Lactobacillus rhamnosus YZULr026 capable of efficiently degrading purine and application of lactobacillus rhamnosus YZULr026
A technology of Lactobacillus rhamnosus and purine, which is applied to the strain of Lactobacillus rhamnosus and its application field in degrading purine, can solve the problem that the strains of purine substances are rare, etc., and achieve the ability to quickly and efficiently degrade guanine, good acid resistance, Effect of reducing guanine absorption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
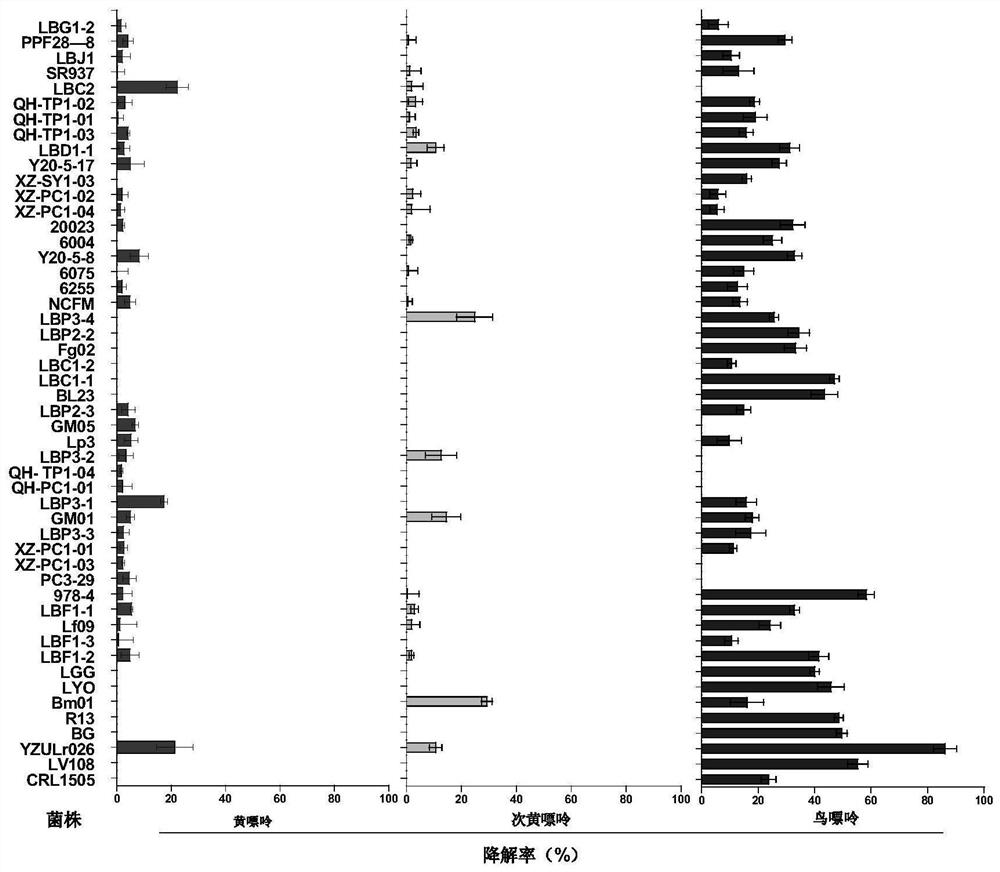
[0035] Screening of high-efficiency purine-degrading strains
[0036] 50 strains isolated from Tibetan traditional fermented food (yogurt, kimchi, sweet fermented grains) were activated 3 times, inoculated with MRS medium, anaerobically cultured at 37°C for 24 hours, and 1 mL of 1×10 7 The CFU / mL bacterial culture solution was centrifuged at 8000r / min at 4°C for 10min to collect the bacterial cells. And resuspend and wash the bacteria 2-3 times with 1mL sterile ultrapure water. Add 750 μL of 20 μg / mL xanthine, hypoxanthine, and guanine reaction solution to the bacteria respectively, add ultrapure water to the control sample, vortex and mix well, incubate at 37°C and 120r / min for 2 hours, and use liquid chromatography to determine Purine content in the sample.
[0037] The specific measurement process is as follows: sampling after cultivation, centrifugation at 8000r / min, 4°C for 10min to take the supernatant; filtering the supernatant through a 0.22μm microporous membrane, a...
