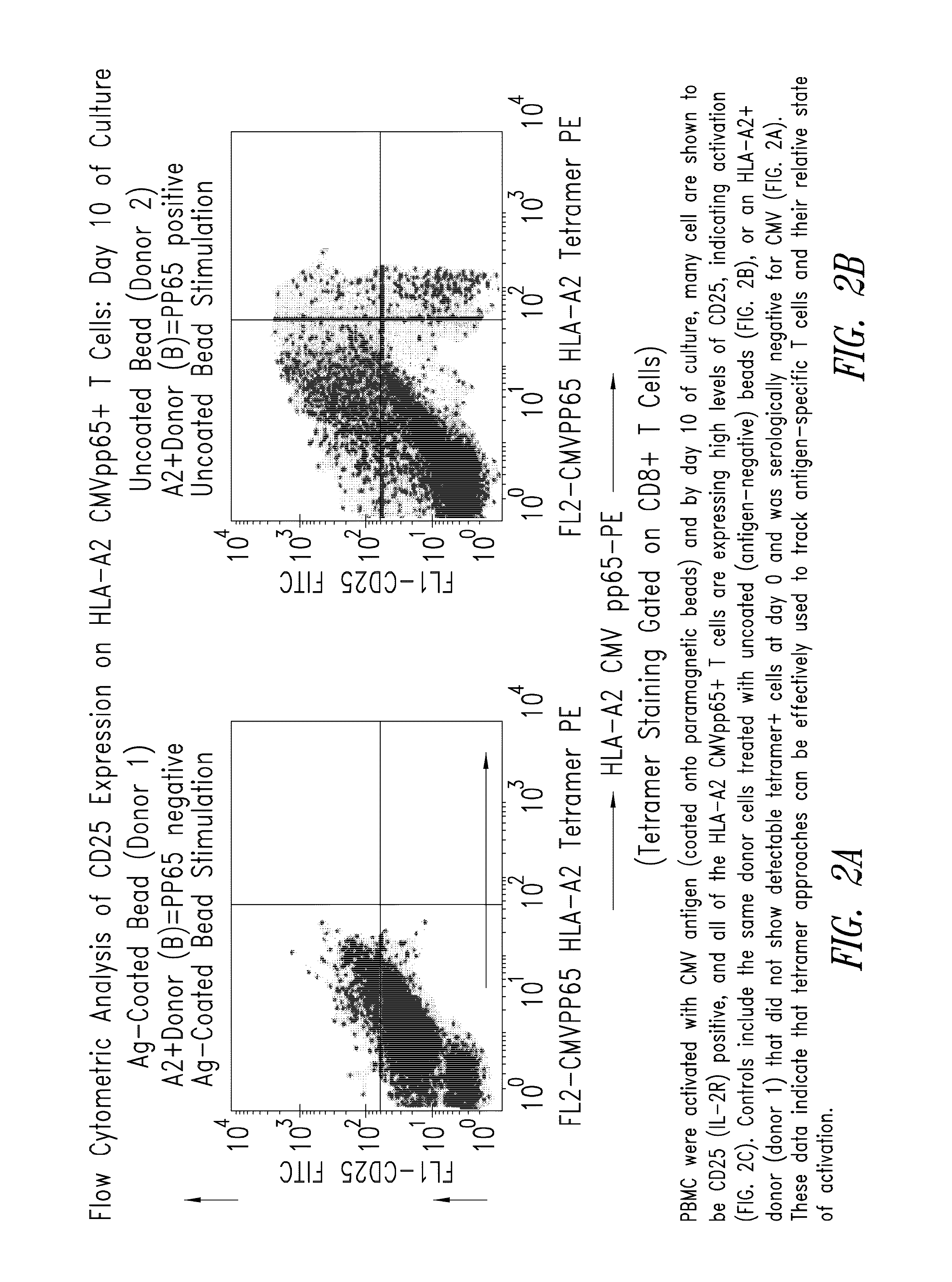
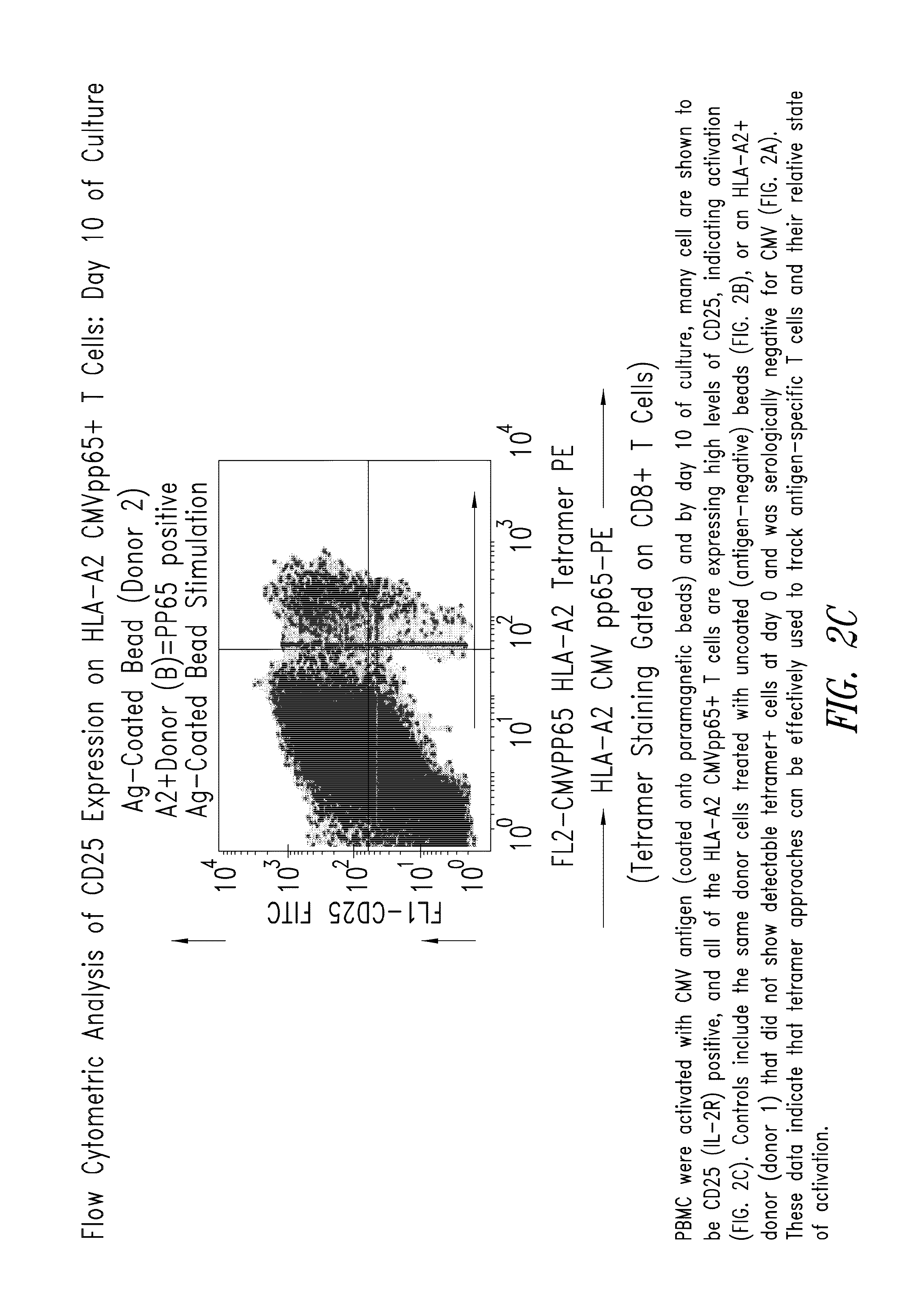
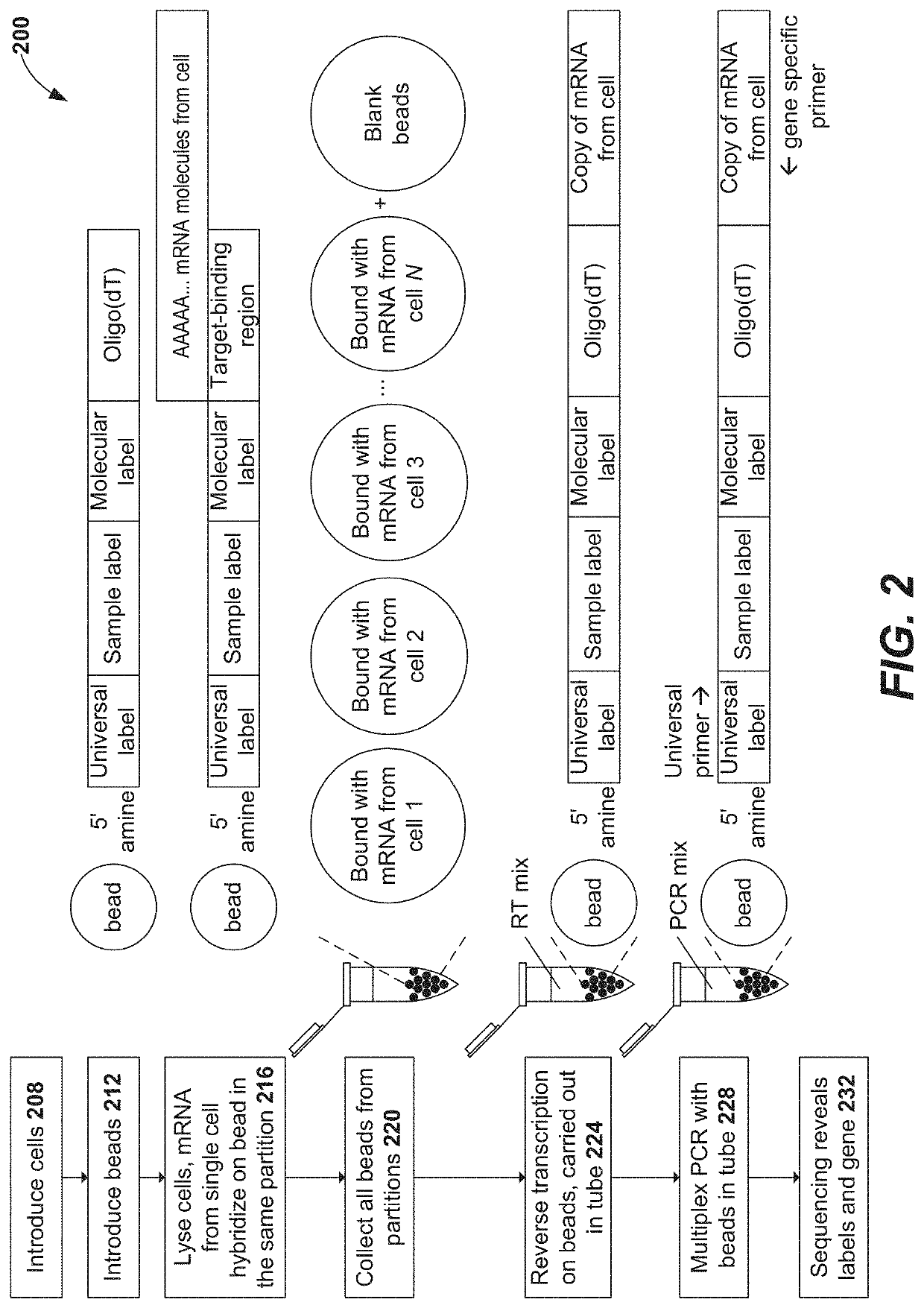
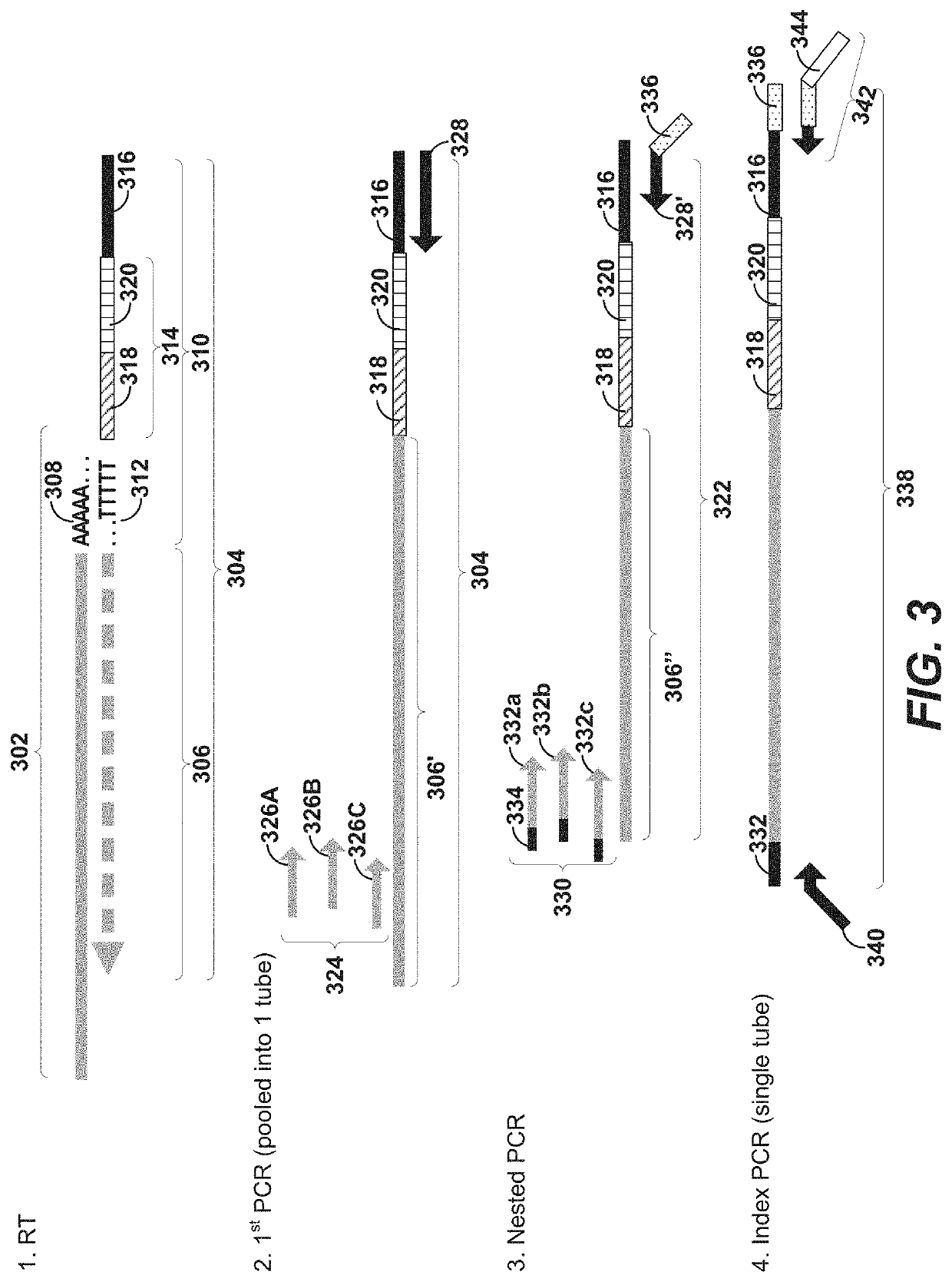
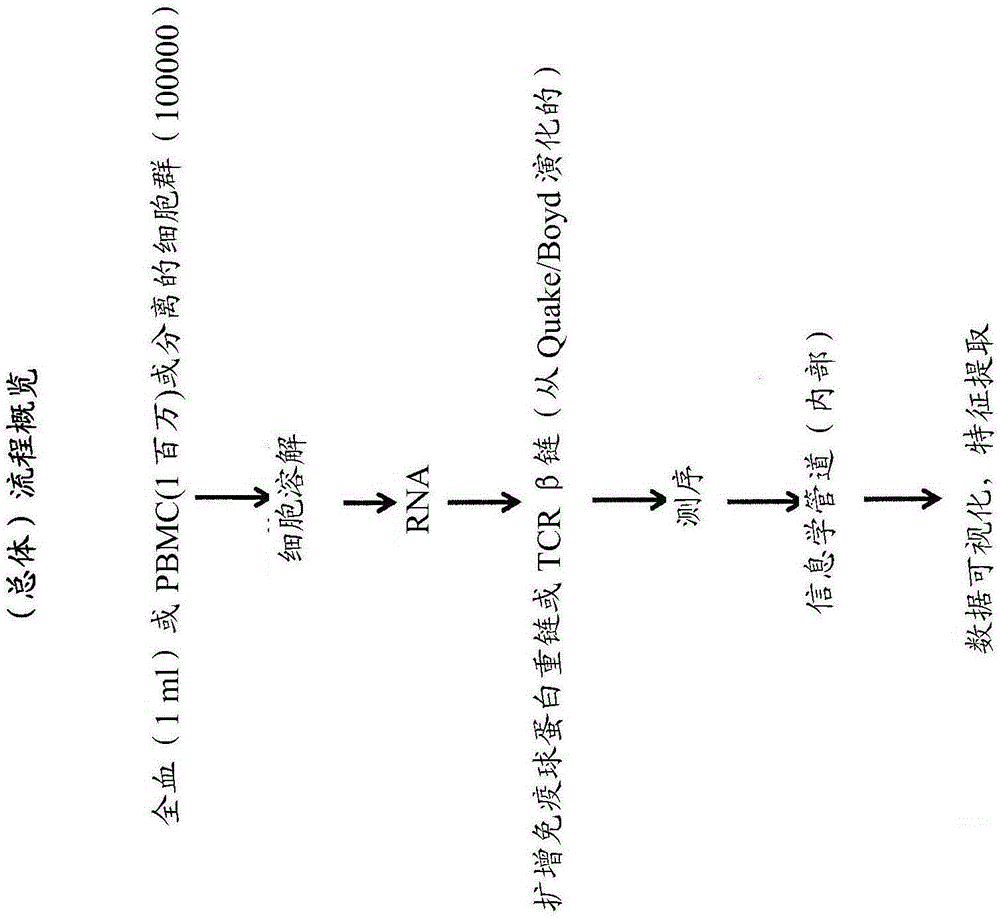Patents
Literature
87 results about "Immune repertoire" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
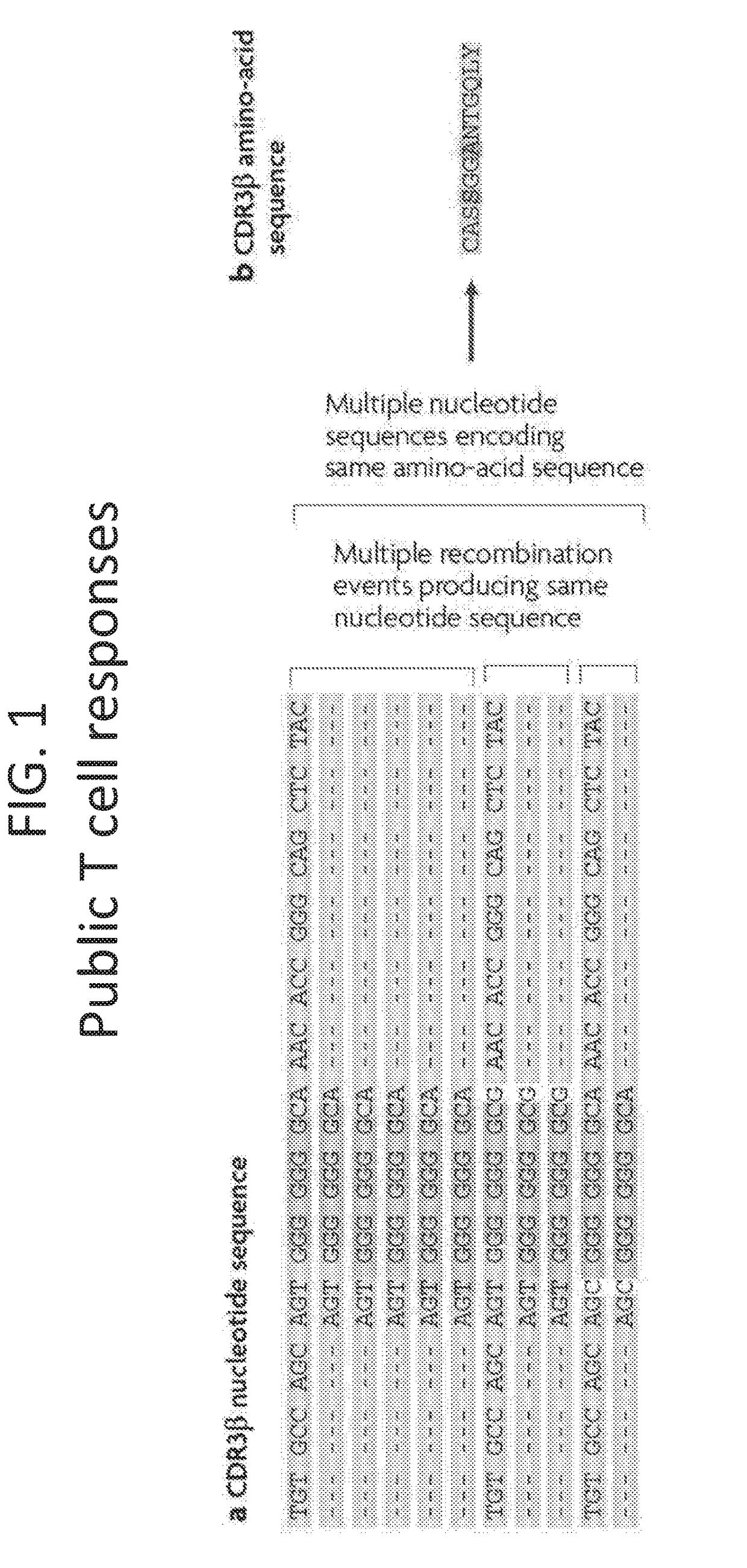
The immune repertoire, is defined as, the number of different sub-types an organism's immune system makes, of any of the 6 key types of protein, either immunoglobulin or T cell receptor.
