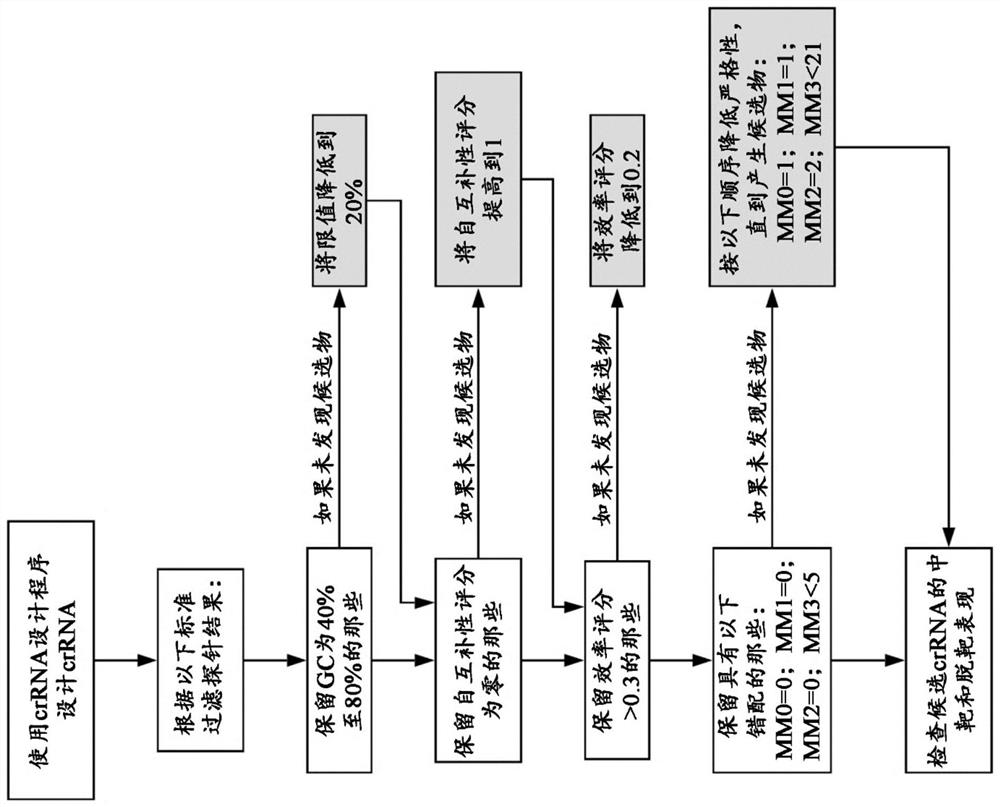
Patents
Literature
54 results about "Genome structure" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genome Structure. The human genome comprises of roughly 3.2 billion base pairs across 46 chromosomes. However, only a small fraction of this sequence codes for functional genes (roughly 1.5% of the genome) The remainder is made up of repeating elements, pseudogenes, microsatellites and transposons.
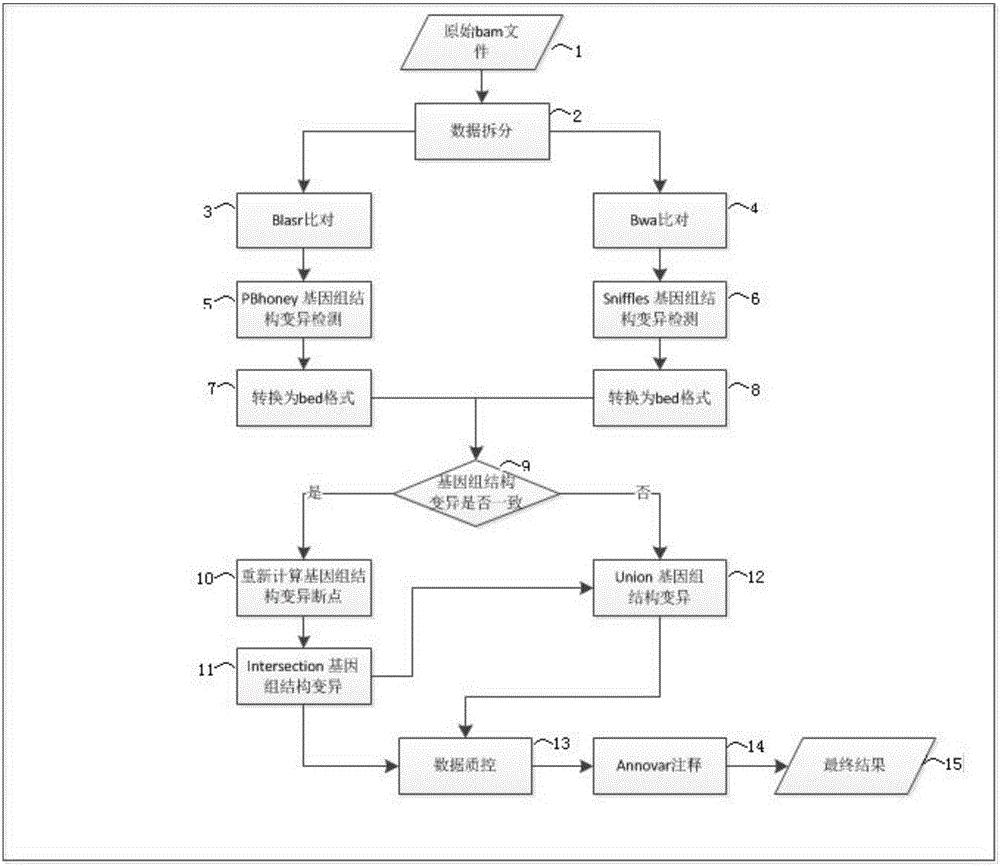
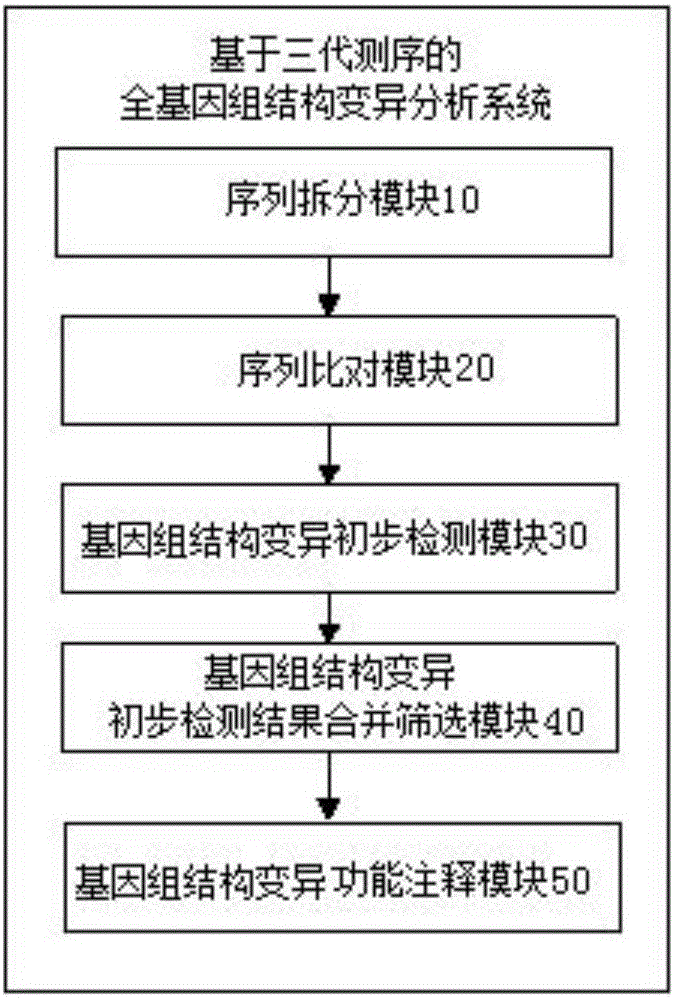
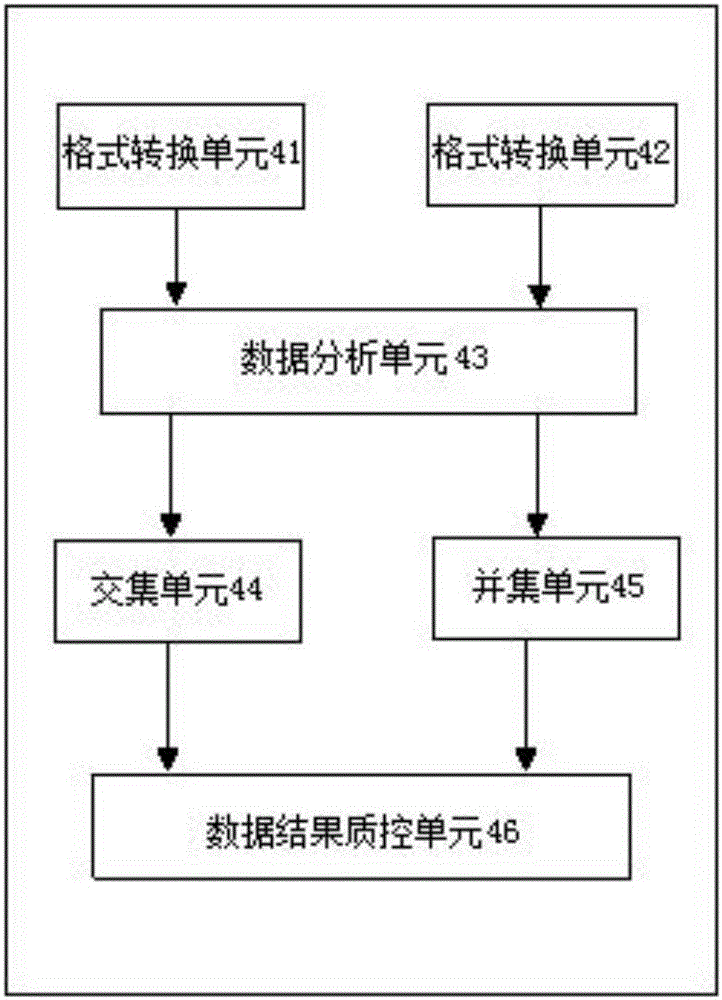
Three-generation sequencing-based whole genome structure variation analysis method and system
The invention discloses a three-generation sequencing-based whole genome structure variation analysis method and system. The method comprises the steps of 1) performing sequence splitting; 2) performing sequence comparison; 3) performing genome structure variation preliminary detection; 4) combining and screening genome structure variation preliminary detection results; and 5) performing genome structure variation function annotation. The system comprises a sequence splitting module, a sequence comparison module, a genome structure variation preliminary detection module, a genome structure variation preliminary detection result combining and screening module and a genome structure variation function annotation module. According to the method and the system, by integrating existing three-generation genome structure variation detection technologies PBhoney and Sniffles, the accuracy and sensitiveness of genome structure variation detection under low coverage degree can be effectively improved, and the reliability of the detection results is ensured while the detection cost is reduced.
Owner:BEIJING GRANDOMICS BIOTECH
Extraction of duck enteritis virus genom DNA and sequence thereof
InactiveCN101182526AHigh homologyShort copy cycleFermentationGenetic engineeringPUC19Escherichia coli
The invention discloses a DNA extraction method of Duck Enteritis Virus genome and the genome sequence. Firstly, the pyrolysis of chicken embryo fibroblast which is infected with DEV is processed by hypertonic buffer; then the nuclease is added for the digestion of cell DNA; and the virus genome DNA is obtained through phenol chloroform extracting method. The obtained sample is broken randomly into 2kb to 2.5kb fragments; the pUC19 vector is linked to transform host E. coli and construct genomic library. The positive clones in the library are randomly sequenced; the total sequence length has 6 times coverage rate for the virus genome. The sequence result is assembled and clustered to obtain the whole DEV genome sequence with the length of156512bp. The analysis shows that the genome structure is the same with that of the members of Varicella virus in Alpha- herpesus subfamily, which provides theoretical basis for the classification of DEV.
