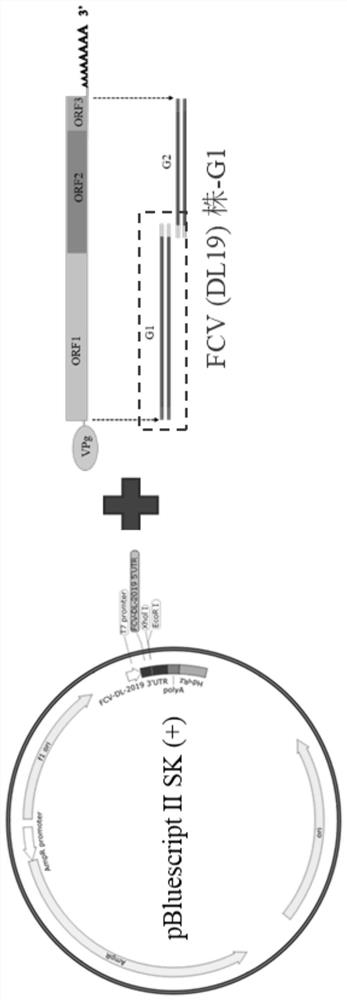
Feline calicivirus reverse genetic platform and construction method thereof
A feline calicivirus and reverse genetic technology, applied in the field of molecular biology, can solve the problems of time-consuming, labor-intensive and complicated process of PCR cloning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1 sequence analysis and the design of primer
[0026] According to the FCV (DL19) strain sequence, DNA MAN and DNA Star were used to design primers, and the online biological software NovoPro (https: / / www.novopro.cn / tools / rest_summary.html) was used to limit the FCV (DL19 strain) sequence 4505 XholⅠ restriction sites were selected for homology arm primer design. The primers were synthesized by Beijing Huada Biotechnology Co., Ltd., and the primer sequences are shown in Table 1.
[0027] Table 1 FCV genome primer sequences
[0028]
[0029]
Embodiment 2
[0030] The extraction of embodiment 2 viral total RNA and the acquisition of cDNA
[0031] The extraction of total virus RNA was carried out according to the instructions of Trizol Reagent, and the specific steps were as follows:
[0032] 1) Take out the sample stored in the -80°C refrigerator, and wait for the sample to completely melt and return to room temperature;
[0033] 2) Add chloroform according to 200 μL chloroform / mL Trizol, vortex and oscillate for 15 seconds to fully mix the sample, and let stand at room temperature for 3 minutes;
[0034] 3) After centrifugation at 12,000rpm for 15min at 4°C, the sample was divided into three layers, and the uppermost aqueous phase containing RNA (about 450μL) was slowly drawn into a new 1.5mL EP tube, and pre-cooled isopropanol was added at a ratio of 1:1. , mix the liquid gently and place it on ice for 10 minutes;
[0035] 5) Take out the mixed liquid and centrifuge at 12,000 rpm at 4°C for 10 minutes. After discarding the su...
Embodiment 3
[0044] Example 3 Expression of eGFP recombinant vector construction
[0045] In order to further verify whether the pBluescript II SK (+) modified vector system is available, the whole eGFP genome was inserted after the pBluescript II SK (+) modified vector FCV 5'UTR to construct a recombinant vector expressing eGFP, and to verify the rescue system minigenome system.
[0046] Expression of eGFP recombinant vector transfection:
[0047] Inoculate BHK-T7 into a 6-well cell plate, and when the cells cover about 80% of the bottom of the well, use Lipofectamine 2000 to transfect the constructed pBluescript II SK(+)-eGFP, and observe the eGFP gene expression under a fluorescent microscope 48 hours after transfection Condition.
[0048]The constructed pBluescript II SK(+)-eGFP was transfected with Lipofectamine 2000, and 48 hours after transfection, the expression of the green fluorescent protein reporter gene was observed under a fluorescent microscope. As shown in Figure 2, the ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com