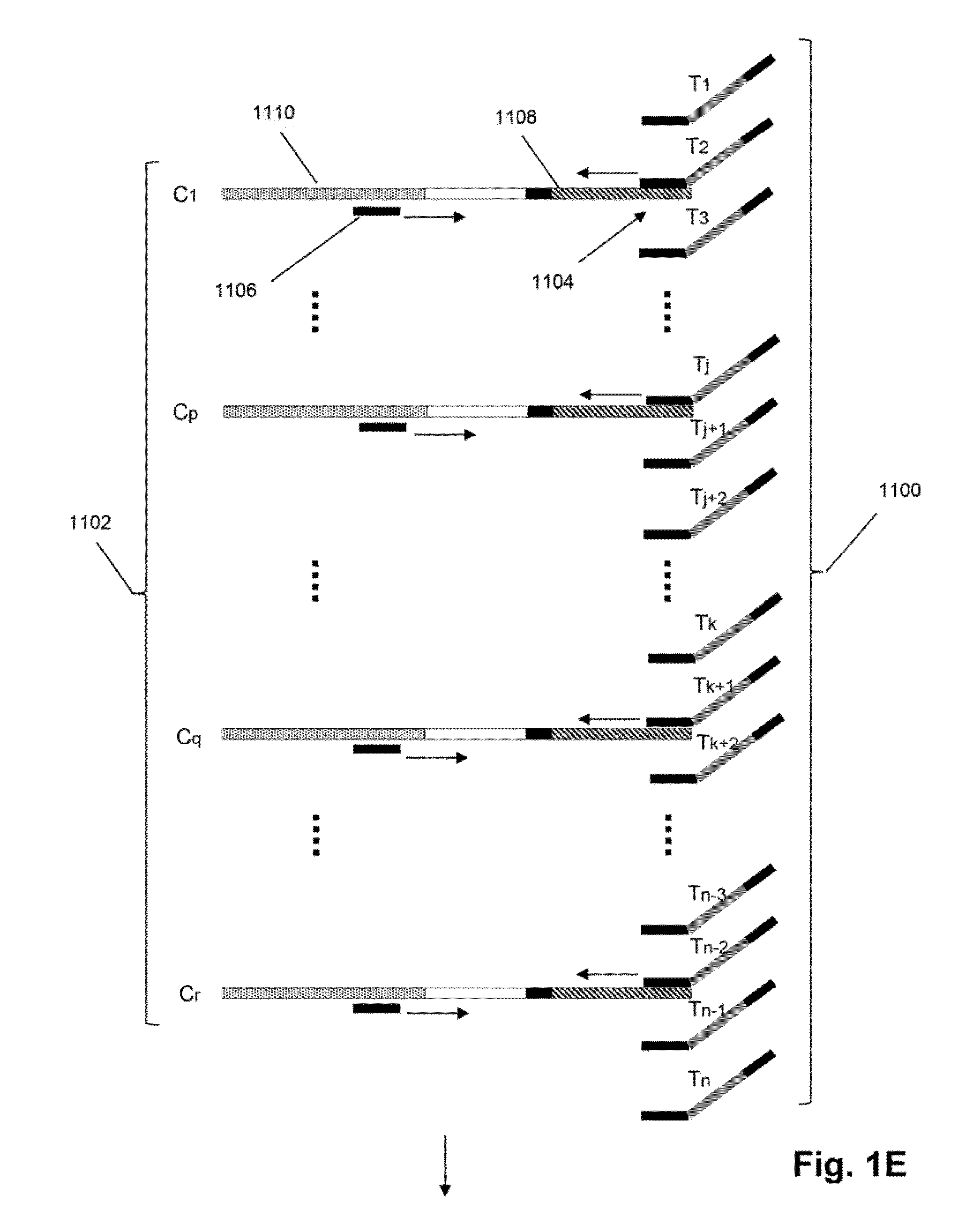
Single cell bar-coding for antibody discovery
Provided herein are methods and composition for immune repertoire sequencing and single cell barcoding for heavy and IgL pairing if antibodies.
Owner:ABVITRO LLC
Single cell bar-coding for antibody discovery
Owner:ABVITRO LLC
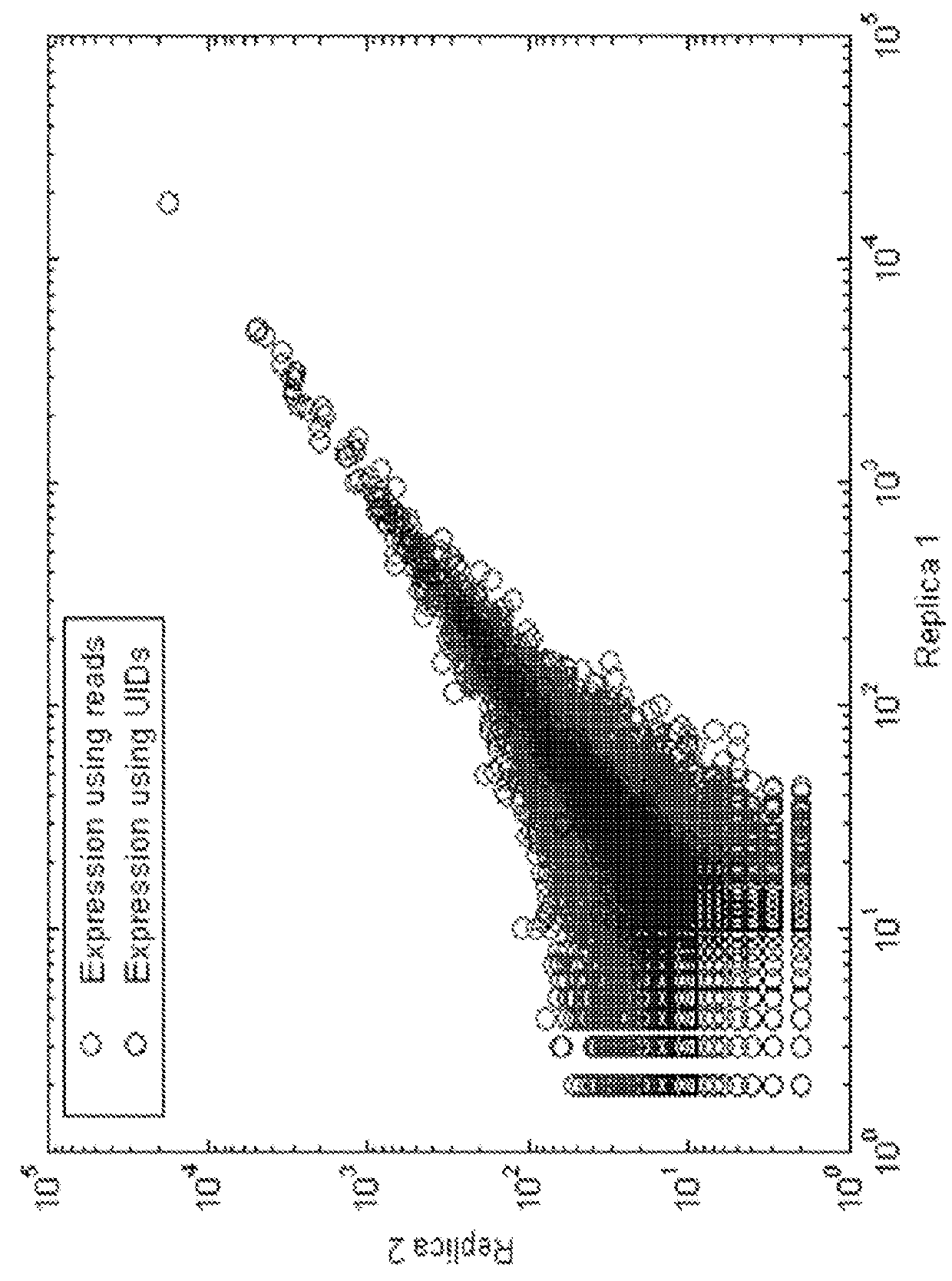
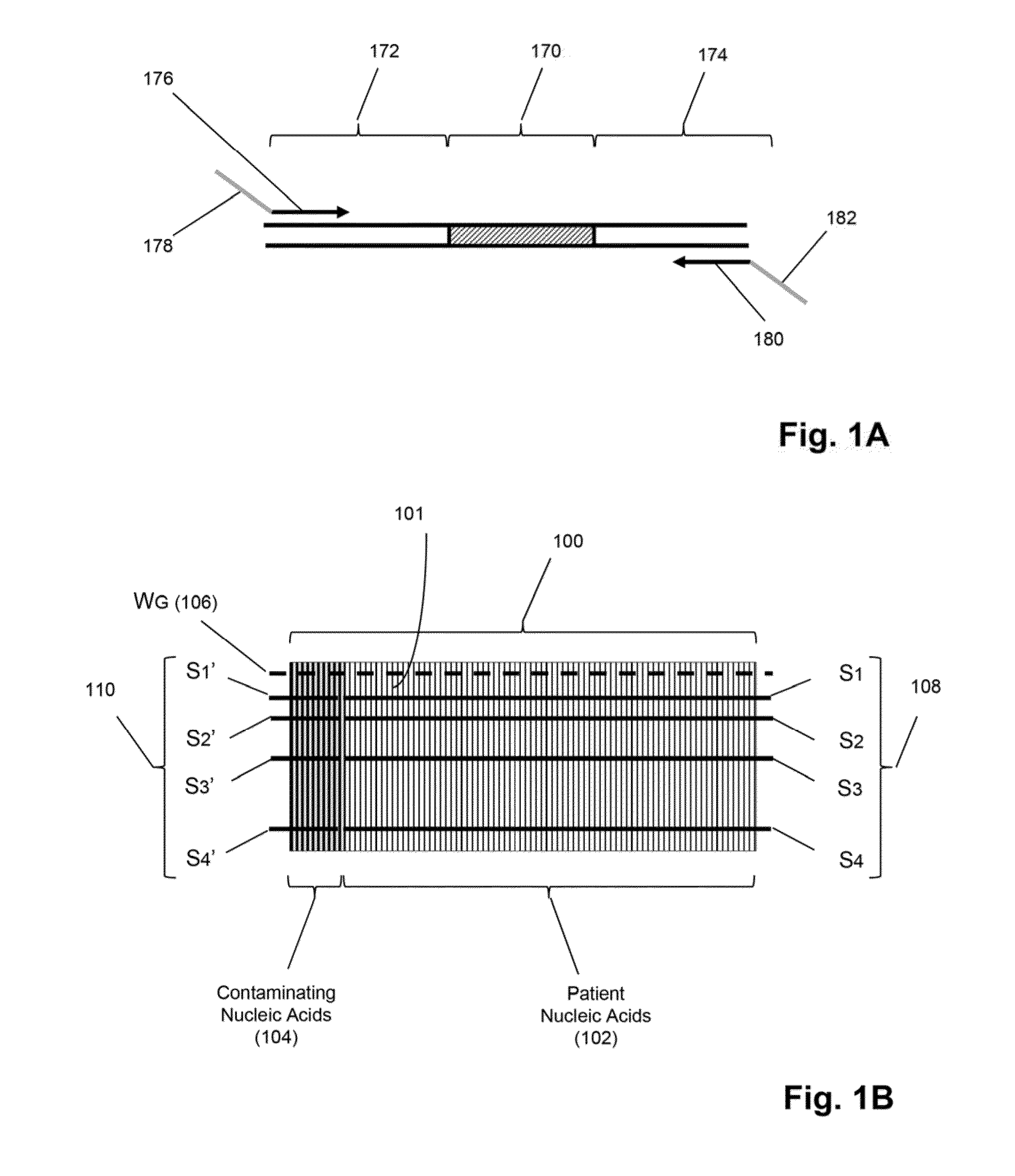
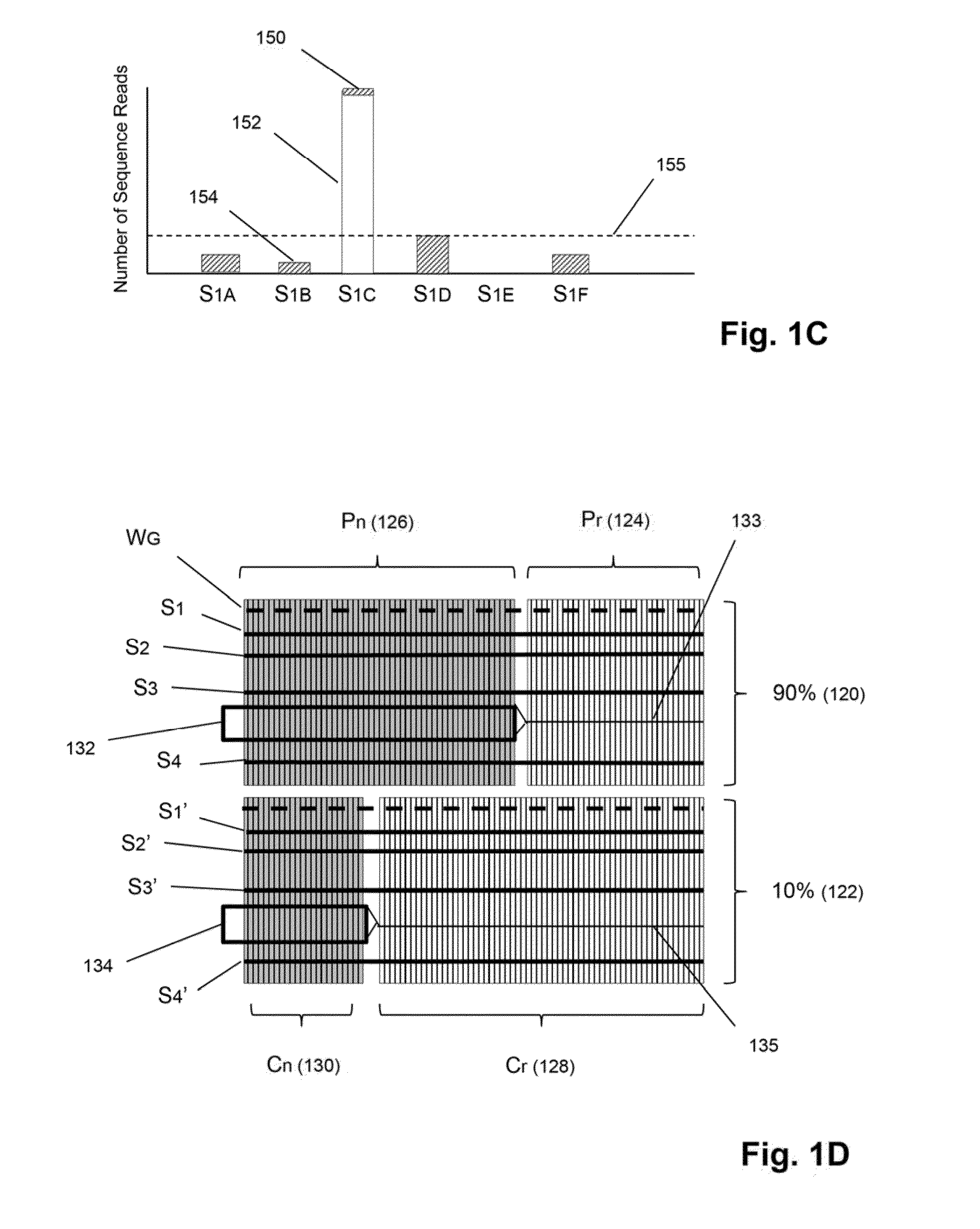
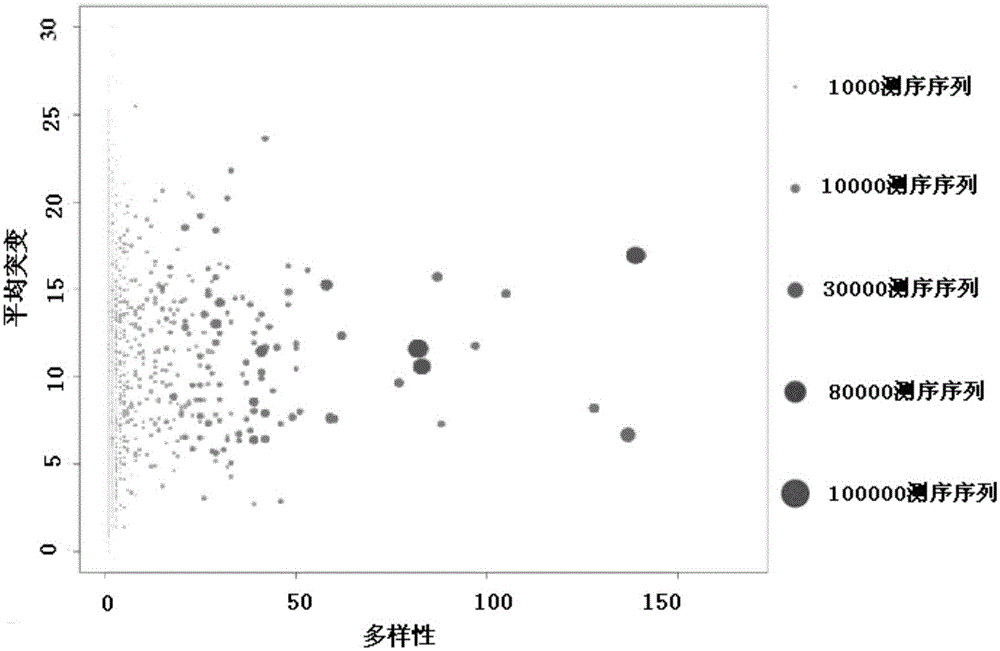
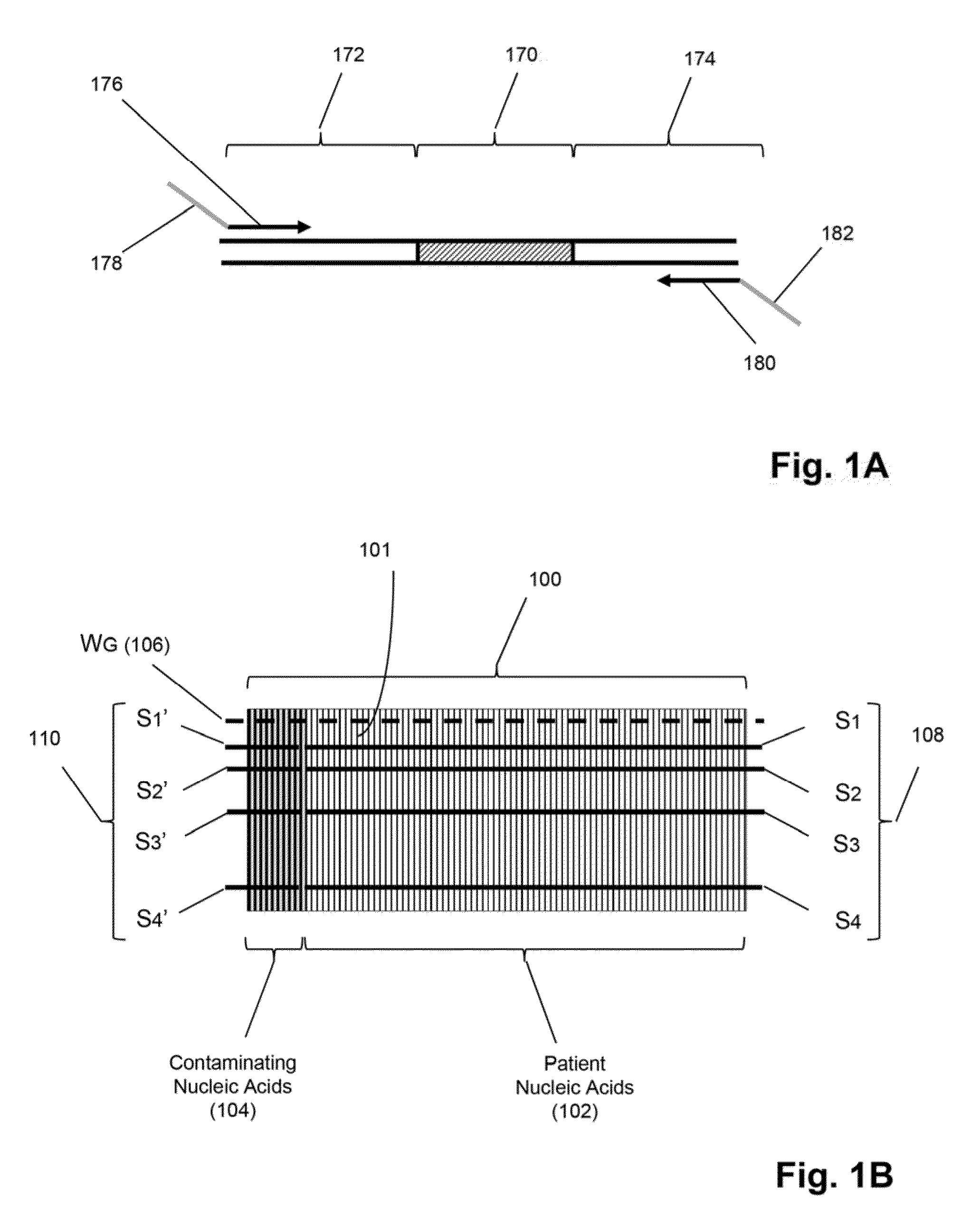
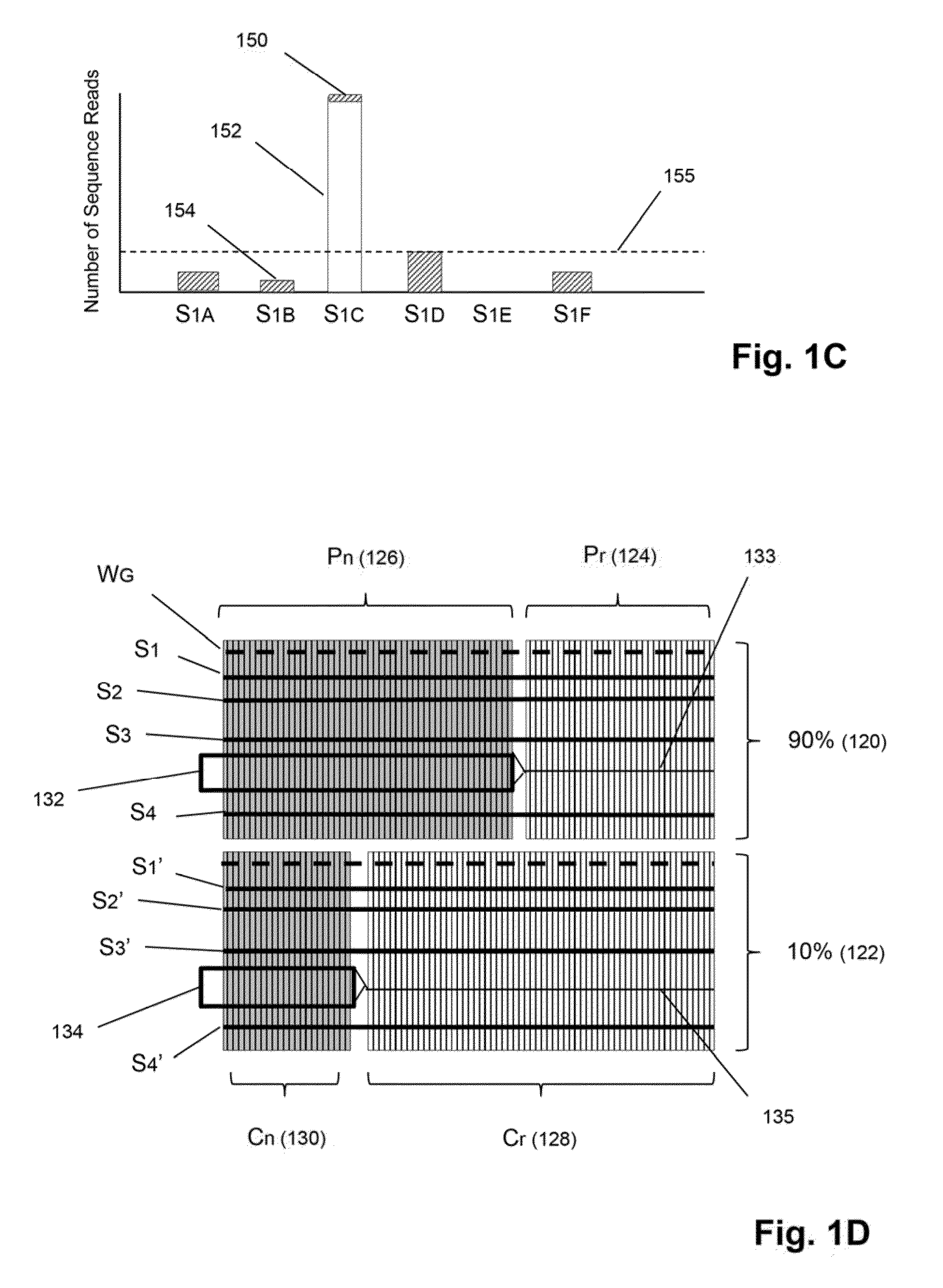
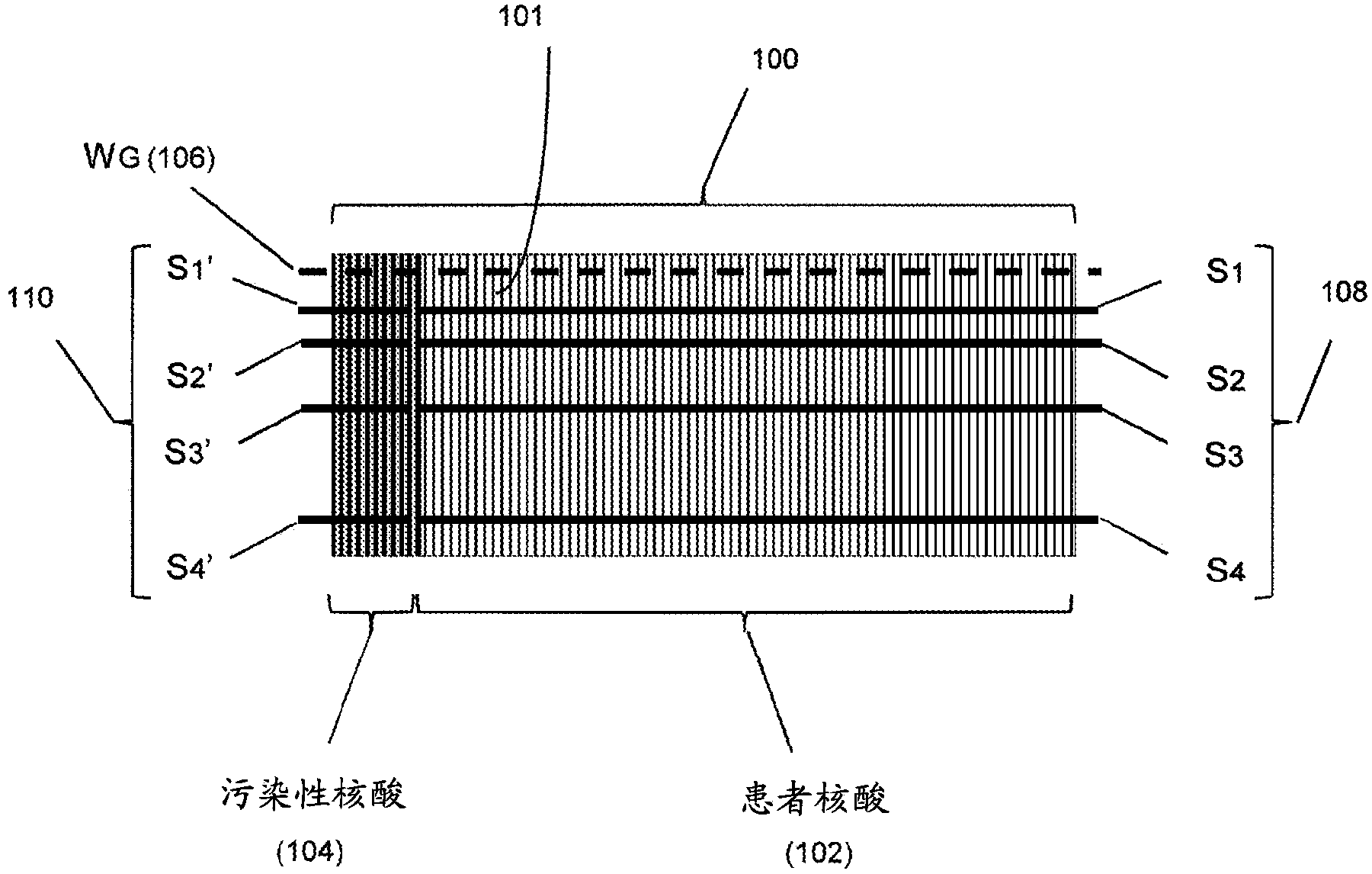
Detection and quantification of sample contamination in immune repertoire analysis
The invention is directed to methods for detecting and quantifying nucleic acid contamination in a tissue sample of an individual containing T cells and / or B cells, which is used for generating a sequence-based clonotype profile. In one aspect, the invention is implemented by measuring the presence and / or level of an endogenous or exogenous nucleic acid tag by which nucleic acid from an intended individual can be distinguished from that of unintended individuals. Endogenous tags include genetic identity markers, such as short tandem repeats, rare clonotypes or the like, and exogenous tags include sequence tags employed to determine clonotype sequences from sequence reads.