Owner:POULTRY INST SHANDONG ACADEMY OF AGRI SCI
Method and test kit for determining genome instability on basis of next-generation sequencing technology
ActiveCN111676277AHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementBiostatisticsGenomic StabilityGenome instability
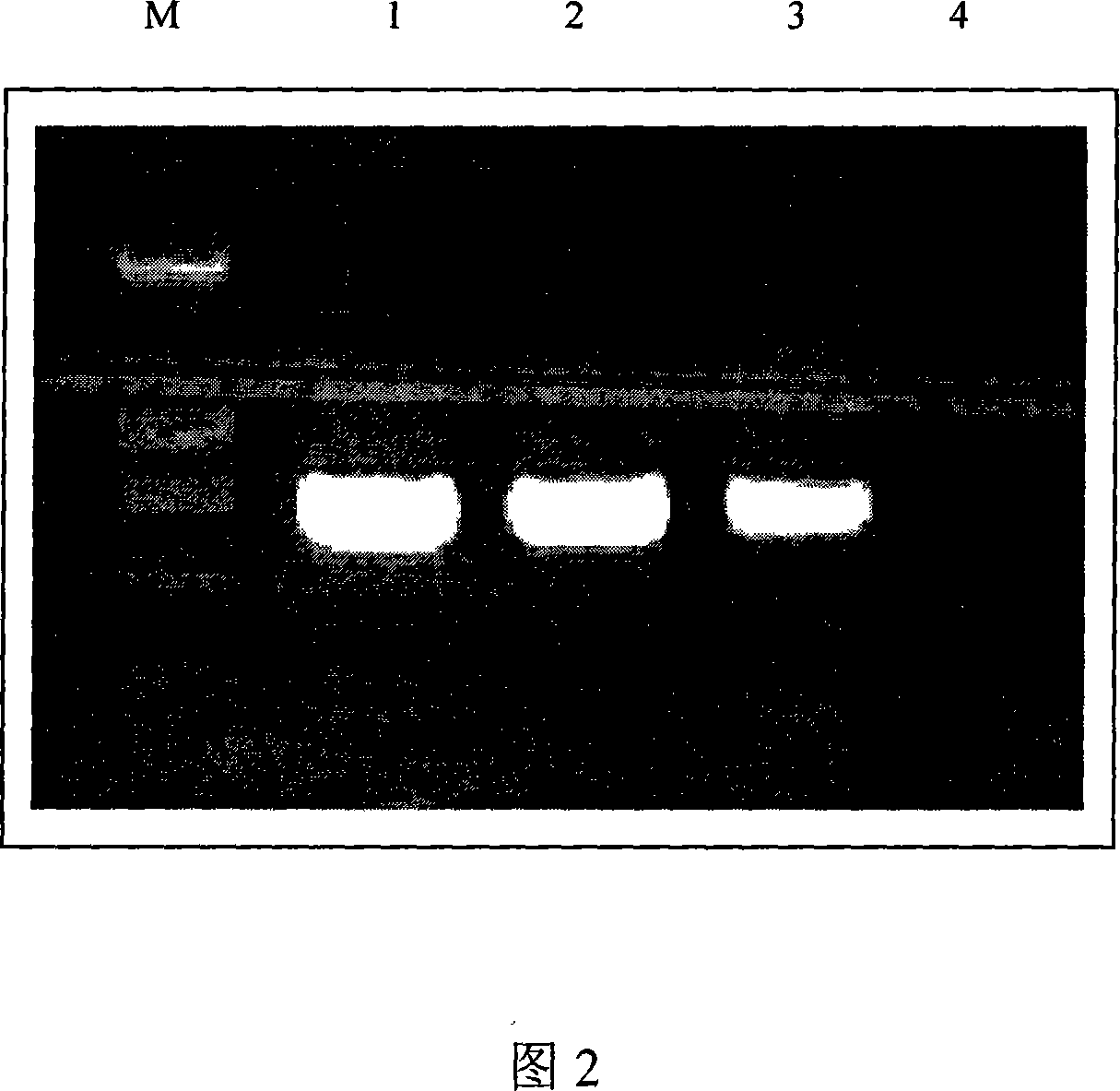
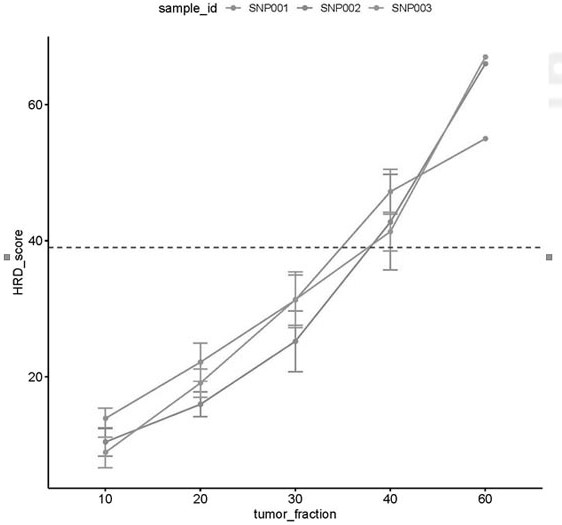
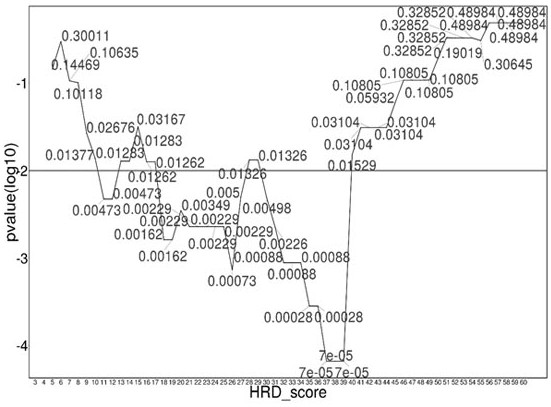
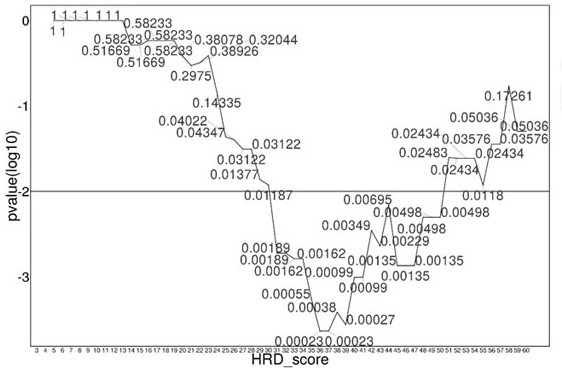
The invention relates to a method and a test kit for determining genome instability on the basis of a next-generation sequencing technology, belonging to the technical field of molecular detection. Inorder to overcome the defect that genome structure variation is not considered during detection of homologous recombination defects in the prior art, the invention provides the method and the test kit for determining genome instability on the basis of the next-generation sequencing technology. The method comprises the following steps: breaking a to-be-detected gene, then adding an A connector fora corresponding PCR reaction, carrying out hybridization capture through a designed probe, then amplifying a captured DNA library, sequencing the library, and carrying out evaluating through a software so as to determine the homologous recombination defect state. Compared with the prior art, the method provided by the invention can comprehensively evaluate the HRD state, is reliable in data results and is applicable to popularization.
Owner:臻和(北京)生物科技有限公司 +1
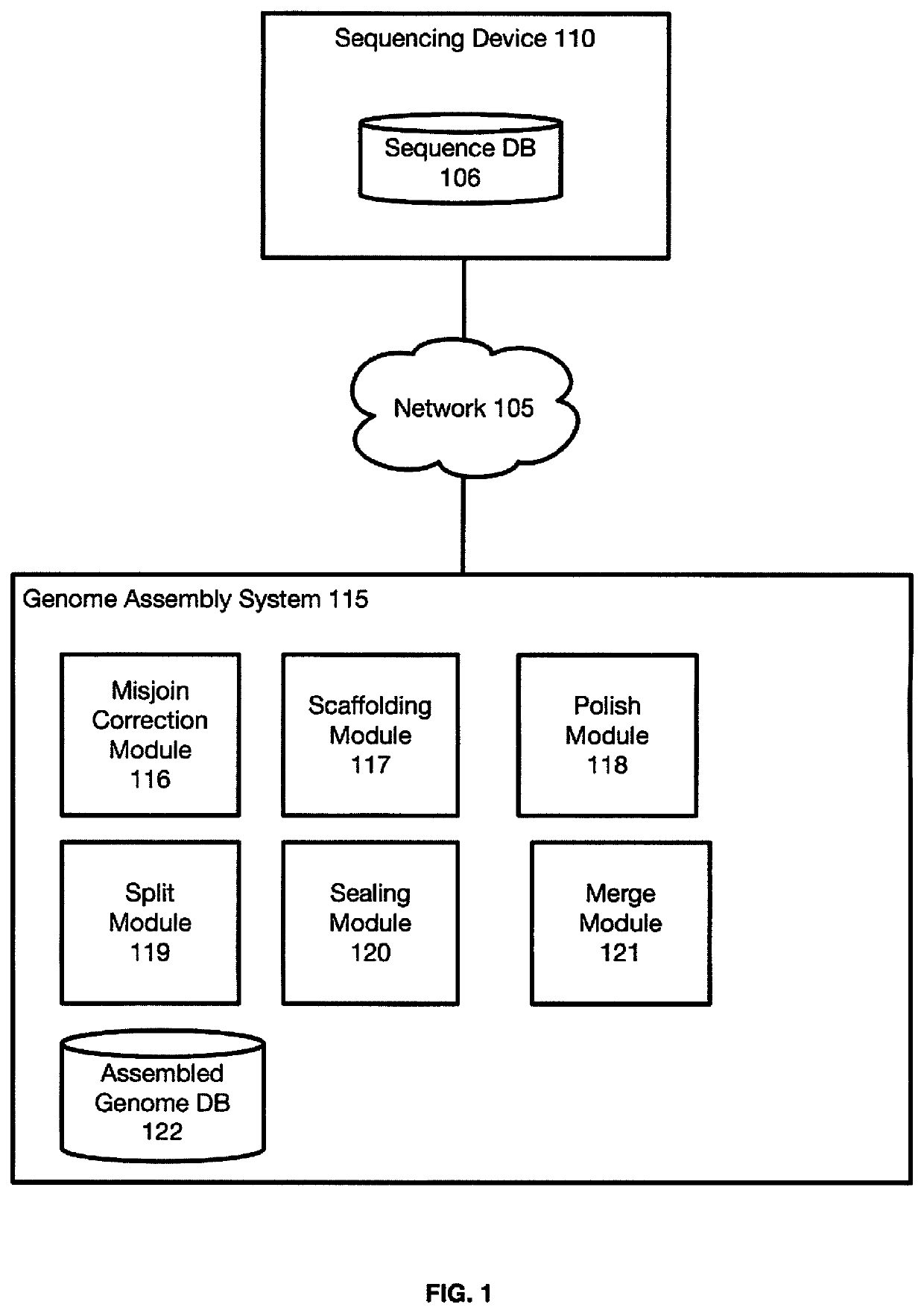
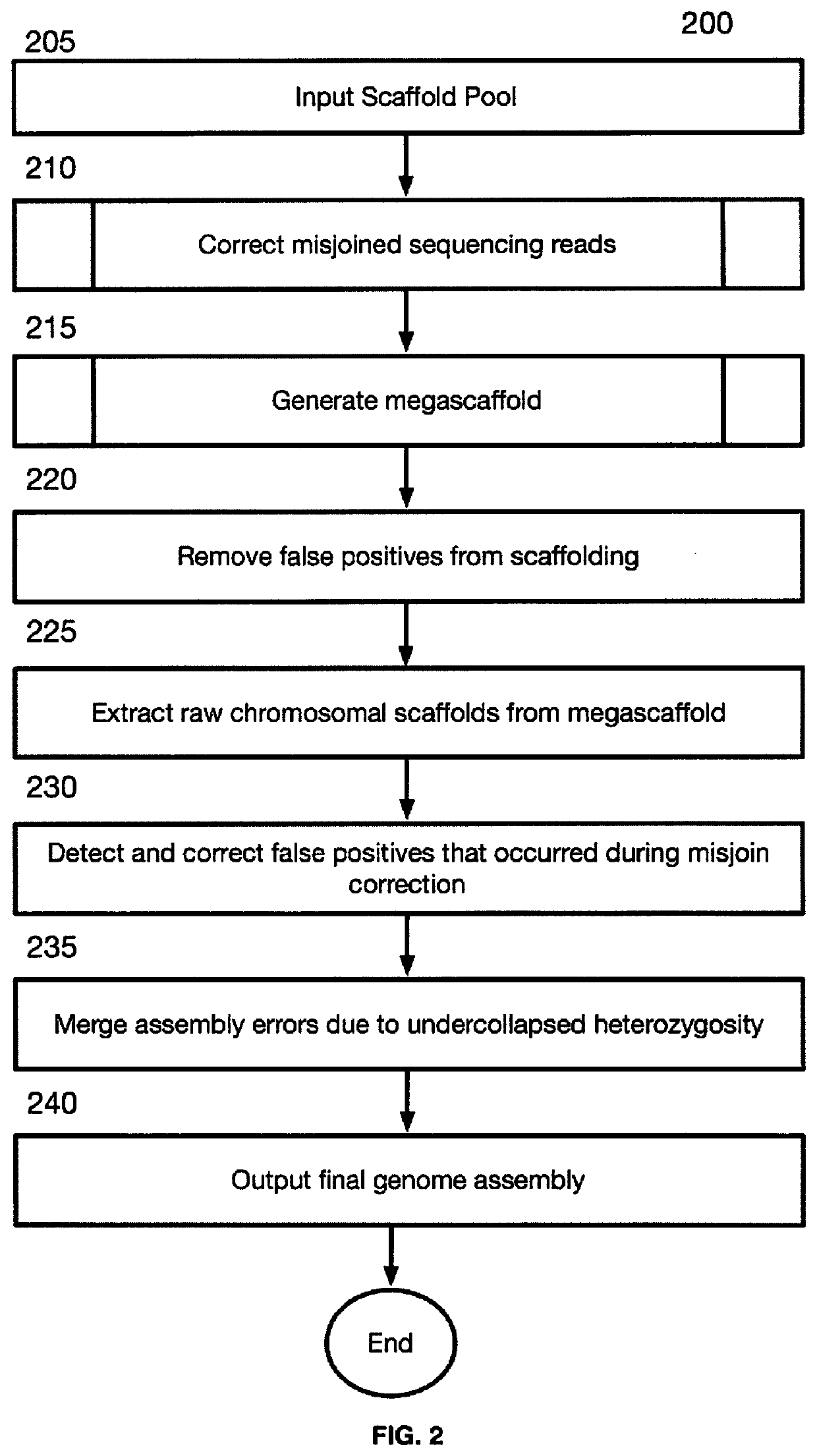
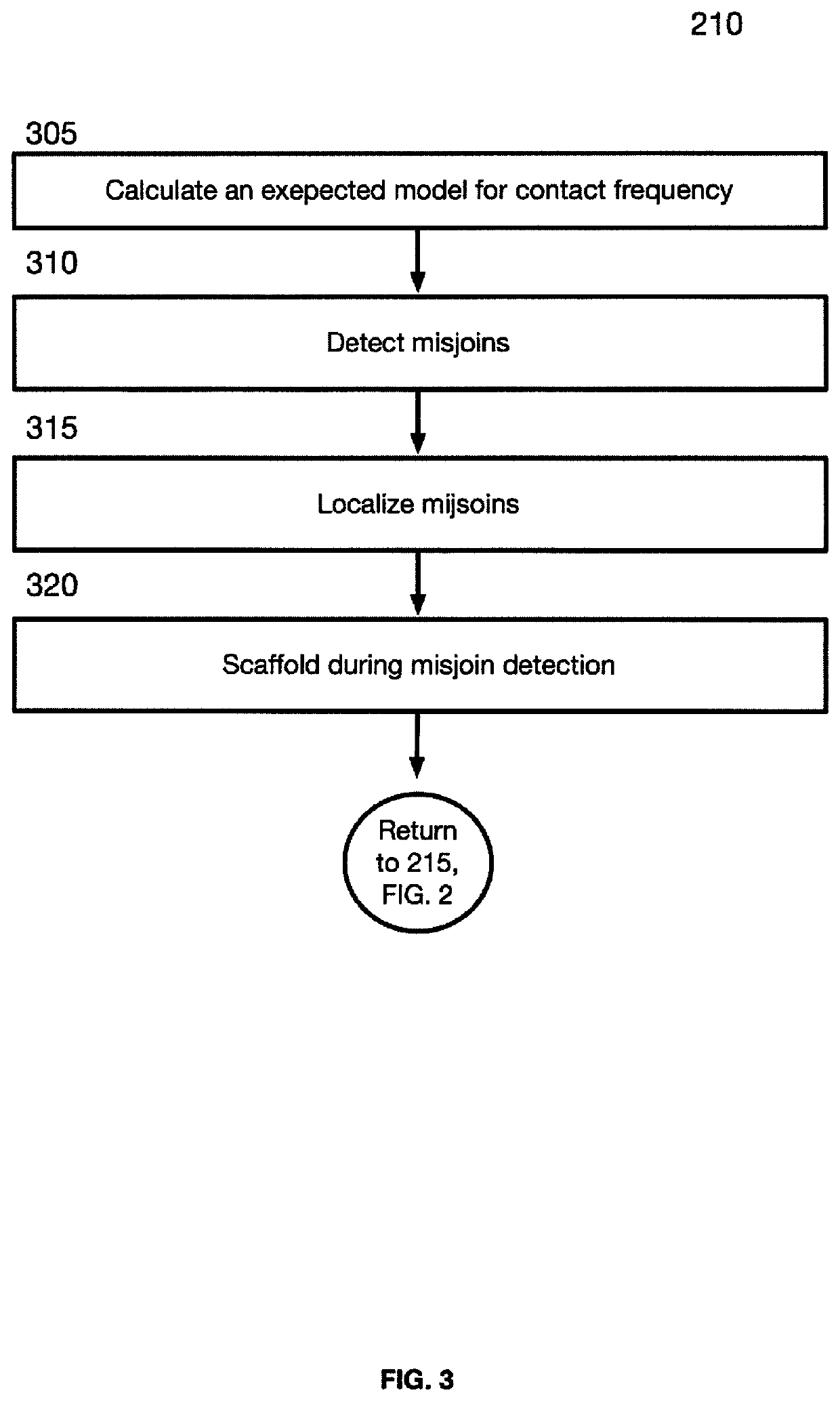
Linear genome assembly from three dimensional genome structure
PendingUS20190385703A1Misassembly detectionEasy to assembleMicrobiological testing/measurementSequence analysisChromatin LoopNucleic acid sequencing
Embodiments provide a method for sequencing and assembling long DNA genomes comprising generating a 3D contact map of chromatin loop structures in a target genome, the 3D contact map of chromatin loop structures defining spatial proximity relationships between genomic loci in the genome, and deriving a linear genomic nucleic acid sequence from the 3D map of chromatin loop structures
Owner:BAYLOR COLLEGE OF MEDICINE +1
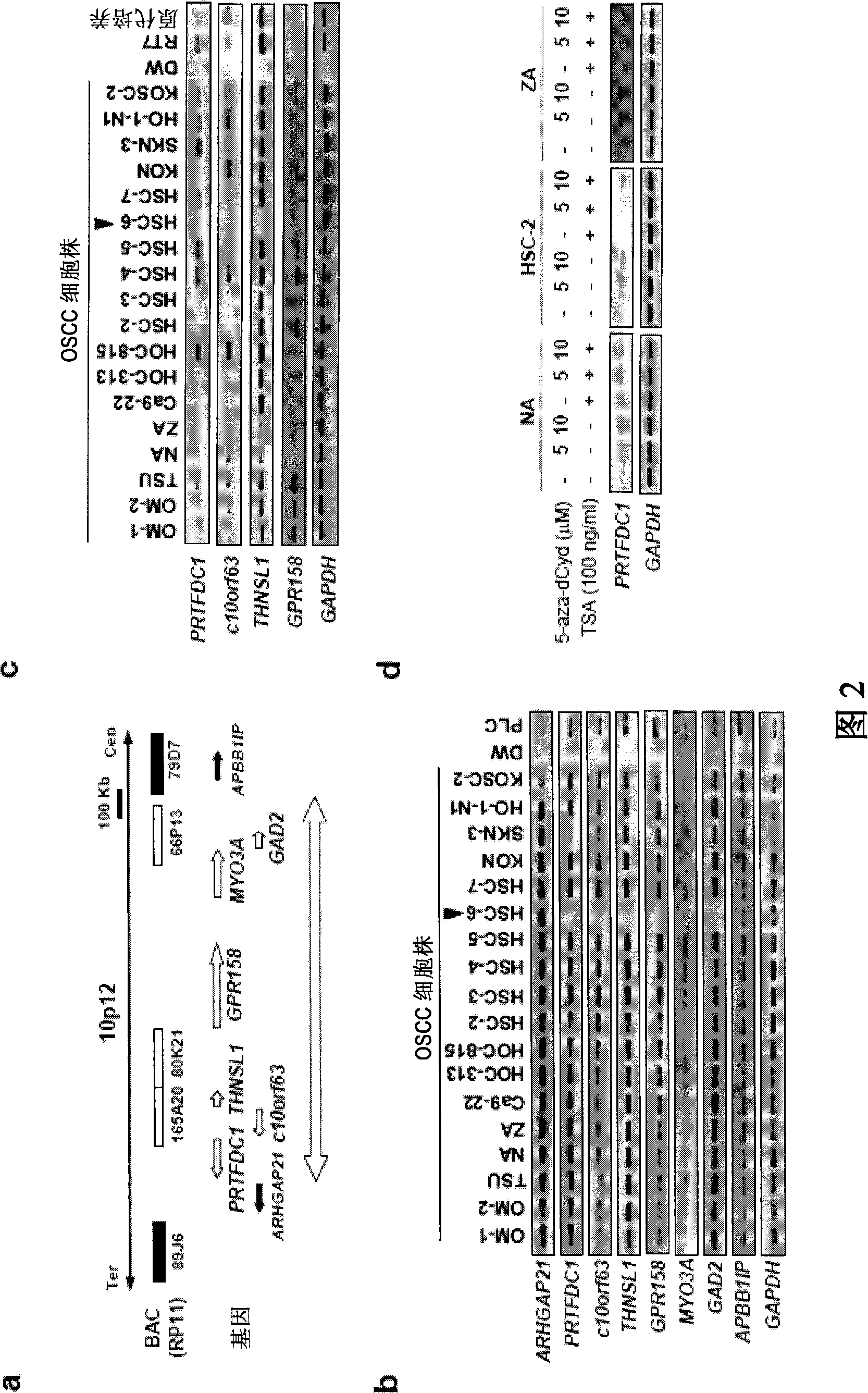
Method for detecting oral squamous-cell carcinoma and method for suppressing the same
InactiveCN101314794APrevent proliferationOrganic active ingredientsPeptide/protein ingredientsAcinar cell carcinomaSmall-cell carcinoma
An object of the present invention is to provide a method for detecting the oral squamous-cell carcinoma including the vicious degree through the change of the genome structure in the oral squamous-cell carcinoma as the index. The invention uses the genome structure in the oral squamous-cell carcinoma as the index, for detecting the oral squamous-cell carcinoma including the vicious degree. In addition, in the oral squamous-cell carcinoma, through recovering the deactivated gene, the multiplication of the oral squamous-cell carcinoma can be restrained.
Owner:FUJIFILM CORP +1
Bacillus subtilis mutant strain
