A Benchmark Set-Based Method for Genome Structural Variation Performance Detection
A detection method and technology for structural variation, applied in the fields of genomics, proteomics, instruments, etc., can solve the problems of lack of detection methods for variation identification results, insufficient detection methods for genome structure variation, etc., to achieve convenient data processing and analysis, fast Detection method, the effect of speeding up the pace
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
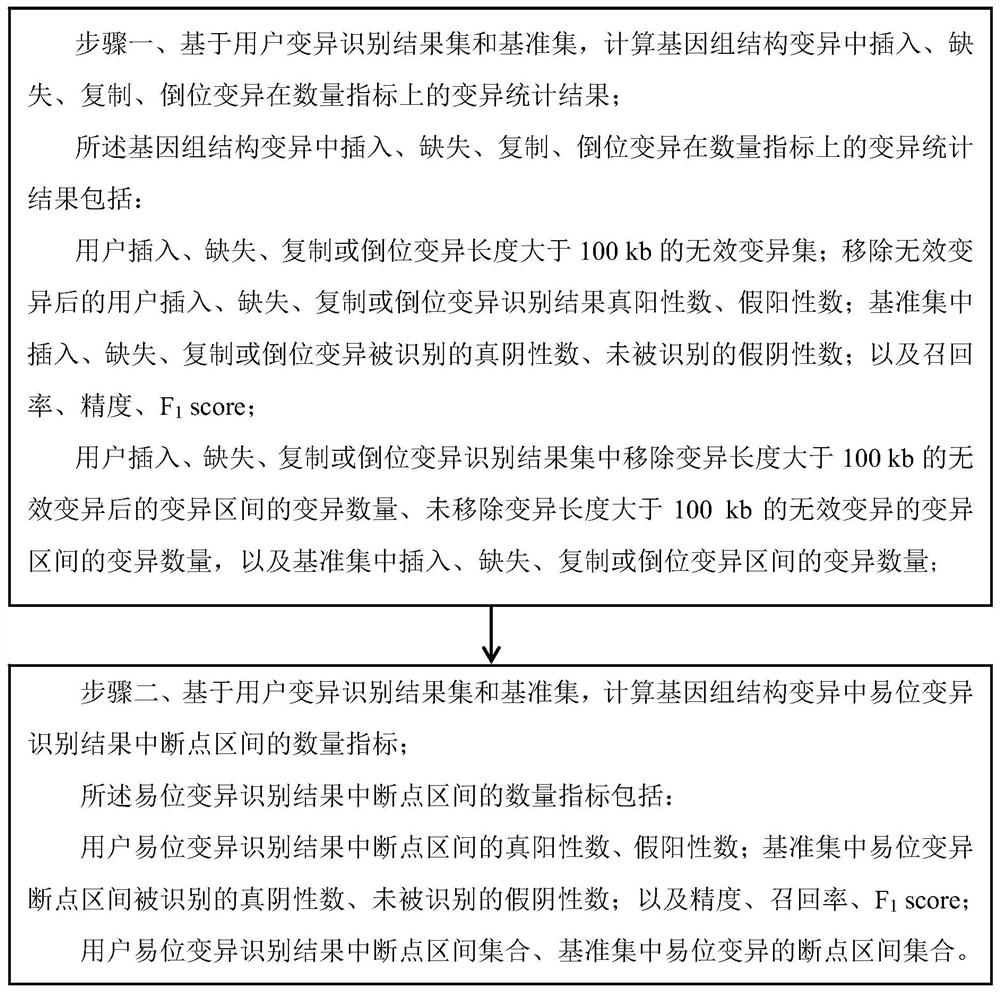
[0044] Embodiment 1: The specific process of a benchmark set-based genome structure variation performance detection method in this embodiment is as follows:
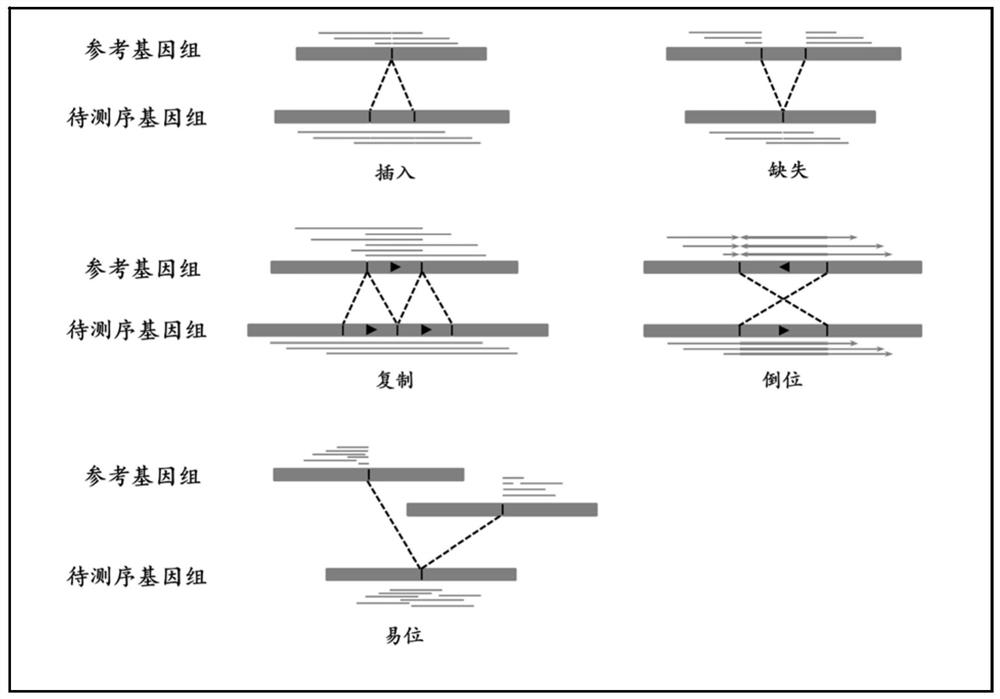
[0045] The present invention proposes a public genome structure variation performance detection method, and conducts a more systematic and detailed analysis on the structural variation of the common types of insertion, deletion, duplication, inversion and translocation in the genome.
[0046] According to whether the data used is simulated data or real data, the detection methods of genome structure variation performance can be divided into: simulated data and real data.
[0047] On the simulated data, due to the benchmark set of structural variation, this type of genomic structural variation performance detection method is suitable for objective analysis and comparison of different structural variation performance detection methods.
[0048] On real sequencing data, due to the lack of a benchmark set of structural varia...
specific Embodiment approach 2
[0058] Embodiment 2: The difference between this embodiment and Embodiment 1 is that in step 1, based on the user variation identification result set and the benchmark set, the insertion, deletion, duplication, and inversion variation in the genome structure variation are calculated on the quantitative index The statistical results of variation of , and output to the terminal screen; the specific process is: The specific process is:
