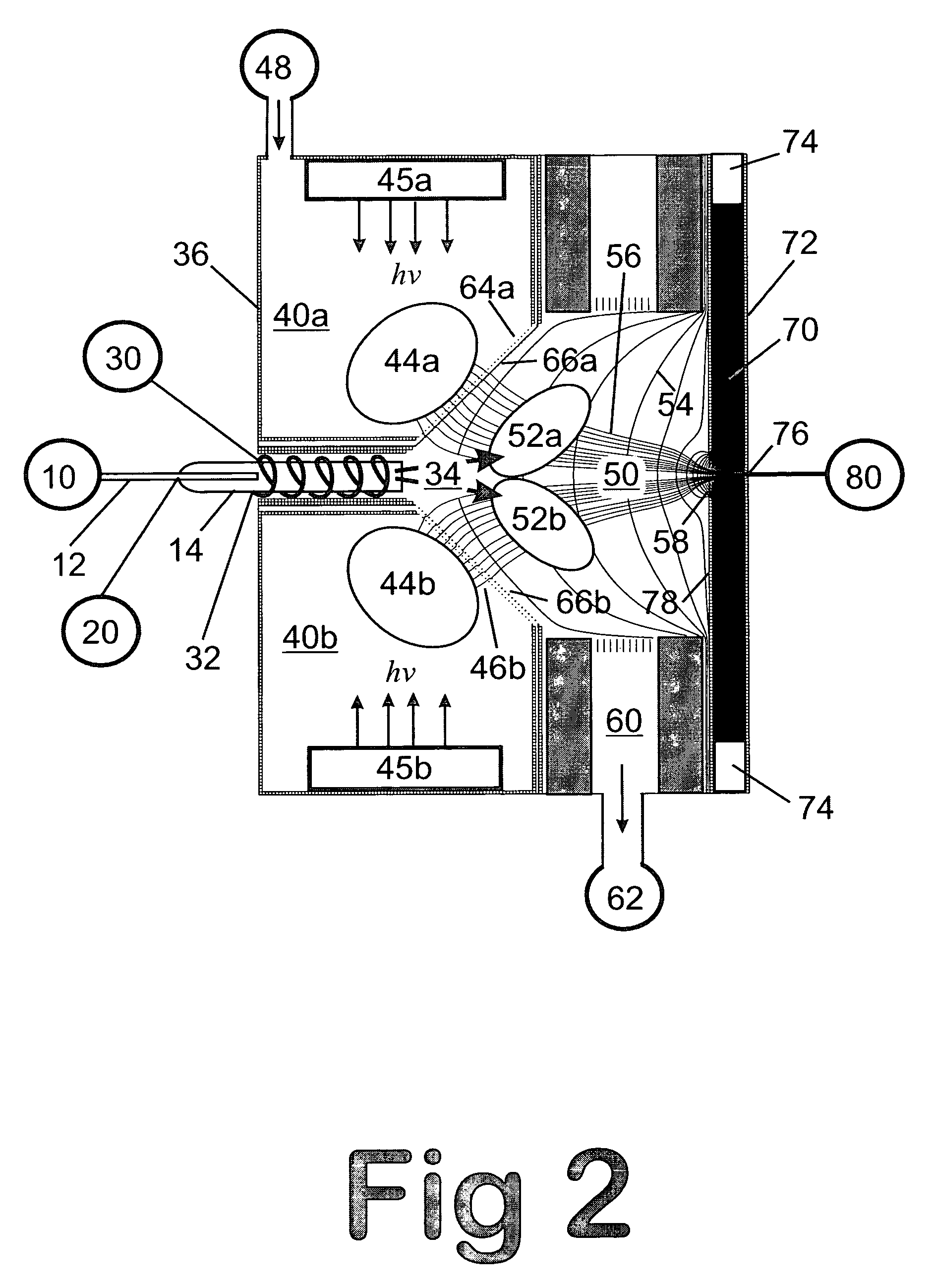
A mass spectrum is an intensity vs. m/z (mass-to-charge ratio) plot representing a chemical analysis. Hence, the mass spectrum of a sample is a pattern representing the distribution of ions by mass (more correctly: mass-to-charge ratio) in a sample.
Light-Emitting Element, Light-Emitting Device, Electronic Appliance, and Lighting Device
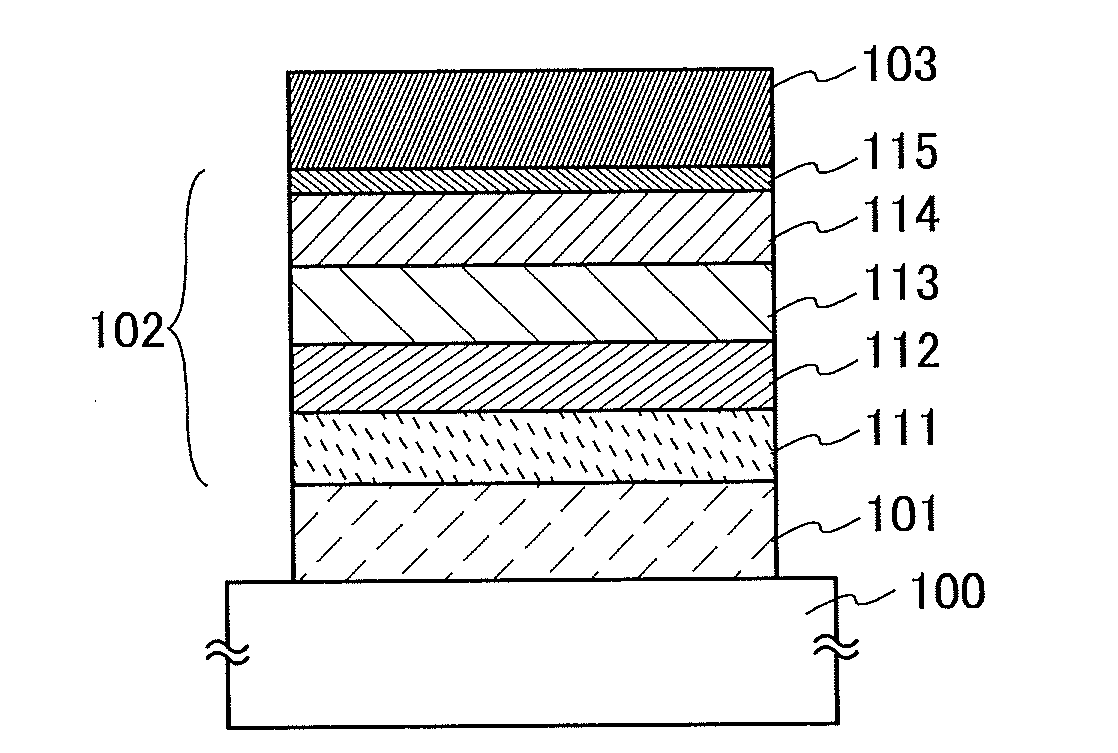
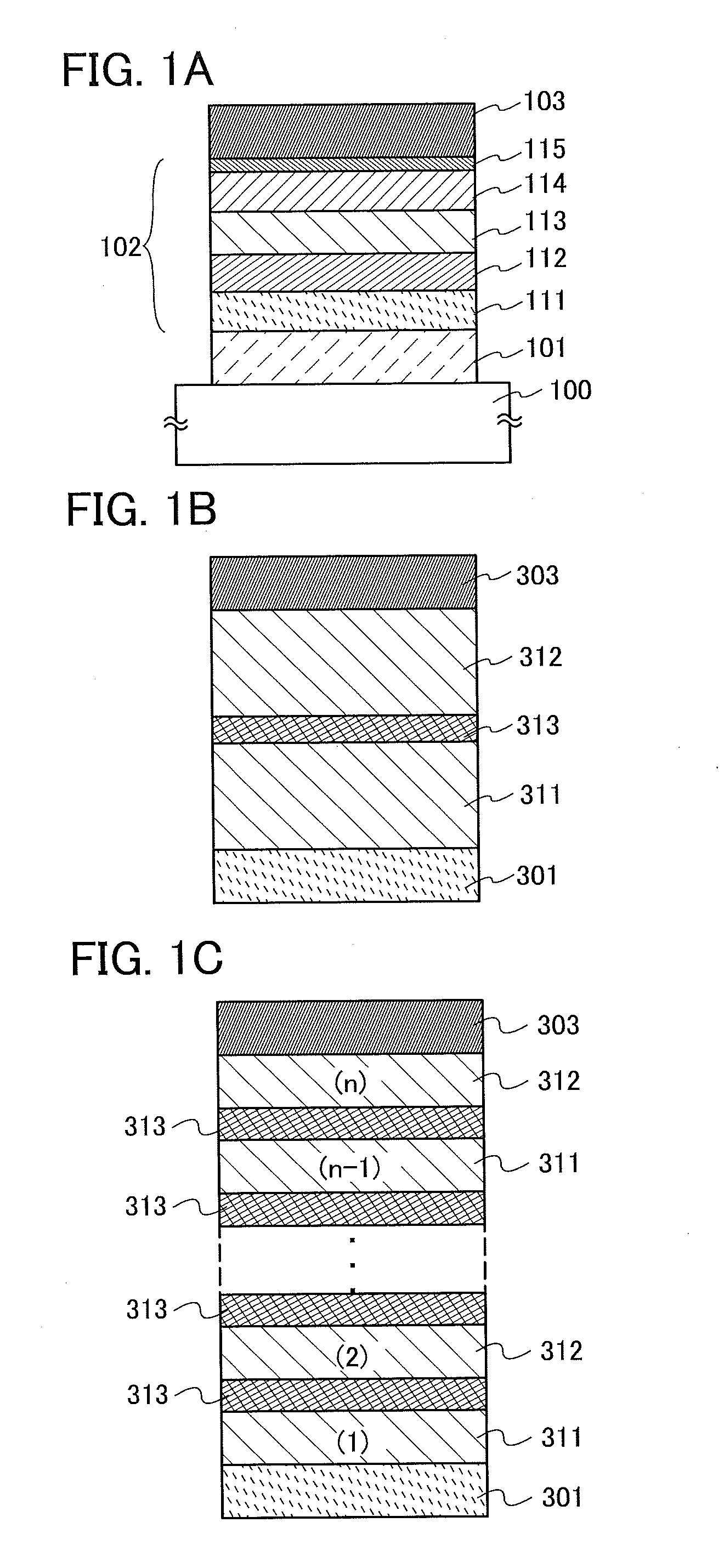
InactiveUS20140183503A1Reduce the driving voltageImprove current efficiencyOrganic chemistrySolid-state devicesLow voltageQuinoline
Disclosed is a light-emitting element having high emission efficiency, capable of driving at low voltage, and showing a long lifetime. The light-emitting element contains a compound between a pair of electrodes, and the compound is configured to give a first peak of m / z around 202 and a second peak of m / z around 227 in a mass spectrum. The first and second peaks are product ions of the compound and possess compositions of C16H9 and C17H10N, respectively, which are derived from a dibenzo[f,h]quinoline unit.
Owner:SEMICON ENERGY LAB CO LTD
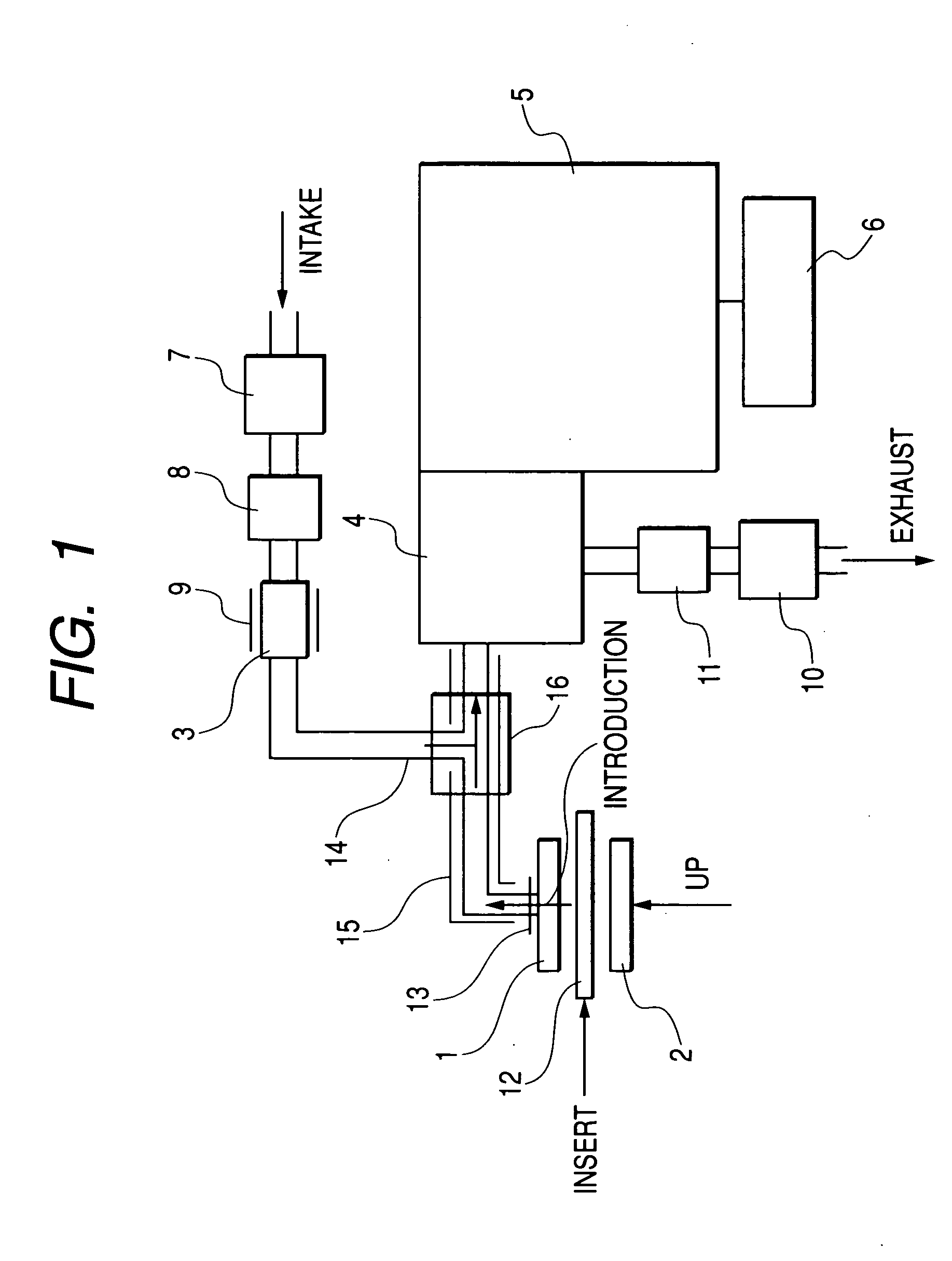
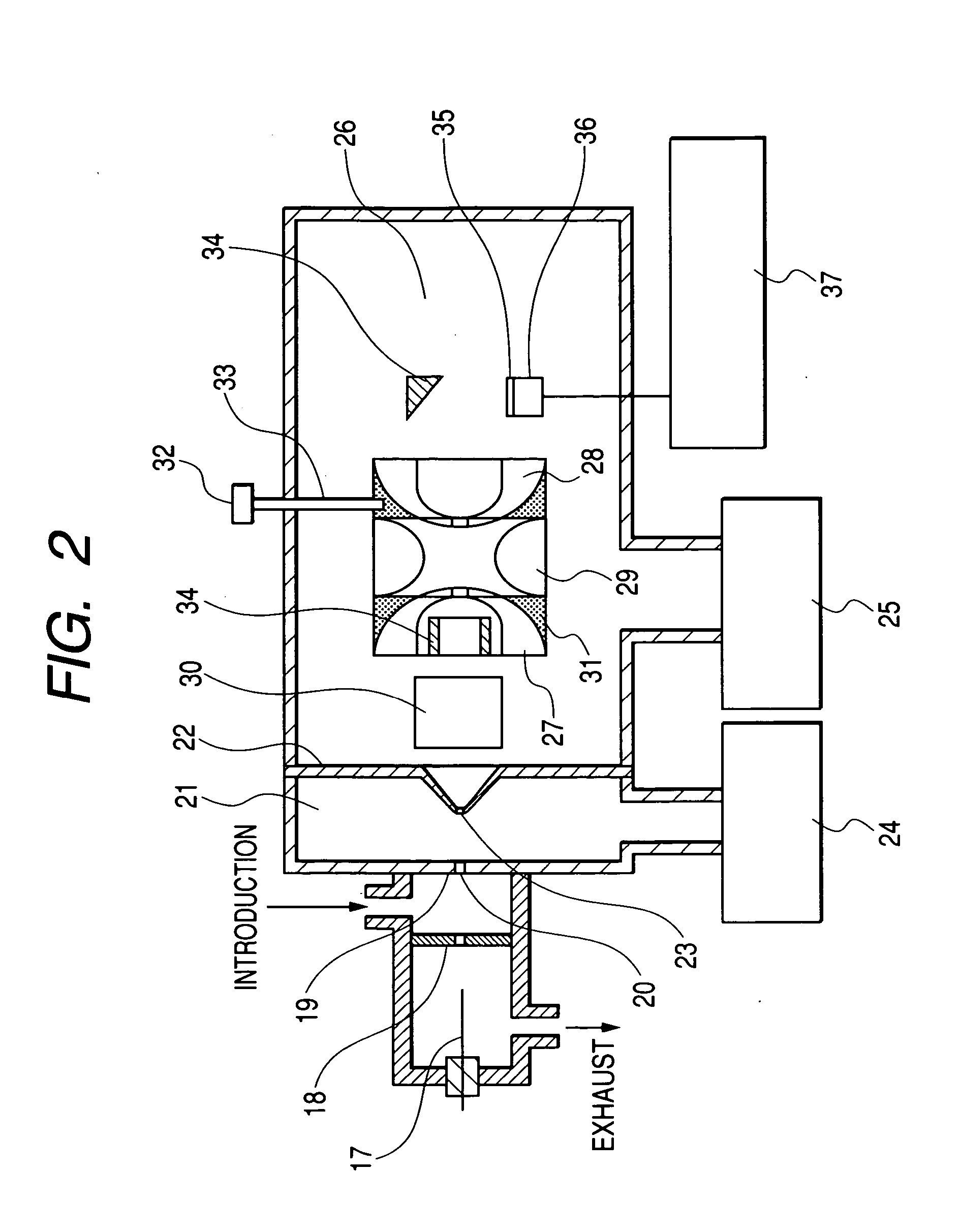
Apparatus for detecting chemical substances and method therefor
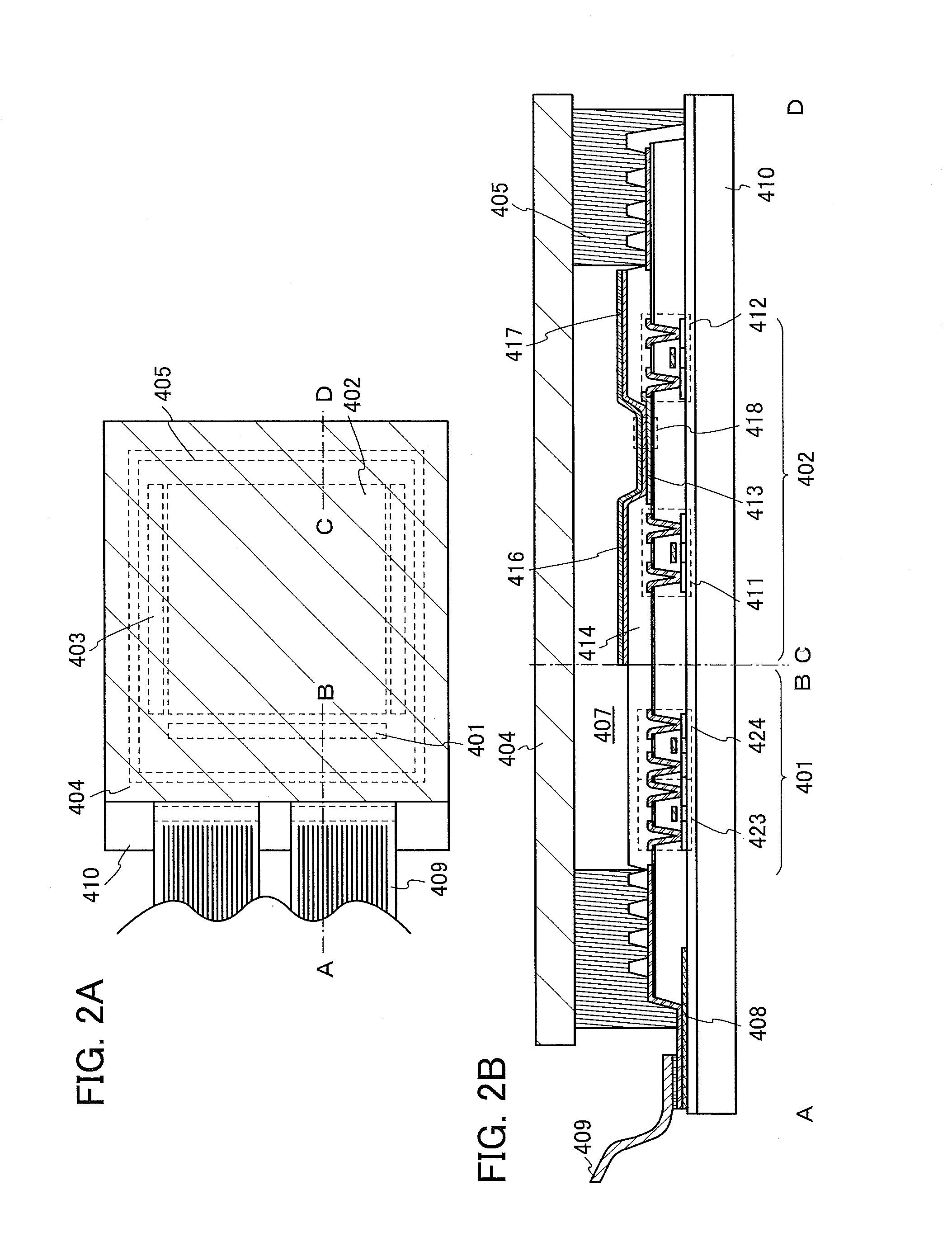
ActiveUS20050061964A1High detection sensitivityPrevent false detectionStability-of-path spectrometersAnalysis using chemical indicatorsNon detectionData treatment
An apparatus for detecting chemical substances which is high in sensitivity and selectivity is provided. An organic acid or an organic acid salt is used to generate an organic acid gas from an organic acid gas generator 3 to be mixed with a sample gas for introduction into an ion source 4 for ionization, thereby obtaining a mass spectrum by a mass analysis region 5. A data processor 6 determines the detection or non-detection of a specific m / z of an organic acid adduct ion obtained by adding a molecule generated from the organic acid to a molecule with specific m / z generated from a target chemical substance to be detected based on the obtained mass spectrum. When there is an ion peak with the m / z of the organic acid adduct ion, the presence of the target chemical substance to be detected is determined, and an alarm is sounded. False detection can be prevented.