Novel Bacillus subtilis mutant strains having good productivity of various enzymes are provided through extensive analysis of strains that are derived from Bacillus subtilis via gene disruption. The Bacillus subtilis mutant strains according to the present invention have genomic structures prepared by deletion of regions listed in the columns for deficient regions. Each of these Bacillus subtilis mutant strains exerts significantly improved secretory productivity of a protein when a gene encoding such a secretory target protein is introduced so that it can be expressed, compared with a case in which the same gene is introduced into a wild-type strain.
Owner:KAO CORP
Isolation and identification of Yunnan tomato leaf curl viral genome and agrobacterium tumefaciens-mediated infective clone construction
InactiveCN102703435AAvoid pitfalls in inoculation methodsReduce dependenceMicrobiological testing/measurementFermentationRestriction enzyme digestionGenomic DNA
The invention discloses isolation and identification of Yunnan tomato leaf curl viral genome and agrobacterium tumefaciens-mediated infective clone construction thereof. Total DNA of a genome of an infected tomato is extracted, virus total genomic DNA is cloned by using PCR (Polymerase Chain Reaction), and a total genomic sequence of a Yunnan tomato leaf curl virus is obtained through sequence measurement. 1.4 direct repeated genomes are obtained through methods of PCR and restriction enzyme digestion and are inserted into a plant expression carrier pBinPLUS with high replication capacity, a recombinant carrier is imported with agrobacterium tumefaciens strains EHA105 with strong infection capacity through an electroporation method, therefore, the agrobacterium tumefaciens-mediated infective clone has high-efficiency infection capacity to a host plant. The invention provides a matured method and system for researching interaction among the host plant, a virus infecting medium and a virus, field detection, genome structure and function research of the Yunnan tomato leaf curl virus.
Owner:ZHEJIANG UNIV
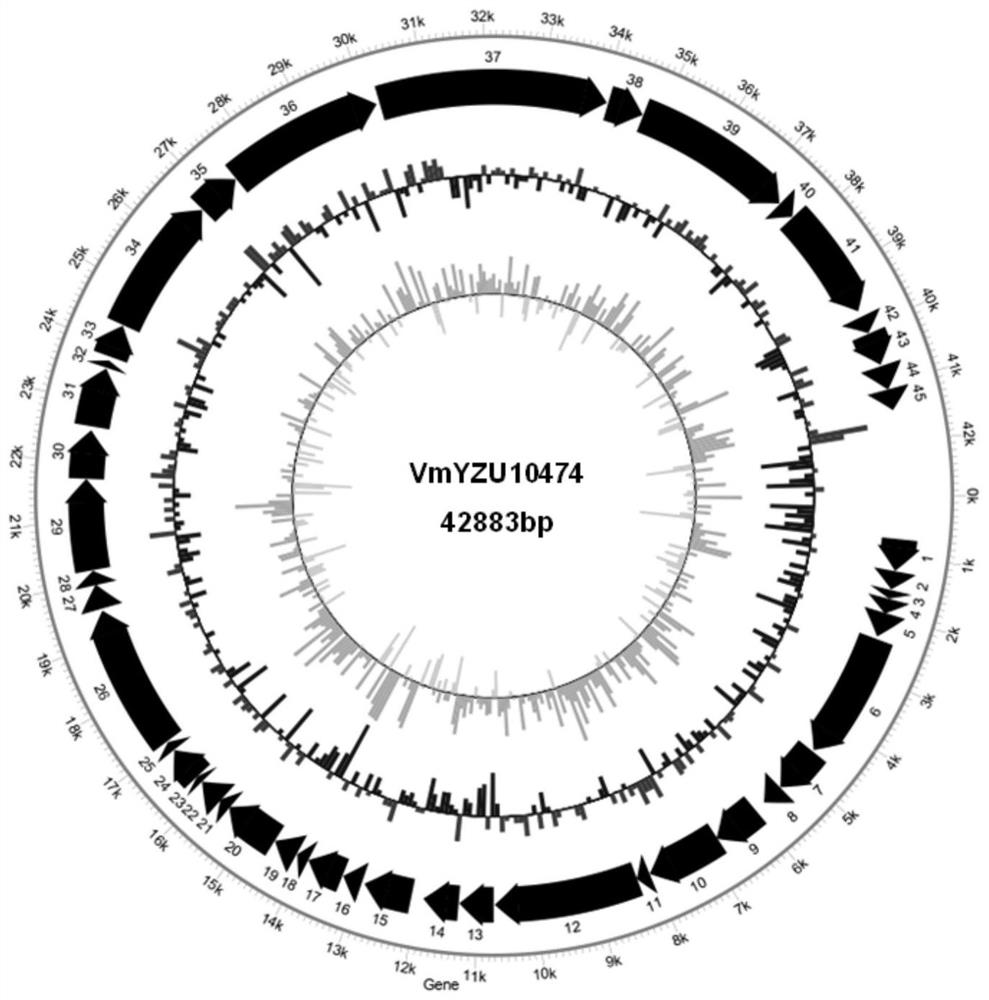
Pathogenic vibrio phage VmYZU10474 and application thereof
ActiveCN112063593AEfficient crackingUnique shapeBiocideMicroorganism based processesBiotechnologyDisease
The invention discloses a pathogenic vibrio phage VmYZU10474 and application thereof. The phage is identified to have unique genome structure characteristics, belongs to a novel phage, and has been preserved in China Center for Type Culture Collection on August 12, 2020 with the preservation number of CCTCC NO: M 2020417. The phage capable of efficiently cracking vibrio is obtained, can inhibit pathogenic vibrio in various matrixes, such as mimicus vibrio, vibrio parahaemolyticus, vibrio alginolyticus and vibrio fluvialis, and can be applied to preparation of bacteriostatic agents for controlling pathogenic vibrio pollution in aquatic products and environments thereof. The prepared bacteriostatic agents can effectively control growth of pathogenic vibrio in culture media, fish juice and aquatic product samples, and meanwhile can effectively inhibit pathogenic vibrio pellicle formation on solids such as equipment and vessels; and in addition, the pathogenic vibrio phage VmYZU10474 is simple to prepare and convenient to use, and the risk that pathogenic vibrio is spread and causes food-borne diseases can be reduced.