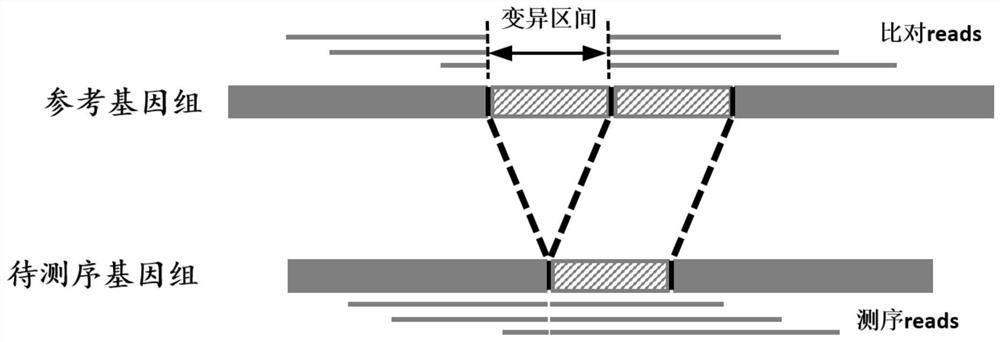
[0059] Due to the identification results of existing identification methods (such as Sniffles, nextSV, PBHoney, SMRT-SV, etc.), there are usually some variations with too large interval lengths. Such variations have no obvious significance due to their too large lengths. Therefore, in SV_STAT In the method, this type of variation is considered to be invalid variation, and these variations need to be removed before the performance detection of genomic structural variation to obtain more objective results. The higher the number of such invalid vari...
specific Embodiment approach 3
[0075] Embodiment 3: This embodiment is different from Embodiment 1 in that: the number of true positives and false positives in the identification results of user insertions, deletions, duplications or inversions after removing invalid variants is calculated; , number of true negatives identified for deletion, duplication or inversion variants, number of false negatives not identified; and recall, precision, F 1 score; the specific process is:
[0076] (1) Traverse S 1 , remove the invalid mutations whose mutation length is greater than 100kb, and obtain the mutation set S' whose mutation length meets the requirements 1 ;
[0077] (2) Statistics S' 1 with S 2 In the two sets, the number of mutations corresponding to each mutation length Size in 0≤mutation length≤2kb is stored in the form of two-tuple (Size,Num);
[0078] (3) Set S' 1 Each mutated Region in i with S 2 Each mutated Region in j Perform a pairwise comparison to the Region i and Region j Calculate the o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com