Owner:ADAPTIVE BIOTECH
Sequence-based measures of immune response
InactiveUS20140356339A1Evaluate effectivenessBiocideMicrobiological testing/measurementTreatment successImmunotherapy
The invention is directed to methods of measuring an immune response by comparing sequence-based clonotype frequency data from successively measured clonotype profiles. In particular, the invention includes immunotherapies of cancers, such as lymphomas, that include sensitive pre- and post-vaccination sequence-based measurements of changes in a patient's immune repertoire, thereby providing a sensitive measure of the likelihood of treatment success.
Owner:ADAPTIVE BIOTECH
Method of determining clonotypes and clonotype profiles
ActiveUS9043160B1Efficiently carrying out sequence comparisonQuick sortingDigital data processing detailsMicrobiological testing/measurementObservational errorNucleic acid sequencing
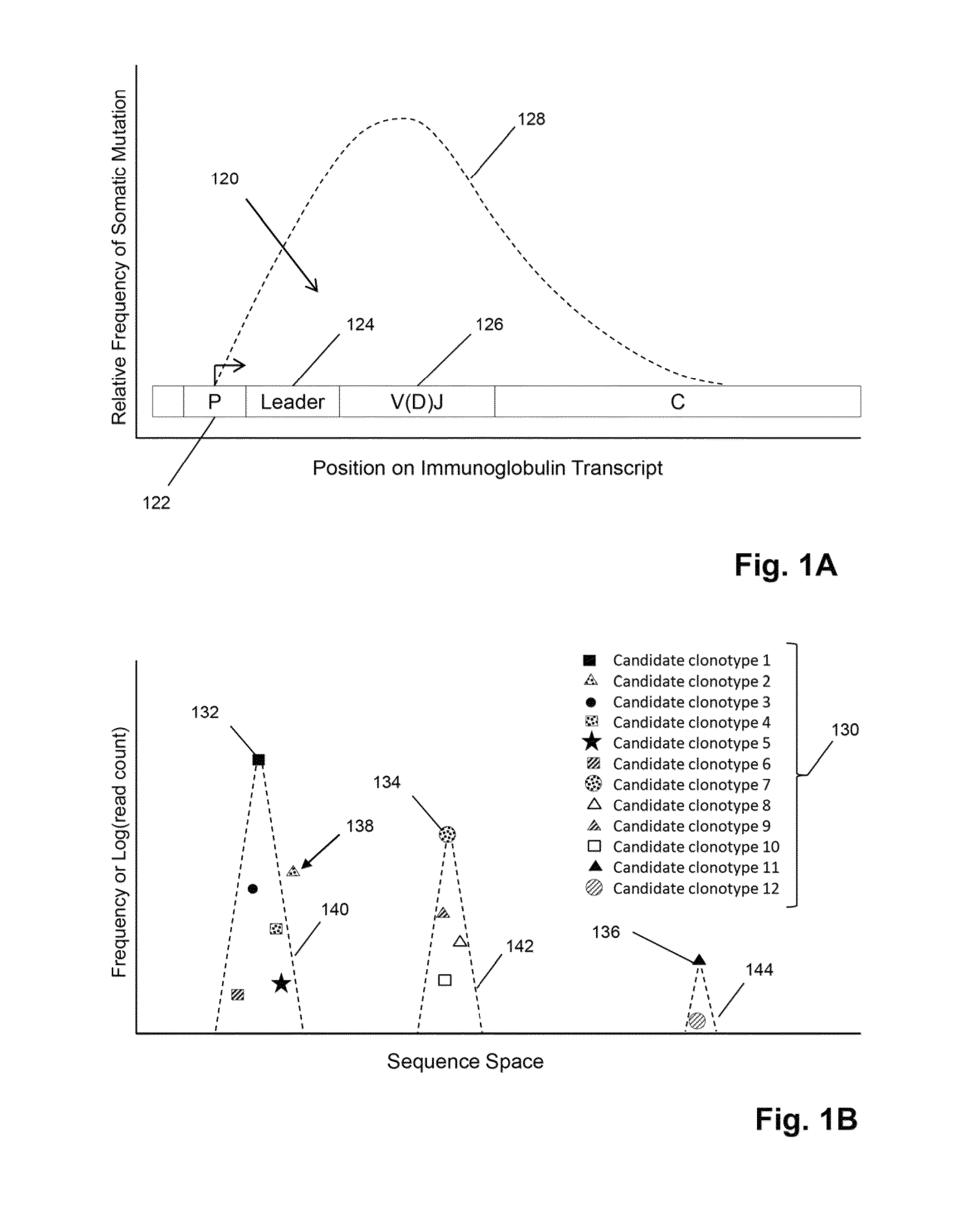
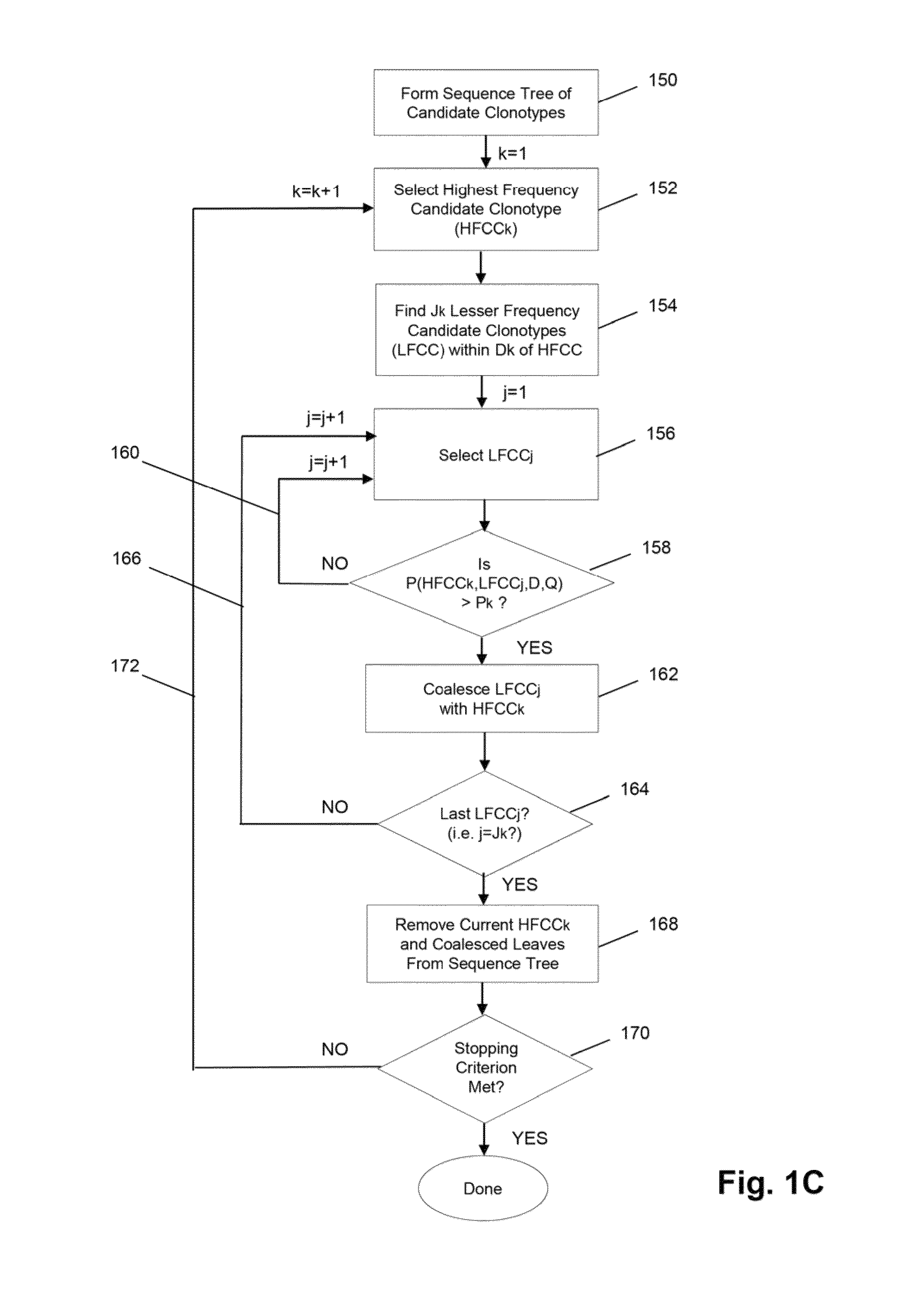
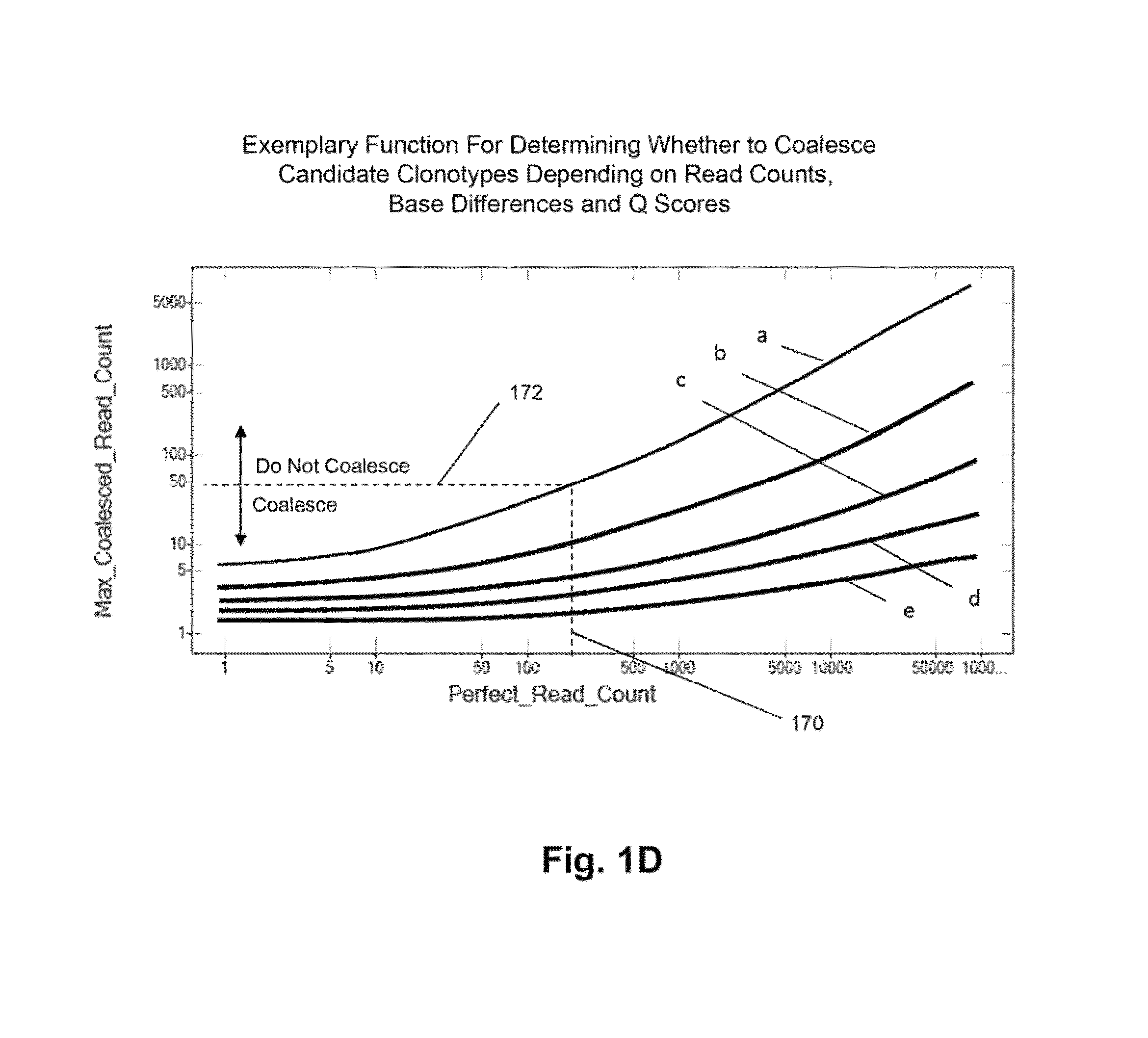
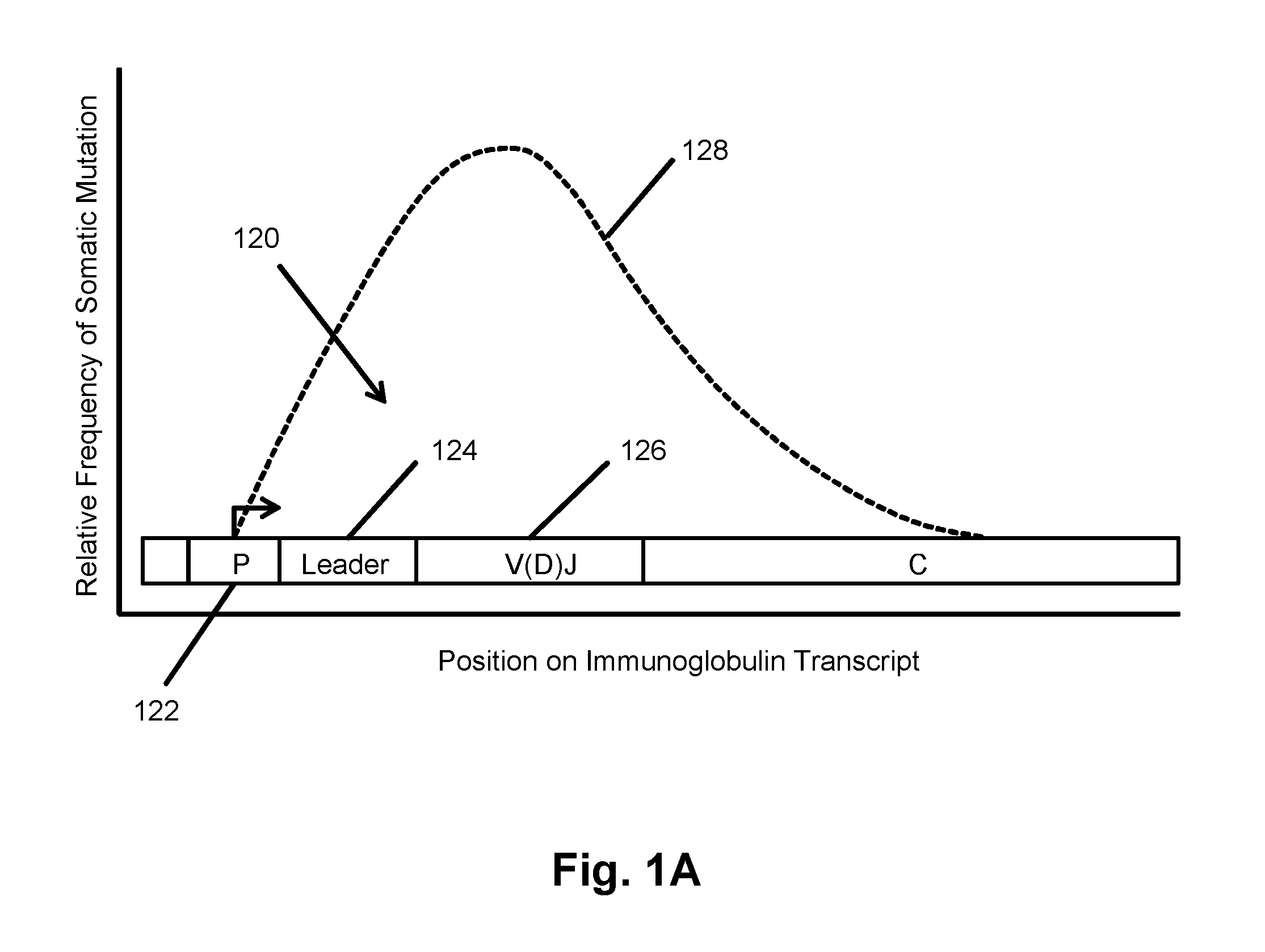
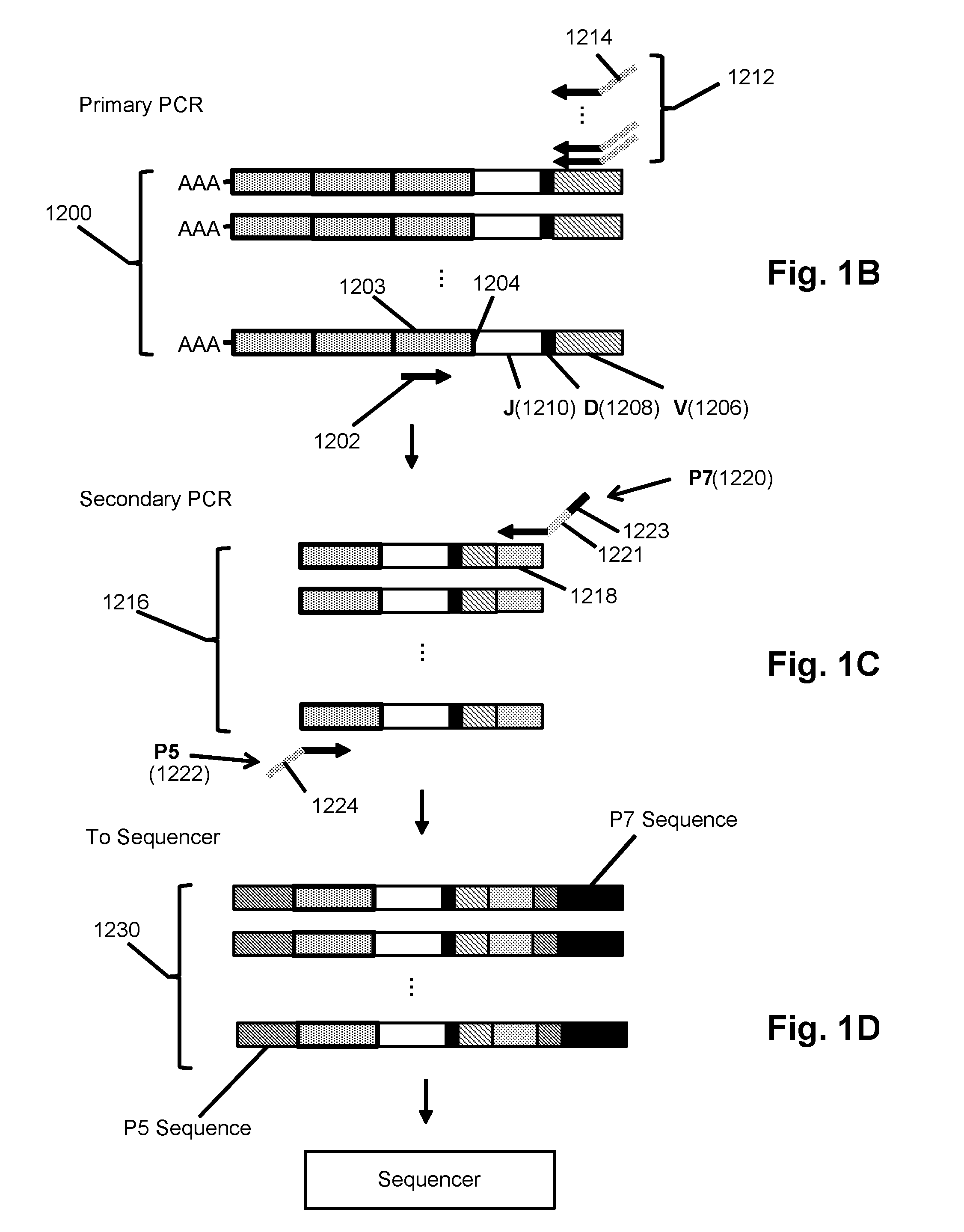