Owner:YANGZHOU UNIV
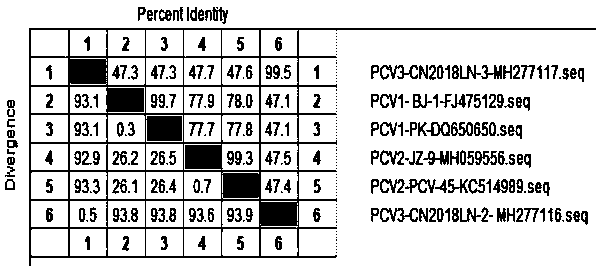
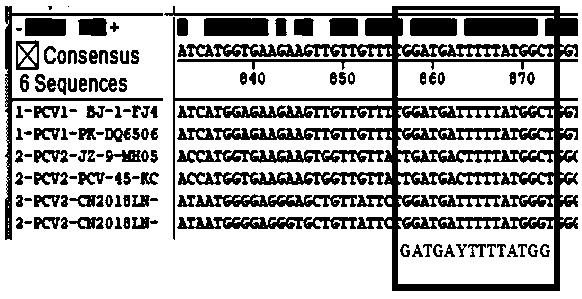
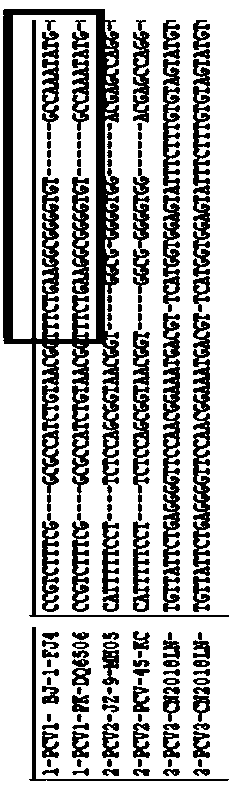
Primer group for identifying porcine circovirus types 1, 2 and 3 and application of primer group
InactiveCN109762943ACondition optimization is simpleEasy to observe the resultsMicrobiological testing/measurementDNA/RNA fragmentationPorcine circovirus type 1Genotype
The invention provides a primer group for identifying porcine circovirus types 1, 2 and 3 and application of the primer group. The sequences of the primer group are shown in SEQ ID NO.1 -4. On the basis of the genome structure characteristics of the three genotypes (PCV1, PCV2 and PCV3) of porcine circovirus, genome specific conserved areas of the three genotypes (PCV1, PCV2 and PCV3) of porcine circovirus are selected as upstream primer design areas, and downstream primers are designed separately in differential areas of genomes generated according to the three genotypes (PCV1, PCV2 and PCV3). The primer is simple in condition optimization, convenient and high in reaction speed, and a result is observed conveniently; an assembled kit can be used for rapid differential diagnosis of the three genotypes of porcine circovirus (PCV1, PCV2 and PCV3).
Owner:INST OF ANIMAL HUSBANDRY & VETERINARY FUJIAN ACADEMY OF AGRI SCI
Genome structure variation annotation method
The present disclosure relates to methods of annotating genomic structural variations and predicting fusion protein formation. The disclosure also provides a system and device for annotating genomic structural variations and predicting fusion protein formation, and a computer-readable media .
Owner:GUANGZHOU BURNING ROCK DX CO LTD
Model-independent genome structure variation detection system and method
ActiveCN111583996ALow detection sensitivityReduce error rateBiostatisticsProteomicsData miningStructural variant
The invention provides a model-independent genome structure variation detection system and method, wherein a model-independent structure variation detection theory is used as a core, and structure variation detection without depending on any variation model is achieved through a variation signal extraction module, a frequent maximum subgraph mining module and a classification module. According tothe system, a frequent variation pattern mining module is used for capturing the characteristics of structural variation left on a genome, and judging a potential structural variation region only by mining abnormal points in a large amount of normal data; and according to different genome disturbance modes of different variation types, different arrangement sequences of variation signals are further caused, and the different variation types are classified on the basis of the different arrangement sequences in combination with a deep learning model with a memory function. According to the invention, the system does not depend on any variation model, so that the variation detection sensitivity and error rate are greatly reduced; and the system is suitable for detection of complex variation types, and an additional structural variation model does not need to be established.
Owner:XI AN JIAOTONG UNIV
Method for acquiring Minxian county Chinese Gentian chloroplast genome sequence by utilizing transcriptome data
InactiveCN110021356ACan judge the qualityMicrobiological testing/measurementSequence analysisContigFiltration
The invention discloses a method for utilizing transcriptome data to acquire Minxian county Chinese Gentian chloroplast genome sequence. The method comprises the steps that transcriptome sequencing raw data of Minxian county Chinese Gentian is acquired through a next-generation sequencing technique, chloroplast gene reads is screened out through filtration and comparison with the known Chinese Gentian chloroplast genome, the screened reads are assembled into contigs, the contigs are built into a local blast database to be compared with reference sequence, ordering and splicing are conducted according to the comparison result, the newly assembled sequence is compared with other chloroplast genome sequences after every 5-6kb is spliced to check the splice sequence, the sequences are assembled into a ring according to the overlapping areas at the head and tail, the gaps are filled through PCR and a first-generation sequencing technique, and the Minxian county Chinese Gentian chloroplast genome sequence is finally obtained. The method is beneficial to understand the important information such as Minxian county Chinese Gentian chloroplast genome structure, and further develops the valueof transcriptome data.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
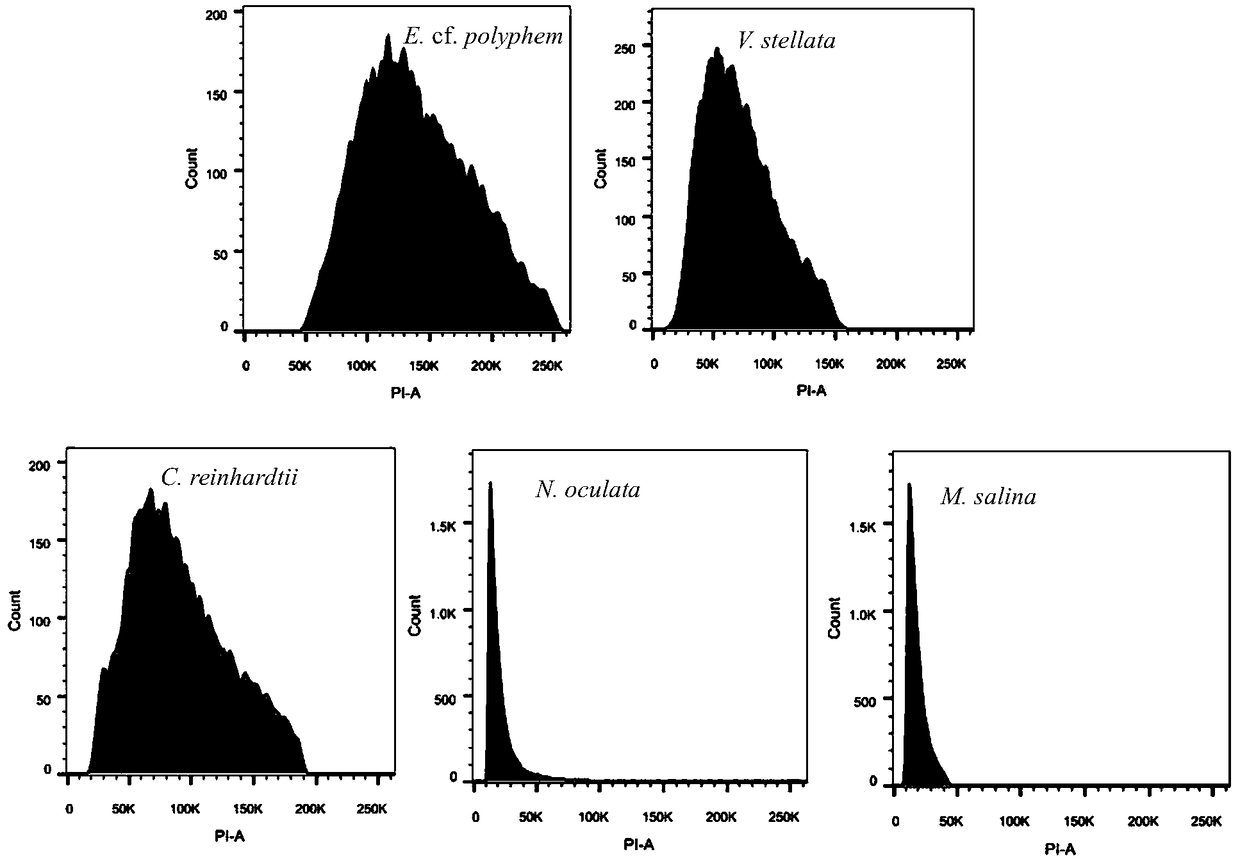
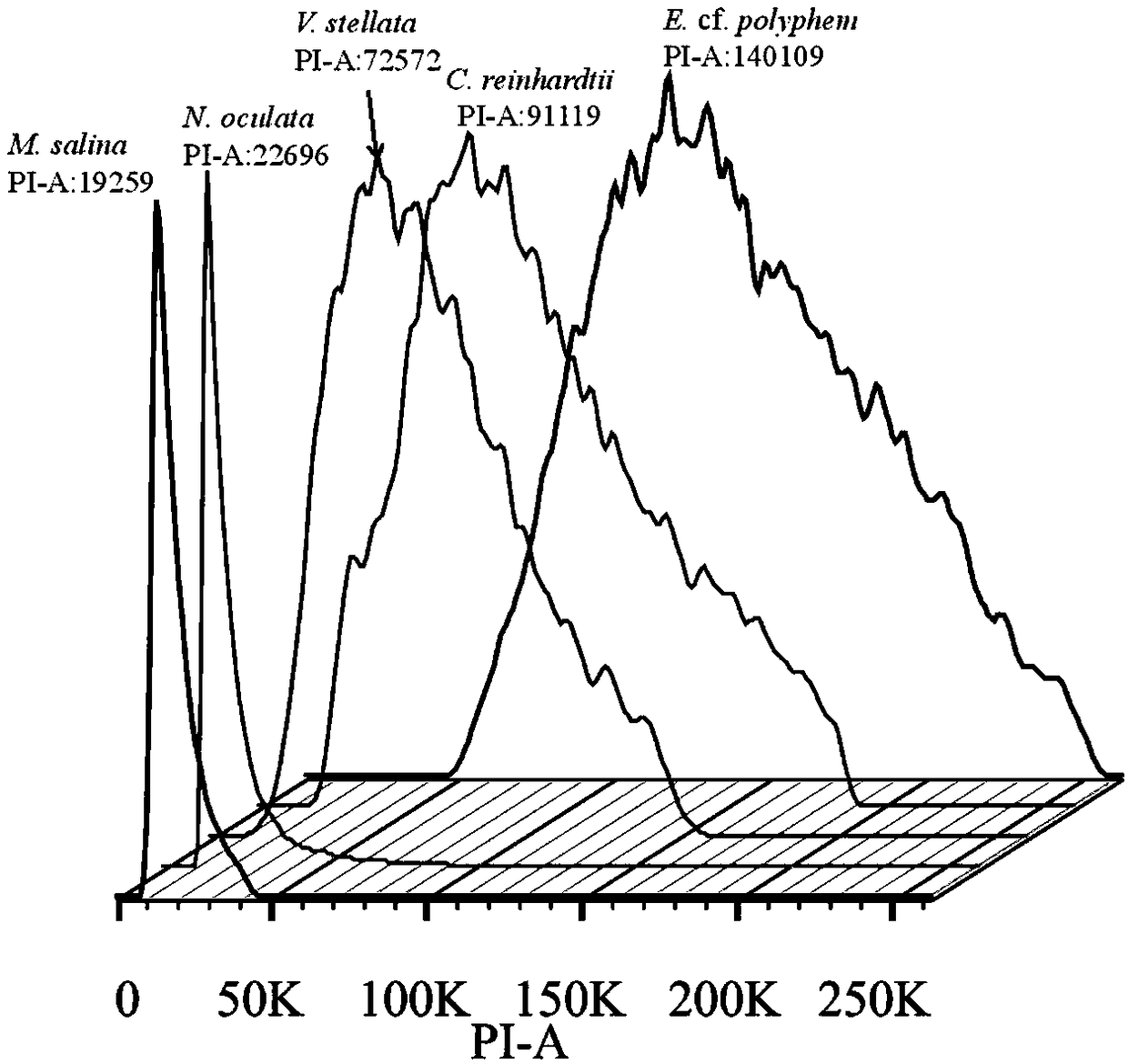
Method for determining size of single-cell eukaryotic algae genome
ActiveCN109295185AEasy to operateImprove accuracyMicrobiological testing/measurementStainingCell wall
The invention discloses a method for determining size of single-cell eukaryotic algae genome. The method is based on the characteristics of single-cell eukaryotic algae, optimizes an extraction and staining method of the nucleus, and obtains the more accurate flow cytometry analysis result, combined with a high-throughput second-generation illumina sequencing technology, the genome size of algae cells is comprehensively evaluated, and the characteristics of the genome can be clarified. The technical method optimizes the flow cytometry analysis of traditional plants, removes the interference ofalgal cytochrome and cell wall by using methanol, greatly improves the dyeing effect of PI, and improves the accuracy of flow cytometry analysis, and combined with a current leading high-flux sequencing technology, the genome structure characteristics of the to-be-tested species are comprehensively analyzed, which can provide a theoretical basis for a genetic structure research of subsequent species and an assembly strategy of whole genome sequencing.
