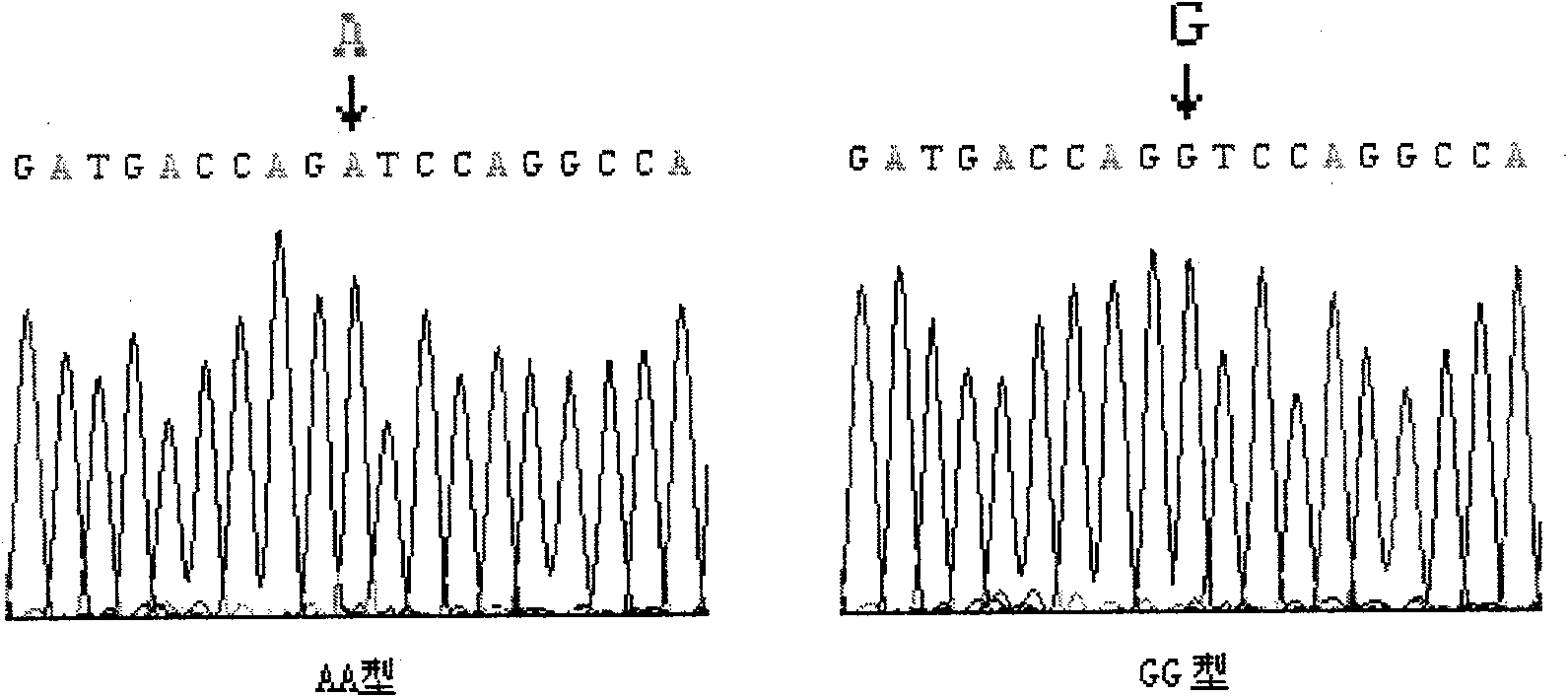
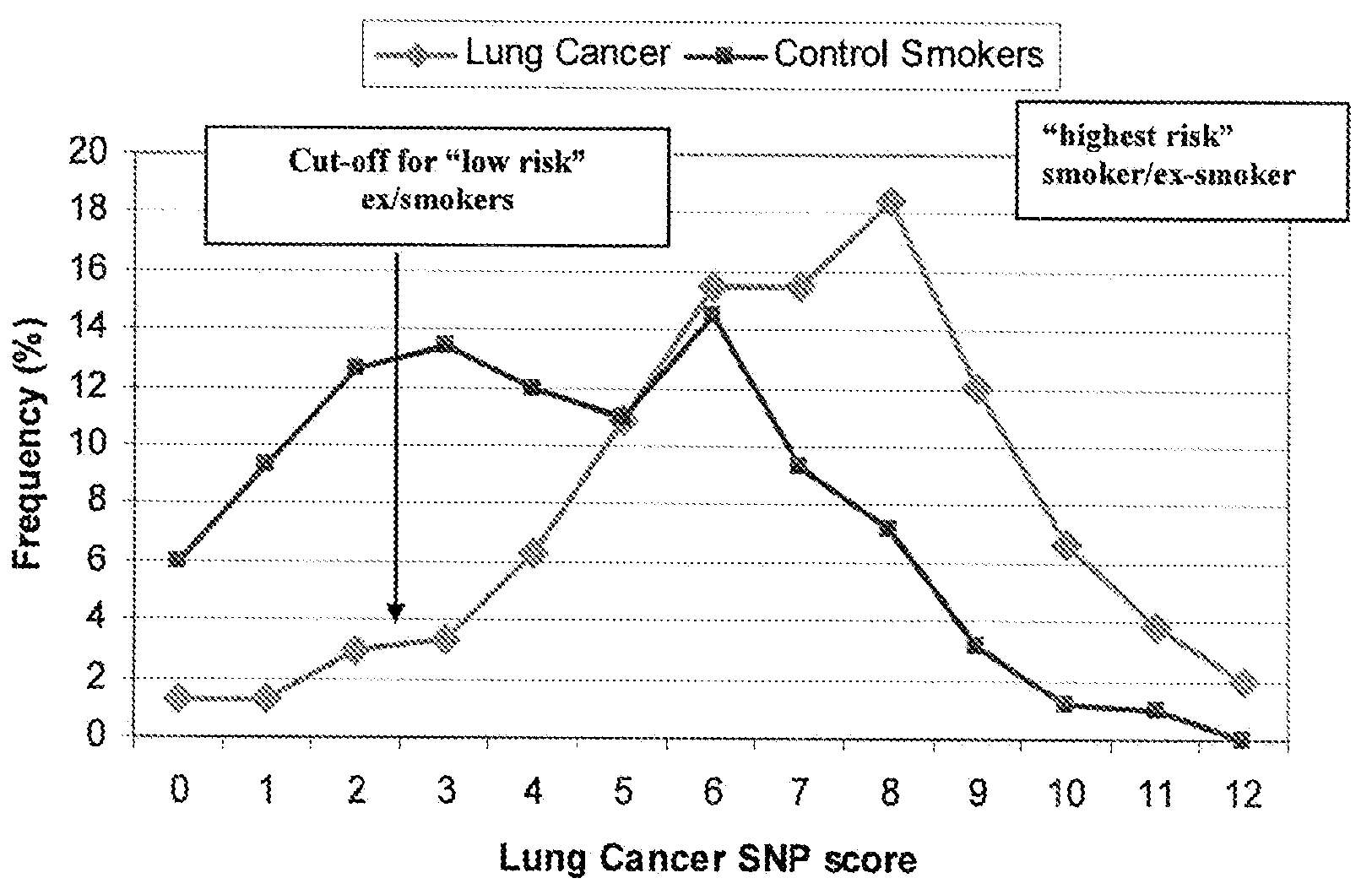
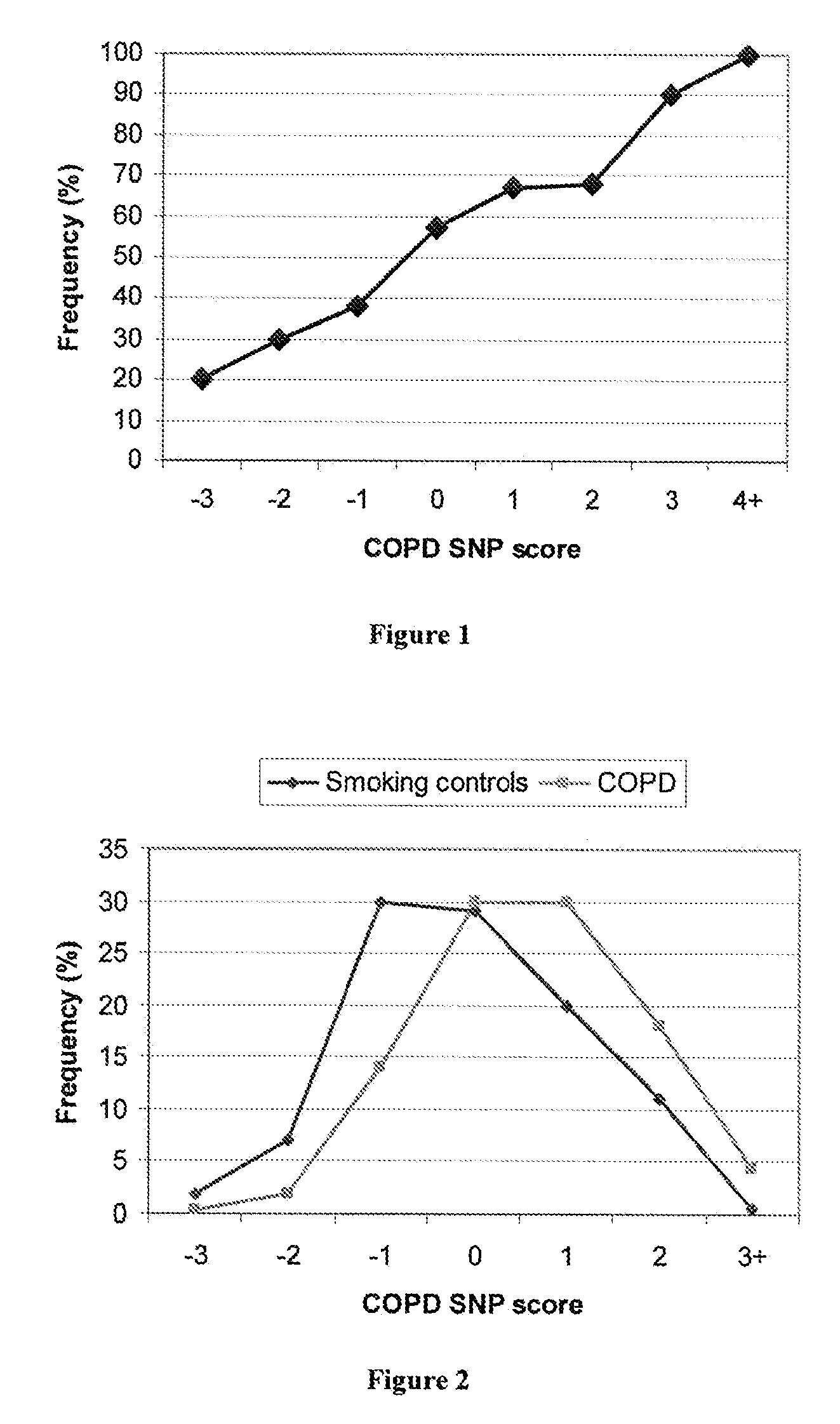
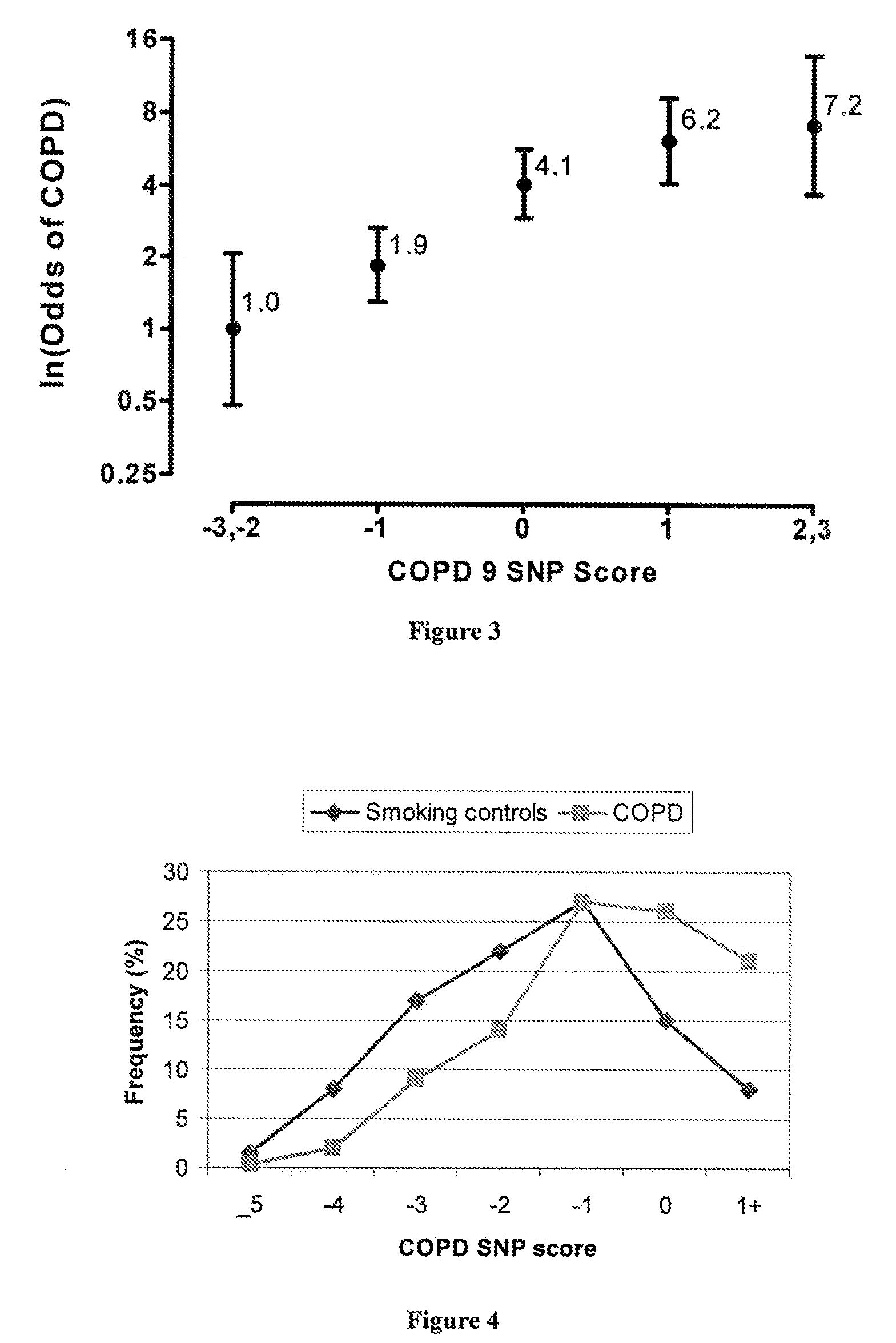
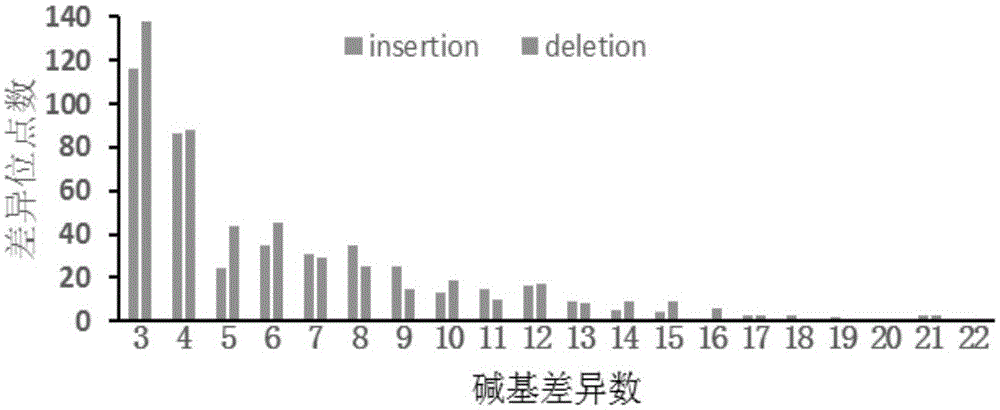
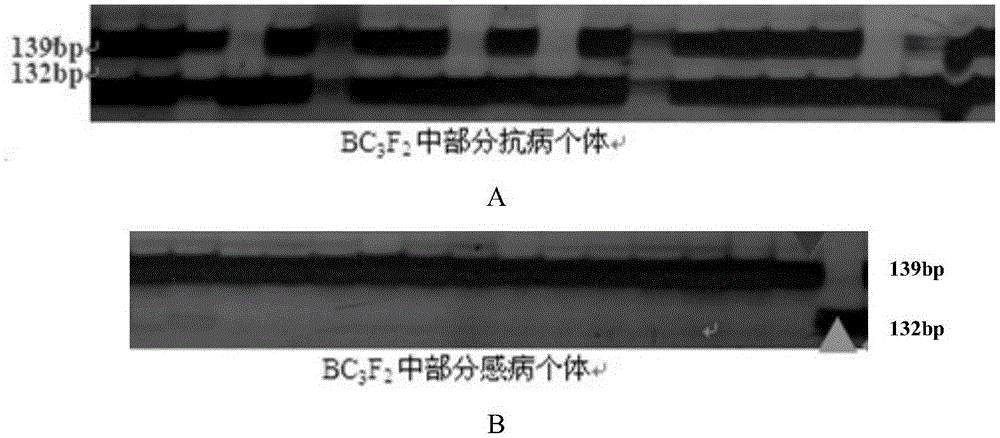
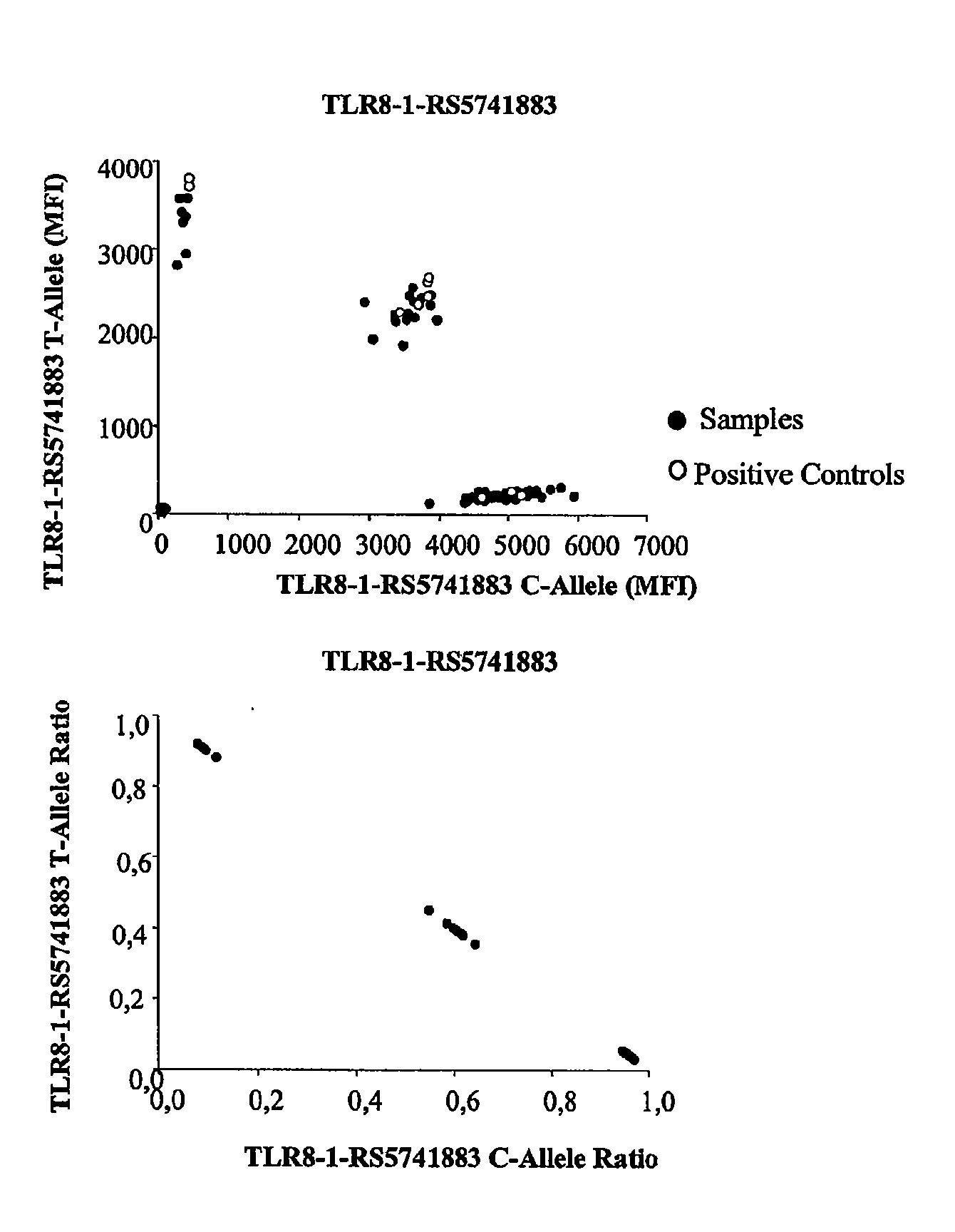
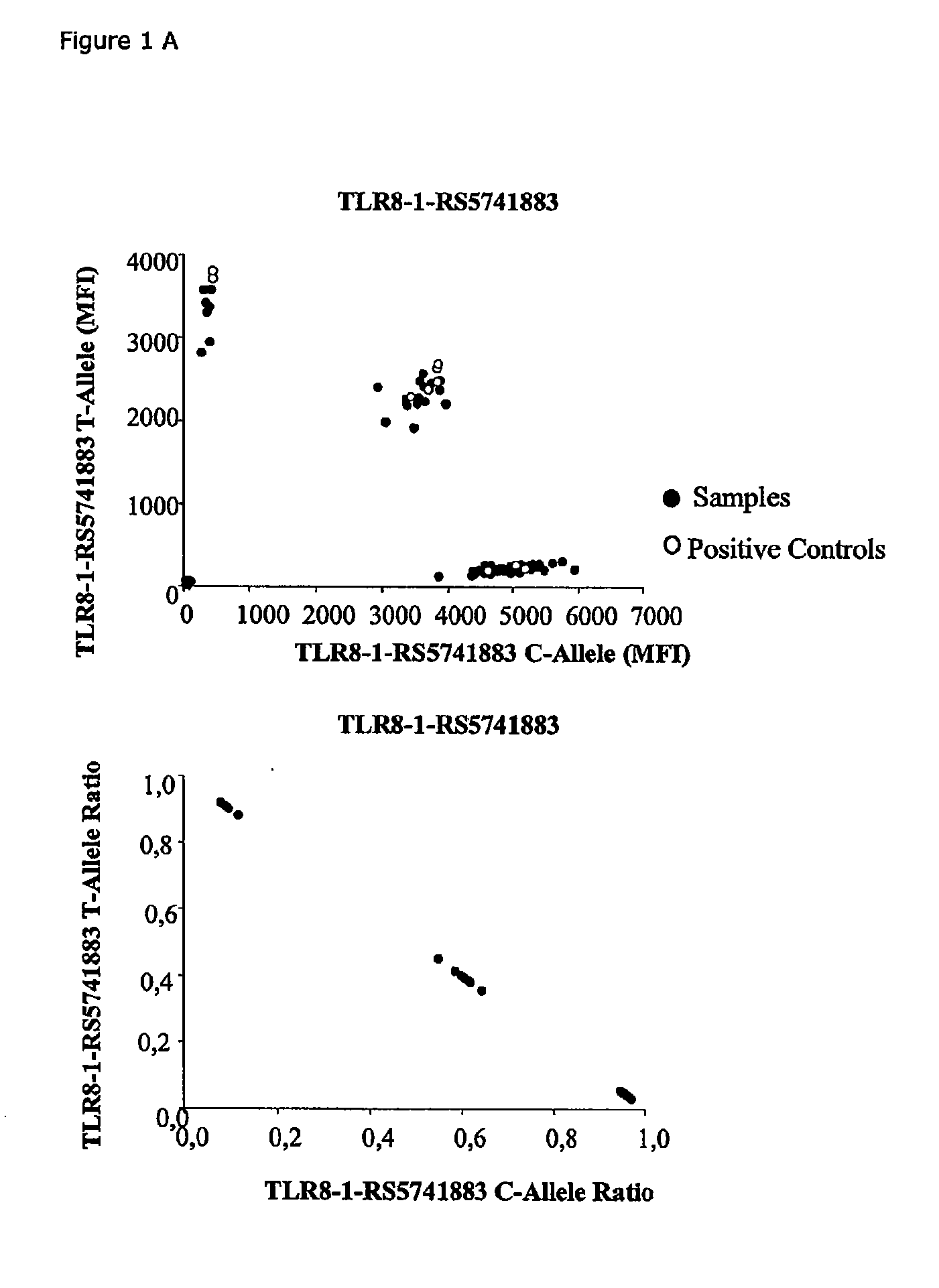
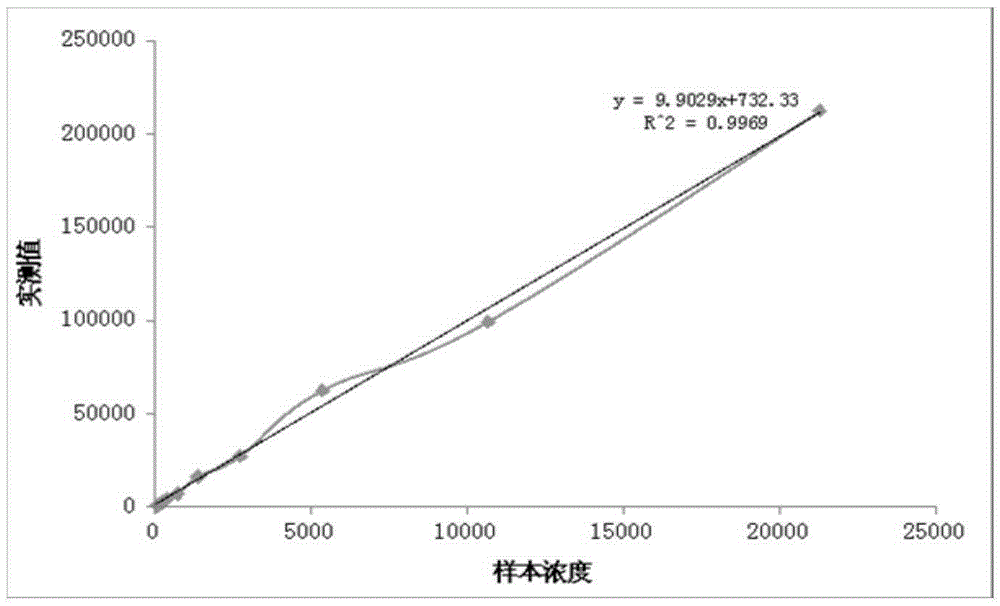
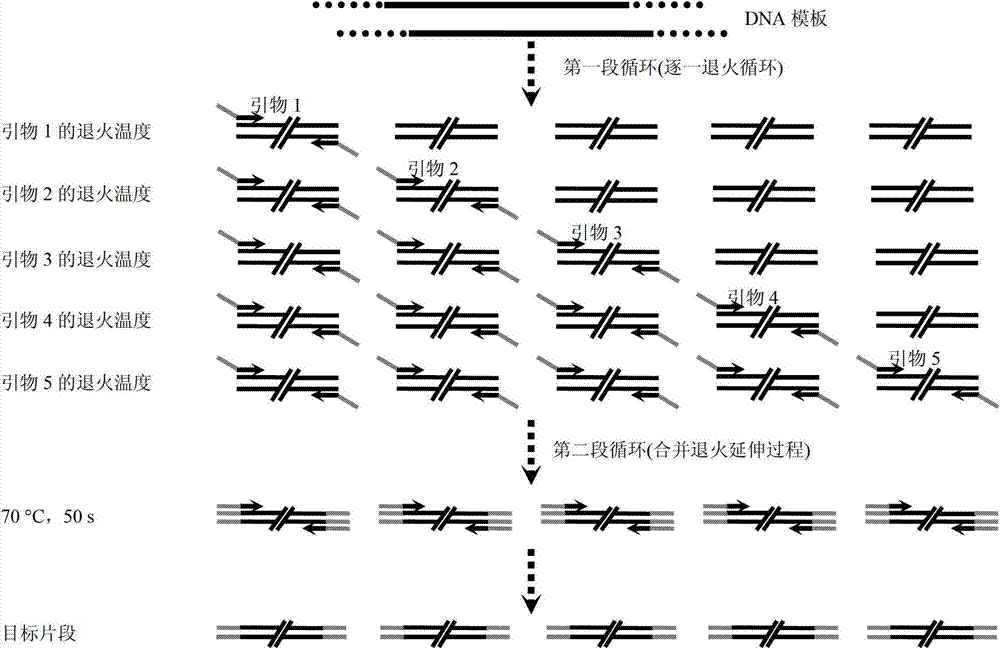
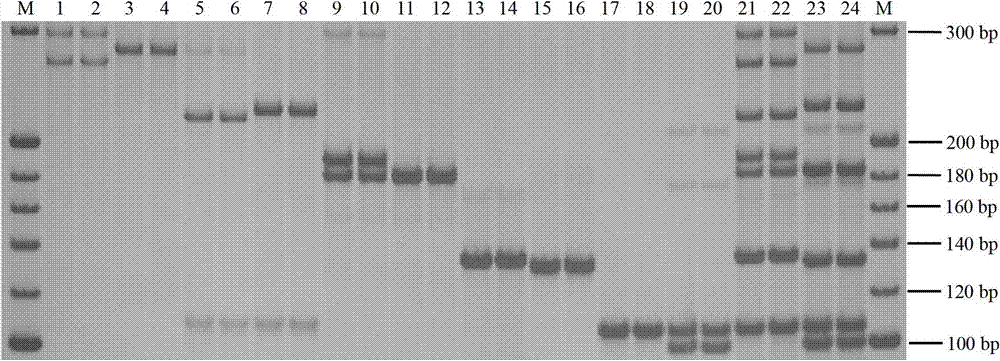
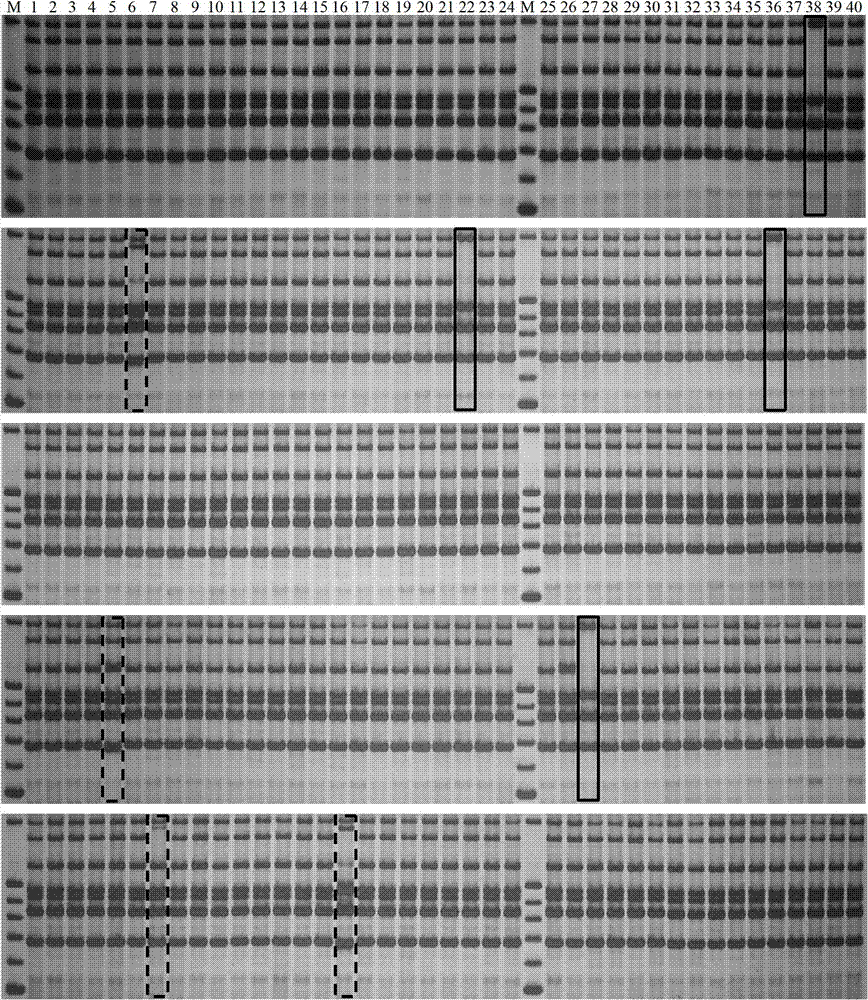
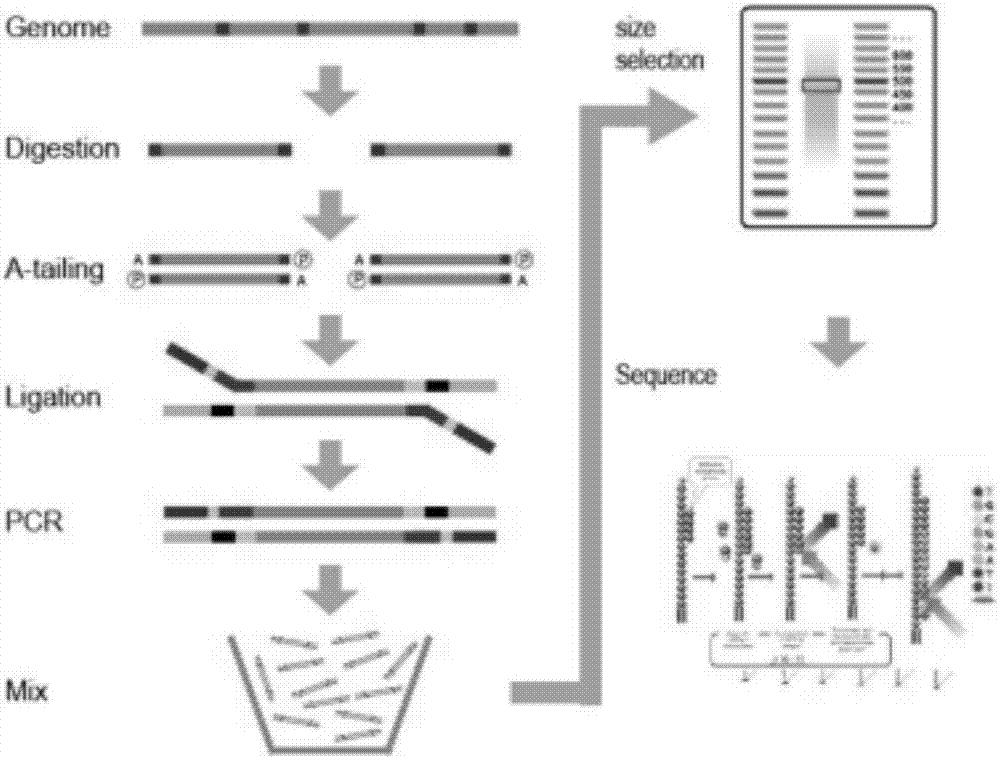
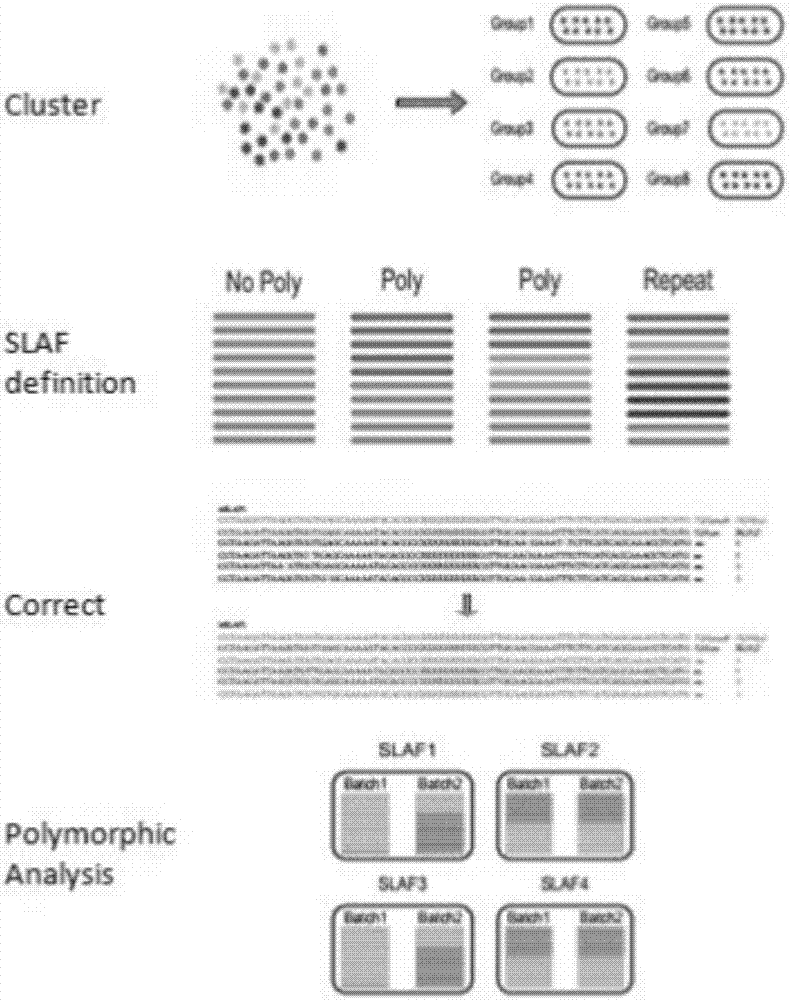
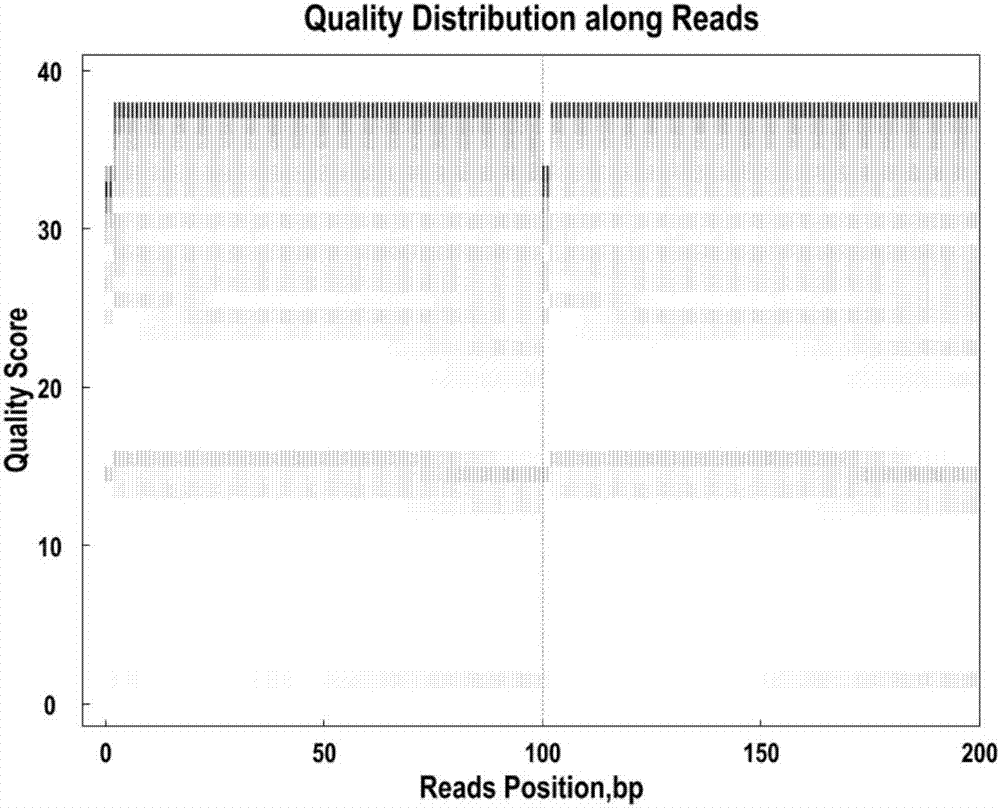
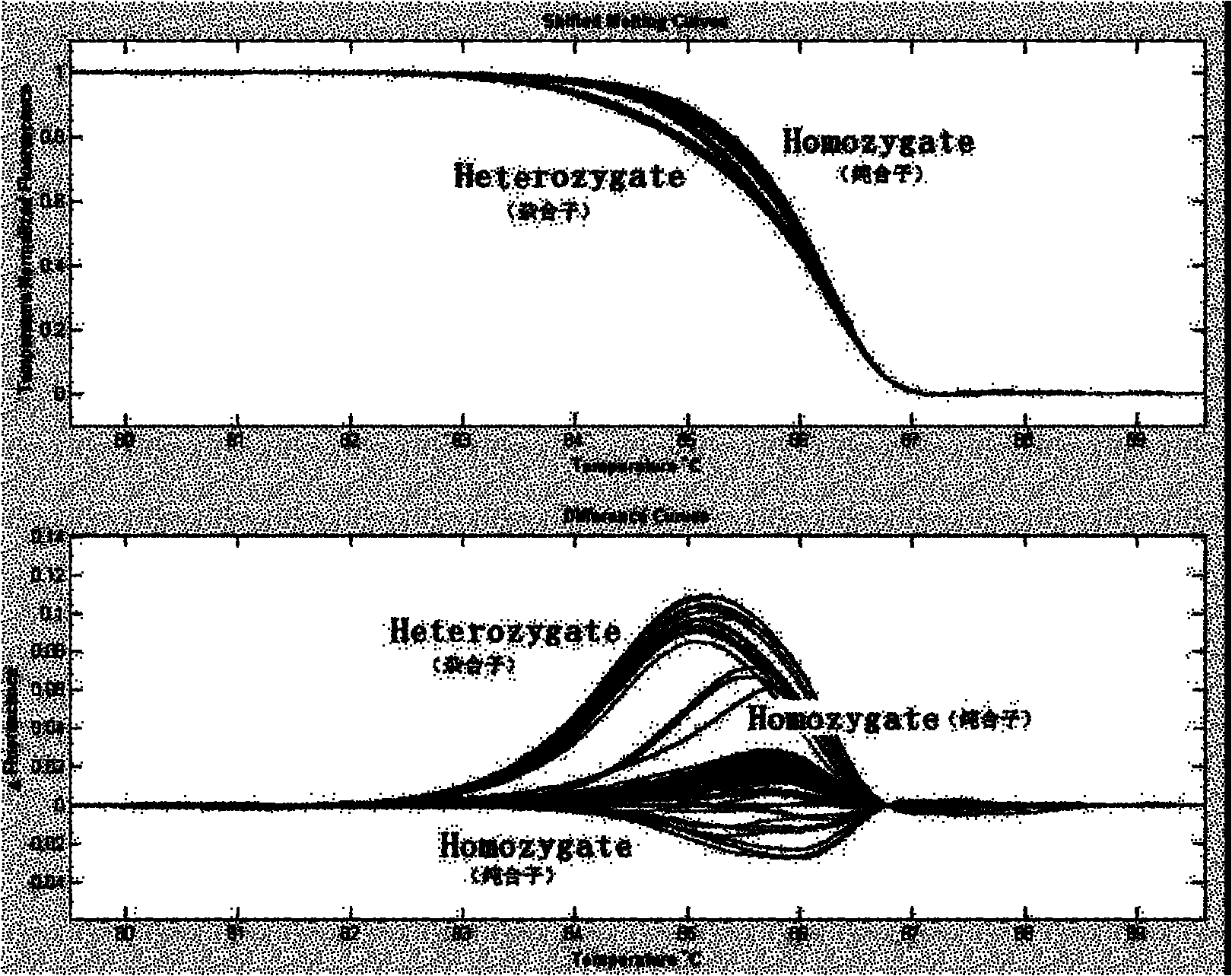
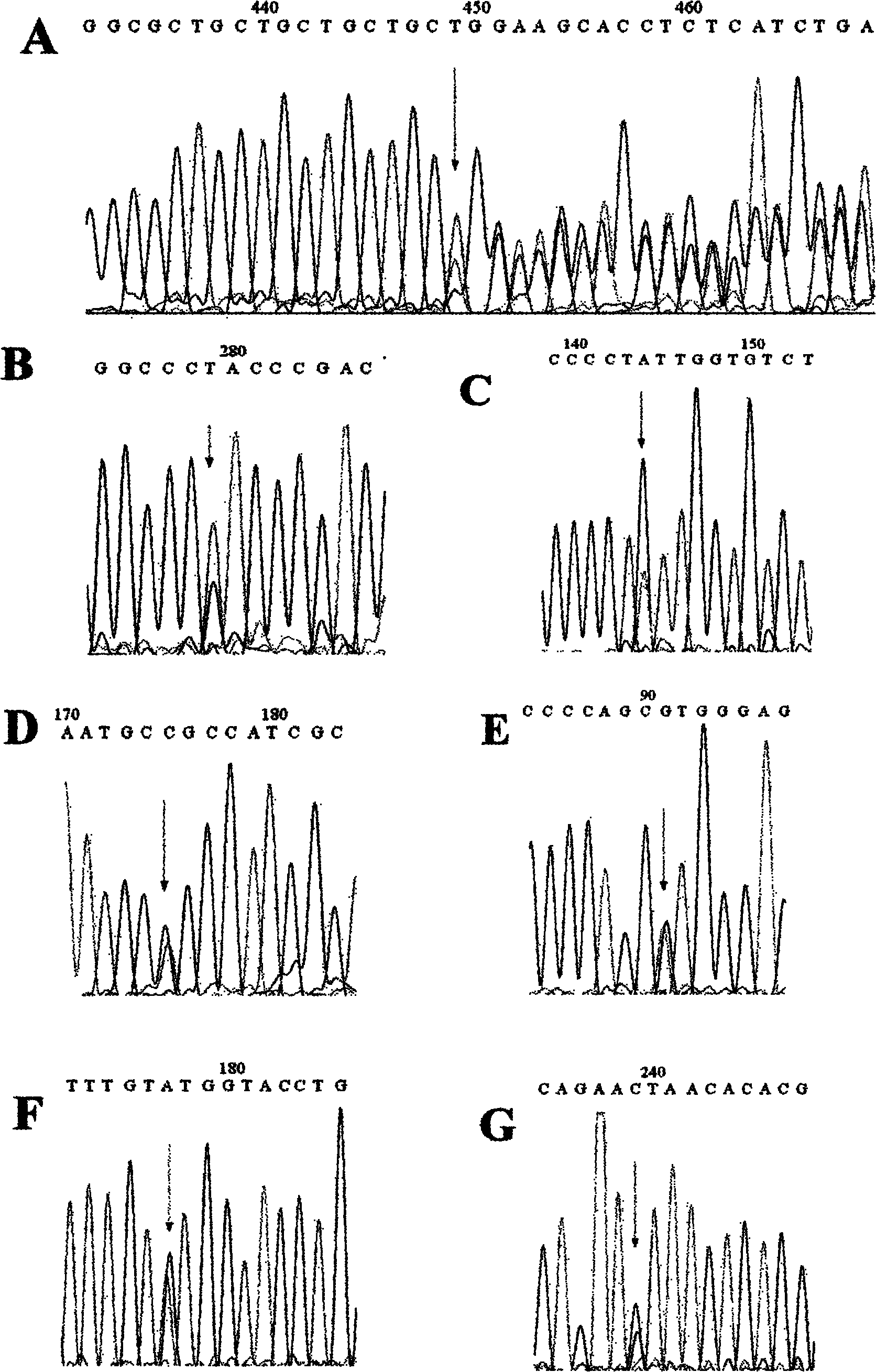
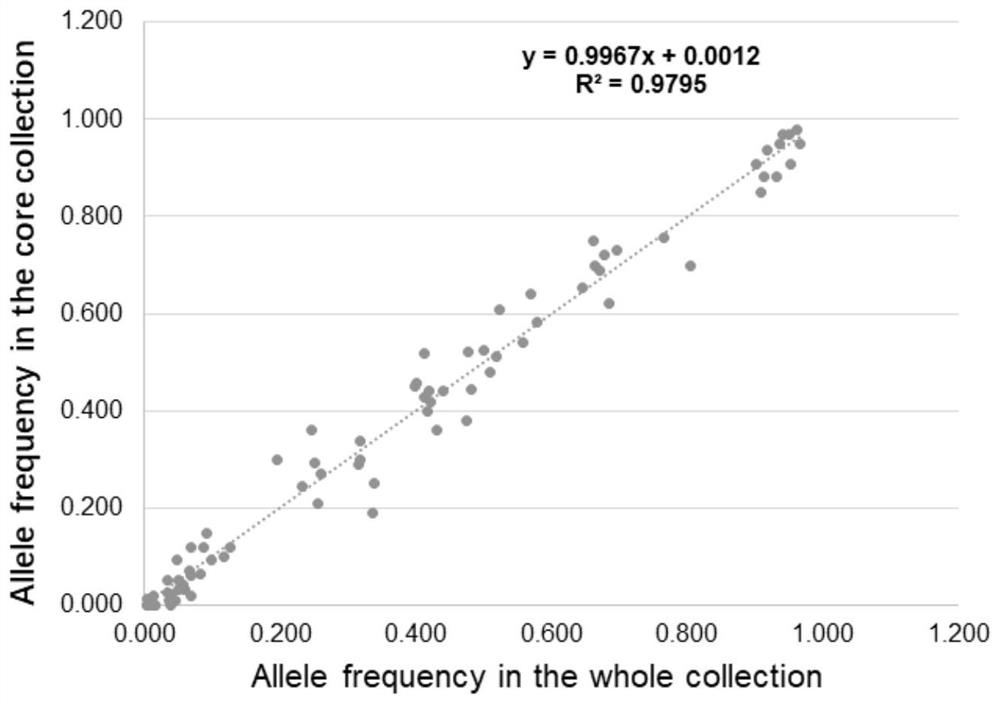