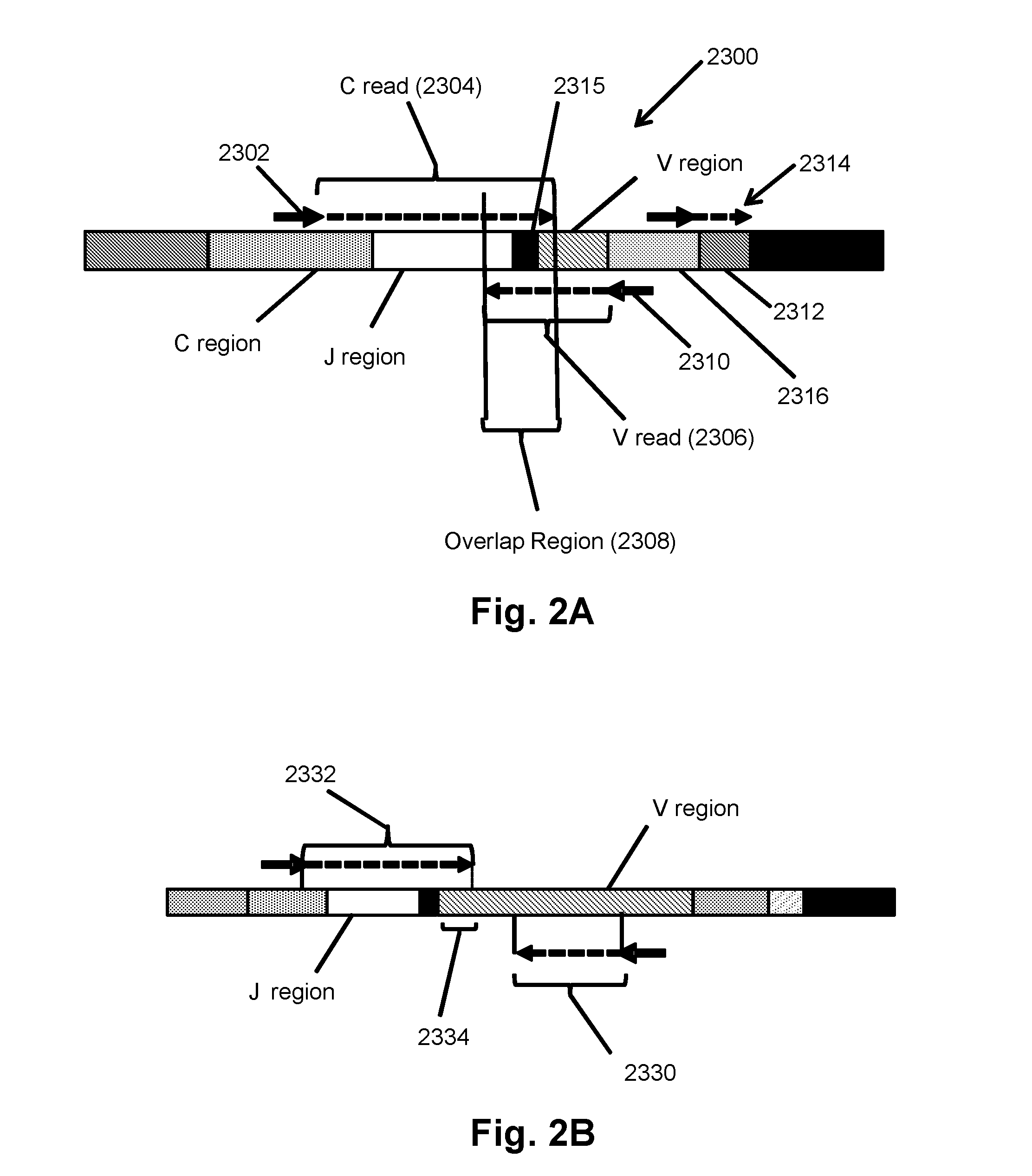
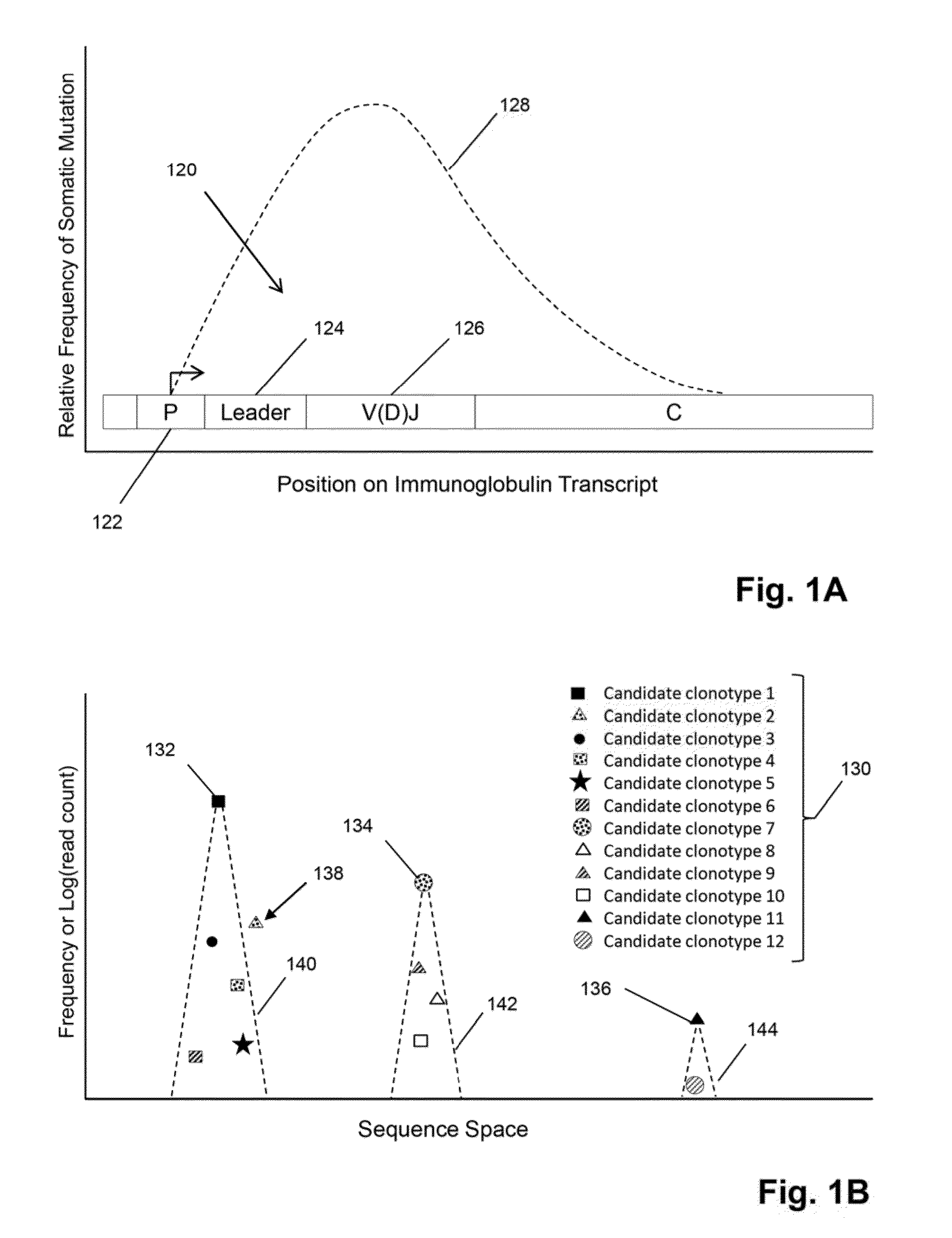
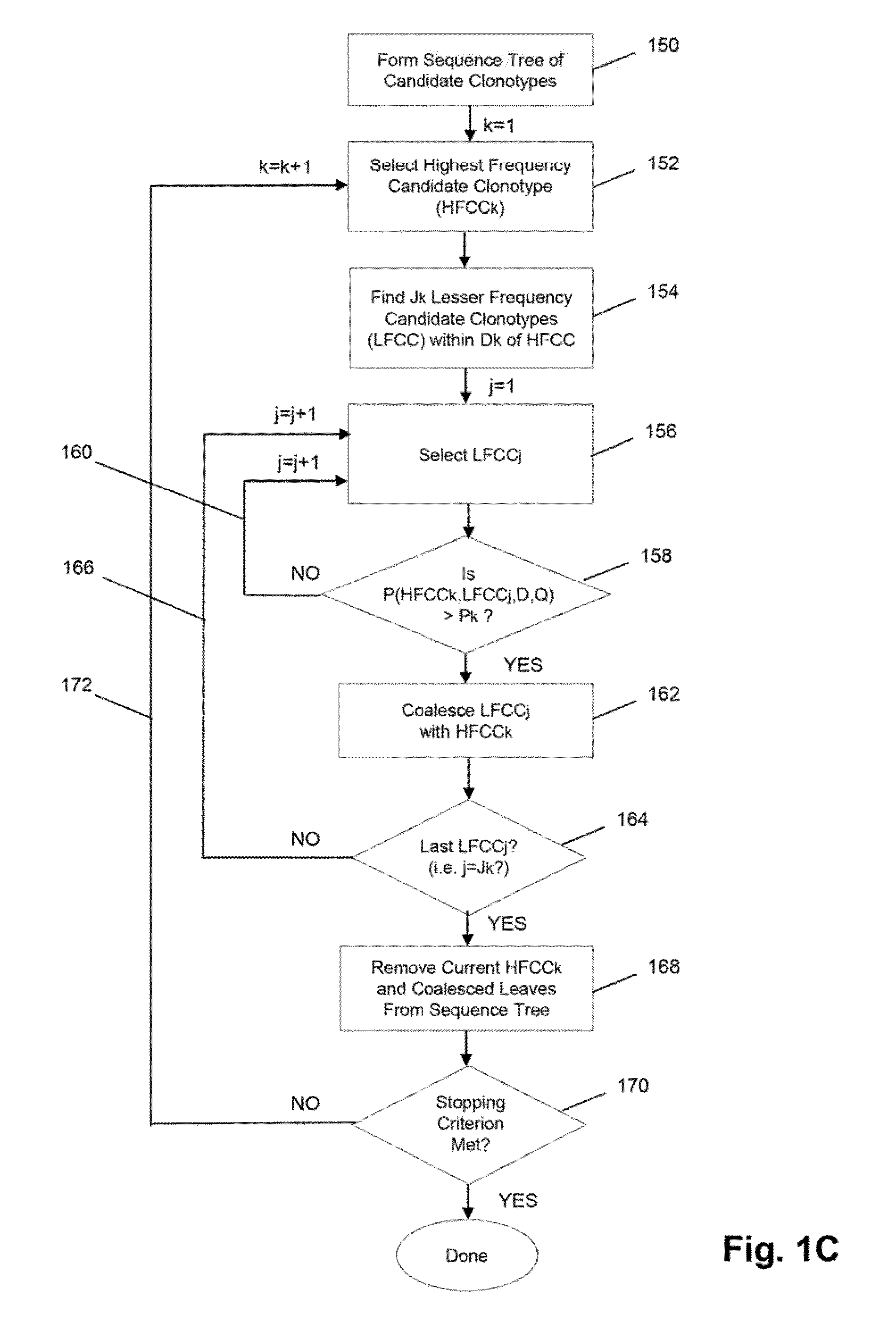
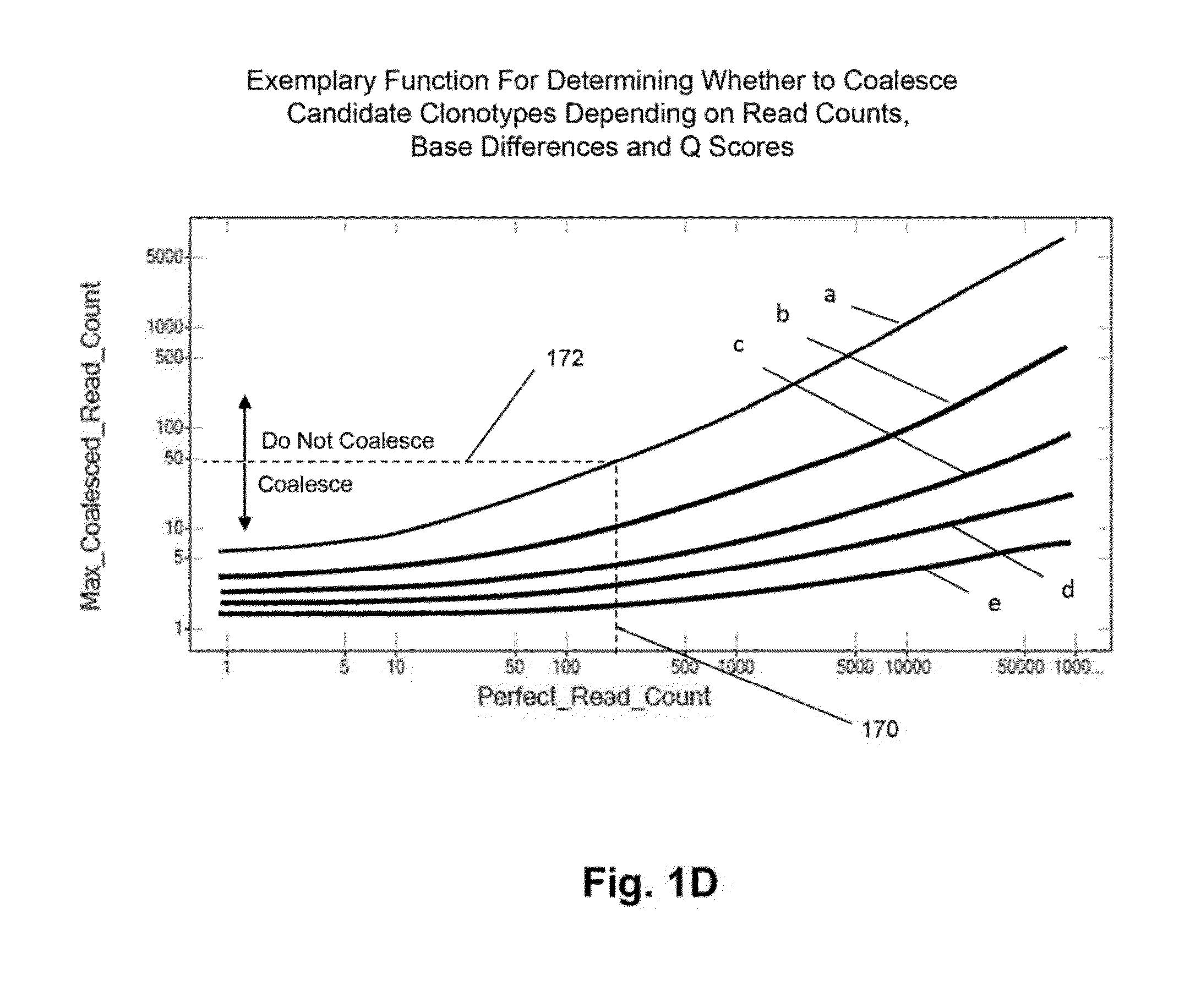
The invention is directed to methods for determining clonotypes and clonotype profiles in assays for analyzing immune repertoires by high throughput nucleic acid sequencing of somatically recombined immune molecules. In one aspect, the invention comprises generating a clonotype profile from an individual by generating sequence reads from a sample of recombined immune molecules; forming from the sequence reads a sequence tree representing candidate clonotypes each having a frequency; coalescing with a highest frequency candidate clonotype any lesser frequency candidate clonotypes whenever such lesser frequency is below a predetermined value and whenever a sequence difference therebetween is below a predetermined value to form a clonotype. After such coalescence, the candidate clonotypes is removed from the sequence tree and the process is repeated. This approach permits rapid and efficient differentiation of candidate clonotypes with genuine sequence differences from those with experimental or measurement errors, such as sequencing errors.