Owner:HITACHI LTD
Mass spectrometry with segmented RF multiple ion guides in various pressure regions
InactiveUS7034292B1Reduce lossesEliminate and reduce numberIsotope separationSpectrometer combinationsFourier transform on finite groupsMass analyzer
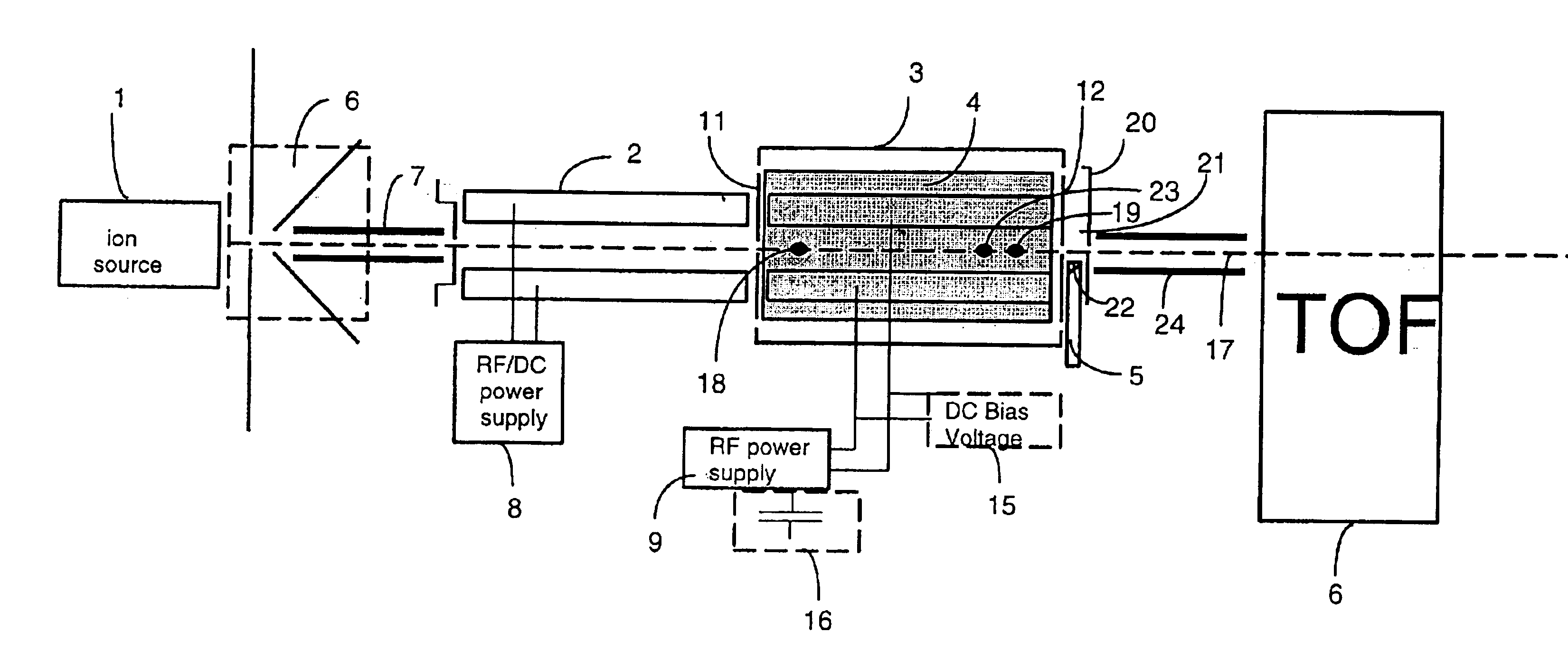
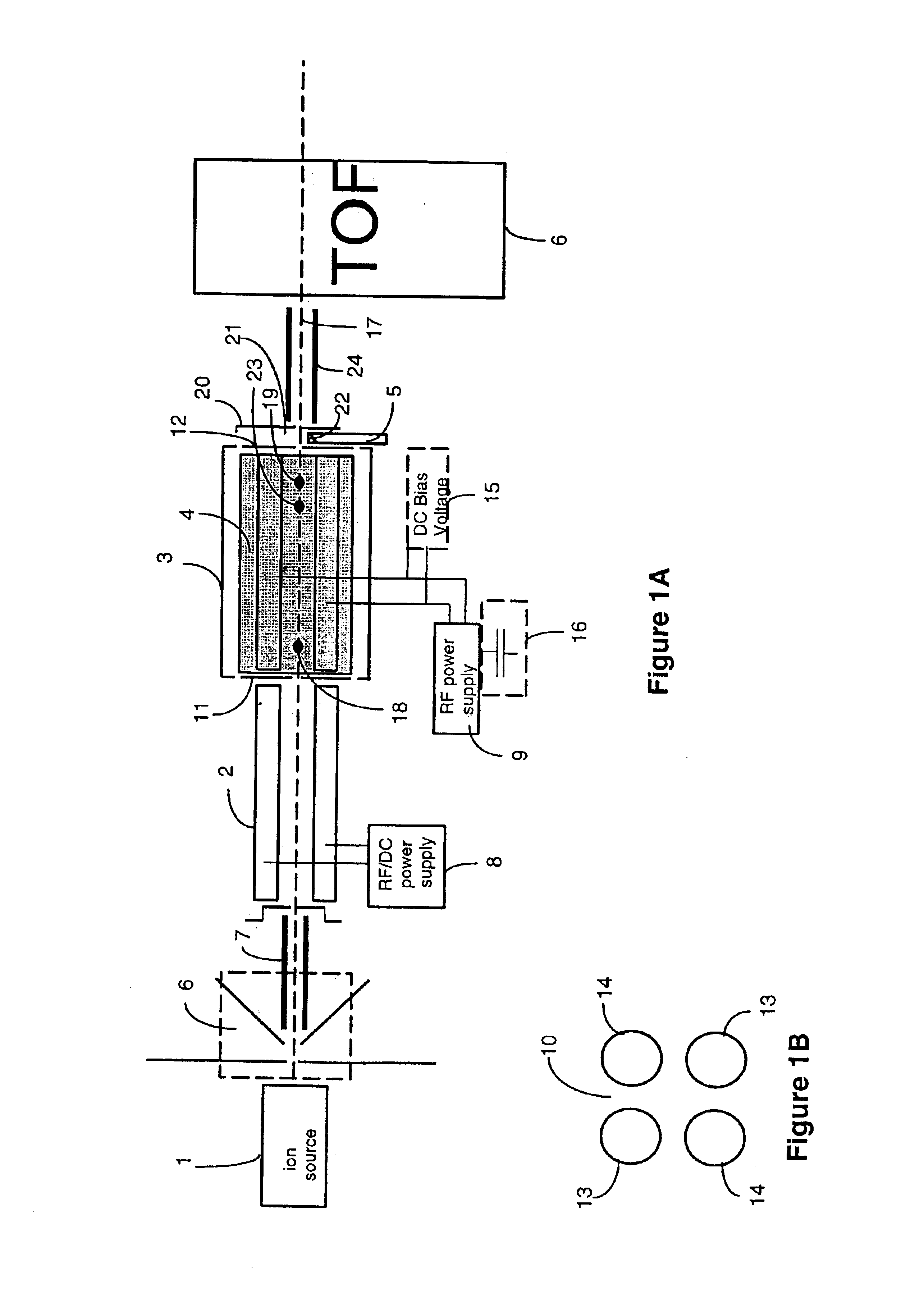
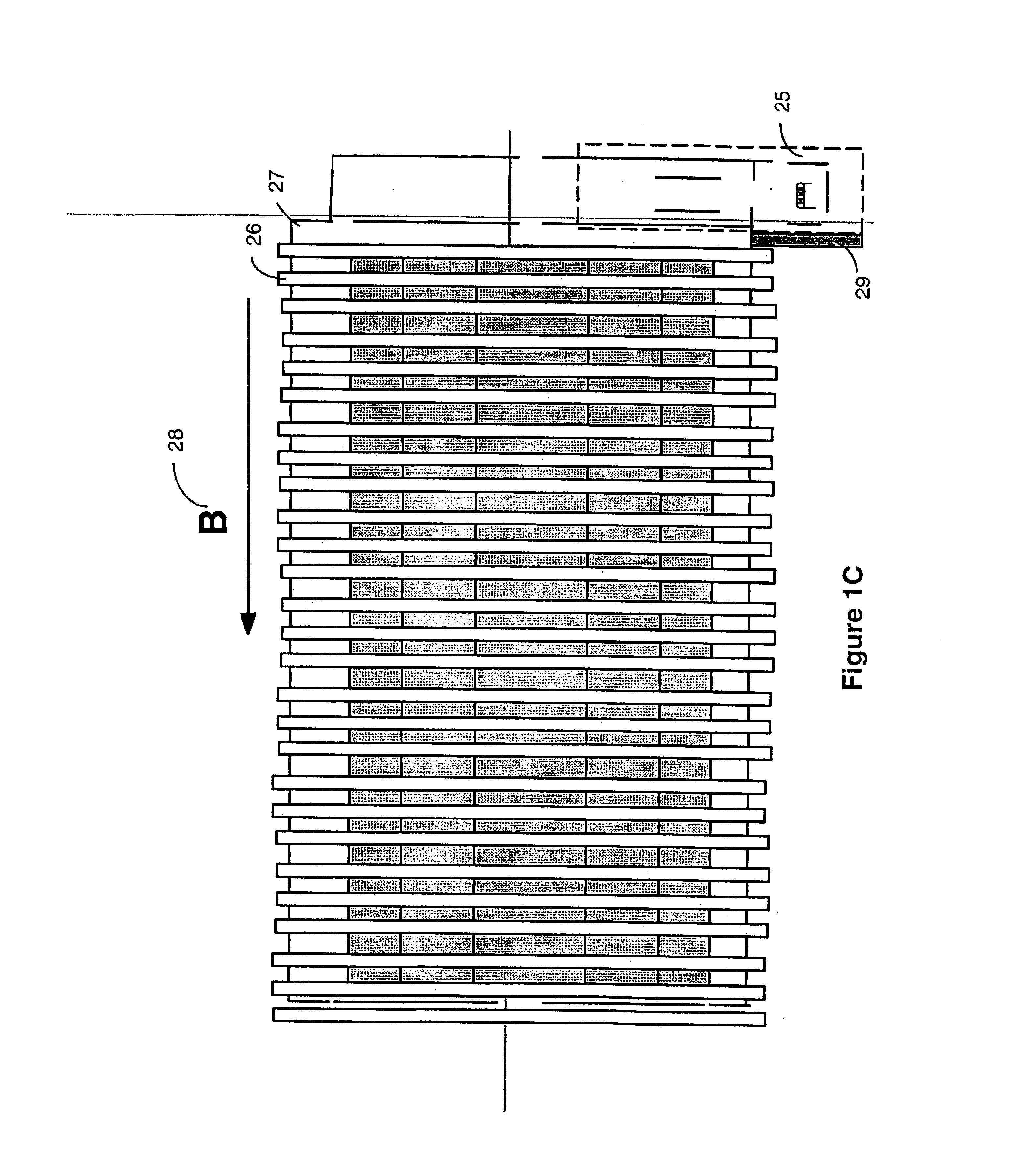
A mass spectrometer is configured with individual multipole ion guides, configured in an assembly in alignment along a common centerline wherein at least a portion of at least one multipole ion guide mounted in the assembly resides in a vacuum region with higher background pressure, and the other portion resides in a vacuum region with lower background pressure. Said multipole ion guides are operated in mass to charge selection and ion fragmentation modes, in either a high or low pressure region, said region being selected according to the optimum pressure or pressure gradient for the function performed. The diameter, lengths and applied frequencies and phases on these contiguous ion guides may be the same or may differ. A variety of MS and MS / MSn analysis functions can be achieved using a series of contiguous multipole ion guides operating in either higher background vacuum pressures, or along pressure gradients in the region where the pressure drops from high to low pressure, or in low pressure regions. Individual sets of RF, + / −DC and resonant frequency waveform voltage supplies provide potentials to the rods of each multipole ion guide allowing the operation of ion transmission, ion trapping, mass to charge selection and ion fragmentation functions independently in each ion guide. The presence of background pressure maintained sufficiently high to cause ion to neutral gas collisions along a portion of each multiple ion guide linear assembly allows the conducting of Collisional Induced Dissociation (CID) fragmentation of ions by axially accelerating ions from one multipole ion guide into an adjacent ion guide. Alternatively ions can be fragmented in one or more multipole ion guides using resonant frequency excitation CID. A multiple multipole ion guide assembly can be configured as the primary mass analyzer in single or triple quadrupole mass analyzers with or without mass selective axial ejection. Alternatively, the multiple multipole ion guide linear assembly can be configured as part of a hybrid Time-Of-Flight, Magnetic Sector, Ion Trap or Fourier Transform mass analyzer.
Owner:PERKINELMER U S LLC
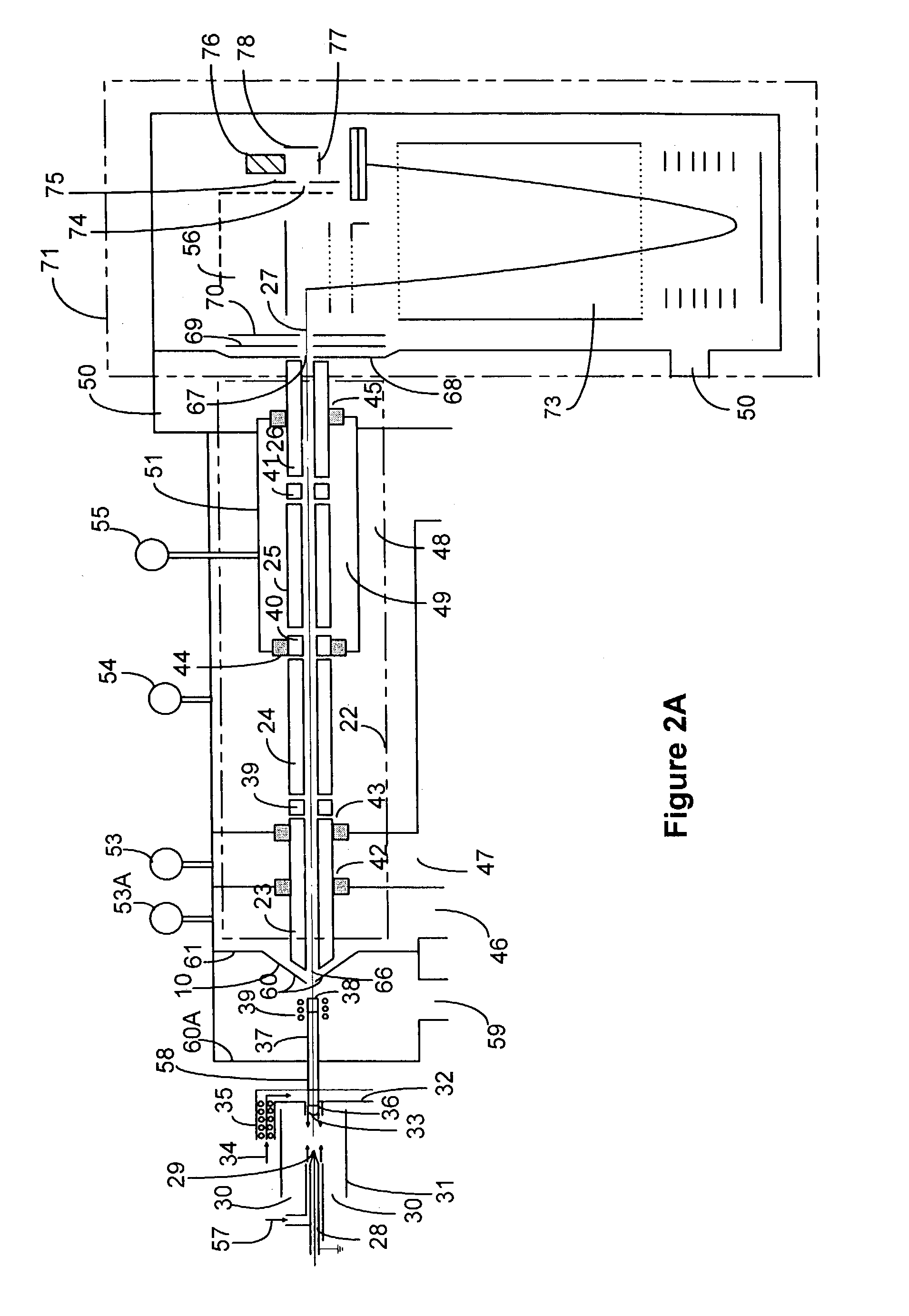
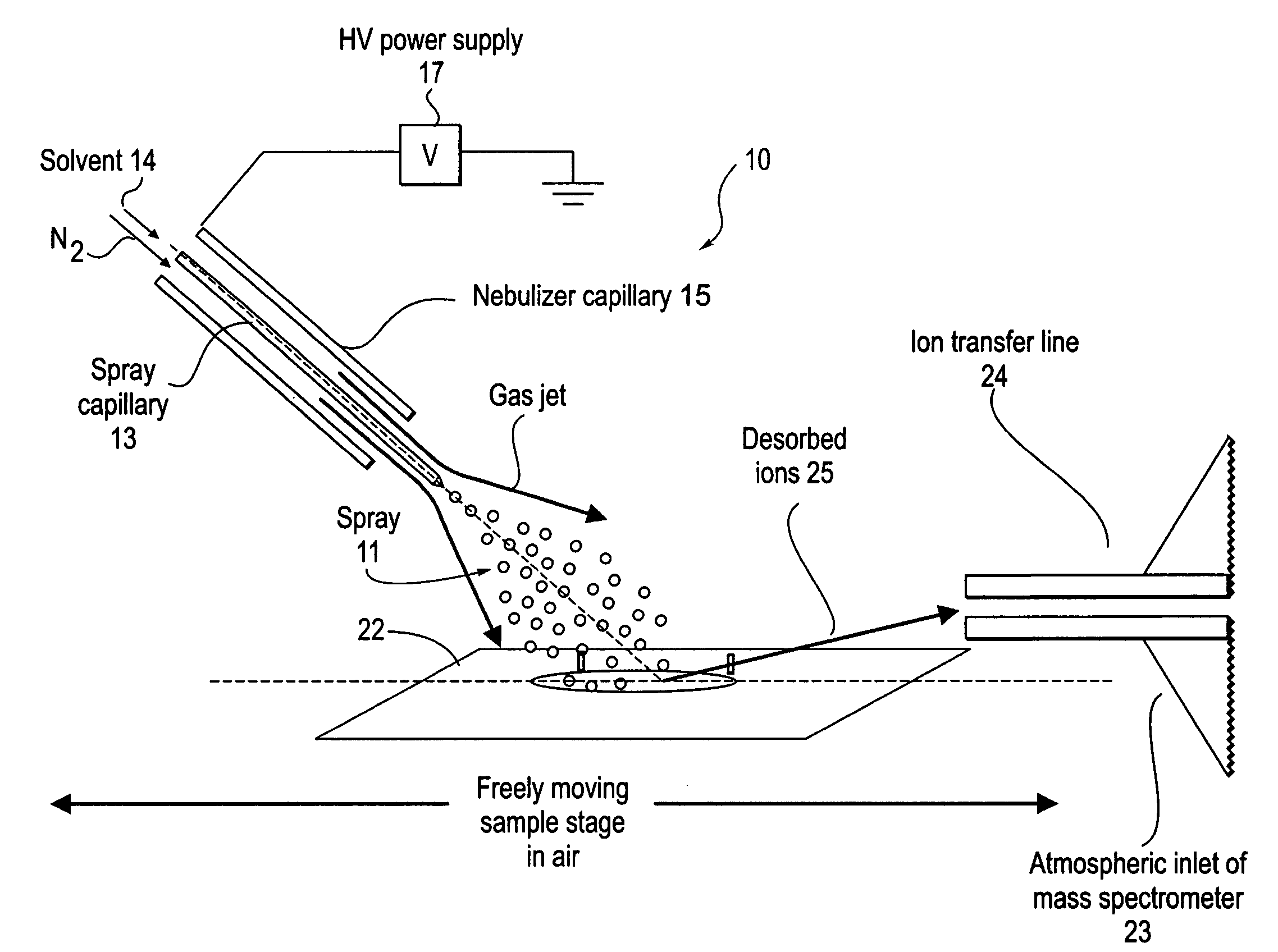
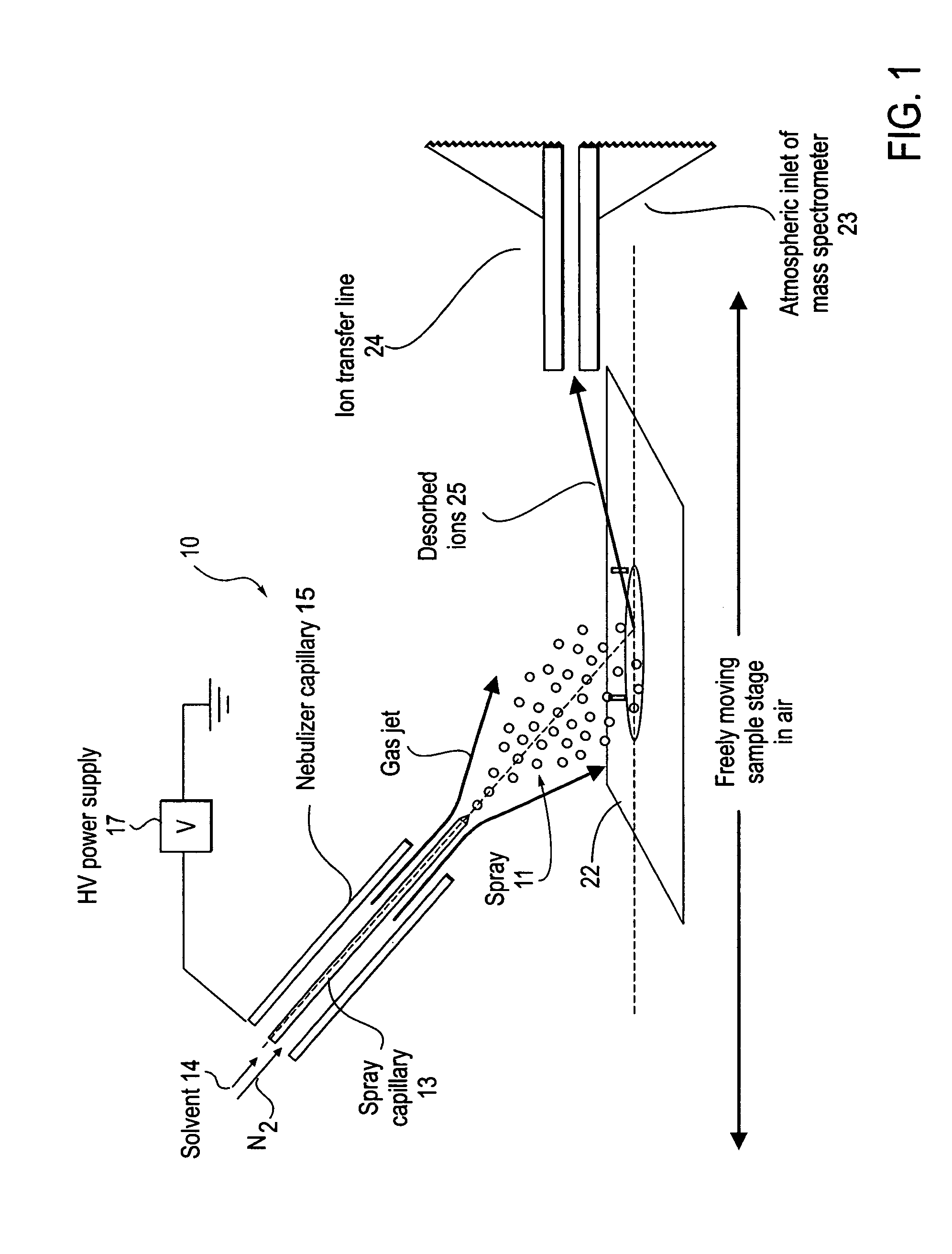
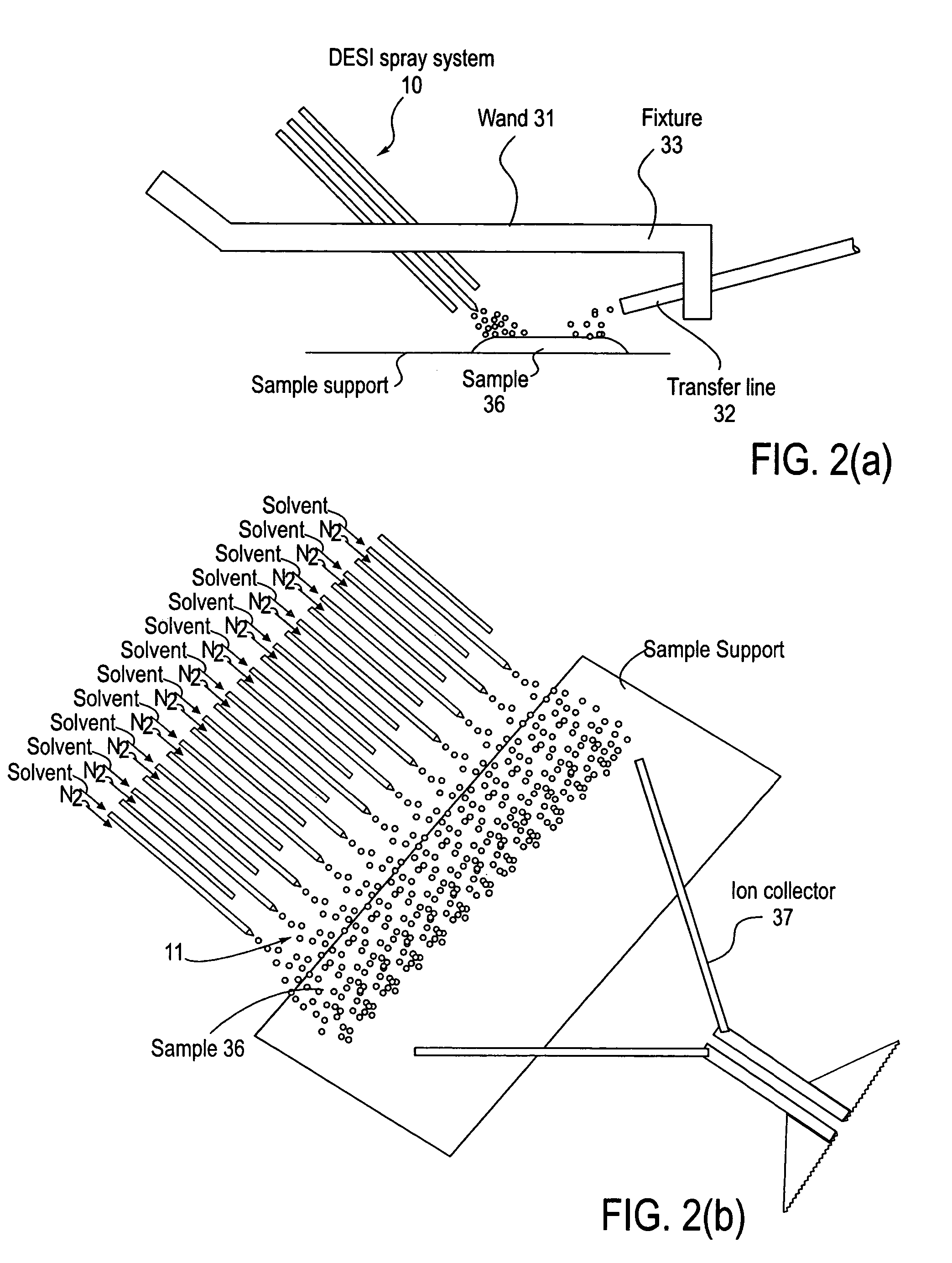
Method and system for desorption electrospray ionization
ActiveUS7335897B2Samples introduction/extractionMaterial analysis by optical meansLycopeneElectrospray ionization
A new method and system for desorption ionization is described and applied to the ionization of various compounds, including peptides and proteins present on metal, polymer, and mineral surfaces. Desorption electrospray ionization (DESI) is carried out by directing charged droplets and / or ions of a liquid onto the surface to be analyzed. The impact of the charged particles on the surface produces gaseous ions of material originally present on the surface. The resulting mass spectra are similar to normal ESI mass spectra in that they show mainly singly or multiply charged molecular ions of the analytes. The DESI phenomenon was observed both in the case of conductive and insulator surfaces and for compounds ranging from nonpolar small molecules such as lycopene, the alkaloid coniceine, and small drugs, through polar compounds such as peptides and proteins. Changes in the solution that is sprayed can be used to selectively ionize particular compounds, including those in biological matrices. In vivo analysis is demonstrated.
Owner:PURDUE RES FOUND INC
IR-MALDI mass spectrometry of nucleic acids using liquid matrices
InactiveUS6706530B2Rapidly and accurately massOptimize allocationNanostructure manufactureSamples introduction/extractionMass Spectrometry-Mass SpectrometryMatrix-assisted laser desorption/ionization
Mass spectrometry of large nucleic acids by infrared Matrix-Assisted Laser Desorption / Ionization (MALDI) using a liquid matrix is reported.
Owner:AGENA BIOSCI
Volatile matrices for matrix-assisted laser desorption/ionization mass spectrometry
InactiveUS6104028AEasy to spreadReduce formationSamples introduction/extractionWithdrawing sample devicesThermal ionization mass spectrometryRoom temperature
A sample preparation method is disclosed for volatilization and mass spectrometric analysis of nonvolatile high molecular weight molecules. Photoabsorbing molecules having significant sublimation rates at room temperature under vacuum, and preferably containing hydroxy functionalities, are disclosed for use as matrices in matrix-assisted laser desorption / ionization mass spectrometry. The samples are typically cooled in the mass spectrometer to temperatures significantly below room temperature.