Owner:JINAN UNIVERSITY
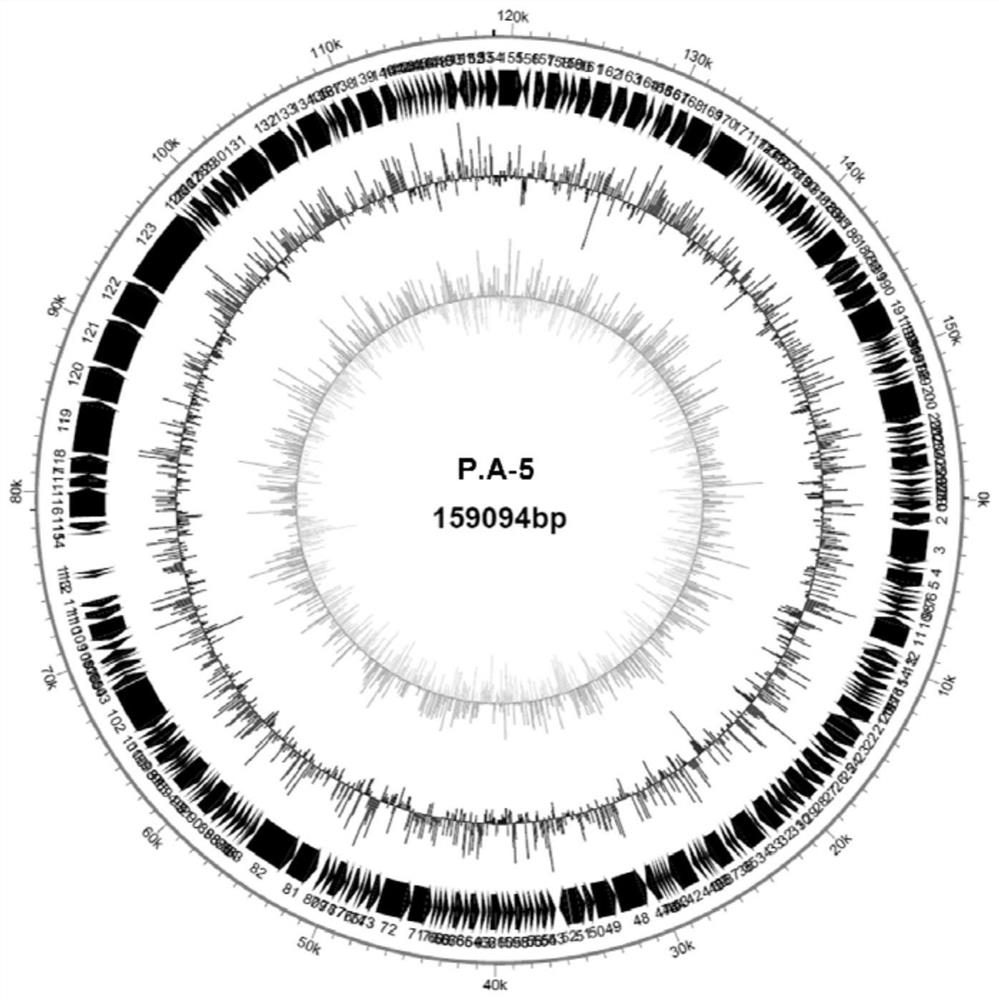
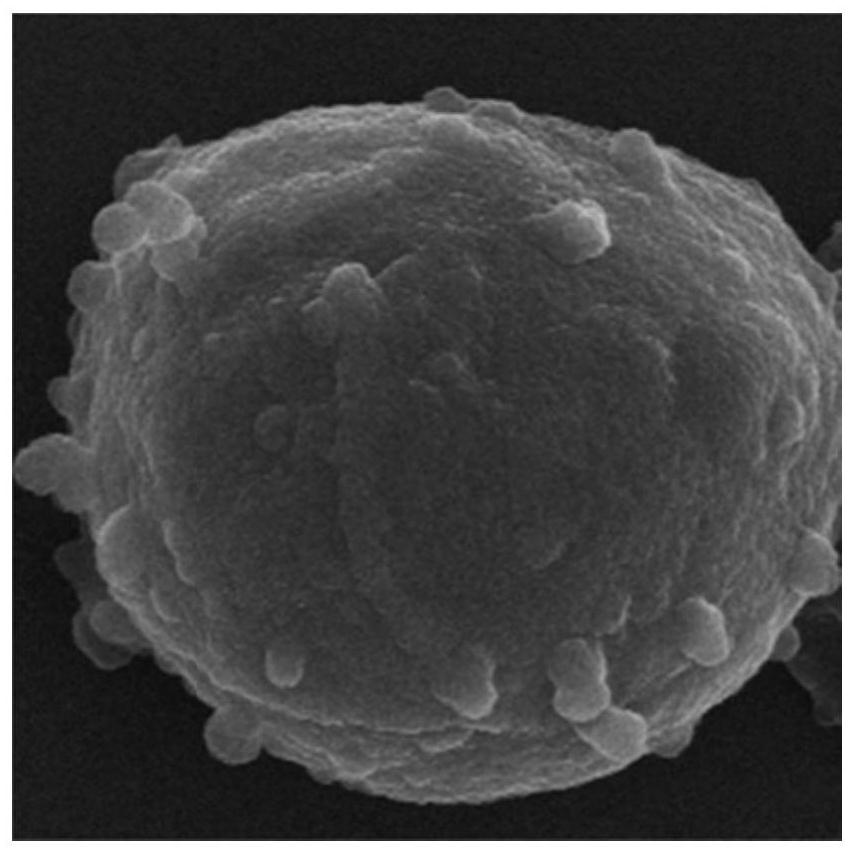
Enterobacter hormaechei bacteriophage YZU.P.A-5 and application thereof
ActiveCN112094820AInhibition formationGrowth inhibitionBiocideOrganic detergent compounding agentsBiotechnologyEnterobacteriales
The invention discloses enterobacter hormaechei bacteriophage YZU.P.A-5 and application thereof. The bacteriophage is identified to have unique genome structure characteristics, and belongs to a novelkind of bacteriophage; and the bacteriophage has been preserved in China Center for Type Culture Collection since June 15, 2020 under the preservation number of CCTCC NO: M 2020206. The bacteriophagedisclosed by the invention can efficiently crack enterobacter hormaechei so as to effectively inhibit enterobacter hormaechei in various matrices; and thus, the bacteriophage can be applied to preparation of a bacterial depressant for controlling enterobacter hormaechei pollution. The bacterial depressant prepared by the bacteriophage can effectively control growth of enterobacter hormaechei in culture media and food matrices, as well as effectively inhibit formation of enterobacter hormaechei mycoderm on surfaces of equipment, utensils and raw material solids; and moreover, the bacterial depressant is simple to prepare and convenient to use, as well as capable of reducing risks of enterobacter hormaechei propagation and food-borne diseases.
Owner:YANGZHOU UNIV
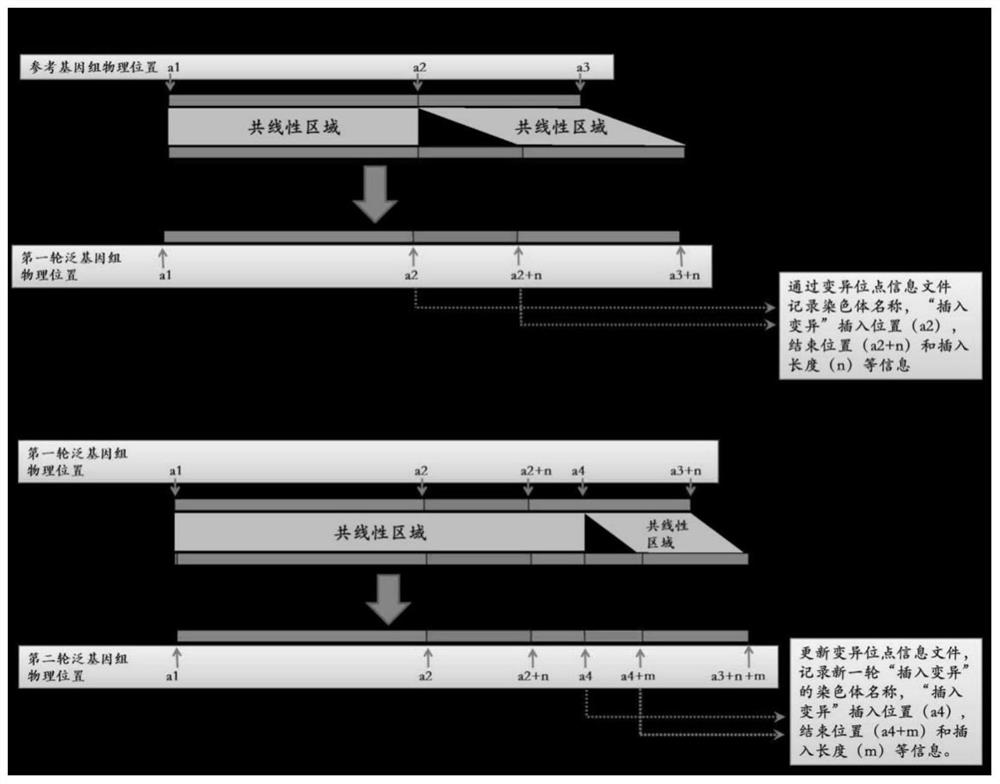
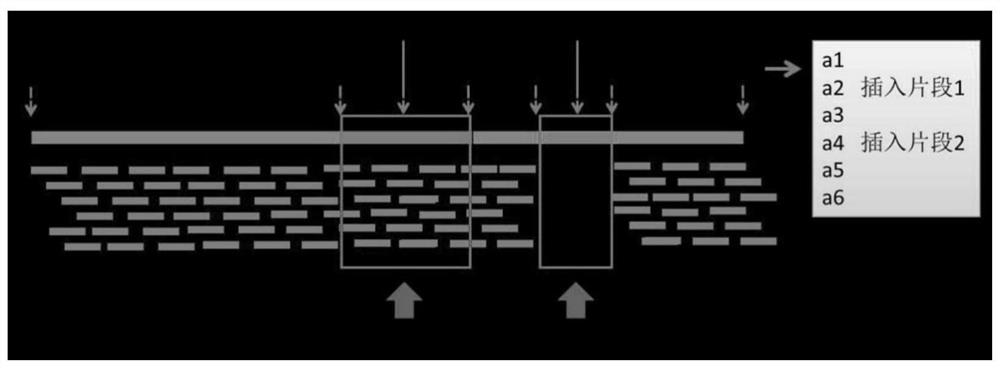
Generic genome construction method and corresponding structural variation mining method thereof
ActiveCN113571131AQuality improvementAchieve large-scale applicationBiostatisticsProteomicsTheoretical computer scienceEngineering
The invention belongs to the technical field of genome data analysis, and particularly relates to a generic genome construction method and a corresponding structural variation mining method thereof, structural variation obtained through genome comparison is put back to a linear genome, meanwhile, a structural variation site information file is added, and an efficient analysis generic genome which is linearized in the form and considers various structure variation forms is constructed. The generic genome not only can capture more brand-new structural variations which are not found by a reference genome, but also better displays the captured structural variations by combining a linearization method with a variation site information file, so that the constructed generic genome is easier to understand and analyze, and is more beneficial to subsequent application; according to the method and the process for analyzing the genome structure variation based on the generic genome and the second-generation sequencing data, a complete program code is compiled, and the efficient and accurate mining of the structure variation based on the relatively low-cost second-generation sequencing data is realized.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI
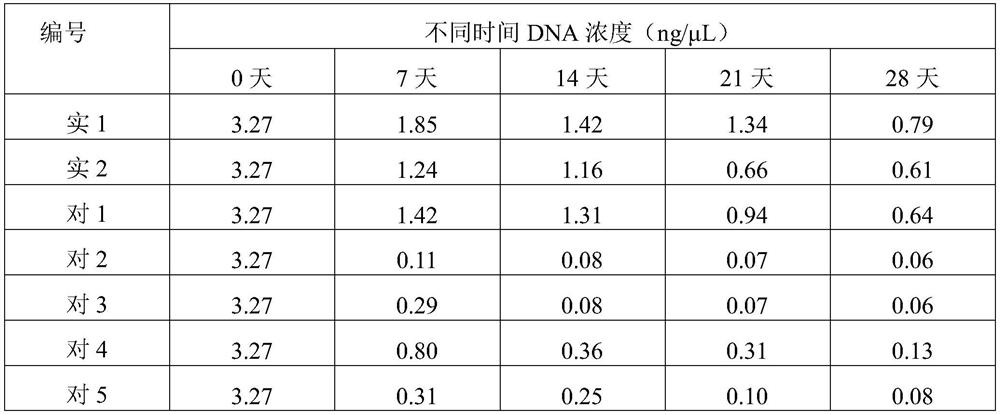
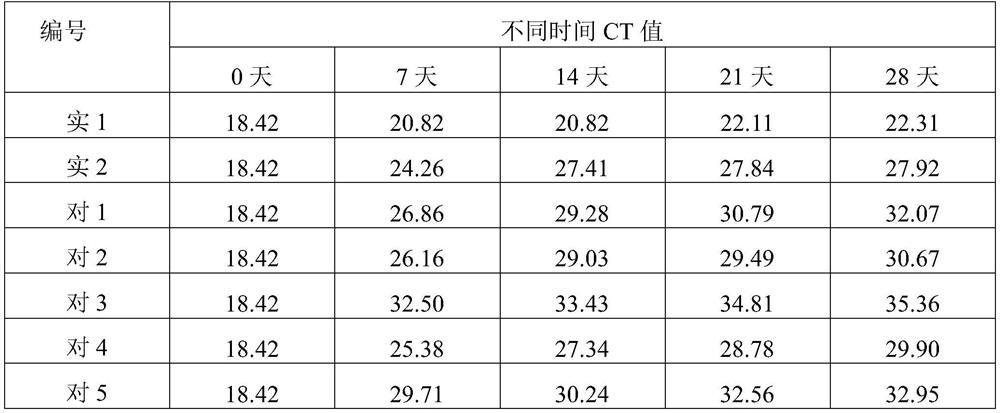
Whole blood genome DNA preservative as well as preparation method and application thereof
PendingCN112575063AGuaranteed stabilityAvoid lossMicrobiological testing/measurementAnticoagulantGenomic information
The invention discloses a whole blood genome DNA preservative as well as a preparation method and application thereof, belonging to the field of biotechnology. The whole blood genome DNA preservativecomprises the following components in parts by weight: 1 to 200 parts of an anticoagulant, 1 to 200 parts of a blood cell stabilizer, 1 to 200 parts of a reactive cell encapsulant, 0.1 to 200 parts ofa pH regulator, 0.1 to 200 parts of an ion concentration regulator and 500 to 1000 parts of RNase-free purified water. According to the whole blood genome DNA preservative, the integrity of a genomeDNA structure and the stability of physicochemical properties can be ensured for a long time at room temperature, and the loss or change of genome information is avoided; besides, the storage time andlong-distance transportation of the genome sample at room temperature can be prolonged. The genome DNA long-term normal-temperature preservation and extraction method can provide powerful guarantee for disease prevention, disease treatment, forensic criminal investigation and the like.