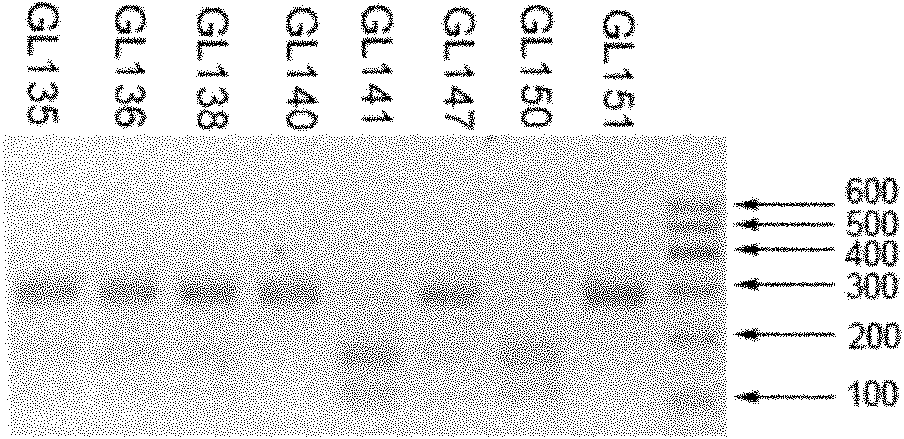Patents
Literature
79 results about "Polymorphism analysis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Restriction Fragment Length Polymorphism (RFLP) is a molecular method of genetic analysis that allows individuals to be identified based on unique patterns of restriction enzyme cutting in specific regions of DNA. Also referred to as RFLP Analysis, the technique takes advantage of the polymorphisms in individual people's genetic codes.
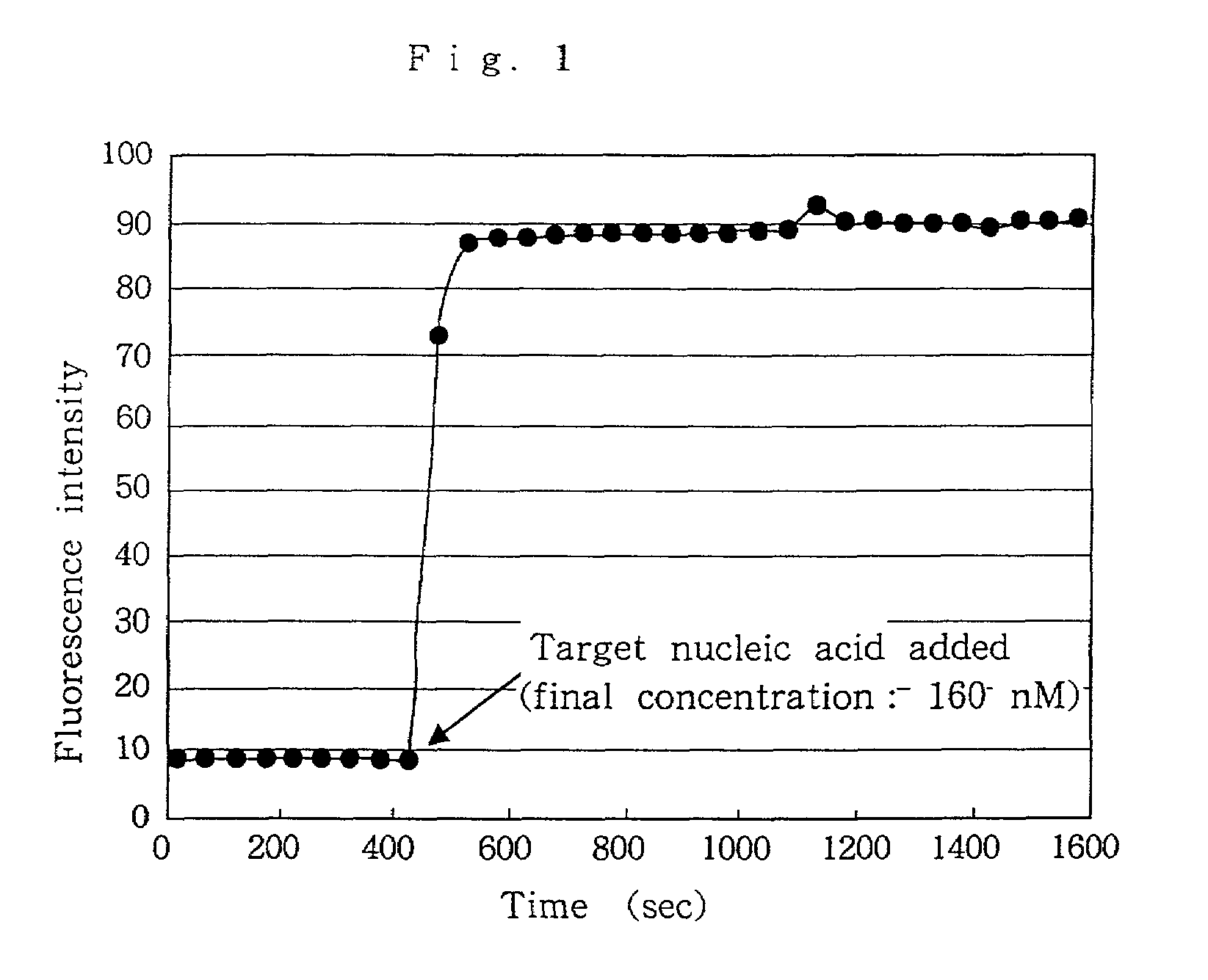
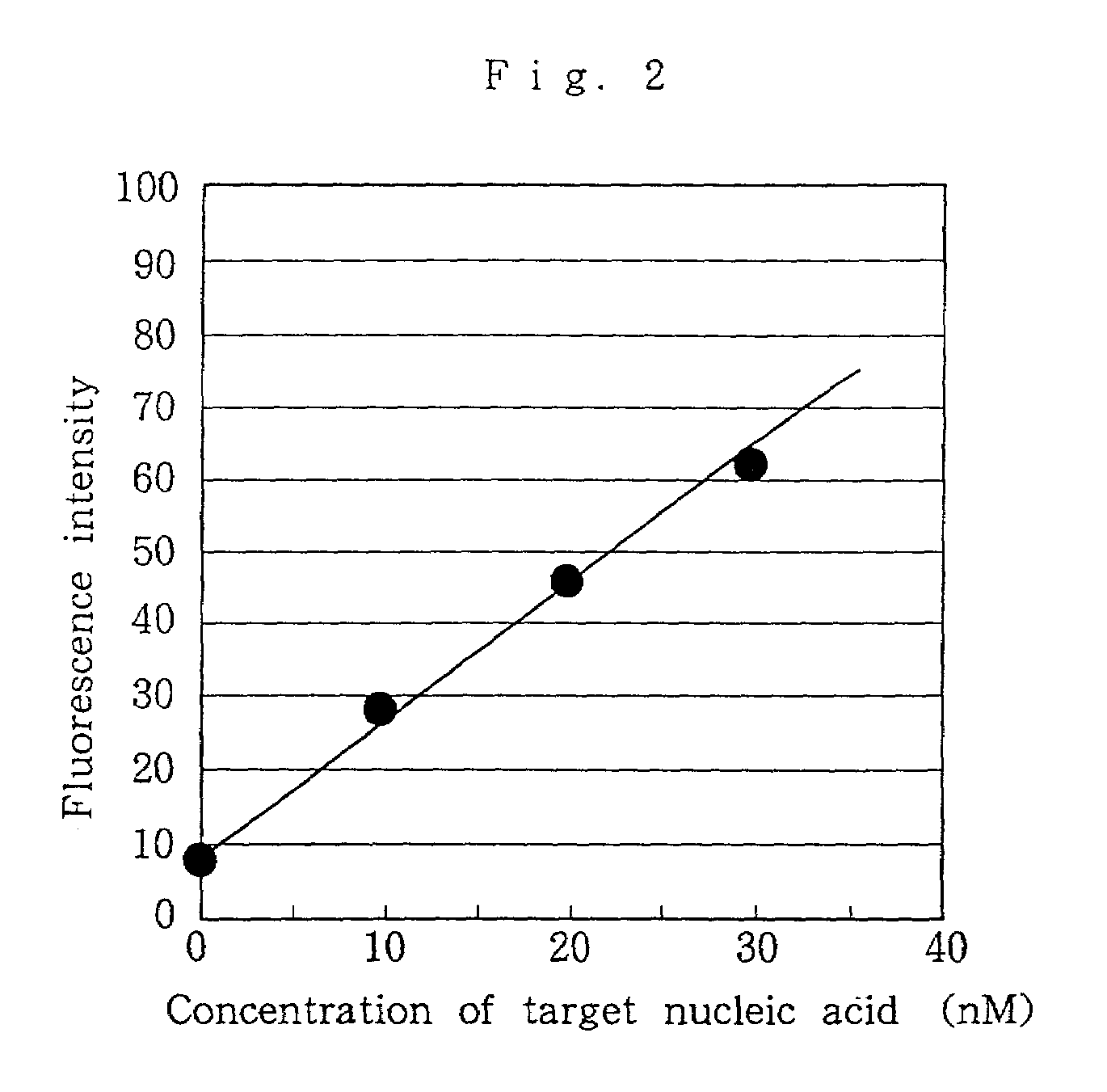
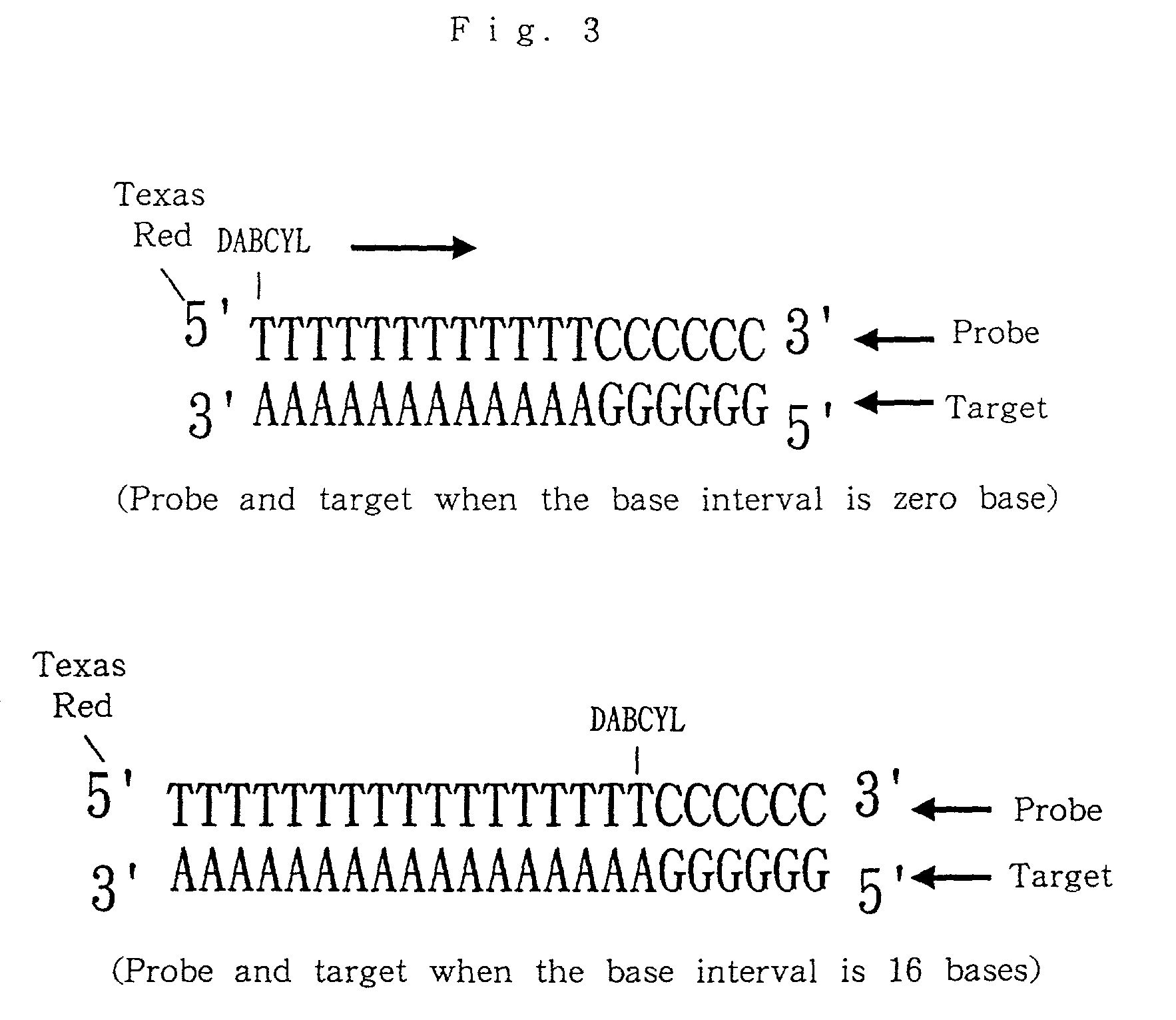
Nucleic acid probes, method for determining concentrations of nucleic acid by using the probes, and method for analyzing data obtained by the method
InactiveUS7354707B2Reduce measurementEasy to measureMonoazo dyesSugar derivativesFluorescenceNucleic Acid Probes
Nucleic acid probes are provided, each of which is formed of a single-stranded oligonucleotide which can hybridize to a target nucleic acid and is labeled with a fluorescent dye or with a fluorescent dye and a quencher substance. The nucleic acid probes can be easily designed, permit determination, polymorphous analysis or real-time quantitative PCR of nucleic acids in short time, and are not dissociated during reactions. Nucleic acid determination methods, polymorphous analysis methods and real-time quantitative PCR methods, which make use of the nucleic acid probes, are also provided.