Owner:AGENA BIOSCI
Mass spectrometer system
ActiveUS20050063864A1Reduce adverse effectsSamples introduction/extractionIsotope separationStructure analysisSpectroscopy
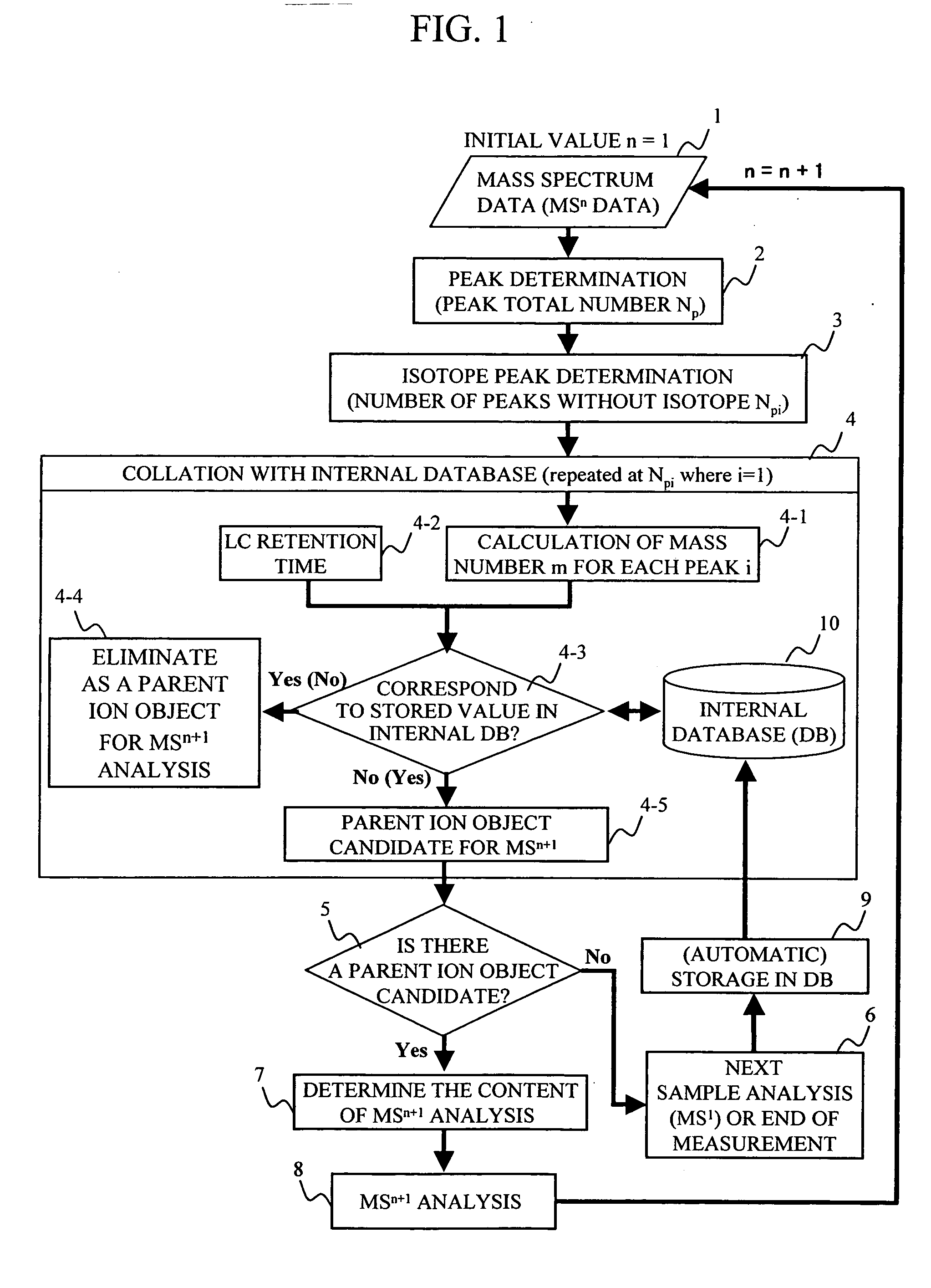
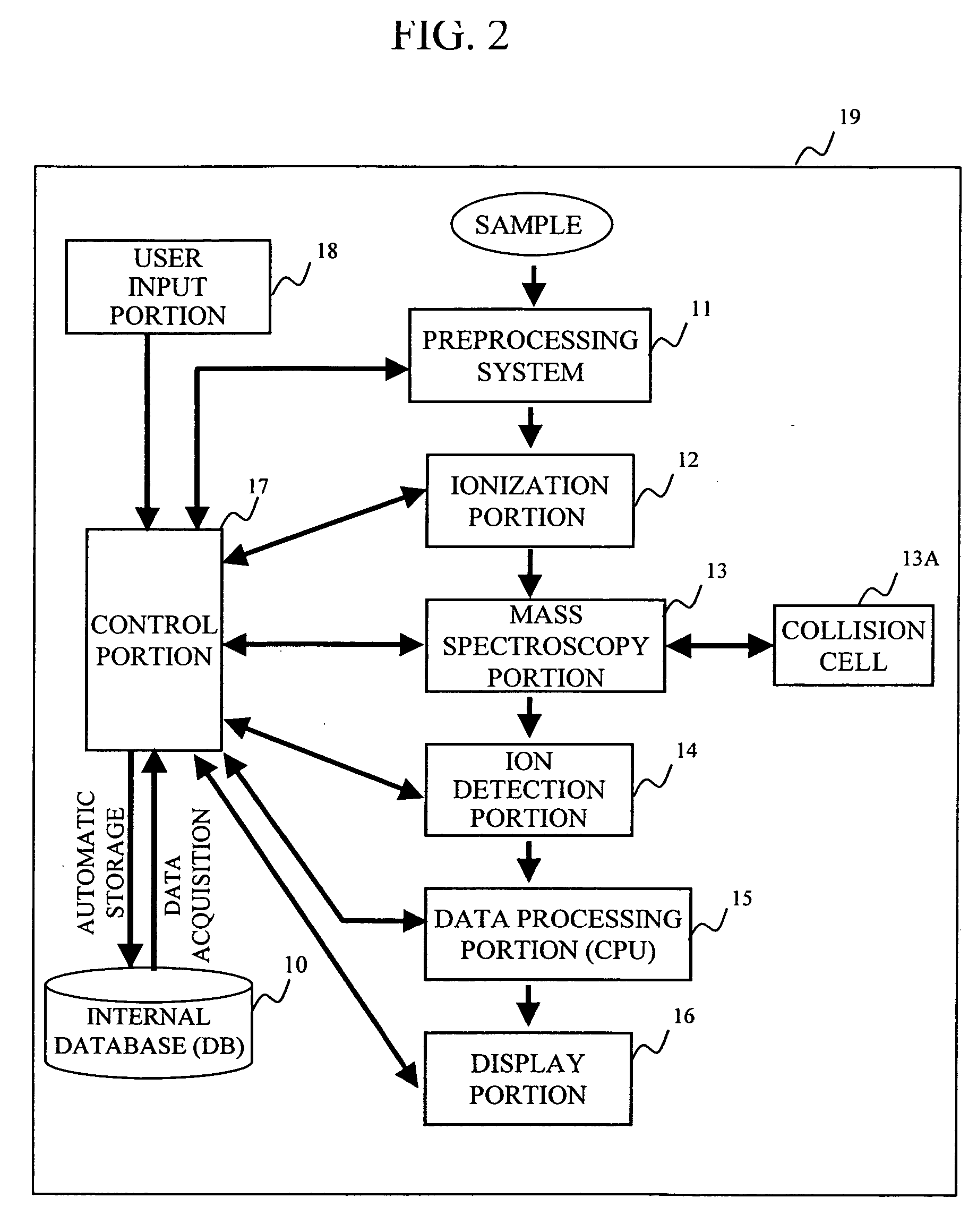
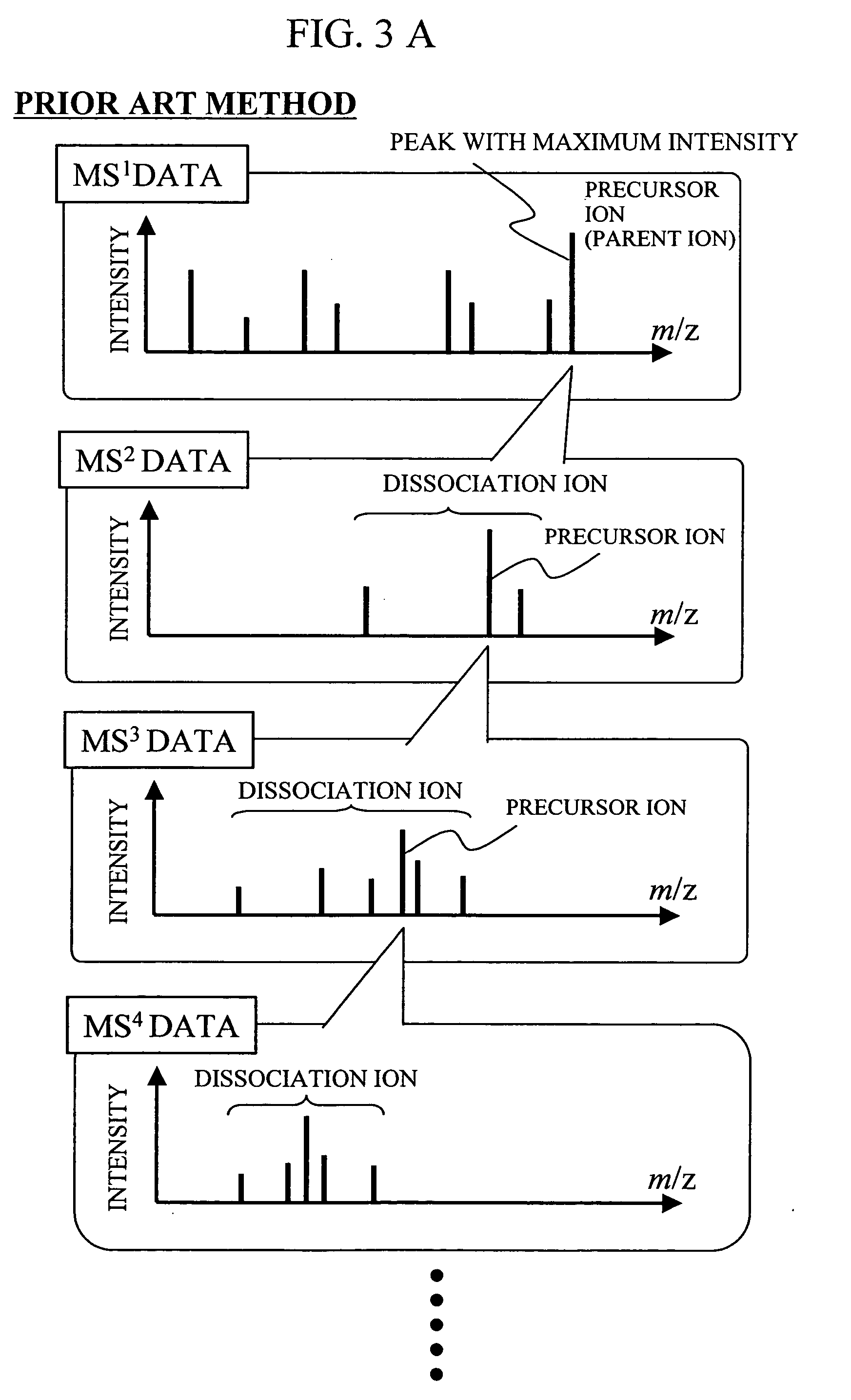
During the structural analysis of a protein or peptide by tandem mass spectroscopy, a peptide ion derived from a protein that has already been measured and that is expressed in great quantities is avoided as a tandem mass spectroscopy target. A peptide derived from a minute amount of protein, which has heretofore been difficult to analyze, can be automatically determined as a tandem mass spectroscopy target within the real time of measurement. Data concerning a protein that has already been measured and a peptide derived from the protein is automatically stored in an internal database. The stored data is collated with measured data with high accuracy to determine an isotope peak. In this way, the process of selecting a peptide peak that has not been measured as the target for the next tandem analysis can be performed within the real time of measurement and a redundant measurement of peptides derived from the same protein can be avoided. The information contained in the MSn spectrum is effectively utilized in each step of the MSn involving a multi-stage dissociation and mass spectroscopy (MSn), so that the flows for the determination of the next analysis content and the selection of the parent ion for the MSn+1 analysis, for example, can be optimized within the real time of measurement and with high efficiency and accuracy. Thus, a target of concern to the user can be subjected to tandem mass spectroscopy without wasteful measurement.
Owner:HITACHI HIGH-TECH CORP
Obtaining tandem mass spectrometry data for multiple parent ions in an ion population
ActiveUS7157698B2Improve performanceImprove ejection efficiencyStability-of-path spectrometersTime-of-flight spectrometersNarrow rangeIon trap mass spectrometry
This invention relates to tandem mass spectrometry and, in particular, to tandem mass spectrometry using a linear ion trap and a time of flight detector to collect mass spectra to form a MS / MS experiment. The accepted standard is to store and mass analyze precursor ions in the ion trap before ejecting the ions axially to a collision cell for fragmentation before mass analysis of the fragments in the time of flight detector. This invention makes use of orthogonal ejection of ions with a narrow range of m / z values to produce a ribbon beam of ions that are injected into the collision cell. The shape of this beam and the high energy of the ions are accommodated by using a planar design of collision cell. Ions are retained in the ion trap during ejection so that successive narrow ranges may be stepped through consecutively to cover all precursor ions of interest.
Owner:THERMO FINNIGAN
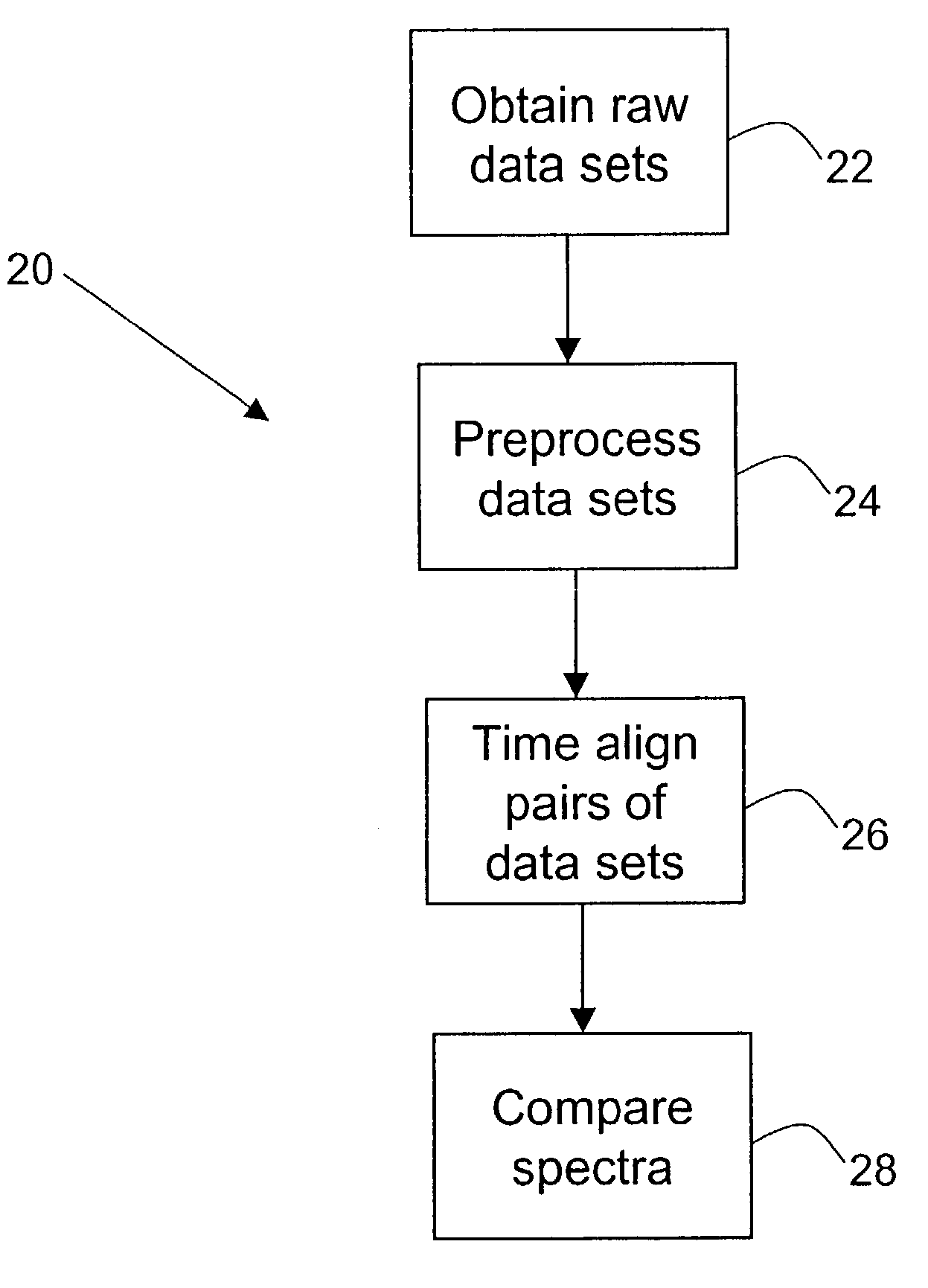
Methods for time-alignment of liquid chromatography-mass spectrometry data
Nonlinear retention time variations in chromatography-mass spectrometry data sets are adjusted by time-alignment methods, enabling automated comparison of spectra for differential phenotyping and other applications.
Owner:CAPRION PROTEOMICS INC
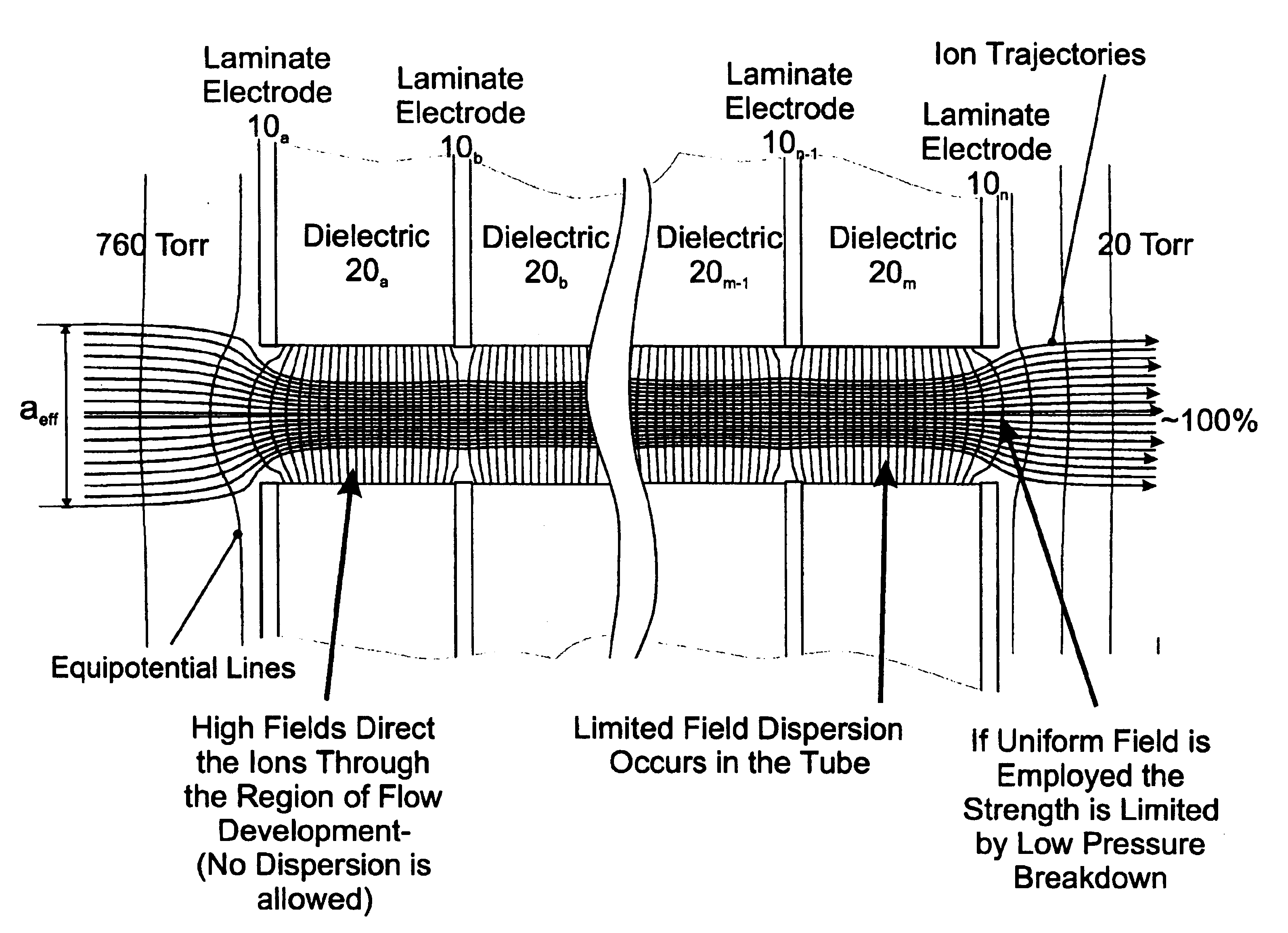
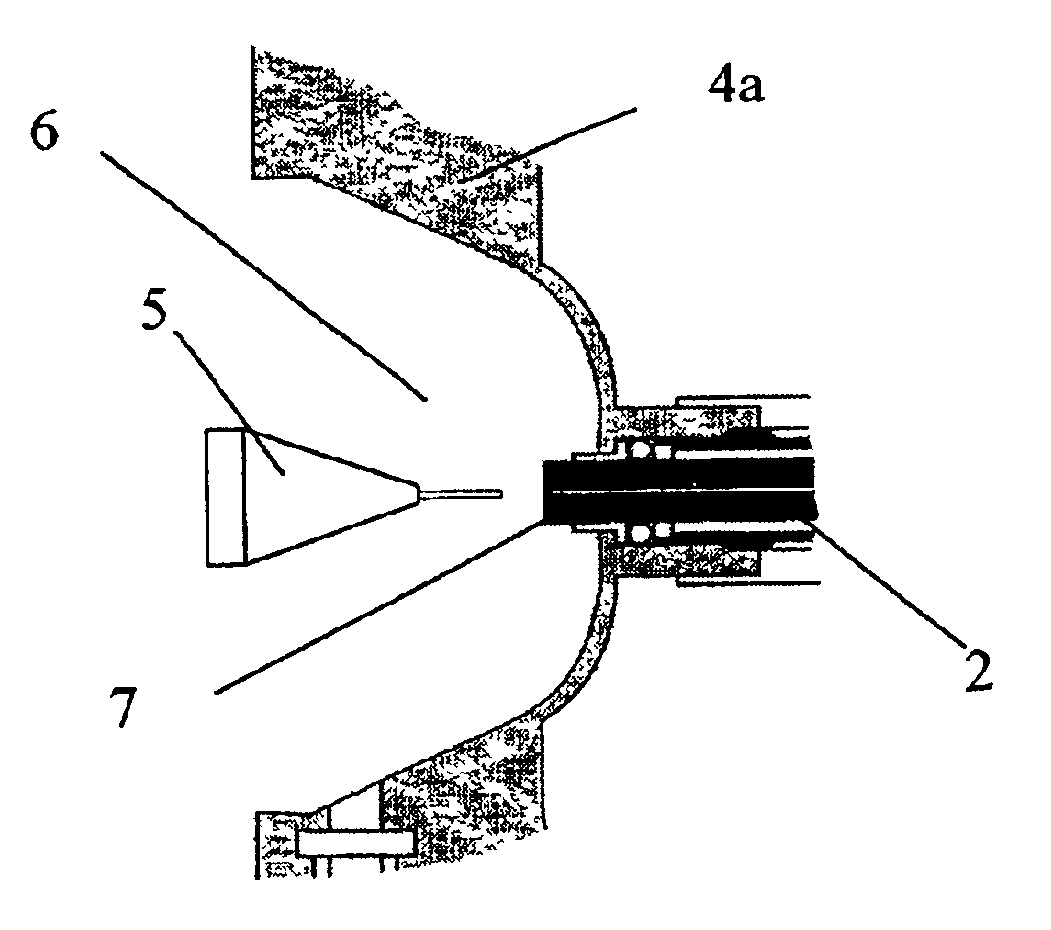
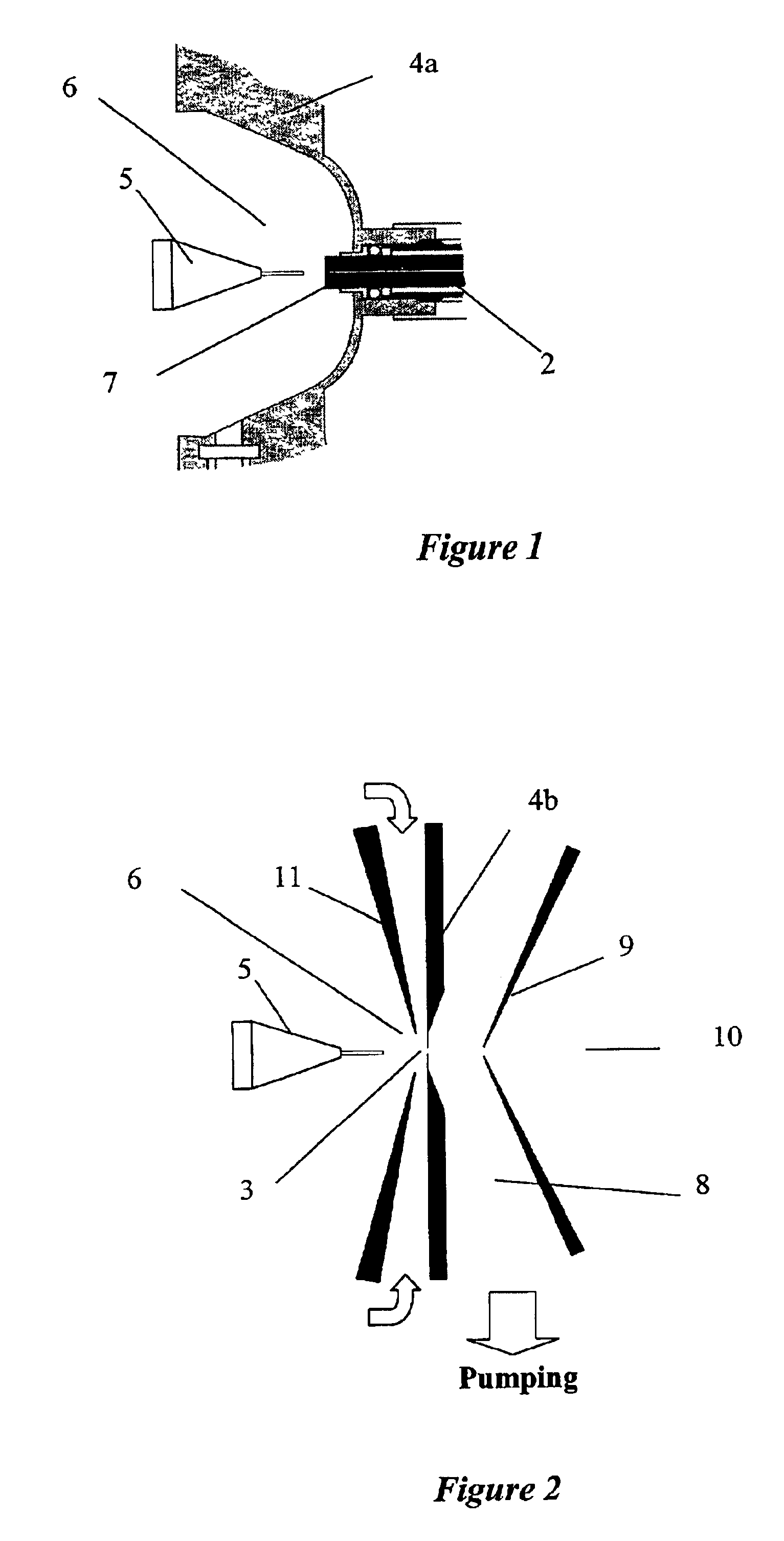
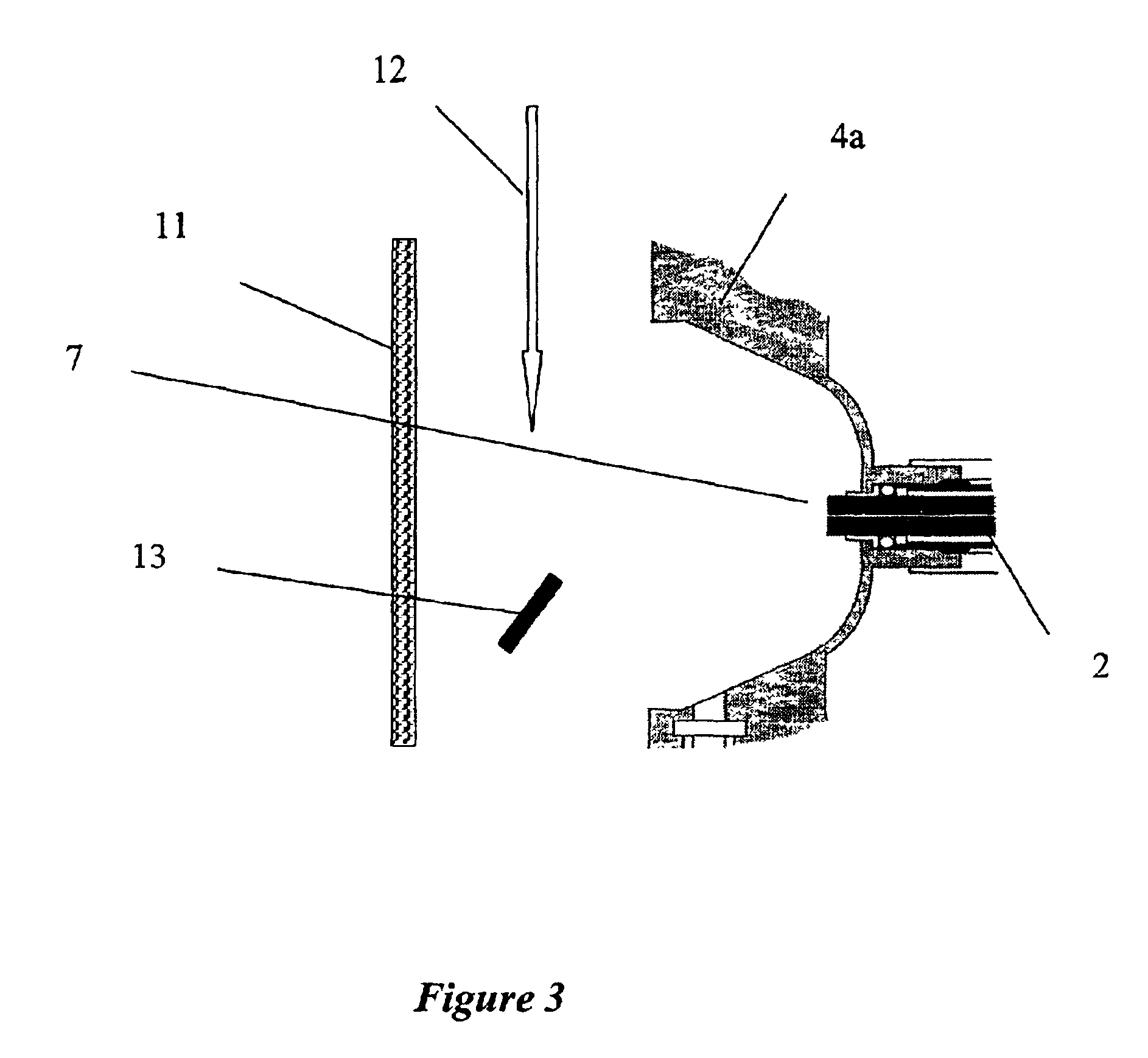
Laminated tube for the transport of charged particles contained in a gaseous medium