Owner:SOUTH CHINA UNIV OF TECH
Genome DNA room-temperature preservation card as well as manufacturing method and application thereof
The invention discloses a genome DNA room-temperature preservation card and a manufacturing method and application thereof, and belongs to the technical field of biology. The genome DNA room-temperature preservation card comprises a medium with liquid absorption performance and a cell preservation liquid premixing agent. According to the genome DNA room-temperature preservation card, the integrity of a genome DNA structure and the stability of physicochemical properties can be ensured for a long time at room temperature, and the loss or change of genome information is avoided; and the storage time of the genome sample at room temperature can be prolonged, and long-distance transportation can be realized. The long-term normal-temperature preservation and extraction method of the genome DNA can provide powerful guarantee for mutation monitoring, disease prevention and disease treatment of somatic cells.
Owner:SOUTH CHINA UNIV OF TECH
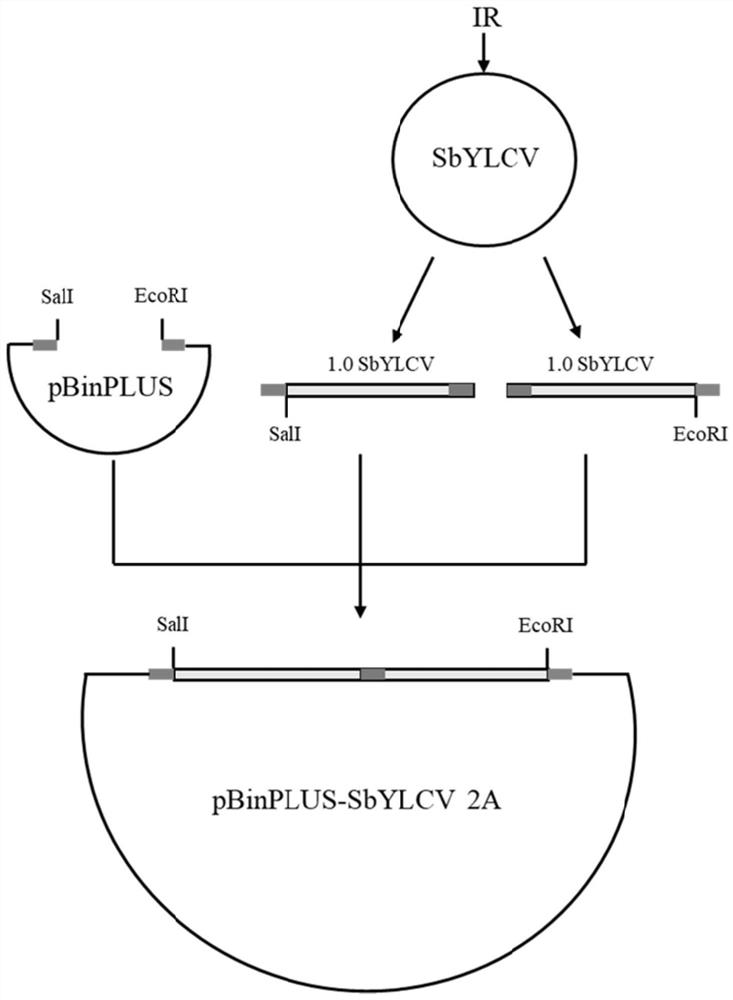
Soybean yellow leaf curl virus infectious cloning vector as well as construction method and application thereof
The invention discloses a soybean yellow leaf curl virus infectious cloning vector as well as a construction method and application thereof. The method specifically comprises the following steps: on the basis of obtaining the complete sequence of the genome of the soybean yellow leaf curl virus, constructing two forward repeated genomes of the soybean yellow leaf curl virus on a binary expression vector pBinPLUS by a gene recombination method; transforming the recombinant vector into an agrobacterium strain EHA105 through electric shock, so that the obtained virus vector can successfully infect plants such as soybeans and nicotiana benthamiana. The invention provides a mature system for researching the genome structure and function of the virus and the interaction between the virus and the host.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Salmonella detection method and application thereof
InactiveCN108531551ALow costEasy to detectMicrobiological testing/measurementAgainst vector-borne diseasesConserved sequencePolystyrene
The invention relates to a salmonella detection method. The method includes the steps of: 1, preparing a probe molecule: according to the genome structure and conserved sequence of salmonella, selecting the invA gene of salmonella as the detection target fragment, according to the salmonella invA gene sequence disclosed by GenBank:EU348365, designing a probe molecule, which has a sequence of 5'-GCGAGTGGTAAATTATCGATGAAGT-3', with the 5' end being labeled with a reporter fluorophore; and 2, mixing a polystyrene nanosphere solution with the probe molecule prepared by step 1, then adding a to-be-detected sample, and performing fluorescence detection.
Owner:江西华牧生物科技有限公司
Lactobacillus rhamnosus YZULr026 capable of efficiently degrading purine and application of lactobacillus rhamnosus YZULr026
ActiveCN112725216AImprove acid resistanceGood resistance to bile saltsBacteriaLactobacillusBiotechnologyPurinergic Effects
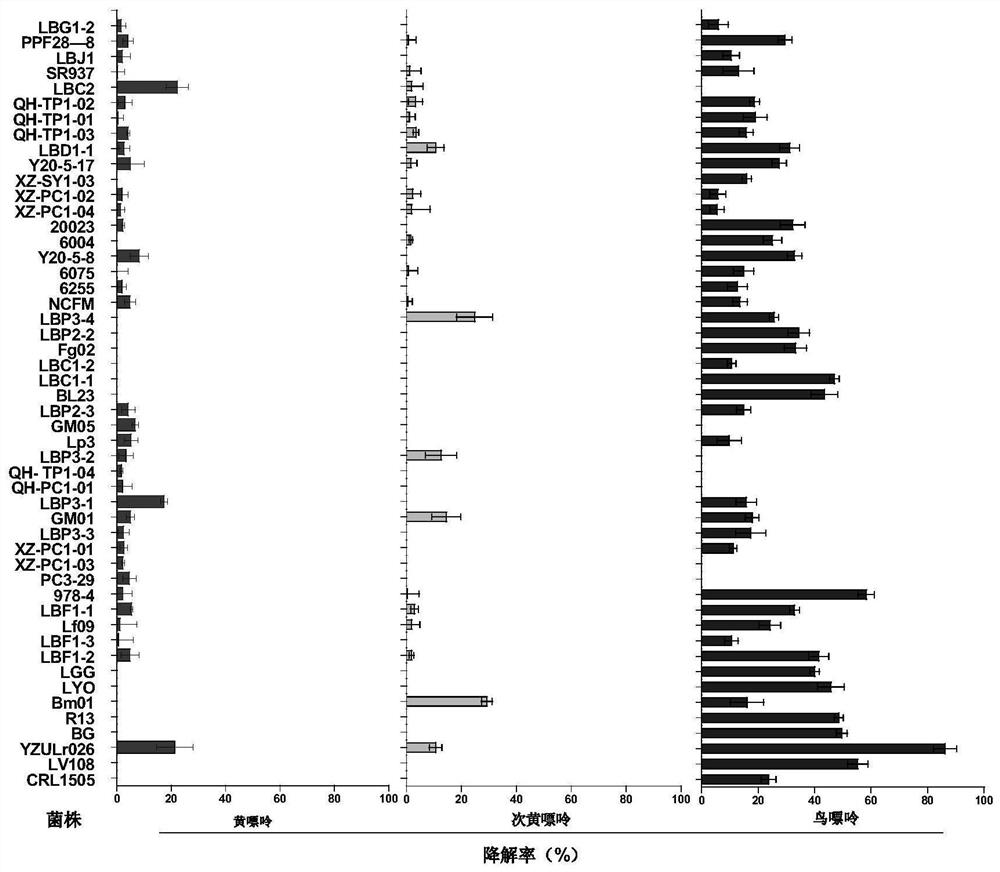
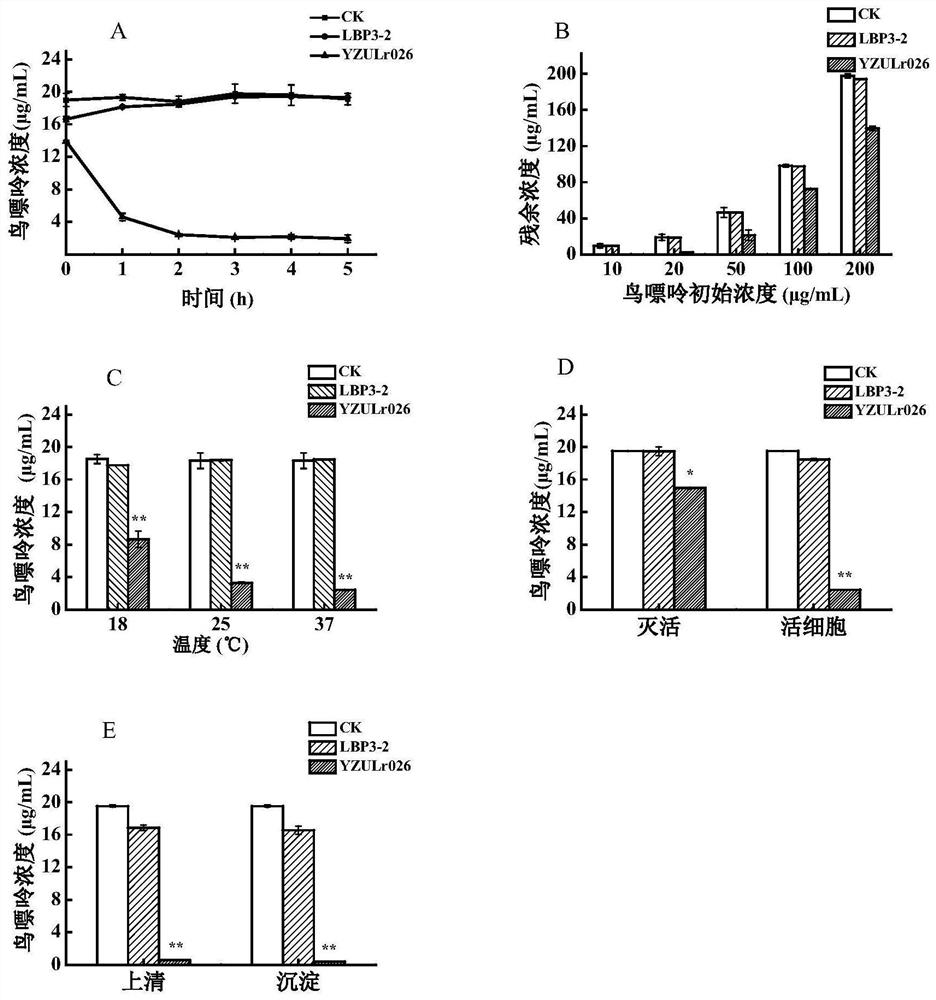
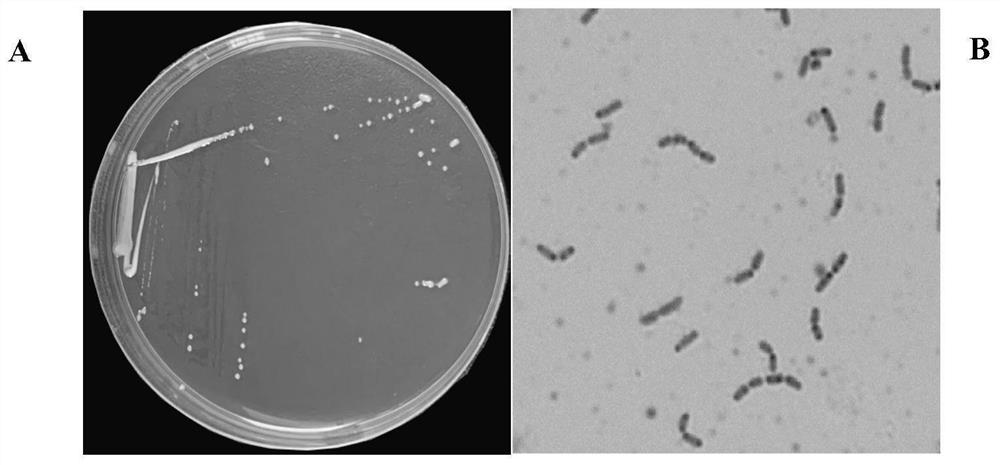
The invention discloses lactobacillus rhamnosus YZULr026 capable of efficiently degrading purine and an application of the lactobacillus rhamnosus YZULr026, the lactobacillus rhamnosus YZULr026 is preserved in China Center for Type Culture Collection (CCTCC), the preservation number is CCTCC NO: M 2020359, and the preservation date is July 27th, 2020. The lactobacillus rhamnosus YZULr026 disclosed by the invention has good acid resistance and bile salt resistance; has efficient purine degradation capacity, and the guanine degradation rate reaches up to 87.85% after the strain and guanine are incubated in a 37 DEG C pure water system for 2 hours; YZULr026 has a unique genome structure, the highest homology of a gene sequence Gene264 for encoding alpha galactosidase and a published sequence is 93.38%, and 28 base differences exist; the strain YZULr026 is used for preparing a microbial preparation, and has a wide application prospect when being used for reducing the purine content in food in vitro and vivo and preventing hyperuricemia.