Owner:NAT INST OF ADVANCED IND SCI & TECH +1
Authentication method of cattle CAPN1 gene as longisimus dorsi tenderness molecular marker and application
InactiveCN101624598AMicrobiological testing/measurementGenetic engineeringAgricultural sciencePolymorphism analysis
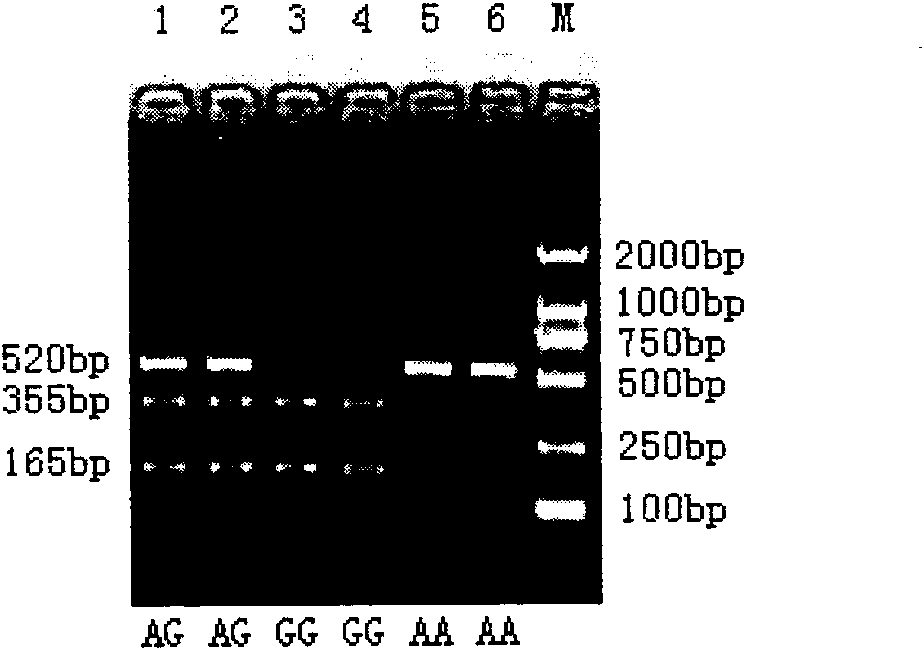
The invention belongs to the technical field of animal gene engineering, relating to clone of cattle longisimus dorsi tenderness related CAPN1 gene segment and an application thereof in cattle marker assisting selection. The obtained CAPN1 gene segment has the nucleotide sequence length of 520bp and comprises full fourteenth exons. A base mutation from A to G exists at the 166bp of the segment, which results in the generation of PsuI-RFLP restriction enzyme polymorphism. The mutation site is taken as a molecular marker, the restriction enzyme PsuI is used for detecting the mutation site, and association analysis can be carried out on the detection result and the cattle longisimus dorsi tenderness. Analysis result shows that striking difference exists among the longisimus dorsi tenderness of different genotype individuals, thus providing a theoretical basis for the seed selection and breeding of cattle. The invention is characterized by obtaining the CAPN1 gene part segment related to the cattle longisimus dorsi tenderness and providing the molecular marker and the application for the marker assisting selection of cattle by using a PCR-RFLP method to carry out polymorphism analysis on the segment.
Owner:JILIN UNIV
Methods of Analysis of Polymorphisms and Uses Thereof
InactiveUS20080195327A1Reduce riskReduce development riskMicrobiological testing/measurementBiological testingDiseaseGenetics
The present invention provides methods for the assessment of a subject's suitability for an intervention in respect of one or more diseases. The methods are dependant on the results of at least one genetic analysis, in particular genetic analyses that are predictive of predisposition to one or more diseases, including one or more genetic analyses of genetic polymorphisms associated with one or more diseases.
Owner:SYNERGENZ BIOSCIENCE LTD
Molecular marker of brassica napus anti-clubroot gene and application of molecular marker to anti-clubroot breeding
ActiveCN105063033AImprove breeding efficiencyShorten the breeding processMicrobiological testing/measurementDNA/RNA fragmentationRe sequencingAgricultural science
The invention discloses a molecular marker of a brassica napus anti-clubroot gene PbBa8.1 and a method for applying the molecular marker to anti-clubroot breeding through re-sequencing. The DNA molecular marker, capable of distinguishing anti-clubroot brassica napus and brassica napus being not resistant to clubroot, is obtained by conducting polymorphic analysis on sequences, around a PbBa8.1 locus, of two parents, and carrying out primer development design by utilizing the flanking sequences of an Indel locus. Primers, namely F A08-300: 5'-GTAGTGCGGGCCACAAAAT-3' and R A08-300: 5'-CACAATGGAGTGTTGAAATTCACT-3', aimed at the molecular marker are designed, and can be used for improving the anti-clubroot gene breeding speed and identification efficiency. The method is easy to carry out, high in repeatability, operability and identification speed, and low in detection cost, time consumption and labor cost, and has the advantages that the workload of selecting an anti-clubroot singleplant through phenotype identification in a field is greatly reduced.
Owner:HUAZHONG AGRI UNIV
Prognostic method for the determination of the suitability of biopharmaceutical treatment
InactiveUS20100311052A1Microbiological testing/measurementSkeletal disorderPolymorphism analysisDisease cause
The invention refers to a method for the prognosis of a disease in a subject by the administration of a biopharmaceutical treatment in a subject suffering from, or likely to suffer from the disease, the method involving the analysis of SNP polymorphisms in the subjects pattern recognition receptor genes (PRRs), eg the analysis of polymorphisms' with the purpose of predicting the response of anti-TNFx antibody therapy in rheumatoid arthritis patients. Also the response to Beta-interferon in multiple sclerosis patients may be predicted. The genes whose polymorphisms are analysed may be TLRs, NOD-like receptors or retinoic acid-inducible gene I-like receptors (RLR).
Owner:BIOMONITOR
Establishing method of cymbidium microsatellite labels, core fingerprint label database and kit
InactiveCN104293778ALarge amount of informationImprove versatilityMicrobiological testing/measurementDNA preparationMicrosatelliteCymbidium ensifolium
The invention discloses an establishing method of cymbidium microsatellite labels, a core fingerprint label database and a kit. The method comprises the following steps: exploiting SSR primers based on Cymbidium ensifolium transcription group information, and carrying out versatility and polymorphism analysis on labels to obtain 49 cymbidium polymorphism labels. The labels have high polymorphism and good repeatability, and are very suitable for the genetic study of cymbidium plants. Additionally, the core fingerprint label database is constructed according to the distinguishing ability of the labels in cymbidium species. The SSR fingerprint label database obtained in the invention has the characteristics of stable and reliable labels, small quantity and strong cymbidium species distinguishing ability, can maximally reflect the interspecific difference (not intraspecies difference) of the cymbidium species, is convenient for realizing statistics, and can be used for identifying cymbidium main species.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
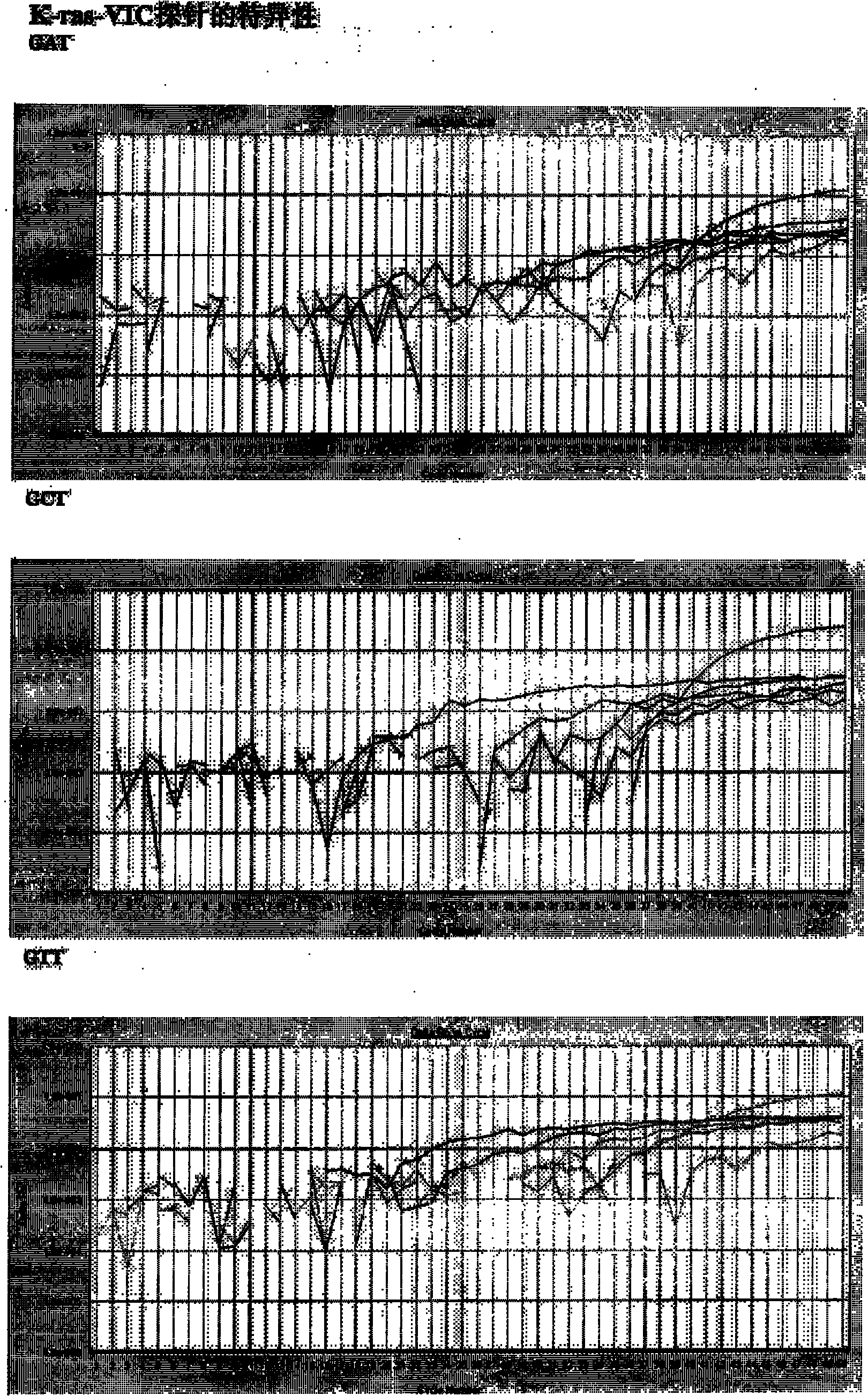
Double fluorescent marker probe real-time quantitative detection method for K-ras gene 12 codon mutation and application
InactiveCN102041313ASimple and fast operationIncreased sensitivityMicrobiological testing/measurementFluorescence/phosphorescenceWild typeTarget peptide
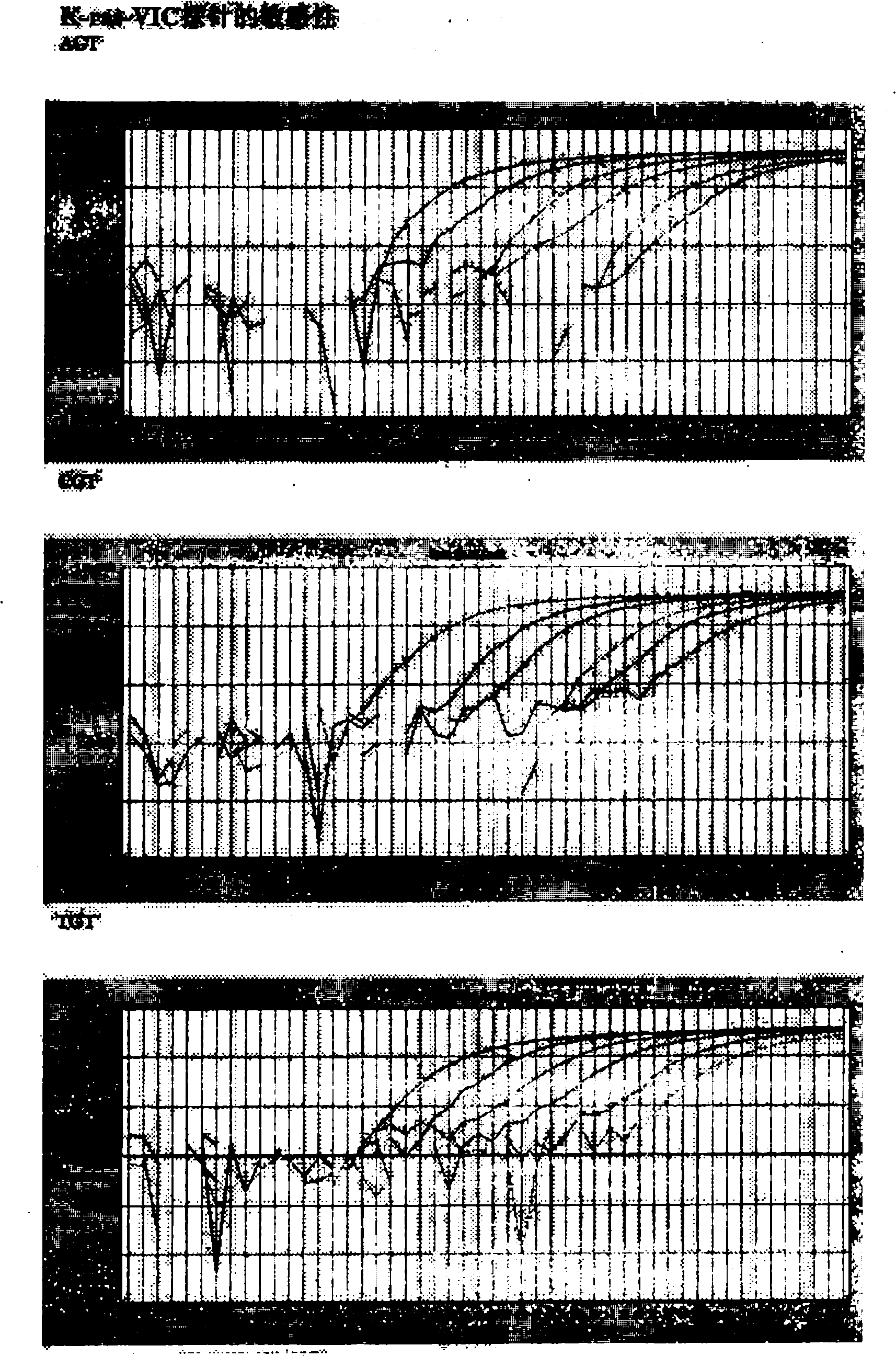
The invention belongs to the technical field of biology. K-ras gene mutation plays an important role in occurrence and development of pancreatic cancer, but the current universal K-ras gene mutation detection methods comprise a limited fragment length polymorphism analysis method and an amplification blocked mutation system method, and the two methods have low specificity and cannot qualitativelyand quantitatively detect the mutation of K-ras genes at the same time. The invention aims to provide a double fluorescent marker probe real-time quantitative detection method for K-ras gene 12 codonmutation with operation convenience, high sensitivity and high specificity. The method comprises the following steps of: designing wild K-ras gene 12 codon-targeted peptide nucleic acid (PNA) and corresponding mutation detection probes, namely a K-ras-FAM Tagman MGB probe and a K-ras-VIC Tagman MGB probe; performing real-time quantitative polymerase chain reaction (PCR) detection on a plasmid standard substance of known mutation quantity by using the PNA and the probes to acquire a standard curve and a fluorescent type; and extracting and purifying sample DNA, measuring the concentration, performing real-time quantitative PCR detection, and judging K-ras gene mutation quantity and mutation type according to the standard curve and the fluorescent type.
Owner:SECOND MILITARY MEDICAL UNIV OF THE PEOPLES LIBERATION ARMY
Identification method taking growth hormone receptor gene as Chinese grassland red bull excellent slaughter trait molecular marker and application thereof
The invention belongs to the technical field of animal genetic engineering, and particularly relates to a growth hormone receptor gene taken as a Chinese grassland red bull excellent slaughter trait molecular marker and an application thereof. The length of a nucleotide sequence of an obtained GHR genetic segment is 489bp and the nucleotide sequence is a part of an exon. A base mutation exists at a 289-position basic group to cause the generation of restrictive fragment length polymorphism. The mutant site is taken as the molecular marker and detected by a PCR-RFLP (polymerase chain reaction-restrictive fragment length polymorphism) method using restriction enzyme BstACI, and detection results are associated with the Chinese grassland red bull slaughter traits so as to provide theoretical basis for selection breeding of Chinese grassland red bull. The invention is characterized in that partial segment of GHR gene associated with the Chinese grassland red bull excellent slaughter traits is obtained, and polymorphism analysis is conducted to the segment for providing the molecular marker for marker assisted selection of the Chinese grassland red bull.