InactiveUS6943347B1Reduce gas loadControl flowElectron/ion optical arrangementsIsotope separationGas phaseMass Spectrometry-Mass Spectrometry
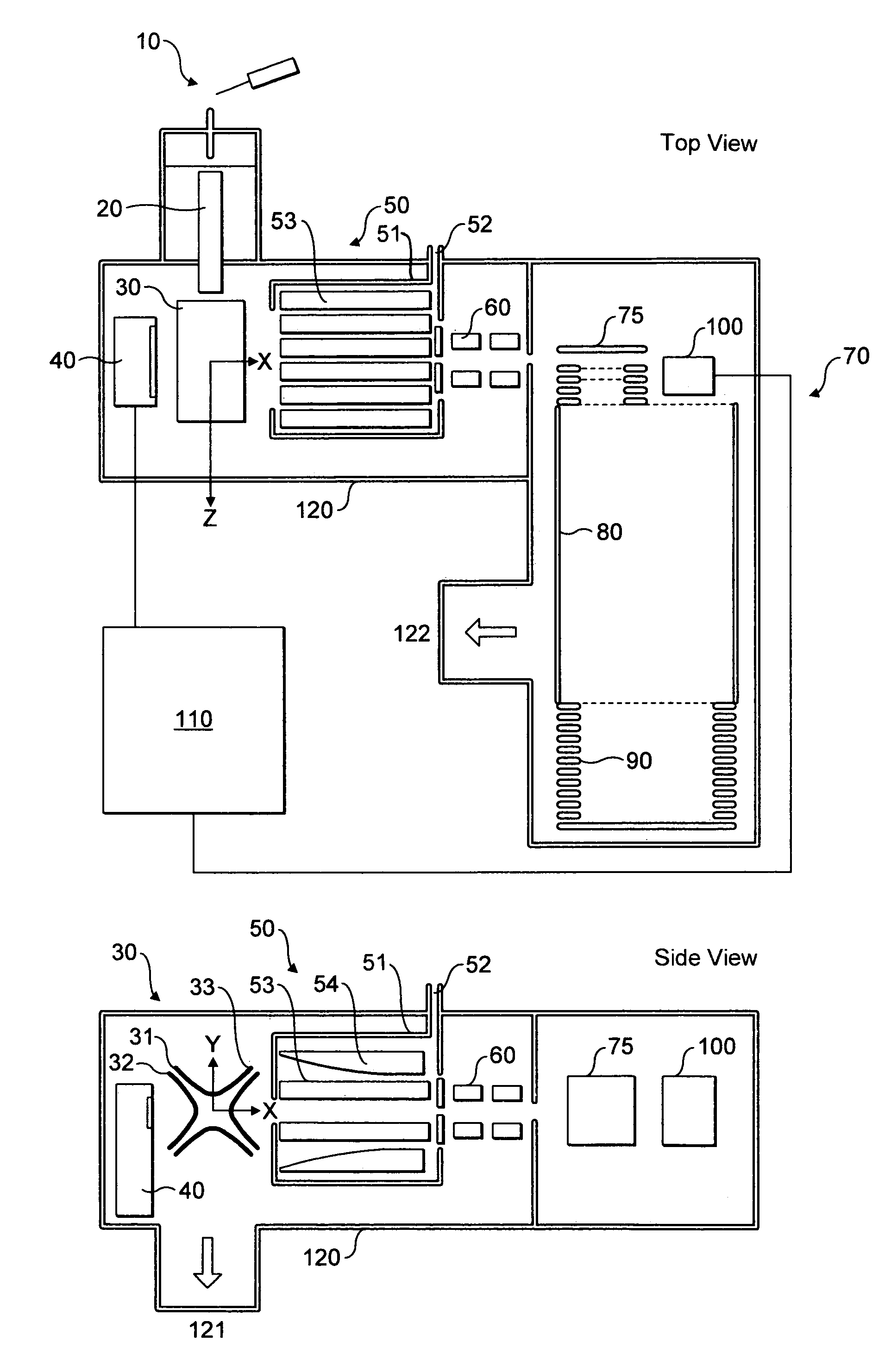
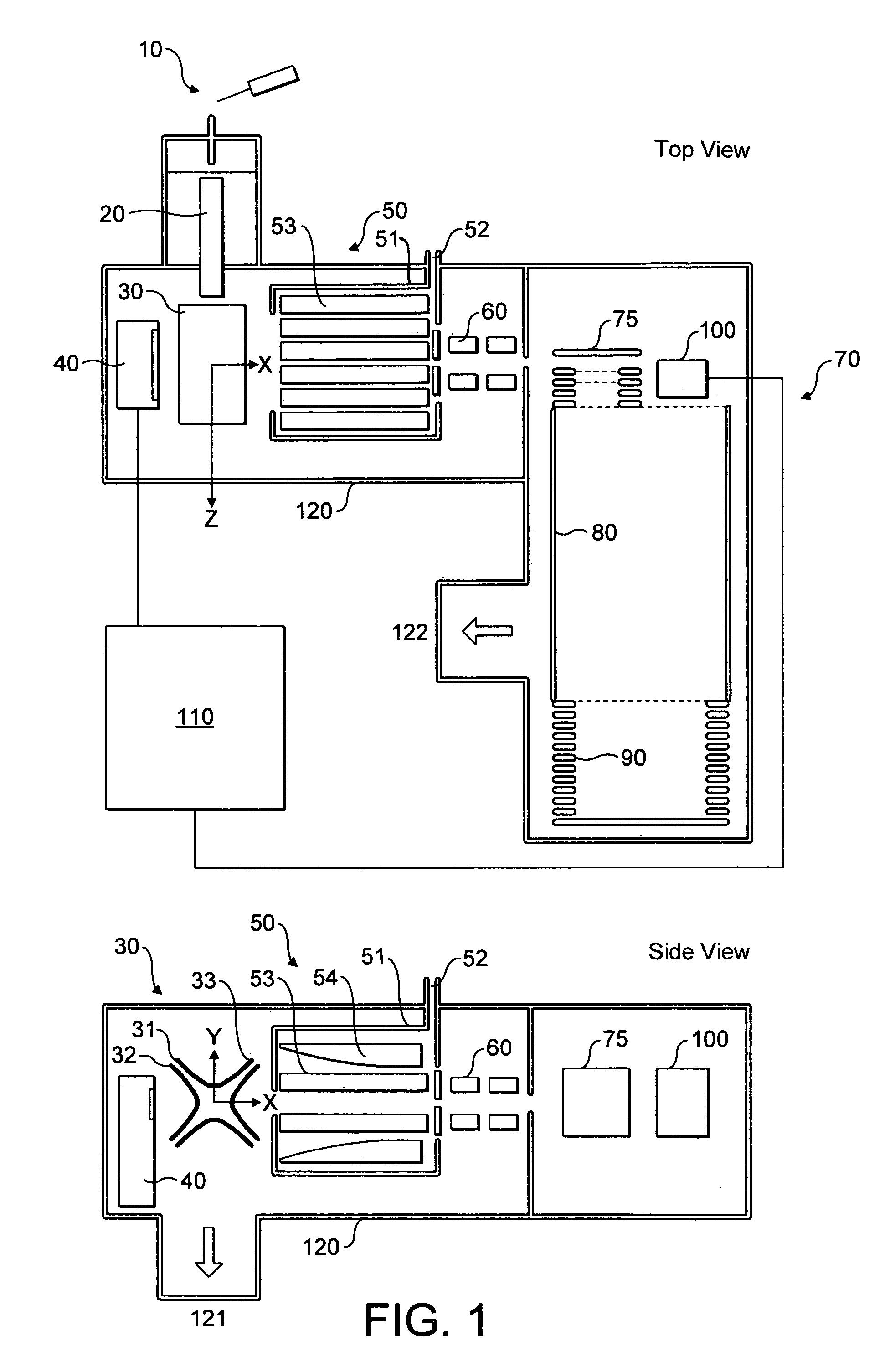
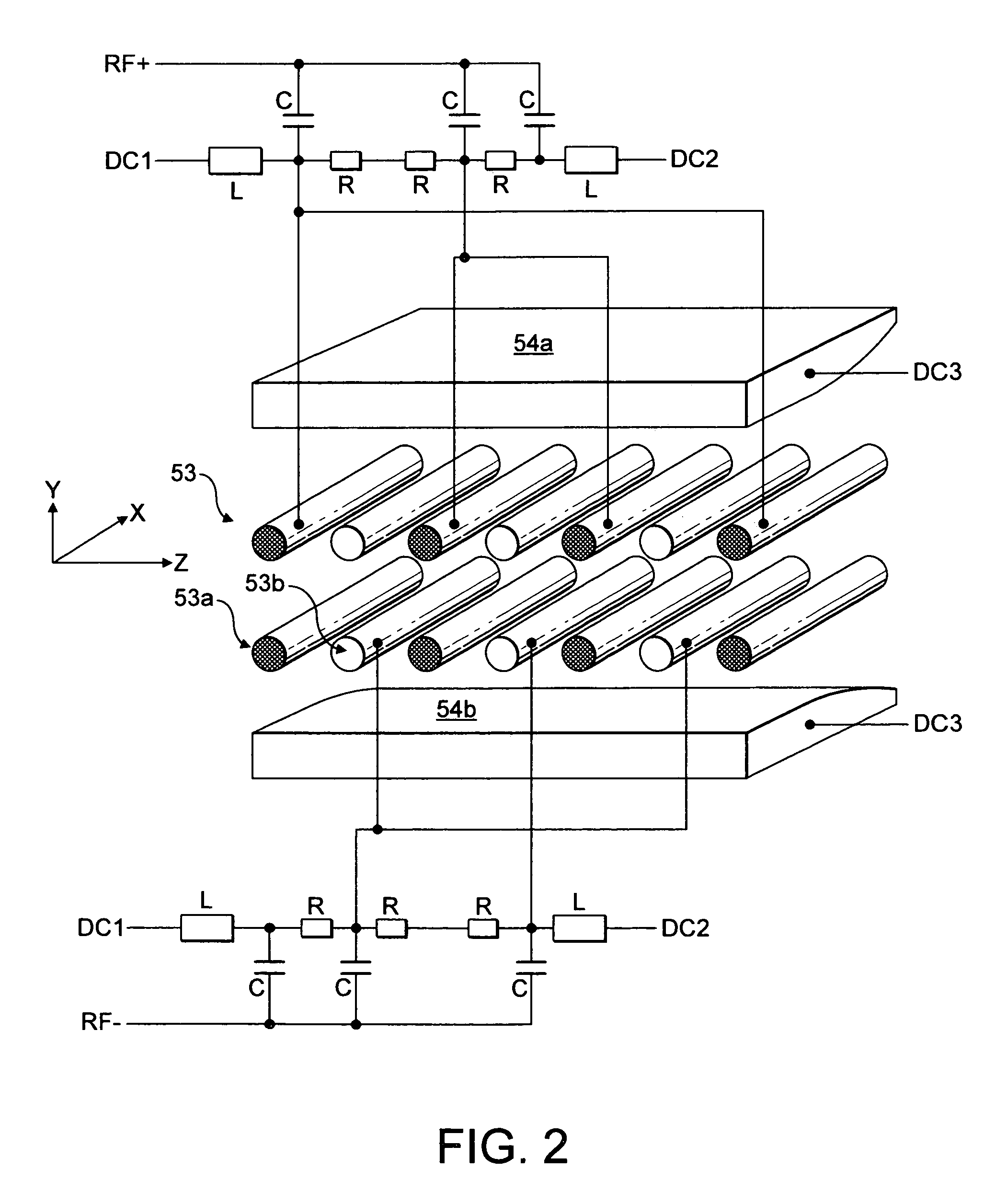
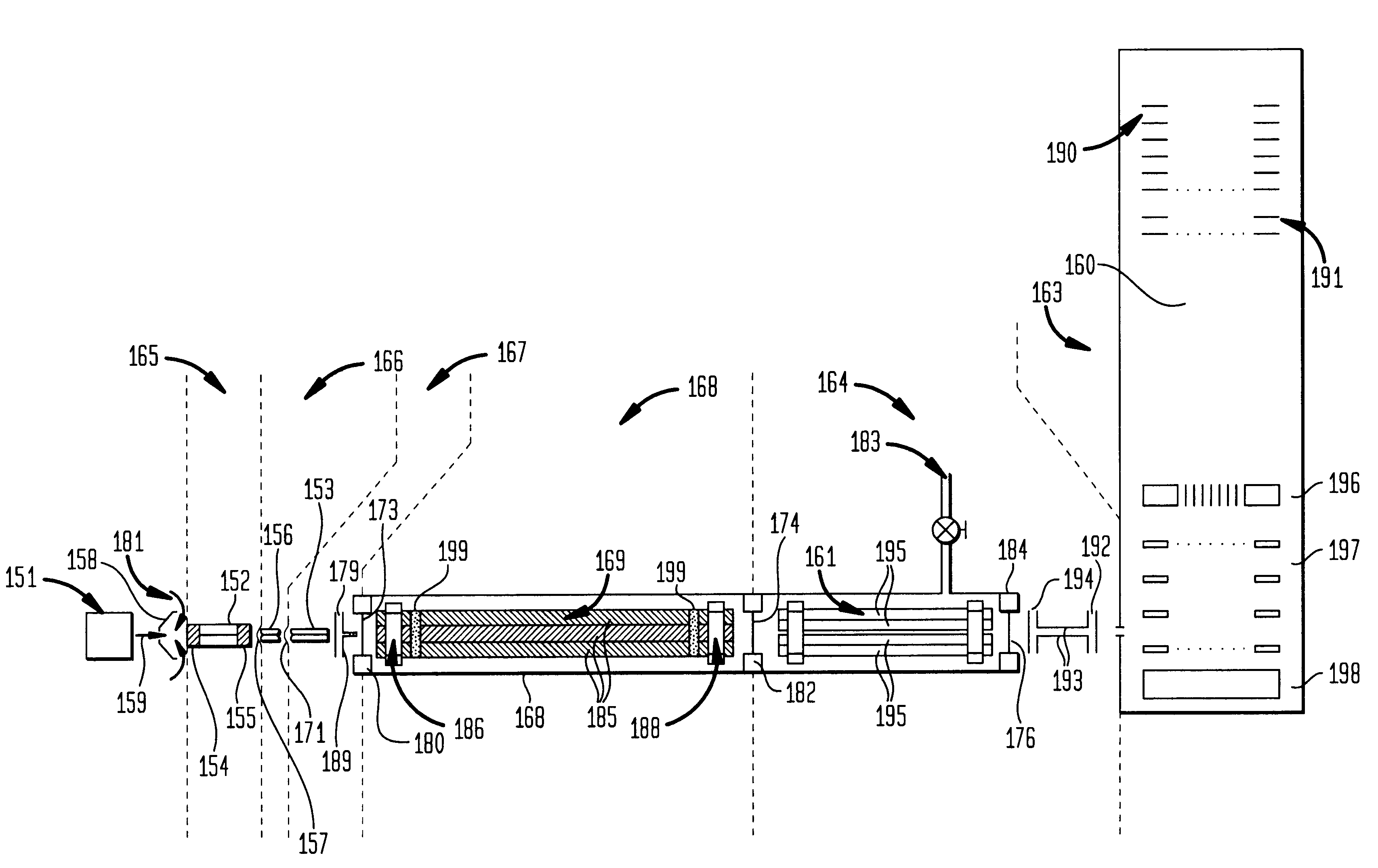
An improved tube for accepting gas-phase ions and particles contained in a gas by allowing substantially all the gas-phase ions and gas from an ion source at or greater than atmospheric pressure to flow into the tube and be transferred to a lower pressure region. Transport and motion of the ions through the tube is determined by a combination of viscous forces exerted on the ions by the flowing gas molecules and electrostatic forces causing the motion of the ions through the tube and away from the walls of the tube. More specifically, the tube is made up of stratified elements, wherein DC potentials are applied to the elements so that the DC voltage on any element determines the electric potential experience by the ions as they pass through the tube. A precise electrical gradient is maintained along the length of the stratified tube to insure the transport of the ions. Embodiments of this invention are methods and devices for improving the sensitivity of mass spectrometry or ion mobility spectrometers when coupled to atmospheric and above atmospheric pressure ionization sources. An alternate embodiment of this invention applies an AC voltage to one or more of the conducting elements in the laminate.
Owner:CHEM SPACE ASSOIATES
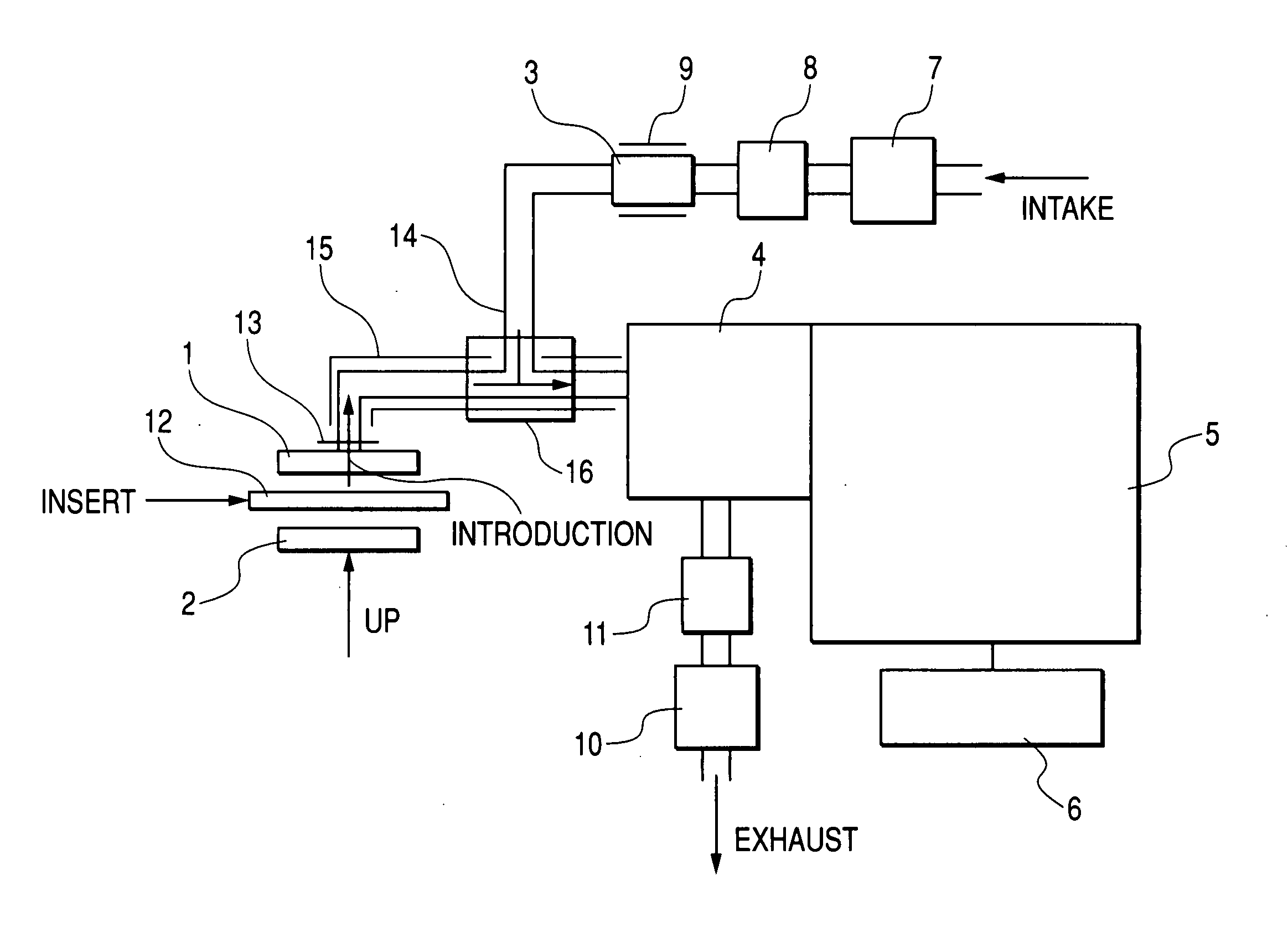
Laser ablation flowing atmospheric-pressure afterglow for ambient mass spectrometry
ActiveUS20090272893A1Time-of-flight spectrometersIsotope separationMass Spectrometry-Mass SpectrometryMass analyzer
Disclosed is an apparatus for performing mass spectrometry and a method of analyzing a sample through mass spectrometry. In particular, the disclosure relates to an apparatus capable of ambient mass spectrometry and mass spectral imaging and a method for the same. The apparatus couples laser ablation, flowing atmospheric-pressure afterglow ionization, and a mass spectrometer.
Owner:INDIANA UNIV RES & TECH CORP
Laser ablation feedback spectroscopy
InactiveUS20050061779A1Precise depth controlReduce the amount requiredWelding/soldering/cutting articlesMetal working apparatusMass spectrometryAblation plasma
Methods, for use with a laser ablation or drilling process, which achieve depth-controlled removal of composite-layered work-piece material by real-time feedback of ablation plasma spectral features. The methods employ the use of electric, magnetic or combined fields in the region of the laser ablation plume to direct the ablated material. Specifically, the electric, magnetic or combined fields cause the ablated material to be widely dispersed, concentrated in a target region, or accelerated along a selected axis for optical or physical sampling, analysis and laser feedback control. The methods may be used with any laser drilling, welding or marking process and are particularly applicable to laser micro-machining. The described methods may be effectively used with ferrous and non-ferrous metals and non-metallic work-pieces. The two primary benefits of these methods are the ability to drill or ablate to a controlled depth, and to provide controlled removal of ablation debris from the ablation site. An ancillary benefit of the described methods is that they facilitate ablated materials analysis and characterization by optical and / or mass spectroscopy.