Owner:ADAPTIVE BIOTECH
Monitoring transformation of follicular lymphoma to diffuse large b-cell lymphoma by immune repertoire analysis
InactiveUS20140349883A1Raise the possibilityMicrobiological testing/measurementLibrary screeningProgenitorSomatic cell
The invention is directed to a method of prognosing in an individual a transformation from follicular lymphoma to diffuse large B-cell lymphoma (DLBCL) by measuring changes and / or lack of changes in certain groups of related clonotypes, referred to herein as “clans,” in successive clonotype profiles of the individual. A clan may arise from a single lymphocyte progenitor that gives rise to many related lymphocyte progeny, each possessing and / or expressing a slightly different immunoglobulin receptor due to somatic mutation(s), such as base substitutions, inversions, related rearrangements resulting in common V(D)J gene segment usage, or the like. A higher likelihood of transformation from follicular lymphoma to DLBCL is correlated with the persistence of clans in successive clonotype profiles whose clonotype membership fails to undergo diversification over time.
Owner:ADAPTIVE BIOTECH
Method of determining clonotypes and clonotype profiles
ActiveUS20150167080A1Quick sortingEfficiently carrying out sequence comparisonsMicrobiological testing/measurementLibrary member identificationAssayNucleic acid sequencing
Owner:ADAPTIVE BIOTECH
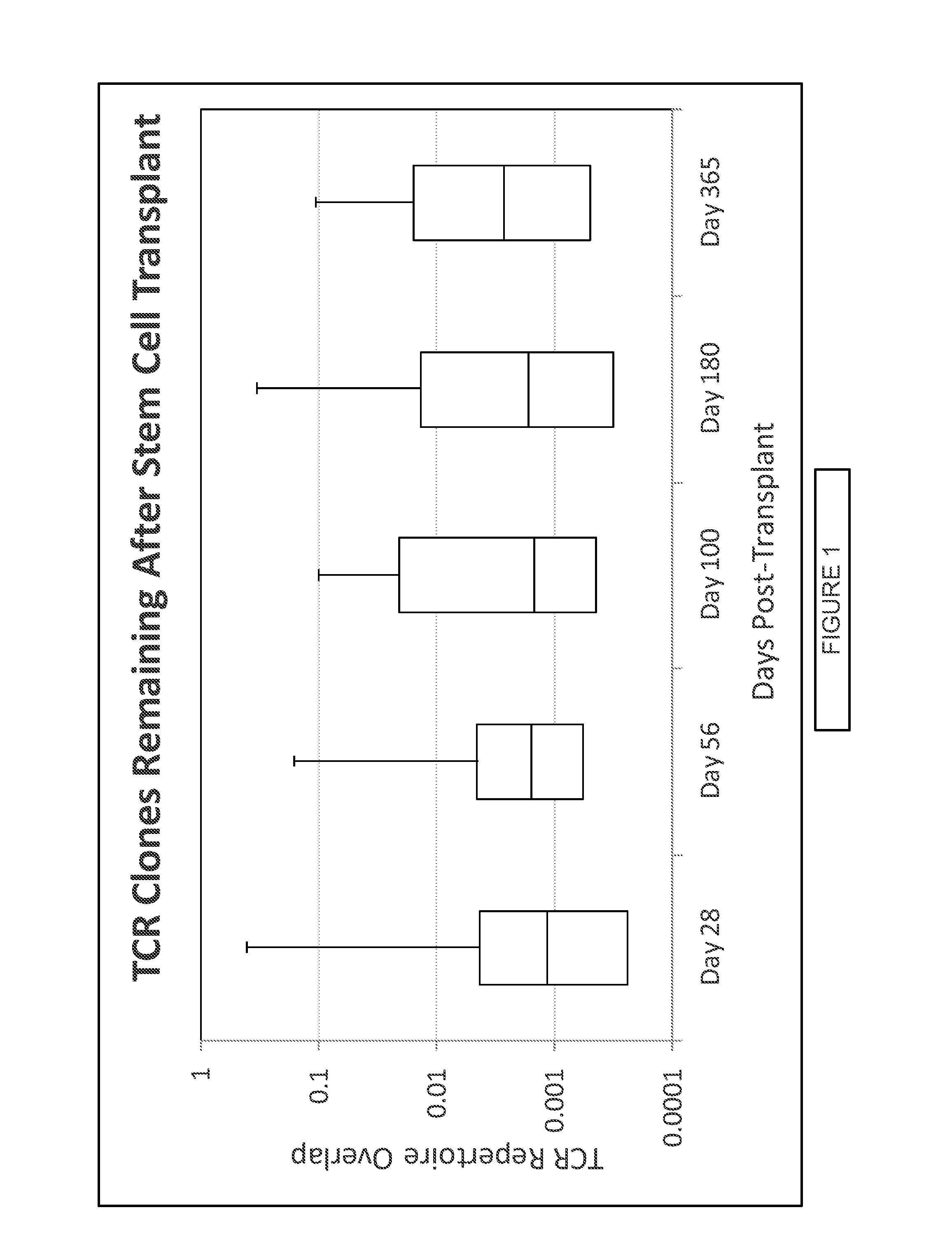
Compositions and methods for eliminating undesired subpopulations of T cells in patients with immunological defects related to autoimmunity and organ or hematopoietic stem cell transplantation
InactiveUS20050084967A1Reduce riskReduce severityMicrobiological testing/measurementArtificial cell constructsAutoimmune responsesHematopoietic stem cell transplantation
The present invention relates generally to methods for stimulating T cells, and more particularly, to methods to eliminate undesired (e.g. autoreactive, alloreactive, pathogenic) subpopulations of T cells from a mixed population of T cells, thereby restoring the normal immune repertoire of said T cells. The present invention also relates to compositions of cells, including stimulated T cells having restored immune repertoire and uses thereof.
Owner:LIFE TECH CORP
Method for detecting BCR and TCR immune repertoire in blood plasma cfDNA
ActiveCN105087789AEliminate errorsImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationInformation analysisSubcloning
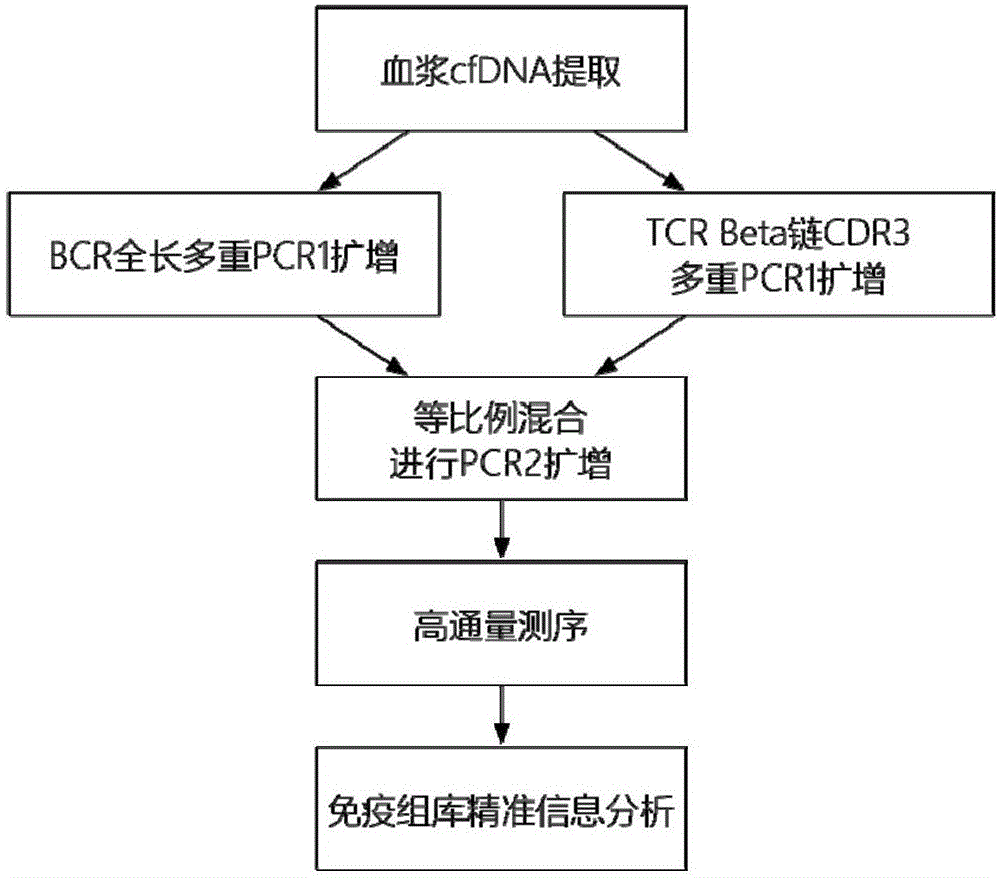
The invention provides a method for detecting BCR and TCR immune repertoire in blood plasma cfDNA. The method includes: blood plasma cfDNA extraction, BCR H chain overall length multiple PCR amplification, TCR beta chain CDR3 multiple PCR amplification, PCR amplification after the products of the two previous amplifications are mixed in an equal-volume manner, high-throughput sequencing, and immune repertoire precise information analysis. The primers of the three amplifications are respectively shown in SEQ ID No. 1-28, SEQ ID No. 29-77 and SEQ ID No. 78-79. The invention further provides a kit, which contains the primers, for detecting the BCR and TCR in the blood plasma cfDNA. By the method and the kit which are promising in application prospect in the field of non-invasive detection, the BCR and TCR in the blood plasma cfDNA can be detected at the same time, precise information analysis and precise quantification of immune repertoire subcloning are achieved.
Owner:BEIJING GENEPLUS TECH +2
Detection and quantification of sample contamination in immune repertoire analysis
The invention is directed to methods for detecting and quantifying nucleic acid contamination in a tissue sample of an individual containing T cells and / or B cells, which is used for generating a sequence-based clonotype profile. In one aspect, the invention is implemented by measuring the presence and / or level of an endogenous or exogenous nucleic acid tag by which nucleic acid from an intended individual can be distinguished from that of unintended individuals. Endogenous tags include genetic identity markers, such as short tandem repeats, rare clonotypes or the like, and exogenous tags include sequence tags employed to determine clonotype sequences from sequence reads.
Owner:ADAPTIVE BIOTECH
Methods for diagnosing infectious disease and determining HLA status using immune repertoire sequencing
Methods are provided for predicting a subject's infection status using high-throughput T cell receptor sequencing to match the subject's TCR repertoire to a known set of disease-associated T cell receptor sequences. The methods of the present invention may be used to predict the status of several infectious agents in a single sample from a subject. Methods are also provided for predicting a subject's HLA status using high-throughput immune receptor sequencing.
Owner:ADAPTIVE BIOTECH +1
Primers for immune repertoire profiling
ActiveUS20210355484A1Immunoglobulin superfamilyMicrobiological testing/measurementSingle cell transcriptomeNucleotide
Disclosed herein include systems, methods, compositions, and kits for immune repertoire profiling. There are provided, in some embodiments, primer panels enabling the determination of the nucleotide sequence of the complete variable region of nucleic acids encoding mouse B cell receptor (BCR) and T cell receptor (TCR) polypeptides. In some embodiments, the method comprises single cell transcriptomic analysis.
Owner:BECTON DICKINSON & CO
Oligonucleotides and beads for 5 prime gene expression assay
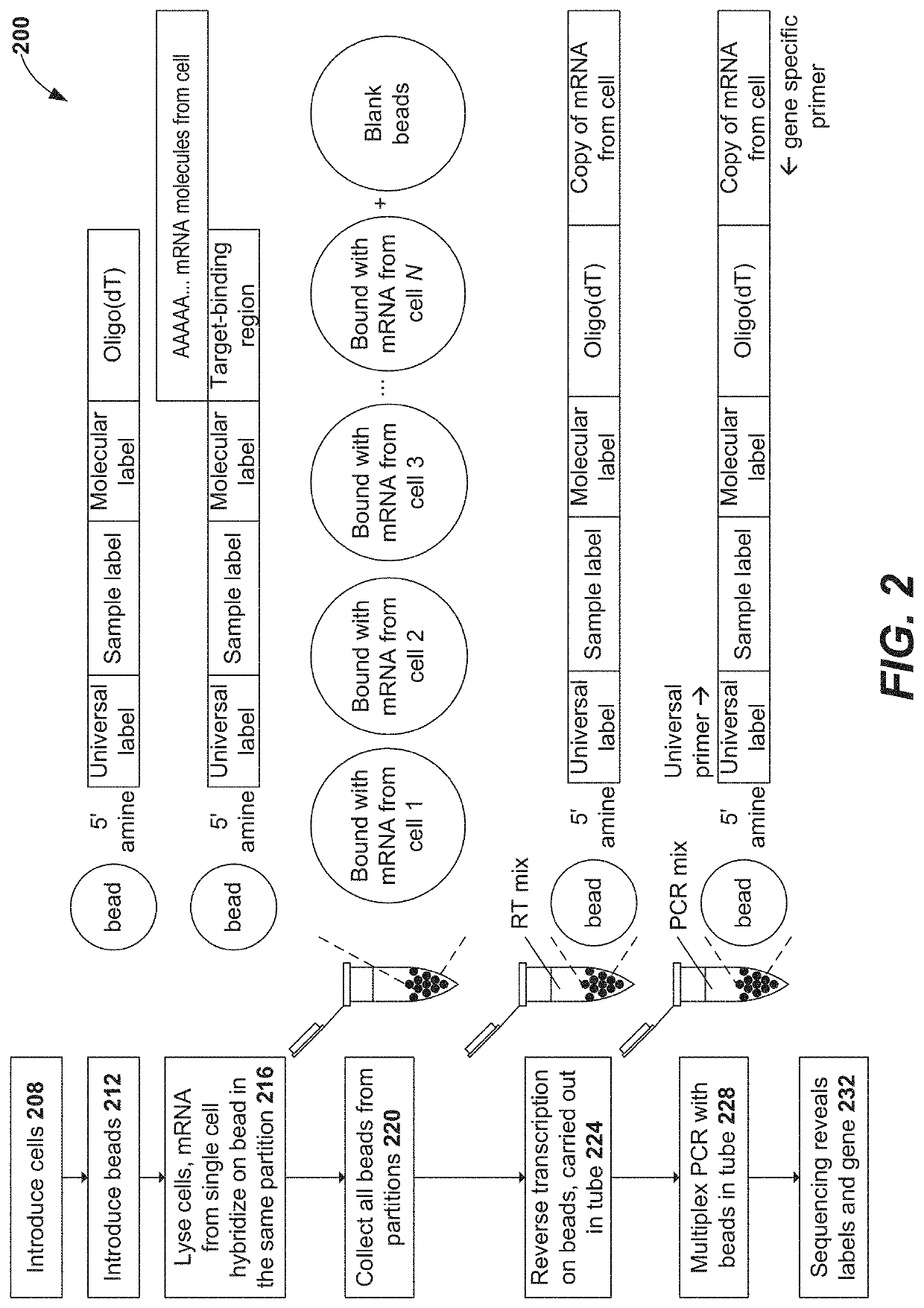
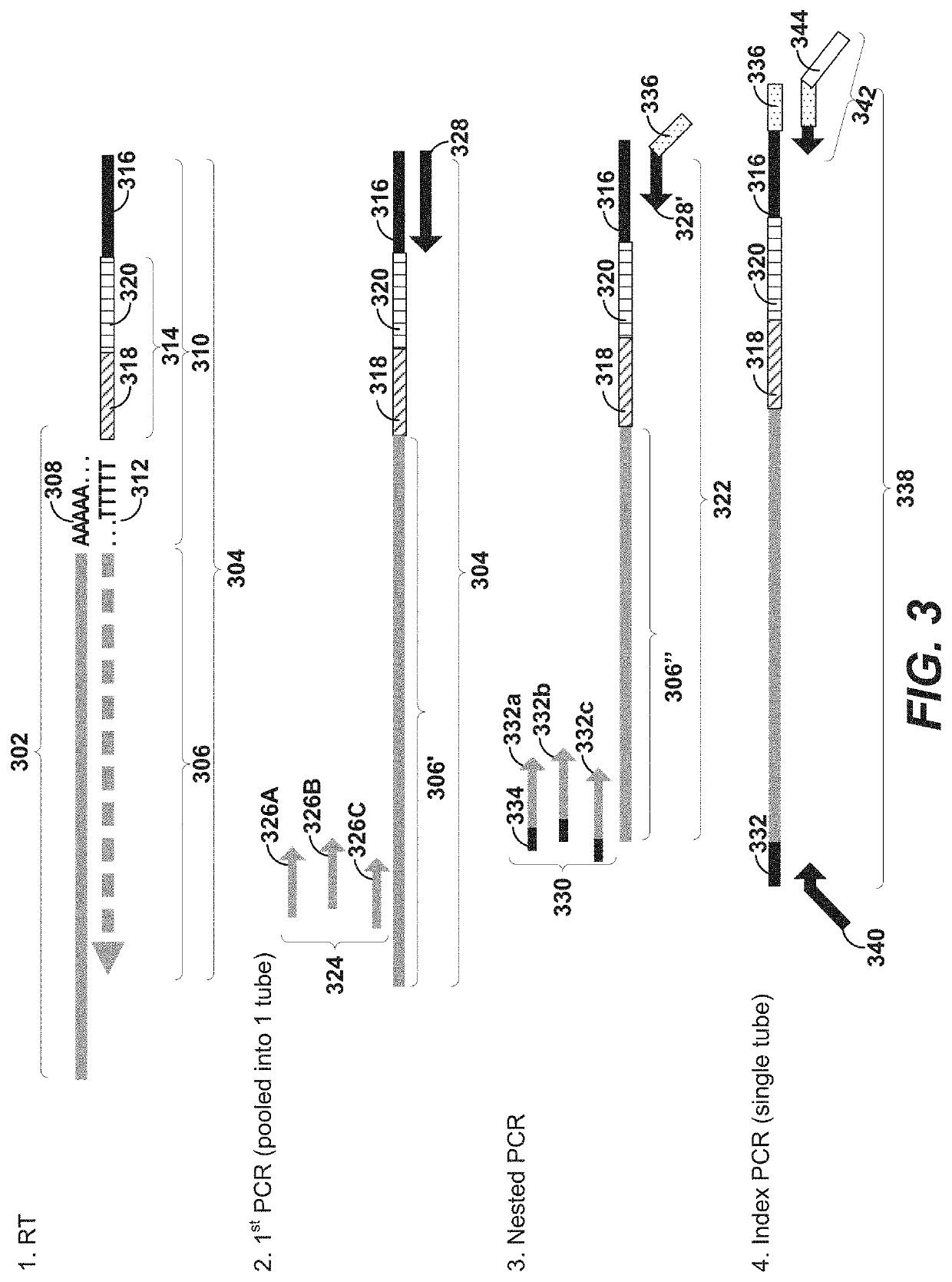
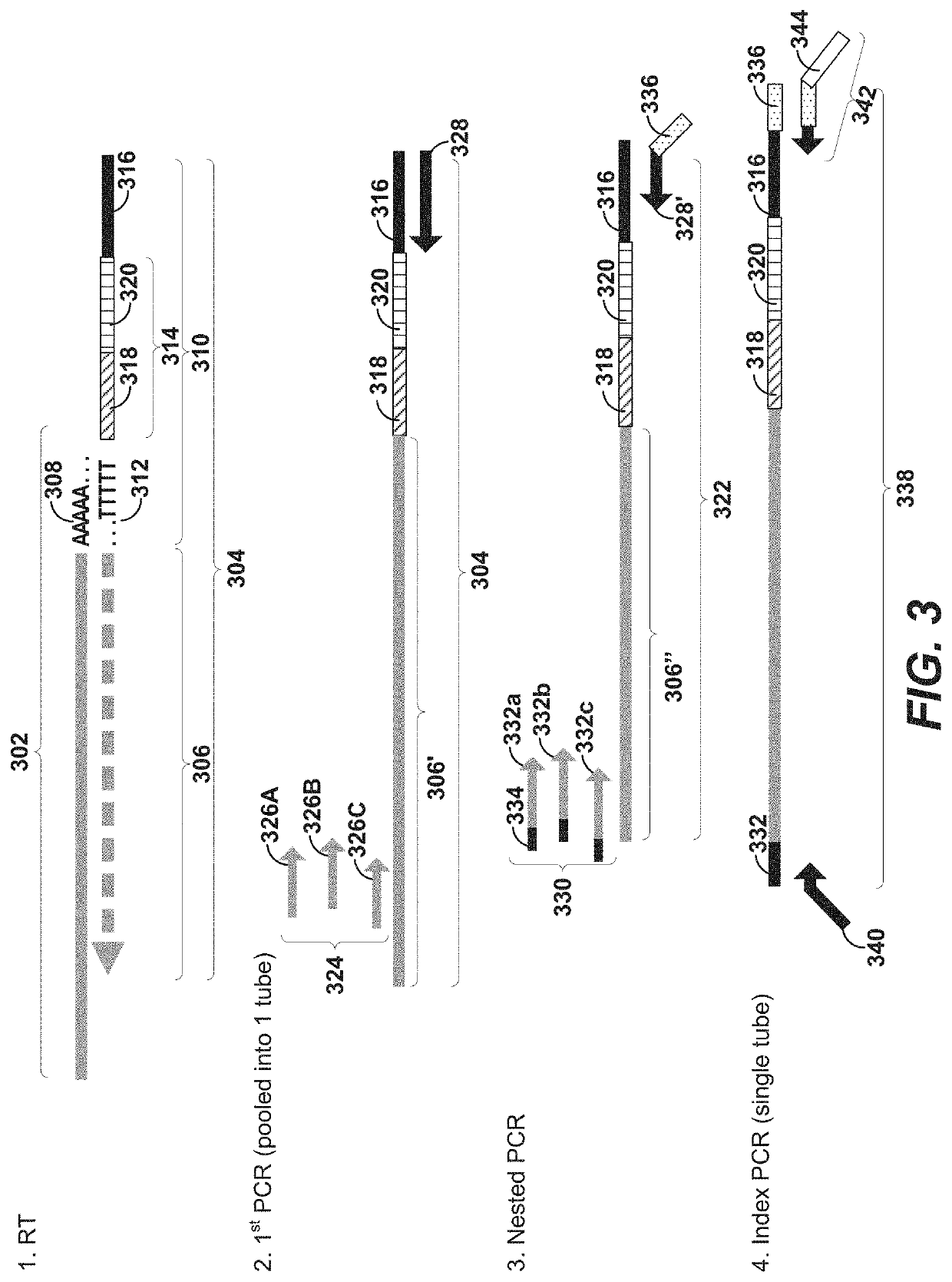
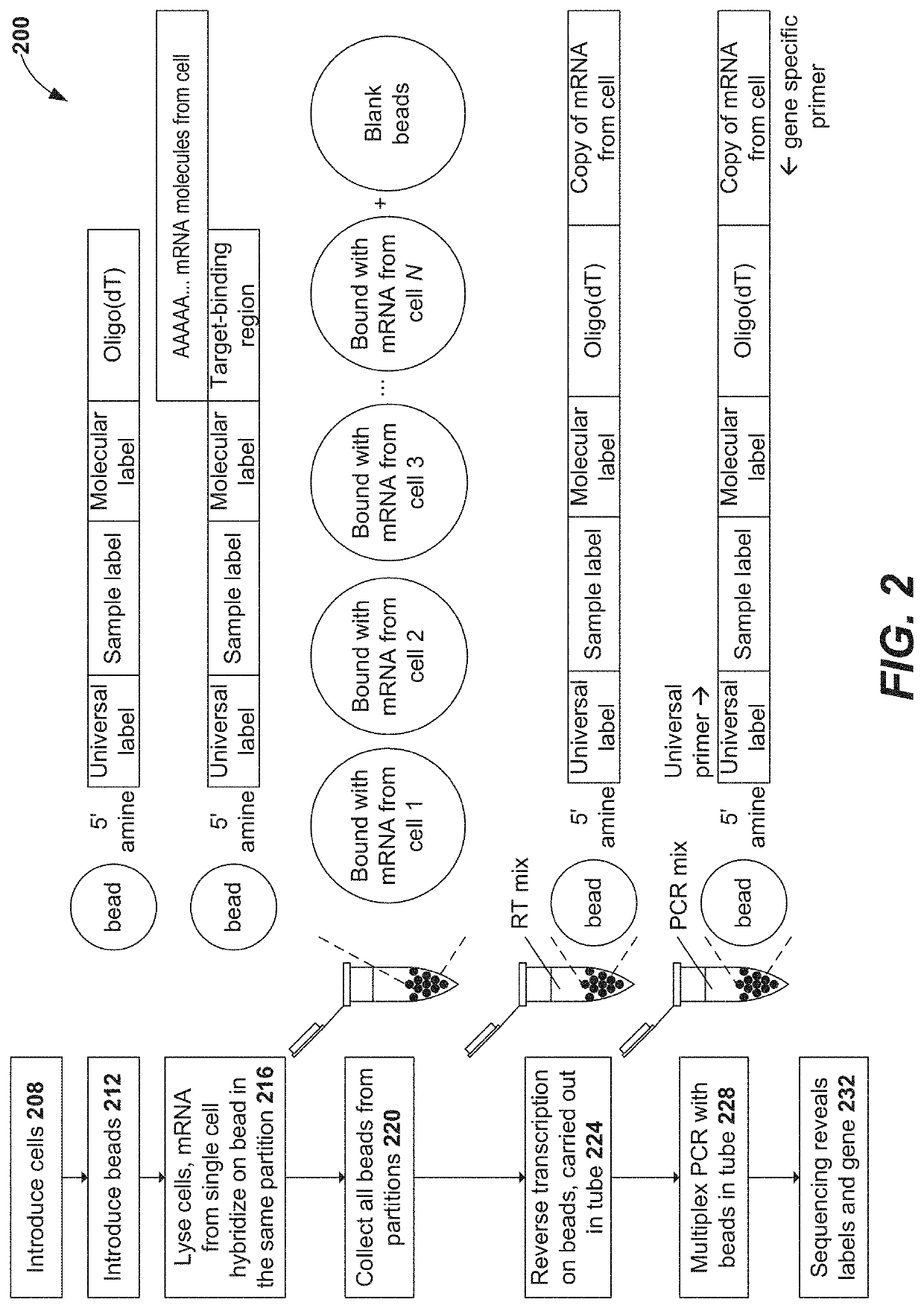
Disclosed herein include systems, methods, compositions, and kits for 5′-based gene expression profiling. Some embodiments provide synthetic particles (e.g., beads) associated with a first plurality of oligonucleotide barcodes and a second plurality of oligonucleotide barcodes. In some embodiments, nucleic acid targets (e.g., mRNAs) are initially barcoded on the 3′ end with the first plurality of oligonucleotide barcodes and subsequently barcoded on the 5′ end following a template switching reaction and intermolecular hybridization with the first plurality of oligonucleotide barcodes and extension. Immune repertoire profiling methods are also provided in some embodiments.