Owner:JILIN UNIV +1
Novel cucumber SNP molecular marker
InactiveCN104988142AHelp with positioningMicrobiological testing/measurementDNA/RNA fragmentationPolymorphism analysisCell stress
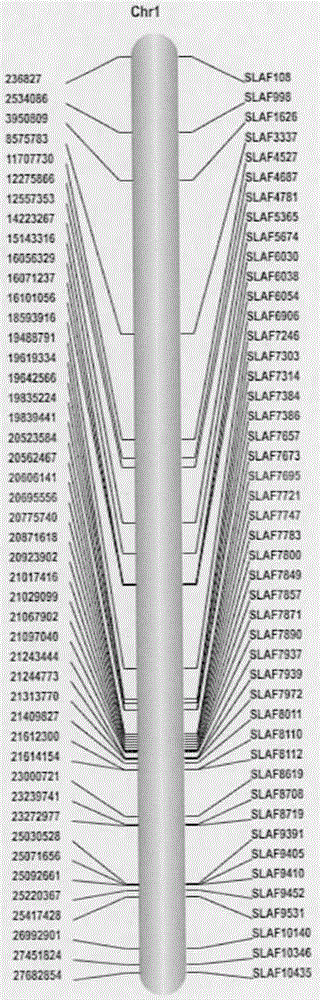
The invention discloses a novel cucumber SNP molecular marker. The mark number of the novel cucumber SNP molecular marker is SLAF7695. According to the novel cucumber SNP molecular marker, the simplified genome depth sequencing technology is utilized, 73100 SLAF labels are obtained totally, 5355 polymorphic markers of three types including SNP, EPSNP and INDEL are obtained through polymorphic analysis, and 140 different markers closely related to cucumber powdery mildew resistance are obtained and mainly concentrated on the first chromosome and the sixth chromosome. Two candidate areas related to powdery mildew resistance characters are successfully located, seven special markers closely linked with cucumber powdery mildew resistance are obtained, 28 genes and related annotation information are obtained through gene annotation, and five genes are related to the functions of defense response, harm response, toxin catabolism, cell stress response and the like. The novel cucumber SNP molecular marker provides a theoretical foundation for exploration of cucumber powdery mildew resisting mechanism and molecular marker assisted selective breeding.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
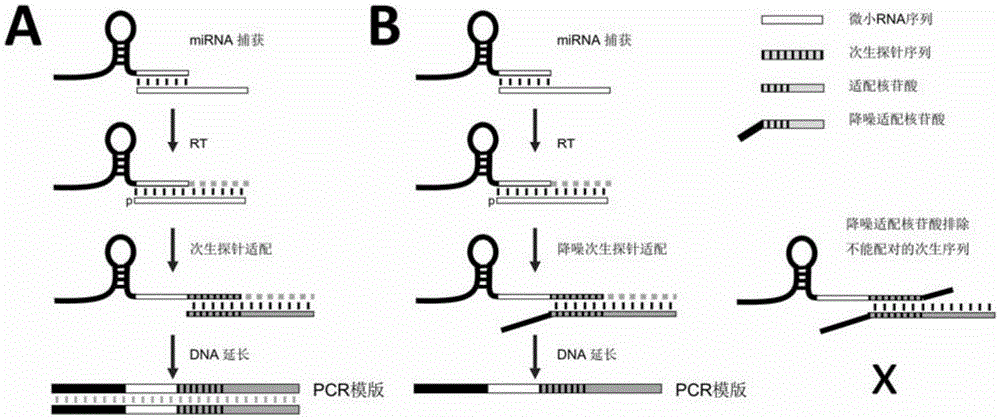
Method for directly detecting miRNA in absolute quantification mode
ActiveCN105331695AAvoid interferenceReduce Reactive Background SignalMicrobiological testing/measurementFluorescenceMagnetic bead
Owner:CHENGDU NUOEN BIOLOGICAL TECH
Universal multiple PCR (Polymerase Chain Reaction) method
InactiveCN102776172AImprove versatilityImprove featuresMicrobiological testing/measurementDNA preparationGeneticsPcr method
The invention relates to a universal multiple PCR (Polymerase Chain Reaction) method. The universal multiple PCR method comprises the steps of: increasing a universal joint sequence to decrease difference of annealing temperature between primers, and increasing the annealing temperature (about 70 DEG C) of the jointed primers to be close to extension temperature; combining annealing and extensionphases; shortening PCR time; and improving the specificity and the yield of a PCR product by using two sections of circulation modes according to the gradual annealing circulation of primer annealingtemperatures from high to low. By using the universal multiple PCR method, the universality of multiple PCR can be remarkably improved, the universal multiple PCR method can be applied to many fields, such as seed purity identification, variety authenticity identification, polymorphism analysis, mutation analysis, quantitative analysis and species identification.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
Method for identifying genetic relationship by developing SNP molecular marker of miscanthus with SLAF-seq technology
The invention discloses a method for identifying the genetic relationship by developing an SNP molecular marker of miscanthus with the SLAF-seq technology. The method comprises the following steps: (1) designing a marker development scheme; (2) selecting an optimal digestion scheme; (3) respectively extracting DNA of genomes for digesting, constructing an SLAF-seq sequencing library, and sequencing; (4) developing an SLAF marker for a single sample, then performing similarity clustering on all SLAF markers to obtain an SLAF marker of the cluster; (5) performing allele number, genetic sequence difference and other polymorphic analysis on the cluster SLAF marker, wherein SNP is taken as a polymorphic marker; (6) coding the genotype for parent, representing a character by a genotype, detecting the daughter genotype after the separation type is determined, and representing the SNP coding genotype with a corresponding character; and (7) selecting SNP marker with few sample loss, extracting all SNP sites, calculating the daughter SNP anomalous typing ratio, determining the genetic relationship between daughter and parent according to whether the daughter SNP anomalous typing ratio is higher than the threshold value or not.
Owner:HUNAN AGRICULTURAL UNIV
Epithelial type cadherin expressed gene CDH1 mutation detection kit and application thereof
The invention relates to a CDH1 mutation detection kit and an application thereof. The kit mainly comprises the following main ingredients: (1) 19 pairs of CDH1 gene PCR amplifying and sequencing primers; and (2) PCR reagent for amplification. The PCR amplification product obtained by the kit can screen gene micromutation by a high-resolution dissolved curve process or carries out polymorphism analysis on restrictive segment length. The kit has high-efficiency mutation detection and functional evaluation actions. The invention is used for CDH1 gene mutation detection.
Owner:NANJING UNIV
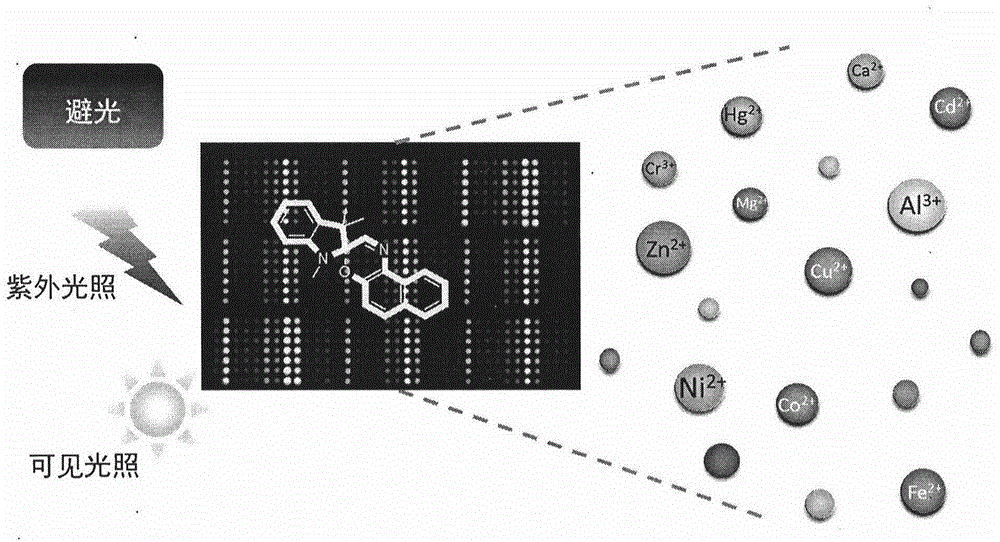
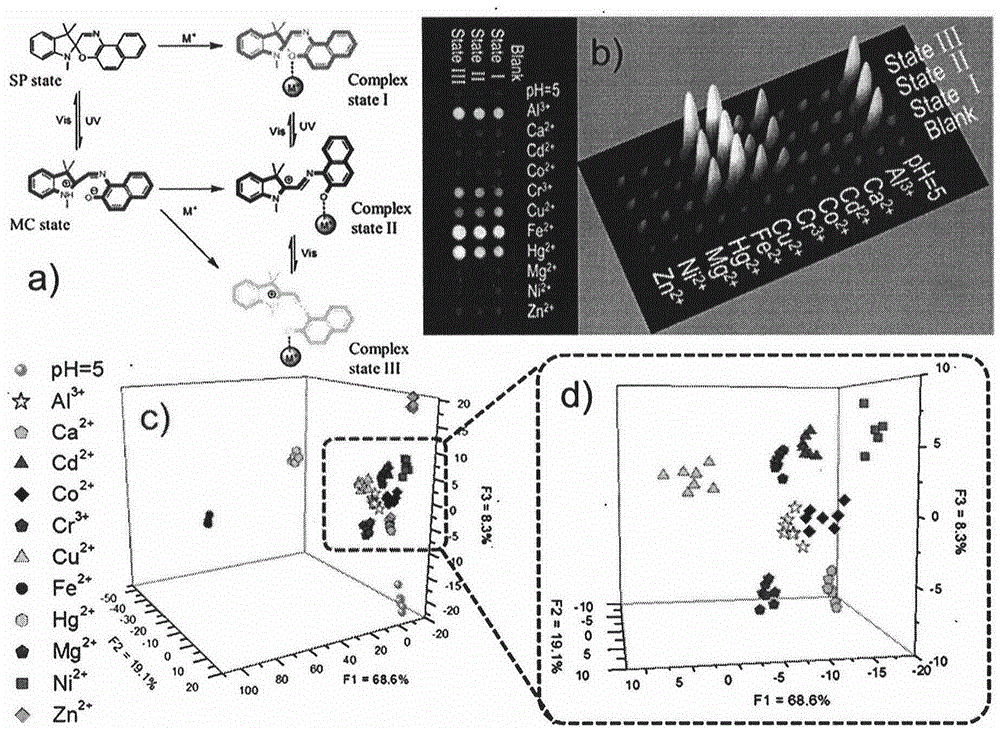
Photochromic dynamic multi-substrate detection microchip and polymorphism analysis method
InactiveCN104437688ARich chemical informationAvoid multipleLaboratory glasswaresFluorescence/phosphorescenceSensor arrayFluorescence
The invention belongs to the field of photochromic material and multi-substrate analysis. Photochromic molecules are introduced into a sensor array so as to prepare an efficient universal multi-substrate detection analysis microchip. According to the invention, photochromic molecules, namely spiropyrane / spirooxazine, are utilized, the response optical difference of the microchip to various metal cations under different photostimulation conditions (light resistance, ultraviolet light illumination and visible light illumination) is analyzed comprehensively, and an array chip consisting of a single spiropyrane or spirooxazine compound is designed. Fluorescence and absorption chemical information is recorded via a plurality of channels, and difference analysis and identification can be carried out on different kinds of metal cations as multiple as possible by utilizing high-flux statistics methods such as principal component analysis (PCA), hierarchic classification analysis (HCA) and linear difference analysis (LDA) and the like. According to the invention, the single chemical sensor is used for detecting and analyzing multiple substrates by constructing the photochromic dynamic multi-substrate detection microchip, and the product has broad-spectrum universality and great operability for multi-substrate recognition and detection at various complex environments.
Owner:INST OF CHEM CHINESE ACAD OF SCI
Method for selecting goose carcass trait microsatellite molecular marker
InactiveCN102719549AHigh precisionEasy to operateMicrobiological testing/measurementMicrosatellitePolymorphism analysis
The invention discloses a method for selecting goose carcass trait microsatellite molecular marker and belongs to the technical field of poultry breeding. The method comprises the processes of: (1) designing specific primers of goose RY 36-site, wherein an upstream primer is 5'-TCCCAGCTTCACTCCTTTTC-3', and a downstream primer is 5'-GTGGTGTTCTTGCGGTGTAG-3'; (2) measuring goose carcass trait; (3) carrying out a PCR (polymerase chain reaction) amplification reaction; (4) carrying out polymorphism analysis of a PCR amplification product, and (5) selecting a superior goose parent individual body. According to the invention, goose carcass trait selection is carried out according to the genotype, the accuracy is high and the operation is simple. The carcass trait selection is begun at an earlier period of goose life; a generation interval is shortened; the breeding process of the goose is accelerated; the selection time can be shortened by about three months in each generation, and the method can be generalized and applied in the field of poultry breeding.
Owner:ZHEJIANG ACADEMY OF AGRICULTURE SCIENCES
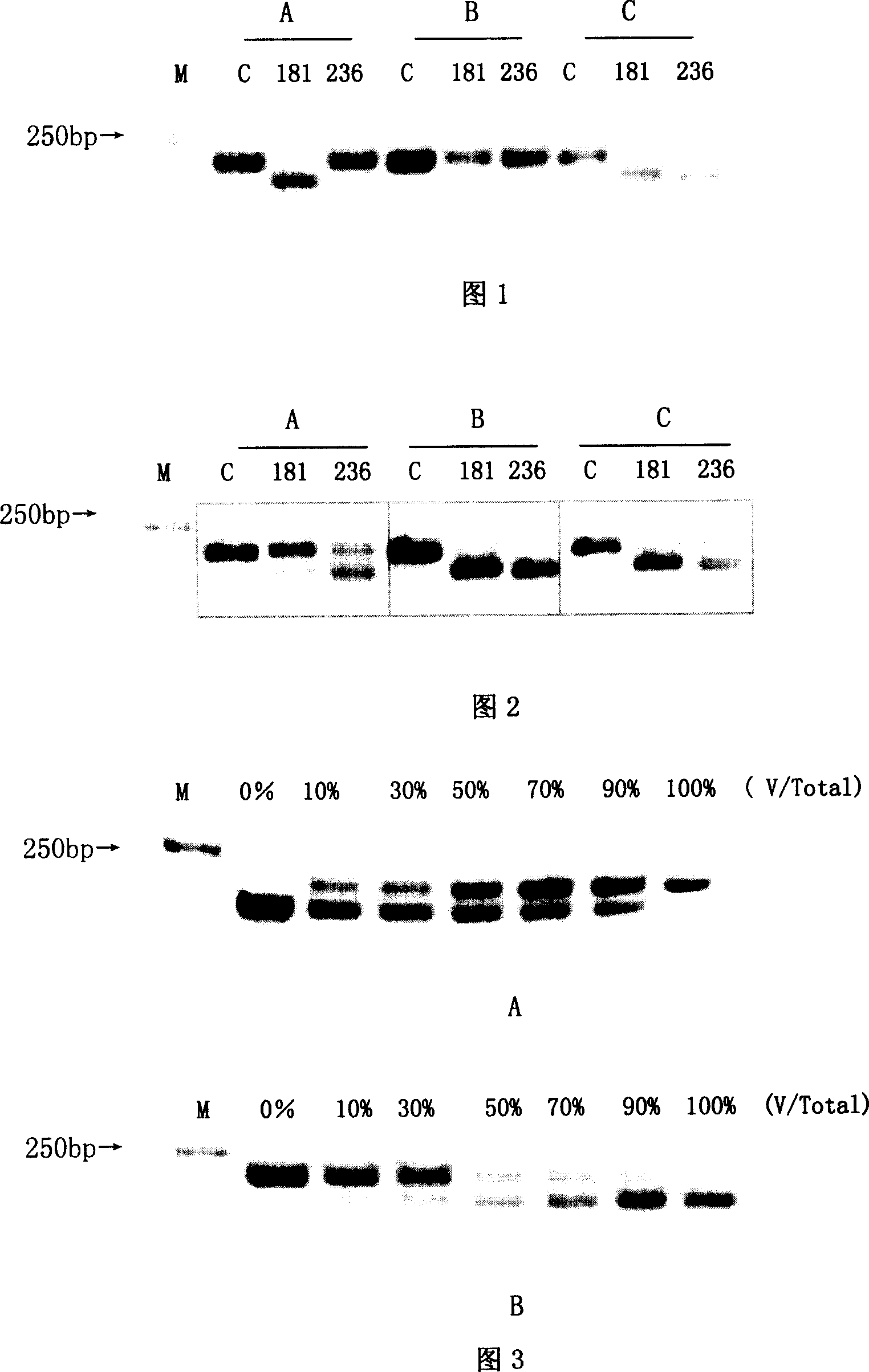
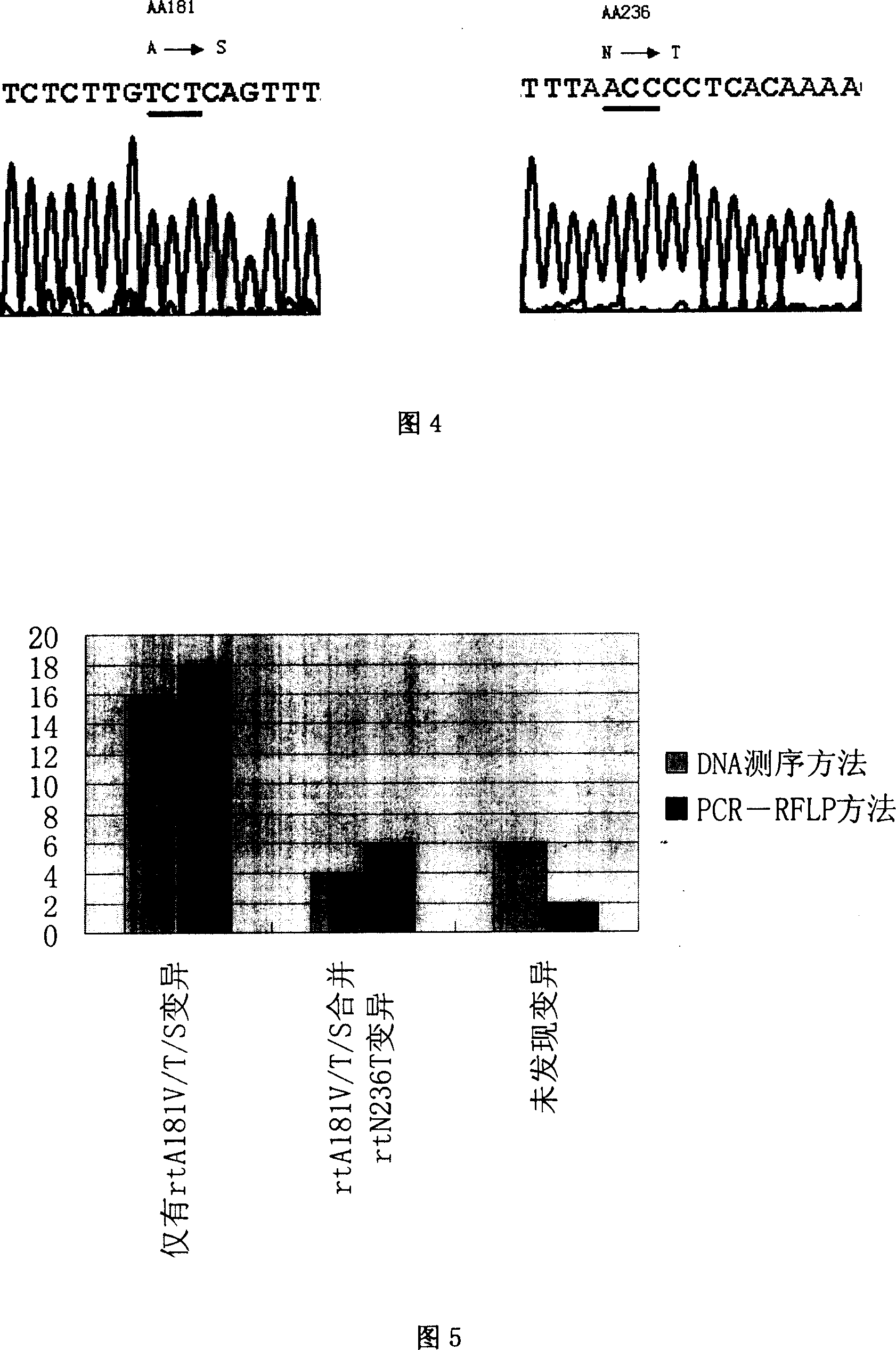
Method for measuring drug fast variation of hepatitis b virus to Adefovir
InactiveCN101004415AIncreased sensitivityHigh sensitivityMicrobiological testing/measurementMaterial analysis by electric/magnetic meansElectrophoresisNucleotide variation
A method for determining adfuwei tolerance variation of hepatitis B virus includes designing specific primer, carrying out amplification by set of PCR means, using BgII and BseDI to carry out enzyme cut reaction electrophoresis on amplified product then carrying out restrictive section length polymorphism analysis.