Owner:BLUMENFELD WALTER +2
Chemical detection method and system
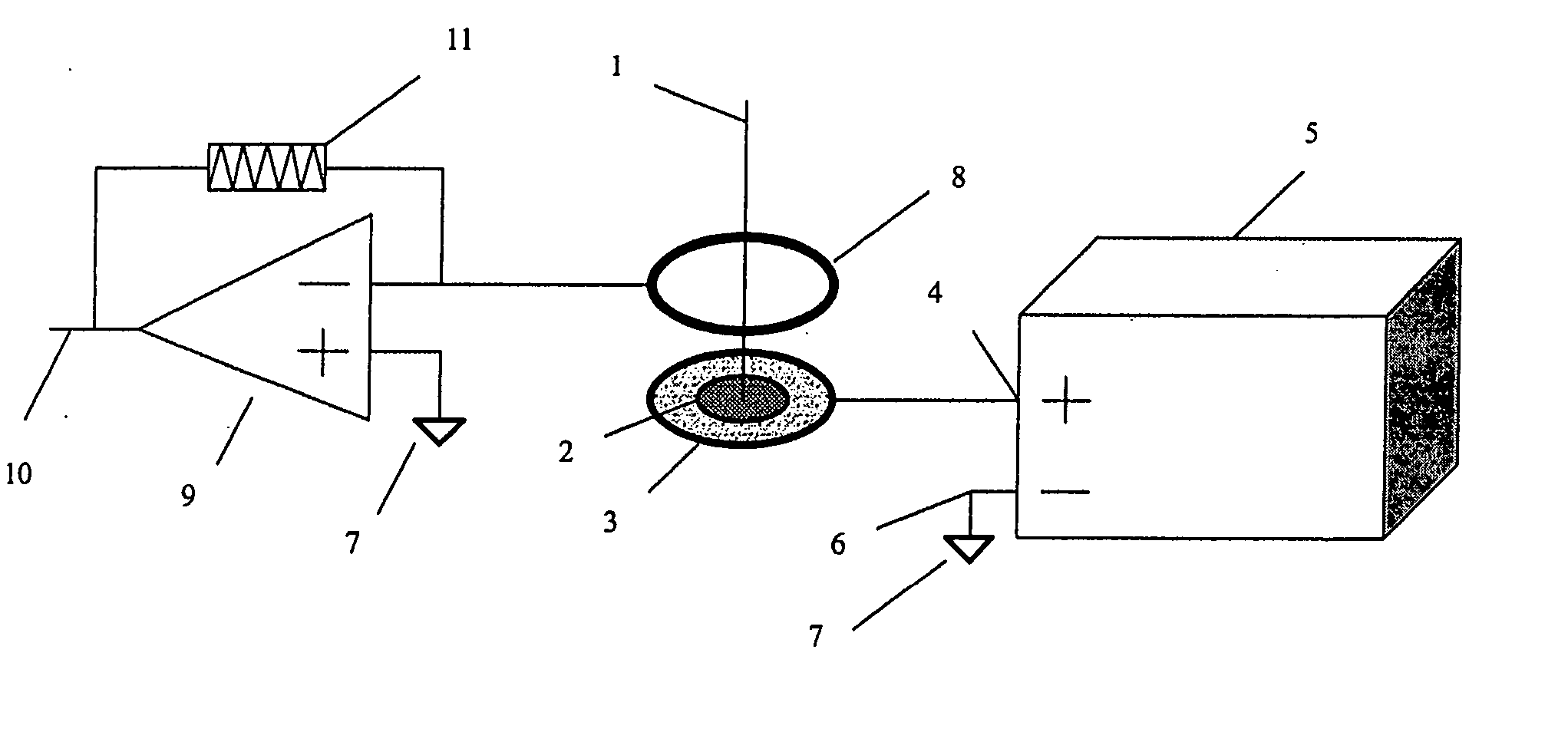
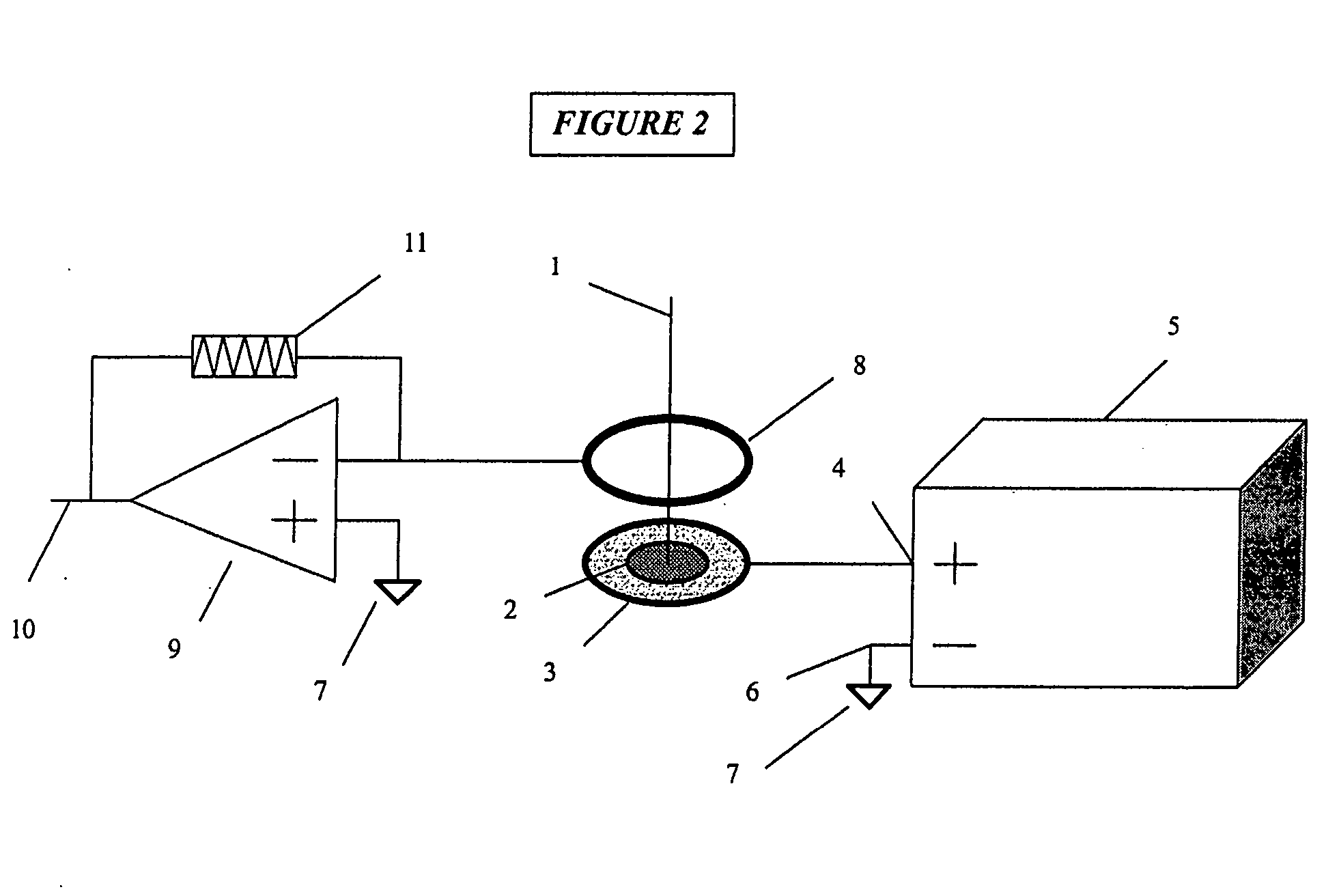
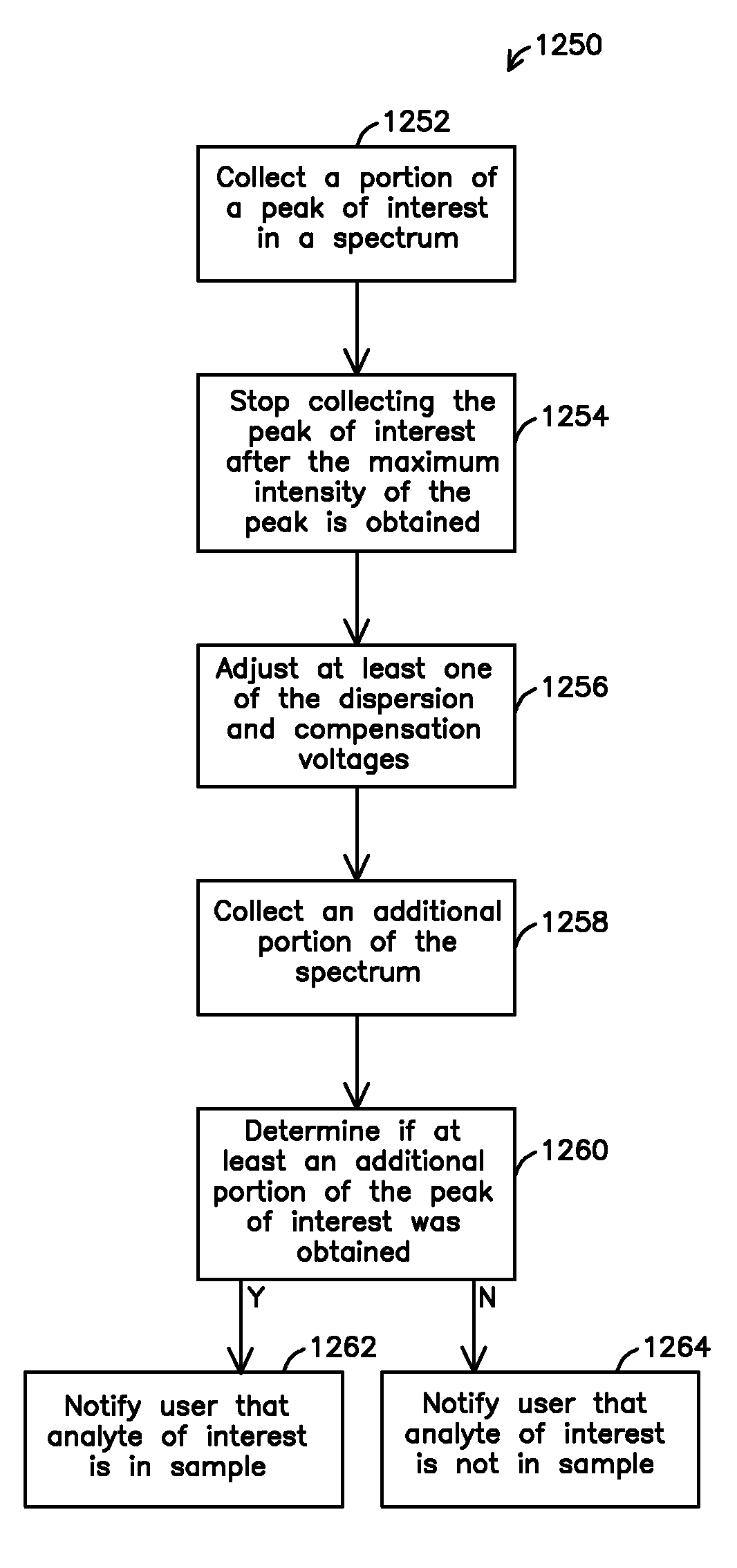
InactiveUS8067731B2Material analysis by electric/magnetic meansIsotope separationAnalyteIon-mobility spectrometry
A system and method for detecting an analyte of interest in a sample is provided. The method includes passing a set of ions obtained from the sample through an ion mobility spectrometer to filter out ions that are not ions of interest and to generate an ion mobility spectrum. A mass spectrum of at least some of the ions is generated using a mass spectrometer. The method also includes determining that the analyte of interest is in the sample when peaks of interest are found in one or more of the ion mobility spectrum and the mass spectrum, and the peaks of interest follow a predetermined pattern of peaks associated with the analyte of interest or are confirmed by ion mobility spectrometry.
Owner:TELEDYNE DETCON INC
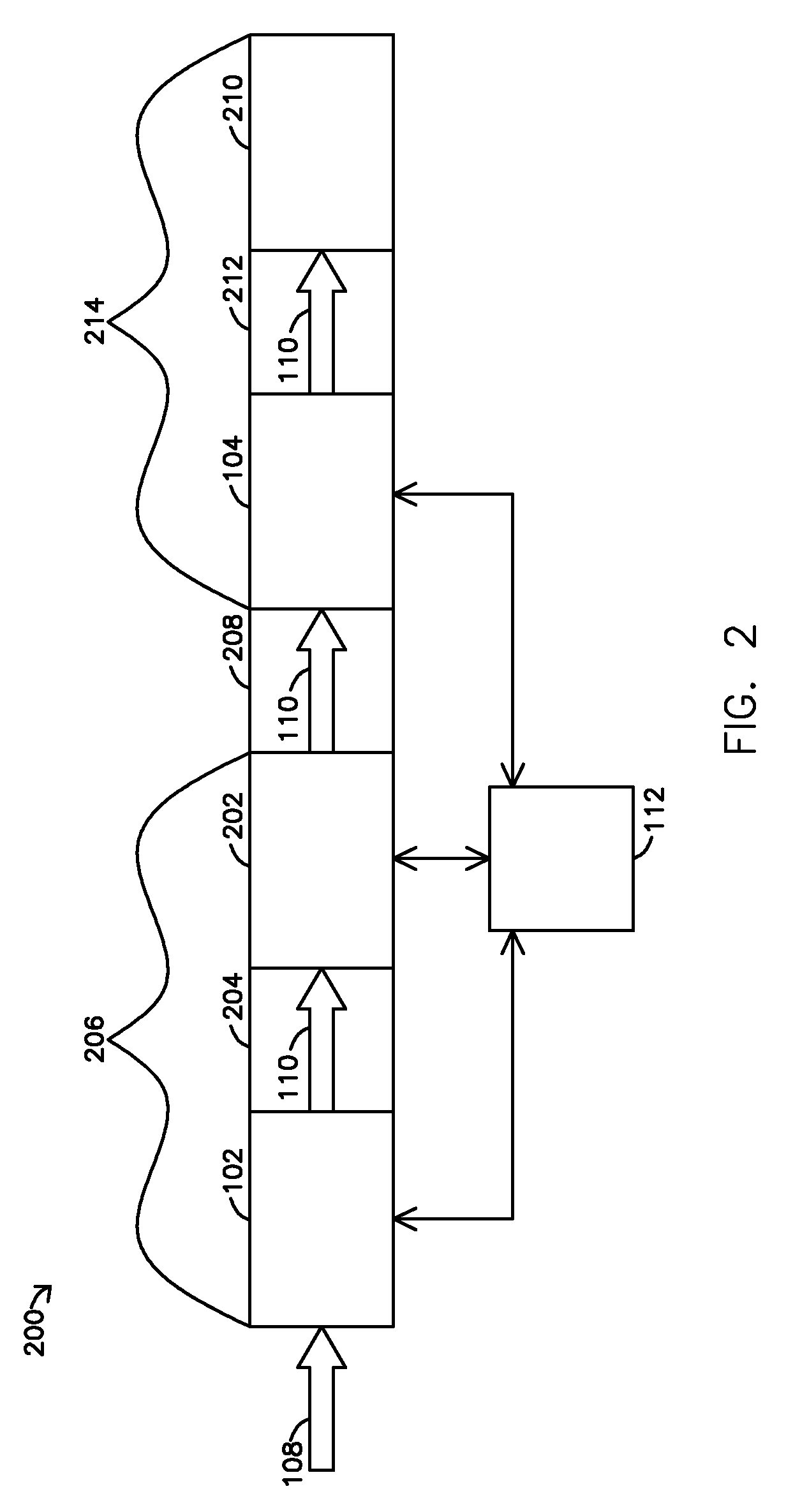
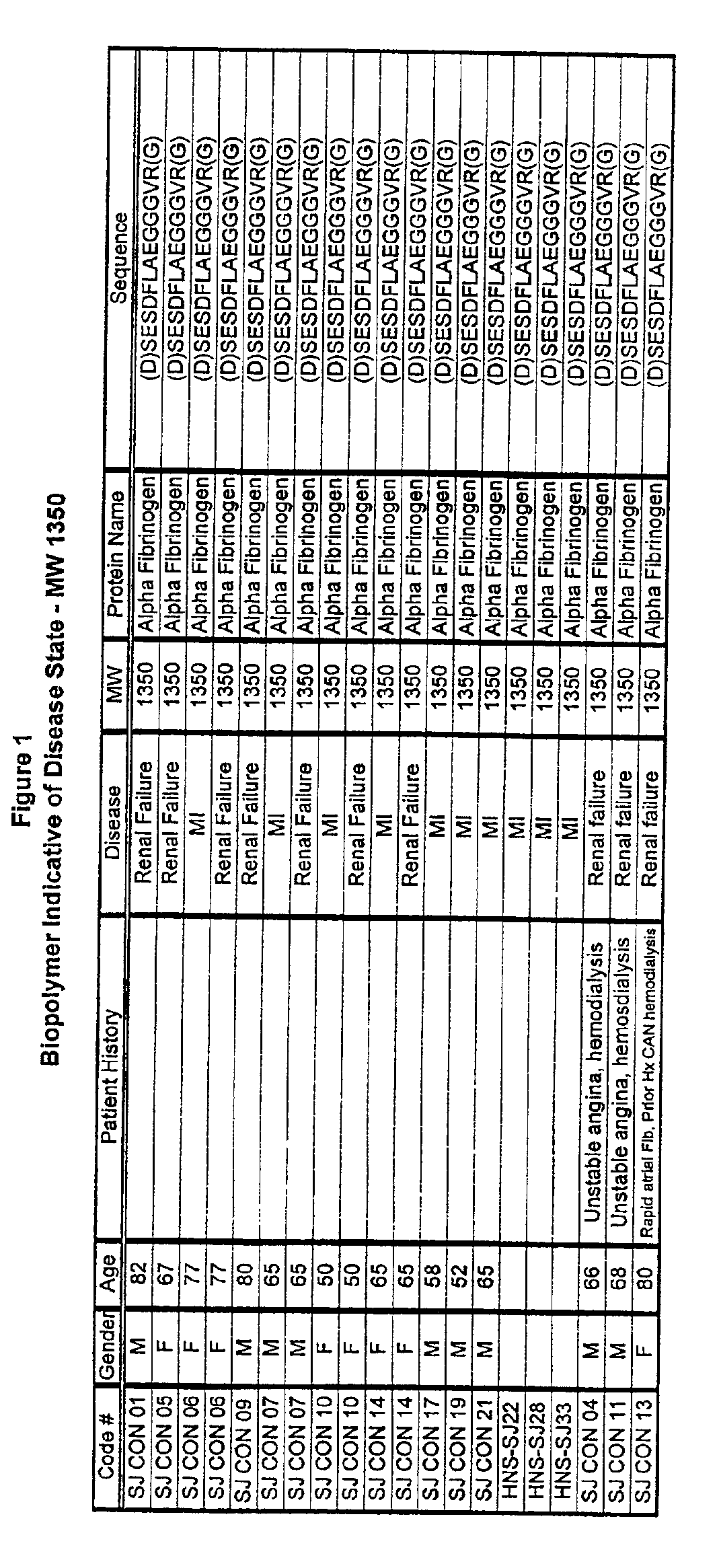
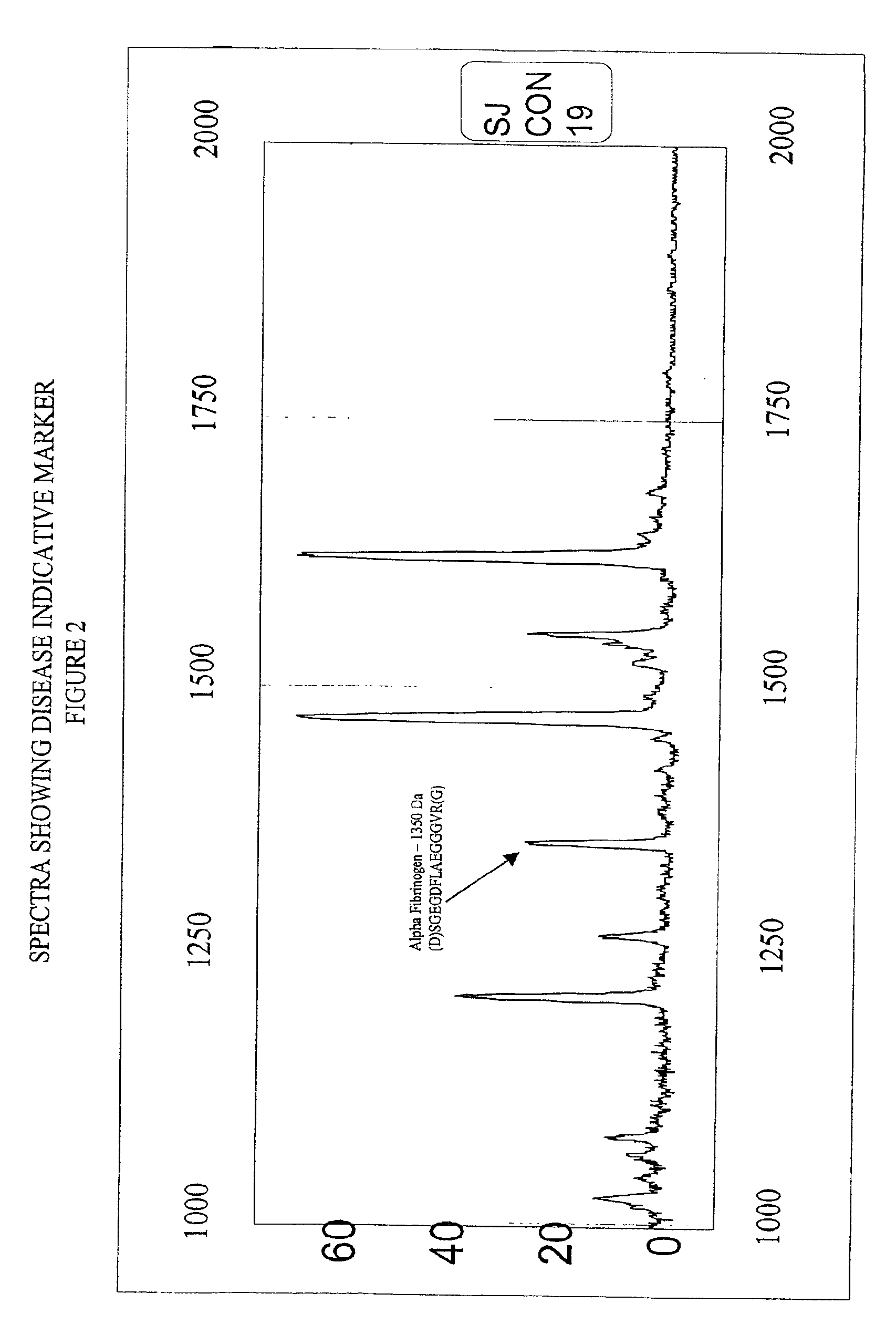
Biopolymer marker indicative of disease state having a molecular weight of 1350 daltons
InactiveUS6890763B2Maximize diversityThwart these maladiesAnalysis using chemical indicatorsPeptide/protein ingredientsDiseaseBiopolymer
The instant invention involves the use of a combination of preparatory steps in conjunction with mass spectroscopy and time-of-flight detection procedures to maximize the diversity of biopolymers which are verifiable within a particular sample. The cohort of biopolymers verified within such a sample is then viewed with reference to their ability to evidence at least particular disease state; thereby enabling a diagnostician to gain the ability to characterize either the presence or absence of at least one disease state relative to recognition of the presence and / or the absence of the biopolymer.
Owner:NANOGEN INC
Remote reagent chemical ionization source
InactiveUS7095019B1Facilitate efficient sample ionizationEasy to collectTime-of-flight spectrometersIsotope separationChromatographic separationGas phase
An improved ion source and portable analyzer for collecting and focusing dispersed gas-phase ions from a reagent source at atmospheric or intermediate pressure, having a remote source of reagent ions generated by direct or alternating currents, separated from a low-field sample ionization region by a stratified array of elements, each element populated with a plurality of openings, wherein DC potentials are applied to each element necessary for transferring reagent ions from the remote source into the low-field sample ionization region where the reagent ions react with neutral and / or ionic sample forming ionic species. The resulting ionic species are then introduced into the vacuum system of a mass spectrometer or ion mobility spectrometer. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when gas and liquid chromatographic separation techniques or probes containing samples are coupled to atmospheric and intermediate pressure photo-ionization, chemical ionization, and thermospray ionization sources.
Owner:CHEM SPACE ASSOCS
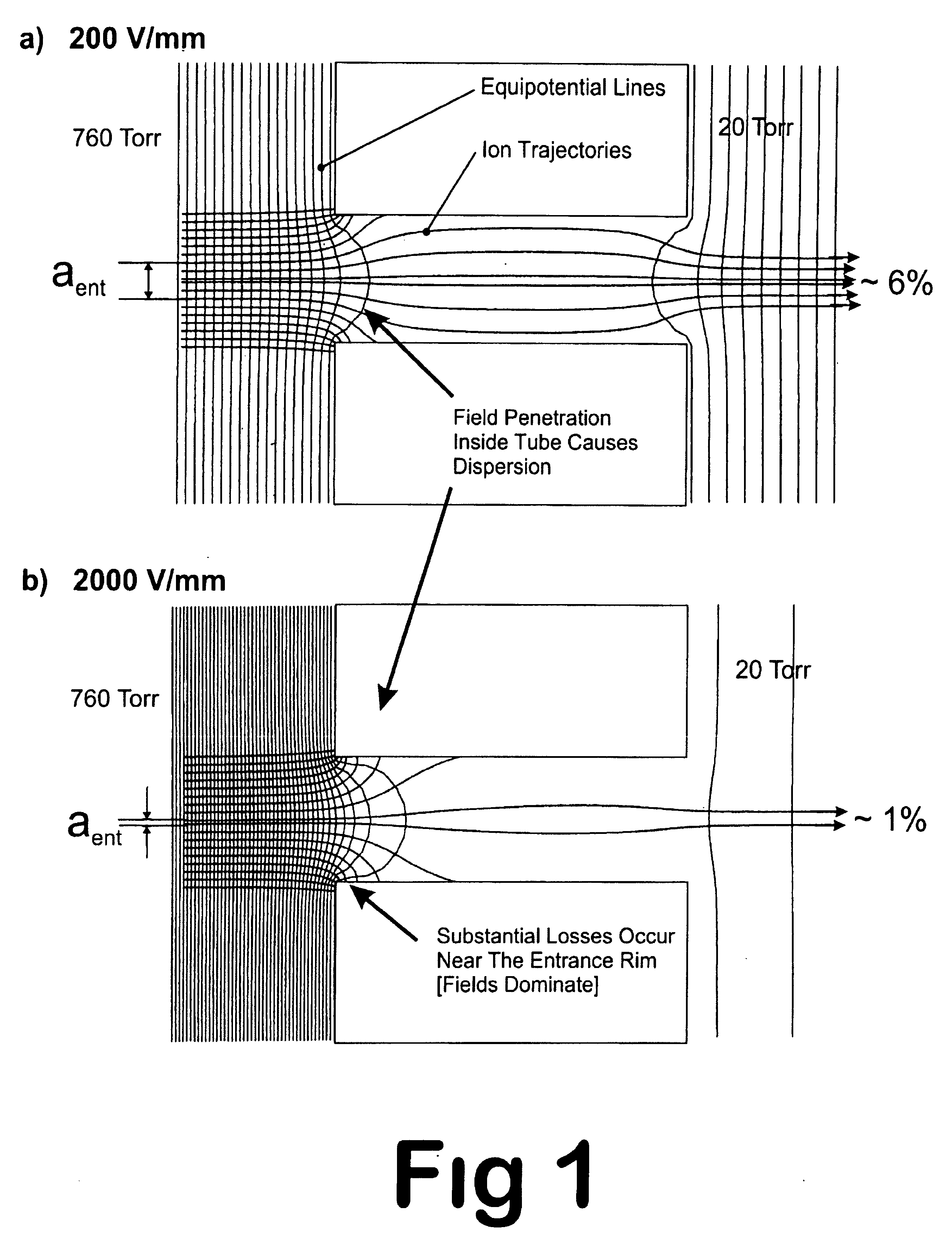
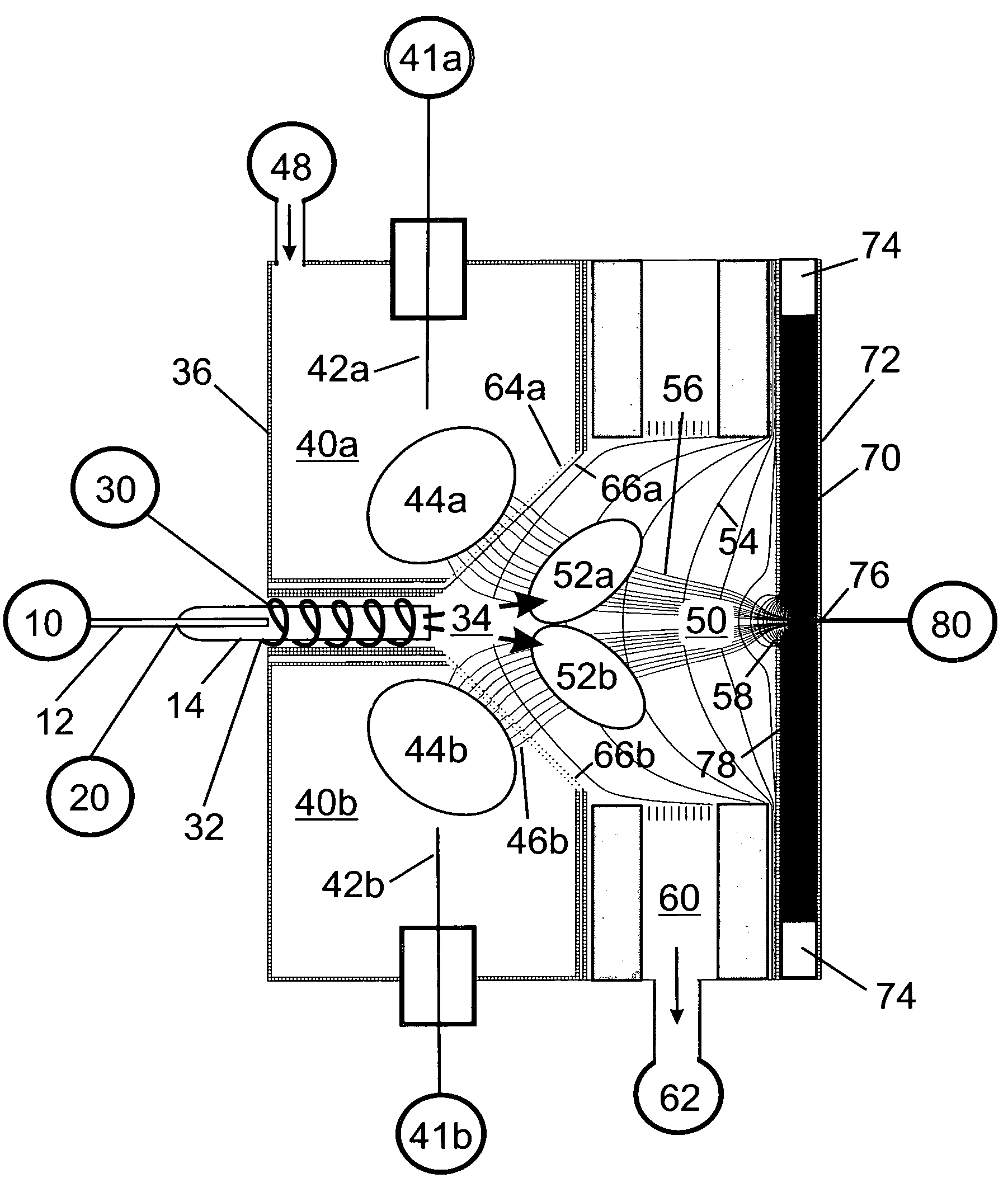
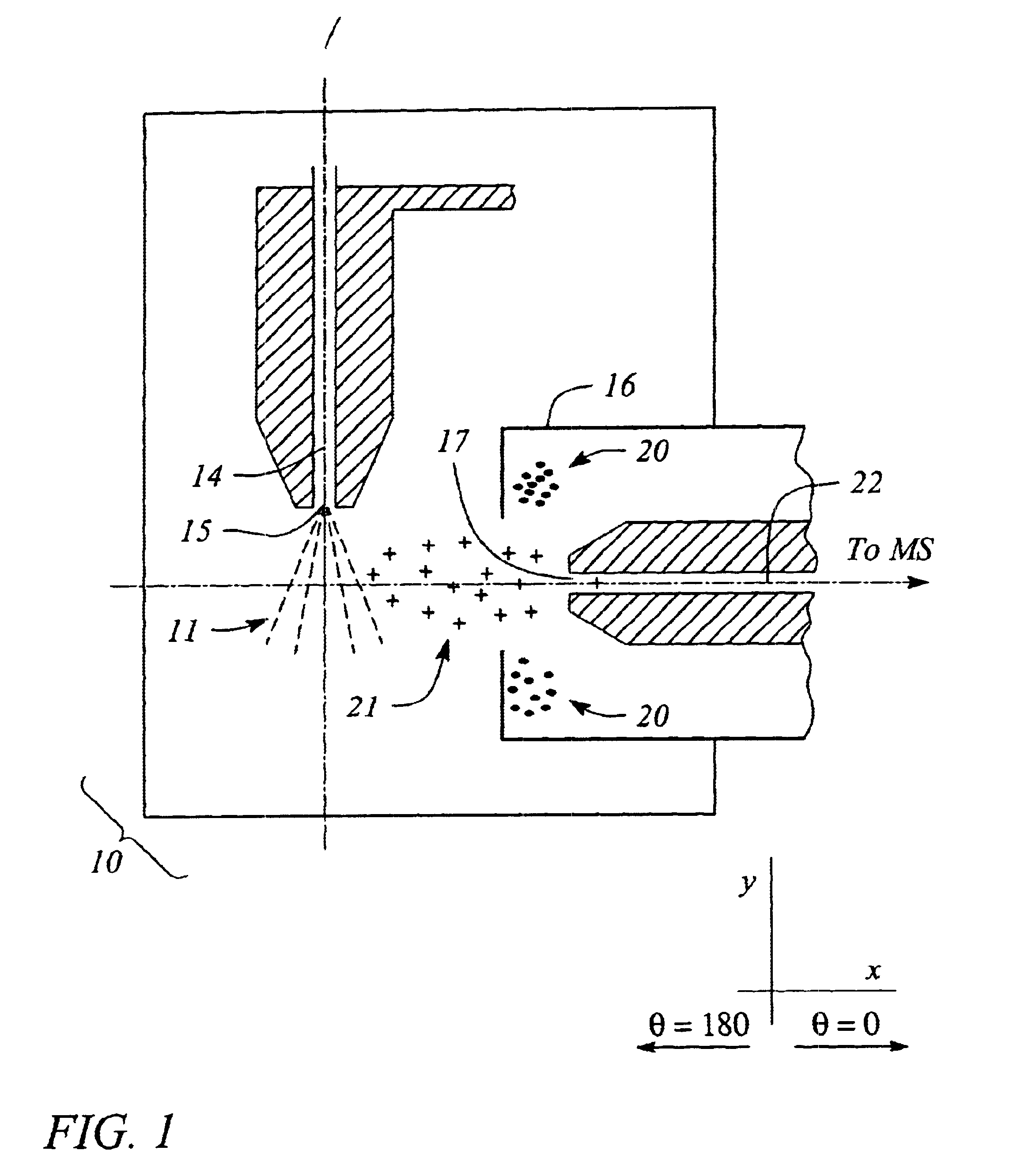
Apparatus and method for focusing ions and charged particles at atmospheric pressure
InactiveUS6744041B2Samples introduction/extractionIsotope separationInductively coupled plasmaElectrospray
Improvements have been made for collection and focusing of ions generated from atmospheric pressure sources such as electrospray, atmospheric pressure chemical ionization, inductively coupled plasma, discharge, photoionization and atmospheric pressure matrix assisted laser desorption ionization. A high transmission electro-optical surface is placed between the source regions and the focusing regions to optimize the field geometries and strengths in each respective region. Compression ratios of greater than 5000 are capable of transferring virtually all ions from large volume dispersive ion regions into ion beam cross-sections of less than 1 mm. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when coupled to atmospheric pressure ionization sources.
