Method for determining size of single-cell eukaryotic algae genome
A measurement method and genome technology, applied in the field of microalgae species, can solve the problems of reducing the fluorescence intensity of PI and affecting the accuracy of genome measurement by flow cytometry, achieving high operability and accuracy, and improving the effect of staining
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
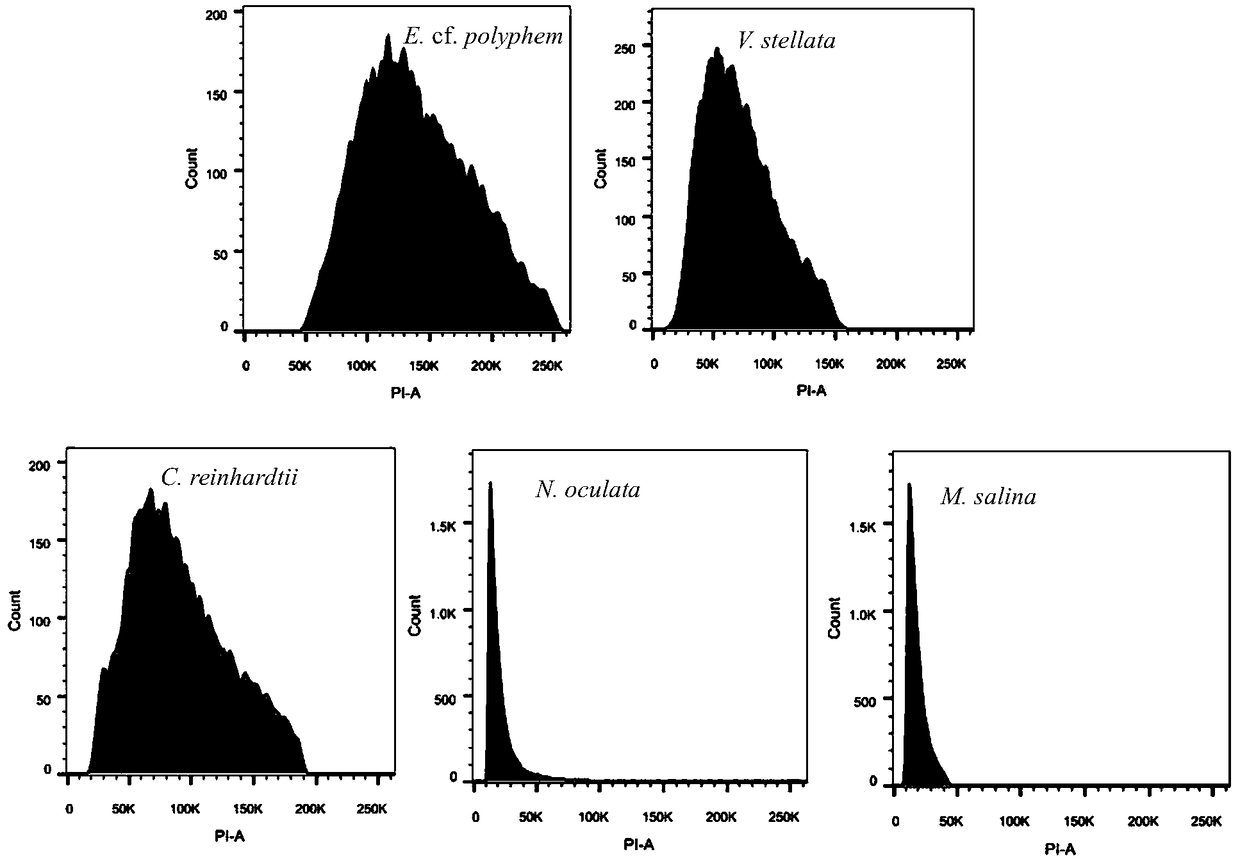
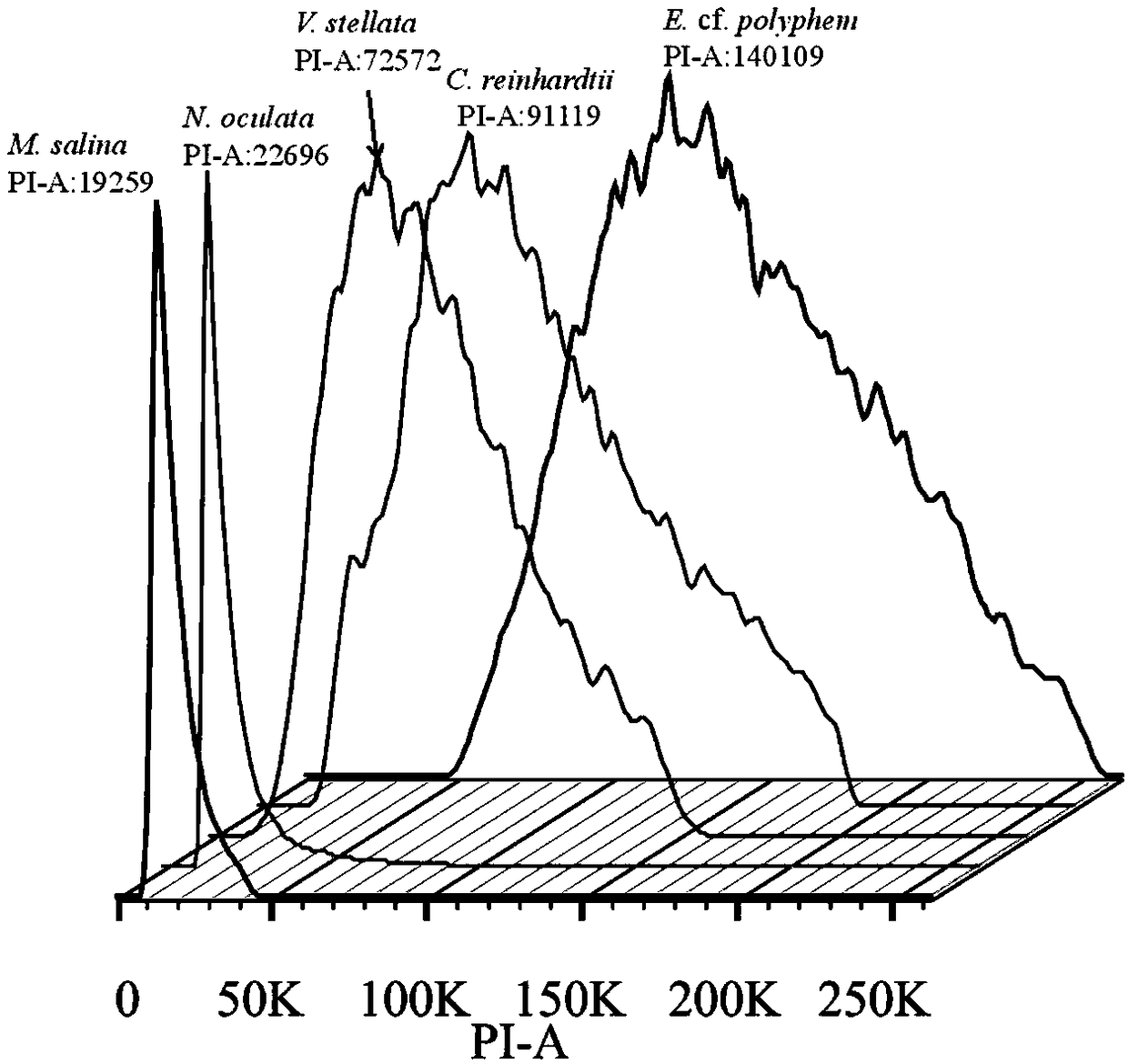
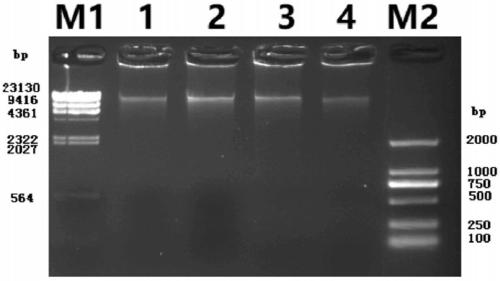
[0046] Taking two unicellular eukaryotic microalgae with unknown genomes, E.cf.polyphem and V.stellata as examples, and Chlamydomonas reinhardtii (C.reinhardtii) as examples The first internal reference, N.oculata and M.salina were the second internal reference, and the genomes of the three known species were mutually calibrated. 5 microalgae cultures (about 50mL, 1×10 5 cells / mL) after centrifugation, add 50mL methanol, magnetically stir, 1-2h each time, centrifuge at 3000rpm to remove the supernatant, repeat the extraction 3-5 times until the cells become white; add 5mL Lysisbuffer LB01 to re- Suspend the cells, process and lyse the nuclei for 0.5h, obtain the cell nucleus suspension, and keep it on ice for later use; take 1mL of the cell nucleus suspension, add 50μL PI (1mg / mL), 50μL RNase (1mg / mL) and 5μL β-mercaptoethanol, mix well Stain in the dark for 0.5 h at 4°C; take 300 μL of the stained cell suspension, add it to a flow-type special round-bottom test tube (Fluores...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com