Owner:YANGZHOU UNIV
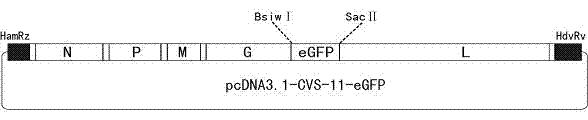
Recombinant virus of standard attack strain CVS-11 and preparation method thereof
InactiveCN102864126AStrong specificityImprove economyMicroorganism based processesViruses/bacteriophagesAntigenStaining
The invention provides a recombinant virus of standard attack strain CVS-11 and a preparation method thereof. The invention establishes a reverse genetic manipulation system for rabies virus international standard attack strain CVS-11, provides an effective platform for further modifying or reconstructing the CVS-11 genome structure, and lays foundation for researching pathopoiesis mechanism, antigen site, virulence determined site and the like of rabies virus on the molecular level. On the basis of the CVS-11 reverse genetic manipulation system, the invention establishes a recombinant virus rCVS-11-eGFP for saving international standard attack strain CVS-11. The rCVS-11-eGFP expresses eGFP in the proliferation process, and can be used for directly observing the proliferation state and diffusion of the virus. By utilizing the characteristics of no toxicity, capability of self-luminescence without dependence on any accessory factor or substrate, and the like, the rCVS-11-eGFP is used as a detection antigen for determining the anti-rabies virus neutralizing antibody titer, and no operations, such as stain fixation and the like, are needed in the result determination process, so that the invention is economic and convenient.
Owner:MILITARY VETERINARY RES INST PLA MILITARY MEDICAL ACAD OF SCI
Staphylococcus aureus phage SapYZUalpha and application thereof
The invention discloses a staphylococcus aureus bacteriophage SapYZUalpha and application thereof, the staphylococcus aureus bacteriophage SapYZUalpha is identified to have unique genome structural characteristics, belongs to a novel bacteriophage and is preserved in the China Center for Type Culture Collection, the preservation time is January 5, 2022, and the preservation number is CCTCC NO: M2022023. The bacteriophage capable of efficiently cracking staphylococcus aureus can effectively inhibit staphylococcus aureus in various matrixes, and can be applied to preparation of a bacteriostatic agent for controlling staphylococcus aureus pollution. The bacteriostatic agent prepared by the invention can effectively control the growth of staphylococcus aureus in a culture medium and a food matrix, can effectively inhibit the formation of staphylococcus aureus pellicles on the surfaces of equipment, vessels and raw material solids, is simple to prepare and convenient to use, and can reduce the propagation of staphylococcus aureus and the risk of causing food-borne diseases.
Owner:YANGZHOU UNIV
A method and kit for determining genome instability based on next-generation sequencing technology
ActiveCN111676277BHigh detection sensitivityImprove capture efficiencyMicrobiological testing/measurementBiostatisticsGenomic StabilityGenome instability
Owner:臻和(北京)生物科技有限公司 +1
A kind of attenuated mutant strain of foot-and-mouth disease virus and its preparation method and application
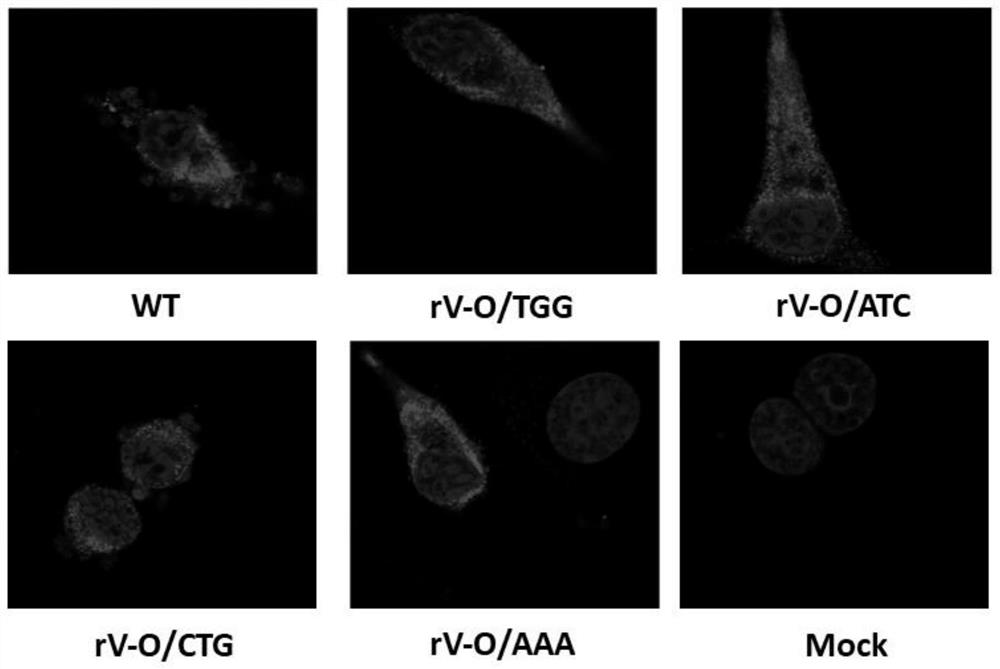
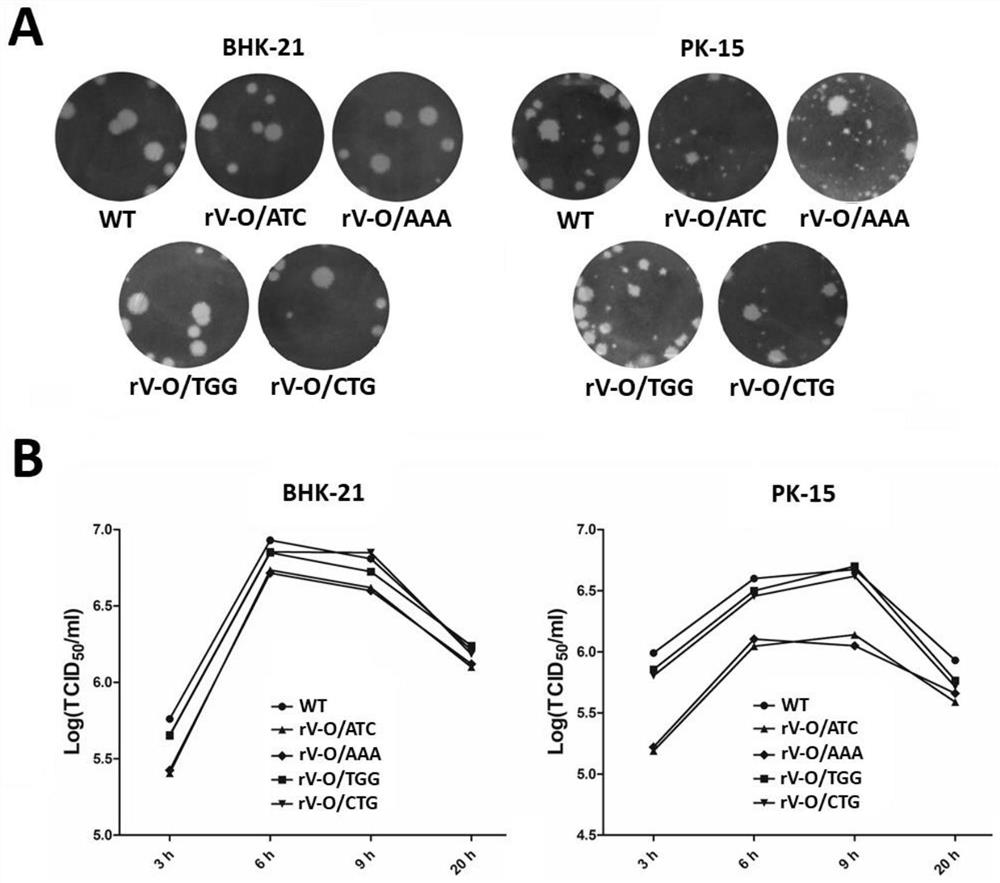
ActiveCN111944770BHigh genetic stabilityReduce pathogenicitySsRNA viruses positive-senseViral antigen ingredientsDiseaseVaccine antigen
The invention provides a foot-and-mouth disease virus attenuated mutant strain and its preparation method and application, belonging to the technical field of biotechnology and biological products. The attenuated mutant strain of foot-and-mouth disease virus is to mutate the Lb AUG in the parental foot-and-mouth disease virus leader protein gene to ATC or AAA. The level is reduced, the pathogenicity to suckling mice is significantly weakened, and this mutation type can be inherited stably. Based on the genome structure of the OZK / 93‑08 vaccine strain, the invention mutates the second start codon AUG in the lead protein gene to ATC or AAA, which can significantly attenuate the O-type foot-and-mouth disease virus. The method of the attenuated vaccine strain will provide a safer platform for the production of the FMD O-type inactivated vaccine antigen.
Owner:LANZHOU INST OF VETERINARY SCI CHINESE ACAD OF AGRI SCI
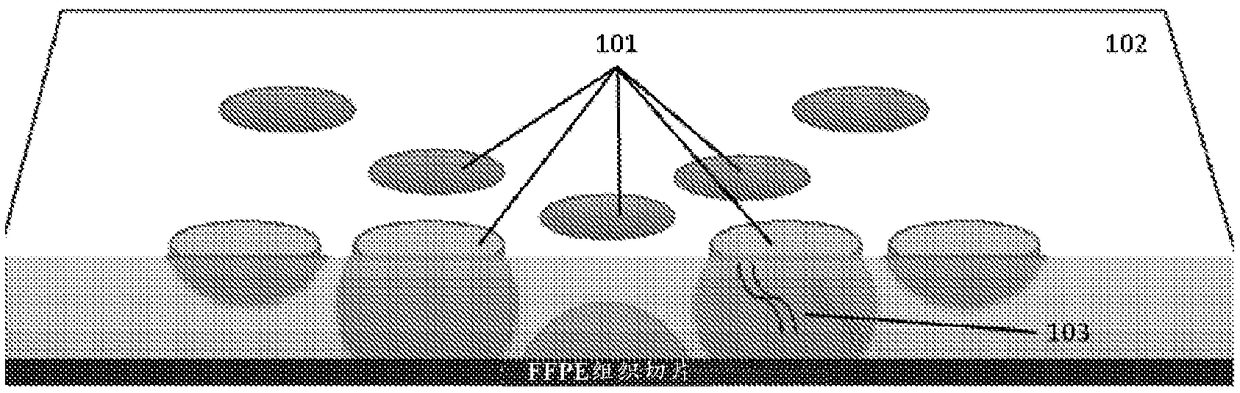
Recovering long-range linkage information from preserved samples
The disclosure provides methods to isolate genome or chromosome level structural information from preserved samples. In some cases, samples preserved under conditions where long-range nucleic acid information is believed to be irreparably lost, such as FFPE samples, are treated to recover nucleic acid-protein complexes stabilized as part of the sample preservation process. The complexes are processed so as to recover information regarding which nucleic acids are bound to a common complex, and the information is used to recover genomic structural information.