Owner:SHEEHAN EDWARD W +1
Remote reagent chemical ionization source
InactiveUS6888132B1Time-of-flight spectrometersIsotope separationChromatographic separationGas phase
An improved ion source for collecting and focusing dispersed gas-phase ions from a reagent source at atmospheric or intermediate pressure, having a remote source of reagent ions separated from a low-field sample ionization region by a stratified array of elements, each element populated with a plurality of openings, wherein DC potentials are applied to each element necessary for transferring reagent ions from the remote source into the low-field sample ionization region where the reagent ions react with neutral and / or ionic sample forming ionic species. The resulting ionic species are then introduced into the vacuum system of a mass spectrometer or ion mobility spectrometer. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when gas and liquid chromatographic separation techniques are coupled to atmospheric and intermediate pressure photo-ionization, chemical ionization, and thermospray ionization sources.
Owner:CHEM SPACE ASSOIATES
Apparatus and method for analyzing samples in a dual ion trap mass spectrometer
InactiveUS6627883B2Stability-of-path spectrometersIsotope separationIon trap mass spectrometryMemory effect
The present invention is an improved apparatus and method for mass spectrometry using a dual ion trapping system. In a preferred embodiment of the present invention, three "linear" multipoles are combined to create a dual linear ion trap system for trapping, analyzing, fragmenting and transmitting parent and fragment ions to a mass analyzer-preferably a TOF mass analyzer. The dual ion trap according to the present invention includes two linear ion traps, one positioned before an analytic quadrupole and one after the analytic multipole. Both linear ion traps are multipoles composed of any desired number of rods-i.e. the traps are quadrupoles, pentapoles, hexapoles, octapoles, etc. Such arrangement enables one to maintain a high "duty cycle" while avoiding "memory effects" and also reduces the power consumed in operating the analyzing quadrupole.
Owner:BRUKER SCI LLC
Laminated lens for introducing gas-phase ions into the vacuum systems of mass spectrometers
InactiveUS6949740B1Time-of-flight spectrometersElectron/ion optical arrangementsGas phaseMass Spectrometry-Mass Spectrometry
An improved lens for collecting and focusing dispersed charged particles or ions having a stratified array of elements at atmospheric or near-atmospheric pressure, each element having successively smaller apertures forming a tapered terminus, wherein the electrostatic DC potentials are applied to each element necessary for focusing ions through the stratified array for introducing charged particles and ions into the vacuum system of a mass spectrometer. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when coupled to both high and low electrostatic field atmospheric pressure ionization sources.
Owner:CHEM SPACE ASSOIATES
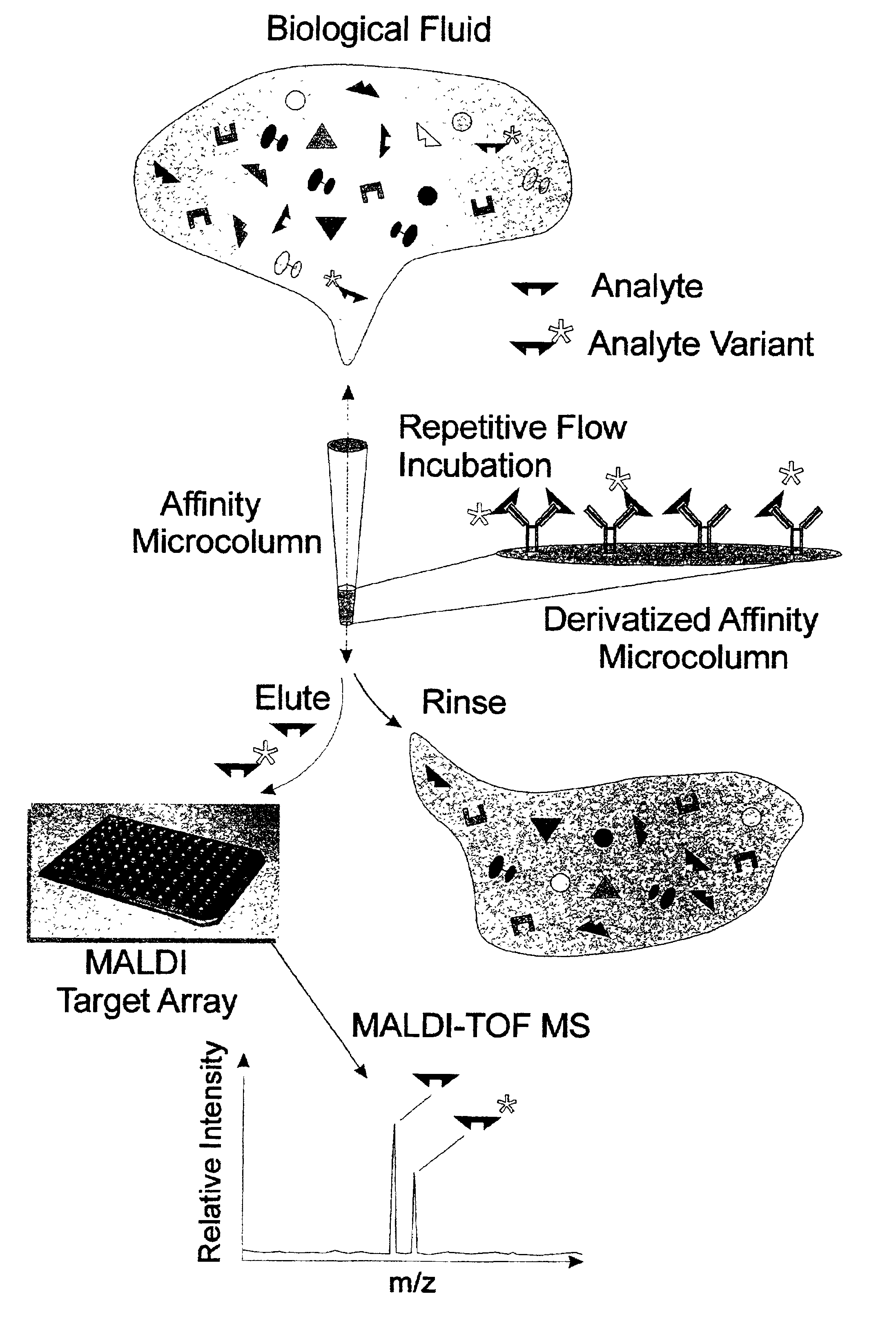
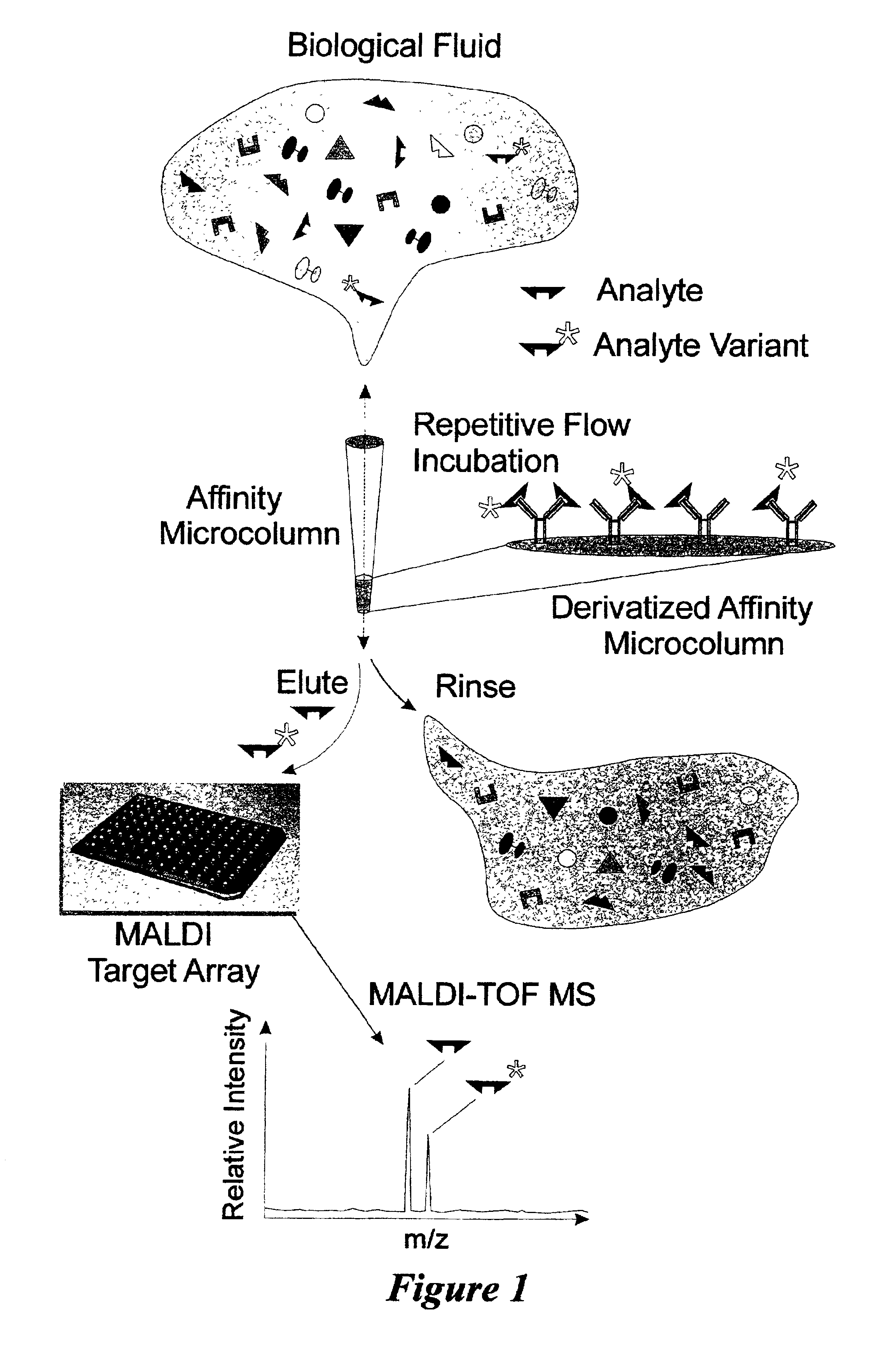
Integrated high throughput system for the mass spectrometry of biomolecules
InactiveUS6783672B2Bioreactor/fermenter combinationsMaterial nanotechnologyDead volumeMass Spectrometry-Mass Spectrometry
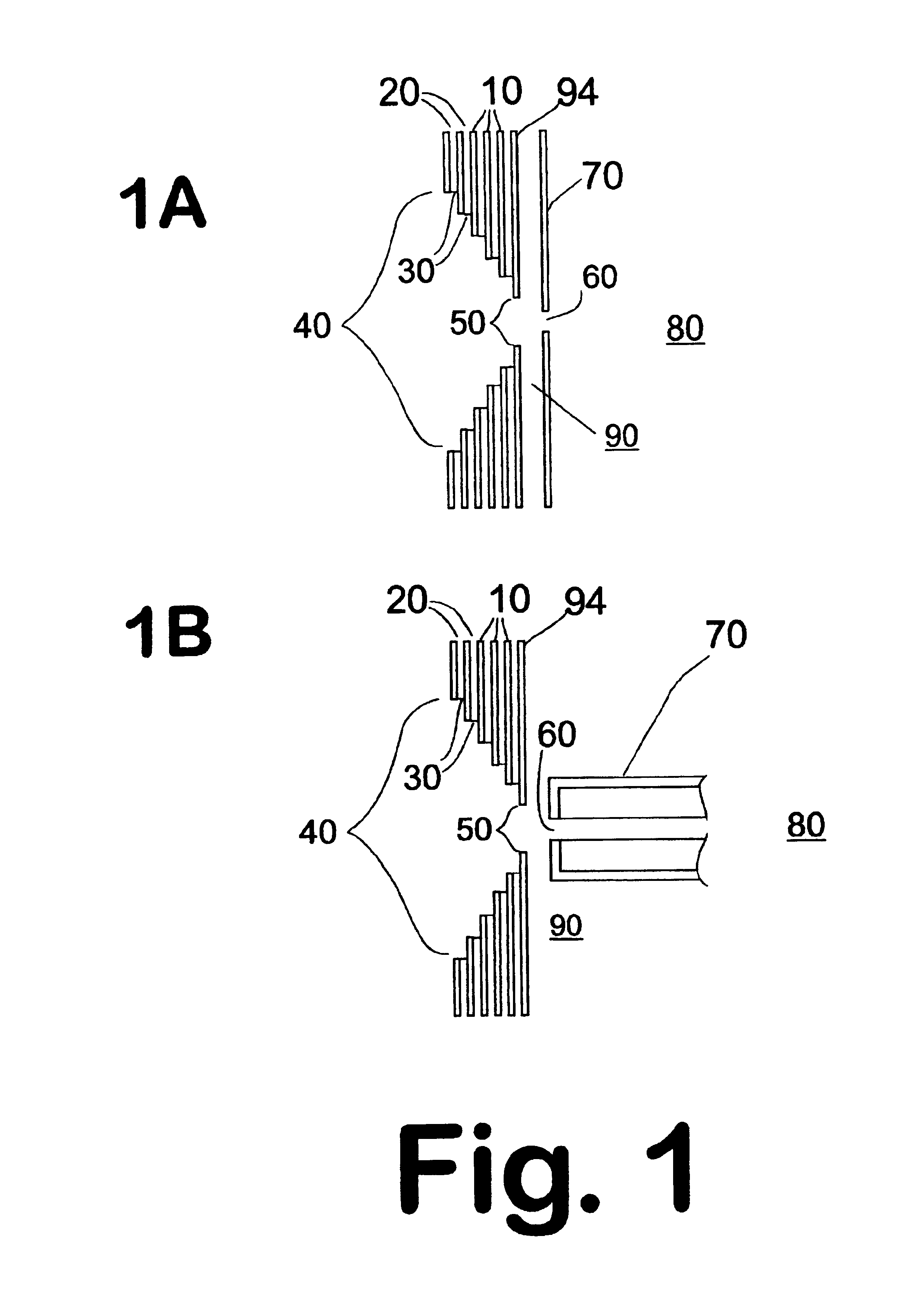
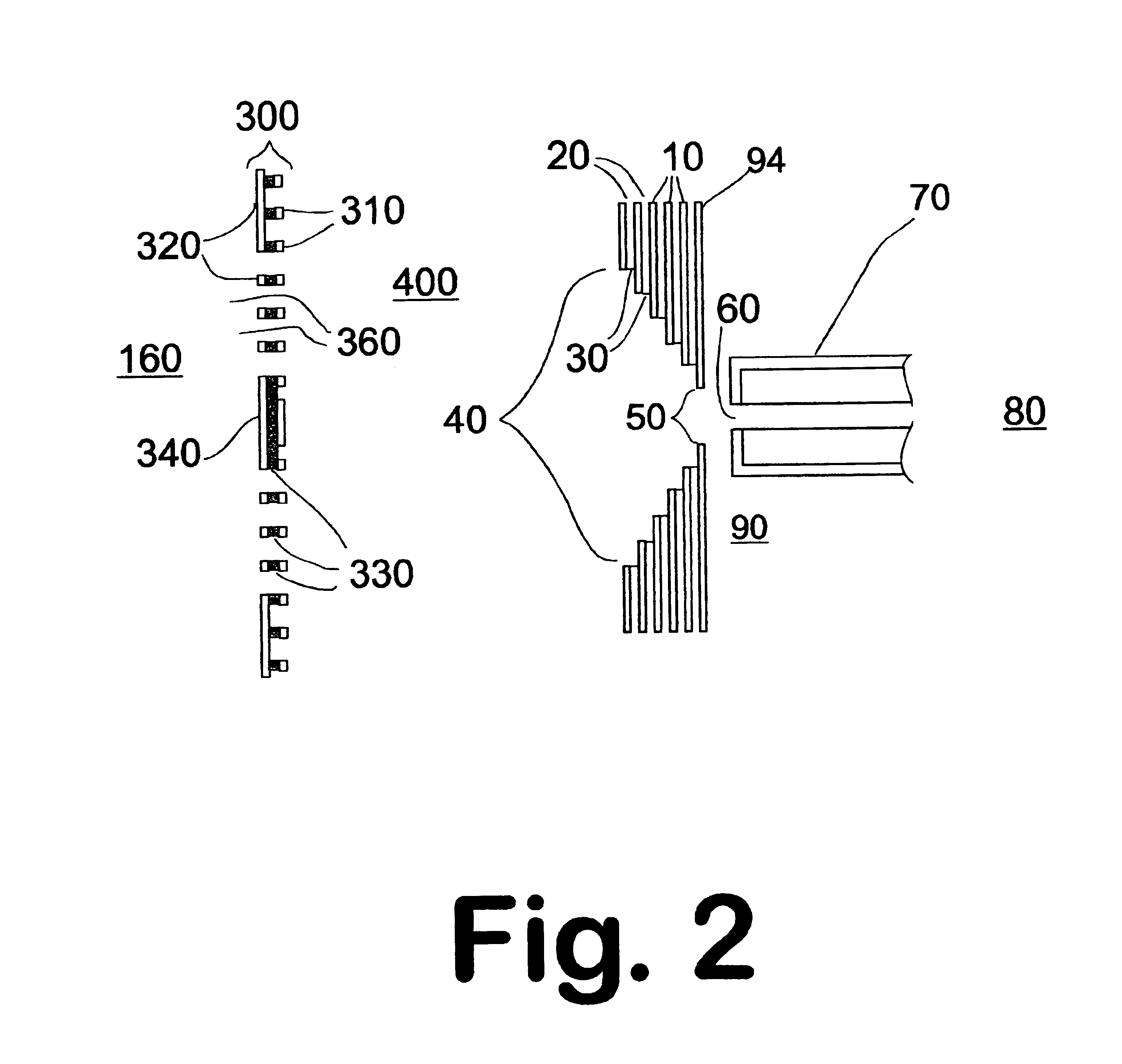
Described is an affinity microcolumn comprising a high surface area material, which has high flow properties and a low dead volume, contained within a housing and having affinity reagents bound to the surface of the high surface area material that are either activated or activatable. The affinity reagents bound to the surface of the affinity microcolumn further comprise affinity receptors for the integration into high throughput analysis of biomolecules.