Owner:BECTON DICKINSON & CO
Methods for eliminating at least a substantial portion of a clonal antigen-specific memory T cell subpopulation
InactiveUS8617884B2Reduce riskReduce severityMammal material medical ingredientsArtificial cell constructsCentral Memory T-CellPartial cloning
The present invention relates generally to methods for stimulating T cells, and more particularly, to methods to eliminate undesired (e.g., autoreactive, alloreactive, pathogenic) subpopulations of T cells from a mixed population of T cells, thereby restoring the normal immune repertoire of said T cells. The present invention also relates to compositions of cells, including stimulated T cells having restored immune repertoire and uses thereof.
Owner:LIFE TECH CORP
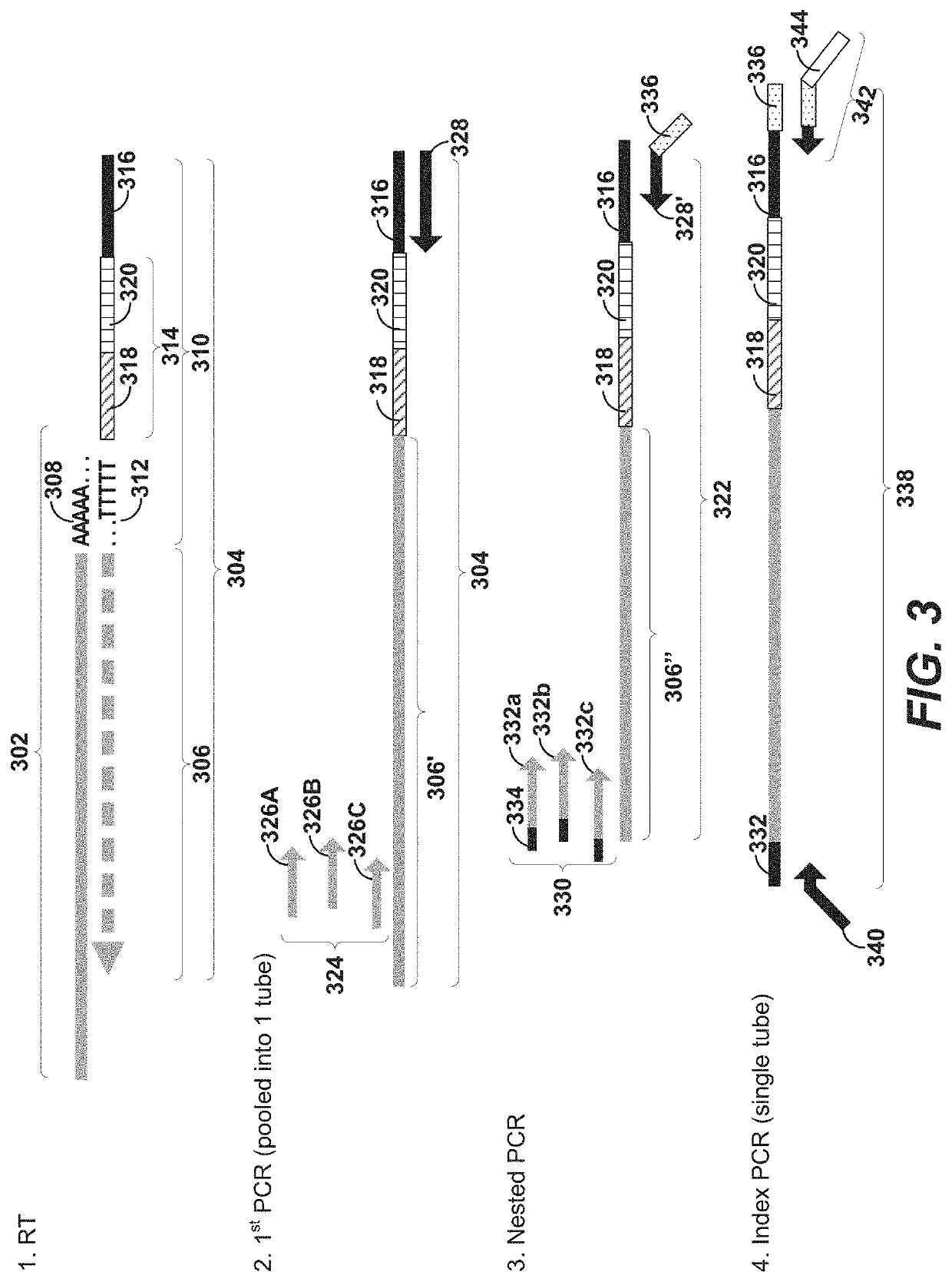
Using random priming to obtain full-length v(d)j information for immune repertoire sequencing
ActiveUS20210139970A1Microbiological testing/measurementDNA preparationGenomic cloneTemplate switching
Disclosed herein include systems, methods, compositions, and kits for labeling nucleic acid targets in a sample. In some embodiments, nucleic acid targets (e.g., mRNAs) are initially barcoded on the 3′ end and are subsequently barcoded on the 5′ end following a template switching reaction and intermolecular and / or intramolecular hybridization and extension. 5′- and / or 3′-barcoded nucleic acid targets can serve as templates for amplification reactions and / or random priming and extension reactions to generate a sequencing library. The method can comprise generating a full-length sequence of the nucleic acid target by aligning a plurality of sequencing reads. Immune repertoire profiling methods are also provided in some embodiments.
Owner:BECTON DICKINSON & CO
Primer group for developing rhesus T cell immune repertoire, high-flux sequencing method and application of method
InactiveCN104673892AFull lengthRich quantityMicrobiological testing/measurementDNA/RNA fragmentationForward primerV region
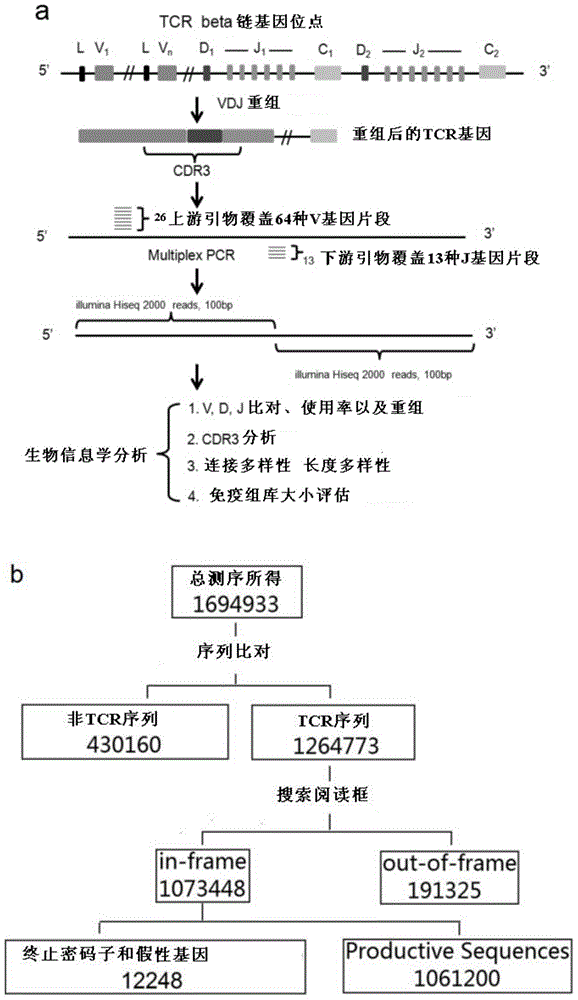
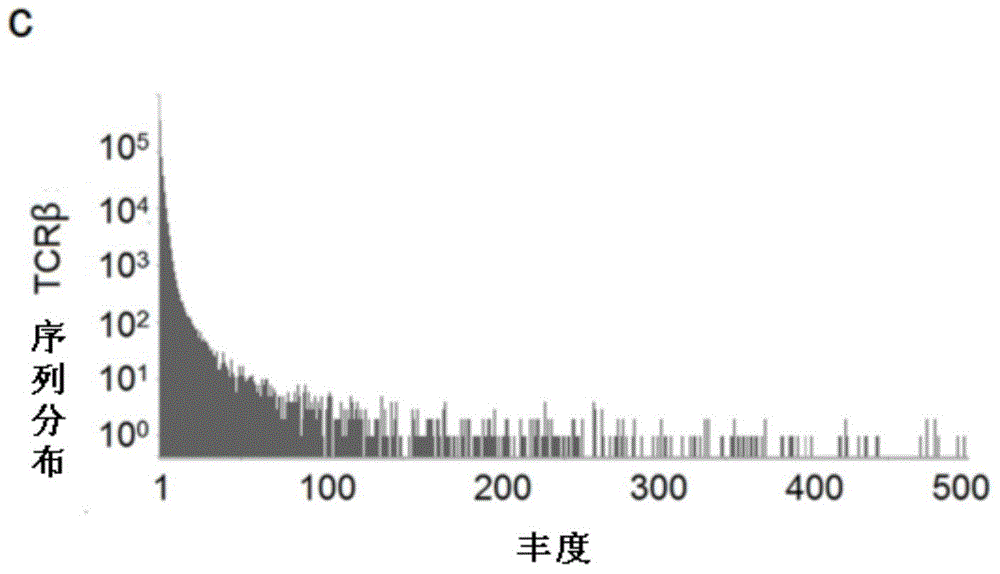
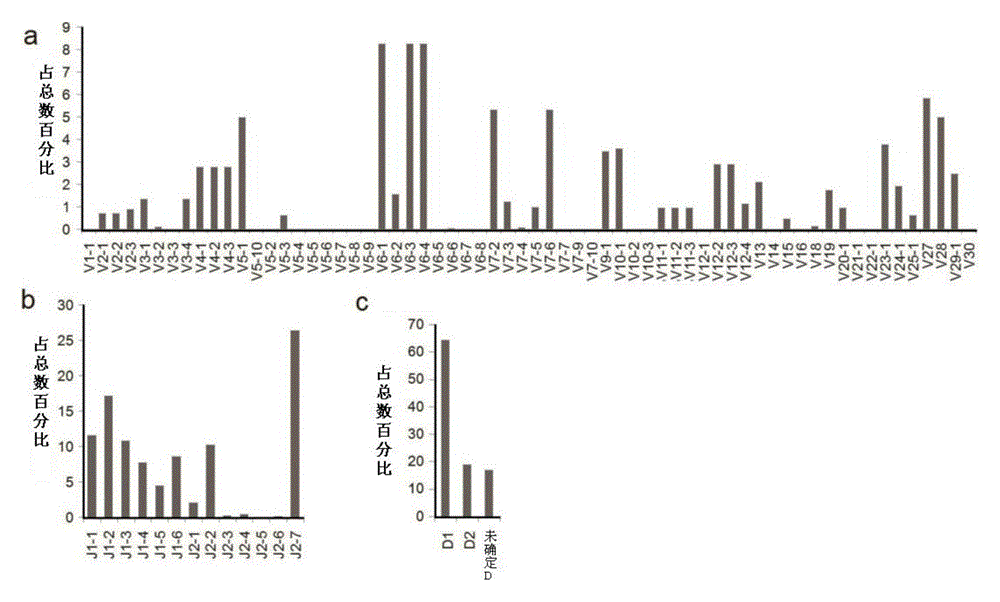
The invention provides a primer group for developing a rhesus T cell immune repertoire, a high-flux sequencing method and application of method. The method comprises the following steps: setting 26 forward primer sequences in a variable region, namely a V region aiming at rhesus TCR; setting 13 reverse primer sequences in a joining region, namely a J region aiming at TCR; performing multiple PCR amplification on the TCR sequence with the magnitude order of about 10<7> in peripheral blood or other immunologic tissues, preparing a high-flux sequencing library, sequencing, and performing comprehensive analysis on the rhesus TCR gene immune repertoire by virtue of bioinformatics. According to the technical scheme, the primer group has significance for constructing a rhesus analysis model, contributes to promoting the application of the rhesus model in research on autoimmune system diseases and infectious diseases and also contributes to deeply and widely recognizing the T cell mediated diseases by the humans.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA
High-throughput nucleotide library sequencing
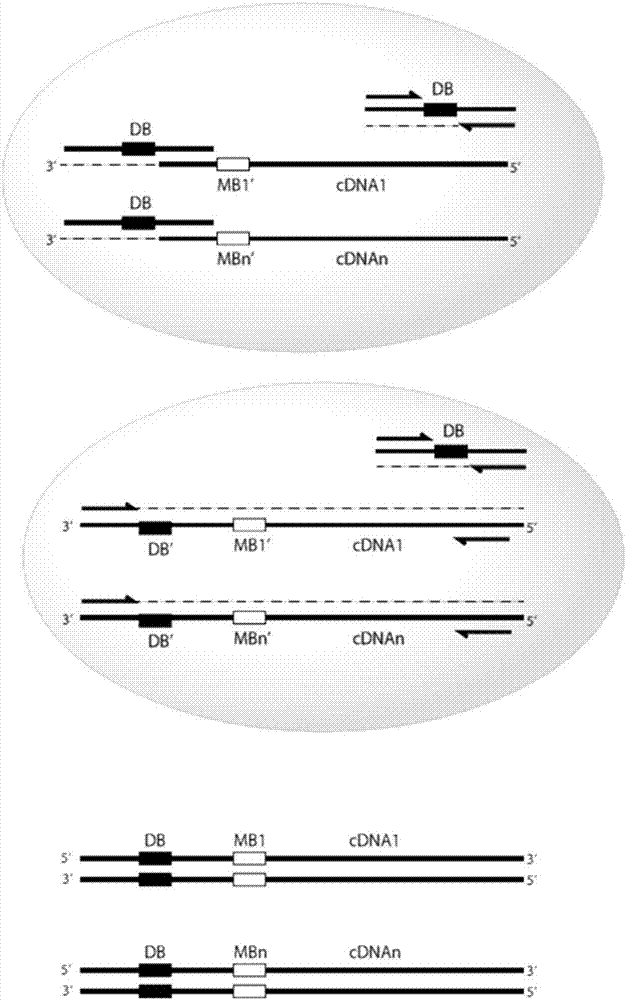
Provided herein are methods and composition for immune repertoire sequencing and single cell barcoding. The methods and compositions can be used to pair any two sequences originating from a single cell, such as heavy and light chain antibody sequences, alpha and beta chain T-cell receptor sequences, or gamma and delta chain T-cell receptor sequences, for antibody and T-cell receptor discovery, disease and immune diagnostics, and low error sequencing.