Owner:AFFILIATED HUSN HOSPITAL OF FUDAN UNIV
Molecular marker method for identifying pepper anti-epidemic disease character
InactiveCN101487048ASimple stepsThe steps are stable and reliableMicrobiological testing/measurementGenomic DNAPolymorphism analysis
The invention discloses a molecular marking method that is used for differentiating pepper disease resisting characteristics, which comprises extraction of pepper genomic DNA, PCR argumentation and polymorphism analysis of a specific enzyme incision product, and is characterized in that a pair of nucleotide molecules with a specified sequence that consists of a certain number of basic groups are artificially synthesized according to a plant disease resistance gene sequence and used as a primer, PCR reaction is realized to obtain a molecular marker, and pepper disease resistant materials and pepper disease susceptible materials can be identified through the size of the specific enzyme incision product in the marker. The molecular marking method can be used for disease resistance filtering in pepper seedling or adult phases and can greatly accelerate the process of disease-resistant pepper breeding.
Owner:NORTHWEST A & F UNIV
Paternity test method for goats and microsatellite primer and kit thereof
InactiveCN102465181AEasy to operateLow costMicrobiological testing/measurementDNA/RNA fragmentationDNA paternity testingElectrophoresis
The invention belongs to the technical field of research on molecular genetics and particularly relates to a method for utilizing a microsatellite marking technology to perform a paternity test on goats and a microsatellite primer and a kit thereof. The invention discloses a paternity test method for the goats, which comprises the following steps of: (1) extracting a blood sample from a goat breed and performing DNA (Deoxyribonucleic Acid) extraction and detection; (2) selecting a microsatellite locus: selecting the microsatellite locus with a stable PCR (Polymerase Chain Reaction) augmenting result and a clear strip as well as a polymorphism but without non-specific augmentation; (3) utilizing each microsatellite locus to perform PCR augmentation on different goat DNA templates, and performing electrophoresis detection on augmenting fragments; and (4) performing genetic polymorphism analysis on the microsatellite locus. The invention also discloses the microsatellite primer and the kit used for the paternity test method for the goats. The paternity test method for the goats, provided by the invention, is simple in operation, high in speed, low in cost, and high in accuracy.
Owner:TONGJI UNIV
SSR primer group developed based on multiple transcriptome sequences of color group zantedeschia aethiopica and acquisition method and application
ActiveCN113151545AAccelerate cultivationImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyZantedeschia aethiopica
The invention discloses an SSR primer group developed based on multiple transcriptome sequences of color group zantedeschia aethiopica and acquisition method and application. The method comprises the following steps of firstly, screening polymorphic SSR primers from transcriptome splicing sequences of three varieties of the color group zantedeschia aethiopica by utilizing a CandiSSR method, determining hundreds of polymorphic SSR primers after experimental verification, and then carrying out polymorphic analysis on more than 160 parts of color group zantedeschia aethiopica varieties by utilizing 35 pairs of screened SSR primers with high polymorphism. In addition, on the basis of the research, 160 core germplasm of the color group zantedeschia aethiopica is constructed through a maximization strategy, finally, 30 optimal representative parents of the color group zantedeschia aethiopica are screened out, and good help is provided for screening of core parents of the color group zantedeschia aethiopica, improvement of breeding efficiency and accelerated cultivation of new varieties with proprietary intellectual property rights in the future.
Owner:BEIJING ACADEMY OF AGRICULTURE & FORESTRY SCIENCES
SSR (Simple Sequence Repeat) primesr of platygaster robiniae buhl and duso and application thereof to population genetic diversity analysis
ActiveCN108588234AMicrobiological testing/measurementDNA/RNA fragmentationPolymorphism analysisPopulation evolution
The invention discloses SSR (Simple Sequence Repeat) primers of platygaster robiniae buhl and duso and application thereof to population genetic diversity analysis. The invention designs according tomitochondrial genome data of the platygaster robiniae buhl and duso to obtain more than 130 thousand of SSR sequences and screens out 37 pairs of SSR specific primers which are relatively stable and have excellent polymorphism from the SSR sequences. The invention further discloses application of the 37 pairs of SSR specific primers in the aspects of population genetic diversity analysis, population evolution analysis and polymorphic analysis of the platygaster robiniae buhl and duso, construction of fingerprint spectrum of the platygaster robiniae buhl and duso and the like. The SSR primers and the application thereof have the important value for developing natural enemy class group population genetic diversity research of the platygaster robiniae buhl and duso, mastering the source and the population differentiation condition of the platygaster robiniae buhl and duso and the like.
Owner:INST OF FOREST ECOLOGY ENVIRONMENT & PROTECTION CHINESE ACAD OF FORESTRY
SSR DNA markers for jute expressed sequence tags
InactiveCN103937786AMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversityMicrobiology
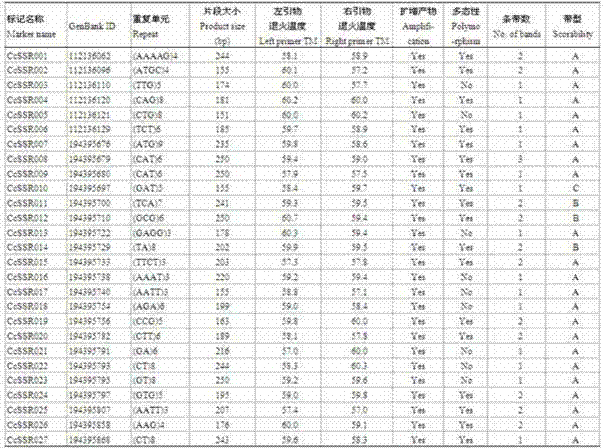
The invention relates to a molecular marker technology, specifically to SSR DNA markers for jute expressed sequence tags. A preparation method for the markers comprises the following steps: developing novel SSR sites; amplifying primer sequences of the SSR sites; and carrying out polymorphic analysis on the SSR sites. The invention is characterized in that jute expressed sequence tags are downloaded from the GenBank common database, 66 novel SSR markers of jute are separated and identified, 10 SSR markers with rich polymorphism are selected from the 66 novel SSR markers, and the 10 SSR markers comprise CcSSR008, CcSSR013, CcSSR016, CcSSR024, CcSSR029, CcSSR040, CcSSR049, CcSSR050, CcSSR052 and CcSSR053. The invention provides 66 novel jute SSR sites and the primer sequences and an amplification method used for amplifying the 66 SSR sites, which can be used for analysis of genetic diversity of different species of jute, construction of a genetic linkage map, molecular marker-assisted breeding and other research.
Owner:FUJIAN AGRI & FORESTRY UNIV
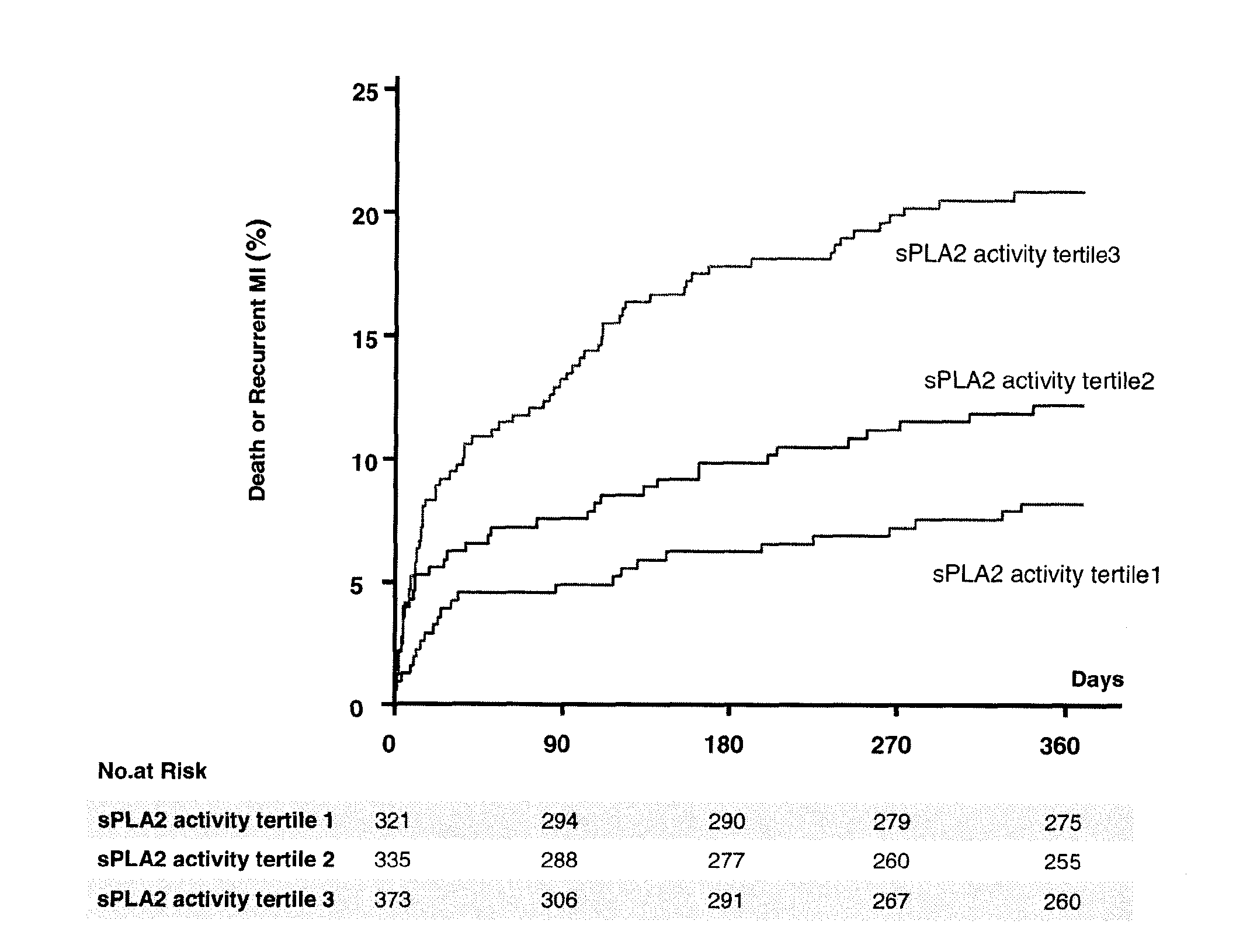
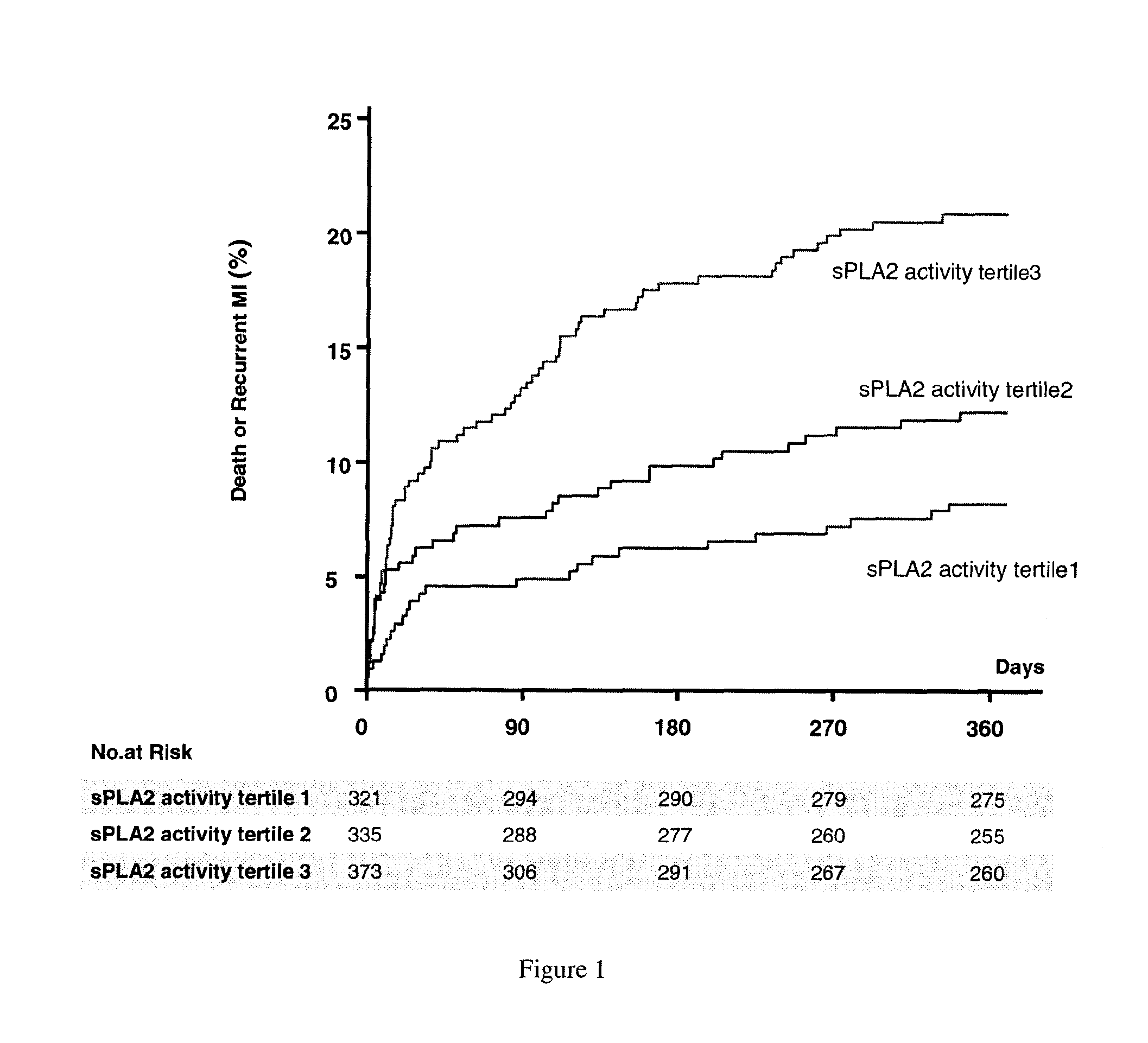
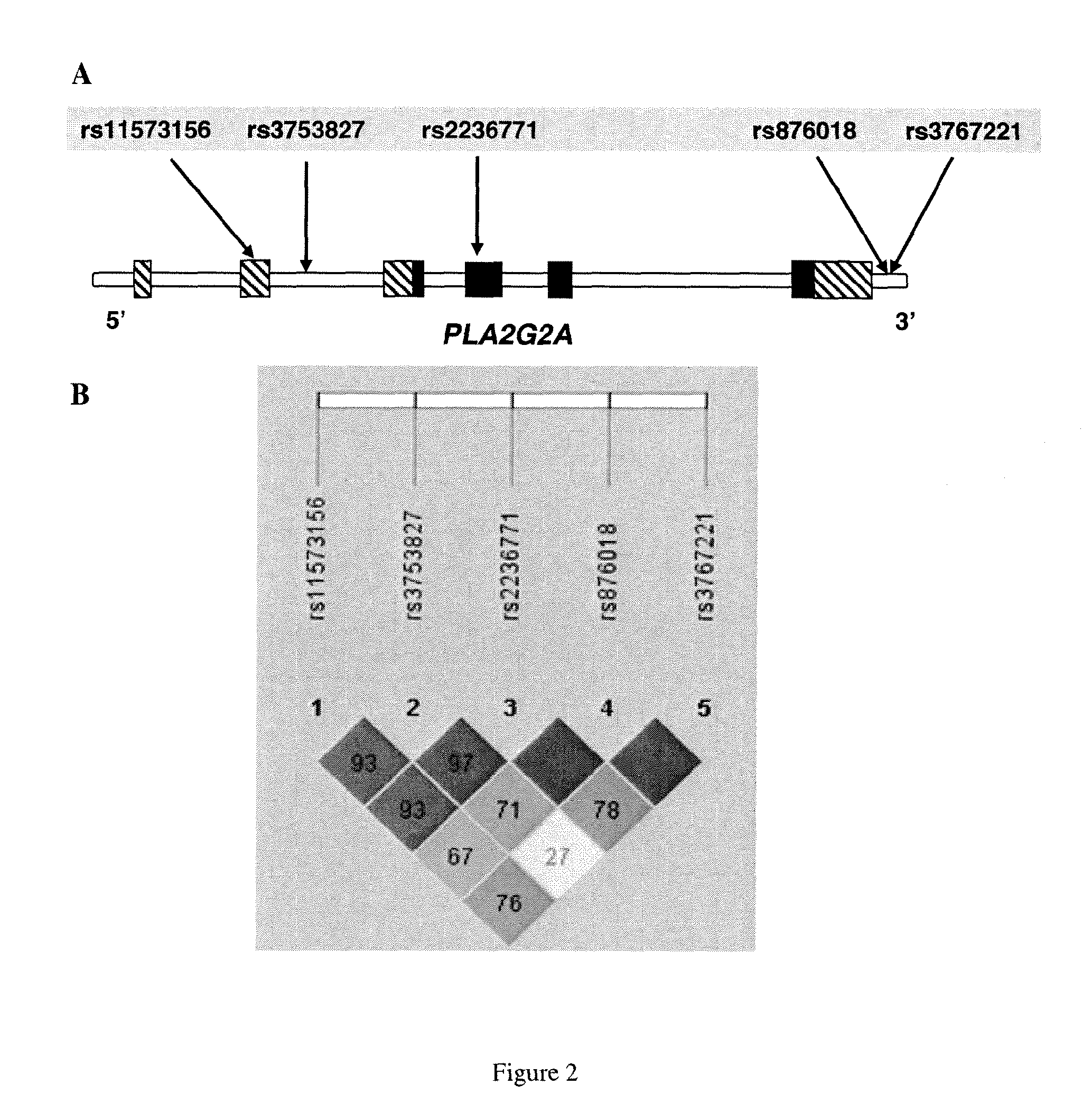
sPLA2 IIA Polymorphism Analysis for the Diagnosis/Prognosis of a Cardiovascular Disease/Event
InactiveUS20120225427A1Reduce riskReduce in quantityMicrobiological testing/measurementNucleotideIncreased risk
The invention relates to a method of identifying a subject having or at risk of having or developing a cardiovascular disease and / or a cardiovascular event, comprising determining, in a sample obtained from said subject, the presence or absence of a variant allele of nucleotide polymorphism (SNP) of the sPLA2 type IIA nucleic acid, wherein the SNP is selected from the group consisting of rs11573156 and rs2236771, wherein the presence of the minor allele (G) of SNP rs11573156 indicates an increased risk of having or being at risk of having or developing a cardiovascular disease and / or cardiovascular event, and the presence of the minor allele (C) of SNP rs2236771 indicates a decreased risk of having or being at risk of having or developing a cardiovascular disease and / or cardiovascular event.
