Multi-omics integration accurate pig breeding method
A technology for piglets and aspects, applied in the field of multi-omics integrated precision breeding of reproductive efficiency, can solve problems such as insufficient understanding and understanding, incomplete pig reproductive efficiency, etc., and achieve the effect of accelerating the breeding process, accelerating genetic progress, and promoting breeding selection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
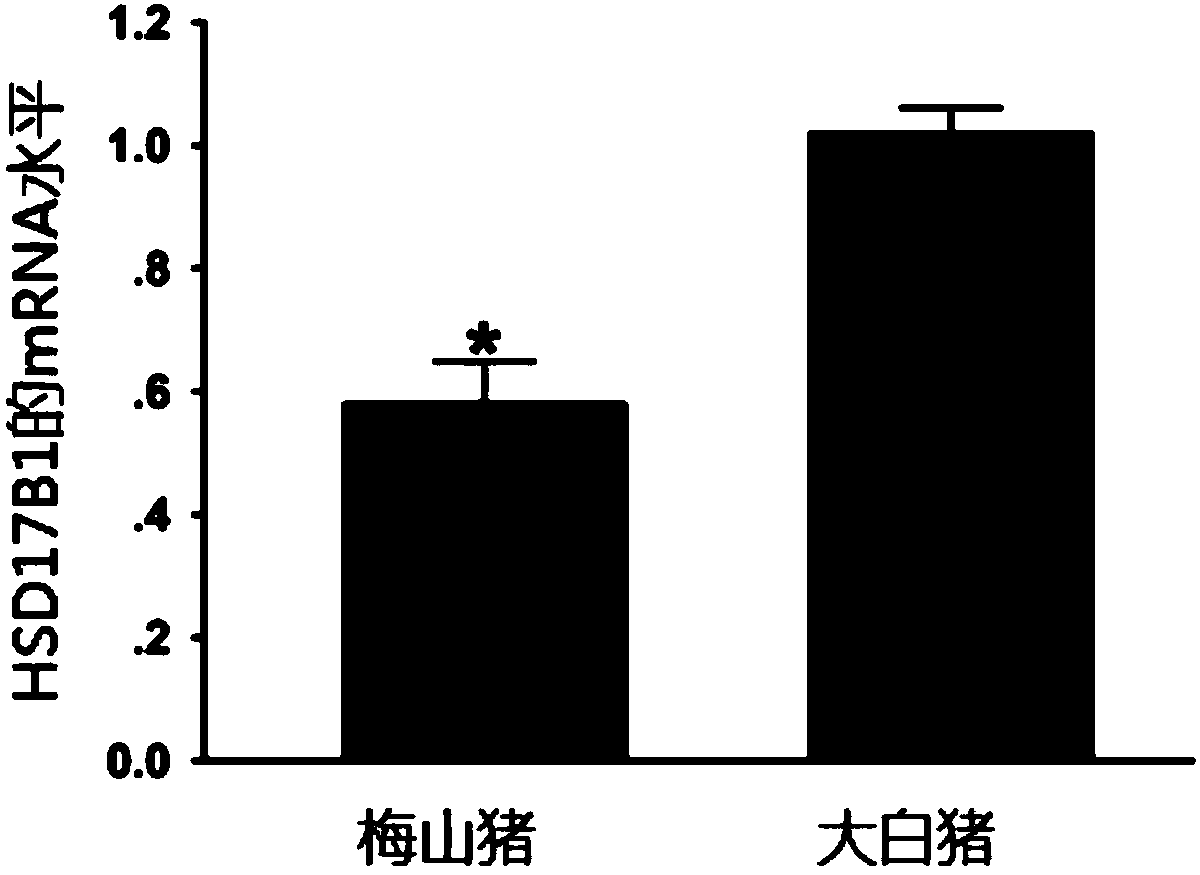
[0087] Example 1 Detection of HSD17B1 gene expression
[0088] 1. Collection of experimental samples
[0089] The large white pigs and Meishan pigs used to detect the expression of the HSD17B1 gene came from the pig farm of the Beijing Institute of Animal Husbandry and Veterinary Medicine, Chinese Academy of Agricultural Sciences, Wuqing District, Tianjin. The ovaries of sows at 49 days of gestation were collected, and the samples were stored in liquid nitrogen to extract total RNA. Three individuals of each species were collected as biological replicates.