Owner:DOVETAIL GENOMICS
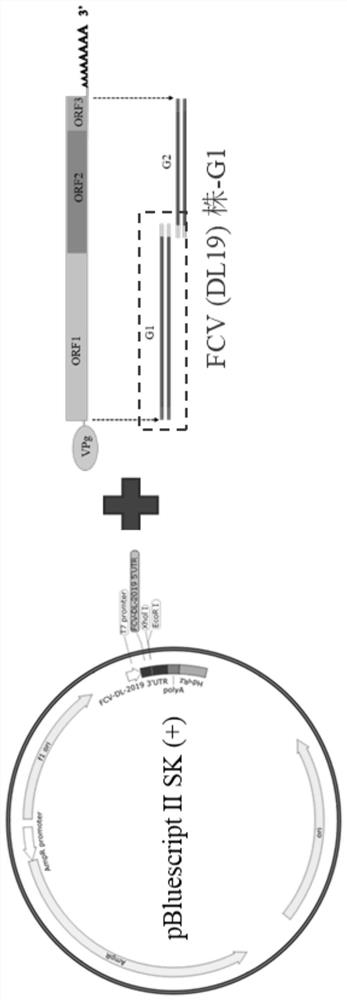
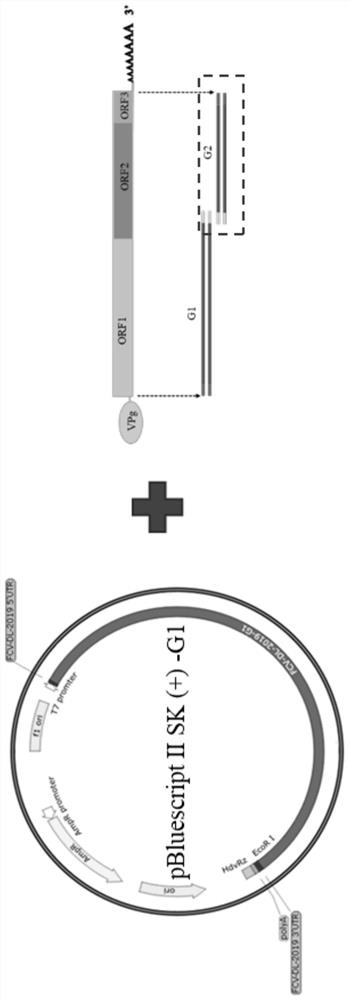
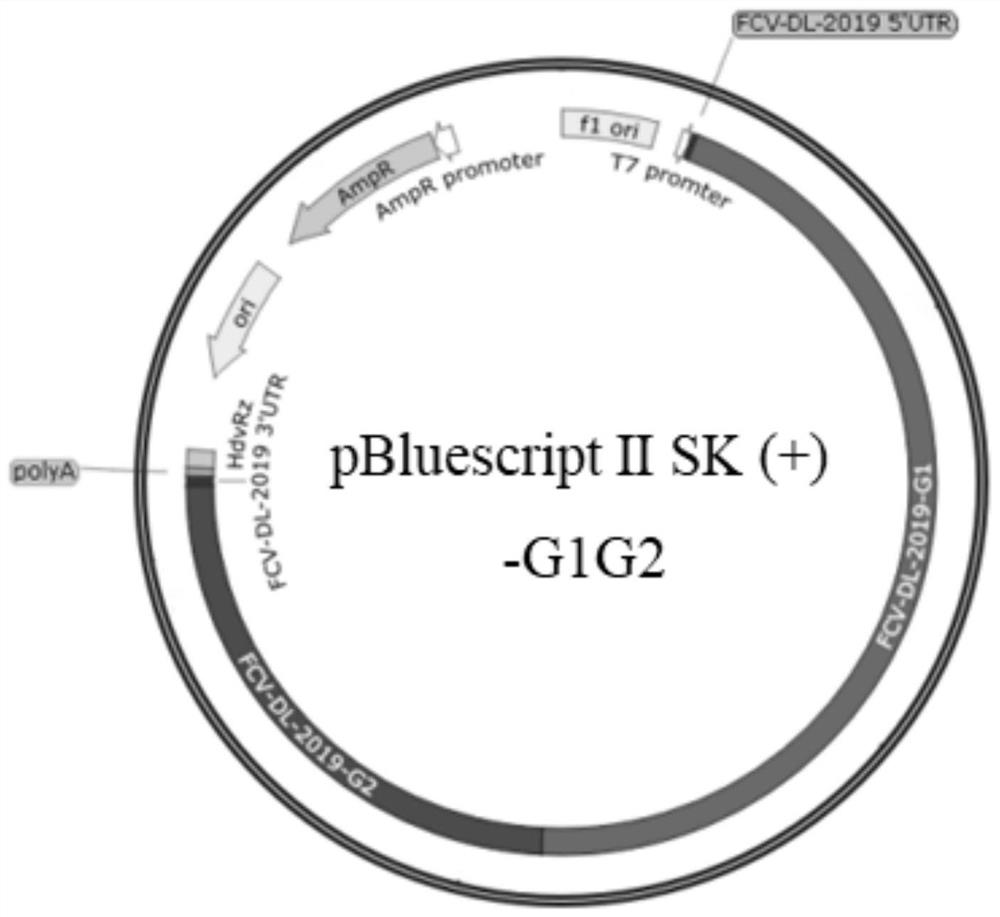
Feline calicivirus reverse genetic platform and construction method thereof
PendingCN113493804ASsRNA viruses positive-senseVirus peptidesFeline calicivirus infectionGenome structure
The invention provides a feline calicivirus reverse genetic platform and a construction method thereof. A feline calicivirus DL19 strain genome is divided into two segments to be connected to a modified pBluescript II SK (+) vector, a feline calicivirus DL19 strain full-length infectious clone plasmid capable of serving as the reverse genetic platform is constructed, and a foundation is laid for subsequent virus rescue and genome structure research.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
cDNA full-length sequence of zebrafish tmem132e gene and application thereof
InactiveCN104818282AUnderstanding Genome StructureFermentationAnimals/human peptidesOpen reading frameNucleotide
Belonging to the technical field of genetic engineering, the invention specifically relates to the cDNA full-length sequence of zebrafish tmem132e gene and application thereof. The invention provides the full-length sequence of zebrafish tmem132e gene, the DNA full length is 3222bp, the cDNA full-length sequence contains 3219bp open reading frame, and the nucleotide sequence is shown as SEQ ID NO.1. The invention also provides a zebrafish tmem132e gene encoded protein molecule. With the zebrafish tmem132e gene provided by the invention, purified protein or oligonucleotide with bioactivity can be artificially synthesized according to the gene sequence or can be obtained by genetically modified biosynthesis, thus being conducive to understanding the genomic structure of the zebrafish tmem132e gene and analyzing the gene expression and protein functions.
Owner:SHANDONG UNIV
Identification of genomic structural variants using long read sequencing
Provided herein are systems and methods for detecting genomic structural variants using non-applied gene editing sample preparation followed by long read sequencing.
Owner:BLACK HAWK GENOMICS LLC
Infective cloning vector of rape mosaic virus as well as construction method and application of infectious cloning vector
ActiveCN114369617ASsRNA viruses positive-senseMicrobiological testing/measurementPathogenicityGenetic engineering
The invention belongs to the technical field of gene engineering, and particularly relates to a rape mosaic virus infectious cloning vector as well as a construction method and application thereof. The technical problem to be solved by the invention is to provide a new choice for screening resistant varieties. According to the technical scheme, the infectious cloning vector for the rape mosaic virus is derived from a strain Agrobacterium tumefaciens YoMV-GD, and the preservation number of the infectious cloning vector is GDMCC No: 62229. A new strain of rape mosaic virus is screened, a rape mosaic virus infectious cloning vector is constructed, the vector is converted into agrobacterium, and then the rape mosaic virus can be inoculated by utilizing an agrobacterium injection method. And a foundation is laid for screening rape mosaic virus resistant materials and varieties and researching a rape mosaic virus genome structure, pathogenicity and interaction with hosts.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
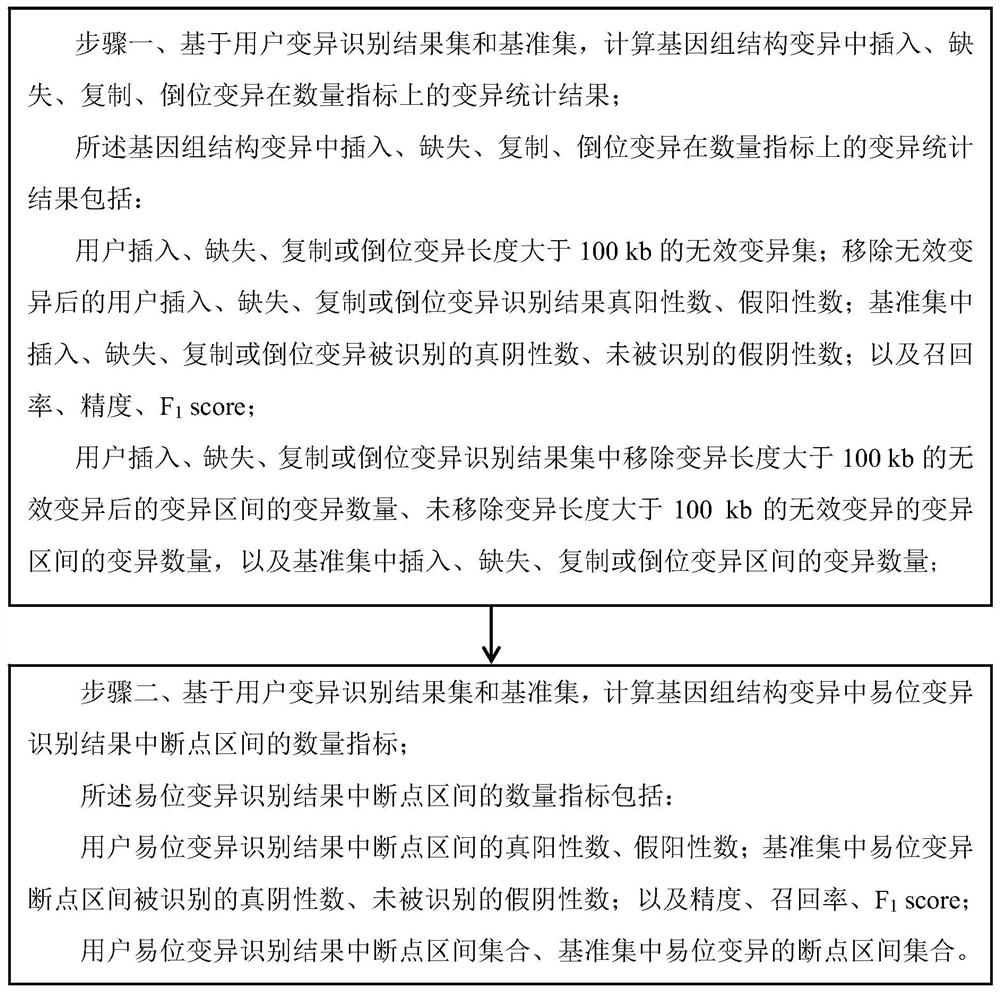
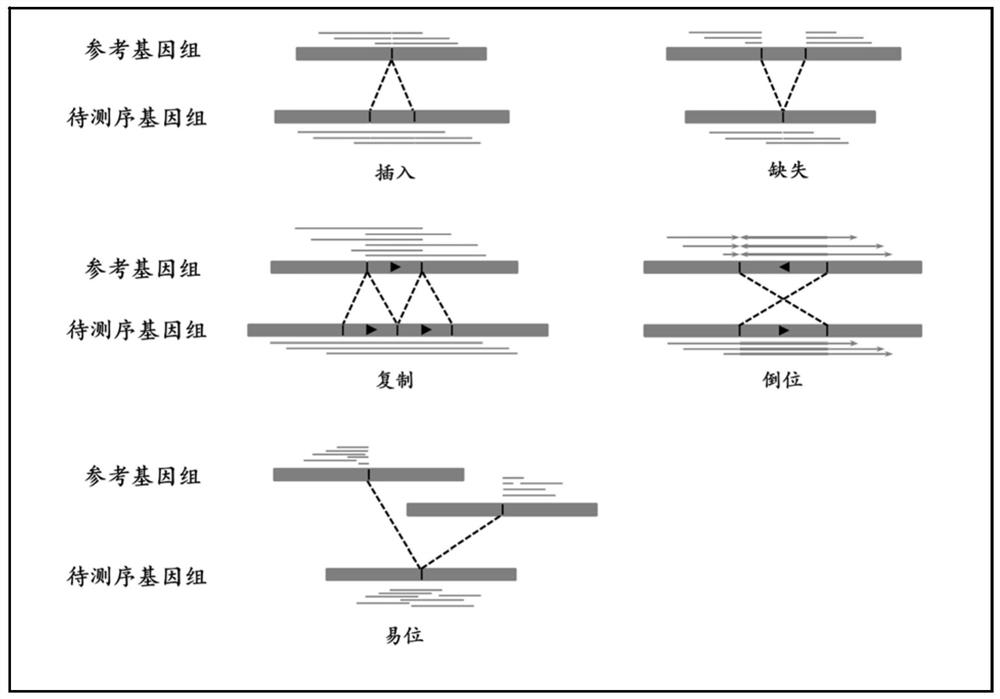
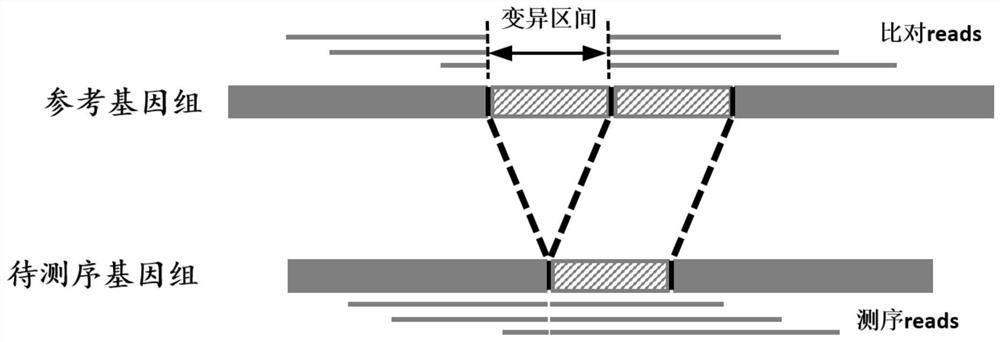
A Benchmark Set-Based Method for Genome Structural Variation Performance Detection
ActiveCN113628680BEasy to detect and analyzeDetailed analysisProteomicsGenomicsData miningStructural variant
The invention relates to a genome structure variation performance detection method based on a benchmark set, and the invention relates to a genome structure variation performance detection method based on a benchmark set. The purpose of the present invention is to solve the problems that the existing genome structure variation detection methods are not comprehensive enough and lack a public variation recognition result detection method. The specific process of a genome structure variation performance detection method based on a benchmark set is as follows: step 1, based on the user variation identification result set and the benchmark set, calculate the variation statistics of insertion, deletion, duplication, and inversion variation in quantitative indicators in the genome structure variation Result; step 2, based on the user variation identification result set and the reference set, calculate the quantity index of the break point interval of the translocation variation identification result in the genome structure variation. The invention is used in the field of genome structure variation performance detection.
Owner:HARBIN NORMAL UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com