Embodiment 2
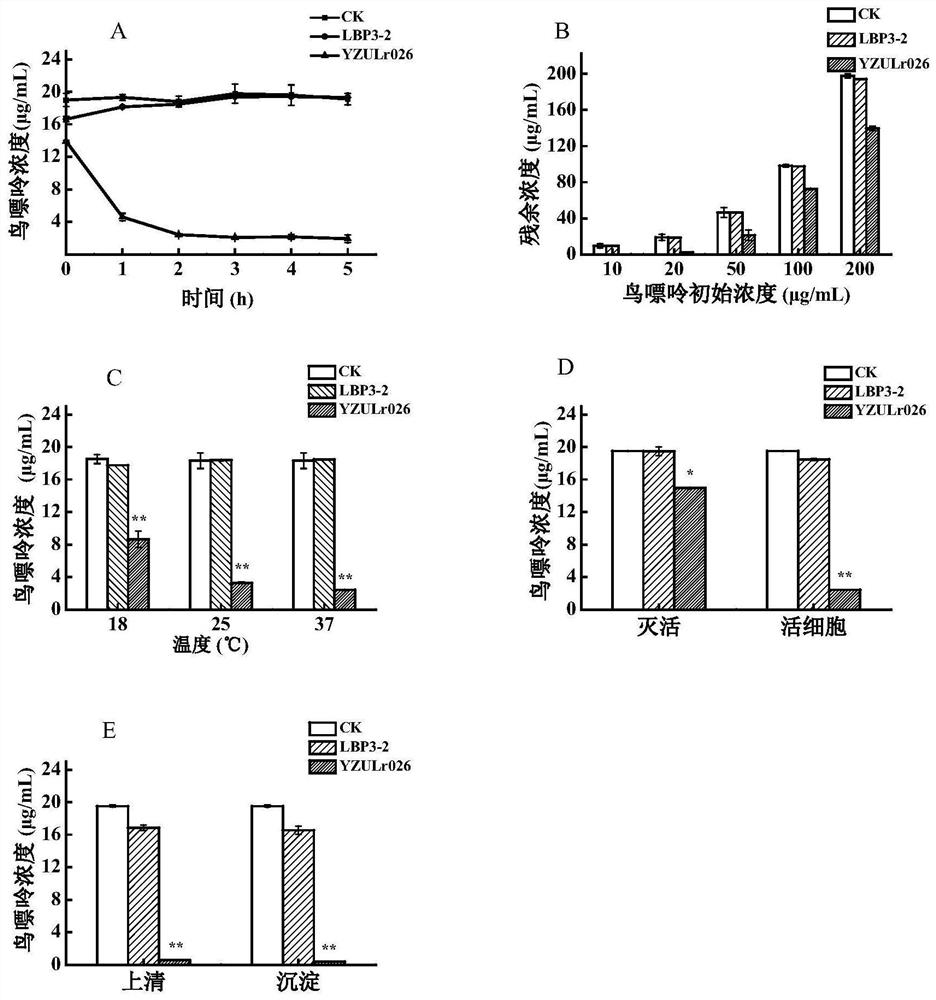
[0040] Degradation characteristics of guanine in strain YZULr026
[0041] 100 μL of strain YZULr026 preservation solution (logarithmic phase) was inoculated into 5 mL of MRS liquid medium, cultured anaerobically at 37 °C for 24 h, and passed three times in a row. The culture solution of the activated strain was inoculated in fresh MRS liquid medium with an inoculum size of 2% by volume, and cultured anaerobically until the stationary phase. Adjust the concentration of YZULr026 bacterial solution to 10 7 CFU / mL Take 1mL in a centrifuge tube, centrifuge at 8000r / min, 4°C for 10min, collect the bacteria, wash 2-3 times with ultrapure water. (1) Add 750 μL of 20 μg / mL purine reaction solution to the cells, vortex and mix well, incubate at 37°C and 120 r / min for 5 h, and take samples for analysis at 0, 1, 2, 3, 4, and 5 h; (2) Add purine reaction solutions with concentrations of 10, 20, 50, 100, and 200 μg / mL to the bacteria, vortex and mix; incubate at 37°C for 2 hours, and take...
Embodiment 3
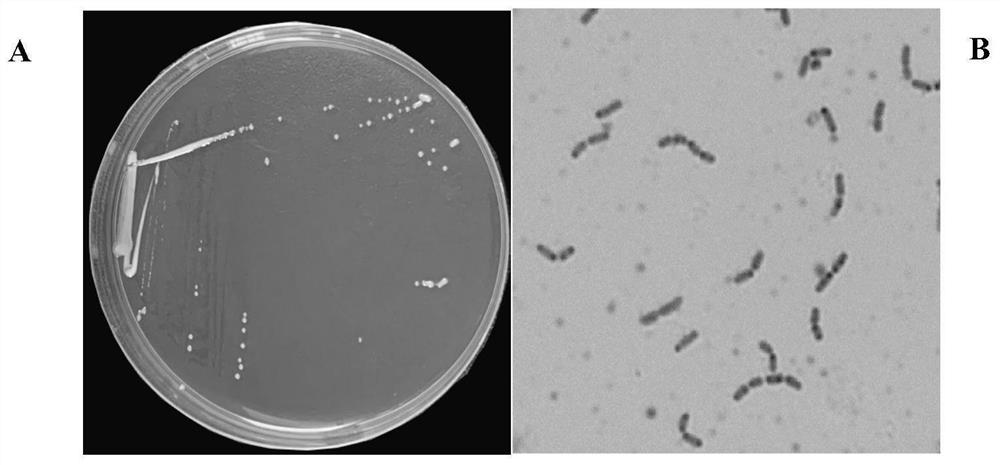
[0044] Physiological and biochemical identification of strain YZULr026
[0045] The obtained strain YZULr026 was inoculated with MRS solid medium by streaking, anaerobically cultured at 37°C for 24 hours, picked a single colony smear on the plate, fixed, added crystal violet staining solution dropwise, stained for 1min, washed with water; added iodine solution dropwise, React for 1 min, wash with water; add 95% ethanol dropwise to decolorize for 15-20 s, wash with water; counterstain with safranin for 1 min, wash with water; examine under microscope and record the cell morphology of the strain. At the same time, single colonies were picked for catalase test, indole, hydrogen sulfide and ammonia, nitrate reduction, arginine hydrolysis, and sugar fermentation tests.
[0046] Implementation results show that the strain is a Gram-positive bacillus ( image 3 ), catalase negative, does not produce indole, hydrogen sulfide and ammonia, does not reduce nitrate, does not hydrolyze ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com