[0090] 2. HSD17B1 gene expression detection
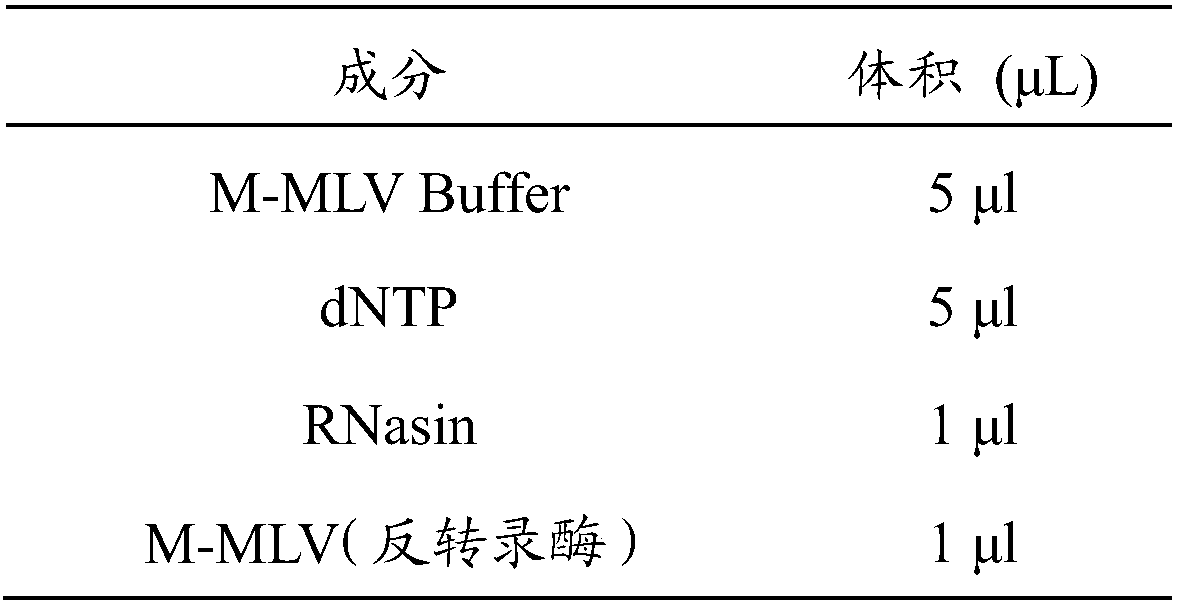
[0091] 2.1 Extraction of total RNA from ovarian tissue
[0092] The glassware and tweezers used in the RNA extraction process need to be dry-baked at 200°C for 4 hours to inactivate RNase. Eppendorf tubes and pipette tips need to use RNase inactivation products. The experimenters need to wear masks and latex gloves during the entire experiment.
[0093] (1) Sample collection: Place the ...
Embodiment 2
[0119] Example 2 Obtaining SNP molecular markers
[0120] 1. Collection of experimental samples
[0121] 375 large white pigs from a certain pig farm in Shandong were used for SNP typing of HSD17B1 gene. Collect ear tissue samples one by one, put them in 75% alcohol and store at -20°C, and prepare DNA.
[0122] 2. SNP typing of HSD17B1 gene
[0123] 2.1 Extraction and detection of pig genomic DNA
[0124] (1) Cut an appropriate amount of pig ear tissue, cut it into pieces, and put it into a 1.5 mL Axgen tube.
[0125] (2) Take a 50 mL BD centrifuge tube, mix the proteinase K with a final concentration of 0.4 mg / mL and the lysis buffer uniformly, and add 0.5 mL of the lysis buffer to a 1.5 mL centrifuge tube containing pig ear tissue for lysis.
[0126] (3) The centrifuge tube is evenly placed on the shaking plate of the thermostatic hybridization furnace in parallel (seal the tube lid tightly to avoid liquid leakage). Store at 55°C for more than 6 hours (full mixing of samples during di...
Embodiment 3
[0161] Example 3 Kit for detecting relevant SNP sites
[0162] The SNP site detection kit of the present invention includes the following components:
[0163] (1), dNTP (2.5mM);
[0164] (2), 5×PCR buffer;
[0165] (3) FastPfuDNA polymerase (250U);
[0166] (4), SNaPshot mixture;
[0167] (5), MgSO 4 Solution (50mM);
[0168] (6), 6×DNA Loading Buffer;
[0169] (7), PCR amplification primers (P 1 And P 2 ): Its nucleotide sequence is shown in SEQ ID No. 4 and SEQ ID No. 5;
[0170] (8) Extension primer: its nucleotide sequence is shown in SEQ ID No. 6;
[0171] (9) Exonuclease (250U);
[0172] (10), shrimp-alkaline phosphatase (100U);
[0173] (11) Buffer for purification:;
[0174] (12), ddNTP;
[0175] (13) Positive control specimen for homozygous mutation of SNP site: a plasmid containing pig rs323407415 site artificially synthesized as G base;
[0176] (14) Positive control specimen of heterozygous mutation at SNP site: a mixture of two plasmids containing artificially synthesized A and G bas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com