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM)
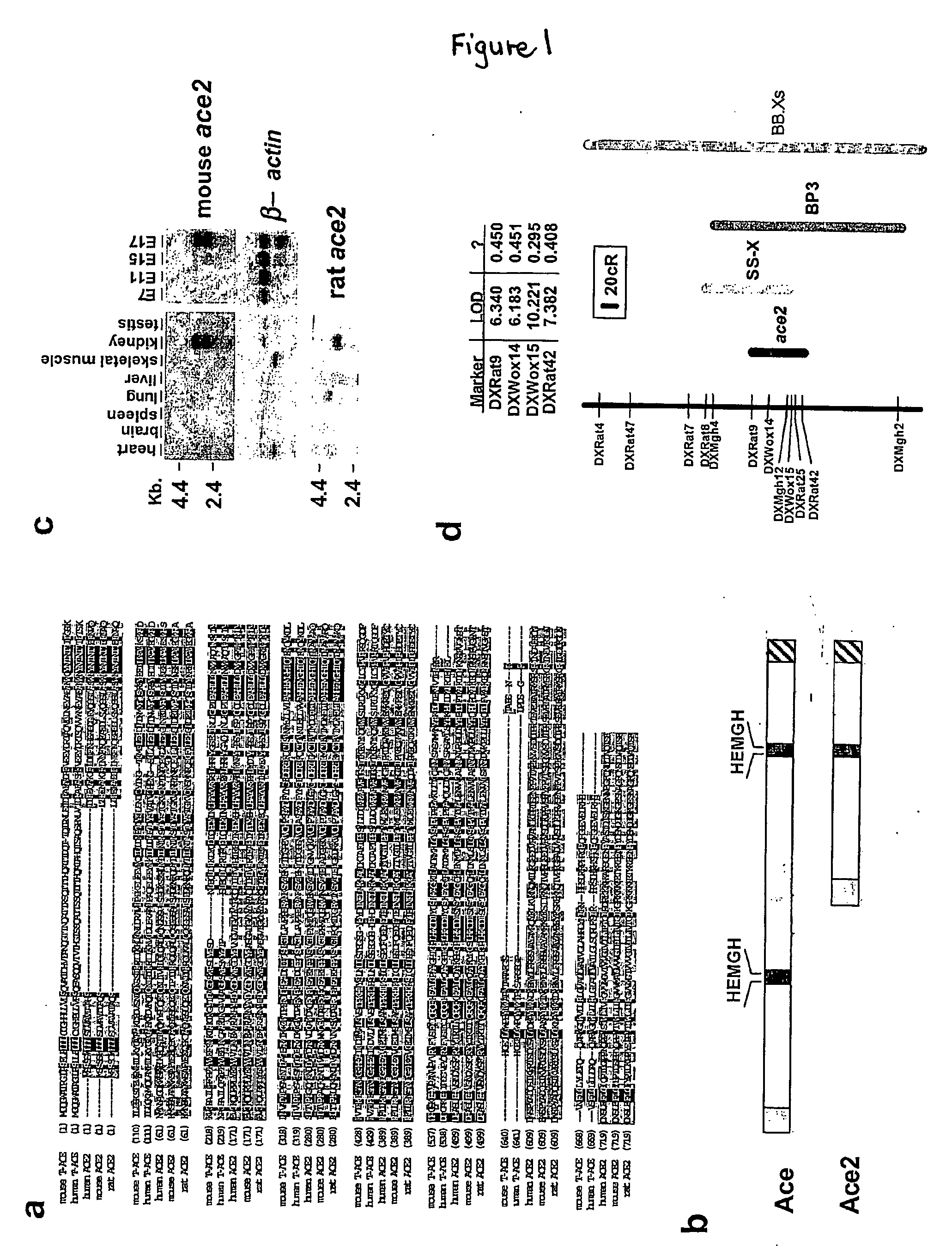
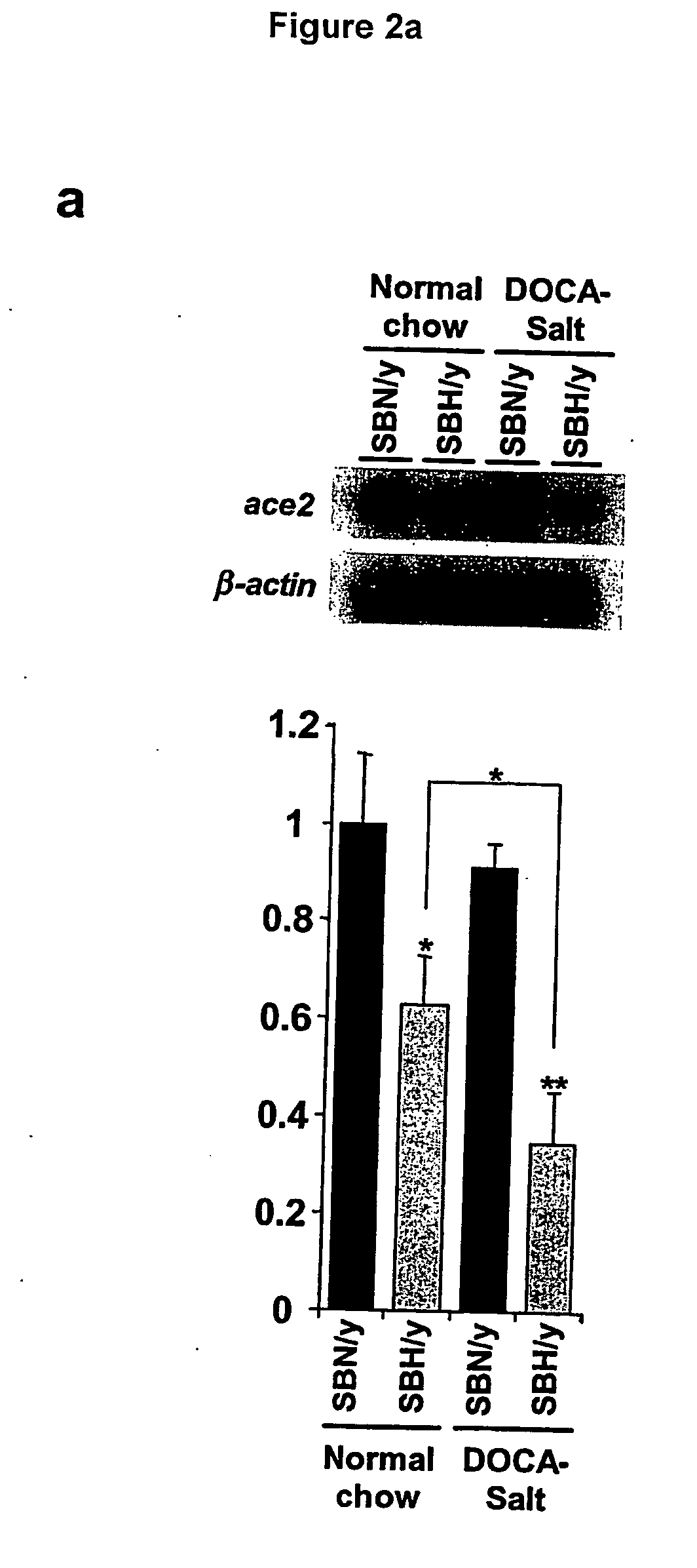
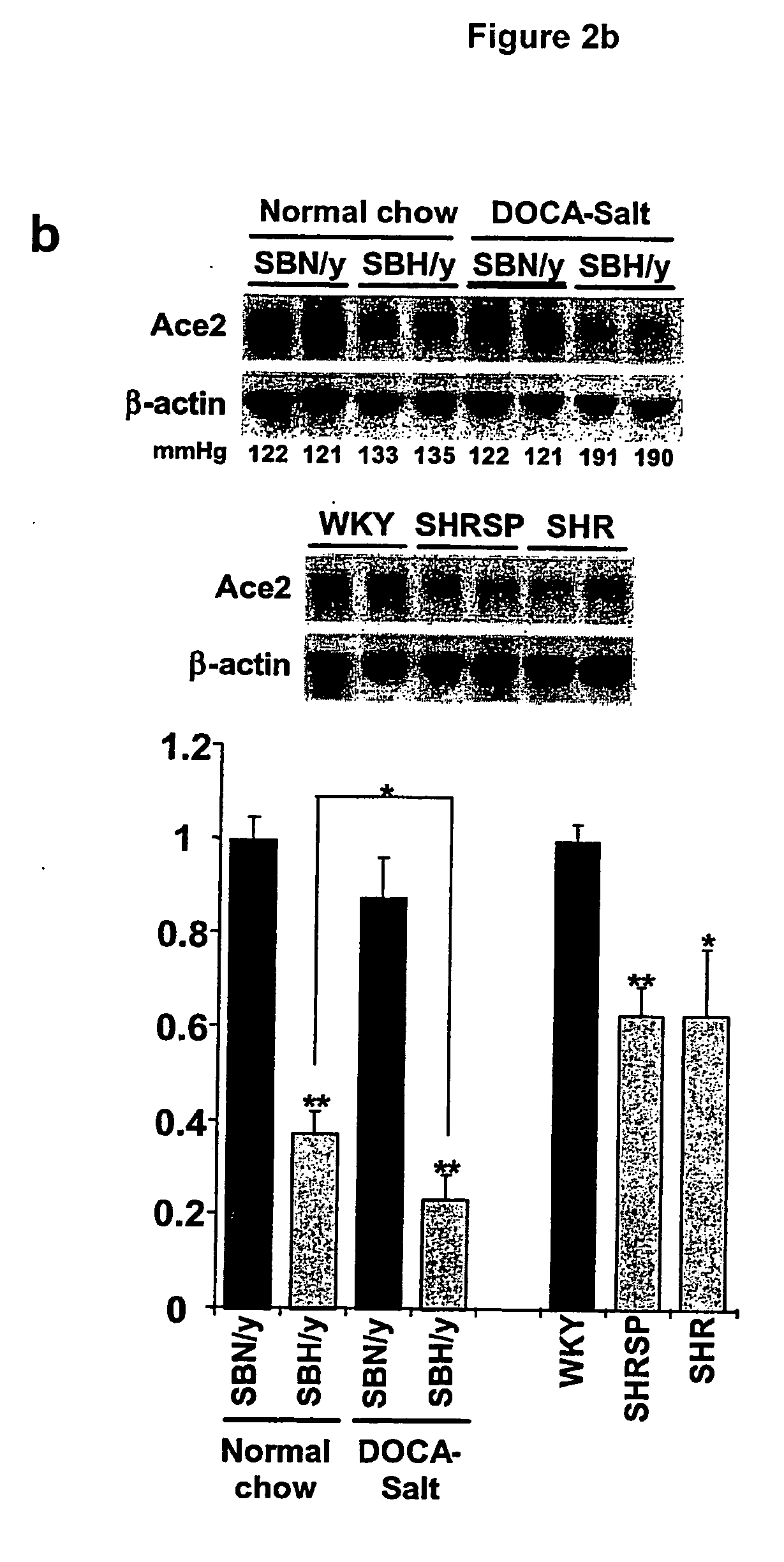
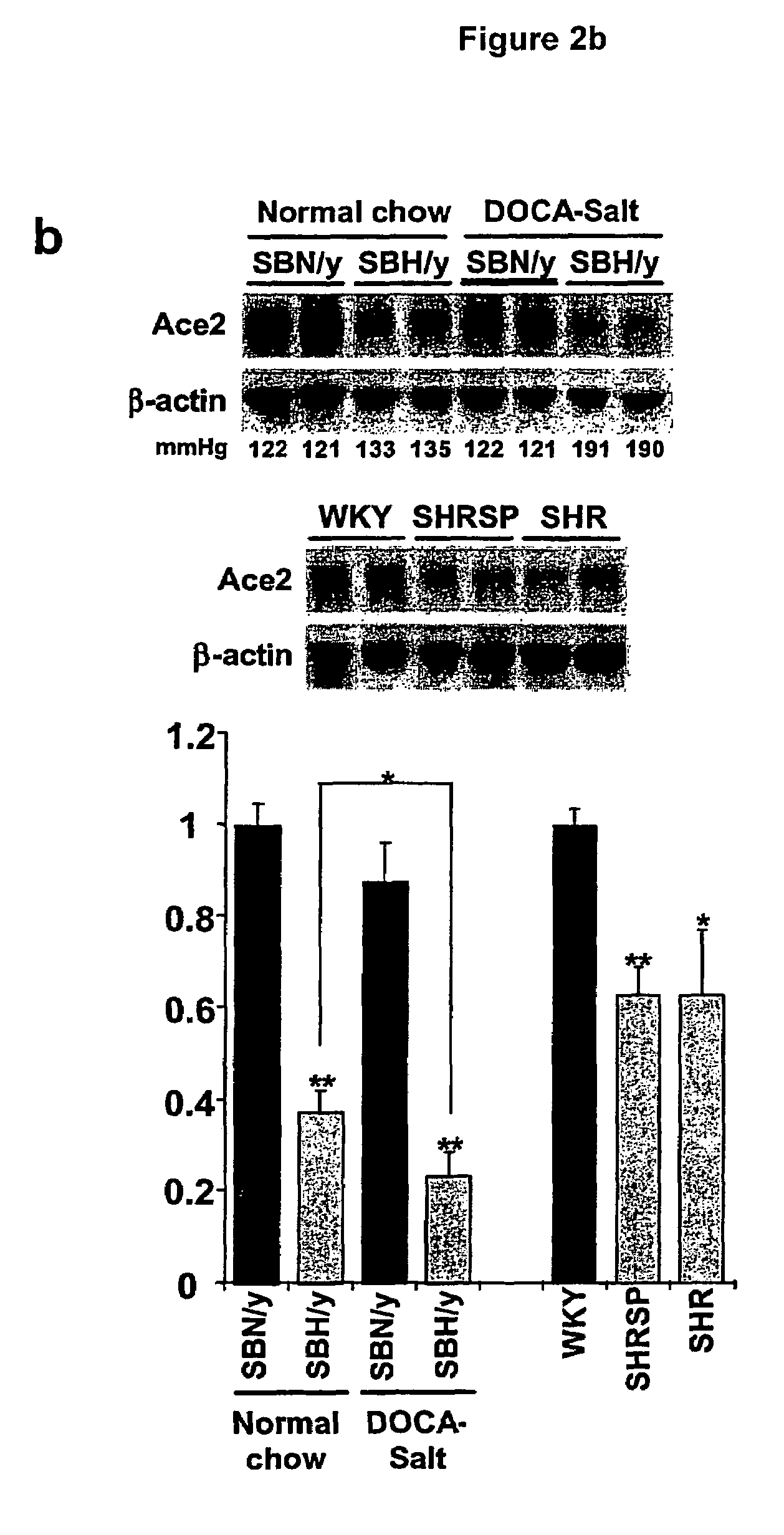
Ace2 activation for treatment of heart, lung and kidney disease and hypertension
InactiveUS20050251873A1Prevents and treat hypertensionPrevents and treat and cardiacBiocidePeptide/protein ingredientsVascular diseaseNephrosis
The invention relates to ACE2 activating compounds for prevention and treatment of cardiovascular disease, kidney disease, lung disease and hypertension. The invention also includes methods of diagnosing cardiovascular disease, kidney disease, lung disease and hypertension by measuring ACE2 expression or nucleotide polymorphism analysis.
Owner:HEALTH NETWORK UNIV +1
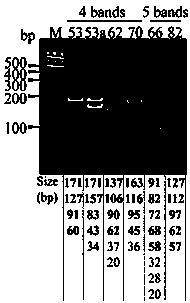
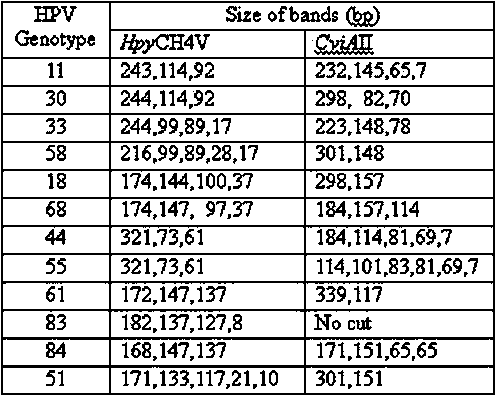
Method and kit for detecting HPV (Human Papillomavirus) genotype
ActiveCN103361442APrecise screeningIntuitively determinedMicrobiological testing/measurementMicroorganism based processesHuman papillomavirusA-DNA
The invention discloses a method and a kit for detecting an HPV (Human Papillomavirus) genotype. The method for detecting the HPV genotype comprises the following steps of: A, extracting DNA (Deoxyribonucleic Acid); b, carrying out PCR (Polymerase Chain Reaction) on a DNA template, and amplifying to obtain a PCR product of an HPV-infected DNA sample; and C, carrying out restricted segment length polymorphism analysis through restriction endonuclease. The method is low in detection cost and can be used for rapidly and intuitively determining the type of the infected HPV according to a special HPV type atlas.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
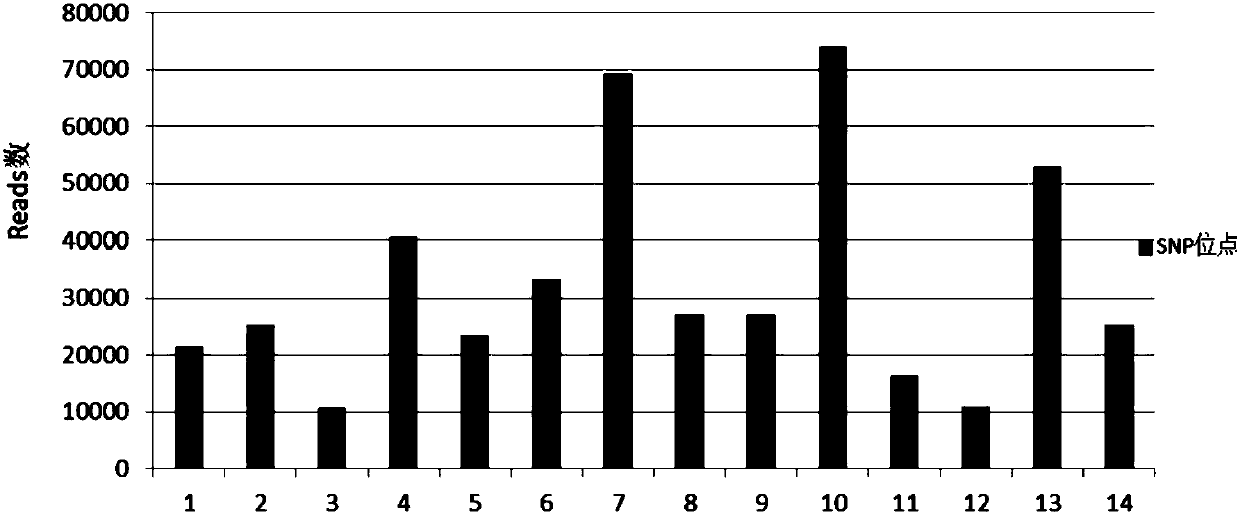
A method and a kit for detecting nucleic acid sample pollution and application of the method
ActiveCN108823296AGuarantee the quality of inspectionEasy to operateMicrobiological testing/measurementPolymorphism analysisBiology
The application discloses a method and a kit for detecting nucleic acid sample pollution and application of the method. The method for detecting the nucleic acid sample pollution includes the following steps: PCR amplification is performed on a nucleic acid sample to be measured by adopting a SNP detection primer, mononucleotide polymorphism of the nucleic acid sample to be measured is analyzed, whether pollution of different individual sources is existed in the nucleic acid sample to be measured or not is judged according to a mononucleotide polymorphism analysis result; and the SNP detectionprimer is composed of at least an analysis primer pair of 14 SNP sites. The specificity of each individual SNP is creatively utilized and is used as specific identification information of the nucleicacid sample to be detected, if SNP information of the detected nucleic acid sample to be measured conforms to the self SNP information, the result that the nucleic acid sample to be detected is not polluted is judged, if not, and the result that the nucleic acid sample to be detected has pollution is judged. Whether the nucleic acid sample to be measured exists pollution or not can be accuratelyjudged by the detection method.
Owner:BGI GENOMICS CO LTD +2
Method for developing ramie genome SNP marks on large scale and primer developed by method
InactiveCN104212898AHigh speedLow costMicrobiological testing/measurementDNA/RNA fragmentationEnzyme digestionSmall fragment
The invention discloses a method for developing ramie genome SNP marks on a large scale and a primer developed by the method. The method particularly comprises the following steps: (1) extracting DNAs of two ramie varieties; (2) carrying out enzyme digestion on two ramie variety genomes by using RsaI enzyme, and thus obtaining small fragments of 214-314bp; (3) sequencing the small fragments obtained by enzyme digestion, and thus obtaining SLAF labels; (4) carrying out polymorphic analysis as to the SLAF labels obtained by the two ramie varieties according to the number of alleles and the difference between the gene sequences, and thus searching the SLAF labels with SNP; and (5) carrying out primer design on the searched SLAF labels, and detecting to be genome SNP marks. Compared with a traditional method for developing molecular markers, the method is feasible, simple and convenient to operate, high in efficiency, and low in cost, and a solid foundation is established for ramie molecular biology and genetics by obtaining of the ramie genome SNP marks on a large scale.
Owner:INST OF BAST FIBER CROPS CHINESE ACADEMY OF AGRI SCI
Construction method for breeding pedigree of lutraria sieboldii
ActiveCN108450378AImprove growth traitsMeet parent number requirementsClimate change adaptationPisciculture and aquariaDiseaseOpen water
The invention discloses a construction method for a breeding pedigree of lutraria sieboldii. The lutraria sieboldii is a rare bivalve shellfish, and excessive breeding causes problems such as germplasm degeneration and frequent diseases; and pedigree breeding becomes a first choice for industrial development, but because of a subtidal open water area breeding environment of the lutraria sieboldii,complex resources, and a large number of individuals to be marked, common marking and molecular marking are difficult, and an effective pedigree breeding mechanism is not established in China. The method disclosed by the invention adopts the technical steps of artificial directional insemination, breeding, pedigree individual marking and cultivation to solve the difficult problem of pedigree construction of pedigree breeding of the lutraria sieboldii, can quickly realize the pedigree breeding by combining genetic polymorphism analysis, and provides technical references for pedigree breeding of the lutraria sieboldii and related subtidal bivalve shellfish.