Owner:ABVITRO LLC
Method and system for processing immune repertoire sequencing data of samples
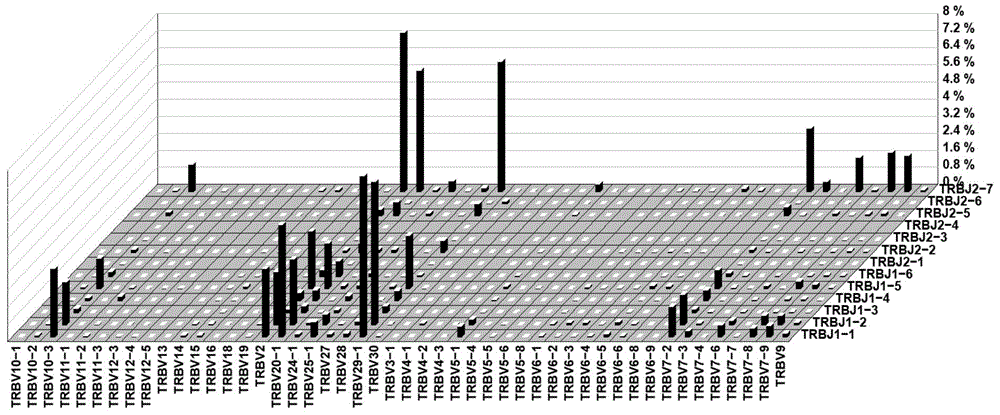
ActiveCN106156536AEffectively reflect the V-J pairing situationIntuitive reflection of V-J pairingSpecial data processing applicationsRepeatabilityComputer science
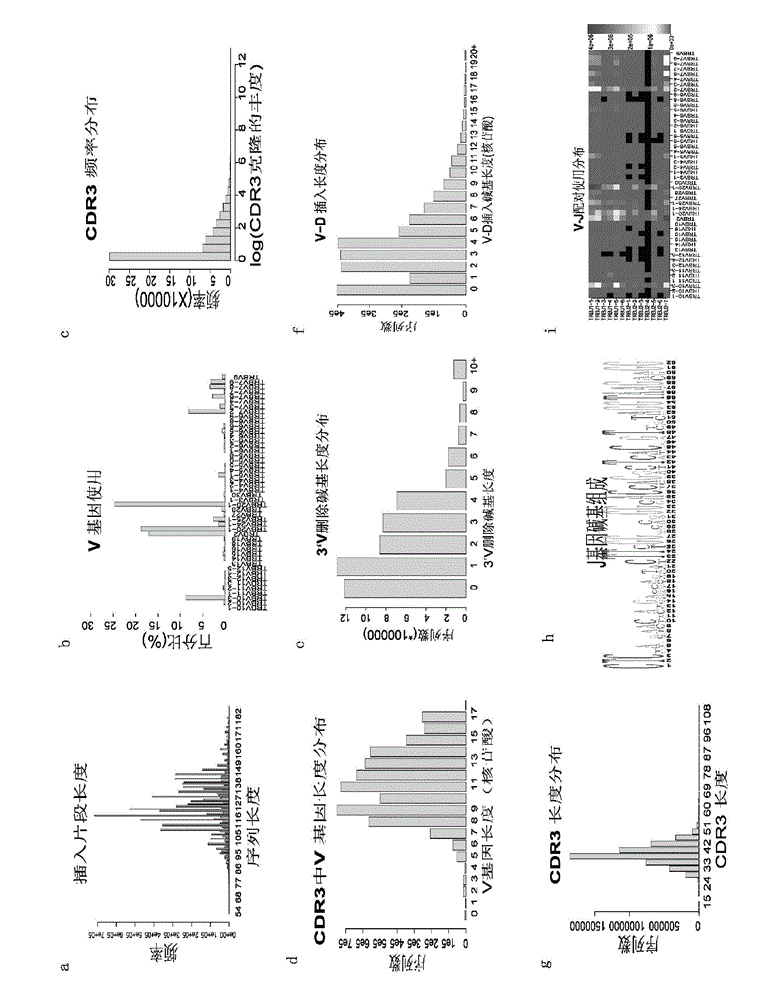
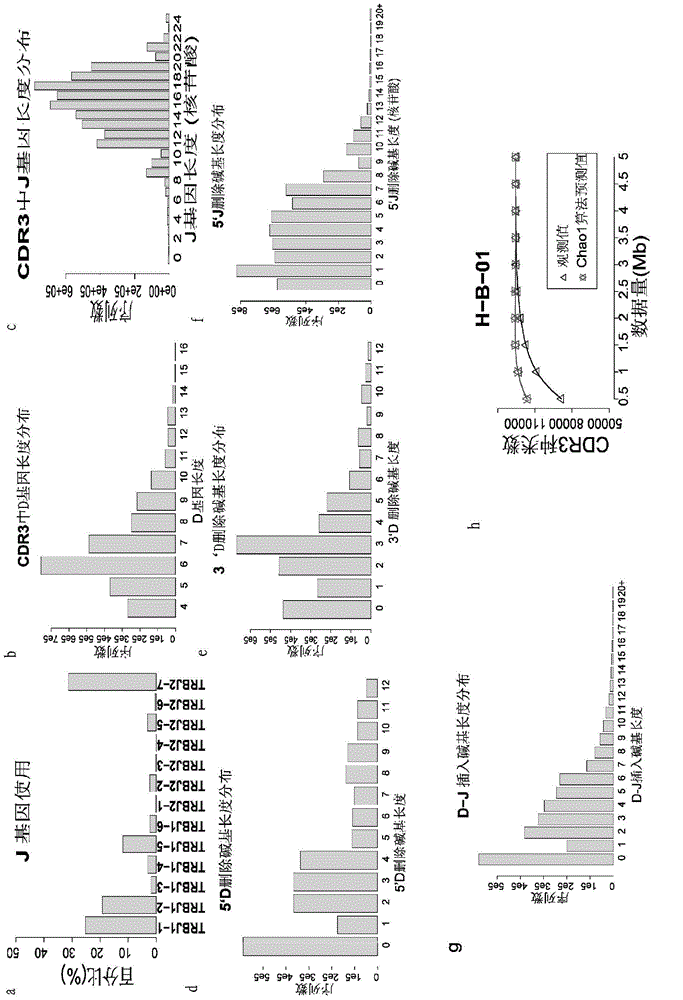
The invention discloses a method and system for processing immune repertoire sequencing data of samples. The method includes the steps that 1, data filtering processing is carried out on the immune repertoire sequencing data of the samples; 2, sequence splicing processing is carried out on the sequencing data obtained after data filtering processing; 3, local comparison is carried out on the sequencing data obtained after sequence splicing processing; 4, re-comparison is carried out on local comparison results; 5, the result with the highest score is screened out of the re-comparison results, and filtering is carried out; 6, error correction processing is carried out on the final comparison result; 7, sequence structure determination and translation are carried out based on the final comparison result obtained after error correction processing. By means of the method, analysis of a large quantity of immune repertoire sequencing data can be effectively achieved, analysis of TCR data and analysis of BCR data can be achieved at the same time, the PCR and sequencing errors can be effectively handled, the accuracy is high, the repeatability is good, and thus the whole condition of an immune repertoire can be truly and effectively reflected.
Owner:BGI SHENZHEN CO LTD
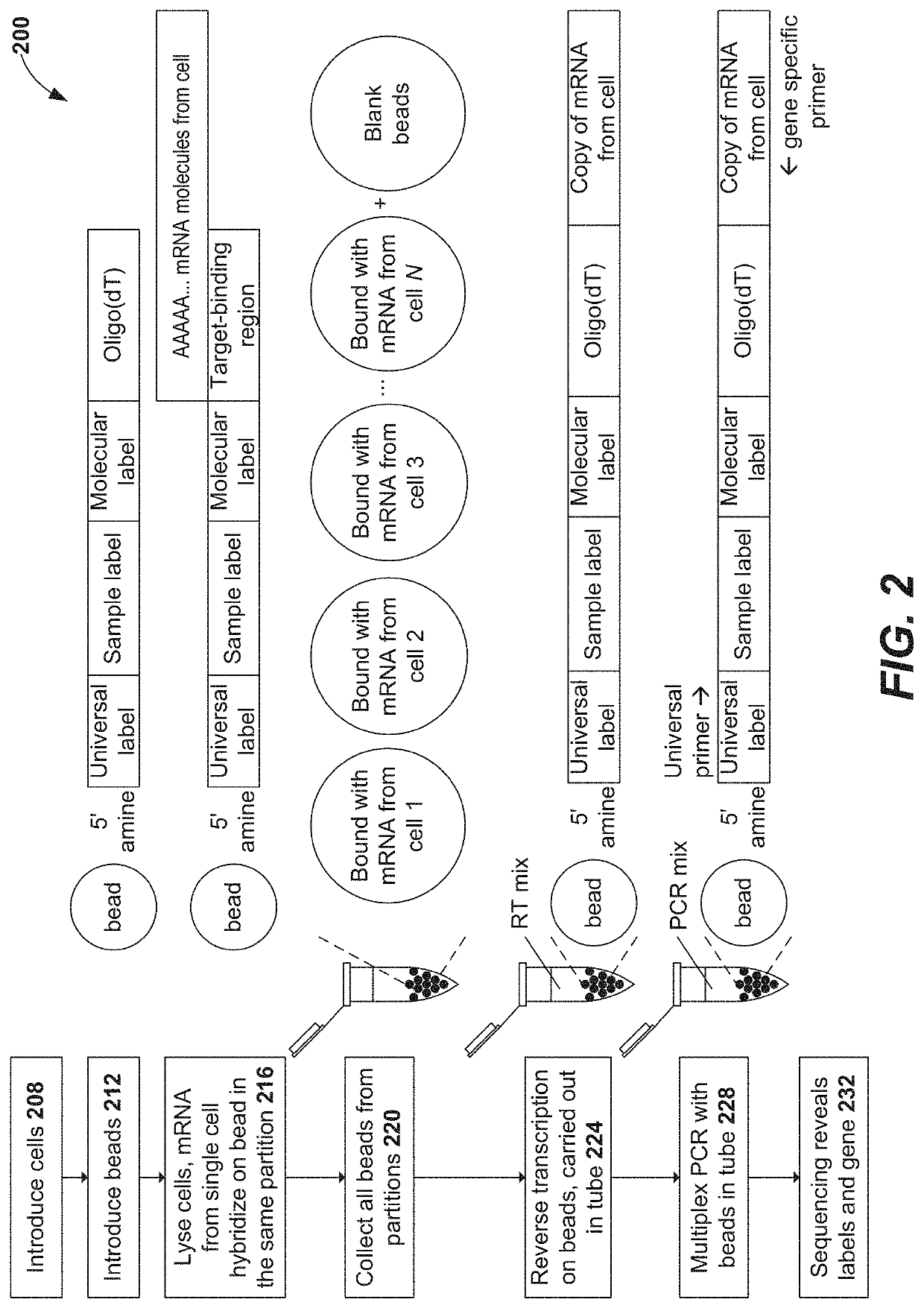
Compositions and methods for immune repertoire sequencing
ActiveUS20180208984A1Immunoglobulin superfamilyMicrobiological testing/measurementB-cell receptorClonal diversity
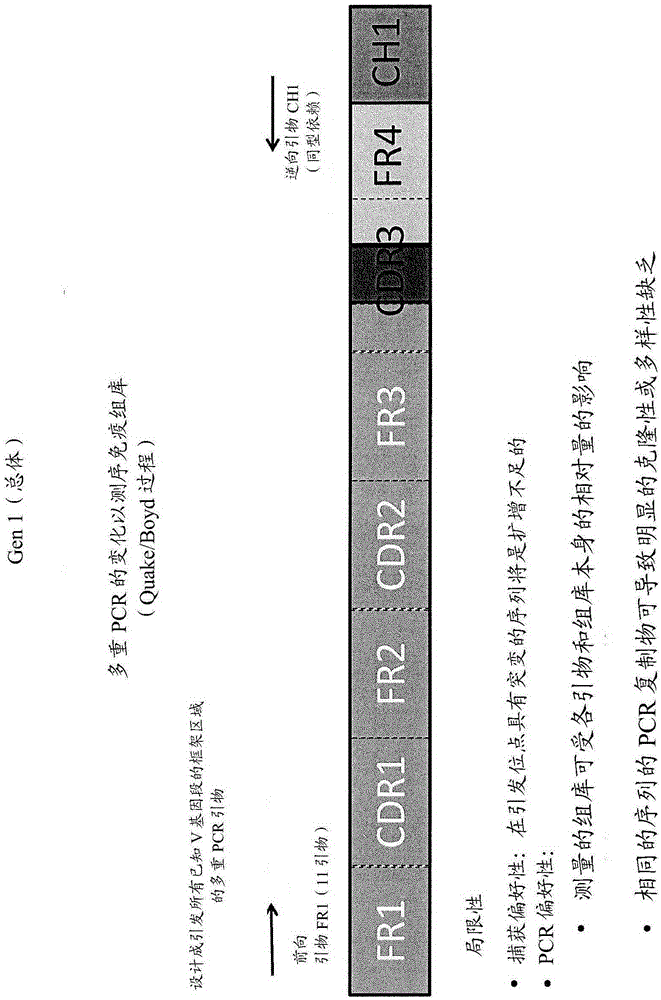
The present disclosure provides methods, compositions, kits, and systems useful in the determination and evaluation of the immune repertoire. In one aspect, target-specific primer panels provide for the effective amplification of sequences of T cell receptor and / or B cell receptor chains with improved sequencing accuracy and resolution over the repertoire. Variable regions associated with the immune cell receptor are resolved to effectively portray clonal diversity of a biological sample and / or differences associated with the immune cell repertoire of a biological sample.
Owner:LIFE TECH CORP
Mesophilic DNA polymerase extension blockers
PendingUS20210238661A1Reduce generationMicrobiological testing/measurementGenomic cloneOligonucleotide
Disclosed herein include systems, methods, compositions, and kits for 5′-based gene expression profiling and for whole transcriptome analysis (WTA) with random priming and extension (RPE). Blocker oligonucleotides capable of specifically binding to a portion of an oligonucleotide barcode are provided in some embodiments. The blocker oligonucleotides can reduce of the generation of undesirable extension products, such as, for example, the extension products of random primers hybridized to a portion of the oligonucleotide barcode. Immune repertoire profiling methods are also provided in some embodiments.
Owner:BECTON DICKINSON & CO
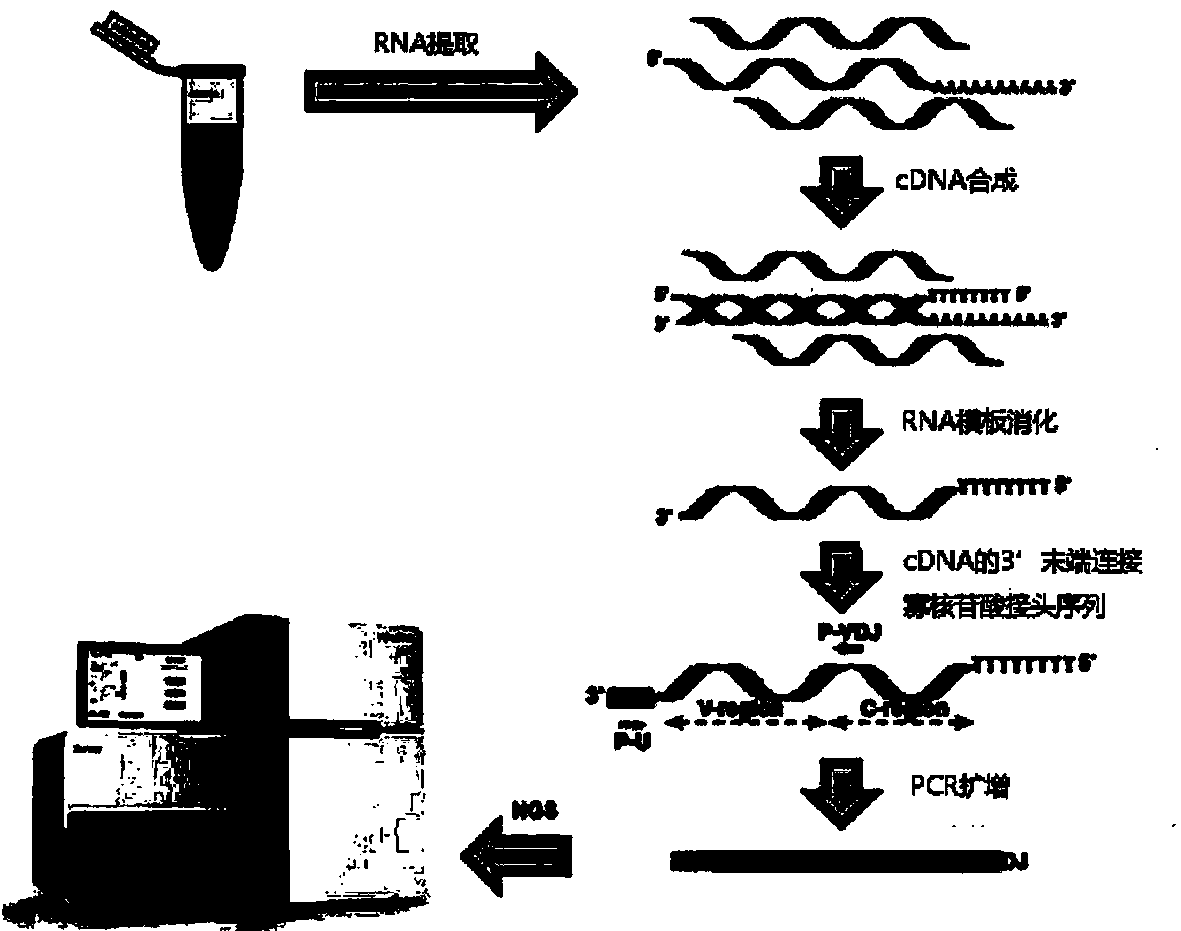
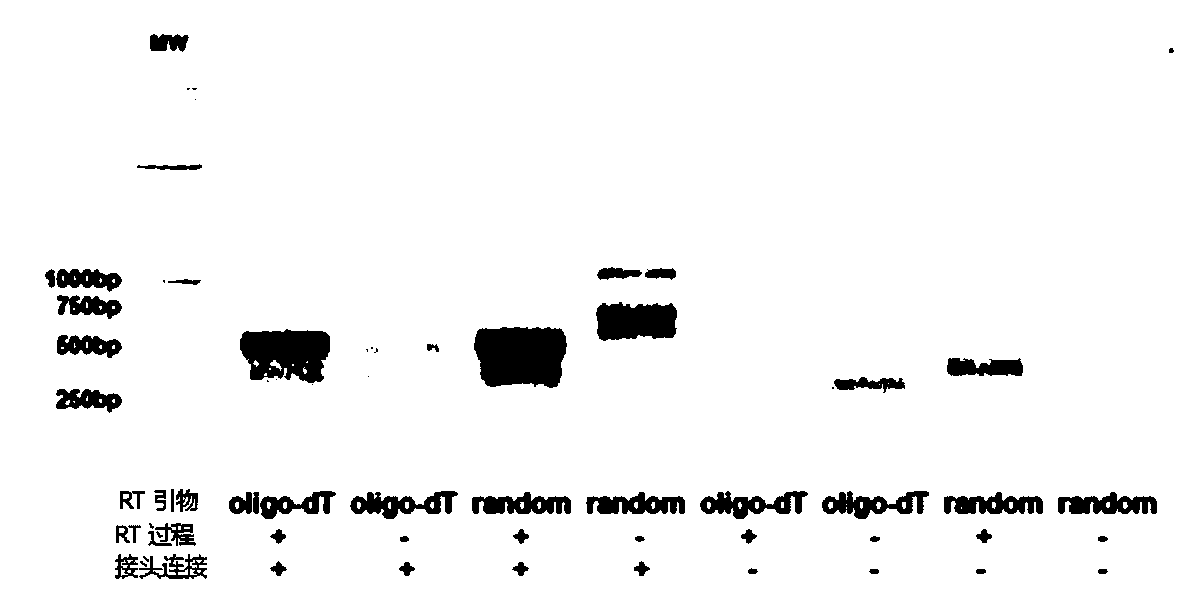
Whole blood immune repertoire detection method based on high-flux sequencing technology
ActiveCN104263818ALow costReduce sequencing timeMicrobiological testing/measurementComplementary deoxyribonucleic acidT cell
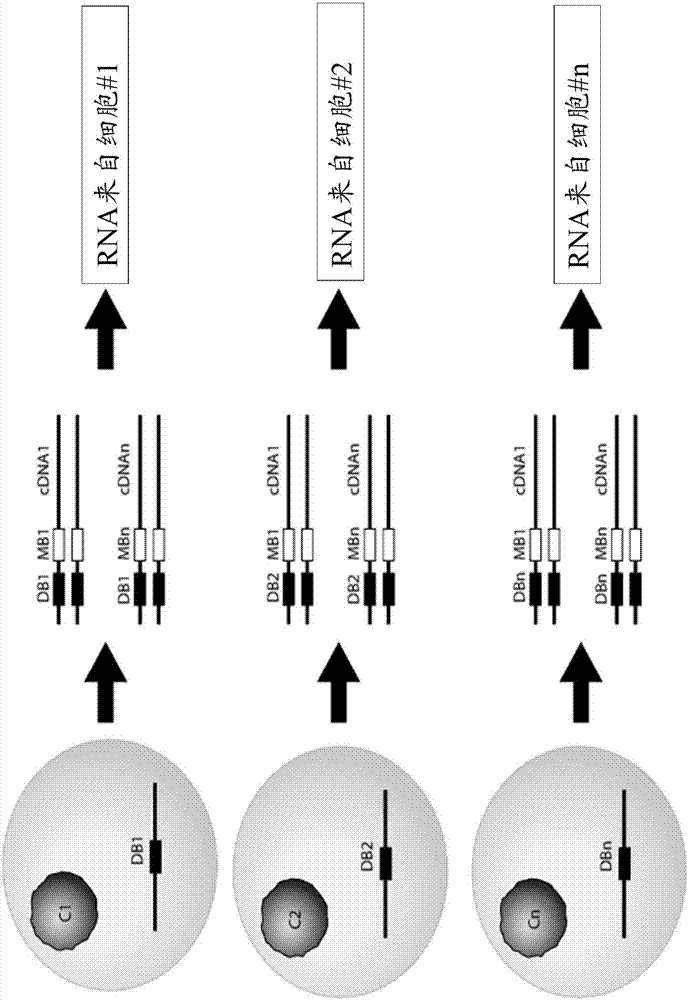
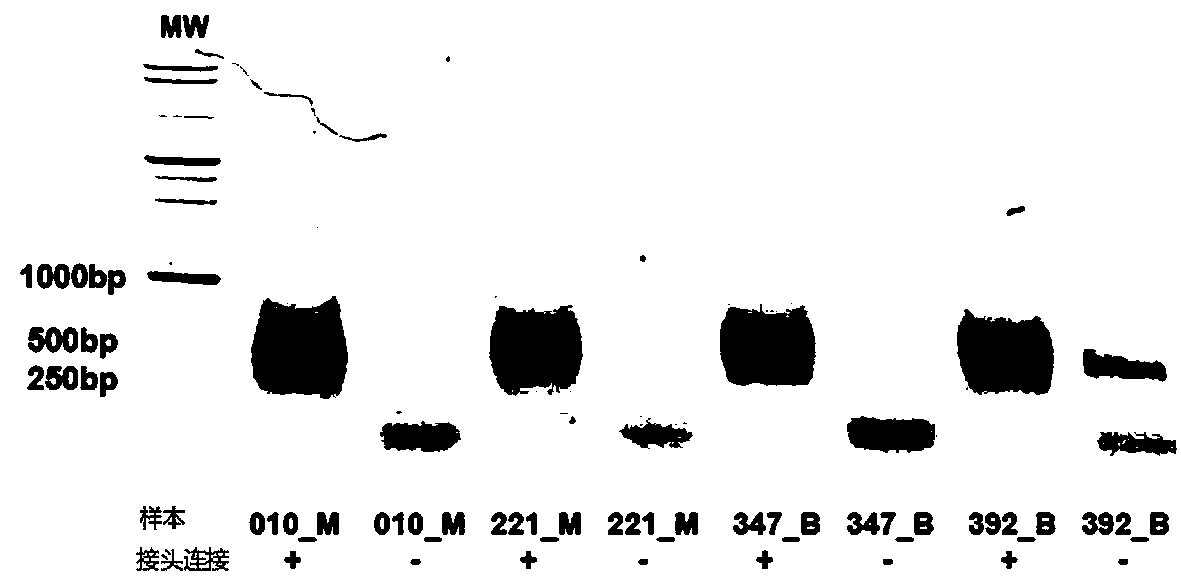
The invention provides a whole blood immune repertoire detection method based on high-flux sequencing technology. The method comprises the following steps: (1) directly extracting RNA (ribonucleic acid) from a whole blood sample; (2) carrying out reverse transcription on the RNA to generate single chain cDNA (complementary deoxyribonucleic acid), connecting an oligonucleotide joint sequence to the tail end of the single chain cDNA, carrying out PCR (polymerase chain reaction) reaction on the single chain cDNA subjected to oligonucleotide joint sequence connection by using a universal primer and a special targeted primer P-VDJ (of which the sequence is GAC CTC GGG TGG GAA CAC) as a special primer pair for PCR reaction, and amplifying the immune receptor genome; and (3) carrying out high-flux sequencing. The method of directly extracting the RNA from the whole blood sample is more suitable for clinic; the single target specific primer is utilized to uniformly amplify the variable region of the subtype T cell receptor gene; and a Hi-SEQ 2500 of the Illumina corporation and a bar code are utilized for sequencing, thereby ensuring the lower cost, the sequencing time of less than 48 hours and favorable sequencing depth.
