A method and a kit for detecting nucleic acid sample pollution and application of the method
A nucleic acid sample and kit technology, applied in the field of nucleic acid sample contamination detection, can solve problems such as cross-contamination and sample contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
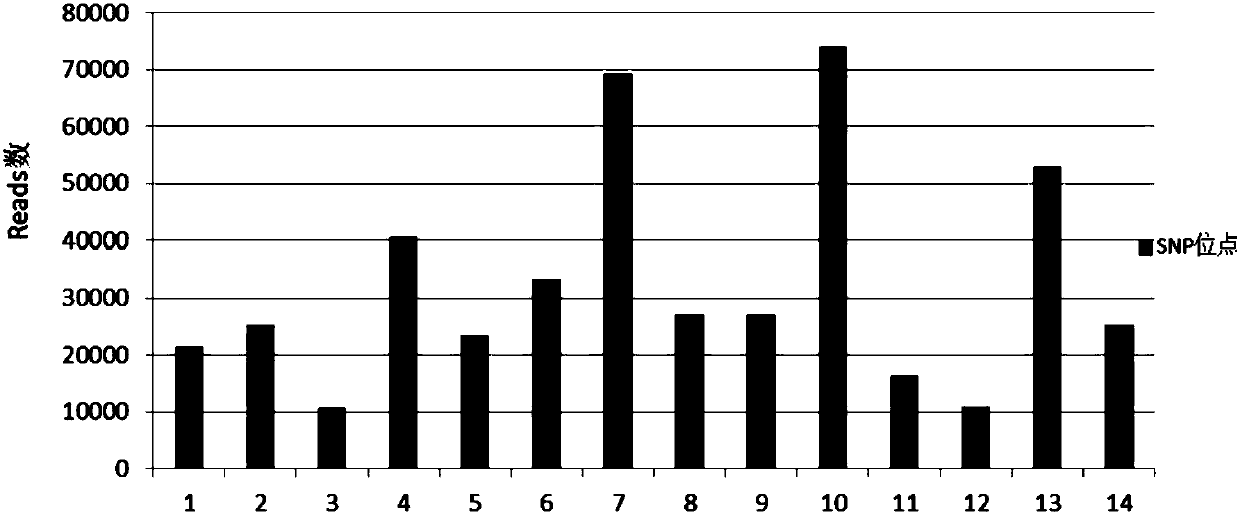
[0050] 1. Screening of SNP loci
[0051] Not all SNP sites are suitable for the detection of immune repertoire contamination samples. First, it must be ensured that the primers for amplifying the SNP sites, the multiple PCR primers of the immune repertoire, and the CDR3 region of BCR / TCR have no more than 4 bp contiguousness. Base identical or complementary pairing, to avoid the impact of SNP site detection on the multiple PCR amplification of immune repertoire.
[0052] In this example, the primers of the existing 21 SNP sites were first compared, analyzed and screened, and then the primers screened out by the comparative analysis were screened through specific tests to use specific SNP site primers as SNP site primers for this example. sample detection method. The sequence comparison analysis in this example uses the sequence comparison software independently developed by BGI, or uses sequence comparison software such as DNAman. The primers for the 21 SNP sites used for scr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com