Owner:广西海洋研究所有限责任公司
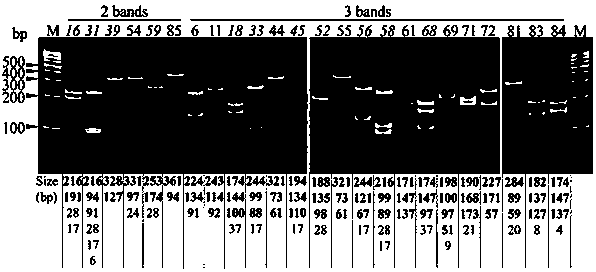
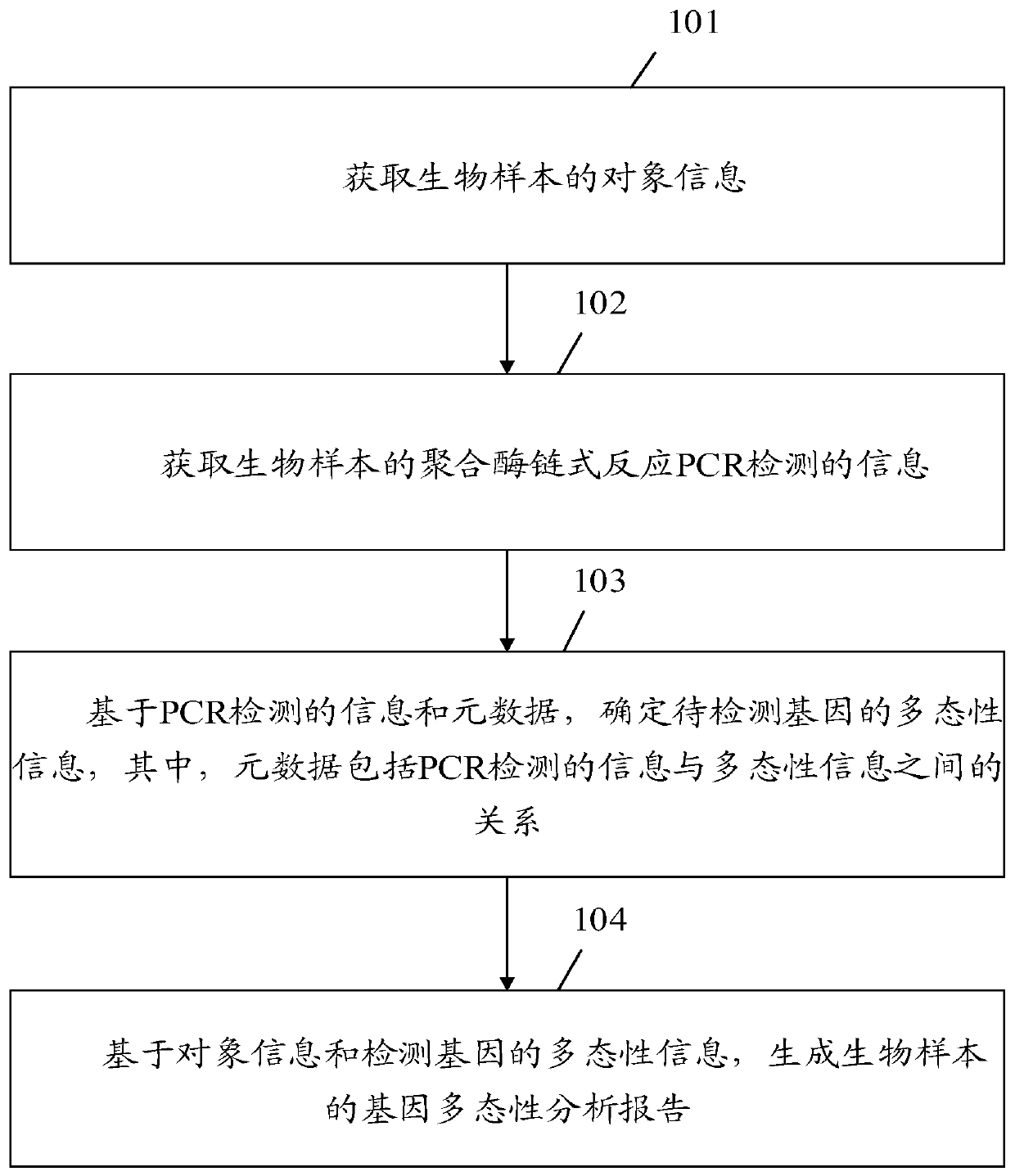
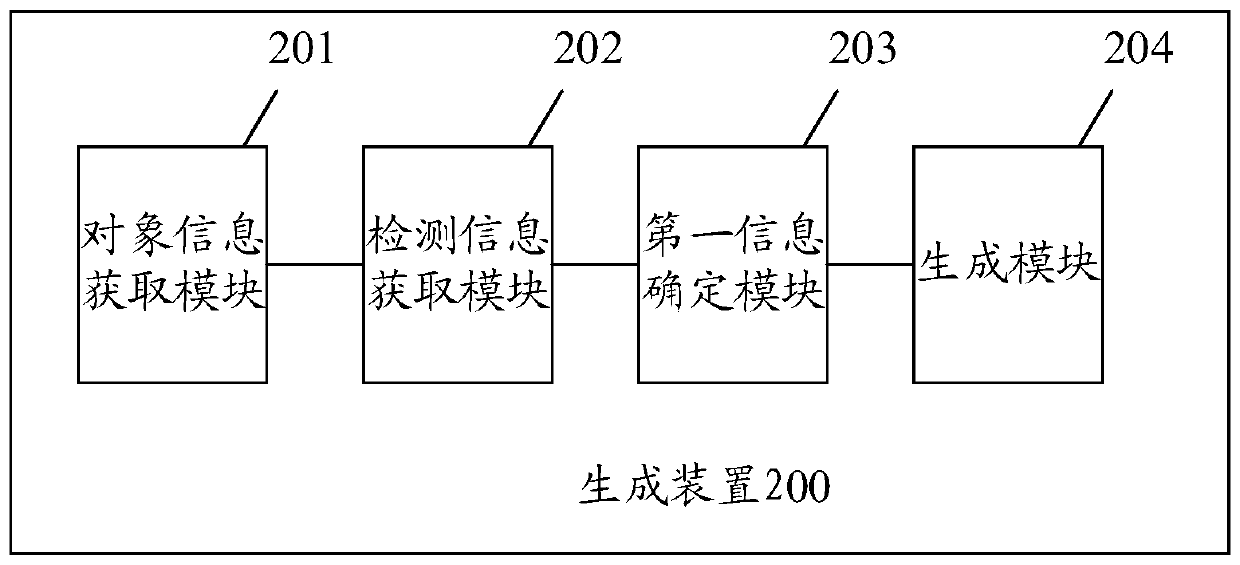
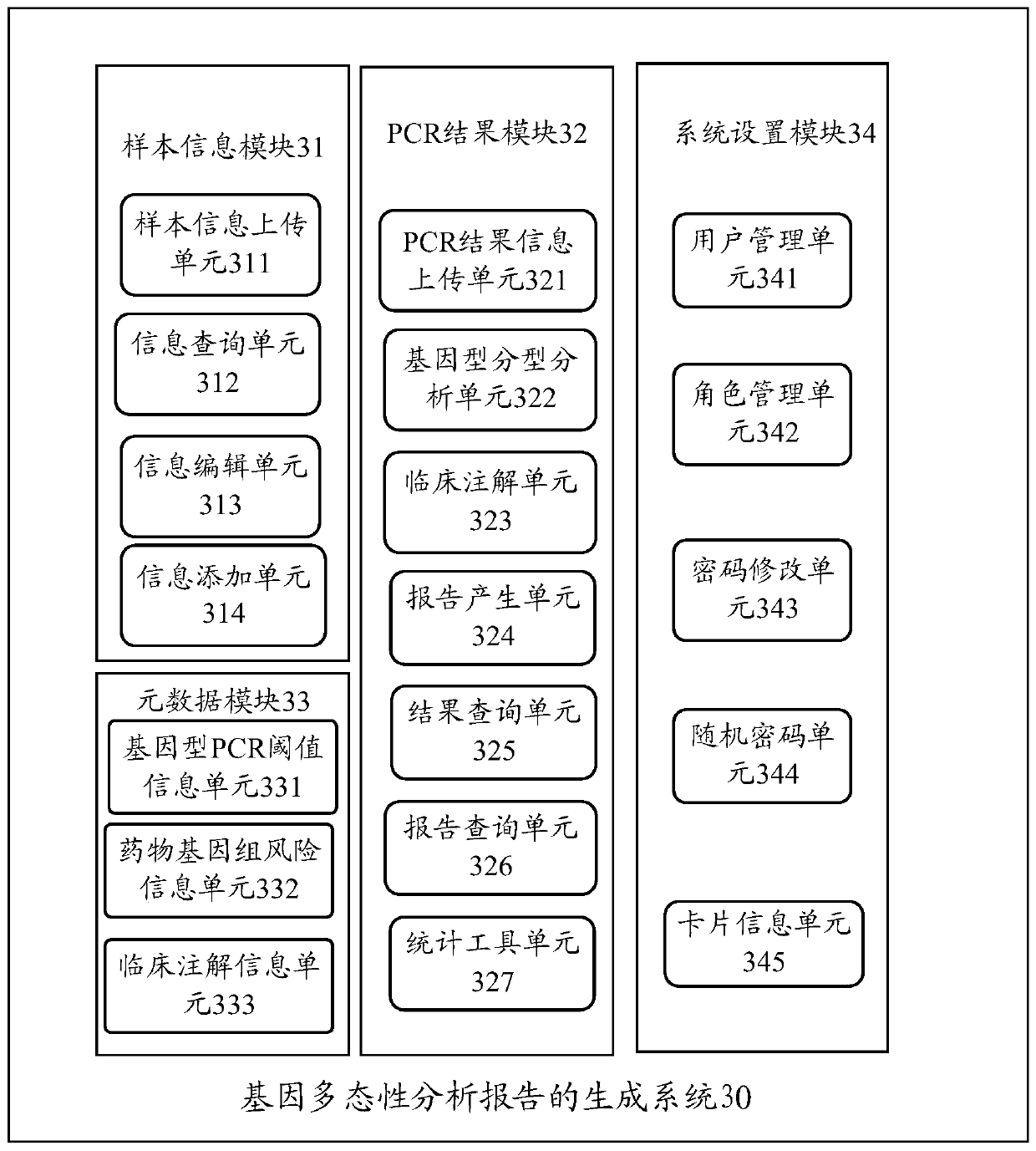
Gene polymorphism analysis report generation method and apparatus, equipment and storage medium
ActiveCN111564178AImprove work efficiencyReduce error rateData visualisationBiostatisticsPolymorphism analysisBioinformatics
Embodiments of the invention disclose a gene polymorphism analysis report generation method and apparatus, equipment and a computer storage medium. The method comprises the steps of acquiring object information of a biological sample; acquiring information about polymerase chain reaction (PCR) detection of the biological sample; on the basis of the PCR detection information and metadata, determining polymorphism information of a detection gene, wherein the metadata comprises relation between the PCR detection information and the polymorphism information; and generating a gene polymorphism analysis report of the biological sample based on the object information and the polymorphism information of the detection gene. Therefore, the analysis of multiple gene polymorphisms of multiple biological samples can be quickly completed according to a PCR detection result, so the error rate of interpreting the PCR detection result is reduced; and the gene polymorphism analysis report of the biological sample can be quickly generated, so the working efficiency of generating the analysis report is improved, and the defect of overlong consumed time is avoided.
Owner:北京圣维尔医学检验实验室有限公司
ACE2 activation for treatment of heart, lung and kidney disease and hypertension
The invention relates to ACE2 activating compounds for prevention and treatment of cardiovascular disease, kidney disease, lung disease and hypertension. The invention also includes methods of diagnosing cardiovascular disease, kidney disease, lung disease and hypertension by measuring ACE2 expression or nucleotide polymorphism analysis.
Owner:HEALTH NETWORK UNIV +1
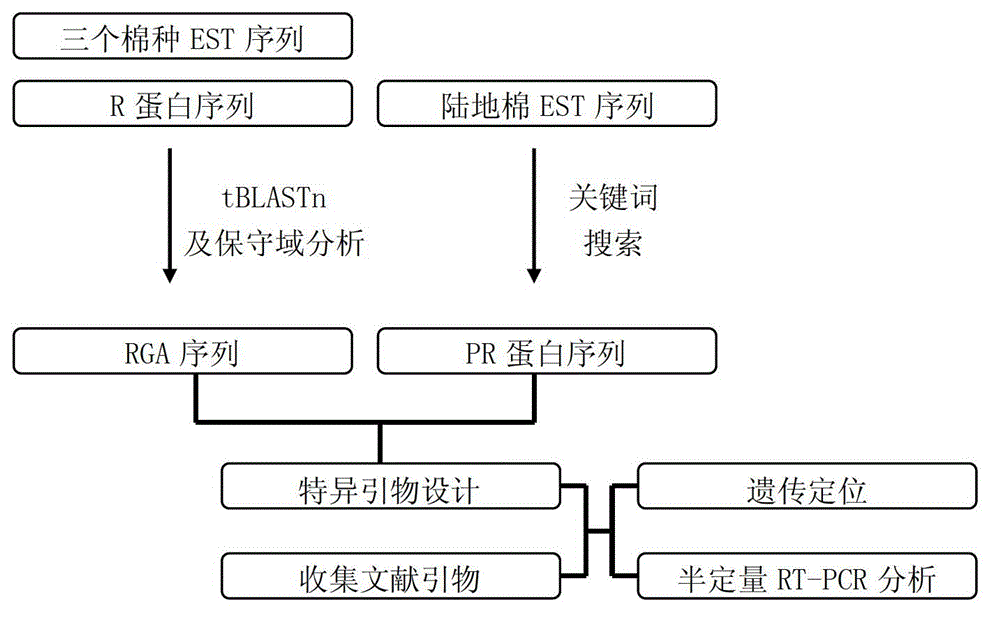
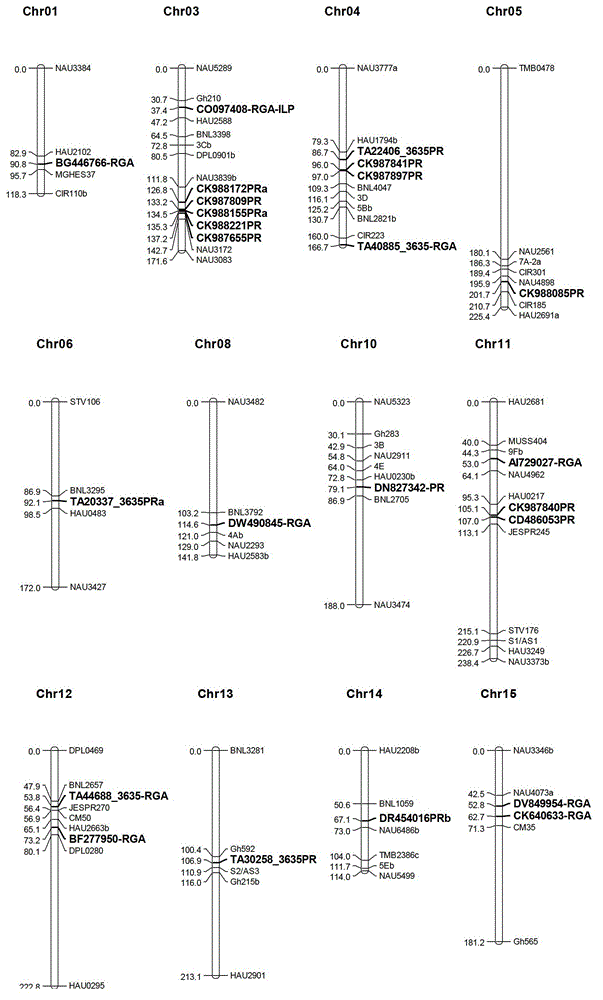
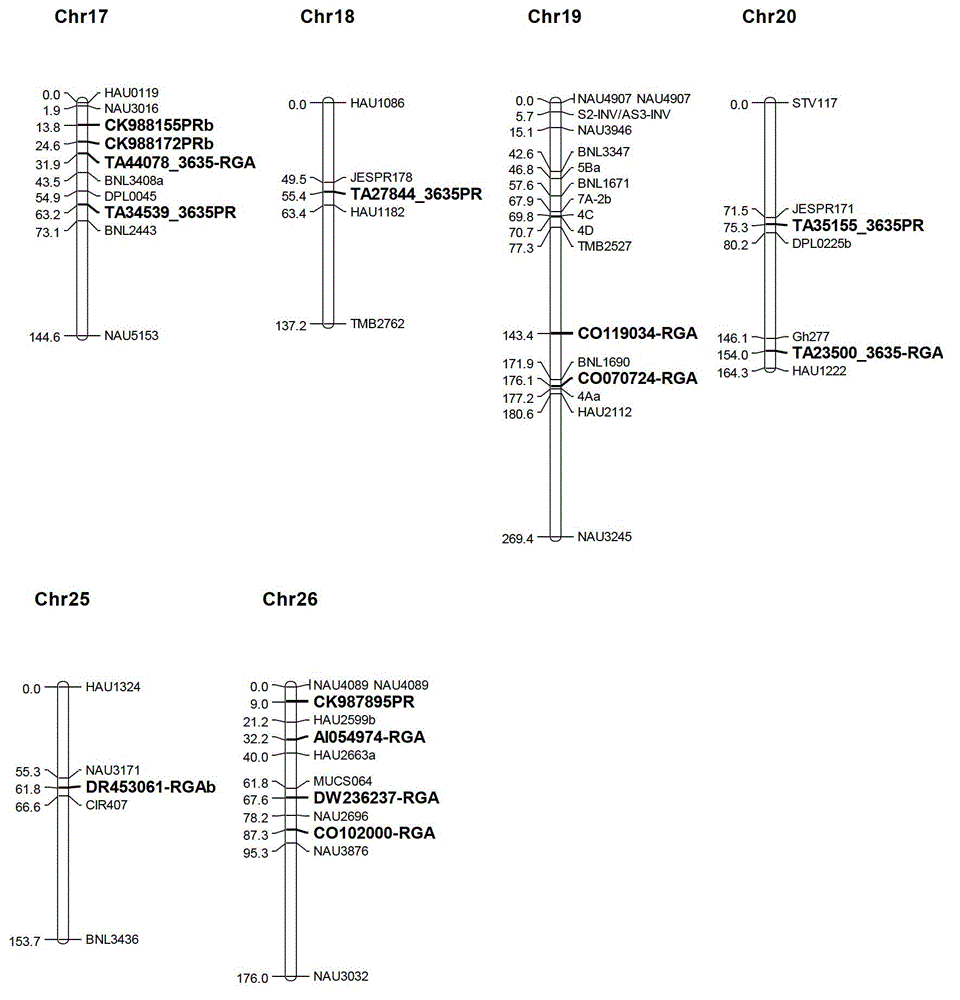
Marking method for resistance gene homologues based on EST(Expressed Sequence Tag) data mining
InactiveCN102747154AAccelerate the breeding process for disease resistanceIncrease the number ofMicrobiological testing/measurementSpecial data processing applicationsSingle-strand conformation polymorphismR protein
The invention discloses a marking method for resistance gene homologues based on EST data mining. The method comprises the following steps: (1) sequence mining and primer designing; (2) RGA sequence mining and primer designing, wherein 96 R protein sequences of different species are collected through literature, 196 R protein sequences of Arabidopis thaliana are downloaded at the same time, and EST data of three cotton species are downloaded from TIGR; and (3) genetic mapping of developed markers, wherein a BC1 population between Gossypium hirsutum and G. barbadense is used, then polymorphism is screened by using the parent of Hubei cotton-22 and 3-79 under the condition of electrophoresis of single-strand conformation polymorphism, screened polymorphism markers are used for polymorphism analysis of the BC1 population, and obtained data are introduced into JoinMap3.0 for construction. The method provided in the invention has the advantages of low cost, no need for cost for amplification of degenerate primers and considerable monoclonal sequencing, a greater number of obtained sequences, more abundant sequence varieties and capacity of providing effective molecular markers for molecular breeding.
Owner:HUAZHONG AGRI UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com