Owner:INTRINSIC BIOPROBES
Protein Microscope
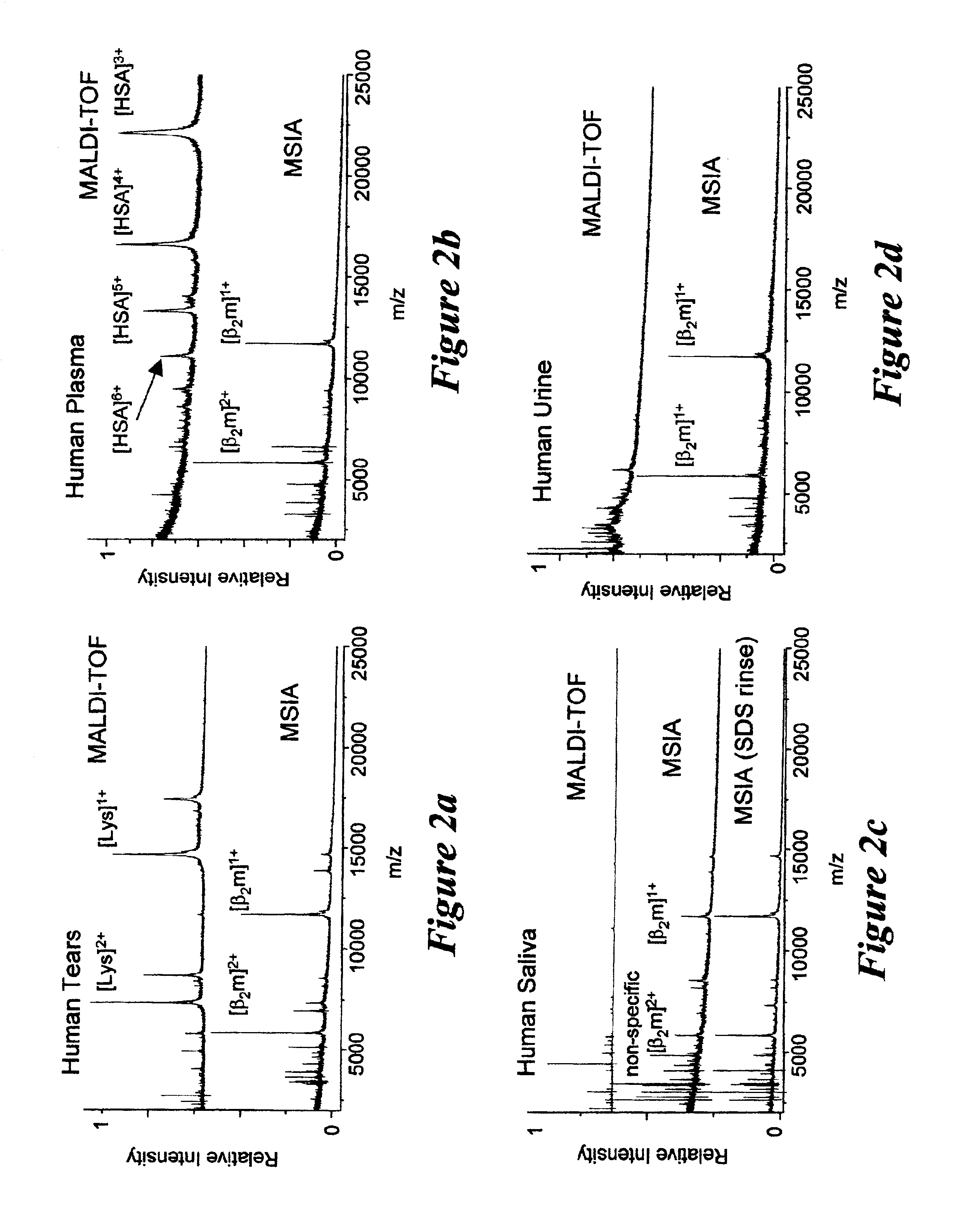
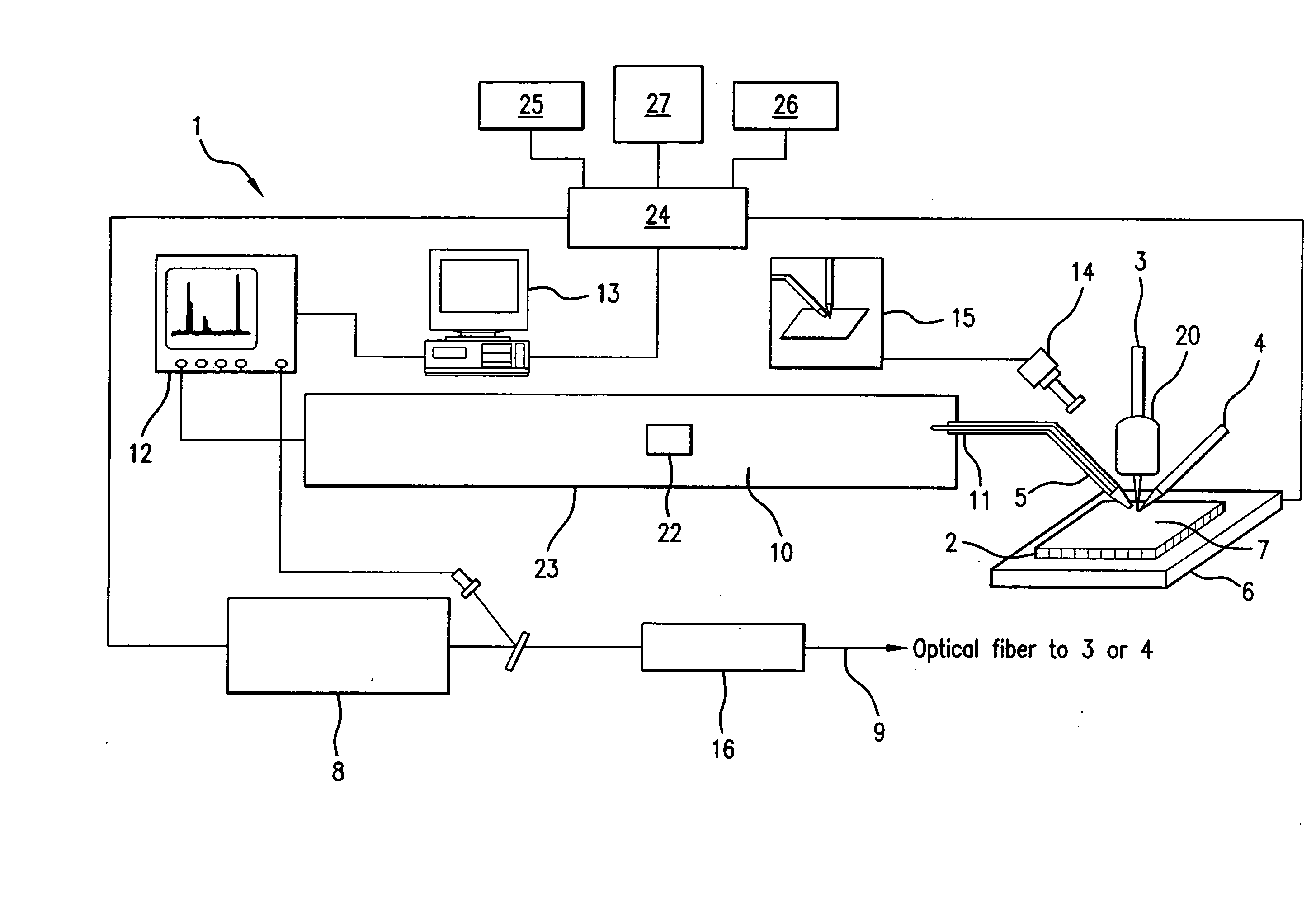
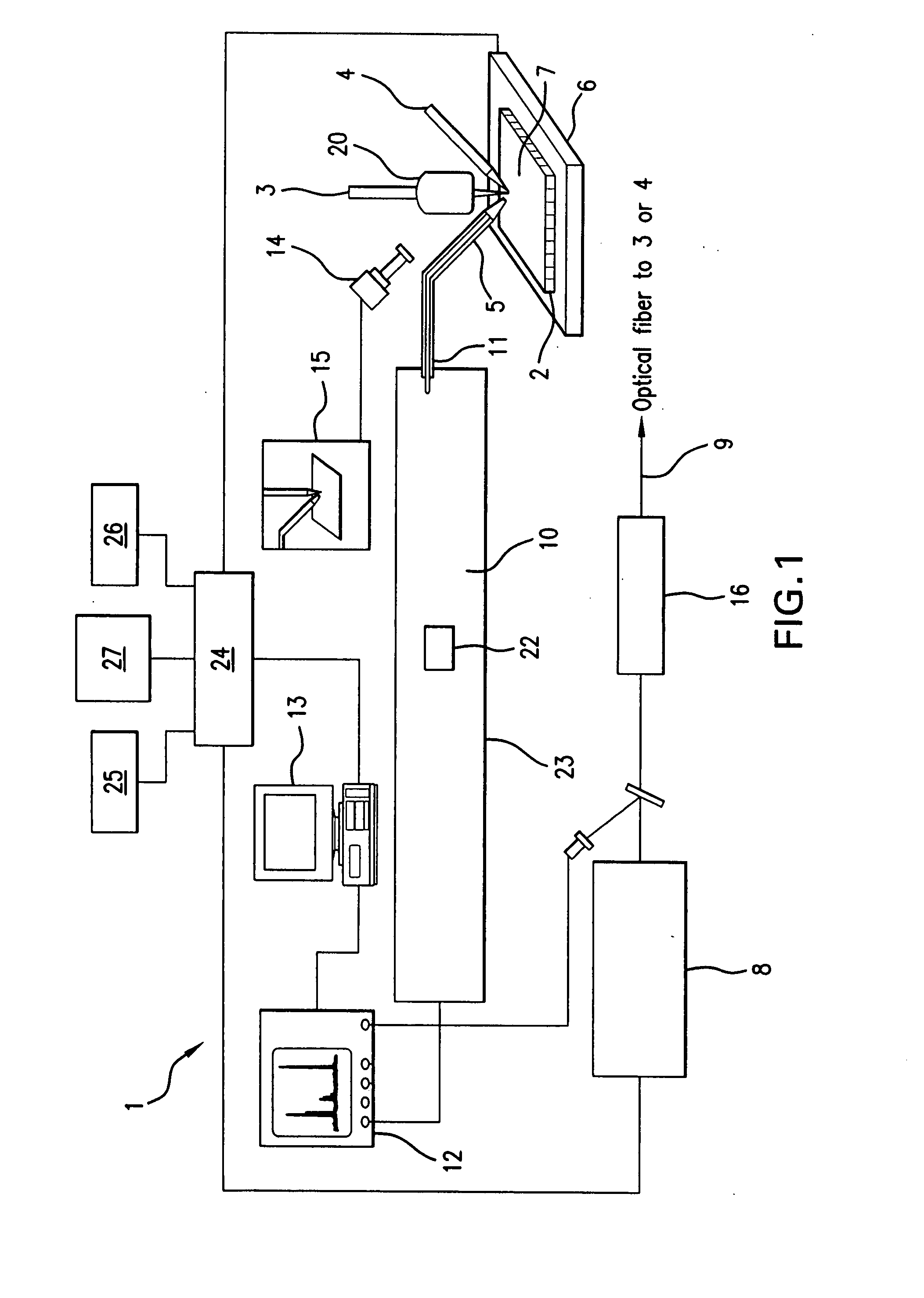
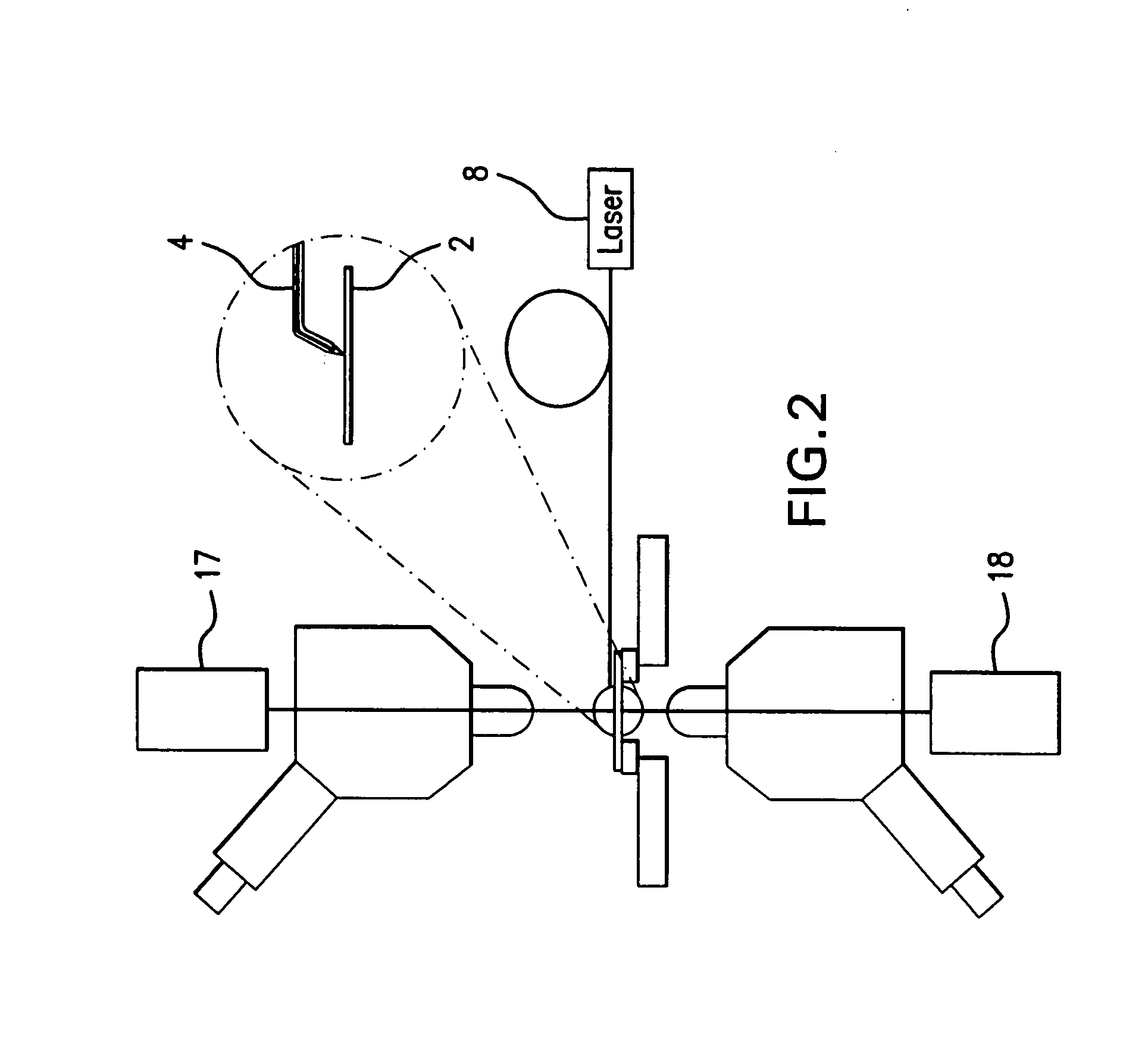
A system and method for analyzing and imaging a sample containing molecules of interest combines modified MALDI mass spectrometer and SNOM devices and techniques, and includes: (A) an atmospheric-pressure or near-atmospheric-pressure ionization region; (B) a sample holder for holding the sample; (C) a laser for illuminating said sample; (D) a mass spectrometer having at least one evacuated vacuum chamber; (E) an atmospheric pressure interface connecting said ionization region and said mass spectrometer; (F) a scanning near-field optical microscopy instrument comprising a near-field probe for scanning the sample; a vacuum capillary nozzle for sucking in particles which are desorbed by said laser, the nozzle being connected to an inlet orifice of said atmospheric pressure interface; a scanner platform connected to the sample holder, the platform being movable to a distance within a near-field distance of the probe; and a controller for maintaining distance information about a current distance between said probe and said sample; (G) a recording device for recording topography and mass spectrum measurements made during scanning of the sample with the near-field probe; (H) a plotting device for plotting said topography and mass spectrum measurements as separate x-y mappings; and (I) an imaging device for providing images of the x-y mappings.
Owner:GEORGE WASHINGTON UNIVERSITY
Remote reagent chemical ionization source
InactiveUS7253406B1Improve collection efficiencyAccurate shapeTime-of-flight spectrometersIsotope separationChromatographic separationGas phase
An improved ion source for collecting and focusing dispersed gas-phase ions from a reagent source at sub-atmospheric or intermediate pressure, having a remote source of reagent ions separated from a low-field sample ionization region by a barrier, comprised of alternating laminates of metal and insulator, populated with a plurality of openings, wherein DC potentials are applied to each metal laminate necessary for transferring reagent ions from the remote source into the low-field sample ionization region where the reagent ions react with neutral and / or ionic sample forming ionic species. The resulting ionic species are then introduced into the vacuum system of a mass spectrometer or ion mobility spectrometer. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when gas and liquid chromatographic separation techniques are coupled to sub-atmospheric and intermediate pressure photo-ionization, chemical ionization, and thermal-pneumatic ionization sources.
Owner:CHEM SPACE ASSOCS
Methods of identification of biomarkers with mass spectrometry techniques
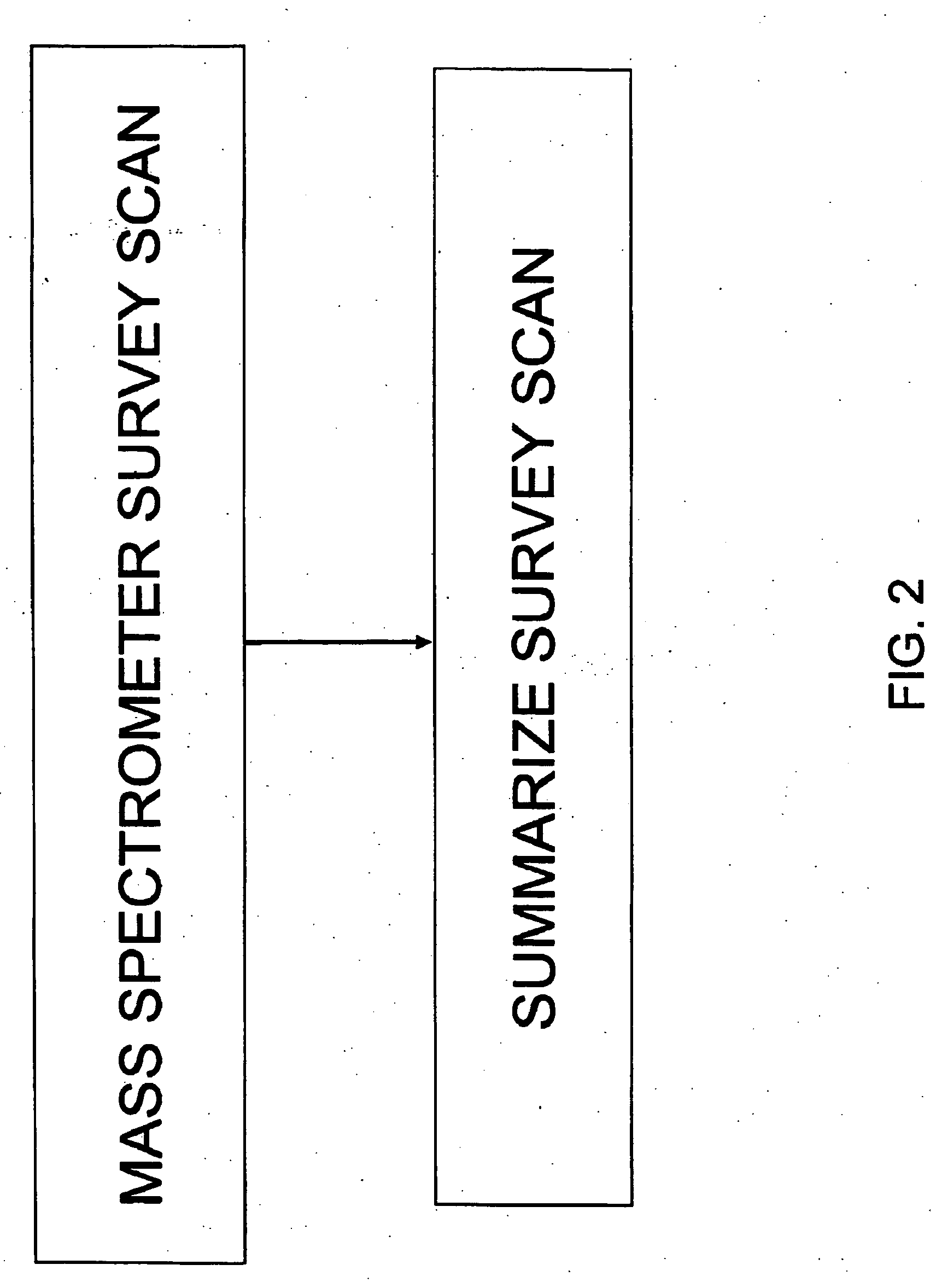
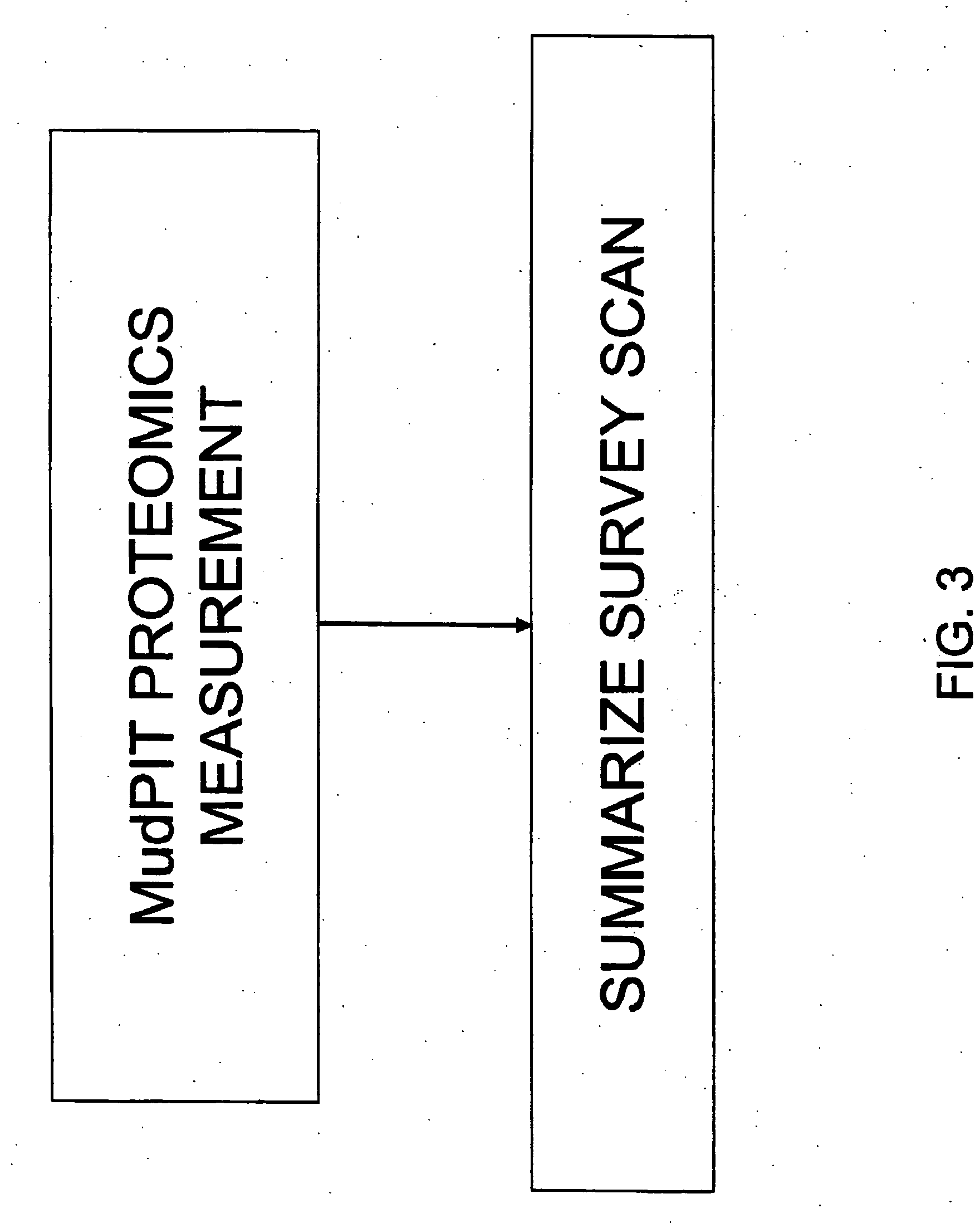
InactiveUS20060172429A1Avoid problemsReduce the impactDisease diagnosisBiological testingMass Spectrometry-Mass SpectrometryDiagnoses diseases
The present invention provides methods for identifying various biological states. Methods for diagnosis of diseases, in particular cardiovascular and brain diseases, are provided herein. One aspect of the invention is the analysis of lipoprotein complexes with summary survey scan mass spectrum for the analysis of biological states. Another aspect of the invention is the use of matrix assisted laser desorption ionization (MALDI) mass spectrometer to analysis lipoprotein complexes for the diagnosis of cardiovascular and brain diseases. Yet another aspect of the invention is a method of diagnosis of brain diseases by evaluating the characteristics of lipoprotein complexes.
Owner:INSILICOS
Ion sampling for APPI mass spectrometry
InactiveUS6653626B2Component separationSamples introduction/extractionMass Spectrometry-Mass SpectrometryPhysical chemistry
An atmospheric pressure ion source, e.g. for a mass spectrometer, that produces ions by atmospheric pressure photoionization (APPI). It includes a vaporizer, a photon source for photoionizing vapor molecules upon exit from the vaporizer, a passageway for transporting ions to, for example, a mass spectrometer system, and a means for directing the ions into the passageway. The center axis of the vaporizer and the center axis of the passageway form an angle that may be about 90 degrees. Included in the invention is a method for creating ions by atmospheric pressure photoionization along an axis and directing them into a passageway oriented at an angle to that axis.
Owner:AGILENT TECH INC
Linear ion trap with an imbalanced radio frequency field
ActiveUS7582864B2High resolutionStability-of-path spectrometersIsotope separationIon trap mass spectrometryIon beam
An imbalanced radio frequency (RF) field creates a retarding barrier near the exit aperture of a multipole ion guide, in combination with the extracting DC field such that the barrier provides an m / z dependent cut of ion sampling. Contrary to the prior art, the mass dependent sampling provides a well-conditioned ion beam suitable for other mass spectrometric devices. The mass selective sampling is suggested for improving duty cycle of o-TOF MS, for injecting ions into a multi-reflecting TOF MS in a zoom mode, for parallel MS-MS analysis in a trap-TOF MS, as well as for moderate mass filtering in fragmentation cells and ion reactors. With the aid of resonant excitation, the mass selective ion sampling is suggested for mass analysis.
Owner:LECO CORPORATION
Remote reagent ion generator
InactiveUS7569812B1Significant excessIncrease percentageBioreactor/fermenter combinationsBiological substance pretreatmentsChromatographic separationGas phase
An improved ion source and means for collecting and focusing dispersed gas-phase ions from a remote reagent chemical ionization source (R2CIS) at atmospheric or intermediate pressure is described. The R2CIS is under electronic control and can produce positive, negative, or positive and negative reagent ions simultaneously. This remote source of reagent ions is separated from a low-field sample ionization region by a stratified array of elements, each element populated with a plurality of openings, wherein DC potentials are applied to each element necessary for transferring reagent ions from the R2CIS into the low-field sample ionization region where the reagent ions react with neutral and / or ionic sample forming sample ionic species. The resulting sample ionic species are then introduced into a mass spectrometer, ion mobility spectrometer or other sensor capable of detecting the sample ions. Embodiments of this invention are methods and devices for improving sensitivity of mass spectrometry when gas and liquid chromatographic separation techniques are coupled to atmospheric and intermediate pressure photo-ionization, chemical ionization, and thermospray ionization sources; and improving the sensitivity of chemical detectors or probes.