Owner:武汉希望组生物科技有限公司
Methods of sequencing the immune repertoire
ActiveCN105189748ASequential/parallel process reactionsMicrobiological testing/measurementNon invasiveImmune repertoire
The invention provides a non-invasive technique for the detection and quantification of the immune repertoire, in a biological sample containing a plurality of distinct cell populations. Methods are conducted using sequencing technology to detect and enumerate immune repertoire within a heterogeneous biological sample.
Owner:LINEAGE BIOSCI
Immunocompetence assessment by adaptive immune receptor diversity and clonality characterization
InactiveUS20160002731A1Microbiological testing/measurementLibrary member identificationImmunocompetenceImmune receptor
Owner:ADAPTIVE BIOTECH +1
Method for constructing high-throughput sequencing library of immune repertoire
InactiveCN108893464AAmplify full-length sequenceGuarantee the success rate of the experimentMicrobiological testing/measurementLibrary creationImmune repertoireConstant region
The invention discloses a library construction scheme for introducing UM, namely molecular bar codes, based on a principle of 5'-RACE. The scheme specifically comprises the following steps: performingreverse transcription by designing single primers in a constant region, performing equivalent amplification to obtain all types of TCR / BCR CDR whole gene sequences, and introducing UMI, so that errors introduced in the subsequent PCR (Polymerase Chain Reaction) or sequencing process can be corrected. According to the method disclosed by the invention, the experimental requirements on library construction of trace samples as low as 100 cells can be met, and the TCR / BCR CDR whole gene sequences can be equivalently amplified.
Owner:广州华银医学检验中心有限公司
Compositions and methods for eliminating undesired subpopulations of t cells in patients with immunological defects related to autoimmunity and organ or hematopoietic stem cell transplantation
InactiveUS20090148404A1Reduce riskReduce severityBiocidePeptide/protein ingredientsAutoimmune responsesHematopoietic stem cell transplantation
The present invention relates generally to methods for stimulating T cells, and more particularly, to methods to eliminate undesired (e.g., autoreactive, alloreactive, pathogenic) subpopulations of T cells from a mixed population of T cells, thereby restoring the normal immune repertoire of said T cells. The present invention also relates to compositions of cells, including stimulated T cells having restored immune repertoire and uses thereof.
Owner:LIFE TECH CORP
Immune Repertoire Sequence Amplification Methods and Applications
ActiveUS20180073014A1Immunoglobulin superfamilyMicrobiological testing/measurementHeavy chainPairing
The present invention relates generally to the field of immune binding proteins and method for obtaining immune binding proteins from genomic or other sources. The present invention also relates to nucleic acids encoding the immune binding proteins in which the natural multimeric association of chains is maintained in the nucleic acids and the immune binding proteins made therefrom. For example nucleic acids encoding antibodies that are amplified from a B-cell using the methods of the invention maintain the natural pairing of heavy and light chains from the B-cell. This maintenance of pairing (or multimerization) produces libraries and / or repertoires of immune binding proteins that are enriched for useful binding molecules.
Owner:AUGMENTA BIOWORKS INC
Immune repertoire biological information analysis process based on molecular markers
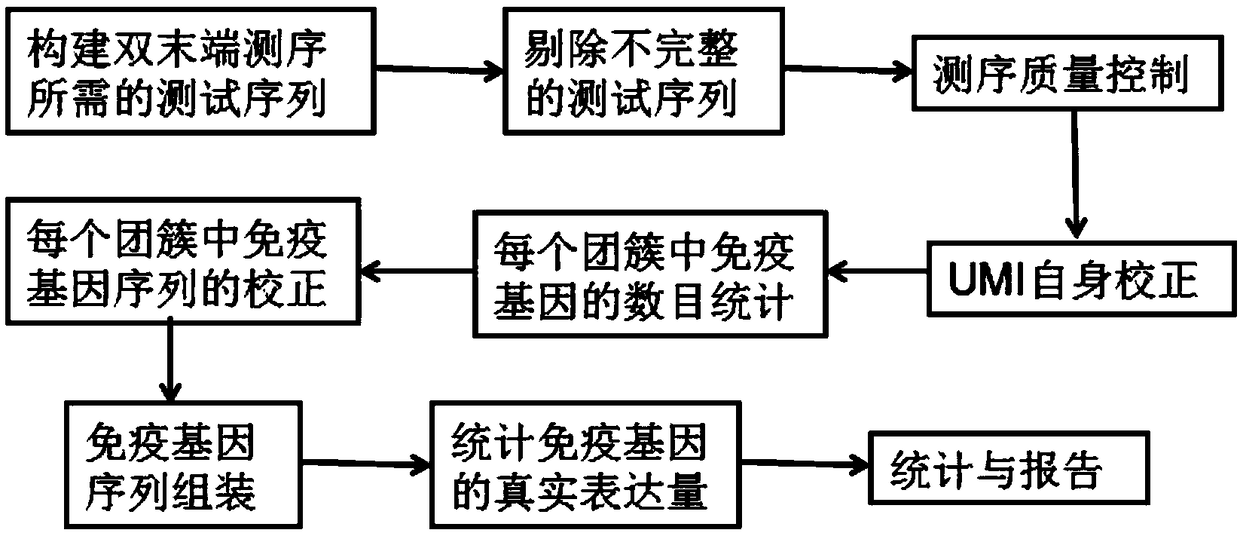
ActiveCN108804874AImprove accuracySpecial data processing applicationsInformation analysisStatistical analysis
The invention discloses an immune repertoire biological information analysis process based on molecular markers. In the process, single-chain immune globulin genes with the same sequence are selected,and primers and UMI sequences with the same structure are added to 3' ends or 5' ends of the selected single-chain immune globulin genes; after addition is completed, PCR amplification is performed on the immune genes to obtain a test sequence, and the test sequence is filtered; after filtering, the UMI sequence carried by the immune genes is corrected by establishing a directed acyclic tree, then the immune genes marked by the corrected UMI sequence are corrected, and the corrected immune genes are assembled; and finally the assembled immune genes are subjected to statistical analysis and reporting. According to the process, through correction of UMI and correction of the immune genes, the influences of amplification errors and sequencing errors on immune gene sequencing are effectivelyremoved, and the accuracy of sequencing data is improved.
Owner:广州华银医学检验中心有限公司
Method for establishing analysis immune library spectra based on random primer barcode technology
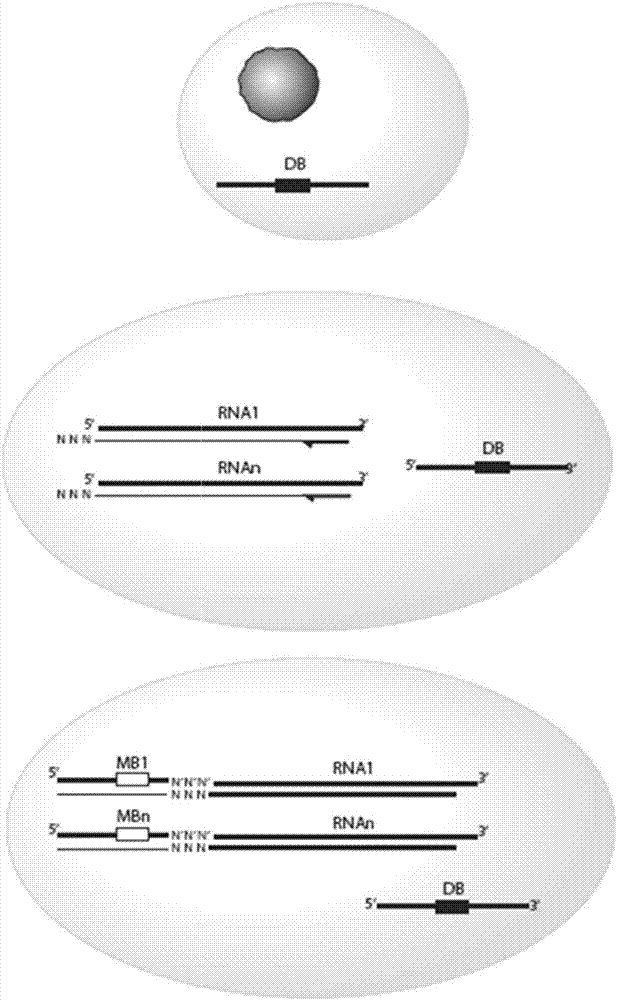
InactiveCN103436523AReduce skewReduce mistakesMicrobiological testing/measurementDNA preparationSequencing dataT-cell receptor
The invention aims at the medical biotechnology, relates to a high-throughput sequencing method based on a primer barcode technology and an application thereof in study of immune genomics, and specifically relates to a T-cell high-throughput sequencing method based on a marking method of cDNA molecules. Specifically, the method comprises the following steps of extracting RNA from a sample, performing reverse transcription on the RNA to obtain cDNA, designing a specific primer according to the obtained cDNA, constituting a random primer barcode by a downstream primer of the designed specific primer, a random barcode and a sequencing joint in a series manner, adding excessive obtained random primer barcode in the obtained cDNA molecules, carrying out high-throughput sequencing by using the sequencing joint in the random primer barcode, carrying out VDJ comparison on sequencing data; analyzing a CDR3 sequence and establishing immune library spectra. The method reduces deflection and error generated in a PCR amplification process. The number of the cDNA molecules can be quantized according to the specific primer barcode corresponding to a given cDNA sequence.
Owner:ARMY MEDICAL UNIV
Detection and quantitation of sample contamination in immune repertoire analysis
The invention is directed to methods for detecting and quantifying nucleic acid contamination in a tissue sample of an individual containing T cells and / or B cells, which is used for generating a sequence-based clonotype profile. In one aspect, the invention is implemented by measuring the presence and / or level of an endogenous or exogenous nucleic acid tag by which nucleic acid from an intended individual can be distinguished from that of unintended individuals. Endogenous tags include genetic identity markers, such as short tandem repeats, rare clonotypes or the like, and exogenous tags include sequence tags employed to determine clonotype sequences from sequence reads.
Owner:SEQUENTA
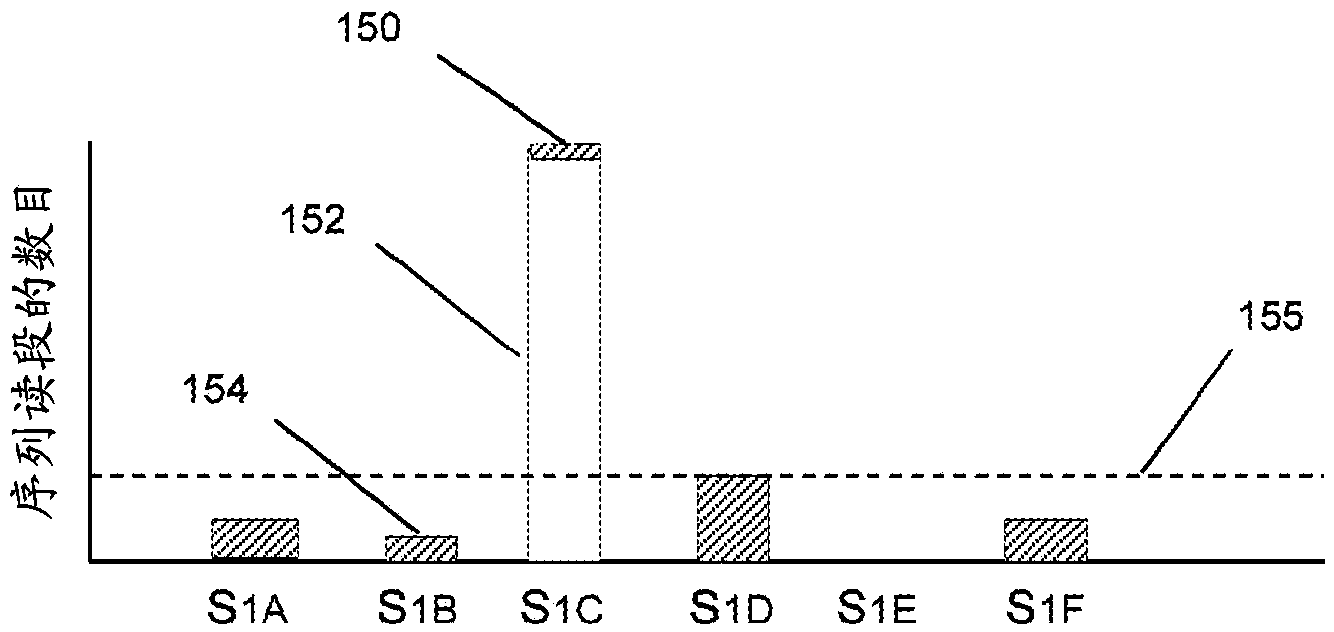
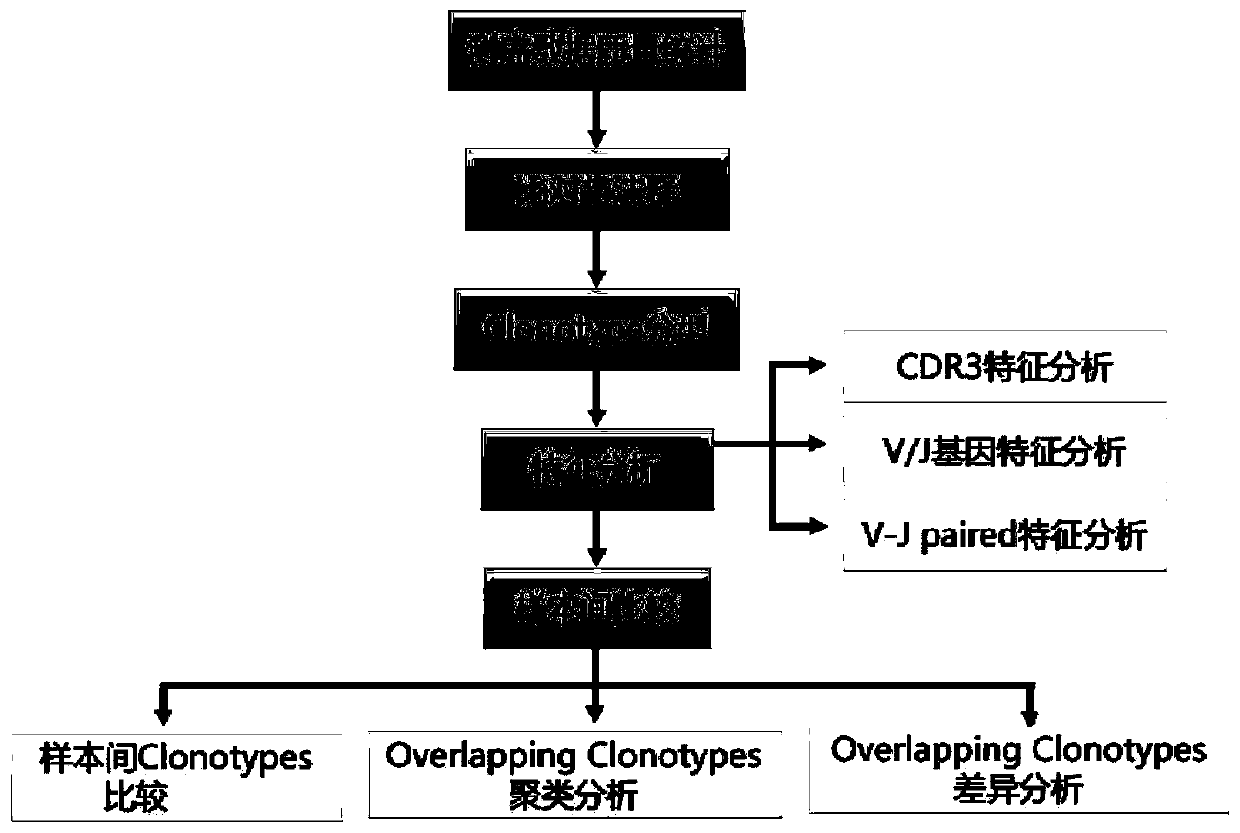
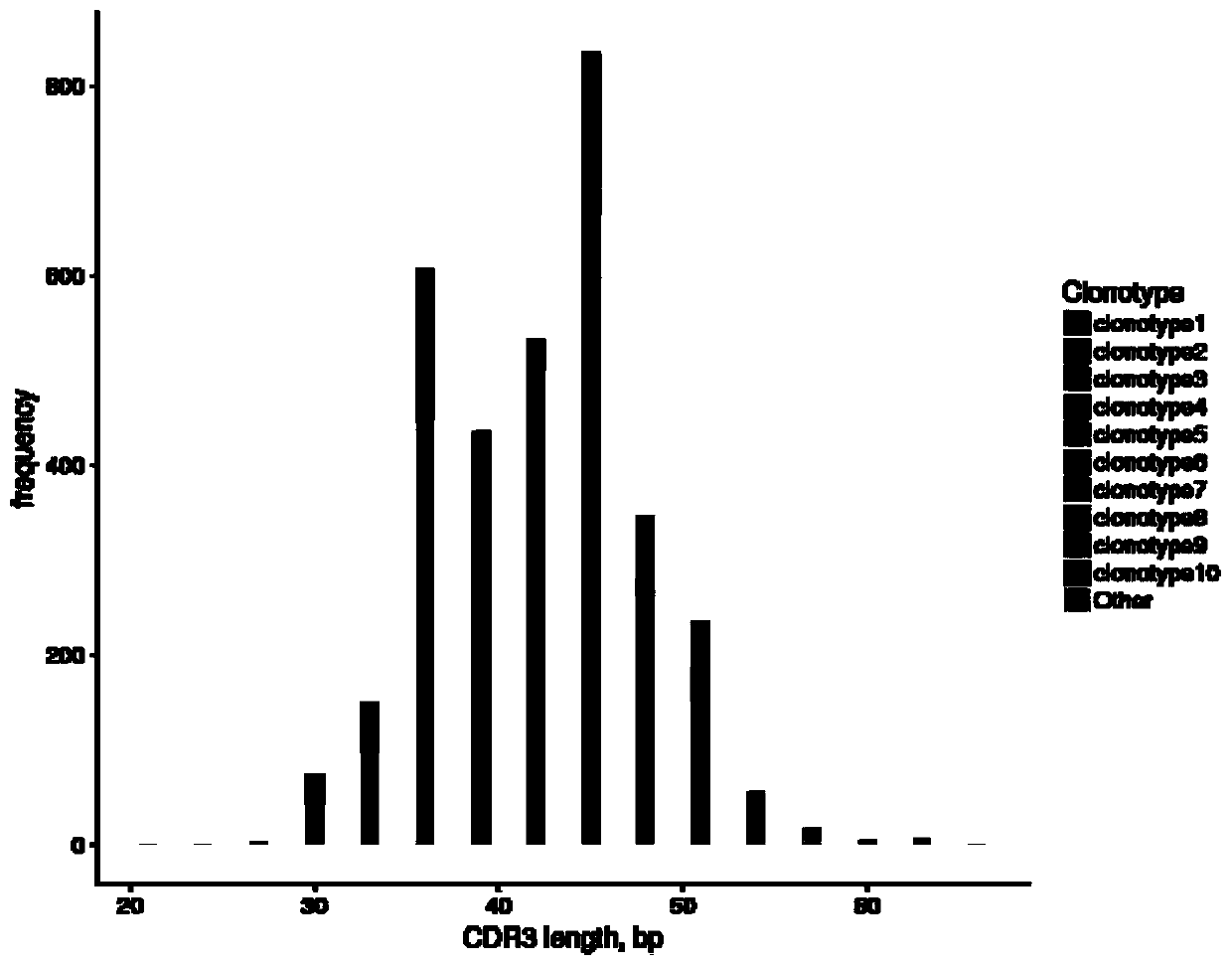
Method for analyzing single-cell immune repertoire sequencing data
ActiveCN109979528AThe analysis process is completeIncrease contentProteomicsGenomicsOriginal dataOrganism
The invention discloses a method for analyzing single-cell immune repertoire sequencing data. The method comprises the following steps of S1, performing data quality statistics and basic quality control on original data; S2, performing sequence alignment and noting on high-quality reads which are obtained in the step S1 and a reference V(D)J fragments sequence, thereby obtaining a Consensus sequence; S3, performing Clonotype typing on a CDR3 amino acid sequence in the Consensus which is obtained through splicing in the step S2; S4, performing CDR3, V / J gene and V-J paired characteristic analysis on the Clonotype in the sample in the step S3; and S5, combining the clonotypes in different samples, and performing multiple-sample analysis. The method has advantages of integral analysis process, abundant content, visual result displaying and comprehensive displaying of immune repertoire information, thereby making a user comprehensively understand and evaluate the immune repertoire condition and more accurately reflecting the immune function in an organism.
Owner:GUANGZHOU GENE DENOVO BIOTECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com