Owner:LEIDOS
Method for separation, characterization and/or identification of microorganisms using mass spectrometry
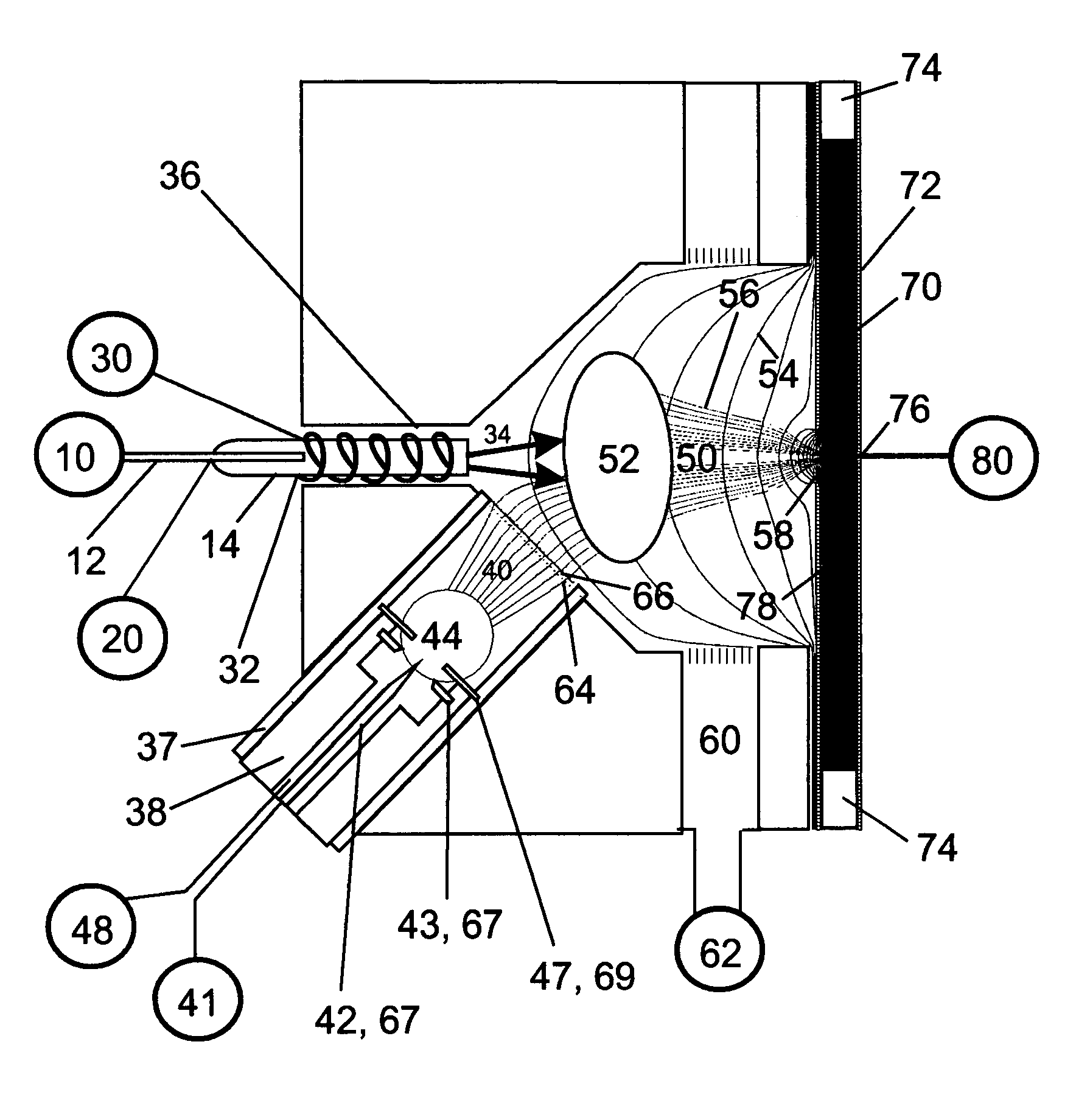
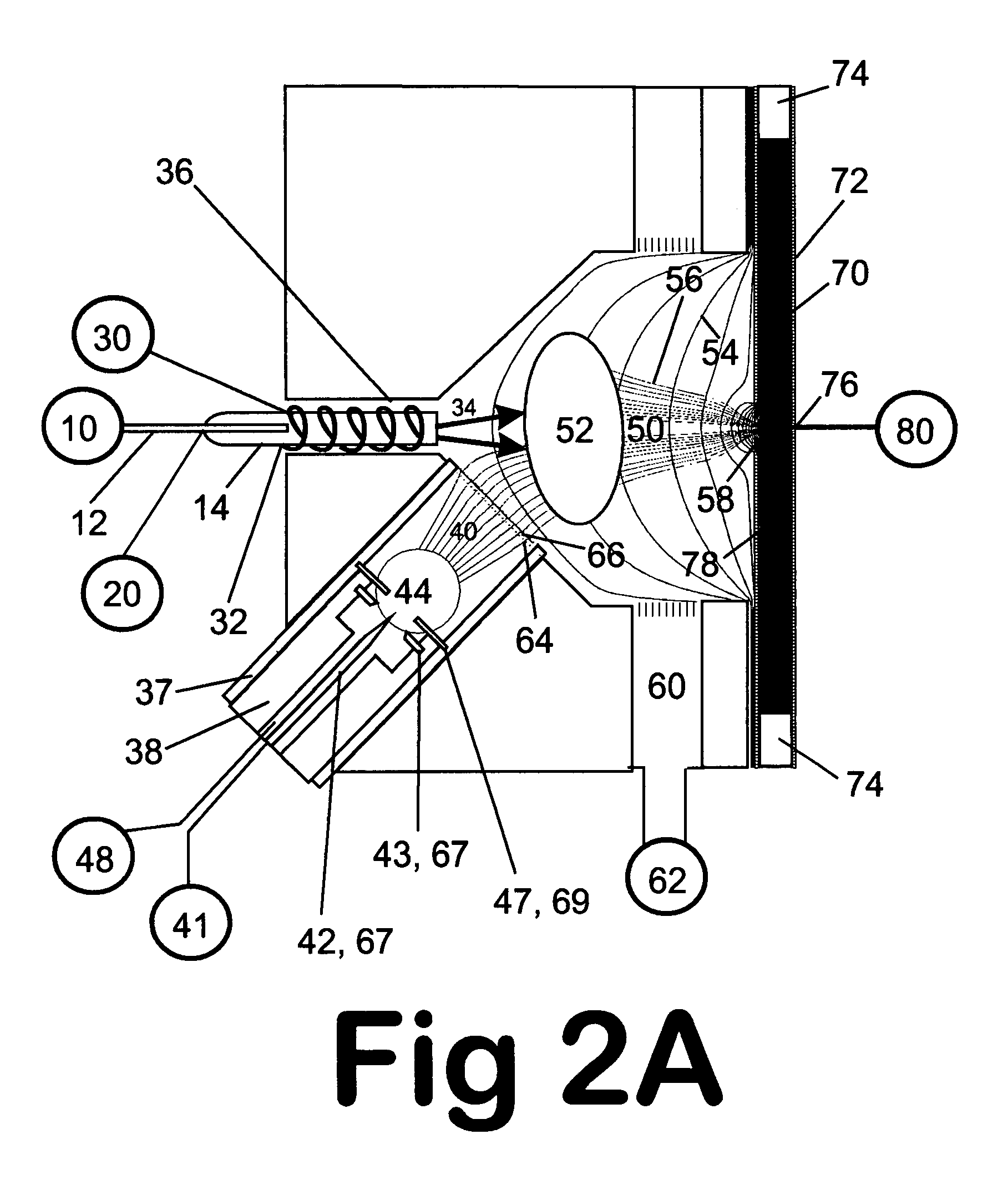
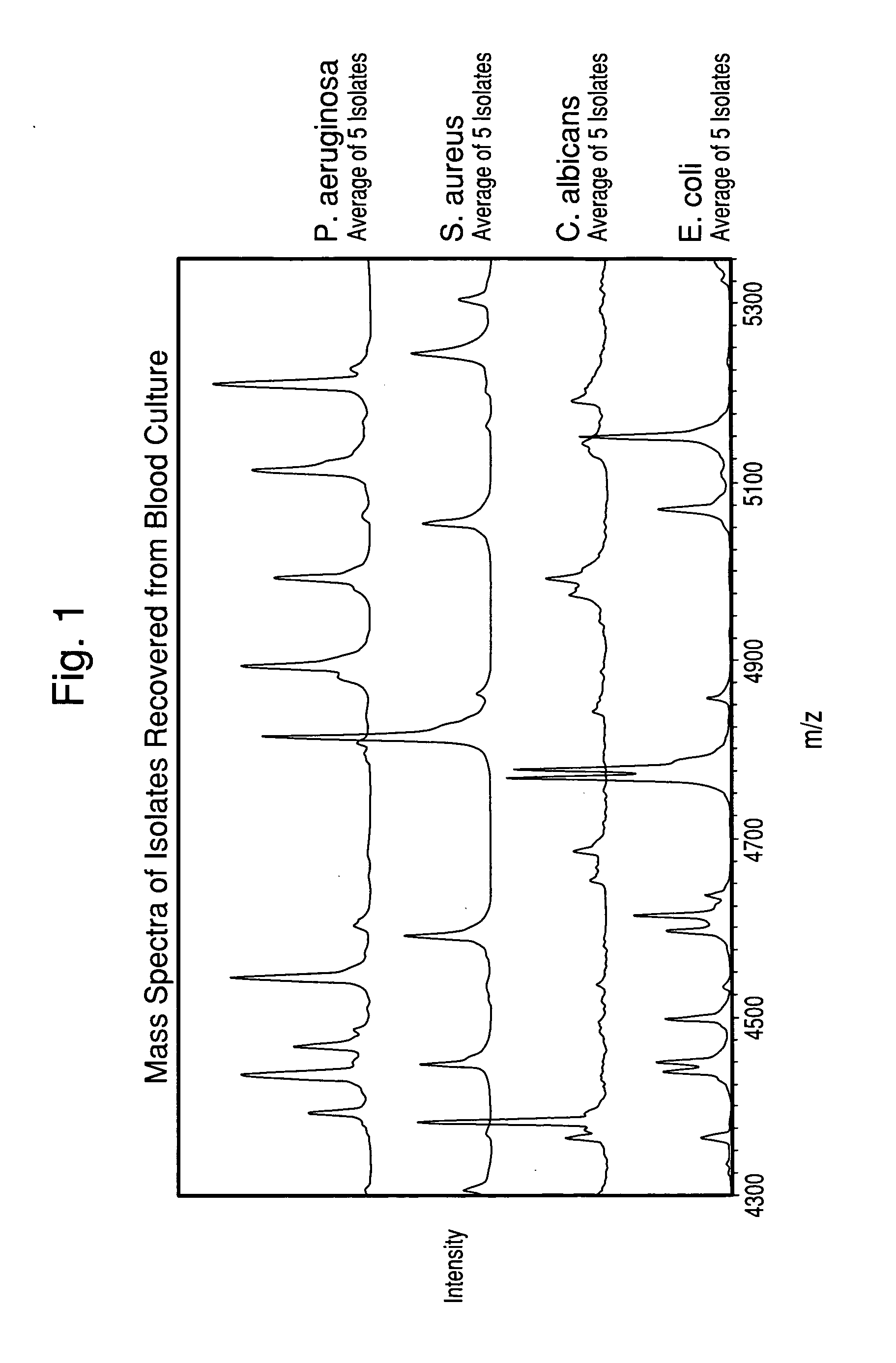
ActiveUS20100120085A1Rapid diagnosisRapid characterizationParticle separator tubesMicrobiological testing/measurementMicroorganismLysis
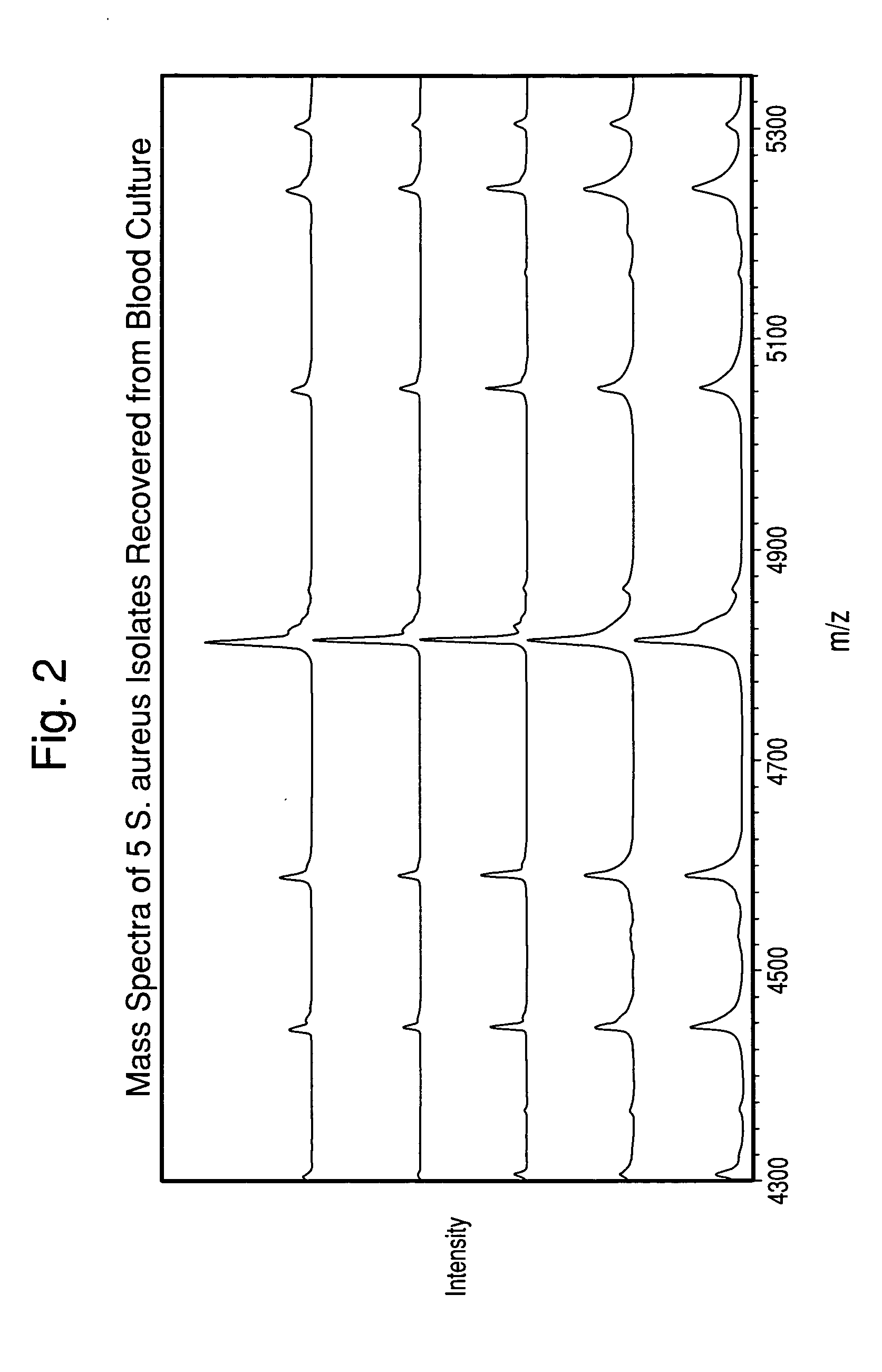
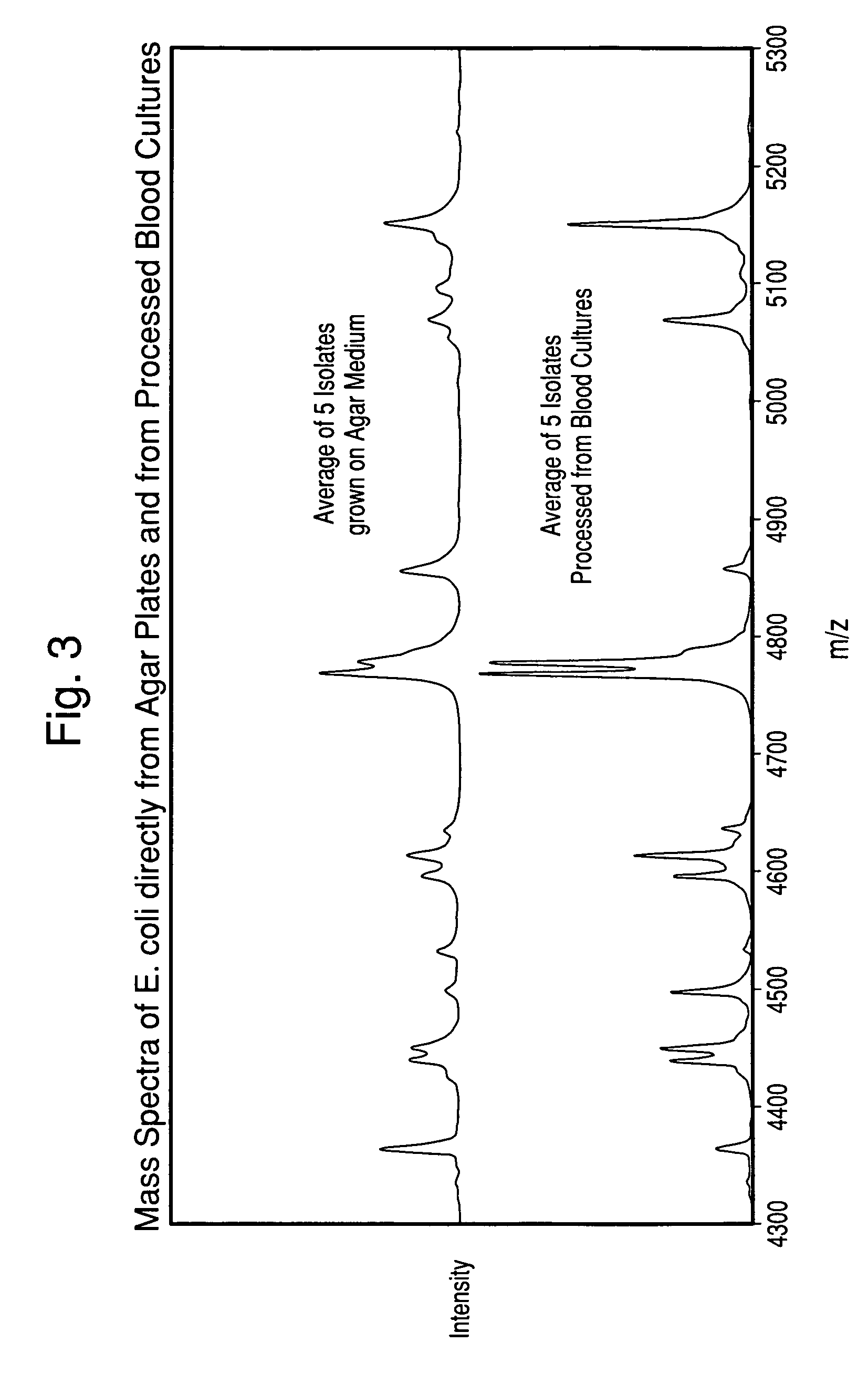
The present invention is directed to a method for separating, characterizing and / or identifying microorganisms in a test sample. The method of the invention comprises an optional lysis step for lysing non-microorganism cells that may be present in a test sample, followed by a subsequent separation step. The method may be useful for the separation, characterization and / or identification of microorganisms from complex samples such as blood-containing culture media. The invention further provides for interrogation of the separated microorganism sample by mass spectrometry to produce a mass spectrum of the microorganism and characterizing and / or identifying the microorganism in the sample using the mass spectrum obtained.
Owner:BIOMERIEUX INC
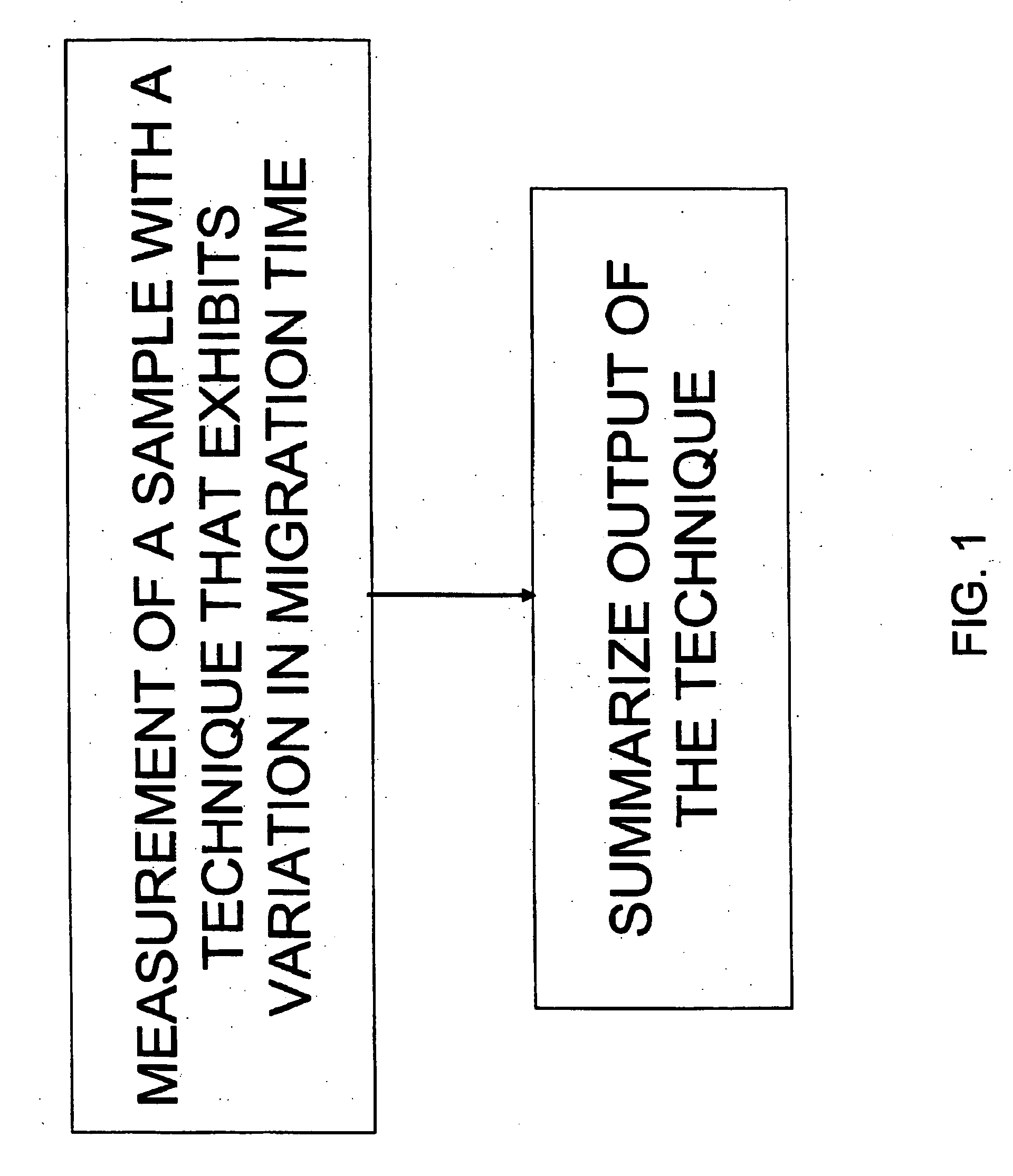
Methods for accurate component intensity extraction from separations-mass spectrometry data
InactiveUS20050255606A1Accurate representationBiological testingRecognisation of pattern in signalsMass Spectrometry-Mass SpectrometryUltimate tensile strength
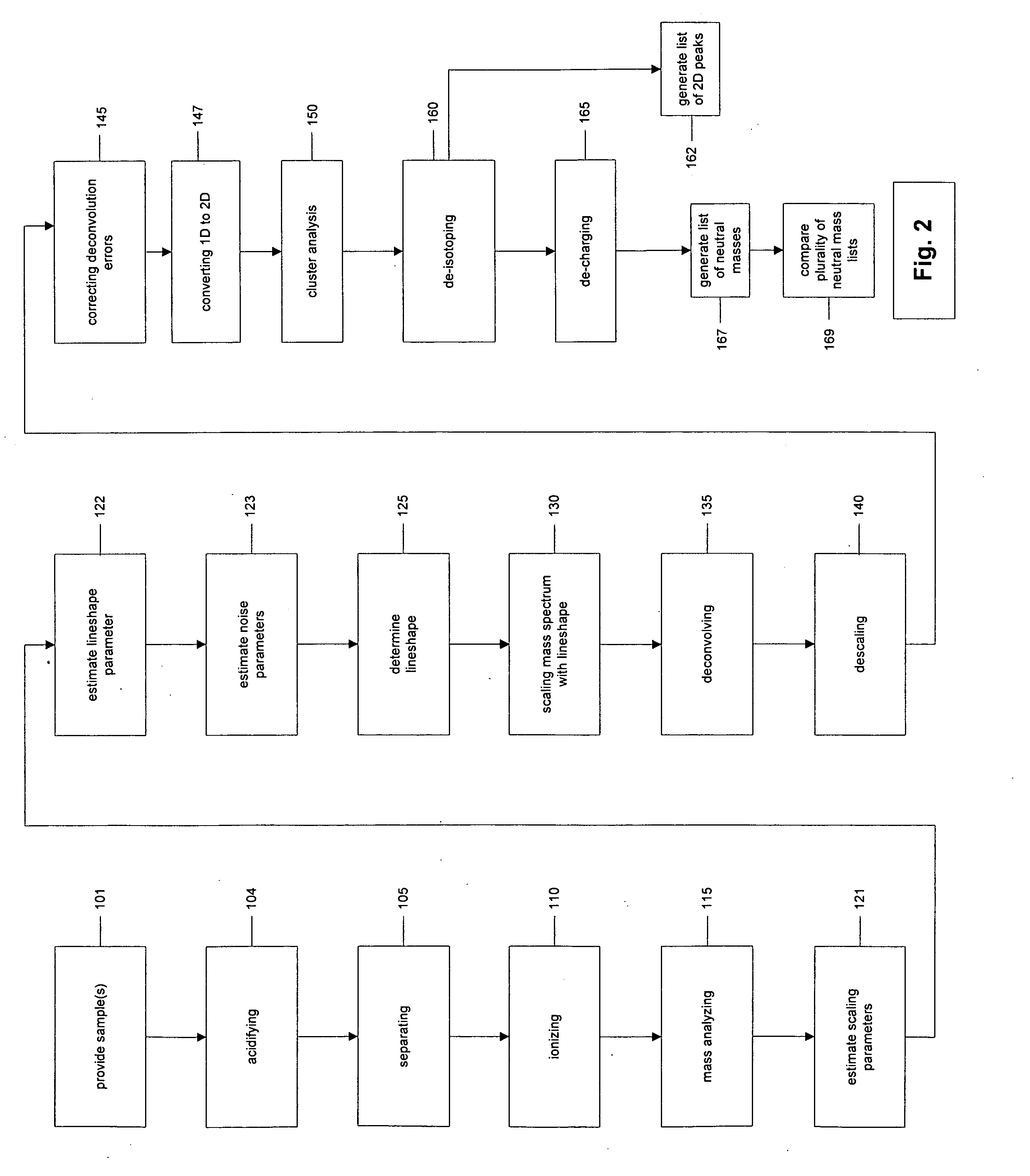
The present invention discloses methods for deconvolving and converting 1D mass spectra to 2D mass spectrum in order to obtain migration time centers and total intensities of the neutral mass envelopes of 2D spectra. The present invention also discloses devices that include a preparation / separation unit coupled to a mass spectrometer unit, and a computer unit capable of deconvolving mass spectra and calculating neutral mass envelopes.
Owner:V&M TCA LP +1
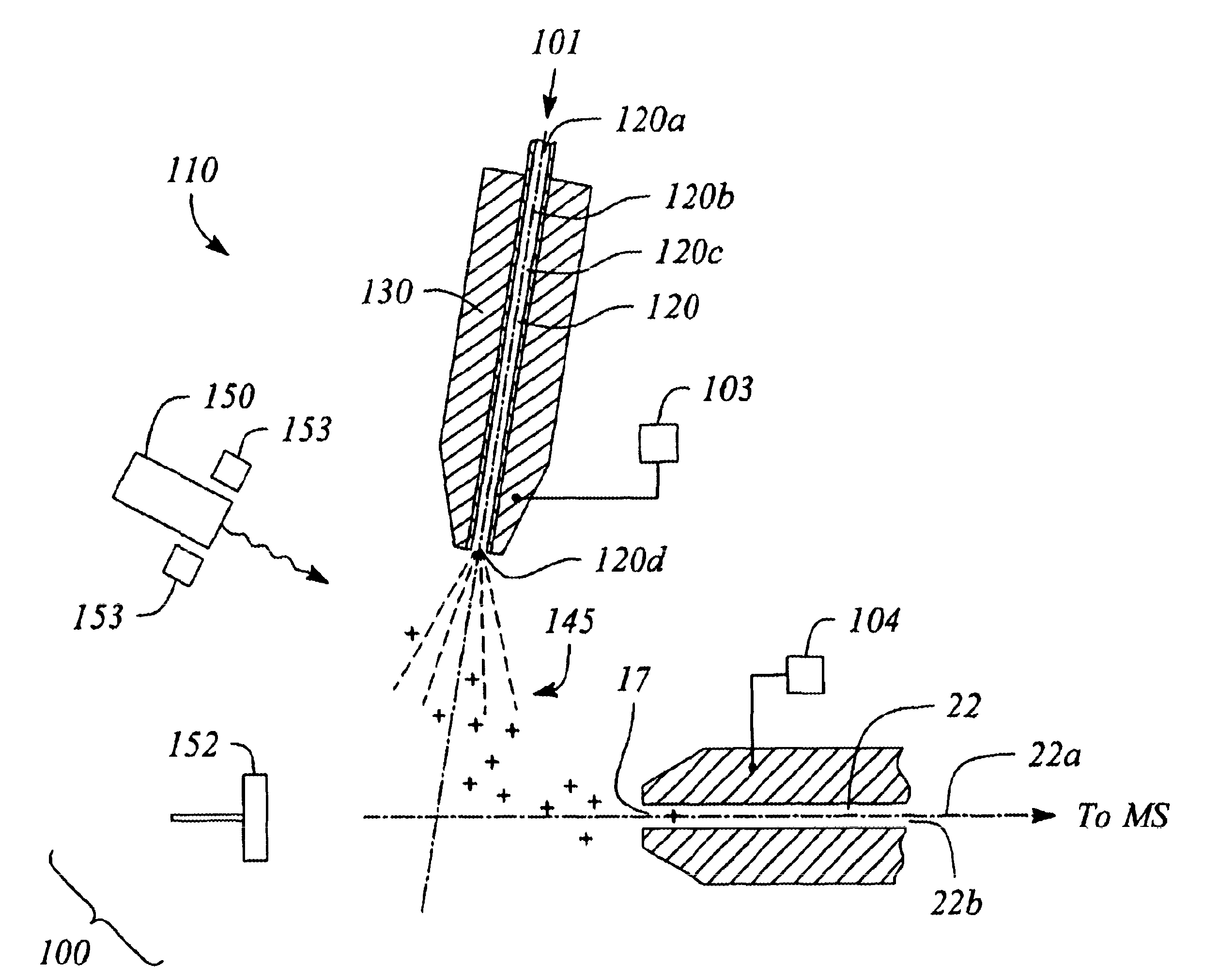
Capillary ion delivery device and method for mass spectroscopy
InactiveUS6806468B2Time-of-flight spectrometersSamples introduction/extractionMass Spectrometry-Mass SpectrometrySpectroscopy
Owner:SCI & ENG SERVICES
Fragmentation methods for mass spectrometry
InactiveUS6919562B1Shorten speedMinimizing velocityIsotope separationMass spectrometersMass Spectrometry-Mass SpectrometryMass analyzer
Apparatus and methods are provided that enable the interaction of low-energy electrons and positrons with sample ions to facilitate electron capture dissociation (ECD) and positron capture dissociation (PCD), respectively, within multipole ion guide structures. It has recently been discovered that fragmentation of protonated ions of many biomolecules via ECD often proceeds along fragmentation pathways not accessed by other dissociation methods, leading to molecular structure information not otherwise easily obtainable. However, such analyses have been limited to expensive Fourier transform ion cyclotron resonance (FTICR) mass spectrometers; the implementation of ECD within commonly-used multipole ion guide structures is problematic due to the disturbing effects of RF fields within such devices. The apparatus and methods described herein successfully overcome such difficulties, and allow ECD (and PCD) to be performed within multipole ion guides, either alone, or in combination with conventional ion fragmentation methods. Therefore, improved analytical performance and functionality of mass spectrometers that utilize multipole ion guides are provided.
Owner:PERKINELMER U S LLC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com